Abstract
Purpose
To ask whether the expression of immune markers and interferon signaling in tumor biopsies changes during concurrent chemoradiotherapy (CCRT).
Experimental design
Tumor biopsies and peripheral mononuclear blood cells (PMBCs) before and immediately after 20Gy/10 fractions (F) of radiation treatment (RT) from 30 cervical cancer patients receiving CCRT were evaluated by IHC and RT-qPCR for immune markers and correlated with the short-term response.
Results
Tumor immune response to radiation before and after 10F RT as reflected by CD8+ T cell infiltration had substantial heterogeneity with increases, decreases and no change all evident. Increases in CD8+ T cells during CCRT correlated with the presence of nuclear IRF1 in tumor cells (r = 0.68, P < 0.0001) and the patient short-term response (P < 0.01). Similarly, in a subset of patients (approx. 40%) PD-L1 positivity in tumor cells increased, which also correlated with nuclear IRF1 staining (r = 0.48, P < 0.01). Patients with augmented PMBC interferon (IFN) signature expression after 10F had a significantly higher probability of PD-L1 induction (83% vs 7%, P < 0.0001). Most patients exhibited abundant expression of SERPINB9 and CD47 in tumor cells, and tumor infiltration by CD68+ cells. SERPINB9 expression correlated with STAT1 signaling in tumor cells.
Conclusions
CCRT leads to differential tumor immunogenicity and IFN signaling in cervical cancer patients, suggesting radiation induction of immunity is limited to a subset of patients and may reflect the heterogeneity of intratumoral induction of IFNs.
Keywords: Cervical cancer, Chemoradiotherapy, Interferon, Tumor immunogenicity, Heterogeneity
Introduction
Immunotherapy using immune checkpoint blockade (ICB) has emerged as the fourth main therapeutic modality for cancer. ICB depends on pre-existing anti-tumor immunity so that inhibition of immune checkpoints can release an effective immune response against the tumor. The immunogenicity of a tumor is thought to be determined by a number of factors, including the number of somatic mutations that might generate neoantigens, the presentation of these neoantigens, the expression of immune checkpoints in the tumor, and the infiltration and status of tumor-specific CD8+ T cells. The key etiological factor for cervical cancer (CC) is persistent human papillomavirus (HPV) infection, present in 85% of invasive CC (1). Despite HPV vaccination, CC is still the 4th leading cause of cancer-related deaths in women. HPV positive tumors actively transcribe HPV-related genes associated with their neoantigen landscape (2). CC is one of the cancer types carrying a high genomic mutation burden (3). These data suggest that CC could often be immunogenic.
Radiation therapy (RT) is one of the main methods of cancer treatment. In addition to directly killing cancer cells, ionizing radiation (IR) has the potential to trigger an anti-tumor immune response. Many mechanisms including the generation of damage associated molecular patterns (DAMPs) and neoantigens, activation of dendritic cells, and production of interferon (IFN) have been identified as potential contributors to a RT-stimulated immune response (4–7). Patients with locoregionally advanced CC are routinely treated with conventionally fractionated external beam RT (EBRT) in combination with concurrent chemotherapy followed by brachytherapy with the 5-year survival very dependent on stage; the survival of the local, regional and distant stages is approximately 87%, 49% and 11%, respectively (8). More recently, clinical evidence suggests that CC also responds to ICB further supporting the presence of tumor immunogenicity. Programmed cell death-1 (PD-1) blockade (Pembrolizumab) has now been approved by the FDA for treatment of recurrent or metastasized CC with positive programmed death-ligand 1 (PD-L1) expression in tumors (9). Furthermore, cytotoxic T-lymphocyte-associated protein 4 (CTLA-4) blockade after chemoradiotherapy (CRT) in curative-intent treatment is effective in CC (10). Combining αPD-L1/PD-1 with CRT in treatment of CC is under evaluation in a number of clinical trials in various setting (11). One question that arises is whether treatment with RT and chemotherapy alters the immune response in CC.
If RT augments tumor immunogenicity and alters the microenvironment in favor of an inflammatory immune response, ICB would be more effective when used in conjunction with RT (12). In preclinical models, combining RT and ICB has a synergistic effect, which is dependent on the proinflammatory effect of RT (13–15). Promising results have been demonstrated in a few clinical trials examining ICB in combination with stereotactic body RT (SBRT) in metastatic tumors, or with conventional CRT in locally advanced diseases (16–18). Intrigued by these positive findings, clinical investigators have launched hundreds of trials to examine the efficacy of various combination between ICB and RT in a number of cancer diseases.
Despite this enthusiasm, only a small proportion of patients have been shown to benefit from adding ICB to RT. In the PACIFIC-1 trial, administration of antibodies against PD-L1 (αPD-L1, durvalumab) after definitive CRT resulted in significant improvement of both objective responses and 2-year overall survival rates in patients with stage III non-small cell lung cancer (NSCLC) (18). However, the majority (70%) of patients did not display improved response from the PD-L1 blockade. The mechanisms underlying lack of response in these patients are still not fully understood.
The proinflammatory effect of RT and its synergistic effect in combination with ICB depends on generation of an IFN response in tumors (4,19). IR and other means of generating DNA damage result in cellular synthesis of IFNs through the cGAS-STING-TBK1-IRF pathway (4,5,20). Both mice and cancer cells lacking IFN signaling fail to elicit an anti-tumor immune response in tumors after RT (4,21). IFN regulates many immune mediators through STAT1 and IRF signaling. IFN signaling has a pivotal role in IR-induced dendritic cell activation and T cell priming (4,7,21,22). However persistent IFN signaling provokes cancer cell-intrinsic resistant mechanisms, including upregulation of PD-L1, SERPINB9, and CASPASE9, protecting cancer cells from T cell-mediated cytotoxicity (14,23,24). IFNs may also suppress anti-tumor immunity by recruiting Tregs and myeloid cells into the tumor microenvironment (25,26). Together, these data suggest that the IFN response and its effect on the expression of key immune mediators dictate the immunomodulatory effect of RT on the tumors.
RT could also lead to immunosuppression due to induction of lymphopenia, which is commonly found in patients receiving RT and is a poor prognostic factor (27,28).
Because of the relatively accessible location of CC, we initiated a study examining biopsies before and during CRT. Tumor biopsies from the primary site and PMBCs were obtained from 30 CC patients before and immediately after 10F EBRT when the total dose is likely to trigger an immune response yet with sufficient tumor available for assessment in most patients. We focused on infiltration of CD8+ T cells and macrophages, STAT1- or IRF1-mediated IFN responses and a variety of IFN-responsive molecules in tumor cells that are critical for the anti-tumor immune response. We found wide variation in the extent of infiltration by CD8+ T cells and in their response during therapy with increases, no change and decreases all evident. However, increases of CD8+ T cells during therapy roughly correlated with IFN signaling and tumor response at the completion of therapy. We also examined an IFN gene signature, which has shown excellent predictive value for both local and systemic IFN response in autoimmune diseases (29), in PMBCs as a surrogate for evaluation of IFN response in local tumors. These data suggest that the immune and IFN response of CC to therapy is highly variable and that IFN signaling in PMBCs could be a surrogate biomarker.
Materials and Methods
Study design
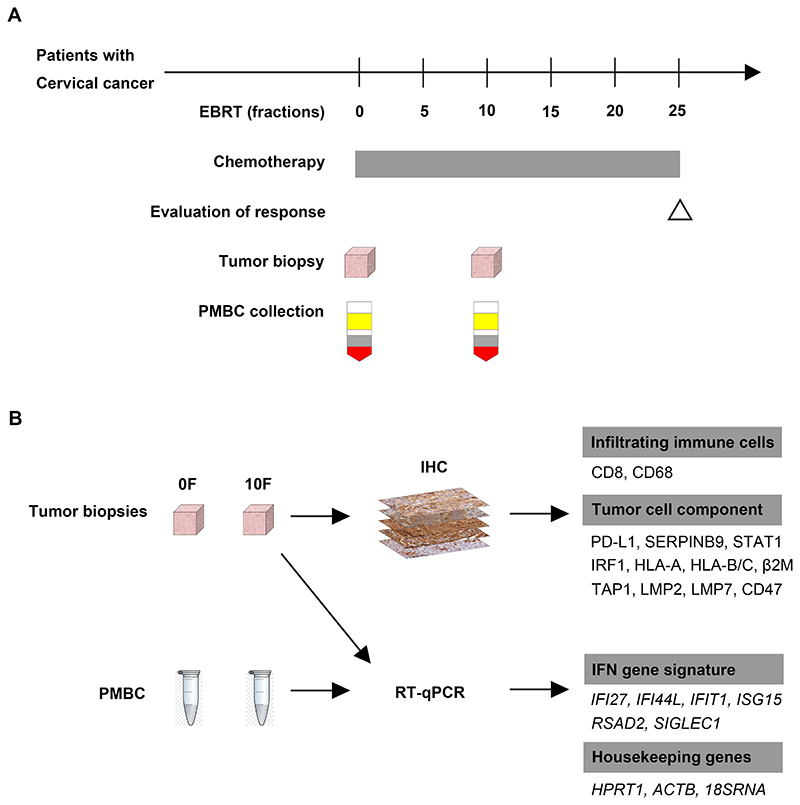
A prospective observation study was launched in 2018 to explore potential biomarkers to predict CC response to CCRT. The study design is shown in Figure 1. Briefly, patients diagnosed with primary squamous cell carcinoma (SCC) of the cervix were subjected to CCRT. Tumor biopsies at primary sites and PMBCs collection were performed at baseline before treatment and within 12h after 10F RT. The expression of biomarkers in these samples was analyzed as described below. This study is in compliance with the declaration of Helsinki. It was approved by the clinical study ethic committee in Cancer Hospital of Shantou University Medical College and registered on Clinicaltrial.gov (NCT03744819). Written informed consent was obtained from all enrolled patients.
