Abstract
The heart muscle diseases hypertrophic (HCM) and dilated (DCM) cardiomyopathies are leading causes of sudden death and heart failure in young otherwise healthy individuals. We conducted genome-wide association studies (GWAS) and multi-trait analyses in HCM (1,733 cases), DCM (5,521 cases), and nine left ventricular (LV) traits in 19,260 UK Biobank participants with structurally normal hearts. We identified 16 loci associated with HCM, 13 with DCM, and 23 with LV traits. We show strong genetic correlations between LV traits and cardiomyopathies, with opposing effects in HCM and DCM. Two-sample Mendelian randomization supports a causal association linking increased contractility with HCM risk. A polygenic risk score (PRS) explains a significant portion of phenotypic variability in carriers of HCM-causing rare variants. Our findings thus provide evidence that PRS may account for variability in Mendelian diseases. More broadly, we provide insights into how genetic pathways may lead to distinct disorders through opposing genetic effects.
Cardiomyopathies are heritable heterogenous diseases characterized by changes in myocardial structure and function. We sought to better understand the genetic underpinnings of HCM and DCM as well as their relation to myocardial traits in the general population (Fig. 1). HCM has a prevalence of 0.2%1 and has been classically considered a Mendelian disease. However, genetic testing identifies a causal rare variant in less than half of cases2, and data from both families and population cohorts support reduced penetrance and variable expressivity3, suggesting a complex genetic architecture. Here, we performed a meta-analysis of three new GWAS comprising 1,733 unrelated HCM cases and 6,628 controls of European ancestry from the Netherlands, United Kingdom and Canada (Supplementary Table 1). Of the cases, 641 (37%) carried pathogenic or likely pathogenic variants in established HCM disease genes (Supplementary Table 2). Analysis of SNP-based heritability (h 2 SNP) using GREML4 demonstrated that a significant portion of HCM liability is attributed to common genetic variation (Supplementary Table 3), with h 2 SNP estimates in meta-analyses ranging from 0.12 (GREML, fixed-effects; P = 8 × 10-6) to 0.29 (GREML-LDMS5, random-effects; P = 9 × 10-3). The GWAS summary meta-analysis results of 6,530,233 variants with a minor allele frequency (MAF) ≥ 0.01 are shown as Manhattan and quantile-quantile (QQ) plots in Figure 2a. The wide association signal observed on chromosome 11 tagging recurrent MYBPC3 founder variants disappeared when restricting the analysis to the 1,445 HCM cases without such variants (Supplementary Fig. 1). Using a conservative threshold of P < 1 × 10-8 to account for multiple testing, a total of six loci were significantly associated with HCM (Table 1, Supplementary Table 4 and Supplementary Fig. 2), of which five are new and one, on chromosome 18 near FHOD3, has been previously published6. Importantly, two of the new HCM loci (chromosome 1 near HSPB7 and chromosome 10 near BAG3) have been previously associated with DCM at genome-wide statistical significance, but with an opposite direction of effect7–10. Specifically, the published DCM lead risk alleles at both loci (rs10927875-C and rs2234962-T)10 were protective for HCM in the present study (odds ratio, OR (95% confidence interval, CI), 0.80 (0.74-0.87) and 0.71 (0.64-0.77), respectively). Recently, both loci were also found to be associated with left ventricular ejection fraction (LVEF, a volume-based assessment of LV contractility) in the general population11, for whom DCM risk alleles decrease LVEF (contractility), while HCM risk alleles increase LVEF (contractility). This indicates that genetic loci underlying variability of LV function in the general population may be differentially involved in susceptibility to HCM and DCM.
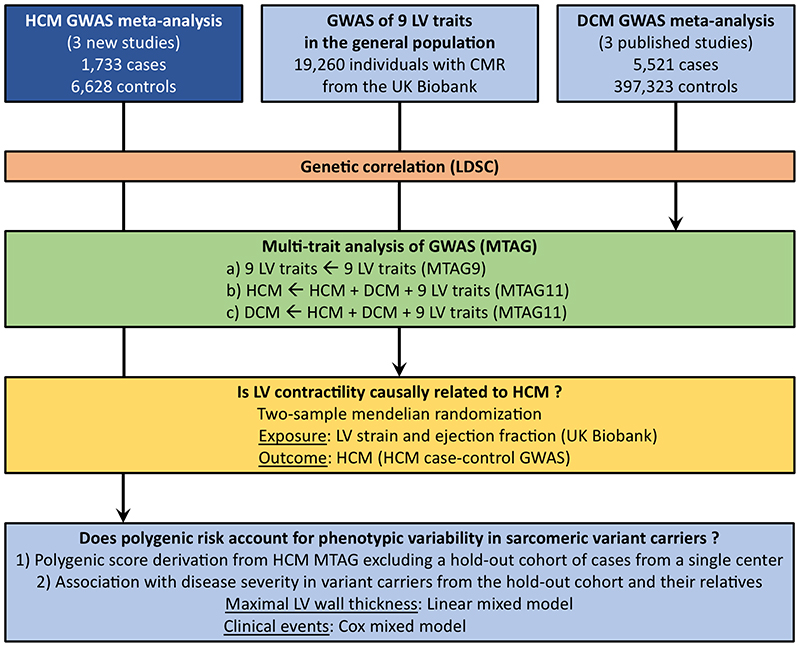
Figure 1. Study flowchart.
CMR, cardiac magnetic resonance; DCM, dilated cardiomyopathy; HCM, hypertrophic cardiomyopathy; LV, left ventricle/ventricular; LDSC, LD score correlation; MTAG, multi-trait analysis of GWAS.
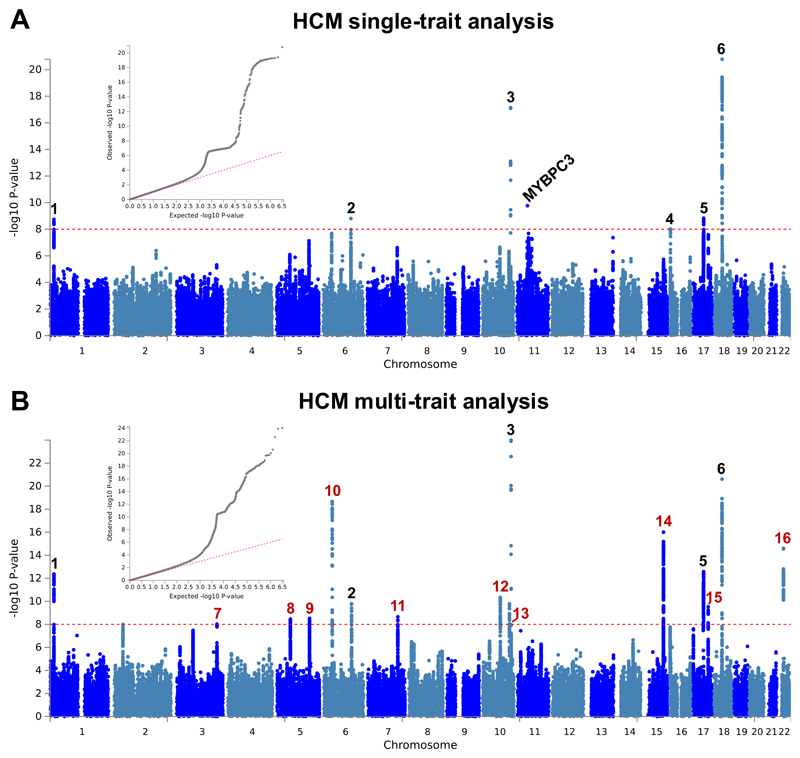
Figure 2. Summary results of the hypertrophic cardiomyopathy (HCM) single trait GWAS and multi-trait analysis.
a,b, Single trait analysis (a) consisted of a fixed effects meta-analysis of case-control GWAS using a frequentist test, and multi-trait analysis results (b) were obtained using MTAG for HCM, including GWAS for dilated cardiomyopathy (DCM) and nine left ventricular (LV) traits. Summary statistics shown as Manhattan plots with red dashed line showing the genome-wide significance threshold of P = 1 × 10-8. Quantile-quantile (QQ) plots are shown as inserts in corresponding panels. Genomic inflation (λ) = 1.081 (single-trait) and 1.082 (MTAG). Six association signals were identified in single trait analysis (a), and an additional 10 signals were identified in multi-trait analysis (b). The wide signal on chromosome 11 tags founder MYBPC3 pathogenic variants. Locus #4 was only significant in the single-trait analysis and did not replicate in an independent HCM GWAS. Numbering of signals is as shown in Table 1 and Supplementary Table 4, where red numbers refer to signals reaching genome-wide significance only in the multi-trait analysis.
Table 1. Lead SNPs and effect estimates for genome-wide significant loci (P < 1 × 10-8) in the HCM single-trait and multi-trait analyses.
| Locus | Lead SNP | GRCh37 | Nearest gene | RA | NRA | RAF | OR | 95%CI | P | P (MTAG11) | P replication |
|---|---|---|---|---|---|---|---|---|---|---|---|
| HCM1 | rs10927886 | 1:16339313 | HSPB7 | G | C | 0.41 | 1.28 | 1.18-1.38 | 1.8E-09 | 7.4E-13 | 2.5E-13 |
| HCM2 | rs12212795 | 6:118654308 | SLC35F1 | C | G | 0.06 | 1.69 | 1.43-2.01 | 1.6E-09 | 1.7E-10 | 6.7E-11 |
| HCM3 | rs17099139 | 10:121419487 | BAG3 | G | C | 0.29 | 1.46 | 1.34-1.59 | 7.2E-18 | 1.0E-24 | 1.3E-22 |
| HCM4* | rs9928278 | 16:2152651 | PKD1 | C | T | 0.18 | 1.45 | 1.28-1.65 | 9.5E-09 | 5.9E-07 | 2.7E-01 |
| HCM5 | rs1378358 | 17:44787312 | NSF | T | C | 0.23 | 1.34 | 1.22-1.47 | 1.5E-09 | 4.7E-13 | 4.7E-08 |
| HCM6 | rs503274 | 18:34253745 | FHOD3 | C | T | 0.31 | 1.52 | 1.40-1.66 | 1.7E-21 | 2.4E-21 | 4.3E-19 |
| HCM7 (MTAG11) | rs9647379 | 3:171785168 | FNDC3B | C | G | 0.42 | 1.22 | 1.12-1.33 | 6.8E-06 | 9.5E-09 | 2.4E-06 |
| HCM8 (MTAG11) | rs2191445 | 5:57011469 | ACTBL2 | T | A | 0.78 | 1.29 | 1.17-1.43 | 8.0E-07 | 3.5E-09 | 2.6E-05 |
| HCM9 (MTAG11) | rs4385202 | 5:138743256 | DNAJC18 | A | G | 0.31 | 1.25 | 1.15-1.37 | 6.0E-07 | 3.0E-09 | 7.3E-05 |
| HCM10 (MTAG11) | rs66761782 | 6:36636080 | CDKN1A | C | T | 0.26 | 1.29 | 1.18-1.41 | 3.4E-08 | 2.1E-19 | 1.3E-08 |
| HCM11 (MTAG11) | rs60871386 | 7:128430437 | CCDC136 | T | G | 0.10 | 1.43 | 1.25-1.64 | 3.4E-07 | 2.2E-09 | 1.4E-04 |
| HCM12 (MTAG11) | rs3740293 | 10:75406141 | SYNPO2L | C | A | 0.15 | 1.33 | 1.19-1.49 | 4.8E-07 | 4.6E-11 | 8.5E-06 |
| HCM13 (MTAG11) | rs11196078 | 10:114487812 | VTI1A | A | G | 0.26 | 1.26 | 1.15-1.38 | 5.2E-07 | 1.6E-10 | 6.7E-07 |
| HCM14 (MTAG11) | rs11073729 | 15:85350081 | ZNF592 | C | T | 0.46 | 1.20 | 1.11-1.30 | 4.2E-06 | 9.9E-17 | 3.6E-07 |
| HCM15 (MTAG11) | rs9892651 | 17:64303793 | PRKCA | T | C | 0.59 | 1.25 | 1.16-1.36 | 2.8E-08 | 3.0E-10 | 2.4E-09 |
| HCM16 (MTAG11) | rs2186370 | 22:24171305 | SMARCB1 | A | G | 0.22 | 1.21 | 1.09-1.34 | 3.5E-04 | 2.5E-15 | 5.5E-15 |
Locus HCM4 is not replicated. Abbreviations: 95%CI, 95% confidence interval; GRCh37, genomic position in GRCh37; MTAG11, Multi-Trait Analysis of GWAS summary statistics from HCM, DCM, and nine quantitative LV traits; NRA, non-risk allele; OR, odds ratio for each risk allele in the single trait HCM analysis; P, single trait analysis P value; P (MTAG11), multi-trait analysis P value for HCM; P replication, P value in the replication dataset; RA, risk allele; RAF, risk allele frequency.
