Abstract
Sequencing of RNA 3’ ends uncovered numerous sites that do not correspond to termination sites of known transcripts. Through their 3’ untranslated regions, protein-coding RNAs interact with RNA-binding proteins and microRNAs, which regulate many properties, including RNA stability and subcellular localization. Here we present the ‘terminal exon characterization’ (TEC) tool (http://tectool.unibas.ch), applicable to RNA sequencing data from any species for which a genome annotation that includes sites of RNA cleavage and polyadenylation is available. We describe hundreds of novel isoforms and cell type-specific terminal exons in human cells. Ribosome profiling data indicate that many of these isoforms are translated. Applying TECtool to single cell sequencing data we find that the newly identified isoforms have typical per-cell abundance, but are expressed in subpopulations of cells. Thus, TECtool enables identification of novel isoforms in well studied cell systems and in rare cell types.
Introduction
Most eukaryotic transcripts undergo maturation through 3’ end cleavage and polyadenylation (CPA). The 3’ untranslated regions (3’ UTRs) of protein-coding messenger RNAs (mRNAs) interact with RNA-binding proteins (RBPs) 1 and microRNAs (miRNAs), which control diverse aspects of gene expression 2. Global changes in 3’ UTR length have been observed during immune responses 3, development 4, and in cancers 5. The initial view was that 3’ UTR shortening serves to counteract the repressive effect of miRNAs in proliferating cells 3,6. However, subsequent studies found largely similar decay rates of long and short 3’ UTR isoforms 7,8, leaving the role of 3’ UTR length changes still unclear. Evidence is accumulating that 3’ UTR-located sequence elements, particularly uridine(U)-rich, regulate many aspects of gene expression, from alternative polyadenylation in the nucleus to the subcellular localization of mRNAs and proteins in the cytoplasm 9–11.
In spite of many efforts to catalog human and mouse transcript isoforms 12–15, sequencing of RNA 3’ ends continues to uncover novel polyadenylation (poly(A)) sites (PAS), many outside of annotated exons 12–15. The presence of well-characterized poly(A) signals indicates that these PAS are genuine 9, yet little is known about their regulation and functions. In a recent study 9 we identified 108’932 PAS in genomic regions annotated as introns in the GENCODE transcript annotation version 19 16. In contrast to the more studied tandem PAS in 3’ UTRs, whose variable processing leads to changes in 3’ UTR length, the use of ‘intronic’ PAS can alter both the encoded protein isoforms and the 3’ UTRs and thereby the interactomes of the corresponding transcripts. To expand genome annotations with transcripts that end at currently ‘intronic’ PAS, we have developed a computational terminal exon characterization (TEC) tool.
Results
Prevalent pre-RNA processing at ‘intronic’ poly(A) sites
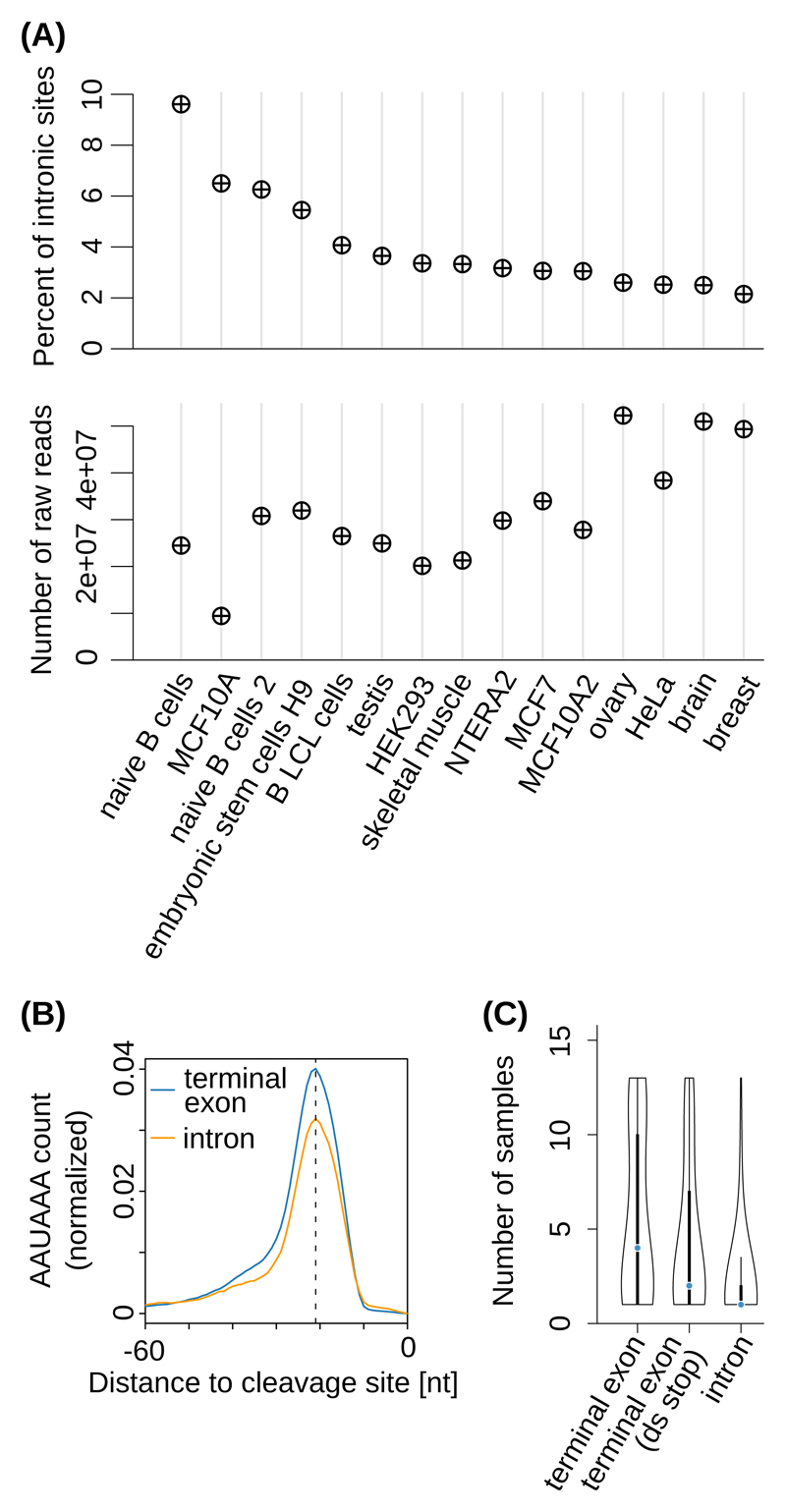
A large proportion of PAS reproducibly identified from ~200 distinct human and mouse 3’ end sequencing samples, are located in genomic regions currently annotated as intronic 9. Representing up to ~10% of the PAS identified in individual tissues (unrelated to sequencing depth, Figure 1A and Supplementary Figure 1A), ‘intronic’ PAS have canonically-positioned polyadenylation signals (~21 nucleotides (nts) upstream of cleavage sites, Figure 1B and Supplementary Figure 1B), but a more specific tissue distribution compared to the PAS in annotated terminal exons (Figure 1C and Supplementary Figure 1C).
