Abstract
Lipoteichoic acids (LTA) are essential cell wall components in Gram-positive bacteria, including the human pathogen Staphylococcus aureus, contributing to cell adhesion, cell division and antibiotic resistance. Genetic evidence suggested that LtaA is the flippase that mediates the translocation of the lipid-linked-disaccharide that anchors LTA to the cell membrane, a rate-limiting step in S. aureus LTA biogenesis. Here, we present the structure of LtaA, describe its flipping mechanism, and show its functional relevance for S. aureus fitness. We demonstrate that LtaA is a proton-coupled antiporter-flippase that by sensing environmental pH contributes to S. aureus survival under physiological acidic conditions. Our results reveal the first known example of a proton dependent flippase and give foundations for the development of novel strategies to counteract S. aureus infections.
The Gram-positive pathogen Staphylococcus aureus is one of the leading causes of nosocomial infections around the globe1,2. Multiple strains have acquired resistance to clinically used antibiotics1,2, being methicillin-resistant S. aureus (MRSA) one of the most successful modern pathogens1–4. The cell wall of Gram-positive bacteria is primarily composed of peptidoglycan (PG), wall-associated proteins, capsular polysaccharides, and teichoic acids (TA)5,6. TA are divided in lipoteichoic acids (LTA) and wall-teichoic acids (WTA)5–7, both of which are necessary for proper localization of the cell wall elongation and division machinery, contribute to immune evasion by concealing cell wall epitopes, prevent recognition and opsonization by antibodies and have been shown to be important for adhesion, colonization and biofilm formation8–12. By altering TA composition, Gram-positive bacteria are able to resist the action of hydrophobic antibacterial agents, cationic antimicrobial peptides, β-lactam, and glycopeptide antibiotics7,13,14.
From the five types of LTA known to date5,15, the best characterized is type I LTA from S. aureus, which is recognized by Toll-like receptor 2 and has been associated with various inflammatory diseases ranging from minor skin diseases to severe sepsis16,17. S. aureus LTA are composed of a polymer of 1,3-glycerol-phosphate repeat units attached to C-6 of the non-reducing glucosyl of the anchor lipid-linked-disaccharide gentiobiosyl-diacylglycerol (anchor-LLD) embedded in the extracellular side of the plasma membrane5–7,18. Synthesis of the anchor-LLD occurs at the cytoplasmic leaflet of the membrane19–22. Thus, translocation of anchor-LLD across the plasma membrane is a requisite for extracellular assembly of LTA7,19,23 (Fig. 1A), thus constituting a rate-limiting step in LTA synthesis since it regulates the pool of precursor anchor-LLD available. This translocation reaction is presumed to be catalyzed by the flippase LtaA, although its flippase activity has not been demonstrated. LtaA is found in all known S. aureus strains and also in closely related Staphylococcus species19. Deletion of the ItaA gene in S. aureus leads to attenuated virulence during animal infection and alterations in LTA composition, such as anchoring to phosphatidylglycerol and longer polymer length7,19. Despite the clear relevance of LtaA in LTA biogenesis, remarkably little is known about its mechanism and function, in part due to the lack of structural information and the absence of comprehensive functional characterization in vitro and in vivo.
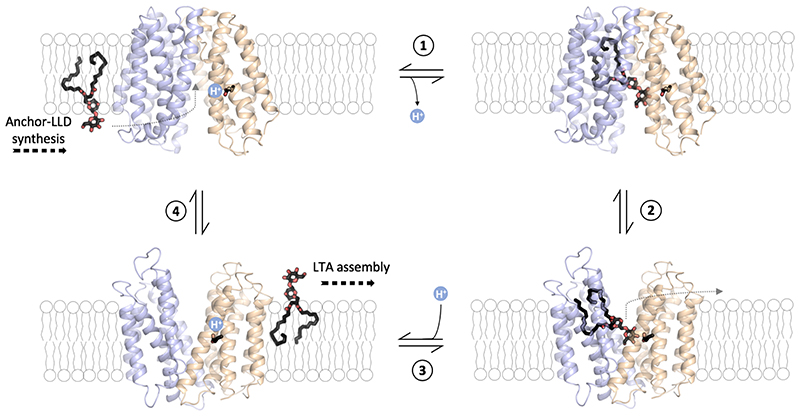
Figure 1. LtaA-catalyzed anchor-LLD flipping.
A. Lipoteichoic acid synthesis pathway in Staphylococcus aureus 5. YpfP synthesizes the LTA anchor-LLD on the cytoplasmic leaflet of the membrane, LtaA performs anchor-LLD flipping towards the extracellular side of the membrane where the LtaS polymerase assemble the 1,3-glycerol-phosphate polymer on anchor-LLD. The structure of the periplasmic domain of LtaS (PDB: 4UOO)22 is shown. PG: Phosphatidylglycerol; DAG: diacylglycerol; Glc: glucose; GroP: glycerol-phosphate; anchor- LLD: gentiobiosyl-diacylglycerol. Figure adapted from Percy and Gründling, 2014 5. B. Scheme of flipping assay. NBD-Anchor-LLD lipids (red spheres) are irreversibly reduced (black spheres) by dithionite (dit.). In protein-free liposomes only outer-leaflet fluorophores are reduced. In proteoliposomes containing LtaA (yellow boxes), a larger portion of the fluorophores are reduced due to exchange. Full fluorescence quenching will be achieved after prolonged incubation (dashed arrow). C. Flipping of NBD-Anchor-LLD by LtaA. Representative traces shown are from protein-free liposomes, proteoliposomes containing either LtaA or a functionally unrelated transporter (bacterial choline transporter) (n ≥ 3). Asterisk marks addition of dithionite. Source data for C are available with the paper online.
LtaA is a member of the Major Facilitator Superfamily (MFS) of transporters19. The MFS superfamily, ubiquitously distributed in all kingdoms of life, consists of 74 families classified based on their type of substrate24. In bacteria, MFS transporters are mainly used for the uptake of nutrients and extrusion of deleterious compounds. Whereas in humans, they are implicated in the transport of a wide range of toxins and drugs, and their malfunction leads to multiple diseases25–27. Currently, only six different MFS families have been structurally characterized28–34. However, there are no reports that describe the structure of flippases in any of the 74 MFS families.
To elucidate the mechanism of LtaA-catalyzed anchor-LLD flipping, we determined the structure of S. aureus LtaA, characterized its transport properties by in vitro flipping assays, and investigated its function and relevance in live S. aureus cells. The structure of LtaA revealed a 12 transmembrane helices (TM) MFS-fold and displays an outward-facing conformation. The central cavity of LtaA shows a unique amphiphilic architecture never seen before in any of the structurally characterized MFS transporters. We show that LtaA is an antiporter-flippase that couples anchor-LLD translocation to proton antiport, which confers LtaA pH-sensing properties used by S. aureus to cope with acidic stress found under normal physiological conditions.
Results
LtaA displays in vitro flipping activity
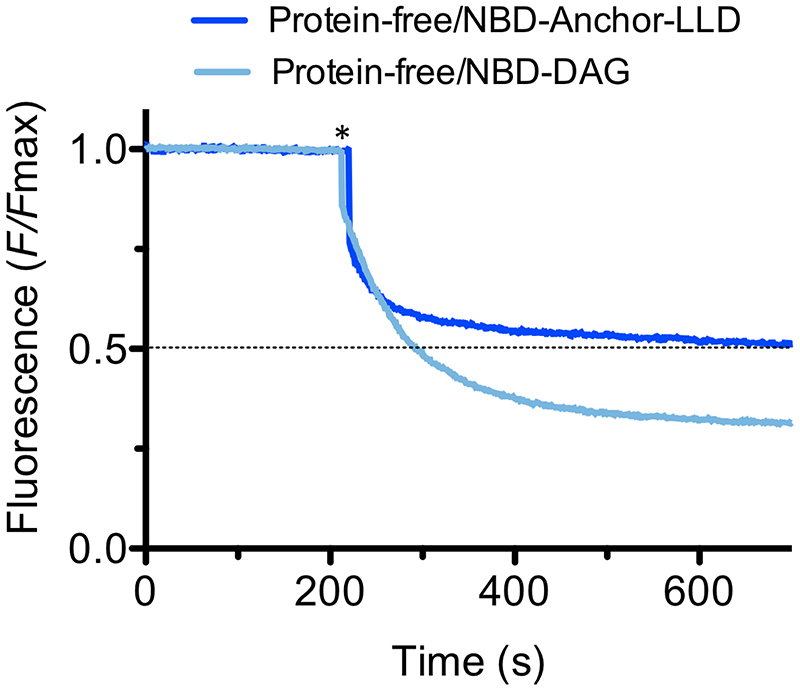
To characterize the flipping mechanism of LtaA in vitro, it is essential to establish reliable measurements of lipid translocation activity of LtaA wild type (WT) and variants. Lipid translocation has been investigated for other lipid transporters using fluorescently labelled substrates that when exposed on the outer leaflet of proteoliposomes are quenched by sodium dithionite, a membrane-impermeable reducing agent35–37. Because a suitable fluorescently labeled lipid substrate was so far not available for LtaA, we performed the chemoenzymatic synthesis of a nitrobenzoxadiazole (NBD) labeled anchor-LLD (NBD-anchor-LLD) using recombinantly expressed and purified YpfP 20,21. In S. aureus, YpfP synthesizes the native LtaA substrate during LTA assembly (Fig. 1A). In an in vitro reaction, purified YpfP catalyzed the reaction between UDP-Glc and NBD-labelled diacylglycerol (NBD-DAG) to produce NBD-anchor-LLD. Tandem MS/MS, parallel-reaction monitoring (PRM) High resolution LC-MS, HPLC-MS, and 1D 1H NMR spectroscopy, confirmed the presence of NBD-anchor-LLD in enriched extracts (Supplementary Fig. 1–8, Supplementary Table 1-6).
For the functional flipping assay, we incorporated NBD-anchor-LLD into protein-free liposomes or into LtaA proteoliposomes. Upon addition of dithionite, the fluorescence of protein-free liposomes plateaus at about 50% of the total fluorescence (Fig. 1B,C), consistent with the expectation that NBD-anchor-LLD distributes symmetrically between the two leaflets and that it does not show spontaneous transbilayer diffusion, due to its prominent hydrophilic disaccharide headgroup. In contrast, in LtaA-proteoliposomes, the NBD-anchor- LLD fluorescence plateaus at about 35% of the initial intensity, clearly demonstrating that LtaA possesses NBD-anchor-LLD flipping activity (Fig. 1B,C).
To verify that the flipping activity detected was LtaA-specific, we performed the same type of experiment but with a functionally unrelated protein (choline transporter) reconstituted in proteoliposomes (Fig. 1C). In this experiment, quenching by dithionite led to a fluorescence plateau of 50%, validating that the flipping activity observed in LtaA proteoliposomes is catalyzed by LtaA. Furthermore, control experiments with protein-free liposomes containing the precursor molecule NBD-DAG plateau at about 30%, (Extended Data Fig. 1) consistent with spontaneous transbilayer diffusion of NBD-DAG due to its prominent hydrophobicity. Together, these experiments demonstrate that LtaA displays anchor-LLD flippase activity.
