Abstract
Locus-specific interrogation of target genes employing functional probes such as proteins and small molecules is paramount in decoding the molecular basis of gene function and designing tools to modulate its downstream effects. In this context, CRISPR-based gene editing and targeting technologies have proved tremendously useful, as they can be programmed to target any gene of interest by simply changing the sequence of the single guide RNA (sgRNA). Although these technologies are widely utilized in recruiting genetically encoded functional proteins, display of small molecules using CRISPR system is not well developed due to the lack of adequate techniques. Here, we have devised an innovative technology called sgRNA-Click (sgR-CLK) that harnesses the power of bioorthogonal click chemistry for remodeling guide RNA to display synthetic molecules on target genes. sgR-CLK employs a novel posttranscriptional chemoenzymatic labeling platform wherein a terminal uridylyl transferase (TUTase) was repurposed to generate clickable sgRNA of choice by site-specific tailoring of multiple azide-modified nucleotide analogues at the 3’ end. The presence of a minimally invasive azide handle assured that the sgRNAs are indeed functional. Notably, an azide-tailed sgRNA targeting the telomeric repeat served as a Trojan horse on the CRISPR-dCas9 system to guide synthetic tags (biotin) site-specifically on chromatin employing copper-catalyzed or strain-promoted click reactions. Taken together, sgR-CLK presents a significant advancement on the utility of bioorthogonal chemistry, TUTase, and the CRISPR toolbox, which could offer a simplified solution for site-directed display of small molecule probes and diagnostic tools on target genes.
Introduction
The bacterial adaptive defense system, clustered regularly interspaced short palindromic repeats (CRISPR)-CRISPR-associated proteins (Cas), is in the spotlight for its ability to serve as a powerful genome targeting and editing tool.1,2 The ease-of-use and flexibility to target any genomic loci by simply changing the sequence of the single guide RNA (sgRNA) sets this technology apart from other traditionally used technologies like zinc finger nucleases (ZFNs) and transcription activator-like effector nucleases (TALENs), which require extensive protein engineering to target specific DNA bases.3–5 The catalytically inactive mutant of Cas9, also called dead Cas9 (dCas9), has further harnessed the utility of this genome editing tool for localized delivery of functional tags to enable epigenome editing, transcriptional activation/repression, baseediting, chromatin pull-down, and gene visualization.6–13 These applications greatly rely on dCas9 derivatized with genetically encoded fusion proteins (e.g., eGFP, APOBEC, VP64) and epitope tags (e.g., Flag and SunTag).14–18 Similarly, engineered sgRNA scaffolds with recruitment sequences for RNA-binding proteins or aptamers have also been used to regulate as well as visualize specific genes.19–22 While tools based on dCas9-sgRNA have been widely utilized in delivering functional proteins, locus-specific delivery of synthetic molecules such as drugs, photoactivatable groups, pull-down probes, and epigenomic and transcription modulators remains a technical challenge due to the lack of adequate labeling technologies.23,24 Therefore, development of practical labeling technologies to recruit such small molecule probes will not only allow the profiling of interaction partners of the target gene but also would profoundly expand the therapeutic and diagnostic potential of the CRISPR system.25–27
Toward this direction, methods including maleimide chemistry, postsynthetic modification of a HaloTag and incorporation of unnatural amino acids have been used to derivatize dCas9.28–30 More recently, expression platforms composed of peptide-conjugated dCas9 and ligation modules have been used to (i) introduce small molecule epigenetic baits using split intein and (ii) transfer biotin using biotin ligase for chromatin capture.23,31,32 However, engineering dCas9 with small molecules using expression platforms is complex, and further, the efficiency of conjugation is dependent on the functional moiety tagged on the ligation module.33 On the other hand, covalent derivatization of sgRNA is envisioned as a powerful alternative to localize small molecule functional tags to gene loci.34,35 For example, sgRNA tagged with a fluorescent dye, generated by splint-ligating a chemically synthesized RNA oligonucleotide, was used in establishing a CRISPR-based FISH assay to visualize gene loci in fixed cells.28 However, except for a very few examples, accessing long and structured RNAs like sgRNA labeled with a wide range of functional tags remains a major challenge due to known drawbacks in conventional chemical and enzymatic labeling approaches. Further, dCas9 and sgRNA conjugation strategies rely on functional tagging prior to the formation of the active ternary complex (dCas9:sgRNA:target DNA).20 Hence, attaching synthetic probes in this way could potentially perturb the ribonucleoprotein (RNP) complex formation or its binding efficiency to the target DNA sequence.34 In this regard, we hypothesized that bioorthogonal click reaction would serve as a powerful tool in installing noninvasive reactive handles on sgRNA, which could be further functionalized chemo-selectively upon formation of the ternary complex for downstream applications.36,37
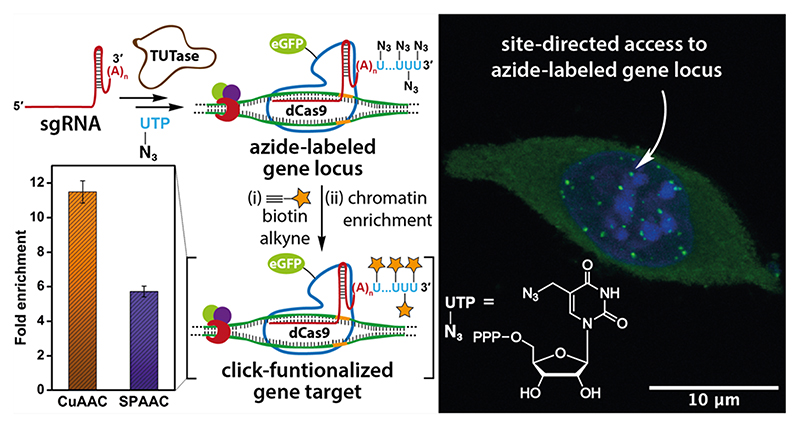
Here, we describe the development of an innovative labeling technology to display minimally perturbing clickable groups (e.g., azide) at a genetic locus of interest by repurposing terminal uridylyl transferase (TUTase) to incorporate azide-modified UTP analogues site-specifically at the 3’ end of sgRNAs. An azide-tailed sgRNA constructed using TUTase served as a Trojan horse on the CRISPR-dCas9 system to direct functional tags on telomeres. Indeed, in situ click reactions performed on the target-bound ternary complex using biotin-alkynes showed significant enrichment of the telomeric DNA repeat upon pull-down. This technique, which we named sgRNA-Click (sgR-CLK), is modular as any sgRNA of choice can be engineered to deploy the desired synthetic probes, thereby offering new means to interrogate gene loci of interest.
Results And Discussion
Blueprint of sgR-CLK
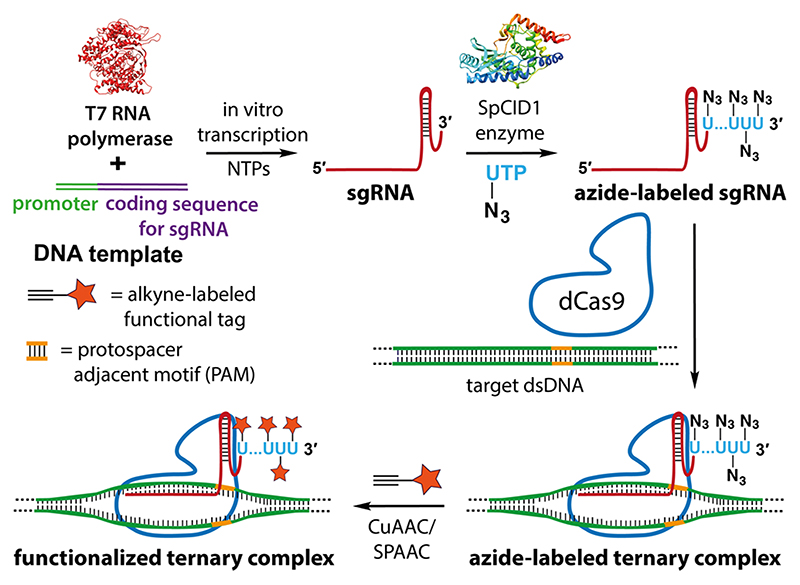
Our strategy to construct clickable sgRNA so as to display functional tags on a specific gene target using the CRISPR system is based on the following consideration. Among various clickable groups, azide is structurally minimally perturbing and can be used in different bioorthogonal reactions like copper-catalyzed azide—alkyne cycloaddition (CuAAC), strain-promoted azide—alkyne cycloaddition (SPAAC), and Staudinger ligation reactions to install variety of functionalities on biomolecules.38,39 However, chemical labeling of RNA with azide groups is not straightforward, as azide-modified phosphoramidites are not stable under solid-phase oligonucleotide synthesis conditions.40 Further, indiscriminate body labeling of sgRNA utilizing RNA polymerases would perturb the formation of an active CRISPR RNP complex.41 To overcome this, we decided to repurpose the RNA tailing ability of a TUTase (SpCID1) in site-specifically derivatizing sgRNA of choice with multiple azide groups at the 3’ end employing azide-modified UTP analogues. An sgRNA thus obtained would enable the display of desired functional tags on the target gene by either posthybridization (Figure 1) or prehybridization click reaction (Figure S1) with appropriate alkyne-labeled tags. In fact, our results demonstrated that posthybridization click strategy is the method of choice for installing synthetic cargos.
Figure 1.
sgR-CLK design to display functional tags on a specific gene. In vitro transcribed CRISPR sgRNA is terminal uridylated with an azide-labeled nucleotide analogue using SpCID1. Azide-labeled sgRNA can be further used to generate a functionalized ternary complex by posthybridization click reaction with an alkyne counterpart containing the desired function tag.
