Abstract
Background & Aims
Ultrasound (US)-based screening has been recommended for patients with an increased risk of hepatocellular Carcinoma (HCC). US analysis is, however, limited in patients who are obese or have small tumors. Addition of measurement of serum level of alpha-fetoprotein (AFP) to US analysis can increase detection of HCC. We analyzed data from patients with chronic liver disease, collected over 15 years in an HCC surveillance program, to develop a model to assess risk of HCC.
Methods
We collected data from 3450 patients with chronic liver disease undergoing US surveillance in Japan from March 1998 through April 2014 and followed for a median 8.83 years. We performed longitudinal discriminant analysis of serial AFP measurements (median number of observations/patient, 56; approximate every 3 months) to develop a model to determine risk of HCC. We validated the model using data from 2 cohorts of patients with chronic liver disease in Japan (404 and 2754 patients) and 1 cohort in Scotland (1596 patients).
Results
HCC was detected in 413 patients (median tumor diameter, 1.8 cm), during a median follow-up time of 6.60 years. In the development dataset, the model identified patients who developed HCC with an area under the curve of 0.78; it correctly identified 74.3% of patients who did develop HCC, and 72.9% of patients who did not. Overall, 73.1% of patients were correctly classified. The model could be used to assign patients to a high-risk group (27.5 HCCs/1000 patient-years) vs a low-risk group (4.9 HCCs/1000 patient-years). Similar performance was observed when the model was used to assess patients with cirrhosis. Analysis of the validation cohorts produced similar results.
Conclusions
We developed and validated a model to identify patients with chronic liver disease who are at risk for HCC based on change in serum level of AFP over time. The model could be used to assign patients to high-risk vs low-risk groups, and might be used to select patients for surveillance.
Keywords: liver cancer, stratification, prognostic factor, biomarker
Hepatocellular carcinoma (HCC) is the third most common cause of cancer-related death worldwide and the leading cause of death in patients with cirrhosis, the setting within which most HCCs arise.1 Since potentially curative therapies are only available for patients with early stage HCC, ultrasound-based screening (USS) is widely recommended although its impact on overall survival remains controversial. USS does however, have severe limitations. Recent studies suggest that its sensitivity for early H2–4 However, USS surveillance may be enhanced by addition of the serum biomarker AFP 4
Japan has implemented a rigorous surveillance program resulting in clear evidence that the stage at which HCC is detected has decreased and that overall survival has been improved. The Japanese surveillance program relies on regular US surveillance with AFP and two additional biomarkers, AFP–L3 and DCP, both of which have FDA-approval for HCC screening. 5–7
Here, we have analysed a unique dataset of patients with chronic liver disease, prospectively followed-up for over 15 years within a rigorous HCC surveillance program using a recently developed statistical methodology (longitudinal discriminant analysis (LoDA))8, 9, with the aim of stratifying patients according to their risk of developing HCC. This could identify a subgroup for more intensive monitoring with the aim of early diagnosis.
The use of longitudinal AFP measurements in screening for HCC has been considered previously with encouraging results.10, 11 However, these models were developed only in patients with hepatitis-C, and have only been validated internally. Here we examine multiple disease aetiologies over long time periods and offer an externally validated tool that is ready for practical application.
Materials and Methods
Model derivation dataset
The model described here was built on a total sample of 3450 patients undergoing surveillance at Ogaki municipal hospital in Japan. Patients were recruited between March 1998 and April 2014 with a median follow up of 8.83 years. AFP levels above 20ng/ml or a positive USS triggered a diagnostic workup for HCC. Of those undergoing surveillance, 413 developed HCC (12%). The diagnosis of HCC was made according to EASL guidelines,12 but it should be noted that diagnosis of HCC was established histologically in more than 99% of cases because of the very high rate of resection. Median maximum tumour size at HCC diagnosis was 2cm, 84.2% were within Milan criteria and 99.76% (412 out of 413) underwent treatment with curative intent.
Fifty four percent of the patients in the Ogaki cohort were HCV seropositive, 23.3% seropositive HBV, 21.8% had ‘other’ forms of chronic disease (Table 1). Of the 1876 (54.4%) patients who had hepatitis C, 998 (53%) were successfully treated with various anti-viral therapies, and only 83 (8.3%) of this subgroup went on to develop HCC. By contrast, 175 (25.8%) of the 679 patients who received no treatment for hepatitis C, and 62 (31.2%) of the 199 patients whose treatment failed, developed HCC. Our preliminary analysis showed that the great majority of cases in which Fib-4 was > 3.25 had severe fibrosis or cirrhosis as assessed histologically. Henceforth we regraded patients with Fib-4 > 3.25 as ‘cirrhotic’.
