Abstract
Acute myeloid leukemia (AML) is a heterogeneous clonal disorder with a poor clinical outcome. Previously we showed that overproduction of reactive oxygen species (ROS), arising from constitutive activation of NOX2 oxidase, occurs in >60% of AML patients and that ROS production promotes proliferation of AML cells. We show here that the process most significantly affected by ROS overproduction is glycolysis. Whole metabolome analysis of 20 human primary AML showed that blasts generating high levels of ROS have increased glucose uptake and correspondingly increased glucose metabolism. In support of this, exogenous ROS increased glucose consumption whilst inhibition of NOX2 oxidase decreased glucose consumption. Mechanistically, ROS promoted uncoupling protein 2 (UCP2) protein expression and phosphorylation of AMPK, upregulating the expression of a key regulatory glycolytic enzyme, 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase (PFKFB3). Overexpression of PFKFB3 promoted glucose uptake and cell proliferation, whilst downregulation of PFKFB3 strongly suppressed leukemia growth both in vitro and in vivo in the NSG model. These experiments provide direct evidence that oxidase-derived ROS promotes the growth of leukemia cells via the glycolytic regulator PFKFB3. Targeting PFKFB3 may therefore present a new mode of therapy for this disease with a poor outcome.
Keywords: Acute Myeloid Leukemia, Reactive Oxygen Species, Metabolism, Glycolysis, Proliferation
Introduction
Reactive oxygen species (ROS) are a heterogeneous group of molecules and free radicals generated as a by-product of mitochondrial oxidative phosphorylation and deliberately generated via nicotinamide adenine dinucleotide phosphate (NADPH) oxidase (NOX) family proteins (1). In particular, NOX2 is expressed in the plasma membrane of hematopoietic cells that generates superoxide. Superoxide rapidly dismutates to hydrogen peroxide (H2O2), a relatively long-lived, mildly reactive molecule that traverses biological membranes and mediates redox signaling in both autocrine and paracrine fashion (2). The capacity of H2O2 to reversibly oxidise cysteine residues in regulatory domains or active sites of proteins is believed to underlie its biological effects (3). Indeed, H2O2 plays an integral role in hematopoiesis both through direct and indirect regulation of gene expression (4).
Excessive production of ROS is a common feature of cancer. In leukemia, ROS are known to cause DNA damage (5) and also promote proliferation (6–8). We previously showed that >60% of AML patients exhibited elevated levels of extracellular superoxide and H2O2 which correlated with NOX2 expression (6). We also found that RAS (which is both directly and indirectly activated in AML (9)) was able to drive the production of NOX2-derived ROS in normal hematopoietic stem / progenitor cells (HSPC) and using this model we were able to show that RAS-induced ROS production contributed to the pro-proliferative effects of this oncogene (7). Despite this, the underlying mechanism through which ROS promote proliferation in cancer remains unclear. Using this model, we show for the first time that ROS particularly impacts on genes associated with the glycolytic pathway, with a key glycolytic regulator, PFKFB3, acting as an important mediator of ROS. Correspondingly, we show that ROS also promotes glycolysis in cell lines and AML patient blasts. Furthermore, myeloid leukemia cells exhibit dependency on PFKFB3 both for their growth and survival. Given the frequently elevated levels of ROS in primary AML, these data provide a plausible mechanism for the enhanced glycolysis seen in AML and suggest that a therapeutic opportunity exists in which agents inhibiting PFKFB3 could be used to treat this disease.
Materials and Methods
Key resources
All reagents and key resources are provided in Supplemental material.
Primary cell material and cell culture
For whole cell metabolomics, a subset of bone marrow samples (n=20) from AML patients who had enrolled in UK MRC/NCRI AML clinical trials at point of diagnosis, before treatment and obtained informed written consent from patients in accordance with the 1964 Declaration of Helsinki, was used. Control mononuclear cells were isolated from peripheral blood of human male/female volunteers (n=6).
Human neonatal cord blood was obtained from healthy full-term pregnancies at the University Hospital Wales, Cardiff, UK. These were obtained with informed consent and with approval from the South East Wales Research Ethics Committee in accordance with the 1964 Declaration of Helsinki. Human CD34+ cells (>95% pure, which constitute a mixed progenitor blood cell population) were isolated, cultured and transduced with retroviral vectors based on the PINCO backbone harboring either a GFP or DsRED selectable marker as previously described (10). Transduced human CD34+ hematopoietic progenitor cells were cultured in supplemented IMDM as previously described (10) containing 20% v/v FCS; supplemented with 5 ng/mL human (hu) IL-3, huG-CSF and huGM-CSF and 20 ng/mL huSCF. For microarray studies, on day 5 of culture (post CD34+ isolation), cells were washed in PBS and resuspended in supplemented IMDM in the presence or absence of 100nM diphenyleneiodonium (DPI) for 18 h prior to Affymetrix microarray (n=4). RNA was extracted using Trizol® as previously described (11). Due to the high frequency of retroviral transduction (~70%), enrichment of transduced cells was unnecessary.
Cell lines were purchased from ATCC or ECACC and cultured according to recommended conditions at 37°C, 5% CO2 for all experiments. All lines are maintained at ≤ 20 passage from receipt. The genetic identity of the cell lines was confirmed by short tandem repeat (STR) at purchase. Monthly monitoring for Mycoplasma contamination was performed and confirmed using the MycoAlert Detection Kit (Sigma). Mice were bred and maintained at Cardiff University (UK) and were cared for in accordance with Institutional Animal Care and Use Committee guidelines. NOD-SCID IL2Rγ(-/-)(NSG) female mice were sub-lethally irradiated with 200cGy total body irradiation 24h before inoculation of THP-1 cells via tail-vein injection. Transplanted cells were analysed using hCD45-FITC, hCD33-APC and mCD45-PerCP-Cy5.5 by flow cytometry. For ex vivo analysis of mouse MRP8 N-RASG12D leukemia (12), bone marrow was harvested from the tibias and glucose uptake was measured using 2-NBDG as described below.
Detection of superoxide
Following gene transduction, the indicated cell cultures were adjusted for viable cell number and superoxide measurement was carried out using the chemiluminescent probe Diogenes™ (Geneflow, U.K.). Briefly, cells were resuspended in their conditioned medium to a density of 1x106 cells/mL and 150 μL aliquots were assayed in triplicate in FluoroNunc Maxisorp 96 well plates (Thermo-Fisher Scientific, Loughborough, UK). Diogenes (50 μL) was added immediately prior to recording chemiluminescence as previously described (7).
Determination of glucose and lactate
Supernatant from the culture media was filtered using Microcon-10 kDa centrifugal filter units (Merck-Millipore, Feltham, UK) at 12,782 × g for 30 min. The levels of D-glucose and L-Lactate were measured by fluorimetry using a glucose and L-Lactate assay kit (Abcam, Cambridge, UK) coupled with a Chameleon Hidex fluorescent plate reader (Ex/Em 535/590 nm), according to the manufacturer’s instructions. Briefly, samples were diluted with proprietary glucose or lactate buffer to a volume of 50μL and added in triplicate to a black 96 well flat bottomed microclear plate (Greiner Bio-One, Stonehouse, UK). Glucose or lactate buffer containing proprietary glucose or lactate probe (0.8% v/v) and proprietary glucose or lactate enzyme mix (0.8% v/v) were added (50μL) to each well and left to incubate in the dark at RT for 30 min. Fluorescence was measured (Ex/Em 535/590nm) and compared with glucose or lactate standards assayed in duplicate on the same plate.
To determine cellular glucose uptake at the individual cell level, the glucose bioprobe 2-NBDG (Life Technologies, U.K.) was employed in conjunction with flow cytometry. Cells were washed twice in PBS then treated with 2-NBDG (10μM) or PBS alone (to establish a background control) followed by incubation for 10 min (37°C, 5% CO2) and two washes in ice cold PBS. Cells were immediately analysed by flow cytometry using an Acurri C6 flow cytometer. 2-NBDG emits fluorescence at a wavelength of 542nM. Having excluded cell debris based on FSC/SSC, the median glucose uptake per cell of the samples was established by subtracting the median value of fluorescence of the background control cells from the median value of fluorescence of the cells treated with 2-NBDG. In some experiments, cells were treated with PEGylated catalase (300 mU/mL) for 24 h at 37°C, prior to analysis of glucose uptake.
Expression analysis
Transduced CD34+ cells were washed in PBS and resuspended in supplemented IMDM in the presence or absence of 100nM diphenyleneiodonium (DPI) for 18 h prior to RNA isolation (n=4) as previously described (11). RNA was hybridized to Affymetrix GeneChip® Human Exon 1.0ST Array for whole-transcript expression analysis. Data were analysed using Partek Genomics Suite (v6.6; Partek, MO, USA). Data analysis of CEL files are described in Supplemental Methods; data available at https://www.ebi.ac.uk/arrayexpress (accession number e-mexp-583). Gene Ontology (GO) enrichment analysis was undertaken using Metacore® (Clarivate Analytics, U.K.).
Detection of each protein was determined by western blot using antibodies described in Supplemental Methods, in conjunction Amersham ECL™ Advance/Prime Western Blotting Detection Kit (GE Healthcare U.K.).
