Abstract
Protein O-GlcNAcylation is a reversible post-translational modification of serines/threonines on nucleocytoplasmic proteins. It is cycled by the enzymes O-GlcNAc transferase (OGT) and O-GlcNAc hydrolase (O-GlcNAcase or OGA). Genetic approaches in model organisms have revealed that protein O-GlcNAcylation is essential for early embryogenesis. Drosophila melanogaster OGT/supersex combs (sxc) is a polycomb gene, null mutants of which display homeotic transformations and die at the pharate adult stage. However, the identities of the O-GlcNAcylated proteins involved, and the underlying mechanisms linking these phenotypes to embryonic development, are poorly understood. Identification of O-GlcNAcylated proteins from biological samples is hampered by the low stoichiometry of this modification and limited enrichment tools. Using a catalytically inactive bacterial O-GlcNAcase mutant as a substrate trap, we have enriched the O-GlcNAc proteome of the developing Drosophila embryo, identifying, amongst others, known regulators of Hox genes as candidate conveyors of OGT function during embryonic development.
Introduction
O-GlcNAcylation, the addition of a single O-linked β-N-acetylglucosamine (O-GIcNAc) to serine or threonine residues on target proteins, is a post-translational modification of nucleocytoplasmic proteins regulated by two enzymes, O-GlcNAc transferase (OGT) and O-GlcNAcase (OGA)1. The donor substrate for protein O-GlcNAcylation is UDP-GlcNAc, produced from the glycolytic metabolite fructose-6-phosphate through the hexosamine biosynthetic pathway. Protein O-GlcNAcylation is a dynamic and reversible modification and is responsive to alterations in nutrient status and cellular stimuli1 and has been implicated in a broad range of cellular process including gene expression, protein trafficking and degradation, stress response1 and autophagy2. Alterations in tissue specific protein O-GlcNAcylation profiles have been linked to a number of human pathologies including diabetes, cancer, cardiovascular disease and neurodegenerative disorders1. In addition, using genetic approaches, it has been demonstrated that OGT, and by extension, protein O-GlcNAcylation, has a critical role in embryonic development in animals3–6, although the mechanisms underpinning this remain largely unclear.
An attractive model organism to begin to dissect the links between protein O-GlcNAcylation and metazoan development is the fruit fly Drosophila melanogaster. Flies that lack zygotic expression of OGT/sxc, but retain maternally contributed OGT protein and transcripts, die at the late pupal pharate adult stage with distinct homeotic transformations3. Flies lacking both zygotic and maternal OGT/sxc arrest development at the end of embryogenesis and show homeotic transformations in the embryonic cuticle3. Studies employing ChIP experiments have shown that O-GlcNAc is highly enriched at polycomb responsive elements (PREs) in Hox and other gene clusters in Drosophil a 7,8. The transcription factor Polyhomeotic (Ph) is a polycomb group protein known to be O-GlcNAcylated7. It has been shown that O-GlcNAcylation of Ph prevents its aggregation, and is required for the formation of functional, ordered assemblies of the protein9. OGT/sxc null mutants recapitulate some of the developmental phenotypes of Ph null mutants10. Other studies in flies have described the association of O-GlcNAc with cellular processes like glucose-insulin homeostasis11, circadian rhythm12–14, temperature stress during development15, FGF signaling16 and autophagy17. It is therefore clear that protein O-GlcNAcylation is involved in several processes in the fly in addition to Ph-dependent Hox gene repression. Our discovery that protein O-GlcNAcylation is dynamic during Drosophila embryogenesis18 led us to pursue the proteomics-based identification of the modified proteins to aid the understanding of the mechanisms responsible for the OGT/sxc null phenotypes. While many proteomics studies have focused on the identification of O-GlcNAcylated proteins in mammalian cells and tissues, there is only a single study reporting O-GlcNAcylated proteins from Drosophila S2 cells with no site assignments19.
Identification of native O-GlcNAcylated proteins by mass spectrometry is hampered by the fact that the O-GlcNAc moiety is labile and lost during standard collision induced dissociation (CID) peptide backbone fragmentation20. Additionally, given the sub-stoichiometric nature of O-GlcNAc, enrichment of modified proteins is required before they can be identified using mass spectrometry. Derivatization of modified substrates by BEMAD (β-elimination followed by Michael addition of DTT) and chemoenzymatic/metabolic labeling approaches have been used for the enrichment/site mapping of O-GlcNAcylated proteins (reviewed by 20). With the advent of electron transfer dissociation (ETD) fragmentation, in which O-GlcNAc is not labile20, strategies for the capture of native O-GlcNAcylated proteins/peptides, such as lectin weak affinity chromatography using wheat germ agglutinin (WGA)21,22 or immunoprecipitation with the anti-O-GlcNAc antibody CTD110.623, have been employed for site mapping O-GlcNAcylated substrates. There are however, a number of limitations associated with these enrichment methods. In addition to O-GlcNAc, O-phosphate groups and O-glycans are also susceptible to BEMAD and rigorous optimization of reaction conditions and the use of appropriate controls such as phosphatase treatment are required to eliminate false positive identifications24. This is also true of chemoenzymatic and metabolic labeling methods, which can lead to the derivatization and enrichment of off- target glycans and other chemical groups. The drawback of using WGA affinity chromatography is its millimolar affinity for GlcNAc20–22. Although possessing much improved affinity for O-GlcNAc, the anti-O-GlcNAc antibody CTD 110.6, like WGA, has been shown to recognize terminal GlcNAc residues in other glycans25,26, making it somewhat non-specific as a bait. Additionally, given that it is raised against a specific immunogen from the C-terminal domain of RNA pol II27, it is possible that regardless of its specificity, CTD 110.6 does not recognize all O-GlcNAc sites. There is thus a need for novel strategies for the native enrichment of O-GlcNAcylated proteins/peptides.
We previously observed that a bacterial orthologue of the eukaryotic OGAs, Clostridium perfringens NagJ (CpOGA), shares 51% sequence similarity with human OGA (hOGA) and possesses remarkable catalytic activity on human O-GlcNAcylated proteins28. We recently demonstrated that an inactive mutant of this enzyme (CpOGAD298N), which retains the ability to bind to O-GlcNAcylated peptides (Fig. 1a) with affinities down to the nM range, could be used for the detection of O-GlcNAc proteins18. Here, we demonstrate that CpOGAD298N is a powerful new tool for the enrichment of O-GlcNAcylated proteins from Drosophila embryos, and use mass spectrometry to identify the first O-GlcNAc proteome associated with embryonic development. We reveal a range of previously unknown O-GlcNAc proteins with established links to homeotic and non-homeotic phenotypes as candidate conveyors of the Drosophila OGT/sxc catalytic null phenotype.