Azide-Tailed sgRNAs Using TUTase
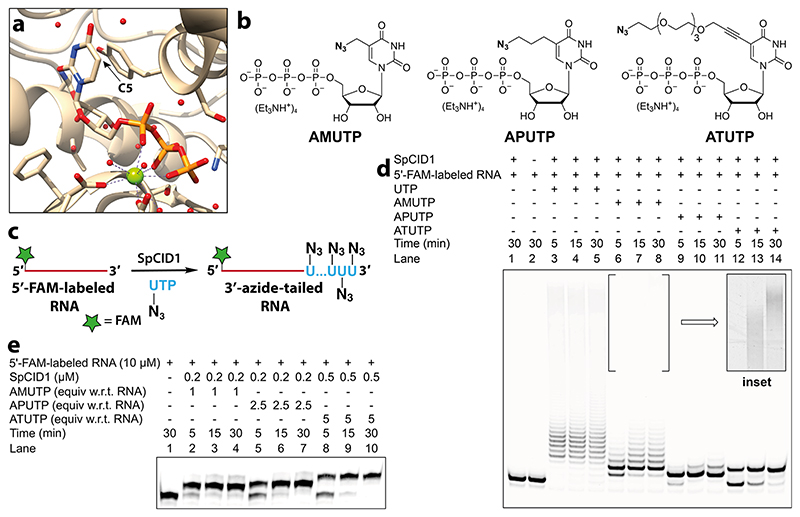
The prerequisite to establish sgR-CLK would require TUTase to efficiently incorporate azide-modified UTP analogues into highly structured sgRNA sequences. TUTases play a key role in epitranscriptomics wherein these enzymes add multiple uridylates at the 3’ end of RNA, which signals RNA biogenesis, processing, degradation, or stabilization.42,43 In the family of TUTases, a cytoplasmic TUTase from Schizosaccharomyces pombe known as caffeine-induced death suppressor 1 (SpCID1) is an important posttranscriptional RNA modifying enzyme.44,45 Careful examination of the crystal structure of SpCID1 in the presence of UTP revealed that the C5-position of uridine does not make contacts with neighboring amino acid residues (Figure 2a).46 This observation suggested that the enzyme might be promiscuous to C5-modified UTP analogues. Therefore, we synthesized azide-modified UTP analogues, namely, AMUTP, APUTP, and ATUTP, with increasing linker length having a methyl, propyl, and tetraethylene glycol spacer, respectively, between the azide group and nucleobase (Figure 2b).47,48 We then cloned SpCID1 gene from S. pombe fission yeast and expressed the recombinant protein in E. coli (see Supporting Information for details, Figure S2-S5, Table S1). The efficacy of SpCID1 to incorporate the nucleotide analogues was evaluated by using a model RNA oligonucleotide sequence bearing a fluorescein (FAM) label at the 5’ end (Figure 2c and Table 1).
Figure 2. SpCID1, a TUTase enzyme, efficiently incorporates C5-azide-modified nucleotides at the 3’ end of an RNA oligonucleotide.
(a) Crystal structure of SpCID1 bound to UTP (PDB 4FH5) generated using UCSF Chimera software.46 C5 position of uridine triphosphate is shown using an arrow. (b) Structure of azide-modified UTP analogues used in terminal uridylation reaction. (c) Scheme showing 3’-terminal uridylation of model 5’-FAM-labeled RNA with modified UTP analogues. (d) Image of PAGE resolved reaction products of terminal uridylation reaction of 5’- FAM-labeled RNA (10 μM) in the presence of UTP/modified UTPs (500 μM) using SpCID1 (0.5 μM). Inset: Processive incorporation of AMUTP was evident upon adjusting the brightness and contrast of the gel image. See Figure S6 for the complete gel picture in these image settings. (e) Gel image showing optimized reaction conditions to effect single nucleotide incorporation.
Table 1. Sequence of Model RNA Oligonucleotide and sgRNAs.
| RNAa | Sequence |
|---|---|
| 5 ‘-FAM-labeled RNA | 5’ FAM-UUACCAUAGAAUCAUGUGCCAUACAUCA 3’ |
| sgRNA 1 | 5’GGGUUAGGGUUAGGGUUAGGGUUAGUUUAAGAGCUAUGCUGGAAACAGCAUAGCAAGUUUA AAUAAGGCUAGUCCGUUAUCAACUUGAAAAAGUGGCACCGAGUCGGUGCUUUAAAAAAAAAAAAAAA3’ |
| sgRNA 1’ | 5’GUUAGGGUUAGGGUUAGGGUUGUUUAAGAGCUAUGCUGGAAACAGCAUAGCAAGUUUAAAUAAGGCUAGUCCGUUAUCAACUUGAAAAAGUGGCACCGAGUCGGUGCUUUAAAAAAAAAAAAAAA 3’ |
| sgRNA 2 | 5’GGCGAGGGCGAUGCCACCUAGUUUAAGAGCUAUGCUGGAAACAGCAUAGCAAGUUUAAAUAAGGCUAGUCCGUUAUCAACUUGAAAAAGUGGCACCGAGUCGGUGCUUUAAAAAAAAAAAAAAA 3’ |
Sequence of sgRNA targeting the protospacer region is represented in blue, and adenylate overhang is represented in red.
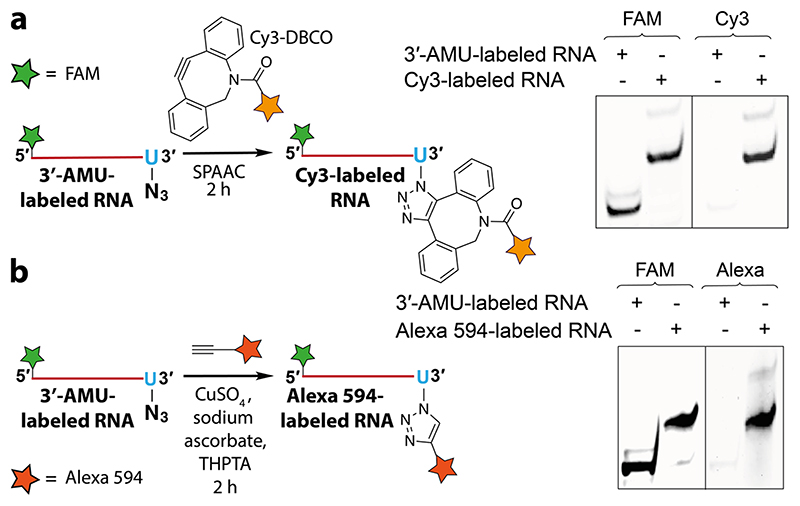
SpCID1 was incubated with 5’-FAM-labeled RNA in the presence of UTP or azide-modified UTPs. Reaction aliquots at different time intervals were resolved by polyacrylamide gel electrophoresis (PAGE) under denaturing conditions and the reaction products were imaged using a fluorescence scanner (Figure 2d). A biphasic incorporation pattern corresponding to a distributive and processive addition of UTP, consistent with a literature report, was observed (lane 3—5).49 Distributive enzymatic UMP addition involves the dissociation of enzyme from substrate RNA after every successive UMP addition giving rise to RNA oligonucleotides containing few added nucleotides (6-9 nucleotides). However, in the processive phase the enzyme adds several nucleotides to the tailed RNA substrates produced in the distributive phase, which is thought to be a result of increase in binding affinity of enzyme to uridylated RNA.49 Remarkably, all the nucleotide analogues served as good substrates in the terminal uridylation reaction. Notably, the analogues were predominantly incorporated via distributive enzyme incorporation, wherein the number of additions is dependent on the length of the linker. While AMU was incorporated multiple times (lane 6-8), APUTP and ATUTP produced RNA oligonucleotides containing one or two nucleotide analogues (lane 9-14). Further optimization by varying enzyme-nucleotide stoichiometry yielded singly modified RNA oligonucleotide as the major product, which was characterized by mass analysis (Figure 2e, Figure S7, S8 and Table S2). AMU-labeled RNA oligonucleotide was readily functionalized by SPAAC and CuAAC reactions using Cy3-DBCO strained alkyne and Alexa 594-alkyne, respectively (Figure 3). It is worth mentioning here that RNA containing an azide label at the 3′ end can be used to generate RNA with terminally or internally labeled biophysical probes by click reaction or ligation followed by click reaction.50 Recently, chemically synthesized crRNAs containing chemoselective reactive handles have been used in stitching tracrRNA to yield full-length sgRNA or donor DNA to facilitate homology directed repair.51,52 However, this strategy is not suitable for introducing functional tags on sgRNA unlike our method, which introduces multiple free azide labels on sgRNA for further functionalization. Hence, in the present study, we preferred to synthesize sgRNAs containing multiple AMU residues at its 3’ end utilizing its minimally perturbing nature, which in turn may not affect the formation of the active CRISPR complex for subsequent display of multiple functional tags by click chemistry.
Figure 3. AMU-labeled RNA oligonucleotide is compatible for click functionalization. The gels were scanned at FAM and Cy3 wavelengths.
(a) Scheme and gel image of SPAAC reaction between 3’-AMU-labeled RNA and Cy3-DBCO. (b) CuAAC reaction between 3’-AMU-labeled RNA and Alexa 594 alkyne. Structures of alkyne substrates are provided in Figure S9.
As a study system to establish sgR-CLK, we designed sgRNAs with a protospacer targeting telomere repeat region and eGFP gene. We used telomere as the target since CRISPR-based tools developed to visualize and pull-down specific genes commonly use this sequence as a model. Guide RNA against eGFP was used as a non-targeting sequence. We initially used a conventional guide RNA design13 to synthesize sgRNA targeting the telomere region by in vitro transcription reaction using T7 RNA polymerase and template CT1 (Table S1). Upon incubation with SpCID1 and AMUTP, we observed no detectable incorporation of the nucleotide analogue (data not shown). This is possibly because the trans-acting CRISPR RNA (tracrRNA) region of sgRNA being RNA pol III transcribed, ends with a strong hairpin loop structure at its 3’ end (Figure S10a).53 This feature is also observed in the crystal structure of guide RNA bound to Cas9 and target DNA.54 In vitro experiments have revealed that SpCID1 requires a ~13 nucleotide stretch of single stranded RNA for efficient binding and uridylation.44 Possibly due to this reason the enzyme failed to incorporate AMUTP into the strongly structured 3’ end of sgRNA.
To circumvent this problem, we took the cue from the ability of SpCID1 to bind and uridylate unstructured poly(A) tail region of mRNA.45 Additionally, poly(A) and poly(U) tracts at the 3’ end of guide RNA has been reported to increase the stability and efficiency of CRISPR systems.55,56 On the basis of these key observations, we decided to reconfigure sgRNA to contain a poly(A) tail at the 3’ end with the view that it would facilitate efficient incorporation of AMUTP. This notion was also supported by secondary structure prediction, which suggested that extension of sgRNA with a 15-mer homopolymer of adenosine preserved the overall structure of the guide RNA and introduced an unstructured 3’-overhang (Figure S10b).
