Abstract
Successful treatment of tuberculosis (TB) depends on the eradication of its causative agent Mycobacterium tuberculosis (Mtb) in the host. However, the emergence of phenotypically drug-resistant Mtb in the host environment tempers the ability of antibiotics to cure disease. Host immunity produces diverse microenvironmental niches that are exploited by Mtb to mobilize adaptation programs. Such differential interactions amplify pre-existing heterogeneity in the host–pathogen milieu to influence disease pathology and therapy outcome. Therefore, comprehending the intricacies of phenotypic heterogeneity can be an empirical step forward in potentiating drug action. With this goal, we review the interconnectedness of the lesional, cellular, and bacterial heterogeneity underlying phenotypic drug resistance. Based on this information, we anticipate the development of new therapeutic strategies targeting host–pathogen heterogeneity to cure TB.
Antimicrobial Resistance (AMR) in Diverse Infection Landscapes
TB remains the number one infectious killer globally. TB treatment is unusually long (minimum of 6-9 months) for an infectious disease and requires the use of a cocktail of antibiotics. A critical impediment to antimicrobial treatment is that the causative agent, Mtb, becomes phenotypically resistant once it is inside its human host.
Genetic AMR emerges when bacteria mutate to gain the heritable ability to multiply in the presence of antibiotics. By comparison, phenotypic AMR is a more fledgling concept, although it was reported as early as World War II when the wonder drug penicillin failed against persistent bacterial infections [1,2]. It is defined as a non-heritable, physiological adaptation that allows bacteria to survive under extreme antibiotic exposure and outgrow once growth-permissive conditions are reinstated. Recent guidelines clarify differences between genetic AMR and two major phenotypic AMR forms known as ‘persistence’ and ‘tolerance’ [3,4]. Of these, the former is an attribute of a smaller fraction of non-replicating bacteria while the latter is a feature of the majority of actively dividing bacteria in an isogenic population [3,4]. ‘Occult’ Mtb fractions that tolerate antibiotic exposure and promote infection relapse have been reported in rabbit cavity caseum and sputum of TB patients [5,6]. Multiple stochastic and deterministic triggers induce a spectrum of responses from a clonal Mtb population during infection, indicating that both host and pathogen drive phenotypic AMR and heterogeneous outcomes of disease [7,8]. Uncovering the mechanisms underlying the generation of phenotypic variability can facilitate the development of novel antibiotics or repurposing of existing molecules to reduce therapy time. This review attempts to delineate phenotypic variations in granulomatous lesions, host immune cells, and Mtb that dictate the emergence of phenotypic AMR.
Lesional Heterogeneity
A History of ‘Vanished’ Mtb
In the late 1950s, Walsh McDermott and his colleagues extensively studied the outcome of prolonged drug treatment in mice and reported that antibiotics could kill actively growing Mtb and render the remaining bacilli ‘sterile’ [9,10]. The sterile fraction lacks the capacity to divide and remains undetectable in tissues. However, a natural relapse of infection was observed in one-third of the animals after cessation of antibiotic therapy, which could be accelerated by immune suppression. The resurgence of ‘vanished’ Mtb was not due to genetic mutations. Moreover, spleens showed higher bacterial load than lungs, indicating a role for the local immune environment in infection relapse. The isoniazid (Inh)–pyrazinamide (Pza) combination was more efficacious than Inh-streptomycin, suggesting the importance of a specific mix of antibiotics in eradicating Mtb. Early Inh activity targets replicating Mtb and the residual non-replicating bacteria are targeted by Pza to achieve near eradication [11]. Combining Inh and Pza with rifampicin (Rif), which targets house-keeping transcriptional functions in replicating and non-replicating Mtb, enhances the sterilizing activity of the drug regimen [12]. Differential activity of drugs against phenotypic Mtb variants is a likely explanation for the underlying antagonism between ethambutol (Emb) and Rif when they are administered together [11]. In support of this, the early bactericidal activity of antibiotics to decrease the Mtb load per milliliter of human sputum during the first 2 days of treatment is reported to be variable [13]. However, the sterilizing activities of antibiotics to clear persisting Mtb over a longer period of exposure are often similar [13]. These studies underscore the importance of studying the mechanisms underlying phenotypic diversity in the Mtb population during infection. In this context, bacilli present in TB patients’ sputum provide exciting and tractable opportunities to investigate the basis of phenotypic heterogeneity and disease transmission.
Out and About: Occult Mtb in Sputum
Active TB disease manifests in only a small fraction of the entire diseased population. The mechanisms of disease transmission via aerosols, however, remain poorly understood. Recently, a cell-wall-associated polyketide (sulfolipid-1) in virulent Mtb strains such as H37Rv and Erdman, has been reported to serve as a nociceptive stimulant for the host’s cough reflex and aid in bacterial transmission through aerosols [14]. Interestingly, H37Rv and Erdman are more resistant to oxidative stress and hypoxia than ancient lineages in animal models of infection [15,16]. However, the transmissibility of strains can vary even within lineages depending on the initial host–pathogen interaction, as demonstrated in a household contact study of TB patients [17]. Mtb sampled through bronchoalveolar lavage and sputum from nasopharyngeal airways is, therefore, essential to understanding of the role of phenotypic diversity and AMR during transmission [18,19].
Sputum bacilli were classically believed to be actively replicating, by virtue of their presence in aerated cavities [20]. However, transcriptome analysis of Mtb in sputum shows similarity with stationary-phase axenic cultures, indicating that growth-variable bacterial fractions might be expectorated by patients [6]. Analysis of sputum from active TB or HIV-TB co-infected patients indicates that not all persistent Mtb can be cultured effectively, indicating the presence of differentially culturable tubercle bacteria (DCTB) [21]. Resuscitation-promoting factors (RPFs), a class of cell wall hydrolyzing enzymes secreted by Mtb, have been reported to aid the revival of non-replicating bacteria and can be harvested from axenic culture filtrate to stimulate the growth of DCTB [22]. Interestingly, sputum from a single patient can harbor non-DCTB cells and culture filtrate-independent, RPF-dependent, and RPF-independent DCTB, indicating extensive heterogeneity in sputum bacilli. DCTB loads vary greatly in absolute numbers among patients; however, HIV-TB co-infected patients with low CD4+ T cell counts yield a lower load in general, indicating a putative link between distinct immune pressures and DCTB generation [21].
Drug tolerance is a common feature of Mtb retrieved from the cavity caseum of infected rabbits and DCTB. However, while the former is unresponsive to most antibiotics except rifamycins, RPF-dependent DCTB show high phenotypic resistance to Rif, Inh, and streptomycin [5,22,23]. Phenotypic AMR in DCTB can be recapitulated in vitro by prolonged sequential exposure to starvation (>2 weeks) and high concentrations (>10 μM) of Rif [24]. This seems intuitive, as Rif penetrates the caseous core of lesions and could alter the Mtb transcriptome to induce persistence [25]. Therefore, it is important to examine phenotypic AMR in the context of TB lesions/granulomas, which are highly dynamic and shaped by both immune signaling and the pathogen.
In Granulomas, the Host Immune System Trusts
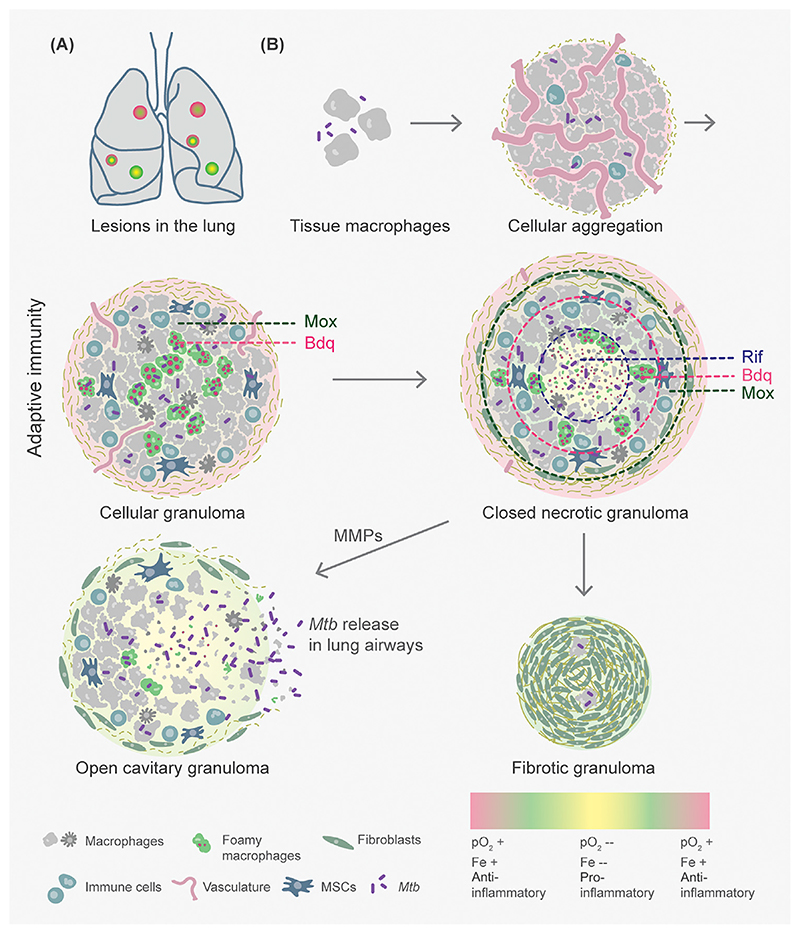
The systemic host immune response to Mtb infection is manifested by the formation of immune cell aggregates or granulomas in infected tissues such as the lung (Figure 1). Immense variability in granuloma make-up has been documented across active, latent, and reactivated disease groups, in humans and cynomolgus macaques [14]. [18F] fluorodeoxyglucose positron emission tomography coupled with CT reveals the coexistence of a range of TB lesions, often within the same lung lobe of a single subject, with no significant correlation with disease presentation [25,26]. Analysis of proteins expressed across multiple granuloma types – cellular, caseous, and open cavitary – through quantitative mass spectrometry indicates distinct temporal variations that occur during lesional progression [26]. The influx and egress of phagocytes continue to occur at a basal level even from latent granulomas, explaining the potential for reactivation of infection in certain subjects [27]. Laser-guided microdissection of tissue in Mtb-infected rabbits and resected human lungs coupled with mass spectrometry, immunohistochemistry, and imaging demonstrates that a third level of functional heterogeneity exists within a granuloma in the form of spatial delineation of inflammatory signals [26]. The core mounts a more proinflammatory response, including the expression of interferon-gamma (IFNγ), tumor necrosis factor-alpha (TNFα), and their downstream bactericidal effectors, such as reactive oxygen species (ROS), cathelicidin, and eicosanoids. More necrosis occurs here than the adjoining areas where tissue damage is limited by a heightened anti-inflammatory response, such as cyclooxygenase-mediated prostanoid biosynthesis [26]. In TB lesions of macaques, this signature closely relates to the cellular arrangement, such that proinflammatory enzymes [nitric oxide (NO) synthases] have higher expression in epithelioid macrophages and neutrophils of the necrotic core compared with the peripheral ring and non-infected tissue, in that order [28]. Transcriptomics of Mtb sampled from peripheral cellular aggregates and caseating necrotic zones of murine granulomas suggest that fatty acids, iron limitation, hypoxia, and NO prevail in the necrotic core [29] (Figure 1). Taking these findings together, a high degree of granuloma variability indicates that local immune responses are likely to shape the pathogen’s physiology, drug permeability, and disease control in individual lesions.
