Summary
The INK4a/ARF locus encodes important cell-cycle regulators p14ARF, p15INK4b, and p16INK4a. The neighboring gene desert to this locus is the most reproducible GWAS hotspot that harbors one of the densest enhancer clusters in the genome. However, how multiple enhancers that overlap with GWAS variants regulate the INK4a/ARF locus is unknown, which is an important step in linking genetic variation with associated diseases. Here, we show that INK4a/ARF promoters interact with a subset of enhancers in the cluster, independent of their H3K27ac and eRNA levels. Interacting enhancers transcriptionally control each other and INK4a/ ARF promoters over long distances as an interdependent single unit. The deletion of even a single interacting enhancer results in an unexpected collapse of the entire enhancer cluster and leads to EZH2 enrichment on promoters in an ANRIL-independent manner. Dysregulated genes genome-wide mimic 9p21-associated diseases under these scenarios. Our results highlight intricate dependencies of promoter-interacting enhancers on each other.
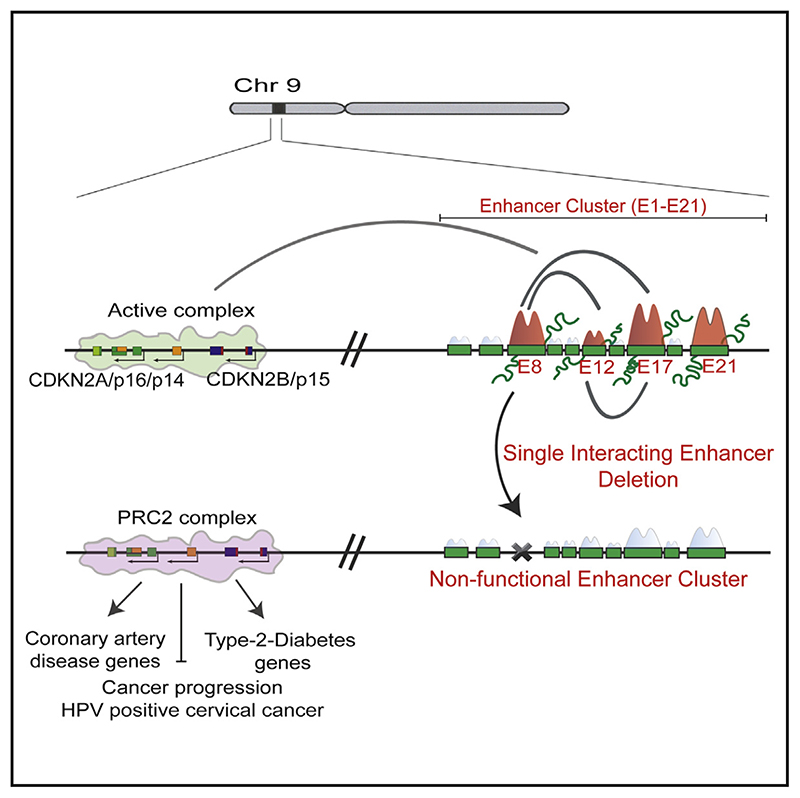
Graphical abstract.
Introduction
The INK4a/ARF locus codes for critical cell-cycle regulators, namely, p14ARF, p15INK4b, and p16INK4a, from two coding genes: CDKN2A (p14ARF and p16INK4a) and CDKN2B (p15INK4b). The locus also harbors a long noncoding RNA, CDKN2BAS (ANRIL), at its 3’ end (Figure 1A; Aguilo et al., 2011; Kim and Sharpless, 2006; Sherr, 2012). Together these proteins inhibit cyclin-dependent kinases (CDKs) to regulate the cell cycle (Baker et al., 2016; Sherr, 2012; Zhang et al., 1998). Due to these inhibitory roles, the locus is either methylated or deleted in 90% of the tumors (Gonzalez and Serrano, 2006; Yap et al., 2010), except for a few types of cancers such as breast, prostate (Drak Alsibai et al., 2019; Pare et al., 2016), non-small cell lung (Drak Alsibai et al., 2019), and human papillomavirus (HPV) positive cancers such as head/neck, and cervical cancers (Kanao et al., 2004; Vazquez-Vega et al., 2013). Notably, ~90% of the cervical tumors are HPV positive.
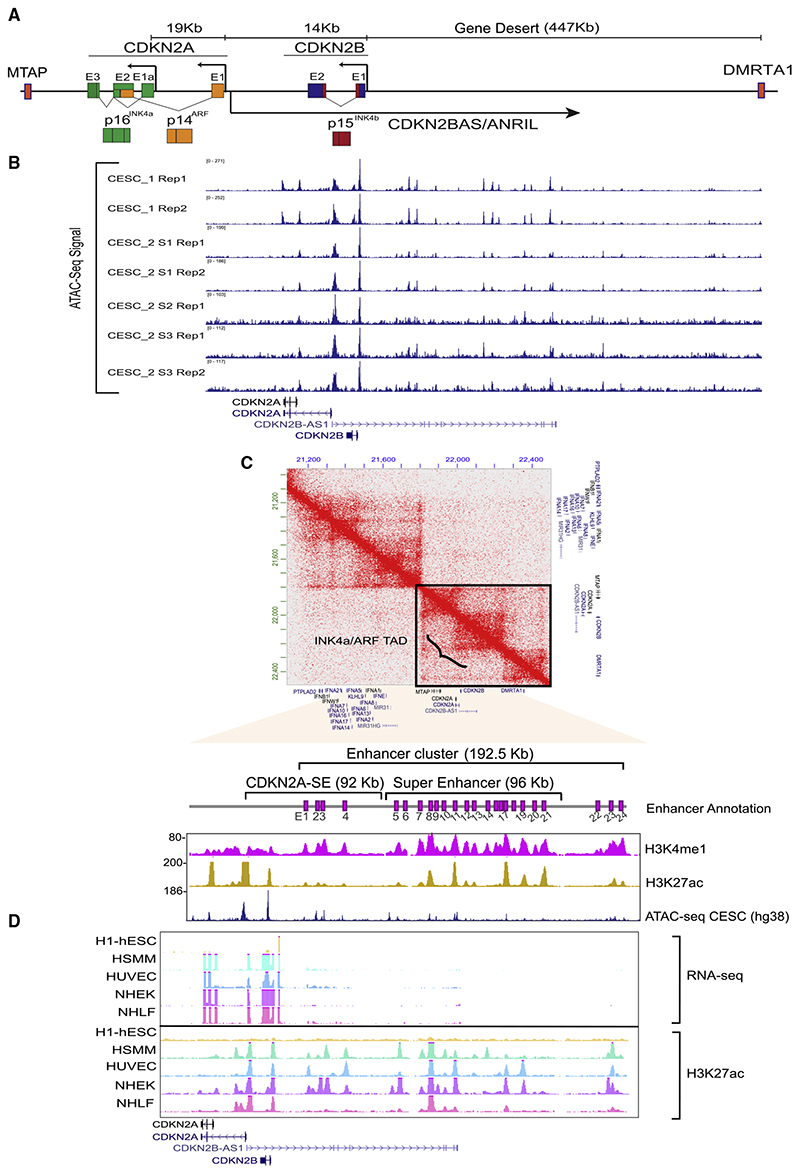
Figure 1. Expression of INK4a/ARF genes is correlated with the presence of an enhancer cluster in the neighboring gene-desert region.
(A) Schematic representing the INK4a/ARF locus. CDKN2A and 2B code for p16INK4a, p14ARF, and p15INK4b.
(B) IGV browser snapshot showing the ATAC-seq signal on INK4a/ARF promoters and the neighboring gene-desert region in cervical cancer tumors from two patients in the TCGA cohort.
(C) Hi-C heatmap depicting the TAD structure at the INK4a/ARF locus in HeLa cell line. The zoom-in region shows the INK4a/ARF promoters and enhancer cluster depicted by pink boxes (E1–E24). These are overlaid by H3K4me1 and H3K27ac (hg19) tracks in HeLa and ATAC-seq (hg38) from cervical tumor.
(D) UCSC genome browser snapshots showing the RNA-seq signal on the locus across different cells. The RNA-seq is overlaid by H3K27ac ChIP-seq.
Apart from cancers, the activation of INK4a/ARF genes is the hallmark of senescence and aging (Baker et al., 2016; Carrasco-Garcia et al., 2015; Jeck et al., 2012; Matheu et al., 2009). Hence, this locus is the most reproducible genome-wide association study(GWAS) hotspot associatedwith various age and lifestyle-related diseases, such as coronary artery disease, type 2 diabetes, Alzheimer disease, and atherosclerosis,among others (Gonzá lez-Navarro et al., 2013; Harismendy et al., 2011; Jeck et al., 2012; Helgadottir et al., 2008; Samani et al., 2007). Despite the tremendous importance of this locus in disease pathologies, its transcriptional regulation in associated cancers and senescence is poorly understood.
Most studiescarriedouton INK4a/ARF transcriptional regulation have focused on promoter-driven mechanisms (Aloia et al., 2015; Hirosue et al., 2012; Lazorthes et al., 2015; Zhang et al., 2019). However, disease-associated variants identified in GWAS studies lie in the gene-desert region adjacent to CDKN2A/2B genes. This hints toward a plausible regulation of the locus through the gene desert. The deletion of the gene-desert region in mouse and induced pluripotent stem cells (iPSCs), and their subsequent differentiation into relevant cell type has shown genome-wide alterations in coding genes associated with coronary artery disease and atherosclerosis (Kojima et al., 2020; Lo Sardo et al., 2018; Visel et al., 2010) suggesting that the gene desert regulates the INK4a/ARF locus. However, functional roles of the regulatory elements in these large, deleted regions remain unknown. We have previously shown that the INK4a/ARF gene desert harbors several enhancers with unexplored potential of regulating this locus (Harismendy et al., 2011). How these multiple enhancers that overlap with GWAS variants transcriptionally regulate this multigene locus is unknown, an important step toward understanding the biological relevance of these variants. Importantly, understanding the regulatory configurations of the individual enhancers within this enhancer cluster (super-enhancer), such as co-operative, hierarchical, mutual, and redundant/non-redundant is crucial in linking the disease-associated variants in this region with pathologies. The knowledge gained will form the basis of designing therapies to modulate the expression of INK4a/ARF genes in aging-related pathologies and cancers.
Here, we show that this gene-desert region is one of the densest enhancer clusters in the genome with 24 constituent enhancers (E1–E24). Super Enhancer (SE) calling identifies a super-enhancer using long-distance cutoffs. The presence of this enhancer cluster in the gene-desert region is tightly correlated with transcriptional activation of the INK4a/ARF locus in various cell types including HPV positive cervical tumors and senescent cells. Using a chromosome conformation capture technique (4C), we observed that the promoter of CDKN2A physically interacts with five enhancers (E5, E8, E12, E17, and E19) that were spread far apart within the cluster. These enhancers exhibited varying levels (very low to the highest) of functional enhancer marks such as H3K27ac, PolII, p300, eRNA, and STARR sequencing (STARR-seq) signal and were comparable to some of the non-interacting enhancers. Importantly, individual deletion of any of the three interacting enhancers (E8, E12, and E17) silenced the transcription of INK4a, ARF, and INK4b promoters, whereas deletion of E21 that is a non-interacting enhancer mildly affected INK4b and ARF expression but not INK4a. Unexpectedly, upon perturbation of even a single promoter-interacting enhancer, we observed loss of H3K27ac, eRNA expression on intact enhancers, and their interaction with promoters and other enhancers. We also observed the loading of the PRC2 complex subunit EZH2 and a subsequent increase in H3K27me3 levels on promoters upon enhancer perturbations. The deletion of any single enhancer severely hampers proliferation, clonogenicity, and migration of HPV-positive cells. Notably, global gene-expression profiling upon enhancer deletions exhibits GO terms that are related to the 9p21 disease association such as coronary artery disease and type 2 diabetes. Our results identify an enhancer network constituted by a subset of enhancers that is dependent on each enhancer for the target gene regulation.
