Abstract
Lysosomal cell death is triggered by lysosomal membrane permeabilization (LMP) and subsequent release of lysosomal hydrolases from the lysosomal lumen into the cytosol. Once released to the cytosol, the lysosomal cathepsin proteases act as executioner proteases for the subsequent cell death - either autonomously without caspase activation or in concert with the classical apoptotic machinery. Lysosomal cell death usually remains functional in apoptosis-resistant cancer cells and thus holds great potential as a therapeutic strategy for circumventing apoptosis deficiency in cancers. Notably, lysosomal cell death also plays an important role in normal physiology, e.g. during the regression of the mammary gland. Here we present four complementary methods for functional quantification and visualization of LMP during the onset of death: (1) enzymatic activity measurements of released lysosomal hydrolases in the cytosol after digitonin extraction, (2) direct visualization of LMP by monitoring the release of fluorescent dextran from lysosomes into the cytosol, (3) immunocytochemistry to detect cathepsins released into the cytosol and, (4) detection of the translocation of galectins from the cytosol to damaged lysosomes. The methods presented here can ideally be combined as needed to provide solid evidence for LMP after a given cytotoxic stimuli.
Keywords: Lysosomal membrane permeabilization, LMP, Death pathway, Method, Lysosomal cell death, Cathepsins, Galectins
Introduction
Lysosomes are acidic vesicles (pH 4-5) found in all mammalian cells except for mature erythrocytes. Their main function is the disposal and re-cycling of worn-out and damaged cellular macromolecules and organelles as well as the digestion of extracellular and foreign materials delivered to them by endo- and phagocytosis (Saftig and Klumperman 623-35;Pryor and Luzio 615-24;Kolter and Sandhoff 1700-12). The digestion of the cargo is brought about by over 50 lysosomal hydrolases including proteases, glycosidases, phosphatases, sulfatases, nucleases and lipases. Most lysosomal hydrolases are optimally active in the acidic environment of the lysosomal lumen but many of them can also function in neutral pH, although with less efficacy. Accordingly, lysosomal hydrolases have been assigned important extralysosomal functions e.g. in programmed cell death, as discussed below, and in cell membrane repair and tissue remodelling (Kroemer and Jaattela 886-97;Vasiljeva and Turk 380-86;Gerasimenko, Gerasimenko, and Petersen R971-R974).
The role of lysosomes and lysosomal hydrolases in cell death was introduced by a Belgian scientist Christian de Duve who was awarded the Nobel Prize for his discovery and characterization of lysosomes (DE DUVE et al. 604-17;de 391-97). Due to the potent hydrolytic capacity of lysosomal hydrolases, de Duve defined lysosomes as “suicide bags” that can cause cell death and tissue damage upon rupture and subsequent release of lysosomal hydrolyses to the cytosol. This definition triggered an intensive search for pharmaceutical agents that either stabilize or destabilize lysosomal membranes for the treatment of degenerative disorders and cancer, respectively. As a result, hydrocortisone and cholesterol were identified as lysosome stabilizing agents (de 391-97), whereas many amines with long hydrophobic chains and pK values around 5 - 9 were identified as lysosomotropic detergents with potential applications in cancer therapy (Firestone, Pisano, and Bonney 1130-33). Interest in lysosomal cell death pathways waned, however, rapidly. de Duve speculated that this was largely due to the lack of assays that could differentiate lysosomal rupture that causes cell death from postmortal alterations in autolytic cells as well as the fear that lysosomotropic detergents would be equally toxic to normal and transformed cells (de 391-97). Consequently, novel more sensitive assays to study lysosomal membrane permeabilization (LMP) (see below) and emerging data indicating that cancer cell lysosomes are less stable than normal lysosomes were needed to initiate a new wave of interest in lysosomal cell death in the beginning of the 21st century (Kroemer and Jaattela 886-97).
Whereas the importance of LMP in cell and tissue autolysis during uncontrolled necrosis was well established already in the 1970’s (de 391-97), it took a long time to recognize its role in the more controlled events of programmed cell death (PCD). One of the reasons for this delay was the fact that methyl ketone peptide inhibitors (e.g. zVAD-fmk) commonly used to assess the role of caspases in PCD also inhibit several lysosomal cathepsins that function as effectors of lysosomal cell death (Schotte et al. 117-21;Foghsgaard et al. 999-1010). Additionally, the lysosomal involvement in PCD was overlooked in some studies, because the lysosomal ultrastructure appears intact even after lysosomal hydrolases have leaked into the cytosol (Brunk, Neuzil, and Eaton 91-97). However, studies with agents that directly disrupt the integrity of lysosomal membrane finally proved that LMP can not only trigger uncontrolled necrosis but also apoptosis or apoptosis-like PCD (Brunk et al. 616-26;Kagedal et al. 335-43;Cirman et al. 3578-87;Boya et al. 1323-34). A quantitative relationship between the amount of lysosomal rupture and the mode of cell death might explain the different morphological outcomes following LMP. According to this model, limited release of lysosomal contents to the cytosol triggers apoptosis or apoptosis-like PCD, while massive lysosomal rupture results in rapid cellular necrosis (Brunk et al. 616-26;Kagedal et al. 335-43)(13, 14). The abundant cysteine cathepsins B and L and aspartate cathepsin D are among the best-defined effector molecules in LMP-induced apoptosis and apoptosis-like PCD (Stoka, Turk, and Turk 555-60;Kirkegaard and Jaattela 746-54)(17, 18). It should, however, be emphasized that their inhibition often provides only partial protection against LMP-induced cell death. Thus, more studies are clearly needed to define the roles of other lysosomal hydrolases (e.g. lipases and phosphatases), lysosome-derived second messengers (e.g. Ca2+ and reactive oxygen species (ROS)) as well as LMP-associated dysfunction of lysosomes and acidification of the cytosol in LMP-induced death.
The distinctive characteristic of LMP is the translocation of soluble hydrolases from the lysosomal lumen to the cytosol but the exact mechanism responsible for LMP is still speculative. Whether special channels are involved in the relocation of lysosomal proteins or whether transient pores of a certain size are formed in the membrane remains to be established. Alternatively, ROS can directly damage the lysosomal membranes leading to a less controlled leakage of lysosomal hydrolases from the affected lysosome. The susceptibility of the individual lysosomes to ROS-induced LMP may be controlled by the spatial distribution of lysosomes in relation to the source of ROS such as damaged mitochondria (Boya and Kroemer 6434-51), or by variations in their content of iron which catalyzes the production of highly reactive pro-oxidants from hydrogen peroxide via Fenton reactions (Link, Pinson, and Hershko 127-34;Kurz, Eaton, and Brunk 1686-97).
Besides their role in death caused by direct lysosomal disrupters, LMP and lysosomal hydrolases also participate in the execution of cell death induced by an array of classic apoptotic stimuli such as death receptor activation, p53 and cytotoxic drugs not primarily designed to target lysosomes (Kroemer and Jaattela 886-97). Importantly, cancer cells with defects in their apoptosis machinery are still able to undergo lysosomal cell death. Rapidly dividing cancer cells are especially dependent on effective lysosomal function, and dramatic changes in lysosomal volume, composition and cellular distribution occur during transformation and cancer progression (Kallunki T;Moin et al. 1093-99;Palermo and Joyce 22-28)(20-22). The realization that these changes which promote invasive growth simultaneously sensitize cells to LMP and lysosomal cell death has initiated a new wave of interest in lysosomes as targets for cancer therapy. Given the potentially fatal outcome of LMP, it is not surprising that cancer cells have developed various strategies to counteract it such as the up-regulation of cytosolic protease inhibitors (Silverman et al. 480-87;Suminami, Nawata, and Kato 488-93) or the translocation of cytosolic heat shock protein 70 (Hsp70) to the lysosomal lumen where it stabilizes the lysosomal membranes by enhancing the activity of acid sphingomyelinase (Nylandsted et al. 425-35;Kirkegaard et al. 549-53). In addition to the growing interest in lysosomal cell death among cancer researchers, it recently entered a broader scene when Christine Watson and co-workers demonstrated the first physiological role for LMP-dependent death during the regression of the mammary gland after involution (Kreuzaler et al. 303-09)(19).