Figure 1. Lesional Heterogeneity Modulates Mycobacterium tuberculosis (Mtb) Survival, Tissue Homeostasis, and Drug Penetration.
(A) Diverse granuloma types – cellular (red), necrotic (yellow), and cavitary (green) – coexist in the lungs of an infected host. (B) As few as two to five Mtb bacilli in aerosol droplets are adequate to infect a susceptible host. Aggregation of differentially activated macrophages and neutrophils, followed by necroptosis, leads to granuloma formation. Cellular granulomas can then morph into a closed lesion, infiltrated by epithelioid and foamy macrophages and mesenchymal stem cells (MSCs) and contained by a peripheral cuff of lymphocytes. Closed granulomas undergo macrophage necrosis and caseation in their core leading to the release of intracellular Mtb. Matrix metalloproteinases (MMPs) induce the degradation of connective tissue, which allows the expulsion of the liquefied contents into adjoining airways to form an aerobically exposed open cavity. By contrast, repopulation with fibroblasts and collagenous material may form a fibrotic granuloma primarily containing latent Mtb. Inter- and intralesional diversity arises through differences in vasculature, oxygen availability (pO2), iron (Fe), and inflammation. These can influence Mtb physiology and drug distribution in distinct structural regions of a granuloma leading to phenotypic antimicrobial resistance (AMR). Mox, moxifloxacin; Bdq, bedaquiline; Rif, rifampicin.
Mtb-Lesion Variability Foils Drug Action
Arising from a single bacillus, each TB lesion bears a finite bacterial load depending on the balance between bactericidal and tissue damage-limiting immune responses [25]. Inside a lesion, Mtb is known to exist at multiple, distinct locations in a mosaic of immune cells and signaling – intracellularly or extracellularly among macrophages, neutrophils, and mesenchymal stem cells (MSCs) or in the caseous core (Figure 1). Variations arise in the Mtb transcriptome during the process of granuloma formation, particularly as a function of bacterial location, and overlap with more than one stress-responsive regulon of Mtb [30,31]. Through changes in the expression of virulence factors, Mtb is likely to influence immunopathology for its survival and spread. For example, the two-component system PhoPR and a redox-sensitive transcription factor, WhiB3, regulate the production of immunomodulatory lipids and proteins (Esx proteins) to coordinate TB pathology in animals and human granuloma formation in vitro [32–34]. Mtb also induces host matrix metalloproteinases leading to active caseation to worsen tissue pathology [35].
The bacillary burden of granulomas in an individual nonhuman primate varies from 100 to 1 million [25]. This variation in bacterial growth, coupled with diversity in granuloma structures, presents diffusion barriers to antibiotics. Matrix-assisted laser desorption/ionization mass spectrometry imaging estimates greater penetration and accumulation of Rif and Pza in the necrotic core, consistent with their ability to target non-replicating bacteria [36,37]. By contrast, moxifloxacin (Mox) concentrates more in the peripheral cellular ring and minimally in the caseum, which possibly explains the failure of a Mox-containing regimen to shorten therapy duration in clinical trials [31]. Additionally, Mox accumulation occurs significantly in foamy macrophages, where quiescent, phenotypically resistant Mtb resides [31]. Correlative live, ion, and electron microscopy suggests that lipid droplets (LDs) in foamy macrophages accumulate varying levels of the newest lipophilic anti-TB drug, bedaquiline (Bdq) [38,39] (Figure 1). Since Mtb exists in a variety of intracellular compartments, Bdq accumulates heterogeneously in the Mtb population within foamy macrophages [38]. This, coupled with the longer half-life of Bdq (6 months), indicates that Bdq stored in LDs might remain obtainable even after cessation of treatment. Clearly, permeation barriers posed by lesional and cellular heterogeneity contribute to the differential efficacy of anti-TB drugs during infection.
Cellular Heterogeneity
Immunity through Macrophage Diversity
Macrophage–Mtb interactions can influence infection by having multiple outcomes, such as bacterial killing, persistence, and macrophage death [40]. Macrophages possess an unusual capacity to polarize towards a specific phenotype based on microenvironmental cues [41]. The traditional definitions of classically (M1) and alternatively (M2) activated macrophages, derived from a common M0 ground state, present an overly simplistic picture of macrophage polarization [42]. IFNγ, lipopolysaccharide (LPS), granulocyte-macrophage colony stimulating factor (GM-CSF), TNFα, or their combinations induce the M1 phenotype, which expresses proinflammatory cytokines [interleukin (IL)-1, IL-12, IL-6, and TNFα], ROS, and reactive nitrogen intermediates to control Mtb replication. The growth-permissive M2 phenotype appears in response to IL-4 and/or IL-13 and is involved in tissue repair and homeostasis while maintaining an antiinflammatory phenotype (IL-10, transforming growth factor β, and IL-1Ra secretion). Interestingly, studies using Salmonella enterica serovar Typhimurium (STm) reveal that M1 and M2 polarized macrophages harbor non-growing and actively growing STm, respectively, and yet an intermediate macrophage phenotype retains STm in a non-growing but metabolically active state [43]. Consistent with this, unconventional stimuli such as high-density lipoproteins, cyclodextrins, and glucocorticoids induce a spectrum of activation states between the M1 and M2 extremes [44].
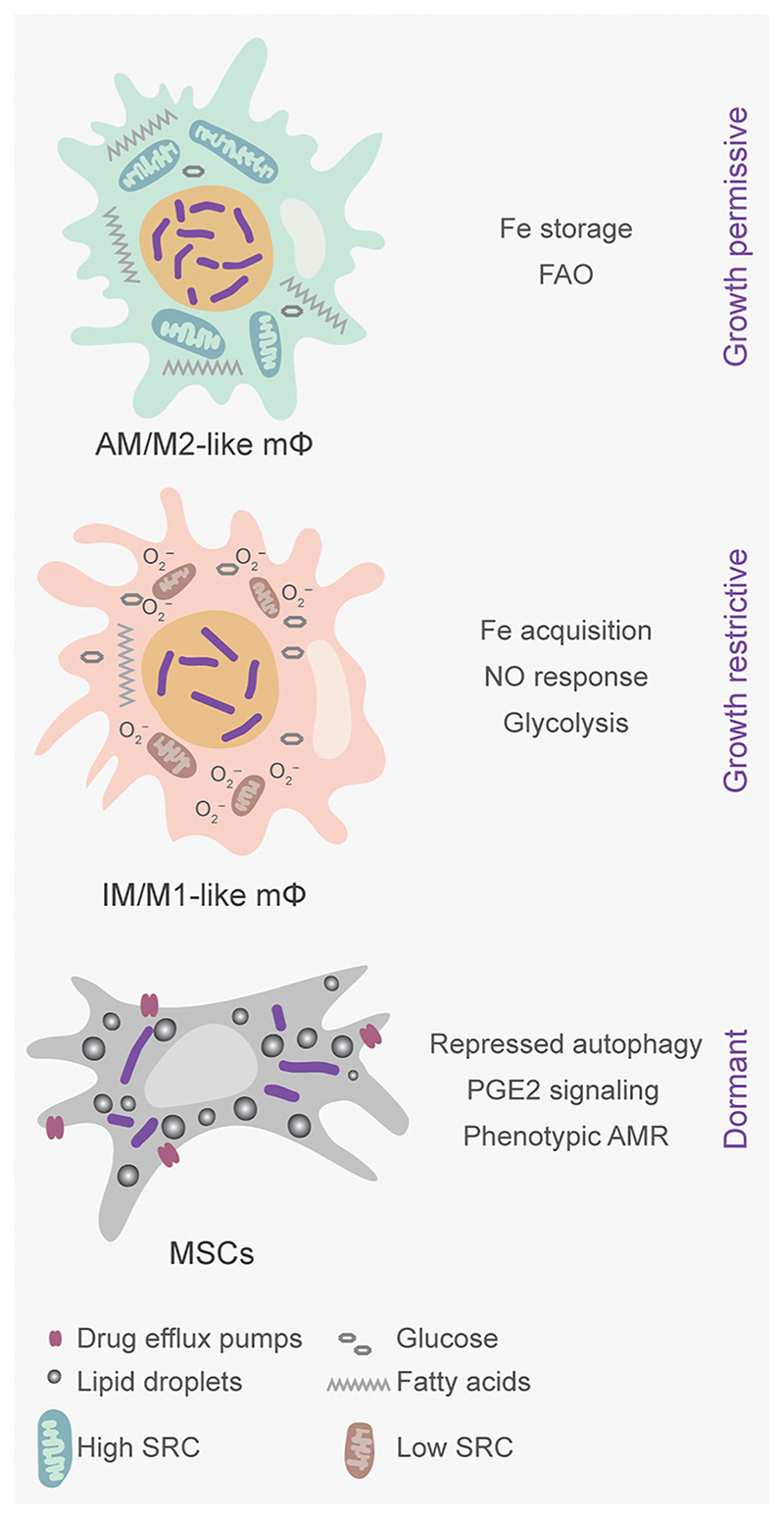
In addition to M1/M2, differences in ontogeny generate distinct alveolar macrophage (AM) and interstitial macrophage (IM) lineages and further contribute to the diversity in host response towards Mtb infection [45,46]. Lung tissue-resident AMs differentiate from fetal liver-derived monocytes and are self-renewing, whereas IMs are recruited from circulating blood monocytes in response to Mtb’s invasion in the lungs. Using fluorescent reporters of Mtb’s replication rate (SSB-GFP) and stress response (hspX’::GFP), AMs have been determined to be more growth permissive than the growth-restrictive IMs [47] (Figure 2).
