Abstract
Anti-EGFR antibodies such as cetuximab are active against KRAS/NRAS wild-type colorectal cancers (CRC) but acquired resistance invariably evolves. Which mutational mechanisms enable resistance evolution and whether adaptive mutagenesis, a transient cetuximab-induced increase in mutation generation, contributes in patients is unknown. Here, we investigate this in exome sequencing data of 42 baseline and progression biopsies from cetuximab treated CRCs. Mutation loads did not increase from baseline to progression and evidence for a contribution of adaptive mutagenesis was limited. However, the chemotherapy-induced mutational signature SBS17b was the main contributor of specific KRAS/NRAS and EGFR driver mutations that are enriched at acquired resistance. Detectable SBS17b activity before treatment predicted for shorter progression free survival and for the evolution of these specific mutations during subsequent cetuximab treatment. This suggests that chemotherapy mutagenesis can accelerate resistance evolution. Mutational signatures may be a new class of cancer evolution predictor.
The anti-EGFR antibody (EGFR-AB) cetuximab is active against many KRAS/NRAS wild-type metastatic colorectal cancers (CRCs)1,2. However, resistance invariably evolves within several months. Darwinian selection of subclones that harbor mutations in KRAS, NRAS and EGFR is among the commonest mechanisms of acquired resistance3-6. Pre-treatment biomarkers that can predict the time to resistance evolution and the specific resistance mechanism that will evolve have not been identified7,8.
Mutation generation is central to resistance evolution, and mutational signature analysis can be used to dissect cancer mutational processes9,10. Yet, how the activity of specific mutational signatures enables or constrains the evolution of cetuximab resistance in CRCs is unknown. Resistance evolution may furthermore be influenced by the timing of specific mutational processes. The pre-existing drug resistance model assumes that such mutations are already present in small subclones before EGFR-AB exposure, making the evolution of acquired resistance inevitable (Fig. 1A)11. Recently, a model of ‘adaptive mutagenesis’ has been proposed in which cetuximab treatment triggers a transient down–regulation of mismatch repair (MMR) and homologous recombination (HR) DNA repair proteins and increased expression of low-fidelity DNA polymerases, which together promote mutation generation in CRC cells12. Such drug-induced mutagenesis could increase the probability of resistance mutation acquisition during treatment (Fig. 1A). Importantly, these are preclinical observations and it is unknown how prevalent cetuximab-induced mutagenesis is in patients13 and whether it impacts the acquisition of common resistance mutations. More generally, it remains undetermined whether any specific mutational signatures change through cetuximab treatment and which signatures generate the majority of resistance mutations in the clinic.
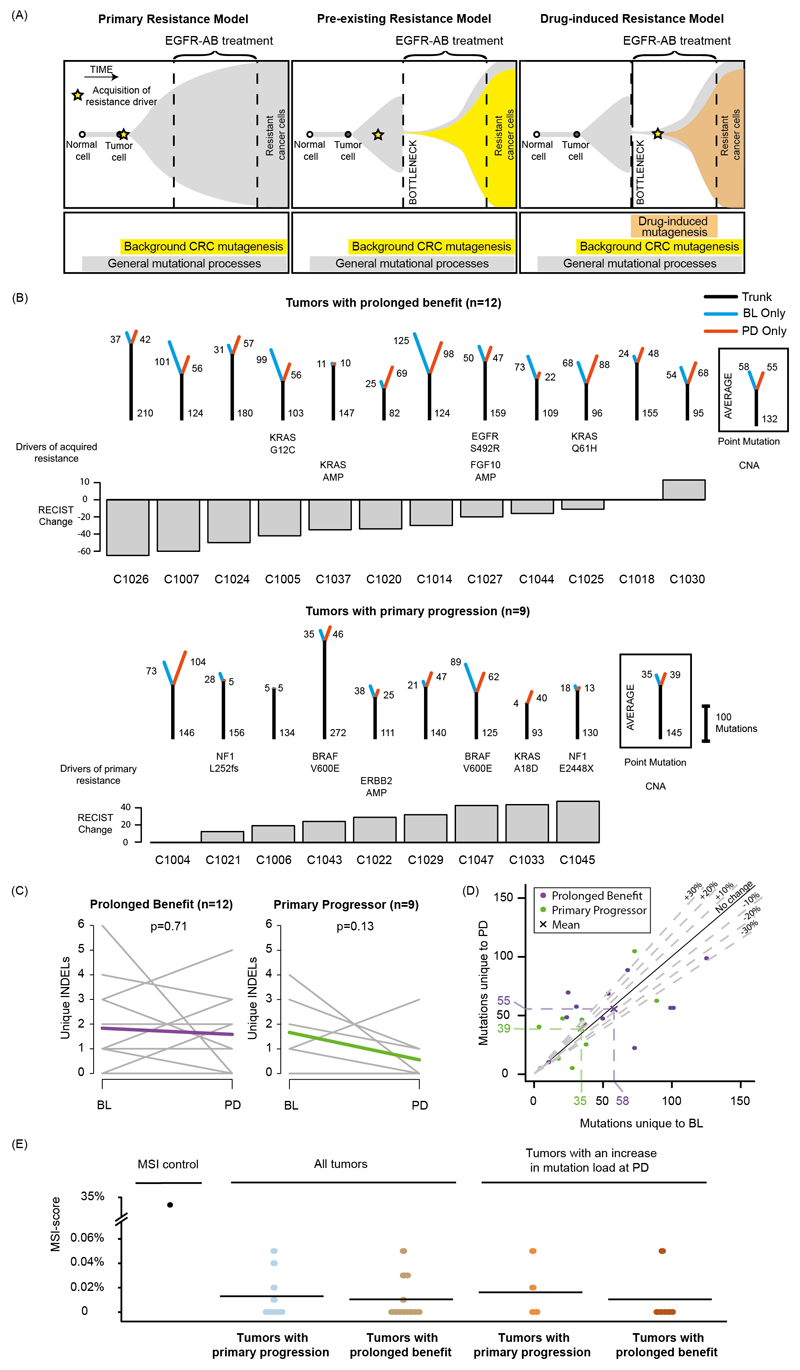
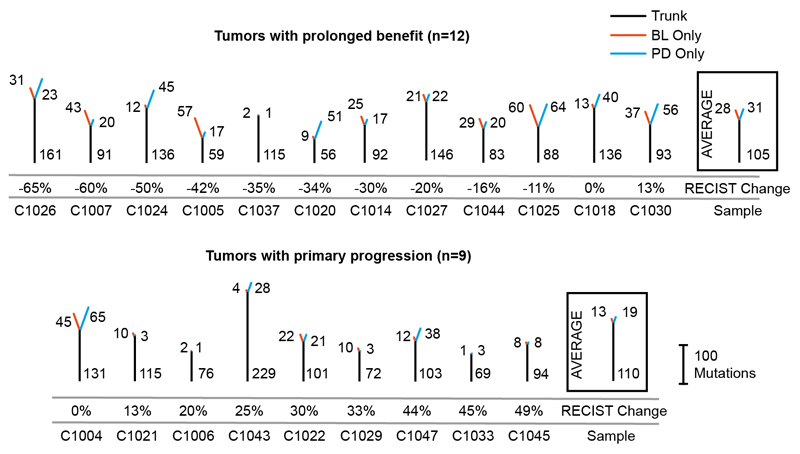
Fig. 1. Cetuximab resistance models and analysis of mutation loads in 21 tumors treated with single-agent cetuximab.
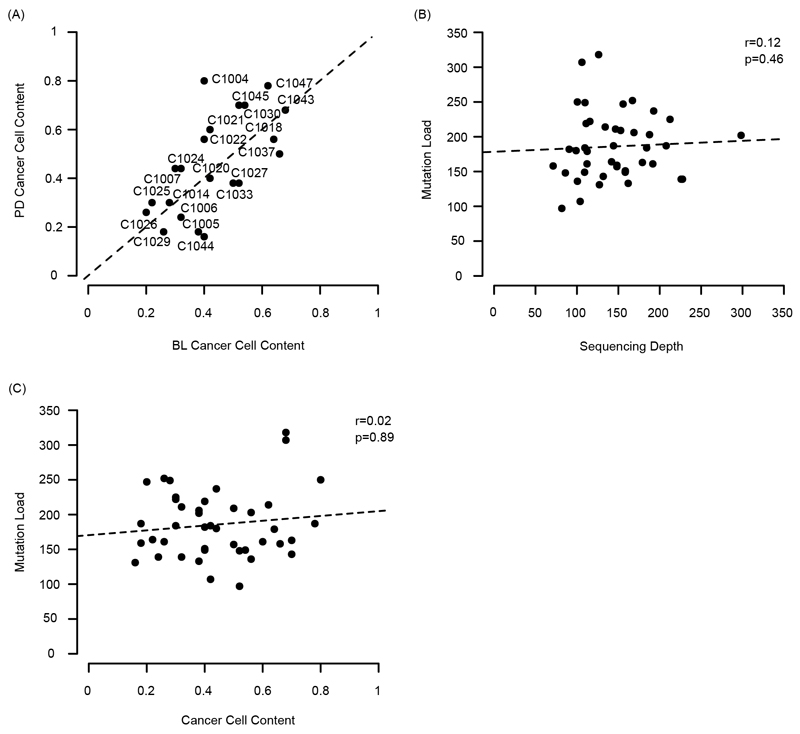
(A) Models of primary and acquired cetuximab resistance and their relationship to mutation signature activity. (B) Mutation trees for 21 tumors from the Prospect-C trial. Grouped into cases with prolonged benefit and primary progression. Numbers next to the trunk or the branches indicate the number of somatic mutations. Cetuximab resistance driver mutations and copy number aberrations (CNA) identified in 3 are shown. The RECIST change indicates the change of the sum of radiological tumor measurements based on RECIST criteria from BL to the time of best response. (C) Change of the unique INDEL numbers from BL to PD. Colored lines show the mean. The p-values were calculated with a paired t-test. (D) Unique mutation loads for each tumor at BL vs. PD. The dashed lines indicate a relative increase or decrease by 10%, 20% or 30%. (E) Microsatellite length variability analysis with the MSIsensor algorithm. MSI-scores indicate the percentage of microsatellite and homopolymer loci with an increased read length variability at PD compared to BL. Horizontal bars show the mean MSI-score for each group. The MSI-score of the only MMR-deficient tumor from the Prospect-C trial (which has not been included in any other analyses as no paired PD sample was available) in comparison to the matched blood sample is shown as a control for correct MSI detection.
