Abstract
CpG islands (CGIs) represent a widespread feature of vertebrate genomes, being associated with ~70% of all gene promoters. CGIs control transcription initiation by conferring nearby promoters with unique chromatin properties. In addition, there are thousands of distal or orphan CGIs (oCGIs) whose functional relevance is barely known. Here we show that oCGIs are an essential component of poised enhancers (PEs) that augment their long-range regulatory activity and control the responsiveness of their target genes. Using a knock-in strategy in mouse embryonic stem cells (ESCs), we introduced PEs with or without oCGIs within topologically associating domains (TADs) harboring genes with different types of promoters. Analysis of the resulting cell lines revealed that oCGIs act as tethering elements that promote the physical and functional communication between PEs and distally located genes, particularly those with large CGI clusters in their promoters. Therefore, by acting as genetic determinants of gene-enhancer compatibility, CGIs can contribute to gene expression control under both physiological and potentially pathological conditions.
Introduction
Enhancers are a heterogeneous group of distal cis-regulatory elements containing clusters of transcription factor binding sites (TFBS) that control gene expression in a distance- and orientation-independent manner1. The regulatory properties of enhancers have been mostly investigated using transgenic reporter assays2 in which enhancer activity is evaluated by measuring the capacity to activate transcription of a reporter gene from a minimal promoter. In these assays, the investigated sequences are placed at short distances from the reporter genes and using a limited set of minimal promoters. On the other hand, insulators prevent enhancers from ectopically activating non-target genes3. In vertebrates, insulators are typically bound by CTCF, which, together with Cohesin, can form long-range chromatin loops that demarcate the boundaries of regulatory domains and limit enhancer activity4. Current models of enhancer function implicitly assume that enhancers and genes can effectively communicate with each other, regardless of distance or sequence composition, as far as they are located within the same regulatory domain3. However, recent studies show that the disruption of regulatory domains does not always lead to changes in gene expression or enhancer-gene communication5–9. Similarly, enhancers and their developmental target genes can reside within the same regulatory domains together with “bystander” genes that are not responsive to the enhancers10. Therefore, additional factors, such as the type of core promoter elements11,12, contribute to enhancer responsiveness. However, it is currently unknown whether other genetic factors (e.g. distance or enhancer sequence composition) can also contribute to such responsiveness, which is essential to understand the pathological consequences of human structural variation13.
We previously showed that PEs control the induction of major neural genes upon mouse ESC differentiation14. Before becoming active in anterior neural progenitors (AntNPCs), PEs are already bookmarked in ESCs with unique chromatin and topological features, including binding by polycomb-group protein complexes (PcG) and pre-formed contacts with their target genes14,15. PEs have a distinctive genetic composition that includes not only clusters of TFBS but also nearby CGIs14. CGIs are a prevalent feature of vertebrate gene promoters, providing them with a permissive chromatin state that facilitates transcription initiation16. However, only half of the CGIs found in the mouse and human genomes are associated with promoters (pCGIs)16,17, while the other half, known as oCGIs, remains poorly studied. oCGIs have been proposed to act as alternative gene promoters18 or highly active enhancers with limited tissue specificity17,19,20. Nevertheless, the mechanisms whereby oCGIs might contribute to transcriptional regulation remain unknown. Here we show that oCGIs act as long-range potentiators of PEs, enabling the functional communication between PEs and developmental genes with CpG-rich promoters. Therefore, our work uncovers CGIs as major determinants of enhancer-gene compatibility and provides important insights into how gene expression programs are specifically and precisely deployed during development.
Results
Genetic properties of PE-associated oCGIs
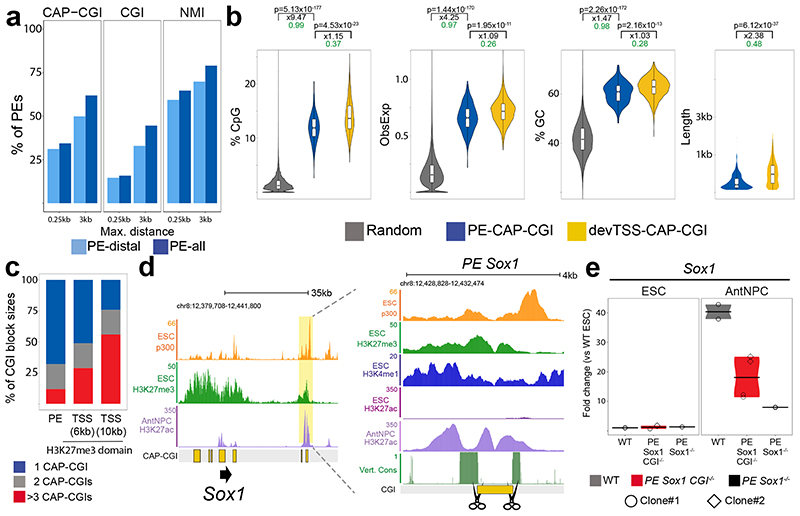
PEs identified in mouse ESCs are commonly located in proximity to computationally predicted CGIs14. However, computational models underestimate the abundance of CGIs, especially those distally located from transcription start sites (TSS)21. Using biochemically identified CGIs ((i) CGIs identified by CXXC affinity purification and deep sequencing (CAP-seq) (i.e. CAP-CGIs)18 and (ii) non-methylated islands (NMIs)21) we found that ~60-80% of PEs are located within 3 kb of a CAP-CGI or a NMI, respectively (Fig. 1a). In comparison to the CAP-CGIs located in proximity of the TSS of developmental genes, those associated with PEs were shorter and had lower CpG density (Fig. 1b). Moreover, PEs tend to be associated with single CAP-CGIs, whereas developmental gene promoters frequently contain clusters of two or more CAP-CGIs22 (Fig. 1c, Extended Data Fig. 1a). Here we use the term oCGI regardless of whether these sequences are identified computationally or experimentally, although many of the PE oCGIs display lower GC content and CpG ratios than the classically defined CGIs (Fig. 1b).
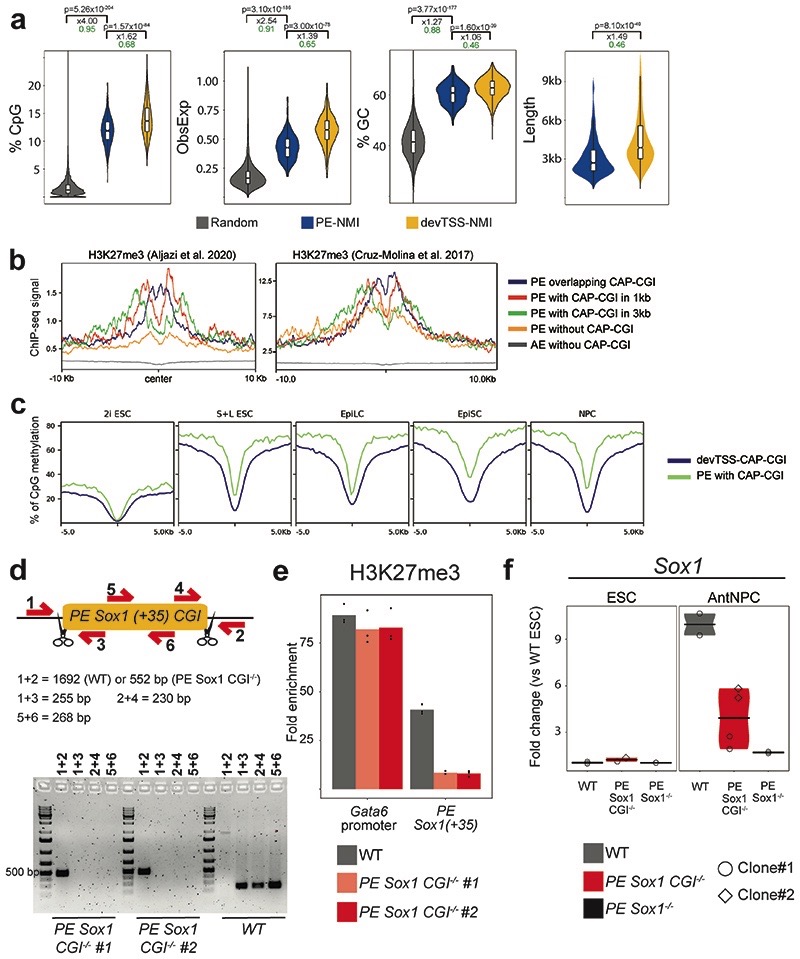
Fig. 1. Genetic properties and functional relevance of orphan CGIs associated with PEs.
a, Percentage of PEs within the indicated maximum distances (0.25 kb or 3 kb) of a CGI identified by CAP-seq18 (left), a NMI21 (middle) or a computationally defined CGI (right). b, Comparison of the CpG%, observed/expected CpG ratio, GC% and sequence length between random regions (n = 436,000), CAP-CGIs associated with PE-distal (PE-CAP-CGI; n = 276) and CAP-CGIs associated with the TSS of developmental genes (devTSS-CAP-CGI; n = 1,926) (Methods). P values were calculated using unpaired two-sided Wilcoxon tests with Bonferroni correction for multiple testing; black numbers indicate median fold-changes; green numbers indicate non-negligible Cliff’s delta effect sizes. The center line of the violin plot represents the median, the boxes encompass the interquartile range and the whiskers extend to the minimum and maximum. c, Percentage of CAP-CGI block sizes (1, 2 or ≥3 CAP-CGIs) associated with PE-distal (n = 253) or the TSS of developmental genes (devTSS; n = 1,522 with at least one CAP-CGI in <3 kb. The devTSS were classified in two groups based on the length of the H3K27me3 domains associated with them (>6 kb (n = 1,522) and >10 kb (n = 599)). d, Left panel: ChIP-seq data14 from ESCs (p300 and H3K27me3) and AntNPCs (H3K27ac) at the Sox1 locus. The PE Sox1(+35) is highlighted in yellow. Right panel: close-up view of the PE Sox1(+35) with additional epigenomic and genomic data, including a computationally defined CGI. Vert. Cons. = vertebrate PhastCons. e, Sox1 expression was investigated by RT-qPCR in cells that were either WT, homozygous for a deletion of the PE Sox1(+35) CGI (PE Sox1 CGI-/-) or homozygous for a deletion of the complete PE Sox1(+35) 14 (PE Sox1-/-). N = 2 independent PE Sox1 CGI-/- ESC clones (circles and diamonds) and n = 1 PE Sox1-/- clone were studied. For each ESC clonal line, n = 2 replicates of the AntNPC differentiation were performed. Expression values were normalized to two housekeeping genes (Eef1a and Hprt) and are presented as fold-changes with respect to WT ESCs. The colored area of the violin plot represents the expression values distribution and the center line represents the median. N = 1 independent biological replicate of this experiment is shown in Extended Data Figure 1f.
pCGIs serve as recruitment platforms for proteins that can modify chromatin (e.g. PcG, TET1)23,24. Consequently, pCGIs are hypomethylated and enriched in H3K27me314,15. Analysis of publically available data14,25–28 showed that PEs associated with CAP-CGIs are also hypomethylated and more enriched in H3K27me3 than PEs or active enhancers (AE) not linked to CAP-CGIs (Extended Data Fig. 1b-c). Therefore, PEs are pervasively found in proximity of CGIs, which in turn might endow them with unique chromatin features.
oCGIs are necessary for PE regulatory function
To start evaluating the regulatory role of oCGIs in the context of PEs, we generated mouse ESC lines with a homozygous deletion of the oCGI associated with PE Sox1(+35), a PE that controls the expression of Sox1 in neural progenitors14 (Fig. 1d; Extended Data Fig. 1d). The oCGI deletion severely reduced H3K27me3 levels around PE Sox1(+35) in ESCs (Extended Data Fig. 1e). Next, we measured Sox1 expression in WT, PE Sox1(+35)CGI-/- and PE Sox1(+35)-/- ESCs as well as upon their differentiation into AntNPCs. In ESCs, neither the deletion of the oCGI nor of the whole PE Sox1(+35) affected Sox1 expression (Fig. 1e; Extended Data Fig. 1f). However, in AntNPCs the oCGI deletion reduced Sox1 expression by >2-fold (Fig. 1e; Extended Data Fig. 1f), thus suggesting that oCGIs might positively influence the cis-activation capacity of PEs14.
Dissection of PE regulatory logic by genetic engineering
The functional assessment of PE oCGIs using loss-of-function approaches has certain limitations: (i) oCGIs can be difficult to delete individually, as they frequently overlap with nearby TFBS (Extended Data Fig. 2a); (ii) PE target genes typically display complex regulatory landscapes in which multiple enhancers control gene expression29, thus potentially masking the regulatory function of individual oCGIs; (iii) the loss of CGI-bound proteins (e.g. PcG) can elicit global molecular changes that indirectly alter PE loci.
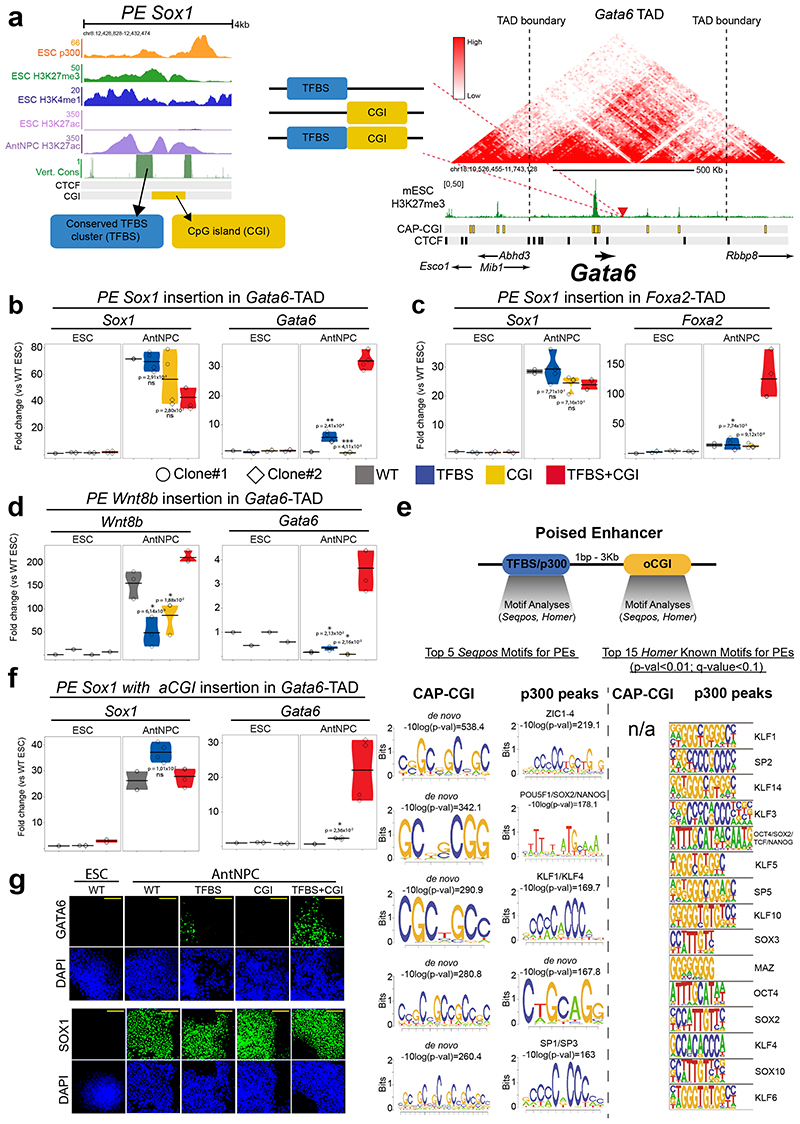
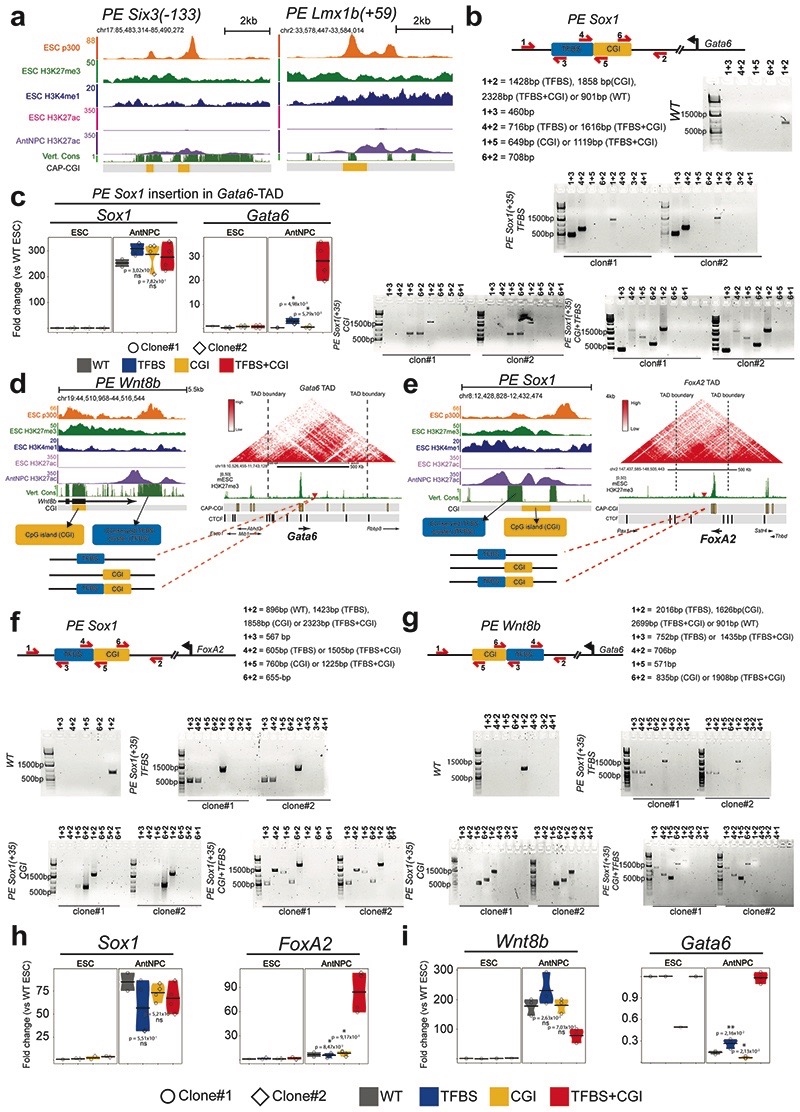
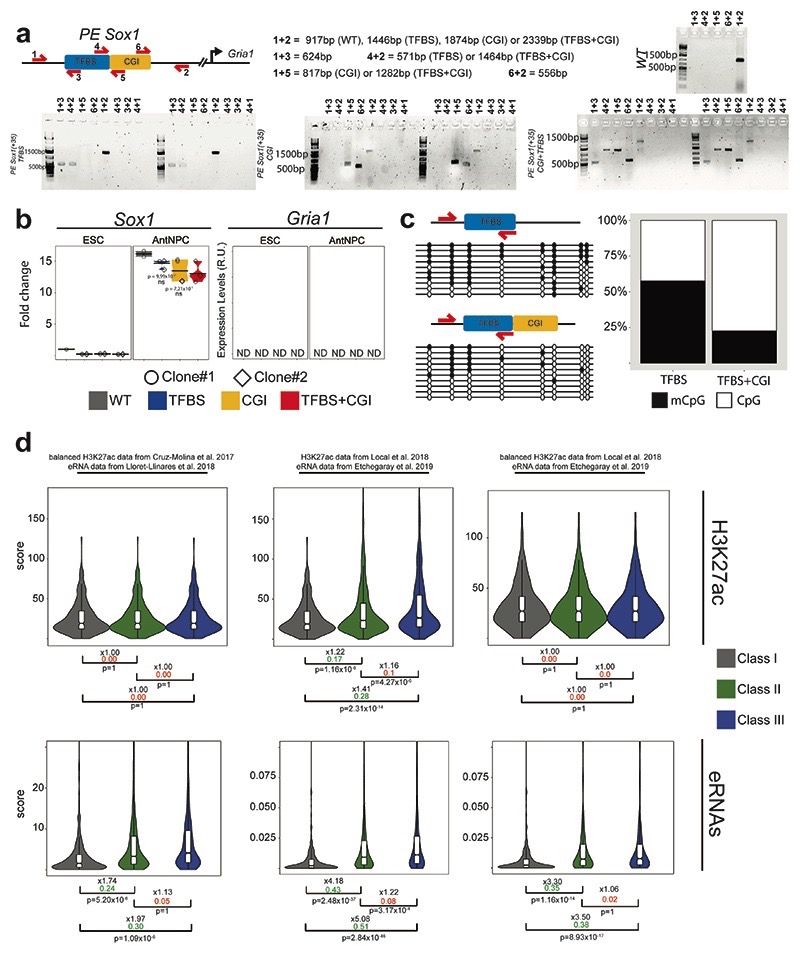
To systematically dissect the contribution of oCGIs to the regulatory function of PEs, we designed a genetic engineering approach to generate ESCs in which the components of selected PEs are modularly inserted (i.e. TFBS, oCGI or TFBS+oCGI) into a fixed genomic location32,33 (Fig. 2a). We reasoned that by selecting insertion sites located within TADs containing developmental genes not expressed in ESCs or AntNPCs and, thus, without active enhancers in these cell types, any changes in the expression of the selected genes could be attributed solely to the inserted PE sequences. To implement this approach, we initially inserted the PE Sox1(+35) components (i.e. PE Sox1(+35)TFBS, PE Sox1(+35)CGI or PE Sox1(+35)TFBS+CGI) approximately 100 kb downstream of Gata6 (Fig. 2a), a gene with multiple CGIs around its promoter region and lowly expressed in both ESCs and AntNPCs. The selected insertion site was not conserved and was not close to any CTCF binding site, thus minimizing the risk of disrupting any regulatory element. Using this strategy, we established two homozygous ESC clones for each of the PE Sox1(+35) inserts described above (Extended Data Fig. 2b). Next, we measured Gata6 expression in the previous ESC lines and upon their differentiation into AntNPCs. In ESCs none of the engineered PE Sox1(+35) combinations affected Gata6 expression (Fig. 2b; Extended Data Fig. 2c). Strikingly, upon differentiation into AntNPCs, Gata6 was strongly induced in cells with the TFBS+CGI insertion (~50-fold vs. WT). In contrast, cells with the TFBS displayed considerably milder Gata6 induction (~7-fold vs. WT), while the CGI had no effect on Gata6 expression (Fig. 2b; Extended Data Fig. 2c).
