Abstract
The lysosomes have definitely polished their status inside the cell. Being discovered as the last resort of discarded cellular biomass, the steady rising of this versatile signaling organelle is currently ongoing. This review discusses the recent data on the unconventional functions of lysosomes, focusing mainly on the less studied lysosomes residing in the cellular periphery. We emphasize our discussion on the emerging paths the lysosomes have taken in promoting cancer progression to metastatic disease. Finally, we address how the altered cancerous lysosomes in metastatic cancers may be specifically targeted and what are the pending questions awaiting for elucidation.
Introduction
Originally described by Christian de Duve in 1950s as membrane-enclosed hydrolase-containing vesicles, lysosomes are known today as the major recycling stations of most eukaryotic cells [1]. Within lysosomes, at least 60 hydrolases are responsible for recycling the majority of cellular macromolecules, which are delivered to them by autophagy, endocytosis and phagocytosis [2,3]. The maturation to acidic lysosomes starts in early endosomes (EE), which change their lipid and protein composition during maturation to late endosomes (LE) and lysosomes that reside mainly in the perinuclear area of the cell. The vacuolar H+-adenosine triphosphatase (V-ATPase) at the limiting membrane gradually acidifies the maturing endosomes, ultimately resulting in a pH of 4.5-5 in lysosomes, an environment where most of the lysosomal hydrolases work best [2,4,5]. The homeostasis of the cells’ nutrient balance and timely degradation of worn-out organelles by lysosomal activities is essential for maintaining cellular integrity. Accordingly, the lysosomal biogenesis is tightly controlled by the metabolic status of the cell, and the lysosomal membrane serves as a platform for signaling molecules that regulate not only lysosome’s own function but also the metabolism of the entire cell [6,7]. When cells are fed and lysosomes produce high amounts of amino acids, a central regulator of cellular and organismal growth, mTORC1 kinase complex, is activated at the lysosomal membrane via a recently discovered mechanism involving lysosomal amino acid transporter SLC38A9, Rag GTPase and Ragulator [8–10] (for detailed review on mTORC1 see [11]. Activated mTORC1 then attenuates lysosomal biogenesis by keeping TFEB (transcription factor EB), the master regulator of lysosomal genes, inactive at the lysosomal membrane [12,13]. Vice versa, when nutrients are sparse and lysosomal amino acid flux reduced, mTORC1 is inhibited and TFEB released to enter the nucleus to activate the transcription of lysosomal genes [7]. Moreover, various stresses that trigger lysosomal calcium release through multiple channels, e.g. Mucolipin 1, can reverse the inhibitory effect of mTORC1 on TFEB and lysosomal biogenesis [6]. Thus, lysosomes are emerging as the major signaling hubs that gather information on cellular nutrient status and forward it to signaling networks that dictate cellular metabolism.
Misrouting the lysosomes from their normal intracellular paths may contribute to various pathologies, such as cancer progression and neuropathologies [5,14–16], and the peripheral positioning of lysosomes is emerging as a critical factor in determining their targeted functions [10,17,18]. Here we discuss the recent data supporting the idea that lysosomal subpopulations at the cancer cell edges have special functions in the regulation of cell adhesion dynamics [17,19], exocytosis, invasion [20–22], signaling and possibly cell death (Figure 1).
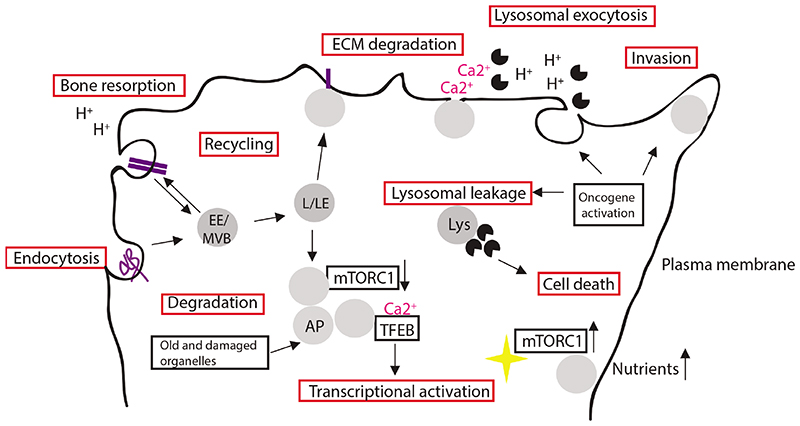
Figure 1.
Lysosomal functions in cancer cells. Endocytosis targets extracellular matrix (ECM) components, cell surface receptors and cell-cell adhesion components to lysosomal degradation and/or recycling back to plasma membrane (PM). The worn-out cellular macromolecules and damaged organelles are targeted to autophagosomes (AP). When nutrients are available, mTORCl kinase is activated at the lysosomal membrane in the cell periphery; during low nutrient conditions, kinase activity is lowered and lysosomes accumulate in juxtanuclear compartment. Lysosomal calcium release via MCOLN1 activates TFEB via calcineurin phosphatase activity and induces TFEB translocation to nucleus to activate transcription. Bone resorption by enzymes in osteoclasts involves polarized accumulation of lysosomes and lysosomal acidification of facilitate bone resorption. In lysosomal exocytosis Involving calcium flux, lysosomes fuse with the plasma membrane and release hydrolases to the cell exterior. Oncogenic activation leads to lysosomal localization to the cellular periphery, increased release of hydrolases to the extracellular compartment which enables ECM degradation and invasion; oncogenic activation can lead to lysosomal membrane destabilization and to altered cell death responses.