Metabolomics
Metabolomic analysis of AML patient blast samples or Mv4;11 was carried out by Metabolon™ (http://www.metabolon.com/). Data was generated using ultra-high performance liquid chromatography-tandem mass spectroscopy (UPLC-MS/MS) and gas-chromatography mass-spectroscopy (GC-MS). Peripheral blood and bone marrow samples collected from a random cohort of AML patients were counted and analysed for viability using 7-AAD. Only those samples with a cell count greater than 30 million and a viability greater than 80% were sent for analysis by Metabolon™ (n=20). Additionally, mononuclear cells isolated from healthy individuals (n=6) were also sent to Metabolon™ as a comparative control. Diogenes™ analysis of AML blasts stratified the patient samples into ROSHigh (above median) and ROSLow (below median). Raw data was extracted, peak identified and quality control processed using proprietary Metabolon™ hardware, software and biochemical library database. Following normalisation to Bradford protein concentration, log transformation and imputation of missing values with the minimum observed value for each compound, Welch’s unequal variance two sample t-test was performed to identify significant differences between the experimental groups. To account for a potentially high false discovery rate (as a consequence of multiple comparisons), a q-value was also calculated, where a lower q-value is an indication of higher confidence in the result.
Protein expression analysis of genes identified by microarray analysis
Detection of each protein (see key resources table) was determined using a monoclonal or polyclonal antibody in conjunction with an anti-mouse or anti-Rabbit HRP linked secondary antibody and Amersham ECL™ Advance/Prime Western Blotting Detection Kit (GE Healthcare UK) according to the manufacturer’s instruction. In the case of NOX2 or glucose transporter cell surface protein expression, PE conjugated antibody to NOX2 epitope or an indirect stain coupled with anti-mouse IgG-APC was used and protein expression was determined by flow cytometry.
Flow cytometric and data analysis
Flow cytometric data were acquired using an Accuri C6 cytometer (BD, U.K.). Data analysis was performed using FCS express v6 (DeNovo Software). The threshold for GFP positivity was determined from the autofluorescence of GFP/DsRED negative cells in mock transduced cultures. Significance of difference was tested using Minitab software version 19 (Minitab Inc, PA) all analyses except those provided by Metabolon assays (see above). Appropriate statistical tests used are labelled in figure legends. To better understand variations between samples, principal component analysis (PCA) or hierarchical clustering using distance Pearson Correlation was employed to provide a global analysis of how closely related or otherwise any given sample is (mRNA or biochemical).
Results
NOX-derived ROS promotes transcriptional change in N-RASG12D expressing hematopoietic progenitor cells and AML patient blasts
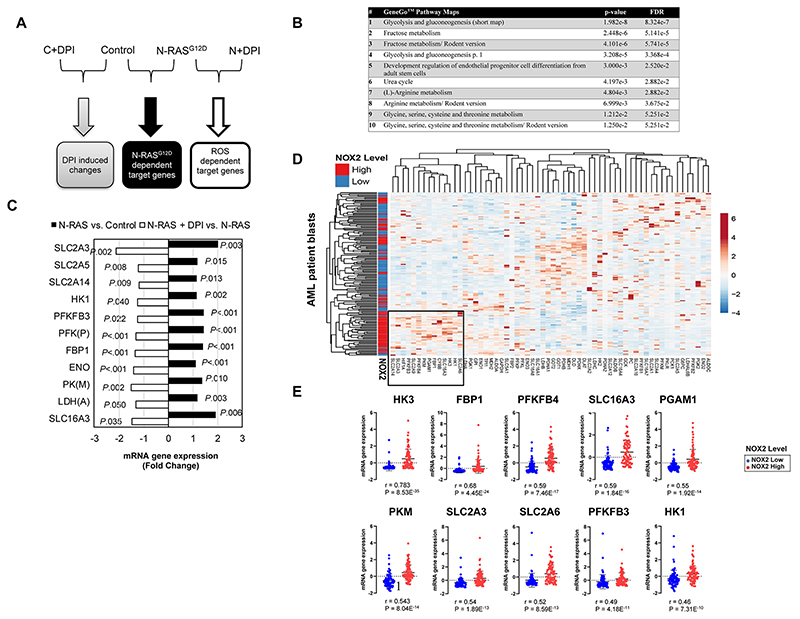
We previously showed that expression of N-RASG12D in HSPC strongly promotes ROS production through activation of NOX oxidases leading to increased proliferation (7). We used this model of ROS overproduction in primary hematopoietic CD34+ cells (Supplemental Fig. S1A-E) to investigate the changes in gene expression mediated by ROS. We reasoned that we could enrich for ROS target genes by looking for gene changes which were absent in mutant N-RASG12D cells treated with the NOX inhibitor, DPI (Fig. 1A). N-RASG12D significantly changed the expression of 305 genes in HSPC (p<0.05) (Supplemental Table S1) of which 24 were specifically attributed to ROS production. Metacore™ pathway analysis identified glycolysis as the most dysregulated pathway (Fig. 1B). ROS significantly impacted the expression of 18% of genes involved in carbohydrate metabolism (Fig. 1C). To examine this in AML patient blasts, we analyzed the TCGA database of 161 AML patients using cBioPortal (13, 14). Hierarchical clustering in Fig. 1D shows that patients with high NOX2 (CYBB) expression (which we have previously shown to correlate with ROS production (7)) clustered together based on correlative expression of genes involved in carbohydrate metabolism suggesting changes in glucose utilization compared to those patients with lower NOX2 expression; more than half the ROS regulated genes observed above are seen in this cluster. Fig. 1E shows the ROS-responsive genes that significantly correlated with NOX2 expression in AML blasts. Data derived from TCGA (15) supports the overexpression of NOX2 mRNA in AML (Supplemental Fig. S2A). Increased NOX2 expression also showed a trend towards poor prognosis (P=0.075; Supplemental Fig. S2B). We did not observe any significant association of expression of genes identified in Fig.1E with AML cytogenetics or survival. Taken together, these data show that NOX inhibition upregulates the expression of genes involved in carbohydrate metabolism.
Figure 1. NOX-derived ROS promotes transcriptional change in N-RASG12D expressing HSPC and AML patient blasts and identifies the glycolytic pathway as a major target of ROS.
(A) Summary flow diagram showing the strategy for changes in mRNA expression analysed by Affymetrix microarray. Four treatment conditions were employed for the examination of the effect of N-RASG12D and ROS on mRNA gene expression: CD34+ HSPC infected with control vector (‘C’) or N-RASG12D (‘N’), incubated in the presence or absence of 100nM DPI for 24 h to determine the ROS-specific gene expression profile (n=4). We examined changes that were only co-directional (i.e. they were similarly dysregulated in each replicate). (B) Significantly changing GeneGo™ Maps in human HSPCs as a response to changes in exposure to ROS. (C) Statistically significant changes in mRNA showing the impact of DPI (unfilled bars) on N-RASG12D dependent target gene expression (filled bars). Non-specific effects of DPI were excluded as described in (A). Only genes involved in carbohydrate metabolism using the Human Exon 1.0ST Full Probe Set list were analysed. Statistically significant gene changes in Control cells treated with DPI compared to untreated control were excluded from the final list. Data represents mean fold change of n=4; P value calculated by 2-way ANOVA with Bonferroni multiple testing correction. (D) Hierarchical clustering of patient AML mRNA expression z-Scores based on RNA Seq V2 RSEM. NOX2 high / low expressing blasts was defined as, above and below the median expression intensity of NOX2 (aka CYBB). Genes involved in glycolysis and carbohydrate transport are shown (as defined by Affymetrix Netaffx™ gene annotation software under advanced pathway searches); AML patient blasts (n=160). Boxed section shows cluster of genes associated with NOX2 high AML blasts. (E) Expression of genes from AML patient blasts significantly correlating with NOX2 (CYBB, R>0.5) expression. R; Spearmans Correlation co-efficient. FDR, False Discovery Rate; GLUT3 (SLC2A3), glucose transporter 3; GLUT5 (SLC2A5), glucose transporter 5; GLUT6 (SLC2A6), glucose transporter 6; GLUT14 (SLC2A14), glucose transporter 14; HK, hexokinase; PFKFB, 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase; PFK(P). phosphofructokinase platelet; FBP1, fructose-1,6-bisphosphatase 1; ENO, enolase; PK(M), pyruvate kinase muscle; LDH(A), lactate dehydrogenase A; MCT4 (SLC16A3), monocarboxylate transporter 4.