Table 1. Characteristics of patients included in the analysis for the main Ogaki dataset, and the Kindai, Red Cross and Edinburgh external validation datasets.
| Variable | Ogaki | Kindai | Red Cross | Edinburgh | ||||
|---|---|---|---|---|---|---|---|---|
| Non HCC | HCC | Non HCC | HCC | Non HCC | HCC | Non-HCC | HCC | |
| Number of Patients | 3037 (88%) | 413 (12%) | 338 (84%) | 66 (16%) | 2413 (88%) | 341 (12%) | 1509 (95%) | 87 (5%) |
| Follow up visits | 72206 | 8646 | 3824 | 1001 | 61561 | 8371 | 14845 | 1068 |
| Follow Up length (Years) | 9.29 (5.99,13.32) | 6.6 (4.65,9.64) | 2.47 (1.04,4.63) | 2.38 (1.13,5.15) | 9.07 (5.85,12.36) | 6.15 (4.11,8.35) | 4.69 (2.38,8.95) | 5.79 (2.85,9.11) |
| Males (%) | 1465 (48.2%) | 251 (60.8%) | 154 (45.6%) | 37 (56.1%) | 1062 (44.0%) | 182 (53.4%) | 860 (57%) | 64 (73.6%) |
| Aetiology HBV:HCV:Both:Other (%) | 749:1556:11:721 (24.7,51.2,0.4,23.7) | 55:320:6:32 (13.3,77.5,1.5,7.7) | 117:148:2:71 (34.6,43.8,0.6,21.0) | 4:53:3:6 (6.1,80.3,4.5,9.1) | 502:1362:15:367 (20.8,56.4,0.6,15.2) (167 unknown) | 34:261:5:41 (10.0,76.5,1.5,12.0) | 157:401:5:946 (10.4,26.6,0.3,62.7) | 4:30:0:53 (4.6,34.5,0,60.9) |
| Maximum Tumor Size 1:2:3:?4cm, (%) | N/A | 92:176:84:61 (22.3,42.6,20.3,14.8) | N/A | 20:32:5:9 (30.3,48.5,7.6,13.6) | N/A | 82:161:56:42 (24.0,47.2,16.4,12.4) | N/A | |
| Number of nodules 1:2:?3, % | N/A | 285:76:52 (69.0,18.4,12.6) | N/A | 53:7:6 (80.3,10.6,9.1) | N/A | 233:65:43 | N/A | |
| Age at first screening (Years) | 60.09 (50.61,67.72) | 64.47 (57.91,69.26) | 62.00 (49.33,70.50) | 66.89 (60.02,72.88) | 60.83 (51.21,68.71) | 65.68 (60.51,71.09) | 51 (42,61) | 59.05 (50.33,66.32) |
| AFP ng/mL | 2.4 (1.5,4) | 7.4 (3.5,19.1) | 3 (2,4) | 10 (4,22) | 4 (2.8,6.4) | 8.6 (4.9,18.75) | 4.84 (2.42,6.05) | 6.05 (3.63,13.31) |
| DCP ng/mL | 0.2 (0.16,0.25) | 0.22 (0.16,0.31) | 0.2 (0.17,0.26) | 0.23 (0.18,0.31) | ||||
| FIB-4 | 2.07 (1.37,3.34) | 4.75 (3.06,7.52) | 2.45 (1.40,4.68) | 4.31 (3.02,6.02) | ||||
| First recorded FIB-4 <1.45:1.45-3.25:>3.25:NA, % |
1102:1292:643:0 (36.3,42.5,21.2,0) |
15:134:262:2 (3.6,32.5,63.4,0.5) |
113:136:78:11 (33.4,40.2,23.1,3.3) |
2:17:47:0 (3.0,25.8,71.2,0) |
||||
| AFP.L3 % | 0.5 (0,0.5) | 0.5 (0,1.1) | 0.5 (0.5,0.5) | 11.2 (0.5,12.2) | ||||
| GALAD | -3.3 (-4.47,-2.13) | -1.66 (-2.78,-0.34) | -3.67 (-4.84,-2.15) | -1.45 (-3.36,-0.23) | ||||
1. Continuous variables are presented as median with interquartile range.
2. Greyed out cells in Edinburgh and Red Cross cohorts did not have available data.
Model validation datasets
External validation was performed in three additional cohorts, two from Japan and one from Scotland. The Kindai cohort consists of 404 patients recruited between January 2000 and July 2017 at the Kindai University hospital in Osaka of whom 66 (16%) developed HCC, during a median follow up of 2.42 years. The Osaka Red Cross Hospital cohort comprised 2754 patients recruited between January 2004 and May 2018 of whom 341 (12%) developed HCC during this period of observation, with a median follow up of 8.63 years.
We also validated our model using a UK-based dataset from the Edinburgh region of Scotland. This dataset comprised 1596 patients, 87 (5%) of whom developed HCC whilst under active HCC surveillance between January 2009 and December 2016 (median follow-up 4.77 years). These patients were all cirrhotic. Details of this cohort have been previously published and the relevant dataset (used here) is publicly available.13
The Red Cross and Kindai cohorts are part of the Japanese surveillance programme, but represent independent institutions. All Japanese patients in this study underwent surveillance according to the Japanese Liver Cancer Group guidelines14 involving ultrasound examination and biomarker analysis. The median size of tumour detected was 1.8 cm, (90%< 3cm). The Edinburgh cohort was included to assess the utility of a model developed in a Japanese cohort in predicting patients at risk of developing HCC within a western population, containing only cirrhotic patients.
Statistical Methods
To visualize changes in AFP levels over time, we plotted time backwards from the diagnosis of HCC or the last sample and presented smoothed mean profiles with 95% confidence intervals (using generalized additive models) separately for patients who developed HCC and those who did not.
