Abstract
Herpes simplex virus type 1 (HSV-1) is a leading cause of infectious blindness. Current treatments for HSV-1 do not eliminate the virus from the site of infection or latent reservoirs in the trigeminal ganglia. Here, we target HSV-1 genomes directly using mRNA-carrying lentiviral particles that simultaneously deliver SpCas9 mRNA and viral gene-targeting gRNAs (designated HSV-1-erasing lentiviral particles, HELP). We show that HELP efficiently blocks HSV-1 replication and the occurrence of herpetic stromal keratitis (HSK) in three different infection models. HELP was capable of eliminating the virus reservoir via retrograde transportation from corneas to trigeminal ganglia. Additionally, HELP inhibited viral replication in human-derived corneas without causing off-targeting effects as determined by whole genome sequencing. These results support the potential clinical utility of HELP for treating refractory HSK.
Herpes simplex virus type I (HSV-1) is among the most common human viruses with 50-80% of the world population being seropositive1. It belongs to the alpha subfamily of herpesviruses, which are enveloped viruses carrying double-stranded DNA, and capable of establishing latent infections in sensory neurons2. HSV-1 infection can cause a wide variety of diseases including herpes simplex encephalitis, which has a high mortality if untreated3. HSV-1 infection in the cornea can cause herpetic stromal keratitis (HSK), which is the leading factor for infectious blindness4. After primary infection and production replication in corneal epithelium, HSV-1 is transported through ophthalmic nerves in a retrograde direction to trigeminal ganglia (TG) where the virus establishes a latent reservoir for lifetime5. Under certain conditions, including immunosuppression, the latent viruses in the TG can be re-activated, leading to the recurrence and aggravation of the disease. Typical blinding HSK develops subsequent to infection in the eye, at which point virus can often not be detected6. Most of the tissue damage occurring in human corneas during HSK is immune-mediated rather than direct viral cytopathic effect7. Globally, it is estimated that 1.5 million episodes of ocular HSV each year and 40,000 people get visual disability4.
Despite the high prevalence, currently, no vaccine is available for HSV infection8, 9. The first-line treatment option for HSV-1 infection is acyclovir (ACV). This compound was developed nearly half a century ago and analogues have subsequently been made, all targeting viral DNA polymerase. In specific patient groups, including immunocompromised and individuals receiving chronic antiviral prophylaxis drug resistance occurs frequently 10–12. Alternative strategies including small molecules that inhibit viral helicase-primase, antibodies, and peptides are still under development13–15. Recently, Jaishankar et al. reported BX795-a commonly used inhibitor of TANK-binding kinase 1- blocks HSV-1 infection in vivo by targeting Akt phosphorylation in the infected cells16. However, none of these strategies can remove the existing virus and modulate its reservoir in TG- therefore, incapable of preventing recurrence.
CRISPR targets genomes directly and has been very successful in treating genetic diseases in preclinical studies17–22. About two years ago, Food and Drug Administration (FDA) approved
CRISPR for phase I/II trial to treat β-thalassemia, sickle cell disease, and LCA-10 (ClinicalTials.gov: NCT04208529; NCT03745287; NCT03872479). Its therapeutic potential on infectious diseases is promising. Dash et. al demonstrated viral clearance in latent infectious reservoirs in HIV-1 infected humanized mice by combing antiviral prodrugs and CRISPR23.
However, to the best of our knowledge, no investigational new drug (IND) application has been registered for infectious diseases. These reflect the challenge of delivering CRISPR to infection sites, and especially to viral reservoirs24. One study delivered an HSV-1 targeting endonuclease using AAV in a mouse model of latent HSV infection, however, neither revealed a detectible loss of viral genome nor therapeutic efficacy25. Recently, the same group showed detectable elimination of latent genomes and therapeutic efficacy by using improved AAV vector and dual-meganuclease targeting the HSV genome26. So far, the anti-HSV activity of CRISPR has only been characterized in vitro and no existing studies have shown the therapeutic efficacy of CRISPR against HSK in vivo 27, 28.
In this study, we developed an HSV-1-erasing lentiviral particle (HELP) and showed its therapeutic efficacy in three different HSK models and human-derived corneas. Furthermore, we found HELP was capable of modulating the HSV-1 reservoir in the TG. The corneas were maintained in a healthy status after intracorneal injection of HELP as shown by a variety of clinical relevant assays. The Cas9 expression from HELP only lasted for 3 days in vivo and no off-targeting was detected in the coding regions of the murine and human genome. Taken together, our study supports clinical translation of HELP for treating refractory HSK, which has been resistant to conventional drugs and corneal transplantations.
Results
HELP blocks HSV-1 replication in vitro
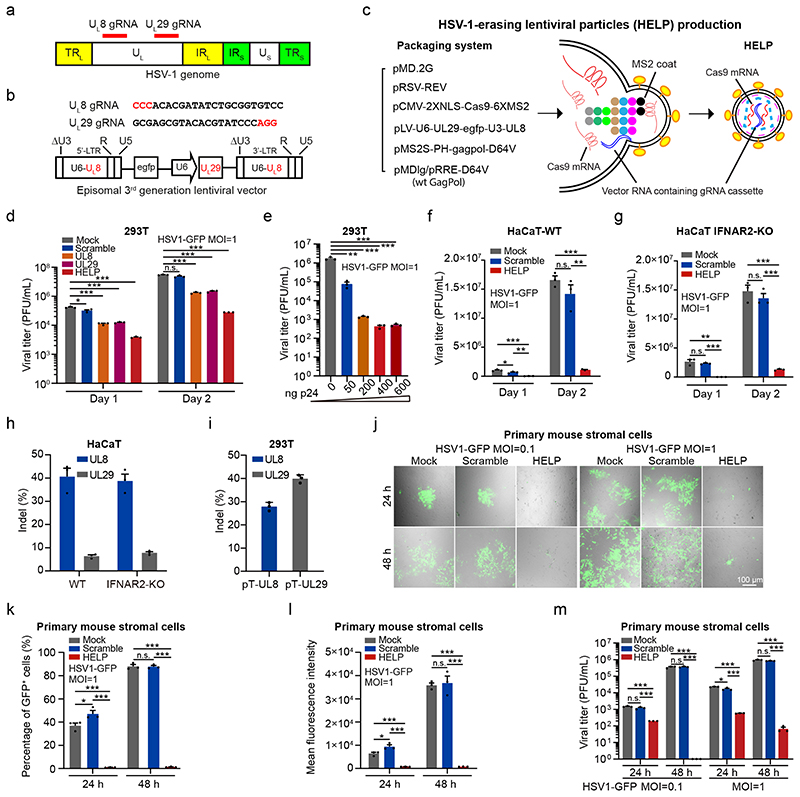
In this study, we designed a gRNA expression cassette targeting two essential genes of HSV-1 - UL8 and UL29 29, 30, simultaneously, and co-packaged with SpCas9 mRNA to mRNA-carrying lentiviral particle (mLP) via the specific binding of pac site-containing SpCas9 mRNA to bacteriophage-derived MS2 coat locating in the N-terminus of lentiviral Gag and GagPol polyproteins (Fig. 1a-c). The MS2 coat protein specifically recognizes and interacts with the pac site-containing SpCas9 mRNA, and co-packages them into the lentiviral particle during viral assembly. As the resulting lentiviral particle was designed to destroy the genome of HSV-1, we therefore designate it HSV-1-erasing lentiviral particles (HELP). The gRNA expression cassette is reverse transcribed and maintained as circular episomal DNA as an integration-defective lentiviral vector (IDLV) (Fig. 1b). As the UL8 gRNA is cloned into the ΔU3 region of the long terminal region (LTR), it will be copied from 3’-LTR to 5’-LTR during reverse transcription (Fig. 1b). We produced the HELP by co-transfection of 6 plasmids to 293T cells and harvested the particles by ultracentrifugation (Fig. 1c). As controls, we also produced mLP expressing single gRNA-either UL8, UL29, or scramble (non-targeting gRNA). To verify whether HELP was indeed capable of inhibiting HSV-1, 293T cells were transduced with HELP for 24 h and infected with HSV-1 (HSV1-GFP). The supernatants were harvested 1 day and 2 days after HSV-1 infection, respectively, and subjected to virus yield assay. We found inhibitory effects for all viral gene-targeting mLPs with the UL8/UL29 co-targeting HELP the most efficient (Fig. 1d and Supplementary Fig. 1). The averaged copy number of Cas9 mRNA in each HELP was 3.5 (Supplementary Fig. 2). Additionally, we conducted a dose-response experiment for HELP, which showed an increased level of virus inhibition and saturated at 400 ng p24 (Fig. 1e). We therefore chose HELP in all the subsequent experiments.
Figure 1. HELP blocks HSV-1 replication in vitro.
a, Schematic representation of the HSV-1 genome and gRNA loci. TRL, terminal repeat long; IRL, internal repeat long; UL, unique long; IRS, internal repeat short; TRS, terminal repeat short; US, unique short. b, The gRNA sequences and expression cassettes for HELP. c, Schematic illustration of HELP production. Coloured dots represent the main components of lentiviral Gag and GagPol polyproteins. Gag is composed of matrix (MA), capsid (CA), nucleocapsid (NC), whereas Pol consists of protease (PR), reverse transcriptase (RT) and integrase (IN). d-g, The anti-viral activity of HELP in cell lines. d, Mock vs. Scramble, UL8, UL29 and HELP, P=0.0220, 0.0003, 0.0003 and 0.0002, respectively, on day 1; P<0.0001 on day 2. e, Mock vs. 50 ng, P=0.0011; P=0.0009 for others. f, HELP vs. Mock and Scramble, P=0.0003 and 0.0019 on day 1; P<0.0001 and P=0.0011 on day 2. Mock vs. Scramble, P=0.0420 on day 1. g, HELP vs. Mock and Scramble, P=0.0013 and P<0.0001 on day 1; P=0.0002 and 0.0001 on day 2. h and i, TIDE analysis of indels in HSV1 genome and plasmid DNA. Viral DNA was from day 2 samples in f and g. j-m, The antiviral activity in primary mouse corneal stromal cells by confocal microscopy (j), flow cytometry (k and l) and PFU (m). k, Mock vs. HELP, P=0.0001 at 24 h; P<0.0001 for others. l, *P=0.0380, ***P=0.0005, ***P=0.0004, ***P<0.0001 and ***P=0.0003, left to right. m, ***P<0.0001, ***P=0.0010, ***P=0.0002, ***P<0.0001, *P=0.0236, ***P<0.0001, ***P=0.0006, ***P<0.0001, ***P<0.0001, left to right. Images are representative of three independent biological replicates in one experiment (j). Scale bars, 100 μm. The gating strategy is provided in Supplementary Fig. 20. Data and error bars represent mean ± SEM from three biologically independent replicates. Unpaired two-tailed Student’s t-tests. n.s.=non-significant.
