Abstract
Oncoproteins of the MYC family are major drivers of human tumorigenesis. Since a large body of evidence indicates that MYC proteins are transcription factors, studying their function has focused on the biology of the downstream target genes. Detailed studies of MYC-dependent changes in RNA levels have provided contrasting models of the oncogenic activity of MYC through either enhancing or repressing the expression of specific target genes, or as a global amplifier of transcription. In this review, we first overview the biochemistry of MYC proteins and summarize what is known (and unclear) about MYC target genes. We then discuss recent progress in defining the MYC and MYCN interactomes and how this information affects central concepts of MYC biology, focusing on mechanisms by which MYC proteins modulate transcription. MYC proteins promote transcription termination upon stalling of RNA Polymerase II and we propose that this mechanism enhances the stress resilience of basal transcription. Furthermore, MYC proteins co-ordinate transcription elongation with DNA replication and cell cycle progression. Finally, we argue that the mechanism by which MYC proteins regulate the transcription machinery is likely to promote tumorigenesis independently of global or relative changes in the expression of their target genes.
Introduction
Since the initial discovery of viral MYC oncogenes in several chicken retroviruses, a large effort has been made in many laboratories to understand the biology of the three cellular MYC oncoproteins: MYC, MYCN and MYCL (for a comprehensive review, see REF.1). It is now unequivocally clear that MYC proteins are major drivers of human tumorigenesis. This assertion rests on several major arguments. First, in the majority of all human tumor types, the expression of MYC proteins is deregulated and enhanced relative to the corresponding normal tissue, and high MYC expression often correlates with poor prognosis. This deregulation is not only due to chromosomal translocations or copy number changes involving MYC genes, but also due to MYC being downstream of multiple oncogenic signaling pathways. For example, the WNT signaling pathway is deregulated in colorectal tumors and invariably this deregulation results in very high MYC levels in colon tumors. Second, activation of MYC in tissue culture is sufficient to induce many properties of tumor cells; for example, activation of MYC induces cell proliferation and cell growth in the absence of external growth factors. Conversely, inhibition of MYC almost invariably suppresses cell proliferation2. Third, genetically defined mouse tumor models show that manipulation of MYC levels dramatically changes tumor incidence and development. On one hand, depletion of MYC in various tumor models abolishes tumorigenesis3 and on the other hand, expression of MYC above physiological levels induces tumor development or strongly accelerates tumorigenesis in multiple tissues. Although all MYC proteins are oncogenic, expression of different MYC paralogues results in tumors with distinct characteristics and dependencies in medulloblastoma4,5 and lung cancer6, indicating that MYC, MYCN and MYCL have unique features. The properties of MYCL, which is best known for its involvement in small cell lung carcinoma 7,8, have been less well studied than those of the other two paralogues. In this Review, we focus on MYC and MYCN.
It has also been established that MYC proteins are a valid target for tumor therapy, since many tumor cells continue to depend on enhanced expression of MYC, and reversion of MYC activity eradicates tumor growth9. This is not only true for tumors driven by MYC transgenes. For example, inhibition of MYC by expression of a dominant negative allele induces tumor regression in mice with established lung adenocarcinoma driven by mutations in RAS and TP53 genes and with glioma10,11. Furthermore, MYC inhibition is tolerated for an extended period of time in adult animals, although MYC is essential during embryogenesis12. When MYC is inactivated, both cell-autonomous processes such as induction of senescence and non-cell autonomous processes such as a collapse of the tumor vasculature contribute to tumor regression13,14. Recent data show that tumor regression following acute deprivation of MYC expression depends on the interaction of tumor cells with the host immune system and argue that MYC controls the synthesis of cytokines that mediate the interaction of tumor cells with NK cells and with B-lymphocytes and T-lymphocytes15-17.
In parallel to the analysis of the oncogenic properties of MYC proteins, a large effort has been made studying the main biochemical properties of these proteins1. However, our current mechanistic understanding of the oncogenic function of MYC, and hence of ways to target this function, lags behind the study of other oncoproteins. In this Review we first summarize the key findings that have generated a seeming paradox in our understanding of MYC function and discuss the progress made in analyzing the interactomes of MYC family proteins and the mechanisms by which they control transcription. We then discuss the evidence showing that MYC proteins can help free promoters from stalling RNA polymerase II (Pol II) complexes and co-ordinate transcription elongation with cell cycle progression, suggesting that they promote cell proliferation and possibly tumorigenesis by altering basic transcription mechanisms in addition to changing target gene expression. We propose that these new data offer a way forward towards a better understanding of the tumor-promoting and oncogenic effects of MYC and also towards the development of novel therapies targeting MYC function.
[H1] Canonical functions of MYC proteins
MYC proteins are part of a network of interacting transcription factors1,18. They form an obligatory heterodimeric DNA binding complex with a partner protein named MAX. MAX proteins can form homodimers, which are largely inert in transcription regulation, or heterodimers with any one of four MXD (MAX dimerization) proteins. Heterodimers of MAX with the MXD proteins, MGA1 or MNT recruit histone deacetylase complexes, through their interaction with a scaffold protein SIN3A. The interaction between MYC, MAX and other members of this network is mediated by a conserved domain in each of the proteins, which encompasses a helix-loop-helix (HLH) domain and a leucine zipper (Figure 1). The highly conserved domains of the dimer-forming proteins19 place the HLH-adjacent basic region of each member of the network in a manner that leads the dimers to bind to an Enhancer-box’ (E-box) sequence with the consensus CAC(G/A)TG. This sequence is enriched in MYC binding sites on chromatin, but many of the binding sites show substantial deviation from this sequence or no similarity to it, suggesting that other factors strongly affect chromatin association of MYC20 (see below). A large body of evidence obtained from genetic models shows that heterodimerization with MAX is essential for MYC function21 and MAX heterodimerization with MNT or MGA1 suppresses cell growth22. The balance within this network is frequently perturbed in human tumors by changes in gene copy number21: whereas MYC predominantly undergoes amplification, MNT and MGA undergo focal deletions, which is consistent with their proposed functions as inducer or suppressors of cell growth, respectively. At high MYC levels, the zinc finger protein MIZ1 binds to the carboxy-terminal of MYC and is recruited to the MYC target sites on DNA, thereby blocking MYC-dependent transcription activation5,23.
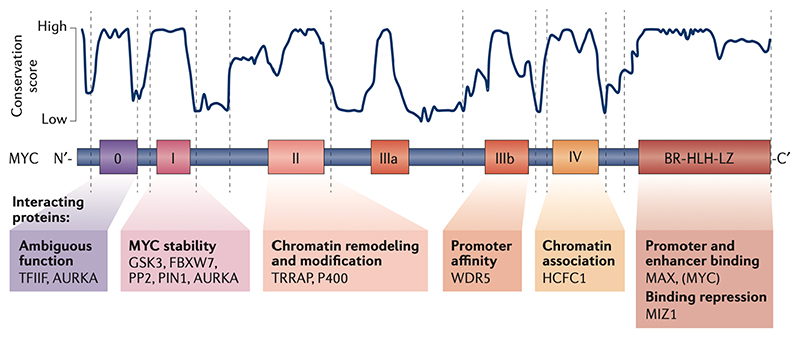
Figure 1. Protein domains of MYC and their canonical function.
Amino acid sequence analysis identified different degrees of conservation within MYC proteins. While the C-terminal domain responsible for DNA binding is entirely conserved only small stretches – the MYC boxes – show a high degree of conservation in the remaining part of the protein. The conservation score shown in the figure was calculated as described in REF 126. Function definitions of MYC boxes 0–IV are based on deletion and/or point mutations. Examples of proteins that interact with the relevant MYC boxes are indicated. AURKA, Aurora kinase A; BR, basic region; FBXL3, F-box and leucine rich repeat protein 3; FBXW7, F-box and WD repeat domain containing 7; GSK3,glycogen synthase kinase 3; HCFC1, host cell factor C1; MAX MYC associated factor X; MIZ1,MYC interacting zing finger protein 1; P400, E1A binding protein p400 ; PIN1, peptidylprolyl cis/trans isomerase NIMA-interaction 1; PP2A, serine/threonine protein phosphatase 2A; TFIIF, general transcription factor IIF subunit 1; TRRAP, transformation/transcription domain associated protein; WDR5, WD repeat domain 5.
Apart from the DNA binding and dimerization domain, the overall sequence homology of the trans-regulatory domains is not particularly high among the three MYC isoforms, but it includes relatively short stretches of amino acids that show very high sequence conservation (Figure 1). These sequences are termed MYC boxes and the current consensus defines six of them. Since their numbering changed with the discovery of new conserved elements, they are now numbered 0, I, II, IIIa, IIIb and IV. Some of the functions and interacting proteins of the MYC boxes are well described. For example, MYC box I controls proteasome-mediated degradation of MYC proteins24. It constitutes a phospho-degron [G] consisting of either three (in MYCN: T58, S62 and S64) or four (in MYC: T58, S62, S64 and S67) closely spaced and highly conserved serine and threonine residues, which are all phosphorylated in vivo. Among them, the best understood modification is the phosphorylation of T58 by the kinase GSK3. This phosphorylation is required for binding to the ubiquitin ligases FBXW7 and FBXL3 and suppresses the interaction with ubiquitin carboxyl-terminal hydrolase 11 (USP11)25. Phosphorylation of S62 by cyclin-dependent kinases and potentially also by MAP kinases primes the phosphorylation of T58 by GSK326. The kinases that phosphorylate S64 (of MYC and MYCN) and S67 (of MYC) and whether they also control protein degradation are unknown. MYC box I is also recognized by serine/threonine-protein phosphatase 2A and by the peptidyl-prolyl cis/trans isomerase PIN127.