Fig. 2. Modular engineering of PEs reveals major regulatory functions for orphan CGIs.
a, Strategy to insert the PE Sox1(+35) components into the Gata6-TAD. Left: epigenomic and genetic features of the PE Sox1(+35). The oCGI is not evolutionary conserved. Middle: the three combinations of PE Sox1(+35) modules inserted into the Gata6-TAD. Right: TAD in which Gata6 is located (i.e. Gata6-TAD)30,31. The red triangle indicates the integration site of the PE Sox1(+35) modules approximately 100 kb downstream of Gata6. b-d and f, The expression of Gata6 (b, d and f), Foxa2 (c), Sox1 (b, c and f) and Wnt8b (d) was measured by RT-qPCR in ESCs and AntNPCs that were either WT or homozygous for the insertion of the different PE Sox1(+35) (b-c) or PE Wnt8b(+21) (d) modules. In (f), the PE Sox1(+35)TFBS was inserted alone or in combination with an artificial CGI into the Gata6-TAD. For the cells with the PE insertions, n = 2 independent clonal cell lines (circles and diamonds) were studied in each case. For each cell line, n = 2 replicates of the AntNPC differentiation were performed. Expression values were normalized to two housekeeping genes (Eef1a and Hprt) and are presented as fold-changes with respect to WT ESC. N = 1 independent biological replicate of these experiments is shown in Extended Data Figure 2. In (b-d and f), the expression differences between AntNPCs with the TFBS+CGI module and AntNPCs with the other PE modules were calculated using two-sided non-paired t-tests (*** fold-change >2 & P <0.0001; ** fold-change >2 & P <0.001; * fold-change >2 & P <0.05; ns: not significant; fold-change <2 or P >0.05). The colored area of the violin plot represents the expression values distribution and the center line represents the median. e, TF motif analyses using Homer32 and Seqpos33 for PEs with a CAP-CGI within less than 3 kb and that do not overlap with the p300 peaks defining the PEs14. Motif analyses were performed separately for the CAP-CGIs and the p300 peaks. g, Immunofluorescence assays for GATA6 and SOX1 in WT ESCs or AntNPCs that were either WT or homozygous for the insertion of the different PE Sox1(+35) modules in the Gata6-TAD. Scale bar = 100 μm.
oCGIs amplify PE regulatory activity
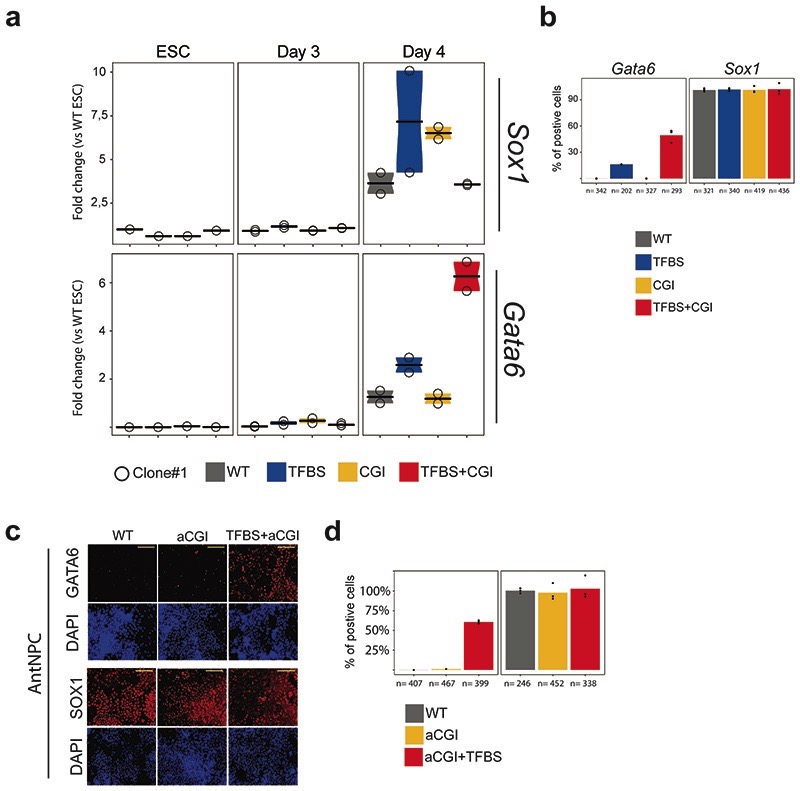
To evaluate whether the previous observations could be generalized, we generated two additional groups of transgenic ESC lines: (i) PE Sox1(+35) components were inserted within the Foxa2-TAD (~100 kb downstream of Foxa2 TSS, which contains several CGIs and is inactive in ESCs and AntNPCs); (ii) PE Wnt8b(+21) 14 components were inserted within the Gata6-TAD (~100 kb downstream of Gata6-TSS) (Extended Data Fig. 2d-g). Importantly, the TFBS+CGI inserts were able to strongly induce gene expression in AntNPCs (Fig. 2c,d; Extended Data Fig. 2h,i), while the TFBS or the oCGI alone lead to either no or minor gene inductions, respectively (Fig. 2c,d; Extended Data Fig. 2h,i).
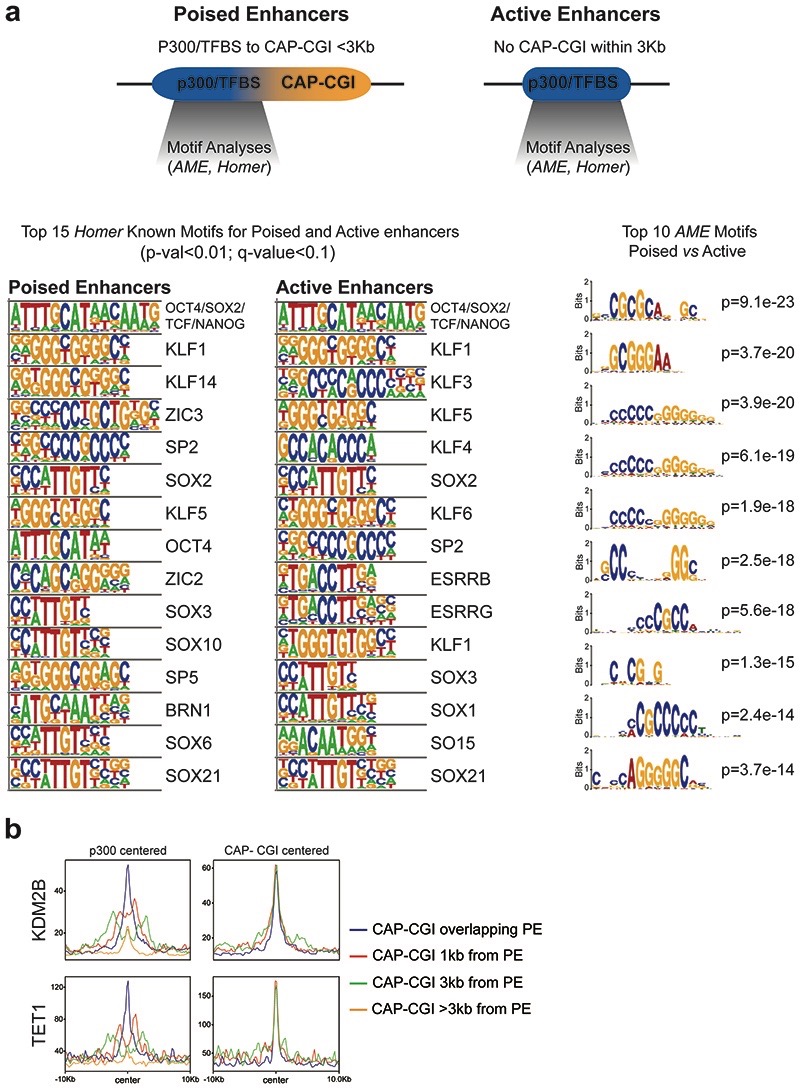
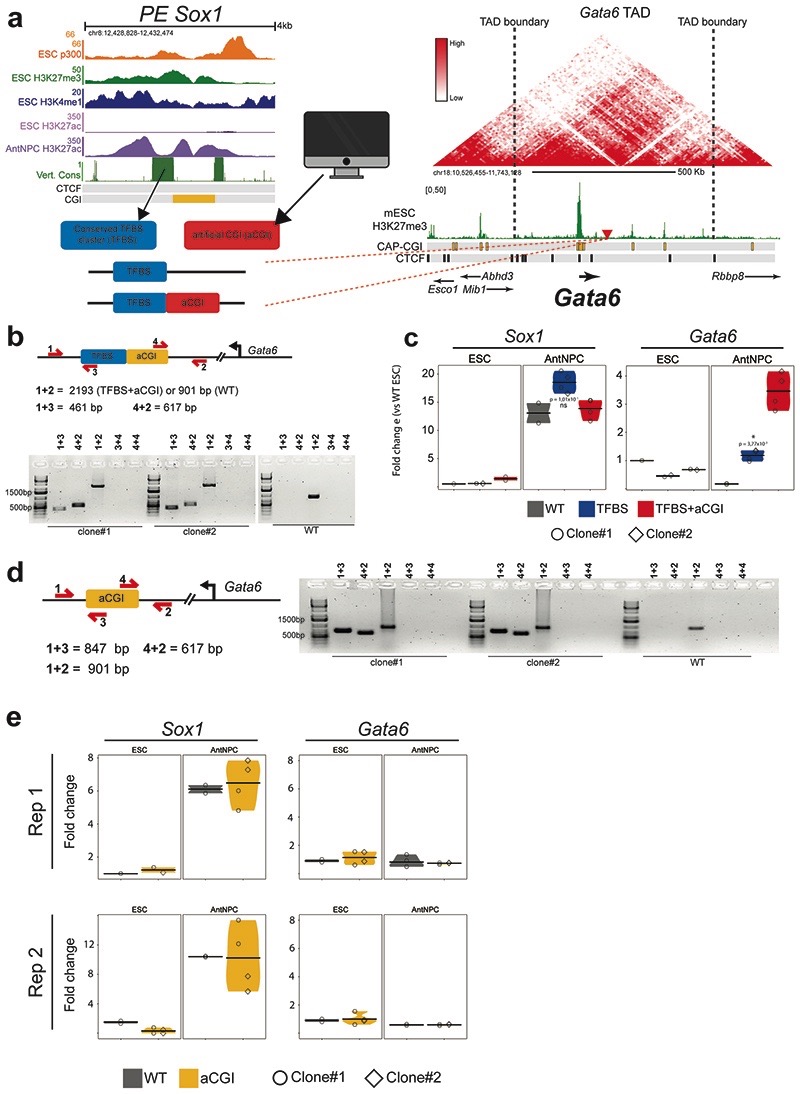
Next, we investigated whether the boosting capacity of the oCGIs could be attributed to other type of regulatory information beyond their CpG-richness (e.g. TF binding sites). In silico motif analyses using as input either the CAP-CGIs or the TFBS/p300 peaks from PEs in which these elements do not overlap (Fig. 2e) showed that p300 peaks, but not CAP-CGIs, were strongly enriched in binding motifs for pluripotency TFs14 (e.g. OCT4, SOX2, NANOG). Similarly, differential motif analyses revealed that CpG-rich motifs were strongly overrepresented among PEs compared to active enhancers (Extended Data Fig. 3a)14. CGIs can serve as recruitment platforms for proteins containing ZF-CxxC domains (e.g. KDM2B, TET1)16,34, which could contribute to the unique chromatin features of PEs (Extended Data Fig. 1b,c). Analysis of KDM2B and TET1 ChIP-seq data generated in ESCs35,36 showed that the binding of these proteins to PEs was positively correlated with the presence of nearby CAP-CGIs (Extended Data Fig. 3b). Next, we designed an artificial CGI (aCGI; Methods) and inserted it alone or together with the PE Sox1(+35)TFBS at the Gata6-TAD (Extended Data Fig. 4a,b,d). Notably, the TFBS+aCGI considerably increased Gata6 expression in AntNPCs compared to the TFBS (Fig. 2f; Extended Data Fig. 4c), whereas the insertion of the aCGI alone did not alter Gata6 expression (Extended Data Fig. 4e). Although we cannot completely dismiss that some oCGIs contain relevant binding sites for tissue-specific TFs, our results indicate that the CpG-richness of the oCGIs is important to increase the regulatory activity of PEs.
The boosting properties of oCGIs might be attributed to a premature induction of the target gene, an increase in the number of cells in which the target gene becomes induced and/or an increase in the expression levels within individual cells. To address this, we focused on those cell lines containing the different PE Sox1(+35) components inserted within the Gata6-TAD. Upon differentiation of the TFBS+CGI ESCs into AntNPCs, Gata6 did not become induced until day 4, thus matching the expression dynamics of Sox1 (the endogenous target of PE Sox1(+35)) and arguing against premature gene induction due to the presence of the oCGI (Extended Data Fig. 5a). Next, we performed immunofluorescence assays to visualize GATA6 and SOX1 proteins in WT and Gata6-TAD cells. SOX1 became strongly and homogeneously induced in AntNPCs derived from all the evaluated cell lines37 (Fig. 2g; Extended Data Fig. 5b-d). Notably, GATA6 was also induced in ~50% and ~60% of the AntNPCs derived from TFBS+CGI or TFBS+aCGI ESCs, respectively (Fig. 2g; Extended Fig. 5c-d). In contrast, the TFBS resulted in noisier and more heterogeneous GATA6 expression, while no GATA6 could be detected in cells having the CGI or aCGI alone. These results suggest that oCGIs increase the number of cells in which the PE target genes get induced, thus potentially leading to high gene expression precision38.