Historically, lysosomal membrane permebilization was assessed using a β-glycerophosphate substrate that does not readily penetrate the lysosomal membrane unless the permeability is altered. The degree of membrane permeability was visualized by Gomori acid phosphatase staining. Based on this approach, Bitensky and co-workers utilized this concept and developed a “lysosomal fragility test” to estimate LMP and suggested the method as a sensitive measure of early cell injury (BITENSKY 193-96). During the last decade, additional methods have been introduced including enzymatic quantification of released lysosomal proteases in the cytosol (Method 1), fluorescent dextran release (Method 2) and immunocytochemistry (Method 3, 4). To quantify LMP by measuring the amount of lysosomal hydrolases (e.g. lysosomal cysteine cathepsins) released into the cytosol, we have developed an assay presented in Method 1. The assay is based on the extraction of cytosol by digitonin followed by measurement of lysosomal hydrolase activities in the extracted cytosol and total cellular lysate with a fluorigenic substrate. LMP can also be monitored by time lapse imaging of cells in which the lysosomes are loaded with fluorescent dextran as described in Method 2. This approach allows the researcher to visualize the LMP process in real time by following the release of dextran into the cytosol and makes it possible to determine the membrane pore sizes by applying different sizes of dextran. Furthermore, with the improvement of flourochromes and antibody technology for immunocytochemistry, LMP is today often monitored simply by staining for lysosomal cathepsin proteases. In healthy cells these are localized in the lumen of lysosomes resulting in a punctate staining pattern, whereas an LMP-inducing insult results in the release of cathepsins and a diffuse staining pattern throughout the cytosol (Nylandsted et al. 425-35). The advantage of this method is that co-staining of other relevant death components (e.g. activated Bax, activated caspases or cytochrome c release) can be used to address the order of events in the death pathway of interest.
Finally, we have recently developed an assay based on the translocation of the sugar-binding proteins galectin-1 and -3 to damaged lysosomes (Method 4). Galectins are normally found in the cytosol and bound to glycans on the cell surface but localize to the lysosomes after LMP. This is thought to be a consequence of their gaining access to the glycocalyx that lines the inside of the lysosomal membrane. Galectin translocation can be detected by immunocytochemistry, as described here, or using fluorescent constructs. The galectin translocation assay is highly sensitive and allows quantification of LMP from the early stage of the death process. It is also the only assay that can detect individual damaged lysosomes. We hope that the protocols presented here and the constant development of better methods to detect and quantify LMP will attract more researchers to this exciting field which still has many unanswered questions.
Methods
Method 1. Quantification of Cathepsin and β-N-Acetyl-Glucosaminidase Release into the Cytosol by Enzymatic Activity Measurement
This protocol takes advantage of digitonin, a detergent that creates pores in cellular membranes by replacing cholesterol. The difference in cholesterol content between the plasma membrane (high) and lysosomal membrane (low) allows the titration of a digitonin concentration that permeabilizes the plasma membrane but leaves lysosomal membranes intact. The extent of LMP can then be determined by measuring the activity of lysosomal hydrolases (e.g. cysteine cathepsins or β-N-acetyl-glucosaminidase (NAG)) in the digitonin-extracted cytosol and comparing it to the total cellular activity. Caspase activity can be measured in parallel in whole-cell extract if desired.
Points of attention
Digitonin extraction is performed on a rocking table with adjustable lifting frequency (15 min extraction; lifting frequency 50-60/min, digitonin concentration 15-25 μg/ml). The cathepsin activity is normalized to Lactate dehydrogenase (LDH) which is a cytosolic protein. Alternatively, cathepsin activity levels can be normalized to cytosolic protein levels by using a dedicated protein determination kit.
When using treatments that influence cellular cholesterol content or have detergent-like properties as e.g. cationic amphiphilic drugs (Petersen et al. 379-93), one should be aware that such treatments may interfere with the digitonin extraction procedure. In such cases, alternative approaches should be used to estimate LMP, e.g. those described in Method 2-4.
Materials
Reagents
β-N-acetyl-glucosaminidase (NAG) reaction buffer (NAG RB)
0.2 M sodium citrate buffer, pH 4.5, containing 300 μg/ml 4-methylumbelliferyl-2-acetamido-2-deoxy-β-d-glucopyranoside (Sigma-Aldrich). The NAG reaction buffer is aliquoted and stored at -20°C.
Caspase reaction buffer (Caspase RB)
100 mM Hepes, 20% glycerol, 0.5 mM EDTA, 0.1% CHAPS, 5 mM dithiothreitol (DTT), and 0.5 mM pefabloc, pH 7.5. Substrate: 50 μM Ac-DEVD–7-amino-trifluoromethylcoumarin (AFC) (BIOMOL Research Laboratories, Inc.).
Prepare the reaction buffer freshly by adding pefabloc 1:200 (100 mM stock), DTT 1:125 (1 mM stock) and Ac-DEVD-AFC 1:400 (20 mM stock) to the premade buffer Caspase RB.
Cathepsin reaction buffer (Cathepsin RB)
50 mM sodium acetate, 4 mM EDTA, 8 mM DTT, and 0.5 mM pefabloc, pH 6.0. Substrate: 50 μM zFR-AFC (Enzyme System Products).
The reaction is made freshly by adding pefabloc 1:200 (100 mM stock), DTT 1:125 (1 mM stock) and zFR-AFC 1:400 (20 mM stock) to the premade sodium acetate/EDTA buffer.
Digitonin extraction buffer (DE buffer)
250 mM sucrose, 20 mM Hepes, 10 mM KCl, 1.5 mM MgCl2, 1 mM EDTA, 1 mM EGTA, and 0.5 mM pefabloc, pH 7.5.
Add fresh pefablock 1:200 (100 mM stock) before extraction.
Digitonin stocks (Sigma)
5 and 50 μg/μl digitonin dissolved in H2O. Store at RT.
Lactate dehydrogenase (LDH) cytotoxicity detection kit (Roche)
Equipment
24-well cell culture plates
Absorbance plate reader (e.g. VersaMax plate reader, Molecular Devices)
Rocking table with adjustable lifting frequency
Spectrofluorometer (e.g. Spectramax Gemini fluorometer, Molecular Devices)
Vacuum suction pump
Time frame
-
-
The optimization of digitonin extraction takes about 1h.
-
-
The procedure for measuring LMP by digitonin extraction takes 2 - 2.5 h for two 24-well plates.
Protocol
Determination of the optimal digitonin concentration for the extraction of lysosome-free cytosol
The optimization procedure described below is necessary to determine the optimal digitonin concentration that only permebilizes the plasma membrane with minimal impact on the lysosomal membrane. It must be performed for each cell line separately and should be done regularly because reagents (e.g. digitonin stocks) and cellular conditions may change over time (Fig. 1). It should be noted that the ability of digitionin to permeabilize cellular membranes depends not only on digitonin concentration but also on the total amount of digitonin / cell and thus the DE buffer should always be used in the same volume / cell.
Seed 5x104 cells per well in a 24-well plate and let cells adhere overnight. Set up at least 12 wells for the optimization.
The day after, prepare 200 μl digitonin extraction buffer (DE buffer) per well (+20 μl to adjust for pipetting loss). The conditions can be scaled up or down accordingly if using e.g. 6- or 96-well plates.
Heat up the digitonin stocks (50 and 5 μg/μl stocks) to 80 °C for 5-10 min to dissolve any precipitates. Digitonin usually precipitates and needs to be re-dissolved by heating and occasional mixing by vortexing.
Make dilutions of digitonin in DE buffer with pefablock i.e.: 0, 5, 10, 15, 20, 25, 30, 35, 40, 45, 50 and 200 μg/ml digitonin. 200 μg/ml digitonin is used for complete permeabilization of cells.