Both MYC box 0 and MYC box I bind to Aurora-A kinase, which stabilizes MYC and MYCN because it antagonizes the binding of FBXW728,29. MYC box II interacts with transformation/transcription domain-associated protein (TRRAP), which is a scaffold protein of several large protein complexes involved in chromatin remodeling and histone acetylation, such as the transcription-promoting histone acetyltransferase complex NuA430,31. TRRAP associates with multiple other proteins such as the helicase P400. MYC box IIIb binds to WDR5, which facilitates histone H3 Lys4 (H3K4) methylation32. Since H3K4 methylation is a mark of active promoters, the interaction of MYC with WDR5 is thought to increase the affinity of MYC for active promoters. In addition, MYC box 0 interacts with the Pol II basal transcription factor IIF (TFIIF)33, and MYC box IV interacts with the transcription co-regulator HCFC134.
MYC proteins are intrinsically disordered and only fold when complexed with other proteins. Crystallography structures have been resolved for complexes of MYC or MYCN with MAX35, WDR532, TBP36 and Aurora-A28. Intrinsically disordered proteins may associate with each other or with partner proteins to form liquid–liquid phase-separated condensates37, and it is possible that MYC participates in the formation of such condensates through the same C-terminal domains involved in transcription regulation.
[H2] The target genes of MYC
The assumption emerging from the biochemical characterization of MYC proteins, such as their functioning as part of a DNA binding complex and their ability to transactivate reporter genes is that they exert their biological effects regulating a set of target genes. Over the past two decades, a large body of work using a wide range of methods identified MYC target genes, which are defined as genes that are either activated or repressed by MYC1,38. Although there is no consensus on the identity of the target genes, a few conclusions can be drawn from the prevailing studies:
-
(i)
Genes transcribed by all three RNA polymerases can be regulated by MYC proteins39,40. Also, different RNA species (including mRNAs, long non-coding RNAs, tRNAs and microRNAs) can be regulated by MYC. The effects of MYC on genes transcribed by Pol I and Pol III are generally positive (transcription activation). By contrast, MYC-binding to promoters transcribed by Pol II can lead to either activation or repression of transcription. Sequencing of nascent transcripts shows that both activation and repression occur at the stage of transcription, not at later steps such as mRNA processing41.
-
(ii)
The number of MYC regulated genes transcribed by Pol II is large and the biological processes defined by these genes encompass virtually every anabolic and growth-promoting process, including those which drive all hallmarks of tumorigenesis. However, the studies identifying target genes have not yet led to the definition of crucial oncogenic processes by which MYC transforms cells.
-
(iii)
MYC and MYCN regulate distinct gene expression programmes. Most studies involving experimentally-manipulated MYC expression or using an inducible MYC-estrogen receptor allele42 concluded that a common set of MYC target genes exists, which is involved in protein translation and purine biosynthesis43-47. By contrast, direct target genes of MYCN that are expressed at higher levels in MYCN-amplified neuroblastoma are most strongly enriched with genes expressed in the S phase and G2 phase of the cells cycle, when MYCN expression is highest48,49 (see below). Similarly, a study probing for a MYC-dependent growth-stimulating agent that acts in the G2 phase of hepatocytes concluded that a master transcription factor of G2 progression, FOXM1, is a crucial target of MYC-induced cell growth50, which is consistent with the mitosis-promoting cyclin B1 being a direct target of MYC51. In a different experimental setup, such as cells that have been exposed to DNA damage (an anti-mitogenic stimulus), transcriptional responses to MYC are very different52. Hence, the identity of the genes regulated by MYC appears to be largely determined by the experimental conditions and cell types used; the only genes that are observed as MYC targets in most studies are those transcribed in the G1 phase of unperturbed growing cells. Even in a single-cell system, different MYC levels regulate different sets of target genes53.
-
(iv)
MYC represses expression of the CDKN2B (p15Ink4b)54,55 and CDKN1A (p21Cip1)47,56,57 genes encoding cyclin-dependent kinase inhibitors and consequently reverses the growth arrest induced by DNA damage and by transforming growth factor β (TGFβ). MYC also represses expression of multiple genes involved in cell adhesion58,59. The use of an interactingdeficient MYC mutant and of a conditional MYC deletion showed that interaction with MIZ1 mediates repression of these genes by MYC60. The set of repressed genes is even more variable than that of MYC-activated genes and includes many weakly expressed or tissuespecific genes. Both direct gene repression mechanisms, such as recruitment of histone demethylases61 or the sequestration of general transcription factors62 and indirect mechanisms63, contribute to the repression of specific sets of target genes.
-
(v)
Almost all the effects of MYC on the expression of specific target genes are weak and often are below twofold even when MYC levels are manipulated to increase over several orders of magnitude. Although interference with the expression of MYC target genes can abolish MYC-driven tumorigenesis, there are very few studies that show that a two-fold change in target gene expression is relevant: examples are 60S ribosomal protein L24 and ornithine decarboxylase, which are both haplo-insufficient for supporting MYC-induced lymphomagenesis, but not for normal growth64,65.
[H2] Current models of MYC function
MYC and MYCN bind to the promoters of virtually all genes that are active in a given cell population as well as to multiple enhancers23,41,66-68. They bind chromatin in complex with MAX. MYC–MAX binding sites can be both 5’ or 3’ of the transcription start site. Binding sites are enriched in E-boxes, but the distribution of MYC and MYCN cannot be explained by their DNA binding specificity alone20,69. Rather, interaction of MYC or MYCN with other proteins enhances their specific DNA interactions, for example with promoters. Examples of co-operative binding include WDR532 and, in Drosophila melanogaster, with RNA polymerase II-associated factor 1 homolog (PAF1)70. Relative MYC occupancy correlates with promoter activity67,71. Related proteins such as the MXD proteins and MGA1 show very similar association patterns with chromatin, which is consistent with the notion that they are part of a network. High levels of MYC can recruit also MIZ1 to many of its target sites23. Attempts to correlate chromatin binding of MYC with gene regulation provided different models of MYC function.
[H3] The specific-gene regulation model
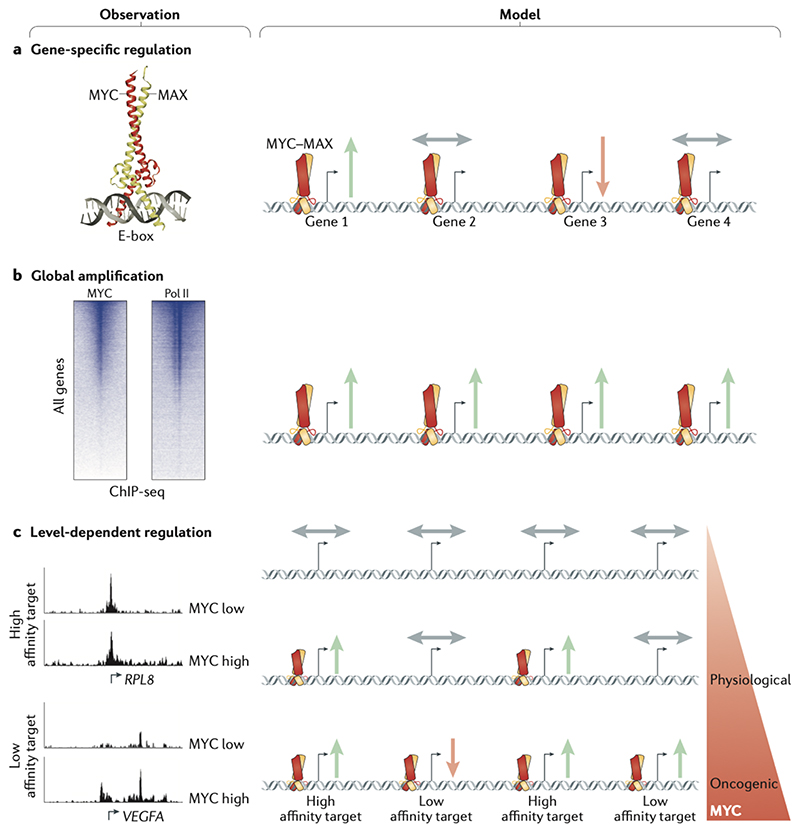
Comparison of the gene expression data discussed above with the chromatin binding data of MYC proteins — specifically, with the genome-wide binding of promoters by MYC proteins — produce a paradox, since in each experimental system or condition, although all active genes are bound by MYC proteins, only a small subset of the genes responds to changes in MYC levels23,41,53 (Figure 2a). For example, a recent careful study carried out in B cells showed that the number of MYC-regulated genes during stimulation of primary B cells is at least an order of magnitude smaller than the number of MYC-bound genes72. Without exception, therefore, these comparisons yield the conclusion that the great majority of MYC binding to promoters does not produce changes in steady-state mRNA levels of the downstream gene43.
Figure 2. Models of gene regulation by MYC.
(a) Specific-gene regulation: Based on the study and structural elucidation of the Enhancerbox (E-box) binding by MYC, specific MYC-induced target genes were discovered and characterized. (b) Global gene activation: Genome-wide chromatin immunoprecipitation sequencing (ChIP-seq) studies of MYC identified that MYC binds at the promoters of most RNA polymerase II (Pol II)-bound and expressed genes. Accordingly, Myc induces a global increase in cellular mRNA levels in some biological systems. (c) Gene-specific affinity: Although MYC binding is detectable at most promoters, promoter affinities for MYC differ widely. Genes with high promoter–MYC affinity are bound and upregulated by MYC at physiological levels, but are not further induced at oncogenic levels, as binding is already saturated. Instead, oncogenic MYC upregulates low affinity target genes. The affinity model can reconcile the seemingly opposing specific gene and global gene models. Green arrows indicate activation, red arrow indicates repression and grey arrows indicate no regulation of genes. RPL8, 60S ribosomal protein L8; VEGFA, vascular endothelial growth factor A.