The HSV-1 infection is sensitive to type I interferons (IFNs) induced by pathogen-associated molecular patterns (PAMPs) even in the absence of gene editing (Supplementary Fig. 3)31. To exclude the necessity of type I IFNs, here, we evaluated the antiviral activity of HELP in both wild-type and IFNAR2 knockout HaCaT cells. We found that HELP, but not the scrambled control, significantly inhibited HSV-1 in both cell lines (Fig. 1f, g). Furthermore, we analysed the UL8 and UL29 loci and found indel frequency about 40% for UL8 while only 7% for UL29 on average (Fig. 1h). The indels on UL29 were relatively low. As ICP8 (encoded by UL29) plays multifunctional roles in the viral life cycle including viral DNA synthesis, we reasoned mutations on UL29 make the virus unable to replicate and tend to be underestimated30. Indeed, when using plasmids containing UL8 and UL29 sequence as the targets, we obtained even higher indels for UL29 than UL8 gRNA (Fig. 1i). Notably, the antiviral activity of HELP is under-estimated using PCR-based indel analysis, since not all the cleavage outcomes, e.g. unrepaired double-strand breaks or large deletions, can be amplified (Supplementary Fig. 4). Also, we found HELP did not provoke innate immune sensing in contrast to HSV-1 strains, which were all sensed by THP-1 derived macrophages at MOI=1 and induced moderate, but significant IFN response (Supplementary Fig. 5). Together, these data suggest the HELP inhibits HSV-1 through DNA disruption, but not type I IFN-dependent innate immune response.
The corneal stroma is highly linked to keratitis recurrence32. The stroma is rich with nerve trunks that originate from the trigeminal ganglion where HSV-1 maintains latency33. Therefore, we explored whether HELP was functional in primary corneal stromal cells from mice. The primary stromal cells were transduced with a non-GFP version HELP for 24 h and then infected with HSV1-GFP. We found that HELP potently suppressed GFP expression as well as viral replication using both low and high MOI on either 24 h or 48 h post-infection, while the scrambled control did not show any protective effects (Fig. 1j-m).
HELP blocks HSV-1 infection of corneas and neurons in the prevention model
Persisting nuclease expression may bring additional risks. From a safety perspective, transient nuclease exposure is desired for CRISPR therapeutics. However, it is unclear if transient Cas9 expression can control HSK as HSV-1 propagates fast (about 18 h for the lytic replication cycle). It is difficult for the CRISPR machinery to remove every single HSV-1 genome. On the other hand, HSV-1 encounters harsh antiviral responses in vivo. Here, we hypothesize that reducing the viral load to a certain level is sufficient to control the virus in vivo. To verify this, we performed dose-response experiments of HSV-1 infection on scarified corneas of mice. Indeed, only when HSV-1 load was over 2x104 plaque-forming units (PFU), did the viability, weight change, and symptoms of keratitis developed (Supplementary Fig. 6).
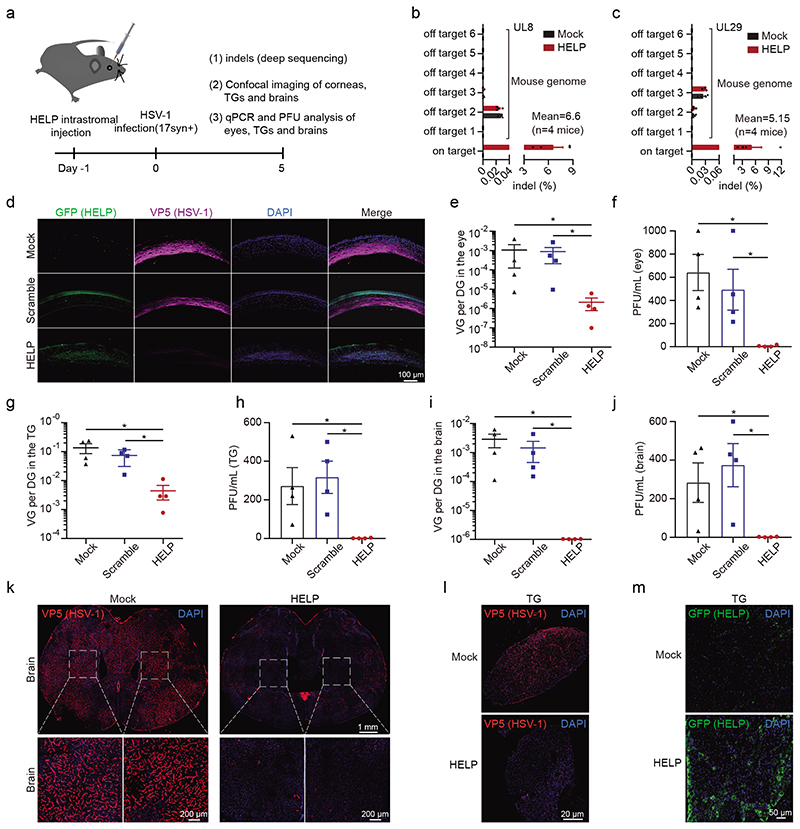
We then set out to investigate the potential of HELP as a novel HSK therapeutic in vivo. To identify the kinetics of HSV-1 infection in our HSK model, we visualized HSV-1 using confocal imagining and found the virus progressively disseminated from surficial to the deeper side of corneal stroma during the time course from 12 h to 8 days post-infection (Supplementary Fig. 7). The experimental set-up is illustrated in Fig. 2a. HELP was administrated by intrastromal injection to corneas 1 day before HSV-1 17syn+ infection (Supplementary Fig. 8). We first deep-sequenced the on-target activity of HELP on the HSV-1 genome and the off-target effects on the murine genome. The indels induced by HELP were approximately 7% on UL8 loci and 5% for UL29 loci while no off-targets were found for both gRNA (Fig. 2b and Fig. 2c). Notably, the Cas9 expression only lasted for 3 days both in vitro and in vivo, which might minimize the off-target activity of HELP (Supplementary Fig. 9). Next, we performed confocal imaging to access HSV-1 replication and HELP distribution in the corneas of mice, which was represented by viral capsid protein VP5 and GFP, respectively. We found HSV-1 was replicating actively in the corneal stroma in the mock and scrambled controls, while it was hardly detectable after HELP treatment (Fig. 2d). Accordingly, HELP was evenly distributed in all corneal structures-from epithelium and stroma to the endothelium (Fig. 2d). To assess whether HELP treatment blocks the transmission of HSV-1 from corneal epithelium to the peripheral and central nervous system (CNS), eyes, TGs, and brain samples from all the infected mice were harvested and examined for the copy number of HSV-1 genome and infectious viruses. In all samples, the viral load was significantly reduced after HELP treatment (Fig. 2e-2j). Additionally, we performed confocal imaging of the whole brain and TG. In agreement with the qPCR and PFU analysis, we found HELP diminished HSV-1 to an almost undetectable level in both the brain and TG (Fig. 2k-2l). Tissue distribution is an important safety index for in vivo gene therapy. Therefore, we evaluated the dissemination of HELP in the whole body and found HELP was highly restricted to the eyes, not other organs, including reproductive organs (Supplementary Fig. 10). Interestingly, we also detected HELP in the TG though it was factually injected in the corneas supporting retrograde delivery of CRISPR from neuron termini in the corneas to neuron body in the TG (Supplementary Fig. 10). This finding was further strengthened by the detection of HELP in the TG by confocal imaging (Fig. 2m).
Figure 2. HELP blocks HSV-1 infection of corneas and neurons in a prevention model.
a, Flow chart for evaluating antiviral effects of HELP in vivo. 100 ng p24 HELP, scramble mLP, or 2 μL PBS (Mock) were injected into corneas of mice by intrastromal injection. After 24 h, the mice were infected with HSV-1 17syn+ (2×106 PFU/eye). b and c, Deep sequencing analysis of on-target effects in HSV-1 and off-target effects in the mouse genome. n=4 mice. d, Confocal imaging of HSV-1 and HELP in corneas. Mouse corneal sections were incubated with both anti-GFP (HELP) and anti-HSV-1 (VP5) antibodies. Scale bars, 100 μm. e-j, qPCR and PFU analysis of HSV-1 dissemination in the eyes, trigeminal ganglia and brains. The abundance of HSV-1 shown as the viral genome (VG) per diploid genome (DG). n=4 mice. P=0.0286 in e-j. k and l, Confocal analysis of HSV-1 presence in the whole brain and TG. Scale bars, 1 mm, 200 μm and 20 μm, respectively. m, Confocal analysis of HELP presence in the TG after intracorneal injection. Scale bars, 50 μm. Data and error bars represent mean ± SEM. Unpaired two-tailed Mann-Whitney-tests. The experiments were repeated twice with similar results.
