Abstract
Repurposing cationic amphiphilic drugs (CADs) for cancer treatment is emerging as an attractive means to enhance the efficacy of chemotherapy. Many commonly used CADs, including several cation amphiphilic antihistamines and antidepressants, induce cancer-specific, lysosome-dependent cell death and sensitize cancer cells to chemotherapy. CAD-induced inhibition of lysosomal acid sphingomyelinase is necessary, but not sufficient, for the subsequent lysosomal membrane permeabilization and cell death, while other pathways regulating this cell death pathway are largely unknown. Prompted by significant changes in the expression of genes involved in Ca2+ and cyclic AMP (cAMP) signaling pathways in CAD resistant MCF7 breast cancer cells, we identified here an early lysosomal Ca2+ release through P2X purinerigic receptor 4 (P2RX4) and subsequent Ca2+- and adenylyl cyclase 1 (ADCY1)-dependent synthesis of cAMP as a signaling route mediating CAD-induced lysosomal membrane permeabilization and cell death. Importantly, pharmacological and genetic means to increase cellular cAMP levels either by activating cAMP-inducing G-protein coupled receptors (GPR3 or ß2 adrenergic receptor) or ADCY1, or by inhibiting cAMP-reducing guanine nucleotide-binding protein G(i) subunit α2, C-X-C motif chemokine receptor type 4 or cAMP phosphodiesterases, sensitized cancer cells to CADs. These data reveal a previously unrecognized lysosomal P2RX4- and ADCY1-dependent signaling cascade as a pathway essential for CAD-induced lysosome-dependent cell death, and encourage further investigations to find the most potent combinations of CADs and cAMP-inducing drugs for cancer therapy.
Keywords: adenyl cyclase 1, calcium, cationic amphiphilic drugs, cyclic AMP, lysosome, P2RX4, signaling
Introduction
Lysosomes are acidic organelles originally described as cellular waste baskets and recycling centers responsible for the digestion and reuse of cargo delivered to them mainly by endocytosis and autophagy. In recent years, lysosomes have emerged as important signaling platforms that regulate cell metabolism, ion homeostasis, motility and survival in response to the metabolic status of the cell [1–4]. In line with their role in such essential cellular processes, reduced lysosomal function is associated with severe degenerative diseases [5–7], while lysosomal activation is a common feature of aggressive cancers, where lysosomes promote cancer metabolism and invasion [8–10]. The activation of lysosomes during malignant transformation does not come without a price. The cancer-associated changes in lysosomal composition result in reduced lysosomal membrane stability, thereby sensitizing cells to lysosome-dependent cell death [11]. This frailty can be targeted by cationic amphiphilic drugs (CADs) that induce lysosome-dependent death of cancer cells regardless of their origin, and sensitize them to chemotherapy in vitro and in various murine cancer models in vivo [12–19]. Suggestive of their efficacy also in human cancer, recent pharmaco-epidemiological studies have reported a reduced incidence of glioma and colorectal cancer among users of tricyclic antidepressants [20], and an association between postdiagnostic use of cationic amphiphilic antihistamines and reduced cancer mortality as compared with similar use of antihistamines that do not classify as CADs [18]. Finally, the promising data is emerging from numerous clinical trials testing the anti-cancer activity of chloroquine and its derivatives [21].
The CAD family comprises a broad spectrum of compound classes, including dozens of approved drugs that are used to treat a wide range of diseases including allergies, heart diseases. psychiatric disorders and infections [22]. Common to all CADs is their amphiphilic nature that is brought about by a hydrophobic ring structure and a hydrophilic side chain containing cationic amine groups. Due to their chemical structure, CADs diffuse freely to the acidic lysosomes, where they are protonated and trapped resulting in up to 1000-fold accumulation in this compartment [23]. Inside the lysosomes, CADs incorporate to luminal membranes where they function as effective inhibitors of acid sphingomyelinase and other lysosomal lipases [24]. The subsequent alterations in the lipid composition lead to increased permeability of lysosomal membranes, and eventually to lysosome-dependent cell death mediated by lysosomal hydrolases leaking into the cytosol [14, 22]. Cancer cells are especially sensitive to the accumulation of sphingomyelin [14, 25, 26], which may, at least partially, explain the cancer-specific toxicity of CADs [14]. Other cellular pathways regulating CAD-induced lysosome-dependent cell death are, however, largely unknown.
Prompted by the apparent anti-cancer activity of CADs, we are presently preparing for clinical trials to test whether repurposing of safe and inexpensive antihistamines for cancer therapy would be warranted. In order to identify optimal chemotherapies and other treatments for the combination therapy with CADs, it is essential to know the signaling pathways that either promote or inhibit CAD-induced cell death. To gain insight into such pathways, we compared transcriptomes of control and CAD resistant MCF7 breast cancer cells. Prompted by the abundance of genes involved in G-protein coupled receptor (GPCR) signaling among the differentially expressed genes, we then undertook a detailed study of the role of GPCR-activated signaling pathways in CAD-induced cell death and investigated the possibilities to exploit these pathways in order to improve the anti-cancer activity of CADs.
Materials and Methods
Reagents
Details of chemicals, reagents, kits, siRNAs, plasmids, primer and antibodies used are listed in Table S1. All kits were used according to the manufacturer’s instructions and drugs were reconstituted in DMSO unless otherwise specified.
Cell culture
Cell lines used included an S1 subclone of MCF7 human ductal breast carcinoma cells [27] and human A549 non-small cell lung carcinoma and HeLa cervix carcinoma cells from the ATCC. Cells were authenticated by sequencing (MCF7) or by short tandem repeat analysis, and used within 6 months after thawing. Cells were cultured in Dulbecco’s Modified Eagle’s medium, supplemented with 6% heat-inactivated fetal calf serum penicillin and streptomycin. CAD resistant cells were established by culturing them with increasing concentration of siramesine for six months, and the resistant cells were resensitized by culturing them without siramesine for three months. Their authenticity as MCF7 cells was confirmed by sequencing after the long-term culture. All cells were cultured at 37 °C in a humidified atmosphere of 5% CO2. Cells were regularly tested and found negative for mycoplasma using Venor®GeM Classic PCR kit.
RNA sequencing
Total RNA was isolated using Nucleospin RNA II kit. The 100 bp reads, paired end RNA sequencing (40 million reads/sample) by Illumina Hiseq2000 was performed by BGI HongKong Co., Ltd. Reads were aligned to the human genomic sequence and quantified using the CLC genomic workbench software. Reads were mapped with a minimum of 50 bases of consecutive match allowing for up to 3 mismatches. Mapped reads were used to calculate the relative expression levels of the identified genes, expressed in reads per kilobase of exon per million mapped sequenced reads (RPKM).
Transfections
Plasmid transfections were performed with Lipofectamine LTX with PLUS Reagent. To obtain stable clones overexpressing the protein of interest, cells were cultured in the appropriate selection medium and single cell cloned. siRNA transfections were performed as reverse transfections using 20 nM siRNA and RNAiMAX.
Cell Death
Subconfluent cells were treated as indicated and cell death was measured after 10 min incubation with propidium iodide (0.2 μg/mL) and Hoechst-33342 (2.5 μg/mL) at 37 °C using Celigo® Imaging Cytometer (Nexcelom Bioscience) according to the manufacturer’s instructions.
Calcium
To measure relative levels of cytosolic free Ca2+ ([Ca2+]c), cells were stained with 3 μM Fluo-4-AM for 25 min, washed twice with Dulbecco’s phosphate-buffered saline (DPBS), resuspended in DPBS without Ca2+/Mg2+ (if not otherwise indicated) plus 20mM HEPES and maintained at 37°C while treating the cells as indicated and analyzing by BD FACSVerse flow cytometer (FL-1 channel).
cAMP measurements
To analyze relative intracellular cAMP levels ([cAMP]i), we transfected MCF7 cells with Flamindo2 plasmid (a gift from Tetsuya Kitaguchi [28]) and created a single cell clone with relatively high expression. For the cAMP analysis, MCF7-Flamindo2 cells were resuspended in DPBS without Ca2+/Mg2+ plus 20 mM HEPES and maintained at 37°C during the indicated treatment and analysis by BD FACSVerse flow cytometer (FL-1 channel). Absolute cAMP levels were quantified with the competitive immunoassay using HitHunter® cAMP-HS+ kit according to manufacturer’s instructions. Luminescence was measured with the PerkinElmer’s EnVision 2105 multimode plate reader. Values were interpolated with the standard curve using GraphPad prism.
Lysosomal pH
To estimate the relative lysosomal pH, subconfluent cells were loaded with 5 μg/ml pH-sensitive PhRodo® Green coupled to 10 kDa dextran and 2.5 mg/ml pH-insensitive tetramethylrhodamine (TMR) coupled to 70 kDa dextran for 18 h, washed and chased in fresh medium for 5 h. Medium was changed to Thermo Fisher Scientific Live Cell Imaging Solution supplemented with 20 mM HEPES before image acquisition by LSM700 confocal laser scanning microscope (Zeiss). Images were analyzed using ImageJ software.
Volume of acidic compartment
Cells were stained with 50 nM LysoTracker green DND-26 in DMEM in an incubator (37°C, 5% CO2) for 30 min, washed once with DPBS and re-suspended in fresh pre-warmed medium without phenol red before the analysis of 10000 cells / sample by BD FACSVerse flow cytometer using FL-1 channel.