To assess a patient’s risk of developing HCC, we applied a longitudinal discriminant analysis (LoDA) approach.8, 9 Changes over time in log(AFP) were modelled using linear mixed models, with separate models for patients who developed HCC during surveillance and those who did not, and were adjusted for age and gender. Full details of the linear mixed models are given in the Supplementary material. The LoDA approach then calculated a probability that a new patient would develop HCC by assessing which of the two average profiles (HCC/Non-HCC) the patient was closest to. The marginal profile of a patient’s biomarker values was used in order to assess similarity to patients in each group.15
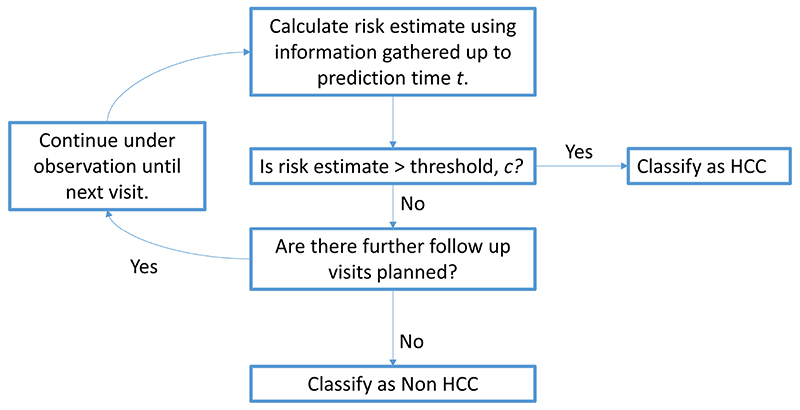
Predictions were dynamically updated each time a new measurement was available for a patient. We adopted the classification scheme shown in Figure 1 to classify patients. At each visit, the patient’s risk of developing HCC was calculated using the current visit and all previously known data from that patient. If this risk was greater than a given threshold (chosen by a ROC analysis), then the patient was classified as likely to develop HCC and prediction would stop for this patient. As long as a patient’s risk of developing HCC remained below the threshold, their risk was updated at their next follow up visit.
Figure 1. Allocation Scheme for sequential HCC risk assessment.
The longitudinal serum biomarker AFP was log-transformed for this analysis due to extreme skewness. Leave-one-out cross-validation was used to assess the predictive accuracy of this model. The model was externally validated in the Kindai, Red Cross and Edinburgh datasets, by fitting the model to all patients in the Ogaki dataset and then predicting the risk of developing HCC for each patient in the three validation cohorts using the allocation scheme in Figure 1. Calculations were performed in R using the mixAK package.16
Prediction accuracy was measured using sensitivity, specificity, positive predictive value (PPV) and negative predictive value (NPV). Since we test our prediction model at successive visits (until a patient is classified as high risk, or all available visits are considered) we report a patient-level sensitivity as the proportion of cases with at least one positive screening test.10 Similarly, we report a patient-level specificity as the proportion of non-cases who have no positive tests whilst under observation. Patient level PPV (proportion of patients predicted to be high risk who went on to develop HCC) and NPV (proportion of patients who never had a positive screening test who did not develop HCC) were also reported. We also calculated the mean lead-time, defined as the average time before actual diagnosis at which an HCC patient was correctly identified. The leadtime gives us an indication of how long before diagnosis patients can be identified as “at-risk”.
Results
Exploratory Plots
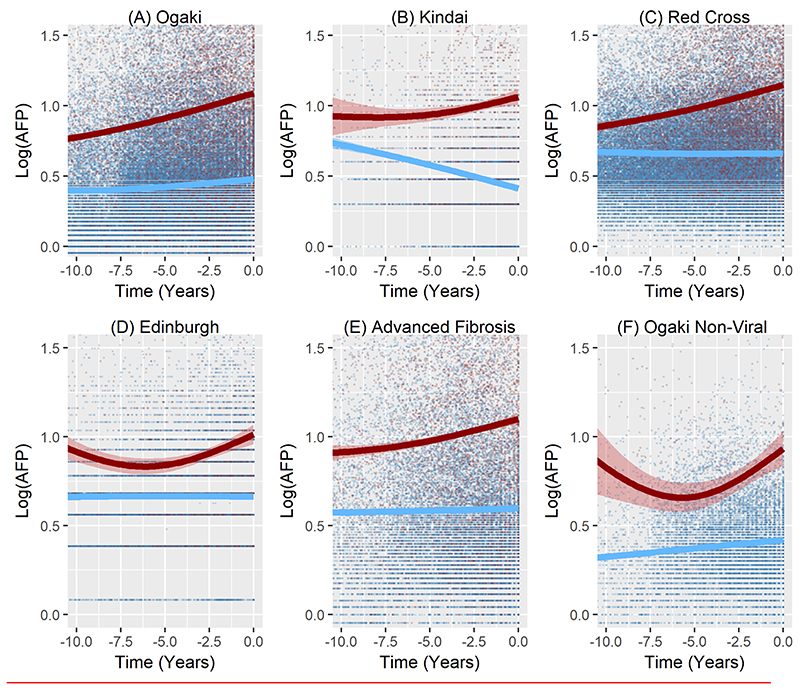
AFP was elevated, on average, among those destined to develop HCC for up to 15 years before the tumour was diagnosed clinically in all four cohorts. (Fig 2 A-D). Figure 2(E) shows that, on average, there is no significant AFP rise in the 796 patients with cirrhosis who did not develop HCC.
