Abstract
Interferon lambda 3 (IFN-λ3 or IFNL3, formerly IL28B), a type III interferon, modulates immune responses during infection/inflammation. Several human studies have reported an association of single nucleotide polymorphisms (SNP) in the IFNL3 locus with expression level of IFNL3. Previous genetic studies, in the context of hepatitis C virus infections, had predicted three regulatory SNPs: rs4803219, rs28416813 and rs4803217 that could have functional/causal roles. Subsequent studies confirmed this prediction for rs28416813 and rs4803217. A dinucleotide TA-repeat variant (rs72258881) has also been reported to be regulating the IFN-λ3 promoter. In this study, we tested all these genetic variants using a sensitive reporter assay. We show that the minor/ancestral alleles of both rs28416813 and rs4803217, together have a strong inhibitory effect on reporter gene expression. We also show an interaction between the two principal transcription factors regulating IFNL3 promoter: IRF7 and NF-kB RelA/p65. We show that IRF7 and p65 physically interact with each other. By using a transient ChIP assay, we show that presence of p65 increases the promoter occupancy of IRF7, thereby leading to synergistic activation of the IFNL3 promoter. We reason that, in contrast to p65, a unique nature of IRF7 binding to its specific DNA sequence makes it more sensitive to changes in DNA phasing. As a result, we see that IRF7, but not p65-mediated transcriptional activity is affected by the phase changes introduced by the TA-repeat polymorphism. Overall, we see that three genetic variants: rs28416813, rs4803217 and rs72258881 could have functional roles in controlling IFNL3 gene expression.
Keywords: IFNL3, Gene expression, IFNL locus genetics, rs12979860, rs8099917, IFNL4, rs28416813, rs4803217, rs368234815, rs117648444, IFN-λ3, IFN-λ4, IFN-λ
1. Introduction
Genome-wide association studies (GWAS) carried out in the year 2009 to identify the genetic determinants behind response to interferon (IFN) alpha-based (conventional) therapy in chronic hepatitis C virus (HCV) infections has remained one of the most successful GWAS carried out on a human complex genetic disease [1–3]. This exercise also helped to revive a new field in innate immune biology, that of the IFN lambda (IFN-λ) or the Type III IFN family. It comprises of four members in humans (IFN-λ1, 2, 3 and 4), all located in tandem on chromosome 19 in a region spanning ~55 kb [4]. The first three members which are closer in homology to each other were discovered in 2003 [5–6], while the fourth member, a recent addition, was discovered in the context of follow-up studies that were conducted to identify the causal variants behind the HCV GWAS [7]. Until the discovery of IFN-λ4 [7] by P-Olsson et al., (2013), it was widely believed that the causal variants behind the HCV GWAS were somehow regulating the expression and/or activity of IFN-λ3 which happened to be the gene nearest to the strongest signalgenerating single nucleotide polymorphisms (SNP) rs12979860 and rs8099917 in the GWAS [8]. However, the discovery of IFN-λ4 has to a large extent, successfully answered many questions about the causal mechanism behind IFN-λ locus variants and HCV persistence following therapy [9]. While several lines of evidence have surfaced that implicates IFN-λ4 as the prime causal mechanism behind persistence of HCV following either the conventional (IFN a-Ribavirin) or the new age (direct-acting antiviral) therapy [9–12], it is still not clear how spontaneous clearance of HCV infections is genetically regulated by the IFN-λ locus. In what is referred to as the IFN-λ4-HCV paradox, expression of a relatively active form of IFN-λ4 is detrimental for the clearance of HCV following conventional or even the new age therapy [13]. This is due to a saturation of expression of IFN-stimulated genes (ISG) in HCV-infected liver expressing an active IFN-λ4 that allows the virus to persist and resist the therapy. But this cannot convincingly explain the genetic link to spontaneous clearance of HCV wherein some individuals but not others are able to clear the virus without the need for any therapy. Interestingly, this phenotype is also strongly associated with the IFN-λ locus genetic variants [14–17], but whether IFN-λ4 can be responsible for this is not obvious. In fact, a recent genetic study has shown that spontaneous HCV clearance cannot be entirely explained by IFN-λ4, implying that other mechanisms related to rs8099917 and another SNP rs4803221 may be in operation [18]. In addition, the genetic association of IFN-λ locus variants has extended beyond HCV infections to include a wide variety of infectious and inflammatory diseases [19–20]. In these diseases it is not clear if IFN-λ4 is causal or other genetic variants in the region that could be influencing IFN-λ3 expression could be causal. In fact, a recent study based on functional and genetic evidence found that it is not IFN-λ4 but IFN-λ3 that may be responsible for severe fibrosis in chronic HCV infections, thereby putting the onus back on IFN-λ3 regulatory SNPs [21].
This makes us to look for the ‘other’ variants in the region that have functional roles, perhaps the ones that may be affecting IFN-λ3. In fact, two previous genetic studies, the first one utilizing a recombination mapping technique [22] identified ‘four causal SNPs’ that could be responsible for spontaneous clearance of HCV infections [22–23]. These SNPs are: rs4803219, rs28416813, rs8103142 and rs4803217. These variants were also shown to have better predictive value of response to therapy in chronic HCV infections [22,24]. Among the four SNPs, rs8103142 is a non-synonymous SNP that has no effect on the activity of the IFN-λ3 protein, at least in vitro [25–27], and hence can be ruled out from the list of causal SNPs. The remaining three SNPs are situated in regulatory regions of IFN-λ3 gene and two of them, rs28416813 and rs4803217 have been documented to potentially affect IFN-λ3 gene transcription or translation [28–29]. Numerous studies have reported an association of IFN-λ locus variants with expression level of IFN-λ3 both in HCV and non-HCV contexts [2–3,30–39]. However, some have also reported of not seeing any such association [1,24]. Therefore, to understand the influence of this evolutionarily important innate immune gene locus and its variation on human disease and health conditions, it is imperative to further the research in this area to better explain the relationship between genetic variants at IFN-λ3 locus and its expression.
We were the first to show an evidence for a causal SNP that could be behind HCV GWAS [28]. We showed that rs28416813 due to its proximity to an NF-κB binding site in the IFN-λ3 promoter region influenced transcription in reporter assays. Later, in the same year rs4803217, present in the IFN-λ3 3’UTR region, was also shown to have a functional role in influencing the IFN-λ3 mRNA stability by interfering with the AU-rich element mediated decay (AMD) pathway [29]. A report also tested for the effect of rs4803219 on IFN-λ3 promoter function but found ‘little’ evidence to implicate this SNP as causal [26]. However, in a natural context, due to the phenomenon of linkage disequilibrium (LD) all the above three SNPs are thought to be highly correlated to each other [4] and therefore, the combined effect of these SNPs on gene expression could be important, that remains to be studied. Moreover, another variant, a TA dinucleotide polymorphism rs7225881 has also been reported to be affecting the IFN-λ3 promoter [26] and at least two genetic studies have found evidence for association of this variant with response to therapy in chronic HCV infections and/or spontaneous clearance [40–41]. In the present study, we have included all the above variants and tested for their influence on reporter gene expression.
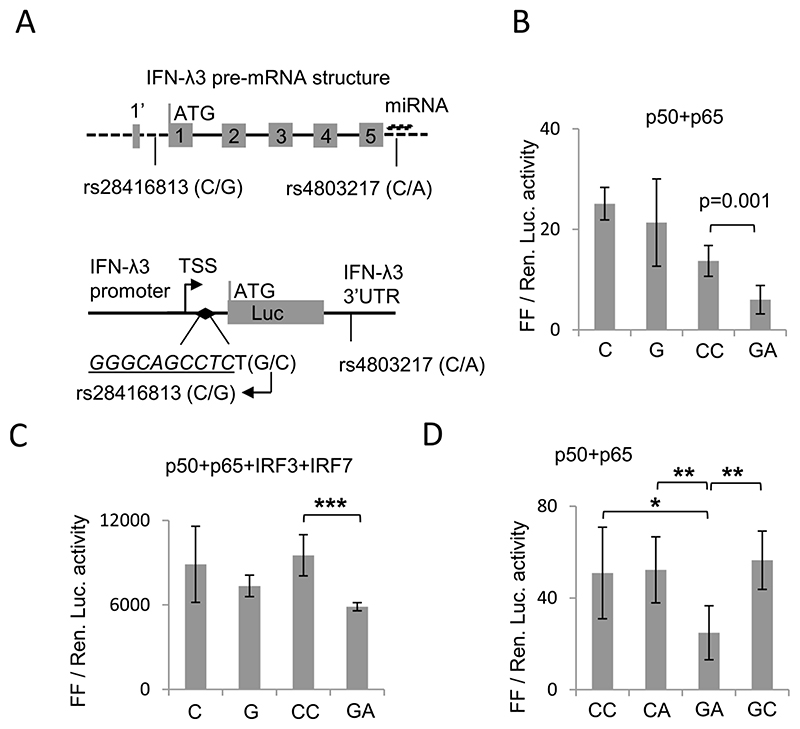
The IFN-λ3 gene, like the other IL10 cytokine superfamily members to which it belongs [5–6], was initially predicted to have five exons but later studies showed that it may have a split 1st exon, with a short (first three amino acids) and a long arm [42] (Fig. 1A). We have previously identified the SNP rs28416813 [28] to be placed very close (one basepair apart) to an NF-κB binding site characterized by Osterlund et al [43] in 2007 (Fig. 1A lower panel). This NF-κB binding site is downstream of the transcription start site (TSS) and is located 37 bp upstream of the translation start site, within the 5’UTR, if IFN-λ3 is considered as having the conventional five exons (Fig. 1A upper panel). However, if IFN-λ3 has a split first exon, then the SNP rs28416813 will be present in the first intron of IFN-λ3 gene. In either case, the significance of the SNP on the IFN-λ3 promoter activity would remain unaffected by either definition of the gene/mRNA structure of IFN-λ3 since transcription precedes splicing. In fact, a previous study tested the effect of rs28416813 on splicing rather than on transcription and found no role for the SNP [26].
Fig. 1. Effect of SNPs rs28416813 and rs4803217 on reporter gene expression.
A) (Top) Structure of the pre-mRNA of IFN-λ3 showing presence of 5 exons. The first exon is split such that the first three amino acids (1’) are encoded in the shorter arm and the remaining amino acids are encoded by the longer arm. The dark line shows introns while dashed lines show UTRs. The SNP rs4803217 overlaps the miRNA binding sites identified in Ref. no. (29); miRNA is shown above the SNP. Depending on whether IFN-λ3 has a split first exon or not, the SNP rs28416813 is either present in the 5’UTR or in the first intron of IFN-λ3. (bottom) Schematic showing the positions of SNPs rs28416813 and rs4803217 within the constructs used in the study. A 1.4 kb DNA fragment upstream of the start codon of IFN-λ3 gene (as shown in top panel) containing the SNP rs28416813 at 37 bp upstream of the start codon and one base pair away from a NF-κB binding site (inverted filled diamond, sequence in italics and underlined) is shown. The 154 bp IFN-λ3 3’UTR is shown downstream of the Luciferase (Luc) gene carrying the SNP rs4803217. The top arrow shows transcription start site (TSS). B) Ancestral alleles G and A at rs28416813 and rs4803217 significantly decrease reporter gene expression. The construct p1.4kbIFNL3 or p1.4kbIFNL3-3’UTR were transfected in HEK293T cells. The experiments were carried out in 96-well plates. The data is from six separate experiments representing three different plasmid preparations. The mean value from all six experiments is shown, while the error bars depict SD. C) Similar to B, except that additional TFs IRF3 and IRF7 were overexpressed and experiments were carried out in 12-well plates. D) The non-ancestral alleles at rs28416813 and rs4803217 have dominant effects on each other while ancestral alleles at both rs28416813 and rs4803217 in combination have a negative effect on reporter gene expression. The experiments were carried out in 96-well plates seeded with HEK293T cells and transfected with p1.4kbIFNL3-3’UTR carrying different allele combinations at the two SNP positions rs28416813 (C/G) and rs4803217 (C/A) respectively as shown. The data is from six separate experiments representing three different plasmid preparations. The mean value from all six experiments is shown, while the error bars depict SD. For B, C and D two tailed student t-test for independent means was used for calculating statistical significance; *p < 0.05, **p < 0.01; ***p < 0.001. All the Luciferase ratio values from each individual experiment are shown in Suppl. Table 1.
In our previous study [28] where we used 1.4 kb IFN-λ3 promoter to characterize rs28416813, we have used the reverse orientation in naming the alleles at rs28416813 due to a discrepancy between the direction of the coding strand and naming convention followed by dbSNP (where naming is based on the complementary reverse strand (Fig. 1A); ex. the two alleles at SNP rs4803217 are shown as C > A, even though the allele G is in the coding strand); the reverse scheme of naming the alleles is also followed by the two reports that have shown the functional significance of the 3’UTR SNP rs4803217 [29,44]. However, in the current study, to maintain uniformity, we have retained the conventional method of naming the alleles at a given SNP position in the same scheme as defined in dbSNP [45]. For ex. rs28416813 (C), means we are referring to the ‘G’ allele in the coding strand and vice versa (Fig. 1A). We have used different lengths of the IFN-λ3 promoter (1.4 kb, 0.5 kb and 4 kb) in this study to characterize its activity in the context of different functional genetic variants and to report on a novel interaction between IRF7 and NF-kB RelA/p65 that leads to transcriptional synergy.