CAD uptake
Cells (0.5 x 106) treated with 15 μM ebastine were suspended in 500 μl distilled H2O and total protein concentration was measured using Pierce BCA Protein Assay Kit. Aliquots of 20 μl were extracted using 200 μl of acetonitrile (ACN), adding 8 μl of internal standard (1 μM terfenadine) for the quantitation. Samples were shaken for 10min at 4°C using an Eppendorf ThermoMixer C (Eppendorf Instrument GmbH, Hamburg, Germany) followed by centrifugation at 10000 rpm for 5 min at 4°C for protein precipitation. Supernatant was transferred into a 1.5 ml Eppendorf tube and the solvent was evaporated. Sample were resuspended in the adequate solvent for the liquid chromatography coupled to mass spectrometry (LC-MS) starting conditions. The quantitation was performed using a (U) HPLC UltiMate 3000 RSLCnano System interfaced on-line to quadrupole-orbitrap mass spectrometer Q-Exactive (both from Thermo Fisher Scientific) using a Triart C18 reverse phase column 150 x 0.5, 1/16”, 5 μm particle size (YMC CO., LTD, Japan).
Parallel reaction monitoring for the specific fragments 167.0858 (ebastine) and 436.3002 (terfenadine) was used for the quantitation of ebastine. The data analysis was performed using TraceFinder (Thermo Fisher Scientific) and Prism GraphPad. All solvents for the extraction were HPLC grade.
Microscopy
For immunocytochemistry, cells grown on coverslips and fixed in 4% paraformaldehyde in Dulbecco’s phosphate-buffered saline (DPBS) for 15 min were permeabilized with 0.2% triton X-100 in DPBS for 15 min and blocked in 5% goat serum in DPBS for 30 min before staining with indicated primary antibodies (Table S1) followed by appropriate AlexaFluor™488- or AlexaFluor™594–coupled secondary antibodies. Nuclei were labeled with 5 mg/ml Hoechst 33342 and coverslips were mounted with Prolong Gold Antifade mounting medium. Images were acquired on LSM700 with a 63X Carl Zeiss oil-objective and the Zeiss Zen software. For P2RX4 and LAMP1 colocalization, cells were transfected with the P2X4-pHluorin123 (a gift from Baljit Khakh, Addgene plasmid # 52926) [29] and LAMP1-mCherry (a gift from Michael Davidson Addgene plasmid # 55073) plasmids for 24 h, after which the medium was replaced with Live Cell Imaging Solution supplemented with 20 mM HEPES and the images were acquired as above.
RNA isolation, reverse transcription and real-time PCR
Cells were lysed after 72 hrs of ADCY1 siRNA silencing and RNA was isolated using Nucleospin™ Total RNA Isolation Kit (Macherey - Nagel) according to kit protocol. Complementary DNA (cDNA) was synthesized from 1 μg RNA using TaqMan™ Reverse transcription Kit (Roche Diagnostics) following manufacturer’s protocol. Reverse transcription was performed on a Gradient thermocycler (Biometra). Quantitative Realtime PCR (qPCR) analysis was performed with SYBR Green QPCR Master Mix (Agilent Technologies) on a 7500 Fast Real-Time PCR System (Applied Biosystems), using 0.4 μM primers (ADCY1 forward primer: 5’-TGG TCA CCT TCG TGT CCT AT -3’, reverse primer: 5’-ACC CCA TAC ATG TTC ACA CC -3’) according to manufacturer’s protocol. The ΔCt method was used to calculate mRNA levels. Expression levels of ADCY1 were normalized to expression levels of the housekeeping gene β-actin.
Immunoblotting
Proteins separated by gradient SDS-PAGE (4–15%) and transferred to a nitrocellulose membrane following standard protocols were detected using primary antibodies and appropriate Horseradish peroxidase-conjugated secondary antibodies listed in Table S1. The blots were developed using Clarity™ Western ECL (BioRad), and images were acquired by the luminescent image analyzer LAS-4000 mini (Fujifilm, GE Healthcare).
Statistical analysis
Data are presented as mean + SD from at least three independent experiments, unless indicated otherwise. Statistical analysis was performed using two-tailed, homoscedastic Student’s t tests in order to evaluate null hypotheses.
Results
Characterization of CAD-resistant MCF7 cells
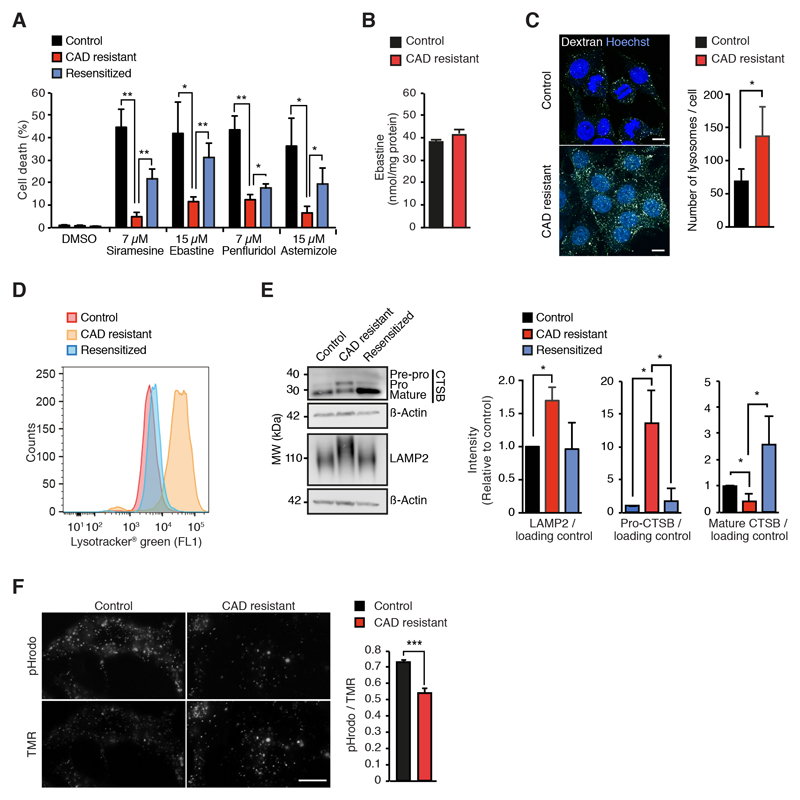
In order to test whether cancer cells can acquire resistance to CAD-induced cytotoxicity, we cultured MCF7 human mammary adenocarcinoma cells with increasing concentrations of siramesine, an experimental antidepressant that has served as a prototype CAD in our previous studies [12, 14, 30]. After six months, the resistant cells were able to grow in the presence of 8 μM siramesine, but could not adjust to higher siramesine concentrations. In addition to siramesine, these cells showed reduced sensitivity towards other tested CADs with cancer specific cytotoxicity, e.g. antihistamines ebastine and astemizole and antipsychotic penfluridol (Fig. 1A). Notably, the resistant phenotype was partially reversed when cells were cultured in the absence of CADs for three months (Fig. 1A). Using liquid chromatography and mass spectrometry (LC/MS)-based quantification of cellular ebastine, we confirmed that the resistance was not associated with reduced CAD uptake (Fig. 1B). Compared to the control and resensitized cells, CAD resistant cells had, however, clearly enlarged acidic compartment associated with an increase in the number of lysosomes and higher levels of lysosome-associated membrane glycoprotein LAMP2 (Figs. 1C, 1D and 1E). In line with the significantly increased lysosomal pH in CAD resistant cells (Fig. 1F), their pH-dependent maturation of cathepsin B was also significantly reduced (Fig. 1E). The enlargement of the lysosomal compartment did not appear to be due to the transcriptional up-regulation of lysosomal genes as mRNA levels of LAMP1 and LAMP2 as well as those of all subunits of the vacuolar H+-ATPase (V-ATPase), most lysosomal hydrolases and key transcriptional regulators of lysosomal biogenesis (i.e. transcription factors E3 (TFE3) and EB (TFEB)) were not significantly upregulated in CAD resistant cells as analyzed by whole transcriptome sequencing (Figs. S1A and S1B; Gene Expression Omnibus (GEO) data set, accession # GSE130363). The mRNA expression of TFEB was, in fact, significantly down-regulated in the resistant cells (Fig. S1B). The other differentially expressed lysosome-associated genes (≥2-fold change, p < 0.05) encoded for ATP binding cassette subfamily B member 6 (ABCB6), Ca2+-activated potassium channel subunit α1 (KCNMA1), H+/Cl- exchange transporters 4 (CLCN4), hematopoietic progenitor cell antigen CD34 (CD34) and antigen-presenting glycoprotein (CD1D) that were down-regulated, and cathepsin O (CTSO), H+/Cl- exchange transporter 5 (CLCN5) and HLA class II histocompatibility antigen γ-chain (CD74) that were up-regulated in CAD-resistant cells (Fig. S1B). Taken together, these data demonstrate that cancer cells can acquire partial and reversible resistance to CADs upon long-term exposure and that the acquired resistance is not associated with reduced uptake of the drugs or major changes in the expression of lysosomal genes. On the other hand, the resistant phenotype could be partially explained by the dilution of the lysosomal CAD concentration to the increased volume of the less acidic lysosomes and their higher expression of LAMP2 protein, which is known to protect lysosomes against CAD-induced membrane permeabilizion [11]. The mechanism by which the CAD resistant cells have increased their lysosomal volume remains to studied, whereas the increase in LAMP2 could be caused by its longer half-life in less acidic lysosomes with lower levels of active cathepsin B as demonstrated previously [11].
Figure 1. CAD resistance is associated with altered lysosomal compartment.
(A) CAD-induced cell death of control, CAD resistant and re-sensitized MCF7 cells treated for 48 h with DMSO, 7 μM siramesine, 15 μM ebastine, 7 μM penfluridol or 15 μM astemizole determined by propidium iodine and Hoechst-33342 staining employing Celígo® Imaging Cytometer.
