Abstract
Background
Past history of gallstones is associated with increased risk of gallbladder cancer (GBC) in observational studies. We conducted complementary observational and Mendelian Randomization (MR) analyses to determine whether history of gallstones is causally related to development of GBC in an Indian population.
Methods
To investigate associations between history of gallstones and GBC, we used questionnaire and imaging data from a GBC case-control study conducted at Tata Memorial Hospital, Mumbai (cases=1170; controls=2525). We then used 26 genetic variants identified in a genome-wide association study of 27,174 gallstones cases and 736,838 controls of European ancestry in a Mendelian randomization approach to assess causality. The association of these genetic variants with both gallstones and GBC was examined in the GBC case-control study. Various complementary MR approaches were used to evaluate the robustness of our results in the presence of pleiotropy and heterogeneity, and to consider the suitability of the selected SNPs as genetic instruments for gallstones in an Indian population.
Results
We found a strong observational association between gallstones and GBC using self-reported history of gallstones (OR=4.5, 95%CI=3.5-5.8) and with objective measures of gallstone presence using imaging techniques (OR=2.0, 95%CI=1.5-2.7). We found consistent causal estimates across all MR techniques, with odds ratios for GBC in the range of 1.3-1.6.
Conclusion
Our findings indicate a causal relationship between history of gallstones and increased risk of GBC, albeit of a smaller magnitude to those found in observational analysis.
Impact
Our findings emphasise the importance of gallstone treatment for preventing GBC in high risk individuals.
Keywords: Gallstones, Gallbladder cancer, GWAS, Mendelian Randomization
Introduction
Although gallbladder cancer (GBC) is rare in most parts of the world, it is more common in certain populations, including some parts of India, Chile and Mexico (1). While the aetiology of GBC remains poorly understood, increased risk of GBC has been associated with Salmonella Typhi infection (2), possession of certain genetic variants (3), obesity (4) and high consumption of mustard oil (5). In addition, a personal history of gallstones has long been found to be strongly related to GBC risk (6).
While a causal effect of gallstones on GBC is probable, variation in estimates of risk have been observed in previous studies, most likely due to differences in study design and the methods used to collect information on gallstones (7). In particular, case-control studies with questionnaire-based retrospective ascertainment of history of gallstones could result in differential misclassification and a biased estimate of risk, since cases might report history of gallstones more frequently than controls as they undergo various investigative procedures, including ones that may uncover silent gallstones. The case-control design also raises the issue of temporality, i.e. it cannot be determined whether the gallstone occurred prior to, or after, the development of GBC.
It is of clinical importance to confirm causality for the association of gallstones with GBC, which could direct intervention strategies in countries with high risk of GBC, since treatment for gallstones is considerably lower in cost, complexity and risk compared to treatment of GBC. In order to better elucidate the role of gallstones history in the aetiology of GBC, and to determine the magnitude of any causal effect, we examined potential biases in the estimation of GBC risk using both self-report and objective measures of gallstone history (from imaging techniques) in a large Indian case-control study of GBC. In addition, we used genetic variants strongly associated with risk of gallstones (8) to estimate the causal effect of gallstones on risk of GBC within a Mendelian randomization (MR) framework. MR is a technique that uses genetic variants as unconfounded proxies for an exposure of interest, which are less susceptible to problems of measurement error and reverse causation, as a method to ascertaining better evidence for causality (9).
Materials and Methods
Study Design
A case-control study was conducted at the Tata Memorial Hospital (TMH), Mumbai from 2010 to 2015 in order to evaluate the relationship between lifestyle and genetic risk factors and GBC. Details of the methodology are published elsewhere. (10) In brief, a total of 1,170 GBC cases and 2,525 visitor controls were enrolled into the study. All cases of GBC (International Classification of Diseases for Oncology Version 3 [ICD-O-3] site code C23) were microscopically confirmed. Controls were recruited from friends, neighbours, colleagues, in-laws, spouses and relatives (other than first-degree relatives) visiting Tata Memorial Hospital. Controls were frequency-matched to cases on age (± 10 years), gender, and region. Matching by geographical region (north, north-east, and west, central and south) was conducted using reported place of current residence at the time of enrolment. We obtained written informed consent from all study participants before enrolment and obtained ethical approval from all relevant local and hospital-based institutional review boards.
A structured questionnaire was used to obtain self-reported history of gallstones along with other lifestyle and environmental risk factors. In order to procure more objective measures of gallstone history, we examined the case records of all GBC cases for imaging techniques such as Ultrasonography (USG), Magnetic resonance imaging (MRI), computed tomography (CT) scan and accordingly classified them as being with or without gallstones. As the visitor controls that were used for the observational study did not undergo similar kinds of imaging investigations, we used breast cancer cases as a second control group as a comparator for this aspect of the study (11). Breast cancer cases were recruited as a control group as virtually all breast cancer cases undergo imaging evaluation (particularly USG) and thus it is possible to look for evidence of the presence of gallstones in their case records. This allowed an additional comparison of female GBC cases with female breast cancer cases as a control for the presence of gallstones confirmed by imaging. Breast cancer cases were enrolled during the same study period using the same questionnaire and case control study design as used for the GBC study.