Figure 2. Tissue-Specific Immune Cell Lineages Compound Phenotypic Variations in Mycobacterium tuberculosis (Mtb) during Infection.
Other than classically activated macrophages, ontologically distinct lineages of alveolar macrophages (AMs) and interstitial macrophages (IMs) as well as mesenchymal stem cells (MSCs) are known to be infection permissive in hosts. Fetal-origin AMs exhibit an M2-like phenotype with a growth-permissive environment for Mtb, whereas bone-marrow-derived IMs exhibit M1-like behavior and restrict Mtb growth. Self-renewing MSCs harbor Mtb in a drug-tolerant state and accumulate lipid droplets, which are metabolized by the bacteria for long-term persistence. FAO, fatty acid oxidation; PGE2, prostaglandin E (2); AMR, antimicrobial resistance; SRC, spare respiratory capacity.
Metabolic Heterogeneity Impacts Cellular Immunity
Enormous metabolic plasticity within macrophage types influences their antimicrobial capacity and could impact anti-TB drugs’ efficacy. IMs utilize glycolysis to meet their bioenergetic demands, whereas AMs prefer fatty acid oxidation (FAO) and maintain high spare respiratory capacity (SRC) during infection [47] (Figure 2).
The link between immunometabolism and Mtb is clarified by studies showing enhanced bacterial survival on inhibition of glycolysis, whereas blocking FAO results in decreased survival in mice. Mechanistically, inhibition of FAO reverses electron flow at the NADH:ubiquinone oxidoreductase (respiratory complex I) to generate mitochondrial ROS (mtROS), which suppresses Mtb by elevating NADPH oxidase activity and xenophagy [48]. In line with these findings, mitochondria of Mtb-infected macrophages prefer fatty acids as oxidizable substrates rather than glucose to sustain bioenergetics [49]. Mtb also utilizes host-derived fatty acids for the biosynthesis of cell wall-associated polyketide lipids [50]. Interestingly, these polyketide lipids affect the cell cycle of Mtb-infected and bystander macrophages by inhibiting the G1-to-S phase transition, further linking the fatty acid metabolism of host and/or pathogen in the promotion of phenotypic plasticity within macrophages [51]. Last, anti-TB drugs (Inh and Pza) perturb the host metabolism by inducing mtROS, which further contributes to lethality against Mtb during infection [52]. Thus, the modulation of metabolism, the cell cycle, and mtROS represents an interesting avenue for the development of host-directed therapeutic (HDT) strategies against TB.
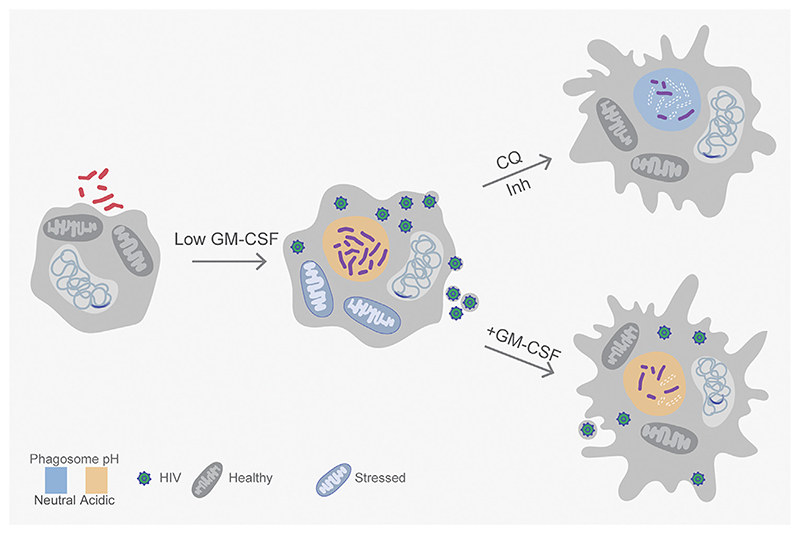
While evidence directly linking immunometabolism to phenotypic AMR is lacking, phagosomal acidification has recently been shown to induce drug tolerance in Mtb in naïve macrophages infected with Mtb alone or co-infected with HIV-1 [53]. However, in contrast to other bacteria, the pH-responsive drug-tolerant fraction of Mtb is fully replicative inside naïve macrophages [53]. IFNγ-mediated immune activation of macrophages further limits the antimycobacterial activity of drugs by inducing a non-growing but metabolically active state in Mtb during infection [54,55]. Contrary to naïve macrophages, NO stress rather than phagosomal acidification drives bacterial phenotypic AMR in activated macrophages [55]. These findings go against the dogma that the primary goal of acidic pH and NO is to suppress Mtb. Here a more sophisticated model emerges, according to which host-generated stresses facilitate the selection of Mtb phenotypes that are difficult to eradicate with anti-TB drugs. Along with IFNγ, GM-CSF signaling promotes nod-like receptor family pyrin domain containing 3-mediated IL-1β upregulation and phagosomal maturation in a subset of macrophages that harbor dead bacteria [56]. Further, GM-CSF signaling is suppressed in macrophages harboring live Mtb, and chemical inhibition of GM-CSF production limits the antimycobacterial capacity of macrophages. Moreover, dampened GM-CSF signaling accelerates the intraphagosomal growth of Mtb during HIV-TB co-infection, whereas exogenous GM-CSF supplementation inhibits the same [57]. Since macrophages co-infected with HIV and Mtb display increased oxidative stress and render Mtb phenotypically resistant to drugs, the contribution of GM-CSF signaling to these processes is likely to be important [53,58] (Figure 3).
Figure 3. Mitochondrial Activity, pH, and Cytokines Influence Survival and Phenotypic Antimicrobial Resistance (AMR) during HIV–Tuberculosis (TB) Co-infection.
Infection of macrophages harboring the latently integrated genome of HIV-1 with Mycobacterium tuberculosis (Mtb) leads to viral reactivation and bacterial proliferation. Phagosomal acidification in co-infected macrophages contributes to redox heterogeneity and phenotypic tolerance towards isoniazid (Inh) in Mtb. Co-infected macrophages experience proton leak from stressed mitochondria and non-mitochondrial reactive oxygen species (ROS), which lead to the reactivation of HIV. Neutralizing phagosomal pH using chloroquine (CQ) subverts redox heterogeneity and Inh tolerance in Mtb inside co-infected macrophages. Phagosomal maturation is controlled heterogeneously within a macrophage population, through active granulocyte-macrophage colony stimulating factor (GM-CSF) signaling and interleukin-1β (IL-1β) induction, to limit Mtb growth. HIV-TB co-infected macrophages can suppress GM-CSF signaling to prevent cytokine-mediated control of Mtb and HIV, whereas exogenous GM-CSF supplementation can reverse this effect.
Besides macrophages, phenotypic AMR has been reported in non-replicating Mtb cells isolated from MSCs often recruited to granulomas [59,60]. Mtb affects several processes in MSCs, such as the accumulation of LDs, autophagy subversion, elevated drug efflux pumps, and prostaglandin E(2) signaling to slow replication and reduce killing by drugs [61] (Figure 2). Perturbation of lipid metabolism using triacsin C and induction of autophagy by the mTOR inhibitor rapamycin potentiate the activity of Inh and clear non-replicating bacilli in MSCs [61,62]. Together, diverse host factors such as activation status, NO, acidic pH, immunometabolism, and MSCs are important determinants of heterogeneity in Mtb’s physiology and, subsequently, therapy outcome.
