Summary
DNA methylation controls eukaryotic gene expression and is extensively reprogrammed to regulate animal development. However, whether developmental methylation reprogramming regulates genes during the diploid life cycle of flowering plants is presently unknown. Here we report a distinctive RNA-directed DNA methylation (RdDM) activity in the Arabidopsis thaliana male sexual lineage that regulates gene expression in meiocytes. Loss of sexual lineage-specific RdDM causes mis-splicing of the MPS1 gene, thereby disrupting meiosis. Our results establish a regulatory paradigm in which de novo methylation creates a cell-lineage-specific epigenetic signature that controls gene expression and contributes to cellular function in flowering plants.
Cytosine methylation is an ancient DNA modification catalyzed by methyltransferases that are conserved across eukaryotes, including plants and animals1. Cytosine methylation in the CG dinucleotide context is maintained by DNA Methyltransferase 1 (Dnmt1, called MET1 in plants), which recognizes hemimethylated CG dinucleotides and adds a methyl group to the unmethylated cytosine during DNA replication2,3. Plant methylation also commonly occurs in the CHG and CHH (H=A, C, or T) contexts, maintained by the CHROMOMETHYLASE3 (CMT3) and CMT2 enzymes, respectively4,5. Establishment of de novo methylation is catalyzed by Dnmt3 and its plant DRM homologues2,3. DRM enzymes (DRM1 and 2 in Arabidopsis thaliana) are part of the RNA-directed DNA methylation (RdDM) pathway, which typically targets transposons and methylates cytosines regardless of sequence context6. In RdDM, 24-nucleotide (nt) small RNAs (sRNAs), which are produced from transcripts created by plant-specific RNA polymerase IV and RNA-dependent RNA polymerase 2 (RDR2), guide DRM methyltransferases to DNA via association with a homologous transcript generated by RNA polymerase V and the DRD1 chromatin remodeling protein5,6.
DNA methylation patterns are faithfully replicated during cell division, which allows methylation to carry epigenetic information within cellular lineages2,3. In the complex genomes of flowering plants and vertebrates, methylation heritably silences transposons, thereby maintaining genome integrity and transcriptional homeostasis2,3. Consistent with this function, DNA methylation of regulatory sequences, especially those near transcriptional start sites, is strongly associated with gene silencing5,7.
In addition to its homeostatic function, DNA methylation can be reprogrammed during development to regulate gene expression. In mammals, this phenomenon has been observed in a number of tissues and cellular lineages, and appears to be a common regulatory mechanism8–14. In plants, gene expression in the transient extra-embryonic endosperm tissue is controlled by active DNA demethylation, which occurs in the central cell (a companion cell of the egg) that gives rise to the endosperm15,16. A similar active demethylation process also occurs in the vegetative cell, a terminally differentiated companion cell of the sperm15,17,18. Beyond the gamete companion cells, although intriguing examples of altered methylation levels and patterns in different cell types17–20 and during responses to biotic and abiotic stimuli21–25 have been described, whether gene expression is controlled by developmental reprogramming of DNA methylation in plants is unknown.
To investigate this question, we analyzed DNA methylation in the male sexual lineage of Arabidopsis thaliana. This allowed us to uncover a sexual-lineage specific DNA methylation signature deposited by the RdDM pathway. We further demonstrated that this de novo methylation regulates gene expression and splicing, and is required for normal meiosis, providing a compelling link between DNA methylation reprogramming, gene expression and developmental outcome. The RdDM pathway is widely present in plant tissues, and therefore has the potential to regulate the development of many cell types and tissues.
Male meiocytes feature a typical germline methylome with high CG and low CHH methylation
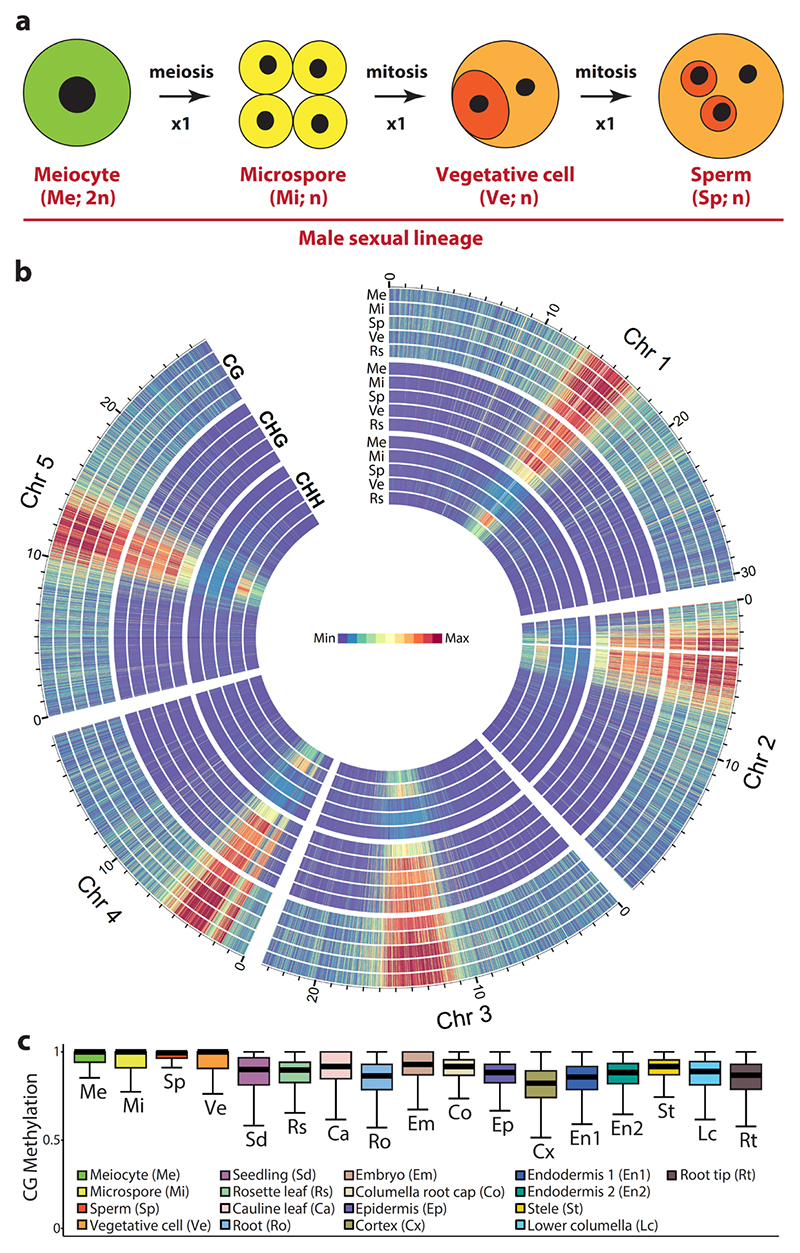
In Arabidopsis and other flowering plants, the male sexual lineage initiates as diploid meiocytes, which give rise to haploid microspores via meiosis26 (Fig. 1a). The microspores subsequently divide mitotically to produce the vegetative and generative cells (Fig. 1a). The generative cell enters one more round of mitosis to generate two sperm cells, which are engulfed within the vegetative cell in the mature pollen grain (Fig. 1a). To comprehensively understand DNA methylation reprogramming within the entire lineage, we generated a genome-wide methylation profile for male Arabidopsis thaliana meiocytes (Supplementary Table 1), which we compared to those of the microspore, sperm and vegetative cell17,18.
Figure 1. Male meiocytes exhibit high CG/CHG and low CHH methylation.
a, Model of male sexual lineage development in Arabidopsis thaliana. n, the number of chromosomes in the haploid genome. b, Heat maps showing CG, CHG and CHH methylation of the male sexual lineage comprising the meiocyte (Me), microspore (Mi), sperm (Sp) and vegetative cell (Vg), in comparison to rosette leaf (Rs). Methylation is calculated and presented in 10 kb windows, with the maximum set at the highest value among the five tissues for each context. The region enriched with mitochondria sequences on Chromosome 2 (3.23 to 3.51 Mb) is removed. c, Box plots demonstrating CG methylation for individual CG sites located within annotated transposons, and with methylation greater than 50% and at least 10 informative sequenced cytosines. Each box encloses the middle 50% of the distribution, with the horizontal line making the median, and vertical lines marking the minimum and maximum values that fall within 1.5 times the height of the box.
