Abstract
Loss-of-function mutations of EZH2, the enzymatic component of PRC2, have been associated with poor outcome and chemotherapy resistance in T-cell acute lymphoblastic leukemia (TALL). Using isogenic T-ALL cells, with and without CRISPR/Cas9-induced EZH2-inactivating mutations, we performed a cell-based synthetic lethal drug screen. EZH2 deficient cells exhibited increased sensitivity to structurally diverse inhibitors of CHK1, an interaction that could be validated genetically. Furthermore, small molecule inhibition of CHK1 had efficacy in delaying tumor progression in isogenic EZH2 deficient, but not EZH2 wild-type T-ALL cells in vivo, as well as in a primary cell model of PRC2 mutant ALL. Mechanistically, EZH2 deficiency resulted in a gene expression signature of immature T-ALL cells, marked transcriptional upregulation of MYCN, increased replication stress, and enhanced dependency on CHK1 for cell survival. Lastly, we demonstrate this phenotype is mediated through de-repression of a distal PRC2-regulated MYCN enhancer. In conclusion, we highlight a novel and clinically exploitable pathway in high-risk EZH2 mutated T-ALL.
Keywords: EZH2, Synthetic lethal screening, T-ALL, CHK1, MYCN
Introduction
Loss-of-function mutations and deletions of Enhancer of Zeste (Drosophila) homologue 2 (EZH2), the enzymatic component of polycomb repressor complex 2 (PRC2), are recurrently found in T-ALL and have been strongly associated with the pathogenesis of T-ALL (1,2). Alterations of the core components of PRC2, including EZH2, EED and SUZ12 are particularly frequent in the early T cell precursor ALL subtype (ETP-ALL) (1,3), a subgroup characterized by aberrant expression of myeloid and stem cell markers (4).
EZH2 catalyzes the trimethylation of histone H3 on Lys27 (H3K27me3), an epigenetic mark associated with transcriptional repression of genes involved in differentiation and development (5). Importantly, work in mice has shown that Ezh2 is required for normal B and T cell maturation (6), and biallelic deletion of Ezh2 leads to highly penetrant spontaneous T-ALL (7). Moreover, recent work associate loss of EZH2 with poor prognosis in T-ALL and acute myeloid leukemia (AML), mediated through an increase in chemoresistance (8–10), emphasizing the need to develop novel therapies for this high-risk subgroup of patients.
Synthetic lethal screening has shown great promise in identifying novel therapeutic targets in tumors deficient in a variety of tumor suppressors (11). Perhaps the best characterized synthetic lethal interaction, targeting the DNA repair pathway with PARP inhibitors in BRCA2 deficient cells (12,13), has translated into clinically meaningful benefits in patients with breast and ovarian cancer (14,15). Identification of synthetic lethal interactions arising from epigenetic deficiencies in cancer cells also offers promising therapeutic possibilities, such as the sensitization to WEE1 inhibition observed in tumors with SETD2 loss (16).
We hypothesized that EZH2 deficiency would render T-ALL cells dependent on distinct survival pathways that could be exploited to provide a novel, less-toxic and more efficacious therapeutic strategy for high-risk T-ALL. Through a targeted drug screen in isogenic T-ALL cells, we discovered EZH2-deficient T-ALL cells are highly sensitive to inhibition of the replication stress checkpoint kinase CHK1. Mechanistically, loss of EZH2 was associated with marked upregulation of MYCN, a significant increase in replication stress and dependency on CHK1 for cell survival.
Results
CHK1 inhibitor MK8776 selectively kills EZH2 deficient T-ALL cells
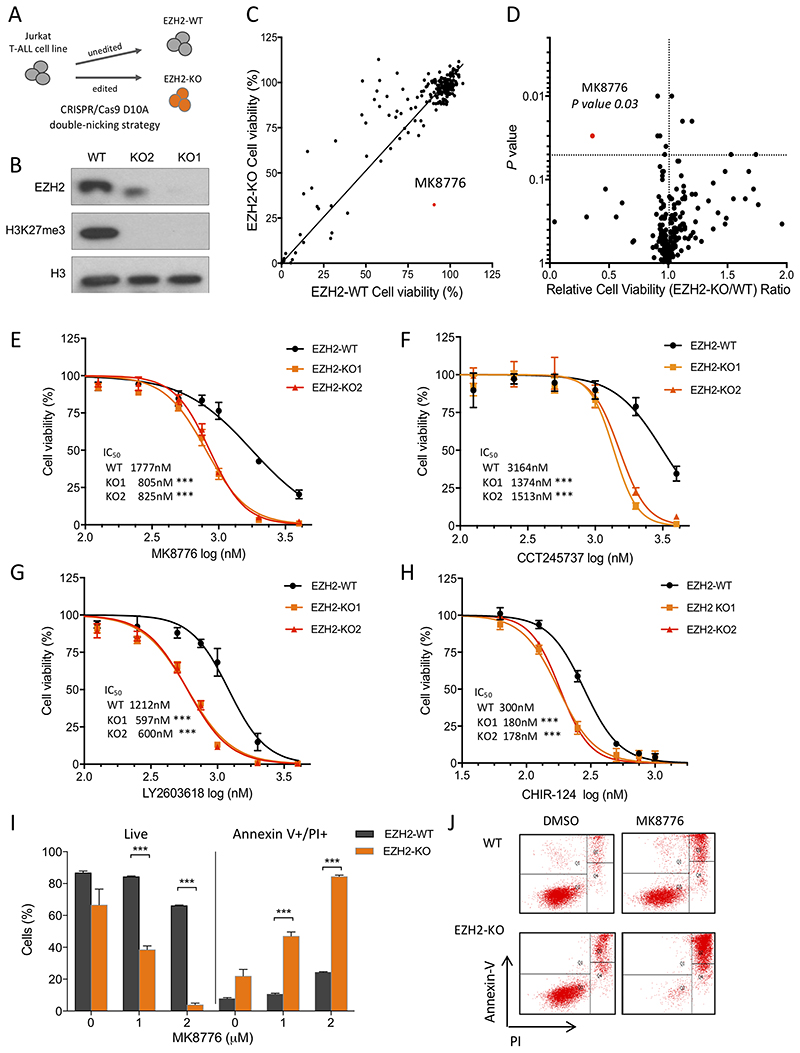
To investigate synthetic lethal interactions that could be exploited in PRC2 deficient T-ALL, we generated isogenic Jurkat T-ALL cells using a double-nicking CRISPR/Cas9 strategy targeting exon 2 of EZH2, an approach that minimizes off-target effects (17). We generated two clones with bialellic frameshift mutations in EZH2 (EZH2-KO1 and EZH2-KO2), that resulted in loss of EZH2 protein expression and a global absence of its functional mark H3K27me3 (Figures 1A and B; supplementary Figure S1A and B). EZH2 deficient clones had similar growth kinetics to their wild-type counterparts (supplementary Figure S1C, solid lines).
Figure 1. Jurkat T-ALL cells with CRISPR/Cas9-induced EZH2 mutations exhibit increased sensitivity to CHK1 inhibition.
A) Schematic diagram of the CRISPR/Cas9 double-nicking strategy used to obtain EZH2-knockout (KO) clones. B) Western blot from nuclear extracts prepared from Jurkat EZH2-WT and EZH2-KO cells, to confirm EZH2 deletion and global H3K27me3 abrogation. Total H3 is shown as loading control. C) Cell based drug screening was performed to test 219 small molecule inhibitors, incubating cells at 1μM dose of each compound for 72 hours. Cell viability of EZH2-WT vs EZH2 KO1 cell lines is shown as determined by CellTiter Glo assay (mean of two independent experiments, constrain linear regression curve is shown). D) Volcano plot representing the relative cell viability (EZH2-KO/WT) versus P value obtained from two-tail Student’s t-test. CHK1 inhibitor MK8776 highlighted in red. E-H) Dose-response of Jurkat EZH2-WT, EZH2-KO1 and EZH2-KO2 cells to four structurally different CHK1 inhibitors: (E) MK8776; (F) CCT245737; (G) LY2603618; (H) CHIR-124. Cells were incubated with drug for 72 hrs and then cell viability determined using the CellTiter Glo assay. Results shown are the mean ± SD from two independent experiments performed in triplicate; *** P < 0.001 by twoway ANOVA multiple comparisons test. I) Percentage of live (Annexin V-/PI-) and apoptotic (Annexin V+/PI+) cells as determined by flow cytometry following incubation with increasing doses of MK8776 (as indicated) for 48 hr, in EZH2-WT and EZH2-KO Jurkat cells performed in triplicate. Significance of difference was assessed by Student’s t-test; *** P value < 0.001. J) Annexin-V versus PI intensity FACS plots from one representative sample of each cell line and condition (DMSO or 2μM MK8776 treatment for 48 hrs).
We assessed the relative sensitivity of EZH2-KO (EZH2-KO1 cells, unless specified) and EZH2-WT cells to an in-house curated library of 219 commercially available compounds targeting specific chromatin modifying enzymes, cell cycle and DNA repair processes, including small-molecule inhibitors that are already in clinical use for the treatment of cancer (supplementary Table 1). The majority of drugs had similar effects on cell viability in both EZH2-WT and -KO cells (Figure 1C and D, and supplementary Table 2). However, MK8776, a CHK1 inhibitor that has been tested in early phase clinical trials in solid tumors (18), demonstrated a significant differential reduction on cell viability in EZH2-KO cells compared to WT cells (P = 0.03 by two-tailed Student’s t-test; Figure 1C and D). By evaluating the dose-response to MK8776, we observed that both EZH2-KO1 and EZH2-KO2 Jurkat clones had a significant reduction of the IC50 compared to EZH2-WT Jurkat cells (IC50 805nM, 825nM vs 1777nM respectively, P < 0.0001 by two-way ANOVA test; Figure 1E). Both EZH2 deficient clones displayed a consistent and similar sensitization to three structurally distinct CHK1 inhibitors, CCT245737, LY2603618 and CHIR-124, validating the specificity of this interaction (Figure 1F–H and supplementary Figure S2A). Importantly, the differential reduction in cell viability on CHK1 inhibition was mediated through a dose-dependent increase in apoptosis, as confirmed by Annexin V-PI staining (84% vs 25% Annexin V+/PI+ apoptotic cells in EZH2 deficient vs wild-type Jurkat cells; Figures 1I and J; supplementary Figure S2B).
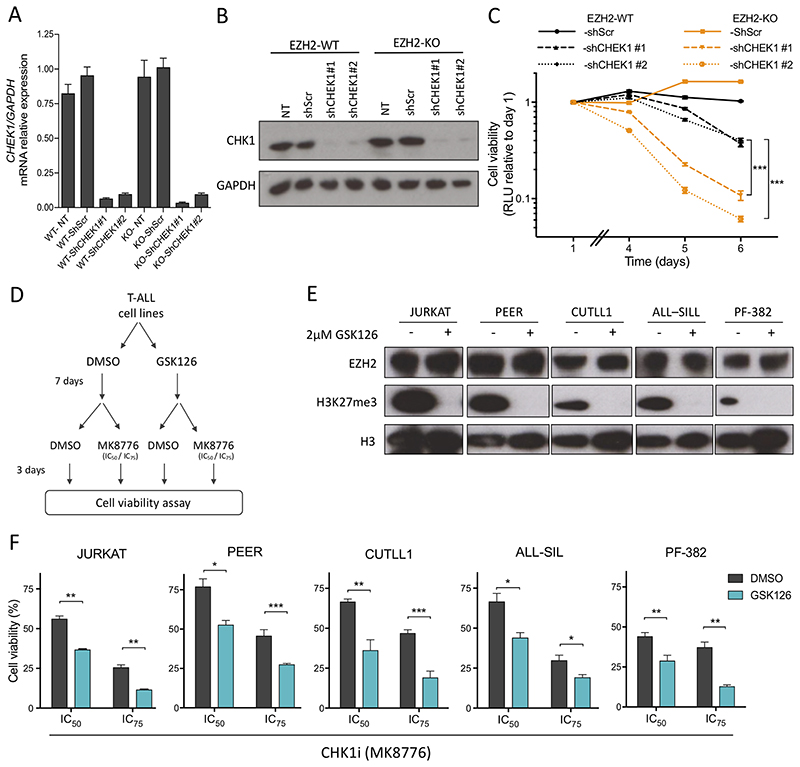
To further establish that the observed effects were specifically due to inhibition of CHK1, and not off-target drug activity, we transduced EZH2-KO and EZH2-WT Jurkat cells with two independent CHEK1 targeting shRNAs and confirmed equivalent knockdown by Q-PCR and western blotting (Figure 2A and B). Consistent with the effects we had observed with small molecule CHK1 inhibitors, shRNA knockdown of CHEK1 had more pronounced effects on impairing cell growth of EZH2 deficient cells as compared to their wild-type isogenic counterpart (Figure 2C).
Figure 2. The interaction of EZH2 activity and CHK1 sensitivity can be recapitulated both genetically and pharmacologically in genetically distinct T-ALL cell lines.
A-B) mRNA expression (A), and protein expression (B) of CHEK1 in Jurkat EZH2-WT and EZH2–KO cells, 72 hrs post infection with lentiviral shRNA constructs targeting CHEK1 (shCHEK1 #1 and #2) or scramble control (shScr). C) Growth curve of EZH2-WT and EZH2-KO Jurkat cells transduced with shRNAs targeting CHEK1 as measured by CellTiterGlo assay. Relative luminescent units (RLU) as compared to day 1. D) Schematic diagram of experimental design to test sensitization of T-ALL cell lines to CHK1 inhibitor, following EZH2 pharmacological inhibition with GSK126. E) Western blot assessing H3K27me3 abrogation in T-ALL cell lines, after seven days of GSK126 (2μM) pre-treatment. F) T-ALL cell lines pre-incubated for 7 days with GSK126, or DMSO control, were subsequently treated with MK8776 at their predetermined IC50 and IC75 for 72 hrs, and cell viability assessed using CellTiter Glo. Bar graphs represent mean of three independent experiments in triplicates. In (C) and (F) significance of difference was assessed by Student’s t-test; * P value <0.05; ** P value <0.01; *** P value < 0.001.
