Abstract
MYC is implicated in the development and progression of Pancreatic cancer, yet the precise level of MYC deregulation required to contribute to tumour development has been difficult to define. We used modestly elevated expression of human MYC, driven from the Rosa26 locus, to investigate the pancreatic phenotypes arising in mice from an approximation of MYC trisomy. We show that this level of MYC alone suffices to drive pancreatic neuroendocrine tumours, and to accelerate progression of KRAS-initiated precursor lesions to metastatic pancreatic ductal adenocarcinoma. Our phenotype exposed suppression of the Type I Interferon pathway by the combined actions of MYC and KRAS and we present evidence of repressive MYC/MIZ1 complexes binding directly to the promoters of type I Interferon regulators IRF5, IRF7, STAT1 and STAT2. De-repression of Interferon regulators allows pancreatic tumour infiltration of B and NK cells, resulting in increased survival.
Keywords: Pancreatic Cancer, PDAC, PNET, KRAS, MYC, Immuno-oncology, Innate Immunity, Interferon, Natural Killer cells
Introduction
Cancers of the pancreas are projected to become the 2nd most frequent cause of cancer-related mortality worldwide by 2030 (1). Ductal adenocarcinoma (PDAC) comprises up to 95% of pancreatic cancers, while neuroendocrine tumours (PNET) and other exocrine tumours account for remainder https://www.pancreaticcancer.org.uk/; https://seer.cancer.gov/statistics/. The 5-year survival rate for PDAC is extremely low at 3-5% and, although that for PNET is considerably higher (30-50%), neither cancer responds effectively to current treatment modalities (2). There is therefore a pressing need to further our understanding of the underlying biology of these cancers.
Activating mutations in KRAS occur in up to 93% of PDAC, while the majority of KRAS wild-type cases bear alternative genetic alterations predicted to result in ectopic RAS->ERK pathway activity, implying a critical requirement for this pathway in PDAC (3, 4). Experiments in genetically altered mice have however demonstrated that KRas activation is alone insufficient for PDAC development: CRE-dependent activation of the endogenously expressed lsl-KRasG12D allele (5) alone gives rise predominantly to low-grade pancreatic intraepithelial neoplasms (PanINs), a subset of which may spontaneously progress to PDAC through the acquisition of additional mutations (6–8). Recent genomic characterisation of cell lines generated from such spontaneously progressing tumours revealed increased K-RasG12D allelic dosage as one of the most frequently recurring genetic alterations associated with progression (9), suggesting that the volume of signalling flux through the RAS pathway is rate-limiting for PDAC progression, as was recently shown for KRAS-driven lung cancer (10–13). Interestingly, cell lines that lacked increased KRasG12D allelic dosage contained amplifications of alternative oncogenes that may serve as potentially rate-limiting RAS effectors, including MYC, YAP1 and NFκB (9, 14). MYC in particular is widely reported to be overexpressed in PDAC, with a propensity for low-level amplification suggesting that subtle changes in MYC expression may suffice to impact on PDAC development (15, 16). Accordingly, expression of murine c-Myc from the Rosa26 locus was recently shown to accelerate progression to PDAC, however the use of murine c-Myc cDNA in the transgene precluded a precise determination of the level of MYC deregulation required for tumour progression (17). Conversely, deletion/depletion of endogenous c-Myc profoundly delayed progression of PDAC driven by KRasG12D and loss of p53 (18, 19), while RNAi-mediated depletion of c-MYC in human PDAC cells suppressed proliferation in vitro and xenograft tumourigenesis in vivo (20), consistent with the hypothesis that MYC is a critical rate-limiting effector of KRAS in this cancer.
Here, we directly investigated the level of MYC deregulation required for pancreatic tumour development using conditional expression of human c-MYC from the Rosa26 locus at levels that approximate expression of endogenous c-Myc. We show that modestly deregulated expression of MYC alone suffices to drive PNET development and dramatically accelerates progression to metastatic PDAC when combined with KRas activation. In PDAC, the combination of MYC & KRasG12D suppresses tumour infiltration of effector immune populations, as recently reported (21). Mechanistically, we reveal that MYC and KRasG12D cooperatively regulate gene expression, with particular convergence on repression of the Type I Interferon pathway. We show that targeted suppression of the MYC/Miz1 transcriptional repressor complex (22) restores interferon-related gene expression and consequent B and NK cell-mediated immune surveillance. Our data reveal a novel mechanism of immune evasion driven by 2 of the most commonly dysregulated oncogenes in human cancer.
Results
Rosa26-driven MYC is expressed at near physiological levels
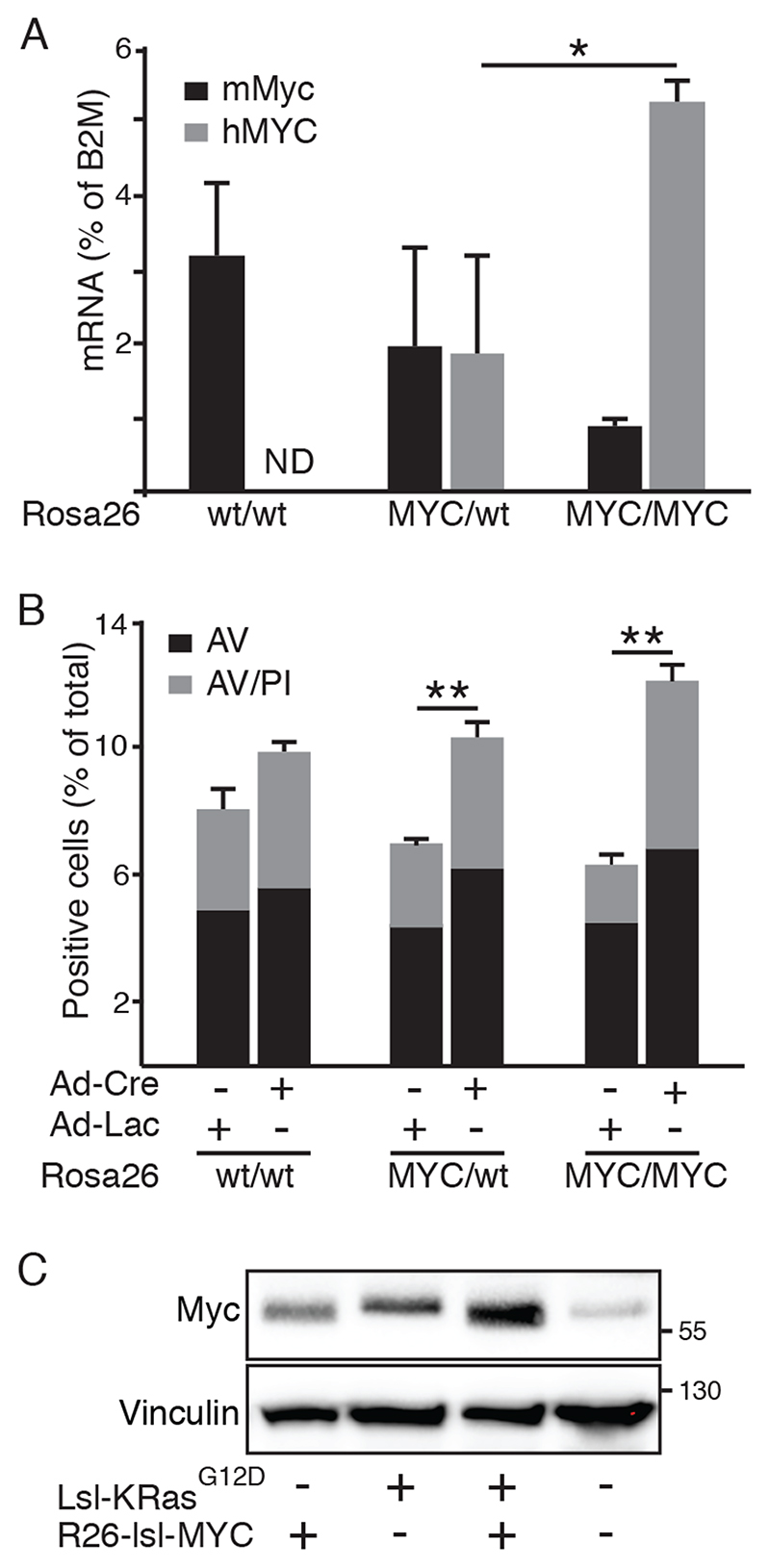
In order to define the level of MYC deregulation required for pancreatic tumour development, we used Rosa26DM-lsl-MYC mice wherein the human c-MYC cDNA, preceded by a floxed translational stop cassette, was inserted into the murine Rosa26 locus by homologous recombination, as previously described (23). Use of the human cDNA facilitated the distinction between endogenously expressed murine c-Myc and Rosa26-driven MYC. RT-PCR demonstrated Cre-dependent expression of human MYC in Rosa26DM-lsl-MYC/+ and Rosa26DM-lsl-MYC/lsl-MYC-derived mouse embryo fibroblasts (MEFs) within 24hrs of infection with Adeno-Cre. Expression of one copy of human MYC mRNA was comparable to murine c-Myc, while 2 copies yielded modestly supraphysiological expression (Fig. 1A). Consistent with previous reports that MYC regulates its own transcription (24), Rosa26MYC/MYC MEFs exhibited reduced expression of endogenous c-Myc and were modestly sensitised to apoptosis upon serum withdrawal (Fig. 1B). At the protein level, activation of Rosa26DM-lsl-MYC/+ drove modestly higher expression of MYC compared with wild-type (wt) MEFs. Notably, Cre-dependent activation of endogenously expressed lsl-KRasG12D also drove higher expression of MYC protein, comparable to that arising from Rosa26MYC/+, and expression was higher again upon combined activation of both alleles (Fig. 1C).
Figure 1. Rosa26-driven MYC is expressed at near physiological levels.
A) RT-PCR comparison of Rosa26-driven MYC with endogenously expressed c-Myc in Rosa26wt/wt, Rosa26DM-lsl-MYC/+ and Rosa26DM-lsl-MYC/lsl-MYC MEFs, 24hrs after infection with Adeno-Cre. N=3, * denotes p<0.05; ANOVA and post-hoc Tukey test. ND = not detected. B) FACS analysis of Annexin V (AV), propidium iodide (PI) labelling of MEFs, infected as per (A) with Adeno-Cre or Adeno-LacZ and cultured overnight in 0.2% FBS. N=3, ** denotes p<0.01; ANOVA with post-hoc Tukey test. C) Immunoblot of MYC expression in wild-type, Rosa26DM-lsl-MYC/+, lsl-KRasG12D and Rosa26DM-lsl-MYC/+;lsl-KRasG12D (double positive) MEFs 24 hrs after Adeno-Cre infection. Image is representative of >3 experiments.