2. Experimental procedures
2.1. Plasmids, mutations, enzymes, cell lines and reagents
The plasmids encoding transcription factors (TF) NF-κB p50, p65, IRF3 and IRF7 were all purchased from Invivogen (San Diego, CA, USA; the IRF3 and IRF7 genes also have a HA tag); they were all present in the pUNO vector background. The phosphomimic versions (constitutively active) of IRF3 and IRF7 (IRF3-SA and IRF7-SA) were gifts from Siddharth Balachandran, Fox Chase Cancer Centre, PA, USA. The construct p1.4kbIFNL3 has been described in detail before (28; referred to as p1.4Il28B in Ref. [28]). Briefly, genomic DNA isolated from whole blood from HCV-infected patients was used as template to amplify the 1.4 kb IFN-λ3 promoter-5’ UTR region in a PCR reaction using the primers: For-5’ GATATCGGTACCCAGTGGAATTCAGGGCAAATTAC-3’ OH; Rev-5’ GATATCAAGCTTGTGTCACAGAGAGAAAGGGAGCT-3’ OH. The PCR fragment was cloned in to pGL3basic vector (Promega, Madison, WI, USA) at KpnI and HindIII restriction sites. The mutation from C to G allele at rs28416813 was introduced by site-directed mutagenesis. The IFN-λ3 3’UTR containing the alternate alleles at the SNP site rs4803217 were cloned at the XbaI site downstream of the Luciferase gene coding sequence in to the p1.4kbIFNL3 construct described above. For the cloning of IFN-λ3 3’UTR fragment, two single stranded DNA molecules were synthesized from IDT Technologies, Coralville, IA, USA containing the entire 3’UTR of IFN-λ3 gene along with the two alleles at rs4803217. This DNA was used as a template to amplify the 3’UTR in a PCR reaction that was used for cloning in to the p1.4kbIFNL3 constructs with two different alleles at rs28416813 to obtain p1.4kbIFNL3 promoter (C or G at rs28416813)-Luciferase-IFNL3 3’UTR (C or A at rs4803217) construct. The T allele at rs4803219 was inserted in to the p1.4kbIFNL3 promoter (C allele at rs28416813)-Luciferase-IFNL3 3’UTR (A allele at rs4803217) construct by subcloning a DNA fragment derived from an HCV-infected patient DNA IFN-λ3 promoter clone that had the T allele at rs4803219 and no other changes in sequence elsewhere in the construct; the fragment was exchanged between the two plasmids at Hind III and Bsu361 restriction sites. All the constructs were confirmed for correctness by DNA sequencing. The details of the HCV-infected patient cohort including ethics approvals are provided elsewhere [10]. All the DNA oligonucleotides used in the study were custom synthesized from IDT; the probes used for electrophoresis mobility shift assays (EMSA) were synthesized by Sigma-Aldrich (St. Louis, MO, USA). p0.5kbIFNL3 was constructed in pGL3basic vector (Promega) in between Kpn I and Hind III restriction sites by subcloning from p1.4kbIFNL3 construct using the primers-for: GATATCGGTACCCCCCTGAGTCTCCATCAGTTTCTCTTT; rev 5’-GATATCAAGCTTGTGTCACAGAGAGAAAGGGAGCT. The ~ 0.5 kb fragment containing p300 binding site (GGGAGTG) was amplified by PCR, using primers: For 5’-GATATCACGCGTTCGGG-GAGCTCCCTGGTTCA-3’ OH and Rev 5’-GATATCA-GATCTGAAGCTGGCGGGGGAGAGGG-3’ OH and cloned in to pGL3Promoter vector (Promega) at MluI and BglII restriction sites. The details of the 4 kb IFN-λ3 promoter construct (p4kbIFNL3) are described in Ref. [10]. The plasmid encoding p300 gene was purchased from Addgene, USA.
All mutations were introduced by PCR-based methods or with site-directed mutagenesis kit (New England Biolabs, Ipswich, MA, USA). NF-κBa (TGGGCTTTCCCAG) was mutated to TTTTGCGGACGCC; NF-κBb (GGGACTGCCC) was mutated to AGCGGCTCGC; NF-κBc (GGGCAGCCTC) was mutated to TTACTGGCGC; IRFa (GTTTCTCTTT) was mutated to GCCCCTCCCC; IRFb (GTTTTCACTTTTC) was mutated to GTCTCCACCTCTC; p300 binding site (GGGAGTG) was mutated to TTCTCTA. The 5 bp insertion had CTACA and the 10 bp insertions had the sequence CTACATCAGC. All mutations were confirmed by DNA sequencing. HEK293T cells were from ATCC (Manssas, VA, USA); they were propagated in DMEM with 10% FBS, glutamax, penicillin and streptomycin (Life Tech., Rockville, MD, USA). All other reagents used were molecular biology grade or higher purchased from Sigma-Aldrich. All the enzymes including the 5X master mix with Taq DNA polymerase used for PCR were from New England Biolabs (Ipswich, MA, USA).
2.2. Expression and purification of recombinant p50 protein
Full-length NF-κB p50 gene (Invivogen) of ~ 1.3 kb was cloned in to pET32a expression plasmid (Novagen, Madison, WI, USA) at EcoRI and HindIII restriction sites. The clone was confirmed by DNA sequencing. Expression of recombinant proteins in pET32a vector gives a fusion recombinant protein with an additional 17 kDa S-tag at the N-terminus (Novagen). Expression was induced from BL21DE3 (Novagen) cells carrying the plasmid by 0.5 mM IPTG at 37 °C overnight in 1 L cultures. Next day, cells were collected by centrifugation at 6000 rpm for 10 min at 4 °C and were resuspended in lysis buffer having 50 mM Tris (pH-7.5), 150 mM NaCl, 1 mM PMSF, 20 mM imidazole and 10% glycerol. The cells were sonicated on ice at 40% amplitude for 5 min with 5 sec on-5 sec off cycles using an ultrasonic processor (Cole-Parmer, India). The cell debris was removed by centrifugation of the lysate at 4000 rpm for 10 min at 4 °C and supernatant was collected. The supernatant was further clarified through a 0.45 um syringe filter (Millipore, USA). Then the soluble fraction was bound to preequilibrated Histrap column (GE Healthcare, USA), washed and eluted in buffer having 50 mM Tris(pH-7.5),150 mM NaCl, 1 mM PMSF, 500 mM imidazole and 10% glycerol. The eluted proteins were checked on SDS-PAGE gels and the fractions containing the recombinant protein were then concentrated and desalted by using Centricon filter units (Amicon, USA) into a buffer containing 50 mM Tris (pH-7.5), 20 mM NaCl and 10% glycerol. The protein was stored at – 20 °C until further use.
2.3. EmSA
EMSA was performed with purified recombinant p50 protein and Cy5 labeled oligonucleotide probes. Oligonucleotide probes used in the EMSA were the following: probe C-For: 5’ Cy5-CCCCBrdUGCCCT-CAGTGGGCAGCCTCTGCATTCCCBrdUCAGCTCCCTTTCTCTCTGTGA-3’ OH; Probe C-Rev: 5’ TCACAGAGAGAAAGGGAGCTGAGGGAATGCA-GAGGCTGCCCACTGAGGGCAGGGG-3’ OH; Probe G-For: 5’ Cy5 CCCCBrdUGCCCTCAGTGGGCAGCCTCTCCATTCCCBrdU-CAGCTCCCTTTCTCTCTGTGA-3’OH; Probe G-Rev: 5’-TCACAGAGA-GAAAGGGAGCTGAGGGAATGGAGAGGCTGCCCACTGAGGGCAGGGG-3’ OH; C-oligo NF-κB -For: 5’-CCGCTGGGACTTTCCAGGA-3’ OH; C-oligo NF-κB -Rev: 5’-TCCTGGAAAGTCCCAGCGG-3’ OH. Probes were annealed at 90 °C and allowed for slow cooling at room temperature overnight. For each binding reaction 1x binding buffer was used containing 50 mM Tris (pH 8.0), 200 mM KCl, 5 mM EDTA, 5 mM DTT and 0.25 μg/microliter BSA (w/v). A typical binding reaction was set up in 20 μl volume and contained 1 μl of labelled probes (5 uM), 6 μl of recombinant protein and 1 μl of cold competitor (5 uM C-oligo). DNA binding reaction was performed on ice for 30 min. 1 pl of 6x DNA loading dye was added to each reaction and run on 4% native PAGE for 50 min at 4 °C. The gel was imaged using Biorad Chemidoc MP imaging system (Universal hood III).
2.4. Luciferase assays
HEK293T cells were seeded at a density of 18x103 cells/well in COSTAR (Corning, NY, USA) 96-well plates (white, clear bottom) (some experiments were undertaken in 12-well plates, as shown in the figure legends). After cells reached a confluency of ~ 70%, they were transfected with plasmids at the indicated concentrations using Lipofect-amine LTX&PLUS reagent (Invitrogen). The IFN-λ3 promoter constructs with or without the IFN-λ3 3’UTR (referred to as p1.4kbIFNL3-3’UTR or p1.4kbIFNL3/p0.5kbIFNL3/p4kbIFNL3 respectively) were transfected at 10 ng/well (100 ng/well for 12-well plates) and the TFs were at 25 ng/well (50 ng/well for 12-well plates); the transfection control plasmid expressing Renilla luciferase (pRLTK) was at 0.01 ng/well (0.1 ng/well for 12-well plates). pUNO vector was used to keep the DNA concentration equal in all the wells. For stimulation of the RLR pathway, HCV NS5B gene at 50 ng/well and RIG-I gene at 10 ng/well were cotransfected as described previously [28]. In experiments with p300 coactivator, 50 ng/well of the plasmid carrying the p300 gene was used. After incubating the cells for 24 hr at 37 °C and at 5% CO2, they were lysed, and Luciferase assays were carried out in the GLOMAX 96-well plate reader with dual-injection system (Promega). The ratio of Firefly Luciferase to Renilla Luciferase was calculated and plotted. All constructs with Luciferase reporter genes carrying different alleles, haplotypes and mutations were quantified using Nanodrop (ThermoFischer, Waltham, MA) by taking the average of several measurements and also the plasmids were run on EtBr-stained gels to make sure that equal amounts of plasmids were used in transfection experiments.
2.5. Virus infection and polyI:C treatment
Rotavirus strain SA11 (RV-SA11) was a kind gift from Dr. Mamta Chawla-Sarkar, National Institute of Cholera and Enteric Diseases, Kolkata, India. 0.09X106 HEK293T cells were seeded in 12-well plates. On next day after reaching 70-80% confluency cells were transfected with Luciferase constructs (p1.4IFNL3-3’UTR construct at 100 ng/well; 0.1 ng/well of pRLTK) using Lipofectamine LTX Plus reagent (Invitrogen). Virus infection was carried out after 24 h. First, the virus was activated by mixing 0.8ul acetylated trypsin and 800ul RV-SA II (1ul/1mL) and incubated in a 37 °C water bath for 1 h. After that 800ul of the activated virus was added to 4 mL MEM media (LifeTech.) and mixed well. Then complete media was removed from wells and washed with 100ul 1x DPBS (LifeTech.) & 300ul of the virus containing MEM media was added to each well and incubated for 1 h in 37 °C incubator. After 1 h the virus containing media was removed from each well and washed with 100ul 1x DPBS and 1 mL MEM media was added to each well and the plate was placed in 37 °C incubator for 6 h. All the processes were carried out for mock wells but without virus. After 6 h, media was removed from all the wells and washed with 100ul 1x DPBS and cells were lysed with PLB buffer (Promega). 20ul of each sample was used to measure Luciferase activity.
For polyI:C treatment, HEK293T cells were seeded in 12-well plates; at 70–80% confluency TFs p50, p65, IRF3 and IRF7 at 50 ng/well, pRLTK at 0.1 ng/well, 100 ng/well of luciferase constructs (p1.4IFNL3-3’ UTR) were transfected. After 24 h, 2.5 ug/well polyI:C (Sigma) was added to media. Luciferase assays were carried out as above after 8 h of further incubation.
2.6. Transient chromatin immunoprecipitation (ChIP)
HEK293T cells in 100 mm dishes at a seeding density of 4X106 were used. Plasmids used for the experiments are pUNO empty vector (as vector control), IRF7-HA in pUNO and p65 in pUNO background. 5ug of each plasmid was used per dish, total DNA amount was kept constant by using pUNO in case of single plasmid transfection. In all the dishes 1ug of p0.5kbIFNL3 construct was also co-transfected. Transfection was performed by using Poly (ethyleneimine) solution (1 ug/ul) (PEI, branched, Cat# 408727-100ML, Sigma) in a ratio of 1:3 (DNA amount: PEI). SimpleChIP® Enzymatic Chromatin IP Kit (Magnetic Beads) (Cat#90035, Cell Signaling Technology) is used for the entire procedure and standard protocol given by the manufacturer was followed. For chromatin immunoprecipitation anti IRF-7 antibody (Cat#ABF130, Sigma-Aldrich) was used. The following primers were used in PCR and qPCR: Vector specific Forward: 5’-GGTGCCAGAACATTTCTCTA-3’ and Reverse: 5’-GGCTTTACCAACAGTACCGG — 3’; IRFa, NF-KBa Reverse: 5’-AGCCTCAGGTAAGACACCG - 3’; NF-κBc Forward: 5’-AAAGACCAGA-GATCAGGAAT-3’; NF-κBa, IRFb, NF-κBb, NF-κBc Forward: 5’-CCTCCAGCTGCTCATCTGGC -3’ and IRFa, NF-κBa, IRFb, NF-κBb Reverse: 5’ - GCTTTTATCCCTGACAGA — 3’. qPCR data was obtained by using Bio-Rad iTaq™ Universal SYBR Green supermix (California, USA) in CFX-96 Real-Time System (Bio-Rad, USA). Data was analysed by using % Input method (thermofischer.com).