HELP suppresses HSV-1 associated disease pathologies in the prevention model
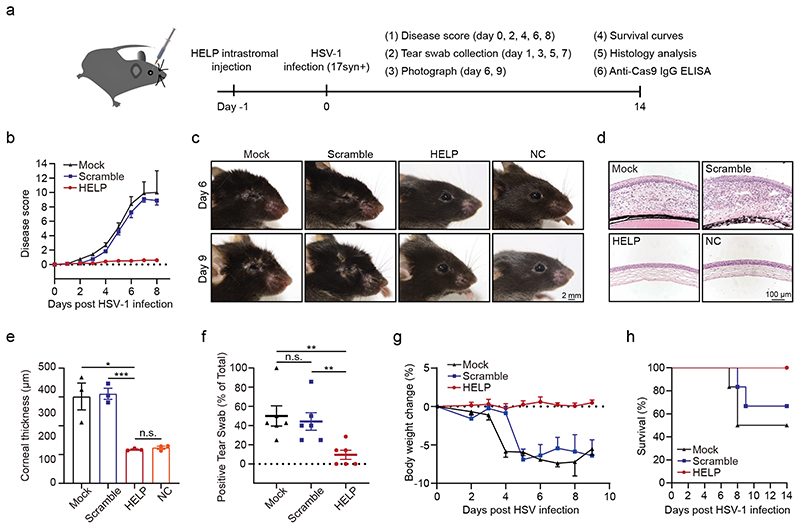
To determine the disease development and therapeutic efficacy, we monitored the clinical signs of acute ocular herpes infection and scored them in a blinded fashion (Fig. 3a). Importantly, mice treated with HELP did not show any disease progress (n=6 mice), while the mock-treated and scramble gRNA-treated eyes developed severe signs of ocular infection (Fig. 3b and Fig. 3c). Next, we performed histological staining to examine the pathology of the eye. The mock- and scramble gRNA-treated eyes presented with irregular stroma matrix and increased corneal thickness, typical signs of acute infection (Fig. 3d and 3e). We further found HSV-1 infection in the corneas induced significant type I IFN response, while HELP transduction did not elicit such a response (Supplementary Fig. 11). Clinical HSK is the result of excessive corneal virus-induced inflammation mediated by infiltration of inflammatory cells, including T cells (both CD4+ and CD8+), polymorphonuclear leukocytes, macrophages34, 35. Indeed, HSV-1 infection provoked corneal expression of the inflammatory molecules IL6, CCL2, and CXCL10, which were blocked after HELP treatment (Supplementary Fig. 12). Using immunohistochemistry, we showed HSV-1 infection led to CD4+ and CD8+ cells infiltration in the corneal stroma for the mock and the scramble groups, but HELP treatment prevented T cells infiltration (Supplementary Fig. 13). Additionally, we stained the corneal sections with two additional markers, CD11b and F4/80, to visualize the myeloid-derived cells and macrophages, respectively. We observed CD11b and F4/80 positive cells in non-therapeutic groups in contrast to the HELP treated mice and the non-infected control (Supplementary Fig. 13). We also noted that PD-L1 was upregulated in the epithelium and stroma of untreated mice after HSV-1 infection consistent with previous observations (Supplementary Fig. 13)36. Increased local PD-L1 expression may inhibit virus clearance by immune cells highlighting the importance of direct DNA degradation by CRISPR. To access the secreted virus, the eye swabs were titrated every other day post-infection (dpi). HELP treatments significantly reduced the viral presence in the eyes (Fig. 3f). In addition, bodyweights were recorded every other day. No loss of bodyweight was observed for HELP treated mice, while it was evident for the mock and scrambled control (Fig. 3g). Notably, all mice survived in the HELP treated groups and no relapse of HSK for the HELP treated mice was found during the 3-month follow-up (Fig. 3h and Supplementary Fig. 14).
Figure 3. HELP suppresses HSV-1-associated disease pathologies in the prevention model.
a, Flow chart for evaluating antiviral effects of HELP in vivo. 100 ng p24 HELP, scramble mLP or 2 μL PBS (Mock) were injected into corneas. After 24 h, the mice were infected with HSV-1 17syn+ (2×106 PFU/eye). b, Ocular disease scores (0 to 4, 4 being severe) in mice. n=6 mice. c, Photographs of the right eyes of differently treated mice on 6 dpi and 9 dpi. Each image is a representative of 3 mice in one experiment. NC, non-treated control. Scale bars, 2 mm. d, Corneal histology of eyes on 14 dpi. Each image is a representative of 3 mice in two independent experiments (c, d). Scale bars, 100 μm. e, Thickness of the corneas assessed from histology. n=3 mice. HELP vs. Mock and Scramble, P=0.0168 and 0.0006. f, Secreted HSV-1 assessed from the swabs of eyes. Tear swabs from each mouse was collected at 1, 3, 5, 7 dpi during the experiment. The percentage of HSV-1 positive swabs was recorded. n=6 mice. Mock vs. HELP, P=0.0056; Scramble vs. HELP, P=0.0072. g, Bodyweight. n=6 mice. h, Kaplan-Meier survival curves. n=6 mice. Data and error bars represent mean ± SEM. Unpaired two-tailed Student’s t-tests. n.s.=non-significant.
Eye health after HELP treatment in the prevention model
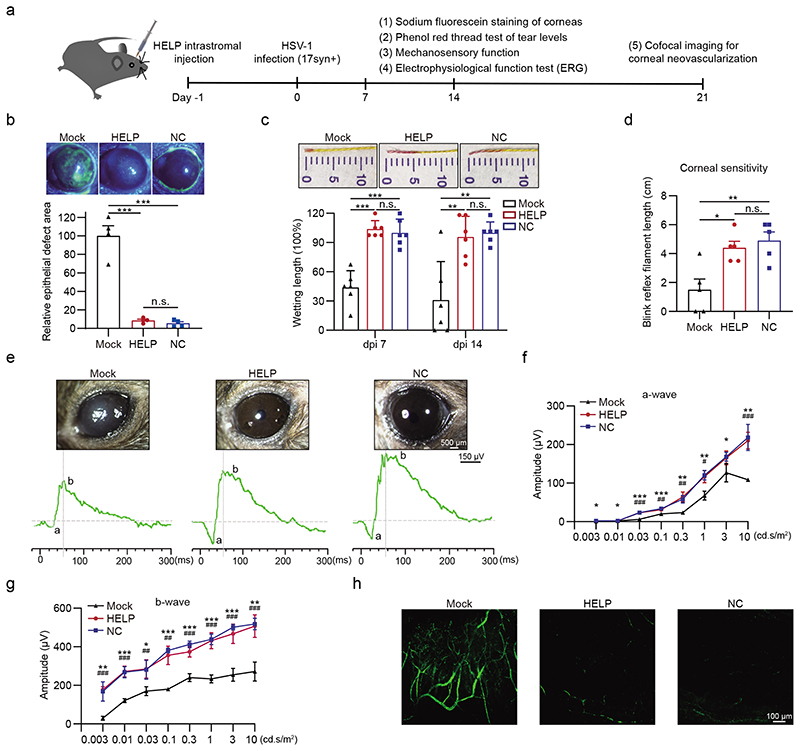
Subsequently, we thoroughly analysed the corneal health using clinical relevant indexes (Fig. 4a). From the lesion perspective, we assessed the epithelial layers using sodium fluorescein, which stains damaged epithelial cells. We found the HELP-treated corneas were significantly protected from HSV-1 infection (Fig. 4b). As reduced tears have been shown in the HSK, we therefore accessed the tear secretion levels of mice using the phenol red thread test. We found HELP treatment significantly protected the infected corneas from desiccation (Fig. 4c). HSV-1 infection often causes denervation of the cornea with a significant loss of sensory fibers. Next, we measured the mechanosensory function of the corneas using an esthesiometer and showed the sensory function of corneas was preserved (Fig. 4d). We further determined the effects of HELP on visual function by performing the full-field electroretinography (ERG). Waveforms were evaluated for the negative a-wave (photoreceptor responses) and the positive b-wave (cone and rod system responses) amplitudes (Fig. 4e). As shown in Fig. 4f-g, the amplitudes of the a-wave and b-wave were significantly reduced in the mock-treated eyes (a-wave: 23.36 ± 2.4 μV vs. 58.11 ± 10 μV, P<0.01; b-wave: 240.12 ± 20.49 μV vs. 412.3 ± 17.38 Mv, P<0.001; n=5 mice) compared with non-infected control eyes (NC) in 3 cd.s/m2flash stimulus (mix cone-rod response). The effect of the a-wave and b-wave amplitude after HELP application were significantly larger in 3 cd.s/m2flash stimulus compared with mock-treated eyes (a-wave: 64.33 ± 13.02 μV vs. 23.36 ± 2.4 Mv, P<0.01; b-wave: 374.44 ± 27.5 μV vs. 240.12 ± 20.49 Mv, P<0.001; n=5 mice). These results confirm that experimental HSK in mice leads to serious deficits in visual function likely due to reduced transparent corneas and lower intensity of light stimulus received by the retina, but not the direct retinal damage (Supplementary Fig. 15). The visual deficits could be prevented by the administration of HELP. Neovascularization is a hallmark of herpetic stromal keratitis37. We therefore assessed for the degree of corneal neovascularization by whole mount FITC-Dextran staining (Fig. 4h). Indeed, HSV-1 infection induced evident neovascularization in the corneas, which was inhibited by HELP treatment.
Figure 4. Eye health after HELP treatment in the prevention model.
a, Flow chart for evaluating eye health in the prevention model. 100 ng p24 HELP or 2 μL PBS (Mock) were delivered into corneas 24 h before the mice were infected with HSV-1 17syn+ (2×106 PFU/eye). b, Sodium fluorescein staining of mice corneas. The defect area of HELP and NC was normalized to Mock. n=4 mice. Mock vs. HELP and NC, P=0.0002 and 0.0001. c, Phenol red thread test of the wettability of the tear fluid. n=6 mice. Mock vs. HELP, P<0.0001 on dpi 7 and P=0.0054 on dpi 14; Mock vs. NC, P=0.0001 on dpi 7 and P=0.0021 on dpi 14. d, Measuring the mechanosensory function of the corneas by esthesiometer. n=5 mice. Mock vs. HELP, P=0.0104; Mock vs. NC, P=0.0074. e-g, Change in ERG amplitudes of treated eyes. Corneal graphs and traces of the a-waves and b-waves (e). Quantitative analysis of a-wave and b-wave amplitude (f and g). n=5 mice. NC vs. Mock (*), P=0.02, 0.01, 0.00002, 0.0006, 0.0026, 0.0036, 0.03 and 0.002; HELP vs. Mock (#), P=0.0002, 0.0092, 0.0018, 0.0114 and 0.0005 for ascending cd.s/m2(f). NC vs. Mock (*), P=0.0040, 0.0003, 0.0200, 0.00001, 0.00001, 0.0001, 0.0001 and 0.0015; HELP vs. Mock (#), P=0.0001, 0.0002, 0.0047, 0.0011, 0.0007, 0.0004, 0.0005 and 0.0005 for ascending cd.s/m2(g). h, Confocal microscopy imaging of neovascularization in the corneas. Data and error bars represent mean ± SEM. Unpaired two-tailed Student’s t-tests. n.s.=non-significant. Each image is a representative of 3 mice in one experiment (b, e, h).
