Abstract
The CD2-CD58 recognition system promotes adhesion and signaling and counters exhaustion in human T cells. We found that CD2 localized to the outer edge of the mature immunological synapse (IS), with cellular or artificial APC, in a pattern we refer to as a “CD2 corolla”. The corolla captured engaged CD28, ICOS, CD226 and SLAM-F1 costimulators. The corolla amplified active phosphorylated Src-family kinases (pSFK), LAT and PLC-γ over T cell receptor (TCR) alone. CD2-CD58 interactions in the corolla boosted signaling by 77% compared to central CD2-CD58 interactions. Engaged PD-1 invaded the CD2 corolla and buffered CD2 mediated amplification of TCR signaling. CD2 numbers and motifs in its cytoplasmic tail controlled corolla formation. CD8+ tumor infiltrating lymphocytes displayed low expression of CD2 in the majority of colorectal, endometrial and ovarian cancer patients. CD2 down-regulation may attenuate anti-tumor T cell responses with implications for checkpoint immunotherapies.
Introduction
Human (h)CD2 and its ligand CD58 were identified by monoclonal antibodies that inhibit T cell killing of alloreactive target cells1. All mature human T cells express CD2, with higher expression on memory T cells2, whereas CD58 is ubiquitous. CD2 and CD58 interact through their extracellular immunoglobulin(Ig)-like domains, creating a complex of 13 nm length between cells, similar to the TCR-pMHC complex and are predicted to function side by side to the exclusion of the CD45 phosphatase3. The CD2 cytoplasmic domain is highly conserved and includes a number of polyproline motifs that mediate Src-family kinase (SFK) activation4 and recruitment of CD2-associated protein (CD2AP)5. CD2-CD58 interactions accumulate in the immunological synapse (IS) and contribute to sustained high intracellular calcium levels6. The IS can be divided into central, peripheral and distal supramolecular activation complexes (c, p and dSMACs, respectively)7. The cSMAC includes formation of synaptic ectosomes, extracellular vesicles that are transferred to the antigen-presenting cells (APC)8. In T cells interacting with CD58-presenting supported lipid bilayers (SLBs), CD2-CD58 interactions form microdomains that coalesce into single contacts with a density of ~800 molecules/μm2, 9. These microdomains are enriched in signaling molecules like phosphorylated lymphocyte-specific protein tyrosine kinase (Lck), and Linker for activation of T cells (LAT), and partially exclude CD4510. Recruitment of engaged CD2 to the IS has been documented, but not mapped at the level of SMACs11.
The CD2-CD58 pathway has been implicated in disease. Genetic variants of CD2 or CD58 associate with multiple sclerosis12 and rheumatoid arthritis13. During in vitro chronic activation of human CD8+ T cells, CD2 ligation prevents development of an exhausted CD127lo Programmed cell death-1 (PD-1)hi phenotype14. The related CD2 transcriptional signature has been associated with better viral clearance, but more severe autoimmunity. In the context of cancer, loss of CD58 expression has been reported mainly in blood malignancies15. In a murine MCA 38 colon adenocarcinoma model, the tumor infiltrating lymphocytes (TILs) displayed reduced CD2, CD8b and LFA-1 expression and defective cytotoxicity16.
We hypothesized that CD2 expression levels affect its function. We first investigated the properties of the CD2-CD58 interaction upon antigen recognition and IS formation. Our study identified the number of CD2 molecules per cell as controlling its location in the IS and costimulatory efficiency. These findings provided a framework to interpret the significance of CD2 expression levels in different contexts, and led us to profile CD2 surface expression of CD8+ TILs in cancer. CD8+ TILs exhibited a lower number of CD2 molecules per cell across multiple cancer types. Finally, we found a weak, but highly significant, negative correlation between an exhaustion gene signature and CD2 expression in publically available singe-cell RNAseq data. We propose that low CD2 impairs CD8+ TILs and hence increasing CD2 expression provides an attractive angle to improve TIL function.
Results
A unique distal “CD2 corolla” formed by CD2-CD58 interactions in the IS
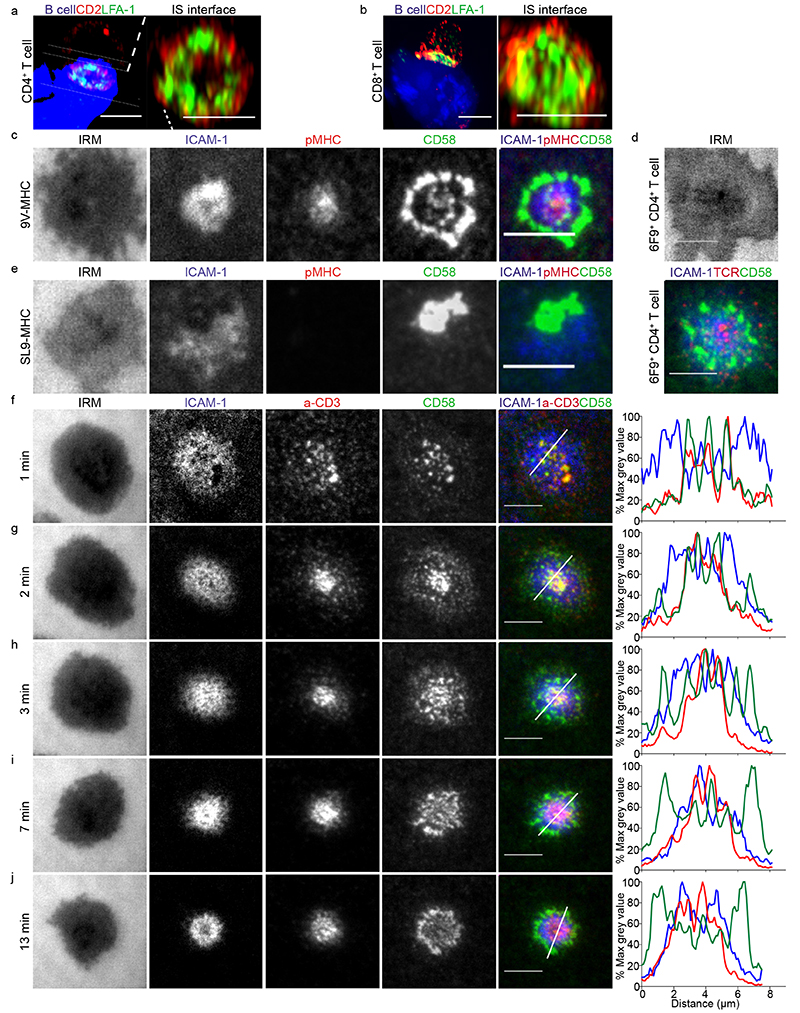
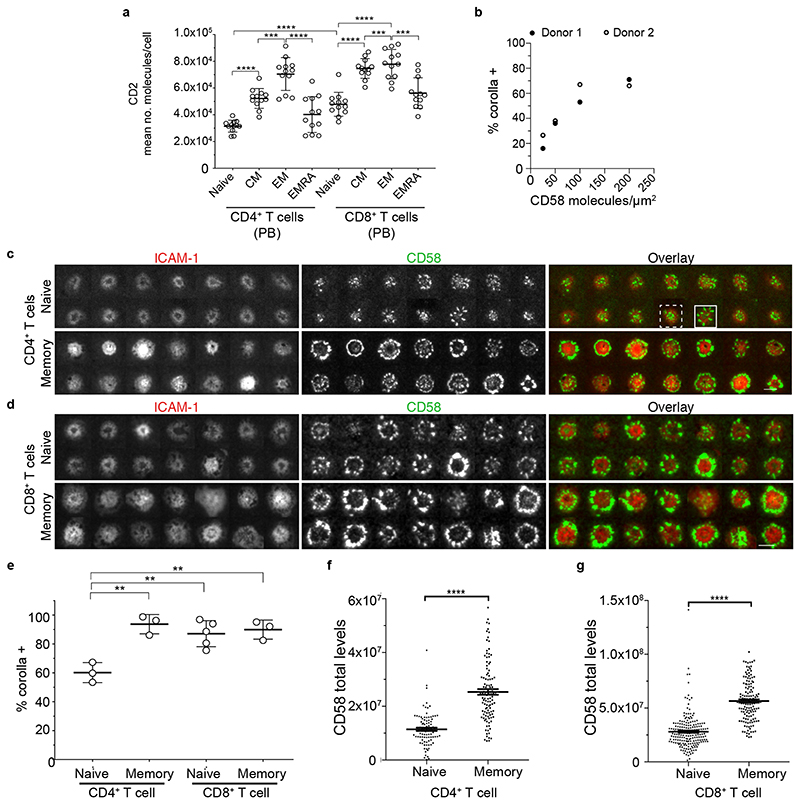
We set out to determine the localization of CD2 in the IS. Human CD4+ and CD8+ T cells, isolated from peripheral blood, were transfected with 6F9 or 1G4 TCR17, and conjugated to MAGE-A3243-258 or NY-ESO-9V157-165 peptide-loaded CF996 EBV-transformed B cells, respectively. At 15 min, 72% of cell conjugates imaged had a CD2 signal and this showed an accumulation in the interface between the T cell and the B cell. CD2 and LFA-1 staining revealed the presence of a strong CD2 signal in the distal regions of the cell-cell interface, outside the LFA-1-enriched pSMAC, and sometimes a weaker CD2 signal in the cSMAC (Fig. 1a-b and Supplementary Fig. 1a). We referred to this unexpected peripheral pattern as a “CD2 corolla” when the distal signal exceeded the central accumulation by at least 2-fold. At 2-5 mins, CD2 signal was in smaller clusters throughout the interface (Supplementary Fig. 1b).
Fig. 1. A unique ring pattern, “corolla”, formed by CD2-CD58 interactions in the IS.
a-b) 3D rendering images (IMARIS software) of T:B cell conjugates and enlarged 1 μm thick slice (white solid lines) of the T:B cell IS interface. 6F9+ CD4+ T cells (a) and 1G4+ CD8+ T cells (b), were conjugated with EBV-transformed B cells (blue-volumetric dye) pulsed with 1 μM MAGE-3A243-258 or 1μM NY-ESO-9V157-165 peptide, respectively, for 25-30 min. CD2 (red) and LFA-1-α-subunit (green) staining shown. Images were captured on an Airyscan confocal microscope (ZEISS). Representative images are shown from two independent experiments (n=25 conjugates). c) 1G4+ CD8+ T cells, fixed 15 min post-incubation on ICAM-1 (200/μm2), CD58 (200/μm2) reconstituted SLBs, in addition to NY-ESO-9V/HLA-A2 (30/μm2). Cells were imaged with TIRFM. d) 6F9+ CD4+ T cells, treated the same as in (c) but on MAGE-A3/HLA-DP4 (30/μm2) instead. 6F9+ TCR CD4+ T cells were stained with a fluorescently labeled anti-mouse TCRβ Fab (H57 clone) prior to SLB incubation. e) Same as in (c) but T cells were incubated on GAG-SL9/HLA-A2 (30/μm2) instead. f-j) CD4+ T cells, incubated on ICAM-1 (200/μm2), anti-CD3 Fab (30/μm2), CD58 (200/μm2) reconstituted SLBs. Cells were imaged at 4s intervals with TIRFM. Representative images within 13 min from initial contact are shown. Histograms depict the intensity profiles on the diagonal white lines in overlay images. Raw pixel intensity signal normalized to maximum intensity pixel of each channel.
A representative experiment of three independent experiments with either CD4+ or CD8+ T cells is shown. IRM, interference reflection microscopy, signal shows the spreading phase of the cell. Scale bar, 5 μm.
To gain a deeper understanding of the dynamics of the CD2 corolla we used SLB reconstituted with fluorescent pMHC, ICAM-1 and CD58. The physiological density of CD58 for incorporation into SLB was determined to be 200 molecules/μm2 (Supplementary Fig.1c and Supplementary Table 1). Antigen specific T cells were incubated on the SLB for 15 mins, fixed and imaged by total internal reflection fluorescence microscopy (TIRFM). 1G4 and 6F9 TCR transfected T cells formed IS with CD2 corolla on SLB presenting the specific pMHC (Fig. 1c, d). The majority of CD2-CD58 interactions were localized in the dSMAC in an array of flower petal-like microdomains that reconstituted the corolla pattern observed in the cell-cell conjugates, with weaker signals in the cSMAC. We extended the definition of the corolla to include the dSMAC:cSMAC ratio of greater than 2:1 and a clear gap between the corolla and cSMAC. 1G4 TCR transfected CD8+ T cells accumulated CD2-CD58 interactions in a single trailing domain on SLB presenting the non-stimulatory GAG-SL9-HLA-A2 (Fig. 1e). These data indicate that productive TCR engagement is required for corolla formation.