[H3] The global gene activation model
A significant change in our understanding of MYC-mediated gene regulation was brought about by the finding that MYC proteins can enhance the overall rate of transcription genome-wide, and that the global increase in mRNA levels during the mitogenic stimulation of early B-cells depends on MYC66,67. The resulting model has been termed the ‘global amplifier’ model of MYC function (Figure 2b); it postulates that binding of MYC to all active promoters is productive because it enhances the rate of overall transcription, and that increasing overall transcription is the central oncogenic function of MYC. In this view, the normalization algorithms used in gene expression studies mask a global MYC-dependent increase in transcription and give the wrong impression that the expression of most genes does not change73. Consistent with this model, MYC-driven and MYCN-driven tumors are highly dependent on cyclin-dependent kinase 7 (CDK7) and CDK9, which globally drive transcription74,75. Global and MYC-dependent increases in the total amount of mRNA in a cell have been reported following mitogenic stimulation of primary B-cells and correlate with cell growth76. However, MYC-dependent changes in global RNA or mRNA levels or in transcription rates in most cells are, like the changes in expression of specific genes discussed above, small and highly dependent on culture conditions76, and occur long after MYC activation41,72 or not at all23.
[H3] The gene-specific affinity model
Several studies have shown that different levels of MYC proteins result in different transcriptional outputs (Figure 2c). For example, B-cells engineered to express either low levels, physiological levels or lymphoma-specific (high) levels of MYC have very different gene expression profiles53. As described above, the affinity of complexes of MYC and MAX for their binding sequence is low, hence the affinity for specific sites is mainly dictated by protein-protein interactions20. Consequently, promoters differ in the level of MYC that is required for their activation and even a globally-binding transcription factor can regulate functionally different sets of genes at physiological and oncogenic expression levels69. This model may be able to reconcile the seemingly opposing observations of global DNA-binding and gene-specific regulation by MYC without the need to invoke productive and non-productive modes of DNA binding69.
The relative merits of each model have been exhaustively discussed and have led to technically detailed debates about the normalization required to account for changes in cell size, the timing of gene-specific versus global changes in gene expression, the distinction of primary versus secondary effects and different peak calling algorithms43,72,73,77. In the absence of decisive evidence for either model, the extensive discrepancy between universal binding and the small effects on either relative or absolute mRNA levels of most genes raises the possibility that MYC has additional functions at core promoters, which are independent of altering steady-state mRNA levels.
[H1] Functions of MYC at promoters
The analysis of the mechanism of MYC function has not received a similar level of attention as the detailed analyses of MYC-dependent gene expression. The assumption, therefore, that to control gene expression, MYC proteins use mechanisms that are similar to those used by other transcription factors, and that the unique role of MYC proteins in oncogenesis is based on their ability to regulate a unique set of target genes has not been scrutinized. By contrast, recent analyses of proteins that interact with MYC and MYCN show that they engage in unexpected protein interactions and that the identity of these proteins is compatible with MYC functionality that differs substantially from the canonical models described above.
[H2] The interactomes of MYC and MYCN
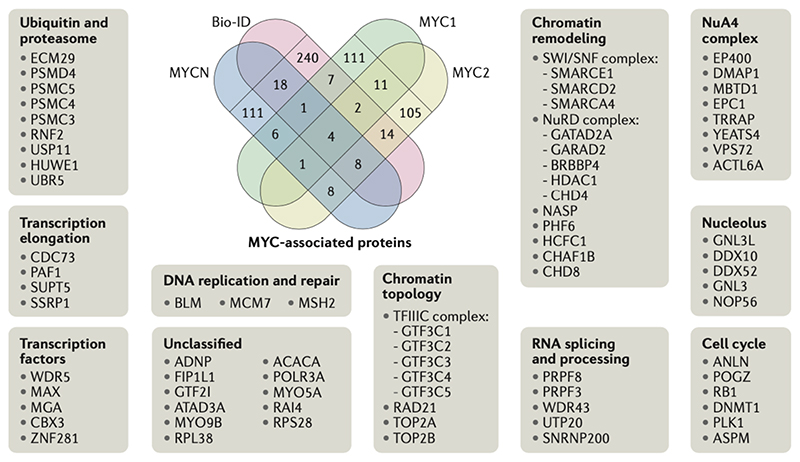
Over the past years, a number of proteomic analyses of the MYC and MYCN interactomes have been published33,62,78. These analyses are based on mass spectrometry of immunoprecipitated MYC or MYCN complexes solubilized from chromatin, or of proteins recovered by Bio-ID techniques [G], which label proteins that are in the vicinity of MYC or MYCN in living cells. Though these interactomes are not identical, they show a significant degree of overlap and have helped to define a core set of MYC-associated proteins (Figure 3). Although any significance cut-off in the absence of clear functional data is arbitrary, a census of the eighty most consistently MYC-bound and MYCN-bound proteins in the overlap of four recently published interactomes yields the following conclusions:
-
(i)
It is very likely that MYC and MYCN engage in several compositions of protein–protein complexes rather than form a single defined complex. As discussed above, the DNA binding affinity of MYC–MAX is low and MYC proteins depend on chromatin-bound factors like WDR5 for recognizing target promoters. At present, therefore, it is unknown which of these complexes recruit MYC to promoters and enhancers and which proteins are effectors that mediate the function of MYC.
-
(ii)
The analyses confirm previously known interactions of MYC with chromatin proteins, for example with MAX, WDR5, the NuA4 complex and P400. Chromatin remodeling factors such as SMARCA4 (of the SWI/SNF complex), transcription elongation factors such as SSRP1 (of the FACT (facilitates chromatin transcription) complex) and SPT5 (of the DSIF complex) and nucleolar ribosome biogenesis factors such as NOP56 are also detected (Figure 3). Conversely, a number of potential MYC-interacting proteins that have been used to argue for major functions of MYC at core promoters transcribed by Pol II and Pol III, are either not present, or present only at low levels. Examples of such proteins are YY-1, TFII-I, TFIIH, mediator-complex subunits and CDK9.
-
(iii)
The ubiquitin ligases HUWE1 und UBR5 and the de-ubiquitylating enzyme USP7 are found in the interactomes. HUWE1 is bound to the proteasome complex79 and proteasome subunits are also found in the interactomes, which is consistent with the observation that MYC can recruit the proteasome to promoters80.
-
(iv)
The analysis points to new functions of MYC proteins in chromosome topology. Chromatin architectural proteins, including the condensin-associated TFIIIC complex, Topoisomerases 2A and 2B, and several SMC (structural maintenance of chromosomes) proteins (for example, RAD21), interact with MYC and MYCN. The presence in the interactomes of DNA replication proteins and of the mitotic kinase PLK1, support previously suspected roles of MYC proteins in cell cycle progression and DNA replication81,82. Furthermore, multiple RNA-binding proteins, in particular proteins involved in splicing, are found in the interactomes; some of these interactions are likely to be mediated by nascent RNA.
Figure 3. MYC-associated proteins.
Schematic of the overlap of recently published MYC and MYCN interacting proteins33,62,78,87. Proteins appearing in at least two data sets are presented in the surrounding boxes and were used for functional annotation. Bio-ID techniques label proteins that are in close proximity to a protein of interest fused to a biotin ligase and was used in 33. The other datasets used immunopurification of MYC complexes:, MYC1 refers to 62, MYC2 to 78 and MYCN to87.
Although these analyses clearly show that MYC proteins interact with multiple chromatinbound proteins, they do not support the notion that their interactions with the core transcription machineries of the three RNA polymerases define the sole or even the major biochemical function of MYC proteins. Instead, the interactomes are equally compatible with other functional descriptions of MYC. For example, the NuA4 complex, which is assumed to be crucial for MYC-dependent transcription activation, also has an important role in the repair of DNA double-stranded breaks (DSBs)83. Deleting an allele of one of its catalytic subunits, TIP60 in a MYC-driven lymphoma model, has no effect on MYC-dependent gene expression, but causes severe DNA damage84. Like NuA4, the ATPase p400, the ubiquitin ligases HUWE1 and UBR5, the de-ubiquitylase USP7, the PAF1 transcription complex, the SMC RAD21– cohesin complex and the histone methyltransferase G9A have all been implicated in cell responses to DNA damage and in DNA repair.
[H2] Hand-over regulation of transcription
Many recent studies show that MYC and MYCN globally affect the function of Pol II. In some experimental systems, the strongest global effect is the ability of MYC and MYCN to induce the release of Pol II from promoter-proximal pausing [G] into productive transcription elongation23,85. Many other effects of MYC and MYCN on Pol II have been documented, ranging from modulating Pol II binding to promoters23,86, promoter escape87 and effects on the processivity and directionality62 of transcription.
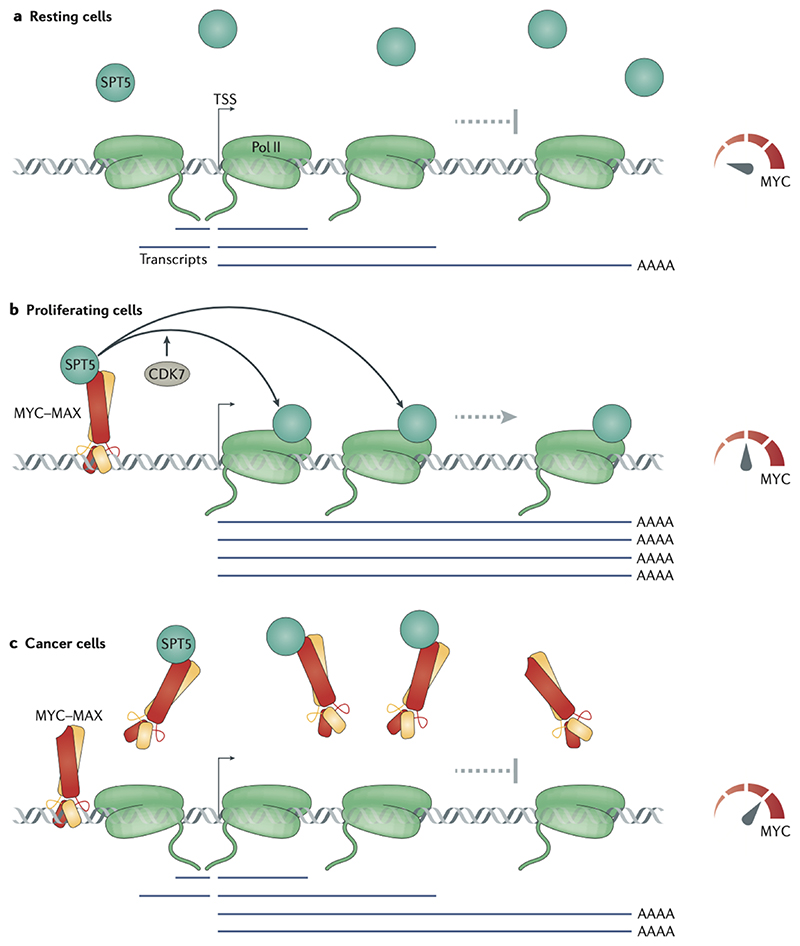
The mechanisms underlying these observations are only beginning to emerge. A number of recent papers support a handover mechanism, in which MYC recruits factors to the promoter and onto Pol II to promote transcription. MYC interacts directly with the transcription pausing and elongation complex DSIF (consisting of SPT4 and SPT5) and transfers SPT5 onto Pol II in a CDK7-dependent manner. Consequently, in absence of MYC, Pol II largely engages in abortive transcription few kilobases downstream from the promoter-proximal pause-site and thus generates short (1–4 kb) nascent mRNA transcripts in both sense and anti-sense directions. MYC therefore can switch transcription from a non-productive mode (including antisense, abortive, slow and pervasive transcription) to a productive mode (directional, sense and fast transcription)62 (Figure 4a,b). Although non-productive transcription at low MYC expression levels is seemingly wasteful, such abortive transcriptional activity could prevent heterochromatinization of promoters and ensure that they remain responsive to stimulatory signals, especially in resting cells88.