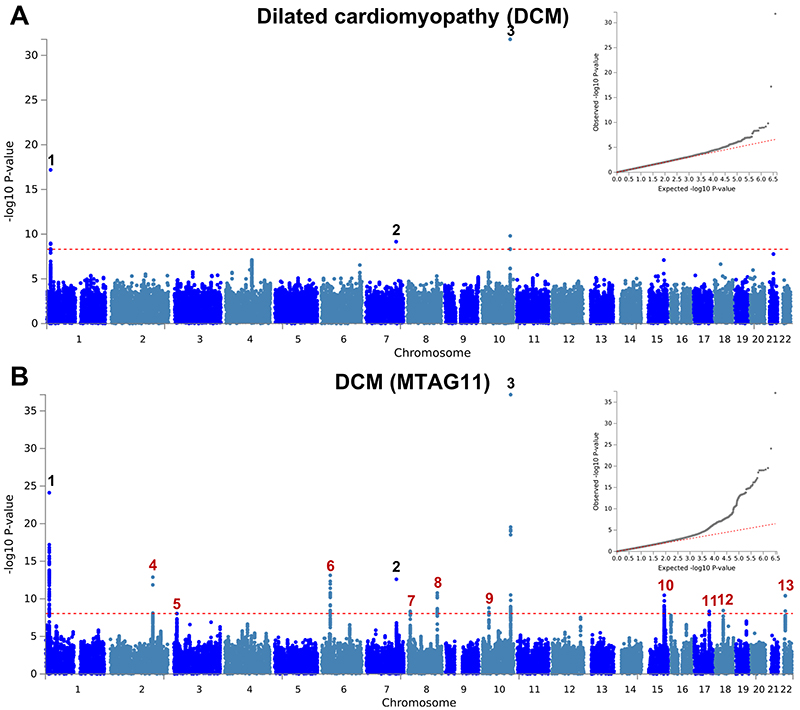
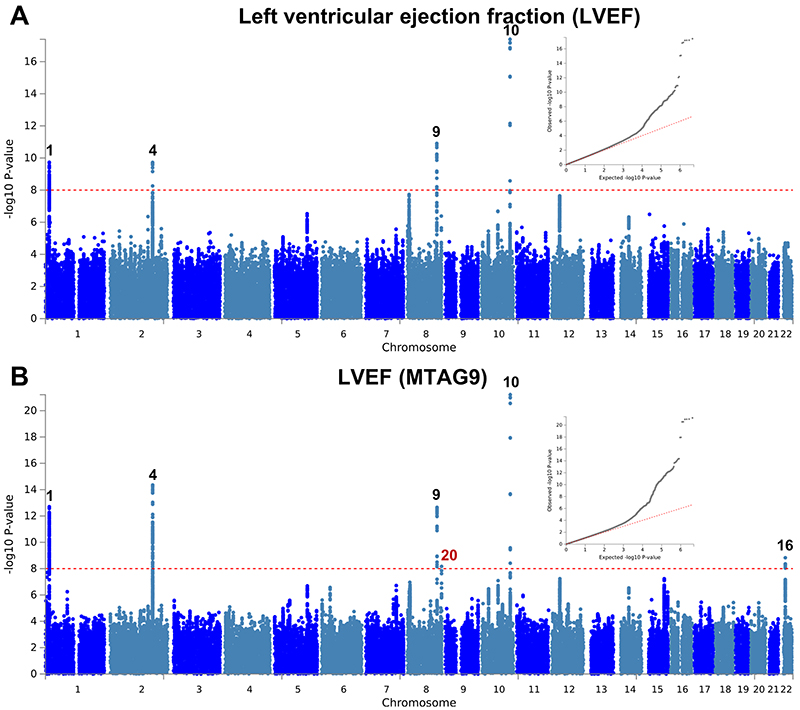
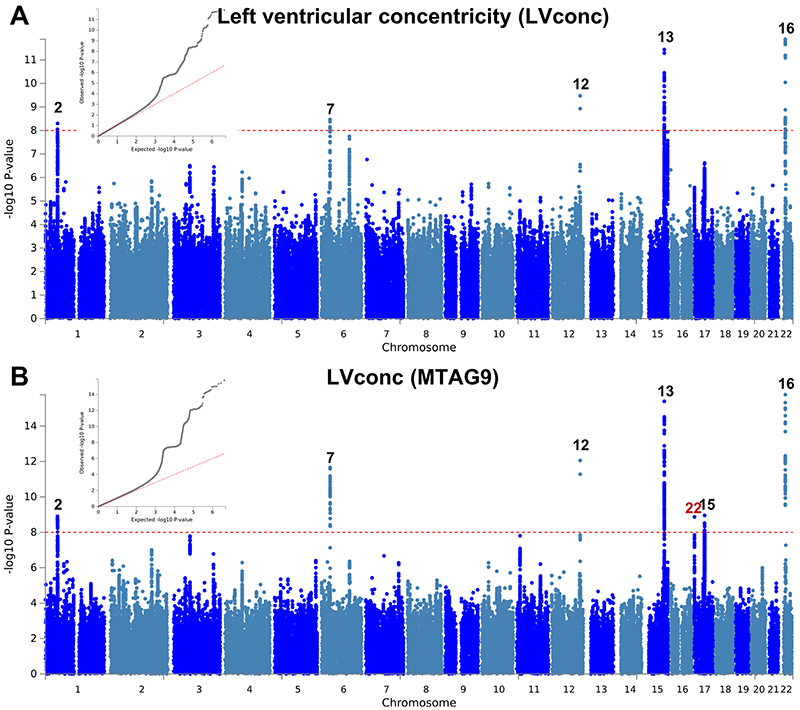
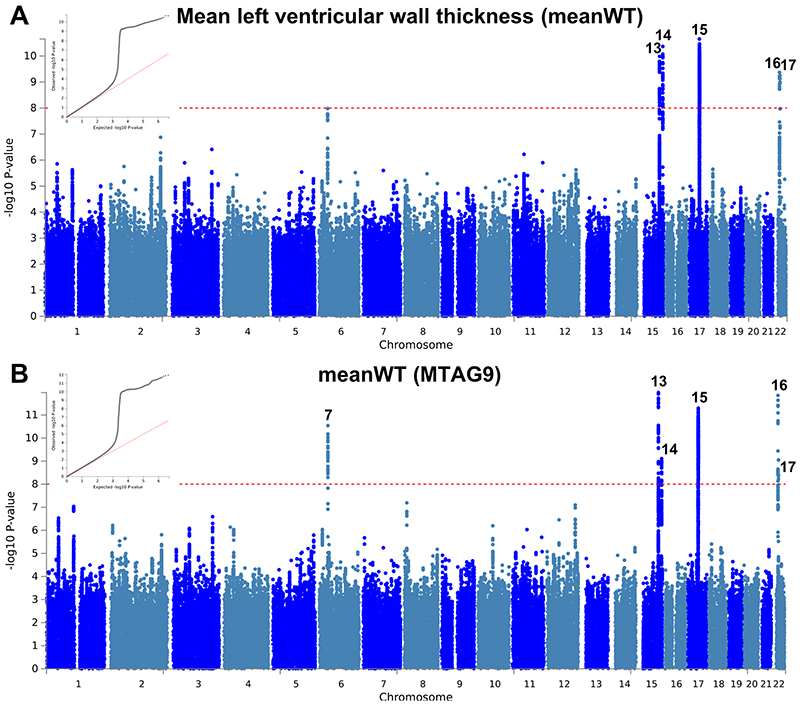
The observation that two loci with opposite directions of effects in HCM and DCM are also associated with LVEF in the general population motivated us to further explore such relationships between HCM, DCM, and LV traits in the general population. We performed a meta-analysis of three published DCM case-control genetic association studies7–9 totaling 5,521 cases and 397,323 controls (Supplementary Table 5), as well as GWAS of nine LV traits derived from cardiac magnetic resonance (CMR) imaging in a cohort of 19,260 participants in UK Biobank without structural heart disease (Supplementary Table 6 and Supplementary Figs. 3-5). Compared to published GWAS on LV traits11, our analysis adds new phenotypes including mean LV wall thickness (meanWT) and measures of myocardial deformation (i.e. strain), which are proposed as more direct markers of contractility than volumetric assessments (LVEF)12. Many LV trait pairs were correlated phenotypically (Supplementary Fig. 6). The Manhattan and QQ plots of the DCM meta-analysis and nine LV traits GWAS are presented in panel a of Extended Data Figures 1-10. A total of three loci in the DCM (all previously published7,8; Supplementary Table 7 and Supplementary Fig. 7) and 17 loci in any of the nine LV traits meta-analyses reached a P-value threshold of 1 × 10-8 (Supplementary Table 8).
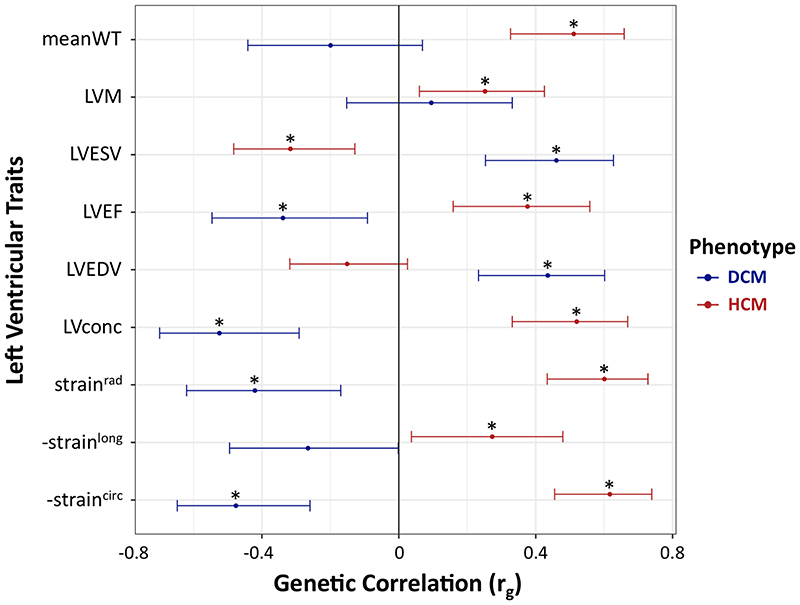
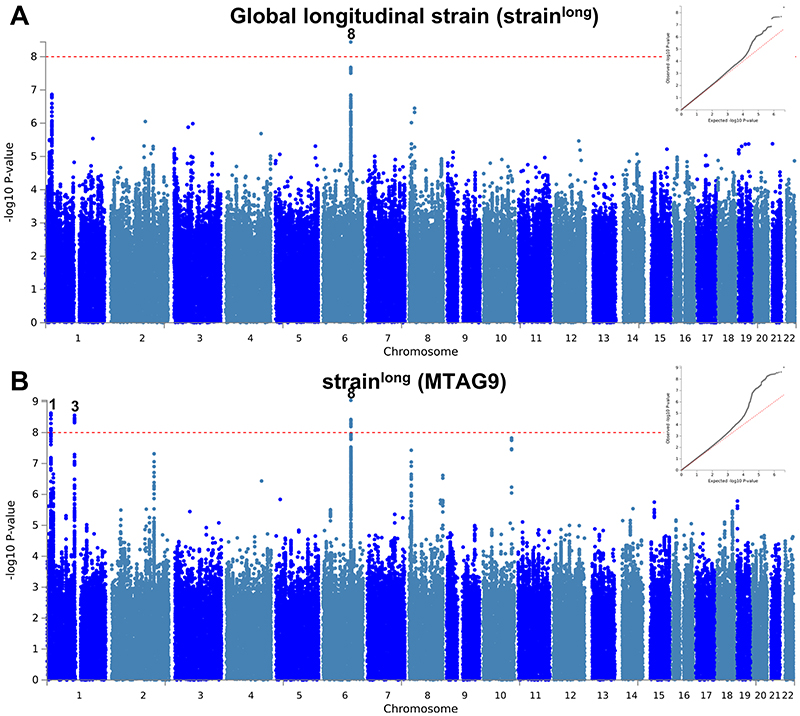
Genetic correlations between LV traits in the general population, HCM and DCM were assessed using LD score regression13, 14 (Supplementary Table 9). The results highlight the divergent relationships of LV traits with DCM and HCM (Fig. 3). HCM showed a positive genetic correlation with mean LV wall thickness (meanWT; r g = 0.51, P = 9 × 10-6), while DCM was positively correlated with LV end-diastolic (LVEDV) and end-systolic (LVESV) volumes (r g = 0.43, P = 2 × 10-4 and r g = 0.46, P = 1 × 10-4, respectively). Decreased LV contractility is a hallmark of DCM, and we observed a negative genetic correlation between DCM and LV contractility, whether assessed using a volumetric measure, LVEF (r g = -0.35, P = 9 × 10-3), or using global LV strain measured in any direction: circumferential (-straincirc; r g = -0.48, P = 2 × 10-4), radial (strainrad; r g = -0.42, P = 3 × 10-3) and longitudinal (-strainlong; r g = -0.27, P = 0.05) (note that for strainlong and straincirc, increasingly negative values reflect higher strain/contractility). Remarkably, all four of these contractility parameters were positively correlated with HCM; increases in contractility are correlated with increased HCM risk (r g ranging from 0.27 for -strainlong (P = 0.03) to 0.62 for -straincirc (P = 1 × 10-7)).