Results
Expression of INK4a/ARF genes is correlated with the presence of an enhancer cluster in the neighboring gene-desert region
The INK4a/ARF locus codes for three cell-cycle regulators from CDKN2A and CDKN2B genes, where CDKN2A alone harbors two promoters, one for INK4a and the other for ARF (Figure 1A). The adjacent gene-desert region is a known GWAS hotspot (Figure 1A). To understand the relevance of the gene desert in INK4a/ARF regulation, we began with analyzing the relative expression of CDKN2A/2B in tumors of different origins. Cervical tumors displayed the highest levels of CDKN2A and CDKN2B (Figures S1A and S1B). Indeed, most of the cervical tumors (~90%) are HPV positive and exhibit elevated levels of p16INK4a (Kanao etal.,2004;Vazquez-Vega etal.,2013).Inorder to understand why the expression of CDKN2A and CDKN2B is high in these cervical tumors, we interrogated the open chromatin signature by ATAC sequencing (ATAC-seq) as a proxy for active cis-regulatory elements. The promoters (INK4a, ARF, and INK4b) and the neighboring gene-desert region that harbors disease-associated variants identified in several GWAS (Figure1A) showed highly open chromatin features suggesting these elements are potential regulatory elements (Figure 1B). To explore these regulatory elements and their mechanisms, we chose HPV positive cervical tumor cells HeLa, as a model that has high levels of p16INK4a. Towards that, we first interrogated the topologically associating domain (TAD) structure at this locus using Hi-C data (Rao et al., 2014). All three INK4a/ARF genes, CDKN2BAS, and neighboring gene MTAP were present within the same TAD (Figure 1C). CDKN2A/2B promoters were well within the TAD, whereas the promoter of MTAP was located at the 5′ boundary of the TAD, and the 3′ boundary was located ~269 kb downstream of CDKN2A promoter (Figure 1C). We have previously shown the presence of several H3K4me1-marked enhancers in this gene-desert region (Harismendy et al., 2011). We observed that these enhancers were within the same TAD and overlapped with the open chromatin features of cervical tumors, pointing toward conserved transcriptional regulation of this locus in HeLa and cervical tumors (Figure 1B).
Enhancers that respond to signaling and development cues are present in active, primed, or poised states (Creyghton etal., 2010; Ostuni etal.,2013, Raisner etal.,2018).In this regard, we noticed that this enhancer cluster in HeLa consists of 24 H3K4me1-marked enhancers (E1–E24) (Figure 1C, magenta annotation). Out of these, 15 enhancers were co-marked by H3K27ac and H3K4me1 (active enhancers), and the remaining enhancers were primed (only H3K4me1). Next, to test the association of these enhancers and INK4a/ARF expression, we chose stem cells and various differentiated primary cell types since the regenerative potential of stem cells is attributed to the silencing of the INK4a/ARF locus. In contrast, the locus is active in differentiated cells (Li et al., 2009). Corroborating with these facts, we observed that, in hESCs, INK4a, ARF, and INK4b did not express and interestingly the H3K27ac mark was absent on promoters and the neighboring gene-desert region (Figure 1D). As opposed to stem cells, we noticed the expression of these genes in several cell types spanning different lineages (Figure 1D). Besides transcription, we also observed several H3K27ac-marked regions overlapping with the promoters and the gene-desert region in these cells. These data strongly link the presence of an enhancer cluster with the expression of the INK4a/ARF locus in cervical tumors, and other lineages.
Promoters of INK4a/ARF interact with multiple, but not all, enhancers in the cluster irrespective of their functional marks
Next, to dissect this enhancer cluster functionally, we first compared the number of H3K27ac-marked enhancers in INK4a/ ARF TAD with other TADs in the genome. INK4a/ARF TAD was in the top 30 densest TADs out of 2,740 TADs in terms of the number of H3K27ac-marked enhancers per TAD in HeLa cells (Figure 2A). Clusters of enhancers grouped within a certain distance and with the presence of Med1 or H3K27ac above a threshold are defined as super-enhancers (Hnisz et al., 2013; Whyte et al., 2013). To test whether these enhancers in the gene desert indeed form a super-enhancer, we analyzed H3K27ac chromatin immunoprecipitation sequencing (ChIP-seq) using the ROSE algorithm (Hnisz et al., 2013). Data suggested that, although the enhancer cluster in INK4a/ARF TAD is one of the 719 super-enhancers observed in HeLa cells, it is not one of the highest-ranking SE (Figure 2B). We noticed 17 (E5–E21) out of 24 enhancers in the cluster were called as SE using unusually long-distance cutoff of 20 kb, and only three enhancers were called as SE at a 12 kb distance as the enhancers - were well spread out in the gene desert. However, the remaining enhancers could still be functional; thus, we disregarded the SE calling and focused on all 24 enhancers present in this locus to assess their regulatory potential in an unbiased manner.
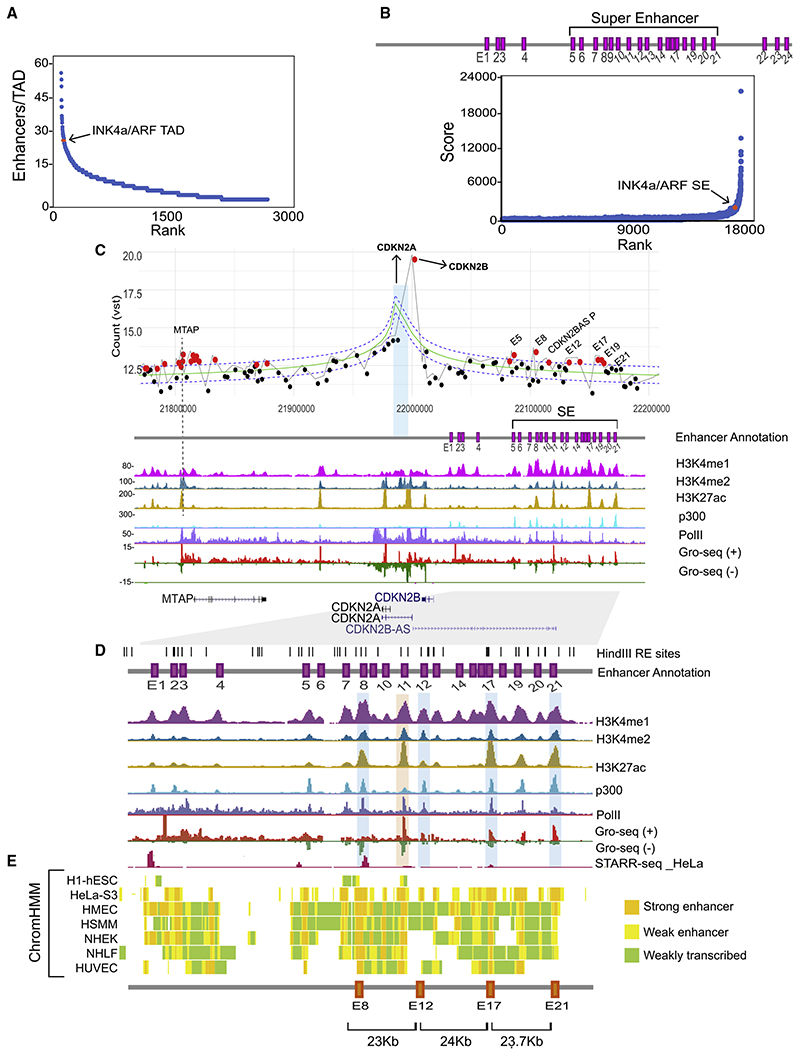
Figure 2. Promoters of INK4a/ARF interact with multiple, but not all, enhancers in the cluster irrespective of their functional marks.
(A) Ranking of TADs based on number of H3K27ac peaks.
(B) Plot showing INK4a/ARF enhancer cluster contains a super-enhancer overlaid by enhancer annotations (purple boxes E1–E24) with the super-enhancer marked (E5–E21).
(C) 4C plot showing the interactions from CDKN2A viewpoint; red dots represent the significant interactions. The 4C plot is overlaid with ChIP-seq and GRO-seq tracks in HeLa.
(D) The UCSC browser shot shows the zoomed region from E1–E21. Tracks are overlaid with STARR-seq track in HeLa..
(E) ChromHMM profiles at the region from E1–E21 across different cell types and distances among E8, E12, E17, and E21 are shown.
Since active enhancers regulate their target promoters via looping (Li et al., 2013), we performed 4C experiments at CDKN2A promoter viewpoint in HeLa. The analyzed contact maps exhibited several high-frequency interactions in both replicates. Interestingly, all interactions originating from CDKN2A promoter were directed either toward the MTAP promoter and its gene body (5′ of TAD) or with several enhancers within the enhancer cluster in the gene-desert region. E22–E24 were within the TAD boundary, but, apart from these, the remaining enhancers within clusters that showed high-frequency interactions were E5, E8, E12, E17, and E19 (Figures 2C and S1C). The sixth region that exhibited looping with CDKN2A promoter was the terminator of CDKN2BAS (ANRIL); thus, we did not regard it as an enhancer (Figure 2C). Interacting enhancers were marked with varying levels of H3K4me1, H3K4me2, H3K27ac, p300, PolII, and STARR-seq signal (Arnold et al., 2013; Muerdter et al., 2018; Figures 2C and 2D, browser shot of ChIP-seq). The presence of PolII prompted us to check the eRNA-expression signature on these enhancers as functional enhancers transcribe eRNAs by virtue of PolII loading (Kim et al., 2010; Li et al., 2013; Mikhaylichenko et al., 2018; Wu et al., 2014). We found that all PolII-enriched enhancers transcribed bidirectional eRNAs, but their expression varied (Figures 2C and 2D, eRNA track). Apart from these enhancers and MTAP, a strong interaction of CDKN2A promoter with CDKN2B promoter was also observed. These data suggest that a multi-promoter:enhancer network exists within the TAD and not all enhancers are part of this network. Further, interacting enhancers exhibit varying levels (very low to the highest levels) of functional enhancer marks, but these marks were not different from non-interacting enhancers (E21) within the cluster (Figure 2D).
We noticed that E8 and E17 consistently featured as active enhancers in other cell types as well, as revealed by their ChromHMM profiles (Figure 2E). These cell types included HSMM and HUVEC that are relevant cell types to coronary artery disease and atherosclerosis (Figure 2E). p14ARF, p15INK4b, and p16INK4a proteins are the key drivers of senescence and stem cell regeneration (Carrasco-Garcia et al., 2015; Li et al., 2009; Sherr, 2012. We observed that the locus is already transcriptionally active inthe young IMR90 fibroblast; however, as it gets highly upregulated upon replicative senescence, an accompanying gain of H3K27ac occurred on the super-enhancer in the cluster (Figure S1D). Importantly, the eRNA expression was robustly induced on E12, E17, E19, and E21 enhancers upon onset of senescence (Figure S1D). All of this evidence strongly suggests that these interacting (E5, E8, E12, E17, and E19) and non-interacting (E21) enhancers have functional potential. Interestingly, all these enhancers were also part of the super-enhancer called using the ROSE algorithm but using unusually long-distance cutoff (Figure 2B).
Each promoter-interacting enhancer fully regulates INK4a/ARF genes
To test whether these enhancers were functional and also whether the levels of these marks correlated with their relative enhancer activity, we blocked three promoter-interacting enhancers, namely, E8, E12, and E17, that exhibited low, medium, and high levels of functional marks and one non-interacting enhancer, E21 that had high levels of these features. These four enhancers had the following distribution of functional features: E8 (interacting enhancer with high H3K27ac, PolII, eRNA, and a very high STARR-seq signal; this enhancer was active in all cell types studied using ChromHMM [Figures 2D and 2E]), E17 (interacting enhancer with highest H3K27ac, moderate PolII, eRNA expression, and STARR-seq signal; similar to E8 this enhancer was also present in all cell types tested using ChromHMM and was robustly induced upon senescence [Figure S1D]), E12 (interacting enhancer with very low H3K27ac, moderate PolII, low eRNA levels, and no STARR-seq signal that was active in HeLa and HSMM but weakly active in HUVEC and HMEC [Figures 2D and 2E]), and also E21 (non-interacting enhancer but with very high levels of H3K27ac, PolII, eRNA and no STARR-seq signal) (Figure 2D). We did not block E5 and E19 enhancers as they exhibited eRNA levels comparable to E8 and E21, respectively (Figure 2D).