2.7. Bimolecular fluorescence complementation assay (BiFC)
For BiFC, IRF7 gene was cloned in Flag-GFP11-pDEST-N vector and p65, IRF3 genes were cloned in HA-GFP1-10-pDEST-C vector (both the plasmids were a gift from Maria Vartiainen (Addgene plasmid # 118366; http://n2t.net/addgene:118366; RRID: Addgene_118366). HEK293T cells were seeded in 12-well plates at a density of 0.1X106. 250 ng each of the plasmids encoding different TFs were transfected. Transfection was performed by using Poly(ethyleneimine) solution (1 ug/ul) (PEI, branched, Cat# 408727-100ML, Sigma) in a ratio of 1:3 (DNA amount: PEI). After 24 h, transfected cells were harvested carefully and re-seeded in 96-well plate at a density of 18,000 cells/well. Next day, images were taken from live cells using EnSight multimode plate reader (PerkinElmer, USA) using GFP filter. Quantification and image analysis were performed in Kaleido data acquisition and analysis software integrated in the machine.
2.8. Co-immunoprecipitation and western blot
HEK293T cells were transfected with 6 ug of plasmids encoding p65, IRF3-HA and IRF7-HA genes in 100 mm dishes. Transfection was performed using Poly(ethyleneimine) (PEI) solution (1 ug/ul) (PEI, branched, Cat.# 408727-100ML, Sigma) in a ratio of 1:3 (DNA amount : PEI). For lysis and immunoprecipitation (IP), Pierce Magnetic HA-Tag IP/Co-IP kit was used. Cells were lysed with 500 μl of IP Lysis buffer supplemented with protease inhibitor cocktail and 100 mM PMSF. Lysates were incubated with beads at RT for 30 min. After performing IP, proteins were eluted from the beads with 60 μl of 2X Laemmli buffer by boiling for 10 min.
Western blot of both the input and IP samples were performed after running the input or immunoprecipitated protein on 10% SDS-PAGE gels. After the transfer, the blot was probed with Anti-NF-κB p65 primary antibody (Anti-NF kB p65, CT, polyclonal antibody, Cat.# 06-418 by Merck-Millipore). Secondary antibody incubation was performed for 1 hr using HRP conjugated Anti-Rabbit secondary antibody (Cat.# 12-348 by Merck-Millipore). Blots were developed and imaged using Biorad Chemidoc MP imaging system (Universal hood III). To check for endogenous p65 expression, HEK293T cells were seeded in 100 mm dishes and then transfected with plasmids pUNO empty vector, IRF3-HA, IRF7-SA and p65 (all in pUNO background). 6 ug each of respective plasmids were used per dish; pUNO vector was used to keep total DNA concentration as a constant in all dishes. Transfection was performed using PEI as above in a ratio of 1:3 (DNA amount: PEI). Western blots were carried out to probe for endogenous or exogenous p65 as above.
2.9. Data analysis
All comparisons of transcription activity between various constructs were performed using one-tailed or two-tailed Student-t test for independent means. A p-value of 0.05 was considered significant. All Luciferase experiments carried out to measure IFN-λ3 promoter activity involving the different genetic variants were carried out as three biological replicates (i.e., three separate plasmid preparations were used after normalizing the concentration of plasmids carrying alternate alleles or haplotypes), wherein plasmids were transfected in single, duplicate or triplicate wells of HEK293T cells for each biological replicate plasmid sample (as mentioned in the figure legends). The ratio values of each FF/Ren. Luc activity reading shown in different figures in this work are tabulated in Suppl. Table 1. Mean values of luciferase gene expression data form nine constructs were used to generate a dendrogram in the clustering analysis. Clustering the data with function “heatmap” was carried out by using R programming (R core team 2019).
3. Results
3.1. Effects of IFN-λ3 SNPs rs28416813 and rs4803217 on reporter gene expression
The SNP rs4803217 (C/A) which overlaps with a miRNA binding site within the IFN-λ3 3’UTR was shown previously to interfere with AMD pathway [29] and to affect mRNA stability [44]. Both reports showed that allele A reduced reporter gene expression compared to allele C by 30-50%. Similarly, our previous report on rs28416813 (C/G) showed that the G allele decreased reporter gene expression by 20–30% compared to the C allele [28]. The A allele at rs4803217 and the G allele at SNP rs28416813 happen to be the ancestral alleles [4] (Fig. 1A). The ancestral alleles at both the SNP sites are expected to be together in human haplotypes due to prevalent LD in majority of the world populations [4]. Therefore, to test the effect of both the SNPs on gene expression, we cloned the IFN-λ3 3’UTR containing the two alternate alleles (C/A) at rs4803217, downstream of the Luciferase gene in our IFN-λ3 promoter clones containing the C and G alleles at rs28416813 (Fig. 1A lower panel).
We transfected HEK293T cells with equal amounts of expression plasmids coding for the NF-κB components p50 (NF-κB1) and p65 (RelA) along with the constructs carrying either variant alleles only at rs28416813 (C or G) or the variant haplotypes at rs28416813 and rs4803217 (CC or GA for the two SNPs respectively) (Fig. 1B). In this experiment we see an insignificant effect of the G allele at rs28416813 compared to the C allele; however, we see a significant decrease in reporter gene activity when both ancestral alleles G and A at SNPs rs28416813 and rs4803217 respectively were present together compared to the non-ancestral alleles (Fig. 1B). Similar result was seen when IRF3 and IRF7 TFs were co-overexpressed along with p50 and p65 (Fig. 1C). Next, to determine the effect of each of the alleles at the two SNP positions individually, we made constructs carrying the other combinations of the ancestral and non-ancestral alleles at the two SNP positions and repeated the experiments with p50 and p65 (Fig. 1D). From this analysis, we see that the major/non-ancestral alleles at the two SNP positions have a dominant effect on each other in keeping the reporter gene expression high, while the presence of both the ancestral alleles simultaneously was necessary to keep the reporter gene expression significantly low. In other words, each SNP shows the effect only in the minor/ancestral allele background of the other SNP. We confirmed that the minor/ancestral alleles present together at both SNP positions showed a significant decrease in reporter gene expression compared to the major/non-ancestral alleles in a Rotavirus infection model system (Suppl. Fig. 1).
3.2. Molecular characterization of the differential allelic effects of the SNP rs28416813
Next, we wanted to further explore the molecular mechanism behind the SNP rs28416813 affecting transcription. We carried out EMSA with fluorescently labelled probes to test for the binding affinity of the two alleles at rs28416813 for NF-κB (Suppl. Fig. 2). We expressed a fulllength NF-κB p50 protein in E.coli and purified it to homogeneity (Suppl. Fig. 2A) and used it in EMSA. The results of EMSA show that the C allele binds more recombinant p50 than the G allele at rs28416813 in the probe. Further, a cold oligonucleotide that had the consensus NF-κB binding DNA sequence competed out the labelled probes for p50 binding showing the specificity of the interaction. We tested the binding of p50 with both probes at increasing concentration of the recombinant protein (Suppl. Fig. 2B). While the C allele showed a higher binding with increasing concentration of p50, the G allele on the other hand showed lower binding throughout and seems to saturate at higher protein concentration. These results show the higher binding affinity for the C allele at rs28416813 with NF-κB p50. We attempted to similarly express and purify the full-length NF-κB p65 in E.coli, but the recombinant protein was unstable and could not be purified.
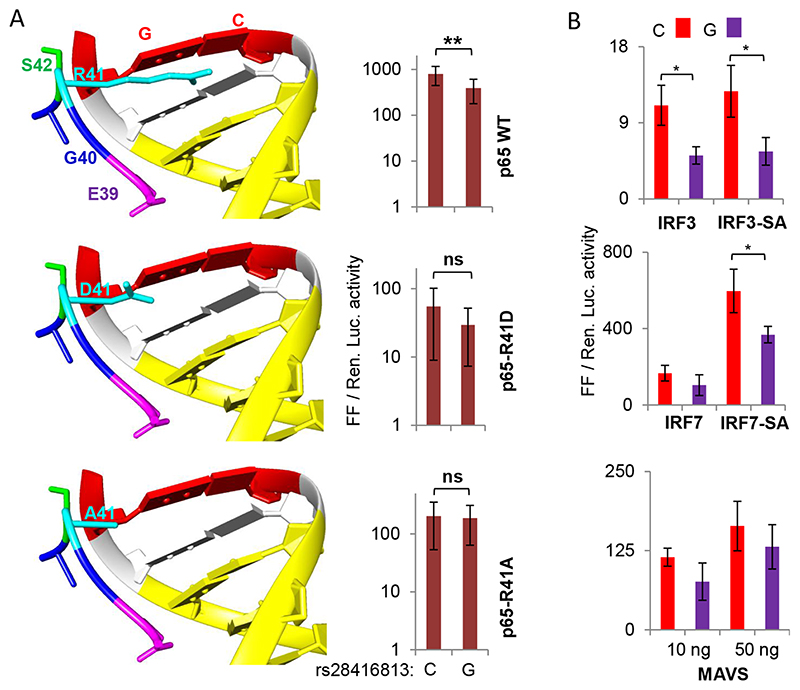
Fig. 2.
A) (Left) R41 of NF-κB interacts with SNP rs28416813. The models show cocrystal structures of NF-κB p50-p65 heterodimers and their cognate DNA together (PDB ID2O61 visualized in UCSF CHIMERA). The peptide shown is that of p65 carrying the wt R41 and the mutants D41 and A41 along with some nearby residues. The NF-κB DNA binding site (10 bp) is shown in yellow and the site at equivalent position to that of the SNP rs28416813 is in red (12th bp). (Right) Equal amounts of plasmids encoding NF-κB p50 and either WT or mutant p65 were used as TFs to stimulate the IFN-λ3 promoter. The construct p1.4kbIFNL3 was used in the experiments with C or G alleles at the SNP rs28416813; the experiments were carried out on HEK293T cells grown in 96-well plates. The data is drawn from nine experiments representing three different plasmid preparations. The mean value from all nine experiments is shown, while the error bars depict SD. B) Overexpression of phosphomimic (constitutively active) versions of IRF3 and IRF7 and the mediator MAVS also reveals differences in transcriptional activity of p1.4kbIFNL3 carrying C or G alleles at rs28416813. The experiments were carried out on HEK293T cells grown in 96-well plates. The data is from three experiments performed with three different plasmid preparations. The mean value from the three experiments is shown, while the error bars depict SD. For both A and B, two tailed student t-test for independent means was used for calculating statistical significance; *p < 0.05, **p < 0.01; ***p < 0.001. All the Luciferase ratio values from each individual experiment are shown in Suppl. Table 1.
We, therefore, tested the interaction of NF-κB p65 with the SNP rs28416813 in a more physiological condition (Fig. 2A). We had predicted that the residue R41 of NF-κB p65 may be interacting with the G-C base pair in the DNA oligonucleotide representing the SNP rs28416813 in the cocrystal structures of the NF-κB p50-p65-DNA complex [28] (12th base pair from the beginning of the NF-κB DNA binding site in the cocrystal structure represents the position equivalent to the SNP position of rs28416813 in the natural IFN-λ3 5’UTR DNA) (ref [28] and Fig. 2A, left). A closer look at the cocrystal structure of NF-κB p65 and the DNA oligo shows that R41 even though has an interaction (<3 A) with the Guanine base situated at a similar position to that of the SNP rs28416813, it is also possible that R41 may have a salt bridge interaction with a nearby residue E39 (Fig. 2A left). To test this possibility, we mutated R41 to D41 and A41 and subjected the constructs to luciferase assay as before along with WT NF-κB p50 and D41 or A41 NF-κB p65. The change from R41 to D41 in p65 drastically reduced the expression of the reporter gene suggesting that the 41st residue is an important one possibly for maintaining the stability of the NF-κB p50-p65 heterodimer and its function as a TF; the change from Arginine to Alanine at the 41st residue partially restored the WT activity of p65 (Fig. 2A, right). Importantly when the mutant p65 were used, we saw that the difference in luciferase activity between the C allele and G allele at rs28416813 lost significance especially with the A41 mutation. These results suggest that our prediction about the R41 residue of p65 contacting the SNP site may be correct.
After establishing this, we wanted to see how the two alleles of the SNP respond in absence of overexpressed p50 and p65 in HEK293T cells. For this purpose, we overexpressed IRF3 or IRF7 either in their unstimulated forms or as their ‘phosphomimic’ constitutively active or superactive (SA) forms (IRF3-SA and IRF7-SA) (Fig. 2B) along with the p1.4kbIFNL3 constructs carrying the two alleles C and G of rs28416813 (Fig. 2B). While there was little difference in promoter activity while overexpressing IRF3 or IRF3-SA, the phosphomimic version of IRF7 showed significantly increased activity of the promoter. But surprisingly, the C and G alleles of rs28416813 showed significant differences in the reporter gene expression when the phosphomimic version of IRF3 and IRF7 were overexpressed as TFs in absence of any exogenous p50 or p65. This would suggest that either there is a yet unidentified IRF binding site overlapping or in the vicinity of the SNP site or the IRFs were activating the endogenous NF-κB components leading to an effect on the reporter gene expression due to the latter’ s differential binding to the NF-κB site near the SNP site (Fig. 2A left). Overexpressing the mediator protein MAVS (mitochondrial antiviral signalling protein) that activates endogenous IRF3/7 and NF-κB [46] also had a similar but insignificant effect on the two alleles of the SNP (Fig. 2B). These results are indeed surprising as there has been no reported IRF-binding site near the SNP rs28416813 [43] suggesting the alternative possibility of the endogenous NF-κB being activated by the mediator MAVS or the IRFs. Since there would be substantial amount of IFN production due to the activity of the overexpressed IRFs, it is possible that endogenous NF-κB is activated by the IFNs. In that case, the endogenous NF-κB present at relatively lower amounts should compete with the overexpressed IRFs such that their effect on the SNP would be revealed. This is possible when there is some interaction between NF-κB and the IRFs.