Dynamics of CD2 corolla formation
In order to use untouched CD4+ and CD8+ T cells isolated from human blood we employed SLB containing fluorescent anti-CD3 Fab in place of specific pMHC to engage polyclonal TCR. Within seconds of contact with SLB T cells initiated contacts through a few points that triggered rapid spreading and developed into microclusters with co-localized anti-CD3 and CD58 (Fig. 1f and Supplementary movies 1 and 2). At 2 min, microclusters with anti-CD3 and CD58 moved towards the center; microdomains of CD58 enrichment appeared in the dSMAC (Fig. 1g). At 3 min, anti-CD3 and CD58 in the forming cSMAC were still equal, while in the dSMAC CD58 microdomains grew (Fig. 1h). At 7 min, CD58 interactions were reduced in the anti-CD3 rich cSMAC and CD2-CD58 interactions increased in the dSMAC. At 13 min, the characteristic >2:1 (dSMAC:pSMAC) ratio of CD58 intensity was established; anti-CD3 microclusters continued to form in the dSMAC and appeared to move through the “fluid” CD58 microdomains of the corolla (Fig. 1h, Supplemental Fig.1d, Supplementary movies 1 and 2). These data suggest that CD2-CD58 dynamics are very different from other costimulatory receptors like CD28 and Inducible T cell costimulator (ICOS), which localize to the cSMAC in SLB lacking CD588.
The CD2 corolla organizes costimulatory interactions
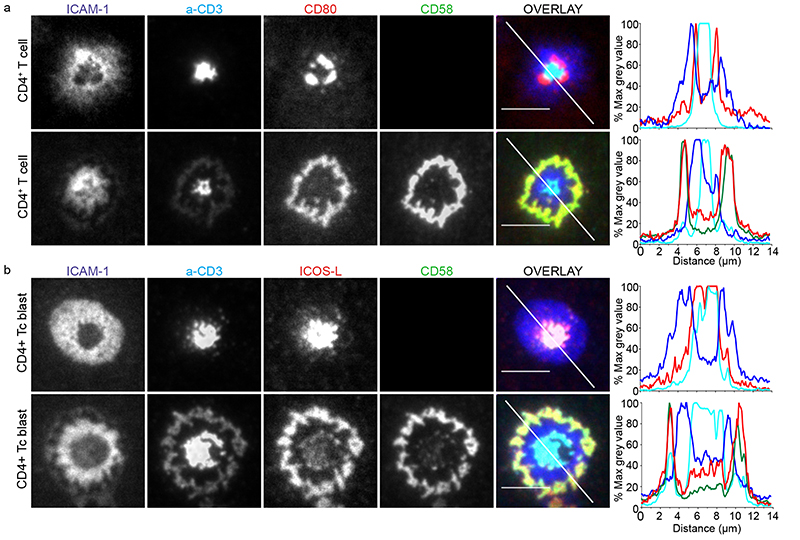
We next asked what happens when CD2 and different co-stimulators are engaged in the same IS. In order to track ligated CD28 in the presence and absence of CD2 corolla formation, fluorescent CD80 was added to the SLB. In the absence of CD58, mean of 98.4(±1.1)%, (±standard deviation or SD)% of CD4+ T cells displayed an annular ring of CD28-CD80 interactions within the cSMAC (Fig. 2a). In the presence of CD58, CD28-CD80 interactions were recruited to the corolla in 90.8±6.6% of CD4+ T cells (Fig. 2a, bottom panel, and Supplementary movie 3). Similar results were obtained with CD8+ T cells for which 99.7±0.4% were corolla positive. This phenotype was reproduced in an antigen-specific SLB system, using 1G4 TCR transfected CD8+ T cells for which ~98% (n=2 donors) were corolla positive (Supplementary Fig. 2a). In order to track ligated ICOS, we incorporated fluorescent ICOSL in the SLB. ICOS-ICOSL interactions were detected within the core of the cSMAC (Fig. 2b, top row), consistent with recent reports that they are sorted into TCR+ synaptic ectosomes in the cSMAC, which may have terminated ICOS signaling8. Addition of CD58 to the SLB redirected the ICOS-ICOSL interactions to the CD2 corolla and away from synaptic ectosomes in the cSMAC (Fig. 2b, bottom row), which may have sustained ICOS signaling.
Fig. 2. The corolla organizes multiple costimulatory receptor interactions.
a) Human CD4+ T cells incubated on ICAM-1 (200/μm2), anti-CD3 Fab (30/μm2), CD80 (100/μm2) without (top) or with CD58 (200/μm2) (bottom) reconstituted SLBs and fixed at 15 min. b) Activated human CD4+ T cell blasts incubated on ICAM-1 (200/μm2), anti-CD3 Fab (30/μm2), ICOS-L (100/μm2) without (top) or with CD58 (200/μm2) (bottom) reconstituted SLBs and fixed 15 min. Histograms depict the intensity profiles on the diagonal white lines in overlay images. Raw pixel intensity signal normalized to maximum intensity pixel of each channel. Cells were imaged with TIRFM and representative images are shown from four independent experiments. Scale bar, 5 μm.
We tested two additional co-stimulators in the presence of CD80 or CD58. The poliovirus receptor (PVR) binds CD226 CD96, and TIGIT on T cells; the first co-stimulatory for LFA-1 and the latter two co-inhibitory18, 19. Consistent with this, PVR accumulated in the pSMAC with ICAM-1 (Supplementary Fig. 2b); without any changes to the CD80 and CD3-enriched cSMAC. Inclusion of CD58, instead of CD80 led to significant redistribution of PVR into the corolla (Supplementary Fig. 2c). SLAM-F1 is a homophilic adhesion molecule20. SLAM-F1 interactions were co-localized with CD28-CD80 interaction in the cSMAC (Supplementary Fig. 2d). When CD80 was exchanged for CD58, SLAM-F1 strongly accumulated in the corolla (Supplementary Fig. 2e). We conclude that CD2 reorganizes diverse costimulatory receptor-ligand pairs to the corolla.
CD2 corolla boosts CD2 dependent TCR signal amplification
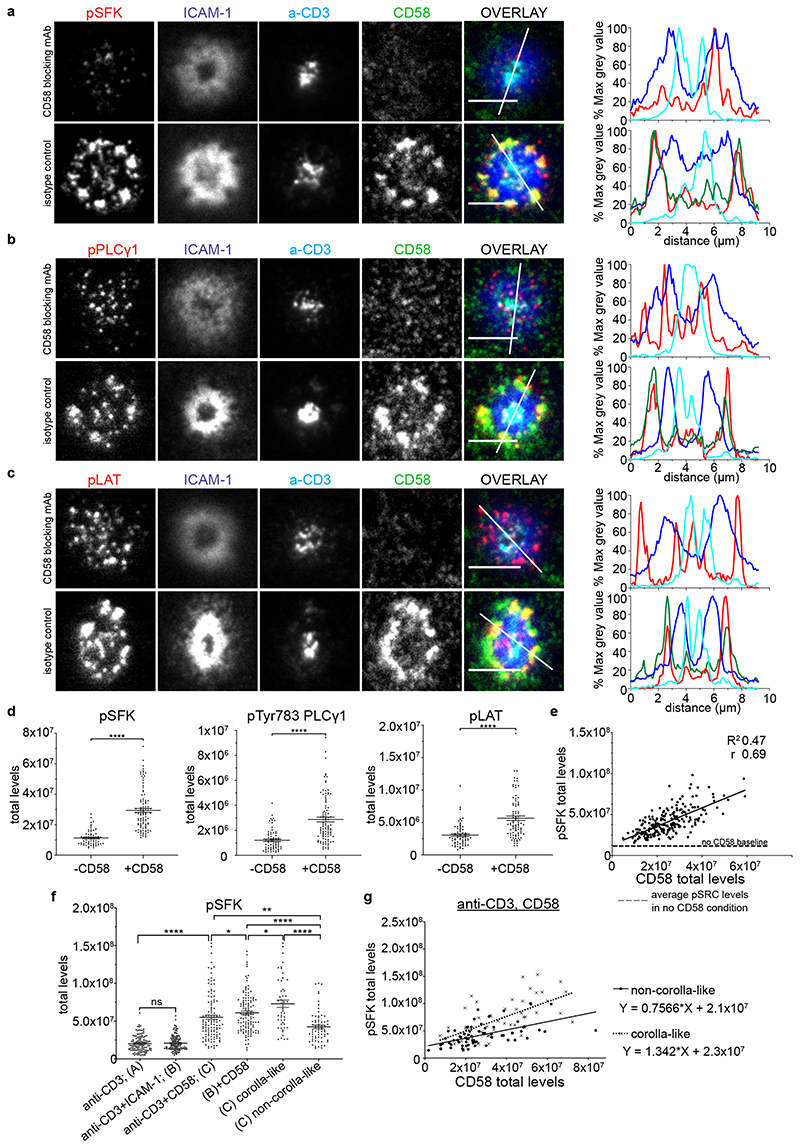
We next sought to investigate the implications of CD2 organization for its co-stimulatory function. T cells formed IS on SLB containing anti-CD3 and ICAM-1 ± CD58, fixed after 15 minutes and stained with antibodies to active phosphorylated SFK (pSFK), phosphorylated PLCγ1 (pPLCγ1) and phosphorylated LAT (pLAT). All three markers were enriched in the corolla formed in the presence of CD58 compared to patterns of microcluster localization in the absence of CD58 (Fig. 3a-c). Quantification of the total pSFK, pPLCγ1 and pLAT signals revealed 2-3-fold increase in total signal in the presence of CD2-CD58 interactions compared to their absence (Fig. 3d). As expected, the CD45 phosphatase was partly excluded from the CD2 corolla based on experiments with fluorescent anti-CD45 Fab and TIRFM (Supplementary Fig.3a). CD2 ligation either reduced 14% (n=5, independent donors) or increased 75% (n=2) the accumulation of anti-CD3 in the cSMAC (Supplementary Fig. 3b,c). In addition, CD2 ligation reduced 25% (n=4), increased 47% (n=6), or did not alter (n=1) the accumulation of ICAM-1 in the pSMAC (Supplementary Fig. 3d, e). The amount of CD58 in the IS correlated with total pSFK (Fig. 3e). The y-intercept of the regression line corresponds to the mean level of pSFK observed in the absence of CD58. These data suggest that CD2-CD58 interactions linearly amplify TCR proximal signals. The slope of this line represents the amplification factor.
Fig. 3. CD2 corolla boosts CD2-dependent TCR signal amplification.
Representative images of CD8+ T cells (n=3 independent experiments) incubated on ICAM-1 (200/μm2), anti-CD3 Fab (30/μm2) and CD58 (200/μm2) reconstituted SLBs, fixed 15 min post-incubation, followed by permeabilization, blocking and intracellular staining for phosphorylated Src kinases, pSFK (a), pTYR783PLCγ1, pPLCγ1 (b) or pTYR171LAT, pLAT in (c). SLBs were previously incubated with blocking anti-CD58 mAb (TS2/9 clone) or an isotype control. Same results were obtained when CD58 was omitted instead of using CD58 blocking mAb. Cells were imaged with TIRFM. Scale bar, 5 μm. Histograms depict the intensity profiles on the diagonal white lines in overlay images. y-axis; raw pixel intensity signal normalized to maximum intensity pixel of each channel. The CD58 signal was omitted in CD58 blocking conditions for clarity. d) Levels of pSFK, pPLCγ1 and pLAT for an average of 80 cells/condition shown in (a-c). ****, p value <0.0001 with unpaired two-tailed Mann Whitney test. A representative of three experiments is shown. Error bars, Mean±SEM. e) pSFK and CD58 levels in single cell synapses for CD8+ T cells treated as in (a); a representative experiment out of three. There is positive correlation between the two parameters with Pearson r 0.69; R2 0.47; 95% CI; ****, p<0.0001; y-intercept 1.32x107, slope 1.123; - - - -, dashed line represents the mean pSFK levels (1.15x107) from a population of single T cells (n=213 cells), in the absence of CD58. f) Levels of pSFK from T cells incubated on four different ligand SLB compositions based on anti-CD3 Fab (A) and anti-CD3, ICAM-1 (B) ±CD58, (n=121,145, 119, 124 cells, respectively and one out of three independent donors shown) reconstituted SLBs, fixed 15 min post-incubation, followed by permeabilization, blocking and intracellular staining for phosphorylated Src kinases (pSFK) and imaged with TIRFM. Error bars, Mean±SEM. *, p<0.05; **, p<0.007; ****, p<0.0001. with unpaired two-tailed Mann Whitney test. g) Comparison of the slopes of pSFK vs CD58 for non-corolla-like and corolla-like synapses in CD8+ T cells treated on anti-CD3, CD58 reconstituted SLBs, as in (f). A representative donor out of three; n=70 and 51 cells per category; statistically significant difference in the slopes, p=0.023. All three donors showed the same trend and two of them had significant p values (0.023, 0.0072, 0.0861) for the difference in the slopes of the relationship between CD58 and pSFK.
