Abstract
The HORMA domain protein REV7, also known as MAD2L2, interacts with a variety of proteins and thereby contributes to the establishment of different complexes. With doing so, REV7 impacts a diverse range of cellular processes and gained increasing interest as more of its activities became uncovered. REV7 has important roles in translesion synthesis and mitotic progression, and acts as a central component in the recently discovered shieldin complex that operates in DNA double-strand break repair. Here we discuss the roles of REV7 in its various complexes, focusing on its activity in genome integrity maintenance. Moreover, we will describe current insights on REV7 structural features that allow it to be such a versatile protein.
Keywords: REV7, HORMA, genome maintenance, translesion synthesis, shieldin
REV7: a versatile protein
Rev7 was initially identified in yeast (S. cerevisiae)[1–3]. Human REV7 was also named MAD2L2 (for MAD2-Like2), MAD2β or MAD2β, based on homology with MAD2[4–6]. These different names reflect the two distinct biological roles that for the first decades after its discovery were recognized as its main activities. First, REV7 was found to compose DNA polymerase ζ (Polζ) with REV3 and facilitate bypass of DNA lesions through translesion synthesis (TLS, see Glossary) together with REV1[7]. Second, homology with the spindle assembly checkpoint (SAC) protein MAD2 sparked exploration of a potential role of REV7 in mitotic regulation, revealing that REV7 interacts with the anaphase-promoting complex/cyclosome (APC/C)-cofactor CDH1[8, 9]. The actions of REV7 appeared not restricted to TLS or mitotic control. Various approaches found a wide variety of proteins to interact with REV7. Following that, not only knowledge on REV7 function in mitosis and TLS expanded, but also important novel roles of REV7 were discovered. REV7 is now also recognized as important for genome integrity by functioning in DNA double-strand break (DSB) repair as the first identified member of the shieldin complex, further consisting of SHLD1, SHLD2 and SHLD3[10–12]. REV7 itself does not harbor recognizable enzymatic activity. How it operates in distinct protein complexes and is regulated among different functions are intriguing questions. Here we discuss the current understanding of REV7 function, including the structural features of REV7 that allow it to be such a versatile protein.
Glossary.
- DNA double-strand break (DSB)
a DNA lesion in which the two strands of the DNA are no longer physically connected. DSBs can be caused by a variety of sources, including IR and collapsed replication forks and are lethal to the cell if left unrepaired.
- DNA repair pathway choice
DNA DSBs can be repaired through NHEJ and HR. Which of these two pathways is employed at the break site depends on multiple factors, and is commonly referred to as DNA repair pathway choice.
- Fanconi Anemia (FA)
the FA pathway specifically participates in DNA-crosslink repair. Mutations in FA-genes, such as FANCD1 (BRCA2), FANCS (BRCA1) and FANCR (RAD51), can result in a complex genetic disease, called Fanconi Anemia, with patients being characterized by congenital abnormalities, bone-marrow failure and increased cancer susceptibility[111].
- Interstrand crosslinks (ICLs)
DNA lesions that are particularly toxic to the cell as they covalently link the two DNA strands, giving rise to problems during replication and transcription.
- Homologous recombination (HR)
an error-free repair process for DNA DSBs. HR requires 5’ to 3’ resection of DNA at the break, followed by strand invasion into the intact sister chromatid, which is then used as a template for DNA synthesis. HR is restricted to the S/G2 phase of the cell cycle.
- Non-homologous end-joining (NHEJ)
a DSB repair process in which two DNA ends are ligated together after minimal processing and without guidance from a correct DNA sequence, risking introduction of small insertions or deletions. NHEJ can operate at any phase of the cell cycle.
- Shieldin
a newly identified protein complex consisting of REV7, SHLD1, SHLD2 and SHLD3. Shieldin is recruited to DNA DSBs downstream of 53BP1 and RIF1 and functions to counteract end-resection, thereby promoting repair through NHEJ.
- Spindle assembly checkpoint (SAC)
An active signal from improperly attached kinetochores that prevents anaphase onset until the microtubules from opposite spindle poles are properly attached to sister kinetochores on replicated chromosomes. The SAC thereby ensures correct chromosome segregation. This is enabled by the mitotic checkpoint complex (MCC) consisting of MAD2, BUBR1, BUB3 and CDC20. Upon SAC silencing, the MCC disassembles, releasing CDC20 through opening of the MAD2-safety belt. CDC20 then activates the APC/C to facilitate anaphase onset.
- Anaphase-promoting complex/cyclosome (APC/C)
E3 ubiquitin ligase that facilitates the timely ubiquitin-mediated proteosomal degradation of cell cycle regulators essential for proper cell cycle progression. CDC20 and CDH1 interact with APC/C to activate its E3 ligase activity towards different substrates at different stages during mitosis. The activity of the APC/CCDC20 is tightly regulated by the SAC to control the onset of anaphase.
- Translesion synthesis (TLS)
facilitates DNA synthesis past a DNA replication blocking lesion by incorporating a nucleotide opposite the lesion. This requires a two-step mechanism; 1) an inserter polymerase incorporates a nucleotide opposite the lesion and 2) an extender polymerase adds a few nucleotides extending from the mismatched base pair, after which replication will be restarted by replicative polymerases[47]. This can be performed error-free or in an erroneous manner.
REV7 is a HORMA domain protein
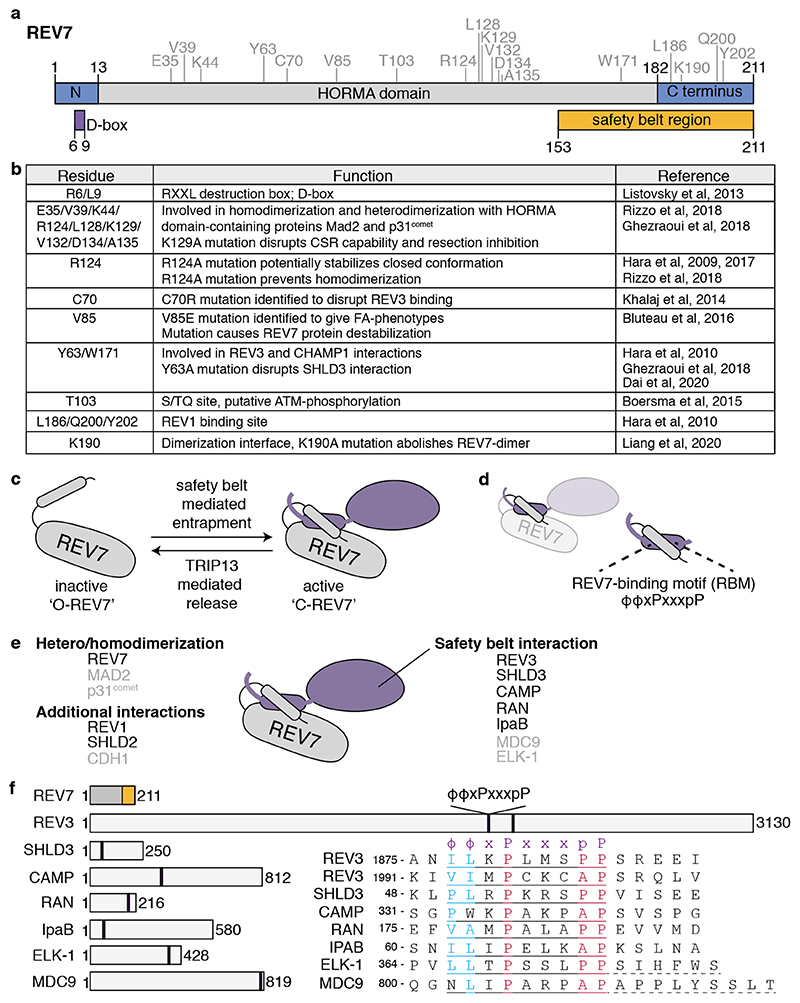
Human REV7 consists of 211 amino acids that mostly contribute to a HORMA domain (Figure 1a,b), defined by sequence conservation among yeast Hop1, Rev7 and Mad2[13, 14]. The HORMA domain is critical for REV7 activity in distinct complexes by enabling REV7 to entrap interacting peptides, dimerize, and engage in additional protein interactions.
Figure 1. REV7 is a HORMA domain containing protein.
a. REV7 domain structure. Important structural features are indicated below, such as the destruction box (D-box) and the safety belt region (a.k.a. seatbelt). REV7 amino acid residues that have been described to be important for REV7 structure and function are indicated above in grey and are shown in (b). b. Table showing the different residues important for REV7 function. c. Schematic representation on how REV7 entraps peptides through its safety belt. Upon entrapment, REV7 undergoes a conformational switch from inactive, open-REV7 (‘O-REV7’) to active closed-REV7 (‘C-REV7’). The reverse reaction is mediated by the action of TRIP13, which thereby facilitates partner release. d. The REV7 safety belt entraps a REV7 Binding Motif (RBM) on its partner. e. Schematic representation of the different interaction-surfaces used by REV7 to interact with other proteins, also indicating what proteins have already been identified to interact with REV7 through each domain. REV7 dimerization appears essential for the interaction with SHLD2, and thereby could provide REV7 with an extra interaction-surface. For proteins indicated in black, the crystal structure in complex with REV7 has been described. f. Schematic representation of REV7 and proteins for which an RBM has been described, with their sizes in amino acids. The amino acids of the respective RBMs and their relative position within the protein are indicated. The RBM is defined as ϕϕxPxxxpP, in which ϕ denotes an aliphatic residue[15]. For ELK-1 and MDC9 additional residues are needed for the interaction with REV7, as indicated with dashed underline[15].
