Abstract
Purpose
Therapies directed to specific molecular targets are still unmet for triple-negative breast cancer (TNBC) patients. Deubiquitinases (DUBs) are emerging drug targets. The identification of a highly active DUBs in TNBC may lead to novel therapies.
Experimental Design
Using DUB activity probes, we profiled global DUB activities in 52 breast cancer cell lines and 52 patients’ tumor tissues. To validate our findings in vivo, we employed both zebrafish and murine breast cancer xenograft models. Cellular and molecular mechanisms were elucidated using in vivo and in vitro biochemical methods. A specific inhibitor was synthesised and its biochemical and biological functions were assessed in a range of assays. Finally, we used patient sera samples to investigate clinical correlations.
Results
Two DUB activity profiling approaches identified UCHL1 as being highly active in TNBC cell lines and aggressive tumors. Functionally, UCHL1 promoted metastasis in zebrafish and murine breast cancer xenograft models. Mechanistically, UCHL1 facilitates TGFβ signaling-induced metastasis by protecting TGFβ type I receptor and SMAD2 from ubiquitination. We found that these responses are potently suppressed by the specific UCHL1 inhibitor, 6RK73. Furthermore, UCHL1 levels were significantly increased in TNBC patient sera, and highly enriched in sera exosomes as well as TNBC cell conditioned media. UCHL1 enriched exosomes stimulated breast cancer migration and extravasation, suggesting that UCHL1 may act in a paracrine manner to promote tumor progression.
Conclusion
Our DUB activity profiling identified UCHL1 as a candidate oncoprotein that promotes TGFβ-induced breast cancer metastasis and may provide a potential target for TNBC treatment.
Keywords: breast cancer, UCHL1, TGFβ/SMAD signaling, DUB inhibitor, exosomes
Introduction
Breast cancer is the most frequently diagnosed cancer in women (1), approximately 90% of breast cancer-related deaths are due to metastasis (2). During the metastasis process, epithelial-mesenchymal transition (EMT) plays an important role, which can be induced by the secreted cytokine transforming growth factor-β (TGFβ) (3). In the late stage of tumorgenesis, TGFβ stimulates cell invasion and modifies the microenvironment to promote cancer cell intravasation into nearby vessels, and stimulate extravasation into distant tissues and forming tumors-initiating seeds (4). Increasing evidence indicates that tumor cell-derived exosomes can profoundly influence the tumor local and systemic environment by transferring oncogenic cargo molecules (including protein, RNAs and lipids) to stromal or less aggressive tumor cells (5). Proteins that are enriched in circulating exosomes can be readily isolated from cancer patient blood and have been used as blood-based diagnostic and prognostic markers (6). Once metastasis has been triggered, current treatments frequently fail to provide durable responses (7). Therefore, an improved understanding of the underlying molecular and cellular mechanisms of metastasis is needed to better prevent and treat metastatic breast cancer.
As a highly heterogeneous disease, breast cancer can be classified into multiple subtypes with distinct metastatic potential based on genetic and clinical features (8). For instance, basal-like breast cancers are more aggressive than luminal and normal-like breast cancers (9), and estrogen receptor (ER) negative tumors are more aggressive than ER positive ones (10). The most aggressive subtype of breast cancer is triple-negative breast cancer (TNBC), which is defined as lacking expression of ER, progesterone receptor (PR), and human epidermal growth factor receptor 2 (HER2). TNBC subtype accounts for approximately 12% to 17% of breast cancers (11). TNBC remains the most challenging subtype of breast cancer to treat due to a low response rate to chemotherapy and lack of clinically meaningful molecular targets (12). Thus, there is an unmet need for newer molecular targets and effective drugs against these novel targets.
Post-translational modification of proteins by ubiquitination is emerging as a key regulatory mechanism in cell biology for regulating protein degradation and signaling activity (13). Ubiquitination is mediated by ubiquitin E3 ligase enzymes and reverted by deubiquitinases (DUBs). About 100 human DUBs have been identified and some of them play important roles in cancer progression (14). The majority of DUBs have a catalytic cysteine in the activity site of the protease, which render them attractive targets for small-molecule drug discovery screens (15). In recent studies, several independent groups have developed USP7 inhibitors (16–18), and, especially, the inhibitor FT671 showed significant inhibition of medulloblastoma, colorectal and lung tumors growth in mice (19). More than a decade after a Nobel prize was awarded for the discovery of the ubiquitin-proteasome system and clinical approval of proteasome and ubiquitin E3 ligase inhibitors, first-generation DUB inhibitors are now approaching to clinical trials (15). Besides, the development of DUB activity based probes (ABPs) provide very useful tools for monitoring target engagement and facilitate progress in drug discovery of DUBs (15,20).
Motivated to better understand the functional importance of differential DUB activities in breast cancer, we profiled DUB activity in different breast cancer subtypes with DUB ABPs. From these landscape profiles of DUB activities, we identified UCHL1 as being highly active in the more aggressive breast cancer subtype. Functionally, UCHL1 promoted TGFβ-induced breast cancer metastasis, and these responses were mitigated by genetic and pharmacological approaches. Furthermore, UCHL1 levels were significantly increased in exosome fractions of aggressive breast cancer patient sera. In this study, we also explored the function of UCHL1 enriched exosomes in promoting TNBC migration and extravasation.
Materials and Methods
Ethics Statement and Preparation of Clinical Samples
ER positive and negative fresh frozen tumor tissues and sera were randomly selected from the historical tumor biobank at the Erasmus MC Cancer institute. Use of biospecimen for biomarker research has been approved by the Medical Ethics Committee of the Erasmus MC and was performed in accordance to the Code of Conduct of the Federation of Medical Scientific Societies in the Netherlands (http://www.federa.org/). TNBC and control sera samples were collected by the Leiden University Medical Center (LUMC) Surgical Oncology Biobank between October 2002 and March 2013 according to a standardized protocol. This study was approved by the Medical Ethics Committee of the LUMC and was performed in accordance to the Code of Conduct of the Federation of Medical Scientific Societies in the Netherlands (http://www.federa.org/). Sera samples from 10 TNBC patients were selected that had no prior treatment and sera from 25 volunteers were selected as controls. Sera samples from TNBC and controls were stored at -80° C.
Cell Lines and Cell Culture
HEK293T and A549 cells were originally obtained from American Type Culture Collection (ATCC) and cultured in Dulbecco’s modified Eagles’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 100U/ml penicillin-streptomycin (15140122; Gibco). The 52 breast cancer cell lines that were used in this study were cultured in Roswell Park Memorial Institute (RPMI) medium (11875093; Gibco) supplemented with 10% FBS, 100 IU/ml penicillin-streptomycin. All the 52 breast cancer cell lines were molecularly and biochemically characterized and are listed in Supplementary Table S1. All the cells were routinely tested for absence mycoplasma contamination and checked for authenticity by STR profiling.
Zebrafish Extravasation Assay of Human Breast Cancer Cells
Transgenic zebrafish lines Tg (fli1: EGFP) were raised according to standard procedures in compliance with the local Institutional Committee for Animal Welfare of the Leiden University. Zebrafish extravasation assay were prepared as previous described (21). Zebrafish were fixed with 4% paraformaldehyde (PFA) 6 days after injection. Imaging and quantification of the results were carried out on an inverted SP5 STED confocal microscope (Leica), At least 40 zebrafish were analyzed for each group and 3 representative images were taken. All the experiments were repeated at least three times, and representative results are shown.
Breast Cancer Metastasis Assay in Mice Xenograft Model
Mice were purchased from the animal husbandry center of the Shanghai Institute Cell Biology, Academia Sinica, Shanghai, China. For the intracardial injection, five-weeks-old female BALB/c nude mice were anesthetized with isoflurane and single-cell suspension of MDA-MB-231 BM Luc (100.000 /100ul PBS) cells or MDA-MB-436 Luc (300.000 /100ul PBS) cells were inoculated into the left heart ventricle according to the method described by Arguello et al (22). Ten mice were injected in each group. Bioluminescent imaging was used to verify successful injection and to monitor the outgrowth of metastasis weekly. Mice experiments were approved by the Zhejiang University Animal Welfare Committee.
6RK73 Synthesis
The 6RK73 compound was synthesized according to a reported procedure (23). NMR and analytical LC-MS analysis was performed to confirm the nature and purity of the compound.
Exosome Isolation from Cell and Sera
Exosome isolation were performed as previous described (24,25). Supernatants from cell and sera samples from patients were concentrated by 100 K NMWL centrifugal filtration (UFC910024; Millipore) at 4 °C 10 x 103g and washed twice with PBS. Exosomes were recovered from the concentrated supernatant by ultracentrifugation at 100 x 103g for 17 hours at 4 °C. Exosome pellets were resuspended in ice-cold PBS at 4 °C. The concentration of exosomal proteins was quantified using DC protein assay (Pierce).
Statistical Analysis
Statistical analysis was performed using Prism 8 software (GraphPad La Jolla, USA). Numerical data from triplicates are presented as the mean ± SD, except for analysis of Zebrafish experiments where a representative result is expressed as mean ± SEM. The significance of differences between two independent subjects was determined using the unpaired Student’s t test. Two-way analysis of variance (ANOVA) has been used to analysis multiple subjects. The Kaplan-Meier method was used to evaluate metastasis free survival of mice between two groups. P value are indicated by asterisks in the figures: *, P < 0.05, **, P < 0.01, ***, P < 0.001 and ****, P < 0.0001. Differences at P =0.05 and lower were considered significant.
See supplementary information for additional descriptions regarding methods that were used.