Figure 1. Study design.
(A): Timeline of the clinical study.
(B): The assessment of patient samples.
Patient eligibility criteria
The inclusion criteria were as follows: (1) Pathological proven diagnosis of primary cervical SCC; (2) Patient will receive CCRT; (3) The primary tumor was accessible for biopsy during the course of CCRT. The exclusion criteria included: (1) History of autoimmune diseases; (2) History of anti-tumor immunotherapy; (3) Prior RT that would result in overlapping of planned RT fields; (4) Concomitant immunotherapy during the course of RT; (5) Contraindications for biopsy.
Treatment
Enrolled patients were subjected to CCRT. RT consisted of EBRT (median dose 46Gy/23F) to the whole pelvis followed by EBRT boost (median dose 14Gy/7F) to positive lymph nodes (LNs) and brachytherapy boost (24-32Gy/4-5 fraction) to the primary tumor and surrounding subclinical disease. During EBRT, patients also received platinum-based concurrent chemotherapy. The chemotherapy regimen consisted of either weekly cisplatin (30-40 mg/m2(body surface area)) or a combination of cisplatin (25mg/m2, day 1-3) and fluorouracil (0.5g/m2, day 1-4) on week 1 and 4.
Clinical evaluation
Patients received the following examination prior to treatment: (1) Tumor biopsy; (2) PMBC collection; (3) other examinations for tumor staging and pre-treatment evaluation. Bone marrow, liver and renal functions were monitored during treatment. Immediately after 10F of RT, another tumor biopsy and PMBC collection was obtained. The short-term tumor response to CCRT in patients was assessed after the whole course of pelvic EBRT according to the Response Evaluation Criteria in Solid Tumors (RECIST 1.0 version). Complete regression (CR) is defined as disappearance of cervical lesion as evaluated by the bimanual examination. Partial regression (PR) is defined as at least 30% decrease in the lesion maximum diameter compared to baseline. The evaluation of tumor response was blind to the biomarker assessment.
Sample collection
Each tumor biopsy was divided into two parts; one fixed in 10% formalin for at least 24 hours, paraffin-embedded, and stored at room temperature. 4μm consecutive sections were stored at 4°C. The other part was submerged in RNAlater solution (Thermo Fisher) and stored at −80 °C. Whole blood samples were collected in EDTA tubes. PMBCs were isolated using Ficoll Paque Plus media (GE Healthcare Life Sciences) according to the manufacturer’s protocol. Isolated PMBCs were then submerged in RNAlater solution and stored at −80°C.
IHC staining and evaluation
The antibodies used for IHC staining in this study are listed in Table S1. Tumor sections were deparaffinized using a series of xylene and ethanol and rehydrated with dH2O. Microwave-stimulated antigen retrieval in buffers was performed as recommended by the manufacturers. Staining was as previously described (23). Slides were scanned (Magascanner, KFBIO) at 40X magnification. Images were processed using K-Viewer (KFBIO).
These IHC images were evaluated by two independent researchers. Up to 10 representative areas of tumor islands were selected. PD-L1, SERPINB9, CD47, nuclear IRF1, nuclear STAT1, HLA-A, HLA-B/C, β2M, TAP1, LMP2 and LMP7 stainings were assessed. For each area, the percentage of cancer cells with positive staining (positivity) was measured. The expression intensity was scored as follows: negative, score = 0; weak, score = 1; moderate, score = 2; strong, score = 3 (Representative images in Figure S1). The H score is the product of positivity multiplied by the score. For infiltrating CD8+ T cells or CD68+ cells, the number of immune cells per mm2 was quantified. The mean expression (positivity, H score or density) over multiple areas was used in subsequent analyses. Representative images were selected from various patients or the same patient for illustration in the figures as indicated in the legends. The image of IRF1 staining is the same in Fig. 2G and 5C and the images of PD-L1, STAT1 and HLA-A staining are the same in Figures S1 and S8 allowing direct comparison.
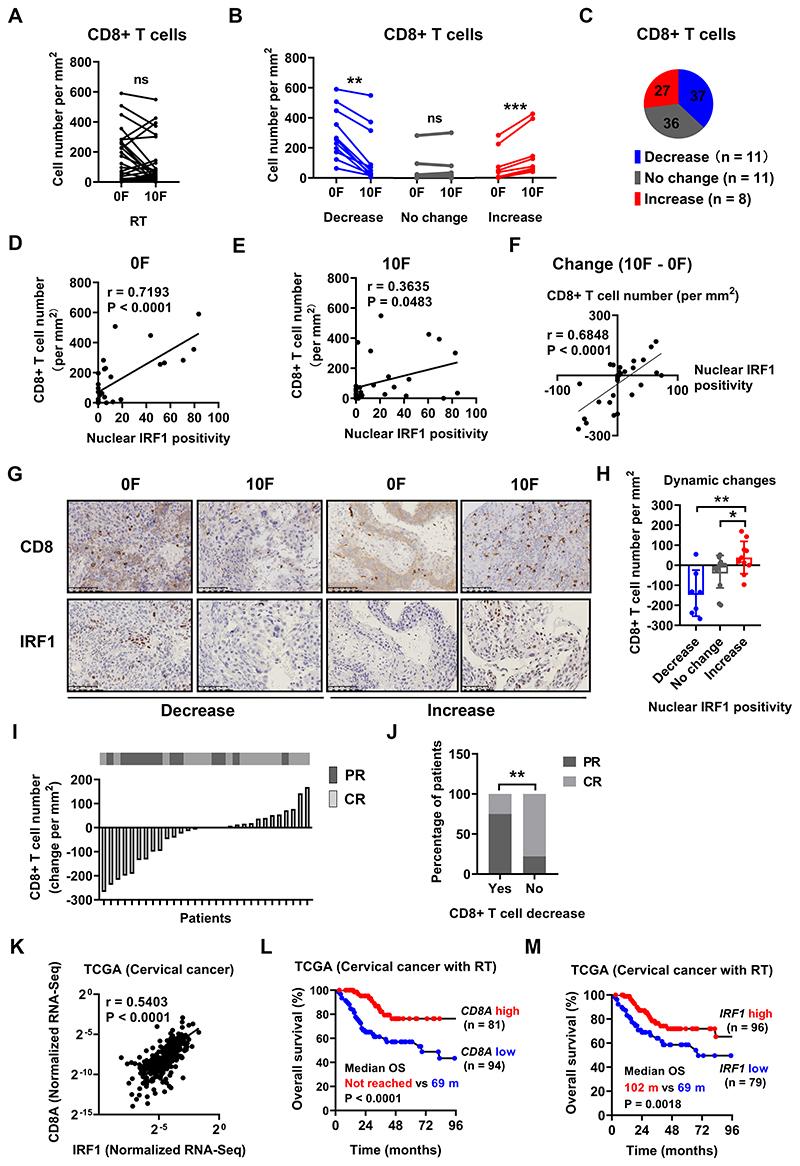
Figure 2. Changes in tumor infiltrating CD8+ T cell number correlate with nuclear IRF1 staining and tumor response to CCRT in CC patients.
Tumor infiltrating CD8+ T cell number before and after 10F RT in aggregate or in subgroups with different dynamic change patterns (decrease, no change and increase) are shown in (A) and (B) respectively. The percentage of various change patterns is summarized in (C). Definition of change patterns for CD8+ T cells: Increase (in red), cell number increased by > 30 per mm; Decrease (in blue), cell number decreased by > 30 per mm; No change (in gray): the difference of cell number ≤ 30 per mm.
The number of tumor infiltrating CD8+ T cells and the expression positivity of nuclear IRF1 in tumor cells before and after 10F RT, and their dynamic changes is plotted in (D), (E) and (F).
(G): Representative immunohistochemical staining of CD8 (upper row) and IRF1 (lower row) before and after 10F RT showing both decreased and increased nuclear IRF1 positivity in tumor cells. (H): Changes of infiltrating CD8+ T cell number after 10F RT in tumors with various dynamic change patterns of nuclear IRF1 positivity in tumor cells.
(I): The change of tumor infiltrating CD8+ T cell number after 10F RT is displayed on the lower panel as a waterfall plot, ranking from the greatest decrease on the left to the greatest increase on the right. The short-term (immediately after external beam radiotherapy) response to concurrent chemoradiotherapy for each individual patient is shown on the upper panel. CR in gray; PR in black.
(J): The percentage of patients with a tumor CR or PR in patients with or without decreased CD8+ T cell number after 10F RT. The cut-off for CD8+ T cell density was set at −19.6 per mm2 which was determined by using the receiver operating characteristic curve method.
