Abstract
Purpose
Patient-derived xenograft (PDX) models accurately recapitulate the tumor of origin in terms of histopathology, genomic landscape, and therapeutic response, but some limitations due to costs associated with their maintenance and restricted amenability for large-scale screenings still exist. To overcome these issues, we established a platform of 2D cell lines (xeno-cell lines, XLs), derived from PDXs of colorectal cancer (CRC) with matched patient germline gDNA available.
Experimental Design
Whole exome and transcriptome sequencing analyses were performed. Biomarkers of response and resistance to anti-HER therapy were annotated. Dependency on the WRN helicase gene was assessed in MSS, MSI-H and MSI-like XLs using a reverse genetics functional approach.
Results
XLs recapitulated the entire spectrum of CRC transcriptional subtypes. Exome and RNA-seq analyses delineated several molecular biomarkers of response and resistance to EGFR and HER2 blockade. Genotype-driven responses observed in vitro in XLs were confirmed in vivo in the matched PDXs. MSI-H models were dependent upon WRN gene expression, while loss of WRN did not affect MSS XLs growth. Interestingly, one MSS XL with transcriptional MSI-like traits was sensitive to WRN depletion.
Conclusion
The XL platform represents a preclinical tool for functional gene validation and proof of concept studies to identify novel druggable vulnerabilities in CRC.
Keywords: Colorectal cancer, preclinical models, EGFR, HER2, resistance
1. Introduction
The knowledge that cancer is a genetic disease and the availability of genomic profiles of tumor samples have paved the way to ‘precision medicine’ in oncology (1).
In the last ten years, major advances in complex genomic technologies have strongly contributed to the mapping of cancer molecular landscape (2). The need to understand the impact of the major drivers of cancer development and progression on therapeutic intervention has thus become of paramount importance. Functional characterization and target validation requires the generation of reliable cellular and animal models to understand the role of genomic alterations on tumor onset and evolution (3). Functional studies have demonstrated that the tumor is often dependent on oncogenic alterations for its growth and maintenance, providing a strong rationale for the development of therapies targeting cancer driver genes (4).
On these premises, modeling disease-associated alterations in models that reflect the clinical features of patients is vital to implement precision medicine. So far, different cellular and animal models have been generated, each presenting both advantages and pitfalls.
Cancer cell lines have been extensively and routinely used for biomarker discovery and drug development. Indeed, the Cancer Cell Line Encyclopedia, the Genomics of Drug Sensitivity in Cancer (GDSC) Sanger Institute project, the National Cancer Institute-60 (NCI-60) cancer cell line screen, and the Cancer Therapeutic Response Portal (CTRP) constitute paradigmatic examples of how coupling cell lines to systematic compound screening can provide an informative clinical platform for pharmacogenomic analyses (5–10). Genomic and transcriptional drift during serial passaging, loss of molecular heterogeneity and lack of the tumor microenvironment represent the three main issues thought to make cell line models divergent from the original tumor. For these reasons, the lack of experimental reproducibility and the clinical relevance of cell lines have repeatedly been questioned (11,12). These drawbacks may play against the recapitulation of the evolutionary nature of cancer itself, thus limiting the translation of the in vitro results to the clinical response of the tumor in the patient. In addition, the unavailability of matched germline tissue has often hampered the confident identification of somatic changes.
To partly obviate these issues and better bridge the bench-to-bedside gap, patient-derived tumor xenografts (PDXs, xenopatients) have been developed. Although these models had already been described in the seventies, they have become popular for assessing genotype-drug correlation response studies in the last decade (13). Comprehensive molecular annotation studies have demonstrated that PDXs maintain the majority of genetic alterations and global pathway activity of primary tumors (14,15). Importantly, the histological structure and intra-tumor clonal heterogeneity are usually preserved (16), although recent literature reports the selection of specific copy number alterations during PDX propagation (17). In colorectal cancer (CRC), seminal studies have already set the stage for pharmacogenomic analyses and association of PDX genotype with targeted therapy response (14,18). PDXs have significantly contributed to shedding light on the mechanisms of resistance and on identification of predictive biomarkers of response, but large-scale screening is actually limited by the costs, size and efforts for animal maintenance and manipulation.
Models that can couple both the ease of handling of cell cultures with the preservation of intra-tumor heterogeneity of PDXs have been proposed. Bruna and colleagues (5) have generated a large biobank of breast cancer PDXs combined with short-term cultures of PDX-derived cells demonstrating that the corresponding cell models could be reliably used to assess drug response and to identify biomarkers of resistance in vitro, as paralleled in vivo. Similar studies have been recently performed also in gastrointestinal and pancreatic cancer (19,20), thus confirming the value and applicability of this approach.
In this work, we describe the establishment of a panel of 29 cell lines obtained from a cohort of 29 CRC PDXs and provide a comprehensive genomic analysis of these models. Importantly, we show that the molecular features of xenopatient-derived cell lines (XLs) closely mimic their matched PDX models. We also show that XLs constitute a valuable model to investigate CRC patient molecular vulnerabilities and provide an effective opportunity to reveal patient-specific drug responses and implement precision oncology in CRC.
2. Materials and Methods
Xenopatient-derived cell line establishment, culture and authentication
For xenopatient generation, fresh surgically resected primary CRC and metastases tissues or biopsies were obtained and collected from consenting patients. All procedures were approved by the Italian Ministry of the Health and the Ethics Committee of the Medical faculty of the University of Rostock, in accordance with generally accepted guidelines for the use of human material.
Tumor specimens were cut into small pieces and either frozen (biobanked) or prepared for implantation, as previously described (14). For tumor implantation, six-week-old female NMRI nu/nu or NOD/SCID mice were used as recipients. All experimental procedures were carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The protocol was approved by the Committee on the Ethics of Animal Experiments of the University of Rostock and the Internal Ethical Committee from Animal Experimentation of the Candiolo Cancer Center, as well as the Italian Ministry of Health.
Established tumors from xenopatients (1500 mm3) were removed and processed for in vitro culture as described below.
PDX tissues were dissociated into single-cell suspension by mechanical dissociation using the Gentle MACS Dissociator (Miltenyi Biotec) and enzymatic degradation of the extracellular matrix using the Human Tumor Dissociation Kit (Miltenyi Biotec) was performed according to the manufacturer’s protocol. Cell suspension was centrifuged three times and the pellet was re-suspended in DMEM/F12 medium containing 10% FBS and 2 mM L-glutamine. The final cell suspension was then filtered through a 70-μm cell strainer (Miltenyi Biotec) and cells that were not filtered out were re-suspended in DMEM/F12 medium containing 10% FBS, 2mM L-glutamine, antibiotics (100 U/ml penicillin and 100 mg/ml streptomycin) and 10 μM ROCK inhibitor Y-27632 (Selleck Chemicals Inc) and cultured on collagen-coated plates (Corning) at 37°C in a humidified atmosphere of 5% CO2. Medium was changed regularly every three days. Growing cell lines were further passaged and were subjected to three sequential cycles of freezing and thawing to ensure the stability of the cell lines. Growing cell lines were stocked at low passages by cryopreservation.
For only one case (CRC0080), the cell line was established as a xenosphere (21) and then adapted to grow in a 2D monolayer. The HROC cell line collection was derived by Michael Linnebacher’s laboratory as previously reported (22).
BT474 and SKBR3 cell lines were purchased from the American Type Culture Collection (ATCC) and maintained in DMEM/F12 and DMEM media, respectively, containing 10% FBS, 2mM L-glutamine, antibiotics (100 U/ml penicillin and 100 mg/ml streptomycin) and cultured at 37°C in a humidified atmosphere of 5% CO2.
Xenopatient-derived cell lines (XLs) were routinely checked for mycoplasma contamination using the Venor GeM Classic Kit (Minerva Biolabs) according to the manufacturer’s protocol. XLs, matched PDXs and patient authentication was performed using the Gene Print 10 System (Promega) through Short Tandem Repeats (STR) at 10 different loci (D5S818, D13S317, D7S820, D16S539, D21S11, vWA, TH01, TPOX, CSF1PO and amelogenin). Amplicons from multiplex PCRs were separated by capillary electrophoresis (3730 DNA Analyzer, Applied Biosystems) and analyzed using GeneMapperID v.3.7 software (Life Technologies).
DNA extraction and MSI analysis
Genomic DNA samples were extracted from each XL and matched PDX using ReliaPrep gDNA Tissue Miniprep System (Promega). Germline genomic DNA obtained from normal tissue (liver) or PBMCs of each patient was used as the reference genome.
The microsatellite instability (MSI) status was evaluated by using the MSI Analysis System kit (Promega) according to manufacturer’s protocol. The analysis requires a multiplex amplification of seven markers including five mononucleotide repeat markers (BAT-25, BAT-26, NR-21, NR-24 and MONO-27) and two pentanucleotide repeat markers (Penta C and Penta D). The products were analyzed by capillary electrophoresis in a single injection using ABI 3730 DNA Analyzer capillary electrophoresis system (Applied Biosystems). The results were analyzed using GeneMapper V5.0 software. Samples with instability in two or more markers were defined as MS-instable (MSI-H). Samples with no detectable alterations were defined as MS-stable (MSS).
MSI events were evaluated by MSIsensor (23). Signatures were calculated using a custom Python script.
Exome sequencing and bioinformatic analysis
Exome sequencing was performed in outsource, while data analysis was performed at the Candiolo Cancer Institute. Each set of DNA samples (PDX, xeno-cell line, germline) was sent to GATC (Kostanz, DE) where library preparation, exome capture, sequencing on Illumina platform and data demultiplexing were performed. Whole-exome sequencing data were analyzed following procedures as previously described (24).
Gene copy number analysis
Real-time PCR was performed with 10 ng of DNA per single reaction using GoTaq QPCR Master Mix (Promega) with an ABI PRISM 7900HT apparatus (Applied Biosytems; primers’ sequences are available on request). Gene copy numbers were normalized to a control diploid cell line, HCEC.
RNA extraction, analysis and identification of cancer-cell intrinsic subtypes
RNA was extracted using miRNeasy Mini Kit (Qiagen), according to the manufacturer’s protocol for both XLs and PDXs. The quantification and quality analysis of RNA was performed on a Bioanalyzer 2100 (Agilent), using RNA 6000 nano Kit (Agilent). Total RNA extracted from XLs was processed for RNA-seq analysis with the TruSeq RNA Library Prep Kit v2 (Illumina) following manufacturer’s instruction and sequenced on a NextSeq 500 system (Illumina). Each fastq file was aligned using MapSplice (25). Version hg19 was used as the genome reference and Gencode v19 was used as the transcriptome reference database and gene quantification was performed with RSEM (26). Assignments of each xeno-cell line to cancer-cell intrinsic subtypes and consensus molecular subtypes were performed as previously described (27,28). Correlation analysis of PDX and matched XL samples was performed with Pearson correlation based on genes with at least one RPM in either datasets for a total of 23 pairs.
Data deposition
WES and RNAseq data from PDXs and matched XLs have been deposited in the European Nucleotide Archive (ENA) with the accession number PRJEB33640.
Drug proliferation assays
CRC XLs were seeded at different densities (5-7 × 103 cells per well) in 100 μl complete growth medium in 96-well plastic culture plates at day 0. The following day, serial dilutions of the indicated drugs were added to the cells in serum-free medium while medium-only was included as controls. Plates were incubated at 37 °C in 5% CO2 for 5 or 6 days, after which cell viability was assessed by measuring ATP content through CellTiter-Glo Luminescent Cell Viability assay (Promega). Luminescence was measured by SPARK M10 (Tecan) plate reader. Treated wells were normalized to untreated/DMSO treated wells.
For long-term proliferation assays, cells were seeded in 24-wells plates (1-2.5 × 104 cells per well) and cultured in the absence and presence of drugs as indicated. Wells were fixed with 4% paraformaldehyde (Santa Cruz) and stained with 1% crystal violet-methanol solution (Sigma-Aldrich) after 2 weeks. All assays were performed independently at least three times.