The ramified F-actin network underlying the IS is established even in conditions where there is no LFA-1-ICAM-1 interaction21. We next asked if the corolla might form in the absence of the LFA-1-ICAM-1 interaction, which would simplify analysis of impact on signaling. To test this, we allowed primary T cells to form IS on SLBs presenting anti-CD3 ± CD58. ICAM-1 removal didn’t impair pSFK signal in response to anti-CD3 stimulation in SLB (Fig. 3f). In the presence of CD58 40% of T cells formed corolla as defined by the ratio of dSMAC:cSMAC signal and a gap between the cSMAC and CD58 accumulation (Supplementary Fig. 3f). We assessed signaling in the structures by quantifying the level of pSFK in the IS. In relation to anti-CD3 alone, SLBs with anti-CD3 and CD58 had 2.7-fold higher pSFK (Fig. 3f). Cells lacking a corolla displayed a 2.2-fold increase, whereas cells with a corolla had a significantly higher, 3.6-fold increase (Fig. 3f). The relationship between CD58 accumulation and pSFK signal showed that the corolla increased the y-intercept (TCR alone pSFK) by only 10%, but the corolla significantly increased the slope by ~77% (Fig. 3g). These results demonstrated that the corolla boosts the amplification of TCR signaling by CD2.
PD-1-PD-L1 interactions buffer signal amplification in corolla
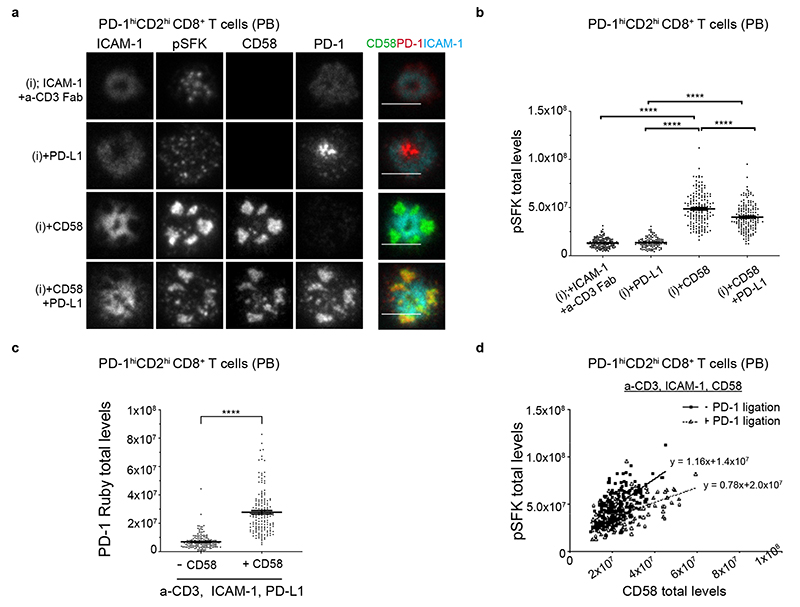
We next investigated the localization of the coinhibitory receptor PD-1 in relation to the CD2 corolla and pSFK levels. We transfected memory CD8+ T cells with PD-1 mRNA to achieve a mean of 1.45±0.56(SD)x104 PD-1 per cell, similar to that used in an earlier study22. We determined the localization of transfected PD-1 and corresponding pSFK levels on SLBs reconstituted with anti-CD3 and ICAM-1 ± PD-L1 ± CD58. PD-1 localized in the cSMAC in the presence of PD-L1 and absence of CD58, whereas PD-1 invaded the corolla in the presence of both CD58 and PD-L1 (Fig. 4a). There was no detectable effect of PD-1 engagement on pSFK in the absence of CD2 ligation. PD-1 engagement in the CD2 corolla reduced the amplification of pSFK by 20% (Fig. 4b). CD58 in the SLB also increased PD-1 accumulation in the IS overall (Fig. 4c). The slope of the CD58 engagement vs pSFK plot decreased by 33% (Fig. 4d). Thus, PD-1 engagement in the CD2 corolla buffers signal amplification.
Fig. 4. Regulation of signaling in the corolla by PD-1 engagement.
a) Representative images of memory CD8+ T cells transfected with TIL-like levels of PD-1-Ruby and incubated on four different ligand compositions based on ICAM-1 and anti-CD3 Fab (i) ±CD58 and ±PD-L1 in SLBs, fixed 15 min post-incubation, followed by permeabilization, blocking and intracellular staining for pSFK and imaged with TIRFM. T cells expressed, on average, 1.5x104 molecules/cell. Overlay image merges signals from ICAM-1, CD58 and PD-1 to assess the localization of CD58 and PD-1 relative to ICAM-1 and to each other. Scale bar, 5 μm.
b) pSFK levels at the IS of single memory PD-1hiCD2hiCD8+ T cells from experiment in (a), n=141, 111, 161, 157 cells per condition. Representative donor out of three is shown. ****, p<0.0001 with non-parametric two-tailed Mann Whitney test. Error bars, Mean±SEM. c) PD-1-Ruby levels accumulating in the synapse of PD-1hiCD2hiCD8+ T cells from experiment shown in (a) in a-CD3 Fab, ICAM-1, PD-L1±CD58 conditions; representative example shown. ****, p<0.0001. Error bars, Mean±SEM d) Plot of CD58 vs pSFK without (n=161 cells) and with PD-L1 (n=157) in SLB from donor in (a); significant difference in slopes, p<0.0001; all three donors showed the same trend and two had significant p values (p<0.0001, p=00.46, p=0.1765) for the difference in the slopes of the two regression lines.
CD2 expression levels determine corolla formation
We next determined the relationship between CD2 levels and corolla formation. The eight major peripheral blood (PB) conventional T cell subsets, naive, central memory (CM), effector memory (EM) and effector memory expressing CD45RA (EMRA) for CD4+ and CD8+ T cells varied in their levels of surface CD2 (Fig. 5a, Supplementary Fig. 4). As expected, memory subsets generally had higher CD2 than naive subsets and CD8+ T cells had more CD2 than CD4+ T cells overall (Fig. 5a and Supplementary Table 2). The EMRA subsets of both CD4+ and CD8+ had intermediate CD2 expression.
Fig. 5. Number of CD2 per human T cell predicts CD58 engagement and corolla formation.
a) After gating on single and live cells, CD4+CD3+ (left) or CD8+CD3+ (right) cells were selected and divided into naive (CD62L+CD45RA+), central memory (CD62L+CD45RA-), effector memory (CD62L-CD45RA-) and effector memory that reexpressed CD45RA (CD62L-CD45RA+) to determine average CD2 absolute number per cell. Error bars, Mean±SD b) The percentage of human CD8+ T cells that formed a CD2 corolla (% corolla+) in experiments on ICAM-1, anti-CD3 presenting SLBs with titrated levels of CD58. Data from 2 donors. c) A random image selection of naive (CD62L+CD45RA-) or memory (CD45RA-) CD4+ T cells incubated on ICAM-1 (200/μm2), anti-CD3 Fab (30/μm2) and CD58 (200/μm2) reconstituted SLBs, fixed at 15 min and imaged with TIRFM; examples of corolla positive (solid line rectangle) and corolla negative (dashed line rectangle) are shown. d) Same as in (c) but with CD8+ T cells. e) Percentage of T cells forming an IS with a CD2 corolla (corolla+) in experiments as in (c-d), was determined for naive and memory T cell subsets. Circles represent donors. **, p value <0.006, unpaired two-tailed t-test with Welch’s correction. For number of cells analyzed/subset see Supplementary Table 6. Error bars, Mean±SD. Quantification (from experiments as in (c-d)) of total CD58 accumulation in the IS of CD4+ (f) (n=91 and 110 cells, respectively) and CD8+ (g) T cell subsets (n=199 and 140, respectively). ****, p value <0.0001 with unpaired two-tailed non-parametric Mann Whitney test. A representative of three experiments is shown. Error bars, Mean±SEM.
Before further exploring the effect of CD2 expression, we tested the effect of titrating CD58 density in the SLB from 25-200/μm2 while maintaining anti-CD3 and ICAM-1 constant. CD2 corolla formation by CD8+ T cells was highly sensitive to CD58 density in the SLB with roughly half-maximal corolla formation at 50 molecules/μm2 (Fig. 5b). For further experiments we continued with 200/μm2 as appropriate to mimic the surface of professional antigen presenting cells (Supplementary Fig.1c and Supplementary Table 1).
We next examined CD2 corolla formation by CD4+ or CD8+ naive (CD62L+, CD45RA+) or pooled central and effector memory (CD62L±, CD45RA-) subsets (Fig. 5c,d). In naive CD4+ T cells, a mean of 60±7(SD)% formed corollas, whereas the remaining cells clustered CD2 within the cSMAC and the inner border of the LFA-1-ICAM-1-enriched pSMAC (Fig. 5c-e). Naive CD8+ T cells were 83±4% corolla positive while memory CD4+ T cells and memory CD8+ T cells were 94%±7% and 90±4% corolla positive, respectively (Fig. 4c-e). Consistent with lower CD2 numbers per cell in naive subsets, we detected lower levels of accumulated CD58 in the IS formed by naive subsets compared to their memory counterparts (Fig. 5f,g).
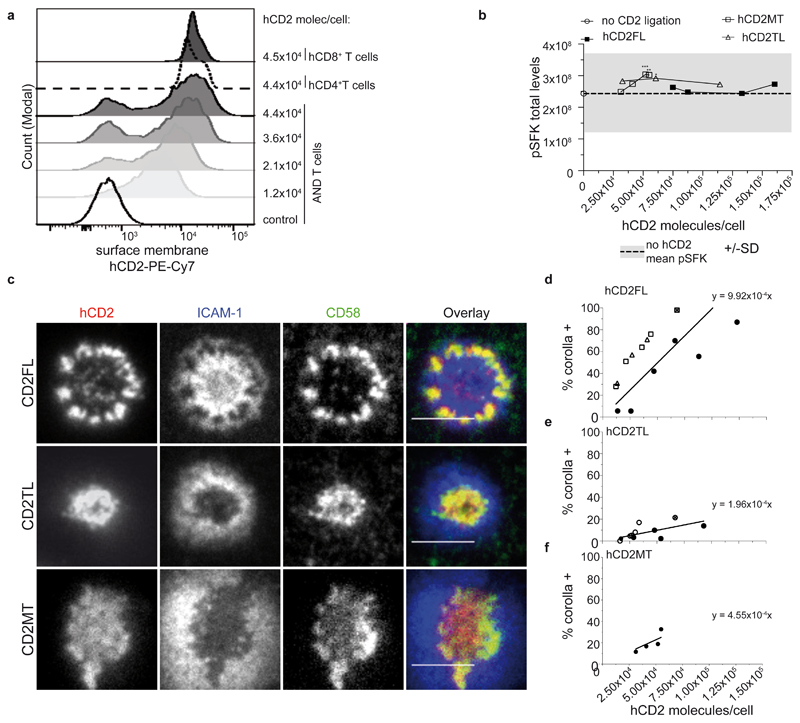
Other differences besides CD2 levels could influence these results. We further tested requirements for CD2 corolla formation in a system where we can express human CD2 (hCD2) at different levels in one cell type and evaluate the effects on corolla formation. As mouse CD2 (mCD2) doesn’t interact with CD58 we expressed Ruby-coupled hCD2 in mouse T cells and tested for corolla formation on SLB with pMHC, ICAM-1 and CD58. We tested “full-length” hCD2 with the intact cytoplasmic domain (hCD2FL), a “tail-less” form of hCD2 with only a 6-residue cytoplasmic domain (hCD2TL), and a “mutated tail” with PR at positions 329 and 330 replaced by AA (hCD2MT), which eliminates CD2AP and F-actin coupling23. The titration of hCD2 expression was achieved by electroporating varying amounts of mRNA encoding hCD2FL, hCD2TL or hCD2MT into mouse AND T cells from transgenic mice that express a TCR specific to Moth Cytochrome C peptide 88-103 in the context of I-Ek 24. The mean number of hCD2 molecules in electroporated AND T cells was quantified by flow cytometry (Fig. 6a, Supplementary Fig.5a). As expected, there was a strong correlation between the hCD2-Ruby and CD58 accumulated in IS at both the single cell (Supplementary Fig.5b) and population levels (Supplementary Fig. 5c). Engaging hCD2 in mouse T cells didn’t enhance pSFK in the IS (Fig. 6b). This system thus provided an opportunity to study the role of hCD2 number per cell on corolla formation without effects of signal amplification. The percentage of AND T cells forming a corolla increased with increasing levels of hCD2FL (Fig. 6c,d and Supplementary Fig. 5d). hCD2TL was 5.1-fold less efficient compared with hCD2FL (Fig. 6e and Supplementary Fig. 5e). hCD2MT was 2.2-fold less efficient than hCD2FL (Fig. 6f and Supplementary Fig. 5f). Collectively, these results suggest the CD2 number per cell, CD58 density on the opposing cell, and interactions with the cytoplasmic tail, including a motif involved in coupling to F-actin, control CD2 corolla formation.