Our aim was to assess the activity of mutational mechanisms in serial biopsies from KRAS/NRAS wild‐type CRC patients who were treated with single-agent cetuximab in a clinical trial. Drug treatment forces the cancer cell population through an evolutionary bottleneck7. We reasoned that this should reveal the mutational signatures operating before or during treatment as these become increasingly clonal and hence detectable by exome sequencing. Cetuximab-induced mutagenesis should increase both, mutation loads and the specific mutational signatures that are characteristic of these mechanisms in patients who benefit (Fig. 1A). In contrast, no changes would be expected in patients with primary progression where cetuximab lacks activity. We furthermore assessed which mutational mechanisms are most relevant for the generation of the hotspot driver mutations that evolve at acquired resistance.
Results
Clinical trial samples
The patient characteristics and biopsy analysis of the Prospect-C phase II trial have been described previously3. Biopsies had been taken at baseline (BL) before cetuximab initiation and at progressive disease (PD) from KRAS/NRAS wild-type CRCs. Paired BL/PD biopsies from 21 patients were successfully analyzed by exome sequencing and had sufficient cancer cell content for bioinformatics analysis (Extended Data Fig. 1A; see Methods for full details). The characteristics of these patients were comparable to the entire population in the Prospect-C trial and in other EGFR-AB trials (Supplementary Table 1). The median sequencing depth of BL (112x) and PD (148x) samples and the median cancer cell content of BL (40%) and PD (44%) samples were similar. Neither sequencing depth nor cancer cell content of samples correlated with the mutation load (Extended Data Fig. 1B,C). There was hence no evidence that sequencing depth or cancer cell content biased the number of detected mutations in BL vs. PD samples. No tumor showed MMR-deficiency at BL3. Progression at or before the first per-protocol CT scan (scheduled at 12 weeks) had been classified as ‘primary progression’ (n=9). The remaining tumors were considered to have obtained ‘prolonged benefit’ (n=12) from treatment3.
Temporal change of mutation loads
Mutation trees were generated to analyze the evolutionary relationship of cancer cells in BL and PD biopsies and changes in the mutation load (Fig. 1B). The trunk represents mutations present in both samples whereas branches indicate mutations unique to BL or PD samples. Truncal mutation loads were similar between tumors with prolonged benefit and primary progression (p=0.53, t‐test). Cancers with prolonged benefit had higher unique mutation numbers compared to primary progressors (mean sum of BL and PD: 113 and 73, respectively, p=0.06, t-test). Although this was not significant, it likely indicates a cetuximab-induced population bottleneck that diminishes treatment-sensitive subclones which are replaced by subclones with distinct mutations at acquired resistance, whereas subclones at BL and PD are more similar in primary progressors.
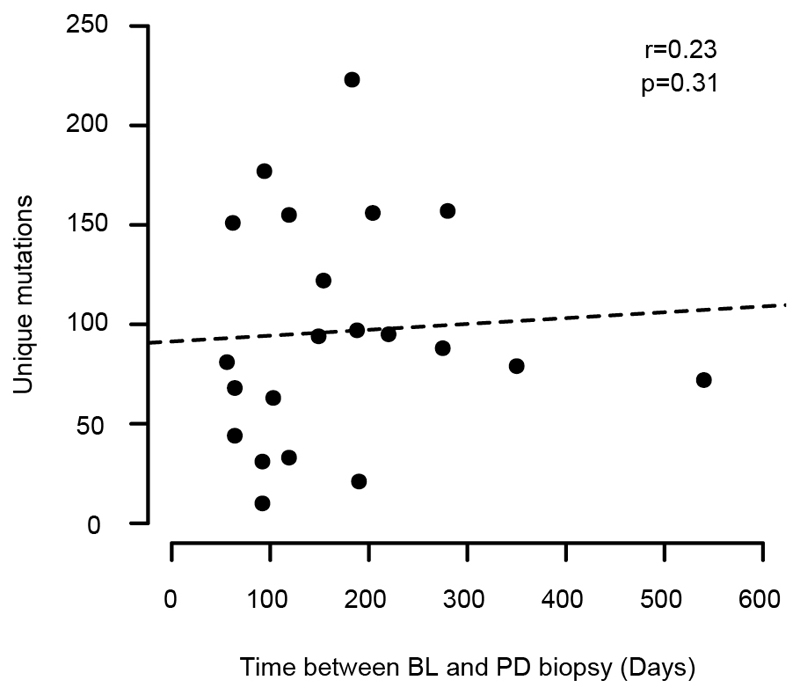
The number of unique mutations did not significantly change from BL to PD in either group (prolonged benefit: p=0.74, primary progression: p=0.62, paired t-test). An increase in the number of small insertions and deletions (INDELs) can be an indicator of acquired MMR-deficiency14 but these did not change significantly from BL to PD (prolonged benefit: p=0.71; primary progression: p=0.13, paired t-test, Fig. 1C). The absence of a population bottleneck in primary progressors is a potential source of bias as these may harbor higher numbers of subclones at PD, leading to higher subclonal mutation loads than in prolonged benefit cases where subclones were pruned. We therefore repeated the analysis by only considering clonal mutations in each sample. This found no significant increase of mutations in tumors with prolonged benefit (p=0.66, Extended Data Fig. 2) or in primary progressors (p=0.20, paired t-test). As mutations accumulate over time, we tested whether the time lapse between BL and PD may influence branch lengths. We found no association between treatment duration and the number of unique mutations (Spearman’s r: 0.23, p=0.31, Extended Data Fig. 3). We furthermore considered that cetuximab-induced mutagenesis may only be active in a subgroup of tumors. 6/12 (50%) cases with prolonged benefit showed an increase of the unique mutation load at PD but also 4/9 (44.4%) of tumors with primary progression (Fig. 1D). Thus, although mutations can increase in individual tumors after treatment, this fraction did not differ between these groups.
Taken together, we found no evidence for a rise in the mutation load through cetuximab treatment. This mirrors results from Russo et al.12 who described only a negligible change in mutation burden in cetuximab treated CRC cell lines analyzed by exome sequencing. Exome sequencing only analyzes ~1-2% of the genome which may be insufficient to detect an increase of mutations across the genome. However, these results show that if drug-induced mutagenesis is active, the impact on the mutation load in the protein-coding genome is small.
Microsatellite tract length variability
Cetuximab-induced mutagenesis increased the accumulation of INDELs in microsatellite tracts in CRC cell lines12. Assessing the length variability of microsatellites showed no increase from BL to PD in tumors with prolonged benefit or with primary progression (Fig. 1E). Restricting the analysis to those tumors with an increase in the unique mutation load at PD also showed no change. We hence found no evidence for a cetuximab-induced increase in microsatellite tract length variability.