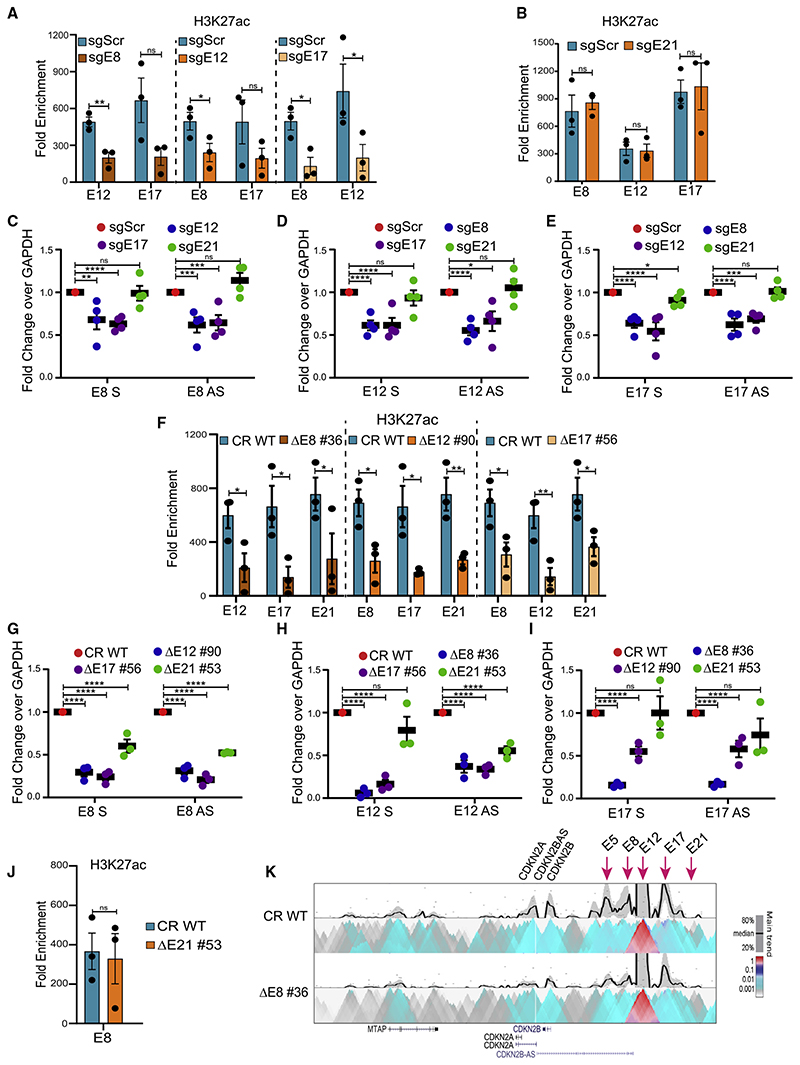
We systematically targeted these four enhancers individually using CRISPRi that employs dCas9-KRAB and specific gRNAs (Figure S2A; Table S2) to mediate repression on targeted regions (Gilbert et al., 2014; Larson et al., 2013). We first confirmed the blocking by observing the gain of H3K9me3 on targeted enhancer using dCas9-KRAB but not by dCas9 alone suggesting the specificity of CRISPRi(Figures S2B and S2C).Further,we observed the loss of H3K27ac (Figures S3A–S3D) and eRNAs (Figures S3E–S3H) on the targeted enhancers. After confirming the blocking of enhancers, we assessed the transcriptional status of INK4a/ARF genes. INK4a, ARF, and INK4b mRNA expression was significantly downregulated upon blocking any of the interacting enhancer (E8, E12, or E17) individually. However, CRISPRi on E21 did not manifest any such effects (Figures 3A–3C). These results point toward the essentiality of E8, E12, and E17 in transcriptional regulation of this locus. Notably, all three enhancers except E21 were promoter-interacting enhancers. Further, to confirm whether the downregulation of gene expression is due to the transcriptional perturbations at the promoter, we extended our analysis to local histone acetylation (H3K27ac) and PolII as a surrogate of promoter function as the levels of H3K27ac are very high on the promoters of INK4a/ARF (Figure 2), and high acetylation on promoters is correlated with the promoter strength (Nicolas et al., 2018; Raisner et al., 2018). H3K27ac and PolII levels were significantly lost on the promoters of INK4a, ARF,andINK4b upon blocking of any interacting enhancer (Figures 3D–3F) but not upon blocking of non-interacting enhancer E21 (Figures 3G–3I). These results corroborate with the downregulation of target genes.
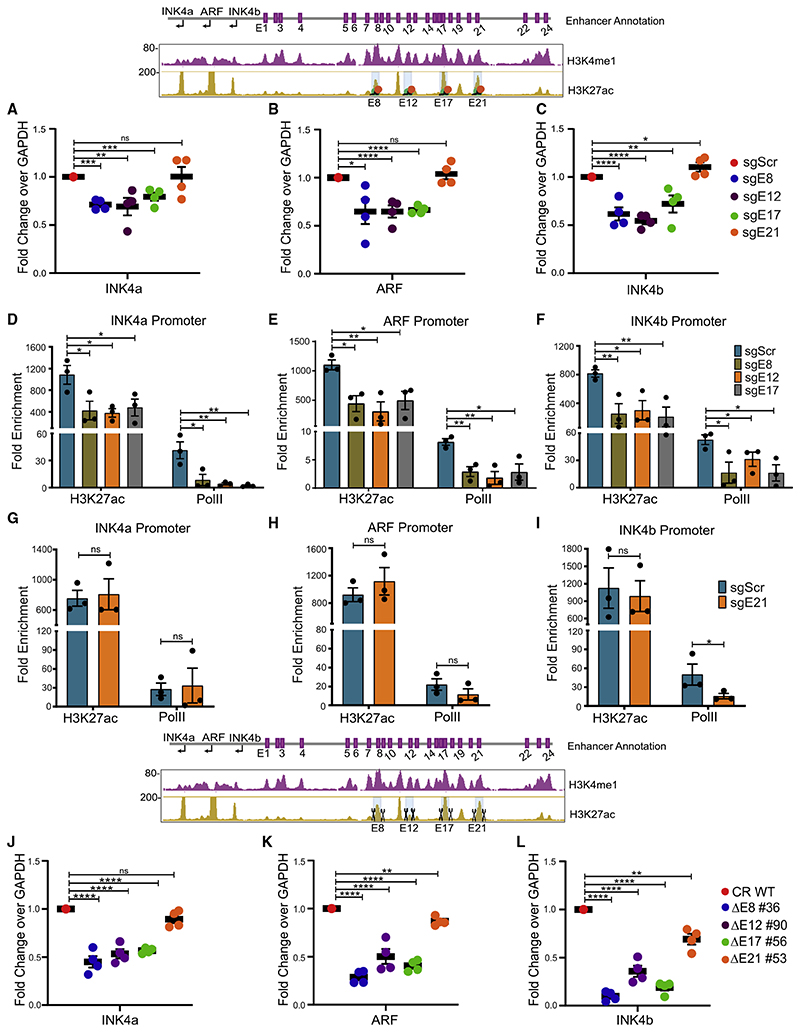
Figure 3. Each interacting enhancer is essential and fully regulates INK4a/ARF genes.
(A–C) mRNA levels of (A) INK4a, (B) ARF, and (C) INK4b genes upon CRISPRi on E8, E12, E17, and E21 (n = 4). Plots are overlaid with a schematic of the INK4a/ ARF locus marked with enhancer annotation, H3K4me1, and H3K27ac tracks, and highlighted enhancers were blocked (orange and green solid circles).
(D–I) H3K27ac and PolII enrichment on promoters (D) INK4a, (E) ARF, and (F) INK4b upon CRISPRi on E8, E12, and E17 individually or E21 (G–I) (n = 3).
(J–L) mRNA levels of (J) INK4a, (K) ARF, and (L) INK4b genes upon deletion of E8, E12, E17, and E21 enhancers (n = 4). Plots are overlaid with histone tracks, and highlighted enhancers were deleted. Statistical significance is determined by unpaired t test (*p < 0.05, **p < 0.01, ***p < 0.005, ****p < 0.001; ns p > 0.05), and error bars denote SEM.
To understand the co-dependence among enhancers and their hierarchy, we created individual enhancer deletions in HeLa using active Cas9 protein and specific gRNAs that target the flanks of the enhancer region (Figure S4A; Table S2). We generated four unique lines carrying a homozygous deletion of individual enhancers, namely, DE8 #36, DE12 #90, DE17 #56, and E21 #53 (Figures S4B–S4I). The expression of INK4a, ARF, and INK4b genes was robustly downregulated upon the deletion of any interacting enhancer individually (Figures 3J–3L). Interestingly, unlike CRISPRi, E21 deletion caused marginal downregulation of INK4b and ARF, whereas the expression of INK4a was unaffected. Further, akin to homozygous deletions, even the heterozygous lines for interacting enhancers exhibited similar effects (Figures S4J–S4L). Intriguingly, akin to CRISPRi, the heterozygous deletion of E21 failed to exert any effects on INK4a/ARF gene regulation (Figure S4M). This suggests that even small perturbations in interacting enhancers cause the severe downregulation of INK4a/ARF genes, but these perturbations in non-interacting enhancers are well tolerated as long as active enhancers are functional to activate the target genes. Notably, deletion of each interacting enhancer displayed an extent of effects on promoters that was the sum of all enhancers. Furthermore, INK4b exhibited almost complete silencing in the deleted lines, suggesting the complete dependence of INK4b for its expression on individual interacting enhancers. To test whether activation of these three enhancers individually upregulates the INK4a expression, we performed CRISPRa in HaCaT cells that are derived from human keratinocytes (epithelial origin similar to HeLa cells). INK4a exhibited significant upregulation upon CRISPRa on E8, E12, or E17 (Figure S3I). Together, these data confirm that the INK4a/ ARF locus is under control of at least these interacting enhancers in one of the densest enhancer clusters and, perturbation of even a single interacting enhancer results in loss of INK4a/ARF promoter activity at a similar extent even though the remaining 23 enhancers in the cluster remain intact. Further, based on these data on interacting enhancers, we believe that other interacting enhancers, E5 and E19, could also be functional.
An interdependent enhancer network operates within the enhancer cluster
The data above suggest that the strength of INK4a/ARF promoters was significantly reduced upon deletion or blocking of even a single interacting enhancer. Even though other interacting enhancers in the cluster were unaltered, they could not rescue the transcription of promoters (Figures 3J–3L). This prompted us to consider whether silencing or deletion of individual enhancer renders the remaining intact enhancers non-functional. Thus, to interrogate whether the interacting enhancers were functionally dependent on each other for their transcriptional activity, we assessed the levels of H3K27ac on interacting enhancers upon CRISPRi or deletions as a proxy of their active status (Nicolas et al., 2018; Raisner et al., 2018). Intriguingly, we observed a drop in H3K27ac enrichment on E12 and E17 upon blocking of E8. Similarly, enrichment of H3K27ac on E8 and E17 was reduced upon E12 blocking, and blocking of E17 resulted in the loss of H3K27ac on E8 and E12 (Figure 4A). However, blocking of E21 had no effect on H3K27ac levels on E8, E12, and E17 (Figure 4B). These data suggest that the intact functional enhancers within the enhancer cluster lose their activity upon blocking of any interacting enhancer.
Figure 4. An interdependent enhancer network operates within the enhancer cluster.
(A) H3K27ac enrichment on intact enhancers in cells carrying CRISPRi on E8, E12, and E17 (A) or E21 (B) (n = 3).
(C–E) Sense and antisense eRNA expression from (C) E8, (D) E12, and (E) E17 upon CRISPRi.
(F) H3K27ac enrichment on enhancers upon deletion of the individual enhancer (n = 3).
(G–I) Sense and antisense eRNA expression from (G) E8, (H) E12, and (I) E17 enhancers upon enhancer deletion (n = 3).
(J) H3K27ac enrichment on E8 upon E21 deletion (n = 3).
(K) 4C heatmap in WT showing the interactions from E12 viewpoint with other enhancers and INK4a/ARF promoters. These interactions are significantly reduced upon E8 deletion (n = 2).
Statistical significance is determined by unpaired t test (*p < 0.05, **p < 0.01, ***p < 0.005, ****p < 0.001; ns p > 0.05), and error bars denote SEM.
Further, we observed that both sense and antisense eRNA expression on unaltered enhancers was downregulated when any interacting enhancer was blocked, but blocking of E21 failed to exert these effects on other intact enhancers tested (Figures 4C–4E). A similar loss of H3K27ac on intact enhancers including E21 was observed upon deleting any interacting enhancer (Figure 4F). Furthermore, robust loss of eRNA levels on other intact interacting enhancers as well as on E21 were also observed (Figures 4G–4I). Significant downregulation of these eRNAs was also observed in another set of clones that were heterozygous for the deletions (Figures S5A–S5C). Interestingly, H3K27ac and eRNA expression remained unaffected on the E8, E12, and E17 enhancers upon blocking of E21, but its complete deletion exhibited effects on eRNA levels on E8 and E12 but not on E17 (Figures 4G–4I). Surprisingly, these effects on the E8 enhancer were not recapitulated by the concomitant loss of H3K27ac (Figure 4J). However, E21 showed loss of H3K27ac and eRNA in the absence of any of the promoter-interacting enhancers (Figures 4F, 4G–4I, and S5D), suggesting E21 is under the control of interacting enhancers in this locus.