Figure 2.
Average changes in Log AFP over the 10 years before HCC detection according to development of HCC in A) Ogaki cohort, B) Kindai Cohort C) Red Cross Cohort, D) Edinburgh Cohort, E) patients who had advanced Fibrosis (defined by Fib-4>3.25) within the first two years of their observations from the Ogaki cohort (262 HCC cases and 796 Non-HCC cases), F) non-viral patients from the Ogaki cohort (32 HCC cases and 721 non-HCC cases).The bands represent 95% confidence intervals around the mean profile.
We also considered the possibility that the rise in AFP in HCC cases is caused by poor response to antiviral treatment resulting in continuing virus-associated inflammatory activity. However, on average, the AFP levels are higher in the patients who go on to develop HCC even in patients with no viral activity (Fig 2(F)), with similar patterns observed in the Edinburgh non-viral cohort, Supplementary Figure 1). Note that the dips in 2(D) and 2(F) are likely due to the smaller number of data points so long before diagnosis, rather than being an actual trend in the AFP profiles. The point of note is that there is a clear separation in average AFP profiles between patients who develop HCC and those who do not, and that is evident a long time before diagnosis.
Predictive Accuracy
A patient was classified as at high-risk of developing HCC if they had a probability of developing HCC (assigned by the LoDA model) of above 25% at any point during their follow up (this threshold was chosen as the threshold closest to the top left corner of a ROC plot). Otherwise, they remained under observation. In the Ogaki cohort this model correctly identified 74.3% of patients who developed HCC (sensitivity), and 72.9% of patients who did not develop HCC (specificity). Overall, this model achieved an AUC of 0.784 (Table 2), with 73.1% of patients correctly identified (PCC). 27.2% of patients predicted to develop HCC actually developed HCC (PPV), whilst 95.4% of patients whose risk of HCC never rose above the optimal threshold of 25%, did not develop HCC (NPV).
Table 2. AFP model performance based on internal validation in the Ogaki cohort, analysis of subgroups within Ogaki, and external validation in the Kindai, Red Cross and Edinburgh cohorts.
| n | HCC | Threshold | Sensitivity | Specificity | PCC | AUC | PPV | NPV | Mean Lead Time (years) | |
|---|---|---|---|---|---|---|---|---|---|---|
| Internal Validation in Ogaki Cohort (Leave-one-out cross-validation) | ||||||||||
| Whole Ogaki Cohort | 3450 | 413 | 0.25 | 0.743 | 0.729 | 0.731 | 0.784 | 0.272 | 0.954 | 5.69 |
| Subgroup Analysis | ||||||||||
| Males | 1716 | 251 | 0.2 | 0.777 | 0.662 | 0.679 | 0.761 | 0.283 | 0.945 | 5.81 |
| Females | 1734 | 162 | 0.25 | 0.796 | 0.743 | 0.748 | 0.807 | 0.242 | 0.973 | 6.05 |
| Aetiology=HBV | 804 | 55 | 0.16 | 0.800 | 0.694 | 0.701 | 0.802 | 0.161 | 0.979 | 5.19 |
| Aetiology=HCV | 1876 | 320 | 0.29 | 0.731 | 0.677 | 0.686 | 0.740 | 0.318 | 0.924 | 5.55 |
| Aetiology=other | 753 | 32 | 0.14 | 0.781 | 0.736 | 0.738 | 0.787 | 0.116 | 0.987 | 6.04 |
| Other Cancers | 657 | 413 | 0.25 | 0.743 | 0.754 | 0.747 | 0.800 | 0.837 | 0.634 | 5.69 |
| Tumour Characteristics | ||||||||||
| Total Tumours = 1 | 413 | 285 | 0.25 | 0.710 | N/A | N/A | N/A | N/A | N/A | 5.97 |
| Total Tumours = 2 | 413 | 76 | 0.25 | 0.803 | N/A | N/A | N/A | N/A | N/A | 5.17 |
| Total Tumours >= 3 | 413 | 52 | 0.25 | 0.854 | N/A | N/A | N/A | N/A | N/A | 4.80 |
| Max Tumour Diameter = 1 | 413 | 92 | 0.25 | 0.761 | N/A | N/A | N/A | N/A | N/A | 6.32 |
| Max Tumour Diameter = 2 | 413 | 176 | 0.25 | 0.693 | N/A | N/A | N/A | N/A | N/A | 6.06 |
| Max Tumour Diameter = 3 | 413 | 84 | 0.25 | 0.786 | N/A | N/A | N/A | N/A | N/A | 5.09 |
| Max Tumour Diameter >= 4 | 413 | 61 | 0.25 | 0.800 | N/A | N/A | N/A | N/A | N/A | 4.56 |
| External validation | ||||||||||
| Kindai | 404 | 66 | 0.25 | 0.682 | 0.882 | 0.849 | 0.831 | 0.529 | 0.934 | 3.37 |
| Kindai (adjusted threshold) | 404 | 66 | 0.13 | 0.818 | 0.760 | 0.770 | 0.831 | 0.400 | 0.955 | 3.39 |
| Red Cross | 2754 | 341 | 0.25 | 0.668 | 0.717 | 0.711 | 0.747 | 0.251 | 0.939 | 8.76 |
| Red Cross (adjusted threshold) | 2754 | 341 | 0.23 | 0.704 | 0.690 | 0.692 | 0.747 | 0.243 | 0.943 | 8.80 |
| Edinburgh | 1596 | 87 | 0.25 | 0.598 | 0.721 | 0.714 | 0.731 | 0.110 | 0.969 | 4.85 |
| Edinburgh (adjusted threshold) | 1596 | 87 | 0.21 | 0.690 | 0.657 | 0.659 | 0.731 | 0.104 | 0.974 | 5.00 |
1. Sensitivity (Specificity) is calculated for each characteristic as the percentage of cases (no cases) correctly predicted to develop (not develop) HCC out of all HCC (non HCC) patients with the given characteristic. Reported threshold is the point closest to the top-left corner on the ROC curve.