Pharmacological inhibition of EZH2 by GSK126 sensitizes T-ALL cells to CHK1 inhibitor
To assess whether our finding could be recapitulated in other T-ALL cell lines, a panel of T-ALL cells representing different genetic subgroups were tested for their response to MK8776 after pre-incubation with GSK126, a specific EZH2 inhibitor (Figure 2D). Notably, incubation of cells with GSK126 alone had no appreciable effect on cell growth (supplementary Figure S1C), EZH2 expression or CHK1 protein levels (supplementary Figure S3A). Following seven days of EZH2 inhibition, a global decrease in H3K27me3 mark in treated cell lines was confirmed by western blotting (Figure 2E). Consistent with findings in our EZH2-KO clones, pre-incubation of Jurkat cells with GSK126 significantly increased their sensitivity to CHK1 inhibition when exposed to MK8776 at doses corresponding their initial IC50 and IC75 (Figure 2F; supplementary Figure S3B). Similar sensitization to MK8776 upon GSK126 pre-incubation was observed in PEER, CUTLL1, ALL-SIL and PF-382 cell lines (Figure 2E and F; supplementary Figure S3B). These data not only support our results in isogenic EZH2-KO Jurkat cells, but also confirms the observed effects are due to EZH2 enzymatic activity, rather than non-canonical EZH2 function.
EZH2-KO cells experience increased levels of replication stress
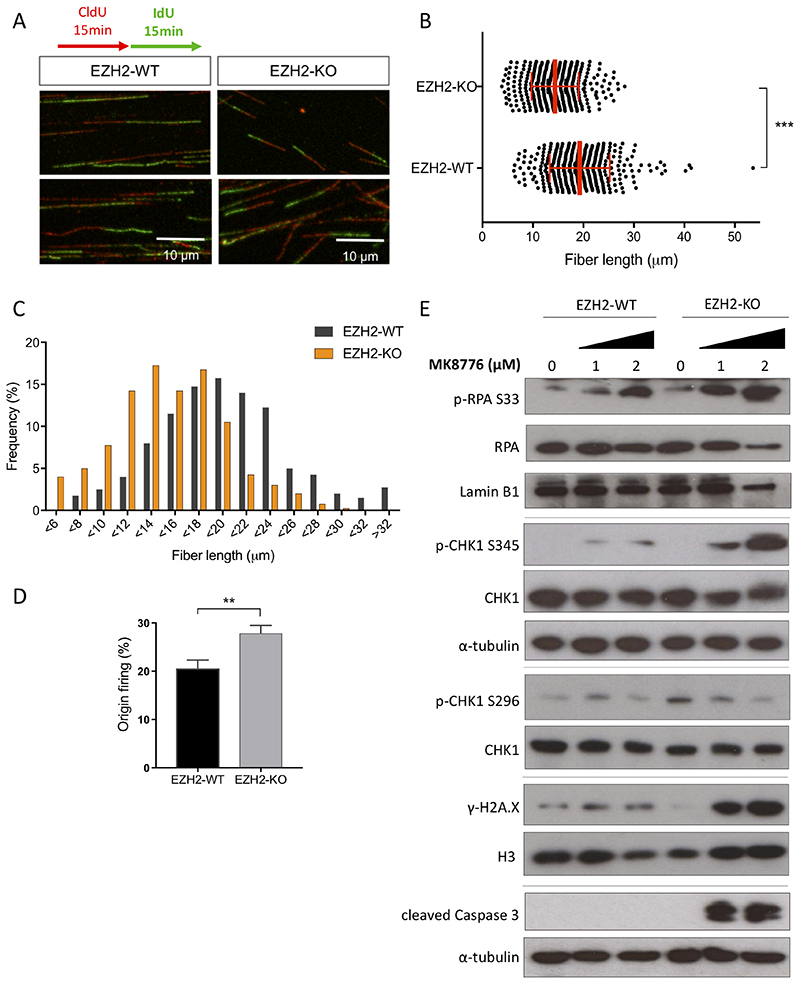
The replication stress checkpoint protein kinase CHK1 has a central role in the cellular response to replication stress (RS) and is essential to prevent replication stress-induced DNA damage (19–23). We hypothesized that EZH2-KO cells might be under higher levels of replication stress with increased dependency on CHK1 function for cell survival. Replication stress is characterized by the slowing and/or stalling of replication forks. To test our hypothesis, we used the DNA fiber assay to study replication fork dynamics in EZH2-WT and EZH2-KO cells at single molecule resolution. DNA replication tracks were labelled with the nucleotide analog CldU (red) followed by the second analog IdU (green; Figure 3A). These nucleotide analogs incorporated during ongoing replication were then visualized by confocal microscopy, and the length of replication tracks (red-green track) was measured. EZH2-KO cells showed a significantly greater population of short tracks in comparison with EZH2-WT cells (Figure 3B; 14.4 ±4.7 vs 19.3 ± 5.9 μm respectively, P <0.001 by two-tailed Student’s t-test) corresponding to a significantly reduced replication fork speed (Figures 3B-C). Moreover, a significantly higher percentage of origin firing (green-red-green tracks) was observed in EZH2 deficient cells compared with control (Figure 3D; P value = 0.003 by two-tailed Student’s t-test).
Figure 3. EZH2 inactivation results in increased DNA replication stress.
A) Representative images of DNA fibers in Jurkat EZH2-WT and EZH2-KO cells. Cells were pulsed with 5-chloro-2’-deoxyuridine (CldU) for 15 minutes following by 5-iodo-2’-deoxyuridine (IdU) for 15 minutes, isolated and DNA spread on slides. The CldU and IdU analogs were visualized with red and green antibodies respectively. B) Quantification of the length of 400 DNA fibers (ongoing structures: red track followed by a green track) measured in total from three independent experiments, in cells treated as in (A). Median and SEM are shown in red. ***P < 0.001 by two-tailed Student’s t-test. C) Replication speed, as indicated by the distribution of the replication length from fibers measured as in (B). D) Percentage of origin firing (green-red-green fibers) in relation to all ongoing fibers (bars represent the average from three independent experiments; **P value < 0.01 by two-tailed Student’s t-tests). E) Western blot of replication stress response proteins, from lysates of cells treated for 24h at concentrations of MK8776 as indicated. Lines indicate separate blots.
Slowing down of replication forks causes a decoupling between the DNA polymerases and the helicase that unwinds DNA, resulting in an accumulation of single stranded DNA (ssDNA). ssDNA is bound and protected by the ssDNA binding protein RPA, which serves as the signal for activation of the replication stress checkpoint protein kinase ATR and its downstream effector CHK1 (24–27). Activation of the replication stress checkpoint can thus be monitored by the phosphorylation levels of RPA, ATR and CHK1, which become phosphorylated upon checkpoint activation. Consistent with previous work showing that CHK1 inhibition increases activation of replication stress markers (28,29), CHK1 inhibition with MK8776 markedly increased phosphorylation of RPA (p-RPA S33) and CHK1 at S345 (pCHK1-S345), with concomitant reduction of CHK1 autophosphorylation point at S296 (p-CHK1 S296) (Figure 3E). This response was more robust in EZH2-KO cells in comparison with WT, suggesting that these cells experience elevated levels of replication stress (Figure 3E). Acute inhibition of CHK1 in cells experiencing replication stress has been shown to cause replication fork collapse whereby replisome components dissociate from the DNA, leading to double strand breaks and increased apoptotic death (30). In line with this, EZH2-KO cells, but not EZH2-WT cells, showed increased double strand breaks as determined by TUNEL (supplementary Figure S4A) and phosphorylation of histone H2AX (γH2AX), with cleavage of caspase 3 after exposure to MK8776 (Figure 3E), further supporting an essential role of the replication checkpoint response for their survival.
Given the close links between the DNA replication stress and DNA damage checkpoints (31), we examined whether inhibition of the checkpoint protein kinases CHK2, ATM and ATR could also induce synthetic lethality in T-ALL cells lacking a functional PRC2. Inhibition of the DNA damage checkpoint kinases, CHK2, by the specific inhibitor CCT241533, or ATM, by Ku-55933, did not differentially affect EZH2-KO cells compared with WT (supplementary Figure S4B and C). In contrast, EZH2 deficient cells were more sensitive to AZ20, a selective inhibitor of the replication stress checkpoint protein kinase ATR (supplementary Figure S4D), indicating that EZH2 loss specifically sensitizes cells to inhibition of the ATR-CHK1 pathway, rather than the CHK2-ATM DNA damage response pathway. Taken together, these data suggest that loss of EZH2 leads to increased levels of replicative stress in T-ALL cells, which results in an increased dependency on the replication stress checkpoint response.
CHK1 inhibition in EZH2-KO cells induces apoptosis in S and G2 phases
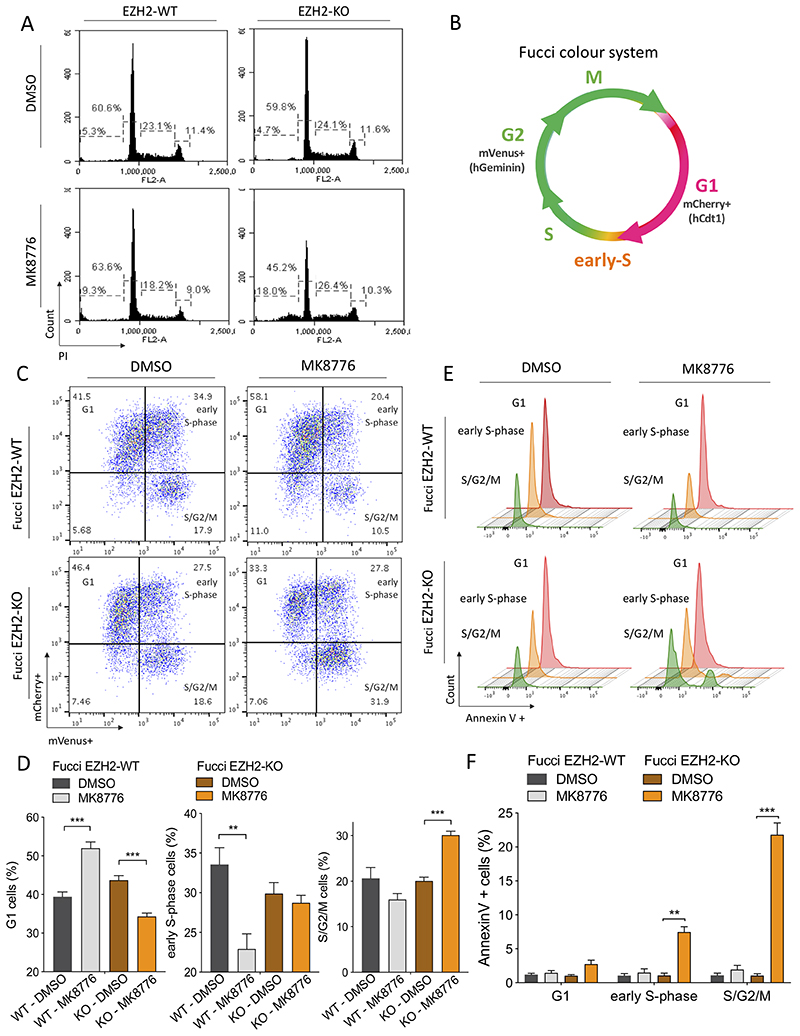
We postulated that interrogation of cell cycle dynamics coupled with apoptotic profiling could provide more insight into the mechanism by which CHK1 inhibition preferentially affects EZH2 deficient cells. Firstly, we analyzed whether lack of EZH2 induces cell cycle alterations in Jurkat cells. Flow cytometric analysis of EZH2-WT and KO cells showed a similar cell cycle pattern, and the percentage of cells at G1, S and G2/M was comparable in both cell lines (Figure 4A, upper panel). However, MK8776 treatment induced an increased proportion of subG1 population in EZH2-KO cells (4.7% in DMSO control vs 18% in MK8776 treated), indicative of cell death, accompanied by a decrease in the G1 population (59.8% in DMSO control vs 45.2% in MK8776 treated) that was not observed in WT control cells (Figure 4A, lower panel).
Figure 4. EZH2 prevents cells from an S-phase/G2 apoptosis upon CHK1 inhibition.
A) Cell cycle analysis using propidium iodide (PI) staining on EZH2-WT and EZH2-KO Jurkat cells treated with 1μM MK8776 or DMSO control for 24 hrs. B) Fucci2 color diagram showing the cell cycle stages that can be distinguished with this system. C) mCherry versus mVenus fluorescence intensity plots obtained from flow cytometry for FucciEZH2-WT and FucciEZH2-KO Jurkat cells, treated with DMSO or MK8776 (2μM) for 20 hrs. One representative image is shown from three independent experiments performed in duplicate. D) Quantification of FucciEZH2-WT and EZH2-KO Jurkat cells in G1, early-S-phase and S/G2/M, incubated with DMSO or MK8876 (2μM) for 20 hrs. Error bars indicate mean ± SD. **P < 0.01; ***P < 0.001 by two-tailed Student’s t-test. E) Representative Annexin V+ flow cytometric histograms of FucciEZH2-WT and FucciEZH2-KO Jurkat cells following DMSO or MK8776 (2μM) treatment, showing apoptotic cells in relation to their stage of the cell cycle. F) Quantification of Annexin V+ cells, in FucciEZH2-WT and FucciEZH2-KO Jurkat cells, following CHK1 inhibition by MK8776 (2μM). Error bars indicate mean ± SD. **P < 0.01; ***P < 0.001 by two-tailed Student’s t-test.