Cetuximab and trastuzumab were obtained from the Pharmacy at Niguarda Cancer Center in Milan, Italy. Lapatinib was purchased from Selleck Chemicals.
RNA interference
A pool of four siRNAs targeting WRN were used (Dharmacon, L-010378-00-0005). A total of 15 XL models classified as MSS, MSI-H and MSI-like, were grown and transfected with siRNA at a final concentration of 20 nM using the RNAiMAX (Invitrogen) transfection reagent following the manufacturer’s instructions. In each experiment, mock control (transfection lipid only), as well as ON-TARGETplus Non-targeting Control Pool (Dharmacon, D-001810-10-05) and polo‐like kinase 1 (PLK1) targeting siRNA (Dharmacon, L-003290-00-0010) were included as negative and positive controls, respectively. After 6 days, cells were collected for CellTiter-Glo assay (Promega). CellTiter-Glo data were read on an Envision Multiplate Reader and data were visualized using GraphPad Prism 7 software. siRNA sequences are the following:
Non-targeting Control Pool: UGGUUUACAUGUCGACUAA, UGGUUUACAUGUUGUGUGA, UGGUUUACAUGUUUUCUGA, UGGUUUACAUGUUUUCCUA
PLK1: GCACAUACCGCCUGAGUCU, CCACCAAGGUUUUCGAUUG, GCUCUUCAAUGACUCAACA, UCUCAAGGCCUCCUAAUAG
WRN: GAUCCAUUGUGUAUAGUUA, GCACCAAAGAGCAUUGUUA, AUACGUAACUCCAGAAUAC, GAGGGUUUCUAUCUUACUA
Lentiviral Transduction of ERBB2-amplified Colorectal XL-cells
The PIK3CA p.H1047R mutation was inserted into pLenti-PIK3CA-myc-DDK vector (Origene) using site directed mutagenesis with the Quick Change II Site-Directed Mutagenesis kit (Agilent) according to manufacturer’s instructions. Primers sequences for PIK3CA site directed mutagenesis were the following: GAAACAAATGAATGATGCACGTCATGGTGGCTGGACAAC (PIK3CA_H1047R_F) and GTTGTCCAGCCACCATGACGTGCATCATTCATTTGTTTC (PIK3CA_H1047R_R). Lentiviral control pLenti-myc-DDK vector was purchased from Origene.
Lentiviral control vectors (pRLL empty and FG12 empty), pRLL-KRAS WT, pRLL-KRAS G13D and FG12-BRAF V600E vectors were previously exploited (29,30).
Lentiviral vector stocks were produced by transient transfection of the transfer plasmids, the packaging plasmids pMDLg/pRRE and pRSV.REV, and the vesicular stomatitis virus (VSV) envelope plasmid pMD2.VSV-G (12, 5, 2.5, and 3 μg, respectively, for 10 cm dishes) in HEK-293T cells. Determination of the viral p24 antigen concentration was done by ALLIANCE HIV-I P24 ELISA 2 PLATE KIT (PerkinElmer Life Science Inc.). Cells were transduced in six-well plates (3 × 105 per well in 2 ml of medium) in the presence of polybrene (8 mg/ml) (Sigma).
Western blotting analysis
Total cellular proteins were extracted by solubilizing the cells in boiling SDS buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, and 1% SDS). Extracts were clarified by centrifugation and protein concentration was determined using the BCA Protein Assay Reagent kit (Thermo). Western blot detection was performed with enhanced chemiluminescence system (GE Healthcare) and peroxidase conjugated secondary antibodies (Amersham). The following primary antibodies were used for western blotting (all from Cell Signaling Technology, except where indicated): anti-phospho-HER2 YY1221/1222); anti-HER2 (Santa Cruz); anti-phospho-EGFR (Y1068); anti-EGFR (clone 13G8, Enzo Life Science); anti-vinculin (Millipore); anti-phospho-p44/42 ERK (Thr202/Tyr204); anti-p44/42 ERK; anti-phospho AKT (Ser473); anti-AKT; anti-PTEN; anti-KRAS (clone 3B10-2F2, Sigma-Aldrich); anti-BRAF; anti-PIK3CA; anti-HSP90 (Santa Cruz); anti-GAPDH (Abcam); anti-ACTIN (Santa cruz); anti-MLH1 (Abcam); anti-MSH2 (Abcam); anti-MSH6 (Abcam); anti-PMS2 (Cell Marque Corporation, USA).
Western blot was employed to validate siRNA-mediated WRN knockdown. Briefly, 1 × 106 cells were seeded in 10 cm well plates and then transfected the day after with the indicated siRNAs, following the procedure presented in the RNA interference section. Seventy-two hours post transfection, cells were lysed with 150 μl RIPA buffer supplemented with protease and phosphatase inhibitors and lysates were used for SDS-PAGE and immunoblot analysis. Primary antibodies against WRN (Cell Signaling Technologies) and anti-β-tubulin (Sigma-Aldrich) were used. Anti-mouse IgG HRP-linked secondary antibody (GE Healthcare, NA931) was used as secondary antibody. Molecular weight markers included: SeeBlue Plus2 Pre-stained Protein Standard (Thermo Fisher Scientific) and Precision Plus Protein Standards (BioRad).
In vivo treatment
Established tumors (average volume 400 mm3) were treated with the following regimens, either single-agent or in combination: lapatinib (Carbosynth) 100 mg/kg daily (vehicle, 0.5% methylcellulose, 0.2% Tween-80); trastuzumab (Roche) 30 mg/kg once weekly (vehicle: physiological saline). Tumor size was evaluated weekly by caliper measurements, and the approximate volume of the mass was calculated using the formula 4/3π (d/2)2 D/2, where d is the minor tumor axis and D is the major tumor axis. Animal procedures were approved by the Ethical Commission of the Candiolo Cancer Institute and by the Italian Ministry of Health.
Statistical Analysis
Results were expressed as means ± standard error of the mean (SEM) or standard deviation (SD) as indicated in the legend. Statistical significance was evaluated by t test or two-way ANOVA using GraphPad Prism software. p<0.05 was considered statistically significant.
3. Results
Establishment of PDX-derived cell lines
We derived primary cell lines from a set of CRC PDX models with the goal of setting up in vitro CRC preclinical models that more closely resembled the features of the patient’s tumor. The clinico-pathological characteristics of patients from whom the PDXs were initially obtained are listed in Supplementary Table S1. The establishment procedure, described in detail in the ‘materials and methods’ section, is schematically represented in Supplementary Fig. S1. Briefly, the PDX tumor is surgically excised and mechanically and enzymatically dissociated to obtain cells that are subsequently cultured in 2D petri dishes. In this work, a total of 29 primary cell lines have been derived from a collection of 29 PDXs established at the Candiolo Cancer Institute (n=17) and at Rostock University (n=12) (14,18,22). Among the 29 PDXs, 10 originated from primary tumors and 19 from liver or peritoneum metastases of 28 CRC patients (two different metastatic lesions - IRCC-5A and IRCC-5B - were obtained from the same patient) (Fig. 1A and Supplementary Fig. S1). To underline the origin of these 2D primary cell lines from xenopatients, we will refer to them as xeno-cell lines (XLs).
Figure 1. Exome sequencing analysis of XLs matches the profile of the corresponding PDX of origin.
A, Site of origin of PDX samples used to generate XLs. B, Relative distribution of microsatellite status in XL-cells. C, Number of MSI events in the 29 PDX-XL pairs, stratified by the length of the repeat units identified by exome analysis. D, Mutational load (number of mutations per Mb) in the 29 PDX-XL pairs, as calculated by exome analysis. E, The counts of non-synonymous mutations (blue bar) and INDELs (red bar) in the 29 PDX-XL pairs, as calculated by exome analysis. F, Mutational spectra in the 29 PDX-XL pairs, as relative proportion of the six different possible base-pair substitutions (nucleotide transitions and trasversions). Microsatellite stable (MSS) and unstable (MSI-H) cases are highlighted in light blue and light red, respectively.
XLs, although presenting cell-specific morphological features, displayed a predominant and typical epithelial pattern characterized by islets of compact or round shaped cells growing predominantly as a monolayer or, in one case (HROC147), as cell-clumps in suspension (Supplementary Fig. S2A). The epithelial phenotype was confirmed by analysis of expression of specific epithelial markers (e.g. E-cadherin, cytokeratin 8, 18 and 19 and EpCAM) with respect to mesenchymal markers (e.g. vimentin, SNAIL and TWIST1) (Supplementary Fig. S2B).
All XLs displayed the ability of long-term propagation (multiple passages) in vitro and their stability was ensured by at least three sequential freezing/thawing cycles prior to bio-banking. Cells were maintained in culture for at least 8 passages prior to nucleic acid extraction.
Xeno-cell lines recapitulate the genomic landscape of matched patient-derived xenografts
To assess if the CRC molecular subtypes that are commonly ascertained in the clinic were preserved in both the PDXs and matched XLs, we performed genomic and transcriptomic analyses systematically comparing the two preclinical platforms.
Initially, we analyzed microsatellite markers and found that 5 out of 29 (17%) PDXs displayed microsatellite instability (MSI-H), while the remaining were microsatellite stable (MSS). The MSI status was maintained in the matched XLs (Fig. 1B).
Transcriptome profiling of XLs and their originating PDXs revealed higher correlation between matched XL-PDX pairs than between unmatched pairs (Kolmogorov-Smirnov, P < 1x10−16, Supplementary Fig. S3A and B).
Next, parallel exome sequencing analyses of XLs and matched PDXs were performed. Importantly, to identify tumor-specific somatic mutations, germline genomic DNA (gDNA) extracted from normal tissues (liver) or blood (PMBCs) of each corresponding patient was used as the reference genome.
Samples were sequenced with high depth and coverage to allow detection of low-frequency somatic mutations in xenopatient tumors and matched XLs despite contamination of human cancer cells with murine infiltrating cells in some samples. The median depth achieved was 143x and the median coverage was 97.8% on the coding regions of the reference genome (assembly hg38) with 90.3% of the bases covered at least 40 times.
A high level of correlation was found between cell lines and matched PDXs regarding the total number of somatic non-synonymous single-nucleotide variants (SNVs) and insertions or deletions (INDELs) (Pearson r=.9870; P<.0001) (Supplementary Fig. S4A). Overall, even taking into consideration genomic variability and tumor evolution that could occur during in vivo passages and in vitro culturing, mutation and copy number profiles detected in PDXs are highly preserved (i.e. shared) in XLs (Supplementary Fig. S4B).
MSI-H models showed a higher number of MSI events compared to MSS cases (median value MSI-H = 7723.5 vs MSS = 263.5) (Fig. 1C). Moreover, the number of MSI events is maintained in each PDX and matched XL (Pearson r = .998), in both MSS and MSI-H cases.
MSI-H cell lines and matched PDXs displayed a hypermutator phenotype with a higher mutational load (number of mutations/Mb) as compared to MSS models (median value MSI-H = 60.5 vs MSS = 5.5) (Fig. 1D), reflecting a higher number of non-synonymous single nucleotide variations (SNVs) (mutation burden) (median value MSI-H = 1691.5 vs MSS = 116) (Fig. 1E). This is consistent with the reported mutation frequency in CRC tissues and cell lines (2,7,31). No MSS model was found to be hypermutated, likely reflecting the lack of mutations in POLE or POLD1, which are known to generate mutations in the absence of MSI-H (2). Moreover, the INDEL burden was higher in MSI-H compared to MSS models (median value MSI-H = 562.5 vs MSS = 7), as previously reported in TCGA cancers and CRC cell lines (2,31) (Fig. 1E and Supplementary Fig. S4C).
Somatic variants within the coding regions in XLs were mostly concordant with the matched PDX specimens for both MSS and MSI-H models, with few exceptions (CRC0078, CRC0740 and CRC0080) displaying a similar or higher number of private mutations (found in XL and/or PDX) with respect to shared mutations (Supplementary Fig. S4D).