Results
DUB activity profiling identified UCHL1 as a highly active DUB in aggressive breast cancer
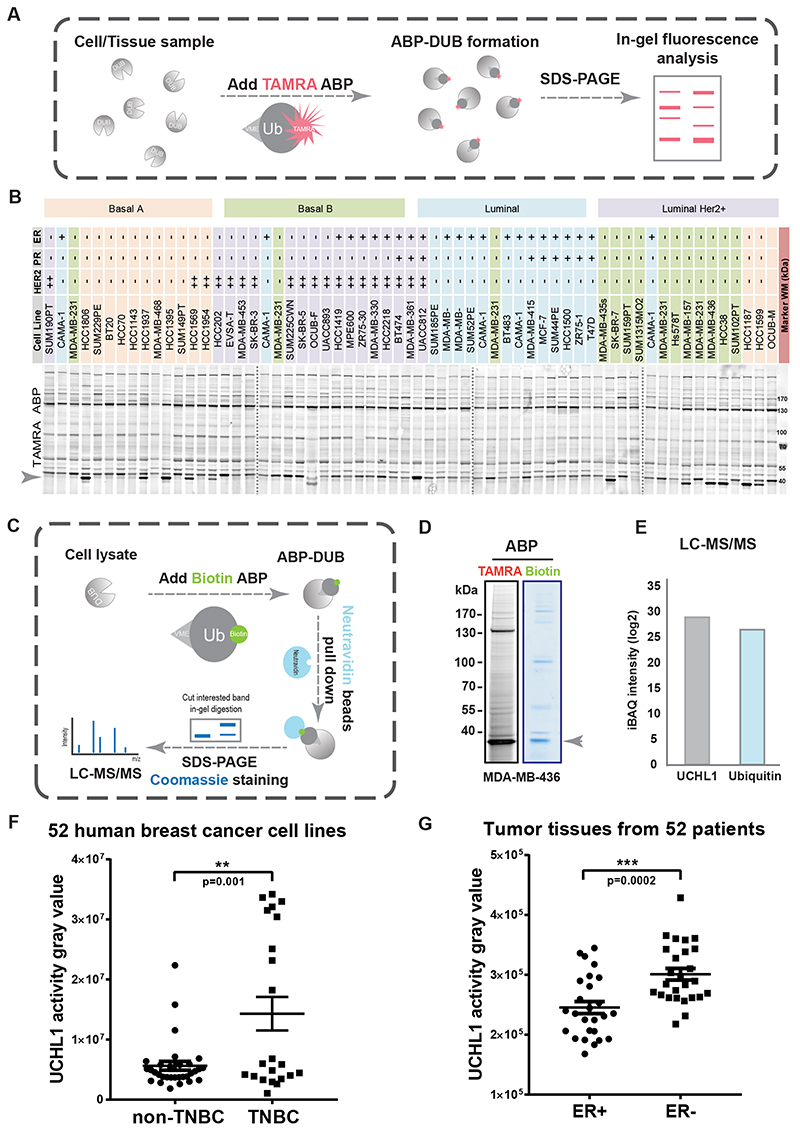
We first established a workflow to systematically determine the differential DUB activities in 52 human breast cancer cell lines and 52 breast cancer patient tumor tissues by using TAMRA-ubiquitin-VME, which is a ubiquitin-based activity probe for cysteine DUBs labeled on the N-terminus with a 5-carboxytetramethylrhodamine (TAMRA) dye and equipped with a reactive C-terminal vinyl methyl ester (VME) warhead (Fig. 1A). Among all the bands that were labelled with TAMRA ABP and visualized by fluorescence scanning, a band on the bottom of the gel displayed large variation in intensity levels between cell lines with representatives for Basal A, Basal B, Luminal, and Luminal HER2+ subtypes (Fig. 1B). To identify the DUB corresponding to this band, we used Biotin-ubiquitin-VME ABP to pull down the protein and identified it by liquid chromatography-tandem mass spectrometry (LC/MS-MS) (Fig. 1C). We performed the DUB identification in MDA-MB-436 cells, which showed strong intensity of the band of interest in the TAMRA and Biotin ABP result (Fig. 1D). The LC/MS-MS identified the DUB as UCHL1, and the Biotin-ubiquitin-VME ABPs were also identified and almost equally enriched with UCHL1 in the samples (Fig. 1E and Supplementary Fig. S1A). Next, we measured the intensities of the UCHL1-corresponding band in the TAMRA ABP profiling results by densitometry to compare UCHL1-corresponding activities between different breast cancer subtypes (Supplementary Table S1); UCHL1 activities were significantly increased in TNBC lines compared to non-TNBC cell lines (Fig. 1F). Next, DUB activity profiling with TAMRA ABP was performed in 26 ER+ and 26 ER- breast cancer patient tumor tissues (Supplementary Fig. S1B), and UCHL1-corresponding activities in ER- patient tumors were significantly higher than the activities in ER+ patient tumors (Fig. 1G and Supplementary Table S2).
Figure 1. DUB activity profiling identified UCHL1 as being selectively highly activated in aggressive breast cancer tumor tissues and cell lines.
A, Schematic overview of DUB activity profiling with TAMRA activity based probe (ABP). B, Atlas of DUB activity in 52 breast cancer cell lines. Four gels were merged together with dashed line in between two gels. C, DUB identification workflow with Biotin ABP. D, TAMRA ABP and Biotin ABP assay in MDA-MB-436 cells. E, LC-MS/MS analysis of in-gel tryptic digestion of excised gel slice indicated in figure 1D. F, UCHL1 activity analysis of 52 breast cancer cell lines. **, P < 0.01, unpaired Student t test. G, UCHL1 activity gravy value analysis of 52 tissues from breast cancer patients. ***, P < 0.001, unpaired Student t test.
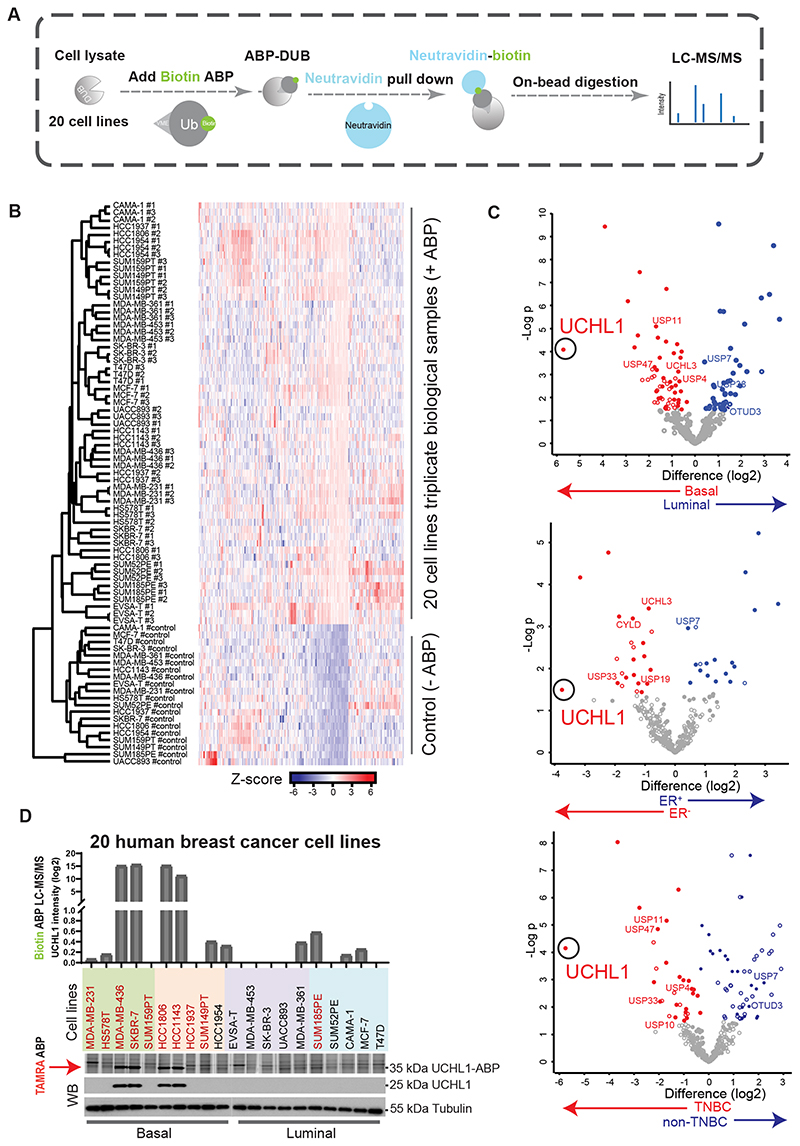
The second parallel DUB activity profiling was performed with Biotin-ubiquitin-VME ABP combined with LC/MS-MS analysis in 20 randomly picked up Basal and Luminal human breast cancer cell lines (Fig. 2A; Supplementary Table S3). All the targets identified by LC/MS-MS were plotted by hierarchical clustering to compare biological replicates (Fig. 2B). Average label-free quantification (LFQ) log2 difference between Basal and Luminal, ER+ and ER-, and TNBC and non-TNBC subtype cell lines revealed that UCHL1 activity was highly enriched in Basal, ER negative and TNBC subgroups (Fig. 2C; Supplementary Table S4). To further validate the Biotin ABP profiling result of UCHL1, we compared UCHL1 activity detected by Biotin ABP and TAMRA ABP profiling, with the UCHL1 protein level measured by Western Blot (WB) in these 20 breast cancer cell lines (Fig. 2D). Both profiling results of UCHL1 activity showed similar results, and the UCHL1 protein level detected by WB was found to be a major determinant for UCHL1 activity level (Fig. 2D). Taken together, both DUB activity profiling methods identified UCHL1 as being highly activated in aggressive breast cancer.
Figure 2. Quantitative DUB activity-based proteomic profiling identified UCHL1 as being selectively highly active in TNBC cell lines.