Bacterial Heterogeneity
Diversity in Axenic Cultures
Host cell heterogeneity and immune pressures have been widely acknowledged to generate adaptive variations among intracellular pathogens such as Listeria monocytogenes, Brucella abortus, Salmonella, and Mtb [55,63–65]. A role reversal, where cell-to-cell variations within a bacterial population drive disparate host immune responses, may yet occur, making phenotypic variability a dynamic bidirectional process.
Heterogeneity can exist within a bacterial population before the invasion of a host cell (Figure 4). Members of the genus Mycobacteria are known to generate asymmetry during cell division [66]. Mycobacterium smegmatis (Msm) and Mtb cells display differences in growth rates that create a wide distribution of cell sizes within microcolonies, as viewed by time-lapse and atomic force microscopy, and generate potential influences on antibiotic susceptibility [67–69]. Rego et al. identified Msm lamA as a ‘divisome’-associated factor involved in maintaining asymmetric cell length and differential antibiotic susceptibility [70]. Transcriptional surges of the catalase-peroxidase KatG in actively dividing Msm can generate bacterial cells with differential susceptibility to Inh, as KatG is required to activate the Inh prodrug [71]. Similarly, phase variation in the expression of glpK, which encodes a glycerol kinase, due to transient frameshift mutations in its hypervariable homopolymeric region induces heterogeneity in colony size, reduces antibiotic efficacy in mice, and may contribute to the emergence of multidrug resistance in clinical Mtb isolates [72,73]. Errors in bacterial translation leading to the generation of multiple ‘flawed’ protein copies and post-translational modifications are also known to enhance phenotypic AMR in Msm [74,75]. Consistent with this, the aminoglycoside kasugamycin can counter Rif-induced indirect tRNA aminoacylation to reduce mistranslation and increase the Rif susceptibility of Mtb in vitro and in mice [76].
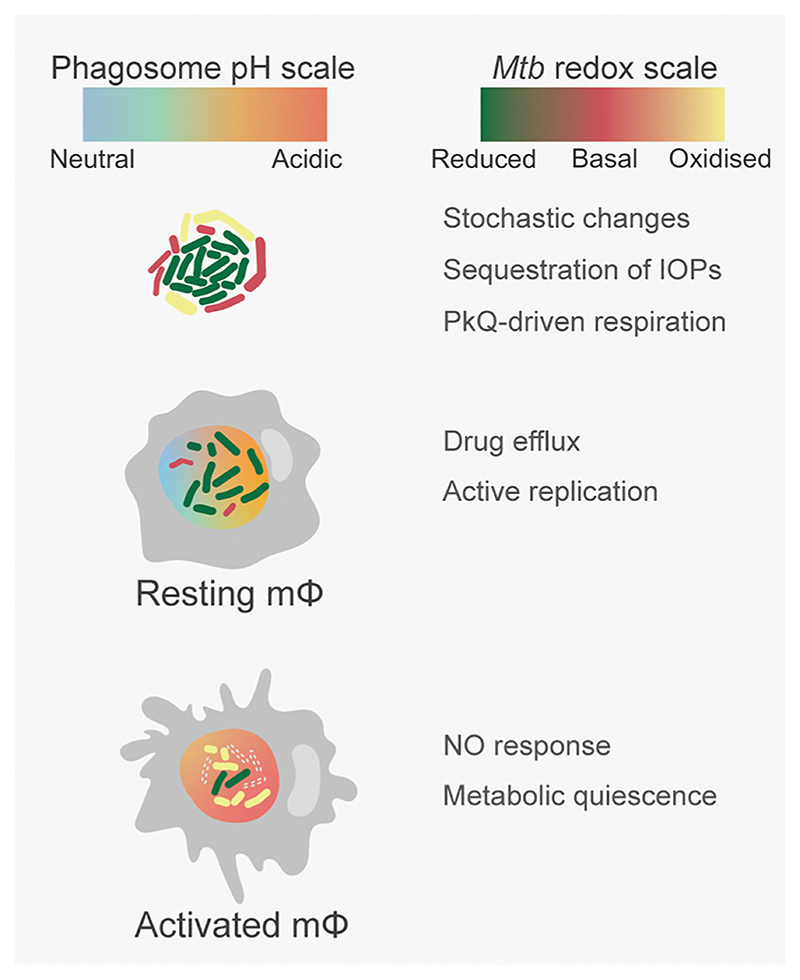
Figure 4. Phagosomal Acidification and Immune Activation Induce Phenotypic Antimicrobial Resistance (AMR).
Both bacterial phenotypic variations and host immune pressures can modulate Mycobacterium tuberculosis (Mtb) proliferation in macrophages. Isogenic Mtb populations demonstrate stochastic heterogeneity in essential cellular processes such as growth, gene expression, and the management of irreversibly oxidized proteins (IOPs). Aggregation into biofilms alters the bacterial redox state with respiration driven by polyketide quinones (PkQs) serving as alternative electron acceptors in a hypoxic niche. Inside naïve macrophages, Mtb faces variable acidification in phagosomes and recalibrates metabolism and efflux activity to cope with metal and redox stress. The consequent generation of redox heterogeneity selects for an actively replicating, drug-tolerant Mtb population. Interferon-gamma (IFNγ) signaling in activated macrophages allows Mtb to be trafficked to highly acidic phagolysosomes with abundant exposure to reactive oxygen species (ROS) and nitric oxide. Enhanced adaptive responses to an increasingly hostile host environment ultimately confer metabolic quiescence on Mtb and further strengthen phenotypic AMR.
Bacterial heterogeneity can be amplified by a prolonged stationary phase and exposure to radical stresses, acidity, and antibiotics during growth. This primes Mtb to maintain cellular and metabolic homeostasis by evoking adaptive mechanisms, which can be indirect contributors to phenotype variation [54,77]. Irreversible oxidation generates misfolded copies of cellular proteins, which interfere with physiological processes. The ClpB chaperone enables the sequestration and partitioning of oxidized protein aggregates between daughter cells of Mtb. Bacterial growth rate is inversely proportional to the amount of oxidized protein aggregates inherited, and this enhances existing asymmetry in growth and drug susceptibility within an isogenic Mtb population [78] (Figure 4). Together these findings point towards a multimorphic existence of Mtb in what should ideally be a synchronized growth condition in axenic cultures.
Looking for Triggers, One Cell at a Time
The immune pressures that Mtb faces on uptake by macrophages directly influence its growth and physiology and amplify in vitro variations (Figure 4). The transcriptome of intramacrophage Mtb simultaneously overlaps with that of Mtb exposed to oxidative and pH stress in synthetic broth, confirming the identities of phagosomal cues that reprogram Mtb to adapt in the host [79,80]. Interestingly, the transcriptome of Mtb exposed to acidic pH, hydrogen peroxide, and NO shows considerable overlap with expression changes in response to frontline anti-TB drugs [55,81]. This suggests that hostile macrophage environments select for bacterial physiologies that tolerate both antibiotics and immune pressures. In this context, a major technological leap has emerged from the development of a genetically encoded biosensor (Mrx1-roGFP2) of the mycobacterial antioxidant buffer mycothiol (MSH). The biosensor reveals marked heterogeneity in the redox potential of MSH (EMSH), with Mtb fractions displaying EMSH-basal, EMSH-reduced, and EMSH-oxidized inside macrophages [82]. EMSH-reduced Mtb is generated in response to phagosomal acidity and is found to be replicative and exceptionally tolerant to anti-TB drugs in macrophages [53,83]. Consequently, alkalization of phagosomal pH by an approved antimalarial drug, chloroquine (CQ), subverts redox heterogeneity and potentiates the efficacy of Inh to eradicate Mtb and reduce post-therapy relapse in animals [53]. These findings reposition CQ as a HDT molecule to increase the efficacy of anti-TB drugs and shorten therapy duration. Further application of Mrx1-roGFP2 has led to the identification of novel inhibitors targeting the redox metabolism of Mtb [84–88].
Until recently, the molecular underpinnings of the link between phagosomal pH, redox physiology, and drug tolerance had been elusive. Using flow cytometry-coupled RNA-seq, we have identified the bacterial determinants of redox-mediated drug tolerance in Mtb [53]. The transcriptome of redox-diverse Mtb indicates that low pH exposes the pathogen to cysteine (CySH) and metal (Fe/Cu) overload. Elevated levels of CySH and metals can lead to ROS generation through metal-catalyzed oxidation of CySH to cystine (CyS2). The drug-tolerant EMSH-reduced Mtb counteracts these stresses by effectively channelizing the flux of CySH into the reverse transsulfuration pathway (H2S gas production), Fe-S cluster biogenesis (suf operon), and MSH generation [53]. Genetic disruption of these mechanisms significantly impairs the ability of Mtb to generate the EMSH-reduced fraction and tolerate antibiotics during infection, suggesting that the management of CySH flux is an important adaptive strategy to protect Mtb from drugs (Figure 5). Similarly, H2S gas coordinates antibiotic tolerance in diverse bacteria by sequestering iron, elevating the activity of antioxidant enzymes, and scavenging ROS [89–91]. Also, altered Fe-S cluster biogenesis influences respiration to affect antibiotic uptake and killing [92,93] Moreover, overwhelming of the CySH pools of Mtb using N-acetylcysteine stimulates respiration, increases ROS, and potentiates the antimycobacterial efficacy of Inh [94]. The enhancing of respiration and ROS has emerged as a prospective strategy to potentiate the action of antibiotics in diverse bacteria [94–97]. Although a few studies have disputed this approach [98,99], it is strongly supported by others demonstrating that bactericidal antibiotics perturb redox physiology, and tolerance depends on the ability of bacteria to counteract ROS [100–103].
