Summary
The MYC oncoprotein globally affects the function of RNA Polymerase II (RNAPII). The ability of MYC to promote transcription elongation depends on its ubiquitylation. Here we show that MYC and PAF1c (Polymerase II associated factor 1 complex) interact directly and mutually enhance each other´s association with active promoters. PAF1c is rapidly transferred from MYC onto RNAPII. This transfer is driven by the HUWE1 ubiquitin ligase and is required for MYC-dependent transcription elongation. MYC and HUWE1 promote histone H2B ubiquitylation, which alters chromatin structure both for transcription elongation and doublestrand break repair. Consistently, MYC suppresses double-strand break accumulation in active genes in a strictly PAF1c-dependent manner. Depletion of PAF1c causes transcription-dependent accumulation of double-strand breaks despite widespread repair-associated DNA synthesis. Our data show that the transfer of PAF1c from MYC onto RNAPII efficiently couples transcription elongation with double-strand break repair to maintain the genomic integrity of MYC-driven tumor cells.
Keywords: MYC, HUWE1, PAF1c, RNAPII, double-strand break repair
Introduction
Deregulated expression of the MYC oncoprotein or one of its paralogues, MYCN and MYCL, drives tumorigenesis in many entities (Dang, 2012). Tumors driven by a number of different oncogenes continuously depend on enhanced MYC expression, suggesting that targeting MYC is a valid approach for tumor therapy (Annibali et al., 2014; Beaulieu et al., 2019; Gabay et al., 2014; Soucek et al., 2013). MYC proteins are transcription factors that bind to virtually all active promoters and many active enhancers (Kress et al., 2015). While MYC generally stimulates the transcription by RNA Polymerases I and III, its effects on expression of individual genes transcribed by RNAPII can be both positive or negative (Herold et al., 2019; Sabo et al., 2014; Tesi et al., 2019; Walz et al., 2014). In addition, MYC-dependent global increases in expression of all mRNAs have been observed (Lin et al., 2012; Nie et al., 2012). Both the general and specific effects of MYC on gene expression are typically weak, raising the possibility that MYC proteins have functions that are independent of altering target gene expression (Baluapuri et al., 2020).
The stepwise assembly of an elongation-competent RNAPII complex is a well-understood process (Cramer, 2019). MYC proteins affect several steps of this process, and their ability to promote transcription elongation is predominant in numerous experiments (Baluapuri et al., 2019; de Pretis et al., 2017; Herold et al., 2019; Rahl et al., 2010; Walz et al., 2014). Several lines of evidence show that MYC not only engages CDK9 (Huang et al., 2014; Rahl et al., 2010), but also the ubiquitin system to promote transcription elongation. First, several MYC-associated ubiquitin ligases are required for MYC-driven gene expression (Adhikary et al., 2005; Kim et al., 2003; von der Lehr et al., 2003). Inhibitors of the HUWE1 ubiquitin ligase abrogate MYC-dependent gene expression in colorectal tumor cells (Peter et al., 2014). Conversely, dephosphorylation of MYCN enables binding of USP11, which leads to transcription termination (Herold et al., 2019). Second, a lysine-free mutant of MYC is capable of recruiting RNAPII to core promoters but fails to promote pause release (Jaenicke et al., 2016). Finally, transcriptional activation by MYC requires ubiquitin-dependent extraction of MYC from chromatin by the p97/VCP complex (Heidelberger et al., 2018).
Ubiquitylation of MYC is required for the transfer of the PAF1c transcription elongation complex from MYC onto RNAPII (Jaenicke et al., 2016). Since MYC-dependent transcriptional elongation also depends on CDK9, our previous data did not distinguish between two possible models: the first model suggests that the actual transfer of PAF1c from MYC onto RNAPII is driven by CDK9, with ubiquitylation of MYC removing non-productive protein complexes from promoters. The other model suggests that the transfer itself is driven by ubiquitylation of MYC. To address the question of how the ubiquitin system promotes MYC dependent transcriptional elongation, we have now analyzed the mechanism of transferring PAF1c from MYC onto RNAPII and the consequences of disrupting this process.
Results
PAF1c binds MYC directly and enhances the association of MYC with active promoters
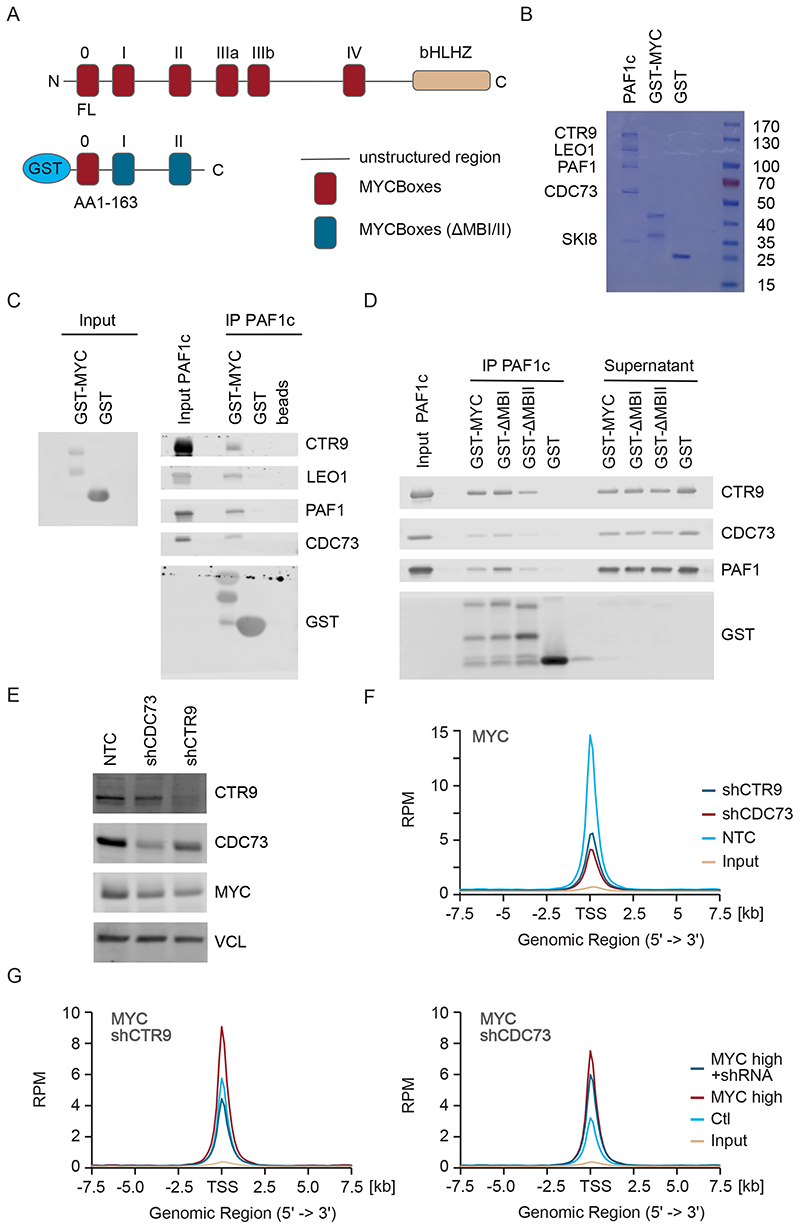
The inhibition of proteasome function in HeLa cells strongly enhances binding of MYC to CTR9 and CDC73, two subunits of the PAF1 complex (PAF1c) (Jaenicke et al., 2016). To test whether this association reflects a direct interaction of MYC with this complex, we incubated a purified glutathione-S-transferase (GST)-MYC fusion protein that encompasses amino acids 1 to 163 of human MYC with purified PAF1c, which has been reconstituted from recombinantly expressed subunits (Vos et al., 2018a) (Figure 1A,B); note that the PAF1c used lacks the RTF1 subunit (Vos et al., 2018a). PAF1c was recovered on beads carrying GST-MYC, but not on beads with an equivalent amount of GST (Figure 1C). Amino acids 1 to 163 of MYC encompass a large part of the transcription regulatory domain and include two of highly conserved MYCBoxes I and II (Baluapuri et al., 2020). Neither MYCBox I nor MYCBox II were required for interaction with PAF1c, consistent with recent Bio-ID data for two PAF1c subunits, CDC73 and PAF1 (Figure 1D) (Kalkat et al., 2018).
Figure 1. Binding of PAF1c to MYC enhances association of MYC with active promoters.
A. Diagram of MYC protein structure showing the position of MYCBoxes and the GST-MYC construct used for pulldown experiments.
B. Coomassie gel showing purified proteins. 10% of input material is shown.
C. Immunoblots of MYC and PAF1c (n=3; in all legends, n indicates the number of independent biological replicates).
D. Immunoblots of pulldown experiment using GST-△MYCBox I and GST-△MYCBox II constructs (n=3).
E. Immunoblot showing levels of CTR9, CDC73 and MYC in U2OS cells after stable expression of constitutive shRNAs. Vinculin (VCL) was used as loading control (n=3).
F. Density plot of MYC centered on the transcription start site (TSS) of 8,437 active promoters in a ChIP-Rx experiment in control U2OS cells or in cells expressing shCTR9 or shCDC73 (n=1). All ChIP-Seq traces show S.E.M. as a shade.
G. Density plot of MYC as in (F) in a ChIP experiment in control U2OS cells or in cells stably expressing MYC and Dox inducible shRNA targeting CTR9 (shCTR9) or shCDC73. (n=1). See also Figure S1.
On chromatin, MYC binds preferentially to active promoters and this preference depends on protein-protein interactions of MYC with promoter-bound factors (Guo et al., 2014; Lorenzin et al., 2016). To determine whether PAF1c is required for the specific binding of MYC to active promoters, we stably expressed shRNAs targeting two subunits of PAF1c, CTR9 and CDC73, in U2OS cells using lentiviral infection. Since the vector used for depletion confers resistance to puromycin, infected cells were selected and used 96 h after infection without further passaging. Each shRNA depleted its target protein by about 80% (Figure 1E and Figure S1A). To precisely evaluate possible changes in chromatin association, we used spike-in ChIP-sequencing (ChIP-Rx) (Orlando et al., 2014). Depletion of CTR9 or CDC73 caused a two-to three-fold decrease in association of MYC with active promoters (Figure 1F and Figure S1B). Subsequent analyses also showed that the decrease in MYC binding upon CTR9 or CDC73 depletion occurred at promoters but not at enhancers (Figure S1C), consistent with previous observations for Drosophila Myc (Gerlach et al., 2017).
Stable depletion of CTR9 or CDC73 also caused a reduction of about 30% in total MYC protein levels, most likely due to a reduction in MYC mRNA levels (Figure 1E and Figure S1A). To rule out the possibility that this decrease accounted for the apparent decrease in MYC binding, we expressed doxycycline (Dox)-inducible shRNAs targeting CTR9 or CDC73 in cells stably expressing ectopic MYC. With around 80,000 molecules of MYC per cell, U2OS cells express relatively low endogenous MYC levels, and stable lentiviral expression raises the number to about 1x106 molecules per cell (Lorenzin et al., 2016). In cells expressing ectopic MYC, addition of Dox led to a 3-fold reduction in levels of the respective target protein but had no effect on MYC levels (Figure S1D-F). Density plots and browser tracks confirmed that depletion of CTR9 and, to a lesser degree, of CDC73 reduced binding of MYC to promoters, while it had little or no effect on MYC binding to enhancers (Figure 1G, Figure S1G-I). We concluded that MYC binds directly to PAF1c and that PAF1c is required for the preferential association of MYC with active promoters.
Rapid transfer of PAF1c from MYC onto RNAPII
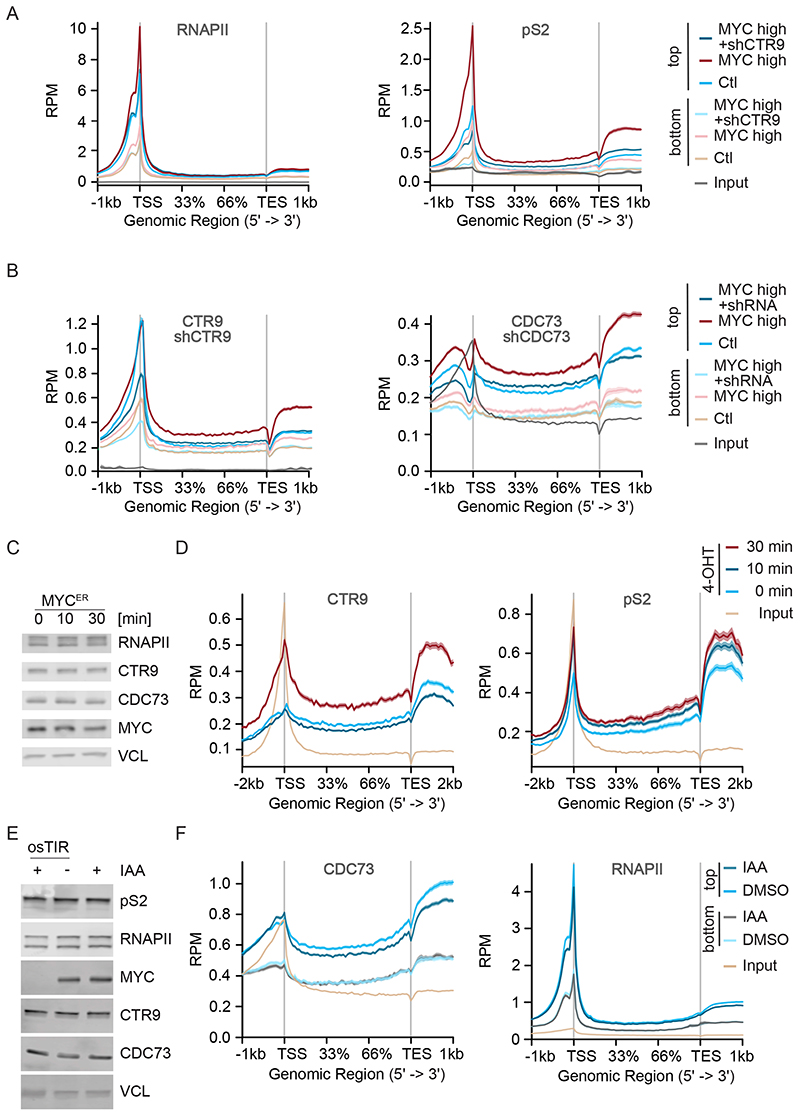
PAF1c travels with elongating RNAPII (Van Oss et al., 2017). To determine whether MYC affects loading of PAF1c onto RNAPII, we analyzed the binding of RNAPII to chromatin by ChIP-sequencing of unperturbed U2OS cells and compared it to cells in which MYC was stably expressed form a lentiviral promoter. Metagene plots showed that ectopic expression of MYC had little effect on chromatin association of total RNAPII, but strongly enhanced the levels of elongating Ser2-phosphorylated RNAPII (pS2 RNAPII) downstream of the start site and in the gene body, consistent with multiple previous data that MYC promotes elongation (Rahl et al., 2010; Walz et al., 2014) (Figure 2A). In parallel, ectopic expression of MYC strongly enhanced the association of both CTR9 and CDC73 with chromatin in gene bodies and depletion of either protein using specific shRNA confirmed the specificity of the ChIP signal (Figure 2B and Figure S2A,B). Stratifying the response for MYC occupancy at the promoter showed that the increase in pS2, CTR9 and CDC73 association with gene bodies was much stronger on genes with strongly MYC-bound (“top”) than on weakly (“bottom”) MYC-bound promoters (Figure 2A,B).
Figure 2. PAF1c is rapidly transferred from MYC on RNAPII.