Structure of S. aureus LtaA
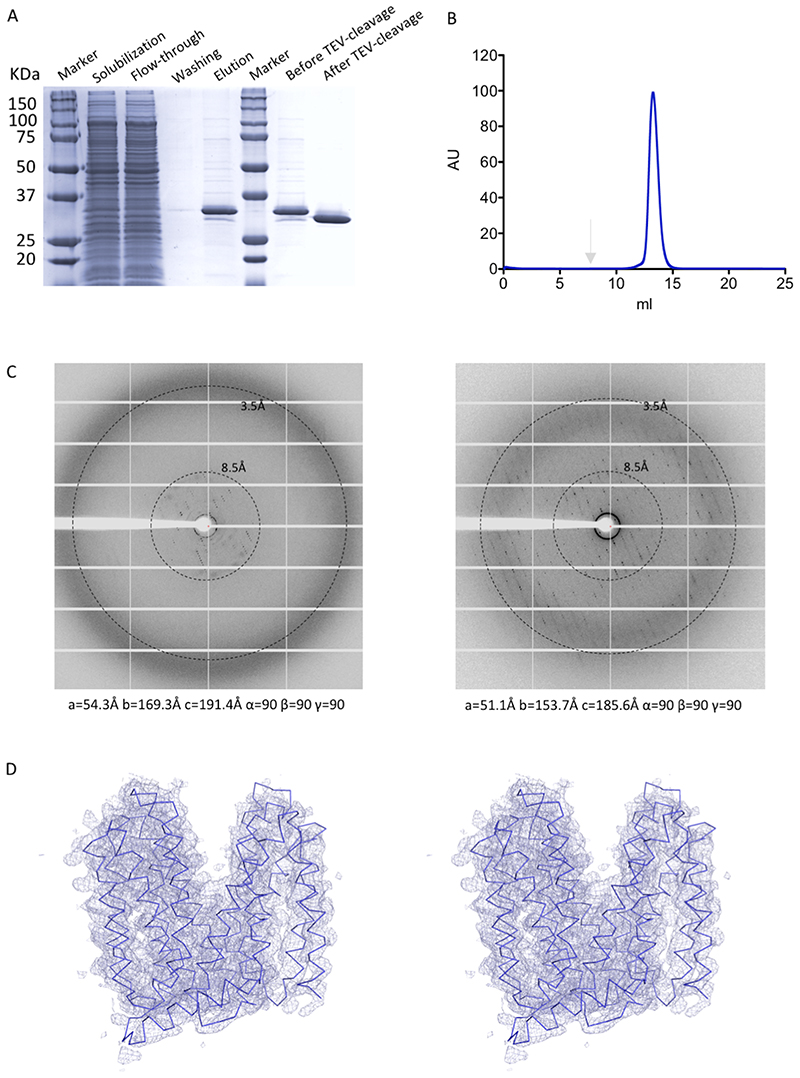
We determined the structure of LtaA from S. aureus at 3.3Å resolution (Fig. 2A and B, Extended Data Fig. 2–4, Table 1). Experimental phases were determined by single-wavelength anomalous diffraction (SAD) from selenomethionine (SeMet) derivative crystals. The register of the resulting model was confirmed by the anomalous densities of 16 SeMet residues along the polypeptide chain (Extended Data Fig. 4). LtaA crystals commonly diffracted X-rays up to 7Å resolution, however by optimizing in situ annealing conditions38 we were able to increase the diffraction resolution up to 3.3 Å, accompanied by an up to 18% reduction of the unit cell volume (Extended Data Fig. 2C).
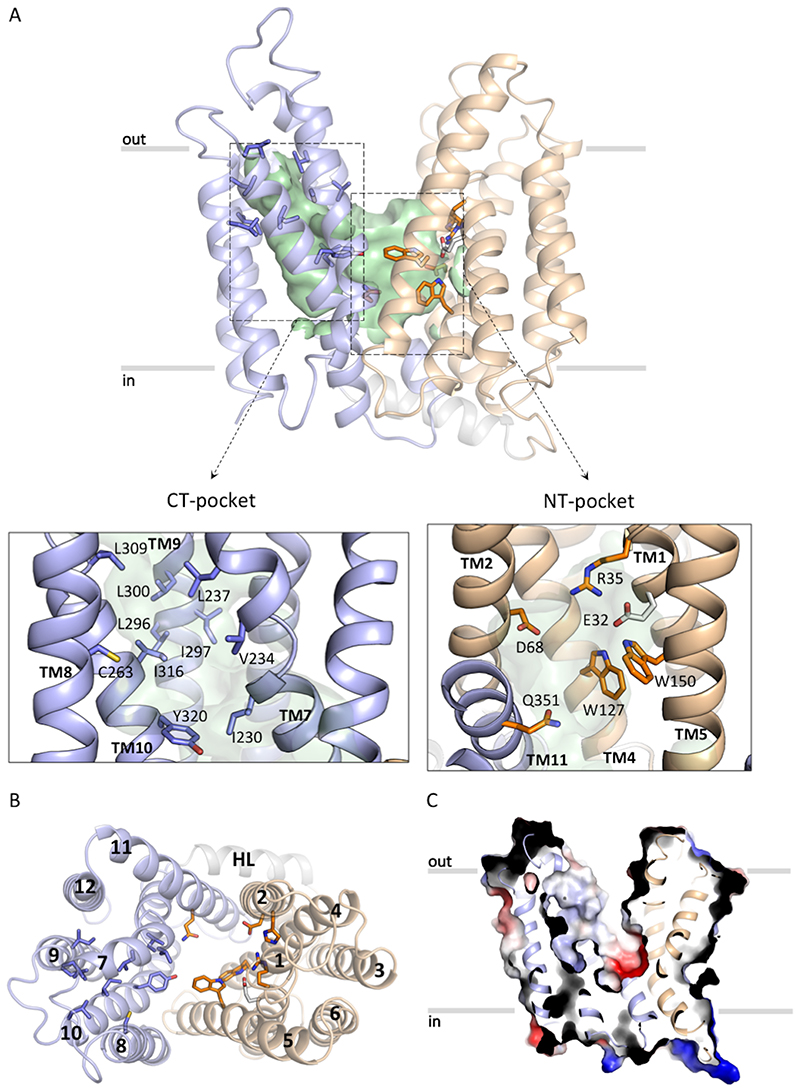
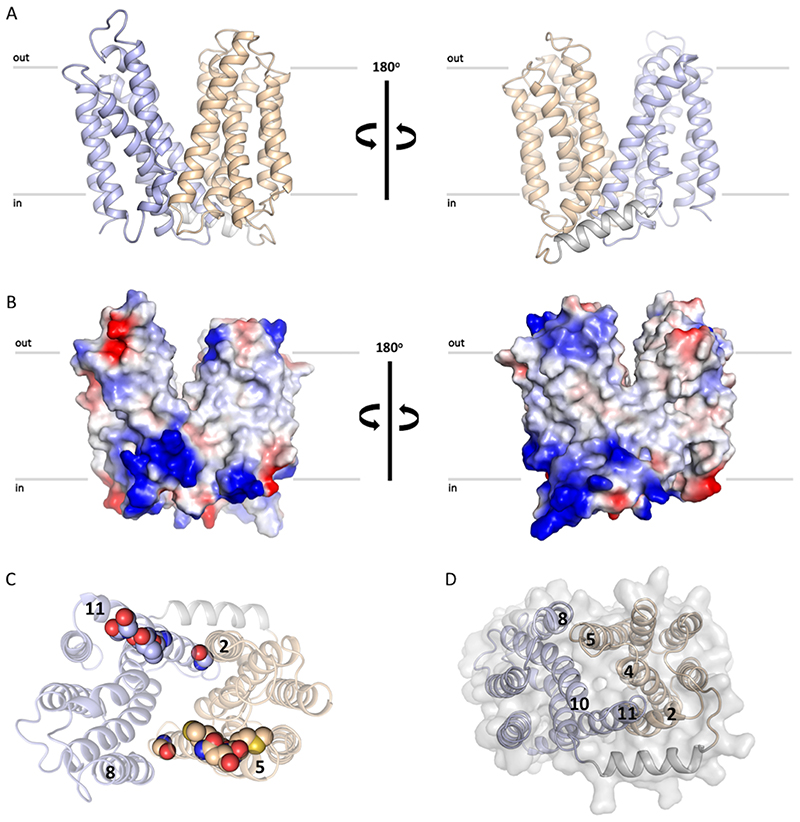
Figure 2. S. aureus LtaA structure.
A. Structure of LtaA showing its central cavity (green surface). CT: C-terminal, NT: N-terminal. The N-terminal domain is shown in light-orange, C-terminal domain is shown in light-blue. Residues forming the hydrophobic C-terminal pocket and the hydrophilic N-terminal pocket are shown. B. Top view of LtaA. Residues in sticks participate in formation of the amphiphilic cavity. C. Vacuum electrostatic surface representation of LtaA showing the internal cavity.
Table 1. Data collection and refinement statistics.
| LtaA (SeMet)a (PDB 6S7V) | |
|---|---|
| Data collection | |
| Space group | C2221 |
| Cell dimensions | |
| a, b, c (Å) | 51.39, 162.47, 191.05 |
| a, ß, γ (°) | 90.0, 90.0, 90.0 |
| Resolution (Å) | 20-3.3 (3.4-3.3)b |
| R meas (%) | 13.5 (153.7) [22.1 (414.8)]c |
| I/σ(I) | 12.76 (1.64) [6.93 (1.36)] |
| CCM (%) | 99.9 (94.4) [100 (86.8)] |
| Completeness (%) | 83.37 (32.3) [98.37 (97.55)] |
| Redundancy | 40.0 (39.0) |
| Refinement | |
| Resolution (Å) | 20-3.3 (3.4-3.3) |
| No. reflections | 12387 [19539] |
| Rwork / Rfree (6%) | 27.05/28.94 |
| No. atoms | |
| Protein | 3001 |
| B factors | |
| Protein | 84.0 |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.006 |
| Bond angles (°) | 0.987 |
Merged from three data-sets collected from one selenomethionine (SeMet) crystal.
Values in parentheses are for highest-resolution shell.
Values in brackets are before anisotropic truncation.
LtaA displays the fold of MFS transporters, which consists of 12 TM divided into two 6TM domains (N- and C-terminal domains) related by a pseudo-rotational two-fold symmetry axis perpendicular to the plane of the membrane (Fig. 2A, Extended Data Fig. 5A). The fold of both domains is similar showing an r.m.s.d. of 3.2Å upon secondary structure superposition. Both domains are connected through a cytoplasmic helical loop, a structural feature observed in many other MFS fold transporters 39,40. The LtaA structure displays an outward-facing state with two prominent lateral hydrophobic entrances (Fig. 2A, Extended Data Fig. 5B). One of the entrances is flanked by TMs 2 and 11, whereas TMs 5 and 8 flank the entrance on the opposite side. TM11 delimiting one of the hydrophobic entrances displays the antiporter motif41 G (G345(X)8G(X)3GP(X)2GG363), whereas on the opposite entrance TM5 displays a similar motif-G-like sequence at the same position (Extended Data Fig. 5C). Access to the central cavity from the cytoplasmic face is sealed off by multiple interactions between TMs 2, 4 and 5, from the N-terminal domain, and TMs 8, 10 and 11, from the C-terminal domain, and cytoplasmic loops connecting TMs 4-5 and TMs 10-11 (Extended Data Fig. 5D).
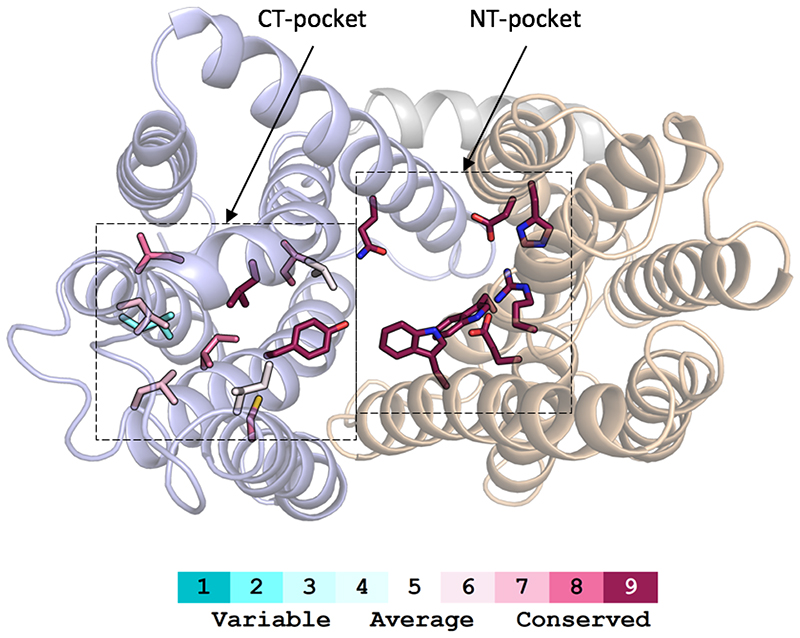
One striking structural feature of LtaA architecture is the presence of a large amphiphilic central cavity whose hydrophilic part is harboured by the N-terminal domain of the flippase with the participation of residues E32, R35, D68, W127, and W150, located in TMs 1, 2, 4 and 5 (Fig. 2A and C). On the other hand, the hydrophobic part of the cavity is harboured by the C-terminal domain of the flippase with the participation of 8 hydrophobic residues located in TMs 7, 8, 9 and 10 (Fig. 2A and C). Such an amphiphilic cavity has not been observed in any other MFS fold transporter described to date. Sequence conservation analysis revealed that residues forming the N-terminal hydrophilic pocket are highly conserved among LtaA homologues found in other Staphylococcus species or further Gram-positive bacteria, whereas residues forming the C-terminal hydrophobic pocket are less conserved, although their positions are prominently occupied by amino-acids with aliphatic side chains (Extended Data Fig. 6, Supplementary Fig. 9).