Genetic data
Participants in the GBC study were genotyped on the HumanOmniExpress-24 version 1.1 IlluminaBeadChip array (Illumina, San Diego, CA, USA) at the Centre for Cancer Epidemiology, Tata Memorial Centre (3). SNPs with a call rate of less than 95% (cut-off level empirically determined), failure to meet Hardy-Weinberg equilibrium at p < 10-6 or a minor allele frequency (MAF) of <0·005 were excluded in quality control procedures. The study also excluded samples with a call rate of less than 90% (cut-off level empirically determined), no intensity, gender discordance (>5% heterozygosity on the basis of the X chromosome SNPs for males or <15% heterozygosity on the basis of the X chromosome SNPs for females), first-degree relatives who were also genotyped in the study on the basis of identity by descent (pi-hat >0·48), and unexpected duplicates. After genotyping, we performed genome-wide imputation to statistically infer untyped variants using IMPUTE2 software version 2.2.2 (12) and version 3 of the 1000 Genomes Project data as the reference set (13). We included 10·4 million SNPs, which were imputed with an INFO score of more than 0·3 and a MAF of more than 0·5%, for analysis. A genome-wide association study (GWAS) of GBC was performed among cases and controls, as previously described (3). The GWAS estimates were adjusted for age, sex, and five significant eigenvectors of the derived principal components.
32 single nucleotide polymorphisms (SNPs) associated with gallstones at a genome-wide level of significance (p<5 x 10-8) have been identified in a previous genome-wide association study (GWAS) (8). Two SNPs (rs756082276, rs756935975) were neither genotyped nor imputed in the GBC study (3). Two SNPs (rs34851490, rs45575636) which were in linkage disequilibrium with other SNPs used in the analysis (r2>0.001) were excluded. Similarly, 2 palindromic SNPs (rs2469991, rs1935) were excluded. The remaining 26 SNPs were used in both one-sample and two-sample MR. All 32 SNPs, their location and nearby gene, with reasons for exclusion are tabulated in Supplementary Table 1.
Statistical Analysis
Observational analysis
The association between gallstones and GBC was initially assessed using self-reported gallstone history. Logistic regression models were adjusted for the following potential confounders: age, current residential region (North, North-East, West, Central and South), education (<5 years, ≥5 years), gender, tobacco-chewing and/or smoking (yes/no), and waist-to-hip ratio (continuous variable), per capita per month mustard oil consumption (continuous), and per capita per week fresh fish consumption (continuous). Mustard oil and fresh fish consumption have been included as potential confounders since both have been associated with gall bladder cancer in the same dataset under investigation (5). To avoid bias due to reporting of gallstones as a consequence of development of cancer, we conducted sensitivity analysis by removing participants with a history of gallstones within one year of diagnosis or interview for cases and controls respectively.
In order to obtain a more objective measure of gallstones, we used information on the presence of gallstones amongst GBC cases confirmed by imaging techniques. The information about the presence of gallstones amongst breast cancer cases was similarly ascertained and breast cancer cases were used as a control. Logistic regression was then used to estimate risk of image-confirmed gallstones on GBC among females only.
The associations obtained were multiplied by 0.693 to observe the change in odds of GBC per two-fold increase (doubling) in the odds of gallstones, to be compared with the results of MR. All observational analyses were performed using the statistical package STATA version 15.0.
Mendelian Randomization analysis
Two-sample MR
We used a two-sample MR approach to estimate the risk of gallstones on the development of GBC, using the TwoSampleMR package in R version 3.5.1 (14). For each of the SNPs robustly associated with gallstones, information on the SNP-exposure (gallstones) and SNP-outcome (GBC) effects were obtained from genome-wide association analyses conducted in separate studies (3, 8). Specifically, we performed a look-up of the 26 SNPs associated with gallstones from the GWAS of GBC (3) and extracted the following summary data for each SNP: the effect estimate (logOR) for GBC per copy of the effect allele and its standard error, the reference allele, and the effect allele along with its frequency. We then combined information on the SNP-gallstone associations from the previous GWAS with information on the SNP-GBC associations and performed MR, using the inverse variance weighted (IVW) approach. For this, we first calculated the causal effect of gallstones on GBC by calculating the SNP-specific Wald ratios (β(outcome~SNP)/β(exposure~SNP) (where β reflects the log odds). Standard errors of the Wald ratios were approximated by the delta method. Wald ratios for each SNP were combined in a fixed effects meta-analysis after weighting each ratio estimate by the inverse variance of their associations with the outcome. We also conducted random effects meta-analysis to allow for heterogeneity in the individual SNP effect estimates. The causal estimate obtained was multiplied by log2(=0.693) to yield the change in log odds of GBC per doubling in the odds of gallstone (15).
In order to account for horizontal pleiotropy, whereby the genetic variants might affect GBC through pathways other than via gallstones, we used different MR methods, namely the weighted median (16), MR Egger (17), and modal estimates (18). The weighted median estimator takes the median value of the Wald ratio over the SNPs, and is robust to outliers as the estimator is consistent for the causal effect if at least half the information in the analysis comes from valid instruments (16). The MR-Egger approach is similar to the IVW approach except that the intercept is not fixed to zero, and so allows for directional pleiotropy. An intercept term that differs from zero is indicative of overall directional pleiotropy (17). The MR-Egger approach will return a valid causal estimate in the presence of directional pleiotropy. The MR modal estimates assume that the most common causal effect is consistent with the true causal effect (18). As each of these methods make different assumptions regarding horizontal pleiotropy, a consistent effect across multiple MR methods strengthens causal evidence (19).
