Abstract
Hepatic fat accumulation is associated with diabetes and hepatocellular carcinoma (HCC). Here we characterize the metabolic response that high fat availability elicits in livers prior to disease development. After a short term on a high fat diet, otherwise healthy mice showed elevated hepatic glucose uptake and increased glucose contribution to serine and pyruvate carboxylase activity compared to control diet mice. This glucose phenotype occurred independently from transcriptional or proteomic programming, which identifies increased peroxisomal and lipid metabolism pathways. High fat diet-fed mice exhibited increased lactate production when challenged with glucose. Consistently, administration of an oral glucose bolus to healthy individuals revealed a correlation between waist circumference and lactate secretion in a human cohort. In vitro, palmitate exposure stimulated production of reactive oxygen species and subsequent glucose uptake and lactate secretion in hepatocytes and liver cancer cells. Furthermore, high fat diet enhanced the formation of HCC compared to control diet in mice exposed to a hepatic carcinogen. Regardless of the dietary background, all murine tumors showed similar alterations in glucose metabolism to those identified in fat exposed non-transformed mouse livers; however, particular lipid species were elevated in high fat diet tumor and non-tumor-bearing high fat diet liver tissue. These findings suggest that fat can induce glucose-mediated metabolic changes in non-transformed liver cells similar to those found in HCC.
Introduction
The tissue environment and nutrient availability are important regulators of cellular processes (1–3). Recent evidence from the study of cancer cells also suggests that nutrient availability can directly modulate cellular metabolism by defining metabolic dependencies (4), metabolic intratumor heterogeneity (5) and the metabolic makeup of metastasis compared to primary tumors (6). These and other publications highlight the importance of studying cellular metabolism in the context of increased nutrient availability.
While many recent nutrient metabolism studies focus on cancer, most nutrients are also processed by normal non-transformed cells. A major difference between normal, non-transformed cells, and cancer cells is that the former are mostly quiescent and only proliferate under defined and regulated circumstances resulting in a different metabolism (7, 8). Normal tissue metabolism has been investigated under disease conditions (9, 10) and has been compared between different tissues (11, 12). With these comparisons, a signaling-independent nutrient availability metabolism link has been observed (13, 14).
Changes in fat availability are common and physiologically relevant. Excess fat is a major cause for the development of insulin resistance, dampening insulin signaling in the liver and skeletal muscles (15, 16). Insulin resistance can also reduce glucose uptake in the liver and skeletal muscles and elevate hepatic gluconeogenesis (15, 16). Importantly, fat can also directly alter the metabolism of non-transformed cells in the liver (17). Moreover, high fat diet (HFD) feeding in mice and rats has been found to alter gluconeogenesis (18) and mitochondrial lipid metabolism, stimulating reactive oxygen species (ROS) (19–21) and endoplasmic reticulum (ER) stress (22), ultimately culminating in DNA damage and cell death (23).
Here, we investigate the in vivo metabolic changes that occur in normal non-transformed liver cells upon increased fat availability before the detection of a fat-promoted oncogenic transformation. We discover that fat rewires glucose metabolism in normal, non-transformed livers and that this fat-induced metabolism is similar to the metabolic state of liver cancers.
Methods
Detailed methods are provided in the supplement.
Animal studies
All experimental animal procedures were approved by the Institutional Animal Care and Research Advisory Committee of the KU Leuven, Belgium. C57BL/6N mice were obtained from the KU Leuven animal laboratory. 2-week old mice were injected i.p. with diethylnitrosamine (DEN, 25 mg/Kg) in PBS (vehicle, 3.17 mg/ml). 4 weeks later, mice were randomized into two groups: control diet (CD, E15742-33 ssniff Spezialdiäten GmbH) or high fat diet (HFD, S8655-E220 sniff Spezialdiäten GmbH). The energy balance between fat/protein/carbohydrates is 13%/27%/60% and 60%/20%/20% for CD and HFD, respectively. Humane endpoints were observed: 20% increase in abdomen diameter and hard mass detection via palpation, loss of ambulation, loss of skin elasticity, labored respiration, or weight loss over 20 % of initial body weight.
Mouse surgery and metabolic infusions
Mice were implanted with a jugular vein catheter and recovered for 1 week. Mice were fasted for 6-hours, then 500 mg/ml 13C6-glucose was infused at a rate of 30 mg/kg/min (14-week old mice) or 7.5 mg/kg/min (35-week old mice). After 6-hours, mice were sacrificed for blood and tissue collection for further analysis.
Glucose and insulin tolerance tests
Mice were fasted overnight, then injected with 2mg/kg of glucose (ipGTT) or 1 U/kg of insulin (ipITT) or gavaged with 2g/kg 13C6-glucose (oGTT). Blood glucose was measured using a glucometer (OneTouch Verio) at multiple time points. In the oGTT, blood was collected in heparin coated capillary tubes to analyze for 13C lactate enrichment.
Metabolic response of H4IIEC3 cells to fatty acids and cell treatments
H4IIEC3, HUH7, and PLC cells were purchased from ATCC, and HHL5 cells were provided by Dr. Arvind Patel (24). Cells were cultured in DMEM media containing 10% fetal bovine serum and 1% penicillin/streptomycin, and incubated at 37°C and 5% CO2. Cells were maintained for a maximum of 4 weeks (approximately 12 splits) before discarding and thawing a new stock. Prior to use, cells were split once (at minimum) following thawing and mycoplasma testing was conducted (MycoAlert Mycoplasma Detection Kit, Lonza). 5x105 cells/well were seeded in a 6-well plate and adhered overnight. 0.4 mM of palmitate-BSA complex or ethanol-BSA control in DMEM media with 13C6-glucose was applied for 8-hours. Etomoxir, NAC, mitoTEMPO, ATZ, and H2O2 were tested by adding the corresponding treatment at the same moment as palmitate-BSA, at final concentrations of 10 μM and 5 mM, 10 mM, 20 mM, and 2 mM, respectively. For catalase stimulation, cells were treated with 5mM Na-butyrate or 100μM WY-14643 for 24h prior to treating the cells with palmitate and ATZ, as described above. Details on stock preparations are available in methods supplement.
Metabolite quenching and extraction
For in vitro cells, media was aspirated from the wells, quickly washed in 0.9% NaCl, and snap frozen in liquid nitrogen. Metabolites were extracted with methanol and water (5:3, 800 μL/well) containing internal standards for organic acids and amino acids (glutarate and norvaline, respectively). 500 μl of chloroform containing heptadecanoic acid (internal standard) was added, vortexed at 4°C for 5 min and centrifuged at 15000 RPM for 5 min. For plasma or tissue metabolite extraction, we performed the above procedure directly in 10 to 20 μl of plasma or in ~10 mg tissue that was pulverized using a liquid nitrogen-chilled cryomill (Retsch). Polar metabolites (upper phase) and fatty acids (lower phase) were collected and dried in a vacuum concentrator (4°C, overnight), and stored in -80°C until further processing by GC/MC or LC/MS.
Oral glucose tolerance test in healthy human volunteers
All human experiments were performed with informed written consent in compliance with Japan’s Ethical Guidelines for Epidemiological Research, and the study was approved by the Ethics Committee of the Faculty of Medicine of the University of Tokyo (10264-(4)). Volunteers participated in the experiment after 10-hours fasting. A glucose solution (Torelan G, 75g in 225ml; AJINOMOTO) was consumed within 1 min. Blood samples were obtained from the cutaneous vein of the forearm at baseline (−10, 0 min) and at 10, 20, 30, 45, 60, 75, 90, 120, 150, 180, 210, 240 min after ingestion. Samples were centrifuged for 10 min at 4°C at 1,380 g, then stored at −80°C. Plasma glucose was measured by the enzymatic method (IATRO LQ GLU). Lactate was measured by the enzymatic method (Determiner LA). Further data analysis methods are detailed in supplemental methods.
Measurement of glucose uptake and reactive oxygen species production in cell lines
Glucose uptake and ROS production were measured in 96-well plates. 7.5x104 cells/well were seeded and adhered overnight. 0.4 mM of palmitate-BSA or control-BSA complex was added to each well (200 μl/well) for 8-hours. Media was removed from the wells, and washed with PBS. 2-deoxy-2-[(7-nitro-2,1,3-benzoxadiazol-4-yl)amino]-D-glucose (2-NBDG, 600 μM, Cayman Chemical) or 2’,7’-dichlorodihydrofluorescein diacetate (CM-H2DCFDA, 5 μM) (ThermoFisher) in serum-free media (and glucose-free media for 2-NBDG) was applied to cells and incubated for 1-hour. Media was removed, and cells washed 3 times with PBS. Intracellular fluorescence was then measured with a Victor X2 plate reader (Perkin Elmer) with an excitation frequency of 485 nm and an emission frequency of 535 nm, and reads were normalized to cell counts.
RNA Isolation for qPCR
Total RNA was isolated with PureLink RNA Mini Kit (Life Technologies) or extracted using TRIazol (Life Technologies) with chloroform and propanol for phase separation and RNA isolation. RNA quality and quantity were measured using a NanoDrop One Microvolume UV-Vis Spectrophotometer (Thermo Scientific). For qPCR, RNA (1 μg) was reverse transcribed into cDNA using a High-Capacity cDNA Reverse Transcription Kit (Life Technologies). qPCR and RNA sequencing were performed as described in supplemental methods.
RNA Sequencing
RNA from liver tissues were extracted using TRIazol (Life Technologies) and quality and quantity were measured (NanoDrop One Microvolume UV-Vis Spectrophotometer, Thermo Scientific). RNAseq libraries were prepared from 1 μg of total RNA per sample (KAPA Stranded mRNA Sequencing Kit, Roche) and poly-A containing mRNA was purified using oligo(dT) magnetic beads and fragmented into 200–500 bp pieces using divalent cations at 94°C for 8 min. The cleaved RNA fragments were copied into first strand cDNA. After second strand cDNA synthesis, fragments were A-tailed and indexed adapters were ligated. PCR was used to purify and enrich products, creating the final cDNA library. After quantification with qPCR, the resulting libraries were sequenced on a HiSeq4000 (Illumina) using a flow cell generating 1x50bp single-end reads. Details on transcriptome analysis are in the supplemental methods. Transcriptomic data is available at the NIH GEO repository, accession code GSE165752.