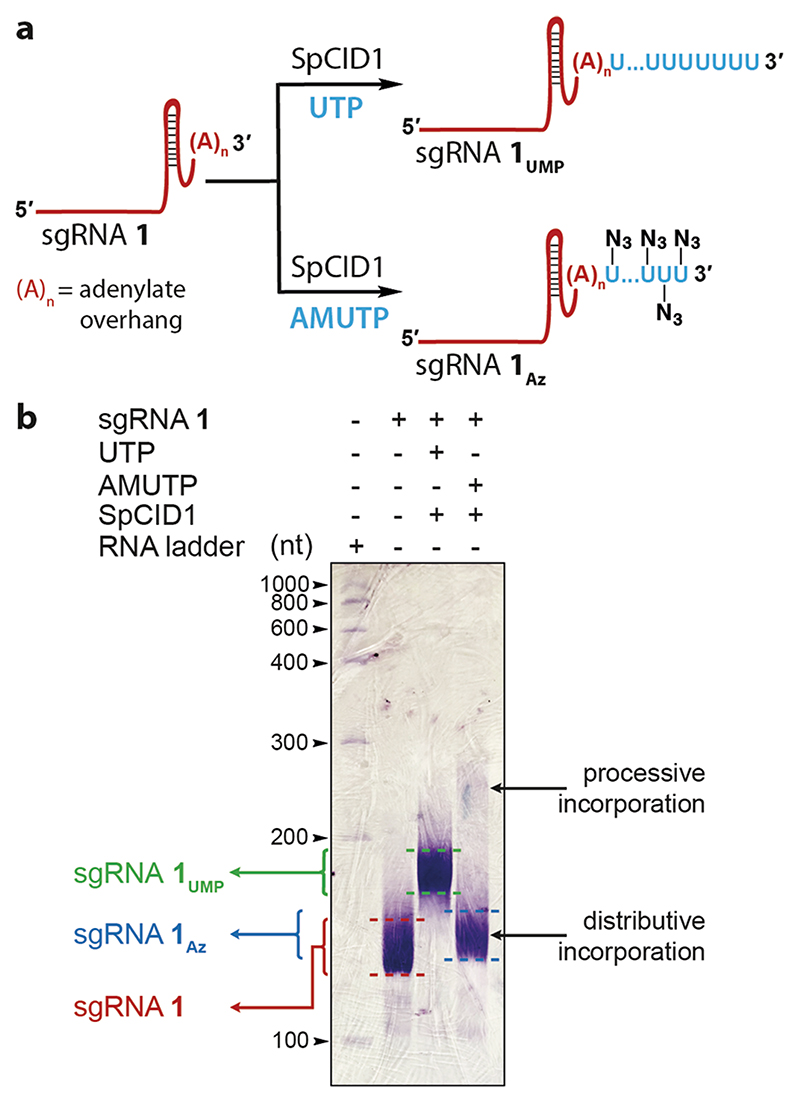
PCR amplified dsDNA templates were in vitro transcribed in the presence of natural NTPs to synthesize poly(A) tail-containing sgRNA 1 and 2, targeting the telomeric repeat and eGFP gene, respectively (Table 1 and Table S1). In order to check if the envisioned sgRNA design was compatible for terminal uridylation, sgRNA 1 was reacted with AMUTP or UTP in the presence of SpCID1, and products were resolved by denaturing PAGE followed by RNA staining (Figure 4). sgRNA 1 showed a slightly diffused band, which can be attributed to transcripts produced with small variations in the number of adenylate residues added at the 3’ end due to known transcript slippage. Rewardingly, bands of lower mobility indicated that the overhang efficiently assisted the terminal uridylation in the presence of natural (1UMP) and modified (1Az) UTPs. Similar to uridylation with the model RNA, AMUTP incorporation was found to be biphasic, prominently favoring distributive addition. For downstream applications, the azide-labeled sgRNAs (1Az and 2Az were synthesized in large-scale by terminal uridylation of sgRNA 1 and 2 and isolated in good yields (Table 1 and Table S3).
Figure 4. SpCID1 efficiently uridylates poly(A) tail-containing sgRNA in the presence of UTP and AMUTP.
(a) Scheme showing uridylation reaction setup. (b) Uridylated sgRNA products were resolved by PAGE under denaturing conditions and were stained using Stains-All reagent.
Azide-Labeled sgRNAs Are Compatible for Click Functionalization
Prehybridization click-conjugation of fluorescent and pull-down tags onto sgRNAs was achieved by reacting 1Az and 2Az with strained alkynes Cy3-DBCO and biotin-DBCO (Figure S9 and Figure S11a). The reaction products were analyzed by imaging the gel at Cy3 wavelength and or using Stains-All reagent. The staining step revealed the click functionalization of sgRNAs wherein the product bands migrated slowly compared to the azide-labeled sgRNAs produced by distributive and processive incorporation of AMUTP (Figure S11 and Table S3). In addition, fluorescence image of the gel confirmed the Cy3 labeling of RNAs.
Modified sgRNAs Are Functional
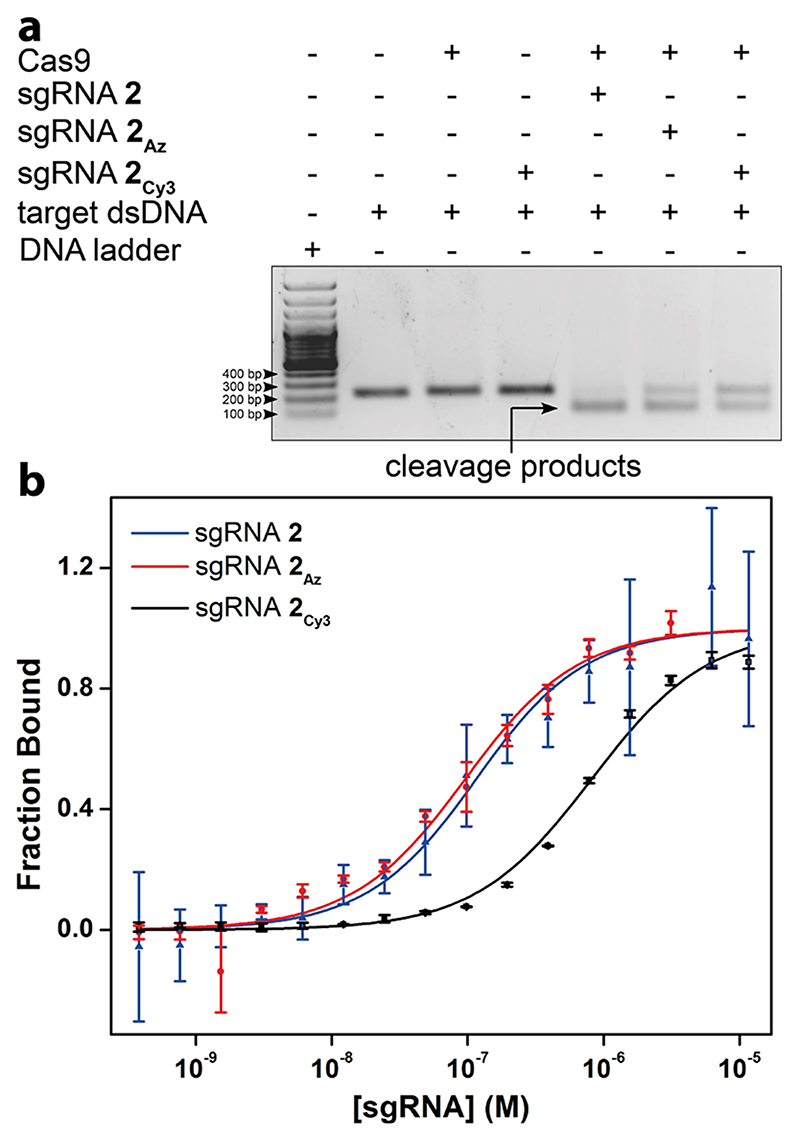
To evaluate whether the modified sgRNAs are functional, we performed cleavage reactions using nuclease protein, Cas9 on a target dsDNA (216 bp) corresponding to a segment of eGFP gene. 1:1 molar equivalence of Cas9 and unlabeled sgRNA 2, azide-labeled (2Az), or Cy3-conjugated sgRNA (2Cy3) were incubated at room temperature to form the ribonucleoprotein (RNP) complex. The target dsDNA was added to the complex in the binding buffer, incubated for 30 min at 37 °C, and resolved by agarose gel. The sgRNAs cleaved the dsDNA to produce two fragments of very similar length expected for this target (~110 and ~106 bp, Figure 5a). While sgRNA 2 produced almost complete cleavage under the reaction conditions, we observed a progressive reduction in cleavage efficiency in the presence of azide-labeled and click functionalized sgRNAs, which is possibly due to differences in binding affinity of modified sgRNAs to Cas9 or RNP complex to target DNA sequence.
Figure 5.
(a) Modified sgRNA-Cas9 complexes cleave target dsDNA. The target dsDNA in the presence of RNP complex produced two fragments of very similar length consistent with the site of cleavage. (b) Curve fits for the binding of unlabeled-(2), AMU-labeled (2Az), and Cy3 click-labeled (2 Cy3) sgRNAs to dCas9-eGFP derived from MST. Values are denoted as mean ± s.d. for n = 2 independent experiments.
In the CRISPR editing or targeting process, sgRNA initially binds to Cas9/dCas9 to form an RNP complex, and subsequently this binary complex binds to the target dsDNA.57,58 The major structural changes in the protein complex occur during the formation of sgRNA-Cas9/sgRNA-dCas9 RNP as compared to the RNP-target dsDNA ternary complex.59 Therefore, any inhibition of RNP complex formation would prevent binding to the target dsDNA and its function. First we determined the efficiency of formation of the RNP complex by microscale thermophoresis (MST). dCas9-eGFP fusion protein was titrated with increasing concentration of sgRNA 2, azide-labeled sgRNA 2Az, or Cy3-labeled sgRNA 2Cy3 and the binding was monitored by MST using the eGFP tag. A similar apparent dissociation constant (Kd) of 98.1 ± 22.1 nM and 87.4 ± 19.8 nM was observed for the sgRNA 2 and sgRNA 2Az (Figure 5b). The Cy3-labeled sgRNA 2Cy3 showed significantly decreased binding affinity (Kd = 799.5 ± 87.6 nM) as compared to the unlabeled and azide-labeled sgRNAs. These results indicate that sgRNA containing a minimally perturbing azide tail does not hamper the binary complex formation as opposed to Cy3-clicked sgRNA.
Further, the ability of an azide-ffinctionalized sgRNA to form a CRISPR ternary complex was investigated by titrating RNP complexes made of dCas9-eGFP and control unlabeled sgRNA 2 or azide-labeled sgRNA 2Az with the target dsDNA (corresponding to eGFP sequence) and recording MST. The dissociation constants for the ternary complexes composed of RNP complexes from control unlabeled (148.7 ± 7.2 nM) and azide-labeled (142.1 ± 12 nM) sgRNAs were very similar (Figure S12). dCas9-eGFP alone and an RNP complex made of a nontargeting sgRNA (also azide-labeled) showed poor binding affinity for the target dsDNA (Kd = 1.17 ± 0.02 μM and 1.17 ± 0.19 μM, respectively). These results indicate that azide-labeled sgRNA, much like native sgRNA, forms a strong ternary complex specifically with the target DNA.