For the graphical representation of the effect of each SNP on GBC, we constructed a forest plot of Wald ratios, with summary effect estimates derived using the MR-Egger and IVW approaches, as well as a scatter plot of the SNP associations with both gallstones and GBC. For assessing asymmetry of SNP-specific causal estimates and thus directional pleiotropy, we provide a funnel plot for visual assessment.
To further detect and correct causal estimates for potential violation of the MR assumptions, we assessed heterogeneity in the causal effects estimated by each SNP by generating Cochran’s Q statistics (20). We performed Radial MR (21) and the MR Pleiotropy Residual Sum and Outlier (MR-PRESSO) test (22) to detect and correct for influential outliers. Both approaches can be used to identify outliers which may represent strong and potentially highly pleiotropic instruments. Using Radial MR, SNPs with the largest contribution to Cochran’s Q for heterogeneity were identified removed and the data reanalysed. We also plotted a Galbraith radial plot for straightforward detection of outliers and influential data points. Three parts of the MR-PRESSO test were also conducted: 1) the MR-PRESSO global test which identifies horizontal pleiotropy, 2) the outlier corrected causal estimates which correct for detected horizontal pleiotropy and 3) the MR-PRESSO distortion test which determines if causal estimates differ after adjustment for outliers. We also performed the contamination mixture method for MR which can i) identify subgroups of genetic variants with similar causal estimates and therefore similar mechanisms of action, and ii) obtain robust causal estimates in the presence of invalid instruments (23).
One-sample MR
One assumption of two-sample MR is that the exposure and outcome datasets are homogeneous with respect to the underlying populations (24). As the GWAS for gallstones was conducted in a European population and the GWAS for GBC in an Indian population, we carried out further analyses to assess the suitability of the selected SNPs as genetic instruments for gallstones in the Indian population.
We investigated associations of the previously identified gallstone SNPs in relation to self-reported history of gallstones in our study. We also performed a one-sample MR (25) using data on SNPs, gallstones, and GBC status for all participants in our case-control study to address the issue of population heterogeneity. For this, we generated a genetic risk score (GRS) for gallstones, calculated as the sum of the number of gallstone-increasing alleles for each of the individuals in our study (26).
To estimate the risk of GBC using a one-sample MR approach, we used the two–stage least squares method (27, 28). In the first stage, the self-reported history of gallstones (exposure) was regressed on the GRS. The predicted values of the gallstones were taken from the first-stage regression model. In the second stage, the GBC (outcome) was regressed over the predicted values of the exposure by logistic regression. We calculated the F-statistic of the GRS on gallstone to assess instrument strength. To test the MR assumption that genetic variants should not be associated with confounders of exposure-outcome relation, we investigated associations between GRS and potential confounders (age, gender, residential region, waist-to-hip ratio, mustard oil consumption, and fresh fish consumption).
Results
Conventional epidemiologic analyses
The overall prevalence of self-reported gallstone history was 40.0% among cases and 2.1% among controls. Gallstone prevalence was higher in the north and north-eastern geographical regions compared to other geographical regions in India (Table 1). The OR per doubling of exposure to gallstones based on self-reported gallstone history and GBC was observed to be 10.0 (95%CI=8.0-12.8) (Table 2). After removing participants with history of gallstones one year prior to diagnosis/interview, the OR was attenuated but still large (OR=4.5; 95%CI=3.5-5.8). The objective measure of gallstone history with 580 female GBC cases and 787 female breast cancer cases treated as “controls” revealed a similar increase in the risk of developing GBC, albeit of a much smaller magnitude (OR=2.0; 95%CI=1.5-2.7).
Table 1. Prevalence of gallstones among gallbladder cancer (GBC) case-control study participants.
| Variable | Presence of gallstone | ||
|---|---|---|---|
| GBC (n=1,170) N (%) | Controls (n = 2,525) N (%) | ||
| Self-reported gallstone using stringent definition b | 145 (12.4) | 54 (2.1) | |
| Self-reported gallstone historya | 468 (40.0) | 60 (2.4) | |
| Gender | Male | 120 (25.6) | 13 (21.7) |
| Female | 348 (74.4) | 47 (78.3) | |
| Geographical regions | North | 207 (44.2) | 16 (26.7) |
| North East | 181 (38.7) | 28 (46.7) | |
| South | 1 (0.2) | 1 (1.6) | |
| West | 47 (10.0) | 12 (20.0) | |
| Central | 32 (6.8) | 3 (5.0) | |
aSelf-Reported Gallstone; reported by study participant as either present/not present.
bSelf reported gallstone using stringent definition; Gallstone history was ascertained using definition of self-reported gallstone; however those gallstones diagnosed within a year prior to the date of diagnosis of gallbladder cancer for cases or within a year prior to the date of interview for controls were excluded from the analysis
Table 2. Odds Ratio (OR) and 95% Confidence Interval (CI) for GBC in relation to gallstone using self-reported and image verified gallstone.
