Abstract
Telomerase is expressed in the majority (>85%) of tumours, but has restricted expression in normal tissues. Long-term telomerase inhibition in malignant cells results in progressive telomere shortening and reduction in cell proliferation. Here we report the synthesis and characterisation of radiolabeled oligonucleotides that target the RNA subunit of telomerase, hTR, simultaneously inhibiting enzymatic activity and delivering radiation intracellularly. Oligonucleotides complementary (match) and non-complementary (scramble or mismatch) to hTR were conjugated to diethylenetriaminepentaacetic dianhydride (DTPA), allowing radiolabeling with the Auger electron-emitting radionuclide indium-111 (111In). Match oligonucleotides inhibited telomerase activity with high potency which was not observed with scramble or mismatch oligonucleotides. DTPA-conjugation and 111In-labeling did not change telomerase inhibition. In telomerase-positive cancer cells, unlabeled match oligonucleotides had no effect on survival, however, 111In-labeled match oligonucleotides significantly reduced clonogenic survival and upregulated the DNA damage marker γH2AX. Minimal radiotoxicity and DNA damage was observed in telomerase-negative cells exposed to 111In-Match oligonucleotides. Match oligonucleotides localised in close proximity to nuclear Cajal bodies in telomerase-positive cells. In comparison to match oligonucleotides, 111In-Scramble or 111In-Mismatch oligonucleotides demonstrated reduced retention and negligible impact on cell survival. This study indicates the therapeutic activity of radiolabeled oligonucleotides that specifically target hTR through potent telomerase inhibition and DNA damage induction in telomerase-expressing cancer cells, and paves way for the development of novel oligonucleotide radiotherapeutics targeting telomerase-positive cancers.
Keywords: Telomerase, Radionuclide therapy, Indium-111, oligonucleotide, DNA damage
Introduction
Human telomeres consist of repetitive TTAGGG nucleotide sequences at the end of chromosomal DNA. These structures protect chromosomes from fusion, deterioration and recognition by DNA damage response proteins. In normal somatic cells, telomeres erode during each cell division as a consequence of the inability of unidirectional DNA synthesis to completely replicate the linear chromosomes in the so-called end replication problem (1, 2). When telomeres shorten to a critical length, cells undergo senescence or apoptosis, limiting the number of possible cell divisions (1, 2). In the case of cancer, cellular processes are activated to maintain telomere length, providing the cell with unrestricted cell division potential (3). In the majority of malignancies (>85%), telomere maintenance is the result of the upregulation of telomerase, a protein that has a restricted expression profile in normal tissue (4). Human telomerase is a ribonucleoprotein minimally composed of human telomerase reverse transcriptase (hTERT), a catalytic subunit, and a human telomerase RNA component (hTR), a template sequence-containing subunit encoding the telomeric repeats (5). The enzyme acts by catalysing the de novo addition of hexanucleotide telomeric repeats to the chromosome-ends, resulting in cell immortality (6). In addition, extra-telomeric properties of telomerase have also been uncovered. hTERT can influence tumour development, oncogenesis and inflammation via NF-κB and Wnt/β-catenin pathways (7). hTR has been shown to protect against apoptosis and oxidative stress (8, 9), and to activate DNA-dependent kinase (DNA-PKcs) to phosphorylate hnRNPA1, which is critical for capping telomeres (10, 11).
As a consequence of its differential expression in cancer versus normal tissue and its role in the maintenance of the malignant phenotype, telomerase is regarded as an attractive cancer therapeutic target and several telomerase-inhibition strategies have been developed. In particular, oligonucleotides complementary to the hTR template sequence act as catalytic inhibitors of telomerase and have shown promise in pre-clinical investigations (12–14). The use of oligonucleotides as therapeutic agents has been potentiated by the modulation of nuclease susceptibility, cell permeability and target affinity while oligonucleotide-based inhibitors offer exquisite specificity of action and high binding affinity, owing to the necessary formation of specific base-pairing during hybridisation (15). One such inhibitor (GRN163L, Imetelstat) is undergoing clinical trials in patients with a variety of cancer indications and positive outcomes have been reported in patients with essential thrombocytopenia and myelofibrosis (16, 17). Interestingly, it has been demonstrated that hTR inhibition results in chemo- and radiosensitisation (18–22), which is attributed to both telomere length-dependent and -independent mechanisms (23).
Based on these observations, we hypothesised that targeting hTR with a radiolabeled complementary oligonucleotide would provide a largely unexplored and potent approach for the targeted radiotherapy of telomerase-positive cancer. The localisation of telomerase in the nucleus creates an opportunity for targeting with short-range (nm scale) Auger electron-emitting radionuclides, such as Indium-111 (111In). High linear energy transfer (LET) Auger electrons are highly toxic when emitted in close proximity to DNA, but have little effect outside this range, thereby providing a potentially highly selective therapy with a low side-effect profile (24). In this study, we describe the design and biological characterisation of 111In-labeled oligonucleotides that target hTR, thereby presenting a novel strategy for targeting telomerase-positive tumours by combining selective telomerase inhibition and molecular radiotherapy in a single agent.
Materials And Methods
Reagents
All chemicals were purchased from Sigma-Aldrich, unless otherwise stated.
Cell culture
MDA-MB-435 (human melanoma), SKBR3 (human breast cancer), U2OS (osteosarcoma) and WI38 (human fibroblasts) cells were obtained from the American Type Culture Collection (ATCC). The MDA-MB-231/H2N human breast cancer cell line was a gift from Dr Robert Kerbel (University of Toronto). Cells were maintained in Dulbecco’s Modified Eagle’s Medium supplemented with 10% foetal bovine serum (FBS; Gibco) and 1% penicillin/streptomycin/glutamate. Cell line authentication (STR profiling) confirmed the identity of the cells used in this work. Cells were regularly checked for Mycoplasma infection and were not used beyond passage 20.
Oligonucleotide synthesis and labeling
2’ O-methyl RNA (2’OMeRNA) oligonucleotides, with or without 5’-amino linkers, were synthesised and HPLC-purified by Sigma-Aldrich. Sequences: Match 5’-CAGUUAGGGUUAG-3’, Scramble 5’-GCAGUGUGAUGAU-3’, Mismatch 1 5’-CAGUUACGCUUAG-3’, and Mismatch 2 5’-CAGAUACGCUUAG-3’. As a control, phosphorothioate (PS) DNA oligonucleotides were similarly obtained. Sequences: PS Match 5’-CAGTTAGGGTTAG-3’, PS Mismatch 5’-CAGTTAGAATTAG-3’ (mismatched bases are underlined).
For diethylenetriaminepentaacetic dianhydride (DTPA) conjugation, oligonucleotides with 5’-amino linkers were reconstituted in sodium bicarbonate buffer (0.1 M, pH 8.3). DTPA was combined with oligonucleotides in 20-fold molar excess and incubated at room temperature (RT) for 60 min. Fractions were separated by size exclusion chromatography (SEC) on a P4-column (BioRad) with sodium citrate buffer (0.1 M, pH 5). Oligonucleotide concentration was determined by measuring absorbance at 260 nm using a NanoDrop spectrometer (Thermo Fisher Scientific). Radiolabeling was performed in sodium citrate buffer by the addition of 0.2 MBq of [111In]InCl3 (subsequently referred to as 111InCl3) followed by incubation at RT for 60 min. Fractions were separated on a P4-column and gamma-counted for 60 seconds using a Wizard scintillation counter (Perkin-Elmer). Oligonucleotides were radiolabeled to the desired molar activity and the radiolabeling efficiency determined by instant thin layer chromatography (ITLC) in sodium citrate buffer (0.1 M, pH 5).
Oligonucleotides were linked to Cy3 for confocal microscopy studies. Oligonucleotides with 5’-amino linkers were reconstituted in sodium bicarbonate buffer (0.1 M, pH 8.3) to a final concentration of 220 μM. Monoreactive functional Cy3 dye (GE Healthcare) was reconstituted in DMSO and added to the oligonucleotide, before incubation for 60 minutes at RT. Reaction products were separated into 50 μL PBS fractions using P4 SEC, as before. Fractions were analysed using a NanoDrop spectrometer (Thermo Fisher Scientific) at 260 and 550 nm.
Telomeric repeat amplification protocol
The telomeric repeat amplification protocol (TRAP) assay (EMD-Millipore) was setup according to manufacturer’s instructions, with an optimised PCR cycle (30 °C, 30 min; 36 cycles of 94 °C / 30 sec; 53.5 °C / 30 seconds; 72 °C / 60 seconds; of 72 °C / 3 minutes), using 200 ng of protein from MDA-MB-435 cell lysate, unless otherwise stated. For inhibition experiments, inhibitor or control was added immediately prior to the telomerase extension step (30 °C, 30 min). Samples were analysed using a fluorescence plate reader (Tecan). The determined telomerase activity was normalised to untreated control.
For analysis of 111In-labeled oligonucleotides, the TRAP assay was modified for the accommodation of radioactive samples. In this system, DTPA-conjugated oligonucleotides were radiolabeled to a molar activity of 27 MBq/nmol and pre-incubated with cell lysate for 24 hours. Radiolabeled inhibitors were combined with MDA-MB-435 lysate at 27 ng/μL protein concentration in a total volume of 60 μL. Following incubation for 24 hours to allow for radioactive decay, inhibitor-enzyme complexes (7.5 μL) were added to 48 μL of TRAP PCR mix to give 200 ng of lysate protein and the final concentration of inhibitor as indicated. Data were fitted using fixed-slope non-linear regression and compared using an exact-sum-of-squares F-test and subject to one-way ANOVA with post hoc Tukey test.