3.3. Interaction between NF-κB and IRFs on the IFNL3 promoter
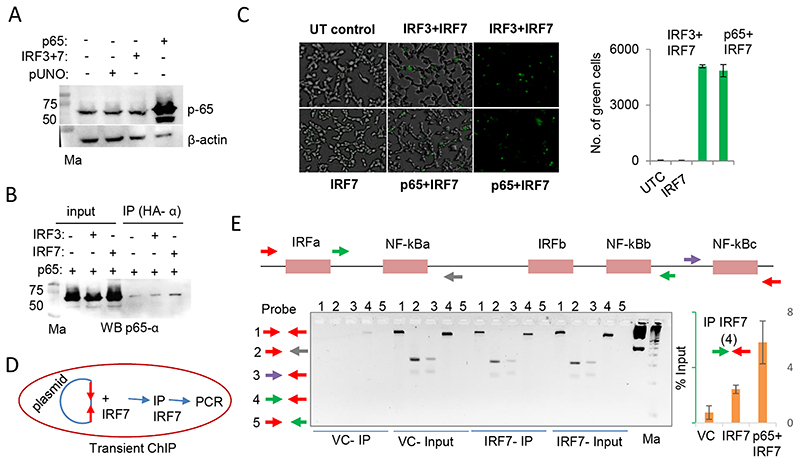
We checked by western blot to see if p65 was endogenously expressed in unstimulated HEK293T cells (Fig. 3A). We saw that there was significant amount of p65 that was endogenously expressed in the HEK293T cells without stimulation and this level was unaffected by overexpression of both IRF3 and IRF7. An interaction between NF-κB p65 and IRF3 has been previously reported [47]. However, there has been no reports to the best of our knowledge to show that NF-κB interacts with IRF7. We wanted to examine this further and used two different techniques for this purpose: co-immunoprecipitation (co-IP) and bimolecular fluorescence complementation assay (BiFC) [48]. Using co-IP we saw that IRF7 and p65 were present in a complex in HEK293T cells. The interaction between IRF7 and p65 seems to be stronger than that between IRF3 and p65 (Fig. 3B). Using BiFC we saw that the interaction between IRF7 and p65 was as efficient as that between the known interactors IRF3 and IRF7 [49] (Fig. 3C). These results show that IRF7 and p65 physically interact with each other. However, we were interested to see if there is an interaction between the two TFs on the IFN-λ3 promoter itself. For this, we used a transient chromatin immunoprecipitation (ChIP) assay [50] (Fig. 3D). There are three known NF-κB and two known IRF binding sites that have been defined for the IFN-λ3 promoter within 500 bases of the proximal promoter [43,51]; hence we used the p0.5kbIFNL3 construct that we are using for reporter gene expression in this study (Fig. 3E top and see later). First, we standardized the transient ChIP assay by overexpressing IRF7 as TF and transfecting p0.5kbIFNL3 construct in HEK293T cells (Fig. 3E bottom left); after pulling out the promoter bound IRF7 complexes and reversing the crosslinks we used five different primer sets to amplify different regions of the construct by PCR; we found that primer set 4 to be ideal considering the length and coverage of the TF binding sites and the input to IP ratio. In the next round of transient ChIP, we overexpressed IRF7 alone and in combination with p65 in HEK293T cells and pulled down the complexes of plasmid DNA-bound IRF7 and p65 by using the IRF7 specific antibody. We found that the 479 bp amplicon generated by the primer set 4 to be suitable for quantifying the input DNA by qPCR (Suppl. Fig. 3). Using qPCR with the primer set 4 we quantified the amount of plasmid construct DNA that was pulled down in the ChIP reaction. The results showed that there was more than a two-fold enrichment of promoter-bound IRF7 when p65 was co-overexpressed (Fig. 3E bottom right). These results show that there is a cooperative interaction between p65 and IRF7 on the promoter DNA such that the promoter occupancy of IRF7 is increased in presence of p65. We were interested to see the functional impact of such a cooperation on the transcriptional activity of the IFN-λ3 promoter.
Fig. 3. NF-κB p65 interacts with IRF7 and increases its promoter occupancy.
A) Western blot (WB) showing endogenous expression of p65 in unstimulated HEK293T cells. The overexpressed TFs in different lanes are shown on the top of the blot. B) Co-IP experiment showing the physical interaction between IRF7 and NF-κB p65. The HA tags present in the IRF3 and IRF7 constructs were used to pull down the complexes before probing for p65 using a specific antibody. The blot shown is a representative of two independent Co-IP experiments. C) Bimolecular Fluorescence Complementation Assay (BiFC) shows that IRF7 interacts with p65. UT-untransfected control. The left and middle images show merged brightfield and fluorescence views; the right image is only from the fluorescence view. The quantification shown on the right graph was carried out with Kaleido (PerkinElmer). D) Schematic showing the flow of transient ChIP experiment. The p0.5kbIFNL3 construct was co-transfected along with IRF7 and ChIP was performed with anti-IRF7 antibody. E) (Top and bottom left) Standardization of primer sets for transient ChIP. After pulling down the complexes and reversing the cross-links PCR was performed with the DNA obtained using 4 different primer set combinations. The top shows the schematic of the five different IRF or NF-κB binding sites on the p0.5kbIFNL3 promoter construct. Each arrow depicts a primer. The red arrows are specific for primers entirely binding in the pGL3basic plasmid DNA sequence; the remaining arrows show primers that bind within the 0.5 kb promoter encompassing different TF binding regions. The gel picture shows the PCR products run on a 2.5% EtBr-stained agarose gel. First marker (Ma) is 1 kb ladder (GeneRuler 1 kb DNA Ladder, ready to use, Cat#SM0313,ThermoFisher Scientific, USA); second marker is 50 bp ladder (TrackIt™ 50 bp DNA Ladder, Cat#10488-043, ThermoFisher Scientific, USA). (bottom right) qPCR showing the enrichment of IRF7-bound DNA which is increased greater than 2-fold when p65 is co-transfected with IRF7. The primer set 4 was used for the qPCR (the forward primer binds downstream of NF-xBa site and the reverse primer binds within the plasmid DNA sequence). The mean values from triplicate experiments are shown, error bars depict SD. VC-vector control.
3.4. Characterization of a synergistic cooperativity between TFs at the IFN-73 promoter
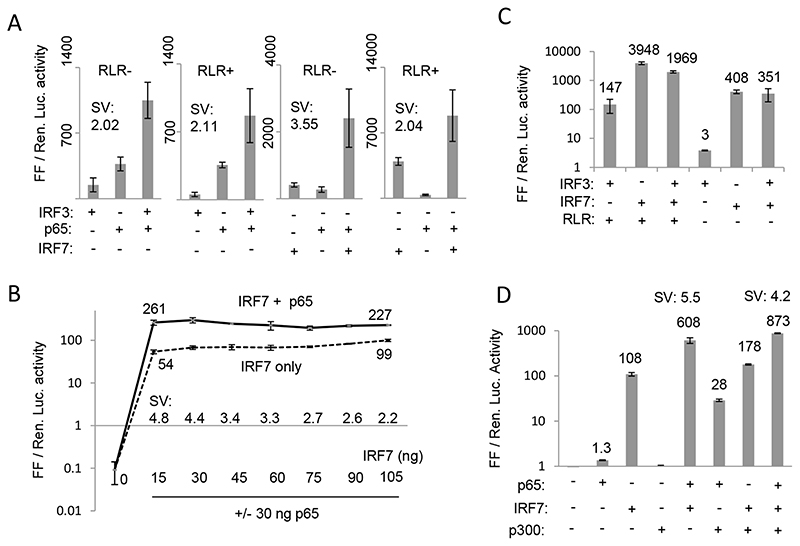
The well-characterized IFN-β promoter is known to assemble an enhanceosome comprising of several TFs which can interact with the coactivator p300 protein and synergistically activate transcription [52]. It is not known if such a mechanism also exists for the IFN-λ3 promoter. IRF and NF-κB TFs are known to be functional at the IFN-λ3 promoter [43,51]. We decided to test the most important viral-associated TFs: IRF3 and 7 and NF-κB p50 and p65 for any synergistic transcriptional cooperation at the IFN-λ3 promoter. We either co-overexpressed IRF3, IRF7 and the NF-κB p50 and p65 components individually or together along with the IFN-λ3 promoter construct (p1.4kbIFNL3 not including the 3’ UTR) that contained the C allele at rs28416813. Since NF-κB p50 does not have a transactivating domain, co-overexpression of this TF did not induce any luciferase expression and when present together with NF-κB p65 it decreased the luciferase activity by ~ 50% compared to the activity when NF-κB p65 was present alone (Suppl. Fig. 4). Therefore, we used only NF-κB p65 and IRF3 and IRF7 in our experiments on synergy (Fig. 4). A synergy value (SV) was defined as the ratio of luciferase activity when two or more TFs were present together in the assay to the sum of the luciferase expression activity of the individual TFs. An SV of more than 1 suggests a synergistic cooperation between the TFs. Fig. 4A shows that both combinations of IRF3 + p65 and IRF7 + p65 showed synergistic cooperation on transcription. To mimic viral infection, we used the HCV RNA-dependent RNA polymerase (RdRp) that can synthesize small RNA molecules and stimulate the RIG-I-like receptor (RLR) pathway as shown previously (Fig. 4A; RLR + or RLR-) [53]. Using this method of RLR pathway stimulation we saw that IRF7 increased luciferase activity by ~ 10 fold compared to the activity when RLR pathway was not stimulated (Fig. 4A); a comparable level of stimulation of IRF7 activity has also been shown upon Sendai virus infection in a previous report on the type I IFN promoter activity suggesting that HCV RdRp could efficiently mimic viral infection by optimally stimulating IRF7 [49]. The results show that the maximal SV could be achieved with IRF7 + p65 combination in absence of RLR pathway stimulation (Fig. 4A). We saw a consistent level of SV of more than 3.5-4.0 in multiple independent experiments (not shown).
Fig. 4. Transcriptional synergy at the IFN-λ3 promoter.
A) Synergy between IRF3/7 and NF-κB p65 with RLR pathway stimulated (RLR +) or not (RLR-). The RLR pathway was stimulated when indicated by co-overexpression of HCV NS5B and RIG-I genes as described before (53). B) p65 enhances IRF7-mediated transcription at the IFN-λ3 promoter. Synergy between IRF7 and p65 at increasing concentration of IRF7 when p65 is held at a constant concentration (30 ng) is shown. C) No evidence of synergy between IRF3 and IRF7 either with or without (+/-) RLR pathway stimulation. D) CBP/p300 has no role in transcriptional synergy at the IFN-λ3 promoter. Synergy between IRF7 and p65 with the p1.4kbIFNL3 promoter clone is shown in presence or absence o of overexpressed p300. All experiments in A, B, C and D were done as three separate experiments with a single plasmid preparation. The results show mean values from the three replicates with error bars showing SD. The mean values are depicted near the curve or above the respective histograms in B, C and D. SV = synergy value defined as ratio of mean Luciferase expression value when both TFs are present together to that of the sum of mean Luciferase expression values when the two TFs are present individually. An SV of more than 1 suggests a synergistic cooperation between the TFs. All the Luciferase ratio values from each individual experiment are shown in Suppl. Table 1.
Next, to further characterize this novel observation, we transfected increasing amount of the IRF7 expression plasmid while keeping a constant (30 ng) concentration of the p65 expression plasmid (Fig. 4B) and tested for IFN-λ3 promoter activity; a control consisted of IRF7 plasmid transfected alone without p65. This experiment would tell us if the synergy we observe between IRF7 and p65 is an artifact of the experiment because of a mere additive effect between the TFs that may be slightly amplified and interpreted as synergy. In fact, the results show contrary evidence. While an increase to 7-fold in IRF7 concentration (15 to 105 ng) could not achieve any significant increase in transcription (mean luciferase expression increased to 99 from 54), maximum SV was seen with just 15 ng of IRF7 and 30 ng of p65. This suggested that it was not a deficiency of IRF7 at the IFN-λ3 promoter that was compensated for by p65 in enhancing transcription and therefore was not a mere additive effect between two TFs. In fact, an increasing IRF7 concentration had a shallow increase in transcription (mean values 54 to 99) in absence of any synergy, while synergy concomitantly decreased from 4.8 to 2.2 suggesting clearly that there was a deficiency of p65 to cooperate with increasing concentration of IRF7 and therefore led to a decrease in synergy. This would suggest that IRF7 binding to the promoter may be the rate-limiting step in transcription from the IFN-λ3 promoter that can be overcome by presence of p65 in agreement with our results from the transient ChIP experiment (Fig. 3E) wherein we saw that promoter occupancy of IRF7 is enhanced by p65. Furthermore, a similar synergistic interaction to enhance transcription was absent between IRF3 and IRF7 (Fig. 4C) even though the two TFs are known to physically interact with each other [49]. These results clearly establish that there is a synergistic cooperation between the TFs IRF7 and p65, possibly due to better promoter occupancy of the former in presence of latter.