| Gallstone History | Self-reported gallstone history a | Self-reported gallstone using stringent definition b | Gallstone history as determined by imaging techniques c | ||||||
|---|---|---|---|---|---|---|---|---|---|
| N(Cases | Controls) | OR(95%CI)d | Pd | N(Cases | Controls) | OR(95%CI)d | Pd | N(Cases | Controls) | OR(95%CI)e | Pe | |
| Never | 693|2455 | Reference | 693|2455 | Reference | 580|787 | Reference | |||
| Ever | 468|60 | 10.0 | ≤0.001 | 145|54 | 4.5 | ≤0.001 | 208|58 | 2.0 | ≤0.001 |
| (8.0-12.8) | (3.5-5.8) | (1.5-2.7) | |||||||
Abbreviations: CI, Confidence Interval; OR, Odds ratio
aSelf reported Gallstone; as per reported by study participant either present/not present.
bSelf reported gallstone using stringent definition; Gallstone history was ascertained using definition of self-reported gallstone; however those gallstones diagnosed within a year prior to the date of diagnosis of gallbladder cancer for cases or within a year prior to the date of interview for controls were excluded from the analysis
c Gall stone history as determined by imaging techniques (USG, MRI and CT scans) and using breast cancer cases as a control group.
d Adjusted for age (continuous), education (less than 5 years schooling, ≥5 year of education), current residential region (north, north-east, central, west, south), gender, waist-to-hip ratio (continuous), tobacco chewing and tobacco smoking (yes/no), per capita per month mustard oil consumption (continuous), per capita per week fresh fish consumption (continuous)
e Same as d; except not adjusted for gender and additionally adjusted for the number of full-term pregnancies
Missing values were excluded from analysis
Genetic associations
The genetic variants used for MR were obtained from a GWAS of gallstones conducted in Europeans. A comparison between European and Indian populations with respect to allele frequencies, risk of developing gallstones and GBC for the genetic variants was made and results are shown in Supplementary Table 1. The allele frequencies between the two populations were similar, although with striking differences for some SNPs (e.g. for rs601338, rs1260326, rs174567, rs2469991, rs2290846, where the difference in minor allele frequency was >15%). The risk for developing gallstones and GBC were in broadly the same direction for the SNPs in the Indian population (consistently increased risk for 80% of SNPs in relation to gallstones and 70% SNPs in relation to GBC). When assessed individually, one SNP (rs11887534) showed an increased risk for gallstones and two SNPs (rs11887534, rs686030) showed an increased risk for GBC in the Indian population which surpassed Bonferroni correction (p<0.002).
Two-sample MR
When 26 SNPs were used to estimate risk between gallstones and GBC, the results of MR were similar when we compared causal effects using the IVW and pleiotropy-robust methods, and ranged from OR=1.34-1.62 (Table 3, Supplementary Figures 1-3). The analysis of the intercept in the MR-Egger test did not provide strong evidence for directional pleiotropy (Egger Intercept=-0.014, p=0.77) (Table 3, Supplementary Figures 1-3). However, the Cochrane’s Q test indicated heterogeneity in the individual SNP effects (QIVW=72.95, p=1.4x10-6 and QMR Egger=72.69, p=8.0x10-7). The Radial MR method indicated 5 outlier SNPs (Supplementary Table 2, Supplementary Figure 4) contributing to heterogeneity in the causal estimates. We repeated the analysis using the various MR methods after removing these outlier SNPs and IVW and MR-Egger effect estimates were largely unchanged (Table 3). The MR-PRESSO analysis also provided no strong evidence of distortion in causal estimates after adjustment for outliers (p=0.303) (Table 3). In addition, the contamination mixture method did not identify any clusters of variants representing distinct causal mechanisms.
Table 3. Estimates from various Mendelian randomization methods for the association between gallstone and gallbladder cancer.
| Method | OR | 95% LCI | 95% UCI | P value |
|---|---|---|---|---|
| MR estimates (SNPs=26) | ||||
| MR Egger | 1.62 | 1.00 | 2.63 | 0.062 |
| Weighted median | 1.49 | 1.15 | 1.94 | 0.003 |
| Inverse variance weighted | 1.53 | 1.16 | 2.02 | 0.003 |
| Simple mode | 1.59 | 0.96 | 2.65 | 0.084 |
| Weighted mode | 1.34 | 1.09 | 1.65 | 0.011 |
| Test for directional horizontal pleiotropy test | ||||
| Egger intercept (SE) | -0.014 (0.046) | 0.774 | ||
| MR estimates After removing outlier SNPs(n=21) | ||||
| MR Egger | 1.54 | 1.12 | 2.13 | 0.016 |
| Weighted median | 1.60 | 1.22 | 2.10 | ≤0.001 |
| Inverse variance weighted | 1.50 | 1.23 | 1.83 | ≤0.001 |
| Simple mode | 1.86 | 1.07 | 3.24 | 0.040 |
| Weighted mode | 1.62 | 1.24 | 2.11 | 0.002 |
| Test for directional horizontal pleiotropy test after removing outlier SNPs(n=21) | ||||
| Radial Egger intercept (SE) | -0.007(0.028) | 0.814 | ||
| MR-PRESSO raw estimates(SD) | 1.53(0.21) | 0.006 | ||
| MR-PRESSO outlier corrected estimate(SD) | 1.39(0.14) | 0.003 | ||
| Distortion coefficient | 0.303 | |||
| Contamination mixture method | 1.57 | 1.00 | 1.99 | 0.050 |
Abbreviations: LCI, Lower Confidence Interval; NCI, Upper Confidence Interval; OR, Odds ratio
One-sample MR
Since the genetic variants were determined from a population of non-Indian ancestry (European ancestry), we performed one-sample MR approach using a genetic risk score (GRS) derived in the Indian case-control in order to confirm that the estimates in the two-samples MR analyses were not affected by subtle differences in the genetic architecture of the different populations.