Internalisation of radiolabeled oligonucleotides
Cells were seeded in a 24-well plate (2 x 104 cells/well). Oligonucleotides were combined with Transfast transfection reagent (Tfx) (Promega) according to the manufacturer’s protocol in antibiotic-free medium to a concentration of 220 nM and incubated for 15 minutes at RT. Cell-medium was removed, replaced with transfection complexes and incubated for the indicated time. Following incubation, medium was aspirated and retained. Cells were washed twice with PBS, and the washes combined with the medium to constitute the free-fraction. Cell membranes were washed using glycine buffer (0.1 M, pH 2.5) at 4 °C for 6 min and re-washed with PBS. To lyse the cells, sodium hydroxide (0.1 M) was added and plates incubated for 20 min at RT. The internalised fraction was collected and combined with two PBS washes. Fractions were counted using a Wizard gamma-counter (Perkin-Elmer).
Clonogenic assay
Cells were seeded in a 24-well plate (2 x 104 cells/well). Cells were transfected with 250 μL of 220 nM oligonucleotide for 2.5 h before addition of 250 μL medium and incubation for a further 24 h. Cells were harvested with trypsin (Gibco) following washing in PBS. Harvested cells were counted before plating in six-well plates at a density sufficient to give > 50 colonies for counting. Untreated cells were typically seeded at 750 cells/well. Colonies were grown for > 7 days, washed in PBS and stained with 1 % methylene blue (Alfa Aesar) in 50 % methanol (Thermo Fisher Scientific). Colonies containing > 50 cells were counted. The surviving fraction (SF) was calculated using the plating efficiency (PE) of untreated cells. Data were fitted using the linear-quadratic function and curves were compared using an exact-sum-of-squares F-test. Differences between treatment groups were analysed with a two-way ANOVA.
Cell viability assay
WI38 cells were seeded in a 24-well plate (1 x 104 cells/well). Cells were transfected with 250 μL of oligonucleotide (220 nM) for 2.5 h before addition of 250 μL medium and incubation for a further 24 h. At 24 h, the solution was removed and replaced with fresh medium. After 72 h, 0.5 mg/mL MTT (thiazolyl blue tetrazolium bromide) dissolved in serum-free medium was added for 120 minutes and the formazan product eluted using DMSO. Absorbance at 540 nm was quantified by plate reader (reference wavelength 650 nm) and the relative cell viability of cell populations determined as a fraction of mock-treated control.
Confocal microscopy
Cells were seeded in 8-well chamber slides (Thermo Fisher Scientific) (2 x 104 cells/well). Cells were transfected for the indicated time with 220 nM oligonucleotide in 100 μL, with an additional 100 μL of medium added after 2.5 h. A 137Cs-irradiator was used to deliver 4 Gy of γ-radiation to induce γH2AX foci, as a positive control for staining. Irradiated cells were incubated at 37 °C for 1 h to allow for foci induction. Immunostaining (anti-γH2AX 50-636-KC, Millipore and anti-coilin ab87913, Abcam) and confocal image acquisitions were performed as described previously (25). Intranuclear DNA damge foci were counted manually in > 100 cells per treatment. Nuclear Cy3 mean intensity was quantified from > 50 cells per treatment. Groups were compared by one-way ANOVA with a post hoc Tukey test. Images represent several fields of view acquired using a 40x/1.2 or 63x/1.4 objective lens, with 4x digital zoom.
Statistical analyses
Data were plotted as the mean ± standard deviation (SD) of the stated number of replicates, unless otherwise indicated. Statistical significance was determined using Graphpad Prism 6.03 software and is indicated as follows: * P < 0.05; ** P < 0.01; *** P < 0.001 (two-sided).
Results
Synthesis and stability of 111In-labeled oligonucleotides
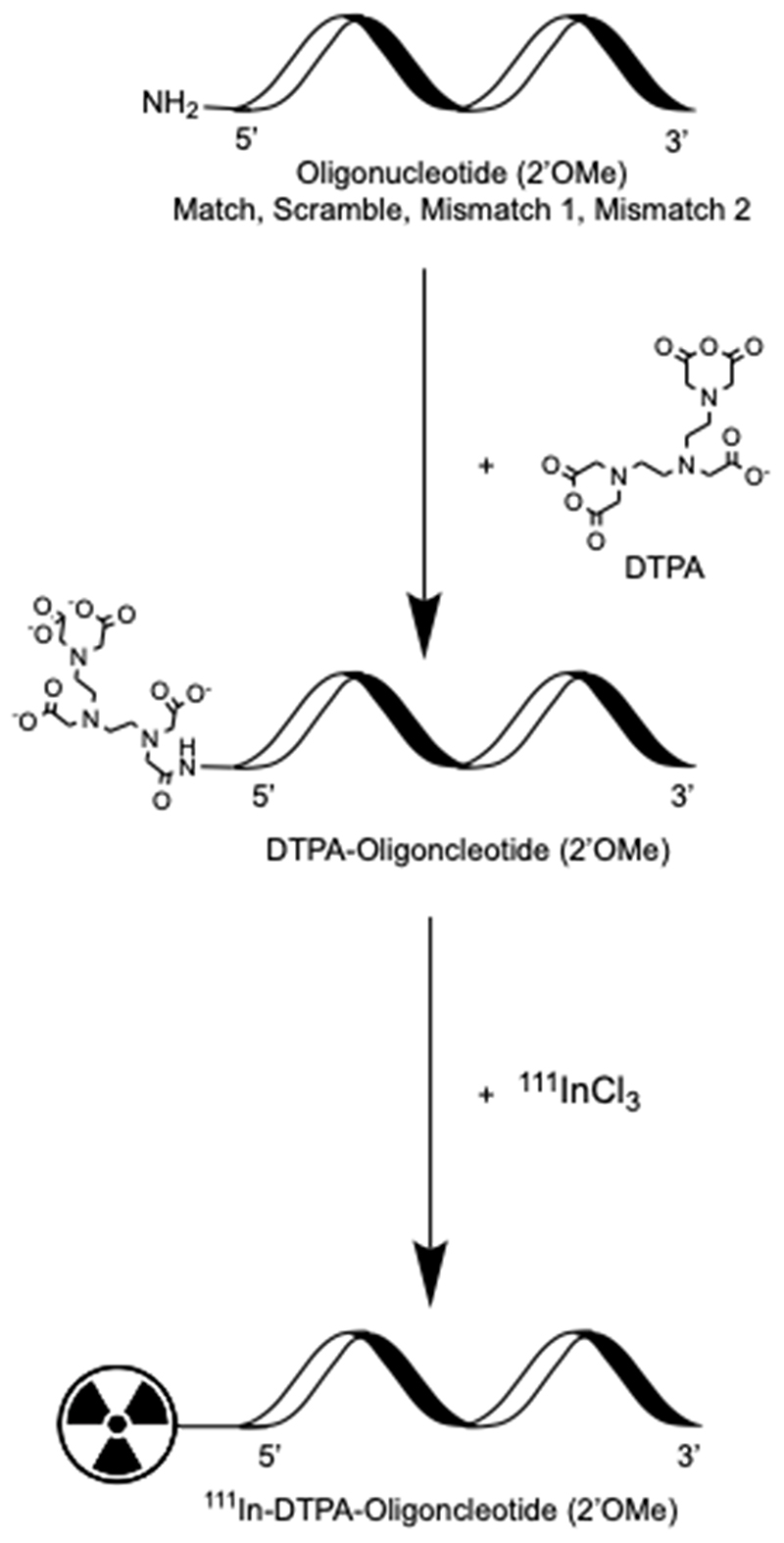
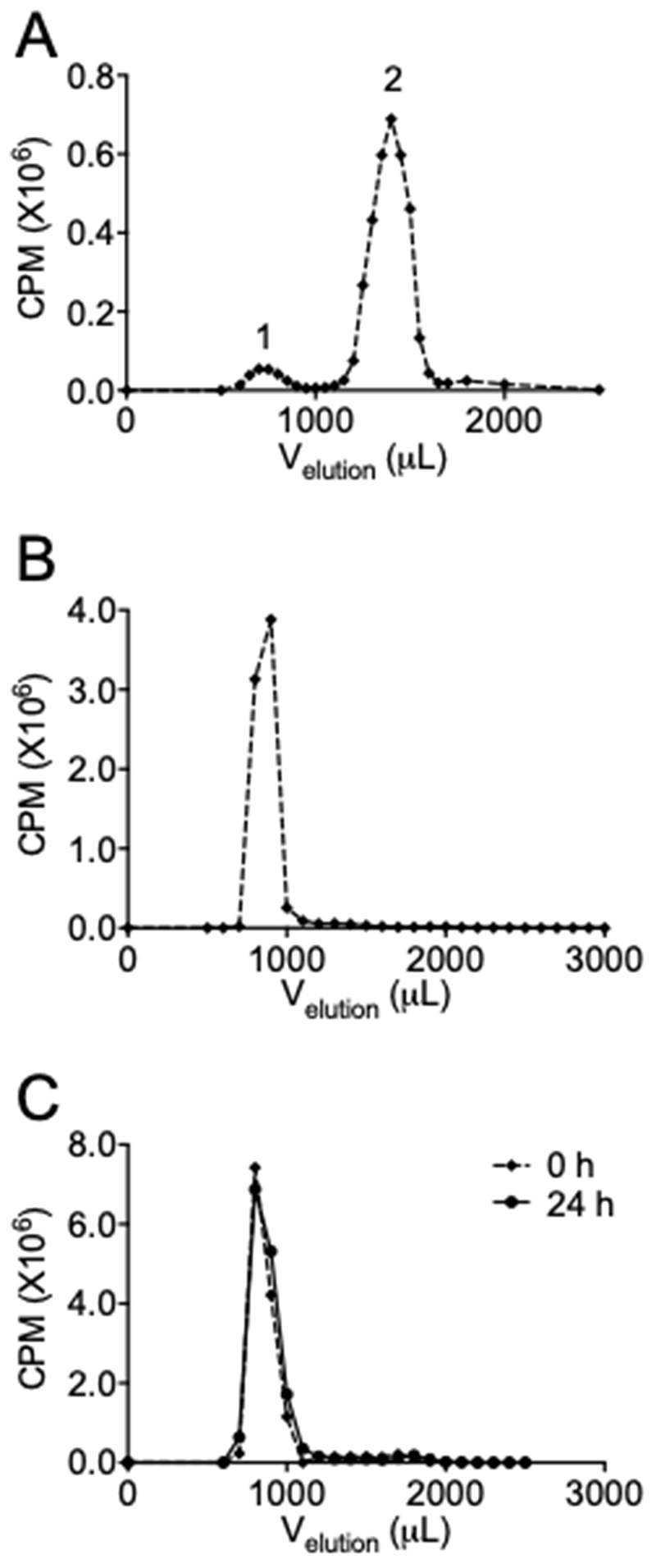
2’OMeRNA oligonucleotides were selected based on their resistance to nucleases and serum stability (26). Amino-modified 2’OMeRNA oligonucleotides containing a sequence complementary to the template region of hTR were conjugated to DTPA to enable subsequent labeling with 111In (Figure 1). The region of complementarity between the Match oligonucleotide and hTR is depicted in Table 1, which despite some overlap is distinct from the GRN163L site. To ensure sequence-specificity, several non-complementary control oligonucleotides (Mismatch 1, Mismatch 2, Scramble) were generated and labeled. Oligonucleotides were purified (or confirmed pure) by HPLC and DTPA conjugation was confirmed by mass spectrometry (Supplementary Figure S1), and the conjugation reaction products were diagnostically radiolabeled (0.2 MBq) before separation by SEC. Analysis of the reaction products by gamma-counting demonstrated the elution of two distinct radiopeaks, at 700 μL (peak 1) and 1400 μL (peak 2) (Figure 2A). Unreacted oligonucleotide was found to elute at 700 μL by absorbance measurement at 260 nm, and radiolabeled, unreacted DTPA at 1400 μL by gamma-counting. Reinjection of the fractions constituting peak 1 on a second SEC column verified the presence of a single radiospecies (Figure 2B). As expected, both the amino-modification and conjugation of DTPA were found to be necessary for oligonucleotide radiolabeling (Supplementary Figure S2).