Additionally, we examined if the intrastromal injection of HELP induces Cas9-specific IgG in the bloodstream. We did not observe significantly higher Cas9-specific IgG for both HELP and scrambled control than the mock (n=5 mice, non-significant, Student’s t-tests). In contrast, when HELP was injected via the footpad route, it provoked significantly higher anti-Cas9 IgG in the sera (n=3 mice, P<0.001, Student’s t-tests) (Supplementary Fig. 16). Interestingly, ocular infection of HSV-1 induced high titers of anti-HSV-1 neutralizing antibodies, which was absent after HELP treatment (Supplementary Fig. 17). Taken together, these results suggest that the administration of HELP significantly reduced the manifestation of disease severity during ocular HSV-1 infection.
HELP cures HSK in the therapeutic and recurrent models
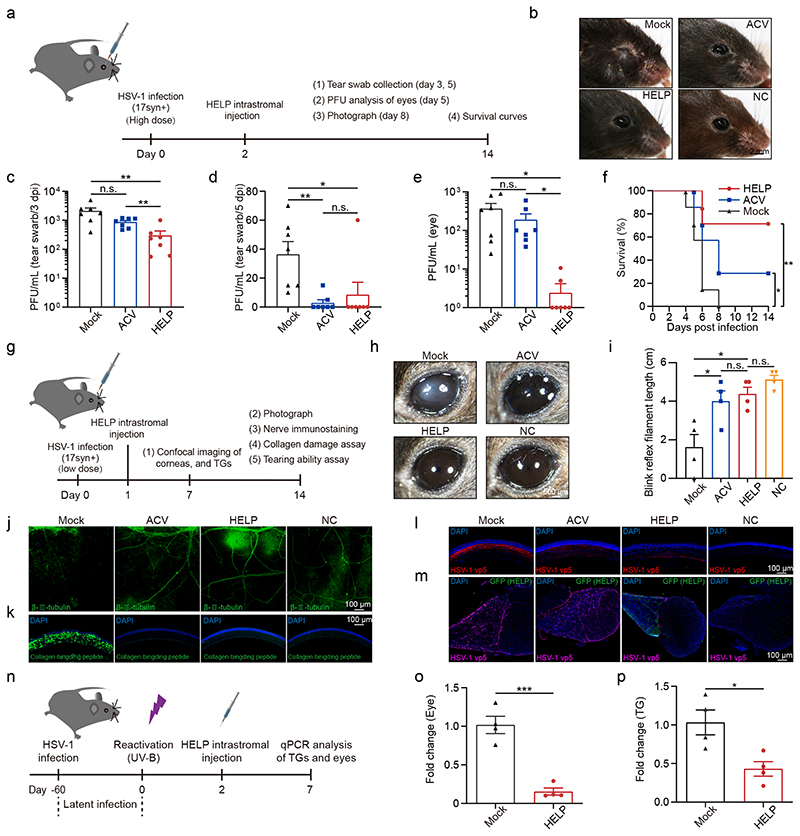
To better mimic the therapeutic process of HSK, we performed HELP administration after HSV infection and evaluated the therapeutic efficacy. We initially inoculated HSV-1 at a dose of 2×106 PFU/eye and treated the mice with HELP or ACV, which was used as a positive control (Fig. 5a). We found that both ACV and HELP treatment inhibited the lesions in the eyelids (Fig. 5b). Both ACV and HELP significantly reduced the secretion of infectious HSV-1 progenies in the tear swabs (Fig. 5c-d). Interestingly, only HELP significantly reduced the levels of infectious HSV-1 in the eyes, suggesting the unique advantage of CRISPR in eliminating viruses (Fig. 5e). While all mice from the Mock group died, both HELP and ACV treatment significantly augmented the survival rates with HELP showing superior effect compared to ACV (Fig. 5f). Next, we lowered the dose of HSV-1 to 5×104 PFU/eye (Fig. 5g). In addition, we injected HELP 1 day instead of 2 days after HSV-1 infection (Fig. 5g). The corneas of Mock developed symptoms of HSK 14 days after infection while both ACV and HELP treatment prevented the disease progress (Fig. 5h). Next, we evaluated the mechanosensory function of the corneas and found it was maintained for both ACV and HELP, but not for the Mock (Fig. 5i). Additionally, we performed confocal microscopy analyses of the cornea by staining of sensory fibers and damaged collagen fibers, thus evaluating for corneal health. We found loss of beta III tubulin and appearance of peptides positive collagens for the Mock, which was absent in both ACV and HELP groups (Fig. 5j-k). Furthermore, we assessed the HSV-1 distribution in the corneas and TGs, respectively. We found both ACV and HELP blocked HSV-1 replication in the corneas with HELP showing superior efficiency (Fig. 5l). Strikingly, while ACV failed to modulate the HSV-1 reservoir in the TG, HELP diminished the HSV-1 to an almost undetectable level in the TG likely via retrograde transportation (Fig. 5m).
Figure 5. HELP cures HSK in the therapeutic and recurrent models.
a, Flow chart for evaluating the antiviral effects of HELP in the HSK therapeutic model. Mice were infected with HSV-1 17syn+ (2×106 PFU/eye). After 48 h, 100 ng p24 HELP or 2 μL PBS (Mock) was administrated. ACV was added topically to both eyes every day for five days. b, Photographs of the eyes of differently treated mice. c and d, Infectious units in tear swabs on 3 dpi and 5 dpi. n=7 mice. c, Mock vs. HELP, P=0.0083; ACV vs. HELP, P=0.0060. d, Mock vs. ACV, P=0.0031; Mock vs. HELP, P=0.0431. e, Plaque assay for HSV-1 in the eyes. n=7 mice. Mock vs. HELP, P=0.0197; ACV vs. HELP, P=0.0360. f, Kaplan-Meier survival curves. n=6 mice. Mock vs. HELP and ACV, P=0.0076 and 0.0297. g, Flow chart for evaluating the antiviral effects of HELP in the HSK therapeutic model. HSV-1 was used 5×104 PFU/eye. h, Photographs of the differently treated eyes. Scale bars, 500 μm. i, Measuring the mechanosensory function of the corneas by esthesiometer. n=4 mice. Mock vs. HELP and ACV, P=0.0127 and 0.0349. j and k, Confocal images of sensory fibers and damaged collagen in corneas. Scale bars, 100 μm. l and m, Confocal images of HSV-1 (VP5) in the corneas and HELP (anti-GFP) in the TG. Scale bars, 100 μm. n, Flow chart for evaluating antiviral activity using a recurrent HSK model. Mice were infected with HSV-1 17syn+ (2×105 PFU/eye). o and p, Virus load in the eyes and TGs detected by qPCR. n=4 mice. o, P=0.0004; p, P=0.0179. Data and error bars represent mean ± SEM. Unpaired two-tailed Student’s t-tests. n.s.=non-significant. Each image is a representative of 4 mice (b, h) or 2 mice (j-m) in one experiment.
To strengthen the notion that HELP can modulate the HSV reservoir, we adopted a recurrent HSK model in which eyes were infected with HSV-1 to establish latency before HELP treatment (Fig. 5n). We reactivated the mice survived from the acute infection by UV-B irradiation of the eyes 60 days after HSV-1 inoculation. We then treated the eyes with HELP by intrastromal injection and quantified the HSV-1 genome seven days later. Indeed, we found that HELP significantly decreased virus load in the eyes, which migrated from TG by anterograde transportation (Fig. 5o). In agreement with Fig. 5m, we found a significantly reduced level of HSV-1 in the TG (Fig. 5p).
HELP eliminates HSV-1 in tissue culture of human corneas
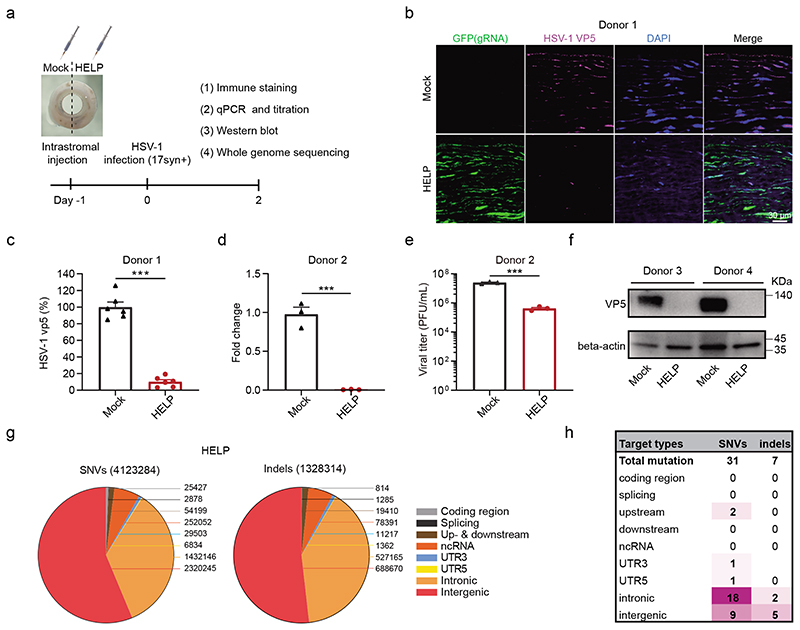
Having established the therapeutic efficacy of HELP in the murine models, we sought to investigate the antiviral potential of HELP in human corneas (Fig. 6a). One human cornea was evenly divided into two halves and injected with either 15 μL HELP (equal to 1.5 μg p24) or PBS, respectively, for confocal imaging. We found that HELP was evenly spread in the stroma and potently inhibited HSV-1 (17syn+) replication as manifested by reduced VP5 compared to mock controls (Fig. 6b-c). With cornea from another donor, we found HELP treatment significantly diminished both the genome of HSV-1 and viral titers (Fig. 6d-e). Additionally, we showed that VP5 protein was hardly detectable by Western blotting after HELP treatment (Fig. 6f). To examine whether HELP causes off-target effects in the human genome, we performed whole genome sequencing (WGS) on a human cornea which was evenly divided for HELP and PBS injection, respectively. We analysed the single-nucleotide variants (SNVs) and indels at an average depth of 51 and 45 fold for HELP and mock, respectively, including coding region, splicing, up- and downstream, non-coding RNA, 5’- and 3’- UTR, intronic and intergenic region. In total 4,123,284 SNVs and 1,328,314 indels were detected in HELP and 3,726,678 SNVs and 981,253 indels were detected in mock (Fig. 6g and Supplementary Fig. 18-19). We then filtered the SNVs and indels using 707 Cas-OFFinder predicted off-target sites and the mock sequence from WGS38. In total, we found 31 SNV and 7 indel mutations (Supplementary Table 1-2), but none of them localized in the coding region indicating no functional off-targets in HELP treated human cornea (Fig. 6h).