A, Schematic overview of quantitative DUB activity profiling with Biotin ABP. B, Heatmap depicting sample clustering considering Z-score of proteins identified by LC-MS/MS after Biotin-ABP profiling. The tree indicates Euclidean distances between samples. C, Scatterplots depicting statistical differences between cell lines grouped by tumor subtype. A filled dot indicates that a protein is statistically significantly different between any of the groups of study and the control sample set. An empty dot indicates that there is no statistically significant difference between any of the groups of study and the control sample group. Location of UCHL1 is marked with a circle. D, Biotin ABP, TAMRA ABP and WB analysis of UCHL1 in 20 breast cancer cell lines, TNBC cell lines were highlighted with red color. Two blots were merged together with a grey line in between two blots. Same blot was used for UCHL1 and Tubulin (loading control).
UCHL1 promotes breast cancer metastasis in xenograft models
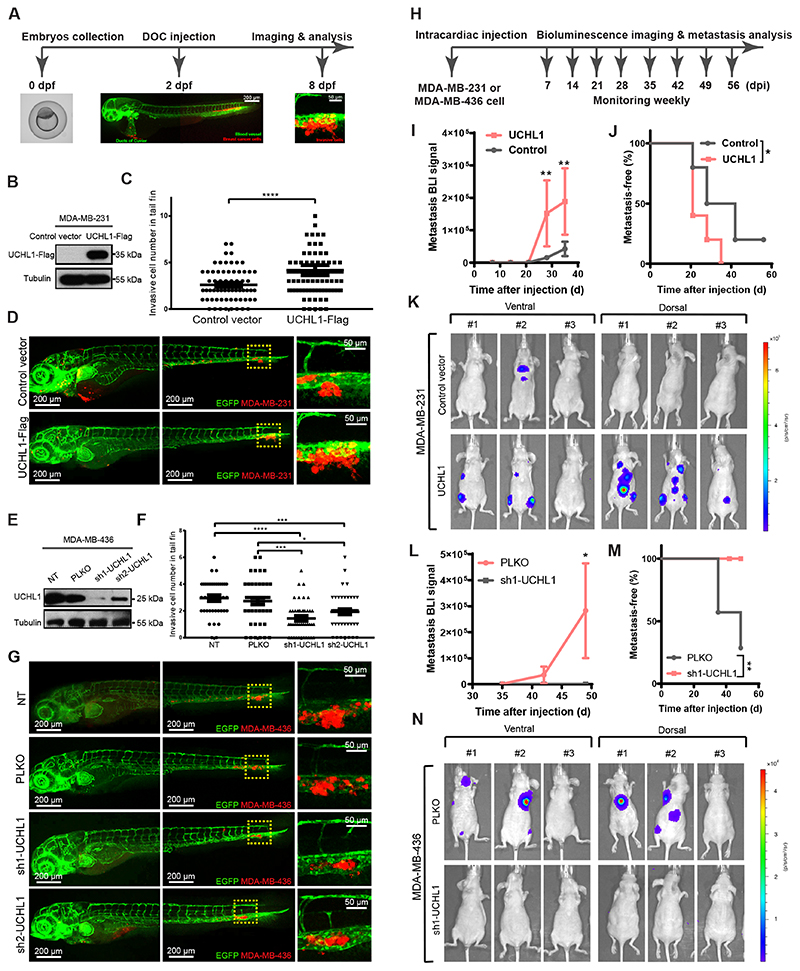
To explore the role of UCHL1 activity in breast cancer metastasis, we first analyzed the effect of its misexpression in breast cancer cells on extravasation in a zebrafish breast cancer xenograft model (Fig. 3A). First, we overexpressed UCHL1 in mCherry-expressing MDA-MB-231 cells, which has a low endogenous UCHL1 expression/activity level as determined by WB and TAMRA ABP assays (Fig. 3B and Supplementary Fig. S2A). Injection of the same number of cells into the circulation of zebrafish embryos revealed after 6 days significantly increased number of invasive cells in the UCHL1-Flag group compared to the vector control group (Fig. 3C and D). The proliferation of both cell lines when grown on plastic showed no significant difference (Supplementary Fig. S2B). Next, we knocked down UCHL1 in mCherry-expressing MDA-MB-436 cells, which have high endogenous UCHL1 expression/activity level by using two independents short hairpin RNAs (shRNAs). The knockdown efficiency was validated by WB and TAMRA ABP assays (Fig. 3E and Supplementary Fig. S2C). The UCHL1 knockdown groups revealed not only less invasive cells but also a weaker metastatic phenotype (cells were unable to extravasate into zebrafish tail fin and formed clusters in between the blood vessels) compared with a non-targeting (NT) shRNA and empty vector (PLKO) control groups (Fig. 3F and G). The proliferation was not affected by UCHL1 depletion (Supplementary Fig. S2D).
Figure 3. UCHL1 promotes breast cancer metastasis in zebrafish and mice xenograft models.
A, Workflow of breast cancer extravasation experiment in a zebrafish model. The blood vessels and cancer cells are fluorescently labelled in green and red, respectively. B, UCHL1 overexpressing and control vector expressing MDA-MB-231 cell lines were established and validated by WB. Same blot was used for UCHL1 and Tubulin (loading control). C, Analysis of invasive cell numbers of control and UCHL1 groups in zebrafish metastasis experiment. ****, P < 0.0001, unpaired Student t test. D, Representative images of zebrafish from the control and the UCHL1 group with zoom-in of invasive cells on the right panel. E, Two UCHL1 shRNA knock down MDA-MB-436 cell lines and two control cell lines PLKO (empty vector) and NT (non-target) were established and validated by WB. Same blot was used for UCHL1 and Tubulin (loading control). F, Analysis of invasive cell numbers of each group in zebrafish metastasis assay. *, P < 0.05, ***, P < 0.001 and ****, P < 0.0001, two-way analysis of variance (ANOVA). The location of nuclear and plasma membrane are indicated with a dashed line. G, Representative images from 4 groups with zoom-in on the right panel. H, Workflow of breast cancer metastasis experiment in mouse model. I, Bioluminescence imaging (BLI) signal of metastasis of control and UCHL1 overexpression in MDA-MB-231 cells were measured at indicated times. **, P < 0.01, two-way ANOVA. J, Percentage of metastasis-free mice in each group followed in time. *, P < 0.05, Log-rank test. K, BLI signal of metastasis of 3 representative mouse images with both ventral and dorsal side from each group at day 35 after injection. L, BLI signal of metastasis of PLKO and sh1-UCHL1 MDA-MB-436 cells were measured at indicated times. *, P < 0.05, two-way ANOVA. M, Percentage of metastasis-free mice in each group followed in time. **, P < 0.01, Log-rank test. N, BLI signal of metastasis of 3 representative mouse images with both ventral and dorsal side from each group at day 49 after injection.
Next, to further confirm that UCHL1 promotes breast cancer metastasis, we used a mouse breast cancer xenograft model in which we intracardially injected breast cancer cells stably expressing firefly luciferase, into female BALB/c athymic nude mice. Bioluminescent images (BLI) were taken every week to monitor colonization in different organs after successful injection (Fig. 3H). UCHL1-overexpressing MDA-MB-231 cells exhibited significantly increased metastasis in different organs 35 days after injection (Fig. 3I and K) and shorter metastasis-free survival periods than the empty-vector control group (Fig. 3J). Furthermore, nude mice were intracardially injected with luciferase-labelled PLKO control and sh1-UCHL1 knockdown MDA-MB-436 cells. The PLKO group showed metastasis in different organs at 49 days after injection (Fig. 3L and N), and shorter metastasis-free survival periods than sh1-UCHL1 group (Fig. 3M). Altogether, the mice and zebrafish results confirm that UCHL1 promotes breast cancer invasion and metastasis.