Deregulated MYC drives pancreatic tumour development and progression
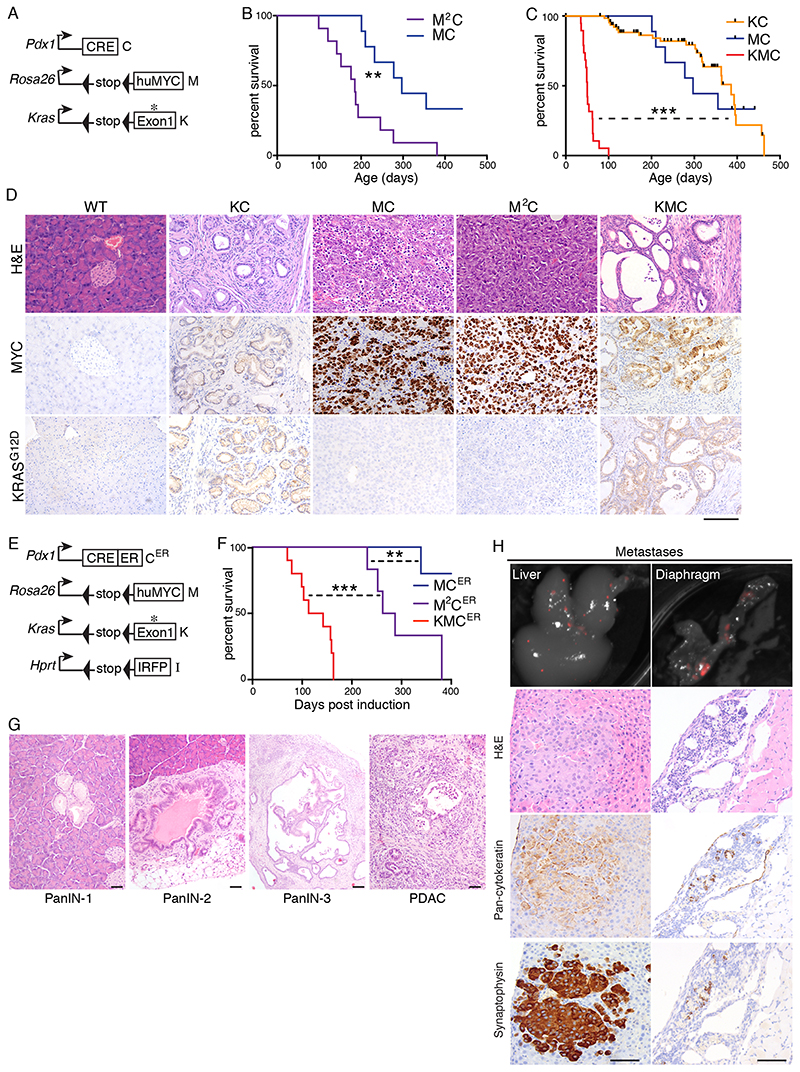
RAS pathway activity elevates MYC expression in pancreatic cancer, primarily via suppression of MYC protein turnover, and MYC was previously shown using RNAi to be required for proliferation of PDAC cells in culture (20, 25, 26). Use of the PICKLES database of CRISPR-mediated essentiality screening (27) confirmed the requirement for MYC to sustain the fitness of all PDAC lines examined, including several lines that do not require KRAS (Fig. S1A). We asked if MYC expressed from the Rosa26 locus is sufficient for tumour formation. We interbred Rosa26DM-lsl-MYC mice (M) with pancreas-specific Pdx1-Cre mice (C), with and without the lsl-KRasG12D allele (K), and aged mice until humane clinical end-points were reached. Pdx1-Cre positive mice carrying one (MC) or two (M2C) copies of Rosa26DM-lsl-MYC developed pancreatic tumours requiring euthanasia at a median age of 297 or 180 days, respectively. The combination of lsl-KRasG12D and one copy of Rosa26DM-lsl-MYC (KMC) yielded a dramatically accelerated tumour phenotype requiring euthanasia at a median age of just 50 days, as compared with KC mice that have a median survival >350 days (Fig. 2A-C). In contrast with KC mice, which developed mostly ductal epithelial PanIN lesions with infrequent progression to PDAC, as previously reported (6), MC and M2C pancreata showed no evidence of ductal epithelial phenotypes and instead presented with neuroendocrine tumours (PNET) characterised by densely populated cells that stained strongly for the neuroendocrine marker synaptophysin and faintly positive for cytokeratin (Fig. 2D & S1B). This PNET phenotype was largely masked upon inclusion of the lsl-KRasG12D allele: rapidly arising tumours in KMC mice displayed a predominantly ductal adenocarcinoma phenotype (PDAC), complete with characteristic desmoplastic stroma, while <5% of tissue area exhibited PNET features (Fig. S1B, C). PNET regions in KMC tumours expressed KRasG12D (Fig. S1D), indicating that the persistence of this phenotype does not arise from failure to activate the lsl-KRasG12D allele in a subset of pancreatic tumour-initiating cells.
Figure 2. Pancreatic cancer phenotypes induced by activation of Rosa26-driven MYC with and without KRasG12D .
A) Schematic of alleles used in panels B-D. B) Overall survival of MC (N=9) and M2C (N=11) mice. Mantel Cox logrank test (panels B, C, F). C) Overall survival of KC (N=65) versus MC (from (B)) and KMC (N=19) mice. Black hash marks indicate mice that were euthanised for reasons unrelated to pancreatic cancer. D) Representative images show tumour histology (H&E) and immunohistochemical detection of total MYC and KRasG12D expression in end-stage tumours of mice of the indicated genotypes. Scale bar = 100μm. E) Schematic of alleles used in panels F-H. F) Overall survival of MCER (N=5), M2CER (N=6) and KMCER (N=10) mice measured in days post induction with Tamoxifen. G) Representative H&E images of ductal tumour progression in KMCER mice. Scale bars = 100μm. H) Top panels show Licor PEARL fluorescent imaging of IRFP-expressing metastases in KMCER mice. Left panels show liver metastases, right panels show diaphragm metastases. Lower panels show H&E and IHC for pan-cytokeratin and synaptophysin. Scale Bars = 100μm.
Sporadic adult activation of Rosa26DM-lsl-MYC and lsl-KRasG12D drives metastatic cancer
The Pdx1 promoter is active from embryonic day 8.5 (28), raising the possibility that blockade of pancreatic progenitor cell differentiation might explain the MYC-dependent neuroendocrine and accelerated PDAC phenotypes. To address this, we substituted the constitutively active Pdx1-Cre allele with a Pdx1-Cre-ER allele (CER) which expresses an inactive Cre:Estrogen Receptor ligand-binding domain fusion protein that can be activated by the synthetic ligand Tamoxifen (29). KMCER mice were additionally interbred with mice carrying a Hprt-lsl-IRFP Cre-reporter allele (30), to facilitate imaging of tumour populations (Fig. 2E). Cre-ER was transiently activated in mice aged 5-6 weeks by 3 days of Tamoxifen injection, and mice were aged to clinical end-point. As was found using the constitutively active Cre, all M2CER mice developed neuroendocrine tumours, reaching median end-point at 275 days from the date of induction, although the majority of MCER mice failed to develop symptomatic disease within 400 days. KMCER mice all developed PDAC, reaching median end-point at 128 days post-induction (Fig. 2F). Periodic sampling of KMCER pancreatic tissue following Cre-ER activation showed temporal progression of incipient tumours from PanIN-1 through to PDAC development (Fig 2G). Furthermore, the IRFP allele revealed metastases in 6/10 mice, of which 4 had liver metastases while 4 had diaphragm metastases, including 2 mice with both (Fig. 2H). As observed with KMC tumours, KMCER tumours contained regions (approx. 15% of tumour area) of PNET (Fig. S1C). All liver metastases histologically resembled the PNET phenotype and all stained strongly for synaptophysin. In contrast, diaphragm metastases all histologically resembled PDAC, but also contained discrete regions where cells stained strongly for either cytokeratin or synaptophysin, suggestive of phenotypic plasticity in the metastatic population (Fig. 2H). Accordingly, synaptophysin/cytokeratin double positive cells could be identified in the ductal epithelium of primary tumours (Fig. S1E), as previously reported using the embryonically active Pdx1-Cre allele (17). The accelerated tumour phenotypes observed upon MYC deregulation can thus arise in fully developed adult tissues.
Endogenous c-Myc is required for KMC tumour development
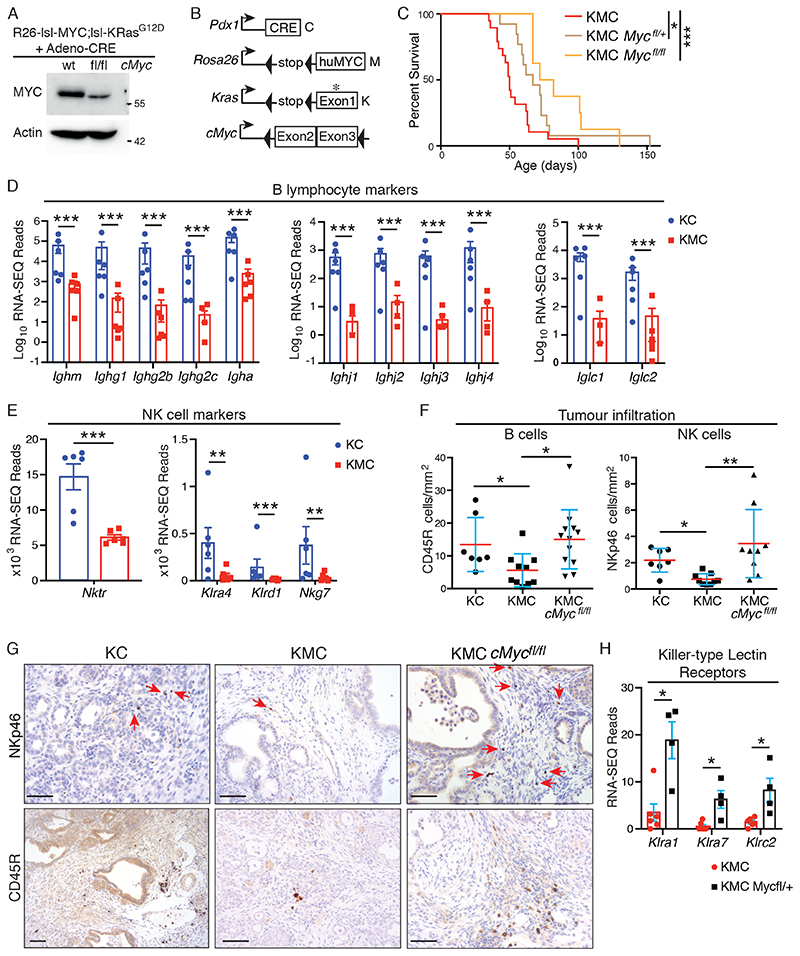
Given the modest expression of Rosa26-driven MYC in MEFs, we asked if autochthonously expressed c-Myc contributes meaningfully to the total pool of MYC protein in the KMC model. Note that while KRas activation does not impact upon transcription from the Rosa26 locus, which we previously showed is refractory to growth factor signalling (31), post-translation regulation of MYC protein by RAS pathway activation should affect murine and human MYC protein equally (32). Accordingly, deletion of floxed murine c-Myc in MEFs reduced the total pool of MYC protein by approximately 50%, despite concurrent activation of Rosa26DM-lsl-MYC and lsl-KRasG12D alleles (Fig. 3A). KMC mice interbred with c-Mycfl/fl mice were aged until humane end-points were reached. KMC mice heterozygous for floxed c-Myc showed significantly increased survival and survival was increased further in homozygous c-Mycfl/fl mice (Fig. 3B, C). Nevertheless, all mice developed PDAC. Examination of murine c-Myc by in situ hybridization (ISH) revealed a mosaic pattern of continued expression of murine c-Myc in much of the ductal epithelium of all KMC-Mycfl/fl tumours examined, indicating escape from Cre-mediated deletion and suggesting a selective pressure to retain expression of endogenous c-Myc despite the presence of Rosa26-driven MYC (Fig. S2A, B). This MYC dose-dependence was also evident in human PDAC cell lines, wherein depletion of MYC suppressed cell proliferation (Fig. S2C), consistent with previous reports (20). These data strongly suggest that the level of MYC expression is rate-limiting for pancreatic tumour progression and, in KMC mice, both Rosa26-driven MYC and endogenously expressed c-Myc functionally contribute to the total pool of MYC protein.