Figure 3. Genetic correlation between left ventricular traits, hypertrophic cardiomyopathy, and dilated cardiomyopathy.
Hypertrophic cardiomyopathy (HCM, red bars) and dilated cardiomyopathy (DCM, blue bars) show strong genetic correlations with quantitative cardiac left ventricular (LV) traits measured in the general population, but with opposite effects. Center values are the estimated genetic correlation (r g), and error bars indicate 95% confidence intervals. Samples sizes for included GWAS are as follows: 1,733 cases and 6,628 controls for HCM; 5,521 cases and 397,323 controls for DCM; and 19,260 for LV traits. Asterisks identify significant genetic correlations with a Benjamini–Hochberg false discovery rate (FDR) < 0.05. Data shown correspond to that in Supplementary Table 9. DCM, dilated cardiomyopathy; straincirc, strainlong and strainrad, global circumferential, longitudinal and radial strain, respectively (measures of contractility based on myocardial deformation); HCM, hypertrophic cardiomyopathy; LV, left ventricular; LVconc, LV concentricity (defined as LVM/LVEDV); LVEDV, LV end-diastolic volume; LVEF, LV ejection fraction (a volumetric measure of contractility); LVESV, LV end-systolic volume; LVM, LV mass; meanWT; mean LV wall thickness. Since straincirc and strainlong are always negative values, -straincirc and -strainlong are plotted to facilitate interpretation of effect direction.
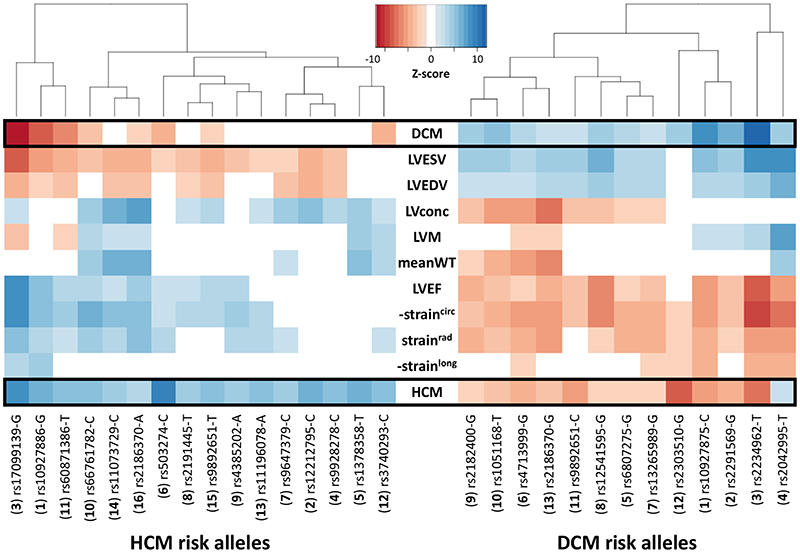
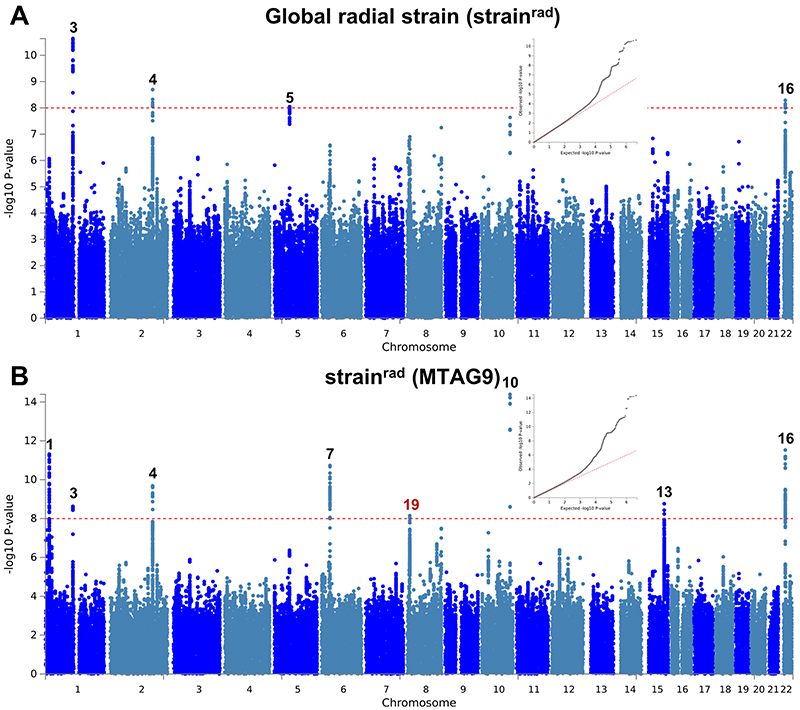
We then performed multi-trait analysis of GWAS summary statistics (MTAG)15 to increase power for discovery of novel loci. Two analyses were performed: (i) MTAG of nine LV traits (referred to as MTAG9) to uncover novel loci associated with LV traits, and (ii) MTAG of HCM, DCM and nine LV traits (henceforth MTAG11) to uncover loci associated with DCM and HCM. The corresponding Manhattan and QQ plots appear as panel b in Figure 2 and Extended Data Figures 1-10. MTAG9 uncovered 6 additional genetic loci associated with LV traits (Supplementary Table 8). MTAG11 uncovered an additional 10 HCM (Table 1, Supplementary Table 4 and Supplementary Fig. 2) and 10 DCM loci (Supplementary Table 7 and Supplementary Fig. 7). Supplementary Tables 4, 7, and 8 tabulate relationships for all traits with all HCM, DCM and LV trait lead SNPs, respectively, highlighting the cross-trait single SNP level correlation. In particular, 8 of the 16 HCM-associated loci also showed significant association with DCM using a Benjamini-Hochberg false discovery rate (FDR) < 0.05, where all 8 lead SNPs showed opposite directions of effect in HCM vs. DCM (Supplementary Table 4). Similarly, all 13 DCM loci were also associated with HCM at an FDR < 0.05 (Supplementary Table 7), with all loci showing an opposite effect in HCM vs. DCM except locus DCM4 near TTN. At this locus, the DCM risk allele also increases risk for HCM. We hypothesize that this unique concordant HCM/DCM effect may be attributable to pleiotropic effects of that locus on LV structure/function, where the HCM and DCM risk increasing allele reduces LV contractility parameters but also increases LV hypertrophy (LV mass and meanWT; Supplementary Table 7). Figure 4 displays a heatmap representation of the direction and strength of effect of all 16 HCM risk alleles and 13 DCM risk alleles in all nine LV traits, HCM and DCM. Many HCM risk alleles are also associated with reduced risk of DCM, and, in the general population, increased LV contractility (LVEF and strain) and decreased LV volumes.
Figure 4. Cross-trait associations of hypertrophic and dilated cardiomyopathy loci.
Heatmap of cross-trait associations of the 16 hypertrophic cardiomyopathy (HCM, left side) and 13 dilated cardiomyopathy (DCM, right) risk variants in HCM, DCM and nine LV traits in the general population. The dbSNP ID and risk alleles are shown on the x-axis, with the corresponding locus number in parenthesis (corresponding to numbering in Fig. 2, Table 1 and Supplementary Table 4 for HCM, and Extended Data Fig. 1 and Supplementary Table 7 for DCM). Variants sorted along the x-axis using Euclidean distance and complete hierarchical clustering (dendrogram on top). Effect of the HCM or DCM risk alleles shown as a colormap of Z-scores (legend), where positive values (concordant effect) are in shades of blue, and negative values (discordant effect) are in shades of red. Only associations with FDR < 0.05 are shown. HCM and DCM loci show many and reciprocal cross-trait associations. Since straincirc and strainlong are negative values, we show -straincirc and -strainlong to facilitate interpretation of effect direction. Lookup in DCM was performed using SNP proxies to maximize sample size, as shown in Supplementary Table 4. Note that the DCM risk allele rs2042995-T also increases risk of HCM, potentially through pleiotropic effects (decreased contractility and increased LV wall thickness). LV traits are as defined in the legend of Figure 3.
Replication of HCM loci was tested in an independent dataset16 comprised of 2,694 cases with HCM included from the Hypertrophic Cardiomyopathy Registry (HCMR)17 or the NIHR Bioresource for Rare Disease (BRRD)18, and 47,486 controls without HCM included from the UK Biobank or BRRD. Of the 16 HCM loci, 15 (all except HCM4) were replicated at P < 0.003 (Table 1 and Supplementary Table 10).
The correlation between increased contractility and HCM risk led us to test the hypothesis that increased contractility is causally associated with HCM. We tested such potential causality between increased LV contractility and HCM using two-sample Mendelian randomization (MR), where the exposure variables were LV contractility measures (-straincirc, strainrad, -strainlong and LVEF; all strongly correlated, with overlap in associated loci) and the outcome was HCM. Genetic instruments for the exposures were selected using two approaches: (i) independent SNPs reaching P < 5 × 10-8 in the single trait analysis, and (ii) those reaching P < 5 × 10-8 in MTAG9 analysis (Supplementary Table 11). The results of the primary inverse-variance weighted (IVW) analysis and sensitivity analyses including leave-one-out analyses are presented in Supplementary Table 12 and Supplementary Figures 8 and 9. Although heterogeneity and the limited number of SNPs in the exposure-outcome effects are limitations, the findings of the main (IVW) and sensitivity analyses support a causal relation between increased LV contractility and increased HCM risk. Both SNP selection approaches demonstrate a significant effect of -straincirc on the odds of HCM (OR 1.89 (1.32-2.70), P = 5 × 10-4 and OR 1.94 (1.43-2.63), P = 2 × 10-5, for approaches i and ii, respectively), while the effect of strainrad and LVEF on HCM risk was only significant using the latter more SNP inclusive approach (ii) in the IVW model (OR 1.38 (1.18-1.62), P = 8 × 10-5 and OR 1.37 (1.10-1.69), P = 4 × 10-3, for strainrad and LVEF, respectively). The effect of strainlong on HCM was not significant. Notably, the magnitude of HCM risk increase with increased contractility is important, e.g., each unit (1%) increase in LVEF and -straincirc increases the risk of HCM by 37% and 89%, respectively (Supplementary Table 12). To place this in context, the standard deviations of LVEF and straincirc in the UK Biobank are 5.5% and 3.1%, respectively.
We performed MAGMA19 gene-based analyses using the HCM and DCM MTAG11 summary statistics. Not surprisingly, gene property analysis for tissue specificity identified muscle (heart and skeletal) as significantly associated with both HCM and DCM (Supplementary Fig. 10). Similarly, MAGMA gene-set analysis identified significantly associated gene sets related to muscle contraction for cellular components (e.g. I Band, contractile fibre), biological processes (e.g. myofibril assembly, sarcomere organization) and molecular functions (actin binding) (Supplementary Tables 13 and 14). Individual HCM, DCM and LV traits loci were annotated with proxy coding variants, significant expression (eQTL) and splice (sQTL) quantitative trait loci in skeletal and heart muscle, and chromatin interactions using Hi-C data obtained in left ventricular tissue (Supplementary Tables 15-18). The established Mendelian cardiomyopathy genes BAG3 (in loci HCM3, DCM3, LV10), ALPK3 (HCM14, DCM10, LV13), FHOD3 (HCM6, DCM12), TTN (DCM4, LV4), FLNC (HCM11, DCM2) and PLN (HCM2, LV8), which directly overlap associated loci (defined with r 2 > 0.6 from the lead SNP), are highly plausible candidates for the functional effects of variation at the corresponding loci. The involvement of FHOD3 and FLNC is further supported by eQTL effects, and involvement of PLN, ALPK3 and TTN is supported by evidence for Hi-C chromatin interactions between the association loci and the gene promoter. Notably, two loci overlap genes that play key roles in cardiomyocyte calcium handling related with muscle contraction (PLN and CASQ2; each supported by eQTL effects). Other candidate genes that emerge based on annotation and prior knowledge include GATA4 (DCM7, LV19)20, PRKCA (HCM15, DCM11)21, HSPB7 (HCM1, DCM1, LV1)22 and TMEM43 (DCM5)23. In aggregate, candidate genes at associated loci suggest susceptibility mechanisms involving regulation of sarcomere assembly, homeostasis, and calcium handling in cardiomyocytes.
