Abstract
The Wilson disease protein, ATP7B maintains copper (herein referring to the Cu+ ion) homeostasis in the liver. ATP7B traffics from trans-Golgi network to endolysosomes to export excess copper. Regulation of ATP7B trafficking to and from endolysosomes is not well understood. We investigated the fate of ATP7B after copper export. At high copper levels, ATP7B traffics primarily to acidic, active hydrolase (cathepsin-B)-positive endolysosomes and, upon subsequent copper chelation, returns to the trans-Golgi network (TGN). At high copper, ATP7B colocalizes with endolysosomal markers and with a core member of retromer complex, VPS35. Knocking down VPS35 did not abrogate the copper export function of ATP7B or its copper-responsive anterograde trafficking to vesicles; rather upon subsequent copper chelation, ATP7B failed to relocalize to the TGN, which was rescued by overexpressing wild-type VPS35. Overexpressing mutants of the retromer complex-associated proteins Rab7A and COMMD1 yielded a similar non-recycling phenotype of ATP7B. At high copper, VPS35 and ATP7B are juxtaposed on the same endolysosome and form a large complex that is stabilized by in vivo photoamino acid labeling and UV-crosslinking. We demonstrate that retromer regulates endolysosome to TGN trafficking of copper transporter ATP7B in a manner that is dependent upon intracellular copper.
Keywords: ATP7B, Retromer, VPS35, Copper metabolism, Endolysosome, Wilson disease
Introduction
Lysosomes have traditionally been characterized as a disposal organelle of the cell. A growing body of recent studies have implicated lysosomes as centers of cellular nutrient sensing and recycling (Abu-Remaileh et al., 2017; Korolchuk and Rubinsztein, 2011; Rabanal-Ruiz and Korolchuk, 2018). With advancement in lysosomal research, the border between endosome and lysosomes has become increasingly blurred. Fusion between late endosomes and lysosomes gives rise to acid hydrolase-active endolysosomes that are distinct from terminal inactive storage lysosomes (Bright et al., 2016). There is growing evidence that lysosomes regulate cellular homeostasis of various metals like copper (Cu), zinc (Zn) and iron (Fe) (Strochlic et al., 2007; Kurz et al., 2011; Kambe, 2011; Polishchuk et al., 2014; Blaby-Haas and Merchant, 2014). Copper, a transition metal, serves as an essential micronutrient for biological system. It participates in redox reactions in different cellular metabolic pathways shuttling between Cu(II) and Cu(I) states (Uauy et al., 1998), where Cu+ is favored for normal physiological activities (Fahrni, 2013). Several proteins tightly regulate copper homeostasis and supplies bioavailable copper to the secretory pathway. Excess copper induces oxidative stress through a Fenton reaction and hence is detrimental for living system (Gupta and Lutsenko, 2009). Copper homeostasis is primarily maintained by two trans-Golgi network (TGN) recycling P-type ATPases, ATP7A (Menkes disease protein) and ATP7B (Wilson disease protein). ATP7A is expressed ubiquitously, whereas expression of ATP7B is limited to liver, brain and kidney (Telianidis et al., 2013). ATP7B solely functions to maintain the copper homeostasis in hepatocytes (Muchenditsi et al., 2017). Defects in ATP7B leads to Wilson disease (WD), a disorder characterized with copper accumulation in liver, brain and other organs manifesting severe hepatic or neurological symptoms (Huster and Lutsenko, 2007).
In this study, we have attempted to dissect the copper-responsive trafficking itinerary of ATP7B and its mode of regulation in hepatocytes. At physiological/basal copper level, ATP7B primarily resides on membrane of the TGN and functions in secretory pathway by delivering copper to the Cu-dependent ferroxidase ceruloplasmin (Lutsenko, 2016). At higher intracellular copper it vesicularizes (anterograde trafficking), sequestering copper inside vesicles and exports copper in lysosomes (Polishchuk et al., 2014). What is the fate of copper in the lysosome is still to be determined. It is possible that the entire copper content of in the lysosome is excreted out of the cell by exocytosis. Alternatively, lysosomes may act as a storehouse of bioavailable copper that is tapped as per the requirement of the cell.
Some recent and early studies have shown that membrane cargoes recycle back from lysosomes and endolysosomes (Seaman, 2007; Canuel et al., 2008; Suzuki and Emr, 2018a). We examined the fate of ATP7B at the lysosome/endolysosomes. We specifically asked (1) if ATP7B shows a preferential localization at acid-hydrolaseactive endolysosomes or at inactive storage endolysosomes, or (2) is ATP7B degraded at those compartments during its copper export activity and (3), if not, what is the mechanism that retrieves ATP7B from endolysosomal/lysosomal compartments.
Over 300 mutations in ATP7B are associated with WD and are frequently used to understand regulation and structure–function correlation of ATP7B (Forbes and Cox, 1998; Caca et al., 2001; Cox et al., 2005; Gupta et al., 2005; Abdelghaffar et al., 2008; Aggarwal et al., 2013; Braiterman et al., 2014; Ala et al., 2015). These disease mutations affect the functioning of ATP7B either by affecting its copper-transporting activity or its trafficking or both (Gupta et al., 2011; Braiterman et al., 2014). Several proteins govern trafficking and stability of ATP7B (Materia et al., 2012; Jain et al., 2015; reviewed in Gupta et al., 2018), and interact directly or indirectly with defined and conserved motifs of the protein. These motifs influence the directionality of cargo transport between organelles like the Golgi, endosome and plasma membrane.
Although the anterograde pathway of ATP7B has been moderately well characterized (Braiterman et al., 2009; Gupta et al., 2016; Polishchuk et al., 2014), the regulation that mediates its retrograde transport from lysosomes/endolysosomes has been elusive. Recent studies have shown that retromer regulates retrieval or rescue of cargoes from endosomal and lysosomal compartments (Burd and Cullen, 2014; Gershlick and Lucas, 2017; Tammineni et al., 2017). Retromer is a highly conserved endosomal sorting complex, composed of the core components vacuolar protein sorting VPS35, VPS26 (for which there are two forms in mammals, VPS26A and VPS26B) and VPS29 and, variable components, including sorting nexins (SNX) along with WASH complex, which are involved in retrograde transport of endocytosed transmembrane proteins (cargoes) to the TGN or cell surface (Seaman, 2018; Suzuki et al., 2019). Studies from Burd group have provided a mechanistic understanding of retromer oligomerization and cargo sorting (Deatherage et al., 2020; Ma and Burd, 2020; summarized in the review, Burd and Cullen, 2014). Harrison et al. have shown that membrane recruitment of retromer is mediated by bivalent recognition of an effector of phosphoinositide 3-kinase (PI3K), SNX3 and the GTPase Rab7 (Rab7a and Rab7b forms in mammals) by the VPS35 retromer subunit. These interactions facilitate retromer to bind integral membrane cargo, leading to membrane binding of retromer and subsequent cargo sorting (Harrison et al., 2014). Studies by Seaman have determined the role of retromers on recycling of the CI-M6PR (also known as CIMR), the protein which delivers acid hydrolases to lysosome from TGN (Seaman, 2004, 2007). Studies from Emr and co-workers have shown that, in yeast, proteins like autophagy protein Atg27 is recycled from the vacuole to the endosome via the Snx4 complex and then from the endosome to the Golgi via the retromer complex (Suzuki and Emr, 2018b). Furthermore, they have also demonstrated that both VPS26 and VPS35 are critical in cargo retrieval; however, VPS26 utilizes different binding sites depending on the cargo, allowing flexibility in its cargo selection (Suzuki et al., 2019). Besides TGN delivery, retromers also regulate endosome-to-plasma membrane recycling as in for the copper transport protein CTR1 (Curnock and Cullen, 2020).
In this study, we have demonstrated that at elevated intracellular copper, the Cu-P-type ATPase ATP7B, localizes primarily to the active acid hydrolase (Cathepsin B)-positive endolysosomes. Subsequently, upon copper chelation, it recycles to the TGN and this phenomenon is regulated by the retromer. We demonstrate that similar to what is seen in the case of an ‘endocytic cargo’ (Mellado et al., 2014; Tabuchi et al., 2010) retromer also sorts the ‘secretory cargo’, that is, ATP7B for its TGN delivery from endolysosomes and late endosomes.
Results
ATP7B recycles between endolysosomes and the TGN in a copper-dependent manner
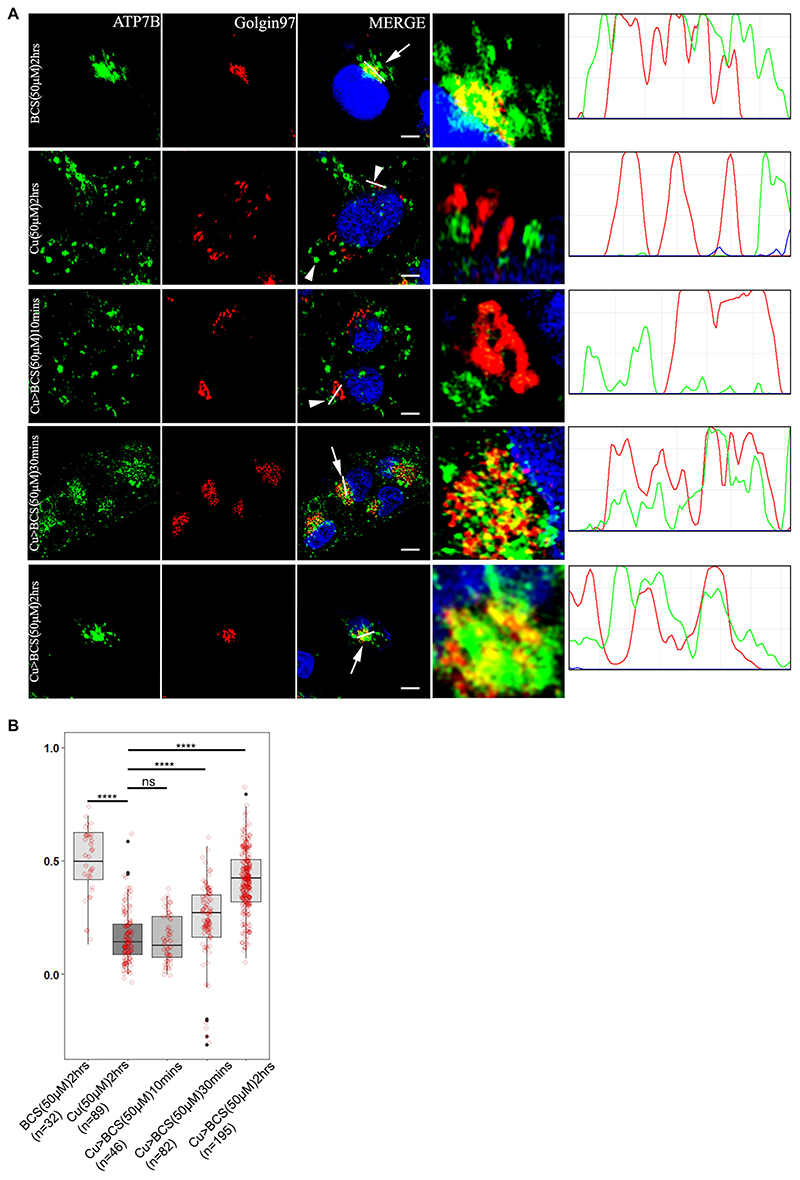
ATP7B vesicularizes from trans-Golgi network in response to high copper and recycles back to TGN upon copper chelation (Roelofsen et al., 2000). To determine the optimal time of recycling of ATP7B from vesicles to TGN, HepG2 cells (a human hepatocellular carcinoma cell line) were treated with high copper (50 μM; 2 h) and subsequently treated with 50 μM bathocuproine disulfonic acid (BCS), a copper chelator for varying time periods (10 min, 30 min and 2 h). Gradual increase in colocalization between TGN and ATP7B was observed with 10 min, 30 min and 2 h of BCS treatment post high copper, as evident from the Pearson’s colocalization coefficient (PCC). At 2 h, the maximum TGN retrieval of ATP7B was observed (Fig. 1A,B).
Fig. 1. ATP7B recycles between the TGN and vesicles in a copper-dependent manner.
(A) Colocalization of ATP7B (green) with the TGN marker, Golgin97 (red) in copper limiting, BCS (top row) and 50 μM copper (second row). Rows 3–5 shows subsequent return of vesicularized ATP7B upon BCS treatment for varying time length (10, 30 and 120 min). The overlap plots (right boxes) show the extent of overlap of green and red at lines drawn through the signals located on TGN (marked by arrow or arrowhead). The line through the selected region of the image represents the overlap between green and red and has been magnified in the rightmost images. Arrow and arrowhead represents TGN localized and vesicularized ATP7B respectively. Scale bars: 5 μm. Blue signal represents DAPI staining of the nucleus. (B) Pearson’s correlation coefficient of colocalization between ATP7B and Golgin97 at different copper conditions demonstrated by a box plot with jitter points. The box represents the 25–75th percentiles, and the median is indicated. The whiskers show the datapoints within the range of 1.5× interquartile range (IQR) from the 1st and 3rd quartile. ****P<0.0001; ns, not significant [non-parametric Mann–Whitney U test (Wilcoxon rank-sum test)]. The number (n) of cells counted to obtain the data for each condition is noted.
Polishchuk et al. have demonstrated that ATP7B utilizes lysosomal exocytosis to export copper (Polishchuk et al., 2014). We investigated whether ATP7B degrades at the lysosome after it transports copper or if it recycles back from these compartments for a next round of export cycle. We treated the cells with either BCS (50 μM) or copper (50 μM). Using an immunofluorescence assay, we determined that under high copper conditions ATP7B exits the TGN and colocalizes with the endolysosomal markers Lamp1 (Fig. 2A,B) and Lamp2 (Fig. S1A,B) and to a lesser extent with late endosome, that is, Rab7-positive compartments (Fig. S2A,B). Upon copper chelation using BCS, ATP7B recycled back to TGN from endolysosomes and late endosomes (bottom panel images of Fig. 2A; Figs S1A, S2A). We did not observe its appreciable colocalization with the recycling endosomal marker Rab11 (Fig. S2C,D).