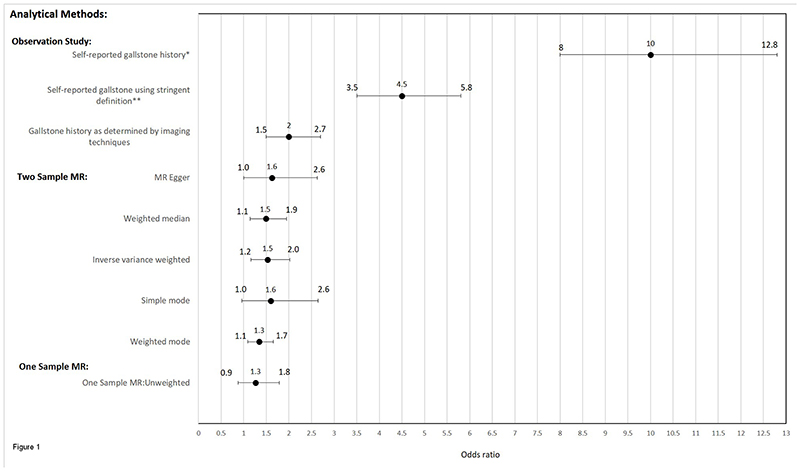
Using a GRS, the risk of developing gallstones was found to increase with an increasing number of risk alleles (OR=1.03; p=0.034, per allele increase), suggesting that these SNPs contributed to risk of developing gallstones in Indians as well as Europeans. The GRS was a strong instrument for the presence of gallstones (F-statistic=11.62, r2=0.024). None of the potential confounders for GBC considered in this study were associated with the GRS (Supplementary Table 3). In two-stage least squares analysis, using the GRS as an instrument for gallstones, we observed an OR of 1.26 (95%CI=0.88-1.79) per doubling of exposure to gallstones. Findings from the one-sample MR were largely consistent with those from the two-sample MR, although confidence intervals crossed the null (Figure 1).
Figure 1. Summary of results from analytical methods assessing gallstones and gallbladder cancer used in this study.
Odds ratios and confidence intervals shown represent the risk of GBC per doubling in liability to gallstone formation
*Self-reported gallstones; reported by study participant as either present or not present
**Self-reported gallstones using stringent definition of self-report, where those diagnosed within a year prior to the date of diagnosis of gallbladder cancer for cases or within a year prior to the date of interview for the controls were excluded from the analysis
Discussion
We consistently observed an increased risk of GBC for individuals with a history of gallstones using various methods to assess personal history of gallstones. Estimates varied in line with the robustness of the exposure data (i.e. measurement of the presence of gallstones) and risk of bias in case-control analysis, as has previously observed (7, 29, 30). The association remained although attenuated when we conducted analysis using more stringent and more objective assessments of the presence of gallstones. Furthermore, evidence of a causal effect was also provided by Mendelian randomization, which is less susceptible to issues of confounding, reverse causation and measurement error, although estimates were of a smaller magnitude to those found in the observational analysis. The effect estimates obtained from both imaging techniques (OR=2.0, 95%CI=1.5-2.7) and MR analysis (ORs ranging from 1.3-1.6) were similar, and largely consistent with findings from a prospective cohort study of screen-detected gallstone disease and gastrointestinal cancer (HR=1.41, 95% CI 1.01, 1.97) (30) as well as a cohort study of self-reported gallstones and gallbladder cancer (HR=3.10, 95% CI 1.55, 6.19) (31).
The use of the MR approach is particularly useful in this setting, since genetic variants robustly associated with liability to form gallstones can be used to appraise their causal role in GBC development. In the absence of any reliable, unbiased objective measures of the presence or absence of actual gallstones, inference can be made about their likely role in GBC. Genetic proxies for gallstones have been identified in several large genome wide association studies (8) and these genetic proxies were tested for association in 1,042 GBC cases and 1,709 controls. We applied several MR methods that rely on different underlying assumptions to evaluate the causal relationship between liability to form gallstones and risk of GBC. We observed consistent estimates of increased risk of GBC using these different MR approaches. An increased risk of GBC was similarly observed when we conducted one-sample MR using a GRS.
