Abstract
Adoptive transfer of antitumor cytotoxic T cells is an emerging form of cancer immunotherapy. A key challenge to expanding the utility of adoptive-cell therapies is how to enhance the survival and function of the transferred T cells. Immune-cell survival requires adaptation to different microenvironments, and particularly to the hypoxic milieu of solid tumors. The hypoxia-inducible factor (HIF) transcription factors are an essential aspect of this adaptation. In this study, we undertook experiments to define structural determinants of HIF that potentiate antitumor efficacy in cytotoxic T cells. We first created retroviral vectors to deliver ectopic expression of HIF1α and HIF2α in mouse CD8+ T cells, together or individually, and with or without sensitivity to the oxygen-dependent HIFα inhibitors Von Hippel-Lindau (VHL) and Factor Inhibiting HIF (FIH). HIF2α, but not HIF1α, drove broad transcriptional changes in CD8+ T cells, resulting in increased cytotoxic differentiation and cytolytic function against tumor targets. A specific mutation replacing the hydroxyl group–acceptor site for FIH in HIF2α gave rise to the most effective antitumor T cells after adoptive transfer in vivo. In addition, co-delivering an FIH-insensitive form of HIF2α with an anti-CD19 chimeric antigen receptor greatly enhanced cytolytic function of human CD8+ T cells against lymphoma cells both in vitro and in a xenograft adoptive transfer model. These experiments point to a means to increase the antitumor efficacy of therapeutic CD8+ T cells via ectopic expression of the HIF transcription factor.
Keywords: hypoxia, CD8+ T cells, adoptive cell transfer, immunotherapy, CAR
Introduction
CD8+ T cells are the cytotoxic arm of the adaptive immune system and play an essential role in antitumor immunity. One key immunotherapeutic strategy is the redirection of T cells against tumor antigens by retroviral gene transfer of defined T-cell receptors (TCR), or chimeric antigen receptors (CAR), which can be adoptively transferred into cancer patients, and mediate tumor regression in some forms of malignancies (1–3).
The process by which TCR/CAR T cells are manufactured can be used to perform additional genetic interventions to improve the safety and antitumor efficacy of the T-cell product. Such interventions include CRISPR-mediated disruption of the endogenous TCR loci (4), disruption of the gene encoding the inhibitor checkpoint protein PD-1 (5) and co-delivery of genes encoding various cytokines (6), chemokines or their receptors (7,8), as well as suicide genes (9). Transcription factors are able to exert large phenotypic changes and guide T-cell differentiation (10) making them an appealing target in TCR/CAR T-cell immunotherapy.
The hypoxia-inducible transcription factors (HIF) are primary regulators of the transcriptional response to hypoxia (11,12). HIFs function as heterodimers composed of an alpha (HIF1α or HIF2α) and a beta subunit (ARNT; also known as HIF-1β) and their function is efficiently inhibited by oxygen. In the presence of oxygen, HIFα subunits are hydroxylated at two conserved prolines by prolyl-hydroxylases (PHD), leading to recognition and ubiquitination by the Von Hippel-Lindau (VHL) protein and subsequent proteasomal degradation (13,14). The second oxygen-sensitive mechanism utilizes the Factor Inhibiting HIF (FIH) enzyme, which hydroxylates a conserved asparagine residue in HIFα subunits, blocking association with the p300/CBP coactivator and thus HIF transcriptional activity (15,16). These repressive mechanisms are reduced or eliminated in low-oxygen environments leading to HIF stabilization, translocation to the nucleus and induction of a hypoxia-response gene expression program.
HIF1α and HIF2α have an identical core-binding motif and overlap significantly in their target gene–binding distribution but HIF1α typically binds close to the transcription start site whereas HIF2α is often found further up- or downstream (17–19). Although HIFs consistently drive expression of genes involved in glycolysis, their overall DNA-binding pattern is highly context specific, varying with cell type, external stimuli and epigenetic landscape (17,18).
Low-oxygen availability in tumors and in inflamed tissues has historically been characterized as immunosuppressive (20–22) but a number of studies show that CD8+ T cells differentiate more efficiently into cytotoxic T cells when cultured in low oxygen (23,24), suggesting that T-cell differentiation is functionally linked to decreased oxygen availability.
This hypothesis was further supported by genetic deletion of Vhl (24) or the genes encoding the three PHD isoforms (25), which caused constitutive accumulation of HIF1α and HIF2α in T cells and increased cytotoxic differentiation as well as improved rejection of primary and metastatic tumors. Ablation of HIF activity via deletion of Arnt (26) or Hif1a (27) had the opposite effect, resulting in poorer cytotoxic differentiation, and reduced tumor rejection after adoptive transfer.
Based on these studies, we hypothesized that engineering CD8+ T cells to enhance HIF expression would result in T cells with enhanced antitumor properties. To test this hypothesis, we designed retroviral vectors to ectopically express HIF1α and HIF2α, alone or in combination, and with or without susceptibility to oxygen-dependent regulation by either VHL- or FIH-dependent mechanisms. We screened these HIF variants in CD8+ T cells to determine whether they would enhance in vivo antitumor efficacy. Our results demonstrate the potential for engineering HIF activity as a way to boost T-cell function in adoptive T-cell immunotherapy.
Materials and Methods
Animals
C57BL/6J (CD45.2) animals were purchased from Janvier Labs. Donor TCR-transgenic OT-I mice (catalogue #003831, The Jackson Laboratory) were crossed with mice bearing the CD45.1 congenic marker (catalogue #002014, The Jackson Laboratory) or TdTomato dLck Cre reporter mice (catalogue #007914 and #012837, The Jackson Laboratory). NSG (NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ) mice were purchased from The Jackson Laboratory (catalogue #005557).
Cell lines
HEK293 cells were a gift from Prof. Dantuma (Karolinska Institute, Stockholm) and cultured between June 2015 and September 2020. MC38 colon cancer cells were a kind gift from Dr Asis Palazon (University of Cambridge) and cultured between February 2015 and October 2020. B16-F10 melanoma cells were purchased from ATCC (CRL-6475) and cultured between August 2016 and November 2020. Lewis lung carcinoma cells (LLC) were purchased from ATCC (CRL-1642) and cultured between September 2016 and October 2020. Raji-GFP-Luc cells were purchased from Biocytogen (B-HCL-010), cultured between January 2019 and October 2020 and regularly monitored for GFP expression by flow cytometry. All adherent cell lines were cultured in DMEM high glucose, pyruvate (Thermo Fisher, #11995065) supplemented with 10% fetal bovine serum (Thermo Fisher, #10270106), 100 U/ml penicillin and 100 μg/ml streptomycin (Thermo Fisher, #15140122). All non-adherent cell lines were cultured in RPMI1640 (Thermo Fisher, #21875034) supplemented with 10% fetal bovine serum, 100 U/ml penicillin, 100 μg/ml streptomycin and 55 μM 2-mercaptoethanol (Thermo Fisher, #21985023). Cell lines were typically passaged 3-4 times between thawing and experimental use. All cell lines were mycoplasma-tested using MycoAlertTM Mycoplasma Detection Kit, Lonza (#LT07-118) in November 2015, except for cells obtained directly from ATCC, which were not tested. Cell lines were not authenticated.
Vectors
DNA encoding a codon-optimized polycistronic peptide composed of mouse Thy-1.1 (AAR17087.1), mouse HIF1α (NP_034561.2; P402A, P577A, N813A) and mouse HIF2α (NP_034267.3; P404A, P530A, N851A) interspersed with picornavirus P2A (GSGATNFSLLKQAGDVEENPGP) and furin (RAKR) cleavage sequences was synthesized by Gene Art (Thermo Fisher). Cell surface and nuclear localization peptides from Thy-1.1 and HIF sequences, respectively, were maintained to ensure proper subcellular localization. Quick Change II Site-directed mutagenesis (Agilent, #200523) was used to revert mutated sites to the native sequence. DNA encoding a codon-optimized polycistronic peptide composed of eGFP (ADQ43426.1), anti-human CD19 CAR and human HIF2α (NP_001421.2, N847A) interspersed with picornavirus P2A and furin cleavage sequences was synthesized by Gene Art (Thermo Fisher). The coding sequences were cloned into the gamma retroviral vector pMP71, a gift from Christopher Baum (MHH, Hannover). Helper vector pCL-Eco (for ecotropic infection) was a gift from Inder Verma (Addgene plasmid #12371) and pCL-Ampho (for amphotropic infection) was purchased from Novus (#NBP2-29541). DNA encoding a codon-optimized polycistronic peptide composed of chicken ovalbumin (OVA; P01012.2), eGFP (ADQ43426.1) and neomycin phosphotransferase (NeoR; CAD21956.1) interspersed with P2A and furin cleavage sites was synthesized by Gene Art (Thermo Fisher) and cloned under control of the SV40 promoter in the transposon vector pT2/BH, a gift from Perry Hackett (Addgene plasmid #26556). pCMV-SB11 encoding the sleeping beauty transposase was a gift from Perry Hackett (Addgene plasmid #26552). All protein sequences and accession numbers are available in Supplementary Material.