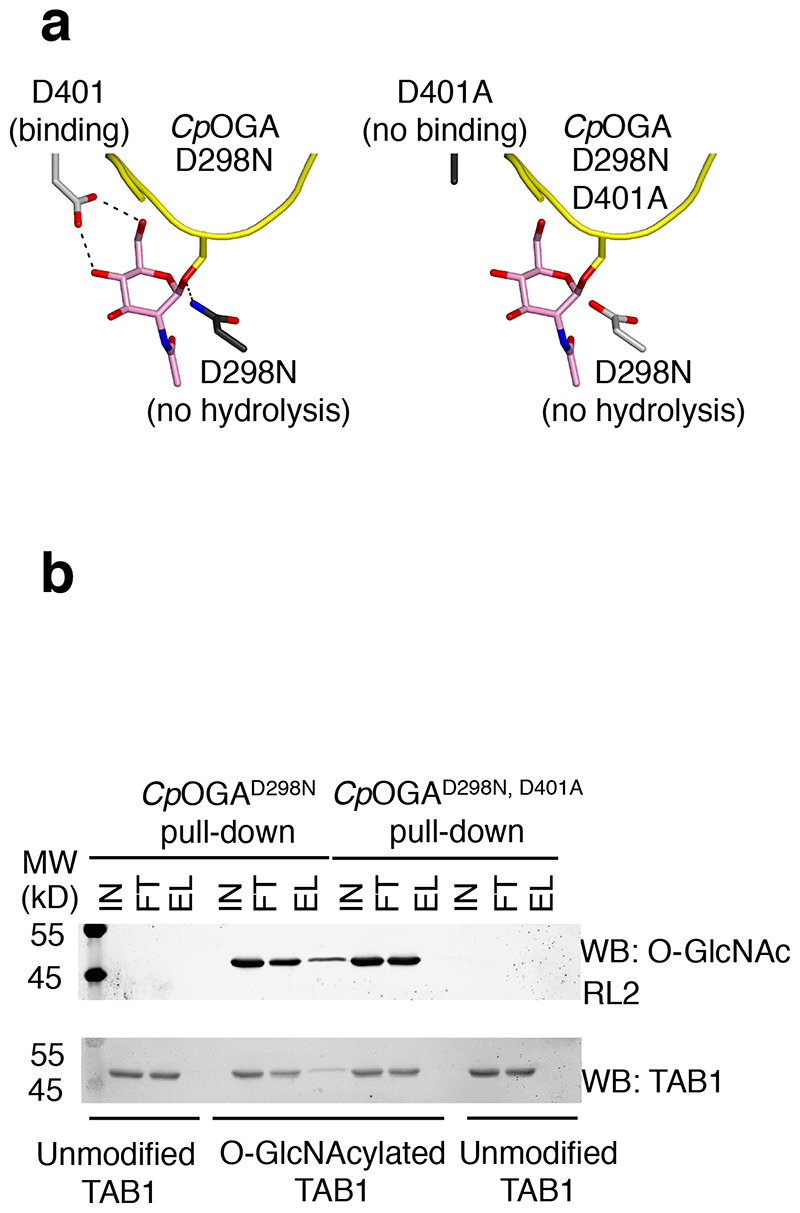
Figure 1. A point mutant of CpOGA can be exploited as a substrate trap for the enrichment of O-GlcNAcylated proteins.
(a) The inactive mutant CpOGAD298N can bind to substrate proteins (substrate is shown as a yellow cartoon, with GlcNAc depicted with pink sticks) but cannot hydrolyse GlcNAc therefore trapping O-GlcNAc modified proteins. The double mutant CpOGAD298N,D401A cannot bind O-GlcNAcylated proteins and therefore cannot act as a substrate trap
(b) Unmodified or O-GlcNAcylated TAB1 was incubated with Halo-CpOGAD298N coupled covalently to HaloLink beads. Pull down using the binding-deficient mutant CpOGAD298N,D401A was included to test the specificity of the pull down. Input, flow-through and elution fractions were blotted and probed with the antibodies mentioned. Elutions were performed by boiling the beads with sample buffer. TAB1 was pulled down in an O-GlcNAc specific manner by CpOGAD298N but not the control probe as evidenced by the presence of modified but not unmodified TAB1 in the elution fractions from CpOGAD298N.
Results
A tool for the enrichment of O-GlcNAcylated proteins
Our earlier work on the elucidation of the catalytic mechanism of OGA, using the bacterial enzyme CpOGA as a model, revealed a number of conserved amino acids in the active site involved in catalysis28,29. In particular, Asp298 (equivalent to Asp175 in hOGA) was identified as the catalytic acid that protonates the glycosidic bond, and Asp401 (equivalent to Asp285 in hOGA) was identified as being involved in hydrogen bonding required for the anchoring of the GlcNAc moiety in the active site through its O4 and O6 hydroxyl groups (Fig. 1a). The D298N mutant of CpOGA was catalytically impaired (8100-fold decrease in kcat compared to wild type enzyme) with negligible effect on the substrate K M, while the D401A mutant demonstrated loss of binding to the model substrate 4-methylumbelliferyl-GlcNAc (4MU-GlcNAc) (5-fold increase in K M, 2400-fold decrease in kcat)28. Having previously shown that CpOGAD298N (but not the binding-deficient CpOGAD401A or the double mutant CpOGAD298N,D401A) can be used as a probe for the specific detection of O-GlcNAcylated proteins in both human and Drosophila cell/tissue lysates18, we wanted to evaluate the feasibility of using it as a substrate trap to pull down O-GlcNAcylated proteins.
To this end, we first carried out a proof of principle experiment. Halo-tagged CpOGAD298N, or the double mutant CpOGAD298N,D401A as a negative control, were covalently coupled to HaloLink™ (Promega) beads and incubated with unmodified or in vitro O-GlcNAcylated recombinant TAB1 (transforming growth factor beta-activated kinase 1 binding protein 1) (Fig. 1b and Supplementary Results, Supplementary Fig. 1), a protein whose O-GlcNAcylation has previously been demonstrated to modulate innate immune signaling downstream of the IL-1 receptor30. Elution of enriched TAB1 from the mutant CpOGA beads was achieved by boiling the beads with sample buffer (see online methods). CpOGAD298N, but not the double mutant, was successful in pulling down O-GlcNAcylated but not unmodified TAB1, showing that the pull down occurred in an O-GlcNAc specific and CpOGA active-site dependent manner (Fig. 1b). The affinity of CpOGAD298N for glycosylated TAB1 was therefore sufficient for it to pull down the modified substrate, suggesting that it might be suitable for the enrichment of O-GlcNAcylated proteins from more complex samples such as cell/tissue lysates.
Prior to applying it to enrich for O-GlcNAcylated proteins from cell/tissue lysates, we wished to further dissect the substrate specificity of CpOGAD298N. It is evident from our previous work that CpOGAD298N is a specific detector of O-linked GlcNAc in HEK293 cell lysates as well in lysates of Drosophila S2 cells and embryos; PNGaseF treatment of lysates does not result in any visible alteration of signal obtained using CpOGAD298N as a probe for detection by Far Western blotting18. To investigate whether CpOGAD298N would bind to N-GlcNAc moieties in lysates resulting from endogenous ENGase activity, we performed a fluorescence polarization assay we previously described18, using an N-GlcNAcylated synthetic peptide derived from Cathepsin D. It appears that the conformation of the sugar/peptide backbone in a short peptide containing an O-linked GlcNAc moiety vs. an N-linked GlcNAc moiety affects CpOGAD298N binding, as no detectable binding was observed when up to 2.5 mM of the N-linked GlcNAc containing peptide derived from Cathepsin D (SYLN(GlcNAc)VTR)31 was used (Supplementary Fig. 2). In contrast, CpOGAD298N binds to an O-GlcNAc peptide derived from dHCF (VPST(GlcNAc)MSAN) with an affinity of 36 μM (highest concentration of peptide used - 2.7 mM)18. SPR experiments to determine differences in the binding to GlcNAc vs. GIcNAc(βl-4)GlcNAc reveal that CpOGAD298N binds the latter with a 20-fold lower affinity (29 μM vs. 590 μM) (Supplementary Fig. 3), suggesting that the mutant protein would have poor affinity for terminal GlcNAc moieties on extended glycan structures and would therefore preferentially bind to O-GlcNAc.