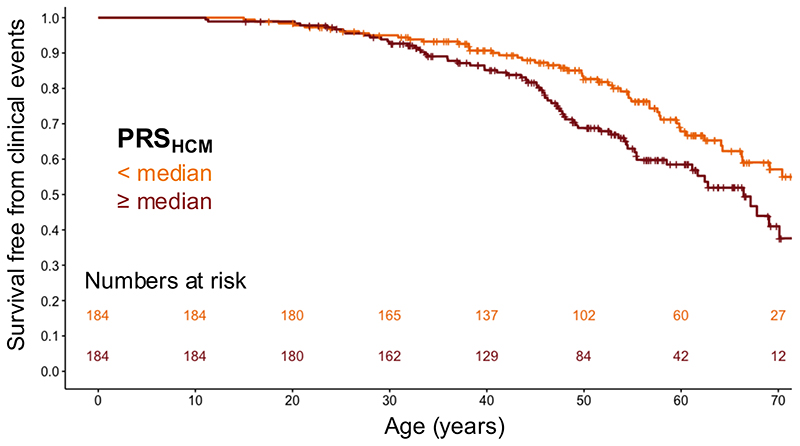
HCM attributed to rare disease-causing sarcomeric variants is characterized by variable disease severity. We investigated whether common variants could explain such phenotypic variability. We first derived a polygenic risk score (PRSHCM; Supplementary Table 19) from an HCM GWAS meta-analysis excluding a hold-out cohort of cases with sarcomeric variants from a single (Dutch) centre. We then assessed the association of PRSHCM with HCM expression and severity in the hold-out cohort and their family members (368 carriers of pathogenic or likely pathogenic sarcomeric variants, Supplementary Table 20). The results are shown in Supplementary Table 21. PRSHCM was associated with maximal left ventricular wall thickness (maxLVWT) indexed to body surface area (BSA), where each standard deviation (SD) increase in the PRSHCM is associated with a 0.7 mm/m2 increase in maxLVWT (P = 1 × 10-4), corresponding to a clinically relevant 1.4 mm absolute increase in maxLVWT for an average BSA of 1.95 m2 (cohort mean, Dutch population). PRSHCM was also associated with adverse clinical events (a composite of septal reduction therapy, cardiac transplantation, sustained ventricular arrhythmia, sudden cardiac death, appropriate defibrillator therapy, or atrial fibrillation/flutter), where each SD increase in PRSHCM was associated with a 28% relative risk increase in adverse clinical events (hazard ratio 1.28, 95% CI 1.06-1.54; P = 9 × 10-3). Figure 5 shows the event-free survival in sarcomeric variant carriers stratified by PRSHCM above vs. below the median.
Figure 5. A polygenic risk score for HCM stratifies event-free survival in carriers of disease-causing variants in sarcomere-encoding genes.
Kaplan-Meier curves showing survival free from adverse clinical events (composite of septal reduction therapy, cardiac transplantation, sustained ventricular arrhythmia, sudden cardiac death, appropriate implantable cardioverter defibrillator [ICD] therapy or atrial fibrillation/flutter) in sarcomeric (likely) pathogenic variant carriers stratified by polygenic score (PRSHCM) below (dark orange) vs. above (dark red) the median. Numbers at risk in each group along the time scale shown at the bottom of the plot. Ticks along the survival curves represent subject censoring. Two-sided log-rank test P = 0.032 (Cox proportional hazard analysis P = 9 × 10-3; see Supplementary Table 21).
Several novel observations emerge from this work: (i) by conducting the first well-powered GWAS in HCM and the largest GWAS meta-analysis in DCM, we identified 15 novel loci associated with HCM, of which 14 replicate in an independent cohort, and 7 novel loci for DCM, bringing the total number of loci to 16 and 13, respectively; (ii) we identified a total of 23 loci for LV traits and extended the study of these traits to include LV strain (13 loci) and mean LV wall thickness (6 loci); (iii) we demonstrate for the first time a direct genetic correlation between LV traits and susceptibility to HCM and DCM with opposing direction of effect, indicating shared pathways for these disorders; (iv) by using MR, we demonstrate that increased cardiac contractility plays an etiologic role in HCM. The demonstration of causal common variant effects on HCM through increased contractility broadens the applicability of therapeutic strategies targeting contractility as has been proposed for rare variants in sarcomere genes24; and; (v) we provide the first evidence that a polygenic score based on common HCM susceptibility variants may explain inter-individual differences in HCM disease severity among carriers of rare disease-causing variants. This work constitutes a proof-of-principle for potential use of polygenic risk scores in HCM risk stratification, to be assessed in future purposely designed and adequately powered studies. More broadly, this work demonstrates that the same genetic pathways may lead to distinct disorders through opposing genetic effects.
Methods
The overall study design and flowchart are shown in Figure 1 and described in detail below and in the Supplementary Note. All human subjects provided written informed consent, and all studies had received approval from the appropriate ethical review boards (see Reporting Summary).
GWAS of hypertrophic cardiomyopathy
Case inclusion
Unrelated cases with hypertrophic cardiomyopathy (HCM) were included from cardiovascular genetics referral centres (Supplementary Table 1). Cases were included if they had a clinical diagnosis of HCM according to current diagnostic criteria3: left ventricular (LV) wall thickness (LVWT) of ≥ 15 mm, ≥ 13 mm in presence of family history of HCM, or Z-score > 2 in children, where LV hypertrophy is not solely explained by loading conditions. Cases were excluded if they had syndromic HCM (e.g. Noonan syndrome spectrum), metabolic disease (e.g. Fabry) or had > 1 sarcomeric pathogenic or likely pathogenic variants (homozygous, compound heterozygous or digenic). The maximal LVWT was collected from chart review of cases using the most recent cardiac imaging report available, prior to septal reduction therapy or cardiac transplantation, if performed. Because cases were referred from multiple centres for cardiogenetic evaluation, imaging data were not available for standardized re-measurements in most cases. Cases underwent targeted sequencing of genes associated with HCM, as per local practice at the time of analysis. Rare variants detected through sequencing in each of the contributing cohorts of this study were centrally assessed for pathogenicity according to the American College of Medical Genetics and Genomics and the Association for Molecular Pathology (ACMG/AMP) guidelines25, using an adapted version of the CardioClassifier resource26 (details provided in the Supplementary Note, and classification results in Supplementary Table 2).
GWAS analysis design
Quality control (QC) and case-control association analysis were performed in three strata (Netherlands, NL; Royal Brompton & Harefield Hospitals, RBH; and Canada, CAN) followed by meta-analysis. See Supplementary Note for details regarding each stratum, including enrolling centres, DNA genotyping, QC and imputation.
Association analysis
The association of alternate allele dosage with HCM was performed for each of the three strata using a frequentist test in an additive model implemented in SNPTEST (v2.5.2 for the CAN and NL strata, v2.5.4 for the RBH stratum), correcting for the first three genotypic principal components. The results of the three strata were then combined using an inverse variance weighted fixed-effect meta-analysis, performing meta-analysis heterogeneity analysis, implemented in METAL27 (version released on 2011-03-25). SNPs that were missing in any of the three strata, as well as those with a heterogeneity test P < 0.05, were excluded. The stringent exclusion P value threshold for meta-analysis heterogeneity test was necessary to exclude most common SNPs that tag one of the population-specific founder HCM-causing pathogenic variants (e.g. NM_000256.3[MYBPC3]: c.2373dup [p.Trp792fs] in Dutch; NM_000364.4[TNNT2]: c.881G>A [p.Trp294Ter] in French Canadians). A subgroup analysis restricted to HCM cases without founder variants in MYBPC3 was performed. For the sake of this analysis, a founder variant was defined as a rare variant classified as pathogenic or likely pathogenic that was observed at least 10 times in the case cohort combining all strata. Using this definition, HCM cases carrying the following MYBPC3 variants (ENST00000545968) were excluded from this analysis: c.2373dupG, c.2827C>T, c.2864_2865delCT, c.3776delA, c.481C>T, c.551dupT, c.927-2A>G. Variant c.551dupT is a French Canadian founder while all other are Dutch founders. The summary results of this sub-analysis are shown in Supplementary Figure 1. The results of the main HCM case-control meta-analysis are shown in Figure 2, Table 1 and Supplementary Table 4. QQ plots of each stratum and forest plots for lead SNPs at all significant loci are shown in Supplementary Figure 2.
Analysis of heritability attributable to common variants
We used the generalized restricted maximum likelihood (GREML) approach of GCTA (version 1.92.4 beta)4,28 to estimate how much of the variance in HCM susceptibility could be attributed to common genetic variants (SNP-based heritability, h 2 SNP). The analysis was performed by stratum (NL, RBH, CAN), followed by a fixed-effects and random-effects meta-analysis using the meta package (v.4.9-9) in R version 3.6.0. Prior to heritability analyses, we performed additional stringent post-imputation QC as suggested29, using hard call genotypes (genotype probability, GP > 0.9) and excluding SNPs with missing rate > 0.01, minor allele frequency (MAF) < 0.05, Hardy-Weinberg test P < 0.05 and phenotype biased missingness P < 0.05, as well as samples with missing rate > 0.01, and excluded regions in the genome that tag founder HCM-causing rare variants in HCM (chr11:42008264-65380094 [MYBPC3] in NL and CAN, and chr1: 196816127-204926893 [TNNT2] in CAN). We then generated a genetic relationship matrix (GRM) and excluded distantly related individuals (proportion identity by descent, IBD > 0.05). We estimated h 2 SNP on the liability scale assuming a prevalence of 0.2%3 with the first 10 genotypic principal components and sex as covariates. In addition to standard GREML, we also performed GREML with linkage disequilibrium and MAF stratification (GREML-LDMS)5 with 200-kb segmentation, stratification of SNPs in four sets by LD scores of individual SNPs in R followed by GRM estimation per SNP stratum and REML analysis with multiple GRMs. To estimate the h 2 SNP specific to the previously published locus (near FHOD3), the 15 novel loci and the rest of the genome, we also performed partitioned heritability analysis using GREML, using the same QC and prevalence described for overall h 2 SNP. The genome was partitioned into three segments (FHOD3 locus, 15 novel loci, and rest of genome). Loci were defined based on the lead SNP ± 500 kb. Analysis was also performed by stratum, followed by an inverse variance weighted meta-analysis using a fixed-effects and random-effects models. The results of h 2 SNP estimation are shown in Supplementary Table 3.
HCM GWAS replication dataset
Replication of HCM loci reaching the significance threshold was tested in an independent dataset comprised of 2,694 cases with HCM included from the Hypertrophic Cardiomyopathy Registry (HCMR)17 or the NIHR Bioresource for Rare Disease (BRRD)18, and 47,486 controls without HCM included from the UK Biobank or BRRD. The detailed methodology is described in Harper et al.16 Of the 2,780 HCM cases included in Harper et al.16, 86 are overlapping with the present discovery dataset and have been excluded from the replication dataset. The results of this replication analysis and a fixed effects model meta-analysis combining the discovery and replication results are shown in Supplementary Table 10.
Meta-analysis of association studies in dilated cardiomyopathy
A meta-analysis of three published case-control association studies7–9 of dilated cardiomyopathy (DCM) was performed. The included studies are described in the Supplementary Note and in Supplementary Table 5. A fixed-effects meta-analysis was performed using METAL27 (version released on 2011-03-25). Study weighting was performed using the case sample size. The results of the DCM meta-analysis are shown in Extended Data Figure 1 and Supplementary Table 7. QQ plots for each study and forest plots for the lead SNPs at all significant loci are shown in Supplementary Figure 7.
GWAS of cardiac magnetic resonance-derived left ventricular traits
UK Biobank (UKBB) study population
The UKBB is an open-access population cohort resource that has recruited half a million participants in its initial recruitment phase, from 2006-2010. At the time of analysis, robust cardiac magnetic resonance (CMR) imaging data was available from 26,523 individuals in the imaging substudy. The UKBB CMR acquisition protocol has been described previously30. In brief, images were acquired according to a basic cardiac imaging protocol using clinical 1.5 Tesla wide bore scanners (MAGNETOM Aera, Syngo Platform VD13A, Siemens Healthcare, Erlangen, Germany) in three separate imaging centers. Extensive clinical and questionnaire data and genotype are available for these individuals. Clinical data were obtained at the time of the imaging visit. These included sex (31), age (21,003), weight (21,002), height (50), SBP (4,080), DBP (4,079), self-reported non-cancer illness code (20,002), and ICD10 codes (41,270). The mean age at the time of CMR was 63 ± 8 (range 45-80), and 46% of participants were male. Cohort anthropometrics, demographics and comorbidities are reported in Supplementary Table 6. Exclusion criteria for the UKBB imaging substudy included childhood disease, pregnancy and contraindications to MRI scanning. For the current analysis, we also excluded, by ICD-10 code and/or self-reported diagnoses, any subjects with heart failure, cardiomyopathy, a previous myocardial infarction, or structural heart disease. We also excluded those with uncontrolled hypertension (defined by systolic or diastolic blood pressure > 180 mmHg or > 110 mmHg, respectively, at time of imaging visit) or with extremes of body mass index (BMI < 16 or > 40). We restricted our analysis to Caucasians, defined by genotype as described (http://www.ukbiobank.ac.uk/wp-content/uploads/2014/04/UKBiobank_genotyping_QC_documentation-web.pdf). After phenotyping, we also excluded subjects with mean wall thickness > 13 mm in any of the 16 American Heart Association (AHA) left ventricular (LV) segments31, and subjects with outlying (>3 SD from mean) LV mass, LV volumes or LV ejection fraction. The UKBB received National Research Ethics Approval (REC reference 11/NW/0382). All participants provided informed consent (https://biobank.ctsu.ox.ac.uk/crystal/docs/Consent.pdf). The present study was conducted under terms of UKBB access approval 18545.