Figure 4. A handover-model of promoter-proximal function of MYC proteins.
(a) In resting cells, which do not express MYC, the transcription elongation factor SPT5 is insufficiently recruited to RNA polymerase II (Pol II), which loses directionality and processivity, resulting in increase in the levels of antisense and abortive transcripts. (b) In growing cells, MYC is expressed, binds SPT5 and recruits it to promoters. The transfer of SPT5 from MYC to Pol II depends on cyclin-dependent kinase 7 (CDK7).Pol II associated with transcription elongation factors engages in productive (fast, processive and directional) transcription elongation and produces full-length mRNAs. (c) In cancer cells expressing high levels of MYC, a considerable fraction of SPT5 is sequestered by soluble MYC, and transcription is decreased at known MYC-repressed genes. TSS, transcription start site.
Similarly, a ubiquitin-ligase dependent handover of the PAF elongation complex by MYC primes Pol II for elongation89. By contrast, the analysis of MYC and MYCN interactomes has not provided clear insight into the interactions through which MYC enhances the binding of Pol II at core promoters or promotes Pol II pause-release. Specifically, CDK9 is very scarce in the interactomes, despite some evidence of in vitro interactions of MYC with the CDK9-interacting protein cyclin T190. Recent work has argued that MYC binds CDK9 indirectly through TFIIF33, but whether CDK9 is thus activated by MYC to support Pol II pause-release remains to be demonstrated.
The hand-over mechanism of transcription regulation by MYC proteins has two functionally relevant consequences. Firstly, the high levels of MYC found in tumor cells sequester SPT5 in non-functional complexes and therefore reduce transcription elongation62 (Figure 4c).
Handover of SPT5 is therefore a mechanism of both transcription activation and repression. Hence, oncogenic MYC levels are not ‘more of the same’, but suppress transcription of antiproliferative and cytokine genes by sequestering SPT562. Secondly, MYC-dependent handover of elongation factors can prevent the response of Pol II to stress conditions that limit transcription elongation in normal cells. One such example is glutamine starvation, which lowers the levels of ribonucleotides and blocks transcription in cells by downregulating MYC levels through a translation control element in the MYC mRNA 3’ untranslated region (3’UTR). In this manner, even tumor cells that express high level of MYC mRNA are protected from apoptosis91. By contrast, cells that express MYC constructs lacking the 3’UTR maintain transcription elongation in the presence of low nucleotide levels, which leads to MYC- dependent stalling of Pol II and R-loop [G] formation in multiple gene bodies91. R-loops are stable hybrids of DNA and nascent RNA that threaten genome stability by promoting collisions between the transcription and replication machineries92. This correlates closely with the well-characterized sensitization of cells to glutamine-starvation-induced apoptosis by such MYC constructs (termed ‘glutamine addiction’). Notably, just as MYC recruits elongation factors to Pol II, it may recruit DNA repair factors to promoter-proximal DSBs.
[H1] New models of MYC function
In parallel to the identification of novel interaction partners, a number of recent studies have identified effects of MYC or MYCN on Pol II that may be crucial for its function in proliferating cells, but are expected to have only minor effects on the expression of individual MYC target genes and on the overall rate of transcription.
[H2] Resilience to transcription stress
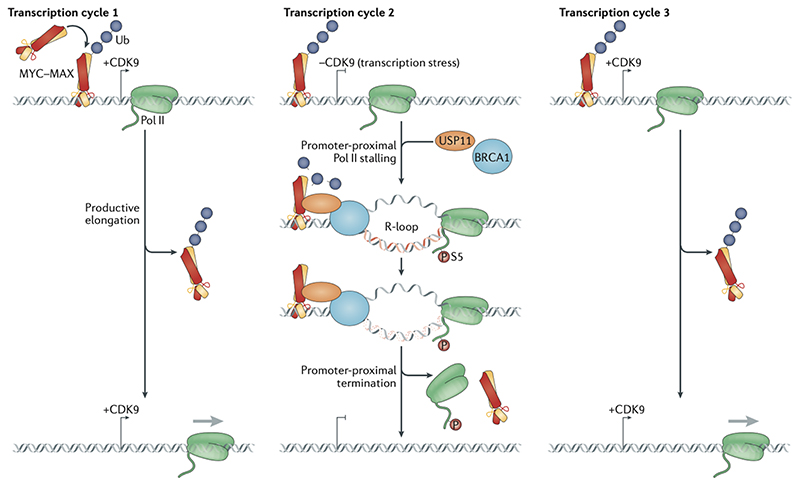
Activation of MYCN in neuroblastoma universally promotes Pol II pause-release from all active promoters, irrespective of whether mRNA levels change in response to MYCN activation; instead, the observed changes in mRNA levels are due to gene-specific differences in promoter-proximal transcription termination25. The critical switch between elongation and termination is phosphorylation of MYCN at T58. This residue promotes MYCN turnover, as its dephosphorylation causes MYCN to bind the de-ubiquitylase USP11–USP7. MYCN de-ubiquitylation and hence stabilization by USP11–USP7 enables it to recruit the tumor suppressor BRCA1to promoters (Figure 5). In turn, BRCA1 recruits mRNA decapping complexes that promote promoter-proximal transcription termination. BRCA1 recruitment is strongly enhanced by compounds that stall Pol II at the promoter, suggesting that this regulatory mechanism prevents accumulation of non-productive Pol II complexes.
Figure 5. MYC binding at core promoters.
Model of global and possibly crucial functions of MYC proteins at all active promoters with very small effects on steady-state mRNA levels. MYC ubiquitylation allows cyclin-dependent kinase 9 (CDK9)-driven RNA polymerase II (Pol) II to generate the mRNA (Transcription Cycle # 1). However, in response to transcription stress, which induces or augments promoter-proximal Pol II stalling and R-loop formation, BRCA1 is recruited to the R-loop with the help of USP11, which de-ubiquitylates MYC, thereby allowing it to interact with BRCA1 (Transcription cycle # 2). This promotes the resolution of the R-loop and dissociation of the Pol II machinery, which makes the promoter ready for a new cycle where Pol II can again transcribe in a CDK9 driven manner (Transcription cycle # 3).
A major consequence of the failure to recruit BRCA is the accumulation of promoter-proximal R-loops, which also accumulate in BRCA1-deficient mammary carcinomas. Suppression of promoter-proximal R-loop formation is one function of the PAF1 complex93; hence, recruitment of elongation factors by MYC may have a similar function. Intriguingly, the co-repressor protein that is recruited to promoters by MXD proteins, SIN3A, also suppresses co-transcriptional R-loop accumulation through its association with THO complex subunit 1, which binds to spliced mRNA and promotes its nuclear export94. It is possible, therefore, that MYC and MXD proteins promote different pathways that limit R-loop accumulation at core promoters. Exactly by which DNA structure(s) and stress signal(s) BRCA1 is recruited to R-loops is unclear. As BRCA1 interacts with Pol II, its recruitment could simply be mediated by Pol II. Alternatively, as active transcription exerts a strong torsional stress on DNA, topoisomerases, which introduce transient DNA breaks to relieve torsional stress (followed by re-ligation), are required to enable transcription95. Indeed, promoter-proximal DSBs occur frequently during the rapid stimulation of transcription in response to mitogenic signals and ligand-dependent activation of nuclear receptors96,97. Consequently, DSBs that are formed by topoisomerase inhibition (that is, by preventing DNA re-ligation by the topoisomerase) occur predominantly in promoter-proximal positions98, and it is possible that BRCA1 recruitment is part of a MYCN-mediated pathway that restricts transcription in the presence of transcription-induced DSBs.
These findings suggest that MYC and MXD proteins may both limit R-loop formation and clear stalled Pol II from promoters, potentially explaining the apparent discrepancy between the widespread occupancy of active promoters by MYC proteins and the lack of apparent functional consequences of MYC binding (Figure 5). R-loop formation following Pol II stalling and — even more so — DNA damage at promoters are likely to be stochastic events, since the fraction of promoters that break during each transcription cycle is thought to be small. If MYC clears a promoter from stalled Pol II and enables transcription restart by another Pol II molecule, it is required at each active promoter, but not necessarily for each round of transcription and hence its effects on steady-state mRNA levels would be small. In this model, MYC proteins productively engage with a promoter even without causing changes in gene expression.