To determine how the substrate trap compares to previously published enrichment methods applied to lysates of a single cell line32,33, pull downs were also performed from HeLa cell lysates. Lysates were incubated for 90 min at 4 °C with Halo-tagged CpOGAD298N or the control mutant CpOGAD298N,D401A covalently coupled to saturation to HaloLink beads (schematic in Fig. 2a). To ensure that the eluents contained O-GlcNAcylated proteins captured specifically by the CpOGAD298N active site, elution was achieved by displacement with a molar excess of the OGA inhibitor Thiamet G34 (Fig. 2a), which retains binding to the inactive CpOGAD298N mutant (with a Kd of 688 nM, Supplementary Fig. 4). The pull down performed with CpOGAD298N, but not that performed with the CpOGAD298N,D401A negative control, resulted in an overall qualitative enrichment of O-GlcNAcylated proteins as visualized by Western blotting of samples using the RL2 antibody (representative blot in Fig. 2b and Supplementary Figs. 5-6), suggesting that this approach is a suitable enrichment method for complex samples. To identify the O-GlcNAcylated proteins enriched, three independent replicate pull downs were performed, including negative controls with CpOGAD298N,D401A. Eluates from these pull downs were processed and subjected to mass spectrometry. A total of 915 protein accessions were identified from the HeLa eluates, of which 859 were significantly enriched (4-fold, p < 0.05) in the CpOGAD298N mutant pull down compared to the control CpOGAD298N,D401A pull down (Supplementary Dataset 1). Bona fide O-GlcNAcylated substrates, such as the histones H2A, H2B, H3 and H435,36, c—Rel37, CREB38, CK2α39,40, TAB130 and OGT21,41 were among the proteins identified, thus validating the enrichment method. In contrast, a previously published study33 identified 199 significantly enriched proteins from HeLa cells using a tagging via substrate (TAS) approach, whereby a cell permeable azide modified analog of UDP-GlcNAc is used for the metabolic labeling of OGT substrates, which are then chemoselectively enriched. 49 of the significantly enriched proteins identified by us were also identified by that study33 (Supplementary Table 1). We identified 550 high confidence O-GlcNAc peptide sequence matches in 3 replicate MS analyses (3 with ETD site assignments). These resulted in a total of 61 high confidence O-GlcNAc peptides being identified that mapped to 29 of the 859 identified proteins (Supplementary Dataset 2 and Supplementary Table 2). This represents 3.3% of significantly enriched proteins on which O-GlcNAc sites were mapped, and is comparable to the 3.8% of significantly enriched proteins (using a metabolic labeling/chemoselective capture approach coupled to BEMAD) from denatured HEK293 cell lysates on which a previous study mapped O-GlcNAc sites32. Interestingly, 373 significantly enriched proteins identified by us from HeLa cells were also identified in that study in HEK293 cells with a large number of substrates unique to both studies (Supplementary Table 3). The prime advantage of enrichment using CpOGAD298N lies in the fact that no derivatization of O-GlcNAc moieties is required prior to enrichment unlike in metabolic (for cell lines) or chemoenzymatic (for tissue samples) labeling - it is a one-step method. Also, unlike WGA, CpOGAD298N possesses better affinity for O-GlcNAc, potentially enabling the enrichment and identification of a larger number of substrates.
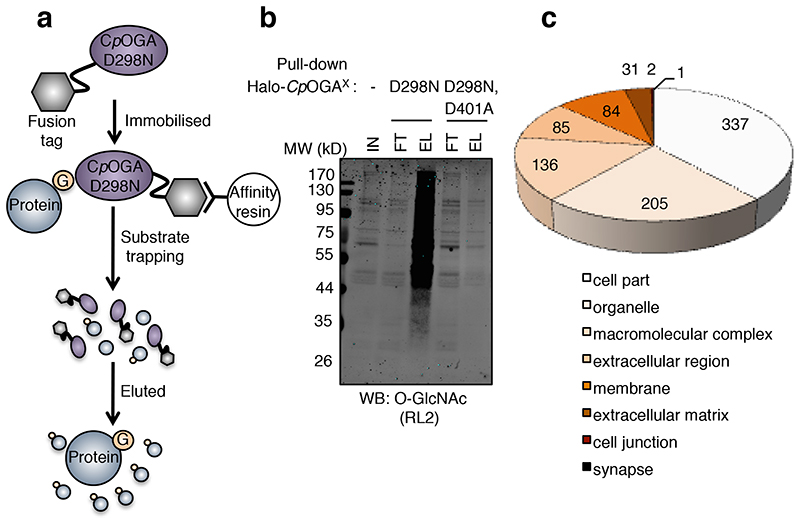
Figure 2. Pull down of O-GlcNAcylated proteins by CpOGAD298N.
(a) Schematic of the CpOGAD298N enrichment method. Halo-tagged CpOGA mutants covalently coupled to
HaloLink beads were used to pull down O-GlcNAcylated proteins. Elution of proteins from the beads was achieved by using a molar excess of the OGA inhibitor Thiamet G. Eluted proteins were concentrated using a spin concentrator and processed for mass spectrometry.
(b) Pull down from Drosophila embryo lysates using CpOGAD298N, but not the control mutant, results in the enrichment of O-GlcNAcylated proteins detected in the elution fractions.
(c) Cellular localization of proteins identified by CpOGAD298N. Cellular component analysis of all proteins identified by CpOGAD298N.
Enrichment of O-GlcNAc proteins from Drosophila embryos
We next used CpOGAD298N to enrich O-GlcNAcylated proteins from Drosophila embryo lysates in an attempt to begin to identify the O-GlcNAc proteome responsible for the sxc null phenotypes. A total of 3558 proteins accessions (isoforms of proteins and redundant entries with unique Uniprot accessions contribute to this number) were identified (Supplementary Dataset 3), of which 2358 were significantly enriched (4-fold, p < 0.05) in the CpOGAD298N mutant pull down compared to the control CpOGAD298N,D401A pull down (Supplementary Dataset 3).
2044 of the 2358 proteins enriched were recognised by PANTHER42, which was used for Gene Ontology (GO) analysis of the data and 881 cellular component hits were obtained. The majority (678) of the hits are nucleocytoplasmic (cell part, organelle and macromolecular complexes in the nucleus and cytoplasm), with 84 proteins being classified as membrane proteins, 116 as secreted or extracellular matrix proteins, 2 as synaptic proteins and 1 as a cell junction protein (Fig. 2c). Significantly enriched (p < 0.05, Bonferroni correction for multiple testing applied) GO cellular compartment terms are detailed in Supplementary Table 4.
Protein class analysis (performed using PANTHER) revealed that nucleic acid binding proteins represent the largest protein class identified, with 14% (289 out of 2136: 2044 recognised proteins with 2136 protein class hits) of proteins belonging to this class, most of these involved in RNA transport and processing (Fig. 3a). Transcription factors represent 4% of classified proteins and include Dp, Taf6, Cand1, fkh and T-related protein (byn orthologue). In mouse synaptic membranes, kinases have been shown to be more frequently O-GlcNAcylated than other protein classes in general (16% versus 10%, p < 3.6 × 10-4)22. In contrast, kinases and phosphatases combined comprise only 5% (~ 2.5% each) of classified proteins in our dataset and do not display a statistically significant overrepresentation (Fig. 3a, Supplementary Table 5). Protein kinases identified include the Akt-1, Cdk7, Cdc2 and Abl orthologues, while protein phosphatases identified include the PP2A 55 kDa subunit and Ptp4E, among others. While histones themselves are absent from the dataset, the HDACs Rpd3 and HDAC3 are present. The putative HAT Enok is also present, as is the bromodomain containing homeotic protein female sterile (fs(1)h- Brd2 orthologue). Significantly enriched (p < 0.05, Bonferroni correction for multiple testing applied) protein classes along with fold enrichment values are listed in Supplementary Table 5.
Figure 3. Protein class grouping of proteins identified by CpOGAD298N and example ETD fragmentation spectra for HexNAc modified peptides from Host Cell Factor (HCF) and Nucleoporin 153 (Nup153).
(a) Protein classes represented by identified proteins. Uniprot accessions of significantly enriched proteins (in CpOGAD298N pulldown vs. control pulldown) provided in Supplementary Dataset 3 were used as input for analysis on PANTHER database.
(b) and (c) Example ETD fragmentation spectra for HexNAc modified peptides from Host Cell Factor (HCF) (b) and Nucleoporin 153 (Nup153) (c). One peptide each from Host Cell Factor and Nucleoporin 153kD are shown. Peptide fragments were assigned using Mascot and Proteome Discoverer 2.0. Signals of charged reduced species of the precursor and neutral losses associated with it in the spectrum were filtered out. For clarity, only c[+1], red, and z[+1], blue, ions are annotated.. The sequence relevant to each ion is shown, lower case “s”/”t” indicate the HexNAc modified residues.