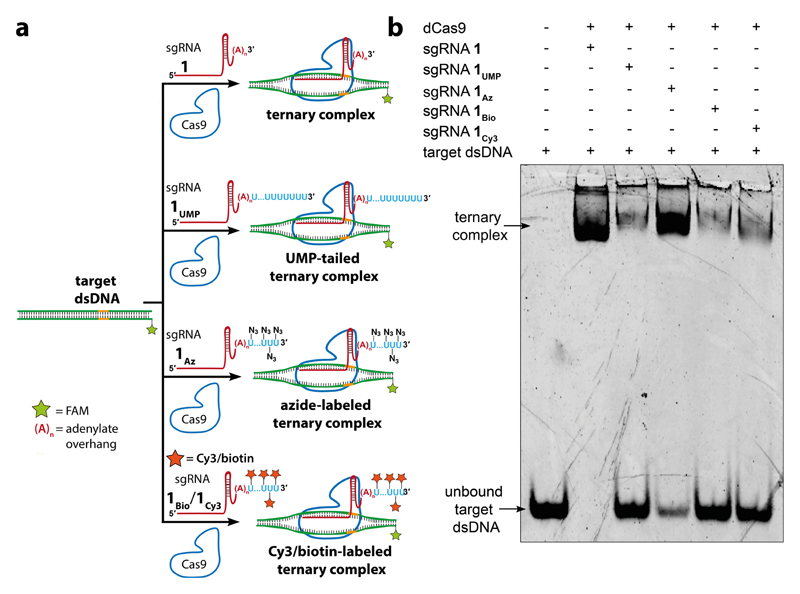
In order to determine whether the posthybridization or prehybridization click labeling approach is best suited for displaying functional tags on the target gene, in this case telomeres, we additionally studied the formation of ternary complex by electrophoretic mobility shift assay (EMSA). sgRNAs (unlabeled 1, UMP-tailed 1UMP, azide-labeled 1Az, biotin-conjugated 1Bio, or Cy3-conjugated 1Cy3) were incubated with dCas9 for 10 min and then with the target dsDNA in a binding buffer (Figure 6a). The dsDNA contained a short telomeric repeat sequence, and one of the strands was labeled with a FAM probe. The samples were resolved by native PAGE and imaged using a gel scanner at FAM and Cy3 wavelengths (Figure 6b and Figure S13). The unlabeled sgRNA 1 showed complete binding to form the ternary complex evident from the formation of a band displaying decreased migration as compared to the dsDNA. On the contrary, UMP-tailed sgRNA showed poor binding. The azide-labeled sgRNA 1Az showed excellent binding of ~85% with a much lower amount of the unbound fraction. It is to be noted that since AMUTP is incorporated with less efficiency compared to natural UTP in terminal uridylation reaction, there is a lesser number of AMUs at the 3’ end of 1Az as compared to Us in 1UMP (vide supra, Figure 4b). Hence, UMP-tail on sgRNA could have hindered its binding to dCas9. Therefore, we presume that the lesser incorporation efficiency of AMUTP has in a way assisted the efficient binding of 1Az to dCas9. However, clicked sgRNAs (1Bio and 1Cy3) showed poor binding efficiency (20—25%) to form the ternary complex potentially due to the introduction of bulky tags (Figure 6b). The formation of a small percentage of Cy3-conjugated ternary complex was also evident upon imaging the gel at Cy3 wavelength (Figure S13). Collectively, minimally invasive nature of the azide label complemented by these results indicate that posthybridization click functionalization approach, i.e., functionalizing ternary complex, would be the method of choice for the chemical display of functional tags on gene of interest.
Figure 6. Azide-labeled sgRNA forms a ternary complex with dCas9 and target DNA efficiently.
(a) Telomeric repeat-containing dsDNA target was incubated with dCas9 and sgRNA 1, UMP-tailed sgRNA 1UMP, azide-labeled sgRNA 1Az, biotin-conjugated sgRNA 1Bio, or Cy3-conjugated sgRNA Icy3. The formation of ternary complex (dCas9-sgRNA-dsDNA) was studied by PAGE under native conditions. (b) Fluorescence image of the gel at FAM wavelength. EMSA results suggest that posthybridization click functionalization is the most favorable approach to display functional tags on target gene.
Azide-Tailed sgRNA Displays the Clickable Tags on Target Gene Locus
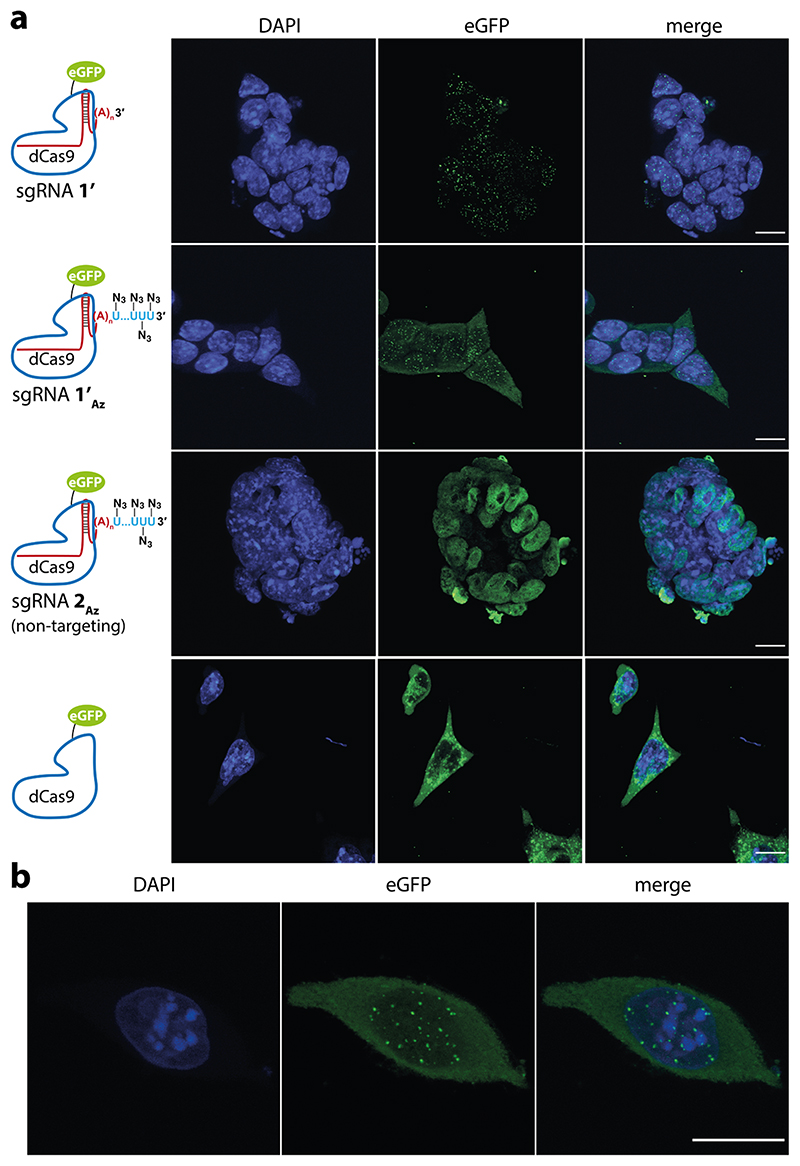
The localization of azide-labeled sgRNA on the telomeric region of mouse embryonic stem cells (mESCs) was confirmed by CRISPR-FISH.28 Fixed and permeabilized mESCs were incubated with an RNP complex composed of either unlabeled sgRNA 1’, azide-labeled sgRNA 1’A (targeting telomeric region) or azide-labeled sgRNA 2Az (targeting eGFP) and dCas9-eGFP fusion protein (Table 1). sgRNAs, 1’ and 1’Az, were designed to match a validated protospacer sequence used to target telomeric repeat in cells.60,61 Nuclear puncta corresponding to telomeres were visualized for both 1’ and 1’Az, which is consistent with the telomere localization pattern established using fluorescent dCas9 fusion proteins (Figure 7a, see Figure 7b for single cell image).13,28,61,62 In a control experiment, nontargeting sgRNA 2Az showed a fluorescence signal, which was nonspecifically distributed all throughout the nucleus. Further, incubation with dCas9-eGFP alone resulted in aggregates mostly localized outside the nucleus. Collectively, these observations confirmed that the RNP complex made of azide-tailed sgRNA can be efficiently used to localize clickable groups on the target gene.
Figure 7. Azide-tailed sgRNA 1’Az localizes on telomeric region.
(a) mESCs were treated with dCas9-eGFP alone or its RNP complex with unlabeled sgRNA 1’ (targeting telomere), azide-labeled sgRNA 1’Az (targeting telomere) and azide-labeled sgRNA 2Az (targeting eGFP: nontargeting control). The cells were imaged in DAPI and eGFP channels, and maximally projected Z-stacks are shown. First two rows: RNP complex of 1’ and 1’Az directs the localization to telomeric regions visualized as nuclear puncta. Third row: RNP complex of 2Az (nontargeting) and dCas9-eGFP nonspecifically distributes throughout the nucleus with no observable puncta. Last row: incubation with dCas9-eGFP alone resulted in aggregates, which were mostly localized outside the nucleus (scale bar, 10 μm). (b) Maximally projected Z-stack image showing nuclear puncta localized to telomeres in a single embryonic stem cell obtained using RNP complex of 1’Az and dCas9-eGFP (scale bar, 10 μm). The experiments were performed as n = 2 biological replicates.
