Abstract
Coenzyme Q (CoQ) is an essential player in the respiratory electron transport chain and is the only lipid-soluble antioxidant synthesized endogenously in mammalian and yeast cells. In humans, genetic mutations, pathologies, certain medical treatments, and aging, result in CoQ deficiencies, which are linked to mitochondrial, cardiovascular, and neurodegenerative diseases. The only strategy available for these patients is CoQ supplementation. CoQ supplements benefit a small subset of patients, but the poor solubility of CoQ greatly limits treatment efficacy. Consequently, the efficient delivery of CoQ to the mitochondria and restoration of respiratory function remains a major challenge. A better understanding of CoQ uptake and mitochondrial delivery is crucial to make this molecule a more efficient and effective therapeutic tool. In this study, we investigated the mechanism of CoQ uptake and distribution using the yeast Saccharomyces cerevisiae as a model organism. The addition of exogenous CoQ was tested for the ability to restore growth on non-fermentable medium in several strains that lack CoQ synthesis (coq mutants). Surprisingly, we discovered that the presence of CoQ biosynthetic intermediates impairs assimilation of CoQ into a functional respiratory chain in yeast cells. Moreover, a screen of 40 gene deletions considered to be candidates to prevent exogenous CoQ from rescuing growth of the CoQ-less coq2Δ mutant, identified six novel genes (CDC10, RTS1, RVS161, RVS167, VPS1, and NAT3) as necessary for efficient trafficking of CoQ to mitochondria. The proteins encoded by these genes represent essential steps in the pathways responsible for transport of exogenously supplied CoQ to its functional sites in the cell, and definitively associate CoQ distribution with endocytosis and intracellular vesicular trafficking pathways conserved from yeast to human cells.
Keywords: Coenzyme Q, ubiquinone, transport, uptake, endocytosis, CoQ6 rescue, Saccharomyces cerevisiae
Introduction
Coenzyme Q (CoQ) is essential for energy production in mitochondria. It functions as an electron carrier in the respiratory chain, moving electrons from Complex I (NADH oxidase in Saccharomyces cerevisiae), II, and several other enzymes, to Complex III [1, 2]. In this manner, CoQ participates in multiple cellular pathways including β-oxidation and de novo synthesis of pyrimidines. In addition, CoQH2 (the reduced or hydroquinone form) has an important role as an antioxidant, protecting DNA, lipids, and proteins from oxidative stress [3, 4]. CoQ is composed of a benzoquinone ring connected to a polyisoprenoid side chain, whose length is species-specific [1, 2]. Six isoprene subunits are present in Saccharomyces cerevisiae (CoQ6), eight in Escherichia coli (CoQ8), and nine and ten (CoQ9 and CoQ10) in rodents and humans, although CoQ9 predominates in rodents and CoQ10 is most prevalent in humans [1, 2]. Almost all cells have the capacity to produce CoQ, and at least 13 genes are necessary for its endogenous production in yeast (see Fig. 1) [1, 2, 5].
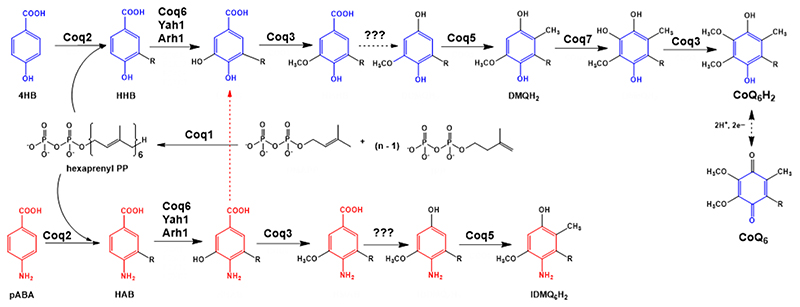
Figure 1. Coenzyme Q biosynthetic pathway in S. cerevisiae.
Adapted from [1]. At least 13 proteins are necessary for efficient CoQ biosynthesis in S. cerevisiae (Coq1-11, Yah1 and Arh1) [1]. The polyprenyl diphosphate is produced by a hexaprenyl diphosphate synthase (Coq1) [6]. Coq2 mediates the condensation of the isoprenoid tail with the aromatic ring precursor, generating a membrane-bound CoQ intermediate. In yeast, both 4HB and pABA can be used as ring precursors for CoQ6 biosynthesis [7, 8]. Intermediates originating from 4HB are depicted in blue, while intermediates originating from pABA are depicted in red. Several Coq proteins (Coq6, Coq3, Coq5 and Coq7) modify the hexaprenylated CoQ precursor to form the final product. Other Coq proteins (Coq4, Coq8, Coq9, CoQ10 and Coq11) are essential for efficient CoQ production. For simplicity, only the relevant intermediates to understand further analysis have been named. 4HB = 4-hydroxybenzoic acid; pABA = para-aminobenzoic acid; HHB = 3-hexaprenyl-4-hydroxybenzoic acid; HAB = 3-hexaprenyl-4-aminobenzoic acid; DMQH2 = demethoxy-QH2; IDMQH2 = 4-imino-demethoxy-QH2; CoQ6H2 = reduced coenzyme Q6H2; CoQ6 = oxidized coenzyme Q6.
In humans, insufficient CoQ10 can result in profound deficits in mitochondrial function [9, 10]. Mutations in genes that participate directly in CoQ10 biosynthesis lead to primary CoQ10 deficiencies, rare conditions that severely affect multiple organ systems, such as the central and peripheral nervous systems, kidney, skeletal muscle, heart, or sensory systems, in highly variable manners [11]. CoQ10 content may also be decreased in other conditions attributed to secondary deficiencies, in which depletion of CoQ10 is caused by mutations in genes unrelated to CoQ10 biosynthesis including genes involved in mitochondrial myopathies or in the mitochondrial DNA depletion syndrome [12, 13]. Secondary deficiencies can also result from non-genetic conditions such as fibromyalgia, clinical treatments (e.g. hypercholesterolemia treatment with statins) [14, 15], environmental toxins, metabolic disorders, aging [16], and age-related diseases [2, 4]. Secondary deficiencies are more common than primary deficiencies and reflect the diversity of biological functions and metabolic pathways that involve CoQ10 [11, 12].
To date, the only treatment to ameliorate the symptoms of human CoQ10 deficiencies is CoQ10 supplementation. At high doses, CoQ10 supplements increase CoQ10 levels in all tissues, especially with certain formulations, and are beneficial for various pathophysiological conditions [11, 17]. For example, the Q-SYMBIO study demonstrated that CoQ10 supplementation improves heart failure symptoms with a significant reduction in major adverse cardiovascular events and mortality [4, 18]. Other studies reported that CoQ10 supplementation could stop the progression of encephalopathy [19] and ameliorate the symptoms of renal disease in patients with defects in CoQ2, CoQ6, or ADCK4 (the human homolog of yeast Coq8) [20, 21]. However, bioavailability studies show that the response of individuals to CoQ10 supplementation is inconsistent, and treatment failure is common [1, 17]. The absorption of CoQ10 is slow and limited due to its high molecular weight and negligible aqueous solubility, making delivery by oral supplementation inefficient [11, 17, 22]. Thus, developing therapies for CoQ10 deficiencies remains a challenge. Current efforts are focused on new delivery strategies for CoQ [23–25], as well as on alternative approaches using compounds that enhance endogenous CoQ10 synthesis in CoQ10-deficient patients [1, 5, 11, 26–29].
In order to improve the therapeutic efficacy of CoQ10 supplementation, it is crucial to define the mechanisms responsible for the uptake and distribution of CoQ within cells. However, complete understanding of these mechanisms remains elusive. In mammalian cells and yeast, CoQ is synthesized in the inner membrane and matrix of mitochondria, yet is found in all cellular membranes, indicating the existence of a transport mechanism from the mitochondria to the rest of the cell [30–32]. Conversely, transport to the mitochondria can also occur from the extracellular environment through the plasma membrane as shown by uptake and assimilation of exogenously supplied CoQ in studies with yeast [33, 34] and mice [35], as well as through successful CoQ10 supplementation in patients with CoQ10 deficiencies [36]. Bidirectionality of CoQ transport, from the extracellular media to the mitochondria and from the mitochondria to other cellular membranes, has been attributed to endomembrane trafficking, yet the pathways and mechanisms remain obscure [32, 33].
In the yeast S. cerevisiae, CoQ6 is not essential for viability as strains that lack CoQ6 (coq mutants) can grow by fermentation on glucose without the involvement of the respiratory chain. However, coq mutants are unable to grow on non-fermentable carbon sources unless they are supplemented with exogenous CoQ, thereby restoring mitochondrial oxidative phosphorylation [34, 37, 38]. In this study, we investigated the mechanism of CoQ uptake and intracellular distribution in yeast. To do this, we interrogated a series of ORFΔcoq2Δ double mutants for their ability to grow in a non-fermentable glycerol-based medium (designated YPG) in the presence of exogenous CoQ6. These mutants harbor the deletion of one gene necessary for CoQ biosynthesis (coq2Δ) in conjunction with the deletion of another gene hypothesized to be involved with CoQ transport. The inability of some of these ORFΔcoq2Δ double mutants to recover growth in YPG even in the presence of CoQ6 allowed us to identify six genes that are important for CoQ6 trafficking, highlighting key components of the intracellular CoQ transport pathway. Intriguingly, our results showed that single coqΔ mutants differ in their ability to be rescued with exogenous CoQ6 and demonstrated that the presence of CoQ6-hexaprenylated biosynthetic intermediates impairs the assimilation of exogenous CoQ6 in yeast cells.
Materials and Methods
Coenzyme Q isoforms
CoQ2 was obtained from Cayman Chemical, CoQ6 from Avanti, and CoQ4 and CoQ10 from Millipore-Sigma. Each was dissolved in ethanol and the concentration was calculated using the absorbance at 275 nm and the corresponding molar extinction coefficients (13,700 M-1cm-1 for CoQ2; 14,200 M-1cm-1 for CoQ4; 14,700 M-1cm-1 for CoQ6 and 15,200 M-1cm-1 for CoQ10).
Determination of CoQ6 movement in mixed vesicle populations
The methodology used was a combination of previously described methods [39–41]. Briefly, LUVETs (large unilamellar vesicles produced by extrusion) were created from 25 mg/mL L-α-phosphatidylcholine (PC) from soybean (Type III-S, Millipore-Sigma) in chloroform, supplemented with 1-pyrene dodecanoic acid (Pyr12, Santa Cruz Biotechnology) to achieve a final concentration of 4 μM. The mixture was evaporated until dry under a stream of nitrogen gas, re-hydrated with MBSE buffer (0.15 M NaCl/ 0.01 M MOPS pH7/ 0.1 M EDTA) to a final PC concentration of 1 mg/mL, and subjected to ten freeze-thaw cycles in a solid CO2/ethanol bath. The multilamellar vesicles were extruded through two polycarbonate filters in tandem using the Avestin Lipofast system (Avestin, Inc). A second population was created as described above, with the addition of CoQ2, CoQ4, CoQ6, or CoQ10 to the chloroform to achieve a final concentration of 200 μM after re-hydration. To monitor the ability of the different CoQ isoforms to move between vesicles, Pyr12 quenching was monitored. Two mL of the initial LUVETs population was placed in a stirred cuvette in a Photon Technology International QuantaMaster Spectrofluorimeter (Horiba) (λex 346 nm and λem 377 nm) at 30 °C. After 90 s, 0.5 mL of a CoQ-containing vesicle population was added and the fluorescence was monitored for a total of 10 min. CoQ collisionally quenches Pyr12 fluorescence if both are present in the same vesicle and are capable of physical interaction [39, 42]. On mixing the two populations of vesicles, a further decrease in the fluorescence signal can only occur if the CoQn isoform is able to move from vesicle to vesicle. This is because the gain in fluorescence in the CoQn vesicle population from CoQ loss is less than the loss in fluorescence in the previously CoQn-less population from the gain of CoQ (for a discussion see [39]). Data are expressed as the ratio of fluorescence at a given point in time (I) to initial fluorescence just before the addition of the second population of vesicles (I 0).
Yeast strains, growth media, and verification of the mutants
S. cerevisiae strains used in this study are described in Table S1. Standard growth media for yeast included YPD (1% Bacto yeast extract, 2% Bacto peptone, 2% dextrose) and YPG (1% Bacto yeast extract, 2% Bacto peptone, 3% glycerol). Solid plate medium contains additional 2% Bacto agar. To select for mutants that contain a kanMX cassette, YPD + 200 mg/L G418 (Santa Cruz Biotechnology, Inc) plates were used. To select for ORFΔcoq2Δ mutants obtained from a previously created library [39], synthetic dextrose media without arginine and histidine (SD –His –Arg) (20 g/L glucose, 1.7 g/L yeast nitrogen base without amino acids and ammonium sulfate, 1 g/L monosodium glutamic acid, and 2 g/L of amino acid supplement lacking histidine and arginine) plates were used. These SD –His –Arg plates were supplemented with 200 mg/L G418, 70 mg/L canavanine (Millipore-Sigma) and 100 mg/L nourseothricin sulfate (ClonNat) (Gold Biotechnology). For mutants of new creation (see method below), SD –Leu (0.18% Difco yeast nitrogen base without amino acids and ammonium sulfate, 0.5% (NH4)2SO4, 0.14% NaH2PO4, 2% dextrose and amino acid supplement lacking leucine) plate medium was used. For plasmid selection (see below), SD –Ura (0.18% Difco yeast nitrogen base without amino acids and ammonium sulfate, 0.5% (NH4)2SO4, 0.14% NaH2PO4, 2% dextrose and amino acid supplement lacking uracil) liquid and plate media were used.