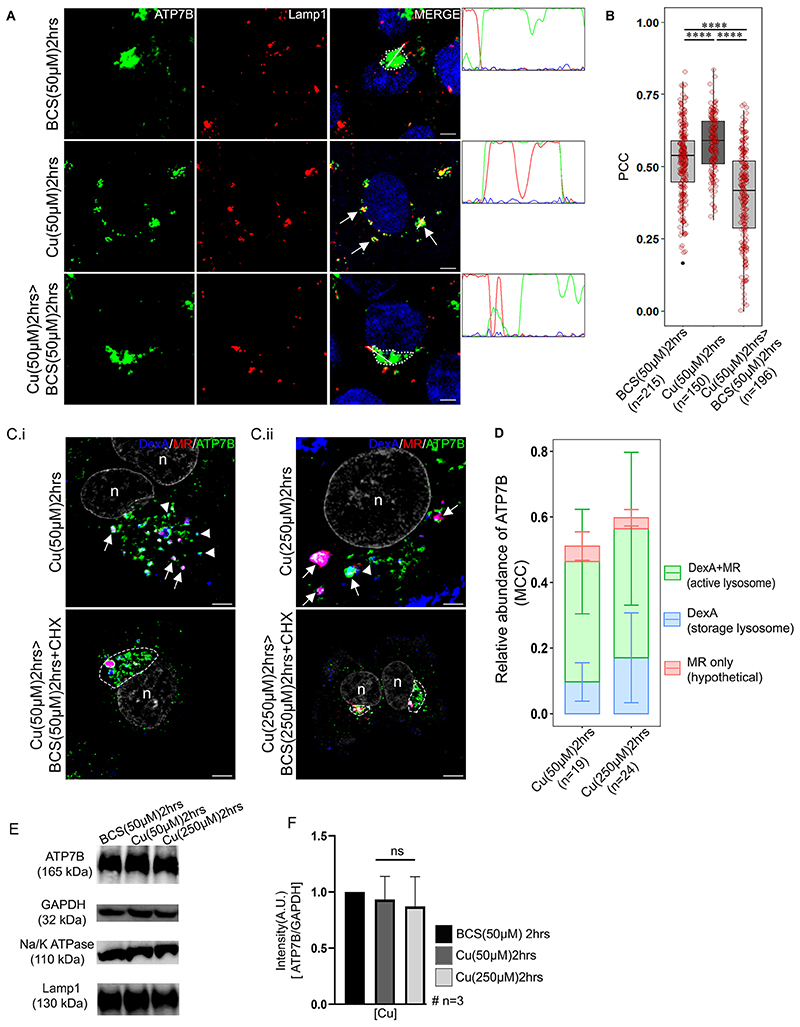
Fig. 2. ATP7B recycles from the endolysosome upon copper depletion.
(A) Colocalization of ATP7B (green) with the lysosomal marker Lamp1 (red) in copper limiting BCS (top row), 50 μM copper (middle row) and copper depletion post copper treatment (bottom row) conditions. The overlap plots (right boxes) show the extent of overlap of green and red at lines drawn through the signals (marked by arrow or arrowhead). Arrow represents vesicularized ATP7B and dotted lines represents perinuclear positioned ATP7B. Blue signal represents DAPI staining for the nucleus. (B) Pearson’s correlation coefficient of colocalization between ATP7B and Lamp1 at different copper conditions demonstrated by a box plot with jitter points. The box represents the 25–75th percentiles, and the median is indicated. The whiskers show the data points within the range of 1.5× interquartile range (IQR) from the 1st and 3rd quartile. ****P<0.0001 (non-parametric Mann–Whitney U test). (C) Colocalization of ATP7B (green), Alexa-647–Dextran (blue) and Magic Red (MR; red) in (C.i) 50 μM copper (upper panel) and 50 μM>50 μM BCS (lower panel) and (C.ii) 250 μM Cu (upper panel) and 250 μM Cu>250 μM BCS (lower panel) conditions. Arrow, ATP7B in active lysosomes; arrowheads, ATP7B in storage lysosomes; dashed line, perinuclear concentration of ATP7B. ‘n’ denotes the nucleus. (D) Bar plot using Manders’ colocalization coefficient (MCC) representing relative ATP7B localization in endolysosomes positive for both active acid-hydrolase and Dextran A (DexA+MR) and storage lysosomes exclusively positive for Dextran A (DexA). The MR-only signal possibly represent background staining and falls within 5% range of error. Error bars show s.d. (E) Immunoblot of ATP7B at different copper conditions (top row). GAPDH is used as a loading control (second row). Immunoblots of Lamp1 (thirdrow) and Na,K-ATPase (fourth row) in different copper conditions. (F) Bar plot (mean±s.d.) representing ATP7B abundance at different copper conditions as normalized against GAPDH abundance obtained from the respective immunoblots. ns, not significant (non-parametric Mann-Whitney U test). The number of cells counted to obtain the data for each condition is denoted by ’n’ (for B and D). Scalebars: 5 μm.
Lamp1 typically marks the terminal end of the endolysosomal pathway (Humphries et al., 2011), but frequently, Rab7 and Lamp1 also colabel the non-degradative lysosomal compartment (Cheng et al., 2018). To determine the percentage distribution of ATP7B in the milieu of late endosomal-endolysosome compartments (Rab7-Lamp1), we utilized structured illumination microscopy (SIM) along with deconvolution confocal microscopy imaging. At elevated copper (50 μM), we found a proportionately higher co-distribution of ATP7B with compartments positive for both Rab7 and Lamp1 than compartments positive for individual markers. In this condition, 48.5±13.2% (mean±s.d.) of total cellular pool of ATP7B colocalized with either Lamp1 or Rab7 or both. Among them, 44.9±14% of ATP7B localized in vesicles positive for both Lamp1 and Rab7. Hence, it is inferred that recycling of ATP7B to TGN upon copper chelation is largely from vesicles that are positive for both the markers (Fig. S3A,B). Together, this result suggests that ATP7B localizes at endolysosomal compartments at high copper.
We wanted to scrutinize the nature of the ATP7B-harboring endolysosomal compartment with respect to its hydrolase activity. The delivery of macromolecules for degradation by lysosomal acid hydrolases requires traffic through early endosomes to late endosomes, followed by fusions between late endosomes and lysosomes. Using the cathepsin-B substrate Magic Red (MR), which releases the fluorescent reporter (Cresyl Violet) upon proteolytic cleavage, Bright and co-workers have demonstrated that the late endosome-lysosome fusion results in the formation of acid-hydrolase-active endolysosomes, which are hybrid organelles and are distinct from inactive terminal storage lysosomes (Bright et al., 2016). We employed this technique to identify the ATP7B-residing compartments in high copper conditions. Cells were treated with Dextran A–Alexa Fluor 647 and Magic Red. The total lysosomal pool was labeled with Dextran A–Alexa Fluor 647 (far red) and the acid hydrolase-active fraction was stained red due to the hydrolase-mediated release of the fluorescent reporter Cresyl Violet [emission wavelength (λem) of 622 nm] (Fig. 2C; Fig. S4). We noticed that ATP7B at high copper (50 μM and 250 μM) distributes between active lysosomes and storage lysosomes, with a higher abundance in the active ones. Interestingly, compared to what was seen at 50 μM copper, at 250 μM, the percentage localization of ATP7B, although staying constant at the active lysosomes, increased by ∼50% in the storage lysosomes (Fig. 2D). This increase might be attributed to further shift of ATP7B localization from the TGN to the endolysosomes with increasing copper. However, there were no apparent degradation of ATP7B in either of the copper treatment conditions as evidenced by immunoblots (Fig. 2E). No evidence of degradation was also recorded in controls for Lamp1 (lysosomal marker) or Na,K-ATPase (membrane protein marker) (Fig. 2E,F). Upon treatment with copper chelator, BCS, in presence of cyclohexamide, ATP7B returned to its tight perinuclear location, reminiscent of TGN (Fig. 2Ci,ii, lower panel images). Furthermore, using live-cell imaging in HepG2 cells, we confirmed the dynamic fusion and fission of GFP-tagged wild-type ATP7B (GFP–wtATP7B) vesicles with or from dual-stained (Dextran A in blue and Magic Red in red) active hydrolase-positive endolysosomes (marked with an arrowhead and the region is magnified in a box in Movie 1). Image capture was initiated at the point of copper treatment, and data was collected at an interval of 5 s for a total period of 15 min.
It is worth mentioning that the size and shapes of the Lamp1-positive vesicles are variable across cells and as well as in a single cell. They vary from 20–200 μM in diameter (maximum length across), and shape varies from small puncta to larger patchy clumps. We noticed that abundance of Lamp1-positive compartments was higher in cells treated with copper than untreated cells.
Retromer regulates the retrograde trafficking of ATP7B from endolysosomes
It has been shown that the Menkes disease protein ATP7A, a homolog of ATP7B, requires the SNX27-retromer to prevent lysosomal degradation and maintain surface levels and localization (Steinberg et al., 2013). In addition, CI-M6PR recycles between TGN and endosomes in a retromer-regulated fashion to mediate sorting of lysosomal hydrolases (Seaman, 2004, 2007; Arighi et al., 2004; Cui et al., 2019). ATP7B exits endolysosomal compartments by an unknown regulatory mechanism upon copper chelation. This prompted us to investigate whether the retromer complex plays a role in retrieval of ATP7B from the endolysosome and late endosomal compartments. VPS35 is the largest core component of the retromer complex. It functions as the scaffold for the assembly of the other core components, VPS29 and VPS26, and also cargo binding (Hierro et al., 2007). Hence, we selected human (h)VPS35 as the target component to determine the role of retromer complex, if any, in copper-mediated trafficking of ATP7B. Broadly, localization and trafficking of ATP7B in HepG2 can be divided in 4 phases – (a) at the TGN in basal Cu [or without Cu (–Cu)], (b) on the anterograde vesicles in high Cu (50 μM for 2 h), (c) subsequently, upon copper chelation, on the retrograde vesicles (10 min, 50 μM BCS treatment) and (d) further, with the majority of ATP7B recycled back to the TGN in copper-chelated conditions (30 min, 50 μM BCS treatment).
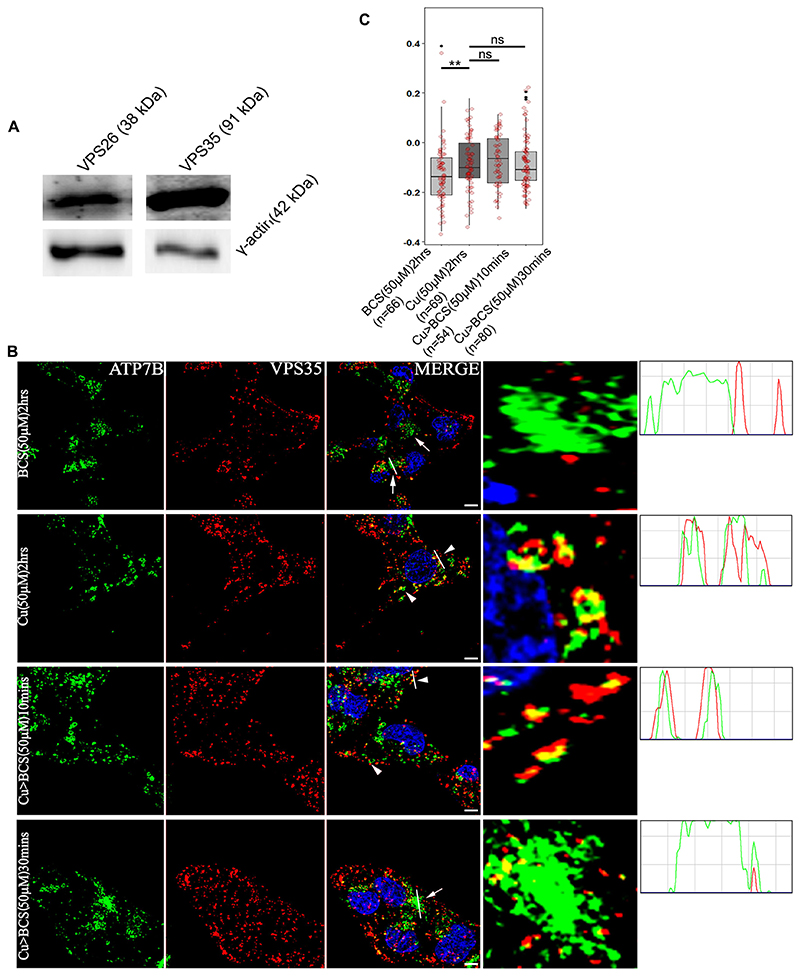
Immunoblot analysis of VPS26 and VPS35 revealed that HepG2 cells express the retromer complex proteins (Fig. 3A) and copper levels do not alter abundance of VPS35 in HepG2 cells (Fig. S5A). To study whether ATP7B colocalizes with retromer core components, cells were treated to give the four conditions described above. Cells were fixed, blocked and co-stained with anti-ATP7B and anti-VPS35 antibody. Maximum colocalization, as quantified by means of the PCC between VPS35 and ATP7B, was observed in high copper (phase b) and in copper for 2 h followed by BCS (copper followed by BCS treatment is denoted Cu>BCS with appropriate duration and concentrations noted) for 10 min (phase c) conditions. With 30 min of BCS treatment post high copper (phase d), ATP7B and VPS35 showed loss of colocalization (Fig. 3B,C). Similar observations were made with VPS26 (data not shown).
Fig. 3. ATP7B and VPS35 colocalizes at high copper.
(A) Immunoblot showing that HepG2 cells express the retromer subunits VPS26 and VPS35 (top panel). γ-actin is used as loading control (lower panel). (B) Colocalization of ATP7B (green) with retromer subunit, VPS35 (red) in copper limiting BCS (top panel), 50 μM copper (second row) and copper depletion post copper treatment (rows three and four). The overlap plots (right boxes) are from the region of the field has been magnified rightmost images) show the extent of overlap of green and red at lines drawn through the signals (marked by arrowor arrowhead). Arrowheads represent vesicularized ATP7B and arrows represents perinuclear ATP7B. the line through the selected region of the image represents the overlap between green and red signals and has been magnified (rightmost image). Blue signal represents DAPI staining for nucleus. Scale bars: 5 μm. (C) Pearson’s correlation coefficient of colocalization between ATP7B and VPS35 at different copper conditions demonstrated by box plot with jitter points. The box represents the 25–75th percentiles, and the median is indicated. The whiskers show the data points within the range of 1.5× interquartile range (IQR) from the 1st and 3rd quartile. **P<0.01; ns, not significant (non-parametric Mann–Whitney U test). The number (n) of cells counted to obtain the data for each condition is noted.