CD8+ T-cell sourcing, activation and restimulation
CD8+ T cells from female and male mice were purified from spleens by CD8α positive selection magnetic bead sorting (Miltenyi, #130-117-044) and activated in RPMI1640 supplemented with 10% fetal bovine serum, 100 U/ml penicillin, 100 μg/ml streptomycin and 55 μM 2-mercaptoethanol (Thermo Fisher, #21985023), with 2 μg/ml Concanavalin A (Sigma, #C5275) and 10 ng/ml recombinant human IL7 (R&D Systems, #207-IL-005) or anti-mouse CD3/CD28 dynabeads (Thermo Fisher, #11453D) at a 1:1 cell-to-bead ratio for 24 hours before transduction. Whole OT-I splenocytes or CD8α-purified OT-I cells were activated with 0.1-1 μg/ml of the OVA-derived peptide SIINFEKL (ProImmune, #P093-0A-E) 24 hours before transduction. Transduced CD8+ T cells were restimulated with anti-mouse CD3/CD28 dynabeads or SIINFEKL. Transduced CD8+ T cells were expanded in the presence of 10 U/ml recombinant human IL2 (Sigma, #11011456001). Human CD8+ T cells were purified from donor PBMCs (National Health Service Blood and Transport, United Kingdom, or Karolinska Hospital Hematology Service, Sweden) by either negative or positive CD8 magnetic bead sorting (Miltenyi, #130-104-075, #130-04-201) and activated in complete RPMI supplemented with 30 U/ml IL-2 with anti-human CD3/CD28 dynabeads at a 1:1 cell-to-bead ratio. In some experiments, cultures were split 1 day after transduction into two sub-cultures treated with 10 μM PT2977 (HIF2α inhibitor; MedChemExpress, #HY-125840) or an equivalent volume of DMSO.
Retroviral transductions
Sub-confluent HEK293 cultures were transfected using FugeneHD (Promega, #E2311/2) with 2.5 μg HIF-encoding vectors and 1.5 μg helper vector pCL-Eco or pCL-Ampho, for generation of mouse- or human-tropic retroviral particles, respectively. Supernatant media containing retroviral particles was harvested 48 hours after transfection and used fresh or stored at -80˚C. Retroviral supernatants were spun onto Retronectin-coated wells (Takara) at 2000 xg for 2 hours at 32˚C and replaced with activated mouse polyclonal, mouse OT-I, or human CD8+ T cells in fresh RPMI supplemented with 10 (mouse) or 30 (human) U/ml IL2. Fresh media was added every 2-3 days. For long-term expansion, transduced cells were restimulated weekly with anti-CD3/CD28 dynabeads.
Flow cytometry
Single-cell suspensions were stained with Near-IR Dead Cell Stain Kit (Thermo Fisher, #L10119) in PBS at room temperature followed by antigen surface staining with fluorochrome-labeled monoclonal antibodies in PBS supplemented with 2% BSA and 2 mM EDTA at 4˚C, followed by cytoplasmic and/or nuclear antigen stain when required (fluorochrome-labeled monoclonal antibodies listed in Supplementary Table 1). Staining of cytoplasmic and nuclear antigens was performed using the Fixation/Permeabilization kit (BD Biosciences, #554714) and the Transcription Factor buffer set (BD Biosciences, #562574), respectively, according to the manufacturers’ protocols. To measure IFN□ secretion, OVA-reactive T cells were incubated overnight in complete RPMI supplemented with 100 ng/ml SIINFEKL and treated with Golgi Stop (BD Biosciences, #554724) 4 hours before intracellular staining and flow cytometry analysis. For proliferation assays, cells were loaded with CellTrace Violet (Thermo Fisher, #C34557) according to manufacturer’s instructions. Samples were analyzed using a FACSCanto II flow cytometer (BD Biosciences).
RNA sequencing
OT-I cells were expanded for 5 days in the presence of IL2 after transduction and sorted on Thy-1.1 surface expression on a BD FACSAria Fusion cell sorter (BD Biosciences) directly into RLT Plus lysis buffer (Qiagen, #1053393). Total RNA was subjected to quality control with Agilent Tapestation according to the manufacturer’s instructions. To construct libraries suitable for Illumina sequencing the Illumina TruSeq Stranded mRNA Sample preparation protocol, which includes cDNA synthesis, ligation of adapters and amplification of indexed libraries, was used (Illumina, #20020594). The yield and quality of the amplified libraries were analysed using Qubit by Thermo Fisher and the Agilent Tapestation. The indexed cDNA libraries were normalised and combined and the pools were sequenced on the Nextseq 550 for a 50-cycle v2.5 sequencing run generating 75 bp single-end reads. Basecalling and demultiplexing was performed using CASAVA software with default settings generating Fastq files for further downstream mapping and analysis. RNA-seq data are deposited in the NCBI’s Gene Expression Omnibus database under GEO accession number GSE166758.
Western blotting
Nuclear protein extracts (15-20 μg) from HEK293 cells transfected with HIF-encoding vectors or from Thy-1.1–purified CD8+ T cells transduced with HIF-encoding vectors were prepared with the NE-PER nuclear extraction kit (Thermo Fisher, #78833) probed with polyclonal antibodies against HIF1α (NB-100-449 or NB-100-105 Novus Biologicals), HIF2α (AF2997, R&D Systems), Lamin B (sc-6217, Santa Cruz), and Histone 3 (4499S, CST) and detected using infra-red labeled secondary antibodies in an Odyssey imaging system (LI-COR).
Generation of OVA-expressing cell lines
B16-F10, LLC, and MC38 were co-transfected using FugeneHD (Promega, #E2311/2) with the transposon vector pT2 encoding OVA, eGFP and neomycin phosphotransferase, and the vector encoding transposase SB11. Three days later 400 mg/ml geneticin (Thermo Fisher, #10131027) was added to culture media to select for transgene-expressing cells. Successful integration was confirmed by analyzing eGFP fluorescence by flow cytometry. Limiting dilution was used to derive monoclonal OVA-expressing lines for each cell line. OVA presentation was confirmed by flow cytometry using a PE-labeled antibody against surface SIINFEKL bound to H-2Kb (clone 25-D1.16, BioLegend).
In vitro cytotoxicity assay
OVA-expressing GFP-positive B16F10, LLC, or MC38 cells were seeded in black polystyrene 96 well plates (Costar, #3603), 1x104 cells per well. Transduced OT-I cells were enriched by magnetic bead sorting against Thy-1.1 (Miltenyi, #130-121-273) and added to tumor cells to a final T cell-to-target ratio of 1:1. Endpoint cytotoxicity was assessed after 20 hours of co-culture. Wells were washed 3 times with PBS to remove T cells and the number of remaining target cells was determined by culturing with 10 μg/ml resazurin (Sigma, #R7017) and measuring fluorescence signal using 540/25 nm excitation wavelength and 620/40 nm emission wavelength in a Synergy 2 multi-mode microplate reader (BioTek). Cytotoxicity was calculated relative to wells with no T cells added. For real-time cytotoxicity, plates containing tumor cells were placed in Incucyte S3 Live-Cell Analysis System (Essen Bioscience, #4647) and imaged once per hour until 24 or 48 hours. OT-I cells were added 4 hours after tumor cell seeding. Images analysed with IncuCyte software and cell density determined by detection of GFP+ cell events.
To determine cytotoxicity of CAR-transduced human T cells, Raji (GFP+CD19+, target) were co-cultured with varying ratios of GFP+CD8+ T cells. After 20h co-culture cytotoxicity was assessed by flow cytometry and the ratio of Raji cells to CountBright Absolute counting beads (Thermo Fisher, #C36950) used to calculate cytotoxicity. To determine specific cytotoxicity, data was normalised to the cytotoxicity of VC-transduced CD8+ T cells of the respective donor.
Adoptive cell transfer of mouse CD8+ T cells
8 to 12-weeks old female C57BL/6J were inoculated subcutaneously with 5×105 B16-F10-OVA and conditioned 4 days later with peritoneal injection of 300 mg/kg cyclophosphamide (Sigma, #C0768). On day 8, 0.5-1×106 Thy-1.1–purified transduced OT-I CD8+ T cells were peritoneally injected. Animals were assigned randomly to each experimental group. Tumor volume was measured every 2-3 days with electronic calipers until day 60. Peripheral blood was collected from the tail vein at day 15 and analyzed by flow cytometry. Tumor volume was calculated using the formula where a is the length and b is the width of the tumor.