In general, the allelic frequencies of alterations shared between XLs and corresponding PDXs were significantly correlated (Pearson r =.8233) (Supplementary Fig. S4E).
Despite the global genetic correlation between the sets of models, we also detected several divergent mutations in either XLs (n = 4019) or PDXs (n = 1885) (private mutations). The putative biological significance in cancer of these private mutations was assessed based on Cancer Gene Census and data reported from CRC analysis (32,33). Only 7.1% (285/4019) divergent mutations found in XLs affected cancer-related genes. Similarly, pan cancer-related genes that were discordant in the PDXs represented 6.7% (126/1885) (Supplementary Table S2). The majority of these discordant mutations in cancer-related genes are found in MSI-H models (97/126 = 77.0% in PDX; 222/285 = 77.9% in XL), as expected due to the hypermutated phenotype of these models. The remaining discordant mutations affecting pan cancer-related genes (n = 92) were identified in MSS models (PDX = 22 SNVs + 7 INDELs; XL = 50 SNVs + 13 INDELs). Of note, among 411 private alterations found in pan cancer-related genes, only 92 affected CRC-related genes (CRC_CGC): 23 in PDXs and 69 in XLs. Twenty-six (13 SNVs + 13 INDELs) out of 92 were found in MSS models (Supplementary Table S2) and the number of private variants occurring in CRC-related genes for each MSS XL or PDX ranged from zero to three. We found that the majority of private alterations have lower variant allele frequency (VAF) compared to shared variants, suggesting that the majority of discordant mutations found in the matched models might be of subclonal origin (Supplementary Fig. S4F). Interestingly, also a subgroup of shared alterations ranged with frequencies between 25% and 50% (Supplementary Fig. S4F), thus endowing the polyclonal architecture characterizing PDXs and retained in matched XLs as a unique model to investigate on heterogeneity and clonal evolution in CRC.
While the SNVs spectrum was maintained between XLs and matched PDXs, it differed significantly between the MSI-H and MSS cases, with an increased proportion of C>G/G>C transversions (about 5-fold) and a decrease in T>C/A>G transitions in the MSS cases (about 1.3-fold, Fig. 1F). Interestingly, MSI-H models present a SNV spectrum typically associated with defective DNA mismatch repair and found in MSI-H CRCs (34) (Supplementary Fig. S5).
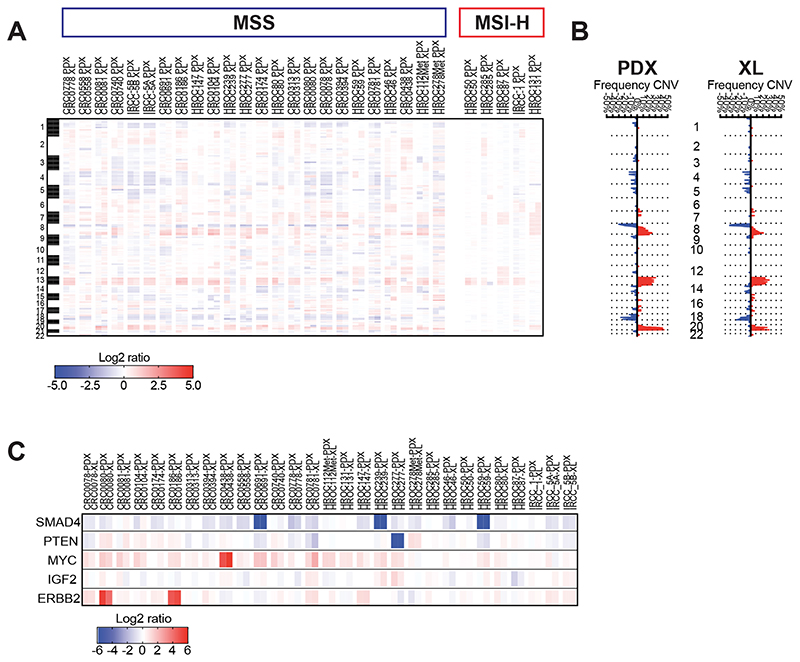
Finally, exome analysis revealed that the copy number profile of each PDX is also maintained in the corresponding derived cell line (Pearson correlation: 0.82 ≤ r ≤ 0.98). As expected, MSS PDXs and matched XLs displayed a higher level of loss of heterozygosity (LOH) or copy-number variations than MSI-H models, which exhibited a more stable CNV profile (Fig. 2A). In MSS cases, the most common deleted chromosomal arms were 8p, 17p (including TP53), and 18q (including SMAD4), and the most common gained regions were chromosome 7, 8q (including MYC), 13, and 20q (Fig. 2B and 2C).
Figure 2. Comparison of copy-number variation unveils gain or loss of selected genome segments in PDX-XL matched models.
A, Heat map comparing copy-number aberrations between matched PDX and XL models in log2 scale, as assessed by whole exome sequencing (WES). Red colors indicate gains, while blue colors indicate losses. The band on the left represents chromosomes 1-22. B, Plot of frequency of gains and losses for each position per group (PDX and XL) along the genome, as assessed by WES. Each horizontal dotted line represents chromosomal boundaries. C, Heat map comparing copy-number between matched PDX and XL models in log2 scale for the indicated genes.
CRC pathway alterations are conserved in XLs
Integration of genomic data allowed for the identification of deregulated signaling pathways, and the genes most commonly altered in CRC (2) were all represented in our panel of PDXs and matched XLs (Fig. 3A). In particular, recurrent alterations in the DNA mismatch repair (MMR), Wnt, MAPK, PI3K/PTEN, TGF-β and p53 pathways were identified (Fig. 3A).
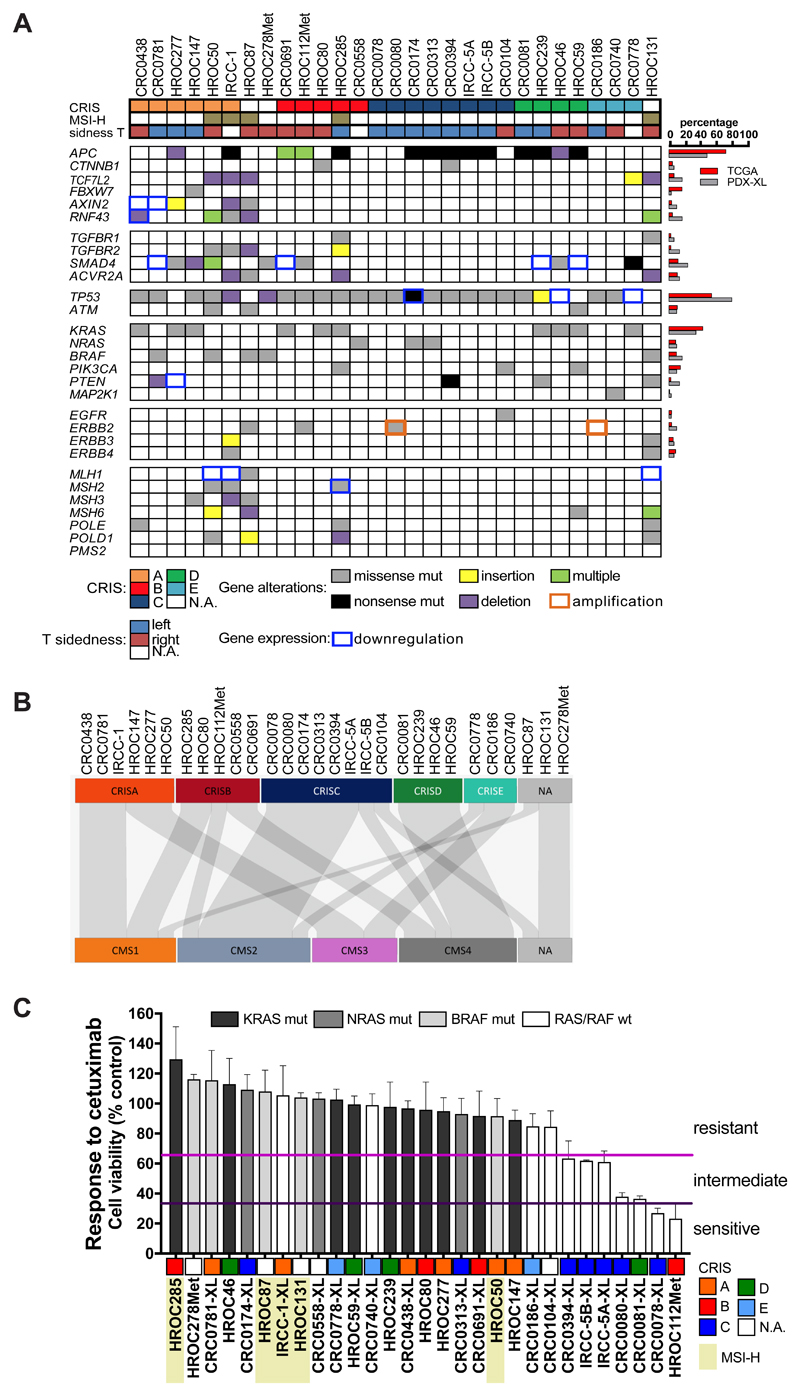
Figure 3. Genomic landscape of deregulated signaling pathways in PDX-XL pairs and response to cetuximab treatment.
A, Landscape of the gene alterations found in the PDX-XL pairs; missense mutations (gray boxes), nonsense mutations (black boxes), insertion (yellow boxes) and deletion (violet boxes) as well as co-occurrence of multiple alterations (green boxes) are displayed. Orange squared boxes indicate gene amplification, while blue squared boxes indicate gene downregulation. Genes are grouped according to signaling pathways; cases are grouped according to transcriptional CRIS classification of XLs (as assessed in (27)). Primary tumor site (right and left) for each cell line are also reported. MSI-H cases are also shown. Right panel, Percentage of gene alterations in PDX-XL pairs (gray bars) are compared to percentage of cases reported in TCGA CRC cases (red bars). B, Caleydo view of correspondences between the CRIS vs CMS class assignments of XLs. C, Bar graph representing the effect of cetuximab treatment normalized on control untreated cells for the XLs. Viability was assessed by measuring ATP content after 6 days of treatment with cetuximab at the clinically relevant dose of 10 ug/ml. Data represent the average of value obtained from at least three independent experiments, error bars represent SD. Based on their response to cetuximab, cell lines were subdivided in three different groups: resistant (first tertile, range 100-66%), intermediate (second tertile, range 65-34%) and sensitive (third tertile, range 33-1%). MSI-H cells are highlighted in light yellow and for each cell line its CRIS classification is reported.
Mutations in MMR genes were found in MSI-H tumors. The alterations detected were missense mutations in MSH2, MSH3, MSH6 and POLE/POLD1 genes, as well as MLH1 expression downregulation (Fig. 3A). Interestingly, three MSS cases displayed alterations in MSH3, MSH6 and POLE DNA repair associated genes in both PDXs and matched XLs. These mutations were not reported previously, nor did they fall in regions important for protein activity, suggesting that these genes are not functionally affected by these mutations.
The Wnt pathway is frequently altered (90%) in CRC. The most frequent alterations include inactivating mutations in APC as well as activating mutations in CTNNB1 (2). Other variants affecting this pathway are inactivating mutations targeting regulators of the Wnt pathway, such as TCF7L2, FBXW7, AXIN2 and RNF43 genes.
APC mutants were identified in 14 out of 29 cases (48.3%), while other alterations were found in the same pathway in APC wild-type cases, such as a FBXW7 alteration (p.R465H) in one case, and AXIN2 downregulation or mutation (p.G495A) in three cases. Similarly, RNF43 alterations were identified in one MSS APC wild-type cell line and in four out of five microsatellite unstable cases, highlighting the association of these alterations with the MSI-H phenotype, as previously reported (35). Of interest, multiple alterations affecting RNF43 were detected in two APC wild-type cases.