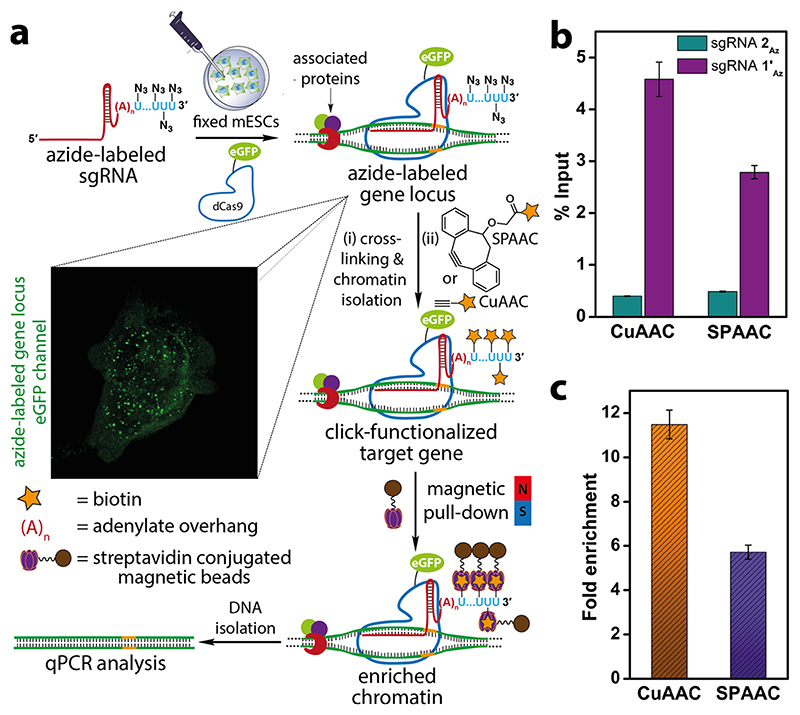
sgR-CLK Enables Site-Directed Functional Tagging of Target Gene
To evaluate the compatibility of azide tags localized on the telomeric region for click-functionalization we performed CuAAC and SPAAC reactions on the isolated chromatin to enrich the target gene (Figure 8a). Fixed and permeabilized mESCs were incubated with azide-labeled sgRNA 1’Az and dCas9-eGFP complex. The localization of azide labels on the telomere region was initially confirmed by the presence of nuclear puncta in the eGFP channel (Figure 8a). The cells were then cross-linked, and the nuclear isolate was sheared until chromatin fragments of the range of 200—500 bp were obtained. CuAAC reaction was performed on chromatin using a biotin-alkyne in the presence of a catalytic system formed by CuSO4, sodium ascorbate, and THPTA. In case of SPAAC reaction, chromatin was first treated with iodoacetamide to sequester protein thiols on chromatin so as to reduce nonspecific enrichment resulting from the reaction between the strained biotin-alkyne substrate and thiols.63 Further, strained alkyne system, sDIBO was preferred in this reaction as DBCO-based alkynes are known to show nonspecific reaction with cellular thiols (Figure S9).63 After click reaction, unreacted biotin substrate was removed and biotinylated-chromatin was incubated with streptavidin-coated magnetic beads. The captured chromatin was eluted, de-cross-linked, and the isolated genomic DNA fragments were subjected to qPCR using telomeric primers as previously described.64 In parallel, a control azide-labeled sgRNA 2Az was subjected to the same experimental procedure to account for the background enrichment from nontarget sgRNA and click reaction. sgR-CLK in the presence of 1’Az revealed significant enrichment of the telomere region by both CuAAC and SPAAC reactions (Figure 8b). On the other hand, chromatin captured using nontargeting sgRNA 2Az labeled by click reactions showed very low enrichment. Further, fold enrichment of the telomeric repeat region calculated for sgRNA 1’Az upon normalizing with sgRNA 2Az was found to be nearly 12 and 6 while employing CuAAC and SPAAC reactions (Figure 8c). Notably, CuAAC reaction exhibited better pull-down efficiency as compared to SPAAC reaction, possibly due to better accessibility and reactivity of the terminal alkyne substrate as compared to the bulky cyclooctyne counterpart.65 These results clearly validate the key concept of this study, i.e., directing multiple clickable groups to a specific region of chromatin, which enables the display of synthetic recruits by click chemistry.
Figure 8. Site-directed functionalization of telomeres by sgR-CLK was confirmed by ChIP-qPCR.
(a) Schematic diagram illustrating the steps involved in chromatin capture by in situ click reaction performed on the target-bound ternary complex using biotin-alkynes. While CuAAC reaction was performed using a biotin substrate containing a terminal alkyne, SPAAC reaction was performed using biotin-conjugated to a strained alkyne (sDIBO). See Figure S9 for complete structure of alkyne substrates. Biotinylated chromatin was enriched using streptavidin beads and subjected to qPCR analysis. (b) qPCR analysis of enriched chromatin obtained by CuAAC and SPAAC reactions using telomere-targeting sgRNA 1’Az and control nontargeting sgRNA 2Az. The enrichment is expressed relative to respective inputs after click reaction step. (c) A plot showing the fold enrichment of telomere DNA using sgRNA 1’Az normalized over sgRNA 2Az for CuAAC and SPAAC reactions (see Experimental Section for details). Values in b and c are denoted as mean ± s.d. performed as n = 3 technical replicates.
Conclusions
In this work, we have developed a novel CRISPR-based technique called sgR-CLK to recruit small-molecule functional tags on the target gene by combining the power of bioorthogonal azide–alkyne click chemistry and our ability to engineer sgRNAs using a uridylyl transferase enzyme (SpCID1). The azide-labeled sgRNAs accessed via this approach were functional and enabled locus-specific display of multiple azide groups on the target telomeric loci. Further, click reaction performed on the azide handles enabled the sitespecific display of biotin tags on chromatin, resulting in significant fold-enrichment of telomeres upon chromatin capture in comparison to other CRISPR-based telomere capture tools. Currently used strategies to tag dCas9 with synthetic molecules employing thiol-maleimide chemistry require selective mutation of cysteine residues on the protein to prevent nonspecific ligation.66 In this context, sgR-CLK using azide–alkyne cycloaddition chemistry is highly sitespecific and bioorthogonal. Alternatively, site-specific enzymatic ligation strategies use dCas9 fused to large enzyme tags like Halo (~33 kDa) or SNAP (~20 kDa) to facilitate the recruitment of the small molecules.28 However, introducing an array of small molecules by these approaches would require fusion of multiple enzyme tags, which could potentially affect the RNP complex formation and, hence, compromise its targetability. Although the trans-splicing strategy using split intein system has been used in the multimeric display of probes, it requires elaborate setup and custom synthesis of individual intein fragment-conjugated to small molecules.23 In comparison, our technology provides an easy platform to install multiple minimally invasive azide handles compatible for introducing functional molecules on a target gene by click chemistry, which could enhance the efficacy and specific action of the molecule.
sgR-CLK has several additional advantages. First, it is modular on two counts. Our design enables insertion of multiple azide groups at the 3’ end of any sgRNA sequence. TUTase can be used to incorporate other minimally perturbing bioorthogonal reactive groups such as alkyne, vinyl, and cyclopropene, which would allow introduction of a range of functions by using a cognate reaction partner.67,68 Hence, a combination of these two features can be conceivably used to prepare an array of clickable sgRNAs targeting the same or different gene loci to either achieve a robust functional output at a designated genomic site or interrogate multiple genes in parallel.69 On the whole, sgR-CLK is anticipated to open up new experimental strategies for gene-specific display of chemical cargos for various applications.
Experimental Section
Detailed procedures for cloning, expressing SpCID1, single nucleotide incorporation, and click labeling of RNA are described in the Supporting Information.
Uridylation of RNA Oligonucleotide Using SpCID1
Model 5’-FAM-labeled RNA oligonucleotide (10 μM, Table 1) was incubated with UTP, AMUTP, APUTP, or ATUTP (500 μM) in the presence of Tris-HCl buffer (10 mM, pH 7.9 at 25 °C), NaCl (50 mM), MgCl2(10 mM), DTT (2 mM), RiboLock RNase inhibitor (1 U/μL), and 1 μL of SpCID1 (10.25 pmol) in a final volume of 20 μL. After 5, 15, and 30 min, 5 μL aliquots of reaction mixture (50 pmol of RNA oligonucleotide) were mixed with 15 μL of denaturing loading buffer (7 M urea in 10 mM Tris-HCl, 100 mM EDTA, 0.05% bromophenol blue, pH 8) and heat-denatured at 75 °C for 3 min. Five μL of the sample was loaded on to 20% denaturing polyacrylamide gel, electrophoresed, and imaged using Typhoon gel scanner at FAM wavelength.
Constructing sgRNAs Containing 3’-Adenylate Overhang
Forward (CFP1, CFP1’ or CFP2) and reverse CRP primers (1 μM) were incubated with sgRNA templates CT1, CT1’ or CT2 (44 nM) in EmeraldAMP GT PCR master mix in a final volume of 25 μL. PCR conditions: heat denaturation at 94 °C for 1 min, 35 cycles of (denaturing: 94 °C for 20 s, annealing: 64 °C for 20 s, extension: 68 °C for 30 s), final extension at 68 °C for 5 min. Multiple small-scale PCRs were performed to isolate dsDNA templates required for large-scale transcription reaction. The amplicons were purified using NucleoSpin Gel and PCR Clean-up kit. PCR with CT1, CT1’ and CT2 yielded dsDNA templates for the synthesis of sgRNAs 1, 1’, and 2. For primer and template sequences see Table S1.
In vitro transcription reactions were performed using ATP, GTP, CTP, and UTP (2 mM), RiboLock RNase inhibitor (0.4 U/μL), respective dsDNA templates (300 nM) and T7 RNA polymerase (3.2 U/μL) in 40 mM Tris-HCl (pH 7.9), 10 mM MgCl2, 20 mM DTT, 10 mM NaCl, and 1 mM spermidine in a final reaction volume of 250 μL. The reaction was incubated at 37 °C for 12 h. 3’-Adenylate sgRNAs 1, 1’, and 2 were isolated by MEGAclear Transcription Clean-Up kit using the manufacturer’s protocol. A typical 250 μL transcription reaction yielded 2-3 nmol of the sgRNAs.
Azide-Tailed sgRNAs Using SpCID1
In vitro transcribed sgRNA 1 (10 μM, 200 pmol) was incubated with natural UTP or AMUTP (500 μM) in the presence of Tris-HCl buffer (10 mM, pH 7.9 at 25 °C), NaCl (50 mM), MgCl2 (10 mM), DTT (2 mM), RiboLock RNase inhibitor (1 U/μL), and SpCID1 (20.5 pmol) in a final volume of 20 μL. The reaction was incubated at 37 °C for 30 min and further heat-denatured by incubating at 75 °C for 3 min. The reaction mixture (150 pmol) was resolved in 8.5% denaturing polyacrylamide gel. Bands corresponding to UMP-tailed sgRNA 1UMP and azide-labeled sgRNA 1Az were visualized by Stains-All reagent (Figure 4).
Multiple such small-scale (20 μL, 200 pmol of sgRNA) reactions were performed with 1, 1’, and 2 and the reactions were pooled together and the product sgRNAs 1Az, 1’Az, and 2Az were precipitated by addition of 1 volume of 5 M ammonium acetate and 10 volume of ethanol followed by incubation for 2 h at -20 °C. The samples were centrifuged at 15 000 rpm for 15 min and RNA pellets obtained were washed with chilled 75% ethanol in water and sgRNAs 1Az, 1’Az, and 2Az thus obtained were dissolved in autoclaved water. See Table S3 for yields.