UCHL1 facilitates TGFβ signaling-induced TNBC migration and extravasation by protecting TβRI and SMAD2 from ubiquitination
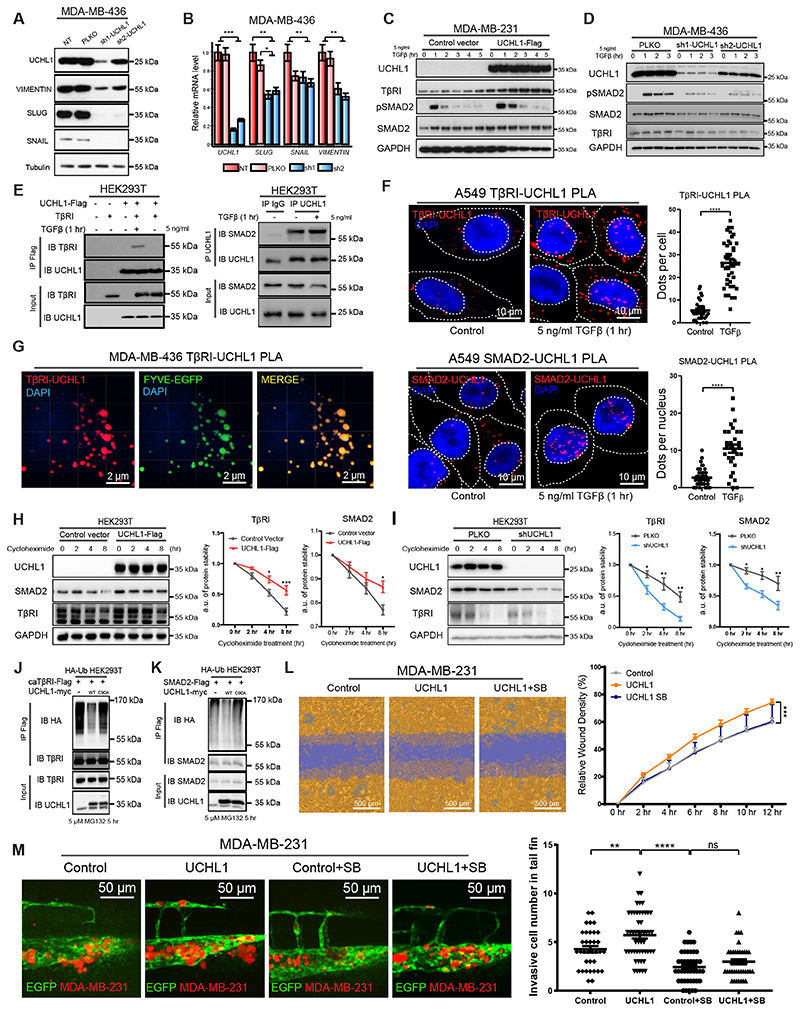
Next, we investigated the underlying mechanism by which UCHL1 promoted breast cancer metastasis. Since EMT plays an important role during breast cancer metastasis (3), we firstly tested the effect of UCHL1 depletion in MDA-MB-436 cells on the levels of several mesenchymal markers. Knockdown of UCHL1 significantly decreased VIMENTIN, SNAIL and SLUG expression both at the RNA and protein level (Fig. 4A and B). In addition, qPCR results showed a modest decrease of β-CATENIN, ZEB1 and ZEB2 expression upon UCHL1 depletion (Supplementary Fig. S3A). Since TGFβ is a key activator of EMT, we next examined whether UCHL1 can control TGFβ signaling. Indeed, ectopic expression of UCHL1 in MDA-MB-231 cells (low endogenous UCHL1 activity) promoted TGFβ-induced pSMAD2 levels, and this coincided with increased TGFβ type I receptor (TβRI) and SMAD2 levels (Fig. 4C). Knockdown of UCHL1 in MDA-MB-436 cells (high endogenous UCHL1 activity) suppressed pSMAD2, TβRI and SMAD2 levels (Fig. 4D). Besides, ectopic expression of UCHL1 in HEK293T cells upregulated the TGFβ-induced SMAD3/4 driven transcriptional CAGA12-luc response, whereas knockdown of UCHL1 decreased this effect significantly (Supplementary Fig. S3C). To investigate whether UCHL1 interacts with TβRI, we performed immunoprecipitation (IP) of UCHL1 followed by WB for TβRI using HEK293T cell lysates. We observed that Flag-tagged UCHL1 interacted with both overexpressed and endogenous TβRI upon TGFβ treatment (Fig. 4E and Supplementary Fig. S3D). Besides, we found that recombinant UCHL1 preferentially binds to ubiquitinated TβRI. This post-translational modification of TβRI is triggered by TGFβ treatment (Supplementary Fig. S3E). We also found in IP-WB experiments that UCHL1 interacts with SMAD2 in HEK293T cells. This endogenous interaction was not TGFβ dependent (Fig. 4E). As UCHL1 is a small protein in which either deletion of N- or C-terminal sequences will result in loss of solubility and misfolding (26), it is not possible to make truncated versions to study the responsible domain of UCHL1 for the interaction with TβRI and SMAD2. We therefore resorted to investigate the effect of specific amino acid mutations in UCHL1 on the interaction of UCHL1 with TβRI or SMAD2. These mutations were previously shown to interfere with UCHL1 DUB function (26,27). Our results showed that the catalytic triad mutants (C90S, C90A, D176N, H161D and H161Y), ubiquitin binding mutant (D30K), and double mutant (D30K & D176N) which are defective in DUB activity as measured by TAMRA-ABP assay, still interact with TβRI or SMAD2. However, the S18Y mutant of UCHL1 retained DUB activity in TAMRA-ABP assay but demonstrated a decreased interaction with TβRI or SMAD2 (Supplementary Fig. S4A and B). These results suggest the N-terminal region in vicinity of Serine18 in UCHL1 plays an important role in the interaction with TβRI or SMAD2. To further validate and investigate the endogenous interactions and subcellular localization of TβRI-UCHL1 and SMAD2-UCHL1, we performed proximity ligation assays (PLA) in A549 cells. A549 cells were chosen as they contain a large cytoplasm unlike MDA MB 436 and HEK293T cells; the large cytoplasm facilitates studies on subcellular distribution. Results showed that the interaction between UCHL1 and TβRI occurs in a ligand dependent manner (Fig. 4F and Supplementary Fig. S3G-H). The interaction between UCHL1 and SMAD2 is not ligand dependent, but we found that the subcellular location of the interaction shifted from cytoplasm to nucleus after TGFβ treatment (Fig. 4F and Supplementary Fig. S3F-H). Importantly, we found that the interaction between UCHL1 and TβRI occurs in the early endosome. The PLA signals for UCHL1-TβRI co-localized to a large extent with the early endosome marker FYVE-EGFP (Fig. 4G and supplementary video).
Figure 4. UCHL1 regulates mesenchymal phenotype of breast cancer cells and promotes TGFβ/SMAD signaling induced breast cancer extravasation.
A, WB analysis of mesenchymal markers in UCHL1 shRNA knock down MDA-MB-436 cells. Same blot was used for UCHL1, SLUG and Tubulin (loading control). VIMENTIN and SNAIL blotting results were obtained from another blot using the same corresponding cell lysates. B, qPCR analysis of mesenchymal markers in UCHL1 shRNA knock down MDA-MB-436 cells. **, P < 0.01, ***, P < 0.001, two-way ANOVA C, WB analysis of TβRI, SMAD2 and TGFβ-induced pSMAD2 in control and UCHL1 overexpressed MDA-MB-231 cells. Same blot was used for TβRI and GAPDH (loading control). UCHL1 and pSMAD2 blotting results were obtained from another blot using the same corresponding cell lysates. SMAD2 result were obtained from another blot using the same corresponding cell lysates. D, WB analysis of TβRI, SMAD2 and pSMAD2 in PLKO and UCHL1 shRNA knockdown MDA-MB-436 cells. Same blot was used for UCHL1, pSMAD2 and GAPDH (loading control). TβRI and SMAD2 results were derived from another two blots using the same corresponding cell lysates. E, The Interaction of UCHL1 with TβRI was detected by immunoprecipitation (IP) of Flag-tagged UCHL1 and immunoblotting (IB) for TβRI in HEK293T cells (left). The endogenous interaction of UCHL1 with SMAD2 was detected by IP of endogenous UCHL1 and IB for SMAD2 in HEK293T cells (right). IP results were obtained from same blot. Input results were from another blot using the same corresponding cell lysates as used for IP. F, PLA of TβRI-UCHL1 and SMAD2-UCHL1 in A549 cells treated with or without 5 ng/ml TGFβ for 1 hour. Representative images are shown in the left panel and signal analysis are shown in the right panel. ****, P < 0.0001, unpaired Student t test. G, PLA of TβRI-UCHL1 in MDA-MB-436 cells transfected with early endosome marker FYVE-EGFP and treated with 5 ng/ml TGFβ for 1 hour. H, Expression levels of TβRI and SMAD2 were analysed by IB in UCHL1 overexpressed and control HEK293T cells treated with 10 μg/ml cycloheximide for the indicated times. WB results are shown in the left panel, and quantification of protein stability of TβRI and SMAD2 are shown in the two panels on the right. Same blot was used for TβRI and GAPDH (loading control). UCHL1 and SMAD2 blotting results were derived from another blot using the same corresponding cell lysates. I, Expression levels of TβRI and SMAD2 were analysed by IB in PLKO and shUCHL1 HEK293T cells treated with 10 μg/ml cycloheximide for the indicated times. WB results are shown in the left panel, protein stability analysis of TβRI and SMAD2 are shown in the right panel. Same blot was used for TβRI and GAPDH (loading control). UCHL1 and SMAD2 results were derived from another blot using the same corresponding cell lysates. J, Ubiquitination of TβRI was detected by IP of Flag-tagged constitutively active TβRI (caTβRI) from HA-Ub transfected HEK293T cells with WT-UCHL1-myc or C90A-UCHL1-myc overexpression. IP results were obtained from same blot. Input results were obtained from another blot using the same corresponding cell lysates. K, Ubiquitination of SMAD2 was detected by IP of Flag-tagged SMAD2 from HA-Ub transfected HEK293T cells with WT-UCHL1-myc or C90A-UCHL1-myc overexpression. IP results were obtained from same blot. Input results were obtained from another blot using the same corresponding cell lysates. L, Real-time scratch assay results of Control, UCHL1 and UCHL1+SB MDA-MB-231 cells. Representative scratch wounds are shown at the end time point of the experiment (left). The region of the original scratch is coloured in purple and the area of cell is coloured in yellow. Relative wound density (closure) were plotted at indicate times (right). M, In vivo zebrafish extravasation assay of UCHL1 overexpressed and control vector expressed MDA-MB-231 cells treated with or without TβRI kinase inhibitor SB-431542. SB groups zebrafish were treated with 5 μM inhibitor in the egg water for 6 days after injection, and refreshed every other day. **, P < 0.01, ****, P < 0.0001, two-way ANOVA. Representative images from 4 groups with zoom in of the tail fin area are shown in the left panel. Analysis of invasive cell number of Control, Control+SB, UCHL1 and UCHL1+SB groups in zebrafish extravasation assay are shown in the right panel.