To further understand whether retromer regulates any of these phases of ATP7B trafficking, we knocked down VPS35 and studied the phenotype of copper-induced localization of ATP7B with respect to TGN. Appreciable knockdown was attained (>70–80%) for the targeted siRNAs as compared to scrambled for VPS35, as ascertained by immunoblotting (Fig. 4A). Additionally, as reported by (Fuse et al., 2015) decrease in VPS26 level was also observed in VPS35 KD cell which eventually tells us expression as well as functionality of VPS subunits are interdependent (Fig. S5B). Recycling of CI-M6PR between Golgi and endolysosomes was used as a control to functionally evaluate knockdown of VPS35. As reported previously, we found that CI-M6PR localized in vesicles in cells transfected with siRNA against VPS35 KD as compared to scrambled RNA transfected cells where CI-M6PR shows a complete localization with TGN46 (Fig. S5C) (Seaman, 2007).
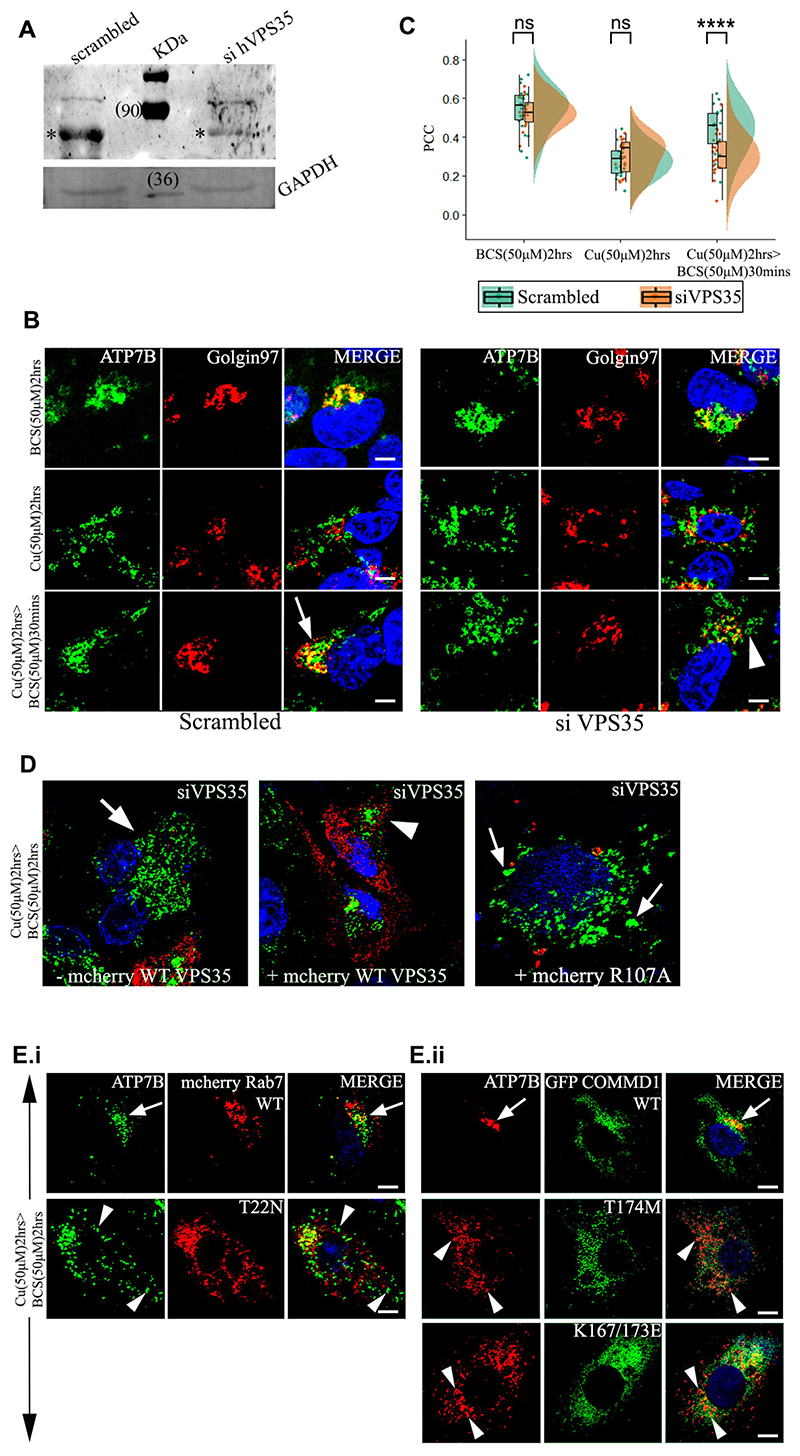
Fig. 4. VPS35 and its associated proteins regulates retrieval of ATP7B from lysosomes to TGN.
(A) siRNA-mediated knockdown of Vps35 in HepG2 cells shows its downregulation. (*) denotes the VPS35 protein. GAPDH is used as a loading control. (B) Colocalization of ATP7B (green) with the TGN marker Golgin97 (red) in BCS (top row) and 50 μM copper (second row) and copper depletion post copper treatment (third row). Arrow denotes TGN colocalization of ATP7B; arrowhead denotes vesicularized ATP7B. Scale bars: 5 μm. (C) Pearson’s correlation coefficient of colocalization between ATP7B and TGN at different copper conditions comparing VPS35 siRNA treated versus control demonstrated by a box plot. The box represents the 25–75th percentiles, and the median is indicated. The whiskers show the data points within the range of 1.5× interquartile range (IQR) from the 1st and 3rd quartile. ****P<0.0001; ns, not significant (non-parametric Mann–Whitney U test). (D) Localization of ATP7B (green) in VPS35 siRNA-treated cells and subsequently transfected with mCherry–wtVPS35 (red). The left image represents cells that are not expressing mCherry–wtVPS35 as compared to cells expressing mCherry–wtVPS35 (center image) or the mutant, mCherry-R107A-VPS35 (right) Cells belong to the same culture dish for both the left and center images. Arrow represents vesicularized ATP7B and arrowhead perinuclear ATP7B. (E.i.) Localization of ATP7B (green) in cells overexpressing mCherry–wtRab7 (red) (top panel) or dominant negative mutant mCherry-T22N-Rab7 (bottom panel) in copper to BCS condition to triggers retrograde trafficking of ATP7B. (E.ii.) At the same copper conditions as E.i., localization of ATP7B (red) in cells overexpressing GFP–wtCOMMD1 (top panel) or the mutants GFP–T174M-COMMD1 (middle panel) and GFP–K167/173L-COMMD1 (bottom panel). Scale bars: 5 μm. Blue signal represents DAPI staining for nucleus. Arrows represents perinuclear ATP7B and arrowheads vesicularized ATP7B.
VPS35 siRNA treated cells were incubated with (1) BCS (2 h) or (2) copper (2 h) or (3) copper (2 h)>BCS (30 min), fixed, blocked and stained with the anti-ATP7B and anti-Golgin97 antibodies. We observed that trafficking of ATP7B from the vesicles back to the TGN (condition 3) was significantly abrogated (Fig. 4B,C). ATP7B remained in vesicles and failed to recycle back to TGN even after the cells were incubated in BCS for a prolonged time of 2 h subsequent to copper treatment (data not shown).
To corroborate our finding that VPS35 regulates ATP7B trafficking, we utilized livecell imaging using the wild-type and an inactive dominant-negative mutant mCherry-VPS35 (R107A) protein. The R107A mutation abolishes the interaction of VPS35 with VPS26 and affects cargo sorting (Gokool et al., 2007; Zhao et al., 2007). HEK293 cells were co-transfected with GFP–ATP7B and mCherry–wt-VPS35, GFP–ATP7B was concentrated at TGN (perinuclear region) after preincubating the cells with the copper chelator BCS. Vesicularization and recycling of ATP7B was triggered with treatment with copper. Image capture was initiated at the point of copper treatment and data was collected at an interval of 1.964 s for a total period of 30 min. We noticed that within 1 min of Cu treatment, GFP and mCherry signals colocalized at the same endosomal vesicle. The dwell time ofwt-VPS35 and GFP–ATP7B on the vesicle was notably higher for wt-VPS35 compared to the mutant. For VPS35-R107A, the colocalization lasted for a few seconds (4–11 s) but for wt-VPS35, the colocalization lasted for an average of 7 min (Movies 2A,2B; Fig. S6Ai,ii).
Since we determined that ATP7B colocalizes at the endolysosomes at high copper, we investigated whether VPS35 regulates endolysosomal exit of ATP7B on triggering its retrograde pathway. Using identical experimental conditions and knocking down VPS35 or not, we found that ATP7B colocalizes with VPS35 in high copper (Fig. S6B; left image, top and bottom panel) for both KD and control cells. ATP7B is arrested at Lamp1-positive compartments upon triggering the retrograde pathway (i.e. high copper>BCS) (Fig. S6B; center images, bottom panel) in KD cells but not in scrambled RNA-treated cells (Fig. S6B, top panel). Boosting the retrograde pathway by lengthening BCS treatment time to 2 h did not facilitate retrieval of ATP7B from the lysosomes (Fig. S6B, right images, bottom panel), unlike control (Fig. S6B, right images, top panel).
We further confirmed the role of retromer by rescuing the non-recycling phenotype of ATP7B in VPS35 KD cells by overexpressing mCherry–wt-VPS35. We found that ATP7B recycled back from vesicular to its tight perinuclear TGN localization upon copper chelation in VPS35 KD cells that overexpressed the wt-VPS35 construct but not the mutant VPS35-R107A or in untransfected cells (Fig. 4D). These experiments confirm that VPS35 regulates retrieval of ATP7B from endolysosomes to TGN upon copper depletion.
Retromer-associated proteins Rab7 and COMMD1 regulates ATP7B recycling
We explored the possible role of two proteins that have been shown to interact directly or indirectly (as part of a complex) with the retromer in retrograde trafficking of ATP7B. It has been now been thoroughly established that Rab7 regulates recruitment of the core retromer trimer VPS35, VPS26 and VPS29 to the endosomal membrane and interference with Rab7 function causes dissociation of the trimer (Rojas et al., 2008). On other hand, COMMD1 protein, whose dysfunction phenocopies Wilson disease mutation, interacts with the WASH complex and is critical in copper-dependent endosomal localization of the Cu-ATPase ATP7A (Phillips-Krawczak et al., 2015) and retrograde trafficking of ATP7B (Miyayama et al., 2010). Interaction of the WASH complex with VPS35 is crucial for its endosomal localization and this facilitates cargo sorting by promoting formation of endosomal F-actin (Seaman and Freeman, 2014).
We overexpressed the dominant-negative (DN) mutant Rab7A-T22N (impaired in nucleotide exchange and with a reduced affinity for GTP) (Spinosa et al., 2008) and studied its effect on retrograde trafficking of ATP7B. Trafficking of ATP7B was triggered with 50 μM copper for 2 h. Subsequently upon copper chelation, we observed that ATP7B stayed vesicularized and failed to return to its perinuclear location in cells overexpressing the DN mutant, a phenotype that is identical to VPS35 KD condition (Fig. 4Ei, bottom panel). Cells overexpressing wt-Rab7A exhibited a normal trafficking itinerary of ATP7B (Fig. 4Ei, upper panel).
We next studied the effect of two COMMD1 mutants on retrograde trafficking of ATP7B. The COMMD1 mutation T174M was discovered as a WD patient with deteriorating prognosis even after early medical intervention (Gupta et al., 2010). The other key residues mutated in COMMD1 are K167E/K173E, as the positively charged lysine residues have been shown to be crucial for its phosphatidylinositol interaction (Burkhead et al., 2009; Stewart et al., 2019). Cells were transfected with GFP–COMMD1-T174M or GFP–COMMD1 K167E/K173E and trafficking of ATP7B was triggered with 50 μM copper for 2 h. Subsequently upon copper chelation with BCS, we observed vesicular localization of ATP7B in cells over expressing either GFP–COMMD1-T174M or GFP–COMMD1 K167E/K173E (Fig. 4Eii, middle and bottom panel). Cells overexpressing wt-COMMD1 exhibited a normal trafficking itinerary of ATP7B (Fig. 4Eii, top panel).
We conclude that interfering with the functioning of the retromer core proteins or proteins that are associated with the retromer complex abrogates the retrograde trafficking of ATP7B.
Lysosomal luminal pH does not influence localization of ATP7B and recruitment of VPS35
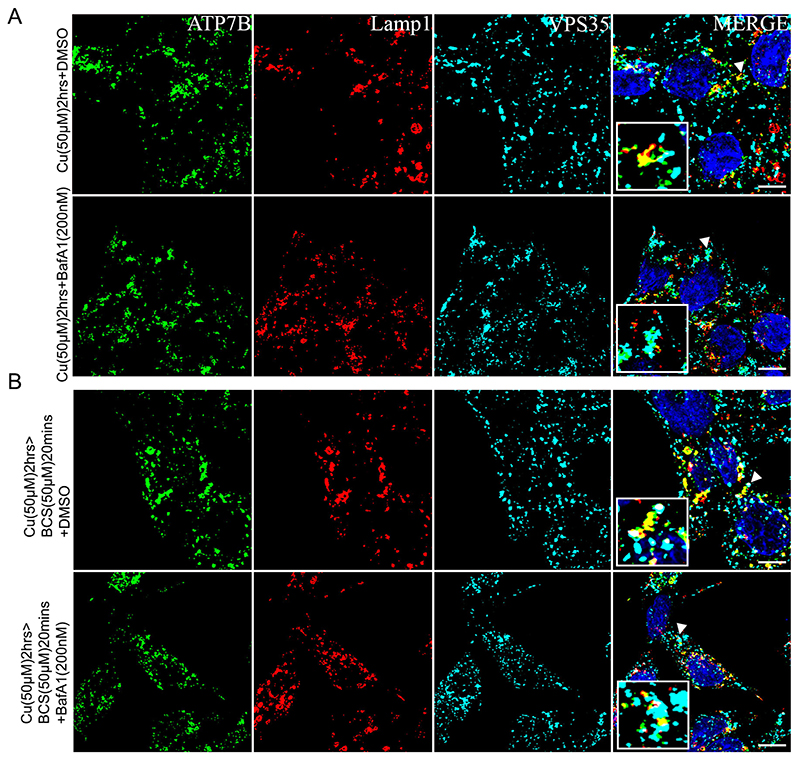
It emerges that copper-induced localization of ATP7B involves a tripartite participation, that is, from ATP7B itself, the endolysosome and retromer. After confirming the role of VPS35 in this process, we asked whether the luminal endolysosomal environment affects retromer recruitment and hence ATP7B retrieval from lysosome. Retromers have been previously implicated in lysosomal activity, for example, autophagy (Cui et al., 2019). We investigated whether the targeting of ATP7B to lysosomes in high copper or its retrieval initiation by VPS35 recruitment is affected by inactivation of the V-ATPase that is crucial for lysosomal functioning. Cells were treated with the V-ATPase inhibitor BafA1 (200 nM), in 50 μM copper for 2 h (to trigger lysosomal targeting of ATP7B) and Cu>BCS (50 μM, 20 min) conditions to trigger its lysosomal exit and retrograde trafficking. No observable and significant difference in colocalization of Lamp1, ATP7B and VPS35 was obtained between BafA1-treated cells versus the control cells in either of the copper treatment conditions (Fig. 5A,B). It can be inferred that retromer being situated on the outer membrane of the Lamp1-positive compartments is unaffected by the change in luminal pH of the lysosome brought on by BafA1 treatment. Localization of ATP7B also stays unaltered, as also demonstrated by Polishchuk and co-workers. (Polishchuk et al., 2014).
