Abstract
Accurately quantifying the proteome remains challenging for large sample series and longitudinal experiments. We report a data-independent acquisition method, Scanning SWATH, that accelerates the mass spectrometric duty cycles, yielding quantitative proteomes in combination with short gradients and high-flow (800 µL/min) chromatography. Exploiting a continuous movement of the precursor isolation window to assign precursor masses to the MS/MS fragment traces, Scanning SWATH increases precursor identifications by ~70% compared to conventional DIA methods on 0.5-5 minute chromatographic gradients. We demonstrate the application of ultra-fast proteomics in drug mode-of-action screening and plasma proteomics. Scanning SWATH proteomes capture the mode-of-action of fungistatic azoles and statins. Moreover, we confirm 43 and identify 11 new plasma proteome biomarkers of COVID-19 severity, advancing patient classification and biomarker discovery. Thus, our results demonstrate a substantial acceleration and increased depth in fast proteomic experiments that facilitate proteomic drug screens and clinical studies.
Introduction
Proteomes bridge between genotype and phenotype, and are important for basic as well as data-driven biology, biotechnology and systems medicine1–3. Indeed, proteins account for more than 2/3rds of drug targets 4. Proteomes are, however, inherently complex, presenting significant analytical challenges 5. Advances in mass spectrometry instrumentation, chromatography, acquisition methods, data analysis strategies, as well as the introduction of ion mobility devices have increased the depth in single-shot proteome measurements 6–12. However, the requirements of quantitative biology, precision medicine, and epidemiology are driving a need to increase throughput, to improve consistency and precision, to facilitate longitudinal studies and to make data acquired across laboratories more comparable 2,13,14. Larger, systematic experiments, and more complete data matrices, would also enable the application of advanced statistical methods, including machine learning, in the analysis of biological and medical problems15–17.
A central challenge is to raise throughput in proteomic measurements without compromising identification numbers, quantitative precision, and data completeness. Here we report a data-independent acquisition (DIA) method, Scanning SWATH, and software algorithms dedicated to analyzing highly complex samples measured with short chromatographic gradients in bottom-up proteomics. First, Scanning SWATH adds a new dimension to the DIA-MS data that is derived from the use of a “sliding” quadrupole (Q1), that was first introduced as part of the “SONAR” method 18,19. In Scanning SWATH, the scanning dimension is exploited to assign precursor masses to MS/MS traces, which is not possible in conventional DIA methods. Scanning SWATH hence combines the advantages of data-dependent and data-independent proteomic acquisition techniques. Second, in Scanning SWATH, Q1 scans are completed in a shorter time than the stepwise acquisition in conventional SWATH 20,21, as there is no need to empty the collision cell between steps.
We demonstrate that the combination of Scanning SWATH with analytical high-flow chromatography (800 µL/min) generates a technology platform for ultra-fast, yet quantitatively precise proteome experiments. This setup allows the processing of several hundred samples per day per mass spectrometer, using chromatographic gradients at a minute or even a sub-minute scale. We show that the enhanced throughput does not come at the expense of quantitative precision. Indeed, we report coefficient of variation (CV) values that are equal or better compared to other proteomic techniques that are of considerably lower throughput. We illustrate the application of ultra-fast proteomics for capturing the response of cytostatic and antifungal drugs (statins, azoles, and antifolates) acting on yeast, for mode-of-action prediction using the proteome, in addition to compound-specific effects. Moreover, we demonstrate that proteomic gradients as fast as 60 seconds allow for classification of disease severity in COVID-19 patients on the basis of their plasma proteomes, and we identify a panel of known and new COVID-19 severity biomarkers.
Results
The scanning quadrupole creates a new data dimension
So far, a disadvantage of SWATH-MS over DDA acquisition techniques has been a lack of precursor mass assignment to the MS/MS traces 20. A continuous movement of the quadrupole in Scanning SWATH results in a time dependency of the fragment signal. The signal of each MS/MS feature first appears and then disappears when the leading margin and the trailing margin of the “sliding” isolation window pass the precursor mass, respectively (Figure 1b). This scanning dimension is complementary to the retention time dimension, as it allows to distinguish co-eluting peptides with different precursor masses and to assign precursor masses to each fragment ion (Figure 1, lower panel).
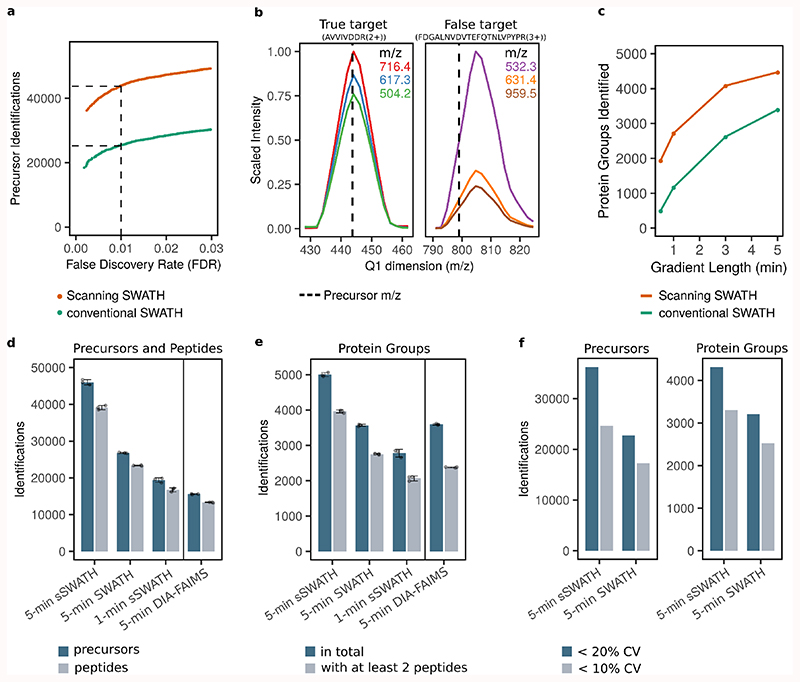
Figure 1. Scanning SWATH replaces the stepwise precursor selection with a continuously moving quadrupole and thereby adds another dimension to the data and shortens duty cycles.
a. In conventional SWATH-MS/DIA-MS, a quadrupole selects a relatively wide mass range, and the detector collects MS/MS spectra for a defined accumulation time. The windows are stepped and are overlapping (to compensate for edge effects20). The collision cell needs to be emptied after each step. b. In Scanning SWATH, the isolation window slides over the precursor mass range and MS/MS spectra are continuously acquired. The continuous movement of the quadrupole results in a time dependency of the fragment intensity. Fragment signals appear when the leading edge of the quadrupole passes the precursor m/z and they disappear when the precursor m/z falls out of the quadrupole isolation window. c. The acquired raw data is sectioned into bins of a defined m/z size. Data from TOF pulses that overlap with a certain m/z bin are summed together and written into the respective bin (e.g. all TOF pulses labeled in red on the diagram are summed together in the respective bin). Therefore, the highest signal for a fragment is in the bin which includes the respective precursor mass. In contrast to conventional SWATH, data from each TOF pulse is written into more than one bin, resulting in a Q1 profile of a triangular shape. d,e. The Q1 profile provides a 4th dimension in the Scanning SWATH data. In conventional SWATH each fragment mass (mass dimension) has a certain intensity (intensity dimension) that is measured along the chromatographic time (retention time dimension). e. In Scanning SWATH data, each fragment gives rise also to a Q1 profile (Q1 dimension) f. Different fragments from the same precursor show correlating Q1 profiles (e.g. green, orange and purple fragments). The apex of the Q1 profile corresponds to the precursor mass and thus fragments from different precursors can be distinguished (e.g. green, orange and purple fragments belong to a different precursor than the pink fragment).
To make the scanning dimension exploitable, the acquired Scanning SWATH data is written into defined m/z bins by summing up all TOF pulses that overlap with the respective precursor range (Figure 1c). The resulting triangular “Q1 profiles” are then mapped to m/z coordinates by calibrating on known masses and aligning the Q1 profiles with the respective MS1 mass (Methods). To exploit the Q1 profiles in proteomic experiments, we developed open-source algorithms and made them broadly accessible via including them in the open-source DIA-NN software suite 22. DIA-NN makes use of the scanning dimension by calculating 15 scores that assess (i) the similarity of the Q1 profiles of the fragments and the non-fragmented precursor (ii) the Q1 profile shapes, and (iii) the relation of the centroided Q1 profile and the expected precursor mass (Figure S1 and Methods). These scores are then analyzed by a deep neural network classifier used in DIA-NN 22, to assign confidence scores to peptide-spectrum matches and thus obtain q-values (Figure S1).
In order to test to what extent the use of the Q1 dimension improves true precursor identification out of complex DIA spectra, we separated 10 µg of a trypsin-digested human cell lines (K562) on a 5-minute high-flow LC gradient, as introduced recently as part of a high-throughput proteomics workflow 23. We used a 10 m/z Scanning SWATH window size as this yields a good compromise between proteomic depth and quantification precision (Methods, Figure S2a). Moreover, we experimentally determined the false discovery rate using a two-species spectral library method (22,24, Methods). In the 5-minute gradient, Scanning SWATH identified 70% more true positive precursors at 1% FDR than an optimized, conventional SWATH acquisition method 23, run on the same chromatographic gradient and mass spectrometer (Figure 2a). To illustrate the impact of the additional dimension, we highlight the true positive target human precursor-AVVIVDDR(2+), for which the apex of the Q1 profile matches the mass of the precursor in the library and thus increases the confidence in this particular identification (Figure 2b - left panel). On the other hand, the apex of the Q1 profiles that correspond to the extracted fragment masses of a false target (Arabidopsis thaliana precursor- FDGALNVDVTEFQTNLVPYPR(3+)) does not match the respective precursor mass (Figure 2b- right panel). Therefore, this particular false target had a reported q-value above 0.01 (not identified) when analyzed with Scanning SWATH, but was incorrectly called as true positive using conventional SWATH (reported q-value below 0.01). Thus, the use of the Q1 profiles facilitates a better distinction of true targets from interferences.