In addition, a translocation of the Wnt-agonistic RSPO3 with PTPRK was found in one BRAF-mutated APC wild-type case, as assessed by RNA-seq analysis on XLs and validated by quantitative RT-PCR (Fig.3B and Supplementary Fig. S6A and S6B). No RSPO2 gene fusions were identified in our collection of XLs.
TP53 alterations were found in 79.3% of cases (23 out of 29). In addition, alterations in ATM were found in 10.35% (3 out of 29). One of these co-occurred with a TP53 mutation, while the remaining two cases were TP53 wild-type.
The TGF-β signaling pathway is also frequently deregulated in CRC (2). In our dataset, we identified two TGFBR1 mutated cases and four cases carrying TGFBR2 alterations. Consistent with previous findings (36), all of these models were MSI-H. Moreover, alterations in SMAD4 were identified in seven cases, while four other cases (CRC0781-XL, CRC0691-XL, HROC59 and HROC239) did not express SMAD4, as assessed by RNA-seq analysis (Fig. 3A and Supplementary Fig. S6C). Lack of SMAD4 expression was ascribed to LOH in three cases (Fig. 2C). No alterations in SMAD2 and SMAD3 were detected. Three MSI-H cases (IRCC-1-XL, HROC131 and HROC285) displayed deletions in the ACVR2A gene, a TGF-β family member, known as a potential player in CRC (2).
Genetic alterations in RAS-MAPK and PI3K-PTEN pathways are very frequent in CRC. Activating mutations in KRAS - including the less common mutations p.Q61K (exon 3), p.K117N and p.A146V (exon 4) - and in NRAS were identified in 13 out 29 XLs and matched PDXs, thus reflecting the prevalence of RAS mutations found in CRC patients. We identified five models harboring BRAF p.V600E mutations, two derived from MSS metastases and three from primary MSI-H CRC. While alterations in KRAS, NRAS and BRAF displayed a significant pattern of mutual exclusivity, two out of ten KRAS mutated models also exhibited activating mutations in the PIK3CA gene (codon 1047 and 1049). Only one PIK3CA variant (p.H1047R) was identified in a KRAS/NRAS/BRAF wild-type case.
Missense (p.A86E and p.D92Y) as well as nonsense (p.R233*) mutations affecting the PTEN gene were identified in three cases. Although both missense mutations fall in the proximity or within the WPD loop of the catalytic site, only alterations in codon 92 have been reported to fully abrogate PTEN function (37). In addition, western blot analysis highlighted the loss of PTEN expression in two XLs: CRC0394-XL carrying the p.R233* nonsense mutation, and HROC277 in which PTEN expression was consistently decreased at the transcript level due to genetic loss of the PTEN locus (Fig. 2C, Supplementary Fig. S6C and S6D). The PTEN p.K342N variant found at high VAF in the MSI-H HROC131 line (Supplementary Table S2) does not affect PTEN protein expression (Supplementary Fig. S6D)(38). Since this variant was absent in the corresponding PDX, we speculate that this alteration appeared in XL as either a sampling bias during the establishment of the cell line or as an acquired randomly generated mutation owing to the intrinsic hypermutator phenotype of this MSI-H cell line.
Alternative mechanisms of activation for the PIK3CA pathway include transcriptional upregulation of IGF2 (Supplementary Fig. S6E). In our panel of XLs, we identified seven cases (CRC0081-XL, CRC0438-XL, HROC46, HROC278Met, HROC59, HROC239 and HROC285) overexpressing the IGF2 transcript as assessed by RNA-seq analysis (log2 ratio > 1.5) (Supplementary Fig. S6E). Of note, overexpression of IGF2 was not associated to genomic amplification of the IGF2 gene, consistent with previous reports (Fig. 2C) (2,39).
Evaluation of genetic alterations in receptors of the ERBB family receptors unveiled the presence of an EGFR extracellular mutation (p.G465E) in a PIK3CA-mutated case (CRC0104-XL). Of interest, this EGFR mutation was previously found to be associated with acquired resistance to anti-EGFR therapies (40,41). Indeed, its corresponding xenopatient was established from a liver metastasis of a patient who had received cetuximab treatment within six months prior to resection (18).
Another clinically actionable ERBB family member frequently altered in cancer is ERBB2 (also known as HER2). ERBB2 gene amplification was identified in two MSS wild-type RAS/BRAF cases (CRC0080-XL and CRC0186-XL) (Supplementary Fig. S7A). Interestingly, CRC0186-XL showed a remarkable increase in ERBB2 gene copy number (n = 33.2), while CRC0080-XL displayed a less pronounced increase (n = 6.1), comparable to that present in the breast cancer cell lines BT474 and SKBR3 (n = 6.5 and n = 5.6, respectively) (Supplementary Fig. S7B). In these cell lines, ERBB2 gene copy number gain led to protein overexpression (Supplementary Fig. S7C). CRC0080-XL also carried an ERBB2 activating mutation (p.L866M) affecting the intracellular kinase domain of the receptor (42) (Fig. 3A).
Mutations in the extracellular region of the ERBB2 receptor (p.S310F and p.L465V) were identified in two additional xenopatients and matched cell lines (HROC87 and HROC112Met, respectively). Of note, ERBB2 p.S310F was co-present in a BRAF p.V600E mutated case. Similarly, in another BRAF mutated case (HROC131), we identified occurrence of the ERBB3 p.V104M variant (Fig. 3A).
Gene expression profiling revealed that molecular subtypes found in XLs correlated with transcriptional classes identified by the recently described CRC intrinsic subtype (CRIS) analysis (27) (Fig. 3A and 3B). In total, 26 out of 29 XLs were significantly assigned to a specific CRIS class (Supplementary Table S3). In particular, MSI-H and KRAS/BRAF mutant cells were enriched in CRIS-A, while CRIS-C included KRAS/BRAF-WT lines. In addition, two cell lines that displayed TGF-β pathway alteration clustered in CRIS-B and IGF-2 overexpressing cells were included in CRIS-D group (Fig. 3 and Supplementary Table S3). A similar distribution in genetic alterations was also found for the consensus molecular subtypes (CMS). MSI and BRAF mutational status were enriched in CMS1, and KRAS mutant lines were partially depleted in CMS2.
Associations between CRIS and CMS transcriptional classes are also evident. Indeed CRIS-A samples are assigned to CMS1 and CMS3, while CRIS-C is predominantly composed of CMS2. Interestingly, in the context of stroma-depleted samples, CRIS-D samples are also classified as CMS4, likely due to the similarities in stem cells rewiring program in intrinsic cancer cell intrinsic traits – i.e. LGR5 pathway – captured by both classifiers (Fig. 3B).
Sensitivity to EGFR blockade is maintained in xeno-derived cell lines
Anti-EGFR monoclonal antibodies are approved to treat metastatic CRC and achieve a 10% objective response rate in unselected patients treated with antibody monotherapy (43). Clinical benefit of EGFR blockade increases up to 25% in cases when tumors are stratified for wild-type KRAS, NRAS and BRAF (43). To assess if XLs could recapitulate this genotype-drug response, and therefore could be exploited as a reliable platform for drug testing, we checked the entire cell platform for cetuximab sensitivity.
Based on the level of response to cetuximab, the cell collection was classified in three different groups: sensitive (n = 2), intermediate (n = 5) and resistant (n = 22) (Fig. 3C). As observed in the clinic, KRAS, BRAF or NRAS mutations, as well as other genetic alterations (i.e. MAP2K1 p.K57N and EGFR p.G465E), are associated with resistance to EGFR blockade in CRC cells (Fig. 3D) (18,40,44,45). Similarly, ERBB2-amplified CRC0186-XL was refractory to cetuximab treatment, confirming previous findings obtained by preclinical and clinical data from the same patient (14,18). Conversely, the other ERBB2-amplified and mutated CRC0080-XL cell line displayed higher sensitivity to cetuximab treatment.
Integration of multi-layer data also revealed a positive correlation with the transcriptional classes obtained from the cancer cell intrinsic classifier (Supplementary Table S3). As previously reported, CRIS-A resulted enriched in both MSI-H and KRAS/BRAF mutated cases (IRCC-1-XL, HROC50, HROC277, HROC147, CRC0438-XL and CRC0781-XL) with marked resistance to cetuximab treatment (Fig. 3B and 3C), and CRIS-C depleted for KRAS mutant cells (IRCC-5A-XL, IRCC-5B-XL, CRC0078-XL, CRC0080-XL and CRC0394-XL), showing sensitivity to anti-EGFR treatment. Confirming previous reports (27,46), both CRIS-C and CMS2 transcriptional classes are enriched for cetuximab responsive models (Supplementary Table S4).
Expression analysis identifies therapeutic targets in xeno-derived cell lines
RNA-seq data analysis unveiled potentially actionable targets in the XL platform. Since receptor tyrosine kinases (RTKs) are often implicated in cancer and are ideally suited for pharmacological inhibition, the analysis was focused on this class of proteins. Overall, 30 distinct RTKs passed the filtering process during RNA-seq analysis due to their expression level in our XL collection. Outlier expression analysis identified ten (log2 ratio ≥ 3.5) overexpressed receptor tyrosine kinase genes including ERBB2, AXL, FGFR1, NTRK1, NTRK2, ROR2, RET, EPHA4, EPHB1 and EPHB6 in individual XLs (Fig. 4A).
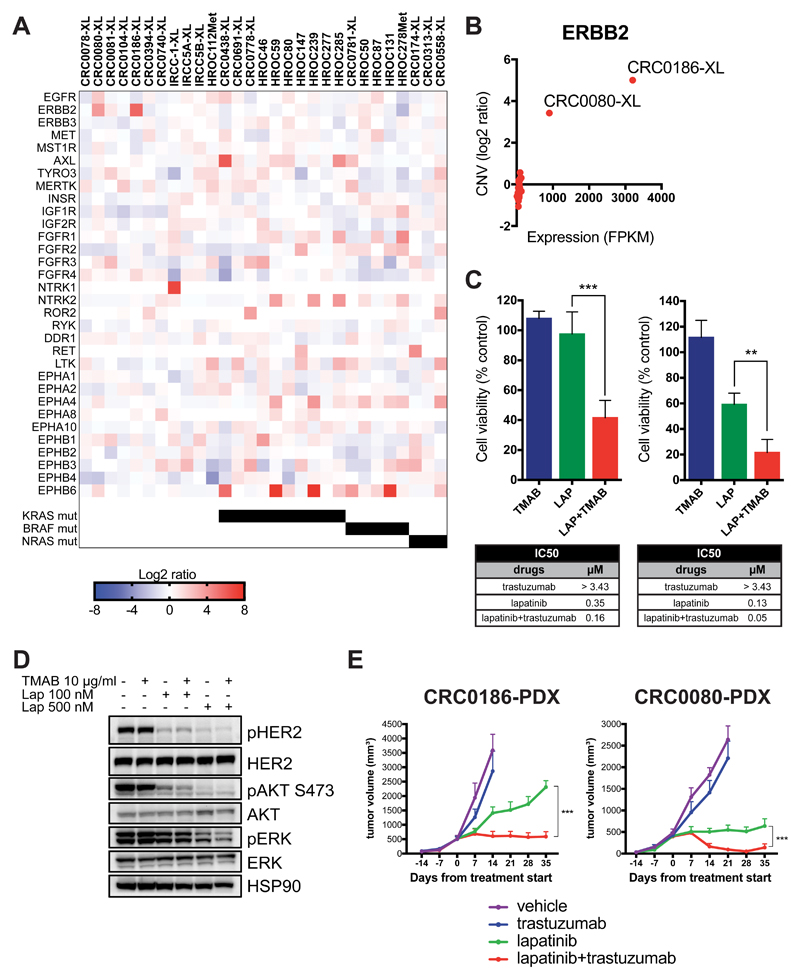
Figure 4. Identification of overexpressed receptor tyrosine kinases (RTKs) in CRC XLs and effects of HER blockade in ErBB2-amplified cell models.