LtaA displays high selectivity towards the headgroup of its substrate
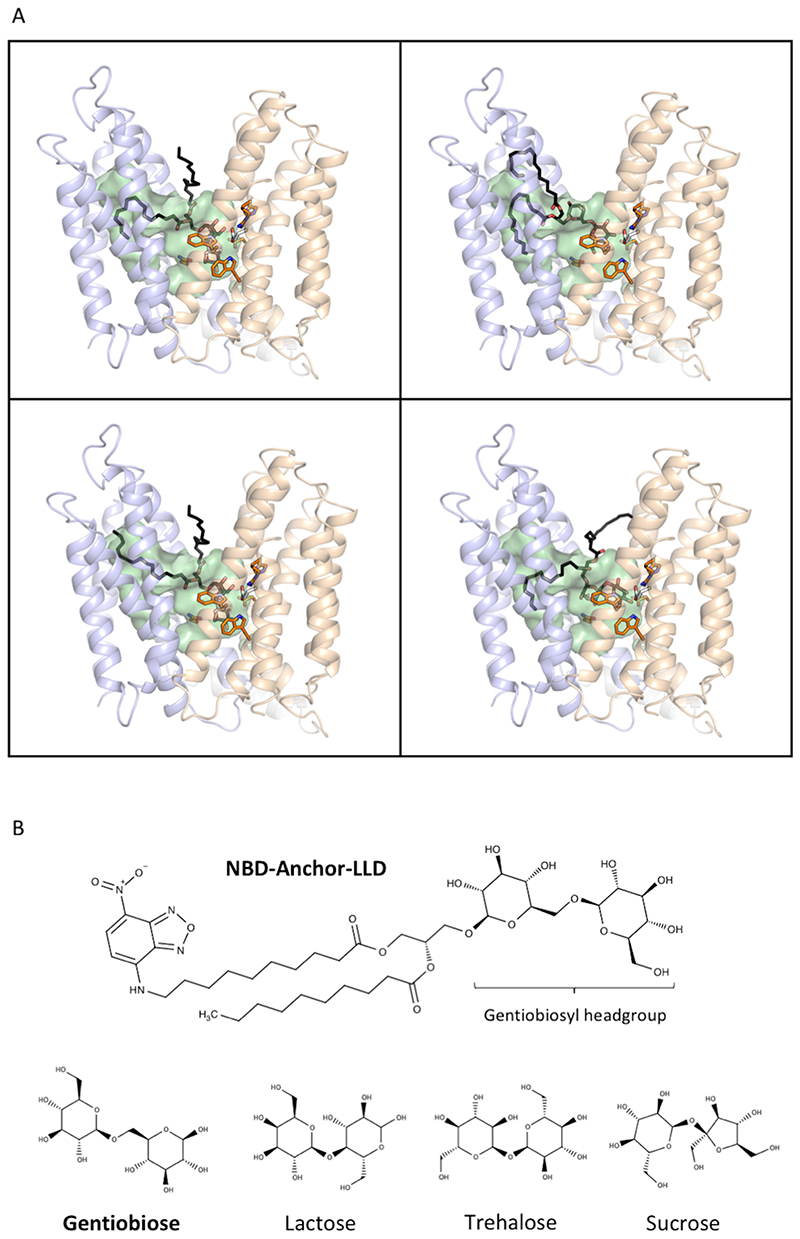
We used computational docking to test whether the binding of anchor-LLD to outward-facing LtaA was feasible. The ligand docking analysis suggests that the gentiobiosyl headgroup of the anchor-LLD is preferentially accommodated into the N-terminal hydrophilic pocket, whereas the diacylglycerol aliphatic tails are docked with one or both chains into the C- terminal hydrophobic pocket in multiple conformations (Fig. 3A, Extended Data Fig. 7A). This, together with the high sequence conservation of the residues forming the hydrophilic pocket led us to hypothesize that LtaA might display high selectivity towards the anchor-LLD headgroup. To test this hypothesis, we assessed flipping activity in the presence of different disaccharide compounds (Fig. 3B-C, Extended Data Fig. 7B). If LtaA displays high selectivity towards the disaccharide headgroup of its substrate, one would expect to see reduced flipping activity in the presence of an excess of gentiobiose (β-D-Glc-(1,6)-D-Glc), a disaccharide with the same chemical composition and conformation as the anchor-LLD headgroup. Indeed, we found that increasing concentrations of gentiobiose inhibit LtaA- catalyzed flipping activity, whereas other disaccharides, i.e. lactose (β-D-Gal-(1,4)-D-Glc), sucrose (α-D-Glc-(1,2)-β-D-Fru), and trehalose (α-D-Glc-(1,1)-α-D-Glc) showed no inhibitory effect at a similar concentration (Fig. 3C). These results indirectly but strongly suggest that LtaA displays high selectivity towards the headgroup of its substrate.
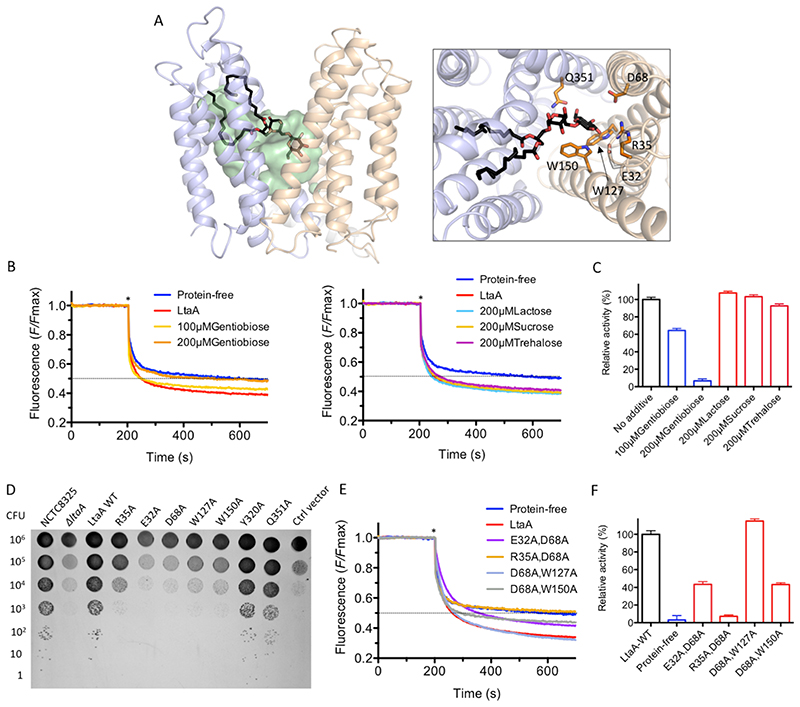
Figure 3. Amphiphilic cavity characterization.
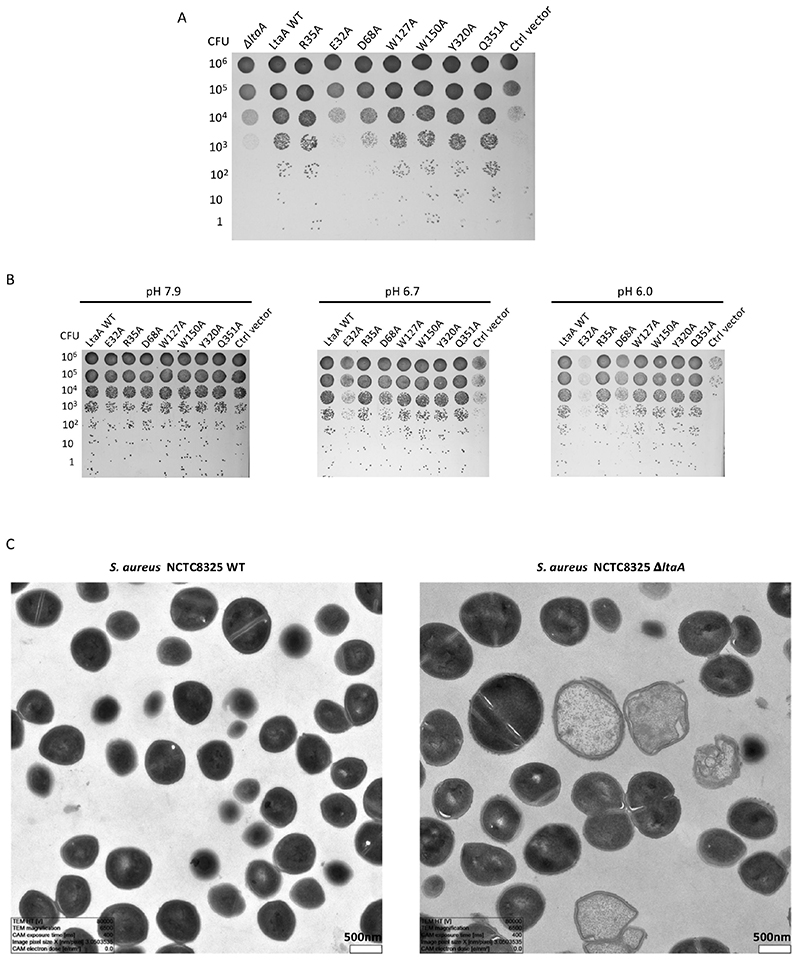
A. A model of lipid-linked-disaccharide docked into the amphiphilic cavity of LtaA (see also Extended Data Fig. 7A). The lipid tails length corresponds to C16 chains19. Right inlet, Top view of anchor-LLD binding pocket and residues coordinating its gentiobiosyl headgroup. B. Representative traces of LtaA catalyzed flipping in the presence of different disaccharides at given concentrations (n ≥ 3). C. Relative flipping activity of assays showed in B. Error bars indicate the s.d. of technical replicates, n ≥ 3. D. Cells growth on IPTG-free C+Y agar plates at 37°C and in the presence of 5% CO2. ΔltaA represents the S. aureus NCTC8325 ΔltaA mutant; LtaA WT represents ΔltaA mutant complemented with wild type ltaA on a multicopy vector (pLOW); Ctrl vector indicates ΔltaA mutant complemented with pLOW carrying a nonrelated gene (dcas9); the other labels represent ΔltaA mutant complemented with ltaA with corresponding point mutations. E. Representative traces of flipping activity of LtaA-WT and variants. F. Relative flipping activity of assays showed in E. Asterisk marks addition of dithionite. Error bars indicate the s.d. of technical replicates, n ≥ 3. Source data for B-C and E-F are available with the paper online.
LtaA N-terminal hydrophilic pocket is crucial for flipping activity
Next, we assessed the functional role of residues in the highly conserved N-terminal hydrophilic pocket. According to our docking model, this pocket might accommodate the anchor-LLD headgroup. First, we constructed a S. aureus NCTC 8325 ΔltaA mutant, which we then complemented with an ectopic copy of the ItaA gene carrying single point mutations and evaluated growth on agar plates at 37°C and in the presence of 5% CO2 (Fig. 3D). Strikingly, while ItaA was shown not to be essential under laboratory conditions19, we find that non-complemented ΔltaA mutants are non-viable in the presence of 5% CO2 (see below) demonstrating that LtaA is crucial for S. aureus fitness. Mutants Y320A and Q351A rescued the growth defect of the ΔltaA strain, whereas mutants E32A, R35A, D68A, W127A, and W150A did not. Notably, under over-expression conditions, the mutants R35A, D68A, W127A and W150A, but not E32A rescued the growth defect (Extended Data Fig. 8 and 9), indicating that E32 plays a key role in LtaA activity.