The positioning of lysosomes to the cellular periphery
All cells, except for mature erythrocytes, contain lysosomes/LE (from here onward referred to as lysosomes) of sizes ranging from 100 to at least 500 nm [3]. They can be divided into two spatially distinct pools: the main one concentrated juxtanuclearly around the microtubule-organizing center (MTOC) and the other peripherally delineating the plasma membrane [23]. Especially the peripheral population of lysosomes has recently attracted the attention of cancer researchers due to its dramatic increase upon malignant transformation and emerging involvement in migration and invasion ([2,24] and see below). Lysosomes of the juxtanuclear and peripheral pools interchange by travelling along the fast two-way microtubule (MT) tracks or by low-speed diffusion [23]. Microtubules emanating from the MTOC grow at their plus-end towards the cellular periphery (anterograde movement), and lysosomal anterograde traffic is regulated by several MT-associated kinesin motor proteins, e.g. kinesin-1 (KIF-1), as well as Arf GTPase Arl8b and its effector SKIP [25–27]. Also RhoA, PI3Ks and protein complex FYCO are implicated to assist in anterograde lysosomal traffic [28,29]. A multisubunit protein complex BORC (consisting of myrlysin, lyspersin, diaskedin, snapin, BLOS1, BLOS2 and KXD1) recruits Arl8b to the lysosomal membrane and drives KIF-1-dependent motility towards cell periphery [17,27]. In contrast, dynein-dynactin motor complex directs the traffic towards the MT minus-ends (retrograde movement). GTPase Rab7 that is specifically associated with the lysosomes, recruits dynein motors via its effector RILP and Rabip4’ and AP-3 [30–32]. Additionally, Huntingtin (Htt) and GTPases Rab34 and Rab36 assist in the retrograde motility [33–35]. Interestingly, inhibition of BORC complex causes dissociation of Arl8b from lysosomes, prevents KIF-1-dependent lysosomal trafficking to the cell periphery and attenuates cell spreading and motility but has no profound effect on degradative functions in the remaining juxtanuclear pool. This indicates that a subset of peripheral lysosomes may be allocated to specifically control adhesion formation and metastatic processes in migratory cells.
How malignant transformation interferes with lysosomal trafficking is as yet largely unknown. One explanation may be the increased acidity of tumor environment, a known trigger of peripheral distribution and exocytosis of lysosomes [2]. On the other hand, studies in ErbB2-transformed cancer cells suggest that the transformation-associated increase in the activity of luminal proteases, especially cysteine cathepsins B and L, promotes the peripheral localization of lysosomes [21,22]. Supporting this hypothesis, increased cathepsin expression in response to overexpression of TFEB has a similar effect on lysosomal localization [36]. How the intraluminal protease activity connects to lysosomal trafficking is unclear, but one possibility is that luminal proteases degrade lysosomal transmembrane proteins from the inside thereby altering their association with trafficking proteins (Table 1).
Table 1. Genes and pharmacological agents that alter the subcellular localization of lysosomes.
| Treatment | Lysosomal localization | LMP | Physiological effect | References |
|---|---|---|---|---|
| ErbB1/2 inhibition (Lapatinib) ERBB2 RNAi CTSB/CTSL RNAi | Perinuclear | ND | Attenuated invasion | [21,22] |
| Cationic amphiphilic drugs Siramesine | Perinuclear | ND + | Expansion of lysosomal compartment | [67]; Unpublished observations |
| BORCS5 (BORC complex protein) KO, ARL8B RNAi | Perinuclear | ND | Reduced cell adhesion and motility, Lysosome traffic | [17,27] |
| KIF2A, KIF1B RNAi, Starvation | Perinuclear | ND | mTORC activation, autophagy modulation | [10] |
| KIF20A, KIF25 RNAi, KIF5B RNAi | Peripheral/protrusions | + | Accumulation of autophagosomes | [57,68] |
| DHC, Htt RNAi, RUFY1, δ-adaptin RNAi | Peripheral/protrusions | ND | Exaggerated protrusions, Lysosomal detachment from MTs | [32,33] |
| RAB7 shRNA | ND | ND | Hydrolase secretion, increased invasion | [69] |
| TPM2, MYH1 RNAi | Peripheral/protrusions | + | Cell death | [57] |
| TMEM106b shRNA | Peripheral | + | Increased lysosomal axonal motility | [16] |
| Cambinol (HDAC inhibitor), Troglizatone (PPARγ agonist), EIPA, (sodium-proton exchanger inhibitor) | Perinuclear | ND | Attenuated invasion | [28,31], Dykes et al, 2015 |
Lysosomal exocytosis and extracellular matrix degradation
During cancer invasion, the cells in the tumor mass pave their way through the microenvironment by forming actin-rich adhesions (invadopodia) to connect the cells to their surroundings and to create the forces needed for the invasion. During invasion, the extracellular matrix (ECM) components need to be degraded efficiently, which can occur both outside and inside of the cells [37]. The extracellular degradation is brought about by a hierarchic activation of proteolytic enzymes (e.g. matrix metalloproteins and plasminogen) secreted by both cancer and stromal cells. The cathepsins trafficked to plasma membrane lipid rafts or secreted via exocytosis of peripheral lysosomes augments this process [20,37,38]. A possible explanation to the ability of lysosomal cathepsins to degrade ECM in the non-favorable