Pathway analysis (performed using PANTHER) identified 33 of 2044 mapped protein accessions (~ 1.6%) as functioning in the Wnt signaling pathway (Supplementary Fig. 7). Examples of the Wnt signaling proteins identified are cadherin-87A, the acetyltransferase Neijre (CREB-binding protein/CBP orthologue), the HDAC Rpd3, the mor orthologue, CK1, and the helicase domino. Other proteins in the dataset are involved in pathways such as the ubiquitin proteasome pathway, DNA replication, apoptosis and cytoskeletal regulation by Rho GTPase (Supplementary Fig. 7). Interestingly, many proteins involved in these pathways are also implicated in the pathogenesis of Huntington’s and Parkinson’s disease. Mutations in huntingtin for example, affect its interaction with hits like CBP43.
Identification of O-GlcNAc proteins linked to development
We next examined the O-GlcNAc sites on enriched proteins. In the CpOGAD298N pull downs we identified, in three experiments, a total of 268 high confidence O-GlcNAc peptide sequence matches (32 with ETD site assignments) (Supplementary Dataset 4 and Supplementary Tables 6-7); ETD fragmentation spectra for two HexNAc peptides are shown in Fig. 3b-c). These resulted in a total of 52 high confidence O-GlcNAc peptides being identified (Supplementary Dataset 4) that were mapped on a total of 43 proteins (Supplementary Table 7). In contrast, only 3 HexNAc peptide sequence matches were identified in the CpOGAD298N,D401A pull downs (none of which with ETD site assignments) (Supplementary Table 6).
The majority of the high confidence O-GlcNAc sites are on nuclear/nucleocytoplasmic proteins. Tay (AUTS2 - like protein), Grunge (Gug - atrophin orthologue), myopic (mop - HDPTP orthologue), and lingerer (lig - UBA domain containing protein) are examples of bona fide O-GlcNAcylated proteins identified in this study and many of these are conserved across evolution. Our dataset also includes the nuclear pore proteins (Nups) (recently reviewed44), Ataxin-2 (Atx2)45, CF11970 (NFRKB orthologue)46 and HCF47, which have previously been shown to be O-GlcNAcylated in other organisms although the role of the modification on these proteins is as yet not understood. GO analysis using STRING48 to determine the biological processes associated with these O-GlcNAcylated proteins categorizes 19 of the 42 proteins mapped as being involved in anatomical structure development and morphogenesis, with four (if, Gug, tay and LanA) amongst those specifically associated with appendage development/morphogenesis, a process clearly affected in OGT null mutant flies given the phenotypes observed (e.g. the homeotic transformation of antennae to prothoracic legs and wings to haltere-like structures3). Interestingly, 11 hits are classified as being involved in nervous system development and include Atx-2 and Iswi.
Improper O-GlcNAc modification of Ph, one of the most prominent substrates of OGT in Drosophila has been proposed to be responsible for the OGT/sxc phenotypes via misexpression of Hox genes7,9. Nevertheless, numerous transcription factors and cell signalling molecules have been identified in this study as being O-GlcNAc modified. These data therefore suggest the possibility that some of the phenotypes associated with the lack of OGT activity may be downstream of hypo-O-GlcNAcylation of one of the non-Ph OGT substrates. Site mapping confirmation in this study establishes Gug as a genuine OGT substrate. We also identified O-GlcNAcylated peptides from Gug in immunoprecipitates obtained from embryo lysates using the anti-O-GlcNAc antibody RL2 but not an isotype control antibody, thereby orthogonally confirming its modification status (Supplementary Fig. 8a shows the EThcD fragmentation spectrum for one of the HexNAc peptides identified). Gug is a nuclear receptor corepressor, which was identified in a screen designed to identify regulators of one of the other O-GlcNAc proteins in the embryo O-GlcNAc proteome, teashirt49. Since then the functions of Gug in transcriptional regulation of EGF receptor signalling50, as a co-repressor for Even skipped51, Tailless52 and Cubitus interruptus53 have been established, outlining its multiple roles during embryonic development. One of the other identified O-GlcNAc modified substrates is mop (also orthogonally verified as being modified using an anti-O-GlcNAc antibody, EThcD fragmentation spectrum in Supplementary Fig. 8b). A protein associated with intracellular vesicles, mop, was found to be essential for transit of ubiquitylated EGF receptor to lysosomes54. In addition, mop is also involved in distribution of integrins during oogenesis55, endocytosis and activation of the Toll56, Wnt/Wingless57, Frizzled58 and Yorkie59 pathways also affecting respective downstream signalling.
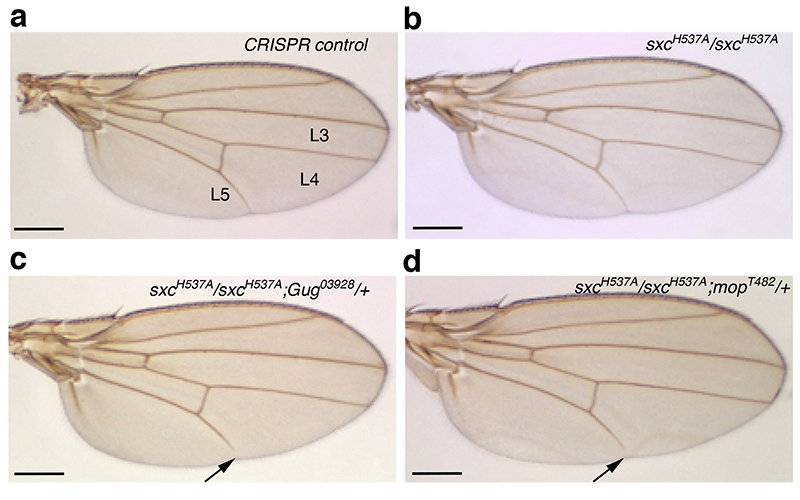
To investigate how reduced O-GlcNAc modification of two of these OGT substrates, Gug and mop, affects their function, genetic interaction experiments were performed. We used an OGT catalytic hypomorphic allele, OGT/sxcH537A (henceforth represented as sxcH537A), that we have generated using CRISPR gene editing (Mariappa et al., Under revision, J. Biol. Chem.). This ensured that any potential genetic interaction we observed was a consequence of reduced OGT catalytic activity and therefore decreased O-GlcNAc modification of Gug and mop. Recessive lethal alleles Gug03928 (P element insertion)50 and mopT482 (Q1968Stop)54 were crossed into either homozygous or heterozygous sxcH537A background. CRISPR control (Cr Control) flies were generated from the BL51323 stock used for CRISPR injections and subjected to the same crossing scheme as the sxcH537A mutant lines. None of the Cr Control (Fig. 4a, Supplementary Fig. 9), OGT/sxcH537A homozygotes (Fig. 4b, Supplementary Fig. 9) or heterozygotes for Gug03928 and mopT482 (Supplementary Fig. 9) displayed wing vein deposition defects. About 2% and 1% of sxcH537A/+;Gug03928/+ and sxcH537A /+;mopT482/+ double heterozygotes had a short L5 longitudinal wing vein that did not reach the wing margin (Supplementary Fig. 9, Supplementary Table 8). This phenotype was enhanced on further reduction in OGT activity in flies homozygous for the sxcH537A allele and heterozygous for either Gug03928 and mopT482; to 14% in sxcH537A;Gug03928/+ flies and 8% sxcH537A;mopT482/+ flies (Fig. 4c-d and Supplementary Table 8). More of the sxcH537A/sxcH537A;Gug03928/+ flies (5%) had the short L5 wing vein defect in both the wings as compared to the sxcH537A/sxcH537A;mopT482/+ flies (0.6%, Supplementary Table 8). These data establish a genetic interaction between the hypomorphic OGT/sxc allele and alleles of two of the OGT substrates Gug and mop. Given that both Gug50 and mop54 have roles in EGF signalling-dependent wing vein specification, O-GlcNAc modification of these two proteins could potentiate their function in EGF signalling.