(B) Mean ebastine concentrations in lipid extracts of control and CAD resistant MCF7 cells treated with 15 μM ebastine for 1 h were measured by LC/MS.
(C) Representative images (left) and quantification (right) of AF594-dextran-positive lysosomes in control and CAD resistant MCF7 cells loaded with AF594-dextran for 16 h and chased for 4 h in the absence of AF594-dextran. Cells were counter-stained with Hoechst 33342. Scale bar, 10 μm.
(D) Representative flow cytometer profiles of control, CAD resistant and resensitized MCF7 cells stained with Lysotracker® Green.
(E) Representative immunoblots (left) and quantification of indicated protein ratios in lysates of control, CAD resistant and re-sensitized MCF7 cells (right).
(F) Representative images (left) and mean intensities (right) of a pH-sensitive (pHrodo; fluorescence induced by low pH) and pH-insensitive lysosomal probes in control and CAD resistant cells. Scale bar, 10 μm.
Error bars, SD of ≥ 3 independent experiments. * P < 0.05, ** P < 0.01, *** P < 0.001 as analyzed by 2-tailed, homoscedastic student’s t-test.
In order to identify targetable pathways associated with CAD resistance, we analyzed the transcriptomes of control and CAD resistant MCF7 cells in further detail. Gene ontology analysis revealed that cellular functions involving G-protein coupled receptor (GPCR) activity, Ca2+ binding, and ion homeostasis were enriched among the differently expressed genes (Fig. S1C). Motivated by the abundance of drugs regulating GPCR pathways, and the fact that most CADs have been originally designed as GPCR agonists or antagonists, we focused on these pathways. Among the differentially expressed GPCR-associated genes were i) GPCRs, i.e. GPR3, C-X-C motif chemokine receptor type 4 (CXCR4) and calcitonin receptor (CALCR), ii) GPCR ligand adrenomedullin 2 (ADM2) that can activate CALCR [31], iii) cAMP modifying enzymes, i.e. adenylyl cyclase 1 (ADCY1) and phosphodiesterase 7B (PDE7B)), iv) regulators of cAMP-dependent protein kinase, i.e. type I-β and type II-α regulatory subunits of cAMP-dependent protein kinase (PRKAR1B and PRKAR2A) and cAMP-dependent protein kinase inhibitor α (PKIA), iv) cAMP-regulated ion channels of hyperpolarization-activated cyclic nucleotide-gated K+ and Na+ channel family (HCN2, HCN3 and HCN4), and v) Ca2+-responsive proteins such as calmodulin (CALM3), Ca2+/calmodulin dependent protein kinase I (CAMK1), calmodulin-regulated spectrin-associated protein 1 (CAMSAP1) and KCNMA1 (Fig. S1D). In light of the reported role of arrestins in GPCR recycling and activation of mitogen-activated protein kinases (MAPKs) [32, 33], it is also worth noting the significant changes in arrestin C (ARR3) and MAPK13 mRNA levels (Fig. S1D). And finally, Ca2+-activated proteins being highly enriched among the genes with altered expression, it was also interesting to note that P2X purinergic receptor 4 (P2RX4), which has been recently identified as an important regulator of lysosomal Ca2+ stores and lysosomal fusion events [34, 35], was significantly down-regulated in CAD resistant cells (Fig. S1D). These data strongly suggest that GPCR-activated cAMP and Ca2+ signaling pathways modulate cellular response to CADs.
CADs trigger a rapid increase in intracellular Ca2+ and cAMP levels in sensitive target cells
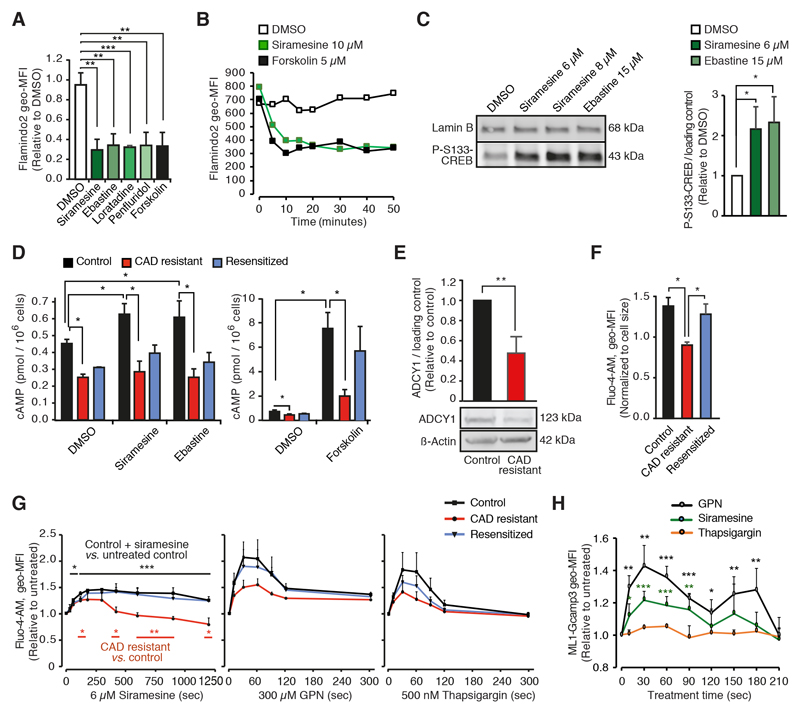
Prompted by the gene ontology data presented above, we analyzed whether CADs increased intracellular cAMP ([cAMP]i) or cytosolic free Ca2+ ([Ca2+]c) levels. To monitor the [cAMP]i, we transfected MCF7 cells with Flamindo2, a yellow fluorescent cAMP indicator whose fluorescence intensity is reduced by cAMP binding by approximately 75% [28]. Forskolin, a potent activator of adenylyl cyclase, reduced the Flamindo2 fluorescence intensity of the MCF7-Flamindo2 cells by approximately 70% (Figs. 2A, 2B and S2A). At concentrations that kill approximately 50% of cells (LC50), all tested CADs (siramesine, ebastine, loratadine and penfluridol) induced a similar rapid decrease in Flamindo2 fluorescence intensity as forskolin (Figs. 2A, 2B and S2A). The siramesine-induced reduction was concentration-dependent and detectable even at sub-lethal concentrations (0.75 - 3 μM; Fig. S2B). CAD-induced cAMP response was accompanied by increased phosphorylation of Ser-133 (S133) in cAMP-responsive element-binding protein 1 (CREB) suggestive of the activation of cAMP-dependent protein kinase (also known as protein kinase A; Fig. 2C). The ability of CADs to induce maximal reduction in Flamindo2 fluorescence intensity suggested that relatively low [cAMP]i could saturate the cAMP binding capacity of Flamindo2. Thus, we used HitHunter® competitive immunoassay to determine the accurate [cAMP]i. CADs at LC50 induced approximately 40% increase in [cAMP]I, whereas forskolin-induced increase was almost 20-fold (Fig. 2D and S2C). Notably, CAD resistant MCF7 cells with significantly reduced expression of the Ca2+/calmodulin-dependent ADCY1 mRNA and protein had significantly lower basal [cAMP]i than parental cells, and they failed to accumulate cAMP in response to either CADs or forskolin (Figs. 2D, 2E and S1D).
Figure 2. CAD resistance is associated with reduced intracellular cAMP and Ca2+ levels.
(A) Geometric mean fluorescence intensities (geo-MFIs) in MCF7-Flaminodo2 cells treated with DMSO, 6 μM siramesine, 15 μM ebastine, 40 μM loratadine, 8 μM penfluridol or 5 μM forskolin for 20 min in the absence of extracellular Ca2+ were analyzed by flow cytometry. The values are presented relative to DMSO-treated cells. Representative flow cytometer profiles are shown in Fig. S2A.
(B) Kinetics of geo-MFIs in MCF7-Flamindo2 cells treated as indicted in the absence of extracellular Ca2+ were analyzed as in (A). Data from one representative experiment is shown (N = 3).
(C) Representative immunoblots of P-S133-CREB and lamin B (loading control) in lysates of MCF7 cells treated as indicated for 2 h (left) and quantification of band intensities (P-S133-CREB/loading control; right).
(D) cAMP levels in control, CAD resistant and re-sensitized MCF7 cells treated for 20 min with DMSO, 10 μM siramesine or ebastine, 8 μM penfluridol or 2 μM forskolin in the absence of extracellular Ca2+ were analyzed by HitHunter® immunoassay. Standard curve for cAMP luminescence is shown in Fig. S2C.
(E) Representative immunoblots of ADCY1 and ß-actin (loading control) in lysates of control and CAD resistant MCF7 cells (top) and quantification of band intensities (ADCY1/loading control; bottom).
(F and G) Fluo-4-AM geo-MFIs in indicated cells left untreated (F) or treated with 6 μM siramesine (G, left), 300 μM GPN (G, middle) or 500 nM thapsigargin (G, right) in the absence of extracellular Ca2+ for indicated times were analyzed by flow cytometry.
(H) ML1-Gcamp3 geo-MFIs in MCF7 cells transfected with ML1-Gcamp3 for 72 h prior to the treatments with 6 μM siramesine, 300 μM GPN or 50 nM thapsigargin in the absence of extracellular Ca2+. The values are normalized to cell size (F) or relative to untreated cells (G). The ability of 50 nM thapisgargin to induce Ca2+ release and lysosomal localization of ML1-Gcamp3 are shown in Figs. S2G and S2H, respectively.
Error bars, SD of ≥ 3 independent experiments. * P < 0.05, ** P < 0.01, *** P < 0.001 as analyzed by 2-tailed, homoscedastic student’s t-test.