extralysosomal pH can be drawn from the biology of the osteoclast-mediated bone resorption (see review [39]). To efficiently resorb bone minerals, osteoclasts use their invadopodia-like, actin- and αvβ3-integrin-containing structures, podosomes, to adhere tightly to the collagen-rich bone surface. To create an acidic enclosed environment for the resorption, a ruffled membrane is formed by fusion of lysosome-related organelles to the plasma membrane. V-ATPase and the ClC-7 chloride channel ensure that sufficient H+ and Cl- ions and low pH can be created in the sealed zone to potentiate the matrix-degrading activities of bone-degrading hydrolases such as cathepsin K. It seems reasonable to envision that the targeted delivery of lysosomes via microtubules enables exocytosis and the required acidic local hub, inside which the ECM degradation is efficiently executed [40]. To support this, the acidifying action of V-ATPase is instrumental in cancer cell invasion [41]. Additionally, cells have specific surface receptors dedicated to internalization of ECM components. Endo180, dystroglycan and integrin α5β1 reroute collagens, laminin and fibronectin, respectively, to lysosomal degradation thus facilitating cell invasion into the ECM [42–44]. Invasive potential is further facilitated by lysosomal targeting of cell-cell adhesion components E-cadherin and junctional adhesion molecule (JAM) [24]. Lysosomally degraded proteins provide essential nutrients and energy for the invading cell and proinvasive endosomal trafficking may be linked to cells’ nutrient status and mTORC1 activation [45,46]. In this context, it is also interesting to note that invadopodia are especially rich in lysosomes [21,22]. Thus, the local activation of mTORC1 in response to the production of amino acids in these lysosomes may initiate signaling cascades that facilitate the function of invadopodia. This hypothesis is supported by the reported increase in mTORC1 activity upon peripheral localization [10], as well as the dependence of invasion on peripheral lysosomal localization [21].
Lysosomes in adhesions
In addition to their role in extracellular matrix degradation, peripheral lysosomes have recently emerged as cytoplasmic regulators of actin-based cell adhesion structures such as focal adhesions, the sites of ECM degradation [17,19,47]. The continuous formation and dissolution of cellular adhesions is needed to allow efficient cell motility [48]. Some indication to lysosomal function related to cellular adhesions can be drawn from a recent study showing that a subpopulation of LAMTOR2/3-expressing lysosomes move in Arl8b- and KIF-1-dependent manner to cell periphery where they target focal adhesions [19]. More specifically, this lysosome population facilitates the disassociation of the adhesion-stabilizing protein IQGAP1 from mature focal adhesions thereby promoting cell motility [19]. Lysosomes can regulate invasion also by controlling the traffic of integrins [49]. For example, the lysosome-targeted active integrin α5β1 is not necessarily degraded in lysosomes but can be recycled back to the plasma membrane at the rear end of the invading cell where it facilitates the release of the cell from the matrix and promotes invasion [49]. This recycling pathway depends on lysosomal chloride channel CLIC3 whose expression correlates with lymph node metastasis and poor prognosis in pancreatic carcinoma. Lysosomal cysteine cathepsins can also modulate focal adhesion molecules, e.g. focal adhesion kinase (FAK), Src and integrin activator talin, thereby promoting tumor growth and invasiveness [50–53]. It is as yet unclear whether these substrates enter the lysosomes to be degraded or whether cathepsins are released from the lysosomes to modulate adhesions intracellularly [24] (Figure2).
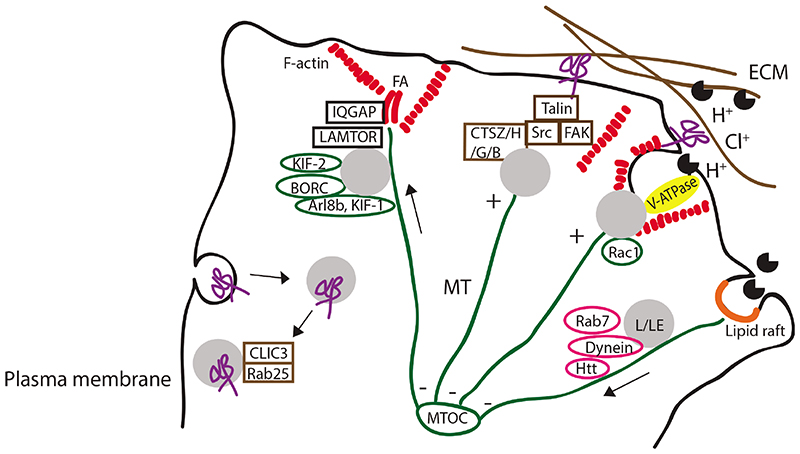
Figure 2.
Lysosomes in cellular adhesions and In invasion. Lysosomes/late endosomes (L/LE) reach the periphery via MT plus –end mediated traffic and with the help of kinesins, Arl8b, BORC and other effector molecules (RhoA, PI3Ks, FYCO, not shown in figure). BORC regulates Arl8b recruitment to lysosomal membranes and drives KIF-l-dependent motility. At focal adhesions (FA), the LAMTOR2/3 in late endosomes facilitates the removal of IQ.GAP from the adhesions thus promoting cell motility. Cathepsins Z, H, B and G activate focal adhesion molecules FAK, Src and integrin-binding protein talin: Cathepsin Z and H promote thereby tumor invasion into extracellular matrix (ECM) whereas cathepsin B promotes invasion in rheumatoid arthtritis and neutrophil cathepsin G affects myocyte survival. In osteoclasts, the Racl-assisted traffic of lysosomes to actin-rich integrin adhesions creates an acidic microenvironment, involving V-ATPase and chloride channels, inside which cathepsin K and matrix metalloproteinases degrade the bone matrix. Cathepsin B localization to and release from lipid rafts promotes cancer progression. Lysosomal traffic towards MTOC is driven by dynein-dynactin complex, Rab7 GTPase and several other molecules (RILP, Rabip4’, AP-3, Huntingtin (Htt) and GTPases Rab34 and Rab36, not shown in figure). Additionally, integrin uptake and release from lysosomes back to the plasma membrane, with the help of Rab25 GTPase and CLIC3, contributes to carcinoma invasion.