Figure 3. MYC expression levels modulate effector immune cell infiltration in PDAC.
A) Genetic deletion of endogenous c-Myc in Rosa26DM-lsl-MYC;lsl-KRasG12D MEFs. Lysates were prepared 24hrs after Adeno-Cre infection. Representative of 2 independent experiments. B) Schematic of alleles used in panels C-G. C) Overall survival of KMC mice with 0 (N=19, from Fig. 2C), 1 (N=13) or 2 (N=8) copies of c-Mycfl. Mantel Cox Logrank test. D) Normalised RNA-SEQ reads of indicated B cell markers in KC (N=6) and KMC (N=6) pancreatic tumours. Note that samples with zero reads are absent from log-scale graphics (pertains only to KMC samples). Adjusted P values were generated in R (panels D, E, H). E) Normalised RNA-SEQ reads of indicated NK cell markers in KC (N=6) and KMC (N=6) pancreatic tumours. F) Quantification of tumour infiltrating NK (NKp46+) and B (CD45R+) cells in KC (N=7), KMC (N=10) and KMC-cMycfl/fl (N=9 & 12, respectively) pancreatic tumours. Kruskal-Wallis and Dunn’s multiple comparison test. G) Representative images of IHC staining for NKp46+ NK cells (upper panels) and CD45R+ B cells (lower panels), as per (F). Scale bars = 100μm. H) RNA-SEQ reads of NK cell killer-type lectins in KMC (N=6) versus KMC-Mycfl/+ (N=4) tumours. For all panels, ns = not significant; *** denotes p<0.001, **denotes p<0.01 and * denotes p<0.05.
MYC suppresses immune cell infiltration in PDAC
To gain insight into the functional roles of MYC in PDAC, we performed bulk-tumour RNA-SEQ on 6 KC, 6 KMC, and 4 KMC-Mycfl/+ end-stage tumours. Note that homozygous KMC-Mycfl/fl mice were omitted from RNA-SEQ analysis given that the failure of Cre recombinase to efficiently delete both copies of endogenous c-Myc would likely confound interpretation. Metacore GeneGO analysis revealed a pronounced reduction in immune cell-related gene expression in KMC relative to KC tumours (Figure S3). Specifically, we found reduced expression of T cell markers, including CD3, CD4 & CD8 (Fig. S4A); B cell immunoglobulin genes, including multiple constant heavy and light chains along with joining regions; and NK cell genes, including Natural Killer Triggering Receptor, Nktr, and Killer-type lectin receptors (Fig. 3D, E). IHC for markers of B (CD45R) and NK (NKp46) cells confirmed their reduced presence in KMC PDAC (Fig. 3F, G). Reduction of total MYC by deletion of endogenous c-Myc reversed the exclusion of B and NK cells but did not significantly influence T cell infiltration (Fig. 3F, G & S4B, C). RNA-SEQ analysis showed increased expression of multiple killer-type lectin receptors, indicative of NK cell activation and memory (33), upon reduction of MYC in KMC tumours (Fig. 3H). Taken together with Figures 1 & 2, these data show that subtle differences in MYC expression profoundly modulates long-term immune cell infiltration in PDAC, extending a recent report of acute immune modulation by MYC using a tamoxifen-dependent variant allele, Rosa26-lsl-MycERT2 (21).
Suppression of the Type I Interferon pathway by MYC and KRAS
MYC was previously shown to modulate the immune landscape in KRAS-driven lung adenocarcinoma via Ccl9-dependent recruitment of pro-tumour macrophages and Il23-dependent exclusion of T, B & NK cells (34). Expression of Ccl9 was not significantly altered in our RNA-SEQ datasets and we found no difference in the number of macrophages present in end-stage KMC versus KMC-Mycfl/fl tumours (Fig. S4D, E). Functional IL23 comprises a heterodimer of p19 and p40 subunits, encoded by IL23a and IL12b, respectively (35). Although Il23a expression was higher in KMC versus KC tumours and significantly reduced in KMC-Mycfl/+ tumours, its dimerizing partner Il12b showed opposite regulation (Fig. S4F), strongly suggesting that Il23 does not account for the immunosuppressive phenotype driven by MYC in PDAC. Expression of Gas6, recently reported to be induced upon acute MYC activation in PanIN lesions (21), was unchanged in end-stage tumours (Fig. S4G). We therefore sought an alternative explanation for MYC-driven immunosuppression.
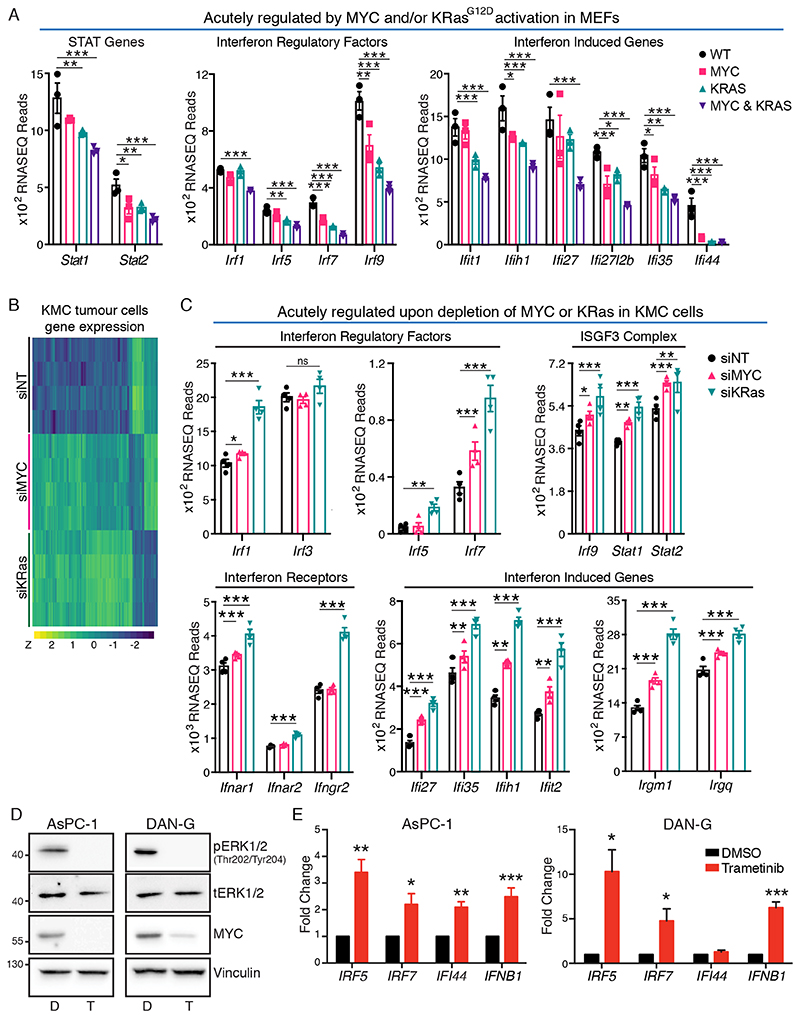
Type I Interferon signalling via JAK/STAT was amongst the most strongly suppressed pathways (ranked 6th) in KMC relative to KC tumours (Fig. S3). Taking advantage of our ability to acutely induce expression of floxed MYC and KRasG12D in otherwise wild-type primary cells, we asked which pathways were regulated upon acute activation of MYC and/or KRas in MEFs. Early passage MEFs carrying Rosa26-lsl-MYCDM, lsl-KRasG12D, or both, were infected with Adeno-CRE overnight in full growth media and their gene expression was compared with that of similarly infected wild-type littermate MEFs. Consistent with positive regulation of MYC by activated KRas (Fig. 1), we found a pronounced overlap in the transcriptomic impact of activation of either oncogene alone (Fig. S5A-C & Table S1-S3). Similar to our comparison between KMC and KC tumours, GeneGO analysis identified Type I Interferon signalling via JAK/STAT as the most significantly downregulated pathway upon combined activation of MYC and KRas alleles (Fig. S5D). Interrogation of the data at the single gene level showed reduced expression of multiple Interferon-induced genes; multiple Interferon Regulatory Factors; and genes encoding STAT1 and STAT2, which combine with IRF9 to form the ISGF3 complex (36). Although several of these effects were evident with activation of either MYC or KRasG12D alone, regulation was most pronounced upon combined oncogene activation (Fig. 4A, S5D).
Figure 4. MYC and KRasG12D suppress the Type I Interferon pathway.