Figure 4. OGT catalytic activity potentiates the function of its substrates Grunge and myopic.
Genetic interaction between OGT/sxcH537A and Gug03928 or mopT482 alleles was assessed in the adult wing. In the Cr control (a), OGT/sxcH537A (b) homozygotes or Gug03928 or mopT482 heterozygotes have a complete L5 longitudinal wing vein that reaches the wing margin. In OGT/sxcH537A/OGT/sxcH537A;Gug03928/+ (c) or OGT/sxcH537A/OGT/sxcH537A;mopT482/+ (d) flies, 14% and 8% of the flies, respectively, have a shorter L5 wing vein. Fewer of the double homozygotes, OGT/sxcH537A/+;Gug03928/+ or OGT/sxcH537A/+;mopT482/+ display this phenotype as demonstrated by the quantification in (e). Arrows in (c) and (d) point to the short L5 wing vein phenotype.
Discussion
Unlike OGT knockout mice, which do not survive beyond the single cell stage60, OGT null flies develop to the pharate adult stage and display the hallmark phenotypes of mutants of polycomb group (PcG) proteins3. This, in addition to the relatively rapid generation time and amenability to genetic manipulation, renders Drosophila melanogaster an attractive model organism in which to dissect the role of O-GlcNAc on proteins, particularly in the context of early development. Targeted investigation of all known members of the PcG has led to the identification of polyhomeotic (ph) as a key OGT substrate from this class of proteins7. The O-GlcNAcylation of ph has been suggested to be important in preventing its self-aggregation9. The discovery that the phenotypes of OGT null mutants resemble a less severe version of the phenotypes of the ph null mutant has led to the suggestion that the loss of O-GlcNAc on ph is the key driver of the manifestation of the defects exhibited by OGT null flies10. Ph is not, however, the sole OGT substrate in Drosophila, and the role of O-GlcNAc on a handful of other proteins has been studied in the fly11–14,16,17. Nevertheless, it is not understood how the O-GlcNAc proteome maps to processes that are critical for development in both Drosophila and vertebrates.
We previously described CpOGAD298N as a versatile and specific tool for the detection of O-GlcNAc in mammalian and Drosophila cell lysates, and used it to demonstrate that protein O-GlcNAcylation is dynamic during Drosophila embryogenesis18. We have now successfully deployed CpOGAD298N for the enrichment of O-GlcNAcylated proteins from Drosophila embryos and have discovered novel substrates of OGT in the fly. Interestingly, genetic interactions of a hypomorphic OGT/sxc allele with lethal recessive alleles of two of the bona fide substrates, Gug and mop, lead to a similar phenotype wherein the L5 wing vein is short. Reduced deposition of wing vein material is observed in mop mutant wings, possibly affecting EGF signalling54. Conversely, EGF signalling dependent wing vein deposition is enhanced in a Gug mutant background50. It is possible that O-GlcNAc modification of Gug and mop could affect their role in EGF signalling via mechanisms that will need to be further investigated. Nevertheless, given the role of both these proteins in numerous other cell signalling and transcriptional control events, O-GlcNAcylation of Gug or mop could be modulating one/multiple such downstream events. In addition, we have also observed genetic interaction between sxcH537A with a Hcf null allele with respect to specification of the thoracic scutellar bristles (Mariappa et al., Under revision, J. Biol. Chem.), thus underlining the multiple roles that can be ascribed to O-GlcNAcylated substrates.
The identification and validation of proteins like Gug and mop as bona fide OGT substrates, and the determination of O-GlcNAc sites on them, paves the way for future studies aimed at investigating the effect of O-GlcNAc on these proteins and the processes they regulate. While some of these hits could contribute to the homeotic transformations observed in OGT/sxc null flies, others might reveal novel, potentially conserved functions of O-GlcNAc through the identification of subtler phenotypes in non-lethal OGT/sxc mutants.
Online methods
Drosophila embryos
Embryos from w1118 wild type flies were used. Fly stocks were maintained by flipping vials once every ten days. Embryos (0-16 h) were collected on apple juice agar plates at 25 °C overnight. For embryo collections, flies were assigned from vials in a rack in random order to three separate cages to represent three biological replicates. Collected embryos were dechorionated with bleach and snap frozen in dry ice and stored at -80 °C until they were processed. Samples were collected over time and on independent occasions in this manner till enough material was obtained for further processing. Lysates were prepared as described below. Bradford assay or Pierce 660 nm protein assay was used to quantify cell lysates.
Cell culture
HeLa cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM; Gibco) supplemented with 10% fetal bovine serum (FBS), L-glutamine, and penicillin streptomycin at 37 °C with humidified air at 5% CO2. Cells were plated on 10 cm dishes and grown to 80% confluence prior to harvesting.
Protein expression and purification
Plasmids containing N-terminally Halo-tagged CpOGA (31-618) were transformed into E. coli BL21-Gold (DE3) pLysS cells (Agilent). Cells were grown overnight at 37 °C in Luria-Bertani medium containing 50 μg/ml Kanamycin (LB-Kan) and used at 10 mL/L to inoculate of fresh LB-Kan. Cells were grown to an OD600 of 0.6-0.8, transferred to 18 °C and induced with 250 μM IPTG and harvested after 16 h by centrifugation for 30 min at 3500 rpm (4 °C). Cell pellets were resuspended in 10-20 mL of 50 mM Tris, 250 mM NaCl at pH 7.5 (lysis buffer) supplemented with protease inhibitors (1 mM benzamidine, 0.2 mM PMSF and 5 μM leupeptin), DNAse and lysozyme prior to lysis. Cells were lysed using a continuous flow cell disrupter (Avestin, 3 passes at 20 kpsi) and the lysate was cleared by centrifugation (30 min, 15,000 rpm, 4 °C). Supernatants were collected and loaded onto a HisTrap HP column (GE Healthcare Life Sciences) charged with NiSO4 and pre-equilibrated with lysis buffer. The column was washed with 10 column volumes of lysis buffer. Proteins were eluted with a linear gradient of imidazole (0-500 mM) over 20 column volumes. Late elution fractions were pooled and dialysed into 1 x TBS and snap frozen with a final concentration of 20% glycerol and stored at -80 °C until use. Untagged proteins used for fluorescence polarization and surface plasmon resonance experiments were prepared as described previously18.
Fluorescence Polarimetry
Experiments were performed as described before18. Briefly, to avoid receptor depletion, reaction mixtures for competition binding experiments contained 5 nM fluorescent probe, 7 nM of CpOGAWT (receptor)/20 nM CpOGAD298N (receptor) and a range of concentrations of ligands. Reactions were allowed to stand at room temperature for 10 min. Highest amount of fluorescent probe bound to CpOGAD298N in the absence of competing ligands was arbitrarily set as 100%. EC50 values were determined by fitting non-linear regression curves with Prism (GraphPad) and converted to Kd as described before18. All experiments were performed in triplicate.
Surface Plasmon Resonance
Experiments were performed as described before18. Briefly, biotinylated proteins were captured on a neutravidin surface prepared on high capacity amine sensor chip of a Mass-1 instrument (Sierra Sensors) at densities 3,600-3,900 RU. Ligands were injected over captured proteins at a flow rate of 30 μL min-1 in running buffer (25 mM Tris pH 7.5, 150 mM NaCl, 0.05% Tween20), with each compound injected in duplicates in concentration series adjusted specifically around their affinities. Association was measured for 60 s and dissociation for 120 s. All data were double referenced for blank injections of buffer and biotin-blocked Streptavidin surface. Data processing and analysis were performed using Analyser 2 (Sierra Sensors) and Scrubber 2 (BioLogic Software).