Tumor-infiltration assessment
8 to 12-weeks old female C57BL/6J were inoculated subcutaneously with 5×105 B16-F10-OVA and conditioned 11 days later with peritoneal injection of 300 mg/kg cyclophosphamide. On day 14, 0.5-1×106 Thy-1.1–purified transduced OT-I CD8+ T cells were intraperitoneally injected. Animals were assigned randomly to each experimental group. On day 19, tumors were dissected, digested with 1 mg/ml Collagenase Type IV (Life Technologies, #17104-019) and 20 U/ml DNAse I (Sigma, #D5025), and processed in a GentleMACS dissociator (Miltenyi). The tumor single cell suspensions were stained with fluorochrome-labeled antibodies and analysed by flow cytometry.
Adoptive cell transfer of human CD8+ T cells
13-week old female NSG mice were injected intravenously with 1×106 Raji-GFP-Luc and injected 4 days later with 2.5×105 GFP+ human CD8+ T cells transduced with vectors encoding GFP alone (VC), GFP + CD19CAR, or GFP + CD19CAR + HIF2α (PPA), 5 days after transduction. Animals were assigned randomly to each experimental group. Baseline tumor burden was measured 1 day before T-cell injection and 1-2 times per week until day 45 using an IVIS-SpectrumCT In Vivo imaging system (Perkin Elmer). Bioluminescent signal was assessed by injecting 150 mg/kg XenoLight D-Luciferin (Perkin Elmer, #122799) intraperitoneally followed by sedation with isoflurane. Bioluminescent signal was quantified using Aura Imaging Software (Spectral Instruments Imaging). Peripheral blood was collected from the tail vein at day 10 post T-cell transfer and analyzed by flow cytometry.
Statistics
Statistical analyses were performed with Prism 8 software (GraphPad). Statistical tests used are stated in figure legends. Statistical significance was set at p<0.01 and denoted in figures as α.
Study approval
All animal experiments were approved by the regional animal ethics Committee of Northern Stockholm, Sweden, under Ethical Permit number 5261-2020.
Results
Generation of retroviral vectors for ectopic expression of HIF1α and HIF2α in CD8+ T cells
To test whether ectopic expression of HIFs can increase the antitumor function of cytotoxic CD8+ T cells we generated retroviral vectors encoding murine HIF1α and HIF2α. HIF protein stability and transcriptional activity is regulated by oxygen at the post-translational level (Figure 1A). In the presence of oxygen, PHD isoforms hydroxylate conserved proline residues, leading to recognition and ubiquitination by the VHL protein, thus targeting HIFs for proteasomal degradation. FIH hydroxylates a conserved asparagine, which blocks recruitment of the coactivator p300/CBP and impairs HIF transcriptional activity. To address the role of HIF regulation we employed site-directed mutagenesis (Figure 1B) to convert conserved proline (P402/P577 in mouse HIF1α; P405/P530 in mouse HIF2α) and asparagine (N813 in mouse HIF1α and N851 in mouse HIF2α) residues into alanine residues. These alanine residues in the resultant vectors cannot be hydroxylated, and thus prevent proline or asparagine hydroxylation and inhibition of HIF accumulation or activity.
Figure 1. Design and validation of retroviral vectors to drive ectopic expression of HIF proteins in mouse CD8+ T cells.
(A) HIF transcription factors are post-translationally regulated. Oxygen-dependent hydroxylation at conserved proline (P) residues by PHD results in VHL)-mediated proteasomal degradation. Hydroxylation at a conserved asparagine (N) residue by FIH prevents recruitment of the coactivator p300/CBP resulting in inhibited transcriptional activity. Once released from VHL/PHD and FIH repression, HIF proteins heterodimerize with ARNT, translocate to the nucleus, bind hypoxia-responsive elements (HRE) and initiate transcription of target genes.
(B) Mutation of key amino-acid residues modulates HIF regulation. Mutation of conserved prolines (P402, P577 in mouse HIF1α; P405, P530 in mouse HIF2α) and of conserved asparagine (N813 in mouse HIF1α and N851 in mouse HIF2α) into alanine (A) prevents hydroxylation by PHD and FIH, respectively.
(C) Retroviral vector design for ectopic HIF expression. After genomic integration, the retroviral long terminal repeat (LTR) promoter drives expression of a polycistronic peptide containing Thy-1.1 (THY), HIF1α and HIF2α interspersed with furin cleavage sites and self-cleaving picornavirus 2A sites. Post-translational processing results in separation of the elements. Surface and nuclear localization sequences target Thy-1.1 to the cell surface and HIF isoforms to the nucleus, respectively.
(D) Nuclear extracts from HEK cells transfected with vectors encoding HIF1α alone, HIF-2α alone or both probed for mouse HIF1α, HIF2α and Lamin B. Vector control (VC) encodes Thy-1.1 alone.
(E) CD8+ T-cell transduction scheme. Primary CD8+ T cells were purified from mouse (C57BL/6J) splenocytes and activated by TCR triggering for 24 hours before transduction with retroviral particles. Transduced T cells were expanded in the presence of IL2 for further 3-5 days before subsequent analysis.
(F) Example of CD8+ T-cell transduction. Representative flow cytometry plot showing retrovirally (RV)-transduced cells expressing Thy-1.1 on the cell surface (red box).
(G) Nuclear extracts from Thy-1.1+CD8+ T cells transfected with vectors encoding HIF1α or HIF2α probed for mouse HIF1α, HIF2α and Histone 3. Vector control (VC) encodes Thy-1.1 alone.
The retroviral vectors designed to carry out these experiments encoded a polycistronic peptide composed of Thy-1.1 (transduction marker), HIF1α, and/or HIF2α (Figure 1C). These elements were interspersed with furin and self-cleaving picornavirus 2A sites that enabled post-translational separation of Thy-1.1 and HIF proteins. Subcellular localization signal peptides trafficked Thy-1.1 to the cell surface, and HIFs to the nucleus. The polycistronic nature of the constructs ensured equimolar production of all proteins, while retroviral vector integration in the T-cell genome ensured constitutive and heritable transgene expression.
A library of vectors was generated encoding both HIF1α and HIF2α together, HIF1α alone, or HIF2α alone, and in every mutational arrangement; these are denoted henceforth as PPN (hydroxylated by VHL and FIH), PPA (hydroxylated by VHL only), AAN (hydroxylated by FIH only), and AAA (no hydroxylation by either VHL or FIH). An empty vector control (VC) was generated that encodes Thy-1.1 alone. The ability of these vectors to deliver HIF1α and HIF2α protein to the nucleus was confirmed in nuclear extracts of transfected HEK cells (Figure 1D). Ectopic expression of HIF isoforms was achieved with every mutational combination. As expected, mutation of proline residues resulted in increased HIF protein accumulation due to the absence of VHL-driven proteasomal degradation.
During retroviral transduction primary mouse CD8+ T cells were activated 24 hours before transduction with retroviral particles, and expanded for a further 3 to 5 days in the presence of IL2 (Figure 1E). Transduced cells expressing Thy-1.1 on the cell surface were identified and characterized by flow cytometry (Figure 1F) or purified using magnetic bead sorting for subsequent analysis. Ectopic expression of HIF1α and HIF2α was confirmed in nuclear extracts of transduced CD8+ T cells (Figure 1G). Having validated these vectors, we next proceeded to investigate the effect of ectopic HIF expression on CD8+ T-cell gene expression, effector differentiation, and antitumor function.
Ectopic HIF2α expression dramatically alters the CD8+ T-cell transcriptome
To assess the impact of ectopic HIF expression on global gene expression we performed transcriptomic analyses of CD8+ T cells transduced with either HIF1α or HIF2α. To increase homogeneity in these analyses we transduced CD8+ T cells from the transgenic OT-I mouse strain in which all T cells express the same MHC class I–restricted TCR specific for a fragment of OVA (Figure 2A). Five days after transduction, Thy-1.1+ OT-I cells were sorted by flow cytometry for RNA purification followed by RNA sequencing (RNA-seq). In total, 12,155 individual transcripts were detected and mapped (Figure 2B). Average transcript frequency and rank distribution was similar to previously published transcriptomic analyses of CD8+ T cells (28,29), including high levels of expression of genes defining CD8+ T cell identity such as granzyme B (Gzmb), Cd8a, Cd3g, perforin (Prf1) and Ifng.
Figure 2. Effects of ectopic expression of HIF1α or HIF2α on the CD8+ T-cell transcriptome.
(A) OVA-specific OT-I splenocytes were activated with an H-2Kb–restricted OVA peptide (SIINFEKL) for 24 hours before transduction with HIF1α- or HIF2α-encoding retroviral vectors. After 5 days of expansion in the presence of IL2, live CD8+Thy-1.1+ cells were sorted by flow cytometry followed by RNA extraction and RNA-seq (n = 3 independent transductions per vector).