(K) mRNA expression correlation between IRF1 and CD8A in the TCGA dataset of patients with cervical cancer. N = 294.
Patients from the TCGA dataset who had received RT (N = 175) were stratified into subgroups according to their tumor mRNA expression level (High in red, low in blue) of CD8A or IRF1. OS curves of these subgroups of patients and comparison of median OS (in months (m)) are plotted in (L) and (M).
Data in bar charts shows the mean ± SD. Mean comparisons were performed using the paired Student’s t test in (A), the Wilcoxon matched-pairs signed rank test in (B), and 1-way ANOVA with Tukey’s multiple-comparisons test in (H). Ratio comparison in 2 groups (J) was performed by using the Chi-square test. The correlation between positivity of various markers was evaluated by Pearson’s correlation (K). Survival curves for different groups of patients were generated using the Kaplan-Meier method (L and M). The log-rank test was used to compare the median overall survival time (* P < 0.05, **P < 0.01).
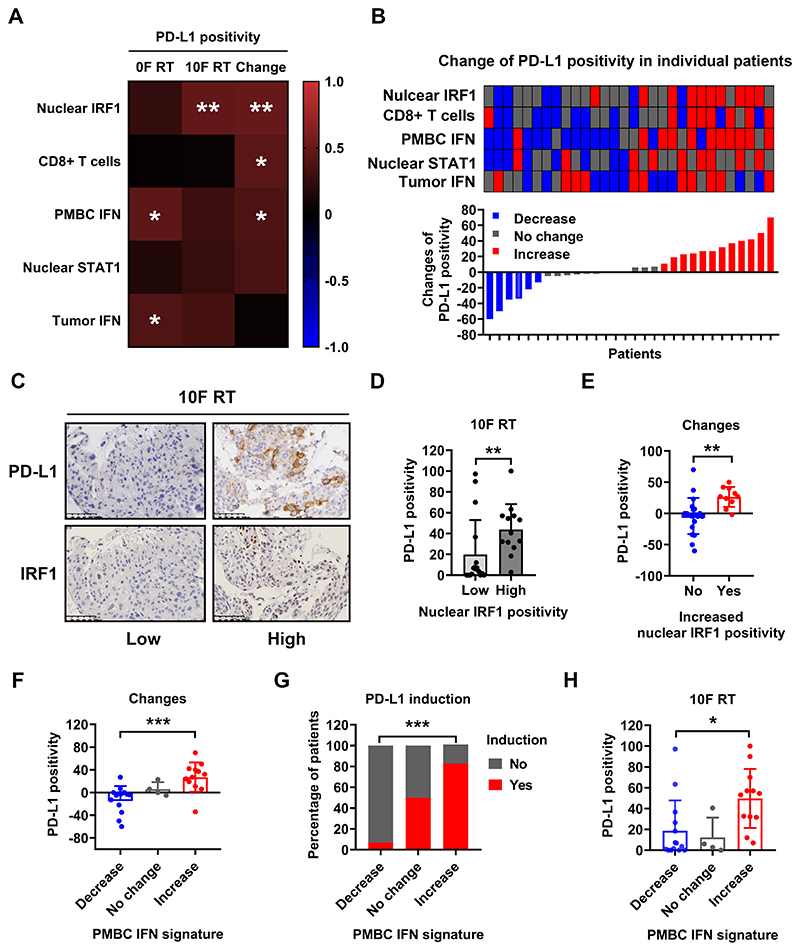
Figure 5. PD-L1 expression positivity correlates with tumor cell nuclear IRF1 staining and the IFN gene signature expression of PMBCs during CCRT.
(A): Heatmap of Pearson correlation coefficients between the expression of PD-L1 (positivity in tumor cells before and after 10F RT and their dynamic change) and various markers.
(B): The change of PD-L1 positivity in tumor cells after RT is displayed on the lower panel as a waterfall plot, ranking from the greatest decrease on the left to the greatest increase on the right. The dynamic change patterns of expression positivity or level of indicated markers in each patient is shown as a heatmap on the upper panel (decrease in blue, no change in gray and increase in red).
(C) Representative immunohistochemical section staining (low vs high) of PD-L1 (top) and IRF1 (bottom) in tumor biopsies from patients after 10F RT. The image of IRF1 staining on the right is the same as that shown in Figure 2G
(D): Quantification of PD-L1 positivity in tumor cells with low (≤ 10%) or high (>10%) nuclear IRF1 positivity in tumor biopsies from patients after 10F RT.
(E): Changes of PD-L1 positivity in tumor cells with or without increased nuclear IRF1 activity in tumor biopsies.
The change of tumor cell PD-L1 positivity, the percentage of patients with PD-L1 induction (increased positivity in tumor cells), and the tumor cell PD-L1 positivity after 10F RT in patients with different change patterns of PMBC IFN signature are summarized in (F), (G) and (H), respectively.
Data in bar charts represent mean ± SD. Mean comparison was performed by using the Mann-Whitney test (2 groups) or 1-way ANOVA with Tukey’s multiple-comparisons test (more than 2 groups). Ratio comparison was performed by using the Chi-square test. The correlation between positivity of various markers was evaluated by Pearson’s correlation (*P < 0.05, ** P < 0.01, *** P < 0.001. P > 0.05 was not shown in heatmaps).
RT-qPCR
After removal of RNAlater, tumor samples and PMBCs were homogenized in Trizol (Thermo Fisher) at 4°C using a magnetic bead homogenizer. The procedures for RNA isolation and RT-qPCR have been described (23). The IFN gene signature used was as described in Rice, R.I, et al and consists of IFI27, IFI44L, IFIT1, ISG15, RSAD2 and SIGLEC1 except that the expression of SIGLEC1 was not robust in these samples and was excluded from analysis (29). Genes of the DDR signature include BBC3, DDB2, EI24, FBXO22, FDXR, GRAD45A, PCNA, SESN1 and TRIAP1. These genes were selected from a previous report excluding genes that could be upregulated by IFNs (http://www.interferome.org/interferome/home.jspx) (30). Of a macrophage signature consisting of ARG1, TNF, CXCL9, IFNG, IL1B, IL10, IL4, IL12B, NOS2 and TGFB1, only IL-1B, TNF, CXCL9 and TGFB showed sufficient robustness in all samples (31). Housekeeping genes were 18sRNA, HPRT1 and ACTB. The primer pairs used are in Table S2. All genes displayed single peak on melting curve analysis after the amplification of qPCR (Figure S2).
Knockout of IRF1 in tumor cell line by using CRISPR/Cas9
IRF1 knockout in HT29 cells (a human colorectal cancer cell line, RRID: CVCL_0320) using CRISPR/Cas9 was as described (20).
TCGA data analysis
TCGA gene expression data (RNA-seq V2 RSEM) of CD8A, IRF1, HPRT1 and ACTB and clinical information in cervical SSC (n = 294) was downloaded via cBioPortal (http://www.cbioportal.org/) (RRID: SCR_014555). The expression of CD8A and IRF1 was normalized to HPRT1 and ACTB. Pearson correlation coefficients (r) are indicated. The predictive capacity of tumor expression of CD8A or IRF1 in patient OS was evaluated by using receiver operating characterisitc (ROC) analysis. Their optimal cut-off was determined using the Youden index. Patients who received RT were divided into subgroups (high vs low) according to the expressoin level of CD8A or IRF1. The survival curves for subgroups of patients were generated using the Kaplan-Meier method. The log-rank test was used to compare the median survival time.
Statistical methods
This study aimed to enroll at least 30 patients with evaluable paired samples. All values represent means (M) and standard deviations (SD). Mean comparisons were performed using the paired Student’s t test in paired groups with normal distribution (evaluated using D’Agostino & Pearson omnibus normality test), the Wilcoxon matched-pairs signed rank test in paired groups with uncertain distribution normality, the Mann-Whitney test in 2 independent groups with uncertain distribution normality and the 1-way ANOVA with Tukey’s multiple-comparisons test in 3 groups. Ratio comparison in 2 groups was performed by using the Chi-square test. The correlation between the expression of markers was evaluated by Pearson’s correlation. P value: NS > 0.05; * P < 0.05; ** P < 0.01; *** P < 0.001; **** P < 0.0001. All graphs were plotted using Graphpad Prism 8 (RRID: SCR_002798). ROC analysis was performed using IBM SPSS Statistics 23 (RRID: SCR_002865).
Results
Patient clinical characteristics
Between September 2018 and May 2020, 58 consecutive patients with primary cervical SCC were evaluated. Of those, 48 were enrolled in this prospective trial (flow diagram in Figure S3). All 48 patients completed the full course of CCRT. 41 had tumor biopsies and PBMC collection before and after 10F RT. Tumor biopsies from 11 patients contained only necrotic or inflammatory tissues or were of poor quality, preventing further assessment. The remaining 30 patients were included as are summarized in Table S3. The majority (90%) had locally advanced disease. Following EBRT, 57% (17/30) of patients had a CR, while the remaining patients had a PR. The number of leukocytes (neutrophils, lymphocytes and monocytes) in the peripheral blood was reduced after EBRT (Figure S4).