Proteomic analysis
The iST sample preparation kit (PreOmics, Germany) was used to isolate peptides from liver tissue. Purified peptides were re-dissolved in 20 μl loading solvent A (0.1% TFA in water/ACN (98:2, v/v)) and the peptide concentration was determined on a Lunatic instrument (Unchained Lab). 2μg peptides were injected for LC-MS/MS analysis on an Ultimate 3000 RSLCnano system in-line connected to a Q Exactive HF BioPharma mass spectrometer (Thermo). Detailed proteomics methods and analysis are in the supplemental methods. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD023848.
Lipidomics analysis
Lipids were extracted from liver tissues by mixing 700 μL in water, homogenizing (Precellys, Bertin) with 800 μL 1 N HCl:CH3OH 1:8 (v/v), 900 μL CHCl3, 200 μg/ml of the antioxidant 2,6-di-tert-butyl-4-methylphenol (BHT; Sigma Aldrich) and 3 μL of SPLASH® LIPIDOMIX® Mass Spec Standard (#330707, Avanti Polar Lipids). After vortexing and centrifugation, the lower organic fraction was collected and evaporated using a Savant Speedvac spd111v (Thermo Fisher Scientific) at room temperature and the remaining lipid pellet was stored at - 20°C under argon. Just before mass spectrometry analysis, lipid pellets were reconstituted in 100% ethanol. Lipid species were analyzed by liquid chromatography electrospray ionization tandem mass spectrometry (LC-ESI/MS/MS) on a Nexera X2 UHPLC system (Shimadzu) coupled with hybrid triple quadrupole/linear ion trap mass spectrometer (6500+ QTRAP system; AB SCIEX). Detailed methods and data analysis are available in the supplemental methods. Lipidomics data is deposited at the NIH Common Fund’s National Metabolomics Data Repository (NMDR) website, the Metabolomics Workbench, https://www.metabolomicsworkbench.org where it has been assigned Project ID (PR001085).
Statistical analysis
For testing when two variables are present (HFD and DEN), two-way ANOVA was used to measure variances, and data from all 4 groups are plotted separately with reported statistics for a diet effect (unless noted otherwise). p-values for main effects and other statistical tests is represented in the figure when possible. Two-tailed unpaired Student’s T-test was used for comparisons between two different groups. One-way ANOVA with Tukey’s multiple comparisons test was used for comparisons of multiple independent variables. Areas under the curve for kinetic measurements were calculated using the lowest value as the baseline. Correlations between waist circumference and lactate levels were determined by calculating the Pearson coefficient. Outlier detection was conducted via ROUT test (Q=1%) and Grubbs tests (α=0.05). Statistical testing and figure generation was conducted in GraphPad Prism 8. For all tests p≤0.05 was considered significant. Bar graphs are represented ± standard deviation. Box plots are represented with the middle line as the median, the box ranging from 25th to 75th quartile and whiskers going from maximum to minimum. Schematic figures were generated using Biorender.com.
Results
To address the question of how the in vivo nutrient environment impacts the metabolism of normal and non-transformed livers before tumor development, we investigated mice on a high fat, normal glucose diet compared to control diet. Specifically, we injected two-week old C57BL/6N mice with phosphate buffered saline (vehicle) or diethylnitrosamine (DEN), a carcinogen which induces hepatocellular carcinoma (HCC) (25). At six weeks of age, mice were subdivided into two additional cohorts, and fed either an HFD or control diet (Fig. S1a; Table S1). After eight weeks on the different diets, we observed an approximate 50% increase in weight and fat mass (measured by dual X-ray absorptiometry (DEXA)) in HFD mice compared to control diet mice (Fig. S1b,c). Since obesity can result in the development of insulin resistance and diabetes, which induces multiple changes in whole body metabolism (26), we tested for signs of insulin resistance by performing intraperitoneal glucose and insulin tolerance tests. We observed that mice on HFD showed a trend toward higher basal blood glucose levels compared to control diet mice (Fig. S1d). Yet, the area under the curve for blood glucose in response to glucose or insulin was not significantly different between groups (Fig. S1d–g). Thus, we concluded that the mice were not yet insulin resistant or glucose intolerant but showed (considering the higher basal glucose levels) a trend towards altered insulin signaling after eight weeks on HFD compared to control diet.
Despite no visible signs of cancer onset at this timepoint (Fig. S2a), we performed a Ki-67 staining, a marker for proliferation. We found that only a small cell fraction (less than 4%) stained positive for Ki-67 (Fig. S2b,c), meeting the expectations of non-transformed livers (27). Moreover, there was no significant difference in Ki-67 positive liver cells across the different cohorts (Fig. S2b,c). HFD and DEN exposure can lead to liver inflammation and immune cell infiltration (28). To address this possibility, we stained liver sections with the murine macrophage marker F4/80 and myeloid cell markers Ly6C/Ly6G (Fig. S2d–g). At this early time point, F4/80 staining was unchanged upon HFD or DEN exposure (Fig. S2d,f). However, Ly6C/Ly6G staining was elevated in the HFD DEN condition and strongly inhibited in control diet DEN condition (Fig. S2e,g), indicative of a pro-inflammatory state within the HFD, DEN-injected livers (29). Thus, we concluded that, at this moment, the livers of these mice showed no sign of hepatocarcinogenesis but showed some signs of inflammation when comparing diet changes in DEN injected animals but not in vehicle injected animals.
Normal, non-transformed liver cells metabolically respond to changes in the in vivo fat availability
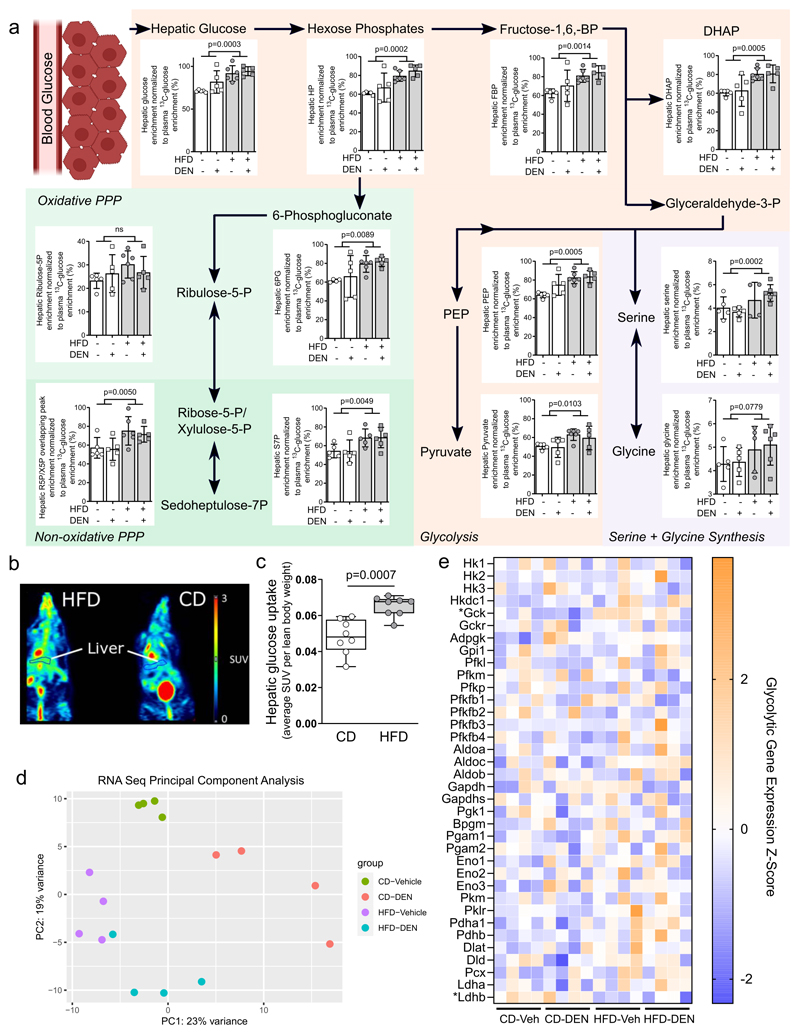
Next, we determined in vivo liver metabolism using 13C tracer analysis (30) in mice after eight weeks of HFD feeding. Specifically, following a six hour fast mice were infused with 13C6-glucose over the course of approximately six hours. Subsequently we measured the 13C enrichment of hepatic metabolites and normalized them to plasma glucose enrichment (31)(Table S2). Additionally, we analyzed hepatic metabolite abundance as well as plasma metabolite 13C enrichment (Tables S3–5). While plasma metabolite levels (Table S5) and hepatic metabolite abundances (Table S3) were changed across central metabolism in mice challenged with fat, we strikingly discovered a large and consistent rewiring of glucose metabolism in the liver of HFD-fed mice based on the 13C tracer data (Table S2). In particular, we found that hepatic 13C enrichment from glucose was increased in all measured glycolytic metabolites and in many branch pathway metabolites (pentose phosphate pathway and serine biosynthesis) upon high fat availability (Fig. 1a; Table S2). To further verify the HFD-induced changes in liver glucose metabolism, we measured hepatic uptake of 18F-fluorodeoxyglucose (18F-FDG) in mice on high fat or control diet using positron emission tomography (PET). We found hepatic glucose uptake increased approximately 35% in HFD-fed mice compared to control (Fig. 1b,c). Thus, we concluded that liver glucose uptake and its metabolism is increased by fat in non-transformed liver cells supporting metabolic pathways downstream of glycolysis.