[H2] Transcription–replication coordination
Both MYC and MYCN are expressed in the S phase and in G2 phase of the cell cycle99,100 and the roles of MYC in mitosis are well-documented101. In this section and in the next, we discuss roles of MYC and MYCN in cell-cycle-dependent control of transcription that are not related to the well-documented roles of MYC and MYCN during early growth-factor-dependent stimulation of cell proliferation. Multiple mechanisms have been identified that link the expression and stability of MYC proteins with progression through the cell cycle. First, both MYC and MYCN are target genes of the E2F family of transcription factors, and their mRNA expression is regulated in a cell-cycle-dependent manner. Second, two cell cycle regulated kinases, PLK1 and Aurora-A control the turnover of MYC and MYCN82. Aurora-A sterically impedes the interaction of MYCN with the ubiquitin ligase SCFFbw7, because it binds adjacently to the MYC degron28. Consequently, MYCN protein levels increase in parallel with those of Aurora-A at the beginning of S phase and are maintained at high levels during G2 phase100. PLK1 phosphorylates SCFFbw7 and promotes its auto-ubiquitylation and proteasomal degradation102. As a result, PLK1, like Aurora-A, blocks MYCN degradation. PLK1 also binds MYC and phosphorylates it at both S62 and S279; the latter phosphorylation protects MYC from proteasomal degradation upon recovery from an S-phase arrest and is required for the resumption of cell cycle progression82,103.
During the S phase of the cell cycle, transcription elongation by Pol II can impede the progress of DNA replication forks, leading to replication–transcription conflicts [G] 104. In particular, transcription-induced R-loops are major impediments to replication forks and can cause head-to-head collisions between Pol II and DNA polymerases92. MYC and MYCN restriction of R-loop formation will therefore diminish replication–transcription conflicts. For example, PAF1 not only restricts promoter-proximal R-loop formation but is also required to remove Pol II from promoters during DNA replication to avoid such conflicts105.
A number of observations indicate that MYC and MYCN have a role in co-ordination of transcription with DNA replication and in preventing replication–transcription conflicts. Aurora- A associates with MYCN specifically during S phase and competes with many MYCN coactivators for binding to MYCN. Disruption of the Aurora-A–MYCN complex restores the binding of MYC co-activators and transcription elongation, which potently activates the kinase ATR through the stalling of DNA replication forks, arguing that the Aurora-A–MYCN association prevents conflict between transcription and replication87. The interaction of MYC with MIZ1 appears to have a similar function. Previous work had established that the interaction of MYC with MIZ1 facilitates the recovery of cells from an ultraviolet-light-induced S-phase arrest and that MIZ1 interacts with components of the S-phase checkpoint machinery46. In several leukemia models, disruption of MIZ1 function or of the interface of MYC with MIZ1 strongly delays lymphomagenesis. This correlates closely with a dramatically enhanced expression of canonical MYC target gene sets and genes expressed in S phase, and also with potent engagement of the ATR-dependent replication checkpoint and with enhanced sensitivity to replication inhibition106. Collectively, the data argue that MIZ1 restricts MYC function during S phase and thus decreases replication stress. Partial depletion of MIZ1 or preventing its interaction with MYC is tolerated by normal tissues, but strongly delays tumorigenesis as shown for pancreatic carcinoma23, lymphoma60, medulloblastoma5 and hepatocellular carcinoma107. Finally, yeast cells escape transcription–replication conflicts at least in part by detaching genes that are close to the nuclear envelope, so that nascent mRNAs are rapidly released from chromatin in order to allow the passage of replication forks108. Intriguingly, MYC proteins have recently been shown to control the association of nascent transcription complexes with nuclear pores, suggesting that they may function in a similar manner109.
At each gene, a collision between Pol II and a replication fork occurs no more than once during a cell cycle. Although transcription is not generally coordinated with the cell cycle, the chance that a collision occurs at a given promoter in a cell is therefore low, as is the formation of promoter-proximal R-loops and DSBs. Collision are also not expected to occur at the same promoters in different cells. Nevertheless, failure to any resolve replication–transcription conflict leads to S-phase checkpoint activation and cell cycle. As we discussed in the previous section, one potential reason for the widespread presence of MYC at active promoters is the need to be able to rapidly respond to replication–transcription conflicts.
[H2] Beyond S phase: G2 phase and mitosis
A number of different observations suggest that the MYC proteins have a specific role during G2 phase and mitosis. First, phosphorylation of MYCN at S62, which primes the phosphorylation of T58 by GSK3 and subsequently MYCN degradation by the ubiquitin ligaseFBXW7, is carried out by cyclin-dependent kinase 1 (CDK1) and hence occurs in G2 and mitosis in primary neuroblasts, thereby coupling MYCN degradation with progression through mitosis110. Interaction with Aurora-A can delay degradation of MYCN until mitotic exit, when Aurora-A itself is degraded111. In combination with E2F-dependent transcription of MYCN, this mechanism sets a window of MYCN expression between S phase and mitotic exit. Although MAP kinase and CDK2 (rather than CDK1) phosphorylate MYC at S62112,113, PLK1 associates with both MYC and MYCN and controls their stability and possibly their transcriptional functions during G2 phase and mitosis82,102,114.
Second, deregulated MYC or MYCN expression enhances the sensitivity of cells to, and alters cell fate in response to mitotis perturbation101,115. In response to disruption of the mitotic spindle, enhanced expression of MYC causes cells to either undergo apoptosis or to become polyploid by replicating their DNA without subsequently undergoing cell division. Consequently, inhibitors of the mitotic kinases CDK1 and Aurora-B have been proposed as therapeutic targets in MYC-driven tumours116,117. One possible mechanistic interpretation of these findings is that MYC and MYCN couple the structural changes that chromatin undergoes during entry into mitosis with Pol II function, for example by maintaining transcription competency of active promoters during the progressive chromatin condensation that takes places in mitosis. Recruitment of BRCA1 by MYCN occurs in both S phase and G2 phase, suggesting that MYCN can terminate transcription also in G2 phase, when there are no conflicts with DNA replication25. Notably, functions of MYC during mitosis, but not other functions of MYC, depend on protein SUMOylation, suggesting that this modification has an important role in mitosis118.
[H1] Oncogenesis ‘without’ target genes?
The unequivocal demonstration that MYC is part of a network of interacting transcription factors has been a major breakthrough in the field. However, the implicit assumption that MYC biology can only be explained either as changes in the expression of target genes or as altered global transcription rates has led to a conceptual standstill in understanding by which major mechanism or pathway MYC proteins promote tumorigenesis. It remains possible of course that either gene-specific or global changes in transcription contribute to or account for MYC- driven tumorigenesis. However, there are no experimental data that systematically test the hypothesis that MYC proteins transform cells because they either (i) induce changes in the expression of a small group of targets with a circumscribed oncogenic function; (ii) induce changes in the expression of large groups of functionally diverse target genes; or (iii) induce weak increases in the global transcription rate. Importantly, the experimental strategies of blunting MYC function using inhibitors that target specific target genes (such as inhibitors of polyamine metabolism), or strategies that globally dampen transcription rates (such as inhibitors of CDK7 and CDK9) have not yet yielded convincing data demonstrating that they specifically target tumors with high MYC or MYCN levels, nor have they reached advanced stages in clinical trials.
Furthermore, recent analyses not only reveal new and unexpected mechanisms, by which MYC proteins control transcription, but also show that MYC-associated proteins are not only related to transcription. The assembly of elongation-competent polymerase complexes and the co-ordination of transcription with mRNA processing are likely to be perturbed at many stages in tumor cells, due to their aneuploidy and the resulting imbalance in the expression of different proteins of the same complex, insufficient amounts of nucleotides and elevated nucleotide oxidation. Moreover, disruption of DNA-damage checkpoints in cancer cells will hamper the co-ordination of transcription elongation with DNA replication. This reduced coordination will increase Pol II stalling, R-loop formation and genome instability owing to replication-transcription conflicts92,119. Importantly, DNA replication stress, which activates the ATR-dependent replication checkpoint and thereby apoptosis or cell cycle arrest, is a barrier to tumorigenesis that inhibits the progression of pre-neoplastic lesions to neoplasia, arguing that mechanisms that relieve replication stress are highly oncogenic120. At the same time, the overall high transcription rate associated with rapid cell growth will aggravate the topological stress associated with transcription and enhance the likelihood of promoter-proximal DSBs. We propose that the oncogenic ability of MYC proteins to enhance transcription-stress resilience of tumor cells is independent of either specific or global changes in gene expression. In our view, the observed MYC-dependent changes in gene expression may be an indirect and context-dependent outcome of this function.
Our model raises two predictions. First, that many phenotypes of MYC activation are likely manifestations of stalling of Pol II, of uncoupling of deregulated nascent transcripts from subsequent processing, or uncoupling of transcription from DNA replication, rather than manifestations of deregulation of hitherto elusive target genes. For example, the ability of MYC to promote cell growth in suboptimal conditions may result from maintaining transcription competence in such conditions. MYC-driven sensitization to apoptosis may result from Pol II stalling following perturbation of cellular nucleotide pools rather than from changes in the relative expression of pro-apoptotic and anti-apoptotic proteins. Conversely, it seems possible that the link between MYC and tumour escape from immune–system surveillance is due to perturbations in transcription and RNA processing in cells over-expressing MYC, which would limit the accumulation of double-stranded RNA that is recognized by pattern recognition receptors [G].
The second prediction is that there are many hitherto unexplored ways to target MYC for tumor therapy. We predict that disrupting the mechanisms that enable MYC to clear promoters from stalled Pol II and disable the coupling of transcription elongation with DNA replication would be therapeutically relevant for MYC- driven and MYCN-driven tumors. Examples include inhibition of the spliceosome and the splicing-associated kinase CLK2, which impairs the survival and tumorigenicity of MYC-dependent cancers121,122, the inhibition of the NUAK1/ARK5 kinase, which controls protein phosphatase 1 complexes that couple transcriptional elongation to spliceosome assembly123,124, as well as the targeting of a MYCN-dependent DNA repair pathway in neuroendocrine prostate carcinoma125. In our view, the systematic analysis of the functions of MYC-interacting proteins in combination with finding new ways to target the stability of proteins will pave the way to rational targeting of MYC functions for tumour therapy.
Glossary.
- Phospho-degron
A short amino acid motif that is recognized by a ubiquitin ligase and promotes the degradation of the protein. Ubiquitin ligases often recognize degrons upon phosphorylation of a critical residue, hence the term phospho-degron.