Next, we examined several of the mutants tested for their ability to rescue ΔltaA, for their capacity to perform flipping of NBD-anchor-LLD. In vitro flipping assays with LtaA double mutants that combine the five mutations that showed the stronger impact on S. aureus growth, revealed that the mutant R35A/D68A is not active and that the relative activity of mutants E32A/D68A and D68A/W150A is about half of that of LtaA-WT (Fig. 3E and F). This agrees with our in vivo assays that revealed that the mutants E32A, R35A, D68A, W127A, and W150A display a strong growth defect of the ΔltaA strain (Fig. 3D). Taken together, these results indicate that slight modifications of the activity of LtaA are sufficient to perturb the assembly line of LTA and in consequence the fitness of S. aureus.
LtaA is a proton-coupled antiporter flippase
Charged residues located in substrate binding sites of secondary transporters have been frequently associated with proton coupling because they may undergo pKa shifts as a consequence of structural changes during the transport cycle39,42. LtaA possesses three candidates for such pH-coupling residues, E32, R35, and D68, which are all located in the N- terminal hydrophilic part of the central amphiphilic cavity (Fig. 2A). We therefore explored if anchor-LLD flipping could be energized by a proton-gradient by measuring LtaA catalyzed NBD-anchor-LLD flipping in proteoliposomes (Fig. 4A, Extended Data Fig. 10). In the absence of a proton gradient, quenching of the NBD-anchor-LLD plateau to about 35% of the total fluorescence. Strikingly, if we impose an inward proton gradient (low pHout / high pHin), larger quenching of NBD-anchor-LLD is observed. On the contrary, if we impose an outward proton gradient (high pHout / low pHin), less quenching of NBD-anchor-LLD is observed, relative to no gradient conditions (Fig. 4A). As controls, we carried out the same type of experiment but with protein-free liposomes (Fig 4B) and with a functionally unrelated protein reconstituted in proteoliposomes (Extended Data Fig. 10A), with the result that NBD- anchor-LLD quenching plateaued at about the same fluorescence level, independent of the pH gradient. In summary, these results reveal that LtaA performs vectorial flipping of anchor- LLD opposite to the direction of the proton gradient.
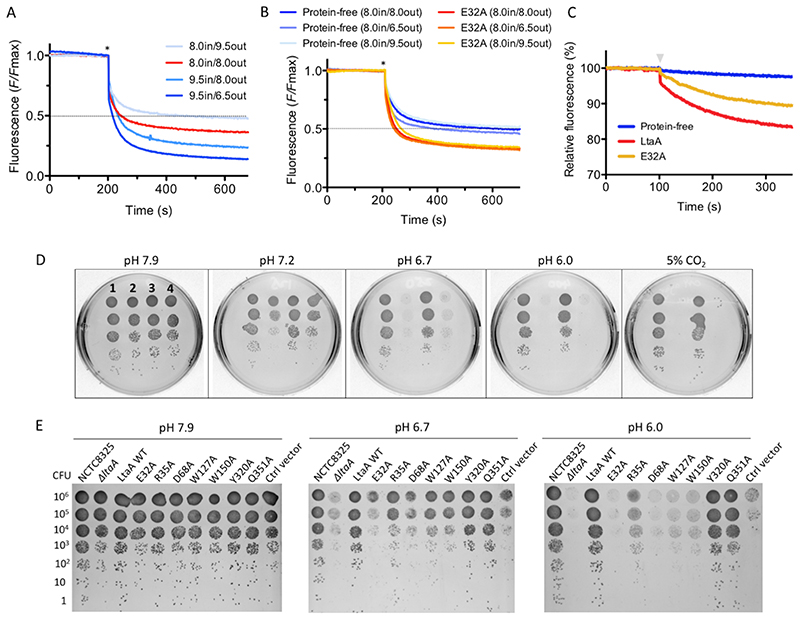
Figure 4. LtaA proton coupling and S. aureus ΔltaA growth under acidic conditions.
A and B. Representative traces of flipping assays with protein-free liposomes, LtaA-WT and LtaA- E32A in the presence of different proton gradients, in and out denote pH of buffer inside and outside of liposomes, respectively (n ≥ 3). Asterisk marks addition of dithionite. C. Protons transport assay. Representative time courses are shown (n ≥ 3). Proteoliposomes and protein- free liposomes containing 100 mM KCl were diluted in buffer containing 10 mM KCl and 9- amino-6-chloro-2-methoxyacridine (ACMA). H+ influx was initiated by establishing a membrane potential by addition of the potassium ionophore valinomycin (gray triangle). D. Cells growth in IPTG-free C+Y agar plates under different pH at 37°C in absence of CO2 (ambient conditions) or in the presence of 5% CO2 (initial pH 7.9). 1 indicates S. aureus NCTC8325 WT, 2 indicates ΔltaA mutant, 3 indicates ΔltaA mutant complemented with wild type ltaA on a multicopy vector (pLOW), 4 indicates ΔltaA mutant complemented with pLOW carrying a functionally unrelated gene (dcas9). E. S. aureus NCTC8325 ΔltaA mutant complemented with ltaA carrying corresponding point mutations. Cells growth on IPTG-free C+Y agar plates under different pH at 37°C. LtaA WT represents ΔltaA mutant complemented with wild type ltaA; Ctrl vector indicates ΔltaA mutant complemented with a functionally unrelated gene (dcas9). Source data for A-C are available with the paper online.
Protons transport mediated by LtaA was demonstrated using a fluorescence assay43 where proteoliposomes loaded with 100 mM KCl (pH 7.3) were diluted 20-fold into an assay buffer containing 10 mM KCl (pH 7.3). Under these conditions, if LtaA performs H+ transport, the addition of the K+ selective ionophore valinomycin will drive H+ influx and cause quenching of the fluorophore 9-amino-6-chloro-2-methoxyacridine (ACMA). The robust fluorescence decrease observed upon the addition of valinomycin reflects H+ influx into the proteoliposomes driven by the membrane potential (Fig. 4C). A control experiment with protein-free liposomes showed little fluorescence change (Fig. 4C).
We performed flipping assays and proton transport assays with the single point mutant LtaA- E32A reconstituted in proteoliposomes (Fig. 4B and C). This residue was selected as the best candidate to undergo protonation and deprotonation due to its theoretical pKa-value of 7.8 in the current outward-facing structure, compared to residues R35 and D68 with pKa values of 14 and 3.4, respectively. Our results show that the E32A mutation is insensitive to transmembrane proton gradients, maintaining basal activity (Fig. 4B). Consistent with this is the observed decrease of H+ influx driven by the membrane potential (Fig. 4C). In summary, these data establish that LtaA works as a proton-coupled antiporter-flippase and that residue E32 plays a crucial role in proton coupling, presumably by being involved in proton transport. The importance of proton coupling for LtaA function in vivo becomes evident as pointed out by the strong growth defect of the S. aureus ΔltaA strain expressing LtaA-E32A (Fig. 3D, Extended Data Fig. 9A).
LtaA is essential to combat acid stress
In S. aureus cells, synthesis of anchor-LLD by the glycosyltransferase YpfP occurs at the cytoplasmic leaflet without interruption as long as the levels of DAG and UDP-Glc in the cell are preserved20,21. Under these conditions, healthy S. aureus displays an outward anchor-LLD gradient that would suffice to drive LtaA-facilitated flipping. Thus, we wondered whether the proton coupling activity of LtaA could play an important role in the survival of S. aureus under physiological conditions encountered at the human nasopharynx, mucous membranes and on the skin, which present mild acidic environments (5.0<pH<6.5) 44–46.
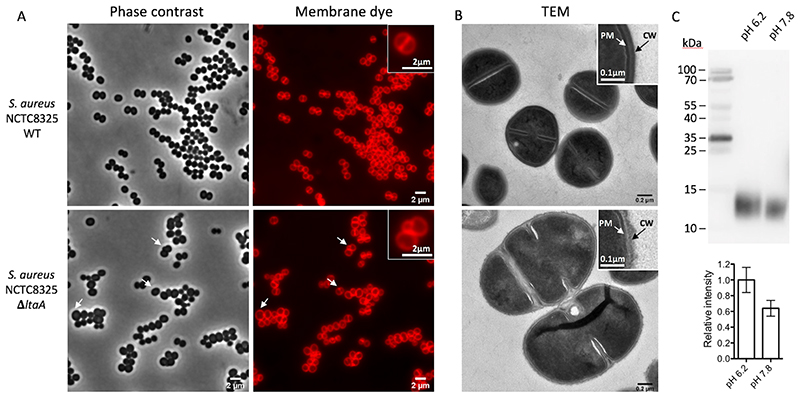
To evaluate this hypothesis, we investigated the growth characteristics of S. aureus NCTC 8325 ΔltaA on plates at 37°C under different pH conditions in the absence of CO2 (ambient conditions) (Fig. 4D). We found that at high pH the ΔltaA mutant does not show growth defect compared to wild type or ltaA-complemented S. aureus, in line with previous findings19. However, very strong growth retardation was observed in the ΔltaA mutant at low pH, while complementation with ltaA-WT restored normal growth. Alternatively, the presence of 5% CO2, which acidifies the medium to about pH 6.0 due to CO2/HCO3 equilibria, equivalently suffices to promote a strong growth defect of the ΔltaA mutant (Fig. 4D). In the same way, point mutations of the proton coupling E32 residue and others forming the N- terminal hydrophilic pocket showed a strong growth defect at low pH (Fig. 4E), consistent with their important role in the coordination of the anchor-LLD headgroup and E32 essential role in proton transport as shown above. The E32A mutant does not recover the growth defect at low pH even under over-expression conditions (Extended Data Fig. 9B). Fluorescence microscopy and transmission electron microscopy (TEM) of S. aureus NCTC 8325 WT and ΔltaA grown at 37°C in LB medium at pH 6.5 or grown in the presence of 5% CO2 (medium pH 6.5) showed that the ltaA deletion mutant display aberrant cell morphologies including enlarged cells, defects in the formation and localization of the division septum, and abnormal cell wall shape (Fig. 5A-B, Extended Data Fig. 9C).
Figure 5. Morphology of S. aureus NCTC8325 WT and ΔltaA mutant, and LTA abundance.
A. Phase contrast and fluorescence images. Bacteria were grown to mid-exponential phase in LB medium at 37°C with 5% CO2 causing acidification of the medium. As membrane dye, Nile red was used. White arrows point to cells with aberrant morphology. B. Transmission electron microscopy (TEM) images. PM indicates plasma membrane. CW indicates cell wall. Low magnification images are shown in Extended Data Fig. 9C. C. Detection of LTA by immunoblotting. S. aureus NCT8325 were cultured in LB medium buffered with PBS to different pH (6.2 and 7.8). Cell lysates were normalized based on optical density. Samples were separated by 12% SDS-PAGE and LTA detected by LTA (polyglycerol-phosphate)- specific primary antibody. Histogram shows relative amounts of LTA determined from band intensities (n=4). Error bars indicate s.d. of technical replicates. Source data for C are available with the paper online.
Our results attribute an essential role to LtaA in the survival of S. aureus under acidic conditions. This is highly relevant since such conditions are encountered in the most common niches of this bacteria in the human body44–47. Thus, the development of drugs targeting LtaA might lead to novel therapies for the treatment of S. aureus infections.