To investigate whether CAD resistance or treatment was associated with changes in Ca2+ signaling, we loaded the cells with green-fluorescent Ca2+ indicator, Fluo-4-AM, whose fluorescence intensity reflects the free [Ca2+]c [36]. CAD resistant cells had significantly lower basal levels of the free [Ca2+]c than control or resensitized cells (Figs. 2F), and siramesine treatment triggered a rapid and long-lasting increase in the free [Ca2+]c in control and resensitized cells, while the response was reduced and significantly shorter in CAD resistant cells (Fig. 2F). CAD resistant cells responded with lower Ca2+ peaks than control cells also to glycyl-L-phenylalanine-beta-naphthylamide (GPN) and thapsigargin (Fig. 2F), which increase the free [Ca2+]c by permeabilizing the lysosomes and by inhibiting the Ca2+ uptake by endoplasmatic reticulum (ER), respectively [37, 38]. The CAD-induced increase in the free [Ca2+]c could result from the entry of the extracellular Ca2+, leakage of Ca2+ from the intracellular stores, e.g. lysosomes or ER, or reduced capacity of cells to pump Ca2+ out of the cytosol. Addition of extracellular Ca2+ to the assay increased the free [Ca2+]c, but had only a minimal accelerating effect on siramesine-induced Ca2+ peak (Fig. S2D), whereas both GPN and thapsigargin effectively inhibited the Ca2+-mobilizing effect of siramesine (Fig. S2E). As expected due to the tight coupling of lysosomal and ER Ca2+ pools [39], GPN treatment also reduced ER Ca2+ stores, and vice versa, thapsigargin reduced lysosomal Ca2+ stores (Fig. S2F). Therefore, it was not possible to determine whether the CAD-mobilized Ca2+ originated from one or both of the studied Ca2+ stores. To test the role of lysosomes in this process more directly, we took advantage of a genetically encoded Ca2+ indicator Gcamp3, which consists of green fluorescent protein (GFP), calmodulin, and a peptide sequence from myosin light chain kinase fused to the lysosomal Ca2+ channel mucolipin 1 (ML1) [40, 41]. Transiently expressed ML1-Gcamp3 co-localized with Lysotracker® Red-labelled lysosomes and reacted to GPN-induced lysosomal Ca2+ release but not to thapsigargin-induced Ca2+ release from the ER (Figs. 2H, S2G and S2H). This lysosome-specific Ca2+ indicator revealed a rapid and significant increase in the lysosome-proximal free [Ca2+]c after addition of CADs to MCF7 cells and HeLa cervix carcinoma cells (Figs. 2H and S2I). Notably the siramesine-induced lysosome-proximal free [Ca2+]c signal detected by ML1-GCamp3 in MCF7 cells peaked already around 30 seconds and disappeared by 2 minutes, whereas the global cytoplasmic signal detected by Fluo-4-AM peaked around 4 minutes and lasted over 20 min (Figs. 2F and 3H).
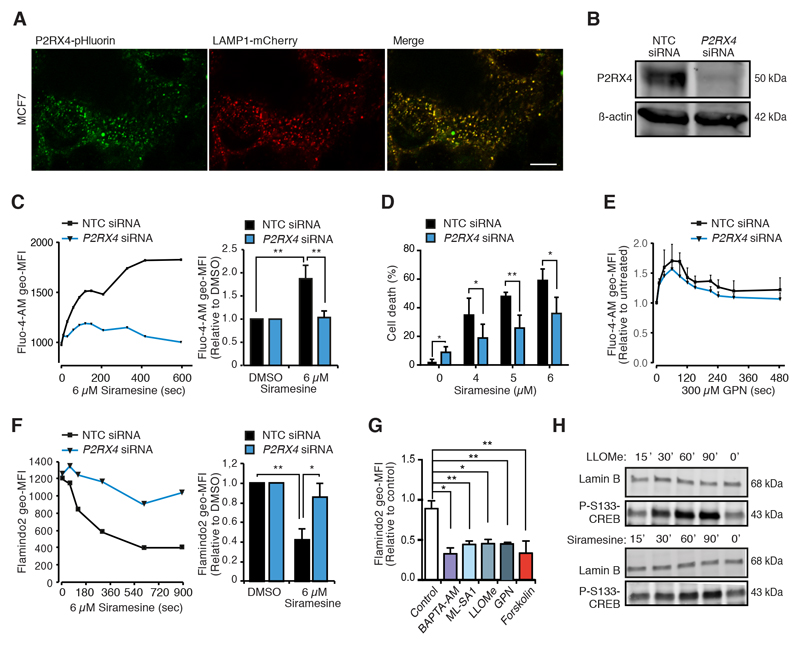
Figure 3. P2RX4 is required for CAD-induced Ca2+ and cAMP responses and cell death.
(A) Representative confocal images of MCF7 cells transfected with P2RX4-pHluorin [29] and LAMP1-mCherry for 72 h. Scale bar, 10 μm.
(B) Representative immunoblots of P2RX4 and β-actin (loading control) in lysates of MCF7 cells treated with non-targeting control (NTC) or P2RX4 siRNAs for 72 h.
(C) Fluo-4-AM geometric mean fluorescence intensities (geo-MFIs) in MCF7 cells treated with indicated siRNAs for 72 h and with 6 μM siramesine in the absence of extracellular Ca2+ for the last 10 min. The values are means of actual geo-MFIs (left) or geo-MFIs relative to untreated samples (right).
(D) Cell death of MCF7 cells treated with indicated siRNAs for 72 h and with indicated concentrations of siramesine for the last 48 h.
(E) Fluo-4-AM geo-MFIs in MCF7 cells treated with indicated siRNAs for 72 h and with 300 μM GPN in the absence of extracellular Ca2+ for the last 8 min. The values are means of geo-MFIs relative to untreated samples.
(F) Flamindo2 geo-MFIs in MCF7-Flamindo2 cells treated with indicated siRNAs for 72 h and with 6 μM siramesine in the absence of extracellular Ca2+ for the last 15 min. The values are presented as means of actual geo-MFIs (left) or geo-MFIs relative to untreated samples (right).
(G) Normalized geometric MFIs in MCF7-Flamindo2 cells treated with DMSO (control), 10 μM BAPTA-AM, 20 μM ML-SA1, 2 mM LLOMe, 200 μM GPN or 5 μM forskolin in the absence of extracellular Ca2+ for 20 min.
(H) Representative immunoblots of P-S133-CREB and lamin B (loading control) in lysates of MCF7 cells treated with 2 mM LLOMe or 6 μM siramesine for indicated times.
Error bars, SD of ≥ 3 independent experiments. * P < 0.05, ** P < 0.01 as analyzed by 2-tailed, homoscedastic student’s t-test.
Taken together, these data indicate that CADs trigger a rapid and transient increase in the lysosome-proximal free [Ca2+]c followed by a long-lasting increase in cytosolic free [Ca2+]c and an increase in the [cAMP]i. The reduced capacity of CAD resistant cells to mobilize Ca2+ from the intracellular stores and to accumulate cAMP suggests that these events contribute to CAD-induced cytotoxcity.
P2RX4 mediates CAD-induced lysosomal Ca2+ release
In our search for the mechanism of CAD-induced lysosomal Ca2+ release, we focused on the P2RX4 purinenergic receptor, whose expression was significantly reduced in CAD resistant MCF7 cells (Fig. S1D). This channel appeared as a good candidate because of its reported abundance in lysosomal membranes, where it forms functional, ATP-activated cation channels with high permeability for Ca2+ upon neutralization of lysosomal pH [34, 35, 42], and its predominantly lysosomal localization in MCF7 cells (Fig. 3A). Indeed, siRNA-mediated depletion of P2RX4 protein inhibited the siramesine-induced increase in the free [Ca2+]c and reduced the subsequent cell death significantly (Figs. 3B-D). Notably, P2RX4 depletion did not have a significant effect on the increase in the free [Ca2+]c in response to GPN-mediated permeabilization of lysosomal membranes (Fig. 3E), indicating that it did not alter lysosomal Ca2+ stores. The depletion of P2RX4 in MCF7-Flamindo2 cells abolished the ability of siramesine to reduce Flamindo2 fluorescence (Fig. 3F), suggesting that a Ca2+-dependent adenylyl cyclase is activated in response to the siramesine-induced lysosomal Ca2+ release. Confirming the ability of increased Ca2+ permeability of the lysosomal membrane to serve as a trigger for the cAMP accumulation, lysosomal detergents, L-leucyl-L-leucine methyl ester (LLOMe) and GPN, as well as ML-SA1, a synthetic agonist of lysosomal Ca2+ channel ML1, whose activation, contrary to that of P2RX4, requires acidic lysosomal pH [43], reduced the Flamindo2 fluorescence intensity and induced the phosphorylation of S133-CREB (Figs. 3G, 3H and S3A). It was unfortunately not possible to further validate the necessity of cytosolic Ca2+ in CAD-induced cAMP accumulation by chelating it with BAPTA-AM because BAPTA-AM itself increased the [cAMP]i and S133-CREB phosphorylation (Figs. 3G and S3A). Importantly, P2RX4 depletion affected neither the uptake of CADs nor their localization to lysosomes as analyzed by flow cytometry and confocal microscopy of MCF7 cells treated with sunitinib and SU11652 (Figs. S3B and S3C), fluorescent CADs that induce similar lysosome-dependent cancer cell death as the CADs studied here [44]. Furthermore, mass spectrometry-based analyses revealed similar accumulation of ebastine in P2RX4-depleted and control cells (Fig. S3D). Taken together, these data suggest that CADs, which are known to increase lysosomal pH [12], activate pH sensitive P2RX4 channels to release Ca2+ from lysosomes to the cytosol, and that this Ca2+ release is required for the subsequent cAMP accumulation and cell death.