The transcriptional coactivator CBP/p300 increases the transcription efficiency of individual TFs at the interferon beta promoter/enhancer site by interacting with p65 through a synergism-specific domain [54]. Therefore, we overexpressed p300 to see if such an effect can be seen with the IFN-λ3 promoter (Fig. 4D). Surprisingly, while we saw an ~ 21-fold increase in p65-mediated transcription and an ~ 1.6-fold increase in IRF7-mediated transcription, there was no positive effect on synergistic activation (SV 5.5 in absence of p300 and 4.2 in presence; Fig. 4D). This would suggest that the IFN-λ3 promoter is regulated differently than interferon beta promoter/enhancer at least in terms of requirement of CBP/p300.
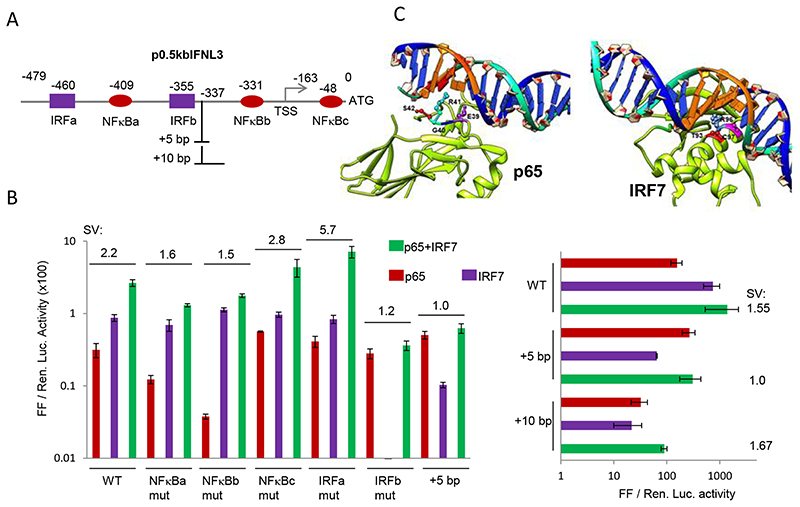
A previous report has characterized the IFN-λ3 promoter and defined two IRF binding sites and two NF-κB binding sites located within the 0.5 kb of the promoter [51] (Fig. 5A). We tested the shortened IFN-λ3 promoter (p0.5kbIFNL3) for synergy and saw that the 0.5 kb promoter retained synergy albeit at a lower level (SV of 2.2; Fig. 5B). To examine if the synergistic activity can be disrupted by interfering with the binding of TFs, we systematically mutated the four TF sites defined by the previous study [51] and included the additional NF-κB site (NF-κBc) present in close proximity to the SNP rs28416813 [28,43], for the mutational analysis (Fig. 5A and B). To ensure maximal loss of TF binding, we introduced mutations to disrupt the entire binding site rather than introducing just point mutations (refer to Material and Methods). Mutational analysis of the three NF-κB binding sites showed that NF-κBb was the most important site for p65 binding followed by NF-κBa. NF-κBc was not needed for p65-mediated transcription, in fact we saw an increase in transcription while it was disrupted suggesting a possible negative regulation of transcription by this site which is present downstream of the TSS. While mutations at both NF-κBa and b affected transcription when p65 was transfected alone, both mutations decreased synergy between p65 and IRF7 compared to the WT p0.5kbIFNL3 promoter construct; disrupting NF-κBc site, however, had a small positive effect on synergy (Fig. 5B). These results suggest that even though NF-κBc is dispensable for transcriptional activity of the IFN-λ3 promoter, its presence, especially close to a SNP site, may be used to regulate transcription. Similarly, mutational analysis of the two IRF binding sites showed that IRFa site was not needed for IRF7 binding but IRFb was most important as there was a complete loss of transcription mediated by IRF7 by the IRFb mutation. In agreement with this result, IRFb mutation had the maximal disruption on synergy (SV of 1.2) compared to all the five mutations. Interestingly, while disrupting IRFa site did not adversely affect either p65 or IRF7-mediated transcription individually, it increased the synergy between them (from 2.2 to 5.7).
Fig. 5. Characterization of transcriptional synergy between IRF7 and NFkB p65.
A) Schematic showing the different IRF and NF-κB binding sites as defined by a previous study (51) along with the NF-κBc site that was characterized by Osterlund et al., (2007) (43) within 0.5 kb of the IFN-λ3 promoter; the numbers show the beginning of each of the site and the site of insertion of 5 bp or 10 bp. B) Mutational analysis of the p0.5kbIFNL3 construct shown in (A). The results show mean values from three separate experiments carried out with a single plasmid preparation with error bars showing SD. SV (synergy value) is defined in Fig. 4. The wt and the mutated sequences are shown in Materials and Methods section; All the Luciferase ratio values from each individual experiment are shown in Suppl. Table 1. C) The nature of DNA binding of NF-κB p65 and IRF7 are distinctly different. Models showing cocrystal structures of TFs NF-κB p65 and IRF7 in their DNA bound forms. It is clear from the structural models that p65 binds to DNA mainly through loops in its structure and does not involve any stable structures while IRF7 has a short helix compactly placed inside the major groove with several amino acids contacting the bases inside the groove. This observation for p65 has also been noted by others who reported the structures (61) (PDB ID2O61 visualized in UCSF CHIMERA).
Interfering with the ‘DNA helix phase’ can influence TF binding and/ or their interactions with other TFs [54]. This can be achieved by inserting 5 bp in the DNA to break the phase or 10 bp to restore it [54]. To test this possibility, we introduced a 5 bp DNA fragment between IRFb and NF-κBb, the two critical binding sites for the two TFs (Fig. 5A). This disruption of the DNA helix by ~ half turn did not affect p65-mediated transcription but substantially decreased IRF7-mediated transcription, suggesting that the IRF7 is more sensitive to changes in DNA phasing (Fig. 5B). Importantly, synergy was completely abolished by this insertion (SV of 1). When we attempted to bring back the DNA helix phasing by introducing an additional 5 bp to the previous insertion, the individual transcriptional activity of both IRF7 and p65 decreased, but importantly, synergy was restored (Fig. 5B, right). Synergy was most negatively affected by two changes: 1) mutation of the IRFb site and 2) disruption of the DNA helix phase by a 5 bp insertion; most interestingly, these two changes were the only ones that affected IRF7-mediated transcription negatively (Fig. 5B left) suggesting that synergy is likely a reflection of enhanced IRF7 mediated transcriptional activity at the IFN-λ3 promoter. Since disruption of DNA helical phase phenocopied IRFb binding site mutation, this would suggest that the 5 bp insertion that led to change in the DNA phase likely affected IRF7 binding. Interestingly, p65 was unaffected by the 5 bp insertion (Fig. 5B) again implying that its binding to the promoter was not adversely affected by the change in DNA phasing. Our results suggest that there could be different effects of DNA phasing on p65 and IRF7. We explored to see if the mode of interaction adopted by the two TFs when they bind to their cognate binding sites are different (Fig. 5C). A closer look at the cocrystal structures of p65-DNA and IRF7-DNA shows that the two TFs have very different binding tendencies to their DNA partners. While p65 has only loops in its structure contacting the DNA binding site with no stable secondary structures like helices or sheets inside the major groove, IRF7 interacts with its DNA binding site through a stable helix compactly packed inside the major grove of the DNA binding site with several residues having important contact points within the groove (Fig. 5C). This discrepancy in binding to their cognate DNA may be affecting their efficiency in driving the promoter when DNA phasing is changed.
3.5. Effect of IFN-λ3 promoter SNP rs4803219 and the TA dinucleotide repeat polymorphism rs7225881 on reporter gene expression
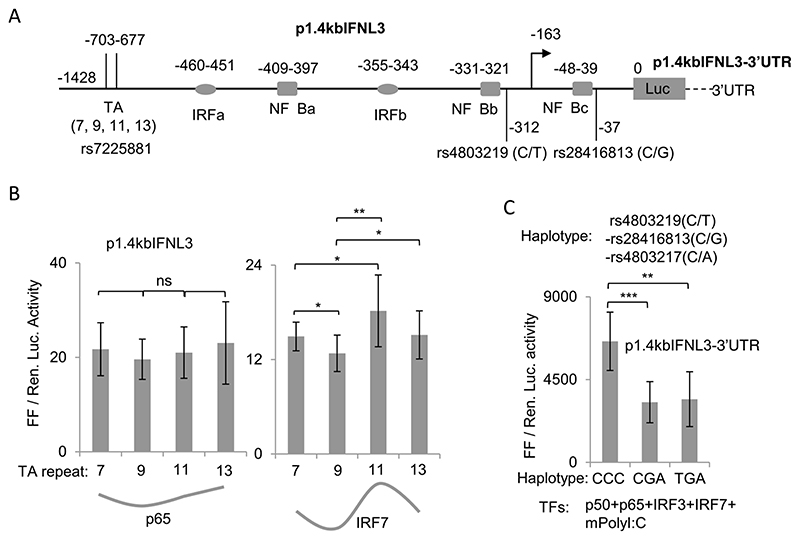
The effect on transcription due to changes in DNA phasing could be important at the IFN-λ3 promoter as this effect may be seen naturally as part of genetic variation linked with the promoter. A TA dinucleotide repeat polymorphism (rs7225881) has been reported to be present around 700 bp upstream to IFN-λ3 gene (Fig. 6A). A previous report showed that increase in the number of TA repeats at the polymorphic site increased reporter gene expression [26], and at least two genetic studies have shown that the TA repeat polymorphism associates with either response to therapy in chronic HCV infections or spontaneous clearance of HCV [40–41]. Therefore, we included different number of TA repeats into our promoter construct at the TA variant site (Fig. 6A). Our original p1.4kbIFNL3 construct was re-sequenced and found to have 7 TA repeats; we introduced 2, 4 and 6 repeats in to the p1.4kbIFNL3 construct to have additional promoter constructs with 9, 11 and 13 TA repeats (Fig. 6A). We conducted luciferase assays with these constructs in presence of NF-κB p65 and IRF7 to assess the effect of TA repeat number variation on transcription driven by these two TFs (Fig. 6B). As per our prediction based on results from our 5 bp and 10 bp insertion (Fig. 5B) and the mode of TF binding (Fig. 5C), we saw that the TA repeats affected IRF7 more than p65-driven transcription of the IFN-λ3 promoter (Fig. 6B). Moreover, the effect on IRF7-driven transcription by the TA repeats was ‘cyclical’. Clearly the TA repeats were affecting transcription possibly by changing the DNA helical phasing in the promoter downstream and the phase changes that occurred at the TF binding sites affected IRF7 binding more than p65 binding. Such a cyclic effect on transcription is also known to be caused by another TA repeat polymorphism present at 550 bp upstream in the NADPH oxidase gp91phox promoter [55].
Fig. 6. Effect of rs7225881 and rs4803219 on IFN-λ3 promoter activity.
A) Schematic of the p1.4kbIFNL3 construct used in the study showing the location of the SNPs rs4803219 and rs28416813 and the TA dinucleotide repeat polymorphism rs7225881 in the context of the different IRF7 and NF-κB TF binding sites. The exact location of each binding site from the ATG start site is shown above them. B) Effect of TA repeat-length on the IFN-λ3 promoter activity when tested against p65 or IRF7. The TA repeats were cloned at the site rs7225881 as shown in (A) in the p1.4IFNL3 construct carrying the C allele at rs28416813; this construct did not include the IFN-λ3 3’UTR. The data was drawn from 9 to 10 separate experiments representing three different plasmid preparations transfected in to HEK293T cells grown in 96-well plates. The histograms show the mean value of all the 9–10 experiments, while error bars are depicting SD. The grey line below the histograms shows the trend in mean Luciferase expression values for the different construct transfected with different TFs. C) The ancestral alleles of the three SNPs rs4803219, rs28416813 and rs4803217 significantly decrease transcription from the IFN-λ3 promoter. All four TFs as shown were transfected in to HEK293T cells grown in 12-well plates and further stimulated by polyI:C as described in the methods section. p1.4kbIFNL3-3’ UTR construct carrying the different haplotypes at the three SNP positions were used. The data is from six separate experiments representing three different plasmid preparations. The histograms show mean values from the six experiments with error bars showing SD. For B and C one tailed student t-test for independent means was used for calculating statistical significance; *p < 0.05, **p < 0.01; ***p < 0.001. All the Luciferase ratio values from each individual experiment for B and C are shown in Suppl. Table 1.
One of the three SNPs that were predicted to have regulatory functions on IFN-λ3 expression is rs4803219 [22,23]. We observe that rs4803219 is close to (8 base pair apart) the NF-κBb site in the IFN-λ3 promoter (Fig. 6A). Since we saw that NFκ-Bb was very important for p65-mediated transcription (Fig. 5B), it is possible that the SNP rs4803219 may also have an influence on NF-κB binding at this site and thereby could influence transcription. Therefore, we inserted the mutation at rs4803219 in to our IFN-λ3 promoter-3’UTR construct and tested for its effect (Fig. 6C). All four TFs: p50, p65, IRF3 and IRF7 were overexpressed and the cells were also stimulated by the RNA mimic polyI:C added into the medium (mpolyI:C). Our results show that the SNP rs4803219 may not have any significant effect on transcription from the IFN-λ3 promoter, at least in the conditions tested (Fig. 6C).
Therefore, we conclude that three genetic variants at the IFN-λ3 gene may be having functional roles: rs28416813 in the 5’UTR, rs4803217 in the 3’UTR and the TA repeat variant (rs7225881) in the promoter may be influencing IFN-λ3 gene expression based on a comprehensive analysis that we have carried out in this study.