HORMA proteins share a safety belt mechanism
HORMA domains consist of a ‘core region’ and a C-terminal ‘safety belt’ that can entrap a short peptide of an interacting protein[14]. Mechanistically this involves a topological change that converts the HORMA protein from an ‘open’ to ‘closed’ form in which the safety belt wraps around the ligand[14]. For REV7, this short peptide was defined as ϕϕxPxxxpP (Figure 1c-f), ‘ϕ‘ indicating aliphatic and proline ‘p’ being less important than ‘P’[15, 16]. This ‘REV7-binding motif’ (RBM) was initially described in REV3, but also exists in several other REV7-interacting proteins, including the chromosome alignment-maintaining phosphoprotein CAMP, the small GTPase RAN, IpaB and SHLD3 (Figure 1, Table 1)[15, 17–21]. Structural analysis of human REV7 in complex with a fragment of REV3, CAMP, RAN, IpaB or SHLD3 indeed showed their entrapment through the REV7 safety belt[17, 18, 22, 23]. Peptide-bound REV7 thereby resembles MAD2 in its closed conformation (C-MAD2)[17, 22]. The opposite, release of the interactor, involves conversion of the closed to an open safety belt. In this process, best described for MAD2, the AAA+ ATPase TRIP13 promotes the opening of C-MAD2 to open (O-)MAD2. This releases CDC20 from MAD2 which then activates the APC/C to facilitate progression into anaphase[24]. TRIP13 also acts towards the HORMA domain protein HORMAD (Hop1)[14, 24]. Although it was suggested earlier that the REV7 structure is dynamic, only recently also the ‘open’ conformation of REV7 was described[17, 25, 26]. Like for MAD2, conversion of closed to open REV7 requires TRIP13 and is facilitated by the p31comet adaptor protein, itself a HORMA domain protein[26–28]. However, while p31comet serves to bridge MAD2 and TRIP13, the mechanistic role of p31comet in the REV7-TRIP13 context is not yet clear and might differ, as TRIP13 and REV7 can interact without p31comet ([26], de Krijger et al., unpublished).
Table 1.
All interactions with REV7 confirmed by other assays, beyond mass spectrometry. Species refers to REV7 (H, human; M, mouse; Y, yeast). For each interactor the presence of a verified (by mutagenesis, ‘confirmed’) or potential RBM is indicated. RBMs are defined as ϕϕxPxxxpP[15], in which ϕ denotes an aliphatic residue. For the selection of potential RBMs, the absence of one out of two aliphatic residues is tolerated. In addition, an alanine (A) at the position of the second proline (p) is allowed. MS, mass spectrometry; CoIP, co-immunoprecipitation; Y2H, yeast-two-hybrid experiments; GST, pull-down experiments using GST-tagged proteins; FRET, fluorescence resonance energy transfer.
| Protein | Gene names | Full protein name | Pathway | Species | Detection method | (potential) RBM at aa: | Reference |
|---|---|---|---|---|---|---|---|
| 53BP1 | TP53BP1 | TP53-binding protein 1 | DNA damage; NHEJ | H | MS - CoIP | Not found | [112] |
| 9-1-1 clamp |
DDC1
RAD17 MEC3, PIP3, PSO9 |
DNA damage checkpoint protein 1 DNA damage checkpoint control protein Rad17 DNA damage checkpoint control protein Mec3 |
DNA damage checkpoint | Y | Y2H - GST - CoIP | - | [113] |
| ADP/E3-11.6K | - | Adenovirus death protein | Adenoviral infection; Cell lysis | H | Y2H - GST - CoIP | Not found | [73] |
| APC3 | CDC27, ANAPC3, D0S143OE, D17S978E | Cell division cycle protein 27 homolog | Mitotic regulation; APC/C | H | CoIP | 308-316 | [74] |
| CAMP | CHAMP1, C13orf8, CAMP, CHAMP, KIAA1802, ZNF828 | Chromosome alignment-maintaining phosphoprotein 1 | Mitosis; Chromosome segregation | H | MS - CoIP - structural data | 204-212, 297-305, 333-341 (confirmed), 531-539 | [17, 85] |
| CDC20 | CDC20 | Cell division cycle protein 20 homolog | Mitotic regulation; APC/C | H | CoIP | 16-24 | [9, 74] |
| CDH1 | FZR1, CDH1, FYR, FZR, KIAA1242 | Fizzy-related protein homolog | Mitotic regulation; APC/C | H/X | GST - CoIP | Not found | [8, 9, 74, 81, 83] |
| CDK1 | Cdk1, Cdc2, Cdc2a, Cdkn1 | Cyclin-dependent kinase 1 | Cell cycle regulator | M?1 | CoIP | Not found | [79] |
| CLTA | CLTA | Clathrin light chain A | Mitosis; Stabilization of kinetochore fibers | H | Y2H - GST - CoIP | 61-69 | [78, 114] |
| CST-complex |
CTC1, C17orf68 STN1, OBFC1 TEN1, C17orf106 |
CST-complex subunit CTC1, STN1 and TEN1 | DNA damage; NHEJ; Telomere maintenance |
H | CoIP - Y2H | Not found | [67] |
| DDK |
DBF4, DNA52 CDC7, OAF2 |
DDK kinase regulatory subunit DBF4 Cell division control protein 7 |
TLS | Y | CoIP | - | [90] |
| ELK-1 | ELK1 | ETS domain-containing protein Elk-1 | Transcription factor | H | Y2H - GST - CoIP | 366-374 | [15, 115] |
| G9A | Ehmt2, Bat8, G9a, Ng36 | Histone-lysine N-methyltransferase EHMT2 | Epigenetics; HMT | M? | CoIP | 249-257 (M) | [79] |
| GLP | Ehmt1, Euhmtase1, Glp, Kmt1d | Histone-lysine N-methyltransferase EHMT1 | Epigenetics; HMT | M? | CoIP | 562-570 (M) | [79] |
| HCCA2 | YY1AP1, HCCA2, YY1AP | YY1-associated protein 1 | Transcription | H | Y2H - GST - CoIP | Not found | [106] |
| HR23B | RAD23B | UV excision repair protein RAD23 homolog B | DNA repair; NER | H | MS - CoIP | Not found | [89] |
| IpaB | ipaB | Invasin IpaB (Shigella flexneri) | Bacterial infection; Shigella Effector | H | Y2H - GST - CoIP - structural data | 62-70 (confirmed) | [18, 74] |
| MAD2 | MAD2L1, MAD2 | Mitotic spindle assembly checkpoint protein MAD2A | Mitosis; SAC | H | Y2H - GST | Not found | [6, 32] |
| MDC9 | ADAM9, KIAA0021, MCMP, MDC9, MLTNG | Disintegrin and metalloproteinase domain-containing protein 9 | Metalloprotease-disintegrin | H | Y2H - GST | 802-810 | [5, 15] |
| NCOA3 | NCOA3, AIB1, BHLHE42, RAC3, TRAM1 | Nuclear receptor coactivator 3 | Transcriptional coactivator | H | MS - CoIP | Not found | [103] |
| p31(comet) | MAD2L1BP, CMT2, KIAA0110 | MAD2L1-binding protein | Mitosis/HORMA domain | H | Y2H - CoIP | Not found | [27, 32] |
| Pol31 | POL31, HUS2, HYS2, SDP5 | DNA polymerase delta small subunit | TLS; Polymerase ζ subunit | Y | Structural data | - | [33] |
| Pol32 | POL32 | DNA polymerase delta subunit 3 | TLS; Polymerase ζ subunit | Y | GST - structural data | - | [33, 116] |
| PRCC | PRCC, TPRC | Proline-rich protein PRCC | H | Y2H - CoIP - FRET | 45-53, 57-65, 62-70, 72-80, 296-304, 371-379, 376-384 | [102] | |
| PRDX2 | PRDX2, NKEFB, TDPX1 | Peroxiredoxin-2 | Antioxidant; radiosensitivity | H | MS - CoIP | Not found | [98] |
| RAN | RAN, ARA24 | GTP-binding nuclear protein Ran | Cell cycle regulator | H | Y2H - GST – CoIP - structural data | 177-185 (confirmed) | [18, 75] |
| REV1 | REV1, REV1L | DNA repair protein REV1 | TLS | Y/M/H | Y2H - GST - CoIP - structural data | Not found | [15, 22, 30, 34, 50, 117, 118] |
| REV3 | REV3L, POLZ REV3 | DNA polymerase zeta catalytic subunit | TLS; Polymerase ζ subunit | Y/M/H | Y2H - GST - CoIP - structural data | 1877-1885 (confirmed), 1993-2001 (confirmed) | [6, 15, 22, 30, 33, 34, 36, 44, 50, 53] |
| SHLD1 | SHLD1, C20orf196, RINN3 | Shieldin complex subunit 1 | DNA damage; NHEJ | H/M | Y2H - GST - MS - CoIP | Not found | [19–21, 62–65] |
| SHLD2 | SHLD2, FAM35A, RINN2 | Shieldin complex subunit 2 | DNA damage; NHEJ | H/M | Y2H - GST - MS - CoIP - structural data | Not found | [19–21, 25, 62–65] |
| SHLD3 | SHLD3, FLJ26957, RINN1 | Shieldin complex subunit 3 | DNA damage; NHEJ | H/M | Y2H - MS - CoIP - structural data | 50-58 (confirmed) | [19–21, 23, 25] |
| SIM2 | SIM2, BHLHE15 | Single-minded homolog 2 | Transcriptional regulator | H/Rat | Y2H - CoIP | 519-527, 575-583, 581-589 | [114] |
| TCF4 | TCF7L2, TCF4 | Transcription factor 7-like 2 | WNT signalling; transactivation | H | Y2H - GST - CoIP | Not found | [104, 105] |
| TFII-I | GTF2I, BAP135, WBSCR6 | General transcription factor II-I | Transcription factor | H | MS - GST - CoIP | Not found | [46] |
| TRIP13 | TRIP13, PCH2 | Pachytene checkpoint protein 2 homolog | Structural regulation, DNA repair | H | MS – CoIP – structural data | Not found | [26, 28] |
M? indicates that the study was performed in mouse cells but uncertain is if the exogenous REV7 cDNA used was from mouse origin.
TRIP13 and p31comet thus seem important for regulating REV7 interactions and thereby formation of different REV7-complexes, such as shieldin and REV1-Polζ[26, 27]. TRIP13 might additionally contribute to REV7 recycling. Also, part of the REV7 safety belt region was suggested to undergo slight structural rearrangements when binding different interactors, thereby potentially further facilitating interaction specificity[18]. Finally, the position of the RBM(s) within the interacting protein might impact the catalytic assembly of REV7-complexes. Motifs at the extremities of a protein might allow an interactor to ‘simply’ slide into the closed safety belt. Opposingly, internally positioned motifs might require more complex catalytic assembly, as recently shown for MAD2-interactions[29]. RBMs are located at the extremities of REV7-interactors, except in REV3 and CAMP (Figure 1f). This potentially reflects the necessity to regulate certain interactions at specific subcellular locations or throughout the cell cycle. For instance, CAMP-REV7 interaction might be restricted to mitosis, and therefore require additional regulation, while SHLD3-REV7 interaction and shieldin-mediated DSB repair are required throughout the cell cycle.