The MR findings supporting a causal role for gallstones in GBC has strong biological plausibility. Of note, many of the genetic variants identified to proxy the presence of gallstones are associated with cholesterol metabolism (8). Cholesterol metabolism is known to play a fundamental role in gallstone formation. In addition, some variants (rs56398830 and rs55971546) are located proximal to the bile acid transporter, SLC10A2 gene. The main function of these transporters is to reabsorb bile salts from the terminal ileum into ileocytes, after which the bile salts are transported back to the liver through the entero-hepatic circulation. As some of the identified variants relates to gallstone risk through pathways which are modifiable (cholesterol levels and bile salts) and relate to genetic liability to gallstones, any strategy to intervene on these pathways has the potential to reduce GBC risk. For example, previous studies have demonstrated a reduction in gallstone disease and biliary tract cancer among long-term users of statins (32–34). However, it should be acknowledged that targeting a single pathway such as cholesterol or bile salts is unlikely to be entirely sufficient for minimising risk and the contamination mixture method did not identify any clusters of variants representing distinct causal pathways.
Several limitations in the study require acknowledgement. While the use of breast cancer cases as “controls” provided us with an opportunity to compare cases and controls who have gone through similar imaging technique procedures, this approach has some limitations. The breast cancer cases might have come from a different source population to the cases. Further, it is possible that some risk factors are common between GBC and breast cancer which might attenuate effect estimates, although we adjusted for additional confounders such as adiposity in the analysis. In addition, even with more objective measure of gallstones, we cannot completely rule out misclassification in measurement. The imaging techniques may not completely capture gallstone history and may detect silent gallstones which are not present years later (35). This is an important consideration since a long duration of gallstones may be necessary to induce chronic trauma to the mucosa, which initiates the sequence of pathological changes resulting in cancer development (36).
While MR offers several advantages to observational analysis, it also relies of various assumptions and any violation of these may bias causal estimates. The core MR assumptions are that: 1) the genetic instrument is strongly associated with the exposure; 2) the genetic instrument is independent of confounding factors and 3) the genetic instrument is only related to the outcome via the exposure of interest (i.e. no horizontal pleiotropy) (9). The genetic variants used in MR may be imperfect at capturing gallstone formation. In particular, since the SNPs were identified in a large GWAS of gallstones among individuals of European ancestry, they may not provide an adequate instrument for gallstone formation in Indians. We carried out analyses to assess the suitability of the selected SNPs as genetic instruments for gallstones in the Indian population. Although the risk for developing gallstones was in the same direction for 80% of SNPs, when assessed individually just one SNP (rs11887534) showed a strong association with gallstones in Indians. While we demonstrated adequate instrument strength of the genetic variants when combined into an GRS in this Indian population (F-stat>10), the identification of genetic variants robustly related to gallstone formation among Indians would be of particular use for confirming the effects observed. With respect to the second assumption, we demonstrated that this genetic instrument was not strongly associated with any potential confounders, unlike self-reported gallstone history. The use of complementary MR approaches allowed us to assess the third assumption and infer that any pleiotropic effects do not seriously distort effect estimates. Using Radial plots, we were able to identify and then remove 5 outlier SNPs, potentially involving pleiotropic pathways to GBC which did not involve gallstone formation. These variants have been most consistently associated with blood metabolite levels and measures of cholesterol, as identified in PhenoScanner (37). Two of these SNPs were found in the HNF4 region which is associated with diabetes, while two variants were found in the FUT2 and FUT6 regions, which are responsible for glycosylation in the gastrointestinal tract (38). We repeated the MR analysis after removing these outliers and the results were broadly consistent, in term of the direction and strength of association observed.
Implications of findings
The consistency of the association between history of gallstones and GBC using a range of analytical methods leads us to believe that this association reflects a causal effect of gallstones on GBC. The findings provide important evidence that is otherwise not feasible to ascertain given the challenges inherent in conducting a randomised controlled trial of gallstone treatment in populations at risk of GBC. Our findings have major implications in developing preventive strategies for GBC, including the early detection and/or prophylactic treatment of gallstones in high risk individuals. As ultrasonography may be difficult to conduct in the field for identification of gallstones especially in resource-poor settings, a panel of SNPs associated with gallstone formation could potentially be used as triage for identifying liability to form gallstones. This could be used to plan interventional strategies to reduce the risk of gallstone formation and consequently reducing the risk of GBC.
Supplementary Material
Acknowledgements
S. Mhatre and R. P. Dikshit are supported by the The Tata Memorial Centre, Department of Biotechnology (DBT-COE grant number BT/01CEIB/09/V/06). R. C. Richmond is a de Pass VC research fellow at the University of Bristol. R. C. Richmond, G. Davey Smith and C. L. Relton are supported by a Cancer Research UK (C18281/A19169) programme grant (the Integrative Cancer Epidemiology Programme) and are part of the Medical Research Council Integrative Epidemiology Unit at the University of Bristol supported by the Medical Research Council (MC_UU_00011/1 and MC_UU_00011/5) and the University of Bristol. G. Davey Smith and C. L. Relton are supported by the National Institute for Health Research (NIHR) Bristol Biomedical Research Centre which is funded by the National Institute for Health Research (NIHR) and is a partnership between University Hospitals Bristol NHS Foundation Trust and the University of Bristol. Department of Health and Social Care disclaimer: The views expressed are those of the authors and not necessarily those of the NHS, the NIHR or the Department of Health and Social Care.