Figure 1. Cell type-dependent usage of ‘intronic’ poly(A) sites.
(A) Top panel: Percentage of ‘intronic’ PAS in individual samples obtained with the 3’-Seq protocol 17. Bottom panel: corresponding sequencing depths. (B) Position-dependent frequency of the canonical poly(A) signal (‘AAUAAA’), dashed line at -21 nts) upstream of ‘intronic’ poly(A) sites (orange) and of poly(A) sites from annotated terminal exons (blue) from the study introduced in (A). (C) Distribution of the number of distinct samples in which individual PAS were observed, for PAS from terminal exons with no stop codon annotated downstream (‘terminal exon’, 26894 PAS), from annotated terminal exons located upstream of an annotated stop codon in the corresponding gene (‘terminal exon (ds stop)’, 3430 PAS), and from genomic regions currently annotated as intronic (‘intron’, 3937 PAS). Black boxes indicate the interquartile range (IQR) with the blue line corresponding to the median, whiskers corresponding to 1.5 times the IQR from the hinge, and densities extending to the most extreme values.
TECtool identifies terminal exons from RNA-sequencing data
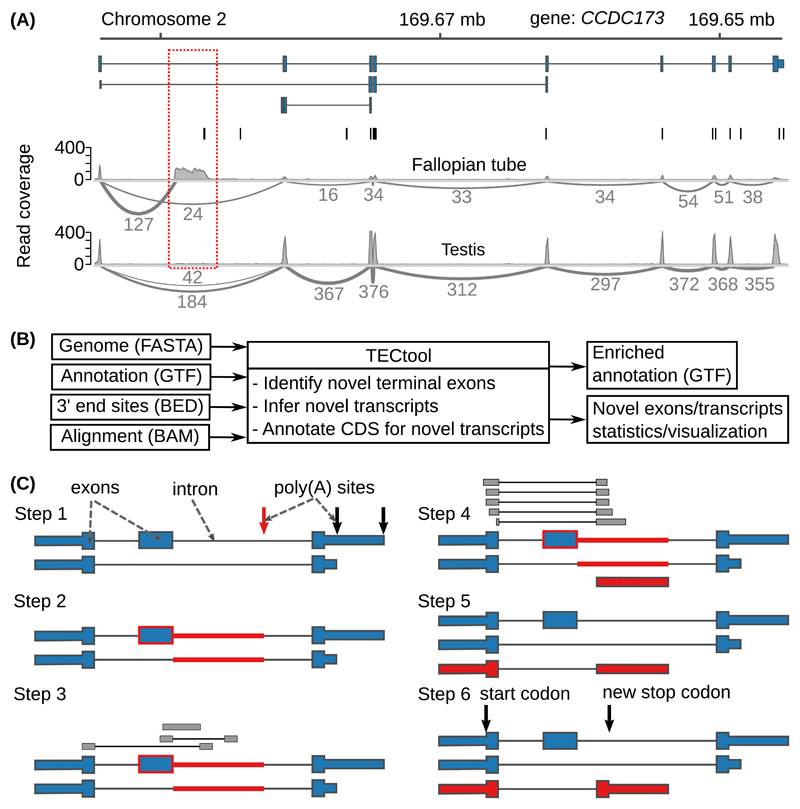
3’ end sequencing data remain relatively scarce. However, public databases contain many RNA sequencing (RNA-seq) data sets, from a wide range of cell types, that provide evidence for yet unannotated transcript isoforms (Figure 2A and Supplementary Figure 2). The terminal exon characterization (TEC) tool that we present here identifies terminal exons and transcript isoforms ending at ‘intronic’ PAS (Figure 2B-C). Based on alignments of RNA-seq reads resulting from single or paired-end sequencing (Supplementary Figure 3), TECtool trains a model (Supplementary Figure 4) to distinguish terminal exons from internal exons and background regions, using a variety of features that reflect differences in the coverage of these regions by RNA-seq reads (Supplementary Figure 5). It then uses the model to predict novel terminal exons, corresponding transcripts and their putative coding regions. TECtool can also be applied to data from unstranded protocols (e.g. Illumina TruSeq RNA v2). In this case, it does not predict terminal exons that overlap with annotated exons encoded on the opposite strand. To analyze data from single cells, where most transcripts are only sparsely covered by reads, we have designed a TECtool workflow that initially pools the reads to infer novel transcripts, and then quantifies the abundance of these transcripts in individual cells (Supplementary Figure 6).
Figure 2. Example and model to identify novel 3’ UTR isoforms.