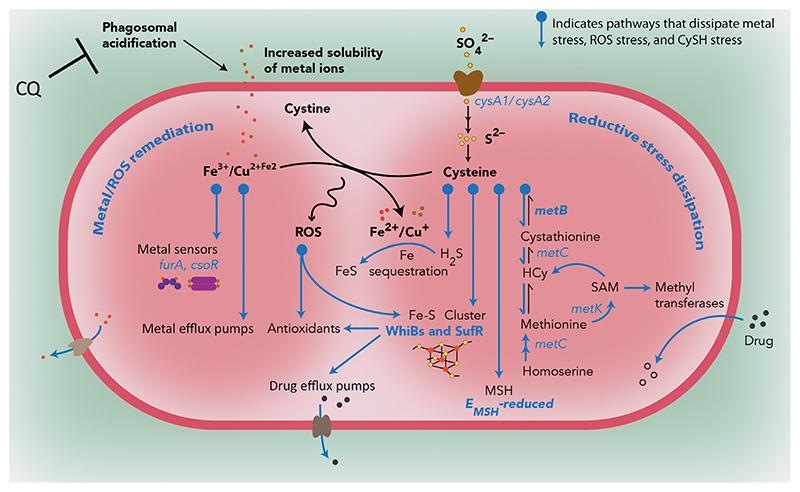
Figure 5. Redox-Altered Mycobacterium tuberculosis (Mtb) Inside Naïve Macrophages Acquires Drug Tolerance through Dissipation of Metal and Reductive Stress.
Exposure to acidic pH in macrophages leads to perturbation of redox homeostasis and the appearance of redox-altered populations of Mtb. Acidity in phagosomes induces the accumulation of soluble forms of metal ions (Fe/Cu) as well as the redox-active amino acid cysteine (CySH). The former can catalyze the generation of reactive oxygen species (ROS) by either driving the Fenton reaction or oxidizing CySH to cystine. Transcriptional profiling of intramacrophage Mtb fractions reveals that drug-tolerant EMSH-reduced Mtb are particularly efficient in dissipating reductive and metal stress during infection. This Mtb fraction channels the flux of CySH into the reverse transsulfuration pathway leading to H2S generation (MetB), Fe-S cluster biogenesis (SufR), and mycothiol (MSH) production. Induction of these pathways affords protection from oxidative stress by increased expression of redox- and acid-sensitive transcription factors (WhiB family and Suf system) that regulate Mtb’s antioxidant response, consume excess CySH and Fe, and induce the activity of metal and drug efflux pumps. Additionally, S-adenosylmethionine (SAM)-dependent methyltransferases are found to be highly expressed in reduced Mtb, where they can inactivate antibiotics by N-methylation. Blocking phagosomal acidification with lysosomotropic agents such as chloroquine (CQ) reverses intramacrophage redox heterogeneity and reduces isoniazid (Inh) or rifampicin (Rif) tolerance in Mtb-infected macrophages as well as in chronically infected animals. From [53], reprinted with permission from AAAS.
The connection between redox and respiration has also been evident in biofilms where mycobacterial cells sustain respiration by producing alkyl benzoquinones, exhibit redox diversity, and tolerate multiple antibiotics [104]. Since environmental cues such as those encountered in vivo (e.g., thiol reductive stress, hypoxia), and exposure to leukocyte lysates induce biofilm-like growth of Mtb, potentiating metabolism and respiration in biofilms can aid in the targeting of highly tolerant bacteria during infection and the development of effective adjuvants for therapy [104–106]. For example, a TCA cycle metabolite (fumarate) stimulates the central metabolism, respiration, and proton motive force of highly tolerant biofilms of Pseudomonas aeruginosa. As a consequence, fumarate increases the uptake of aminoglycosides and killing [107]. However, a similar approach in the case of Mtb would require a comprehensive understanding of host and pathogen metabolism during infection. To begin to address this, a recent study has delineated host–pathogen metabolic heterogeneity using dual RNA-seq of infected AMs and IMs [108]. In particular, glucose and nitrogen metabolism and Fe uptake appear to be highly induced in Mtb derived from IMs, driven by elevated NO levels and iron depletion. By contrast, growth-permissive AMs, known for their dependence on FAO for metabolic needs, induce fatty acid uptake pathways in resident Mtb. Together, the characterization of bacterial heterogeneity during infection can help to decipher the effects of nutrient and antibiotic availability in lesional environments and cellular immune signaling on Mtb physiology. The information derived thereof will hold immense potential for the characterization of phenotypic tolerance and its consequences for successful chemotherapy and AMR.
Concluding Remarks and Perspective
The relative contributions of host and pathogen components is still in need of considerable investigation to identify the source of phenotypic diversity (see Outstanding Questions). Phenotypic AMR in a replication-competent and metabolically active intramacrophage Mtb population has only recently been acknowledged. This area requires the active development of tools, such as noninvasive biosensors, to quantify physiological perturbations and gain insights into mechanisms other than redox metabolism and drug efflux. From the host’s perspective, the impacts of ontogeny, immunometabolism, and HIV co-infection need to be assessed individually to identify novel host-directed targets for therapy. Table 1 summarizes the currently available tools to quantify host and bacterial phenotypic diversity and its consequence for antibiotic therapy design. Chronic TB infections stem from the dynamic balance maintained between hosts and Mtb. It is essential to comprehend the mechanistic alterations involved during this interaction for effective disease management.
Outstanding Questions.
Despite successful systemic control, TB disease progresses at individual foci due to the failure of local immune responses. How can local immune responses be mapped to granuloma diversity in the tissue microenvironment during disease progression and therapy?
Does Mtb exhibit stochastic heterogeneous expression of virulence factors to drive variable responses within individual infected host immune cells and granulomas?
How can the distribution and sequestration of anti-TB drugs at a subcellular level in the infected tissues be quantified?
What is the relative contribution of bacterial heterogeneity that develops in response to host immune pressures or is pre-existing within subpopulations to the emergence of phenotypic AMR in vivo?
Is it possible to eradicate infection by developing antimicrobials that target phenotypic AMR arising from both growth-independent (e.g., nonreplicating but metabolically active) and growth-dependent (e.g., efflux pump activity) mechanisms?
How can differences among macrophage types (AMs, IMs, M1, and M2) and within a macrophage subcategory (AMs/IMs) influence bacterial heterogeneity and phenotypic AMR?
Can immunomodulatory therapy be used to mobilize the more potent subset of host immune cells to appropriate locations for the targeting of drug-tolerant Mtb during infection?
How can the central carbon metabolism, respiration, and redox homeostasis of Mtb be perturbed in the complex environment of immune cells and tissues to improve antibiotic efficacy?
Is it possible to image and capture heterogeneity in the physiology of the Mtb population present in human sputum and/or in biofilms in vivo to understand the requirement for prolonged chemotherapy?
Table 1. Tools That Can Be Applied to Study Bacterial, Cellular, and Lesional Heterogeneity and Their Influence on Phenotypic AMRa .
| Tool | Description | Key finding | Refs |
|---|---|---|---|
| Reporter strains | |||
| hspx’::GFP | hspX promoter-driven GFP expression | Higher NO stress in IMs and neutrophils | [47] |
| SSB-GFP | Binds single DNA strands and marks active replisomes | IMs are growth restrictive while AMs are growth permissive | [47] |
| Mrx1-roGFP2 | Reduction-oxidation-sensitive GFP linked to MSH-dependent oxidoreductase (Mrx1) | Intramacrophage redox diversification of replicating Mtb mediates drug tolerance Acidic pH, NO, and antibiotics perturb redox homeostasis in Mtb | [53,82] [34,83,85,86] |
| Live-dead reporter | Constitutive, long-lived RFP; Tet-inducible promoter-coupled GFP to assess transcriptional activity in bacteria | Heterogeneous GM-CSF signaling regulates Mtb survival in TB and HIV-TB co-infection | [56] |
| Transcriptomics (coupled with flow cytometry) | Analyze and sort cell/bacterial subpopulations to quantify messenger and noncoding RNA | Intramacrophage Mtb senses host cues to exhibit phenotypic AMR Simultaneous host and pathogen profiling to identify ontogeny-based regulation of pathogen burden in AMs and IMs Regulatory RNA MrsI mediates iron-sparing response in iron-limited Mtb Analysis of cell-to-cell heterogeneity |
[53,55] [47,108] [109] [40,110] |
| Microscopy (confocal, ion, electron, atomic force, microfluidics-coupled time lapse) | Image and quantify host–pathogen crosstalk Analyze inter- and intracellular changes in real time Drug permeation and sequestration |
LDs bind Bdq and facilitate drug trafficking inside Mtb Neutrophils, AMs, and IMs have different capacities to limit Mtb proliferation HMDMs display heterogeneity in Mtb control Asymmetric cell division promotes phenotypic AMR in axenic Mtb cultures Stochastic transcriptional pulsing generates Mtb variants Redox-altered Mtb fractions reside in specific subcellular compartments |
[38] [47] [56] [67,69] [71–73] [82,83] |
| Small-molecule inhibitors (etomoxir, trimetazidine, 2-deoxyglucose, glibenclamide, lysosomotropic agents) | Low-molecular-weight compounds allow metabolic manipulations in host and/or pathogen | FAO and glycolysis regulate Mtb survival in vivo
GM-CSF signaling controls Mtb burden in HMDMs Blocking phagosomal acidification subverts redox-related drug tolerance |
[47,48] [56] [53,83] |
| Proteomics | Protein quantification during host–pathogen interaction |
Mtb WhiB3 differentially regulates host cell cycle Mtb-infected macrophages transfer host proteins to reactivate HIV in bystander cells Antibiotic exposure rewires Mtb metabolic path ways Post-translational modifications occur in host proteins on Mtb infection |
[51] [58] [111] [112] |
| Metabolomics | Metabolite quantification during host-pathogen interaction | Elevated intracellular levels of NAD and glutathione in infected macrophages Perturbation in amino acid and phospholipid metabolism in infected mouse lungs | [113] [114] |
| Seahorse extracellular flux analysis | Profiling of metabolic activity of cells in real time | Immunometabolism of AMs and IMs regulates Mtb growth Mtb infection induces quiescence and dependency on exogenous fatty acids in macrophages Screening drug candidates in Mtb |
[108] [49] [115] |
| Laser capture microdissection | Acquisition of specific cell types or spatial regions from complex granulomas | Spatiotemporal differences in inflammatory signaling within granulomas Nutrient and drug penetration vary in lesions |
[26] [31,37] |
Abbreviations: HMDM, human monocyte-derived macrophage; hspX, heat shock protein X; Mrx1, mycoredoxin-1; roGFP2, redox-sensitive GFP; SSB, single-strand binding protein.