To analyze this in finer detail, we made use of the fluorescent ubiquitination-based cell cycle indicator system (Fucci2), based on the expression of two chimeric proteins (mCherry-hCdt1 and mVenus-hGeminin) that accumulate reciprocally in the nuclei of transduced cells during the cell cycle (32). In this system, the red fluorescence of mCherry becomes detectable in early G1 phase, the mVenus fluorescence appears in early S-phase and disappears rapidly in late M phase, and during the G1/S transition both fluorophores are present (Figure 4B). When Fucci-EZH2-WT Jurkat cells were incubated with MK8776 we observed an increased percentage of cells at G1, accompanied by a decreased percentage of cells at early S-phase, indicative of G1 phase arrest. Interestingly, MK8776 did not induce this effect in EZH2-KO cells which instead accumulated in S/G2/M (Figure 4C and D). Importantly, we observed that upon exposure to the CHK1 inhibitor, there was a significant increase in apoptosis in Fucci-EZH2-KO cells but not in WT cells, with Annexin V+ staining occurring in early S-phase, at a time-point when replication forks are first formed, and then notably in cells in S/G2/M, where we also observed increased cleaved Caspase 3 (Figure 4E and F; supplementary Figure S5A).
Together with our previous data, these results are consistent with the notion that upon CHK1 inhibition, EZH2-KO cells are unable to prevent excessive levels of replication stress, leading to apoptosis in S/G2/M. In contrast, the lower levels of replication stress experienced by EZH2-WT cells upon inactivation of the replication checkpoint response allows cells to enter M phase, probably with small under-replicated regions, which can be detected and repaired by the DNA damage checkpoint in G1 (33–35).
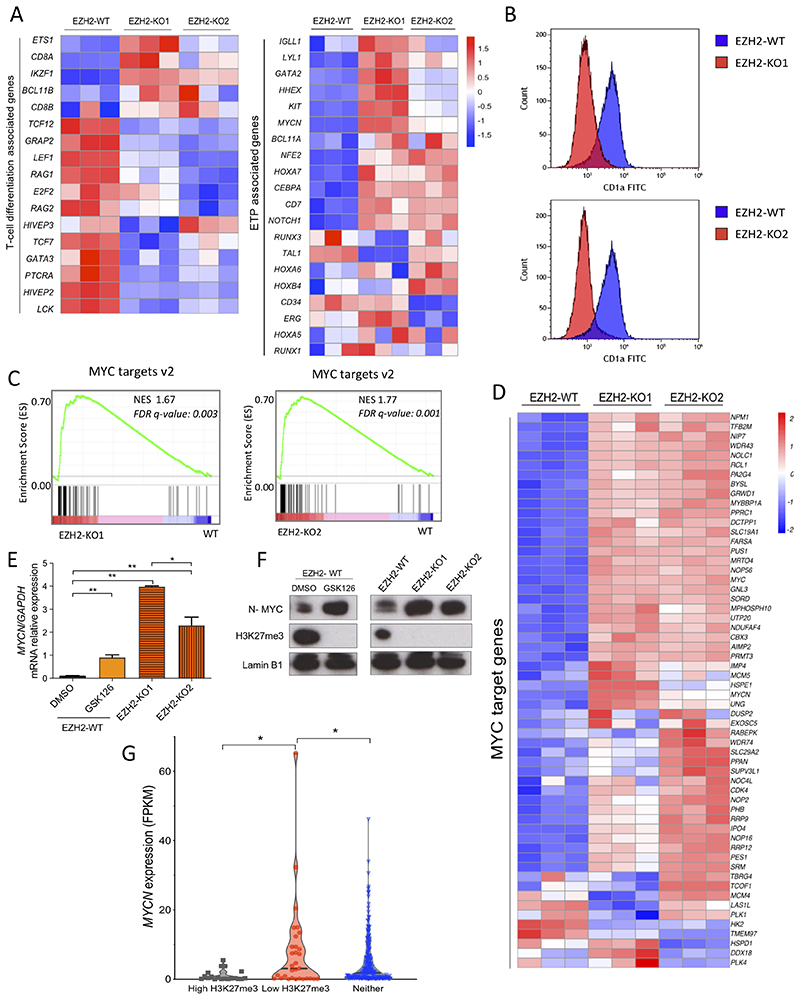
EZH2 loss leads to an ETP-ALL signature and increased expression of MYCN
Given the role of the epigenetic mark H3K27me3 in transcriptional repression, we next evaluated whether the observed phenotype in EZH2-KO cells was due to changes in gene expression. Through RNA sequencing (RNA-seq) of EZH2-WT, EZH2-KO1 and EZH2-KO2 Jurkat cells, we observed a marked downregulation of genes associated with T-cell differentiation (36,37) such as RAG1, PTCRA and LCK (Figure 5A, left panel), and these changes were also observed phenotypically as indicated by downregulation of the cortical cell surface marker CD1a when analyzed by flow cytometry (Figure 5B). Moreover, gene set enrichment analysis (GSEA) (38) in EZH2-KO cells versus EZH2-WT control showed significant enrichment of genes upregulated in ETP-ALL as compared to more mature T-ALL cases (supplementary Figure S6A) (1), with notable upregulation of early thymic genes such as LYL1, HHEX, KIT and MYCN (Figure 5A, right panel). When these gene expression data were compared with those from normal thymic subtypes (37) using principal component analysis (PCA), EZH2 deficiency reflected the disparity between less differentiated thymic subsets (Thy 1-3) as compared to more mature ones (Thy 4-6) at a global transcriptional level (principal component 2, supplementary Figure S6B). We hypothesized that a similar mechanism may occur in T-ALL patients, whereby loss-of-function mutations in PRC2 are acquired at a relatively advanced thymic developmental stage but cause reprogramming to give an ETP immunophenotype. To explore this possibility we analyzed the V(D)J recombination status of the T-cell receptor γ (TRG) gene in a published dataset of pediatric ETP-ALL (1). Recombination of TRG occurs at the pro-T stage of T-cell development (39), such that cells that have undergone biallelic deletion at the TRG locus have matured well beyond the ETP stage to the cortical stage of thymic development (supplementary Figure S6C) (40). Remarkably, of the 22 cases of ETP-ALL that also had loss-of-function mutations in PRC2 members, 13 (59%) had biallelic TRG deletions (1), consistent with our hypothesis (supplementary Figure S6D).
Figure 5. EZH2 loss leads to an ETP-ALL signature and increased expression of MYCN.
A) Differential mRNA expression heatmap of T-cell differentiation and ETP associated genes (as per Casero et al., 2015 and Ha et al., 2017) from RNA-seq data of EZH2-WT versus EZH2-KO1 and EZH2-KO2 Jurkat cells (n=3). B) Flow cytometric histograms for the cortical thymic marker CD1a in both EZH2-KO1 and EZH2-KO2 Jurkat cell lines in comparison with EZH2-WT isogenic control cells. C) Gene set enrichment analysis (GSEA) of MYC target genes v2 from EZH2-KO1 and EZH2-KO2 versus EZH2-WT Jurkat cells. NES, normalized enrichment score; FDR q-value, false discovery rate q value. D) Heatmap visualization of the upregulation of MYC target genes v2 (as per the Molecular Signatures Database) in EZH2-KO1 and EZH2-KO2 Jurkat cells as compared to Jurkat EZH2-WT cells. E) MYCN mRNA expression (relative to GAPDH) analyzed by Q-PCR of Jurkat EZH2-WT cells treated with EZH2 inhibitor GSK126 (2μM) or DMSO (control) for 7 days and compared with Jurkat EZH2-KO1 and EZH2-KO2 clones. **P < 0.01 by two-tailed Student’s t-test. F) Western blot of lysates of MYCN, H3K27me3 and Lamin B1 loading control from cells treated as in (E). G) MYCN expression in FPKM from the pediatric and young adult T-ALL patients cohort reported in Liu et al., 2017. Patients were sub grouped according to the presence of mutations (identified by exome sequencing) or deletions (based on DNA copy number analysis of SNP array data) potentially leading to low H3K27me3 levels (Low H3K27me3, n = 31), mutations or deletions leading to high H3K27me3 (High H3K27me3, n = 19) or neither of them (Neither, n = 210). Statistical significance was assessed by two-tailed Student’s t-test. *P value < 0.05.
Strikingly, GSEA using the MSigDB Hallmark gene set collection (41) identified MYC target genes ("MYC target v2’) as significantly enriched in both EZH2-KO clones compared to the parental line, indicating that genetic inactivation of EZH2 results in accentuated expression of these genes (Figure 5C and D). While we observed a modest 2-fold increase in MYC expression (supplementary Figure S6E), there was a more striking upregulation of MYCN, which was 18 and 10-fold upregulated in EZH2-KO1 and EZH2-KO2 cells respectively by RNA-seq (1.5 vs 28.1 and 15.6 FPKM respectively; supplementary Figure S6E). Furthermore, upregulation of MYCN could be recapitulated in parental cells treated with the EZH2 inhibitor GSK126 (Figure 5E) and this was confirmed at the protein level by western blotting (Figure 5F). Altogether, these data suggest that loss-of-function mutations in EZH2 acquired during intrathymic T cell maturation may cause epigenetic reprograming, shaping their transcriptional pattern towards a less differentiated phenotype, with derepression of early thymic genes such as MYCN.
To ascertain whether an inverse association is observed between H3K27me3 levels and MYCN expression in patient samples, we analyzed gene expression and mutational data from a large cohort of pediatric T-ALL patients (3). Patients with mutations predicted to lower H3K27me3 levels had significantly higher levels of MYCN expression as compared to those with mutations predicted to lead to high H3K27me3 (7.9 vs 1.1 FPKM respectively; P value = 0.029 by two-tailed Student’s t-test). Furthermore, patients with low H3K27me3 had a trend towards higher MYCN expression than those patients who did not have a defined mutation in PRC2 components ("neither group’, which likely represent patients with a wide variety of H3K27me3 levels; 7.9 vs 4.8 FPKM; P value = 0.046; Figure 5G and supplementary Table 3). This result is supported by previous work in transgenic mice, where there is approximately 4-fold upregulation of Mycn in Ezh2 KO T helper CD4+ cells compared with their WT counterpart (supplementary Figure S6F) (42).
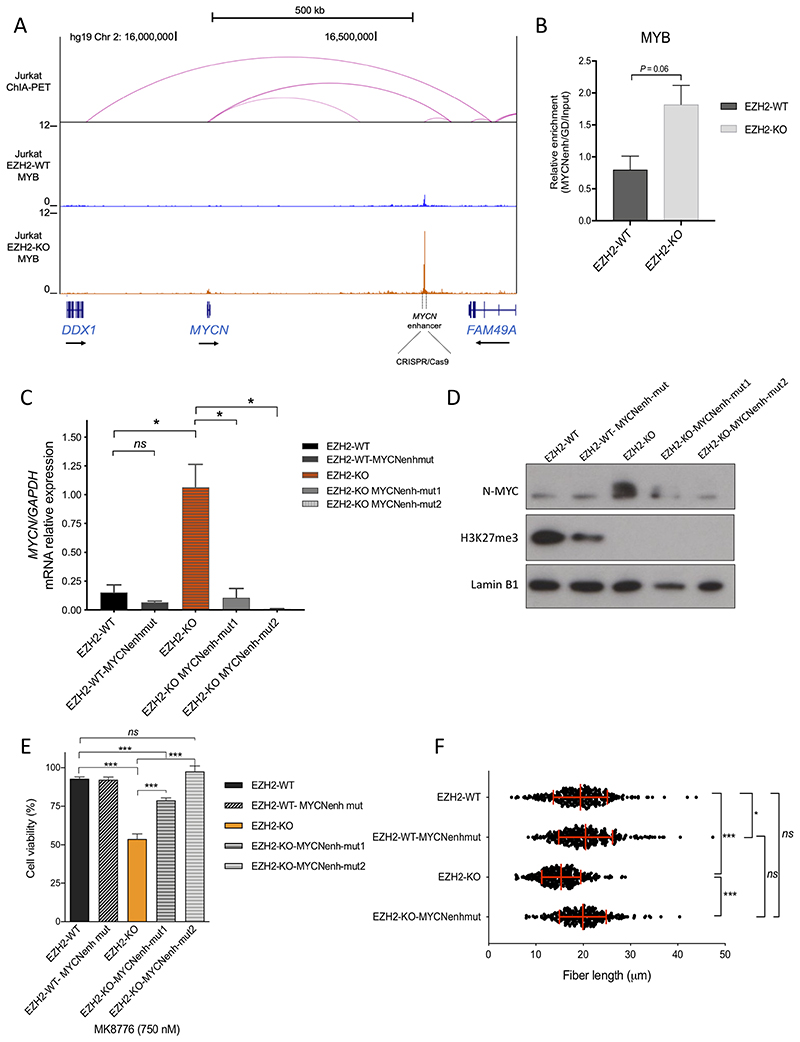
Activation of a distal MYCN enhancer contributes to CHK1 sensitivity in EZH2 deficient cells
To evaluate the contribution of MYCN to CHK1 dependence in EZH2-KO cells, we first attempted to knockdown MYCN with shRNAs. However, we were unable to expand EZH2-deficient cells after MYCN shRNA knockdown, consistent with the oncogene addiction model (43).
Given we had previously identified MYB as a key regulator of the majority of active enhancers in T-ALL cells (44), we interrogated MYB binding around the MYCN locus by ChIP-seq. Within the MYCN topologically associated domain (TAD), as defined in Jurkat cells by ChIA-PET for the cohesin subunit SMC1A (45), we noted a site of MYB occupancy +540 Kb distal to the MYCN transcriptional start site that appeared significantly enriched in EZH2-KO as compared to EZH2-WT cells (Figure 6A), and we were able to validate differential MYB binding by ChIP-qPCR (Figure 6B). To ascertain whether increased MYB enrichment at this site was responsible for MYCN upregulation, we performed CRISPR/Cas9 genome editing to delete approximately 400 bp of this region, by targeting two guide RNAs flanking either side of the MYB binding peak (supplementary Figure S7A and B). While there was no significance difference in MYCN expression upon MYCN enhancer deletion in EZH2-WT clones, there was a striking downregulation of MYCN mRNA and protein levels in EZH2-KO-MYCN-enhancer deleted clones (Figures 6C and D), confirming this site acts as a MYCN enhancer in these cells.