oCGIs do not increase the local activation of PEs
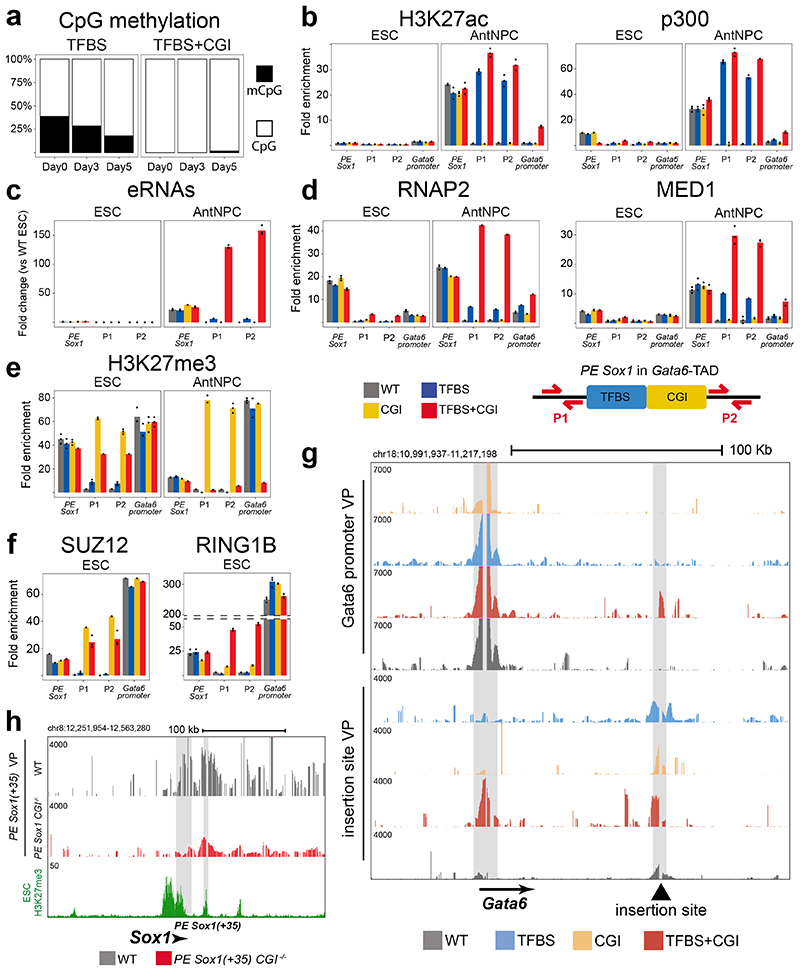
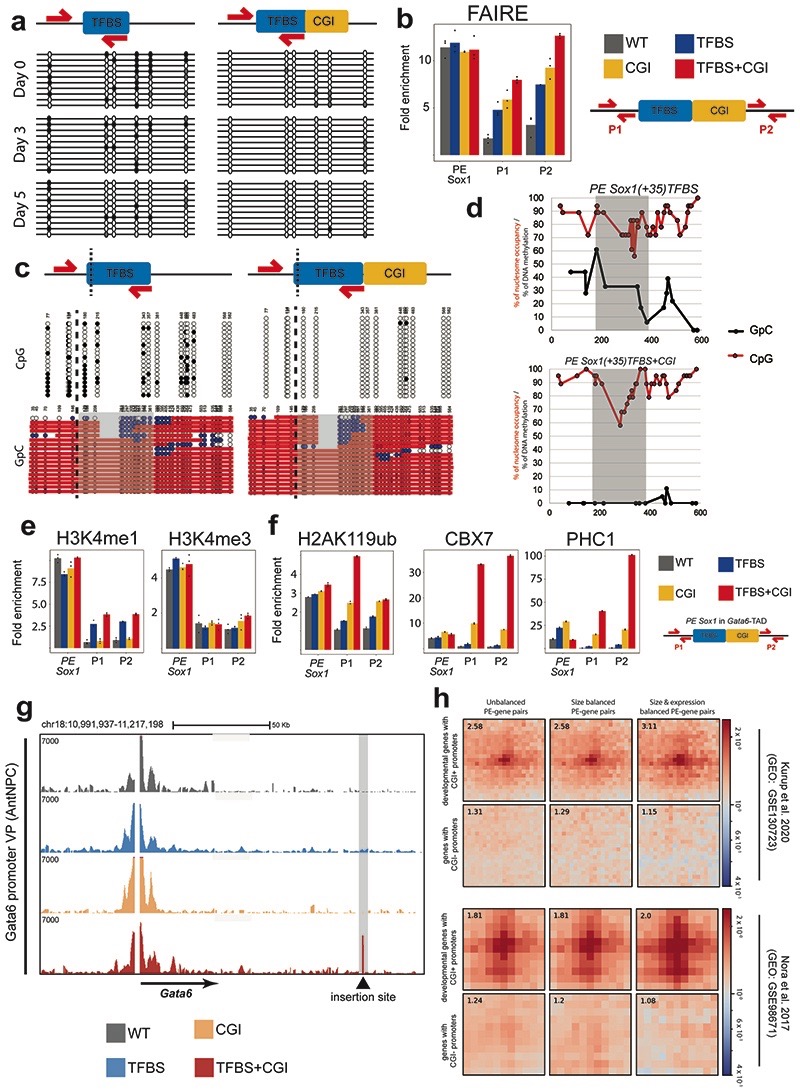
To investigate the mechanisms whereby oCGIs potentiate the regulatory function of PEs, we focused on the ESC lines in which the PE Sox1(+35) components were inserted within the Gata6-TAD. pCGIs are typically devoid of CpG methylation and display low nucleosomal density, which might provide a chromatin environment permissive for TF binding and transcription initiation39,40. Bisulfite sequencing experiments in TFBS+CGI and TFBS ESCs showed that the TFBS sequences acquired intermediate CpG methylation levels when inserted alone, while becoming completely unmethylated when combined with the oCGI (Fig. 3a; Extended Data Fig. 6a). In contrast, FAIRE assays showed that the oCGI only moderately increased chromatin accessibility whether inserted alone or in conjunction with the TFBS (Extended Data Fig. 6b). To simultaneously measure nucleosome occupancy and CpG methylation at the inserted TFBS with single-DNA molecule resolution41, we also performed NOME-PCR assays. These experiments confirmed that oCGIs protect nearby TFBS from CpG methylation without a major impact on chromatin accessibility (Extended Data Fig. 6c,d). Furthermore, upon differentiation into AntNPCs, the TFBS got progressively demethylated in the TFBS cells (Fig. 3a; Extended Data Fig. 6a), suggesting that, even in the absence of an oCGI, TFs can access and activate PEs in AntNPCs42. To test this prediction, we performed ChIP-qPCR experiments to measure p300 binding and H3K27ac levels, two major hallmarks of active enhancers15, around the inserted PE Sox1(+35) constructs. Interestingly, in AntNPCs the PEs containing the TFBS alone or together with the oCGI became strongly and similarly enriched in H3K27ac and p300 (Fig. 3b). Therefore, the boosting capacity of the oCGIs cannot be simply attributed to their local chromatin effects.
Fig. 3. Characterization of the epigenetic, topological and regulatory features of the PE Sox1(+35) modules engineered within the Gata6-TAD.
a, Bisulfite sequencing analyses in ESCs (Day 0) and AntNPCs (Day 5) differentiated from cell lines with the PE Sox1(+35)TFBS or PE Sox1(+35)TFBS+CGI modules inserted in the Gata6-TAD. DNA methylation levels were measured using a forward bisulfite primer upstream of the insertion site and a reverse primer inside the TFBS module (Methods). b, H3K27ac and p300 levels at the endogenous PE Sox1(+35), the Gata6-TAD insertion site (P1 and P2 primer pairs) and the Gata6 promoter were measured by ChIP-qPCR in ESCs (left) and AntNPCs (right) that were either WT (gray) or homozygous for the insertion of the different PE Sox1(+35) modules. ChIP-qPCR signals were normalized against two negative control regions (Supplementary Data 1). The bars display the mean of n = 3 technical replicates (black dots). c, eRNA levels at the endogenous PE Sox1(+35) and the Gata6-TAD insertion site (P1 and P2 primer pairs) were measured by RT-qPCR in ESCs (left) and AntNPCs (right) that were either WT (gray) or homozygous for the insertions of the different PE Sox1(+35) modules. Expression values were normalized to two housekeeping genes (Eef1a and Hprt) and are presented as fold-changes with respect to WT ESCs. The bars display the mean of n = 3 technical replicates (black dots). d-f, RNAP2 and MED1 (d), H3K27me3 (e) or SUZ12 and RING1B (f) levels were measured by ChIP-qPCR as described in (b). g, 4C-seq experiments were performed using the Gata6 promoter (upper panels) or the Gata6-TAD insertion site (lower panels) as viewpoints in ESCs that were either WT (grey) or homozygous for the insertions of the different PE Sox1(+35) modules. h, 4C-seq experiments were performed using the PE Sox1(+35) as a viewpoint in ESCs that were either WT or homozygous for the deletion of PE Sox1(+35) CGI (PE Sox1 CGI-/-). The genomic location of PE Sox1(+35) and Sox1 are highlighted in grey.
oCGIs increase PE-target gene communication
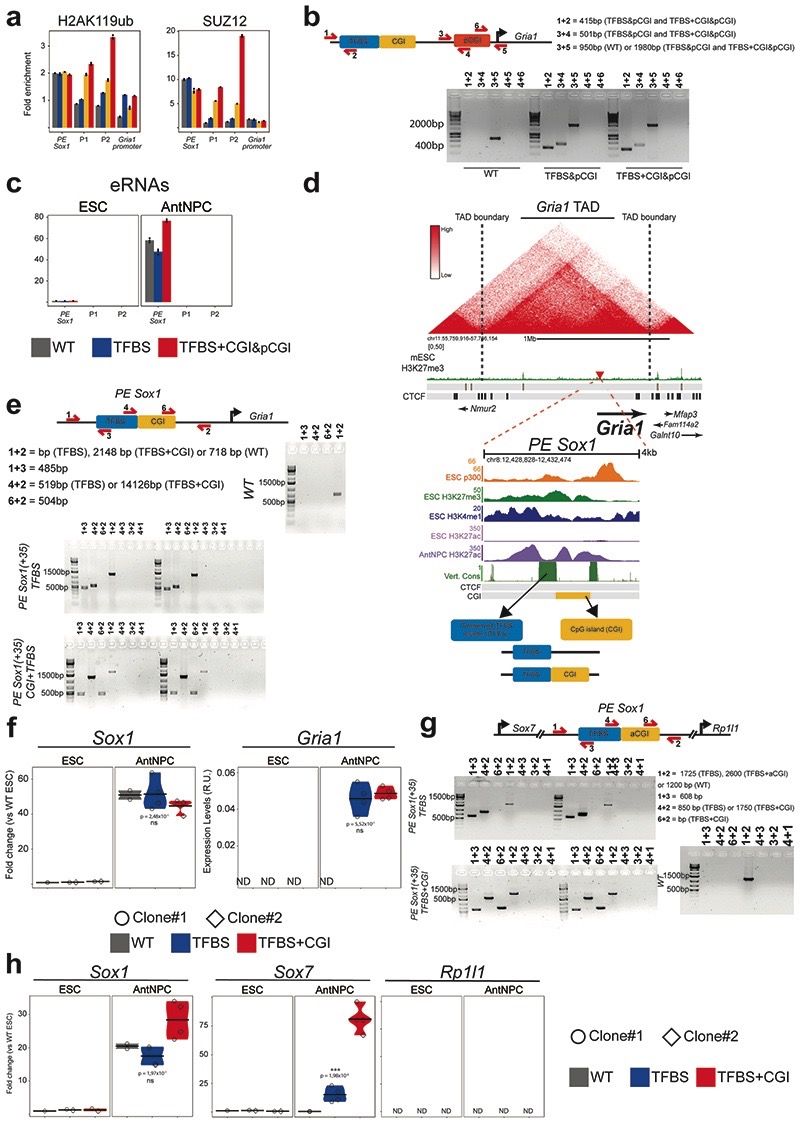
Another distinctive hallmark of active enhancers is the production of short bidirectional transcripts termed enhancer RNAs (eRNAs)43. Remarkably, eRNA levels in AntNPCs were >20-fold higher around the TFBS+CGI insert in comparison with the TFBS alone (Fig. 3c). Moreover, upon AntNPC differentiation, the TFBS+CGI insert became highly enriched in RNA polymerase II (RNAP2) and Mediator (Fig. 3d). In contrast, the binding of these proteins to the TFBS and CGI inserts was either considerably weaker or undetectable, respectively (Fig. 3d). Similarly, the recruitment of RNAP2 and Mediator to the Gata6 promoter was also stronger in AntNPCs with the TFBS+CGI insert (Fig. 3d).
In their inactive state, PEs are enriched in histone modifications (i.e. H3K27me3 and H3K4me1) and protein complexes (e.g. PcG) implicated in the establishment of long-range chromatin interactions14,15,44,45. Therefore, oCGIs could be implicated in the establishment of PEs’ unique chromatin signature, facilitating the physical communication between PEs and their target genes. To investigate this possibility, we performed ChIP for H3K4me1, H3K4me3 and H3K27me3/PcG in the ESC lines containing the different PE Sox1(+35) components within the Gata6-TAD (Fig. 3e; Extended Data Fig. 7a). H3K4me1 was weakly enriched around the PE Sox1(+35) inserts containing the TFBS with or without the oCGI, while no enrichment was observed for the oCGI insert alone (Extended Data Fig. 7a). On the other hand, H3K4me3 was not enriched in any of the evaluated ESC lines (Extended Data Fig. 7a), indicating that oCGIs do not adopt the same chromatin state as pCGI. Most interestingly, H3K27me3, H2AK119ub and additional PcG subunits (i.e. SUZ12, CBX7, PHC1 and RING1B) were strongly enriched around the PE Sox1(+35) inserts containing the oCGI (Fig. 3e-f; Extended Fig. 7b). Intriguingly, PRC1 recruitment (i.e. CBX7, PHC1 and RING1B) was considerably stronger for the TFBS+oCGI insert than for the oCGI alone (Fig. 3f; Extended Data Fig. 7b).
Since PcG can mediate long-range homotypic interactions between distal PcG-bound loci44–49, we investigated the three-dimensional organization of the Gata6 locus in our engineered ESC lines. 4C-seq experiments using either the Gata6 promoter or the PE Sox1(+35) insertion site as viewpoints revealed strong PE-Gata6 contacts only in the TFBS+CGI cells (Fig. 3g). The lack of PE-gene contacts in cells with the CGI alone could be attributed to the weaker recruitment of PRC1 to the insert in these cells50,51 (Extended Data Fig. 7b). Furthermore, the strong interactions between the TFBS+CGI insert and the Gata6 promoter were also observed upon differentiation into AntNPCs (Extended Data Fig. 7c). Although the TFBS insert alone did not significantly contact with the Gata6 promoter in AntNPCs (Extended Data Fig. 7c), Gata6 was induced, albeit weakly, in these cells (Fig. 2b). This could be explained by the more transient and/or heterogeneous interactions between Gata6 and the PE in the absence of the oCGI and/or by the capacity of enhancers to induce gene expression without getting into close proximity of their target genes3,52,53. Next, to evaluate whether oCGIs are important for PE-gene contacts in an endogenous genomic context, we performed 4C-seq experiments in the ESCs in which the oCGI associated with PE Sox1(+35) was deleted. Importantly, the deletion of this oCGI reduced the interactions between Sox1 and PE Sox1(+35) (Fig. 3h).
Overall, our data suggest that oCGIs increase the functional communication between PEs and their target genes by bringing them into close spatial proximity.
CpG-poor promoters do not show responsiveness to distal PEs
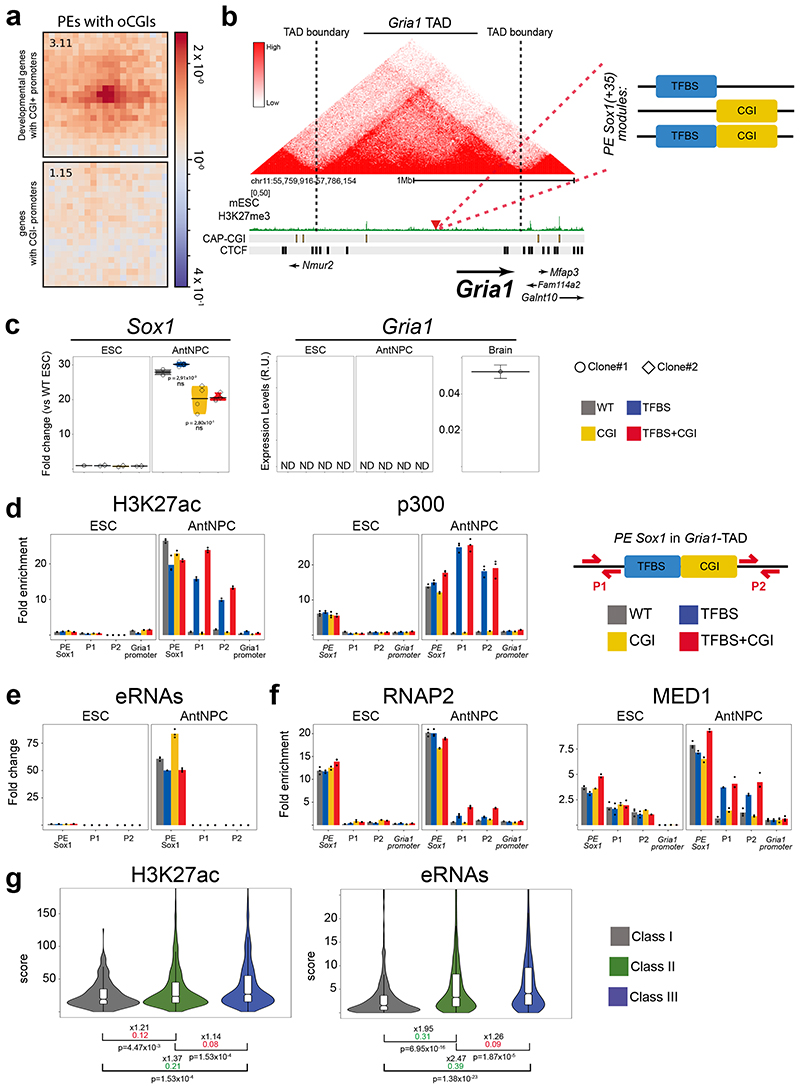
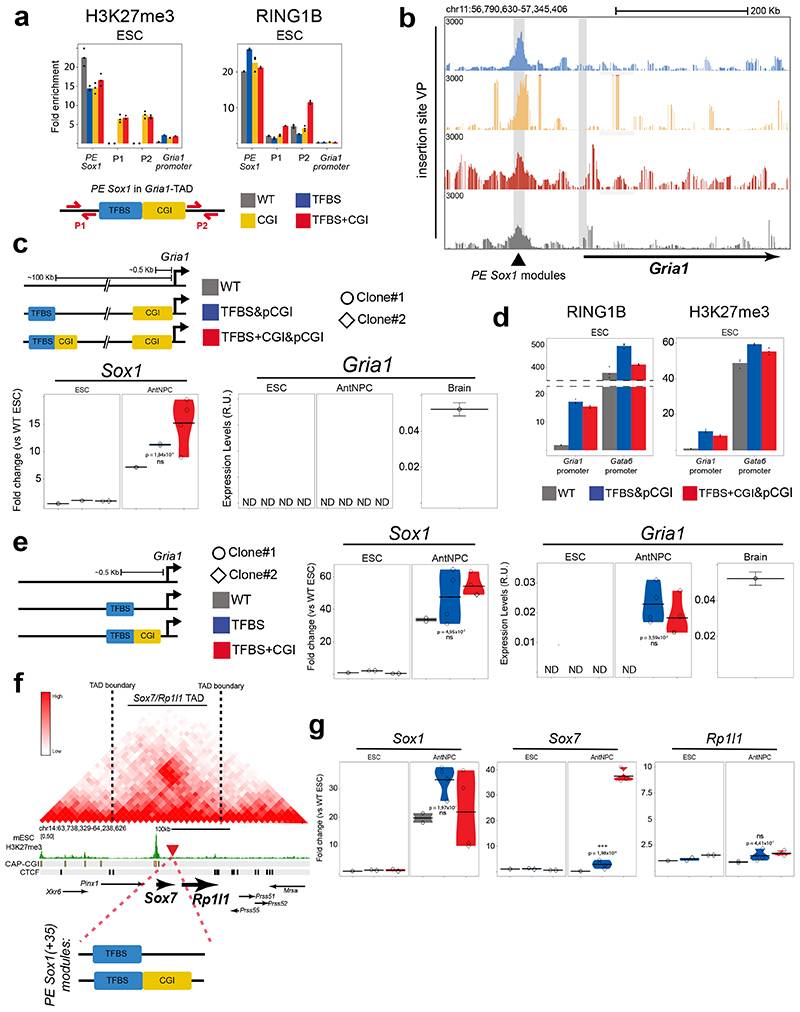
Developmental genes, such as those regulated by PEs14,15, contain large CGI clusters around their promoters, whereas tissue-specific genes tend to have CpG-poor promoters22. PE and their target genes could spatially segregate from genes with CpG-poor promoters by engaging into active (i.e. transcription factories) or inactive (i.e. polycomb bodies) homotypic chromatin interactions depending on their transcriptional state54,55. Therefore, the responsiveness of developmental genes to PEs could depend not only on the presence of PE-oCGIs, but also on CGIs located at the target gene promoters. In agreement with this hypothesis, analysis of Hi-C data generated in ESCs7,56 showed that PEs strongly interact with developmental genes with CGI-rich promoters located in the same TADs, but not with genes with CGI-poor promoters (Fig. 4a; Extended Data Fig. 7d). To test whether CpG-poor promoters are responsive to PEs, we inserted the PE Sox1(+35) components into the Gria1-TAD, approximately 100 kb upstream of the Gria1-TSS (Fig. 4b; Extended Data Fig. 8a). Similarly to Gata6 and Foxa2, Gria1 is not expressed in either ESCs or AntNPCs. However, the Gria1 promoter does not contain CGIs and is not bound by PcG but fully DNA methylated instead (Fig. 4b). Remarkably, upon differentiation of the Gria1-TAD cell lines, none of the PE Sox1(+35) inserts was able to induce Gria1 expression (Fig. 4c; Extended Data Fig. 8b). To gain mechanistic insights into this lack of responsiveness, we measured DNA methylation, H3K27ac, p300, RNAP2, MED1 and eRNA levels around the inserted PE Sox1(+35) constructs. Similarly to what we observed within the Gata6-TAD, the TFBS became demethylated in ESCs, albeit partially, when combined with the oCGI (Extended Data Fig. 8c). Furthermore, upon differentiation into AntNPCs, the TFBS+CGI and TFBS inserts became strongly and similarly enriched in H3K27ac and p300 (Fig. 4d). However, in contrast to what we observed in the Gata6-TAD, we did not detect eRNAs around any of the PE Sox1(+35) inserts (Fig. 4e). Congruently, the recruitment of RNAP2 and MED1 to the PE Sox1(+35) was weak regardless of whether the TFBS were alone or with the oCGI (Fig. 4f). In addition, RNAP2 and MED1 were not recruited to the Gria1 promoter, thus in agreement with the lack of Gria1 induction observed upon differentiation of the Gria1-TAD ESC lines (Fig. 4f).