Changes in tumor infiltrating CD8+ T cells: correlation with nuclear IRF1 staining and tumor response.
There was a wide range of CD8 T cell densities in the tumors as well as divergent responses after radiation (Figure 2A). The patients could be divided into subgroups (decrease, no change and increase) based upon the change in CD8 T cell density within the tumor after RT (Figure 2B, C). We evaluated the relationship between CD8+ T cell density and a series of immunologically relevant markers (Figure S5, S6). There was a strong correlation between the number of tumor-infiltrating CD8+ T cells and nuclear IRF1 staining in tumor cells (Figure 2D, E). The change of CD8+ T cells during CCRT also correlated with nuclear IRF1 in tumor cells (Figure 2F, G). The number of tumor infiltrating CD8+ T cells increased by approximately 28 cells /mm2 (median) after RT in patients with tumor cells in which nuclear IRF1 positivity was also upregulated in comparison to tumors with no significant change in CD8+ T cell density after RT (0 cells/mm2) in which there was also no significant change of IRF1 positivity. Those tumors with a decrease (on average of 135 cells/mm2) also had reduced nuclear IRF1 in tumor cells (Figure 2H). These data suggest that IRF1-mediated signaling in tumor cells influences the infiltration and/or survival of CD8+ T cells into tumors during CCRT.
The change in tumor infiltrating CD8+ T cells after 10F RT correlated with patient short-term response to CCRT. PR was associated with change of CD8+ T cell density in tumors, change of LMP7, tumor size and platelet-neutrophil-ratio at baseline (Table S4). However, only change of CD8+ T cells stood out as an independent factor associated with the risk of PR in multivariate analysis (Table S5, S6, S7). We ranked patients according to the change of T cell number in their tumors in parallel with their short-term response (Figure 2I). The majority of patients with PR clustered with decreased CD8+ T cell number after CCRT. Patients with decreased numbers of tumor infiltrating CD8+ T cells after CCRT had significantly higher risk (75% vs 22%, P = 0.0080) of PR compared to patients without (Figure 2J). Consistently, significant correlation was found between the expression of CD8A and IRF1 in cervical cancer in the TCGA database (Figure 2K). More importantly, in patients treated with RT, high expression of CD8A was associated with better OS (Figure 2L). A similar result was found in comparing patients with high vs low expression of IRF1 in their tumors (Figure 2M). Macrophages were assessed by CD68 and showed an increase with CCRT in 52% of patients (Figure S7 A–C). The macrophages in irradiated tumors were polarized towards the M1 phenotype in 53% of patients and M2 in 26% of patients after RT (Figure S7 D–H). Thus, macrophages also had a heterogenous response. Taken together, these data suggest that CD8+ T cell-mediated immunity has a role in tumor response to CCRT in cervical cancer.
IFN gene expression signature
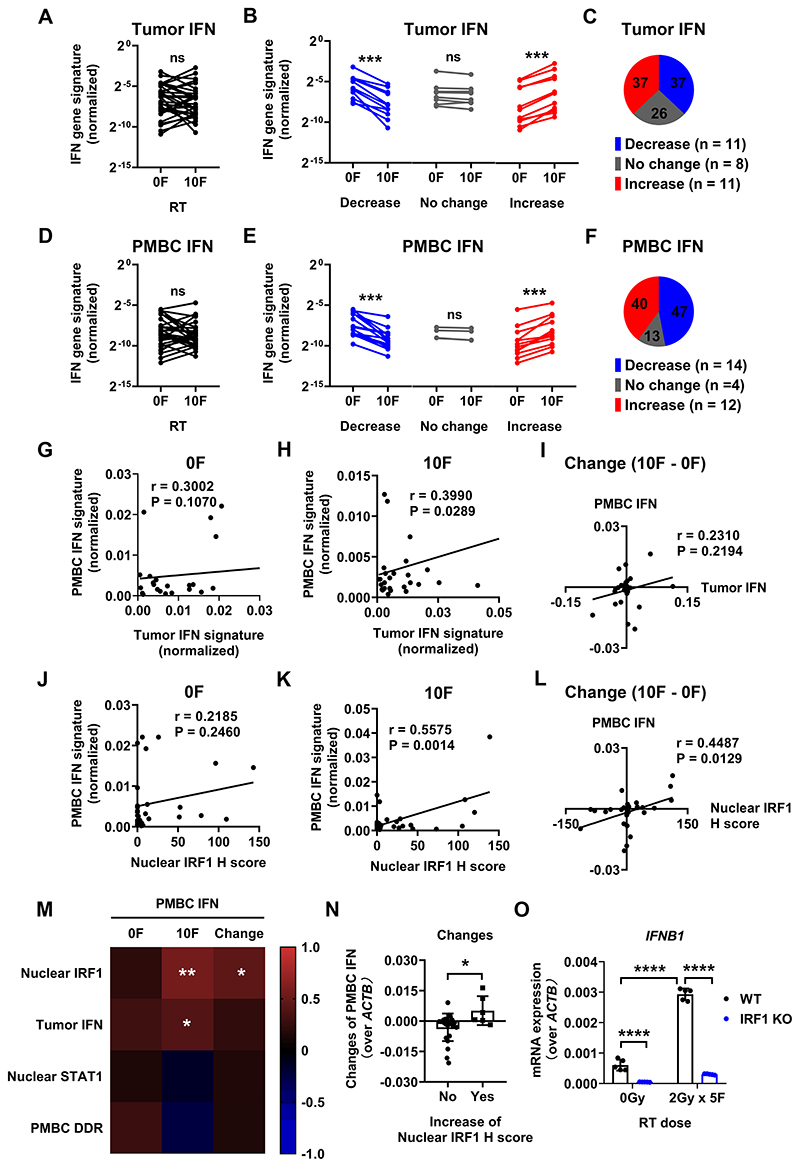
There was similar heterogeneity in the IFN gene expression signature. Overall, there was no average difference in the IFN gene expression signature before and after 10F RT in both the tumors and the PMBCs (Figure 3A, D). However, substantial changes were found in subgroups of patients (Figure 3B, E). The IFN gene signature mainly either increased or decreased after 10F RT in both tumors and PMBCs and these responses were correlated. The baseline IFN gene signature showed a trend of correlation (Figure 3C, F).
Figure 3. IFN gene signature expression signature during CCRT in tumors and PMBCs.
IFN gene signature expression in both tumors and PMBCs before and after 10F RT are summarized in (A) and (D).
The comparisons of IFN signature (both in tumors and PMBCs) before and after 10F RT with different dynamic change patterns (decrease, no change and increase) are summarized in (B) and (E). The case numbers (N) for each change pattern are as indicated.
Definition of change patterns for IFN gene signature: Increase (in red), gene expression after 10F RT increased by > 0.5 fold compared to that before; Decrease (in blue), gene expression after 10F RT decreased by > 0.5 fold compared to that before; No change (in gray): the difference of gene expression after RT ≤ 0.5 fold.
The percentages of various dynamic change patterns of CD8+ T cells and IFN gene signature are summarized in (C) and (F).
PMBC and tumor IFN gene signature expression before and after 10F RT, and their dynamic changes are plotted in (G-I).
The expression of PMBC IFN gene signature and nuclear IRF1 expression H score in tumor cells before and after 10F RT, and their dynamic changes are plotted in (J-K).
(M): Heatmap of Pearson correlation coefficients between the expression of PMBC IFN signature and various markers.
(N): Change of PMBC IFN gene signature in patients with or without increased nuclear IRF1 positivity in tumor cells.
(O) The mRNA expression of IFNB1 (normalized to ACTB) in WT vs IRF1 KO HT29 cells (a human colorectal cancer cell line) after mock or 2Gy x 5F irradiation. The KO of IRF1 was performed using the CRISPR/Cas9 methodology.
The bar charts show the mean ± SD. Mean comparisons were performed using the paired Student’s t test in paired groups with normal distribution (A and D), the Wilcoxon matched-pairs signed rank test in paired groups with uncertain distribution normality (B and E), the Mann-Whitney test in 2 groups with uncertain distribution normality (H), the 1-way ANOVA with Tukey’s multiple-comparisons test in 3 groups (I). The expression correlation between markers was evaluated by Pearson’s correlation (ns: P > 0.05, *P < 0.05, ** P < 0.01, *** P < 0.001, **** P < 0.0001. P > 0.05 was not shown in heatmaps).
The IFN response in PMBCs could be triggered by direct exposure of PMBCs as they circulate in the radiation field. Alternatively, the PMBCs might be exposed to IFN generated by the tumor in response to radiation. Direct exposure would be predicted to lead to DDR expression. To distinguish these possibilities, we analyzed the correlation between the IFN gene signature and a DDR signature in PMBC and several other markers in tumors. (Figure 3M). No correlation was found between the PMBC IFN and DDR signatures suggesting that IFN signaling in the PMBCs was not due to direct exposure to radiation.