Figure 2. Scanning SWATH improves peptide identification in short gradients.
a. 10 µg human cell line digest was acquired with a Scanning SWATH method (10 m/z window) and a conventional stepped SWATH method 23 using a 5-minute high-flow (800 µL/min) LC gradient. The data were analyzed with DIA-NN using a two-species library, that contains human and Arabidopsis thaliana precursors, to validate the FDR experimentally (methods) 8,22. b. left panel: Q1 profiles of fragments corresponding to a true-positive target precursor (human) with a mass of 443.8 m/z (AVVIVDDR(2+)). b. right panel: Q1 profile of fragments corresponding to a false target (Arabidopsis thaliana precursor) with the precursor mass 799.1 m/z (FDGALNVDVTEFQTNLVPYPR(3+)). c. Number of protein groups identified (1% FDR) in a K562 cell lysate with Scanning SWATH and conventional stepped SWATH, using 5, 3, 1 minute and 30 second chromatographic gradients and adjusted duty cycles (Table S1, S2, S3). d. Number of precursors (peptides ionized to a specific charge) and peptides (stripped sequences) identified (1% FDR) in human cell lysates measured with different acquisition schemes and platforms. A K562 digest was analyzed with 5 and 1-minute gradient Scanning SWATH (“5-min sSWATH” and “1-min sSWATH”) and 5-minute conventional stepped SWATH (“5 min SWATH”). 10 µg was injected for the 5-minute gradients and 5 µg for the 1-minute gradient. To put the results into context, we compared them to a publicly available 5-minute gradient human cell line (HeLa) DIA dataset as recorded with an Evosep One system coupled to an Orbitrap Exploris 480 with FAIMS (“5-min DIA-FAIMS”) (PXD016662)25. Project-specific libraries and the same software settings were used for raw data analysis (Methods). Data are presented as mean +/- standard deviation (n=3 replicate injections). e. The number of protein groups (1% FDR) with at least one or two peptide identifications, respectively. Data are presented as mean +/- standard deviation (n=3 replicate injections). f. Number of precursors (left) and protein groups (right) quantified with a coefficient of variation (CV) below 20% and below 10% in triplicate injections. CV values were calculated from n = 3 replicate injections.
Scanning SWATH records precise proteomes with short gradients
In order to increase the throughput capacity in proteomics, we have recently introduced protocols that use high-flow liquid chromatography with flow rates of several hundred microliters per minute, and with gradient lengths of 5 minutes and faster 23. With only 3-minute overheads between injections, that include all washing and equilibration steps, 5-minute high-flow LC gradients facilitate a proteomic throughput of 180 samples/day 23. We have further shown that high-flow chromatography offers also other benefits to proteomic experiments. It provides high peak capacities, reduces carry-over, and improves longitudinal chromatographic and electrospray stability, factors that lead to an increased quantification precision and data completeness 23.
To test to which extent Scanning SWATH facilitates proteome experiments with fast, high-flow chromatographic gradients, we ran linear gradients with 5, 3, and 1 minute as well as 30-second length at a flow rate of 800 µL/min. We acquired the data with correspondingly adjusted duty cycles (see Methods) and injected 5 µg K562 tryptic digests. With Scanning SWATH cycle times as short as 280 ms (30-second gradient), the method recorded on average >3 data points per peak at full width at half maximum (FWHM), which corresponds to > 5 points per peak width at base (1.7*FWHM 26), which is sufficient for precise quantification in DIA-MS experiments 11,27. The 30-second gradient quantified 1,937 protein groups at 1% FDR. The identification numbers increased to 2,720 protein groups with a 60-second chromatographic gradient (cycle time of 310 ms), and to 4,470 protein groups in 5 minutes (cycle time of 520 ms) when 5 µg human cell lysate digest was injected (Figure 2c).
To determine the dependency on sample amount injected, we measured a dilution series of K562 tryptic digests, ranging from 10 µg to 250 ng, with 5-minute conventional SWATH and 5-minute Scanning SWATH. Scanning SWATH shows overall higher ID numbers, even at lower injection amounts (Figure S3ab). We have further measured a dilution series with micro-flow liquid chromatography (5 µl/min and 20-min gradients). Here, Scanning SWATH identifies 4,896 human protein groups from 1.25 µg of tryptic digest (Figure S3c).
To put the performance of Scanning SWATH into context, we illustrate a benchmark in which we compare the 5-minute Scanning SWATH runs (“5-min sSWATH”) with 1-minute Scanning SWATH (“1-min sSWATH”) and with 5-minute conventional stepped SWATH (“5-min SWATH”) on the same LC-MS setup. This benchmark confirmed an increase of 70% in precursor identifications (peptides ionized to a specific charge) compared to stepped SWATH on the same instrument setup on the 5-minute high-flow rate gradient (46,009 vs 26,795) (Figure 2d). We further compare these results with a reference dataset that has been recorded with an Orbitrap Exploris 480 instrument (Thermo) with FAIMS interface and with 5-minute separations on an Evosep One LC system (“5-min DIA-FAIMS”) 25. If analyzed with the same software and software settings to render the results comparable, the 5-min Evosep-DIA-FAIMS study quantified 15,576 precursor, which is less than the Scanning SWATH precursor identifications in 1/5th of the gradient time (60-second experiment, 19,401 precursors) (Figure 2d). The Scanning SWATH experiment identified 5,004 protein groups (corresponding to 4,394 unique proteins, detected with proteotypic peptides), while conventional stepped SWATH and DIA-FAIMS identified with a comparable gradient length 3,568 and 3,594 protein groups, respectively (Figure 2e). Out of these, 3,962, 2,753, and 2,380 protein groups are identified with at least 2 peptides in 5-minute Scanning SWATH, 5-minute conventional SWATH, and 5-minute DIA-FAIMS, respectively.
Scanning SWATH also improves quantification precision, expressed as a median coefficient of variation (CV) values of 6.4% and 8.8% for all protein groups and precursors quantified, respectively. When comparing on the same set of protein groups quantified in both Scanning- and conventional SWATH runs, Scanning SWATH yielded median CV values of 4.1% compared to 4.9% for conventional SWATH (Figure S2b and Figure S2c for protein groups and precursors, respectively). In absolute numbers, Scanning SWATH quantified 4,317 protein groups (out of 5,004 identified) with CV < 20% and 3,308 with CV < 10%, while conventional SWATH quantifies 3,208 and 2,527 with CV < 20% and CV < 10%, respectively (Figure 2f - left panel for precursors and right panel for protein groups).
The quantification precision obtained with the Evosep-DIA-FAIMS using the 5-minute gradients in the reference dataset 25 is substantially lower compared to the performance of Scanning SWATH (Figure S2b and Figure S2c for protein groups and precursors). However, we would like to highlight caveats in directly comparing the quantification and identification performance of the high-flow rate LC Scanning SWATH to the Evosep-Exploris DIA data 25. While conventional SWATH and Scanning SWATH experiments are directly comparable (only the acquisition method differs), the Evosep-Exploris and the high-flow qTOF platform differ in multiple design parameters 25, so that the acquisitions differed not only in flow rate and scan mode but also in precursor range, cycle time (1 sec for the Exploris) and in the fact that the Evosep system uses as separate solid-phase extraction (SPE) tip for each sample23,28. Furthermore, the FAIMS-DIA reference data used a non-commercial mammalian cell line (HeLa) tryptic digest generated with an in-house protocol. We have no influence on the design elements, but we would like to note that they affect both depth and quantification precision, and that therefore, the direct comparison is compromised by the fact that the platforms differ in multiple parameters.
Scanning SWATH captures drug responses
If proteomics can be run at high-throughput, it has the potential to be used in drug screens as a comprehensive phenotypic readout. Proteomic screening strategies are particularly attractive for those medical applications where classic screening approaches have yielded only a few hits, such as in the development of antifungals 29,30. Despite 1.6 million people dying each year from invasive fungal infections, immense screening efforts have so far only yielded three classes of clinically applicable antifungals (azoles, echinocandins, and polyenes) 31,32. To test whether a high-throughput proteomic approach could be used for exploratory drug screening, we applied high-flow Scanning SWATH to measure proteomes of the single cellular fungus Saccharomyces cerevisiae treated with 16 different drugs from 3 different drug classes (antifolates, statins, and azoles - Table S4) at 10 µM working concentration, which is a typical screening concentration in large-scale experiments. These specific classes were chosen because of their clinical availability, well-characterized mode-of-action, documented resistance mechanisms and, considering azoles, their successful application in antifungal therapies 33–35. Samples were measured in quadruplicates and pooled sample were repeatedly injected for quality control. Using 5-minute high-flow gradients (7.3 minutes including column wash and equilibration steps, see Table S5), the measurement of 103 proteomes (samples plus controls) was completed within 13.5 hours. On average 1,980 unique proteins (1% FDR) (Figure S4a) were identified per run. The yeast proteome that is less complex than the mammalian proteome, was hence captured at a depth that is comparable to previous yeast proteomic experiments using SWATH-MS with several times longer gradients and nano- or micro-flow rate chromatography 17,36. The proteins were quantified with a median coefficient of variation of 8% for instrument control samples (injections of the same sample across the measurements) (Figure S4b). High-flow Scanning SWATH hence facilitated the acquisition of the proteomes not only in a much shorter time, but indeed yielded also substantial gain of precision compared to previous yeast DIA proteomic studies that required weeks to months to process a similar number of samples 17,37,38.
A principal component analysis (PCA) of the differentially expressed proteins across each drug class revealed a clear separation of the antifolate and statin/azole drug classes (Figure 3a). Furthermore, the proteomes reflect the drug potency within the classes, which is in turn reflected by the number of differentially expressed proteins (Figure S4c). For instance, the 10 µM treatment with atorvastatin had a much more prevalent impact on the proteome with 287 proteins differentially expressed, compared to a treatment with lovastatine which resulted in 38 differentially expressed proteins (Figure S4c). Furthermore, mapping the affected pathways consistently identified the mode-of-action related to the drug class, primarily resulting in the upregulation of associated enzymes of the inhibited pathway (Figure 3b). For example, azoles elicit a strong upregulation of multiple proteins involved in the ergosterol biosynthesis pathway (Figure 3d, Figure S4e), most notably in the azole target lanosterol 14-alpha demethylase (gene product of ERG11) itself which is in agreement with previous findings. Consistent with this, a response in the same pathway was also observed in statin-treated cells (Figure 3d, Figure S4d), however squalene monooxygenase (gene product of ERG1), which is significantly upregulated upon treatment with statins, was found to be downregulated in azoles (Figure 3d, Figure 3e). Thus, although statins and azoles target the same pathway, the proteomes reveal class-dependent signatures that permit differentiation of these two drug classes. Indeed, the antifolate methotrexate induced an upregulation in proteins primarily related to nucleotide, purine, and pyrimidine biosynthesis; a clear and different response compared to statins/azoles, linking dihydrofolate reductase (gene product of DHFR1) inhibition to nucleotide starvation and cessation of DNA replication 40. The structurally related antifolate pralatrexate instead induces a change in a broader range of pathways additional to nucleotide biosynthesis despite its lower potency (Figure 3b). Each of these drug treatments therefore shows a specific proteome response and drugs from the same class show similar patterns.