Fig. 6. CD2 expression determines corolla formation independent of signaling.
a) The histograms show an example of AND T cells transfected with different levels of human CD2 (hCD2FL; full length protein) and compared to CD2 levels found in PB T cells from healthy individuals. Control cells were stained untransfected AND T cells (empty solid line histogram). b) Mean pSFK levels of AND T cells expressing either human CD2 full length (hCD2FL), hCD2p.240_351del (hCD2TL), or hCD2 p.PR329AA (hCD2MT) at different levels, on ICAM-1 (200/μm2), MCC-I-Ek (30/μm2) and CD58 (200/μm2) reconstituted SLBs, stained for pSFK and imaged with TIRFM. Dashed line represents mean pSFK levels AND T cells on ICAM-1, pMHC to which rest of conditions were compared to. *, p<0.05; **, p=0.0012, ***, p=0.001 with unpaired two-tailed non-parametric Mann Whitney test. For number of cells per data point see Supplementary Table 6. c) Representative images of AND T cells expressing either hCD2FL, hCD2MT or hCD2TL incubated on ICAM-1 (200/μm2), MCC-I-Ek (30/μm2) and CD58 (200/μm2) reconstituted SLBs, fixed 15 min post-incubation and imaged with TIRFM. Scale bar, 5 μm. d-f) The percentage of hCD2FL-, hCD2TM- or hCD2MT-expressing AND T cells that formed a CD2 corolla (% corolla+) in experiments such as in (c) over a range of hCD2FL (d) n= 4 independent experiments, hCD2TM expression (e) n=3 and hCD2MT (f) n=1. At least 30 cells were considered/data point. 95% CI of regression lines for FL, 7.15x10-4 to 12.69x10-4, line is significantly non-zero, F=59.76, p<0.0001; for TM 1.17x10-4 to 2.75x10-4, line is significantly non-zero, F=30.83, p=0.0002, MT, 2.62x10-4 to 6.48x10-4, line is significantly non-zero, F=56.21, p=0.0049. Each different symbol represents one data point from an individual experiment. See statistical analysis section for slope comparison test.
CD8+ TILs from cancer patients have heterogeneous CD2 expression
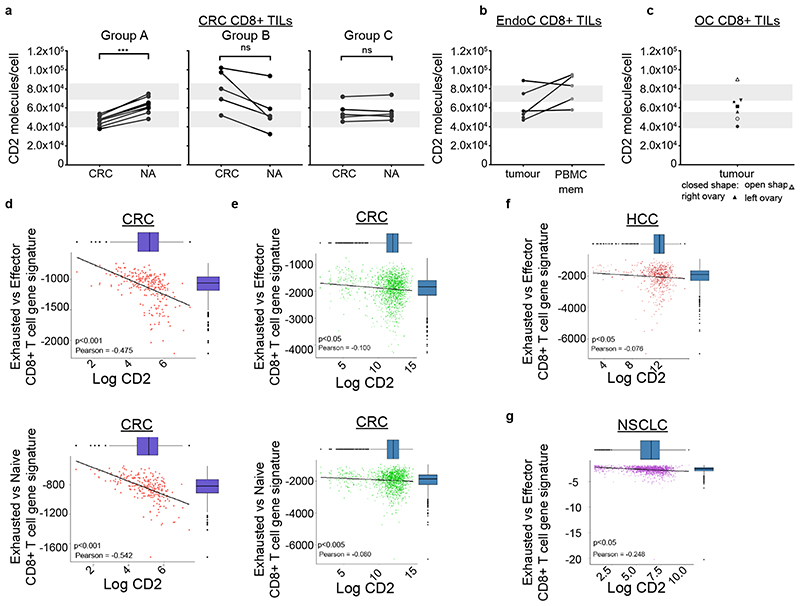
Since CD2 number per T cell was the most observable parameter that controls corolla formation we next investigated CD2 expression in human CD8+ and CD4+ TILs. CRC is paradoxical in that CD8+ T cell infiltration predicts outcome over a wide range of CRC subtypes25, and yet, only a small proportion of CRC with deficient mismatch repair respond to checkpoint therapies26. This suggests that other parameters besides CTLA-4, CD80, PD-1 or PD-L1 status control CD8+ T cell responses in CRC. Single cell suspensions obtained from primary CRC tissue resections were profiled using flow cytometry to investigate CD2 in the two main T cell compartments (CD4+ and CD8+) (Supplementary Fig. 6a).
We determined the number of CD2 molecules per cell in TILs and “normal adjacent” tissue (NA) from nineteen CRC patients (Supplementary Table 3a), for CD8+ (Fig. 7a) and CD4+ (Supplementary Fig. 6b). Based on the number of CD2 per cell of CD8+ TILs relative to NA T cells we stratified CRC patients into three groups (Fig. 7a, Supplementary Table 4). Group A (n=8 patients) had lower CD2 expression on TIL compared to NA, Group B (n=5) had higher levels of CD2 on TIL than NA and Group C (n=6) had no significant difference in CD2 levels between TIL and NA. Our calibration of CD2 number per cell enabled comparison to PB subsets, which revealed that 15 patients had CD8+ TILs with low numbers of CD2 per cell, which mimic the naïve PB CD8+ T cells with regards to CD2 expression. However, these were not naïve T cells as 90% were positive for CD45RO (data not shown). The CD2lo phenotype described for the CD8+ TILs wasn’t observed in the CD4+ T cells (Supplementary Fig. 6b). The CD2lo phenotype was also detected in Endometrial (EndoC) and Ovarian cancers (OC) (Supplementary Table 5). Four out of six EndoC patients were CD2lo in CD8+ TILs, but not in CD4+ TILs (Fig. 7b and Supplementary Fig. 6c). For comparison, we also determined CD2 levels in matched blood memory T cells from EndoC patients whenever available which had CD2 expression similar to healthy individuals. Two out of five OC patients tested, exhibited the CD2lo phenotype in CD8+ TILs but not in CD4+ TILs (Fig. 7c, Supplementary Fig. 6d). One patient had an intermediate CD2 phenotype. As with CRC and EndoC, CD4+ TILs did not exhibit a CD2lo phenotype in OC.
Fig. 7. CD8+ TILs from cancer patients can express considerable low levels of CD2.
a) The mean number of CD2 molecules/cell per CRC patient, for the CD8+ TILs from tumor (CRC) compared to matched normal adjacent tissue (NA). Patients were grouped in three separate groups (Group A, n=8; Group B, n=5; Group C, n=6) based on CD2 level change of tumor T cells relative to NA. ***, p=0.0007 with unpaired two-tailed t test with Welch’s correction. b) The mean number of CD2 molecules/cell per endometrial cancer (EndoC) patient (n=6 independent patients), for the CD8+ TILs compared to memory CD8+ T cells in matched patient PBMCs (one donor without matched PBMCs). c) The mean number of CD2 molecules/cell per ovarian cancer (OC) patient (n=5 independent patients) in CD8+ TILs.
For a-c, grey bars represent the average CD2 levels (mean±SD) expressed in memory (top) and naive (bottom) CD3+CD8+ T cells found in blood from healthy individuals. d) The correlation between exhausted CD8+ T cell gene signature and CD2 expression from whole transcriptomic analysis of CRC patients from The Cancer Genome Atlas (TCGA) is shown (n=255, independent patients) for (top) “exhausted vs effector CD8+ T cell gene signature” and (left) “exhausted vs naive CD8+ T cell gene signature”. Pearson r -0.475, -0.542, respectively; p<0.001 for both e) The correlation between exhausted CD8+ T cell gene signature and CD2 expression from publically available single cell transcriptomic dataset from CRC patients. The correlation was investigated in both exhaustion gene signatures (GSE 41867, top panel - exhausted vs. effector CD8 T cells, Pearson r -0.100, p<0.05, left panel-exhausted vs. naive CD8 T cells, Pearson r -0.542; p<0.005). f,g) The correlation between exhausted CD8+ T cell gene signature and CD2 expression from publically available single cell transcriptomic dataset from hepatocellular carcinoma (HCC) patients (Pearson r -0.076, p<0.05) and non-small cell lung cancer (NSLCC) patients (Pearson r -0.248, p<0.05). The correlation was investigated in both exhaustion gene signatures (signatures (GSE 41867, exhausted vs. effector CD8 T cells shown only). Statistical analyses for (d-g) see section “Data analysis - CD8 exhausted T cell subpopulations” in Methods.
Exhaustion negatively correlates with CD2 expression in CD8+ TILs
Attenuated function of CD8+ TILs in tumors has often been attributed to an “exhaustion” gene signature. We next investigated whether CD2 mRNA levels correlated with exhaustion. A CD8+ T cell exhaustion transcriptomic signature negatively correlated with bulk CD2 mRNA expression in 255 CRC patients in the TCGA cohort (https://www.cancer.gov/tcga) (Fig. 7d) and this was validated in 148 stage III CRCs in the METALLIC cohort (Supplementary Fig. 6e). Further analysis in the TCGA cohort revealed that this negative correlation was independent of microsatellite instability, which correlates with CD8+ T cell infiltration in CRC27. Bulk transcriptomic data has the limitation that correlations potentially could have arisen through expression of exhaustion associated genes and CD2 by different cells, and so we next revisited this in publically available single cell RNAseq datasets.
Single cell transcriptomic profiles for CD8+ TILs from patients with CRC (Fig. 7e), hepatocellular carcinoma (HCC) (Fig. 7f), non-small cell lung carcinoma (NSCLC) (Fig. 7g) and melanoma (Supplementary Fig. 6f) from the National Center for Biotechnology Information Gene Expression Omnibus were interrogated (Supplementary Table 7)28, 29, 30. Linear regression demonstrated that the T cell exhaustion gene signature was significantly negatively correlated with CD2 expression in CRC (Fig. 7e). This result was obtained for both gene signatures of “Exhausted CD8 T cell vs. Effector” (Fig. 7e, left) and “Exhausted CD8 T cell vs. Naïve CD8” (Fig.7e, right). Significant correlations were also observed in HCC (Fig. 7f), NSCLC (Fig. 7g) and melanoma (Supplementary Fig. 6f). To confirm this trend, linear regression was performed with all scRNAseq samples combined, following correction for each sequencing run, further reinforcing the weak (~-0.2) but highly statistically significant negative correlation between the “Exhausted CD8 T cell vs. Effector” (Pearson r -0.242, p=3.68x10-9) and “Exhausted CD8 T cell vs. Naïve CD8” (Pearson r -0.231, p= 4.37x10-8) gene signatures and CD2 mRNA (Supplementary Table 8).
Discussion
Using model systems for T cell activation (T:APC cell conjugates and T:SLB hybrid junctions) we revealed unexpected properties of CD2 that contribute to its function as an adhesion receptor, signal amplifier and signal integrator. Our observations that CD2-CD58 and TCR-pMHC interactions are initially colocalized fit with expectations based on extracellular domain size and functional synergy3. Segregation of ligated CD2 and TCR at later times had been reported in the SLB system10, 31, but our observations in T:APC conjugates verify that this is the physiologically relevant distribution in the mature IS. We use the SLB system to show that engagement of sufficient CD2 molecules with CD58 results in a unique protein rearrangement in the IS, the “corolla” pattern in the dSMAC, which boosts the amplification of TCR signaling by CD2 and uniquely changes the localization of classical costimulatory or checkpoint interactions with CD80, ICOSL, PVR, SLAM-F1 and PD-L1. We speculate that the CD2 corolla boost is based on exploiting dynamic F-actin structures in the dSMAC32, 33. TCR microclusters appear to traverse the corolla as discrete structures, but phosphorylated SFK, LAT and PLC-γ fill the corolla. Similarly, invasion by the inhibitory receptor PD-1 checks, but doesn’t turn off CD2 mediated amplification. We have not directly examined if being captured in the corolla impacts signals generated by CD80 receptors (CD28 or CTLA-4), ICOS, PVR receptors (CD226, CD96, TIGIT) or SLAM-F1. Forcing distal CD28 engagement through microcontact printing of anti-CD28 antibodies was shown to boost CD28-mediated enhancement of IL-2 production34. Moreover, CD2 corolla could function as an expanded receptive area of the IS across which a T cell can extend its scanning activity of the APC surface35.
Corolla formation displays a number of unexpected features – notably the persistence of thousands of CD2-CD58 interactions in the dSMAC in the face of centripetal F-actin flow. CD2-CD58 interactions are transient with a half-life of only a fraction of a second and turnover rapidly in contact areas36. Due to the high lateral mobility of CD2, most CD2 molecules on the cell can diffuse into the IS and engage CD58 over minutes. Since CD2-CD58 interactions maintain a density of ~800 μm-2 as the number of interactions increases, cells with 4x104 CD2 molecules will have a total CD2 microdomain area approaching 50 μm2, which is about half the area of a mature IS9. Dynamics and size may contribute to the apparent fluidity of the corolla and its resistance to central transport. We also found that CD2 corolla can form even in the absence of ICAM-1, and preliminary computational simulations predict that repulsion from large, non-ligated molecules like CD45 may also promote the corolla pattern37.