Highlights.
Heterogeneity in host-pathogen interactions introduces incredible diversity in TB lesions that leads to heterogeneous drug distribution and selects for Mtb phenotypes with reduced susceptibility to drugs.
Immune signaling, ontogeny, and HIV co-infection dictate Mtb’s survival and phenotypic antimicrobial resistance (AMR) in the host.
Intraphagosomal Mtb faces low pH and nitric oxide and the metabolic adaptations required to survive under these conditions lead to phenotypic AMR.
Metabolic adaptations under low pH generate redox heterogeneity in a replicating Mtb population with drug-tolerant bacteria exhibiting elevated mycothiol, Fe-S clusters, and H2S gas.
The link between acidic pH, metabolism, and phenotypic AMR opens novel approaches for therapy; for example, phagosomal de-acidification by chloroquine subverts redox heterogeneity and eradicates drug-tolerant Mtb.
Acknowledgments
The work was supported by the following Wellcome Trust/DBT India Alliance Grant: IA/S/16/2/502700 (A.S.), and in part by Department of Biotechnology (DBT) Grant BT/PR11911/BRB/10/1327/2014, BT/PR5020/MED/29/1454/2012 (A.S.) and the DBT-IISc Partnership Program (22-0905-0006-05-987-436) and Infosys Foundation. A.S. is a senior fellow of the Wellcome Trust/DBT India Alliance. R.M., V.Y., and M.G. gratefully acknowledge CSIR-India and IISc for fellowships.
References
- 1.Hobby GL, et al. Observations on the mechanism of action of penicillin. Proc Soc Exp Biol Med. 1942;50:281–285. [Google Scholar]
- 2.Bigger JW. Treatment of staphylococcal infections with penicillin by intermittent sterilisation. Lancet. 1944;244:497–500. [Google Scholar]
- 3.Balaban NQ, et al. Definitions and guidelines for research on antibiotic persistence. Nat Rev Microbiol. 2019;17:441–448. doi: 10.1038/s41579-019-0196-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schrader SM, et al. Biology of antimicrobial resistance and approaches to combat it. Sci Transl Med. 2020;12:eaaz6992. doi: 10.1126/scitranslmed.aaz6992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sarathy JP, et al. Extreme drug tolerance of Mycobacterium tuberculosis in caseum. Antimicrob Agents Chemother. 2018;62:e02266–17. doi: 10.1128/AAC.02266-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Garton NJ, et al. Cytological and transcript analyses reveal fat and lazy persister-like bacilli in tuberculous sputum. PLoS Med. 2008;5:e75. doi: 10.1371/journal.pmed.0050075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dhar N, et al. Phenotypic heterogeneity in Mycobacterium tuberculosis . Microbiol Spectr. 2016 doi: 10.1128/microbiolspec.TBTB2-0021-2016. Published online November 11, 2016. [DOI] [PubMed] [Google Scholar]
- 8.Cadena AM, et al. Heterogeneity in tuberculosis. Nat Rev Immunol. 2017;17:691–702. doi: 10.1038/nri.2017.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McCune RM, Jr, et al. The fate of Mycobacterium tuberculosis in mouse tissues as determined by the microbial enumeration technique. II. The conversion of tuberculous infection to the latent state by the administration of pyrazinamide and a companion drug. J Exp Med. 1956;104:763–802. doi: 10.1084/jem.104.5.763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McCune RM, et al. Microbial persistence i The capacity of tubercle bacilli to survive sterilization in mouse tissues. J Exp Med. 1966;123:445–468. doi: 10.1084/jem.123.3.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McCune RM, et al. Microbial persistence Ii Characteristics of the sterile state of tubercle bacilli. J Exp Med. 1966;123:469–486. doi: 10.1084/jem.123.3.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brindle R, et al. Serial counts of Mycobacterium tuberculosis in sputum as surrogate markers of the sterilising activity of rifampicin and pyrazinamide in treating pulmonary tuberculosis. BMC Pulm Med. 2001;1:2. doi: 10.1186/1471-2466-1-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jindani A, et al. Bactericidal and sterilizing activities of antituberculosis drugs during the first 14 days. Am J Respir Crit Care Med. 2003;167:1348–1354. doi: 10.1164/rccm.200210-1125OC. [DOI] [PubMed] [Google Scholar]
- 14.Ruhl CR, et al. Mycobacterium tuberculosis sulfolipid-1 activates nociceptive neurons and induces cough. Cell. 2020;181:293–305.:e211. doi: 10.1016/j.cell.2020.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Saelens JW, et al. Mycobacterial evolution intersects with host tolerance. Front Immunol. 2019;10:528. doi: 10.3389/fimmu.2019.00528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bottai D, et al. TbD1 deletion as a driver of the evolutionary success of modern epidemic Mycobacterium tuberculosis lineages. Nat Commun. 2020;11:684. doi: 10.1038/s41467-020-14508-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Verma S, et al. Transmission phenotype of Mycobacterium tuberculosis strains is mechanistically linked to induction of distinct pulmonary pathology. PLoS Pathog. 2019;15:e1007613. doi: 10.1371/journal.ppat.1007613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garcia BJ, et al. Sputum is a surrogate for bronchoalveolar lavage for monitoring Mycobacterium tuberculosis transcriptional profiles in TB patients. Tuberculosis (Edinb) 2016;100:89–94. doi: 10.1016/j.tube.2016.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fennelly KP, et al. Cough-generated aerosols of Mycobacterium tuberculosis: a new method to study infectiousness. Am J Respir Crit Care Med. 2004;169:604–609. doi: 10.1164/rccm.200308-1101OC. [DOI] [PubMed] [Google Scholar]
- 20.Young DB, Duncan K. Prospects for new interventions in the treatment and prevention of mycobacterial disease. Annu Rev Microbiol. 1995;49:641–673. doi: 10.1146/annurev.mi.49.100195.003233. [DOI] [PubMed] [Google Scholar]
- 21.Chengalroyen MD, et al. Detection and quantification of differentially culturable tubercle bacteria in sputum from patients with tuberculosis. Am J Respir Crit Care Med. 2016;194:1532–1540. doi: 10.1164/rccm.201604-0769OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mukamolova GV, et al. Resuscitation-promoting factors reveal an occult population of tubercle bacilli in sputum. Am J Respir Crit Care Med. 2010;181:174–180. doi: 10.1164/rccm.200905-0661OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Turapov O, et al. Phenotypically adapted Mycobacterium tuberculosis populations from sputum are tolerant to first-line drugs. Antimicrob Agents Chemother. 2016;60:2476–2483. doi: 10.1128/AAC.01380-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saito K, et al. Rifamycin action on RNA polymerase in antibiotic-tolerant Mycobacterium tuberculosis results in differentially detectable populations. Proc Natl Acad Sci U S A. 2017;114:E4832–E4840. doi: 10.1073/pnas.1705385114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lin PL, et al. Sterilization of granulomas is common in active and latent tuberculosis despite within-host variability in bacterial killing. Nat Med. 2014;20:75–79. doi: 10.1038/nm.3412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marakalala MJ, et al. Inflammatory signaling in human tuberculosis granulomas is spatially organized. Nat Med. 2016;22:531–538. doi: 10.1038/nm.4073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cardona PJ. A dynamic reinfection hypothesis of latent tuberculosis infection. Infection. 2009;37:80–86. doi: 10.1007/s15010-008-8087-y. [DOI] [PubMed] [Google Scholar]
- 28.Mattila JT, et al. Microenvironments in tuberculous granulomas are delineated by distinct populations of macrophage subsets and expression of nitric oxide synthase and arginase isoforms. J Immunol. 2013;191:773–784. doi: 10.4049/jimmunol.1300113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Timm J, et al. Differential expression of iron-, carbon-, and oxygen-responsive mycobacterial genes in the lungs of chronically infected mice and tuberculosis patients. Proc Natl Acad Sci U S A. 2003;100:14321–14326. doi: 10.1073/pnas.2436197100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fenhalls G, et al. In situ detection of Mycobacterium tuberculosis transcripts in human lung granulomas reveals differential gene expression in necrotic lesions. Infect Immun. 2002;70:6330–6338. doi: 10.1128/IAI.70.11.6330-6338.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Blanc L, et al. High-resolution mapping of fluoroquinolones in TB rabbit lesions reveals specific distribution in immune cell types. eLife. 2018;7:e41115. doi: 10.7554/eLife.41115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Steyn AJ, et al. Mycobacterium tuberculosis WhiB3 interacts with RpoV to affect host survival but is dispensable for in vivo growth. Proc Natl Acad Sci U S A. 2002;99:3147–3152. doi: 10.1073/pnas.052705399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Walters SB, et al. The Mycobacterium tuberculosis PhoPR two-component system regulates genes essential for virulence and complex lipid biosynthesis. Mol Microbiol. 2006;60:312–330. doi: 10.1111/j.1365-2958.2006.05102.x. [DOI] [PubMed] [Google Scholar]