Place cell plate on ice and remove medium using a vacuum suction pump; remove only medium for 6 wells at a time to avoid cells drying out. Add 200 μl of digitonin dilutions per well. Make sure to add DE buffer to the side of the well to avoid flushing off attached cells.
Start the timer when DE buffer is added to each set of wells (i.e. 6 wells). Note: The time is critical to ensure that each well is extracted equally.
Incubate cells for 15 min on a rocking table (lifting frequency approx. 50-60 /min) and in the meanwhile mark a 96-well plate appropriately.
Then, transfer 180 μl of extract to the 96-well plate and place it on ice.
The cathepsin reaction is started by mixing 50 μl of extract with 50 μl cathepsin reaction buffer (Cathepsin RB) in a black 96 well plate. Pre-incubate the plate at 30 °C for 5 min in the plate reader before reading is started. The kinetics of cathepsin activity (i.e. Vmax of the liberation of AFC; excitation, 400 nm; emission, 489 nm) are measured for 20 min at 30 °C in a fluorometer.
While the cathepsin measurement is running set up an LDH assay to measure plasma membrane permeabilization by placing 30 μl extract into a 96-well plate and letting it equilibrate to room temperature for 5-10 min.
Add 30 μl of mixed LDH reagent per well and let the reaction run for 2-10 min before stopping it with 20 μl 1 M HCL. Make sure that all samples have equal reaction time before ending the reaction typically when some samples are medium to intense red.
Measure LDH activity at OD 490 nm in an absorbance microplate reader.
To determine the optimal digitonin concentration for cytosolic extraction the cathepsin release and LDH raw values are compared in a graph. The concentration that gives the best possible permeabilizion of the plasma membrane (LDH release) with minimal cathepsin release from the lysosomes is optimal (Fig. 1).
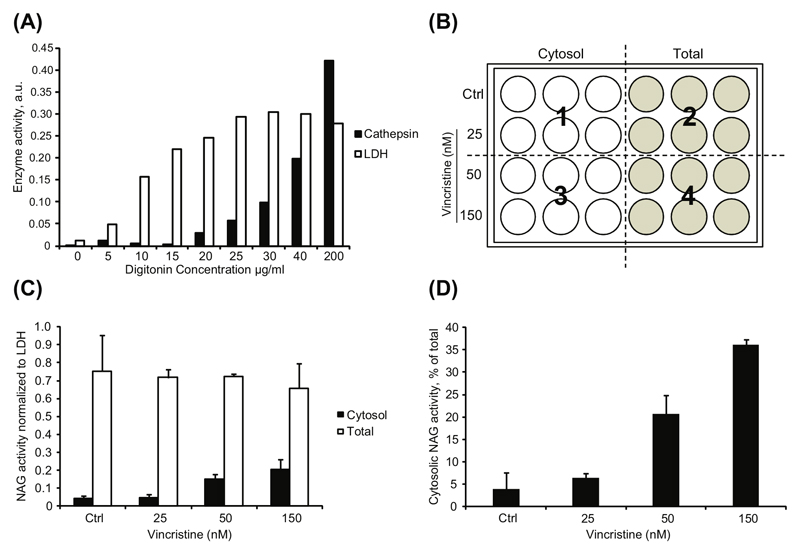
Figure 1.
(A) Example of digitonin optimization for cytosolic extraction of mouse embryo fibroblasts (MEFs). Cells (5x104 per well) were extracted for 10 min on a rocking table (lifting frequency 110/min) with indicated digitonin concentrations and the level of plasma membrane permeabilization (LDH activity) and lysosome permeabilization (Cathepsin activity) was measured. A digitonin concentration of 15-17 μg/ml is optimal for these cells. (B) Exemplified LMP extraction setup in 24-well plate formats with the vinca alkaloid vincristine as LMP inducer, which depolymerises microtubules, aggregates lysosomes and triggers LMP (Groth-Pedersen et al. 2217-25). Cells are seeded in triplicates for cytosolic and total cellular extraction respectively and digitonin is added to sets of six wells (marked 1-4) and timed, to ensure that all wells are extracted equally. (C) A representative example of lysosomal NAG release obtained from HeLa cells treated for 48 h with vincristine. Results are presented as NAG values normalized to LDH for cytosol and total digitonin extraction respectively or (D) presented as % cytosolic release of total cellular NAG activity.
Measurement of LMP
The level of LMP as a response to the given treatment can now be measured by digitonin extraction of the cytosolic fraction followed by the hydrolase activity measurements. The following protocol is used for this:
-
14.
Seed cells at a density that will on the day of analysis result in the density used in the digitonin optimization experiment (approximately 5x104 cells). For each cellular condition (e.g. siRNA or cytotoxic compound), cells are seeded in triplicates (24-well plate) and parallel triplicate wells for whole-cell cellular cathepsin measurements are included (Fig. 1).
-
15.
Shortly before the LMP measurement, dilute digitonin in the DE buffer with pefablock for cytosolic and whole-cell lyate extraction. In the example here, we use 17 μg/ml and 200 μg/ml digitonin for cytosolic and total cell extractions in murine embryonic fibroblasts, respectively (Fig. 1).
-
16.
Place cell plates on ice and remove medium with a vacuum suction pump. Remove only medium from maximum of 6 wells at a time to avoid the drying of the cells. Add 200 μl of digitonin dilutions per well. Make sure to add DE buffer to the side of the well to avoid flushing off attached cells.
-
17.
Start the timer when the DE buffer is added to each set of wells (i.e. 6 wells). Note: the timing is critical to ensure that each well is extracted for equal time. Mark the time e.g. on the plate lid for each set. Swift pipetting is necessary to avoid too much variation between wells.
-
18.
Incubate the plates on ice for 15 min on a rocking table (lifting frequency approx. 50-60 /min). Note: the optimal time and lifting frequency may vary between cell lines.
-
19.
Transfer 180 μl of extract from each well into a 96-well plate on ice. At this point, the cytosolic/total extracts can be used to measure lysosomal cysteine cathepsin, NAG, LDH and caspase 3-like activities.
-
20.
The cathepsin reaction is started by mixing 50 μl of extract with 50μl cathepsin RB in a black 96-well plate.
-
21.
The caspase 3-like activity is measured by incubating 50 μl of the total extract with 50 μl of Caspase RB and can be measured on the same plate as the cathepsin reaction provided that the substrates are coupled to the same fluorescent marker (e.g. AFC).
-
22.
Pre-incubate the reaction plate at 30° C for 5 min in the plate reader before starting the analysis. The kinetics of the enzyme activity (Vmax of the liberation of AFC; excitation, 400 nm; emission, 489 nm) are measured for 20 min at 30° C in a fluorometer (45 sec interval).
-
23.
To measure lysosomal NAG release, 30 μl extract from each well is mixed with 100 μl NAG RB in a black 96-well plate and pre incubated for 3-5 min at 30 °C in the plate reader at 30° C. The Vmax of the liberation of methylumbelliferyl (excitation, 356 nm; emission, 444 nm) is measured for 20 min at 30° C with a fluorometer (45 sec interval).
-
24.
LDH activities in the extracts are used as internal standards to which the hydrolase activities are normalized. Transfer 30 μl from each extract into a 96-well plate and let it equilibrate to room temperature for 5-10 min. Add 30 μl of mixed LDH reagent per well and let the reaction run for 2-10 min before stopping it with 20 μl 1 M HCL. Measure the LDH activity at OD 490 nm in an absorbance plate reader. Make sure that all samples have equal reaction time before ending the reaction typically when the samples with highest LDH content are medium to intense red. If the treatment of the cells alters the cellular LDH activity, hydrolase activities can be normalized to total protein content using a commercial kit dedicated for this purpose.
Data analysis
In order to normalize the hydrolase levels to an internal standard, all values are related to the corresponding LDH value from the same well and mean values are calculated for cytosolic and total protease levels respectively (Fig. 2A). The percentage of released enzyme activity is calculated by relating the LDH-corrected cytosolic activity of the lysosomal hydrolase measured to the corresponding LDH-corrected total cellular activity: Cytosolic Activity / Total Activity x100 (Fig. 1B).