(A) ‘Sashimi plots’ 18 of RNA-seq reads mapped to a region within the Coiled-coil Domain Containing 173 (CCDC173) gene locus, with the annotated ENSEMBL transcripts (blue), the PAS annotated in the PolyAsite atlas (vertical black lines, http://polyasite.unibas.ch) and densities of RNA-seq reads (gray) from fallopian tube and testis samples. The novel terminal exon is marked by the red dashed box, gray arcs indicate putative splice junctions, and numbers on the arcs indicate supporting reads (for clarity, only splice junctions supported by at least 10% of the maximum number of split reads between two exons in the genomic locus are shown, see also Supplementary Figure 2A). (B) Flow of the data through TECtool (input and output file formats are indicated in parentheses). (C) Outline of the main computational steps: Step 1 - Selection of PAS located within regions that with respect to the input annotation (see ‘Annotation (GTF)’ in (B)), are ‘intronic’ (red arrow), and not exonic, intergenic or antisense (black arrows). Step 2 - Identification of the ‘feature’ region of the putative novel terminal exon (red line), extending from the ‘intronic’ poly(A) site up to the closest annotated exon upstream (blue box with red border). Step 3 - Identification of reads that map uniquely to the feature region. Step 4 - Definition of terminal exon boundaries (red box), given by a splice site at the 5’ end - inferred from split reads -, and the ‘intronic’ poly(A) site at the 3’ end. Classification of putative terminal exons is done with a Bayes classifier. Step 5 - The newly identified terminal exons are linked to upstream exons to which they were found to be spliced based on split reads, to generate novel isoforms. Step 6 - Prediction of protein coding regions in newly identified transcripts.
TECtool reproducibly and accurately identifies novel transcripts
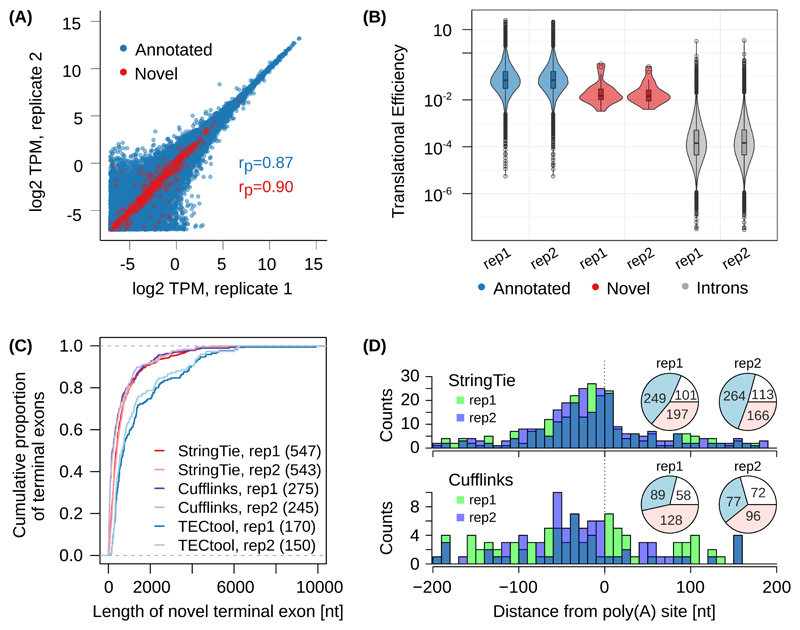
To evaluate TECtool, we took advantage of extensive datasets generated from human embryonic kidney (HEK) 293 cells. The ‘support level’ annotation of individual transcripts in ENSEMBL 19 provides a natural way to validate the tool, as we can determine whether isoforms that are predicted as novel relative to the annotation with the strongest experimental support (transcript support level (TSL) 1) are present in the annotation with more limited experimental evidence. From two biological replicates of RNA-seq in HEK 293 cells 20 TECtool identified 327 and 337 terminal exons (510 in total and 154 in common) that were novel with respect to TSL1 annotation. 321 of the 510 overlapped with terminal exons from the TSL1-5 annotation, and both annotated and novel transcripts had very reproducible expression in the two replicates (Supplementary Figure 7A). Repeating the inference starting from the known TSL1-5 transcripts, we obtained 170 and 150 novel terminal exons in the two replicates (250 total, 70 common), similar in properties to transcripts identified starting from TSL1 annotation (Figure 3A). These results show that TECtool identifies many novel terminal exons even from a highly studied cell line such as HEK 293.
Figure 3. Evaluation of TECtool’s performance.
(A) Scatter plot of estimated expression levels of already annotated transcripts (ENSEMBL v87, transcript support level 1-5 (TSL1-5), blue, 168'726 transcripts) and of transcripts ending at TECtool-identified terminal exons (red, 842 novel transcripts), in biological replicates of RNA-seq from HEK 293 cells (rP indicate the corresponding Pearson correlations). (B) Translational efficiencies computed for annotated terminal exons, novel terminal exons and intronic regions (two-tailed t-test p-values for pairwise comparisons of regions based on TSL1-5, novel versus intron replicate 1 (rep1): 2.1e-16; replicate 2 (rep2): 5.4e-18, and annotated versus novel rep1: 1.4e-5; rep2: 8.6e-7). The numbers of annotated, novel and introns were in rep1: 16068, 24, and 64455, and in rep2: 15772, 25, and 63932. Boxes indicate the interquartile range (IQR) with the line corresponding to the median, whiskers correspond to the most extreme value that is within 1.5 times the IQR from the hinge and outliers beyond this range are shown as individual points. (C) Cumulative distribution of the length of novel terminal exons identified by TECtool, StringTie and Cufflinks in the two replicate RNA-seq data sets, relative to the TSL1-5 annotation. The number of novel terminal exons identified by each tool is indicated in parentheses. (D) Distance between experimentally determined PAS from the PolyAsite atlas 9 and the 3’ ends of novel transcripts identified by StringTie (top panel) and Cufflinks (bottom panel). Pie-charts show the number of 3’ ends of novel transcripts that have an experimentally determined PAS within +/-200 nts (blue), or have experimentally determined PAS farther away but in the same intron (red) or do not have any experimentally observed PAS in the respective intron (white).
Ribosome profiling data from HEK 293 cells 21 revealed that novel terminal exons had much higher translational efficiency compared to intronic sequences, but lower than already annotated terminal exons (Figure 3B and Supplementary Figure 7B). The ribosome footprint density peaked around stop codons, whether already annotated or predicted in the novel terminal exon isoforms (Supplementary Figure 7C). These results indicate that TECtool-predicted isoforms are sufficiently stable to undergo translation.