Fig. 6. MYCN overexpression contributes to CHK1 sensitivity in EZH2 deficient cells.
A) ChIP-seq tracks for MYB at the MYCN locus in EZH2-WT and EZH2-KO Jurkat cells. Dotted lines flanking the MYB peak at +540 Kb downstream MYCN, indicate the targeted region (aprox. 400 bp) deleted using CRISPR/Cas9 RNP strategy. Y-axis values are reads per million mapped reads. The cohesin (SMC1) ChIA-PET interactions from Hnisz et al., 2016 are displayed above the ChIP-seq profiles (intensity of pink arc represents interaction score). B) ChIP-Q-PCR analysis of MYB enrichment at the MYCN enhancer locus. Bars represent the mean and SEM from two independent ChIP-Q-PCR experiments. P = 0.06 by two-tailed Student’s t-test. C) MYCN mRNA expression relative to GAPDH in EZH2-WT, EZH2-WTMYCNenhancer-mutant, EZH2-KO, and EZH2-KO-MYCNenhancer-mutant cells, from Q-PCR analysis performed in duplicates, *P< 0.05 by two-tailed Student’s t-test from two independent experiments. D) Western blot showing N-MYC and H3K27me3 expression in lysates from EZH2-WT, EZH2-WT-MYCNenhancer-mutant, EZH2-KO, and EZH2-KO-MYCNenhancer-mutant cells. Lamin B1 is shown as loading control. E) Cell viability assay using CellTiter Glo, after 72 hours incubation with MK8776 (750nM) in EZH2-WT, EZH2-WT-MYCNenhancer-mutant, EZH2-KO, and EZH2-KO-MYCNenhancer-mutant cells. Bar graphs represent mean ±SEM of two independent experiments performed in triplicates. ***P < 0.001 by two-tailed Student’s t-test. F) Quantification of the length of 300 DNA fibers (ongoing structures: red track followed by a green track) measured in total from two independent experiments, in EZH2-WT, EZH2-WT-MYCNenhancer-mutant, EZH2-KO1, and EZH2-KO-MYCNenh-mut2 cells, treated as in Figure 3A. Median and SEM are shown in red. ***P < 0.001, *P < 0.05 by two-tailed Student’s t-test.
To further evaluate the contribution of MYCN levels to CHK1 dependency, we then tested the sensitivity of these cells to CHK1 inhibition. Deletion of the MYCN enhancer in EZH2-KO cells partially rescued cell viability when incubated with MK8776 at a dose surrounding the IC50 (Figure 6E), confirming that increased expression of MYCN through activation of the +540 kb enhancer plays a predominant role in sensitization to CHK1 inhibition. We next evaluated the replication fork speed in MYCN-enhancer deleted cells. Confirming our initial observation, EZH2-KO cells had significantly shorter DNA fiber length compared to EZH2-WT (P < 0.0001, Figure 6F). However, EZH2-KO-MYCN-enhancer deleted cells showed a significant recovery of replication fork speed, similar to that observed in EZH2-WT control cell lines (P = 0.25, Figure 6F). Taken together, these results support the hypothesis that MYCN mediates enhanced replication stress in EZH2-deficient cells.
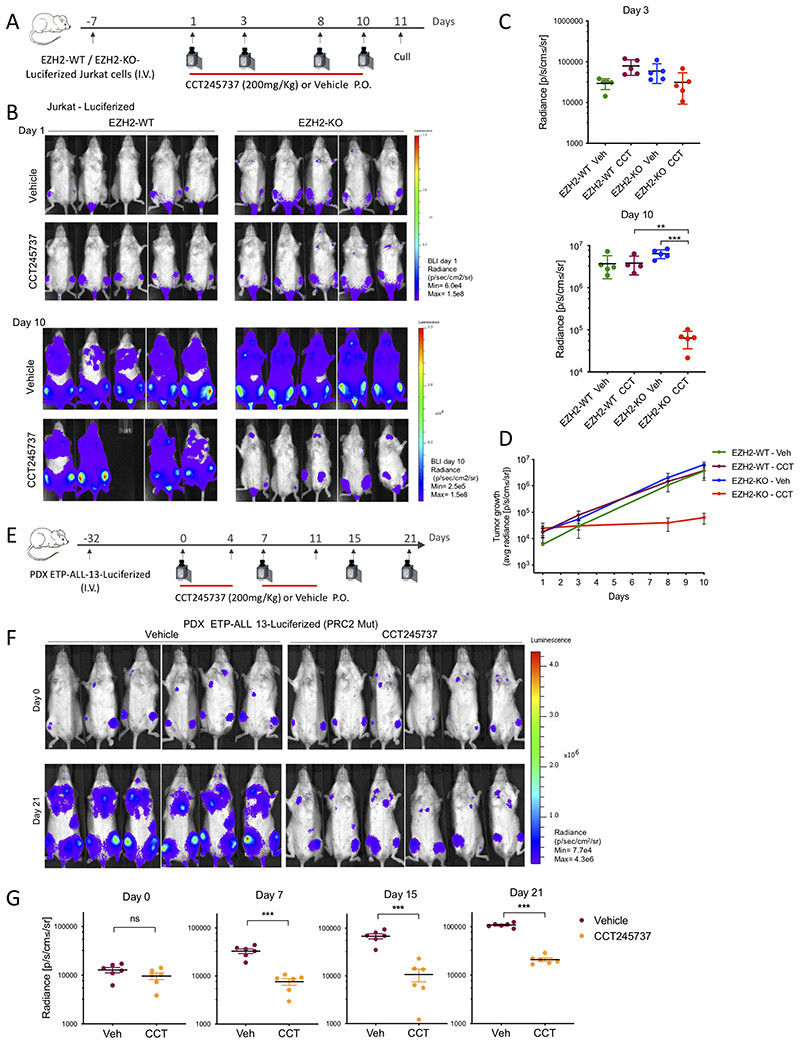
CHK1 inhibitor CCT245737 reduces tumor growth of PRC2-mutated T-ALL in vivo
To test whether our observations would translate into a meaningful benefit in vivo, we used the potent CHK1 inhibitor CCT245737, because of its excellent oral bioavailability and significant antitumor activity as a single-agent in an Eμ-Myc driven mouse model of B-cell lymphoma (46). We verified target engagement of CCT245737 in vivo using tumor homogenates tested for CHK1 autophosphorylation at S296 by ELISA and western blotting, as it is the most sensitive and specific biomarker of CHK1 kinase activity (46) (supplementary Figure S8A and B).
To generate an in vivo model, EZH2-WT and EZH2-KO Jurkat cells were transduced to stably express firefly luciferase, engrafted and harvested from the bone marrow of NSG female mice, and tested for their response to CCT245737 ex vivo (EZH2-WT/KO-luciferized Jurkat cells; supplementary Figure S8C). Intravenously injected cells in NSG female mice were allowed to grow for 8 days, at which point engraftment in bone marrow was confirmed in all mice through bioluminescent imaging (BLI) (Figure 7A). Mice were treated daily with oral CCT245737 or vehicle control for 10 days and tumor burden was reassessed by BLI. Strikingly, CCT245737 treatment significantly reduced leukemia growth in mice engrafted with EZH2-KO cells, with a two-log reduction in average radiance compared to vehicle treated (P < 0.001 by two tailed Student’s t-test) (Figure 7B–D). In contrast, CHK1 inhibitor treatment of tumors generated from EZH2-WT cells did not have any significant effect on tumor growth (Figure 7B–D).
Fig. 7. CHK1 inhibitor reduced tumor growth of PRC2-mutated T-ALL.
A) Schematic diagram of the Jurkat in vivo experiment protocol. Female NSG mice were I.V. injected with EZH2-WT- or EZH2-KO-Luciferized Jurkat cells, and at 8 days post-injection animals were administered CCT245737 (200mg/Kg P.O.) or vehicle daily for 10 days. Mice were culled 24h after the last treatment. Red bar represents CCT245737 or vehicle treatment days. Camera, BLI imaging; I.V., intravenous tail vein injection; P.O., oral dosing. B) BLI of mice at days 1 and 10 of treatment. BLI Day 1 radiance (p/sec/cm2/sr) scale: min= 6.4e4, max= 1.5e8. BLI Day 10 radiance (p/sec/cm2/sr) scale: min= 2.5e5, max= 1.5e8. p, photon; sr, steradian. n = 5 mice per group C) Radiance from individual mice at Day 3 (upper panel) and Day 10 (lower panel) following CCT245737 (200mg/kg) or Vehicle treatment. ***P < 0.001; **P = 0.002 by Student’s t-test. D) Tumor growth observed during the treatment period in mice engrafted with EZH2-WT- or EZH2-KO-Luciferised Jurkat cells. Each point represents the average radiance ± SEM from each group, at the indicated time points. E) Schematic diagram of the PDX experiment protocol. Female NSG mice were I.V. injected with PDX ETP-ALL-13-Luciferized cells, and at 32 days post-injection animals were administered CCT245737 (200mg/Kg P.O.) or vehicle daily for 2 weeks, 5 days per week. Red bar represents CCT245737 or vehicle treatment days. Camera, BLI imaging; I.V., intravenous tail vein injection; P.O., oral dosing. F) BLI of mice injected with patient derived xenograft (PDX) ETP-ALL 13-Luciferized tumor cells, at day 0 and day 21 after beginning of treatment. BLI radiance (p/sec/cm2/sr) scale: min= 7.7e4, max= 4.3e6. p, photon; sr, steradian. n = 6 mice per group G) Radiance from individual mice at days 0, 7, 15 and 21 following CCT245737 (200mg/kg) or Vehicle treatment. ***P < 0.001 by Student’s t-test.
To assess whether this differential effect could translate into a therapeutic window, we treated a separate cohort of mice and monitored their survival rate. In line with clinical observations, EZH2-KO cells were highly aggressive, leading to significantly shorter survival rates compared to their EZH2-WT counterparts when treated with vehicle alone (P<0.001). CCT245737 treatment resulted in a survival benefit only in mice bearing EZH2-KO tumors (supplementary Figure S8D, right panel, P < 0.001) while no differences in survival was observed in mice engrafted with EZH2-WT cells (supplementary Figure S8D, left panel, P value 0.98). This confirms that the sensitization we had observed in vitro translates to an in vivo benefit, at tolerated doses and in a more relevant tissue microenvironment.
To further evaluate the therapeutic potential of targeting CHK1 in a model of endogenous PRC2 deficiency, we next tested an ETP-ALL patient derived xenograft (PDX) carrying an inactivating mutation in the PRC2 component EED (47), and transduced to stably express firefly luciferase, ETP-ALL-13-Luciferized (see Methods). Importantly, ETP-ALL-13 expresses similar levels of MYCN when compared with EZH2-KO Jurkat cells (supplementary Figure S8E). We injected PRC2-mutant ETP-ALL-13-Luciferized into NSG mice and upon confirmation of engraftment (4.5 weeks post-injection), started treatment with CCT245737 or vehicle (5 days per week for two weeks, starting at Day 0) (Figure 7E). CCT245737 treatment significantly inhibited tumor growth, as observed by reduction of average radiance compared to the vehicle treated group (P < 0.001 by two tailed Student’s t-test on Days 7, 15 and 21) (Figures 7F and G).Taken together, these data identify CHK1 inhibition as an exploitable therapeutic approach to target PRC2 deficient T-ALL associated with high levels of MYCN and replication stress.
Discussion
One of the main obstacles in cancer treatment is the inability to selectively and effectively target cancer cells, while leaving healthy cells unharmed. The discovery of novel synthetic lethal interactions offers the potential of exploiting a previously unappreciated therapeutic window, enabling personalized treatment for cases with specific genetic alterations. The data presented here demonstrates that EZH2 activity, or H3K27me3 status, is a key molecular determinant for MYCN expression, and thereby CHK1 inhibitor sensitivity in T-ALL. Our findings fit a model whereby loss-of-function mutations in EZH2, the enzymatic component of PRC2, lead to increased expression of the oncogenic transcription factor MYCN, causing oncogene-induced replication stress, rendering cells more dependent on CHK1 activity for replication fork integrity. Overexpression or constitutive activation of oncogenes such as MYC or MYCN is one of the most relevant causes of oncogene-induced replication stress in human cancer (48–51). Previous studies have indicated that over-replication, an increase in the number of active replication forks, is the most likely mechanism of MYC-induced replication stress (52,53). Moreover, DNA replication deregulation due to increased expression of MYC proteins has been proposed to mediate the sensitivity to CHK1 inhibition in murine models of MYC-driven lymphoma and MYCN-driven neuroblastoma, a finding that has prompted testing of these agents in clinical trials for MYCN-amplified neuroblastoma (54–56).
We found that genetically inactivating EZH2 in T-ALL cells led to marked changes in its gene expression pattern, with downregulation of expression of T-cell differentiation associated genes and concomitant up-regulation of ETP signature genes. This finding is in line with previous work that associate EZH2 loss with stem-cell-associated transcriptional programs (57), and in correlation with the high frequency of genetic lesions in PRC2 components observed in the ETPALL subtype of T-ALL (1,3). Furthermore, these results suggest that PRC2 loss-of-function mutations acquired in T-cell lineage cells of a relatively advanced thymic developmental stage may cause epigenetic deregulation leading to aberrant expression of myeloid and stem cell markers characteristic of less differentiated subtypes. Therefore, the increased frequency of PRC2 mutations within the ETP-ALL subgroup of T-ALL might be a consequence of such epigenetic and transcriptional reprogramming, explaining the observation that these phenotypically immature ALL cases have frequently undergone the types of T-cell receptor rearrangement characteristic of more differentiated T-ALL subtypes (1,4,58).
Although the exact mechanism of cellular reprogramming that occurs with PRC2 loss will require further study, the enrichment of MYB binding we observed at the +540 kb MYCN enhancer in EZH2-KO cells suggests that PRC2 is involved in maintaining repression of key regulatory sites normally silenced during T-cell development. Using CRISPR/Cas9 engineering, we were able to demonstrate the importance of the +540 kb MYB-bound MYCN enhancer on regulation of MYCN expression and replication stress, in turn enabling us to conclude that MYCN overexpression is the major contributor to CHK1 sensitivity.