A. Metagene plot of total RNAPII or pS2 RNAPII in a ChIP experiment in control U2OS cells or in cells stably expressing MYC and Dox inducible shCTR9; metagene plots of 7,479 most strongly MYC-bound (“top”) or 5,768 weakly MYC-bound (“bottom”) genes are shown (see STAR methods). Input shows 17,697 genes. Chromatin of six independent experiments was pooled for ChIP-sequencing.
B. Metagene plot of CTR9 or CDC73 binding to chromatin in a ChIP experiment, in control U2OS cells or in cells stably expressing MYC and Dox inducible shCTR9 or shCDC73; metagene plots of top or bottom MYC-bound genes. Input shows 17,697 genes. Chromatin of six independent experiments was pooled for ChIP-sequencing.
C. Immunoblot of U2OS cells expressing a MYCER chimeric protein with or without MYC activation upon addition of 4-OHT (200 nM) for 10 min or 30 min.
D. Metagene plot of CTR9 and pS2 in a ChIP-Rx experiment in U2OS MYCER cells treated as described in (C). The plot shows profiles of the top 4000 MYC bound genes (n=2).
E. Immunoblot of K562-MYC-AID erythroleukemia cells (n=2). Indole-3-acetic acid (IAA: 100 μM) was added for 30 min. Vinculin (VCL) was used as loading control (n=3).
F. Metagene plot of CDC73 or total RNAPII in a ChIP-Rx experiment in K562-MYC-AID cells treated as in (E). Metagene plots of top or bottom MYC-bound genes (n=2). Input shows 17,697 genes. See also Figure S2.
Importantly, while depletion of CTR9 had no obvious effect on distribution of total RNAPII, it reverted the MYC-dependent increase in pS2 association with chromatin in promoter-proximal regions, gene bodies and transcription end site regions, demonstrating that transfer of PAF1c is required for MYC-driven transcription elongation (Figure 2A,B).
To test how quickly PAF1c is transferred from MYC onto RNAPII, we used U2OS cells that carry a hormone-inducible MYCER chimeric protein and harvested cells 10 and 30 minutes after addition of 200nM 4-OHT. Activation of MYC had no effect on total levels of CTR9, CDC73, RNAPII or pS2 RNAPII at these short time points (Figure 2C). ChIP-Rx experiments revealed a strong increase in MYC association with promoters (Figure S2C) and an increase in association of pS2 RNAPII with chromatin in gene bodies downstream of the TSS at 10 min after MYC activation (Figure 2D, Figure S2D) and a further increase at 30 min. This data is consistent with previous data showing that MYC promotes pause release of RNAPII (Rahl et al., 2010; Walz et al., 2014). We did not detect an increase in association of PAF1c with gene bodies 10 min after MYC activation, but a strong increase at 30 min, demonstrating that MYC-dependent PAF1c transfer is delayed relative to pause release (Figure 2D, Figure S2D).
Conversely, we wanted to know whether the high levels of MYC found in human tumor cells are rate-limiting for the association of PAF1c with RNAPII. Since U2OS cells express relative low levels of endogenous MYC, we used K562 cells, whose endogenous MYC has been replaced by a chimeric protein, in which MYC is fused to an auxin-induced degron (AID) (Muhar et al., 2018). In these cells, addition of indole-3 acetic acid (IAA) induces rapid degradation of the MYC AID chimera (Figure 2E and Figure S2E). Importantly, addition of IAA for 30 minutes led to a significant decrease in association of CDC73 with gene bodies (Figure 2F, left panel). At this early time point, this decrease was not secondary to a decrease in RNAPII association with chromatin in promoter or gene body regions (Figure 2F, right panel). As before, the extent of decrease in CDC73 correlated with MYC occupancy of the promoter (Figure 2F). We concluded that MYC recruits PAF1c to active promoters and that PAF1c is rapidly transferred from MYC onto RNAPII following MYC-dependent pause-release.
HUWE1 drives the transfer of PAF1c from MYC onto RNAPII
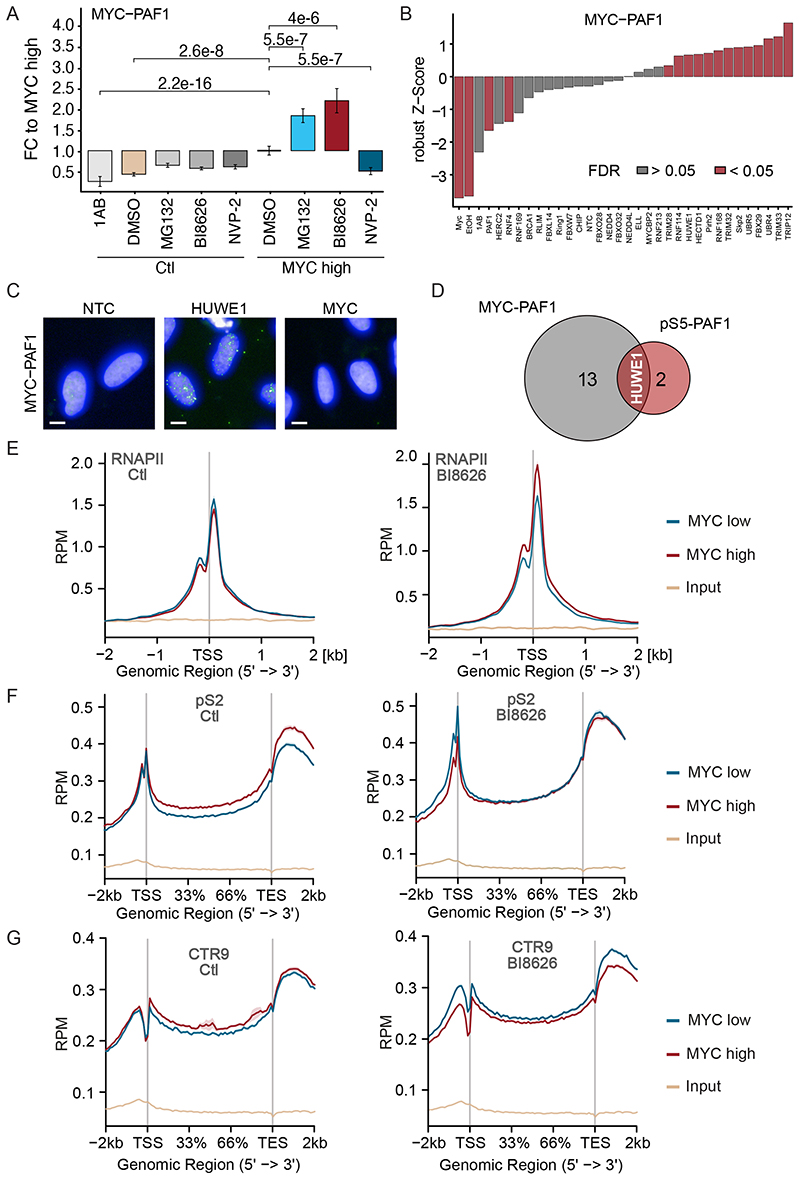
We next used proximity ligation assays (PLAs) with PAF1 and MYC antibodies to understand how PAF1c is transferred from MYC onto RNAPII. Controls established that the signal obtained with PAF1c and MYC antibodies was specific, since it did not occur when only one antibody was added and dependent on Dox-inducible induction of MYC (Figure 3A). Consistent with our previous experiments, inhibition of the proteasome by MG132 strongly enhanced the PLA signal between MYC and PAF1 (Figure 3A) (Jaenicke et al., 2016). In contrast, inhibition of CDK9 using a specific inhibitor, NVP-2 (Olson et al., 2018), decreased the PLA signal between MYC and PAF1 (Figure 3A). This is consistent with observations that CDK9 phosphorylates the C-terminal domain (CTD) of RNAPII and the NELF complex and causes it to dissociate from RNAPII (Fujinaga et al., 2004). Dissociation of NELF frees the interaction surface for PAF1c on RNAPII (Vos et al., 2018a; Vos et al., 2018b), arguing that inhibition of CDK9 precludes PAF1c from interacting with MYC at core promoters.
Figure 3. The HUWE1 ubiquitin ligase drives the transfer of PAF1c from MYC onto RNAPII.
A. Box plot of proximity ligation assays (PLAs) between MYC and PAF1 in U2OS cells expressing Dox inducible MYC. Where indicated (“MYC high”) Dox (1 μg/ml) was added for 24 h, MG132 (20 μM), BI8626 (10 μM) and NVP2 (1 μM) was added for 4 h (n=3). 1AB refers to control samples containing solely the anti-PAF1 antibody.
B. Results of a siRNA screen targeting MYC-associated ubiquitin ligases using PLAs between MYC and PAF1 as readout (n=10).
C. Representative micrographs. Nuclei were stained with Hoechst; bright dots indicate proximity of MYC with PAF1. Scale bar: 10 μm
D. Venn diagram of siRNAs significantly enhancing the proximity between the corresponding proteins.
E. Metagene plot of total RNAPII in a ChIP-Rx experiment in U2OS cells expressing Dox inducible MYC. The plots show metagene profiles of all active promoters (n=17,674) with or without addition of Dox in control (DMSO-treated) cells or cells exposed to BI8626 (n=2).
F. Metagene plot of pS2 RNAPII in a ChIP-Rx experiment. Plots and conditions are as in (E) (n=2).
G. Metagene plot of CTR9 in a ChIP-Rx experiment. Plots and conditions are as in (E) (n=2). See also Figure S3.
To better understand the dependency of PAF1c transfer on the ubiquitin system, we screened siRNAs targeting all ubiquitin ligases that have been reported in the literature to associate with MYC or MYCN using proximity of MYC with the PAF1c subunit PAF1 as readout (Figure 3B and Figure S3A). Depletion of several ligases significantly enhanced the proximity of MYC with PAF1 in cells. To focus on core promoters, we performed a second screen using the proximity of PAF1 with Ser5-phosphorylated (pS5) RNAPII, which is strongly enriched at core promoters (Figure S3B). This showed that siRNA-mediated depletion of three ubiquitin ligases significantly enhanced the extent of PAF1c association with RNAPII (Figure S3B). The overlap of both screens identified the ubiquitin ligase HUWE1 as a significant hit of both screens (Figure 3C,D). HUWE1 has been shown to bind to MYC and be required for transcriptional activation by MYC (Adhikary et al., 2005; Baluapuri et al., 2019; Peter et al., 2014).
In order to rapidly perturb HUWE1 function and study its impact on MYC and RNAPII function, we used a previously characterized inhibitor of HUWE1, BI8626 (Peter et al., 2014). This inhibitor blocks the activity of HUWE1, but not that of other HECT-domain ubiquitin ligases, and blocks MYC-dependent transcriptional activation in colorectal cancer cells. We confirmed that BI8626 enhances the MYC/PAF1-PLA signal to a similar degree as depletion of HUWE1 (Figure 3A). To better characterize its mode of action in cells and to ascertain the specificity of BI8626, we performed ubiquitin remnant profiling (Kim et al., 2011; Xu et al., 2010) in U2OS cells, in which we depleted HUWE1 using an siRNA, and compared this to inhibitor-treated cells. We were able to identify a total of 1,825 ubiquitylated sites across four individual experiments. The changes in ubiquitylation in response to depletion of HUWE1 highly correlated with those of HUWE1 inhibition (Figure S3C, left) and very few changes in ubiquitylation at individual sites differed between inhibitor and siRNA (Figure S3C, right). A notable exception was HUWE1 itself, since multiple ubiquitylation sites on HUWE1 decreased in abundance upon depletion but not upon inhibition of HUWE1, suggesting that the corresponding ubiquitylation sites are targeted by other ligases (Figure S3C, right). We concluded that the HUWE1 inhibitor targets a spectrum of ubiquitylation sites that are largely direct or indirect targets of HUWE1.
While the impact of HUWE1 depletion on steady-state levels of soluble MYC is weak, ubiquitylation by HUWE1 extracts MYC from chromatin via the p97 ATPase (Heidelberger et al., 2018). Consistent with this, incubation of U2OS cells with BI8626 or the VCP inhibitor NMS-873 (Magnaghi et al., 2013) enhanced MYC occupancy at the Nucleolin (NCL) promoter (Figure S3D), while neither depletion nor inhibition of HUWE1 affected levels of soluble MYC (Figure S3E). To globally test the effects of HUWE1 inhibition on MYC levels and on RNAPII function, we performed ChIP-Rx sequencing from U2OS cells that carry Dox-inducible MYC at physiological levels of MYC or after induction of MYC by addition of Dox for 24 hours. Metagene plots of all expressed genes showed that inhibition of HUWE1 caused a small increase in MYC-association at promoters at physiological MYC level and a much larger increase in cells expressing ectopic MYC (Figure S3F). Induction of ectopic MYC expression in control cells had no obvious effect on chromatin association of total RNAPII but caused a significant accumulation of RNAPII close to the transcription start site when HUWE1 was inhibited (Figure 3E). Consistent with multiple previous data, induction of MYC caused a large increase in pS2 RNAPII within the gene body and the transcription end site in control cells (Figure 3F). Inhibition of HUWE1 abrogated the MYC-dependent increase in elongation. Instead, induction of MYC rather caused a small decrease in association of pS2 RNAPII with promoter-proximal regions in the presence of BI8626 (Figure 3F). Finally, induction of MYC increased association of CTR9 with gene bodies in control cells, but decreased CTR9 association with chromatin in the gene body upon HUWE1 inhibition (Figure 3G). We concluded that blockade of HUWE1 abolishes the ability of MYC to promote transcription elongation and exposes an ability of MYC to retain RNAPII close to the promoter. Notably, the phenotype is similar to that observed in response to expression of a lysine-free mutant of MYC, supporting the notion that a decrease in ubiquitylation of MYC itself is critical for the effects of HUWE1 inhibition (Jaenicke et al., 2016).
MYC and HUWE1 promote global histone H2B ubiquitylation
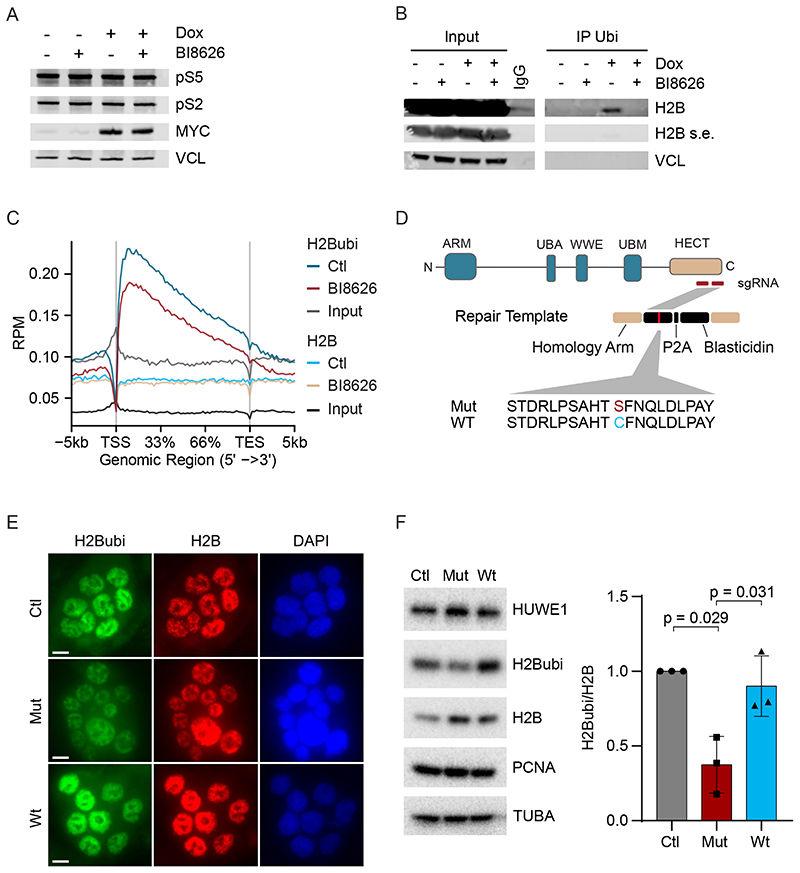
We performed two experiments to confirm that MYC and HUWE1 do not act upstream of NELF to promote the transfer of PAF1c onto RNAPII. First, immunoblots did not reveal a global HUWE1-dependent change in phosphorylation of RNAPII at serines 2 and 5 (Figure 4A). Second, we generated NELF ChIP-Seq data and used them to show that MYC promotes transcription elongation and promotes the transfer of CTR9 both on genes with NELF-bound promoters on genes that do not have NELF bound at their promoters (Figure S4A,B). We concluded that HUWE1 does not promote RNAPII pause release via CDK9 and NELF. Since PAF1c does not bind to RNAPII when NELF is bound, we suggest that MYC together with HUWE1 stimulates elongation from promoters after CDK9-dependent dissociation of NELF. This interpretation is consistent with observations that NELF acts upstream of the actual release of RNAPII into the gene body (Aoi et al., 2020).