Contrary to the speculation of DNA demethylation in male meiocytes27, we found meiocyte methylation resembles that of microspores and sperm, with high levels of CG and CHG methylation in transposons (Fig. 1b,c and Supplementary Fig. 1). This is consistent with robust transposon silencing in the germline, an essential function for ensuring genetic integrity across generations16,28. In the CHH context, the microspore and sperm cells of the germline have low levels of methylation compared to somatic tissues and, especially, vegetative cells17,18,28 (Fig. 1b and Supplementary Fig. 1). However, the male meiocyte has even lower CHH methylation than microspore and sperm cells (Fig. 1b and Supplementary Fig. 1). Low levels of CHH methylation in microspore and sperm were proposed to result from lack of methylation maintenance during meiotic divisions17. Our results demonstrate this is not the case, and that CHH methylation instead undergoes an overall increase during the development of the male sexual lineage.
Hypermethylated loci are observed in the male sexual lineage
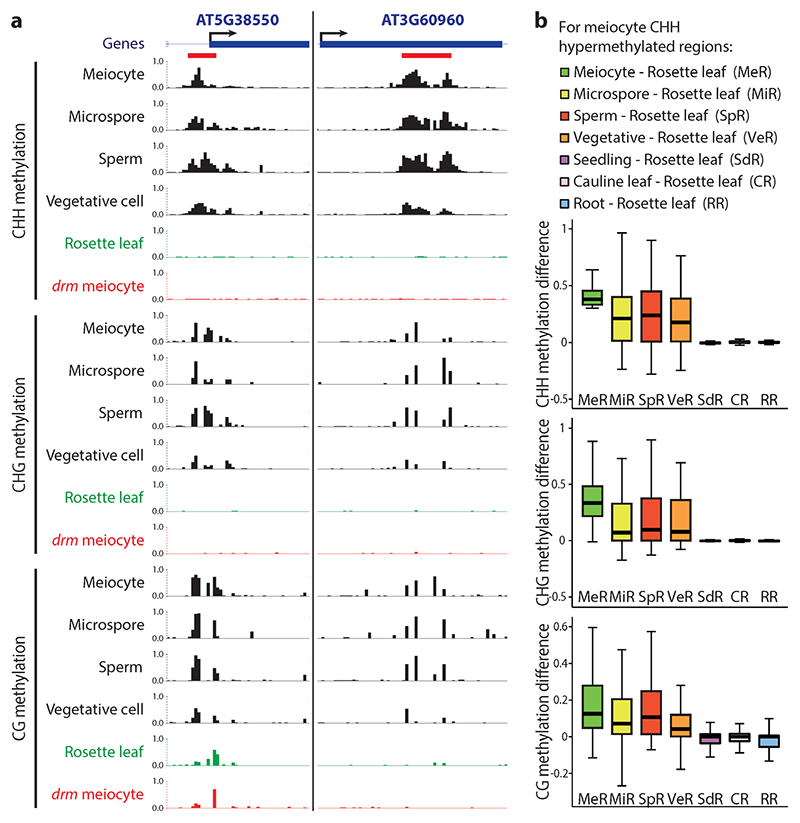
Comparison of DNA methylation patterns between male sex cells and somatic tissues (seedlings, rosette leaves, cauline leaves, roots) revealed regions that are strongly hypermethylated in sex cells (Fig. 2a and Supplementary Fig. 2). Furthermore, loci hypermethylated in one male sex cell type tend to be hypermethylated in other sex cells (Fig. 2a,b and Supplementary Fig. 2). This hypermethylation is most prominent in the CHH context, but encompasses other contexts as well (Fig. 2a,b and Supplementary Fig. 2), so that the same locus is often hypermethylated at CG, CHG and CHH sites (Fig. 2a and Supplementary Fig. 2). Among the 1301 loci we identified that are consistently differentially methylated between sex cells and somatic tissues, the overwhelming majority (1265; 97%) are hypermethylated in sex cells (Supplementary Data 1). These sexual-lineage hypermethylated loci (SLHs) are typically small (529 nucleotides on average; Supplementary Data 1), altogether encompassing 0.6% of the nuclear genome.
Figure 2. Sexual-lineage hypermethylated loci (SLHs) in Arabidopsis.
a, Snapshots of cytosine methylation in wild-type male sex cells (black), rosette leaf (green) and drm1drm2 (drm) mutant meiocyte (red) at two SLH examples. SLHs (refer to Supplementary Data 1 for a full list) are underlined in red. Methylation patterns in other somatic tissues and drm sex cells are shown in Supplementary Fig. 2. b, Box plots showing absolute methylation difference between specific cells/tissues and rosette leaf for 50 bp windows that are CHH hypermethylated in meiocytes in comparison to rosette leaves.
Sexual-lineage hypermethylated loci (SLHs) are caused by RdDM
The SLHs resemble targets of the small RNA-directed DNA methylation (RdDM) pathway, which establishes and maintains methylation in all sequence contexts, but is particularly important for CHH methylation of relatively small loci3,5,6. To test whether RdDM is responsible for SLHs, we analyzed the methylomes of meiocytes, sperm and vegetative cells with mutations in both DRM1 and DRM2, as well as the methylomes of sperm and vegetative cells with a mutation in RDR2 (Supplementary Table 1). In all examined mutant sex cells, SLHs are extensively hypomethylated in all sequence contexts (Figs 2a and 3a; Supplementary Fig. 2), demonstrating that SLHs are a product of RdDM. As expected of RdDM targets in the sexual lineage, SLHs are associated with the 24-nt sRNAs that guide RdDM at levels similar to those of other RdDM target loci in pollen, but not in shoots (Fig. 3b). The vast majority of SLHs (99.4%; 1257 loci) have significantly less methylation in drm1drm2 mutant sex cells in comparison to those of wild type (none in the reverse comparison; both P < 0.001; Fisher’s exact test); this slightly reduced group of SLHs is used for subsequent analyses (Supplementary Data 1). Collectively, our results demonstrate that RdDM is the underlying mechanism producing SLHs.
Figure 3. Sexual-lineage hypermethylated loci (SLHs) are produced by RdDM.
a, Box plots showing the absolute methylation at SLHs in the drm1drm2 (drm) and rdr2 mutants in comparison to wild type. b, Box plots demonstrating the abundance of 24 nucleotide (nt) small RNA in pollen or shoot at SLHs and non-SLH RdDM target loci. *P < 0.001; Kolmogorov-Smirnov test. c, Box plots showing the absolute methylation at SLH and non-SLH 50 bp windows in ros1dml2dml3 (rdd) mutant rosette leaf, wild-type rosette leaf, and wild-type male sex cells (meiocyte, microspore and sperm).
In Arabidopsis, RdDM is counteracted by active DNA demethylation3, suggesting that somatic demethylation may contribute to the distinct patterns of RdDM in male sex cells and somatic tissues. To test this possibility, we examined available methylation data for rosette leaves with mutations in the three demethylases expressed in somatic tissues: ROS1, DML2 and DML3 29. CG methylation at SLHs in these rdd mutants is indeed much higher than in wild-type control leaves, but not as high as in wild-type sex cells (Fig. 3c). CHG methylation in rdd leaves is also higher than in wild-type, but substantially lower than in sex cells (Fig. 3c), and CHH methylation is only slightly higher in rdd leaves compared to wild-type and much lower than in sex cells (Fig. 3c). Given the known ability of RdDM-established CG methylation, but not CHH methylation, to be maintained in the absence of RdDM6, our data suggest that active demethylation removes some of the CG (and CHG) methylation that is induced by RdDM in the sexual lineage and maintained in somatic tissues.