Fig. 4. Genes with CpG-poor promoters do not show long-range responsiveness to PEs.
a, Pile-up plots showing average Hi-C interactions in ESCs56 between PE-distal and developmental genes with CGI-rich promoters (n = 401 PE-gene pairs) or genes with CGI-poor promoters (n = 900 PE-gene pairs) (Methods). b, Strategy to insert the PE Sox1(+35) components into the Gria1-TAD30,31. c, Gria1 and Sox1 expression was measured by RT-qPCR in ESCs and AntNPCs with the indicated genotypes as in Fig. 2 (n = 1 independent biological replicate is shown in Extended Data Fig. 8b). Gria1 was also measured in the mouse brain to illustrate the quality of the RT-qPCR primers. Gria1 expression values are presented as arbitrary units (R.U.) since it was not detectable (ND) except in the brain. For Sox1, expression differences between AntNPCs with the TFBS+CGI module or the other PE modules were calculated using two-sided non-paired t-tests (ns: not significant; fold-change <2 or P >0.05). d, H3K27ac and p300 levels at the endogenous PE Sox1(+35), the Gria1-TAD insertion site (P1 and P2) and the Gria1 promoter were measured by ChIP-qPCR in cells with the indicated genotypes. ChIP-qPCR signals were calculated as described in Figure 3. e, eRNA levels at the endogenous PE Sox1(+35) and the Gria1-TAD insertion site (P1 and P2) were measured by RT-qPCR in cells with the indicated genotypes. RT-qPCR signals were calculated as described in Figure 3. f, RNAP2 and MED1 levels were measured by ChIP-qPCR as in (d). g, Violin plots showing H3K27ac and eRNA levels for active enhancers classified into three categories: Class I (active enhancers in TADs containing only poorly expressed genes; n = 271); Class II (active enhancers in TADs with at least one highly expressed gene); n = 2,566; Class III (active enhancers whose closest genes in the same TAD is highly expressed; n = 1,294) (see Methods). P values were calculated using two-sided unpaired Wilcoxon tests with Bonferroni correction for multiple testing; the numbers in black indicate median fold-changes; the colored numbers correspond to negligible (red) and non-negligible (green) Cliff’s delta effect sizes. The violin box graphs were calculated as in Figure 1.
Our results indicate that H3K27ac and eRNA production can be uncoupled from each other and represent different steps during PE activation (Fig. 3b,c; Fig. 4d,e). Namely, the accumulation of H3K27ac might occur as PEs become locally activated, while the production of eRNAs, which is coupled with gene transcription, could signify the functional activation of the PEs43,57. To assess if these observations could be generalized, we compared eRNA production between three classes of active enhancers using nascent transcriptomic and epigenomic data generated in ESCs14,58–60: (I) enhancers located in TADs containing only poorly expressed genes; (II) enhancers located in TADs with at least one highly expressed gene; (III) enhancers whose closest gene within the same TAD is highly expressed (Methods). Interestingly, Class I enhancers showed ~2 and 2.5-fold lower eRNA levels than Class II and Class III enhancers, respectively, while H3K27ac levels were similar among the three enhancer groups (Fig. 4g; Extended Data Fig. 8d). These results suggest that enhancer and gene transcription are frequently coupled and mutually dependent on each other43,61.
Promoters with large CGI clusters are responsive to PEs
The experiments within the Gata6-TAD suggest that the responsiveness to PEs involves the physical proximity between PEs and their target genes, which in ESCs is likely to be mediated by PcG present at both PEs and promoters14,62 (Fig. 3g). ChIP experiments in the Gria1-TAD ESC lines revealed that PcG were recruited to the PE Sox1(+35) inserts containing an oCGI (Fig. 5a; Extended Data Fig. 9a), albeit not as strongly as for the Gata6-TAD (Fig. 3e,f). Furthermore, the Gria1 promoter, which does not contain pCGIs, was not bound by PcG (Fig. 5a; Extended Data Fig. 9a). Accordingly, 4C-seq analyses showed that none of the inserted PE Sox1(+35) constructs were able to interact with the Gria1 promoter (Fig. 5b). In principle, the addition of pCGIs to the Gria1 promoter could increase PcG recruitment and, consequently, the physical and functional communication with the distal PE Sox1(+35) constructs. To test this prediction, we introduced one of the Gata6 pCGI into the Gria1 promoter in those ESC lines containing either the TFBS+CGI or TFBS inserts 100 kb away from the Gria1-TSS (Fig. 5c; Extended Data Fig. 9b). Upon differentiation into AntNPCs, the addition of the pCGI did not result in detectable Gria1 mRNA or eRNAs around the PE inserts, suggesting that a single CGI is not sufficient to trigger the long-range responsiveness to PEs (Fig. 5c; Extended Data Fig. 9c). Interestingly, in comparison to the PcG levels observed for promoters with large CGI clusters (e.g. Gata6), the insertion of a single pCGI into the Gria1 promoter led to relatively mild PcG recruitment (Fig. 5d). This could explain, at least partly, the lack of Gria1 responsiveness to the distal PE Sox1(+35). Alternatively, the Gria1 promoter might contain core-promoter elements that are not responsive to developmental enhancers11,12,63. To evaluate this possibility, we generated ESC lines in which the PE Sox1(+35)TFBS or PE Sox1(+35)TFBS+CGI constructs were integrated 380 bp upstream of the Gria1-TSS (Fig. 5e; Extended Data Fig. 9d,e). Remarkably, both the TFBS+CGI and TFBS inserts were able to strongly induce Gria1 expression upon differentiation into AntNPCs (Fig. 5e; Extended Data Fig. 9f). These results show that the Gria1 promoter can respond to the PE Sox1(+35) and suggest that the boosting effect of the oCGI might be lost when the PEs are located close to gene promoters.
Fig. 5. Promoters with large CGI clusters are particularly responsive to distal PEs.
a, H3K27me3 and RING1B levels at the endogenous PE Sox1(+35), the Gria1 TAD insertion site (P1 and P2) and the Gria1 promoter were measured by ChIP-qPCR in cells with the indicated genotypes. ChIP-qPCR signals were calculated as in Figure 3. b, 4C-seq experiments were performed using the Gria1-TAD insertion site as a viewpoint in ESCs with the indicated genotypes. c, ESC clonal lines with homozygous insertions of PE Sox1(+35)TFBS or PE Sox1(+35)TFBS+CGI 100 kb upstream of the Gria1-TSS (Fig. 4b), respectively, were used to insert a Gata6-pCGI immediately upstream of the Gria1-TSS. Gria1 and Sox1 expression was measured by RT-qPCR in cells with the indicated genotypes. For the PE Sox1(+35)TFBS cells, a single clone was used, while for the PE Sox1(+35)TFBS+CGI cells, n = 2 independent clonal lines (circles and diamonds) were studied. For each cell line, n = 2 replicates of the AntNPC differentiation were performed. The mouse brain expression values are the same as in Figure 4c. d, RING1B and H3K27ac levels at the Gria1 and Gata6 promoter were measured by ChIP-qPCR in ESCs with the indicated genotypes. ChIP-qPCR signals were calculated as in Figure 2. e, Gria1 and Sox1 expression was measured by RT-qPCR in ESCs and AntNPCs that were WT or homozygous for the indicated PE Sox1(+35) modules inserted 380 bp upstream of the Gria1 TSS (an independent biological replicate is shown in Extended Data Fig. 9e). For cells with the PE module insertions, two different clonal lines (circles and diamonds) were studied in each case. f, Strategy to insert the PE Sox1(+35) components into the Sox7/Rp1l1-TAD. The red triangle indicates the integration site located in between Sox7 and Rp1l1. g, Sox1, Sox7 and Rp1l1 expression was measured by RT-qPCR in cells with the indicated genotypes. For cells with the PE insertions, n = 2 independent clonal lines (circles and diamonds) were studied in each case. In (c, e and g), the expression differences between AntNPCs with TFBS+CGI or TFBS were calculated using two-sided non-paired t-tests (*** fold-change >2 & P <0.0001; ns: not significant; fold-change <2 or P >0.05).
When inserted into the Gria1-TAD, the PE Sox1(+35)TFBS+CGI did not acquire the same chromatin state as within the Gata6-TAD (i.e. lower PRC1 levels and higher DNA methylation (Fig. 5a; Extended Data Fig. 8c)). Therefore, the constitutive heterochromatin environment of the Gria1-TAD might result in chromatin and/or topological properties at the PE insertion site that somehow compromise the regulatory function of the oCGI. To investigate this and further assess whether developmental genes with large CGI clusters in their promoters are particularly responsive to PEs, we inserted the PE Sox1(+35) components into the Sox7/Rp1l1-TAD, right between Sox7 and Rp1l1 (24 kb from Sox7 and Rp1l1 TSSs) (Fig. 5f; Extended Data Fig. 9g). Sox7 and Rp1l1 are both inactive in ESCs and AntNPCs, but differ in their type of promoter (Fig. 5f): Sox7 is an endodermal regulator whose promoter contains a large CGI cluster and is strongly bound by PcG, while Rp1l1 is specifically expressed in mature rod cells and its promoter does not contain CGIs and is not bound by PcG. Remarkably, upon AntNPC differentiation, none of the PE Sox1(+35) inserts was able to induce Rp1l1 expression (Fig. 5g; Extended Data Fig. 9h). In stark contrast, Sox7 was strongly induced by the TFBS+CGI, while the TFBS alone led to a milder gene induction (Fig. 5g; Extended Data Fig. 9h). Together with our experiments in other TADs, these results strongly indicate that developmental genes with large CGI clusters in their promoters are particularly responsive to distal PEs.
CGIs and TAD boundaries control gene expression specificity
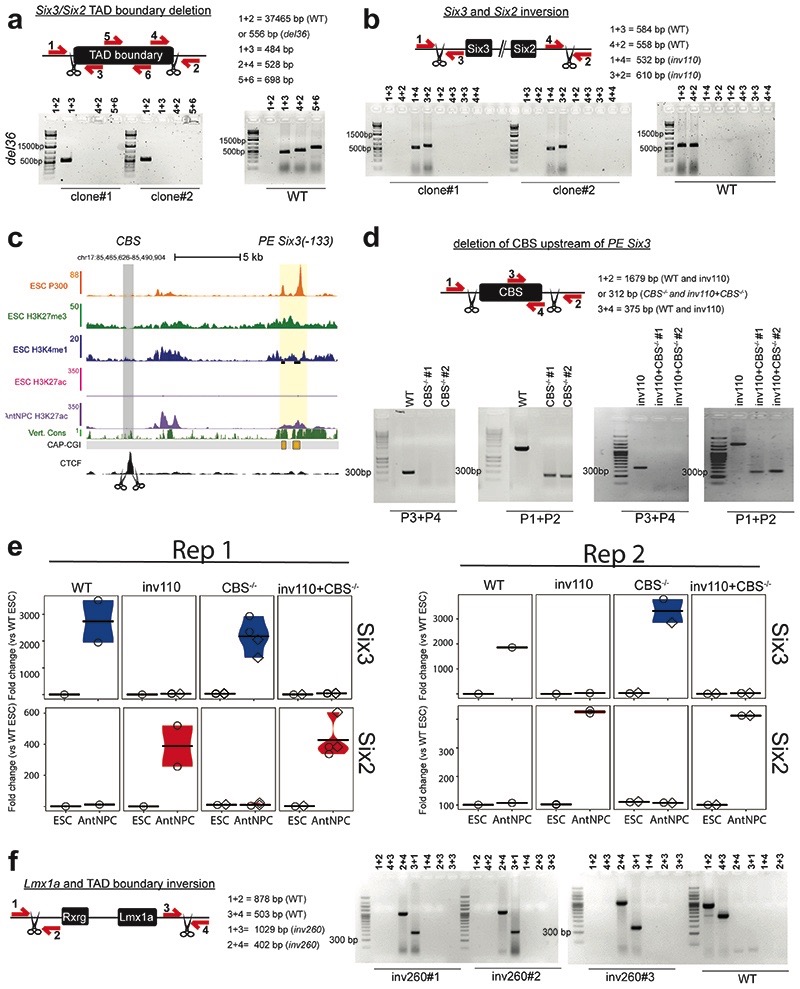
Our data suggest that, in addition to TAD boundaries, the interactions between PE-associated oCGIs and pCGI clusters proximal to developmental genes might contribute to gene expression specificity during embryogenesis. To test this prediction, we genetically engineered two different loci: Six3/Six2 and Lmx1a/Lrrc52/Mgst3 (Fig. 6a,d).
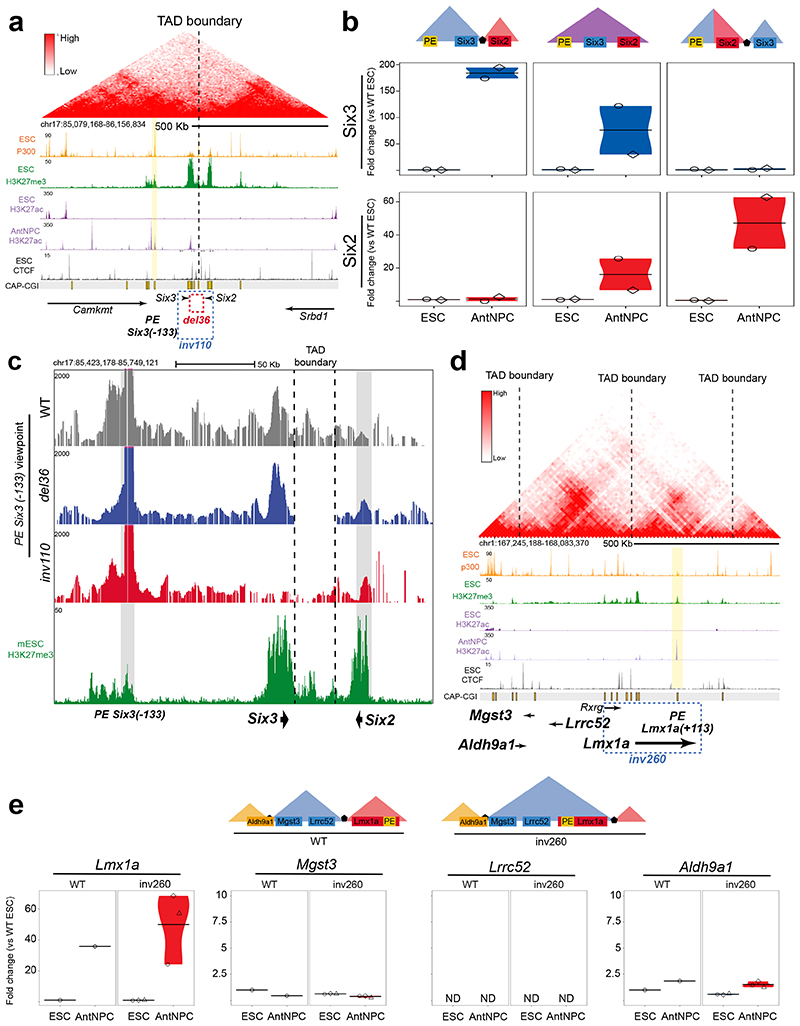
Fig. 6. oCGIs and TAD boundaries enable PEs to specifically induce their target genes.
a, The TADs in which Six3 and Six2 are located (i.e. Six3-TAD and Six2-TAD) are shown according to publically available Hi-C data30,31. Below the Hi-C data, several epigenomic and genetic features of the Six3-TAD and the Six2-TAD are shown. The dotted rectangles indicate the location of the 36-kb deletion (red) and 110-kb inversion (blue) engineered in ESCs. b, The expression of Six3 (blue) and Six2 (red) was measured by RT-qPCR in ESCs and AntNPCs that were either WT, homozygous for the 36-kb deletion (del36) or homozygous for the 110-kb inversion (inv110). For each of the engineered structural variants, n = 2 clonal cell lines were generated and independently differentiated into AntNPCs. Expression values were calculated as described in Figure 2. c, 4C-seq experiments were performed using the PE Six3(-133) as viewpoint in ESCs with the indicated genotypes. d, The TADs in which Lmx1a, Lrrc52 and Mgst3 are located are shown according to publically available Hi-C data30,31. Below the Hi-C data, several epigenomic and genetic features of the corresponding TADs are shown. The dotted rectangle indicates the location of the 260-kb inversion (inv260) engineered in ESCs. e, The expression of Lmx1a, Mgst3, Lrrc52 and Aldh9a1 was measured by RT-qPCR in cells with the indicated genotypes. For the inv260, n = 3 clonal cell lines were generated and independently differentiated into AntNPCs. Expression values were calculated as in Figure 2.
We first focused on the Six3/Six2 locus (Fig. 6a): (i) Six3 and Six2 are contained within two neighboring TADs separated by a conserved TAD boundary64,65; (ii) Six3 and Six2 display mutually exclusive expression patterns during embryogenesis (e.g. Six3 in brain; Six2 in facial mesenchyme)64; (iii) the Six3-TAD contains a PE (i.e. PE Six3(-133)) that controls the induction of Six3 in AntNPCs without any effects on Six2 14; (iv) in ESCs, the PE Six3(-133) strongly interacts with Six3 but not with Six2 14, although both genes contain multiple pCGIs. Next, we generated ESCs with either a 36-kb deletion spanning the Six3/Six2-TAD boundary (del36) or a 110-kb inversion that places Six3 within the Six2-TAD and vice versa (inv110) (Fig. 6a; Extended Data Fig. 10a,b). Upon differentiation into AntNPCs, Six2 was strongly induced in del36 and inv110 cells (~12- and ~35-fold vs. WT, respectively), while Six3 expression was dramatically reduced in inv110 cells (~77-fold vs. WT) and mildly affected in del36 cells (~2.5-fold vs. WT) (Fig. 6b). In agreement with these gene expression changes, 4C-seq experiments in WT, del36 and inv110 ESCs showed that both the boundary deletion and the inversion resulted in increased interactions between Six2 and PE Six3(-133) (Fig. 6c). Furthermore, a CTCF binding site (CBS) immediately upstream of the PE Six3(-133) could also contribute to the long-range communication with Six3/Six2 (Fig. 6a, Extended Data Fig. 10c). However, the deletion of this CBS did not have any major impact on Six3 or Six2 expression in either WT or inv110 AntNPCs, respectively (Extended Data Fig. 10c-e). Altogether, these results indicate that Six3 and Six2, whose promoters have large CGI clusters, are responsive to the PE Six3(-133) and, potentially, to other enhancers located within the Six3-TAD14.