Figure 4. HUWE1 and MYC control global histone H2B ubiquitylation.
A. Immunoblot of U2OS cells with or without induction of MYC in the presence or absence of BI8626 (n=2).
B. Immunoprecipitation using an anti-ubiquitin antibody (FK2) from U2OS cells. Input shows 1% of the material used in the immunoprecipitation (n=2). A shorter exposure (s.e.) was chosen to visualize total H2B levels.
C. Metagene plot of a ChIP-Rx experiment of ubiquitylated H2B (“H2Bubi”) and H2B in U2OS cells expressing Dox inducible MYC after addition of in DMSO-treated cells or cells exposed to BI8626. The metagene plot shows the profile of all active promoters (n=2).
D. Diagram illustrating knock-in mutagenesis strategy towards the catalytic cysteine of HUWE1.
E. Immunofluorescence of H2Bubi levels in wild-type HCT116 cells (“Ctl”) and in cells upon bi-allelic replacement of the catalytic cysteine of HUWE1 with serine (“Mut”). As control, HCT116 cells, in which a repair template with cysteine (“Wt”) was used, are shown (n=3). Scale bar: 10 μm
F. Left: Immunoblot (see Figure 5D for description) (n=3). Right: Quantification of the results. Tubulin (TUBA) was used as loading control. Data show mean +/- S.D. (n=3). See also Figure S4.
RTF1, a subunit of PAF1c, interacts directly with and activates the BRE1A/B (RNF20/40) ubiquitin ligase, which mono-ubiquitylates histone H2B at K120 (Kim et al., 2009; Van Oss et al., 2016), suggesting that HUWE1 and MYC act upstream of histone H2B ubiquitylation to modulate RNAPII function. To test this hypothesis, we precipitated lysates of U2OS cells expressing Dox-inducible MYC before and after incubation with a HUWE1 inhibitor with an anti-ubiquitin antibody. Under these conditions, induction of MYC increased ubiquitylation of histone H2B and this was suppressed by inhibition or siRNA-mediated depletion of HUWE1 (Figure 4B and Figure S4C). ChIP-Rx sequencing confirmed that histone H2B ubiquitylation at expressed genes is globally suppressed upon HUWE1 inhibition, while total levels of H2B remain unaffected (Figure 4C and S4D). HUWE1-dependent ubiquitylation of MYC was detectable on paused but not on non-paused genes (Figure S5A) and occurred both on NELF-bound and non NELF-bound genes (Figure S5B), supporting the view that ubiquitylation of MYC contributes to MYC-dependent pause release and occurs independent of the CDK9-dependent NELF release from RNAPII (Aoi et al., 2020; Jaenicke et al., 2016). To confirm that these effects reflect an on-target activity of HUWE1 inhibitors, we generated an HCT116 cell line, which carries a bi-allelic knock-in replacing the cysteine residue of HUWE1 that forms a thioester with ubiquitin with a serine (Figure 4D). Both immunofluorescence (Figure 4E) and immunoblots of cell lysates (Figure 4F) showed a significant decrease in histone H2B ubiquitylation. We concluded that HUWE1 and MYC globally control H2B ubiquitylation at active genes.
MYC-promotes double-strand repair in transcribed regions
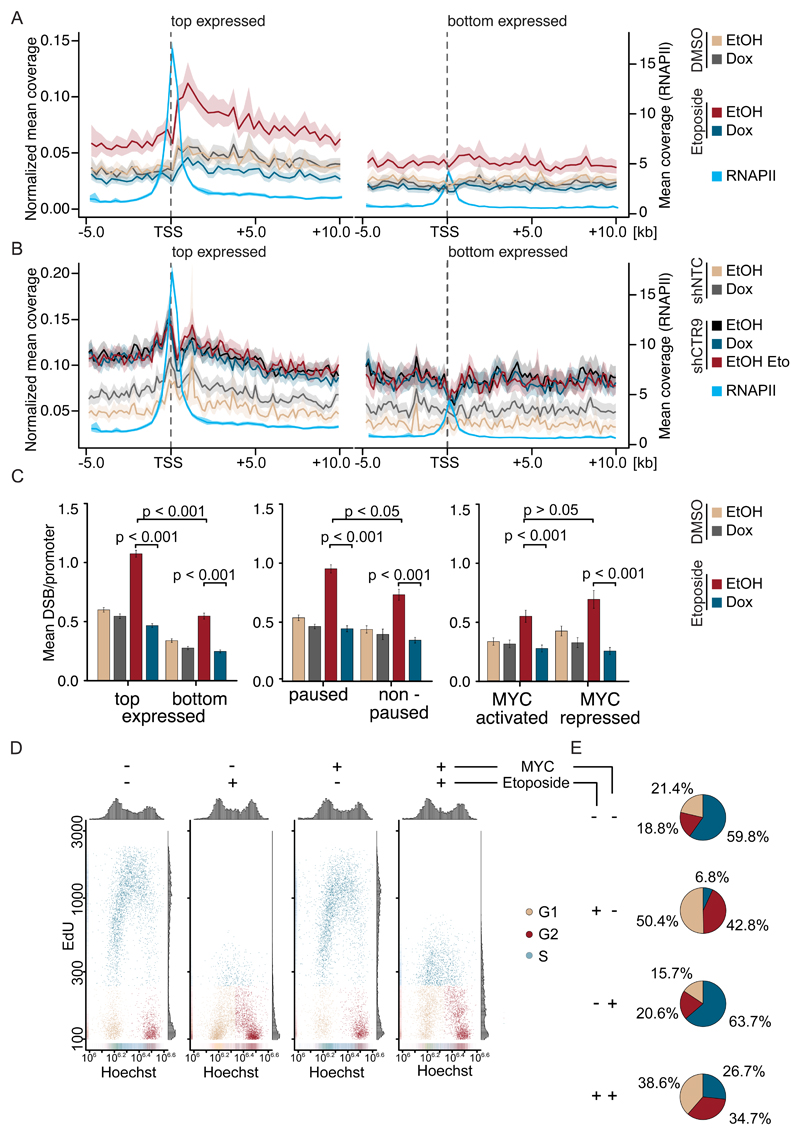
Ubiquitylation of histone H2B supports both transcriptional elongation (Fuchs et al., 2014) and the opening of chromatin for the repair of double-strand breaks (Moyal et al., 2011; Nakamura et al., 2011). Transcription-dependent breaks occur due to torsional stress that can be relieved by recruitment of topoisomerases; inhibition of topoisomerase II therefore induces doublestrand breaks both in gene bodies and, most strongly, downstream of active promoters (Gothe et al., 2019; Kouzine et al., 2013; Singh et al., 2020). To test whether MYC enhances DNA repair, we blocked topoisomerase II using a specific inhibitor, etoposide, and asked whether induction of MYC suppresses the formation of double-strand breaks using BLISS sequencing (Breaks Labeling In Situ and Sequencing) (Yan et al., 2017).
Consistent with previously published observations (Madabhushi et al., 2015), a limited number of double-strand breaks were detectable downstream of the transcription start site of actively transcribed genes, but not at weakly expressed genes (Figure 5A). Addition of etoposide strongly increased the number of promoter-proximal breaks on active genes. Induction of MYC by Dox had only a small effect on the number of breaks in control cells, but suppressed the accumulation of etoposide-induced double-strand breaks at active promoters and gene bodies. Conversely, depletion of MYC in K562-AID cells enhanced the accumulation of breaks after etoposide treatment at active promoters; notably, depletion of MYC in the absence of etoposide actually decreased double-strand frequency most likely due to a decreased overall transcription rate (Figure S5C). Depletion of CTR9 caused a strong increase in double-strand breaks around the TSS and in gene bodies of actively transcribed, but not of weakly expressed, genes and addition of etoposide did not lead to a further increase in double-strand break accumulation (Figure 5B). Importantly, induction of MYC was unable to revert the shCTR9-dependent increase double strand breaks (Figure 5B).
Figure 5. MYC-promotes double-strand repair at active promoters.
A. Density plot showing normalized mean coverage and estimated confidence interval of double-strand breaks (BLISS8) and total RNAPII (ChIP-Rx) around the transcription start sites of top (n=3954; left) and bottom (n=3012; right) expressed genes in U2OS cells expressing Dox inducible MYC. Shown is the merge of biological triplicates.
B. Density plot of double-strand breaks (BLISS8) and total RNAPII (ChIP-Rx) of U2OS cells expressing stable shCTR9 or non-targeting control. Shown is the merge of biological triplicates.
C. Stratification of double-strand breaks by promoter features. Data are mean +/- S.E.M.
D. Cell cycle distribution and EdU incorporation in U2OS cells expressing Dox inducible MYC. Dox and etoposide were added as described (n=3). 5,000 cells are shown per condition.
E. Pie chart visualizing the cell cycle distribution of U2OS treated as described in (D). See also Figure S5.
The effects of MYC and etoposide were strongest at highly expressed genes (Figure 5A) and neither MYC nor etoposide caused significant changes in double-strand breaks in heterochromatin (Figure S5D). While the total number of breaks per promoter correlated with overall gene expression, MYC reverted the etoposide-induced increase in breaks on both highly and weakly expressed genes, on genes which showed pausing of RNAPII and on non-paused genes, and on both MYC activated and MYC repressed genes, arguing that the effects of MYC on the accumulation of double-strand breaks are independent of effects on gene expression (Figure 5C). MYC also reverted the etoposide-induced increase uniformly when promoters were stratified by the number of breaks (Figure S5E). To test whether MYC has an effect on cellular responses to etoposide, we performed cell cycle analyses of U2OS cells that were exposed to etoposide for 3 h both with and without induction of MYC. Consistent with multiple previous data, etoposide strongly suppressed the DNA synthesis in control cells (Figure 5D). While induction of MYC had only a small effect on the cell cycle distribution of U2OS cells in the absence of etoposide (Walz et al., 2014), it restored DNA synthesis in etoposide-treated cells to a significant degree (Figure 5D,E). While the experiment does not distinguish between replicative DNA synthesis and DNA synthesis associated with DNA repair, the data show that MYC promotes the repair of promoter-proximal DNA breaks and alleviates the inhibition of DNA synthesis in response to topoisomerase inhibition.
PAF1c suppresses transcription-dependent DNA damage
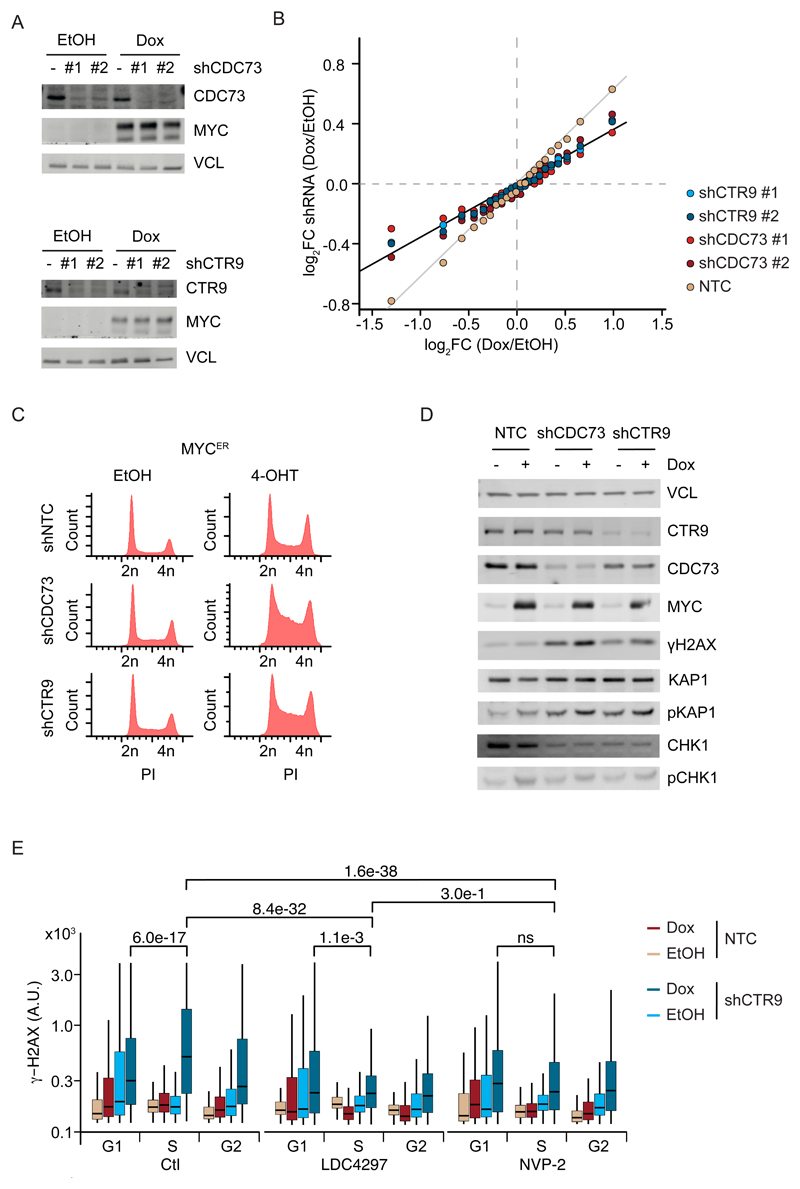
To assess the specific contribution of PAF1c to MYC-dependent cellular phenotypes, we depleted CTR9 or CDC73 by stable expression of two different shRNAs each and found that depletion of either protein retarded the proliferation of U2OS cells with or without induction of MYC (Figure 6A and Figure S6A). RNA sequencing confirmed that MYC exerted both weakly positive and negative effects on large groups of genes that have been previously described in this and multiple other systems (Figure 6B) (see Introduction). Note that the induction of MYC in the exponentially growing U2OS cells used here does not alter the total amount of mRNA per cell (Walz et al., 2014). Depletion of CTR9 or CDC73 attenuated the effects of MYC on both induced and repressed genes, consistent with the decrease in chromatin association of MYC with promoters observed in CTR9- and CDC73-depleted cells (Figure 6B).
Figure 6. MYC induces rampant DNA damage in the absence of PAF1c.
A. Immunoblot of MYC in U2OS cells expressing Dox inducible MYC and non-targeting shRNA, shCDC73 (top) or shCTR9 (bottom) (n=4).
B. Summary of RNA sequencing experiments from the cells described in (A). Genes were sorted in 23 bins (see STAR methods). The plot shows the change in gene expression observed for each bin upon expression of the indicated shRNAs. Values are average of four biological replicates using two different shRNAs each for CTR9 and CDC73.
C. FACS-profile of propidium-iodide stained U2OS-MYCER cells expressing shCTR9 or shCDC73 upon addition of 4-OHT (200 nM) for 24 h (n=2).