As discussed above, microtubule dynamics can promote focal adhesion turnover [54] by serving as highways that deliver lysosomes and proteins essential for focal adhesion disassembly to the areas close to the plasma membrane. It is, however, unclear how the cargo hop off the tubulin tracks to reach the actin-based adhesions? Although some molecules connecting actin and tubulin networks have been discovered [54], only a few clues regarding the trafficking of lysosomes from tubulin to actin are available [33,55]. Once the lysosomes have reached the cellular periphery, local Ca2+ fluxes and F-actin formation, however, influence their localization and movement [26,56]. Furthermore, in the absence of tropomyosin 2 (TPM2), which can link actin with focal adhesions, lysosomes cluster in the periphery and lose their membrane integrity [57]. Thus, correct connection to cytoskeleton may in addition to trafficking regulate lysosomal function and stability.
Lysosomal membrane permeabilization
Cancer cell lysosomes tend to get hyperactive when fulfilling the needs of the demanding cancer microenvironment. They need to upregulate exocytosis, ingest excessive amounts of engulfed ECM and adhesion molecules, repair damaged membranes and move around the crowded cell [2,3]. This busy lysosomal behavior is associated with an increased vulnerability of the lysosomal membrane, a frailty that can be targeted by several cationic amphiphilic drugs (CADs) that induce lysosomal damage preferentially in cancer cells [58]. CADs include hundreds of pharmacologic agents used to treat a plethora of common diseases [59]. Due to their chemical structure, they accumulate up to 1000-fold inside acidic lysosomes, where their incorporation into lumenal membranes interferes with the function of several lysosomal lipases, especially acid sphingomyelinase [60]. Cancer cells are especially sensitive to the subsequent accumulation of sphingomyelin, which may explain why CADs that are effective acid sphingomyelinase inhibitors are especially potent in killing cancer cells. In addition to the altered sphingolipid metabolism, increased cathepsin activity, misregulated cholesterol and free fatty acid metabolism, increased size, high lysosomal iron content and reduced pH also contribute to reduced lysosomal membrane stability in cancer cells [5,61–63].
Upon lysosomal membrane rupture, the exposed beta-galactoside attracts the diffusely localized sugar-binding galectins to the damaged membrane [64,65]. This biology of galectin recruitment can be used as a sensitive antibody- or fluorescent protein-based detection tool to study the early enzymatic leakage from lysosomes [65]. Due to its extreme sensitivity, this method has revealed that minor lysosomal leakage is not necessarily lethal and opened the intriguing possibility that the intracellular release of lysosomal enzymes may serve yet unknown physiological functions, for example in dissolving the focal adhesions from the cytosolic side [24,65]. Indeed, we have detected galectin-positive “leaky” lysosomes in protrusions of invasive breast carcinoma cells in optimal growth conditions (unpublished observations). And, in neuronal cells, the depletion of lysosomal transmembrane protein TMEM106B causes lysosomes to disperse in the distal periphery, which sensitizes them to oxidative stress in contrast to the more resistant juxtanuclearly positioned TMEM106B-expressing lysosomes [16]. These observations together with the findings where depletion of cytoskeleton-associated molecules alters lysosomal localization and membrane integrity [57] raise intriguing questions regarding the membrane stability in peripherally localized lysosomes.
Targeting the peripherally localized lysosomes
The finding that lysosomes travel to invasive protrusions of cancer cells is fascinating, especially considering the observed altered lysosomal membrane stability in these structures ([57] and our unpublished observations). By identifying the alterations in these “invasive tips” it is intriguing to envision the scenario that the peripherally localized subpopulation of lysosomes could be specifically targeted. Thus, combined targeting of MTs and lysosomes in cancer cells might be beneficial as exemplified by the ability of MT-stabilizing chemotherapeutic drugs (vinca alkaloids and taxanes) as well as siRNAs targeting MT motors to sensitize cells to the lysosomal cell death induced by a lysosome-destabilizing siramesine [57,66].
Conclusion and perspectives
Here we have highlighted the recent evidence that places lysosomes in key position to regulate cell adhesion and motility, the fundamental phenomena in cancer invasion. From the cancer progression point of view, the peripheral localization of lysosomes might serve to build local acidic environment for the ingestion and/or modulation of components of the adhesion machinery. What is still an open question is the way MTs and actin filaments cooperate in this process and the signaling hubs they serve at their tips. It also remains unclear whether the subcellular localization and association to cytoskeleton alters the lysosomal membrane stability, and whether the peripheral pool of lysosomes differs from that in the juxtanuclear region what comes to membrane composition and hydrolytic capacity. Answers to these questions will hopefully open new possibilities in targeting the invasion-associated lysosomes specifically.
Acknowledgements
This work was supported by the European Research Council (LYSOSOME), Danish National Research Foundation (CARD) and the Danish Cancer Society. We are grateful to Drs Kenji Maeda and Elena Favaro for critical comments on this manuscript.