Abbreviations
- GBC
gallbladder cancer
- GWAS
genome-wide association studies
- MR
Mendelian randomization
- OR
odds ratio
- PRESSO
Mendelian Randomization Pleiotropy Residual Sum and Outlier
- SNP
single nucleotide polymorphism
Footnotes
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed by the authors.
Authors’ contributions
Conception and design: S. Mhatre, R. C. Richmond, G. Davey Smith, C. L. Relton, R. P. Dikshit
Statistical analysis: S. Mhatre and R. C. Richmond
Acquisition of data: S. Mhatre, R. Badwe, M. Goel, S. Patkar, S. V. Shrikhande, P. S. Patil, R. P. Dikshit
Interpretation of data: All authors
Writing initial draft of manuscript: S. Mhatre, R. C. Richmond, G. Davey Smith, C. L. Relton, R. P. Dikshit
Critical review of manuscript: All authors
References
- 1.Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136(5):E359–86. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 2.Tewari M, Mishra RR, Shukla HS. Salmonella typhi and gallbladder cancer: report from an endemic region. Hepatobiliary Pancreat Dis Int. 2010;9(5):524–30. [PubMed] [Google Scholar]
- 3.Mhatre S, Wang Z, Nagrani R, Badwe R, Chiplunkar S, Mittal B, et al. Common genetic variation and risk of gallbladder cancer in India: a case-control genome-wide association study. Lancet Oncol. 2017;18(4):535–44. doi: 10.1016/S1470-2045(17)30167-5. [DOI] [PubMed] [Google Scholar]
- 4.Hsing AW, Sakoda LC, Rashid A, Chen J, Shen MC, Han TQ, et al. Body size and the risk of biliary tract cancer: a population-based study in China. Br J Cancer. 2008;99(5):811–5. doi: 10.1038/sj.bjc.6604616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mhatre S, Rajaraman P, Chatterjee N, Bray F, Goel M, Patkar S, et al. Mustard oil consumption, cooking method, diet and gallbladder cancer risk in high- and low-risk regions of India. Int J Cancer. 2020 doi: 10.1002/ijc.32952. [DOI] [PubMed] [Google Scholar]
- 6.Lowenfels AB, Lindstrom CG, Conway MJ, Hastings PR. Gallstones and risk of gallbladder cancer. J Natl Cancer Inst. 1985;75(1):77–80. [PubMed] [Google Scholar]
- 7.Randi G, Franceschi S, La Vecchia C. Gallbladder cancer worldwide: geographical distribution and risk factors. Int J Cancer. 2006;118(7):1591–602. doi: 10.1002/ijc.21683. [DOI] [PubMed] [Google Scholar]
- 8.Ferkingstad E, Oddsson A, Gretarsdottir S, Benonisdottir S, Thorleifsson G, Deaton AM, et al. Genome-wide association meta-analysis yields 20 loci associated with gallstone disease. Nat Commun. 2018;9(1):5101. doi: 10.1038/s41467-018-07460-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davey Smith G, Hemani G. Mendelian randomization: genetic anchors for causal inference in epidemiological studies. Hum Mol Genet. 2014;23(R1):R89–98. doi: 10.1093/hmg/ddu328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mhatre SS, Nagrani RT, Budukh A, Chiplunkar S, Badwe R, Patil P, et al. Place of birth and risk of gallbladder cancer in India. Indian J Cancer. 2016;53(2):304–8. doi: 10.4103/0019-509X.197723. [DOI] [PubMed] [Google Scholar]
- 11.Nagrani RT, Budukh A, Koyande S, Panse NS, Mhatre SS, Badwe R. Rural urban differences in breast cancer in India. Indian J Cancer. 2014;51(3):277–81. doi: 10.4103/0019-509X.146793. [DOI] [PubMed] [Google Scholar]
- 12.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5(6):e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Genomes Project C, Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, et al. A global reference for human genetic variation. Nature. 2015;526(7571):68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hemani G, Zheng J, Elsworth B, Wade KH, Haberland V, Baird D, et al. The MR-Base platform supports systematic causal inference across the human phenome. Elife. 2018;7 doi: 10.7554/eLife.34408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Burgess S, Labrecque JA. Mendelian randomization with a binary exposure variable: interpretation and presentation of causal estimates. Eur J Epidemiol. 2018;33(10):947–52. doi: 10.1007/s10654-018-0424-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bowden J, Davey Smith G, Haycock PC, Burgess S. Consistent Estimation in Mendelian Randomization with Some Invalid Instruments Using a Weighted Median Estimator. Genet Epidemiol. 2016;40(4):304–14. doi: 10.1002/gepi.21965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bowden J, Davey Smith G, Burgess S. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int J Epidemiol. 2015;44(2):512–25. doi: 10.1093/ije/dyv080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hartwig FP, Davey Smith G, Bowden J. Robust inference in summary data Mendelian randomization via the zero modal pleiotropy assumption. Int J Epidemiol. 2017;46(6):1985–98. doi: 10.1093/ije/dyx102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hemani G, Bowden J, Davey Smith G. Evaluating the potential role of pleiotropy in Mendelian randomization studies. Hum Mol Genet. 