Many SLHs are novel RdDM targets specific to the sexual lineage
As a manifestation of RdDM activity in the sexual lineage, SLHs can simply be a product of increased RdDM activity at canonical targets, an expansion of RdDM into novel targets, or both. As CHH/G methylation levels are indicators of RdDM activity, we separated SLHs into two groups based on the level of CHH/G methylation in somatic tissues: canonical SLHs (724 loci), which have CHH/G methylation in the soma, and sexual-lineage specific methylated loci (SLMs; 533 loci), which lack CHH/G methylation in somatic tissues (Supplementary Data 1).
To further evaluate the cell/tissue specificity of SLMs, we examined the root cap columella cell, which has high levels of RdDM-associated CHH methylation19. Whereas canonical SLHs show methylation in all sequence contexts in columella cells (Fig. 4a), SLMs have little CHH/G methylation in columella (Supplementary Fig. 3). Furthermore, whereas 76% (551/724) of the canonical SLHs overlap with published columella differentially methylated regions (DMRs)19, most SLMs (88%, 469 sites; Supplementary Data 1) do not overlap with columella DMRs and show no CHH/G methylation in columella (Fig. 4b). Examination of DNA methylation in the embryo – another tissue with reported CHH hypermethylation20 – revealed that although canonical SLHs are methylated (Fig. 4a), the 469 SLMs that do not overlap with columella DMRs show no CHH/G methylation in embryo (Fig. 4b). We use this group of highly specific SLMs for all subsequent analyses (Supplementary Data 1).
Figure 4. SLMs are novel RdDM targets specific to the sexual lineage.
a,b, Box plots showing the absolute methylation at canonical SLHs (a) and SLMs (b) in somatic tissues (Sd, seedling; Rs, rosette leaf; Ca, cauline leaf; Ro, root), sex cells (Me, meiocyte; Mi, microspore; Sp, sperm), columella root cap (Co) and embryo (Em). c, Box plots illustrating CG methylation at SLMs in wild-type (WT) seedling (refer to b for other somatic tissues), and seedlings and sex cells from RdDM mutants: drm1drm2 (drm) and rdr2. d, Scatter plot showing the linear correlation between CG methylation in WT (y axis) and drd1 mutant (x axis) roots at SLMs (Pearson’s R = 0.80). e, Scatter plot showing the linear correlation between average CG methylation in WT somatic tissues (cauline leaf, rosette leaf, root and seedling; y axis) and that in drm mutant sex cells (meiocytes, sperm and vegetative cell; x axis) at SLMs (Pearson’s R = 0.76). f, Box plots demonstrating the absolute CG methylation at SLMs in WT seedling (same data as used in b and c), and published data† 30 including WT control seedlings (Sd†), metl mutant, and MET1 reintroduction lines (T-METla T2 and T-MET1b T5).
Although SLMs lack CHH/G methylation in somatic tissues, some CG methylation is present (Fig. 4b and Supplementary Fig. 2d,e). This remnant CG methylation can either be induced by sexual-lineage specific RdDM and maintained in somatic tissues by MET1, or result directly from somatic RdDM activity. To distinguish between these hypotheses, we analyzed SLM CG methylation in RdDM mutant somatic tissues, which showed similar overall levels to wild-type somatic tissues (Fig. 4c). Furthermore, SLM CG methylation in RdDM (drd1, drm2 and rdr2) mutant somatic tissues correlates with that in wild-type (Pearson’s R = 0.80, 0.58 and 0.70, respectively; Fig. 4d and Supplementary Fig. 4), demonstrating that RdDM is not required to maintain somatic CG methylation at SLMs.
The hypothesis that CG methylation at SLMs is initiated by RdDM in sex cells and is maintained at lower levels by MET1 in the absence of RdDM in somatic cells makes several predictions. First, MET1 should be able to maintain CG methylation in sex cells without RdDM at levels similar to wild-type somatic tissues. Indeed, SLM CG methylation in drm1drm2 mutant sex cells is similar to wild-type somatic tissues (Fig. 4c and Supplementary Fig. 2d,e) and strongly correlates with that in wild-type somatic tissues (Pearson’s R = 0.76; Fig. 4e). Second, somatic CG methylation at SLMs should be MET1-dependent, which it is (Fig. 4f). Finally, CG methylation at SLMs should be reestablished after it is erased, because CG methylation is known to be reestablished at loci that are targeted by RdDM in a manner that is not dependent on preexisting CG methylation (i.e. at loci where RdDM still works in met1 mutants)30. Indeed, somatic CG methylation at SLMs is restored to wild-type levels through introduction of functional MET1 into met1 mutant plants (Fig. 4f). Taken together, our analyses demonstrate that SLMs are products of sexual-lineage specific RdDM activity, which establishes methylation in all sequence contexts. In somatic tissues, residual CG methylation at SLMs is maintained by MET1 in the absence of RdDM.
RdDM-induced sexual-lineage specific methylation regulates gene expression in meiocytes
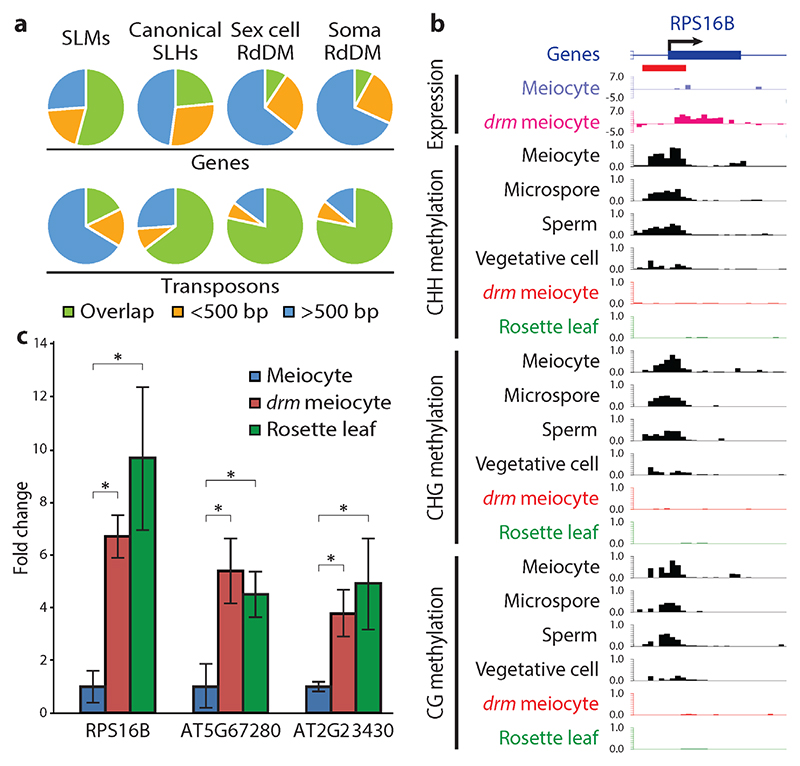
Because SLMs are not targeted by RdDM outside the sexual lineage, we analyzed whether they resemble conventional RdDM loci by comparing the proximity of SLMs, canonical SLHs, and other RdDM targets to genes and transposons. Canonical SLHs correspond mostly to transposons, but overlap genes more frequently than other RdDM targets (Fig. 5a). Furthermore, canonical SLHs are more likely to overlap annotated transposons than randomly selected sets of loci that are comparably located in relation to genes throughout the genome, but are less likely to overlap transposons than other RdDM target loci (Supplementary Table 2). Surprisingly, the majority of SLMs overlap genes (Fig. 5a), and are even slightly less likely to overlap annotated transposons than random control loci (Supplementary Table 2). These results indicate that canonical SLHs are an extension of conventional, transposon-targeted RdDM, which is consistent with their methylation in some somatic cell types, whereas SLMs are an aggressive expansion of RdDM into genes.
Figure 5. SLMs target genes and regulate gene expression in meiocytes.
a, Pie charts illustrating percentages of SLMs, canonical SLHs and other RdDM target loci overlapping (green), within 500 bp (yellow), and more than 500 bp from (blue) genes or transposons (numbers shown in Supplementary Table 2). b, Snapshots of transcription (in log2RPKM) and DNA methylation (similar to Fig. 2a), at the RPS16B gene drm, drm1drm2; SLM is underlined in red. c, Quantitative RT-PCR showing the expression of three SLM-regulated genes. *P < 0.02 (t-test).