References
- [1].de Duve C. The lysosome turns fifty. Nat Cell Biol. 2005;7:847–849. doi: 10.1038/ncb0905-847. [DOI] [PubMed] [Google Scholar]
- [2].Kallunki T, Olsen OD, Jaattela M. Cancer-associated lysosomal changes: friends or foes? Oncogene. 2013;32:1995–2004. doi: 10.1038/onc.2012.292. [DOI] [PubMed] [Google Scholar]
- [3].Xu H, Ren D. Lysosomal physiology. Annu Rev Physiol. 2015;77:57–80. doi: 10.1146/annurev-physiol-021014-071649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Eskelinen EL, Tanaka Y, Saftig P. At the acidic edge: emerging functions for lysosomal membrane proteins. Trends Cell Biol. 2003;13:137–145. doi: 10.1016/s0962-8924(03)00005-9. [DOI] [PubMed] [Google Scholar]
- [5].Appelqvist H, Waster P, Kagedal K, Ollinger K. The lysosome: from waste bag to potential therapeutic target. JMol Cell Biol. 2013;5:214–226. doi: 10.1093/jmcb/mjt022. [DOI] [PubMed] [Google Scholar]
- [6].Medina DL, Di Paola S, Peluso I, Armani A, De Stefani D, Venditti R, Montefusco S, Scotto-Rosato A, Prezioso C, Forrester A, et al. Lysosomal calcium signalling regulates autophagy through calcineurin and TFEB. Nat Cell Biol. 2015;17:288–299. doi: 10.1038/ncb3114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Settembre C, De Cegli R, Mansueto G, Saha PK, Vetrini F, Visvikis O, Huynh T, Carissimo A, Palmer D, Klisch TJ, et al. TFEB controls cellular lipid metabolism through a starvation-induced autoregulatory loop. Nat Cell Biol. 2013;15:647–658. doi: 10.1038/ncb2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Wang S, Tsun ZY, Wolfson RL, Shen K, Wyant GA, Plovanich ME, Yuan ED, Jones TD, Chantranupong L, Comb W, et al. Metabolism. Lysosomal amino acid transporter SLC38A9 signals arginine sufficiency to mTORCl. Science. 2015;347:188–194. doi: 10.1126/science.1257132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Rebsamen M, Pochini L, Stasyk T, de Araujo ME, Galluccio M, Kandasamy RK, Snijder B, Fauster A, Rudashevskaya EL, Bruckner M, et al. SLC38A9 is a component of the lysosomal amino acid sensing machinery that controls mTORC1. Nature. 2015;519:477–481. doi: 10.1038/nature14107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Korolchuk VI, Saiki S, Lichtenberg M, Siddiqi FH, Roberts EA, Imarisio S, Jahreiss L, Sarkar S, Futter M, Menzies FM, et al. Lysosomal positioning coordinates cellular nutrient responses. Nat Cell Biol. 2011;13:453–460. doi: 10.1038/ncb2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Shimobayashi M, Hall MN. Making new contacts: the mTOR network in metabolism and signalling crosstalk. Nat Rev Mol Cell Biol. 2014;15:155–162. doi: 10.1038/nrm3757. [DOI] [PubMed] [Google Scholar]
- [12].Sardiello M, Palmieri M, di Ronza A, Medina DL, Valenza M, Gennarino VA, Di Malta C, Donaudy F, Embrione V, Polishchuk RS, et al. A gene network regulating lysosomal biogenesis and function. Science. 2009;325:473–477. doi: 10.1126/science.1174447. [DOI] [PubMed] [Google Scholar]
- [13].Ferron M, Settembre C, Shimazu J, Lacombe J, Kato S, Rawlings DJ, Ballabio A, Karsenty G. A RANKL-PKCbeta-TFEB signaling cascade is necessary for lysosomal biogenesis in osteoclasts. Genes Dev. 2013;27:955–969. doi: 10.1101/gad.213827.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Steffan JJ, Williams BC, Welbourne T, Cardelli JA. HGF-induced invasion by prostate tumor cells requires anterograde lysosome trafficking and activity of Na+-H+ exchangers. J Cell Sci. 2010;123:1151–1159. doi: 10.1242/jcs.063644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Erie C, Sacino M, Houle L, Lu ML, Wei J. Altered lysosomal positioning affects lysosomal functions in a cellular model of Huntington’s disease. Eur J Neurosci. 2015;42:1941–1951. doi: 10.1111/ejn.12957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Stagi M, Klein ZA, Gould TJ, Bewersdorf J, Strittmatter SM. Lysosome size, motility and stress response regulated by fronto-temporal dementia modifier TMEM106B. Mol Cell Neurosci. 2014;61:226–240. doi: 10.1016/j.mcn.2014.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Pu J, Schindler C, Jia R, Jarnik M, Backlund P, Bonifacino JS. BORC, a multisubunit complex that regulates lysosome positioning. Dev Cell. 2015;33:176–188. doi: 10.1016/j.devcel.2015.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Garg S, Sharma M, Ung C, Tuli A, Barral DC, Hava DL, Veerapen N, Besra GS, Hacohen N, Brenner MB. Lysosomal trafficking, antigen presentation, and microbial killing are controlled by the Arf-like GTPase Arl8b. Immunity. 2011;35:182–193. doi: 10.1016/j.immuni.2011.