(B) Violin plot representing transcript frequency in Log2 counts per million (CPM) of 12155 mapped transcripts. Solid vertical line: median. Dashed vertical line: quartiles. Red circles represent transcripts defining CD8+ T-cell identity.
(C) Mean-difference plots showing Log2 fold change of transcripts in HIF1α- and HIF2α-transduced relative to vector control (VC)-transduced CD8+ T cells. Pink and green circles: differentially expressed transcripts as defined by a false discovery rate (FDR) < 0.01 and Log2 fold change >1 or <−1. Grey circles: non differentially expressed transcripts.
(D) Bar chart summarizing total number of up- and down-regulated transcripts. Values over bars: total number of differentially expressed genes.
(E) Scattered dot plot showing absolute Log2 fold change of differentially expressed transcripts in each transduction. Lines: median and interquartile range. Values over plots: median Log2 fold change. α, P < 0.01; Kruskal-Wallis with Dunn’s multiple comparison test.
(F) Heatmap representing correlation in transcript frequency between HIF1α and HIF2α-transduced CD8+ T cells. Values in boxes: Spearman’s rank correlation coefficient.
We next compared transcript frequency of HIF-transduced CD8+ T cells with vector control (VC)-transduced CD8+ T cells (Figure 2C). Ectopic expression of VHL-sensitive (PPN and PPA) HIF1α had minimal impact on gene expression, with only 5 and 11 differentially expressed genes, respectively (Figure 2D). Ablation of VHL control over HIF1α (AAN and AAA) significantly altered expression of 437 and 445 genes, respectively, suggesting that high levels of HIF1α protein are required to elicit transcriptional changes in this context. Indeed, we were only able to detect nuclear HIF1α protein in extracts from CD8+ T cells if VHL regulation was absent (AAN and AAA) (Figure 1G).
Ectopic expression of HIF2α resulted in a significantly greater number of differentially expressed genes relative to expression of HIF1α, irrespective of regulatory status (Figure 2C, 2D). HIF2α altered the expression of 1300, 743, 1491 and 2353 genes in the PPN, PPA, AAN and AAA formats, respectively. Furthermore, the magnitude of changes in gene expression was generally higher in HIF2α-expressing CD8+ T cells than in those expressing HIF-1α (Figure 2E).
There was high transcriptional similarity amongst cells expressing ectopic HIF1α, irrespective of susceptibility to VHL or FIH inhibition (Figure 2F). The transcriptional changes elicited by HIF2α were also similar amongst the PPN, PPA, AAN and AAA variants. However, HIF1α and HIF2α appear to alter CD8+ T-cell transcription differently, as shown by low correlation coefficients when comparing differentially expressed genes between the two HIF isoform mutational groups (Figure 2F).
Ectopic HIF2α altered the expression of several genes involved in CD8+ T-cell function (Figure 3A). Notably, HIF2α increased expression of Prf1 and Gzmb, which encode two highly expressed proteins essential for cytolytic function. Ectopic HIF2α also increased expression of genes encoding co-stimulators such as CD30 (also known as TNFRSF88), 4-1BB (also known as TNFRSF9), OX40 (also known as TNFRSF4) and ICOS and co-inhibitors such as LAG3 and CTLA-4. Reduced expression of the gene encoding IFNγ, a cytokine that promotes antitumor function by inducing antigen presentation in cancer cells, as well as the genes encoding its receptor (Ifngr1 and Ifngr2) was detected in HIF2α-expressing cells. Expression of the genes encoding the alpha (CD25) and beta chains (CD12) of the IL2 receptor was increased, suggesting a greater responsiveness to IL2. Genes encoding protein involved in T-cell trafficking such as integrins and the bone marrow–homing receptor CXCR4 (30) were also induced by ectopic HIF2α expression.
Figure 3. Effects of ectopic expression of HIF1α or HIF2α in genes involved in CD8+ T-cell function.
(A) Heatmaps showing Log2 fold change of transcripts involved in functional aspects of CD8+ T cells.
(B) Expression of differentiation markers determined by flow cytometry in CD8+ T cells transduced with vectors encoding HIF-1α and HIF-2α, HIF-1α alone or HIF-2α alone (day 3 to 5 post-transduction). Data expressed as Log2 fold change of median fluorescence intensity (MFI) relative to VC-transduced cells. Each data point represents an independent transduction (n=4-24). Results are pooled from a minimum of two independent experiments. α, P < 0.01; one-way ANOVA with Dunnett’s multiple comparison test relative to VC. Histograms are representative flow cytometry plots for each parameter and are pre-gated on live, singlet, CD8+Thy-1.1+ events.
The broad transcriptional changes induced by HIF-2α can be partially explained by altered expression of a wide array of transcriptional factors and transcriptional modulators (Supplementary Figure 1A and B). While few transcriptional modulators were altered by HIF1α, ectopic expression of HIF2α significantly altered expression of 77, 40, 98, and 135 transcriptional modulator genes in the PPN, PPA, AAN and AAA mutational variants respectively, thus amplifying the transcriptional impact of HIF2α. Notably, HIF2α decreased expression of genes encoding transcription factors associated with naive/memory CD8+ T cells as well as the exhausted state (Tcf7 (31,32), Tox (33), Nr4a1, Nr4a3 (34) and Eomes (35)). Transcripts associated with a response to hypoxia (Supplementary Figure 1C) and induction of glycolysis (Supplementary Figure 1D) were also upregulated in CD8+ T cells expressing HIF2α, but not in those expressing HIF1α.
HIF2α drives phenotypical changes in CD8+ T cells
We next sought to confirm if the transcriptional changes induced by ectopic HIF expression translated to alterations in protein expression. For that we transduced CD8+ T cells with vectors encoding HIF1α and HIF2α together, HIF1α alone, or HIF2α alone and analysed their phenotype by flow cytometry (Figure 3B). In line with the RNA-seq results described above, expression of the co-inhibitors LAG3 and CTLA-4 was augmented by HIF2α, particularly in the absence of VHL inhibition (AAN and AAA). Ectopic co-expression of HIF1α and HIF2α yielded similar results to HIF2α alone. Increased expression of the IL2 receptor α and β chains, CD25 (Figure 3B) and CD122 (Supplementary Figure 2A), was also confirmed at the protein level as was expression of the chemokine receptor CXCR4 (Supplementary Figure 2A). Likewise, expression of GZMB, ICOS and 4-1BB, key components of the cytotoxic function of CD8+ T cells, was increased by ectopic expression of HIF2α (Figure 3B). Notably, GZMB expression was only significantly increased when HIF2α was susceptible to VHL regulation (PPN and PPA). Suppression of the transcription factor EOMES by HIF2α was also confirmed by intranuclear flow cytometry (Figure 3B). Of the assayed proteins, only CD147 (also known as Basigin), a known direct HIF1α target (36), was augmented by HIF-1α (Figure 3B). The increased expression of GZMB, LAG3, CTLA-4, CD25 and 4-1BB and suppression of EOMES was confirmed in CD8+ T cells transduced with HIF2α and cultured over a period of 21 days with weekly restimulation (Supplementary Figure 2B and C), confirming the long-term stability of the phenotypes conferred by the integrated retroviral vectors. Altogether, these data show that ectopic HIF2α expression, but not HIF1α expression, was able to substantially alter the phenotype of CD8+ T cells and that the magnitude of those changes was greater in the absence of inhibition by VHL.
Ectopic expression of VHL-insensitive HIF2α alters CD8+ T-cell phenotype
During long-term culture of HIF-transduced CD8+ T cells (Supplementary Figure 2B) the frequency of cells expressing VHL-insensitive HIF2α (AAN and AAA) diminished progressively over time (Figure 4A) whereas the frequency of the remaining transduced populations increased slightly over 21 days in culture. This was due to reduced proliferation of VHL-insensitive HIF2α-transduced CD8+ T cells (Figure 4B, 4C and Supplementary Figure 3A) and not due to increased apoptosis (Supplementary Figure 3B).
Figure 4. Ectopic expression of VHL-insensitive HIF2α reduces CD8+ T-cell proliferation and drives TCR loss.
(A) Fitness of HIF-transduced CD8+ T cells over time. CD8+ T cells were transduced with HIF1α and HIF2α-coding vectors and cultured for 21 days in the presence of IL2. Cells were restimulated with CD3/CD28 beads on days 7 and 14. Fitness was calculated as the difference in % of Thy-1.1+ cells in culture relative to day 7 (Δ% Thy-1.1+). VC: vector control.
(B) Proliferation of HIF2α-transduced CD8+ T cells. Cells were loaded with CellTrace Violet (CTV) proliferation dye 6 days after transduction and were restimulated with αCD3/CD28 beads for 3 days. Proliferation was determined by CTV dilution in flow cytometry. Left: representative histograms pre-gated on live, singlet, CD8+. Thy-1.1+ events. Right: summary data showing CTV mean fluorescence intensity (MFI). n = 7 independent transductions. Lines: median and interquartile range.