Given the role of CHK1 in mediating cell cycle arrest during the DNA-damage response, CHK1 inhibitors have undergone clinical testing in certain solid tumors in combination with DNA-damaging agents such as gemcitabine and radiotherapy (59,60). This mechanism of action seems distinct from the single agent efficacy of CHK1 inhibition in MYC-driven models, where CHK1 activity is required to tolerate high levels of oncogenic replication stress. While CHK1 inhibitors have shown efficacy as monotherapy in preclinical studies of T-ALL (61), the biomarkers identifying those cases most likely to respond have remained elusive (62). Our data suggests a clinically exploitable therapeutic window in T-ALL cases with the highest levels of replication stress, occurring through EZH2-deficiency, low H3K27me3 and high MYCN. These results indicate that CHK1 inhibitors should be considered for clinical testing in such T-ALL cases, which currently do poorly with standard chemotherapy approaches.
Materials and Methods
Experimental Models and Subject Details
Patient-Derived Xenograft
The patient-derived xenograft ETP-ALL-13 (SJTALL004) was established from a 3-year old male, and carries a mutation in the PRC2 complex gene EED (EED S259F) that impairs trimethylation of lysine 27 in Histone 3 (47).
Cell line cultures
All T-ALL cell lines used in this study (Jurkat, PEER, CUTLL1, ALL-SIL, PF-382) were obtained from DSMZ and ATCC, and cultured at 37 °C with 5% CO2 in RPMI-1640 medium supplemented with 10% v/v fetal bovine serum (FBS) and penicillin (100 units/ml)/streptomycin (0.1 mg/ml). HEK-293T cells were maintained in Dulbecco’s modified Eagle’s medium supplemented with 10% FBS, L-glutamine, and penicillin/streptomycin. All cell lines were routinely tested for mycoplasma using EZ-PCR Mycoplasma Detection Kit (Biological Industries) and the identity of T-ALL cell lines used in this study were verified by short tandem repeat analysis using the PowerPlex 1.2 system (Promega) in June 2017. Experiments were performed within four weeks after fresh viable cells were thawed.
Animal model
All animal protocols were approved by the UK Home Office. Mice were maintained in individually ventilated cages in a specific-pathogen-free facility. Female NSG mice were obtained from Charles River, and assigned randomly to control and experimental groups. All mice had ad libitum access to feed.
Generation of isogenic EZH2-KO cells by CRISPR-Cas9 double-nicking strategy
The px335-CRISPR-Cas9-D10A plasmid was obtained from Addgene (Cat. 42335) and modified to express BFP or GFP by the UCL-CI Genomics and Genome Engineering Core Facility. A pair of gRNAs targeting exon 2 of EZH2, obtained from MIT CRISPR design tool (http://crispr.mit.edu/) (gRNA #1: ACACGCTTCCGCCAACAAACTGG, quality score: 84; gRNA #2: TGCGACTGAGACAGCTCAAGAGG, quality score: 74) were each cloned into either BFP-px335 or GFP-px335 respectively. The plasmids were electroporated into Jurkat cells using AMAXA Nucleofector (Lonza), and BFP/GFP double positive cells were single cell sorted by FACS (BD FACSAria™ Fusion, BD Biosciences) into 96 well plates and incubated under standard tissue culture conditions in RPMI supplemented with 10% FBS. Once the clones had grown to confluency, genomic DNA was extracted using the QuickExtract DNA Extraction solution (Epicentre) following manufacturer’s instructions and clones were screened for mutations by genomic PCR and Sanger sequencing. Briefly, PCR was performed using primers located approximately 150 bp on each side of the target site (Fw: 5’-TGATTGTTAGTTTGCTGCGGA; Rv: 5’ CAGATCAAGAACCTAAGCTTCCA). The PCR product was cleaned by Qiagen spin column and sequenced by Sanger sequencing. Oligos cloned into BFP-px335, Ex2_EZH2 G1-UP: 5’-CACCGACACGCTTCCGCCAACAAAC, Ex2_EZH2 G1-DOWN: 5’-AAACGTTTGTTGGCGGAAGCGTGTC. Oligos cloned into GFP-px335, Ex2_EZH2 G2-UP: 5’-CACCGTGCGACTGAGACAGCTCAAG, Ex2_EZH2 G2-DOWN: 5’-AAACCTTGAGCTGTCTCAGTCGCAC.
Genomic validation of CRISPR EZH2 knockout cells
The CRISPR knockout cells were validated at the DNA level, analyzing the mutations on both alleles by TOPO-cloning. Briefly, genomic DNA was extracted from Jurkat parental and Jurkat CRISPR EZH2-knockout cells using Qiagen DNeasy kit, and the targeted EZH2 region amplified by PCR using the screening primers described above. PCR products were subcloned into a TOPO vector using TOPO-TA cloning kit (Invitrogen) and transformed into competent cells. Six colonies from each cell line were mini-prep and sequenced separately by Sanger sequencing. Sequences were aligned using Geneious R9 multiple sequence alignment tool. The amino acid translation was also checked and compared with the transcript reference sequence obtained from Ensemble (www.ensemble.org).
Synthetic lethal drug screening of small molecule inhibitors
An in-house curated epigenetic library (Selleckchem; Cat. L1900) of 219 commercially available compounds specifically targeting chromatin modifying enzymes, cell cycle and DNA repair processes, including small-molecule inhibitors already in clinical use in cancer chemotherapy, was obtained at a stock concentration of 10mM in DMSO (supplementary Tables 1 and 2). Cell based drug screening was performed in 96 well plates, incubating the cells with a working concentration of 1 μM of each compound for 72 hours. Cell viability was measured using CellTiterGlo (Promega) as per manufacturer instructions. VarioSkan plate reader (ThermoScientific) was used to determine relative luminescence units (RLU) and cell viability was calculated relative to vehicle treated samples.
Cell Viability Assays
T-ALL cell lines were cultured (20,000cells/well) in 96 well white plates for luminescence assays (CulturePlateTM, PerkinElmer) and treated with the indicated range of doses of CHK1 inhibitor for 72 hrs. Cell viability was determined using CellTiter-Glo Luminescent Viability Assay (Promega) as directed and luminescence measured using Varioskan LUX Microplate Reader (Thermo Scientific).
Apoptosis analysis using Annexin V/ PI staining by flow cytometry
The FITC Annexin V/Dead Cell Apoptosis Kit (Invitrogen; Cat. V13242) with FITC annexin V and PI for flow cytometry was used as per manufacturer’s instructions. Briefly, after incubation of cells with MK8776 or DMSO (control) as indicated in Figure legends, cells were washed in PBS and resuspended in 1X annexin-binding buffer (100 μl per assay). 5μl of FITC annexin V and 1μl of 100μg/ml PI working solution were added to each cell suspension and incubated for 15 minutes. After this time, 400μl of 1X annexin-binding buffer was added and samples were kept on ice until analyzed by flow cytometry.
Cell Cycle analysis by PI staining using flow cytometry
After incubation of cells with MK8776 or DMSO (control) as indicated in Figure legends, cells were washed in PBS and resuspended to fix in 1 ml of cold 70% methanol (SIGMA). Methanol solution was added drop wise to the pellet while vortexing, to ensure fixation of all cells and minimize clumping. Following fixation overnight at −20 °C, cells were washed twice in PBS and treated with RNase A (SIGMA) by adding 50μl of 0.1mg/ml solution to pelleted cells, and 300 μl of 50μg/ml propidium iodide (PI) solution. Cells were incubated for 5 minutes at room temperature and cell cycle was analyzed by flow cytometry.
Western blot
Protein lysates were prepared from 4×106 cells, in either Nuclear Extraction Reagents (Pierce Biotechnology) or pre-chilled RIPA buffer (150 mM NaCl, 50 mM Tris-HCl pH 8, 0.5% sodium deoxycholate, 0.1% SDS, 1% Triton X-100). Buffers were supplemented with protease and phosphatase inhibitors (Sigma; Roche). Protein concentrations were determined using BCA Protein Assay kit (Pierce). Cell lysates were separated by SDS-PAGE electrophoresis (NuPAGE; 4%–12% Bis‐ Tris gels or NuPAGE; 3-8% Tris-Acetate Protein Gels; Invitrogen). Spectra Multicolor Broad Range Protein Ladder was used as a size marker. Proteins were transferred to a 0.22μm PVDF membrane (Maine Manufacturing), pre-activated with 100% Methanol, using the Trans–Blot SD semi–dry Transfer Cell (Bio–Rad). Membranes were blocked with 5% bovine serum albumin (BSA) in PBS 0.1% Tween 20 (PBS-T) and incubated overnight with a primary antibody in 2.5% BSA or milk at 4°C. Membranes were then washed in PBS-T, before probing with the appropriate HRP-conjugated secondary antibody in 2.5% BSA or milk- PBS-T for 1 hr. Membranes were washed and developed on X ray film using either Pierce ECL western blotting substrate (Thermo Scientific) or Luminata Forte Western HRP Substrate (Millipore). In some cases, membranes were stripped using Restore PLUS Western Blot Stripping Buffer (Thermo Scientific) and re-probed with another primary antibody.
The following antibodies purchased from Cell Signaling were used in this study: EZH2 (Cat. 5246), Tri-Methyl-Histone H3 (Lys27) (Cat. 9733), Histone H3 (Cat. 4499), CHK1 (Cat. 2360), GAPDH (Cat. 5174), Lamin B1 (Cat. 12586), N-Myc (Cat. 9405), Phospho-CHK1 S345 (Cat. 2348), Phospho-CHK1 S296 (Cat. 90178), Phospho Histone H2A.X S139 (Cat. 9718), Cleaved Caspase3 (Cat. 9664), RPA32 (Cat. 2208). Phospho RPA32 (S33) antibody was purchased from Bethyl (Cat. A300-246A) and monoclonal Anti-α-Tubulin antibody was purchased from Sigma Aldrich (Cat. T9026).
ELISA assay to validate CCT245737 target engagement in vivo
ELISA assay for phospho-CHK1 S296 and S317 were performed using PDX ETP-ALL13 tumor homogenates, from mice at 13 weeks post-IV injection of tumor cells. Spleen tissue was harvested 4 hours following CCT245737 or Vehicle P.O. dosing as indicated. ELISA assay was based on commercially available kit PathScan Phospho-CHK1 (S317) Sandwich ELISA (Cell Signaling Technology), and was used as previously reported (46). For phospho-CHK1 S296 detection the commercial phospho-CHK1 S317 antibody was replaced with phospho-CHK1 S296 (Cat. 90178). Tumor homogenates were added to 96 well plates (50μg protein) and quantification of the colorimetric readout was determined at 450nm using a Multiskan FC (Thermo Scientific) plate reader. Confirmation of phospho-CHK1 S296 levels in homogenates was also performed by western blotting using the same antibody.
shRNA knockdown using Lentivirus
The target sequences of CHEK1 shRNAs used (shCHEK1 #1: 5’-GAAGGCAATATCCAATATTTA; and shCHEK1 #2: 5’-GTGGTTTATCTGCATGGTATT) were obtained from the Broad Institute Genetic Perturbation Platform (GPP web portal https://portals.broadinstitute.org/gpp/public/). Sequences were cloned into pLKO.1 vector (63) and lentiviral particles were produced as previously described (64). Briefly, lentivirus was generated by transfecting H293T cells with each pLKO-1 shRNA vector and the packaging plasmids VsVg and psPAX2 using GeneJuice (Merck-Millipore) as per manufacturer instructions. The virus–containing medium was collected at 48h post transfection, filtered through a 0.45μM nitrocellulose membrane filter and stored at 4°C. 8 x 105 target Jurkat cells were resuspended in virus media, polybrene (Millipore) was added at 8μg/ml and spinfection was carried out at 2,500rpm for 90minutes at 37°C. Following overnight incubation, cells were resuspended in fresh complete medium and plated in 96 well plates to assess cell viability. For protein and mRNA expression analysis, cells were harvested 72hours after spinfection.
Quantitative Real-Time Polymerase Chain Reaction (Q-PCR)
Total RNA was extracted with RNeasy Mini Kit (Qiagen) as per manufacturer’s protocol and concentrations were measured on a Nanodrop 1000 spectrophotometer (Thermo Scientific). For two-step Q-PCR, cDNA was synthesized initially using Omniscript RT Kit (Qiagen) and 500ng input RNA was used for each reaction. FastStart Universal SYBR Green Master (ROX) mix was used as per manufacturer’s protocol and samples were run on a Mastercycler epgradient S thermocycler (Eppendorf). Normalized expression ratios were calculated by the relative standard curve method whilst using GAPDH as the endogenous reference mRNA. Primer pairs for CHEK1 were Fw: 5’-ATATGAAGCGTGCCGTAGAC and Rv: 5’-TGCCTATGTCTGGCTCTATTC. Primer pairs for the housekeeping gene GAPDH were Fw: 5’-TGCACCACCAACTGCTTAGC and Rv: 5’-GGCATGGACTGTGGTCATGAG. Primers used for MYCN were Fw: 5’-GACTGTAGCCATCCGAGGAC, Rv: 5’-AATGTGGTGACAGCCTTGGT.
Generation of Jurkat-Fucci cells
The CSII-EF-MCS vectors encoding mCherry-hCdt1(30/120) and mVenus-hGem(1/110) were obtained from Atsushi Miyawaki at the RIKEN BioResource Research Center. Both vectors were co-transfected with the packaging plasmids psPAX2 (Addgene, Cat. 12260) and VSV-G (Addgene, Cat. 14888) into 293T cells to produce lentiviral particles. Viral solutions were prepared and used for transduction into EZH2-WT and EZH2-KO1 Jurkat cells. Stably transformed cells were established by FACS, and mCherry+/mVenus+ double-positive cells were bulk sorted (BD FACSAria™ Fusion, BD Biosciences) to ensure a homogeneous population expressing both cell cycle indicators.