Temporal change of mutational signatures
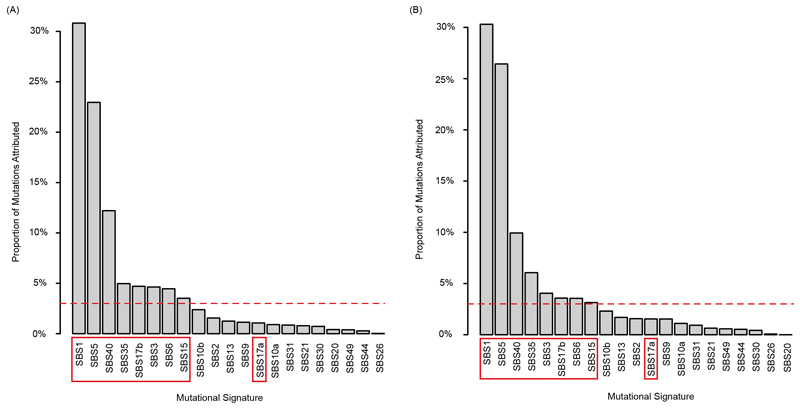
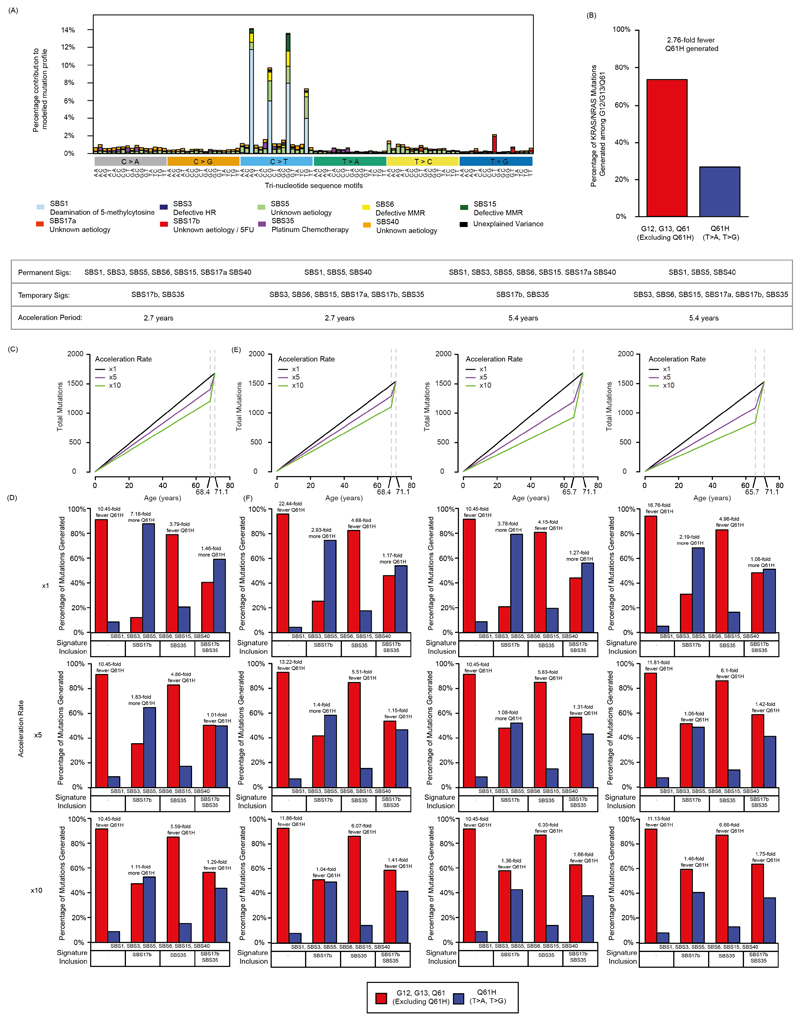
Mutational signature analysis9 should reveal changes in the activity of mutagenic processes independent of mutation loads. All single nucleotide substitutions and the two flanking bases were analyzed, corresponding to 96 tri-nucleotide sequence motifs. Individual tri-nucleotide motifs only showed small changes from BL to PD without obvious differences between tumors with prolonged benefit vs. primary progression (Fig. 2A). We next assigned these mutations to individual mutational signatures15. To limit the impact of signature bleeding, which can lead to the misassignment of mutations to signatures with high similarity16, we included only a) signatures that were detectable in a large series of CRC samples (Extended Data Fig. 4; SBS1, SBS5 and SBS40 which are clock-like based on their relatively constant rate over time17, SBS15 which is typical for CRCs with MMR-deficiency18, SBS17b which can be present in CRCs that were treated with 5-fluorouracil (5-FU) chemotherapy19,20 and SBS17a which remains of uncertain aetiology although oxidative damage has been suggested to contribute to SBS17a/SBS17b21), b) additional signatures of mutational processes that were reported to increase through cetuximab-induced mutagenesis by Russo et al.12 (HR‐deficiency signature SBS3, MMR‐deficiency signature SBS69,22) and c) the platinum chemotherapy signature SBS35 as all tumors had received chemotherapy.
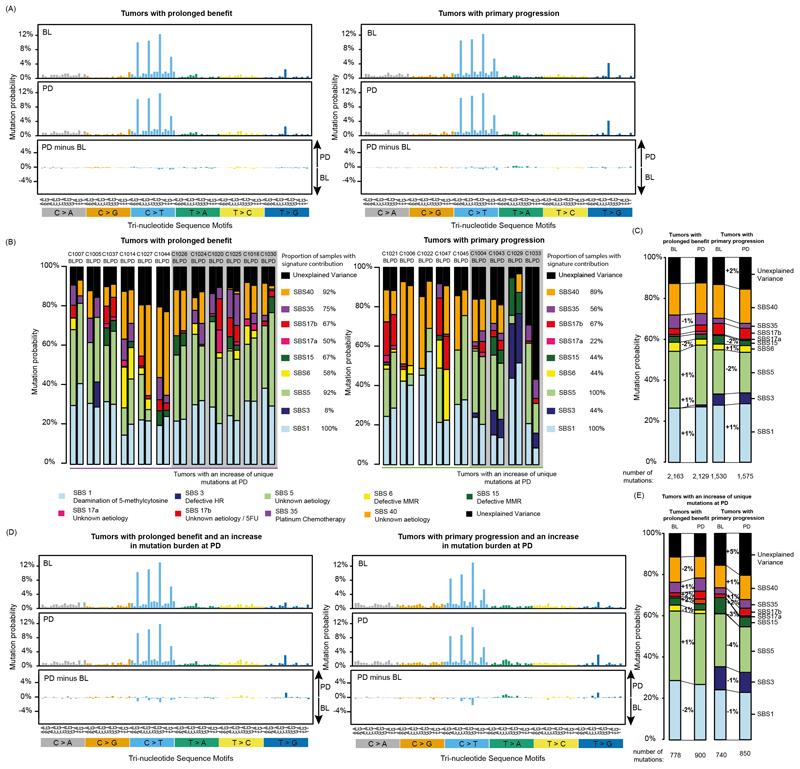
Fig. 2. Mutational signatures in tumors treated with cetuximab.
(A) 96 tri-nucleotide motif plot of all single base substitutions prior to cetuximab treatment (BL) and at progression (PD). The bottom panel shows the difference between BL and PD. (B) Attribution of single base substitutions to mutational signatures shows the contribution of each signature to individual samples at BL and PD. (C) Signature contribution for the combined group of cases with prolonged benefit or primary progression. (D) Mutational signatures in tumors where an increase in unique mutation burden was found at PD. (E) Mutational signature contribution for the combined group of cases with prolonged benefit or primary progression which also showed an increase in unique mutation burden.
SBS1 and signatures with a broad range of substitution motifs (SBS5, SBS40) were most abundant (Fig. 2B,C). The platinum signature SBS35 and the 5-FU-associated signature SBS17b, which is characterized by a unique predominance of T>G mutations in a CTT context, were the next most abundant. SBS1, SBS5 and SBS40 were active in most samples whereas SBS35 and SBS17b were only detected in a subset.
We investigated whether any of the signatures increased with cetuximab treatment in the prolonged benefit group. SBS1 and SBS5 both showed small (1%) increases from BL to PD (Fig. 2C). The HR-deficiency signature SBS3 also showed a 1% increase but this was driven by a single case (C1005, Fig. 2B). Focusing only on the 6 tumors in the prolonged benefit group that showed an increase in the unique mutation load revealed the largest rise for SBS17a and SBS17b (+2% each, Fig. 2D,E) but this was driven by a single tumor (C1020, Fig. 2B). SBS17b also rose by 2% among the 4 tumors with primary progression that showed a mutation increase at PD and a single case (C1004) showed a relatively large increase in SBS17a and SBS17b. Thus, neither SBS17a nor SBS17b appear to be specifically promoted by cetuximab.