CpOGAD298N pull downs
Halo-tagged CpOGA proteins were purified as described above and coupled to Magne™HaloTag® Beads (Promega) as per the manufacturer’s instructions. Briefly, Magne™HaloTag® Beads (Promega) were equilibrated with 50 mM Tris pH 7.5, 150 mM NaCl (wash buffer) supplemented with 0.05% Tween-20 (binding buffer). The binding capacity of the beads for tagged CpOGA was determined to be 8 mg per mL of settled beads. Beads were coupled to saturation with CpOGA for 90 min h at 4 °C then washed extensively with wash buffer and stored on ice. Halo-CpOGA beads were prepared freshly for each experiment.
For the TAB1 pull down experiment, in vitro O-GlcNAcylation of TAB1 was performed by incubating 24 μg (18.2 μM) of TAB1 with 10 μg (4.1 μM) hOGT and 10 mM UDP-GlcNAc in a final volume of 30 μL for 3 h at 37 °C. The reaction was stopped by the addition of a final concentration of 20 mM of UDP. ‘Unmodified TAB1’ was the product of reactions containing all components except for UDP-GlcNAc. The reactions were split in four equal volumes (containing 3 μg of total TAB1 each), of which two were retained for loading as input and one each of the remainder of the two loaded onto 50 μL of a 20% slurry of HaloLink beads coupled to saturation with CpOGAD298N or CpOGAD298N,D401A . The incubation with beads was performed in a total volume of 200 μL made up with binding buffer for 1 h at 4 °C. The flow-through was collected, beads washed 3 times with wash buffer and bound protein eluted using 200 μL of 10 mM Tris pH 6.8, 4% SDS, 200 mM DTT by boiling for 2 min. The ‘input’ fractions were also made up to a volume of 200 μL and 8 μL of all fractions were subjected to SDS PAGE and Western blotting. The antibodies used were Anti-TAB1 (C25E9 - Cell Signaling, 1:5000) and anti-O-GlcNAc RL2 (ab2739 - Abcam, 1:3000 or 1:1000)
Drosophila embryo lysates and HeLa lysates were prepared with RIPA buffer (50 mM Tris pH 7.5, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, 150 mM NaCl, 2 mM EDTA and 50 mM NaF). For each replicate experiment, protein lysates were split in half to carry out pull downs with either CpOGAD298N or the control CpOGAD298N,D401A. 7 mg of lysates were incubated with 200 μL of settled HaloLink beads coupled to saturation with CpOGAD298N or CpOGAD298N, D401A for 90 min at 4 °C. The flow through was collected and the beads washed extensively with wash buffer. Bound proteins were eluted by incubating the beads 2 x for 30 min at 4 °C with 250 μL wash buffer supplemented with 3 mM Thiamet G. The eluents were concentrated using a 10 kDa molecular weight cut off spin concentrator and ~2 μg set aside for Western blotting and the rest prepared for mass spectrometry analysis as below. Experiments were performed in triplicate.
RL2 immunoprecipitation
5 mg of embryo lysates prepared as described above were incubated for 3 h at 4 °C with 5 μg of RL2 or Mouse normal IgG1 (Cell Signaling) antibody bound to Protein G dynabeads (Invitrogen) as per the manufacturer’s instructions. The flow through was collected and incubated with freshly coupled RL2/IgG1 (5 μg) dynabeads overnight at 4 °C. The immunoprecipitates were washed several times with 500 μL of 1 X TBS containing 0.02% Tween 20 per wash and eluted by boiling the beads for 5 min in 50 mM Tris pH 6.8 containing 4% SDS and 200 mM DTT. Eluates were processed for mass spectrometry as described below.
Sample preparation for mass spectrometry
Samples were run halfway down precast NuPAGE 4-12% Bis-Tris gels (Invitrogen) and stained in clean plastic containers with InstantBlue (Expedeon) Coomassie stain then de-stained using mass spec grade water (VWR). Each lane on the gel was excised into up to 0.5 cm X 0.5 cm sections and then further diced into 1 mm cubes using a clean scalpel. The excised gel pieces were de-stained till colourless using 50% methanol, rinsed with 50% acetonitrile and subsequently with 50% acetonitrile in 50 mM ammonium bicarbonate buffer (wash buffer). In-gel reduction was performed by incubating gel pieces in 10 mM DTT made in 50 mM ammonium bicarbonate for 20 min at RT, then alkylated by adding 50 mM iodoacetamide made in 50 mM ammonium bicarbonate buffer for 30 min at RT in the dark. The gel pieces were then washed several times with wash buffer and dehydrated by incubating for 10 min at RT in 100% acetonitrile. Gel pieces were then swelled with enough 25 mM triethylammonium bicarbonate buffer to cover them and subjected to enzymatic digestion using Trypsin (mass spec grade, Promega) at 5 μg per mL of triethylammonium bicarbonate buffer at 30 °C for 16 h. The solution containing liberated peptides was then collected and more peptides extracted from the gel pieces using 50% acetonitrile containing 2.5% formic acid. Peptides were pooled and dried in a SpeedVac and stored at -80 °C until MS analysis.
Mass spectrometry and data analysis
HCD and ETD mass spectrometry analysis (or EThcD for RL2 immunoprecipitates) was performed by LC-MS- MS on a Fusion ion trap-orbitrap hybrid mass spectrometer (Thermo Scientific) coupled to a U3000 RSLC HPLC (Thermo Scientific). 50%/10% of the Drosophila embryo samples/HeLa samples were injected. Peptides were trapped on a nanoViper Trap column, 2 cm x 100 μm C18 5 μm 100 Å (Thermo-Fisher, 164564) then separated on a 50 cm EasySpray column (Thermo, ES803) equilibrated with a flow of 300 nl/min of 3% Solvent B [Solvent A was 2% acetonitrile, 0.1% formic acid, 3% DMSO in H2O; Solvent B was 80% acetonitrile, 0.08% formic acid, 3% DMSO in H2O]. The elution gradient was as follows, Time (min): Solvent B (%); 0:3, 5:3, 55:25, 74:40, 74.5: 99, 79.5:99, 80:3, 90:3. Data were acquired in the data-dependent mode, automatically switching between MS and MS-MS acquisition. MS full scan spectra were acquired in the orbitrap with S-lens RF level of 60 %, resolution of 120000 (scan range m/z 400-1600), with a maximum ion injection time of 50 ms, and AGC setting of 400000 ions. HCD normalized collision energy was set to 30% and fragment ions were detected in the linear ion trap using 1 microscan, with a maximum injection time of 250 ms and AGC setting of 100 ions. ETD MS2 analyses were triggered by the presence of product ions with m/z 204.0867 (HexNAc oxonium) and/or 138.0545 (HexNAc fragment) and detected in the Ion Trap, AGC Target 10000 and maximum injection time of 105 ms. EThcD reactions were triggered as for ETD or by the presence of the 366.1396 HexNAcHex ion, and detected in the orbitrap (resolution of 30000, scan range m/z 120-2000) using 1 microscan, AGC setting of 300000 ions and maximum injection time of 150 ms. Data files were analysed for HexNAc peptides by Proteome Discoverer 2.0 (Thermo), using Mascot 2.4.1 (Matrix Science), and searched against the Uniprot_DROME database or the Uniprot_HUMAN database as appropriate. Allowance was made for fixed, (carbamidomethyl (C)), and variable modifications (oxidation (M), dioxidation (M), phospho (S/T) and HexNAc (S/T)). Protein abundance analysis was performed using MaxQuant 1.5.1.7 and data was further analysed using the Perseus software package; significant proteins were identified using a two-tailed t-test (p < 0.05).