2. PCC=Probability of correct classification, AUC=Area under ROC curve, PPV=Positive Predictive Value, NPV=Negative Predictive Value.
3. Lead time is defined as the average length of time before diagnosis at which patients who developed HCC were correctly predicted to develop HCC.
4. External Validation assessed the ability of the Ogaki model to predict HCC using the threshold chosen from internal validation of the main Ogaki cohort (0.25). The predictive ability of the model if each cohort was allowed their unique threshold (adjusted threshold) is also provided.
5. The “Other cancers” group consisted of 244 patients who did not develop HCC but developed other cancers whilst under observation.
In the Ogaki cohort, 67.2% of patients never rose above the threshold of 0.25. Patients in this group were observed for a total of 21550.93 patient years with 106 HCC cases (incidence rate of 4.92 cases per 1000 patient years). This compares to an incidence rate of 27.5 cases per 1000 patient years among those in the high-risk group, and to an incidence rate of 12.62 for the entire Ogaki cohort. Patients who were correctly identified as HCC cases by the model were identified on average 5.69 years before their actual diagnosis.
By contrast, if patients were risk stratified based on presence of advanced fibrosis, then this approach would achieve a sensitivity of 88.9% but a specificity of only 55.1% in the Ogaki cohort. This is in comparison to a sensitivity of 74.3%, and specificity of 72.9% using the AFP LoDA model with a cut-off chosen by ROC analysis.
Stratifying based on fibrosis would correctly identify an extra 60 HCC cases than the LoDA model, but at the expense of 50.1% of the population being put into the “high-risk group” (Compared with 33% in the LoDA approach).
Subgroup Analyses
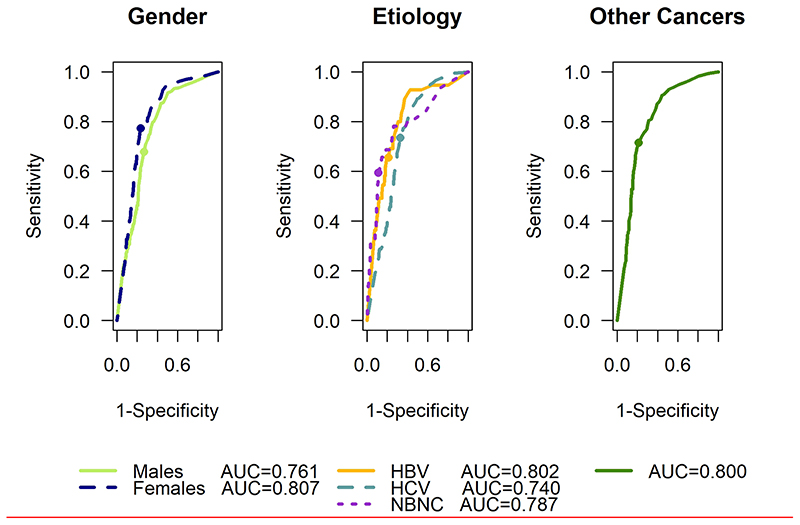
The predictive accuracy of the model for various patient subgroups in the Ogaki cohort is given in Table 2 and ROC curves are shown in Figure 3. Our model performed well regardless of sex, aetiology, and ultimate tumour features with further details given in the supplementary material. Particularly noteworthy is the ability of the model to predict HCC in patients who have neither Hepatitis B, Hepatitis C or alcohol related liver disease, (most likely non-alcoholic-fatty liver disease) and go on to develop HCC.
Figure 3. ROC curves for subgroup analysis of Ogaki cohort showing model performance by gender (left panel) and aetiology (centre panel) to predict HCC. The right panel shows the model performance to discriminate between HCC and other cancers.