ROS promotes functional changes in glucose uptake
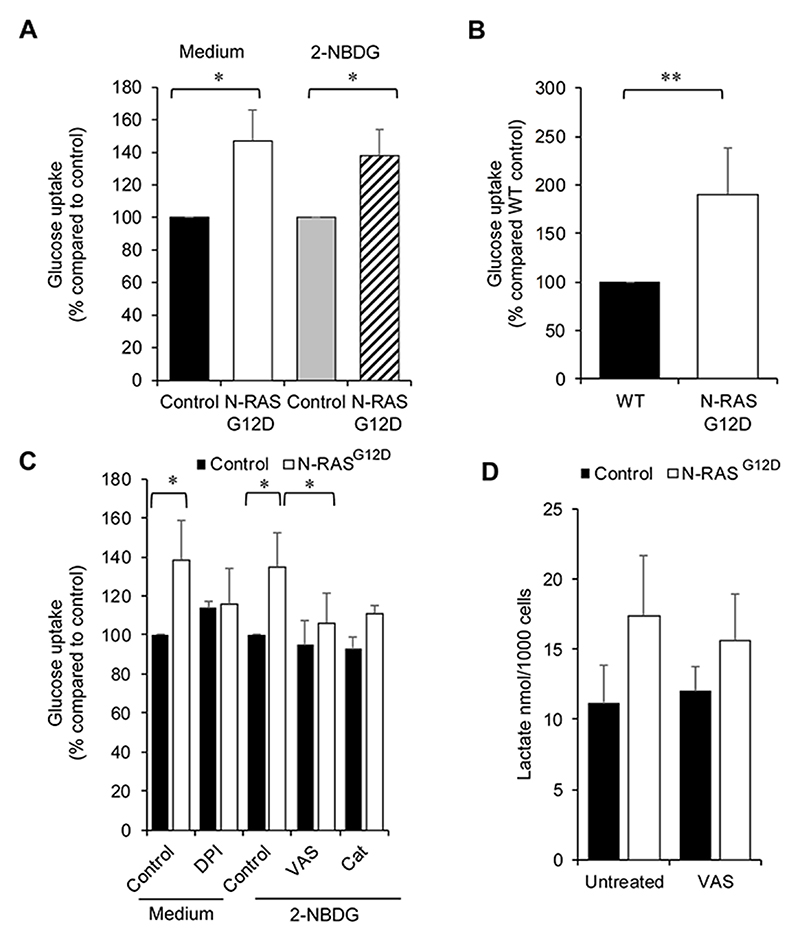
Increased aerobic glycolysis is a common feature of cancerous cells with concomitant increases in cellular glucose uptake and lactate secretion (16). In order to assess whether transcriptional changes observed above resulted in functional glycolytic changes, glucose uptake and lactate secretion were measured. The level of glucose taken up by N-RASG12D HSPC was significantly more (48±19%) when compared to control (Fig. 2A) when analysing glucose remaining in the culture media. To confirm that these changes in cellular glucose consumption occurred at the single cell level, the fluorescent glucose bioprobe 2-NBDG was employed (Supplemental Fig. S3). N-RASG12D increased glucose uptake by 36±15% compared to controls (Fig. 2A). To support these in vitro data, glucose uptake was assayed ex vivo in bone marrow cells harvested from secondary transplants of transgenic mice expressing N-RASG12D (12). A significant 90±48% increase in glucose uptake was observed in N-RASG12D mice compared to wild type control (Fig. 2B). These data demonstrate that expression of mutant N-RASG12D increases glucose uptake both in vitro and ex vivo.
Figure 2. ROS promotes glucose uptake.
(A) ‘Medium’ - following transduction, N-RASG12D were cultured for 3 days without growth factors, glucose present in the media was assayed using a fluorometric glucose kit (n=4) (see methods) and normalised to empty-vector control. Glucose uptake in the cell is inversely proportional to the glucose remaining in the media. ‘2-NBDG’ - glucose uptake using the fluorogenic substrate 2-NBDG (normalised to control) in N-RASG12D HSPC (n=4) cultured as above. (B) Glucose uptake (normalised to Wild Type (WT) control) ex vivo in N-RASG12D bone marrow compared to WT control cells (n=8). (C) N-RASG12D HSPCs (day 5 post infection) were treated with 100nm DPI and cultured for 24 h without growth factors. Glucose in culture media (normalised to untreated control) was assayed (n=3). Glucose uptake in the cell is inversely proportional to the glucose remaining in the media. N-RASG12D HSPCs were treated with 5μM VAS-2870 (VAS; n=3) or 300mU/mL PEGylated catalase (Cat; n=2) for 24 h and glucose uptake assayed using 2-NBDG (normalised to PEG-treated control). (D) Concentration of extracellular L-lactate in culture media of transduced CD34+ cells (cultured as above) treated with 5 μM VAS-2870 as above (n=4). Data represents mean±1SD. * denotes p<0.05 and ** p<0.001 analysed by one sample t-test.
To assess whether increases in glucose uptake were mediated by NOX-derived ROS, we examined the effect of the NOX inhibitor, DPI, on N-RASG12D HSPC. NOX inhibition reduced glucose uptake by 20% compared to untreated cells (Fig. 2C) when analysing glucose remaining in the culture media. Similar data were obtained with the alternative NOX inhibitor, VAS-2870 (17). Treatment with PEGylated catalase (which catabolises the destruction of H2O2 at the plasma membrane (18)) also reverted the increase in glucose uptake (Fig. 2C). In contrast, treatment of control HSPC with DPI or VAS-2870 had no significant effect on glucose uptake. To determine whether these changes in glucose uptake resulted in increases in extracellular lactate production, levels of L-lactate in the culture supernatant were assayed. Interestingly, no significant changes in lactate secretion (see below) was observed in cells expressing N-RASG12D compared to control (Fig. 2D).
Overproduction of ROS is associated with changes in glucose utilization in primary AML blasts
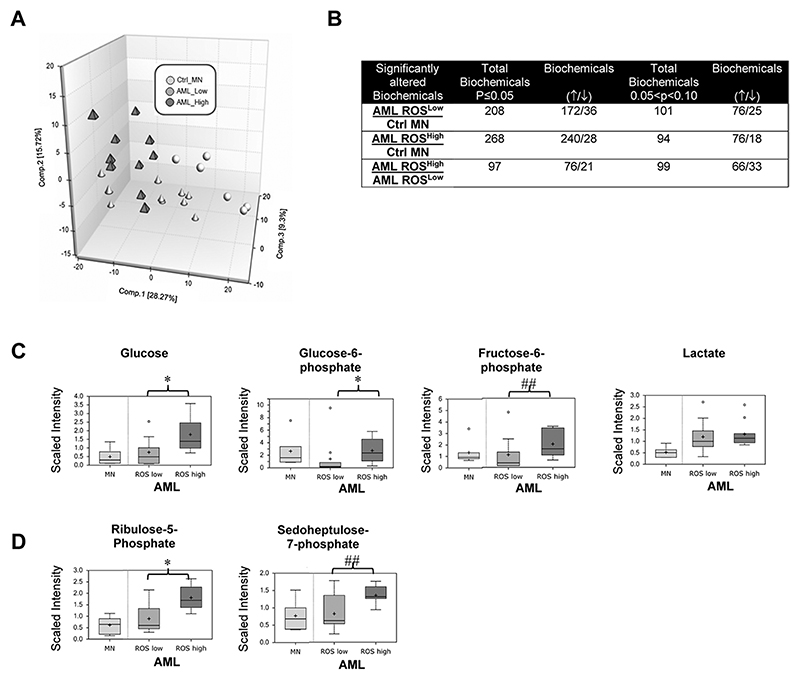
The above data suggests increased glucose uptake is at least in part mediated by production of NOX-derived ROS. To establish evidence for this in primary AML, we stratified AML blasts according to extracellular ROS production (ROSHigh and ROSLow; Supplemental Fig. S4A-B) and analysed the global biochemical metabolomic profile. Using this approach, 444 named metabolites were identified which distinguished ROSHigh, ROSLow and control MNC by PCA (Fig. 3A). A summary of the biochemicals that achieved statistical significance (p≤0.05) or approached it (0.05<p<0.10), is shown in Fig. 3B and in full in Supplemental Table S2. Random Forest (RF) analysis of the cellular metabolic profiles resulted in 85% predication accuracy in differentiating ROSHigh and ROSLow groups (Supplemental Fig. S5). Among the 30 top-ranking metabolites resulting from the RF analyses were biochemicals spanning different pathways, but primarily limited to those associated with nucleotides and lipid metabolism. Polyunsaturated fatty acids and lipid-related changes in n3 and n6 polyunsaturated fatty acids showed significant accumulations within the AMLHigh compared to the AMLLow samples which would be indicative of increased uptake (Supplemental Table S2). Interesting, within the AMLHigh samples, lysolipids, monoacylglycerols and glycerol were consistently and significantly higher in relation to AMLLow ROS (see discussion).
Figure 3. Overproduction of ROS is associated with changes in glucose utilizations in primary AML blasts.
Data from global biochemical profiling of AML blasts stratified according to extracellular ROS production was performed. (A) Principal components analysis of global biochemical profiling of AML cells with high and low ROS production; ROSLow, n=10; ROShigh, n=10. Also shown are normal human mononuclear control cells (Ctrl_MN: n=4). (B) Summary of the numbers of biochemicals that achieved statistical significance (*p≤0.05), as well as those approaching significance (##0.05<p<0.10) analysed by Welch’s two sample t-test. Levels of biochemicals normalised to total protein in (C) glycolysis and (D) Pentose Phosphate Pathway (PPP) are shown.
Focussing specifically on detected metabolites within the glycolytic pathway, we found, glucose, glucose-6-phosphate and fructose-6-phosphate levels were significantly higher within the ROSHigh blasts compared to ROSLow blasts indicating that increased glucose utilisation correlates with elevated ROS (Fig. 3C). In agreement with our in vitro HSPC model, changes in lactate were not significantly different between the ROS high and ROS low groups but were higher than the control MNC (Fig. 3C). This suggests that whilst glucose utilisation is increased in primary AML, higher levels of ROS are consistent with increased levels of glycolytic intermediates. Indeed, metabolites associated with the pentose phosphate pathway (PPP) including the isobaric compound ribulose/xylulose 5-phosphate and sedoheptulose-7-phosphate were also higher in ROSHigh vs ROSLow samples (Fig. 3D). We next set out to determine whether the addition of exogenous H2O2 to the ROSLow AML cell line, Mv4;11 could itself promote changes in glucose metabolism. Similarly to primary AML-ROShigh we observed an increase in glucose consumption (Fig. 4) and biochemicals associated with the PPP (Supplemental Fig. S6A and B). Taken together these data suggests increased glucose utilization by PPP, potentially driving nucleotide biosynthesis and NAD(P)H generation within ROSHigh blasts.