D. Immunoblot of U2OS cells expressing Dox inducible MYC and non-targeting shRNA or shCDC73 or shCTR9 (n=3).
E. Quantitative immunofluorescence of γ-H2AX in U2OS control cells (n=3) or cells treated with the CDK7 inhibitor LCD4297 or the CDK9 inhibitor NVP-2 for 3h. Cells were stratified for their cell cycle position by Hoechst staining. Shown is the mean intensity of γ-H2AX for at least 800 cells. See also Figure S6.
The attenuation of MYC-dependent gene expression upon PAF1c depletion raised the expectation that depletion of either PAF1c subunit would also have mild effects on MYC-dependent cell cycle progression. Induction of MYC activity in U2OS-MYCER cells by addition of 4-OHT or induction of MYC expression by Dox led to small increases in the proportion of cells in the S-phase of the cell cycle (Figure 6C and Figure S6B). In contrast, induction of MYC led to a much larger accumulation of cells in the S-phase of the cell cycle upon depletion of CTR9 or CDC73, indicating a strong delay in DNA replication (Figure 6C and Figure S6B). To understand this observation, we used antibodies that indicate activation of the ATM or the ATR kinase, reflecting double-strand breaks and replication stress, respectively (Figure 6D, Figure S6C). While induction of MYC had little effect on the activity of these kinases under control conditions, depletion ofCTR9 or CDC73 increased phosphorylation of H2AX and KAP1 at S824 and additional induction of MYC led to a further increase in phosphorylation of both proteins. Both sites are phosphorylated by the ATM kinase. In contrast, we did not observe an effect of MYC induction after CTR9 or CDC73 depletion on phosphorylation of CHK1 (S345) or RPA S33, which are target sites of the ATR kinase, or on phosphorylation of RPAS4/8, a target site of DNA-PK (Figure 6D and Figure S6C). We noted that the MYC-stimulated increase H2AX phosphorylation in CTR9-depleted cells was highest in S-phase, suggesting that PAF1 complexes have a role in preventing replication-transcription conflicts (Figure 6E). Incubation of cells with specific inhibitors of CDK7 (LDC4297;(Hutterer et al., 2015)) or CDK9 (NVP-2;(Olson et al., 2018)) for three hours strongly attenuated the induction of DNA damage in CTR9-depleted cells, demonstrating that DNA damage is caused by the residual transcription in these cells (Figure 6E and Figure S6D).
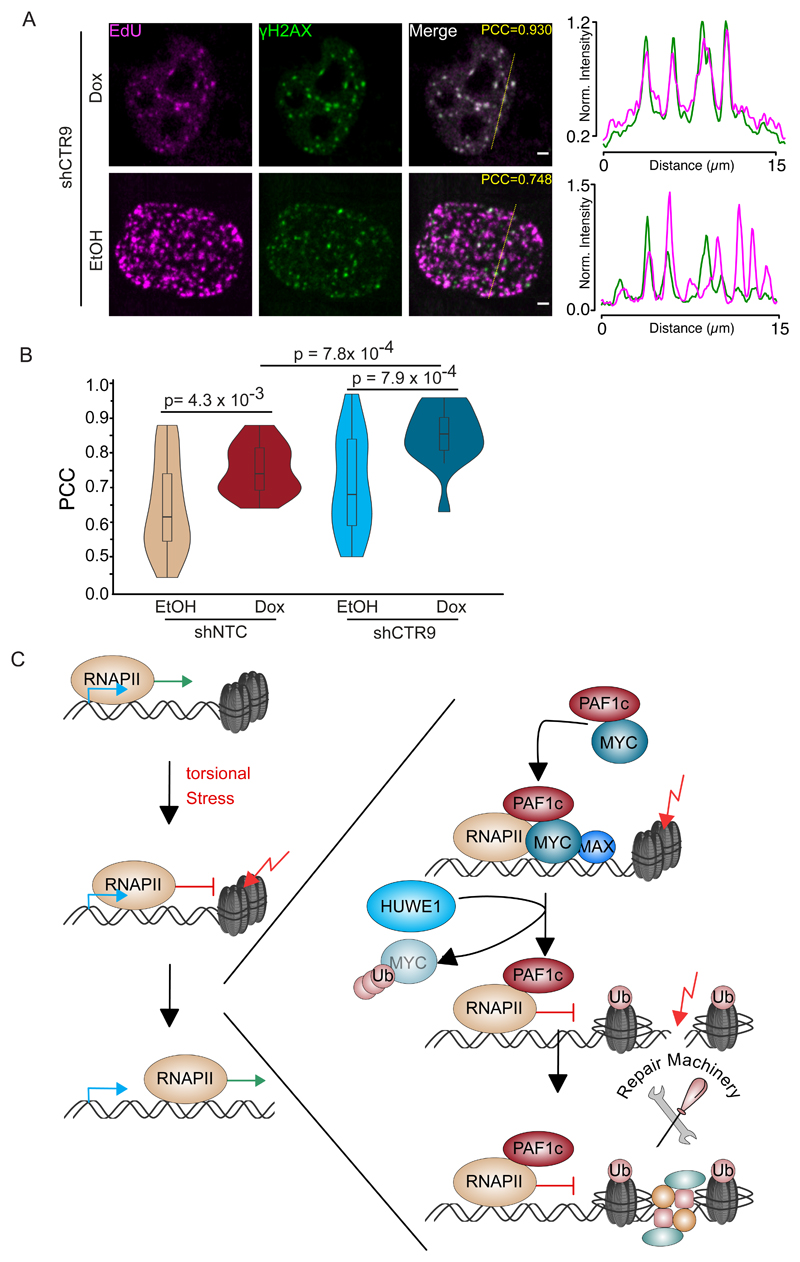
Parallel FACS analysis of propidium iodide-stained control cells, in which nascent DNA had been labelled with a pulse of BrdU revealed a normal profile with well-separated BrdU-positive and -negative populations (Figure S6E). Depletion of CTR9 or CDC73 led to a marked reduction in overall DNA synthesis and apparent DNA synthesis in cells with a 2n and a 4n DNA content; this phenotype was aggravated by the expression of high MYC levels (Figure S6E). The observation suggests that the DNA synthesis may be due to the repair of doublestrand breaks rather than normal replication (Orthwein et al., 2015). In strong support, confocal microscopy revealed a large increase in colocalization of γ-H2AX foci with EdU incorporation, indicative of DNA repair-associated DNA synthesis (Figure 7A,B and Figure S7A). We concluded that PAF1c has moderate effects on MYC-dependent gene expression but coordinates elongation with DNA repair to suppress transcription-induced DNA damage (Figure 7C).
Figure 7. Repair-associated DNA synthesis upon PAF1 depletion.
A. Confocal images of U2OS cells showing EdU incorporation immediately adjacent to γ-H2AX foci in U2OS cells upon constitutive expression shCTR9 and induction of MYC for 24 h. Insert shows Pearson´s correlation constant (“PCC”). Scale bars represent 2 μm.
B. Violin Plot of Pearson correlation coefficients for co-localization of DNA synthesis and γ-H2AX for each of the experimental conditions (n=17-21 cells).
C. Model of our findings. See also Figure S7.
Discussion
Both ubiquitylation of MYC and CDK9 are required for MYC-driven transcriptional elongation (Bywater et al., 2020; Huang et al., 2014; Jaenicke et al., 2016; Rahl et al., 2010). The CDK9-dependent dissociation of NELF frees the interaction surface for PAF1c on RNAPII (Vos et al., 2018a; Vos et al., 2018b) and there is a second, PAF1c-dependent pause site downstream of the NELF-dependent site (Aoi et al., 2020). Here we showed that HUWE1-dependent ubiquitylation of MYC and transfer of PAF1c control elongation at a step downstream of the CDK9-dependent release of RNAPII from NELF-inhibition. Both in flies and in humans, binding of MYC to PAF1c at promoters has moderate effects on gene expression, raising the question what is principal biological function may be (Gerlach et al., 2017).
Actively transcribed genes are particular susceptible to DNA damage. To maintain genomic stability, dedicated mechanisms ensure that DNA repair is particularly effective at transcribed genes (Gregersen and Svejstrup, 2018; Lans et al., 2019). The transcription process itself causes torsional stress that is a major cause for double-strand breaks in genes (Kouzine et al., 2013) and the relief of torsional stress is critical for transcription elongation (Bunch et al., 2015; Ju et al., 2006; Puc et al., 2015). Several mechanisms counteract transcription-induced torsional stress: for example, phosphorylated RNAPII stimulates the activity of topoisomerase I (Baranello et al., 2016). Also, both MYC and MYCN interact with topoisomerases I and II (Baluapuri et al., 2019; Buchel et al., 2017). Nevertheless, double-strand breaks are enriched downstream of active promoters in proliferating cells (Chiarle et al., 2011; Gothe et al., 2019; Klein et al., 2011). Here we show that MYC promotes the repair of DNA breaks via the HUWE1-dependent transfer of PAF1c from MYC to RNAPII. PAF1c in turn directly activates the BRE1A/B (RNF20/40) ligase that mono-ubiquitylates H2B (Van Oss et al., 2016). This histone modification alters chromatin structure and promotes the repair of double-strand breaks bynon-homologous end joining or homologous recombination (Moyal et al., 2011; Oliveira et al., 2014). Since MYC suppresses accumulation of double-strand breaks on genes independently of changes in steady-state mRNA levels, our observations can provide a plausible explanation for the pervasive presence of MYC proteins at virtually all active promoters (Figure 7C).
In addition to topoisomerases, two partner proteins of MYC and MYCN, the TRRAP-containing NuA4 complex (Jacquet et al., 2016; Kim et al., 2010; Murr et al., 2006) and the p400 helicase (Courilleau et al., 2012; Frank et al., 2003; Fuchs et al., 2001) have direct functions in double-strand break repair. Furthermore, the neuronal MYC paralogue, MYCN, recruits the BRCA1 protein, a central scaffold protein of multiple complexes involved in homologous recombination (Venkitaraman, 2014), to active promoters (Herold et al., 2019). A hallmark of DNA repair proteins is that their association with RNA polymerases, replication forks or chromatin is transient and regulated by protein ubiquitylation. Consistently, several MYC-associated ubiquitin ligases (Gudjonsson et al., 2012; Qiao et al., 2020) as well as MYC-associated ubiquitin-specific proteases (Herold et al., 2019; Knobel et al., 2014; Orthwein et al., 2015; Popov et al., 2007; Sondalle et al., 2019; Sun et al., 2015; Zhang et al., 2006) regulate the stability or interactions of DNA repair proteins. We propose, therefore, that MYC proteins engage multiple complexes that suppress double-strand break accumulation at active promoters and MYC-associated ubiquitylation and de-ubiquitylation reactions dynamically control their interactions with the transcription machinery.
Depletion of PAF1c induces transcription-dependent DNA damage, arguing that the residual transcription that occurs in depleted cells is highly DNA damaging. This increase was highest in S-phase, consistent with a previous report that shows that PAF1c is critical for preventing transcription-replication conflicts (Poli et al., 2016). Such conflicts can be co-directional or head-on with conflicts being largely head-on, since origins of replication are localized at active promoters (Chen et al., 2019). Co-directional conflicts activate ATM, but not ATR (Hamperl et al., 2017), arguing that PAF1c prevents the accumulation of breaks that occur during co-directional conflicts. It is striking that a second transcription factor that engages PAF1c in a similar manner is β-catenin, which drives the oncogenic growth of colorectal tumor cells; like MYC, β-catenin is also targeted by the HUWE1 ligase (Dominguez-Brauer et al., 2017; Mosimann et al., 2006; Moyal et al., 2011). We hypothesize, therefore, that targeting PAF1c and its role for repair of transcription-induced double-strand breaks will open a wide therapeutic window for colorectal and other MYC-driven tumors.
Star Methods
Resource Availability
Lead Contact
Further information and requests for resources and reagents should be directed to the lead contact, Martin Eilers (martin.eilers@biozentrum.uni-wuerzburg.de).
Materials Availability
Unique and stable reagents generated in this study are available upon request.
Data and Code Availability
The ChIP-Rx, BLISS and RNA sequencing data are deposited at the GEO (Gene Expression Omnibus) database (GEO accession: GSE150217). Original image data have been deposited to Mendeley Data: 10.17632/n4rr8ck4w3.1
Experimental Model And Subject Details
Cell cultures, primary cells, viral strains
HEK293TN, NIH3T3 and U2OS cells were grown in DMEM (Sigma-Aldrich and Thermo Fisher Scientific). K562 were grown in RPMI 1640 supplemented with 4 mM Glutamine. Medium was supplemented with 10% fetal calf serum (Biochrom and Sigma-Aldrich) and penicillin–streptomycin (Sigma-Aldrich). All cells were routinely tested for mycoplasma contamination.
Cell line manipulation and generation
Lentiviral packaging plasmids psPAX2 (Addgene 12260) and pMD2.G (Addgene 12259) were used to generate stable cell lines. Lentivirus production was carried out in HEK293 cells and cell-free, virus-containing supernatant was used for infections. If not specified otherwise inhibitor treatment of U2OS cells was as follows: Doxycycline (24h, 1 μg/ml), NVP-2 (3h, 1 μM), LDC4297 (0.5 μM, 3 h), BI8626 (10 μM, 4h), NMS-873 (5 μM, 4 h). BrdU (10 μM) or EdU (10 μM) treatment was for 30-60 min. Transfection with siRNA was performed using the RNAiMAX reagent (Thermo Fischer Scientific) according to the manufacturer’s protocol. Cells were collected 40 h after transfection.
Method Details
General Cloning
GST-MYC1-163 was cloned as described (Baluapuri et al., 2019). GST-MYC1-163 ΔMBI and GST-MYC1-163 ΔMBII were cloned by PCR amplification using the primers MYC-f (pGex4T3) and MYC-r (pGex4T3) and inserted into pGex4T3 using EcoRI and XhoI restriction sites.
shRNA Experiments
U2OS cells were infected with lentiviral supernatants in the presence of polybrene (4 μg/ml) or protamine sulfate (5 μg/ml) for 24 h. Medium (1:1, v/v) was added for 24 h. Cells were selected for 24 h with puromycin (2 μg/ml) and afterwards plated for the experiment. shRNAs against CTR9 and CDC73 were selected as described (Fellmann et al., 2013) and lentivirally transduced into the cell genome.
For Figure 1G, Figure 2B, Figure S1E, F,H and I and Figure S2A and B shRNA mirE3 against CDC73 was induced by doxycycline for 48 h. For Figure 1G, Figure 2A, and Figure S1D,F,H and I and Figure S2A and B shRNA mirE3 against CTR9 was induced by doxycycline for 48 h. For Figure 1E,F, Figure 6A-D, Figure S1 A,B and C and Figure S6A,B,C and E shRNA mirE3 against CDC73 was constitutively expressed. For Figure 6A,B shRNA mirE4 against CDC73 was constitutively expressed. For Figure 1E,F, Figure 5B, Figure 6A-E, Figure 7A, Figure S1 A,B and C and Figure S6A-E and Figure S7A shRNA mirE3 against CTR9 was constitutively expressed. For Figure 6A,B shRNA mirE5 against CTR9 was constitutively expressed.