REV7 homodimerization
MAD2 homodimerization and heterodimerization with p31comet are important for its SAC function, involving incorporation of MAD2 into the mitotic checkpoint complex (MCC)[24]. The formation of an O-MAD2:C-MAD2 dimer is an important step for the catalysis of the MCC although eventually only C-MAD2 is incorporated[14]. REV7 can also homodimerize[14, 30–32], as well as heterodimerize with p31comet and MAD2 (Figure 1e)[6, 32]. Dimerization depends on 9 residues within one particular surface of REV7 and is essential for its TLS function; REV3 contains two RBMs that are both important for DNA damage tolerance towards cisplatin and UV, and REV7-residues mediating its dimerization are essential in survival upon cisplatin treatment (Figure 2a,g)[16, 32]. Indeed, in vitro the two RBMs of one REV3 can simultaneously bind two REV7 molecules, forming a REV3-tethered homodimer, in which the two REV7 molecules in closed conformation (C-REV7) dimerize through the canonical HORMA domain dimerization-interface[16, 32].
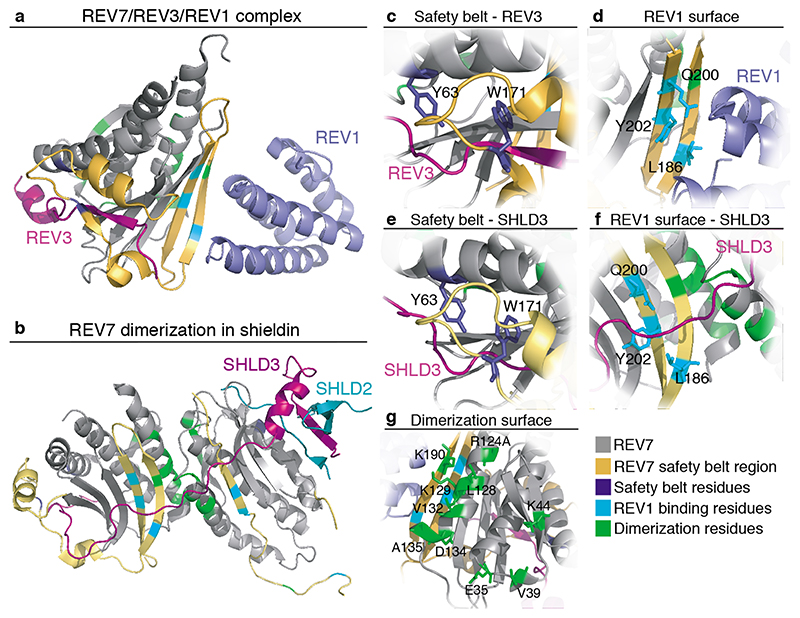
Figure 2. Crystal structure of REV7, highlighting residues important for REV7 interactions.
a. Structure (PDB: 3VU7[34]) of human REV7 (R124A) shown in grey, entrapping a fragment of human REV3 (residues 1847-1898, purple) together with REV1 (residues 1140-1251, light purple). The REV7 safety belt (yellow) spans residues 153-211. Residues important for the safety belt interaction (dark blue), REV1 interaction (cyan) or contributing to the dimerization-surface (green) are indicated[22, 25, 32]. b. Structure of human SHLD3-C-REV7-O-REV7-SHLD2 complex, containing a REV7-dimer. PDB: 6KTO[25]. SHLD3 fragment (residues 1-64) is shown in pink, SHLD2 fragment (residues 1-52) in blue and REV7 in grey, with residues indicated as in a. c. Y63 and W171 residues important for REV7 safety belt interactions (dark blue) d. L186, Q200 and Y202 residues described to mediate REV1 interaction (cyan). e. Y63 and W171 residues important for REV7 safety belt interactions (dark blue) when interacting with SHLD3 within the shieldin complex. f. SHLD3 binds over the REV1 interaction-surface. Residues important for the REV1 interaction indicated as in d. when REV7 forms a dimer within the shieldin complex. g. E35, V39, K44, R124, L128, K129, V132, D134, A135 and K190 residues were identified as critical for REV7 dimerization (green)[25, 32].
Initially, two potential RBMs were identified in SHLD3, but only one appeared essential[20, 23]. This raised the question whether homodimerization of REV7 is important in shieldin. Recent biochemical and structural insights shed light on this, showing that REV7 interacts with SHLD3 as a dimer through the originally identified RBM and a novel region in SHLD3, and in a manner critical for its role in DSB repair ([25], de Krijger et al, unpublished). Within shieldin, REV7 utilizes the conventional HORMA dimer-interface centered around the REV7 αC Helix, similar to how MAD2 dimerizes, head-to-head oriented (Figure 2b,g). Moreover, REV7 appears to form an O-REV7:C-REV7 dimer[25]. Whether this mode of O/C-dimerization contributes to the catalysis of the complex is still unclear since unlike MAD2 in the MCC, both REV7 molecules are incorporated in shieldin and important for its function. Recently, a detailed S. cerevisiae Polζ-structure revealed Rev3/Pol31/Pol32 in complex with two Rev7 molecules in a surprising head-to-tail arrangement, different from other HORMA domain protein dimers[33]. It therefore seems that REV7 can dimerize in both a head-to-tail and head-to-head orientation. This might be determined by additional protein interactions such as by Pol31, Pol32, REV1 or SHLD2/3[33]. Alternatively, this could reflect a difference between yeast and human REV7-containing complexes. The orientation of the REV7 dimer could in turn impact additional protein interactions centered around the dimer.
Although REV7 can still bind peptides through its safety belt in the presence of a dimer-disrupting mutation (REV7R124A)[17, 19, 20, 22, 23, 31], REV7 dimerization might stabilize existing protein interactions, or create a new protein interaction-surface. While the latter appears not the case for REV7-REV1 interaction in TLS, as REV1 binding is unaffected when using a dimerization-defective REV7 mutant and mediated by only one of the REV7 molecules, it does seem to apply to REV7-SHLD2/3 interaction (see below)[22, 25, 32, 33]. Interestingly, the more recent insights on the functional importance of REV7 dimerization indicate that structural and biochemical studies, facilitated by using dimerization-impaired REV7R124A instead of wild-type REV7[31], were on partially defective REV7.
Additional REV7 interaction-surfaces
Aside from the safety belt and dimerization-domain, REV7 contains additional binding-surfaces, allowing multiple interactions simultaneously (Figure 1e). This is illustrated by Polζ and shieldin where a REV7 dimer can engage with REV3 or SHLD3, and simultaneously with REV1 or SHLD2 (Figure 2a,b). Moreover, it has become clear that REV7 dimerization in shieldin creates an opportunity for SHLD3 and SHLD2 to interact with an open REV7. In this dimer SHLD3 binds partially over the REV1 interaction-surface, indicating that SHLD3 and REV1 cannot participate in the same REV7-complex simultaneously[25]. It therefore seems that REV7 has at least four surfaces for protein interaction; a ‘safety belt’, a ‘dimerization-domain’, a ‘REV1 binding-domain’ and an interaction-surface exposed by a REV7 dimer (Figure 2a–g). Finally, in yeast Polζ, Pol31 and Pol32 form separate interactions with a Rev3-bound Rev7 dimer, indicating additional REV7 interaction-surfaces[33]. In TLS, REV7-REV3 bind first and REV1 subsequently interacts with the closed REV7, indicating a certain order of events is needed for protein interactions surrounding REV7[22, 34]. Also important to consider is that mutations disturbing REV7 safety belt interactions (Y63A, W171A and potentially C70R, Figure 2c,e) likely impact multiple REV7 functions. Table 1 summarizes the different REV7 protein interactions, for only a subset the REV7 interaction-surface is known.
REV7 contributes to genome integrity in multiple ways
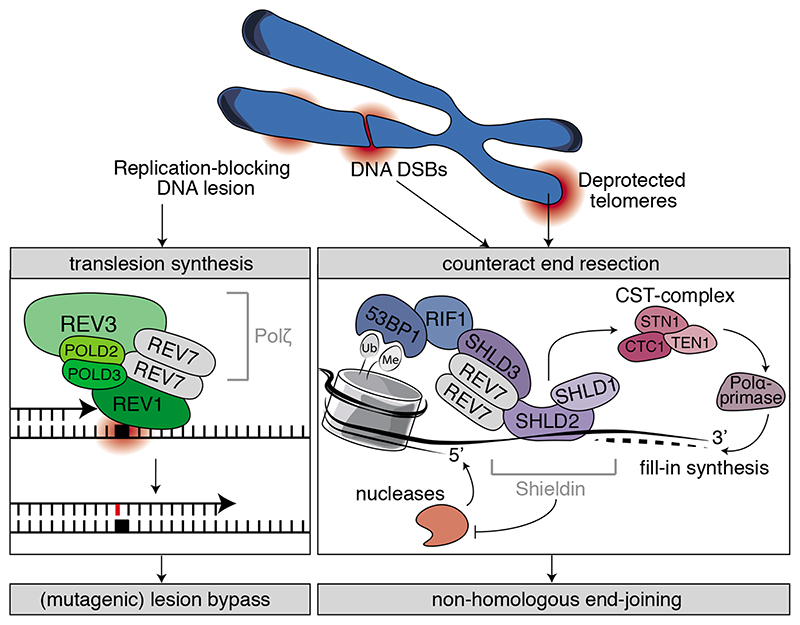
DNA constantly endures various types of damage that require resolution to maintain genome integrity. Chemicals or UV radiation cause adducts to the DNA or covalently link the two DNA strands, thereby obstructing DNA replication. Moreover, collapsed replication forks, ionizing radiation (IR) or other sources can cause DSBs that are lethal if unrepaired[35]. REV7 plays an important role in the replication bypass of DNA lesions as well as in DSB repair by functioning in two distinct protein complexes; Polζ and shieldin (Key Figure).
Key Figure. REV7 and genome integrity.
REV7 contributes to genome integrity through performing a central structural role in two distinct protein complexes: Polζ and shieldin. It functions in TLS together with REV3, POLD2, POLD3 and REV1 to facilitate (mutagenic) lesion bypass. In the shieldin complex, REV7 interacts with a second set of proteins, SHLD3, SHLD2 and SHLD1 that are recruited downstream of 53BP1 and RIF1. The shieldin complex counteracts 5’ end-resection and thereby promotes non-homologous end-joining.
REV7 as subunit of Polζ
Rev1, Rev3 and Rev7 were originally identified in S. cerevisiae through screening for defective mutagenesis-induced locus reversion[1–3]. It is now well established that REV1, REV3 and REV7 operate together in TLS and are conserved from yeast to human. The catalytic subunit REV3 and accessory subunit REV7 together form Polζ[36]. Two subunits of Polδ, POLD2 (yeast Pol31) and POLD3 (yeast Pol32), interact with Polζ via the REV3 C-terminal domain (CTD), increasing its TLS activity[33, 37–40]. Polζ has a crucial role in lesion bypass with the REV1 DNA polymerase[7, 41, 42], in which REV7 and REV3 bind each other first and REV7 subsequently interacts with the REV1-CTD[22, 30, 34, 43, 44]. REV7 thereby plays an important structural role in Polζ-REV1 complex formation.