Figure 3. The proteome response in drug-treated Saccharomyces cerevisiae captured with 5-minute Scanning SWATH.
Prototrophic Saccharomyces cerevisiae (S288c background) yeast cells were grown in minimal media and treated with 10 µM of the indicated drug. 5 µg peptides were injected and analyzed with Scanning SWATH and 5-minute water-to-acetonitrile chromatographic gradients (800 µL/min flow rate). a. Principal component analysis separates the samples according to drug class as well as potency. Proteins that are differentially expressed in at least one of the drug classes (compared to DMSO) were considered (two-sided t-test, adjusted p-value < 0.01, Benjamini-Hochberg multiple testing correction 41). The quantities were log2 transformed and centered. Drugs that have > 20 differentially expressed proteins are shown. b. Pathway enrichment of proteomic data identifies the target pathways. Pathway enrichment among differentially expressed proteins (two-sided t-test, adjusted p-value < 0.01, Benjamini-Hochberg multiple testing correction 41) was conducted using hypergeometric testing. c. The proteome responses are drug class-specific. Differentially expressed proteins in at least one drug class are illustrated as a heatmap. Clustering was performed row-wise but not column-wise. Drugs that have > 20 differentially expressed proteins are shown. d. Differential protein expression varies by drug class, and identifies the targeted pathways for azoles (left panel) and statins (right panel). Significance (-log10(adjusted p-value)) was calculated with a t-test (two-sided) and is plotted as a function of the log2 fold-changes (ratio of expression levels in treated and DMSO-treated cells). Proteins in the cholesterol pathway that have an adjusted p-value < 0.01 are highlighted and are labelled with the respective gene name. The Benjamini-Hochberg procedure was used for multiple testing correction 41. e. Treatments with azoles and statins result in down- and upregulation of the Squalene monooxygenase (gene product of ERG1), respectively. The expression levels are given as fold changes (ratio of expression levels in treated and DMSO-treated cells). The boxes show the first and third quartile as well as the median (middle) and the whiskers extend to the most extreme data point, which is no more than 1.5 times the interquartile range from the box. n = 5 azoles, n = 7 statins.
COVID-19 severity classification with 1-minute gradients
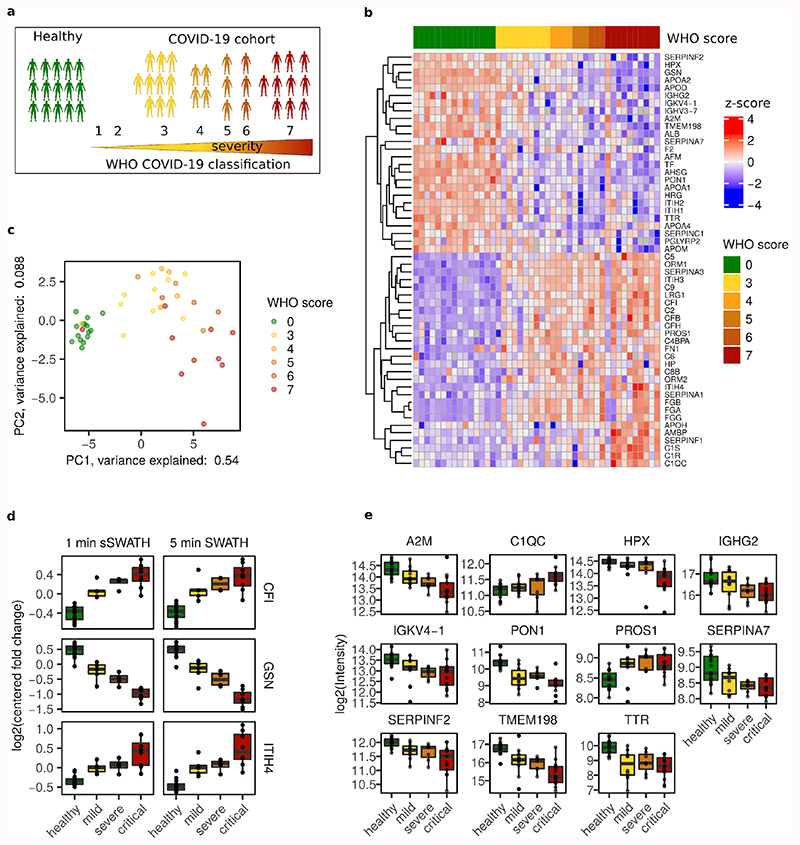
In addition to drug-screening approaches, clinical proteomics is a major application for fast proteomic methods. Applied on blood plasma or serum samples, proteomics can i) classify patients, ii) identify biomarkers, and iii) provide molecular signatures for diagnostic and prognostic models 23,24,45,46. Fast proteomic methods are specifically relevant during pandemics, as they can rapidly assess pathomechanisms in an unbiased fashion 23,47–50. To demonstrate the application of ultra-fast proteomic methods for plasma proteomics, we processed non-depleted citrate plasma samples from a cohort of 30 COVID-19 patients hospitalized at Charité Universitätsmedizin Berlin between 1st and 26th March 2020 51, and compared their plasma proteomes to those of 15 healthy individuals. The cohort was relatively well-balanced between individuals suffering from COVID-19 with different levels of severity (Table S6 and Figure 4a), as graded according to the WHO ordinal outcome scale of clinical improvement (score 3 = hospitalized, no oxygen therapy, score 4 = oxygen by mask or nasal prongs, score 5 = non-invasive ventilation or high-flow oxygen, score 6 = intubation and mechanical ventilation, score 7 = ventilation and additional organ support - pressors, RRT, ECMO) 52. We analyzed the citrate plasma samples using Scanning SWATH in conjunction with 60-second water to acetonitrile chromatographic gradients (3.5 minutes total runtime, Table S7). We identified on average around 2,600 precursors (Figure S5a) corresponding to 180 protein groups (Figure S5b). The achieved proteomic depth is hence comparable to precursor identification numbers we have previously acquired with 5-times longer LC gradients and conventional SWATH 23. 47 of the quantified proteins are FDA approved biomarkers 53, indicating the information-richness of the highly abundant plasma protein fraction (Table S8). Median protein coefficients of variation were 4.4 % and 6.6% for instrument QC (repeat injection of a pooled digest) and process QC (separately prepared digests from the same commercial plasma sample), respectively (Figure S5c). This indicates that the short measurement times do not compromise quantification precision. In parallel, we compared the 1-minute Scanning SWATH to a conventional SWATH method that uses 5-minute chromatographic gradients 23. The same cohort was measured with both methods on two different qTOF instruments, and we compared the relative abundance changes measured. Both methods result in similar quantitative changes but Scanning SWATH achieves this with a 5 times shorter gradient than the conventional SWATH method (Figure 4d for selected proteins, Figure S5d for all proteins). We further compared the relative abundance changes of the different precursors (most abundant and 2nd most abundant) from the same protein across all COVID-19 plasma samples (Figure S5e). Although not all peptides derived from the same protein are expected to correlate with one another (i.e. due to differential post-translational modification) 37,54–56, we obtained a linear correlation for the majority of peptide quantities that were assigned to the same protein (Figure S5e).
Figure 4. 1-minute gradients and Scanning SWATH identify biomarkers that classify COVID-19 patients.
a. Plasma samples were taken from 30 hospitalized COVID-19 patients of different severity, as well as 15 healthy individuals. b. Plasma proteomes classify COVID-19 patients according to the severity. Centered and standardized quantities (z-scores) for 54 proteins that are significantly differentially expressed depending on COVID-19 severity are illustrated on a heatmap (Kendall's Tau test for the Theil-Sen trend estimator, adjusted p-value < 0.01, Benjamini-Hochberg for multiple testing 41). Clustering was performed row-wise but not column-wise. Labels indicate the corresponding gene names. c. Principal component analysis separates patients according to their severity. Proteins found significantly differentially expressed depending on severity were considered. d. The 1-minute Scanning SWATH method gives similar quantities as conventional SWATH with 5 times shorter gradients. Boxplots comparing 5-minute conventional SWATH 23 with 1-minute Scanning SWATH quantifying the COVID-19 severity biomarkers as a function of COVID-19 severity. Plots are labelled with gene names that encode the respective proteins: CFI (Complement factor I), GSN (Gelsolin) and ITIH4 (Inter-alpha-trypsin inhibitor heavy chain H4). The intensities were normalized to the mean values of each protein. n = 15 healthy patients, n = 5 mild patients, n = 4 severe patients, n= 8 critical patients. e. COVID-19 severity biomarkers, that have to our knowledge not been associated to COVID-19 severity by proteomics before. Plots are labelled with gene names that encode the respective proteins: A2M (Alpha-2-macroglobulin), C1QC (Complement C1q subcomponent subunit C), HPX (Hemopexin), IGHG2 (Immunoglobulin heavy constant gamma 2), IGKV4-1 (Immunoglobulin kappa variable 4-1), PON1 (Serum paraoxonase/arylesterase 1), PROS1 (Vitamin K-dependent protein S), SERPINA7 (Thyroxine-binding globulin), SERPINF2 (Alpha-2-antiplasmin), TMEM198 (Transmembrane protein 198), TTR (Transthyretin). Protein quantities (Log2 transformed) are plotted as a function of COVID-19 severity. The boxes in d. and e. show the first and third quartile, the median (middle) and the whiskers extend to the most extreme data point, which is no more than 1.5 times the interquartile range from the box. n = 15 healthy patients, n = 10 mild patients, n = 7 severe patients, n= 13 critical patients (Table S6).