Deletions in each and every strain (both ORF and COQ2 deletions) were confirmed by PCR, using primer pairs A - KanB or D - KanC. Primer A and D are specific for each ORF while KanB and KanC are located whithin the kanMX cassette. Primers were obtained from the Saccharomyces Genome Deletion Project database (http://www-sequence.stanford.edu/group/yeast_deletion_project/deletions3.html). Tm ranged between 57 and 62 °C. The rho status of the cells was also confirmed using JM6 and JM8 as rho0 test strains [34]. Additionally, prior to the initiation of CoQ6 rescue experiments, we confirmed the ability of each of the corresponding single selected ORFΔ mutants to grow on YPG. The deletion of a single gene could compromise the ability of a mutant to grow on YPG if the gene is involved in respiratory activity. Demonstrated growth of the single ORFΔ on YPG ensures that recovery of respiration of the corresponding ORFΔcoq2Δ mutants with exogenous CoQ6 is theoretically possible. Any single mutant unable to grow on YPG automatically eliminated the corresponding ORFΔcoq2Δ from the study.
Generation of double deletion mutants
Some non-available mutants of interest for the study were created de novo using a PCR-based gene deletion strategy [43]. In all cases, the ORF of interest was replaced with a LEU2 auxotrophic marker amplified from the pRS305 vector using primers Leu forward: 5’-TGCCCTCCTCCTTGTCAATA-3’ and Leu reverse: 5 ‘-GGCGCCTGATTCAAGAAATA-3’, to which homologous sequences to the upstream of ORF start codon and downstream of the ORF stop codon were attached. Details about these homologous sequences are described in Table S2. Following PCR clean-up (Thermo Fisher Scientific), the PCR products were used to transform corresponding ORFΔ mutant strain using the lithium acetate/single-stranded carrier DNA/Polyethylene Glycol method [44], and transformed cells were selected twice on SD –Leu plates. Genomic DNA isolation following by PCR and DNA sequencing (Laragen, Inc.) were used to verify the correct integration of the auxotrophic marker at the correct gene locus. In these verification steps, specific primers A and D from the Saccharomyces Genome Deletion Project database were used, together with two internal primers inside the LEU vector (5’-CCATCACCATCGTCTTCCTT-3’ and 5’-CTGTGGGTGGTCCTAAATGG-3’) for DNA sequencing.
Generation of rho negative yeast
To generate rho0 BY4741 yeast, cells were incubated on YPD media plus 25 μg/mL ethidium bromide for two subsequent overnight periods. After that, cells were diluted (1:20,000 from a 1 A600/mL solution) and plated on YPD. After 1-2 days, cells were replica plated to YPG plate medium to test their ability to respire. Colonies unable to grow on YPG were considered rho0. To further confirm the absence of mtDNA, two distant mitochondrial DNA loci, COX3 and OLI1, were checked by PCR (COX3-F: 5’-GGTAATATGAATATGGTATATTTAGC-3’; COX3-R: 5’ -GTTACAGTAGCACCAGAAGATAATAAG-3’; OLI1-F: 5’-ATGCAATTAGTATTAGCAGCTAAAT-3’; OLI1-R: 5’-CCGAATAATAATAAGAATGAAACCATTA-3’) [45]. The ERG2 locus was included as a positive control for a nuclear gene, using erg2A and erg2D primers from the Saccharomyces Deletion Project Database.
Exogenous CoQ rescue assays
Yeast mutants were grown overnight in YPD at 30 °C 250 rpm. The next day, the A600nm/mL was measured and yeast strains were sub-inoculated into tubes containing 5 mL of YPG supplemented with CoQ (2 μM of CoQ6 or 100 μM of CoQ2) or with the corresponding amount of ethanol at a cell density of 0.1 A600nm/mL. In all experiments the amount of ethanol added to the culture (containing CoQ or as vehicle) was less than 1% of the culture volume. Yeast were allowed to grow at 30 °C 250 rpm for 7 days. To construct the growth curves (see examples in Fig. S4), A600nm measurements were taken at approximately 0, 7, 24, 30, 48, 54, 72, 78, and 144 hours. An aliquot of each culture (200 μL) was transferred to a 96-well plate and A600nm was measured in a TECAN M1000 plate reader. 1/10 dilutions were made if necessary. With the exception of Fig. S4, CoQ rescue is always represented as a percentage of a designated control, considered as 100% and represented as a dashed line in the graphics. A600nm from the last time point, 144 h, was used to calculate the degree of rescue.
Determination of CoQ6 and CoQ6 intermediates
To determine the content of CoQ6 and CoQ6 intermediate in coqΔ mutants, 15 mL of yeast grown on YPD were harvested at approximately 1 A600nm/mL. Lipid extractions and the following analysis of CoQ content in a 4000 QTRAP linear MS/MS spectrometer (Applied Biosystems) were performed as described [46]. Aliquots of the culture were saved prior to lipid extraction to determine protein concentration using Bradford assay [47].
Immunoblot analysis
Three mL of yeast grown on YPD were harvested at approximately 1 A600nm/mL. Pellets were resuspended in 100 μL 2M lithium acetate and incubated on ice for 5 min. Samples were centrifuged and pellets were resuspended in 100 μL 0.4M NaOH, then incubated on ice for 5 min. After this step, a 10 μL aliquot of each sample was saved for further protein quantification. Samples were resuspended in 100 μL 1X SDS sample buffer (50 mM Tris pH 6.8, 10% glycerol, 2% SDS, 0.1% bromophenol blue, and 1.33% β-mercaptoethanol) and boiled for 5 min. After centrifugation, supernatants were moved to clean tubes and stored at −20 °C until further use. 30-50 μg of protein was loaded in individual lanes and separated by SDS gel electrophoresis on 12% Tris-glycine polyacrylamide gels. Proteins were subsequently transferred to 0.45 μm Immobilon-P membrane (Bio-Rad). Blots were stained with Ponceau S dye for visualization of protein lanes and assessment of equal protein loading. After blocking (blocking buffer: 0.5% BSA, 0.1% Tween 20, 0.02% SDS in phosphate-buffered saline), blots were incubated with 1:1,000 primary anti-Porin 1 (obtained from Dr. Carla Koehler, University of California, Los Angeles) and 1:20,000 secondary IRDye 680LT goat anti-rabbit IgG antibody (LiCOR). Immunoblot images were visualized with a LiCOR Odyssey infrared scanner (LiCOR), and relative protein levels were quantified by band densitometry using both Image Studio Lite 5.2 and ImageJ.
Complementation of ORFΔcoq2Δ mutants with yeast COQ2
To re-introduce COQ2 in the ORFΔcoq2Δ double mutants, the vector pRSQ2 was constructed. The sequence of yeast COQ2 plus 350 bp of the upstream sequence was cloned into the vector pRS316 [48] using the BamHI restriction site. Then, the empty vector, as well as pRSQ2, were each used to transform coq2Δ, cdc10Δcoq2Δ, rts1Δcoq2Δ, rvs161Δcoq2Δ, rvs167Δcoq2Δ and nat3Δcoq2Δ. Transformation of WT and the corresponding single ORFΔ with pRS316 served as controls. Cells containing the plasmid were double selected on SD −Ura plates. The ability of the ORFΔcoq2Δ to recover their respiratory capacity after COQ2 was analyzed using spot plate assays. Briefly, plasmid-containing cells were grown overnight in SD −Ura at 30 °C 250 rpm. The next morning, A600nm/mL was measured, cells were diluted to 0.2 A600nm/mL in PBS, and a series of 5-fold dilutions were prepared. 2 μL of each dilution were plated onto YPD, YPG and SD −Ura plates. Plates were incubated at 30 °C and after 3-4 days, pictures were taken.
Statistical analysis
Data shown in this work represent mean ± standard deviation (SD) from at least three biological replicates. Statistical analyses and graphics were performed with Graphpad Prism 7 (Graphpad Software Inc., San Diego, CA, USA). When a specific mutant is compared to WT or to a designated normalization variable (specific coqΔ + CoQ6 or ORFΔ) a paired t-test analysis was performed. When comparing two specific mutants between each other, an unpaired t-test analysis was used. Significant differences were referred as *p<0.05, **p<0.01, ***p<0.001, and ****p<0.0001.
Results
1. CoQ6 does not move passively between non-contiguous membranes
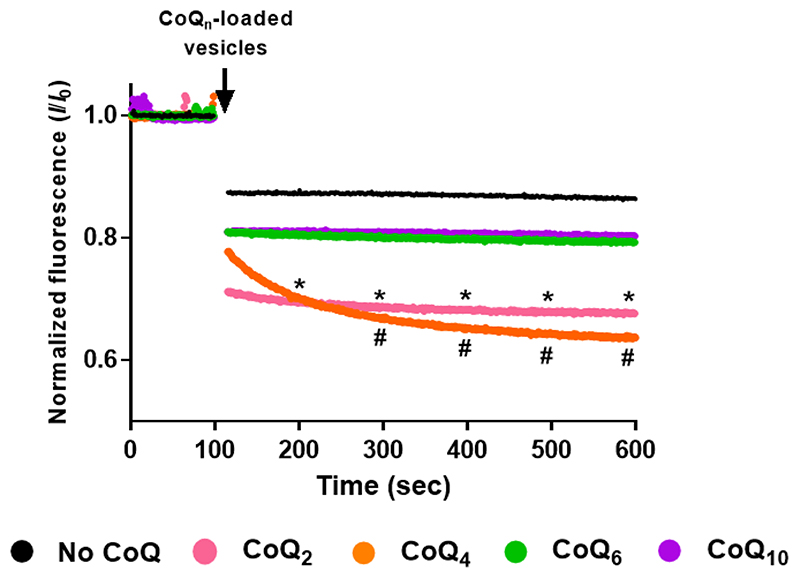
Growth of coqΔ strains on YPG with exogenous CoQ6 [33, 34] indicated that a sufficient quantity of the supplied CoQ6 reached the mitochondrial inner membrane. However, the uptake of a particular CoQ isoform depends on the length of the isoprenoid chain. A previous study assessed the ability of different CoQ isoforms to diffuse between non-contiguous membranes [39]. Short CoQ isoforms such as CoQ2 and CoQ4 that are more water soluble, freely exchanged between phospholipid bilayers, while a long CoQ isoform (CoQ9) did not due to its extreme hydrophobicity, and thus required a mechanism of transport [39]. The behaviour of the S. cerevisiae isoform CoQ6 has not been evaluated. Therefore, to assess the ability of CoQ6 to move between non-contiguous membranes, fluorescence quenching experiments were performed with two populations of phosphatidylcholine vesicles. Both populations contained equal concentrations of the fluorophore 1-pyrene dodecanoic acid (Pyr12), but only the second population contained the specified isoform of CoQn. In this second population, the presence of CoQn fully quenches Pyr12 fluorescence by collisional quenching. On mixing the two populations of vesicles, a further decrease in the fluorescence signal can only occur if the CoQn isoform is able to move from vesicle to vesicle [39, 49]. Results showing the behavior of CoQ6 in comparison with CoQ2, CoQ4 and CoQ10 are depicted in Fig. 2. When the population of CoQ-loaded vesicles was added, the fluorescence decreased dramatically when CoQ2 or CoQ4 were present, while fluorescence was maintained when the vesicles were loaded with either CoQ6 or CoQ10. These results indicated that CoQ2 and CoQ4 were able to move rapidly between the different vesicles, in agreement with previous results [39], while CoQ10 and CoQ6 were non-diffusible. The inability of CoQ6 to move between synthetic phospholipid membranes supports the idea that a transport/distribution mechanism is needed due to its negligible water solubility.
Figure 2. CoQ6 and CoQ10 isoforms fail to move freely between phosphatidylcholine vesicles.
The membrane behavior of different CoQn isoforms was analyzed using a fluorescence-quenching assay. An initial population of vesicles containing 4 μM Pyr12 was mixed with 500 μL of a second population of vesicles that contained 4 μM Pyr12 and either a control, with no addition (black), or a CoQ isoform: CoQ2 (pink), CoQ4 (orange), CoQ6 (green) or CoQ10 (purple). Fluorescence was monitored for 600 seconds and data points were collected every second. The data are expressed as the relative amount of fluorescence at a given point in time (I) to the initial fluorescence just before the addition of the CoQn-loaded vesicles (IO). Dots represent the average, at each point, of at least three independent experiments. Standard deviations have been omitted for clarity in this figure but they are provided in Fig. S1. At intervals of 100 sec, the (*) symbol represents significant differences (p<0.05) between CoQ2 and the no-CoQ condition, while (#) represents differences between CoQ4 and the no-CoQ condition.