Figure 1. Preparation of radiolabeled oligonucleotides.
Amino-modified 2’OMeRNA oligonucleotides of sequence complementary (Match) and non-complementary (Scramble, Mismatch) to hTR were conjugated to the chelator DTPA, which allowed for site-specific radiolabeling with Auger electron-emitting 111In.
Table 1. 2’O-MeRNA oligonucleotide sequences. The template region of hTR is shown in bold and mismatched bases are underlined. For reference, the sequence of GRN163L (Imetelstat) is also shown.
| hTR template (3’-5’) | GUCAAUCCCAAUCUGUU | - | - |
|
| |||
| GRN163L | TAGGGTTAGACAA | - | - |
| Match | CAGUUAGGGUUAG | 2’OMeRNA | Amine (AmC6) |
| Mismatch 1 | CAGUUACGCUUAG | 2’OMeRNA | Amine (AmC6) |
| Mismatch 2 | CAGAUACGCUUAG | 2’OMeRNA | Amine (AmC6) |
| Scramble | GCAGUGUGAUGAU | 2’OMeRNA | Amine (AmC6) |
Figure 2. Synthesis and stability of radiolabeled oligonucleotides.
(A) Oligonucleotides modified with a 5’-amino-linker were reacted with cyclic DTPA, radiolabeled with 0.2 MBq of 111In and separated by SEC. Peak 1 represents radiolabeled oligonucleotides, peak 2 unreacted DTPA. (B) SEC of peak 1. A single peak represents the existence of a single radiospecies. (C) Oligonucleotides were radiolabeled to 27 MBq/nmol and subjected to SEC immediately (92%) or after 24 hours (94%) to assess radiolysis of the construct. CPM – counts per minute.
To test the stability of the radiolabeled construct, the SEC profile was analysed following incubation for 0 or 24 hours. A single radiopeak was observed for both time points (Figure 2C). This peak accounted for 92 and 94% of the radioactivity at 0 and 24 hours respectively, determined by area under the curve analysis. These results indicated the short-range Auger electrons do not induce significant autoradiolysis of oligonucleotide constructs over this time period. High radiolabeling efficiency (>90%) was confirmed by ITLC and molar activities up to 27 MBq/nmol were employed for subsequent in vitro experiments.
hTR-targeted 111In-labeled oligonucleotides inhibit telomerase activity
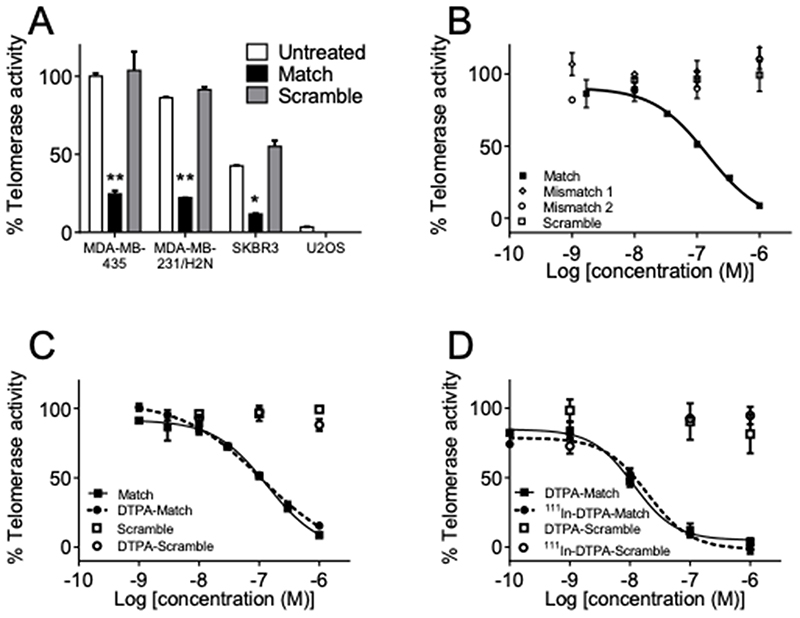
To assess telomerase inhibition by 111In-labeled oligonucleotides, relative telomerase activity was determined by a polymerase chain reaction (PCR) based telomeric repeat amplification protocol (TRAP) assay in a panel of cancer cell lines. MDA-MB-435 melanoma cells were found to have the highest telomerase activity, with MDA-MB-231/H2N and SKBR3 breast cancer cells expressing 86.3 ± 0.5 and 42.6 ± 0.4 % of that activity, respectively (Figure 3A). The osteosarcoma cell line, U2OS, was found to lack detectable telomerase activity in the TRAP assay, as described previously (27).
Figure 3. Oligonucleotides inhibit telomerase activity in a sequence-dependent manner.
(A) Telomerase activity was assayed in MDA-MB-435, MDA-MB-231/H2N, SKBR3 and U2OS cell lysate following treatment with 300 nM Match or Scramble oligonucleotide. Data were compared using one-way ANOVA with post hoc Tukey test. (B) Telomerase activity of MDA-MB-435 cell lysate after treatment with increasing concentration of Match, Mismatch 1, Mismatch 2, and Scramble oligonucleotides. (C) Telomerase activity of MDA-MB-435 cell lysate after treatment with increasing concentration of Match and Scramble oligonucleotides following DTPA-conjugation. (D) Telomerase activity of MDA-MB-435 cell lysate after pre-incubation with 111In-DTPA-Match and 111In-DTPA-Scramble (27 MBq/nmol) in the modified TRAP assay. Data were fitted using fixed-sloped non-linear regression and compared by F-test. ** P < 0.01, * P < 0.05. n=2, data represents three independent repeat experiments.
In each cell line, the effect of non-radiolabeled 2’OMeRNA Match oligonucleotide on telomerase activity was tested immediately following addition of the construct. Following normalisation to untreated control, inhibition of 75, 74 and 72 % was recorded in MDA-MB-435, MDA-MB-231/H2N and SKBR3 lysates incubated with Match oligonucleotide at a concentration of 300 nM, respectively (Figure 3A). In contrast, Scramble showed no inhibitory activity. The amplification of a pre-elongated control substrate, TSR8, was not affected by Match (Supplementary Figure S3), confirming inhibition was occurring during telomerase-dependent substrate elongation (28). Further characterisation showed that Match inhibited the telomerase activity of MDA-MB-435 lysate in a dose-dependent manner with maximum inhibition of 91% at a concentration of 1 μM and an IC50 of 147.4 nM (95% CI: 91.1–238.3 nM) (Figure 3B). DTPA-conjugation did not significantly alter the IC50 (101.4 nM, 95% CI: 75.9–135.6 nM) of Match (P > 0.05) (Figure 3C). Furthermore, in a TRAP assay modified for inclusion of radioactive samples, no difference in inhibitory potency was observed following radiolabeling with 111In (Figure 3D). In this modified assay where, as reported previously, pre-incubation with inhibitor leads to a perceived increase in potency (29), the IC50 values were found to be 19.6 nM (95% CI: 11.5–33.3 nM) for 111In-DTPA-Match (subsequently referred to as 111In-Match) and 11.5 nM (95% CI: 7.7–17.2 nM) for DTPA-Match (P > 0.05). Inhibition of > 96% was achieved with 111In-Match at a concentration of 1 μM. The IC50 of Match recorded in the modified TRAP assay was in agreement with previously published data, particularly where pre-incubation was explicitly employed (30). In all cases, disruption of oligonucleotide complementarity to hTR (Scramble, Mismatch 1, Mismatch 2) abrogated inhibitory activity, irrespective of modification with DTPA (Figure 3B, C, Supplementary Figure S4). Importantly, radiolabeling did not confer inhibitory capability on control oligonucleotides, with 111In-DTPA-Scramble (subsequently referred to as 111In-Scramble) lacking inhibitory activity (Figure 3D), confirming the preservation of sequence-specific telomerase inhibition.