Figure 6. HELP eliminates HSV-1 in tissue culture of human corneas.
a, Flow chart for evaluating antiviral effects of HELP in human corneas. 1.5 μg p24 HELP or 15 μL PBS (Mock) were injected into the corresponding punches derived from the same human cornea. After 24 h, the corneal punches were infected with HSV-1 17syn+ (2×106 PFU/piece). b, Confocal analysis of the distribution of HSV-1 and HELP in the human cornea. GFP is indicative of the presence of HELP and VP5 for HSV-1. Scale bar, 30 μm. c, Percentage of VP5+ cells presented in (b). Data shown as the percentage of mock-treated tissues. n=6 biologically independent samples. P<0.0001. d, qPCR analysis the fold change of HSV-1 genome. n=3 biologically independent samples. P=0.0005. e, Titration of supernatants from human corneal cultures. n=3 biologically independent samples. P=0.0003. f, Western blot analysis of VP5 protein. The experiment was repeated twice with similar results. g, Identification of SNV and indel mutations in the HELP treated corneal punch at WGS level. Valid sequencing data were aligned to Human Genome version 19 (HG19). h, Summary of unique SNV and indel mutations. Data and error bars represent mean ± SEM. Unpaired two-tailed Student’s t-tests.
Discussion
In this study, we show that transient gene editing via mRNA-based CRISPR delivery is sufficient to achieve therapeutic efficacy against HSK in vivo, and block HSV-1 replication in human corneas. Modulating the virus reservoir is essential to prevent HSK from recurrence. Our study further provides evidence of HSV elimination in the reservoir by HELP via retrograde transportation from the cornea to TG.
The administration of HELP was through intrastromal delivery which has been frequently used in clinical practice to deliver bevacizumab to the corneal stroma of patients with HSK to prevent corneal neovascularization39, 40. In addition, the intrastromal injection of antibiotics is a routine procedure to treat patients with fungal keratitis41, 42. Using a package of evaluation methods
including sodium fluorescein staining, phenol red thread test, esthesiometer, ERG, FITC-Dextran staining, beta III tubulin, and peptides staining, we showed healthy corneal status after CRISPR therapy suggesting the intrastromal injection of HELP is practical for clinical translation.
HSK is thought to be the result of HSV-1 induced corneal infiltration of a cocktail of inflammatory cells consisting of T cells (both CD4+ and CD8+), macrophages, and polymorphonuclear leukocytes. Corneas removed from patients requiring corneal transplants due to HSK contained both CD4+ and CD8+ T cells43, 44. Consistently, our study also showed infiltration of both CD4+ and CD8+ cells to corneas of HSV infected mice. Moreover, we also found the infiltration of myeloid-derived cells and macrophages in the HSV-1 infected corneas (Supplementary Fig. 13). Notably, the infiltration of inflammatory cells was prevented after HELP administration.
In humans, HSK often develops after virus replication can be detected, and may develop after most of the replicating viruses are cleared7. Asymptomatic virus shedding has been reported in tears of healthy individuals though it is relatively rare45. As HELP is an antiviral therapy, the optimal efficacy of HELP treatment may link to the time point of administration, which has to been investigated by clinical testing. From a clinical perspective, HELP may first be applied to patients with acute corneal perforation or corneal graft failure due to the recurrence of virus in combination with keratoplasty. Given the safety and efficacy shown in the most serious HSK, HELP may also be extended to the early stage HSK as a first-line choice to cure or prevent the recurrence of HSK by eliminating the virus in the corneas and TG.
The HELP used in the current study is coated by VSV-G envelope protein. Lentiviral vector pseudotyped by VSV-G is capable of retrograde transportation potentially mediated by cytoplasmic dynein as has been shown previously46, 47. Our study has demonstrated the possibility to reduce HSV-1 in the TG, however, the efficiency may further be enhanced by coating HELP with derivatives of rabies virus glycoprotein46, 48.
While our manuscript was under revision49, Aubert et al. reported that AAV delivery of meganucleases was also capable of modulating the latent HSV-1 in vivo 26. The study, however, largely focused on analysis of HSV-1 genome and did not include a disease model. Therefore, it is difficult to predict the therapeutic benefits. Notably, the study hardly detected gene editing of HSV with CRISPR - likely, due to the large size of Cas9 and low expression of Cas9 and gRNA from the single-stranded AAV in vivo 26. In contrast, HELP is able to overcome the size limitation of AAV and co-packaging both SpCas9 and gRNA to efficiently eliminate HSV genome in vivo - 1 to 2 logs higher than meganuclease in diminishing the virus genome. Importantly, HELP is mRNA-based and promoter-less for Cas9. On the contrary, AAV will likely persist for a long time in the non-dividing neurons and require a strong promoter for efficient nuclease expression - therefore, is accompanied by long-term safety risk from a clinical perspective.
In conclusion, the efficacy and safety profile of HELP shown in our study strongly supports further clinical testing of these lentiviral particles targeting HSV-1 by CRISPR. Additionally, since the gRNAs of HELP target the genome of the virus instead of humans, this may accelerate the clinical translation. Our study may also facilitate the development of CRISPR therapeutics targeting other viruses such as human papillomavirus (HPV) or inherited diseases.
Methods
Cell cultures and HSV-1 propagation
293T, Vero, HaCaT and HaCaT IFNAR2-KO cells were cultured in DMEM (Gibco). THP1 cells were cultured in RPMI 1640 (Gibco). DMEM and RPMI 1640 media were supplemented with 10% fetal bovine serum (Gibco) and 1% penicillin/streptomycin (P/S) (Thermo Fisher Scientific). All used cell lines were obtained from the laboratory of Soren Riis Paludan and not authenticated in our laboratory. None of the cell lines were listed in the database of commonly misidentified cell lines maintained by ICLAC. Primary mouse corneal stromal cells were digested from corneal stromal tissues and maintained in MEM (Gibco) supplemented with 1% P/S and 10% fetal bovine serum. Human cornea tissues were maintained in MEM (Gibco) supplemented with 1% P/S and 10% fetal bovine serum. All cells were cultured at 37°C and 5% (vol/vol) CO2. HSV-1 strains including 17syn+, McKrae, F, and HSV1-GFP (HSV-1 KOS strain expressing GFP driven by the CMV promoter)31 were propagated in and titrated on Vero cells. Only 17syn+ was used for in vivo study. All cell lines were tested negative for mycoplasma contamination.
Production of mLPs
293T cells were seeded in 15-cm dishes at a density of 107/dish 24 h before calcium phosphate transfection. 24 h after transfection, the media were refreshed, and the supernatants were harvested 48 h and 72 h post-transfection before passing through a 0.22-μm filter (Millipore) and ultracentrifugation at RPM 25,000 at 4°C for 2 h. Pellets were re-suspended in PBS and stored at −80°C. To produce ‘all-in-one’ mLPs, 293T cells were transfected with 9.07 μg pMD.2G, 7.26 μg pRSV-Rev, 15.74 μg pMDlg/pRRE-D64V, 15.74 μg pMS2M-PH-gagpol-D64V, 31.46 μg pCMV-Cas9-6XMS2 and 31.46 μg pLV-egfp-U3-osp-gRNA with corresponding gRNA sequence. To produce HELP, pLV-U6-UL29-egfp-U3-UL8 was used as the gRNA producing plasmid. To produce non-GFP version HELP, pLV-U6-UL29-U3-UL8 was used. The plasmids are deposited in Addgene.
HSV-1 plaque assay
HSV-1 plaque assays were performed in triplicates for each biological sample. 1.5×105 Vero cells were seeded in a 12-well plate in the complete DMEM and infected the following day with various dilutions of HSV stocks or culture supernatants. Two hours after infection, cells were overlaid with 1% agarose (Sangon) solution. After incubation for 3 days, cells were fixed with 4% formaldehyde and stained using 1% crystal violet solution at room temperature for 2 hours. After 3 washes with PBS, plates were allowed to dry and the numbers of plaques were counted. Viral titers were calculated as PFU/mL.
Infection and transduction of cells
For 293T, HaCaT, HaCaT-IFNAR2-KO, THP-1, and primary mouse corneal stromal cells, 4×104 cells were seeded in a 48-well plate well and transduced with 400 ng mLP on the following day. The media were refreshed 12 hours post-infection (hpi). After 24-h transduction, the cell infected with HSV1-GFP at an MOI of 1. The cells and supernatants were harvested at 24 and 48 hpi for flow cytometry and plaque assay, respectively. The DNA isolated from the cell lysates using the viral DNA extraction kit (TaKaRa) was used to determine the cleavage activity of HSV-1 genomes by Sanger sequencing and TIDE 2.0.1 analysis. The used primers are shown in Supplementary Table 3.
Flow cytometry analysis
The primary mouse corneal stromal cells were seeded at a density of 4×104/well on day 1 and transduced with UL29-UL8 co-targeting HELP or scrambled control (non-GFP version) on day 2. The cells were then infected with HSV1-GFP on day 3. The GFP signals were determined by flow cytometry (BD LSRFortessa, BD Biosciences). Data were collected by BD FACSDiva 7 and analysed by FlowJo 7.6 for the percentage of GFP+ cells and mean fluorescence intensity on day 4 and 5. Gating strategies were shown in Supplementary Fig. 20.