Since single interacting enhancer deletions caused the eRNA and H3K27ac loss on all other intact enhancers, we asked whether these effects on intact enhancers are the result of enhancer:enhancer interactions. Toward this, we performed the 4C assay on the E12 enhancer as a viewpoint and observed its interactions with promoters of INK4a/ARF. E12 also exhibited physical proximity with E5, E8, and E17. These data validate that indeed these enhancers form a network with the promoters as also revealed by 4C at CDKN2A viewpoint (Figures 2C and 4K, top heatmap panel). Further, to test whether the downregulation of INK4a/ARF genes upon deletion of the interacting enhancer is due to the loss of the enhancer:promoter interaction and whether the enhancer:enhancer interaction is also altered, we extended 4C assays at the E12 enhancer upon E8 enhancer deletion. As expected, we found a weaker interaction between E12 and INK4a/ARF promoters. Interestingly, we also observed weaker interaction of E12 with E5 and E17 (Figure 4K, bottom heatmap panel). These data suggest that, upon single enhancer deletions, other interacting enhancers lose their physical proximity with the target promoters and with each other.
Though, we did not monitor the levels of H3K27ac and eRNA on all intact enhancers in the cluster, the fact that none of the intact enhancers could rescue the activity of promoters in deletion lines strongly indicates that all the intact enhancers lose their activity upon deletion of even a single functional enhancer.
Together, these data suggest that functional enhancers within a super-enhancer form a single functional unit with the target promoters where they regulate each other just the way they regulate the target promoters. Further, the non-interacting enhancers, even with comparable levels of H3K27ac, have partial effects on transcriptional activity, but they are regulated by interacting enhancers.
Loss of a single enhancer results in EZH2 enrichment at the INK4a/ARF promoters
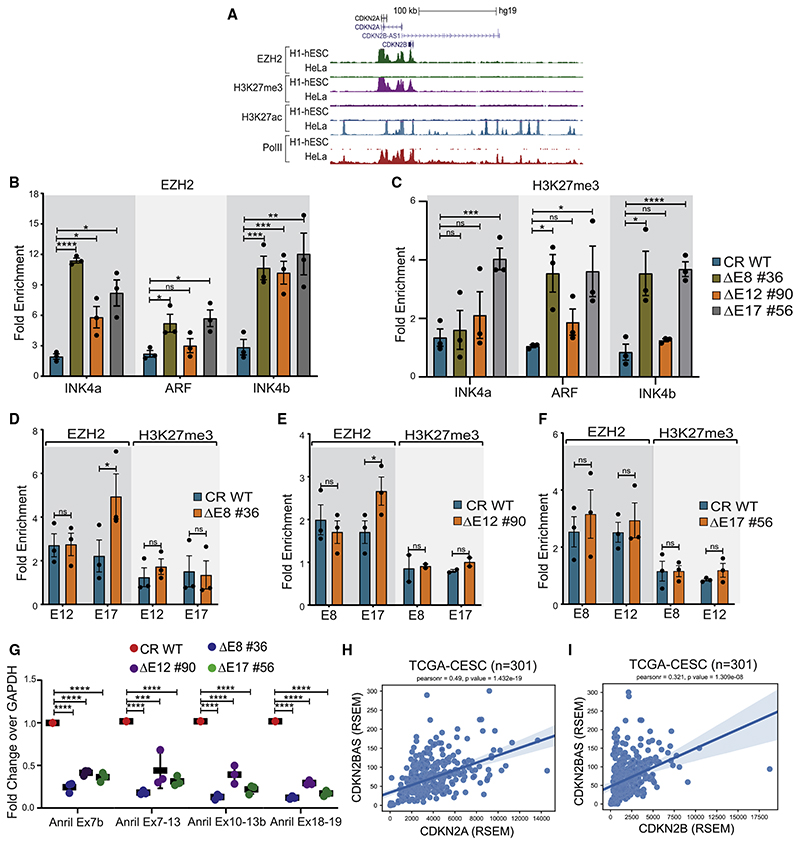
The INK4a/ARF locus is suppressed by loading of Polycomb on its promoters in stem cells and many tumors (Aguilo et al., 2011; Gamell et al., 2017; Li et al., 2009; Mosteiro et al., 2018; Yap et al., 2010; Zhang et al., 2019). Hence, we interrogated whether the promoter silencing upon enhancer deletions is due to the loading of EZH2 (polycomb complex subunit) on promoters. Toward this, we first compared the EZH2 enrichment on the INK4a/ARF locus in HeLa, where the locus is highly active, with hESCs, where the locus is completely silent (Figure 1D). The EZH2 ChIP-seq showed its expected loading on INK4a/ARF and INK4b promoters in stem cells and a complete absence in HeLa cells (Figure 5A). Importantly, the gene-desert region in stem cells did not exhibit EZH2 occupancy, suggesting the recruitment of EZH2 on promoters is independent of the gene-desert region in stem cells where the enhancer cluster is absent and transcriptional state of the INK4a/ARF locus is silent (Figures 1D, 2E, and 5A).
Figure 5. Loss of a single enhancer results in EZH2 enrichment at the INK4a/ARF promoters.
(A) UCSC genome browser snapshot at the INK4a/ARF locus shows ChIP-seq tracks for EZH2 and H3K27me3, H3K27ac, and PolII in hESCs and HeLa.
(B) EZH2 enrichment on INK4a, ARF, and INK4b promoters upon deletion of E8, E12, and E17 enhancers (n = 3).
(C) H3K27me3 enrichment on INK4a, ARF, and INK4b promoters upon deletion of E8, E12, and E17 enhancers (n = 3).
(D–F) EZH2 and H3K27me3 enrichment on (D) E12 and E17 upon E8 deletion (n = 3), (E) E8 and E17 upon E12 deletion (EZH2 n = 3 and H3K27me3 n = 2), and (F) E8 and E12 upon E17 deletion (n = 3).
(G) Expression of different isoforms of ANRIL upon deletion of E8, E12, and E17 enhancer (n = 3).
(H and I) Pearson correlation plots depicting positive correlation between (H) CDKN2A and CDKN2BAS, and (I) CDKN2B and CDKN2BAS in cervical cancer tumors from TCGA cohort; Pearson correlation value and p value, respectively, are shown at top of each plot. Statistical significance is determined by unpaired t test (*p < 0.05, **p < 0.01, ***p < 0.005, ****p < 0.001, ns p > 0.05), and error bars denote SEM.
After confirming the absence of EZH2 on promoters and the enhancer cluster in HeLa cells, we next investigated whether INK4a/ARF promoters gain EZH2 in the absence of enhancer activity. We focused on interacting enhancers as they exhibited effects on all INK4a/ARF genes. Interestingly, we observed a gain of EZH2 on INK4a, ARF, and INK4b promoters in the absence of E8, E12, or E17 enhancers individually (Figure 5B). Since the polycomb complex exerts its effects by methylating histone H3 at lysine 27, we found the higher levels of this mark on these promoters upon deletion of E8 and E17 enhancers (Figure 5C).
Surprisingly, except for the gain of EZH2 on E17, we did not notice EZH2 gain on other interacting enhancers upon deletion of any enhancer (Figures 5D–5F). Corroborating with the EZH2 levels, we did not observe a gain of H3K27me3 on these enhancers including the E17 enhancer (Figures 5D–5F). These results explain that enhancers do not deliver EZH2 to promoters after losing their activity, but one of the indirect mechanisms by which enhancers exert their function on the INK4a/ARF promoters is by protecting the promoters from EZH2 loading or from active suppression of the TAD.
Next, we tested whether the enrichment of EZH2 on promoters upon enhancer deletion is due to the higher expression of ANRIL, as ANRIL directly recruits polycomb on CDKN2A/2B promoters, and its elevated expression is associated with the silencing of the INK4a/ARF locus (Yap et al., 2010). Strikingly, we observed a loss of ANRIL expression in these lines suggesting that ANRIL itself is a transcriptional target of the enhancer cluster. Thus, EZH2 loading on INK4a/ARF promoters in the absence of enhancers was independent of ANRIL (Figure 5G; Figures S5E–S5G). A positive correlation between CDKN2A and CDKN2B with ANRIL is reported (Drak Alsibai et al., 2019; Lo Sardo et al., 2018) and was also observed in cervical cancer tumors (Figures 5H and 5I).
Perturbation in the enhancer network affects the cancerous properties of HeLa cells
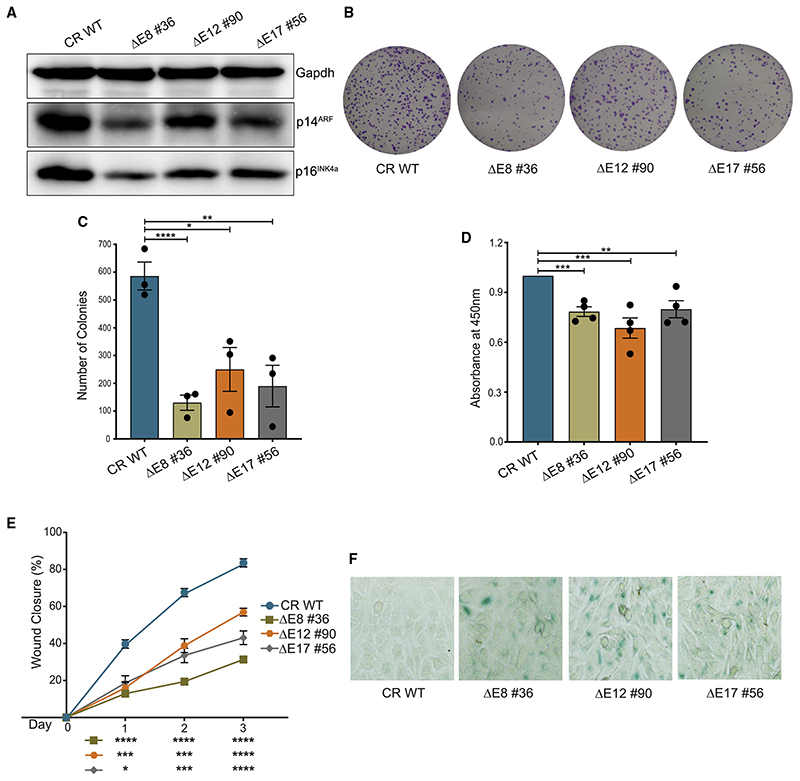
Small interfering RNA (siRNA)-mediated targeting of p16INK4a in HPV positive cancer cells is associated with tumor-growth inhibition (McLaughlin-Drubin et al., 2013; Pauck et al., 2014; Zhang et al., 2014, Kanao et al., 2004). Thus, we interrogated whether the hallmarks of cancer such as cell proliferation and colony-formation potential are affected in HeLa cells in the absence of these enhancers. We first validated the reduction in protein levels of p14ARF and p16INK4a in enhancer-deleted lines (Figure 6A). Not surprisingly, the enhancer deletion resulted in a severe loss of colonies (Figures 6B and 6C). Similarly, the proliferation rate (Figure 6D) and the cell migration were also affected in these cells (Figure 6E). Further, we noticed heightened senescence-associated β-galactosidase (SA-β-gal) staining upon deletion of these enhancers (Figure 6F). These observations suggest that the loss of enhancers in the INK4a/ARF locus alters the cancerous properties of HeLa and induces senescence.
Figure 6. Perturbation in the enhancer network affects the cancerous properties of HeLa.
(A) Immunoblots show the protein levels of p14ARF and p16INK4a upon deletion of E8, E12, and E17. Gapdh was used as a loading control (n = 2).
(B) Representative images of clonogenicity assay.
(C) Quantification of colonies using ImageJ upon deletion of E8, E12, and E17 enhancers (n = 3).
(D) Bromodeoxyuridine (BrdU) cell-proliferation assay in the absence of individual enhancers (n = 4).
(E) The percentage of wound closure in WT and upon deletion of E8, E12, and E17 (n = 2).
(F) Representative images of β-galactosidase activity in WT and enhancer deletions (n = 2). Statistical significance is determined by unpaired t test (*p < 0.05, **p < 0.01, ***p < 0.005, ****p < 0.001; ns p > 0.05), and error bars denote SEM.
Genes dysregulated upon enhancer deletions corroborate with disease association of 9p21 locus 9p21 locus is the most reproducible GWAS locus associated with type 2 diabetes and coronary artery disease across different races and ethnicities (Deloukas et al., 2013; González-Navarro et al., 2013; Harismendy et al., 2011; Helgadottir et al., 2008; Jeck et al., 2012; Nikpay et al., 2015; Samani et al., 2007). The GWAS variants associated with these diseases are located in the gene desert starting from CDKN2B promoter till the 3’ boundary region of the TAD (Figure 1A). However, the causal SNPs in this vast region are largely unknown.