2. Yeast mutants harboring deletions in distinct COQ genes show different degrees of rescue in response to exogenous CoQ6
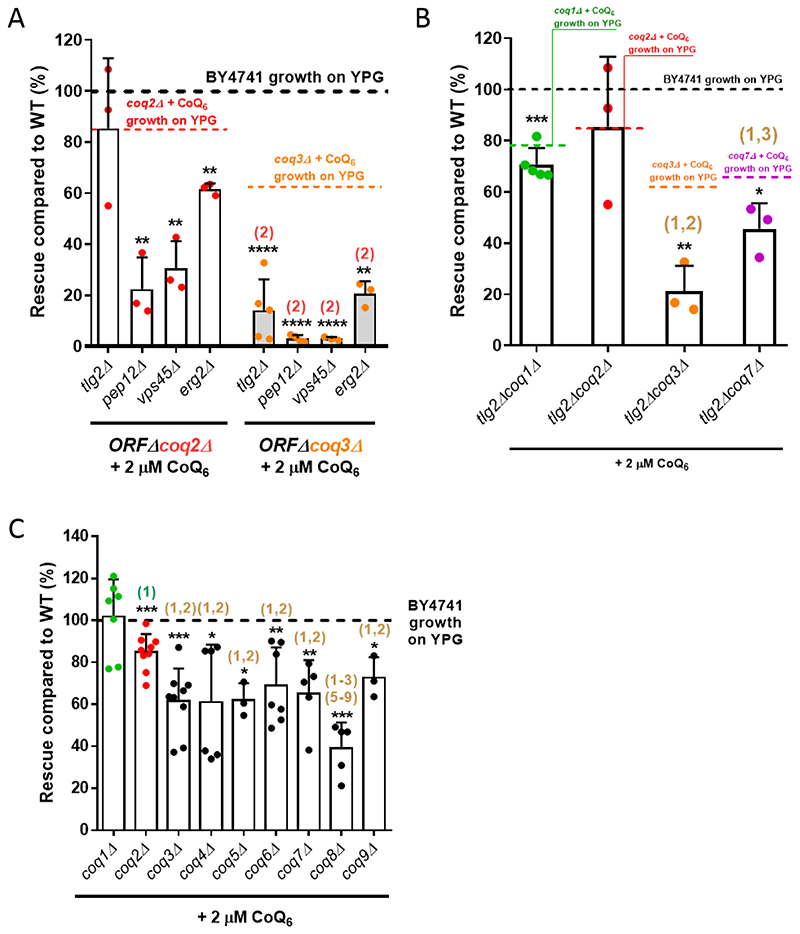
Previous work indicated that at least four genes of the endocytic pathway are required for rescue of S. cerevisiae by exogenous CoQ6 [33]. In that study, ERG2, TLG2, PEP12, or VPS45 deleted in combination with a COQ3 deletion inhibited growth of the resulting double mutants on YPG supplemented with CoQ6. CoQ6 is produced in the mitochondrial inner membrane by a large multiprotein complex (the CoQ synthome) formed by several Coq proteins (Coq3-Coq9, Coq11) [1]. The absence of one Coq polypeptide can result in the loss of other Coq proteins [1, 50]. Given the effects of distinct COQ gene deletions on the CoQ synthome composition, we tested whether there is a difference in the ability of CoQ6 to rescue strains carrying different COQ deletions in combination with the endocytic pathway mutations. Deletions of either COQ2 or COQ3 were combined with erg2Δ, tlg2Δ, pep12Δ, or vps45Δ, and the resulting strains were assessed for the degree of rescue by CoQ6 (Fig. 3A, Table S3). When normalized to the wild-type parental strain (WT), rescue of the four ORFΔcoq2Δ double mutants by exogenous CoQ6 was more robust than rescue of the ORFΔcoq3Δ counterparts. Similar results were observed if the degree of rescue was compared to the corresponding positive control (either coq2Δ + CoQ6 or coq3Δ + CoQ6), instead of to WT growth (Fig. 3A, Table S3). These results suggested that coq3△ cells are sensitized to the effects of endocytic pathway mutations on CoQ6 rescue compared to coq2Δ cells. This was most apparent in the case of tlg2Δ, which severely inhibited rescue in coq3Δ cells but had no effect on rescue in coq2Δ cells. It thus seemed possible that the presence of different CoQ6 synthesis intermediates might influence the rescue with exogenous CoQ6. In a coq3Δ mutant, the Coq2 polypeptide is present and active, resulting in the production of two hexaprenylated CoQ6 intermediates: 3-hexaprenyl-4-hydroxybenzoic acid (HHB), and 3-hexaprenyl-4-aminobenzoic acid (HAB) (Fig. 1) [1]. On the other hand, in a coq2Δ mutant, CoQ biosynthesis is blocked at the hexaprenyl transfer step, and neither HHB nor HAB can be formed.
Figure 3. Yeast mutants harboring deletions in distinct COQ genes show different degrees of rescue in response to exogenously added CoQ6.
(A) Yeast coq2Δ mutants harboring additional deletions in tlg2Δ, pepl2Δ, vps45Δ or erg2Δ showed a more pronounced YPG growth rescue by CoQ6 than do the comparable coq3Δ double mutants. (B) The tlg2ΔcoqlΔ, tlg2Δcoq3Δ and tlg2Δcoq7Δ double mutants presented an impaired degree of rescue as compared to WT. The decrease of rescue was more pronounced in tlg2Δcoq3Δ and tlg2Δcoq7Δ, but especially in the tlg2Δcoq3Δ mutant. (C) Yeast mutants harboring deletions in either the COQ1 or COQ2 gene showed more robust rescue in response to exogenous CoQ6 treatment than do the other single coq null mutants (coq3Δ - coq9Δ). In all cases, columns represent the degree of rescue (in %) ± SD of a strain compared to WT, which is defined as 100% and represented as a dashed line. In A and B, the average of rescue of the specific positive controls (coq1Δ + CoQ6, coq2Δ + CoQ6, coq3Δ + CoQ6, or coq7Δ + CoQ6), which were analyzed in parallel with the samples, is represented as a dashed line. Three or more independent rescue experiments were performed for every strain. Asterisks on top of the columns represent significant differences when compared to WT (*p<0.05, **p<0.01, ***p<0.001, and ****p<0.0001). Statistically significant differences between a specific coqΔ mutant (or ORFΔcoqΔ mutant) and one of its counterparts (another coqΔ or ORFΔcoqΔ mutant) are denoted with numbers in parentheses on top of the columns. (1) represents differences comparing to coqlA, (2) represents differences comparing to ORFΔcoq2Δ (panel A) or coq2Δ (panel B-C), (3) represents differences comparing to coq3Δ, etc. Statistical comparisons not detailed in the figure are provided in Table S3.
To further examine whether the accumulation of early CoQ6 synthesis intermediates influences the degree of rescue by exogenous CoQ6, we focused on a particular endocytosis-defective mutant, tlg2Δ, and constructed COQ1 and COQ7 deletion strains. Coq1 synthesizes the CoQ polyisoprenoid tail, so its deletion prevents the formation of any hexaprenylated CoQ precursors (Fig. 1) [1]. Coq7 is a hydroxylase responsible for catalyzing the penultimate step of the CoQ biosynthetic pathway. However, because it is also a required structural component of the CoQ synthome, its deletion leads to the accumulation of HHB and HAB [1]. As shown in Fig. 3B, the degree of rescue of either tlg2Δcoq3Δ or tlg2Δcoq7Δ was significantly different from the mutants carrying tlg2Δcoq1Δ or tlg2Δcoq2Δ. It is notable that the tlg2Δcoq3Δ exhibits the most severely impaired degree of rescue (Fig. 3B, Table S3). The same general trend was observed when the degree of rescue is compared to each of the corresponding positive controls (e.g. coq1Δ + CoQ6; coq2Δ + CoQ6; etc.), instead of to WT growth (Fig. 3B, Table S3). These results suggest that the accumulation of CoQ hexaprenylated intermediates can affect the restoration of respiratory chain function by exogenous CoQ6.
We then assessed the degree of rescue of each of the coq1Δ to coq9Δ single mutants in presence of exogenous CoQ6 (Fig. 3C). The coq1Δ mutant grew to WT levels under these conditions (Fig. 3C, Table S3A). Although exogenous CoQ6 does not completely restore the coq2Δ growth to WT levels, the response of the coq2Δ mutant is significantly better as compared to each of the other mutants, from coq3Δ to coq9Δ (Fig. 3C, Table S3). Interestingly, the coq8Δ mutant is more impaired than each of the other mutants, except for CoQ4Δ (Fig. 3C, Table S3). This finding might suggests an active role of Coq8 in CoQ6 transport. As coq3Δ - coq9Δ mutants are reported to accumulate hexaprenylated CoQ6 intermediates, these results further support the hypothesis that the presence of such intermediates inhibits rescue of respiratory defective cells by CoQ6.
3. The accumulation of early CoQ6 hexaprenylated intermediates impairs CoQ6 rescue
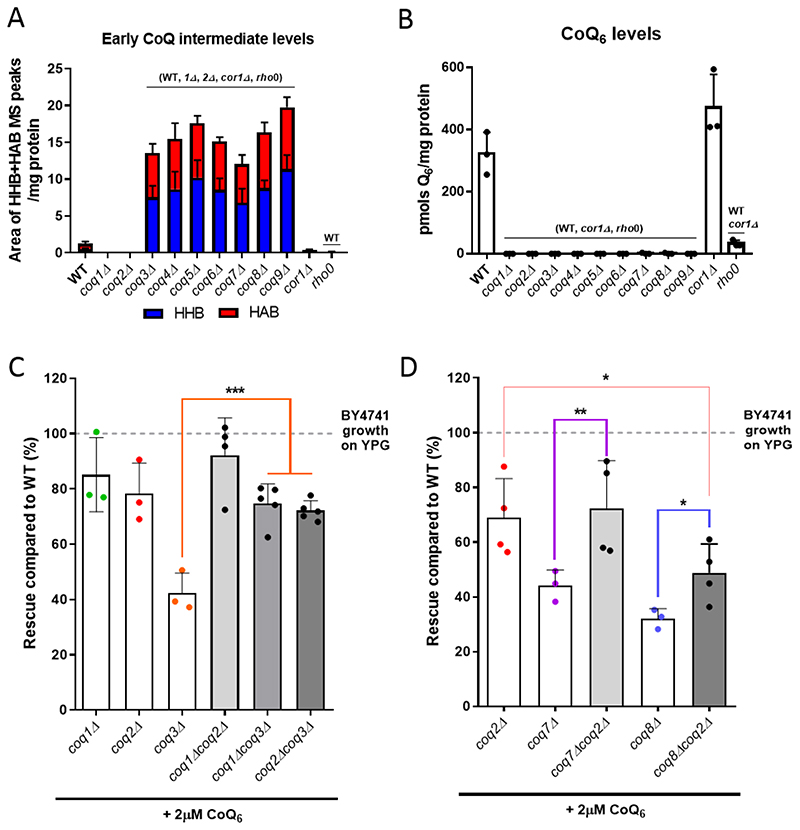
The presence of HHB and HAB in coq3Δ-coq9Δ mutants has been described previously [1, 2, 5], but a direct comparison of intermediates between the different strains is not available. We now show this comparison in Fig. 4A. These experiments revealed the loss of CoQ6 in each of the coqΔ mutant strains (Fig. 4B) and a similar accumulation of HHB and HAB in coq3Δ - coq9Δ mutants (Fig. 4A, Fig. S2). None of the coqΔ mutants accumulated late-stage CoQ6 intermediates (Fig. S2). In all cases, several controls were included: WT, which defines normal content of CoQ6 and CoQ6 biosynthetic intermediates in yeast cells; corlΔ, as an example of a mutant with normal CoQ6 content but an inability to respire due to the deletion of COR1, an essential component of complex III; and a rho0 mutant on the same genetic background, to evaluate the absence of mitochondrial DNA (mtDNA) on CoQ6 content. The absence of mtDNA can be considered a secondary CoQ deficiency [2, 51], and accordingly a dramatic decrease of CoQ6 is observed (Fig. 4B). Additionally, to investigate whether altered mitochondrial mass might affect the degree of rescue of coq3Δ-coq9Δ mutants, we measured levels of the porin 1 polypeptide as a mitochondrial mass marker. Mitochondrial mass was not altered in any of the nuclear mutants (Fig. S3).
Figure 4. The elimination of HHB and HAB intermediates augments the degree of rescue with exogenous CoQ6.
(A) Each of the coq mutants, from coq3Δ to coq9Δ, accumulated early CoQ6 intermediates (HHB and HAB). The amount of HHB and HAB was calculated as the area of the MS peak/mg protein. (B) CoQ6 was not detected in any of the coqΔ mutants. CoQ6 (pmol/mg protein) was determined based on a CoQ6 standard curve as described in [46]. (C) Additional deletions of either COQ1 or COQ2 restored the deficient CoQ6-rescue of a coq3Δ mutant, but do not affect the phenotype observed in coq1Δ or coqΔ2 strains. (D) Similar results were observed when COQ2 is deleted in a coq7Δ strain, but only a partial improvement was observed in a coq8Δ mutant. In A and B, columns represent mean ± SD and strains named in parentheses on top of the columns denote statistically significant differences (p<0.01). In C and D, columns represent the degree of rescue (in %) ± SD of a strain in comparison to WT (considered 100% and represented as a dashed line). Asterisks denote significant differences as compared to the single coqΔ strain (*p<0.05, **p<0.01, and *** p<0.001). In all cases, three or more independent biological replicates were performed.
If accumulation of hexaprenylated CoQ6 intermediates HAB and HHB caused by coq3Δ - coq9Δ mutations is responsible for impaired rescue by exogenous CoQ6, then deletion of either COQ1 or COQ2, which prevent the prenylation, should improve growth of coq3Δ - coq9Δ in the presence of CoQ6. As predicted, deletion of COQ1 or COQ2 in a coq3Δ mutant provided a degree of CoQ6 rescue comparable to either the coq1Δ and coq2Δ single mutant, and approximately double that of a coq3Δ mutant (Fig. 4C). Similarly, deletion of COQ2 in either a coq7Δ or a coq8Δ mutant enhanced rescue by exogenous CoQ6 as compared with the rescue of either the coq7Δ or coq8Δ single mutants (Fig. 4D). Interestingly, the rescue of the coq8Δcoq2Δ mutant was still lower than the rescue of a single coq2Δ strain (Fig. 4D). Together, our data indicate that the accumulation of HHB and HAB intermediates in coq3Δ-coq9Δ mutants impairs rescue by exogenous CoQ6.