A, Heat map showing the relative expression (log2 ratio FPKM) of receptor tyrosine kinases (RTKs) as identified by RNA-seq analysis of 29 XLs. RTKs listed here (n=32) represent the only members of this class of protein passing the filtration process during RNA-seq analysis. KRAS, BRAF and NRAS mutational status is also indicated. B, Scatter plot displaying the correlation between absolute ERBB2 expression (FPKM, x-axis) and relative ERBB2 copy-number variation (CNV, log2ratio) in the 29 XLs, as assessed by RNA-seq and WES analyses, respectively. ERBB2-overexpressing and amplified XLs (CRC0080-XL and CRC01086-XL) are indicated. C, Short-term viability assay showing the effects of single dose of trastuzumab (TMAB, 25 μg/mL), lapatinib (LAP, 120 nM), and combination of lapatinib and trastuzumab (LAP+TMAB) on ERBB2-amplified CRC0186-XL (left panel) and CRC0080-XL (right panel). Viability was assessed by measuring ATP content after 6 days of treatment. Data represent the values obtained from at least three independent experiments in triplicate, normalized on control DMSO-treated cells; error bars indicate SD (lapatinib vs lapatinib+trastuzumab in CRC0186-XL: ***, P = 0.001; lapatinib vs lapatinib+trastuzumab: **, P = 0.0036 in CRC0080-XL; unpaired Student’s t test). IC50 values were calculated from dose inhibition curves using GraphPad Prism software. D, Activation status/phosphorylation of HER2 receptor and downstream pathway in dose-response experiments in CRC0186-XL. Cells were treated with the indicated concentrations of trastuzumab (TMAB), lapatinib (Lap), or the combination of both drugs for 4 hours. Cell extracts were immunoblotted with the indicated antibodies. HSP90 was used as loading control. E, In vivo tumor growth curves of CRC0186-PDX (n=6) and CRC0080-PDX (n=6) treated with the indicated modalities. Data represent the mean of tumor volume at each time point; error bars indicate S.E.M. (lapatinib vs lapatinib+trastuzumab: ***, P < 0.001 by two-way Anova Test). Response curves for both models were previously reported in (49) and are reshown here for comparative purposes.
Among these, only two RTKs were overexpressed in RAS/RAF wild-type cells (“WT-specific” RTKs), namely NTRK1 (TRKA) and ERBB2 (HER2). Integrated analysis showed that the observed overexpression is associated with different molecular alterations, namely gene translocation (NTRK1) or gene amplification (ERBB2) (Fig. 3A, 4B and Supplementary Fig. S6F, S7A and S7B).
In the TRKA overexpressed case, we identified a genetic rearrangement involving exon 10 or 11 of LMNA and exon 11 of NTRK1 genes (47). The two distinct splice variants encoding exons 1-10 or 1-11 of LMNA gene fused to exons 11-16 of NTRK1 gene reported in the donor patient were identified by RNA-seq analysis in the matched XL (Supplementary Fig. S6F). Remarkably, the pharmacological response of these xeno-cells resembled the one observed in the corresponding PDX upon entrectinib treatment (47).
In order to validate ERBB2 amplification as an actionable target, we first evaluated biochemical pathway activation. In contrast to CRC0186-XL, CRC0080-XL showed lower level of HER2 protein, but still an active downstream pathway due to the presence of the L866M mutation (Supplementary Fig. S7C).
This analysis confirmed that overexpression and constitutive signaling, as well as downstream pathway activation of HER2, paralleled the extension of ERBB2 gene amplification.
To validate overexpressed ERBB2 as putative target for pharmacological inhibition (14), we challenged our in vitro models with rational combinations of drugs. We initially tested the effect of drugs (lapatinib and trastuzumab) that have previously shown activity on HER2 positive metastatic CRC patients (14,48). While monotherapy showed no or mild effects, both lines (CRC0080-XL and CRC0186-XL) displayed significant sensitivity to the combinatorial treatment (Fig. 4C) in short-term proliferation assays. We performed western blot analysis showing that the biological effect of the drugs on ERBB2-amplified CRC cell lines is matched by the quenching of HER2 biochemical downstream signals (Fig. 4D). Of note, similar results were also obtained in vivo, when the corresponding ERBB2-amplified xenopatients were challenged with the same treatments for 28 - 35 days (49) (Fig. 4E), thus confirming the XL as a valuable preclinical model to assess drug response and to dissect biochemical pathway activation.
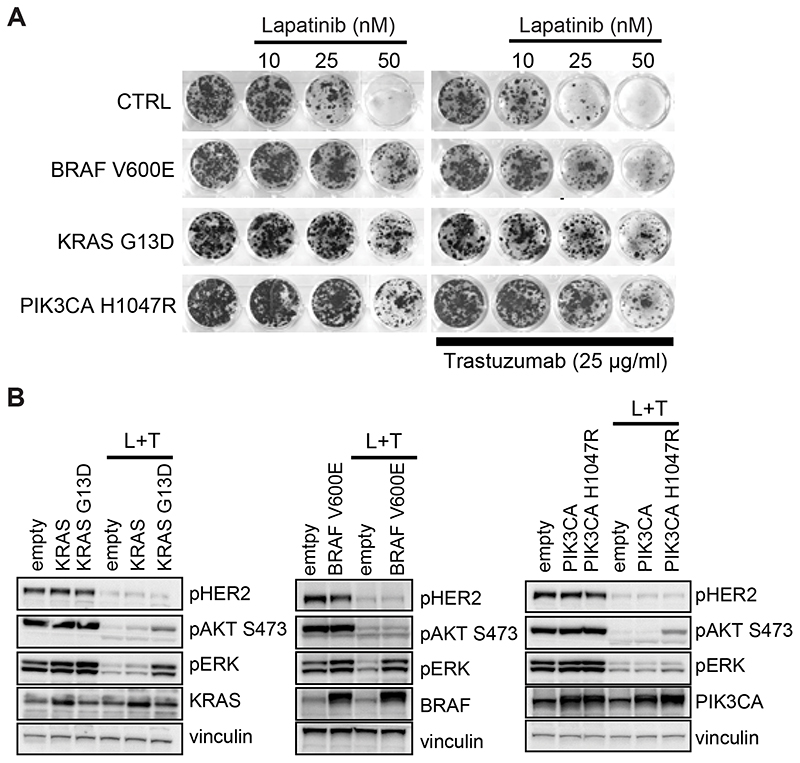
Combinatorial treatment with lapatinib and trastuzumab of HER2+ patients has been exploited in the recently completed proof-of-concept clinical trial named HERACLES (48). Our previous work based on liquid biopsy analysis has shown that 32 out of 35 patients treated with anti-HER2 inhibitors relapsed after initial response revealing the presence of molecular mechanisms potentially responsible for resistance (primary or acquired) to HER2 blockade (50). In particular, either KRAS or BRAF mutated clones were preferentially found in patients who were initially refractory to anti-HER2 treatment (primary resistance), while PIK3CA mutations were detected at progression in patients who had responded to lapatinib+trastuzumab combinatorial treatment and then relapsed (acquired resistance). In order to validate these genetic players as putative drivers of resistance, we transduced HER2+ CRC XL-cells by means of lentiviral vectors and performed pharmacological and biochemical analysis of treated cells. We found that overexpression of mutant alleles of KRAS, BRAF and PIK3CA conferred resistance to combinatorial treatment (Fig. 5A and Supplementary Fig. S7D), due to sustained ERK and/or AKT activation (Fig. 5B and Supplementary Fig. S7E). Consistent with this data, the combination of lapatinib and trastuzumab was ineffective in lines lacking ERBB2 amplification and naturally bearing RAS/BRAF/PIK3CA mutations (Supplementary Fig. S8).
Figure 5. Constitutive expression of mutant RAS/BRAF/PIK3CA mediates resistance to lapatinib+trastuzumab combinatorial treatment in ErBB2-amplified cells.
A, Long-term viability assay showing the effect of different indicated doses of lapatinib in absence or in presence of doses of 25 μg/ml of trastuzumab on ERBB2-amplified CRC0186-XL transduced with lentiviral vector overexpressing BRAF V600E, KRAS G13D or PIK3CA H1047R mutants. Cells transduced with empty lentiviral vector were used as control (CTRL). Cell viability was assessed by crystal violet staining after 15 days of treatment. Representative wells were photographed and reported. B, Activation status/phosphorylation of HER2 receptor and downstream pathway in CRC0186-XL transduced with lentiviral vector overexpressing the indicated alleles. Cells transduced with empty lentiviral vector were used as control. Cells were treated with the combination of 500 nM of lapatinib and 10 μg/ml of trastuzumab (L+T) for 4 hours. Cell extracts were immunoblotted with the indicated antibodies. Vinculin was used as loading control.
WRN is a target in MSI and in selected MSI-like CRC XL cells
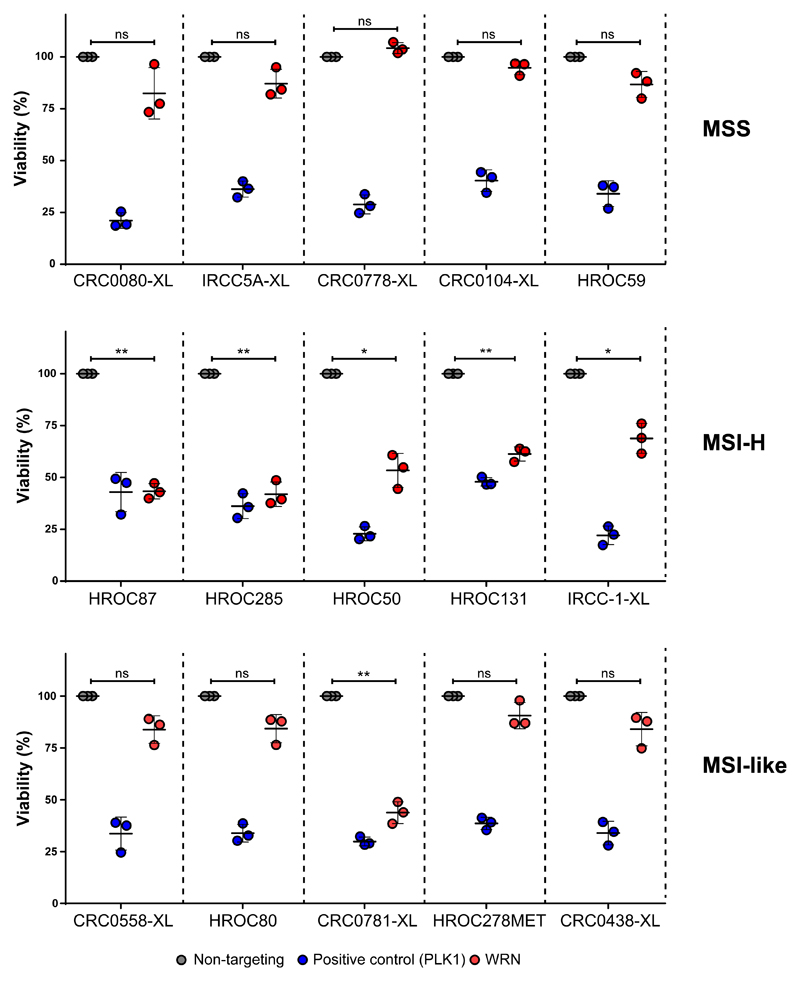
We then exploited our XL-cell models to explore novel dependencies in CRC. We focused on the WRN gene which encodes a RecQ helicase involved in Werner syndrome. MSI-H CRCs were recently found to have a synthetic-lethal dependency on the WRN gene for cell fitness and viability, while MSS CRCs were not dependent on WRN expression (51–54). It is presently unknown whether MSI-like CRC cells, which are genetically MSS but are transcriptionally defined as MSI-H (55), also rely on WRN gene expression for their survival.