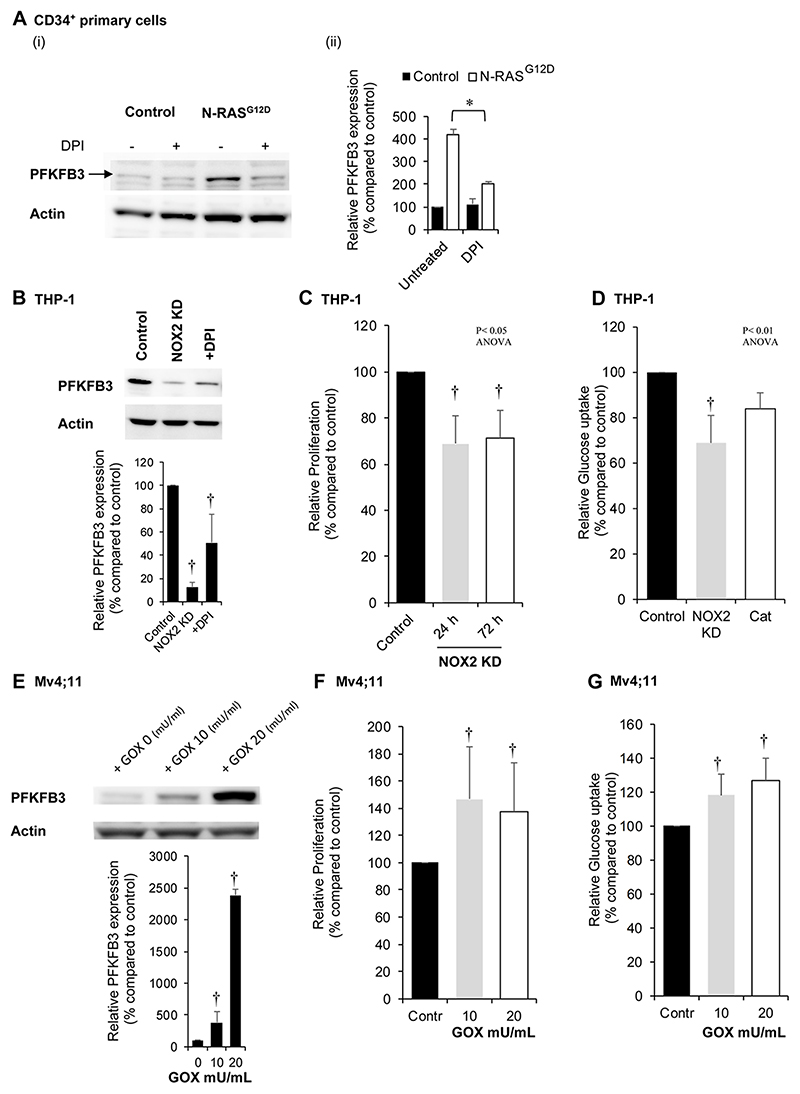
Figure 4. PFKFB3 protein expression correlates with levels of NOX2 derived extracellular ROS.
(A) Human CD34+ HSPC control and N-RASG12D (day 5 post transduction) were cultured for 24 h in cytokine free media in the presence or absence of DPI (100nM) followed by whole cell protein extraction. (i) Example western blot of PFKFB3 protein expression. (ii) Relative protein expression (as measured by pixel densitometry of equivalent regions of interest (ROI) between different samples on the same blot then normalised to control) of PFKFB3 (n=3). (B) Western blot showing PFKFB3 protein expression levels in THP-1 cells with NOX2 knocked down (KD) or cells treated with DPI (100nM) for 24 h compared to control (non-mammalian shRNA) cells. Lower panel showing relative protein expression of PFKFB3 compared to control (n=3). (C) Percentage proliferative change (normalised to control) in THP-1 cells with NOX2 KD (n>3) over 72 h. (D) Glucose uptake in single cell analysis using 2-NBDG (normalised to control) in THP-1 cells with NOX2 KD (n=4) or THP-1 cells treated with 300 mU/mL PEGylated catalase (Cat; n=6). (E) Western blot showing PFKFB3 protein expression in Mv4;11 cells treated with glucose oxidase (GOX) (10 and 20mU/mL for 24 h), which catalyses production of hydrogen peroxide (H2O2) in cell culture media; (imitating the effect of NOX2-generated ROS production). Lower panel showing relative protein expression of PFKFB3 compared to control (n=3). Correlation of PFKFB3 overexpression in Mv4;11 cells treated with GOX for 24 h on (F) proliferation and (G) glucose uptake using 2-NDBG (n=5). Actin was used as a loading control. Data represents mean±1SD. † denotes p<0.05 analysed by ANOVA with Tukey’s honestly significance difference. * denotes p<0.05 analysed by one sample t-test.
PFKFB3 is a ROS responsive target that mediates changes in glucose utilization
The data above suggest that ROS-induced changes in mRNA expression of genes of glycolysis (Fig. 1C) was associated with altered glucose utilization (Fig. 3). To validate these findings, we surveyed the expression of these ROS-responsive genes at the protein level (Supplemental Fig. S7A-D). ROS induced changes at the protein level, only occurred in the expression of the regulatory glycolytic enzyme 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase 3 (PFKFB3)(Fig. 4A and Supplemental Fig. S7A). PFKFB is a bifunctional enzyme with both kinase and phosphatase activity that regulates the glycolytic pathway (19). To support this data, we inhibited NOX derived ROS production in the ROSHigh AML cell line, THP-1, using NOX2 knock-down (Supplemental Fig. S8A-C). In line with our previous data (7), NOX inhibition with DPI suppressed proliferation of N-RASG12D cells. As shown in Fig. 4B, loss of NOX2 protein expression and ablation of superoxide production in these cells reduced the expression of PFKFB3 compared to control cells with a concomitant reduction in proliferation by 30±12% when compared to control (Fig. 4C). Glucose uptake was similarly reduced by 25±17% (Fig. 4D). We next set out to determine whether the addition of exogenous H2O2 to the ROSLow AML cell line, Mv4;11 cells, could itself promote PFKFB3 protein expression. As predicted, we observed a dose-dependent increase in PFKFB3 protein expression (Fig. 4E) with concomitant increases in proliferation (Fig. 4F and Supplemental Fig. S9) and glucose uptake (Fig. 4G).
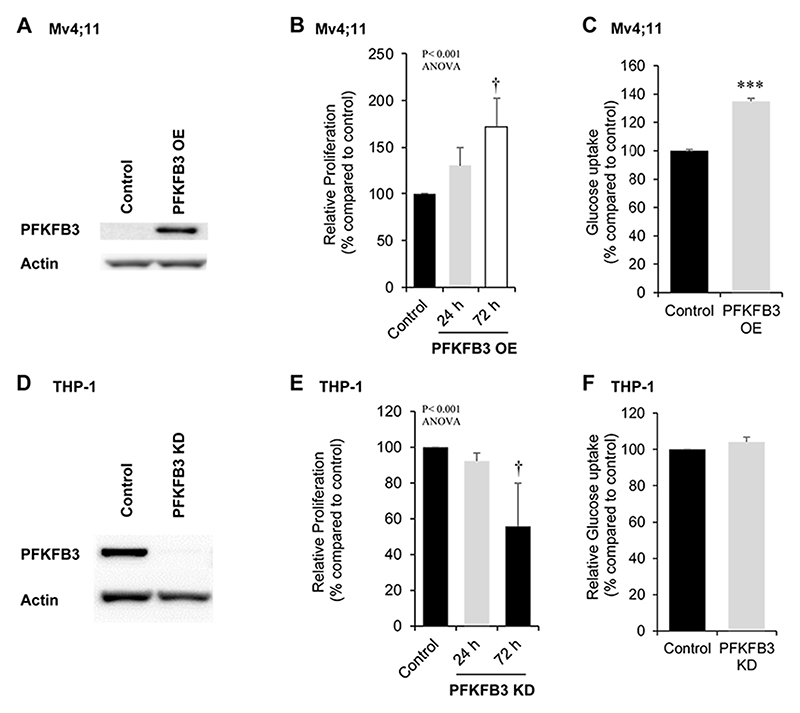
To investigate whether PFKFB3 itself could mediate these phenotypic changes, we overexpressed PFKFB3 in Mv4;11 cells (PFKFB3-OE; Fig. 5A). Cells overexpressing PFKFB3 showed a 72±30% increase in proliferation (at 72 h) compared to control cells (Fig. 5B) and correspondingly showed an increased proportion of cells in S+G2M phase of the cell cycle (Supplemental Fig. S10A). The levels of glucose uptake in PFKFB3-OE cells were significantly more (35±2%) compared to control cells (Fig. 5C). To determine the impact of decreased expression of PFKFB3 in ROS generating cells, PFKFB3 was knocked-down in THP-1 cells (Fig. 5D). Knock-down of PFKFB3 resulted in reduced proliferation of these cells (Fig. 5E) and a reduction in the percentage of cells in cycle (Supplemental Fig S10B), though without detectable change in glucose uptake in this context (Fig. 5F). These experiments provide the first direct evidence that PFKFB3 controls the growth of leukemia cells which is consistent with cells producing high levels of ROS.