Annexin V and cleaved Caspase-3 analysis of Jurkat-Fucci cells
For Annexin V staining, following overnight (18 hrs) incubation of cells with MK8776 (2μM) or DMSO (control), cells were collected and washed with PBS, stained using Pacific Blue-Annexin V (BioLegend; Cat. 640917) and analyzed by flow cytometry using a BD FACSymphonyTM cytometer (BD Biosciences). 10,000 events were recorded, and doublets were excluded. For cleaved-Caspase-3, cells were fixed with 3% methanol-free formaldehyde (Thermo Scientific Pierce) for 10 minutes, and permeabilized with NP40 (0,2% in PBS) for 10 minutes. Following a PBS wash, cells were incubated with a V450 rabbit anti-Active-caspase-3 antibody (BD Horizon) and analyzed by flow cytometry using BD FACSymphonyTM.
TUNEL assay
Detection of DNA break sites in cells was achieved by Terminal Deoxynucleotide Transferase dUTP Nick End Labelling or TUNEL using APO-BrdU TUNEL assay Kit (Invitrogen) following manufacturer’s instructions. Briefly, following incubation with MK8776 or DMSO at indicated times, cells were washed in PBS and resuspended in 1% (w/v) paraformaldehyde in PBS, placed on ice for 15 minutes. Following a wash with PBS, cells were resuspended in ice-cold 70% ethanol and stored in −20C freezer until TUNEL staining. Control and experimental samples were washed and incubated with DNA-labeling solution 2 hours at 37C. Following washes with rinse buffer, cells were incubated with antibody solution for 30 minutes at room temperature, and propidium iodide/RNase A staining buffer was added for another 30 minutes. Cells were analyzed by flow cytometry using BD FACSymphonyTM.
DNA fiber experiments
Exponentially growing Jurkat cells were pulse labelled with 25 μM CldU followed by 250 μM IdU for 15 min each. Labelling reaction was stopped by placing cells on ice. Cells were pelleted, washed in cold PBS immediately and resuspended in PBS at a final concentration of 100 x 104. 2μl of cell suspension was placed on each slide and allowed to partially dry for maximum 5 min, afterwards 7μl of spreading buffer (200mM Tris pH 7.4; 50mM EDTA; 0.5% SDS) was added to the cells and incubated for 2 minutes, then the slides were tilted to allow the stretching of DNA fibers along the slide. After drying for 2 min at room temperature, slides were fixed in Methanol/Acidic acid (3:1 volume) for 10 minutes, air-dried and placed either at 4°C for storage, or treated for immunofluorescence detection. From this time onwards, the slides were protected from direct light. Slides were denatured by incubation at room temperature in freshly made 2.5 M HCl for maximum 1hour 15min. After repeated wash in PBS, slides were incubated in blocking buffer (PBS, 1% BSA and 0.1% Tween) for 30 min. Incubation with primary antibodies rat anti BrdU (AbD SeroTec; Cat. OBT0030G), and mouse anti BrdU (Becton Dickinson; Cat. 347580), 1:750 and 1:100 in blocking buffer respectively, was carried out overnight at 4°C. Washes in PBS were followed by an incubation with 4% Formaldehyde for 10 min. Slides were washed in PBS and a final wash in blocking buffer. Incubation with secondary antibodies Alexafluor 488-conjugated goat anti-mouse IgG (Molecular Probes) and Alexafluor 555-conjugated goat anti-rat IgG (Molecular Probes), 1:500 in blocking buffer, was carried out for 1 hour at room temperature. Slides were washed in PBS and mounted with mounting medium (SIGMA; Cat. F6182). Imaging was performed using a Leica TCS SPE microscope with a 63X objective. The lengths of CldU (AF 555, red) and IdU (AF 488, green) labelled patches were measured using the ImageJ software (http://rsb.info.nih.gov/ij/). Origin structures (green-red-green) were quantified as a percentage of all on-going structures (red-green).
RNA sequencing
Jurkat EZH2-WT, EZH2-KO1 and EZH2-KO2 RNA was extracted from 1,000,000 cells per sample using the RNeasy Mini Kit (Qiagen). For library preparation, samples were processed using the KAPA mRNA HyperPrep Kit (Cat. KK8580) according to manufacturer’s instructions. Briefly, mRNA was isolated from total RNA using Oligo dT beads to pull down poly-adenylated transcripts. The purified mRNA was fragmented using chemical hydrolysis and primed with random hexamers. Strand-specific first strand cDNA was generated using Reverse Transcriptase in the presence of Actinomycin D. The second cDNA strand was synthesised using dUTP in place of dTTP, to mark the second strand. The resultant cDNA was then “A-tailed” at the 3’ end to prevent self-ligation and adapter dimerisation. Truncated adaptors, containing a T- overhang are ligated to the A-Tailed cDNA. Successfully ligated cDNA molecules were then enriched with limited cycle PCR (13 cycles using 500ng of total RNA). High fidelity polymerase was used in the PCR to make the library strand specific. Samples were sequenced on the NextSeq 500 instrument (Illumina) using a 43bp paired end run. Run data were demultiplexed and converted to Fragments Per Kilobase of sequence per Million mapped reads (FPKM) files using Illumina’s BaseSpace RNA-Seq Alignment and Cufflinks Assembly & DE applications.
RNA-seq analysis
Gene set enrichment analysis was performed using GSEA software (v 3.0) using the Hallmark gene sets obtained from the Molecular Signatures Database (MSigDB v4.0) (41). ETP-ALL gene sets were derived from the top 500 upregulated genes in ETP-ALL relative to non-ETP T-ALL samples (1,65). Heatmaps were generated in R with the pheatmap package. The FPKM gene expression data is scaled by row for the plot. For PCA analysis Casero et al. data (37) was downloaded from the GEO public repository (Accession GSE69239) as was processed by the authors. Both their data and ours were normalised using FPKM. Data for PCA contains only the genes that are expressed in at least one sample on both datasets. PCA was generated in R with the prcomp package.
For analysis of the T-ALL patients cohort (3), FPKM matrix was obtained from their published supplementary materials. The relevant RNAseq sheet was exported as a tab delimited file for analysis within R. Patients were sub grouped according to the presence of mutations (based on the sequence of mutations identified by exome sequencing) or deletions (based on DNA copy number analysis of SNP array data) potentially leading to low H3K27me3 levels (Low H3K27me3), mutations or deletions leading to high H3K27me3 (High H3K27me3) or neither of them (Neither) (supplementary Table 3). A Student’s T-test comparing the subgroups was performed using GraphPad Prism v7. The violin plots were then generated using the ggplots2 package.
ChIP-sequencing
For MYB ChIP-seq a ChIPmentation protocol was adapted from (66). Briefly, protein A Dynabeads (Invitrogen) were washed with PBS/BSA 0.5% using lo-bind tubes (Eppendorf). After washing, Dynabeads were resuspended in PBS/BSA 0.5%, 2μg of antibody added (MYB Millipore Mouse IgGak 1μg/μl) and incubated on an orbital shaker for 2.5hours at 4°C. 5 million cells were harvested per IP and washed twice in PBS. Cells were double cross linked in PBS with 2mM disuccinimidyl glutarate (Sigma, Cat. 80424) for 45 mins, with the addition of formaldehyde to 1% w/v final for the final 15 mins. Cross linking was quenched with glycine to a final concentration of 0.125M. Cells were washed with ice cold PBS plus 1X complete proteinase inhibitor (Sigma, Cat. 11836145001) and 5mM sodium butyrate (Sigma, Cat. B5887) and resuspended in 100μl sonication buffer (Tris pH 8.0 10mM, NaCl 100mM, EDTA 1mM, EGTA 0.5mM, sodium deoxycholate 0.1%, 0.5% sodium N-lauryl sarcosine, plus proteinase inhibitors and butyrate). Cells were sonicated in a Diagenode Bioruptor Pico® using lo-bind tubes (Eppendorf). Sonicated material was centrifuged at 14000g at 4°C for 10mins and supernatants transferred to a fresh tube. 2% of the sonicated chromatin was removed for input controls. The samples were incubated at 65°C overnight in elution buffer (Tris pH8.0 50mM, EDTA 10mM, SDS 1%) with the addition of proteinase K (20μg/μl) and acidified with 10μl 3M sodium acetate prior to purification over a MinElute® column (Qiagen). Sonication efficiency was assessed by 2% agarose gel and eluted DNA quantified by Qubit™ (Invitrogen). The antibody-Dynabead conjugates were washed 3 times in PBS/BSA. The remainder of the sonicated chromatin was adjusted to a volume of 100μl, 10μl of 10% triton X-100 was added and then 10μl of washed beads added per IP. The chromatin-bead mix was incubated overnight at 4°C on an orbital shaker. The following day, the antibody-bead-chromatin mix was washed 3 times in wash buffer (Hepes pH 7.6, EDTA 1mM, LiCl 0.5M. sodium deoxycholate 0.7%, NP-40 1%), followed by 2 washes in 10mM Tris pH8.0 before resuspension in 25μl of tagmentation reaction mix (10mM Tris pH8.0, 5mM MgCl2, 1μl Nextera tagmentation enzyme (Illumina, Cat. FC-121-1030)). Chromatin was tagmented for 10mins at 37°C in a thermocycler, and reaction was stopped by addition of ChIP dilution buffer (Tris pH8.0 10mM, NaCl 100mM, EDTA 1mM, EGTA 0.5mM, sodium deoxycholate 0.1%, N-lauryl-sarcosine 0.5%, SDS 0.1%), followed by two further washes in ChIP dilution buffer. Beads were then washed twice with low salt TE (Tris pH8.0 10mM, EDTA 1mM, NaCl 50mM) before decrosslinking in 48μl elution buffer supplemented with 2μl proteinase K (20μg/μl) incubation at 55°C for 1 hour and 65°C overnight. All buffers prior to this were supplemented by proteinase inhibitor and sodium butyrate as above. The next day, beads were magnetised, and supernatants saved to fresh lo-bind tubes. The remaining beads were further digested in 19μl elution buffer with 1μl proteinase K (20μg/μl) at 55°C for one hour. The supernatants were pooled, acidified with sodium acetate and purified over a MinElute® column (Qiagen) into 22μl Qiagen elution buffer EB. 2μl of each IP was test-amplified by real time Q-PCR in a 10μl reaction (2xKAPA HiFi ready mix (pre-heated to 98°C for 30s), Primer Ad1-Ad2.1 each to final concentration of 0.75 μM, 1:10000 dilution of SYBR dye (Thermo, Cat. S7563)) using the following programme (72°C 5mins, 98°C 30s; 25 cycles of 98°C 10s, 63°C 30s, 72°C 30s; 72°C 1min). Fluorescence was plotted using a linear scale against cycle number, and the number of cycles that resulted in 50% total amplification chosen for amplification of the bulk ChIP DNA. The remaining DNA was amplified in the following conditions (25μl KAPA HiFi 2x master mix (pre-activated), 20μl ChIP DNA, Nextera primer Ad1/Ad2.x at 0.75 μM, were Ad2 primers were indexed per sample in a 50μl reaction volume) and cycled as above to the number of cycles judged from the above Q-PCR. Input controls were tagmented in the following conditions (2.5ng input DNA, 1μl tagmentation enzyme, 1μl 5x tagmentation buffer (Tris pH8.0 50mM, MgCl2 25mM) in a total volume of 5μl at 55°C for 5 mins. Tagmentation was stopped by the addition of 1μl 0.6% SDS at room temperature for 5mins. Inputs were amplified in the following conditions (25μl KAPA HiFi 2x master mix (pre-activated), 1.5μl Ad1 primer, 1.5μl Ad2.x index primer, 25μl, 6μl tagmented DNA in a 50μl reaction) for 12 cycles as above. The resulting PCR products were purified using 90μl magnetic SPRI beads (Agencourt AMPure XP Beckman Coulter, Cat. A63881) and after two 80% ethanol washes were resuspended in 50μl water. Size separation was performed by incubation with 32.5μl SPRI beads, transfer of supernatant to a new tube and then addition of 12.5μl fresh SPRI beads. Following two further 80% ethanol washes the size selected libraries were eluted into 15μl water. Libraries were checked and quantified by Qubit™ (Invitrogen) and Tapestation (Agilent). Normalised libraries were pooled and sequenced using a NextSeq® (Illumina) 75bp paired end short read sequencer using standard protocols.
ChIP-seq analysis
ChIP sequencing data was aligned to the hg19 human assembly using bwa-mem (v0.7.15). BigWig files for viewing ChIP alignments were created using deeptools bamCoverage (v3.0.2) with bin height scaled to expected read counts using the Counts Per Million mapped reads (CPM) method (67). For Figure 6A, Cohesin (SMC1) ChIA-PET interactions in Jurkat (two replicates) from (45) were visualized on the WashU Genome Browser (68). Only the high confidence interactions are displayed between paired-end tags (PET) peaks (3 PET, > 5kb, FDR < 0.01), with an interaction score (number of PETs) threshold set at 5.