The cytoplasmic domain of CD2 promoted corolla formation in a setting in which signal amplification at the level of pSFK was disabled. The lack of hCD2 dependent enhancement of pSFK signaling in mouse T cells was surprising, but may be due to a dominant effect of endogenous mCD2 cytoplasmic domains, which have been shown to participate in signaling reactions even when not engaged38. The cytoplasmic tail also links CD2 via CD2AP and CIN85 adaptor proteins to other signaling proteins and to the actin cytoskeleton39. The hCD2MT does not bind CD2AP and had compromised corolla formation, consistent with collective association of the fluid CD2 corolla microdomains to lamellipodial F-actin in the dSMAC.
In light of the importance of the number of CD2 to scaling of TCR signaling we propose that the CD2lo phenotype we have observed in CD8+ TILs renders them less responsive to tumor antigens and may compromise tumor clearance. It has been shown that PD-1-mediated inhibition preferentially targets CD28-mediated costimulation22. We have determined that, even in the absence of CD28 engagement, PD1 can also buffer CD2-mediated costimulation. The amount of PD-1 expression regulates the magnitude of PD-1-mediated inhibition of T cell activation and cytokine production40, hence PD-1 acts as a quantitative checkpoint in the context of CD2. Similarly, we have shown that the number of CD2 per cell regulates corolla formation, which boosts CD2-mediated enhancement of TCR signaling. Hence the conditions that create the CD2lo phenotype take away a key IS organizer and check signal amplification. An important future goal would be to understand how the corolla influences the interaction of PD-1 with CD28 and if any emergent signaling networks with unique outputs are generated by combining costimulators and checkpoints in the corolla.
In addition, CD28-CD8+ T cells are enriched in the periphery of older adults and are CD2hi similar to effector memory CD8 T cells in blood41; it was reported that CD28-CD8+ T cells use CD2 as their primary costimulatory pathway during T cell activation42. CD8+ TILs in CRC patients were often CD28-, but could at the same time exhibit the CD2lo phenotype. CD4+ TILs, did not suffer the same loss of CD28 as CD8+ TILs. This highlights further the potential impact that the CD2lo phenotype can have on CD8+ TIL function. Decrease in CD2 expression on non-functional TILs was also reported in the MCA38 mouse model16. CD4 T cells did not display low CD2 and it will be important to investigate CD2 levels specifically in tumor associated regulatory T cells as elevated CD58 has been associated with greater Treg function12.
Our results suggest that this CD2lo phenotype is also detectable at the mRNA level, but this does not exclude other post-translational regulation. Why TILs from some CRC patients retain high levels of CD2 expression is yet to be addressed. It will be important to monitor CD2 levels in CRC patients treated with anti-PD-1 mAbs as lowered CD2 expression might offset the benefit of PD-1 targeted therapy. Finally, the CD2lo phenotype could be an indicator of poor prognosis or a predictor of poor response of CRC patients to certain immunotherapies and could be, by itself, an interesting target for immunotherapy, perhaps through manipulation of transcription factors controlling CD2 expression. Engineering T cells to bolster CD2 expression might make them more resilient in adoptive immunotherapy of solid tumors.
Methods
Reagents
RPMI 1640 (31870074), Dynabeads Human T-Activator CD3/CD28 for T cell expansion and Activation kit (1132D) were purchased from Thermofisher Scientific. Hyclone Fetal bovine serum was obtained from Fisher Scientific. Poly-L-lysine solution 0.1% (w/v) in H2O was obtained from Sigma (25988-63-0). The following anti-human antibodies were purchased from Biolegend, CD62L (DREG-56), CCR7 (G043H7), CD3 (UCHT-1), CD4 (A161A1), CD8 (HIT8a), CD2 (RPA2.10), CD28 (CD28.2), PD-1 (EH12.2H7), CD11a (TS2/4), mouse IgG1 isotype control (MOPC-21), CD58 (TS2/9), mouse IgG1 isotype control (MG1-45; 401402), CD45 (HI30), CD127 (A019D5), HLA-A2 (BB7.2), CD45RO (UCHL1). The following anti-human antibodies were purchased from BD Bioscience, CD45RA (HI100), CD127 (HIL-7R-M21), CD58 (1C3), CD2 (CD2.1) clone was a gift of D. Olive (Aix Marseille University). The following anti-human antibodies were purchased from eBioscience, CD8 (OKT8) and CD28 (CD28.2). Flow cytometry mAbs were used between 1-5ug/ml depending on how many cells were staining (i.e. between 5x103- 1x106). The following antibodies were purchased from Cell Signaling, pPLCγ1(pY783), pLAT(pY171), pSFK (pY416) (D49G4) and used at dilutions recommended by the company. The following anti-mouse antibodies were purchased from Biolegend, CD4 (RM4- 5), CD3 (145-2C11), TCRβ (H57-97). TCRβ (H57-97) Fab was obtained from Bio X Cell.
Plasmids and constructs
For 1G4 and 6F9 TCR please see Abu-Shah et al 17. The human CD2FL and CD2TL constructs were provided by S. Davis (University of Oxford) and were then cloned into the pGEM-GFP-64A (gift of J. Riley, U. Pennsylvania, USA) backbone vector. The mutant CD2MT insert was designed in silico in house and synthesized by Thermofisher. The human PD-1 construct was provided by P. A. van der Merwe (University of Oxford) and cloned into pGEM-GFP-64A. Protein expression in 293 HEK cells utilized either the pHR lentiviral vector (S. Davis) or pOPINTTGneo (R. Owens, Protein Production UK, Harwell, UK).
Fluorescently labeled Fab and extracellular domains (ECDs)
UCHT1 Fab’2 was generated by Papain digestion of UCHT1 IgG (Bio X Cell, Lebanon, NH). UCHT1 Fab were made by reduction and alkylation of Fab’2 with maleimide-PEG2-biotin and Alexa Fluor™ 488 or 568 NHS Ester at fluorophore:proteins ratio between 1:1 to 2:145. Other proteins used in preparation of ligand-presenting SLBs were all complete ECDs with a C-terminal 12His tag. These were generated in S9 cells (ICAM-1) or 293 HEK cells (CD58, CD80 and others) and purified by Ni2+ affinity chromatography and gel filtration. ICAM-1 was labelled with Alexa Fluor™ 405 NHS Ester and other proteins were labelled with the Alexa Fluor™ dye-maleimide indicated on a free cystein added between the ECD and the 12His tag (CD58-AF488, CD58-AF568, CD80-AF568, CD80-AF647, ICOS-L-AF647, PD-L1-AF647, PVR-AF647, SLAM-F1-AF647).
Isolation of T cell subsets, cell culture and T cell blast generation
Human T cell subsets were isolated from leukapheresis products (NHS Blood and Transplant/ London-Fulham Research Ethics Committee/ Barlow House/ 3rd floor, 4 Minshull Street/ Manchester/ M1 3DZ/ phone 0207 1048021) using negative selection kits from STEMCELL Technologies. Rosette Sep (Stemcell Technologies) was used to enrich for total CD8+ or CD4+ T cells. Naïve and memory cells were isolated using EasySep Enrichment kits (negative selection, Stemcell Technologies). Flow cytometric analysis based on CD4/CD8 and CD62L and CD45RA was used to confirm purity is above >92%. Human T cells were cultured in resting conditions, with no IL-2, at 37°C, 5% CO2, in RPMI 1640 (Roswell Park Memorial Institute) medium (Life Technologies) supplemented with 10% FBS, 5% Penicillin-Streptomycin, Penstrep (Gibco), 1x MEM Non-Essential Amino Acids Solution (Thermo Fisher Scientific), 10mM HEPES (Life Technologies), 1mM Sodium Pyruvate (Thermo Fisher Scientific). T cells were cultured at 2x106 cells/ml for a maximum of 3 days. Human T cells isolated were also used to generate T cells blasts using the Dynabeads Human T-Activator CD3/CD28 for T cell expansion and Activation kit (Life Technologies) as per company’s protocol. Human T cells blasts were ready to be used in experiments on Days 6-7 of expansion protocol. No IL-2 was supplemented in media the day before an experiment.
AND Mouse T cells (AND T cell blasts) were generated from spleen and lymph nodes of TcrAND B10.Br mice; mice were housed under pathogen-free conditions in the Kennedy Institute of Rheumatology Animal Facility in accordance with local and Home Office regulations. The mice were housed in individually ventilated cages (IVC) with corn cob bedding. 12 hours of Light/dark cycle with half an hour of dim light period in place. Appropriate environmental enrichment is provided: Enviro-dri, and housing tunnel, cage balcony and chew blocks. The temperature is maintained at 21 degrees +/- 2 and the humidity 55% +/- 10. The protocols were reviewed by local Veterinary Surgeon (Vet) and Named Animal Care and Welfare Officer (NACWO) before being reviewed and approved by “ Animal Welfare and Ethical Review Body (AWERB). All procedures were conducted in accordance with the UK Scientific Procedures Act of 1986 and overlooked by University of Oxford Department of Biomedical Services, Clinical medicine ORC (old road campus).
TcrAND.B10 (JAX stock #002761) and B10.Br (JAX stock #000465) mice were mated, and their F1 progeny was used for experiments at 6-10weeks of age. Briefly, spleen and lymph nodes from F1 TcrAND B10.Br mice were crashed over 70 μm strainer to generate single cell suspensions and following red blood cell lysis, incubated with 1 μM moth cytochrome c, MCC, or, T102S peptide (variant of MCC) and 50 U recombinant human IL-2 in High Glucose (25mM) Dulbecco’s Modified Eagle Medium supplemented with 0.116g/L L-Arg-HCl, 0.036g/L L-asparagine, 2g/L NaHCO3, 2.385 g/L HEPES, 1mM sodium pyruvate, 1.5 mM L-glutamine, Pen/Strep and 50 μM 2-mercaptoethanol at 37°C, 5% CO2 incubator to expand AND T cells. AND T cells were used for experiments on day 6-7 of culture; cell culture was enriched (>95%) in AND CD4+ T cells. No IL-2 was supplemented in media the day before an experiment. AND T cells were also frozen on day 6-7 to be used in future experiments when needed i.e. they would be thawed and rested overnight in 50 U IL-2 before being used in SLBs or for mRNA electroporation transfections.
Cell line culture
The CF996 EBV-transformed B cell line (Sigma Aldrich), was maintained in supplemented RPMI-1640 medium with 10 % FBS at cell densities of 0.75-1x106 cells/ml in a 37°C, 5% CO2 incubator. Trombelli cell line (a kind gift from Prof. V. Cerundolo, University of Oxford), was grown in supplemented RPMI-1640 as above. None of the above cell lines were tested for mycoplasma while in our cultures nor any signs of aberrant cell culture were observed suggestive of any infection. Antibiotics and antifungals were used where necessary.
Generation of monocyte-derived DCs
Monocyte-derived Dendritic cells were prepared as described17. In brief, human monocytes were isolated from leukapheresis products using the Rosette Human Monocyte Enrichment Cocktail kit (Stem Cell). Purified cells were cultured in 2 ml/well in 12 well plates for at least 4 hr at 37°C to allow for adhesion and enrichment of monocytes. Culture supernantant was resuspended and removed. Cells were washed twice with fresh media before adding culture media supplemented with 0.1 μg/ml, granulocytemonocyte colony stimulating factor, GM-CSF (Immunotools), and 0.05 μg/ml IL-4 (Peprotech). Cells were incubated for 24 hr in the above conditions at a 37°C and 5% CO2 incubator. Culture supernatant was supplemented the day after with a final concentration of 0.1 μg/ml prostaglandin E, 0.04 μg/ml TNF-α, 0.02 μg/ml IL-1β and 0.02 μg/ml IFNγ. This 24 hr incubation was used to activate the cells.
RNA transfection of human and mouse T cells
RNA constructs were prepared from DNA constructs in pGEM plasmids linearized with SpeI restriction enzyme at 37°C for 12 hrs. DNA precipitation, in vitro RNA transcription and in vitro poly-adenylation were performed exactly as described in the protocol of the mMESSAGE mMACHINE T7 ULTRA Transcription kit (Thermofisher Scientific). RNA was purified as per instructions of MEGAclear Transcription Clean-up kit (Thermofisher Scientific). RNA aliquots of 5 μg per PCR/vial were prepared and stored at - 80°C. RNA was only thawed once and used immediately. TCR RNA was prepared as described17.
Freshly isolated T cells (human or mouse) or T cells in culture (human or mouse) were harvested and washed three times in Opti-MEM media (LifeTechnologies). Depending on the expression levels to be achieved and the number of available cells, T cells were resuspended between 1x106-2.5x106 per 100 μl. Each 100 μl cell suspension was mixed with the RNA preparation and transferred to a Gene Pulser/Micropulser electroporation cuvette, 0.2 cm gap (BIO-RAD). Cells were electroporated at 300V, 2ms in an ECM 830 Square Wave Electroporation System (BTX). Cells were harvested using 0.5 ml of pre-warmed culture media at 37°C and cultured at ~1-2x106 cells/ml in warm media. Cells were incubated for at least 12 hr in 37°C, 5% CO2 incubator before testing expression levels and transfection efficiency. For TCR RNA transfection we used 5 μg of each TCRα and β chains and 5 μg of CD3-ζ chain for 2-5x106 T cells17. BTX cuvettes, 2mm gap (VWR) were used for TCR transfection. Two TCR constructs were used: the 6F9 TCR that recognizes an HLA-DPB1*04:01-restricted MAGE-A3 melanoma antigen43 and the 1G4 TCR that recognizes an HLA-A*0201-restricted NY-ESO-1 cancer-testis antigen44.