4. Genes that diminish growth response to exogenous CoQ6
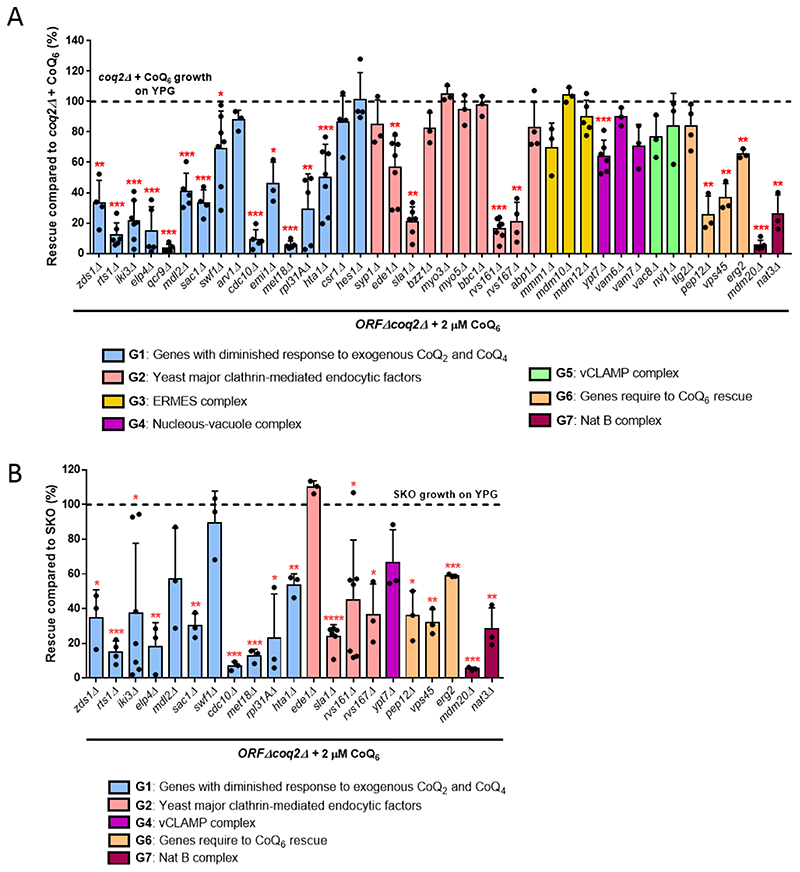
To identify genes involved in CoQ6 uptake and trafficking to mitochondria, we used a library of double ORFΔcoq2Δ mutants, and tracked their ability to recover growth on YPG when supplemented with exogenous CoQ6. This library was previously developed to identify proteins or pathways that might be involved in CoQ2 or CoQ4-dependent growth on non-fermentable substrates [39]. Based on the results described above, the use of coq2Δ in the library avoids potential complications from accumulation of HHB and HAB. Rescue with exogenous CoQ6 requires vigorous aeration of liquid cultures, which does not occur in high throughput multi-well assays [52]. Therefore, we selected 48 candidate genes that previous literature suggests may be involved in CoQ6 uptake/distribution [32, 33, 39]. The genes can be grouped into seven categories (Table 1): 16 genes described to have diminished response to exogenous CoQ2 and CoQ4 [39] plus two extra genes involved in phospholipid and sterol transport (CSR1 and HES1) with limited growth on exogenous CoQ2 and CoQ4; 15 genes implicated in yeast clathrin-mediated endocytosis [53]; four genes that assemble the endoplasmic reticulum (ER)-mitochondria encounter structure (ERMES) [54]; two genes that encode the nucleus-vacuole junction (NVJ); three genes that represent the vacuole-mitochondria patch (vCLAMP) [55, 56]; four endocytosis genes already described to play a role in CoQ6 transport [33]; and two genes that form the N-terminal acetyltransferase B (Nat B) complex. Some ORFΔcoq2Δ double mutants were not recoverable from the library (see Table 1), so a total of 40 mutants were ultimately analyzed.
Table 1. Yeast gene candidates selected for screening.
| Group 1 | Genes with diminished response to exogenous CoQ2 and CoQ4 [39] | ZDS1, RTS1, IKI3, ELP4, QCR9, ACO1, MDL2, SAC1, SWF1, ARV1, CDC10, KCS1, EMI1, MET18, RPL31A, HTA1, CSR1, HES1 |
| Group 2 | Yeast major clathrin-mediated endocytic factors [53] | SYP1, EDE1, CHC1, CLC1, SLA2, END3, SLA1, VRP1, BZZ1, MYO3, MYO5, BBC1, RVS161, RVS167, ABP1 |
| Group 3 | ER-mitochondria encounter structure (ERMES) | MMM1, MDM10, MDM12, MDM34 |
| Group 4 | Nucleous-vacuole junction (NVJ) | VAC8, NVJ1 |
| Group 5 | Vacuole-mitochondria patch (vCLAMP) | YPT7, VAM6, VAM7 |
| Group 6 | Genes previously described to be required for CoQ6 rescue [33] | TLG2, PEP12, ERG2, VPS45 |
| Group 7 | Nat B complex | MDM20, NAT3 |
Genes depicted in light grey represent the ORFΔcoq2Δ double mutants that were not recoverable for the ORFΔcoq2Δ library.
The ability of each ORFΔcoq2Δ double mutant to recover respiratory growth on YPG in the presence of 2 μM CoQ6 was monitored for seven days. In all experiments, WT, coq2Δ and cor1Δ strains were included as controls in the presence and absence of CoQ6. This screen identified 23 mutants that showed significantly diminished rescue compared to the positive control (coq2Δ + 2 μM CoQ6) (Fig. 5A), indicating that the presence of these particular ORFs may be necessary for optimal CoQ6 rescue. A representative ORFΔcoq2Δ mutant growth curve from each group is displayed in Fig. S4. Almost all double mutants from the heterogeneous Group 1 had diminished ability to be rescued by CoQ6, in agreement with the deficient response to addition of CoQ2 or CoQ4 [39]. Four affected candidates were yeast endocytic factors (Group 2), while six other members of this group retained the ability to be rescued with exogenous CoQ6. This result suggests that the entire endocytic pathway may not be necessary for CoQ6 transport, or that alternative or redundant mechanisms of CoQ6 transport may compensate for the deletion of a particular gene. A strong deficiency in rescue was also observed in three of the four gene deletions previously studied [33] (Group 6) and in the members of the Nat B complex. Interestingly, the groups representing the connections between organelles were in general not affected. These results may either exclude gene products involved in organelle connections as participants in the mechanism of CoQ6 transport, or suggest that CoQ6 transport has more than one path and so the elimination of one organelle connection is not sufficient to produce a defective phenotype.
Figure 5. Two rounds of screening identified 17 ORFΔcoq2Δ double mutants to have a diminished response to exogenous CoQ6.
(A) First, the degree of rescue of the ORFΔcoq2Δ candidates was calculated in comparison with the positive control: coq2Δ + CoQ6 . Of a total of 40 candidates, 23 showed a significantly decreased ability to rescue in presence of exogenous CoQ6. (B) Since a single deletion can compromise the ability of a mutant to grow on YPG, the degree of rescue of the previous significant candidates was calculated in comparison to the corresponding ORFΔ in the second phase of the screening. The emilΔcoq2Δ and qcr9Δcoq2Δ mutants were eliminated for further analysis due to special phenotypes (see main text). 17 candidates display impaired rescue in response to treatment with exogenous CoQ6. In both cases, columns represent the average degree of rescue (in %) ± SD of mutants growing on YPG in presence of 2 μM CoQ6 for 7 days. Three or more replicas were included for all candidates. The normalization variable is considered 100% and represented as a dashed line. Asterisks denote the different degrees of significance: *p<0.05, **p<0.01, ***p<0.001, and ****p<0.0001.
Deletions of certain genes may not completely abolish the ability of mutants to grow on YPG, but can still compromise growth. To examine this, a second round of screening was performed and the ability of the ORFΔcoq2Δ mutants to grow in the presence of exogenous CoQ6 was compared with the corresponding single ORFΔ. We analyzed the 23 candidates identified in Fig. 5A for their degree of rescue when compared to the single ORFΔ. It should be noted that two double mutants were excluded: qcr9Δcoq2Δ and emilΔcoq2Δ. The growth of the single qcr9Δ in YPG was highly inconsistent, for reasons that are unclear. In the case of the emilΔcoq2Δ mutant, the phenotype observed was unlikely to be related to CoQ6 transport, and is being investigated separately. Of the remaining 21 ORFΔcoq2Δ mutants, 17 candidates were significantly different from the corresponding ORFΔ (Fig. 5B), suggesting they are involved in CoQ6 transport. The four ORFΔ that were eliminated were each rescued to their maximum potential level, indicating that the slow YPG growth is unrelated to CoQ6 transport. A good example of this phenomenon can be observed in Fig. S4B, where the growth of ede1Δcoq2Δ + CoQ6 is similar to the growth of ede1Δ, and both display decreased growth as compared to the positive control.
5. Comparison of the CoQ2 and CoQ6 rescue phenotypes identifies six candidates necessary for CoQ6 transport
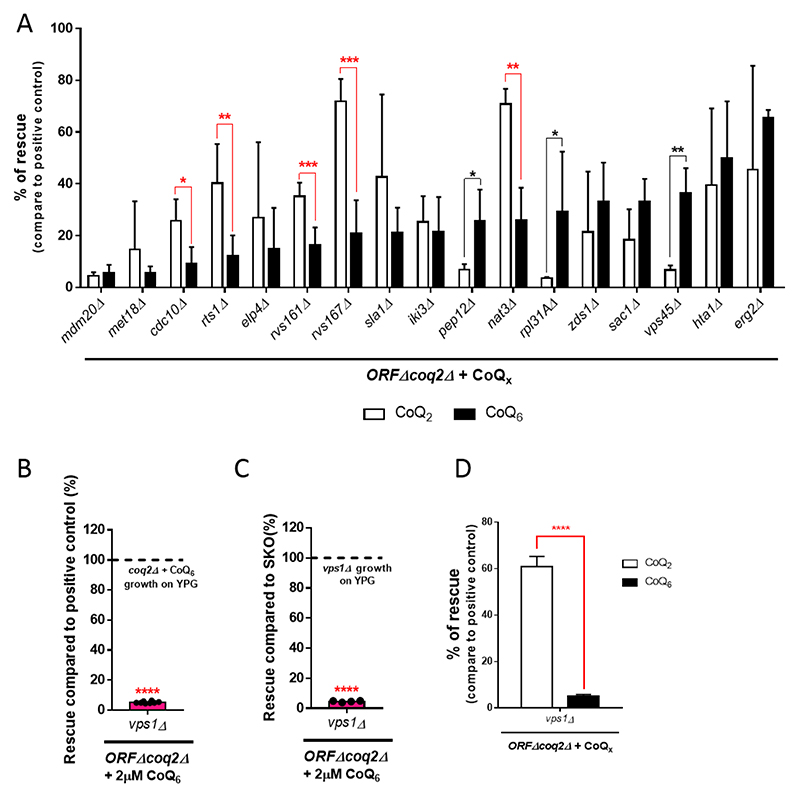
In order to identify the most relevant genes in the transport of CoQ6, we analyzed the rescue phenotype of the 17 previously identified candidates in response to exogenous CoQ2. We hypothesized that if the growth defect of the ORFΔcoq2Δ mutants is in fact due to the uptake and/or distribution of CoQ6, a shorter, diffusible isoform of CoQ such as CoQ2 should allow for a better degree of rescue than CoQ6. In this scenario, CoQ2 will be able to reach the mitochondria of the mutants even when the mechanisms of transport are impaired. It is important to note that the degree of rescue of the coq2Δ mutant in the presence of CoQ2 was significantly lower than that observed in the presence of CoQ6 (Fig. S5A). It is possible that the more rapid diffusion of CoQ2 between membranes due to its far lower hydrophobicity limits its accumulation in the mitochondria where it must partition to recover respiratory function.
Comparison between CoQ2 and CoQ6 rescue phenotypes of each of the 17 ORFΔcoq2Δ mutants is depicted in Fig. 6A. The CoQ2 rescue phenotype of the mutants on their own can be observed in Fig. S5B. Of the 17 ORFΔcoq2Δ mutants analyzed, eight showed statistically significant differences between CoQ2 and CoQ6 rescue. However, only five ORFΔcoq2Δ candidates fit the criterion of having a degree of CoQ2 rescue significantly greater than that by CoQ6. Thus, these five genes (CDC10, RTS1, RVS161, RVS167, and NAT3) seem good candidates to encode proteins that are required for CoQ6 uptake and/or distribution to mitochondria within yeast cells. The known functions of these proteins are summarized in Table 2. In Fig. S6, we showed that the transformation of the final five ORFΔcoq2Δ candidates with the plasmid pRSQ2 (containing the COQ2 gene) restores the growth of the ORFΔ on YPG plates.
Figure 6. Comparison of the CoQ2 and CoQ6 rescue phenotypes identifies six genes necessary for CoQ6 transport.