- Bio-ID techniques
Allow the systematic identification in living cells of proteins that are in close proximity to a protein of interest fused to a biotin ligase.
- Promoter-proximal pausing
A transcription regulatory step, whereby RNA polymerase II pauses about 80 nucleotides downstream of the transcription start site.
- R-loop
A three-stranded nucleic acid structure consisting of an RNA–DNA hybrid and the corresponding single strand of DNA.
- Replication–transcription conflicts
Collusions of the transcription and replication machineries during S phase, which could result in DNA damage and genome instability.
- Pattern recognition receptors
Cellular innate immunity factors that recognize molecules that are typical of pathogens.
Acknowledgements
We thank many colleagues in the field for many fruitful discussions and apologize to the many colleagues whose work we did not quote due to lack of space. The work in the authors’ laboratories is funded by grants from the European Research Council (AuroMYC, TarMyc) and the German Research Council (DFG).
Footnotes
Author contributions
The authors contributed equally to all aspects of the article.
Competing interests
The authors declare no competing interests.
Peer review information
Nature Reviews Molecular Cell biology thanks William Tansey and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Dang CV. MYC on the path to cancer. Cell. 2012;149:22–35. doi: 10.1016/j.cell.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mateyak MK, Obaya AJ, Adachi S, Sedivy JM. Phenotypes of c-Myc-deficient rat fibroblasts isolated by targeted homologous recombination. Cell Growth Differ. 1997;8:1039–1048. [PubMed] [Google Scholar]
- 3.Sansom OJ, et al. Myc deletion rescues Apc deficiency in the small intestine. Nature. 2007;446:676–679. doi: 10.1038/nature05674. [DOI] [PubMed] [Google Scholar]
- 4.Kawauchi D, et al. A mouse model of the most aggressive subgroup of human medulloblastoma. Cancer cell. 2012;21:168–180. doi: 10.1016/j.ccr.2011.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vo BT, et al. The Interaction of Myc with Miz1 Defines Medulloblastoma Subgroup Identity. Cancer cell. 2016;29:5–16. doi: 10.1016/j.ccell.2015.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dammert MA, et al. MYC paralog-dependent apoptotic priming orchestrates a spectrum of vulnerabilities in small cell lung cancer. Nat Commun. 2019;10:3485. doi: 10.1038/s41467-019-11371-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Makela TP, Saksela K, Evan G, Alitalo K. A fusion protein formed by L-myc and a novel gene in SCLC. The EMBO journal. 1991;10:1331–1335. doi: 10.1002/j.1460-2075.1991.tb07652.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nau MM, et al. L-myc, a new myc-related gene amplified and expressed in human small cell lung cancer. Nature. 1985;318:69–73. doi: 10.1038/318069a0. [DOI] [PubMed] [Google Scholar]
- 9.Gabay M, Li Y, Felsher DW. MYC activation is a hallmark of cancer initiation and maintenance. Cold Spring Harbor perspectives in medicine. 2014;4 doi: 10.1101/cshperspect.a014241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Soucek L, et al. Modelling Myc inhibition as a cancer therapy. Nature. 2008;455:679–683. doi: 10.1038/nature07260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Annibali D, et al. Myc inhibition is effective against glioma and reveals a role for Myc in proficient mitosis. Nat Commun. 2014;5:4632. doi: 10.1038/ncomms5632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dubois NC, et al. Placental rescue reveals a sole requirement for c-Myc in embryonic erythroblast survival and hematopoietic stem cell function. Development (Cambridge, England) 2008;135:2455–2465. doi: 10.1242/dev.022707. [DOI] [PubMed] [Google Scholar]
- 13.Sodir NM, et al. Endogenous Myc maintains the tumor microenvironment. Genes & development. 2011;25:907–916. doi: 10.1101/gad.2038411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu CH, et al. Cellular senescence is an important mechanism of tumor regression upon c-Myc inactivation. Proc Natl Acad Sci U S A. 2007;104:13028–13033. doi: 10.1073/pnas.0701953104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Casey SC, et al. MYC regulates the antitumor immune response through CD47 and PD-L1. Science (New York, N Y) 2016;352:227–231. doi: 10.1126/science.aac9935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kortlever RM, et al. Myc Cooperates with Ras by Programming Inflammation and Immune Suppression. Cell. 2017;171:1301–1315.:e1314. doi: 10.1016/j.cell.2017.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Topper MJ, et al. Epigenetic Therapy Ties MYC Depletion to Reversing Immune Evasion and Treating Lung Cancer. Cell. 2017;171:1284–1300.:e1221. doi: 10.1016/j.cell.2017.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Conacci-Sorrell M, McFerrin L, Eisenman RN. An overview of MYC and its interactome. Cold Spring Harbor perspectives in medicine. 2014;4:a014357. doi: 10.1101/cshperspect.a014357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mathsyaraja H, et al. Max deletion destabilizes MYC protein and abrogates Emicro-Myc lymphomagenesis. Genes Dev. 2019;33:1252–1264. doi: 10.1101/gad.325878.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guo J, et al. Sequence specificity incompletely defines the genome-wide occupancy of Myc. Genome biology. 2014;15 doi: 10.1186/s13059-014-0482-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schaub FX, et al. Pan-cancer Alterations of the MYC Oncogene and Its Proximal Network across the Cancer Genome Atlas. Cell Syst. 2018;6:282–300.:e282. doi: 10.1016/j.cels.2018.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hurlin PJ, et al. Deletion of Mnt leads to disrupted cell cycle control and tumorigenesis. EMBO J. 2003;22:4584–4596. doi: 10.1093/emboj/cdg442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Walz S, et al. Activation and repression by oncogenic MYC shape tumour-specific gene expression profiles. Nature. 2014;511:483–487. doi: 10.1038/nature13473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Farrell AS, Sears RC. MYC degradation. Cold Spring Harb Perspect Med. 2014;4 doi: 10.1101/cshperspect.a014365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Herold S, et al. Recruitment of BRCA1 limits MYCN-driven accumulation of stalled RNA polymerase. Nature. 2019;567:545–549. doi: 10.1038/s41586-019-1030-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Welcker M, et al. The Fbw7 tumor suppressor regulates glycogen synthase kinase 3 phosphorylation-dependent c-Myc protein degradation. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:9085–9090. doi: 10.1073/pnas.0402770101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yeh E, et al. A signalling pathway controlling c-Myc degradation that impacts oncogenic transformation of human cells. Nature cell biology. 2004;6:308–318. doi: 10.1038/ncb1110. [DOI] [PubMed] [Google Scholar]
- 28.Richards MW, et al. Structural basis of N-Myc binding by Aurora-A and its destabilization by kinase inhibitors. Proc Natl Acad Sci U S A. 2016;113:13726–13731. doi: 10.1073/pnas.1610626113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dauch D, et al. A MYC-aurora kinase A protein complex represents an actionable drug target in p53-altered liver cancer. Nat Med. 2016;22:744–753. doi: 10.1038/nm.4107. [DOI] [PubMed] [Google Scholar]
- 30.Zhang N, et al. MYC interacts with the human STAGA coactivator complex via multivalent contacts with the GCN5 and TRRAP subunits. Biochim Biophys Acta. 2014;1839:395–405. doi: 10.1016/j.bbagrm.2014.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McMahon SB, Van Buskirk HA, Dugan KA, Copeland TD, Cole MD. The novel ATM-related protein TRRAP is an essential cofactor for the c-Myc and E2F oncoproteins. Cell. 1998;94:363–374. doi: 10.1016/s0092-8674(00)81479-8. [DOI] [PubMed] [Google Scholar]
- 32.Thomas LR, et al. Interaction with WDR5 Promotes Target Gene Recognition and Tumorigenesis by MYC. Molecular Cell. 2015;58:1–13. doi: 10.1016/j.molcel.2015.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kalkat M, et al. MYC Protein Interactome Profiling Reveals Functionally Distinct Regions that Cooperate to Drive Tumorigenesis. Mol Cell. 2018;72:836–848.:e837. doi: 10.1016/j.molcel.2018.09.031. [DOI] [PubMed] [Google Scholar]
- 34.Thomas LR, et al. Interaction of MYC with host cell factor-1 is mediated by the evolutionarily conserved Myc box IV motif. Oncogene. 2016;35:3613–3618. doi: 10.1038/onc.2015.416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nair SK, Burley SK. X-ray structures of Myc-Max and Mad-Max recognizing DNA. Molecular bases of regulation by proto-oncogenic transcription factors. Cell. 2003;112:193–205. doi: 10.1016/s0092-8674(02)01284-9. [DOI] [PubMed] [Google Scholar]
- 36.Wei Y, et al. Multiple direct interactions of TBP with the MYC oncoprotein. Nat Struct Mol Biol. 2019;26:1035–1043. doi: 10.1038/s41594-019-0321-z. [DOI] [PubMed] [Google Scholar]
- 37.Hnisz D, Shrinivas K, Young RA, Chakraborty AK, Sharp PA. A Phase Separation Model for Transcriptional Control. Cell. 2017;169:13–23. doi: 10.1016/j.cell.2017.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Eilers M, Eisenman RN. Myc’s broad reach. Genes & development. 2008;22:2755–2766. doi: 10.1101/gad.1712408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Grandori C, et al. c-Myc binds to human ribosomal DNA and stimulates transcription of rRNA genes by RNA polymerase I. Nature cell biology. 2005;7:311–318. doi: 10.1038/ncb1224. [DOI] [PubMed] [Google Scholar]