Next, we focused on the Lmx1a/Lrrc52/Mgst3 locus (Fig. 6d). Lmx1a and Lrrc52/Mgst3 are located in neighboring TADs separated by a strong TAD boundary. The three genes have different types of promoters15,22 and expression patterns in ESCs and AntNPCs14. Lmx1a, a developmental gene with a large CGI cluster in its promoter, is bound by PcG in ESCs and induced in AntNPCs. Lrrc52, a tissue-specific gene without CGIs, is not bound by PcG in ESCs and is inactive in ESCs and AntNPCs. Mgst3, an ubiquitously expressed gene with a single and short CGI centered on its TSS, is not bound by PcG and is active in both ESCs and AntNPCs. The Lmx1a-TAD contains a PE (i.e. PE Lmx1a(+113) that becomes active in AntNPCs and that presumably contributes to Lmx1a induction in these cells. Considering all this, we generated two ESCs lines with a 260-kb inversion that places Lmx1a and PE Lmx1a(+113) within the Lrrc52/Mgst3-TAD (inv260) (Fig. 6d; Extended Data Fig. 10f). Notably, neither Lrrc52 nor Mgst3 were induced upon differentiation of the inv260 ESCs into AntNPCs (Fig. 6e). These results indicate that tissue-specific and housekeeping genes without large CGI clusters in their promoters are not responsive to distal PEs.
Overall, our data suggest that PEs specifically execute their regulatory functions due to the combined effects of TAD boundaries, which provide insulation, and homotypic interactions between oCGIs and pCGIs, which confer enhancer responsiveness (Fig. 7).
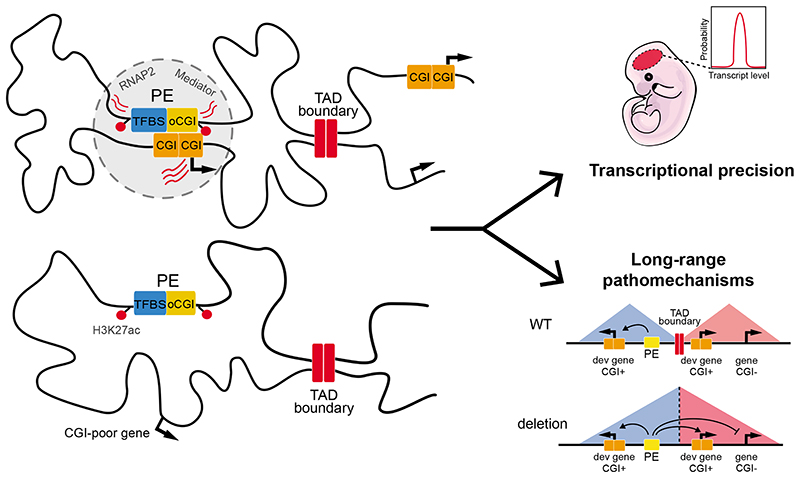
Fig. 7. Proposed model for the role of oCGI as amplifiers of PE regulatory activity and determinants of PE-gene compatibility.
The presence of oCGIs increases the physical communication of PEs with their target genes due to homotypic chromatin interactions between oCGIs and promoter-proximal CGI clusters. Consequently, the oCGIs can increase the number of cells and alleles in which the PEs and their target genes are in close spatial proximity (i.e. permissive regulatory topology) both during pluripotency and upon differentiation. This will ultimately result in a timely and homogenous induction of PE target genes once the PEs become active (i.e. increase transcriptional precision). In addition, the compatibility and responsiveness between PE and their target genes depends on the presence of oCGIs at the PEs and of the pCGI clusters at the target genes. Therefore, the oCGI can increase the specificity of PEs by enabling them to preferentially communicate with their CpG-rich target genes while still being insulated by TAD boundaries. These PE-gene compatibility rules may improve our ability to predict and understand the pathomechanisms of human structural variants.
Discussion
Deciphering the factors that control enhancer-promoter compatibility is a major challenge in the enhancer field66. According to current models, insulator proteins demarcate TAD boundaries and restrict enhancers to act upon genes located within their same TADs13,67,68. Nonetheless, enhancers do not promiscuously activate all the genes present within a TAD5,8,10,67,69, suggesting that additional factors control enhancer responsiveness. Massively parallel reporter assays in Drosophila showed that enhancer responsiveness is determined by the sequence composition of core promoters12,70. We now show that, in the context of PE loci, such responsiveness is also dependent on distal genetic elements, namely oCGIs, which serve as tethering elements that allow PEs to preferentially activate promoters containing large CGI clusters (Fig. 7). Although CGIs are considered a vertebrate-specific genomic feature, regulatory sequences with similar tethering functions have been also described in invertebrates71–73.
Our data suggest that the role of oCGIs as potentiators of PEs regulatory function does not involve the local activation of PEs but rather the establishment of long-range interactions with developmental genes (Fig. 7). In pluripotent cells, these PE-gene interactions are likely to be mediated by PRC1 complexes recruited to both oCGIs and pCGIs14,24,50,74,75. Intriguingly, our data suggest that the binding of PRC1 to the PEs is increased by the combination of TFBS and oCGIs. While the importance of CGIs as PcG recruitment platforms is well established24,75, how the TFBS can contribute to PRC1 recruitment is still an open question. Furthermore, our experiments in the Gria1-TAD suggest that a single pCGI is not sufficient to enable the long-range communication with PEs. This could be explained, at least partly, by the low levels of PRC1 recruitment that a single CGI can confer in comparison to the large CGI clusters associated with developmental gene promoters. Genetic engineering experiments whereby multiple and long CGIs are inserted in CpG-poor promoters will be required to assess if these genetic features are sufficient to increase the long-range responsiveness to PEs. Regardless, once recruited, PcG complexes might keep PEs and their target genes close together during pluripotent cell differentiation, ensuring that PEs uniformly induce their target genes as they become active. Then, once RNAs are produced at both PEs and their target genes, this would result in PcG eviction76. Although PRC1 might also contribute to PE-gene communication once PEs become active51, additional proteins are likely to be involved in the maintenance of such contacts77. Interestingly, upon PE activation the oCGIs increase the loading of Mediator and RNAP2 to both PEs and their target genes (Fig. 3d), suggesting that oCGIs might favor the formation of phase-separated transcriptional condensates78. Once PEs are active, multivalent interactions occurring within these condensates could robustly maintain PE-gene communication78. According to our analyses, the regulatory function of the PE-associated oCGIs could be primarily attributed to their CpG-richness. Namely, oCGIs can serve as recruitment platforms for ZF-CXXC proteins that, as part of major complexes (e.g. PcG, TrxG, Mediator), can facilitate the physical and functional communication between PEs and their target genes24,79,80. In addition, TFs with CG-rich binding sites (e.g. Sp1)81,82 might be also recruited to oCGIs and thereby contribute to PE-gene communication. Lastly, some oCGIs might contain binding sites for tissue-specific TFs that are important for the regulatory activity of PEs83.
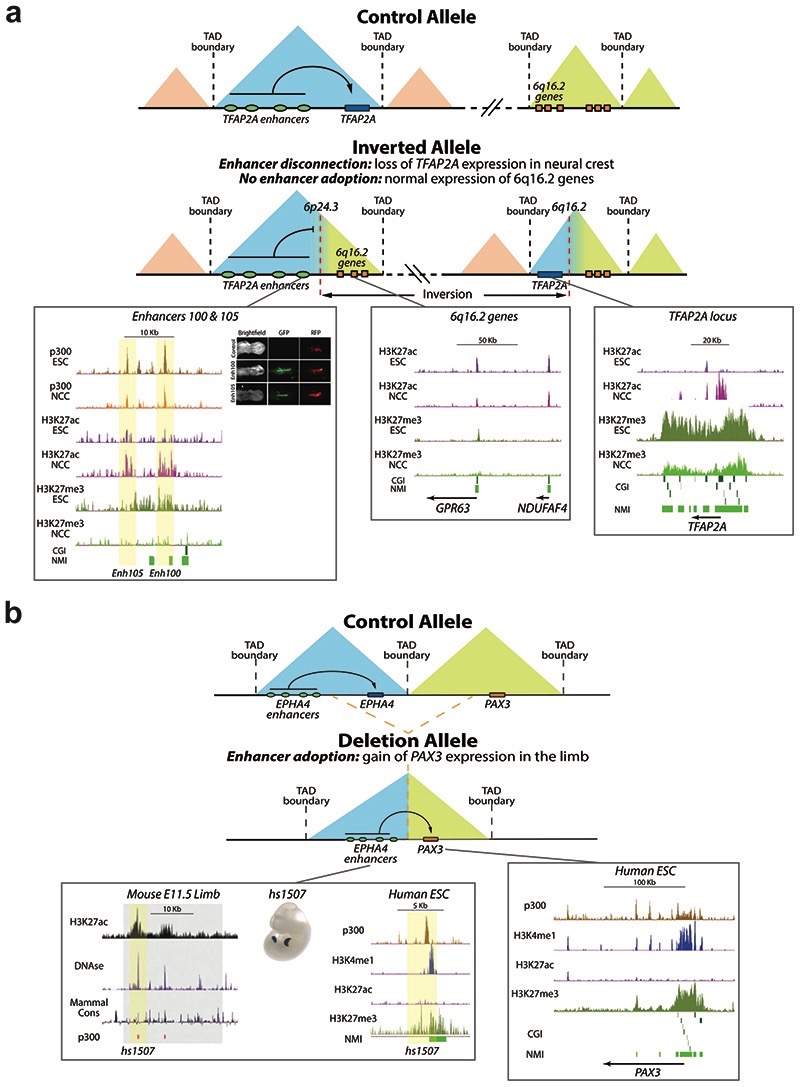
We propose a model whereby the precise and specific induction of certain developmental genes is achieved through the combination of CGI-mediated long-range chromatin interactions and the insulation provided by TAD boundaries (Fig. 7). As illustrated by the TFAP2A and EPHA4 loci, the function of CGIs as determinants of enhancer-gene compatibility can help understanding why only some structural variants that disrupt TAD organization lead to enhancer adoption and major changes in gene expression (Extended Data Fig. 11)5,9,67. Therefore, our findings may have important medical implications, as they could improve our ability to predict the pathological consequences of human structural variation13 (Fig. 7; Extended Data Fig. 11).
Methods
Cell lines and differentiation protocol
E14Tg2a (E14) mouse ESCs were cultured on gelatin-coated plates using Knock-out DMEM (Life Technologies, 10829018) supplemented with 15% FBS (Life Technologies, 10082147) and LIF. For the AntNPC differentiation86, ESCs were plated at 12,000 cells/cm2 on gelatin-coated plates and grown for three days in N2B27 medium supplemented with 10 ng/ml bFGF (Life Technologies, PHG0368) without serum or LIF. Subsequently, cells were grown for another two days in N2B27 medium without bFgf (D3–D5). From D2-D5 the N2B27 medium was supplemented with 5 mM Xav93987 (Sigma, 284028-89-3). N2B27 medium: Advanced Dulbecco’s Modified Eagle Medium F12 (Life Technologies, 21041025) and Neurobasal medium (Life Technologies, 12348017) (1:1), supplemented with 1× N2 (Life Technologies, 17502048), 1× B27 (Life Technologies, 12587010), 2 mM L-glutamine (Life Technologies, 25030024), 40 mg/ml BSA (Life Technologies, 15260037), 0.1 mM 2-mercaptoethanol (Life Technologies, 31350010).
RNA isolation, cDNA synthesis and RT-qPCR
Total RNA was isolated using Innuprep RNA mini kit (Analytik Jena, 845-KS-2040250). cDNA was generated using ProtoScript II First Strand cDNA Synthesis Kit (New England Biolabs, E6560L). RT-qPCRs were performed on the Light Cycler 480II (Roche) using Eef1a1 and Hptr as housekeeping genes. For each sample, RT-qPCRs were performed as technical triplicates using primers listed in Supplementary Data 1.
ChIP
5 × 107 (p300/RNAP2/MED1/PcG ChIPs) or 1 × 107 (histone ChIPs) cells were crosslinked with 1% formaldehyde for 10 min at room temperature (RT) and quenched with 0.125 M glycine for 10 min. Cells were washed and resuspended sequentially in three lysis buffers (Buffer 1: 50 mM HEPES, 140 mM NaCl, 1 mM EDTA, 10% glycerol, 0.5% NP-40, 0.25% TX-100; Buffer 2: 10 mM Tris, 200 mM NaCl, 1 mM EDTA, 0.5 mM EGTA; Buffer 3: 10 mM Tris, 100 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, 0.1% Na-deoxycholate, 0.5% N-lauroylsarcosine) to isolate chromatin. Chromatin was sonicated for 15 cycles (20 s on, 30 s off, 25% amplitude) using an EpiShear probe sonicator (Active Motif). Sonicated chromatin was incubated overnight at 4°C with 3 μg antibody for histones or 10 μg antibody for other proteins. Next, 50 μl of protein G magnetic beads (Invitrogen, 10004D) were added and incubated for four hours at 4°C. Magnetic beads were washed and the chromatin eluted, followed by de-crosslinking and DNA purification. The ChIP and input DNAs were analyzed by qPCR using two mm10 intergenic regions as negative controls (chr2:73,030,265-73,030,373; chr6: 52,339,345-52,339,505). The qPCRs for each sample were performed as technical triplicates. All antibodies and primers used in ChIP-qPCR experiments are listed in Supplementary Data 1.
Bisulfite sequencing
Bisulfite conversion of 400 ng of genomic DNA was performed using the EZ DNA Methylation Kit (Zymo Research, D5001). The investigated sequences were amplified by PCR using EpiTaq polymerase (Takara Bio, R110B) and primers described in Supplementary Data 1. PCR products were cloned into the pGEM-T vector (Promega, A1360) and sequenced with the M13 reverse primer.
Immunofluorescence
Cells were fixed for 10 min in 3.7% paraformaldehyde at RT, permeabilized with 0.1% Triton X-100 for 15 minutes at RT and blocked in PBS with 5% BSA for 1 hour at RT. Cells were incubated with primary antibodies (anti-GATA6 (AF1700, R&D systems, 8 μl/ml) or anti-SOX1 (AF3369, R&D systems, 8 μl/ml)) in blocking solution overnight at 4°C, rinsed and incubated with secondary antibodies (Fig. 2g: donkey anti-goat IgG Alexa Fluor Plus 488 (A32814, Invitrogen, 1 μl/ml); Extended Data Fig. 5c: donkey anti-goat IgG Alexa Fluor Plus 594 (A32758, Invitrogen, 1 μl/ml)) in blocking solution for 30 minutes at RT. Nuclei were stained with DAPI (Sigma, 28718-90-3) and mounting with anti-fading mounting medium (Life Technologies, P10144).
4C-seq
1 × 107 cells were crosslinked with 1% formaldehyde for 20 minutes and quenched with 0.125 M glycine for 10 minutes. Cells were washed with PBS and resuspended in lysis buffer (50 mM Tris–HCl pH 7.5, 150 mM NaCl, 5 mM EDTA, 0.5% NP-40, 1% TX-100 and 1× protease inhibitors) during 10 minutes on ice. Following centrifugation, nuclei were re-suspended in 0.5 ml of 1.2× restriction buffer with 0.3% SDS and incubated at 37°C/900 rpm for 1 hour. Triton X-100 was added to a concentration of 2% followed by 1 hour incubation at 37°C/900 rpm. Next, chromatin was digested overnight at 37°C/900 rpm with 400U of NlaIII (R0125L, NEB). NlaIII was inactivated by adding SDS to a concentration of 1.6% and incubating for 20 minutes at 65°C/900 rpm. The digested chromatin was mixed with 6.125 ml of 1.15× ligation buffer (50 mM Tris-HCl pH 7.6, 10 mM MgCl2, 1 mM ATP, 1 mM DTT). Triton X-100 was added to a concentration of 1% and the solution was incubated for 1 hour at 37°C while shaking gently. The digested chromatin was ligated with 100U of T4 DNA ligase (15224-041, Life Technologies) for 8 hours at 16°C, followed by RNase A treatment (Peqlab, 12-RA-03) for 45 minutes at 37°C. Subsequently, chromatin was de-crosslinked with 300 mg of Proteinase K (Peqlab, 04-1075) and incubated at 65°C overnight. DNA was purified by phenol/chloroform extraction and ethanol precipitation, re-suspended in 100 ml of water and digested with 50U of DpnII (R0543M, NEB) at 37°C overnight. DNA samples were purified by phenol/chloroform extraction and ethanol precipitation and resuspended in 500 μl of H2O. 200U of T4 DNA Ligase were added into a final volume of 14 ml 1× Ligation Buffer, followed by overnight incubation at 16°C. DNA samples were subjected to phenol/chloroform extraction and ethanol precipitation, re-suspended in 100 μl of water and column-purified (28104, QIAgen). The resulting DNA products were amplified by inverse PCR using primers located within selected viewpoints (Supplementary Data 1) and the expand long template PCR system (11681842001, Roche) (94°C 2 min, 30× [94°C 10 s, 60°C 1 min, 68°C 3 min], 68°C 5 min).
oCGI deletion using CRISPR-Cas9
To generate the deletion of the PE Sox1(+35)CGI, a pair of sgRNAs flanking the oCGI were designed with Benchling’s CRISPR toolkit (www.benchling.com) (Supplementary Data 1). For each sgRNA, two oligonucleotides were synthesized (IDT), annealed and cloned into a CRISPR-Cas9 expression vector (pX330-hCas9-long-chimeric-grna-g2p; Leo Kurian’s laboratory). ESCs were transfected with the pair of gRNAs-Cas9 expressing vectors using Lipofectamine (Thermo Scientific, L3000001). After 16 hours, puromycin selection was performed for 48 hours. Surviving cells were isolated in 96-well plates by serial dilution and clones with the deletion were identified by PCR using primers listed in Supplementary Data 1. The presence of the deletion was confirmed by Sanger sequencing.
Homology-dependent knock-in
Knock-In of PE modules was performed as previously described in88. Briefly, a sgRNA was designed for the insertion site of interest and cloned in the CRISPR-Cas9 expression described above. Then, the cassette-vector was generated by ligating: (i) 300-bp homology arms flanking the insertion site; (ii) construct of interest; and (iii) cloning vector. The resulting cassette-vector was used as a template for amplifying the knock-in donor (left homology arm+construct+right homology arm) by PCR (Supplementary Data 2). The resulting PCR product was column-purified (28104, QIAgen). ESCs were transfected with the sgRNA-Cas9 expressing vector and the knock-in donor using Lipofectamine (Thermo Scientific, L3000001). After 16 hours, puromycin selection was performed for 48 hours. Surviving cells were isolated in 96-well plates by serial dilution and clones with insertions were identified by PCR using the primers listed in Supplementary Data 1. The PE insertions were confirmed by Sanger sequencing.