With regards to expressing the different forms of human CD2 (hCD2FL, hCD2TM, hCD2MT) in AND T cells, we found that the amounts needed to get a similar expression level of the mutants to the hCD2FL were significantly higher. We tried to rule out any technical factors that might result in the above variability. There is also the potential of an underlying biological factor controlling this that we do not necessarily understand at this point.
Supported Lipid Bilayer (SLB) in IBIDI chambers, immunostaining and TIRFM Imaging
SLB were prepared as described previously in45, 46. Briefly, glass coverslips (Nexterion) were cleaned in piranha solution, rinsed in water, dried, plasma cleaned and mounted onto six-channel chambers (Ibidi). Small unilamellar liposomes were prepared using an extruder (Avestin, 100 nm pore size filter) using 18:1 DGS-NTA(Ni), NTA-lipids, (Stratech, Avanti Polar Lipids, 790404C-AVL) that bind HIS-tag proteins, CapBio 18:1 Biotinyl Cap PE (870282C-AVL), CapBio-lipids, and 18:1 (Δ9-Cis) PC, DOPC-lipids, (850375C-AVL). Lipids were prepared in stock concentrations of 25% (4mM) NTA-lipids, 4mM CapBio-lipids and 4mM DOPC-lipids. For experiments NTA-lipids were used at a final concentration of 12.5% and CapBio-lipids were used at a concentration determined to give 30 molecules/μm2 of monobiotinylated unlabeled, AF568 or AF647-conjugated anti-CD3 Fab or monobiotinylated unlabeled peptide MHC. Channels in Ibidi chamber were covered with liposome mixture and after a 20 min incubation at RT that gives rise to the SLB, they were washed with human serum albumin (HSA)-supplemented HEPES buffered saline (HBS) supplemented with 1 mM CaCl2 and 2 mM MgCl2, referred to as HBS/HSA. SLBs were blocked with 5% casein in PBS containing 100 μM NiSO4, to saturate NTA sites and washed with HBS/HSA. Unlabeled/labeled streptavidin was added to channels coupling of biotinylated proteins to cap-biotinyl-containing lipids. Combination of the following reagents: Biotinylated UCHT1 Fab (30 molecules/μm2), peptide-HLA-A2, HLA-DP4 (30 molecules/μm2), ICAM-1-AF405 (200 molecules/μm2), CD58-AF488 (200 molecules/μm2), CD80-AF647 (200 molecules/μm2) ICOSL-AF647 (100 molecules/μm2), PD-L1-AF647(100 molecules/μm2), moth cytochrome c peptide (MCC)- I-EK (0.1 or 30 molecules/μm2), were then incubated onto the SLBs at specific concentrations to achieve the indicated molecule densities. Channels were washed and kept in HBS/HSA until ready to be used. 6F9 TCR transfected human CD4+ T cells or AND T cells were stained with anti-TCRβ (H57) Fab to allow tracking of TCR during imaging. For fixed experiments, cells were allowed to interact with SLBs for 15 min at 37°C, before a 10 min fixation with 2% PFA in PHEM buffer at 37°C. Cells were permeabilized with 0.1% Triton X-100 in HBS/HAS for 2 min at RT where required. Cells were blocked in 5% Casein containing 5% Donkey serum for 60 min at RT. Cells were stained overnight at 4°C with primary anti-pSFK, anti-PLCγ1, anti-pLAT prepared in 5% casein containing 5% donkey serum followed by washing and incubation in secondary donkey anti-rabbit antibody for 45 min at RT. Cells were immediately imaged or fixed as above for 5 min at RT and stored at 4°C. Imaging was performed on an Olympus IX83 inverted microscope equipped with a TIRF module and using the Olympus cellSens v1.15 (build 14760) software either at room temperature for fixed samples or at 37°C for live cell imaging. For live cell imaging, cells were imaged every ~4.5 seconds for various durations depending on the experiment. The instrument was equipped with an Olympus UAPON 150x TIRF N.A 1.45 objective, 405 nm, 488 nm, 561 nm and 640 nm laser lines and Photomertrics Evolve delta EMCCD camera. In live imaging experiments, SLBs were transferred to a pre-heated incubator on top of the TIRF microscope and cells were added in the well.
Preparation of SLBs for high throughput imaging in 96-well plate
96-well glass bottom plates (Brooks Life Science Systems) were incubated with 1% Hellmanex III in 50% isopropanol overnight at room temperature (RT). The wells were then rinsed with 5 ml milliQ water and dried by vacuum suction. To remove residual water, the plates were centrifuged upside down for 1 min at 200 rpm and dried with nitrogen gas. The wells were then cleaned with 3M NaOH for 1 hour at RT, rinsed with 5 ml milliQ water and dried as previously described. 50μl 12.5% NTA lipids in DOPC were then loaded per well and the plate incubated for 20 min at RT. Each well of the plate was then manually washed with 2 ml 0.1% BSA in HBS (20 mM HEPES, 137 mM NaCl, 1.7 mM KCl, 0.7 mM Na2PO4, 5 mM glucose, 2 mM MgCl2, 1 mM CaCl2, pH 7.2), incubated in 400 μl blocking solution (2% BSA in HBS) and put on a plate shaker for 3 min at 300 rpm. The plates were then incubated for 20 min at RT before they were washed with 2 ml 0.1% BSA in HBS.
Each well was loaded with 100 μl of a protein mix containing one of the three following combinations: 1. ICAM-1-AF405, UCHT1-AF488 and CD80-AF568, 2. ICAM-1-AF405, UCHT1-AF488, CD58-AD568 and SLAMF1-AF647 or 3. ICAM-1-AF405, UCHT1-AF488, CD58-AF568 and PVR-AF647. The protein concentrations were calibrated by flow cytometry to yield a density of 200 molecules/μm2 ICAM-1, 30 molecules/μm2 UCHT1, and 200 molecules/μm2 CD80, CD58, SLAMF1 and PVR. Proteins and lipids were then mixed for 3 min at 300 rpm and incubated for 30 min at RT. Each well was subsequently washed with 2 ml 0.1% BSA in HBS. After adding the cells to the bilayers the plates were centrifuged at 600 rpm for 1 min and incubated for 15 minutes at 37°C. Cells were fixed with 200 μl 4% PFA for 10 min at RT then the wells were washed with 2 ml 0.1% BSA in HBS and. Following fixation, the cells were imaged with an IN Cell Analyzer 6000 (GE Healthcare) with a 40x (0.75 NA) objective.
Cell conjugates and imaging
CF996 EBV-transformed B cells were stained with 0.5 μM cell trace violet (CTV, Thermofisher) by incubation at 37°C, 5% CO2 for 2-5min or 10-15 min. 10% FBS was added to quench dye and cells washed twice in media. MAGE-A3243-258 or NY-ESO-9V157-165 loaded CF996 EBV-transformed B-cells were conjugated with TCR transfected CD4+ and CD8+ T cells at 1:1 ratio, in round bottom tubes for 25-30 min at 37°C. Conjugates were transferred onto a poly-L-lysine coated μ-slide 8-well glass bottom chambers and fixed with 4% paraformaldehyde for 10 min at 37°C. Washing steps with PBS/0.5%BSA were performed in-between each of the following steps. Cells were washed, permeabilized for 4-5 min with 0.1% Triton-X100 prepared in PBS/0.5%BSA for 10 min and blocked with PBS/5%BSA/5% donkey serum for 1hr at RT. Cells were washed and incubated with unlabeled, non-blocking mouse anti-CD2 (CD2.1) in PBS/5%BSA, overnight at 4°C. Cells were washed and incubated in secondary antibody, AF647-conjugated donkey anti-mouse (Jackson ImmunoResearch Laboratories) for 45 min at RT. Cells were washed and fixed with 2% PFA for 5 min at RT. Cells were washed and incubated in FITC-conjugated anti-CD11a (TS2/4) in PBS/0.5%BSA for 1hr, at RT. Cells were washed and fixed wit 2% PFA for 5 min at RT. Finally, cells were loaded with Vectashield antifade mounting medium (H-1000) to minimize the movement of cell conjugates during imaging. Imaging was performed on a Zeiss inverted LSM 880 AiryScan confocal microscope in xyz dimensions in the Airyscan mode. We acquired the Airyscan data using the ZEISS ZEN software used to operate the microscope and the Airyscan settings were used in the automated mode. The instrument was equipped with a Plan Apochromat, 63x, NA 1.40 oil objective.
Flow cytometry analysis to determine ligand and receptor expression levels
1x106 blood mononuclear cells, PBMCs, 1×106 cells from single cell suspensions of human tissue (tumor or NA) or 105 cells (T cells, CF996 B-cell line, monocyte or DCs) were stained in 96-well U-bottom plates. For staining PBMCs, single cell suspensions from tissue, monocytes, DC and B cells, an Fc block step was performed by re-suspending cells in 50μl of Fc block prepared in PBS/0.5%BSA (room temperature) using 5μl of Human Trustain FcX (Biolegend) per sample i.e. 5μl/50μl of PBS/0.5%BSA. Cell were incubated at room temperature for 10 min. Cells were stained with an antibody mix prepared in ice cold PBS/0.5%BSA (and Fc block where appropriate) and incubated on ice, in the dark, for 15-20 min. Cells were then ready to be analyzed. Cells were analyzed on an LSRII or Fortessa machine using FACSDIVA 8.01 software. Analysis of acquired data was performed on FlowJo 10.4.2 and GraphPad Prism 7. For absolute quantification of CD58, CD2 and PD-1 expression, the Quantum Simply Cellular antimouse IgG kit was used (Bangs Laboratories, see further details in next section). We counted two CD2 molecules for each anti-CD2 IgG detected as we have previously found that the anti-CD2 mAbs bind bivalently at 10 μg/ml46. We applied the same for CD58 and PD-1 quantification. For CD58 densities on the B-cell line, we determined a mean surface area 277.6 μm2 using the CASY cell counter and analyzer.
Determining absolute molecular densities on SLB-coated silica beads and cells
Silica microspheres (SS05003/SS05N) (Bangs Laboratories) were used for SLB preparation. In order to simulate the surface area used in our SLBs prepared in 6-well chamber IBIDI slides, we used the number of beads that gives us the same surface area as the surface area covered by the SLBs in Ibidi channels. Silica bead vial was vortexed briefly to uniformly re-suspend silica beads. The required volume of beads was transferred to a clean 1.5ml Eppendorf tube and 1 ml of PBS was added to wash the beads by spinning beads down. This was repeated twice. Beads were then incubated in the required mixture of lipids as described in ‘Supported Lipid Bilayer (SLB) in IBIDI chambers, immune-staining and TIRFM Imaging’ and the same steps were performed from this point onwards. If titration of biotinylated proteins was used, cap-bio lipids were titrated instead of the protein itself, hence silica bead with different lipid mixtures were prepared and processed separately. If biotinylated proteins were titrated, streptavidin, at saturating concentrations, was added to the silica beads for 10 min at RT. His-tag proteins were serially diluted and added in 100 μl of a protein of interest. Proteins were incubated with silica beads for 20 min at RT on a plate shaker to avoid beads from settling to bottom of well. If proteins were already fluorescently-tagged with a known dye-protein ratio then the SLB-covered beads were resuspended in 200μl of flow cell buffer and run on a flow cytometry machine (LSII or FORTESSA) along with MESF standard beads, Quantum Alexa Fluor 488 MESF or Quantum Alexa Fluor 647 MESF (Bangs Laboratories). If unlabeled proteins were tested, silica beads were stained with saturating concentration of antibodies against the specific ligand reconstituted on the SLB-coated beads. Silica beads with no protein or DOPC-covered silica beads were prepared as controls. MESF standard beads were used to translate the mean fluorescence intensity of protein-coated SLB-silica beads to number of dye per bead. Then using the dye-protein ratio of the ligand tested, we calculated the mean number of ligands per beads. The surface area of a bead was known hence the density of ligands was calculated by dividing the mean number of ligands per bead by the surface area of the bead. If ligands were detected with anti-ligand mAb, then, we used either the dye-protein ratio of the mAb and assumed a 1:2 ratio for mAb:ligand to determine the mean number of ligand per bead. All calculations were performed in Microsoft Excel. If the dye-protein ratio of antibody was not known, the Quantum Simply Cellular anti-mouse IgG kit was used to make a calibration standard curve (Bangs Laboratories, 815).