We then looked for a correlation between tumor cell response to radiation and the PMBC IFN gene signature. We used nuclear IRF1 as a marker for tumor cell response to radiation perhaps by IFN generation. Nuclear IRF1 in tumor cells correlates with the PMBC IFN signature during CCRT (Figure 3K). The dynamic changes of these two markers also displayed significant correlation (Figure 3L). Upregulation of nuclear IRF1 staining in tumor cells was associated with induction of the IFN gene signature in PMBCs after 10F RT (Figure 3N). These data are consistent with the notion that DNA damage inducing treatments, RT and chemotherapy induce IFNs in cancer cells in an IRF-dependent manner (20,32). It is striking, however that this occurred in only a subset of the patients. IRF1 mediates type III IFN induction in HT29, a human colorectal cancer cell line, after IR (20). Similarly, induction of IFNB1 by IR requires intact IRF1 signaling in cancer cells (Figure 3O). Collectively, these results suggest that IFN induction mediated by IRF1 in tumor cells make a substantial contribution to the IFN response in circulating PMBCs during CCRT.
Immune marker expression in tumor cells
We assessed the expression of some IFN-responsive markers, which could affect tumor immune response, including PD-L1, SERPINB9, IRF1, STAT1, HLA-A, HLA-B/C, β2M, LMP2, LMP7 and TAP1(Figure 4A, B, D). PD-L1 expression in tumor cells (>1%) was found in 83.3% of patients both before after 10F RT. While there was a trend of an increase in the average positivity of PD-L1 across the whole cohort immediately after 10F RT, the difference was not statistically significant (30% vs 24%, P > 0.05). The majority of tumor samples had tumor-cell expression of SERPINB9, with an average positivity of 56% before treatment and 47% during CCRT.
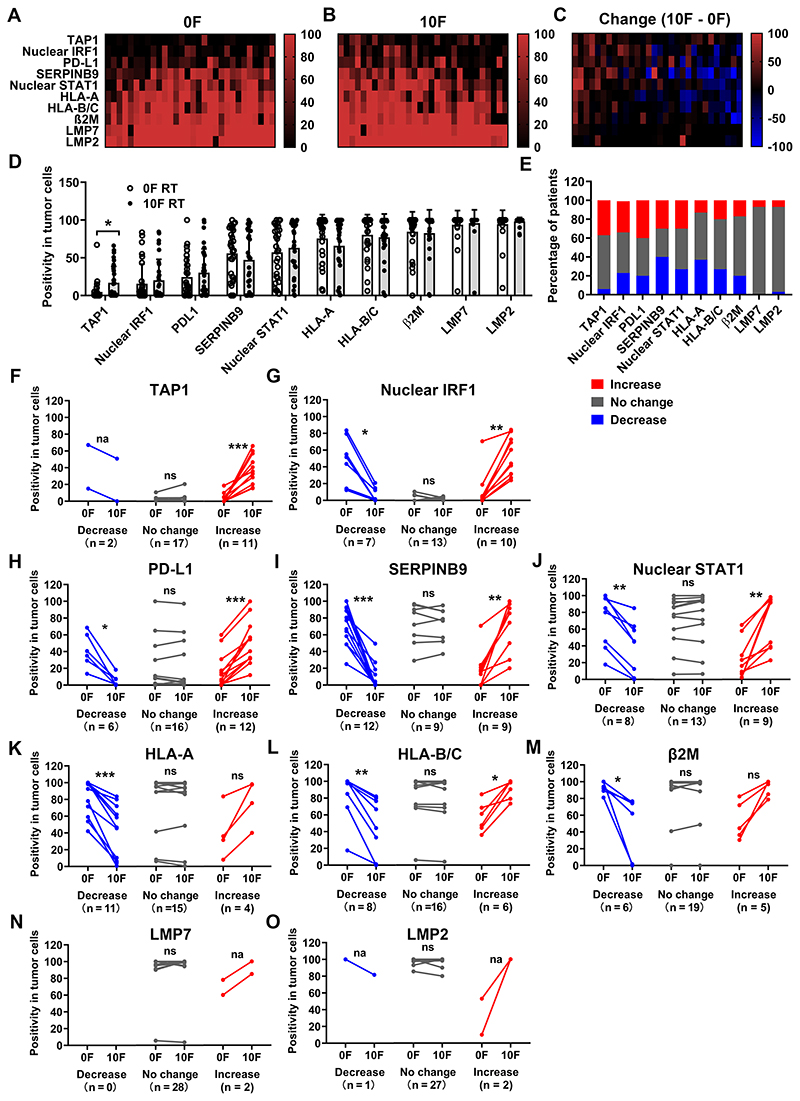
Figure 4. Immune marker expression positivity in tumor cells before and during CCRT.
Heatmaps of expression positivity of indicated immune markers in tumor cells before (A) and after 10F RT (B), and their dynamic changes (C). Patients were ranked according to their mean changes of positivity from the highest on the left to the lowest on the right. Markers were ranked according to their mean positivity of the whole cohort of patients from the lowest on the top to the highest on the bottom. The vertical color bars display the range of values. N = 30.
(D): Summary of the positivity of these markers in tumor cells before and after 10F RT.
(E): Summary of the dynamic change (10F vs 0F) patterns (increase, no change and decrease) of these markers. Increase (in red): positivity increased by > 10%; Decrease (in blue): positivity decreased by > 10%; No change (in gray): Changes of positivity ≤ 10%.
Comparisons of marker expression positivity before and after 10F RT in subgroups with different dynamic change patterns (decrease, no change and increase) are summarized in (F-O). The markers and case numbers (N) for each change pattern are indicated.
Mean comparisons in (D) (paired groups with normal distribution) were performed by using the paired Student’s t test. Mean comparison in (F-O (paired groups with uncertain distribution normality) was performed by using the Wilcoxon matched-pairs signed rank test.
ns: P > 0.05; * P < 0.05; ** P < 0.01; *** P < 0.001; na: the comparison is not applicable.
P values are not shown if they > 0.05 in (D).
Both STAT1 and IRF1 are important transcriptional factors for mediating the IFN response. Their presence in the nucleus could reflect activation of signaling pathways leading to IFN transcription. Nuclear STAT1 was found in 57% of tumor cells before treatment, while only 16% of displayed nuclear IRF1 staining. Both markers showed a trend of increasing during CCRT.
The antigen presentation machinery (APM) consists of a variety of components, including molecules involved in protein degradation (e.g., LMP2 and LMP7), peptide transportation (e.g., TAP1) and presentation on the cellular surface (e.g. HLA-A, HLA-B, HLA-C and β2M). The majority of tumor cells, both before and after 10F RT, expressed these components with the exception of TAP1, which was found in only 5% of tumor cells at baseline. Upon RT, the average positivity of TAP1 in tumor cells increased to 17%. The dynamic change of TAP1 expression correlated with that of nuclear IRF1 (r = 0.41, P < 0.05). No significant change was noted in the other APM components examined.
Many of the markers in individual patients exhibited patterns of change similar to the infiltration of CD8 T cells (increase, no change and decrease) after 10F RT (representative images in Figure S8, heatmap graph in Figure 4C, summary of change pattern proportion in Figure 4E). The comparisons of marker expression in the different subgroups of patients are shown in Figure 4F–O with similar results with the H score (Figure S9). Markers with both significant increases or decreases in subgroups of patients included nuclear IRF1, PD-L1, SERPINB9, nuclear STAT1 and HLA-B/C. Around 40% of patients exhibited upregulation of PD-L1 expression (17% vs 50%, P < 0.001) in tumor cells, while 20% of patients showed decreases (41% vs 6%, P < 0.05) (Figure 4H). CD47 (a protein often expressed by tumor cells that inhibits phagocytosis by macrophages) was found in the majority of tumor cells with variable staining intensity at baseline and sustained changes after irradiation in a subgroup of patients (Figure S10) (33). Significant induction of TAP1 (3% vs 38%, P < 0.001) was found in 37% of patients, while the tumors the other patients did not show substantial changes (Figure 4F). Significant downregulation of HLA-A and β2M was observed in 37% and 20% of patients, respectively (Figure 4K, M). The expression of LMP2 and LMP7 was unresponsive to RT and was found in approximately 95% of cancer cells before and after RT (Figure 4N, O). Overall, CCRT led to expression changes of key immune markers involved in immune surveillance that might affect a therapy-induced anti-tumor immune response.