In total, 54 of the quantified proteins were significantly up- or downregulated (adjusted p-value < 0.01) depending on the COVID-19 disease severity (Table S9). They characterize the host response to the infection, as illustrated by a heatmap grouping the patients/individuals according to their severity (WHO score, Figure 4b). Additionally, a principal component analysis allows a separation of the patients according to their severity in the first principal component, indicating that the 1-minute Scanning SWATH runs capture clinical classifiers for COVID-19 severity. Out of the 54 significantly changed proteins, 43 proteins have been previously related to COVID-19 severity 23,47,49. We have further identified 11 proteins that are changing significantly and, to our knowledge, have not been associated with COVID-19 severity before (Figure 4e). Several of these belong to the acute phase response and the complement cascade, that are involved in the antiviral host response. For example, we detected an upregulation of Alpha-2-macroglobulin (gene product of A2M) as well as Alpha-2-antiplasmin (gene product of SERPINF2) and downregulation of Vitamin K-dependent protein S (gene product of PROS1), highlighting the role of coagulation in COVID-19 infections. Alpha-2-macroglobulin and Alpha-2-antiplasmin are protease inhibitors and inactivate thrombin 57 and plasmin 58, respectively, whilst Vitamin K-dependent protein S is an anticoagulate plasma protein. Moreover, we detect an upregulation of Complement C1q subcomponent subunit C (gene product of C1QC) and downregulation of Immunoglobulin kappa variable 4-1 (gene product of IGKV4-1), both involved in the activation of the classical complement pathway and thus the innate immune response. We found a downregulation of Thyroxine-binding globulin (gene product of SERPINA7) and Transthyretin (gene product of TTR), both involved in binding and transport of thyroid hormones, which is consistent with previous studies that found associations of thyroid dysfunction and the severity of COVID-19 infection59,60. Further, we observed downregulation of Serum paraoxonase/arylesterase 1 (gene product of PON1), which is associated with the cholesterol-carrying high-density lipoprotein (HDL), a modulator of innate immune response and inflammation 61–64. This agrees with previous studies that related downregulation of Serum paraoxonase/arylesterase 1 with other inflammatory and infectious diseases 65,66. Equally, dysregulation of HDL related proteins (like apolipoproteins (APOs)) in severe COVID-19 patients have been previously reported 23,47. Further, Hemopexin (gene product of HPX), a heme-binding and transporting protein, is downregulated. This could be due to its role in iron homeostasis, which is known to play a role in viral infections 67–69. Proteome experiments that use LC gradients as fast as 60 seconds are hence able to capture known and novel information in major infectious disease, including COVID-19, while allowing the measurement of hundreds of thousands of samples.
Discussion
Bottom-up proteomics has become popular in part because it substantially increases the number of proteins that can be studied in parallel in biological samples7,8,71,72. More recently, the proteomic field has sought to increase throughput and data robustness. High-throughput proteomics benefits from recent developments in sample preparation, chromatography, data acquisition, and data analysis. Automation and 96-well plate-based sample processing allow the preparation of hundreds of samples per day and reduce batch effects that limit large-scale and longitudinal experiments 23,72–77. Fast, efficient, and robust chromatographic separations have been achieved by replacing traditional nano-flow LC78,79 with setups that use higher flow rates. This ranges from micro-flow LC systems (5-50 µl/min)24,38,80, to LC devices with preformed gradients 25,81. More recently, we have introduced proteome experiments that make use of high-flow liquid chromatography (800 µl/min). In 5-minute chromatographic gradients, these allowed up to 180 proteome injections/day on a single LC-MS instrument, while increasing robustness, cost-effectiveness and quantification precision in longitudinal proteome experiments 23. The development of algorithms to deconvolute complex spectra resulting from fast chromatographic measurements is ongoing, but several major steps have been recently achieved, and have increased proteomic depth as well as quantification precision in conjunction with the fast chromatographic methods 22,82–84.
Missing so far have been mass spectrometric acquisition modes that are specifically designed for the challenges of complex samples analyzed in a short period of time. Here we demonstrate that the precise acquisition of proteomes in short gradients is facilitated by Scanning SWATH. This method, which requires a fast scanning qTOF, but no proprietary reagents, adds an additional scanning dimension to the raw data, which increases depth and true positive precursor identification. Scanning SWATH further shortens mass spectrometric duty cycles and allows narrow precursor isolation windows. Scanning SWATH hence brings the requirement for ‘deep’ proteomes and high throughput closer together.
We have benchmarked the platform’s identification and quantification performance on a human cell line digest that is commercially available (Promega) and thus facilitates comparability across laboratories. Further, we show the application of this technology in yeast antifungal drug screens, and in COVID-19 patient classification based on plasma samples. In human cell lysates we achieve the quantification of 1,937 protein groups in conjunction with a chromatographic gradient as fast as 30 seconds and show that with 60 seconds to 5-minute gradients, at least 70% more precursors are quantified compared to previous methods. Despite this high-throughput, the quantification precision is comparable if not better to the most recent achievements in human and yeast samples, even if similar chromatographic setups, sample preparation, and instruments are used 17,23,38. Indeed, achieving a median CV of 6.4% in quantifying the protein groups in a cell lysate indicates that, despite the high-throughput, the combination of high-flow chromatography and Scanning SWATH is among the most precise proteomic methods currently available.
The ideal balance between throughput and proteomic depth in a proteomic experiment is determined by the scientific question asked. Scanning SWATH has benefits for both; it facilitates faster chromatographic gradients and better measurement precision in high-throughput applications, but it also improves proteomic depth with longer gradients. With a 30-minute gradient on our high-flow system, conventional stepped SWATH and Scanning SWATH identify 5,958 and 6,564 protein groups, respectively (Figure S6). Scanning SWATH is most advantageous over existing methods in conjunction with fast chromatographic gradients and in the analysis of complex samples, wherein the assignment of precursor masses to MS/MS traces has large benefits in improving true positive precursor identification. Scanning SWATH combined with 5-minute LC gradients measures protein group intensities across 4 orders of magnitude (Figure S7a) and the intensity values are in agreement with those obtained using ten times longer gradients (Figure S7b). This setup allows measuring several hundreds of proteomes per day on a single LC-MS instrument 23.
The optimal injection amount is less a consequence of the Scanning SWATH method, but depends on the applied chromatographic flow rate. The amount of sample injected for the 5-minute gradient methods was 5 µg to 10 µg, which is an accessible protein amount with conventional digestion protocols 17,23,85,86. For instance, the digestion of just 5 µL of blood plasma would allow ten injections on the high-flow LC system.
We also note that in contrast to conventional SWATH, wherein different precursor isotopologues might fall into different windows, Scanning SWATH can preserve the isotopic patterns of the fragments as there are no “quadrupole edges” in Scanning SWATH raw data. This might have future applications. For instance, one could exploit this feature to improve SILAC experiments, where it has been noted before that in conventional SWATH experiments problems might occur when one of the precursor distributions (e.g. light) is split between 2 windows but the other (e.g. heavy) is not 20.
Methods
Materials
Water (LC-MS Grade, Optima; 10509404), Acetonitrile (LC-MS Grade, Optima; 10001334), Methanol (LC-MS Grade, Optima, A456-212) and Formic acid (LC-MS Grade, Thermo Scientific Pierce; 13454279) were purchased from Fisher Chemicals. Human cell lysate (MS Compatible Human Protein Extract, Digest, V6951) and Trypsin (Sequence grade, V511X) were purchased from Promega. DL-Dithiothreitol (BioUltra, 43815), Iodoacetamide (BioUltra, I1149) Ammonium Bicarbonate (Eluent additive for LC-MS, 40867), Yeast Nitrogen Base Without Amino Acids (Y0626) and glass beads (acid washed, 425-600 µm, Sigma, G8772) were purchased from Sigma Aldrich. Urea (puriss. P.a., reag. Ph. Eur., 33247H) and Acetic Acid (Eluent additive for LC-MS, 49199) were purchased from Honeywell Research Chemicals. Rosuvastatin Calcium (S2169), Fluvastatin Sodium (S1909), Pyrimethamine (S2006), Pitavastatin Calcium (S1759), Pemetrexed Disodium Hydrate (S7785), Pravastatin Sodium (S3036), Clotrimazole (S1606), Miconazole Nitrate (S1956), Lovastatin (S2061), Ketoconazole (S1353), Atorvastatin (S2077), Methotrexate Disodium (S5097), Simvastatin (S1792), Uniconazole (S3660), Itraconazole (S2476) were purchased from Selleck chemicals and Pralatrexate (A4350) was purchased from APExBIO. Control samples for SARS-CoV-2 study were prepared from commercial Human Plasma (EDTA, Pooled Donor, Genetex GTX73265).
Clinical samples of COVID-19 patients
Sampling was performed as part of the Pa-COVID-19 study, a prospective observational cohort study assessing pathophysiology and clinical characteristics of patients with COVID-19 at Charité Universitätsmedizin Berlin 51. All patients with SARS-CoV-2 infection proven by positive PCR from respiratory specimens and willing to provide written informed consent are eligible for inclusion. Exclusion criteria are refusal to participate in the clinical study by patient or legal representative or clinical conditions that do not allow for blood sampling. The study assesses epidemiological and demographic parameters, medical history, clinical course, morbidity and quality of life during the hospital stay of COVID-19 patients. Moreover, serial high-quality bio-sampling consisting of various sample types with deep molecular, immunological and virological phenotyping is performed. Treatment and medical interventions follow the standard of care as recommended by current international and German guidelines for COVID-19. The severity of illness in the present study follows the WHO ordinal outcome scale 52. The Pa-COVID-19 study is carried out according to the Declaration of Helsinki and the principles of Good Clinical Practice (ICH 1996) where applicable and was approved by the ethics committee of Charité-Universitätsmedizin Berlin (EA2/066/20).
Sample preparation
The human cell lysate was obtained commercially (Promega) and was dissolved 0.1% formic acid. Plasma samples were prepared as previously described 23.
The yeast samples for the drug response measurements were prepared and digested as follows: The auxotrophic Saccharomyces cerevisiae strain BY4741 (∆his3, ∆leu2, ∆ura3, ∆met15) was rendered prototrophic via genomic knock-in of the missing genes. This prototrophic, wild-type strain was grown on agar plates containing synthetic minimal (SM) media for 3 days. Subsequently, colonies were inoculated in SM liquid media (25 ml) and incubated at 30°C for 1 day. The yeast culture was transferred to 96 deep well plates and drugs were added to achieve a working concentration of 10 μM (1 ml total volume/well). The yeast culture was incubated at M (1 ml total volume/well). The yeast culture was incubated at 30°C and was grown overnight to the exponential phase. Cells were pelleted by centrifugation at 3220 rcf for 5 minutes, the supernatant was discarded and plates were stored at −80°C until further processing.