To generalise these findings using an alternative oligonucleotide chemistry, we employed phosphorothioate (PS) DNA oligonucleotides as an additional control (29, 31). However, PS oligonucleotides were found to inhibit telomerase in a sequence-independent fashion (Supplementary Figure S5) and so were unsuitable for therapeutic development. This finding is in agreement with published work (29, 32).
Delivery of 111In-labeled oligonucleotides to cancer cells
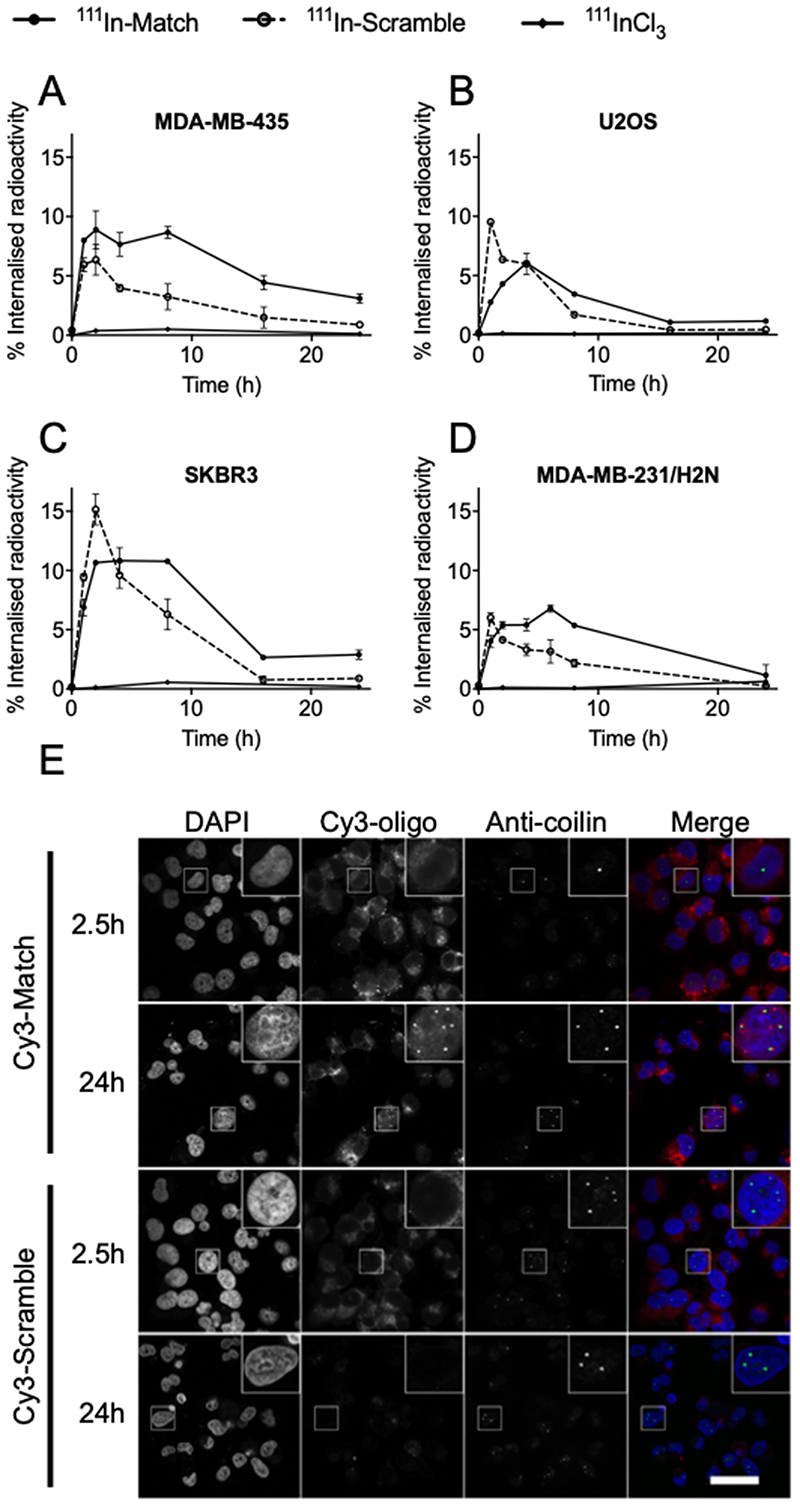
To facilitate in vitro cellular characterisation, 111In-Match and 111In-Scramble were introduced into cancer cells using liposomal transfection reagents. For each cell line, the transfection conditions were optimised to approximately equalise the peak delivery of 111In-Match and 111In-Scramble. Examining the uptake of oligonucleotides over a 24 h time period, maximum internalisation occurred after approximately 4 hours of treatment in MDA-MB-435, U2OS, SKBR3, and MDA-MB-231/H2N cells (Figure 4A–D). The peak uptake of 111In-Match achieved was 8.9 ± 1.6, 6.1 ± 0.1, 10.8 ± 1.1 and 6.8 ± 0.3 % of total administered radioactivity in MDA-MB-435, U2OS, SKBR3, and MDA-MB-231/H2N cells, respectively. Interestingly, while the maximum uptake for cell lines was comparable, 111In-Match was retained to a far greater extent than 111In-Scramble in the telomerase-positive cell lines (Figure 4A, C, D); an effect not apparent in telomerase-negative U2OS cells where both sequences demonstrated comparable temporal uptake profiles (Figure 4B).
Figure 4. Cellular uptake and subcellular distribution of hTR-targeting oligonucleotides.
(A-D) 250 μL of 111In-oligonucleotides (220 nM, 3.6 MBq/nmol) or 111InCl3 were combined with transfection reagent and incubated with (A) MDA-MB-435, (B) U2OS, (C) SKBR3 and (D) MDA-MB-231/H2N cells for the indicated time. Cells were washed and lysed, and the proportion of internalised radioactivity determined by gamma-counting. n=6, data from three independent repeat experiments. (E) Subcellular localisation of fluorophore-labeled oligonucleotide inhibitors of telomerase in MDA-MB-435 cells. Cy3-labeled Match and Scramble oligonucleotides (220 nM) were transfected into MDA-MB-435 cells for 2.5 or 24 hours before fixation. Cells were counterstained for the Cajal body protein coilin, and DNA (DAPI). Images represent data from three independent repeat experiments. Scale bar 40 μm.
hTR-targeted oligonucleotides enter the nucleus and associate with Cajal bodies
As telomerase is predominantly localised to the nucleus (1) and Auger electron-emitting radionuclides exert their therapeutic effects most effectively when in close proximity to DNA, the subcellular localisation of the oligonucleotides was investigated using fluorophore-labeled constructs in MDA-MB-435 cells. Cy3-labeled oligonucleotides were synthesised and purified by SEC (Supplementary Figure S6). Following transfection for 2.5 hours, a robust Cy3 signal was observed primarily in the cytoplasm of cells treated with Cy3-Match or Cy3-Scramble oligonucleotides (Figure 4E). In contrast, 24 hours after treatment, Cy3-Match oligonucleotides were retained, with a focal nuclear distribution apparent in approximately 30% of cells, while Cy3-Scramble treated cells showed a substantial decrease in signal intensity, which remained predominantly cytoplasmic (Figure 4E). Consistent with the uptake of radiolabeled oligonucleotides, a reduced signal was recorded in cells treated with label alone (Cy3), transfected unlabeled oligonucleotides or non-transfected Cy3-oligonucleotides (Supplementary Figure S7). Quantification of the oligonucleotide-Cy3 signal confirmed the preferential nuclear retention of Match over Scramble constructs after 24 hours (Supplementary Figure S8).
Previous studies have reported that hTR localises to subnuclear Cajal bodies during telomerase biosynthesis (33). Importantly, the punctate nuclear oligonucleotide signal that was observed in cells incubated with Cy3-Match for 24 hours was found to associate with the periphery of coilin, a protein marker of Cajal bodies (Figure 4E) (34). Such an association was absent in telomerase-negative U2OS cells (Supplementary Figure S9). These findings suggest telomerase-targeted oligonucleotides localise to and are retained at the nuclear sites of telomerase assembly.
hTR-targeted 111In-labeled oligonucleotides reduce the survival of telomerase-positive cancer cells
Next, the potential of 111In-Match to selectively reduce the survival of telomerase-positive cancer cells was assessed by clonogenic assay. MDA-MB-435 and U2OS cell lines were employed as telomerase-positive and telomerase-negative cells, respectively, while treatment with 111In-labeled non-complementary Scramble or Mismatch oligonucleotides were included to provide an indication of sequence-specificity. Transfection conditions were optimised to provide comparable levels of cellular uptake for each individual oligonucleotide; approximately 5-10% internalised radioactivity (Supplementary Figure S10).