FAIRE (Formaldehyde-Assisted Isolation of Regulatory Elements)
Chromatin was sonicated as described for ChIP and then subject to three rounds of phenol/chloroform purification followed by ethanol precipitation89. The FAIRE and input DNA was analyzed by qPCR using two mm10 intergenic regions as negative controls (chr2:73,030,265-73,030,373; chr6:52,339,345-52,339,505) and the primers listed in Supplementary Data 1.
NOMe-PCR
Nuclei extraction and M.CviPI treatment were performed as described previously90. Briefly, isolated nuclei were incubated with 200 U of M.CviPI (NEB, M0227L) for 15 min at 37 °C. Then, bisulfite conversion was performed using the EZ DNA Methylation Kit (Zymo Research, D5001) and the converted DNA was amplified by PCR. PCR products were cloned into the pGEM-T vector (Promega, A1360) and sequenced with the M13 reverse primer. NOMe-PCR data was analyzed with the NOMePlot web app tool (http://www.landeiralab.ugr.es/software)90.
Computational and Statistical Analyses
Statistics and Reproducibility
Immunofluorescence assays and genotyping of all the ESC lines were independently performed twice with similar results.
For RT-qPCR measurements in transgenic cell lines, expression levels were measured in two independent biological replicates. In each of these biological replicate experiments, two different clonal cell lines for each of the investigated genotypes were generally studied (unless stated otherwise), and for each clonal cell line two replicates of the AntNPC differentiation were measured. The statistical significance of the expression differences was calculated between AntNPCs with the TFBS+CGI module and AntNPCs with the other PE modules whenever the number of biological replicates n ≥ 3.
Analyses of qPCR data
RT-qPCR: relative gene expression levels were calculated with the 2ΔCt method using Eef1a and Hprt as housekeeping genes. Primers can be found in Supplementary Data 1.
ChIP-qPCR: for each sample, signals were calculated as % of input using technical triplicates and normalized to the average signals obtained in the same sample for two negative control regions (Chr2_neg and Chr6_neg; Supplementary Data 1).
aCGI design
The aCGI was designed by randomly incorporating nucleotides into an 800-bp sequence with a 50% higher chance of incorporating C or G rather than A or T. These GC-rich sequences were filtered to fulfil the Gardiner-Gardner criteria (i.e. observed/expected ratio of CpGs >0.6 and CG% > 50%)91. Then, the resulting CGIs were analyzed with the EMBOSS Cpgplot 92 and only those sequences with high GC% along the whole sequence were selected as possible candidates for synthesis. Finally, the sequence with lowest complexity was ordered as a gBlock from IDT (Supplementary Data 2).
4C-seq analysis
4C-seq samples were sequenced on an Illumina HiSeq 2500 sequencer, generating single reads of 74 bases in length. Reads were assigned to samples based on their first 10 bases, the primer sequences were removed from the reads and the remaining sequences were trimmed to 36 bases/read. These 36 bases were aligned to the mm10 reference genome using the HISAT2 aligner56. From these alignments, the reads per NlaIII restriction fragment were quantified using bedtools 93. Then, the reads mapping to the viewpoint as well as the preceding and following restriction fragments were removed. Finally, the resulting bedgraph files were normalized as RPM (reads per million) considering the total number of mappable reads left for each sample. These normalized bedgraph files were used for downstream visualization of the 4C-seq data.
Gene Annotation
The RefSeq gene annotation was downloaded from UCSC Table Browser94 and used for the different analyses described in this work.
ChIP-seq and PRO-seq pre-processing steps
ChIP-seq or PRO-seq fastq files read quality was assessed with FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/) and MultiQC 95.
For ChIP-seq data, the removal of read adapters and low quality filtering was done with trimmomatic 96.
For PRO-seq data, adapter removal was performed with cutadapt 1.18 97, filtering for a minimum of 15 bases (adapter sequence: TGGAATTCTCGGGTGCCAAGG). In addition, reads mapping to mouse rDNA repeats (BK000964.3) were discarded.
For both data types, reads were mapped to the mm9 reference genome with Bowtie2 98. For ChIP-seq samples duplicated reads were discarded with SAMtools99.
Genetic properties of CGIs
Data retrieval and pre-processing
PE coordinates were downloaded from14. Only PEs >2.5 kb away from any TSS (PE-all) were considered. PEs >10 kb away from any TSS are referred to as PE-distal.
NMI coordinates were obtained from21. CAP-CGI coordinates were obtained from18. Computational CGIs (GC content >50%; Length >200 bp; CpG Observed to expected ratio >0.6) were retrieved from the UCSC browser.
H3K27me3 ChIP-seq data generated in ESCs (GSE89209; H3K27me3: SRR4453259, Input: SRR4453262) were used to call H3K27me3 peaks using MACS2 100 with broad peak calling mode. Peaks with a fold-enrichment >3 and q-value <0.1 were considered. Subsequently, peaks within 1 kb of each other were merged using bedtools, and associated with a protein coding gene when overlapping a TSS. Lastly, the knee of the size distribution of the H3K27me3 peaks associated with genes was determined with findiplist() (inflection R package; https://cran.r-project.org/web/packages/inflection/vignettes/inflection.html). Upon curvature analysis, genes with a H3K27me3 peak >6 kb were considered as developmental genes (devTSS).
NMIs and CAP-CGIs were associated with PE-distal or devTSS if located <3 kb away from them. In addition, to create a group of random regions, each region associated with a PE-distal was randomly relocated along the genome 1,000 times (maintaining its size).
Sequence Composition
To retrieve DNA sequences, BSgenome 101 and the unmasked mm9 genome were used. The length, G+C percentage, CpG percentage and CpG observed/expected ratio was calculated for each region. The CpG% was calculated as the ratio of CpG dinucleotide counts with respect to half the total region length. The CpG observed/expected ratio was calculated as described in91.
CGI Block Sizes
All CAP-CGIs <3 kb from the region of interest (PE or TSS) were obtained, with smaller and larger CGIs coordinates constituting the CGI block initial limits. If another CAP-CGI was encountered in the next 5 kb from the CGI block limits, it was added to it, and the CGI block limits were expanded taking into account the newly included CGI. The second step was recursively applied until no CGI was found in the next 5 kb.
Comparison of eRNA levels between different classes of active enhancers
Data retrieval and pre-processing
Gene expression levels (FPKMs) and active enhancer coordinates from WT ESCs were obtained from14. To avoid confounding effects between transcripts produced by enhancers or genes, only active intergenic enhancers located >10 kb from any TSS and >20 kb from any transcription termination site (TTS) were considered102.
For the analyses presented in Figure 4g and Extended Figure 7d (left), the H3K27ac ChIP-seq fastq files were retrieved from GEO (GSM2360929; sample ID: SRR4453258) and pre-processed as indicated above. For the analyses presented in Extended Figure 7d (middle and right), two H3K27ac bigWig files were downloaded from GEO (GSM2808655 and GSM2808669).
For Figure 4g and Extended Figure 7d (left), PRO-seq fastq files were obtained from GEO (GSE115713; sample IDs: SRR7300121, SRR7300122) and the two replicates were combined and pre-processed as described above. For Extended Figure 7g (middle and right), two PRO-seq bigWig files (one for each DNA strand) were obtained from GEO (GSE130691).
TAD maps from ESCs were retrieved from31. For Figure 4g and Extended Figure 7d (left): mESC_Dixon2012-raw_TADs.txt. For Extended Figure 7d (middle and right): mESC.Bonev_2017-raw.domains.
H3K27ac & PRO-seq enhancer levels quantification
Figure 4g and Extended Figure 7d (left): H3K27ac and PRO-seq reads with a mapping quality <10 were discarded using SAMtools99. Next, bigwig files were generated with deepTools103 using bamCoverage (RPGC normalization) and then used to calculate the H3K27ac and PRO-seq enhancer mean scores with computeMatrix from deepTools. For H3K27ac and PRO-seq, the signals were calculated using a ±1-kb or ±0.5-kb window from the enhancer midpoints, respectively.
Extended Figure 7d (middle and right): H3K27ac and PRO-seq mean signals for the enhancers were calculated with the bigWigAverageOverBed UCSC binary tool. PRO-seq signals for each enhancer from the two different strands were averaged and the same was done for the signals coming from different H3K27ac replicates.
Active enhancers classification
Three groups of AEs were defined: (I) enhancers located in TADs only containing poorly expressed genes (<0.5 FPKM); (II) enhancers located in TADs with at least one gene with >10 FPKM; (III) enhancers whose closest gene within the same TAD has >10 FPKM.
Balancing of H3K27ac levels within enhancer classes
Enhancers with similar H3K27ac levels belonging to the three enhancer classes were selected by applying the nearest neighbor matching method (without replacement and ratio = 1) using MatchIt (https://cran.r-project.org/web/packages/MatchIt/MatchIt.pdf) and considering the enhancer group (I) as the treatment condition.
Cliff’s delta effect size estimator
Cliff’s delta104,105 was used to quantify the differences between groups of genomic regions. This measure is robust to skewed signal distributions106. Cliff’s delta, was estimated using the cliff.delta() function from the R package effsize (https://cran.r-project.org/web/packages/effsize/index.html). Differences between groups with |delta| < 0.147 can be considered as negligible and |delta| ≥ 0.147 as non-negligible.
Hi-C analyses
Pre-processing
GSE130723: the.hic files for two Hi-C replicates (GSM3752487, GSM3752488) generated in ESCs were downloaded. The.hic format was converted to.cool format using a 5-kb matrix resolution with the hic2cool software (https://github.com/4dn-dcic/hic2cool).
GSE98671: the.cool format files for two untreated ESC Hi-C replicates (GSM2644945, GSM2644946) at a 20-kb matrix resolution were downloaded.
For both datasets the corresponding replicates in.cool format were merged with cooler merge 107 and normalized with cooler balance 107.
Definition of PE-gene pairs
Group A: When a PE-distal was found in a TAD with a devTSS, both coordinates were selected to define a PE-gene pair. Only devTSS with a CAP-CGI in <3 kb were considered.
Group B: CGI-poor TSS do not have a CAP-CGI in <3 kb and are not enriched in H3K27me3 (H3K27me3 ChIP-seq peaks described above). When a PE-distal was found in a TAD with a CGI-poor TSS, both coordinates were selected to define a PE-gene pair.
Two additional filters were applied: (i) PE-gene pairs were balanced to compare groups of PE-gene pairs without significant differences in their linear genomic sizes. PE-gene pairs with similar lengths were selected by applying the nearest neighbor matching method (without replacement and ratio = 1) using MatchIt (https://cran.r-project.org/web/packages/MatchIt/MatchIt.pdf) and considering Group A as the treatment condition; (ii) only TSSs of genes with expression <1 FPKM were considered.
We used TADs defined in mESC_Dixon2012-raw_TADs.txt 30.
Pile-up plot generation
The pile-up plots for the GSE130723 and GSE98671 Hi-C datasets were generated with coolpup.pyc 108 using a padding of ±50 kb or ±100 kb, respectively.
TF Motif Analyses
The genomic coordinates of PEs and AEs were defined by p300 peaks identified in ESCs14. and located >2.5 kb away from any RefSeq TSS.
CAP-CGI vs. p300 peaks
Among the previously reported PEs14, we only considered those with a CAP-CGI in <3 kb and that did not overlap with the p300 peaks defining the PEs. Then, motif analyses were performed separately for the CAP-CGIs and the p300 peaks associated with the selected PEs using Homer 32 and Seqpos 33.
Homer: input regions (p300 peaks or CAP-CGIs) were analyzed with the following parameters: -size given -mset vertebrates.
SeqPos: Curated cistrome motif database and de novo motif searches were used. The species list parameter was used to filter the results considering both Homo sapiens and Mus musculus. All other parameters were used with the default settings
PEs vs. AEs
We considered PEs with a CAP-CGI in <3 kb and AEs without a CAP-CGI in <3 kb. The motif composition of PEs and AEs was analyzed using two different tools:
-
(i)
Homer32 was utilized to analyze each enhancer group separately with the same parameters described above.
-
(ii)
AME84 was used to perform a differential motif enrichment analysis between PEs and AEs. The Eukaryote DNA & Vertebrates motif database was used. All other parameters were used with default settings.
Whole-genome bisulfite sequencing (WGBS) analysis
For WGBS analyses we used public data from 2i ESCs26, day 2 epiblast-like cells (EpiLCs)27, epiblast stem cells (EpiSCs)27, serum+LIF ESCs (GSE82125) and neural progenitor cells (NPCs) (GSE82125). The adapters were trimmed with Trim Galore and mapped to the mm10 reference genome using Bismark-v0.16.1 109 and bowtie2-2.2.9 110. For each cell type, the CpG methylation levels were estimated with the Bismark methylation extractor, considering only CpGs with a coverage of 3-100 reads. For visualization of CpG methylation levels around pCGI and oCGI, the average CpG methylation signal was visualized with deeptools-3.3.1 103.
ChIP-seq profile plots
PE classification
PE-distal were separated in four groups: (i) PEs with overlapping TFBS/p300 and CAP-CGIs; (ii) PEs with TFBS/p300 separated by 1 bp to 1 kb from a CAP-CGI; (iii) PEs with TFBS/p300 separated by 1-3 kb from a CAP-CGI; (iv) PEs without CAP-CGIs in 3 kb. The coordinates of AEs without CAP-CGI in < 3 kb were also considered.
Datasets Used
H3K27me3: GSE157748 (Extended Data Fig. 1b left) and GSE89209 (Extended Data Fig. 1b right) H3K27me3 ChIP-seq datasets from ESCs were used. For GSE89209, the fastq file SRR4453259 was processed as described in previous sections. For GSE157748, bigwig files (GSM4774518, GSM4774519) were downloaded and combined using bigWigMerge and bedGraphToBigWig UCSC tools 111.
TET1: GSE104067 was used. The bigwig files of three untreated ESC replicates (GSM2788888, GSM2788889, GSM2788890) were downloaded and combined using bigWigMerge and bedGraphToBigWig UCSC tools111.
KDM2B: GSE126862 was used. The bigwig file with all the merged untreated ESC replicates (GSE126862_KDM2AB_CXXCfl_KDM2B_UNT_mm10_downsampled_merged.bw) was downloaded. Bigwig coordinates were converted from mm10 to mm9 with CrossMap 112.
Plots Generation
Profiles plots were generated using computeMatrix and plotProfile from deepTools103.
Extended Data
Extended Data Fig. 1. Genetic and epigenetic features of the oCGIs associated with PEs.
a, Comparison of CpG%, observed/expected CpG ratio, GC% and sequence length between random regions (n=436000), NMIs associated to PE-distal (PE-NMIs; n=345) and NMIs associated to the devTSS (devTSS-NMIs; n=1476) (Methods). The p-values were calculated using two-sided unpaired Wilcoxon tests with Bonferroni correction for multiple testing; black numbers indicate median fold-changes; green numbers indicate non-negligible Cliff Delta effect sizes. The coloured area of the violin plot represents the expression values distribution and the center line represents the median. b, H3K27me3 ChIP-seq levels14,24 around: PE-distal with overlapping TFBS/p300 peaks and CAP-CGIs (n=135), PE-distal with TFBS/p300 peaks separated by 1bp-1kb from CAP-CGIs (n=65), PE-distal with TFBS/p300 peaks separated by 1-3kb from CAP-CGIs (n=53), PE-distal without CAP-CGIs within 3kb (n=254) and AEs without CAP-CGI within 3kb (n=8115). c, % of CpG methylation at CAP-CGI associated with PE-distal (PE-CAP-CGI; n=276) and CAP-CGI associated with the TSS of developmental genes (devTSS-CAP-CGI; n=1926) in the indicated cell types (Methods). d, For the identification of the PE Sox1(+35)CGI deletion, primer pairs flanking each of the deletion breakpoints (1+3 and 4+2), located within the deleted region (5+6) or amplifying a large or small fragment depending on the absence or presence of the deletion (1+2) were used. e, H3K27me3 levels at PE Sox1(+35) were measured by ChIP-qPCR in WT ESCs and in n=2 independent PE Sox1(+35)CGI -/- ESCs clones using primers adjacent to the deleted region. The bars display the mean of n=3 technical replicates (black dots). f, Independent biological replicate for the data presented in Fig. 1d. Sox1 expression was investigated by RT-qPCR in ESCs and AntNPC with the indicated genotypes. N=2 independent PE Sox1 CGI -/- ESC clones (circles and diamonds) and n=1 PE Sox1 -/- clone were studied. For each cell line, n=2 replicates of the AntNPC differentiation were performed. Expression values were normalized to two housekeeping genes (Eef1a and Hprt) and are presented as fold-changes with respect to WT ESCs. The coloured area of the violin plot represents the expression values distribution and the center line represents the median.
Extended Data Fig. 2. Modular engineering of PEs modules within the Gata6-TAD and FoxA2-TAD.
a, Epigenomic and genomic features of two previously characterized PEs4 (PE Six3(-133); PE Lmx1b(+59)) in which the oCGIs overlap with conserved sequences bound by p300 and, thus, likely to contain relevant TFBS. b, The different PE Sox1(+35) insertions were identified using primer pairs flanking the insertion borders (1+3 and 4+2; 1+5 and 6+2; 1+3 and 6+2), amplifying potential duplications (4+3, 3+2 and 4+1; 6+5, 5+2 and 6+1) and amplifying a large or small fragment depending on the absence or presence of the insertion (1+2), respectively. The PCR results obtained for WT ESCs and for two ESC clonal lines with homozygous insertions of the PE Sox1(+35) modules in the Gata6-TAD are shown. c, Independent biological replicate for the data presented in Fig. 2b. d-e, Strategy used to insert the PE Wnt8b(+21) (d) or the PE Sox1(+35) (e) components into the Gata6-TAD (d) or Foxa2-TAD (e), respectively. The right panels shows the TADs in which Gata6 (d) or Foxa2 (e) are included according to publically available Hi-C data30,31, with the red triangle indicating the integration site of the PE modules, approximately 100 Kb downstream of Gata6 (d) or Foxa2 (e). f-g, For identifying the successful insertion of the different PE Sox1(+35) (f) or PE Wnt8b(+21) (g) modules, primer pairs flanking the insertion borders (1+3 and 4+2; 1+5 and 6+2; 1+3 and 6+2), amplifying potential duplications (4+3, 3+2 and 4+1; 6+5, 5+2 and 6+1) and amplifying a large or small fragment depending on the absence or presence of the insertion (1+2), respectively, were used. The PCR results obtained for two ESC clonal lines with homozygous insertions of the indicated PE modules in the Foxa2-TAD (f) or Gata6-TAD (g), respectively, are shown. h-i, Independent biological replicates for the data shown in Fig. 2c (h) and Fig. 2d (i). In (c), (h) and (i), the expression differences between AntNPCs with the TFBS+CGI module and AntNPCs with the other PE modules were calculated using two-sided non-paired t-tests (**: foldchange>2 & p<0.001; *: foldchange> 2 & p<0.05; ns: not significant; fold-change<2 or p>0.05).