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Schiefermeier N, Scheffler JM, de Araujo ME, Stasyk T, Yordanov T, Ebner HL, Offterdinger M, Munck S, Hess MW, Wickstrom SA, et al. The late endosomal p14-MP1 (LAMTOR2/3) complex regulates focal adhesion dynamics during cell migration. J Cell Biol. 2014;205:525–540. doi: 10.1083/jcb.201310043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Bian B, Mongrain S, Cagnol S, Langlois MJ, Boulanger J, Bernatchez G, Carrier JC, Boudreau F, Rivard N. Cathepsin B promotes colorectal tumorigenesis, cell invasion, and metastasis. Mol Carcinog. 2015 doi: 10.1002/mc.22312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Brix DM, Rafn B, Bundgaard Clemmensen K, Andersen SH, Ambartsumian N, Jaattela M, Kallunki T. Screening and identification of small molecule inhibitors of ErbB2-induced invasion. Mol Oncol. 2014;8:1703–1718. doi: 10.1016/j.molonc.2014.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Rafn B, Nielsen CF, Andersen SH, Szyniarowski P, Corcelle-Termeau E, Valo E, Fehrenbacher N, Olsen CJ, Daugaard M, Egebjerg C, et al. ErbB2-driven breast cancer cell invasion depends on a complex signaling network activating myeloid zinc finger-1-dependent cathepsin B expression. Mol Cell. 2012;45:764–776. doi: 10.1016/j.molcel.2012.01.029. [DOI] [PubMed] [Google Scholar]
- [23].Matteoni R, Kreis TE. Translocation and clustering of endosomes and lysosomes depends on microtubules. J Cell Biol. 1987;105:1253–1265. doi: 10.1083/jcb.105.3.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Olson OC, Joyce JA. Cysteine cathepsin proteases: regulators of cancer progression and therapeutic response. Nat Rev Cancer. 2015;15:712–729. doi: 10.1038/nrc4027. [DOI] [PubMed] [Google Scholar]
- [25].Bagshaw RD, Callahan JW, Mahuran DJ. The Arf-family protein, Arl8b, is involved in the spatial distribution of lysosomes. Biochem Biophys Res Commun. 2006;344:1186–1191. doi: 10.1016/j.bbrc.2006.03.221. [DOI] [PubMed] [Google Scholar]
- [26].Granger E, McNee G, Allan V, Woodman P. The role of the cytoskeleton and molecular motors in endosomal dynamics. Semin Cell Dev Biol. 2014;31:20–29. doi: 10.1016/j.semcdb.2014.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Rosa-Ferreira C, Munro S. Arl8 and SKIP act together to link lysosomes to kinesin-1. Dev Cell. 2011;21:1171–1178. doi: 10.1016/j.devcel.2011.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Steffan JJ, Snider JL, Skalli O, Welbourne T, Cardelli JA. Na+/H+ exchangers and RhoA regulate acidic extracellular pH-induced lysosome trafficking in prostate cancer cells. Traffic. 2009;10:737–753. doi: 10.1111/j.1600-0854.2009.00904.x. [DOI] [PubMed] [Google Scholar]
- [29].Pankiv S, Alemu EA, Brech A, Bruun JA, Lamark T, Overvatn A, Bjorkoy G, Johansen T. FYCO1 is a Rab7 effector that binds to LC3 and PI3P to mediate microtubule plus end-directed vesicle transport. J Cell Biol. 2010;188:253–269. doi: 10.1083/jcb.200907015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Cantalupo G, Alifano P, Roberti V, Bruni CB, Bucci C. Rab-interacting lysosomal protein (RILP): the Rab7 effector required for transport to lysosomes. EMBO J. 2001;20:683–693. doi: 10.1093/emboj/20.4.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Steffan JJ, Cardelli JA. Thiazolidinediones induce Rab7-RILP-MAPK-dependent juxtanuclear lysosome aggregation and reduce tumor cell invasion. Traffic. 2010;11:274–286. doi: 10.1111/j.1600-0854.2009.01012.x. [DOI] [PubMed] [Google Scholar]
- [32].Ivan V, Martinez-Sanchez E, Sima LE, Oorschot V, Klumperman J, Petrescu SM, van der Sluijs P. AP-3 and Rabip4’ coordinately regulate spatial distribution of lysosomes. PLoS One. 2012;7:e48142. doi: 10.1371/journal.pone.0048142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Caviston JP, Zajac AL, Tokito M, Holzbaur EL. Huntingtin coordinates the dynein-mediated dynamic positioning of endosomes and lysosomes. Mol Biol Cell. 2011;22:478–492. doi: 10.1091/mbc.E10-03-0233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Kasmapour B, Cai L, Gutierrez MG. Spatial distribution of phagolysosomes is independent of the regulation of lysosome position by Rab34. Int J Biochem Cell Biol. 2013;45:2057–2065. doi: 10.1016/j.biocel.2013.07.003. [DOI] [PubMed] [Google Scholar]
- [35].Chen L, Hu J, Yun Y, Wang T. Rab36 regulates the spatial distribution of late endosomes and lysosomes through a similar mechanism to Rab34. Mol Membr Biol. 2010;27:23–30. doi: 10.3109/09687680903417470. [DOI] [PubMed] [Google Scholar]