Next, we investigated whether UCHL1 can stabilize TβRI and SMAD2 protein levels. We examined the stability of TβRI or SMAD2 in the presence of cycloheximide upon misexpression of UCHL1 in HEK293T cells. Results demonstrated that UCHL1 overexpressing cells showed longer protein half-lives of TβRI and SMAD2 than control cells, while UCHL1 knockdown cells showed shorter protein half-lives of TβRI and SMAD2 than the PLKO cells (Fig. 4H and I). The mRNA level of TβRI and SMAD2 were not significantly different between UCHL1 overexpressing and knock down cells, compared to their control cells (Supplementary Fig. S4C and D). Thereafter, we investigated whether UCHL1 protects TβRI and SMAD2 from ubiquitination. We tested the ubiquitination of constitutively active TβRI (caTβRI) and SMAD2 with overexpression of wild type (WT) and catalytic inactive mutant (C90A) UCHL1. Results showed that only WT UCHL1, but not C90A UCHL1 mitigates caTβRI and SMAD2 ubiquitination (Fig. 4J and K). In addition, we found that UCHL1 mainly regulates lysine 48-linked poly-ubiquitination of TβRI and SMAD2 (Supplementary Fig. S4E and F). Besides, we found that recombinant UCHL1 protein is able to deubiquitinate TβRI and SMAD2 in vitro directly, and N-ethylmaleimide (NEM) treatment blocked this process by inhibiting UCHL1 DUB activity (Supplementary Fig. S4H and I).
To investigate whether UCHL1-induced metastasis is dependent on its ability to potentiate TGFβ signaling, we employed the selective TβRI /SMAD signaling inhibitor SB431542 (SB) to block TGFβ receptor signaling in migration and extravasation assays. In a scratch assay, SB treatment blocked the ability of UCHL1 to promote MDA-MB-231 cell migration (Fig. 4L). Results of the extravasation assay showed that SB also blocked the stimulatory effect of UCHL1 on MDA-MB-231 cell extravasation in an in vivo zebrafish xenograft model (Fig. 4M). Besides, we performed functional rescue experiments. Ectopic expression of TβRI, SMAD2 or SMAD3 mimicked the promoting effect of UCHL1 on migration of MDA-MB-231 cells as measured by real-time imaging system (Supplementary Fig. S5A). In addition, we found that overexpression of TβRI, SMAD2 or SMAD3 partially compensated the inhibitory effect of UCHL1 knock down in MDA-MB-436 cells in a migration assay (Supplementary Fig. S5B). Altogether, these results demonstrate that UCHL1 facilitates TGFβ signaling-induced TNBC migration and extravasation by protecting TβRI and SMAD2 from ubiquitination. Besides, we found that DUB activity of UCHL1 is required for the metastasis-promoting activity of UCHL1. When we overexpressed C90A catalytically inactive mutant of UCHL1 in MDA-MB-231 cells, UCHL1 lost its promotion function and showed slightly dominant-negative regulation of migration and extravasation (Supplementary Fig. S5C and D).
UCHL1 activity inhibitor antagonizes TGFβ/SMAD signaling and inhibits breast cancer migration and extravasation
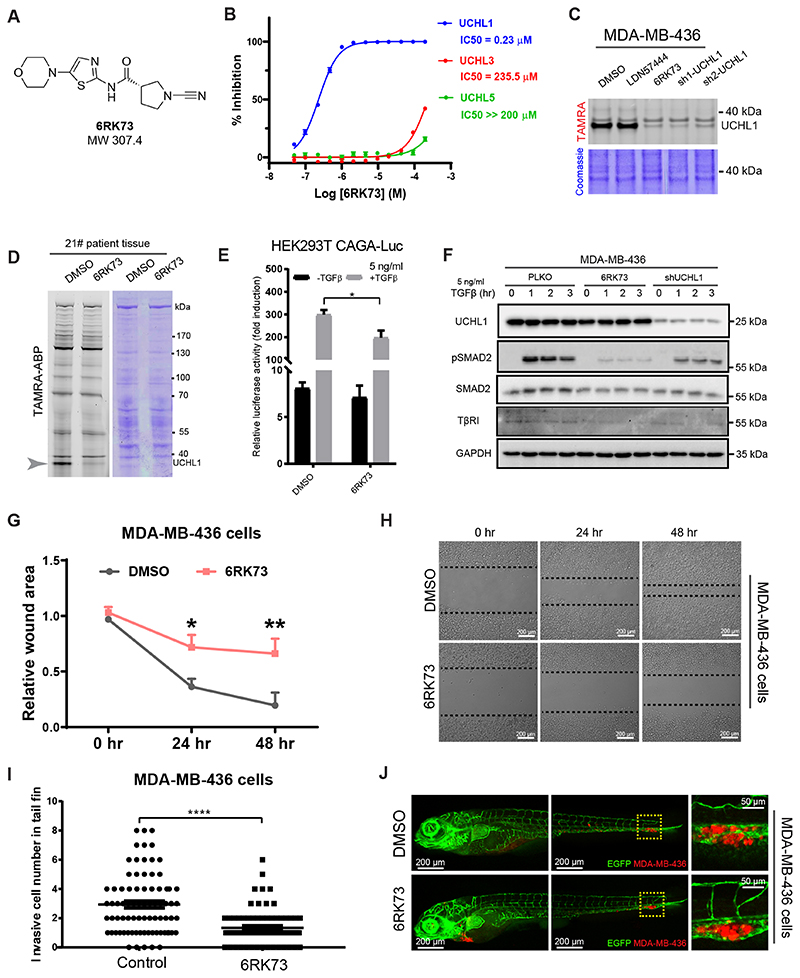
In order to study the effect of UCHL1 activity inhibition on the TGFβ pathway and breast cancer metastasis we turned to a recently reported panel of UCHL1 inhibitors and decided to synthesize and characterize one of the most potent ones (23). This compound, 6RK73 covalently binds to UCHL1 (Fig. 5A and Supplementary Fig. S6A), showed excellent inhibitory potency towards UCHL1 based on an in vitro half-maximum inhibitory concentration (IC50) assay, and a high selectivity over other DUBs including its closest family members UCHL3 and UCHL5 (Fig. 5B). We examined its effect in living cells, and we took along the reversible competitive UCHL1 inhibitor LDN57444. 6RK73 showed more potent inhibition than LDN57444 on UCHL1 activity in MDA-MB-436 cells by TAMRA ABP assay; the inhibitory efficiency of 6RK73 was comparable to genetic knockdown of UCHL1 (Fig. 5C). Next, we used 6RK73 to test its specificity against UCHL1 activity on all the DUBs by performing a TAMRA ABP assay in patient tumor specimen, only UCHL1 band decreased among all the DUBs detected (Fig. 5D). We can conclude that 6RK73 displays a potent and specific inhibitory effect on UCHL1 both in vitro and in vivo.
Figure 5. UCHL1 activity inhibitor antagonizes TGFβ signaling and inhibits breast cancer migration and extravasation.
A, Chemical structure of the selective covalent UCHL1 activity inhibitor 6RK73. B, IC50 analysis of 6RK73. C, TAMRA ABP analysis of UCHL1 reversible activity inhibitor LDN67444 and covalent activity inhibitor 6RK73 in MDA-MB-436 cells, 5 μM LDN57444 or 6RK73 was added to the cells overnight. TAMRA and Coomassie results were obtained from the same gel. D, TAMRA ABP analysis of 6RK73 in 21# patient specimen, 5 μM 6RK73 was added in the lysate for 30 min. TAMRA and Coomassie results were obtained from the same gel. E, CAGA12-Luc reporter analysis of 6RK73 in HEK293T cells, 5 μM 6RK73 was added to the cells overnight. *, P < 0.05, two-way ANOVA. F, WB analysis of TβRI, SMAD2 and pSMAD2 in MDA-MB-436 cells treated with or without 5 μM 6RK73 overnight. Same blot was used for UCHL1, pSMAD2 and GAPDH (loading control). TβRI and SMAD2 were obtained from another two blots using the same corresponding cell lysates. G, In vitro scratch wound healing assay of MDA-MB-436 cells treated with and without 5 μM 6RK73 for 48 hr, time-lapse imaging was performed every hour. Relative wound area was analysed for each group at indicate times. *, P < 0.05, **, P < 0.01, two-way ANOVA. H, Representative images of cells from DMSO and 6RK73 groups. I, In vivo zebrafish extravasation assay of MDA-MB-436 cells, in which the injected zebrafish were treated with or without 6RK73 for 6 days. 5 μM 6RK73 was added in the egg water and refreshed every other day. Invasive cell number were analysed for DMSO and 6RK73 groups. ****, P < 0.0001, unpaired Student t test. J, Representative images of zebrafish from DMSO and 6RK73 groups with zoom in of invasive cells are shown in the right panel.
To investigate whether 6RK73 can inhibit TGFβ signaling, we performed a CAGA12-Luc transcriptional reporter assay in HEK293T cells. Treating cells with 6RK73 inhibited the TGFβ/SMAD-induced transcriptional response (Fig. 5E). Moreover, 6RK73 treatment of MDA-MB-436 cells displayed strong inhibition of the TGFβ-induced pSMAD2 and pSMAD3, and a decrease of TβRI and total SMAD protein levels; the inhibitory efficiency of 6RK73 was stronger than the effect observed after shRNA-mediated UCHL1 knockdown (Fig. 5F and Supplementary Fig. S6B). Furthermore, we tested the effect of 6RK73 on migration of MDA-MB-436 cells. 6RK73 treated MDA-MB-436 cells migrated significantly slower than the DMSO control group (Fig. 5G and H). To study 6RK73 function in extravasation, we used the MDA-MB-436 cells injected zebrafish xenograft treated with DMSO or 6RK73 that was added in the egg water surrounding the zebrafish embryos. Extravasation of the cells in 6RK73-treated zebrafish were potently inhibited (Fig. 5I and J). Taken together, 6RK73 showed specific inhibition of UCHL1 activity and TGFβ/SMAD2 and SMAD3 signaling, and potent inhibition of breast cancer migration and extravasation.
UCHL1+ exosomes upregulate TGFβ signaling and serves as blood-based biomarker for aggressive breast cancer.