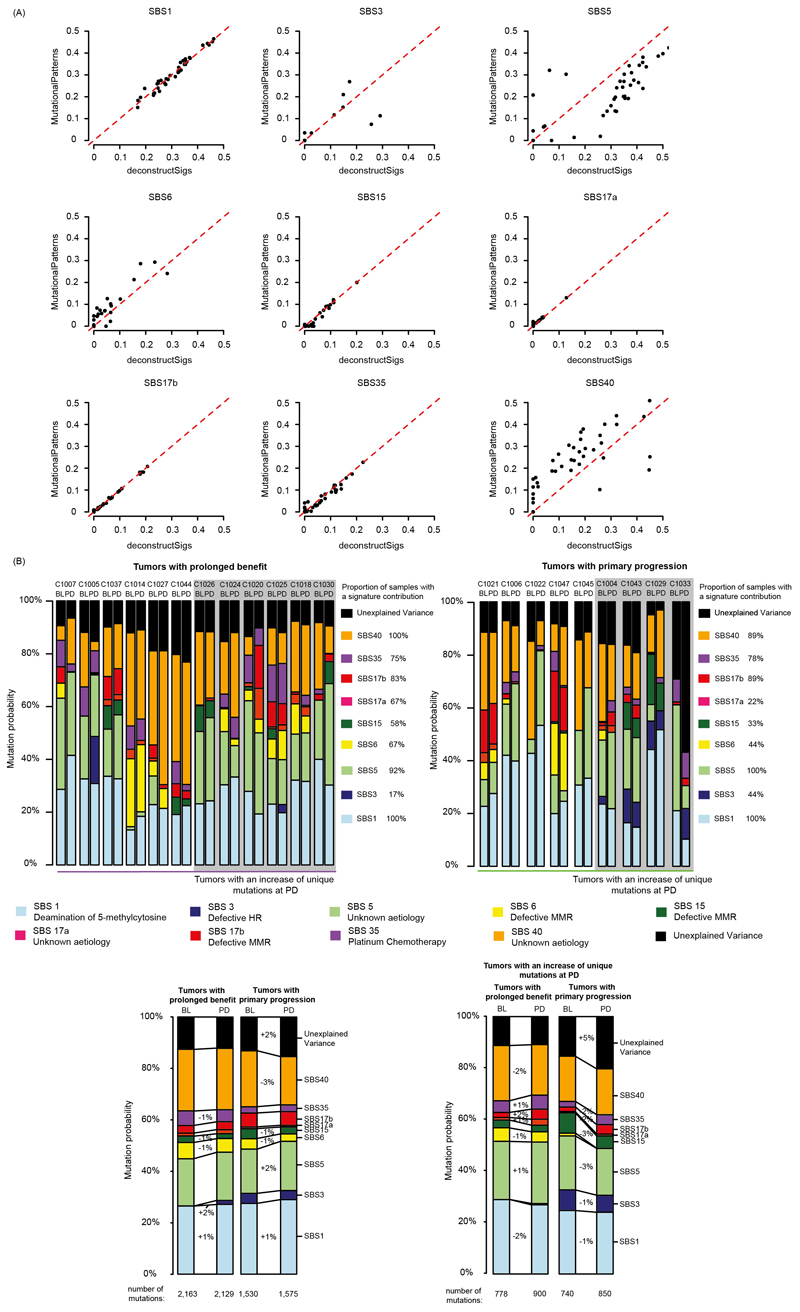
To ascertain our results, we repeated the mutational signature analysis with a second independent method which applies a non-negative least squares approach to signature fitting23 instead of the iterative linear regression method24 used for Fig. 2. Signature SBS40 was more and SBS5 less abundant with this approach. All other signatures showed a high level of agreement (Extended Data Fig. 5A). Comparison of signature abundance at BL and PD in cases with prolonged benefit vs. primary progression (Extended Data Fig. 5B) supported the same conclusions as the analysis in Fig. 2.
Taken together, no signature noticeably increased at PD in the prolonged benefit group despite a median cetuximab treatment duration of 26wks (range: 18-96). Signatures that would be expected to increase most strongly through cetuximab-induced mutagenesis in the prolonged benefit group showed only a 1% increase which was driven by a single case (SBS3: HR-deficiency), remained unchanged (SBS15: MMR-deficiency) or even decreased (SBS6: MMR-deficiency). These results are inconsistent with a major contribtution of drug-induced mutagenesis to exonic mutations in CRC patients.
SBS17b disproportionally contributes to driver mutations enriched at acquired resistance
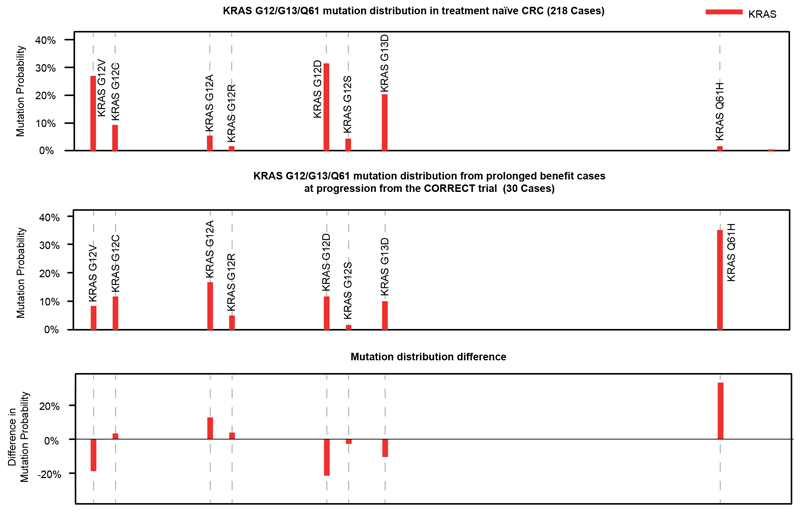
KRAS/NRAS and EGFR mutations are the commonest genetic mechanisms of acquired cetuximab resistance in CRC3-6. Mutations in these genes at acquired resistance differ from those in treatment-naïve CRCs: EGFR mutations at acquired resistance disrupt cetuximab binding epitopes and do not occur in untreated CRCs as they provide no fitness advantage in the absence of treatment25. Furthermore, comparing biopsy- and ctDNA-sequencing results of CRCs with acquired cetuximab resistance3,5,26 to biopsy sequencing data of KRAS/NRAS mutant treatment–naïve CRCs27 showed that KRAS/NRAS codon 12/13 mutations were 1.7-fold lower and codon 61 mutations 4.2-fold higher in tumors with acquired resistance compared to tumors with expected primary resistance. Q61H mutations showed the largest increase (11.8-fold, Fig. 3A). Analysis of the CORRECT trial28 even showed a 21.1-fold increase of KRAS Q61H mutations at acquired cetuximab resistance compared to treatment-naïve KRAS mutant CRCs (Extended Data Fig. 6). Motivated by the observation that signature contributions varied between tumors in the Prospect-C trial, we questioned whether signature activity before cetuximab initiation influences which resistance driver mutations evolve at acquired resistance.
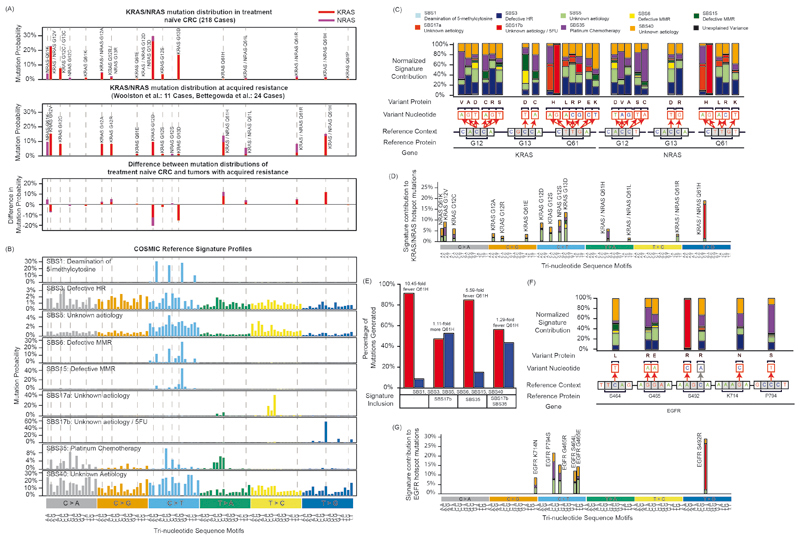
Fig. 3. Relationship of mutational signatures to specific KRAS/NRAS and EGFR mutations.
(A) KRAS/NRAS codon 12/13/61 mutation frequency in treatment naïve CRCs from the TCGA Pan-Cancer study versus those identified in CRCs with acquired EGFR-AB resistance3,5,26. (B) Sigprofiler exome SBS reference profiles (syn11967914.3) of all active signatures included in the analyses of the Prospect-C cohort. (C) Relative contribution of each of the signatures in B corresponding to the indicated KRAS/NRAS mutations when an equal number of mutations is generated with each signature. All reference contexts in the figure show the main genomic strand. (D) Modelling of the relative contribution of each of the signatures in B to all indicated KRAS/NRAS mutations when the observed mutational signature distribution at BL in cases with prolonged benefit is taken into account. (E) Modelled contribution of chemotherapy related mutation signatures (SBS17b, SBS35) to KRAS/NRAS Q61H mutations vs. all other hotspot mutations. Results presented are from a model that assumes a 10x acceleration in mutation accumulation of signatures SBS1, SBS3, SBS5, SBS6, SBS15 and SBS40 between diagnosis and BL biopsy. (F) Repeat of the analysis in panel C for EGFR mutations. (G) Repeat of the analysis in panel D for EGFR mutations.
We first compared KRAS/NRAS mutation profiles in CRC (Fig. 3A) to the published15 mutational signature profiles (Fig. 3B). SBS3, SBS5 and SBS40 overlapped with most hotspot mutations. The remaining signatures only overlapped with a few KRAS/NRAS mutations, indicating that the activity of these signatures could influence the probability that specific mutations are generated and thereby account for genetically distinct evolutionary outcomes. We hence calculated the probability for each signature to generate specific KRAS/NRAS mutations (Fig. 3C). Intriguingly, SBS17b showed a strong preference to create KRAS/NRAS Q61H mutations and almost exclusively generated the T>G mutation that was most enriched at acquired cetuximab resistance. The platinum signature SBS35 also overlapped with a KRAS/NRAS Q61H mutation (T>A) that is enriched at acquired resistance. Thus, SBS17b and SBS35 activity could critically influence the probability that these mutations evolve.