Discussion
Taken together, our biochemical and structural studies suggest a functional cycle in which LtaA performs anchor-LLD translocation energized by proton antiport (Fig. 6). The mechanism likely follows in its structural conformations the classic antiporter alternating access cycle48,49. However, due to the amphiphilic nature of the substrate, the conformational and energy landscape of lipid flipping by LtaA might look very different to that of canonical transporters of water-soluble substrates. Specifically, substrate recognition, loading into the central cavity, and release are expected to entail different mechanisms. In a first step, deprotonated LtaA in an inward-facing conformation recognizes and binds the anchor-LLD in its central cavity positioning the lipid tails at the C-terminal hydrophobic pocket and the disaccharide headgroup at the N-terminal hydrophilic pocket. Recognition and extraction of the anchor-LLD from the pool of surrounding lipids are facilitated by specific binding of the gentiobiosyl headgroup which allows extracting the lipid out of the membrane. Subsequently, LtaA will change its conformation to an outward-facing state promoting substrate release into the membrane. We suggest that the prominent lateral entrances observed in the crystal structure might facilitate the toppling of the anchor-LLD back into the bilayer. After substrate release in the outward facing state, residue E32 will become protonated, facilitating the transition to an inward-facing state, where it will then undergo deprotonation. This mechanism explains the stimulation of transport by a proton gradient, while in the absence of it, LtaA performs antiport driven by the outward directed anchor-LLD gradient maintained by the activity of YpfP in S. aureus cells. In the absence of a proton gradient, anchor-LLD translocation will happen at a slower rate in the cell, in full agreement with our in vitro flipping assays.
Figure 6. LtaA anchor-LLD flipping mechanism.
1 Binding of lipid-linked-disaccharide (black and red sticks) in the central cavity of LtaA in inward-facing conformation (modelled conformation) and deprotonation of E32. 2 Transition to outward-facing state (structure determined in this study). 3 Substrate release into the membrane and protonation of E32. 4 Transition to inward-facing state (modeled conformation). The N-terminal domain is shown in light-orange, C-terminal domain is shown in light-blue. The inward-facing model of LtaA was constructed by rigid body alignment of the N-terminal domain (TM1-6) and the C- terminal domain (TM7-12) to those of inward-facing LacY (PDB: 2CFQ)60.
The LtaA flipping mechanism proposed here has certain similarities to the flipping mechanism proposed for the lipid-II flippase MurJ50,51. MurJ belongs to the multidrug/oligosaccharyl-lipid/polysaccharide (MOP) family of transporters, and plays an important role in peptidoglycan assembly52. Similar to the C-terminal hydrophobic pocket observed in LtaA, in MurJ a hydrophobic groove formed by C-terminal TM helices 13 and 14 contributes to binding of the undecaprenyl lipid tail of lipid-II. These similarities suggest that secondary transporter flippases might share a mechanism where part of the internal cavity specializes in binding specific lipid moieties, facilitating in this way loading and translocation of amphiphilic molecules. Thus, the amphiphilic cavities observed in LtaA and MurJ might constitute a general structural signature that may contribute to the identification of other flippases in silico. The architecture of the outward-open LtaA structure provides the basis to understand anchor-LLD recognition, binding and release into the membrane. These results together with the apparent high selectivity of LtaA for the anchor-LLD headgroup and inhibition of the flipping activity by gentiobiose provide the structural basis to design inhibitors targeting LTA assembly.
The mechanism by which LtaA contributes to S. aureus survival under acidic conditions is intimately related to its capacity to couple anchor-LLD flipping to proton transport. This mechanism makes LtaA a “pH sensing” flippase that by increasing anchor-LLD transport under low pH conditions, it enlarges the population of LTA at the outer leaflet of the plasma membrane (Fig. 5C). Besides the already known important functions of LTA for cell division and protection against environmental threats5,7–9, we hypothesize that increased amounts of LTA in the cell wall will likely provide an efficient way to buffer against acidification thanks to the high negative charge of the LTA backbone polymer. Notably, the description of a pH sensing mechanism in a lipid transporter represents a fundamental conceptual advance in the field of lipid transport, with LtaA being the first “sensor-flippase” described.
Membrane transport proteins performing unidirectional active translocation of lipids (flipping) generate membrane asymmetry 50,51,53–57. On the other hand, membrane proteins that perform passive bi-directional translocation of lipids (scrambling) disrupt the membrane asymmetry36,37,58. Lipid flipping energized by ATP-hydrolysis has been well characterized for multiple protein families53,55–57, our results identify LtaA as a new class of flippase that energizes lipid translocation by coupling to a transmembrane proton gradient. These findings will potentiate the search of other ion-coupled flippases, not only in Gram-positive bacteria but in all prokaryotes. For example, it has been shown that the lipid-II flippase MurJ requires a membrane potential for its function59. In this case, in addition to driving export of lipid-II due to its intrinsic net negative charge, a membrane potential might drive lipid-II flipping by transport of a coupling ion, but this remains to be shown.
Methods
LtaA expression and purification
LtaA was overexpressed in E. coli BL21-Gold (DE3) (Stratagene) cells. Cells were transformed with a modified pET-19b vector (Novagen) carrying the gene encoding S.aureus LtaA with an N-terminal His10 affinity tag. Cells were grown at 37°C in Terrific Broth medium supplemented with 1% glucose (w/v) and induced with 0.2mM Isopropyl β-D-1-thiogalactopyranoside (IPTG). Cells were harvested and resuspended in 50mM Tris-HCl, pH 8.0; 500mM NaCl; 5mM β-mercaptoethanol; 0.5mM PMSF, disrupted and membranes were collected by ultracentrifugation. Membranes containing LtaA were solubilized in 50mM Tris-HCl, pH 8.0; 200mM NaCl; 20mM Imidazole; 15% glycerol (v/v); 5mM β-mercaptoethanol; 1% Lauryl Maltose Neopentyl Glycol (w/v) (LMNG, Anatrace); 1% N-dodecyl- β -D-maltopyranoside (w/v) (DDM, Anatrace) for 2h at 4°C. After centrifugation, the supernatant was loaded onto a NiNTA superflow affinity column (Qiagen), washed with 50mM Tris-HCl, pH 8.0; 200mM NaCl; 50mM Imidazole; 10% glycerol (v/v); 5mM β-mercaptoethanol; 0.02% LMNG and 0.02% DDM and then washed a second time with the same buffer only containing 0.02% LMNG. Elution was performed in the same buffer containing 200mM Imidazole. Imidazole was removed by desalting using a PD-10 column (GE Healthcare). The His10 affinity tag was removed by overnight treatment with Tobacco Etch Virus (TEV) protease. TEV protease was later removed by passing through Ni-NTA affinity column. LtaA was further purified by size exclusion chromatography (SEC) in buffer 10 mM Tris-HCl pH 8.0, 150mM NaCl, 0.02% LMNG (Superdex 200 Increase 10/300 GL, GE Healthcare). The main peak was collected and the buffer exchanged to 10 mM Tris-HCl pH 8.0, 150 mM NaCl, 0.1% Cymal-7 (Anatrace) using a PD-10 desalting column61.
Seleno-methionine derivative production
E. coli BL21-Gold (DE3) (Stratagene) cells carrying the LtaA expression vector were grown in TB-glucose media at 37 °C until OD600 0.5-1.0. These were then used to inoculate a preculture of M9 media supplemented with vitamin B1 hydrochloride, cells were then grown until OD600 0.5 and used to inoculate 2L of M9 media supplemented with vitamin B1 hydrochloride. Cells were grown overnight at 37°C until OD600 ~ 0.9, followed by the addition of an amino-acids/Seleno-methionine (SeMet) cocktail and incubation for 30 minutes. SeMet-LtaA expression was induced with 0.2 mM IPTG and 90 minutes incubation.
LtaA crystallization
LtaA was concentrated up to 6.0mg/ml using a 30kDa MWCO VivaspinTM20 concentrator (GE healthcare). After extensive optimization of crystallization conditions, LtaA crystals were obtained at 16°C using sitting-drop vapor diffusion. Plate shaped crystals were obtained in reservoir conditions containing 30-70 mM magnesium acetate, 80-120 mM glycine pH 9.5 and 30-34% PEG 300. Crystals appeared after 3–4 days and matured to full size within 1 week. Crystals were dehydrated and cryoprotected by gently increasing PEG 300 concentration in the drop followed by flash freezing by immersion in liquid nitrogen61.
In situ annealing and data collection
LtaA crystals diffracted X-rays up to about 6-7Å resolution in general. Performing in situ annealing led to X-rays diffraction to higher resolution (3.8Å to 3.3Å, depending on the crystal)61. In situ annealing was performed by blocking the cold nitrogen stream with a thin film while the crystal was mounted on the goniometer. Annealing time, thickness of the film, and distance of the cryo-stream to the crystal were critical parameters to optimize. The X-ray diffraction patterns displayed in Extended Data Fig. 2C show an example of diffraction before and after annealing.
Enhancement in X-rays diffraction resolution was accompanied by a 10% to 18% reduction of the unit cell volume. The best data-sets were collected from crystals exposed to X-rays only after annealing. Annealing time of about 10 seconds produced the best results. Three data-sets collected from one LtaA crystal were merged and used to determine the structure. This LtaA crystal showed anisotropic diffraction up to 3.3Å and belonged to the space group C2221, with unit cell constants a=51.39Å, b=162.47Å, c=191.05Å and α=90°, ß=90°, γ=90° (Table 1). Data were processed and merged with XDS62 and anisotropic scaling/ellipsoid truncation was performed. Resolution limits after ellipsoid truncation were a*=3.0Å, b*=4.0Å, and c*=3.5Å. We used Karplus CC* (Pearson correlation coefficient) based data cutoff approach to determine the usable resolution of the data-sets63. The resolution limit was set taking into account a CC1/2>~40% based on data merging statistics and a CC* analysis against unmerged intensities in Phenix package64 satisfying Karplus CC* against CCwork and CCfree criteria, as well as, Rfree of the highest resolution shell against the refined structure being less than or equal to ~50% (Table 1). A second criterion for limiting the resolution was the overall completeness percentage observed after anisotropic ellipsoid truncation, which was kept above 80%. Diffraction data was collected at the beamline X06SA at the Swiss Light Source (SLS, Villigen).
Structure determination
The structure of LtaA was solved by single-wavelength anomalous diffraction (SAD). SeMet positions were found using SHELX65 and refined using CCP4i2 programs66, CRANK267, and PHASER68. Solvent flattening and density modification were performed using PARROT69. The resulting phases and electron density maps were used to build an initial model. The anomalous densities of 16 SeMet residues along the polypeptide chain aided in tracing the correct amino acids register in the resulting model (Extended Data Fig. 4). Tracing of TM helices was facilitated by placing fractions of a LtaA homology model generated by Swissmodel70 using as reference model the structure of the MFS transporter YajR71 (PDB: 3WDO). Model building was performed in Coot72. Subsequently, multiple rounds of molecular replacement combined with single-wavelength anomalous diffraction (MR-SAD) in CRANK267 contributed to further improvement of the initial phases and electron density maps, facilitating the improvement of the model. Multiple rounds of refinement and model building were then performed using Phenix64 and Coot72. Map sharpening was used to facilitate model building. X-ray data and refinement statistics are given in Table 1. The final refined structure has Rwork=27.05% and Rfree=28.94%, with 93.35% of residues in the Ramachadran favored region; 6.38% in Ramachadran allowed; and 0.27% as Ramachandran outliers. Molecular graphics were created in PyMOL73. Surface electrostatics were calculated with the APBS PyMOL plugin.
YpfP expression and purification
YpfP with an N-terminal His10 affinity tag was overexpressed in E. coli BL21-Gold (DE3) (Stratagene) cells by IPTG induction. Cells were disrupted by sonication and YpfP was purified using affinity chromatography. YpfP was desalted in buffer 50 mM Tris-HCl pH 8.0, 200 mM NaCl, 10 % Glycerol, concentrated to 2.4 mg/ml, flash frozen in liquid nitrogen and stored at -80°C.
Synthesis of NBD-anchor-LLD
Synthesis of fluorescently labelled NBD-anchor-LLD was performed following a modification of the protocol described by Kiriukhin et. al. 2001 and Jorasch et al. 1998 20,21. Purified YpfP was incubated with 2mM UDP-Glucose (Sigma) and 2mM l-NBD-decanoyl-2-decanoyl-sn-Glycerol (Cayman) at 30°C for about 16 hours. The reaction product was separated using thin-layer chromatography (TLC) in a solvent mixture consisting of chloroform:methanol:water (65:25:4, vol/vol/vol)19 (Supplementary Figure 1). NBD-lipids were visualized in a fluorescence scanner (Amersham Typhoon Imaging System). The main product was extracted from the silica using a 50:50 mixture of chloroform:methanol, followed by evaporation of solvents. The extracted product was resuspended in buffer 50mM Tris-HCl pH 8.0, 200mM NaCl, 3%Glycerol.