LV trait phenotyping
A description of CMR image analysis appears in the Supplementary Note and Supplementary Figure 3. We included nine LV phenotypes for GWAS analyses: end-diastolic volume (LVEDV), end-systolic volume (LVESV), ejection fraction (LVEF), mass (LVM), concentricity (LVconc = LVM/LVEDV), mean wall thickness (meanWT), as well as global peak strain in radial (strainrad), longitudinal (strainlong) and circumferential (straincirc) directions. The mean and SD of all nine LV phenotypes overall and stratified by sex are shown in Supplementary Table 6. The distributions of raw measures of these nine phenotypes are shown in Supplementary Figure 4. Despite non-normal distribution of some of the raw LV phenotypes, the residuals from our regression model including covariates (as defined below) approximated to normal distributions for all phenotypes (Supplementary Fig. 5). Therefore, the primary analysis was conducted using raw, non-normalized phenotypes.
LV trait genome-wide association analyses
A description of genotyping, imputation and QC appears in the Supplementary Note. The GWAS model included age, sex, height, weight, and mean arterial pressure (MAP) as covariates. We performed a meta-analysis comprised of two strata. Subjects recruited and imaged in the North of England (Cheadle, Newcastle) were treated as the first stratum (n = 15,215 after exclusions) and those recruited in the South of England (Reading) comprised the second stratum (n = 4,045). For each stratum, BOLT-LMM32 (v2.3.2) was used to construct mixed models for association with around 9.5 million directly genotyped and imputed SNPs. A high-quality set of directly genotyped model SNPs was selected to account for random effects in the genetic association analyses. These were selected by MAF (> 0.001), and LD-pruned (r 2 < 0.8) to create an optimum SNP set size of around 500,000. The model was then applied to the > 9.8 million imputed SNPs passing quality control and filtering. Inverse-variance weighted meta-analysis was carried out with summary statistics from both strata using METAL27 (version released on 2011-03-25). The results of the LV traits GWAS are shown in Supplementary Table 8 and Extended Data Figures 2–10.
GWAS statistical significance threshold
We accounted for multiple testing to define the P value threshold for genome-wide statistical significance in the HCM, DCM, and LV traits GWAS. As expected, LV trait pairs are phenotypically correlated (Supplementary Table 9). Supplementary Figure 6 displays a phenotypic correlation heatmap using absolute values with dendrograms constructed using Euclidean distance and complete hierarchical clustering. Cutting the dendrogram at a height of 1.5 results in three LV phenotype clusters (Supplementary Fig. 6): (i) a contractility cluster comprising strainlong, straincirc, strainrad and LVEF; (ii) a LV volume cluster comprising LVEDV and LVESV; and (iii) a LV hypertrophy cluster comprising LVM, meanWT and LVconc. The genome-wide significance threshold for the GWAS of LV traits, HCM and DCM was therefore set to P < 1 × 10-8 (5 × 10-8/5; accounting for three LV clusters + DCM + HCM).
Pairwise genetic correlation
We performed pairwise genetic correlation between HCM, DCM and the nine LV traits using LDSC (Version 1.0.1)13,14. For each GWAS, we first reformatted summary statistics using the “munge_sumstats.py” command, filtering for the HapMap3 SNPs with corresponding alleles using the “--merge-alleles w_hm3.snplist” flag, as recommended. The HapMap3 SNPs were downloaded from “https://data.broadinstitute.org/alkesgroup/LDSCORE/w_hm3.snplist.bz2”. We then assessed genetic correlation for each of the 55 pairs (HCM, DCM and nine LV traits) using the “ldsc.py –rg” command and pre-computed LD scores from the European 1000 Genomes Project dataset which were downloaded from “https://data.broadinstitute.org/alkesgroup/LDSCORE/eur_w_ld_chr.tar.bz2”. We did not constrain the single-trait and cross-trait LD score regression intercepts. The results of the genetic correlation analyses are shown in Figure 3 and Supplementary Table 9.
Multi-trait analysis of GWAS
We performed multi-trait analysis of GWAS summary statistics using MTAG (version 1.0.8)15 to increase power for discovery of genetic loci associated with HCM, DCM and LV traits. MTAG uncovers genetic loci associated with a phenotype when the standard single-trait GWAS is underpowered. By definition, it will uncover new genetic loci whenever these are associated with the other phenotypes included. It may theoretically fail to identify phenotype-specific loci. Two MTAG analyses were performed, one only including the summary statistics of the nine LV traits (referred to as “MTAG9”), and another one including the summary statistics of the nine LV traits, HCM and DCM (referred to as “MTAG11”). Specifically, MTAG9 was used for additional locus discovery for the nine LV traits, and MTAG11 was used for additional locus discovery for HCM and DCM. Only SNPs included in all meta-analyses (i.e. HCM, DCM and LV traits) were used in MTAG. The coded/non-coded alleles were aligned for all 11 studies prior to MTAG, and multi-allelic SNPs were removed. All summary statistics refer to the positive strand of GRCh37 and, as such, ambiguous/palindromic SNPs (having alleles A/T or C/G) were not excluded. Regression coefficients (beta) and their standard errors were used for MTAG9, and Z-scores were used for MTAG11 (since regression coefficients and standard errors were not directly available in all included DCM studies). The results of the multi-trait analyses are shown as panel b in Figure 2 and Extended Data Figures 1–10, as well as Supplementary Tables 4, 7, and 8.
Two sample Mendelian randomization
We assessed whether increased contractility is causally linked to increased HCM risk using two sample Mendelian randomization (MR) using LV contractility parameters as exposure variables and HCM as an outcome. Two sample MR was performed using the TwoSampleMR (MRbase) package33 (version 0.4.25) in R version 3.6.0. Four exposure variables corresponding to measures of LV contractility were used separately: LVEF as a volumetric marker of contractility, and global strain (circumferential (straincirc), radial (strainrad) and longitudinal (strainlong)) as contractility markers based on myocardial tissue deformation. We used two approaches for instrument SNP selection: (i) P < 5 × 10-8 in the single LV trait analysis; and (ii) P < 5 × 10-8 in the MTAG9 analysis (i.e. excluding HCM and DCM). Only independent SNPs (using r 2 < 0.01 in the European 1000 Genomes population) were included. The outcome summary statistics were those of the single trait HCM case-control meta-analysis. Insertions/deletions and palindromic SNPs with intermediate allele frequencies (MAF > 0.42) were excluded, and other SNPs in the same locus were included only if reaching the P value threshold for instrument SNP selection. The SNPs included in MR analyses and their effects in the exposure and outcome studies are shown in Supplementary Table 11. Inverse variance weighting (IVW) was used as a primary analysis. We used three additional methods as sensitivity analyses: Weighted median34, weighted mode35 and robust adjusted profile score (RAPS)36. RAPS was used with the default parameters (over.dispersion = TRUE and loss.function = “tukey”). MR Egger37 was not used given the limited number of SNP instruments. Cochran’s Q statistics were calculated to investigate heterogeneity between SNP causal effects using IVW. Evidence of directional pleiotropy was also assessed using the MR-Egger intercept. Mean F-statistics were calculated to assess the strength of the genetic instruments used. Leave-one-out analysis was also performed to ensure the SNP causal effects are not driven by a particular SNP. We also performed a secondary analysis using the generalized summary-data-based Mendelian randomization (GSMR) implemented in GCTA (version 1.92.4 beta)38. LD estimation was performed using the European samples from the 1000 Genomes Project reference dataset. As for the analysis using TwoSampleMR, instrument SNPs were selected using two approaches (see i and ii above). The default parameters were used, with the following exceptions: the r 2 clumping threshold was changed to 0.01, and the minimum number of SNPs required was changed to 5. Removal of pleiotropic SNPs was performed as suggested using the GSMR-implemented HEIDI outlier algorithm with default parameters38. The results of the MR are shown in Supplementary Table 12, with effect plots and leave-one-out analyses shown in Supplementary Figures 8 and 9, respectively.
Genome-wide visualization and annotation
Summary statistics for all single-trait and multi-trait analyses were uploaded to FUMA (Functional Mapping and Annotation of GWAS, v1.3.5)39 for visualization and genome-wide analyses. Manhattan and quantile-quantile plots were constructed. Gene-set and tissue expression analyses were performed using MAGMA19 v1.07, as implemented in FUMA. We used Gene Ontology (GO) gene sets from the Molecular Signatures Database (MSigDB, v6.2)40 for the gene-set analysis, and the Genotype-Tissue Expression project (GTEx41, version 8) for the tissue specificity analysis. The results of MAGMA analyses using the HCM and DCM MTAG summary statistics are shown in Supplementary Tables 13 and 14 (gene-set analyses) and Supplementary Figure 10 (tissue specificity analyses).
Locus annotation
All loci associated with HCM, DCM and LV traits in the single trait or multi-trait analyses were annotated using lookup for: (i) proxy coding variants (Supplementary Table 15), (ii) cis-expression quantitative trait loci (eQTL, Supplementary Table 16), (iii) cis-splice quantitative trait loci (sQTL, Supplementary Table 17), and (iv) contact with gene promoters using Hi-C (Supplementary Table 18). We also assessed whether GWAS loci co-localize with genes associated with Mendelian cardiomyopathy and performed cross-traits lookups for all HCM, DCM and LV traits loci (Fig. 4 and Supplementary Tables 4, 7, and 8). See Supplementary Note for methodological details.
Association of an HCM polygenic risk score (PRSHCM) with phenotypic manifestation in sarcomeric variant carriers
We sought to assess whether a common variant polygenic risk score derived from the HCM case-control GWAS (PRSHCM) accounts for phenotypic variability and severity in pathogenic or likely pathogenic variant carriers.
Study population
In this analysis, we included probands and family members that carry pathogenic or likely pathogenic variants associated with HCM from the Erasmus Medical Center (EMC). All variants were centrally curated as described in the Supplementary Note, and only subjects that carry (likely) pathogenic variants were included (see list in Supplementary Table 2). Homozygous carriers and those carrying multiple pathogenic or likely pathogenic variants were excluded. Clinical data including maximal left ventricular wall thickness on cardiac imaging and time of clinical events were retrieved from an ongoing registry42,43, including all HCM patients and their relatives at the EMC. Missing data were collected from chart review. The baseline characteristics of the study population are shown in Supplementary Table 20.
Derivation of an HCM polygenic risk score (PRSHCM)
The PRSHCM was derived from an independent GWAS, excluding HCM cases with (likely) pathogenic variants from EMC. This was done to ensure that the PRS is derived from an independent cohort. Specifically, a Dutch HCM case-control GWAS was repeated after excluding 161 cases from EMC cases that carry (likely) pathogenic variants, followed by a meta-analysis combined with the RBH HCM GWAS and the CAN HCM GWAS, followed by MTAG11 as described above. Lead SNPs reaching the genome-wide significance threshold (P < 5 × 10-8) were included in the PRSHCM. Those SNPs included in the PRSHCM and their corresponding weights are shown in Supplementary Table 19.
Calculation of PRSHCM
All EMC samples that carry pathogenic or likely pathogenic HCM variants underwent array genotyping on the Illumina GSA. Quality control (QC) was performed as described in the Supplementary Note for the Dutch HCM GWAS, except for identify by descent (IBD) analysis, where only duplicate (or twin) samples were excluded. No sample was excluded for relatedness, which was accounted for using a GRM as described in the statistical analyses paragraph below. Imputation and post-imputation QC were performed also as described for the GWAS in the Supplementary Note. PRSHCM was calculated by summing the products of each lead risk allele dosage by the corresponding regression coefficient in the derivation study (Supplementary Table 19) using Plink 2.0, followed by scaling to a mean of 0 and SD of 1.