The median length of TECtool-predicted terminal exons in the two HEK 293 RNA-seq samples was 732 and 632 nts, respectively, larger than the median length of terminal exons predicted by StringTie 22 (380 and 412 nts, respectively) and Cufflinks 23 (199 and 232 nts, respectively), the two currently most accurate transcript reconstruction methods 24 (Figure 3C). TECtool did not predict any exon shorter than 50 nts, in contrast to StringTie (3.5% and 3.9% of terminal exons in the two replicates, respectively) and especially Cufflinks (21.5% and 22.4%, respectively). This is a reflection of transcript reconstruction tools being largely unable to correctly determine transcript 3’ ends, where the coverage by RNA-seq reads is reduced. Consistent with an accurate assignment of polyadenylation sites, only TECtool-predicted terminal exons had the canonical poly(A) signal (‘AAUAAA’) at the expected position, ~21 nts upstream of PAS (Supplementary Figure 7D). In fact, only a minority of ‘intronic’ terminal exons predicted by Cufflinks (32.4% and 31.4%) and StringTie (45.5% and 48.6%) had experimentally-identified intronic PAS in the region +/-200 nts around their 3’ end (Figure 3D). Even when we defined unique terminal exons solely by their splicing-determined 5’ end, TECtool made more reproducible predictions from replicate samples (40% of the union of predicted exons were identified from both replicates, compared to 30% for StringTie and 18% for Cufflinks) (Supplementary Figure 7E,F), while its predictions were largely not covered by the other tools (58 or 63% of its predicted novel exons). Thus, TECtool identifies with high reproducibility many exons not found by transcript reconstruction methods, having the unique advantage of accurately annotating transcript 3’ ends. TECtool further predicts the coding region of novel transcripts, facilitating downstream analyses of encoded proteins.
Lagarde et al. 25 recently sequenced samples from four tissues, in parallel on both short and long read sequencing platforms, allowing us to further validate TECtool-generated transcript models. Even though full-length RNA Capture Long Sequencing (CLS) primarily captures highly expressed transcripts (Supplementary Figure 8), ~8% of the novel transcripts predicted by TECtool from short read sequencing were also identified by long read sequencing (44, 5, 0, and 1 of the 464, 88, 63 and 20 novel transcripts predicted from testis, brain, heart and liver samples, respectively). Thus, CLS validates highly expressed TECtool-predicted transcripts and altogether, our analysis shows that TECtool can substantially improve transcriptome annotation.
TECtool identifies cell type-specific isoforms
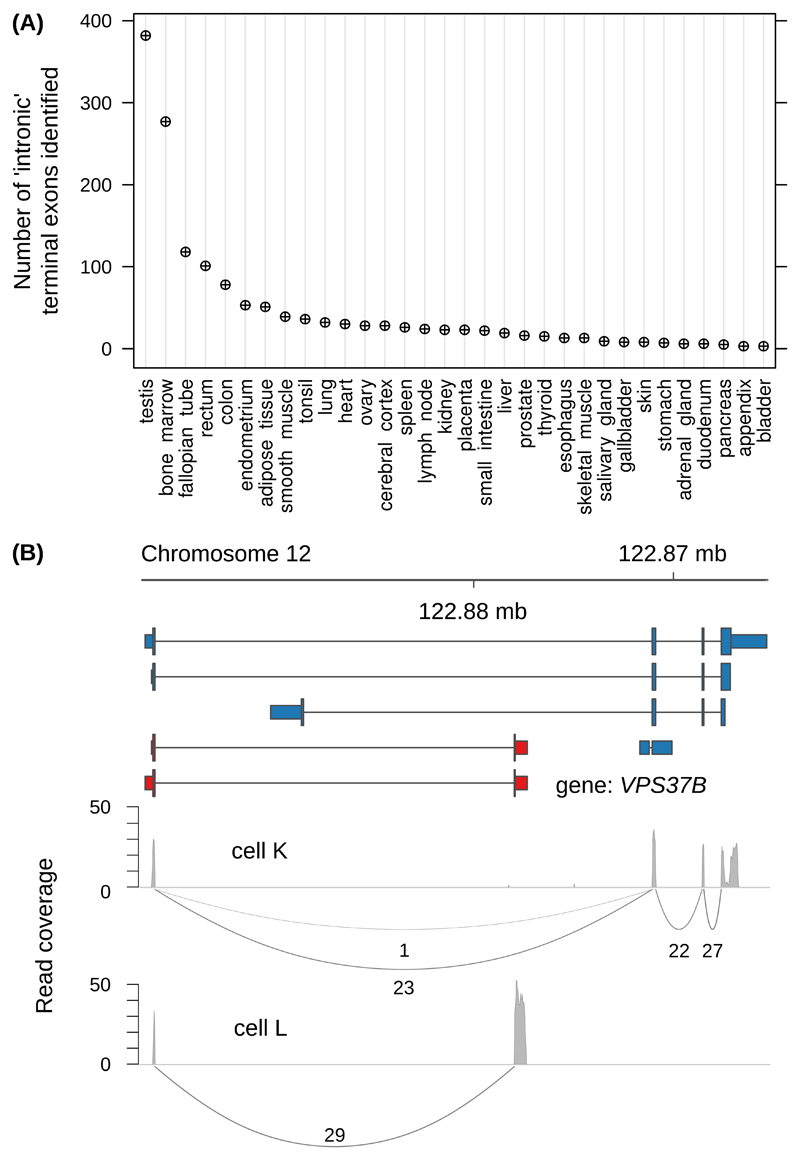
Turning to an RNA-seq data set that covered 32 human tissues 26, TECtool identified hundreds of novel terminal exons, primarily from testis and bone marrow samples (Figure 4A). This was not a mere reflection of the library sizes (Supplementary Figure 9). Furthermore, many novel isoforms were the most expressed transcripts of their corresponding genes (Supplementary Figure 10), indicating a special relevance of intronic polyadenylation in these tissues.
Figure 4. TECtool identifies novel isoforms with cell type-specific expression.
(A) Number of novel terminal exons identified by TECtool in at least one sample from the indicated tissues. (B) VPS37B gene locus with the ENSEMBL-annotated transcripts (blue), novel transcripts predicted by TECtool (red), and Sashimi 18 plots of RNA-seq read densities (gray) from two single T cells (labeled as cell K and cell L).