200 μl 7M Urea/ 100 mM ammonium bicarbonate and glass beads (~100 mg/well) were added to the frozen pellet. Subsequently, the plates were sealed (Cap mats, Spex, 2201) and lysed in a bead beater for 5 minutes at 1500rpm (Spex Geno/Grinder). After 1 minute centrifugation at 4000 rpm, 20 μl 55mM DL-Dithiothreitol was added (final concentration 5 mM), mixed and the samples were incubated for 1h at 30°C. Subsequently, 20 μl 120 mM Iodoacetamide was added (final concentration 10 mM) and incubated 30 minutes in the dark at room temperature. 1 ml 100mM ammonium bicarbonate was added, centrifuged 3 minutes at 4000 rpm and 230 μl were transferred to prefilled trypsin plates (9 μl 0.1 μg/μl trysin). After incubation of the samples for 17 hours at 37°C, 24 μl 10% formic acid was added. The digestion mixtures were cleaned-up using C18 96-well plates (96-Well MACROSpin C18, 50-450 μL, The Nest Group, SNS SS18VL). For the solid phase extraction, 1-minute centrifugation steps at the described speeds (Eppendorf Centrifuge 5810R) were applied to push the liquids through the stationary phase. A liquid handler (Biomek NXP) was used to pipette the liquids onto the material in order to make four 96-well plates/batch feasible. The plates were conditioned with methanol (200 μl, centrifuged at 50g), washed twice with 50% ACN (200 μl, centrifuged at 50g and flow-through discarded), equilibrated three times with 3% ACN, 0.1% FA (200 μl, centrifuged at 50g/80g and 100 g respectively and flow-through discarded). Then 200 μl of digested samples were loaded (centrifuged at 100g), washed three times with 3% ACN, 0.1% FA (200 μl, centrifuged at 100g). After the last washing step, the plates were centrifuged another time at 180g before the peptides were eluted in 3 steps (twice with 120 μl and once with 130 μl) 50% ACN (180g) into a collection plate (1.1 ml, Square well, V-bottom). The collected material was completely dried on a vacuum concentrator (Eppendorf Concentrator Plus) and redissolved in 40 μl 3% ACN, 0.1% formic acid before transferred into a 96-well plate (700 μl round, waters, 186005837). QC samples for repeat injections were prepared by pooling 3 μl of each digested sample. All pipetting was done with a liquid handling robot (Biomek NXP automated liquid handler), shaking was done with a thermomixer (Eppendorf Thermomixer C) after each step and for incubation a Memmert IPP55 incubator was used.
Liquid chromatography-mass spectrometry
Liquid chromatography was performed on an Agilent Infinity II ultra-high-pressure system coupled to a Sciex TripleTOF 6600. The peptides were separated in reversed-phase mode using an InfinityLab Poroshell 120 EC-C18 at a column temperature of 30°C. The dimensions of the columns were 2.1 × 30 mm, 1.9 μm particles for the yeast drug screen and 2.1 × 50 mm, 1.9 μm particles for all other measurements. For the K562 benchmarks a gradient was applied which ramps from 3% B to 36% B in 5 min (Buffer A: 1% acetonitrile and 0.1% formic acid; Buffer B: acetonitrile and 0.1% formic acid) with a flow rate of 800 µL/min. For washing the column, the flow rate was increased to 1 ml/min and the organic solvent was increased to 80% B in 0.5 minutes and was kept for 0.2 minutes at this composition before going back to 3% B in 0.1 minutes. Subsequently the column was equilibrated for 2.1 minutes (Table S10). An IonDrive Turbo V Source was used with ion source gas 1 (nebulizer gas), ion source gas 2 (heater gas) and curtain gas set to 50, 40 and 25. The source temperature was set to 450 and the ion spray voltage to 5500V.
For comparing different gradient length (0.5, 1, 3 and 5 minutes) we applied linear gradients ramping from 3% B to 36% B (Buffer A: 1% acetonitrile and 0.1% formic acid; Buffer B: acetonitrile and 0.1% formic acid) with a flow rate of 800 µL/min. For the Scanning SWATH and conventional stepped SWATH the duty cycles were adjusted accordingly (Table S1 and S3). For conventional SWATH this was done by adjusting the number of variable windows to reach cycle times comparable to the Scanning SWATH duty cycles (Table S2 and S3). For this particular comparison, the accumulation times of the MS1 scan was 10 ms and of the MS/MS scans 25 ms.
The 1-minute gradients for the measurement of the COVID-19 patient samples were slightly adjusted. 3 µg of the digested proteins were injected and we applied linear ramping from 3% B to 15% B (Buffer A: 1% acetonitrile and 0.1% formic acid; Buffer B: acetonitrile and 0.1% formic acid) in 0.1 minutes followed by linear ramping from 15 % B to 40 % B in 0.9 minutes (Table S7 for detailed gradient parameters).
For the yeast drug screen we reduced the column length to 3 cm (InfinityLab Poroshell 120 EC-C18, 2.1 × 30 mm, 1.9 μm particles) and increased the flow rate during the column wash up to 2.3 ml/min, which reduced the method overhead times to 140 seconds (Table S5 for detailed gradient parameters).
Scanning SWATH settings and operation
The Scanning SWATH runs were acquired with a Scanning SWATH beta version. If not mentioned otherwise, the following settings were applied in the Scanning SWATH runs: The precursor isolation window was set to 10 m/z and a mass range from 400 m/z to 900 m/z was covered in 0.5 seconds. These settings provided a compromise between identification and quantification performance. We optimised the window size on yeast (S. cerevisiae) whole proteome tryptic digests and a 5-minute high-flow water-to-acetonitrile gradient where we tested window sizes ranging from 3 m/z to 20 m/z, covering a precursor range from 400 m/z to 900 m/z. The best results in terms of identifications and quantitative precision were achieved with window sizes of 10 m/z (Figure S2a). Reducing the window size further would result in even higher identification numbers due to less interference but the resulting shorter effective accumulation times would lower the quantitative precision. The raw data was binned in the quadrupole or precursor dimension into 2 m/z bins, providing a resolution in the Q1 dimension that allows the effective use of the Q1 scores. The MS1 scan was omitted for the benchmarks and the data was acquired in high sensitivity mode.
The instrument control software calculates an RF/DC ramp which is applied to quadrupole filter 1. The ramp is calculated from the experiment start transmission mass, stop transmission mass, transmission width, and cycle time. The calculation uses previously acquired calibrations to calculate ramps for mass DACS and resolution DACS. The quadrupole start mass is calculated as experiment start mass minus transmission width, and, the quadrupole stop mass is experiment stop mass plus transmission width. This allows for correct precursor profiles of all fragments at the boundaries of the experimental mass range. Collision energy is calculated using the +2 Rolling Collision energy equation based on the center masses for each transmission window. This results in a small collision energy spread depending on the width of the transmission window relative to the range being scanned. In these experiments the effect is typical around 1 eV spread for a given precursor.
The instrument acquisition software organizes ion detection responses into calculated 2 m/z precursor isolation bins given the current TOF pusher pulse number relative to the start of the scan applying the Scanning SWATH offset curve described above. The 2 m/z precursor isolation bins are organized in the data file as adjacent experiments allowing for the extraction of precursor profiles for any given fragment in a given cycle by tracing fragment response across experiments as well as normal chromatographic profiles across cycles.
The bins-to-sum (consolidation of data points in time-of-flight dimension) was set to 4 (4× 25ps = 100ps) for the K562 benchmark experiment and 8 (8× 25ps = 200ps) for all other experiments.
Scanning SWATH calibration was obtained while processing each sample file from the sample data itself. An automated algorithm finds the maximum residual precursors for each transmission window across the entire sample. This results in several accurate mass TOF measurements paired with the centroid of the quadrupole mass traces per quadrupole transmission region of which there are usually 10 or more per 100 da. For example if the algorithm used the best 3 residual precursors across the LC for a given transmission region and, the scan range was 500 da with a transmission width of 10 then there would be 500/10 * 3 = 150 calibration point pairs consisting of quadrupole mass and TOF accurate mass. Since it can occur that an intense peak within the quadrupole transmission region is not in fact a residual precursor a selection algorithm filters out points using an outlier rejection algorithm which considers local variance. Typically a point is evaluated relative to its neighbors in a 50-100 da region. Once a multi-point calibration curve is obtained the calibration is applied to the data by updating the begin and end mass region defined in the header from each experiment stored such that the center is calculated from the calibration function while maintaining continuity of boundaries in adjacent experiments.
Matching precursors to MS/MS fragment traces in DIA-NN
DIA-NN takes full advantage of the 4th dimension in Scanning SWATH data. In DIA-NN, a set of scores is calculated for each precursor-spectrum match (PSM), to distinguish true signals from noise using linear classifiers and an ensemble of deep neural networks. DIA-NN also selects the ‘best’ fragment ion per PSM, as the one with the clearest signal, with other fragment ions then being assessed by comparing their MS2 traces to those of the best fragment22. Scores specifically related to Q1 profile assessment have now been added to DIA-NN algorithms. The Q1 profiles are extracted at the apex of the respective elution peak and the following scores are calculated. (i) Scores that reflect how similar are the Q1 profiles of the fragments and the non-fragmented precursor to the Q1 profile of the best fragment. One score is calculated as the sum of correlations between Q1 profiles of the fragments and the Q1 profile of the ‘best’ fragment, as designated by DIA-NN during candidate elution peak identification 22. The other score is the correlation of the Q1 profile of the non-fragmented precursor and the Q1 profile of the ‘best’ fragment. (ii) Scores that reflect how well the Q1 profile shapes match the expected triangular shape. For each fragment, a score is calculated, with values between 0 and 1, reflecting whether its Q1 profile increases monotonically to the left from the apex. These scores are then multiplied by the correlation between elution profiles of the fragments and the ‘best’ fragment, and summed across all the fragments. A similar sum is calculated reflecting whether the Q1 profiles are monotonically decreasing to the right from the apex. (iii) The difference between the centroid of the Q1 profile of the ‘best’ fragment and the library precursor mass is calculated. DIA-NN calculates the scores listed in (i) - (iii) at 3 different scales by using 3, 7 or 11 bins closest to the Q1 profile apex, respectively, yielding 3 × (2 + 2 + 1) = 15 scores total (see Figure S1 for more details on the algorithm).