Study endpoints
Two primary endpoints were predefined. The first primary endpoint was maximal left ventricular wall thickness (maxLVWT) at last available transthoracic echocardiogram (TTE) or CMR. MaxLVWT is routinely assessed in clinical practice as a measure of HCM severity and for risk stratification of life-threatening ventricular arrhythmias3. For subjects that had cardiac transplantation and/or septal reduction therapy to relieve LV obstruction, the last available CMR or TTE prior to cardiac transplantation and/or septal reduction therapy was used. Considering the higher accuracy of CMR to assess LVWT in all LV segments, maxLVWT from CMR was used whenever available unless TTE was performed more than 5 years after last CMR. To account for body size, a determinant of LVWT in the general population44 and in HCM45, maxLVWT was indexed to body surface area (BSA) calculated using the DuBois formula (0.007184 x height (cm)0.725 x weight (kg)0.425). The second primary endpoint was time to first adverse clinical event (a composite of invasive septal reduction therapy, cardiac transplantation, sustained ventricular arrhythmia, sudden cardiac death, appropriate defibrillator therapy or atrial fibrillation/flutter). The components of this composite endpoint were also assessed as secondary endpoints. As a sensitivity analysis, we also performed an analysis for the primary outcomes restricted to non-probands.
Statistical analyses
A GRM was estimated using GCTA (version 1.92.4 beta) and used to account for the between-sample relatedness. The association of PRSHCM with maxLVWT indexed to BSA (in mm/m2) was performed using a mixed linear model integrating the GRM as a random effect. Neither sex, nor rare variant type (MYBPC3 truncation vs. others) were associated with maxLVWT and were therefore not included as covariates. The association of PRSHCM with the composite primary clinical events endpoint and secondary endpoints were assessed using a Cox proportional hazards mixed effects model integrating the GRM as a random effect. Since biological male sex was significantly associated with increased risk for clinical events, it was added as a fixed effect covariate. Time 0 was set to birth in the Cox model to maximize statistical power by including events that occurred at the time of first medical encounter. Given the genetic nature of our exposure factor, all study subjects are exposed since birth. Nevertheless, there is a possibility of selection bias in our cohort, since study subjects have to reach the age of inclusion. Study subjects were censored at the time of last clinical follow-up. For analyses of secondary endpoints that do not include cardiac transplantation, study subjects were also censored at the time of cardiac transplantation. In addition to PRSHCM, we also assessed the association of a genome-wide score (PRSAF) derived from a large atrial fibrillation meta-analysis and validated by Khera et al.46 with atrial fibrillation within the study population. Mixed effects analyses of PRSHCM with maxLVWT was performed using the lmekin and mixed effects analyses of PRSHCM and PRSAF with clinical events was performed using the coxme function, both from the coxme package v2.2-14 in R version 3.6.0. The statistical significance threshold was set to P < 0.025 for the primary endpoints (0.05/2 primary endpoints) and P < 0.05 for hypothesis-generating secondary endpoints. The results of the primary, secondary and sensitivity analyses are shown in Supplementary Table 21. Kaplan-Meier curves stratified by PRSHCM above or below the median are shown in Figure 5.
Extended Data
Extended Data Fig. 1. Manhattan and QQ plots of DCM GWAS and MTAG.
a,b, Summary results of the dilated cardiomyopathy (DCM) GWAS meta-analysis of 5,521 cases and 397,323 controls shown as Manhattan plots for the single trait (a) and the multi-trait analyses (MTAG; b). Single trait analysis (a) consisted of a fixed effects meta-analysis of case-control GWAS using summary statistics of three previously published DCM GWAS, and multi-trait analysis results (b) were obtained using MTAG for DCM, including GWAS for hypertrophic cardiomyopathy (HCM) and nine left ventricular (LV) traits. Red dashed line shows the significance threshold of P = 1 × 10-8. Quantile-quantile (QQ) plots shown as inserts in corresponding panels. Genomic inflation (λ) = 1.028 (single-trait) and 1.049 (MTAG). Numbering of signals as shown in Supplementary Table 7. Black numbers refer to loci reaching the statistical significance threshold in single trait analysis, while red numbers refer to loci only reaching statistical significance in the multi-trait analysis. The low density of association signals in the single trait analysis (a) is attributable to the inclusion of a large sample size study that used a low density array (Illumina Infinium HumanExome BeadChip; Supplementary Table 5).
Extended Data Fig. 2. Manhattan and QQ plots of LV ejection fraction GWAS and MTAG.
a,b, Summary results of the left ventricular ejection fraction (LVEF) GWAS in the UK Biobank (n = 19,260) shown as Manhattan plots for the single trait (a) and the multi-trait analyses (MTAG; b). Single trait analysis (a) consisted of a fixed effects meta-analysis of case-control GWAS using a linear mixed model (BOLT-LMM), and multi-trait analysis results (b) were obtained using MTAG including summary statistics for all nine left ventricular (LV) traits. Red dashed line shows the significance threshold of P = 1 × 10-8. Quantile-quantile (QQ) plots shown as inserts in corresponding panels. Genomic inflation (λ) = 1.041 (single-trait) and 1.049 (MTAG). Numbering of loci as shown in Supplementary Table 8. Black numbers refer to loci reaching the statistical significance threshold in any single trait analysis, while red numbers refer to loci only reaching statistical significance in the multi-trait analysis.
Extended Data Fig. 3. Manhattan and QQ plots of LV concentricity GWAS and MTAG.
a,b, Summary results of the left ventricular concentricity index (LVconc) GWAS in the UK Biobank (n = 19,260) shown as Manhattan plots for the single trait (a) and the multi-trait analyses (MTAG; b). LVconc is defined as the ratio of left ventricular mass to the left ventricular end-diastolic volume. Single trait analysis (a) consisted of a fixed effects meta-analysis of case-control GWAS using a linear mixed model (BOLT-LMM), and multi-trait analysis results (b) were obtained using MTAG including summary statistics for all nine left ventricular (LV) traits. Red dashed line shows the significance threshold of P = 1 × 10-8. Quantile-quantile (QQ) plots shown as inserts in corresponding panels. Genomic inflation (λ) = 1.06 (single-trait) and 1.084 (MTAG). Numbering of signals as shown in Supplementary Table 8. Black numbers refer to loci reaching the statistical significance threshold in any single trait analysis, while red numbers refer to loci only reaching statistical significance in the multi-trait analysis.
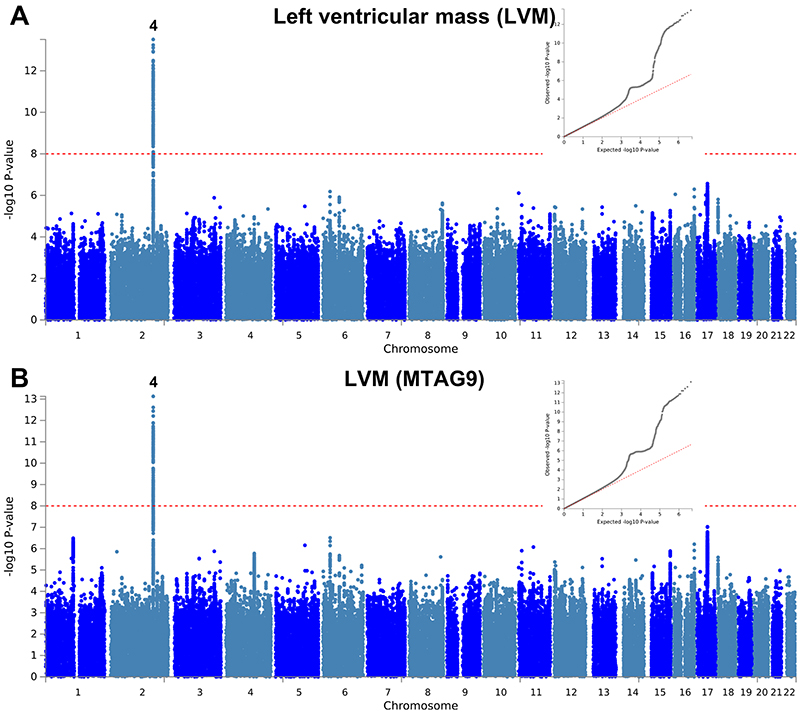
Extended Data Fig. 4. Manhattan and QQ plots of LV mass GWAS and MTAG.
a,b, Summary results of the left ventricular mass (LVM) GWAS in the UK Biobank (n = 19,260) shown as Manhattan plots for the single trait (a) and the multi-trait analyses (MTAG; b). Single trait analysis (a) consisted of a fixed effects meta-analysis of case-control GWAS using a linear mixed model (BOLT-LMM), and multi-trait analysis results (b) were obtained using MTAG including summary statistics for all nine left ventricular (LV) traits. Red dashed line shows the significance threshold of P = 1 × 10-8. Quantile-quantile (QQ) plots shown as inserts in corresponding panels. Genomic inflation (λ) = 1.081 (single-trait) and 1.071 (MTAG). Numbering of signals as shown in Supplementary Table 8.
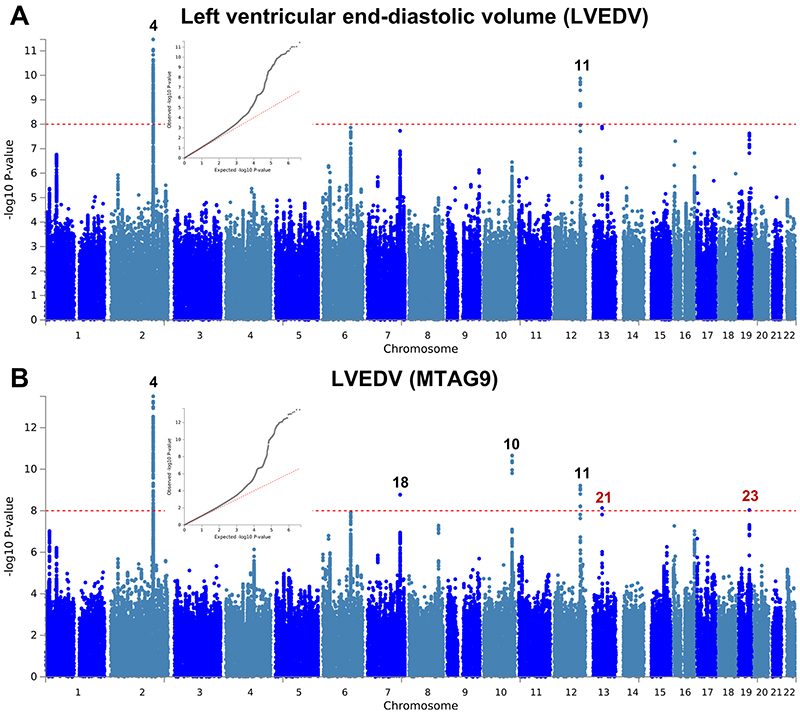
Extended Data Fig. 5. Manhattan and QQ plots of LV end-diastolic volume GWAS and MTAG.
a,b, Summary results of the left ventricular end-diastolic volume (LVEDV) GWAS in the UK Biobank (N=19,260) shown as Manhattan plots for the single trait (a) and the multi-trait analyses (MTAG; b). Single trait analysis (a) consisted of a fixed effects meta-analysis of case-control GWAS using a linear mixed model (BOLT-LMM), and multi-trait analysis results (b) were obtained using MTAG including summary statistics for all nine left ventricular (LV) traits. Red dashed line shows the significance threshold of P = 1 × 10-8. Quantile-quantile (QQ) plots shown as inserts in corresponding panels. Genomic inflation (λ) = 1.076 (single-trait) and 1.078 (MTAG). Numbering of signals as shown in Supplementary Table 8. Black numbers refer to loci reaching the statistical significance threshold in any single trait analysis, while red numbers refer to loci only reaching statistical significance in the multi-trait analysis.
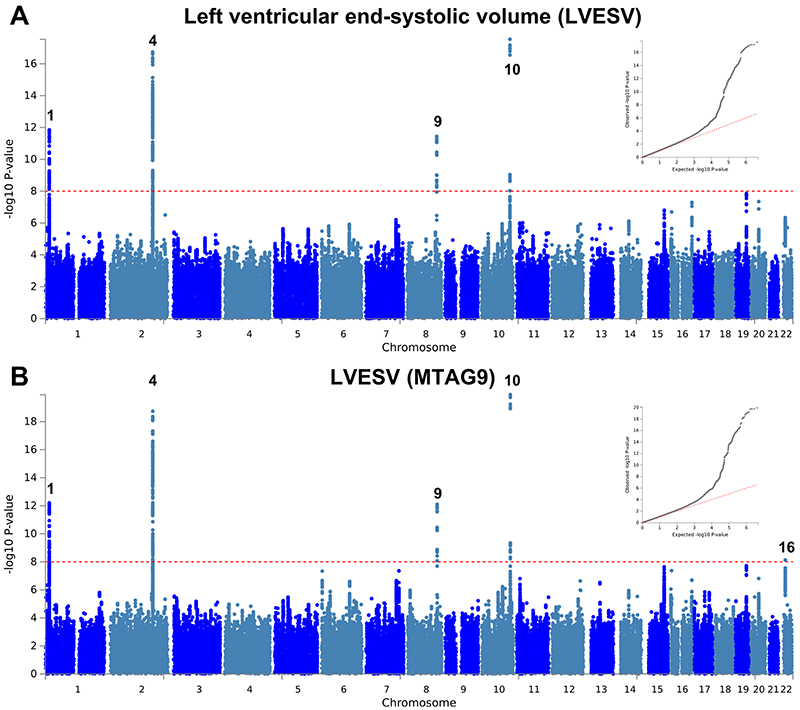
Extended Data Fig. 6. Manhattan and QQ plots of LV end-systolic volume GWAS and MTAG.