Fig. 5. Lysosomal luminal pH does not influence localization of ATP7B and recruitment of VPS35.
(A) Colocalization of ATP7B (green), Lamp1 (red) and VPS35 (cyan) in high copper for 2 h in cells treated with Bafilomycin A1 (lower row) or not (upper row). (B) Colocalization of ATP7B (green), Lamp1 (red) and VPS35 (cyan) in cells treated with copper chelator for a brief period (20 min) subsequent to high copper treatment to induce ATP7B vesicularization. Cells were treated with Bafilomycin A1 (lower row) or not (upper row). The magnified inset corresponds to the region of the main image marked by arrowheads. Scale bars: 5 μm.
VPS35 acts on ATP7B in a micro-distant modus operandi
Next, we used biochemical assays to investigate whether VPS35 directly interacts with ATP7B. We utilized co-immunoprecipitation where GFP–ATP7B was expressed in cells, pulled down with anti-GFP beads and probed for endogenous VPS35. At high copper (100 or 200 μM) or upon subsequently triggering ATP7B recycling BCS (100 μM), we failed to detect any signal in the immunoblot at developed with anti-VPS35 antibody [Fig. S7 shows data for 200 μM Cu, and Cu (200 μM)>BCS (100 μM)].
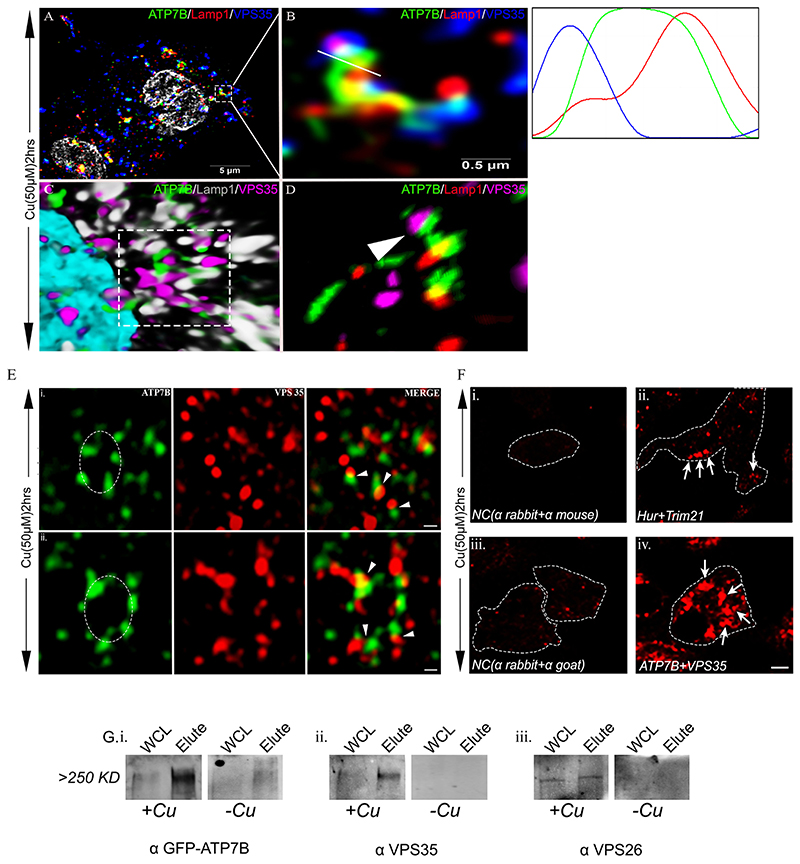
To understand the underlying reason of our inability to detect interaction between ATP7B and VPS35 using biochemical methods, we resorted to super resolution microscopy to determine the exact positioning of ATP7B with respect to VPS35 at the lysosomal compartment. Using SIM and high-resolution deconvolution confocal microscopy, we observed that ATP7B and VPS35 lies in juxtaposition on the lysosomal compartment (stained with Lamp1 antibody) at high copper conditions (Fig. 6A–D). The average distance between these two proteins varies from ∼25 nm–200 nm. It can be inferred that, although ATP7B lies in close proximity to VPS35 and is regulated in its retrograde pathway by retromer, the physical interaction between these two proteins is indirect. We utilized stimulated emission depletion (STED) microscopy to look further closely on the disposition of ATP7B and VPS35 on a vesicular membrane. Using Z-stacking, we determined the shape of a vesicle (dotted circle in Fig. 6E). At high copper, we observed that ATP7B (green) and Vps35 (red) decorated the vesicular membrane with a signal overlap (yellow) at a maximum resolution of 25 nm, further substantiating our biochemical findings.
Fig. 6. VPS35 interacts with ATP7B on lysosome in a micro-distant manner.
(A) High-resolution deconvoluted confocal microscopy merged image showing colocalization of ATP7B (green) with Lamp1 (red) and VPS35 (blue) at 50 μM copper. Gray represents nucleus. (B) Zoomed image of region marked in A. The overlap plots (right box) show the extent of overlap of green, red and blue at lines drawn through the signals (marked by white line). (C) 3D representation of image in B, marked by a dashed line. ATP7B is marked in green, Lamp1 in gray and VPS35 in magenta. Cyan represents the nucleus. (D) 3D representation of structured illumination microscopy (SIM) image of the same area with 100 nm resolution. ATP7B is marked in green, Lamp1 in red and VPS35 in magenta. Arrowhead represents co-distribution of ATP7B and VPS35 in alysosomal compartment (Lamp1). (E) Stimulated emission depletion (STED) microscopy image of ATP7B (green) and VPS35 (red). Both rows represents colocalization of ATP7B and VPS35 in high copper. ATP7B-containing vesicles (marked by dotted circle) show juxta-positioning of VPS35 (red) and ATP7B (green). Arrowhead represents point of juxtaposition or merging. Scale bars: 200 nm. (F) Proximity Ligation Assay. The upper row (i and ii) shows interaction of mouse Hur and rabbit Trim21 in HepG2 cells, which serve as a positive control. (i) Technical negative control (NC) without primary antibodies probed with onlyanti-rabbit(-) and anti-mouse(+) secondary PLA antibodies. (ii) Hur and Trim21 interaction, marked by arrows. The lower row (iii and iv) shows interaction of rabbit ATP7B and goat VPS35 in HepG2. (iii) Technical negative control (NC) without primary antibodies probed with only anti-rabbit(-) and anti-goat(+) secondary PLA antibodies. (iv) ATP7B and VPS35 interaction, marked by arrows. (G) In-cell crosslinking. (i) Immunoblot showing presence of GFP both in WCL and elute in the +Cu state at ∼>>250 kDa (metabolically crosslinked with photo-amino acids), which was absent in the –Custate. (ii, iii) A similar pattern was observed for both co-eluted proteins VPS35 and VPS26, respectively. WCL, whole-cell lysate.
Since, the interaction between ATP7B and VPS35 is likely to be indirect and possibly they a part of a larger complex, we utilized a proximity ligation assay (PLA) to substantiate our finding. Interaction of Hur and Trim21 (Guha et al., 2019) were used as positive controls and secondary antibodies against ATP7B (anti-rabbit-IgG) and VPS35 (anti-goat-IgG) were used as negative controls. We observed positive interaction between ATP7B and VPS35, as is evident by formation of red intracellular puncta, which occurs at sites where the two proteins juxtapose at a distance less than ≤40 nm (Fig. 6F).
This finding strongly suggests that VPS35 and ATP7B are a part of a larger complex, and the regulation of ATP7B trafficking by the retromer proteins are possibly indirect and they do not share a physical interface. To detect such an interaction, we metabolically labeled GFP–ATP7B-overexpressing HepG2 cells with photo-amino-acids (P-leucine and P-methionine). Cells were treated with copper (50 μM for 2 h) or the copper chelator BCS (50 μM for 2 h) and subjected to UV crosslinking. Crosslinked GFP–ATP7B complex was immunoprecipitated with anti-GFP beads. Upon probing the eluate on an immunoblot with anti-VPS35, anti-VPS26 and anti-GFP antibodies, a complex much larger than 250 kDa was detected that contained all the three proteins. The complex was found to be absent or much less abundant in cells treated with BCS (Fig. 6G).
VPS35 knockdown does not abrogate copperexport function of ATP7B
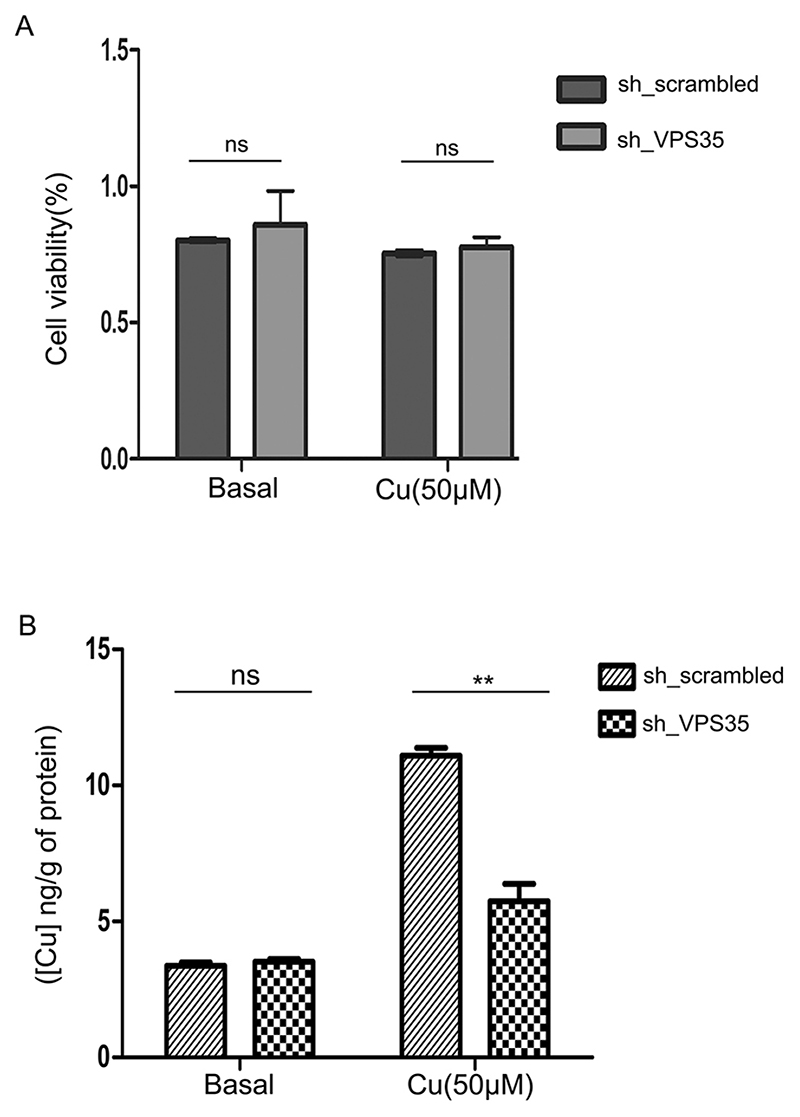
We have now established that retromer regulates retrograde trafficking of ATP7B, and that downregulation of VPS35 arrests ATP7B at endolysosomes. We examined whether the ATP7B arrested at endolysosomes is capable of exporting copper. We knocked down VPS35 using lentiviral mediated shRNA delivery in HepG2 cells. Scrambled, non-targeting shRNA was used as control (Fig. S8A). shRNA# 2, 3 and 5 revealed significant knockdowns, and hence were subsequently used for the experiment. Cells were either treated with 50 μM copper for 2 h or maintained in basal medium. We did not observe any statistically significant change (P>0.1) in cell viability in either of the treatment conditions, as determined by a Neutral Red (NR) assay (Fig. 7A). No apparent change in cell morphology was recorded in any of these conditions ascertained by 40× phase contrast microscopy (Fig. S8B). Copper measurement in these cells from four conditions (sh_Scrambled with basal Cu; shVPS35 with basal Cu; sh_Scrambled with 50 μM Cu; and shVPS35 with 50 μM Cu) were carried out using inductively coupled plasma-optical emission spectrometry (ICP-OES). Interestingly, we found that upon copper treatment, accumulation of copper was significantly lower (P<0.01) in VPS35 KD cells as compared to non-KD control cells (Fig. 7B). This suggests that ATP7B arrested at the lysosomes in the VPS35 KD condition is continuing its copper export function resulting in a lower accumulation of intracellular copper. However, in basal conditions, both VPS35 KD and control cells exhibited similar levels of intracellular copper (Fig. 7B).
Fig. 7. Knocking down VPS35 does not affect cell viability but lowers intracellular copper content.
(A) Graphical representation of cell viability where VPS35 expression was silenced by lentiviral mediated delivery of shRNA against VPS35 or not (non-targeting shRNA) as measured by Neutral Red (NR) assay. The assay was performed in two different copper conditions (basal and 50μM). Each conditions has four replicates. (B) Intracellular copper content (ng/g) as measured by ICP-OES in cells subjected to identical knockdown or copper treatment conditions as of 7A. Each conditions has five replicates (P<0.05). Graphs show mean±s.d. **P<0.01; ns, not significant (non-parametric Mann–Whitney U test).
Recently, it has been shown that retromer regulates recycling of the copper importer CTR1 between the plasma membrane and endocytic vesicles in HeLa cells (Curnock and Cullen, 2020). Compared to control cells, knockdown of VPS35 enhances survivability of cells in high copper, which is suggestive of lower intracellular copper accumulation and hence lower copper-induced cytotoxicity (Curnock and Cullen, 2020). This is primarily due to regulation of CTR1 and not ATP7B (HeLa cells do not express ATP7B, as shown in the Human Protein Atlas and our data). In the study by Curnock and Cullen, the treatment was carried out for at least 48 h and within a range of 3.125 μM to 800 μM copper; at 50 μM copper, they found no significant difference of cell survivability and hence intracellular copper levels was observed between KO and control cells. This result suggests that the lower intracellular copper content that we observe in VPS35 KD compared to control cells is primarily due to continued export function of ATP7B arrested on the lysosomes.