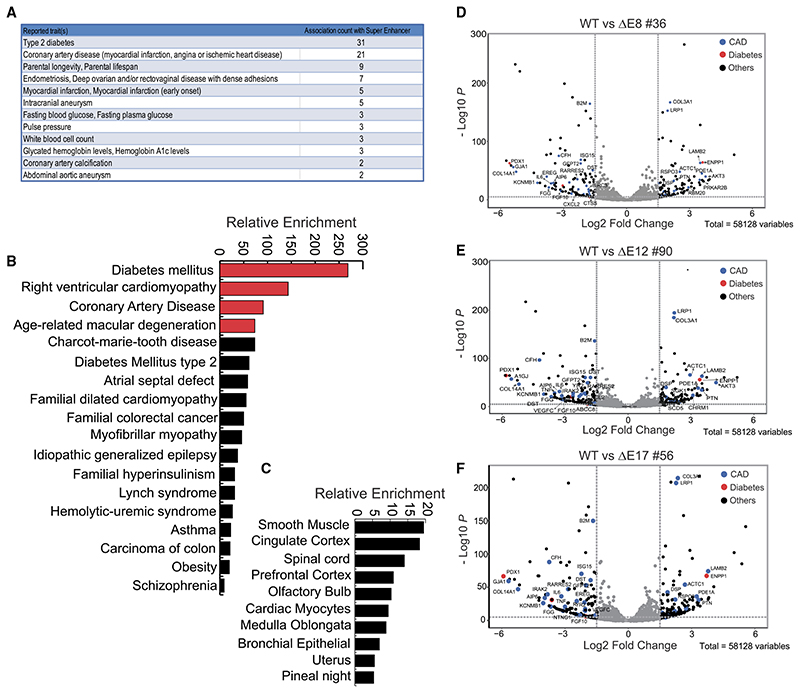
We hypothesized that, since the identified enhancer network is responsible for transcriptional regulation of this locus, its loss of function should mimic the transcriptional scenarios linked with several diseases with which this locus is associated. Though HeLa is not the relevant cell type, E8 and E17 enhancers were commonly active in cells of various lineages including endothelium and smooth muscle cells, as seen in Figure 2E and Figure S1D; thus, the regulation by these enhancers is likely similar in these tissues. Toward this, we first identified the disease association of an entire super-enhancer (96 kb) that spans from E5 to E21 using the GWAS catalog tool with default parameters (Welter et al., 2014). We noted the variants that are present in the SE are associated with type 2 diabetes, coronary artery disease, lifespan, endometrial cancer, etc. (Figure 7A). This suggests that the SE alone harbors the variants associated with diseases with which the entire gene desert between CDKN2B promoter to the 3’ boundary of TAD is associated (Figure 1A).
Figure 7. Dysregulated genes upon enhancer deletions corroborate with disease association of 9p21 locus.
(A) Chart from GWAS catalog tool shows the disease terms of GWAS variants and their counts present within the enhancer region (E8–E21).
(B) Graph from Enrichr tool shows the relative enrichment of disease terms based on dysregulated genes upon the E8, E12, and E17 enhancer deletions.
(C) Graph from Enrichr tool shows the relative enrichment of dysregulated genes, also expressed in various cell types.
(D–F) Volcano plots show the log2 fold changes versus –log10p values of genes comparing WT with (D) E8, (E) E12, and (F) E17 deletions.
Next, we tested whether genes dysregulated upon deletion of individual enhancers indeed corroborate with the diseases with which this super-enhancer is linked. We performed total RNA sequencing (RNA-seq) in two replicates in the wild-type (WT), E8, E12, and E17 deletions. The positional clustering and MA plots exhibited high correlation among various samples (Figures S7A–S7C). We noticed the downregulation of CDKN2A intronic region (transcriptional status) and exonic region (mature transcript), CDKN2B, CDKN2BAS (ANRIL), and MTAP (Figures S6A–S6D), whereas the transcriptional status of another large region on chr9 remained unaffected (Figure S6E).
Next, we identified approximately 200 differentially regulated genes comparing the WT and E8 deletion (Figure S7D). Not surprisingly, we found these genes being similarly affected in E12 and E17 as well (Figures S7C and S7D), suggesting the targets of these enhancers (directorindirect) arecommon. Approximately 100 genes were downregulated and other 100 were upregulated among dysregulated genes (Figure S7D). Interestingly, the enriched disease terms with these genes were diabetes mellitus, cardiomyopathy, coronary artery disease, and age-related macular degeneration (Figure 7B). Further, most dysregulated genes were expressed in smooth muscle cells and cortex (Figure 7C). These differentially regulated genes compared to WT cells revealed that some of the highly dysregulated genes are indeed associated with CAD and T2D (Figures 7D–7F), and they are well-known candidates with important functions. For example, PDX1 an important receptor for insulin, was significantly downregulated in all deletion lines. COL3A1, which is frequently altered or mutated in aortic and arterial aneurism, was one of the genes with the highest fold upregulation and lowest p value. Similarly, LRP1 (LDL receptor protein 1), an important member in clearance of apoptotic cells and lipid metabolism, was highly upregulated to the similar levels as COL3A1. Endothelial function in INK4a/ARF-deleted mice has been associated with dysregulated levels of interleukin-6 (IL-6) (Mosteiro et al., 2018, Yang et al., 2012). Similarly, IL-6 and IL-8 have been shown tobe regulated by ANRIL (Zhou et al., 2016). In corroboration with this, we observed robust downregulation of IL-6 and IL-8 in our RNA-seq data. These data suggest that the perturbed enhancers are indeed functional enhancers; hence, the genetic variation in them is potentially causal.
Discussion
Using a series of enhancer blockings and deletions, we observed that the INK4a/ARF locus is under the regulation of a few enhancers within a dense enhancer cluster. These enhancers act on the promoters as an interdependent network that is non-redundant for transcriptional activity. Loss of even a single promoter-interacting enhancer in the network results in inactivation of the entire enhancer cluster, EZH2 loading on INK4a/ARF promoters, and their downregulation. Further, global gene-expression profiles upon these deletions display effects that are relevant to the diseases with which the entire 9p21 locus is associated.
CDKN2A promoter interacts with only a subset of enhancers in the cluster
The gene desert harbors 24 enhancers; three enhancers are present within the TAD boundary, and, out of the remaining 21 enhancers, only five enhancers loop with CDKN2A promoter. These enhancers were within a SE that constitutes 17 enhancers (Figure 2B). These data suggest that SE can be used to call the functional regions; however, only a subset of enhancers even within SE loop with promoters and are functional. The levels of H3K27ac alone did not predict the looping potential of enhancers as the interacting enhancer E12 had the lowest H3K27ac, whereas non-interacting enhancer E21 exhibited very high levels. Similarly, the levels of PolII and eRNA expression also varied greatly on these enhancers. These data suggest that high enrichment of H3K27ac and robust eRNA induction on enhancers are not good predictors for interaction with promoter. Similar non-reliance on H3K27ac for functionality predictions has been previously reported (Zhang et al., 2020). Within this framework, the chromatin interactions between enhancers and promoters remained the best indicator of functional enhancers. These functional differences among enhancers independent of H3K27ac levels could arise from differential protein complexes loaded on to the enhancers (Li et al., 2013, Bose et al., 2017; Mousavi et al., 2013; Zhao et al., 2019).
The data also raise the question, even though only a subset of enhancers within the super-enhancer are functionally relevant for the target gene activation, then why do enhancer clusters and SEs contain multiple enhancers? It is suggested that these non-functional enhancers may still play a role in keeping the environment favorable such as open chromatin around functional enhancers, maintaining activator concentration and not allowing the spread of H3K27me3 within the super-enhancer. Partial effects of E21 deletion on INK4a/ARF transcription could be due to its potential role in maintaining the co-activator concentrations within SE.
Loss of H3K27ac levels indicate the enhancer function loss but not the extent
We observed the similar extent of H3K27ac loss on intact enhancers upon CRISPRi or deletion (Figures 4A and 4F). However, the loss of eRNA on intact enhancers was much more profound upon deletions (Figures 4C–4E and 4G–4I). Accordingly, the suppression of genes was robust upon deletions (Figures 3A–3C and 3J–3L). Similarly, E8 exhibited downregulation of eRNA (Figure 4G) upon E21 deletion but did not show any alteration in H3K27ac (Figure 4J). These data indicate that loss of H3K27ac is a good indicator of loss of function but not the extent, and it is not the cause of enhancer or promoter activity, rather the outcome of transcriptional activity. In agreement with this, the mutation at K27 of histone H3 has been shown to have no effects on enhancer functions (Zhang et al., 2020). However, in other cases, the CBP/p300 that catalyzes acetylation at H3K27 plays a critical role in enhancer function (Raisner et al., 2018). Further, high levels of H3K27ac could be an outcome of eRNA presence at these enhancers, again suggesting its presence as the result of enhancer activity (Pnueli et al., 2015).
An interdependent enhancer network operates within enhancer cluster
We observed the downregulation of eRNA, loss of PolII, and H3K27ac on intact enhancers upon deletion of any interacting enhancer out of three enhancers tested. Thus, apart from expected enhancer:promoter network, we unexpectedly observed the enhancer:enhancer network in which these enhancers, though located far apart, depend on each other for their absolute functional activity in regulating the target gene. Such effects are likely due to their close physical proximity in 3D nuclear space such that the functional enhancers form a hub or cluster with the target promoters (Liu et al., 2014). Collectively, functional enhancers within a super-enhancer can form an equal-weighted network in which target genes cannot express in the absence of even a single enhancer. The enhancers are non-redundant in these clusters as opposed to earlier observations genomewide (Moorthy et al., 2017).
The loss of function by a mutation in any enhancer within equal-weight networks would result in dysregulation of all enhancers in the network thereby leading to promoter dysregulation. Perhaps, the INK4a/ARF locus being the most reproducible locus across GWAS is the result of such enhancer dependencies where variation in any enhancer would result in a complete collapse of transcriptional output of the locus.
EZH2 gain on promoters is independent of ANRIL
The antisense noncoding RNAfrom this locus, ANRIL, represses the promoter of INK4a by direct loading of polycomb complexes (Yap etal., 2010). However, upon the deletion of enhancers, even ANRIL expression was downregulated (Figure 5G) suggesting that INK4a/ARF genes and ANRIL are co-regulated by the enhancers in a gene desert. Further, EZH2 was present on INK4a/ARF promoters in spite of the absence of ANRIL transcription suggesting its ANRIL independence. We also observed a positive correlation between ANRIL and CDKN2A/2B expression in cervical tumors (Figures 5H and 5I). These positive correlations have been shown across the tumors types (Drak Alsibai et al., 2019; Lo Sardo et al., 2018; Visel et al., 2010).
A possible mechanism of EZH2 presence on promoters in the absence of enhancer and promoter transcription could be due to the lack of EZH2 sequestration from promoters by direct binding of promoter-driven transcripts with EZH2 (Beltran et al., 2016; Cifuentes-Rojas et al., 2014; Kaneko et al., 2014; Wang et al., 2017). However, the active promoters in the presence of enhancers have been shown to actively engage in such exclusion of EZH2. Moreover, combination of transcription loss and reduced H3K27ac at promoters may trigger the robust loading of EZH2, which is sufficient to maintain H3K27me3 (Figure 5A; Hosogane et al., 2016; Lavarone et al., 2019; Saxena et al., 2017).
Our results provide a template for studies in other systems for such enhancer dependencies, and the identified functional enhancers in the INK4a/ARF locus open up new avenues to understand the functional relevance of the 9p21 gene desert in several pathologies.