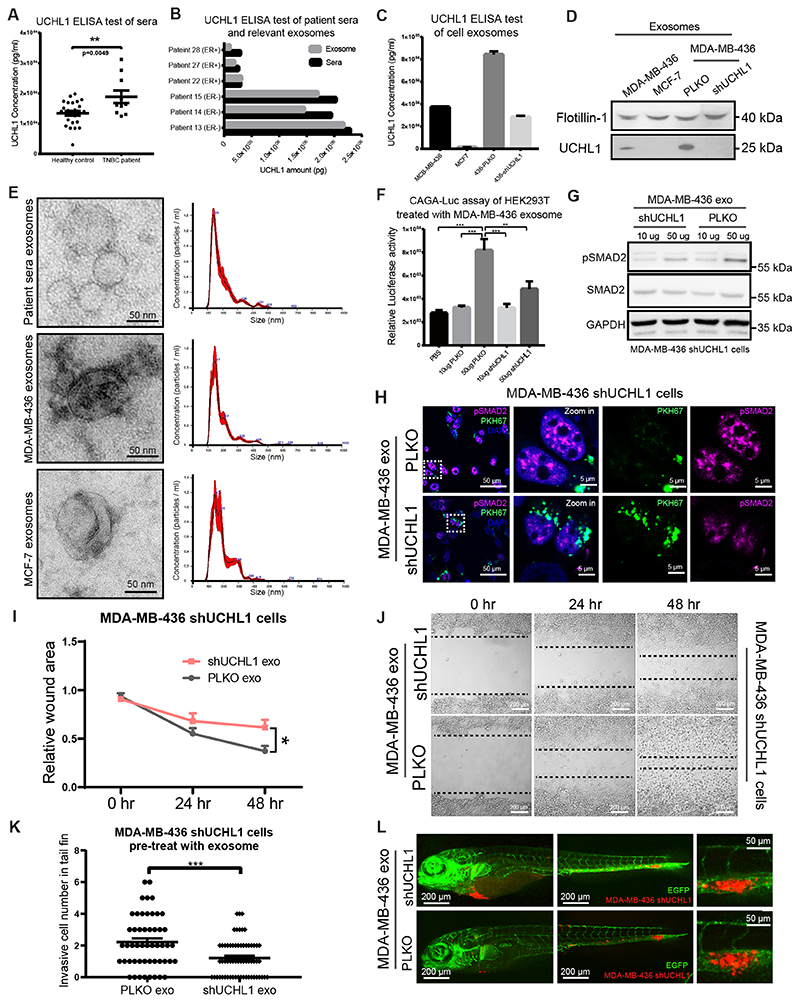
Clinically, UCHL1 has successfully been used as blood biomarker for traumatic brain injury and concussion (28). However, it is unknown whether UCHL1 has a role and/or is present in breast cancer patient sera. Therefore, we performed a UCHL1 enzyme-linked immunosorbent assay (ELISA) on sera samples collected from TNBC patients and healthy donors. Interestingly, UCHL1 protein levels in the TNBC group are significantly higher as compared to healthy controls (Fig. 6A). In addition, UCHL1 levels are also higher in ER negative patient sera than in sera from ER positive cases (Supplementary Fig. S6C). Nevertheless, UCHL1 is not a secreted protein, and this raised the question on how UCHL1 can be found in the blood circulation. A recent study demonstrated that cancer overexpressed proteins can be packaged in exosomes and enter circulation which is useful for minimally-invasive cancer detection (29). We hypothesized that UCHL1 overexpressed in aggressive breast cancer may be secreted via exosomes by cells and that circulated in the blood of patients via exosomes. To further verify this hypothesis, we isolated exosomes from breast cancer patient sera by differential ultracentrifugation (24), and found that the total amount of UCHL1 in sera is highly enriched in the isolated exosomes fraction (Fig. 6B). Next, we isolated exosomes from conditioned media of MDA-MB-436 (TNBC) and MCF-7 (non TNBC) cell cultures. ELISA results showed that the UCHL1 levels were higher in exosomes isolated from MDA-MB-436 cells than from MCF-7 cells. Moreover, UCHL1 level significantly decreased in exosomes isolated from MDA-MB-436 cells that were depleted for UCHL1 compared with PLKO cells (Fig. 6C). WB analysis of UCHL1, and that of a common exosomal marker protein Flotillin-1, showed lower UCHL1 levels in MCF7 exosomes than MDA-MB-436 exosomes, and a decreased UCHL1 level in shUCHL1 MDA-MB-436 exosomes (Fig. 6D). Exosomes are extracellular vesicles with a diameter of 50-200 nm (30). To further characterize the exosomes we isolated them from sera and cell conditioned media, we performed transmission electron microscopy (TEM) imaging to show the shape and size of these purified exosomes, and used nanoparticle tracking analysis (NTA) to determine their concentrations and size distributions. Results showed that both sera and cell samples displayed exosome-typical size and morphology by TEM analysis, and were enriched in the size from 100 nm to 200 nm vesicles by NTA analysis (Fig. 6E). Taken together, UCHL1 levels were significantly increased in TNBC patient sera, and highly enriched in exosomes of aggressive tumor bearing patient sera and TNBC cell conditioned media.
Figure 6. UCHL1+ exosomes upregulate TGFβ signaling and serves as blood-based biomarker for aggressive breast cancer.
A, ELISA analysis of UCHL1 levels in serum samples from healthy donors and TNBC patients. **, P < 0.01, unpaired Student t test. B, ELISA analysis of UCHL1 level in serum and relative exosomes from 6 breast cancer patients. C, ELISA analysis of UCHL1 level in exosomes from breast cancer cell lines. D, WB analysis of exosomes maker Flotillin-1 and UCHL1 in exosomes from four breast cancer cell lines. Same blot was used for UCHL1 and Flotillin-1 (loading control). E, TEM imaging of exosomes from patient serum and breast cancer cell lines (left). Nanoparticle tracking analysis (NTA) of relative exosomes (right). X-axis represents exosomes size distribution. Y-axis shows the concentration of exosomes. F, CAGA12-Luc transcriptional reporter analysis of HEK293T cells treated with UCHL1 high or low exosomes isolated from PLKO or shUCHL1 MDA-MB-436 cells. **, P < 0.01, ***, P < 0.001, two-way ANOVA. G, WB analysis of SMAD2 and pSMAD2 in MDA-MB-436 UCHL1 stable known down cells treated with exosomes isolated from PLKO or shUCHL1 MDA-MB-436 cells. Same blot was used for pSMAD2 and GAPDH (loading control). SMAD2 blotting results were obtained from another blot using the same corresponding cell lysates. H, Immunofluorescence staining of pSMAD2 in MDA-MB-436 UCHL1 stable known down cells treated with exosomes isolated from PLKO or shUCHL1 MDA-MB-436 cells labelled with green PKH67 exosomes dye. I, In vitro scratch wound healing assay of shUCHL1 MDA-MB-436 cells pre-treated with exosomes isolated from PLKO or shUCHL1 MDA-MB-436 cells for 24 hr, time-lapse imaging was carried out for 48 hours, images were taken every hour. Relative wound areas were analysed for each group at indicate times. *, P < 0.05, two-way ANOVA. J, Representative images of cells from PLKO and shUCHL1 groups. K, In vivo extravasation assay of zebrafish injected with shUCHL1 MDA-MB-436 cells pre-treated with exosomes isolated from PLKO or shUCHL1 MDA-MB-436 cells for 24 hr. Invasive cell number were analysed for PLKO and shUCHL1 groups. ***, P < 0.001, unpaired Student t test. L, Representative images of zebrafish from PLKO and shUCHL1 groups with zoom in of invasive cells on the right panel.
To further investigate whether UCHL1+ exosomes regulate TGFβ/SMAD signaling, we first tested the effect of the PLKO and shUCHL1 MDA-MB-436 exosomes on the CAGA12-Luc transcriptional reporter activity in HEK293T cells. Treatment of UCHL1 containing exosomes resulted in higher luciferase signal in HEK293T cells than exosomes in which UCHL1 was depleted (Fig. 6F). Consistent with this finding, pSMAD2 levels were also increased upon treatment with control exosomes from MDA-MB-436 cells but not by exosomes depleted of UCHL1 as analyzed by WB in MDA-MB-436 cells with UCHL1 knockdown (Fig. 6G). To further validate these results, we labelled the exosomes that were isolated from PLKO and shUCHL1 MDA-MB-436 cells with a fluorescent lipid dye (PKH67), and thereafter added them to MDA-MB-436 cells with UCHL1 knockdown. After the exosomes were taken up by the cells, we performed immunofluorescent (IF) staining of pSMAD2. Confocal microscopy imaging revealed that the pSMAD2 levels were higher in the cells which were treated with PLKO MDA-MB-436 cells exosomes than shUCHL1 MDA-MB-436 cells exosomes (Fig. 6H). To further evaluate the biological function of UCHL1+ exosomes, we examined their effect on the migration of MDA-MB-436 cells with UCHL1 knockdown. Cells treated with PLKO MDA-MB-436 exosomes migrated more than exosomes depleted for UCHL1 (Fig. 6I and J). The potential function of UCHL1+ exosomes was further validated in a zebrafish xenograft model by injecting MDA-MB-436 cells with UCHL1 knockdown. The cells pre-treated with PLKO MDA-MB-436 exosome showed more invasion and stronger extravasation phenotype than the cells pre-treated with shUCHL1 MDA-MB-436 exosome (Fig. 6K and L). Taken together, donor cells highly active for UCHL1 can via exosome transfer upregulate TGFβ/SMAD signaling in recipient cells and promote their migration and extravasation.
Discussion
Large-scale conventional genomic and proteomic profiling have been performed in breast cancer (31), and a growing numbers of DUBs have been uncovered to be aberrantly expressed in breast cancer (32). However, there is still very little knowledge on the overall activities of DUBs in breast cancer. Thus, we performed activity profiling studies using ABPs on DUBs in human breast cancer cell lines and patient tumor tissues to study its activity-related biological function in different subtypes of breast cancer. UCHL1 was identified as the most specific highly active DUB in the TNBC subtype, and targeting of its activity mitigated TNBC cell migration and metastasis.