Generation of isogenic EZH2-KO-MYCN enhancer mutant cells by CRISPR-Cas9 RNP strategy
To delete the putative MYCN enhancer in EZH2-WT and EZH2-KO1 Jurkat cells, the Alt-R CRISPR RNP system from IDT was used which is comprised of three components: crRNA, tracrRNA, Cas9. We used the crRNA:tracrRNA duplex, in which crRNA must be used in conjunction with tracrRNA to form a functional gRNA duplex. In the crRNA:tracrRNA duplex, tracrRNA serves as a universal 67mer tracrRNA that is attached to a fluorophore-ATTO550, while the crRNA provides sequence specificity against the target gene. We designed two guide RNAs (crRNA) flanking the observed MYB peak (Chr2: 16,438,737-16,439,346; hg19) using MIT CRISPR design tool (http://crispr.mit.edu/) (Guide #1: GGCTTTGCTCAATGTGGTAC; Guide #2: TAATCCCCAAAAACTCCATC). The crRNAs were incubated with tracrRNA at 95°C, for 5 minutes to form a duplex. This was subsequently incubated with recombinant Cas9 protein at room temperature for 20 minutes to form the RNP complex. The RNP complex was then delivered to Jurkat EZH2-KO cells by electroporation using AMAXA nucleofector I (Lonza). Following electroporation, cells were recovered in 2 ml pre-warmed RPMI complete medium and were incubated at 37°C overnight. The following day, electroporated cells were single cell sorted into 96-well plates using BD FACS Aria™ Fusion flow cytometer. The electroporation efficiency was determined by the percentage of ATTO 550 positive cells, which was about 80%. 96-well plates were incubated at 37°C for three weeks to allow formation of colonies. Genomic DNA from clones was extracted using Quick DNA extract solution (Epicentre), and PCR screening was performed to amplify across the gRNA break sites (primer pairs used Fw: 5’- GAAGTGGTTTCCCCACCACA; Rv: 5’-TAGCCCTAGTTGAAGCCCCA) products were run on 1.5% agarose gel. The expected PCR product size was 604 bp in Jurkat parental lines with intact MYCN enhancer. The CRISPR/Cas9-induced excision of the enhancer was expected to result in a 144 bp PCR product. Sanger sequencing of the PCR products of EZH2-KO-MYCNenh-mut2 showed the upper band (~604bp) contained an inversion of the WT sequence, resulting in a compound heterozygous clone.
In vivo experiments
For the in vivo study using isogenic cell lines, Jurkat EZH2-WT and EZH2-KO cells were firstly transduced to stably express firefly luciferase and engrafted once in female NSG mice by intravenous (I.V.) injection of 2.5 million cells. After 3 weeks post-injection, luciferase expressing Jurkat cells were harvested from the bone marrow of mice and selected in puromycin containing medium (EZH2-WT- and EZH2-KO-luciferized Jurkat cells).
For in vivo administration of CCT245737 (APExBIO; Cat. B8223), female NSG mice were I.V. injected with 2.5 million EZH2-WT- or EZH2-KO-luciferized Jurkat cells. 100 mg/ml stock of CCT245737 prepared in DMSO (SIGMA) was diluted 1:10 before dosing into pre-filtered PTW (20% PEG400 (Qiagen), 5% Tween80 (SIGMA), in sterile water), and administered daily by oral gavage at the indicated doses and points. Control mice received the same volume of diluted DMSO in PTW (Vehicle).
Engraftment and tumor burden were assessed by bioluminescent imaging (BLI), with mice anesthetized and imaged using a Xenogen IVIS Imaging System (Perkin Elmer Life Sciences) following intraperitoneal injection of D-luciferin (VWR; Cat. HISM1-360223-200).
For the PDX in vivo model, ETP-ALL-13 cells harvested from the spleen of an engrafted mouse, were transduced to stably express firefly luciferase and engrafted back in female NSG mice by I.V. injection of 4 million cells. After 8 weeks post-injection, luciferase expressing ETP-ALL-13 cells (ETP-ALL-13-Luciferized-mCherry+) were harvested from the spleen of a mouse and sorted by flow cytometry to ensure a homogeneous population expressing luciferase was obtained. ETP-ALL-13-Luciferized cells were then expanded in two NSG female mice before using them to test CCT245737. For in vivo administration of CCT245737 in this model, female NSG mice were I.V. injected with ~0.4 million ETP-ALL-13-luciferized cells. Four weeks post-injection, engraftment was confirmed by BLI (as described above) and 200mg/Kg CCT245737, prepared in DMSO and diluted 1:10 into pre-filtered PTW before dosing, was administered for 10 days by oral gavage (5 consecutive days per week, for 2 weeks). Control mice received a similar volume of vehicle.
Quantification and Statistical Analysis
Details of statistical tests used are indicated in the Figure Legends. Statistical analyses were performed using GraphPad Prism v7 or Microsoft Excel. Student’s T- test was used to compare experimental groups as indicated in the Figure legends. Survival curves were compared using the log-rank test. Sensitivity of experimental groups to CHK1 inhibitors in in vitro dose-response assays were compared using the two-way ANOVA (multiple comparison) test. Numbers of replicates for each experiment are indicated in the Figure Legends.
Supplementary Material
Statement of significance.
Loss-of-function mutations of PRC2 genes are associated with chemotherapy resistance in TALL, yet no specific therapy for this aggressive subtype is currently clinically available. Our work demonstrates that loss of EZH2 activity leads to MYCN-driven replication stress, resulting in increased sensitivity to CHK1 inhibition, a finding with immediate clinical relevance.
Acknowledgments
Authors acknowledge University College London (UCL) Genomics team for their support with RNA-sequencing, UCL-Cancer Institute Flow Cytometry Core Facility and UCL-Biological Services (Mr. G. Martin) for technical assistance, Dr. Atsushi Miyawaki from RIKEN Brain Science Institute for kindly providing the Fucci2 plasmids. Dr. David Teachey, Dr. Adam Mead and Dr. Chris Booth for providing the PDX ETP-ALL-13, and Prof. Richard Jenner, Dr Alejandro Gutierrez and Dr Alex Kentsis for insightful discussions about this work.
Financial support
This work was funded by Blood Cancer UK (M.R.M. and S.R.), Freemason’s Grand Charity (M.M.), the Hospital Saturday Fund, UCL/UCLH Biomedical Research Centre and Cancer Research UK (CRUK) Funding. T.E.L. was funded by The Prince Fund. T.R.D.S. was funded by the Carol’s Smile Charity. N.F. was funded by Great Ormond Street Hospital Children’s Charity and a Cancer Research UK Clinical PhD studentship. S.A. was funded by Hardy Keinan Fellowship. S.E.R. was funded by a UK NIHR Clinical Lectureship. J.A.G.A. was funded by the CRUK-UCL Centre Award. R.d.B and C.B. were supported by core funding to the MRC-UCL University Unit and funding by R.d.B.’s Cancer Research UK Programme Foundation Award. M.R.M. was funded by a Blood Cancer UK Bennett Fellowship.
Footnotes
Declaration of interests: The authors declare no competing interests.
Author contributions: T.E.L. designed and performed experiments, analyzed and interpreted data and wrote the manuscript. T.R.D. performed experiments and analyzed data. S.R., M.M., N.F., S.E.R. and S.A. provided methodological and technical support, performed experiments and analyzed data. B.P. provided essential reagents and advice. R.G. and E.N. provided technical support. J.A.G.A., J.H. and S.H. helped with data analysis. D.C.L. provided funding and supervised the study. C.B. and R.d.B provided advice and technical support, helped with data analysis, and edited the manuscript. M.R.M. conceptualized and designed experiments, provided funding, supervised the overall study, analyzed data and wrote the manuscript.
Data and materials availability
RNA-seq data and MYB ChIP-seq datasets generated in this publication have been deposited in NCBI’s Gene Expression Omnibus (69) and are accessible through GEO Series accession number GSE127261 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi’acc=GSE127261), and GSE147198 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi’acc=GSE147198) respectively. Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Marc R. Mansour (m.mansour@ucl.ac.uk).
References
- 1.Zhang J, Ding L, Holmfeldt L, Wu G, Heatley SL, Payne-Turner D, et al. The genetic basis of early T-cell precursor acute lymphoblastic leukaemia [Internet] Nature. 2012;481:157–63. doi: 10.1038/nature10725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ntziachristos P, Tsirigos A, Vlierberghe P Van, Nedjic J, Trimarchi T, Flaherty MS, et al. Genetic inactivation of the polycomb repressive complex 2 in T cell acute lymphoblastic leukemia. Nat Med. 2012;18:298–303. doi: 10.1038/nm.2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu Y, Easton J, Shao Y, Maciaszek J, Wang Z, Wilkinson MR, et al. The genomic landscape of pediatric and young adult T-lineage acute lymphoblastic leukemia. Nat Genet. 2017;49:1211–8. doi: 10.1038/ng.3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Coustan-Smith E, Mullighan CG, Onciu M, Behm FG, Raimondi SC, Pei D, et al. Early T-cell precursor leukaemia: a subtype of very high-risk acute lymphoblastic leukaemia. Lancet Oncol. 2009;10:147–56. doi: 10.1016/S1470-2045(08)70314-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Morey L, Helin K. Polycomb group protein-mediated repression of transcription. Trends Biochem Sci [Internet] 2010;35:323–32. doi: 10.1016/j.tibs.2010.02.009. Available from: http://linkinghub.elsevier.com/retrieve/pii/S0968000410000307. [DOI] [PubMed] [Google Scholar]
- 6.Su I, Dobenecker M-W, Dickinson E, Oser M, Basavaraj A, Marqueron R, et al. Polycomb group protein ezh2 controls actin polymerization and cell signaling. Cell [Internet] 2005;121:425–36. doi: 10.1016/j.cell.2005.02.029. [DOI] [PubMed] [Google Scholar]
- 7.Simon C, Chagraoui J, Krosl J, Gendron P, Wilhelm B, Lemieux S, et al. A key role for EZH2 and associated genes in mouse and human adult T-cell acute leukemia. Genes Dev. 2012;26:651–6. doi: 10.1101/gad.186411.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ariёs IM, Bodaar K, Karim SA, Chonghaile TN, Hinze L, Burns MA, et al. PRC2 loss induces chemoresistance by repressing apoptosis in T cell acute lymphoblastic leukemia. J Exp Med [Internet] 2018:jem.20180570. doi: 10.1084/jem.20180570. Available from: http://www.jem.org/lookup/doi/10.1084/jem.20180570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Göllner S, Oellerich T, Agrawal-Singh S, Schenk T, Klein H-U, Rohde C, et al. Loss of the histone methyltransferase EZH2 induces resistance to multiple drugs in acute myeloid leukemia. Nat Med [Internet] 2017;23:69–78. doi: 10.1038/nm.4247. Available from: http://www.nature.com/articles/nm.4247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bond J, Labis E, Marceau-Renaut A, Duployez N, Labopin M, Hypolite G, et al. Polycomb repressive complex 2 haploinsufficiency identifies a high-risk subgroup of pediatric acute myeloid leukemia. Leukemia [Internet] 2018;32:1878–82. doi: 10.1038/s41375-018-0187-9. Available from: http://www.nature.com/articles/s41375-018-0187-9. [DOI] [PubMed] [Google Scholar]
- 11.Ashworth A, Lord CJ. Synthetic lethal therapies for cancer: what’s next after PARP inhibitors? Nat Rev Clin Oncol [Internet] 2018;15:564–76. doi: 10.1038/s41571-018-0055-6. [DOI] [PubMed] [Google Scholar]
- 12.Bryant HE, Schultz N, Thomas HD, Parker KM, Flower D, Lopez E, et al. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature [Internet] 2005;434:913–7. doi: 10.1038/nature03443. Available from: http://www.nature.com/articles/nature03443. [DOI] [PubMed] [Google Scholar]
- 13.Farmer H, McCabe N, Lord CJ, Tutt ANJ, Johnson DA, Richardson TB, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature [Internet] 2005;434:917–21. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- 14.Drew Y, Ledermann J, Hall G, Rea D, Glasspool R, Highley M, et al. Br J Cancer [Internet] Vol. 114. Nature Publishing Group; 2016. Phase 2 multicentre trial investigating intermittent and continuous dosing schedules of the poly (ADP-ribose) polymerase inhibitor rucaparib in germline BRCA mutation carriers with advanced ovarian and breast cancer; pp. 723–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Robson M, Im S-A, Senkus E, Xu B, Domchek SM, Masuda N, et al. Olaparib for Metastatic Breast Cancer in Patients with a Germline BRCA Mutation. N Engl J Med [Internet] 2017;377:523–33. doi: 10.1056/NEJMoa1706450. Available from: http://www.nejm.org/doi/10.1056/NEJMoa1706450. [DOI] [PubMed] [Google Scholar]
- 16.Pfister SX, Markkanen E, Jiang Y, Sarkar S, Woodcock M, Orlando G, et al. Cancer Cell [Internet] Vol. 28. Elsevier Ltd; 2015. Inhibiting WEE1 Selectively Kills Histone H3K36me3-Deficient Cancers by dNTP Starvation; pp. 557–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ran FA, Hsu PD, Lin C-Y, Gootenberg JS, Konermann S, Trevino AE, et al. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell [Internet] 2013;154:1380–9. doi: 10.1016/j.cell.2013.08.021. Available from: http://www.ncbi.nlm.nih.gov/pubmed/23992846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Daud AI, Ashworth MT, Strosberg J, Goldman JW, Mendelson D, Springett G, et al. Phase I dose-escalation trial of checkpoint kinase 1 inhibitor MK-8776 as monotherapy and in combination with gemcitabine in patients with advanced solid tumors. J Clin Oncol. 2015;33:1060–6. doi: 10.1200/JCO.2014.57.5027. [DOI] [PubMed] [Google Scholar]
- 19.Feijoo C, Hall-Jackson C, Wu R, Jenkins D, Leitch J, Gilbert DM, et al. Activation of mammalian Chk1 during DNA replication arrest: a role for Chk1 in the intra-S phase checkpoint monitoring replication origin firing. J Cell Biol [Internet] 2001;154:913–23. doi: 10.1083/jcb.200104099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maya-Mendoza A, Petermann E, Gillespie DAF, Caldecott KW, Jackson DA. Chk1 regulates the density of active replication origins during the vertebrate S phase. EMBO J [Internet] 2007;26:2719–31. doi: 10.1038/sj.emboj.7601714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu Q, Guntuku S, Cui XS, Matsuoka S, Cortez D, Tamai K, et al. Chk1 is an essential kinase that is regulated by Atr and required for the G(2)/M DNA damage checkpoint. Genes Dev [Internet] 2000;14:1448–59. [PMC free article] [PubMed] [Google Scholar]