Protein expression, purification and in vitro pulldown
PAF1c (CTR9, Leo1, Paf1, CDC73, WDR61) was expressed and purified as described (Vos et al., 2018a). pGex4T3 plasmids (GST, GST-MYC1-163, GST-MYC1-163ΔMBI and GST-MYC1-163ΔMBII) were transformed into BL21 E. coli and preculture was incubated overnight. LB-media was inoculated until an OD600 of 0.5. Overexpression was induced with 100 mM IPTG for 6 h. Bacteria were pelleted and lysed in STE buffer (150 mM NaCl, 10 mM Tris/HCl pH 8, 1 mM EDTA, 0.5 mM TCEP, protease inhibitors (Sigma)). Lysate was sonicated for three times 1 min (1 s pulse on, 1 s pulse off) and centrifuged. Washed Sepharose beads (GE Healthcare/VWR International) were incubated with lysate for 1 h at 4 °C. After coupling, beads were washed with STE buffer. GST and GST-MYC1-163 were of the same preparation as described in Baluapuri et al, 2019. For in vitro pull-down, GST or GST-MYC1-163 as well as GST-MYC1-163ΔMBI and GST-MYC1-163ΔMBII coupled beads were washed with pull-down buffer (100 mM NaCl, 20 mM Na-HEPES pH 7.5, 4% glycerol, 3 mM MgCl2, 1 mM 1,4-Dithiothreitol, 300 ng/ml BSA) and incubated with PAF1c overnight at 4 °C on a rotating wheel. After pull-down, beads were washed with pull-down buffer and NETN buffer (20 mM Tris pH 8, 100 mM NaCl, 1 mM EDTA, 0.5% NP-40). Pull-down was eluted from beads in 2x Laemmli sample buffer (20 mM Tris pH 6.8, 4% SDS, 0.02% bromophenol blue, 13.4% glycerol, 2 mM 1,4-Dithiothreitol) at 95 °C for 5 min.
Immunoblot
Cells were lysed in RIPA lysis buffer (50 mM HEPES pH 7.9, 140 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% SDS, 0.1% sodium deoxycholate) containing protease and phosphatase inhibitors (Sigma-Aldrich) and incubated for 20 min at 4 °C with rotation. The lysate was cleared by centrifugation and protein concentration was determined using the BCA assay. The cell lysate (same number of cells or amount of protein) was separated by BisTris-PAGE and transferred to PVDF membranes (Millipore). Membranes were blocked for 1 h and probed using antibodies against total RNAPII (Santa Cruz Biotechnology, sc-55492, sc-17798), pS2-Pol II (Abcam, ab24758), MYC (Abcam, ab32072), CTR9 (Bethyl Laboratories, A301-395), CDC73 (Bethyl Laboratories, A300-171A), PAF1 (Abcam, ab20662), Leo1 (Novus Biologicals, NB600-276), GST (GE Healthcare/Sigma-Aldrich, GE27-4577-01), Vinculin (Sigma, V9131), CDK2 (Cell Signaling Technology, sc-163), CHK1 (Santa Cruz Biotechnology, sc-7898), phospho-Chk1 (Cell Signaling, 2348), phospho-Histone H2A.X (Cell Signaling, 2577), phospho-KAP1 (phospho S824, Abcam, ab70369), KAP1 (Bethyl Laboratories, A300-274A), Histone H2B (Abcam, ab1790), Ubiquityl-Histone H2B (Cell Signaling Technology, 5546), HUWE1 (Abcam, ab70161), mono- & polyubiquitinylated conjugates (FK2, Enzo Life Sciences, BML-PW8810-0100). For visualization the LAS3000 or LAS4000 Mini (Fuji) or Odyssey CLx Imaging System (LICOR Biosciences) were used. Quantification was performed using Image Studio (LI-COR Biosciences, version 5.2.5).
Immunoprecipitation
Cells were resuspended in lysis buffer (20 mM HEPES pH 7.9, 180 mM NaCl, 1.5 mM magnesium dichloride, 10 % glycerol, 0.2 % NP-40) supplemented with a cocktail of protease and phosphatase inhibitors (Sigma-Aldrich) and 10 mM N-Ethylmaleinimid. After brief sonication, samples were incubated on ice for 30 min with 10 U Benzonase and cleared by centrifugation. Dynabeads (20 μl of Protein A/G beads, Thermo Fisher Scientific) were preincubated, overnight at 4 °C with rotation, in the presence of 5 g/l BSA and 3 μg antibody targeting mono- & polyubiquitinylated conjugates (Enzo Life Sciences, BML-PW8810-0100). Co-immunoprecipitation was carried out in lysis buffer with an adjusted amount of lysate according to protein concentration and incubated for 12 h at 4 °C. Elution of dynabeads was performed by heating in 1.5x Laemmli sample buffer (15 mM Tris pH 6.8, 3% SDS, 0.015% bromophenol blue, 10% glycerol, 1.5 mM 1,4-Dithiothreitol) for 5 min at 95 °C. Samples were analyzed by immunoblotting.
Chromatin IP without or with reference exogenous genome spike-in (ChIP, ChIP-Rx)
For each ChIP or ChIP-Rx sequencing experiment, 5x107 cells per immunoprecipitation condition were fixed with formaldehyde (final concentration, 1%) for 5-10 min at room temperature. Fixation was stopped by adding 125 mM glycine for 5 min. Cells were harvested in ice-cold PBS containing protease and phosphatase inhibitors (Sigma-Aldrich). All further used buffers also contained protease and phosphatase inhibitors. As exogenous control (spike-in), murine NIH 3T3 cells were added at a 1:10 cell ratio during cell lysis. Cell lysis was carried out for 20 min in lysis buffer I (5 mM PIPES pH 8.0, 85 mM KCl, 0.5% NP-40) and nuclei were collected by centrifugation at 1500 rpm for 20 min at 4 °C. Crosslinked chromatin was prepared in lysis buffer II (10 mM Tris pH 7.5, 150 mM NaCl, 1 mM EDTA, 1% NP-40, 1% sodium deoxycholate, 0.1% SDS) and fragmented by sonication (total duration, 20 min with 10 s pulses and 45 s pausing) or by using the Covaris Focused Ultrasonicator M220 for 50 min per ml lysate. Fragment size of 150-300 bp was validated by agarose gel electrophoresis. Chromatin was centrifuged for 20 min at 14,000 rpm at 4 °C before IP. For each IP reaction, 100 μl Dynabeads Protein A and Protein G (Thermo Fisher Scientific) were pre-incubated overnight with rotation in the presence of 5 mg/ml BSA and 15 μg antibody (total Pol II (Santa Cruz Biotechnology, sc-17798), pS2-Pol II (Abcam, ab24758), MYC (Abcam, ab32072), CTR9 (Bethyl Laboratories, A301-395), CDC73 (Bethyl Laboratories, A300-171A), Histone H2B (Abcam, ab1790), Ubiquityl-Histone H2B (Cell Signaling Technology, 5546), NELFC (Cell Signaling Technology, 12265S). Chromatin was added to the beads, and IP was performed for at least 6 h at 4 °C with rotation. Beads were washed three times each with washing buffer I (20 mM Tris pH 8.1, 150 mM NaCl, 2 mM EDTA, 1% Triton X-100, 0.1% SDS), washing buffer II (20 mM Tris pH 8.1, 500 mM NaCl, 2 mM EDTA, 1% Triton X-100, 0.1% SDS), washing buffer III (10 mM Tris pH 8.1, 250 mM LiCl, 1 mM EDTA, 1% NP-40, 1% sodium deoxycholate; including a 5 min incubation with rotation), and TE buffer (Thermo Fisher Scientific). Chromatin was eluted twice by incubating with 150 ml elution buffer (100 mM NaHCO3, 1% SDS) for 15 min with rotation. Input samples and eluted samples were de-crosslinked overnight. Protein and RNA were digested with proteinase K and RNase A, respectively. DNA was isolated by phenol-chloroform extraction and ethanol precipitation and analyzed by qPCR using StepOnePlus Real-Time PCR System (Thermo Fisher Scientific) and SYBR Green Master Mix (Thermo Fisher Scientific) or sequencing on the Illumina Next-Seq500.
For ChIP or ChIP-Rx sequencing, DNA was quantified using the Quant-iT PicoGreen dsDNA assay (Thermo Fisher Scientific). DNA library preparation was done using the NEBNext ChIP-Seq Library Prep Master Mix Set for Illumina (New England Biolabs) or NEBnext Ultra II DNA Library Prep Kit (New England Biolabs) following manufacturer's instructions. Quality of the library was assessed on the Fragment Analyzer (Agilent) using the NGS Fragment High Sensitivity Analysis Kit (1-6,000 bp; Agilent). Finally, libraries were subjected to cluster generation and base calling for 75 cycles on Illumina NextSeq500 platform.
Proximity Ligation Assay
2500 U2OS cells expressing doxycycline-inducible MYC were seeded per well in a 384 well format (PerkinElmer) and allowed to settle overnight. Where indicated, cells were treated with doxycycline (1 μg/ml, 24 h) or equal amounts of ethanol. 4 h before fixation with 4% paraformaldehyde, the indicated inhibitors or equal amounts of DMSO were added if indicated. Fixed cells were permeabilized with 0.3% Triton X-100, washed in PBS, and blocked (5% BSA in PBS) for 60 min. Cells were incubated overnight at 4 °C with primary antibodies against MYC (Santa Cruz Biotechnolog, sc-42), PAF1 (Abcam, ab20662), pS5-RNAPII (Biolegend, 904001) in 5% BSA in PBS. Cells were treated for 1 h at 37 °C with plus (Sigma-Aldrich, DUO92002) and minus (Sigma-Aldrich, DUO92004) probes directed at rabbit and mouse antibodies, respectively, and ligated for 30 min at 37 °C. Next, in situ PCR amplification was done with Alexa 488-conjugated oligonucleotides (Sigma-Aldrich, DUO92014) for 2 h at 37 °C. Samples were counter-stained with Hoechst 33342 (Thermo Fisher Scientific). Image acquisition was done using the Operetta CLS High-Content Analysis System with 40x magnification (PerkinElmer) and were processed using Harmony High Content Imaging and Analysis Software (PerkinElmer) and R. Wells with focus error were discarded.
siRNA screen
1,250 U2OS cells expressing doxycycline-inducible MYC were seeded per well in a 384 well format (PerkinElmer) and allowed to settle for 10 h. Transfection was performed using RNAiMAX reagent (Thermo Fisher Scientific) according to manufacturer’s protocol using a pool of 4 siRNAs against each listed E3-ligase from ubiquitin conjugation libraries (Dharmacon, GU-105635, GU-105625, GU-105615). 16 h post transfection 1 μg/ml doxycycline or equal amounts of ethanol were added for 24 h. Where indicated, MG132 (20 μM, Calbiochem / Merck) was added 4 h before fixation with 4% paraformaldehyde. Proximity Ligation Assay was performed as described. The readout parameter for statistical analysis was foci/nucleus as produced by the Harmony High Content Imaging and Analysis Software (PerkinElmer). Statistical analysis was performed in R by calculating the fold change to the non-targeting siRNA of the respective replicate and applying Welch’s t-test over all replicates for each siRNA to the non-targeting control with subsequent correction for multiple testing using Benjamini and Hochberg's FDR method. To reduce the influence of outliers, generated by transfection, Proximity Ligation Assay and image acquisition, a modified Z-score ("robust Z-score") (Iglewicz and Hoaglin, 1993) was calculated as follows:
BLISS8
The original BLISS protocol was adapted and modified from (Yan et al., 2017). For experiments in U2OS cells expressing doxycycline inducible MYC, cells were plated in a 24-well plate (Greiner) and incubated with ethanol or doxycycline (1 μg/ml) from the following day onwards for 24 h. Where indicated, etoposide was added (3 h, 25 μM). For Figure 5B U2OS cells expressing doxycycline-inducible MYC were infected with lentiviral supernatants in the presence of 4 μg/ml polybrene or protamine sulfate (5 μg/ml) for 24 h. Medium (1:1, v/v) was added for 24 h. Cells were selected for 24 h with puromycin (2 μg/ml) and afterwards 20,000 control cells and 30,000 cells expressing a constitutive active shCTR9 were plated for the experiment. Cells were incubated with ethanol or doxycycline (1 μg/ml) for 24 h. Cells were fixed by addition of paraformaldehyde directly to the media to a final concentration of 3.7%, washed with PBS and either stored at 4 °C or directly processed. For experiments in K562-AID cells, cells were transferred to 24-well plate 12 h before fixation and spun down 5 min preceding and during fixation. Lysis was performed by incubation in lysis buffer 1 (10 mM Tris-HCl, 10 mM NaCl, 1 mM EDTA, 0.2% Triton X-100, pH 8) for 1 h at 4 °C, brief rinsing in PBS and incubation in lysis buffer 2 (10 mM Tris-HCl, 150 mM NaCl, 1 mM EDTA, 0.3% SDS, pH 8) for 1 h at 37 °C. Following rinsing in PBS, cells were equilibrated in CutSmart buffer (New England Biolabs) previous to restriction enzyme digestion using AsiSi (New England Biolabs) according to manufacturer’s protocol. Following rinsing in PBS and equilibrating the cells in CutSmart buffer, blunting of double-strand breaks using Quick Blunting Kit (New England Biolabs) following manufacturer’s protocol was performed. Sense and antisense adapter-oligos were annealed by heating them for 5 min at 95 °C, followed by a gradual cooldown to 25 °C over a period of 45 min. Consecutive to equilibration in CutSmart buffer (New England Biolabs) and T4 Ligase buffer (New England Biolabs) annealed adapters were dispensed on samples and ligated using T4 DNA Ligase (New England Biolabs) using manufacturer’s recommendations for 16 h at 16 °C. Excessive Adapters were removed by repeated rinsing in a high-salt wash buffer (10 mM Tris-HCl, 2M NaCl, 2 mM EDTA, 0.5% Triton X-100, pH 8). Genomic DNA was extracted in DNA extraction buffer (1% SDS, 100 mM NaCl, 50 mM EDTA, 10 mM Tris-HCl, pH 8) supplemented with Proteinase K (1 mg/ml, Roth) for 16 h in a thermoshaker at 55 °C. DNA was isolated by phenol-chloroform extraction and isopropanol precipitation, resuspended in TE buffer and sonicated using the Covaris Focused Ultrasonicator M220 for 1 to 2 min to achieve a fragment size of 300-500 bp. Fragment size was assessed on the Fragment Analyzer (Agilent) using the NGS Fragment High Sensitivity Analysis Kit (1-6,000 bp; Agilent). The DNA was concentrated using Agentcourt AMPure XP Beads (Beckman Coulter), transcribed into RNA and DNA digested using MEGAscript™ T7 Transcription Kit (Thermo Fischer Scientific) following manufacturer’s recommendations. A two-sided RNA cleanup with a ratio of 0.4 followed by 0.2x was performed using Agencourt RNAClean XP Beads (Beckman Coulter). RNA concentration was assessed on the Fragment Analyzer (Agilent) by using Standard Sensitivity RNA Analysis Kit (Agilent). Library preparation was performed by ligating the RA3 adapter to the samples with a T4 RNA Ligase 2 (New England Biolabs) supplemented with Recombinant Ribonuclease Inhibitor (Thermo Fischer Scientific). Samples were reverse transcribed using SuperScript III Reverse Transcriptase kit (Thermo Fischer Scientific) and library indexing and amplification performed using NEBNext High-Fidelity 2X PCR Master Mix (New England Biolabs) with RP1- and desired RPI-primer with 18 (Figure 5A), 19 cycles (Figure 5B) or 17 cycles (Figure S5C) with half of the amount of the prepared library. The libraries were cleaned up using Agentcourt AMPure XP Beads (Beckman Coulter), quality, quantity and fragment size assessed on the Fragment Analyzer (Agilent) using the NGS Fragment High Sensitivity Analysis Kit (1-6,000 bp; Agilent) and subsequently subjected to Illumina NextSeq 500 sequencing, according to manufacturer’s instructions. Adapters and oligos were custom synthesized and Unique Molecular Identifiers (UMIs) generated by random incorporation of the four standard dNTPs using the ‘Machine mixing' option.