(A) The degree of CoQ2 and CoQ6 rescue, compared to the corresponding positive control (coq2Δ + CoQ2 or coq2Δ + CoQ6), are represented side by side (from low to high degree of CoQ6 rescue). Five candidates (denoted with red lines) fulfilled the “CoQ6 rescue < CoQ2 rescue” criteria and have been identified as involved in CoQ6 transport. This panel merges data from Figure 5A and Figure S5B to allow for a straightforward comparison. (B) The degree of rescue of vps1Δcoq2Δ was observed to be dramatically low when compared to the positive control. (C) Similar results were observed when the rescue of the vps1Δcoq2Δ mutant was compared to vps1Δ. (D) The degree of rescue with the addition of exogenous CoQ2 was superior to the one observed with the addition of CoQ6. Degree of rescue is referred to the corresponding positive control (coq2Δ + CoQ2 or coq2Δ + CoQ6). In all cases, columns represent the average degree of rescue (in %) ± SD of mutants growing on YPG in presence of CoQ for 7 days. At least three replicas were included in all determinations. The normalization variable is considered 100% and represented as a dashed line. Asterisks denote: *p<0.05, **p<0.01, ***p<0.001, and ****p<0.0001.
Table 2. Genes identified to be related to CoQ6 transport.
| ORFΔ (protein name) | ORF description (www.yeastgenome.org) | Human homolog |
|---|---|---|
| YCR002C (Cdc10) | Subunit of the septin complex with GTPase activity. Localizes to the bud neck septin ring, and can function as scaffolds for recruiting cell division factors and as barriers to prevent diffusion of specific proteins. | SEPT9 |
| YOR014W (Rts1) | Protein phosphatase regulator involved in protein dephosphorylation, mitotic spindle orientation checkpoint, and septin ring organization and disassembly. | PPP2R5C |
| YCR009C (Rvs161) | N-BAR domain protein that interacts with Rvs167 and regulates polarization of the actin cytoskeleton, endocytosis, cell polarity, cell fusion and viability following starvation or osmotic stress. | BIN3 |
| YDR388W (Rvs167) | N-BAR domain protein with roles in endocytic membrane tabulation, constriction and exocytosis. Interacts with Rvs161 to regulate actin cytoskeleton, endocytosis and viability following starvation or osmotic stress. | BIN1, AMPH [57–59] |
| YPR131C (Nat3) | Catalytic subunit of the NatB N-terminal acetyltransferase complex, which is in charge of the N-acetylation of the amino-terminal methionine residues of all proteins beginning with Met-Asp or Met-Glu and of some proteins beginning with Met-Asn or Met-Met (approximately 20% of proteins in yeast). | NAA20 |
| YKR001C (Vps1) | Dynamin-like GTPase required for vacuolar sorting, protein retention in Golgi apparatus, peroxisome organization and fission, endocytosis, and actin cytoskeleton organization. | Dynamin [57, 60] |
Unless otherwise is indicated, the closest human homolog genes were located in the HomoloGene tool from the NCBI database.
Two of the five identified proteins, Rvs161 and Rvs167, function together in the vesicle scission step of the clathrin-mediated endocytosis pathway. Multiple studies have identified Vps1 as a third player in this mechanism [61, 62]. Moreover, Vps1 and Rvs167 have been reported to function together during endocytosis [63]. VPS1 was not part of our initial selection of candidates, but due to its functional relationship with Rvs161 and Rvs167, we hypothesized that it might also function in CoQ6 transport. The degree of rescue with exogenous CoQ6 of the vps1Δcoq2Δ mutant, in comparison to the positive control and also with vps1Δ, is represented in Fig. 6B-C. In both cases, the degree of CoQ6 rescue is dramatically impaired. In addition, the degree of rescue with exogenous CoQ2 was analyzed, and the comparison with the CoQ6 rescue phenotype showed that the “CoQ2 rescue > CoQ6 rescue” criteria is fulfilled (Fig. 6D). These results confirmed VPS1 as an additional gene required for CoQ6 transport.
Discussion
CoQ10 is among the most widely used dietary and nutritional supplements on the market, ranging from an over-the-counter supplement to more specific administration for particular disorders [64]. Numerous disease processes associated with CoQ10 deficiency may benefit from CoQ10 supplementation, including primary and secondary CoQ10 deficiencies, mitochondrial diseases, fibromyalgia, cardiovascular disease, neurodegenerative diseases, cancer, diabetes mellitus, infertility, and periodontal disease [17, 65, 66]. However, CoQ10 is not an efficient oral supplement. Its high lipophilicity and poor solubility lead to low bioavailability due to limited absorption into systemic circulation, making the effective delivery of CoQ10 to the mitochondria a major challenge [23, 67]. Understanding the mechanism(s) used to uptake and distribute CoQ10 within cells is of great relevance to help CoQ10 become an efficient therapy. Towards this end, the studies in yeast reported here uncovered an unexpected inhibitory effect of prenylated CoQ intermediates on exogenous CoQ assimilation and identified six genes required for functional rescue of CoQ-deficiency by exogenous supplementation with the normal cellular form of CoQ. While we have utilized respiration as a means to identify genes that transport CoQ, CoQ supplementation via these transport pathways could offer antioxidant protection against mitochondrial-derived oxidants.
Multiple lines of evidence support the existence of one or more import pathways for exogenous CoQ6 and CoQ10, as well as an intracellular distribution mechanism for de novo synthesized CoQ from the mitochondria to other cellular membranes [30–34]. Results presented in Fig. 2 establish the necessity of transport systems for movement of CoQ10 and CoQ6 between membranes, supporting the view that CoQ transport requires the endomembrane system [32, 33]. To address the mechanism of transport, we adopted a genetic strategy in which gene deletions were screened for effects on the ability of exogenous CoQ6 to rescue the growth of a coqΔ strain deficient in CoQ synthesis (ref 30). During the course of these experiments, we observed differential responses of coqΔ mutants to exogenous CoQ6 due to reduced rescue in strains (coq3Δ-coq9Δ) that accumulate early-stage prenylated CoQ6 intermediates, HHB and HAB (Figs. 3 and 4). This likely also explains the previous observation that exogenous decylubiquinone blunts rescue by exogenous CoQ2 in coq2Δ mutants [39].
The exact reason why the presence of early CoQ6 hexaprenylated intermediates prevents the rescue with exogenous CoQ6 is not clear, but different hypotheses can be proposed: CoQ6 intermediates could (i) produce a certain level of toxicity that affects the normal operation of the cells; (ii) mimic the behavior of detergents by trapping CoQ6 in micelles and prevent its correct delivery to the mitochondrial inner membrane; or (iii) impede incorporation of CoQ6 into the mitochondrial inner membrane by saturating the bilayer or transport proteins with polyisoprenoids; or (iv) interact with CoQ-utilizing proteins and prevent the proper function of imported exogenous CoQ6. It is also possible that the slight rescue deficiency observed in coq2Δ (Fig. 3C) is due to the presence of hexaprenyl-diphosphate in the membranes. The accumulation of CoQ prenylated intermediates is not as well explored in human cells as it is in yeast cells. However the accumulation of DMQ10 has been reported in fibroblasts derived from patients with inherited COQ4, COQ7, and COQ9 deficiencies [64, 68, 69], as well as in fibroblasts where COQ3, COQ5, or COQ6 were knocked down with siRNAs [64]. DMQ is considered a late-stage CoQ intermediate and it is not clear whether this precursor negatively affects CoQ rescue. However, if this is the case, patients with CoQ10 deficiencies that accumulate DMQ10 might not respond well to CoQ10 supplementation and alternative treatments, such as the induction of internal CoQ10 synthesis, should be considered instead. Based on our results, in addition to CoQ10 intermediates serving as a potentially important biomarker for the diagnosis of CoQ10 deficiency disorders [64], they may also impact selection of an appropriate treatment.
Our use of coq2Δ cells to screen for genes involved in CoQ6 transport avoids complications that could arise from accumulation of prenylated CoQ6 intermediates. This was evident in our analyses of TLG2, previously identified as required for CoQ6 uptake using coq3Δ cells. In our experiments, whereas deletion of TLG2 in a coq3Δ strain impaired rescue by exogenous CoQ6, the same deletion in a coq2Δ mutant had no significant impact on rescue, indicating that TLG2 does not play a significant role in CoQ6 transport in cells that do not accumulate HHB and HAB (Fig. 3).
Comparison of rescue by CoQ6 and CoQ2 provided a way to distinguish mutations that specifically impeded CoQ6 transport (preferentially rescued by CoQ2) from those that generally prevented utilization of CoQ. For example, in addition to TLG2, three other genes were identified using ORFΔcoq3Δ mutants: ERG2, PEP12 and VPS45 (30). Unlike deletion of TLG2, deletion of these genes in coq2Δ strains still reduced CoQ6 rescue, though not to the extent observed with coq3Δ cells. However, the coq2Δ strains harbouring deletions of ERG2, PEP12 or VPS45 were not preferentially rescued by CoQ2 (Fig. 6A), suggesting that defective transport of CoQ6 is not the underlying issue. Like the erg2Δcoq2Δ mutant, many other ORFΔcoq2Δ strains in Fig. 6A responded similarly to exogenous CoQ2 and CoQ6, indicating that the defect in these strains is also more likely to be related to CoQ utilization. Notably, pep12Δcoq2Δ and vps45Δcoq2Δ, as well as rpl31AΔcoq2Δ, responded better to CoQ6 than to CoQ2. The basis for this phenotype remains to be determined, but may be due to lower levels of delivery to the vacuole and degradation.
The Coq8 polypeptide is a member of an ancient atypical kinase family [70], with several conserved kinase motifs that are essential for CoQ biosynthesis [71, 72]. Unique characteristics have been attributed to Coq8 like the fact that its overexpression in coqΔ mutants augments the levels of several Coq polypeptides, stabilizes CoQ synthome formation and restores the formation of CoQ domains [73–77]. We find it intriguing that the coq8Δ mutant yeast also displayed an unique phenotype with regard to rescue by exogenous CoQ6. Its degree of rescue is lower as compared to the other coqΔ mutants (coq3Δ, coq5Δ - coq7Δ and coq9Δ; Fig. 3C), and the additional deletion of COQ2 only partially augmented rescue (Fig. 4D). Together, these observations might suggest an active role of Coq8 in CoQ6 transportation. Such a role is in agreement with previous studies, where Coq8 has been proposed to function as a chaperone that facilitates the partial extraction of lipophilic CoQ intermediates out of the inner mitochondrial membrane and into the aqueous matrix environment promoting the enzymatic reactions catalyzed by other Coq proteins [78].
Due to their endosymbiotic origin, mitochondria are not embedded within the vesicular trafficking system [79]. Instead, they are connected to other cellular organelles by multiple membrane contact sites [79, 80]. Recently, it has been described that the CoQ synthome is localized to distinct puncta within the inner mitochondrial membrane, termed CoQ domains [76]. The CoQ domains are non-randomly localized to adjacent ER-mitochondria contact sites [76, 81]. Moreover, it was observed that the disruption of ERMES alters the distribution of CoQ6 and its precursors [81]. These observations suggest a role for ER-mitochondria contacts in CoQ domain formation and CoQ6 production and distribution from mitochondria to the rest of the cell. Our screen to identify genes involved in CoQ6 transport revealed that ORFΔcoq2Δ strains missing an ERMES gene were not affected in their ability to be rescued by exogenous CoQ6. Deletion of genes belonging to the vCLAMP or the NVJ families also had no significant impact on rescue by exogenous CoQ6. These results may suggest that exogenous CoQ6 is reaching the mitochondria by a mechanism unrelated to these organelle connections, perhaps by means of the novel clathrin-mediated vesicle transport from the plasma membrane to the mitochondria [82], or via a still-unknown transport process. It is also possible that transport of CoQ6 through organelle connections occurs via redundant steps. It has been shown previously that for phospholipid exchange, mitochondria are dependent on at least one functional contact site with the endomembrane system: ERMES or vCLAMP, whose functions are tightly co-regulated [83]. In this case, the loss of one system elicits a compensatory response in the other, and loss of both is lethal. Thus, the cell is able to adapt and, in the absence of one structure, an increase in the other serves as a “backup” path for small-molecule transport [83].
We identified six ORFΔcoq2Δ double mutants that are compromised in their CoQ6 rescue, and perform better when diffusible CoQ2 is supplied instead (Figs. 5 and 6). These six ORFs (CDC10, RTS1, RVS161, RVS167, NAT3, and VPS1) encode proteins that are necessary for CoQ6 transport and identify essential steps that cannot be compensated by other proteins or pathways. Three of these proteins (Rvs161, Rvs167, and Vps1) are directly involved in the endocytosis process at the plasma membrane. Rvs161 and Rvs167 are Bin/amphiphysin/Rvs (BAR) domain proteins that form a heterodimeric protein complex capable of deforming the membrane, and are necessary for invagination and scission of vesicles in the final step of clathrin-mediated endocytosis [84]. In mammalian cells, in addition to Rvs161/167 homologues, the GTPase dynamin (Vps1 human homolog) is essential for scission of clathrin-coated vesicles. In yeast, the role of Vps1 in yeast endocytosis has been controversial [84], but recent reports demonstrated that Vps1 functions with the Rvs161/167 heterodimer to facilitate scission and release of the vesicles [61–63, 85]. Indeed, Vps1 and Rvs167 were found to physically interact with each other [63]. The rvs161Δ and rvs167Δ mutants have a specific defect: a significant fraction of the endocytic patches begins to be internalized and move inward from the cell surface but are then retracted toward the cortex [86]. This defective phenotype might impair the uptake of exogenous CoQ6, resulting in the poor rescue phenotype observed in rvs161Δcoq2Δ and rvs167Δcoq2Δ double mutants (Fig. 5 and 6A). Invaginations do not form correctly in the absence of Vps1 either [61]. Vps1 plays a significant role in endosomal trafficking, and the vps1Δ mutant accumulates endosomes in the cytoplasm [60]. Perhaps these attributes of the vps1Δ mutant account for the extremely deficient rescue phenotype of the vps1Δcoq2Δ double mutant (<5% of the positive control) (Fig. 6B).