Novel isoforms are expressed in subsets of single cells
Single cell RNA sequencing allows one to assess whether a low average expression of a particular transcript results from ‘transcriptional noise’, affecting all cells, or from highly specific expression in rare cell types. Applying TECtool to a recently published single cell RNA-seq data set of 201 T cells 27, we found that the distribution of novel isoform expression levels in individual cells is within the range of already annotated isoforms. Once transcripts reach an average expression of 1-2 reads per million per cell (considering only reads that splice into the 5’ splice site of the terminal exon), they start to be detected in multiple cells (Supplementary Figure 11A). However, multiple isoforms with distinct terminal exons are rarely present in a cell at the same time (Supplementary Figure 11B). Thus, rather than being co-expressed with the more abundant annotated isoforms, novel isoforms appear to be expressed in subsets of cells, at a per-cell level similar to that of annotated transcripts. Examples of isoform switching between individual cells are shown in Figure 4B and Supplementary Figure 11C-D. These results illustrate the potential of TECtool to improve the characterization of transcript isoforms expressed in individual cells, thereby enabling the characterization of rare cell types.
Discussion
Following the initial assembly of the human genome 28,29, full-length RNAs and expressed sequence tags were used to annotate gene structures 16,30. However, many transcripts that are specific to cell types or conditions remain uncharacterized, even though targeted sequencing of RNA 3’ ends hints to their existence 9,13. While analysis of RNA-seq data increasingly involves transcript reconstruction, the accuracy of the approach is limited by alignment errors, intron retention events and 3’ end bias when poly(A) selection is performed 24,31. Terminal exons are especially problematic, because the transcript coverage by RNA-seq reads decreases towards the 3’ end. Here we have demonstrated that the accuracy of isoform annotation can be substantially improved by incorporating experimentally identified polyadenylation sites in transcript reconstruction. The approach can be applied to any RNA-seq data set from a species for which polyadenylation sites have been mapped, including human and mouse (Supplementary Figure 12). Like transcript reconstruction methods, TECtool relies on high-quality RNA-seq data, from samples with minimal RNA degradation and little bias in coverage along transcripts. We have obtained good results with samples for which transcript integrity scores 32 were greater than 0.8. To enable the analysis of samples that may have insufficient coverage and training examples, TECtool also provides the option to analyze new data sets with a model build from samples with deep coverage and high RNA integrity, such as the HEK 293 RNA-seq data sets that we have used in this study.
Although 3rd generation sequencing technologies have made the full-length sequencing of RNAs more common, the capture of low-abundance transcripts remains very limited. Making use of extensive short read sequencing data available from cell populations and especially from single cells, TECtool supports identification of even relatively rare transcripts. The tool is fully automated and easy to use. It does not require any customized input files or specific parameters, as it trains its own classifier based on the input data.
Online Methods
Datasets
Table 1. Data sets used in this study.
| Dataset | Dataset reference | Downloaded from | Dataset use |
|---|---|---|---|
| 3’end sequencing | 9 | (http://polyasite.unibas.ch) | Figures 1,2;Suppl. Figs. 1,2,12 |
| RNA-seq in HEK 293 | 20 | GEO database 33: GSE56010 | Figure 3 Suppl. Fig. 7 |
| Ribosome profiling in HEK 293 | 21 | GEO database 33: GSE73136 | Figure 3, Suppl. Fig. 7 |
| RNA-seq in tissues from Protein Atlas | 26 | ArrayExpress database 34: E-MTAB-2836 | Figures 2A and 4A, Suppl. Figs. 2A,B, 9 and 10 |
| RNA-seq and PacBio reads in 4 different tissues | 25 | GEO data base 33: GSE93848 | Suppl. Fig. 2C, Suppl. Fig. 8 |
| Single-cell data | 27 | GEO data base 33: GSE85527 | Figure 4B, Suppl. Fig. 11 |
| Mouse data | 35 | GEO data base 33: GSE52260 | Suppl. Fig. 4B, 12 |
Poly(A) sites
The genome coordinates of the poly(A) sites from the recently published atlas 9 were converted to the GRCh38 genome assembly version with liftOver 36.
Analysis of ‘intronic’ poly(A) sites identified by 3’ end processing
The locations of all poly(A) sites were associated with the set of transcripts of support levels 1-5 from the ENSEMBL gene annotation version 87 30. Pre-mRNA 3’ end processing sites inferred from the samples that were part of two 3’ end sequencing studies, utilizing either the 3’-Seq 37 or the SAPAS 38 protocol, were intersected with the annotated PAS. PAS with expression of at least five reads per million in individual samples were identified. PAS from introns, terminal exons or terminal exons located upstream of an annotated stop codon in the same gene were identified based on the ENSEMBL annotation.
TECtool
TECtool is implemented as open source Python (version 3.4 and higher) software that can be obtained from http://tectool.unibas.ch. It depends on the packages HTSeq version 0.9.1 39, Bedtools version 2.26.0 40, Pybedtools version 0.7.10 41, pyfasta version 0.5.2, numpy version 1.13 42, scipy version 0.19 43 and scikit-learn version 0.19.0 44, pandas version 0.2 45 and progress version 1.3.
Inputs, outputs and user options
TECtool requires the following inputs (Figure 2B): (1) a file containing all chromosomes in fasta format, (2) a file with the corresponding annotation in ENSEMBL GTF format 30, (3) a file with genome coordinates of 3’ end processing sites (in BED format) and (4) a file containing spliced alignments of RNA-seq reads to the corresponding genome (in BAM format, sorted by coordinates and indexed). For human and mouse, downloadable files of poly(A) sites can be found on the website of the ‘PolyAsite’ atlas (http://polyasite.unibas.ch, 9). The output of TECtool (Figure 2B) is an augmented annotation file (in GTF format), containing the input as well as the newly annotated transcripts. Additional files, summarizing the features of annotated and newly identified exons, that are generated during the run, are also provided (in tab-delimited format). The tool requires that the sequencing direction is specified (as forward/unstranded) for the reads in the BAM file using the --sequencing_direction flag. Other implemented options allow the specification of the number of spliced reads required to support a novel exon, or whether to enforce the use of specific features in training the model and predicting new terminal exons. The tool can also be run with a user-specified, pre-trained model (TECtool options: --use_precalculated_training_set, --training_set_directory), that the user would need to obtain in a preliminary run with a dataset with good transcript coverage by reads. This may be useful when the coverage of annotated exons in the input RNA-seq data is low and therefore too little data is available in order to train an appropriate model.