To address this issue, we selected a subgroup of 15 XLs, including 5 MSI-H (HROC50, IRCC-1-XL, HROC87, HROC285, HROC131), 5 MSS with predicted MSI-like features (CRC0438-XL, CRC0781-XL, HROC80, CRC0558-XL and HROC278Met) and 5 MSS (CRC0080-XL, IRCC-5A-XL, HROC59, CRC0778-XL, CRC0104-XL), recapitulating the CMS and CRIS transcriptional subgroups.
The MSI-like phenotype of the selected PDX-derived cell lines were defined as follows: i) classification into the CMS1 subgroup (CRC0438-XL, CRC0781-XL, HROC80) or ii) classification through the algorithm described by Tian and colleagues (55) (CRC0558-XL and HROC278Met).
The panel of 15 XL-cell lines was subjected to siRNA-mediated WRN knockdown to assess their dependency on WRN. Effective targeting of WRN was verified by Western blot (Supplementary Figure S9A).
As expected, WRN depletion selectively reduced the fitness of all 5 MSI-H cells, whereas none of the MSS models were sensitive (Figure 6). Four of the MSI-like XLs were insensitive to WRN knockdown; notably however one model (CRC0781-XL) was highly sensitive to WRN depletion. To confirm that CRC0781-XL was indeed MSS we repeated the microsatellite and MMR protein analysis, which confirmed that this model is MSS both genetically and biochemically (Supplementary Figure S9B, C).
Figure 6. siRNA-mediated WRN depletion assay in 15 selected XLs.
Dot plots represent the normalized viability data obtained upon siRNA-mediated WRN depletion (red dots) in 5 MSS (upper panel), 5 MSI (middle panel) and 5 MSI-like phenotypes (lower panel). Non-targeting (grey dots) and PLK1 (blue dots) siRNAs were used as negative and positive controls, respectively. Dots represent mean ±SD of 3 independent experiments with 5 technical replicates each. Statistical significance was calculated comparing WRN siRNA vs non-targeting siRNA: ns, not significant; *p< 0.05; **p< 0.01; ***p< 0.001 (Welch’s t test).
4. Discussion
Human cancer cell lines represent a tool extensively exploited in the last half century to study the biology of different tumor types and for extensive drug screening efforts. Exceptional efforts, such as the generation of the Cancer Cell Line Encyclopedia (7), have been pivotal for unveiling novel genomic predictors of drug sensitivities in 36 tumor types, thus coupling cell line genomic annotation to individual pharmacological profiles.
We have previously described a bowel cancer cell-line collection including 151 established cell lines (56). This platform can recapitulate the majority of CRC molecular and transcriptional subtypes previously defined in patients. Genomic analysis has allowed for the identification of novel outliers in response to selected targeted agents. Yet infrequent molecular subtypes or tumors carrying rare genetic lesions may be under-represented or completely absent in the above mentioned CRC cell lines collection. For instance, while ERRB2-amplified cell line models have been widely employed for breast and gastric cancer studies (57,58), none of the models in the 151-CRC cell collection were found to carry this alteration. In this work, we provide an accurate characterization of two novel cancer cell lines of colorectal origin bearing ERRB2 amplification – which could be used in the future to further dissect the role of ERBB2 oncogenic signaling in the context of gut pathophysiology. Testing drug-genotype correlations in the pertinent tissue is necessary since the same molecular alteration could result in dissimilar responses to the same drug, as exemplified by the discrepant anti-proliferative effects of BRAF inhibitors in BRAF mutant melanoma vs colorectal cancer (59).
Cell lines may have intrinsic limitations that hamper representation of the whole tumor disease in the patient. Common immortalized cell lines have been adapted to growing on plastic for decades and this may have caused genetic drifts and the acquisition of phenotypic features different from original cancers in patients. It is now evident that CRC is indeed a heterogeneous disease and that CRC development and progression relies on continuous evolution of multiple clones under environmental and pharmacological pressure (60). For this reason, precision medicine, which is based on the design of personalized treatments, requires a repertoire of preclinical models that mirror the in vivo tumor more closely than conventional cancer cell lines. Derivation of long-term culture directly from tumor samples has proven to be difficult and inefficient, at least in CRC, not only because of contaminations but also due to failure to adhere to the culture dish or loss of proliferative capacity after few passages (61). Establishment of short-term tumor cell culture has proven to be successful (62), but long-term experiments as well as gene editing by lentiviral transduction for experimental needs might not be feasible.
Establishment of 3D cultures (organoids, tumoroids, spheroids, etc.) has provided significant advancements for the identification of personalized therapeutic options by integrating detailed genomic information with high-throughput pharmacological screenings for patients with advanced disease (63–67). These types of cultures have been able to provide important correlations between tumor genomics and response to chemotherapy or targeted drugs within short intervals from their derivation. One of the main limitations for organoid generation is due to the necessity of acquiring a significant amount of fresh tissue with viable tumor cells, and is often not possible when only very small tissue biopsies are available. In our work we have chosen to use small amounts of available tissue material to generate PDXs in order to expand the quantity of tumor tissue for subsequent preclinical model derivation for genomic and drug-response studies.
Generation of PDX models has contributed to the identification and validation of biomarkers of response and resistance to current anticancer therapies (18). However, this model presents some important shortcomings such as long time for establishment and expansion, cumbersome manipulation and cost-ineffective experiments, especially if used on a large scale.
Here we present a complementary approach based on PDX-derived cell lines (XLs). We have established 29 cell lines from PDXs originally derived from samples of primary or metastatic CRC tissue. Initially, to assess whether passage from 3D/in vivo to 2D/in vitro growing condition did not select for genomic alterations that may compromise the biology of the tumor, we performed genetic and transcriptomic analyses to compare matched samples. Whole exome sequencing confirmed preservation of the PDX main genetic features in 2D-derived cell lines. High concordance in genetic variation was evident between the two platforms when considering cancer-related genes, although we identified a variable range of private variants in each PDX-XL pair. Although the majority of private alterations was found in MSI-H models, likely due to their hypermutator phenotype (68), a small number of private mutations in CRC-related genes were also identified at subclonal levels in MSS PDX or XL models. This variation could either reflect the enrichment/depletion of a subclonal population or the acquisition of additional mutations during derivation or propagation of the XL-cells with respect to the matched xenopatient. Our results are in line with recent findings unveiled by barcoding experiments showing that cell line evolution may occur as a result of positive clonal selection under culture conditions (12). Few shared variants were also identified at subclonal levels in the MSS models, thus fueling heterogeneity in these CRC models. This highlights PDXs and matched XLs as unique preclinical tools to investigate in vitro and in vivo clonal evolution in response to therapies in CRC.
Although a certain degree of drifting may occur, gene expression profiles of matched XLs and PDX models were highly correlated, suggesting that in vitro propagation may have a minimal impact on transcriptional traits. The molecular classification of cell lines based on transcriptional traits did reproduce the pattern observed in the original tumor, and conserved the distinct genetic, molecular and pharmacological characteristics. Indeed, despite the small size of our dataset, we could confirm the association between CRIS-C assignment and higher probability of response to anti-EGFR treatment, as well as the association between CRIS-A and MSI status (27).
Of note, comparing the classification of CMS and CRIS in the context of cell models we unveiled correspondence in classification of CMS4 and CRIS-D, likely emerging due to absence of stromal cells in the cancer cells profiles and similarities in intrinsic traits, such as the LGR5 stem-like signature detected in both subtypes (28,69).
We next assessed whether these preclinical CRC models could represent a reliable tool for biomarker validation and pharmacological screening. The identification of genomic players that may confer sensitivities or resistance to selected therapies is of paramount importance for the design of effective therapeutic strategies. As a proof of concept, we have shown two independent examples in which the results observed with the XLs highly mirrored what was previously observed in PDXs. We focused our attention on anti-HER therapies, which are approved or used as investigational agents for patients with RAS wild-type and HER2-overexpressing CRC tumors. When we challenged XLs with the anti-EGFR monoclonal antibody cetuximab, we observed a distribution of responses that closely correlated with genetic and non-genetic biomarkers of resistance identified in the corresponding PDXs (18).
As a second paradigmatic example, we have identified in our cohort two XLs harboring HER2 receptor overexpression due to ERBB2 gene amplification. When these cells where challenged with a rational combination of HER inhibitors, we observed that cell proliferation was severely impaired, as previously seen with the matched PDXs treated with the same drugs. This further confirms that the XL platform is a valuable tool for genotype-drug correlation studies. Moreover, we have validated molecular determinants of resistance to dual HER2 blockade in CRC, as emerged from the HERACLES trial (50), suggesting that combinatorial treatment with other targeting agents might be beneficial for HER2+ patients (49).
We have exploited our XL collection to assess if WRN dependency, which has been recently reported as a new synthetic lethality trait in MSI-H cancers (51–54), could also apply to MSI-like colorectal tumors. MSI-like CRC models are genetically MSS, but bear intrinsic transcriptional features that resemble MSI-H cancers (55). MSI-like CRC have limited therapeutic options and are presently excluded from immune based regimens but might benefit from alternative therapy regimens. We therefore hypothesized that they could be reliant upon the WRN gene. By means of a siRNA-mediated WRN depletion assay, we discovered that MSI-like CRCs are generally not dependent on WRN downregulation. Interestingly however we found one MSI-like XL cell line (CRC0781-XL) sensitive to WRN suppression. Future analysis should be performed to uncover the mechanistic basis of this findings using larger cohort of MSI-like CRC models.
In summary, we have established a compendium of 29 XLs that substantially recapitulate the genomic features of matched PDXs with the great advantage of providing ease of handling and amenability for high-throughput compound screening. This may significantly accelerate the translation of drug-response information from in vitro testing to clinical practice.
Another notable advantage provided by this approach is the possibility of generating cell lines with genotypes that are either missing or rare in common commercial repositories (i.e. ERBB2-amplified or NRAS Q61-mutated cell lines), as well as increasing the number of MSS CRC cell lines, which are typically underrepresented in existing cell line collections (31,56). We proposed that generation of a large cohort of XLs will provide a novel tool to help researchers better understand and model the molecular mechanisms that contribute to CRC heterogeneity. In addition, we believe that our XL collection offers significant advantages with respect to commercially available cells for several reasons. First, the PDX-XL pairs provide a new strategy for pharmacogenomics studies: XLs could be exploited as an in vitro surrogate of matched PDXs in biomarker-based preclinical drug screening studies, and the selected drug-target can be further validated in the corresponding PDX model. Second, clinical and pharmacological will be available for the majority of the patients, thus making this cell line collection a unique annotated tool to perform screenings with other agents of interest in the context of chemo-resistance/sensitivity. Moreover, the subclonal architecture identified in PDXs and XLs provides opportunities to investigate how heterogeneity and evolution affect response to therapeutic agents in CRC models closely paralleling the clinical setting. Third, for all patients from whom matched normal genomic DNA is provided, comparison between normal and XL samples will enable the confident identification of somatic alterations. Fourth, some of the patient models have matched PBMCs and tumor infiltrating lymphocytes, thus rendering this XL-collection a unique resource to perform co-culture assays or to study genotype-drug correlations that may be dependent upon the immune microenvironment.
In conclusion, we present a novel preclinical platform that – owing to its reliability, large-scale use and proximity to original patient tumors – could offer easier and more rapid access to the dissection of genetic and patient-selective therapeutic vulnerabilities, thus leading to the design of novel effective treatment strategies.
Supplementary Material
Translational Relevance.
Progress in the development of effective cancer treatments is limited by the availability of tumor models. For decades, established cancer cell lines represented the mainstay preclinical tumor model, but these tools have limitations, such as limited capacity to recapitulate inter- and intra-tumor heterogeneity, adaptation to grow in two-dimensional cultures and the lack of interaction with the microenvironment. To overcome these restraints, patient-derived tumor xenografts (PDXs) have been developed in recent years. Although this model mirrors histological and molecular features of the patient’s tumor, PDXs also have limitations including maintenance costs and unsuitability for large-scale screenings. Here we describe a novel platform of PDX-derived cell lines (xeno-cell lines, XLs) that retain the pharmacogenomic profile of the sample of origin and offer advantages such as reduced working costs and ease of handling. In conclusion, we provide a preclinical model resource for large-scale functional gene validation and assessment of novel therapeutic strategies in colorectal cancer.