HPLC-MS product analysis
Saples of NBD-DAG (Cayman), completed reaction, and extracted product from preparative TLC (NBD-DAG-Glc2) were subjected to HPLC-MS analysis using a ReproSil-Pur ODS-3 column (Dr. Maisch) and a flow rate of 1 ml/min with a solvent gradient elution of 0.1% Acetic acid (Eluent A) against Acetonitril with 0.1% Formic acid (Eluent B). The gradient consisted of 95% of eluent A to 95% of eluent B over a range of 22 minutes. The eluted species were analysed on a Bruker microTOF electrospray ionization mass spectrometer (Bruker Daltonics) in positive ion mode with the following settings; capillary voltage: 4500V, nebulizer pressure: 2 bar, drying gas: 9 l/min at 220°C, mass range 50 – 2000 m/z.
Tandem MS/MS and parallel reaction monitoring (PRM) LC-MS analysis
Pure NBD-DAG (Cayman) and extracted NBD-DAG-Glc2 (10 pmol/ul in 30% acetonitrile/69.9% water/0.1% formic acid) were prepared and analyzed by high-resolution mass spectrometry on a Q-Exactive mass spectrometer equipped with a Heated Electrospray Ionization (HESI-II) Probe (both Thermo Fisher Scientific) with direct infusion. For both molecules, full and tandem mass spectra were acquired at a resolution of 140,000 FWHM (at 200 m/z). The three most intense fragment ions were manually selected for quantitative parallel reaction-monitoring (PRM) MS analysis74. The setup of the μRPLC-MS system was as described previously75. The extracted ion chromatograms (XICs) were obtained using the most intense fragments selected before and employed for quantification (see Supplementary Notes).
NMR product analysis
NBD-DAG and DGDG were dissolved in CDCl3 at a concentration of 100μM and 75μM, respectively. NBD-anchor-LLD was extracted from TLC plates and dissolved in CDCl3 for NMR analysis. The experiments were recorded at 298K on a Bruker Ascend 600 MHz spectrometer running Topspin 3.2 equipped with a cryogenically cooled triple-resonance probe, using a 16 ppm spectral width, 30° flip angle, 1.5 s relaxation delay, and 2.7 s acquisition time. All data were processed with Bruker TOPSPIN-NMR software (version 3.2, Bruker).
Formation of LtaA proteoliposomes
We reconstituted LtaA in unilamellar liposomes prepared by extrusion through polycarbonate filters (400 nm pore size) from a 3:1 (w:w) mixture of E. coli polar lipids and L-α-phosphatidylcholine (Avanti). Liposomes were diluted in buffer containing 20mM Tris-HCl pH 8.0, 150mM NaCl and 2mM β-mercaptoethanol. After saturation with DDM (Anatrace), liposomes were mixed with purified protein at a 50:1 (w/w) lipid/protein ratio. Removal of detergent was performed by incubation with BioBeads (Biorad). Proteoliposomes containing a final concentration of 20mg/ml lipids, 7.8μM LtaA were centrifuged, washed and resuspended before being used for NBD-anchor-LLD reconstitution or stored at -80°C after freezing in liquid nitrogen. Before doing flipping assays, proteoliposomes were thawed and β-mercaptoethanol was removed by cycles of freeze/thaw and washing upon ultracentrifugation. NBD-anchor-LLD was then incorporated into proteoliposomes performing freeze/thaw cycles and extrusion through a polycarbonate filter (400 nm pore size). LtaA/NBD-anchor-LLD proteoliposomes were then immediately used for flipping assays. Protein-free proteoliposomes were prepared in the same way but without the addition of protein.
In vitro flipping assay
LtaA/NBD-anchor-LLD proteoliposomes, protein-free NBD-anchor- LLD liposomes and control transporter NBD-anchor-LLD proteoliposomes were diluted in buffer 20mM Tris-HCl pH8.0, 150mM NaCl before extrusion through polycarbonate filters (400 nm pore size). For sodium- and pH-gradient experiments, proteoliposomes were subjected to freeze/thaw cycles and resuspended in two steps: (i) in 200μl of the same “internal buffer” before extrusion; (ii) dilution in 800μl of “external buffer” before starting fluorescence recording. This was performed in order to minimize the effect of pH equilibration inside and outside proteoliposomes. Internal and external buffer compositions were 20mM of either Tris pH 8.0, Mes pH 6.5, or Glycine pH 9.5, together with 150mM NaCl or 150mM KCl. Flipping of the NBD-anchor-LLD was measured by determining the percentage of NBD-fluorescence that is quenched by 5 mM sodium dithionite (Sigma) added after 200 seconds of starting fluorescence recording. 100 seconds before finishing data recording 0.5% Triton X100 was applied to permeabilize the liposomes, making all NBD- anchor-LLD molecules accessible to dithionite reduction. Fluorescence was recorded at 20°C using a Jasco Fluorimeter. Excitation and emission wavelengths were 470 and 535 nm, respectively. For analysis, the fluorescence intensity was normalized to F/Fmax. Relative flipping activities were calculated as follows, Rel. act. = 100 x [(F/Fmax)i - (F/Fmax)liposomes]/[(F/Fmax)wt - (F/Fmax)liposomes], where i correspond to each respective treatment/mutants, liposomes correspond to liposomes without protein, wt correspond to wild-type LtaA proteoliposomes, and F/Fmax values correspond to the observed plateau for each recording. Curves were plotted using GraphPad Prism 5. Time courses of the dithionite- induced fluorescence decay in liposomes were repeated at least 3 times for each individual experiment.
Protons transport assay
Purified LtaA WT or the variant LtaA-E32A were reconstituted into POPE:POPG (3:1) liposomes at a protein to lipid ratio of 1:50 (w/w). Synthetic anchor- LLD, produced from 1,2-Dimyristoyl-sn-Glycerol (Avanti) and 2 mM UDP-Glucose as described above, was incorporated into proteoliposomes performing freeze/thaw cycles. Proteoliposomes and protein-free liposomes were resuspended in 5 mM Tris-HCl/HEPES pH 7.3, 100 mM KCl and extruded through a polycarbonate filter (400 nm pore size). After brief sonication, proteoliposomes were 20-fold diluted in buffer containing 5 mM Tris-HCl/HEPES pH 7.3, 10 mM KCl, 90 mM NaCl, and 0.5 μM 9-amino-6-chloro-2-methoxyacridine (ACMA). Fluorescence was measured at 20°C using a Jasco Fluorimeter. Excitation and emission wavelengths were 410 nm and 480 nm, respectively. After the fluorescence signal was stable, H+ influx was initiated by establishing a membrane potential by the addition of the potassium ionophore valinomycin (5 nM). Time courses of the proton transport assays were repeated at least 3 times for each individual experiment.
Mutagenesis
LtaA mutants were generated using Q5® Site-Directed Mutagenesis Kit (NEB). The sequence of the resulting constructs was confirmed by DNA sequencing (Microsynth).
Sequence conservation analysis
The sequence conservation analysis shown in Extended Data Fig. 6 and Supplementary Fig. 9 were computed using the ConSurf server76. Briefly, 76 LtaA homologues found in related Staphylococcus species or other Gram-positive bacteria were selected from a protein sequence BLAST search on the NCBI public database using S. aureus LtaA protein sequence as a query. We then generated a multiple sequence alignment using the HHMER algorithm provided by ConSurf, with conservation scores plotted in PyMOL73.
Docking of anchor-LLD
Docking of anchor-LLD (1,2-dihexadecanoic-3-O-(β -D- glucopyranosyl-1→6-O- β -D-glucopyranosyl-sn-Glycerol) to the LtaA structure was done with Autodock Vina77. Initial anchor-LLD coordinates were generated from 2D geometry in Phenix (eLBOW)64, and stereochemistry was corrected in Phenix (REEL)64, with reference to the X-ray crystal structures of ligands PDB:6GB and PDB:DDR. Docking was carried out over a search space of 60 × 54 × 36 Å covering the entire amphiphilic central cavity.
Construction of mutants in Staphylococcus aureus NCTC8325
Strains, plasmids and oligos used in this study are listed in Supplementary Table 7. A vector for efficient allelic replacement, pMAD78, was used for knockout of ltaA in Staphylococcus aureus NCTC8325. Specifically, the upstream and downstream of the ltaA coding region were amplified from genomic DNA by primer pairs OVL2253-OVL 2254 and OVL2255-OVL2256, respectively. A spectinomycin resistance cassette was amplified with oligos OVL2257 and OVL2258 from plasmid pMAD-int2-luc-spc-gfp79. Then the three fragments were assembled by gibson assembly to produce the fragment upstream-spectinomycin-downstream. The gibson assembly product was then used as template for amplification of the ltaA replacement fragment with oligos OVL2259 and OVL2260 containing NcoI and BamHI sites. The ltaA replacement fragment and vector pMAD-int2-luc-spc-gfp were both ligated to produce the ltaA knockout plasmid pMAD-ΔltaA construct, followed by transformation into E. coli strain IM08B80. The previously described gene deletion method for the temperature sensitive vector pMAD78 was used for the construction of the ltaA deletion strain in NCTC8325 with pMAD- ΔltaA. The final ΔltaA genotype of the mutant was confirmed by both PCR and Sanger sequencing.
S. aureus phenotypic assay
Bacteria cells were grown in 3 ml of Luria-Bertani (LB) medium at 37°C with 200 rpm shaking to OD600=0.3. For the complementary strains with pLOW vector, a final concentration of 5μg/ml of erythromycin was added to the medium. The bacterial culture was then serially diluted. 10μl of the original and its serial dilutions were spotted onto C+Y agar plates at different pH, without or complemented with 0.1mM IPTG. In the absence of IPTG, the pLOW vector provides a mild protein expression level79, as indicated by growth restoration of the ΔltaA mutant by ltaA-WT and single point mutants in absence of IPTG, whereas in the presence of IPTG it provides protein overexpression79. The plates were incubated at 37°C with or without 5%CO2 overnight. The images of the plates were captured by BioRad Gel Doc XR+ imaging system.
S. aureus complementation assays
The pLOW vector was used for the construction of ltaA complementary strains. The ltaA fragment was amplified from genomic DNA with oligos OVL2243 and OVL2244 containing SalI and NotI digestion sites (Supplementary Table 7). The amplified ltaA fragment was cloned into the vector pLOW-dCas979 to produce pLOW- ltaA. This was then transformed into E. coli IM08B. Point mutations were introduced by PCR mutagenesis (Supplementary Table 7). Later ltaA variatns were cloned into pLOW with the same method described above. The pLOW vector carrying ltaA or variants were then introduced into NCTC8325 ΔltaA strain by electroporation transformation with erythromycin selection (5μg/ml) on LBA plates as described above (see Supplementary Notes).
S. aureus fluorescent microscopy
LB medium pre-warmed at 37°C in the presence of 5% CO2 overnight (causing acidification to about pH 6.5) or LB medium with low pH (adjusted by dissolving LB Broth powder in PBS pH 6.5) were used to grow cells for fluorescence microscopy experiments. Fluorescence microscopy was performed using a Leica DMi8 microscope with a 100x phase contrast objective (NA 1.40) with a SOLA Light Engine (Lumencor) light source. A chroma cube nr (Quad=Chroma 89000, mCT_LP=49017) was used. For nile red staining, light was filtered through external excitation filter 545/25 nm (Chroma ET545/25x), and the external filter ET605/70 nm was used for emission. An exposure time of 800 ms with 100% of light from SOLA Light Engine was used for capturing images. The images were obtained with LasX software (Leica) and processed with ImageJ (https://imagej.nih.gov) (see Supplementary Notes).