- [36].Medina DL, Fraldi A, Bouche V, Annunziata F, Mansueto G, Spampanato C, Puri C, Pignata A, Martina JA, Sardiello M, et al. Transcriptional activation of lysosomal exocytosis promotes cellular clearance. Dev Cell. 2011;21:421–430. doi: 10.1016/j.devcel.2011.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Sevenich L, Joyce JA. Pericellular proteolysis in cancer. Genes Dev. 2014;28:2331–2347. doi: 10.1101/gad.250647.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Cavallo-Medved D, Dosescu J, Linebaugh BE, Sameni M, Rudy D, Sloane BF. Mutant K-ras regulates cathepsin B localization on the surface of human colorectal carcinoma cells. Neoplasia. 2003;5:507–519. doi: 10.1016/s1476-5586(03)80035-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Kikuta J, Ishii M. Osteoclast migration, differentiation and function: novel therapeutic targets for rheumatic diseases. Rheumatology (Oxford) 2013;52:226–234. doi: 10.1093/rheumatology/kes259. [DOI] [PubMed] [Google Scholar]
- [40].Macpherson IR, Rainero E, Mitchell LE, van den Berghe PV, Speirs C, Dozynkiewicz MA, Chaudhary S, Kalna G, Edwards J, Timpson P, et al. CLIC3 controls recycling of late endosomal MT1-MMP and dictates invasion and metastasis in breast cancer. J Cell Sci. 2014;127:3893–3901. doi: 10.1242/jcs.135947. [DOI] [PubMed] [Google Scholar]
- [41].Cotter K, Stransky L, McGuire C, Forgac M. Recent Insights into the Structure, Regulation, and Function of the V-ATPases. Trends Biochem Sci. 2015;40:611–622. doi: 10.1016/j.tibs.2015.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Melander MC, Jurgensen HJ, Madsen DH, Engelholm LH, Behrendt N. The collagen receptor uPARAP/Endo180 in tissue degradation and cancer (Review) Int J Oncol. 2015;47:1177–1188. doi: 10.3892/ijo.2015.3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Lobert VH, Brech A, Pedersen NM, Wesche J, Oppelt A, Malerod L, Stenmark H. Ubiquitination of alpha 5 beta 1 integrin controls fibroblast migration through lysosomal degradation of fibronectin-integrin complexes. Dev Cell. 2010;19:148–159. doi: 10.1016/j.devcel.2010.06.010. [DOI] [PubMed] [Google Scholar]
- [44].Leonoudakis D, Huang G, Akhavan A, Fata JE, Singh M, Gray JW, Muschler JL. Endocytic trafficking of laminin is controlled by dystroglycan and is disrupted in cancers. J Cell Sci. 2014;127:4894–4903. doi: 10.1242/jcs.152728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Rainero E, Howe JD, Caswell PT, Jamieson NB, Anderson K, Critchley DR, Machesky L, Norman JC. Ligand-Occupied Integrin Internalization Links Nutrient Signaling to Invasive Migration. Cell Rep. 2015 doi: 10.1016/j.celrep.2014.12.037. [DOI] [PubMed] [Google Scholar]
- [46].Commisso C, Davidson SM, Soydaner-Azeloglu RG, Parker SJ, Kamphorst JJ, Hackett S, Grabocka E, Nofal M, Drebin JA, Thompson CB, et al. Macropinocytosis of protein is an amino acid supply route in Ras-transformed cells. Nature. 2013;497:633–637. doi: 10.1038/nature12138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Wang Y, McNiven MA. Invasive matrix degradation at focal adhesions occurs via protease recruitment by a FAK-p130Cas complex. J Cell Biol. 2012;196:375–385. doi: 10.1083/jcb.201105153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Kim DH, Wirtz D. Focal adhesion size uniquely predicts cell migration. FASEB J. 2013;27:1351–1361. doi: 10.1096/fj.12-220160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Dozynkiewicz MA, Jamieson NB, Macpherson I, Grindlay J, van den Berghe PV, von Thun A, Morton JP, Gourley C, Timpson P, Nixon C, et al. Rab25 and CLIC3 collaborate to promote integrin recycling from late endosomes/lysosomes and drive cancer progression. Dev Cell. 2012;22:131–145. doi: 10.1016/j.devcel.2011.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Tong B, Wan B, Wei Z, Wang T, Zhao P, Dou Y, Lv Z, Xia Y, Dai Y. Role of cathepsin B in regulating migration and invasion of fibroblast-like synoviocytes into inflamed tissue from patients with rheumatoid arthritis. Clin Exp Immunol. 2014;177:586–597. doi: 10.1111/cei.12357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Rafiq K, Guo J, Vlasenko L, Guo X, Kolpakov MA, Sanjay A, Houser SR, Sabri A. c-Cbl ubiquitin ligase regulates focal adhesion protein turnover and myofibril degeneration induced by neutrophil protease cathepsin G. J Biol Chem. 2012;287:5327–5339. doi: 10.1074/jbc.M111.307009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Akkari L, Gocheva V, Kester JC, Hunter KE, Quick ML, Sevenich L, Wang HW, Peters C, Tang LH, Klimstra DS, et al. Distinct functions of macrophage-derived and cancer cell-derived cathepsin Z combine to promote tumor malignancy via interactions with the extracellular matrix. Genes Dev. 2014;28:2134–2150. doi: 10.1101/gad.249599.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Jevnikar Z, Rojnik M, Jamnik P, Doljak B, Fonovic UP, Kos J. Cathepsin H mediates the processing of talin and regulates migration of prostate cancer cells. J Biol Chem. 2013;288:2201–2209. doi: 10.1074/jbc.M112.436394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Yue J, Xie M, Gou X, Lee P, Schneider MD, Wu X. Microtubules regulate focal adhesion dynamics through MAP4K4. Dev Cell. 2014;31:572–585. doi: 10.1016/j.devcel.2014.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Sun Y, Buki KG, Ettala O, Vaaraniemi JP, Vaananen HK. Possible role of direct Rac1-Rab7 interaction in ruffled border formation of osteoclasts. J Biol Chem. 2005;280:32356–32361. doi: 10.1074/jbc.M414213200. [DOI] [PubMed] [Google Scholar]