In this work, two different ABP-based DUB activity profiling methods were performed. Each profiling method has its own advantages and drawbacks. The TAMRA ABP profiling method is a simple, fast and convenient method, which allow us to achieve a snapshot of the DUB activity landscape with a very small amount of protein, whereas the Biotin ABP profiling coupled to mass spectroscopy analysis method is a more laborious requiring a larger amount of protein, but enables for the identification of the DUBs in a quantitative manner. Both DUB activity profiling methods identified UCHL1 as the most specific highly active DUB in the TNBC subtype. In the Biotin ABP profiling, other DUBs such as USP4, were previously reported to promote breast cancer metastasis that were detected in the Biotin ABP profiling to be highly active in TNBC group (33). OTUD3 that displayed selective high activity in non-TNBC was found formerly to act as a suppressor in breast cancer tumorigenesis and metastasis (34) (Fig. 2C). There are some other interesting hits for which still little is known about their function in breast cancer that can be studied in the future (Supplementary Table S3 and S4). Although there are several reports that UCHL1 may possibly act as a tumor suppressor in breast cancer pathogenesis, most evidence supports its role as a positive regulator of tumorigenesis (35) (36). These differences may be attributed to differential action of UCHL1 in different breast cancer subtypes.
Functionally, we observed that UCHL1 promoted breast cancer migration, extravasation and metastasis both in zebrafish and mice xenograft models. Mechanistically, UCHL1 facilitates TGFβ/SMAD2 and SMAD3 signaling and TGFβ-induced TNBC migration and extravasation by protecting TβRI and SMAD2/3 from ubiquitination. Next, we found that UCHL1 mainly regulates lysine 48-linked ubiquitination of TβRI and SMAD2/3 (Supplementary Fig. S4E-G). The interaction of TβRI with UCHL1 was found to be ligand dependent. UCHL1 interacted more efficiently with ubiquitylated TβRI, and TGFβ triggers the ubiquitination of TβRI. The latter may thus contribute to the ligand-induced interaction between TβRI and UCHL1. The interaction between UCHL1 and TβRI occurs in early endosomes, where activated TGFβ receptor complexes promote SMAD dependent signaling responses (37). Although our results point to a pivotal role for UCHL1 in stimulating breast cancer extravasation by regulating TGFβ signaling, we do not preclude that UCHL1 may also promote invasion and metastasis by targeting other signaling proteins. Previous studies showed that UCHL1 can also regulate protein kinase B (AKT) and hypoxia-inducible factor 1 α (HIF1α) signaling (36) (38). However, upon shRNA-mediated knockdown in MDA-MB-436 cells we were unable to detect changes in AKT and HIF1α protein levels (Supplementary Fig. S3B), suggesting a context-dependent role for UCHL1 in breast cancer.
When comparing the UCHL1 inhibitor LDN57444 with 6RK73, LDN57444 is a reversible and competitive inhibitor of UCHL1 activity (IC50 = 0.88 μM) (39), whereas 6RK73 is a covalent irreversible inhibitor of UCHL1 activity (IC50 = 0.23 μM) (Fig. 5B and Supplementary Fig. S6A). LDN57444 also inhibit UCHL3 activity (IC50 = 25 μM) (39), whereas, 6RK73 showed almost no inhibition of UCHL3 (IC50 = 236 μM) (Fig. 5B). Besides, 6RK73 displayed a potent inhibition of breast cancer extravasation in zebrafish (Fig. 5I and J), and this result is reminiscent to the inhibitory effect observed upon genetic UCHL1 depletion (Fig. 3F and J). Clinically, high UCHL1 expression is also associated with many other types of cancers including lung, colorectal, and pancreatic (40). Thus, 6RK73 may provide a new choice for the development of a clinical drug for targeting UCHL1 activity in the treatment of aggressive breast cancer and other UCHL1 overactive cancers.
In addition, UCHL1 was found to be highly enriched in TNBC patient sera compared with samples from healthy individuals. More importantly, we found that UCHL1 was specifically enriched in exosomes from aggressive breast cancer patient sera and TNBC cell conditioned medium. In this respect, our finding that UCHL1 and TβRI colocalize in early endosomes is of interest as early endosomes are precursor vesicles for exosomes (41). Another group, also recently detected UCHL1 in the exosomes of breast cancer patient sera, and high UCHL1 levels were found to be correlated with chemotherapy resistance phenotype (42). We found that UCHL1+ exosomes upregulated TGFβ/SMAD signaling and promoted migration and extravasation of the recipient breast cancer cells. This suggests that UCHL1 may act in cancer cells in both cell autonomous and paracrine manners to stimulate tumorigenesis.
Altogether, our results demonstrate the important roles for UCHL1 in breast cancer migration and extravasation by upregulating TGFβ signaling and highlight a potential novel therapy for cancer treatment by targeting UCHL1. UCHL1-containing exosomes also have the potential to be a blood-based biomarker for early diagnosis of aggressive breast cancer. The selective, potent and covalent UCHL1 activity inhibitor 6RK73 may open new avenues for therapeutic intervention in breast cancer and beyond.
Supplementary Material
Translational Relevance.
Metastasis is the leading cause of breast cancer-associated death. Triple-negative breast cancer remains the most challenging subtype to treat. Deubiquitinases (DUBs) are emerging drug targets in cancer treatment. To discover new DUB targets, we profiled global DUB activities in 52 human breast cancer cell lines and 52 patients’ tumor tissue samples. Two independent DUB activity profiling approaches identified UCHL1 as being highly active in TNBC cell lines and aggressive tumors. Mechanistically, UCHL1 facilitate TGFβ signaling-induced metastasis by restricting ubiquitination of TGFβ type I receptor and its downstream effector SMAD2. We further found UCHL1 covalent activity inhibitor 6RK73 can be used as a potential drug to specifically inhibit UCHL1 activity in breast cancer. Furthermore, we observed that TNBC patient sera contains high UCHL1 levels, which may represent a blood-based biomarker for early diagnosis of metastasis. In sum, our study has identified UCHL1 as a potential target for TNBC treatment.
Acknowledgments
We are grateful to Hiroshi Harada for the pcDNA4-UCHL1-myc construct and Andrew Hinck for recombinant TGFβ3. We thank Hans van Dam and David Baker for critical reading of the manuscript, and our lab members for valuable discussion. We are grateful to Kiki. M. H. Vangangelt for the collection of sera samples in LUMC. We thank Marcel Smid, Fuyu Cai and Fons J. Verbeek for bioinformatic analysis. We are grateful to Marie-José Goumans, Rubina Baglio and Michiel Pegtel for expert advice on exosome preparation, and Luis J. Cruz and Fabio Baldazzi for NTA analysis. We thank Annelies van der Laan and Lennard Voortman for confocal imaging technical assistance, Midory Thorikay for technical assistance, and Martijn Rabelink for shRNA constructs and determining of lentiviral titers.
Financial support
This work is supported by the Cancer Genomics Centre Netherlands, NWO (VICI grant 724.013.002 to H. Ovaa) and ERC (grant 310913 to A.Vertegaal). S. Liu is supported by China Scholarship Council. P.V. Iyengar is supported by the European Union Horizon 2020 Research and Innovation Program with a Marie Skłodowska‐Curie Individual Fellowship (786880).