S. aureus transmission electronic microscopy
The same bacterial cells for fluorescent microscopy were used for transmission electronic microscopy. 50nm ultrathin sections of cells fixed in agarose were cut on a Leica Ultracut (Leica Mikrosysteme) and picked up on a copper slot grid 2x1mm coated with a polystyrene film. Sections were poststained with 4% uranyl acetate. Micrographs were collected with a transmission electron microscope Philips CM100 (Thermo Fisher Scientific) at an acceleration voltage of 80 kV with a TVIPS TemCam-F416 digital camera (TVIPS). The final images were processed with ImageJ (https://imagej.nih.gov) (see Supplementary Notes).
Immunoblotting detection of LTA in Staphylococcus aureus NCTC8325 at different pH
LB media with different pH were prepared by dissolving LB powder (DifcoTM LB Broth, BD) in PBS followed by pH adjustment, and then sterilized by filtering with 0.2 μM membranes. The sample preparation and western blotting of LTA were performed based on the previous studies of Gründling et al. 19,22. Briefly, S. aureus cells were cultured in LB at 37°C, collected and resuspended in 500 μl of TBS buffer (20 mM Tris-Cl, pH 7.4, 150 mM NaCl). Bacteria were lysed by bead beater in 4 runs of 30 seconds per cycle with 6 m/s. The cell lysate was separated on 12% SDS-PAGE followed by western blot analysis. LTA was detected with LTA (polyglycerolphosphate)-specific primary antibody (clone 55, HyCult Biotechnology) and antimouse IgG (H+L) HRP antibody (Promega) as the secondary antibody. The blots were developed with SuperSignalTM west pico plus chemiluminescent substrate (Thermo Fisher Scientific), and the images were obtained with a Fusion FX7 imaging system (Witec). The relative amount of LTA was determined from the band intensities of western blots (n=4) using ImageJ (https://imagej.nih.gov).
Preparation of S. aureus membranes for LC-MS analysis
S. aureus cells were grown in 4 ml of Luria-Bertani (LB) medium at 37°C and 200 rpm shaking to OD600=0.4. For the complementary strains with pLOW vector, a final concentration of 5 μg/ml of erythromycin and 0.1 mM IPTG were added to the medium. Cells were harvested by centrifugation and resuspended in 10 mM Tris-HCl pH 8.0, 1 mM EDTA with lysostaphin, followed by a 0.5 h incubation at 37°C. Cells were further subjected to sonication. Membranes were isolated and resuspended in 100 mM Tris-HCl pH 8.5, 5% SDS and 10 mM tris(2-carboxyethyl) phosphine (TECP). Samples were incubated at 37°C for 1h after the addition of chloroacetamide to reduce and alkylate disulfides. Samples were then loaded on an S-Trap™ Micro Spin column (Protifi). On-column peptide digestion was performed by adding trypsin and incubated at 47°C for 1h. Digested peptides were collected by passing 50 mM triethylammonium bicarbonate (TEAB) buffer, 0.2 % formic acid (w/v) in distillated water, and 0.2 % formic acid (w/v) in 50% acetonitrile (v/v) through the column and dried in a SpeedVac (Labconco). Dried peptides were re-suspended in 0.1% formic acid (w/v) with iRT normalization peptide mix (Biognosys) and stored at -20 °C.
Targeted PRM-LC-MS analysis of LtaA and variants in S. aureus membranes
In a first step, parallel reaction-monitoring (PRM) assays74 for all possible peptides of LtaA being 6 to 25 amino acid long comprising double and triple charged precursor ions were generated. In total, 5 peptides were found to match the length criteria leading to a total of 10 PRM assays. These were applied to identify LtaA in membrane fractions of wild-type S. aureus. The setup of the μRPLC-MS system was as described previously75. Mass spectrometry analysis was performed on a Q-Exactive mass spectrometer equipped with a nanoelectrospray ion source (both Thermo Fisher Scientific). Each MS1 scan was followed by high-collision-dissociation (HCD) of the 10 LtaA precursor ions in PRM mode using a global isolation mass list. Using strict identification criteria, three peptides ions of LtaA, LTNYNTRPVK (2+ and 3+ ion) and MQDSSLNNYANHK (2+) could be confidently identified and were used for label-free PRM quantification. To control for variation in sample amounts, the total ion chromatogram (only comprising peptide ions with two or more charges) of each sample was determined by label- free quantification using Progenesis QI (version 2.0, Waters) and used for normalization. The integrated peak areas of the 3 peptide ions quantified by PRM were summed and employed for LtaA quantification (see Supplementary Notes).
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Extended Data
Extended Data Fig. 1.
Extended Data Fig. 2.
Extended Data Fig. 3.
Extended Data Fig. 4.
Extended Data Fig. 5.
Extended Data Fig. 6.
Extended Data Fig. 7.
Extended Data Fig. 8.
Extended Data Fig. 9.
Extended Data Fig. 10.
Supplementary Material
Acknowledgments
We thank the staff at the PX beamline of the Swiss Light Source, Switzerland. We thank G. Cebrero and N. Bärland for providing a control transporter sample. We thank J. Daraspe and M. Rengifo for contributing to TEM images acquisition. We thank U. Lanner, A. Schmidt, and T. Müntener for contributing to HPLC-MS and PRM-MS studies. This work was supported by the Swiss National Science Foundation (SNSF) (PP00P3_170607 to C.P and 31003A_172861 to J.W.V.). Further funding came from a JPIAMR grant (40AR40_185533 to J.W.V.) and ERC consolidator grant 771534-PneumoCaTChER (to J.W.V). E.L. was funded by the Biozentrum International PhD Program.
Footnotes
Author Contributions
B.Z. performed purification and crystallization of LtaA. C.P. assisted B.Z. during data collection, structure determination and docking analysis. B.Z., E.L. and C.P. established and performed in vitro flipping assays. C.P., B.Z. and E.L. analyzed the structural and in vitro functional data. E.L. performed reaction products characterization. X.L. and E.L. performed experiments in live cells and together with C.P. and J.W.V. analyzed in vivo data. G.M. and S.H. performed NMR analysis. C.P. conceived the project and wrote the manuscript with input from all authors.
Competing interests
The authors declare no competing interests.
Data availability
Atomic coordinates have been deposited in the Protein Data Bank under accession code PDB 6S7V. Source data for Figures 1C, 3B-C, 3E-F, 4A-C, 5C and Extended Fig. 1 are available with the paper online.
References
- 1.Rivera AM, Boucher HW. Current concepts in antimicrobial therapy against select gram-positive organisms: methicillin-resistant Staphylococcus aureus, penicillin-resistant pneumococci, and vancomycin-resistant enterococci. Mayo Clin Proc. 2011;86:1230–43. doi: 10.4065/mcp.2011.0514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cosgrove SE, et al. The impact of methicillin resistance in Staphylococcus aureus bacteremia on patient outcomes: mortality, length of stay, and hospital charges. Infect Control Hosp Epidemiol. 2005;26:166–74. doi: 10.1086/502522. [DOI] [PubMed] [Google Scholar]
- 3.Turner NA, et al. Methicillin-resistant Staphylococcus aureus: an overview of basic and clinical research. Nat Rev Microbiol. 2019;17:203–218. doi: 10.1038/s41579-018-0147-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown S, et al. Methicillin resistance in Staphylococcus aureus requires glycosylated wall teichoic acids. Proc Natl Acad Sci U S A. 2012;109:18909–14. doi: 10.1073/pnas.1209126109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Percy MG, Grundling A. Lipoteichoic acid synthesis and function in gram positive bacteria. Annu Rev Microbiol. 2014;68:81–100. doi: 10.1146/annurev-micro-091213-112949. [DOI] [PubMed] [Google Scholar]
- 6.Brown S, Santa Maria JP, Jr, Walker S. Wall teichoic acids of gram-positive bacteria. Annu Rev Microbiol. 2013;67:313–36. doi: 10.1146/annurev-micro-092412-155620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xia G, Kohler T, Peschel A. The wall teichoic acid and lipoteichoic acid polymers of Staphylococcus aureus. Int J Med Microbiol. 2010;300:148–54. doi: 10.1016/j.ijmm.2009.10.001. [DOI] [PubMed] [Google Scholar]
- 8.Reichmann NT, et al. Differential localization of LTA synthesis proteins and their interaction with the cell division machinery in Staphylococcus aureus. Mol Microbiol. 2014;92:273–86. doi: 10.1111/mmi.12551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sewell EW, Brown ED. Taking aim at wall teichoic acid synthesis: new biology and new leads for antibiotics. J Antibiot (Tokyo) 2014;67:43–51. doi: 10.1038/ja.2013.100. [DOI] [PubMed] [Google Scholar]
- 10.Lee JH, et al. Surface Glycopolymers Are Crucial for In Vitro Anti-Wall Teichoic Acid IgG-Mediated Complement Activation and Opsonophagocytosis of Staphylococcus aureus. Infect Immun. 2015;83:4247–55. doi: 10.1128/IAI.00767-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gautam S, Kim T, Lester E, Deep D, Spiegel DA. Wall teichoic acids prevent antibody binding to epitopes within the cell wall of Staphylococcus aureus. ACS chemical biology. 2016;11:25–30. doi: 10.1021/acschembio.5b00439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bucher T, Oppenheimer-Shaanan Y, Savidor A, Bloom-Ackermann Z, Kolodkin-Gal I. Disturbance of the bacterial cell wall specifically interferes with biofilm formation. Environ Microbiol Rep. 2015 doi: 10.1111/1758-2229.12346. [DOI] [PubMed] [Google Scholar]
- 13.Campbell J, et al. Synthetic lethal compound combinations reveal a fundamental connection between wall teichoic acid and peptidoglycan biosyntheses in Staphylococcus aureus. ACS Chem Biol. 2011;6:106–16. doi: 10.1021/cb100269f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peschel A, et al. Inactivation of the dlt operon in Staphylococcus aureus confers sensitivity to defensins, protegrins, and other antimicrobial peptides. J Biol Chem. 1999;274:8405–10. doi: 10.1074/jbc.274.13.8405. [DOI] [PubMed] [Google Scholar]
- 15.Reichmann NT, Grundling A. Location, synthesis and function of glycolipids and polyglycerolphosphate lipoteichoic acid in Gram-positive bacteria of the phylum Firmicutes. FEMS Microbiol Lett. 2011;319:97–105. doi: 10.1111/j.1574-6968.2011.02260.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hong SW, et al. Lipoteichoic acid of Streptococcus mutans interacts with Toll-like receptor 2 through the lipid moiety for induction of inflammatory mediators in murine macrophages. Mol Immunol. 2014;57:284–91. doi: 10.1016/j.molimm.2013.10.004. [DOI] [PubMed] [Google Scholar]
- 17.Kang SS, Sim JR, Yun CH, Han SH. Lipoteichoic acids as a major virulence factor causing inflammatory responses via Toll-like receptor 2. Arch Pharm Res. 2016;39:1519–1529. doi: 10.1007/s12272-016-0804-y. [DOI] [PubMed] [Google Scholar]
- 18.Fischer W, Koch HU, Rosel P, Fiedler F, Schmuck L. Structural requirements of lipoteichoic acid carrier for recognition by the poly(ribitol phosphate) polymerase from Staphylococcus aureus H. A study of various lipoteichoic acids, derivatives, and related compounds. J Biol Chem. 1980;255:4550–6. [PubMed] [Google Scholar]
- 19.Grundling A, Schneewind O. Genes required for glycolipid synthesis and lipoteichoic acid anchoring in Staphylococcus aureus. J Bacteriol. 2007;189:2521–30. doi: 10.1128/JB.01683-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jorasch P, Wolter FP, Zahringer U, Heinz E. A UDP glucosyltransferase from Bacillus subtilis successively transfers up to four glucose residues to 1,2-diacylglycerol: expression of ypfP in Escherichia coli and structural analysis of its reaction products. Mol Microbiol. 1998;29:419–30. doi: 10.1046/j.1365-2958.1998.00930.x. [DOI] [PubMed] [Google Scholar]