A) RNA-SEQ analysis, of indicated gene expression in Rosa26wt/wt (WT), Rosa26DM-lsl-MYC/+(MYC), lsl-KRasG12D (KRAS) and Rosa26DM-lsl-MYC/+;lsl-KRasG12D (MYC & KRAS) MEFs, 24hrs after infection with Adeno-Cre. Adjusted P values were generated in R (panels A, C). B) Heatmap of significant gene expression changes induced by depletion of MYC and KRas in KMC-derived cultured PDAC cells compared to non-targeting control, analysed by RNA-SEQ. N=4 biological replicates. C) RNA-SEQ analysis of the indicated genes upon depletion of MYC or KRas in KMC PDAC cells, as per (B). D) Reduced expression of MYC in human PDAC cells treated with Trametinib (T), compared with DMSO vehicle (D). Representative of 3 individual experiments. E) RT-PCR analysis of Interferon-related gene expression in human PDAC cell lines treated +/- Trametinib. Mean and SEM shown for N=3 biological replicates. For all panels, *** denotes p<0.001, **denotes p<0.01, * denotes p<0.05, ns = not significant.
We next compared the transcriptomic impact of acute depletion of MYC or KRas from KMC-derived tumour cell lines. As was observed in MEFs, we found a pronounced overlap in significantly regulated genes altered in murine PDAC cells upon depletion of either MYC or KRas (Fig. 4B & S5E). Notably, the Type I interferon pathway was again amongst the topmost regulated pathways in both instances (Fig. S5F), with increased expression of the ISGF3 complex subunits, multiple Irfs, Interferon receptors, and Interferon-induced genes, detected upon depletion of either oncogene. These effects were typically stronger upon KRas depletion and some Irf genes (eg. Irf5) were regulated by KRas but not by MYC in this analysis, perhaps reflecting the limits of transcript detection at the sequencing read-depth used (Fig. 4C). To extend these data to the human setting, we treated PDAC cell lines with the MEK1/2 inhibitor Trametinib, which acutely reduced expression of c-MYC downstream of KRAS (Fig. 4D). In both AsPC-1 and DAN-G cells Trametinib increased expression of IRF5, IRF7, IFNb1 and a representative Interferon-inducible gene, IFI44 (Fig. 4E). We conclude from these experiments that MYC and KRAS cooperate to suppress the Type I Interferon pathway.
The role of MIZ1 in suppression of Interferon regulators by MYC
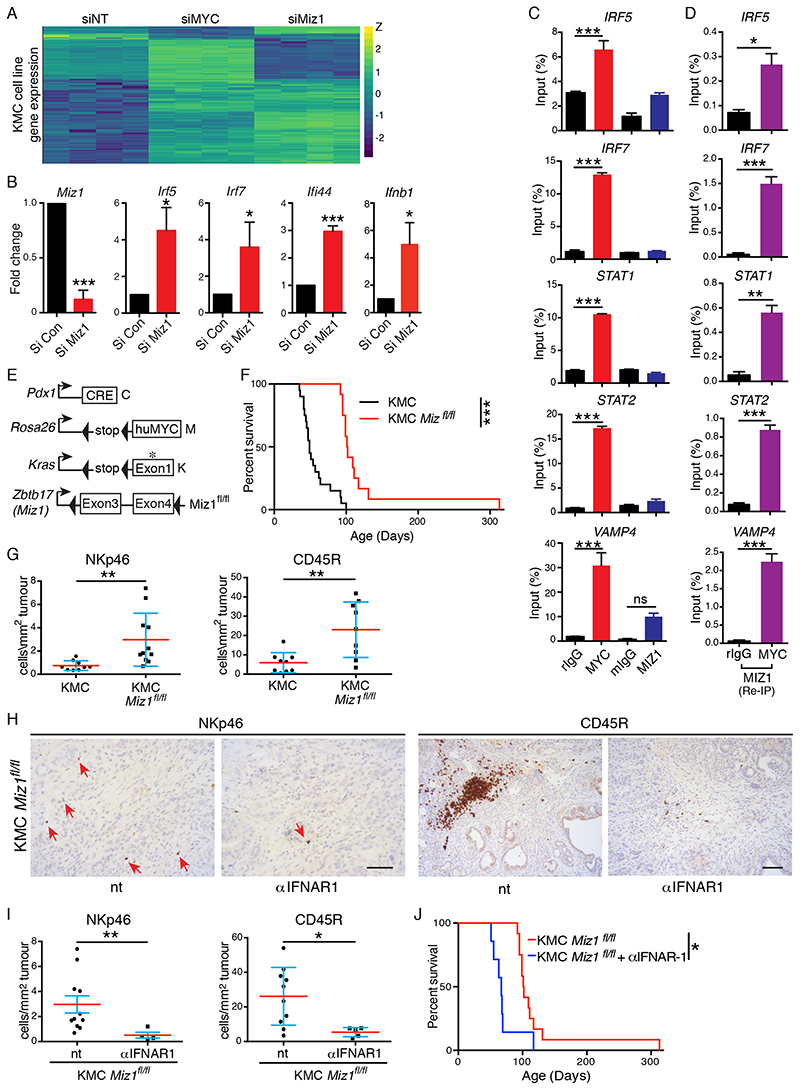
In many instances MYC-dependent transcriptional repression involves binding to the MYC-interacting zinc finger protein MIZ1, encoded by ZBTB17 (18, 37). We therefore investigated the acute impact of Miz1 depletion in KMC PDAC cells in vitro. Transcriptomic analysis revealed that depletion of Miz1 upregulated approximately 60% of the same genes upregulated upon depletion of MYC from KMC cells (Fig. 5A & Table S4). Using Q-PCR we determined that depletion of Miz1 or MYC specifically increased expression of Irf5, Irf7, Ifnb1 and Ifi44 (Figs. 5B, S6A), as was found upon Trametinib treatment of human PDAC cells. Chromatin immunoprecipitation (ChIP) analyses showed MYC binding to the promoters of STAT1, STAT2, IRF5 and IRF7 in DAN-G cells (Fig. 5C). ChIP-re-ChIP using an anti-MIZ1 antibody showed efficient recovery of the same IRF & STAT promoter fragments initially precipitated using anti-MYC but absent from control IgG precipitates, strongly suggesting that MYC and MIZ1 form repressive complexes on the STAT1, STAT2, IRF5 and IRF7 promoters (Fig. 5D & S6B), as previously reported for the Stat1 promoter in murine KPC PDAC cells (18). To investigate the functional role of Miz1 in vivo we used Miz1ΔPOZ mice (38) in which the MYC-interacting POZ domain of Miz1 is flanked by LoxP sites (Fig. 5E). Efficient deletion of the Miz1 POZ domain in KMC-Miz1fl/fl mice was verified by ISH & significantly extended the lifespan of tumour-bearing mice (Figs. 5F & S6C). PDAC subtype analysis performed as per Bailey et al. (3) revealed that deletion of either c-Myc or Miz1 significantly increased the immunogenic subtype signature but did not significantly alter any of the other subtypes (Fig. S6D). As was observed upon deletion of endogenous c-Myc, deletion of Miz1 restored tumour infiltration of NK and B cells (Fig. 5G). Antibody-mediated blockade of the Type I Interferon receptor, IFNAR, suppressed NK and B cell infiltration and negated the survival benefit of Miz1 deletion in tumour-bearing KMC mice (Fig. 5H-J), implicating Interferon signalling in lymphocyte recruitment and in the survival benefit observed upon Miz1 deletion.
Figure 5. Transcriptional repression of the Type I Interferon pathway by MYC/Miz1.
A) Heatmap of gene expression changes induced by RNAi-mediated depletion of MYC or Miz1 in KMC-derived cultured PDAC cells compared to non-targeting (NT) control, analysed by RNA-SEQ. N=4 biological replicates. B) RT-PCR analysis of Interferon-related gene expression in KMC tumour cells upon siRNA mediated depletion of Miz1 compared to non-targeting control. Mean & SEM of 3 independent experiments shown. T test. C) Chromatin-IP analysis of MYC and Miz1 binding to the promoters of the indicated genes in human DAN-G PDAC cells compared with a known MYC/Miz1 target, VAMP4. Mean and SEM of technical replicates from 1 of 3 independent experiments shown. ANOVA and Tukey’s multiple comparison test. D) Re-CHIP analysis of Miz1 binding to anti-MYC-precipitated promoter regions. Mean and SEM of technical replicates from 1 of 3 independent experiments shown. T test. E) Schematic of alleles used in panels F-J. F) Overall survival of KMC (N=19, from Fig. 2C) and KMC-Miz1fl/fl (N=12) mice. *** denotes p<0.001; Mantel Cox Logrank test (panels F, J). G) Quantification of tumour infiltrating NK (NKp46+) and B (CD45R+) cells in KMC and KMC Miz1fl/fl pancreatic tumours. Mann-Whitney Test (panels G, I). H) IHC detection of tumour infiltrating NK and B cells after 3 weeks of IFNAR-1 blockade in KMC-Miz1fl/fl PDAC. Scale bars = 100μm. I) Quantification of tumour infiltrating NK (NKp46+) and B (CD45R+) cells in untreated (N=11 & 10, respectively; from panel G) versus IFNAR1 antibody treated (3 weeks treatment, N=5) KMC-Miz1fl/fl PDAC. J) Overall survival of KMC-Miz1fl/fl mice treated with (N=7) or without (N=12, from panel F) anti-IFNAR1 blocking antibody. For all panels, *** denotes p<0.001, **denotes p<0.01, * denotes p<0.05, ns = not significant.