In Vitro Cleavage Assay
A 216 bp dsDNA template corresponding to a segment of eGFP gene required for cleavage assay was generated by PCR using forward (eGFPF) and reverse (eGFPR) primers (0.5 μM). The primers in the presence of dNTPs (0.2 mM) were incubated with linearized eGFP-N1 plasmid (100 ng) and Taq DNA polymerase (2.5 U) in 1× PCR buffer in a final volume of 50 μL. PCR conditions: heat denaturation at 95 °C for 5 min, 35 cycles of (denaturing: 95 °C for 30 s, annealing: 58 °C for 30 s, extension: 72 °C for 30 s) and final extension at 72 °C for 5 min. The amplicons were purified by using PCR Clean-up kit.
A 1:1 solution (1.25 μM) of sgRNA 2, azide-labeled sgRNA 2Az, or Cy3-labeled sgRNA 2Cy3 and Cas9 was incubated at room temperature for 10 min. eGFP dsDNA was mixed with the above RNP complexes in 20 mM HEPES (pH 7.5), 100 mM KCl, 5 mM MgCl2, 1 mM DTT to make a final volume of 15 μL. The final concentration of dsDNA and RNP complex was 50 nM and 250 nM, respectively. The reaction was performed for 30 min at 37 °C. Proteinase K (40 μg) was added to the reaction mix and incubated at 55 °C for 30 min. Proteinase K was deactivated by incubating at 70 °C for 10 min, and RNase A (10 μg) was added. DNA gel loading buffer was added, and the samples were loaded on a 2% agarose gel, resolved, and stained using EtBr (Figure 5a).
MST Analysis to Quantify RNP Complex
70 dCas9-eGFP (180 nM) in 20 mM HEPES (pH 7.5), 150 mM KCl, 10 mM MgCl2 and 1 mM DTT was incubated with varying concentrations of sgRNA 2, azide-labeled sgRNA 2Az, or Cy3-labeled sgRNA 2Cy3 (0.4 nM to 12.5 μM) in a final volume of 10 μL for 10 min at room temperature. The samples (~5 μL) were loaded onto standard treated quartz capillary and excited at 60% LED power and 40% MST power at 25 °C using blue filter, and MST was recorded. The experiments were performed individually as two replicates. Data was analyzed using MST analysis software to determine bound protein fraction with respect to sgRNA concentration.
MST Quantification of Ternary Complex
70 A 29 bp target dsDNA corresponding to eGFP sequence was prepared by annealing ssDNAs (TS3 and TS4) at 95 °C for 5 min followed by slowly cooling to 25 °C (Table S1). RNP complexes consisting of sgRNA 2, azide-labeled sgRNA 2Az, azide-labeled sgRNA 1’Az (360 nM, nontargeting), and dCas9-eGFP (360 nM) in 20 mM HEPES (pH 7.5), 150 mM KCl, 10 mM MgCl2, and 1 mM DTT were generated by incubating at 25 °C for 10 min. The RNP complexes (360 nM) or dCas9-eGFP (360 nM) alone were incubated with varying concentrations of target dsDNA (0.9 nM to 30 μM) in 20 mM HEPES (pH 7.5), 150 mM KCl, 10 mM MgCl2 in a final volume of 10 μL for 30 min at 37 °C. The samples (~5 μL) were loaded onto standard treated quartz capillary and excited at 40% LED power and 40% MST power at 25 °C using blue filter, and MST was recorded. The experiments were performed individually as two replicates. Data was analyzed using MST analysis software to determine the bound dCas9 or RNP fraction with respect to dsDNA concentration.
EMSA to Estimate Ternary Complex Formation
sgRNA 1, 1UMP, 1AZ, 1BIO, or 1Cy3 (2 μM) was incubated with dCas9 (2.5 μM) in the presence of RiboLock RNase inhibitor (4 U/μL) in a total volume of 5 μL at room temperature for 10 min. Five μL solution of target dsDNA (0.2 μM, formed by annealing TS1 and TS2 DNA oligonucleotides, Table S1) and DTT (20 μM) was added to the individual RNP complex in binding buffer (20 mM HEPES (pH 7.5), 100 mM KCl, 5 mM MgCl2, 1 mM DTT, 1 mg/mL BSA, 0.1% Triton X-100, 5% glycerol). The samples were incubated for 30 min at 37 °C. The samples (0.5 pmol DNA) were resolved by native PAGE (8% gel containing 10 mM MgCl2) at 4 °C in 1× TBE buffer supplemented with 10 mM MgCl2. The gel was imaged using Typhoon gel scanner at FAM wavelength (Figure 6b).
Cellular Localization of Azide-Labeled sgRNA on Telomeres
mESCs (R1/E) were seeded in coverslips and cultured in DMEM in the presence of leukemia inhibiting factor (LIF) at 37 °C and 5% CO2 to maintain cells in its pluripotent state.71 Postseeding (24 h), the media was removed and cells were washed with 1× PBS. The cells were then fixed in prechilled 50% acetic acid in methanol at -20 °C for 20 min. The cells were washed three times with 1 × PBS for 5 min with gentle shaking. Further, the cells were permeabilized by incubating in hybridization buffer (20 mM HEPES (pH 7.5), 150 mM KCl, 10 mM MgCl2, 1 mM DTT, 2% BSA, 5% glycerol and 0.1% Triton-X) at 37 °C for 30 min. dCas9-eGFP (200 nM) and sgRNA 1’, sgRNA 1’Az, or sgRNA 2Az (200 nM) and RiboLock RNase inhibitor (1.6 U/μL) were incubated for 20 min at room temperature in hybridization buffer in a final volume of 50 μL. The cells were incubated with the respective RNP complex or dCas9-eGFP alone in a humid chamber for 4 h at 37 °C and then washed three times with hybridization buffer for 5 min. The cells were stained with 5 μg/mL DAPI and the coverslip was mounted on glass slides for imaging. The cells were visualized in Leica TCS SP8 confocal microscope in DAPI and eGFP channels. Images were deconvoluted on Leica LAS X software using blind deconvolution and Z-stacks were projected (maximum intensity) using Fiji image analysis software (Figure 7).
Chromatin Capture Using sgR-CLK
mESCs (R1/E) were seeded in 10 cm dish and cultured in DMEM in the presence of leukemia inhibiting factor (LIF) at 37 °C and 5% CO2. At around 70% confluency, cells were fixed and permeabilized as above. dCas9-eGFP (200 nM), azide-labeled sgRNA 1’Az or 2Az (200 nM), and SUPERaseIn RNase Inhibitor (0.1 U/μL) were incubated in hybridization buffer for 20 min at room temperature in a final volume of 1 mL for the formation of RNP complex. The cells were incubated with the above solution in a humid chamber for 1.5 h at 37 °C and then washed with 1× PBS for 5 min. Cells were cross-linked with 1% formaldehyde in DMEM for 10 min at room temperature with gentle shaking. Further, glycine (125 mM) was added and incubated for 5 min. Cells were harvested and washed twice with 1× protease inhibitor cocktail (PIC) in 1× PBS. For nuclear isolation, cells were resuspended in 2 mL of PBS followed by the addition of 2 mL of nuclear isolation buffer (40 mM Tris-HCl pH 7.5, 1.28 M sucrose, 20 mM MgCl2 and 4% Triton X-100) in a final volume of 10 mL. Cell suspension was incubated on ice for 20 min with frequent mixing. The nuclear isolate was centrifuged at 2500g for 15 min at 4 °C. The nuclear pellet obtained was resuspended in 1 mL RNA immunoprecipitation buffer (25 mM Tris HCl pH 7.5, 150 mM KCl, 5 mM EDTA pH 8.0, 0.5 mM DTT, 0.5% Igpal, 0.1 U/μL SUPERaseIn RNase Inhibitor, 1× PIC) and sonicated for 4 cycles of 15 min (30 s on and 30 s off) at 4 °C in a Bioruptor Plus sonication device. After sonication, the fragment size (200—500 bp) was confirmed by agarose gel electrophoresis. Further, the nuclear debris were centrifuged at 13 300 rpm for 10 min, and the supernatant was transferred to a fresh vial to afford the chromatin. For SPAAC reaction, chromatin was treated with 50 mM iodoacetamide in a final volume of 1 mL for 60 min at room temperature. Samples were buffer-exchanged to 1× PBS using 10 kDa cutoff columns at 4 °C. SPAAC reaction on chromatin was performed with 100 μM biotin-sDIBO alkyne in a final volume of 500 μL having 0.1 U/μL SUPERaseIn RNase Inhibitor. CuAAC reaction on chromatin was performed with 100 μM biotin-alkyne in the presence of 1 mM CuSO4, 1 mM sodium ascorbate, 1 mM THPTA. Both the reactions were performed for 1 h at room temperature. The excess biotin was removed by buffer exchanging to 1 × PBS using 10 kDa size cutoff columns to a final volume of 1 mL.
Meanwhile, 50 μL of Dynabeads MyOne Streptavidin C1 magnetic beads for each sample of the click reaction was washed thrice with 1× PBS. 5% volume inputs (50 μL) were taken out, stored at —20 °C, and remaining samples were treated with 50 μL of Dynabeads, 0.1 U/ μL SUPERaseIn RNase Inhibitor and incubated overnight with gentle rotation at 4 °C. The immobilized samples were washed with 1 mL of each wash buffer for 5 min at 4 °C with gentle rotation (once with low salt buffer: 20 mM Tris HCl (pH 8.0), 150 mM NaCl, 2 mM EDTA, 1% Triton X-100, 0.1% SDS; once with high salt buffer: 20 mM Tris HCl (pH 8.0), 500 mM NaCl, 2 mM EDTA, 1% Triton X-100, 0.1% SDS; once with LiCl buffer: 10 mM Tris HCl (pH 8.0), 1 mM EDTA (pH 8.0), 250 mM LiCl, 1% sodium deoxycholate, 1% Igpal; twice with TE buffer: 10 mM Tris HCl (pH 8.0), 1 mM EDTA).