(C) Fitness of HIF2α-transduced cells after restimulation. Fitness was calculated as the difference in % of Thy-1.1+ cells in culture between restimulated and unstimulated cultures (Δ% Thy-1.1+). n = 3 independent transductions. Lines: median and interquartile range.
(D) Mean-difference plots showing Log2 fold change of TCR chain-coding transcripts in HIF2α-transduced relative to VC-transduced CD8+ T cells. Green circles: differentially expressed transcripts as defined by a false discovery rate (FDR) < 0.01 and Log2 fold change >1 or <−1. Grey circles: non differentially expressed transcripts. Trbv12-1 codes the Vβ5 segment of the OT-I TCRβ chain.
(E) Expression of TCR Vα2 and TCR Vβ chains, and CD3 determined by flow cytometry in OT-I CD8+ T cells transduced with vectors encoding HIF1α and HIF2α, HIF1α alone or HIF2α alone (day 3 to 5 post-transduction). Data expressed as Log2 fold change of MFI relative to VC-transduced cells. Each data point represents an independent transduction (n=4-24). Results are pooled from a minimum of two independent experiments.
(F) Surface expression of OT-I TCR chains in HIF2α-transduced OT-I cells on day 4 post-transduction. Flow cytometry zebra plots pre-gated on live, singlet, CD8+ events showing surface expression of OT-I TCR Vα2 and TCR Vβ5 chains in transduced (Thy-1.1+; top row) and non-transduced (Thy-1.1−; bottom row). Values are the percentage of events within the double-negative quadrant.
(G) Frequency of TCR-negative cells. n=5 independent transductions. Lines: median and interquartile range.
(H) Surface expression of CD3 and the constant region of the TCRβ chain in HIF2α-transduced polyclonal and OT-I CD8+ T cells. Flow cytometry zebra plots pre-gated on live, singlet, CD8+, Thy-1.1+ events. Values are the percentage of events within the double-negative quadrant.
(I) Frequency of TCR-negative cells in HIF2α-transduced CD8+ T cells cultured with DMSO or 10 μM PT2977 (HIF-2α inhibitor). n=5 independent transductions. Lines: median and interquartile range.
α, P < 0.01; one-way ANOVA with Dunnett’s multiple comparison test relative to VC.
Transcriptomic analysis revealed downregulation of genes encoding TCR chains in HIF2α-transduced cells, particularly when VHL regulation was absent (Figure 4D). OT-I cells transduced with VHL-insensitive (AAN and AAA) HIF2α had reduced surface expression of TCR alpha (Vα2) and TCR beta (Vβ5) chains (Figure 4E), whereas VHL-sensitive HIF2α or any form of HIF1α had no effect on TCR expression. Surface expression of CD3, the signalling component of the TCR complex, was also reduced (Figure 4E). Loss of TCR chains occurred synchronously, resulting in TCR loss in over 50% of AAN and AAA HIF2α-expressing cells (Figure 4F and 4G) and occurred progressively after transduction (Supplementary Figure 3C). TCR loss was observed in polyclonal CD8+ T cells to the same extent as in OT-I cells (Figure 4H and 4I). Interestingly, the magnitude of TCR loss was influenced by the type of in vitro stimulus used to activate T cells on the day prior to transduction, with concanavalin A (ConA) stimulation resulting in low TCR loss and OVA-antigen stimulation resulting in greater TCR loss (Supplementary Figure 3D). Culturing HIF2α-transduced T cells with the HIF2α inhibitor PT2977 (37) completely abrogated TCR loss (Figure 4I and Supplementary Figure 3D) and augmented expression of LAG3 and CD25 (Supplementary Figure 3E). PT2977 also reversed the proliferative disadvantage of ectopic HIF2α expression (Supplementary Figure 3E).
Production of IFNγ after TCR triggering with OVA was also reduced in OT-I cells transduced with VHL-insensitive HIF2α (AAN and AAA) (Figure 5A and 5B), a likely consequence of reduced TCR expression, since stimulation with PMA and ionomycin, which bypasses TCR signaling, resulted in unchanged IFNγ secretion (Supplementary Figure 2C).
Figure 5. Ectopic expression of HIF2α enhances cytotoxicity against tumor cells.
(A) IFNγ secretion determined by intracellular cytokine flow cytometry in OT-I CD8+ T cells transduced with vectors encoding HIF1α and HIF2α, HIF1α alone or HIF2α alone and restimulated for 4 hours with 1 μM OVA (SIINFEKL) peptide. Values are the percentage within the IFNγ+ gate. Pre-gated on live, singlet, CD8+. Thy-1.1+ events
(B) Summary data expressed as % IFNγ+ cells. Each data point represents an independent transduction (n=3-7). Results are pooled from a minimum of two independent experiments.
(C) Real-time cytotoxicity assay. Upper row: B16F10-OVA, MC38-OVA and LLC-OVA cell density over time after addition of HIF2α-transduced OT-I CD8+ T cells. Tumor cells were seeded 5 hours prior. Bottom row: endpoint cytotoxicity. n=6-12 replicate wells. Grey horizontal area: interquartile range of no T-cell control.
(D) Real-time cytotoxicity assay with DMSO- or PT2977-treated HIF2α-transduced OT-I CD8+ T cells. Left: B16F10-OVA cell density over time after addition of T cells. Right: endpoint cytotoxicity. n=6-12 replicate wells. Grey horizontal area: interquartile range of no T cell control.
α, P < 0.01; one-way ANOVA with Dunnett’s multiple comparison test relative to VC.
To assess how the phenotypic and functional changes caused by ectopic HIF2α expression affect cytotoxicity against tumor cells, we co-cultured transduced OT-I cells with OVA-expressing B16-F10 melanoma, LLC or MC38 colon cancer cells and monitored cell growth in real-time (Figure 5C). Ectopic HIF2α expression increased cytotoxicity against B16-F10, LLC and MC38 cells, but more so when VHL regulation was retained (PPN and PPA). Noticeably, OT-I cells expressing VHL-insensitive HIF2α (AAN and AAA) performed worse than controls in restraining B16-F10 growth, but this defect was reversed when T cells were cultured with the HIF2α inhibitor PT2977 (Figure 5D). Killing of B16-F10 cells was similar whether co-culture occurred at atmospheric oxygen conditions (21% O2) or at physiologically relevant oxygen tensions (5% and 1% O2) (Supplementary Figure 3F). This result can be partly attributed to increased GZMB levels (Figure 3B) and to largely unchanged proliferation rates, TCR expression and IFNγ secretion in the HIF2α PPN and PPA groups (Figures 4 and 5). Overall, these results reveal that ectopic expression of highly stable HIF2α protein (AAN and AAA) in CD8+ T cells leads to a deleterious phenotype, whereas expression of VHL-sensitive (PPN and PPA) HIF2α potentiates in vitro cytotoxicity against tumor cells.
In vivo efficacy of adoptively transferred CD8+ T cells ectopically expressing FIH-insensitive HIF2α
In therapy, adoptively transferred CAR/TCR engineered T cells must engraft, proliferate, traffic to tumor sites and eliminate cancer cells to deliver therapeutic benefit. To test the antitumor function of HIF-transduced CD8+ T cells we employed a mouse model of adoptive cell therapy (ACT) (Figure 6A). Wild-type mice were inoculated with OVA-expressing B16-F10 melanoma cells, followed by lymphodepleting chemotherapy and adoptive transfer of HIF1α- or HIF2α-transduced OT-I cells (Supplementary Figure 4A). A week after T-cell injection, peripheral blood was harvested, and the frequency of transduced OT-I cells assessed by flow cytometry (Figure 6B and Supplementary Figure 4B). The engraftment of T cells expressing VHL- and FIH-sensitive (PPN) HIF2α was significantly increased, whereas cells expressing VHL-insensitive (AAN and AAA) HIF2α were nearly absent in peripheral blood. The frequency of T cells expressing VHL- and FIH-insensitive (AAA) HIF1α was also reduced relative to the control group.
Figure 6. Adoptive transfer of CD8+ T cells ectopically expressing FIH-insensitive HIF2α results in delayed tumor growth and increased survival rate.
(A) ACT model. C57BL/6J mice were injected subcutaneously with 5×105 OVA-expressing B16-F10 (B16-F10-OVA) and 4 days later were lymphodepleted with 300 mg/kg cyclophosphamide. On day 8, 0.5-1 × 106 HIF-transduced (Thy-1.1 enriched) OVA-specific OT-I CD8+ T cells were adoptively transferred into tumor-bearing mice. Peripheral blood was sampled at day 15 and analysed by flow cytometry. Tumor growth was monitored every 2-3 days until day 60.
(B) Frequency of HIF-transduced OT-I cells per million PBMCs in peripheral blood. n = 8-13 animals pooled from two independent experiments. Grey horizontal line: median of VC group. α, P < 0.01; one-way ANOVA with Dunnett’s multiple comparison test relative to VC.