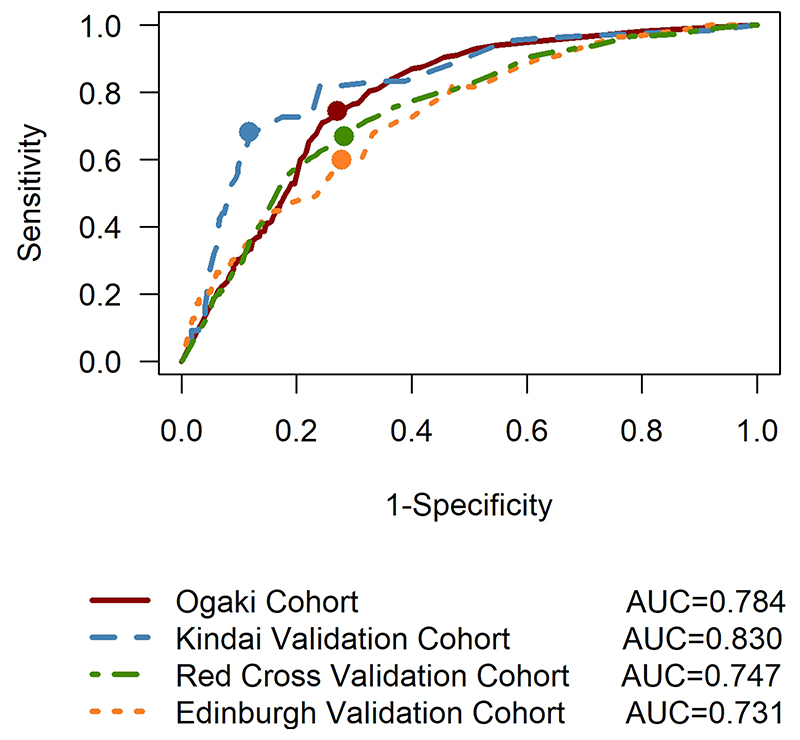
External Validation
In both of the independent Japanese cohorts our model was able to identify a group of patients with a high-risk of developing HCC substantially before clinical appearance of HCC (Table 2 and Figure 4). The changes over time in AFP, and the classification accuracy were similar in the Edinburgh cohort to those seen in Japan (Figure 2(C)). Incidence rates and additional details regarding the external validation are provided in the supplementary material.
Figure 4. ROC curves for the model developed with the Ogaki cohort using the Ogaki cohort (internal validation) and the Kindai, Red Cross and Edinburgh cohorts (external validation). The dots represent the values of sensitivity and specificity for the chosen threshold of 0.25.
Surveillance of only cirrhotic patients
In the West, only patients with cirrhosis are considered for HCC surveillance programmes. As a sensitivity analysis, we developed our model only including patients once they developed cirrhosis, and achieved broadly similar accuracy results (See supplementary material).
Using other longitudinal biomarkers
It is possible that the predictive accuracy of our model could be improved by including additional longitudinal markers such as DCP, platelet count or ALT. As an example, we considered a trivariate MGLMM in each group consisting of longitudinal observations of log(AFP), log(ALT) measurements and platelet counts.
This model achieved a higher sensitivity (81.4%) and NPV (96.5) but lower specificity (69.9%) and PPV (26.9%) and with an increased AUC of 0.817. DeLong’s test for correlated ROC curves showed a statistically significant improvement in the AUC of the trivariate model (p<0.001).
Discussion
In this study, we have applied a recently developed longitudinal discriminant analysis approach that takes account of correlation between AFP values for the same patient within a flexible model, and also allows patient specific predictions of risk to be made. These can be updated over time, allowing the potential for personalised decision making.
Our AFP model is able to identify patients at high-risk of developing HCC in cohorts that may be considered appropriate for HCC surveillance. The model validates well, not just in two Japanese cohorts, but also in a UK-based cohort, implying the generalisability of the model. Patients who developed HCC were identified as being at high-risk, 3 to 5 years before clinical detection emphasising the potential of the model for risk stratification.
In contradistinction to Western guidelines, Japanese guidelines14 define the high risk population who should undergo HCC surveillance for HCC on the basis of aetiology, rather than degree for fibrosis. Hence all patients who have chronic HBV or HCV infection are considered as ‘high-risk’, (regardless of the presence or absence of cirrhosis) and offered surveillance.
The most striking of these findings is that average AFP levels tend to be elevated and start to rise, in those who go on to develop HCC, more than 15 years before diagnosis. By any standard, diagnosis is considered early with a median tumour size of < 2cm at the time of detection, so this tool allows the potential for closer follow up long before tumours are visible.
It is unlikely that the early changes observed in AFP represent early/sub-clinical HCC. Much more plausible is that we are detecting the population of patients with chronic liver disease that are at risk of HCC development. The model presented in this paper requires only a record of all available AFP values for a patient, their sex, and their age at their first surveillance visit. Using this information the model provides a prediction of the patient’s risk of developing HCC. This risk can be updated each time new information is available for the patient. Our model achieves good accuracy and identifies patients who develop HCC with an accuracy of around 75%. The incidence of HCC, as measured per 1000 years of patient follow-up is about 5-fold higher in the high-risk group than the low-risk group.
Regarding the practical implementation of the model, we have developed a web-based tool which acts as a calculator of a patient’s risk of developing HCC. A preliminary version of this can be accessed at https://biostats.liv.ac.uk/olc/. A clinician could enter a patient’s age at first surveillance and sex, as well as all available AFP history, and the tool would calculate a probability that the patient is at high-risk of developing HCC.