The chromatin from each sample was eluted from the beads by incubating with 500 μL of elution buffer (100 NaHCO3, 1% SDS) at room temperature for 15 min. Input samples were also treated with 500 μL of elution buffer under same conditions. The eluted chromatin samples were de-cross-linked by incubating in 200 mM NaCl at 65 °C overnight. Each sample was treated with proteinase K (20 μg), 10 μL of 0.5 M EDTA (pH 8.0) and 20 μL of 1 M Tris HCl (pH 7.5) at 42 °C for 60 min for digesting the proteins. Genomic DNA fragments from each sample were isolated using PCR-clean up kit and subjected to qPCR.
qPCR to Estimate Telomere DNA Enrichment
To 2 μL of 1% of input or 2 μL of pull-down samples corresponding to azide-labeled sgRNA 1’Az and 2Az obtained by SPAAC and CuAAC reactions were added 7.5 μL TB Green Premix Ex TaqII and 1 μL TeloF and TeloR primer (0.5 μM each, Table S1). The final volume was adjusted to 15 μL with water. qPCR conditions: heat denaturation at 95 °C for 1 min, 40 cycles of (denaturing: 95 °C for 10 s, annealing: 52 °C for 30 s, extension: 72 °C for 30 s), final extension at 72 °C for 5 min. The Ct values for input were adjusted to 100%, and then pull-down values were normalized with 100% input and represented as enrichment relative to respective inputs for sgRNA 1’Az and sgRNA 2Az for CuAAC and SPAAC reactions, respectively. Further, the fold enrichment for telomere-pull down with sgRNA 1’Az over nontargeting sgRNA 2AZ was calculated for both the click reactions using the equation given below. Reactions were performed in triplicate.
Supplementary Material
Acknowledgements
We thank Dr. Gayathri Pananghat and Dr. Amrita Hazra for their help in cloning and protein purification experiments. We thank Rupam Bhattacharjee for his help in the mass analysis of RNA oligonucleotides. J.T.G. thanks IISER Pune and Wellcome Trust-DBT India Alliance for graduate research fellowship. D.C. acknowledges a research grant from DBT (GAP 0175). This work was supported by Wellcome Trust-DBT India Alliance senior fellowship (IA/S/16/1/502360) to S.G.S.
Footnotes
The authors declare no competing financial interest.
Contributor Information
Jerrin Thomas George, Department of Chemistry, Indian Institute of Science Education and Research (IISER), Pune 411008, India.
Mohd. Azhar, Chemical and Systems Biology Unit, Council of Scientific and Industrial Research-Institute of Genomics & Integrative Biology, New Delhi 110025, India
Meghali Aich, Genomics and Molecular Medicine Unit, Council of Scientific and Industrial Research-Institute of Genomics & Integrative Biology, New Delhi 110025, India.
Dipanjali Sinha, Chemical and Systems Biology Unit, Council of Scientific and Industrial Research-Institute of Genomics & Integrative Biology, New Delhi 110025, India.
Uddhav B. Ambi, Department of Chemistry, Indian Institute of Science Education and Research (IISER), Pune 411008, India
References
- (1).Doudna JA, Charpentier E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346:1258096. doi: 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]
- (2).Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genomee Engineering. Cell. 2014;157:1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Sander JD, Joung JK. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat Biotechnol. 2014;32:347–355. doi: 10.1038/nbt.2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Raper AT, Stephenson AA, Suo Z. Sharpening the scissors: Mechanistic details of CRISPR/Cas9 improve functional understanding and inspire future research. J Am Chem Soc. 2018;140:11142–11152. doi: 10.1021/jacs.8b05469. [DOI] [PubMed] [Google Scholar]
- (5).Kim H, Kim J-S. A guide to genome engineering with programmable nucleases. Nat Rev Genet. 2014;15:321–334. doi: 10.1038/nrg3686. [DOI] [PubMed] [Google Scholar]
- (6).Chen T, Gao D, Zhang R, Zeng G, Yan H, Lim E, Liang F-S. Chemically controlled epigenome editing through an inducible dCas9 system. J Am Chem Soc. 2017;139:11337–11340. doi: 10.1021/jacs.7b06555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Chavez A, Scheiman J, Vora S, Pruitt BW, Tuttle M, P R Iyer E, Lin S, Kiani S, Guzman CD, Wiegand DJ, Ter-Ovanesyan D, et al. Highly efficient Cas9-mediated transcriptional programming. Nat Methods. 2015;12:326–328. doi: 10.1038/nmeth.3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Didovyk A, Borek B, Tsimring L, Hasty J. Transcriptional regulation with CRISPR-Cas9: principles, advances, and applications. Curr Opin Biotechnol. 2016;40:177–184. doi: 10.1016/j.copbio.2016.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Gaudelli NM, Komor AC, Rees HA, Packer MS, Badran AH, Bryson DI, Liu DR. Programmable base editing of A•T to G•C in genomic DNA without DNA cleavage. Nature. 2017;551:464–471. doi: 10.1038/nature24644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Molla KA, Yang Y. CRISPR/Cas-mediated base editing: Technical considerations and practical applications. Trends Biotechnol. 2019;37:1121–1142. doi: 10.1016/j.tibtech.2019.03.008. [DOI] [PubMed] [Google Scholar]
- (11).Tsui C, Inouye C, Levy M, Lu A, Florens L, Washburn MP, Tjian R. dCas9-targeted locus-specific protein isolation method identifies histone gene regulators. Proc Natl Acad Sci U S A. 2018;115:E2734–E2741. doi: 10.1073/pnas.1718844115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Qiu W, Xu Z, Zhang M, Zhang D, Fan H, Li T, Wang Q, Liu P, Zhu Z, Du D, Tan M, et al. Determination of local chromatin interactions using a combined CRISPR and peroxidase APEX2 system. Nucleic Acids Res. 2019;47:e52. doi: 10.1093/nar/gkz134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Chen B, Gilbert LA, Cimini BA, Schnitzbauer J, Zhang W, Li G-W, Park J, Blackburn EH, Weissman JS, Qi LS, Huang B. Dynamic imaging of genomic loci in living human cells by an optimized CRISPR/Cas system. Cell. 2013;155:1479–1491. doi: 10.1016/j.cell.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Wang H, La Russa M, Qi LS. CRISPR/Cas9 in Genome Editing and Beyond. Annu Rev Biochem. 2016;85:227–264. doi: 10.1146/annurev-biochem-060815-014607. [DOI] [PubMed] [Google Scholar]
- (15).Fujita T, Fujii H. Efficient isolation of specific genomic regions and identification of associated proteins by engineered DNA-binding molecule-mediated chromatin immunoprecipitation (en-ChIP) using CRISPR. Biochem Biophys Res Commun. 2013;439:132–136. doi: 10.1016/j.bbrc.2013.08.013. [DOI] [PubMed] [Google Scholar]
- (16).Williams P, Li L, Dong X, Wang Y. Identification of SLIRP as a G quadruplex-binding protein. J Am Chem Soc. 2017;139:12426–12429. doi: 10.1021/jacs.7b07563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Pflueger C, Tan D, Swain T, Nguyen T, Pflueger J, Nefzger C, Polo JM, Ford E, Lister R. A modular dCas9-SunTag DNMT3A epigenome editing system overcomes pervasive off-target activity of direct fusion dCas9-DNMT3A constructs. Genome Res. 2018;28:1193–1206. doi: 10.1101/gr.233049.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Papikian A, Liu W, Gallego-Bartolomé J, Jacobsen SE. Site-specific manipulation of Arabidopsis loci using CRISPR-Cas9 SunTag systems. Nat Commun. 2019;10:729. doi: 10.1038/s41467-019-08736-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Zalatan JG, Lee ME, Almeida R, Gilbert LA, Whitehead EH, La Russa M, Tsai JC, Weissman JS, Dueber JE, Qi LS, Lim WA. Engineering complex synthetic transcriptional programs with CRISPR RNA scaffolds. Cell. 2015;160:339–350. doi: 10.1016/j.cell.2014.11.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Adli M. The CRISPR tool kit for genome editing and beyond. Nat Commun. 2018;9:1911. doi: 10.1038/s41467-018-04252-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Qin P, Parlak M, Kuscu C, Bandaria J, Mir M, Szlachta K, Singh R, Darzacq X, Yildiz A, Adli M. Live cell imaging of low-and non-repetitive chromosome loci using CRISPR-Cas9. Nat Commun. 2017;8:14725. doi: 10.1038/ncomms14725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Shechner DM, Hacisuleyman E, Younger ST, Rinn JL. Multiplexable, locus-specific targeting of long RNAs with CRISPR-Display. Nat Methods. 2015;12:664–670. doi: 10.1038/nmeth.3433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Liszczak GP, Brown ZZ, Kim SH, Oslund RC, David Y, Muir TW. Genomic targeting of epigenetic probes using a chemically tailored Cas9 system. Proc Natl Acad Sci U S A. 2017;114:681–686. doi: 10.1073/pnas.1615723114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).David Y, Muir TW. Emerging chemistry strategies for engineering native chromatin. J Am Chem Soc. 2017;139:9090–9096. doi: 10.1021/jacs.7b03430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Anders L, Guenther MG, Qi J, Fan ZP, Marineau JJ, Rahl PB, Lovén J, Sigova AA, Smith WB, Lee TI, Bradner JE, et al. Genome-wide localization of small molecules. Nat Biotechnol. 2014;32:92–96. doi: 10.1038/nbt.2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Kellner MJ, Koob JG, Gootenberg JS, Abudayyeh O, Zhang F. SHERLOCK: nucleic acid detection with CRISPR nucleases. Nat Protoc. 2019;14:2986–3012. doi: 10.1038/s41596-019-0210-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Guk K, Keem JO, Hwang SG, Kim H, Kang T, Lim E-K, Jung J. A facile, rapid and sensitive detection of MRSA using a CRISPR-mediated DNA FISH method, antibody-like dCas9/sgRNA complex. Biosens Bioelectron. 2017;95:67–71. doi: 10.1016/j.bios.2017.04.016. [DOI] [PubMed] [Google Scholar]
- (28).Deng W, Shi X, Tjian R, Lionnet T, Singer RH. CASFISH: CRISPR/Cas9-mediated in situ labeling of genomic loci in fixed cells. Proc Natl Acad Sci U S A. 2015;112:11870–11875. doi: 10.1073/pnas.1515692112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Hemphill J, Borchardt EK, Brown K, Asokan A, Deiters A. Optical Control of CRISPR/Cas9 Gene Editing. J Am Chem Soc. 2015;137:5642–5645. doi: 10.1021/ja512664v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Koopal B, Kruis AJ, Claassens NJ, Nobrega FL, van der Oost J. Incorporation of a synthetic amino acid into dCas9 improves control of gene silencing. ACS Synth Biol. 2019;8:216–222. doi: 10.1021/acssynbio.8b00347. [DOI] [PubMed] [Google Scholar]