3.6. Characterization of a CBP/p300 binding site in the distal region of IFN-λ3 promoter and natural variation in the IFN-λ3 promoter DNA derived from human samples and its effect on transcription
The transcriptional coactivator p300 functions by mediating protein-protein interactions to enhance transcription from enhancers and promoters. However, p300 is also known to bind to DNA to perform its enhancer functions. The p300 binding site GGGAGTG has been previously characterized [56] and known to have enhancer functions [57]. Interestingly, while scanning for different TF binding sites in the IFN-λ3 promoter we came across a p300 DNA binding site within the body of the IFN-λ4 gene which is situated in the distal IFN-λ3 promoter region (Suppl. Fig. 5A). A similar site was observed ~ 21 kb away at a similar position in the IFN-λ2 promoter. We were interested to test if this binding site is functionally important; we therefore cloned a 500 bp DNA fragment encompassing the p300 DNA binding site in to a pGL3promoter vector (Promega) which has an SV40 promoter that could report on any enhancer activity present in the cloned fragment. Indeed, upon p300 overexpression, we saw an enhancer activity present within this fragment and mutational analysis of the p300 DNA binding site confirmed this observation (Suppl. Fig. 5B). We next used a 4 kb IFN-λ3 promoter (carrying C allele at rs28416813) construct that included the proposed p300 DNA binding site and subjected to luciferase assays in presence and absence of co-overexpressed p300 protein to check for synergy between IRF7 and p65 (Suppl. Fig. 5C). Interestingly, we saw that the SV value was higher (greater than7.0) with the p4kbIFNL3 construct than that observed with p1.4kbIFNL3 construct (3.5–5.5; Fig. 4), which in turn was higher than that seen for the p0.5kbIFNL3 construct (1.55–2.2; Fig. 5B). However, we only saw a modest increase in synergy (SV 7.4 from 7.2) from the p4kbIFNL3 construct in presence of p300 (Suppl. Fig. 5C). Similarly, we had seen that there was no further stimulatory effect of p300 overexpression on synergy between the TFs in the p1.4kbIFNL3 construct also, even though it significantly increased transcription from p65 (Fig. 4D). Therefore, it is unlikely that p300 has any role in mediating the synergy between IRF7 and p65 at the IFN-λ3 promoter, unlike its role on the interferon beta promoter/enhancer [54].
We wanted to examine the natural genetic variation that may be present in the IFN-λ3 promoter and its possible influence on the promoter activity in the context of our results obtained so far. We amplified the 1.4 kb IFN-λ3 promoter and 5’UTR DNA from five HCV-infected patients from our cohort (described in Ref. [10]) and cloned it in pGL3basic (Promega) vectors. Among the five patients (A-E), four (A-D) were responders to the conventional IFN-a-Ribavirin therapy and one (E) was a non-responder. The other details of this patient cohort are provided elsewhere [10]. After cloning we isolated plasmids derived from each patient sample randomly from colonies and subjected to DNA re-sequencing of the entire cloned promoter-5’UTR DNA fragment. We found that four patients (A-D) were heterozygous at various positions and one patient (E) was a homozygote from this limited analysis (Suppl. Fig. 6A). Sequencing results showed the following: a) we saw variation at five SNP sites defined in the dbSNP (including rs4803219 and rs28416813); b) nineteen variations were observed that have not been reported in dbSNP, all of them seem to be rare variants and c) high level of variation was present at the TA dinucleotide repeat polymorphism (7, 11, 12 and 13 TA repeats). We saw that rs28416813 was maximally correlated to rs4803219 and another SNP in the region rs75158108 (Suppl. Fig. 6B) but not with other SNPs. None of the rare variants showed any significant influence on transcription (Suppl. Fig. 6C). The SNP rs28416813 seem to have a strong effect on transcription but confounding effects from other SNPs like rs4803219 (Suppl. Fig. 6D), rs75158108 and the TA repeat polymorphism could not be verified in our limited experiments carried out from a small sample size.
4. Discussion
The initial GWAS for HCV identified the IFN-λ locus to be behind response to therapy in chronic HCV infections [1–3], but later studies also showed the same set of variants to be responsible for spontaneous clearance of HCV [14–17]. The search for the possible causal variants was taken up by several groups around the world including those that conducted the GWAS [1–3]. Ge et al., (2009) [1] in their GWAS report on response to therapy in chronic HCV infections, had also performed fine-mapping at the IFN-λ locus and identified two potential causal SNPs rs28416813 and rs8103142; SNP rs28416813 was also identified by fine-mapping in the Tanaka et al, study [3]. The most significant study conducted to identify candidate causal SNPs/variants responsible for spontaneous clearance of HCV infections, happens to be that conducted by Rauch and others (2011) [22] and de lulio and others [23]. In these studies, two important conclusions were drawn: 1) The SNPs rs4803219, rs28416813, rs8103142 and rs4803217 are likely to be causal variants responsible for spontaneous viral clearance in both a single-source (exposed to a single HCV genotype) and a multiple-source (exposed to multiple HCV genotypes) HCV cohort; 2) The candidate causal SNPs are highly correlated to the SNP rs12979860. The studies suggested that the four identified SNPs within and around the IFN-λ3 gene were ideal to be included for further functional analysis. The study also found no evidence for any structural (copy number) variants that could be causal. These four candidate SNPs also show a better predictive value for response to conventional therapy against HCV [24]. Interestingly, a study also showed that three out of the four candidate causal SNPs (rs28416813, rs8103142 and rs4803217) were among the five SNPs that were under strong selection pressure in the Asian population [58].
When several reports were emerging about association of IFN-λ locus genetic variants with expression of IFN-λ3 gene/protein in human samples, it was thought that a genetically controlled IFN-λ3 gene expression could be the causal mechanism behind the HCV GWAS [8]. During this time, a novel type III IFN, IFN-λ4 was identified and this discovery has convincingly explained the genetic association of IFN-λ locus variants with response to therapy in chronic HCV infections [7]. However, the expression of a novel IFN-λ4 gene that could somehow be responsible for non-clearance of HCV infections at the acute phase without any therapy is not sufficiently convincing to explain spontaneous viral clearance, especially considering a recent report [18] that shows the possibility of variants other than those linked with IFN-λ4 expression and activity, like rs368234815 and rs117648444 (P70S), could have independent roles in spontaneous HCV clearance. Therefore, we need further research to understand how the IFN-λ locus variants control spontaneous clearance after an acute HCV infection.
Furthermore, even in HCV clearance after therapy, the role of IFN-λ3 gene cannot be undermined given its immunological significance. Out of the four candidate causal SNPs identified by Rauch and others (2010) [22] and di Iulio and others (2011) [23], two SNPs rs28416813 and rs4803217 situated in the 5’UTR and 3’UTR genes respectively seem to have potential roles in regulating transcription and translation of IFN-λ3 gene [28–29]. The two non-ancestral alleles in these two SNPs could increase IFN-λ3 gene expression while the ancestral allele background gives rise to IFN-λ4 gene [19]. Therefore, the effect of genetic variants influencing IFN-λ genes seem to be functioning reciprocally: an increase in IFN-λ3 gene expression by presence of non-ancestral alleles at SNPs that regulate its expression would mean no IFN-λ4 gene being expressed since the non-ancestral allele disrupts its open reading frame [7] and when IFN-λ4 gene is expressed by the ancestral allele, the expression of IFN-λ3 gene is expected to be low. Given this scenario, even though strong evidence has emerged (including from our group [10]), implicating IFN-λ4 gene in regulating HCV viral diversity [12] and clearance following conventional therapy, the role of IFN-λ3 cannot be completely ruled out, especially since the protective non-ancestral alleles have been found to potentially increase IFN-λ3 gene expression [28–29,44]. Moreover, the natural suppressive control on IFN-λ4 expression [59] and a lack of evidence so far for its expression in multiple cells and tissues, puts the onus back on IFN-λ3 regulatory SNPs, especially to understand the wide variety of health and disease states that the IFN-λ locus has shown genetic association with recently [19–20,60].
In this study, firstly, we show that the previously described functional genetic variants of the IFN-λ3 gene: rs28416813 and rs4803217 together can have a strong effect on gene expression (Fig. 1). It seems that the ancestral alleles need to be present simultaneously at both these SNP positions to cause a significant decrease in gene expression (Fig. 1D); since the two SNPs are in strong LD in most world populations, this result could be important in future genetic association studies involving the IFNL locus variants. Next, we revisit our previous work [28] and provide additional evidence for a role of the SNP rs28416813 in controlling transcription at the IFN-λ3 promoter (Figs. 1, 2 and Suppl. Fig. 2). Secondly, we test all the three regulatory SNPs-rs4803219, rs28416813 and rs4803217-predicted by Rauch and others (2010) [22] for their combined effect on reporter gene expression (Fig. 6C). Thirdly, during our study we have observed an interaction between the TFs IRF7 and p65 at the IFN-λ3 promoter. We show that both these TFs not only show physical interaction but also the promoter occupancy of IRF7 increases in presence of p65 (Fig. 3E). This interaction has a functional implication at the IFN-λ3 promoter in terms of a synergistic effect on transcription (Fig. 4). We define this synergistic effect by carrying out a systematic mutational analysis of TF binding sites (Fig. 5). Fourthly, we identify that IRF7 is particularly sensitive to changes in DNA phasing, likely due to its unique nature of binding to DNA (Fig. 5B and C). Fifthly, we show that this sensitivity of IRF7 towards DNA binding may have physiological significance as a particular genetic variant present upstream in the promoter could be responsible for changes in DNA phasing and this effect may be naturally present in the human population. Indeed, we see that IRF7 but not p65 shows significant differences in promoter activation when provided with IFN-λ3 promoter containing different number of TA repeats (Fig. 6B).
In our previous study [28], we had included a nearby SNP rs71356849 due to its physical proximity (two base pairs apart) to the candidate causal SNP rs28416813 in our EMSA analysis using nuclear lysates from p50 and p65 overexpressed cells and also with a recombinant p50 protein. To the best of our knowledge, no study has reported any genetic association with any phenotype with the SNP rs71356849 and we too did not see its presence in our study population, hence we have not studied this variant any further. In the present study, our EMSA (involving only rs28416813) results clearly show that the C allele at rs28416813 has a strong affinity for the full-length p50 homodimer compared to the G allele, which happens to be the risk allele in case of HCV infections (Suppl. Fig. 2). We were not able to do a similar EMSA with a full-length p65, but we were able to address the interaction of the SNP with p65 in a more physiological setting (Fig. 2A). The NF-κB TF functions as a homo or a heterodimer of five different proteins, among which p50 and p65 form the most stable dimer [61]. The p50 homodimer can bind to the same DNA binding site as that of the p50-p65 heterodimer, but with lesser affinity [61]. While p50 homodimer is a transcriptional repressor, p50-p65 is a transcriptional activator [62]. The exact dimer combinations involving p50 and p65 that are formed after activation of the classical NF-κB pathway is difficult to predict and it is possible that both homo and heterodimers are important in different physiological and pathological conditions [63–64]. NF-κB TF is not only activated during viral infections but can be an important TF during a variety of immune stimuli including during inflammation and development [62]. For instance, TNF-a that functions by activating the classical NF-κB pathway is not only a cytokine that functions during infection and inflammation but has an important role in development of several immune cells [62]. Therefore, the regulation of IFN-λ3 gene expression by the SNP rs28416813 through binding of NF-κB components could have a deep impact not only on disease but also on health.
Transcriptional synergy between TFs is common in regulating expression of cytokine and chemokine genes [54,64–65]. The IFN-β enhanceosome is a supreme example of such a phenomenon [52]. However, the synergy we report at the IFN-λ3 promoter seems unlike that at the IFN-β enhanceosome and is more like the one reported for the RANTES and IL10 genes [64–65]. Multiple lines of evidence in our study establish that the binding of IRF7 to the promoter is a rate-limiting step in activation of the IFN-λ3 promoter. Firstly, we see that a 7-fold increase in IRF7 concentration causes only a modest 1.8-fold increase in transcription activity when the TF is present alone (Fig. 4B, dose response curve); while presence of p65 at only 30 ng concentration increased the transcription activity of IRF7 at its lowest concentration tested, by 4.8-fold. Secondly, our results from the transient ChIP reaction clearly shows that IRF7 promoter occupancy is enhanced by p65 (Fig. 3E). Thirdly, we see that IRF7-mediated transcription is more affected when DNA helical phase is disrupted (Fig. 5B and 6B). Furthermore, the synergy efficiency seems to depend on the length of the promoter construct (Figs. 4, 5 and Suppl. Fig. 5), the longer the promoter, the more is the synergy value; this would suggest that there is some correlation between the length of the promoter DNA used in our experiments and the rate-limiting nature of IRF7 binding to the DNA. Therefore, enhancing the promoter occupancy of IRF7 (and/or IRF3) at the IFN-λ3 promoter may be the mechanism behind transcriptional synergy at the IFN-λ3 promoter (Fig. 7A). NF-κB p65, by binding to its cognate binding sites around the IRF binding sites may ‘open’ the major groove within the IRF binding site through allosteric mechanisms, so that IRF3/7 can better access their binding surface. Although, we also see that IRF7 can physically interact with p65 (Fig. 3B and C; and IRF3 can also interact with p65, Ref. 47), we believe that, unlike the case with the interferon beta promoter/enhancer, this mechanism of achieving synergy, i.e., by enhancing the recruitment of the basal transcriptional apparatus by protein-protein contacts between IRFs and NF-κB at the IFN-λ3 promoter is less likely (Fig. 7B). This is primarily because of the distantly placed IRF and NF-κB binding sites at the IFN-λ3 promoter (within ~ 500 bp; Fig. 5A) compared to that of the interferon beta promoter/enhancer (within 55 bp) [52].