PD-L1 expression correlates with tumor cell nuclear IRF1 staining and the IFN gene signature of PMBCs during CCRT
PD-L1 expression on cancer cells is an indicator of tumor response to αPD-L1/PD-1 antibodies in some situations. The expression positivity of PD-L1 in tumor cells correlated with the IFN gene signature in tumors (r = 0.39, P < 0.01) and PMBCs (r = 0.45, P < 0.01) before CCRT (Figure 5A and Figure S11). Following 10F RT, PD-L1 positivity showed a significant correlation (r = 0.48, P < 0.01) with nuclear IRF1 positivity (Figure S11) and a trend of association with others, including the nuclear STAT1 and IFN gene signature in tumors or in PMBCs. Patients with high (> 10%) nuclear IRF1 positivity in tumor cells had more than 2 times higher expression positivity of PD-L1 (44% vs 20%, P < 0.01) in tumor cells than those with low nuclear IRF1 positivity (representative images in Figure 5C and quantification in Figure 5D). The dynamic change of PD-L1 positivity was associated with nuclear IRF1 positivity in tumor cells, tumor infiltrating CD8+ T cell number and the PMBC IFN gene signature (Figure 5A). Consistently, augmented PD-L1 expression was associated with upregulation of nuclear IRF1, CD8+ T cell number, the PMBC IFN signature and to lesser extent, nuclear STAT1 positivity (heatmap graph in Figure 5B). Patients with increased IRF1 positivity or the PMBC IFN signature had greater upregulation of PD-L1 than patients without (Figure 5E, F). The probability of PD-L1 induction is significantly higher in patients with augmentation of their PMBC IFN signature than patients without (83% vs 7%, P < 0.0001). Indeed, following 10F RT, the average tumor cell PD-L1 positivity was 50% in patients with an increased PMBC IFN gene signature compared to 19% in patients with decreases (P < 0.05, Figure 5H). The H score also correlated with nuclear IRF1 H score, CD8+ T cell number and the PMBC IFN signature (Figure S12). Patients with an increased PMBC IFN gene signature had significantly higher risk of PD-L1 H score upregulation in tumor cells than patients without (50% vs 7%, P < 0.05). In summary, the expression of PD-L1 in tumor cells is correlated with IRF1-mediated signaling, numbers of tumor infiltrating CD8+ T cells and the PMBC IFN gene signature.
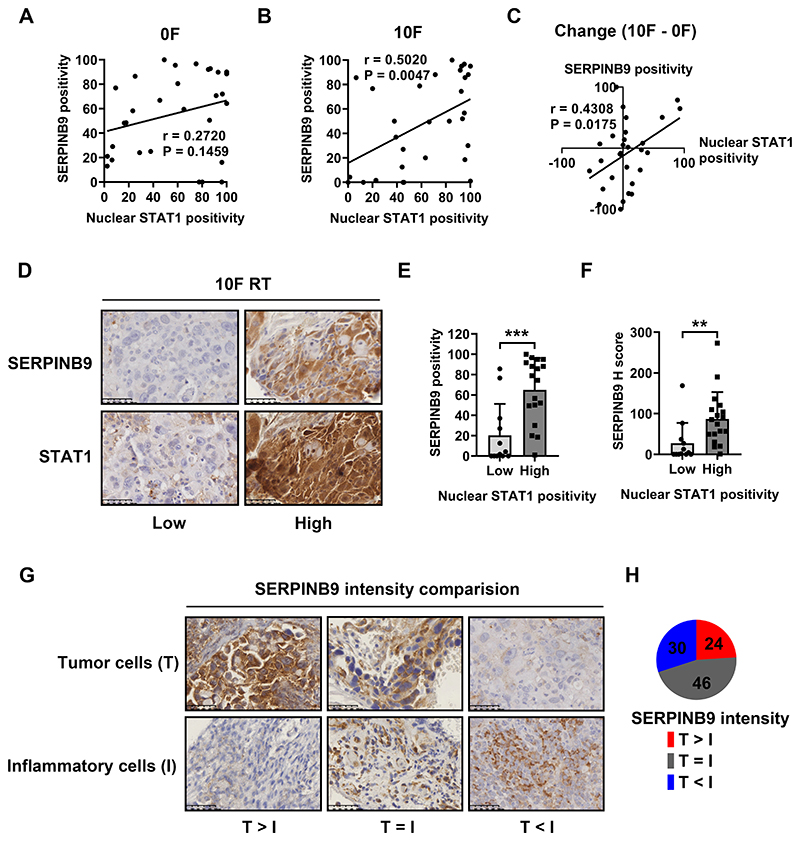
SERPINB9 expression correlates with STAT1-mediated signaling in tumor cells
SERPINB9 is an emerging target for cancer immunotherapy. It mediates resistance to radiation-induced immunity and to ICB in both experimental settings and retrospective clinical data (23,34,35). SERPINB9 expression correlates with STAT1-mediated signaling in tumor cells in biopsies from CC patients (Figure 6A–C). High nuclear STAT1 positivity is associated with greater expression of SERPINB9 in tumor cells after 10F RT (representative images in Figure 6D, quantification in Figure 6E, F). SERPINB9 expression was also found in immune cells, including T cells and dendritic cells. We compared the expression intensity of SERPINB9 between these two cellular components in patients and categorized them into 3 subgroups (representative images in Figure 6G, quantification in Figure 6H). Greater expression of SERPINB9 in cancer cells than inflammatory cells was found in 24% of patients, while 30% of patients displayed higher staining intensity in inflammatory cells than in cancer cells. The majority (46%) of patients had comparable expression levels of SERPINB9 in both cellular components.
Figure 6. SERPINB9 expression correlates with the STAT1-mediated signaling in tumor cells.
Expression positivity of SERPINB9 and nuclear STAT1 in tumor cells before and after 10F RT, and their dynamic changes are plotted in (A), (B) and (C).
(D): Representative immunohistochemical section staining (low vs high) of SERPINB9 (top) and nuclear STAT1 (bottom) in tumor biopsies from patients after 10F RT.
SERPINB9 expression (positivity and H score) in tumor cells with low (≤ 50%) or high (> 50%) nuclear STAT1 positivity after 10F RT was summarized in (E) and (F).
(G): Representative immunohistochemical staining of tissue sections with various SERPINB9 expression differences between tumor cells (T, on the top) and inflammatory cells (I, on the bottom) from patients after 10F RT. T > I, T = I or T < I: the median expression intensity of SERPINB9 in tumor cells is greater, equal to or weaker than that of surrounding inflammatory cells.
(H): Percentages of patients with different expression patterns of SERPINB9 in tumor cells and inflammatory cells after 10F RT.
Data in bar charts represent mean ± SD. Mean comparison in 2 groups was performed by using the Mann-Whitney test. The correlation between positivity of various markers was evaluated by Pearson’s correlation (**P < 0.01, *** P < 0.001).
In addition, STAT1-mediated signaling was associated with expression of APM components in tumor cells, in particular HLA-A, HLA-B/C, β2M and LMP7 (Figure S13).
Discussion
Here we assessed evidence for an immune response and IFN signaling in biopsies before and during therapy of CC, a cancer often immunogenic due to HPV. Most notably, we found substantial heterogeneity in CD8+ T cell infiltration and in IFN signaling after 10F of CCRT with increases, no change and decreases all represented.
OS of CC patients, who received RT, correlates strongly with tumor CD8A expression in the TCGA database and in this study patients with decreased tumor infiltrating CD8+ T cells during CCRT had a significantly higher risk of PR than of CR. Low CD8+ T cell infiltration also correlates with increased risk of relapse (36,37). Therefore, our results suggest that the response of tumor infiltrating CD8+ T cells in CC treated with CCRT contributes to therapeutic efficacy. Whether this early CD8 T cell response reflects long-term local tumor control remains to be determined. The heterogenous T cell response we observed might be related to timing as the infiltration of CD8+ T cells probably varies during treatment with diminished levels at least in mice, early after radiation followed by expansion (23,38). Dorta-Estremera et al. found an initial decline of CD8+ T cell number in the first week, which was followed by variable expansion later in irradiated cervical cancers consistent with the results here (39).
Activation of IFN signaling after RT has been observed in many murine tumor models (4,21,40). However, this activation did not occur in many of our CC patients. Only one third of patients showed augmented IFN gene expression or nuclear STAT1 and IRF1 staining after 10F RT. Lack of response might be due to deficiencies of key mediators in IFN signaling. cGAS-STING or IFNAR1 signaling is frequently suppressed in tumors (41–43). In addition, mutation of these mediators is possible and has been shown for STING (44). Additionally, HPV-derived E7 inhibits the cGAS-STING pathway in human cervical cancer cells (45). Here only 16% of CC cells expressed IRF1, a key transcription factor in IFN induction, a limitation that might contribute to the impaired IFN response. Activation of negative feedback loops in response to IFNs can inhibit IFN-mediated signaling (40,46). Further necrosis in tumors following CCRT could affect the overall IFN response. Still it is striking that a positive IFN response to CCRT was found in less than 40% of these patients as compared to results using animal models. These data highlight the danger of extrapolation from animal model-derived results.
The benefit from adding ICB to RT may depend on the expression of immune checkpoints. 40% of patients in this study displayed PD-L1 induction in tumor cells during CCRT. The combination of CD47-SIRPα blockade and RT could be synergistic resulting in enhanced anti-tumor T cell immunity (33,47). We separated patients into subgroups based on changes of PD-L1 positivity, CD8+ T cell number, CD47 H score and CD68+ cell number (Figure S14). If cancers with increases of either the immune checkpoint (PD-L1 or CD47) or the effector cells (CD8+ or CD68+ cells) would benefit from blockade of CD47 or PD-L1 then CCRT might benefit from additional ICB in 59% of patients: 11% from αPD-L1/PD-1, 26% from αCD47/SIRPα, and 22% from both (Figure S14).
SERPINB9 was frequently expressed in the samples we examined, validating it as a potential target for cancer immunotherapy (23). Consistently, emerging data suggests that SERPINB9 can protect cancer cells and immunosuppressive cells from Granzyme B-mediated killing, which can be targeted simultaneously in vivo via a small molecule SERPINB9 inhibitor (48). TAP1 is one component of APM frequently deficient in cervical cancer cells and its deficiency can mediate tumor evasion (49,50). Very few patients enrolled in this study exhibited TAP1 expression in tumor cells at baseline. However after 10F, 37% of tumors displayed significant induction of TAP1 in an IRF1-dependent manner suggesting its induction might contribute to anti-tumor immunity.