We therefore modelled the KRAS/NRAS mutation distribution that would be generated in prolonged benefit cases based on the observed signature contribution at BL (Supplementary Table 2, Extended Data Fig. 7A). Despite the higher activity of SBS1, SBS5 and SBS40 (together accounting for 70% of mutations, Fig. 2C), SBS17b was the largest contributor of KRAS/NRAS Q61H mutations (65% of all Q61H mutations, Fig. 3D). SBS35 generated the second highest proportion of Q61H mutations (13% of all Q61H mutations), although it contributed more codon 12 mutations than codon 61 mutations. Codon 12 and codon 13 mutations were most likely to be generated by the clock-like signatures SBS5/SBS40.
To further substantiate whether the chemotherapy-induced signatures SBS17b (5-FU) and SBS35 (platinum) can explain the strong enrichment of Q61H mutations among KRAS/NRAS mutations at acquired cetuximab resistance (Extended Data Fig. 7B), we modelled the distribution of KRAS/NRAS codon 12, 13 and 61 mutations that would be expected in the presence or absence of these signatures. Tumors harbor higher numbers of mutations corresponding to the clock-like signatures (SBS1, SBS5, SBS40) than to SBS17b and SBS35 but the former are active over the lifetime of the patient, whereas the chemotherapy-induced signatures SBS17b and SBS35 are acquired over a much shorter period of time. In addition, even signatures that are active over the patients lifetime can accelerate up to ~10-fold once a cancer is established due to increased proliferation and genomic instability29. Thus, the signature composition we observe at BL may not be reflective of the true activity of the signatures at the biopsy timepoint. We therefore estimated the contemporaneous activity of each mutational signature by taking into account the time period over which it is likely active and a range of acceleration rates.
Our model assumes that SBS1, SBS5 and SBS40 have a constant mutation rate from birth until diagnosis (median 68.4y) followed by a period of acceleration from the time of diagnosis to biopsy (median 2.7y, Extended Data Fig. 7C). Chemotherapy-induced signatures (SBS17b, SBS35) were assumed active only after cancer diagnosis. The temporal varibility of SBS3, SBS6, SBS15 and SBS17a is poorly understood but they are not known to increase through chemotherapy treatment. They were therefore modelled analogous to SBS1, SBS5 and SBS40.
The model shows that in the absence of the SBS17b and SBS35, KRAS/NRAS Q61H mutations are generated with a 10.45-fold lower probability than all other KRAS/NRAS 12/13/61 hotspot mutations taken together (Fig. 3E). The likelihood of generating a Q61H mutation increases when the platinum signature SBS35 is added, but still remains 5.59-fold lower than all other hotspot mutations. However, when the SBS17b signature is added in the model, Q61H becomes the predominant KRAS/NRAS mutation (1.11-fold higher probability than all other KRAS/NRAS mutations taken together). When both signatures are added together, Q61H mutations are 1.29-fold lower than all other hotspot mutations. The slightly lower enrichment is explained by the generation of additional codon 12/13 mutations by SBS35. Our simplified model hence demonstrates that SBS17b signature activity and to a smaller extent also SBS35 are able to explain the inflated frequency of KRAS/NRAS Q61H mutations at acquired cetuximab resistance (Extended Data Fig. 7B).
We next varied several model assumptions to assess whether this would change these conclusions. Firstly, the tumor is likely to be present several months to years prior to diagnosis. Therefore, we considered an extended period of tumor growth (twice the time from diagnosis: 5.4y). Secondly, it is unclear whether SBS3, SBS6, SBS15 and SBS17a are acquired over the patients lifetime. We hence assessed whether restricting their activity only to the growth phase (equivalent to SBS17b and SBS35 modelling) impacts the results. We finally tested additional acceleration factors (1x,5x). All models showed a consistant increase in the likelihood of KRAS/NRAS Q61H generation with the inclusion of SBS35 and SBS17b (Extended Data Fig. 7D–F) and a dominant role of SBS17b as the leading contributor of Q61H mutations.
We next investigated how mutational signatures influence EGFR mutations (Fig. 3F). Similar to what we found for KRAS/NRAS Q61H mutations, the EGFR S492R A>C mutation, which is common at acquired resistance25,30 was almost exclusively generated by SBS17b. When the signature contributions at BL in tumors with prolonged benefit was taken into account, SBS17b was the major signature generating this mutation (Fig. 3G).
This indicates that SBS17b and SBS35 activity are sufficient to explain the predominant evolution of KRAS/NRAS Q61H and EGFR S492R mutations at acquired resistance in tumors where these signatures are active.
SBS17b signature activity as a predictor of mutation evolution and progression free survival
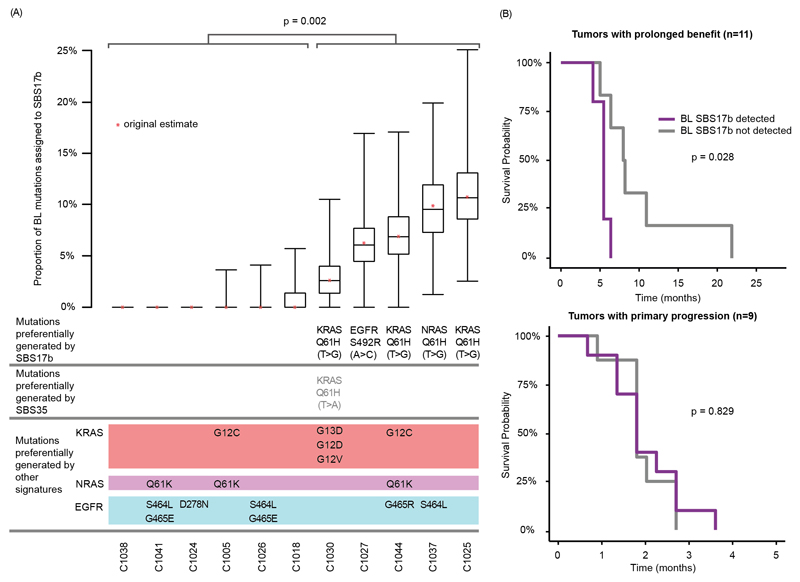
To substantiate the relevance of the SBS17b signature in patients, we investigated in the Prospect-C trial whether SBS17b activity in BL samples can predict the evolution of specific drivers at acquired resistance and of progression free survival (PFS). SBS17b was detectable in five cases at BL and a bootstrap analysis confirmed the stability of the signature attribution (Fig. 4A). KRAS/NRAS Q61H T>G mutations evolved in four of these and an EGFR S492R A>C mutation in one. No KRAS/NRAS Q61H or EGFR S492R mutations were identified in tumors without a detected SBS17b activity. This statistically significant enrichment (p=0.002, Fisher’s exact test) suggests that SBS17b activity canalizes the evolution of these resistance driver mutations. Furthermore, SBS17b predicted for a significantly shorter PFS in the prolonged benefit group but not in primary progressors (p=0.028, log rank test, Fig. 4B).
Fig. 4. Association of detected SBS17b at BL with specific KRAS/NRAS and EGFR mutation evolution at the time of acquired resistance and with progression free survival.
(A) SBS17b signature contribution calculated from whole exome mutation analysis of BL biopsies for all prolonged benefit cases with available ctDNA sequencing vs. resistance driver mutations in KRAS/NRAS and EGFR that were detected at PD in ctDNA. The stability of the SBS17b attributions was assessed by bootstrap analysis based on 1,000 replicates. Signature decomposition was then calculated for each replicate and the 25th, 50th and 75th percentiles presented. Statistical significance was assessed with the Fisher’s exact test. (B) Kaplan-Meier analysis of progression free survival for tumors with and without a detected SBS17b contribution at BL. Statistical significance was assessed with the Log-rank test.
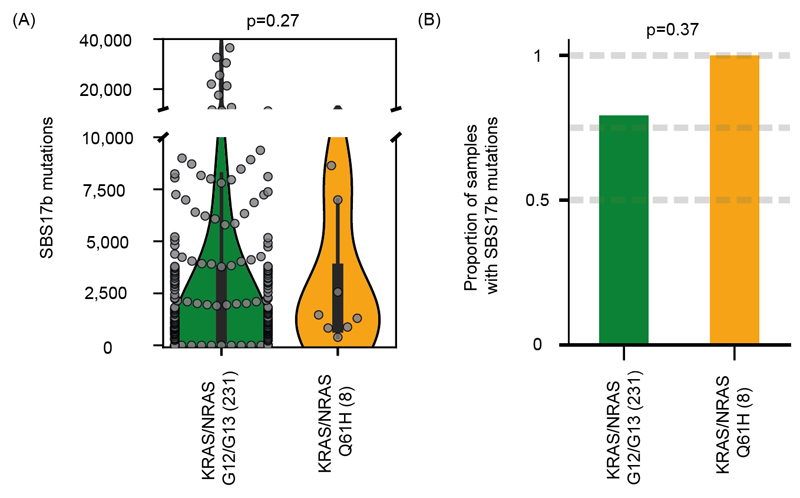
We finally investigated the relationship of SBS17b with KRAS/NRAS Q61H mutations in an independent cohort of 239 chemotherapy treated CRC samples with KRAS/NRAS G12/G13 or Q61H mutations20,31. Only 8 tumors harbored Q61H mutations and all had a detectable SBS17b activity compared to 79% of tumors with KRAS/NRAS codon 12/13 mutations (Extended Data Fig. 8A,B). Firm conclusions cannot be drawn because of the small number of Q61H mutations but the results do not contradict the notion that Q61H mutations predominantly occur through signature SBS17b.