- [56].Jung SR, Seo JB, Shim D, Hille B, Koh DS. Actin cytoskeleton controls movement of intracellular organelles in pancreatic duct epithelial cells. Cell Calcium. 2012;51:459–469. doi: 10.1016/j.ceca.2012.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Groth-Pedersen L, Aits S, Corcelle-Termeau E, Petersen NH, Nylandsted J, Jaattela M. Identification of cytoskeleton-associated proteins essential for lysosomal stability and survival of human cancer cells. PLoS One. 2012;7:e45381. doi: 10.1371/journal.pone.0045381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Petersen NH, Olsen OD, Groth-Pedersen L, Ellegaard AM, Bilgin M, Redmer S, Ostenfeld MS, Ulanet D, Dovmark TH, Lonborg A, et al. Transformation-associated changes in sphingolipid metabolism sensitize cells to lysosomal cell death induced by inhibitors of acid sphingomyelinase. Cancer Cell. 2013;24:379–393. doi: 10.1016/j.ccr.2013.08.003. [DOI] [PubMed] [Google Scholar]
- [59].Kornhuber J, Tripal P, Reichel M, Muhle C, Rhein C, Muehlbacher M, Groemer TW, Gulbins E. Functional Inhibitors of Acid Sphingomyelinase (FIASMAs): a novel pharmacological group of drugs with broad clinical applications. Cell Physiol Biochem. 2010;26:9–20. doi: 10.1159/000315101. [DOI] [PubMed] [Google Scholar]
- [60].Kolzer M, Werth N, Sandhoff K. Interactions of acid sphingomyelinase and lipid bilayers in the presence of the tricyclic antidepressant desipramine. FEBS Lett. 2004;559:96–98. doi: 10.1016/S0014-5793(04)00033-X. [DOI] [PubMed] [Google Scholar]
- [61].Sargeant TJ, Lloyd-Lewis B, Resemann HK, Ramos-Montoya A, Skepper J, Watson CJ. Stat3 controls cell death during mammary gland involution by regulating uptake of milk fat globules and lysosomal membrane permeabilization. Nat Cell Biol. 2014;16:1057–1068. doi: 10.1038/ncb3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Fehrenbacher N, Bastholm L, Kirkegaard-Sorensen T, Rafn B, Bottzauw T, Nielsen C, Weber E, Shirasawa S, Kallunki T, Jaattela M. Sensitization to the lysosomal cell death pathway by oncogene-induced down-regulation of lysosome-associated membrane proteins 1 and 2. Cancer Res. 2008;68:6623–6633. doi: 10.1158/0008-5472.CAN-08-0463. [DOI] [PubMed] [Google Scholar]
- [63].Eriksson I, Joosten M, Roberg K, Ollinger K. The histone deacetylase inhibitor trichostatin A reduces lysosomal pH and enhances cisplatin-induced apoptosis. Exp Cell Res. 2013;319:12–20. doi: 10.1016/j.yexcr.2012.10.004. [DOI] [PubMed] [Google Scholar]
- [64].Paz I, Sachse M, Dupont N, Mounier J, Cederfur C, Enninga J, Leffler H, Poirier F, Prevost MC, Lafont F, et al. Galectin-3, a marker for vacuole lysis by invasive pathogens. Cell Microbiol. 2010;12:530–544. doi: 10.1111/j.1462-5822.2009.01415.x. [DOI] [PubMed] [Google Scholar]
- [65].Aits S, Kricker J, Liu B, Ellegaard AM, Hamalisto S, Tvingsholm S, Corcelle-Termeau E, Hogh S, Farkas T, Holm Jonassen A, et al. Sensitive detection of lysosomal membrane permeabilization by lysosomal galectin puncta assay. Autophagy. 2015;11:1408–1424. doi: 10.1080/15548627.2015.1063871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Groth-Pedersen L, Ostenfeld MS, Hoyer-Hansen M, Nylandsted J, Jaattela M. Vincristine induces dramatic lysosomal changes and sensitizes cancer cells to lysosome-destabilizing siramesine. Cancer Res. 2007;67:2217–2225. doi: 10.1158/0008-5472.CAN-06-3520. [DOI] [PubMed] [Google Scholar]
- [67].Funk RS, Krise JP. Cationic amphiphilic drugs cause a marked expansion of apparent lysosomal volume: implications for an intracellular distribution-based drug interaction. Mol Pharm. 2012;9:1384–1395. doi: 10.1021/mp200641e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Cardoso CM, Groth-Pedersen L, Hoyer-Hansen M, Kirkegaard T, Corcelle E, Andersen JS, Jaattela M, Nylandsted J. Depletion of kinesin 5B affects lysosomal distribution and stability and induces peri-nuclear accumulation of autophagosomes in cancer cells. PLoS One. 2009;4:e4424. doi: 10.1371/journal.pone.0004424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Alonso-Curbelo D, Riveiro-Falkenbach E, Perez-Guijarro E, Cifdaloz M, Karras P, Osterloh L, Megias D, Canon E, Calvo TG, Olmeda D, et al. RAB7 controls melanoma progression by exploiting a lineage-specific wiring of the endolysosomal pathway. Cancer Cell. 2014;26:61–76. doi: 10.1016/j.ccr.2014.04.030. [DOI] [PubMed] [Google Scholar]