- 34.Mehta M, et al. Mycobacterium tuberculosis WhiB3 maintains redox homeostasis and survival in response to reactive oxygen and nitrogen species. Free Radic Biol Med. 2019;131:50–58. doi: 10.1016/j.freeradbiomed.2018.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brace PT, et al. Mycobacterium tuberculosis subverts negative regulatory pathways in human macrophages to drive immunopathology. PLoS Pathog. 2017;13:e1006367. doi: 10.1371/journal.ppat.1006367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Prideaux B, et al. The association between sterilizing activity and drug distribution into tuberculosis lesions. Nat Med. 2015;21:1223–1227. doi: 10.1038/nm.3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zimmerman M, et al. Spatial quantification of drugs in pulmonary tuberculosis lesions by laser capture microdissection liquid chromatography mass spectrometry (LCM-LC/MS) J Vis Exp. 2018;134:57402. doi: 10.3791/57402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Greenwood DJ, et al. Subcellular antibiotic visualization reveals a dynamic drug reservoir in infected macrophages. Science. 2019;364:1279–1282. doi: 10.1126/science.aat9689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tiberi S, et al. Bedaquiline in MDR/XDR-TB cases: first experience on compassionate use. Eur Respir J. 2014;43:289–292. doi: 10.1183/09031936.00122313. [DOI] [PubMed] [Google Scholar]
- 40.Avraham R, et al. Pathogen cell-to-cell variability drives heterogeneity in host immune responses. Cell. 2015;1621:1309–1321. doi: 10.1016/j.cell.2015.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Benoit M, et al. Macrophage polarization in bacterial infections. J Immunol. 2008;181:3733–3739. doi: 10.4049/jimmunol.181.6.3733. [DOI] [PubMed] [Google Scholar]
- 42.Martinez FO, Gordon S. The M1 and M2 paradigm of macrophage activation: time for reassessment. F1000Prime Rep. 2014;6:13. doi: 10.12703/P6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stapels DAC, et al. Salmonella persisters undermine host immune defenses during antibiotic treatment. Science. 2018;362:1156–1160. doi: 10.1126/science.aat7148. [DOI] [PubMed] [Google Scholar]
- 44.Xue J, et al. Transcriptome-based network analysis reveals a spectrum model of human macrophage activation. Immunity. 2014;40:274–288. doi: 10.1016/j.immuni.2014.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guilliams M, et al. Alveolar macrophages develop from fetal monocytes that differentiate into long-lived cells in the first week of life via GM-CSF. J Exp Med. 2013;210:1977–1992. doi: 10.1084/jem.20131199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yona S, et al. Fate mapping reveals origins and dynamics of monocytes and tissue macrophages under homeostasis. Immunity. 2013;38:79–91. doi: 10.1016/j.immuni.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huang L, et al. Growth of Mycobacterium tuberculosis in vivo segregates with host macrophage metabolism and ontogeny. J Exp Med. 2018;215:1135–1152. doi: 10.1084/jem.20172020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chandra P, et al. Inhibition of fatty acid oxidation promotes macrophage control of Mycobacterium tuberculosis . mBio. 2020;11:e01139–20. doi: 10.1128/mBio.01139-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cumming BM, et al. Mycobacterium tuberculosis induces decelerated bioenergetic metabolism in human macrophages. eLife. 2018;7:e39169. doi: 10.7554/eLife.39169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Singh A, et al. Mycobacterium tuberculosis WhiB3 maintains redox homeostasis by regulating virulence lipid anabolism to modulate macrophage response. PLoS Pathog. 2009;5:e1000545. doi: 10.1371/journal.ppat.1000545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cumming BM, et al. Mycobacterium tuberculosis arrests host cycle at the G1/S transition to establish long term infection. PLoS Pathog. 2017;13:e1006389. doi: 10.1371/journal.ppat.1006389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kim JJ, et al. Host cell autophagy activated by antibiotics is required for their effective antimycobacterial drug action. Cell Host Microbe. 2012;11:457–468. doi: 10.1016/j.chom.2012.03.008. [DOI] [PubMed] [Google Scholar]
- 53.Mishra R, et al. Targeting redox heterogeneity to counteract drug tolerance in replicating Mycobacterium tuberculosis . Sci Transl Med. 2019;11:eaaw6635. doi: 10.1126/scitranslmed.aaw6635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Manina G, et al. Stress and host immunity amplify Mycobacterium tuberculosis phenotypic heterogeneity and induce nongrowing metabolically active forms. Cell Host Microbe. 2015;17:32–46. doi: 10.1016/j.chom.2014.11.016. [DOI] [PubMed] [Google Scholar]
- 55.Liu Y, et al. Immune activation of the host cell induces drug tolerance in Mycobacterium tuberculosis both in vitro and in vivo . J Exp Med. 2016;213:809–825. doi: 10.1084/jem.20151248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bryson BD, et al. Heterogeneous GM-CSF signaling in macrophages is associated with control of Mycobacterium tuberculosis . Nat Commun. 2019;10:2329. doi: 10.1038/s41467-019-10065-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Patel NR, et al. HIV impairs TNF-α mediated macrophage apoptotic response to Mycobacterium tuberculosis . J Immunol. 2007;179:6973–6980. doi: 10.4049/jimmunol.179.10.6973. [DOI] [PubMed] [Google Scholar]
- 58.Tyagi P, et al. Mycobacterium tuberculosis reactivates HIV-1 via exosome-mediated resetting of cellular redox potential and bioenergetics. mBio. 2020;11:e03293–19. doi: 10.1128/mBio.03293-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Das B, et al. CD271+ bone marrow mesenchymal stem cells may provide a niche for dormant Mycobacterium tuberculosis . Sci Transl Med. 2013;5:170ra113. doi: 10.1126/scitranslmed.3004912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Raghuvanshi S, et al. Mycobacterium tuberculosis evades host immunity by recruiting mesenchymal stem cells. Proc Natl Acad Sci U S A. 2010;107:21653–21658. doi: 10.1073/pnas.1007967107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jain N, et al. Mesenchymal stem cells offer a drug-tolerant and immune-privileged niche to Mycobacterium tuberculosis . Nat Commun. 2020;11:3062. doi: 10.1038/s41467-020-16877-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fatima S, et al. Mycobacterium tuberculosis programs mesenchymal stem cells to establish dormancy and persistence. J Clin Invest. 2020;130:655–661. doi: 10.1172/JCI128043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rengarajan M, Theriot JA. Rapidly dynamic host cell heterogeneity in bacterial adhesion governs susceptibility to infection by Listeria monocytogenes. Mol Biol Cell. 2020;31:2097–2106. doi: 10.1091/mbc.E19-08-0454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zai X, et al. Relative quantitative proteomic analysis of Brucella abortus reveals metabolic adaptation to multiple environmental stresses. Front Microbiol. 2017;8:2347. doi: 10.3389/fmicb.2017.02347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Helaine S, et al. Internalization of Salmonella by macrophages induces formation of nonreplicating persisters. Science. 2014;343:204–208. doi: 10.1126/science.1244705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kieser KJ, Rubin EJ. How sisters grow apart: mycobacterial growth and division. Nat Rev Microbiol. 2014;12:550–562. doi: 10.1038/nrmicro3299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Aldridge BB, et al. Asymmetry and aging of mycobacterial cells lead to variable growth and antibiotic susceptibility. Science. 2012;335:100–104. doi: 10.1126/science.1216166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Richardson K, et al. Temporal and intrinsic factors of rifampicin tolerance in mycobacteria. Proc Natl Acad Sci U S A. 2016;113:8302–8307. doi: 10.1073/pnas.1600372113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hannebelle MTM, et al. A biphasic growth model for cell pole elongation in mycobacteria. Nat Commun. 2020;11:452. doi: 10.1038/s41467-019-14088-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rego EH, et al. Deletion of a mycobacterial divisome factor collapses single-cell phenotypic heterogeneity. Nature. 2017;546:153–157. doi: 10.1038/nature22361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wakamoto Y, et al. Dynamic persistence of antibiotic-stressed mycobacteria. Science. 2013;339:91–95. doi: 10.1126/science.1229858. [DOI] [PubMed] [Google Scholar]