(C) B16-F10-OVA tumor growth after ACT. B16-F10-OVA tumor volume measured until day 60 after mice received VC-HIF1α- or HIF2α-transduced OT-I cells on day 8. Thin lines: tumor growth from individual animals. Thick line: centered sixth order polynomial curve. Shaded area: 99% confidence level interval. n = 9-25 animals per group pooled from two (HIF1α) or four (HIF2α) independent experiments.
(D) Survival curves for tumor growth shown in (D). Threshold for survival was set at 200 mm3. Grey line: no ACT. Black line: ACT of VC-transduced OT-I. Pink or green lines: ACT of HIF-1α- or HIF-2α-transduced OT-I, respectively. α, P < 0.01; log-rank (Mantel-Cox) test relative to VC.
In this model, transfer of VC-transduced OT-I CD8+ T cells delayed tumor growth relative to the untreated (no ACT) group and increased median survival from 22 to 29 days. However, there were no complete regressions in this group (Figure 6D and Supplementary Figure 4C). Adoptive transfer of T cells expressing VHL-sensitive, but FIH-insensitive (PPA) HIF2α significantly delayed tumor growth, boosted median survival to 34 days, and fully eliminated tumors in 23% of the treated animals (Figure 6D and Supplementary Figure 4C). In contrast, animals receiving T cells expressing VHL-insensitive but FIH-sensitive (AAN) HIF2α performed significantly worse than controls, resulting in median survival times comparable to those of the untreated group (23 days). Likewise, T cells expressing VHL- and FIH-insensitive (AAA) HIF1α failed to protect animals from tumor growth, while the FIH-insensitive (PPA) HIF1α variant resulted in complete tumor clearance in 15% of animals; albeit without significantly delaying tumor growth (Figure 6D and Supplementary Figure 4C). These tumor rejection rates did not correlate with a differential ability to infiltrate tumor sites, since cells expressing VHL-sensitive HIF2α (PPN and PPA) infiltrated tumors at the same rate as controls, while cells expressing VHL-insensitive HIF2α (AAN and AAA) failed to infiltrate the tumor site (Supplementary Figure 4D-F). Infiltration of endogenous CD8+ T cells was unaltered (Supplementary Figure 4G). These experiments reveal an unexpected role of the FIH hydroxylation site on the HIF2α protein for the in vivo antitumor function of CD8+ T cells, and indicate an important role for oxygen-mediated transcriptional control of HIF2a that is qualitatively different from the role played by the VHL pathway.
Antitumor efficacy of human CD8+ T cells co-expressing a CAR and FIH-insensitive HIF2α
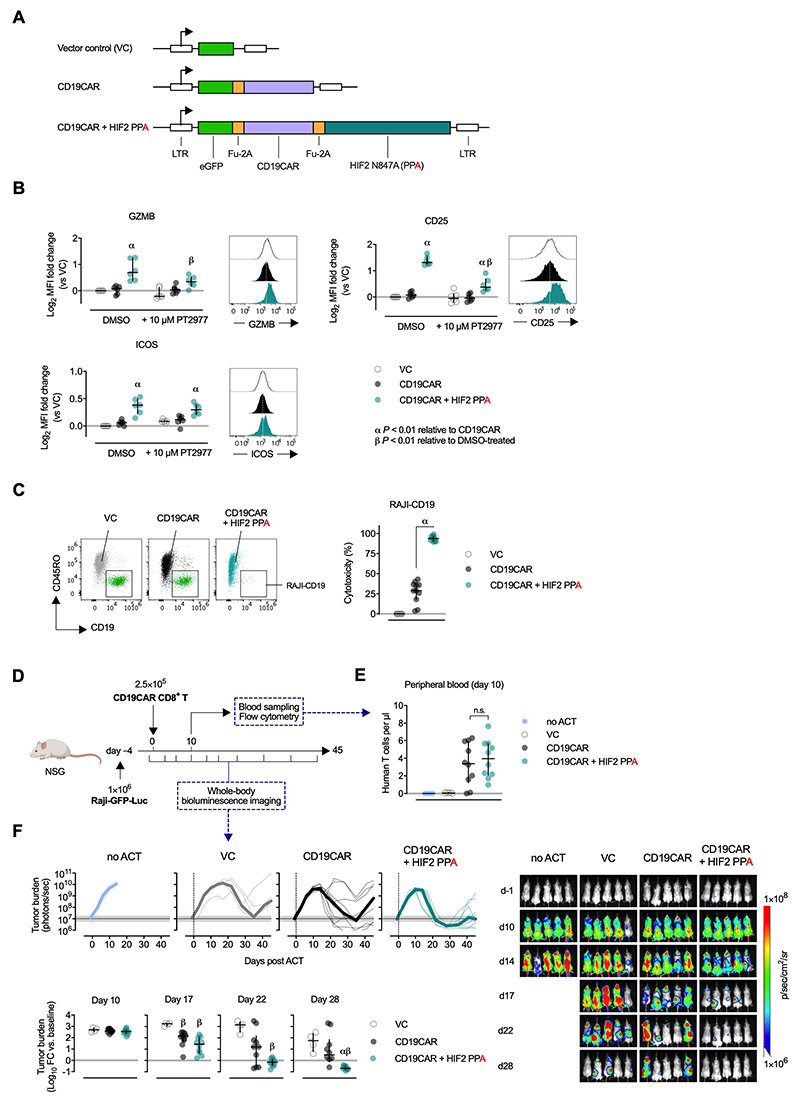
To test whether HIF2α expression could improve the antitumor function of human therapeutic T cells we designed a vector encoding an anti-CD19 CAR (CD19CAR) alone or in combination with FIH-insensitive HIF2α (N847A) (Figure 7A and Supplementary Figure 5A). CD8+ T cells purified from multiple human PBMC transduced with CD19CAR and HIF2α had increased expression of GZMB, CD25, ICOS, CCR7, 4-1BB and CD95/FAS relative to cells expressing CD19CAR alone (Figure 7B and Supplementary Figure 5B). Inhibition of HIF2α activity with PT2977 reduced expression of GZMB and CD25, although not entirely to baseline level, but did not alter ICOS expression (Figure 7B). Cytolytic function of CAR-transduced CD8+ T cells against Raji (CD19+) lymphoma cells was significantly increased when HIF2α was co-expressed (Figure 7C and Supplementary Figure 5C). Next, we performed a xenograft ACT using immunodeficient NSG mice. Animals were injected with 1 × 106 luciferase-expressing CD19+ Raji cells followed by 2.5 × 105 CD19CAR (or VC) CD8+ T cells 4 days later (Figure 7D and Supplementary Figure 5D). Tumor burden was assessed by total body luminescence before and after ACT. Peripheral blood analysis 10 days after T-cell transfer revealed successful and equal engraftment of both CD19CAR and CD19CAR+HIF2α T cells (Figure 7E and Supplementary Figure 5D). Notably, the frequency of circulating Raji cells was lower in mice injected with HIF2α-expressing T cells (Supplementary Figure 5E). Tumor burden increased rapidly in the first days after engraftment and by day 14 all animals that did not receive T cells had to be sacrificed due to hind limb paralysis (Figure 7F). An acute drop in tumor burden occurred after day 14 in mice injected with CD19CAR T cells. Reduction of tumor burden was significantly more pronounced in mice receiving CD19CAR+HIF2α T cells, with tumor regressing under baseline levels more rapidly than with cells expressing the CAR vector alone (Figure 7F). A delayed antitumor effect of the VC-transduced control cells was also observed, likely due to a polyclonal TCR-driven response.
Figure 7. FIH-insensitive HIF2α increases cytotoxicity of human CD8+ CAR-T cells against lymphoma cells.
(A) Retroviral vector design for co-expression of an anti-CD19 CAR and FIH-insensitive (N847A; PPA) human HIF2α. LTR: long terminal repeats. Fu-2A: furin and picornavirus 2A self-cleaving sequence.
(B) Expression of GZMB, CD25 and ICOS on GFP+ human CD8+ T cells 6 days after transduction with vector control (VC), CD19CAR, or CD19CAR+HIF2. Cells were cultured with DMSO or 10 μM PT2977 (HIF2α inhibitor) from day 1 post-transduction. n = 6 donors. Mean Fluorescence Intensity (MFI) is normalized to DMSO-treated VC-transduced cells of respective donors. Lines: median and interquartile range.
(C) Cytotoxicity of VC- or CAR-transduced human CD8+ T cells against GFP+CD19+ RAJI lymphoma cells after 20 hours of co-culture at a 1:1 effector-to-target ratio. Representative flow cytometry showing transduced (GFP+, CD45RO+) CD8+ T cells and RAJI targets (GFP+, CD19+) after gating on live, singlet GFP+ events. Summary data of specific cytotoxicity of 11 donors. Lines: median and interquartile range.