Drosophila genetics
The following fly stocks were obtained from Bloomington Drosophila Stock Centre: Gug03928/TM3, Sb1,Ser1 and mopT482/TM6B, Tb1. The catalytically hypomorphic OGT/sxcH537A flies were generated using CRISPR/Cas9 gene editing (Mariappa et al., Under revision, J. Biol. Chem.). The BL51323 Vasa::Cas9 stock used for the CRISPR injections were crossed with the balancer stocks to eliminate the Vasa::Cas9 containing X chromosome similar to the mutant flies to derive the CRISPR control (Cr control) stock. To derive double heterozygotes Cr control virgins were crossed with OGT/sxcH537A/OGT/sxcH537A;Gug03928/TM6 or OGT/sxcH537A/OGT/sxcH537A;mopT482/TM6 flies. To derive Gug03928or mopT482 Cr control virgins were crossed with Gug03928/TM3, Sb1, Ser1 or mopT482/TM6B, Tb1 flies, respectively. Wing phenotypes of flies of the various genotypes were assessed using a Motic SMZ microscope. Wing preparations were made by dissecting whole wings from the flies and transferring them into isopropanol for 24 h. The wings were then mounted in DPX Mounting medium (Sigma) and imaged with a Leica E24 HD dissection microscope.
Supplementary Material
Acknowledgements
This work is funded by a Wellcome Trust Senior Investigator Award (110061) to D.M.F.v.A. M.T. is funded by a MRC grant (MC_UU_12016/5). R.W is funded by a Royal Society Research Grant. We thank Julien Peltier for help with mass spectrometry and Olawale Raimi for help with protein purification.
Footnotes
Author contributions
N.S., R.W. and D.M.F.v.A conceived the study; N.S., R.W., and D.M. performed experiments; D.G.C., R.G. and M.T. performed mass spectrometry; A.T.F. performed molecular biology; T.A. and I.H.N. performed SPR; N.S., D.G.C., and M.T. analysed mass spectrometry data; D.M. analysed genetics data and N.S., R.W., DM., and D.M.F.v.A. interpreted the data and wrote the manuscript with input from all authors.
Conflict of interest
The authors declare that there are no conflicts of interest.
References
- 1.Hart GW, Slawson C, Ramirez-Correa G, Lagerlof O. Cross talk between O-GlcNAcylation and phosphorylation: roles in signaling, transcription, and chronic disease. Annual review of biochemistry. 2011;80:825–58. doi: 10.1146/annurev-biochem-060608-102511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Guo B, et al. O-GlcNAc-modification of SNAP-29 regulates autophagosome maturation. Nature Cell Biology. 2014;16:1215–26. doi: 10.1038/ncb3066. [DOI] [PubMed] [Google Scholar]
- 3.Ingham PW. A gene that regulates the bithorax complex differentially in larval and adult cells of Drosophila. Cell. 1984;37:815–23. doi: 10.1016/0092-8674(84)90416-1. [DOI] [PubMed] [Google Scholar]
- 4.Ingham PW. Genetic control of the spatial pattern of selector gene expression in Drosophila. Cold Spring Harbor Symposia on Quantitative Biology. 1985;50:201–8. doi: 10.1101/sqb.1985.050.01.026. [DOI] [PubMed] [Google Scholar]
- 5.Webster DM, et al. O-GlcNAc modifications regulate cell survival and epiboly during zebrafish development. BMC developmental biology. 2009;9:28. doi: 10.1186/1471-213X-9-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kenwrick S, Amaya E, Papalopulu N. Pilot morpholino screen in Xenopus tropicalis identifies a novel gene involved in head development. Developmental dynamics : an official publication of the American Association of Anatomists. 2004;229:289–99. doi: 10.1002/dvdy.10440. [DOI] [PubMed] [Google Scholar]
- 7.Gambetta MC, Oktaba K, Muller J. Essential role of the glycosyltransferase sxc/Ogt in polycomb repression. Science. 2009;325:93–6. doi: 10.1126/science.1169727. [DOI] [PubMed] [Google Scholar]
- 8.Sinclair DA, et al. Drosophila O-GlcNAc transferase (OGT) is encoded by the Polycomb group (PcG) gene, super sex combs (sxc) Proceedings of the National Academy of Sciences of the United States of America. 2009;106:13427–32. doi: 10.1073/pnas.0904638106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gambetta MC, Muller J. O-GlcNAcylation Prevents Aggregation of the Polycomb Group Repressor Polyhomeotic. Developmental cell. 2014;31:629–39. doi: 10.1016/j.devcel.2014.10.020. [DOI] [PubMed] [Google Scholar]
- 10.Gambetta MC, Muller J. A critical perspective of the diverse roles of O-GlcNAc transferase in chromatin. Chromosoma. 2015 doi: 10.1007/s00412-015-0513-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sekine O, Love DC, Rubenstein DS, Hanover JA. Blocking O-linked GlcNAc cycling in Drosophila insulin-producing cells perturbs glucose-insulin homeostasis. The Journal of biological chemistry. 2010;285:38684–91. doi: 10.1074/jbc.M110.155192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Diernfellner AC, Brunner M. O-GlcNAcylation of a circadian clock protein: dPER taking its sweet time. Genes and Development. 2012;26:415–6. doi: 10.1101/gad.188524.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim EY, et al. A role for O-GlcNAcylation in setting circadian clock speed. Genes and Development. 2012;26:490–502. doi: 10.1101/gad.182378.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaasik K, et al. Glucose sensor O-GlcNAcylation coordinates with phosphorylation to regulate circadian clock. Cell Metabolism. 2013;17:291–302. doi: 10.1016/j.cmet.2012.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Radermacher PT, et al. O-GlcNAc reports ambient temperature and confers heat resistance on ectotherm development. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:5592–7. doi: 10.1073/pnas.1322396111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mariappa D, et al. Protein O-GlcNAcylation is required for fibroblast growth factor signaling in Drosophila. Science signaling. 2011;4:ra89. doi: 10.1126/scisignal.2002335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Park S, et al. O-GlcNAc modification is essential for the regulation of autophagy in Drosophila melanogaster. Cellular and Molecular Life Sciences. 2015:1–11. doi: 10.1007/s00018-015-1889-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mariappa D, et al. A mutant O-GlcNAcase as a probe to reveal global dynamics of protein O-GlcNAcylation during Drosophila embryonic development. The Biochemical journal. 2015;470:255–62. doi: 10.1042/BJ20150610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sprung R, et al. Tagging-via-substrate strategy for probing O-GlcNAc modified proteins. Journal of proteome research. 2005;4:950–7. doi: 10.1021/pr050033j. [DOI] [PubMed] [Google Scholar]
- 20.Ma J, Hart GW. O-GlcNAc profiling: from proteins to proteomes. Clinical proteomics. 2014;11:8. doi: 10.1186/1559-0275-11-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alfaro JF, et al. Tandem mass spectrometry identifies many mouse brain O-GlcNAcylated proteins including EGF domain-specific O-GlcNAc transferase targets. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:7280–5. doi: 10.1073/pnas.1200425109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Trinidad JC, et al. Global Identification and Characterization of Both O-GlcNAcylation and Phosphorylation at the Murine Synapse. Molecular & Cellular Proteomics. 2012;11:215–229. doi: 10.1074/mcp.O112.018366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zachara NE, Molina H, Wong KY, Pandey A, Hart GW. The dynamic stress-induced “O-GlcNAc-ome” highlights functions for O-GlcNAc in regulating DNA damage/repair and other cellular pathways. Amino Acids. 2011;40:793–808. doi: 10.1007/s00726-010-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wells L, et al. Mapping sites of O-GlcNAc modification using affinity tags for serine and threonine post-translational modifications. Molecular and Cellular Proteomics. 2002;1:791–804. doi: 10.1074/mcp.m200048-mcp200. [DOI] [PubMed] [Google Scholar]
- 25.Reeves RA, Lee A, Henry R, Zachara NE. Characterization of the specificity of O-GlcNAc reactive antibodies under conditions of starvation and stress. Analytical biochemistry. 2014;457:8–18. doi: 10.1016/j.ab.2014.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ogawa M, et al. GTDC2 modifies O-mannosylated alpha-dystroglycan in the endoplasmic reticulum to generate N-acetyl glucosamine epitopes reactive with CTD110.6 antibody. Biochemical and biophysical research communications. 2013;440:88–93. doi: 10.1016/j.bbrc.2013.09.022. [DOI] [PubMed] [Google Scholar]