Following normalisation to untreated control, the non-radiolabeled Match oligonucleotide (i.e. 0 MBq/nmol) was shown to have no effect on the survival of cancer cells (SF >0.8, Figure 5), a result in line with the notion that long-term treatment with existing telomerase inhibitors is necessary to lead to telomere erosion sufficient for therapeutic effect (1). This finding was confirmed in MDA-MB-435 cells treated with different concentrations of radiolabeled and non-labeled oligonucleotide (Supplementary Figure S11). A significant effect on clonogenic survival was observed only following treatment with radiolabeled Match (27 MBq/nmol), with non-labeled Match showing no therapeutic activity over this timeframe.
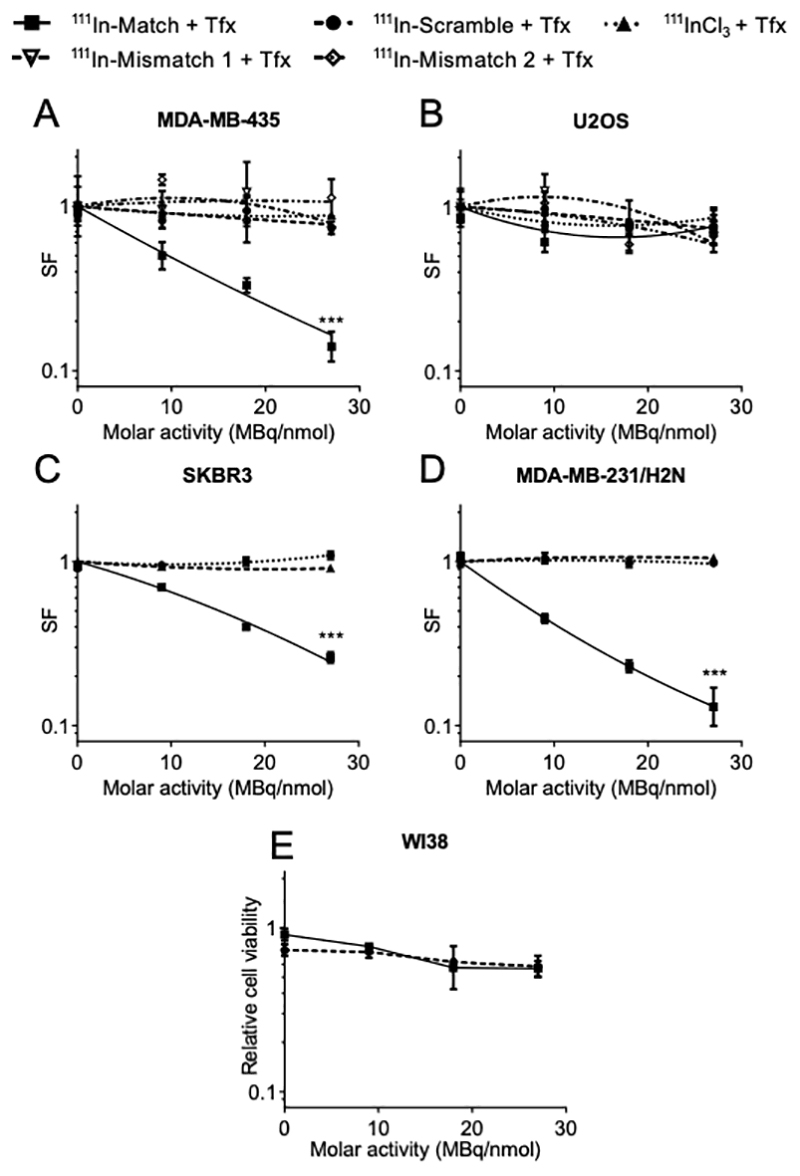
Figure 5. Radiolabeled oligonucleotide inhibitors of telomerase reduce clonogenic survival in a specific manner.
Oligonucleotides (220 nM in 250 μL) were radiolabeled to the indicated molar activity and transfected into (A) MDA-MB-435, (B) U2OS, (C) SKBR3 and (D) MDA-MB-231/H2N cells for 24 hours before plating for colony formation in the clonogenic assay. Colonies of > 50 cells were counted and the surviving fraction (SF) calculated. (E) Normal fibroblasts (WI38) were transfected with oligonucleotides (220 nM in 250 μL) radiolabeled to the indicated molar activity for 24 hours and cell viability determined 72 hours later. Data were normalised to untreated control. Treatment groups were compared using two-way ANOVA and F-test of linear quadratic fitting. *** P < 0.001, n=6, data from at least two independent repeat experiments.
As shown in Figure 5A, 111In-Match caused a significant reduction in the clonogenic survival of telomerase-positive MDA-MB-435 cells in an 111In-dependent manner, where treatment with the highest molar activity (27 MBq/nmol) resulted in a surviving fraction (SF) of 0.14 ± 0.03. In contrast, 111In-Scramble, 111In-Mismatch 1 and 111In-Mismatch 2 sequences had minimal impact on MDA-MB-435 survival even at high molar activities (SF >0.7 at 27 MBq/nmol). Likewise, 111InCl3 in the presence of transfection agent demonstrated no therapeutic activity. Importantly, 111In-Match did not significantly reduce the survival of telomerase-negative U2OS cells, with SF of 0.73 ± 0.16 at the maximum molar activity of 27 MBq/nmol (Figure 5B) but exhibited therapeutic activity towards two additional telomerase-positive cancer cell lines, SKBR3 and MDA-MB-231/H2N (Figure 5C, D). 111In-Match in the absence of liposomal transfection reagent failed to significantly reduce clonogenic survival at the equivalent levels of radioactivity, confirming the requirement for intracellular delivery of 111In for therapeutic activity (Supplementary Figure S12).
The potential for off-target toxicity of these novel radiopharmaceuticals was further investigated by measurement of the viability of treated normal fibroblasts. These cells cannot be analysed by clonogenic assay as they do not form colonies. In WI38 cells, shown to lack telomerase activity and exhibiting similar uptake of transfected radiopharmaceuticals (Supplementary Figure S13), oligonucleotides radiolabeled to 27 MBq/nmol had minimal effect on cell viability (Figure 5E). Furthermore, the modest reduction in viability occurred in an oligonucleotide sequence-independent manner, indicating a minimal and non-specific effect on normal cells.
Long-term telomerase inhibition is known to induce downregulation of hTERT and associated downstream cellular effects, for example those leading to altered regulation of the pro-apoptotic pathway (35). However, in the short timescale necessary for reduction in clonogenic survival following treatment with radiolabeled oligonucleotides, no effect on hTERT expression was observed (Supplementary Figure S14). These results indicate that hTR-targeted radiolabeled oligonucleotides induce rapid 111In-mediated radiotoxicity selectively towards telomerase-positive cancer cells.
hTR-targeted 111In-labeled oligonucleotides induce DNA damage specifically in telomerase-positive cancer cells
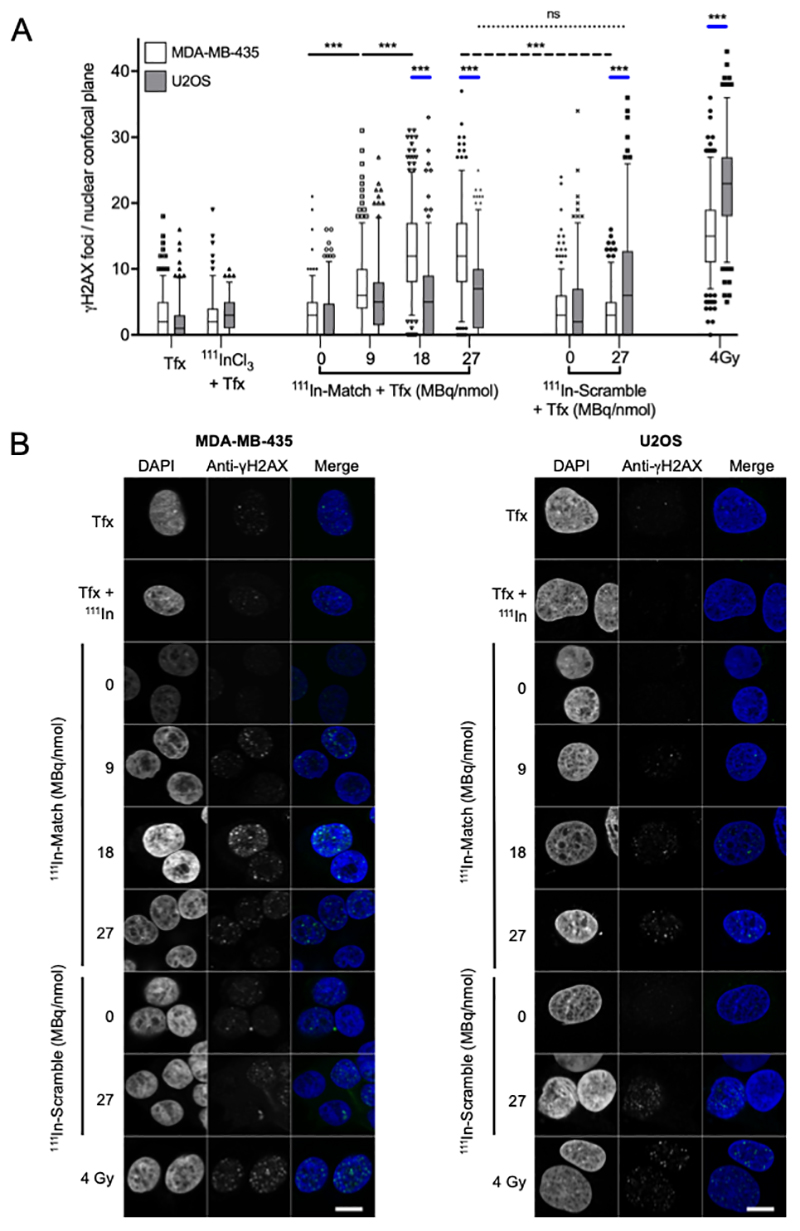
To further explore the preferential activity of 111In-labeled oligonucleotides towards telomerase-positive cancer cells, the upregulation of γH2AX was investigated. γH2AX is a protein involved in the recruitment of DNA damage repair factors and is a surrogate marker of DNA double-strand breaks (36). In MDA-MB-435 cells, a molar activity-dependent increase in γH2AX foci number was observed following treatment with 111In-Match (Figure 6A, B). Treatment with Match labeled to 0, 9, 18 and 27 MBq/nmol led to upregulation of γH2AX to 3.1 ± 3.1, 7.4 ± 5.3, 12.5 ± 6.5, and 12.7 ± 6.9 foci per nuclear confocal plane (NCP), respectively. In each case, the number of foci was significantly higher than in cells treated with Tfx alone (3.0 ± 3.3 foci/NCP) and Tfx + 111InCl3 (3.1 ± 2.5 foci/NCP). Notably, the effect of 111In-Scramble labeled to 27 MBq/nmol (3.6 ± 3.4 foci/NCP) was not significantly different from control groups, confirming the sequence-specific effect of 111In-Match.