Summary results of the left ventricular end-systolic volume (LVESV) GWAS in the UK Biobank (n= 19,260) shown as Manhattan plots for the single trait (a) and the multi-trait analyses (MTAG; b). Single trait analysis (a) consisted of a fixed effects meta-analysis of case-control GWAS using a linear mixed model (BOLT-LMM), and multi-trait analysis results (b) were obtained using MTAG including summary statistics for all nine left ventricular (LV) traits. Red dashed line shows the significance threshold of P = 1 × 10-8. Quantile-quantile (QQ) plots shown as inserts in corresponding panels. Genomic inflation (λ) = 1.069 (single-trait) and 1.081 (MTAG). Numbering of signals as shown in Supplementary Table 8.
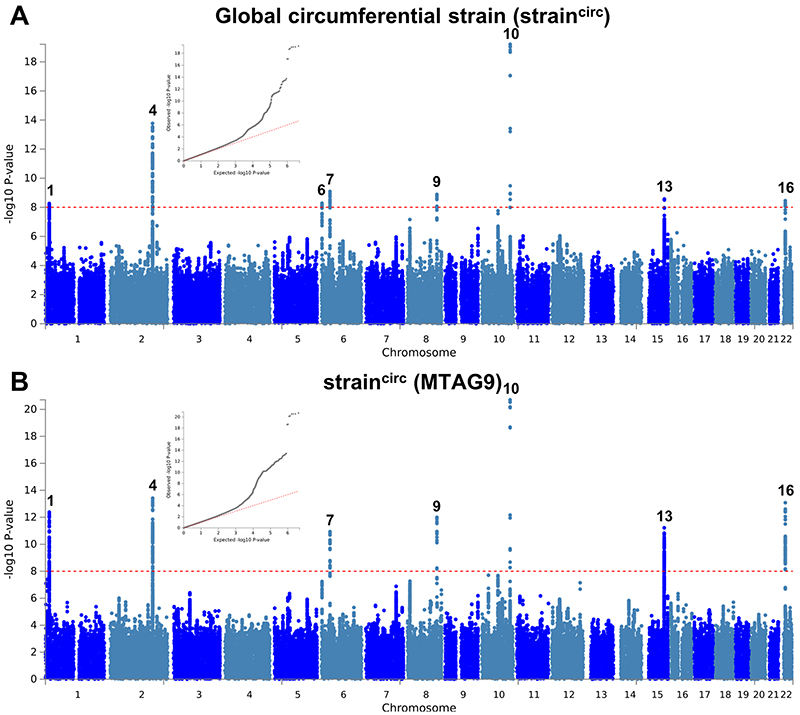
Extended Data Fig. 7. Manhattan and QQ plots of LV global circumferential strain GWAS and MTAG.
a,b, Summary results of the left ventricular global circumferential strain (straincirc) GWAS in the UK Biobank (N=19,260) shown as Manhattan plots for the single trait (a) and the multi-trait analyses (MTAG; b). Single trait analysis (a) consisted of a fixed effects meta-analysis of case-control GWAS using a linear mixed model (BOLT-LMM), and multi-trait analysis results (b) were obtained using MTAG including summary statistics for all nine left ventricular (LV) traits. Red dashed line shows the significance threshold of P = 1 × 10-8. Quantile-quantile (QQ) plots shown as inserts in corresponding panels. Genomic inflation (λ) = 1.046 (single-trait) and 1.061 (MTAG). Numbering of signals as shown in Supplementary Table 8.
Extended Data Fig. 8. Manhattan and QQ plots of LV global radial strain GWAS and MTAG.
a,b, Summary results of the left ventricular global radial strain (strainrad) GWAS in the UK Biobank (n = 19,260) shown as Manhattan plots for the single trait (a) and the multi-trait analyses (MTAG; b). Single trait analysis (a) consisted of a fixed effects meta-analysis of case-control GWAS using a linear mixed model (BOLT-LMM), and multi-trait analysis results (b) were obtained using MTAG including summary statistics for all nine left ventricular (LV) traits. Red dashed line shows the significance threshold of P = 1 × 10-8. Quantile-quantile (QQ) plots shown as inserts in corresponding panels. Genomic inflation (λ) = 1.049 (single-trait) and 1.057 (MTAG). Numbering of signals as shown in Supplementary Table 8. Black numbers refer to loci reaching the statistical significance threshold in any single trait analysis, while red numbers refer to loci only reaching statistical significance in the multi-trait analysis.
Extended Data Fig. 9. Manhattan and QQ plots of LV global longitudinal strain GWAS and MTAG.
a,b, Summary results of the left ventricular global longitudinal strain (strainlong) GWAS in the UK Biobank (n = 19,260) shown as Manhattan plots for the single trait (a) and the multi-trait analyses (MTAG; b). Single trait analysis (a) consisted of a fixed effects meta-analysis of case-control GWAS using a linear mixed model (BOLT-LMM), and multi-trait analysis results (b) were obtained using MTAG including summary statistics for all nine left ventricular (LV) traits. Red dashed line shows the significance threshold of P = 1 × 10-8. Quantile-quantile (QQ) plots shown as inserts in corresponding panels. Genomic inflation (λ) = 1.040 (single-trait) and 1.059 (MTAG). Numbering of signals as shown in Supplementary Table 8.
Extended Data Fig. 10. Manhattan and QQ plots of LV mean wall thickness GWAS and MTAG.
a,b, Summary results of the mean left ventricular wall thickness (meanWT) GWAS in the UK Biobank (n = 19,260) shown as Manhattan plots for the single trait (a) and the multi-trait analyses (MTAG; b). Single trait analysis (a) consisted of a fixed effects meta-analysis of case-control GWAS using a linear mixed model (BOLT-LMM), and multi-trait analysis results (b) were obtained using MTAG including summary statistics for all nine left ventricular (LV) traits. Red dashed line shows the significance threshold of P = 1 × 10-8. Quantile-quantile (QQ) plots shown as inserts in corresponding panels. Genomic inflation (λ) = 1.065 (single-trait) and 1.072 (MTAG). Numbering of signals as shown in Supplementary Table 8.
Supplementary Material
Acknowledgements
R.T. received funding from the Canadian Heart Rhythm Society (George Mines Award), the European Society of Cardiology (Research fellowship grant), the Philippa and Marvin Carsley Cardiology Chair at Université de Montréal and the Canadian Institutes of Health Research (funding reference number 169063), and is currently a clinical research scholar of the Fonds de Recherche du Québec—Santé (reference number 135055). C.F. received funding from a British Heart Foundation Clinical Research Training Fellowship FS/15/81/31817. X.X. is currently a post-doc scientist funded by the Medical Research Council London Institute of Medical Sciences. A.R.H. received support from the Medical Research Council Doctoral Training Partnership. R.W. received support from an Amsterdam Cardiovascular Sciences fellowship. The study was supported by the Netherlands Cardiovascular Research Initiative (CVON PREDICT 2012-10, DOSIS 2014-40, eDETECT 2015-12 and PREDICT2 2018-30). W.P.t.R. is supported by a Postdoctoral Fellowship CURE-PLaN (Netherlands Heart Institute) from the Leducq Foundation. J.A.O. received support from the Amsterdam UMC’s PhD scholarship. A.d.M. received support from the National Institute for Health Research Biomedical Research Centre based at Imperial College Healthcare NHS Trust and Imperial College London; the Medical Research Council, UK; the Academy of Medical Sciences (SGL015/1006) and a Mason Medical Research Trust grant. J.H.V. received funding from the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation programme (grant agreement n° 772376 - EScORIAL). E.K. received support from the German Center for Cardiovascular Research (DZHK) Rotation Grant. F.M. is supported by a post-doctoral research fellowship from the University of Florence and by the European Union Horizon 2020 framework programme (SILICOFCM, GA 777204). H.S. received support from grants from the Japan Society for the Promotion of Science (18K15410). D.S.-L. received support from UK Med-Bio. P.R. received support from the Federation Française de Cardiologie. P.R. and F.A. were supported by Ligue contre la cardiomyopathie. E.V. and P.C. received support from the CONNY-MAEVA charitable foundation and GenMed LABEX. R.A.H. is supported by the Jacob J. Wolfe Distinguished Medical Research Chair, the Edith Schulich Vinet Research Chair in Human Genetics, and the Martha G. Blackburn Chair in Cardiovascular Research, and has received operating grants from the Canadian Institutes of Health Research (Foundation award) and the Heart and Stroke Foundation of Canada (G-18-0022147). M.-P.D. holds a Canada Research Chair in Precision Medicine Data Analysis. J.-C.T. holds the Canada Research Chair in personalized medicine and the University of Montreal endowed research chair in atherosclerosis and is the principal investigator of the Montreal Heart Institute André and France Desmarais hospital cohort funded by the Montreal Heart Institute Foundation. R.T.L. is supported by a UK Research and Innovation Rutherford Fellowship. F.W.A. is supported by the UCL Hospitals NIHR Biomedical Research Centre. F.W.A. and P.C. received support from European Union’s Horizon 2020 research and innovation program under the ERA-NET Co-fund action no. 680969 (ERA-CVD DETECTIN-HF), jointly funded by the Dutch Heart Foundation (2016T096) and Netherlands Organisation for Health Research and Development (ZonMw). P.J.R.B. is supported by a Health Innovation Challenge Fund award from the Wellcome Trust and Department of Health, UK [HICF-R6–373]. D.P.O’R. is funded by the Medical Research Council, National Institute for Health Research Biomedical Research Centre based at Imperial College Healthcare NHS Trust and Imperial College London, and a British Heart Foundation Program Grant (RG/19/6/34387). K.J.H.V. receives support from the Foundation Volksbond Rotterdam. M.T. receives support from Monat foundation. G.L. is funded by the Montreal Heart Institute Foundation, the J.C. Edwards Foundation and the Canada Research Chair Program. Y.M.P. is supported by the Netherlands Heart Foundation (CVON PRIME). B.M. received support from DZHK, the German Ministry of Education and Research (CaRNAtion), Informatics for Life (Klaus Tschira Foundation) and the European Union (FP7 BestAgeing), and was supported by an excellence fellowship of the Else Kröner Fresenius Foundation. P.C. received funding from PROMEX charitable foundation. I.C. receives support from the Netherlands Cardiovascular Research Initiative: CVON2017-10 DOLPHIN-GENESIS. H.W. have received support from the Wellcome Trust core award [090532/Z/09/Z], the BHF Centre of Research Excellence, Oxford [RE/13/1/30181], the National Institute for Health Research (NIHR) Oxford Biomedical Research Centre and a National Heart, Lung, and Blood Institute grant [U01HL117006-01A1]. P.M.M. acknowledges generous personal and research support from the Edmond J Safra Foundation and Lily Safra, an NIHR Senior Investigator Award and the UK Dementia Research Institute. J.S.W. was supported by Wellcome Trust [107469/Z/15/Z]; British Heart Foundation [SP/17/11/32885; RE/18/4/34215]; Medical Research Council (UK); NIHR Royal Brompton Cardiovascular Biomedical Research Unit; NIHR Imperial College Biomedical Research Centre; a Health Innovation Challenge Fund award from the Wellcome Trust and UK Department of Health [HICF-R6-373]. C.R.B. acknowledges the support from the Netherlands Organization for Scientific Research (VICI fellowship, 016.150.610) and the Leducq Foundation (project 17CVD02). This research has been in part conducted using the UK Biobank Resource under Application Number 18454, and the Genotype-Tissue Expression (GTEx) Project supported by the Common Fund of the Office of the Director of the National Institutes of Health, and by NCI, NHGRI, NHLBI, NIDA, NIMH, and NINDS. This study was supported in part by a kind donation from family and friends of Jean-Paul Balkestein who died at 32 years of age from hypertrophic cardiomyopathy.
Footnotes
Author contributions
R.T., C.F., A.M.C.V., J.M.V., W.B., N.L., P.E., E.V., M.W.T.T., J.P.v.T., P.J.R.B., S.A.C., S.K.P., J.v.d.V., K.J.H.V., G.L., B.M., P.C., I.C., M.M., A.A.M.W., H.W., P.M.M., J.S.W., and C.R.B. conceived or designed elements of the study. R.T., C.F., X.X., A.M.C.V., A.R.H., R.H., K.K.B., R.W., E.T.H., W.P.t.R., R.J.B., H.G.v.V., M.A.v.S., J.M.V., J.A.O., W.B., A.d.M., L.B., J.C.K., J.H.V., E.K., A.P., A.J.B., N.W., F.M., G.S., H.S., D.S.-L., P.R., F.A., E.V., P.L., T.M., A.T., D.M., R.A.H., J.D.R., J.A., M.-P.D., J.C.-T., G.G., P.L.L., P.G., J.-C.T., S.M.B., R.T.L., F.W.A., P.J.R.B., S.A.C., S.K.P., D.P.O’R., M.T., G.L., Y.M.P., B.M., P.C., R.A.d.B., M.M., A.A.M.W., H.W., J.S.W., and C.R.B. acquired, analyzed or interpreted data. R.T., C.F., X.X., R.W., J.A.O., W.B., J.S.W., and C.R.B. drafted the manuscript. All authors critically revised the manuscript for important intellectual content and approved the final version.