The common occurrence of SLMs in genes and the well-demonstrated role of DNA methylation in suppressing gene expression3 suggested that SLMs may repress gene expression in the sexual lineage. To test this hypothesis, we analyzed mRNA levels in drm1drm2 mutant meiocytes and wild-type controls by RNA-seq. Expression of meiosis-associated genes is substantially enriched in our data compared with published meiocyte RNA-seq results (Supplementary Data 2)31,32, suggesting high meiocyte purity. Among the 47 genes with a greater than four-fold change in expression between wild-type and drm1drm2 meiocytes, all of which are activated in drm1drm2 meiocytes, seven overlap an SLM and one has an SLM within 20 bp (Fig. 5b,c, Supplementary Fig. 5 and Supplementary Data 3), a much higher fraction (17%) than expected by random chance (Fisher’s exact test, P = 1.40x10-8), as only 0.9% of nuclear genes are within 20 bp of an SLM. Furthermore, all four of the SLM-associated genes that are overexpressed in drm1drm2 meiocytes and significantly differentially expressed between meiocytes and leaves are suppressed in meiocytes compared to leaves (Fig. 5c and Supplementary Data 3). Expression levels of these genes in leaves are not elevated by RdDM mutation (Supplementary Data 3). These data indicate that RdDM-mediated SLMs specifically regulate gene expression in meiocytes.
Pre-tRNA genes encoding specific anti-codons are hypermethylated in the male sex lineage
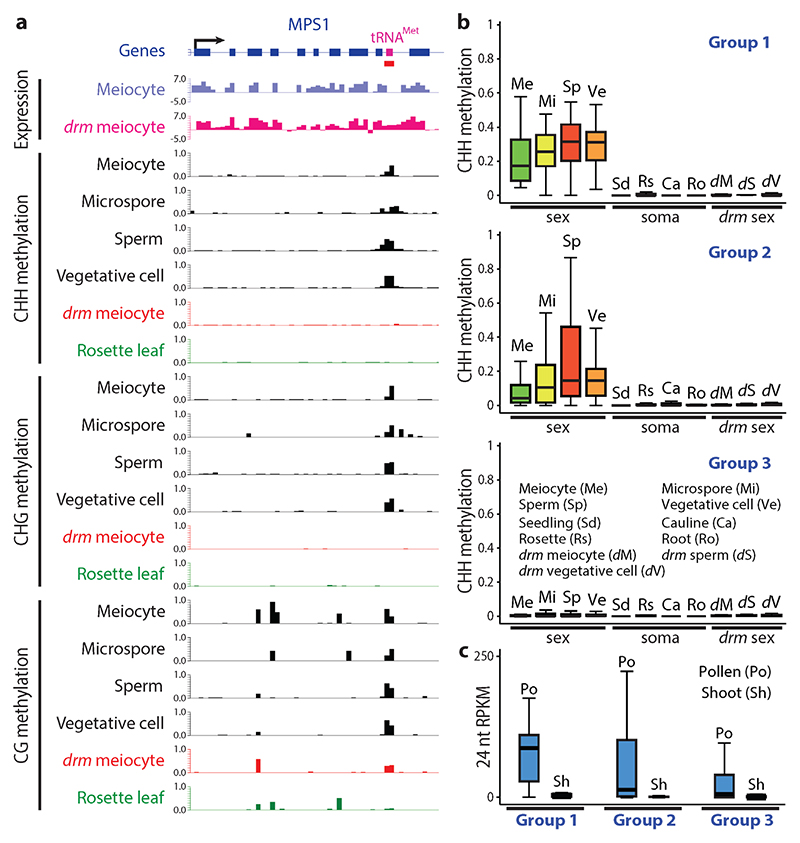
Among the genes containing SLMs, an unexpected group is comprised of pre-tRNA genes. 24 pre-tRNA loci overlap SLMs, with preference for specific anti-codons: for example, 75% and 21% of the phenylalanine and methionine pre-tRNA genes are covered by SLMs (Fig. 6a, Supplementary Fig. 6 and Supplementary Table 3a); numbers that are substantially higher than expected by random chance (both P < 2.63x10-6, Fisher’s exact test). As our criteria for calling SLMs are very stringent, we performed a genome-wide analysis to specifically detect sexual lineage hypermethylation of pre-tRNA genes. We found an additional set of 60 pre-tRNAs with significantly more CHH and CHG methylation in at least two of the sex cells in comparison to somatic tissues, and in wild-type sex cells in comparison to drm1drm2 sex cells (both P < 0.001, Fisher’s exact test; Fig. 6b, Supplementary Fig. 7 and Supplementary Table 3b). Together, the 84 hypermethylated loci include 100%, 75%, 73% and 42% of the phenylalanine, valine, cysteine and methionine pre-tRNA genes, respectively (Supplementary Table 3b). Consistently, 24-nt sRNAs are enriched at these pre-tRNA genes in pollen, but not in shoots (Fig. 6c). The preferential hypermethylation of certain pre-tRNA genes, together with the recent discovery of small tRNA fragments in Arabidopsis pollen33, suggest that tRNA biology may have interesting aspects that are particular to sex cells.
Figure 6. Pre-tRNA genes are hypermethylated in the male sexual lineage.
a, Snapshots of transcription and DNA methylation (similar to Fig. 5b) at the methionine pre-tRNA locus (magenta box) in the last intron of the MPS1 gene. drm, drm1drm2; SLM is underlined in red. Methylation patterns in other somatic tissues and drm sex cells are shown in Supplementary Fig. 6b. b, Box plots showing the absolute CHH methylation at three groups of pre-tRNA genes in sex cells (Me, meiocyte; Mi, microspore; Sp, sperm; Ve, vegetative cell), somatic tissues (Sd, seedling; Rs, rosette leaf; Ca, cauline leaf; Ro, root), and drm mutant sex cells (dM, drm meiocyte; dS, drm sperm; dV, drm vegetative cell). Group 1, the 24 pre-tRNA genes that overlap SLMs; Group 2, the additional 60 genes hypermethylated in the sexual lineage by RdDM; Group 3, the remaining 605 nuclear pre-tRNA genes. c, Box plots demonstrating the abundance of 24 nucleotide (nt) small RNA in pollen (Po) or shoot (Sh) at the abovementioned three groups of pre-tRNA genes.