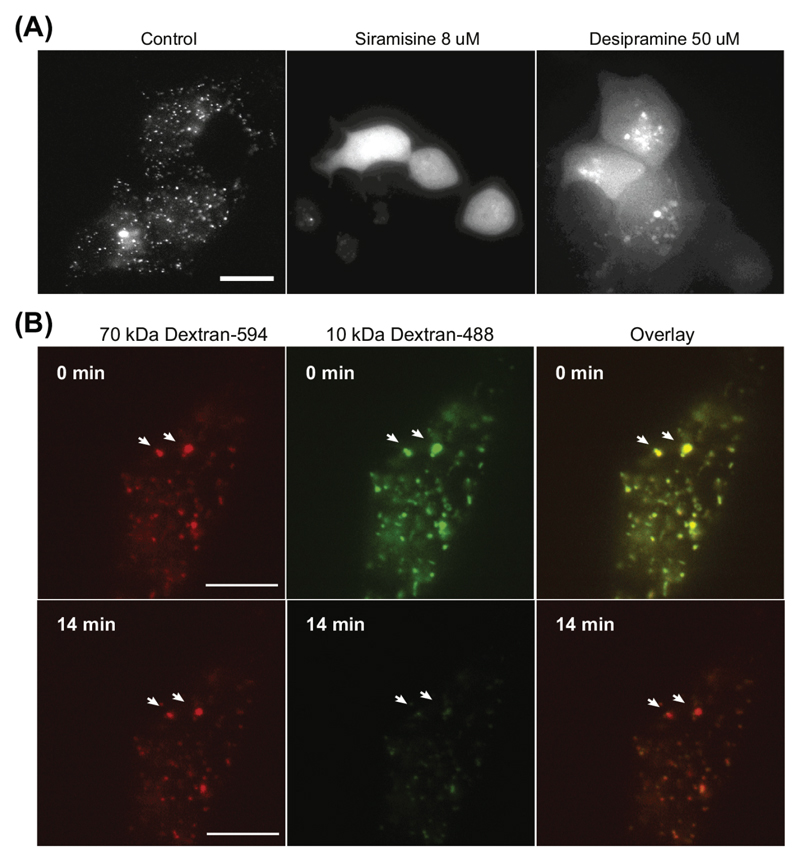
Figure 2. LMP visualized by fluorescent dextran release.
(A) Representative pictures of MCF-7 breast carcinoma cells loaded with 10 kDa Alexa Fluor®488 dextran and treated for 20 h with indicated cationic amphiphilic drugs that induce LMP (Petersen et al. 379-93). (B) Pictures obtained from a time-lapse movie of HeLa cells loaded with 10 kDa Alexa Fluor®488 and 70 kDa Alexa Flour®594 dextran and incubated with hydrogen peroxide for 20 min. Only 10 kDa dextran is released to the cytosol after 14 min H2O2 exposure indicating that the pores formed in the membrane are restricting 70 kDa dextran to be released. Scale bars: 20 μM. Unpublished data.
Troubleshooting
Problem
A common difficulty associated with LMP measurements is digitonin over-extraction resulting in high cytosolic cathepsin/NAG background levels in control cells (more than 10%).
Solution
This can be adjusted by further finetuning the digitonin extraction (Ad. 1-13) with a narrower digitonin concentration range to determine the optimal concentration. In addition, the cell density is critical to achieve the best digitonin/cell ratio for cytosolic extraction and this is optimized by keeping the digitonin concentration constant and varying the cell density. Finally, the optimization of the extraction time may be helpful in some cells.
Problem
As mentioned above (see Points of attention), cytotoxic stimuli that have a direct impact on lipids in the plasma or lysosomal membrane may interfere with the digitonin extraction procedure giving misleading results.
Solution
LMP should be assessed by using Method 2-4.
Problem
Due to a shorter half-life of cathepsins in the cytosol as compared to the lysosome, the obtained values are, in fact, lower than the actual release of the hydrolases. This may become a problem especially in long-term experiments and should be considered if the total cathepsin/LDH ratios decline extensively.
Solution
The cytosolic half-life of NAG is longer than that of cathepsins and thus NAG measurements can give more accurate values in long-term assays. Additionally, determination of the kinetics of the LMP by multiple measurements at different time points after the stimulus may help the interpretation of the results.
Method 2. LMP Visualized by Release of Fluorescent Dextran to the Cytosol
LMP can be monitored by taking advantage of the steady endocytic capacity of cells, which allows the loading of fluorescent dextran into lysosomes. The method presented here can be used to monitor LMP simply by observing the translocation of lysosomally localized dextran into the cytosol after an LMP insult. In healthy cells fluorescent dextran appears in punctuate structures inside lysosomes, whereas after LMP, a diffuse staining pattern throughout the cytosol is observed (Fig. 2A). The method allows following the LMP process in real time using time-lapse imaging. It can also be used to determine the dimensions of pores formed in the membrane during LMP by size exclusion using dextrans of different sizes and colours e.g. by comparing the release of 10, 70 and 500 kDa Dextran in different colours (Bidere et al. 31401-11). Exposure of HeLa cells to H2O2 creates pores in the lysosomal membrane of a size that only allows 10 kDa dextran to be released and retains 70 kDa dextran in the lumen as shown in Figure 2B.
Furthermore, flourescein isothiocyanate (FITC) conjugated to dextran (Ex 488 nm) can be used to monitor early changes in lysosomal pH during the LMP process. The fluorescence intensity of FITC-dextran is dramatically reduced in normal lysosomes at acidic pH 4-5. Thus, neutralization of lysosomes upon LMP results in up to 8-fold increase in fluorescence intensity.
Materials
Reagents
Alexa Fluor® 488/594 dextran 10 or 70 kDa (Life Technologies/Molecular Probes), Anionic fixable (Other dextran sizes are commercially available e.g. 500 kDa). Stock solution: 5 μg/μl in serum-free medium (store at 4 °C).
Flourescein isothiocyanate (FITC) conjugated to dextran (70 kDa) (Life Technologies/Molecular Probes). Stock solution: 5 μg/μl in serum-free medium (store at 4 °C).
Equipment
Glass chamber slides for live cell imaging (e.g. Lab-Tek Chambered coverglass, Nalge Nunc International)
Glass coverslips
Inverted fluorescence microscope or/and confocal microscope
Time frame
Loading cells with fluorescent dextran including a 2 h chase period can be done within 4-8 h even though longer (16 h) incubation is recommended to increase the number of stained lysosomes.
Protocol
The following guide applies to adherent cells for visualizing LMP by dextran release:
Seed cells in chamber slides or on glass coverslips and allow cells to adhere.
Add fluorescent dextran to the medium in the concentration 50-200 μg/ml of e.g. Alexa Fluor® 488- or 594-Dextran (10 kDa) or 75-100 μg/ml FITC-Dextran.
Incubate for 2 – 16 h. The optimal dextran concentration and incubation time varies between cell lines and should be optimized. For HeLa and MCF-7 cells, 50-100 μg/ml Alexa Fluor® 488/594-Dextran (10 kDa) for 3-6 h works well.
Alternatively, cells can be loaded faster by incubating them directly in the dextran stock solution i.e. 5 μg/μl for 1-2 h. To save the valuable dextran, the stock solution can be collected and re-used.
After dextran loading, wash cells twice in PBS and chase for 2 h in fresh medium. Dextran uptake into lysosomes can after this point be inspected live using an inverted fluorescence microscope.
Cells with loaded lysosomes can subsequently be treated with LMP-inducing cytotoxic stimuli.
After proper incubation time, inspect cells under a fluorescence microscope. The degree of LMP (dextran release into the cytosol) can be imaged and quantified either manually or by using dedicated imaging software). When using FITC-dextran to monitor LMP-associated pH changes, include a positive control, e.g. concanamycin A, which inhibits the activity of the lysosomal V-H+-ATPase pump and increases lysosomal pH and FITC fluorescence up to 8 fold normally without causing LMP.