Star★Methods
Key Resources Table
| REAGENT OR RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-H3K27ac | Abcam | ab4729 |
| Anti PolII | Santa Cruz | sc-899 |
| Anti-EZH2 | Active Motif | 39875 |
| Anti-H3K27me3 | Millipore | 07-449 |
| Anti-p14 | Santa Cruz | sc-53639 |
| Anti-p16 | Cell Signaling | 4824S |
| Anti-Gapdh | Santa Cruz | sc-32233 |
| Chemicals, peptides, and recombinant proteins | ||
| Trizol | Ambion | 15596018 |
| Lipofectamine 2000 | Invitrogen | 11668-019 |
| Protein G Beads | Invitrogen | 10004D |
| ABI PowerUp SYBR Green Master Mix | Applied Biosystems | A25742 |
| SuperScript IV First-Strand Synthesis System | Invitrogen | 18091050 |
| DMEM | GIBCO | 10569-010 |
| FBS | GIBCO | 26140079 |
| Penicillin-Streptomycin | GIBCO | 15070-063 |
| Opti-MEM | GIBCO | 31985-070 |
| 0.25% Trypsin-EDTA (1X) | GIBCO | 25200-072 |
| T4 DNA Ligase | New England Biolabs | M0202M |
| DpnII | New England Biolabs | R0543M |
| HindIII | New England Biolabs | R0104M |
| Puromycin | GIBCO | A11138-03 |
| Hexadimethrine Bromide (Polybrene) | Sigma | H9268 |
| ECL Western Blotting Detection Reagent | GE Healthcare | RPN2106 |
| BrdU Cell Proliferation Assay Kit | Cell Signaling | 6813S |
| Senescence ß-Galactosidase Staining Kit | Cell Signaling | 9860S |
| Deposited data | ||
| RNA-Seq data upon enhancer deletions | This Study | GEO: GSE153410 |
| Experimental models: cell lines | ||
| HeLa Cell Line | ATCC | N/A |
| 293FT | ATCC | N/A |
| Software and algorithms | ||
| ImageJ | Software | https://imagej.nih.gov/ij/download.html |
| Graphpad Prism 8 | Software | https://www.graphpad.com/scientific-software/prism/ |
| Adobe Illustrator | Software | https://www.adobe.com/in/products/illustrator.html |
| HOMER | Software | http://homer.ucsd.edu/homer/ |
| Bowtie2 | Software | http://bowtie-bio.sourceforge.net/bowtie2/index.shtml |
| Juicer | Software | https://github.com/aidenlab/juicer |
| Juicebox | Software | https://github.com/aidenlab/Juicebox |
| ROSE (Rank Ordering of Super-enhancers) algorithm | Software | https://github.com/stjude/ROSE |
| FourCSeq | Software | https://bioconductor.org/packages/release/bioc/html/FourCSeq.html |
| 4Cseqpipe | Software | http://compgenomics.weizmann.ac.il/tanay/?page_id=367 |
| hisat2 | Software | http://daehwankimlab.github.io/hisat2/ |
| Samtools | Software | http://samtools.sourceforge.net/ |
| HTSeq pipeline | Software | https://htseq.readthedocs.io/en/master/ |
| DESeq2 | Software | https://bioconductor.org/packages/release/bioc/html/DESeq2.html |
| EnhancedVolcano | Software | https://github.com/kevinblighe/EnhancedVolcano |
Resource Availability
Lead contact
The lead contact, Dimple Notani (dnotani@ncbs.res.in), should be contacted for requests regarding resources, data, and reagents used in the study.
Materials availability
Cell lines generated in this study can be requested from the lead contact.
Experimental Model And Subject Details
HeLa and 293FT cell lines were obtained from ATCC. Both the cell lines were maintained in DMEM supplemented with 10% FBS at 37° C and 5% CO2.
Method Details
Cell Culture
HeLa cells were obtained from ATCC and were maintained in Dulbecco’s Modified Eagle’s medium (DMEM) supplemented with 10% Fetal Bovine Serum and 1% Penicillin/Streptomycin. The cells were maintained in humidified environment at 37°C in presence of 5% CO2. HeLa cells were passaged every third day.
HiC analysis
HiC forward and reverse end reads were trimmed and aligned separately. The raw reads were mapped to hg19 assembly using bowtie 2. The HOMER program makeTagDirectory was first used to create tag directories with tbp 1. Data was further processed by HOMER in order to remove small fragment and self-ligations using makeTagDirectory with the following options:-removePEbg -removeSpikes 10000 5. Next, findTADsAndLoops.pl was used to obtain overlapping TADs, produced at a 20kb resolution with 40kb windows. H3K27ac peaks intersecting with the identified TADs were counted and ranked to obtain the enhancers per TAD slope. The HiC datasets were analyzed using the Juicer pipeline for visualization. The .hic file generated from the juicer pipeline was then visualized using Juicebox. The contact maps were generated using Balanced normalization (Knight-Ruiz balancing algorithm).
Super-enhancer calling
Super-enhancers were identified using the ROSE (Rank Ordering of Super-enhancers) algorithm (https://bitbucket.org/young_computation/) using the aligned ChIP-seq reads as input with parameters-s 12500.
Circular chromosome conformation capture (4C)
4C was performed as per the protocol described in (van de Werken etal., 2012) with minor variations. HeLa cells were fixed with 1.5% fresh formaldehyde for 10 mins at room temperature and quenched with glycine (125mM) for 5 mins. The cells were washed thrice with ice-cold PBS and scraped, pelleted and stored at -80°C. Lysis buffer [Tris-Cl pH 8.0 (10mM), NaCl (10mM), NP-40 (0.2%), PIC (1X)] was added to the pellets and homogenized by Dounce homogenizer (20 strokes with pestle A followed by 20 strokes with pestle B). HindIII (400U, NEB) was used for the 3C digestion and T4 DNA ligase was used for ligation along with ligation mix [1% Triton X-100, 1X Ligation buffer (Tris-Cl pH 7.5 (50mM), MgCl2 (10mM), DTT (10mM)), BSA (0.0105mg/ml), ATP (0.105mM). The ligated samples were purified by PCI, followed by ethanol precipitation. The pellet was dissolved in 1XTE (pH 8.0) to obtain the 3C library. DpnII (50U, NEB) was used for 4C digestion and the samples were ligated, purified and precipitated similar to the 3C library to obtain the 4C library. The 4C library was treated with RNaseA to remove any trace of contaminating RNA and purified by the QIAquick PCR purification kit. The library was then subjected to PCR using the oligos designed for the CDKN2A viewpoint (Table S1). The PCR amplicon were purified using the same kit and subjected to next-generation sequencing with Illumina HiSeq2500 using 50bp single-end reads. Number of reads in each replicate are mentioned in Table S3.
4C-seq analysis
The sequenced reads were aligned to hg19 assembly using default Bowtie2 options. The output BAM file was used as the input for the FourCSeq pipeline. The first and second restriction site sequence was provided along with the primer sequence in the metadata. The viewpoint information is also provided. The reference genome is then in-silico digested to obtain the reference fragments. Reads mapping exactly to the fragment ends are then counted. This data is then plotted after smoothening and calculating Z-scores to detect interactions. Interacting regions are defined with the following thresholds: a fragment must have z-scores larger than 3 and an adjusted p value of 0.01 (Figure 2C; Figure S1 C).
Beside FourCseq pipeline, 4Cseqpipe (http://compgenomics.weizmann.ac.il/tanay/?page_id=367) was also used to process the sequenced data in (Figure 4K). 4C-seq images were generated using truncated mean at a 10kb resolution (Figure 4K).
Chromatin immunoprecipitation
Cells were crosslinked with 1% formaldehyde (Sigma-F8775) at room temperature for 10 mins with constant shaking. Glycine was added to a final concentration of 125 mM to quench the formaldehyde for 5 mins. Cells were washed thrice with 1X ice cold PBS. Cells were scraped in 3 mL of 1X PBS and pelleted down at 2K rpm for 5 mins at 4°C. Cells were gently resuspended in L1A Buffer (10 mM HEPES/KOH pH 7.9, 85 mM KCl, 1 mM EDTA pH 8.0). Nuclei were isolated by adding L1B buffer (10 mM HEPES/KOH pH 7.9, 85mM KCl, 1 mM EDTA pH 8.0,1% NP40) to the cells, resuspended and incubated on ice for 10 minutes. Nuclei were obtained by centrifuging at 3.5Krpmfor5 mins. Nuclear lysis buffer (L2) (50 mM Tris-HCl pH 7.4,1% SDS, 10 mM EDTA pH 8.0) supplemented with 1X PIC was added to the nuclear pellet and incubated on ice for 10 mins. Samples were sonicated using Diagenode biorupter with a setting of 30 Sec ON and 30 Sec OFF for 25 cycles. Cell lysate was cleared by centrifuging samples at 12K rpm for 12 mins. 100 ug of sheared chromatin was taken for each IP. Sheared chromatin was diluted by adding Dilution Buffer (DB) (20 mM Tris-HCl pH 7.4, 100 mM NaCl, 2 mM EDTA pH 8.0, 0.5% Triton X-100) supplemented with 1X PIC in 1:1.5 (1 volume of sheared chromatin and 1.5 volumes of Dilution Buffer) ratio. 5% of diluted chromatin was set aside as Input. 1 ug of antibody was added to immunoprecipitate the DNA and incubated on rocking platform for overnight at 4°C. Protein G Dynabeads (Invitrogen-140004D) were prepared by blocking in 1% BSA prepared in 1X PBS at 4°C for 1 hour followed by washing with 1X PBS. Immunoprecipitated DNA was collected by adding 15 ul of BSA blocked beads to each sample and incubated at 4°C for 4 hours. Beads were collected, flowthrough was discarded and 600 ul of Wash Buffer I (20 mM Tris-HCl pH 7.4,150 mM NaCl, 0.1% SDS, 2 mM EDTA pH 8.0,1% Triton X-100) was added. Washings were carried out at 4°C on a rocking platform. Washings were sequentially repeated with Wash Buffer II (20 mM Tris-HCl pH 7.4, 500 mM NaCl, 2 mM EDTA pH 8.0,1% Triton X-100), Wash Buffer III (10 mM Tris-HCl pH 7.4, 250 mM LiCl, 1% NP40, 1% Sodium Deoxycholate, 1 mM EDTA pH 8.0 and 1XTE (10 mM Tris pH 8.0,1mM EDTA pH 8.0). Immunoprecipitated DNA was eluted by adding 200 ul of Elution Buffer (100 mM NaHCO3,1% SDS) for 45 mins at 37°C in a thermomixer with rpm of 1200. Eluate was transferred to fresh tubes and 14 ul of 5M NaCl was added and kept overnight at 65°C. Immunoprecipitated DNA was purified by Phenol:chloroform:isoamyl alcohol (Ambion-AM9732) method, followed by ethanol precipitation. The final dried DNA pellet was dissolved in 100 ul of 1X TE. The ChIP data was plotted as the fold enrichment of percent input. First, percent inputs were calculated for beads and antibody and then values obtained from beads were divided from antibody values to get the fold enrichment.
RNA Isolation and cDNA synthesis
Cells were lysed with 1 mLof Trizol reagent (Invitrogen-15596018). 1/5th volume of chloroform was added to samples, briefly vortexed and centrifuged at 12K rpm for 12 mins. The aqueous phase was carefully collected and transferred to fresh tubes. 0.7 volumes of isopropanol were added to samples and incubated at room temperature for 10 mins to precipitate the RNA. The samples were centrifuged at 12K rpm for 12 mins, supernatant was discarded without disturbing the pellet. The pellet obtained was washed with 75% ethanol. Pellet was allowed to dry for 20 mins and dissolved in RNase free water. RNA obtained was treated with ezDNase (Invitogen-117660) to remove the traces of contaminating DNA. 1 ug of RNA was taken for the cDNA synthesis. cDNA was synthesized with Superscript IV (Invitrogen-18091050) cDNA kit.
CRISPR Cas9 mediated deletion
gRNAs were designed with https://zlab.bio/guide-design-resources tool. gRNAs were selected based on the highest score with a least number of off targets. All the gRNAs were cloned in pgRNA humanized vector (#44248) a gift from Stanley Qi between BstX1 and Xho1 restriction sites. gRNAs were co-transduced with lenti-Cas9 vector (#52962) a gift from Feng Zhang. Cells were selected for pgRNA humanized vector with puromycin (3ug/ml) for 48 hours. The remaining cells were seeded in a 96 well plate such that each well gets a single cell. Wells with single cells were identified under microscope and allowed to grow till colonies appeared. Media was changed after every fifth day. The cells were trypsinized and half of the cells were taken for the surveyor assay. Surveyor assay was carried out by PCR-based method.
CRISPRi
For carrying out CRISPRi, gRNAs were designed to target the core ofthe enhancers. For each enhancer two gRNAs were designed. Lentiviruses were made carrying dCas9-KRAB (#99372), a gift from Kristen Brennand and enhancer specific gRNAs. At the time of transduction, cells were around 75% confluent. Viral soup supplemented with 8ug/ml of polybrene was added to cells. Infection was stopped after 16 hours of transduction. Cells carrying the vectors were selected with 3ug/ml of puromycin for 72 hours.
Lentiviral transduction
HEK293FT cells were seeded in culture dishes coated with poly D lysine. Transfection of lentiviral packaging plasmid like pCMV-VSV-G (#8454), a gift from Bob Weinberg lab along with the plasmid of interest was carried out by Lipofectamine 2000. Media was changed after 6 hours. Viral soup was collected after 48 hours and 72 hours, pooled together, filtered and finally added to cells along with 8ug/ml of polybrene. Infection was stopped after 16 hours of transduction.