Figure 5. Effect of PFKFB3 overexpression and knock down on proliferation and glucose uptake in leukemia cells.
(A) Western blot analysis of PFKFB3 protein comparing control and PFKFB3-overexpression (OE) Mv4;11 cells. Actin was used as a loading control. (B) Percentage proliferation (normalised to control) of Mv4;11 PFKFB3 over-expressed (OE) cells compared to control (n>3). (C) Glucose uptake in Mv4;11 PFKFB3-OE cells compared to control following 72 h (n=3). Glucose was assayed in the culture media and glucose uptake in the cell is inversely proportional to the glucose remaining in the media. The concentration of glucose in the culture media (starting concentration 25nmol/μL) of PFKFB3-OE cells after 24 h was 15nmol/μL compared with 23nmol/μL in control cells. (D) Western blot analysis of PFKFB3 protein comparing control (non-mammalian targeting shRNA control) and PFKFB3 knock down (KD) in THP-1 cells. Actin was used as a loading control. (E) Percentage proliferation (normalised to control) of THP-1 PFKFB3-KD cells compared to control (n>3). (F) Glucose uptake in single cell analysis using 2-NBDG (normalised to control) of THP-1 PFKFB3-KD cells compared to control following 24h growth (n=5). Data represents mean±1SD. † denotes p<0.05 analysed by ANOVA with Tukey’s honestly significance difference. ***p<0.005 denotes statistical significance calculated by Student’s t-test calculated.
ROS induced changes in PFKFB3 expression are mediated via UCP/p-AMPK
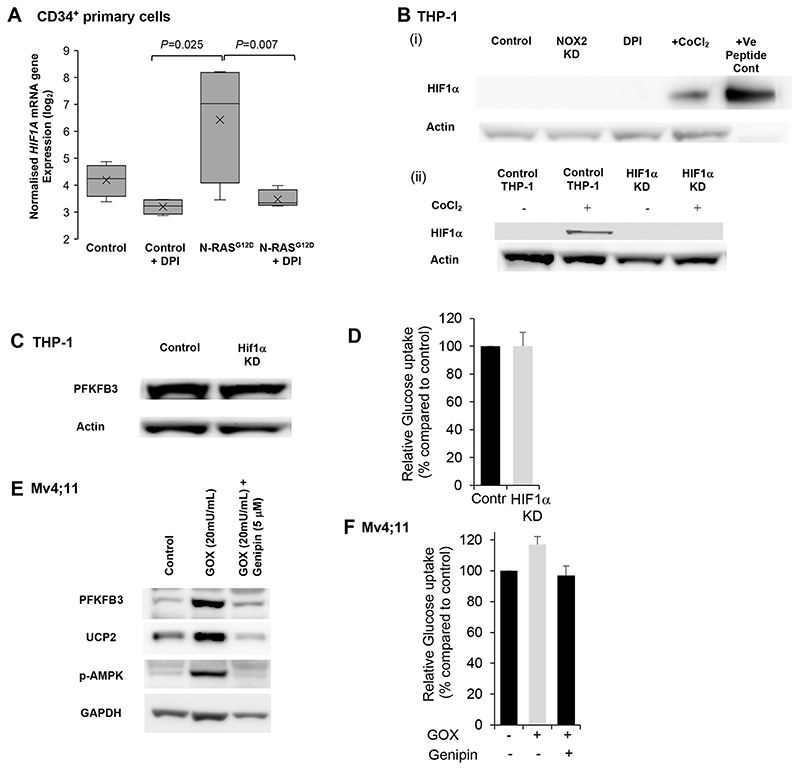
To determine the mechanism of ROS induced PFKFB3 expression we analysed two potential mechanisms. Firstly, HIF-1α has been shown to induce increased PFKFB3 mRNA expression (20). Analysing our transcriptome data revealed that HIF-1α is a strongly ROS-responsive gene (Fig. 6A). However, we were unable to detect expression of HIF1-α protein regardless of NOX2 status (Fig. 6B). Furthermore, HIF1α knock-down (Fig. 6B) did not change PFKFB3 protein expression (Fig. 6C) or glucose uptake (Fig. 6D). Collectively, these data do not therefore support a role of HIF1-α in mediating glycolytic changes in these cells.
Figure 6. ROS induced changes in PFKFB3 expression is mediated via UCP/p-AMPK.
(A) Normalised log2 expression of HIFA (transcript ID 3567728) mRNA in control HSPC, DPI, N-RASG12D HSPC and N-RASG12D HSPC treated with DPI (100nM). Box plots represent median quartile ranges, x represents mean value (n=4). p-Value calculated by 2-way ANOVA with Bonferroni multiple testing correction. (B)(i) Western blot showing expression of HIF-1α in control THP-1, THP-1 NOX2-KD and THP-1 cells treated with DPI as (A). As positive controls, THP-1 cells were treated with CoCl2 as indicated (an inhibitor of HIF-1α degradation (50)). HIF-1α recombinant protein were also immunoblotted. Actin was used as loading control. (B)(ii) Western blot showing expression of HIF-1α, comparing control THP-1 (non-mammalian shRNA target) and THP-1 HIF-1α knocked down (KD). Cells were also untreated or treated with CoCl2 as above. (C) Western blot showing expression of PFKFB3 in THP-1 cells knocked down with HIF1-α. (D) Glucose uptake (normalised to control) of THP HIF-1α KD cells (n=3). Data represents mean±1SD. (E) Immunoblot showing PFKFB3, UCP2 and p-AMPK expression upon 1 h pre-treatment of Mv4;11 cells with the UCP2 inhibitor Genipin (5μM) followed by GOX treatment for 24 h. (F) Glucose uptake using 2-NBDG (normalised to untreated cells; control) of Mv4;11 cells treated with GOX (20mU/mL) and / or Genipin (5μM) (n=5). Data represents mean±1SD. ***p<0.005 and §p<0.01 denotes statistical significance calculated by Student’s t-test calculated.
Superoxide levels are sensed by uncoupling protein 2 (UCP2) which drives an adaptive response to protect against oxidative stress including the activation of AMP-activated protein kinase (AMPK) (21) leading to increased production of PFKFB3 (22). In accord with this we observed that treatment of the ROSLow AML cell line, Mv4;11, with exogenous H2O2 increased both PFKFB3 expression and AMPK phosphorylation (Fig. 6E). This induction was inhibited by Genipin (which specifically inhibits UCP2 expression (23)) decreasing both p-AMPK and PFKFB3 levels (Fig. 6D) and concomitant reduction in glucose consumption (Fig. 6F). To confirm, changes in p-AMPK/mTOR signalling, we next examined downstream signalling of mTOR by western blot. As shown in Supplemental Fig. S11A and B phosphorylation of S6 kinase is decreased following treatment with H2O2; an effect reversed when UCP2 is inhibited. These data support a role for UCP2 having a pivotal role in mediating ROS-induced changes in the expression of PFKFB3.
Targeting PFKFB3 reduces glucose uptake and cell proliferation in AML cells
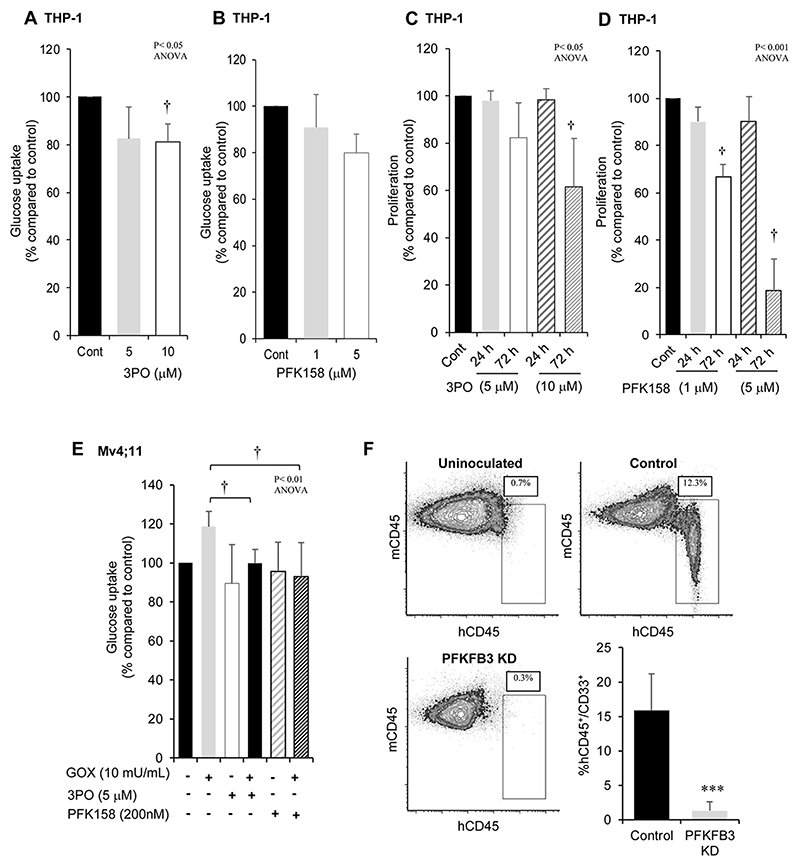
We next wanted to establish whether chemical inhibition of PFKFB3 could have the potential for a therapeutic impact in the treatment of AML. Having established appropriate inhibitory concentrations of two chemical inhibitors of PFKFB3, 3-(3-pyridinyl)-1-(4-pyridinyl)-2-propen-1-one (3PO) (24) and the more specific PFK158 (25) (Supplemental Fig. S12A-C). We next treated ROSHigh cells (THP-1) with 3-PO or PFK158 which resulted in a reduction in glucose uptake (Fig. 7A and B). Correspondingly, treatment with 3PO resulted in a significant dose dependent decrease in proliferation of THP-1 cells (Fig. 7C) whilst treatment with the more specific PFK158 (25) also significantly reduced proliferation at doses >500nM (Fig. 7D). We next investigated whether PFKFB3 inhibition could suppress the effects of exogenous ROS on the ROSLow cell line Mv4;11. No change in viability in Mv4;11 (or THP-1) cells treated with 3PO, PFK158 or H2O2 were observed. Treatment of Mv4;11 cells with H2O2 resulted in a 20±9% increase in glucose uptake compared with control (as expected), whilst combined treatment with H2O2 and 3PO or with PFK158 inhibitor ablated the response to peroxide (Fig. 7E). Treatment with these inhibitors alone did not alter glucose uptake when compared to control cells.