Cells loaded with anionic fixable Alexa Fluor® 488/594 can be fixated in 4% paraformaldehyde in PBS and then imaged/estimated using a fluorescence or confocal microscope. However, the fixation should be carefully optimized to achieve good results.
Troubleshooting
Problem
In cases where the degree of LMP triggered by a particular cytotoxic insult is relatively weak it might be difficult to recognize/monitor LMP by microscopy using the dextran release procedure.
Solution
Increasing the concentration of dextran and/or applying sensitive imaging software dedicated to measure differences in cellular fluorescence (e.g. MetaMorph software) might prove advantageous.
Method 3. LMP Visualized by Cathepsin Immunocytochemistry
The protocol presented here is based on immunocytochemical staining of the lysosomal protease cathepsin L that visualizes its translocation from the lysosomal lumen to the cytosol upon LMP. Alternatively, staining for cathepsin B can be used in cells with low cathepsin L expression. In healthy cells cathepsins will appear in localized punctuate structures inside lysosomes whereas LMP will cause their release and result in a diffuse staining pattern throughout the cytoplasm. LMP can be triggered upstream, downstream or independently of the classical apoptotic death pathway involving mitochondrial outer membrane permeabilization (MOMP). Co-staining with antibodies recognizing e.g. the active form of Bax will allow this method to be used to address the order of events in death signalling between LMP and MOMP.
Materials
Reagents
0.2% Triton X-100 in PBS
4% paraformaldehyde in PBS
Ice cold methanol (-20 °C)
Alexa Fluor® 488-conjugated anti-mouse secondary antibody
Alexa Fluor® 594-conjugated anti-rabbit secondary antibody
Anti-human cathepsin L mouse antibody (BD Transduction Laboratories, cat. no. 611084)
Anti-human Bax rabbit antibody, active conformation (Cell Signaling, cat. no. 2772)
Immunoflourescence buffer-1 (IF-Buffer-1)
1% BSA, 0.3% triton X-100 in PBS (Store at -20 °C)
Immunoflourescence buffer-2 (IF-Buffer-2)
0.25% BSA, 0.1% Triton X-100 in PBS (Store at 4 °C)
Immunoflourescence buffer-3 (IF-Buffer-3)
0.05% Tween-20 in PBS (Store at 4 °C)
Mounting medium
Equipment
Glass coverslips
24-well cell culture plates
Glass slides
Flourescence or confocal microscope
Fine tweezers
Time frame
The immunocytochemisty immune-staining procedure can be performed within 4-5 h.
Protocol
Seed cells on coverslips at 25-50% confluency depending on the length of the following treatment period. Usually 2.5x104 cells for coverslips that fit into a 24-well plate using 48 h treatment.
When cells have firmly adhered to the coverslip start the treatment by adding the cytotoxic agent (compound or siRNA) for the desired length of time. Remember to include a positive control for LMP e.g. L-leucyl-L-leucine-methyl ester (2-6 h treatment with 1-2.5 mM suitable for most cell lines).
Wash cells twice in PBS and fixate in 4% paraformaldehyde for 20 min at RT or 3 min in ice-cold methanol. If using treatments that cause early detachment (e.g. cytoskeleton disrupting drugs like vincristine) cells can be centrifuged onto glass slides (Cytospin: 600 x g for 5 min) before fixation.
Wash once in PBS and permeabilize cells in 0.2% Triton X-100 for 2 min. After this point cells can stored at 4 °C in PBS for later staining.
Wash twice in PBS and block cells by incubating coverslips in IF-Buffer-1 containing 5% fetal calf serum for 20 min. At this point coverslips can gently be transferred from the dish to a plane area wrapped in para film for subsequent staining to minimize antibody use.
Wash once in IF-Buffer-1 and overlay coverslips with approximately 100 μl of cathepsin L/Bax antibody (dilution 1:350) mixture in IF-Buffer-1 for 1 h at RT. If incubating longer ensure that the antibody solution doesn’t dry out e.g. by placing the coverslips in a humidified chamber.
Wash the coverslips 3x5 min in IF-Buffer-2.
Incubate cells on coverslips with Alexa Fluor®488/594-conjugated secondary antibodies (1:1000 dilution) in IF-Buffer-2 (approx. 100 μl) for 1 h at RT in the dark.
Wash 3x5 min in IF-Buffer-3.
Mount coverslips after a brief wash in water (to get rid of salt from the washing solution)- dry off excess liquid on a paper towel and mount on glass slides (cell side down) with a drop of antifade mounting medium (store slides at 4 °C).
Let the mounting medium solidify (usually after 2-3 h) before the slides are examined by fluorescence or confocal microscopy.
Cells are analysed for vesicular versus diffuse staining of lysosomal cathepsin L and absence versus presence of active Bax by imaging.
Data analysis
For each experiment one hundred cells are randomly chosen and the number of cells containing released cathepsin L and active Bax translocated to mitochondria out of the total cells are counted for each condition (Groth-Pedersen et al. 2217-25). Dedicated imaging software e.g. ImageJ can be used here to quantify cathepsin /Bax translocation from captured pictures.
Troubleshooting
Problem
Cathepsin L/Bax staining looks unspecific with too high background.
Solution
Make sure to use fresh 4% paraformaldehyde (or thaw a fresh stock solution from -20 °C) for fixation and in general prepare reagents fresh. Optimize the fixation procedure further by reducing the fixation time, changing the permeabilization agent to saposin or combining 4% paraformaldehyde fixation with subsequent methanol permeabilization/fixation.
Method 4. Detection of Damaged Lysosomes by Galectin-1 and -3 Translocation
The protocol presented here is based on the detection of the sugar-binding proteins galectin-1 and -3 which translocate from the cytosol to lysosomes upon the permeabilization of the lysosomal membrane, regardless of the mode of damage (unpublished data). This can be visualized as a change in staining pattern from a diffuse cytosolic staining to a dotted staining after immunostaining (Figure 4). After screening and pilot experiments using galectin staining alone, it should be verified that galectin dots indeed label lysosomes since galectins also translocate to damaged endo- or phagosomes. This can e.g. be done by co-staining with lysosomal membrane proteins such as LAMPs or LIMPs. If desired, the staining can also be combined with staining for cathepsins or the active form of Bax (see Method 3).
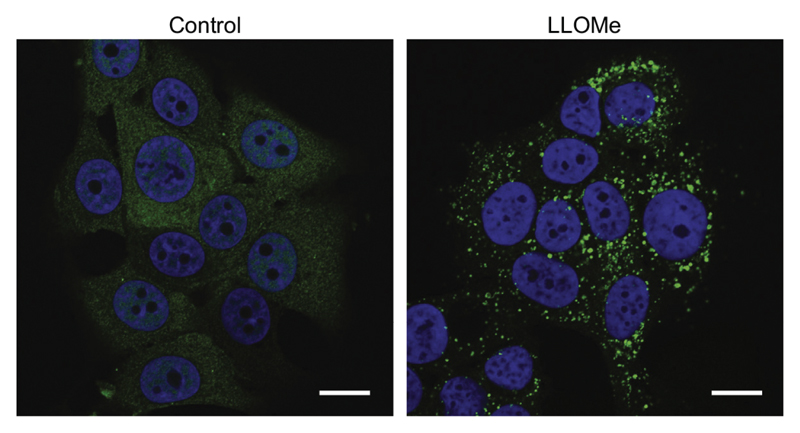
Figure 4. LMP visualized by galectin-3 translocation.
MCF-7 cells were treated with 2 mM L-leucyl-L-leucine-methyl ester (LLOMe) for 2 h to induce LMP and stained for Galectin-3. Nuclei were labelled with Hoechst 33342. Unpublished data.