Extended Data Fig. 3. PEs are enriched in CpG-rich motifs and are bound by CxxC-domain containing proteins.
a, Comparison of the TF motifs enriched in either PEs with a CAP-CGI in <3kb and active enhancers without CAP-CGIs in <3kb. Motif enrichment analyses were performed with Homer 32 (left) and AME 84 (right). b, ChIP-seq signals for KDM2B36 (upper panel) and TET135 (lower panel) are shown around: PE-distal with overlapping TFBS/p300 peaks and CAP-CGIs (n=135), PE-distal with TFBS/p300 peaks separated by 1bp-1kb from CAP-CGIs (n=65), PE-distal with TFBS/p300 peaks separated by 1-3kb from CAP-CGIs (n=53) and PE-distal without CAP-CGIs within 3kb (n=254). ChIP-seq profile plots were generated using either the p300 peaks (left) or the CAP-CGIs (right) associated with the PEs as midpoints.
Extended Data Fig. 4. Engineering of ESC lines containing the PE Sox1(+35) TFBS and an artificial CGI within the Gata6-TAD.
a, Strategy used to insert the PE Sox1(+35)TFBS alone or together with an aCGI into the Gata6-TAD. The upper left panel shows the epigenomic and genetic features of the PE Sox1(+35). The lower left panel shows the PE Sox1(+35) modules inserted into the Gata6-TAD. The right panel shows the Gata6-TAD according to publically available Hi-C data44,45. The red triangle indicates the integration site of the PE Sox1(+35) modules approximately 100 Kb downstream of Gata6. b, For the identification of the PE Sox1(+35)TFBS+aCGI insertion, primer pairs flanking the insertion borders (1+3 and 4+2), amplifying potential duplications (4+3 and 4+4) and amplifying a large or small fragment depending on the absence or presence of the insertion (1+2), respectively, were used. The PCR results obtained for two ESC clonal lines with homozygous insertions of PE Sox1(+35)TFBS+aCGI in the Gata6-TAD are shown. c, Independent biological replicate for the data presented in Fig. 2f. The expression differences between AntNPCs with the TFBS+CGI module and AntNPCs with the other PE modules were calculated using two-sided non-paired t-tests (*: foldchange> 2 & p<0.05; ns: not significant; fold-change<2 or p>0.05). d, For the identification of the aCGI insertion alone, primer pairs flanking the insertion borders (1+3 and 4+2), amplifying potential duplications (4+3 and 4+4) and amplifying a large or small fragment depending on the absence or presence of the insertion (1+2), respectively, were used. The PCR results obtained from two ESC clonal lines with heterozygous insertions of aCGI in the Gata6-TAD are shown. e, The expression of Gata6 and Sox1 was measured by RT-qPCR in cells that were either WT or heterozygous for the aCGI insertion in the Gata6-TAD (two different clones; circles and diamonds). For each cell line, n=2 replicates of the AntNPC differentiation were performed. The results obtained in n=2 independent biological replicates are presented in each panel (Rep1 and Rep2).
Extended Data Fig. 5. Gata6 expression patterns in cell lines with the PE Sox1(+35) modules inserted within the Gata6-TAD.
a, Gata6 and Sox1 expression was measured by RT-qPCR in ESCs and at intermediate stages of AntNPC differentiation (Day 3 and Day 4). The analysed cells were either WT or homozygous for the insertions of the different PE Sox1(+35) modules within the Gata6-TAD. For the cells with the PE module insertions, n=1 clonal cell line was studied. For each cell line, n=2 replicates of the AntNPC differentiation were performed. Expression values were normalized to two housekeeping genes (Eef1a and Hprt) and are presented as fold-changes with respect to WT ESCs. b, Quantification of cells expressing GATA6 or SOX1 according to immunofluorescence assays as the ones shown in Fig 2g. The analysed cells were either WT of homozygous for the insertions of the different PE Sox1(+35) modules within the Gata6-TAD. c, The expression patterns of GATA6 (upper panel) and SOX1 (lower panel) were investigated by immunofluorescence in WT ESCs or AntNPCs that were either WT, homozygous for the insertion of the PE Sox1(+35)TFBS+aCGI in the Gata6-TAD or heterozygous for the insertion of the aCGI alone in the Gata6-TAD. Nuclei were stained with DAPI. Scale bar = 100μm. d, Quantification of cells expressing GATA6 or SOX1 according to the immunofluorescence assays described in (c). In (b) and (d), the bars display the mean of n=3 technical replicates (black dots).
Extended Data Fig. 6. Epigenetic and topological characterization of the Gata6-TAD cell lines.
a, Bisulfite sequencing data presented in Fig. 3a for the indicated Gata6-TAD cell lines. The circles correspond to individual CpG dinucleotides located within the TFBS module. Unmethylated CpGs are shown in white, methylated CpGs in black and not-covered CpGs in gray. b, Chromatin accessibility at the endogenous PE Sox1(+35) and the Gata6-TAD insertion site (P1 and P2) were measured by FAIRE-qPCR in cells with the indicated genotypes. c, DNA methylation and nucleosome occupancy at the TFBS were simultaneously analyzed by NOME-PCR in the indicated Gata6-TAD ESC lines. In the upper panels, the black and white circles represent methylated or unmethylated CpG sites, respectively. In the lower panels, the blue or white circles represent accessible or inaccessible GpC sites for the GpC methyltransferase, respectively. Red bars represent inaccessible regions large enough to accommodate a nucleosome. The dotted line indicates where the TFBS starts. The grey shaded area represents a nucleosome-depleted region. d, Scatter plots showing population-averaged nucleosome occupancy (red) and DNA methylation (black) levels within the TFBS in the indicated Gata6-TAD ESC lines. The grey shaded area represents a nucleosome depleted region. e-f, H3K4me1, H3K4me3, H2AK119ub, CBX7 and PHC1 levels at the endogenous PE Sox1(+35) and the Gata6-TAD insertion site (P1 and P2) were measured by ChIP-qPCR in cells with the indicated genoytpes. ChIP-qPCR signals were calculated as described in Fig. 3. g, 4C-seq experiments were performed using the Gata6 promoter as a viewpoint in AntNPC with the indicated genotypes. h, Pile-up plots showing average Hi-C7,56 signals in ESC between two groups of PE-gene pairs: PEs and developmental genes with CGI-rich promoters; PEs and genes with CGI-poor promoters. For each PE-gene pair, both the PE and the gene were located within the same TAD. Left panels include all the considered PE-gene pairs (n=401 pairs for developmental genes; n=900 for CGI-poor promoters; middle panels includes PE-gene pairs with the same genomic size in the two groups (n=401 pairs); right panels consist of PE-gene pairs with the same genomic size and genes with expression levels <1 FPKM9 (n=290 pairs) (Methods).
Extended Data Fig. 7. Generation of cell lines with engineered PE Sox1(+35) modules within the Gria1-TAD and global characterization of H3K27ac and eRNA levels at active enhancers.
a, ESC clonal lines with insertions of the different PE Sox1(+35) modules were identified using primer pairs flanking the insertion borders (1+3 and 4+2; 1+5 and 6+2; 1+3 and 6+2), amplifying potential duplications (4+3, 3+2 and 4+1; 6+5, 5+2 and 6+1) and amplifying a large or small fragment depending on the absence or presence of the insertion (1+2), respectively. The PCR results obtained for WT ESCs or two ESC clonal lines with homozygous insertions of the different PE Sox1(+35) modules in the Gria1-TAD are shown. b, Independent biological replicate for the data presented in Fig. 4b. The expression differences between AntNPCs with the TFBS+CGI module and AntNPCs with the other PE modules were calculated using two-sided non-paired t-tests (ns: not significant; fold-change<2 or p>0.05). c, Bisulfite sequencing analyses of ESC lines with the indicated PE Sox1(+35) modules inserted in the Gria1-TAD. The circles correspond to individual CpG dinucleotides located within the TFBS: unmethylated CpGs (white), methylated CpGs (black) and not-covered CpGs (gray) are shown. The plot on the right summarizes the DNA methylation levels measured within the TFBS in the indicated ESC lines. d, Active enhancers (AEs) identified in ESCs based on the presence of distal H3K27ac peaks were classified into three categories (Methods): Class I (AEs in TADs containing only poorly expressed genes; n=271(left); n=340 (middle, right); Class II (AEs in TADs with at least one highly expressed gene; n=271(left); n=2353(middle); n=340(right)); Class III (AEs whose closest genes in the same TAD is highly expressed; n=271(left); n=1262(middle); n=340(right)). The violin plots show the H3K27ac and eRNA levels in ESC for each AE category. P-values were calculated using unpaired Wilcoxon tests with Bonferroni correction for multiple testing; the numbers in black indicate the median fold-changes between the indicated groups; the coloured numbers correspond to Cliff Delta effect sizes: negligible (red) and non-negligible (green). In the left and right panels, eRNA levels for the three enhancers classes are compared after correcting for H3K27ac differences (Methods).
Extended Data Fig. 8. Generation and characterization of cell lines with PE insertions at the Gria1 and Sox7/Rp1l1 TADs.
a, H2AK229ub and SUZ12 levels at the endogenous PE Sox1(+35), the Gria1 promoter and the Gria1-TAD insertion site (P1 and P2; Fig. 4d) were measured by ChIP-qPCR in ESCs with the indicated genotypes. ChIP-qPCR signals were calculated as in Fig. 3. b, ESC clonal lines in which a pCGI was inserted 380bp upstream of the Gria1-TSS in cells with the indicated PE Sox1(+35) modules 100Kb upstream from Gria1 were identified using the indicated primer pairs. PCR results for clonal ESC lines with the indicated double homozygous insertions are shown. c, eRNA levels at the endogenous PE Sox1(+35) and the Gria1-TAD insertion site (P1 and P2) were measured by RT-qPCR in cells with the indicated genotypes. Expression values were calculated as in Fig. 3. d, Strategy to insert the indicated PE Sox1(+35) modules 380bp upstream (red triangle) of the Gria1-TSS. e, ESC clonal lines with the PE Sox1(+35) modules 380bp upstream of the Gria1-TSS were identified using the indicated primer pairs. PCR for ESC clonal lines with homozygous insertions of the indicated PE Sox1(+35) modules are shown. f, Independent biological replicate for the data presented in Fig. 5e. g, ESC clonal lines with the PE Sox1(+35) modules within the Sox7/Rp1l1-TAD were identified using primers flanking the insertion borders (1+3 and 4+2; 1+3 and 6+2), amplifying potential duplications (4+3, 3+2 and 4+1) and amplifying a large or small fragment depending on the absence or presence of the insertion (1+2), respectively. PCR results for ESC clonal lines with homozygous insertions of the indicated PE Sox1(+35) modules are shown. h, Independent biological replicate for the data presented in Fig. 5g. In (a) and (c), the bars display the mean of n=3 technical replicates (black dots). In (f) and (h), the expression differences between AntNPCs with the TFBS+CGI module or the other PE modules were calculated using two-sided non-paired t-tests (***: foldchange> 2 & p<0.0001; ns: not significant; fold-change<2 or p>0.05).
Extended Data Fig. 9. Generation of ESC lines with structural variants.
a, ESC lines with the Six3/Six2 TAD boundary deletion were identified using primers flanking the deleted region (1+3 and 4+2), amplifying the deleted fragment (5+6) and amplifying a large or small fragment depending on the absence or presence of the deletion (1+2), respectively. The PCR results for two ESC clonal lines with 36Kb homozygous deletions (del36) are shown. b, ESC lines with the Six3/Six2 inversion were identified using primer pairs flanking the inverted region (1+3, 4+2, 1+4 and 3+2) and amplifying potential duplications (4+3, 3+3 and 4+4). The PCR results for two ESC clonal lines with 110Kb homozygous inversions (inv110) are shown. c, Epigenomic and genetic features of a CTCF binding site85 (CBS; highlighted in grey) located upstream of the PE Six1(-133) (highlighted in yellow). d, ESC lines with the CBS deletion were identified using primers flanking the deleted region (1+2) or located in the CBS (3+4). The PCR results for two ESC clonal lines with homozygous CBS deletions are shown. e, The expression of Six3 and Six2 was measured by RT-qPCR in cells with the indicated genotypes. For each of the engineered structural variants, n=2 independent clonal cell lines were generated (circles and diamonds). In each plot, the number of circles and/or diamonds correspond to the number AntNPC differentiations performed. The results obtained in n=2 independent biological replicates are presented in each panel (Rep1 and Rep2). Expression values are presented as fold-changes with respect to WT ESCs. f, ESC lines with the Lmx1a-TAD boundary inversion were identified using primers flanking the inverted region (1+3, 4+2, 1+4 and 3+2) and amplifying potential deletions (1+4) or duplications (4+3, 3+3 and 4+4). The PCR results for three ESC clonal lines with 260 Kb homozygous inversions (inv260) are shown.
Extended Data Fig. 10. Examples of human congenital diseases caused by structural variants that disrupt developmental loci with PE-associated oCGIs.
a, Upper panel: heterozygous inversion in a patient with Branchio-oculo-facial syndrome (BOFS)5. Lower panel: epigenomic and genetic features of TFAP2A neural crest (NC) cognate enhancers (left), 6q16.2 genes (middle) and TFAP2A (right). In the lower left panel, enhancer reporter assays in chicken embryos are shown for two representative TFAP2A enhancers5. Computational CGI and NMIs are represented as green rectangles. The inversion places one TFAP2A allele into a novel TAD and impairs its normal expression in NC cells due to the physical disconnection from its enhancers. TFAP2A has a promoter with a large CGI cluster and marked with a broad H3K27me3 domain in ESCs. Some TFAP2A NC enhancers are associated with oCGIs and marked with H3K27me3 in ESCs. Moreover, this inversion places genes originally found within the 6q16.2 locus in proximity of the TFAP2A NC enhancers within a shuffled domain. The promoters of these 6q16.2 genes (i.e GPR63 and NDUFAF4) contain a short CGI centered on their TSSs. In agreement with our findings, none of the 6q16.2 genes is responsive to the TFAP2A NC enhancers5. b, Upper panel: deletion found in families with brachydactyly involving a TAD boundary located between the EPHA4 and the PAX3 loci67. Lower panel: epigenomic and genetic features of the Epha4 cognate enhancers in the mouse E11.5 limb (left) and in human ESCs (right). Representative reporter assay in E11.5 mouse embryos for the hs1507 element is shown in the middle67. The deletion includes EPHA4, a gene highly expressed in the developing limb, and the TAD boundary separating the EPHA4 and PAX3 TADs. As a result, enhancers that control EPHA4 expression in the limb establish ectopic interactions with PAX3 (i.e. enhancer adoption) and strongly induce its expression in the limb. PAX3 promoter contains a large CGI cluster and is marked with H3K27me3 in ESCs, while one of the major EPHA4 enhancers (hs1507) is associated with an oCGI and is marked with H3K27me3 in ESCs. The high responsiveness of PAX3 to the EPHA4 enhancers is in agreement with our findings.
Supplementary Material
Acknowledgements
We thank the Rada-Iglesias laboratory members for insightful comments and critical reading of the manuscript. Tomas Pachano is supported by a doctoral fellowship from the DAAD (Germany). Víctor Sánchez-Gaya is supported by a doctoral fellowship from the University of Cantabria (Spain). Work in the Rada-Iglesias laboratory was supported by the EMBO Young Investigator Programme, CMMC intramural funding (Germany), the German Research Foundation (DFG) (Research Grant RA 2547/2-1), “Programa STAR-Santander Universidades, Campus Cantabria Internacional de la convocatoria CEI 2015 de Campus de Excelencia Internacional” (Spain), the Spanish Ministry of Science, Innovation and Universities (Research Grant PGC2018-095301-B-I00 and RED2018-102553-T REDEVNEURAL 3.0) and the European Research Council (ERC CoG “PoisedLogic”). The Landeira laboratory is funded by grants of the Spanish Ministry of Science and Innovation (BFU2016-75233-P and PID2019-108108GB-I00) and the Andalusian Regional Government (PC-0246-2017).
Footnotes
Author Contributions
Conceptualization: T.P., A.R.-I.; Experimental investigation: T.P., T.E., M.M.-F., H.G.A., P.R., M.M.-S., E.H.; Data analysis: T.P., V.S.-G., T.B.; Writing, Review & Editing: T.P., A.R.-I.; Resources: S.C.-M., W.F.J.v.I., D.L., A.R.-I.; Supervision and Funding Acquisition: A.R.-I..
Competing Interests
The authors declare no competing interests.
Data Availability
All the generated 4C-seq data generated in this study are available through GEO (GSE156465). All the generated transgenic ESC lines are available upon request.