Figure 7. Targeting PFKFB3 reduces glucose uptake and cell proliferation in AML cells.
Glucose uptake using 2NBDG (normalised to untreated control) in THP-1 cells lines treated with (A) 3PO or (B) PFK158 for 24 h. Vehicle control for 3PO and PFK158 was DMSO 0.05% and DMSO 0.01% respectively. Data represents mean±1SD (n>3). Proliferation (normalised to control) in THP-1 cells, seeded at 4 x 105/mL treated with (C) 3PO or (D) PFK158. Vehicle control for 3PO, DMSO 0.05% (n=6). Vehicle control for PFK158, DMSO 0.01% (n=6). (E) Glucose uptake using 2NBDG (normalised to untreated control) in Mv4;11 cells treated with inhibitors to PFKFB3 and/or incubated with 10 mU/mL GOX (source of H2O2) (n=3). (F) Representative flow cytometric bivariate plots from bone marrow harvested from tibias and femurs of adult NSG mice (7-10 weeks old) sub-lethally irradiated with 200cGy total body irradiation 24 h before injection of control THP-1 cells or THP-1 cells where PFKFB3 was KD. Human cells were distinguished from mouse cells using hCD45-FITC, hCD33-APC and mCD45-PerCP-Cy5.5 antibodies. Uninoculated NSG mice were used to control for the analysis of THP-1 engraftment (n=4) and analysed at week 6. Data represents mean±1SD. † denotes p<0.05 and represent significantly different groups from control, analysed by ANOVA with Tukey’s honestly significance difference. ***p<0.005 denotes statistical significance calculated by Student’s t-test calculated.
Finally, we investigated whether loss of PFKFB3 also affected leukemia growth in vivo. Knock-down of PFKFB3 expression in THP-1 cells resulted in a significant reduction (83%) of leukemia cell growth, supporting a role of PFKFB3 in the proliferation of AML cells in vivo (Fig. 7F). Overall these experiments provide the first direct evidence that oxidase-derived ROS promotes the growth of leukemia cells via PFKFB3 expression and suggests a potential ROS-dependent mechanism for these changes.
Discussion
Whilst increased levels of ROS produced by AML blasts or tumor cells have been shown to induce double strand breaks (26), they also promote proliferation (6, 27). Here we present evidence that the proliferative response to ROS is supported by changes in glucose uptake and altered glucose metabolism. Furthermore, we demonstrate that AML cells exhibit enhanced PFKFB3 expression and that inhibitors that target the function of this protein (or its upstream pathway UCP2/AMPK) may provide a tractable therapeutic target in AML.
ROS are now recognized as important secondary messengers, serving as critical cell signaling molecules through the capacity of H2O2 to reversibly oxidise cysteine residues (28). Using a primary cell model for ROS production, we investigated the effect of ROS on gene transcription and found that changes in mRNA expression were associated with enzymes involved in glucose metabolism. Specifically, we identified several gene changes associated with glycolysis coupled with increased cellular glucose uptake. This is consistent with many solid tumor models that shift energy production from oxidative phosphorylation toward the less efficient glycolytic pathway, a phenomenon known as the Warburg effect (29). Indeed, our previous data have shown no significant differences in mitochondrial ROS were observed in mutant Ras expressing CD34+ cells compared to controls (7). Using inhibitors to NOX2, we were able to attribute the effects of observed increases in glucose uptake to the production of extracellular NOX2-derived ROS. Additionally, we found leukemic cell lines increased glucose uptake in response to exogenous ROS or showed decreased uptake following NOX2 knock-down. We show for the first time that the effect of ROS on glucose uptake is mediated by PFKFB3 (see below). However, pro-glycolytic effects of ROS on other genes have been observed (30) and is consistent with increased glycolysis in solid tumor models associated with GLUT upregulation (31).
In the context of primary AML, we previously demonstrated that >60% of patients exhibited high levels of ROS which correlated with NOX2 expression (6). Here, AML blasts stratified according to level of extracellular ROS production were analysed by global biochemical metabolomic profiling. Several hundred metabolites were identified which distinguished patients according to ROS production including metabolite levels within the glycolytic pathway (glucose, glucose-6-phosphate and fructose-6-phosphate) suggesting that the impact of ROS is to increase the levels of glycolytic intermediates, primarily those in the early part of the glycolytic pathway. In support of this, metabolites associated with the PPP such as sedoheptulose-7-phosphate and the isobaric compounds ribulose/xylulose 5-phosphate were also elevated in ROSHigh samples. These data may be indicative of a reprogramming of metabolic pathways, where increased glucose consumption, is metabolised via the PPP, resulting in increased generation of NADPH (to counter oxidative stress) and biosynthetic precursors such as nucleotides, necessary for cell growth and DNA repair. Interestingly, metabolomic profiling of serum from AML patients has also revealed distinct increases in the glycolytic metabolic (32, 33). Additionally, NOX2 expression has previously been demonstrated to regulate self-renewal of leukemic stem cells (34). Using a murine model of leukemia, Adane et al showed that suppression of NOX2 expression led to increased fatty acid oxidation and potential limiting of substrates passing through glycolysis. Our study supports this notion, where we also observed significant changes associated with lipid metabolism in human AML; lipid-related changes in n3 and n6 polyunsaturated fatty acids showed significant accumulations within the AMLHigh in relation to the AMLLow samples. The higher levels of monoacylglycerols and glycerol in the AMLHigh samples may also be an indicator of increased lipolysis to support free fatty acid levels. Taken together, these data show that NOX2 derived ROS impact on glucose metabolism, an effect consistent with that seen in solid tumors (35).
We identified significant ROS induced changes in mRNA and in protein of the regulatory glycolytic enzyme PFKFB3. PFKFB3 mRNA expression has been shown to be upregulated in several solid tumors including colon, breast, prostate, ovary, thyroid and head and neck squamous cell carcinoma (36, 37). Whilst PFKFB3 mRNA is elevated in AML we did not observe any significant association of expression with particular AML subgroups or clinical outcome. PFKFB is a bifunctional enzyme, which catalyses both forward and reverse reaction of F-6-P to F-2,6-BP (19). In turn, F-2,6-BP is a powerful allosteric activator of PFK which catalyses F-6-P to F-1,6-BP, a rate limiting step in glycolysis. The PFKFB3 isoform contains a lysine and serine at position 79 and 80 respectively (38) resulting in increased kinase activity, which is 740 times greater than other PFKFB isoforms, making it a powerful driver of glycolysis (39). Here we also showed ROS dependent changes in PFKFB3 expression in AML lines with constitutive NOX2 activity/ROS production coupled with suppression of reduction in proliferation and glucose uptake upon ROS inhibition. Overexpression/knock-down of PFKFB3 generated the predicted changes in glucose uptake and cell proliferation in vitro whilst knock-down of PFKFB3 strongly suppresses leukemia growth in vivo (though we have not confirmed what the mechanistic basis of the effects are, in vivo data).
It has previously been established that ROS can regulate HIF-1α in a non-hypoxic pathway (40). Further, stabilisation of HIF-1α has been associated with increased expression of glycolytic genes, including PFKFB3 (41, 42). Our data showed increased expression of HIF-1α mRNA correlated with increased ROS levels. However, immunoblotting showed that HIF-1α was not expressed at detectable levels and furthermore knock-down of the mRNA for this protein did not result in any changes in glucose uptake. ROS has also previously been shown to activate mitochondrial proteins (UCP2) to regulate the leak of protons across the inner membrane, resulting in poor fuel conversion efficiency and a more pro-glycolytic phenotype including AMPK activation (21). We show that inhibition of UCP2 led to decreased p-AMPK and PFKFB3 levels. Further, analysis of the mTOR pathway showed decreased S6-Kinase expression in response to ROS which was reverted upon UCP inhibition. AMPK is a master regulator of cellular energy homeostasis, upregulating catabolic metabolic processes including increased glycolytic flux and protects against ROS accumulation by increasing NADPH production (see below) (17). AMPK has previously been shown to be activated via ROS (43), in addition to regulating glycolysis and PFKFB3 expression in a phosphorylation dependent manner in cancer cells (22, 44). Domenech et al., have shown that mitotic arrest of cancer cells leads to altered energy requirements through switching to a more glycolytic phenotype and increased AMPK phosphorylation (22). This suggests that PFKFB3 could also be increased through changes to cell cycle or autophagy. Metabolomic data generated as part of this study also indicated increased levels of ROS correlated with an increase in fatty acid metabolites (Supplemental Table S2). Interestingly, changes in expression of UCP2 has been linked with 2-3 fold elevation of plasma fatty acids reviewed in (45). Further, activation of fatty acid metabolism by AMPK (46) is interesting given that ROS activation of AMPK has also been shown to potentially influence and maintain HSC (47). Conversely, inhibition of mitochondrial fatty acid oxidation induces loss of HSC maintenance (48).