Materials
Reagents
4% paraformaldehyde in PBS
50 mM ammonium chloride in PBS
mouse anti-galectin-3 antibody (BD Biosciences, cat. no. 556904)
rabbit anti-galectin-1 antibody (Abcam, cat. no. ab25138)
Alexa 488 or 594-donkey anti-mouse antibody (Life Technologies, cat. no. A21202 or A21203)
Alexa 488 or 594-donkey anti-rabbit antibody (Life Technologies, cat. no. A21206 or A21207) goat serum
Hoechst 33342 (25 mg/ml in PBS)
Immunoflourescence buffer-1 (IF-Buffer-1)
1% BSA, 0.3% Triton X-100 in PBS (Store at -20 °C)
Immunoflourescence buffer-2 (IF-Buffer-2)
0.25% BSA, 0.1% Triton X-100 in PBS (Store at 4 °C)
Immunoflourescence buffer-3 (IF-Buffer-3)
0.05% Tween-20 in PBS (Store at 4 °C)
Anti-fade mounting medium
Equipment
24-well cell culture plates
Glass coverslips
Glass slides
Fine tweezers
Flourescence or confocal microscope
Time frame
This immunostaining procedure is normally performed over 2 days, followed by an extra day for letting the slides dry.
Protocol
Place coverslips in 24-well culture plates and seed cells so they are 50-70% confluent at the time of treatment (e.g. 2 x104 cells for U2-OS and 5x104 cells for MCF7 for treatment on the next day). Alternatively, cells can be seeded in microscopy slides with removable chambers.
When cells have firmly adhered (usually the next day) subject them to the treatment of interest. Remember to include a positive control for LMP e.g. L-leucyl-L-leucine-methyl ester (2-6 h treatment with 1-2.5 mM suitable for most cell lines.
Remove medium and fixate cells in 4% paraformaldehyde for 10 min at RT. Make sure coverslips do not dry out at this or any other subsequent step. If required, cells can also be fixed in ice-cold methanol for 2 min.
Wash once in PBS. After this point cells can stored at 4 °C in PBS for later staining.
Transfer coverslips to a flat area (e.g. a large Petri dish) covered with Parafilm and incubate 10 min in ammonium chloride solution. For this and all subsequent steps 90-100 μl of the solution is pipetted onto each cover slip. The Parafilm ensures that the liquid forms a drop and does not run off. After the incubation, the solution is carefully removed with vacuum suction.
Wash twice in PBS and then permeabilize and block by incubating in IF-Buffer-1 containing 5% goat serum for 20 min.
Dilute galectin-1 (1:1000) and/or galectin-3 antibodies (1:50 – 1:100) in IF-Buffer-1 and incubate coverslips with antibody solutions overnight at 4 °C in a humidified chamber. The choice of galectin depends on the expression level in the cell line of interest (e.g. galectin-1 is best for HeLa and U2-OS whereas galectin-3 is best for MCF7). For pilot experiments it is best to use both.
Wash 3x5 min in IF-Buffer-2.
Dilute Alexa Fluor®488/594-conjugated secondary antibodies 1:1000 in IF-Buffer-2 and incubate coverslips with antibody solutions for 1 h at RT in the dark. Centrifuge secondary antibodies 2 min at 20000xg before diluting and then take from the top of the solution to eliminate fluorchrome aggregates which can be mistaken for galectin dots.
Wash 3x5 min in IF-Buffer-3 and once briefly in PBS.
Incubate coverslips 10 min in Höchst 33342 (1:5000) to label nuclei.
Wash twice in PBS.
Mount coverslips onto glass slides – add a small drop of mounting medium (~7 μl) to the slide, remove excess liquid from the coverslip by placing its edge on a paper towel and then put it on the slide with the cell side downwards.
Let the mounting medium solidify for 1 day at RT in the dark. Afterwards, store slides at -20 °C until analysis.
Examine slides in a fluorescence or confocal microscope.
Data analysis
Both the number of galectin dots per cell and the percentage of cells scoring positive for galectin dots can be quantified by manual counting or automated image analysis. Quantification should be performed on randomly selected fields chosen in the Hoechst channel to avoid bias.
Alternative assay with fluorescent constructs
Even though staining for endogenous galectins as described here is preferable in most cases, the galectin translocation assay can also be performed by expressing fluorescently labelled galectin-3 in the cells. This can be used if endogenous galectin levels are too low or for live cell microscopy.
Fluorescent constructs with galectin-1 should be avoided if possible as the tag may interfere with its translocation due to the small size of this protein.
Troubleshooting
Problem
No galectin dots detected in L-leucyl-L-leucine-methyl ester positive control.
Solution
Extend treatment time and/or concentration of L-leucyl-L-leucine-methyl ester. Ensure that the galectin for which staining is performed is expressed and exclude that the observed staining is not due to unspecific background (e.g. by using galectin siRNA). If expression levels are too low, use galectin-1 instead of galectin-3 and vice versa or transfect cells with fluorescently labelled galectin-3.
Discussion
The four complementary methods presented here should provide a solid foundation for studying lysosomal cell death.
We have applied the digitonin extraction protocol for LMP quantification (Method 1) to various cell lines treated with a range of different cytotoxic stimuli and it performs well for most standard cancer cell lines including MCF-7, HeLa, U2OS (Foghsgaard et al. 999-1010;Nylandsted et al. 425-35;Groth-Pedersen et al. 2217-25). However, for other cell lines including immortalized mouse embryonic fibroblasts (MEFs) the cholesterol content, membrane topography and hence digitonin extraction time and lifting frequency of the rocking table may be different and should be optimized (e.g. MEFs: 10 min extraction; lifting frequency 110/min, digitonin concentration 17 μg/ml) (Gyrd-Hansen et al. 7880-91). Further, as mentioned previously digitonin-based extraction can only be used with cytotoxic treatments that do not interfere with membrane cholesterol content since this severely affects the assay. This is evident when treating cells with mild detergents like cationic amphiphilic drugs (CADs) which accumulate in lysosomes and induce LMP by inhibiting acidic sphingomyelinase activity (Pedersen et al). In such cases, LMP can be measured by monitoring Galactin translocation to permeabilized lysosomes (Method 4) (Fig. 4), by fluorescent dextran release from lysosomes (Method 2) (Fig. 2A) or/and by cathepsin immunocytochemistry (Method 3). In principle, it is possible to follow LMP in real time by time-lapse video microscopy of dextran release or translocation of fluorescently-tagged galectins. However, since lysosomes are highly dynamic organelles that regularly move in and out of the focus plane and many LMP-inducing insults only induce LMP in a fraction of lysosomes it can be challenging to capture LMP events in real time. Still, if using potent LMP inducers, e.g. O-methyl-serine dodecylamide hydrochloride (MSDH), L-leucyl-L-leucine-methyl ester (LLOMe) that accumulates in lysosomes and has detergent-like properties (Li et al. 35-39) or hydrogen peroxide it is possible to capture the process in real time as shown in Figure 2B. The advantage of monitoring LMP by dextran release is that LMP pore size can be defined by using different sizes of fluorophore coupled dextran (Fig. 2B).
Immunocytochemistry is useful for assessing LMP and mitochondrial outer membrane permebilization (MOMP) concurrently under various cellular conditions, as presented in Method 3. LMP can, dependent on the cytotoxic stimuli and cell type, be activated in a MOMP- and apoptosome-independent manner which still involves caspase-9 activation (Gyrd-Hansen et al. 7880-91) or be induced before MOMP as in HeLa cells treated with the microtubule depolymerizing drug vincristine (Groth-Pedersen et al. 2217-25). It can even be triggered independently of the intrinsic apoptosis pathway as seen after Hsp70 depletion (Nylandsted et al. 7871-76;Nylandsted et al. 425-35). The advantage of this procedure is that Co-staining for LMP and apoptosis markers at different time points can be used to address the sequence of events in the death pathway of interest and thereby establish the initiating mechanism and secondary effects. The method can also be adjusted by using antibodies to other abundant luminal lysosomal proteins (e.g. cathepsin B or NAG) as needed and other relevant death components (e.g. activated caspases, activated Bax, or cytochrome c release).