Interferon signalling to NK and B cells is mediated by CXCL13
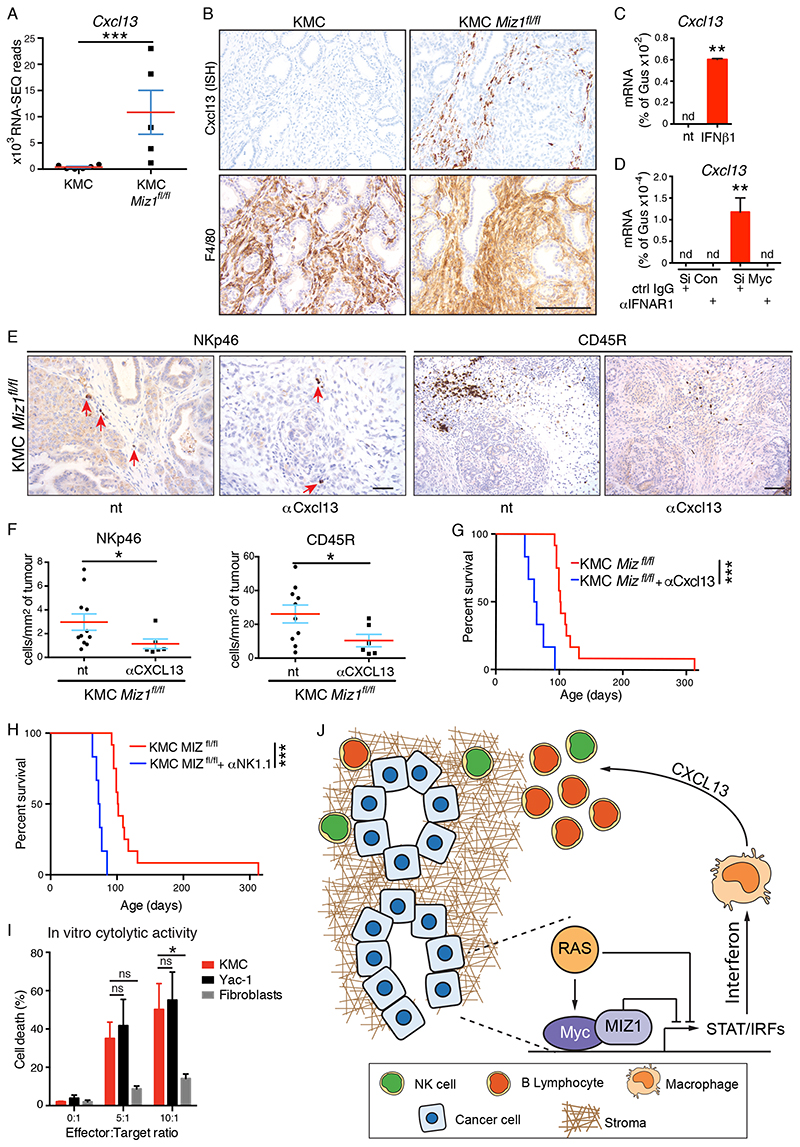
Type I Interferons were recently shown to induce expression of the B cell chemokine, CXCL13 (39). RNA-SEQ analysis showed extremely high expression of Cxcl13 in KMC-Miz1fl/fl tumours, as compared with KMC tumours that express very little Cxcl13 (Fig. 6A). ISH analysis of KMC-Miz1fl/fl tumours showed Cxcl13 expression was restricted to a subset of F4/80-positive macrophages (Fig. 6B). Notably, only macrophages in close proximity to areas of ductal tumour epithelium stained positive for Ccxl13 mRNA, suggesting paracrine signalling from tumour epithelium to adjacent macrophages. A similar, albeit weaker, pattern of Cxcl13 expression in F4/80-positive macrophages adjacent to ductal epithelium was observed in KMC-Mycfl/fl tumours but was absent from KMC tumours (Figs. 6A, S6E). We used exogenous mouse IFNβ1 treatment of bone marrow-derived macrophages to confirm that Interferon can stimulate Cxcl13 expression in macrophages (Fig. 6C). Next, we cultured macrophages with conditioned media harvested from KMC tumour cells treated with non-targeting or MYC-depleting siRNA and found that only media from MYC-depleted tumour cells could stimulate Cxcl13 expression. Importantly, pre-treatment of macrophages with IFNAR1 blocking antibody completely abrogated Cxcl13 induction (Fig. 6D), confirming that Type I Interferon released from MYC-depleted tumour cells drives Cxcl13 production in macrophages. Antibody-mediated depletion of Cxcl13 from KMC-Miz1fl/fl mice suppressed infiltration of both B and NK cells and negated the survival benefit of Miz1 deletion (Fig. 6E-G), whereas isotype control antibody had no effect (Fig. S6F). Depletion of NK cells likewise negated the survival benefit, strongly suggesting that they actively restrain the tumour phenotype (Fig. 6H, S6G). Accordingly, co-culture of KMC tumour cells with in vitro-activated splenocytes enriched for NK cells showed that KMC tumour cells are efficiently killed by NK cells, equivalent to the established NK target cell line, YAC1 (40), in contrast with primary fibroblasts that are largely resistant to NK-mediated killing (Fig. 6I). Taken together, these data delineate a pathway by which KRAS and MYC cooperate to evade NK-mediated anti-tumour activity in PDAC through suppression of the Type I Interferon pathway (Fig. 6J).
Figure 6. Cxcl13 mediates Interferon signalling to B and NK cells in PDAC.
A) Normalised RNA-SEQ reads of Cxcl13 expression in KMC (N=6) and KMC Miz1fl/fl (N=5) pancreatic tumours. B) In situ hybridization (ISH) analysis of Cxcl13-expressing cells in KMC and KMC-Miz1fl/fl tumours. Lower panels show IHC for F4/80. Scale bar = 100μm. C) RT-PCR analysis of Cxcl13 gene expression in bone marrow-derived macrophages upon treatment (10ng/ml for 4hrs) with recombinant mouse IFNβ1. Mean and SEM of technical replicates from 1 of 3 independent experiments. T test; nd = none detected. D) RT-PCR analysis of Cxcl13 gene expression in bone marrow-derived macrophages after 24hrs treatment with conditioned media from MYC-depleted or control KMC tumour cells. Macrophages were pre-treated (20μg/ml, overnight) with IFNAR1 blocking antibody or isotype control, where indicated. Mean and SEM of technical replicates from 1 of 3 independent experiments. ANOVA and Tukey post-hoc test; nd = none detected. E) IHC detection of tumour infiltrating NK and B cells after 3 weeks blockade of Cxcl13 in KMC-Miz1fl/fl PDAC. Scale bars = 100μm. F) Quantification of tumour infiltrating NK (NKp46+) and B (CD45R+) cells in untreated (N=11 & 10, respectively, from Figure 5G) and anti-Cxcl13-treated (N=6) KMC-Miz1fl/fl PDAC. Mann Whitney test. G) Overall survival of KMC-Miz1fl/fl mice treated with (N=6) or without (N=12, from Fig. 5F) anti-Cxcl13 blocking antibody for 3 weeks. Mantel Cox logrank test (G, H). H) Overall survival of KMC-Miz1fl/fl mice treated with (N=6) or without (N=12, from Fig. 5F) anti-NK1.1 depleting antibody for 4 weeks. I) FACS analysis of Zombie NIR™ labelling of target cells (KMC, Yac-1, Fibroblasts) co-cultured with IL-2 stimulated, NK cell enriched splenocytes for 4hrs. Mean and SEM of 3 independent experiments shown. 2-way ANOVA and Tukey’s multiple comparisons test. J) Model showing the mechanism of MYC/Miz1-dependent immune evasion in PDAC. For all panels, *** denotes p<0.001, **denotes p<0.01, * denotes p<0.05, ns = not significant.
Discussion
The evasion of anti-tumour immunity has emerged in recent years as a major hallmark of cancer that presents genuine therapeutic opportunities through the targeted suppression of immune-evasion mechanisms, often with long-term patient benefit. Most efforts to date have focussed on mechanistic evasion of T lymphocyte-mediated tumour immunity, with targeted blockade of CTLA4 and PD1/PD-L1 establishing a paradigm for successful therapeutic exploitation of immuno-oncology. Here we present evidence of a novel mechanism of evasion of innate tumour immunity achieved through cooperation of the frequently dysregulated oncogenes MYC and KRAS via suppression of the Type I Interferon pathway.
We identified the Type I Interferon pathway as a major target of oncogene-mediated suppression through the unbiased analysis of multiple RNA-SEQ datasets: comparing end-stage pancreatic tumours driven by KRasG12D alone with those driven by the combination of KRasG12D & MYC; comparing KMC-derived tumour cells depleted of MYC, Miz1 or KRas with mock-depleted parental cells; comparing MEFs upon acute activation of lsl-KRasG12D, Rosa26DM-lsl-MYC, or both, with wild-type littermate controls. In all such comparisons the Type I Interferon pathway ranked amongst the pathways most significantly regulated. Although our mechanistic analysis here was performed exclusively in the context of pancreatic cancer, the conservation of this regulation in MEFs suggests that suppression of Type I Interferons may be a general feature of cancers wherein MYC and KRAS are dysregulated. For instance, a strong negative correlation between tumours with an IRF8/9 gene signature and those with a MAX gene signature (MAX being the obligate heterodimerising partner of MYC) was recently reported in Mesothelioma (41). Moreover, the Type I Interferon gene cluster is syntenic with CDKN2A/B on Chromosome 9 and is frequently co-deleted with the latter in a spectrum of human cancers: Across multiple cancers, patients with co-deletion show significantly reduced overall survival compared with those that retain the Interferon gene cluster, providing further evidence of an anti-tumour role for this pathway and a selective pressure for cancers to reduce its activity (42).
Mechanistically we present evidence of repressive MYC/Miz1 complexes binding to the promoters of STAT1, STAT2, IRF5 and IRF7. These data are complemented by a previous report of MYC/Miz1 binding to the Stat1 promoter in KPC (KRasG12D;Tp53R172H) murine pancreatic tumour cells (18) and MYC repression of STAT2 in breast cancer cells (43). Recent work has shown that basal expression of many Interferon-stimulated genes is regulated by STAT2/IRF9 complexes independently of IFNAR signalling. Upon Interferon-dependent activation of IFNAR, signalling through JAK family kinases leads to STAT1 binding to STAT2/IRF9 to form the ISGF3 complex (36), driving expression of, amongst other genes, IRF7, which in turn is strictly required for IFNα/β production (44). Note that other STAT family members may have very different effects, as recently evidenced by STAT6 driving expression of c-Myc during PDAC progression in response to Th2-derived interleukins (45). MYC/Miz1 thus attenuates the Type I Interferon cascade at multiple points that would limit both basal and Interferon-stimulated gene expression. The effects of KRAS on this pathway appear to be largely a consequence of MYC stabilisation, at least in PDAC cells (25), however, signalling cross-talk between the RAS and JAK-STAT pathways likely contributes to some of the MYC-independent effects observed, or indeed to the stronger effects of KRAS modulation in these systems (46). Similarly, Miz1 likely has additional roles in PDAC beyond its function as a MYC-corepressor, evidenced by divergent regulation of some 40% of MYC-regulated genes upon acute depletion of Miz1 in KMC cells. The fact that Miz1 could be efficiently deleted in ductal tumour epithelium, unlike endogenous Myc, argues that its role in PDAC is more limited and likely explains the stronger impact of the floxed Miz1 genotype upon Cxcl13 induction in vivo, as compared with the floxed c-Myc allele.
We demonstrate that de-repression of the Type I Interferon pathway in PDAC tumour cells stimulates production in nearby macrophages of the canonical B cell chemokine Cxcl13, resulting in tumour infiltration of B and NK cells. Although the recruitment of NK cells may well be an indirect consequence of B cell recruitment, as suggested by recent data (21), evidence is emerging of a direct role for CXCL13 in NK cell recruitment and a subset of NK cells express the CXCL13 receptor, CXCR5 (47). Depletion of Cxcl13 or blockade of the Type I Interferon receptor, IFNAR, each suppressed B and NK cell infiltration and reversed the survival benefit of Miz1 deletion in KMC tumours. Using co-culture experiments we showed that NK cells can efficiently kill KMC tumour cells, and that their depletion also negated the survival benefit observed upon Miz1 deletion. As is the case with cytotoxic T cells, NK cells express a variety of activating and repressing surface markers that are amenable to extrinsic modulation. As such, targeted activation of NK cells is emerging as an attractive therapeutic option, particularly in tumours with low mutation burden or lack of PD-L1 expression (48).