Upon replication fork stalling at a DNA lesion, REV1 or POLD2/POLD3 recruit Polζ to mono-ubiquitinated PCNA[39, 42, 45]. Also the TFII-I transcription factor has been suggested to contribute to Polζ recruitment by bridging PCNA-REV7 interaction[46]. Polζ has no proofreading capacity by lacking 3’-5’ exonuclease activity, but is highly proficient in extending from a mismatched base pair, while the role of REV1 is primarily structural and for recruitment[36, 42, 47]. Polζ-REV1 is responsible for the majority of spontaneous and UV-induced mutagenesis[48]. Indeed, depletion of REV1, REV3 or REV7 reduces mutagenesis and increases sensitivity to DNA damage-inducing drugs, UV and IR[41, 48–53].
REV7 in ICL-repair and Fanconi Anemia
Through TLS, REV7 also aids repair of intra- and interstrand crosslinks (ICLs), as Polζ by itself, or with REV1, is important for DNA extension past an ICL-lesion[54, 55]. Hence, in various cell systems, absence of either REV1, REV3 or REV7 strongly increases sensitivity to the DNA-crosslinking agent cisplatin[41, 52, 56–58]. Moreover, bi-allelic mutations in REV7, causing undetectable levels of REV7 protein, were identified in a patient presenting with characteristics of Fanconi Anemia (FA)[59]. FA is a rare genetic disease associated with sensitivity to DNA-crosslinking agents and defects in a cluster of FA genes that are responsible for DNA repair via homologous recombination (HR). Re-expressing REV7 in patient cells reversed FA-phenotypes such as chromosome breakage and proliferation defects[59]. Accordingly, REV7 was included in the FA gene-family as FANCV. Further supporting that the Polζ role of REV7 contributes to FA, mice harboring a REV7 C70R mutation, that abolishes Polζ activity, displayed FA-phenotypes and hypersensitivity to ICL-induction[60]. However, roles of REV7 in mitotic regulation or DSB repair might also contribute to cell cycle defects and unresolved DNA damage in FA cells.
REV7 at the core of shieldin
Two different functional genetic screens independently identified REV7 as an important regulator of DSB repair by non-homologous end joining (NHEJ) and HR[10, 11]. These two main and opposing repair pathways differ in their requirements for DSB end processing. NHEJ can only occur at minimally processed breaks, while HR requires 5’ DNA end-resection to generate 3’ single-stranded DNA tails for DNA strand invasion. Hence, regulation of end-resection is critical to DNA repair pathway choice. Central here are 53BP1-RIF1 and BRCA1, that respectively inhibit or promote end-resection[61]. REV7 facilitates NHEJ by counteracting 5’ end-resection downstream of 53BP1-RIF1[10, 11]. Moreover, in a BRCA1-deficient background, REV7-loss causes resistance to PARP inhibitors due to restoration of end-resection and HR[11]. Observations that REV7 acts independent from REV1/REV3 in NHEJ, already suggested that REV7 operates in different protein complexes to facilitate TLS and inhibit resection[10, 11]. Indeed, multiple laboratories independently identified that in DSB repair REV7 associates with three previously uncharacterized proteins: SHLD1, SHLD2 and SHLD3, in a complex called ‘shieldin’[12, 19–21, 62–65]. As for 53BP1-RIF1, depletion of any shieldin-member impairs NHEJ and causes PARP-inhibitor resistance in BRCA1-deficient human and mouse cells[10, 11, 19–21, 61–65]. SHLD3 interacts directly with REV7[19–21, 23, 25]. SHLD3-REV7 then interacts with SHLD2 and SHLD1, of which SHLD2 has single-stranded DNA binding motifs. Shieldin also recruits the CTC1-STN1-TEN1 (CST)-complex to DSBs, with depletion of CST-members resembling functional loss of shieldin[66, 67]. CST is well known for promoting fill-in synthesis by Polα-primase upon over-resection of telomeric DNA ends after replication. At DSBs this activity could counteract resection, thereby limiting 3’ overhang formation[67]. Shieldin might thus operate in two ways: by recruiting fill-in activities that counteract resection and by hindering nucleases from performing resection.
Paradoxically, REV7 also appears to contribute to late steps of HR. Both in yeast and chicken DT40 cells, REV1-Polζ has been implicated in (error-prone) DNA repair synthesis at DSBs[56, 57, 68, 69]. Also, in a HR-reporter system in human cells REV7 contributed to repair[53]. At first glance this seems difficult to reconcile with REV7-loss restoring HR in BRCA1-deficient cells, but on the other hand might explain why HR is only partially restored in this setting[11]. How REV7 acts in DNA repair might be context-dependent and dictated by the type of damage (UV, ICLs, DSBs), protein complex (shieldin versus REV1-Polζ), or cell cycle stage.
REV7 and the response to pathogens
Within Polζ and shieldin, REV7 contributes to different biological processes that enable cells to recognize and combat pathogens. First, somatic hypermutation of immunoglobulin genes involves the generation of mutations and Polζ-mediated extension from the mismatched base pair[70–72]. Secondly, immunoglobulin class-switch recombination (CSR) requires shieldin-promoted joining of induced DSBs by NHEJ[10, 11, 20, 72]. Besides in shieldin and Polζ, additional protein interactions connect REV7 to the cellular response to foreign elements: REV7 interacts with adenovirus death protein (ADP) and decreases its efficiency to lyse human cells[73], while binding of the Shigella bacterial effector IpaB to REV7 interferes with its regulation of the APC/C[74].
Additional roles of REV7
Besides contributing to genome integrity maintenance, REV7 has been implicated in additional cellular processes. Structural features shared between REV7 and MAD2 and localization of REV7 at the mitotic spindle and at mitotic chromosomes in human cells provided starting points for studying the role of REV7 in mitosis[17, 75–78]. In addition, Rev7-knockout mice revealed a role for REV7 in chromatin regulation[79, 80].
REV7 in mitosis
During the SAC, MAD2 operates in the MCC by binding the primary APC/C-coactivator CDC20, thereby preventing APC/CCDC20-activation[14]. Upon correct kinetochore-microtubule attachment, the SAC is inactivated through the disassembly of the MCC. This releases CDC20 to promote APC/CCDC20 activity, which initiates anaphase onset. Hence, cells lacking MAD2 lack a functional SAC. REV7 was found to interact with the second APC/C-coactivator CDH1[8, 9, 81], and sequester it away from the APC/C[81]. REV7 itself is degraded by APC/CCDC20 in early anaphase, which releases CDH1 and increases APC/CCDH1 activity to promote timely mitotic progression[81]. Therefore, unlike MAD2, REV7 is not essential for the SAC, but rather seems to fine-tune correct mitotic timing to prevent premature mitotic exit[57, 77, 81, 82]. This function of REV7 is hijacked during infection with Shigella through its effector IpaB[74]. IpaB interaction with REV7 releases CDH1, causing unscheduled activation of the host-APC/CCDH1. CDH1 lacks an RBM but was recently found to interact with REV7 in a manner resembling REV1[15, 83]. REV7 also directly interacts with RAN, clathrin light chain A (CLTA) and CAMP[17, 18, 75, 78, 84, 85]. RAN, CLTA and CAMP function in microtubule organization and co-localize with REV7 at the mitotic spindle[75, 78, 85]. Although the exact role of REV7 in this context is unclear, its localization to the spindle and mitotic chromosomes seems dependent on CAMP and important for correct recruitment of RAN and CLTA in mitosis[17, 77, 78, 85].
In various cellular conditions, REV7 depletion increases mitotic aberrations, including lagging chromosomes and anaphase bridges[76–78, 81, 82]. How much this relates to the ability of REV7 to regulate APC/C-activity, localize at the mitotic spindle and mitotic chromosomes, or interact with various mitotic proteins is still unclear. Also the critical roles for REV7 in TLS and DSB repair add to the complexity in understanding these mitotic phenotypes arising with compromised REV7, as also unrepaired or improperly repaired lesions can contribute to problems in mitosis. Moreover, REV1 was found to undergo REV7-dependent proteasomal degradation via the APC/C, suggesting an interplay between APC/C function and TLS[86]. Though further exploring the mitotic roles of REV7 is interesting and important, clearly attributing mitotic phenotypes to specific functions and interactions of REV7 might be challenging in the light of such interplays between different REV7 activities.
REV7 in development and control of chromatin
Rev7 is highly expressed in testes of mice[60, 79, 80]. While mice heterozygous for Rev7 are healthy, homozygous knockout mice are born in sub-Mendelian ratios with underdeveloped gonads and infertility, or show complete embryonic lethality, depending on genetic background[20, 79, 80]. Indeed, primordial germ cells (PGC’s), normally giving rise to oocytes and sperm cells, are lost through apoptosis in Rev7-deficient embryos[79, 80]. This is accompanied by accumulation of DNA damage in PGC’s and surrounding somatic cells and seems related to REV7’s role in Polζ, as mice containing a REV3-REV7 interaction and Polζ-activity disrupting mutation (Rev7 C70R) also show defective PGC development[60, 80]. However, since a REV7C70R mutation potentially also impairs other (safety belt) interactions, disrupted TLS might not be the only explanation for how Rev7-deficiency impairs PGC development. Particularly relevant in this regard seems a REV7 effect on PGC epigenetic reprogramming and chromatin regulation. Upon development, PGC’s arrest in G2 and undergo major epigenetic reprogramming. However, Rev7 -/- PGC’s fail to arrest in G2 and show dysregulated H3K9me2 and H3K27me3 chromatin marks[79]. Although requiring further characterization, potential explanations may lie in the associated observations that REV7 can bind G9A and GLP H3K9me2-methyltransferases, and Cdk1[79]. REV7 is also important for maintenance of pluripotent mouse embryonic stem cells (ESC’s) and PGC-like cells (PGCLCs)[87, 88]. In absence of REV7, ESC’s, normally characterized by open chromatin, display increased heterochromatin, dysregulation of H3K9me2 and H3K27me3, spontaneous epithelial differentiation and MAPK-pathway hyperactivity[87, 88]. Moreover, Rev7 -/- ESC’s show suppression of Dppa3, a regulator of DNA-methylation, and changes in global DNA-methylation[88].