Cdc10 was also identified in our screens as a protein involved in CoQ6 transport (Fig. 5 and 6A). Cdc10 is one of the four core septin proteins in S. cerevisiae, a group of GTPases that assemble into filaments and higher-order structures [87–89]. In yeast cells, septin filaments assemble at the bud neck in a ‘septin ring’, which is essential in cytokinesis during which it acts as a scaffold to recruit cytokinetic factors to the site of cell division [88, 90]. In addition to scaffolding, the other main function of septins is to serve as cortical barriers to prevent the free diffusion of membrane proteins between different compartments [88, 90]. Septins may play a role in endocytosis since they interact with a subset of endocytic proteins, including Vps1, and the accessory proteins Syp1 and Sla2 [91]. SEPT9, the human homolog of Cdc10, has been implicated in endosomal sorting [92]. Interaction partners previously identified suggested a role for SEPT9 in vesicle transport to and from the plasma membrane [93]. The other three core septins (Cdc3, Cdc11 and Cdc12) are encoded by essential genes and therefore the null mutants cannot be interrogated [94]. Whether the involvement of Cdc10 (or potentially other septins) in CoQ6 uptake/transport is related to the role of septins in endocytosis or a different septin function, will require further experimentation.
The remaining two proteins identified as necessary for CoQ6 transport, Rts1 and Nat3, may function as regulators. Rts1 is one of the two regulatory subunits of protein phosphatase 2A (PP2A), a major class of serine/threonine protein phosphatases that play an important role in many biological processes [95, 96]. The regulatory subunit is thought to determine substrate specificity, targeting PP2ARts1 for regulation of the mitotic spindle orientation checkpoint, cell size control, and septin ring organization and disassembly during cytokinesis [96]. Phosphoproteomic analyses in rts1Δ cells reveals 156 hyperphosphorylated proteins, highlighting the multitude of PP2ARts1 complex targets [97]. The cytokinesis defect observed in rts1Δ cells is in agreement with the role of PP2ARts1 in the regulation of septin dynamics, possibly by dephosphorylating some factor(s) at the bud neck [98]. PP2ARts1 reverses phosphorylation of (at least) septin Shs1 and contributes to timely septin disassembly after cytokinesis [90]. Yet-to-be-identified relationships between septins and PP2ARts1 are possible as suggested by data from other fungal species. AspB (the CDC3 ortholog in Aspergillus fumigatus) is dephosphorylated in a PP2A-dependent manner [99]. To the best of our knowledge, there is no direct association between PP2ARts1 and Cdc10, although Rts1 has a strong potential to serve as a regulator during CoQ6 transport even if it is not related with septins. Almost all endocytic proteins participating in clathrin-mediated endocytosis are phosphorylated [100], including Rvs161, Rvs167, and Vps1 [100–102], and are excellent targets for phospho/dephosphorylation regulation.
The final gene influencing CoQ6 uptake or transport is Nat3, the catalytic subunit of the NatB complex. Together with its auxiliary subunit (Mdm20), Nat3 mediates the N-terminal acetyl transfer of about 20% of proteins in yeast and humans [103–105]. In yeast, there are four functional NATs, and each has distinct substrate specificity [105, 106]. Could N-acetylation modifications play a role in some of the proteins involved in CoQ6 transport? This seems less likely given that mdm20Δcoq2Δ had profound rescue defects in response to both exogenous CoQ2 and CoQ6 (Fig. 6A), excluding MDM20 from consideration as an ORF related with CoQ6 transport. Thus, we believe that Nat3 and CoQ6 may be connected through an unknown, but NatB-independent, function of this protein. Although nat3Δ and mdm20Δ share several phenotypes, nat3Δ mutants possess additional unique phenotypes [104]. Moreover, Mdm20 has recently been related with several cellular functions independent of Nat3 [107, 108], supporting the idea that Nat3 participates in additional pathways, either by itself or in conjunction with another auxiliary protein. It is also possible that the deficient rescue displayed by mdm20Δcoq2Δ (Fig. 6A) is a consequence of multiple defects in this strain, and that N-acetylation modifications somehow regulate CoQ6 transport.
Conclusion
Our studies have revealed deleterious effects of CoQ6 hexaprenylated intermediates on rescue with exogenous CoQ6. Additionally, our research defines six novel proteins as necessary for appropriate CoQ6 uptake and transport, and strongly connects CoQ6 distribution with endocytosis and membrane trafficking pathways.
Acknowledgements
We thank the UCLA Molecular Instrumentation Core proteomics facility, and Dr. Yu Chen, for the use of the QTRAP4000 for lipid analysis. We thank undergraduate UCLA researcher Peter Lee for the creation of the pRSQ2 plasmid. We thank Kelly Quinn and Christopher P. Kampmeyer for their participation in the early stages of the project.
Funding Sources
This work was supported by the National Science Foundation Grant MCB-1330803 (CFC), the National Institutes of Health Grant T32 GM 007185 Ruth L. Kirschstein National Service (MCB), the Whitcome Individual Predoctoral Fellowship (MCB), and a UCLA Summer Undergraduate Research Fellowship Department of Chemistry & Biochemistry (MK). Work in the MPM laboratory is supported by the Medical Research Council UK (MC_U105663142) and by a Wellcome Trust Investigator award (110159/Z/15/Z).
Abbreviations
- CoQn
coenzyme Q containing a polyisoprenyl tail of n isoprene units
- CoQ6
oxidized coenzyme Q6
- CoQ6H2
reduced coenzyme Q6H2
- Coq1
polypeptide encoded by the COQ1 gene in Saccharomyces cerevisiae
- coqlΔ
yeast mutant harboring a deletion in the COQ1 gene
- DMQH2
demethoxy-QH2
- ERMES
endoplasmic reticulummitochondria encounter structure
- HAB
3-hexaprenyl-4-aminobenzoic acid
- 4HB
4-hydroxybenzoic acid
- HHB
3-hexaprenyl-4-hydroxybenzoic acid
- IDMQH2
4-imino-demethoxy-QH2
- LUVET
large unilamellar vesicles produced by extrusion
- mtDNA
mitochondrial DNA
- NatB
N-acetyltransferase B complex
- NVJ
nucleus vacuole junction
- ORFΔcoq2Δ
yeast double mutant harboring a gene deletion in a designated open reading frame plus a deletion in COQ2
- pABA
para-aminobenzoic acid
- PC
phosphatidylcholine
- Pyr12
1-pyrene dodecanoic acid
- SD
synthetic dextrose medium
- vCLAMP
vacuole-mitochondria patch
- WT
wild-type parental yeast strain
- YPD
rich growth medium containing dextrose as a fermentable carbon source
- YPG
rich growth medium contain glycerol as the sole non-fermentable carbon source
Footnotes
Conflict Of Interests
The authors declare no conflict of interest for any of the work presented in this article.
References
- [1].Awad AM, Bradley MC, Fernandez-del-Rio L, Nag A, Tsui H, Clarke CF. Coenzyme Q10 deficiencies: pathways in yeast and humans. Essays Biochem. 2018;62(3):361–376. doi: 10.1042/EBC20170106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Wang Y, Hekimi S. The Complexity of Making Ubiquinone. Trends in endocrinology and metabolism: TEM. 2019;30(12):923–943. doi: 10.1016/j.tem.2019.08.009. [DOI] [PubMed] [Google Scholar]
- [3].Bentinger M, Tekle M, Dallner G. Coenzyme Q-biosynthesis and functions. Biochemical and biophysical research communications. 2010;396(1):74–9. doi: 10.1016/j.bbrc.2010.02.147. [DOI] [PubMed] [Google Scholar]
- [4].Gutierrez-Mariscal FM, Yubero-Serrano EM, Villalba JM, Lopez-Miranda J. Coenzyme Q10: from bench to Clinic in Aging Diseases, a translational review. Critical reviews in food science and nutrition. 2018;59(14):2240–2257. doi: 10.1080/10408398.2018.1442316. [DOI] [PubMed] [Google Scholar]
- [5].Stefely JA, Pagliarini DJ. Biochemistry of Mitochondrial Coenzyme Q Biosynthesis. Trends in biochemical sciences. 2017;42(10):824–843. doi: 10.1016/j.tibs.2017.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Saiki R, Nagata A, Kainou T, Matsuda H, Kawamukai M. Characterization of solanesyl and decaprenyl diphosphate synthases in mice and humans. The FEBS journal. 2005;272(21):5606–22. doi: 10.1111/j.1742-4658.2005.04956.x. [DOI] [PubMed] [Google Scholar]
- [7].Marbois B, Xie LX, Choi S, Hirano K, Hyman K, Clarke CF. para-Aminobenzoic acid is a precursor in coenzyme Q6 biosynthesis in Saccharomyces cerevisiae . The Journal of biological chemistry. 2010;285(36):27827–38. doi: 10.1074/jbc.M110.151894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Pierrel F, Hamelin O, Douki T, Kieffer-Jaquinod S, Muhlenhoff U, Ozeir M, Lill R, Fontecave M. Involvement of mitochondrial ferredoxin and para-aminobenzoic acid in yeast coenzyme Q biosynthesis. Chemistry & biology. 2010;17(5):449–59. doi: 10.1016/j.chembiol.2010.03.014. [DOI] [PubMed] [Google Scholar]
- [9].Yubero D, Montero R, Santos-Ocana C, Salviati L, Navas P, Artuch R. Molecular diagnosis of coenzyme Q10 deficiency: an update. Expert review of molecular diagnostics. 2018;18(6):491–498. doi: 10.1080/14737159.2018.1478290. [DOI] [PubMed] [Google Scholar]
- [10].Yubero D, Allen G, Artuch R, Montero R. The Value of Coenzyme Q10 Determination in Mitochondrial Patients. Journal of clinical medicine. 2017;6(4):1–10. doi: 10.3390/jcm6040037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Alcazar-Fabra M, Trevisson E, Brea-Calvo G. Clinical syndromes associated with Coenzyme Q10 deficiency. Essays in biochemistry. 2018;62(3):377–398. doi: 10.1042/EBC20170107. [DOI] [PubMed] [Google Scholar]
- [12].Desbats MA, Lunardi G, Doimo M, Trevisson E, Salviati L. Genetic bases and clinical manifestations of coenzyme Q10 (CoQ10) deficiency. Journal of inherited metabolic disease. 2015;38(1):145–56. doi: 10.1007/s10545-014-9749-9. [DOI] [PubMed] [Google Scholar]
- [13].Doimo M, Desbats MA, Cerqua C, Cassina M, Trevisson E, Salviati L. Genetics of coenzyme Q10 deficiency. Molecular syndromology. 2014;5(3-4):156–62. doi: 10.1159/000362826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Littarru GP, Langsjoen P. Coenzyme Q10 and statins: biochemical and clinical implications. Mitochondrion. 2007;7(Suppl):S168–74. doi: 10.1016/j.mito.2007.03.002. [DOI] [PubMed] [Google Scholar]
- [15].Passi S, Stancato A, Aleo E, Dmitrieva A, Littarru GP. Statins lower plasma and lymphocyte ubiquinol without affecting other antioxidants and PUFA. BioFactors. 2003;18(1-4):113–24. doi: 10.1002/biof.5520180213. [DOI] [PubMed] [Google Scholar]
- [16].Barcelos IP, Haas RH. CoQ10 and Aging. Biology. 2019;8(2):1–22. doi: 10.3390/biology8020028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Hernández-Camacho JD, Bernier M, López-Lluch G, Navas P. Coenzyme Q10 Supplementation in Aging and Disease. Frontiers in physiology. 2018;9(44):1–11. doi: 10.3389/fphys.2018.00044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Oleck S, Ventura HO. Coenzyme Q10 and Utility in Heart Failure: Just Another Supplement? Current heart failure reports. 2016;13(4):190–5. doi: 10.1007/s11897-016-0296-6. [DOI] [PubMed] [Google Scholar]
- [19].Salviati L, Sacconi S, Murer L, Zacchello G, Franceschini L, Laverda AM, Basso G, Quinzii C, Angelini C, Hirano M, Naini AB, et al. Infantile encephalomyopathy and nephropathy with CoQ10 deficiency: a CoQ10-responsive condition. Neurology. 2005;65(4):606–8. doi: 10.1212/01.wnl.0000172859.55579.a7. [DOI] [PubMed] [Google Scholar]
- [20].Montini G, Malaventura C, Salviati L. Early coenzyme Q10 supplementation in primary coenzyme Q10 deficiency. The New England journal of medicine. 2008;358(26):2849–50. doi: 10.1056/NEJMc0800582. [DOI] [PubMed] [Google Scholar]
- [21].Acosta MJ, Vazquez Fonseca L, Desbats MA, Cerqua C, Zordan R, Trevisson E, Salviati L. Coenzyme Q biosynthesis in health and disease. Biochimica et biophysica acta. 2016;1857(8):1079–1085. doi: 10.1016/j.bbabio.2016.03.036. [DOI] [PubMed] [Google Scholar]
- [22].Lopez-Lluch G, Del Pozo-Cruz J, Sanchez-Cuesta A, Cortes-Rodriguez AB, Navas P. Bioavailability of coenzyme Q10 supplements depends on carrier lipids and solubilization. Nutrition (Burbank, Los Angeles County, Calif) 2018;57:133–140. doi: 10.1016/j.nut.2018.05.020. [DOI] [PubMed] [Google Scholar]