β-galactosidase staining
Cells were stained for senescence associated β-Galactosidase activity according to the manufacturer’s protocol (Senescence β-Galactosidase Staining from Cell Signaling #9860). Briefly, growth media was removed and cells were washed once with 1X PBS. 1 mL of 1X Fixative solution was added to each well and allowed to fix for 15 mins at room temperature. Fixed cells were washed twice with 1X PBS. To stain the cells, 1ml of β-Galactosidase Staining Solution was added to each well. Plates were carefully sealed with parafilm to avoid evaporation and incubated at 37°C for 48 hours in a dry incubator devoid of CO2. Images were taken with Olympus IX73. ImageJ was used to quantify the cells stained for senescence associated β-Galactosidase activity.
Cell proliferation assay
Cell proliferation assay was performed according to the manufacturer’s protocol (BrdU Cell Proliferation Assay Kit from cell signaling #6813). Briefly, 2500 cells were seeded in 3 wells (triplicates) of 96 well plate. Cells were incubated for 48 hours. 1XBrdU solution was added to each well and incubated for 24 hours. Growth media was removed and 100 ul of Fixing/Denaturing Solution was added to each well for 30 mins at room temperature. Post 30 mins fixing solution was removed and 100 ul of 1X detection antibody solution was added to each well for 1 hour at room temperature. Detection antibody solution was removed, followed by washing each well 3 times with 1X Wash Buffer. 100 ul of 1XHRP-conjugated secondary antibody solution was added for 30 mins at room temperature. Solution was removed followed by washing 3 times with 1X Wash Buffer. 100 ul of TMB substrate was added for around 20 mins and reaction was stopped by adding 100 ul stop solution. Absorbance was measured at 450 nm.
Wound healing assay
Cells were grown to confluence in 6 well plates and two scratches per well were created. Cells were washed with DPBS to remove the cellular debris. After removing debris, images were taken and labeled as A0. Cells were allowed to migrate for 72 hours. The extent of migration was recorded every 24 hours labeled as A1, A2 and A3. Wound closure (2 scratches per replicate) was measured with ImageJ where wound closure (%) = (wounded area after every 24 hours/wounded area at A0) X 100.
Colony formation assay
For colony formation assay, 2000 cells each of CR WT and enhancer knockout lines were seeded in one well of 6 well plate. Cells were allowed to form colonies for 10 days. Growth media was removed and cells were washed twice with 1X PBS. Colonies were fixed with absolute methanol for 10 mins followed by staining with 0.5% crystal violet for 20 mins. Colonies containing at least 50 cells were counted using ImageJ.
RNA-Seq analysis
The raw reads were mapped to hg19 assembly using hisat2 in a strand specific manner. The output BAM file was sorted using Samtools. This was then provided as an input for htseq-count, of the HTSeq pipeline, to count reads in the exonic features. The raw counts from the different datasets were then used as in input in DESeq2 to obtain differentially expressed genes using default thresholds. The volcano plots were plotted using the EnhancedVolcano tool on R.
Gene expression in tumors
Gene expression data from cancer patients belonging to the TCGA cohort were obtained from TCGA Genomic Data Commons (GDC) (https://portal.gdc.cancer.gov/).
ATAC-seq data in cervical tumors
ATAC-seq data of Cervical squamous cell carcinoma and endocervical adenocarcinoma (CESC) tumors from TCGA cohort were obtained from TCGA study (Corces et al., 2018).
Quantification And Statistical Analysis
Statistical significance is determined by unpaired t test (*p < 0.05; **p < 0.01; ***p < 0.005; ****p < 0.001; ns p > 0.05). The error bars denote SEM. The statistical details of experiments can be found in the figure legends. n represents number of biological replicates.
Supplementary Material
Highlight.
Only a few enhancers from the dense multi-enhancer cluster regulate the INK4a/ARF locus
Functional enhancers are not defined by high levels of H3K27ac or eRNAs
Deletion of a single functional enhancer renders the entire SE non-functional
Enhancer activation prevents EZH2 loading onto the INK4a/ ARF promoters
Acknowledgments
We thank Michael Geoff Rosenfeld for the discussions during the initial phase of study. We also thank D.N. lab members and Amanjot Singh for discussions. We acknowledge the support of the Department of Atomic Energy, Government of India, under project no. 12-R&D-TFR-5.04-0800, and intramural funds from NCBS-TIFR (D.N.). We also acknowledge funding support from Welcome-IA: IA/1/14/2/501539 (D.N.). D.N. is a EMBO Global Investigator. U.F. is supported by CSIR-SRF; K.W. and S.S. by SPM-CSIR, India; and A.K.S. and S.M. by NCBS/TIFR graduate program.
Footnotes
Author Contributions
U.F. and D.N. conceived the project, designed the experiments, and wrote the manuscript. U.F. generated most of the data with help from Z.I., K.W., R.S.J., S.M., and S.S. Informatics on data was performed by B.S. The patient data analysis was performed by A.K.S. All authors read and edited the manuscript.
Declaration of Interests
The authors declare no competing interests.
Data and code availability
The NGS datasets (RNA-Seq upon enhancer deletions) supporting the conclusions of this article are available at GEO (https://www.ncbi.nlm.nih.gov/geo/) with the accession numbers GEO: GSE153410.
References
- Aguilo F, Zhou MM, Walsh MJ. Long noncoding RNA, polycomb, and the ghosts haunting INK4b-ARF-INK4a expression. Cancer Res. 2011;71:5365–5369. doi: 10.1158/0008-5472.CAN-10-4379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aloia L, Demajo S, Di Croce L. ZRF1: a novel epigenetic regulator of stem cell identity and cancer. Cell Cycle. 2015;14:510–515. doi: 10.4161/15384101.2014.988022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold CD, Gerlach D, Stelzer C, Boryn qM, Rath M, Stark A. Genome-wide quantitative enhancer activity maps identified by STARR-seq. Science. 2013;339:1074–1077. doi: 10.1126/science.1232542. [DOI] [PubMed] [Google Scholar]
- Baker DJ, Childs BG, Durik M, Wijers ME, Sieben CJ, Zhong J, Saltness RA, Jeganathan KB, Verzosa GC, Pezeshki A, et al. Naturally occurring p16(Ink4a)-positive cells shorten healthy lifespan. Nature. 2016;530:184–189. doi: 10.1038/nature16932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beltran M, Yates CM, Skalska L, Dawson M, Reis FP, Viiri K, Fisher CL, Sibley CR, Foster BM, Bartke T, et al. The interaction of PRC2 with RNA or chromatin is mutually antagonistic. Genome Res. 2016;26:896–907. doi: 10.1101/gr.197632.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bose DA, Donahue G, Reinberg D, Shiekhattar R, Bonasio R, Berger SL. RNA Binding to CBP Stimulates Histone Acetylation and Transcription. Cell. 2017;168:135–149. doi: 10.1016/j.cell.2016.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrasco-Garcia E, Arrizabalaga O, Serrano M, Lovell-Badge R, Matheu A. Increased gene dosage of Ink4/Arf and p53 delays age-associated central nervous system functional decline. Aging Cell. 2015;14:710–714. doi: 10.1111/acel.12343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cifuentes-Rojas C, Hernandez AJ, Sarma K, Lee JT. Regulatory interactions between RNA and polycomb repressive complex 2. Mol Cell. 2014;55:171–185. doi: 10.1016/j.molcel.2014.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corces MR, Granja JM, Shams S, Louie BH, Seoane JA, Zhou W, Silva TC, Groeneveld C, Wong CK, Cho SW, et al. The chromatin accessibility landscape of primary human cancers. Science. 2018;80:362. doi: 10.1126/science.aav1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci USA. 2010;107:21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deloukas P, Kanoni S, Willenborg C, Farrall M, Assimes TL, Thompson JR, Ingelsson E, Saleheen D, Erdmann J, Goldstein BA, et al. CARDIo-GRAMplusC4D Consortium; DIAGRAM Consortium; CARDIOGENICS Consortium; MuTHER Consortium; Wellcome Trust Case Control Consortium Large-scale association analysis identifies new risk loci for coronary artery disease. Nat Genet. 2013;45:25–33. doi: 10.1038/ng.2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drak Alsibai K, Vacher S, Meseure D, Nicolas A, Lae M, Schnitzler A, Chemlali W, Cros J, Longchampt E, Cacheux W, et al. High positive correlations between ANRIL and p16-CDKN2A/p15-CDKN2B/p14-ARF gene cluster overexpression in multi-tumor types suggest deregulated activation of an ANRIL-ARF bidirectional promoter. Noncoding RNA. 2019;5:44. doi: 10.3390/ncrna5030044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamell C, Ginsberg D, Haupt S, Haupt Y. New insights on the regulation of INK4/ARF locus expression. Oncotarget. 2017;8:106147–106148. doi: 10.18632/oncotarget.22258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert LA, Horlbeck MA, Adamson B, Villalta JE, Chen Y, Whitehead EH, Guimaraes C, Panning B, Ploegh HL, Bassik MC, et al. Genome-Scale CRISPR-Mediated Control of Gene Repression and Activation. Cell. 2014;159:647–661. doi: 10.1016/j.cell.2014.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez S, Serrano M. Anew mechanism of inactivation of the INK4/ARF locus. Cell Cycle. 2006;5:1382–1384. doi: 10.4161/cc.5.13.2901. [DOI] [PubMed] [Google Scholar]
- González-Navarro H, Vinué Á, Sanz MJ, Delgado M, Pozo MA, Serrano M, Burks DJ, Andrés V. Increased dosage of Ink4/ Arf protects against glucose intolerance and insulin resistance associated with aging. Aging Cell. 2013;12:102–111. doi: 10.1111/acel.12023. [DOI] [PubMed] [Google Scholar]
- Harismendy O, Notani D, Song X, Rahim NG, Tanasa B, Heintzman N, Ren B, Fu XD, Topol EJ, Rosenfeld MG, Frazer KA. 9p21 DNA variants associated with coronary artery disease impair interferon-g signalling response. Nature. 2011;470:264–268. doi: 10.1038/nature09753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helgadottir A, Thorleifsson G, Magnusson KP, Grétarsdottir S, Steinthorsdottir V, Manolescu A, Jones GT, Rinkel GJE, Blankensteijn JD, Ronkainen A, et al. The same sequence variant on 9p21 associates with myocardial infarction, abdominal aortic aneurysm and intracranial aneurysm. Nat Genet. 2008;40:217–224. doi: 10.1038/ng.72. [DOI] [PubMed] [Google Scholar]
- Hirosue A, Ishihara K, Tokunaga K, Watanabe T, Saitoh N, Nakamoto M, Chandra T, Narita M, Shinohara M, Nakao M. Quantitative assessment of higher-order chromatin structure of the INK4/ARF locus in human senescent cells. Aging Cell. 2012;11:553–556. doi: 10.1111/j.1474-9726.2012.00809.x. [DOI] [PubMed] [Google Scholar]
- Hnisz D, Abraham BJ, Lee TI, Lau A, Saint-André V, Sigova AA, Hoke HA, Young RA. Super-enhancers in the control of cell identity and disease. Cell. 2013;155:934–947. doi: 10.1016/j.cell.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosogane M, Funayama R, Shirota M, Nakayama K. Lack of Transcription Triggers H3K27me3 Accumulation in the Gene Body. Cell Rep. 2016;16:696–706. doi: 10.1016/j.celrep.2016.06.034. [DOI] [PubMed] [Google Scholar]
- Jeck WR, Siebold AP, Sharpless NE. Review: a meta-analysis of GWAS and age-associated diseases. Aging Cell. 2012;11:727–731. doi: 10.1111/j.1474-9726.2012.00871.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanao H, Enomoto T, Ueda Y, Fujita M, Nakashima R, Ueno Y, Miya-take T, Yoshizaki T, Buzard GS, Kimura T, et al. Correlation between p14(ARF)/p16(INK4A) expression and HPV infection in uterine cervical cancer. Cancer Lett. 2004;213:31–37. doi: 10.1016/j.canlet.2004.03.030. [DOI] [PubMed] [Google Scholar]
- Kaneko S, Son J, Bonasio R, Shen SS, Reinberg D. Nascent RNA interaction keeps PRC2 activity poised and in check. Genes Dev. 2014;28:1983–1988. doi: 10.1101/gad.247940.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim WY, Sharpless NE. The regulation of INK4/ARF in cancer and aging. Cell. 2006;127:265–275. doi: 10.1016/j.cell.2006.10.003. [DOI] [PubMed] [Google Scholar]