Figure 1. Fat hyperactivates glucose metabolism.
a) Metabolic differences detected in liver tissue of mice after 8-weeks on CD (CD-vehicle n=5, CD-DEN n=5) or HFD (HFD-vehicle n=7, HFD-DEN n=6) diet normalized to plasma glucose enrichment. Abbreviations: BP, bisphosphate; FBP, fructose-1,6-bisphosphate; DHAP, dihydroxyacetone phosphate; PEP, phosphenolpyruvate; HP, Hexose phosphates; R5P, ribuose-5-phosphate; X5P, Xylulose-5-phosphate; S7P, sedoheptulose-7-phosphate; 6PG, 6-phoshogluconate; PPP, pentose-phosphate pathway. p-value statistics indicate variance caused by diet effect in two-way ANOVA testing.
b,c) 18F-fluorodeoxyglucose positron emission tomography (18F-FDG-PET) of mice after eight weeks of control (n=8) or high fat (n=8) diet. Representative images highlighting the region that was selected to assess hepatic 18F-FDG uptake, normalized to lean weight of each mouse. Statistics: Two-tailed unpaired Student’s T-test.
d) Principle component analysis of transcriptomics dataset from bulk RNA sequencing of liver tissue from CD-vehicle, CD-DEN, HFD-vehicle and HFD-DEN. n=4 per group. X and Y axes indicate the amount of variance accounted for by the first and second components, in percentage.
e) Heat map of genes involved in glycolysis expressed as Z-score. * indicates p-adjusted value < 0.05 from DESeq analysis testing for a diet effect.
To determine whether this glycolytic phenotype was driven by transcriptional changes, we conducted bulk RNA sequencing on the liver tissues (Table S6). We observed a diet effect on sample clustering based on principle component analysis (PCA) (Fig 1d). Using Gene Set Enrichment Analysis (GSEA), we found that HFD exposure enriched gene sets involved in peroxisomes and fatty acid metabolism, and accordingly, several genes involved in fatty acid metabolism were upregulated (Fig. S3a–c). Interestingly, despite the physiological changes to glycolysis, we were unable to detect a strong signature for glycolytic gene expression, with glucokinase (Gck) being the only significantly upregulated glycolytic gene in HFD fed liver tissue (Fig 1e). This observation was verified on the protein level using proteomics (Fig S3d–f). Thus, we concluded that the observed changes in glycolysis and glucose uptake where largely independent of the transcriptome and proteome alterations induced by HFD.
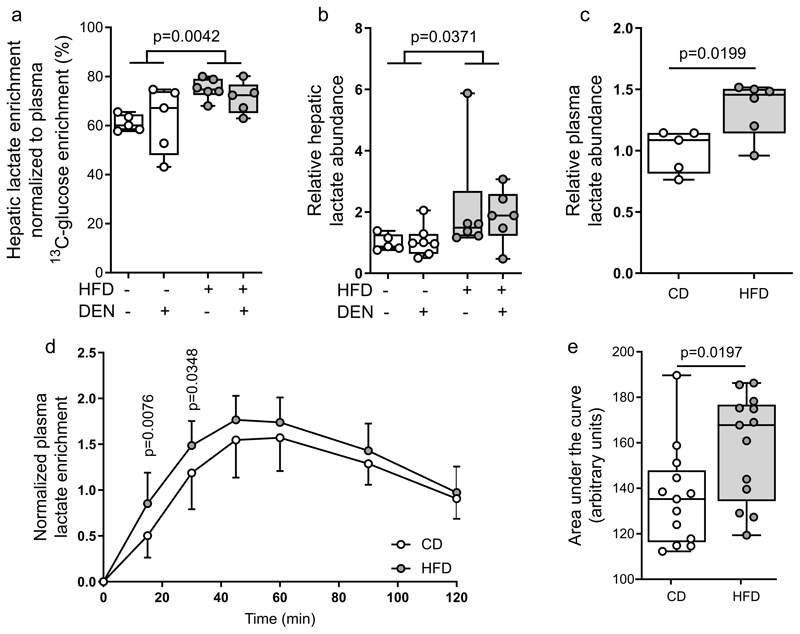
One major fate of glucose metabolism is lactate production. Accordingly, we observed increased 13C enrichment of hepatic tissue lactate, and increased lactate abundance in both plasma and liver tissues in HFD animals compared to controls (Fig. 2a–c). Notably, 13C enrichment of hepatic lactate was similar or slightly lower compared to phosphoenolpyruvate (PEP) (32) (Fig. 1a, 2a; Table S2). Moreover, we excluded that the observed 13C enrichment of hepatic lactate in HFD animals was dependent on an altered Cori cycle (Fig S4). Thus, our data suggest that lactate was in absolute terms produced rather than taken up. To confirm that HFD-fed mice produce more lactate from glucose, we measured the time resolved 13C enrichment of plasma lactate after administering an oral bolus of 13C6-glucose (2 g/kg) HFD and control diet mice. If glycolysis is increased and lactate secretion is upregulated, we expect to see increased 13C enriched lactate released into the plasma. Indeed, we measured faster and more pronounced 13C enrichment dynamics in plasma lactate of HFD compared to control diet mice, an effect that occurred despite a slightly impaired glucose tolerance (Fig. 2d,e; Fig. S4i, j). Taken together, these data are consistent with the notion that fat induces elevated lactate production from glucose in non-transformed mouse livers.
Figure 2. Fat induces lactate production from glucose.
a, b) Hepatic lactate enrichment normalized towards the 13C enrichment of plasma glucose, and hepatic lactate abundance CD (CD-vehicle n=5, CD-DEN n=5) or HFD (HFD-vehicle n=7, HFD-DEN n=6).
c) Plasma lactate abundance of mice after 8-weeks of control (CD-vehicle n=5) or high fat (HFD-vehicle n=6) diet normalized to control. Statistics: Two-tailed unpaired Student’s T-test.
d, e) Time-resolved changes in 13C lactate enrichment and corresponding area under the curve in mouse plasma after eight weeks on CD (n=13) or HFD (n=13) diet in response to oral administration of 2 g/kg of 13C6 glucose. Statistics: Two-way ANOVA with Fisher LSD testing to compare each time point (d), and two-tailed unpaired Student’s T-test (e).
Statistics: Unless noted otherwise, two-way ANOVA, with p-values representing the diet effect.
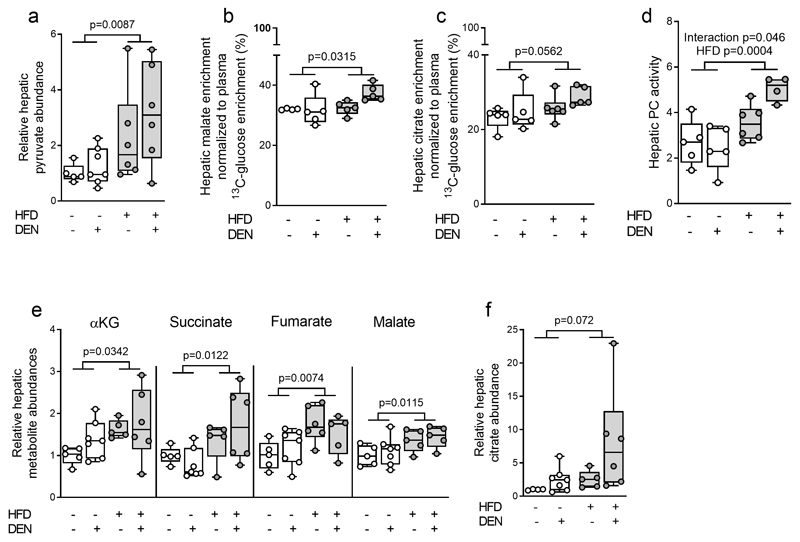
The tricarboxylic acid (TCA) cycle is another major fate of glucose metabolism. Glucose-derived carbon has two major routes to enter the TCA cycle, either via pyruvate carboxylase (PC) or pyruvate dehydrogenase (PDH). Hepatic pyruvate was elevated (Fig. 3a) and malate and citrate exhibited a higher 13C enrichment in HFD liver tissues (Fig. 3b,c). This suggests that PC activity may be increased upon HFD feeding. In line, we found that PC activity (33) was increased upon HFD feeding (Fig. 3d, Table S2). In hepatic metabolism, PC can function in conjunction with phosphoenolpyruvate carboxykinase (PCK1) activity which funnels carbons from the TCA cycle back into phosphoenolpyruvate, which can support gluconeogenesis. Our RNA-Seq data showed downregulation of Pck1, Pck2, and the gluconeogenic enzyme Fbp1 expression while Pcx expression was unchanged in HFD challenged livers (Fig. 1e, Fig. S4h). This suggests that fat may shift the PC to PCK1 balance in favor of TCA cycle fueling. Accordingly, we observed an increase in the abundance of the TCA cycle metabolites α–ketoglutarate, succinate, fumarate and malate, and a trend towards increased citrate abundance in HFD-exposed livers (Fig. 3e,f; Table S3). These data suggest that HFD results in an upregulation of PC-dependent glucose metabolism in mouse liver tissue. Collectively, we found that an increase in fat availability in vivo induces increased glucose uptake and contribution to central metabolism in normal, non-transformed livers.
Figure 3. Fat promotes pyruvate carboxylase activity.
a) Hepatic pyruvate abundance of mice after 8-weeks of control or high fat diet normalized to CD (CD-vehicle n=5, CD-DEN n=7) or HFD (HFD-vehicle n=7, HFD-DEN n=6).
b, c) Hepatic malate and citrate enrichment normalized towards the 13C enrichment of plasma glucose CD (CD-vehicle n=5, CD-DEN n=5) or HFD (HFD-vehicle n=7, HFD-DEN n=6).
d) Pyruvate carboxylase (PC) activity (based on 13C tracer analysis) in mouse livers after eight weeks of CD (CD-vehicle n=5, CD-DEN n=5) or HFD (HFD-vehicle n=6, HFD-DEN n=4).
e, f) Metabolite abundance in liver tissue of mice after eight weeks of CD (CD-vehicle n=5, CD-DEN n=7) or HFD (HFD-vehicle n=7, HFD-DEN n=6) normalized to CD-vehicle. αKG refers to α-ketoglutarate.
Unless noted otherwise, two-way ANOVA.