ELISA
The p24 of mLP was measured using HIV p24 ELISA according to the manufacturer (Beijing Biodragon Immunotechnologies). To detect mouse humoral IgG immune response to Cas9, IgG Mouse ELISA Kit (Abcam) was used following the manufacturer’s protocol with a few modifications. A total of 0.25 μg of recombinant Cas9 proteins (Novoprotein) suspended in PBS were used to coat 96-well ELISA plates and incubated at 4°C for 12 h, then washed three times using 1X Wash Buffer. Plates were blocked with 2% BSA Blocking Solution for 2 h at room temperature, then washed three times. Serum samples were added to each well. The remaining steps were doing as the manufacturer’s protocol. Cas9 Mouse mAb (Cell Signaling Technology) was used to make a standard curve, the dilution gradient was accorded to the instruction of IgG Mouse ELISA Kit.
Western blotting
The human and mouse corneal tissues were ground using TissueLyser with magnetic beads. The corneal suspensions and 293T cells were lysed in RIPA (Beyotime Biotechnology) in the presence of a protease inhibitor (Beyotime Biotechnology) for 30 minutes and incubated with SDS-PAGE sample loading buffer (Beyotime Biotechnology) for 15 minutes at 98°C. The proteins were separated by SDS-polyacrylamide gel electrophoresis and transferred to the PVDF membrane. The membrane was blocked by 5% fat-free milk dissolved in TBS/0.05% Tween-20 for 1 h and was cut off according to the marker to incubate with different primary antibodies overnight at 4°C. The membranes were then incubated with anti-mouse secondary antibodies (1:3000, Cell Signaling Technology, cat:4409) and visualized by hypersensitive ECL chemiluminescence (Beyotime Biotechnology) using gel imaging system (Amersham ImageQuant 680, GE). Beta-actin was used for signal normalization across samples. The primary antibodies used in this experiment were HSV VP5 monoclonal antibody (1:1000, Santa Cruz Biotechnology, cat: sc56989), beta-actin mouse monoclonal antibody (1:3000, Cell Signaling Technology, cat: 3700) and Cas9 mouse monoclonal antibody (1:3000, Cell Signaling Technology, cat: 14697).
Quantitative PCR
Genomic DNA and total RNA from all samples were extracted using Viral DNA/RNA extraction kit (TaKaRa). cDNA was synthesized using the QuantScript RT Kit (TIANGEN) according to the manufacturer’s protocol. qPCR was performed using PowerUp SYBR Green Master Mix (Applied Biosystems) following the manufacturer’s protocol. The qPCR experiments were performed by Real-Time PCR system (LightCycler 96, Roche). To quantify HSV-1 genome in the murine tissues or human corneas, genomic DNA and viral DNA were extracted from the corresponding parts of mice or human corneas and subjected to qPCR to detect HSV-1 (primer Y5/Y6) which was then normalized to mouse GAPDH (SK13/SK14) or human GAPDH (SK55/SK56), respectively. To detect mLP distribution in vivo, genomic DNA was extracted from the eye, trigeminal ganglion, heart, liver, spleen, lung, kidney, and testis. qPCR was performed to detect WPRE (primer SK9/SK10) which was then normalized to GAPDH (SK13/SK14). To detect innate immune response induced by HELP and HSV-1 in mice, total RNA was extracted from eyes. RT-qPCR was performed to detected ISG15 (SK51/SK52), RIG-I (Y7/Y8) and IFNB1 (Y9/Y10) which was then normalized to GAPDH (Y23/Y24). To detect the innate immune response in human cells, RT-qPCR was performed to detected ISG15 (Y11/Y12), RIG-I (Y13/Y14) and IFNB1 (Y15/Y16) which was then normalized to GAPDH (SK55/SK56). To measure the inflammatory molecules are expressed in the cornea following HSV-1 infection. RT-qPCR was performed to detected IL6 (Y17/Y18), CCL2 (Y19/Y20) and CXCL10 (Y21/Y22) which was then normalized to GAPDH (Y23/Y24). To detect the copy number of Cas9 mRNA in each HELP particle, the same amount p24 of HELP and lenti-CRISPR were used to extract total RNA and synthesized to cDNA. RT-qPCR was performed using primer SK11/SK12. Data of HELP was normalized to lenti-CRISPR. The used primer sequences were listed in Supplementary Table 4.
Mice
6-8 weeks old, male, specific-pathogen-free (SPF) C57BL/6J mice were used in this study. Mice were housed in an environmentally controlled room (23 °C, with 55±5% humidity and 12 h/12 h light-dark cycle). The HELP or PBS (Mock) was injected into mice by intrastromal injection under the stereo light microscope (SMZ800N, Nikon). All mice studies have complied with the guidelines of the Institutional Animal Care and Use Committee (IACUC) of the Shanghai Jiao Tong University with approval from the animal Ethics Committee.
Intrastromal injection
The mice were anesthetized and a small intrastromal pocket was carefully created in the mid-peripheral cornea by a 29 G needle. Then a 33 G needle was inserted towards the central cornea, and 2 μL HELP or PBS was injected into the corneal stroma. Both eyes of the mouse were injected in this study. Mice were allocated to the experimental groups and control randomly.
Murine acute HSV-1 infection model
The mice were anesthetized by intraperitoneal injection of 1.25% Avertin. Corneas were scarified with a 3×3 crosshatch pattern. The mice were inoculated with 2×106 PFU (or 5×104) HSV-1 17syn+ on both eyes. The bodyweight and disease scores were measured at the indicated times post-infection. The scoring was performed as blinded study: hair loss (0: none, 1: minimal periocular hair loss, 2: moderate periocular hair loss, 3: severe hair loss limited to periocular, 4: hair loss severe and extensive); hydrocephalus (0: none, 1: minor bump, 2: moderate bump, 3: large bump); symptoms related to neurological disease (0: normal, 1: jumpy, 2: uncoordinated, 3: hunched/lethargic, 4: unresponsive/no movement); eye swell/lesions (0: none, 1: minor swelling, 2: moderate swelling, 3: severe swelling and skin lesions, 4: lesions extensive). Mice were sacrificed at the specified time post-infection. To collect the eye swabs, mouse eyes were gently proptosed, then wiping with a sterile cotton swab (Miraclean Technology) three times around the eye in a circular motion and twice across the center of the cornea in a ‘+’ pattern. The cotton swabs were placed in 1 mL of DMEM containing 2% (vol/vol) FBS, 1% P/S and stored at -80°C until titrated by plaque assay. Corneal graphs were collected using color digital camera (DS-Ri2, Nikon) attached to stereo light microscope (SMZ800N, Nikon) at the indicated time. Sera were collected at 14 dpi to test the mouse humoral IgG specific to SpCas9 by ELISA.
Murine recurrent HSV-1 infection model
The mice were inoculated with 2×105 PFU HSV-1 17syn+ for both eyes on scarified corneas. Mice survived from acute infection were maintained for 60 days and reactivated by UV-B irradiation of the eyes, followed by HELP or PBS (Mock) treatment. The TGs and eyes were collected to quantify HSV-1 DNA by qPCR.
Human corneal HSV-1 infection
The human corneas were obtained from fresh cadavers and supplied by the Eye Bank of Eye, Ear, Nose and Throat Hospital, Fudan University under the approval of the hospital Ethics Committee (EENTIRB-2017-06-07-01). The experiments were conducted according to the Declaration of Helsinki and in compliance with China law. The corneas were evenly divided into two halves. One half was dosed with 15 μL HELP by intrastromal injection (equal to 1.5 μg p24) while the other was dosed with PBS by intrastromal injection as a mock control. The corneas were then infected with 2×106 PFU of HSV-1 17syn+ in MEM medium containing 2% FBS. The media were refreshed 2 hpi with MEM containing 10% FBS and 5% P/S. Two days after HSV-1 infection, the corneas were processed for immunofluorescence imaging, immunoblotting or DNA isolation by the viral DNA extraction kit to determine the viral genomes by qPCR. The supernatants were collected for plaque assay.
Immunofluorescence imaging
For confocal imaging, the 293T and mouse corneal stromal cells were imaged under Laser Scanning Confocal Microscope (A1si, Nikon) at the indicated time. The eyes, trigeminal ganglia, and brains were fixed in 4% PFA overnight at 4°C before transferring to 30% sucrose. The OCT-embedded tissues were sectioned to 10-μm thickness using the freezing microtome (CM1950, Leica) and processed for immunofluorescence. The slides were dried at room temperature for 10 minutes and blocked in blocking buffer with 5% normal goat serum (Solarbio), 1% BSA, 0.3% Triton X-100 in PBS in a humidified box at room temperature for 30 minutes. The slides were incubated with the primary antibody against HSV-1 VP5 (1:200, Santa Cruz Biotechnology, cat: sc56989) or rabbit primary antibody against GFP (1:1000, GeneTex, cat: 113617) in 1% BSA overnight at 4°C. After washing, the slides were incubated with anti-mouse secondary antibody (1:100, Santa Cruz Biotechnology, cat: 516176) or anti-rabbit secondary antibody (1:500, Beyotime Biotechnology, cat: a0468) in 1% BSA for 1 hour. For whole-mount mouse corneal staining, freshly isolated mouse corneal buttons were cut from the eye globe on ice with dissecting microscope. Corneal buttons were fixed 4% PFA in 4 °C overnight, then washed in blocking buffer (1% BSA, 5% goat serum and 0.03% Triton X-100 in 1× PBS) for 1 hour at room temperature. Corneas were incubated with primary anti-mouse anti-beta III-tubulin antibody (1 ug/mL, Abcam, cat: 238697) at 4 °C for 72 h, then washed thoroughly with washing buffer (0.05% Tween20 in PBS). The corneas were incubated with the corresponding secondary antibodies for 2 hours at room temperature and washed and affixed with a coverslip. Fluorescent staining was imaged with Laser Scanning Confocal Microscope (A1si, Nikon). The thickness of Z-stack generated for cornea whole mounts was 19 to 21 slices (4.2-mm step size).