An SLM regulates the splicing of MPS1 and is important for meiosis
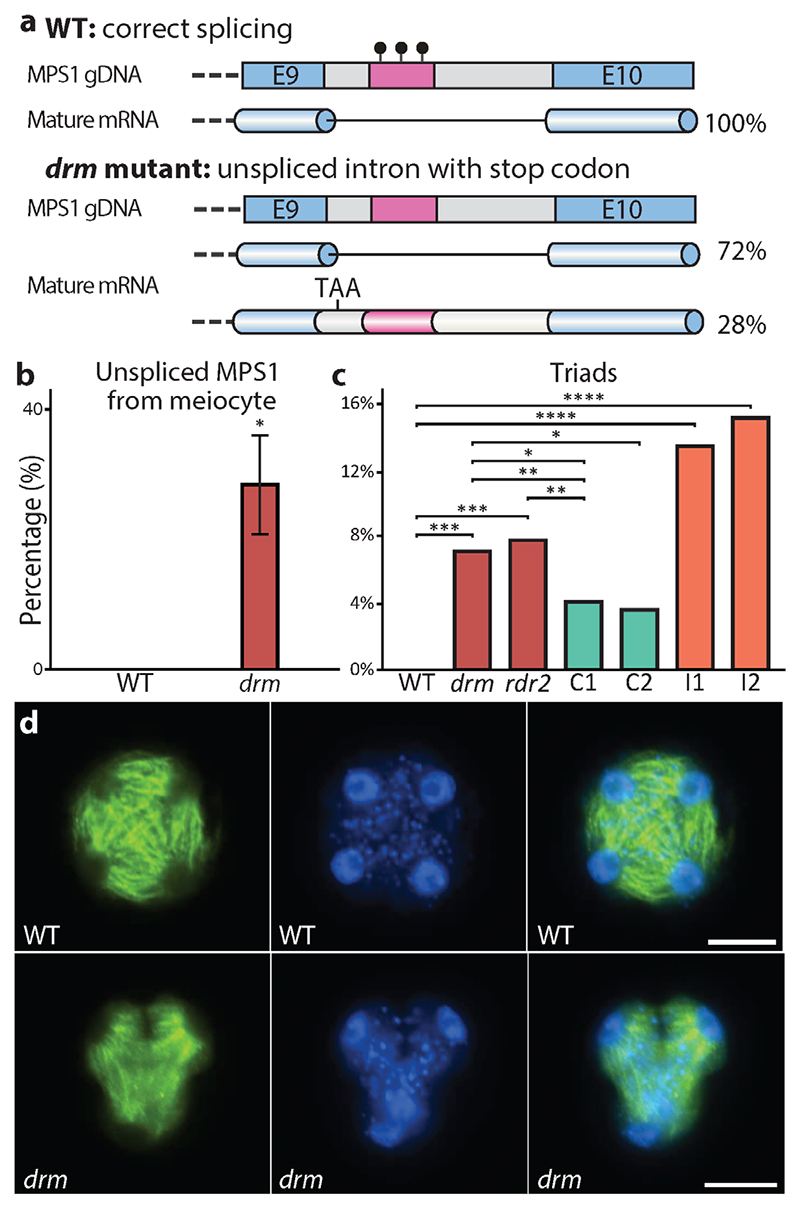
One SLM-covered methionine pre-tRNA gene attracted our attention because it occurs within the last intron (between exons 9 and 10) of another gene called MULTIPOLAR SPINDLE 1 (MPS1; Figs 6a and 7a, and Supplementary Fig. 6b). Given the emerging evidence of DNA methylation’s influence on splicing in plants34,35 and animals36,37, we examined our RNA-seq data to determine if the methylation status of this pre-tRNA locus affects MPS1 splicing. Indeed, we detected cDNA reads that indicate incorrect splicing of MPS1 at the last intron in drm1drm2 mutant meiocytes (Fig. 6a). Quantitative RT-PCR analysis demonstrated that 28% of the mature MPS1 mRNA retains the last intron in drm1drm2 mutant meiocytes, whereas no such retention occurs in wild-type (Fig. 7a,b), confirming that the SLM within the intron is required for correct splicing of the MPS1 transcript.
Figure 7. RdDM is important for the splicing of MPS1 and normal meiosis.
a, Gene model illustrating that the methionine pre-tRNA SLM (magenta bar) located in the last intron of MPS1 affects the splicing of this intron. E, exon; black lollipops, DNA methylation. b, Quantitative RT-PCR showing the percentage of unspliced MPS1 transcript in wild type (WT) and drm1drm2 (drm) mutant meiocytes. *P < 0.02 (t-test). c, Percentage of meiotic triads in WT, drm and rdr2 mutants, two complementation lines (C1 and C2) and two interference lines (I1 and I2) (***P < 1x 10-7, **P < 0.02, *P < 0.05, ****P < 1x 10-14; Fisher’s exact test; C1, 514 observations; C2, 167 observations; for observation numbers of other genotypes refer to Supplementary Fig. 8 legend. d, Spindles (green) and nuclei (blue) of WT (tetrad) and drm (triad) meiotic products at tetrad stage. Scale bars, 10 μm.
We were intrigued by aberrant splicing of MPS1 RNA in meiocytes because this gene is required for Arabidopsis meiosis38,39, and retention of the last intron introduces a premature stop codon (Fig. 7a). Furthermore, one of the described loss-of-function alleles affects splicing between exons 9 and 10, and another is an insertion in the intervening intron, indicating that exon 10 is essential for MPS1 activity39. We therefore analyzed meiotic progression in drm1drm2 and rdr2 mutants. Loss of MPS1 causes polyads – meiotic products numbering other than four38. Consistent with this phenotype, we found a significant occurrence of cellular triads in RdDM mutants (7.1% and 7.8% in drm1drm2 and rdr2, respectively; Fig. 7c,d and Supplementary Fig. 8a-c). We also observed pentads in drm1drm2 and rdr2 mutants (Supplementary Fig. 8d,e), as has been reported for other RdDM mutants40, whereas we did not observe triads or pentads in wild type. Introduction of an MPS1 transgene lacking the last intron into the drm1drm2 background reduced the number of meiotic triads (4.1% and 3.6% for two independent complementation lines; Fig. 7c), but not to the undetectable level of wild-type plants. The persistence of triads suggested that the mis-spliced MPS1 mRNA produces a protein that interferes with meiosis. To test this hypothesis, we introduced an MPS1 transgene with mutations that prevent splicing of the last intron into wild-type plants (Supplementary Fig. 8f). The resulting transgenic plants exhibited a substantially higher percentage of meiotic triads than drm1drm2 or rdr2 mutants (13.4% and 15.1% for two independent interference lines; Fig. 7c and Supplementary Fig. 8g). Our results indicate that loss of methylation at the SLM within the last intron of MPS1 causes intron retention and the production of an aberrant MPS1 protein that interferes with meiosis.
Discussion
Our results reveal the presence of a specific DNA methylation signature in the Arabidopsis male sexual lineage mediated by the RdDM pathway. SLMs suppress gene transcription and promote the splicing of a gene essential for meiosis, and are required for normal meiotic progression. This demonstrates that developmental gene regulation through DNA methylation reprogramming is not confined to gamete companion cells in flowering plants, and can occur through the deposition as well as the removal of methylation. Because RdDM appears to be ubiquitous in plant tissues, modulation of the RdDM pathway that achieves cell-specific methylation patterns can plausibly occur in any cell type. The epigenetic regulatory paradigm we describe here may therefore be broadly applicable to plant development.
SLMs are the product of developmentally orchestrated remodeling of DNA methylation via the RdDM pathway, but the small number of genes directly controlled by SLMs suggest that gene regulation is not the only, and perhaps not the main function of this remodeling. RdDM is known to target transposons2,6,7, there is a characterized tradeoff between methylation of transposons and gene expression41, and transposon suppression should be particularly important in cells that contribute to the next generation26. Therefore, RdDM may be balanced more aggressively in sex cells to ensure transposon silencing, even at the expense of gene expression – a phenomenon that might also occur in the shoot apical meristem, which gives rise to all above-ground plant cell types, including the gametes42. This would explain why we see sexual-lineage specific methylation, but very little if any soma-specific methylation. A more aggressive setting of the self versus non-self threshold in sex cells would also explain why sexual-lineage specific RdDM targets genes with such high frequency. Most SLMs may therefore be functionally neutral, or even slightly deleterious, and are likely evolutionarily transient, but a few, such as the one in MPS1, would be expected to confer a benefit and be retained through selection.
The substantial number (253) of SLMs that overlap genes in the Arabidopsis genome may elucidate a longstanding mystery regarding plant DNA methylation. The genes of flowering plants frequently exhibit CG-specific methylation of unclear origin and function2. This methylation has been hypothesized to arise due to transient RdDM activity43, which would have to occur in cells that contribute to the next generation – a description that fits SLMs. The remaining somatic CG methylation at SLMs (Fig. 4b and Supplementary Fig. 2d,e), which is maintained during somatic development without RdDM (Fig. 4c), provides evidence in support for this hypothesis. SLMs cover only a small fraction of the over 4000 genes with body methylation44,45, indicating that most body methylated genes are not presently targeted by RdDM in the male sexual lineage. However, shifting patterns of SLMs over thousands of generations could have plausibly created the existing gene body methylation pattern due to the strong trans-generational heritability of CG methylation6.
Methods
Isolation of A. thaliana meiocytes, sperm and vegetative cell nuclei
A. thaliana plants of Col-0 ecotype were grown under 16h light/8h dark in a growth chamber (21°C, 70% humidity). Stage 9 flower buds were collected and gently squeezed between a glass slide and coverslip. The released meiocytes (of meiotic prophase I) were examined carefully under a microscope. Clean meiocytes free from any somatic cell debris were transferred to a new slide using capillary glass pipettes, washed by 1 x PBS buffer 3 times, and frozen in liquid nitrogen. Sperm cells and vegetative cell nuclei were isolated as described previously18.