Our model can be used for practical ‘risk stratification’. In particular, patients in the high-risk group (HCC risk > 25%) might be offered more intensive surveillance than those in the lower risk group, along with proposals for lifestyle changes with the aim of reducing HCC risk. The levels of AFP increase markedly two years prior to diagnosis, as seen in Figure 2(a). We have found similar abrupt rise in other tumour biomarkers such as AFP-L3 and DCP (data not shown). Such changes can be captured by the GALAD score, which combines these markers with age and gender.17–19 Thus the former (AFP) is, as presented here, of practical importance in defining the high-risk population for screening whereas the latter changes (via GALAD) offer the opportunity of early diagnosis.
Our preliminary analysis of an expanded biomarker dataset suggests that the predictive accuracy of our model could be improved by including additional longitudinal markers (such as DCP, platelet count or ALT), that have been shown to improve accuracy of diagnostic models for AFP.20 However, further work would be needed to assess the optimal combination of longitudinal markers, and we are currently reviewing models that include DCP, albumin, FIB-4 and markers related to diabetes. Such expansion will require careful balancing of the increased complexity of the model (which limits its practical application) and the performance improvement.Our model could be used to identify a subgroup of patients to be monitored more frequently in order to watch for early signs of sharp increase in AFP, whilst patients deemed to be lower risk could be screened less frequently and have updated predictions of risk of HCC at a future visit. In the high- risk group, models could be developed to identify patients as soon as their AFP profiles show an increase of certain magnitude. It is possible that additional markers such as DCP, AFP-L3, or the composite GALAD score could offer additional help in this early detection stage. Ultrasound screening, in combination with the GALAD score offers the potential for accurate diagnosis once HCC has developed.19 Further studies would be required in order to assess whether our AFP based approach could complement ultrasound screening in a similar manner.
It is important to emphasise that prediction was individualised and a patient was only classified as likely to develop HCC once their personalised risk was higher than the threshold. This means that patients with more unclear status will, in general, only be identified closer to their diagnosis time. This may be considered desirable in order to not proceed with early interventions once there is sufficient confidence in a patient’s classification, hence avoiding unnecessary anxiety and potentially ineffective treatment.
The negative predictive value of our model confirms that as long as a patient’s risk stays below the chosen threshold, they have a low-risk of developing HCC. This information is potentially useful in determining when they should next visit a clinic for a blood sample to be taken. Since most HCC patients are identified a long time before actual diagnosis, those patients who are deemed low-risk at a current visit could be asked to return for samples less frequently. Further work would be required to assess the cost benefit of such an approach and also to determine whether a less frequent surveillance system missed true cases of HCC significantly more frequently than the current system.
A limitation of our model is that patients were classified as non-HCC only at their last recorded clinic visit. Some of these patients may eventually develop HCC under longer follow up and so some of our ‘false-positives’ may in fact be true HCC cases, but we have not observed them for long enough. This limitation means that our predictive accuracy measures are lower bounds, and that sensitivity may in fact be higher, if we had longer follow up.
A further limitation of our model is the fact that the prediction model takes no account of the fact that patients with hepatitis C can now be treated effectively. However, a subgroup analysis of prediction accuracy according to treatment response gave broadly similar results (See Supplementary material).
Although further work is needed to identify the extent to which these results can be applied in the West, our analysis suggests that a model just based on AFP performs well in the Scottish dataset.
Supplementary Material
Need to Know.
Background
Addition of measurement of serum level of alpha-fetoprotein (AFP) to ultrasound analysis can increase detection of HCC.
Findings
The authors developed and validated model to identify patients with chronic liver disease who are at risk for HCC based in change in serum level of AFP over time; it accurately identified 73.1% of patients who did or did not develop HCC.
Implications for patient care
The model could be used to assign patients to groups with high vs low risk for HCC and might be used to select patients for surveillance.
Acknowledgments
We are grateful to Graeme Alexander and Amit Singal for comments on earlier versions of this manuscript. We are also grateful to Kevin Beresford, Keith Kennedy and Robert Sherman for assistance with the technical aspects of developing the online calculator.
Grant Support
DMH and MGF acknowledge the support of the Medical Research Council (Research project MR/L010909/1). DMH was supported by a UKRI Innovation Fellowship, funded by the MRC (Research Project MR/R024847/1). MGF, SB and RK acknowledge the support of the UK EPSRC grant EP/N014499/1. TB is supported by the Wellcome Trust (WT107492/Z/15/Z) and a Scottish Government CSO grant (CGA/17/19).
Abbreviations
- AFP
alpha-fetoprotein
- DCP
des-gamma-carboxy prothrombin
- AFP-L3
lectin-reactive alpha-fetoprotein.
- ALT
Alanine aminotransferase
Footnotes
Disclosures:
All authors have nothing to disclose
Author Contributions:
Study Concept and Design: DMH, MGF, PJ.
Acquisition of data: HT, TT, TKu, SS, NN, MK, TKi, TB.