Footnotes
Authors’ Contributions
Conception and design: P. ten Dijke, S. Liu
Development of methodology: S. Liu, M. van Dinther, R. González-Prieto, H. Zhu
Acquisition of data (provided animals, acquired and managed patients, provided facilities, etc.): S. Liu, R. González-Prieto, M. Zhang, P.P. Geurink, P.V. Iyengar, M. van Dinther, R. Kooij, E. Bos, X. Zhang, S. Le Dévédec, B. van de Water, R.I. Koning, W. Mesker, H. Ovaa, L. Zhang, A. Vertegaal, J.W.M. Martens, P. ten Dijke
Analysis and interpretation of data (e.g., statistical analysis, biostatistics, computational analysis): S. Liu, R. González-Prieto, M. Zhang, P.P. Geurink, P.V. Iyengar, M. van Dinther, R. Kooij, E. Bos, R.I. Koning, H. Zhu, W. Mesker, H. Ovaa, L. Zhang, A. Vertegaal, J.W.M. Martens, P. ten Dijke
Writing, review, and/or revision of the manuscript: S. Liu, R. González-Prieto, P.P. Geurink, J.W.M. Martens, P. ten Dijke
Study supervision: P. ten Dijke
Conflict of interest disclosure statement: All authors, except Huib Ovaa and Bob van de Water, have no conflict of interest. Huib Ovaa is cofounder and shareholder of UbiQ company. Bob van de Water is cofounder and equity option of OcellO BV; equity options VitroScan BV.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 2.Cummings MC, Simpson PT, Reid LE, Jayanthan J, Skerman J, Song S, et al. Metastatic progression of breast cancer: insights from 50 years of autopsies. J Pathol. 2014;232:23–31. doi: 10.1002/path.4288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dongre A, Weinberg RA. New insights into the mechanisms of epithelial–mesenchymal transition and implications for cancer. Nat Rev Mol Cell Biol. 2019;20:69–84. doi: 10.1038/s41580-018-0080-4. [DOI] [PubMed] [Google Scholar]
- 4.Drabsch Y, ten Dijke P. TGF-β signaling in breast cancer cell invasion and bone metastasis. J Mammary Gland Biol Neoplasia. 2011;16:97–108. doi: 10.1007/s10911-011-9217-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Becker A, Thakur BK, Weiss JM, Kim HS, Peinado H, Lyden D. Extracellular Vesicles in Cancer: Cell-to-Cell Mediators of Metastasis. Cancer Cell. 2016;30:836–48. doi: 10.1016/j.ccell.2016.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thery C. Cancer: Diagnosis by extracellular vesicles. Nature. 2015;523:161–2. doi: 10.1038/nature14626. [DOI] [PubMed] [Google Scholar]
- 7.Massagué J, Obenauf AC. Metastatic colonization by circulating tumour cells. Nature. 2016;529:298–306. doi: 10.1038/nature17038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A. 2001;98:10869–74. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leidy J, Khan A, Kandil D. Basal-like breast cancer: update on clinicopathologic, immunohistochemical, and molecular features. Arch Pathol Lab Med. 2014;138:37–43. doi: 10.5858/arpa.2012-0439-RA. [DOI] [PubMed] [Google Scholar]
- 10.Rochefort H, Glondu M, Sahla ME, Platet N, Garcia M. How to target estrogen receptor-negative breast cancer? Endocr Relat Cancer. 2003;10:261–6. doi: 10.1677/erc.0.0100261. [DOI] [PubMed] [Google Scholar]
- 11.Foulkes WD, Smith IE, Reis-Filho JS. Triple-Negative Breast Cancer. N Engl J Med. 2010;363:1938–48. doi: 10.1056/NEJMra1001389. [DOI] [PubMed] [Google Scholar]
- 12.Garrido-Castro AC, Lin NU, Polyak K. Insights into Molecular Classifications of Triple-Negative Breast Cancer: Improving Patient Selection for Treatment. Cancer Discov. 2019;9:176–98. doi: 10.1158/2159-8290.CD-18-1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ciechanover A. The unravelling of the ubiquitin system. Nat Rev Mol Cell Biol. 2015;16:322–4. doi: 10.1038/nrm3982. [DOI] [PubMed] [Google Scholar]
- 14.Liu S, de Boeck M, van Dam H, ten Dijke P. Regulation of the TGF-β pathway by deubiquitinases in cancer. Int J Biochem Cell Biol. 2016;76:135–45. doi: 10.1016/j.biocel.2016.05.001. [DOI] [PubMed] [Google Scholar]
- 15.Harrigan JA, Jacq X, Martin NM, Jackson SP. Deubiquitylating enzymes and drug discovery: emerging opportunities. Nat Rev Drug Discov. 2017;17:57–78. doi: 10.1038/nrd.2017.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kategaya L, Di Lello P, Rougé L, Pastor R, Clark KR, Drummond J, et al. USP7 small-molecule inhibitors interfere with ubiquitin binding. Nature. 2017;550:534–38. doi: 10.1038/nature24006. [DOI] [PubMed] [Google Scholar]
- 17.Lamberto I, Liu X, Seo HS, Schauer NJ, Iacob RE, Hu W, et al. Structure-Guided Development of a Potent and Selective Non-covalent Active-Site Inhibitor of USP7. Cell Chem Biol. 2017;24:1490–500. doi: 10.1016/j.chembiol.2017.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pozhidaeva A, Valles G, Wang F, Wu J, Sterner DE, Nguyen P, et al. USP7-Specific Inhibitors Target and Modify the Enzyme’s Active Site via Distinct Chemical Mechanisms. Cell Chem Biol. 2017;24:1501–12. doi: 10.1016/j.chembiol.2017.09.004. [DOI] [PubMed] [Google Scholar]
- 19.Turnbull AP, Ioannidis S, Krajewski WW, Pinto-Fernandez A, Heride C, Martin ACL, et al. Molecular basis of USP7 inhibition by selective small-molecule inhibitors. Nature. 2017;550:481–6. doi: 10.1038/nature24451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.de Jong A, Merkx R, Berlin I, Rodenko B, Wijdeven RH, El Atmioui D, et al. Ubiquitin-based probes prepared by total synthesis to profile the activity of deubiquitinating enzymes. Chembiochem. 2012;13:2251–8. doi: 10.1002/cbic.201200497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ren J, Liu S, Cui C, ten Dijke P. Invasive Behavior of Human Breast Cancer Cells in Embryonic Zebrafish. J Vis Exp. 2017;122:e55459. doi: 10.3791/55459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Arguello F, Baggs RB, Frantz CN. A murine model of experimental metastasis to bone and bone marrow. Cancer Res. 1988;48:6876–81. [PubMed] [Google Scholar]
- 23.Jones A, Kemp M, Stockley M, Gibson K, Whitlock G, Madin AJW. Novel Compounds. Patent WO 2016/046530 A1. 2016
- 24.Gao L, Wang L, Dai T, Jin K, Zhang Z, Wang S, et al. Tumor-derived exosomes antagonize innate antiviral immunity. Nat Immunol. 2018;19:233–45. doi: 10.1038/s41590-017-0043-5. [DOI] [PubMed] [Google Scholar]
- 25.Xu R, Greening DW, Zhu HJ, Takahashi N, Simpson RJ. Extracellular vesicle isolation and characterization: toward clinical application. J Clin Invest. 2016;126:1152–62. doi: 10.1172/JCI81129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bishop P, Rocca D, Henley JM. Ubiquitin C-terminal hydrolase L1 (UCH-L1): structure, distribution and roles in brain function and dysfunction. Biochem J. 2016;473:2453–62. doi: 10.1042/BCJ20160082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu Y, Fallon L, Lashuel HA, Liu Z, Lansbury PT., Jr The UCH-L1 gene encodes two opposing enzymatic activities that affect α-synuclein degradation and Parkinson’s disease susceptibility. Cell. 2002;111:209–18. doi: 10.1016/s0092-8674(02)01012-7. [DOI] [PubMed] [Google Scholar]
- 28.Papa L, Brophy GM, Welch RD, Lewis LM, Braga CF, Tan CN, et al. Time Course and Diagnostic Accuracy of Glial and Neuronal Blood Biomarkers GFAP and UCH-L1 in a Large Cohort of Trauma Patients With and Without Mild Traumatic Brain Injury. JAMA Neurol. 2016;73:551–60. doi: 10.1001/jamaneurol.2016.0039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Melo SA, Luecke LB, Kahlert C, Fernandez AF, Gammon ST, Kaye J, et al. Glypican-1 identifies cancer exosomes and detects early pancreatic cancer. Nature. 2015;523:177–82. doi: 10.1038/nature14581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mathieu M, Martin-Jaular L, Lavieu G, Théry C. Specificities of secretion and uptake of exosomes and other extracellular vesicles for cell-to-cell communication. Nat Cell Biol. 2019;21:9–17. doi: 10.1038/s41556-018-0250-9. [DOI] [PubMed] [Google Scholar]
- 31.Chakraborty S, Hosen MI, Ahmed M, Shekhar HU. Onco-Multi-OMICS Approach: A New Frontier in Cancer Research. Biomed Res Int. 2018;2018:9836256. doi: 10.1155/2018/9836256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xiao Z, Zhang P, Ma L. The role of deubiquitinases in breast cancer. Cancer Metastasis Rev. 2016;35:589–600. doi: 10.1007/s10555-016-9640-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang L, Zhou F, Drabsch Y, Gao R, Snaar-Jagalska BE, Mickanin C, et al. USP4 is regulated by AKT phosphorylation and directly deubiquitylates TGF-β type I receptor. Nat Cell Biol. 2012;14:717–26. doi: 10.1038/ncb2522. [DOI] [PubMed] [Google Scholar]
- 34.Yuan L, Lv Y, Li H, Gao H, Song S, Zhang Y, et al. Deubiquitylase OTUD3 regulates PTEN stability and suppresses tumorigenesis. Nat Cell Biol. 2015;17:1169–81. doi: 10.1038/ncb3218. [DOI] [PubMed] [Google Scholar]
- 35.Lien HC, Wang CC, Lin CH, Lu YS, Huang CS, Hsiao LP, et al. Differential expression of ubiquitin carboxy-terminal hydrolase L1 in breast carcinoma and its biological significance. Hum Pathol. 2013;44:1838–48. doi: 10.1016/j.humpath.2013.02.006. [DOI] [PubMed] [Google Scholar]
- 36.Hurst-Kennedy J, Chin L-S, Li L. Ubiquitin C-terminal hydrolase L1 in tumorigenesis. Biochem Res Int. 2012;2012:123706. doi: 10.1155/2012/123706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yakymovych I, Yakymovych M, Heldin CH. Intracellular trafficking of transforming growth factor β receptors. Acta Biochim Biophys Sin. 2018;50:3–11. doi: 10.1093/abbs/gmx119. [DOI] [PubMed] [Google Scholar]
- 38.Goto Y, Zeng L, Yeom CJ, Zhu Y, Morinibu A, Shinomiya K, et al. UCHL1 provides diagnostic and antimetastatic strategies due to its deubiquitinating effect on HIF-1α. Nat Commun. 2015;6:6153. doi: 10.1038/ncomms7153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu Y, Lashuel HA, Choi S, Xing X, Case A, Ni J, et al. Discovery of inhibitors that elucidate the role of UCH-L1 activity in the H1299 lung cancer cell line. Chem Biol. 2003;10:837–46. doi: 10.1016/j.chembiol.2003.08.010. [DOI] [PubMed] [Google Scholar]
- 40.Pfoh R, Lacdao IK, Saridakis V. Deubiquitinases and the new therapeutic opportunities offered to cancer. Endocr Relat Cancer. 2015;22:T35–54. doi: 10.1530/ERC-14-0516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McAndrews KM, Kalluri R. Mechanisms associated with biogenesis of exosomes in cancer. Mol Cancer. 2019;18:52. doi: 10.1186/s12943-019-0963-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ning K, Wang T, Sun X, Zhang P, Chen Y, Jin J, et al. UCH-L1-containing exosomes mediate chemotherapeutic resistance transfer in breast cancer. J Surg Oncol. 2017;115:932–40. doi: 10.1002/jso.24614. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.