Altogether this indicates that REV7-loss causes defective epigenetic reprogramming due to aberrant DNA- or histone-methylation. Interestingly, mass spectrometry identified associations of REV7 with multiple chromatin factors of which the functional consequence is unclear and interesting to further explore[84]. Moreover, an intriguing question is to what extent chromatin-associated REV7 activities contribute to its other functions, in particular in TLS and DSB repair.
How is REV7 regulated?
REV7’s roles range from maintaining genome integrity to mitotic progression and chromatin regulation and are enabled by various protein interactions (Figure 3a–h). How specificity towards its different interactors is regulated, how REV7 is directed to each of them at specific subcellular locations, upon different upstream stimuli or throughout the cell cycle are important open questions. These are also interesting from a clinical perspective, especially since REV7 is dysregulated in different cancers and affects the response to cancer treatment (box 1).
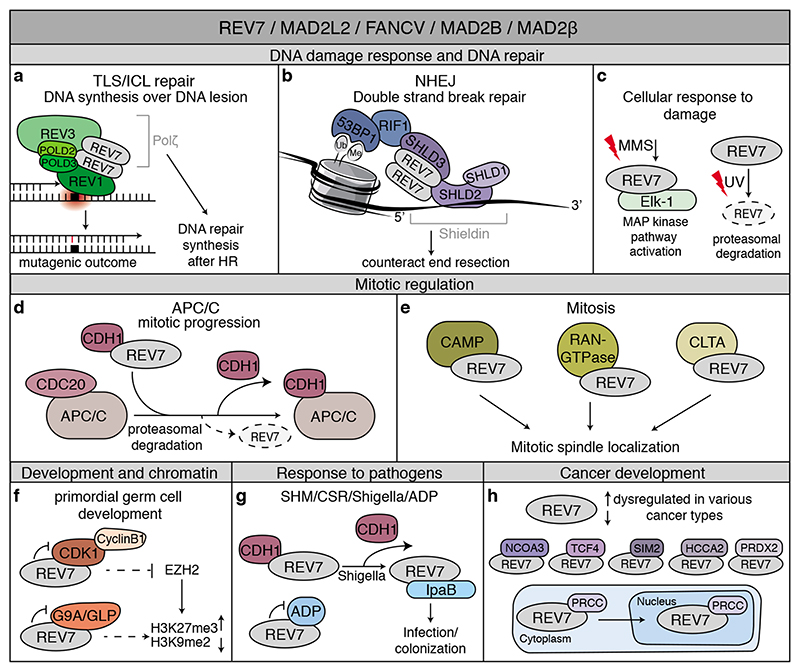
Figure 3. REV7: Jack of many trades.
Overview of the wide range of processes in which REV7 is involved through interactions with various proteins. See text, Table 1 and box 1 for details. a. REV7 functions in TLS by forming Polζ together with REV3, POLD2 and POLD3. Polζ also contributes to DNA repair synthesis after HR. b. The shieldin complex, consisting of REV7, SHLD3, SHLD2 and SHLD1, counteracts resection at DNA DSBs and thereby contributes to NHEJ. c. REV7 contributes to the cellular response to DNA damage. Upon DNA damage exposure by methyl methanesulfonate (MMS), REV7 contributes to MAP kinase pathway activation through the transcription factor Elk-1. Additionally, UV-damage induces degradation of REV7. d. REV7 binds to and sequesters the APC/C-coactivator CDH1, and thereby contributes to the correct timing of mitotic progression. e. REV7 interacts with CAMP, RAN GTPase and CLTA that colocalize with REV7 at the mitotic spindle. f. REV7 contributes to primordial germ cell development and chromatin regulation. In this context, REV7 was found to interact with G9A/GLP methyltransferases and Cdk1. g. REV7 contributes to somatic hypermutation (SHM) of immunoglobulin genes and immunoglobulin class-switch recombination (CSR) through functioning in TLS and shieldin. Additionally, REV7 interacts with IpaB and ADP. Together, REV7 thereby contributes to the cellular response to pathogens. h. REV7 is dysregulated in various cancer types. In addition, REV7 interacts with NCOA3, TCF4, SIM2, HCCA2 and PRDX2 in various cellular contexts or related to cancer development and treatment. The interaction of REV7 with PRCC contributes to REV7 subcellular localization.
Box 1. REV7 in cancer development and treatment.
Aberrant cell cycle regulation and DNA damage resolution can result in genomic instability and tumorigenesis[91]. REV7, significantly impacting both processes, is found dysregulated in different cancer cell lines and tumor tissues[73, 92–98]. Moreover, high REV7 expression is associated with poor prognosis in various cancer types[93, 96, 99, 100]. How REV7 dysregulation is connected to poor prognosis is mostly speculative. Increased REV7 expression (and possibly thereby increased error-prone Polζ activity) might contribute to mutational load, while its upregulation might also be a consequence of cancer cells dealing with increased replication stress, DNA damage or mitotic problems. A role for Polζ in cancer development and treatment has been reviewed elsewhere, and it seems that through its role in Polζ, REV7 operates mostly as a tumor suppressor[42]. Supporting this, female homozygous REV7C70R mice developed ovarian tumors that show accumulation of DNA damage, although Polζ-independent consequences of the C70R mutation cannot be excluded[101]. In line with a tumor suppressive role, FA, now also linked to REV7 loss-of-function, is associated with predisposition to cancer.
Beyond the more established interactions and roles in mitotic control, TLS and DSB repair, REV7 appears to affect cancer development in diverse additional ways via interactions with multiple different factors, including PRCC, NCOA3, TCF4 and HCCA2. In a subset of renal cell carcinoma (RCC), a translocation of PRCC with TFE3 leads to a fusion-protein that disrupts REV7-PRCC interaction and affects REV7’s subcellular localization. Tumor cell lines with this translocation show an impaired mitotic checkpoint, and overexpressing the PRCCTFE3 fusion protein increases chromosome misalignment, which could indicate disturbed REV7 function[78, 102]. On the other hand, REV7 seems to act as a tumor suppressor by promoting p38-mediated degradation of NCOA3, a protein associated with poor prognosis in CRC[103]. REV7 interaction with TCF4, a downstream effector of WNT, reduces TCF4 transcriptional activity, thereby inhibiting TCF4-β-catenin signaling[104, 105]. REV7 might thereby affect TCF4-induced epithelial to mesenchymal transition (EMT), which is characteristic of metastasis. Finally, REV7 interacts and colocalizes with HCCA2, but the functional relevance of this interaction is unclear[106]. Exactly how these different REV7-interactions contribute to cancer development requires further study.
In various cancer cell lines, as well as in mouse models for cancer, REV7 depletion increases sensitivity to DNA damaging treatments, including cisplatin and IR[82, 95, 96, 98, 107]. This is often accompanied with accumulation of DNA damage response proteins in cells, indicative of defective repair[82, 95, 96]. These effects can therefore largely be explained by REV7’s functions in Polζ and shieldin. However, Polζ- or shieldin-independent REV7 functions may also contribute to the observed sensitivity via additional REV7 interactions (table 1). For instance, REV7 interaction with peroxiredoxin 2 (PRDX2) might contribute to radioresistance in esophageal squamous cell carcinoma[98]. It will be interesting to see whether enhanced sensitivity upon REV7 inhibition can be exploited as potential treatment option. On this note, a small molecule inhibitor of REV7 has recently been described that inhibits REV3-REV7 interaction and may sensitize cancer cells to cisplatin [108]. However, as a note of caution, this compound might also inhibit additional interactions that use the REV7-REV3 interaction-interface and thereby affect Polζ-independent REV7 functions, which could affect its usability.
By loss of 53BP1, RIF1, shieldin-components (including REV7) or members of the CST-complex, Brca1-deficient tumors acquire resistance to PARP-inhibition[12]. Interestingly, BRCA1-deficient human and mouse cells that became resistant to PARP-inhibition by loss of 53BP1 or REV7 showed hypersensitivity to the G4-stabilizing compound pyridostatin (PDS) and acquired sensitivity to radiotherapy[109, 110]. Also, BRCA1-deficient human cells lacking SHLD1 or SHLD2, although resistant to PARP-inhibitors, are now hypersensitive to cisplatin treatment[62]. This could therefore provide alternative treatment opportunities in BRCA1-deficient cells that became resistant to PARP-inhibition.
Possibly post-translational modifications (PTMs) provide the cell with means to regulate REV7 activity. REV7 contains a D-box that is recognized by the APC/C, targeting REV7 for ubiquitin-mediated degradation at the end of mitosis[81]. Nevertheless, sufficient levels of REV7 apparently remain to participate in shieldin and facilitate NHEJ in G1. UV-damage, but not DSBs or ICLs, also induces degradation of REV7, which can be prevented by the nucleotide excision repair (NER) protein HR23B[81, 89]. Other than ubiquitination, it is not clear whether REV7 undergoes additional PTMs. Yeast Rev7 was found to be phosphorylated by Dbf4-dependent kinase (DDK) in vitro[90], but so far the in vivo relevance of this observation is unclear. Also, a putative ATM-dependent phosphorylation site (S/TQ) on REV7 appeared unimportant for REV7 function in NHEJ but could still be relevant in other settings[10].
Besides potential regulation through PTMs, REV7 might be heavily regulated at the level of opening and closing of the safety belt. Similar to how MAD2 requires a switch-like conversion to (de)activate the SAC, REV7 function might be silenced via disassembly of REV7-complexes, or dissociation from its substrate or effectors. How could such a switch-like conversion be critical to REV7-controlled processes? It is conceivable that in DNA repair, it facilitates timely dissociation of shieldin when resection is counteracted sufficiently. Disassembly of shieldin, mediated by TRIP13-dependent opening of the REV7 safety belt and release from SHLD3, could thereby allow NHEJ progression and finalize damage resolution. Furthermore, active dissociation of shieldin during S-phase might facilitate the switch from NHEJ to HR by allowing resection to take place in S/G2. In the context of TLS, a switch-like behavior could aid in polymerase switching, or dissociation of Polζ after bypass synthesis. Intriguingly, it was recently found that REV3 can localize to DSBs where it was hypothesized to contribute to fill-in synthesis through Polζ[25]. Although this remains to be further proven, a switch-like behavior might allow REV7 to quickly shuttle between different complexes and move from shieldin to Polζ.