Identification of metabolic differences between normal and malignant tissue creates a therapeutic opportunity for targeting of glycolysis in the treatment of AML; the potential for targeted therapy in AML, through reduction of aberrant metabolic activity via inhibition of 6-phosphogluconate dehydrogenase (6-PGD) function has also recently been shown (32). The therapeutic potential of PFKFB3 inhibition (e.g. PFK158) (25) in cancer is currently undergoing phase I clinical trials (49). Consistent with studies in solid tumors (50, 51), data presented here shows that in ROSHigh AML cells, treatment with PFKFB3 inhibitor significantly reduced glucose uptake and also proliferation both in vitro and in vivo. This supports a previous study which showed chemical inhibition of PFKFB3 in Jurkat T-cell leukemia cells results in decreased proliferation and glucose uptake (24). In myeloproliferative neoplasms expressing JAK2 mutations, PFKFB3 is required for increased growth and metabolic activity, an effect blocked by targeted knock-down of PFKFB3. This study therefore suggested that therapies specifically blocking PFKFB3 activity/expression would be expected to inhibit JAK2/STAT5-dependent malignancies (52).
In conclusion, this and previous data suggest that production of ROS may confer a competitive advantage on premalignant/malignant cells by promoting the proliferation of these cells via changes to carbohydrate metabolism. We show for the first time a link between increased NOX-derived ROS production and increased expression of the key glycolytic regulatory enzyme, PFKFB3. Furthermore, PFKFB3 inhibitors or genetic knock-down established a causal link between ROS production, cellular glucose uptake and PFKFB3 activity.
Supplementary Material
Supplemental Information
Supplemental information is available at Cancer Research website.
Significance.
Findings show that ROS generated by NOX2 in AML cells promotes glycolysis by activating PFKFB3, and suggest PFKFB3 as a novel therapeutic target in AML.
Acknowledgments
We are grateful to Nick Jones (Swansea University, Wales, UK) for critically appraising the manuscript. We would also like to thank Megan Musson CBS, Cardiff University for her technical expertise for Affymetrix Microarray. We are grateful to Prof Tee (Cardiff University) for his expertise in AMPK/mTOR signalling advice. The authors are grateful for support from the NCRI AML trials cell bank and the AML patients for providing primary samples used in this study. This work was supported by grants from Tenovus Cancer Care (A.R), Bloodwise (13029), Medical Research Council (G.L.H.) and Health and Care Research Wales (G.L.H; H07-3-06). NR is a Sêr Cymru II Fellow supported by Welsh Government, European Regional Development Fund (NR; 80762-CU-182).
¥. Funding
This work was supported by grants from Tenovus Cancer Care (A.R), Bloodwise (13029), Medical Research Council (G.L.H.), Health and Care Research Wales (G.L.H; H07-3-06), Cancer Research UK (C7838/A25173) and Sêr Cymru II Fellow supported by Welsh Government, European Regional Development Fund (NR; 80762-CU-182)
Footnotes
Conflict of interest: The authors declare no conflict of interest.
References
- (1).Lambeth JD, Neish AS. Nox enzymes and new thinking on reactive oxygen: a double-edged sword revisited. Annu Rev Pathol. 2014;9:119–45. doi: 10.1146/annurev-pathol-012513-104651. [DOI] [PubMed] [Google Scholar]
- (2).Hole PS, Darley RL, Tonks A. Do reactive oxygen species play a role in myeloid leukemias? Blood. 2011;117:5816–26. doi: 10.1182/blood-2011-01-326025. [DOI] [PubMed] [Google Scholar]
- (3).Bindoli A, Rigobello MP. Principles in redox signaling: from chemistry to functional significance. Antioxid Redox Signal. 2013;18:1557–93. doi: 10.1089/ars.2012.4655. [DOI] [PubMed] [Google Scholar]
- (4).Prieto-Bermejo R, Romo-Gonzalez M, Perez-Fernandez A, Ijurko C, Hernandez-Hernandez A. Reactive oxygen species in haematopoiesis: leukaemic cells take a walk on the wild side. J Exp Clin Cancer Res. 2018;37:125. doi: 10.1186/s13046-018-0797-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Mesbahi Y, Zekri A, Ghaffari SH, Tabatabaie PS, Ahmadian S, Ghavamzadeh A. Blockade of JAK2/STAT3 intensifies the anti-tumor activity of arsenic trioxide in acute myeloid leukemia cells: Novel synergistic mechanism via the mediation of reactive oxygen species. Eur J Pharmacol. 2018;834:65–76. doi: 10.1016/j.ejphar.2018.07.010. [DOI] [PubMed] [Google Scholar]
- (6).Hole PS, Zabkiewicz J, Munje C, Newton Z, Pearn L, White P, et al. Overproduction of NOX-derived ROS in AML promotes proliferation and is associated with defective oxidative stress signaling. Blood. 2013;122:3322–30. doi: 10.1182/blood-2013-04-491944. [DOI] [PubMed] [Google Scholar]
- (7).Hole PS, Pearn L, Tonks AJ, James PE, Burnett AK, Darley RL, et al. Ras-induced reactive oxygen species promote growth factor-independent proliferation in human CD34+ hematopoietic progenitor cells. Blood. 2010;115:1238–46. doi: 10.1182/blood-2009-06-222869. [DOI] [PubMed] [Google Scholar]
- (8).Jayavelu AK, Moloney JN, Bohmer FD, Cotter TG. NOX-driven ROS formation in cell transformation of FLT3-ITD-positive AML. Exp Hematol. 2016;44:1113–22. doi: 10.1016/j.exphem.2016.08.008. [DOI] [PubMed] [Google Scholar]
- (9).Qian X, Nie X, Yao W, Klinghammer K, Sudhoff H, Kaufmann AM, et al. Reactive oxygen species in cancer stem cells of head and neck squamous cancer. Semin Cancer Biol. 2018;53:248–57. doi: 10.1016/j.semcancer.2018.06.001. [DOI] [PubMed] [Google Scholar]
- (10).Tonks A, Pearn L, Tonks AJ, Pearce L, Hoy T, Phillips S, et al. The AML1-ETO fusion gene promotes extensive self-renewal of human primary erythroid cells. Blood. 2003;101:624–32. doi: 10.1182/blood-2002-06-1732. [DOI] [PubMed] [Google Scholar]
- (11).Tonks A, Pearn L, Musson M, Gilkes A, Mills KI, Burnett AK, et al. Transcriptional dysregulation mediated by RUNX1-RUNX1T1 in normal human progenitor cells and in acute myeloid leukaemia. Leukemia. 2007;21:2495–505. doi: 10.1038/sj.leu.2404961. [DOI] [PubMed] [Google Scholar]
- (12).Omidvar N, Kogan S, Beurlet S, le PC, Janin A, West R, et al. BCL-2 and mutant NRAS interact physically and functionally in a mouse model of progressive myelodysplasia. Cancer Res. 2007;67:11657–67. doi: 10.1158/0008-5472.CAN-07-0196. [DOI] [PubMed] [Google Scholar]
- (13).Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:l1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–4. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Ley TJ, Miller C, Ding L, Raphael BJ, Mungall AJ, Robertson A, et al. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013;368:2059–74. doi: 10.1056/NEJMoa1301689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Mikawa T, LLeonart ME, Takaori-Kondo A, Inagaki N, Yokode M, Kondoh H. Dysregulated glycolysis as an oncogenic event. Cell Mol Life Sci. 2015;72:1881–92. doi: 10.1007/s00018-015-1840-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Altenhofer S, Kleikers PW, Radermacher KA, Scheurer P, Rob Hermans JJ, Schiffers P, et al. The NOX toolbox: validating the role of NADPH oxidases in physiology and disease. Cell Mol Life Sci. 2012;69:2327–43. doi: 10.1007/s00018-012-1010-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Beckman JS, Minor RL, Jr, White CW, Repine JE, Rosen GM, Freeman BA. Superoxide dismutase and catalase conjugated to polyethylene glycol increases endothelial enzyme activity and oxidant resistance. J Biol Chem. 1988;263:6884–92. [PubMed] [Google Scholar]
- (19).Ros S, Schulze A. Balancing glycolytic flux: the role of 6-phosphofructo-2-kinase/fructose 2,6-bisphosphatases in cancer metabolism. Cancer Metab. 2013;1:8. doi: 10.1186/2049-3002-1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Yalcin A, Clem BF, Simmons A, Lane A, Nelson K, Clem AL, et al. Nuclear targeting of 6-phosphofructo-2-kinase (PFKFB3) increases proliferation via cyclin-dependent kinases. J Biol Chem. 2009;284:24223–32. doi: 10.1074/jbc.M109.016816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Echtay KS, Roussel D, St-Pierre J, Jekabsons MB, Cadenas S, Stuart JA, et al. Superoxide activates mitochondrial uncoupling proteins. Nature. 2002;415:96–9. doi: 10.1038/415096a. [DOI] [PubMed] [Google Scholar]