Sequencing library construction and analysis
Single-end bisulfite sequencing libraries for Illumina sequencing were constructed using the Ovation Ultralow Methyl-Seq Library Systems (Nugen, #0336) and EpiTect Fast Bisulfite Conversion (Qiagen, #59802) kits according to the kit protocols, except the incorporation of two rounds of bisulfite conversion. Bisulfite libraries were constructed from 2 biological replicates of wild-type (WT) meiocytes, drm1-2drm2-2 (drm1drm2) mutant meiocytes, and rdr2-1 mutant sperm and vegetative cell nuclei. Strand-specific RNA sequencing libraries were prepared using ScriptSeq v2 RNA-Seq (Illumina, #SSV21106) Library Preparation and Ovation RNA-Seq Systems for Model Organisms Arabidopsis (Nugen, #0351) kits, from 3 and 5 biological replicates of WT and drm1drm2 mutant meiocytes, respectively, and 3 and 1 biological replicates of WT and drm1drm2 mutant rosette leaf (from 40-day old plants). Bisulfite sequencing data from WT microspore17, sperm18, vegetative cell18, and embryo18, and drm1drm2 mutant sperm and vegetative cell28 were used. Bisulfite sequencing data from 4 WT somatic tissues (in this section specifically referring to: cauline leaf46, rosette leaf46,47, roots46,48 and seedlings48), rdd mutant rosette leaf29, drm2 and rdr2 mutant seedlings48, and drd1 mutant roots48 were obtained from published sources. Published sRNA data49,50, and bisulfite sequencing data from various root cell types19, and seedlings of met1-1, pMET1::MET1 met1-1 complementation lines (T-MET1a and b) and their WT control plants30 were also used in this study.
Sequencing was performed at the DNA Sequencing Facility of the University of Cambridge Department of Biochemistry, BGI Tech Solutions Ltd. and the Vincent J. Coates Genomic Sequencing Laboratory at UC Berkeley. DNA methylation analysis was performed as previously described18, transcriptome analysis was performed using the Tophat-2.0.10 and Cufflinks-2.2.1 packages, sRNA abundances were calculated using Reads Per Kilobase per Million (RPKM; Fig. 3b) or Reads Per Million (RPM; Fig. 6c) of mapped 24-nt sRNA reads filtered for rRNAs.
Transposon and gene meta analysis (ends analysis)
This was performed as described previously18.
Identification of differentially methylated loci between the sexual lineage and somatic tissues
Fractional methylation in 50 bp windows across the genome was compared between an average of selected sex cells (SexAV) and an average of the four somatic tissues (SomAV) (Diff = SexAV - SomAV). CG and CHG methylation averages in sex cells were calculated using meiocytes, microspores and sperm, and CHH methylation average was calculated using microspores and sperm. We first selected windows meeting the following criterion: Diff_CG > 0 & Diff_CHG > 0 & Diff_CHH > 0 & (Diff_CG + Diff_CHG + Diff_CHH) > 0.3. The selected windows were merged to generate larger SLMs if they occurred within 100 bp. Merged SLMs were retained if they covered at least 100 bp, with significantly different levels of total methylation (Fisher’s exact test P-value < 0.001), having more methylation in all sex cell replicates than all somatic tissues, and met the following criterion: Diff_CG > 0 & Diff_CHG > 0.05 & Diff_CHH > 0.1 & (Diff_CG + Diff_CHG + Diff_CHH) > 0.4. This resulted in the identification of 1265 SLHs (used in Fig. 3a,b). The same criteria, except reversing the relationship between sexual lineage and somatic methylation, were used to identify 36 loci hypomethylated in the sexual lineage. SLHs were further refined by the criterion of having significantly (Fisher’s exact test P-value < 0.001) less methylation in sex cells (meiocyte, sperm and vegetative cell) of drm1drm2 mutant in comparison to those of WT, leaving 1257 loci as a refined list of SLHs for further analyses (used in Fig. 3c). Refer to Supplementary Data 1 for abovementioned lists of loci.
The refined list of SLHs (1257 loci) was then separated into two groups based on the level of CHH/G methylation in somatic tissues: 1) SLMs with CHH and CHG methylation lower than 0.05 and 0.1, respectively, in all 4 somatic tissues (533 loci; Supplementary Data 1; used in Supplementary Fig. 3); 2) canonical SLHs with CHH methylation higher than 0.05 or CHG methylation higher than 0.1, in any of the 4 somatic tissues (724 loci; Supplementary Data 1; used in Fig. 4a). Both groups were analysed for overlap with published columella root cap DMRs (a merged list from reported C and CHH/G DMRs19): 173 and 469 of the canonical SLHs and SLMs have less than 10% overlap, respectively (Supplementary Data 1). The non-overlapping 469 SLMs were used in subsequent analyses (Figs 4b-f, 5 and 6, and Supplementary Figs 4-7) as a refined list of SLMs.
Identification of RdDM targets in sexual lineage and somatic tissues
Fractional methylation differences in 50 bp windows across the genome was compared between an average of RdDM mutants and WT tissues. For RdDM targets in sex cells, differential methylation was calculated using the average methylation in meiocyte, sperm and vegetative cells of WT subtracted by that of the drm1drm2 mutant; for targets in somatic tissues, differential methylation was calculated using the average methylation in the 4 WT somatic tissues subtracted by the average methylation in drm2 and rdr2 mutant seedlings. For both targets, 50 bp windows meeting these criteria were selected: with a differential methylation larger than 0.05, 0.1 and 0.05 in CG, CHG and CHH contexts, respectively, and with a total differential methylation in CG, CHG and CHH contexts exceeding 0.2. Selected windows were subsequently merged to generate larger islands if they occurred within 100 bp. These islands were retained if they are at least 100 bp, with significantly different levels of total methylation (Fisher’s exact test P-value < 0.001), and meet the above-mentioned two criteria, except the 2nd criterion uses a stricter cutoff of 0.3. This resulted in 9051 and 9993 RdDM targets in sex cells and somatic tissues, respectively.
Box plots
All box plots follow this format: each box encloses the middle 50% of the distribution, with the horizontal line making the median, and vertical lines marking the minimum and maximum values that fall within 1.5 times the height of the box. Fig. 2b was generated using 50 bp windows with fractional CHH methylation larger than 0.3 in meiocytes compared to rosette leaf, and at least 20 informative sequenced cytosines in each of the 4 somatic tissues and 4 sex cells (meiocyte, microspore, sperm and vegetative cells). Fig. 3c was generated using 50 bp windows with substantial methylation in either WT sex cells or rosette leaf in the corresponding sequence context (cutoff settings: 0.3, 0.3 and 0.1 in CG, CHG and CHH contexts, respectively), and at least 10 informative sequenced cytosines in each replicate of the WT and rdd rosette leaf sample and 3 sex cells (meiocyte, microspore and sperm).
RT-PCR
100 ng and 500 ng total RNA was reverse transcribed using RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher scientific, #K1621) for quantitative RT-PCR (qRT-PCR; Figs 5c and 7b) and RT-PCR (Supplementary Fig. 8f), respectively. qRT-PCR was performed using SYBR Green (Roche, #4707516001) in triplicate on the LightCycler 480 Real-Time PCR System (Roche) and Cp values were averaged between 3 technical replicates to determine the target/reference ratio. Figs 5c and 7b show the averages of at least 3 biological replicates for each genotype or tissue type. RT-PCR was performed with 46 PCR cycles using primers JW134 and JW141 for MPS1, and 30 cycles using primers PHG34 and PHG101 for the control ACTIN8. ACTIN8 was used as internal control in both RT- and qRT-PCRs, and all primers are listed in Supplementary Table 4.
Meiosis microscopy
Slides were prepared from fixed Arabidopsis buds as previously described51. Cells were visualized and analyzed using a Nikon Ni-E Eclipse fluorescent microscope (Nikon UK Ltd.) with NIS-elements imaging software. Triads were defined as cells with three nuclei that were observed without a boundary of overlap.