Additionally, our work suggests that the IFN signature in PMBCs may reflect that of the tumor and then reflect tumor expression changes in PD-L1. Induction of serum IFNβ correlated with an abscopal response in late-stage NSCLC patients who underwent SBRT in combination with CTLA-4 blockade (17). If the PMBC IFN gene signature mirrors that in the tumor, it could be used to monitor therapy and select patients for combinational therapeutics.
In summary, we demonstrate that CCRT leads to differential tumor immunogenicity in CC patients. Increases in tumor infiltrating CD8+ T cells correlate with IRF1-mediated signaling in tumor cells and short-term response to CCRT. The dynamic and differential tumor immunogenicity in CC patients in response to CCRT suggest that RT induction of immunity is limited to a subset of patients and may reflect the heterogeneity of intratumoral induction of IFN.
Supplementary Material
Translational relevance.
The combination of immune checkpoint blockade (ICB) with radiation therapy for cancer is of great promise, although the extent of anti-tumor immunity that develops in response to irradiation is uncertain. Here we show in cervical cancer patients that increased CD8 T cell infiltration during concurrent chemoradiotherapy (CCRT) is limited to a subset of patients. Similarly, only a subset of patients showed tumoral interferon induction during CCRT, which correlated with CD8 T cell infiltration and only 40% of patients displayed PD-L1 induction in tumor cells during chemoradiotherapy also correlating with interferon signaling. The limitation of induction of anti-tumor immunity and interferon to a subset of patients suggests that the combination of CCRT and ICB will only be effective selectively dependent upon interferon signaling in response to CCRT. Additionally, our results suggest the interferon signature in peripheral mononuclear blood cells could be an easily accessible biomarker to select patients for these combinational therapeutics.
Acknowledgments
This study was funded by Cancer Research UK grant C5255/A15935 (to RJM); Shantou University Medical College Clinical Research Enhancement Initiative, N0201424 (to CC and JC); Collaborative and Creative Center, Molecular Diagnosis and Personalized Medicine, Shantou University, Guangdong, China (to CC); Science and Technology Special Fund of Guangdong Province of China, 2019-132 (to CC); Strategic and Special Fund for Science and Technology Innovation of Guangdong Province of China, 180918114960704 (to CC). We thank Geng Wang from Department of Pathology, Jiongyu Chen, Chaoqun Hong, Jiediao Lin from Oncological Research Lab, Cancer Hospital of Shantou University Medical College and the core facility team in the Central Laboratory of Shantou University Medical College for technical support.
Footnotes
The authors declare no potential conflicts of interest.
References
- 1.de Sanjose S, Quint WG, Alemany L, Geraets DT, Klaustermeier JE, Lloveras B, et al. Human papillomavirus genotype attribution in invasive cervical cancer: a retrospective cross-sectional worldwide study. Lancet Oncol. 2010;11(11):1048–56. doi: 10.1016/S1470-2045(10)70230-8. [DOI] [PubMed] [Google Scholar]
- 2.Qin Y, Ekmekcioglu S, Forget MA, Szekvolgyi L, Hwu P, Grimm EA, et al. Cervical Cancer Neoantigen Landscape and Immune Activity is Associated with Human Papillomavirus Master Regulator. Front Immunol. 2017;8 doi: 10.3389/fimmu.2017.00689. ARTN 689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SAJR, Behjati S, Biankin AV, et al. Signatures of mutational processes in human cancer. Nature. 2013;500(7463):415–21. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Deng L, Liang H, Xu M, Yang X, Burnette B, Arina A, et al. STING-Dependent Cytosolic DNA Sensing Promotes Radiation-Induced Type I Interferon-Dependent Antitumor Immunity in Immunogenic Tumors. Immunity. 2014;41(5):843–52. doi: 10.1016/j.immuni.2014.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harding SM, Benci JL, Irianto J, Discher DE, Minn AJ, Greenberg RA. Mitotic progression following DNA damage enables pattern recognition within micronuclei. Nature. 2017;548(7668):466–70. doi: 10.1038/nature23470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reits EA, Hodge JW, Herberts CA, Groothuis TA, Chakraborty M, Wansley EK, et al. Radiation modulates the peptide repertoire, enhances MHC class I expression, and induces successful antitumor immunotherapy. J Exp Med. 2006;203(5):1259–71. doi: 10.1084/jem.20052494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gupta A, Probst HC, Vuong V, Landshammer A, Muth S, Yagita H, et al. Radiotherapy promotes tumor-specific effector CD8+ T cells via dendritic cell activation. J Immunol. 2012;189(2):558–66. doi: 10.4049/jimmunol.1200563. [DOI] [PubMed] [Google Scholar]
- 8.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70(1):7–30. doi: 10.3322/caac.21590. [DOI] [PubMed] [Google Scholar]
- 9.Chung HC, Schellens JHM, Delord JP, Perets R, Italiano A, Shapira-Frommer R, et al. Pembrolizumab treatment of advanced cervical cancer: Updated results from the phase 2 KEYNOTE-158 study. J Clin Oncol. 2018;36(15) doi: 10.1200/JCO.2018.36.15_suppl.5522. [DOI] [PubMed] [Google Scholar]
- 10.Mayadev JS, Enserro D, Lin YG, Da Silva DM, Lankes HA, Aghajanian C, et al. Sequential Ipilimumab After Chemoradiotherapy in Curative-Intent Treatment of Patients With Node-Positive Cervical Cancer. JAMA Oncol. 2019 doi: 10.1001/jamaoncol.2019.3857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dyer BA, Feng CH, Eskander R, Sharabi AB, Mell LK, McHale M, et al. Current Status of Clinical Trials for Cervical and Uterine Cancer Using Immunotherapy Combined with Radiation. Int J Radiat Oncol Biol Phys. 2020 doi: 10.1016/j.ijrobp.2020.09.016. [DOI] [PubMed] [Google Scholar]
- 12.McLaughlin M, Patin EC, Pedersen M, Wilkins A, Dillon MT, Melcher AA, et al. Inflammatory microenvironment remodelling by tumour cells after radiotherapy. Nat Rev Cancer. 2020;20(4):203–17. doi: 10.1038/s41568-020-0246-1. [DOI] [PubMed] [Google Scholar]
- 13.Dewan MZ, Galloway AE, Kawashima N, Dewyngaert JK, Babb JS, Formenti SC, et al. Fractionated but Not Single-Dose Radiotherapy Induces an Immune-Mediated Abscopal Effect when Combined with Anti-CTLA-4 Antibody. Clinical Cancer Research. 2009;15(17):5379–88. doi: 10.1158/1078-0432.Ccr-09-0265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deng L, Liang H, Burnette B, Beckett M, Darga T, Weichselbaum RR, et al. Irradiation and anti-PD-L1 treatment synergistically promote antitumor immunity in mice. J Clin Invest. 2014;124(2):687–95. doi: 10.1172/JCI67313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Twyman-Saint Victor C, Rech AJ, Maity A, Rengan R, Pauken KE, Stelekati E, et al. Radiation and dual checkpoint blockade activate non-redundant immune mechanisms in cancer. Nature. 2015;520(7547):373–7. doi: 10.1038/nature14292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Luke JJ, Lemons JM, Karrison TG, Pitroda SP, Melotek JM, Zha Y, et al. Safety and Clinical Activity of Pembrolizumab and Multisite Stereotactic Body Radiotherapy in Patients With Advanced Solid Tumors. J Clin Oncol. 2018;36(16):1611–8. doi: 10.1200/JCO.2017.76.2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Formenti SC, Rudqvist NP, Golden E, Cooper B, Wennerberg E, Lhuillier C, et al. Radiotherapy induces responses of lung cancer to CTLA-4 blockade. Nat Med. 2018;24(12):1845–51. doi: 10.1038/s41591-018-0232-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Antonia SJ, Villegas A, Daniel D, Vicente D, Murakami S, Hui R, et al. Overall Survival with Durvalumab after Chemoradiotherapy in Stage III NSCLC. New Engl J Med. 2018;379(24):2342–50. doi: 10.1056/NEJMoa1809697. [DOI] [PubMed] [Google Scholar]
- 19.Formenti SC, Rudqvist N-P, Golden E, Cooper B, Wennerberg E, Lhuillier C, et al. Radiotherapy induces responses of lung cancer to CTLA-4 blockade. Nature medicine. 2018;24(12):1845–51. doi: 10.1038/s41591-018-0232-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen J, Markelc B, Kaeppler J, Ogundipe VML, Cao Y, McKenna WG, et al. STING-Dependent Interferon-lambda1 Induction in HT29 Cells, a Human Colorectal Cancer Cell Line, After Gamma-Radiation. Int J Radiat Oncol Biol Phys. 2018;101(1):97–106. doi: 10.1016/j.ijrobp.2018.01.091. [DOI] [PubMed] [Google Scholar]