- (22).Domenech E, Maestre C, Esteban-Martinez L, Partida D, Pascual R, Fernandez-Miranda G, et al. AMPK and PFKFB3 mediate glycolysis and survival in response to mitophagy during mitotic arrest. Nat Cell Biol. 2015;17:1304–16. doi: 10.1038/ncb3231. [DOI] [PubMed] [Google Scholar]
- (23).Mailloux RJ, Adjeitey CN, Harper ME. Genipin-induced inhibition of uncoupling protein-2 sensitizes drug-resistant cancer cells to cytotoxic agents. PLoS One. 2010;5:e13289. doi: 10.1371/journal.pone.0013289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Clem B, Telang S, Clem A, Yalcin A, Meier J, Simmons A, et al. Small-molecule inhibition of 6-phosphofructo-2-kinase activity suppresses glycolytic flux and tumor growth. Mol Cancer Ther. 2008;7:110–20. doi: 10.1158/1535-7163.MCT-07-0482. [DOI] [PubMed] [Google Scholar]
- (25).Clem BF, O’Neal J, Tapolsky G, Clem AL, Imbert-Fernandez Y, Kerr DA, et al. Targeting 6-phosphofructo-2-kinase (PFKFB3) as a therapeutic strategy against cancer. Mol Cancer Ther. 2013;12:1461–70. doi: 10.1158/1535-7163.MCT-13-0097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Stanicka J, Russell EG, Woolley JF, Cotter TG. NADPH oxidase-generated hydrogen peroxide induces DNA damage in mutant FLT3-expressing leukemia cells. J Biol Chem. 2015;290:9348–61. doi: 10.1074/jbc.M113.510495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Tang CT, Lin XL, Wu S, Liang Q, Yang L, Gao YJ, et al. NOX4-driven ROS formation regulates proliferation and apoptosis of gastric cancer cells through the GLI1 pathway. Cell Signal. 2018;46:52–63. doi: 10.1016/j.cellsig.2018.02.007. [DOI] [PubMed] [Google Scholar]
- (28).Paik JY, Jung KH, Lee JH, Park JW, Lee KH. Reactive oxygen species-driven HIF1alpha triggers accelerated glycolysis in endothelial cells exposed to low oxygen tension. Nucl Med Biol. 2017;45:8–14. doi: 10.1016/j.nucmedbio.2016.10.006. [DOI] [PubMed] [Google Scholar]
- (29).Koppenol WH, Bounds PL, Dang CV. Otto Warburg’s contributions to current concepts of cancer metabolism. Nat Rev Cancer. 2011;11:325–37. doi: 10.1038/nrc3038. [DOI] [PubMed] [Google Scholar]
- (30).Prata C, Maraldi T, Fiorentini D, Zambonin L, Hakim G, Landi L. Nox-generated ROS modulate glucose uptake in a leukaemic cell line. Free Radic Res. 2008;42:405–14. doi: 10.1080/10715760802047344. [DOI] [PubMed] [Google Scholar]
- (31).Macheda ML, Rogers S, Best JD. Molecular and cellular regulation of glucose transporter (GLUT) proteins in cancer. J Cell Physiol. 2005;202:654–62. doi: 10.1002/jcp.20166. [DOI] [PubMed] [Google Scholar]
- (32).Chen WL, Wang JH, Zhao AH, Xu X, Wang YH, Chen TL, et al. A distinct glucose metabolism signature of acute myeloid leukemia with prognostic value. Blood. 2014;124:1645–54. doi: 10.1182/blood-2014-02-554204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Wang Y, Zhang L, Chen WL, Wang JH, Li N, Li JM, et al. Rapid diagnosis and prognosis of de novo acute myeloid leukemia by serum metabonomic analysis. J Proteome Res. 2013;12:4393–401. doi: 10.1021/pr400403p. [DOI] [PubMed] [Google Scholar]
- (34).Adane B, Ye H, Khan N, Pei S, Minhajuddin M, Stevens BM, et al. The Hematopoietic Oxidase NOX2 Regulates Self-Renewal of Leukemic Stem Cells. Cell Rep. 2019;27:238–54. doi: 10.1016/j.celrep.2019.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Shanmugam M, McBrayer SK, Rosen ST. Targeting the Warburg effect in hematological malignancies: from PET to therapy. Curr Opin Oncol. 2009;21:531–6. doi: 10.1097/CCO.0b013e32832f57ec. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Atsumi T, Chesney J, Metz C, Leng L, Donnelly S, Makita Z, et al. High expression of inducible 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase (iPFK-2; PFKFB3) in human cancers. Cancer Res. 2002;62:5881–7. [PubMed] [Google Scholar]
- (37).Li HM, Yang JG, Liu ZJ, Wang WM, Yu ZL, Ren JG, et al. Blockage of glycolysis by targeting PFKFB3 suppresses tumor growth and metastasis in head and neck squamous cell carcinoma. J Exp Clin Cancer Res. 2017;36:7. doi: 10.1186/s13046-016-0481-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Kim SG, Manes NP, El-Maghrabi MR, Lee YH. Crystal structure of the hypoxia-inducible form of 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase (PFKFB3): a possible new target for cancer therapy. J Biol Chem. 2006;281:2939–44. doi: 10.1074/jbc.M511019200. [DOI] [PubMed] [Google Scholar]
- (39).Marsin AS, Bouzin C, Bertrand L, Hue L. The stimulation of glycolysis by hypoxia in activated monocytes is mediated by AMP-activated protein kinase and inducible 6-phosphofructo-2-kinase. J Biol Chem. 2002;277:30778–83. doi: 10.1074/jbc.M205213200. [DOI] [PubMed] [Google Scholar]
- (40).Haddad JJ, Land SC. A non-hypoxic, ROS-sensitive pathway mediates TNF-alpha-dependent regulation of HIF-1alpha. FEBS Lett. 2001;505:269–74. doi: 10.1016/s0014-5793(01)02833-2. [DOI] [PubMed] [Google Scholar]
- (41).Chesney J, Telang S. Regulation of glycolytic and mitochondrial metabolism by ras. Curr Pharm Biotechnol. 2013;14:251–60. doi: 10.2174/1389201011314030002. [DOI] [PubMed] [Google Scholar]
- (42).Yalcin A, Telang S, Clem B, Chesney J. Regulation of glucose metabolism by 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatases in cancer. Exp Mol Pathol. 2009;86:174–9. doi: 10.1016/j.yexmp.2009.01.003. [DOI] [PubMed] [Google Scholar]
- (43).Shi SY, Lu SY, Sivasubramaniyam T, Revelo XS, Cai EP, Luk CT, et al. DJ-1 links muscle ROS production with metabolic reprogramming and systemic energy homeostasis in mice. Nat Commun. 2015;6:7415. doi: 10.1038/ncomms8415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Novellasdemunt L, Bultot L, Manzano A, Ventura F, Rosa JL, Vertommen D, et al. PFKFB3 activation in cancer cells by the p38/MK2 pathway in response to stress stimuli. Biochem J. 2013;452:531–43. doi: 10.1042/BJ20121886. [DOI] [PubMed] [Google Scholar]
- (45).Thompson MP, Kim D. Links between fatty acids and expression of UCP2 and UCP3 mRNAs. FEBS Lett. 2004;568:4–9. doi: 10.1016/j.febslet.2004.05.011. [DOI] [PubMed] [Google Scholar]
- (46).Carracedo A, Cantley LC, Pandolfi PP. Cancer metabolism: fatty acid oxidation in the limelight. Nat Rev Cancer. 2013;13:227–32. doi: 10.1038/nrc3483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Liu X, Zheng H, Yu WM, Cooper TM, Bunting KD, Qu CK. Maintenance of mouse hematopoietic stem cells ex vivo by reprogramming cellular metabolism. Blood. 2015;125:1562–5. doi: 10.1182/blood-2014-04-568949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Ito K, Carracedo A, Weiss D, Arai F, Ala U, Avigan DE, et al. A PML-PPAR-delta pathway for fatty acid oxidation regulates hematopoietic stem cell maintenance. Nat Med. 2012;18:1350–8. doi: 10.1038/nm.2882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Lu L, Chen Y, Zhu Y. The molecular basis of targeting PFKFB3 as a therapeutic strategy against cancer. Oncotarget. 2017;8:62793–802. doi: 10.18632/oncotarget.19513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Zhu W, Ye L, Zhang J, Yu P, Wang H, Ye Z, et al. PFK15, a Small Molecule Inhibitor of PFKFB3, Induces Cell Cycle Arrest, Apoptosis and Inhibits Invasion in Gastric Cancer. PLoS One. 2016;11:e0163768. doi: 10.1371/journal.pone.0163768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).O’Neal J, Clem A, Reynolds L, Dougherty S, Imbert-Fernandez Y, Telang S, et al. Inhibition of 6-phosphofructo-2-kinase (PFKFB3) suppresses glucose metabolism and the growth of HER2+ breast cancer. Breast Cancer Res Treat. 2016;160:29–40. doi: 10.1007/s10549-016-3968-8. [DOI] [PubMed] [Google Scholar]
- (52).Reddy MM, Fernandes MS, Deshpande A, Weisberg E, Inguilizian HV, Abdel-Wahab O, et al. The JAK2V617F oncogene requires expression of inducible phosphofructokinase/fructose-bisphosphatase 3 for cell growth and increased metabolic activity. Leukemia. 2012;26:481–9. doi: 10.1038/leu.2011.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.