- [23].Zaki NM. Strategies for oral delivery and mitochondrial targeting of CoQ10 . Drug delivery. 2016;23(6):1868–81. doi: 10.3109/10717544.2014.993747. [DOI] [PubMed] [Google Scholar]
- [24].Lee JS, Suh JW, Kim ES, Lee HG. Preparation and Characterization of Mucoadhesive Nanoparticles for Enhancing Cellular Uptake of Coenzyme Q10 . Journal of agricultural and food chemistry. 2017;65(40):8930–8937. doi: 10.1021/acs.jafc.7b03300. [DOI] [PubMed] [Google Scholar]
- [25].Sato Y, Yokoyama S, Yamaki Y, Nishimura Y, Miyashita M, Maruyama S, Takekuma Y, Sugawara M. Enhancement of intestinal absorption of coenzyme Q10 using emulsions containing oleyl polyethylene acetic acids. European journal of pharmaceutical sciences : official journal of the European Federation for Pharmaceutical Sciences. 2019;142:105–144. doi: 10.1016/j.ejps.2019.105144. [DOI] [PubMed] [Google Scholar]
- [26].Herebian D, Lopez LC, Distelmaier F. Bypassing human CoQ10 deficiency. Molecular genetics and metabolism. 2018;123(3):289–291. doi: 10.1016/j.ymgme.2017.12.008. [DOI] [PubMed] [Google Scholar]
- [27].Berenguel Hernandez AM, de la Cruz M, Alcazar-Fabra M, Prieto-Rodriguez A, Sanchez-Cuesta A, Martin J, Tormo JR, Rodriguez-Aguilera JC, Cortes-Rodriguez AB, Navas P, Reyes F, et al. Design of High-Throughput Screening of Natural Extracts to Identify Molecules Bypassing Primary Coenzyme Q Deficiency in Saccharomyces cerevisiae . SLAS discovery : advancing life sciences R & D. 2019;21:1–11. doi: 10.1177/2472555219877185. [DOI] [PubMed] [Google Scholar]
- [28].Xie LX, Williams KJ, He CH, Weng E, Khong S, Rose TE, Kwon O, Bensinger SJ, Marbois BN, Clarke CF. Resveratrol and para-coumarate serve as ring precursors for coenzyme Q biosynthesis. Journal of lipid research. 2015;56(4):909–19. doi: 10.1194/jlr.M057919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Fernandez-Del-Rio L, Nag A, Gutierrez Casado E, Ariza J, Awad AM, Joseph AI, Kwon O, Verdin E, de Cabo R, Schneider C, Torres JZ, et al. Kaempferol increases levels of coenzyme Q in kidney cells and serves as a biosynthetic ring precursor. Free radical biology & medicine. 2017;110:176–187. doi: 10.1016/j.freeradbiomed.2017.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Takahashi T, Okamoto T, Mori K, Sayo H, Kishi T. Distribution of ubiquinone and ubiquinol homologues in rat tissues and subcellular fractions. Lipids. 1993;28(9):803–9. doi: 10.1007/BF02536234. [DOI] [PubMed] [Google Scholar]
- [31].Bentinger M, Dallner G, Chojnacki T, Swiezewska E. Distribution and breakdown of labeled coenzyme Q10 in rat. Free radical biology & medicine. 2003;34(5):563–75. doi: 10.1016/s0891-5849(02)01357-6. [DOI] [PubMed] [Google Scholar]
- [32].Fernandez-Ayala DJ, Brea-Calvo G, Lopez-Lluch G, Navas P. Coenzyme Q distribution in HL-60 human cells depends on the endomembrane system. Biochimica et biophysica acta. 2005;1713(2):129–37. doi: 10.1016/j.bbamem.2005.05.010. [DOI] [PubMed] [Google Scholar]
- [33].Padilla-Lopez S, Jimenez-Hidalgo M, Martin-Montalvo A, Clarke CF, Navas P, Santos-Ocana C. Genetic evidence for the requirement of the endocytic pathway in the uptake of coenzyme Q6 in Saccharomyces cerevisiae . Biochimica et biophysica acta. 2009;1788(6):1238–48. doi: 10.1016/j.bbamem.2009.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Santos-Ocana C, Do TQ, Padilla S, Navas P, Clarke CF. Uptake of exogenous coenzyme Q and transport to mitochondria is required for bc1 complex stability in yeast coq mutants. The Journal of biological chemistry. 2002;277(13):10973–81. doi: 10.1074/jbc.M112222200. [DOI] [PubMed] [Google Scholar]
- [35].Wang Y, Hekimi S. Mitochondrial respiration without ubiquinone biosynthesis. Human molecular genetics. 2013;22(23):4768–83. doi: 10.1093/hmg/ddt330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Weber C, Bysted A, Holmer G. Coenzyme Q10 in the diet-daily intake and relative bioavailability. Molecular aspects of medicine. 1997;18(Suppl):S251–4. doi: 10.1016/s0098-2997(97)00003-4. [DOI] [PubMed] [Google Scholar]
- [37].Santos-Ocana C, Villalba JM, Cordoba F, Padilla S, Crane FL, Clarke CF, Navas P. Genetic evidence for coenzyme Q requirement in plasma membrane electron transport. Journal of bioenergetics and biomembranes. 1998;30(5):465–75. doi: 10.1023/a:1020542230308. [DOI] [PubMed] [Google Scholar]
- [38].Jonassen T, Proft M, Randez-Gil F, Schultz JR, Marbois BN, Entian KD, Clarke CF. Yeast Clk-1 homologue (Coq7/Cat5) is a mitochondrial protein in coenzyme Q synthesis. The Journal of biological chemistry. 1998;273(6):3351–7. doi: 10.1074/jbc.273.6.3351. [DOI] [PubMed] [Google Scholar]
- [39].James AM, Cocheme HM, Murai M, Miyoshi H, Murphy MP. Complementation of coenzyme Q-deficient yeast by coenzyme Q analogues requires the isoprenoid side chain. The FEBS journal. 2010;277(9):2067–82. doi: 10.1111/j.1742-4658.2010.07622.x. [DOI] [PubMed] [Google Scholar]
- [40].MacDonald RC, MacDonald RI, Menco BP, Takeshita K, Subbarao NK, Hu LR. Small-volume extrusion apparatus for preparation of large, unilamellar vesicles. Biochimica et biophysica acta. 1991;1061(2):297–303. doi: 10.1016/0005-2736(91)90295-j. [DOI] [PubMed] [Google Scholar]
- [41].James AM, Sharpley MS, Manas AR, Frerman FE, Hirst J, Smith RA, Murphy MP. Interaction of the mitochondria-targeted antioxidant MitoQ with phospholipid bilayers and ubiquinone oxidoreductases. The Journal of biological chemistry. 2007;282(20):14708–18. doi: 10.1074/jbc.M611463200. [DOI] [PubMed] [Google Scholar]
- [42].James AM, Cocheme HM, Smith RA, Murphy MP. Interactions of mitochondria-targeted and untargeted ubiquinones with the mitochondrial respiratory chain and reactive oxygen species. Implications for the use of exogenous ubiquinones as therapies and experimental tools. The Journal of biological chemistry. 2005;280(22):21295–312. doi: 10.1074/jbc.M501527200. [DOI] [PubMed] [Google Scholar]
- [43].Baudin A, Ozier-Kalogeropoulos O, Denouel A, Lacroute F, Cullin C. A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae . Nucleic Acids Research. 1993;21(14):3329–3330. doi: 10.1093/nar/21.14.3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Gietz RD, Woods RA. Yeast transformation by the LiAc/SS Carrier DNA/PEG method. Methods in molecular biology (Clifton, NJ) 2006;313:107–20. doi: 10.1385/1-59259-958-3:107. [DOI] [PubMed] [Google Scholar]
- [45].Dirick L, Bendris W, Loubiere V, Gostan T, Gueydon E, Schwob E. Metabolic and environmental conditions determine nuclear genomic instability in budding yeast lacking mitochondrial DNA. G3 (Bethesda, Md) 2014;4(3):411–23. doi: 10.1534/g3.113.010108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Tsui HS, Pham NVB, Amer BR, Bradley MC, Gosschalk JE, Gallagher-Jones M, Ibarra H, Clubb RT, Blaby-Haas CE, Clarke CF. Human COQ10A and COQ10B are distinct lipid-binding START domain proteins required for coenzyme Q function. Journal of lipid research. 2019;60(7):1293–1310. doi: 10.1194/jlr.M093534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Stoscheck CM. Quantitation of protein. Methods in enzymology. 1990;182:50–68. doi: 10.1016/0076-6879(90)82008-p. [DOI] [PubMed] [Google Scholar]
- [48].Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae . Genetics. 1989;122(1):19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Harnly JM, Bhagwat S, Lin LZ. Profiling methods for the determination of phenolic compounds in foods and dietary supplements. Analytical and bioanalytical chemistry. 2007;389(1):47–61. doi: 10.1007/s00216-007-1424-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Hsieh EJ, Gin P, Gulmezian M, Tran UC, Saiki R, Marbois BN, Clarke CF. Saccharomyces cerevisiae Coq9 polypeptide is a subunit of the mitochondrial coenzyme Q biosynthetic complex. Arch Biochem Biophys. 2007;463(1):19–26. doi: 10.1016/j.abb.2007.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Cotan D, Cordero MD, Garrido-Maraver J, Oropesa-Avila M, Rodriguez-Hernandez A, Gomez Izquierdo L, De la Mata M, De Miguel M, Lorite JB, Infante ER, Jackson S, et al. Secondary coenzyme Q10 deficiency triggers mitochondria degradation by mitophagy in MELAS fibroblasts. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2011;25(8):2669–87. doi: 10.1096/fj.10-165340. [DOI] [PubMed] [Google Scholar]
- [52].Tran UC, Clarke CF. Endogenous synthesis of coenzyme Q in eukaryotes. Mitochondrion. 2007;7(Suppl):S62–71. doi: 10.1016/j.mito.2007.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Boettner DR, Chi RJ, Lemmon SK. Lessons from yeast for clathrin-mediated endocytosis. Nature cell biology. 2011;14(1):2–10. doi: 10.1038/ncb2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Michel AH, Kornmann B. The ERMES complex and ER-mitochondria connections. Biochem Soc Trans. 2012;40(2):445–50. doi: 10.1042/BST20110758. [DOI] [PubMed] [Google Scholar]
- [55].Tamura Y, Endo T. Role of Intra- and Inter-mitochondrial Membrane Contact Sites in Yeast Phospholipid Biogenesis. Advances in experimental medicine and biology. 2017;997:121–133. doi: 10.1007/978-981-10-4567-7_9. [DOI] [PubMed] [Google Scholar]
- [56].Tamura Y, Kawano S, Endo T. Organelle contact zones as sites for lipid transfer. Journal of biochemistry. 2019;165(2):115–123. doi: 10.1093/jb/mvy088. [DOI] [PubMed] [Google Scholar]
- [57].Robertson AS, Smythe E, Ayscough KR. Functions of actin in endocytosis. Cellular and molecular life sciences : CMLS. 2009;66(13):2049–65. doi: 10.1007/s00018-009-0001-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Lombardi R, Riezman H. Rvs161p and Rvs167p, the two yeast amphiphysin homologs, function together in vivo . The Journal of biological chemistry. 2001;276(8):6016–22. doi: 10.1074/jbc.M008735200. [DOI] [PubMed] [Google Scholar]
- [59].Ren G, Vajjhala P, Lee JS, Winsor B, Munn AL. The BAR domain proteins: molding membranes in fission, fusion, and phagy. Microbiology and molecular biology reviews : MMBR. 2006;70(1):37–120. doi: 10.1128/MMBR.70.1.37-120.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Williams M, Kim K. From membranes to organelles: emerging roles for dynamin-like proteins in diverse cellular processes. European journal of cell biology. 2014;93(7):267–77. doi: 10.1016/j.ejcb.2014.05.002. [DOI] [PubMed] [Google Scholar]
- [61].Smaczynska-de R, II, Allwood EG, Aghamohammadzadeh S, Hettema EH, Goldberg MW, Ayscough KR. A role for the dynamin-like protein Vps1 during endocytosis in yeast. J Cell Sci. 2010;123(Pt 20):3496–506. doi: 10.1242/jcs.070508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Wang D, Sletto J, Tenay B, Kim K. Yeast dynamin implicated in endocytic scission and the disassembly of endocytic components. Communicative & integrative biology. 2011;4(2):178–81. doi: 10.4161/cib.4.2.14257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Smaczynska-de R, II, Allwood EG, Mishra R, Booth WI, Aghamohammadzadeh S, Goldberg MW, Ayscough KR. Yeast dynamin Vps1 and amphiphysin Rvs167 function together during endocytosis. Traffic (Copenhagen, Denmark) 2012;13(2):317–28. doi: 10.1111/j.1600-0854.2011.01311.x. [DOI] [PubMed] [Google Scholar]
- [64].Herebian D, Seibt A, Smits SHJ, Bunning G, Freyer C, Prokisch H, Karall D, Wredenberg A, Wedell A, Lopez LC, Mayatepek E, et al. Detection of 6-demethoxyubiquinone in CoQ10 deficiency disorders: Insights into enzyme interactions and identification of potential therapeutics. Molecular genetics and metabolism. 2017;121(3):216–223. doi: 10.1016/j.ymgme.2017.05.012. [DOI] [PubMed] [Google Scholar]
- [65].Shukla S, Dubey KK. CoQ10 a super-vitamin: review on application and biosynthesis. 3 Biotech. 2018;8(5):249. doi: 10.1007/s13205-018-1271-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Garrido-Maraver J, Cordero MD, Oropesa-Avila M, Vega AF, de la Mata M, Pavon AD, Alcocer-Gomez E, Calero CP, Paz MV, Alanis M, de Lavera I, et al. Clinical applications of coenzyme Q10 . Frontiers in bioscience (Landmark edition) 2014;19:619–33. doi: 10.2741/4231. [DOI] [PubMed] [Google Scholar]