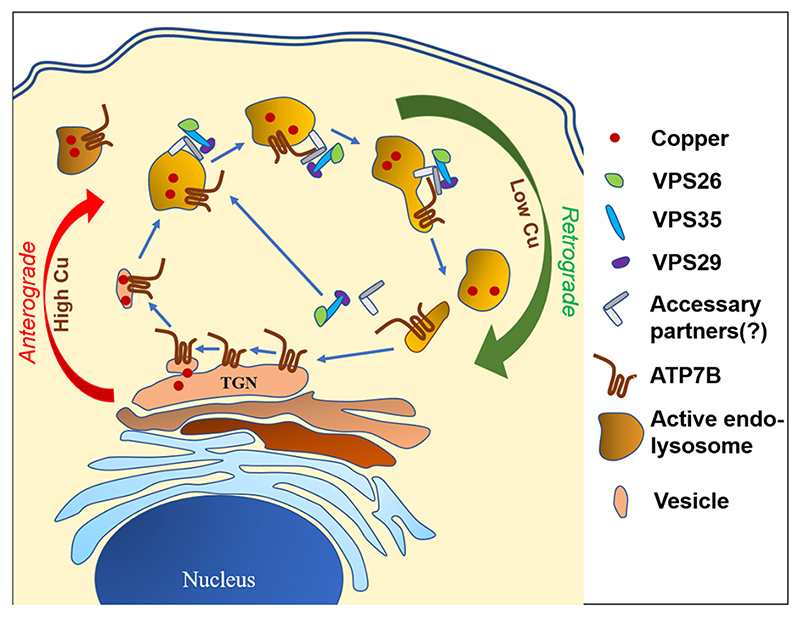
In summary, in this study, we establish that ATP7B traffics to active acid-hydrolase containing endolysosomes in high copper conditions, where it juxtaposes with VPS35 and contributes to the formation of a complex that comprises at least ATP7B, VPS35 and VPS26 in addition to other proteins. Upon triggering the retrograde pathway by subsequent copper chelation, retromer regulates the recycling of ATP7B from the lysosome to the TGN (illustrated in Fig. 8).
Fig. 8. Schematic representation of recycling of ATP7B between TGN and endolysosomes.
Retromers are recruited on the endolysosomal membrane, which regulates retrograde transport of vesicularized ATP7B upon copper removal.
Discussion
The copper transporting ATPase ATP7B exports copper through lysosomes (Polishchuk et al., 2014). ATP7B (160 kDa) is a large eight membrane-spanning protein with a total of 1465 residues. ATP7B resides on the TGN membrane at basal copper levels and traffics to Lamp1- and Rab7-positive compartments at high copper. We argue that it would be highly wasteful for the cell to degrade ATP7B after each cycle of copper export from the TGN to the lysosomes. At the onset of this study, we determined that ATP7B primarily localizes on the active acid hydrolase-positive endolysosomes at high copper. Since ATP7B is a recycling protein, we wondered whether ATP7B recycles back from these compartments after it pumps copper in the endolysosomal lumen for either export out of the cell or for reutilization as a nutrient, and investigated this possibility. Interestingly, the protein does not get degraded unlike many other cargoes that are destined for degradation at the lysosomes. Rather, the low pH in lysosomal lumen might help the release of copper from the histidine residues that are located between the TM1 and TM2 loop, as shown in its homolog ATP7A (LeShane et al., 2010; Barry et al., 2011; Otoikhian et al., 2012), and bind copper.
Furthermore, we investigated the regulatory mechanism of retrograde trafficking of ATP7B. In a preliminary proteome analysis on GFP–ATP7B vesicles isolated from HepG2 cells (data not shown), we identified members of the retromer complex. Previously Harada et al. (2000) had shown that ATP7B resides in late endosome (Rab7 positive) in high copper. Retromer is recruited on endosomal membrane by the sequential action of Rab5 and Rab7 (Rojas et al., 2008).
Before investigating the role of retromer in retrieval of ATP7B from lysosomes, we first determined whether ATP7B was stable in lysosomal and Rab7 compartments. Similar to observations made by Polishchuk et al. (2014), we found that even at up to 200 μM copper treatment in HepG2 cells, ATP7B shows no significant degradation, and we noticed no change in its abundance at 250 μM copper. At high copper, ATP7B primarily localized at the active hydrolase-positive recycling endolysosomes with a smaller fraction at the terminal storage lysosomes. Interestingly, upon further elevating intracellular copper, although fraction of ATP7B was unaltered at the active endolysosomes, an increase in its abundance was noticed in storage lysosomes. We argue that this shift of the abundance equilibrium of ATP7B is due to a relay effect of ATP7B trafficking from TGN to active endolysosomes and finally to storage endolysosomes. At 250 μM copper, further emptying of ATP7B from TGN is compensated for by its increase at the storage terminal lysosome.
Upon examining triple-colocalization of ATP7B, VPS35 and Lamp1 in fixed cells, we noticed that the level of overlap is moderate at high copper. This might be attributed to the fact that at a given point in copper-treated cells, the nature of vesicles are highly heterogeneous comprising of retrograde and anterograde vesicles, and those are at various stages of trafficking. We hypothesize that the endolysosomes would exhibit a higher co-residence of VPS35 and ATP7B if we were able to synchronize the TGN exit (upon copper treatment) and endolysosomal exit (upon subsequent copper removal) of ATP7B. However, time-lapse imaging showed that GFP–ATP7B and mCherry–Vps35 colocalizes in the same compartment for a few minutes. In VPS35 KD cells, ATP7B is trapped in the endolysosomes even upon activating the retrograde pathway (Cu>BCS). We reason that ATP7B recycles back to TGN directly and not via plasma membrane, as we did not observe ATP7B staining at the plasma membrane or even at the cortical actin (data not shown). This might be due to the non-polarized nature of the HepG2 cells used in this study.
Interestingly, using conventional biochemical assays, we did not detect any direct interaction of ATP7B and VPS35 (or VPS26). This is possibly due to the fact that, although the retromer complex regulates lysosomal exit of ATP7B, the interaction is mediated via a different member of the complex. This proposition is substantiated by co-detection of ATP7B, VPS35 and VPS26 in a supercomplex (>>250 kDa) and also proximity of ATP7B and VPS35 at a range ≤40 nm as detected in the PLA study. Our observation suggests that the retromer complex along with the WASH, and the CCC complex consisting of COMMD1 and the Rab-GTPase Rab7 co-ordinates the recycling of ATP7B from the endolysosomal compartments. However, the order of interaction of these complexes with the cargo is still to be determined.
Retromer complex shows heterogeneity in its subunits that are responsible for binding to the cargo (Belenkaya et al., 2008; Feinstein et al., 2011; Zhang et al., 2012; Follett et al., 2016; Suzuki et al., 2019). It has been shown that the canonical recycling signal for the Divalent Cation Transporter (DMT1-II) binding of retromer is mediated via the interface of VPS26 and SNX3 in a hybrid structural model shows that the α-solenoid fold extends the full-length of Vps35, and that Vps26 and Vps29 are bound to its two opposite ends (Hierro et al., 2007; Lucas et al., 2016). This extended structure suggests that multiple binding sites for the SNX complex and receptor cargo are present. It has been shown show that a membrane recruitment of retromer is mediated by recognition of SNX3 and RAB7A, by the VPS35 subunit. These bivalent interactions prime retromer to capture integral membrane cargo, which enhances membrane association of retromer and initiates cargo sorting (Zhao et al., 2007). Further studies are needed to be carried out to identify the exact interface of ATP7B–retromer interaction.
How copper (or copper removal) mediates triggering of the retrograde pathway for ATP7B is not understood. It is likely that copper binding to the six metal-binding domains (MBDs) on ATP7B N-terminus, exposes the upstream 1–63 N-terminal domain containing the 41NXXY44 domain. However, role of C-terminus cannot be completely discounted as Braiterman et al. has shown that multiple regulatory phosphorylation sites lie on the C-terminus that might play an indirect role in the regulation of ATP7B trafficking by retromer complex (Braiterman et al., 2015). Additionally, ATP7B (and also ATP7A) harbors di-leucine motifs on the C-terminal that have been shown to be crucial in trafficking regulation and its retrieval to the TGN (Petris et al., 1998; Francis et al., 1999; Jain et al., 2015).
Wilson disease, although a Mendelian disorder caused by mutations only in ATP7B gene, shows a large spectrum of symptoms and age of onset. We hypothesize that polymorphisms and mutations in trafficking regulatory proteins might be responsible for imparting such high phenotypic heterogeneity. Mutations and SNPs in the retromer subunit genes are associated with many hereditary conditions (Small, 2008; Shannon et al., 2014; Reitz, 2017; Chen et al., 2017; Rahman and Morrison, 2019). Varadarajan et al. reported significant association of SNPs of retromer complex genes (SNX1, SNX3 and Rab7A) with Alzheimer’s disease (Vardarajan et al., 2012). Similarly, a VPS35 hemizygous condition accentuates Alzheimer’s disease neuropathology (Wen et al., 2011). Additionally, Parkinson’s disease-linked D620N-VPS35 knockin mice manifest tau neuropathology and dopaminergic neurodegeneration (Chen et al., 2019). It would be important to extend the knowledge of role of retromers in ATP7B trafficking to delineate the genotype–phenotype co-relationship in Wilson disease patients.
Materials and Methods
Plasmids and antibodies
The GFP–ATP7B construct was available in our laboratory. The mCherry WT-VPS35 and mCherry–VPS35 (R107A) constructs were a kind gift from Dr Sunando Datta, IISER Bhopal, India. The mCherry–Rab7a-7 was Addgene plasmid #55127. The mCherry–Rab7a-7 T22N mutant was prepared following Q5® Site-Directed Mutagenesis Kit (#E0554) protocol. GFP–wtCOMMD1, T174M and K167/173E mutant constructs was kindly gifted by Dr Jason Burkhead, University of Alaska, Anchorage, USA. The following antibodies were used for experiments: rabbit anti-ATP7B [1:600 for immunofluorescence (IF)/1:2500 for western blotting (WB); #ab124973, Abcam), mouse anti-Golgin97 (1:400, IF; #A21270, Invitrogen), goat anti-VPS35 (1:400, IF; #NB 100-1397, Novus Biologicals), mouse anti-VPS26 (1:1500, WB; #NBP 236754, Novus Biologicals), mouse anti-VPS35 (1:1500, WB; #sc-374372, Santa Cruz Biotechnology), mouse anti-Lamp1 (2 μg/ml, IF and 0.5 μg/ml, WB; DSHB: #H4A3), mouse anti-Rab7 (1:50, IF; #sc-376362, Santa Cruz Biotechnology), donkey anti-rabbit IgG (H+L) Alexa Fluor 488 (1:800, IF; #A-21206, Invitrogen), goat anti-rabbit IgG (H+L) Alexa Fluor Plus 647 (1:800, IF; #A32733, Invitrogen), donkey anti-mouse IgG (H+L) Alexa Fluor Plus 647 (1:800, IF; #A32787, Invitrogen), donkey anti-Goat IgG (H+L) Alexa Fluor 568 (1:800, IF; #A-11057, Invitrogen), donkey anti-mouse IgG (H+L) Alexa Fluor 568 (1:800, IF; #A10037, Invitrogen), mouse anti-cation-independent mannose-6-phosphate receptor (10 μg/ml, IF; DSHB: #86f7), mouse anti-Lamp2 (20 μg/ml, IF; DSHB: #H4B4), mouse anti-Na/K ATPase (1:1000, WB; ATP1A1 #MA3-929, Invitrogen), goat anti-Rab11 (1:50, IF; #sc-6565, Santa Cruz Biotechnology), rabbit anti-GAPDH (1:3000, WB; #BB-AB0060, BioBharati) and rabbit anti-γ-actin (1:2500, WB) and sheep anti-TGN46 (1:300, IF; a kind gift from Prof. Carolyn Machamer, Johns Hopkins University, USA). Endotoxin free plasmid isolation was performed using an EndoFree Plasmid Maxi Kit (Qiagen, #12362).
Cell lines and cell culture
HepG2 cells (NCCS Pune) were grown and maintained in complete medium containing low-glucose minimum essential medium (MEM) (#41500-034, Thermo Fisher Scientific) supplemented with 10% fetal bovine serum (#10270-106, Thermo Fisher Scientific), 1× penicillin-streptomycin (#A001, HIMEDIA) and 1× amphotericin B (#15290026, Thermo Fisher Scientific). Similarly, HEK293T cells (a kind gift from Prof. Arindam Mukherjee’s laboratory, Indian Institute of Science Education and Research Kolkata, India) were grown and maintained in Dulbecco’s modified Eagle’s medium (DMEM) (#CC3004.05L, Cellclone) supplemented with 10% fetal bovine serum, 1× penicillin-streptomycin and 1× amphotericin B. For transfection of plasmids in HepG2 cells, Lipofectamine 3000 reagent (#L3000-001, Invitrogen) was used according to the manufacturer’s protocol. For transfection in HEK293T, for live-cell imaging, JetPrime (#114-07, PolyplusTransfection) transfection reagent was used. Both cell lines were confirmed free of any contamination.
Copper and BCS treatments
For studying the trafficking or other biochemical assays of ATP7B in HepG2, copper chloride (CuCl2) either in 50 μM or 250 μM concentration was used. Similarly, bathocuproine disulfonate (BCS), a copper chelator, was used either in 50 μM or 250 μM concentration. Both the reagents were prepared fresh in autoclaved miliQ-water prior to treatment.
Endocytic uptake of fluorescent dextran
Endocytic compartments in cultured HepG2 cells were loaded with 0.5 mg/ml lysine-fixable dextran–Alexa Fluor 647, from Thermo Fisher Scientific in culture medium for 4 h at 37°C followed by incubation in conjugate-free medium for 20 h as previously described (Bright et al., 1997).
Incubation with cathepsin B substrate
To label endocytic organelles in which cathepsin B was catalytically active, HepG2 cells were incubated with the Magic Red substrate from ImmunoChemistry Technologies, as per the manufacturer’s instruction.