Evidence for fat-induced lactate production upon glucose availability in humans
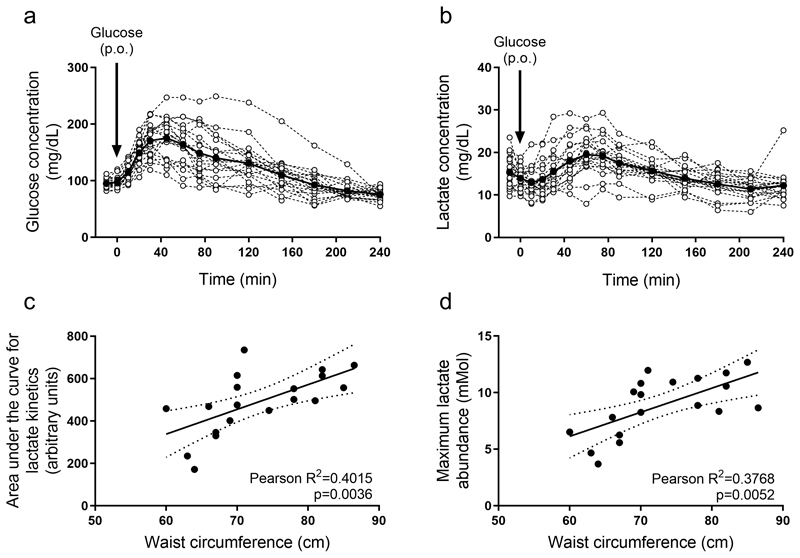
Our mouse data show that high fat availability induces a Warburg-like glucose metabolization in non-transformed mouse liver cells. Thus, we next asked whether there is some indication that similar changes occur in humans. We investigated whether we can observe an increased lactate production upon a glucose bolus in dependence of waist circumference. We analyzed 20 healthy humans of Japanese origin, with different visceral fat content, as determined by waist circumference (Table S7), which is a better predictor than body mass index for fat related liver diseases regardless whether the individual is overweight or has a normal weight (34–36). All individuals received an oral bolus of glucose (75g) under fasting conditions and blood was collected at several time points over a 4-hour period. Using blood glucose levels (Fig. 4a) we calculated the Matsuda index, HOMA-IR, Insulinogenic index, and disposition index (Table S7). All individuals showed normal values for the Matsuda index (below 2.5 units). HOMA-IR levels were also normal, with the exception of one individual showing a slightly higher value (2.8, cutoff 2.5). The insulinogenic index showed considerably more variability, with 7 out of 20 individuals showing a lower value than the standard cutoff rate of 0.4 units, indicating a possible defect with insulin secretion. However, in Japanese cohorts, even normoglycemic patients may show a lower than normal insulinogenic index compared to Caucasian counterparts (37) and, in fact, 19 out of 20 individuals showed a normal disposition index (cutoff ≥1) (Table S7). These results indicate that these individuals do not show any significant levels of insulin resistance.
Figure 4. Evidence for fat-induced lactate production upon glucose availability in humans.
a-b) Glucose and lactate abundance in the blood plasma of healthy human individuals upon oral administration of 75 g glucose (n=20). Black line indicates average over all individuals. Arrows indicate the time at which glucose was consumed. P.O refers to per os.
c-d) Correlation of waist circumference (WC; surrogate of visceral fat) with the area under the curve of the lactate kinetics or the maximum lactate abundance in healthy individuals upon oral administration of 75 g glucose (n=20).
Correlations were calculated by performing linear regression analysis, followed by an F-test to determine significant deviation from a 0-slope line.
In mice, we had observed that the fat-induced glucose metabolism resulted in elevated blood lactate abundance. Therefore, we analyzed the dynamic change in blood plasma lactate abundance in humans after administration of glucose (Fig. 4b). We correlated the area under the curve for lactate abundance over time and the maximum lactate abundance with the corresponding waist circumference of each individual. We expect that if visceral fat induces an increased glucose metabolization in humans, the area under the curve for lactate abundance and the maximum lactate abundance would both correlate with waist circumference. Indeed, we observed the expected correlation (Fig. 4c,d). Thus, we concluded that similarly to mice, humans with elevated visceral fat, as indicated by waist circumference, responded to glucose availability with lactate production.
ROS production may be linked to fat-induced glucose metabolism
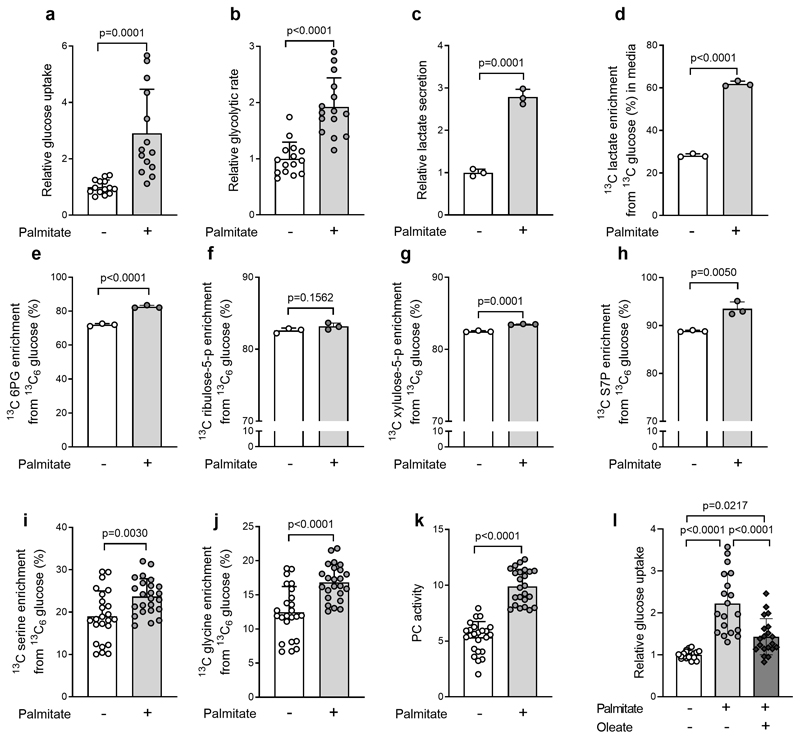
Next, we asked which metabolic changes link fat to an hyperactivated glucose metabolism in hepatic cells. Palmitate is highly abundant in blood plasma and HFD increased its concentration (Fig. S5a). Therefore, we focused on palmitate (0.4mM) and determined its effect on glucose uptake in liver (cancer) cell lines (Fig S5b). All of these cell lines displayed an increased glucose uptake upon palmitate treatment (Fig S5b). We then decided to determine further metabolic parameters in H4IIEC3cells (38). We supplemented H4IIEC3 cells with palmitate and measured glucose uptake, glycolytic flux, lactate production and enrichment from 13C glucose, as well as pentose phosphate pathway usage, serine biosynthesis and serine conversion to glycine and PC activity. In accordance with our in vivo data, we found that these metabolic pathways were highly induced upon palmitate supplementation, with the exception of the non-oxidative branch of the pentose phosphate pathway, which only showed a very minor change (Fig. 5a–k). In line with our in vivo data, we further found that these changes did not require alterations in glycolytic gene expression (Fig S5c).
Figure 5. Fat-induced changes in liver metabolism can be recapitulated at the cellular level.
a, b) Glucose uptake (n=14 for each group) and glycolytic rate (n=15 for each group) in H4IIEC3 cells treated with 0.4 mM palmitate or vehicle for 8-hours normalized to vehicle.
c, d) 13C-labelled lactate secretion into cell medium with following an 8 hour treatment of vehicle or 0.4mM palmitate in 13C6-glucose supplemented DMEM. Lactate secretion was calculated with the assumption of exponential cell growth during the 8 hour treatment phase, normalized to control. n=3 for each condition.
e-k) 6-phosphogluconate (n=3 for each group), ribulose-5-phosphate (n=3 for each group), xylulose 5-phosphate (n=3 for each group) and sedoheptulose-7-phosphate (n=3 for each group), serine synthesis (n=24 for each group) with conversion to glycine (n=24 for each group), and pyruvate carboxylase (PC) activity (n=24 for each group), in H4IIEC3 cells treated with 0.4 mM palmitate or vehicle for 8-hours.
I) Glucose uptake in H4IIEC3cells treated with 0.4mM palmitate and/or oleate for 8 hours normalized to control. n=24 replicates. Statistics: one-way ANOVA with Tukey ‘s multiple comparisons test.
Unless otherwise stated, two-tailed unpaired Student’s T-test, with p-values as indicated. All data are represented as mean ± standard deviation.
In vivo HFD treatment increases also monounsaturated fatty acids such as oleate (Fig. S5d). Therefore, we next combined palmitate and oleate treatment of H4IIEC3 cells and measured glucose uptake. We found that the combination still elevated glucose uptake, yet to a smaller extent, which was an expected outcome (39, 40) (Fig. 5l). Notably, our ability to reproduce the majority of the metabolic rewiring that we detected in vivo upon fat availability in in vitro cultured cells treated with palmitate provides further evidence that the link between fat and glucose metabolism is independent of insulin resistance.
In vivo we had observed that the glycerol-3-phosphate abundance and glycerol-3-phosphate dehydrogenase (Gpd) 2 gene expression were elevated upon fat availability (Table S3, Fig. S6a). Since glycerol-3-phosphate is necessary for building triglycerides, we argued that changes within this metabolic pathway indicate an elevated availability of free fatty acids for oxidation. The mitochondria are a major compartment of fatty acid oxidation. We treated H4IIEC3 cells in the presence of palmitate with etomoxir, which inhibits mitochondrial β-oxidation and measured metabolic pathways. Surprisingly, etomoxir treatment did not alter glucose uptake, and slightly increased serine synthesis with glycine conversion without changing PC activity (Fig. S6b–e). Accordingly, acetyl-CoA abundance, which is a product of mitochondrial β-oxidation was not altered upon palmitate treatment (Fig. S6f). These data suggest that mitochondrial β-oxidation may not be required for the observed hyperactivation of glucose metabolism upon palmitate availability.