Figure 6. γH2AX-induction in MDA-MB-435 and U2OS cell lines following treatment with radiolabeled oligonucleotides.
(A) MDA-MB-435 and U2OS cell lines were treated with radiolabeled oligonucleotides for 6 hours before fixation and staining for γH2AX. For quantification, foci contained within a single confocal plane of > 100 nuclei were counted. (B) Confocal microscopy images of MDA-MB-435 and U2OS cells treated with oligonucleotides/controls. Cells were stained for γH2AX and DNA (DAPI). γ-radiation (4 Gy) was used as a positive control for γH2AX staining. The number of γH2AX foci is statistically significantly greater in MDA-MB-435 cells following treatment with 111In-Match versus 111In-Scramble at 27 MBq/nmol (dashed line). Significant differences in the induction of γH2AX foci between cell lines treated with each construct and 4 Gy are indicated by blue lines. Scale bar 10 μm. Tfx -transfection reagent. Groups were compared using one-way ANOVA with post hoc Tukey test. *** P<0.001, ns -not significant. n>100, data from three independent repeat experiments. Line and box represent median and 25-75th percentile, respectively. Whiskers represent the 5-95th percentile.
In contrast, in telomerase-negative U2OS cells, upregulation of γH2AX foci number was not oligonucleotide-sequence dependent (Figure 6A, B) and both 111In-Match and 111In-Scramble upregulated γH2AX foci number to a modest extent compared to controls but there was no statistically significant difference between the two constructs, and no apparent dependency on molar activity (111In-Match: 6.1 ± 5.6, 6.1 ± 6, 7.2 ± 6 foci/NCP for 9, 18, and 27 MBq/nmol; 111In-Scramble: 8.2 ± 8.3 foci/NCP for 27 MBq/nmol and Tfx and Tfx+111InCl3: 2.0 ± 2.9 and 2.7 ± 3.3 foci/NCP). It was observed that for 111In-Scramble (27 MBq/nmol) and 4Gy γ-radiation there were statistically significantly more foci in U2OS than in MDA-MB-435 cells. We speculate that this may indicate greater DNA repair capacity in MDA-MB-435 compared to U2OS. Despite this, importantly, the induction of γH2AX foci by 111In-Match labeled to specific activities above 9 MBq/nmol was statistically significantly greater in MDA-MB-435 than in U2OS cells (P<0.001). The modest upregulation in foci number in U2OS cells, coupled to the lack of therapeutic activity of the oligonucleotides in these cells, likely reflects the non-specific, sublethal induction of DNA damage following internalisation of radiolabeled oligonucleotides. In U2OS cells, unlabeled Match and Scramble did not elicit a significant effect (2.6 ± 3.8 and 5.5 ± 5.7 foci/NCP, respectively). Together, these results are consistent with a telomerase- and 111In-dependent mechanism of DNA damage induction in telomerase-positive cancer cells.
Discussion
Since the original discovery of telomerase and the observation that it is expressed abundantly in malignant compared to normal cells, telomerase inhibition has been intensively investigated as an anticancer strategy, including in combination with external beam radiation or chemotherapy. It has been demonstrated that telomere shortening caused by down-regulation of telomerase results in radiosensitisation (18-23, 37-39), and, conversely, that increased telomerase expression is linked to apoptosis resistance and enhanced DNA repair (7). Telomerase protects aneuploid cells against replication stress, and promotes genomic stability in cancer cells (40, 41). Moreover, oxidative DNA damage, which is a major determinant of radiation toxicity, stimulates telomerase activity (42). We have shown previously that targeting hTERT with radiolabeled small molecule inhibitors is a viable anticancer strategy in which telomerase inhibition is achieved synchronously with delivery of a high radiation dose (43). Together, these findings provide a strong rationale for combining hTR inhibition with molecular radiotherapy for the treatment of neoplastic disease. Although the metal chelator DTPA has been shown to facilitate the therapeutic labeling of a wide range of biomolecules with the Auger electron-emitting radionuclide, 111In (44), the use of radiolabeled oligonucleotides has not been explored in detail. Yet oligonucleotide-based inhibitors offer high binding affinities and specificity which, if combined with the radiotoxicity of short-range emitters such as 111In, have the potential to generate gene-targeted radiotherapeutics. This concept was proposed by Neumann and co-workers, who examined sequence-specific triplex-forming oligonucleotides to carry 125I for this purpose (45, 46). The use of modified nucleic acids confers improved characteristics on therapeutic oligonucleotides. However, for therapeutic development, the selection of alternative chemistry must be carefully coordinated with the application. This necessitates the inclusion of stringent experimental controls, as exemplified by sequence-independent inhibition of telomerase activity mediated by PS oligonucleotides reported here and previously (32).
We demonstrate that DTPA-conjugation to the 5’-end of 2’OMe-modified oligonucleotides results in site-specific 111In-labeling to high molar activity (27 MBq/nmol). An oligonucleotide complementary to the template region of hTR inhibited telomerase activity in a sequence- and concentration-dependent manner, and inhibitory potency was not compromised by DTPA-conjugation or 111In-labeling. Upon cellular delivery, oligonucleotides targeted to hTR were retained in telomerase-positive cancer cells to a greater extent than the non-targeting control, while clonogenic assay data clearly showed a sequence-specific and dose-dependent effect of radiolabeled oligonucleotides on telomerase-positive cell survival. The rapid radiation-dependent loss of clonogenic survival demonstrated here offers a marked advantage over existing strategies reliant solely on inhibition of telomerase, which require long-term treatment for therapeutic effect (1). Crucially, telomerase-targeted radiolabeled oligonucleotides had little effect on the survival of telomerase-negative osteosarcoma cells and normal fibroblasts, despite internalisation occurring to a similar extent as in telomerase-positive cell lines. The normal fibroblast cell line, WI38, has low expression of hTR (47), and this is a possible explanation for its resistance to 111In-Match. These observations suggest a therapeutic window for the selective targeting of telomerase-positive cancer over normal tissue. At the activities assayed, a small fraction of cells survived treatment with the radiolabeled construct, possibly due to the stochastic nature of radioactive decay and the short range of Auger electrons, the existence of a radioresistant subpopulation of cells, or a failure of delivery. Use of alternative radionuclides and development of an efficient in vivo delivery system will be of future importance to address such questions.
Importantly, sub-cellular localisation studies revealed that 24 hours after treatment the non-hTR targeting oligonucleotides remained mainly cytoplasmic, whereas oligonucleotides complementary to hTR were found within the nucleus of MDA-MB-435 cells in distinct foci in close proximity to Cajal bodies. In agreement with our findings, the nuclear localisation of fluorophore-labeled 2’OMeRNA (48) and peptide nucleic acid (PNA) oligonucleotide telomerase inhibitors, has been reported previously in cancer and immortalised cells (49, 50). Furthermore, unassigned intranuclear foci were observed in a subset of cells (48, 49). The current study shows, for the first time, that oligonucleotide telomerase inhibitors accumulate in close association with nuclear Cajal bodies. Fluorescence in situ hybridisation staining for the RNA component of telomerase, performed post-fixation, revealed a cell cycle-dependent peripheral association between hTR and Cajal bodies (51, 52). This cell cycle-dependency likely explains the presence of oligonucleotide foci in only a proportion, about 30%, of cells in this study. From the perspective of the radiolabeled hTR oligonucleotides, the reported intranuclear accumulation is suggestive of specific target engagement, whilst also having the potential to localise the constructs in proximity to DNA and so maximise the effects of short-range Auger electron molecular radiotherapy.