We found that the Quantum Simply Cellular anti-mouse IgG kit overestimates the antibody binding capacity of the included beads for monoclonal antibodies perhaps due to isotype specific binding by the capture antibodies and hence this resulted in systematic underestimation of specific activity. After communication with the supplier, we were informed the Quantum Simply Cellular anti-mouse IgG kit beads were calibrated using polyclonal mouse IgG. The supplier assumes that monoclonal antibodies of any isotype would be bind with equal capacity to Quantum Simply Cellular anti-mouse IgG beads; but the supplier didn’t confirm this experimentally. After some inconsistent results we decided to experimentally test this. We cross-tested the AF647 MESF beads against the Quantum Simply Cellular anti-mouse IgG kit from the same supplier (Bangs) using samples of directly conjugated Alexa Fluor 647 conjugated mAb of different isotypes with spectroscopically determined dye to protein ratios. The MESF beads were used to generate a standard curve for number of dye molecules and the corresponding mean fluorescent intensity they produce on a flow cytometer. This allowed us to determine the mean number of dye molecules on each bead population of the Quantum Simply Cellular anti-mouse IgG kit. Knowing the mean number of dye molecules on each bead population and the dye to protein ratio of each mAb allowed us to calculate the number of antibodies captured by each Quantum Simply Cellular anti-mouse IgG bead population directly for different IgG isotypes under saturating conditions. We determined that the binding capacity of the Quantum Simply Cellular anti-mouse IgG kit was overestimated by an average of 5.6-fold for any given mouse IgG isotype and we applied the appropriate correction depending upon the isotype to our unknown conjugated mAb for absolute quantification. Given that this is a large correction that will likely impact biological conclusions, we suggest that investigators who must use the Quantum Simply Cellular anti-mouse IgG kit, or similar kits, for absolute quantification perform their own testing for the capacity for relevant isotypes, using this traceable approach. We have previously verified by comparison with binding of radioiodinated antibodies that the 488 and 647 MESF beads are accurate standards.
Processing of CRC, EndoC and OC samples
CRC tissue was collected from colorectal cancer patients that underwent colectomy at the Oxford University Hospitals NHS Foundation Trust. Informed consent was obtained from all patients involved and the samples were collected under the Ethics 11/YH/0020 and16/YH/0247 for the BRC Oxford GI Biobank - supported by NIHR Biomedical Research Center, Oxford. The EndoC and OC cases in this study were recruited under the Gynecological Oncology Targeted Therapy Study 01 (GO-Target-01, research ethics approval #11-SC-0014 supported by the University of Oxford, (Committee: South Central - Berkshir Research Ethic Committee). All participants involved in this study were appropriately informed. In total, we collected uterine curettage samples from six patients with EndoC (Ethical numbers: 15091, 15092, 15096, 15100, 15103,15104) and 5 ovarian and peritoneal biopsies from 5 patients with OC (Ethical numbers: 11581, 11582, 11583, 11584) (Supplementary Table 5).
CRC tumor tissue was cut into small pieces and digested with Collagenase A, 1 mg/ml (Sigma Aldrich) prepared in ADF+++ (DMEM/F12, 200mM GLutamax, 1M HEPES and 1× PenStrep) on a shaker at 37 °C, shaking for 1 hr. At the end of 1 hr incubation, the digested tissue was passed through a 70 μm mesh filter sequentially to generate a single cell suspension. Cells were washed in 1XHBSS/1%PenStrep. Red blood cells lysis was performed and the cells were counted for downstream surface staining and flow cytometric analysis. EndoC and OC samples were processed using the Tumor Dissociation Kit (human, Miltenyi, 130-095-929, without the kit hardware) i.e. in brief, tissue was cut into small pieces, transferred into 10 ml of media, as per kit instructions for soft tissue and incubated on a shaker for 1 hr at at 37 °C. Tissue was then mashed over a 100 μm filter, followed by red blood cell lysis, washing and subsequent processing for staining for flow cytometry analysis.
NCBI Gene Expression Omnibus single cell sequencing data
The NCBI Gene Expression Omnibus data was obtained from the NCBI GEO series outlined in table below. These data are publically available for the scientific research community and comprises the transcriptomic profiles of single T cells from colorectal cancer (mixed cohort of colon and rectal cancer patients), hepatocellular carcinoma and non-small cell lung carcinoma patients in the form of transcript counts data (normalized counts data was available for all datasets except for GSE99254). Un-normalized counts data was normalized by performing a log 2 + 1 transformation in R version 3.6.0. Ethical approval for the data was available from the National Institute of Health (NIH). The European Genome-phenome Archive (EGA) provided raw data for three of the cohorts under accession numbers EGAS00001002430 (GSE99254), EGAS00001002791 (GSE108989) and EGAS00001002072 (GSE98638).
TCGA human cohort data
The human cohort data was obtained from The Cancer Genome Atlas (TCGA), which is publically available for the scientific research community, and comprises the transcriptomic profiles of cancer patients in the form of RSEM count data at the mean. Criteria for inclusion in analyses were CRC patients with colon adenocarcinoma where mRNA sequencing data was available. This also allowed us to directly compare the results with the METALLIC cohort (see below) that consists of solely colon cancer patients. In total, the transcriptomic profiles of 255 colon cancers were taken for downstream analyses. Ethical approval for the data was obtained from the National Institute of Health (NIH). Expression data from the TCGA colorectal cancer cohort was obtained directly from the Broad Institute website (https://gdac.broadinstitute.org).
Metastatic lymph nodes in colorectal cancer (METALLIC) cohort
Metastatic lymph nodes in colorectal cancer (METALLIC) cohort biospecimens were collected from newly diagnosed patients with stage III sporadic colon adenocarcinoma who had undergone surgical resection and had received no prior treatment for their disease at the Oxford University Hospitals between March 2012 to May 2015 (n=148). All patients had given consent for this study and this was performed under REC reference 09/H0606/5+5. The research project was titled “Metastasis Disease Representation - a transcriptomics approach”, and ethical approval for the project was given by Oxford Radcliffe Biobank, Nuffield Division of Laboratory Sciences, University of Oxford and NRES Committee South Central - Oxford C. Cases were staged according to the European Society of Medical Oncology (ESMO) guidelines. Patients were excluded from this analysis if there was a previous history of CRC, any predisposing conditions for CRC (such as IBD, polyposis syndromes), patients presenting with synchronous CRC lesions or if there was evidence of inherited CRC predisposition syndromes.
Data analysis – Data normalization and sequencing run adjustment
Single cell sequencing data from the NCBI gene expression omnibus series GSE108989 (Colorectal)29, GSE72056 (Melanoma)47, GSE98638 (Liver)30 and GSE99254 (Lung)28 were processed and normalized as previously described. In instances where normalized counts data were not available, data were normalized by performing a log 2 + 1 transformation in R version 3.6.0. Prior to analysis of a combined dataset comprising GSE108989 (Colorectal), GSE72056 (Melanoma), GSE98638 (Liver) and GSE99254 (Lung), sequencing run adjustment was performed utilizing the Combat() function of the ‘car’ and ‘sva’ R packages48.
Data analysis - CD8 exhausted T cell subpopulations
Gene expression of different CD8 exhausted T cell subpopulations in the count data from the TCGA and METALLIC cohorts was analyzed using publically available genesets from the molecular signatures database (MSigDB) maintained by the Broad Institute (GSE 41867 genesets). Gene expression of different cell subpopulations was assessed from transcriptomic data by calculating a mean value for expression of each geneset in each sample. Similarly, single cell count data from the NCBI gene expression omnibus series GSE108989 (Colorectal), GSE98638 (Liver) and GSE99254 (Lung) were analyzed using the publically available T cell exhaustion gene signature from the molecular signatures database (MSigDB) maintained by the Broad Institute (GSE 41867). T cell exhaustion signature expression for individual cells was determined from the single cell transcriptomic dataset by calculating a geometric mean value of the exhaustion signature.
Statistical analyses were performed for each CD8 exhausted T cell geneset in R version 3.6.0. Linear regression models, generated using the glm function in R, were utilized for testing for statistical significance between CD8 exhausted T cell geneset expression and CD2 gene expression. Pearson and Spearman correlations were calculated using the corr() function in R. Marginal plots were generated using the ‘ggplot2’ and ‘ggExtra’ r packages (ggMarginal).
Image Analysis
TIRFM image or IN Cell analyzer image analysis was performed in Fiji (ImageJ, version 2.0.0) and analysis presented in GraphPad Prism 7. For quantification analysis, background signal was subtracted from IS image, by acquiring the same square area of a nearby position void of cells; the area was just big enough to fit the synapsed cell and same square area was used for all cells and conditions within the same experiment. In effect the analysis was for the quantification of total signal accumulating in the synapsed cell. For Airyscan© confocal images initial processing was carried out in Zen© 2.1 SP3 software (Zeiss), 3D rendering was performed in Imaris© software (Bitplane), and final panel preparation in Fiji© software (http://Fiji/sc).
Statistical Analysis
All statistical analysis and regression lines were performed using GraphPad Prism 7 (Version 7.0d); the slope of the regression lines was tested for being different than zero using GraphPrism’s default test. Comparison of the slopes of regression lines was performed in GraphPad prism using the built-in option that uses calculations following a method described in Chapter 18 of J Zar, Biostatistical Analysis, 2nd edition, Prentice-Hall, 1984. It is equivalent to analysis of covariance (ANCOVA). The remaining statistical tests are indicated in the figure legends and were two-tailed unless mentioned otherwise in the legend.
For the analysis methods and tools on transcriptomic data see above sections “Data analysis – CD8 exhausted T cell subpopulations”, “Data analysis – CD8 exhausted T cell subpopulations” and Supplementary Table 8.
Supplementary Material
Acknowledgements
We thank S. Davis, P.A. Van Der Merwe, O. Dushek, R. Owens (University of Oxford), for kindly providing plasmids and HLA-A2 pMHC monomers. We also thank M. Dumoux (The Rosalind Frankling Institute) for kindly providing plasmids M. H. Brown and S. Sivakumar for helpful discussion and feedback on experiments and writing of manuscript and all the Dustin lab members for their kind support. S. Balint for his help on some of the experiments. We thank S. Balint for maintaining the TIRF microscope, C. Laggerholm for access to and assistance with the Airyscan confocal microscope, and P. Cespedes for his contribution in methods development for quantification of surface molecules on T cells. The results published here are in part based upon data generated by the TCGA Research Network: https://www.cancer.gov/tcga.
A Kennedy Trust for Rheumatology (KTRR) Prize Studentship supported P.D. An UCB-Oxford Post-doctoral Fellowship supported E.A.S. A grant from The Research Council of Norway in conjunction with Marie Sklodowska-Curie Actions (275466) supported A.K. Wellcome Trust Principal Research Fellowship 100262Z/12/Z and a grant from KTRR supported M.L.D. KTRR supports the Kennedy Institute Microscopy Facility and the Wolfson Foundation supports the Weatherall Institute Microscopy Facility. A collaborative grant from the Human Frontiers Science Program supported E.A.S., A.S. and M.M.H. A.S. was supported by the German Research Foundation (DFG) and Collaborative Research Center (SFB 854) “Molecular Organization of Cellular Communication within the Immune System”. A fellowship from Philippe Foundation partially supported D.D. A Wellcome Trust Senior Research Fellowship 207537/Z/17/Z supported E.A.S., and M.K. NIHR Biomedical Research Centre, Oxford, supports the Oxford Gastro-Intestinal Biobank and the Oxford Inflammatory Bowel Disease Cohort study. The views expressed are those of the authors and not necessarily those of the NHS, the NIHR or the Department of Health. We acknowledge the contribution to this study made by the Oxford Centre for Histopathology Research and the Oxford Radcliffe Biobank, which are supported by the NIHR Oxford Biomedical Research Centre.
Footnotes
Author Contributions
P.D. conceptualized the project, designed and performed experiments, analyzed the data and co-wrote the manuscript. E.A.S. designed, performed and analyzed experiments and co-wrote the manuscript. S.V. prepared and performed experiments and maintained critical infrastructure. K.K. assisted in and acquired confocal microscopy experiments. A.K. performed and analyzed experiments. S.M., M.F., E.M., prepared single cell suspensions from CRC tissue. E.A.S prepared single cell suspensions from EndoC and OC patient samples. S.M. performed transcriptional analysis. P.D., E.A.S, performed the staining and acquisition experiments of CRC tissues. E.A.S performed the staining and acquisition experiments of EndoC and OC tissues. J.A., H.R., S.Y., S.V., V.M., M.K. prepared essential reagents for experiments. V.M. designed image analysis software and trained P.D. in its use. L.L. and T.S. performed transcriptional analysis of CRC, HCC, NSCLC and melanoma cohorts. The O.IBDC.I. provided access to CRC tissue and clinical data. M.M. provided EndoC and OC samples, clinical data and contributed to discussion of patient data. N.W. and A.A.A. provided access to Endo and OC samples. P.D., E.A.S., V.M., D.D., A.S., M.M.H. and M.L.D. made intellectual contributions to the project through regular discussions. D.D. provided training to P.D., conceptualized and designed experiments. M.L.D. supervised the research and facilitated collaboration. P.D. drafted the manuscript and E.A.S., V.M., S.M., D.D., and M.L.D. contributed to writing and editing.