- 22.Patil M, Pabla N, Dong Z. Checkpoint kinase 1 in DNA damage response and cell cycle regulation. Cell Mol Life Sci [Internet] 2013;70:4009–21. doi: 10.1007/s00018-013-1307-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Petermann E, Maya-Mendoza A, Zachos G, Gillespie DAF, Jackson DA, Caldecott KW. Chk1 requirement for high global rates of replication fork progression during normal vertebrate S phase. Mol Cell Biol [Internet] 2006;26:3319–26. doi: 10.1128/MCB.26.8.3319-3326.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Toledo LI, Altmeyer M, Rask M-B, Lukas C, Larsen DH, Povlsen LK, et al. ATR prohibits replication catastrophe by preventing global exhaustion of RPA. Cell [Internet] 2013;155:1088–103. doi: 10.1016/j.cell.2013.10.043. [DOI] [PubMed] [Google Scholar]
- 25.Nam EA, Cortez D. ATR signalling: more than meeting at the fork. Biochem J [Internet] 2011;436:527–36. doi: 10.1042/BJ20102162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zeman MK, Cimprich KA. Causes and consequences of replication stress. Nat Cell Biol. 2014;16:2–9. doi: 10.1038/ncb2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Herlihy AE, de Bruin RAM. The Role of the Transcriptional Response to DNA Replication Stress. Genes (Basel) [Internet] 2017;8:92. doi: 10.3390/genes8030092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Syljuåsen RG, Sørensen CS, Hansen LT, Fugger K, Lundin C, Johansson F, et al. Inhibition of human Chk1 causes increased initiation of DNA replication, phosphorylation of ATR targets, and DNA breakage. Mol Cell Biol [Internet] 2005;25:3553–62. doi: 10.1128/MCB.25.9.3553-3562.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Petermann E, Woodcock M, Helleday T. Chk1 promotes replication fork progression by controlling replication initiation. Proc Natl Acad Sci U S A [Internet] 2010;107:16090–5. doi: 10.1073/pnas.1005031107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Myers K, Gagou ME, Zuazua-Villar P, Rodriguez R, Meuth M. ATR and Chk1 suppress a caspase-3-dependent apoptotic response following DNA replication stress. PLoS Genet [Internet] 2009;5:e1000324. doi: 10.1371/journal.pgen.1000324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cimprich KA, Cortez D. ATR: an essential regulator of genome integrity. Nat Rev Mol Cell Biol [Internet] 2008;9:616–27. doi: 10.1038/nrm2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sakaue-Sawano A, Kobayashi T, Ohtawa K, Miyawaki A. Drug-induced cell cycle modulation leading to cell-cycle arrest, nuclear mis-segregation, or endoreplication. BMC Cell Biol [Internet] 2011;12:2. doi: 10.1186/1471-2121-12-2. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3277280&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Toledo L, Neelsen KJ, Lukas J. Replication Catastrophe: When a Checkpoint Fails because of Exhaustion. Mol Cell [Internet] 2017;66:735–49. doi: 10.1016/j.molcel.2017.05.001. [DOI] [PubMed] [Google Scholar]
- 34.Barr AR, Cooper S, Heldt FS, Butera F, Stoy H, Mansfeld J, et al. DNA damage during S-phase mediates the proliferation-quiescence decision in the subsequent G1 via p21 expression. Nat Commun [Internet] 2017;8:14728. doi: 10.1038/ncomms14728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lukas C, Savic V, Bekker-Jensen S, Doil C, Neumann B, Pedersen RS, et al. 53BP1 nuclear bodies form around DNA lesions generated by mitotic transmission of chromosomes under replication stress. Nat Cell Biol [Internet] 2011;13:243–53. doi: 10.1038/ncb2201. [DOI] [PubMed] [Google Scholar]
- 36.Ha VL, Luong A, Li F, Casero D, Malvar J, Kim YM, et al. The T-ALL related gene BCL11B regulates the initial stages of human T-cell differentiation. Leukemia [Internet] 2017;31:2503–14. doi: 10.1038/leu.2017.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Casero D, Sandoval S, Seet CS, Scholes J, Zhu Y, Ha VL, et al. Long non-coding RNA profiling of human lymphoid progenitor cells reveals transcriptional divergence of B cell and T cell lineages. Nat Immunol [Internet] 2015;16:1282–91. doi: 10.1038/ni.3299. Available from: http://www.nature.com/articles/ni.3299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A [Internet] 2005;102:15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dik WA, Pike-Overzet K, Weerkamp F, de Ridder D, de Haas EFE, Baert MRM, et al. New insights on human T cell development by quantitative T cell receptor gene rearrangement studies and gene expression profiling. J Exp Med [Internet] 2005;201:1715–23. doi: 10.1084/jem.20042524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cieslak A, Le Noir S, Trinquand A, Lhermitte L, Franchini D-M, Villarese P, et al. RUNX1-dependent RAG1 deposition instigates human TCR-δ locus rearrangement. J Exp Med [Internet] 2014;211:1821–32. doi: 10.1084/jem.20132585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liberzon A, Birger C, Thorvaldsdóttir H, Ghandi M, Mesirov JP, Tamayo P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst [Internet] 2015;1:417–25. doi: 10.1016/j.cels.2015.12.004. Available from: https://linkinghub.elsevier.com/retrieve/pii/S2405471215002185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tumes DJ, Onodera A, Suzuki A, Shinoda K, Endo Y, Iwamura C, et al. The polycomb protein Ezh2 regulates differentiation and plasticity of CD4(+) T helper type 1 and type 2 cells. Immunity [Internet] 2013;39:819–32. doi: 10.1016/j.immuni.2013.09.012. [DOI] [PubMed] [Google Scholar]
- 43.Garber K. New Insights Into Oncogene Addiction Found. JNCI J Natl Cancer Inst [Internet] 2007;99:264–9. doi: 10.1093/jnci/djk087. Available from: https://academic.oup.com/jnci/article-lookup/doi/10.1093/jnci/djk087. [DOI] [PubMed] [Google Scholar]
- 44.Mansour MR, Abraham BJ, Anders L, Berezovskaya A, Gutierrez A, Durbin AD, et al. An oncogenic super-enhancer formed through somatic mutation of a noncoding intergenic element. [cited 2014 Nov 15];Science [Internet] 2014 :1–9. doi: 10.1126/science.1259037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hnisz D, Weintraub AS, Day DS, Valton A, Bak RO, Li CH, et al. Activation of protooncogenes by disruption of chromosome neighborhoods. Science (80-) [Internet] 2016;351:1454–8. doi: 10.1126/science.aad9024. Available from: http://science.sciencemag.org/content/351/6280/1454.long. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Walton MI, Eve PD, Hayes A, Henley AT, Valenti MR, De Haven Brandon AK, et al. The clinical development candidate CCT245737 is an orally active CHK1 inhibitor with preclinical activity in RAS mutant NSCLC and E&micro;-MYC driven B-cell lymphoma. Oncotarget [Internet] 2016;7:2329–42. doi: 10.18632/oncotarget.4919. Available from: http://www.oncotarget.com/fulltext/4919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Maude SL, Dolai S, Delgado-martin C, Vincent T, Robbins A, Selvanathan A, et al. Efficacy of JAK / STAT pathway inhibition in murine xenograft models of early T-cell precursor (ETP) acute lymphoblastic leukemia. Blood. 2015;125:1759–68. doi: 10.1182/blood-2014-06-580480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Srinivasan SV, Dominguez-Sola D, Wang LC, Hyrien O, Gautier J. Cdc45 Is a Critical Effector of Myc-Dependent DNA Replication Stress. Cell Rep [Internet] 2013;3:1629–39. doi: 10.1016/j.celrep.2013.04.002. Available from: https://linkinghub.elsevier.com/retrieve/pii/S2211124713001691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Maya-Mendoza A, Ostrakova J, Kosar M, Hall A, Duskova P, Mistrik M, et al. Myc and Ras oncogenes engage different energy metabolism programs and evoke distinct patterns of oxidative and DNA replication stress. Mol Oncol [Internet] 2015;9:601–16. doi: 10.1016/j.molonc.2014.11.001. Available from: http://doi.wiley.com/10.1016/j.molonc.2014.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mazouzi A, Velimezi G, Loizou JI. DNA replication stress: causes, resolution and disease. Exp Cell Res [Internet] 2014;329:85–93. doi: 10.1016/j.yexcr.2014.09.030. [DOI] [PubMed] [Google Scholar]
- 51.Kotsantis P, Petermann E, Boulton SJ. Mechanisms of oncogene-induced replication stress: Jigsaw falling into place. Cancer Discov. 2018;8:537–55. doi: 10.1158/2159-8290.CD-17-1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dominguez-Sola D, Ying CY, Grandori C, Ruggiero L, Chen B, Li M, et al. Nontranscriptional control of DNA replication by c-Myc. Nature [Internet] 2007;448:445–51. doi: 10.1038/nature05953. [DOI] [PubMed] [Google Scholar]
- 53.Blow JJ, Gillespie PJ. Replication licensing and cancer--a fatal entanglement? Nat Rev Cancer [Internet] 2008;8:799–806. doi: 10.1038/nrc2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Murga M, Campaner S, Lopez-Contreras AJ, Toledo LI, Soria R, Montaña MF, et al. Exploiting oncogene-induced replicative stress for the selective killing of Myc-driven tumors. Nat Struct Mol Biol [Internet] 2011;18:1331–5. doi: 10.1038/nsmb.2189. Available from: http://www.nature.com/articles/nsmb.2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cole KA, Huggins J, Laquaglia M, Hulderman CE, Russell MR, Bosse K, et al. RNAi screen of the protein kinome identifies checkpoint kinase 1 (CHK1) as a therapeutic target in neuroblastoma. Proc Natl Acad Sci U S A [Internet] 2011;108:3336–41. doi: 10.1073/pnas.1012351108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ferrao PT, Bukczynska EP, Johnstone RW, McArthur GA. Oncogene [Internet] Vol. 31. Nature Publishing Group; 2012. Efficacy of CHK inhibitors as single agents in MYC-driven lymphoma cells; pp. 1661–72. [DOI] [PubMed] [Google Scholar]
- 57.Danis E, Yamauchi T, Echanique K, Zhang X, Haladyna JN, Riedel SS, et al. Ezh2 Controls an Early Hematopoietic Program and Growth and Survival Signaling in Early T Cell Precursor Acute Lymphoblastic Leukemia. Cell Rep [Internet] 2016;14:1953–65. doi: 10.1016/j.celrep.2016.01.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Neumann M, Coskun E, Fransecky L, Mochmann LH, Bartram I, Sartangi NF, et al. FLT3 mutations in early T-cell precursor ALL characterize a stem cell like leukemia and imply the clinical use of tyrosine kinase inhibitors. PLoS One [Internet] 2013;8:e53190. doi: 10.1371/journal.pone.0053190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rundle S, Bradbury A, Drew Y, Curtin NJ. Targeting the ATR-CHK1 Axis in Cancer Therapy. Cancers (Basel) [Internet] 2017;9:1–25. doi: 10.3390/cancers9050041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dent P, Tang Y, Yacoub A, Dai Y, Fisher PB, Grant S. CHK1 inhibitors in combination chemotherapy: thinking beyond the cell cycle. Mol Interv [Internet] 2011;11:133–40. doi: 10.1124/mi.11.2.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sarmento LM, Póvoa V, Nascimento R, Real G, Antunes I, Martins LR, et al. CHK1 overexpression in T-cell acute lymphoblastic leukemia is essential for proliferation and survival by preventing excessive replication stress. Oncogene [Internet] 2015;34:2978–90. doi: 10.1038/onc.2014.248. [DOI] [PubMed] [Google Scholar]
- 62.Sarmento LM, Barata JT. CHK1 and replicative stress in T-cell leukemia: Can an irreverent tumor suppressor end up playing the oncogene? Adv Biol Regul [Internet] 2016;60:115–21. doi: 10.1016/j.jbior.2015.10.007. Available from: https://linkinghub.elsevier.com/retrieve/pii/S2212492615300336. [DOI] [PubMed] [Google Scholar]
- 63.Moffat J, Grueneberg DA, Yang X, Kim SY, Kloepfer AM, Hinkle G, et al. A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell [Internet] 2006;124:1283–98. doi: 10.1016/j.cell.2006.01.040. [DOI] [PubMed] [Google Scholar]
- 64.Sanda T, Lawton LN, Barrasa MI, Fan ZP, Kohlhammer H, Gutierrez A, et al. Core transcriptional regulatory circuit controlled by the TAL1 complex in human T cell acute lymphoblastic leukemia. Cancer Cell [Internet] 2012;22:209–21. doi: 10.1016/j.ccr.2012.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Booth CAG, Barkas N, Neo WH, Boukarabila H, Soilleux EJ, Giotopoulos G, et al. Ezh2 and Runx1 Mutations Collaborate to Initiate Lympho-Myeloid Leukemia in Early Thymic Progenitors. Cancer Cell [Internet] 2018;33:274–291.:e8. doi: 10.1016/j.ccell.2018.01.006. [DOI] [PubMed] [Google Scholar]
- 66.Schmidl C, Rendeiro AF, Sheffield NC, Bock C. ChIPmentation: fast, robust, low-input ChIP-seq for histones and transcription factors. Nat Methods [Internet] 2015;12:963–5. doi: 10.1038/nmeth.3542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ramírez F, Ryan DP, Grüning B, Bhardwaj V, Kilpert F, Richter AS, et al. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res [Internet] 2016;44:W160–5. doi: 10.1093/nar/gkw257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhou X, Lowdon RF, Li D, Lawson HA, Madden PAF, Costello JF, et al. Exploring long-range genome interactions using the WashU Epigenome Browser. Nat Methods [Internet] 2013;10:375–6. doi: 10.1038/nmeth.2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res [Internet] 2002;30:207–10. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq data and MYB ChIP-seq datasets generated in this publication have been deposited in NCBI’s Gene Expression Omnibus (69) and are accessible through GEO Series accession number GSE127261 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi’acc=GSE127261), and GSE147198 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi’acc=GSE147198) respectively. Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Marc R. Mansour (m.mansour@ucl.ac.uk).