- [67].Beg S, Javed S, Kohli K. Bioavailability enhancement of coenzyme Q10: an extensive review of patents. Recent patents on drug delivery & formulation. 2010;4(3):245–55. doi: 10.2174/187221110793237565. [DOI] [PubMed] [Google Scholar]
- [68].Smith AC, Ito Y, Ahmed A, Schwartzentruber JA, Beaulieu CL, Aberg E, Majewski J, Bulman DE, Horsting-Wethly K, Koning DV, Rodenburg RJ, et al. C. Care4Rare Canada A family segregating lethal neonatal coenzyme Q10 deficiency caused by mutations in COQ9. Journal of inherited metabolic disease. 2018;41(4):719–729. doi: 10.1007/s10545-017-0122-7. [DOI] [PubMed] [Google Scholar]
- [69].Danhauser K, Herebian D, Haack TB, Rodenburg RJ, Strom TM, Meitinger T, Klee D, Mayatepek E, Prokisch H, Distelmaier F. Fatal neonatal encephalopathy and lactic acidosis caused by a homozygous loss-of-function variant in COQ9. European journal of human genetics : EJHG. 2016;24(3):450–4. doi: 10.1038/ejhg.2015.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Stefely JA, Reidenbach AG, Ulbrich A, Oruganty K, Floyd BJ, Jochem A, Saunders JM, Johnson IE, Minogue CE, Wrobel RL, Barber GE, et al. Mitochondrial ADCK3 employs an atypical protein kinase-like fold to enable coenzyme Q biosynthesis. Mol Cell. 2015;57(1):83–94. doi: 10.1016/j.molcel.2014.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Cheng W, Li W. Structural insights into ubiquinone biosynthesis in membranes. Science. 2014;343(6173):878–81. doi: 10.1126/science.1246774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Xie LX, Hsieh EJ, Watanabe S, Allan CM, Chen JY, Tran UC, Clarke CF. Expression of the human atypical kinase ADCK3 rescues coenzyme Q biosynthesis and phosphorylation of Coq polypeptides in yeast coq8 mutants. Biochim Biophys Acta. 2011;1811(5):348–60. doi: 10.1016/j.bbalip.2011.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].He CH, Xie LX, Allan CM, Tran UC, Clarke CF. Coenzyme Q supplementation or over-expression of the yeast Coq8 putative kinase stabilizes multi-subunit Coq polypeptide complexes in yeast coq null mutants. Biochimica et Biophysica Acta (BBA) - Molecular and Cell Biology of Lipids. 2014;1841(4):630–644. doi: 10.1016/j.bbalip.2013.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Xie LX, Ozeir M, Tang JY, Chen JY, Jaquinod SK, Fontecave M, Clarke CF, Pierrel F. Overexpression of the Coq8 kinase in Saccharomyces cerevisiae coq null mutants allows for accumulation of diagnostic intermediates of the coenzyme Q6 biosynthetic pathway. The Journal of biological chemistry. 2012;287(28):23571–81. doi: 10.1074/jbc.M112.360354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Tauche A, Krause-Buchholz U, Rodel G. Ubiquinone biosynthesis in Saccharomyces cerevisiae: the molecular organization of O-methylase Coq3p depends on Abc1p/Coq8p. FEMS Yeast Res. 2008;8(8):1263–75. doi: 10.1111/j.1567-1364.2008.00436.x. [DOI] [PubMed] [Google Scholar]
- [76].Subramanian K, Jochem A, Le Vasseur M, Lewis S, Paulson BR, Reddy TR, Russell JD, Coon JJ, Pagliarini DJ, Nunnari J. Coenzyme Q biosynthetic proteins assemble in a substrate-dependent manner into domains at ER-mitochondria contacts. The Journal of cell biology. 2019;218(4):1353–1369. doi: 10.1083/jcb.201808044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Bradley MC, Yang K, Fernandez-Del-Rio L, Ngo J, Ayer A, Tsui HS, Novales NA, Stocker R, Shirihai OS, Barros MH, Clarke CF. COQ11 deletion mitigates respiratory deficiency caused by mutations in the gene encoding the coenzyme Q chaperone protein Coq10. The Journal of biological chemistry. 2020 doi: 10.1074/jbc.RA119.012420. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Reidenbach AG, Kemmerer ZA, Aydin D, Jochem A, McDevitt MT, Hutchins PD, Stark JL, Stefely JA, Reddy T, Hebert AS, Wilkerson EM, et al. Conserved Lipid and Small-Molecule Modulation of COQ8 Reveals Regulation of the Ancient Kinase-like UbiB Family. Cell chemical biology. 2018;25(2):154–165. doi: 10.1016/j.chembiol.2017.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Ellenrieder L, Rampelt H, Becker T. Connection of Protein Transport and Organelle Contact Sites in Mitochondria. J Mol Biol. 2017;429(14):2148–2160. doi: 10.1016/j.jmb.2017.05.023. [DOI] [PubMed] [Google Scholar]
- [80].Dimmer KS, Rapaport D. Mitochondrial contact sites as platforms for phospholipid exchange. Biochimica et biophysica acta. 2017;1862(1):69–80. doi: 10.1016/j.bbalip.2016.07.010. [DOI] [PubMed] [Google Scholar]
- [81].Eisenberg-Bord M, Tsui HS, Antunes D, Fernandez-Del-Rio L, Bradley MC, Dunn CD, Nguyen TPT, Rapaport D, Clarke CF, Schuldiner M. The Endoplasmic Reticulum-Mitochondria Encounter Structure Complex Coordinates Coenzyme Q Biosynthesis. Contact (Thousand Oaks (Ventura County, Calif)) 2019;2:1–14. doi: 10.1177/2515256418825409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Wei Z, Su W, Lou H, Duan S, Chen G. Trafficking pathway between plasma membrane and mitochondria via clathrin-mediated endocytosis. Journal of molecular cell biology. 2018;10(6):539–548. doi: 10.1093/jmcb/mjy060. [DOI] [PubMed] [Google Scholar]
- [83].Elbaz-Alon Y, Rosenfeld-Gur E, Shinder V, Futerman AH, Geiger T, Schuldiner M. A dynamic interface between vacuoles and mitochondria in yeast. Developmental cell. 2014;30(1):95–102. doi: 10.1016/j.devcel.2014.06.007. [DOI] [PubMed] [Google Scholar]
- [84].Weinberg J, Drubin DG. Clathrin-mediated endocytosis in budding yeast. Trends in cell biology. 2012;22(1):1–13. doi: 10.1016/j.tcb.2011.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Nannapaneni S, Wang D, Jain S, Schroeder B, Highfill C, Reustle L, Pittsley D, Maysent A, Moulder S, McDowell R, Kim K. The yeast dynamin-like protein Vps1:vps1 mutations perturb the internalization and the motility of endocytic vesicles and endosomes via disorganization of the actin cytoskeleton. European journal of cell biology. 2010;89(7):499–508. doi: 10.1016/j.ejcb.2010.02.002. [DOI] [PubMed] [Google Scholar]
- [86].Kaksonen M, Toret CP, Drubin DG. A modular design for the clathrin- and actin-mediated endocytosis machinery. Cell. 2005;123(2):305–20. doi: 10.1016/j.cell.2005.09.024. [DOI] [PubMed] [Google Scholar]
- [87].Mostowy S, Cossart P. Septins: the fourth component of the cytoskeleton. Nature reviews Molecular cell biology. 2012;13(3):183–94. doi: 10.1038/nrm3284. [DOI] [PubMed] [Google Scholar]
- [88].Glomb O, Gronemeyer T. Septin Organization and Functions in Budding Yeast. Frontiers in cell and developmental biology. 2016;4:123. doi: 10.3389/fcell.2016.00123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].McMurray MA, Bertin A, Garcia G, 3rd, Lam L, Nogales E, Thorner J. Septin filament formation is essential in budding yeast. Developmental cell. 2011;20(4):540–9. doi: 10.1016/j.devcel.2011.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Juanes MA, Piatti S. The final cut: cell polarity meets cytokinesis at the bud neck in S. cerevisiae . Cellular and molecular life sciences : CMLS. 2016;73(16):3115–36. doi: 10.1007/s00018-016-2220-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Renz C, Oeljeklaus S, Grinhagens S, Warscheid B, Johnsson N, Gronemeyer T. Identification of Cell Cycle Dependent Interaction Partners of the Septins by Quantitative Mass Spectrometry. PloS one. 2016;11(2):e0148340. doi: 10.1371/journal.pone.0148340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Song K, Russo G, Krauss M. Septins As Modulators of Endo-Lysosomal Membrane Traffic. Frontiers in cell and developmental biology. 2016;4:124. doi: 10.3389/fcell.2016.00124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Hecht M, Rosler R, Wiese S, Johnsson N, Gronemeyer T. An Interaction Network of the Human SEPT9 Established by Quantitative Mass Spectrometry. G3 (Bethesda, Md) 2019;9(6):1869–1880. doi: 10.1534/g3.119.400197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Mela A, Momany M. Septin mutations and phenotypes in S. cerevisiae . Cytoskeleton (Hoboken, NJ) 2019;76(1):33–44. doi: 10.1002/cm.21492. [DOI] [PubMed] [Google Scholar]
- [95].Han Q, Pan C, Wang Y, Wang N, Wang Y, Sang J. The PP2A regulatory subunits, Cdc55 and Rts1, play distinct roles in Candida albicans’ growth, morphogenesis, and virulence. Fungal genetics and biology : FG & B. 2019;131:103240. doi: 10.1016/j.fgb.2019.103240. [DOI] [PubMed] [Google Scholar]
- [96].Arino J, Velazquez D, Casamayor A. Ser/Thr protein phosphatases in fungi: structure, regulation and function. Microbial cell (Graz, Austria) 2019;6(5):217–256. doi: 10.15698/mic2019.05.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Zapata J, Dephoure N, Macdonough T, Yu Y, Parnell EJ, Mooring M, Gygi SP, Stillman DJ, Kellogg DR. PP2ARts1 is a master regulator of pathways that control cell size. The Journal of cell biology. 2014;204(3):359–76. doi: 10.1083/jcb.201309119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Dobbelaere J, Gentry MS, Hallberg RL, Barral Y. Phosphorylation-dependent regulation of septin dynamics during the cell cycle. Developmental cell. 2003;4(3):345–57. doi: 10.1016/s1534-5807(03)00061-3. [DOI] [PubMed] [Google Scholar]
- [99].Vargas-Muniz JM, Renshaw H, Richards AD, Waitt G, Soderblom EJ, Moseley MA, Asfaw Y, Juvvadi PR, Steinbach WJ. Dephosphorylation of the Core Septin, AspB, in a Protein Phosphatase 2A- Dependent Manner Impacts Its Localization and Function in the Fungal Pathogen Aspergillus fumigatus . Frontiers in microbiology. 2016;7:997. doi: 10.3389/fmicb.2016.00997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Lu R, Drubin DG, Sun Y. Clathrin-mediated endocytosis in budding yeast at a glance. J Cell Sci. 2016;129(8):1531–6. doi: 10.1242/jcs.182303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Friesen H, Murphy K, Breitkreutz A, Tyers M, Andrews B. Regulation of the yeast amphiphysin homologue Rvs167p by phosphorylation. Molecular biology of the cell. 2003;14(7):3027–40. doi: 10.1091/mbc.E02-09-0613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Smaczynska-de R, II, Marklew CJ, Allwood EG, Palmer SE, Booth WI, Mishra R, Goldberg MW, Ayscough KR. Phosphorylation Regulates the Endocytic Function of the Yeast Dynamin-Related Protein Vps1. Mol Cell Biol. 2015;36(5):742–55. doi: 10.1128/MCB.00833-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Polevoda B, Cardillo TS, Doyle TC, Bedi GS, Sherman F. Nat3p and Mdm20p are required for function of yeast NatB Nalpha-terminal acetyltransferase and of actin and tropomyosin. The Journal of biological chemistry. 2003;278(33):30686–97. doi: 10.1074/jbc.M304690200. [DOI] [PubMed] [Google Scholar]
- [104].Singer JM, Shaw JM. Mdm20 protein functions with Nat3 protein to acetylate Tpm1 protein and regulate tropomyosin-actin interactions in budding yeast. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(13):7644–9. doi: 10.1073/pnas.1232343100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Aksnes H, Hole K, Arnesen T. Molecular, cellular, and physiological significance of N-terminal acetylation. International review of cell and molecular biology. 2015;316:267–305. doi: 10.1016/bs.ircmb.2015.01.001. [DOI] [PubMed] [Google Scholar]
- [106].Arnesen T, Van Damme P, Polevoda B, Helsens K, Evjenth R, Colaert N, Varhaug JE, Vandekerckhove J, Lillehaug JR, Sherman F, Gevaert K, et al. Proteomics analyses reveal the evolutionary conservation and divergence of N-terminal acetyltransferases from yeast and humans. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(20):8157–62. doi: 10.1073/pnas.0901931106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Yasuda K, Takahashi M, Mori N. Mdm20 Modulates Actin Remodeling through the mTORC2 Pathway via Its Effect on Rictor Expression. PloS one. 2015;10(11):e0142943. doi: 10.1371/journal.pone.0142943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Yasuda K, Ohyama K, Onga K, Kakizuka A, Mori N. Mdm20 stimulates polyQ aggregation via inhibiting autophagy through Akt-Ser473 phosphorylation. PloS one. 2013;8(12):e82523. doi: 10.1371/journal.pone.0082523. [DOI] [PMC free article] [PubMed] [Google Scholar]