Discussion
We showed that KRAS/NRAS Q61H mutations are 11.8-21.1-fold more common at acquired resistance than in treatment-naïve KRAS/NRAS mutant CRCs. A pan-cancer analysis found a higher selective advantage of codon 12/13 vs. codon 61 mutations32, questioning why a less beneficial mutation evolves with a strikingly increased frequency after cetuximab treatment. Q61 mutations have been suggested to have higher oncogenic potential than codon 12/13 mutations when KRAS expression is low and that this explains overrepresentation at acquired resistance33. Yet, there is little evidence for lower KRAS/NRAS expression at acquired resistance. We have now shown that Q61H is predominantly generated by SBS17b which is undetectable in most treatment-naïve CRCs but present in 67% of chemotherapy treated CRCs3,5,26. The platinum signature SBS35 may further contribute. The preferential generation of Q61H mutations by these chemotherapy-induced signatures provides a compellingly simple explanation for the mutation bias between primary and acquired resistance. SBS17b signature activity may also explain the high prevalence of the S492R mutation among EGFR mutations25,34. Prior analyses of large datasets with predominantly treatment-naïve tumors found no link between Signature 17 and KRAS/NRAS Q61 mutations35,36. This is a likely consequence of the low prevalence of Q61 mutations in tumors that have not been treated with EGFR-AB and of Signature 17 in the absence of 5-FU treatment.
Datasets for independent validation of these findings are not available in the public domain but our results are strengthened by the use of data from a prospective trial which limits selection biases and by four independent lines of evidence: We showed that SBS17b disproportionally contributes to KRAS/NRAS Q61H and EGFR S492R mutation generation. Secondly, the observed signature contribution in BL biopsies leads to an excess of KRAS/NRAS Q61H mutations similar to that observed at acquired resistance. Thirdly, we showed that SBS17b at BL correlated with the evolution of KRAS/NRAS Q61H and EGFR S492R mutations in individual patients. Finally, PFS was shorter in patients where SBS17b was detectable at BL, suggesting that this signature increases cancer evolvability during cetuximab treatment. Thus, SBS17b activity may be the first evolutionary biomarker to predict shorter PFS with cetuximab treatment. This hypothesis requires confirmation in future clinical trials. By linking accelerated drug resistance evolution in patients to chemotherapy-induced mutagenesis, our results furthermore highlight opportunities for the development of optimised treatment sequences that restrain cancer evolution.
We found no increase of mutation loads at acquired resistance, nor evidence for cetuximab-mediated MMR-deficiency. We detected a 1% increase in SBS3 mutations in tumors with prolonged benefit. This may be the consequence of reduced HR-fidelity through cetuximab‐induced mutagenesis, however the change was only observed in one patient. We also showed that SBS3 would only contribute minimally to KRAS/NRAS and EGFR mutations (Fig. 3D). Thus, despite the functional evidence for cetuximab-induced mutagenesis in CRC cell lines12, our analysis in patients shows that its contribution to cetuximab resistance evolution is likely small. There are limitations of our analysis. Although it is the largest series of paired biopsies from cetuximab treated CRCs that has been interrogated by exome sequencing, the analysis of further cohorts, ideally by whole-genome sequencing, may strengthen the evidence for drug-induced mutagenesis. Moreover, SBS3 is a ‘broad’ signature with mutation motifs overlapping those of SBS5/SBS40 which may lead to signature bleeding. Using two independent signature assignment algorithms, we demonstrated the largest discrepency in these broad signatures which highlights the technical difficulties of disentangling signatures.
Taken together, this exploratory analysis indicates that chemotherapy-induced mutation signatures can influence and predict the evolution of cetuximab resistance in CRC patients. This defines a strategy for the development of evolutionary biomarkers in precision cancer medicine.
METHODS
Trial Design And Samples
Prospect-C is a single-arm phase II trial that investigated biomarkers of response or resistance to single‐agent cetuximab in KRAS/NRAS wild-type metastatic CRCs (clinicaltrials.gov/ct2/show/NCT02994888). The trial has previously been described in detail3. Patient characteristics are described in Supplementary Table 1. The study was carried out in accordance with the Declaration of Helsinki and approved by the national UK ethics committee (approval number: 12/LO/0914). Written informed consent for trial participation and the molecular analysis of tumor biopsies was obtained from all patients.
Patient Selection
The 21 cases analysed in this study were selected based only on sufficient DNA availability from biopsies and the inferred cancer cell contents. Cancer cell contents were estimated using the variant allele frequency (VAF) of the somatic mutations. Furthermore, we required an adequate cancer cell content to construct the integer copy number profile for clonality assessment. Cancer cell content and the integer copy number profiles have been presented previously3.
Somatic Mutation And Clonality Assessment
Published mutation calls were re-analyzed3,26. A mutation call with variant allele frequency (VAF) less than 5% was considered absent in either paired biopsy. The clonality of somatic variants was assessed as previously described3.
Mutational Signature Analysis
We identified a set of potentially active signatures by comparing with the ‘ColoRect-AdenoCa’ samples from the PCAWG7 TCGA exome cohort (syn11801497.7). This was done by selecting signatures with any non-zero mutation attribution to ensure the widest set of relevant signatures were included at the first stage. We added, if required, a further 6 signatures with aetiology associated to HR-deficiency (SBS3) and MMR-deficiency (SBS6, SBS15, SBS21, SBS26, SBS44) to test the acquired resistance model hypothesis and a further two signatures associated with platinum chemotherapy treatment (SBS31, SBS35) that may be relevant to the samples analysed in our cohort. This resulted in 21 signatures in total.
The single base substitution (SBS) mutation profile for each patient biopsy were fitted to the sigProfiler exome SBS signatures (syn12026190) using whichSignatures in the deconstructSigs24 (v1.8.0) R library. A second method of signature decomposition was applied using the fit_to_signatures function in the MutationalPatterns23 R library (v2.99.7) to assess mutation assignment bias between two independent approaches.
The inclusion of too many signatures would increase the likelihood of misasignment. Therefore, we looked to identify a set of signatures active in the Prospect-C samples for subsequent analysis. We applied a generalized cut-off to discard signatures with insufficient cohort-wide contribution. This required the total assignment of mutations to contribute a minimum of 3% of all SNVs across the cohort to consider the signature active (Extended Data Fig. 4A). This subset was further strengthened when looking just at prolonged benefit PD samples to ensure that potentially relevant signatures that may be involved in resistance driver acquisition were being considered (Extended Data Fig. 4B). Furthermore, despite not achieving the criteria, we included SBS17a due to the inclusion of the ‘connected’ SBS17b signature37.
The observed and reconstructed mutation profiles show residual differences. This error represents an unexplained portion of the mutation profile that is not captured by the signature subset. We estimated the proportion of variance explained by the signature set using a standard coefficient of determination (R2) measure. This was calculated using the computeExplanedVariance function in the decompTumor2Sig38 R library (v2.6.0). The signature weights were subsequently rescaled proportional to the explained variance (R2) of each sample. The remaining variance (1-R2) was considered unexplained.
Microsatellite Tract Length Analysis
MSIsensor39 (v0.6) scan was run on the complete hg19 reference sequence to identify homopolymer and microsatellite regions with a minimum of five consecutive repeats. This identified a total of 23,147,854 regions. Regions were filtered for those located on autosomal chromosomes. MSIsensor msi was run on each BL and PD pair, ensuring that all regions had a minimum of 20X coverage and were located within SureSelect v5 target regions. All microsatellites that showed a significant difference in length distribution were manually reviewed to identify those that showed an increase in the PD sample. The ratio proportion of microsatellites with increased length variability divided by the total number of assessed microsatellites defines the MSI-score.
KRAS, NRAS and EGFR mutation codon biases
Somatic mutation calls from The Cancer Genome Atlas (TCGA) were downloaded from the cBio web portal40,41 by selecting for ‘Colorectal Adenocarcinoma’ in the ‘PanCancer Atlas’. Mutation calls from studies3,5,26 that reported the specific base change alterations in KRAS, NRAS and EGFR mutations in ctDNA were pooled to generate a comparative distribution from CRCs with acquired resistance to EGFR-AB. Only cases with KRAS/NRAS codon 12/13/61 mutations were included and these mutations were assessed. Mutation calls in KRAS were also identified from ctDNA in the CORRECT trial28. Similarly, only KRAS codon 12/13/61 mutations were analyzed.
EGFR mutation calls in 3,5 were used to assess mutation codon biases in EGFR at acquired resistance.
To assess the relevance of mutational signature activity on the generation of KRAS, NRAS and EGFR hotspot mutations we modelled a BL prolonged benefit profile using the deconstructSigs signature weights generated for the corresponding 12 tumors (Supplementary Table 2). The weights were rescaled to sum to the explained variance of the sample (R2) and then multiplied by the corresponding mutation load to generate mutation attributions corresponding to each signature for each tumor. The mutation totals were then summed across the tumors and converted to an overall proportional contribution of each signature.