- 40.Gomez-Roman N, Grandori C, Eisenman RN, White RJ. Direct activation of RNA polymerase III transcription by c-Myc. Nature. 2003;421:290–294. doi: 10.1038/nature01327. [DOI] [PubMed] [Google Scholar]
- 41.Sabo A, et al. Selective transcriptional regulation by Myc in cellular growth control and lymphomagenesis. Nature. 2014;511:488–492. doi: 10.1038/nature13537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Eilers M, Picard D, Yamamoto KR, Bishop JM. Chimaeras of myc oncoprotein and steroid receptors cause hormone-dependent transformation of cells. Nature. 1989;340:66–68. doi: 10.1038/340066a0. [DOI] [PubMed] [Google Scholar]
- 43.Kress TR, Sabo A, Amati B. MYC: connecting selective transcriptional control to global RNA production. Nature reviews Cancer. 2015;15:593–607. doi: 10.1038/nrc3984. [DOI] [PubMed] [Google Scholar]
- 44.Muhar M, et al. SLAM-seq defines direct gene-regulatory functions of the BRD4-MYC axis. Science. 2018;360:800–805. doi: 10.1126/science.aao2793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stine ZE, Walton ZE, Altman BJ, Hsieh AL, Dang CV. MYC Metabolism, and Cancer. Cancer discovery. 2015;5:1024–1039. doi: 10.1158/2159-8290.CD-15-0507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Herold S, et al. Negative regulation of the mammalian UV response by Myc through association with Miz-1. Molecular cell. 2002;10:509–521. doi: 10.1016/s1097-2765(02)00633-0. [DOI] [PubMed] [Google Scholar]
- 47.Seoane J, Le HV, Massague J. Myc suppression of the p21(Cip1) Cdk inhibitor influences the outcome of the p53 response to DNA damage. Nature. 2002;419:729–734. doi: 10.1038/nature01119. [DOI] [PubMed] [Google Scholar]
- 48.Berwanger B, et al. Loss of a FYN-regulated differentiation and growth arrest pathway in advanced stage neuroblastoma. Cancer Cell. 2002;2:377–386. doi: 10.1016/s1535-6108(02)00179-4. [DOI] [PubMed] [Google Scholar]
- 49.Murphy DM, et al. Dissection of the oncogenic MYCN transcriptional network reveals a large set of clinically relevant cell cycle genes as drivers of neuroblastoma tumorigenesis. Mol Carcinog. 2011;50:403–411. doi: 10.1002/mc.20722. [DOI] [PubMed] [Google Scholar]
- 50.Blanco-Bose WE, et al. C-Myc and its target FoxM1 are critical downstream effectors of constitutive androstane receptor (CAR) mediated direct liver hyperplasia. Hepatology. 2008;48:1302–1311. doi: 10.1002/hep.22475. [DOI] [PubMed] [Google Scholar]
- 51.Yin XY, et al. Inverse regulation of cyclin B1 by c-Myc and p53 and induction of tetraploidy by cyclin B1 overexpression. Cancer Res. 2001;61:6487–6493. [PubMed] [Google Scholar]
- 52.Wanzel M, et al. Akt and 14-3-3eta regulate Miz1 to control cell-cycle arrest after DNA damage. Nat Cell Biol. 2005;7:30–41. doi: 10.1038/ncb1202. [DOI] [PubMed] [Google Scholar]
- 53.Yustein JT, et al. Induction of ectopic Myc target gene JAG2 augments hypoxic growth and tumorigenesis in a human B-cell model. Proc Natl Acad Sci U S A. 2010;107:3534–3539. doi: 10.1073/pnas.0901230107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Seoane J, et al. TGFbeta influences Myc, Miz-1 and Smad to control the CDK inhibitor p15INK4b. Nature cell biology. 2001;3:400–408. doi: 10.1038/35070086. [DOI] [PubMed] [Google Scholar]
- 55.Staller P, et al. Repression of p15INK4b expression by Myc through association with Miz-1. Nature cell biology. 2001;3:392–399. doi: 10.1038/35070076. [DOI] [PubMed] [Google Scholar]
- 56.Herold S, et al. Negative regulation of the mammalian UV response by Myc through association with Miz-1. Mol Cell. 2002;10:509–521. doi: 10.1016/s1097-2765(02)00633-0. [DOI] [PubMed] [Google Scholar]
- 57.Wu S, et al. Myc represses differentiation-induced p21CIP1 expression via Miz-1-dependent interaction with the p21 core promoter. Oncogene. 2003;22:351–360. doi: 10.1038/sj.onc.1206145. [DOI] [PubMed] [Google Scholar]
- 58.Inghirami G, et al. Down-regulation of LFA-1 adhesion receptors by C-myc oncogene in human B lymphoblastoid cells. Science. 1990;250:682–686. doi: 10.1126/science.2237417. [DOI] [PubMed] [Google Scholar]
- 59.Gebhardt A, et al. Myc regulates keratinocyte adhesion and differentiation via complex formation with Miz1. J Cell Biol. 2006;172:139–149. doi: 10.1083/jcb.200506057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.van Riggelen J, et al. The interaction between Myc and Miz1 is required to antagonize TGFbeta-dependent autocrine signaling during lymphoma formation and maintenance. Genes & development. 2010;24:1281–1294. doi: 10.1101/gad.585710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tu WB, et al. MYC Interacts with the G9a Histone Methyltransferase to Drive Transcriptional Repression and Tumorigenesis. Cancer Cell. 2018;34:579–595.:e578. doi: 10.1016/j.ccell.2018.09.001. [DOI] [PubMed] [Google Scholar]
- 62.Baluapuri A, et al. MYC Recruits SPT5 to RNA Polymerase II to Promote Processive Transcription Elongation. Mol Cell. 2019;74:674–687.:e611. doi: 10.1016/j.molcel.2019.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kaur M, Cole MD. MYC acts via the PTEN tumor suppressor to elicit autoregulation and genome-wide gene repression by activation of the Ezh2 methyltransferase. Cancer Res. 2013;73:695–705. doi: 10.1158/0008-5472.CAN-12-2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Barna M, et al. Suppression of Myc oncogenic activity by ribosomal protein haploinsufficiency. Nature. 2008;456:971–975. doi: 10.1038/nature07449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.D’Andrea A, et al. The mitochondrial translation machinery as a therapeutic target in Myc-driven lymphomas. Oncotarget. 2016;7:72415–72430. doi: 10.18632/oncotarget.11719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lin CY, et al. Transcriptional amplification in tumor cells with elevated c-Myc. Cell. 2012;151:56–67. doi: 10.1016/j.cell.2012.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nie Z, et al. c-Myc is a universal amplifier of expressed genes in lymphocytes and embryonic stem cells. Cell. 2012;151:68–79. doi: 10.1016/j.cell.2012.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fernandez PC, et al. Genomic targets of the human c-Myc protein. Genes & development. 2003;17:1115–1129. doi: 10.1101/gad.1067003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lorenzin F, et al. Different promoter affinities account for specificity in MYC-dependent gene regulation. eLife. 2016;5 doi: 10.7554/eLife.15161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gerlach JM, et al. PAF1 complex component Leo1 helps recruit Drosophila Myc to promoters. Proc Natl Acad Sci U S A. 2017;114:E9224–E9232. doi: 10.1073/pnas.1705816114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Guccione E, et al. Myc-binding-site recognition in the human genome is determined by chromatin context. Nature cell biology. 2006;8:764–770. doi: 10.1038/ncb1434. [DOI] [PubMed] [Google Scholar]
- 72.Tesi A, et al. An early Myc-dependent transcriptional program orchestrates cell growth during B-cell activation. EMBO Rep. 2019;20:e47987. doi: 10.15252/embr.201947987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Loven J, et al. Revisiting global gene expression analysis. Cell. 2012;151:476–482. doi: 10.1016/j.cell.2012.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Huang CH, et al. CDK9-mediated transcription elongation is required for MYC addiction in hepatocellular carcinoma. Genes Dev. 2014;28:1800–1814. doi: 10.1101/gad.244368.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chipumuro E, et al. CDK7 inhibition suppresses super-enhancer-linked oncogenic transcription in MYCN-driven cancer. Cell. 2014;159:1126–1139. doi: 10.1016/j.cell.2014.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lewis LM, et al. Replication Study: Transcriptional amplification in tumor cells with elevated c-Myc. Elife. 2018;7 doi: 10.7554/eLife.30274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wolf E, Lin CY, Eilers M, Levens DL. Taming of the beast: shaping Myc-dependent amplification. Trends in Cell Biology. 2014 doi: 10.1016/j.tcb.2014.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Heidelberger JB, et al. Proteomic profiling of VCP substrates links VCP to K6-linked ubiquitylation and c-Myc function. EMBO Rep. 2018;19 doi: 10.15252/embr.201744754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Besche HC, Haas W, Gygi SP, Goldberg AL. Isolation of mammalian 26S proteasomes and p97/VCP complexes using the ubiquitin-like domain from HHR23B reveals novel proteasome-associated proteins. Biochemistry. 2009;48:2538–2549. doi: 10.1021/bi802198q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.von der Lehr N, et al. The F-box protein Skp2 participates in c-Myc proteosomal degradation and acts as a cofactor for c-Myc-regulated transcription. Mol Cell. 2003;11:1189–1200. doi: 10.1016/s1097-2765(03)00193-x. [DOI] [PubMed] [Google Scholar]
- 81.Dominguez-Sola D, et al. Non-transcriptional control of DNA replication by c-Myc. Nature. 2007;448:445–451. doi: 10.1038/nature05953. [DOI] [PubMed] [Google Scholar]
- 82.Popov N, Schulein C, Jaenicke LA, Eilers M. Ubiquitylation of the amino terminus of Myc by SCF(beta-TrCP) antagonizes SCF(Fbw7)-mediated turnover. Nature cell biology. 2010;12:973–981. doi: 10.1038/ncb2104. [DOI] [PubMed] [Google Scholar]
- 83.Jacquet K, et al. The TIP60 Complex Regulates Bivalent Chromatin Recognition by 53BP1 through Direct H4K20me Binding and H2AK15 Acetylation. Mol Cell. 2016;62:409–421. doi: 10.1016/j.molcel.2016.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gorrini C, et al. Tip60 is a haplo-insufficient tumour suppressor required for an oncogene-induced DNA damage response. Nature. 2007;448:1063–1067. doi: 10.1038/nature06055. [DOI] [PubMed] [Google Scholar]