Only the monoisotopic fragment masses are used for the Q1 profile assessment as the Q1 profiles of different fragment isotopologues are shifted relative to each other. We have illustrated this for a doubly charged precursor (Figure S8). As one would expect, the Q1 profiles of the +1 13C and the +2 13C fragment isotopologues are shifted by ~0.5 m/z and ~1 m/z to the monoisotopic mass, respectively. Depending on the precursor mass and fragment mass, a small fraction of the monoisotopic fragments might also result from a +1 13C precursor isotope. This does slightly distort the Q1 profile of monoisotopic fragments but as this distortion/shift is in the range of the mass accuracy of the quadrupole, this shift/distortion and its impact is negligible in practice.
Conventional DIA and SWATH runs (for benchmark)
The conventional 5-minute “stepped” SWATH method is based on a previously published method23. To make it comparable to the developed Scanning SWATH method we applied the same 0.5-second duty cycle and the same precursor mass range from 400 m/z to 900 m/z as in the developed Scanning SWATH method. Each duty cycle consists of one MS1 scan with 20 ms accumulation time and 17 MS/MS scans with variable windows (Table S11) and with 25 ms accumulation time.
The DIA-FAIMS data acquired on an Evosep One LC system coupled to an Orbitrap Exploris 480 was downloaded from ProteomeXchange (dataset PXD016662). Triplicate runs with 500 ng HeLa tryptic digests loaded on column (highest load in this dataset), a compensation value of -45 V for FAIMS, a resolving power of 15,000 and a cycle time with 1 second were considered as these runs provided the best identification numbers while maintaining quantitative accuracy 25. The DIA-FAIMS data was analyzed with a project-specific library acquired on the same setup (PXD016662, “5min-library.kit”). For the analysis in DIA-NN, the library was exported from Spectronaut (v. 13.12.200217.43655 (Laika)) with the “Export Spectral Library” function and reannotated with the “Reannotate” function in DIA-NN using the UniProt87 human canonical proteome (3AUP000005640). The DIA-FAIMS data was analyzed with Spectronaut (v. 13.12.200217.43655 (Laika)) and DIA-NN but as the identification numbers were higher with DIA-NN, we used these values for the benchmark.
Data processing and data analysis
Raw data processing was carried out with DIA-NN version 1.7.12 and with default settings in “robust LC (high accuracy)” mode. Protein quantities were obtained using the MaxLFQ algorithm 88 as implemented in DIA-NN (yeast drug screen) or as implemented in the diann R package (https://github.com/vdemichev/diann-rpackage) (all other samples).
The data processing and batch correction for the COVID-19 patient measurements was done as described previously 23. Briefly, the report was filtered at 0.01 precursor-level q-value and 0.05 protein group-level q-value. Intra-batch correction was performed for each peptide precursor separately and based on the sample preparation controls, using linear regression on the injection number. Linear regression was applied only for at least 10 data points. Testing of the relation between log2-transformed protein levels and the WHO severity grade (as classified according to the WHO ordinal scale 52) was performed using Kendall's Tau test as implemented in the EnvStats R package 70 (adjusted p-value < 0.01, Benjamini-Hochberg for multiple testing 41). Proteins were only considered for the differential expression analysis when identified in at least 90% of the individuals/patients.
For the analysis of the yeast drug screen data, proteins were only considered if detected in more than 50% of the samples and samples were removed if they had less than 80% of the maximum identification number across samples. Only proteins identified with proteotypic (i.e. specific) peptides and 1% protein q-value were considered. The differential expression analysis (drug-treated vs DMSO-treated) for the yeast drug screen was done on the log2-transformed protein quantities using a t-test (two-sided), considering proteins that are detected in at least 3 out of the 4 replicates. Benjamini-Hochberg procedure 41 was used for multiple testing correction. Drugs were only considered for the subsequent analysis if they had > 20 differentially expressed proteins and samples treated with folic acid and FIN56 were excluded from the analysis as they do not belong to the three studied drug classes.
Coefficients of variations (CV) were calculated for each protein or precursor as its empirical standard deviation divided by its empirical mean and reported in percent. CV values were calculated for proteins or precursors that were identified in at least 2 replicate measurements. PCA analysis was always performed only on ubiquitously identified proteins: imputation was not used. Heatmaps were generated with the ComplexHeatmap R package and default settings 44. Pathway enrichment was performed with the clusterProfiler R package 42 and wikipathways database 43. Z-scores were calculated by dividing the (centered) protein quantities by their standard deviations. All plots were generated with R (Version 3.6.3)89.
Spectral Libraries
The libraries for the K562 benchmark experiments and for the yeast drug screen were generated from “gas-phase fractionation” runs using Scanning SWATH and small precursor isolation windows. 5 µg of K562 cell lysate (Promega) or 5 µg yeast digests were injected and run on a nanoAcquity UPLC (Waters) coupled to a SCIEX TripleTOF 6600 with a DuoSpray Turbo V source. The peptides were separated on a Waters HSS T3 column (150 mm x 300 µm, 1.8 µm particles) with a column temperature of 35 °C and a flow rate of 5 µL/min. A 55-minute linear gradient ramping from 3% ACN/0.1FA to 40% ACN/0.1% FA was applied. The ion source gas 1 (nebulizer gas), ion source gas 2 (heater gas), and curtain gas set to 15, 20 and 25 respectively. The source temperature was set to 75 and the ion spray voltage to 5500 V. In total 12 injections were run with the following mass ranges: m/z 400-450, m/z 445-500, m/z 495-550, m/z 545- 600, m/z 595-650, m/z 645-700, m/z 695-750, m/z 745-800, m/z 795-850, m/z 845-900, m/ z 895-1000 and m/z 995-1200. The precursor isolation window was set to m/z 1 except for the mass ranges m/z 895-1000 and m/z 995-1200, where the precursor windows were set to m/z 2 and m/z 3, respectively. The cycle time was 3 seconds consisting of high and low energy scan and data was acquired in “high resolution” mode. A spectral library was generated using library-free analysis with DIA-NN directly from these Scanning SWATH acquisitions. For this DIA-NN analysis, MS2 and MS1 mass accuracies were set to 25 ppm and 20 ppm, respectively, and scan window size set to 6.
For the analysis of COVID-19 plasma samples, a project-independent public spectral library 24 was used as described previously 23. Human UniProt 87 isoform sequence database (3AUP000005640) was used to annotate the library. The library was first automatically refined based on the dataset in question at 0.01 global q-value (using the “Generate spectral library” option in DIA-NN). DIA-NN performs such refinement by finding the highest-scoring identification for each library precursor, across all runs in the experiment, and then replacing the library data with the empirically observed spectrum and retention time.
Empirical FDR estimation with two-species library
As false-discovery rate (FDR) calculations are software- and acquisition mode-specific, thus potentially affecting benchmarking results, we also compared Scanning SWATH data to conventional stepped SWATH using the two-species library approach, which estimates true positive calls in an unbiased fashion on the basis of an empirically measured FDR 8,22. We augmented the human library with Arabidopsis thaliana precursors, obtained from ProteomeXchange (dataset PXD012710, Arabidopsis proteome spectral library, “Arabidopsis_Library_TripleTOF5600_Spectronaut.xls”) as negative controls. Peptides that matched to both, the UniProt87 human canonical proteome (3AUP000005640) as well as the UniProt Arabidopsis thaliana canonical proteome (3AUP000005648) were removed from the library. The spectra and retention times in the merged human/Arabidopsis thaliana library were replaced with in silico predicted values whenever possible using the deep learning-based prediction integrated in DIA-NN. The empirical FDR was estimated as previously described 22. In short, the empirical FDR is the ratio of Arabidopsis thaliana precursors identified and human precursors identified multiplied by the ratio of human precursors and Arabidopsis thaliana precursors in the library (only precursors ranging from 400 m/z to 900 m/z were considered).
Supplementary Material
Acknowledgments
We thank Michael Hummel, Denise Treue, Dana Briesemeister, Wolfgang M. Kuebler, Charlotte Thibeault, Moritz Pfeiffer, Stefan Hippenstiel, Andreas Hocke, Christof von Kalle, Martin Witzenrath and Christian Drosten (all Charité Universitätsmedizin Berlin) for their help in providing the clinical samples and data. We thank Lisa Kahl for proofreading the manuscript and Robert King, Robert Lane, Emma Hudson, Nick Morrice and Jean Baptiste Vincendet for their help with the TripleTOF 6600s. This work was supported by the Berlin University Alliance (501_Massenspektrometrie, 501_Linklab, 112_PreEP_Corona_Ralser), by UKRI/NIHR through the UK Coronavirus Immunology Consortium (UK-CIC), the Ministry of Education and Research (BMBF) (01KI20160A, 01ZX1604B, 01KI20337, 01KX2021), the Berlin Institute of Health (BIH) (BIH_PA_COVID19_Ralser), the BBSRC (BB/N015215/1, BB/N015282/1), a Crick Idea to Innovation (i2i) initiative (10658), the Crick LifeArc (1290305) and the Francis Crick Institute which receives its core funding from Cancer Research UK (FC001134), the UK Medical Research Council (FC001134), and the Wellcome Trust (FC001134). The work was further supported by the Ministry of Education and Research (BMBF), as part of the National Research Node ‘Mass spectrometry in Systems Medicine (MSCoresys), under grant agreement 031L0220A. Marco Kreidl was supported by the University of Innsbruck and the Austrian Drug Screening Institute. Leif Erik Sander is supported by the German Research Foundation (DFG, SFB-TR84 114933180) and by the Berlin Institute of Health (BIH), which receives funding from the Ministry of Education and Research (BMBF).
Footnotes
Author contributions
M.R., C.B.M. and V.D. designed and supervised the study; S.T, N.B., M.R., C.B.M. and V.D. conceived the acquisition mode; N.B., S.T., F.W and G.I. implemented the acquisition mode; C.B.M., A.E., M.K, A.F. and J.S.L.Y. carried out the experiments; V.D. developed and implemented the processing algorithms; C.B.M. and V.D designed the experiments, analysed and visualised the data; L.E.S., F. K., L.J. and N.S. conducted clinical assessment and supervised clinical sample handling; K.S.L., M.M., A.Z. helped supervise the project and contributed to the interpretation of the results; M.R., C.B.M., V.D. wrote the paper with contributions from all authors.
Reporting Summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Competing interest
N.B, G.I., F.W and S.T. work for SCIEX. All other authors have no competing interests.
Data availability
The generated data for the benchmarks and the drug response screen in yeast have been deposited to the ProteomeXchange Consortium via the PRIDE 90 partner repository with the dataset identifier PXD023613; previously published data were also used for the benchmarks (PXD016662) and 2-species library generation (PXD012710).