Acknowledgments
The authors thank all the members of Molecular Oncology Laboratory at Candiolo Cancer Institute for critically reading the manuscript and, in particular, Mariangela Russo, Giulia Siravegna, Benedetta Mussolin, Francesca Lodi, Riccardo Volta, and Gaia Grasso for their help with experiments and constructive discussion, Daniela Cantarella from the Oncogenomics laboratory for technical assistance in RNAseq data generation, Francesco Galimi from the Molecular Pharmacology laboratory for providing RNA from PDXs, and Esmée van Vliet from M.J.G.’s lab for assistance with WRN dependency experiments. The authors also thank Dr. Paolo Luraghi for providing CRC0080-XL and for his helpful discussion on cell line derivation. The research leading to these results has received funding from AIRC under MFAG 2017 -ID 20236 project -P.I. Arena Sabrina; the research leading to these results has received funding from Fondazione AIRC under 5 per Mille 2018 - ID. 21091 program - P.I. Bardelli Alberto, G.L. (group leader) Medico Enzo, G.L. Siena Salvatore, G.L. Bertotti Andrea, G.L Trusolino Livio, G.L. Di Nicolantonio Federica. This work was also supported by European Community’s Seventh Framework Programme under grant agreement no. 602901 MErCuRIC(to A. Bardelli); H2020 grant agreement no. 635342-2 MoTriColor (to A. Bardelli and S. Siena); IMI contract no. 115749 CANCER-ID (to A. Bardelli); AIRC IG 2018 - ID. 21923 project – principal investigator A. Bardelli; AIRC IG no. 17707 and IG no. 21407 (to F. Di Nicolantonio); AIRC IG no. 20685 (to S. Siena); Terapia Molecolare Tumori by Fondazione Oncologia Niguarda Onlus (to A. Sartore-Bianchi and S. Siena); Genomic-Based Triage for Target Therapy in Colorectal Cancer Ministero della Salute, Project no. NET 02352137 (to A. Sartore-Bianchi, A. Bardelli, and S. Siena). TRANSCAN-2 JTC 2014 contract no. TRS-2015-00000060 INTRACOLOR (to S. Arena); TRANSCAN-2 JTC 2014 TACTIC (to L. Trusolino); European Research Council Consolidator Grant 724748 - BEAT (to A. Bertotti); AIRC IG n. 18532 (to L. Trusolino); AIRC IG n. 20697 (to A. Bertotti); AIRC-CRUK-FC AECC Accelerator Award contract 22795 (to A. Bardelli); Fondazione Piemontese per la Ricerca sul Cancro-ONLUS 5 per mille 2014 e 2015 Ministero della Salute (to A. Bardelli and E. Medico). Work in M.J. Garnett lab is supported by the Wellcome Trust (grant no: 206194).
Footnotes
Conflicts of Interest: A.B. is an advisory board member for Roche; A.S.B. is an advisory board member for Amgen, Bayer and Sanofi; SS. Siena is a consultant/advisory board member for Amgen, Bayer, BMS, CheckmAb, Celgene, Clovis, Daiichi-Sankyo, Incyte, Merck, Novartis, Roche-Genentech, and Seattle Genetics; potential conflicts of interest were disclosed by the other authors.
References
- 1.Kelloff GJ, Sigman CC. Cancer biomarkers: selecting the right drug for the right patient. Nat Rev Drug Discov. 2012;11(3):201–14. doi: 10.1038/nrd3651. [DOI] [PubMed] [Google Scholar]
- 2.Network TCGA. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487(7407):330–7. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Friedman AA, Letai A, Fisher DE, Flaherty KT. Precision medicine for cancer with next-generation functional diagnostics. Nat Rev Cancer. 2015;15(12):747–56. doi: 10.1038/nrc4015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weinstein IB, Joe A. Oncogene addiction. Cancer Res. 2008;68(9):3077–80. doi: 10.1158/0008-5472.CAN-07-3293. discussion 80. [DOI] [PubMed] [Google Scholar]
- 5.Bruna A, Rueda OM, Greenwood W, Batra AS, Callari M, Batra RN, et al. A Biobank of Breast Cancer Explants with Preserved Intra-tumor Heterogeneity to Screen Anticancer Compounds. Cell. 2016;167(1):260–74.:e22. doi: 10.1016/j.cell.2016.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang W, Soares J, Greninger P, Edelman EJ, Lightfoot H, Forbes S, et al. Genomics of Drug Sensitivity in Cancer (GDSC): a resource for therapeutic biomarker discovery in cancer cells. Nucleic Acids Res. 2013;41(Database issue):D955–61. doi: 10.1093/nar/gks1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483(7391):603–7. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garnett MJ, Edelman EJ, Heidorn SJ, Greenman CD, Dastur A, Lau KW, et al. Systematic identification of genomic markers of drug sensitivity in cancer cells. Nature. 2012;483(7391):570–5. doi: 10.1038/nature11005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Seashore-Ludlow B, Rees MG, Cheah JH, Cokol M, Price EV, Coletti ME, et al. Harnessing Connectivity in a Large-Scale Small-Molecule Sensitivity Dataset. Cancer Discov. 2015;5(11):1210–23. doi: 10.1158/2159-8290.CD-15-0235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ghandi M, Huang FW, Jané-Valbuena J, Kryukov GV, Lo CC, McDonald ER, et al. Next-generation characterization of the Cancer Cell Line Encyclopedia. Nature. 2019;569(7757):503–8. doi: 10.1038/s41586-019-1186-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gillet JP, Varma S, Gottesman MM. The clinical relevance of cancer cell lines. J Natl Cancer Inst. 2013;105(7):452–8. doi: 10.1093/jnci/djt007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ben-David U, Siranosian B, Ha G, Tang H, Oren Y, Hinohara K, et al. Genetic and transcriptional evolution alters cancer cell line drug response. Nature. 2018;560(7718):325–30. doi: 10.1038/s41586-018-0409-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Byrne AT, Alférez DG, Amant F, Annibali D, Arribas J, Biankin AV, et al. Interrogating open issues in cancer precision medicine with patient-derived xenografts. Nat Rev Cancer. 2017;17(4):254–68. doi: 10.1038/nrc.2016.140. [DOI] [PubMed] [Google Scholar]
- 14.Bertotti A, Migliardi G, Galimi F, Sassi F, Torti D, Isella C, et al. A molecularly annotated platform of patient-derived xenografts (“xenopatients”) identifies HER2 as an effective therapeutic target in cetuximab-resistant colorectal cancer. Cancer Discov. 2011;1(6):508–23. doi: 10.1158/2159-8290.CD-11-0109. [DOI] [PubMed] [Google Scholar]
- 15.Julien S, Merino-Trigo A, Lacroix L, Pocard M, Goéré D, Mariani P, et al. Characterization of a large panel of patient-derived tumor xenografts representing the clinical heterogeneity of human colorectal cancer. Clin Cancer Res. 2012;18(19):5314–28. doi: 10.1158/1078-0432.CCR-12-0372. [DOI] [PubMed] [Google Scholar]
- 16.Galimi F, Torti D, Sassi F, Isella C, Corà D, Gastaldi S, et al. Genetic and expression analysis of MET, MACC1, and HGF in metastatic colorectal cancer: response to met inhibition in patient xenografts and pathologic correlations. Clin Cancer Res. 2011;17(10):3146–56. doi: 10.1158/1078-0432.CCR-10-3377. [DOI] [PubMed] [Google Scholar]
- 17.Ben-David U, Ha G, Tseng YY, Greenwald NF, Oh C, Shih J, et al. Patient-derived xenografts undergo mouse-specific tumor evolution. Nat Genet. 2017;49(11):1567–75. doi: 10.1038/ng.3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bertotti A, Papp E, Jones S, Adleff V, Anagnostou V, Lupo B, et al. The genomic landscape of response to EGFR blockade in colorectal cancer. Nature. 2015;526(7572):263–7. doi: 10.1038/nature14969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Damhofer H, Ebbing EA, Steins A, Welling L, Tol JA, Krishnadath KK, et al. Establishment of patient-derived xenograft models and cell lines for malignancies of the upper gastrointestinal tract. J Transl Med. 2015;13:115. doi: 10.1186/s12967-015-0469-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Knudsen ES, Balaji U, Mannakee B, Vail P, Eslinger C, Moxom C, et al. Pancreatic cancer cell lines as patient-derived avatars: genetic characterisation and functional utility. Gut. 2017 doi: 10.1136/gutjnl-2016-313133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Luraghi P, Bigatto V, Cipriano E, Reato G, Orzan F, Sassi F, et al. A molecularly annotated model of patient-derived colon cancer stem-like cells to assess genetic and non-genetic mechanisms of resistance to anti-EGFR therapy. Clin Cancer Res. 2017 doi: 10.1158/1078-0432.CCR-17-2151. [DOI] [PubMed] [Google Scholar]
- 22.Maletzki C, Stier S, Gruenert U, Gock M, Ostwald C, Prall F, et al. Establishment, characterization and chemosensitivity of three mismatch repair deficient cell lines from sporadic and inherited colorectal carcinomas. PLoS One. 2012;7(12):e52485. doi: 10.1371/journal.pone.0052485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Niu B, Ye K, Zhang Q, Lu C, Xie M, McLellan MD, et al. MSIsensor: microsatellite instability detection using paired tumor-normal sequence data. Bioinformatics. 2014;30(7):1015–6. doi: 10.1093/bioinformatics/btt755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Corti G, Bartolini A, Crisafulli G, Novara L, Rospo G, Montone M, et al. A Genomic Analysis Workflow for Colorectal Cancer Precision Oncology. Clin Colorectal Cancer. 2019;18(2):91–101.:e3. doi: 10.1016/j.clcc.2019.02.008. [DOI] [PubMed] [Google Scholar]
- 25.Wang K, Singh D, Zeng Z, Coleman SJ, Huang Y, Savich GL, et al. MapSplice: accurate mapping of RNA-seq reads for splice junction discovery. Nucleic Acids Res. 2010;38(18):e178. doi: 10.1093/nar/gkq622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Isella C, Brundu F, Bellomo SE, Galimi F, Zanella E, Porporato R, et al. Selective analysis of cancer-cell intrinsic transcriptional traits defines novel clinically relevant subtypes of colorectal cancer. Nat Commun. 2017;8:15107. doi: 10.1038/ncomms15107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sveen A, Bruun J, Eide PW, Eilertsen IA, Ramirez L, Murumägi A, et al. Colorectal Cancer Consensus Molecular Subtypes Translated to Preclinical Models Uncover Potentially Targetable Cancer Cell Dependencies. Clin Cancer Res. 2018;24(4):794–806. doi: 10.1158/1078-0432.CCR-17-1234. [DOI] [PubMed] [Google Scholar]
- 29.De Roock W, Claes B, Bernasconi D, De Schutter J, Biesmans B, Fountzilas G, et al. Effects of KRAS, BRAF, NRAS, and PIK3CA mutations on the efficacy of cetuximab plus chemotherapy in chemotherapy-refractory metastatic colorectal cancer: a retrospective consortium analysis. Lancet Oncol. 2010;11(8):753–62. doi: 10.1016/S1470-2045(10)70130-3. [DOI] [PubMed] [Google Scholar]
- 30.Di Nicolantonio F, Arena S, Gallicchio M, Zecchin D, Martini M, Flonta SE, et al. Replacement of normal with mutant alleles in the genome of normal human cells unveils mutation-specific drug responses. Proc Natl Acad Sci U S A. 2008;105(52):20864–9. doi: 10.1073/pnas.0808757105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mouradov D, Sloggett C, Jorissen RN, Love CG, Li S, Burgess AW, et al. Colorectal cancer cell lines are representative models of the main molecular subtypes of primary cancer. Cancer Res. 2014;74(12):3238–47. doi: 10.1158/0008-5472.CAN-14-0013. [DOI] [PubMed] [Google Scholar]