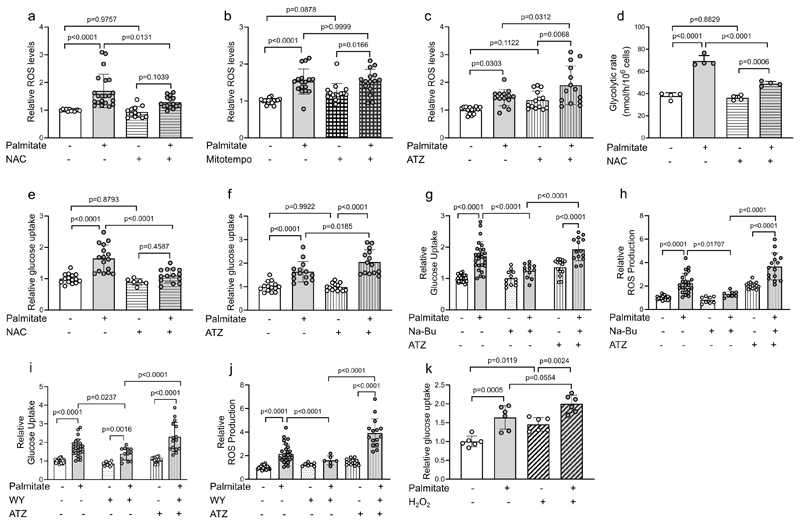
Fatty acids are not only oxidized in the mitochondria, but also in the peroxisomes. We observed that the most strongly enriched gene set in HFD-exposed liver tissue was related to peroxisomes (Fig S3a,c), and one of the few peptides significantly affected by HFD feeding was a peroxisomal β-oxidation enzyme (EHHADH; Fig S3f). Peroxisomes do not possess electron acceptors of the respiratory chain and thus water is used as final electron acceptor, generating the ROS species H2O2, which can be degraded by catalase (41). Therefore, we investigated whether ROS levels increased upon palmitate treatment. Indeed, we found an elevation in ROS with palmitate treatment, an effect partially mitigated with the addition of oleate (Fig S6g). When H4IIEC3cells were treated with palmitate in combination with N-acetyl-L-cysteine (NAC), ROS levels decreased, yet this effect did not occur with the mitochondrial ROS scavenger mitoTEMPO (Fig. 6a, b). Accordingly, the catalase inhibitor 3-amino-1,2,4-triazole (ATZ) further increased ROS levels upon palmitate treatment compared to the control condition (Fig.6c). These data may indicate palmitate-induced ROS generation in the peroxisomes.
Figure 6. ROS production is required for the hyperactivation of glucose metabolism upon palmitate supplementation.
a-c) Reactive oxygen species (ROS) in H4IIEC3 cells treated with 0.4 mM palmitate and/or NAC (5 mM) (n=22, 21, 12, 15 individual replicates for vehicle, palmitate, vehicle + NAC and NAC + palmitate groups, respectively), Mitotempo (n=18, 15, 14 and 15 individual replicates for vehicle, palmitate, mitotempo and mitotempo + palmitate, respectively) or ATZ (20 mM) (n=14 individual replicates for each group) normalized to control.
d) Glycolytic rate in H4IIEC3cells treated with 0.4 mM palmitate and/or NAC (5 mM) (n=4 individual replicates for each group).
e) Glucose uptake in H4IIEC3cells treated with 0.4mM palmitate and/or NAC (5 mM) (n=6 with individual replicates plotted for each group) normalized to control.
f) Glucose uptake in H4IIEC3cells treated with 0.4 mM palmitate and/or 3-ATZ (20 mM) (n=14 individual replicates for each group) normalized to control.
g-j) Relative glucose uptake and ROS production in in H4IIEC3cells pre-treated for 24-hours with sodium butyrate (Na-Bu, 5mM) or WY-14643 (WY, 100μM) followed by 8-hour treatment with 0.4mM palmitate and ATZ (20mM). (n=3-6 with individual replicates plotted for each group) normalized to control. Statistics: two-way ANOVA with Fisher LSD post-hoc testing, with p-values as indicated.
k) Relative glucose uptake in cells treated with 0.4 mM palmitate and/or H2O2 (2 mM) (n=6 with individual replicates plotted) normalized to control.
Unless otherwise noted, one-way ANOVA with Tukey’s multiple comparisons with p-values for multiple comparisons as indicated.
Next, we asked whether ROS production links palmitate to the hyperactivation of glucose metabolism by measuring glycolytic rate and/or uptake in the presence of palmitate upon treatment with NAC or ATZ. Strikingly, NAC dampened the palmitate-induced glycolytic flux and glucose uptake increase by approximately 60% and 50% respectively, while ATZ treatment led to a 68% increase in glucose uptake in the presence of palmitate (Fig. 6d–f). Notably, serine biosynthesis with conversion to glycine was also reduced upon NAC treatment by approximately 44%, whereas PC-activity and oxidative pentose phosphate pathways usage were unchanged (Fig. S6h–k). Additionally, NAC treatment had no effect on gene expression for glycolytic enzymes (Fig. S5c).
Next, we stimulated catalase expression using sodium-butyrate (Na-Butyrate (42)) and the PPARα agonist WY-14643 (43), increasing catalase gene expression by 44% and 72%, respectively (Fig. S6l). We then measured glucose uptake and ROS levels (Fig. 6g–j). We found that catalase stimulation by either compound inhibited palmitate-induced glucose uptake and ROS production, and that each compound on its own had no effect on these measurements (Fig. 6g–j). Next, we used ATZ, which blocked the effect of the catalase stimulators (Fig. 6g–j). Taken together, these data show that modulation in catalase activity altered ROS burden and glucose uptake upon palmitate supplementation.
Finally, we investigated whether ROS are sufficient to cause an increase in glycolysis by treating H4IIEC3C cells with H2O2 (2mM). Indeed, we found that H2O2 increased glucose uptake, and that the combination of H2O2 with palmitate further exacerbated the increase (Fig. 6k). These data indicate that ROS production is linked to the palmitate-induced increase in glucose metabolism in in vitro cultured hepatocyte-like cells.
Metabolic pathways induced by fat in non-transformed mouse livers resemble the metabolic hallmarks of liver cancer
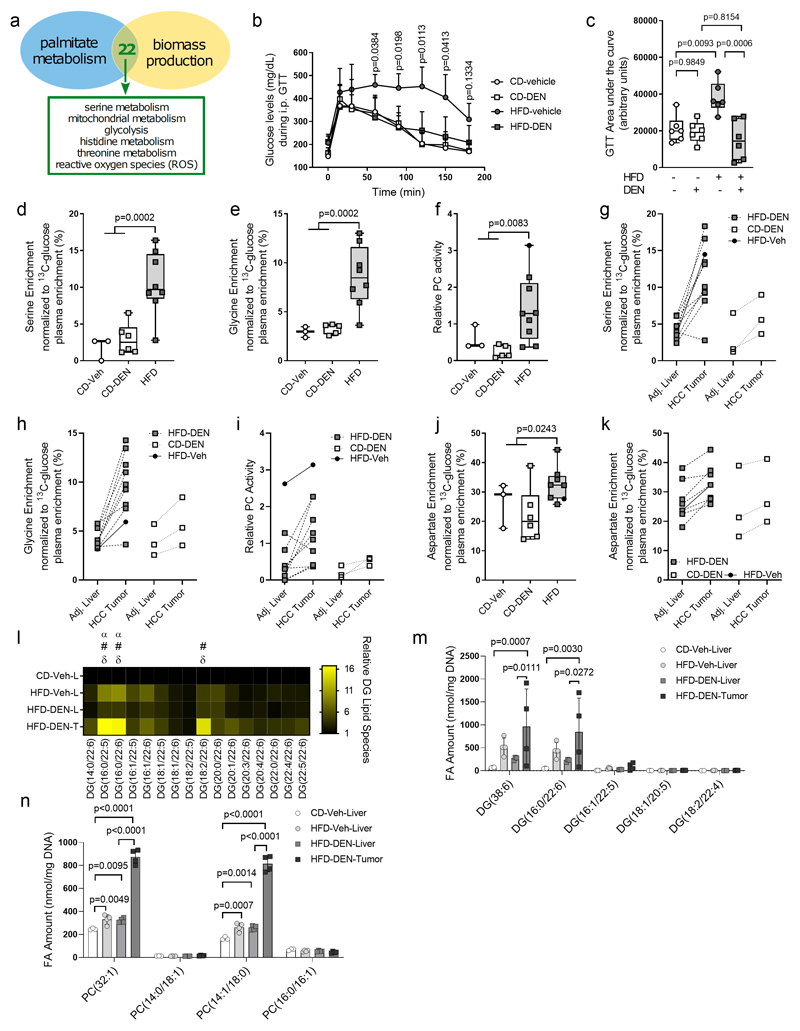
Metabolic rewiring is an essential hallmark of tumor development and progression (6, 44). Therefore, we asked whether the metabolic changes we observed in non-transformed liver cells induced by fat are related to liver cancer, particularly HCC metabolism. Thus, we used a computational modeling approach to predict potential commonalities between the metabolism of hepatocytes (origin of HCC (45)) challenged with palmitate utilization (as surrogate for increased fat availability) or proliferation (a metabolic surrogate for cellular transformation). We applied differential flux-balance analysis (DFA) based on the extended HepatoNet1 model (46), a computational modeling approach to identify metabolites that change most significantly upon a certain perturbation. To extract metabolic commonalities, we overlaid the top 75 most altered metabolites of each perturbation and found that both perturbations had 22 metabolite changes in common (Table S8). Mapping these metabolites onto the metabolic network, we identified glycolysis, mitochondrial metabolism, and serine metabolism as the three highest-ranking metabolic commonalities between palmitate utilization and proliferation-induced metabolic changes (Fig. 7a, Table S8). Thus, this analysis predicts that these three metabolic pathways, which we have shown to be induced in vivo by fat in non-transformed liver cells, could also be a hallmark of HCC.