Cell Cycle Immunofluorescence
For Figure 5D U2OS cells expressing doxycycline inducible MYC were plated in a 96-well plate (Greiner) and incubated with ethanol or doxycycline (1 μg/ml) from the following day onwards for 24 h. Where indicated, etoposide was added (3 h, 25 μM, Sigma-Aldrich). For Figure 6E and Figure S6D U2OS cells expressing doxycycline inducible MYC were infected with lentiviral supernatants in the presence of 4 μg/ml polybrene or protamine sulfate (5 μg/ml) for 24 h. Medium (1:1, v/v) was added for 24 h. Cells were selected for 24 h with puromycin (2 μg/ml) and afterwards plated for the experiment. Cells were pulsed with 10 μM EdU (Jena Bioscience) for 30 minutes and subsequently fixed with 3.7% paraformaldehyde in PBS. After removing paraformaldehyde and washing with PBS, cells were permeabilized with 0.3% Triton X-100 in PBS and blocked with 5% BSA in PBS. Newly synthesized DNA was visualized by performing a copper(I)-catalyzed azide-alkyne cycloaddition (100 mM Tris pH 8.5, 4mM CuSO4, 10mM AFDye 647 Azide (Jena Bioscience), 10mM L-Ascorbic Acid). Samples were stained with primary antibodies against phospho-Histone H2A.X (Cell Signaling, 2577) in 5% BSA in PBS overnight at 4 °C and after rinsing with PBS, incubated with secondary antibody (Thermo Fisher Scientific, A-11029) for 1 h at room temperature. Counter-staining was performed using Hoechst 33342 (Sigma-Aldrich). Images were taken with an Operetta High-Content Imaging System with 20x magnification. Images were processed using Harmony High Content Imaging and Analysis Software and R. Cells were grouped into cell cycle phase according to EdU and Hoechst staining of the control condition.
Confocal Microscopy
Leica SP8 (DM6000) upright microscope was used to scan all cells under 63x GLY objective with HyD detector for EdU channel and PMT for γ-H2AX at 400 Hz scan rate. Stacks with planes 330 nm apart were taken under same gain and laser power values for all conditions.
Image Processing & Colocalization Quantification
3-5 planes of each stack were converted to a single image via maximum intensity projection followed by addition of a Gaussian blur filter with sigma value of 0.8. Composite images of all channels were created, and representative line profiles were generated using “Plot Profile” function in ImageJ over a width of single Gaussian corrected pixel. Pearson’s correlation constant was calculated for brightness and contrast corrected images (EdU - min. values 10-20 & max. values 90-150; pgH2AX - min. values 10 & max. values 90-125) using Coloc 2 plugin in ImageJ (https://imagej.net/Coloc_2).
RNA sequencing
U2OS cells expressing doxycycline inducible MYC were infected with lentiviral supernatants in the presence of 4 μg/ml polybrene or protamine sulfate (5 μg/ml) for 24 h. Medium (1:1, v/v) was added for 24 h. Cells were selected for 24 h with puromycin (2 μg/ml) and afterwards plated for the experiment. Cells were incubated with ethanol or doxycycline (1 μg/ml) for 24 h. Treatment was stopped by adding RLT Buffer (QIAGEN) containing β-Mercaptoethanol according to instruction manual. Total RNA was extracted using RNeasy mini column (QIAGEN) including on-column DNase I digestion. mRNA was isolated using NEBNext Poly(A) mRNA Magnetic Isolation Module (NEB) and library preparation was performed with the NEBNext Ultra RNA Library Prep Kit for Illumina following the instruction manual. Libraries were size selected using Agentcourt AMPure XP Beads (Beckman Coulter), followed by amplification with 12 PCR cycles. RNA quality was assessed on the Fragment Analyzer (Agilent) by using Standard Sensitivity RNA Analysis Kit (Agilent).
Flow cytometry
U2OS cells expressing doxycycline inducible MYC were infected with lentiviral supernatants in the presence of 4 μg/ml polybrene or protamine sulfate (5 μg/ml) for 24 h. Medium (1:1, v/v) was added for 24 h. Cells were selected for 24 h with puromycin (2 μg/ml) and afterwards plated on a 6 cm dish. Cells were labelled with 10 μM 5-Bromo-2’-deoxyuridine (BrdU, Sigma) for 30- 50 min. Both supernatant and cells were harvested. Cell pellets were washed with ice-cold PBS and fixed with ice-cold 80 % ethanol, then incubated at -20 °C overnight. The cells were collected by centrifugation, washed with ice-cold PBS and incubated in 2 M HCl with 0.5 % Triton X-100 for 30 min at room temperature. Cell pellets were neutralized with Natriumtetraborat. Anti-BrdU-FITC antibody (BD Biosciences, BLD-364104) incubation was done in 100 μl 1 % BSA-PBS-T (0.5 % Tween-20 in PBS) for 30 min at room temperature in the dark. Then cells were washed with 1 % BSA-PBS-T and incubated in PBS and 24 ug/ml RNase A (Roche) at 4 °C over night in the dark. Data were acquired using the FACScanto II (BD Biosciences).
Colony formation assay
U2OS cells expressing doxycycline inducible MYC were infected with lentiviral supernatants in the presence of 4 μg/ml polybrene or protamine sulfate (5 μg/ml) for 24 h. Medium (1:1, v/v) was added for 24 h. Cells were selected for 24 h with puromycin (2 μg/ml) and afterwards plated on a 6-well plate. Cells were incubated with ethanol or doxycycline (1 μg/ml) for 24 h. Cells were fixed with 70% ethanol, dried and stained with crystal violet.
Knock-in of the HUWE1 catalytic mutant in HCT116 cells
The HUWE1 repair template included the sequence of human HUWE1 ORF (ENST00000342160.7) that encodes protein residues 4277-4374 of the isoform Q7Z6Z7-1 (with either cysteine or serine at position 4341), the P2A self-cleaving peptide, a blasticidin resistance gene, and homology arms spanning genomic positions chrX:53561159-53561889 and chrX:53559367-53560269. Two sgRNAs against HUWE1 were cloned in the PX459 vector (kind gift from Feng Zhang, Addgene 62988) and co-transfected with the repair template plasmid in the HCT116 cells using the Fugene reagent (Promega). Transfected cells were selected with puromycin and blasticidin (InvivoGen). Individual clones were validated by PCR and Sanger sequencing of genomic DNA and mRNA.
SILAC-based ubiquitin remnant profiling
U2OS SILAC-labeled cells were transfected with siRNAs targeting non-targeting control or HUWE1 as previously described or treated with BI8626 (10 μM, 24 h). Cells were lysed in modified RIPA buffer (50 mM Tris pH 7.5, 150 mM NaCl, 1 mM EDTA, 1% NP-40, 0.1% sodium deoxycholate) supplemented with protease inhibitors (Complete protease inhibitor cocktail tablets, Roche Diagnostics), 1 mM sodium orthovanadate, 5 mM β-glycerophosphate, 5 mM sodium fluoride and 10 mM N-ethylmaleimide. Proteins were digested with endoproteinase Lys-C (Wako Chemicals) and sequencing grade modified trypsin (Sigma-Aldrich). Modified peptide enrichment was done using di-glycine-lysine antibody resin (Cell Signaling Technology, 5562). Peptides were analyzed on a quadrupole Orbitrap mass spectrometer (Q Exactive Plus, Thermo Scientific) equipped with a UHPLC system (EASY-nLC 1000, Thermo Scientific) as described (Kelstrup et al., 2012; Michalski et al., 2012). MaxQuant (development version 1.5.2.8) was used to analyze the raw data files (Cox and Mann, 2008). Parent ion and MS2 spectra were searched against a human protein database obtained from UniProtKB released in May 2016 using Andromeda search engine (Cox et al., 2011). Experimental details were described previously (Heidelberger et al., 2018).
Bioinformatics
Sequencing libraries were subjected to Illumina NextSeq 500 sequencing according to the manufacturer’s instructions. After base calling with Illumina’s FASTQ Generation software v1.0.0 (NextSeq 500 sequencing), high quality PF-clusters were selected for further analyses and sequencing quality was ascertained using FastQC. ChIP samples were mapped to human hg19 and ChIP-Rx samples were mapped separately to the human hg19 and to the murine mm10 genome using Bowtie1 (Langmead et al., 2009) or Bowtie 2 (Langmead and Salzberg, 2012) with default parameters. ChIP samples were normalized to the number of mapped reads in the smallest sample. For ChIP-Rx spike-in normalized reads were calculated by dividing the number of mapped reads mapped to hg19 by the number of reads mapped to mm10 for each sample and multiplying this ratio with the smallest number of reads mapped to mm10 for any sample. RNAseq samples were mapped to hg19 using Bowtie2 (Langmead and Salzberg, 2012) and samples were normalized to the number of mapped reads in the smallest sample. Reads per gene were counted using the “summarizeOverlaps” function from the R package “GenomicAlignments” using the “union”-mode and Ensembl genes. Non- and weakly expressed genes were removed (mean count over all samples <1). Differentially expressed genes were called with edgeR and P-values were adjusted for multiple-testing using the Benjamini-Höchberg procedure. Metagene plots were generated with ngs.plot.r (Shen et al., 2014). Non-scaled density plots were produced with the plotProfile program from the DeepTools suite (Ramirez et al., 2016). Bin plots (Figure 6B) were drawn using R, by ordering 13’337 genes with significant expression in U2OS cells according to their inducibility by overexpressed MYC (Walz et al., 2014) into bins of 600 genes each. For each bin, the average expression ratio in control cells (“EtOH) or cells overexpressing MYC (“Dox) in the presence of the indicated shRNA was plotted on the Y axis, against the average inducibility by overexpressed MYC as derived from Walz et al. (2014) on the X axis (ratio “Dox / EtOH”). MYC-reads in promoter or enhancer regions were determined by processing read-normalized MYC ChIPseq bam-files with the BEDtools intersectBed program (Quinlan and Hall, 2010). Promoter regions were defined here as TSS +/- 1 kb, and enhancer regions were as previously defined (Walz et al., 2014). MYC-activated and -repressed genes were derived from (Lorenzin et al., 2016).
BLISS8 samples were demultiplexed based on their condition-specific barcodes using UMI-tools (Smith et al., 2017), allowing 1 mismatch in the barcode, and separately mapped to hg19
using Bowtie2 (Langmead and Salzberg, 2012) with default parameters. For Figure 5A,B and Figure S5C, respective samples of biological triplicates were merged preceding to mapping and collectively processed. Samples were filtered against an ENCODE Blacklist file to remove regions of high variance in mappability commonly found in satellite, centromeric and telomeric repeats (Amemiya et al., 2019) using bedtools intersect (Quinlan and Hall, 2010). To allow absolute quantification of double-strand breaks and remove PCR-introduced artifacts, duplicated reads were identified based on their UMI, grouped and deduplicated using UMI-tools (Smith et al., 2017) with default parameters. For normalization, deduplicated reads in AsiSI specific restriction sites were counted using countBamInGRanges from the R package exomeCopy. The sample with the smallest number of AsiSI specific reads was divided by the number of respective reads from each sample. Resulting ratio was multiplied by the total amount of deduplicated reads and samples subsequently randomly subsampled to the calculated number of reads. AsiSI specific restriction sites were generated by in silico digestion of the hg19 genome. From the 1,123 predicted restriction sites, sites without mapped reads across all conditions in the respective experiment were dropped. BLISS8 density profiles were generated using the R package metagene2 with the assay parameter ‘ChIPseq’, 150 bp read extension and 50 bins to smoothen the graph. Promoter counts were generated using the R package exomeCopy in the region of 500 bp up- and downstream of the annotated transcriptional start site and divided by the number of genes in the corresponding gene set. Gene sets were generated from RNA sequencing data using RPKM (gene expression) and logCPM (MYC response), or RNAPII ChIP-Rx data using the occupancy in the gene body versus the occupancy in the corresponding promoter region (pausing). Heterochromatic regions in U2OS were identified using a 16-state model (Ho et al., 2014) with chromHMM (Ernst and Kellis, 2012) and published datasets for H3K9me3 (Tasselli et al., 2016), H3K4me3, H3K27me3 (Easwaran et al., 2012),H3K79me2 (Clouaire et al., 2018), H3K36me3 (Wen et al., 2014), H3K4me1, H3K27ac (Walz et al., 2014).
Subsets of genes were derived from our RNAseq data in U2OS as well as from published U2OS ChIPseq data (Lorenzin et al., 2016; 3000 most/least expressed genes passing the minimal expression threshold; MYC-bound: 7684 genes that are expressed in U2OS according to our RNAseq data and contain a MYC-binding site in the region between -1500 and +500 relative to the transcription start site; non MYC-bound: 10013 expressed genes lacking such a MYC binding site; the former group was further subdivided based on the presence of a canonical E-box sequence [CACGTG] within 100 nucleotides of their MYC-binding summit, producing sub-groups of 1169 and 6515 genes, respectively). BLISS8 stratification by expression is based on published K562 polyA-RNAseq data from the ENCODE portal (https://www.encodeproject.org/) with the following identifiers: ENCSR040YBR. The R data set TxDb.Hsapiens.UCSC.hg19.knownGene was subsampled using the selected 5,576 bottom and 5,458 top expressed genes and filtered for a minimum gene length of 1500 bp. Further stratifications are based on respective lists mentioned in this paragraph and adapted as described. Artefacts produced by proximal downstream transcriptional start sites were filtered out. For Figure 5C the number of double stand breaks is presented relative to the number of genes per group, stratified by expression (top n=3,177, bottom n=1,814), pausing of RNAPII (paused n=3414, non-paused n=671), and response to MYC (activated n=340, repressed n=296).
Quantification And Statistical Analysis
General statistics
Statistical significance in Figure 3A and Figure S3A was determined by applying Wilcoxon rank sum test. Statistical analysis in Figure 3B and Figure S3B was performed in R by calculating the fold change to the non-targeting siRNA of the respective replicate and applying Welch’s t- test over all replicates for each siRNA to the non-targeting control with subsequent correction for multiple testing using Benjamini and Hochberg's FDR method. For Figure 4F, and Figure S5E, statistical significance was determined by applying the Welch’s t-test. For Figure 5C significance was calculated applying Wilcoxon rank sum test. Correlation and respective P- value were calculated using the Pearson product-moment correlation coefficient for the shown experiment in Figure S3C (left). Statistical significance in Figure S3C (right) was calculated using the R package limma across all biological replicates and corrected for multiple testing using Benjamini and Hochberg's FDR method. For Figure 6E and Figure S6E Wilcoxon test was used to calculate the significance. To calculate the significance in Figure S6E the different conditions were compared against the reference group (“shNTC EtOH”) within one cell cycle phase. For Figures S1A and S1I the relevant samples were compared using a unpaired t-test.