- 21.Burnette BC, Liang H, Lee Y, Chlewicki L, Khodarev NN, Weichselbaum RR, et al. The efficacy of radiotherapy relies upon induction of type i interferon-dependent innate and adaptive immunity. Cancer Res. 2011;71(7):2488–96. doi: 10.1158/0008-5472.CAN-10-2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vatner RE, Janssen EM. STING, DCs and the link between innate and adaptive tumor immunity. Molecular Immunology. 2019;110:13–23. doi: 10.1016/j.molimm.2017.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen J, Cao Y, Markelc B, Kaeppler J, Vermeer JA, Muschel RJ. Type I IFN protects cancer cells from CD8+ T cell-mediated cytotoxicity after radiation. J Clin Invest. 2019;129(10):4224–38. doi: 10.1172/JCI127458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Han C, Liu Z, Zhang Y, Shen A, Dong C, Zhang A, et al. Tumor cells suppress radiation-induced immunity by hijacking caspase 9 signaling. Nat Immunol. 2020;21(5):546–54. doi: 10.1038/s41590-020-0641-5. [DOI] [PubMed] [Google Scholar]
- 25.Liang H, Deng L, Hou Y, Meng X, Huang X, Rao E, et al. Host STING-dependent MDSC mobilization drives extrinsic radiation resistance. Nat Commun. 2017;8(1):1736. doi: 10.1038/s41467-017-01566-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jacquelot N, Yamazaki T, Roberti MP, Duong CPM, Andrews MC, Verlingue L, et al. Sustained Type I interferon signaling as a mechanism of resistance to PD-1 blockade. Cell Res. 2019;29(10):846–61. doi: 10.1038/s41422-019-0224-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eric Hall AG. Radiobiology for radiobiologist. 7th edion. Lippincott Williams & Wilkins; 2011. pp. 334–5. [Google Scholar]
- 28.Venkatesulu BP, Mallick S, Lin SH, Krishnan S. A systematic review of the influence of radiation-induced lymphopenia on survival outcomes in solid tumors. Crit Rev Oncol Hemat. 2018;123:42–51. doi: 10.1016/j.critrevonc.2018.01.003. [DOI] [PubMed] [Google Scholar]
- 29.Rice GI, Forte GM, Szynkiewicz M, Chase DS, Aeby A, Abdel-Hamid MS, et al. Assessment of interferon-related biomarkers in Aicardi-Goutieres syndrome associated with mutations in TREX1, RNASEH2A, RNASEH2B, RNASEH2C, SAMHD1, and ADAR: a case-control study. Lancet Neurol. 2013;12(12):1159–69. doi: 10.1016/S1474-4422(13)70258-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ghandhi SA, Shuryak I, Morton SR, Amundson SA, Brenner DJ. New Approaches for Quantitative Reconstruction of Radiation Dose in Human Blood Cells. Sci Rep. 2019;9(1):18441. doi: 10.1038/s41598-019-54967-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martinez FO, Gordon S, Locati M, Mantovani A. Transcriptional profiling of the human monocyte-to-macrophage differentiation and polarization: new molecules and patterns of gene expression. J Immunol. 2006;177(10):7303–11. doi: 10.4049/jimmunol.177.10.7303. [DOI] [PubMed] [Google Scholar]
- 32.Brzostek-Racine S, Gordon C, Van Scoy S, Reich NC. The DNA damage response induces IFN. J Immunol. 2011;187(10):5336–45. doi: 10.4049/jimmunol.1100040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Soto-Pantoja DR, Terabe M, Ghosh A, Ridnour LA, DeGraff WG, Wink DA, et al. CD47 in the tumor microenvironment limits cooperation between antitumor T-cell immunity and radiotherapy. Cancer Res. 2014;74(23):6771–83. doi: 10.1158/0008-5472.CAN-14-0037-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jiang P, Gu S, Pan D, Fu J, Sahu A, Hu X, et al. Signatures of T cell dysfunction and exclusion predict cancer immunotherapy response. Nat Med. 2018;24(10):1550–8. doi: 10.1038/s41591-018-0136-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pan D, Kobayashi A, Jiang P, Ferrari de Andrade L, Tay RE, Luoma AM, et al. A major chromatin regulator determines resistance of tumor cells to T cell-mediated killing. Science. 2018;359(6377):770–5. doi: 10.1126/science.aao1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nedergaard BS, Ladekarl M, Thomsen HF, Nyengaard JR, Nielsen K. Low density of CD3+, CD4+ and CD8+ cells is associated with increased risk of relapse in squamous cell cervical cancer. Br J Cancer. 2007;97(8):1135–8. doi: 10.1038/sj.bjc.6604001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nedergaard BS, Ladekarl M, Nyengaard JR, Nielsen K. A comparative study of the cellular immune response in patients with stage IB cervical squamous cell carcinoma. Low numbers of several immune cell subtypes are strongly associated with relapse of disease within 5 years. Gynecol Oncol. 2008;108(1):106–11. doi: 10.1016/j.ygyno.2007.08.089. [DOI] [PubMed] [Google Scholar]
- 38.Dovedi SJ, Cheadle EJ, Popple AL, Poon E, Morrow M, Stewart R, et al. Fractionated Radiation Therapy Stimulates Antitumor Immunity Mediated by Both Resident and Infiltrating Polyclonal T-cell Populations when Combined with PD-1 Blockade. Clin Cancer Res. 2017;23(18):5514–26. doi: 10.1158/1078-0432.CCR-16-1673. [DOI] [PubMed] [Google Scholar]
- 39.Dorta-Estremera S, Colbert LE, Nookala SS, Yanamandra AV, Yang G, Delgado A, et al. Kinetics of Intratumoral Immune Cell Activation During Chemoradiation for Cervical Cancer. Int J Radiat Oncol Biol Phys. 2018;102(3):593–600. doi: 10.1016/j.ijrobp.2018.06.404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vanpouille-Box C, Alard A, Aryankalayil MJ, Sarfraz Y, Diamond JM, Schneider RJ, et al. DNA exonuclease Trex1 regulates radiotherapy-induced tumour immunogenicity. Nat Commun. 2017;8:15618. doi: 10.1038/ncomms15618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xia T, Konno H, Ahn J, Barber GN. Deregulation of STING Signaling in Colorectal Carcinoma Constrains DNA Damage Responses and Correlates With Tumorigenesis. Cell Rep. 2016;14(2):282–97. doi: 10.1016/j.celrep.2015.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Katlinski KV, Gui J, Katlinskaya YV, Ortiz A, Chakraborty R, Bhattacharya S, et al. Inactivation of Interferon Receptor Promotes the Establishment of Immune Privileged Tumor Microenvironment. Cancer Cell. 2017;31(2):194–207. doi: 10.1016/j.ccell.2017.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kitajima S, Ivanova E, Guo S, Yoshida R, Campisi M, Sundararaman SK, et al. Suppression of STING Associated with LKB1 Loss in KRAS-Driven Lung Cancer. Cancer Discov. 2019;9(1):34–45. doi: 10.1158/2159-8290.CD-18-0689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Konno H, Yamauchi S, Berglund A, Putney RM, Mule JJ, Barber GN. Suppression of STING signaling through epigenetic silencing and missense mutation impedes DNA damage mediated cytokine production. Oncogene. 2018;37(15):2037–51. doi: 10.1038/s41388-017-0120-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lau L, Gray EE, Brunette RL, Stetson DB. DNA tumor virus oncogenes antagonize the cGAS-STING DNA-sensing pathway. Science. 2015;350(6260):568–71. doi: 10.1126/science.aab3291. [DOI] [PubMed] [Google Scholar]
- 46.Arimoto KI, Miyauchi S, Stoner SA, Fan JB, Zhang DE. Negative regulation of type I IFN signaling. J Leukoc Biol. 2018 doi: 10.1002/JLB.2MIR0817-342R. [DOI] [PubMed] [Google Scholar]
- 47.Schwartz AL, Nath PR, Allgauer M, Lessey-Morillon EC, Sipes JM, Ridnour LA, et al. Antisense targeting of CD47 enhances human cytotoxic T-cell activity and increases survival of mice bearing B16 melanoma when combined with anti-CTLA4 and tumor irradiation. Cancer Immunol Immunother. 2019;68(11):1805–17. doi: 10.1007/s00262-019-02397-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jiang L, Wang Y-J, Zhao J, Uehara M, Hou Q, Kasinath V, et al. Direct Tumor Killing and Immunotherapy through Anti-SerpinB9 Therapy. Cell. 2020;183(5):1219–33.:e18. doi: 10.1016/j.cell.2020.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cromme FV, Airey J, Heemels MT, Ploegh HL, Keating PJ, Stern PL, et al. Loss of transporter protein, encoded by the TAP-1 gene, is highly correlated with loss of HLA expression in cervical carcinomas. J Exp Med. 1994;179(1):335–40. doi: 10.1084/jem.179.1.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Evans M, Borysiewicz LK, Evans AS, Rowe M, Jones M, Gileadi U, et al. Antigen processing defects in cervical carcinomas limit the presentation of a CTL epitope from human papillomavirus 16 E6. J Immunol. 2001;167(9):5420–8. doi: 10.4049/jimmunol.167.9.5420. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.