ADCY1 mediates CAD-induced cAMP response and cell death
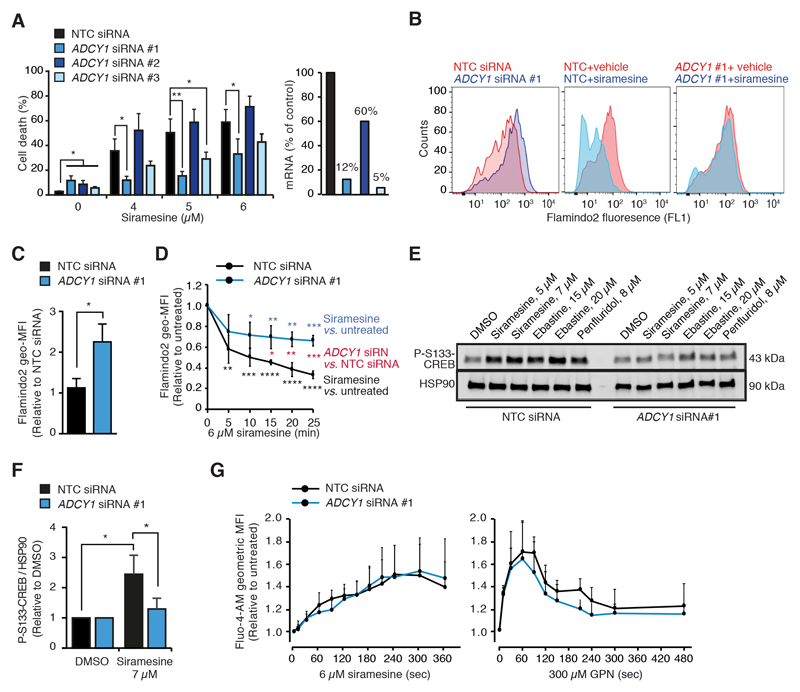
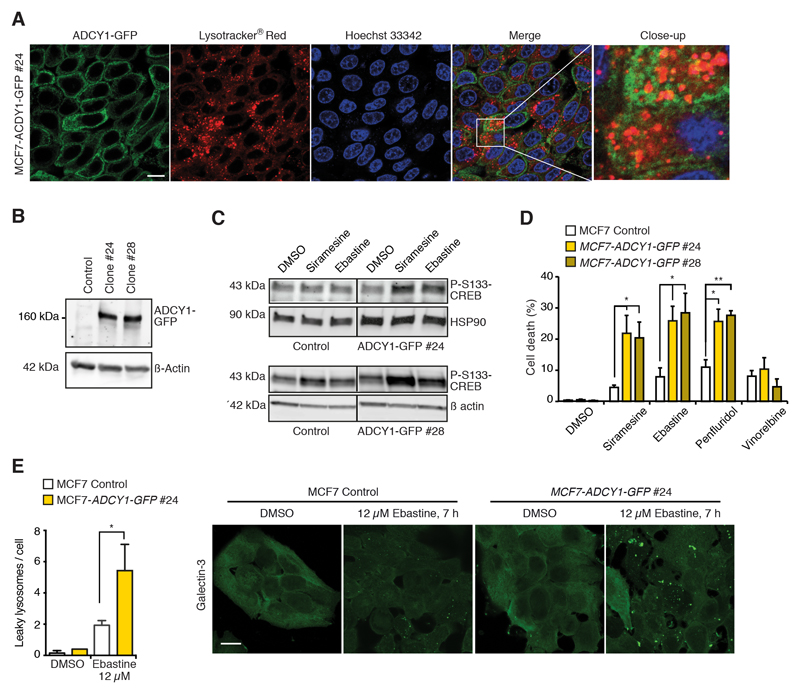
The CAD-induced increase in the [cAMP]i being dependent on Ca2+, we next investigated the role of the Ca2+-activated ADCY1, whose mRNA and protein expression was reduced in CAD resistant cells (Figs. 2E and S1D), in CAD-induced cAMP response and cell death. For this purpose, we treated MCF7 and A549 human non-small-cell lung carcinoma cells with three independent ADCY1 siRNAs, two of which (#1 and #3) effectively reduced ADCY1 mRNA and protein levels (Figs. 4A and S4A). The two effective siRNAs significantly inhibited siramesine-induced cell death in both cell lines (Figs. 4A and S4A). In A549 cells, ADCY1 depletion also reduced astemizole-induced cell death and inhibited the ability of astemizole to sensitize cells to vinorelbine (S4A). The depletion of ADCY1 in MCF7-Flamindo2 cells increased the Flamindo2 fluorescence intensity more than 2-fold implying that ADCY1 is constitutively active in MCF7 cells (Figs. 4B and 4C). It also reduced the CAD-induced cAMP response as indicated by a significantly smaller CAD-induced reduction in Flamindo2 fluorescence and reduced phosphorylation of Ser133-CREB (Figs. 4B-F). ADCY1 depletion affected neither the ability of siramesine or GPN to mobilize Ca2+ from the intracellular stores (Figs. 4G), nor the uptake and lysosomal localization of CADs in MCF7 cells (Figs. S3B-D). Akin to MCF7 cells, the siramesine-induced Ca2+ release was also unaffected by ADCY1 depletion in A549 cells (Fig. S4B). To validate the role of ADCY1 in CAD-induced cell death, we created two MCF7 clones stably expressing ADCY1-GFP fusion protein, majority of which localized to the plasma membrane or structures in the close proximity of the plasma membrane with only minimal co-localization with lysosomes (Figs. 5A and 5B). The functionality of the fusion protein was confirmed by its ability to increase the phosphorylation of S133-CREB induced by low concentrations of CADs (Fig. 5C). Both ADCY1-GFP expressing clones were significantly sensitized to CADs, but not to vinorelbine, an unrelated microtubule destabilizing cancer drug (Fig. 5D). The ADCY1-GFP-associated increase in CAD-induced cell death was preceded by a significant increase in lysosomal membrane permeabilization analyzed by lysosomal galectin-3 puncta assay [45] (Figs. 5E and S4C). Thus, ADCY1 mediates CAD-induced signaling pathways leading to lysosome-dependent cell death without affecting cellular uptake of CADs or CAD-induced increase in the [Ca2+]c.
Figure 4. ADCY1 depletion inhibits CAD-induced cAMP response and cell death.
(A) Cell death of MCF7 cells treated with non-targeting control (NTC) and 3 independent ACDY1 siRNAs for 72 h and with indicated concentrations of siramesine for the last 48 h (left). The efficacy of siRNAs was analyzed by qPCR 48 h after the transfection (right).
(B) Representative flow cytometer profiles for Flaminod2 fluorescence in MCF7-Flamindo2 cells treated with NTC or ADCY1 #1 siRNAs for 48 h. When indicated, cells were treated with vehicle (DMSO) or 6 μM siramesine in the absence of extracellular Ca2+ for the last 20 min.
(C) Relative Flamindo2 geometric mean fluorescence intensities (geo-MFIs) in MCF7-Flamindo2 cells treated with NTC or ADCY1 #1 siRNAs for 72 h.
(D) Kinetics of relative Flamindo2 geo-MFIs in MCF7-Flamindo2 cells transfected as in (C) and treated with 6 μM siramesine in the absence of extracellular Ca2+ for the last 25 min.
(E and F) Representative immunoblots of P-S133-CREB and HSP90 (loading control) in MCF7 cells transfected with siRNAs as in (C) and treated with indicated concentrations of CADs for the last 2 h (E), and mean P-S133-CREB/loading control ratios in indicated samples from 3 independent experiments (F)
(G) Levels of free [Ca2+]c in MCF7 cells treated with NTC or ADCY1 #1 siRNAs for 72 h, and with 6 μM siramesine (left) or with 300 μM GPN (right) in the absence of extracellular Ca2+ for indicated times were analyzed by flow cytometry using Fluo-4-AM probe as a Ca2+ sensor.
Error bars, SD of ≥ 3 independent experiments. * P < 0.05, ** P < 0.01, *** P < 0.001, **** P < 0.0001 as analyzed by 2-tailed, homoscedastic student’s t-test.
Figure 5. Ectopic ADCY1 enhances CAD-induced cAMP response and cell death.
(A) Representative images of MCF7-ADCY-GFP #24 cells stained with Lysotracker® Red and Hoechst 33342. Scale bar, 10 μm.
(B) Representative immunoblots of ADCY1-GFP, ADCY1 and β actin (loading control) in control MCF7 cells and indicated stable ADCY1-GFP expressing clones.
(C) Representative immunoblots of P-S133-CREB and HSP90 or β actin (loading controls) in indicated MCF7 clones treated with DMSO, 4 μM siramesine (top), 6 μM siramesine (bottom) or 10 μM ebastine (top and bottom). Vertical lines indicate a cut in the blot, but both sides originate from the same blot.
(D) Death of indicated MCF7 clones treated with DMSO, 4 μM siramesine, 12 μM ebastine, 6 μM penfluridol or 30 nM vinorelbine for 48 h.
(E) Quantification (left) and representative images (right) of lysosomal galectin-3 puncta in indicated MCF7 clones treated with DMSO or 12 μM ebastine for 7 h and stained for galectin-3 and LAMP2. See Fig. S4C for LAMP2 and merged LAMP2 and galectin-3 stainings. Scale bar, 10 μm.