Code availability
The algorithms are included in the open-source DIA-NN software suite (https://github.com/vdemichev/diann).
References
- 1.Aebersold R, Mann M. Mass-spectrometric exploration of proteome structure and function. Nature. 2016;537:347–355. doi: 10.1038/nature19949. [DOI] [PubMed] [Google Scholar]
- 2.Cox J, Mann M. Quantitative, high-resolution proteomics for data-driven systems biology. Annu Rev Biochem. 2011;80:273–299. doi: 10.1146/annurev-biochem-061308-093216. [DOI] [PubMed] [Google Scholar]
- 3.Duarte T, Spencer C. Personalized Proteomics: The Future of Precision Medicine. Proteomes. 2016;4:29. doi: 10.3390/proteomes4040029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Santos R, et al. A comprehensive map of molecular drug targets. Nat Rev Drug Discov. 2017;16:19–34. doi: 10.1038/nrd.2016.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nilsson T, et al. Mass spectrometry in high-throughput proteomics: ready for the big time. Nat Methods. 2010;7:681–685. doi: 10.1038/nmeth0910-681. [DOI] [PubMed] [Google Scholar]
- 6.Hebert AS, et al. Comprehensive Single-Shot Proteomics with FAIMS on a Hybrid Orbitrap Mass Spectrometer. Anal Chem. 2018;90:9529–9537. doi: 10.1021/acs.analchem.8b02233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hebert AS, et al. The one hour yeast proteome. Mol Cell Proteomics. 2014;13:339–347. doi: 10.1074/mcp.M113.034769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Muntel J, et al. Surpassing 10000 identified and quantified proteins in a single run by optimizing current LC-MS instrumentation and data analysis strategy. Molecular Omics. 2019;15:348–360. doi: 10.1039/c9mo00082h. [DOI] [PubMed] [Google Scholar]
- 9.Meier F, et al. diaPASEF: parallel accumulation-serial fragmentation combined with data-independent acquisition. Nat Methods. 2020;17:1229–1236. doi: 10.1038/s41592-020-00998-0. [DOI] [PubMed] [Google Scholar]
- 10.Meier F, Geyer PE, Virreira Winter S, Cox J, Mann M. BoxCar acquisition method enables single-shot proteomics at a depth of 10,000 proteins in 100 minutes. Nat Methods. 2018;15:440–448. doi: 10.1038/s41592-018-0003-5. [DOI] [PubMed] [Google Scholar]
- 11.Bruderer R, et al. Optimization of Experimental Parameters in Data-Independent Mass Spectrometry Significantly Increases Depth and Reproducibility of Results. Mol Cell Proteomics. 2017;16:2296–2309. doi: 10.1074/mcp.RA117.000314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shishkova E, Hebert AS, Westphall MS, Coon JJ. Ultra-High Pressure (>30,000 psi) Packing of Capillary Columns Enhancing Depth of Shotgun Proteomic Analyses. Anal Chem. 2018;90:11503–11508. doi: 10.1021/acs.analchem.8b02766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen R, Snyder M. Promise of personalized omics to precision medicine. Wiley Interdiscip Rev Syst Biol Med. 2013;5:73–82. doi: 10.1002/wsbm.1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ebhardt HA, Root A, Sander C, Aebersold R. Applications of targeted proteomics in systems biology and translational medicine. Proteomics. 2015;15:3193–3208. doi: 10.1002/pmic.201500004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Costello Z, Martin HG. A machine learning approach to predict metabolic pathway dynamics from time-series multiomics data. npj Systems Biology and Applications. 2018;4 doi: 10.1038/s41540-018-0054-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ahadi S, et al. Personal aging markers and ageotypes revealed by deep longitudinal profiling. Nat Med. 2020;26:83–90. doi: 10.1038/s41591-019-0719-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zelezniak A, et al. Machine Learning Predicts the Yeast Metabolome from the Quantitative Proteome of Kinase Knockouts. Cell Syst. 2018;7:269–283.e6. doi: 10.1016/j.cels.2018.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Juvvadi PR, et al. Scanning Quadrupole Data-Independent Acquisition, Part B: Application to the Analysis of the Calcineurin-Interacting Proteins during Treatment of Aspergillus fumigatus with Azole and Echinocandin Antifungal Drugs. Journal of Proteome Research. 2018;17:780–793. doi: 10.1021/acs.jproteome.7b00499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moseley MA, et al. Scanning Quadrupole Data-Independent Acquisition, Part A: Qualitative and Quantitative Characterization. Journal of Proteome Research. 2018;17:770–779. doi: 10.1021/acs.jproteome.7b00464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ludwig C, et al. Data-independent acquisition-based SWATH-MS for quantitative proteomics: a tutorial. Mol Syst Biol. 2018;14:e8126. doi: 10.15252/msb.20178126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gillet LC, et al. Targeted data extraction of the MS/MS spectra generated by data independent acquisition: a new concept for consistent and accurate proteome analysis. Mol Cell Proteomics. 2012;11:0111.016717. doi: 10.1074/mcp.O111.016717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Demichev V, Messner CB, Vernardis SI, Lilley KS, Ralser M. DIA-NN: neural networks and interference correction enable deep proteome coverage in high throughput. Nat Methods. 2020;17:41–44. doi: 10.1038/s41592-019-0638-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Messner CB, et al. Ultra-high-throughput clinical proteomics reveals classifiers of COVID-19 infection. Cell Systems. 2020 doi: 10.1016/j.cels.2020.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bruderer R, et al. Analysis of 1508 Plasma Samples by Capillary-Flow Data-Independent Acquisition Profiles Proteomics of Weight Loss and Maintenance. Mol Cell Proteomics. 2019;18:1242–1254. doi: 10.1074/mcp.RA118.001288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bekker-Jensen DB, et al. A Compact Quadrupole-Orbitrap Mass Spectrometer with FAIMS Interface Improves Proteome Coverage in Short LC Gradients. Mol Cell Proteomics. 2020;19:716–729. doi: 10.1074/mcp.TIR119.001906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hinshaw JV. How do your peaks measure up? LC GC NORTH AMERICA. 2013;31:860–+. [Google Scholar]
- 27.Doellinger J, Blumenscheit C, Schneider A, Lasch P. Isolation Window Optimization of Data- Independent Acquisition Using Predicted Libraries for Deep and Accurate Proteome Profiling. Anal Chem. 2020;92:12185–12192. doi: 10.1021/acs.analchem.0c00994. [DOI] [PubMed] [Google Scholar]
- 28.Bache N, et al. A Novel LC System Embeds Analytes in Pre-formed Gradients for Rapid, Ultra-robust Proteomics. Mol Cell Proteomics. 2018;17:2284–2296. doi: 10.1074/mcp.TIR118.000853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pais P, et al. Membrane Proteome-Wide Response to the Antifungal Drug Clotrimazole in Candida glabrata: Role of the Transcription Factor CgPdr1 and the Drug:H Antiporters CgTpo1_1 and CgTpo1_2. Molecular & Cellular Proteomics. 2016;15:57–72. doi: 10.1074/mcp.M114.045344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ball B, Bermas A, Carruthers-Lay D, Geddes-McAlister J. Mass Spectrometry-Based Proteomics of Fungal Pathogenesis, Host-Fungal Interactions, and Antifungal Development. Journal of Fungi. 2019;5:52. doi: 10.3390/jof5020052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Berman J, Krysan DJ. Drug resistance and tolerance in fungi. Nat Rev Microbiol. 2020;18:319–331. doi: 10.1038/s41579-019-0322-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Krysan DJ. The unmet clinical need of novel antifungal drugs. Virulence. 2017;8:135–137. doi: 10.1080/21505594.2016.1276692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Whaley SG, Rogers PD. Azole Resistance in Candida glabrata. Curr Infect Dis Rep. 2016;18:41. doi: 10.1007/s11908-016-0554-5. [DOI] [PubMed] [Google Scholar]
- 34.Stylianou M, Kulesskiy E, Lopes JP. Antifungal application of nonantifungal drugs. Antimicrob Agents Chemother. 2014 doi: 10.1128/AAC.01087-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gonen N, Assaraf YG. Antifolates in cancer therapy: Structure, activity and mechanisms of drug resistance. Drug Resist Updat. 2012;15:183–210. doi: 10.1016/j.drup.2012.07.002. [DOI] [PubMed] [Google Scholar]
- 36.Selevsek N, et al. Reproducible and consistent quantification of the Saccharomyces cerevisiae proteome by SWATH-mass spectrometry. Mol Cell Proteomics. 2015;14:739–749. doi: 10.1074/mcp.M113.035550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Alam MT, et al. The metabolic background is a global player in Saccharomyces gene expression epistasis. Nat Microbiol. 2016;1:15030. doi: 10.1038/nmicrobiol.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vowinckel J, et al. Cost-effective generation of precise label-free quantitative proteomes in high-throughput by microLC and data-independent acquisition. Sci Rep. 2018;8:4346. doi: 10.1038/s41598-018-22610-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mazu KT, Bricker AB, Flores-Rozas HY, Ablordeppey S. The Mechanistic Targets of Antifungal Agents: An Overview. Mini Rev Med Chem. 2016;16:555–578. doi: 10.2174/1389557516666160118112103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhao R, Goldman ID. Resistance to antifolates. Oncogene. 2003;22:7431–7457. doi: 10.1038/sj.onc.1206946. [DOI] [PubMed] [Google Scholar]
- 41.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc. 1995 [Google Scholar]
- 42.Yu G, Wang LG, Han Y, He QY. cluster Profiler: an R Package for Comparing Biological Themes Among Gene Clusters. OMICS. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Slenter DN, et al. Wiki Pathways: a multifaceted pathway database bridging metabolomics to other omics research. Nucleic Acids Res. 2018;46:D661–D667. doi: 10.1093/nar/gkx1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gu Z, Eils R, Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics. 2016;32:2847–2849. doi: 10.1093/bioinformatics/btw313. [DOI] [PubMed] [Google Scholar]
- 45.Geyer PE, Holdt LM, Teupser D, Mann M. Revisiting biomarker discovery by plasma proteomics. Mol Syst Biol. 2017;13:942. doi: 10.15252/msb.20156297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wright I, Van Eyk JE. A Roadmap to Successful Clinical Proteomics. Clin Chem. 2017;63:245–247. doi: 10.1373/clinchem.2016.254664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shen B, et al. Proteomic and Metabolomic Characterization of COVID-19 Patient Sera. Cell. 2020;182:59–72.e15. doi: 10.1016/j.cell.2020.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Whetton AD, Preston GW, Abubeker S, Geifman N. Proteomics and Informatics for Understanding Phases and Identifying Biomarkers in COVID-19 Disease. J Proteome Res. 2020 doi: 10.1021/acs.jproteome.0c00326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Overmyer KA, et al. Large-scale Multi-omic Analysis of COVID-19 Severity. medRxiv. 2020 doi: 10.1101/2020.07.17.20156513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mahmud I, Garrett TJ. Mass Spectrometry Techniques in Emerging Pathogens Studies: COVID-19 Perspectives. J Am Soc Mass Spectrom. 2020 doi: 10.1021/jasms.0c00238. [DOI] [PubMed] [Google Scholar]
- 51.Kurth F, et al. Studying the pathophysiology of coronavirus disease 2019: a protocol for the Berlin prospective COVID-19 patient cohort (Pa-COVID-19) Infection. 2020;48:619–626. doi: 10.1007/s15010-020-01464-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.World Health Organisation. WHO R&D Blueprint novel Coronavirus COVID-19 Therapeutic Trial Synopsis. who.int. 2020 https://www.who.int/blueprint/priority-diseases/key-action/COVID-19_Treatment_Trial_Design_Master_Protocol_synopsis_Final_18022020.pdf.