- 32.Futreal PA, Coin L, Marshall M, Down T, Hubbard T, Wooster R, et al. A census of human cancer genes. Nat Rev Cancer. 2004;4(3):177–83. doi: 10.1038/nrc1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lawrence MS, Stojanov P, Mermel CH, Robinson JT, Garraway LA, Golub TR, et al. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature. 2014;505(7484):495–501. doi: 10.1038/nature12912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV, et al. Signatures of mutational processes in human cancer. Nature. 2013;500(7463):415–21. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Giannakis M, Hodis E, Jasmine Mu X, Yamauchi M, Rosenbluh J, Cibulskis K, et al. RNF43 is frequently mutated in colorectal and endometrial cancers. Nat Genet. 2014;46(12):1264–6. doi: 10.1038/ng.3127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Markowitz S, Wang J, Myeroff L, Parsons R, Sun L, Lutterbaugh J, et al. Inactivation of the type II TGF-beta receptor in colon cancer cells with microsatellite instability. Science. 1995;268(5215):1336–8. doi: 10.1126/science.7761852. [DOI] [PubMed] [Google Scholar]
- 37.Rodríguez-Escudero I, Oliver MD, Andrés-Pons A, Molina M, Cid VJ, Pulido R. A comprehensive functional analysis of PTEN mutations: implications in tumor- and autism-related syndromes. Hum Mol Genet. 2011;20(21):4132–42. doi: 10.1093/hmg/ddr337. [DOI] [PubMed] [Google Scholar]
- 38.Han SY, Kato H, Kato S, Suzuki T, Shibata H, Ishii S, et al. Functional evaluation of PTEN missense mutations using in vitro phosphoinositide phosphatase assay. Cancer Res. 2000;60(12):3147–51. [PubMed] [Google Scholar]
- 39.Zanella ER, Galimi F, Sassi F, Migliardi G, Cottino F, Leto SM, et al. IGF2 is an actionable target that identifies a distinct subpopulation of colorectal cancer patients with marginal response to anti-EGFR therapies. Sci Transl Med. 2015;7(272):272ra12. doi: 10.1126/scitranslmed.3010445. [DOI] [PubMed] [Google Scholar]
- 40.Arena S, Siravegna G, Mussolin B, Kearns JD, Wolf BB, Misale S, et al. MM-151 overcomes acquired resistance to cetuximab and panitumumab in colorectal cancers harboring EGFR extracellular domain mutations. Sci Transl Med. 2016;8(324):324ra14. doi: 10.1126/scitranslmed.aad5640. [DOI] [PubMed] [Google Scholar]
- 41.Arena S, Bellosillo B, Siravegna G, Martínez A, Cañadas I, Lazzari L, et al. Emergence of Multiple EGFR Extracellular Mutations during Cetuximab Treatment in Colorectal Cancer. Clin Cancer Res. 2015;21(9):2157–66. doi: 10.1158/1078-0432.CCR-14-2821. [DOI] [PubMed] [Google Scholar]
- 42.Kavuri SM, Jain N, Galimi F, Cottino F, Leto SM, Migliardi G, et al. HER2 activating mutations are targets for colorectal cancer treatment. Cancer Discov. 2015;5(8):832–41. doi: 10.1158/2159-8290.CD-14-1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Misale S, Di Nicolantonio F, Sartore-Bianchi A, Siena S, Bardelli A. Resistance to anti-EGFR therapy in colorectal cancer: from heterogeneity to convergent evolution. Cancer Discov. 2014;4(11):1269–80. doi: 10.1158/2159-8290.CD-14-0462. [DOI] [PubMed] [Google Scholar]
- 44.Siravegna G, Mussolin B, Buscarino M, Corti G, Cassingena A, Crisafulli G, et al. Clonal evolution and resistance to EGFR blockade in the blood of colorectal cancer patients. Nat Med. 2015;21(7):795–801. doi: 10.1038/nm.3870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Russo M, Siravegna G, Blaszkowsky LS, Corti G, Crisafulli G, Ahronian LG, et al. Tumor Heterogeneity and Lesion-Specific Response to Targeted Therapy in Colorectal Cancer. Cancer Discov. 2016;6(2):147–53. doi: 10.1158/2159-8290.CD-15-1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Guinney J, Dienstmann R, Wang X, de Reyniès A, Schlicker A, Soneson C, et al. The consensus molecular subtypes of colorectal cancer. Nat Med. 2015;21(11):1350–6. doi: 10.1038/nm.3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Russo M, Misale S, Wei G, Siravegna G, Crisafulli G, Lazzari L, et al. Acquired Resistance to the TRK Inhibitor Entrectinib in Colorectal Cancer. Cancer Discov. 2016;6(1):36–44. doi: 10.1158/2159-8290.CD-15-0940. [DOI] [PubMed] [Google Scholar]
- 48.Sartore-Bianchi A, Trusolino L, Martino C, Bencardino K, Lonardi S, Bergamo F, et al. Dual-targeted therapy with trastuzumab and lapatinib in treatment-refractory, KRAS codon 12/13 wild-type, HER2-positive metastatic colorectal cancer (HERACLES): a proof-of-concept, multicentre, open-label, phase 2 trial. Lancet Oncol. 2016;17(6):738–46. doi: 10.1016/S1470-2045(16)00150-9. [DOI] [PubMed] [Google Scholar]
- 49.Leto SM, Sassi F, Catalano I, Torri V, Migliardi G, Zanella ER, et al. Sustained Inhibition of HER3 and EGFR Is Necessary to Induce Regression of HER2-Amplified Gastrointestinal Carcinomas. Clin Cancer Res. 2015;21(24):5519–31. doi: 10.1158/1078-0432.CCR-14-3066. [DOI] [PubMed] [Google Scholar]
- 50.Siravegna G, Lazzari L, Crisafulli G, Sartore-Bianchi A, Mussolin B, Cassingena A, et al. Radiologic and Genomic Evolution of Individual Metastases during HER2 Blockade in Colorectal Cancer. Cancer Cell. 2018;34(1):148–62.:e7. doi: 10.1016/j.ccell.2018.06.004. [DOI] [PubMed] [Google Scholar]
- 51.Behan FM, Iorio F, Picco G, Gonçalves E, Beaver CM, Migliardi G, et al. Prioritization of cancer therapeutic targets using CRISPR-Cas9 screens. Nature. 2019;568(7753):511–6. doi: 10.1038/s41586-019-1103-9. [DOI] [PubMed] [Google Scholar]
- 52.Chan EM, Shibue T, McFarland JM, Gaeta B, Ghandi M, Dumont N, et al. WRN helicase is a synthetic lethal target in microsatellite unstable cancers. Nature. 2019;568(7753):551–6. doi: 10.1038/s41586-019-1102-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kategaya L, Perumal SK, Hager JH, Belmont LD. Werner Syndrome Helicase Is Required for the Survival of Cancer Cells with Microsatellite Instability. iScience. 2019;13:488–97. doi: 10.1016/j.isci.2019.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lieb S, Blaha-Ostermann S, Kamper E, Rippka J, Schwarz C, Ehrenhöfer-Wölfer K, et al. Werner syndrome helicase is a selective vulnerability of microsatellite instability-high tumor cells. Elife. 2019;8 doi: 10.7554/eLife.43333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tian S, Roepman P, Popovici V, Michaut M, Majewski I, Salazar R, et al. A robust genomic signature for the detection of colorectal cancer patients with microsatellite instability phenotype and high mutation frequency. J Pathol. 2012;228(4):586–95. doi: 10.1002/path.4092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Medico E, Russo M, Picco G, Cancelliere C, Valtorta E, Corti G, et al. The molecular landscape of colorectal cancer cell lines unveils clinically actionable kinase targets. Nat Commun. 2015;6:7002. doi: 10.1038/ncomms8002. [DOI] [PubMed] [Google Scholar]
- 57.Jernström S, Hongisto V, Leivonen SK, Due EU, Tadele DS, Edgren H, et al. Drug-screening and genomic analyses of HER2-positive breast cancer cell lines reveal predictors for treatment response. Breast Cancer (Dove Med Press) 2017;9:185–98. doi: 10.2147/BCTT.S115600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen CT, Kim H, Liska D, Gao S, Christensen JG, Weiser MR. MET activation mediates resistance to lapatinib inhibition of HER2-amplified gastric cancer cells. Mol Cancer Ther. 2012;11(3):660–9. doi: 10.1158/1535-7163.MCT-11-0754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Prahallad A, Sun C, Huang S, Di Nicolantonio F, Salazar R, Zecchin D, et al. Unresponsiveness of colon cancer to BRAF(V600E) inhibition through feedback activation of EGFR. Nature. 2012;483(7387):100–3. doi: 10.1038/nature10868. [DOI] [PubMed] [Google Scholar]
- 60.Amirouchene-Angelozzi N, Swanton C, Bardelli A. Tumor Evolution as a Therapeutic Target. Cancer Discov. 2017 doi: 10.1158/2159-8290.CD-17-0343. [DOI] [PubMed] [Google Scholar]
- 61.Dangles-Marie V, Pocard M, Richon S, Weiswald LB, Assayag F, Saulnier P, et al. Establishment of human colon cancer cell lines from fresh tumors versus xenografts: comparison of success rate and cell line features. Cancer Res. 2007;67(1):398–407. doi: 10.1158/0008-5472.CAN-06-0594. [DOI] [PubMed] [Google Scholar]
- 62.Lee JK, Liu Z, Sa JK, Shin S, Wang J, Bordyuh M, et al. Pharmacogenomic landscape of patient-derived tumor cells informs precision oncology therapy. Nat Genet. 2018;50(10):1399–411. doi: 10.1038/s41588-018-0209-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pauli C, Hopkins BD, Prandi D, Shaw R, Fedrizzi T, Sboner A, et al. Personalized In Vitro and In Vivo Cancer Models to Guide Precision Medicine. Cancer Discov. 2017;7(5):462–77. doi: 10.1158/2159-8290.CD-16-1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vlachogiannis G, Hedayat S, Vatsiou A, Jamin Y, Fernández-Mateos J, Khan K, et al. Patient-derived organoids model treatment response of metastatic gastrointestinal cancers. Science. 2018;359(6378):920–6. doi: 10.1126/science.aao2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tiriac H, Belleau P, Engle DD, Plenker D, Deschênes A, Somerville TDD, et al. Organoid Profiling Identifies Common Responders to Chemotherapy in Pancreatic Cancer. Cancer Discov. 2018;8(9):1112–29. doi: 10.1158/2159-8290.CD-18-0349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yan HHN, Siu HC, Law S, Ho SL, Yue SSK, Tsui WY, et al. A Comprehensive Human Gastric Cancer Organoid Biobank Captures Tumor Subtype Heterogeneity and Enables Therapeutic Screening. Cell Stem Cell. 2018;23(6):882–97.:e11. doi: 10.1016/j.stem.2018.09.016. [DOI] [PubMed] [Google Scholar]
- 67.Lee SH, Hu W, Matulay JT, Silva MV, Owczarek TB, Kim K, et al. Tumor Evolution and Drug Response in Patient-Derived Organoid Models of Bladder Cancer. Cell. 2018;173(2):515–28.:e17. doi: 10.1016/j.cell.2018.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rospo G, Lorenzato A, Amirouchene-Angelozzi N, Magrì A, Cancelliere C, Corti G, et al. Evolving neoantigen profiles in colorectal cancers with DNA repair defects. Genome Med. 2019;11(1):42. doi: 10.1186/s13073-019-0654-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Isella C, Terrasi A, Bellomo SE, Petti C, Galatola G, Muratore A, et al. Stromal contribution to the colorectal cancer transcriptome. Nat Genet. 2015;47(4):312–9. doi: 10.1038/ng.3224. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.