Error bars, SD of > 3 independent experiments. * P < 0.05, ** P < 0.01 as analyzed by 2-tailed, homoscedastic student’s t-test.
cAMP sensitizes cancer cells to CAD-induced cytotoxicity
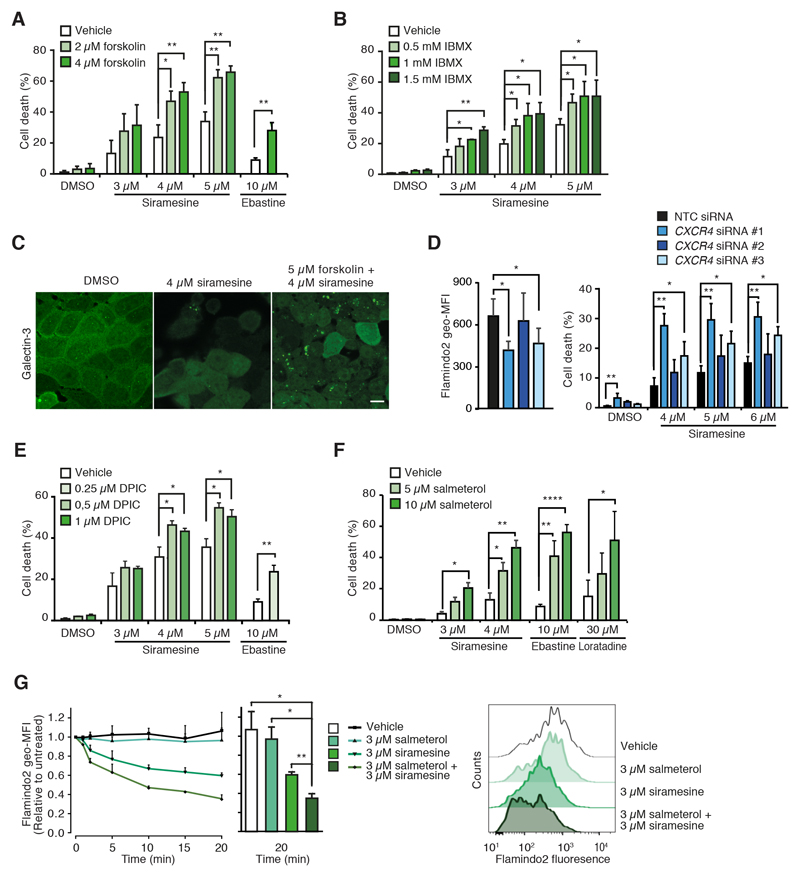
Prompted by the role of ADCY1 in CAD cytotoxicity, we next tested whether known cAMP-inducing drugs would sensitize cells to CADs. Indeed, both activation of cAMP synthesis by forskolin and inhibition of its hydrolysis by phosphodiesterase inhibitor, 3-isobutyl-1-methylxanthine (IBMX) as well as treatment with synthetic cAMP analogues sensitized MCF7 cells to CADs (Figs. 6A, 6B and S5A). In addition to cell death, forskolin increased siramesine-induced lysosomal membrane permabilization as evidenced by the enhanced formation of lysosomal galectin-3 puncta (Figs. 6C and S5B).
Figure 6. cAMP inducers sensitize cancer cells to CAD-induced cytotoxicity.
(A) Cell death of MCF7 cells treated with indicated concentrations of siramesine or ebastine and forskolin for 48 h.
(B) Cell death of MCF7 cells treated with indicated concentrations of siramesine and IBMX for 48 h.
(C) Representative confocal images of MCF7 cells treated as indicated for 16 h and stained for galectin-3 and LAMP2. See Fig. S5B for LAMP2 and merged LAMP2 and galectin-3 stainings. Scale bar, 10 μm.
(D) Flamindo2 geometric mean fluorescence intensities (geo-MFIs; left) and cell death (right) of MCF7-Flamindo2 cells treated with non-targeting control (NTC) or CXCR4 siRNAs for 72 h. For cell death measurements, cells were treated with DMSO or indicated concentrations of siramesine for the last 48 h.
(E and F) Cell death of MCF7 cells treated with indicated combinations of indicated CADs and DPIC (E) or salmeterol (F) for 48 h.
(G) Kinetics of Flamindo2 geo-MFIs in MCF7-Flamindo2 cells treated as indicated for 0-20 min in the absence of extracellular Ca2+ (left) and representative flow cytometry profiles after 20 min treatment (right).
Error bars, SD of ≥ 3 independent experiments. * P < 0.05, ** P < 0.01, **** P < 0.0001 as analyzed by 2-tailed, homoscedastic student’s t-test.
Based on the ability of cAMP to enhance CAD-induced lysosome-dependent cell death, we speculated that the down-regulation of the guanine nucleotide-binding protein G(s)-associated, cAMP-inducing GPCRs (GPR3 [46] and CALCR [47]), and the up-regulation of the guanine nucleotide-binding protein G(i)-associated, cAMP-inhibiting CXCR4 [48] observed in CAD resistant cells (Fig. S1D), contributed to the resistant phenotype. Indeed, two CXCR4 siRNAs that increased [cAMP]i also enhanced siramesine-induced cell death (Fig. 6D), whereas CALCR siRNA-treated cells had a tendency to be more resistant to siramesine than control cells (Fig. S5C). Similarly, effective siRNA-mediated depletion of GNAI2, the major guanine nucleotide-binding protein G(i) subunit in MCF7 cells, increased both cellular cAMP levels and siramesine-induced cell death significantly (Fig. S5D). To study the role of GPR3, we took advantage of a GPR3 agonist, diphenyleneiodonium chloride (DPIC) [49], which sensitized MCF7 cells to CAD-induced cytotoxicity significantly (Figs. 6E). These data raise the possibility that clinically relevant drugs that induce cAMP accumulation, e.g. agonists of ß2 adrenergic receptors that are commonly used to treat asthma and chronic obstructive pulmonary disease, could also sensitize cancer cells to CADs. Supporting this hypothesis, salmeterol, a long-acting ß2 adrenergic receptor agonist, had a synergistic effect with CADs not only on cAMP accumulation but also on cytotoxicity in MCF7 and A549 cells (Figs. 6F, 6G and S5E). Short-acting salbutamol and another long-acting ß2 adrenergic receptor agonist, formoterol, also sensitized MCF7 cells to siramesine, albeit to a much lesser extent than salmeterol (Fig. S5F).
Discussion
Accumulating experimental and epidemiological evidence encourage the development of CADs, especially commonly used and safe antihistamines, as anti-cancer agents [12–19]. In spite of CAD-induced significant reduction in tumor growth observed in various murine cancer models, monotherapy with CADs is, however, not likely to be efficient enough to treat human cancer. In this study, we searched for signaling pathways and putative drug targets that could be exploited to enhance CAD-induced lysosome-dependent cell death. Our approach to identify pathways involved in this process by comparing transcriptomes of CAD sensitive and CAD resistant MCF7 breast cancer cells pointed to GPCR signaling as important regulators of cellular sensitivity to CADs. Motivated by these data and the abundance of existing drugs that regulate GPCR signaling [50, 51], we studied the involvement of the main signaling pathways activated by GPCRs in CAD-induced cell death in further detail.
GPCRs, also known as seven transmembrane receptors, constitute the largest, most widely expressed and most versatile family of cell surface receptors in human genome [32]. The over 800 members of the GPCR family activate a wide array of signaling cascades in response to hormones, cytokines, neurotransmitters, metabolites, drugs, and other stimuli, and regulate cellular functions controlling multiple aspects of cellular physiology and pathology, including tumorigenesis and tumor progression [50, 51]. In resting or native state, GPCRs are bound to a heterotrimeric G protein complex consisting of Gα, Gβ and Gy subunits. Upon activation, GPCRs coupled to different subtypes of Gα either enhance (Gαs) or inhibit (Gαi) adenylate cyclases, or activate phospholipase C (Gαq) or Rho GTPases (Gα12/13), whereas the GβY dimer regulates various phospholipases and ion channels. The rapid CAD-induced accumulation of cellular cAMP in sensitive target cells and the absence of this response in CAD resistant cells suggested that either the activation of Gαs- or inhibition of Gαi-coupled receptors were involved in CAD-induced cytotoxicity (Fig. S6). This was strongly supported by the significantly reduced ability of CADs to trigger the accumulation of cAMP and cell death after depletion of ADCY1, an adenylate cyclase that is activated by Gαs and inhibited by Gαi [52]. The role of cAMP in CAD-induced cytotoxicity was further supported by the death-promoting effect of synthetic cAMP analogues and all tested treatments that increase the [cAMP]i, i.e. activation of adenlate cyclses by forskolin, inhibition of cAMP phosphodiesterases by IBMX, co-treatment with agonists of Gαs-coupled GPR3 or ß2 adrenergic receptors and the depletion of Gαs-coupled CXCR4 as well as the protective effect of the depletion of Gαs-coupled CALCR. It remains, however, unclear which GPCRs are responsible for CAD-induced increase in the [cAMP]i. In this context it is important to note that CADs used in this study function as antagonists of several GPCRs, e.g. histamine 1, cholinergic and adrenergic receptors, at low nanomolar concentrations. Thus, the micromolar concentrations required to trigger the cAMP response and to kill cancer cells are likely to affect multiple GPCRs. Moreover, the altered CAD sensitivity observed upon manipulation of GPR3, CXCR4, CALCR or ß2 adrenergic receptors does not necessarily mean that these receptors are direct targets of CADs. Akin to forskolin and IBMX treatments, their manipulation is likely to sensitize cells to CADs due to their ability to increase the [cAMP]I independent of CADs.