Taken together, these findings demonstrate that the enhanced retention and nuclear localisation of radiolabeled oligonucleotides targeted to hTR results in a telomerase-dependent cytotoxic induction of DNA double-strand breaks by 111In. In addition, the role of telomerase in the DNA damage response and genomic stability offers the opportunity for simultaneous therapeutic exploitation of the reported interaction between telomerase inhibition and ionising radiation. It is also possible that perturbation of hTR function by 111In-Match results in inhibition of DNA-PKcs, a phenomenon that has been reported by others (10). DNA-PK plays a central role in non-homologous end joining (NHEJ), the major pathway for repair of ionising radiation-induced DNA double-strand breaks in human cells. Therefore, through its inhibition of hTR and, perhaps DNA-PKcs, 111In-Match may be impeding the repair of DNA lesions that it is itself causing.
This work supplements the emerging potential of Auger electron-emitting oligonucleotides as molecular radiotherapeutic agents (45, 53, 54). The intrinsic characteristics of oligonucleotide biotherapeutics, including high affinity and specificity of binding and intermediate molecular weight, offer therapeutic advantages over existing approaches. However, a recognised challenge of therapeutic oligonucleotides is that of effective drug delivery (55) and our current studies are aimed at developing nanoparticulate solutions for cancer cell-specific delivery of telomerase-targeted radio-oligonucleotides.
In conclusion, we report for the first time the development of an hTR subunit-targeting nucleic acid radiopharmaceutical for the selective treatment of cancer. The positive results of 111In-Match in telomerase inhibition and clonogenic survival studies provide a strong incentive for further development of telomerase-targeted molecular radiotherapy.
Supplementary Material
Significance.
Findings present a novel radiolabeled oligonucleotide for targeting telomerase-positive cancer cells that exhibits dual activity by simultaneously inhibiting telomerase and promoting radiation-induced genomic DNA damage.
Financial Support
This work was supported by Cancer Research-UK [C5255/A15935]; the Medical Research Council [MC_PC_12004]; the Engineering and Physical Sciences Research Council (EPSRC) Oxford Centre for Drug Delivery Devices [EP/L024012/1]; and the CR-UK/EPSRC Cancer Imaging Centre Oxford [C5255/A16466].
Footnotes
Conflict of Interest Disclosure Statement: KAV is a consultant for Blue Earth Diagnostics Ltd
References
- 1.Sekaran V, Soares J, Jarstfer MB. Telomere maintenance as a target for drug discovery. J Med Chem. 2014;57:521–538. doi: 10.1021/jm400528t. [DOI] [PubMed] [Google Scholar]
- 2.Martínez P, Blasco MA. Telomeric and extra-telomeric roles for telomerase and the telomere-binding proteins. Nat Rev Cancer. 2011;11:161–176. doi: 10.1038/nrc3025. [DOI] [PubMed] [Google Scholar]
- 3.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 4.Shay JW. Role of telomeres and telomerase in aging and cancer. Cancer Discov. 2016;6:584–593. doi: 10.1158/2159-8290.CD-16-0062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sauerwald A, Sandin S, Cristofari G, Scheres SHW, Lingner J, Rhodes D. Structure of active dimeric human telomerase. Nat Struct Mol Biol. 2013;20:454–460. doi: 10.1038/nsmb.2530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schmidt JC, Cech TR. Human telomerase: biogenesis, trafficking, recruitment, and activation. Genes Dev. 2015;29:1095–1105. doi: 10.1101/gad.263863.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li Y, Tergaonkar V. Noncanonical functions of telomerase: implications in telomerase-targeted cancer therapies. Cancer Res. 2014;74:1639–1644. doi: 10.1158/0008-5472.CAN-13-3568. [DOI] [PubMed] [Google Scholar]
- 8.Gazzaniga FS, Blackburn EH. An antiapoptotic role for telomerase RNA in human immune cells independent of telomere integrity or telomerase enzymatic activity. Blood. 2014;124:3675–3684. doi: 10.1182/blood-2014-06-582254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eitan E, Tamar A, Yossi G, Peleg R, Braiman A, Priel E. Expression of functional alternative telomerase RNA component gene in mouse brain and in motor neurons cells protects from oxidative stress. Oncotarget. 2016;7:78297–78309. doi: 10.18632/oncotarget.13049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ting NSY, Pohorelic B, Yu Y, Lees-Miller SP, Beattie TL. The human telomerase RNA component, hTR, activates the DNA-dependent protein kinase to phosphorylate heterogeneous nuclear ribonucleoprotein A1. Nucleic Acids Res. 2009;37:6105–6115. doi: 10.1093/nar/gkp636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sui J, Lin Y-F, Xu K, Lee K-J, Wang D, Chen BPC. DNA-PKcs phosphorylates hnRNP-A1 to facilitate the RPA-to-POT1 switch and telomere capping after replication. Nucleic Acids Res. 2015;43:5971–5983. doi: 10.1093/nar/gkv539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Herbert B, Pitts AE, Baker SI, Hamilton SE, Wright WE, Shay JW, Corey DR. Inhibition of human telomerase in immortal human cells leads to progressive telomere shortening and cell death. Proc Natl Acad Sci U S A. 1999;96:14276–14281. doi: 10.1073/pnas.96.25.14276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Herbert B-S, Gellert GC, Hochreiter A, Pongracz K, Wright WE, Zielinska D, Chin AC, Harley CB, Shay JW, Gryaznov SM. Lipid modification of GRN163, an N3’-->P5’ thio-phosphoramidate oligonucleotide, enhances the potency of telomerase inhibition. Oncogene. 2005;24:5262–5268. doi: 10.1038/sj.onc.1208760. [DOI] [PubMed] [Google Scholar]
- 14.Jafri MA, Ansari SA, Alqahtani MH, Shay JW. Roles of telomeres and telomerase in cancer, and advances in telomerase-targeted therapies. Genome Med. 2016;8:69. doi: 10.1186/s13073-016-0324-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kurreck J. Antisense technologies. Eur J Biochem. 2003;270:1628–1644. doi: 10.1046/j.1432-1033.2003.03555.x. [DOI] [PubMed] [Google Scholar]
- 16.Baerlocher GM, Oppliger Leibundgut E, Ottmann OG, Spitzer G, Odenike O, McDevitt MA, Röth A, Daskalakis M, Burington B, Stuart M, et al. Telomerase inhibitor imetelstat in patients with essential thrombocythemia. N Engl J Med. 2015;373:920–928. doi: 10.1056/NEJMoa1503479. [DOI] [PubMed] [Google Scholar]
- 17.Tefferi A, Lasho TL, Begna KH, Patnaik MM, Zblewski DL, Finke CM, Laborde RR, Wassie E, Schimek L, Hanson CA, et al. A pilot study of the telomerase inhibitor imetelstat for myelofibrosis. N Engl J Med. 2015;373:908–919. doi: 10.1056/NEJMoa1310523. [DOI] [PubMed] [Google Scholar]
- 18.Yu C, Yu Y, Xu Z, Li H, Yang D, Xiang M, Zuo Y, Li S, Chen Z, Yu Z. Antisense oligonucleotides targeting human telomerase mRNA increases the radiosensitivity of nasopharyngeal carcinoma cells. Mol Med Rep. 2015;11:2825–2830. doi: 10.3892/mmr.2014.3105. [DOI] [PubMed] [Google Scholar]
- 19.Wu X, Smavadati S, Nordfjäll K, Karlsson K, Qvarnström F, Simonsson M, Bergqvist M, Gryaznov S, Ekman S, Paulsson-Karlsson Y. Telomerase antagonist imetelstat inhibits esophageal cancer cell growth and increases radiation-induced DNA breaks. Biochim Biophys Acta. 2012;1823:2130–2135. doi: 10.1016/j.bbamcr.2012.08.003. [DOI] [PubMed] [Google Scholar]
- 20.Goldblatt EM, Gentry ER, Fox MJ, Gryaznov SM, Shen C, Herbert B-S. The telomerase template antagonist GRN163L alters MDA-MB-231 breast cancer cell morphology, inhibits growth, and augments the effects of paclitaxel. Mol Cancer Ther. 2009;8:2027–2035. doi: 10.1158/1535-7163.MCT-08-1188. [DOI] [PubMed] [Google Scholar]
- 21.Ji X, Xie C, Fang M, Zhou F, Zhang W, Zhang M, Zhou Y. Efficient inhibition of human telomerase activity by antisense oligonucleotides sensitizes cancer cells to radiotherapy. Acta Pharmacol Sin. 2006;27:1185–1191. doi: 10.1111/j.1745-7254.2006.00417.x. [DOI] [PubMed] [Google Scholar]
- 22.Gomez-Millan J, Goldblatt EM, Gryaznov SM, Mendonca MS, Herbert B-S. Specific telomere dysfunction induced by GRN163L increases radiation sensitivity in breast cancer cells. Int J Radiat Oncol Biol Phys. 2007;67:897–905. doi: 10.1016/j.ijrobp.2006.09.038. [DOI] [PubMed] [Google Scholar]
- 23.Berardinelli F, Coluzzi E, Sgura A, Antoccia A. Targeting telomerase and telomeres to enhance ionizing radiation effects in in vitro and in vivo cancer models. Mutat Res. 2017;773:204–219. doi: 10.1016/j.mrrev.2017.02.004. [DOI] [PubMed] [Google Scholar]