REV7 protein is quite abundant, in sharp contrast to the low abundance of REV7’s safety belt interactors in Polζ and shieldin, REV3 and SHLD3[16, 19, 42]. It thus seems possible for REV7 to engage with most or all of its different interactors in parallel and require little regulation with regard to protein entrapment. In line with this, the interactions between REV3-REV7, and among REV7-shieldin occur independently of DNA damage[19, 39, 63]. Considering the many proteins that need to accumulate at sites of DNA damage, a relatively stable interaction between SHLD3-REV7, and REV3-REV7 might allow protein complexes to assemble faster. If, and how the interactions around the safety belt are regulated through various upstream stimuli requires additional research and will be instrumental in better understanding of REV7’s ability to function in different complexes.
Concluding remarks
Over the past years, REV7 gained increasing interest. This small HORMA domain protein thereby metaphorically evolved from an accessory protein of REV3 to an important mediator of various critical biological processes. REV7 contains a dynamic C-terminal safety belt for protein entrapment and has at least three additional interaction-surfaces, including one mediating homodimerization and one created upon homodimerization, that allow it to interact with multiple proteins simultaneously[16, 25, 32]. It is important to further define how critical REV7-homodimerization is in REV7-mediated processes other than TLS and DSB repair and at which REV7-surface its various interactors bind (see Outstanding Questions). This will not only aid better understanding of the different biological processes involving REV7, but will also be important in developing approaches for regulating the assembly/disassembly and activity of REV7-complexes. Moreover, given the prominent roles of REV7 in the repair of DSBs, TLS and mitotic regulation, further following up on REV7’s impact on tumorigenesis and drug response will also be of great interest. Together, this could lead to improved stratification of patients for specific DNA damaging therapies on the basis of REV7 status or yield opportunities for new therapeutic strategies by modulating REV7-controlled processes.
Outstanding Questions.
-
-
How is REV7 regulated among its various protein interactions?
-
-
To what extent does REV7 protein entrapment in fact require regulation given its relative high abundance versus multiple of its (lowly abundant) critical interactors? Is regulation of REV7-complex formation therefore mostly required at the level of protein release and/or perhaps mostly needed for these low abundance proteins to be able to act at the right time and place, rather than for REV7 itself?
-
-
Why is REV7 promiscuous in its behaviour, while other HORMA domain proteins (like MAD2 and Hop1) are limited in their interactions?
-
-
If REV7 function is silenced via disassembly of REV7-complexes or dissociation from its substrate or effectors, what prevents rebinding?
-
-
To what extent is the ability of REV7 to dimerize important for its function in protein complexes other than Polζ and shieldin?
-
-
Is REV7 function in both shieldin and polζ necessary at DSBs to counteract resection and perform fill-in synthesis? Would this then require partial or complete shieldin-disassembly by release of REV7-protein interactions followed by re-assembly of REV7 with REV3, or is the high abundance of REV7 sufficient to be present and function at DSBs in both complexes in parallel?
-
-
How does the role of REV7 in finalizing HR connect to its role in NHEJ? How can REV7 be needed in HR, while its loss restores HR in a BRCA1-deficient background?
-
-
How do SHLD1/2-deficient cells become hypersensitive to cisplatin? Do shieldin complex members, other than REV7, also function in TLS?
-
-
To what extend is DNA end protection by REV7-shieldin important in the context of replication stress?
-
-
Do roles of REV7 in chromatin regulation also contribute to its roles in TLS, mitosis and DSB repair?
-
-
(How) are the different functions of REV7 connected and coordinated?
Highlights.
For long REV7 was best known for acting in TLS as a component of Polζ. Recent work uncovered that REV7 also acts in DNA repair pathway choice with previously uncharacterized proteins, in a complex called shieldin, to counteract resection at broken DNA ends. Loss of shieldin members in BRCA1-deficient cells confers PARP-inhibitor resistance.
REV7 has a dynamic structure, it contains a ‘safety belt’ to entrap interacting peptides and can thereby switch between open (unbound) and closed (bound) forms.
REV7 homodimerizes, which is critical for its role in genome maintenance.
Analogous to MAD2 and other HORMA proteins, the ATPase TRIP13 was recently found to interact with REV7 and facilitate REV7 partner entrapment and release.
Deregulation of REV7-controlled processes causes disease, such as Fanconi Anemia and cancer, and affects sensitivity to DNA damaging agents.
Acknowledgments
We apologize to authors whose work was not cited owing to space limitations and thank Alex Faesen for comments. Authors were supported by European Research Council (ERC-StG 311565) and Dutch Cancer Society (KWF 10999/2017-1) grants to J.J.L.J.
Footnotes
Declaration of Interests
The authors declare no competing interests.
References
- 1.Lemontt JF. Mutants of yeast defective in mutation induced by ultraviolet light. Genetics. 1971;68(1):21–33. doi: 10.1093/genetics/68.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lawrence CW, et al. REV7, a new gene concerned with UV mutagenesis in yeast. Mol Gen Genet. 1985;200(1):80–5. doi: 10.1007/BF00383316. [DOI] [PubMed] [Google Scholar]
- 3.Torpey LE, et al. Cloning and sequence of REV7, a gene whose function is required for DNA damage-induced mutagenesis in Saccharomyces cerevisiae. Yeast. 1994;10(11):1503–9. doi: 10.1002/yea.320101115. [DOI] [PubMed] [Google Scholar]
- 4.Cahill DP, et al. Characterization of MAD2B and other mitotic spindle checkpoint genes. Genomics. 1999;58(2):181–7. doi: 10.1006/geno.1999.5831. [DOI] [PubMed] [Google Scholar]
- 5.Nelson KK, et al. Evidence for an interaction of the metalloprotease-disintegrin tumour necrosis factor alpha convertase (TACE) with mitotic arrest deficient 2 (MAD2), and of the metalloprotease-disintegrin MDC9 with a novel MAD2-related protein, MAD2beta. Biochem J. 1999;343(Pt 3):673–80. [PMC free article] [PubMed] [Google Scholar]
- 6.Murakumo Y, et al. A human REV7 homolog that interacts with the polymerase zeta catalytic subunit hREV3 and the spindle assembly checkpoint protein hMAD2. J Biol Chem. 2000;275(6):4391–7. doi: 10.1074/jbc.275.6.4391. [DOI] [PubMed] [Google Scholar]
- 7.Lawrence CW. Cellular roles of DNA polymerase zeta and Rev1 protein. DNA Repair (Amst) 2002;1(6):425–35. doi: 10.1016/s1568-7864(02)00038-1. [DOI] [PubMed] [Google Scholar]
- 8.Pfleger CM, et al. Inhibition of Cdh1-APC by the MAD2-related protein MAD2L2: a novel mechanism for regulating Cdh1. Genes Dev. 2001;15(14):1759–64. doi: 10.1101/gad.897901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen J, Fang G. MAD2B is an inhibitor of the anaphase-promoting complex. Genes Dev. 2001;15(14):1765–70. doi: 10.1101/gad.898701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boersma V, et al. MAD2L2 controls DNA repair at telomeres and DNA breaks by inhibiting 5’ end resection. Nature. 2015;521(7553):537–40. doi: 10.1038/nature14216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu G, et al. REV7 counteracts DNA double-strand break resection and affects PARP inhibition. Nature. 2015;521(7553):541–4. doi: 10.1038/nature14328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Setiaputra D, Durocher D. Shieldin - the protector of DNA ends. EMBO Rep. 2019;20(5) doi: 10.15252/embr.201847560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aravind L, Koonin EV. The HORMA domain: a common structural denominator in mitotic checkpoints, chromosome synapsis and DNA repair. Trends Biochem Sci. 1998;23(8):284–6. doi: 10.1016/s0968-0004(98)01257-2. [DOI] [PubMed] [Google Scholar]
- 14.Rosenberg SC, Corbett KD. The multifaceted roles of the HORMA domain in cellular signaling. J Cell Biol. 2015;211(4):745–55. doi: 10.1083/jcb.201509076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hanafusa T, et al. Overlapping in short motif sequences for binding to human REV7 and MAD2 proteins. Genes Cells. 2010;15(3):281–96. doi: 10.1111/j.1365-2443.2009.01380.x. [DOI] [PubMed] [Google Scholar]
- 16.Tomida J, et al. REV7 is essential for DNA damage tolerance via two REV3L binding sites in mammalian DNA polymerase zeta. Nucleic Acids Res. 2015;43(2):1000–11. doi: 10.1093/nar/gku1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hara K, et al. Dynamic feature of mitotic arrest deficient 2-like protein 2 (MAD2L2) and structural basis for its interaction with chromosome alignment-maintaining phosphoprotein (CAMP) J Biol Chem. 2017;292(43):17658–17667. doi: 10.1074/jbc.M117.804237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang X, et al. REV7 has a dynamic adaptor region to accommodate small GTPase RAN/Shigella IpaB ligands and its activity is regulated by RanGTP/GDP switch. J Biol Chem. 2019 doi: 10.1074/jbc.RA119.010123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gupta R, et al. DNA Repair Network Analysis Reveals Shieldin as a Key Regulator of NHEJ and PARP Inhibitor Sensitivity. Cell. 2018;173(4):972–988.:e23. doi: 10.1016/j.cell.2018.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ghezraoui H, et al. 53BP1 cooperation with the REV7-shieldin complex underpins DNA structure-specific NHEJ. Nature. 2018;560(7716):122–127. doi: 10.1038/s41586-018-0362-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Noordermeer SM, et al. The shieldin complex mediates 53BP1-dependent DNA repair. Nature. 2018;560(7716):117–121. doi: 10.1038/s41586-018-0340-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hara K, et al. Crystal structure of human REV7 in complex with a human REV3 fragment and structural implication of the interaction between DNA polymerase zeta and REV1. J Biol Chem. 2010;285(16):12299–307. doi: 10.1074/jbc.M109.092403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dai Y, et al. Structural basis for shieldin complex subunit 3-mediated recruitment of the checkpoint protein REV7 during DNA double-strand break repair. J Biol Chem. 2020;295(1):250–262. doi: 10.1074/jbc.RA119.011464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vader G. Pch2(TRIP13): controlling cell division through regulation of HORMA domains. Chromosoma. 2015;124(3):333–9. doi: 10.1007/s00412-015-0516-y. [DOI] [PubMed] [Google Scholar]