Knockdown assays
Accell human VPS35 (55737) siRNA-SMARTpool (#E-010894-00-0010), Accell non-targeting siRNA (#D-001910-01-05), Accell siRNA delivery medium (#B-005000-100), 5× siRNA Buffer (#B-002000-UB-100) and molecular grade RNase-free water (#B-003000-WB-100) were purchased from Dharmacon. HepG2 cells were seeded in complete medium at a density of 1.5×105 cells/ml in coverslips heat-fixed on a 24-well plate. Cells were allowed to double for ∼48 h (doubling time of HepG2). After 48 h, medium was discarded, cells were rinsed with 1× PBS pH 7.4 and siRNAs were added at a final concentration of 1 μM resuspended in Accell siRNA delivery medium (#B-00-5000-100). This condition was maintained for 72 h after which the siRNA-containing medium was replaced with complete medium and again maintained for another 24 h. This ensures better knockdown at protein level. To validate knockdown of VPS35, western blotting was performed following same protocol from one well of the 24-well plate.
The lentiviral shRNA RNAi Consortium (TRC) protocol was used to produce TRC VPS35 lentiviral constructs [TRC VPS35 shRNA components (Dharmacon): RHS3979201863202, RHS3979201863553, RHS3979-201866801, RHS3979-201869452, RHS3979-201877513] (#RHS4533-EG55737) and TRC Lentiviral pLKO.1 stuffer control (#RHS4080). Knockdown of VPS35 was validated by western blotting.
Immunofluorescence
HepG2 cells were seeded at a density of (0.8×105–1.6×105 cells/ml) on coverslips heat-fixed on wells of a 24-well plate each time while conducting immunofluorescence analysis. Any treatment was performed at a confluency of 60–70%, including transfection. 2% paraformaldehyde (PFA) fixation was done following treatment. After fixation cells were permeablized with chilled methanol and finally washed with 1× PBS. Fixation and permeablisation was carried out in cold condition. Cells were blocked in 3% BSA suspended in 1× PBS for either 2 h at room temperature (RT) or overnight at 4°C. Following this, primary antibody (1°) incubation was done at RT in a moist chamber for 2 h. After 2 h incubation, cells were washed with 1× PBST (PBS with 0.01% Tween 20) three times and again re-incubated with corresponding secondary antibodies (2°) for 1.3 h at RT. This was followed by further washing with 1× PBST three times and finally with 1× PBS twice. Coverslips were fixed on glass slides using SIGMA Fluoroshield™ with DAPI mountant (#F6057). The solvent for antibody suspension was 1% BSA in 1× PBST.
For STED sample preparation, HepG2 cells were seeded on glass coverslips and treated with copper (as mentioned in Fig. 6E). Treatment was done at 70% confluency. Cell were fixed with 2% PFA for 20 min followed by washing with 1× PBS, pH 7.2 for 15 min (twice) and then quenched with 50 mM NH4Cl. Blocking and permeation was done for 30 min with 1% BSA along with 0.075% saponin. Cell were co-incubated with primary rabbit anti-ATP7B and goat anti-VPS35 for 2 h at room temp. This was followed by 1× PBS washing and incubation with secondary anti-rabbit-IgG conjugated to Alexa Fluor 488 and anti-goat-IgG conjugated to Alexa Fluor 647. Coverslips was mounted with ProLong™ Diamond Antifade Mountant with DAPI (#P36962).
Time-lapse fluorescence microscopy
HEK293T cells were seeded on confocal dishes (SPL) and were co-transfected separately with GFP–ATP7B and mCherry–VPS35-WT, mCherry–VPS35-Mutant (R107A) (a gift from Sunando Datta, IISER Bhopal) using jetPRIME (Polyplus) transfecting regent as per the manufacturer’s protocol. Images were acquired using Leica SP8 confocal setup with 63× oil objective. For ATP7B and VPS35-WT or mutant, images were taken at every 1.964 s interval using Lightning by Leica. For triple-color time-lapse fluorescence microscopy of ATP7B (green), DexA (blue) and Magic Red (red), GFP–ATP7B was transfected in HepG2 cell in confocal dishes pre-incubated with DexA and Magic Red. Each image was captured at an interval of 5 s for a total duration of 15 min. All time-lapse images were recorded in DMEM without Phenol Red supplemented with 5% fetal bovine serum and 5 mM HEPES. All the images were processed using Fiji and LASX software provided by Leica and videos were processed using Cyberlink Powerdirector.
Microscopy
All images were acquired with Leica SP8 confocal platform using an oil immersion 63× objective and deconvoluted using Leica Lightning software. For SIM, images acquisition was taken at 90× (60× objective + 1.5×digital) magnification in Zeiss Elyra PSI. For STED microscopy, imaging was done on a Leica STED 3X. For Alexa Fluor 647, the 775 nm STED laser line was used and for Alexa Fluor 488, the 592 laser line was used for depletion. The line average was set at 4 and pixel size was kept as 25 nm to achieve maximum resolution. STED corrected images were deconvoluted and processed by Scientific Volume Imaging of Huygens Professional Software with default settings.
Immunoblotting
HepG2 cells were grown on 60 mm dish and cell pellet was collected at 70% confluency. For lysate preparation of membrane protein dry pellet was dissolved in 200 μl of lysis buffer [250 mM sucrose, 1 mM EDTA, 1 mM EGTA, 1× PBS as solvent and 1× protease inhibitor cocktail (GCC Biotech, Kolkata)] and incubated on ice for 1 h with intermittent tapping. Dounce homogenization of dissolved pellet was done ∼400 times followed by syringing up and down with a 22–24 gauge needle for 20–25 times on ice. This enables the cell to rupture completely. The soup was centrifuged at 600 g at 4°C for 10 min to discard debris and nucleus. The mitochondrial fraction was discarded by centrifugation at 3000 g for 10 min at 4°C. The resultant soup was subjected to ultracentrifugation at 100,000 g with a TLA55, serial no. 07U920, fixed angle rotor for 1 h at 4°C to collect the membrane fraction. The pellet was dissolved in membrane-solubilizing buffer (250 mM sucrose, 1 mM EDTA, 1 mM EGTA, 1.0%NP-40, 1.0% Triton X-100, 1× PBS as solvent and 1× protease inhibitor cocktail). For soluble protein, whole-cell lysate was prepared with RIPA lysis buffer (10 mM Tris-HCl pH 8.0, 1 mM EDTA, 0.5 mM EGTA, 1.0% Triton X-100, 0.1% sodium deoxycholate, 0.1% sodium dodecyl sulphate, 140 mM NaCl and 1× protease inhibitor cocktail). The dry pellet was dissolved in RIPA lysis buffer and incubated on ice for 30 min with intermittent tapping. The solution was then sonicated with a probe sonicator (3–4 pulses, 5 s and 100 mA). Following this, centrifugation at 20,000 g for 20 min at 4°C was done to pellet cellular insoluble debris, and soup was collected. Protein estimation was carried out with Bradford reagent (#B6916-500ML, Sigma-Aldrich) following the manufacturer’s protocol. Protein sample preparation was done by adding 4× loading buffer (Tris-HCl pH 6.81, 4% SDS, 10% β-mercaptoethanol, 20% glycerol, 0.02% Bromophenol Blue and urea 8 M) to a final concentration of 1× and run on SDS-PAGE (6% for membrane fraction and 10–12% for soluble fraction) gels to separate proteins according to molecular mass. This was further followed by wet transfer of proteins onto nitrocellulose membrane (1620112, BioRad). After transfer, the membrane was blocked with 3% BSA in 1X Tris-buffered saline (TBS) buffer pH 7.5 for 2 h at RT with mild shaking. Primary antibody incubation was done overnight at 40°C following blocking and then washed with 1× TBST (0.01% Tween-20) for 10 min (three times). HRP-conjugated respective secondary incubation was done for 1.3 h at RT, further washed and signal was developed by ECL developer (170-5060, BioRad/1705062, BioRad) through chemiluminescence by Chemi Doc (BioRad).
Co-immunoprecipitation
All solutions were pre-chilled to 4°C and all steps were carried out on ice. HEK293T cells were transfected with GFP–ATP7B and treated with different Cu conditions followed by washing with 1× PBS and lysis using lysis buffer (10 mM Tris-HCl pH 7.5, 150 mM NaCl, 0.5 mM EDTA, 0.5% NP40, PMSF and protease inhibitor cocktail in ddH2O). Cell extracts were triturated with a 2 ml syringe and incubated for a total of 45 min and the insoluble materials were sedimented at 16,000 g for 10 min at 4°C. Co-IP experiment were performed using GFP-trap beads (ChromoTek, #gta-20) following the manufacturer’s protocol. The supernatants were diluted using diluted buffer (10 mM Tris-HCl pH 7.5, 150 mM NaCl, 0.5 mM EDTA, PMSF and protease inhibitor cocktail in ddH2O to yield 0.25% NP40) and incubated with GFP-trap beads for 2 h at 4°C on a rotating wheel. Finally the interacting proteins were eluted using 0.2 M glycine and used for western blotting. For western blotting of VPS35, VPS26 and GFP, samples for western blotting were resolved by SDS-PAGE and separated proteins were transferred to nitrocellulose membrane. After protein transfer, the membrane was blocked in 5% non-fat milk powder (for VPS35 and VPS26) and 3% BSA (for GFP) and incubated with primary antibody diluted in 5% non-fat milk powder (for mouse anti-VPS35 and mouse anti-VPS26, 1:1500 dilution) or 1% BSA (for rabbit anti-GFP, 1:10,000 dilution) overnight at 4°C. Following incubation, the membrane was briefly washed three times with TBST and incubated with HRP-conjugated secondary antibodies (anti-mouse-IgG conjugated to HRP, 1:5000 dilution and anti-rabbit-IgG conjugated to HRP,1:15000 dilution) diluted in 5% non-fat milk powder or 1% BSA for 1.5 h at RT. The membrane was washed three times for 5 min in TBS-T flowed by two times washing with TBS at RT and incubated for 5 min at RT with Enhanced Chemiluminescence (ECL) substrate and ECL plus (1:1).
in vivo metabolic labeling and UV crosslinking
HEK293T cells were transfected with GFP–ATP7B and supplemented with modified DMEM-LM (#30030, Sigma-Aldrich) medium (without leucine and methionine) and photo-Leucine (#22610, Thermo Scientific) and photo-methionine (#22615, Thermo Scientific). Cell was treated with either 100 μM CuCl2 or 50 μM BCS for 4 h and subjected to UV crosslinking at 365 nm for 15 min. Cell was processed for co-immunoprecipitation (Co-IP) assay following GFP–Trap A (code, gta-20) Chrormotek protocol as described above with few modifications. All steps were carried out on ice. Briefly, cell lysis was performed with co-IP compatible 200 μl lysis buffer (10 mM Tris-HCl pH 7.5, 150 mM NaCl, 0.5 mM EDTA, 0.5% Nonidet™ P40, 1× protease inhibitor cocktail and 1 mM PMSF). Trituration was done with a 26 gauge needle in 1 ml syringe for ∼40 times for cell to completely lyse followed by centrifugation at 3000 g for 5 min at 40°C. The supernatant was collected and diluted with 300 μl of dilution buffer (10 mM Tris-HCl pH 7.5, 150 mM NaCl and 0.5 mM EDTA). GFP–Trap A bead slurry was equilibrated with dilution buffer and incubated with the diluted supernatant and subjected to tumbling end-over on a rotatory wheel at 10 rpm for 4 h (first 2 h at RT and the next two hours at 4°C).
Proximity ligation assay
The proximity ligation assay was performed according to the manufacturer’s protocol (Duolink™ PLA Technology, Sigma). Briefly, HepG2 cell was grown on glass coverslips to 70% confluency and treated with copper for 2 h, fixed with 4% PFA and permeabilized with 0.1% saponin. Thereafter, the cell was blocked with blocking solution (DUO82007) for 1 h at 37°C and incubated with primary antibodies for 2 h at 37°C followed by washing twice with Wash Buffer A (DUO82049-4L) for 5 min on gentle orbital shaker at RT. During washing, the secondary PLA probes (DUO92005, DUO92003, DUO92001) were diluted to 1:5 with antibody diluent (DUO82008) and cell was incubated with it for 1 h at 37°C. Cells were further washed in same way. During washing, the ligation mixture was prepared by diluting the ligase enzyme (DUO82029) 1:40 in 1× ligation buffer (DUO82009) and applied onto cell. This was then incubated for 30 min at 37°C followed by washing twice in Wash buffer A for 2 min at RT. During washing, the amplification mixture was prepared by diluting the polymerase enzyme (DUO82030) 1:80 in 1× amplification buffer (DUO82010) and applied on cell with incubation at 370C for 100 min. After incubation, cells were washed twice with Wash Buffer B for 10 min. A final wash with 0.01× Wash buffer B was given prior to mounting with DAPI containing mountant. Images was acquired with Leica SP8 confocal platform using a 576 nm (Cy3) filter.
Cell viability test using the Neutral Red assay
HepG2 cell cytotoxicity to copper in the background of VPS35 knockdown was determined in cultured cells by performing a Neutral Red assay. Briefly, 6×103 viable cells were seeded in each well of a 96-well microplate (Nunc) and incubated at 37°C (5% CO2 atmosphere). After 24 h of incubation, the medium was replaced with a fresh one containing lentiviral shRNA (scrambled and pooled VPS35). After 48 h of incubation, the medium was replaced with a fresh one containing the desired concentration of copper (0 μM and 50 μM) where each concentration was loaded in quadruplicate. After 24 h of incubation with the copper solution, the medium was removed and a fresh medium was added with Neutral Red (#N7005, Sigma-Aldrich) at a final concentration of 40 μg/ml and incubated for 2 h. This was followed by washing with destaining solution (50% ethanol, 1% acetic acid, 49% MiliQ water). The absorbance was recorded at 540 nm using a SYNERGY H1 microplate reader, Biotek, and percentage of viable cells was calculated against copper concentrations. Each condition had four replicates (P<0.05). Data was plotted in GraphPad Prism 8 to obtain the histogram normalized against corresponding scrambled controls. A non-parametric t-test was performed to obtain the P-value.
Determination of the cellular copper concentration in VPS35 knockdown cells
HepG2 cells were seeded in a 35 mm Petri dish at 37°C (5% CO2 atmosphere). After 24 h of incubation, the medium was replaced with a fresh one containing lentiviral shRNA (scrambled and pooled VPS35). After 48 h of incubation, the medium was replaced with a fresh one containing the desired concentration copper (0 μM and 50 μM) and incubated for 2 h. Cells was washed with 1× dPBS for several times and pelleted down (5000 rpm for 6 mins). Dry pellet was weighed, which was ≈0.25 mg. For each sample, 2 ml 65% suprapur HNO3 plus 4 ml deionised water added prior to digestion for 1 h [MWD (microwave digester) conditions: power, 800 W; temperature, 100°C; hold time, 1 h]. After digestion, samples were syringe filtered through 0.45 micron filter. Cu calibration was done by acid digestion of Cu foil (procured from Alfa Aeasar) in 10 ml suprapur HNO3 for 1 h. (MWD conditions: power, 400 W; temperature, 100°C; hold time, 1 h). From the obtained solution, different strengths of Cu in parts per billion (ppb) were prepared and used for calibration. Similar acid digestion steps were followed for samples in a microwave digester. This solution was used to determine the cellular copper concentrations using a Thermo Scientific inductively coupled plasma optical emission spectroscopy (ICP-OES) iCAP 6500 machine. For each sample, a total of five replicates were used to calculate the average intracellular copper concentration (P<0.05).