- 24.Cornelissen B, Vallis KA. Targeting the nucleus: an overview of Auger-electron radionuclide therapy. Curr Drug Discov Technol. 2010;7:263–279. doi: 10.2174/157016310793360657. [DOI] [PubMed] [Google Scholar]
- 25.Gill MR, Harun SN, Halder S, Boghozian RA, Ramadan K, Ahmad H, Vallis KA. A ruthenium polypyridyl intercalator stalls DNA replication forks, radiosensitizes human cancer cells and is enhanced by Chk1 inhibition. Sci Rep. 2016;6:31973. doi: 10.1038/srep31973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Geary RS, Norris D, Yu R, Bennett CF. Pharmacokinetics, biodistribution and cell uptake of antisense oligonucleotides. Adv Drug Deliv Rev. 2015;87:46–51. doi: 10.1016/j.addr.2015.01.008. [DOI] [PubMed] [Google Scholar]
- 27.Bryan TM, Englezou A, Dalla-Pozza L, Dunham MA, Reddel RR. Evidence for an alternative mechanism for maintaining telomere length in human tumors and tumor-derived cell lines. Nat Med. 1997;3:1271–1274. doi: 10.1038/nm1197-1271. [DOI] [PubMed] [Google Scholar]
- 28.Piotrowska K, Kleideiter E, Mürdter TE, Taetz S, Baldes C, Schaefer U, Lehr C-M, Klotz U. Optimization of the TRAP assay to evaluate specificity of telomerase inhibitors. Lab Investig J Tech Methods Pathol. 2005;85:1565–1569. doi: 10.1038/labinvest.3700352. [DOI] [PubMed] [Google Scholar]
- 29.Norton JC, Piatyszek MA, Wright WE, Shay JW, Corey DR. Inhibition of human telomerase activity by peptide nucleic acids. Nat Biotechnol. 1996;14:615–619. doi: 10.1038/nbt0596-615. [DOI] [PubMed] [Google Scholar]
- 30.Pitts AE, Corey DR. Inhibition of human telomerase by 2’-O-methyl-RNA. Proc Natl Acad Sci U S A. 1998;95:11549–11554. doi: 10.1073/pnas.95.20.11549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mata JE, Joshi SS, Palen B, Pirruccello SJ, Jackson JD, Elias N, Page TJ, Medlin KL, Iversen PL. A hexameric phosphorothioate oligonucleotide telomerase inhibitor arrests growth of Burkitt’s lymphoma cells in vitro and in vivo. Toxicol Appl Pharmacol. 1997;144:189–197. doi: 10.1006/taap.1997.8103. [DOI] [PubMed] [Google Scholar]
- 32.Matthes E, Lehmann C. Telomerase protein rather than its RNA is the target of phosphorothioate-modified oligonucleotides. Nucleic Acids Res. 1999;27:1152–1158. doi: 10.1093/nar/27.4.1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhu Y, Tomlinson RL, Lukowiak AA, Terns RM, Terns MP. Telomerase RNA accumulates in Cajal bodies in human cancer cells. Mol Biol Cell. 2004;15:81–90. doi: 10.1091/mbc.E03-07-0525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Raska I, Ochs RL, Andrade LE, Chan EK, Burlingame R, Peebles C, Gruol D, Tan EM. Association between the nucleolus and the coiled body. J Struct Biol. 1990;104:120–127. doi: 10.1016/1047-8477(90)90066-l. [DOI] [PubMed] [Google Scholar]
- 35.Rubis B, Holysz H, Gladych M, Toton E, Paszel A, Lisiak N, Kaczmarek M, Hofmann J, Rybczynska M. Telomerase downregulation induces proapoptotic genes expression and initializes breast cancer cells apoptosis followed by DNA fragmentation in a cell type dependent manner. Mol Biol Rep. 2013;40:4995–5004. doi: 10.1007/s11033-013-2600-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem. 1998;273:5858–5868. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- 37.Millet P, Granotier C, Etienne O, Boussin FD. Radiation-induced upregulation of telomerase activity escapes PI3-kinase inhibition in two malignant glioma cell lines. Int J Oncol. 2013;43:375–382. doi: 10.3892/ijo.2013.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Castella M, Puerto S, Creus A, Marcos R, Surralles J. Telomere length modulates human radiation sensitivity in vitro. Toxicol Lett. 2007;172:29–36. doi: 10.1016/j.toxlet.2007.05.012. [DOI] [PubMed] [Google Scholar]
- 39.Goytisolo FA, Samper E, Martín-Caballero J, Finnon P, Herrera E, Flores JM, Bouffler SD, Blasco MA. Short telomeres result in organismal hypersensitivity to ionizing radiation in mammals. J Exp Med. 2000;192:1625–1636. doi: 10.1084/jem.192.11.1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Meena JK, Cerutti A, Beichler C, Morita Y, Bruhn C, Kumar M, Kraus JM, Speicher MR, Wang Z-Q, Kestler HA, et al. Telomerase abrogates aneuploidy-induced telomere replication stress, senescence and cell depletion. EMBO J. 2015;34:1371–1384. doi: 10.15252/embj.201490070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Akiyama M, Yamada O, Kanda N, Akita S, Kawano T, Ohno T, Mizoguchi H, Eto Y, Anderson KC, Yamada H. Telomerase overexpression in K562 leukemia cells protects against apoptosis by serum deprivation and double-stranded DNA break inducing agents, but not against DNA synthesis inhibitors. Cancer Lett. 2002;178:187–197. doi: 10.1016/s0304-3835(01)00838-2. [DOI] [PubMed] [Google Scholar]
- 42.Lee H-T, Bose A, Lee C-Y, Opresko PL, Myong S. Molecular mechanisms by which oxidative DNA damage promotes telomerase activity. Nucleic Acids Res. 2017;45:11752–11765. doi: 10.1093/nar/gkx789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Waghorn PA, Jackson MR, Gouverneur V, Vallis KA. Targeting telomerase with radiolabeled inhibitors. Eur J Med Chem. 2017;125:117–129. doi: 10.1016/j.ejmech.2016.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cornelissen B, Darbar S, Kersemans V, Allen D, Falzone N, Barbeau J, Smart S, Vallis KA. Amplification of DNA damage by a ΓH2AX-targeted radiopharmaceutical. Nucl Med Biol. 2012;39:1142–1151. doi: 10.1016/j.nucmedbio.2012.06.001. [DOI] [PubMed] [Google Scholar]
- 45.Sedelnikova OA, Karamychev VN, Panyutin IG, Neumann RD. Sequence-specific gene cleavage in intact mammalian cells by 125I-labeled triplex-forming oligonucleotides conjugated with nuclear localization signal peptide. Antisense Nucleic Acid Drug Dev. 2002;12:43–49. doi: 10.1089/108729002753670256. [DOI] [PubMed] [Google Scholar]
- 46.Sedelnikova OA, Luu AN, Karamychev VN, Panyutin IG, Neumann RD. Development of DNA-based radiopharmaceuticals carrying Auger-electron emitters for antigene radiotherapy. Int J Radiat Oncol Biol Phys. 2001;49:391–396. doi: 10.1016/s0360-3016(00)01486-3. [DOI] [PubMed] [Google Scholar]
- 47.Atkinson SP, Hoare SF, Glasspool RM, Keith WN. Lack of telomerase gene expression in alternative lengthening of telomere cells is associated with chromatin remodeling of the hTR and hTERT gene promoters. Cancer Res. 2005;65:7585–7590. doi: 10.1158/0008-5472.CAN-05-1715. [DOI] [PubMed] [Google Scholar]
- 48.Beisner J, Dong M, Taetz S, Nafee N, Griese E-U, Schaefer U, Lehr C-M, Klotz U, Mürdter TE. Nanoparticle mediated delivery of 2’-O-methyl-RNA leads to efficient telomerase inhibition and telomere shortening in human lung cancer cells. Lung Cancer. 2010;68:346–354. doi: 10.1016/j.lungcan.2009.07.010. [DOI] [PubMed] [Google Scholar]
- 49.Shammas MA, Simmons CG, Corey DR, Reis RJS. Telomerase inhibition by peptide nucleic acids reverses `immortality’ of transformed human cells. Oncogene. 1999;18:1203069. doi: 10.1038/sj.onc.1203069. [DOI] [PubMed] [Google Scholar]
- 50.Villa R, Folini M, Lualdi S, Veronese S, Daidone MG, Zaffaroni N. Inhibition of telomerase activity by a cell-penetrating peptide nucleic acid construct in human melanoma cells. FEBS Lett. 2000;473:241–248. doi: 10.1016/s0014-5793(00)01540-4. [DOI] [PubMed] [Google Scholar]
- 51.Jády BE, Richard P, Bertrand E, Kiss T. Cell cycle-dependent recruitment of telomerase RNA and Cajal bodies to human telomeres. Mol Biol Cell. 2006;17:944–954. doi: 10.1091/mbc.E05-09-0904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tomlinson RL, Ziegler TD, Supakorndej T, Terns RM, Terns MP. Cell cycle-regulated trafficking of human telomerase to telomeres. Mol Biol Cell. 2006;17:955–965. doi: 10.1091/mbc.E05-09-0903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu X, Wang Y, Nakamura K, Kawauchi S, Akalin A, Cheng D, Chen L, Rusckowski M, Hnatowich DJ. Auger radiation-induced, antisense-mediated cytotoxicity of tumor cells using a 3-component streptavidin-delivery nanoparticle with 111In. J Nucl Med. 2009;50:582–590. doi: 10.2967/jnumed.108.056366. [DOI] [PubMed] [Google Scholar]
- 54.He Y, Panyutin IG, Karavanov A, Demidov VV, Neumann RD. Sequence-specific DNA strand cleavage by 111In-labeled peptide nucleic acids. Eur J Nucl Med Mol Imaging. 2004;31:837–845. doi: 10.1007/s00259-003-1446-0. [DOI] [PubMed] [Google Scholar]
- 55.Juliano RL. The delivery of therapeutic oligonucleotides. Nucleic Acids Res. 2016;44:6518–6548. doi: 10.1093/nar/gkw236. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.