The reference signature profile confers the likelihood of observing a mutation corresponding to each of the 96 trinucleotide mutation motifs if the signature is active. However, the trinucleotide frequencies across the exome are not evenly distributed and so this must be adjusted to assess the likelihood of a specific mutation occurring. We used the function get_context_freq in the SigsPack42 R library to calculate the frequency of each trinucleotide context across the exonic regions and normalized the reference signatures to reflect a profile with even context frequency using the normalize function in SigsPack.
The normalized reference signatures were rescaled using the signature proportions obtained from the BL prolonged benefit tumors to generate a mutation probability profile. The resulting matrix confers the contribution of each individual signature to the overall probability of a mutation occurring at each of the 96 trinucleotide motifs (Extended Data Fig. 7A). The mutation probabilities of KRAS, NRAS and EGFR hotspot mutations observed at acquired resistance were extracted and rescaled proportional to all contexts (Fig. 3D,G).
To assess the impact of the chemotherapy-induced signatures, SBS17b (5-FU) and SBS35 (platinum), on the acquisition of KRAS/NRAS Q61H mutations we calculated the mutation probabilities of trinucleotide contexts corresponding to observed codon 12/13/61 hotspot mutations. The observed mutation signature attributions were adjusted to reflect the time period in which they were likely to be active. For instance, the clock-like signatures (SBS1, SBS5, SBS40) have accumulated mutations over the lifetime of the patient. In contrast, SBS17b and SBS35 are assumed to only be detectable after chemotherapy treatment. As the activity of SBS3, SBS6, SBS15 and SBS17a has not been reported to increase following chemotherapy treatment they were initially modelled as active throughout the patient lifecourse. We modelled a constant mutation accumulation of these signatures from birth to diagnosis, followed by an accelerated mutation accumulation 10 times the rate29 during the tumor growth period from diagnosis to BL biopsy. The attributions of these signatures during the growth phase were combined with the chemotherapy signature attributions to generate an adjusted weight matrix.
We applied the model with and without a zero constraint on the corresponding signature weight. Probabilities calculated for each hotspot mutation context were summed to demonstrate the resulting likelihood of each hotspot mutation. Furthermore, we modelled a range of realistic parameters to reflect the uncertainty of the time of tumor growth from malignant transformation (2.7-5.4yrs), the acceleration of mutation rates during this period (x1, x5, x10) and the time point in which the HR, MMR and SBS17a signatures become active.
The stability of the SBS17b attributions was assessed by bootstrap analysis using the function resample_mut_mat from the MutationalPatterns23 R library. This involves resampling the mutation count matrix using the observed context counts as probabilities. We specified 1,000 bootstrap replicates. Signature decomposition was then calculated for each replicate and percentile descriptives plotted.
Kaplan-Meier Analysis
The survfit function in the Survival (v.2.44-1.1) R library was used to run the Kaplan-Meier analysis. Progression free survival (PFS) was measured from start of treatment to date of progression or death from any cause.
SBS17b signature activity in an independent CRC cohort
The mutation calling from 536 whole-genome sequenced colorectal metastatic samples was obtained from Hartwig Medical Foundation31. A de-novo non-negative matrix factorization based mutational signature extraction was performed using SigProfilerJulia20,43. Two signatures with high cosine similarity to the canonical PCAWG SBS17b15 - related to 5-FU and the canonical signature - were selected. Samples with exposure to any of these signatures were deemed as SBS17b active.
Quantification And Statistical Analysis
All analyses were performed in R (v3.5.0)44. All p-values are two-sided and p<0.05 was considered significant. All t-tests were unpaired unless otherwise stated.
Extended Data
Extended Data Fig. 1. Plots of cancer cell content, sequencing depth and mutation load for the paired BL/PD biopsies from 21 patients in the Prospect-C trial.
(A) Estimated cancer cell contents of paired BL and PD samples. A 1:1 ratio line has been added for reference. (B) Mutation load vs. mean sequencing depth for all BL and PD samples. p-value from Spearman’s test. A linear regression line has been added for reference. (C) Mutation load vs. cancer cell content for all BL and PD samples. p-value from Spearman’s test. A linear regression line has been added for reference.
Extended Data Fig. 2. Clonal mutation trees for 21 tumors from the Prospect-C trial.
Grouped into cases with prolonged benefit and primary progression. The numbers next to the trunk or the branches indicate clonal somatic mutations.
Extended Data Fig. 3. Number of unique mutations detected for each of 21 paired biopsies from the Prospect-C trial vs. time lapse between BL and PD biopsies.
p-value from Spearman’s test. A linear regression line has been added for reference.
Extended Data Fig. 4. Proportion of SBS mutations attributed to each mutational signature.
Signatures were selected using the ‘ColoRect-AdenoCa’ samples from the SigProfiler TCGA whole exome cohort (n=496) (syn11801497.7). All signatures in the cohort with a non-zero mutation attribution were considered along with all MMR-deficiency signatures and platinum treatment signatures. Plots show the cohort wide signature attribution among (A) all 21 Prospect-C samples and (B) only in the PD tumors of the 12 patients with prolonged benefit. The red horizontal dashed line illustrates the 3% threshold used to define signatures as ‘active’ and the red box shows the signatures retained for subsequent analysis. SBS17a and SBS17b are described as ‘connected’ signatures15. SBS17a was retained due to the inclusion of SBS17b despite not reaching the threshold.
Extended Data Fig. 5. Signature attributions based on 21 paired BL/PD biopsies from the Prospect-C trial using MutationalPatterns and deconstructSigs.
(A) Mutation signature attribution using independent decomposition methods (deconstructSigs and MutationalPatterns). (B) Fig. 2 repeated with the ‘fit_to_sigs’ function in MutationalPatterns to assess the variability of estimates between methods.
Extended Data Fig. 6. Mutation frequency profiles of treatment naïve CRCs from the TCGA Pan-Cancer study vs. the KRAS hotspot mutations identified in 28.
The TCGA profile has been adjusted to only consider KRAS mutations that were assessed in the CORRECT trial.
Extended Data Fig. 7. Modelling the impact of mutational signatures on the likelihood of acquired hotspot mutations.
(A) Modelled mutational profile of a BL tumor with prolonged benefit. Exome normalised reference signatures have been scaled by the observed signature exposures of the 12 BL tumors with prolonged benefit to represent a mutation probability at each trinucleotide mutation context. (B) Observed mutation frequencies of KRAS/NRAS Q61H vs. all other KRAS/NRAS hotspot mutations identified in CRCs with acquired EGFR-AB resistance3,5,26. (C) Modelled mutation accumulation of the permanent signatures. A varying acceleration parameter of x1, x5, x10 is applied to the tumor growth period. (D) Impact of SBS17b and SBS35 on the likelihood of generating KRAS/NRAS Q61H mutations vs. all other detected KRAS/NRAS hotspot mutations.
Extended Data Fig. 8. Analysis of an independent cohort of 239 patients with metastatic colorectal cancer and a KRAS/NRAS G12/G13 or Q61H mutation.
(A) Total mutations attributed to SBS17b. Statistical significance was assessed with the Fisher’s exact test. (B) Proportion of tumors with a detectable SBS17b signature activity. Statistical significance was assessed with the Mann-Whitney U test.
Supplementary Material
Acknowledgements
DC received funding from the NIHR Biomedical Research Centre for Cancer at the Institute of Cancer Research and the Royal Marsden Hospital. MG, AW and LJB received funding from the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation programme (grant agreement No. 820137). The paper is dedicated to the memory of Tim Morgan who supported this work with a generous donation.
Footnotes
Author Contributions Statement
MG conceived, funded and supervised the molecular analysis. DC is the chief investigator of the Prospect C trial and funded the trial. NS, IC, SR, and DW recruited trial patients. BG prepared trial samples and NM supervised sequencing. LJB performed ctDNA sequencing and analysis. AW performed the bioinformatics analysis. OP and NL-B provided the analysis of metastatic CRC samples from the Hartwig Medical Foundation. AW and MG performed statistical analysis. AW and MG wrote the manuscript. LJB, OP and NL-B provided feedback. All authors approved the final manuscript.
Competing Interests Statement
IC has consultant/advisory roles with Eli-Lilly, BMS, MSD, Merck KG, Roche, Bayer, and Five Prime Therapeutics. DC receives research funding from Amgen, Sanofi, Merrimack, Astra Zeneca, Celegene, MedImmune, Bayer, 4SC, Clovis, Eli-Lilly, Janssen, and Merck KG. MG and NS receive research funding from Merck KG and BMS.
Data Availability
All analyses were performed on previously published datasets3,5,20,26-28. The datasets can be accessed as described in the primary publications. DNA sequencing data from the Prospect-C trial is deposited in The European Genome-phenome Archive with the accession code EGAS00001003367 (https://www.ebi.ac.uk/ega/studies/EGAS00001003367). As they include exome sequencing data that could potentially permit the re-identification of trial participants, a data sharing agreement is required as stated in the primary publication3.
Code Availability
Custom code to reproduce the mutational signature modelling is freely available on Github (https://github.com/AWoolston/Evolution-of-anti-EGFR-antibody-resistance).