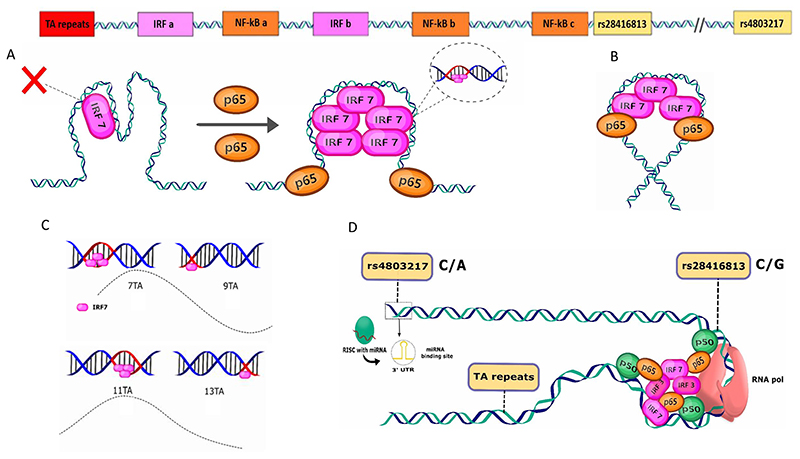
Fig. 7. Schematic representation and summary of the results of this study.
The top panel shows the IFN-λ3 promoter with the different TF binding sites identified in the context of the three functional genetic variants studied. A) Our results show that IRF7 (or IRF3) binding to its DNA is rate-limiting on the IFN-λ3 promoter. Binding of NF-κB p65 (as a hetero or a homodimer) enhances the promoter occupancy of IRF7 possibly by causing allosteric changes to the IRF-binding site DNA and allowing for a better access of IRF7 into the major groove of the DNA thereby overcoming the rate-limiting step in promoter loading. This may occur without any protein-protein contacts between IRF7 and p65; or may involve such contacts in a long-range over the promoter that somehow brings the distantly bound TFs in proximity in the 3-D conformation (B); however, we feel that this model (B) is less likely to lead to promote synergy at the IFN-λ3 promoter. C) The mechanism behind the functioning of the TA repeat polymorphism in affecting IRF7-mediated transcription. The changes in the DNA helix phase disrupts IRF7 binding since IRF7 binding seems to be sensitive to such changes as extensive contacts between a well-structured protein region (in the form of a short alpha-helix) with the nucleotides within the major grove (red) are observed in the co-crystal structures. Every two TA repeats increases the nucleotide length by 4 which is close to half the number of bases that lead to a turn of the DNA helix, thereby disrupting the helix such that bases in the major groove are now in the minor grove and therefore IRF7 finds it difficult to tuck its short alpha-helix into the minor grove in the new phase. This leads to a cyclical effect on transcription (dashed line) as the next two TA repeats can partially restore the helix phase. D) Three functional genetic variants of the IFN-λ3 promoter: rs28416813, rs4803217 and TA-repeat polymorphism (rs7225881). SNP rs28416813 affects NF-κB binding near the basal transcription apparatus (RNA pol) and SNP rs4803217 interferes with miRNA binding and the mRNA stability while present in the 3’UTR of the mRNA. The ancestral alleles of both these SNPs significantly decrease gene expression. The TA-repeat polymorphism affects DNA phasing and therefore likely affects IRF7-mediated transcription.
The TA repeat polymorphism seems to be exploiting the unique nature of IRF7 binding (Fig. 5C) to its DNA within the major groove, which seems to be rate-limiting, to impact on transcription efficiency of the IFN-λ3 promoter by causing DNA helical phase changes (Fig. 7C). Even though we have shown additional evidence for the mechanism behind the SNP rs28416813 affecting IFN-λ3 promoter activity (Suppl. Fig. 2 and Fig. 2A), we could not completely explain the situation wherein the C and G alleles show significant effect on reporter gene expression when only IRF3-SA and IRF7-SA, without any NF-κB TFs, are overexpressed (Fig. 2B). Nonetheless, our study shows that the physiologically relevant haplotype consisting of the ancestral alleles at rs28416813 and rs4803217 can significantly downregulate reporter gene expression in a variety of conditions including during virus infection (Suppl. Fig. 1). It seems that the IFN-λ3 gene expression is regulated by three functional genetic variants at transcription, mRNA stability and translational levels (Fig. 7D). Our study, using in vitro reporter assays shows no evidence for a role for the promoter SNP rs4803219 in affecting transcription, but we do see a functional role for the TA repeat polymorphism (rs72258881). The drawback of the study is that the evidence provided in support of the functional nature of the genetic variants is entirely from in vitro experiments; however, several previous studies have already reported an association of the genetic variants (either the ones studied here or those that are in LD with them) with expression of IFN-λ3 [2,3,30–39]. In conclusion, we show that three functional genetic variants may be regulating IFN-λ3 gene expression and we also show a novel interaction between TFs at the promoter that results in transcriptional synergy. This study further adds to the mechanism of gene regulation at the IFN-λ locus.
Supplementary Material
Acknowledgements
This work was supported by the DBT/Wellcome Trust India Alliance Fellowship (grant number IA/I/17/1/503122) awarded to S.C. SR thanks the UGC for providing the JRF; DGR thanks DST for the JRF; SB thanks DBT for the SRF and AB thanks the DBT for JRF and SRF. The authors thank the Director of NIBMG Prof. Saumitra Das for providing the facilities and for encouragement and support.
Footnotes
CRediT authorship contribution statement
Subhajit Roy: Methodology, Validation, Investigation, Formal analysis, Visualization. Debarati Guha Roy: Methodology, Validation, Investigation, Formal analysis, Visualization. Anand Bhushan: Methodology, Investigation, Resources, Writing - original draft. Seema Bharatiya: Methodology, Resources. Sreedhar Chinnaswamy: Conceptualization, Funding acquisition, Methodology, Investigation, Formal analysis, Project administration, Supervision, Writing - original draft, Writing - review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- [1].Ge D, Fellay J, Thompson AJ, Simon JS, Shianna KV, Urban TJ, et al. Genetic variation in IL28B predicts hepatitis C treatment-induced viral clearance. Nature. 2009;461:399–401. doi: 10.1038/nature08309. [DOI] [PubMed] [Google Scholar]
- [2].Suppiah V, Moldovan M, Ahlenstiel G, Berg T, Weltman M, Abate ML, et al. IL28B is associated with response to chronic hepatitis C interferon-alpha and ribavirin therapy. Nat Genet. 2009;41:1100–1104. doi: 10.1038/ng.447. [DOI] [PubMed] [Google Scholar]
- [3].Tanaka Y, Nishida N, Sugiyama M, Kurosaki M, Matsuura K, Sakamoto N, et al. Genome-wide association of IL28B with response to pegylated interferon-alpha and ribavirin therapy for chronic hepatitis C. Nat Genet. 2009;41:1105–1109. doi: 10.1038/ng.449. [DOI] [PubMed] [Google Scholar]
- [4].Prokunina-Olsson L. Genetics of the human interferon lambda region. J Interferon Cytokine Res. 2019;39(10):599–608. doi: 10.1089/jir.2019.0043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Kotenko SV, Gallagher G, Baurin VV, Lewis-Antes A, Shen M, Shah NK, Langer JA, Sheikh F, Dickensheets H, Donnelly RP. IFN-lambdas mediate antiviral protection through a distinct class II cytokine receptor complex. Nat Immunol. 2003;4(1):69–77. doi: 10.1038/ni875. [DOI] [PubMed] [Google Scholar]
- [6].Sheppard P, Kindsvogel W, Xu W, Henderson K, Schlutsmeyer S, Whitmore TE, Kuestner R, Garrigues U, Birks C, Roraback J, Ostrander C, et al. IL-28, IL-29 and their class II cytokine receptor IL-28R. Nat Immunol. 2003;4(1):63–68. doi: 10.1038/ni873. [DOI] [PubMed] [Google Scholar]
- [7].Prokunina-Olsson L, Muchmore B, Tang W, Pfeiffer RM, Park H, Dickensheets H, Hergott D, Porter-Gill P, Mumy A, Kohaar I, Chen S, et al. A variant upstream of IFNL3 (IL28B) creating a new interferon gene IFNL4 is associated with impaired clearance of hepatitis C virus. Nat Genet. 2013;45(2):164–171. doi: 10.1038/ng.2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Horner SM, Gale M., Jr Regulation of hepatic innate immunity by hepatitis C virus. Nat Med. 2013;19(7):879–88. doi: 10.1038/nm.3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].O’Brien TR, Jackson SS. What Have We Learned from Studies of IFN-λ Variants and Hepatitis C Virus Infection? J Interferon Cytokine Res. 2019;39(10):618–626. doi: 10.1089/jir.2019.0048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Bhushan A, Ghosh S, Bhattacharjee S, Chinnaswamy S. Confounding by Single Nucleotide Polymorphism rs117648444 (P70S) Affects the Association of Interferon Lambda Locus Variants with Response to Interferon-alpha-Ribavirin Therapy in Patients with Chronic Genotype 3 Hepatitis C Virus Infection. J Interferon Cytokine Res. 2017;37(8):369–382. doi: 10.1089/jir.2017.0002. [DOI] [PubMed] [Google Scholar]
- [11].Terczynska-Dyla E, Bibert S, Duong FH, Krol I, Jorgensen S, Collinet E, Kutalik Z, Aubert V, Cerny A, Kaiser L, Malinverni R, et al. Reduced IFNlambda4 activity is associated with improved HCV clearance and reduced expression of interferon-stimulated genes. Nat Commun. 2014;5:5699. doi: 10.1038/ncomms6699. [DOI] [PubMed] [Google Scholar]
- [12].Ansari MA, Aranday-Cortes E, Ip CLC, da Silva Filipe A, Hin LS, Bamford CGG, Bonsall D, Trebes A, Piazza P, Sreenu V, Cowton VM, et al. Interferon lambda 4 impacts the genetic diversity of hepatitis C virus. eLife. 2019;8:e42463. doi: 10.7554/eLife.42463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].O’Brien TR, Prokunina-Olsson L, Donnelly RP. IFN-l4: the paradoxical new member of the interferon lambda family. J Interferon Cytokine Res. 2014;34(11):829–838. doi: 10.1089/jir.2013.0136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Thomas DL, Thio CL, Martin MP, Qi Y, Ge D, O’huigin C, et al. Genetic variation in IL28B and spontaneous clearance of hepatitis C virus. Nature. 2009;461:798–801. doi: 10.1038/nature08463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Tillmann HL, Thompson AJ, Patel K, Wiese M, Tenckhoff H, Nischalke HD, et al. A polymorphism near IL28B is associated with spontaneous clearance of acute hepatitis C virus and jaundice. Gastroenterology. 2010;139(1586-1592):1592e1581. doi: 10.1053/j.gastro.2010.07.005. [DOI] [PubMed] [Google Scholar]
- [16].Sacks-Davis R, Grebely J, Dore GJ, Osburn W, Cox AL, Rice TM, et al. Hepatitis C virus reinfection and spontaneous clearance of reinfection - the InC3 study. J Infect Dis. 2015;212:1407–1419. doi: 10.1093/infdis/jiv220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Aka PV, Kuniholm MH, Pfeiffer RM, Wang AS, Tang W, Chen S, et al. Association of the IFNL4-DeltaG allele with impaired spontaneous clearance of hepatitis C virus. J Infect Dis. 2014;209:350–354. doi: 10.1093/infdis/jit433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Vergara C, Duggal P, Thio CL, Valencia A, O’Brien TR, Latanich R, et al. Multi-ancestry fine mapping of interferon lambda and the outcome of acute hepatitis C virus infection. Genes Immun. 2020;21(5):348–359. doi: 10.1038/s41435-020-00115-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Bhushan A, Chinnaswamy S. Identifying causal variants at the interferon lambda locus in case-control studies: utilizing non-synonymous variant rs117648444 to probe the role of IFN-lambda4. Gene. 2018;664:168–180. doi: 10.1016/j.gene.2018.04.076. [DOI] [PubMed] [Google Scholar]
- [20].Chinnaswamy S. Gene-disease association with human IFNL locus polymorphisms extends beyond hepatitis C virus infections. Genes Immun. 2016;17:265–275. doi: 10.1038/gene.2016.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Eslam M, McLeod D, Kelaeng KS, Mangia A, Berg T, Thabet K, Irving WL, Dore GJ, Sheridan D, Gronbaek H, Abate ML, et al. International Liver Disease Genetics C, IFN-lambda3, not IFN-lambda4, likely mediates IFNL3-IFNL4 haplotype-dependent hepatic inflammation and fibrosis. Nat Genet. 2017;49(5):795–800. doi: 10.1038/ng.3836. [DOI] [PubMed] [Google Scholar]
- [22].Rauch A, Kutalik Z, Descombes P, Cai T, Di Iulio J, Mueller T, et al. Genetic variation in IL28B is associated with chronic hepatitis C and treatment failure: a genome-wide association study. Gastroenterology. 2010;138:1338–1345. doi: 10.1053/j.gastro.2009.12.056. [DOI] [PubMed] [Google Scholar]