Collagen-binding peptides staining
The N terminus 5-FAM labeled collagen-binding peptides (Cys-Gln-Asp-Ser-Glu-Thr-Arg-Thr-Phe-Tyr) were purchased from Sangon Biotech Co, Ltd. The mouse eyes were frozen in OCT (Sakura Finetek) compound and cut to 15-μm thick sections. The sections were blocked with PBS (containing 5% donkey serum) for 30 min at room temperature. Collagen-binding peptides were dissolved in PBS to obtain a final concentration of 50 ng/μL. Before staining, the peptides were heated for 5 min at 80°C water, then immediately incubated on ice. The sections were then stained with peptides solution at 4°C overnight. Following incubation, the sections were rinsed three times with PBS. The samples were analysed using Laser Scanning Confocal Microscope (A1si, Nikon) at 10× magnification.
Evaluation of corneal neovascularization
FITC-Dextran of MW 2×106 (Sigma Aldrich) was diluted in saline to a concentration of 50 mg/mL before injecting (0.15 mL) into the left ventricle of the mouse heart. Eyes were enucleated at 5 min after injection and fixed for 2 h with 4% PFA at 4°C. Corneas were isolated under the Stereo Microscope with 4 radial incisions made to flatten each cornea on a slide. Slides were fixed for 10 min with 4% PFA, washed with PBS. The Cornea was flattened on a glass slide and imaged under Laser Scanning Confocal Microscope (A1si, Nikon).
Phenol red thread test
A 25-mm-long phenol red impregnated thread with 3-mm bent end thread was placed in lower fornix of the mouse eye for 20 s. The phenol red thread changes colours from yellow to red when it contacts with tears. The length of the red portion was measured with a ruler.
Cornea epithelial lesion test
Anesthetized mice were placed on a mice holder and the entire frame of the cornea was visible. A total volume of 4 μL sodium fluorescein (0.5%) was added to the mouse eye. The images were captured in the cobalt blue channel with the surgical microscope (OPMI VISU S8, Carl Zeiss), and the stained area was quantified using Image J 1.52v software.
Electroretinography
Mice were dark-adapted overnight and anesthetized by intraperitoneal injection of 1.25% Avertin. Corneal anesthesia and mydriasis were achieved with 0.5% proparacaine hydrochloride (Alcon-Couvreur, Belgium) and 0.5% tropicamide (Alcon Laboratories, Australia). A thermal plate was used to maintain the body temperature (37°C). The full-field electroretinography (ERG) was recorded from both eyes using an Espion Diagnosys system (Espion E2, Diagnosys LLC). One subdermal needle was inserted into the tail acting as the ground electrode, while another subdermal needle was placed over the nasal bone served as the reference electrode. Electrical signals were recorded with two 3-mm diameter platinum wire loop electrodes placed on the corneal surface. Eyes were lubricated with a 2.5% hydroxypropyl-methylcellulose solution (Gonak, Akorn). Light stimuli were delivered using a ColorDome unit. For dark-adapted responses, ERG was performed to a series of white flash stimuli ranged from 0.003 to 10 cd.s/m2. A total of 10 responses were averaged with interstimulus intervals (ISI) of 10 seconds. Mice were subsequently adapted for a 5-minute period of light adaptation with a white background over which a series of flash stimuli (3, 10, and 30 cd.s/m2) were superimposed (20 responses each, ISI=2 seconds). The band-pass cutoff frequencies were 0.3 and 300 Hz. The data were analysed using Espion software 6.0.54.
Deep sequencing
The top-ranked off-target sites in mouse genome for UL29-targeting and UL8-targeting gRNA were identified by Cas-OFFinder online predictor respectively. The on-target and predicted off-target sites were PCR amplified and pooled by an equal molar ratio for double-end sequencing using Illumina MiSeq at Novogene. Raw data of Next-generation sequencing were analysed by Cas-analyzer (version 2016.12.14). The primer sequences are listed in Supplementary Table 5.
Whole genome sequencing
Genomic DNA from human cornea was isolated using Magen HiPure Blood DNA Mini Kit (AnGen Biotech). The purity, quantity, and size of genomic DNA were assessed by NanoDrop and agarose gel electrophoresis. Genomic DNA was subjected to whole genome DNA library preparation for high-throughput sequencing (Illumina platform) with a mean coverage of 51 and 45 fold for HELP and mock, respectively, in GENEWIZ. The Q30 was set above 85%, and the average error rate was required below 0.1%. Valid sequencing data were aligned to HG19 using Burrows-Wheeler Aligner 0.7.12. All polymorphic SNVs and indel sites in the genome were extracted, and high-confidence SNVs and indel data sets were ultimately obtained and analysed.
Prediction of potential OT sites
The Cas-OFFinder tool was used to find all potential off-target sites based on sequence homology to either the UL8 or UL29 gRNAs, allowing up to 5 mismatches. This resulted in 269 and 438 potential off-target sites being detected for UL8 and UL29, respectively. These potential off-target sites were separated as the coding region, splicing, up- and downstream, ncRNA, 3’-UTR, 5’-UTR, intronic, and intergenic regions. The 100 bp in upstream and downstream of potential off-target sites were used to find the SNVs and indels. The results were further filtered according to identity with the mock and excluded repetitive sequences.
Histology
Mouse eyes were dissected and fixed in paraformaldehyde before embedding in paraffin, sectioning at 10-μm thickness and staining with hematoxylin and eosin. For immunohistochemistry staining, the sections were de-paraffinated and rehydrated followed by incubated with citrate buffer for antigen retrieval. To block endogenous peroxidase activity, the sections were treated with 3% H2O2 for 25 minutes. The sections were then blocked with 3% BSA at room temperature for 30 minutes, followed by incubating with anti-CD4+ (1:100, Servicebio, cat: gb13064), anti-CD8+ (1:1000, Servicebio, cat: gb11068), anti-PD-L1 antibody (1 ug/mL, Abcam, cat: 238697), anti-CD11b antibody (1:500, Servicebio, cat: gb11058), and anti-F4/80 antibody (1:500, Servicebio, cat: gb11027), respectively, at 4°C overnight. The slides were then incubated with an anti-rabbit secondary antibody (1:500, Servicebio, cat: gb23303) or anti-mouse secondary antibody (1:500, Servicebio, cat: gb23301), followed by incubating with the freshly prepared DAB substrate solution to reveal the color of antibody. At last, the tissue slides were counterstained with hematoxylin and blued with ammonia water, and then dehydrated and coverslipped. Images were collected by a fluorescence microscope (Eclipse Ni, Nikon).
Statistics
For in vitro studies, sample sizes were determined by triplicate samples or more for comparisons between one or multiple groups, followed by the statistical test. Each experiment was repeated at least twice and the experimental findings can be reliably reproduced. For in vivo studies, at least four mice were used for each group. Data are presented as mean ± SEM in all experiments. Student’s t-tests, Mann-Whitney-tests or one-way analysis of variance (ANOVA) were performed to determine the P values. The specific statistical method applied, description of replicates can be found in the figure legends. * and # indicates statistical significance (*P< 0.05, **P< 0.01, ***P< 0.001; #P < 0.05, ##P < 0.01, ###P < 0.001; n.s.=non-significant). Statistics were analysed with GraphPad Prism 8.
Supplementary Material
Editorial summary.
CRISPR-Cas9 eliminates herpes simplex virus both in the corneal infection site and in the viral reservoir of the trigeminal ganglia.
Acknowledgements
We thank F. Zhang (MIT, USA) for reading and commenting on our manuscript. The work was supported by grants from the National Natural Science Foundation of China [31971364], Pujiang Talent Project of Shanghai [GJ4150006], Natural Science Foundation of Shanghai [BS4150002], and Startup funding from Shanghai Center for Systems Biomedicine, Shanghai Jiao Tong University [WF220441504] to Y.C.; the National Natural Science Foundation of China [81970766 and 81670818], the Shanghai Rising-Star Program [18QA1401100], the Shanghai Innovation Development Program [2020779], and the Shanghai Key Clinical Research Program [SHDC2020CR3052B] to J.H.. S.R.P. is supported by the European Research Council [ERC-AdG ENVISION; 786602].
Footnotes
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Competing interests
The authors declare no competing interests.
Author contribution
D.Y, S.L., J.H., and Y.C. conceived the study and designed the experiments; D.Y., S.L., D.W., Y.D., H.J., and X.Z. performed the experiments; all the authors analysed the data; D.Y., S.L., and Y.C. wrote the manuscript with the help from all the authors.
Data availability
Data generated or analysed during this study are available from the corresponding author on reasonable request. The deep sequencing and whole genome sequencing data are available at NCBI BioProject. The BioProject IDs are PRJNA668071 and PRJNA668060, respectively.