Selection of ‘intronic’ PAS
In a first step, TECtool uses the provided transcript and PAS annotations of the genome to select candidate ‘intronic’ PAS. These are located within the loci of annotated genes, but outside of annotated exons. When the RNA-seq was not done preserving strand information, TECtool discards PAS that are located in introns of genes that have other exons annotated on the complementary strand.
Identification of candidate novel terminal exon
For each ‘intronic’ PAS, TECtool defines a ‘feature’ region, that extends from the PAS to the closest upstream exon (Figure 2C). The upstream exon is considered the ‘reference’ region. When the upstream exon has multiple possible 5’ ends, the longest exon variant becomes the ‘reference’ region.
Uniquely mapping reads overlapping with the ‘feature’ region, either unspliced or mapping across splice junctions, with the 5’ end in an exon upstream of the candidate ‘intronic’ PAS and the 3’ splice site within the ‘feature’ region, are identified. When the number of such spliced reads surpasses a user-defined lower bound (default: 5 reads), a putative terminal exon is constructed, extending from the 5’ splice site of the spliced reads to the ‘intronic’ PAS. Potential terminal exons that overlap with annotated exons of other genes are not considered.
Collection of training exonic regions
TECtool aims to classify
Terminal exons: unique last exons of annotated transcripts, as defined in the provided annotation file, not including exons that overlap with other exons or that do not have the (user-)defined minimum number of splice-in reads (default: 5 reads).
Internal exons: exons that are neither first nor last exon of an annotated transcript, do not overlap with any other exon and have the (user-)defined minimum number of splice-in reads.
‘Background’ regions: annotated terminal exons that do not overlap with other exons, but have less than the (user-)defined minimum number of splice-in reads.
Feature computation
For each exonic region in the training set (‘object’), TECtool computes the following features (Supplementary Figure 3B-H):
Splicing-in-boundary/all: Counts of reads that splice from an upstream region into the 5’ boundary/anywhere within the entire length of the object.
Splicing-out-boundary/all: Counts of reads that splice from the 3’ boundary/anywhere within the entire length of the object to a downstream region.
Crossing-in/out-boundary: Counts of unspliced reads overlapping the 5’/3’ boundary of the object.
Unspliced-within-boundaries: Counts of unspliced reads that are contained in the object.
Reads-within-gene-loci: Number of reads that map within gene loci.
Union-exon-length: Length of the union exons of the gene.
TECtool then calculates (Supplementary Figure 5):
Reads-out versus reads-in ratio: the ratio of reads splicing out or crossing the 3’ boundary of the object and reads splicing in or crossing the 5’ boundary of the object.
Normalized region expression: ratio between the expression of the object (per kilobase, including splicing-in/out-all, crossing-in/out-boundary, and unspliced-within-boundary reads) and the expression of the corresponding gene (per kilobase, reads-within-gene-loci/length of union-exons).
Object length.
- Entropy efficiency: A measure of the ‘uniformity’ of read coverage along the object, defined as the Shannon entropy of read coverage per position divided by the maximum value it can take based on the object length
where n represents the length of the object and p(xi) the coverage at position i divided by the total coverage along the object EE(x) takes values between 0 and 1. Relative positions of 5% and 95% quantile coverage: where the cumulative distribution of read coverage along the object reaches 5% and 95%.
Splicing-in-all versus 5’ end expression: ratio between the number of reads splicing into the object (see ‘Splicing-in-all’ above) and the mean coverage per position over the first 10 nucleotides of the object.
Splicing-out-all versus 3’ end expression: ratio between the number of reads splicing out from the object (see ‘Splicing-out-all’ above) and the mean coverage per position over the last 10 nucleotides of the object.
Crossing-in versus 5’ end expression: ratio between the number of reads overlapping the 5’ boundary of the object (see ‘Crossing-in-boundary’ above) and the mean coverage per position over the first 10 nucleotides of the object.
Crossing-out versus 3’ end expression: ratio between the number of reads overlapping the 3’ boundary of the object (see ‘Crossing-out-boundary’ above) and the mean coverage per position over the last 10 nucleotides of the object.
Splicing-in-boundary versus 5’ end expression: ratio between the number of reads splicing into the object (see ‘Splicing-in-boundary’ above) and the mean coverage per position over the first 10 nucleotides of the region.
Splicing-out-boundary versus 3’ end expression: ratio between the number of reads splicing out from the object (see ‘Splicing-out-boundary’ above) and the mean coverage per position over the last 10 nucleotides of the region.
Splicing-in-boundary versus Splicing-in-all: ratio between the number of reads splicing into the 5’ boundary of the object (see ‘Splicing-in-boundary’ above) and the number of reads splicing into the object (see ‘Splicing-in-all’ above).
Classifier training and prediction of novel terminal exons
TECtool samples randomly 20% of the training data for validation, and approximates the distributions of all features described above for each region type in the remaining 80% of the training data (Supplementary Figure 4A) using Kernel Density Estimation (KDE). We chose an exponential kernel function, to better approximate drops at the boundary of the empirical distributions. Under the assumption of uncorrelated features, the KDEs represent posterior probabilities to use in the Bayes Classifier. As samples generated with different sequencing protocols typically have different coverage patterns along genes, the features that best distinguish exon types may change from sample to sample. Thus, TECtool uses a forward greedy feature selection, incrementally and greedily adding features that increase the performance (F1-score t-value > 1.37) on the validation data, starting from a core set of features (‘Entropy efficiency’ and ‘Reads-out versus reads-in ratio’). To increase the stability of the model, TECtool trains classifiers on 10 randomized subsets of the training data (1000 objects in each class, or the entire set if smaller than 1000), and then uses each of them to evaluate each candidate terminal exon (Supplementary Figure 4A). The average probabilities of the candidate exon to be terminal, internal, or background, computed over the 10 classifiers, determine the category to which the candidate exon is assigned. When there are multiple putative terminal exons with the same 5’ splice site but different PAS, only the exon with the highest probability of being terminal is reported in the final gtf.