Figure 7. Metabolic pathways induced by fat in non-transformed mouse livers are hallmarks of HCC.
a) In silico predictions of common metabolic pathways between cells supplemented with palmitate and cells maximizing biomass production based on 22 common metabolic intermediates.
b-c), Intraperitoneal glucose tolerance test in mice after 29 weeks on CD (CD-vehicle, n = 6; CD-DEN, n = 6) or HFD (HFD-vehicle, n = 6; HFD-DEN, n = 6) with DEN or vehicle exposure. Data are expressed as the mean of measured blood glucose levels (mg/dL) and the AUC. Statistics, two-way ANOVA followed by Dunnett’s multiple comparisons, with P values for HFD-vehicle compared with CD-vehicle as indicated in B and one-way ANOVA with Tukey’s multiple comparisons test with P values represented in c.
d-f) Serine biosynthesis with conversion to glycine normalized toward the 13C enrichment of plasma glucose and pyruvate carboxylase (PC) activity in normal liver (CD-vehicle, n = 3; CD-DEN, n = 6) or HCC tumor tissue [HFD-vehicle, n = 1 (full-black circle); HFD-DEN, n = 9 (PC activity) or n = 8 (serine biosynthesis with conversion to glycine)] of mice after 29 weeks on CD with DEN or vehicle exposure. Statistics, two-tailed unpaired Student t test comparing CD to HFD.
g-i) Serine biosynthesis with conversion to glycine normalized toward the 13C enrichment of plasma glucose and PC activity in matched pairs of HCC tumor and adjacent normal liver tissue of mice after 29 weeks on CD (n = 3) or HFD (n = 8) with vehicle or DEN exposure. In two HFD mice, two tumors were analyzed and one HFD-vehicle mouse (solid-black dot and line) was included.
j) Aspartate synthesis normalized toward the 13C enrichment of plasma glucose from glucose in normal liver (CD-vehicle, n = 3; CD-DEN, n = 6) or HCC tissue [HFD-vehicle, n = 1 (full-black circle); HFD-DEN, n = 7] of mice after 29 weeks on CD with DEN or vehicle exposure. Statistics, two-tailed unpaired Student t test comparing CD to HFD.
k) Aspartate synthesis normalized toward the 13C enrichment of plasma glucose from glucose in matched pairs of HCC tumor and adjacent normal liver tissue of mice after 29 weeks on CD (n = 3) or HFD (n = 8) with vehicle or DEN exposure. For two HFD-DEN animals, two tumors were analyzed and connected to the same adjacent liver sample and one HFD-vehicle mouse is included (solid-black circle and solid line).
I) Heat map with normalized diacylglycerides (DG) species that contain 22:5 and 22:6 acyl chains in CD-vehicle liver tissue (n = 3), HFD-vehicle liver tissue (n = 4), HFD-DEN liver tissue (n = 3), and HFD-DEN tumor tissue (n = 4), all normalized to CD-vehicle liver tissue. Statistics, two-way ANOVA with Tukey’s multiple comparisons testing. α, significant differences between CD-vehicle liver to HFD-vehicle liver; #, significant differences between CD-vehicle liver and HFD-DEN tumor tissue; δ, significant differences between HFD-DEN liver tissue and HFD-DEN tumor tissue, with P < 0.05 considered significant.
m-n) Total amounts of the sum notations of DG (38:6) and PC (32:1), and the specific lipid species that make up the sum notations in CD-vehicle liver tissue (n = 3), HFD-vehicle liver tissue (n = 4), HFD-DEN liver tissue (n = 4), and HFD-DEN tumor tissue (n = 4).
Statistics, two-way ANOVA with Tukey’s multiple comparisons tests, with P values represented.
It is well established that increased glucose uptake is a hallmark of HCC metabolism (47). Yet, whether serine biosynthesis and mitochondrial PC-activity are elevated in HCC cells is poorly defined. Thus, we measured the activity of these two metabolic pathways in HCC tissues from DEN-injected mice. Following the same experimental design as for our early time point, we extended HFD feeding to 29 weeks. In this setup, we expected that only animals injected with DEN would show substantial HCC development (Fig. S7a) and that mice on HFD would develop insulin resistance. Therefore, we conducted glucose and insulin tolerance tests following long-term HFD exposure. We found that animals on HFD showed impaired insulin response compared to control diet animals with or without DEN injection (Fig. S7b,c). Strikingly, glucose sensitivity was completely normalized in DEN-injected HFD-fed mice and was indistinguishable from all control diet fed animals, in which exposure to DEN had no impact on glucose tolerance (Fig. 7b,c). Comparatively, HFD-fed vehicle-injected mice had severely impaired glucose clearance (Fig. 7b,c). These data suggest that the tumor burden in HFD animals injected with DEN is responsible for significant levels of glucose clearance.
After 29 weeks on HFD, we infused the mice with 13C6-glucose and measured the 13C enrichment of metabolites in hepatic and tumor tissue as described above. At this point, all HFD-fed DEN-injected animals showed massive HCC development (Fig. S7a). All animals injected with DEN on control diet showed signs of HCC development, but only three out of six animals had tumors with a size that allowed resection. Meanwhile, one out of five animals injected with vehicle on HFD developed tumors and none of the vehicle injected animals on control diet showed signs of tumor development.
We then analyzed serine biosynthesis with conversion to glycine and PC-activity in tumor tissues of HFD mice (DEN or vehicle injected) compared to liver tissue of mice on control diet based on the 13C enrichment of metabolites. Both pathways were increased in HFD induced tumors compared to liver tissue of control diet mice (Fig. 7d–f, Table S9). Subsequently, we analyzed matched pairs of tumors and normal adjacent liver tissue from both diets. We observed an elevation in serine biosynthesis with glycine conversion and PC-activity compared to the corresponding adjacent non-transformed liver tissue in 7 out of 8 tissues from HFD-fed mice and, surprisingly, all control diet paired tissues (Fig. 7g–i). In line with the idea that this PC activity fuels anaplerosis required for proliferation, 13C aspartate enrichment was also elevated in all tumors compared to adjacent normal tissue, regardless of diet (Fig. 7j,k). Interestingly, in the presence of tumors, the adjacent liver does not seem to differ any longer between high fat and control diet animals. Therefore, it is tempting to speculate that the tumor educates the metabolism of the organ in which it grows. Collectively, these data demonstrate that glucose uptake, serine biosynthesis with glycine conversion and PC-activity define the in vivo metabolism of HCC regardless whether tumor development and proliferation was accelerated with HFD or not. Moreover, the data show that a large fraction of the glucose metabolism-related rewiring induced by fat in non-transformed and tumor-free livers were induced and further promoted in HCC.
To further explore changes in liver tissue adjacent to growing tumors, we conducted lipidomics analysis (Fig. S7d). We observed that HFD vehicle livers and tumor adjacent HFD DEN livers clustered together, while control diet vehicle livers and HFD DEN tumor tissue clustered separately from the other groups (Fig S7e). This indicates that tumor tissue has a different lipidome compared to its adjacent liver tissue, and that it is influenced by the diet. Further, tumor tissue had 67 lipid species that were significantly different from adjacent liver tissue, with a particular diacylglycerol (DG) and phosphatidylcholine (PC) signature (Fig. 7l–n, Fig. S7f–h, Table S10). Total levels of DG in tumor tissue were elevated compared to control diet livers, but not adjacent HFD DEN-exposed liver tissue (Fig. S7f). Despite no measurable differences in total DG levels between tumor tissue and adjacent HFD DEN liver tissue, tumor tissue had elevated levels of DG species containing 16:0, 22:5, or 22:6 acyl chains when compared to adjacent HFD DEN liver tissue and control diet liver tissue (Fig. 7l). These particular DG species were also increased in the livers of non-tumor bearing HFD vehicle injected mice compared to control diet livers (Fig. 7l). Further, analysis of the sum notation of DG (the sum of all carbons and double bonds in both acyl chains) indicated tumor tissue was enriched in certain DG species, including DG 38:6 and DG 36:6 (Fig. S7g). DG 38:6 was the most abundant of these lipids, and this elevation was found to be driven by DG (16:0/22:6) specifically (Fig 7m). Tumor tissue was also enriched in numerous PC lipids compared to control diet liver tissue (Fig S7h). Tumor tissue was particularly enriched in PC 32:1, and this was found to be driven specifically by PC (14:1/18:0) (Fig. 7n, Fig. S7h). These data indicate that the presence of tumors may alter the HFD induced lipid composition of adjacent liver tissue. Further, while tumors certainly had a particular lipid profile, our analysis showed some indications of HFD priming of the normal liver lipidome towards the lipid state of HCC.
Discussion
Here, we show that normal, non-transformed and tumor-free livers respond to the nutrient fat by inducing an increased glucose metabolism, which is similar to the metabolic state of HCC regardless whether the tumors developed in lean or obese mice.
Previous studies have analyzed the impact of fat-induced insulin resistance on liver metabolism (9) and fatty acid metabolization in vitro (38, 48) and in isolated hepatocytes (20). Complementary to these studies, we provide an in vivo analysis of insulin-independent consequences of fat availability on normal liver metabolism. Moreover, we provide evidence that fat utilization elicits increased glucose metabolism that is mediated by peroxisomal ROS production. Interestingly, this finding is in line with previous work reporting the importance of peroxisomal lipid metabolism in tempering palmitate lipotoxicity (49, 50) and inducing HCC (25). Further, we find that these alterations in glucose metabolism are similar to those found in HCC. Thus, it is tempting to speculate that high fat availability could metabolically prime normal hepatocytes for HCC development. Accordingly, our data may explain the observation that HFD in the presence of glucose accelerates HCC development and progression while HFD in the absence of glucose has no effect aside from the manifestation of obesity and insulin resistance (51). Thus, it may be relevant to investigate sugar reduced diets to decrease the risk of liver cancer in obese patients.
Despite the fact that nutrients are important regulators of in vivo cancer metabolism, we further found that core hallmarks of glucose metabolism in HCC are the same regardless whether the tumor developed in a high fat or lean environment. Interestingly, our data indicate that fat further promotes the activity of metabolic pathways such as serine biosynthesis and PC activity which is rewired upon cellular transformation leading to HCC development. Thus, indicating that targeting these metabolic pathways could have the potential to impair HCC progression.
Using lipidomics, we found elevations in some DG lipids, which are involved in protein kinase C (PKC) activity (52), a protein involved in numerous diseases, including diabetes and cancer. Further, DG have been reported to determine cancer cell sensitivity to FASN inhibitors (53). We also found increases in some PC species in tumor tissue compared to adjacent liver tissue. PC are an important plasma membrane building block and regulator of some cell signaling pathways (54, 55), including PPAR activity. Additionally, we observed lipid species from these two classes to be elevated in HFD vehicle liver tissue compared to control diet liver tissue, mirroring changes in tumor tissue compared to adjacent HFD liver tissue. Thus, it is tempting to speculate that fat is an inducer of a pro-tumor metabolism in non-transformed livers.