- 27.Comer FI, Vosseller K, Wells L, Accavitti MA, Hart GW. Characterization of a mouse monoclonal antibody specific for O-linked N-acetylglucosamine. Analytical biochemistry. 2001;293:169–77. doi: 10.1006/abio.2001.5132. [DOI] [PubMed] [Google Scholar]
- 28.Rao FV, et al. Structural insights into the mechanism and inhibition of eukaryotic O-GlcNAc hydrolysis. The EMBO journal. 2006;25:1569–78. doi: 10.1038/sj.emboj.7601026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schimpl M, Borodkin VS, Gray LJ, van Aalten DM. Synergy of peptide and sugar in O-GlcNAcase substrate recognition. Chemistry & biology. 2012;19:173–8. doi: 10.1016/j.chembiol.2012.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pathak S, et al. O-GlcNAcylation of TAB1 modulates TAK1-mediated cytokine release. The EMBO journal. 2012;31:1394–404. doi: 10.1038/emboj.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang H, Li XJ, Martin DB, Aebersold R. Identification and quantification of N-linked glycoproteins using hydrazide chemistry, stable isotope labeling and mass spectrometry. Nature Biotechnology. 2003;21:660–666. doi: 10.1038/nbt827. [DOI] [PubMed] [Google Scholar]
- 32.Hahne H, et al. Proteome wide purification and identification of O-GlcNAc-modified proteins using click chemistry and mass spectrometry. Journal of Proteome Research. 2013;12:927–36. doi: 10.1021/pr300967y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nandi A, et al. Global identification of O-GlcNAc-modified proteins. Analytical chemistry. 2006;78:452–8. doi: 10.1021/ac051207j. [DOI] [PubMed] [Google Scholar]
- 34.Yuzwa SA, et al. A potent mechanism-inspired O-GlcNAcase inhibitor that blocks phosphorylation of tau in vivo. Nature chemical biology. 2008;4:483–90. doi: 10.1038/nchembio.96. [DOI] [PubMed] [Google Scholar]
- 35.Sakabe K, Wang Z, Hart GW. Beta-N-acetylglucosamine (O-GlcNAc) is part of the histone code. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:19915–20. doi: 10.1073/pnas.1009023107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sakabe K, Hart GW. O-GlcNAc transferase regulates mitotic chromatin dynamics. The Journal of biological chemistry. 2010;285:34460–8. doi: 10.1074/jbc.M110.158170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ramakrishnan P, et al. Activation of the Transcriptional Function of the NF-κB Protein c-Rel by O-GlcNAc Glycosylation. Science Signaling. 2013;6:ra75-ra75. doi: 10.1126/scisignal.2004097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rexach JE, et al. Dynamic O-GlcNAc modification regulates CREB-mediated gene expression and memory formation. Nature chemical biology. 2012;8:253–61. doi: 10.1038/nchembio.770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tarrant MK, et al. Regulation of CK2 by phosphorylation and O-GlcNAcylation revealed by semisynthesis. Nature chemical biology. 2012;8:262–9. doi: 10.1038/nchembio.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lazarus MB, Nam Y, Jiang J, Sliz P, Walker S. Structure of human O-GlcNAc transferase and its complex with a peptide substrate. Nature. 2011;469:564–7. doi: 10.1038/nature09638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Griffin ME, et al. Comprehensive mapping of O-GlcNAc modification sites using a chemically cleavable tag. Molecular Biosystems. 2016;12:1756–1759. doi: 10.1039/c6mb00138f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mi H, Muruganujan A, Casagrande JT, Thomas PD. Large-scale gene function analysis with the PANTHER classification system. Nature Protocols. 2013;8:1551–66. doi: 10.1038/nprot.2013.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cong SY, et al. Mutant huntingtin represses CBP, but not p300, by binding and protein degradation. Molecular and Cellular Neurosciences. 2005;30:560–71. [PubMed] [Google Scholar]
- 44.Li B, Kohler JJ. Glycosylation of the nuclear pore. Traffic. 2014;15:347–61. doi: 10.1111/tra.12150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Teo CF, et al. Glycopeptide-specific monoclonal antibodies suggest new roles for O-GlcNAc. Nature chemical biology. 2010;6:338–343. doi: 10.1038/nchembio.338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang Z, et al. Extensive crosstalk between O-GlcNAcylation and phosphorylation regulates cytokinesis. Science Signaling. 2010;3:ra2. doi: 10.1126/scisignal.2000526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Myers SA, Daou S, Affar el B, Burlingame A. Electron transfer dissociation (ETD): the mass spectrometric breakthrough essential for O-GlcNAc protein site assignments-a study of the O-GlcNAcylated protein host cell factor C1. Proteomics. 2013;13:982–91. doi: 10.1002/pmic.201200332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Szklarczyk D, et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Research. 2015;43:D447–52. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Erkner A, et al. Grunge, related to human Atrophin-like proteins, has multiple functions in Drosophila development. Development. 2002;129:1119–29. doi: 10.1242/dev.129.5.1119. [DOI] [PubMed] [Google Scholar]
- 50.Charroux B, Freeman M, Kerridge S, Baonza A. Atrophin contributes to the negative regulation of epidermal growth factor receptor signaling in Drosophila. Dev Biol. 2006;291:278–90. doi: 10.1016/j.ydbio.2005.12.012. [DOI] [PubMed] [Google Scholar]
- 51.Zhang S, Xu L, Lee J, Xu T. Drosophila atrophin homolog functions as a transcriptional corepressor in multiple developmental processes. Cell. 2002;108:45–56. doi: 10.1016/s0092-8674(01)00630-4. [DOI] [PubMed] [Google Scholar]
- 52.Wang L, Rajan H, Pitman JL, McKeown M, Tsai CC. Histone deacetylase-associating Atrophin proteins are nuclear receptor corepressors. Genes Dev. 2006;20:525–30. doi: 10.1101/gad.1393506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang Z, et al. Atrophin-Rpd3 complex represses Hedgehog signaling by acting as a corepressor of CiR. The Journal of cell biology. 2013;203:575–83. doi: 10.1083/jcb.201306012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Miura GI, Roignant JY, Wassef M, Treisman JE. Myopic acts in the endocytic pathway to enhance signaling by the Drosophila EGF receptor. Development. 2008;135:1913–22. doi: 10.1242/dev.017202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen DY, et al. The Bro1-domain-containing protein Myopic/HDPTP coordinates with Rab4 to regulate cell adhesion and migration. Journal of cell science. 2012;125:4841–52. doi: 10.1242/jcs.108597. [DOI] [PubMed] [Google Scholar]
- 56.Huang HR, Chen ZJ, Kunes S, Chang GD, Maniatis T. Endocytic pathway is required for Drosophila Toll innate immune signaling. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:8322–7. doi: 10.1073/pnas.1004031107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pradhan-Sundd T, Verheyen EM. The role of Bro1-domain-containing protein Myopic in endosomal trafficking of Wnt/Wingless. Dev Biol. 2014;392:93–107. doi: 10.1016/j.ydbio.2014.04.019. [DOI] [PubMed] [Google Scholar]
- 58.Pradhan-Sundd T, Verheyen EM. The Myopic-Ubpy-Hrs nexus enables endosomal recycling of Frizzled. Mol Biol Cell. 2015;26:3329–42. doi: 10.1091/mbc.E15-02-0086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gilbert MM, Tipping M, Veraksa A, Moberg KH. A screen for conditional growth suppressor genes identifies the Drosophila homolog of HD-PTP as a regulator of the oncoprotein Yorkie. Developmental cell. 2011;20:700–12. doi: 10.1016/j.devcel.2011.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shafi R, et al. The O-GlcNAc transferase gene resides on the X chromosome and is essential for embryonic stem cell viability and mouse ontogeny. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:5735–9. doi: 10.1073/pnas.100471497. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.