Novel transcripts and CDS annotation
Having identified putative terminal exons, TECtool constructs putative novel transcripts, starting from annotated transcripts that contain an exon that splices to the novel terminal exon. These transcripts (which we call ‘root transcripts’) and upstream exons are identified based on spliced reads.
In the final step, TECtool annotates the putative protein-coding region in the newly annotated transcripts. When the root transcripts are protein-coding, TECtool uses the already annotated start codon and searches for the first in-frame stop codon. If found, the novel transcript is annotated as protein-coding. When the root transcript had no annotated start codon or when no in-frame stop codon is found, the transcript is classified as non-coding.
Automated analysis of RNA-seq data sets with TECtool
We implemented automated TECtool analyses of standard RNA-seq (Supplementary Figure 3A), as well as single cell RNA sequencing data (Supplementary Figure 6). The analysis flows are implemented in the snakemake framework 46, and the parameters for each type of analysis are specified in a corresponding configuration file. The single cell sequencing data poses the challenge of relative low and highly non-uniform coverage for most genes. Therefore, we initially pool the data from all cells in a sample to identify the terminal exons, which we then quantify in individual cells with a method for transcript isoform quantification.
Analysis of mouse RNA sequencing data
To demonstrate the generality of the tool, we have also applied it to RNA sequencing data from a time series of mouse T cell activation (accession GSE52260). A summary of the results for individual time points is shown in Supplementary Figure 12 together with genome browser screenshots for two individual examples.
Analysis of novel transcript expression in 32 human tissues
We analyzed the mRNA-seq data generated for 32 human tissues 26 with the TECtool version for processing paired-end reads. We merged the enriched annotation files corresponding to replicate samples from the same tissue, to construct tissue-specific annotation files. Estimates of transcript and gene expression levels in each tissue were obtained with Salmon 47.
Visualization of read densities
Sashimi plots 18 were generated with a custom script that is based on the following R libraries: Gviz 48, rtracklayer 49 and GenomicFeatures 50.
Statistics
For the comparison of translation efficiency, two-tailed t-tests were used not treating the two variances as being equal. The number of cases and p-values are given in the legend of Figure 3.
Reporting Summary
Further information on experimental design is available in the Nature Research Reporting Summary linked to this article.
Supplementary Material
Acknowledgements
We thank Martin Jacquot and the members of sciCore for help with the infrastructure that we used to process the data. This work was supported by the Swiss National Science Foundation (Grants #31003A_170216 and #51NF40_141735 (National Center for Competence in Research ‘RNA & Disease’)) and a Marie Curie Initial Training Network (project #607720, RNATRAIN).
Footnotes
Data availability
Accession numbers of all data analyzed in this study are listed in this published article (and its supplementary information files).
Code availability
TECtool is available at http://tectool.unibas.ch.
Author contributions
AJG conceived the study. AJG, FG, MZ designed the study. AJG, FG, AR developed the method. AJG, FG, AR, RS analyzed the data with input from MZ. All authors wrote and approved the manuscript.
Competing interests
The authors declare no competing interests.
References
- 1.Kishore S, Luber S, Zavolan M. Deciphering the role of RNA-binding proteins in the post-transcriptional control of gene expression. Brief Funct Genomics. 2010;9:391–404. doi: 10.1093/bfgp/elq028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hausser J, Zavolan M. Identification and consequences of miRNA--target interactions—beyond repression of gene expression. Nat Rev Genet. 2014;15:599. doi: 10.1038/nrg3765. [DOI] [PubMed] [Google Scholar]
- 3.Sandberg R, Neilson JR, Sarma A, Sharp PA, Burge CB. Proliferating Cells Express mRNAs with Shortened 3’ Untranslated Regions and Fewer MicroRNA Target Sites. Science. 2008;320:1643–1647. doi: 10.1126/science.1155390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lackford B, et al. Fip1 regulates mRNA alternative polyadenylation to promote stem cell self-renewal. EMBO J. 2014;33:878–889. doi: 10.1002/embj.201386537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gruber AJ, et al. Discovery of physiological and cancer-related regulators of 3’ UTR processing with KAPAC. Genome Biol. 2018;19:44. doi: 10.1186/s13059-018-1415-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mayr C, Bartel DP. Widespread Shortening of 3′UTRs by Alternative Cleavage and Polyadenylation Activates Oncogenes in Cancer Cells. Cell. 2009;138:673–684. doi: 10.1016/j.cell.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Spies N, Burge CB, Bartel DP. 3’ UTR-isoform choice has limited influence on the stability and translational efficiency of most mRNAs in mouse fibroblasts. Genome Res. 2013;23:2078–2090. doi: 10.1101/gr.156919.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gruber AR, et al. Global 3’ UTR shortening has a limited effect on protein abundance in proliferating T cells. Nat Commun. 2014;5 doi: 10.1038/ncomms6465. 5465. [DOI] [PubMed] [Google Scholar]
- 9.Gruber AJ, et al. A comprehensive analysis of 3’ end sequencing data sets reveals novel polyadenylation signals and the repressive role of heterogeneous ribonucleoprotein C on cleavage and polyadenylation. Genome Res. 2016;26:1145–1159. doi: 10.1101/gr.202432.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Plass M, Rasmussen SH, Krogh A. Highly accessible AU-rich regions in 3’ untranslated regions are hotspots for binding of regulatory factors. PLoS Comput Biol. 2017;13:e1005460. doi: 10.1371/journal.pcbi.1005460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martin G, Gruber AR, Keller W, Zavolan M. Genome-wide analysis of pre-mRNA 3’ end processing reveals a decisive role of human cleavage factor I in the regulation of 3' UTR length. Cell Rep. 2012;1:753–763. doi: 10.1016/j.celrep.2012.05.003. [DOI] [PubMed] [Google Scholar]