(D) Experimental scheme for ACT of human CD19CAR-transduced CD8+ T cells. Immunocompromised NSG mice were injected with 1×106 luciferase-expressing CD19+ Raji cells followed by (ACT) of 2.5×105 VC-, CD19CAR- or CD19CAR+HIF2-transduced CD8+ T cells 4 days later. Tumor burden assessed by whole-body bioluminescence imaging up to day 45. Adoptive cell engraftment monitored on day 10 by sampling peripheral blood.
(E) Frequency of human CD8+ T cells in peripheral blood on day 10 after ACT. n = 5-10 mice. Lines: median and interquartile range.
α, P < 0.01; one-way ANOVA with Dunnett’s multiple comparison test relative to VC.
β, P < 0.01; one-way ANOVA with Dunnett’s multiple comparison between PT2977- and DMSO-treated counterparts.
(F) Top: Tumour burden after ACT shown as bioluminescent signal (photons/second; thick line: group median; thin lines: individual animals). Horizontal shaded area and line: range ad and median tumor burden at baseline. Bottom: Log10 fold change in tumor burden relative to baseline (median and interquartile range) at different time points. α,β P < 0.01; one-way ANOVA with Tukey’s multiple comparison test relative to CD19CAR or VC, respectively. Right: Representative images of bioluminescent whole-body imaging. Scale ranges from 1×106 to 1×108 photons/sec/cm2 on a logarithmic scale.
These results show that the enhanced antitumor effects of T cells expressing ectopic FIH-insensitive HIF2α can be translated to a preclinical human setting and that this approach has the potential to improve current cancer therapies involving gene-engineered T cells.
Discussion
In this study we set out to determine if ectopic HIF expression can be used to increase the antitumor efficacy of therapeutic CD8+ T cells with the goal of improving ACT protocols for cancer. Retroviral vectors are currently used to deliver ectopic expression of cancer antigen–specific TCRs or CARs to T cells and can be easily modified to deliver additional proteins (7,8). Previous studies employing T-cell specific deletion of VHL (24), PHDs (25), ARNT (26), HIF1α and HIF-2α (27), or hypoxic-priming (23) of CD8+ T cells laid the groundwork for this study by showing that HIF activity drives cytotoxic differentiation and in vivo antitumor function.
Here we took an agnostic approach regarding which of the two main HIFα isoforms and which oxygen-dependent suppression mechanisms could most potently boost antitumor effects of T cells. Previous studies suggested non-degradable HIF1α as the strongest candidate for over-expression, given that VHL- and PHD-null T cells reject tumors more efficiently (24,25), whereas HIF1α-null T cells perform quite poorly in immunotherapy models (27).
Our findings reveal that in an ectopic expression setting, it is HIF2α, and not HIF1α, that is able to most potently boost CD8+ T-cell antitumor cytotoxicity. Ectopic HIF2α was able to elicit broad changes in gene expression by altering the T-cell transcription factor network, irrespective of VHL and FIH suppression. However, the magnitude of change in gene expression was typically higher when HIF2α was resistant to VHL-mediated proteolytic degradation. The transcripts altered by HIF2α extend beyond known HIF target genes. This is likely due to altered expression of a network of transcriptional modulators, many of them known to have implications for CD8+ T-cell differentiation, including Tcf7, Nr4a1, Nr4a3, Tox and Eomes, all of which drive memory and/or exhaustion phenotypes, and are downregulated by HIF2α. Ectopic HIF2α expression does not result in a gene signature that clearly identifies a memory, effector or exhausted population, rather it generates an unconventional phenotype with upregulation of co-stimulators (4-1BB, ICOS), effector proteins (GZMB, Perforin), and also co-inhibitors (CTLA-4, LAG3), as well as downregulation of IFN□.
Although transcriptional changes detected by RNA sequencing were largely replicated at protein level, it was at the functional level that oxygen-dependent regulation of HIF2α proved to be most important. Proteolytic-resistant HIF2α (AAN and AAA) severely impaired T-cell function by restricting proliferation, causing TCR downregulation and decreasing IFN□ secretion. It is worth noting that, unlike other phenotypic changes, proliferation deficiency caused by HIF2α was not observed when HIF1α was co-expressed, suggesting that HIF1α can partially counteract this HIF2α effect. These results are surprising given the greater antitumor function of VHL- and PHD-null T cells reported previously (24,25). Co-culture with a variety of tumor targets consistently revealed that CD8+ T cells expressing VHL-sensitive (PPN and PPA) HIF2α had the highest cytolytic function suggesting that intermediate levels of HIF2α protein offer the most benefit.
Regulation of HIF2α has striking effects in the ACT models we employed. When transferred to melanoma-bearing mice, CD8+ T cells expressing HIF2α proteins that are insensitive to FIH, but sensitive to VHL-mediated post-translational degradation, provided the best protection against tumor growth. Interestingly, FIH-sensitive and VHL-insensitive HIF2α expressing vectors completely inhibit antitumor immunity by T cells. This unexpected role for FIH hydroxylation was difficult to discern outside of the in vivo setting.
Transcriptional analyses showed great similarity between the PPN and PPA groups (0.97 Spearman r) and between the AAN and AAA groups (0.98 Spearman r) despite hydroxylation at N851 being reported as crucial for the transcriptional activity of HIF2α. It appears as though the consequences of mutating N851 in HIF-2α only become apparent once T cells are allowed to differentiate and proliferate in vivo, where they are guided by the varying microenvironments a T cell inhabits and transits through, including the tumor microenvironment.
To translate these findings into a preclinical human setting we generated a vector to co-express a CD19CAR and FIH-insensitive HIF2α (N847A) in CD8+ T cells. Expression of human HIF2α greatly increased the cytotoxic function of CD19CAR T cells against a B-cell lymphoma both in vitro and in an in vivo xenograft ACT model. These results demonstrate that such vectors could represent an important and novel addition to the CAR T-cell armamentarium.
Supplementary Material
Synopsis.
In this study, ectopic expression of FIH-insensitive HIF2α enhances human CD8+ T cell–cytolytic function in vitro and in a xenograft model. Modifying HIF expression may provide a way to enhance the antitumor efficacy of adoptive T-cell therapy.
Acknowledgements
The authors thank Angelika Holler at University College London for providing reagents and discussions, and the Bioinformatics and Expression Analysis (BEA) Core Facility at the Department of Biosciences and Nutrition, which is supported by the board of research at the Karolinska Institute and the research committee at the Karolinska hospital.
Funding info
Funding has been provided by the Swedish Cancer Society (Cancerfonden), the Swedish Childhood Cancer Fund (Barncancerfonden), the Swedish Research Council (Vetenskapsrådet), the Wellcome Trust, the Knut and Alice Wallenberg Foundation, and the Portuguese Foundation for Science and Technology scholarship (SFRH/BD/115612/2016).
Footnotes
Author’s disclosures The authors have declared that no conflict of interest exists.