- [23].di Iulio J, Ciuffi A, Fitzmaurice K, Kelleher D, Rotger M, Fellay J, Martinez R, Pulit S, Furrer H, Gunthard HF, Battegay M, et al. Estimating the net contribution of interleukin-28B variation to spontaneous hepatitis C virus clearance. Hepatology. 2011;53:1446–1454. doi: 10.1002/hep.24263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].de Castellarnau M, Aparicio E, Parera M, Franco S, Tural C, Clotet B, Martinez MA. Deciphering the interleukin 28B variants that better predict response to pegylated interferon- a and ribavirin therapy in HCV/HIV-1 coinfected patients. PLoS ONE. 2012;7:e31016. doi: 10.1371/journal.pone.0031016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Urban TJ, Thompson AJ, Bradrick SS, Fellay J, Schuppan D, Cronin KD, Hong L, McKenzie A, Patel K, Shianna KV, McHutchison JG, et al. IL28B genotype is associated with differential expression of intrahepatic interferon-stimulated genes in patients with chronic hepatitis C. Hepatology. 2010;52:1888–1896. doi: 10.1002/hep.23912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Sugiyama M, Tanaka Y, Wakita T, Nakanishi M, Mizokami M. Genetic variation of the IL-28B promoter affecting gene expression. PLoS ONE. 2011;6:e26620. doi: 10.1371/journal.pone.0026620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Friborg J, Levine S, Chen CQ, Sheaffer AK, Chaniewski S, Voss S, Lemm JA, McPhee F. Combinations of lambda interferon with direct-acting antiviral agents are highly efficient in suppressing hepatitis C virus replication. Antimicrob Agents Chemo. 2013;57:1312–1322. doi: 10.1128/AAC.02239-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Chinnaswamy S, Chatterjee S, Boopathi R, Mukherjee S, Bhattacharjee S, Kundu TK. A single nucleotide polymorphism associated with hepatitis C virus infections located in the distal region of the IL28B promoter influences NF-kappaB-mediated gene transcription. PLoS ONE. 2013;8:e75495. doi: 10.1371/journal.pone.0075495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].McFarland AP, Horner SM, Jarret A, Joslyn RC, Bindewald E, Shapiro BA, Delker DA, Hagedorn CH, Carrington M, Gale M, Jr, Savan R. The favorable IFNL3 genotype escapes mRNA decay mediated by AU-rich elements and hepatitis C virus-induced microRNAs. Nat Immunol. 2013;15(1):72–79. doi: 10.1038/ni.2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Langhans B, Kupfer B, Braunschweiger I, Arndt S, Schulte W, Nischalke HD, Nattermann J, Oldenburg J, Sauerbruch T, Spengler U. Interferon-lambda serum levels in hepatitis C. J Hepatol. 2011;54:859–865. doi: 10.1016/j.jhep.2010.08.020. [DOI] [PubMed] [Google Scholar]
- [31].Rallon NI, Soriano V, Naggie S, Restrepo C, McHutchison J, Vispo E, Benito JM. Impact of IL28B gene polymorphisms on interferon-l3 plasma levels during pegylated interferon a/ribavirin therapy for chronic hepatitis C in patientscoinfected with HIV. J Antimicrob Chemother. 2012;67(5):1246–1249. doi: 10.1093/jac/dkr598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Shi X, Pan Y, Wang M, Wang D, Li W, Jiang T, Zhang P, Chi X, Jiang Y, Gao Y, Zhong J, et al. IL28B genetic variation is associated with spontaneous clearance of hepatitis C virus, treatment response, serum IL-28B levels in chinese population. PLoS ONE. 2012;7(5):e37054. doi: 10.1371/journal.pone.0037054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Bibert S, Roger T, Calandra T, Bochud M, Cerny A, Semmo N, et al. IL28B expression depends on a novel TT/-G polymorphism which improves HCV clearance prediction. J Exp Med. 2013;210:1109–1116. doi: 10.1084/jem.20130012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Murata K, Sugiyama M, Kimura T, Yoshio S, Kanto T, Kirikae I, Saito H, Aoki Y, Hiramine S, Matsui T, Ito K, et al. Ex vivo induc- tion of IFN-lambda3 by a TLR7 agonist determines response to Peg-IFN/ribavirin therapy in chronic hepatitis C patients. J Gastroenterol. 2014;49(1):126–137. doi: 10.1007/s00535-013-0814-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Fukuhara T, Taketomi A, Motomura T, Okano S, Ninomiya A, Abe T, Uchiyama H, Soejima Y, Shirabe K, Matsuura Y, Maehara Y. Variants in IL28B in liver recipients and donors correlate with response to peg-interferon and ribavirin therapy for recurrent hepatitis C. Gastroenterology. 2010;139(5):1577–1585.:1585.e1-e3. doi: 10.1053/j.gastro.2010.07.058. [DOI] [PubMed] [Google Scholar]
- [36].Yoshio S, Kanto T, Kuroda S, Matsubara T, Higashitani K, Kakita N, Ishida H, Hiramatsu N, Nagano H, Sugiyama M, Murata K, et al. Human blood dendritic cell antigen 3 (BDCA3)(+) dendritic cells are a potent producer of interferonlambda in response to hepatitis C virus. Hepatology. 2013;57(5):1705–1715. doi: 10.1002/hep.26182. [DOI] [PubMed] [Google Scholar]
- [37].Li W, Jiang Y, Jin Q, Shi X, Jin J, Gao Y, Pan Y, Zhang H, Jiang J, Niu J. Expression and gene polymorphisms of interleukin 28B and hepatitis B virus infection in a Chinese Han population. Liver Int. 2011;31(8):1118–1126. doi: 10.1111/j.1478-3231.2011.02507.x. [DOI] [PubMed] [Google Scholar]
- [38].Egli A, Levin A, Santer DM, Joyce M, O’Shea D, Thomas BS, Lisboa LF, Barakat K, Bhat R, Fischer KP, Houghton M, et al. Immunomodulatory function of interleukin 28B during primary infection with cytomegalovirus. J Infect Dis. 2014;210(5):717–727. doi: 10.1093/infdis/jiu144. [DOI] [PubMed] [Google Scholar]
- [39].Egli A, Mandal J, Schumann DM, Roth M, Thomas B, Tyrrell DL, Blasi F, Kostikas K, Boersma W, Milenkovic B, Lacoma A, et al. IFN lambda 3/4 locus polymorphisms and IFN lambda 3 circulating levels are associated with COPD severity and outcomes. BMC Pulm Med. 2018;18(1):51. doi: 10.1186/s12890-018-0616-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Thong VD, Wasitthankasem R, Tangkijvanich P, Vongpunsawad S, Poovorawan Y. Prevalence of thymine-adenine dinucleotide repeat, IL28B and IFNL4 in Thai population and correlation with spontaneous clearance and treatment outcome of hepatitis C infection. PLoS ONE. 2015;10:e0125400. doi: 10.1371/journal.pone.0125400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Hiramine S, Sugiyama M, Furusyo N, Uto H, Ido A, Tsubouchi H, et al. A thymine adenine dinucleotide repeat polymorphism near IL28B is associated with spontaneous clearance of hepatitis C virus. J Gastroenterol. 2015;50:1069–1077. doi: 10.1007/s00535-015-1056-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Fox BA, Sheppard PO, O’Hara PJ. The role of genomic data in the discovery, annotation and evolutionary interpretation of the interferon-lambda family. PLoS ONE. 2009;4:e4933. doi: 10.1371/journal.pone.0004933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Osterlund PI, et al. IFN regulatory factor family members differentially regulate the expression of type III IFN (IFN-lambda) genes. J Immunol. 2007;179:3434–3442. doi: 10.4049/jimmunol.179.6.3434. [DOI] [PubMed] [Google Scholar]
- [44].Lu YF, Mauger DM, Goldstein DB, Urban TJ, Weeks KM, Bradrick SS. IFNL3 mRNA structure is remodelled by a functional non-coding polymorphism associated with hepatitis C virus clearance. Sci Rep. 2015;5:16037. doi: 10.1038/srep16037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM, Sirotkin K. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001 Jan 1;29(1):308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Hiscott J, Lin R, Nakhaei P, Paz S. MasterCARD: a priceless link to innate immunity. Trend Mol Med. 2006;12(2):53–56. doi: 10.1016/j.molmed.2005.12.003. [DOI] [PubMed] [Google Scholar]
- [47].Ogawa S, Lozach J, Benner C, Pascual G, Tangirala RK, Westin S, et al. Molecular determinants of crosstalk between nuclear receptors and Toll-like receptors mediating counter-regulation of inflammatory responses. Cell. 2005;12295:707–721. doi: 10.1016/j.cell.2005.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Kerppola T. Design and implementation of bimolecular fluorescence complementation (BiFC) assays for the visualization of protein interactions in living cells. Nature. 2006;1:278–1286. doi: 10.1038/nprot.2006.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Lin R, Mamane Y, Hiscott J. Multiple regulatory domains control IRF-7 activity in response to virus infection. J Biol Chem. 2000;275:34320–34327. doi: 10.1074/jbc.M002814200. [DOI] [PubMed] [Google Scholar]
- [50].Lavrrar JL, Farnham PJ. The use of transient chromatin immunoprecipitation assays to test models for E2F1-specific transcriptional activation. J Biol Chem. 2004;279:46343–46349. doi: 10.1074/jbc.M402692200. [DOI] [PubMed] [Google Scholar]
- [51].Lee HC, Narayanan S, Park SJ, et al. Transcriptional regulation of IFN-k genes in hepatitis C virus-infected hepatocytes via IRF-3$IRF-7$NF-jB complex. J Biol Chem. 2014;289:5310–5319. doi: 10.1074/jbc.M113.536102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Panne D, Maniatis T, Harrison SC. An atomic model of the interferon-β enhanceosome. Cell. 2007;129:1111–1123. doi: 10.1016/j.cell.2007.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Ranjith-Kumar CT, Wen Y, Baxter N, Bhardwaj K, Kao CC. A cell basedassay for RNA synthesis by the HCV polymerase reveals new insights on mechanism of polymerase inhibitors and modulation by NS5A. PLoS ONE. 2011;6(7):e22575. doi: 10.1371/journal.pone.0022575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Merika M, Williams AJ, Chen G, Collins T, Thanos D. Recruitment of CBP/p300 by the IFN beta enhanceosome is required for synergistic activation of transcription. Mol Cell. 1998;1:277–287. doi: 10.1016/s1097-2765(00)80028-3. [DOI] [PubMed] [Google Scholar]
- [55].Uhlemann AC, Szlezak NA, Vonthein R, Tomiuk J, Emmer SA, Lell B, Kremsner PG, Kun JF. DNA phasing by TA dinucleotide microsatellite length determines in vitro and in vivo expression of the gp91phox subunit of NADPH oxidase and mediates protection against severe malaria. J Infect Dis. 2004;189:2227–2234. doi: 10.1086/421242. [DOI] [PubMed] [Google Scholar]
- [56].Rikitake Y, Moran E. DNA-binding properties of the E1A-associated 300-kilodalton protein. Mol Cell Biol. 1992;12:2826–2836. doi: 10.1128/mcb.12.6.2826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Chen H, Hung M-C. Involvement of co-activator p300 in the transcriptional regulation of the HER-2/neu gene. J Biol Chem. 1997;272:6101–6104. doi: 10.1074/jbc.272.10.6101. [DOI] [PubMed] [Google Scholar]
- [58].Manry J, Laval G, Patin E, Fornarino S, Itan Y, Fumagalli M, Sironi M, Tichit M, Bouchier C, Casanova J-L, Barreiro LB, et al. Evolutionary genetic dissection of human interferons. J Exp Med. 2011;208:2747–2759. doi: 10.1084/jem.20111680. phel. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Hong M, Schwerk J, Lim C, Kell A, Jarret A, Pangallo J, Loo YM, Liu S, Hagedorn CH, Gale M, Jr, Savan R. Interferon lambda 4 expression is suppressed by the host during viral infection. J Exp Med. 2016;213(12):2539–2552. doi: 10.1084/jem.20160437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Rugwizangoga B, Andersson ME, Kabayiza J-C, Nilsson MS, Ármannsdóttir B, Aurelius J, Nilsson S, Hellstrand K, Lindh M, Martner A. IFNL4 Genotypes Predict Clearance of RNA Viruses in Rwandan Children With Upper Respiratory Tract Infections. Front Cell Infect Microbiol. 2019;9:340. doi: 10.3389/fcimb.2019.00340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Phelps CB, Sengchanthalangsy LL, Malek S, Ghosh G. Mechanism of KB DNA binding by Rel/NF-KB dimers. J Biol Chem. 2000;275:24392–24399. doi: 10.1074/jbc.M003784200. [DOI] [PubMed] [Google Scholar]
- [62].Hayden MS, Ghosh S. Signaling to NF-kappaB. Genes Dev. 2004;18:2195–2224. doi: 10.1101/gad.1228704. [DOI] [PubMed] [Google Scholar]
- [63].Hayden MS, West AP, Ghosh S. NF-kappaB and the immune response. Oncogene. 2006;25:6758–6780. doi: 10.1038/sj.onc.1209943. [DOI] [PubMed] [Google Scholar]
- [64].Genin P, Algarte M, Roof P, Lin R, Hiscott J. Regulation of RANTES chemokine gene expression requires cooperativity between NF-κB and IFN-regulatory factor transcription factors. J Immunol. 2000;164:5352–5361. doi: 10.4049/jimmunol.164.10.5352. [DOI] [PubMed] [Google Scholar]
- [65].Wang J, Vodovotz Y, Fan L, Li Y, Liu Z, Namas R, Jiang Y. Injury-induced MRP8/MRP14 stimulates IP-10/CXCL10 in monocytes/macrophages. FASEB J. 2015;29:250–262. doi: 10.1096/fj.14-255992. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.