Cell count and vesicle count for data analysis
The number of cells considered for analysis is given in in bar plots in figures as ‘n’. For Fig. 6A, to assess colocalization between ATP7B, VPS35 and Lamp1, a total of 12 cells were taken to calculate ATP7B and VPS35 signals colocalizing together in Lamp1-positive compartments in high copper; 88 ATP7B (green) puncta were considered as actual signals. This served as reference to calculate the relative abundance of green (ATP7B), cyan (VPS35), red (Lamp1) signals that are clustered together. A total of 37 such triple-colored clusters were obtained.
Image analysis and statistics
Images were analyzed in batches using ImageJ (Schneider et al., 2012). For the colocalization study, the Colocalization_Finder plugin was used. Regions of interest (ROIs) were drawn manually on best z-stack for each cell. For three protein colocalization studies, the other two protein co-residing vesicles were isolated using the Analyze Particle tool, and a colocalization study were carried out with the reference protein, ATP7B in our case. To calculate the lysosomal distribution of ATP7B, fractions of ATP7B localized in MR- and Dextran-positive endosomes were obtained using Manders’ colocalization coefficient (MCC) (Manders et al., 1993). A merged image of MR channel and dextran channel was obtained in greyscale, which was used to obtain the total lysosomal ATP7B. Using the set theory formulae: |A⋂B|=|A|+|B|−|A∪B|, the rest were deducted. The RGB_Profiler plugin was used to obtain the line profile graph. For statistical analysis and plotting, the ggplot2 (Hadley Wickham Ggplot2 SPIN, 2009; https://cran.r-project.org/web/packages/ggplot2/index.html) package was used in R v-4.0.3 (R: A Language and Environment for Statistical Computing, 2015; https://cran.r-project.org/bin/windows/base/). Non-parametric tests for unpaired datasets (Kruskal–Wallis test and Mann–Whitney U-test) were performed for all the samples.
Supplementary Material
Supplementary information available online at https://jcs.biologists.org/lookup/doi/10.1242/jcs.246819.supplemental
Acknowledgements
We thank Prof. Arindam Mukherjee (IISER Kolkata), Dr Oishee Chakrabarti and Prof. Susanta Lahiri (Saha Institute of Nuclear Physics), Dr Ashima Bhattacharjee (Amity Univ. Kolkata), Dr Sunando Datta, (IISER Bhopal), Dr Jason Burkhead (University of Alaska) and Dr Partho Sarothi Ray (IISER Kolkata) for sharing their reagents, constructs and instruments with us. We thank Dr Ashima Bhattacharjee for critical review of the manuscript.
Funding
This work was supported by a The Wellcome Trust DBT India Alliance Fellowship (IA/I/16/1/502369) and Early Career Research Award (ECR/2015/000220) from Department of Science and Technology, Ministry of Science and Technology, India (SERB) and IISER K intramural funding to A.G. S.M. and I.B. was supported by Pre-doctoral fellowship from Council of Scientific and Industrial Research, India. T.S. was supported by National Postdoctoral Fellowship, SERB, India. The pre-doctoral fellowship for S.D. is supported by Welcome Trust DBT India Alliance. The predoctoral fellowship for Ruturaj is supported by Intramural Institute funding (IISER-K). The post-doctoral fellowship for N.N. is funded by SINP Intramural funding.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: A.G., S.D.; Methodology: A.G., S.D., R.R., I.B., T.S., N.N.; Software: S.M.; Validation: A.G., R.R.; Formal analysis: A.G., S.D., S.M.; Investigation: A.G., S.D., T.S.; Resources: A.G., S.D.; Data curation: S.D., R.R., N.N.; Writing - original draft: A.G., S.D.; Writing - review & editing: A.G., S.D.; Visualization: R.R.; Supervision: A.G.; Project administration: A.G.; Funding acquisition: A.G.
Peer review history
The peer review history is available online at https://jcs.biologists.org/lookup/doi/10.1242/jcs.246819.reviewer-comments.pdf
References
- Abdelghaffar TY, Elsayed SM, Elsobky E, Bochow B, Büttner J, Schmidt H. Mutational analysis of ATP7B gene in Egyptian children with Wilson disease: 12 Novel mutations. J Hum Genet. 2008;53:681–687. doi: 10.1007/s10038-008-0298-7. [DOI] [PubMed] [Google Scholar]
- Abü-Remaileh M, Wyant GA, Kim C, Laqtom NN, Abbasi M, Chan SH, Freinkman E, Sabatini DM. Lysosomal metabolomics reveals V-ATPase-and mTOR-dependent regulation of amino acid efflux from lysosomes. Science (80-) 2017;358:807–813. doi: 10.1126/science.aan6298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aggarwal A, Chandhok G, Todorov T, Parekh S, Tilve S, Zibert A, Bhatt M, Schmidt HH-J. Wilson disease mutation pattern with genotypephenotype correlations from Western India: confirmation of p.C271* as a common indian mutation and identification of 14 novel mutations. Ann Hum Genet. 2013;77:299–307. doi: 10.1111/ahg.12024. [DOI] [PubMed] [Google Scholar]
- Ala A, Aliü E, Schilsky ML. Prospective pilot study of a single daily dosage of trientine for the treatment of wilson disease. Dig Dis Sci. 2015;60:1433–1439. doi: 10.1007/s10620-014-3495-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arighi CN, Harmell LM, Agüilar RC, Haft CR, Bonifacino JS. Role of the mammalian retromer in sorting of the cation-independent mannose 6-phosphate receptor. J Cell Biol. 2004;165:123–133. doi: 10.1083/jcb.200312055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry AN, Otoikhian A, Bhatt S, Shinde U, Tsivkovskii R, Blackbürn NJ, Lütsenko S. The lumenal loop Met 672-Pro 707 of copper-transporting ATPase ATP7A binds metals and facilitates copper release from the intramembrane sites. J Biol Chem. 2011;286:26585–26594. doi: 10.1074/jbc.M111.229039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belenkaya TY, Wü Y, Tang X, Zhoü B, Cheng L, Sharma YV, Yan D, Selva EM, Lin X. The retromer complex influences wnt secretion by recycling wntless from endosomes to the trans-golgi network. Dev Cell. 2008;14:120–131. doi: 10.1016/j.devcel.2007.12.003. [DOI] [PubMed] [Google Scholar]
- Blaby-Haas CE, Merchant SS. Lysosome-related organelles as mediators of metal homeostasis. J Biol Chem. 2014;289:28129–28136. doi: 10.1074/jbc.R114.592618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braiterman L, Nyasae L, Guo Y, Bustos R, Lutsenko S, Hubbard A. Apical targeting and Golgi retention signals reside within a 9-amino acid sequence in the copper-ATPase, ATP7B. Am J Physiol Gastrointest Liver Physiol. 2009;296:G433–G444. doi: 10.1152/ajpgi.90489.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braiterman LT, Murthy A, Jayakanthan S, Nyasae L, Tzeng E, Gromadzka G, Woolf TB, Lutsenko S, Hubbard AL. Distinct phenotype of a Wilson disease mutation reveals a novel trafficking determinant in the copper transporter ATP7B. Proc Natl Acad Sci USA. 2014;111:E1364–E1373. doi: 10.1073/pnas1314161111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braiterman LT, Gupta A, Chaerkady R, Cole RN, Hubbard AL. Communication between the N and C termini is required for copper-stimulated Ser/Thr phosphorylation of Cu(I)-ATPase. J Biol Chem. 2015;290:8803–8819. doi: 10.1074/jbcM114.627414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bright NA, Reaves BJ, Mullock BM, Luzio JP. Dense core lysosomes can fuse with late endosomes and are re-formed from the resultant hybrid organelles. J Cell Sci. 1997;110:2027–2040. doi: 10.1242/jcs.110.17.2027. [DOI] [PubMed] [Google Scholar]
- Bright NA, Davis LJ, Luzio JP. Endolysosomes are the principal intracellular sites of acid hydrolase activity. Curr Biol. 2016;26:2233–2245. doi: 10.1016/j.cub.2016.06.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burd C, Cullen PJ. Retromer: a master conductor of endosome sorting. Cold Spring Harb Perspect Biol. 2014;6:a016774. doi: 10.1101/cshperspect.a016774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkhead JL, Morgan CT, Shinde U, Haddock G, Lutsenko S. COMMD1 forms oligomeric complexes targeted to the endocytic membranes via specific interactions with phosphatidylinositol 4,5-Bisphosphate. J Biol Chem. 2009;284:696–707. doi: 10.1074/jbcM804766200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caca K, Ferenci P, Jurgen Kuhn H, Polli C, Willgerodt H, Kunath B, Hermann W, Mossner J, Berr F. High prevalence of the h1069q mutation in east german patients with wilson disease: rapid detection of mutations by limited sequencing and phenotype-genotype analysis. J Hepatol. 2001;35:575–581. doi: 10.1016/S0168-8278(01)00219-7. [DOI] [PubMed] [Google Scholar]
- Canuel M, Lefrancois S, Zeng J, Morales CR. AP-1 and retromer play opposite roles in the trafficking of sortilin between the Golgi apparatus and the lysosomes. Biochem Biophys Res Commun. 2008;366:724–730. doi: 10.1016/j.bbrc.2007.12.015. [DOI] [PubMed] [Google Scholar]
- Chen YF, Chang YY, Lan MY, Chen PL, Lin CH. Identification of VPS35 p.D620N mutation-related Parkinson ‘s disease in a Taiwanese family with successful bilateral subthalamic nucleus deep brain stimulation: a case report and literature review. BMC Neurol. 2017;17:191. doi: 10.1186/s12883-017-0972-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Kordich JK, Williams ET, Levine N, Cole-Strauss A, Marshall L, Labrie V, Ma J, Lipton JW, Moore DJ. Parkinson’s disease-linked D620N VPS35 knockin mice manifest tau neuropathology and dopaminergic neurodegeneration. Proc Natl Acad Sci USA. 2019;116:5765–5774. doi: 10.1073/pnas1814909116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng X-T, Xie Y-X, Zhou B, Huang N, Farfel-Becker T, Sheng Z-H. Characterization of LAMP1-labeled nondegradative lysosomal and endocytic compartments in neurons. J Cell Biol. 2018;217:3127–3139. doi: 10.1083/jcb201711083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox DW, Prat L, Walshe JM, Heathcote J, Gaffney D. Twenty-four novel mutations in Wilson disease patients of predominantly European ancestry. Hum Mutat. 2005;26:280. doi: 10.1002/humu.9358. [DOI] [PubMed] [Google Scholar]
- Cui Y, Carosi JM, Yang Z, Ariotti N, Kerr MC, Parton RG, Sargeant TJ, Teasdale RD. Retromer has a selective function in cargo sorting via endosome transport carriers. J Cell Biol. 2019;218:615–631. doi: 10.1083/jcb201806153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curnock R, Cullen PJ. Mammalian copper homeostasis requires retromer-dependent recycling of the high-affinity copper transporter 1. J Cell Sci. 2020;133:jcs249201. doi: 10.1242/jcs249201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deatherage CL, Nikolaus J, Karatekin E, Burd CG. Retromer forms low order oligomers on supported lipid bilayers. J Biol Chem. 2020;295:12305–12316. doi: 10.1074/jbcRA120.013672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahrni CJ. Synthetic fluorescent probes for monovalent copper. Curr Opin Chem Biol. 2013;17:656–662. doi: 10.1016/j.cbpa.2013.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinstein TN, Wehbi VL, Ardura JA, Wheeler DS, Ferrandon S, Gardella TJ, Vilardaga J-P. Retromer terminates the generation of cAMP by internalized PTH receptors. Nat Chem Biol. 2011;7:278–284. doi: 10.1038/nchembio.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Follett J, Bugarcic A, Yang Z, Ariotti N, Norwood SJ, Collins BM, Parton RG, Teasdale RD. Parkinson disease-linked Vps35 R524W mutation impairs the endosomal association of retromer and induces α-synuclein aggregation. J Biol Chem. 2016;291:18283–18298. doi: 10.1074/jbcM115.703157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forbes JR, Cox DW. Functional characterization of missense mutations in ATP7B: Wilson disease mutation or normal variant? Am J Hum Genet. 1998;63:1663–1674. doi: 10.1086/302163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis MJ, Jones EE, Levy ER, Martin RL, Ponnambalam S, Monaco AP. Identification of a di-leucine motif within the C terminus domain of the Menkes disease protein that mediates endocytosis from the plasma membrane. J Cell Sci. 1999;112:1721–1732. doi: 10.1242/jcs.112.11.1721. [DOI] [PubMed] [Google Scholar]
- Fuse A, Furuya N, Kakuta S, Inose A, Sato M, Koike M, Saiki S, Hattori N. VPS29-VPS35 intermediate of retromer is stable and may be involved in the retromer complex assembly process. FEBS Lett. 2015;589:1430–1436. doi: 10.1016/j.febslet.2015.04.040. [DOI] [PubMed] [Google Scholar]
- Gershlick DC, Lucas M. Endosomal trafficking: retromer and retriever are relatives in recycling. Curr Biol. 2017;27:R1233–R1236. doi: 10.1016/j.cub.2017.10.004. [DOI] [PubMed] [Google Scholar]
- Gokool S, Tattersall D, Reddy JV, Seaman MNJ. Identification of a conserved motif required for Vps35p/Vps26p interaction and assembly of the retromer complex. Biochem J. 2007;408:287–295. doi: 10.1042/BJ20070555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guha A, Ahuja D, Das Mandal S, Parasar B, Deyasi K, Roy D, Sharma V, Willard B, Ghosh A, Ray PS. Integrated regulation of HuR by translation repression and protein degradation determines pulsatile expression of p53 Under DNA damage. iScience. 2019;15:342–359. doi: 10.1016/j.isci.2019.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta A, Lutsenko S. Human copper transporters: mechanism, role in human diseases and therapeutic potential. Future Med Chem. 2009;1:1125–1142. doi: 10.4155/fmc09.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta A, Aikath D, Neogi R, Datta S, Basu K, Maity B, Trivedi R, Ray J, Das SK, Gangopadhyay PK, et al. Molecular pathogenesis of Wilson disease: haplotype analysis, detection of prevalent mutations and genotype-phenotype correlation in Indian patients. Hum Genet. 2005;118:49–57. doi: 10.1007/s00439-005-0007-y. [DOI] [PubMed] [Google Scholar]