Microtubule immunolocalization
Anthers were dissected from buds of the correct size (0.4-0.6mm long) and incubated in m-maleimidobenzoyl-N-hydroxysuccinimide ester (1mM in 1 x PBS and 0.05% [v/v] Triton X-100) for 30min52. Anthers were then fixed in 4% paraformaldehyde at 4°C for 1h. After one wash in 1 x PBS, anthers were digested in 0.5% (w/v) cytohelicase (Sigma), 0.3% (w/v) cellulase (Melford) and 0.3% (w/v) pectolyase (MP Biomedicals) in citrate buffer at 37°C for 75min. The enzyme mixture was then replaced with 1.5% Lipsol (Fisher Scientific) and transferred to Superfrost Plus slides (VWR). The digested anthers were spread with a mounted needle and then a coverslip was added. The slide was turned upside down and squashed onto the bench and then transferred to liquid nitrogen for 30s. The cover slip was immediately removed using a razor blade and the slide was allowed to air dry for 20min. An anti-α-Tubulin (Biorad) antibody (1:100 dilution in 1 x PBS and 1% BSA) was directly added to the slide and incubated overnight at 4°C. The slide was washed in 2 x 5min in 1 x PBS 0.1% Triton X-100 and then an anti-rat 488 DyLight (Vector labs) antibody (1:100 dilution in 1 x PBS and 1% BSA) was incubated for 30min at 37°C. The slide was then washed 2 x 5min in 1 x PBS 0.1% Triton X-100 and the excess buffer was allowed to drain off. DAPI in Vectashield (Vector labs) mounting medium was added to the slide.
Construction of MPS1 complementation and interference lines
To construct the MPS1 complementation lines, overlapping PCR was carried out using primers PHG102, PHG103, PHG104 and PHG105 (Supplementary Table 4) to amplify the full MPS1 genomic region covering the promoter, coding region and 3’ region with the exception of the last intron. To construct the MPS1 interference lines with splicing site mutations, DNA was amplified by PCR for the full MPS1 genomic region, using primers PHG102 and PHG103. Two point mutations at the splicing sites of the last intron were created using the QuikChange Lightning Multi Site-Directed Mutagenesis Kit (Agilent, #210515) using primers PHG106 and PHG107. Both entry clones were sequenced and cloned into destination vectors pGWB13-Bar (a modified pGWB13 53 vector with BASTA resistance gene replacing Hygromycin and Kanamycin resistance genes) and pMDC107-NTF (a modified pMDC107 vector with NTF54 replacing mGFP6) via Gateway cloning, and transformed into drm1drm2 mutant and wild-type plants to generate the complementation lines (pGWB13 for C1 and pMDC107 for C2) and interference lines (pGWB13 for I1 and pMDC107 for I2). 5, 2, 5 and 4 T1 generation plants of the C1, C2, I1 and I2 lines, respectively, were used in meiotic phenotype analysis.
Supplementary Material
Acknowledgments
We thank Daniel Zilberman for intellectual contributions to this work and assistance with manuscript preparation. We also thank Caroline Dean, Kirsten Bomblies, Vinod Kumar, Siobhan Brady and Sophien Kamoun for comments on the manuscript, Hugh Dickinson and Josephine Hellberg for developing the meiocyte isolation method, Giles Oldroyd for the pGWB13-Bar vector, Elisa Fiume for the pMDC107-NTF vector, Matthew Hartley, Matthew Couchman and Tjelvar Sten Gunnar Olsson for bioinformatics support, and the John Innes Centre Bioimaging Facility (Elaine Barclay and Grant Calder) for their assistance with microscopy. This work was funded by a Biotechnology and Biological Sciences Research Council (BBSRC) David Phillips Fellowship (BBL0250431) to X.F., a BBSRC grant (BBM01973X1) to J.H., and a Sainsbury PhD Studentship to J.W.
Footnotes
Author Contributions
X.F. designed the study, J.W., H.G., J.Z., B.A., K.F., J.H. and X.F. performed the experiments, J.W., H.G., M.V. and X.F. analyzed the data, X.F. wrote the manuscript.
Sequencing data have been deposited in GEO (GSE86583).
The authors declare no competing financial interests.
References
- 1.Zemach A, Zilberman D. Evolution of eukaryotic DNA methylation and the pursuit of safer sex. Curr Biol. 2010;20:R780–785. doi: 10.1016/j.cub.2010.07.007. [DOI] [PubMed] [Google Scholar]
- 2.Law JA, Jacobsen SE. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat Rev Genet. 2010;11:204–220. doi: 10.1038/nrg2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.He XJ, Chen T, Zhu JK. Regulation and function of DNA methylation in plants and animals. Cell Res. 2011;21:442–465. doi: 10.1038/cr.2011.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Du J, Johnson LM, Jacobsen SE, Patel DJ. DNA methylation pathways and their crosstalk with histone methylation. Nat Rev Mol Cell Biol. 2015;16:519–532. doi: 10.1038/nrm4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pikaard CS, Mittelsten Scheid O. Epigenetic regulation in plants. Cold Spring Harb Perspect Biol. 2014;6:a019315. doi: 10.1101/cshperspect.a019315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Matzke MA, Mosher RA. RNA-directed DNA methylation: an epigenetic pathway of increasing complexity. Nat Rev Genet. 2014;15:394–408. doi: 10.1038/nrg3683. [DOI] [PubMed] [Google Scholar]
- 7.Kim MY, Zilberman D. DNA methylation as a system of plant genomic immunity. Trends Plant Sci. 2014;19:320–326. doi: 10.1016/j.tplants.2014.01.014. [DOI] [PubMed] [Google Scholar]
- 8.Lipka DB, et al. Identification of DNA methylation changes at cis-regulatory elements during early steps of HSC differentiation using tagmentation-based whole genome bisulfite sequencing. Cell Cycle. 2014;13:3476–3487. doi: 10.4161/15384101.2014.973334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dawlaty MM, et al. Loss of Tet enzymes compromises proper differentiation of embryonic stem cells. Dev Cell. 2014;29:102–111. doi: 10.1016/j.devcel.2014.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kubo N, et al. DNA methylation and gene expression dynamics during spermatogonial stem cell differentiation in the early postnatal mouse testis. BMC Genomics. 2015;16:624. doi: 10.1186/s12864-015-1833-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Slieker RC, et al. DNA Methylation Landscapes of Human Fetal Development. Plos Genet. 2015;11:e1005583. doi: 10.1371/journal.pgen.1005583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xie W, et al. Epigenomic analysis of multilineage differentiation of human embryonic stem cells. Cell. 2013;153:1134–1148. doi: 10.1016/j.cell.2013.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cacchiarelli D, et al. Integrative Analyses of Human Reprogramming Reveal Dynamic Nature of Induced Pluripotency. Cell. 2015;162:412–424. doi: 10.1016/j.cell.2015.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Benner C, Isoda T, Murre C. New roles for DNA cytosine modification, eRNA, anchors, and superanchors in developing B cell progenitors. Proc Natl Acad Sci U S A. 2015;112:12776–12781. doi: 10.1073/pnas.1512995112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rodrigues JA, Zilberman D. Evolution and function of genomic imprinting in plants. Genes Dev. 2015;29:2517–2531. doi: 10.1101/gad.269902.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Park K, et al. DNA demethylation is initiated in the central cells of Arabidopsis and rice. Proc Natl Acad Sci U S A. 2016;113:15138–15143. doi: 10.1073/pnas.1619047114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Calarco JP, et al. Reprogramming of DNA methylation in pollen guides epigenetic inheritance via small RNA. Cell. 2012;151:194–205. doi: 10.1016/j.cell.2012.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ibarra CA, et al. Active DNA demethylation in plant companion cells reinforces transposon methylation in gametes. Science. 2012;337:1360–1364. doi: 10.1126/science.1224839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kawakatsu T, et al. Unique cell-type-specific patterns of DNA methylation in the root meristem. Nat Plants. 2016;2:16058. doi: 10.1038/nplants.2016.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hsieh TF, et al. Genome-wide demethylation of Arabidopsis endosperm. Science. 