- (31).Thompson RE, Stevens AJ, Muir TW. Protein engineering through tandem transamidation. Nat Chem. 2019;11:737–743. doi: 10.1038/s41557-019-0281-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Liu X, Zhang Y, Chen Y, Li M, Zhou F, Li K, Cao H, Ni M, Liu Y, Gu Z, Dickerson KE, et al. In situ capture of chromatin interactions by biotinylated dCas9. Cell. 2017;170:1028–1043. doi: 10.1016/j.cell.2017.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Volkmann G, Iwaï H. Protein trans-splicing and its use in structural biology: opportunities and limitations. Mol BioSyst. 2010;6:2110–2121. doi: 10.1039/c0mb00034e. [DOI] [PubMed] [Google Scholar]
- (34).Kelley ML, Strezoska Ž, He K, Vermeulen A, Smith AvB. Versatility of chemically synthesized guide RNAs for CRISPR-Cas9 genome editing. J Biotechnol. 2016;233:74–83. doi: 10.1016/j.jbiotec.2016.06.011. [DOI] [PubMed] [Google Scholar]
- (35).Højfeldt JW, Van Dyke AR, Mapp AK. Transforming ligands into transcriptional regulators: building blocks for bifunctional molecules. Chem Soc Rev. 2011;40:4286–4294. doi: 10.1039/c1cs15050b. [DOI] [PubMed] [Google Scholar]
- (36).Jao CY, Salic A. Exploring RNA transcription and turnover in vivo by using click chemistry. Proc Natl Acad Sci U S A. 2008;105:15779–15784. doi: 10.1073/pnas.0808480105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Anhäuser L, Hüwel S, Zobel T, Rentmeister A. Multiple covalent fluorescence labeling of eukaryotic mRNA at the poly(A) tail enhances translation and can be performed in living cells. Nucleic Acids Res. 2019;47:e42. doi: 10.1093/nar/gkz084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Sletten EM, Bertozzi CR. Bioorthogonal Chemistry: Fishing for Selectivity in a Sea of Functionality. Angew Chem Int Ed. 2009;48:6974–6998. doi: 10.1002/anie.200900942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).El-Sagheer AH, Brown T. Click chemistry with DNA. Chem Soc Rev. 2010;39:1388–1405. doi: 10.1039/b901971p. [DOI] [PubMed] [Google Scholar]
- (40).Wada T, Mochizuki A, Higashiya S, Tsuruoka H, Kawahara S-i, Ishikawa M, Sekine M. Synthesis and properties of 2-azidodeoxyadenosine and its incorporation into oligodeoxynucleotides. Tetrahedron Lett. 2001;42:9215–9219. [Google Scholar]
- (41).Habibian M, McKinlay C, Blake TR, Kietrys AM, Waymouth RM, Wender PA, Kool ET. Reversible RNA acylation for control of CRISPR-Cas9 gene editing. Chem Sci. 2020;11:1011–1016. doi: 10.1039/c9sc03639c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Menezes MR, Balzeau J, Hagan JP. 3’ RNA uridylation in epitranscriptomics, gene regulation, and disease. Front Mol Biosci. 2018;5:1–20. doi: 10.3389/fmolb.2018.00061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Wickens M, Kwak JE. A Tail Tale for U. Science. 2008;319:1344–1345. doi: 10.1126/science.1154946. [DOI] [PubMed] [Google Scholar]
- (44).Yatés LA, Fleurdepine S, Rissland OS, De Colibus L, Harlos K, Norbury CJ, Gilbert RJC. Structural basis for the activity of a cytoplasmic RNA terminal uridylyl transferase. Nat Struct Mol Biol. 2012;19:782–787. doi: 10.1038/nsmb.2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Rissland OS, Norbury CJ. The Cid1 poly(U) polymerase. Biochim Biophys Acta, Gene Regul Mech. 2008;1779:286–294. doi: 10.1016/j.bbagrm.2008.03.003. [DOI] [PubMed] [Google Scholar]
- (46).Lunde BM, Magler I, Meinhart A. Crystal structures of the Cid1 poly (U) polymerase reveal the mechanism for UTP selectivity. Nucleic Acids Res. 2012;40:9815–9824. doi: 10.1093/nar/gks740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Rao H, Tanpure AA, Sawant AA, Srivatsan SG. Enzymatic incorporation of an azide-modified UTP analog into oligoribonucleotides for post-transcriptional chemical functionalization. Nat Protoc. 2012;7:1097–1112. doi: 10.1038/nprot.2012.046. [DOI] [PubMed] [Google Scholar]
- (48).Sawant AA, Tanpure AA, Mukherjee PP, Athavale S, Kelkar A, Galande S, Srivatsan SG. A versatile toolbox for posttranscriptional chemical labeling and imaging of RNA. Nucleic Acids Res. 2016;44:e16. doi: 10.1093/nar/gkv903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Munoz-Tello P, Gabus C, Thore S. A critical switch in the enzymatic properties of the Cid1 protein deciphered from its productbound crystal structure. Nucleic Acids Res. 2014;42:3372–3380. doi: 10.1093/nar/gkt1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Winz M-L, Samanta A, Benzinger D, Jäschke A. Sitespecific terminal and internal labeling of RNA by poly(A) polymerase tailing and copper-catalyzed or copper-free strain-promoted click chemistry. Nucleic Acids Res. 2012;40:e78. doi: 10.1093/nar/gks062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Taemaitree L, Shivalingam A, El-Sagheer AH, Brown T. An artificial triazole backbone linkage provides a split-and-click strategy to bioactive chemically modified CRISPR sgRNA. Nat Commun. 2019;10:1610. doi: 10.1038/s41467-019-09600-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Lee K, Mackley VA, Rao A, Chong AT, Dewitt MA, Corn JE, Murthy N. Synthetically modified guide RNA and donor DNA are a versatile platform for CRISPR-Cas9 engineering. eLife. 2017;6:e25312. doi: 10.7554/eLife.25312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Ui-Tei K, Maruyama S, Nakano Y. Enhancement of single guide RNA transcription for efficient CRISPR/Cas-based genomic engineering. Genome. 2017;60:537–545. doi: 10.1139/gen-2016-0127. [DOI] [PubMed] [Google Scholar]
- (54).Nishimasu H, Ran FA, Hsu PD, Konermann S, Shehata SI, Dohmae N, Ishitani R, Zhang F, Nureki O. Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell. 2014;156:935–949. doi: 10.1016/j.cell.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Mu W, Zhang Y, Xue X, Liu L, Wei X, Wang H. 5’ capped and 3’ polyA-tailed sgRNAs enhance the efficiency of CRISPR-Cas9 system. Protein Cell. 2019;10:223–228. doi: 10.1007/s13238-018-0552-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Bin Moon S, Lee JM, Kang JG, Lee N-E, Ha D-I, Kim DY, Kim SH, Yoo K, Kim D, Ko J-H, Kim Y-S. Highly efficient genome editing by CRISPR-Cpf1 using CRISPR RNA with a uridinylate-rich 3’-overhang. Nat Commun. 2018;9:3651. doi: 10.1038/s41467-018-06129-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Zhu X, Clarke R, Puppala AK, Chittori S, Merk A, Merrill BJ, Simonović M, Subramaniam S. Cryo-EM structures reveal coordinated domain motions that govern DNA cleavage by Cas9. Nat Struct Mol Biol. 2019;26:679–685. doi: 10.1038/s41594-019-0258-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Raper AT, Stephenson AA, Suo Z. Functional insights revealed by the kinetic mechanism of CRISR/Cas9. J Am Chem Soc. 2018;140:2971–2984. doi: 10.1021/jacs.7b13047. [DOI] [PubMed] [Google Scholar]
- (59).Jinek M, Jiang F, Taylor DW, Sternberg SH, Kaya E, Ma E, Anders C, Hauer M, Zhou K, Lin S, Kaplan M, et al. Structures of Cas9 endonucleases reveal RNA-mediated conformational activation. Science. 2014;343:1247997. doi: 10.1126/science.1247997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (60).Gao XD, Tu L-C, Mir A, Rodriguez T, Ding Y, Leszyk J, Dekker J, Shaffer SA, Zhu LJ, Wolfe SA, Sontheimer EJ. C-BERST: defining subnuclear proteomic landscapes at genomic elements with dCas9-APEX2. Nat Methods. 2018;15:433–436. doi: 10.1038/s41592-018-0006-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (61).Acharya S, Mishra A, Paul D, Ansari AH, Azhar M, Kumar M, Rauthan R, Sharma N, Aich M, Sinha D, Sharma S, et al. Francisella novicida Cas9 interrogates genomic DNA with very high specificity and can be used for mammalian genome editing. Proc Natl Acad Sci U S A. 2019;116:20959–20968. doi: 10.1073/pnas.1818461116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (62).Wu X, Mao S, Yang Y, Rushdi MN, Krueger CJ, Chen AK. A CRISPR/molecular beacon hybrid system for live-cell genomic imaging. Nucleic Acids Res. 2018;46:e80. doi: 10.1093/nar/gky304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (63).van Geel R, Pruijn GJM, van Delft FL, Boelens WC. Preventing thiol-yne addition improves the specificity of strain-promoted azide-alkyne cycloaddition. Bioconjugate Chem. 2012;23:392–398. doi: 10.1021/bc200365k. [DOI] [PubMed] [Google Scholar]
- (64).Cawthon RM. Telomere measurement by quantitative PCR. Nucleic Acids Res. 2002;30:e47. doi: 10.1093/nar/30.10.e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (65).Cserép GB, Herner A, Kele P. Bioorthogonal fluorescent labels: a review on combined forces. Methods Appĩ. Fluoresc. 2015;3:042001. doi: 10.1088/2050-6120/3/4/042001. [DOI] [PubMed] [Google Scholar]
- (66).O’Connell MR, Oakes BL, Sternberg SH, East-Seletsky A, Kaplan M, Doudna JA. Programmable RNA recognition and cleavage by CRISPR/Cas9. Nature. 2014;516:263–266. doi: 10.1038/nature13769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (67).Patterson DM, Nazarova LA, Prescher JA. Finding the right (bioorthogonal) chemistry. ACS Chem Biol. 2014;9:592–605. doi: 10.1021/cb400828a. [DOI] [PubMed] [Google Scholar]
- (68).Devaraj NK. The future of bioorthogonal chemistry. ACS Cent Sci. 2018;4:952–959. doi: 10.1021/acscentsci.8b00251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (69).Ford K, McDonald D, Mali P. Functional genomics via CRISPR-Cas. J Mol Biol. 2019;431:48–65. doi: 10.1016/j.jmb.2018.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (70).Seidel SAI, Dijkman PM, Lea WA, van den Bogaart G, Jerabek-Willemsen M, Lazic A, Joseph JS, Srinivasan P, Baaske P, Simeonov A, Katritch I, et al. Microscale thermophoresis quantifies biomolecular interactions under previously challenging conditions. Methods (Amsterdam, Neth) 2013;59:301–315. doi: 10.1016/j.ymeth.2012.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (71).Evans MJ, Kaufman MH. Establishment in culture of pluripotential cells from mouse embryos. Nature. 1981;292:154–156. doi: 10.1038/292154a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.