- 12.Lee JY, Yeh I, Park JY, Tian B. PolyA_DB 2: mRNA polyadenylation sites in vertebrate genes. Nucleic Acids Res. 2007;35:D165–8. doi: 10.1093/nar/gkl870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Derti A, et al. A quantitative atlas of polyadenylation in five mammals. Genome Res. 2012;22:1173–1183. doi: 10.1101/gr.132563.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin Y, et al. An in-depth map of polyadenylation sites in cancer. Nucleic Acids Res. 2012;40:8460–8471. doi: 10.1093/nar/gks637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tian B, Hu J, Zhang H, Lutz CS. A large-scale analysis of mRNA polyadenylation of human and mouse genes. Nucleic Acids Res. 2005;33:201–212. doi: 10.1093/nar/gki158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harrow J, et al. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res. 2012;22:1760–1774. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lianoglou S, Garg V, Yang JL, Leslie CS, Mayr C. Ubiquitously transcribed genes use alternative polyadenylation to achieve tissue-specific expression. Genes Dev. 2013;27:2380–2396. doi: 10.1101/gad.229328.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Katz Y, et al. Sashimi plots: Quantitative visualization of alternative isoform expression from RNA-seq data. bioRxiv. 2014 doi: 10.1101/002576. [DOI] [Google Scholar]
- 19.Kersey PJ, et al. Ensembl Genomes 2016: more genomes, more complexity. Nucleic Acids Res. 2015;44:D574–D580. doi: 10.1093/nar/gkv1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu N, et al. N(6)-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature. 2015;518:560–564. doi: 10.1038/nature14234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Calviello L, et al. Detecting actively translated open reading frames in ribosome profiling data. Nat Methods. 2016;13:165–170. doi: 10.1038/nmeth.3688. [DOI] [PubMed] [Google Scholar]
- 22.Pertea M, et al. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol. 2015;33:290–295. doi: 10.1038/nbt.3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Trapnell C, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hayer KE, Angel P, Lahens NF, Hogenesch JB, Grant GR. Benchmark Analysis of Algorithms for Determining and Quantifying Full-length mRNA Splice Forms from RNA-Seq Data. Bioinformatics. 2015 doi: 10.1093/bioinformatics/btv488. btv488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lagarde J, Uszczynska-Ratajczak B, Carbonell S. High-throughput annotation of full-length long noncoding RNAs with Capture Long-Read Sequencing (CLS) bioRxiv. 2017 [Google Scholar]
- 26.Uhlén M, et al. Proteomics. Tissue-based map of the human proteome. Science. 2015;347 doi: 10.1126/science.1260419. 1260419. [DOI] [PubMed] [Google Scholar]
- 27.Long SA, et al. Partial exhaustion of CD8 T cells and clinical response to teplizumab in new-onset type 1 diabetes. Sci Immunol. 2016;1 doi: 10.1126/sciimmunol.aai7793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Venter JC, et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 29.Lander ES, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 30.Aken BL, et al. The Ensembl gene annotation system. Database. 2016;2016 doi: 10.1093/database/baw093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lahens NF, et al. IVT-seq reveals extreme bias in RNA sequencing. Genome Biol. 2014;15:R86. doi: 10.1186/gb-2014-15-6-r86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gallego Romero I, Pai AA, Tung J, Gilad Y. RNA-seq: impact of RNA degradation on transcript quantification. BMC Biol. 2014;12:42. doi: 10.1186/1741-7007-12-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Barrett T, et al. NCBI GEO: archive for functional genomics data sets--update. Nucleic Acids Res. 2013;41:D991–5. doi: 10.1093/nar/gks1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kolesnikov N, et al. ArrayExpress update--simplifying data submissions. Nucleic Acids Res. 2015;43:D1113–6. doi: 10.1093/nar/gku1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tuomela S, et al. Comparative analysis of human and mouse transcriptomes of Th17 cell priming. Oncotarget. 2016;7:13416–13428. doi: 10.18632/oncotarget.7963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hinrichs AS, et al. The UCSC Genome Browser Database: update 2006. Nucleic Acids Res. 2006;34:D590–8. doi: 10.1093/nar/gkj144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lianoglou S, Garg V, Yang JL, Leslie CS, Mayr C. Ubiquitously transcribed genes use alternative polyadenylation to achieve tissue-specific expression. Genes Dev. 2013;27:2380–2396. doi: 10.1101/gad.229328.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.You L, et al. APASdb: a database describing alternative poly(A) sites and selection of heterogeneous cleavage sites downstream of poly(A) signals. Nucleic Acids Res. 2015;43:D59–67. doi: 10.1093/nar/gku1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Anders S, Pyl PT, Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dale RK, Pedersen BS, Quinlan AR. Pybedtools: a flexible Python library for manipulating genomic datasets and annotations. Bioinformatics. 2011;27:3423–3424. doi: 10.1093/bioinformatics/btr539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Van Der Walt S, Colbert SC. The NumPy array: a structure for efficient numerical computation. Comput Sci Eng. 2011 [Google Scholar]
- 43.Jones E, Oliphant T, Peterson P, et al. SciPy: Open source scientific tools for Python. 2001. 2016 URL http://www.scipy.org.
- 44.Pedregosa F, et al. Scikit-learn: Machine Learning in Python. J Mach Learn Res. 2011;12:2825–2830. [Google Scholar]
- 45.McKinney W. pandas: a foundational Python library for data analysis and statistics. Python for High Performance and Scientific Computing. 2011:1–9. [Google Scholar]
- 46.Köster J, Rahmann S. Snakemake—a scalable bioinformatics workflow engine. Bioinformatics. 2012 doi: 10.1093/bioinformatics/bts480. [DOI] [PubMed] [Google Scholar]
- 47.Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C. Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods. 2017 doi: 10.1038/nmeth.4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hahne F, Ivanek R. Visualizing Genomic Data Using Gviz and Bioconductor. Methods Mol Biol. 2016;1418:335–351. doi: 10.1007/978-1-4939-3578-9_16. [DOI] [PubMed] [Google Scholar]
- 49.Lawrence M, Gentleman R, Carey V. rtracklayer: an R package for interfacing with genome browsers. Bioinformatics. 2009;25:1841–1842. doi: 10.1093/bioinformatics/btp328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lawrence M, et al. Software for computing and annotating genomic ranges. PLoS Comput Biol. 2013;9:e1003118. doi: 10.1371/journal.pcbi.1003118. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.