2009;324:1451–1454. doi: 10.1126/science.1172417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Secco D, et al. Stress induced gene expression drives transient DNA methylation changes at adjacent repetitive elements. Elife. 2015;4 doi: 10.7554/eLife.09343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jiang C, et al. Environmentally responsive genome-wide accumulation of de novo Arabidopsis thaliana mutations and epimutations. Genome Res. 2014;24:1821–1829. doi: 10.1101/gr.177659.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dowen RH, et al. Widespread dynamic DNA methylation in response to biotic stress. Proc Natl Acad Sci U S A. 2012;109:E2183–2191. doi: 10.1073/pnas.1209329109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bilichak A, Ilnystkyy Y, Hollunder J, Kovalchuk I. The progeny of Arabidopsis thaliana plants exposed to salt exhibit changes in DNA methylation, histone modifications and gene expression. PLoS One. 2012;7:e30515. doi: 10.1371/journal.pone.0030515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wibowo A, et al. Hyperosmotic stress memory in Arabidopsis is mediated by distinct epigenetically labile sites in the genome and is restricted in the male germline by DNA glycosylase activity. Elife. 2016;5 doi: 10.7554/eLife.13546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Feng X, Zilberman D, Dickinson H. A conversation across generations: soma-germ cell crosstalk in plants. Dev Cell. 2013;24:215–225. doi: 10.1016/j.devcel.2013.01.014. [DOI] [PubMed] [Google Scholar]
- 27.Kawashima T, Berger F. Epigenetic reprogramming in plant sexual reproduction. Nat Rev Genet. 2014;15:613–624. doi: 10.1038/nrg3685. [DOI] [PubMed] [Google Scholar]
- 28.Hsieh PH, et al. Arabidopsis male sexual lineage exhibits more robust maintenance of CG methylation than somatic tissues. Proc Natl Acad Sci U S A. 2016;113:15132–15137. doi: 10.1073/pnas.1619074114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stroud H, Greenberg MV, Feng S, Bernatavichute YV, Jacobsen SE. Comprehensive analysis of silencing mutants reveals complex regulation of the Arabidopsis methylome. Cell. 2013;152:352–364. doi: 10.1016/j.cell.2012.10.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Catoni M, et al. DNA sequence properties that predict susceptibility to epiallelic switching. Embo J. 2017;36:617–628. doi: 10.15252/embj.201695602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen C, et al. Meiosis-specific gene discovery in plants: RNA-Seq applied to isolated Arabidopsis male meiocytes. BMC Plant Biol. 2010;10:280. doi: 10.1186/1471-2229-10-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang H, Lu P, Wang Y, Ma H. The transcriptome landscape of Arabidopsis male meiocytes from high-throughput sequencing: the complexity and evolution of the meiotic process. Plant J. 2011;65:503–516. doi: 10.1111/j.1365-313X.2010.04439.x. [DOI] [PubMed] [Google Scholar]
- 33.Martinez G, Choudury SG, Slotkin RK. tRNA-derived small RNAs target transposable element transcripts. Nucleic Acids Res. 2017 doi: 10.1093/nar/gkx103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang X, et al. DNA Methylation Affects Gene Alternative Splicing in Plants: An Example from Rice. MolPlant. 2016;9:305–307. doi: 10.1016/j.molp.2015.09.016. [DOI] [PubMed] [Google Scholar]
- 35.Regulski M, et al. The maize methylome influences mRNA splice sites and reveals widespread paramutation-like switches guided by small RNA. Genome Res. 2013;23:1651–1662. doi: 10.1101/gr.153510.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Naftelberg S, Schor IE, Ast G, Kornblihtt AR. Regulation of alternative splicing through coupling with transcription and chromatin structure. Annu Rev Biochem. 2015;84:165–198. doi: 10.1146/annurev-biochem-060614-034242. [DOI] [PubMed] [Google Scholar]
- 37.Lev Maor G, Yearim A, Ast G. The alternative role of DNA methylation in splicing regulation. Trends Genet. 2015;31:274–280. doi: 10.1016/j.tig.2015.03.002. [DOI] [PubMed] [Google Scholar]
- 38.Jiang H, et al. MULTIPOLAR SPINDLE 1 (MPS1), a novel coiled-coil protein of Arabidopsis thaliana, is required for meiotic spindle organization. Plant J. 2009;59:1001–1010. doi: 10.1111/j.1365-313X.2009.03929.x. [DOI] [PubMed] [Google Scholar]
- 39.De Muyt A, et al. A high throughput genetic screen identifies new early meiotic recombination functions in Arabidopsis thaliana. Plos Genet. 2009;5:e1000654. doi: 10.1371/journal.pgen.1000654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Oliver C, Santos JL, Pradillo M. Accurate Chromosome Segregation at First Meiotic Division Requires AGO4, a Protein Involved in RNA-Dependent DNA Methylation in Arabidopsis thaliana. Genetics. 2016;204:543–553. doi: 10.1534/genetics.116.189217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hollister JD, Gaut BS. Epigenetic silencing of transposable elements: a trade-off between reduced transposition and deleterious effects on neighboring gene expression. Genome Res. 2009;19:1419–1428. doi: 10.1101/gr.091678.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Baubec T, Finke A, Mittelsten Scheid O, Pecinka A. Meristem-specific expression of epigenetic regulators safeguards transposon silencing in Arabidopsis. EMBO Rep. 2014;15:446–452. doi: 10.1002/embr.201337915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tran RK, et al. DNA methylation profiling identifies CG methylation clusters in Arabidopsis genes. Curr Biol. 2005;15:154–159. doi: 10.1016/j.cub.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 44.Takuno S, Gaut BS. Body-methylated genes in Arabidopsis thaliana are functionally important and evolve slowly. Mol Biol Evol. 2012;29:219–227. doi: 10.1093/molbev/msr188. [DOI] [PubMed] [Google Scholar]
- 45.Zilberman D, Gehring M, Tran RK, Ballinger T, Henikoff S. Genome-wide analysis of Arabidopsis thaliana DNA methylation uncovers an interdependence between methylation and transcription. Nat Genet. 2007;39:61–69. doi: 10.1038/ng1929. [DOI] [PubMed] [Google Scholar]
- 46.Coleman-Derr D, Zilberman D. Deposition of histone variant H2A.Z within gene bodies regulates responsive genes. Plos Genet. 2012;8:e1002988. doi: 10.1371/journal.pgen.1002988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stroud H, et al. Non-CG methylation patterns shape the epigenetic landscape in Arabidopsis. Nat Struct Mol Biol. 2014;21:64–72. doi: 10.1038/nsmb.2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zemach A, et al. The Arabidopsis nucleosome remodeler DDM1 allows DNA methyltransferases to access H1-containing heterochromatin. Cell. 2013;153:193–205. doi: 10.1016/j.cell.2013.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liang C, Liu X, Sun Y, Yiu SM, Lim BL. Global small RNA analysis in fast-growing Arabidopsis thaliana with elevated concentrations of ATP and sugars. BMC Genomics. 2014;15:116. doi: 10.1186/1471-2164-15-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Martinez G, Panda K, Kohler C, Slotkin RK. Silencing in sperm cells is directed by RNA movement from the surrounding nurse cell. Nat Plants. 2016;2:16030. doi: 10.1038/nplants.2016.30. [DOI] [PubMed] [Google Scholar]
- 51.Armstrong SJ, Sanchez-Moran E, Franklin FC. Cytological analysis of Arabidopsis thaliana meiotic chromosomes. Methods Mol Biol. 2009;558:131–145. doi: 10.1007/978-1-60761-103-5_9. [DOI] [PubMed] [Google Scholar]
- 52.Sonobe S, Shibaoka H. Cortical Fine Actin-Filaments in Higher-Plant Cells Visualized by Rhodamine-Phalloidin after Pretreatment with M-Maleimidobenzoyl N-Hydroxysuccinimide Ester. Protoplasma. 1989;148:80–86. [Google Scholar]
- 53.Nakagawa T, et al. Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J Biosci Bioeng. 2007;104:34–41. doi: 10.1263/jbb.104.34. [DOI] [PubMed] [Google Scholar]
- 54.Deal RB, Henikoff S. The INTACT method for cell type-specific gene expression and chromatin profiling in Arabidopsis thaliana. Nat Protoc. 2011;6:56–68. doi: 10.1038/nprot.2010.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.