References
- 1.June CH, Riddell SR, Schumacher TN. Adoptive cellular therapy: a race to the finish line. Sci Transl Med. 2015;7:280ps7. doi: 10.1126/scitranslmed.aaa3643. [DOI] [PubMed] [Google Scholar]
- 2.Zhang T, Cao L, Xie J, Shi N, Zhang Z, Luo Z, et al. Efficiency of CD19 chimeric antigen receptor-modified T cells for treatment of B cell malignancies in phase I clinical trials: a meta-analysis. Oncotarget. 2015 doi: 10.18632/oncotarget.5582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhao L, Cao YJ. Engineered T Cell Therapy for Cancer in the Clinic. Front Immunol. 2019;10:2250. doi: 10.3389/fimmu.2019.02250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eyquem J, Mansilla-Soto J, Giavridis T, van der Stegen SJC, Hamieh M, Cunanan KM, et al. Targeting a CAR to the TRAC locus with CRISPR/Cas9 enhances tumour rejection. Nature. 2017;543:113–7. doi: 10.1038/nature21405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rupp LJ, Schumann K, Roybal KT, Gate RE, Ye CJ, Lim WA, et al. CRISPR/Cas9-mediated PD-1 disruption enhances anti-tumor efficacy of human chimeric antigen receptor T cells. Sci Rep. 2017;7:737. doi: 10.1038/s41598-017-00462-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yeku OO, Purdon TJ, Koneru M, Spriggs D, Brentjens RJ. Armored CAR T cells enhance antitumor efficacy and overcome the tumor microenvironment [Internet] Scientific Reports. 2017 doi: 10.1038/s41598-017-10940-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Martinez M, Moon EK. CAR T Cells for Solid Tumors: New Strategies for Finding, Infiltrating, and Surviving in the Tumor Microenvironment [Internet] Frontiers in Immunology. 2019 doi: 10.3389/fimmu.2019.00128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brandt LJB, Barnkob MB, Michaels YS, Heiselberg J, Barington T. Emerging Approaches for Regulation and Control of CAR T Cells: A Mini Review [Internet] Frontiers in Immunology. 2020 doi: 10.3389/fimmu.2020.00326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stavrou M, Philip B, Traynor-White C, Davis CG, Onuoha S, Cordoba S, et al. A Rapamycin-Activated Caspase 9-Based Suicide Gene. Mol Ther. 2018;26:1266–76. doi: 10.1016/j.ymthe.2018.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lynn RC, Weber EW, Sotillo E, Gennert D, Xu P, Good Z, et al. c-Jun overexpression in CAR T cells induces exhaustion resistance [Internet] Nature. 2019:293–300. doi: 10.1038/s41586-019-1805-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dengler VL, Galbraith M, Espinosa JM. Transcriptional regulation by hypoxia inducible factors. Crit Rev Biochem Mol Biol. 2014;49:1–15. doi: 10.3109/10409238.2013.838205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schödel J, Oikonomopoulos S, Ragoussis J, Pugh CW, Ratcliffe PJ, Mole DR. High-resolution genome-wide mapping of HIF-binding sites by ChIP-seq [Internet] Blood. 2011:e207–17. doi: 10.1182/blood-2010-10-314427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haase V. The VHL Tumor Suppressor: Master Regulator of HIF. Current Pharmaceutical Design. 2009:3895–903. doi: 10.2174/138161209789649394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maxwell PH, Wiesener MS, Chang GW, Clifford SC, Vaux EC, Cockman ME, et al. The tumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature. 1999;399:271–5. doi: 10.1038/20459. [DOI] [PubMed] [Google Scholar]
- 15.Lando D. FIH-1 is an asparaginyl hydroxylase enzyme that regulates the transcriptional activity of hypoxia-inducible factor [Internet] Genes & Development. 2002:1466–71. doi: 10.1101/gad.991402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mahon PC, Hirota K, Semenza GL. FIH-1: a novel protein that interacts with HIF-1alpha and VHL to mediate repression of HIF-1 transcriptional activity. Genes Dev. 2001;15:2675–86. doi: 10.1101/gad.924501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tausendschön M, Rehli M, Dehne N, Schmidl C, Döring C, Hansmann M-L, et al. Genome-wide identification of hypoxia-inducible factor-1 and -2 binding sites in hypoxic human macrophages alternatively activated by IL-10. Biochim Biophys Acta. 2015;1849:10–22. doi: 10.1016/j.bbagrm.2014.10.006. [DOI] [PubMed] [Google Scholar]
- 18.Mole DR, Blancher C, Copley RR, Pollard PJ, Gleadle JM, Ragoussis J, et al. Genome-wide association of hypoxia-inducible factor (HIF)-1alpha and HIF-2alpha DNA binding with expression profiling of hypoxia-inducible transcripts. J Biol Chem. 2009;284:16767–75. doi: 10.1074/jbc.M901790200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smythies JA, Sun M, Masson N, Salama R, Simpson PD, Murray E, et al. Inherent DNA -binding specificities of the HIF -1a and HIF -2a transcription factors in chromatin [Internet] EMBO reports. 2019 doi: 10.15252/embr.201846401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li Y, Patel SP, Roszik J, Qin Y. Hypoxia-Driven Immunosuppressive Metabolites in the Tumor Microenvironment: New Approaches for Combinational Immunotherapy. Front Immunol. 2018;9:1591. doi: 10.3389/fimmu.2018.01591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Corbet C, Feron O. Tumour acidosis: from the passenger to the driver’s seat [Internet] Nature Reviews Cancer. 2017:577–93. doi: 10.1038/nrc.2017.77. [DOI] [PubMed] [Google Scholar]
- 22.Sitkovsky MV, Kjaergaard J, Lukashev D, Ohta A. Hypoxia-Adenosinergic Immunosuppression: Tumor Protection by T Regulatory Cells and Cancerous Tissue Hypoxia [Internet] Clinical Cancer Research. 2008:5947–52. doi: 10.1158/1078-0432.ccr-08-0229. [DOI] [PubMed] [Google Scholar]
- 23.Gropper Y, Feferman T, Shalit T, Salame T-M, Porat Z, Shakhar G. Culturing CTLs under Hypoxic Conditions Enhances Their Cytolysis and Improves Their Anti-tumor Function. Cell Rep. 2017;20:2547–55. doi: 10.1016/j.celrep.2017.08.071. [DOI] [PubMed] [Google Scholar]
- 24.Doedens AL, Phan AT, Stradner MH, Fujimoto JK, Nguyen JV, Yang E, et al. Hypoxia-inducible factors enhance the effector responses of CD8 T cells to persistent antigen [Internet] Nature Immunology. 2013:1173–82. doi: 10.1038/ni.2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Clever D, Roychoudhuri R, Constantinides MG, Askenase MH, Sukumar M, Klebanoff CA, et al. Oxygen Sensing by T Cells Establishes an Immunologically Tolerant Metastatic Niche. Cell. 2016;166:1117–31.:e14. doi: 10.1016/j.cell.2016.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Finlay DK, Rosenzweig E, Sinclair LV, Feijoo-Carnero C, Hukelmann JL, Rolf J, et al. PDK1 regulation of mTOR and hypoxia-inducible factor 1 integrate metabolism and migration of CD8 T cells [Internet] The Journal of Experimental Medicine. 2012:2441–53. doi: 10.1084/jem.20112607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Palazon A, Tyrakis PA, Macias D, Veliça P, Rundqvist H, Fitzpatrick S, et al. An HIF-1a/VEGF-A Axis in Cytotoxic T Cells Regulates Tumor Progression. Cancer Cell. 2017;32:669–83.:e5. doi: 10.1016/j.ccell.2017.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hukelmann JL, Anderson KE, Sinclair LV, Grzes KM, Murillo AB, Hawkins PT, et al. The cytotoxic T cell proteome and its shaping by the kinase mTOR. Nat Immunol. 2016;17:104–12. doi: 10.1038/ni.3314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hermans D, Gautam S, García-Cañaveras JC, Gromer D, Mitra S, Spolski R, et al. Lactate dehydrogenase inhibition synergizes with IL-21 to promote CD8 T cell stemness and antitumor immunity. Proc Natl Acad Sci U S A. 2020 doi: 10.1073/pnas.1920413117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Khan AB, Carpenter B, Santos E, Sousa P, Pospori C, Khorshed R, Griffin J, et al. Redirection to the bone marrow improves T cell persistence and antitumor functions. J Clin Invest. 2018;128:2010–24. doi: 10.1172/JCI97454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Danilo M, Chennupati V, Silva JG, Siegert S, Held W. Suppression of Tcf1 by Inflammatory Cytokines Facilitates Effector CD8 T Cell Differentiation. Cell Rep. 2018;22:2107–17. doi: 10.1016/j.celrep.2018.01.072. [DOI] [PubMed] [Google Scholar]
- 32.Chen Z, Ji Z, Ngiow SF, Manne S, Cai Z, Huang AC, et al. TCF-1-Centered Transcriptional Network Drives an Effector versus Exhausted CD8 T Cell-Fate Decision. Immunity. 2019:840–55.:e5. doi: 10.1016/j.immuni.2019.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Khan O, Giles JR, McDonald S, Manne S, Ngiow SF, Patel KP, et al. TOX transcriptionally and epigenetically programs CD8 T cell exhaustion. Nature. 2019;571:211–8. doi: 10.1038/s41586-019-1325-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen J, López-Moyado IF, Seo H, Lio C-WJ, Hempleman LJ, Sekiya T, et al. NR4A transcription factors limit CAR T cell function in solid tumours. Nature. 2019;567:530–4. doi: 10.1038/s41586-019-0985-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li J, He Y, Hao J, Ni L, Dong C. High Levels of Eomes Promote Exhaustion of Anti-tumor CD8 T Cells. Front Immunol. 2018;9:2981. doi: 10.3389/fimmu.2018.02981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ke X, Fei F, Chen Y, Xu L, Zhang Z, Huang Q, et al. Hypoxia upregulates CD147 through a combined effect of HIF-1α and Sp1 to promote glycolysis and tumor progression in epithelial solid tumors. Carcinogenesis. 2012;33:1598–607. doi: 10.1093/carcin/bgs196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xu R, Wang K, Rizzi JP, Huang H, Grina JA, Schlachter ST, et al. 3-[(1,2,3)-2,3-Difluoro-1-hydroxy-7-methylsulfonylindan-4-yl]oxy-5-fluorobenzonitrile (PT2977), a Hypoxia-Inducible Factor 2a (HIF-2a) Inhibitor for the Treatment of Clear Cell Renal Cell Carcinoma. J Med Chem. 2019;62:6876–93. doi: 10.1021/acs.jmedchem.9b00719. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.