In addition to cathepsin immunostaining, staining for galectin-1 and -3 can be used to detect LMP as these proteins move from the cytosol to lysosomes upon damage. This assay is extremely sensitive and galectin dots become visible before the cathepsin immunostaining can detect LMP. The formation of dots is less difficult to see than the appearance of a diffuse cytosolic cathepsin staining, making this assay less prone to misinterpretation and easier to quantify in an automated manner. Furthermore, this assay is the only LMP assay that can detect individual damaged lysosomes rather than the response of the entire lysosomal compartment. In contrast to many lysosomal proteins, galectin-3 can also be expressed with fluorescent tags without problems, allowing this assay to be adapted for live cell microscopy. The galectin translocation assay is thus robust and versatile and is therefore recommended as the first assay to use for the detection of LMP.
Lastly, it is important to note that LMP does not equal lysosomal cell death as the cells have a range of defense mechanisms that may allow them to survive LMP up to a certain threshold. In addition, LMP can be both the primary event that initiates cell death or a secondary consequence of the activation of other death pathways such as apoptosis. It is therefore important to always try to examine whether or not the observed LMP actually causes cell death. This is often done using pharmacological cathepsin inhibitors such as Ca-074-Me or z-FA-fmk or by depleting the cells of a specific cathepsin with genetic approaches. While this is a useful strategy it is, however, important to remember that there is redundancy between the different cathepsins and that they are also not the only potentially lethal lysosomal components released upon LMP. Thus, a lack of cell death reduction after cathepsin inhibition/depletion does not necessarily mean that the observed LMP is not causing cell death.
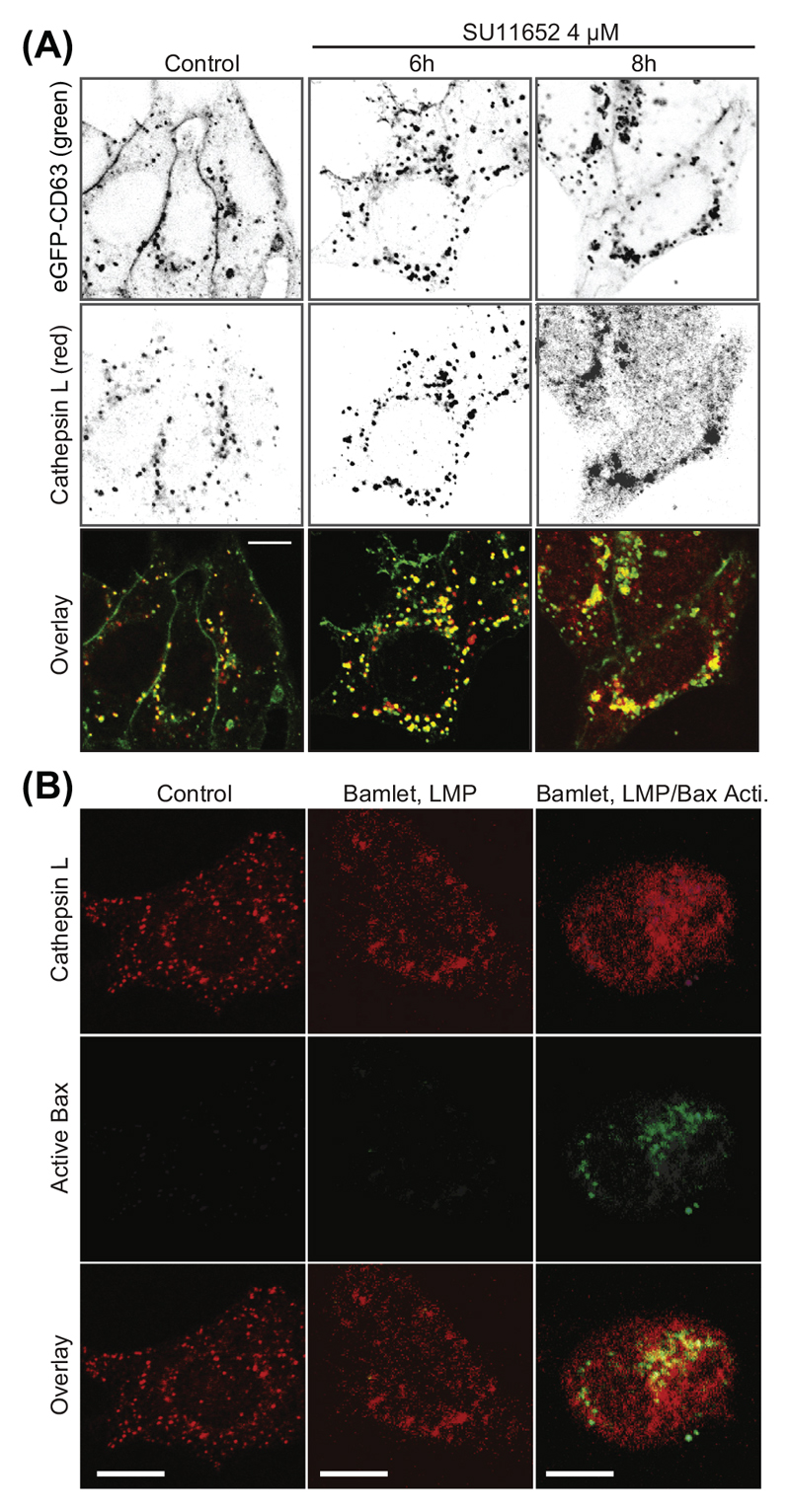
Figure 3. Example of LMP followed by Bax activation in MCF-7 cells induced by a complex of bovine lactalbumin and oleic acid (BAMLET).
Representative confocal pictures of MCF-7 cells treated with 100 μg/ml of BAMLET for 3 h (1 h without serum followed by 2 h with serum) and stained for cathepsin L and the active conformation of Bax (Rammer et al. 24-32). Scale Bars, 20 μm.
Acknowledgment
We thank both present and former colleagues from Unit for Cell Death and Metabolism, Danish Cancer Society Research Center for optimizing and fine tuning the methods presented here. This work was supported by the Danish Cancer Society, the Danish National Research Foundation, the Danish Council for Independent Research in Medical Sciences, the Association for International Cancer Research, theEuropean Research Council, the Swedish Research Council, the Novo Nordisk Foundation, and the Lundbeck Foundation
Contributor Information
Sonja Aits, Email: sonjaa@cancer.dk.
Marja Jäättelä, Email: mj@cancer.dk.