It would be implausible to suggest that the dramatic acceleration of PDAC development observed upon MYC and KRasG12D co-expression is entirely explained by these effects on the immune landscape. Indeed, these are highly pleiotropic oncogenes with broad influence over many cell-intrinsic and cell-extrinsic biological features (21, 45). It is notable however that regulation of the Type I Interferon pathway occurs upon physiological expression of KRasG12D or upon very modest overexpression of c-MYC, suggesting that this regulation may be present from the very outset of tumour initiation. This raises the exciting possibility of early intervention to reactivate Interferon signalling. Although we show that pharmacological inhibition of the RAS pathway can restore Interferon related gene expression in vitro, we would caution against such an approach in vivo given that immune cells use the very same RAS pathway to achieve their own rapid expansion (49). What is clear however is that a very modest increase in MYC suffices to dramatically accelerate KRAS-initiated tumours to metastatic PDAC, as we previously reported for lung adenocarcinoma (12), or indeed to alone drive PNET formation. The different aetiologies of these phenotypes remain to be fully explored.
Methods
Animal Studies
All experiments involving mice were approved by the local animal welfare (AWERB) committee and conducted under UK Home Office licences, PE47BC0BF, 70/7950 and 70/8375. Mice were maintained on a constant 12hr light/dark cycle, fed and watered ad libitum, and all were mixed background (FVBN and C57Bl/6). The following genetically modified mice were described previously: Lsl-KRasG12D (5); Rosa26DM-lsl-MYC (23); Pdx1-Cre (6); Pdx1-CreER (29); c-MycFl (50); Hprt-lsl-IRFP (30); Miz1ΔPOZ (38). All genotyping was performed by Transnetyx Inc.. To induce allele recombination, CreERT2 was activated in the pancreas of 5-6-week old mice by IP injection of 2mg/kg Tamoxifen (dissolved in peanut oil) for 3 consecutive days. For overall survival analysis, cohorts of mice were monitored and euthanized when clinical end-point was reached. Endpoint monitoring was performed by facility staff without knowledge of genotype. For histological analysis, mouse tissues were fixed with 10% neutral buffered formalin overnight. At euthanasia, a small portion of pancreatic tumour was snap frozen for RNA analysis. To determine the influence of Cxcl13, a cohort of randomly selected KMC-Miz1ΔPOZ mice were treated with Cxcl13 blocking antibody (0.5μg, IV, AF470, R&D Systems) or isotype control (Goat IgG, AB-108-C R&D) from 6 weeks of age, twice weekly for 3 weeks, and sacrificed at clinical end-point. To determine the influence of IFN signalling, a cohort of randomly selected KMC-Miz1ΔPOZ mice were treated with IFNAR1 blocking antibody (clone MAR1.5A3, BE0241, BioxCell) IP, 200μg/20g body mass on first dose and 100 μg/20g for the following 6 doses, from 6 weeks of age, once every 3 days for 3 weeks, and sacrificed at clinical endpoint. For NK cell depletion, 6 week old mice were treated with anti-NK1.1 (clone PK136, BP0036, BioxCell), IP, 100ug/20g, 2 doses in the first week and once per week for the following 3 weeks. To verify NK cell depletion following the NK1.1 antibody treatment, blood was sampled by tail bleed from mice prior to and after 2 weeks treatment. Cells were stained with CD45 (10311, Biolegend), CD3 (100327, Biolegend), NK1.1 (108707, Biolegend), NkP46 (137612, Biolegend) & Zombie NIR10311, (423106, Biolegend) after RBC lysis (eBioscience, 00-4333-57) and analysed by flow cytometry (BD Fortessa). FACS profiles were generated and quantified using FlowJo (Tree Star). Rosa26DM-lsl-MYC mice are available from Jaxmice at https://www.jax.org/strain/033805.
Immunohistochemistry and Tissue Analysis
All immunohistochemistry and in situ hybridization staining were performed on 4 μm formalin fixed paraffin embedded sections which had previously been heated to 60°C for 2 hours. Peroxidase blocking was performed for 10 mins in 1% H2O2 diluted in H2O, followed by heat mediated or enzyme mediated antigen retrieval. Non-specific antibody binding was blocked with up to 3% BSA or up to 5% normal goat serum for 1h at RT. The following antibodies were used at the indicated dilution and indicated antigen retrieval method: Synaptophysin (ab8049, 1:50, pH6), CD45R (ab64100, 1:200, ER2 Leica), NKp46 (aF2225, 1:200, pH6), F4/80 (ab6640, 1:100, Enzyme 1 Leica), MYC (ab32072, 1:100, pH6)[Abcam], KRasG12D (CST14429, 1:50, pH9)[Cell Signaling Tech.], pan-Cytokeratin (MS-343, 1:100, pH6)[ThermoFisher]. Nkp46, Pan-cytokeratin and Synaptophysin were stained on a Dako AutostainerLink48, CD45R and F4/80 were stained on the Leica Bond Rx autostainer; KRasG12D and MYC were stained manually. Mouse EnVision (Agilent), Goat ImmPRESS Kit and Rabbit (Vector Labs) were used as secondary antibodies. The HRP signal was detected using liquid DAB (Agilent and Invitrogen). Sections were counterstained with haematoxylin and cover-slipped using DPX mount (Cellpath, UK). In situ-hybridisation detection for Cxcl13 (ACD 406318), Miz1 (ACD 520288) and PP1β (ACD 313918; Advanced Cell Diagnostic, Hayward, CA) mRNA was performed using RNAscope 2.5 LS detection kit (ACD). Detection of murine c-Myc (ACD 712368) was performed using a BaseScope 2.5 LS detection kit (ACD). Both techniques were performed on a Leica Bond Rx autostainer, strictly adhering to ACD protocols. Tumour area was calculated using HaLo software (Indica Labs) as the percent area of pancreas occupied by ductal adenocarcinoma (PDAC) and neuroendocrine tumours (PNET), measured on haematoxylin & eosin (H&E) stained sections. To quantify the immune infiltration, positive cells were counted manually using QuPath cell (https://qupath.github.io/) counter function and normalized to tumour area (PDAC+PNET) calculated as described above.
Cell culture
Human pancreatic cell lines were obtained from ATCC and were maintained in DMEM (MIA PaCa-2) or RPMI (AsPC-1, DAN-G) supplemented with 10% FBS and Pen/Strep. All cell lines were validated using an approved in-house validation service (CRUK-BICR) and tested periodically for mycoplasma. Cell lines were thawed from primary stocks maintained under liquid nitrogen & cultured for a maximum of 8 weeks (< 20 passages), during which time all experiments were performed. Cells were treated with 10nM (DAN-G) or 50nM (AsPC-1) Trametinib (MedChem Express) for 16hrs and used for protein and RNA analysis. Primary mouse embryonic fibroblasts (MEFs) were generated from E13.5 embryos by interbreeding mice carrying Rosa26DM-lsl-MYC, lsl-KRasG12D and c-Mycfl/fl to obtain desired genotypes. MEFs were cultured in DMEM with 10% FBS & Pen/Strep in 3% oxygen. To activate floxed alleles, MEFs were infected with 300 pfu/cell of Adeno-Cre (University of Iowa, vector core facility) and harvested at 24hrs. Mouse pancreatic tumour lines were generated from end-stage KMC mice. Tumours were disintegrated mechanically and cultured in DMEM supplemented with 20% FBS and Pen/Strep. Once established, KMC cells were maintained in DMEM supplemented with 10% FBS and Pen/Strep. For cell death measurements, cells were trypsinized, quenched with 1% BSA followed by replacement of original supernatant, and centrifuged at 300 x G for 5mins; 200μl of Annexin binding buffer (10mM HEPES pH 7.40, 140mM NaCl, 2.5mM CaCl2) and 2μl of Annexin V-APC (Biolegend 640920) were added to the pellet and incubated for 15mins. Propidium iodide (PI, 10μg/ml) was added immediately prior to FACS analysis. For immunoblotting, whole cell lysates were prepared in RIPA buffer (150mM NaCl, 50mM Tris pH 7.5, 1% NP-40, 0.5% Sodium Deoxycholic acid, 1% SDS, plus complete protease and phosphatase inhibitor cocktail) followed by sonication (40% Amp for 5s). MYC (ab32072), Vinculin (ab129002), Histone H2B (ab1790) [Abcam], pERK1/2 (CST 4370), tERK1/2 (CST 4695) [Cell Signaling Tech.], Actin (SC 47778) [Santa Cruz], were used as primary antibodies. Secondary horseradish peroxidase-conjugated antibodies (α-mouse IgG NA931V; α-rabbit NA934V, both GE Healthcare; α -goat IgG, Vector Labs PI-9500) were detected by chemiluminescence (Biorad western blotting substrate 1705060). To deplete MYC in human pancreatic cell lines, following siRNA were used: SiMYC 5 (Qiagen SI00300902), SiMYC 9 (SI03101847). To deplete MYC in mouse pancreatic tumour lines, a combination of mouse and human siRNA were used as follows: Pool 1 (SI01321012, SI00300902), Pool 2 (SI01321012, SI03101847), Pool 3 (SI01320991, SI00300902) (all fromQiagen). To deplete Miz1 in mouse lines SI01320991 (Qiagen) was used. To deplete KRas in mouse lines, SI02742439 (Qiagen) was used.
Bone marrow derived macrophages (BMDM)
Bone marrow cells were isolated from wildtype mice by snipping the end of the femur and centrifuging at 5000 rpm for 1min. Monocytes were differentiated into macrophages by culturing in non-TC treated plates (Corning, 430597) for 6 days in RPMI medium containing M-CSF (10ng/ml). Media containing M-CSF were replaced after 3 days. Where indicated, BMDM’s were treated with recombinant mouse IFN-beta1 (8234-MB, R&D systems) for 4hrs (10ng/ml). KMC tumour cells were used to generate conditioned media. Fresh media (DMEM+20%FBS) were added 24hrs after transfection with siMYC or non-targeting control siRNA. 24hrs later, conditioned media from MYC or non-targeting control depleted cells were collected, centrifuged (1200 rpm, 5mins), supplemented with M-CSF and, where indicated, anti-IFNAR1 blocking antibody (20ng/ml, BE0241, BioxCell) or mIgG (BE0083) prior to BMDM treatment. BMDMs were additionally pre-treated with anti-IFNAR1 or mIgG overnight. BMDM’s were treated with conditioned media for 24hrs. To detect Cxcl13 expression in BMDM’s, cDNA was synthesized with Oligo-dT primer (M510, PromegA), pre-amplified (40nM of each primers, SYBR Green buffer, 1mg/ml BSA, 2.5% glycerol) for GusB and Cxcl13 for 20cycles. Diluted cDNA (1:20 in 10mMTris and 1mM EDTA) was quantified by real time PCR using SYBR Green method (VWR QUNT95072).