- 25.Liang L, et al. Molecular basis for assembly of the shieldin complex and its implications for NHEJ. Nat Commun. 2020;11(1):1972. doi: 10.1038/s41467-020-15879-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Clairmont CS, et al. TRIP13 regulates DNA repair pathway choice through REV7 conformational change. Nat Cell Biol. 2020;22(1):87–96. doi: 10.1038/s41556-019-0442-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sarangi P, et al. p31(comet) promotes homologous recombination by inactivating REV7 through the TRIP13 ATPase. Proc Natl Acad Sci U S A. 2020;117(43):26795–26803. doi: 10.1073/pnas.2008830117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xie W, et al. Molecular mechanisms of assembly and TRIP13-mediated remodeling of the human Shieldin complex. Proc Natl Acad Sci U S A. 2021;118(8) doi: 10.1073/pnas.2024512118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Piano V, et al. CDC20 assists its catalytic incorporation in the mitotic checkpoint complex. Science. 2021;371(6524):67–71. doi: 10.1126/science.abc1152. [DOI] [PubMed] [Google Scholar]
- 30.Murakumo Y, et al. Interactions in the error-prone postreplication repair proteins hREV1, hREV3, and hREV7. J Biol Chem. 2001;276(38):35644–51. doi: 10.1074/jbc.M102051200. [DOI] [PubMed] [Google Scholar]
- 31.Hara K, et al. Purification, crystallization and initial X-ray diffraction study of human REV7 in complex with a REV3 fragment. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2009;65(Pt 12):1302–5. doi: 10.1107/S1744309109046181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rizzo AA, et al. Rev7 dimerization is important for assembly and function of the Rev1/Polzeta translesion synthesis complex. Proc Natl Acad Sci U S A. 2018 doi: 10.1073/pnas.1801149115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Malik R, et al. Structure and mechanism of B-family DNA polymerase zeta specialized for translesion DNA synthesis. Nat Struct Mol Biol. 2020;27(10):913–924. doi: 10.1038/s41594-020-0476-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kikuchi S, et al. Structural basis of recruitment of DNA polymerase zeta by interaction between REV1 and REV7 proteins. J Biol Chem. 2012;287(40):33847–52. doi: 10.1074/jbc.M112.396838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ciccia A, Elledge SJ. The DNA damage response: making it safe to play with knives. Mol Cell. 2010;40(2):179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nelson JR, et al. Thymine-thymine dimer bypass by yeast DNA polymerase zeta. Science. 1996;272(5268):1646–9. doi: 10.1126/science.272.5268.1646. [DOI] [PubMed] [Google Scholar]
- 37.Johnson RE, et al. Pol31 and Pol32 subunits of yeast DNA polymerase delta are also essential subunits of DNA polymerase zeta. Proc Natl Acad Sci U S A. 2012;109(31):12455–60. doi: 10.1073/pnas.1206052109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Baranovskiy AG, et al. DNA polymerase delta and zeta switch by sharing accessory subunits of DNA polymerase delta. J Biol Chem. 2012;287(21):17281–7. doi: 10.1074/jbc.M112.351122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Makarova AV, et al. A four-subunit DNA polymerase zeta complex containing Pol delta accessory subunits is essential for PCNA-mediated mutagenesis. Nucleic Acids Res. 2012;40(22):11618–26. doi: 10.1093/nar/gks948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee YS, et al. Human Pol zeta purified with accessory subunits is active in translesion DNA synthesis and complements Pol eta in cisplatin bypass. Proc Natl Acad Sci U S A. 2014;111(8):2954–9. doi: 10.1073/pnas.1324001111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Makarova AV, Burgers PM. Eukaryotic DNA polymerase zeta. DNA Repair (Amst) 2015;29:47–55. doi: 10.1016/j.dnarep.2015.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Martin SK, Wood RD. DNA polymerase zeta in DNA replication and repair. Nucleic Acids Res. 2019 doi: 10.1093/nar/gkz705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Acharya N, et al. Complex formation with Rev1 enhances the proficiency of Saccharomyces cerevisiae DNA polymerase zeta for mismatch extension and for extension opposite from DNA lesions. Mol Cell Biol. 2006;26(24):9555–63. doi: 10.1128/MCB.01671-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wojtaszek J, et al. Structural basis of Rev1-mediated assembly of a quaternary vertebrate translesion polymerase complex consisting of Rev1, heterodimeric polymerase (Pol) zeta, and Pol kappa. J Biol Chem. 2012;287(40):33836–46. doi: 10.1074/jbc.M112.394841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Garg P, et al. Proliferating cell nuclear antigen promotes translesion synthesis by DNA polymerase zeta. J Biol Chem. 2005;280(25):23446–50. doi: 10.1074/jbc.C500173200. [DOI] [PubMed] [Google Scholar]
- 46.Fattah FJ, et al. The transcription factor TFII-I promotes DNA translesion synthesis and genomic stability. PLoS Genet. 2014;10(6):e1004419. doi: 10.1371/journal.pgen.1004419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Prakash S, et al. Eukaryotic translesion synthesis DNA polymerases: specificity of structure and function. Annu Rev Biochem. 2005;74:317–53. doi: 10.1146/annurev.biochem.74.082803.133250. [DOI] [PubMed] [Google Scholar]
- 48.Lawrence CW, Maher VM. Mutagenesis in eukaryotes dependent on DNA polymerase zeta and Rev1p. Philos Trans R Soc Lond B Biol Sci. 2001;356(1405):41–6. doi: 10.1098/rstb.2000.0746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Diaz M, et al. Decreased frequency and highly aberrant spectrum of ultraviolet-induced mutations in the hprt gene of mouse fibroblasts expressing antisense RNA to DNA polymerase zeta. Mol Cancer Res. 2003;1(11):836–47. [PubMed] [Google Scholar]
- 50.Hirano Y, Sugimoto K. ATR homolog Mec1 controls association of DNA polymerase zeta-Rev1 complex with regions near a double-strand break. Curr Biol. 2006;16(6):586–90. doi: 10.1016/j.cub.2006.01.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McNally K, et al. hRev7, putative subunit of hPolzeta, plays a critical role in survival, induction of mutations, and progression through S-phase, of UV((254nm))-irradiated human fibroblasts. DNA Repair (Amst) 2008;7(4):597–604. doi: 10.1016/j.dnarep.2007.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Neal JA, et al. The role of hRev7, the accessory subunit of hPolzeta, in translesion synthesis past DNA damage induced by benzo[a]pyrene diol epoxide (BPDE) BMC Cell Biol. 2010;11:97. doi: 10.1186/1471-2121-11-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sharma S, et al. REV1 and polymerase zeta facilitate homologous recombination repair. Nucleic Acids Res. 2012;40(2):682–91. doi: 10.1093/nar/gkr769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Raschle M, et al. Mechanism of replication-coupled DNA interstrand crosslink repair. Cell. 2008;134(6):969–80. doi: 10.1016/j.cell.2008.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Budzowska M, et al. Regulation of the Rev1-pol zeta complex during bypass of a DNA interstrand cross-link. EMBO J. 2015;34(14):1971–85. doi: 10.15252/embj.201490878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sonoda E, et al. Multiple roles of Rev3, the catalytic subunit of polzeta in maintaining genome stability in vertebrates. EMBO J. 2003;22(12):3188–97. doi: 10.1093/emboj/cdg308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Okada T, et al. Multiple roles of vertebrate REV genes in DNA repair and recombination. Mol Cell Biol. 2005;25(14):6103–11. doi: 10.1128/MCB.25.14.6103-6111.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hicks JK, et al. Differential roles for DNA polymerases eta, zeta, and REV1 in lesion bypass of intrastrand versus interstrand DNA cross-links. Mol Cell Biol. 2010;30(5):1217–30. doi: 10.1128/MCB.00993-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bluteau D, et al. Biallelic inactivation of REV7 is associated with Fanconi anemia. J Clin Invest. 2016;126(9):3580–4. doi: 10.1172/JCI88010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Khalaj M, et al. A missense mutation in Rev7 disrupts formation of Polzeta, impairing mouse development and repair of genotoxic agent-induced DNA lesions. J Biol Chem. 2014;289(6):3811–24. doi: 10.1074/jbc.M113.514752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Panier S, Boulton SJ. Double-strand break repair: 53BP1 comes into focus. Nat Rev Mol Cell Biol. 2014;15(1):7–18. doi: 10.1038/nrm3719. [DOI] [PubMed] [Google Scholar]
- 62.Dev H, et al. Shieldin complex promotes DNA end-joining and counters homologous recombination in BRCA1-null cells. Nat Cell Biol. 2018;20(8):954–965. doi: 10.1038/s41556-018-0140-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Findlay S, et al. SHLD2/FAM35A co-operates with REV7 to coordinate DNA double-strand break repair pathway choice. EMBO J. 2018 doi: 10.15252/embj.2018100158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tomida J, et al. FAM35A associates with REV7 and modulates DNA damage responses of normal and BRCA1-defective cells. EMBO J. 2018;37(12) doi: 10.15252/embj.201899543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gao S, et al. An OB-fold complex controls the repair pathways for DNA double-strand breaks. Nat Commun. 2018;9(1):3925. doi: 10.1038/s41467-018-06407-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Barazas M, et al. The CST Complex Mediates End Protection at Double-Strand Breaks and Promotes PARP Inhibitor Sensitivity in BRCA1-Deficient Cells. Cell Rep. 2018;23(7):2107–2118. doi: 10.1016/j.celrep.2018.04.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mirman Z, et al. 53BP1-RIF1-shieldin counteracts DSB resection through CST- and Polalpha-dependent fill-in. Nature. 2018;560(7716):112–116. doi: 10.1038/s41586-018-0324-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Holbeck SL, Strathern JN. A role for REV3 in mutagenesis during double-strand break repair in Saccharomyces cerevisiae. Genetics. 1997;147(3):1017–24. doi: 10.1093/genetics/147.3.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rattray AJ, et al. The roles of REV3 and RAD57 in double-strand-break-repair-induced mutagenesis of Saccharomyces cerevisiae. Genetics. 2002;162(3):1063–77. doi: 10.1093/genetics/162.3.1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Diaz M, et al. Evolution and the molecular basis of somatic hypermutation of antigen receptor genes. Philos Trans R Soc Lond B Biol Sci. 2001;356(1405):67–72. doi: 10.1098/rstb.2000.0750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Daly J, et al. Altered Ig hypermutation pattern and frequency in complementary mouse models of DNA polymerase zeta activity. J Immunol. 2012;188(11):5528–37. doi: 10.4049/jimmunol.1102629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yang D, et al. REV7 is required for processing AID initiated DNA lesions in activated B cells. Nat Commun. 2020;11(1):2812. doi: 10.1038/s41467-020-16632-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ying B, Wold WS. Adenovirus ADP protein (E3-11.6K), which is required for efficient cell lysis and virus release, interacts with human MAD2B. Virology. 2003;313(1):224–34. doi: 10.1016/s0042-6822(03)00287-3. [DOI] [PubMed] [Google Scholar]