Analysis and Interpretation of the data: DMH, SB, CED, RSA, RKD MGF, PJ
Drafting of Manuscript: DMH, PJ
Critical Revision of the manuscript for intellectual content: All authors
Statistical Analysis: DMH, MGF
Study Supervision: MGF, PJ
References
- El-Serag HB. Hepatocellular Carcinoma. New England Journal of Medicine. 2011;365:1118–1127. doi: 10.1056/NEJMra1001683. [DOI] [PubMed] [Google Scholar]
- Kansagara D, Papak J, Pasha AS, et al. Screening for hepatocellular carcinoma in chronic liver disease: a systematic review. Annals of internal medicine. 2014;161:261–269. doi: 10.7326/M14-0558. [DOI] [PubMed] [Google Scholar]
- Liver EAFTSOT. EASL-EORTC clinical practice guidelines: management of hepatocellular carcinoma. Journal of hepatology. 2012;56:908–943. doi: 10.1016/j.jhep.2011.12.001. [DOI] [PubMed] [Google Scholar]
- Tzartzeva K, Obi J, Rich NE, et al. Surveillance imaging and alpha fetoprotein for early detection of hepatocellular carcinoma in patients with cirrhosis: a meta-analysis. Gastroenterology. 2018;154:1706–1718.:e1. doi: 10.1053/j.gastro.2018.01.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson P, Berhane S, Kagebayashi C, et al. Impact of disease stage and aetiology on survival in hepatocellular carcinoma: implications for surveillance. British journal of cancer. 2017;116:441. doi: 10.1038/bjc.2016.422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudo M. Japan’s Successful Model of Nationwide Hepatocellular Carcinoma Surveillance Highlighting the Urgent Need for Global Surveillance. Liver Cancer. 2012;1:141–3. doi: 10.1159/000342749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singal AG, El-Serag HB. Hepatocellular carcinoma from epidemiology to prevention: translating knowledge into practice. Clinical gastroenterology and hepatology. 2015;13:2140–2151. doi: 10.1016/j.cgh.2015.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes DM, Komárek A, Czanner G, et al. Dynamic longitudinal discriminant analysis using multiple longitudinal markers of different types. Statistical methods in medical research. 2018;27:2060–2080. doi: 10.1177/0962280216674496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komárek A, Hansen BE, Kuiper EM, et al. Discriminant analysis using a multivariate linear mixed model with a normal mixture in the random effects distribution. Statistics in Medicine. 2010;29:3267–3283. doi: 10.1002/sim.3849. [DOI] [PubMed] [Google Scholar]
- Tayob N, Lok AS, Do K-A, et al. Improved detection of hepatocellular carcinoma by using a longitudinal alphafetoprotein screening algorithm. Clinical Gastroenterology and Hepatology. 2016;14:469–475.:e2. doi: 10.1016/j.cgh.2015.07.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tayob N, Stingo F, Do KA, et al. A Bayesian screening approach for hepatocellular carcinoma using multiple longitudinal biomarkers. Biometrics. 2018;74:249–259. doi: 10.1111/biom.12717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galle PR, Forner A, Llovet JM, et al. EASL clinical practice guidelines: management of hepatocellular carcinoma. Journal of hepatology. 2018;69:182–236. doi: 10.1016/j.jhep.2018.03.019. [DOI] [PubMed] [Google Scholar]
- Bird TG, Dimitropoulou P, Turner RM, et al. Alpha-fetoprotein detection of hepatocellular carcinoma leads to a standardized analysis of dynamic AFP to improve screening based detection. PLoS One. 2016;11:e0156801. doi: 10.1371/journal.pone.0156801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omata M, Cheng A-L, Kokudo N, et al. Asia-Pacific clinical practice guidelines on the management of hepatocellular carcinoma: a 2017 update. Hepatology international. 2017;11:317–370. doi: 10.1007/s12072-017-9799-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes DM, El Saeiti R, García-Fiñana M. A comparison of group prediction approaches in longitudinal discriminant analysis. Biometrical Journal. 2018;60:307–322. doi: 10.1002/bimj.201700013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komárek A, Komárková L. Capabilities of R package mixAK for clustering based on multivariate continuous and discrete longitudinal data. Journal of Statistical Software. 2014;59:1–38. [Google Scholar]
- Berhane S, Toyoda H, Tada T, et al. Role of the GALAD and BALAD-2 serologic models in diagnosis of hepatocellular carcinoma and prediction of survival in patients. Clinical Gastroenterology and Hepatology. 2016;14:875–886.:e6. doi: 10.1016/j.cgh.2015.12.042. [DOI] [PubMed] [Google Scholar]
- Johnson PJ, Pirrie SJ, Cox TF, et al. The detection of hepatocellular carcinoma using a prospectively developed and validated model based on serological biomarkers. Cancer Epidemiology and Prevention Biomarkers. 2014;23:144–153. doi: 10.1158/1055-9965.EPI-13-0870. [DOI] [PubMed] [Google Scholar]
- Yang JD, Addissie BD, Mara KC, et al. GALAD Score for Hepatocellular Carcinoma Detection in Comparison with Liver Ultrasound and Proposal of GALADUS Score. Cancer Epidemiology and Prevention Biomarkers. 2019;28:531–538. doi: 10.1158/1055-9965.EPI-18-0281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Serag HB, Kanwal F, Davila JA, et al. A new laboratory-based algorithm to predict development of hepatocellular carcinoma in patients with hepatitis C and cirrhosis. Gastroenterology. 2014;146:1249–1255.:e1. doi: 10.1053/j.gastro.2014.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.