References
- 1.Spitz F, Furlong EEM. Transcription factors: From enhancer binding to developmental control. Nat Rev Genet. 2012;13:613–626. doi: 10.1038/nrg3207. [DOI] [PubMed] [Google Scholar]
- 2.Kvon EZ. Using transgenic reporter assays to functionally characterize enhancers in animals. Genomics. 2015;106:185–192. doi: 10.1016/j.ygeno.2015.06.007. [DOI] [PubMed] [Google Scholar]
- 3.Furlong EEM, Levine M. Developmental enhancers and chromosome topology. Science. 2018;361:1341–1345. doi: 10.1126/science.aau0320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dixon JR, et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Laugsch M, et al. Modeling the Pathological Long-Range Regulatory Effects of Human Structural Variation with Patient-Specific hiPSCs. Cell Stem Cell. 2019;24:736–752.:e12. doi: 10.1016/j.stem.2019.03.004. [DOI] [PubMed] [Google Scholar]
- 6.Rao SSP, et al. Cohesin Loss Eliminates All Loop Domains. Cell. 2017;171:305–320.:e24. doi: 10.1016/j.cell.2017.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nora P, et al. Targeted Degradation of CTCF Decouples Local Insulation of Chromosome Domains from Genomic Compartmentalization. Cell. 2017;169:930–944. doi: 10.1016/j.cell.2017.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ghavi-Helm Y, et al. Highly rearranged chromosomes reveal uncoupling between genome topology and gene expression. Nat Genet. 2019;51:1272–1282. doi: 10.1038/s41588-019-0462-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kraft K, et al. Serial genomic inversions induce tissue-specific architectural stripes, gene misexpression and congenital malformations. Nat Cell Biol. 2019;21:305–310. doi: 10.1038/s41556-019-0273-x. [DOI] [PubMed] [Google Scholar]
- 10.Kikuta H, et al. Genomic regulatory blocks encompass multiple neighboring genes and maintain conserved synteny in vertebrates. Genome Res. 2007;17:545–555. doi: 10.1101/gr.6086307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arnold CD, et al. Genome-wide assessment of sequence-intrinsic enhancer responsiveness at single-base-pair resolution. Nat Biotechnol. 2016;35:136–144. doi: 10.1038/nbt.3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haberle V, et al. Transcriptional cofactors display specificity for distinct types of core promoters. Nature. 2019;570:122–126. doi: 10.1038/s41586-019-1210-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Spielmann M, Lupiáñez DG, Mundlos S. Structural variation in the 3D genome. Nat Rev Genet. 2018;19:453–467. doi: 10.1038/s41576-018-0007-0. [DOI] [PubMed] [Google Scholar]
- 14.Cruz-Molina S, et al. PRC2 Facilitates the Regulatory Topology Required for Poised Enhancer Function during Pluripotent Stem Cell Differentiation. Cell Stem Cell. 2017;20:1–17. doi: 10.1016/j.stem.2017.02.004. [DOI] [PubMed] [Google Scholar]
- 15.Rada-Iglesias A, et al. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2011;470:279–83. doi: 10.1038/nature09692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deaton AM, Bird A. CpG islands and the regulation of transcription. Genes Dev. 2011;25:1010–1022. doi: 10.1101/gad.2037511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bell JSK, Vertino PM. Orphan CpG islands define a novel class of highly active enhancers. Epigenetics. 2017;12:449–464. doi: 10.1080/15592294.2017.1297910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Illingworth RS, et al. Orphan CpG Islands Identify numerous conserved promoters in the mammalian genome. PLoS Genet. 2010;6:e1001134. doi: 10.1371/journal.pgen.1001134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Steinhaus R, Gonzalez T, Seelow D, Robinson PN. Pervasive and CpG-dependent promoter-like characteristics of transcribed enhancers. Nucleic Acids Res. 2020;48:5306–5317. doi: 10.1093/nar/gkaa223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bogdanović O, et al. Active DNA demethylation at enhancers during the vertebrate phylotypic period. Nat Genet. 2016;48:417–426. doi: 10.1038/ng.3522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Long HK, et al. Epigenetic conservation at gene regulatory elements revealed by non-methylated DNA profiling in seven vertebrates. Elife. 2013;2:1–19. doi: 10.7554/eLife.00348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lenhard B, Sandelin A, Carninci P. Metazoan promoters: Emerging characteristics and insights into transcriptional regulation. Nat Rev Genet. 2012;13:233–245. doi: 10.1038/nrg3163. [DOI] [PubMed] [Google Scholar]
- 23.Williams K, et al. TET1 and hydroxymethylcytosine in transcription and DNA methylation fidelity. Nature. 2011;473:343–349. doi: 10.1038/nature10066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Blackledge NP, et al. Variant PRC1 complex-dependent H2A ubiquitylation drives PRC2 recruitment and polycomb domain formation. Cell. 2014;157:1445–1459. doi: 10.1016/j.cell.2014.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aljazi MB, Gao Y, Wu Y, Mias GI, He J. Cell Signaling Coordinates Global PRC2 Recruitment and Developmental Gene Expression in Murine Embryonic Stem Cells. iScience. 2020;23:101646. doi: 10.1016/j.isci.2020.101646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Habibi E, et al. Whole-genome bisulfite sequencing of two distinct interconvertible DNA methylomes of mouse embryonic stem cells. Cell Stem Cell. 2013;13:360–369. doi: 10.1016/j.stem.2013.06.002. [DOI] [PubMed] [Google Scholar]
- 27.Zylicz JJ, et al. Chromatin dynamics and the role of G9a in gene regulation and enhancer silencing during early mouse development. Elife. 2015;4:1–25. doi: 10.7554/eLife.09571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee SM, et al. Intragenic CpG islands play important roles in bivalent chromatin assembly of developmental genes. Proc Natl Acad Sci U S A. 2017;114:E1885–E1894. doi: 10.1073/pnas.1613300114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bolt CC, Duboule D. The regulatory landscapes of developmental genes. Development. 2020;147:1–7. doi: 10.1242/dev.171736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang Y, et al. The 3D Genome Browser: A web-based browser for visualizing 3D genome organization and long-range chromatin interactions. Genome Biol. 2018;19:1–12. doi: 10.1186/s13059-018-1519-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bonev B, et al. Multiscale 3D Genome Rewiring during Mouse Neural Development. Cell. 2017;171:557–572. doi: 10.1016/j.cell.2017.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Heinz S, et al. Simple Combinations of Lineage-Determining Transcription Factors Prime cis-Regulatory Elements Required for Macrophage and B Cell Identities. Mol Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu T, et al. Cistrome: An integrative platform for transcriptional regulation studies. Genome Biol. 2011;12:R83. doi: 10.1186/gb-2011-12-8-r83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Blackledge NP, Klose R. CpG island chromatin. Epigenetics. 2011;2294:147–152. doi: 10.4161/epi.6.2.13640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Turberfield AH, et al. KDM2 proteins constrain transcription from CpG island gene promoters independently of their histone demethylase activity. Nucleic Acids Res. 2019;47:9005–9023. doi: 10.1093/nar/gkz607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Arab K, et al. GADD45A binds R-loops and recruits TET1 to CpG island promoters. Nat Genet. 2019;51:217–223. doi: 10.1038/s41588-018-0306-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Diez R, Storey KG. Markers in vertebrate neurogenesis. Nat Rev Neurosci. 2001;2:835–839. doi: 10.1038/35097587. [DOI] [PubMed] [Google Scholar]
- 38.Bentovim L, Harden TT, DePace AH. Transcriptional precision and accuracy in development: From measurements to models and mechanisms. Dev. 2017;144:3855–3866. doi: 10.1242/dev.146563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Boyes J, Bird A. DNA methylation inhibits transcription indirectly via a methyl-CpG binding protein. Cell. 1991;64:1123–1134. doi: 10.1016/0092-8674(91)90267-3. [DOI] [PubMed] [Google Scholar]
- 40.Klemm SL, Shipony Z, Greenleaf WJ. Chromatin accessibility and the regulatory epigenome. Nat Rev Genet. 2019;20:207–220. doi: 10.1038/s41576-018-0089-8. [DOI] [PubMed] [Google Scholar]
- 41.You JS, et al. OCT4 establishes and maintains nucleosome-depleted regions that provide additional layers of epigenetic regulation of its target genes. Proc Natl Acad Sci. 2011;108:14497–14502. doi: 10.1073/pnas.1111309108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stadler MB, et al. DNA-binding factors shape the mouse methylome at distal regulatory regions. Nature. 2011;480:490–495. doi: 10.1038/nature10716. [DOI] [PubMed] [Google Scholar]
- 43.Kim T-K, et al. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465:182–187. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mas G, Di Croce L. The role of Polycomb in stem cell genome architecture. Curr Opin Cell Biol. 2016;43:87–95. doi: 10.1016/j.ceb.2016.09.006. [DOI] [PubMed] [Google Scholar]
- 45.Yan J, et al. Histone H3 lysine 4 monomethylation modulates long-range chromatin interactions at enhancers. Cell Res. 2018;28:204–220. doi: 10.1038/cr.2018.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Denholtz M, et al. Long-Range Chromatin Contacts in Embryonic Stem Cells Reveal a Role for Pluripotency Factors and Polycomb Proteins in Genome Organization. Cell Stem Cell. 2013;13:602–616. doi: 10.1016/j.stem.2013.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang J, et al. A protein interaction network for pluripotency of embryonic stem cells. Nature. 2006;444:364–368. doi: 10.1038/nature05284. [DOI] [PubMed] [Google Scholar]
- 48.Pachano T, Crispatzu G, Rada-Iglesias A. Polycomb proteins as organizers of 3D genome architecture in embryonic stem cells. Brief Funct Genomics. 2019;18 doi: 10.1093/bfgp/elz022. [DOI] [PubMed] [Google Scholar]
- 49.Bantignies F, et al. Polycomb-Dependent Regulatory Contacts between Distant Hox Loci in Drosophila. 2007 doi: 10.1016/j.cell.2010.12.026. [DOI] [PubMed] [Google Scholar]
- 50.Isono K, et al. SAM domain polymerization links subnuclear clustering of PRC1 to gene silencing. Dev Cell. 2013;26:565–577. doi: 10.1016/j.devcel.2013.08.016. [DOI] [PubMed] [Google Scholar]
- 51.Loubiere V, Papadopoulos GL, Szabo Q, Martinez AM, Cavalli G. Widespread activation of developmental gene expression characterized by PRC1-dependent chromatin looping. Sci Adv. 2020;6:eaax4001. doi: 10.1126/sciadv.aax4001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Benabdallah NS, et al. Decreased Enhancer-Promoter Proximity Accompanying Enhancer Activation. Mol Cell. 2019:1–12. doi: 10.1016/j.molcel.2019.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lim B, Heist T, Levine M, Fukaya T. Visualization of Transvection in Living Drosophila Embryos. Mol Cell. 2018;70:287–296.:e6. doi: 10.1016/j.molcel.2018.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Beck S, et al. Implications of CpG islands on chromosomal architectures and modes of global gene regulation. Nucleic Acids Res. 2018:1–10. doi: 10.1093/nar/gky147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu S, et al. From 1D sequence to 3D chromatin dynamics and cellular functions: A phase separation perspective. Nucleic Acids Res. 2018;46:9367–9383. doi: 10.1093/nar/gky633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kurup JT, Han Z, Jin W, Kidder BL. H4K20me3 methyltransferase SUV420H2 shapes the chromatin landscape of pluripotent embryonic stem cells. Development. 2020;147:dev188516. doi: 10.1242/dev.188516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Andersson R, Sandelin A, Danko CG. A unified architecture of transcriptional regulatory elements. Trends Genet. 2015;31:426–433. doi: 10.1016/j.tig.2015.05.007. [DOI] [PubMed] [Google Scholar]
- 58.Lloret-Llinares M, et al. The RNA exosome contributes to gene expression regulation during stem cell differentiation. Nucleic Acids Res. 2018;46:11502–11513. doi: 10.1093/nar/gky817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Local A, et al. Identification of H3K4me1-associated proteins at mammalian enhancers. Nat Genet. 2018;50:73–82. doi: 10.1038/s41588-017-0015-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Etchegaray JP, et al. The Histone Deacetylase SIRT6 Restrains Transcription Elongation via Promoter-Proximal Pausing. Mol Cell. 2019;75:683–699. doi: 10.1016/j.molcel.2019.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hirabayashi S, et al. NET-CAGE characterizes the dynamics and topology of human transcribed cis-regulatory elements. Nat Genet. 2019;51:1369–1379. doi: 10.1038/s41588-019-0485-9. [DOI] [PubMed] [Google Scholar]
- 62.Schoenfelder S, et al. Polycomb repressive complex PRC1 spatially constrains the mouse embryonic stem cell genome. Nat Genet. 2015;47:1179–86. doi: 10.1038/ng.3393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Butler JEF, Kadonaga JT. Enhancer – promoter specificity mediated by DPE or TATA core promoter motifs. 2001:2515–2519. doi: 10.1101/gad.924301.protein. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gómez-Marín C, et al. Evolutionary comparison reveals that diverging CTCF sites are signatures of ancestral topological associating domains borders. Proc Natl Acad Sci U S A. 2015;112:7542–7547. doi: 10.1073/pnas.1505463112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.O’Brien LL, et al. Transcriptional regulatory control of mammalian nephron progenitors revealed by multi-factor cistromic analysis and genetic studies. PLoS Genetics. 2018;14 doi: 10.1371/journal.pgen.1007181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Catarino RR, Stark A. Assessing sufficiency and necessity of enhancer activities for gene expression and the mechanisms of transcription activation. Genes Dev. 2018;32:202–223. doi: 10.1101/gad.310367.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lupiáñez DG, et al. Disruptions of topological chromatin domains cause pathogenic rewiring of gene-enhancer interactions. Cell. 2015;161:1012–1025. doi: 10.1016/j.cell.2015.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kragesteen BK, et al. Dynamic 3D chromatin architecture contributes to enhancer specificity and limb morphogenesis. Nat Genet. 2018;50:1463–1473. doi: 10.1038/s41588-018-0221-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li X, Noll M. Compatibility between enhancers and promoters determines the transcriptional specificity of gooseberry and gooseberry neuro in the Drosophila embryo. EMBO J. 1994;13:400–406. doi: 10.1002/j.1460-2075.1994.tb06274.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zabidi MA, et al. Enhancer-core-promoter specificity separates developmental and housekeeping gene regulation. Nature. 2015;518:556–559. doi: 10.1038/nature13994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mahmoudi T, Katsani KR, Verrijzer CP. GAGA can mediate enhancer function in trans by linking two separate DNA molecules. EMBO J. 2002;21:1775–1781. doi: 10.1093/emboj/21.7.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Calhoun VC, Levine M. Long-range enhancer-promoter interactions in the Scr-Antp interval of the Drosophila Antennapedia complex. Proc Natl Acad Sci U S A. 2003;100:9878–9883. doi: 10.1073/pnas.1233791100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Calhoun VC, Stathopoulos A, Levine M. Promoter-proximal tethering elements regulate enhancer-promoter specificity in the Drosophila Antennapedia complex. Proc Natl Acad Sci U S A. 2002;99:9243–9247. doi: 10.1073/pnas.142291299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Boyle S, et al. A central role for canonical PRC1 in shaping the 3D nuclear landscape. Genes Dev. 2020;34:931–949. doi: 10.1101/gad.336487.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Perino M, et al. MTF2 recruits Polycomb Repressive Complex 2 by helical-shape-selective DNA binding. Nat Genet. 2018;50:1002–1010. doi: 10.1038/s41588-018-0134-8. [DOI] [PubMed] [Google Scholar]
- 76.Beltran M, et al. The interaction of PRC2 with RNA or chromatin s mutually antagonistic. Genome Res. 2016;26:896–907. doi: 10.1101/gr.197632.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Crispatzu G, et al. The chromatin, topological and regulatory properties of pluripotency-associated poised enhancers are conserved in vivo. bioRxiv. 2021:2021.01.18.427085. doi: 10.1038/s41467-021-24641-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Shrinivas K, et al. Enhancer Features that Drive Formation of Transcriptional Condensates. Mol Cell. 2019;75:549–561.:e7. doi: 10.1016/j.molcel.2019.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Dimitrova E, et al. FBXl19 recruits CDK-Mediator to CpG islands of developmental genes priming them for activation during lineage commitment. Elife. 2018;7:1–27. doi: 10.7554/eLife.37084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Long HK, Blackledge NP, Klose RJ. ZF-CxxC domain-containing proteins, CpG islands and the chromatin connection. Biochem Soc Trans. 2013;41:727–740. doi: 10.1042/BST20130028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mastrangelo IA, Courey AJ, Wall JS, Jackson SP, Hough PVC. DNA looping and Sp1 multimer links: A mechanism for transcriptional synergism and enhancement. Proc Natl Acad Sci U S A. 1991;88:5670–5674. doi: 10.1073/pnas.88.13.5670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Su W, Jackson S, Tjian R, Echols H. DNA looping between sites for transcriptional activation: Self-association of DNA-bound Sp1. Genes Dev. 1991;5:820–826. doi: 10.1101/gad.5.5.820. [DOI] [PubMed] [Google Scholar]
- 83.Hartl D, et al. CG dinucleotides enhance promoter activity independent of DNA methylation. Genome Res. 2019;29:554–563. doi: 10.1101/gr.241653.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bailey TL, et al. MEME Suite: Tools for motif discovery and searching. Nucleic Acids Res. 2009;37:202–208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Pope BD, et al. Topologically associating domains are stable units of replication-timing regulation. Nature. 2014;515:402–405. doi: 10.1038/nature13986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gouti M, et al. In vitro generation of neuromesodermal progenitors reveals distinct roles for wnt signalling in the specification of spinal cord and paraxial mesoderm identity. PLoS Biol. 2014;12:e1001937. doi: 10.1371/journal.pbio.1001937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Matsuda K, Kondoh H. Dkk1-dependent inhibition of Wnt signaling activates Hesx1 expression through its 5’ enhancer and directs forebrain precursor development. Genes to Cells. 2014;19:374–385. doi: 10.1111/gtc.12136. [DOI] [PubMed] [Google Scholar]
- 88.Yao X, et al. Tild-CRISPR Allows for Efficient and Precise Gene Knockin in Mouse and Human Cells. Dev Cell. 2018;45:526–536.:e5. doi: 10.1016/j.devcel.2018.04.021. [DOI] [PubMed] [Google Scholar]
- 89.Giresi PG, Kim J, McDaniell RM, Iyer VR, Lieb JD. FAIRE (Formaldehyde-Assisted Isolation of Regulatory Elements) isolates active regulatory elements from human chromatin. Genome Res. 2007;17:877–85. doi: 10.1101/gr.5533506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Requena F, et al. NOMePlot: analysis of DNA methylation and nucleosome occupancy at the single molecule. Sci Rep. 2019;9:1–10. doi: 10.1038/s41598-019-44597-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gardiner-Garden M, Frommer M. CpG islands in vertebrate genomes. J Mol Biol. 1987;196:261–282. doi: 10.1016/0022-2836(87)90689-9. [DOI] [PubMed] [Google Scholar]
- 92.Madeira F, et al. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 2019;47:W636–W641. doi: 10.1093/nar/gkz268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Quinlan AR, Hall IM. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Karolchik D, et al. The UCSC table browser data retrieval tool. Nucleic Acids Res. 2004;32:493–496. doi: 10.1093/nar/gkh103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ewels P, Magnusson M, Lundin S, Käller M. a: Summarize analysis results for multiple tools and samples in a single report. Bioinformatics. 2016;32:3047–3048. doi: 10.1093/bioinformatics/btw354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Bolger AM, Lohse M, Usadel B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnetjournal. 2011;17:10–12. [Google Scholar]
- 98.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Feng J, Liu T, Qin B, Zhang Y, Liu XS. Identifying ChIP-seq enrichment using MACS. Nat Protoc. 2012;7:1728–1740. doi: 10.1038/nprot.2012.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Pagès H. R package version 1.56.0BSgenome: Software infrastructure for efficient representation of full genomes and their SNPs. 2020
- 102.Wang J, et al. Nascent RNA sequencing analysis provides insights into enhancer-mediated gene regulation. BMC Genomics. 2018;19:1–18. doi: 10.1186/s12864-018-5016-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ramírez F, et al. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 2016;44:W160–W165. doi: 10.1093/nar/gkw257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Cliff N. Dominance Statistics: Ordinal Analyses to Answer Ordinal Questions. Psychol Bull. 1993;114:494–509. [Google Scholar]
- 105.Macbeth G, Razumiejczyk E, Ledesma RD. Cliff’s Delta Calculator: A non-parametric effect size program for two groups of observations. Univ Psychol. 2011;10:545–555. [Google Scholar]
- 106.Bush SJ, McCulloch MEB, Summers KM, Hume DA, Clark EL. Integration of quantitated expression estimates from polyA-selected and rRNA-depleted RNA-seq libraries. BMC Bioinformatics. 2017;18:1–12. doi: 10.1186/s12859-017-1714-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Abdennur N, Mirny LA. Cooler: Scalable storage for Hi-C data and other genomically labeled arrays. Bioinformatics. 2020;36:311–316. doi: 10.1093/bioinformatics/btz540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Flyamer IM, Illingworth RS, Bickmore WA. Coolpup.py: Versatile pile-up analysis of Hi-C data. Bioinformatics. 2020;36:2980–2985. doi: 10.1093/bioinformatics/btaa073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Krueger F, Andrews SR. Bismark: A flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics. 2011;27:1571–1572. doi: 10.1093/bioinformatics/btr167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kent WJ, Zweig AS, Barber G, Hinrichs AS, Karolchik D. BigWig and BigBed: Enabling browsing of large distributed datasets. Bioinformatics. 2010;26:2204–2207. doi: 10.1093/bioinformatics/btq351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Zhao H, et al. CrossMap: A versatile tool for coordinate conversion between genome assemblies. Bioinformatics. 2014;30:1006–1007. doi: 10.1093/bioinformatics/btt730. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the generated 4C-seq data generated in this study are available through GEO (GSE156465). All the generated transgenic ESC lines are available upon request.