Competing Interests
The authors declare no competing interests
Contributor Information
TGU Investigators:
Philip Allan, Timothy Ambrose, Carolina Arancibia, Adam Bailey, Ellie Barnes, Elizabeth Bird-Lieberman, Jan Bornschein, Oliver Brain, Barbara Braden, Jane Collier, Jeremy Cobbold, Emma Culver, James East, Lucy Howarth, Paul Klenerman, Simon Leedham, Rebecca Palmer, Michael Pavlides, Fiona Powrie, Astor Rodrigues, Jack Satsangi, Alison Simmons, Peter Sullivan, Holm Uhlig, and Alissa Walsh
Data availability
scRNA seq data are available in: see Supplementary Table 7 in NCBI Gene Expression Omnibus single cell sequencing data section. Additional data and information that support the findings of this study are available from the corresponding author upon reasonable request. Source data are provided with this paper.
Code availability
No custom code or mathematical algorithm was used when acquiring or analyzing data included in this study.
References
- 1.Sanchez-Madrid F, et al. Three distinct antigens associated with human T-lymphocyte-mediated cytolysis: LFA-1, LFA-2, and LFA-3. Proc Natl Acad Sci U S A. 1982;79:7489–7493. doi: 10.1073/pnas.79.23.7489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sanders ME, et al. Human memory T lymphocytes express increased levels of three cell adhesion molecules (LFA-3, CD2, and LFA-1) and three other molecules (UCHL1, CDw29, and Pgp-1) and have enhanced IFN-gamma production. J Immunol. 1988;140:1401–1407. [PubMed] [Google Scholar]
- 3.Davis SJ, van der Merwe PA. The kinetic-segregation model: TCR triggering and beyond. Nature immunology. 2006;7:803–809. doi: 10.1038/ni1369. [DOI] [PubMed] [Google Scholar]
- 4.Carmo AM, Mason DW, Beyers AD. Physical association of the cytoplasmic domain of CD2 with the tyrosine kinases p56lck and p59fyn. European journal of immunology. 1993;23:2196–2201. doi: 10.1002/eji.1830230922. [DOI] [PubMed] [Google Scholar]
- 5.Dustin ML, et al. A novel adapter protein orchestrates receptor patterning and cytoskeletal polarity in T cell contacts. Cell. 1998;94:667–677. doi: 10.1016/s0092-8674(00)81608-6. [DOI] [PubMed] [Google Scholar]
- 6.Espagnolle N, et al. CD2 and TCR synergize for the activation of phospholipase Cgamma1/calcium pathway at the immunological synapse. International immunology. 2007;19:239–248. doi: 10.1093/intimm/dxl141. [DOI] [PubMed] [Google Scholar]
- 7.Freiberg BA, et al. Staging and resetting T cell activation in SMACs. Nature immunology. 2002;3:911–917. doi: 10.1038/ni836. [DOI] [PubMed] [Google Scholar]
- 8.Saliba DG, et al. Composition and structure of synaptic ectosomes exporting antigen receptor linked to functional CD40 ligand from helper T-cells. Elife. 2019;8:600551. doi: 10.7554/eLife.47528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dustin ML, Ferguson LM, Chan PY, Springer TA, Golan DE. Visualization of CD2 interaction with LFA-3 and determination of the two-dimensional dissociation constant for adhesion receptors in a contact area. The Journal of cell biology. 1996;132:465–474. doi: 10.1083/jcb.132.3.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kaizuka Y, Douglass AD, Vardhana S, Dustin ML, Vale RD. The coreceptor CD2 uses plasma membrane microdomains to transduce signals in T cells. The Journal of cell biology. 2009;185:521–534. doi: 10.1083/jcb.200809136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zaru R, Cameron TO, Stern LJ, Muller S, Valitutti S. Cutting edge: TCR engagement and triggering in the absence of large-scale molecular segregation at the T cell-APC contact site. J Immunol. 2002;168:4287–4291. doi: 10.4049/jimmunol.168.9.4287. [DOI] [PubMed] [Google Scholar]
- 12.De Jager PL, et al. The role of the CD58 locus in multiple sclerosis. Proc Natl Acad Sci U S A. 2009;106:5264–5269. doi: 10.1073/pnas.0813310106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Raychaudhuri S, et al. Genetic variants at CD28, PRDM1 and CD2/CD58 are associated with rheumatoid arthritis risk. Nature genetics. 2009;41:1313–1318. doi: 10.1038/ng.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McKinney EF, Lee JC, Jayne DR, Lyons PA, Smith KG. T-cell exhaustion, co-stimulation and clinical outcome in autoimmunity and infection. Nature. 2015;523:612–616. doi: 10.1038/nature14468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Abdul Razak FR, Diepstra A, Visser L, van den Berg A. CD58 mutations are common in Hodgkin lymphoma cell lines and loss of CD58 expression in tumor cells occurs in Hodgkin lymphoma patients who relapse. Genes Immun. 2016;17:363–366. doi: 10.1038/gene.2016.30. [DOI] [PubMed] [Google Scholar]
- 16.Koneru M, Monu N, Schaer D, Barletta J, Frey AB. Defective adhesion in tumor infiltrating CD8+ T cells. J Immunol. 2006;176:6103–6111. doi: 10.4049/jimmunol.176.10.6103. [DOI] [PubMed] [Google Scholar]
- 17.Abu-Shah E, D P, Mayya V, Balint S, Kutuzov M, Dushek O, Dustin ML. A tissue-like platform for studying engineered primary human immune cell interactions. bioRxiv. 2019 doi: 10.7554/eLife.48221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vendrame E, et al. TIGIT is upregulated by HIV-1 infection and marks a highly functional adaptive and mature subset of natural killer cells. AIDS. 2020 doi: 10.1097/QAD.0000000000002488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ramsbottom KM, et al. Cutting edge: DNAX accessory molecule 1-deficient CD8+ T cells display immunological synapse defects that impair antitumor immunity. J Immunol. 2014;192:553–557. doi: 10.4049/jimmunol.1302197. [DOI] [PubMed] [Google Scholar]
- 20.Aversa G, Chang CC, Carballido JM, Cocks BG, de Vries JE. Engagement of the signaling lymphocytic activation molecule (SLAM) on activated T cells results in IL-2-independent, cyclosporin A-sensitive T cell proliferation and IFN-gamma production. J Immunol. 1997;158:4036–4044. [PubMed] [Google Scholar]
- 21.Colin-York H, Kumari S, Barbieri L, Cords L, Fritzsche M. Distinct actin cytoskeleton behaviour in primary and immortalised T-cells. J CellSci. 2019;133 doi: 10.1242/jcs.232322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hui E, et al. T cell costimulatory receptor CD28 is a primary target for PD-1-mediated inhibition. Science (New York NY) 2017;355:1428–1433. doi: 10.1126/science.aaf1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Clarkson NG, Brown MH. Inhibition and activation by CD244 depends on CD2 and phospholipase C-gamma1. J Biol Chem. 2009;284:24725–24734. doi: 10.1074/jbc.M109.028209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kaye J, et al. Selective development of CD4+ T cells in transgenic mice expressing a class II MHC-restricted antigen receptor. Nature. 1989;341:746–749. doi: 10.1038/341746a0. [DOI] [PubMed] [Google Scholar]
- 25.Pages F, et al. International validation of the consensus Immunoscore for the classification of colon cancer: a prognostic and accuracy study. Lancet. 2018;391:2128–2139. doi: 10.1016/S0140-6736(18)30789-X. [DOI] [PubMed] [Google Scholar]
- 26.Huyghe N, Baldin P, Van den Eynde M. Immunotherapy with immune checkpoint inhibitors in colorectal cancer: what is the future beyond deficient mismatch-repair tumours? Gastroenterol Rep (Oxf) 2020;8:11–24. doi: 10.1093/gastro/goz061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chirica M, et al. Phenotypic analysis of T cells infiltrating colon cancers: Correlations with oncogenetic status. Oncoimmunology. 2015;4:e1016698. doi: 10.1080/2162402X.2015.1016698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guo X, et al. Global characterization of T cells in non-small-cell lung cancer by single-cell sequencing. Nature medicine. 2018;24:978–985. doi: 10.1038/s41591-018-0045-3. [DOI] [PubMed] [Google Scholar]
- 29.Zhang L, et al. Lineage tracking reveals dynamic relationships of T cells in colorectal cancer. Nature. 2018;564:268–272. doi: 10.1038/s41586-018-0694-x. [DOI] [PubMed] [Google Scholar]
- 30.Zheng C, et al. Landscape of Infiltrating T Cells in Liver Cancer Revealed by Single-Cell Sequencing. Cell. 2017;169:1342–1356.e1316. doi: 10.1016/j.cell.2017.05.035. [DOI] [PubMed] [Google Scholar]
- 31.Grakoui A, et al. The immunological synapse: a molecular machine controlling T cell activation. Science. 1999;285:221–227. [PubMed] [Google Scholar]
- 32.Valitutti S, Dessing M, Aktories K, Gallati H, Lanzavecchia A. Sustained signaling leading to T cell activation results from prolonged T cell receptor occupancy. Role of T cell actin cytoskeleton. The Journal of experimental medicine. 1995;181:577–584. doi: 10.1084/jem.181.2.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kumari S, et al. Actin foci facilitate activation of the phospholipase C-gamma in primary T lymphocytes via the WASP pathway. Elife. 2015;4 doi: 10.7554/eLife.04953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shen K, Thomas VK, Dustin ML, Kam LC. Micropatterning of costimulatory ligands enhances CD4+ T cell function. Proc Natl Acad Sci U S A. 2008;105:7791–7796. doi: 10.1073/pnas.0710295105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cai E, et al. Visualizing dynamic microvillar search and stabilization during ligand detection by T cells. Science. 2017;356 doi: 10.1126/science.aal3118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dustin ML. Adhesive bond dynamics in contacts between T lymphocytes and glass-supported planar bilayers reconstituted with the immunoglobulin-related adhesion molecule CD58. J Biol Chem. 1997;272:15782–15788. doi: 10.1074/jbc.272.25.15782. [DOI] [PubMed] [Google Scholar]
- 37.Siokis A, et al. In silico characterization of mechanisms positioning costimulatory and checkpoint complexes in immune synapses. bioRxiv. 2020 doi: 10.1016/j.isci.2021.103100. 2020.2001.2016.908723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Douglass AD, Vale RD. Single-Molecule Microscopy Reveals Plasma Membrane Microdomains Created by Protein-Protein Networks that Exclude or Trap Signaling Molecules in T Cells. Cell. 2005;121:937–950. doi: 10.1016/j.cell.2005.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hutchings NJ, Clarkson N, Chalkley R, Barclay AN, Brown MH. Linking the T cell surface protein CD2 to the actin-capping protein CAPZ via CMS and CIN85. J Biol Chem. 2003;278:22396–22403. doi: 10.1074/jbc.M302540200. [DOI] [PubMed] [Google Scholar]
- 40.Wei F, et al. Strength of PD-1 signaling differentially affects T-cell effector functions. Proceedings of the National Academy of Sciences of the United States of America; 2013. pp. E2480–2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lo DJ, et al. Selective targeting of human alloresponsive CD8+ effector memory T cells based on CD2 expression. American journal of transplantation : official journal of the American Society of Transplantation and the American Society of Transplant Surgeons. 2011;11:22–33. doi: 10.1111/j.1600-6143.2010.03317.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Leitner J, Herndler-Brandstetter D, Zlabinger GJ, Grubeck-Loebenstein B, Steinberger P. CD58/CD2 Is the Primary Costimulatory Pathway in Human CD28-CD8+ T Cells. J Immunol. 2015;195:477–487. doi: 10.4049/jimmunol.1401917. [DOI] [PubMed] [Google Scholar]
- 43.Yao X, et al. Isolation and Characterization of an HLA-DPB1*04: 01-restricted MAGE-A3 T-Cell Receptor for Cancer Immunotherapy. Journal of immunotherapy (Hagerstown, Md : 1997) 2016;39:191–201. doi: 10.1097/CJI.0000000000000123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li Y, et al. Directed evolution of human T-cell receptors with picomolar affinities by phage display. Nature biotechnology. 2005;23:349–354. doi: 10.1038/nbt1070. [DOI] [PubMed] [Google Scholar]
- 45.Choudhuri K, et al. Polarized release of T-cell-receptor-enriched microvesicles at the immunological synapse. Nature. 2014;507:118–123. doi: 10.1038/nature12951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dustin ML, Starr T, Varma R, Thomas VK. Supported planar bilayers for study of the immunological synapse. Current protocols in immunology. 2007 doi: 10.1002/0471142735.im1813s76. Chapter 18, Unit 18.13. [DOI] [PubMed] [Google Scholar]
- 47.Tirosh I, et al. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science (New York, NY) 2016;352:189–196. doi: 10.1126/science.aad0501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics (Oxford, England) 2007;8:118–127. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
scRNA seq data are available in: see Supplementary Table 7 in NCBI Gene Expression Omnibus single cell sequencing data section. Additional data and information that support the findings of this study are available from the corresponding author upon reasonable request. Source data are provided with this paper.
No custom code or mathematical algorithm was used when acquiring or analyzing data included in this study.