- Kim TK, Hemberg M, Gray JM, Costa AM, Bear DM, Wu J, Harmin DA, Laptewicz M, Barbara-Haley K, Kuersten S, et al. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465:182–187. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima Y, Ye J, Nanda V, Wang Y, Flores AM, Jarr KU, Tsantilas P, Guo L, Finn AV, Virmani R, Leeper NJ. Knockout of the murine ortholog to the human 9p21 coronary artery disease locus leads to smooth muscle cell proliferation, vascular calcification, and advanced atherosclerosis. Circulation. 2020;141:1274–1276. doi: 10.1161/CIRCULATIONAHA.119.043413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson MH, Gilbert LA, Wang X, Lim WA, Weissman JS, Qi LS. CRISPR interference (CRISPRi) for sequence-specific control of gene expression. Nat Protoc. 2013;8:2180–2196. doi: 10.1038/nprot.2013.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavarone E, Barbieri CM, Pasini D. Dissecting the role of H3K27 acetylation and methylation in PRC2 mediated control of cellular identityy. Nat Commun. 2019;10:1679. doi: 10.1038/s41467-019-09624-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazorthes S, Vallot C, Briois S, Aguirrebengoa M, Thuret JY, Laurent GS, Rougeulle C, Kapranov P, Mann C, Trouche D, et al. A vlincRNA participates in senescence maintenance by relieving H2AZ-mediated repression at the INK4 locus. Nat Commun. 2015;6:1–16. doi: 10.1038/ncomms6971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Collado M, Villasante A, Strati K, Ortega S, Canamero M, Blasco MA, Serrano M. The Ink4/Arf locus is a barrier for iPS cell reprogramming. Nature. 2009;460:1136–1139. doi: 10.1038/nature08290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Notani D, Ma Q, Tanasa B, Nunez E, Chen AY, Merkurjev D, Zhang J, Ohgi K, Song X, et al. Functional roles of enhancer RNAs for oestrogen-dependent transcriptional activation. Nature. 2013;498:516–520. doi: 10.1038/nature12210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Legant WR, Chen BC, Li L, Grimm JB, Lavis LD, Betzig E, Tjian R. 3D imaging of Sox2 enhancer clusters in embryonic stem cells. eLife. 2014;3:e04236. doi: 10.7554/eLife.04236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo Sardo V, Chubukov P, Ferguson W, Kumar A, Teng EL, Duran M, Zhang L, Cost G, Engler AJ, Urnov F, et al. Unveiling the Role of the Most Impactful Cardiovascular Risk Locus through Haplotype Editing. Cell. 2018;175:1796–1810.:e20. doi: 10.1016/j.cell.2018.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matheu A, Maraver A, Collado M, Garcia-Cao I, Canamero M, Borras C, Flores JM, Klatt P, Vina J, Serrano M. Anti-aging activity of the Ink4/Arf locus. Aging Cell. 2009;8:152–161. doi: 10.1111/j.1474-9726.2009.00458.x. [DOI] [PubMed] [Google Scholar]
- McLaughlin-Drubin ME, Park D, Munger K. Tumor suppressor p16INK4A is necessary for survival of cervical carcinoma cell lines. Proc Natl Acad Sci USA. 2013;110:16175–16180. doi: 10.1073/pnas.1310432110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikhaylichenko O, Bondarenko V, Harnett D, Schor IE, Males M, Viales RR, Furlong EEM. The degree of enhancer or promoter activity is reflected by the levels and directionality of eRNA transcription. Genes Dev. 2018;32:42–57. doi: 10.1101/gad.308619.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moorthy SD, Davidson S, Shchuka VM, Singh G, Malek-Gilani N, Langroudi L, Martchenko A, So V, Macpherson NN, Mitchell JA. Enhancers and super-enhancers have an equivalent regulatory role in embryonic stem cells through regulation of single or multiple genes. Genome Res. 2017;27:246–258. doi: 10.1101/gr.210930.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosteiro L, Pantoja C, de Martino A, Serrano M. Senescence promotes in vivo reprogramming through p16INK4aand IL-6. Aging Cell. 2018;17:e12711. doi: 10.1111/acel.12711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mousavi K, Zare H, Dell’orso S, Grontved L, Gutierrez-Cruz G, Derfoul A, Hager GL, Sartorelli V. eRNAs promote transcription by establishing chromatin accessibility at defined genomic loci. Mol Cell. 2013;51:606–617. doi: 10.1016/j.molcel.2013.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muerdter F, Boryń ŁM, Woodfin AR, Neumayr C, Rath M, Zabidi MA, Pagani M, Haberle V, Kazmar T, Catarino RR, et al. Resolving systematic errors in widely used enhancer activity assays in human cells. Nat Methods. 2018;15:141–149. doi: 10.1038/nmeth.4534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolas D, Zoller B, Suter DM, Naef F. Modulation of transcriptional burst frequency by histone acetylation. Proc Natl Acad Sci USA. 2018;115:7153–7158. doi: 10.1073/pnas.1722330115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikpay M, Goel A, Won HH, Hall LM, Willenborg C, Kanoni S, Saleheen D, Kyriakou T, Nelson CP, Hopewell JC, et al. A comprehensive 1,000 Genomes-based genome-wide association meta-analysis of coronary artery disease. Nat Genet. 2015;47:1121–1130. doi: 10.1038/ng.3396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostuni R, Piccolo V, Barozzi I, Polletti S, Termanini A, Bonifacio S, Curina A, Prosperini E, Ghisletti S, Natoli G. Latent enhancers activated by stimulation in differentiated cells. Cell. 2013;152:157–171. doi: 10.1016/j.cell.2012.12.018. [DOI] [PubMed] [Google Scholar]
- Pare R, Shin JS, Lee CS. Increased expression of senescence markers p14(ARF) and p16(INK4a) in breast cancer is associated with an increased risk of disease recurrence and poor survival outcome. Histopathology. 2016;69:479–491. doi: 10.1111/his.12948. [DOI] [PubMed] [Google Scholar]
- Pauck A, Lener B, Hoell M, Kaiser A, Kaufmann AM, Zwerschke W, Jansen-Duürr P. Depletion of the cdk inhibitor p16INK4a differentially affects proliferation of established cervical carcinoma cells. J Virol. 2014;88:5256–5262. doi: 10.1128/JVI.03817-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pnueli L, Rudnizky S, Yosefzon Y, Melamed P. RNA transcribed from a distal enhancer is required for activating the chromatin at the promoter of the gonadotropin a-subunit gene. Proc Natl Acad Sci USA. 2015;112:4369–4374. doi: 10.1073/pnas.1414841112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raisner R, Kharbanda S, Jin L, Jeng E, Chan E, Merchant M, Haverty PM, Bainer R, Cheung T, Arnott D, et al. Enhancer Activity Requires CBP/P300 Bromodomain-Dependent Histone H3K27 Acetylation. Cell Rep. 2018;24:1722–1729. doi: 10.1016/j.celrep.2018.07.041. [DOI] [PubMed] [Google Scholar]
- Rao SSP, Huntley MH, Durand NC, Stamenova EK, Bochkov ID, Robinson JT, Sanborn AL, Machol I, Omer AD, Lander ES, Aiden EL. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 2014;159:1665–1680. doi: 10.1016/j.cell.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samani NJ, Erdmann J, Hall AS, Hengstenberg C, Mangino M, Mayer B, Dixon RJ, Meitinger T, Braund P, Wichmann H-E, et al. WTCCC and the Cardiogenics Consortium Genomewide association analysis of coronary artery disease. N Engl J Med. 2007;357:443–453. doi: 10.1056/NEJMoa072366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxena M, Roman AKS, O’Neill NK, Sulahian R, Jadhav U, Shivdasani RA. Transcription factor-dependent ‘anti-repressive’ mammalian enhancers exclude H3K27me3 from extended genomic domains. Genes Dev. 2017;31:2391–2404. doi: 10.1101/gad.308536.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherr CJ. Ink4-Arf locus in cancer and aging. Wiley Interdiscip Rev Dev Biol. 2012;1:731–741. doi: 10.1002/wdev.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Werken HJ, de Vree PJ, Splinter E, Holwerda SJ, Klous P, de Wit E, de Laat W. 4C technology: protocols and data analysis. Methods Enzymol. 2012;513:89–112. doi: 10.1016/B978-0-12-391938-0.00004-5. [DOI] [PubMed] [Google Scholar]
- Vazquez-Vega S, Sanchez-Suarez LP, Andrade-Cruz R, Castellanos-Juarez E, Contreras-Paredes A, Lizano-Soberon M, Garcia-Carranca A, Benitez Bribiesca L. Regulation of p14ARF expression by HPV-18 E6 variants. J Med Virol. 2013;85:1215–1221. doi: 10.1002/jmv.23568. [DOI] [PubMed] [Google Scholar]
- Visel A, Zhu Y, May D, Afzal V, Gong E, Attanasio C, Blow MJ, Cohen JC, Rubin EM, Pennacchio LA. Targeted deletion of the 9p21 non-coding coronary artery disease risk interval in mice. Nature. 2010;464:409–412. doi: 10.1038/nature08801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Paucek RD, Gooding AR, Brown ZZ, Ge EJ, Muir TW, Cech TR. Molecular analysis of PRC2 recruitment to DNA in chromatin and its inhibition by RNA. Nat Struct Mol Biol. 2017;24:1028–1038. doi: 10.1038/nsmb.3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welter D, MacArthur J, Morales J, Burdett T, Hall P, Junkins H, Klemm A, Flicek P, Manolio T, Hindorff L, Parkinson H. The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res. 2014;42:D1001–D1006. doi: 10.1093/nar/gkt1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whyte WA, Orlando DA, Hnisz D, Abraham BJ, Lin CY, Kagey MH, Rahl PB, Lee TI, Young RA. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013;153:307–319. doi: 10.1016/j.cell.2013.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Nord AS, Akiyama JA, Shoukry M, Afzal V, Rubin EM, Pennacchio LA, Visel A. Tissue-specific RNA expression marks distantacting developmental enhancers. PLoS Genet. 2014;10:e1004610. doi: 10.1371/journal.pgen.1004610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Kantrow S, Sai J, Hawkins OE, Boothby M, Ayers GD, Young ED, Demicco EG, Lazar AJ, Lev D, Richmond A. INK4a/ARF [corrected] inactivation with activation of the NF-κB/IL-6 pathway is sufficient to drive the development and growth of angiosarcoma. Cancer Res. 2012;72:4682–4695. doi: 10.1158/0008-5472.CAN-12-0440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap KL, Li S, Munoz-Cabello AM, Raguz S, Zeng L, Mujtaba S, Gil J, Walsh MJ, Zhou MM. Molecular interplay of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a. Mol Cell. 2010;38:662–674. doi: 10.1016/j.molcel.2010.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Xiong Y, Yarbrough WG. ARF promotes MDM2 degradation and stabilizes p53: ARF-INK4a locus deletion impairs both the Rb and p53 tumor suppression pathways. Cell. 1998;92:725–734. doi: 10.1016/s0092-8674(00)81401-4. [DOI] [PubMed] [Google Scholar]
- Zhang CY, Bao W, Wang LH. Downregulation of p16(ink4a) inhibits cell proliferation and induces G1 cell cycle arrest in cervical cancer cells. Int J Mol Med. 2014;33:1577–1585. doi: 10.3892/ijmm.2014.1731. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Hyle J, Wright S, Shao Y, Zhao X, Zhang H, Li C. A cis-element within the ARF locus mediates repression of p16INK4A expression via long-range chromatin interactions. Proc Natl Acad Sci USA. 2019;116:26644–26652. doi: 10.1073/pnas.1909720116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, Zhang Z, Dong Q, Xiong J, Zhu B. Histone H3K27 acetylation is dispensable for enhancer activity in mouse embryonic stem cells. Genome Biol. 2020;21:45. doi: 10.1186/s13059-020-01957-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Zhou J, He L, Li Y, Yuan J, Sun K, Chen X, Bao X, Esteban MA, Sun H, Wang H. MyoD induced enhancer RNA interacts with hnRNPL to activate target gene transcription during myogenic differentiation. Nat Commun. 2019;10:5787. doi: 10.1038/s41467-019-13598-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Han X, Wittfeldt A, Sun J, Liu C, Wang X, Gan LM, Cao H, Liang Z. Long non-coding RNAANRIL regulates inflammatory responses as a novel component of NF-kB pathway. RNA Biol. 2016;13:98–108. doi: 10.1080/15476286.2015.1122164. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The NGS datasets (RNA-Seq upon enhancer deletions) supporting the conclusions of this article are available at GEO (https://www.ncbi.nlm.nih.gov/geo/) with the accession numbers GEO: GSE153410.