In vitro cytotoxicity assay
NK cells were activated ex vivo as previously described (51): Freshly isolated splenocytes of naïve mice at 3x106 cells/mL were treated for 72h with 50 ng/mL of IL-2 (Biolegend, Catalog # 575404). Target cells, stained with a cell trace dye (Thermofisher, Catalog # C34567 or V12883) following manufacturer instructions, were co-culture with different ratios (0, 5 and 10) of IL-2 stimulated splenocytes, enriched for activated NK cells. After 4h co-incubation, cells were harvested, stained with Zombie NIR (Biolegend, Catalog # 423106) and analyzed by flow cytometry (BD Fortessa). FlowJo (Tree Star) software was used for analysis.
Chromatin Immunoprecipitation
Dan-G cells were cross-linked with formaldehyde (1% final conc.) for 10 min at 37°C. Cells were scraped in PBS containing protease inhibitors and centrifuged at 300 x G for 5 min (4°C). Cells were resuspended in lysis buffer 1 (5mM PIPES pH8, 85mM KCl, 0.5% NP40) and incubated on ice for 20 min. After lysis, cells were centrifuged at 300 x G for 5 min (4°C) and the pellets were resuspended in lysis buffer II (RIPA). After 10 min incubation, lysates were sonicated to fragment DNA. Chromatin was precleared and IP’ed with 5μg of Myc (N262, SC764) or Miz1(B10, SC136985) antibody overnight at 4°C followed by incubation with 60μl of Protein G beads (17-0618-01, GE). Rabbit IgG (CS2729) or Mouse IgG (BE0083) was used as antibody controls. DNA was then eluted using elution buffer (1%SDS, 0.1M NaHCO3), de-crosslinked and purified using Qiagen PCR purification kit for Q-PCR analysis. For Re-CHIP, α-MYC or RIgG control IP’ed chromatin was eluted with Re-CHIP elution buffer (1xTE, 2%SDS, 15mM DTT) at 37°C for 30mins, IP’ed with 5 μg of Miz1 (B10, Sc136985) antibody overnight at 4°C. The immunoprecipitated DNA was eluted as described above. The following primers sets were used: IRF7 promoter: gacaccagcctgaccaacatag, acaatcttggcccaccacaac; IRF5 promoter: aagagcaagagttaccaagcga, taaagaacctcaccccagaacc; IRF5 intronic region: ctctggctttctcctgcagacc, cccattgaagccctgggtact; STAT1 promoter: gctggtcgtcactctcacaa, tcgcctactcttaaggggct; STAT2 promoter: tccaggctcctcaagctagt, gcactttctacgaggggagg; VAMP4 promoter: cagtggttgttcctcccta, ccgagccctattcacctaaa.
RNA-SEQ Analysis
Ribosome-depleted total RNA was used for analysis of mouse embryo fibroblast gene expression. Poly-A enriched RNA was analysed for bulk tumour and KMC cell line gene expression. Datasets are available from ArrayExpress under accession numbers E-MTAB 6824 (MEFs); E-MTAB 8792 (KMC cell lines) and E-MTAB 8797 (PDAC tumours). Full details of NGS protocols and downstream analysis are provided in the Supplementary Materials.
Statistical analysis
Raw data obtained from quantitative Real Time PCR, FACS, & growth curves were copied into Excel (Microsoft) or Graphpad prism spreadsheets. All Mean and SEM values of biological replicates were calculated using the calculator function. Graphical representation of such data was produced in Excel or Graphpad Prism. Statistical significance was determined by the Student’s T test. For multiple comparisons, ANOVA was used with a post-hoc Turkey test or post-hoc Fischer LSD test. For non-normally distributed data (eg. survival benefit; immune cell infiltration), Mantel-Cox (2-way comparison) or Kruskal-Wallis (multiple comparison) tests were performed. For RNA-SEQ data, Adjusted P values calculated in R are shown. * denotes P<0.05; ** denotes P<0.01; *** denotes P<0.005.
Supplementary Material
Significance.
We define herein a novel mechanism of evasion of NK cell-mediated immunity through the combined actions of endogenously expressed mutant KRAS and modestly deregulated expression of MYC, via suppression of the Type I Interferon pathway. Restoration of Interferon signalling may improve outcomes for PDAC patients.
Acknowledgements
The authors thank all members of the Murphy lab past and present who contributed to the development of this work. We would like to thank the core services at the CRUK Beatson Institute with particular thanks to the Biological Services Unit and histology core facility. Thanks to Kirsteen Campbell, Florian Bock and the Stephen Tait lab for helpful discussions and to Catherine Winchester for critical reading of the manuscript. Artwork was provided by Sara Zanivan using images from https://smart.servier.com/. Miz1ΔPOZ mice were provide by Martin Eilers.
Financial Support
Funding for this work was provided by Worldwide Cancer Research grant AICR 15-0279; Pancreatic Cancer UK Future Leaders Academy 2017; European Commission Marie-Curie actions PCIG13-GA-2013-618448, SERPLUC and H2020-MCSA-IF-2015-705190-NuSiCC; Cancer Research UK grants A21139 (to O.J.S.), and A23983 (to L.M.C.). N.M received a L’Oreal/UNESCO Women in Science 2018 award. T.M. was funded through a British Lung Foundation studentship (BLF-APHD13-5). Additional support was provided by Wellcome Trust grant 105614/Z/14/Z; CRUK Glasgow Cancer Centre grant A25142 and CRUK Beatson Institute core facilities grant A17196.
Footnotes
Conflicts of Interest
The Murphy lab currently receives funding from Puma Biotechnology and the Merck Group for work unrelated to this manuscript. The Morton lab has received funding from Elstar Therapeutics and UCB for work unrelated to this manuscript. The Sansom lab currently receives funding from Astra Zeneca, Celgene, Novartis and Cancer Research Technologies, and previously received funding from Jansen Pharmaceuticals. Andrew Biankin is a consultant for Celgene, AstraZeneca, Elstar Therapeutics & My Tomorrow; has received honororia &/or travel-related expenses from Celgene, AstraZeneca, Havas Lynx, Roche, Elstar Therapeutics and MyTomorrow; receives royalties from Agilent; and receives research funding from Celgene & Astra Zeneca. The remaining authors declare no potential conflicts of interest.
References
- 1.Rahib L, Smith BD, Aizenberg R, Rosenzweig AB, Fleshman JM, Matrisian LM. Projecting cancer incidence and deaths to 2030: the unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res. 2014;74(11):2913–21. doi: 10.1158/0008-5472.CAN-14-0155. [DOI] [PubMed] [Google Scholar]
- 2.Neoptolemos JP, Kleeff J, Michl P, Costello E, Greenhalf W, Palmer DH. Therapeutic developments in pancreatic cancer: current and future perspectives. Nat Rev Gastroenterol Hepatol. 2018;15(6):333–48. doi: 10.1038/s41575-018-0005-x. [DOI] [PubMed] [Google Scholar]
- 3.Bailey P, Chang DK, Nones K, Johns AL, Patch AM, Gingras MC, et al. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature. 2016;531(7592):47–52. doi: 10.1038/nature16965. [DOI] [PubMed] [Google Scholar]
- 4.Cancer Genome Atlas Research Network. Electronic address aadhe, Cancer Genome Atlas Research N Integrated Genomic Characterization of Pancreatic Ductal Adenocarcinoma. Cancer Cell. 2017;32(2):185–203.:e13. doi: 10.1016/j.ccell.2017.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jackson EL, Willis N, Mercer K, Bronson RT, Crowley D, Montoya R, et al. Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes Dev. 2001;15(24):3243–8. doi: 10.1101/gad.943001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hingorani SR, Petricoin EF, Maitra A, Rajapakse V, King C, Jacobetz MA, et al. Preinvasive and invasive ductal pancreatic cancer and its early detection in the mouse. Cancer Cell. 2003;4(6):437–50. doi: 10.1016/s1535-6108(03)00309-x. [DOI] [PubMed] [Google Scholar]
- 7.Aguirre AJ, Bardeesy N, Sinha M, Lopez L, Tuveson DA, Horner J, et al. Activated Kras and Ink4a/Arf deficiency cooperate to produce metastatic pancreatic ductal adenocarcinoma. Genes Dev. 2003;17(24):3112–26. doi: 10.1101/gad.1158703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Morton JP, Timpson P, Karim SA, Ridgway RA, Athineos D, Doyle B, et al. Mutant p53 drives metastasis and overcomes growth arrest/senescence in pancreatic cancer. Proc Natl Acad Sci U S A. 2010;107(1):246–51. doi: 10.1073/pnas.0908428107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mueller S, Engleitner T, Maresch R, Zukowska M, Lange S, Kaltenbacher T, et al. Evolutionary routes and KRAS dosage define pancreatic cancer phenotypes. Nature. 2018;554(7690):62–8. doi: 10.1038/nature25459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cicchini M, Buza EL, Sagal KM, Gudiel AA, Durham AC, Feldser DM. Context-Dependent Effects of Amplified MAPK Signaling during Lung Adenocarcinoma Initiation and Progression. Cell Rep. 2017;18(8):1958–69. doi: 10.1016/j.celrep.2017.01.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kerr EM, Gaude E, Turrell FK, Frezza C, Martins CP. Mutant Kras copy number defines metabolic reprogramming and therapeutic susceptibilities. Nature. 2016;531(7592):110–3. doi: 10.1038/nature16967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kruspig B, Monteverde T, Neidler S, Hock A, Kerr E, Nixon C, et al. The ERBB network facilitates KRAS-driven lung tumorigenesis. Sci Transl Med. 2018;10(446) doi: 10.1126/scitranslmed.aao2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moll HP, Pranz K, Musteanu M, Grabner B, Hruschka N, Mohrherr J, et al. Afatinib restrains K-RAS-driven lung tumorigenesis. Sci Transl Med. 2018;10(446) doi: 10.1126/scitranslmed.aao2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kapoor A, Yao W, Ying H, Hua S, Liewen A, Wang Q, et al. Yap1 activation enables bypass of oncogenic Kras addiction in pancreatic cancer. Cell. 2014;158(1):185–97. doi: 10.1016/j.cell.2014.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wirth M, Schneider G. MYC: A Stratification Marker for Pancreatic Cancer Therapy. Trends Cancer. 2016;2(1):1–3. doi: 10.1016/j.trecan.2015.12.002. [DOI] [PubMed] [Google Scholar]