References
- 1.Karapetis CS, et al. K-ras mutations and benefit from cetuximab in advanced colorectal cancer. The New England journal of medicine. 2008;359:1757–1765. doi: 10.1056/NEJMoa0804385. [DOI] [PubMed] [Google Scholar]
- 2.Van Cutsem E, et al. Fluorouracil, leucovorin, and irinotecan plus cetuximab treatment and RAS mutations in colorectal cancer. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2015;33:692–700. doi: 10.1200/JCO.2014.59.4812. [DOI] [PubMed] [Google Scholar]
- 3.Woolston A, et al. Genomic and Transcriptomic Determinants of Therapy Resistance and Immune Landscape Evolution during Anti-EGFR Treatment in Colorectal Cancer. Cancer cell. 2019;36:35–50.:e39. doi: 10.1016/j.ccell.2019.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Misale S, et al. Emergence of KRAS mutations and acquired resistance to anti-EGFR therapy in colorectal cancer. Nature. 2012;486:532–536. doi: 10.1038/nature11156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bettegowda C, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med. 2014;6:224ra224. doi: 10.1126/scitranslmed.3007094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Montagut C, et al. Identification of a mutation in the extracellular domain of the Epidermal Growth Factor Receptor conferring cetuximab resistance in colorectal cancer. Nature medicine. 2012;18:221–223. doi: 10.1038/nm.2609. [DOI] [PubMed] [Google Scholar]
- 7.Lipinski KA, et al. Cancer Evolution and the Limits of Predictability in Precision Cancer Medicine. Trends Cancer. 2016;2:49–63. doi: 10.1016/j.trecan.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maley CC, et al. Classifying the evolutionary and ecological features of neoplasms. Nat Rev Cancer. 2017;17:605–619. doi: 10.1038/nrc.2017.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alexandrov LB, et al. Signatures of mutational processes in human cancer. Nature. 2013;500:415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gerlinger M, Swanton C. How Darwinian models inform therapeutic failure initiated by clonal heterogeneity in cancer medicine. British journal of cancer. 2010;103:1139–1143. doi: 10.1038/sj.bjc.6605912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Diaz LA, Jr, et al. The molecular evolution of acquired resistance to targeted EGFR blockade in colorectal cancers. Nature. 2012;486:537–540. doi: 10.1038/nature11219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Russo M, et al. Adaptive mutability of colorectal cancers in response to targeted therapies. Science (New York, NY) 2019;366:1473–1480. doi: 10.1126/science.aav4474. [DOI] [PubMed] [Google Scholar]
- 13.Gerlinger M. Targeted drugs ramp up cancer mutability. Science (New York, NY) 2019;366:1452–1453. doi: 10.1126/science.aaz9900. [DOI] [PubMed] [Google Scholar]
- 14.Kim TM, Laird PW, Park PJ. The landscape of microsatellite instability in colorectal and endometrial cancer genomes. Cell. 2013;155:858–868. doi: 10.1016/j.cell.2013.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alexandrov LB, et al. The repertoire of mutational signatures in human cancer. Nature. 2020;578:94–101. doi: 10.1038/s41586-020-1943-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maura F, et al. A practical guide for mutational signature analysis in hematological malignancies. Nature communications. 2019;10:2969. doi: 10.1038/s41467-019-11037-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alexandrov LB, et al. Clock-like mutational processes in human somatic cells. Nature genetics. 2015;47:1402–1407. doi: 10.1038/ng.3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sveen A, et al. Multilevel genomics of colorectal cancers with microsatellite instability-clinical impact of JAK1 mutations and consensus molecular subtype 1. Genome Med. 2017;9:46. doi: 10.1186/s13073-017-0434-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Christensen S, et al. 5-Fluorouracil treatment induces characteristic T>G mutations in human cancer. Nature communications. 2019;10:4571. doi: 10.1038/s41467-019-12594-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pich O, et al. The mutational footprints of cancer therapies. Nature genetics. 2019;51:1732–1740. doi: 10.1038/s41588-019-0525-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tomkova M, Tomek J, Kriaucionis S, Schuster-Bockler B. Mutational signature distribution varies with DNA replication timing and strand asymmetry. Genome biology. 2018;19:129. doi: 10.1186/s13059-018-1509-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meier B, et al. Mutational signatures of DNA mismatch repair deficiency in C. elegans and human cancers. Genome research. 2018;28:666–675. doi: 10.1101/gr.226845.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Blokzijl F, Janssen R, van Boxtel R, Cuppen E. MutationalPatterns: comprehensive genome-wide analysis of mutational processes. Genome Med. 2018;10:33. doi: 10.1186/s13073-018-0539-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rosenthal R, McGranahan N, Herrero J, Taylor BS, Swanton C. DeconstructSigs: delineating mutational processes in single tumors distinguishes DNA repair deficiencies and patterns of carcinoma evolution. Genome biology. 2016;17:31. doi: 10.1186/s13059-016-0893-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arena S, et al. Emergence of Multiple EGFR Extracellular Mutations during Cetuximab Treatment in Colorectal Cancer. Clinical cancer research : an official journal of the American Association for Cancer Research. 2015;21:2157–2166. doi: 10.1158/1078-0432.ccr-14-2821. [DOI] [PubMed] [Google Scholar]
- 26.Khan KH, et al. Longitudinal Liquid Biopsy and Mathematical Modeling of Clonal Evolution Forecast Time to Treatment Failure in the PROSPECT-C Phase II Colorectal Cancer Clinical Trial. Cancer discovery. 2018;8:1270–1285. doi: 10.1158/2159-8290.CD-17-0891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cancer Genome Atlas Research, N et al. The Cancer Genome Atlas Pan-Cancer analysis project. Nature genetics. 2013;45:1113–1120. doi: 10.1038/ng.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tabernero J, et al. Analysis of circulating DNA and protein biomarkers to predict the clinical activity of regorafenib and assess prognosis in patients with metastatic colorectal cancer: a retrospective, exploratory analysis of the CORRECT trial. The Lancet Oncology. 2015;16:937–948. doi: 10.1016/S1470-2045(15)00138-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gerstung M, et al. The evolutionary history of 2,658 cancers. Nature. 2020;578:122–128. doi: 10.1038/s41586-019-1907-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Price T, et al. Frequency of S492R mutations in the epidermal growth factor receptor: analysis of plasma DNA from patients with metastatic colorectal cancer treated with panitumumab or cetuximab monotherapy. Cancer Biol Ther. 2020;21:891–898. doi: 10.1080/15384047.2020.1798695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Priestley P, et al. Pan-cancer whole-genome analyses of metastatic solid tumours. Nature. 2019;575:210–216. doi: 10.1038/s41586-019-1689-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cannataro VL, Gaffney SG, Townsend JP. Effect Sizes of Somatic Mutations in Cancer. J Natl Cancer Inst. 2018;110:1171–1177. doi: 10.1093/jnci/djy168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ali M, et al. Codon bias imposes a targetable limitation on KRAS-driven therapeutic resistance. Nature communications. 2017;8:15617. doi: 10.1038/ncomms15617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Montagut C, et al. Efficacy of Sym004 in Patients With Metastatic Colorectal Cancer With Acquired Resistance to Anti-EGFR Therapy and Molecularly Selected by Circulating Tumor DNA Analyses: A Phase 2 Randomized Clinical Trial. JAMA oncology. 2018;4:e175245. doi: 10.1001/jamaoncol.2017.5245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Poulos RC, Wong YT, Ryan R, Pang H, Wong JWH. Analysis of 7,815 cancer exomes reveals associations between mutational processes and somatic driver mutations. PLoS genetics. 2018;14:e1007779. doi: 10.1371/journal.pgen.1007779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Temko D, Tomlinson IPM, Severini S, Schuster-Bockler B, Graham TA. The effects of mutational processes and selection on driver mutations across cancer types. Nature communications. 2018;9:1857. doi: 10.1038/s41467-018-04208-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Poetsch AR. The genomics of oxidative DNA damage, repair, and resulting mutagenesis. Comput Struct Biotechnol J. 2020;18:207–219. doi: 10.1016/j.csbj.2019.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kruger S, Piro RM. decompTumor2Sig: identification of mutational signatures active in individual tumors. BMC Bioinformatics. 2019;20:152. doi: 10.1186/s12859-019-2688-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Niu B, et al. MSIsensor: microsatellite instability detection using paired tumor-normal sequence data. Bioinformatics (Oxford, England) 2014;30:1015–1016. doi: 10.1093/bioinformatics/btt755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cerami E, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer discovery. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gao J, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schumann F, et al. SigsPack, a package for cancer mutational signatures. BMC Bioinformatics. 2019;20:450. doi: 10.1186/s12859-019-3043-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Alexandrov LB, Nik-Zainal S, Wedge DC, Campbell PJ, Stratton MR. Deciphering signatures of mutational processes operative in human cancer. Cell reports. 2013;3:246–259. doi: 10.1016/j.celrep.2012.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; Vienna, Austria: 2018. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All analyses were performed on previously published datasets3,5,20,26-28. The datasets can be accessed as described in the primary publications. DNA sequencing data from the Prospect-C trial is deposited in The European Genome-phenome Archive with the accession code EGAS00001003367 (https://www.ebi.ac.uk/ega/studies/EGAS00001003367). As they include exome sequencing data that could potentially permit the re-identification of trial participants, a data sharing agreement is required as stated in the primary publication3.
Custom code to reproduce the mutational signature modelling is freely available on Github (https://github.com/AWoolston/Evolution-of-anti-EGFR-antibody-resistance).