In conclusion, our data show that fat can increase some metabolic hallmarks of liver cancers in normal, non-transformed and tumor-free livers.
Supplementary Material
Significance.
With obesity-induced hepatocellular carcinoma on a rising trend, this study shows in normal, non-transformed livers that fat induces glucose metabolism similar to an oncogenic transformation.
Acknowledgments
We thank, Dr. Arvind Patel for the HHL5 cell line, Theo Killian (CCB-Bioinformatics core) for contributing to the bioinformatics analyses, Thomas Van Brussel and Bram Boeckx (VIB-KU Leuven) for transcriptomics services, Sara Dufour (VIB-Proteomics core) for the proteomics mass spectrometry service, David Nittner (VIB Histology and Imaging Core) for their histology services, Nienke Meeuws for assistance with IHC analysis, and Paul Van Veldhoven and David Cassiman for their valuable suggestions.
JADG, RS, and JFG were/are supported by FWO postdoctoral and KVe by FWO PhD fellowships. TGPG acknowledges funding from the German Cancer Aid (DKH-70114111 and DKH-70112257), the Dr. Leopold and Carmen Ellinger Foundation, the Matthias-Lackas Foundation, the Dr. Rolf M. Schwiete Foundation, the Gert and Susanna Mayer Foundation, the SMARCB1 association, and the Barbara and Wilfried Mohr Foundation. SK was supported by the Creation of Fundamental Technologies for Understanding and Control of Biosystem Dynamics (JPMJCR12W3), CREST, of the Japan Science and Technology Agency (JST) and by the Japan Society for the Promotion of Science (JSPS) KAKENHI Grant Number (17H06300, 17H6299, 18H03979). YK was supported by JSPS KAKENHI Grant Number (18K16578). SMF acknowledges funding from the European Research Council under the ERC Consolidator Grant Agreement n. 771486–MetaRegulation and Marie Curie CIG n. 617727–MetabolismConnect, FWO Odysseus II, FWO Project (G098120N), Fonds Baillet Latour and KU Leuven Methusalem Co-funding.
Funding Acquisition
SMF
The authors declare that all data supporting the findings of this study are available within the article, its extended data files, or from the corresponding author upon reasonable request.
Footnotes
Conflict of Interest
SMF has received funding from Bayer, Merck and Black Belt Therapeutics and has consulted for Fund+. All other authors declare to have no competing interest.
Author Contributions
Conceptualization: LAB, JAGD, RS, SMF
Methodology: JAGD, RS, LAB, DB
Software: FN, JFG, DDB
Validation: JAGD, RS, LAB, DB
Formal Analysis: JAGD, RS, LAB, KVe, KVr, YK, FN, SF, MF, ME, JFG, JD
Investigation: JAGD, RS, LAB, DB, KVe, YK, SF, MF, FN, BH, RV, JFG, RAR, JD, AL, SK, KDB
Resource: JvP, TGPG, DDB, CM, CMD, JZ, JVS, DL, SK, KDB, JVS, DL
Writing: Original Draft SMF
Writing: Review & Editing JAGD, LAB, SMF
Supervision: SMF
Visualization: JADG, LAB
References
- 1.Lundsgaard A-M, Fritzen AM, Kiens B. Molecular Regulation of Fatty Acid Oxidation in Skeletal Muscle during Aerobic Exercise. Trends in Endocrinology & Metabolism. 2018;29(1):18–30. doi: 10.1016/j.tem.2017.10.011. [DOI] [PubMed] [Google Scholar]
- 2.Lorendeau D, Christen S, Rinaldi G, Fendt S-M. Metabolic control of signaling pathways and metabolic auto-regulation. Biology of the Cell. 2015;107(8):251–72. doi: 10.1111/boc.201500015. [DOI] [PubMed] [Google Scholar]
- 3.Rinaldi G, Rossi M, Fendt S-M. Metabolic interactions in cancer: Cellular metabolism at the interface between the microenvironment, the cancer cell phenotype and the epigenetic landscape. WIREs Syst Biol Med. 2017:e1397. doi: 10.1002/wsbm.1397. in press. [DOI] [PubMed] [Google Scholar]
- 4.Vande Voorde J, Ackermann T, Pfetzer N, Sumpton D, Mackay G, Kalna G, et al. Improving the metabolic fidelity of cancer models with a physiological cell culture medium. Science Advances. 2019;5(1):eaau7314. doi: 10.1126/sciadv.aau7314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hensley Christopher T, Faubert B, Yuan Q, Lev-Cohain N, Jin E, Kim J, et al. Metabolic Heterogeneity in Human Lung Tumors. Cell. 2016;164(4):681–94. doi: 10.1016/j.cell.2015.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bergers G, Fendt SM. The metabolism of cancer cells during metastasis. Nature reviews Cancer. 2021 doi: 10.1038/s41568-020-00320-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coloff Jonathan L, Murphy JP, Braun Craig R, Harris Isaac S, Shelton Laura M, Kami K, et al. Differential Glutamate Metabolism in Proliferating and Quiescent Mammary Epithelial Cells. Cell metabolism. 2016;23(5):867–80. doi: 10.1016/j.cmet.2016.03.016. [DOI] [PubMed] [Google Scholar]
- 8.Kalucka J, Bierhansl L, Conchinha NV, Missiaen R, Elia I, Brüning U, et al. Quiescent Endothelial Cells Upregulate Fatty Acid β-Oxidation for Vasculoprotection via Redox Homeostasis. Cell metabolism. 2018;28(6):881–94.:e13. doi: 10.1016/j.cmet.2018.07.016. [DOI] [PubMed] [Google Scholar]
- 9.Kakimoto PA, Kowaltowski AJ. Effects of high fat diets on rodent liver bioenergetics and oxidative imbalance. Redox Biology. 2016;8:216–25. doi: 10.1016/j.redox.2016.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nagarajan SR, Paul-Heng M, Krycer JR, Fazakerley D, Sharland AF, Hoy AJ. Lipid and Glucose Metabolism in Hepatocyte Cell Lines and Primary Mouse Hepatocytes: A comprehensive resource for in vitro studies of hepatic metabolism. American Journal of Physiology-Endocrinology and Metabolism. 2019 doi: 10.1152/ajpendo.00365.2018. [DOI] [PubMed] [Google Scholar]
- 11.Neinast MD, Jang C, Hui S, Murashige DS, Chu Q, Morscher RJ, et al. Quantitative Analysis of the Whole-Body Metabolic Fate of Branched-Chain Amino Acids. Cell metabolism. 2018 doi: 10.1016/j.cmet.2018.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hui S, Ghergurovich JM, Morscher RJ, Jang C, Teng X, Lu W, et al. Glucose feeds the TCA cycle via circulating lactate. Nature. 2017;551(7678):115–8. doi: 10.1038/nature24057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sunny NE, Bril F, Cusi K. Mitochondrial Adaptation in Nonalcoholic Fatty Liver Disease: Novel Mechanisms and Treatment Strategies. Trends in Endocrinology & Metabolism. 2017;28(4):250–60. doi: 10.1016/j.tem.2016.11.006. [DOI] [PubMed] [Google Scholar]
- 14.Goodpaster BH, Sparks LM. Metabolic Flexibility in Health and Disease. Cell metabolism. 2017;25(5):1027–36. doi: 10.1016/j.cmet.2017.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kahn SE, Hull RL, Utzschneider KM. Mechanisms linking obesity to insulin resistance and type 2 diabetes. Nature. 2006;444:840. doi: 10.1038/nature05482. [DOI] [PubMed] [Google Scholar]
- 16.Taniguchi CM, Emanuelli B, Kahn CR. Critical nodes in signalling pathways: insights into insulin action. Nature Reviews Molecular Cell Biology. 2006;7:85. doi: 10.1038/nrm1837. [DOI] [PubMed] [Google Scholar]
- 17.Anstee QM, Reeves HL, Kotsiliti E, Govaere O, Heikenwalder M. From NASH to HCC: current concepts and future challenges. Nature Reviews Gastroenterology & Hepatology. 2019;16(7):411–28. doi: 10.1038/s41575-019-0145-7. [DOI] [PubMed] [Google Scholar]
- 18.Eisenstein AB, Strack I, Steiner A. Increased Hepatic Gluconeogenesis without a Rise of Glucagon Secretion in Rats Fed a High Fat Diet. Diabetes. 1974;23(11):869. doi: 10.2337/diab.23.11.869. [DOI] [PubMed] [Google Scholar]
- 19.Díaz-Rúa R, van Schothorst EM, Keijer J, Palou A, Oliver P. Isocaloric high-fat feeding directs hepatic metabolism to handling of nutrient imbalance promoting liver fat deposition. International Journal of Obesity. 2016;40(8):1250–9. doi: 10.1038/ijo.2016.47. [DOI] [PubMed] [Google Scholar]
- 20.Vial G, Dubouchaud H, Couturier K, Cottet-Rousselle C, Taleux N, Athias A, et al. Effects of a high-fat diet on energy metabolism and ROS production in rat liver. Journal of Hepatology. 2011;54(2):348–56. doi: 10.1016/j.jhep.2010.06.044. [DOI] [PubMed] [Google Scholar]
- 21.Matsuzawa-Nagata N, Takamura T, Ando H, Nakamura S, Kurita S, Misu H, et al. Increased oxidative stress precedes the onset of high-fat diet;induced insulin resistance and obesity. Metabolism - Clinical and Experimental. 2008;57(8):1071–7. doi: 10.1016/j.metabol.2008.03.010. [DOI] [PubMed] [Google Scholar]
- 22.Townsend LK, Medak KD, Peppler WT, Meers GM, Rector RS, LeBlanc PJ, et al. High-saturated-fat diet-induced obesity causes hepatic interleukin-6 resistance via endoplasmic reticulum stress. Journal of Lipid Research. 2019;60(7):1236–49. doi: 10.1194/jlr.M092510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Seki S, Kitada T, Yamada T, Sakaguchi H, Nakatani K, Wakasa K. In situ detection of lipid peroxidation and oxidative DNA damage in non-alcoholic fatty liver diseases. Journal of Hepatology. 2002;37(1):56–62. doi: 10.1016/s0168-8278(02)00073-9. [DOI] [PubMed] [Google Scholar]