- 16.Schleger C, Verbeke C, Hildenbrand R, Zentgraf H, Bleyl U. c-MYC activation in primary and metastatic ductal adenocarcinoma of the pancreas: incidence, mechanisms, and clinical significance. Mod Pathol. 2002;15(4):462–9. doi: 10.1038/modpathol.3880547. [DOI] [PubMed] [Google Scholar]
- 17.Farrell AS, Joly MM, Allen-Petersen BL, Worth PJ, Lanciault C, Sauer D, et al. MYC regulates ductal-neuroendocrine lineage plasticity in pancreatic ductal adenocarcinoma associated with poor outcome and chemoresistance. Nat Commun. 2017;8(1):1728. doi: 10.1038/s41467-017-01967-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Walz S, Lorenzin F, Morton J, Wiese KE, von Eyss B, Herold S, et al. Activation and repression by oncogenic MYC shape tumour-specific gene expression profiles. Nature. 2014;511(7510):483–7. doi: 10.1038/nature13473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saborowski M, Saborowski A, Morris JPt, Bosbach B, Dow LE, Pelletier J, et al. A modular and flexible ESC-based mouse model of pancreatic cancer. Genes Dev. 2014;28(1):85–97. doi: 10.1101/gad.232082.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hayes TK, Neel NF, Hu C, Gautam P, Chenard M, Long B, et al. Long-Term ERK Inhibition in KRAS-Mutant Pancreatic Cancer Is Associated with MYC Degradation and Senescence-like Growth Suppression. Cancer Cell. 2016;29(1):75–89. doi: 10.1016/j.ccell.2015.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sodir NM, Kortlever RM, Barthet VJA, Campos T, Pellegrinet L, Kupczak S, et al. Myc instructs and maintains pancreatic adenocarcinoma phenotype. Cancer Discov. 2020 doi: 10.1158/2159-8290.CD-19-0435. [DOI] [PubMed] [Google Scholar]
- 22.Herold S, Wanzel M, Beuger V, Frohme C, Beul D, Hillukkala T, et al. Negative regulation of the mammalian UV response by Myc through association with Miz-1. Mol Cell. 2002;10(3):509–21. doi: 10.1016/s1097-2765(02)00633-0. [DOI] [PubMed] [Google Scholar]
- 23.Neidler S, Kruspig B, Hewit K, Monteverde T, Gyuraszova K, Braun A, et al. Identification of a Clinically Relevant Signature for Early Progression in KRAS-Driven Lung Adenocarcinoma. Cancers (Basel) 2019;11(5) doi: 10.3390/cancers11050600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Facchini LM, Chen S, Marhin WW, Lear JN, Penn LZ. The Myc negative autoregulation mechanism requires Myc-Max association and involves the c-myc P2 minimal promoter. Mol Cell Biol. 1997;17(1):100–14. doi: 10.1128/mcb.17.1.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vaseva AV, Blake DR, Gilbert TSK, Ng S, Hostetter G, Azam SH, et al. KRAS Suppression-Induced Degradation of MYC Is Antagonized by a MEK5-ERK5 Compensatory Mechanism. Cancer Cell. 2018;34(5):807–22.:e7. doi: 10.1016/j.ccell.2018.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Santana-Codina N, Roeth AA, Zhang Y, Yang A, Mashadova O, Asara JM, et al. Oncogenic KRAS supports pancreatic cancer through regulation of nucleotide synthesis. Nat Commun. 2018;9(1):4945. doi: 10.1038/s41467-018-07472-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lenoir WF, Lim TL, Hart T. PICKLES: the database of pooled in-vitro CRISPR knockout library essentiality screens. Nucleic Acids Res. 2018;46(D1):D776–D80. doi: 10.1093/nar/gkx993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wilson ME, Scheel D, German MS. Gene expression cascades in pancreatic development. Mech Dev. 2003;120(1):65–80. doi: 10.1016/s0925-4773(02)00333-7. [DOI] [PubMed] [Google Scholar]
- 29.Gu G, Dubauskaite J, Melton DA. Direct evidence for the pancreatic lineage: NGN3+ cells are islet progenitors and are distinct from duct progenitors. Development. 2002;129(10):2447–57. doi: 10.1242/dev.129.10.2447. [DOI] [PubMed] [Google Scholar]
- 30.Hock AK, Cheung EC, Humpton TJ, Monteverde T, Paulus-Hock V, Lee P, et al. Development of an inducible mouse model of iRFP713 to track recombinase activity and tumour development in vivo. Sci Rep. 2017;7(1):1837. doi: 10.1038/s41598-017-01741-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Murphy DJ, Junttila MR, Pouyet L, Karnezis A, Shchors K, Bui DA, et al. Distinct thresholds govern Myc’s biological output in vivo. Cancer Cell. 2008;14(6):447–57. doi: 10.1016/j.ccr.2008.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sears R, Nuckolls F, Haura E, Taya Y, Tamai K, Nevins JR. Multiple Ras-dependent phosphorylation pathways regulate Myc protein stability. Genes Dev. 2000;14(19):2501–14. doi: 10.1101/gad.836800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ram DR, Manickam C, Hueber B, Itell HL, Permar SR, Varner V, et al. Tracking KLRC2 (NKG2C)+ memory-like NK cells in SIV+ and rhCMV+ rhesus macaques. PLoS Pathog. 2018;14(5):e1007104. doi: 10.1371/journal.ppat.1007104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kortlever RM, Sodir NM, Wilson CH, Burkhart DL, Pellegrinet L, Brown Swigart L, et al. Myc Cooperates with Ras by Programming Inflammation and Immune Suppression. Cell. 2017;171(6):1301–15.:e14. doi: 10.1016/j.cell.2017.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Oppmann B, Lesley R, Blom B, Timans JC, Xu Y, Hunte B, et al. Novel p19 protein engages IL-12p40 to form a cytokine, IL-23, with biological activities similar as well as distinct from IL-12. Immunity. 2000;13(5):715–25. doi: 10.1016/s1074-7613(00)00070-4. [DOI] [PubMed] [Google Scholar]
- 36.Platanitis E, Demiroz D, Schneller A, Fischer K, Capelle C, Hartl M, et al. A molecular switch from STAT2-IRF9 to ISGF3 underlies interferon-induced gene transcription. Nat Commun. 2019;10(1):2921. doi: 10.1038/s41467-019-10970-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lorenzin F, Benary U, Baluapuri A, Walz S, Jung LA, von Eyss B, et al. Different promoter affinities account for specificity in MYC-dependent gene regulation. Elife. 2016;5 doi: 10.7554/eLife.15161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kosan C, Saba I, Godmann M, Herold S, Herkert B, Eilers M, et al. Transcription factor miz-1 is required to regulate interleukin-7 receptor signaling at early commitment stages of B cell differentiation. Immunity. 2010;33(6):917–28. doi: 10.1016/j.immuni.2010.11.028. [DOI] [PubMed] [Google Scholar]
- 39.Denton AE, Innocentin S, Carr EJ, Bradford BM, Lafouresse F, Mabbott NA, et al. Type I interferon induces CXCL13 to support ectopic germinal center formation. J Exp Med. 2019;216(3):621–37. doi: 10.1084/jem.20181216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nieswandt B, Hafner M, Echtenacher B, Mannel DN. Lysis of tumor cells by natural killer cells in mice is impeded by platelets. Cancer Res. 1999;59(6):1295–300. [PubMed] [Google Scholar]
- 41.Hmeljak J, Sanchez-Vega F, Hoadley KA, Shih J, Stewart C, Heiman D, et al. Integrative Molecular Characterization of Malignant Pleural Mesothelioma. Cancer Discov. 2018;8(12):1548–65. doi: 10.1158/2159-8290.CD-18-0804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ye Z, Dong H, Li Y, Ma T, Huang H, Leong HS, et al. Prevalent Homozygous Deletions of Type I Interferon and Defensin Genes in Human Cancers Associate with Immunotherapy Resistance. Clin Cancer Res. 2018;24(14):3299–308. doi: 10.1158/1078-0432.CCR-17-3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Qiao X, Liu Y, Llamazares Prada M, Mohan AK, Gupta A, Jaiswal A, et al. UBR5 is co-amplified with MYC in breast tumors and encodes an ubiquitin ligase that limits MYC-dependent apoptosis. Cancer Res. 2020 doi: 10.1158/0008-5472.CAN-19-1647. [DOI] [PubMed] [Google Scholar]
- 44.Honda K, Yanai H, Negishi H, Asagiri M, Sato M, Mizutani T, et al. IRF-7 is the master regulator of type-I interferon-dependent immune responses. Nature. 2005;434(7034):772–7. doi: 10.1038/nature03464. [DOI] [PubMed] [Google Scholar]
- 45.Dey P, Li J, Zhang J, Chaurasiya S, Strom A, Wang H, et al. Oncogenic Kras driven metabolic reprogramming in pancreas cancer cells utilizes cytokines from the tumor microenvironment. Cancer Discov. 2020 doi: 10.1158/2159-8290.CD-19-0297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thomas SJ, Snowden JA, Zeidler MP, Danson SJ. The role of JAK/STAT signalling in the pathogenesis, prognosis and treatment of solid tumours. Br J Cancer. 2015;113(3):365–71. doi: 10.1038/bjc.2015.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang CL, Zhang P, Liu RT, Zhang N, Zhang M, Li H, et al. CXCR5-negative natural killer cells ameliorate experimental autoimmune myasthenia gravis by suppressing follicular helper T cells. J Neuroinflammation. 2019;16(1):282. doi: 10.1186/s12974-019-1687-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chiossone L, Dumas PY, Vienne M, Vivier E. Natural killer cells and other innate lymphoid cells in cancer. Nat Rev Immunol. 2018;18(11):671–88. doi: 10.1038/s41577-018-0061-z. [DOI] [PubMed] [Google Scholar]
- 49.Berridge MJ. Lymphocyte Activation in Health and Disease. Crit Rev Immunol. 2017;37(2-6):439–62. doi: 10.1615/CritRevImmunol.v37.i2-6.120. [DOI] [PubMed] [Google Scholar]
- 50.de Alboran IM, O’Hagan RC, Gartner F, Malynn B, Davidson L, Rickert R, et al. Analysis of C-MYC function in normal cells via conditional gene-targeted mutation. Immunity. 2001;14(1):45–55. doi: 10.1016/s1074-7613(01)00088-7. [DOI] [PubMed] [Google Scholar]
- 51.Lafreniere R, Rosenberg SA. Successful immunotherapy of murine experimental hepatic metastases with lymphokine-activated killer cells and recombinant interleukin 2. Cancer Res. 1985;45(8):3735–41. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.