- 53.UVic Genome BC Proteomics Centre. Assays for FDA approved protein biomarkers. 2020 http://mrmassaydb.proteincentre.com/fdaassay/
- 54.Zhang B, Pirmoradian M, Zubarev R, Käll L. Covariation of Peptide Abundances Accurately Reflects Protein Concentration Differences. Mol Cell Proteomics. 2017;16:936–948. doi: 10.1074/mcp.O117.067728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schwarz E, Levin Y, Wang L, Leweke FM, Bahn S. Peptide correlation: a means to identify high quality quantitative information in large-scale proteomic studies. J Sep Sci. 2007;30:2190–2197. doi: 10.1002/jssc.200700190. [DOI] [PubMed] [Google Scholar]
- 56.Dermit M, Meyer JG. Peptide Correlation Analysis (PeCor A) Reveals Differential Proteoform Regulation. J Proteome Res. 2020 doi: 10.1021/acs.jproteome.0c00602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fischer AM, Tapon-Bretaudiere J, Bros A, Josso F. Respective roles of antithrombin III and alpha 2 macroglobulin in thrombin inactivation. Thromb Haemost. 1981;45:51–54. [PubMed] [Google Scholar]
- 58.Wu G, et al. Structural studies of plasmin inhibition. Biochem Soc Trans. 2019;47:541–557. doi: 10.1042/BST20180211. [DOI] [PubMed] [Google Scholar]
- 59.Gorini F, Bianchi F, Iervasi G. COVID-19 and Thyroid: Progress and Prospects. Int J Environ Res Public Health. 2020;17 doi: 10.3390/ijerph17186630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen M, Zhou W, Xu W. Thyroid Function Analysis in 50 Patients with COVID-19: A Retrospective Study. Thyroid. 2020 doi: 10.1089/thy.2020.0363. [DOI] [PubMed] [Google Scholar]
- 61.Gordon SM, Hofmann S, Askew DS, Davidson WS. High density lipoprotein: it’s not just about lipid transport anymore. Trends Endocrinol Metab. 2011;22:9–15. doi: 10.1016/j.tem.2010.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.White R, Giordano S, Datta G. Role of HDL-Associated Proteins and Lipids in the Regulation of Inflammation. In: Isbir T, editor. Advances in Lipoprotein Research. intechopen; 2017. p. 53. [Google Scholar]
- 63.Fotakis P, et al. Anti-Inflammatory Effects of HDL (High-Density Lipoprotein) in Macrophages Predominate Over Proinflammatory Effects in Atherosclerotic Plaques. Arterioscler Thromb Vasc Biol. 2019;39:e253–e272. doi: 10.1161/ATVBAHA.119.313253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Macpherson ME, et al. Impaired HDL Function Amplifies Systemic Inflammation in Common Variable Immunodeficiency. Sci Rep. 2019;9:9427. doi: 10.1038/s41598-019-45861-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Farid AS, Horii Y. Modulation of paraoxonases during infectious diseases and its potential impact on atherosclerosis. Lipids Health Dis. 2012;11:92. doi: 10.1186/1476-511X-11-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bacchetti T, et al. Oxidative stress and psoriasis: the effect of antitumour necrosis factor-α inhibitor treatment. Br J Dermatol. 2013;168:984–989. doi: 10.1111/bjd.12144. [DOI] [PubMed] [Google Scholar]
- 67.Drakesmith H, Prentice A. Viral infection and iron metabolism. Nat Rev Microbiol. 2008;6:541–552. doi: 10.1038/nrmicro1930. [DOI] [PubMed] [Google Scholar]
- 68.Roldan EQ, Biasiotto G, Magro P, Zanella I. The possible mechanisms of action of 4- aminoquinolines (chloroquine/hydroxychloroquine) against Sars-Cov-2 infection (COVID-19): A role for iron homeostasis? Pharmacological Research. 2020;158:104904. doi: 10.1016/j.phrs.2020.104904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Weinberg ED. Iron and infection. Microbiol Rev. 1978;42:45–66. doi: 10.1128/mr.42.1.45-66.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Millard SP. Env Stats, an RPackage for Environmental Statistics. Wiley Stats Ref: Statistics Reference Online. 2014 doi: 10.1002/9781118445112.stat07181. [DOI] [Google Scholar]
- 71.Wilhelm M, et al. Mass-spectrometry-based draft of the human proteome. Nature. 2014;509:582–587. doi: 10.1038/nature13319. [DOI] [PubMed] [Google Scholar]
- 72.Müller JB, et al. The proteome landscape of the kingdoms of life. Nature. 2020 doi: 10.1038/s41586-020-2402-x. [DOI] [PubMed] [Google Scholar]
- 73.Leutert M, Rodríguez-Mias RA, Fukuda NK, Villén J. R2-P2 rapid-robotic phosphoproteomics enables multidimensional cell signaling studies. Mol Syst Biol. 2019;15 doi: 10.15252/msb.20199021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Müller T, et al. Automated sample preparation with SP3 for low-input clinical proteomics. Mol Syst Biol. 2020;16 doi: 10.15252/msb.20199111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Macron C, Nunez Galindo A, Cominetti O, Dayon L. A Versatile Workflow for Cerebrospinal Fluid Proteomic Analysis with Mass Spectrometry: A Matter of Choice between Deep Coverage and Sample Throughput. Methods Mol Biol. 2019;2044:129–154. doi: 10.1007/978-1-4939-9706-0_9. [DOI] [PubMed] [Google Scholar]
- 76.Bennike TB, et al. A Cost-Effective High-Throughput Plasma and Serum Proteomics Workflow Enables Mapping of the Molecular Impact of Total Pancreatectomy with Islet Autotransplantation. J Proteome Res. 2018;17:1983–1992. doi: 10.1021/acs.jproteome.8b00111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fu Q, et al. Highly Reproducible Automated Proteomics Sample Preparation Workflow for Quantitative Mass Spectrometry. J Proteome Res. 2018;17:420–428. doi: 10.1021/acs.jproteome.7b00623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wilson SR, Vehus T, Berg HS, Lundanes E. Nano-LC in proteomics: recent advances and approaches. Bioanalysis. 2015;7:1799–1815. doi: 10.4155/bio.15.92. [DOI] [PubMed] [Google Scholar]
- 79.Gama MR, Collins CH, Bottoli CBG. Nano-liquid chromatography in pharmaceutical and biomedical research. J Chromatogr Sci. 2013;51:694–703. doi: 10.1093/chromsci/bmt023. [DOI] [PubMed] [Google Scholar]
- 80.Bian Y, et al. Robust, reproducible and quantitative analysis of thousands of proteomes by microflow LC-MS/MS. Nat Commun. 2020;11:157. doi: 10.1038/s41467-019-13973-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Müller JB, Meier F, Olsen JV, Vorm O, Mann M. A novel LC system embeds analytes in pre-formed gradients for rapid, ultra-robust proteomics. Molecular & Cellular. 2018 doi: 10.1074/mcp.TIR118.000853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ting YS, et al. PECAN: library-free peptide detection for data-independent acquisition tandem mass spectrometry data. Nat Methods. 2017;14:903–908. doi: 10.1038/nmeth.4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Peckner R, et al. Specter: linear deconvolution for targeted analysis of data-independent acquisition mass spectrometry proteomics. Nat Methods. 2018;15:371–378. doi: 10.1038/nmeth.4643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Heaven MR, et al. micro DIA (μDIA): data-independent acquisition for high-throughput proteomics and sensitive peptide mass spectrum identification. Anal Chem. 2018;90:8905–8911. doi: 10.1021/acs.analchem.8b01026. [DOI] [PubMed] [Google Scholar]
- 85.Vowinckel J, et al. The beauty of being (label)-free: sample preparation methods for SWATH-MS and next-generation targeted proteomics. F1000Res. 2013;2:1–26. doi: 10.12688/f1000research.2-272.v2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wisniewski JR, Zougman A, Nagaraj N, Mann M. Universal sample preparation method for proteome analysis. Nat Methods. 2009;6:359–362. doi: 10.1038/nmeth.1322. [DOI] [PubMed] [Google Scholar]
- 87.Uni Prot Consortium. Uni Prot: a worldwide hub of protein knowledge. Nucleic Acids Res. 2019;47:D506–D515. doi: 10.1093/nar/gky1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Cox J, et al. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed Max LFQ. Mol Cell Proteomics. 2014;13:2513–2526. doi: 10.1074/mcp.M113.031591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.R Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing; Vienna, Austria: 2004. [Google Scholar]
- 90.Perez-Riverol Y, et al. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 2019;47:D442–D450. doi: 10.1093/nar/gky1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The generated data for the benchmarks and the drug response screen in yeast have been deposited to the ProteomeXchange Consortium via the PRIDE 90 partner repository with the dataset identifier PXD023613; previously published data were also used for the benchmarks (PXD016662) and 2-species library generation (PXD012710).
The algorithms are included in the open-source DIA-NN software suite (https://github.com/vdemichev/diann).