KEY RESOURCE TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mono- & polyubiquitinylated conjugates (FK2) | Enzo Life Sciences | BML-PW8810 RRID: AB_10541840 |
| Mouse monoclonal anti-Pol II (A-10) | Santa Cruz Biotechnology | Cat#sc-17798; RRID: AB_677355 |
| Mouse monoclonal anti-Pol II (F-12) | Santa Cruz Biotechnology | Cat#sc-55492 RRID: AB_630203 |
| Rabbit monoclonal anti-MYC (clone Y69) | Abcam | Cat# ab32072 RRID: AB_731658 |
| Mouse Monoclonal anti-MYC (C33) | Santa Cruz Biotechnology | Cat#sc-42 RRID: AB_2282408 |
| Rabbit polyclonal anti-phospho-Ser2-RNAPII | Abcam | Cat# ab5095 RRID: AB_304749 |
| Mouse monoclonal anti-RNAPII | MBL International | Cat# MABI0601 RRID: AB_2728735 |
| Rabbit polyclonal anti-Histone H2B antibody | Abcam | Ab1790 RRID: AB_302612 |
| Rabbit polyclonal anti-HUWE1/Mule antibody | Abcam | Ab70161 RRID: AB_1209511 |
| Rabbit polyclonal anti-CTR9 | Novus Biologicals | NB100-68205 RRID: AB_11002327 |
| Rabbit polyclonal anti-CTR9 | Bethyl Laboratories | A301-395 RRID: AB_960973 |
| Rabbit polyclonal anti-Parafibromin (CDC73) | Bethyl Laboratories | A300-171A RRID: AB_2078660 |
| Rabbit polyclonal anti-PAF1 | Abcam | Ab20662 RRID: AB_2159769 |
| Rabbit polyclonal anti-LEO1 | Novus Biologicals | NB600-276 RRID: AB_2281237 |
| Goat polyclonal anti-GST | GE Healthcare/Sigma- Aldrich | Cat#GE27-4577-01; Lot: 362611 |
| Rabbit monoclonall Ubiquityl-Histone H2B (Lys120) (D11) XP® | Cell Signaling Technology | Cat#5546 RRID: AB_10693452 |
| Mouse monoclonal anti-VCL | Sigma-Aldrich | Cat#V9131 RRID: AB_477629 |
| Mouse monoclonal anti-phospho-Ser5-RNAPII | Biolegend | Cat#904001 RRID: AB_2565036 |
| Rabbit polyclonal anti- CDK2 | Santa Cruz Biotechnology | Cat#sc-163 RRID: AB_631215 |
| Rabbit polyclonal anti-CHK1(FL-476) | Santa Cruz Biotechnology | Cat#sc-7898 RRID: AB_2229488 |
| Rabbit polyclonal anti-phospho-Chk1 (Ser345) (133D3) | Cell Signaling Technology | Cat#2348 RRID: AB_331212 |
| Rabbit polyclonal anti- phospho-Histone H2A.X (Ser 139) | Cell Signaling Technology | Cat#2577 RRID: AB_2118010 |
| Rabbit polyclonal anti-KAP1 (phospho S824) | Abcam | Cat#ab70369 RRID: AB_1209417 |
| Rabbit polyclonal anti- KAP1 | Bethyl Laboratories | Cat#A300-274A RRID: AB_185559 |
| Rabbit monoclonall Anti-TH1L (D5G6W) (NELFC) | Cell Signaling Technology | Cat#12265S Lot:1 RRID: AB_2797862 |
| Mouse monoclonal FITC anti-BrdU (clone 3D4) | Biozol / BioLegend | Cat#364104 RRID: AB_2564481 |
| Donkey polyclonal anti-goat IgG-HRP secondary antibody | Santa Cruz Biotechnology | Cat#sc-2020 RRID: AB_631728 |
| ECL-Anti-rabbit IgG Horseradish Peroxidase | GE Healthcare / Fisher Scientific GmbH | Cat#1079-4347 / GEHENA934 |
| ECL-Anti-mouse IgG Horseradish Peroxidase | GE Healthcare / Fisher Scientific GmbH | Cat#1019-6124 / GEHENA931 |
| IRDye 800CW Donkey anti-Rabbit IgG (H + L) | LI-COR Biosciences | Cat#926-32213 RRID: AB_621848 |
| IRDye 680RD Donkey anti-Mouse IgG (H + L) | LI-COR Biosciences | Cat#926-68072 RRID: AB_10953628 |
| Goat anti-Mouse IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor 488 | Thermo Fisher Scientific | Cat#A-11029 RRID: AB_138404 |
| Goat anti-Mouse IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor 568 | Thermo Fisher Scientific | Cat#A-11004 RRID: AB_2534072 |
| Bacterial and Virus Strains | ||
| pRRL-SFFV-IRES-Hygro | Wiese et al., 2015 | N/A |
| pRRL-SFFV-MYC-IRES-Hygro | Wiese et al., 2015 | N/A |
| pRRL-SFFV-OsTir1_3x_Myc_tag-T2A-eBFP2 | Muhar et al., 2018 | N/A |
| pInducer10 shCDC73-3 | This paper | N/A |
| pInducer10 shCTR9-3 | This paper | N/A |
| pGIPZ shCDC73-3 | This paper | N/A |
| pGIPZ shCDC73-4 | This paper | N/A |
| pGIPZ shCTR9-3 | This paper | N/A |
| pGIPZ shCTR9-5 | This paper | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| MG-132 | Calbiochem / Merck | Cat#474790-20MG |
| HUWE1-Inhibitor BI8626 | ProbeChem | N/A |
| Doxycycline hyclate | Sigma-Aldrich | Cat#D9891-10G |
| Indole-3-acetic acid sodium salt | Sigma-Aldrich | Cat# I5148-2G |
| Hoechst 33342 | Sigma-Aldrich | Cat#B2261-25MG |
| 5-Ethynyl-2'-deoxyuridine (5-EdU) | Jena Bioscience | Cat#CLK-N001-100 |
| AF647-Picolyl-Azide | Jena Bioscience | Cat#CLK-1300-1 |
| Lipofectamine RNAiMAX Transfection Reagent | Thermo Fischer Scientific | Cat#13778-150 |
| Dynabeads Protein A | Thermo Fisher Scientific | Cat#10001D |
| Dynabeads Protein G | Thermo Fisher Scientific | Cat#10003D |
| Dynabeads® MyOne™ Streptavidin T1 | Thermo Fisher Scientific | Cat#65601 |
| Etoposide | Sigma-Aldrich | Cat#E1383 |
| 4-Hydroxytamoxifen | Sigma-Aldrich | Cat#H7904 |
| Opti-MEM I | Thermo Fischer Scientific | Cat#31985-047 |
| Propidium iodide | Sigma-Aldrich | Cat#81845 |
| Protease inhibitor cocktail | Sigma-Aldrich | Cat#P8340 |
| Phosphatase inhibitor cocktail 2 | Sigma-Aldrich | Cat#P5726 |
| Phosphatase inhibitor cocktail 3 | Sigma-Aldrich | Cat#P0044 |
| NuPAGE LDS Sample Buffer (4X) | Sigma-Aldrich | Cat#NP0007 |
| Pierce™ DTT (Dithiothreitol), No-Weigh™ Format | Thermo Fisher Scientific | Cat#20291 |
| Benzonase nuclease purity >99% 25U/μl | Merck Millipore | Cat#70664-3 |
| InstantBlue(TM) Safe Coomassie Stain | Sigma-Aldrich | Cat#ISB1L-1L |
| Proteinase K | Roth | Cat#7528.2 |
| RNase A | Roth | Cat#7156.1 |
| 16% Paraformaldehyde (Formaldehyde) Aqueous Solution, EM Grade | Science Services GmbH | Cat#E15710 |
| Penicillin-Streptomycin | Sigma-Aldrich | Cat#P4333-100ML |
| HiMark pre-stained HMW STD | Thermo Fisher Scientific | Cat#LC5699 |
| UltraPure BSA (50 mg/mL) | Thermo Fisher Scientific | Cat#AM2616 |
| Polybrene | Sigma-Aldrich | Cat#H9268 |
| Protamine sulfate | Sigma-Aldrich | Cat#P3369 |
| Alexa Fluor568 Phalloidin | Thermo Fisher Scientific | Cat#A12380 |
| N-Ethylmaleinimid | Sigma-Aldrich | Cat#E3876 |
| Puromycin | InvivoGen | Cat#70664-3 |
| Hygromycin B Gold solution | InvivoGen | Cat#ant-hg-05 |
| Blasticidin | InvivoGen | Cat#ant-bl-05 |
| CutSmart® Buffer | New England Biolabs | Cat#B7204S |
| T4 DNA Ligase Buffer | New England Biolabs | Cat#B0202S |
| Agencourt AMPure XP Beads | Beckman Coulter | Cat#A63881 |
| Agencourt RNAClean XP Beads | Beckman Coulter | Cat#A63987 |
| LDC4297 | Selleckchem / Biozol | Cat#SEL-S7992 |
| NVP-2 | Tocris/Bio-Techne | Cat#6535/5 |
| Odyssey Blocking Buffer in TBS | LI-COR Biosciences | Cat#927-50000 |
| Glutathione Sepharose 4B | GE Healthcare / VWR International | Cat#17075601 |
| L-Glutathion reduced, cell culture tested | Sigma-Aldrich | Cat#G6013-5G |
| Fugene | Promega | Cat#E2311 |
| Critical Commercial Assays | ||
| Duolink In Situ PLA Probe Anti-Rabbit PLUS, Affinity purified Donkey anti-Rabbit IgG (H+L) | Sigma-Aldrich | Cat#DUO92002 RRID: AB_2810940 |
| Duolink In Situ PLA Probe Anti-Mouse MINUS, Affinity purified Donkey anti-Mouse IgG (H+L) | Sigma-Aldrich | Cat#DUO92004 RRID: AB_2713942 |
| Duolink In Situ Wash Buffers, Fluorescence | Sigma-Aldrich | Cat#DUO82049 |
| Duolink In Situ Detection Reagents Green | Sigma-Aldrich | Cat#DUO92014 |
| NGS Fragment High Sensitivity Analysis Kit, 1-6,000 bp, 500 samples | Agilent | DNF-474-0500 |
| Standard Sensitivity RNA Analysis Kit (15nt), 500 samples | Agilent | DNF-471-0500 |
| RNeasy Mini Kit | Qiagen | Cat#74106 |
| MinElute PCR Purification Kit | Qiagen | Cat#28006 |
| QIAquick PCR Purification Kit | Qiagen | Cat#28106 |
| QIAquick Gel Extraction Kit | Qiagen | Cat#28704 |
| NEBNext Ultra RNA Library Prep Kit for Illumina | New England Biolabs | Cat#E7530S |
| NEBNext Poly(A) mRNA Magnetic Isolation Modul | New England Biolabs | Cat#E7490L |
| NEBNext ChIP-Seq Prep Master Mix Set for Illumina | New England Biolabs | Cat#E6240S |
| NEBNext Ultra II DNA Library Prep | New England Biolabs | Cat# E7103L |
| NEBNext Multiplex Small RNA Library Prep Kit | New England Biolabs | Cat#E7560S |
| NextSeq 500/550 High Output Kit v2 (75cycles) | Illumina | FC-404-2005 |
| Quant-iT Pico Green | Thermo Fischer Scientific | Cat#P7589 |
| AsiSI | New England Biolabs | Cat# R0630 |
| ABsolute QPCR Mix, SYBR Green, no ROX | Thermo Fischer Scientific | Cat#AB-1158/B |
| Quick Blunting Kit | New England Biolabs | Cat#E1201L |
| T4 DNA Ligase, conc. | New England Biolabs | Cat#M0202M |
| T4 RNA Ligase 2, truncated | New England Biolabs | Cat#M0242L |
| NEBNext High-Fidelity 2X PCR Master Mix | New England Biolabs | Cat#M0541L |
| MEGAscript T7 Transcription Kit | Thermo Fischer Scientific | Cat#AM1334 |
| SuperScript III Reverse Transcriptase | Thermo Fischer Scientific | Cat#T18080044 |
| RNaseOUT Recombinant Ribonuclease Inhibitor | Thermo Fischer Scientific | Cat#10777019 |
| ON-TARGETplus Non-targeting Pool | Horizon Discovery Group | Cat#D-001810-10-50 |
| ON-TARGETplus Set of Four siRNA Library- Human Ubiquitin Conjugation Subset 1 | Dharmacon | GU-105615 Lot 11107 |
| ON-TARGETplus Set of Four siRNA Library- Human Ubiquitin Conjugation Subset 2 | Dharmacon | GU-105625 Lot 11108 |
| ON-TARGETplus SMARTpool® siRNA Library - Human Ubiquitin Conjugation Subset 3 | Dharmacon | GU-105635 Lot 11117 |
| PTMScan Ubiquitin Remnant Motif (K-ε-GG) Kit | Cell Signaling Technology | Cat#5562 |
| Deposited Data | ||
| Sequencing Data | This paper | GSE150217 |
| Raw imaging data | This paper | 10.17632/n4rr8ck4w 3.1 |
| Experimental Models: Cell Lines | ||
| NIH 3T3 | ATCC | CVCL_0594 |
| U2OS | ATCC | N/A |
| HEK293TN | ATCC | CRT-11268 |
| U2OS MYC-Tet-On | Walz et al., 2014 | N/A |
| U2OS MYC-ER | Liu et al., 2012 | N/A |
| K562 MYC-Aid | Muhar et al., 2018 | N/A |
| Oligonucleotides | ||
| Primer ChIP_qPCR NCL_f CTACCACCCTCATCTGAATCC | Baluapuri et al., 2019 | N/A |
| Primer ChIP qPCR NCL_r TTGTCTCGCTGGGAAAGG | Baluapuri et al., 2019 | N/A |
| Primer ChIP qPCR NegReg_f TTTTCTCACATTGCCCCTGT | Baluapuri et al., 2019 | N/A |
| Primer ChIP qPCR NegReg_r TCAATGCTGTACCAGGCAAA | Baluapuri et al., 2019 | N/A |
| Primer ChIP qPCR GNL3 f GTGACGCTCGTCAGTGG | Jaenicke et al., 2016 | N/A |
| Primer ChIP qPCR GNL3 r CATATTGGCTGTAGAAGG AAGC | Jaenicke et al., 2016 | N/A |
| Primer ChIP qPCR NPM1 f TTCACCGGGAAGCATGG | Jaenicke et al., 2016 | N/A |
| Primer ChIP qPCR NPM1 r CACGCGAGGTAAGTCTACG | Jaenicke et al., 2016 | N/A |
| Primer ChIP qPCR IFRD1 r CGTGGTTTGGCTACTGAACT | This paper | N/A |
| Primer ChIP qPCR IFRD1 f CCTGTCCCGACACACTCTC | This paper | N/A |
| Primer ChIP qPCR CHMP2A r CAAGGTGGTGTTGGAGACCT | This paper | N/A |
| Primer ChIP qPCR CHMP2A f GGGGATCCCAGAAAGAGAAG | This paper | N/A |
| shCDC73 human mirE3 TGCTGTTGACAGTGAGCGCCAGCGATCTACTCAAG TCAAATAGTGAAGCCACAGATGTATTTGACTTGAGT AGATCGCTGATGCCTACTGCCTCGGA | Fellmann et al., 2013 | N/A |
| shCDC73 human mirE4 TGCTGTTGACAGTGAGCGCCAGGTACATGGTAAAG CATAATAGTGAAGCCACAGATGTATTATGCTTTACC ATGTACCTGTTGCCTACTGCCTCGGA | Fellmann et al., 2013 | N/A |
| shCTR9 human mirE3 TGCTGTTGACAGTGAGCGCTCGGATGAGGATAAAC TTAAATAGTGAAGCCACAGATGTATTTAAGTTTATCC TCATCCGAATGCCTACTGCCTCGGA | Fellmann et al., 2013 | N/A |
| shCTR9 human mirE5 TGCTGTTGACAGTGAGCGAAAGCAACAAAAGAGAA GAAAATAGTGAAGCCACAGATGTATTTTCTTCTCTTT TGTTGCTTCTGCCTACTGCCTCGGA | Fellmann et al., 2013 | N/A |
| MYC-f (pGex4T3) CCCGAATTCGCCCCTCAACGTTAGCTTC | Baluapuri et al., 2019 | N/A |
| MYC-r (pGex4T3) GGGCTCGAGTCAGTTCGGGCTGCCGCTGTCT | Baluapuri et al., 2019 | N/A |
| A1_Bottom [P]GCGTGATGNNNNNNNNGATCGTCGGACTGTAGA ACTCTGAACCCCTATAGTGAGTCGTATTACCGGCCT CAATCGAA | Yan et al., 2017 | N/A |
| A1_Top CGATTGAGGCCGGTAATACGACTCACTATAGGGGT TCAGAGTTCTACAGTCCGACGATCNNNNNNNNCAT CACGC | Yan et al., 2017 | N/A |
| A2_Bottom [P]GGAACGACNNNNNNNNGATCGTCGGACTGTAGA ACTCTGAACCCCTATAGTGAGTCGTATTACCGGCCT CAATCGAA | Yan et al., 2017 | N/A |
| A2_Top CGATTGAGGCCGGTAATACGACTCACTATAGGGGT TCAGAGTTCTACAGTCCGACGATCNNNNNNNNGTC GTTCC | Yan et al., 2017 | N/A |
| A3_Bottom [P]GATCATCANNNNNNNNGATCGTCGGACTGTAGAA CTCTGAACCCCTATAGTGAGTCGTATTACCGGCCTC AATCGAA | Yan et al., 2017 | N/A |
| A3_Top CGATTGAGGCCGGTAATACGACTCACTATAGGGGT TCAGAGTTCTACAGTCCGACGATCNNNNNNNNTGAT GATC | Yan et al., 2017 | N/A |