cAMP is an evolutionarily conserved signaling molecule that serves as an indicator of stress from the bacteria to humans. An increase in the [cAMP]i is translated into a wide range of biological responses by cAMP-induced conformational changes in a set of proteins that contain a cyclic nucleotide-binding domain [53]. Such downstream effectors can be divided into three main signaling pathways that are activated by i) cAMP-dependent protein kinase, ii) exchange factors directly activated by cAMP 1 and 2 (EPAC1 and 2) that serve as guanine nucleotide exchange factors for small G-proteins RAP1 and RAP2 [54], and iii) cAMP-activated ion channels [55]. In accordance with the data presented here, a recent study demonstrated a synergistic anti-cancer effect for the combination of cationic amphiphilic tricyclic antidepressants and ticlopidine, an anticoagulant that elevates the [cAMP]I by inhibiting the Gαi-coupled purinergic receptor P2Y12, in a murine model of glioma [17]. The authors explained this sensitizing effect by cAMP- and EPAC1-mediated enhancement of autophagic flux. This explanation is, however, not likely to account for the sensitizing effect of cAMP in our study because autophagy has a slightly protective effect against siramesine-induced cytotoxicity in MCF7 cells [12]. It should also be noted that after the initial activation of autophagy, all effective CADs neutralize lysosomal pH and thereby act themselves as effective inhibitors of autophagic flux. Our preliminary studies regarding the downstream effectors of cAMP do not support the involvement of EPACs as mediators of cell death. Thus, we are presently investigating the role of cAMP-dependent protein kinase and cAMP-activated ion channels in this process.
Contrary to our study employing adenocarcinoma cells and that of Shchors and coworkers employing glioma cells, CADs have been reported to specifically kill cancer cells of neuroendocrine origin by reducing cellular cAMP levels [16]. Neuroendocrine cells being especially dependent on GPCR-induced cAMP for their survival, the authors suggested that the potent effect of CADs in the induction of cell death in neuroendocrine cancer cells was mediated by CAD-induced disruption of autocrine survival loops between neurotransmitters and their receptors. Thus, when planning therapies combining CADs with cAMP inducing drugs, it should be kept in mind that cAMP may have opposing effects on CAD sensitivity depending on the origin of the cancer cells and their dependence on GPCR signaling.
In addition to the putative role of multiple GPCRs, our data show that lysosomal Ca2+ release through P2RX4 channels contributes to the CAD-induced cAMP accumulation via the activation of Ca2+/calmodulin-dependent ADCY1. Supporting P2RX4 as an essential effector of this process, its depletion in MCF7 cells almost completely inhibited the early CAD-induced increase in the free [Ca2+]c, the subsequent accumulation of cAMP and cell death. In line with the recently demonstrated increase in the Ca2+ permeability of lysosomal P2RX4 upon neutralization of lysosomal lumen [34, 35], the rapid increase in lysosomal pH caused by the accumulation of basic CADs into lysosomes [12], is likely to serve as a stimulus for the opening of lysosomal P2RX4 channels.
Taken together, the data presented here reveal an early CAD-induced signaling pathway consisting of P2RX4-mediated lysosomal Ca2 release, Ca2-dependent activation of ADCY1 and an increase in [cAMP]i as an essential contributor to CAD-induced lysosome-dependent cancer cell death (Fig. S6). It remains to be studied how the activation of this pathway translates to lysosomal leakage. In light of the recently discovered role of P2RX4 channels in lysosomal membrane fusion [42], it is tempting to speculate that lysosomal fusion could promote lysosomal leakage either due to membrane destabilization during the fusion event itself or due to the increased lysosomal size, which reduces lysosomal membrane stability [56]. In spite of the remaining open questions regarding the mechanism by which the CAD-Ca2+-P2XR4-ADCY1 pathway promotes CAD-induced lysosome-dependent cell death, the data presented here open possibilities for the development of new and inexpensive cancer therapies by combining already approved and safe CADs with other well-characterized and safe cAMP-inducing drugs, such as ß2 adrenergic receptor agonists.
Supplementary Material
Acknowledgements
We thank Signe Diness Vindeløv for preparing the CAD resistant cells, Siv A. Hjorth and Thue W. Schwartz for sharing their expertise in cAMP measurements, Michael Davidson, Baljit Khakh, Tetsuya Kitaguchi, Christine Volbracht, Ekkehard Weber and Developmental Studies Hybridoma Bank (developed under auspices of the National Institute of Child Health and Human Resources and maintained by the University of Iowa) for valuable reagents, and Marianne Johansen, Tiina Naumanen Dietrich, Dianna Skousborg Larsen and Louise Vanderfox for technical assistance.
Financial Information
The research reported in this publication was supported by the European Research Council (AdG 340751), Danish Cancer Society (R90-A5783 and R167-A11061), Danish National Research Foundation (DNRF125), and Novo Nordisk Foundation (NNF15OC0016914) to M.J.
Footnotes
AUTHOR CONTRIBUTIONS
A.A. developed and optimized methodology, designed and performed the great majority of experiments and analyzed the data. B.L. acted as a daily co-supervisor of A.A. and performed the analyses of lysosomal pH. J.D.G. and K.M. performed lipid mass spectrometry analyses. M.R. acted as the daily supervisor for A.A and performed all bioinformatics analyses. M.J. designed the overall study, supervised the experiments, assisted in the data analyses and wrote the first draft of the manuscript. All authors contributed to the final text and approved it.
ACCESSION NUMBERS
The GEO accession number for RNA sequencing data is 130363.
Conflict of interest disclosure statement: The authors disclose no potential conflicts of interest.
References
- 1.Bar-Peled L, Sabatini DM. Regulation of mTORCl by amino acids. Trends Cell Biol. 2014 doi: 10.1016/j.tcb.2014.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Settembre C, et al. Signals from the lysosome: a control centre for cellular clearance and energy metabolism. Nat Rev Mol Cell Biol. 2013;14(5):283–96. doi: 10.1038/nrm3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Morgan AJ, et al. Molecular mechanisms of endolysosomal Ca2+ signalling in health and disease. Biochem J. 2011;439(3):349–74. doi: 10.1042/BJ20110949. [DOI] [PubMed] [Google Scholar]
- 4.Liu B, et al. STAT3 associates with vacuolar H(+)-ATPase and regulates cytosolic and lysosomal pH. Cell Res. 2018;28(10):996–1012. doi: 10.1038/s41422-018-0080-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nixon RA. The role of autophagy in neurodegenerative disease. Nat Med. 2013;19(8):983–97. doi: 10.1038/nm.3232. [DOI] [PubMed] [Google Scholar]
- 6.Colacurcio DJ, Nixon RA. Disorders of lysosomal acidification-The emerging role of v-ATPase in aging and neurodegenerative disease. Ageing Res Rev. 2016;32:75–88. doi: 10.1016/j.arr.2016.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Platt FM. Sphingolipid lysosomal storage disorders. Nature. 2014;510(7503):68–75. doi: 10.1038/nature13476. [DOI] [PubMed] [Google Scholar]
- 8.Kallunki T, Olsen OD, Jäättelä M. Cancer-associated lysosomal changes: friends or foes? Oncogene. 2013;32(16):1995–2004. doi: 10.1038/onc.2012.292. [DOI] [PubMed] [Google Scholar]
- 9.Olson OC, Joyce JA. Cysteine cathepsin proteases: regulators of cancer progression and therapeutic response. Nat Rev Cancer. 2015;15(12):712–29. doi: 10.1038/nrc4027. [DOI] [PubMed] [Google Scholar]
- 10.Perera RM, et al. Transcriptional control of autophagy-lysosome function drives pancreatic cancer metabolism. Nature. 2015;524(7565):361–5. doi: 10.1038/nature14587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fehrenbacher N, et al. Sensitization to the lysosomal cell death pathway by oncogene-induced down-regulation of lysosome-associated membrane proteins l and 2. Cancer Res. 2008;68(16):6623–33. doi: 10.1158/0008-5472.CAN-08-0463. [DOI] [PubMed] [Google Scholar]
- 12.Ostenfeld MS, et al. Anti-cancer agent siramesine is a lysosomotropic detergent that induces cytoprotective autophagosome accumulation. Autophagy. 2008;4(4):487–99. doi: 10.4161/auto.5774. [DOI] [PubMed] [Google Scholar]
- 13.Groth-Pedersen L, et al. Vincristine induces dramatic lysosomal changes and sensitizes cancer cells to lysosome destabilizing siramesine. Cancer Res. 2007;67:2217–2225. doi: 10.1158/0008-5472.CAN-06-3520. [DOI] [PubMed] [Google Scholar]
- 14.Petersen NH, et al. Transformation-associated changes in sphingolipid metabolism sensitize cells to lysosomal cell death induced by inhibitors of acid sphingomyelinase. Cancer Cell. 2013;24(3):379–93. doi: 10.1016/j.ccr.2013.08.003. [DOI] [PubMed] [Google Scholar]