2018;27(R2):R195–R208. doi: 10.1093/hmg/ddy163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bowden J, Del Greco MF, Minelli C, Davey Smith G, Sheehan N, Thompson J. A framework for the investigation of pleiotropy in two-sample summary data Mendelian randomization. Stat Med. 2017;36(11):1783–802. doi: 10.1002/sim.7221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bowden J, Spiller W, Del Greco MF, Sheehan N, Thompson J, Minelli C, et al. Improving the visualization, interpretation and analysis of two-sample summary data Mendelian randomization via the Radial plot and Radial regression. Int J Epidemiol. 2018;47(4):1264–78. doi: 10.1093/ije/dyy101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Verbanck M, Chen CY, Neale B, Do R. Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat Genet. 2018;50(5):693–8. doi: 10.1038/s41588-018-0099-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Burgess S, Foley CN, Allara E, Staley JR, Howson JMM. A robust and efficient method for Mendelian randomization with hundreds of genetic variants. Nat Commun. 2020;11(1):376. doi: 10.1038/s41467-019-14156-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Consortium E-I. Burgess S, Scott RA, Timpson NJ, Davey Smith G, Thompson SG. Using published data in Mendelian randomization: a blueprint for efficient identification of causal risk factors. Eur J Epidemiol. 2015;30(7):543–52. doi: 10.1007/s10654-015-0011-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Burgess S, Davey Smith G, Davies NM, Dudbridge F, Gill D, GM M, et al. Guidelines for performing Mendelian randomization investigations. Wellcome Open Research. 2020;4(186) doi: 10.12688/wellcomeopenres.15555.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Burgess S, Thompson SG. Use of allele scores as instrumental variables for Mendelian randomization. Int J Epidemiol. 2013;42(4):1134–44. doi: 10.1093/ije/dyt093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Angrist JD, Imbens GW. Two-stage least squares estimation of average causal effects in models with variable treatment intensity. Journal of the American Statistical Association. 1995;90:431–42. [Google Scholar]
- 28.Burgess S, Small DS, Thompson SG. A review of instrumental variable estimators for Mendelian randomization. Stat Methods Med Res. 2017;26(5):2333–55. doi: 10.1177/0962280215597579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hsing AW, Gao YT, Han TQ, Rashid A, Sakoda LC, Wang BS, et al. Gallstones and the risk of biliary tract cancer: a population-based study in China. Br J Cancer. 2007;97(11):1577–82. doi: 10.1038/sj.bjc.6604047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shabanzadeh DM, Sorensen LT, Jorgensen T. Association Between Screen-Detected Gallstone Disease and Cancer in a Cohort Study. Gastroenterology. 2017;152(8):1965–74.:e1. doi: 10.1053/j.gastro.2017.02.013. [DOI] [PubMed] [Google Scholar]
- 31.Ishiguro S, Inoue M, Kurahashi N, Iwasaki M, Sasazuki S, Tsugane S. Risk factors of biliary tract cancer in a large-scale population-based cohort study in Japan (JPHC study); with special focus on cholelithiasis, body mass index, and their effect modification. Cancer Causes Control. 2008;19(1):33–41. doi: 10.1007/s10552-007-9067-8. [DOI] [PubMed] [Google Scholar]
- 32.Bodmer M, Brauchli YB, Krahenbuhl S, Jick SS, Meier CR. Statin use and risk of gallstone disease followed by cholecystectomy. JAMA. 2009;302(18):2001–7. doi: 10.1001/jama.2009.1601. [DOI] [PubMed] [Google Scholar]
- 33.Liu Z, Alsaggaf R, McGlynn KA, Anderson LA, Tsai HT, Zhu B, et al. Statin use and reduced risk of biliary tract cancers in the UK Clinical Practice Research Datalink. Gut. 2019;68(8):1458–64. doi: 10.1136/gutjnl-2018-317504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Erichsen R, Froslev T, Lash TL, Pedersen L, Sorensen HT. Long-term statin use and the risk of gallstone disease: A population-based case-control study. Am J Epidemiol. 2011;173(2):162–70. doi: 10.1093/aje/kwq361. [DOI] [PubMed] [Google Scholar]
- 35.Schmidt M, Hausken T, Glambek I, Schleer C, Eide GE, Sondenaa K. A 24-year controlled follow-up of patients with silent gallstones showed no long-term risk of symptoms or adverse events leading to cholecystectomy. Scand J Gastroenterol. 2011;46(7-8):949–54. doi: 10.3109/00365521.2011.571710. [DOI] [PubMed] [Google Scholar]
- 36.Shaffer EA. Gallbladder cancer: the basics. Gastroenterol Hepatol (N Y) 2008;4(10):737–41. [PMC free article] [PubMed] [Google Scholar]
- 37.Staley JR, Blackshaw J, Kamat MA, Ellis S, Surendran P, Sun BB, et al. PhenoScanner: a database of human genotype-phenotype associations. Bioinformatics. 2016;32(20):3207–9. doi: 10.1093/bioinformatics/btw373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Velkova A, Diaz JEL, Pangilinan F, Molloy AM, Mills JL, Shane B, et al. The FUT2 secretor variant p.Trp154Ter influences serum vitamin B12 concentration via holo-haptocorrin, but not holo-transcobalamin, and is associated with haptocorrin glycosylation. Hum Mol Genet. 2017;26(24):4975–88. doi: 10.1093/hmg/ddx369. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.