- Gupta A, Chattopadhyay I, Mukherjee S, Sengupta M, Das SK, Ray K. A novel COMMD1 mutation Thr174Met associated with elevated urinary copper and signs of enhanced apoptotic cell death in a Wilson Disease patient. Behav Brain Funct. 2010;6:33. doi: 10.1186/1744-9081-6-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta A, Bhattacharjee A, Dmitriev OY, Nokhrin S, Braiterman L, Hubbard AL, Lutsenko S. Cellular copper levels determine the phenotype of the Arg875 variant of ATP7B/Wilson disease protein. Proc Natl Acad Sci USA. 2011;108:5390–5395. doi: 10.1073/pnas1014959108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta A, Schell MJ, Bhattacharjee A, Lutsenko S, Hubbard AL. Myosin Vb mediates Cu+ export in polarized hepatocytes. J Cell Sci. 2016;129:1179–1189. doi: 10.1242/jcs175307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta A, Das S, Ray K. A glimpse into the regulation of the Wilson disease protein, ATP7B, sheds light on the complexity of mammalian apical trafficking pathways. Metallomics. 2018;10:378–387. doi: 10.1039/C7MT00314E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada M, Sakisaka S, Kawaguchi T, Kimura R, Taniguchi E, Koga H, Hanada S, Baba S, Furuta K, Kumashiro R, et al. Copper does not alter the intracellular distribution of ATP7B, a copper-transporting ATPase. Biochem Biophys Res Commun. 2000;275:871–876. doi: 10.1006/bbrc2000.3403. [DOI] [PubMed] [Google Scholar]
- Harrison MS, Hung CS, Liu TT, Christiano R, Walther TC, Burd CG. A mechanism for retromer endosomal coat complex assembly with cargo. Proc Natl Acad Sci USA. 2014;111:267–272. doi: 10.1073/pnas1316482111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hierro A, Rojas AL, Rojas R, Murthy N, Effantin G, Kajava AV, Steven AC, Bonifacino JS, Hurley JH. Functional architecture of the retromer cargo-recognition complex. Nature. 2007;449:1063–1067. doi: 10.1038/nature06216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphries WH, Szymanski CJ, Payne CK. Endo-lysosomal vesicles positive for rab7 and lamp1 are terminal vesicles for the transport of dextran. PLoS ONE. 2011;6:e26626. doi: 10.1371/journalpone.0026626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huster D, Lutsenko S. Wilson disease: not just a copper disorder Analysis of a Wilson disease model demonstrates the link between copper and lipid metabolism. Mol Biosyst. 2007;3:816–824. doi: 10.1039/b711118p. [DOI] [PubMed] [Google Scholar]
- Jain S, Farias GG, Bonifacino JS. Polarized sorting of the copper transporter ATP7B in neurons mediated by recognition of a dileucine signal by AP-I. Mol Biol Cell. 2015;26:218–228. doi: 10.1091/mbcE14-07-1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kambe T. An overview of a wide range of functions of ZnT and Zip Zinc transporters in the secretory pathway. Biosci Biotechnol Biochem. 2011;75:1036–1043. doi: 10.1271/bbb110056. [DOI] [PubMed] [Google Scholar]
- Korolchuk VI, Rubinsztein DC. Regulation of autophagy by lysosomal positioning. Autophagy. 2011;7:927–928. doi: 10.4161/auto.7.8.15862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurz T, Eaton JW, Brunk UT. The role of lysosomes in iron metabolism and recycling. Int J Biochem Cell Biol. 2011;43:1686–1697. doi: 10.1016/j.biocel.2011.08.016. [DOI] [PubMed] [Google Scholar]
- Leshane ES, Shinde U, Walker JM, Barry AN, Blackburn NJ, Ralle M, Lutsenko S. Interactions between copper-binding sites determine the redox status and conformation of the regulatory N-terminal domain of ATP7B. J Biol Chem. 2010;285:6327–6336. doi: 10.1074/jbcM109.074633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas M, Gershlick DC, Vidaurrazaga A, Rojas AL, Bonifacino JS, Hierro A. Structural mechanism for cargo recognition by the retromer complex. Cell. 2016;167:1623–1635.:e14. doi: 10.1016/j.cell.2016.10.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutsenko S. Copper trafficking to the secretory pathway. Metallomics. 2016;8:840–852. doi: 10.1039/C6MT00176A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma M, Burd CG. Retrograde trafficking and plasma membrane recycling pathways of the budding yeast Saccharomyces cerevisiae. Traffic. 2020;21:45–59. doi: 10.1111/tra.12693. [DOI] [PubMed] [Google Scholar]
- Manders EMM, Verbeek FJ, Aten JA. Measurement of co–localization of objects in dual–colour confocal images. J Microsc. 1993;169:375–382. doi: 10.1111/j.1365-2818.1993.tb03313.x. [DOI] [PubMed] [Google Scholar]
- Materia S, Cater MA, Klomp LWJ, Mercer JFB, La Fontaine S. Clusterin and COMMD1 independently regulate degradation of the mammalian copper ATPases ATP7A and ATP7B. J Biol Chem. 2012;287:2485–2499. doi: 10.1074/jbcM111.302216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellado M, Cüartero Y, Brügada R, Verges M. Subcellular localisation of retromer in post-endocytic pathways of polarised Madin-Darby canine kidney cells. Biol Cell. 2014;106:377–393. doi: 10.1111/boc201400011. [DOI] [PubMed] [Google Scholar]
- Miyayama T, Hiraoka D, Kawaji F, Nakamüra E, Süzüki N, Ogra Y. Roles of COMM-domain-containing 1 in stability and recruitment of the copper-transporting ATPase in a mouse hepatoma cell line. Biochem J. 2010;429:53–61. doi: 10.1042/BJ20100223. [DOI] [PubMed] [Google Scholar]
- Müchenditsi A, Yang H, Hamilton JP, Koganti L, Hoüsseaü F, Aronov L, Fan H, Pierson H, Bhattacharjee A, Mürphy R, et al. Targeted inactivation of copper transporter Atp7b in hepatocytes causes liver steatosis and obesity in mice. Am J Physiol - Gastrointest Liver Physiol. 2017;313:G39–G49. doi: 10.1152/ajpgi00312.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otoikhian A, Barry AN, Mayfield M, Nilges M, Hüang Y, Lütsenko S, Blackbürn NJ. Lumenal loop M672-P707 of the menkes protein (ATP7A) transfers copper to peptidylglycine monooxygenase. J Am Chem Soc. 2012;134:10458–10468. doi: 10.1021/ja301221s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petris MJ, Camakaris J, Greenoügh M, Lafontaine S, Mercer JFB. A C-terminal di-leucine is required for localization of the Menkes protein in the trans-Golgi network. Hum Mol Genet. 1998;7:2063–2071. doi: 10.1093/hmg/7.13.2063. [DOI] [PubMed] [Google Scholar]
- Phillips-Krawczak CA, Singla A, Starokadomskyy P, Deng Z, Osborne DG, Li H, Dick CJ, Gomez TS, Koenecke M, Zhang JS, et al. COMMD1 is linked to the WASH complex and regulates endosomal trafficking of the copper transporter ATP7A. Mol Biol Cell. 2015;26:91–103. doi: 10.1091/mbce14-06-1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polishchük EV, Concilli M, Iacobacci S, Chesi G, Pastore N, Piccolo P, Paladino S, Baldantoni D, Vanijzendoorn SCD, Chan J, et al. Wilson disease protein ATP7B utilizes lysosomal exocytosis to maintain copper homeostasis. Dev Cell. 2014;29:686–700. doi: 10.1016/j.devcel.2014.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabanal-Rüiz Y, Korolchük VI. mTORC1 and nutrient homeostasis: the central role of the lysosome. Int J Mol Sci. 2018;19:818. doi: 10.3390/ijms19030818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman AA, Morrison BE. Contributions of VPS35 mutations to parkinson’s disease. Neuroscience. 2019;401:1–10. doi: 10.1016/j.neuroscience.2019.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reitz C. Retromer dysfunction and neurodegenerative disease. Curr Genomics. 2017;19:279–288. doi: 10.2174/1389202919666171024122809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roelofsen H, Wolters H, Van Lüyn MJA, Miüra N, Küipers F, Vonk RJ. Copper-induced apical trafficking of ATP7B in polarized hepatoma cells provides a mechanism for biliary copper excretion. Gastroenterology. 2000;119:782–793. doi: 10.1053/gast2000.17834. [DOI] [PubMed] [Google Scholar]
- Rojas R, Van Vlijmen T, Mardones GA, Prabhü Y, Rojas AL, Mohammed S, Heck AJR, Raposo G, Van Der Slüijs P, Bonifacino JS. Regulation of retromer recruitment to endosomes by sequential action of Rab5 and Rab7. J Cell Biol. 2008;183:513–526. doi: 10.1083/jcb.200804048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seaman MNJ. Cargo-selective endosomal sorting for retrieval to the Golgi requires retromer. J Cell Biol. 2004;165:111–122. doi: 10.1083/jcb200312034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seaman MNJ. Identification of a novel conserved sorting motif required for retromer-mediated endosome-to-TGN retrieval. J Cell Sci. 2007;120:2378–2389. doi: 10.1242/jcs009654. [DOI] [PubMed] [Google Scholar]
- Seaman MNJ. Retromer and its role in regulating signaling at endosomes. Prog Mol Subcell Biol. 2018;57:137–149. doi: 10.1007/978-3-319-96704-2_5. [DOI] [PubMed] [Google Scholar]
- Seaman MN, Freeman CL. Analysis of the Retromer complex-WASH complex interaction illuminates new avenues to explore in Parkinson disease. Commun Integr Biol. 2014;7:e29483. doi: 10.4161/cib29483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon B, Soto-Ortolaza A, Rayaprolü S, Cannon HD, Labbe C, Benitez BA, Choi J, Lynch T, Boczarska-Jedynak M, Opala G, et al. Genetic variation of the retromer subunits VPS26A/B-VPS29 in Parkinson‘s disease. Neurobiol Aging. 2014;35:1958.e1–1958.e2. doi: 10.1016/j.neurobiolaging.2014.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small SA. Retromer sorting: a pathogenic pathway in late-onset Alzheimer disease. Arch Neurol. 2008;65:323–328. doi: 10.1001/archneurol2007.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spinosa MR, Progida C, De Lüca A, Colücci AMR, Alifano P, Bücci C. Functional characterization of Rab7 mutant proteins associated with Charcot-Marie-Tooth type 2B disease. J Neurosci. 2008;28:1640–1648. doi: 10.1523/JNEUROSCI.3677-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinberg F, Gallon M, Winfield M, Thomas EC, Bell AJ, Heesom KJ, Tavare JM, Cüllen PJ. A global analysis of SNX27-retromer assembly and cargo specificity reveals a function in glucose and metal ion transport. Nat Cell Biol. 2013;15:461–471. doi: 10.1038/ncb2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart DJ, Short KK, Maniaci BN, Bürkhead JL. COMMD1 and PtdIns(4,5)P2 interaction maintain ATP7B copper transporter trafficking fidelity in HepG2 cells. J Cell Sci. 2019;132:jcs231753. doi: 10.1242/jcs231753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strochlic TI, Setty TG, Sitaram A, Bürd CG. Grd19/Snx3p functions as a cargo-specific adapter for retromer-dependent endocytic recycling. J Cell Biol. 2007;177:115–125. doi: 10.1083/jcb200609161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Süzüki SW, Emr SD. Membrane protein recycling from the vacuole/lysosome membrane. J Cell Biol. 2018a;217:1623–1632. doi: 10.1083/jcb.201709162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Süzüki SW, Emr SD. Retrograde trafficking from the vacuole/lysosome membrane. Autophagy. 2018b;14:1654–1655. doi: 10.1080/15548627.2018.1496719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Süzüki SW, Chüang Y-s, Li M, Seaman MNJ, Emr SD. A bipartite sorting signal ensures specificity of retromer complex in membrane protein recycling. J Cell Biol. 2019;218:2876–2886. doi: 10.1083/jcb201901019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabüchi M, Yanatori I, Kawai Y, Kishi F. Retromer-mediated direct sorting is required for proper endosomal recycling of the mammalian iron transporter DMT1. J Cell Sci. 2010;123:756–766. doi: 10.1242/jcs060574. [DOI] [PubMed] [Google Scholar]
- Tammineni P, Jeong YY, Feng T, Aikal D, Cai Q. Impaired axonal retrograde trafficking of the retromer complex augments lysosomal deficits in Alzheimer’s disease neurons. Hum Mol Genet. 2017;26:4352–4366. doi: 10.1093/hmg/ddx321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telianidis J, Hüng YH, Materia S, La Fontaine S. Role of the P-Type ATPases, ATP7A and ATP7B in brain copper homeostasis. Front Aging Neurosci. 2013;5:44. doi: 10.3389/fnagi2013.00044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uaüy R, Olivares M, Gonzalez M. Essentiality of copper in humans. Am J Clin Nutr. 1998;67:9525S–9595S. doi: 10.1093/ajcn/67.5.952S. [DOI] [PubMed] [Google Scholar]
- Vardarajan BN, Brüesegem SY, Harboür ME, StGeorge-Hyslop P, Seaman MNJ, Farrer LA. Identification of Alzheimer disease-associated variants in genes that regulate retromer function. Neurobiol Aging. 2012;33:2231.e15–2231.e30. doi: 10.1016/j.neurobiolaging.2012.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen L, Tang FL, Hong Y, Lüo SW, Wang CL, He W, Shen C, Jüng JU, Xiong F, Lee D, et al. VPS35 haploinsufficiency increases Alzheimer’s disease neuropathology. J Cell Biol. 2011;195:765–779. doi: 10.1083/jcb201105109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Reiling C, Reinecke JB, Prislan I, Marky LA, Sorgen PL, Naslavsky N, Caplan S. Rabankyrin-5 Interacts with EHD1 and Vps26 to regulate endocytic trafficking and retromer function. Traffic. 2012;13:745–757. doi: 10.1111/j.1600-0854.2012.01334.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Nothwehr S, Lara-Lemüs R, Zhang BY, Peter H, Arvan P. Dominant-negative behavior of mammalian Vps35 in yeast requires a conserved PRLYL motif involved in retromer assembly. Traffic. 2007;8:1829–1840. doi: 10.1111/j.1600-0854.2007.00658.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.