- 15.Sukhai MA, et al. Lysosomal disruption preferentially targets acute myeloid leukemia cells and progenitors. J Clin Invest. 2013;123(1):315–28. doi: 10.1172/JCI64180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jahchan NS, et al. A drug repositioning approach identifies tricyclic antidepressants as inhibitors of small cell lung cancer and other neuroendocrine tumors. Cancer Discov. 2013;3(12):1364–77. doi: 10.1158/2159-8290.CD-13-0183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shchors K, Massaras A, Hanahan D. Dual Targeting of the Autophagic Regulatory Circuitry in Gliomas with Repurposed Drugs Elicits Cell-Lethal Autophagy and Therapeutic Benefit. Cancer Cell. 2015;28(4):456–71. doi: 10.1016/j.ccell.2015.08.012. [DOI] [PubMed] [Google Scholar]
- 18.Ellegaard AM, et al. Repurposing Cationic Amphiphilic Antihistamines for Cancer Treatment. EBioMedicine. 2016;9:130–9. doi: 10.1016/j.ebiom.2016.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rebecca VW, et al. A unified approach to targeting the lysosome’s degradative and growth signaling roles. Cancer Discov. 2017 doi: 10.1158/2159-8290.CD-17-0741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Walker AJ, et al. Tricyclic antidepressants and the incidence of certain cancers: a study using the GPRD. Br J Cancer. 2011;104(1):193–7. doi: 10.1038/sj.bjc.6605996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Verbaanderd C, et al. Repurposing Drugs in Oncology (ReDO)-chloroquine and hydroxychloroquine as anti-cancer agents. Ecancermedicalscience. 2017;11:781. doi: 10.3332/ecancer.2017.781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kornhuber J, et al. Functional Inhibitors of Acid Sphingomyelinase (FIASMAs): a novel pharmacological group of drugs with broad clinical applications. Cell Physiol Biochem. 2010;26(1):9–20. doi: 10.1159/000315101. [DOI] [PubMed] [Google Scholar]
- 23.Trapp S, et al. Quantitative modeling of selective lysosomal targeting for drug design. Eur Biophys J. 2008;37(8):1317–28. doi: 10.1007/s00249-008-0338-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kolzer M, Werth N, Sandhoff K. Interactions of acid sphingomyelinase and lipid bilayers in the presence of the tricyclic antidepressant desipramine. FEBS Lett. 2004;559(1-3):96–8. doi: 10.1016/S0014-5793(04)00033-X. [DOI] [PubMed] [Google Scholar]
- 25.Barcelo-Coblijn G, et al. Sphingomyelin and sphingomyelin synthase (SMS) in the malignant transformation of glioma cells and in 2-hydroxyoleic acid therapy. Proc Natl Acad Sci U S A. 2011;108(49):19569–74. doi: 10.1073/pnas.1115484108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Teres S, et al. 2-Hydroxyoleate, a nontoxic membrane binding anticancer drug, induces glioma cell differentiation and autophagy. Proc Natl Acad Sci U S A. 2012;109(22):8489–94. doi: 10.1073/pnas.1118349109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jäättelä M, et al. Bcl-x andBcl-2 inhibit TNF andFas-induced apoptosis and activation of phospholipase A2 in breast carcinoma cells. Oncogene. 1995;10:2297–2305. [PubMed] [Google Scholar]
- 28.Odaka H, et al. Genetically-encoded yellow fluorescent cAMP indicator with an expanded dynamic range for dual-color imaging. PLoS One. 2014;9(6):e100252. doi: 10.1371/journal.pone.0100252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu J, et al. Imaging P2X4 receptor subcellular distribution, trafficking, and regulation using P2X4-pHluorin. J Gen Physiol. 2014;144(1):81–104. doi: 10.1085/jgp.201411169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ostenfeld MS, et al. Effective tumor cell death by sigma-2 receptor ligand siramesine involves lysosomal leakage and oxidative stress. Cancer Res. 2005;65(19):8975–83. doi: 10.1158/0008-5472.CAN-05-0269. [DOI] [PubMed] [Google Scholar]
- 31.Hay DL, et al. Update on the pharmacology of calcitonin/CGRP family of peptides: IUPHAR Review 25. Br J Pharmacol. 2018;175(1):3–17. doi: 10.1111/bph.14075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hilger D, Masureel M, Kobilka BK. Structure and dynamics of GPCR signaling complexes. Nat Struct Mol Biol. 2018;25(1):4–12. doi: 10.1038/s41594-017-0011-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gurevich VV, Gurevich EV. Arrestins: Critical Players in Trafficking of Many GPCRs. Prog Mol Biol Transl Sci. 2015;132:1–14. doi: 10.1016/bs.pmbts.2015.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Qureshi OS, et al. Regulation of P2X4 receptors by lysosomal targeting, glycan protection andexocytosis. J Cell Sci. 2007;120(Pt 21):3838–49. doi: 10.1242/jcs.010348. [DOI] [PubMed] [Google Scholar]
- 35.Huang P, et al. P2X4 forms functional ATP-activated cation channels on lysosomal membranes regulated by luminal pH. J Biol Chem. 2014;289(25):17658–67. doi: 10.1074/jbc.M114.552158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gee KR, et al. Chemical and physiological characterization offluo-4 Ca(2+)-indicator dyes. Cell Calcium. 2000;27(2):97–106. doi: 10.1054/ceca.1999.0095. [DOI] [PubMed] [Google Scholar]
- 37.Rogers TB, et al. Use of thapsigargin to study Ca2+ homeostasis in cardiac cells. Biosci Rep. 1995;15(5):341–9. doi: 10.1007/BF01788366. [DOI] [PubMed] [Google Scholar]
- 38.Haller T, et al. The lysosomal compartment as intracellular calcium store in MDCK cells: a possible involvement in InsP3-mediated Ca2+ release. Cell Calcium. 1996;19(2):157–65. doi: 10.1016/s0143-4160(96)90084-6. [DOI] [PubMed] [Google Scholar]
- 39.Raffaello A, et al. Calcium at the Center of Cell Signaling: Interplay between Endoplasmic Reticulum, Mitochondria, and Lysosomes. Trends Biochem Sci. 2016;41(12):1035–1049. doi: 10.1016/j.tibs.2016.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tian L, et al. Imaging neural activity in worms, flies and mice with improved GCaMP calcium indicators. Nat Methods. 2009;6(12):875–81. doi: 10.1038/nmeth.1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shen D, et al. Lipid storage disorders block lysosomal trafficking by inhibiting a TRP channel and lysosomal calcium release. Nat Commun. 2012;3:731. doi: 10.1038/ncomms1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cao Q, et al. Calcium release through P2X4 activates calmodulin to promote endolysosomal membrane fusion. J Cell Biol. 2015;209(6):879–94. doi: 10.1083/jcb.201409071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cao Q, et al. The lysosomal Ca(2+) release channel TRPML1 regulates lysosome size by activating calmodulin. J Biol Chem. 2017;292(20):8424–8435. doi: 10.1074/jbc.M116.772160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ellegaard AM, et al. Sunitinib and SU11652 Inhibit Acid Sphingomyelinase, Destabilize Lysosomes, and Inhibit Multidrug Resistance. Mol Cancer Ther. 2013;12(10):2018–30. doi: 10.1158/1535-7163.MCT-13-0084. [DOI] [PubMed] [Google Scholar]
- 45.Aits S, et al. Sensitive detection of lysosomal membrane permeabilization by lysosomal galectinpuncta assay. Autophagy. 2015;11(8):1408–24. doi: 10.1080/15548627.2015.1063871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Uhlenbrock K, Gassenhuber H, Kostenis E. Sphingosine 1-phosphate is a ligand of the human gpr3, gpr6 and gpr12 family of constitutively active G protein-coupled receptors. Cell Signal. 2002;14(11):941–53. doi: 10.1016/s0898-6568(02)00041-4. [DOI] [PubMed] [Google Scholar]
- 47.Suzuki H, et al. Calcitonin-induced changes in the cytoskeleton are mediated by a signal pathway associated with protein kinase A in osteoclasts. Endocrinology. 1996;137(11):4685–90. doi: 10.1210/endo.137.11.8895334. [DOI] [PubMed] [Google Scholar]
- 48.Zheng J, et al. Intracellular CXCR4 signaling, neuronal apoptosis and neuropathogenic mechanisms of HIV-1-associateddementia. J Neuroimmunol. 1999;98(2):185–200. doi: 10.1016/s0165-5728(99)00049-1. [DOI] [PubMed] [Google Scholar]
- 49.Ye C, et al. Identification of a novel small-molecule agonist for human G protein-coupled receptor 3. J Pharmacol Exp Ther. 2014;349(3):437–43. doi: 10.1124/jpet.114.213082. [DOI] [PubMed] [Google Scholar]
- 50.Liu Y, et al. G protein-coupled receptors as promising cancer targets. Cancer Lett. 2016;376(2):226–39. doi: 10.1016/j.canlet.2016.03.031. [DOI] [PubMed] [Google Scholar]
- 51.Lappano R, Maggiolini M. G protein-coupled receptors: novel targets for drug discovery in cancer. Nat Rev Drug Discov. 2011;10(1):47–60. doi: 10.1038/nrd3320. [DOI] [PubMed] [Google Scholar]
- 52.Willoughby D, Cooper DM. Organization and Ca2+ regulation of adenylyl cyclases in cAMP microdomains. Physiol Rev. 2007;87(3):965–1010. doi: 10.1152/physrev.00049.2006. [DOI] [PubMed] [Google Scholar]
- 53.Berman HM, et al. The cAMP binding domain: an ancient signaling module. Proc Natl Acad Sci U S A. 2005;102(1):45–50. doi: 10.1073/pnas.0408579102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gloerich M, Bos JL. Epac: defining a new mechanism for cAMP action. Annu Rev Pharmacol Toxicol. 2010;50:355–75. doi: 10.1146/annurev.pharmtox.010909.105714. [DOI] [PubMed] [Google Scholar]
- 55.Biel M. Cyclic nucleotide-regulated cation channels. J Biol Chem. 2009;284(14):9017–21. doi: 10.1074/jbc.R800075200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ono K, Kim SO, Han J. Susceptibility of lysosomes to rupture is a determinant for plasma membrane disruption in tumor necrosis factor alpha-induced cell death. Mol Cell Biol. 2003;23(2):665–76. doi: 10.1128/MCB.23.2.665-676.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.