References
- Bidere N, et al. Cathepsin D triggers Bax activation, resulting in selective apoptosis-inducing factor (AIF) relocation in T lymphocytes entering the early commitment phase to apoptosis. J Biol Chem. 2003;278(33):31401–11. doi: 10.1074/jbc.M301911200. [DOI] [PubMed] [Google Scholar]
- Bitensky L. Modifications to the Gomori acid phosphatase technique for controlled temperature frozen sections. Q fi microsc Sci. 1963;104:193–96. [Google Scholar]
- Ref Type: Generic
- Boya P, et al. Lysosomal membrane permeabilization induces cell death in a mitochondrion-dependent fashion. J Exp Med. 2003;197(10):1323–34. doi: 10.1084/jem.20021952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boya P, Kroemer G. Lysosomal membrane permeabilization in cell death. Oncogene. 2008;27(50):6434–51. doi: 10.1038/onc.2008.310. [DOI] [PubMed] [Google Scholar]
- Brunk UT, et al. Photo-oxidative disruption of lysosomal membranes causes apoptosis of cultured human fibroblasts. Free Radic Biol Med. 1997;23(4):616–26. doi: 10.1016/s0891-5849(97)00007-5. [DOI] [PubMed] [Google Scholar]
- Brunk UT, Neuzil J, Eaton JW. Lysosomal involvement in apoptosis. Redox Rep. 2001;6(2):91–97. doi: 10.1179/135100001101536094. [DOI] [PubMed] [Google Scholar]
- Cirman T, et al. Selective disruption of lysosomes in HeLa cells triggers apoptosis mediated by cleavage of Bid by multiple papain-like lysosomal cathepsins. J Biol Chem. 2004;279(5):3578–87. doi: 10.1074/jbc.M308347200. [DOI] [PubMed] [Google Scholar]
- De Duve C, et al. Tissue fractionation studies. 6. Intracellular distribution patterns of enzymes in rat-liver tissue. Biochem J. 1955;60(4):604–17. doi: 10.1042/bj0600604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Duve C. Lysosomes revisited. Eur J Biochem. 1983;137(3):391–97. doi: 10.1111/j.1432-1033.1983.tb07841.x. [DOI] [PubMed] [Google Scholar]
- Firestone RA, Pisano JM, Bonney RJ. Lysosomotropic agents. 1. Synthesis and cytotoxic action of lysosomotropic detergents. J Med Chem. 1979;22(9):1130–33. doi: 10.1021/jm00195a026. [DOI] [PubMed] [Google Scholar]
- Foghsgaard L, et al. Cathepsin B acts as a dominant execution protease in tumor cell apoptosis induced by tumor necrosis factor. J Cell Biol. 2001;153(5):999–1010. doi: 10.1083/jcb.153.5.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerasimenko JV, Gerasimenko OV, Petersen OH. Membrane repair: Ca(2+)-elicited lysosomal exocytosis. Curr Biol. 2001;11(23):R971–R974. doi: 10.1016/s0960-9822(01)00577-2. [DOI] [PubMed] [Google Scholar]
- Groth-Pedersen L, et al. Vincristine induces dramatic lysosomal changes and sensitizes cancer cells to lysosome-destabilizing siramesine. Cancer Res. 2007;67(5):2217–25. doi: 10.1158/0008-5472.CAN-06-3520. [DOI] [PubMed] [Google Scholar]
- Groth-Pedersen L, et al. Vincristine induces dramatic lysosomal changes and sensitizes cancer cells to lysosome-destabilizing siramesine. Cancer Res. 2007;67(5):2217–25. doi: 10.1158/0008-5472.CAN-06-3520. [DOI] [PubMed] [Google Scholar]
- Gyrd-Hansen M, et al. Apoptosome-independent activation of the lysosomal cell death pathway by caspase-9. Mol Cell Biol. 2006;26(21):7880–91. doi: 10.1128/MCB.00716-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagedal K, et al. Sphingosine-induced apoptosis is dependent on lysosomal proteases. Biochem J. 2001;359(Pt 2):335–43. doi: 10.1042/0264-6021:3590335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallunki T, Olsen OD, Jäättelä M. Cancer-associated lysosomal changes friends or foes. Cancer-associated lysosomal changes: friends or foes? 2012. Oncogene in press. doi: 10.1038/onc.2012.292. [DOI] [PubMed] [Google Scholar]
- Ref Type: Generic
- Kirkegaard T, Jaattela M. Lysosomal involvement in cell death and cancer. Biochim Biophys Acta. 2009;1793(4):746–54. doi: 10.1016/j.bbamcr.2008.09.008. [DOI] [PubMed] [Google Scholar]
- Kirkegaard T, et al. Hsp70 stabilizes lysosomes and reverts Niemann-Pick disease-associated lysosomal pathology. Nature. 2010;463(7280):549–53. doi: 10.1038/nature08710. [DOI] [PubMed] [Google Scholar]
- Kolter T, Sandhoff K. Lysosomal degradation of membrane lipids. FEBS Lett. 2010;584(9):1700–12. doi: 10.1016/j.febslet.2009.10.021. [DOI] [PubMed] [Google Scholar]
- Kreuzaler PA, et al. Stat3 controls lysosomal-mediated cell death in vivo. Nat Cell Biol. 2011;13(3):303–09. doi: 10.1038/ncb2171. [DOI] [PubMed] [Google Scholar]
- Kroemer G, Jaattela M. Lysosomes and autophagy in cell death control. Nat Rev Cancer. 2005;5(11):886–97. doi: 10.1038/nrc1738. [DOI] [PubMed] [Google Scholar]
- Kurz T, Eaton JW, Brunk UT. The role of lysosomes in iron metabolism and recycling. Int J Biochem Cell Biol. 2011;43(12):1686–97. doi: 10.1016/j.biocel.2011.08.016. [DOI] [PubMed] [Google Scholar]
- Li W, et al. Induction of cell death by the lysosomotropic detergent MSDH. FEBS Lett. 2000;470(1):35–39. doi: 10.1016/s0014-5793(00)01286-2. [DOI] [PubMed] [Google Scholar]
- Link G, Pinson A, Hershko C. Iron loading of cultured cardiac myocytes modifies sarcolemmal structure and increases lysosomal fragility. J Lab Clin Med. 1993;121(1):127–34. [PubMed] [Google Scholar]
- Moin K, et al. Tumor cell membrane cathepsin B. Biol Chem. 1998;379(8-9):1093–99. doi: 10.1515/bchm.1998.379.8-9.1093. [DOI] [PubMed] [Google Scholar]
- Nylandsted J, et al. Heat shock protein 70 promotes cell survival by inhibiting lysosomal membrane permeabilization. J Exp Med. 2004;200(4):425–35. doi: 10.1084/jem.20040531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nylandsted J, et al. Selective depletion of heat shock protein 70 (Hsp70) activates a tumor-specific death program that is independent of caspases and bypasses Bcl-2. Pro cNatl Acad Sci USA. 2000;97(14):7871–76. doi: 10.1073/pnas.97.14.7871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palermo C, Joyce JA. Cysteine cathepsin proteases as pharmacological targets in cancer. Trends Pharmacol Sci. 2008;29(1):22–28. doi: 10.1016/j.tips.2007.10.011. [DOI] [PubMed] [Google Scholar]
- Petersen NH, et al. Transformation-associated changes in sphingolipid metabolism sensitize cells to lysosomal cell death induced by inhibitors of acid sphingomyelinase. Cancer Cell. 2013;24(3):379–93. doi: 10.1016/j.ccr.2013.08.003. [DOI] [PubMed] [Google Scholar]
- Pryor PR, Luzio JP. Delivery of endocytosed membrane proteins to the lysosome. Biochim Biophys Acta. 2009;1793(4):615–24. doi: 10.1016/j.bbamcr.2008.12.022. [DOI] [PubMed] [Google Scholar]
- Rammer P, et al. BAMLET activates a lysosomal cell death program in cancer cells. Mol Cancer Ther. 2010;9(1):24–32. doi: 10.1158/1535-7163.MCT-09-0559. [DOI] [PubMed] [Google Scholar]
- Saftig P, Klumperman J. Lysosome biogenesis and lysosomal membrane proteins: trafficking meets function. Nat Rev Mol Cell Biol. 2009;10(9):623–35. doi: 10.1038/nrm2745. [DOI] [PubMed] [Google Scholar]
- Schotte P, et al. Non-specific effects of methyl ketone peptide inhibitors of caspases. FEBS Lett. 1999;442(1):117–21. doi: 10.1016/s0014-5793(98)01640-8. [DOI] [PubMed] [Google Scholar]
- Silverman GA, et al. SCCA1 and SCCA2 are proteinase inhibitors that map to the serpin cluster at 18q21.3. Tumour Biol. 1998;19(6):480–87. doi: 10.1159/000030041. [DOI] [PubMed] [Google Scholar]
- Stoka V, Turk V, Turk B. Lysosomal cysteine cathepsins: signaling pathways in apoptosis. Biol Chem. 2007;388(6):555–60. doi: 10.1515/BC.2007.064. [DOI] [PubMed] [Google Scholar]
- Suminami Y, Nawata S, Kato H. Biological role of SCC antigen. Tumour Biol. 1998;19(6):488–93. doi: 10.1159/000030042. [DOI] [PubMed] [Google Scholar]
- Vasiljeva O, Turk B. Dual contrasting roles of cysteine cathepsins in cancer progression: apoptosis versus tumour invasion. Biochimie. 2008;90(2):380–86. doi: 10.1016/j.biochi.2007.10.004. [DOI] [PubMed] [Google Scholar]