References
- 1.Liesegang TJ. Herpes simplex virus epidemiology and ocular importance. Cornea. 2001;20:1–13. doi: 10.1097/00003226-200101000-00001. [DOI] [PubMed] [Google Scholar]
- 2.Paludan SR, Bowie AG, Horan KA, Fitzgerald KA. Recognition of herpesviruses by the innate immune system. Nat Rev Immunol. 2011;11:143–154. doi: 10.1038/nri2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bradshaw MJ, Venkatesan A. Herpes Simplex Virus-1 Encephalitis in Adults: Pathophysiology, Diagnosis, and Management. Neurotherapeutics. 2016;13:493–508. doi: 10.1007/s13311-016-0433-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Farooq AV, Shukla D. Herpes simplex epithelial and stromal keratitis: an epidemiologic update. Surv Ophthalmol. 2012;57:448–462. doi: 10.1016/j.survophthal.2012.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Crumpacker CS, Schaffer PA. New anti-HSV therapeutics target the helicase–primase complex. Nat Med. 2002;8:327–328. doi: 10.1038/nm0402-327. [DOI] [PubMed] [Google Scholar]
- 6.Remeijer L, et al. Prevalence and Clinical Consequences of Herpes Simplex Virus Type 1 DNA in Human Cornea Tissues. The Journal of Infectious Diseases. 2009;200:11–19. doi: 10.1086/599329. [DOI] [PubMed] [Google Scholar]
- 7.Wang L, Wang R, Xu C, Zhou H. Pathogenesis of Herpes Stromal Keratitis: Immune Inflammatory Response Mediated by Inflammatory Regulators. Frontiers in Immunology. 2020;11 doi: 10.3389/fimmu.2020.00766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Awasthi S, et al. Nucleoside-modified mRNA encoding HSV-2 glycoproteins C, D, and E prevents clinical and subclinical genital herpes. Science Immunology. 2019;4:eaaw7083. doi: 10.1126/sciimmunol.aaw7083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bolland S, Pierce SK. Ups and downs in the search for a Herpes simplex virus vaccine. eLife. 2015;4:e06883. doi: 10.7554/eLife.06883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vadlapudi AD, Vadlapatla RK, Mitra AK. Update on emerging antivirals for the management of herpes simplex virus infections: a patenting perspective. Recent Pat Antiinfect Drug Discov. 2013;8:55–67. doi: 10.2174/1574891x11308010011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jiang YC, Feng H, Lin YC, Guo XR. New strategies against drug resistance to herpes simplex virus. Int J Oral Sci. 2016;8:1–6. doi: 10.1038/ijos.2016.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schaeffer HJ, et al. 9-(2-hydroxyethoxymethyl) guanine activity against viruses of the herpes group. Nature. 1978;272:583–585. doi: 10.1038/272583a0. [DOI] [PubMed] [Google Scholar]
- 13.Koganti R, Yadavalli T, Shukla D. Current and Emerging Therapies for Ocular Herpes Simplex Virus Type-1 Infections. Microorganisms. 2019;7:429. doi: 10.3390/microorganisms7100429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Crute JJ, et al. Herpes simplex virus helicase-primase inhibitors are active in animal models of human disease. Nat Med. 2002;8:386–391. doi: 10.1038/nm0402-386. [DOI] [PubMed] [Google Scholar]
- 15.Kleymann G, et al. New helicase-primase inhibitors as drug candidates for the treatment of herpes simplex disease. Nat Med. 2002;8:392–398. doi: 10.1038/nm0402-392. [DOI] [PubMed] [Google Scholar]
- 16.Jaishankar D, et al. An off-target effect of BX795 blocks herpes simplex virus type 1 infection of the eye. Sci Transl Med. 2018;10:eaan5861. doi: 10.1126/scitranslmed.aan5861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nelson CE, et al. Long-term evaluation of AAV-CRISPR genome editing for Duchenne muscular dystrophy. Nat Med. 2019;25:427–432. doi: 10.1038/s41591-019-0344-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maeder ML, et al. Development of a gene-editing approach to restore vision loss in Leber congenital amaurosis type 10. Nat Med. 2019;25:229–233. doi: 10.1038/s41591-018-0327-9. [DOI] [PubMed] [Google Scholar]
- 19.Beyret E, et al. Single-dose CRISPR-Cas9 therapy extends lifespan of mice with Hutchinson-Gilford progeria syndrome. Nat Med. 2019;25:419–422. doi: 10.1038/s41591-019-0343-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Santiago-Fernandez O, et al. Development of a CRISPR/Cas9-based therapy for Hutchinson-Gilford progeria syndrome. Nat Med. 2019;25:423–426. doi: 10.1038/s41591-018-0338-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee B, et al. Nanoparticle delivery of CRISPR into the brain rescues a mouse model of fragile X syndrome from exaggerated repetitive behaviours. Nat Biomed Eng. 2018;2:497–507. doi: 10.1038/s41551-018-0252-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gao X, et al. Treatment of autosomal dominant hearing loss by in vivo delivery of genome editing agents. Nature. 2018;553:217–221. doi: 10.1038/nature25164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dash PK, et al. Sequential LASER ART and CRISPR Treatments Eliminate HIV-1 in a Subset of Infected Humanized Mice. Nat Commun. 2019;10:2753. doi: 10.1038/s41467-019-10366-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.de Buhr H, Lebbink RJ. Harnessing CRISPR to combat human viral infections. Curr Opin Immunol. 2018;54:123–129. doi: 10.1016/j.coi.2018.06.002. [DOI] [PubMed] [Google Scholar]
- 25.Aubert M, et al. In vivo disruption of latent HSV by designer endonuclease therapy. JCI Insight. 2016;1:e88468. doi: 10.1172/jci.insight.88468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aubert M, et al. Gene editing and elimination of latent herpes simplex virus in vivo. Nat Commun. 2020;11:4148. doi: 10.1038/s41467-020-17936-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van Diemen FR, et al. CRISPR/Cas9-Mediated Genome Editing of Herpesviruses Limits Productive and Latent Infections. PLoS Pathog. 2016;12:e1005701. doi: 10.1371/journal.ppat.1005701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oh HS, et al. Herpesviral lytic gene functions render the viral genome susceptible to novel editing by CRISPR/Cas9. Elife. 2019;8 doi: 10.7554/eLife.51662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weerasooriya S, DiScipio KA, Darwish AS, Bai P, Weller SK. Herpes simplex virus 1 ICP8 mutant lacking annealing activity is deficient for viral DNA replication. Proc Natl Acad Sci U S A. 2019;116:1033–1042. doi: 10.1073/pnas.1817642116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weller SK, Coen DM. Herpes simplex viruses: mechanisms of DNA replication. Cold Spring Harb Perspect Biol. 2012;4:a013011. doi: 10.1101/cshperspect.a013011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reinert LS, et al. Sensing of HSV-1 by the cGAS-STING pathway in microglia orchestrates antiviral defence in the CNS. Nat Commun. 2016;7:13348. doi: 10.1038/ncomms13348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Oral acyclovir for herpes simplex virus eye disease: effect on prevention of epithelial keratitis and stromal keratitis. Herpetic Eye Disease Study Group. Arch Ophthalmol. 2000;118:1030–1036. [PubMed] [Google Scholar]
- 33.Kennedy DP, et al. Ocular herpes simplex virus type 1: is the cornea a reservoir for viral latency or a fast pit stop? Cornea. 2011;30:251–259. doi: 10.1097/ICO.0b013e3181ef241d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Newell CK, Martin S, Sendele D, Mercadal CM, Rouse BT. Herpes simplex virus-induced stromal keratitis: role of T-lymphocyte subsets in immunopathology. J Virol. 1989;63:769–775. doi: 10.1128/jvi.63.2.769-775.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stuart PM, Summers B, Morris JE, Morrison LA, Leib DA. CD8+ T cells control corneal disease following ocular infection with herpes simplex virus type 1. J Gen Virol. 2004;85:2055–2063. doi: 10.1099/vir.0.80049-0. [DOI] [PubMed] [Google Scholar]
- 36.Jeon S, Rowe AM, Carroll KL, Harvey SAK, Hendricks RL. PD-L1/B7-H1 Inhibits Viral Clearance by Macrophages in HSV-1-Infected Corneas. J Immunol. 2018;200:3711–3719. doi: 10.4049/jimmunol.1700417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Biswas PS, Rouse BT. Early events in HSV keratitis—setting the stage for a blinding disease. Microbes Infect. 2005;7:799–810. doi: 10.1016/j.micinf.2005.03.003. [DOI] [PubMed] [Google Scholar]
- 38.Bae S, Park J, Kim J-S. Cas-OFFinder: a fast and versatile algorithm that searches for potential off-target sites of Cas9 RNA-guided endonucleases. Bioinformatics. 2014;30:1473–1475. doi: 10.1093/bioinformatics/btu048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sarah B, Ibtissam H, Mohammed B, Hasna S, Abdeljalil M. Intrastromal Injection of Bevacizumab in the Management of Corneal Neovascularization: About 25 Eyes. Journal of ophthalmology. 2016;2016:6084270-6084270. doi: 10.1155/2016/6084270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Berrozpe-Villabona C, et al. Intrastromal bevacizumab injection for corneal neovascularization in herpetic stromal keratitis. J Fr Ophtalmol. 2015;38:776–777. doi: 10.1016/j.jfo.2015.02.005. [DOI] [PubMed] [Google Scholar]
- 41.Sharma N, et al. Management Algorithm for Fungal Keratitis: The TST (Topical, Systemic, and Targeted Therapy) Protocol. Cornea. 2019;38:141–145. doi: 10.1097/ICO.0000000000001781. [DOI] [PubMed] [Google Scholar]
- 42.Narayana S, et al. Mycotic Antimicrobial Localized Injection: A Randomized Clinical Trial Evaluating Intrastromal Injection of Voriconazole. Ophthalmology. 2019;126:1084–1089. doi: 10.1016/j.ophtha.2019.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Koelle DM, et al. Tegument-specific, virus-reactive CD4 T cells localize to the cornea in herpes simplex virus interstitial keratitis in humans. J Virol. 2000;74:10930–10938. doi: 10.1128/jvi.74.23.10930-10938.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Maertzdorf J, Verjans GM, Remeijer L, van der Kooi A, Osterhaus AD. Restricted T cell receptor beta-chain variable region protein use by cornea-derived CD4+ and CD8+ herpes simplex virus-specific T cells in patients with herpetic stromal keratitis. J Infect Dis. 2003;187:550–558. doi: 10.1086/367991. [DOI] [PubMed] [Google Scholar]
- 45.Okinaga S. Shedding of herpes simplex virus type 1 into tears and saliva in healthy Japanese adults. Kurume Med J. 2000;47:273–277. doi: 10.2739/kurumemedj.47.273. [DOI] [PubMed] [Google Scholar]
- 46.Kobayashi K, et al. Pseudotyped Lentiviral Vectors for Retrograde Gene Delivery into Target Brain Regions. Front Neuroanat. 2017;11:65. doi: 10.3389/fnana.2017.00065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Arriagada G. Retroviruses and microtubule-associated motor proteins. Cell Microbiol. 2017;19:e12759. doi: 10.1111/cmi.12759. [DOI] [PubMed] [Google Scholar]
- 48.Kato S, et al. Enhancement of the transduction efficiency of a lentiviral vector for neuron-specific retrograde gene delivery through the point mutation of fusion glycoprotein type E. J Neurosci Methods. 2019;311:147–155. doi: 10.1016/j.jneumeth.2018.10.023. [DOI] [PubMed] [Google Scholar]
- 49.Yin D, et al. Intracorneal delivery of HSV-targeting CRISPR/Cas9 mRNA prevents herpetic stromal keratitis. bioRxiv. 2020:2020.2002.2008.934125 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data generated or analysed during this study are available from the corresponding author on reasonable request. The deep sequencing and whole genome sequencing data are available at NCBI BioProject. The BioProject IDs are PRJNA668071 and PRJNA668060, respectively.