Competing interests statement
M.-P.D. is author on a patent pertaining to pharmacogenomics-guided CETP inhibition (US20170233812A1), has a minor equity interest in DalCor and has received honoraria from Dalcor and Servier and research support (access to samples and data) from AstraZeneca, Pfizer, Servier, Sanofi and GlaxoSmithKline. J.-C.T. has received research grants from Amarin, AstraZeneca, DalCor, Esperion, Ionis, Sanofi and Servier; honoraria from AstraZeneca, DalCor, HLS, Sanofi and Servier; holds minor equity interest in DalCor; and is an author of a patent on pharmacogenomics-guided CETP inhibition (US20170233812A1). B.M. has received Research Funding from Siemens Healtheneers, Daiichi Sankyo. The UMCG, which employs R.A.d.B., has received research grants and/or fees from AstraZeneca, Abbott, Bristol-Myers Squibb, Novartis, Novo Nordisk, and Roche. R.A.d.B. received speaker fees from Abbott, AstraZeneca, Novartis, and Roche. HW is a consultant for Cytokinetics. P.M.M. receives an honorarium as Chair of the UKRI Medical Research Council Neuroscience and Mental Health Board. He acknowledges consultancy fees from Adelphi Communications, MedScape, Neurodiem, Nodthera, Biogen, Celgene and Roche. He has received speakers’ honoraria from Celgene, Biogen, Novartis and Roche, and has received research or educational funds from Biogen, GlaxoSmithKline and Novartis. He is paid as a member of the Scientific Advisory Board for Ipsen Pharmaceuticals. J.S.W. has received research support and consultancy fees from Myokardia, Inc.
Data availability statement
Data from the Genome Aggregation Database (gnomAD, v2.1) are available at https://gnomad.broadinstitute.org. Data from the UK Biobank participants can be requested from the UK Biobank Access Management System (https://bbams.ndph.ox.ac.uk). Data from the Genotype Tissue Expression (GTEx) consortium are available at the GTEx portal (https://gtexportal.org). Other datasets generated during and/or analyzed during the current study can be made available upon reasonable request to the corresponding authors. Individual level data sharing is subject to restrictions imposed by patient consent and local ethics review boards. Results from meta-analyses of genome-wide association studies reported in this article are available at https://www.heart-institute.nl/gwas and https://data.hpc.imperial.ac.uk [doi.org/10.14469/hpc/7468].
References
- 1.Semsarian C, Ingles J, Maron MS, Maron BJ. New perspectives on the prevalence of hypertrophic cardiomyopathy. J Am Coll Cardiol. 2015;65:1249–1254. doi: 10.1016/j.jacc.2015.01.019. [DOI] [PubMed] [Google Scholar]
- 2.Walsh R, et al. Quantitative approaches to variant classification increase the yield and precision of genetic testing in Mendelian diseases: the case of hypertrophic cardiomyopathy. Genome Med. 2019;11:5. doi: 10.1186/s13073-019-0616-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Elliott PM, et al. 2014 ESC Guidelines on diagnosis and management of hypertrophic cardiomyopathy: the Task Force for the Diagnosis and Management of Hypertrophic Cardiomyopathy of the European Society of Cardiology (ESC) Eur Heart J. 2014;35:2733–2779. doi: 10.1093/eurheartj/ehu284. [DOI] [PubMed] [Google Scholar]
- 4.Yang J, et al. Common SNPs explain a large proportion of the heritability for human height. Nat Genet. 2010;42:565–569. doi: 10.1038/ng.608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang J, et al. Genetic variance estimation with imputed variants finds negligible missing heritability for human height and body mass index. Nat Genet. 2015;47:1114–1120. doi: 10.1038/ng.3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wooten EC, et al. Formin homology 2 domain containing 3 variants associated with hypertrophic cardiomyopathy. Circ Cardiovasc Genet. 2013;6:10–18. doi: 10.1161/CIRCGENETICS.112.965277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aragam KG, et al. Phenotypic refinement of heart failure in a national biobank facilitates genetic discovery. Circulation. doi: 10.1161/CIRCULATIONAHA.118.03577. published online 11 November 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Esslinger U, et al. Exome-wide association study reveals novel susceptibility genes to sporadic dilated cardiomyopathy. PLoS One. 2017;12:e0172995. doi: 10.1371/journal.pone.0172995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meder B, et al. A genome-wide association study identifies 6p21 as novel risk locus for dilated cardiomyopathy. Eur Heart J. 2014;35:1069–1077. doi: 10.1093/eurheartj/eht251. [DOI] [PubMed] [Google Scholar]
- 10.Villard E, et al. A genome-wide association study identifies two loci associated with heart failure due to dilated cardiomyopathy. Eur Heart J. 2011;32:1065–1076. doi: 10.1093/eurheartj/ehr105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aung N, et al. Genome-wide analysis of left ventricular image-derived phenotypes identifies fourteen loci associated with cardiac morphogenesis and heart failure development. Circulation. 2019;140:1318–1330. doi: 10.1161/CIRCULATIONAHA.119.041161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stokke TM, et al. Geometry as a confounder when assessing ventricular systolic function: comparison between ejection fraction and strain. J Am Coll Cardiol. 2017;70:942–954. doi: 10.1016/j.jacc.2017.06.046. [DOI] [PubMed] [Google Scholar]
- 13.Bulik-Sullivan B, et al. An atlas of genetic correlations across human diseases and traits. Nat Genet. 2015;47:1236–1241. doi: 10.1038/ng.3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bulik-Sullivan BK, et al. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet. 2015;47:291–295. doi: 10.1038/ng.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Turley P, et al. Multi-trait analysis of genome-wide association summary statistics using MTAG. Nat Genet. 2018;50:229–237. doi: 10.1038/s41588-017-0009-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harper AR, et al. Common genetic variants and modifiable risk factors underpin susceptibility and expressivity in hypertrophic cardiomyopathy. Nat Genet. doi: 10.1038/s41588-020-00764-0. accepted [CE: add doi for NG-LE54654R1 Watkins] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kramer CM, et al. Hypertrophic Cardiomyopathy Registry: The rationale and design of an international, observational study of hypertrophic cardiomyopathy. Am Heart J. 2015;170:223–230. doi: 10.1016/j.ahj.2015.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Turro E, et al. Whole-genome sequencing of patients with rare diseases in a national health system. Nature. 2020;583:96–102. doi: 10.1038/s41586-020-2434-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.de Leeuw CA, Mooij JM, Heskes T, Posthuma D. MAGMA: generalized gene-set analysis of GWAS data. PLoS Comput Biol. 2015;11:e1004219. doi: 10.1371/journal.pcbi.1004219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oka T, et al. Cardiac-specific deletion of Gata4 reveals its requirement for hypertrophy, compensation, and myocyte viability. Circ Res. 2006;98:837–845. doi: 10.1161/01.RES.0000215985.18538.c4. [DOI] [PubMed] [Google Scholar]
- 21.Braz JC, et al. PKC-alpha regulates cardiac contractility and propensity toward heart failure. Nat Med. 2004;10:248–254. doi: 10.1038/nm1000. [DOI] [PubMed] [Google Scholar]
- 22.Wu T, et al. HSPB7 is indispensable for heart development by modulating actin filament assembly. Proc Natl Acad Sci USA. 2017;114:11956–11961. doi: 10.1073/pnas.1713763114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Merner ND, et al. Arrhythmogenic right ventricular cardiomyopathy type 5 is a fully penetrant, lethal arrhythmic disorder caused by a missense mutation in the TMEM43 gene. Am J Hum Genet. 2008;82:809–821. doi: 10.1016/j.ajhg.2008.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Green EM, et al. A small-molecule inhibitor of sarcomere contractility suppresses hypertrophic cardiomyopathy in mice. Science. 2016;351:617–621. doi: 10.1126/science.aad3456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Richards S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Whiffin N, et al. CardioClassifier: disease- and gene-specific computational decision support for clinical genome interpretation. Genet Med. 2018;20:1246–1254. doi: 10.1038/gim.2017.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet. 2011;88:76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee SH, Wray NR, Goddard ME, Visscher PM. Estimating missing heritability for disease from genome-wide association studies. Am J Hum Genet. 2011;88:294–305. doi: 10.1016/j.ajhg.2011.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Petersen SE, et al. UK Biobank’s cardiovascular magnetic resonance protocol. J Cardiovasc Magn Reson. 2016;18:8. doi: 10.1186/s12968-016-0227-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cerqueira MD, et al. Standardized myocardial segmentation and nomenclature for tomographic imaging of the heart A statement for healthcare professionals from the Cardiac Imaging Committee of the Council on Clinical Cardiology of the American Heart Association. Circulation. 2002;105:539–542. doi: 10.1161/hc0402.102975. [DOI] [PubMed] [Google Scholar]
- 32.Loh PR, et al. Efficient Bayesian mixed-model analysis increases association power in large cohorts. Nat Genet. 2015;47:284–290. doi: 10.1038/ng.3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hemani G, et al. The MR-Base platform supports systematic causal inference across the human phenome. Elife. 2018;7:e34408. doi: 10.7554/eLife.34408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bowden J, Davey Smith G, Haycock PC, Burgess S. Consistent estimation in Mendelian randomization with some invalid instruments using a weighted median estimator. Genet Epidemiol. 2016;40:304–314. doi: 10.1002/gepi.21965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hartwig FP, Davey Smith G, Bowden J. Robust inference in summary data Mendelian randomization via the zero modal pleiotropy assumption. Int J Epidemiol. 2017;46:1985–1998. doi: 10.1093/ije/dyx102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao Q, Wang J, Hemani G, Bowden J, Small DS. Statistical inference in two-sample summary-data Mendelian randomization using robust adjusted profile score. arXiv e-prints. 2018 [Google Scholar]
- 37.Bowden J, Davey Smith G, Burgess S. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int J Epidemiol. 2015;44:512–525. doi: 10.1093/ije/dyv080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhu Z, et al. Causal associations between risk factors and common diseases inferred from GWAS summary data. Nat Commun. 2018;9:224. doi: 10.1038/s41467-017-02317-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Watanabe K, Taskesen E, van Bochoven A, Posthuma D. Functional mapping and annotation of genetic associations with FUMA. Nat Commun. 2017;8:1826. doi: 10.1038/s41467-017-01261-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.GTEx Consortium. Genetic effects on gene expression across human tissues. Nature. 2017;550:204–213. doi: 10.1038/nature24277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van Velzen HG, et al. Effect of gender and genetic mutations on outcomes in patients with hypertrophic cardiomyopathy. Am J Cardiol. 2018;122:1947–1954. doi: 10.1016/j.amjcard.2018.08.040. [DOI] [PubMed] [Google Scholar]
- 43.Vriesendorp PA, et al. Long-term outcomes after medical and invasive treatment in patients with hypertrophic cardiomyopathy. JACC Heart Fail. 2014;2:630–636. doi: 10.1016/j.jchf.2014.06.012. [DOI] [PubMed] [Google Scholar]
- 44.LaBounty TM, Bach DS, Bossone E, Kolias TJ. Indexing left ventricular wall thickness to body surface area improves prognostic value. Echocardiography. 2019;36:824–830. doi: 10.1111/echo.14317. [DOI] [PubMed] [Google Scholar]
- 45.Huurman R, et al. Effect of body surface area and gender on wall thickness thresholds in hypertrophic cardiomyopathy. Neth Heart J. 2020;28:37–43. doi: 10.1007/s12471-019-01349-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Khera AV, et al. Genome-wide polygenic scores for common diseases identify individuals with risk equivalent to monogenic mutations. Nat Genet. 2018;50:1219–1224. doi: 10.1038/s41588-018-0183-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data from the Genome Aggregation Database (gnomAD, v2.1) are available at https://gnomad.broadinstitute.org. Data from the UK Biobank participants can be requested from the UK Biobank Access Management System (https://bbams.ndph.ox.ac.uk). Data from the Genotype Tissue Expression (GTEx) consortium are available at the GTEx portal (https://gtexportal.org). Other datasets generated during and/or analyzed during the current study can be made available upon reasonable request to the corresponding authors. Individual level data sharing is subject to restrictions imposed by patient consent and local ethics review boards. Results from meta-analyses of genome-wide association studies reported in this article are available at https://www.heart-institute.nl/gwas and https://data.hpc.imperial.ac.uk [doi.org/10.14469/hpc/7468].