- 74.Iwai H, et al. A bacterial effector targets Mad2L2, an APC inhibitor, to modulate host cell cycling. Cell. 2007;130(4):611–23. doi: 10.1016/j.cell.2007.06.043. [DOI] [PubMed] [Google Scholar]
- 75.Medendorp K, et al. The mitotic arrest deficient protein MAD2B interacts with the small GTPase RAN throughout the cell cycle. PLoS One. 2009;4(9):e7020. doi: 10.1371/journal.pone.0007020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bhat A, et al. Rev3, the catalytic subunit of Polzeta, is required for maintaining fragile site stability in human cells. Nucleic Acids Res. 2013;41(4):2328–39. doi: 10.1093/nar/gks1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bhat A, et al. Rev7/Mad2B plays a critical role in the assembly of a functional mitotic spindle. Cell Cycle. 2015;14(24):3929–38. doi: 10.1080/15384101.2015.1120922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Medendorp K, et al. The mitotic arrest deficient protein MAD2B interacts with the clathrin light chain A during mitosis. PLoS One. 2010;5(11):e15128. doi: 10.1371/journal.pone.0015128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pirouz M, et al. A critical function of Mad2l2 in primordial germ cell development of mice. PLoS Genet. 2013;9(8):e1003712. doi: 10.1371/journal.pgen.1003712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Watanabe N, et al. The REV7 subunit of DNA polymerase zeta is essential for primordial germ cell maintenance in the mouse. J Biol Chem. 2013;288(15):10459–71. doi: 10.1074/jbc.M112.421966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Listovsky T, Sale JE. Sequestration of CDH1 by MAD2L2 prevents premature APC/C activation prior to anaphase onset. J Cell Biol. 2013;203(1):87–100. doi: 10.1083/jcb.201302060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Cheung HW, et al. Inactivation of human MAD2B in nasopharyngeal carcinoma cells leads to chemosensitization to DNA-damaging agents. Cancer Res. 2006;66(8):4357–67. doi: 10.1158/0008-5472.CAN-05-3602. [DOI] [PubMed] [Google Scholar]
- 83.Pernicone N, et al. CDH1 binds MAD2L2 in a Rev1-like pattern. Biochem Biophys Res Commun. 2020;531(4):566–572. doi: 10.1016/j.bbrc.2020.07.118. [DOI] [PubMed] [Google Scholar]
- 84.Vermeulen M, et al. Quantitative interaction proteomics and genome-wide profiling of epigenetic histone marks and their readers. Cell. 2010;142(6):967–80. doi: 10.1016/j.cell.2010.08.020. [DOI] [PubMed] [Google Scholar]
- 85.Itoh G, et al. CAMP (C13orf8, ZNF828) is a novel regulator of kinetochore-microtubule attachment. EMBO J. 2011;30(1):130–44. doi: 10.1038/emboj.2010.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Chun AC, et al. REV7 is required for anaphase-promoting complex-dependent ubiquitination and degradation of translesion DNA polymerase REV1. Cell Cycle. 2013;12(2):365–78. doi: 10.4161/cc.23214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Pirouz M, et al. Destabilization of pluripotency in the absence of Mad2l2. Cell Cycle. 2015;14(10):1596–610. doi: 10.1080/15384101.2015.1026485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Rahjouei A, et al. MAD2L2 Promotes Open Chromatin in Embryonic Stem Cells and Derepresses the Dppa3 Locus. Stem Cell Reports. 2017;8(4):813–821. doi: 10.1016/j.stemcr.2017.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bhat A, et al. Rev7, the regulatory subunit of Polzeta, undergoes UV-induced and Cul4-dependent degradation. FEBS J. 2017;284(12):1790–1803. doi: 10.1111/febs.14088. [DOI] [PubMed] [Google Scholar]
- 90.Brandao LN, et al. The role of Dbf4-dependent protein kinase in DNA polymerase zeta-dependent mutagenesis in Saccharomyces cerevisiae. Genetics. 2014;197(4):1111–22. doi: 10.1534/genetics.114.165308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–74. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 92.Janoueix-Lerosey I, et al. Gene expression profiling of 1p35-36 genes in neuroblastoma. Oncogene. 2004;23(35):5912–22. doi: 10.1038/sj.onc.1207784. [DOI] [PubMed] [Google Scholar]
- 93.Rimkus C, et al. Expression of the mitotic checkpoint gene MAD2L2 has prognostic significance in colon cancer. Int J Cancer. 2007;120(1):207–11. doi: 10.1002/ijc.22155. [DOI] [PubMed] [Google Scholar]
- 94.Pinto M, et al. Expression changes of the MAD mitotic checkpoint gene family in renal cell carcinomas characterized by numerical chromosome changes. Virchows Arch. 2007;450(4):379–85. doi: 10.1007/s00428-007-0386-7. [DOI] [PubMed] [Google Scholar]
- 95.Zhao J, et al. Mitotic arrest deficient protein MAD2B is overexpressed in human glioma, with depletion enhancing sensitivity to ionizing radiation. J Clin Neurosci. 2011;18(6):827–33. doi: 10.1016/j.jocn.2010.11.009. [DOI] [PubMed] [Google Scholar]
- 96.Niimi K, et al. Suppression of REV7 enhances cisplatin sensitivity in ovarian clear cell carcinoma cells. Cancer Sci. 2014;105(5):545–52. doi: 10.1111/cas.12390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Feng L, et al. Knockdown of REV7 Inhibits Breast Cancer Cell Migration and Invasion. Oncol Res. 2016;24(5):315–325. doi: 10.3727/096504016X14666990347590. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 98.Gu C, et al. REV7 confers radioresistance of esophagus squamous cell carcinoma by recruiting PRDX2. Cancer Sci. 2019;110(3):962–972. doi: 10.1111/cas.13946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Okina S, et al. High expression of REV7 is an independent prognostic indicator in patients with diffuse large B-cell lymphoma treated with rituximab. Int J Hematol. 2015;102(6):662–9. doi: 10.1007/s12185-015-1880-3. [DOI] [PubMed] [Google Scholar]
- 100.Pernicone N, et al. MDA-MB-157 Cell Line Presents High Levels of MAD2L2 and Dysregulated Mitosis. Anticancer Res. 2020;40(10):5471–5480. doi: 10.21873/anticanres.14558. [DOI] [PubMed] [Google Scholar]
- 101.Abbasi A, et al. Lack of Rev7 function results in development of tubulostromal adenomas in mouse ovary. Mol Cell Endocrinol. 2015;412:19–25. doi: 10.1016/j.mce.2015.05.022. [DOI] [PubMed] [Google Scholar]
- 102.Weterman MA, et al. Impairment of MAD2B-PRCC interaction in mitotic checkpoint defective t(X;1)-positive renal cell carcinomas. Proc Natl Acad Sci U S A. 2001;98(24):13808–13. doi: 10.1073/pnas.241304198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Li Y, et al. MAD2L2 inhibits colorectal cancer growth by promoting NCOA3 ubiquitination and degradation. Mol Oncol. 2018;12(3):391–405. doi: 10.1002/1878-0261.12173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Hong CF, et al. MAD2B, a novel TCF4-binding protein, modulates TCF4-mediated epithelial-mesenchymal transdifferentiation. J Biol Chem. 2009;284(29):19613–22. doi: 10.1074/jbc.M109.005017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Yu N, et al. MAD2B acts as a negative regulatory partner of TCF4 on proliferation in human dermal papilla cells. Sci Rep. 2017;7(1):11687. doi: 10.1038/s41598-017-10350-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Li L, et al. Hepatocellular carcinoma-associated gene 2 interacts with MAD2L2. Mol Cell Biochem. 2007;304(1-2):297–304. doi: 10.1007/s11010-007-9512-8. [DOI] [PubMed] [Google Scholar]
- 107.Hatano K, et al. A functional screen identifies miRNAs that inhibit DNA repair and sensitize prostate cancer cells to ionizing radiation. Nucleic Acids Res. 2015;43(8):4075–86. doi: 10.1093/nar/gkv273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Actis ML, et al. Identification of the first small-molecule inhibitor of the REV7 DNA repair protein interaction. Bioorg Med Chem. 2016;24(18):4339–4346. doi: 10.1016/j.bmc.2016.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Zimmer J, et al. Targeting BRCA1 and BRCA2 Deficiencies with G-Quadruplex-Interacting Compounds. Mol Cell. 2016;61(3):449–460. doi: 10.1016/j.molcel.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Barazas M, et al. Radiosensitivity Is an Acquired Vulnerability of PARPi-Resistant BRCA1-Deficient Tumors. Cancer Res. 2019;79(3):452–460. doi: 10.1158/0008-5472.CAN-18-2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Aparicio T, et al. DNA double-strand break repair pathway choice and cancer. DNA Repair (Amst) 2014;19:169–75. doi: 10.1016/j.dnarep.2014.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Simonetta M, et al. H4K20me2 distinguishes pre-replicative from post-replicative chromatin to appropriately direct DNA repair pathway choice by 53BP1-RIF1-MAD2L2. Cell Cycle. 2018;17(1):124–136. doi: 10.1080/15384101.2017.1404210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Sabbioneda S, et al. The 9-1-1 checkpoint clamp physically interacts with polzeta and is partially required for spontaneous polzeta-dependent mutagenesis in Saccharomyces cerevisiae. J Biol Chem. 2005;280(46):38657–65. doi: 10.1074/jbc.M507638200. [DOI] [PubMed] [Google Scholar]
- 114.Meng X, et al. A novel binding protein of single-minded 2: the mitotic arrest-deficient protein MAD2B. Neurogenetics. 2012;13(3):251–60. doi: 10.1007/s10048-012-0333-x. [DOI] [PubMed] [Google Scholar]
- 115.Zhang L, et al. Rev7/MAD2B links c-Jun N-terminal protein kinase pathway signaling to activation of the transcription factor Elk-1. Mol Cell Biol. 2007;27(8):2861–9. doi: 10.1128/MCB.02276-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Gomez-Llorente Y, et al. The architecture of yeast DNA polymerase zeta. Cell Rep. 2013;5(1):79–86. doi: 10.1016/j.celrep.2013.08.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Masuda Y, et al. Structure and enzymatic properties of a stable complex of the human REV1 and REV7 proteins. J Biol Chem. 2003;278(14):12356–60. doi: 10.1074/jbc.M211765200. [DOI] [PubMed] [Google Scholar]
- 118.Guo C, et al. Mouse Rev1 protein interacts with multiple DNA polymerases involved in translesion DNA synthesis. EMBO J. 2003;22(24):6621–30. doi: 10.1093/emboj/cdg626. [DOI] [PMC free article] [PubMed] [Google Scholar]