- 72.Bellerose MM, et al. Common variants in the glycerol kinase gene reduce tuberculosis drug efficacy. mBio. 2019;10:e00663–19. doi: 10.1128/mBio.00663-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Safi H, et al. Phase variation in Mycobacterium tuberculosis glpK produces transiently heritable drug tolerance. Proc Natl Acad Sci U S A. 2019;116:19665–19674. doi: 10.1073/pnas.1907631116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Javid B, et al. Mycobacterial mistranslation is necessary and sufficient for rifampicin phenotypic resistance. Proc Natl Acad Sci usA. 2014;111:1132–1137. doi: 10.1073/pnas.1317580111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sakatos A, et al. Posttranslational modification of a histone-like protein regulates phenotypic resistance to isoniazid in mycobacteria. Sci Adv. 2018;4:eaao1478. doi: 10.1126/sciadv.aao1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chaudhuri S, et al. Kasugamycin potentiates rifampicin and limits emergence ofresistance in Mycobacterium tuberculosis by specifically decreasing mycobacterial mistranslation. eLife. 2018;7:e36782. doi: 10.7554/eLife.36782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Vandal OH, et al. A membrane protein preserves intrabacterial pH in intraphagosomal Mycobacterium tuberculosis . Nat Med. 2008;14:849–854. doi: 10.1038/nmXXXX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Vaubourgeix J, et al. Stressed mycobacteria use the chaperone ClpB to sequester irreversibly oxidized proteins asymmetrically within and between cells. Cell Host Microbe. 2015;17:178–190. doi: 10.1016/j.chom.2014.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Schnappinger D, et al. Transcriptional adaptation of Mycobacterium tuberculosis within macrophages: insights into the phagosomal environment. J Exp Med. 2003;198:693–704. doi: 10.1084/jem.20030846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rohde KH, et al. Linking the transcriptional profiles and the physiological states of Mycobacterium tuberculosis during an extended intracellular infection. PLoS Pathog. 2012;8:e1002769. doi: 10.1371/journal.ppat.1002769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Rohde KH, et al. Mycobacterium tuberculosis invasion of macrophages: linking bacterial gene expression to environmental cues. Cell Host Microbe. 2007;2:352–364. doi: 10.1016/j.chom.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 82.Bhaskar A, et al. Reengineering redox sensitive GFP to measure mycothiol redox potential of Mycobacterium tuberculosis during infection. PLoS Pathog. 2014;10:e1003902. doi: 10.1371/journal.ppat.1003902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mehta M, et al. Mycobacterium tuberculosis WhiB3 responds to vacuolar pH-induced changes in mycothiol redox potential to modulate phagosomal maturation and virulence. J Biol Chem. 2016;291:2888–2903. doi: 10.1074/jbc.M115.684597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tyagi P, et al. Mycobacterium tuberculosis has diminished capacity to counteract redox stress induced by elevated levels of endogenous superoxide. Free Radic Biol Med. 2015;84:344–354. doi: 10.1016/j.freeradbiomed.2015.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mishra S, et al. Efficacy of β-lactam/β-lactamase inhibitor combination is linked to WhiB4-mediated changes in redox physiology of Mycobacterium tuberculosis . eLife. 2017;6:e25624. doi: 10.7554/eLife.25624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Padiadpu J, et al. Identifying and tackling emergent vulnerability in drug-resistant mycobacteria. ACS Infect Dis. 2016;2:592–607. doi: 10.1021/acsinfecdis.6b00004. [DOI] [PubMed] [Google Scholar]
- 87.Libardo MDJ, et al. Phagosomal copper-promoted oxidative attack on intracellular Mycobacterium tuberculosis . ACS Infect Dis. 2018;4:1623–1634. doi: 10.1021/acsinfecdis.8b00171. [DOI] [PubMed] [Google Scholar]
- 88.Palde PB, et al. First-in-class inhibitors of sulfur metabolism with bactericidal activity against non-replicating M. tuberculosis . ACS Chem Biol. 2016;11:172–184. doi: 10.1021/acschembio.5b00517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Shatalin K, et al. H2S: a universal defense against antibiotics in bacteria. Science. 2011;334:986–990. doi: 10.1126/science.1209855. [DOI] [PubMed] [Google Scholar]
- 90.Shukla P, et al. “On demand” redox buffering by H2S contributes to antibiotic resistance revealed by a bacteria-specific H2S donor. Chem Sci. 2017;8:4967–4972. doi: 10.1039/c7sc00873b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Pal VK, et al. Hydrogen sulfide in physiology and pathogenesis of bacteria and viruses. IUBMB Life. 2018;70:393–410. doi: 10.1002/iub.1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ezraty B, et al. Fe-S cluster biosynthesis controls uptake of aminoglycosides in a ROS-less death pathway. Science. 2013;340:1583–1587. doi: 10.1126/science.1238328. [DOI] [PubMed] [Google Scholar]
- 93.Kohanski MA, et al. A common mechanism of cellular death induced by bactericidal antibiotics. Cell. 2007;130:797–810. doi: 10.1016/j.cell.2007.06.049. [DOI] [PubMed] [Google Scholar]
- 94.Vilcheze C, et al. Enhanced respiration prevents drug tolerance and drug resistance in Mycobacterium tuberculosis . Proc Natl Acad Sci U S A. 2017;114:4495–4500. doi: 10.1073/pnas.1704376114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Brynildsen MP, et al. Potentiating antibacterial activity by predictably enhancing endogenous microbial ROS production. Nat Biotechnol. 2013;31:160–165. doi: 10.1038/nbt.2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Grant SS, et al. Eradication of bacterial persisters with antibiotic-generated hydroxyl radicals. Proc Natl Acad Sci U S A. 2012;109:12147–12152. doi: 10.1073/pnas.1203735109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Makafe GG, et al. Quinoline derivatives kill Mycobacterium tuberculosis by activating glutamate kinase. Cell Chem Biol. 2019;26:1187–1194.:e1185. doi: 10.1016/j.chembiol.2019.05.003. [DOI] [PubMed] [Google Scholar]
- 98.Liu Y, Imlay JA. Cell death from antibiotics without the involvement of reactive oxygen species. Science. 2013;339:1210–1213. doi: 10.1126/science.1232751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Keren I, et al. Killing by bactericidal antibiotics does not depend on reactive oxygen species. Science. 2013;339:1213–1216. doi: 10.1126/science.1232688. [DOI] [PubMed] [Google Scholar]
- 100.Dwyer DJ, et al. Antibiotics induce redox-related physiological alterations as part of their lethality. Proc Natl Acad Sci U S A. 2014;111:E2100–E2109. doi: 10.1073/pnas.1401876111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Foti JJ, et al. Oxidation of the guanine nucleotide pool underlies cell death by bactericidal antibiotics. Science. 2012;336:315–319. doi: 10.1126/science.1219192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Martins D, et al. Superoxide dismutase activity confers (p)ppGpp-mediated antibiotic tolerance to stationary-phase Pseudomonas aeruginosa. Proc Natl Acad Sci U S A. 2018;115:9797–9802. doi: 10.1073/pnas.1804525115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Rowe SE, et al. Reactive oxygen species induce antibiotic tolerance during systemic Staphylococcus aureus infection. Nat Microbiol. 2020;5:282–290. doi: 10.1038/s41564-019-0627-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Anand A, et al. Polyketide quinones are alternate intermediate electron carriers during mycobacterial respiration in oxygen-deficient niches. Mol Cell. 2015;60:637–650. doi: 10.1016/j.molcel.2015.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Trivedi A, et al. Thiol reductive stress induces cellulose-anchored biofilm formation in Mycobacterium tuberculosis . Nat Commun. 2016;7:11392. doi: 10.1038/ncomms11392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ackart DF, et al. Expression of antimicrobial drug tolerance by attached communities of Mycobacterium tuberculosis . Pathog Dis. 2014;70:359–369. doi: 10.1111/2049-632X.12144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Meylan S, et al. Carbon sources tune antibiotic susceptibility in Pseudomonas aeruginosa via tricarboxylic acid cycle control. Cell Chem Biol. 2017;24:195–206. doi: 10.1016/j.chembiol.2016.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Pisu D, et al. Dual RNA-seq of Mtb-infected macrophages in vivo reveals ontologically distinct host–pathogen interactions. Cell Rep. 2020;30:335–350.:e334. doi: 10.1016/j.celrep.2019.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Gerrick ER, et al. Small RNA profiling in Mycobacterium tuberculosis identifies MrsI as necessary for an anticipatory iron sparing response. Proc Natl Acad Sci U S A. 2018;115:6464–6469. doi: 10.1073/pnas.1718003115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Saliba AE, et al. Single-cell RNA-seq ties macrophage polarization to growth rate of intracellular Salmonella . Nat Microbiol. 2016;2:16206. doi: 10.1038/nmicrobiol.2016.206. [DOI] [PubMed] [Google Scholar]
- 111.Danelishvili L, et al. Mycobacterium tuberculosis proteome response to antituberculosis compounds reveals metabolic “escape” pathways that prolong bacterial survival. Antimicrob Agents Chemother. 2017;61:e00430–17. doi: 10.1128/AAC.00430-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Budzik JM, et al. Dynamic post-translational modification profiling of Mycobacterium tuberculosis-infected primary macrophages. eLife. 2020;9:e51461. doi: 10.7554/eLife.51461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Vrieling F, et al. Analyzing the impact of Mycobacterium tuberculosis infection on primary human macrophages by combined exploratory and targeted metabolomics. Sci Rep. 2020;10:7085. doi: 10.1038/s41598-020-62911-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Fernandez-Garcia M, et al. Comprehensive examination of the mouse lung metabolome following Mycobacterium tuberculosis infection using a multiplatform mass spectrometry approach. J Proteome Res. 2020;19:2053–2070. doi: 10.1021/acs.jproteome.9b00868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Lamprecht DA, et al. Turning the respiratory flexibility of Mycobacterium tuberculosis against itself. Nat Commun. 2016;7:12393. doi: 10.1038/ncomms12393. [DOI] [PMC free article] [PubMed] [Google Scholar]