- 85.Rahl PB, et al. c-Myc regulates transcriptional pause release. Cell. 2010;141:432–445. doi: 10.1016/j.cell.2010.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.de Pretis S, et al. Integrative analysis of RNA polymerase II and transcriptional dynamics upon MYC activation. Genome Research. 2017;27:1658–1664. doi: 10.1101/gr.226035.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Buchel G, et al. Association with Aurora-A Controls N-MYC-Dependent Promoter Escape and Pause Release of RNA Polymerase II during the Cell Cycle. Cell Rep. 2017;21:3483–3497. doi: 10.1016/j.celrep.2017.11.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chiu AC, et al. Transcriptional Pause Sites Delineate Stable Nucleosome-Associated Premature Polyadenylation Suppressed by U1 snRNP. Mol Cell. 2018;69:648–663.:e647. doi: 10.1016/j.molcel.2018.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Jaenicke LA, et al. Ubiquitin-Dependent Turnover of MYC Antagonizes MYC/PAF1C Complex Accumulation to Drive Transcriptional Elongation. Molecular cell. 2016;61:54–67. doi: 10.1016/j.molcel.2015.11.007. [DOI] [PubMed] [Google Scholar]
- 90.Eberhardy SR, Farnham PJ. Myc recruits P-TEFb to mediate the final step in the transcriptional activation of the cad promoter. The Journal of biological chemistry. 2002;277:40156–40162. doi: 10.1074/jbc.M207441200. [DOI] [PubMed] [Google Scholar]
- 91.Dejure FR, et al. The MYC mRNA 3’-UTR couples RNA polymerase II function to glutamine and ribonucleotide levels. EMBO J. 2017;36:1854–1868. doi: 10.15252/embj.201796662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Crossley MP, Bocek M, Cimprich KA. R-Loops as Cellular Regulators and Genomic Threats. Mol Cell. 2019;73:398–411. doi: 10.1016/j.molcel.2019.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Shivji MKK, Renaudin X, Williams CH, Venkitaraman AR. BRCA2 Regulates Transcription Elongation by RNA Polymerase II to Prevent R-Loop Accumulation. Cell Rep. 2018;22:1031–1039. doi: 10.1016/j.celrep.2017.12.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Salas-Armenteros I, et al. Human THO-Sin3A interaction reveals new mechanisms to prevent R-loops that cause genome instability. EMBO J. 2017;36:3532–3547. doi: 10.15252/embj.201797208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kouzine F, et al. Transcription-dependent dynamic supercoiling is a short-range genomic force. Nat Struct Mol Biol. 2013;20:396–403. doi: 10.1038/nsmb.2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Madabhushi R, et al. Activity-Induced DNA Breaks Govern the Expression of Neuronal Early-Response Genes. Cell. 2015;161:1592–1605. doi: 10.1016/j.cell.2015.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Haffner MC, et al. Androgen-induced TOP2B-mediated double-strand breaks and prostate cancer gene rearrangements. Nat Genet. 2010;42:668–675. doi: 10.1038/ng.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Gothe HJ, et al. Spatial Chromosome Folding and Active Transcription Drive DNA Fragility and Formation of Oncogenic MLL Translocations. Mol Cell. 2019;75:267–283.:e212. doi: 10.1016/j.molcel.2019.05.015. [DOI] [PubMed] [Google Scholar]
- 99.Hann SR, Thompson CB, Eisenman RN. c-myc oncogene protein synthesis is independent of the cell cycle in human and avian cells. Nature. 1985;314:366–369. doi: 10.1038/314366a0. [DOI] [PubMed] [Google Scholar]
- 100.Otto T, et al. Stabilization of N-Myc is a critical function of Aurora A in human neuroblastoma. Cancer Cell. 2009;15:67–78. doi: 10.1016/j.ccr.2008.12.005. [DOI] [PubMed] [Google Scholar]
- 101.Topham C, et al. MYC Is a Major Determinant of Mitotic Cell Fate. Cancer Cell. 2015;28:129–140. doi: 10.1016/j.ccell.2015.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Xiao D, et al. Polo-like Kinase-1 Regulates Myc Stabilization and Activates a Feedforward Circuit Promoting Tumor Cell Survival. Mol Cell. 2016;64:493–506. doi: 10.1016/j.molcel.2016.09.016. [DOI] [PubMed] [Google Scholar]
- 103.Chakraborty AA, Tansey WP. Inference of cell cycle-dependent proteolysis by laser scanning cytometry. Exp Cell Res. 2009;315:1772–1778. doi: 10.1016/j.yexcr.2009.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Hamperl S, Cimprich KA. Conflict Resolution in the Genome: How Transcription and Replication Make It Work. Cell. 2016;167:1455–1467. doi: 10.1016/j.cell.2016.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Poli J, et al. Mec1 INO80 and the PAF1 complex cooperate to limit transcription replication conflicts through RNAPII removal during replication stress. Genes Dev. 2016;30:337–354. doi: 10.1101/gad.273813.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ross J, et al. Deletion of the Miz-1 POZ Domain Increases Efficacy of Cytarabine Treatment in T-and B-ALL/Lymphoma Mouse Models. Cancer Res. 2019;79:4184–4195. doi: 10.1158/0008-5472.CAN-18-3038. [DOI] [PubMed] [Google Scholar]
- 107.Kress TR, et al. Identification of MYC-Dependent Transcriptional Programs in Oncogene-Addicted Liver Tumors. Cancer research. 2016;76:3463–3472. doi: 10.1158/0008-5472.CAN-16-0316. [DOI] [PubMed] [Google Scholar]
- 108.Bermejo R, et al. The replication checkpoint protects fork stability by releasing transcribed genes from nuclear pores. Cell. 2011;146:233–246. doi: 10.1016/j.cell.2011.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Su Y, et al. Post-translational modification localizes MYC to the nuclear pore basket to regulate a subset of target genes involved in cellular responses to environmental signals. Genes Dev. 2018;32:1398–1419. doi: 10.1101/gad.314377.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Sjostrom SK, Finn G, Hahn WC, Rowitch DH, Kenney AM. The Cdk1 complex plays a prime role in regulating N-myc phosphorylation and turnover in neural precursors. Dev Cell. 2005;9:327–338. doi: 10.1016/j.devcel.2005.07.014. [DOI] [PubMed] [Google Scholar]
- 111.Brockmann M, et al. Small molecule inhibitors of aurora-a induce proteasomal degradation of N-myc in childhood neuroblastoma. Cancer Cell. 2013;24:75–89. doi: 10.1016/j.ccr.2013.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Hydbring P, et al. Phosphorylation by Cdk2 is required for Myc to repress Ras-induced senescence in cotransformation. Proc Natl Acad Sci U S A. 2010;107:58–63. doi: 10.1073/pnas.0900121106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Benassi B, et al. c-Myc phosphorylation is required for cellular response to oxidative stress. Mol Cell. 2006;21:509–519. doi: 10.1016/j.molcel.2006.01.009. [DOI] [PubMed] [Google Scholar]
- 114.Tan J, et al. PDK1 signaling toward PLK1-MYC activation confers oncogenic transformation, tumor-initiating cell activation, and resistance to mTOR-targeted therapy. Cancer Discov. 2013;3:1156–1171. doi: 10.1158/2159-8290.CD-12-0595. [DOI] [PubMed] [Google Scholar]
- 115.Li Q, Dang CV. c-Myc overexpression uncouples DNA replication from mitosis. Mol Cell Biol. 1999;19:5339–5351. doi: 10.1128/mcb.19.8.5339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Goga A, Yang D, Tward AD, Morgan DO, Bishop JM. Inhibition of CDK1 as a potential therapy for tumors over-expressing MYC. Nat Med. 2007;13:820–827. doi: 10.1038/nm1606. [DOI] [PubMed] [Google Scholar]
- 117.Ricke RM, Jeganathan KB, Malureanu L, Harrison AM, van Deursen JM. Bub1 kinase activity drives error correction and mitotic checkpoint control but not tumor suppression. J Cell Biol. 2012;199:931–949. doi: 10.1083/jcb.201205115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Kessler JD, et al. A SUMOylation-dependent transcriptional subprogram is required for Myc-driven tumorigenesis. Science. 2012;335:348–353. doi: 10.1126/science.1212728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Chen L, et al. The Augmented R-Loop Is a Unifying Mechanism for Myelodysplastic Syndromes Induced by High-Risk Splicing Factor Mutations. Mol Cell. 2018;69:412–425.:e416. doi: 10.1016/j.molcel.2017.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Bartkova J, et al. Oncogene-induced senescence is part of the tumorigenesis barrier imposed by DNA damage checkpoints. Nature. 2006;444:633–637. doi: 10.1038/nature05268. [DOI] [PubMed] [Google Scholar]
- 121.Hsu TYT, et al. The spliceosome is a therapeutic vulnerability in MYC-driven cancer. Nature. 2015;525:384–388. doi: 10.1038/nature14985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Iwai K, et al. Anti-tumor efficacy of a novel CLK inhibitor via targeting RNA splicing and MYC-dependent vulnerability. EMBO Mol Med. 2018;10 doi: 10.15252/emmm.201708289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Cossa G, et al. Localized inhibition of protein phosphatase 1 by NUAK1 promotes spliceosome activity and reveals a MYC-sensitive feedback control of transcription. Molecular Cell. 2020 doi: 10.1016/j.molcel.2020.01.008. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Liu L, et al. Deregulated MYC expression induces dependence upon AMPK-related kinase 5. Nature. 2012;483:608–612. doi: 10.1038/nature10927. [DOI] [PubMed] [Google Scholar]
- 125.Zhang W, et al. Targeting the MYCN-PARP-DNA Damage Response Pathway in Neuroendocrine Prostate Cancer. Clin Cancer Res. 2018;24:696–707. doi: 10.1158/1078-0432.CCR-17-1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Goldenberg O, Erez E, Nimrod G, Ben-Tal N. The ConSurf-DB: pre-calculated evolutionary conservation profiles of protein structures. Nucleic Acids Res. 2009;37:D323–327. doi: 10.1093/nar/gkn822. [DOI] [PMC free article] [PubMed] [Google Scholar]