- 24.Clayton RF, Rinaldi A, Kandyba EE, Edward M, Willberg C, Klenerman P, et al. Liver cell lines for the study of hepatocyte functions and immunological response. Liver international : official journal of the International Association for the Study of the Liver. 2005;25(2):389–402. doi: 10.1111/j.1478-3231.2005.01017.x. [DOI] [PubMed] [Google Scholar]
- 25.Heindryckx F, Colle I, Van Vlierberghe H. Experimental mouse models for hepatocellular carcinoma research. International journal of experimental pathology. 2009;90(4):367–86. doi: 10.1111/j.1365-2613.2009.00656.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hopkins BD, Goncalves MD, Cantley LC. Obesity and Cancer Mechanisms: Cancer Metabolism. Journal of Clinical Oncology. 2016;34(35):4277–83. doi: 10.1200/JCO.2016.67.9712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.King K-L, Hwang J-J, Chau G-Y, Tsay S-H, Chi C-W, Lee T-G, et al. Ki-67 expression as a prognostic marker in patients with hepatocellular carcinoma. Journal of Gastroenterology and Hepatology. 2009;13(3):273–9. doi: 10.1111/j.1440-1746.1998.01555.x. [DOI] [PubMed] [Google Scholar]
- 28.Park EJ, Lee JH, Yu G-Y, He G, Ali SR, Holzer RG, et al. Dietary and genetic obesity promote liver inflammation and tumorigenesis by enhancing IL-6 and TNF expression. Cell. 2010;140(2):197–208. doi: 10.1016/j.cell.2009.12.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Song P, Zhang J, Zhang Y, Shu Z, Xu P, He L, et al. Hepatic recruitment of CD11b+Ly6C+ inflammatory monocytes promotes hepatic ischemia/reperfusion injury. International journal of molecular medicine. 2018;41(2):935–45. doi: 10.3892/ijmm.2017.3315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Broekaert D, Fendt S-M. Measuring in vivo tissue metabolism using 13C glucose infusions in mice. Methods in Molecular Biology. 2019:1862. doi: 10.1007/978-1-4939-8769-6_5. Metabolic Signaling. [DOI] [PubMed] [Google Scholar]
- 31.Fernández-García J, Altea-Manzano P, Pranzini E, Fendt S-M. Stable Isotopes for Tracing Mammalian-Cell Metabolism. Trends in biochemical sciences. 2020;45(3):185–201. doi: 10.1016/j.tibs.2019.12.002. [DOI] [PubMed] [Google Scholar]
- 32.Faubert B, Li KY, Cai L, Hensley CT, Kim J, Zacharias LG, et al. Lactate Metabolism in Human Lung Tumors. Cell. 2017;171(2):358–71.:e9. doi: 10.1016/j.cell.2017.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Christen S, Lorendeau D, Schmieder R, Broekaert D, Metzger K, Veys K, et al. Breast cancer-derived lung metastasis show increased pyruvate carboxylase-dependent anaplerosis. Cell Reports. 2016;17(3):837–48. doi: 10.1016/j.celrep.2016.09.042. [DOI] [PubMed] [Google Scholar]
- 34.Shen W, Punyanitya M, Chen J, Gallagher D, Albu J, Pi-Sunyer X, et al. Waist Circumference Correlates with Metabolic Syndrome Indicators Better Than Percentage Fat. Obesity (Silver Spring, Md) 2006;14(4):727–36. doi: 10.1038/oby.2006.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bener A, Yousafzai MT, Darwish S, Al-Hamaq AOAA, Nasralla EA, Abdul-Ghani M. Obesity Index That Better Predict Metabolic Syndrome: Body Mass Index, Waist Circumference, Waist Hip Ratio, or Waist Height Ratio. Journal of Obesity. 2013;2013:269038. doi: 10.1155/2013/269038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Campbell PT, Newton CC, Freedman ND, Koshiol J, Alavanja MC, Beane Freeman LE, et al. Body Mass Index, Waist Circumference, Diabetes, and Risk of Liver Cancer for U.S. Adults Cancer research. 2016;76(20):6076. doi: 10.1158/0008-5472.CAN-16-0787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fukushima M, Suzuki H, Seino Y. Insulin secretion capacity in the development from normal glucose tolerance to type 2 diabetes. Diabetes Res Clin Pract. 2004;66(Suppl 1):S37–43. doi: 10.1016/j.diabres.2003.11.024. [DOI] [PubMed] [Google Scholar]
- 38.Egnatchik RA, Leamy AK, Noguchi Y, Shiota M, Young JD. Palmitate-induced Activation of Mitochondrial Metabolism Promotes Oxidative Stress and Apoptosis in H4IIEC3 Rat Hepatocytes. Metabolism. 2014;63(2):283–95. doi: 10.1016/j.metabol.2013.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee H, Lim J-Y, Choi S-J. Oleate Prevents Palmitate-Induced Atrophy via Modulation of Mitochondrial ROS Production in Skeletal Myotubes. Oxid Med Cell Longev. 2017;2017:2739721. doi: 10.1155/2017/2739721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Srivastava S, Chan C, Srivastava S, Chan C. Hydrogen peroxide and hydroxyl radicals mediate palmitate-induced cytotoxicity to hepatoma cells: Relation to mitochondrial permeability transition. Free Radical Research. 2007;41(1):38–49. doi: 10.1080/10715760600943900. [DOI] [PubMed] [Google Scholar]
- 41.Schrader M, Fahimi HD. Peroxisomes and oxidative stress. Biochim Biophys Acta. 2006;1763(12):1755–66. doi: 10.1016/j.bbamcr.2006.09.006. [DOI] [PubMed] [Google Scholar]
- 42.Sauer J, Richter KK, Pool-Zobel BL. Physiological concentrations of butyrate favorably modulate genes of oxidative and metabolic stress in primary human colon cells. The Journal of Nutritional Biochemistry. 2007;18(11):736–45. doi: 10.1016/j.jnutbio.2006.12.012. [DOI] [PubMed] [Google Scholar]
- 43.Shin MH, Lee S-R, Kim M-K, Shin C-Y, Lee DH, Chung JH. Activation of Peroxisome Proliferator-Activated Receptor Alpha Improves Aged and UV-Irradiated Skin by Catalase Induction. PloS one. 2016;11(9):e0162628-e. doi: 10.1371/journal.pone.0162628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fendt SM, Frezza C, Erez A. Targeting Metabolic Plasticity and Flexibility Dynamics for Cancer Therapy. Cancer discovery. 2020;10(12):1797–807. doi: 10.1158/2159-8290.CD-20-0844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mu X, Español-Suñer R, Mederacke I, Affò S, Manco R, Sempoux C, et al. Hepatocellular carcinoma originates from hepatocytes and not from the progenitor/biliary compartment. The Journal of clinical investigation. 2015;125(10):3891–903. doi: 10.1172/JCI77995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pagliarini R, Castello R, Napolitano F, Borzone R, Annunziata P, Mandrile G, et al. In Silico Modeling of Liver Metabolism in a Human Disease Reveals a Key Enzyme for Histidine and Histamine Homeostasis. Cell Reports. 2016;15(10):2292–300. doi: 10.1016/j.celrep.2016.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Elia I, Schmieder R, Christen S, Fendt S-M. Organ-Specific Cancer Metabolism and Its Potential for Therapy. Handbook of Experimental Pharmacology. 2016;233:321–53. doi: 10.1007/164_2015_10. [DOI] [PubMed] [Google Scholar]
- 48.Leamy AK, Hasenour CM, Egnatchik RA, Trenary IA, Yao C-H, Patti GJ, et al. Knockdown of triglyceride synthesis does not enhance palmitate lipotoxicity or prevent oleate-mediated rescue in rat hepatocytes. Biochimica et Biophysica Acta (BBA) - Molecular and Cell Biology of Lipids. 2016;1861(9, Part A):1005–14. doi: 10.1016/j.bbalip.2016.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhu XG, Nicholson Puthenveedu S, Shen Y, La K, Ozlu C, Wang T, et al. CHP1 Regulates Compartmentalized Glycerolipid Synthesis by Activating GPAT4. Molecular Cell. 2019;74(1):45–58.:e7. doi: 10.1016/j.molcel.2019.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Benard O, Lim J, Apontes P, Jing X, Angeletti RH, Chi Y. Impact of high-fat diet on the proteome of mouse liver. The Journal of Nutritional Biochemistry. 2016;31:10–9. doi: 10.1016/j.jnutbio.2015.12.012. [DOI] [PubMed] [Google Scholar]
- 51.Healy ME, Chow JDY, Byrne FL, Breen DS, Leitinger N, Li C, et al. Dietary effects on liver tumor burden in mice treated with the hepatocellular carcinogen diethylnitrosamine. Journal of Hepatology. 2015;62(3):599–606. doi: 10.1016/j.jhep.2014.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mochly-Rosen D, Das K, Grimes KV. Protein kinase C, an elusive therapeutic target? Nature Reviews Drug Discovery. 2012;11(12):937–57. doi: 10.1038/nrd3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Benjamin DI, Li DS, Lowe W, Heuer T, Kemble G, Nomura DK. Diacylglycerol Metabolism and Signaling Is a Driving Force Underlying FASN Inhibitor Sensitivity in Cancer Cells. ACS Chem Biol. 2015;10(7):1616–23. doi: 10.1021/acschembio.5b00240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Furse S, de Kroon AIPM. Phosphatidylcholine’s functions beyond that of a membrane brick. Molecular Membrane Biology. 2015;32(4):117–9. doi: 10.3109/09687688.2015.1066894. [DOI] [PubMed] [Google Scholar]
- 55.Chakravarthy MV, Lodhi IJ, Yin L, Malapaka RR, Xu HE, Turk J, et al. Identification of a physiologically relevant endogenous ligand for PPARalpha in liver. Cell. 2009;138(3):476–88. doi: 10.1016/j.cell.2009.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.