- 21.Kiriukhin MY, Debabov DV, Shinabarger DL, Neuhaus FC. Biosynthesis of the glycolipid anchor in lipoteichoic acid of Staphylococcus aureus RN4220: role of YpfP, the diglucosyldiacylglycerol synthase. J Bacteriol. 2001;183:3506–14. doi: 10.1128/JB.183.11.3506-3514.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grundling A, Schneewind O. Synthesis of glycerol phosphate lipoteichoic acid in Staphylococcus aureus. Proc Natl Acad Sci U S A. 2007;104:8478–83. doi: 10.1073/pnas.0701821104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lu D, et al. Structure-based mechanism of lipoteichoic acid synthesis by Staphylococcus aureus LtaS. Proc Natl Acad Sci USA. 2009;106:1584–9. doi: 10.1073/pnas.0809020106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reddy VS, Shlykov MA, Castillo R, Sun EI, Saier MH., Jr The major facilitator superfamily (MFS) revisited. FEBS J. 2012;279:2022–35. doi: 10.1111/j.1742-4658.2012.08588.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cura AJ, Carruthers A. Role of monosaccharide transport proteins in carbohydrate assimilation, distribution, metabolism, and homeostasis. Compr Physiol. 2012;2:863–914. doi: 10.1002/cphy.c110024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smith DE, Clemencon B, Hediger MA. Proton-coupled oligopeptide transporter family SLC15: physiological, pharmacological and pathological implications. Mol Aspects Med. 2013;34:323–36. doi: 10.1016/j.mam.2012.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Quistgaard EM, Low C, Guettou F, Nordlund P. Understanding transport by the major facilitator superfamily (MFS): structures pave the way. Nat Rev Mol Cell Biol. 2016;17:123–32. doi: 10.1038/nrm.2015.25. [DOI] [PubMed] [Google Scholar]
- 28.Iancu CV, Zamoon J, Woo SB, Aleshin A, Choe JY. Crystal structure of a glucose/H+ symporter and its mechanism of action. Proc Natl Acad Sci U S A. 2013;110:17862–7. doi: 10.1073/pnas.1311485110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Deng D, et al. Molecular basis of ligand recognition and transport by glucose transporters. Nature. 2015;526:391–6. doi: 10.1038/nature14655. [DOI] [PubMed] [Google Scholar]
- 30.Sun L, et al. Crystal structure of a bacterial homologue of glucose transporters GLUT1-4. Nature. 2012;490:361–6. doi: 10.1038/nature11524. [DOI] [PubMed] [Google Scholar]
- 31.Pedersen BP, et al. Crystal structure of a eukaryotic phosphate transporter. Nature. 2013;496:533–6. doi: 10.1038/nature12042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zheng H, Wisedchaisri G, Gonen T. Crystal structure of a nitrate/nitrite exchanger. Nature. 2013;497:647–51. doi: 10.1038/nature12139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yan H, et al. Structure and mechanism of a nitrate transporter. Cell Rep. 2013;3:716–23. doi: 10.1016/j.celrep.2013.03.007. [DOI] [PubMed] [Google Scholar]
- 34.Newstead S, et al. Crystal structure of a prokaryotic homologue of the mammalian oligopeptide-proton symporters, PepT1 and PepT2. EMBO J. 2011;30:417–26. doi: 10.1038/emboj.2010.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Menon I, et al. Opsin is a phospholipid flippase. Curr Biol. 2011;21:149–53. doi: 10.1016/j.cub.2010.12.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brunner JD, Lim NK, Schenck S, Duerst A, Dutzler R. X-ray structure of a calcium-activated TMEM16 lipid scramblase. Nature. 2014;516:207–12. doi: 10.1038/nature13984. [DOI] [PubMed] [Google Scholar]
- 37.Malvezzi M, et al. Ca2+-dependent phospholipid scrambling by a reconstituted TMEM16 ion channel. Nat Commun. 2013;4:2367. doi: 10.1038/ncomms3367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hanson BL, Bunick GJ. Annealing macromolecular crystals. Methods Mol Biol. 2007;364:31–42. doi: 10.1385/1-59745-266-1:31. [DOI] [PubMed] [Google Scholar]
- 39.Nagarathinam K, et al. Outward open conformation of a Major Facilitator Superfamily multidrug/H(+) antiporter provides insights into switching mechanism. Nat Commun. 2018;9:4005. doi: 10.1038/s41467-018-06306-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bibi E, Kaback HR. In vivo expression of the lacY gene in two segments leads to functional lac permease. Proc Natl Acad Sci U S A. 1990;87:4325–9. doi: 10.1073/pnas.87.11.4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Varela MF, Sansom CE, Griffith JK. Mutational analysis and molecular modelling of an amino acid sequence motif conserved in antiporters but not symporters in a transporter superfamily. Mol Membr Biol. 1995;12:313–9. doi: 10.3109/09687689509072433. [DOI] [PubMed] [Google Scholar]
- 42.Smirnova IN, Kasho V, Kaback HR. Protonation and sugar binding to LacY. Proc Natl Acad Sci U S A. 2008;105:8896–901. doi: 10.1073/pnas.0803577105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Feng L, Campbell EB, MacKinnon R. Molecular mechanism of proton transport in CLC Cl-/H+ exchange transporters. Proc Natl Acad Sci U S A. 2012;109:11699–704. doi: 10.1073/pnas.1205764109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.du Plessis JL, Stefaniak AB, Wilhelm KP. Measurement of Skin Surface pH. Curr Probl Dermatol. 2018;54:19–25. doi: 10.1159/000489514. [DOI] [PubMed] [Google Scholar]
- 45.Frank DN, et al. The human nasal microbiota and Staphylococcus aureus carriage. PLoS One. 2010;5:e10598. doi: 10.1371/journal.pone.0010598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Harell M, Mover-Lev H, Levy D, Sade J. Gas composition of the human nose and nasopharyngeal space. Acta Otolaryngol. 1996;116:82–4. doi: 10.3109/00016489609137718. [DOI] [PubMed] [Google Scholar]
- 47.Williams MR, Nakatsuji T, Gallo RL. Staphylococcus aureus: Master Manipulator of the Skin. Cell Host Microbe. 2017;22:579–581. doi: 10.1016/j.chom.2017.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Law CJ, Maloney PC, Wang DN. Ins and outs of major facilitator superfamily antiporters. Annu Rev Microbiol. 2008;62:289–305. doi: 10.1146/annurev.micro.61.080706.093329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jardetzky O. Simple allosteric model for membrane pumps. Nature. 1966;211:969–70. doi: 10.1038/211969a0. [DOI] [PubMed] [Google Scholar]
- 50.Kuk AC, Mashalidis EH, Lee SY. Crystal structure of the MOP flippase MurJ in an inward-facing conformation. Nat Struct Mol Biol. 2017;24:171–176. doi: 10.1038/nsmb.3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zheng S, et al. Structure and mutagenic analysis of the lipid II flippase MurJ from Escherichia coli. Proc Natl Acad Sci U S A. 2018;115:6709–6714. doi: 10.1073/pnas.1802192115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sham LT, et al. Bacterial cell wall. MurJ is the flippase of lipid-linked precursors for peptidoglycan biogenesis. Science. 2014;345:220–2. doi: 10.1126/science.1254522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Timcenko M, et al. Structure and autoregulation of a P4-ATPase lipid flippase. Nature. 2019;571:366–370. doi: 10.1038/s41586-019-1344-7. [DOI] [PubMed] [Google Scholar]
- 54.Hiraizumi M, Yamashita K, Nishizawa T, Nureki O. Cryo-EM structures capture the transport cycle of the P4-ATPase flippase. Science. 2019 doi: 10.1126/science.aay3353. [DOI] [PubMed] [Google Scholar]
- 55.Perez C, et al. Structure and mechanism of an active lipid-linked oligosaccharide flippase. Nature. 2015;524:433–8. doi: 10.1038/nature14953. [DOI] [PubMed] [Google Scholar]
- 56.Mi W, et al. Structural basis of MsbA-mediated lipopolysaccharide transport. Nature. 2017;549:233–237. doi: 10.1038/nature23649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bi Y, Mann E, Whitfield C, Zimmer J. Architecture of a channel-forming O-antigen polysaccharide ABC transporter. Nature. 2018;553:361–365. doi: 10.1038/nature25190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kalienkova V, et al. Stepwise activation mechanism of the scramblase nhTMEM16 revealed by cryo-EM. Elife. 2019;8 doi: 10.7554/eLife.44364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rubino FA, Kumar S, Ruiz N, Walker S, Kahne DE. Membrane Potential Is Required for MurJ Function. J Am Chem Soc. 2018;140:4481–4484. doi: 10.1021/jacs.8b00942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mirza O, Guan L, Verner G, Iwata S, Kaback HR. Structural evidence for induced fit and a mechanism for sugar/H+ symport in LacY. EMBO J. 2006;25:1177–83. doi: 10.1038/sj.emboj.7601028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang B, Perez C. Stabilization and Crystallization of a Membrane Protein Involved in Lipid Transport. Methods Mol Biol. 2020;2127:283–292. doi: 10.1007/978-1-0716-0373-4_19. [DOI] [PubMed] [Google Scholar]
- 62.Kabsch W. Xds. Acta Crystallogr D Biol Crystallogr. 2010;66:125–32. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Karplus PA, Diederichs K. Linking crystallographic model and data quality. Science. 2012;336:1030–3. doi: 10.1126/science.1218231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–21. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sheldrick GM. A short history of SHELX. Acta Crystallogr A. 2008;64:112–22. doi: 10.1107/S0108767307043930. [DOI] [PubMed] [Google Scholar]
- 66.Collaborative Computational Project, N. The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–3. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 67.Skubak P, et al. A new MR-SAD algorithm for the automatic building of protein models from low-resolution X-ray data and a poor starting model. IUCr J. 2018;5:166–171. doi: 10.1107/S2052252517017961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.McCoy AJ, et al. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cowtan K. Recent developments in classical density modification. Acta Crystallogr D Biol Crystallogr. 2010;66:470–8. doi: 10.1107/S090744490903947X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Waterhouse A, et al. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 2018;46:W296–W303. doi: 10.1093/nar/gky427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jiang D, et al. Structure of the YajR transporter suggests a transport mechanism based on the conserved motif A. Proc Natl Acad Sci U S A. 2013;110:14664–9. doi: 10.1073/pnas.1308127110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Fratamico PM, et al. Escherichia coli serogroup O2 and O28ac O-antigen gene cluster sequences and detection of pathogenic E. coli O2 and O28ac by PCR. Can J Microbiol. 2010;56:308–16. doi: 10.1139/w10-010. [DOI] [PubMed] [Google Scholar]
- 74.Peterson AC, Russell JD, Bailey DJ, Westphall MS, Coon JJ. Parallel reaction monitoring for high resolution and high mass accuracy quantitative, targeted proteomics. Mol Cell Proteomics. 2012;11:1475–88. doi: 10.1074/mcp.O112.020131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ahrne E, et al. Evaluation and Improvement of Quantification Accuracy in Isobaric Mass Tag-Based Protein Quantification Experiments. J Proteome Res. 2016;15:2537–47. doi: 10.1021/acs.jproteome.6b00066. [DOI] [PubMed] [Google Scholar]
- 76.Ashkenazy H, et al. ConSurf 2016: an improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Res. 2016;44:W344–50. doi: 10.1093/nar/gkw408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31:455–61. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Arnaud M, Chastanet A, Debarbouille M. New vector for efficient allelic replacement in naturally nontransformable, low-GC-content, gram-positive bacteria. Appl Environ Microbiol. 2004;70:6887–91. doi: 10.1128/AEM.70.11.6887-6891.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Stamsas GA, et al. CozEa and CozEb play overlapping and essential roles in controlling cell division in Staphylococcus aureus. Mol Microbiol. 2018 doi: 10.1111/mmi.13999. [DOI] [PubMed] [Google Scholar]
- 80.Monk IR, Tree JJ, Howden BP, Stinear TP, Foster TJ. Complete Bypass of Restriction Systems for Major Staphylococcus aureus Lineages. MBio. 2015;6:e00308-15. doi: 10.1128/mBio.00308-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Atomic coordinates have been deposited in the Protein Data Bank under accession code PDB 6S7V. Source data for Figures 1C, 3B-C, 3E-F, 4A-C, 5C and Extended Fig. 1 are available with the paper online.