| A4_Bottom [P]GATGTCGTNNNNNNNNGATCGTCGGACTGTAGA ACTCTGAACCCCTATAGTGAGTCGTATTACCGGCCT CAATCGAA | Yan et al., 2017 | N/A |
| A4_Top CGATTGAGGCCGGTAATACGACTCACTATAGGGGT TCAGAGTTCTACAGTCCGACGATCNNNNNNNNACG ACATC | Yan et al., 2017 | N/A |
| A5_Bottom [P]GGATGATGNNNNNNNNGATCGTCGGACTGTAGA ACTCTGAACCCCTATAGTGAGTCGTATTACCGGCCT CAATCGAA | Yan et al., 2017 | N/A |
| A5_Top CGATTGAGGCCGGTAATACGACTCACTATAGGGGT TCAGAGTTCTACAGTCCGACGATCNNNNNNNNCAT CATCC | Yan et al., 2017 | N/A |
| A6_Bottom [P]GCGGTCGTNNNNNNNNGATCGTCGGACTGTAGA ACTCTGAACCCCTATAGTGAGTCGTATTACCGGCCT CAATCGAA | Yan et al., 2017 | N/A |
| A6_Top CGATTGAGGCCGGTAATACGACTCACTATAGGGGT TCAGAGTTCTACAGTCCGACGATCNNNNNNNNACG ACCGC | Yan et al., 2017 | N/A |
| RPI_01 CAAGCAGAAGACGGCATACGAGATCGAGTAATGTG ACTGGAGTTCCTTGGCACCCGAGAATTCCA | This paper | N/A |
| RPI_02 CAAGCAGAAGACGGCATACGAGATTCTCCGGAGTG ACTGGAGTTCCTTGGCACCCGAGAATTCCA | This paper | N/A |
| RPI_03 CAAGCAGAAGACGGCATACGAGATAATGAGCGGTG ACTGGAGTTCCTTGGCACCCGAGAATTCCA | This paper | N/A |
| RPI_04 CAAGCAGAAGACGGCATACGAGATGGAATCTCGTG ACTGGAGTTCCTTGGCACCCGAGAATTCCA | This paper | N/A |
| RPI_05 CAAGCAGAAGACGGCATACGAGATTTCTGAATGTGA CTGGAGTTCCTTGGCACCCGAGAATTCCA | This paper | N/A |
| RPI_06 CAAGCAGAAGACGGCATACGAGATACGAATTCGTG ACTGGAGTTCCTTGGCACCCGAGAATTCCA | This paper | N/A |
| RPI_07 CAAGCAGAAGACGGCATACGAGATAGCTTCAGGTG ACTGGAGTTCCTTGGCACCCGAGAATTCCA | This paper | N/A |
| RPI_08 CAAGCAGAAGACGGCATACGAGATGCGCATTAGTG ACTGGAGTTCCTTGGCACCCGAGAATTCCA | This paper | N/A |
| RPI_09 CAAGCAGAAGACGGCATACGAGATCATAGCCGGTG ACTGGAGTTCCTTGGCACCCGAGAATTCCA | This paper | N/A |
| RPI_10 CAAGCAGAAGACGGCATACGAGATTTCGCGGAGTG ACTGGAGTTCCTTGGCACCCGAGAATTCCA | This paper | N/A |
| RPI_11 CAAGCAGAAGACGGCATACGAGATGCGCGAGAGTG ACTGGAGTTCCTTGGCACCCGAGAATTCCA | This paper | N/A |
| RPI_12 CAAGCAGAAGACGGCATACGAGATCTATCGCTGTG ACTGGAGTTCCTTGGCACCCGAGAATTCCA | This paper | N/A |
| RA3 TGGAATTCTCGGGTGCCAAGG | Illumina | N/A |
| RTP GCCTTGGCACCCGAGAATTCCA | Illumina | N/A |
| RP1 AATGATACGGCGACCACCGAGATCTACACGTTCAG AGTTCTACAGTCCGA | Illumina | N/A |
| SiHUWE1 GAGUUUGGAGUUUGUGAAG[dT][dT] | Heidelberger et al., 2018 | N/A |
| sgRNA HUWE1_1 AAGGCCCTGCCCAACTCCGT | This paper | N/A |
| sgRNA HUWE1_2 CATGCTACTGTTGGCTATCC | This paper | N/A |
| Recombinant DNA | ||
| pGex-4T3 | Pharmacia | N/A |
| pGex-4T3-MYC1–163 | Baluapuri et al., 2019 | N/A |
| pGex-4T3-MYC1–163 DMBI | This Paper | N/A |
| pGex-4T3-MYC1–163 DMBII | This Paper | N/A |
| pInducer10 | Trono Laboratory | N/A |
| pGIPZ | Dharmacon | N/A |
| psPAX2 | Trono Laboratory | Addgene 12260 |
| pMD2.G | Trono Laboratory | Addgene 12259 |
| PX459 | Zhang Laboratory | Addgene 62988 |
| Software and Algorithms | ||
| FASTQ Generation software v1.0.0 | Illumina | http://illumina.com |
| FastQC v0.11.3 | http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ | https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ |
| Tophat v2.1.0 | Kim et al., 2013 | https://ccb.jhu.edu/software/tophat/index.shtml |
| Bowtie v1.2 | Langmead et al., 2009 | http://bowtie-bio.sourceforge.net/index.shtml |
| Bowtie v2.3.2 | Langmead and Salzberg, 2012 | http://bowtie-bio.sourceforge.net/bowtie2/index.shtml |
| MACS v1.4.1 | Zhang et al., 2008 | https://github.com/taoliu/MACS |
| BEDtools v2.26.0 | Quinlan and Hall, 2010 | https://bedtools.readthedocs.io/en/latest/ |
| SAMtools v1.3 | Li et al., 2009 | http://www.htslib.org/ |
| NGSplot v2.61 | Shen et al., 2014 | https://github.com/shenlab-sinai/ngsplot |
| Integrated Genome Browser v9.0.0 | Freese et al., 2016 | https://bioviz.org/ |
| GraphPad Prism v5/6.0 for Mac | GraphPad software | https://www.graphpad.com/scientific-software/prism/ |
| Harmony High Content Imaging and Analysis Software | PerkinElmer | http://www.perkinelmer.de/product/harmony-4-8-office-hh17000001 |
| StepOne software v2.3 | StepOne | https://www.thermofisher.com/de/de/home/technical-resources/software-downloads/StepOne-and-StepOnePlus-Real-Time-PCR-System.html |
| BD FACSDIVA Software v6.1.2 | BD | http://www.bdbiosciences.com/us/instruments/research/software/flow-cytometry-acquisition/bd-facsdiva-software/m/111112/overview |
| EdgeR | Robinson et al., 2010 | https://bioconductor.org/packages/release/bioc/html/edgeR.html |
| R version 3.6.3 | The R Foundation | https://www.R-project.org/ |
| UMI-tools v1.0.0 | Smith et al., 2017 | https://umi-tools.readthedocs.io/en/latest/index.html |
| DeepTools | Ramirez et al., 2016 | https://deeptools.readthedocs.io/en/develop/ |
| Image Studio version 5.2.5 | LI-COR | http://opensource.licor.com/licenses/ImageStudio/index.html |
| MaxQuant version 1.5.2.8 | Cox and Mann, 2008 | https://www.maxquant.org/ |
Supplementary Material
Acknowledgements
This work was supported by grants from the European Research Council (AuroMYC), the German Research Foundation via the Graduate College GRK 2243 and the European Fonds for Regional Development to M.E.. P.B. is supported by the Emmy Noether Program (BE 5342/1-1 and BE5342/1-2). S.M.V. was supported by an EMBO Long-Term Fellowship (ALTF 745-2014). N.P is supported by the DFG Research Unit FOR2314 "Targeting therapeutic windows in essential cellular processes for tumor therapy" (PO 1458/7-1).
Footnotes
Author Contributions
T.E. and D.S. performed ChIP-qPCR, ChIP-Rx, BLISS8, Immunoblot and Cell-Cycle-Immunofluorescence assays. T.E. performed RNAseq, in-vitro pulldown, shRNA and MYC-ER/-AID assays. D.S. performed siRNA-screen, PLA and Immunoprecipitation assays. D.S. and J.B.H. performed diGLY-SILAC. N.P. and V.A. developed and analysed the HUWE1 knock-in cells. A.B. and E.W. performed confocal imaging and image analysis. P.G., T.E. and D.S. performed bioinformatic analyses. P.B., P.G. and M.E. wrote the paper. S.M.V. and P.C. purified and characterized the PAF1 complex, M.M. and J.Z. generated and characterized the MYC AID system.
Declaration of Interests
The authors declare no competing interests.
References
- Adhikary S, Marinoni F, Hock A, Hulleman E, Popov N, Beier R, Bernard S, Quarto M, Capra M, Goettig S, et al. The ubiquitin ligase HectH9 regulates transcriptional activation by Myc and is essential for tumor cell proliferation. Cell. 2005;123:409–421. doi: 10.1016/j.cell.2005.08.016. [DOI] [PubMed] [Google Scholar]
- Amemiya HM, Kundaje A, Boyle AP. The ENCODE Blacklist: Identification of Problematic Regions of the Genome. Scientific reports. 2019;9:9354. doi: 10.1038/s41598-019-45839-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Annibali D, Whitfield JR, Favuzzi E, Jauset T, Serrano E, Cuartas I, Redondo-Campos S, Folch G, Gonzalez-Junca A, Sodir NM, et al. Myc inhibition is effective against glioma and reveals a role for Myc in proficient mitosis. Nat Commun. 2014;5:4632. doi: 10.1038/ncomms5632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoi Y, Smith ER, Shah AP, Rendleman EJ, Marshall SA, Woodfin AR, Chen FX, Shiekhattar R, Shilatifard A. NELF Regulates a Promoter-Proximal Step Distinct from RNA Pol II Pause-Release. Mol Cell. 2020;78:261–274.:e265. doi: 10.1016/j.molcel.2020.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baluapuri A, Hofstetter J, Dudvarski Stankovic N, Endres T, Bhandare P, Vos SM, Adhikari B, Schwarz JD, Narain A, Vogt M, et al. MYC Recruits SPT5 to RNA Polymerase II to Promote Processive Transcription Elongation. Mol Cell. 2019;74:674–687.:e611. doi: 10.1016/j.molcel.2019.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baluapuri A, Wolf E, Eilers M. Target gene-independent functions of MYC oncoproteins. Nature reviews. 2020;21:255–267. doi: 10.1038/s41580-020-0215-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baranello L, Wojtowicz D, Cui K, Devaiah BN, Chung HJ, Chan-Salis KY, Guha R, Wilson K, Zhang X, Zhang H, et al. RNA Polymerase II Regulates Topoisomerase 1 Activity to Favor Efficient Transcription. Cell. 2016;165:357–371. doi: 10.1016/j.cell.2016.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaulieu ME, Jauset T, Masso-Valles D, Martinez-Martin S, Rahl P, Maltais L, Zacarias-Fluck MF, Casacuberta-Serra S, Serrano Del Pozo E, Fiore C, et al. Intrinsic cell-penetrating activity propels Omomyc from proof of concept to viable anti-MYC therapy. Science translational medicine. 2019;11 doi: 10.1126/scitranslmed.aar5012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchel G, Carstensen A, Mak KY, Roeschert I, Leen E, Sumara O, Hofstetter J, Herold S, Kalb J, Baluapuri A, et al. Association with Aurora-A Controls N-MYC-Dependent Promoter Escape and Pause Release of RNA Polymerase II during the Cell Cycle. Cell reports. 2017;21:3483–3497. doi: 10.1016/j.celrep.2017.11.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunch H, Lawney BP, Lin YF, Asaithamby A, Murshid A, Wang YE, Chen BP, Calderwood SK. Transcriptional elongation requires DNA break-induced signalling. Nat Commun. 2015;6:10191. doi: 10.1038/ncomms10191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bywater MJ, Burkhart DL, Straube J, Sabo A, Pendino V, Hudson JE, Quaife-Ryan GA, Porrello ER, Rae J, Parton RG, et al. Reactivation of Myc transcription in the mouse heart unlocks its proliferative capacity. Nat Commun. 2020;11:1827. doi: 10.1038/s41467-020-15552-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YH, Keegan S, Kahli M, Tonzi P, Fenyo D, Huang TT, Smith DJ. Transcription shapes DNA replication initiation and termination in human cells. Nat Struct Mol Biol. 2019;26:67–77. doi: 10.1038/s41594-018-0171-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiarle R, Zhang Y, Frock RL, Lewis SM, Molinie B, Ho YJ, Myers DR, Choi VW, Compagno M, Malkin DJ, et al. Genome-wide translocation sequencing reveals mechanisms of chromosome breaks and rearrangements in B cells. Cell. 2011;147:107–119. doi: 10.1016/j.cell.2011.07.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clouaire T, Rocher V, Lashgari A, Arnould C, Aguirrebengoa M, Biernacka A, Skrzypczak M, Aymard F, Fongang B, Dojer N, et al. Comprehensive Mapping of Histone Modifications at DNA Double-Strand Breaks Deciphers Repair Pathway Chromatin Signatures. Mol Cell. 2018;72:250–262.:e256. doi: 10.1016/j.molcel.2018.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courilleau C, Chailleux C, Jauneau A, Grimal F, Briois S, Boutet-Robinet E, Boudsocq F, Trouche D, Canitrot Y. The chromatin remodeler p400 ATPase facilitates Rad51-mediated repair of DNA double-strand breaks. J Cell Biol. 2012;199:1067–1081. doi: 10.1083/jcb.201205059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- Cox J, Neuhauser N, Michalski A, Scheltema RA, Olsen JV, Mann M. Andromeda: a peptide search engine integrated into the MaxQuant environment. J Proteome Res. 2011;10:1794–1805. doi: 10.1021/pr101065j. [DOI] [PubMed] [Google Scholar]
- Cramer P. Organization and regulation of gene transcription. Nature. 2019;573:45–54. doi: 10.1038/s41586-019-1517-4. [DOI] [PubMed] [Google Scholar]
- Dang CV. MYC on the path to cancer. Cell. 2012;149:22–35. doi: 10.1016/j.cell.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Pretis S, Kress TR, Morelli MJ, Sabo A, Locarno C, Verrecchia A, Doni M, Campaner S, Amati B, Pelizzola M. Integrative analysis of RNA polymerase II and transcriptional dynamics upon MYC activation. Genome Res. 2017;27:1658–1664. doi: 10.1101/gr.226035.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez-Brauer C, Khatun R, Elia AJ, Thu KL, Ramachandran P, Baniasadi SP, Hao Z, Jones LD, Haight J, Sheng Y, et al. E3 ubiquitin ligase Mule targets beta-catenin under conditions of hyperactive Wnt signaling. Proceedings of the National Academy of Sciences of the United States of America. 2017;114:E1148–E1157. doi: 10.1073/pnas.1621355114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Easwaran H, Johnstone SE, Van Neste L, Ohm J, Mosbruger T, Wang Q, Aryee MJ, Joyce P, Ahuja N, Weisenberger D, et al. A DNA hypermethylation module for the stem/progenitor cell signature of cancer. Genome Res. 2012;22:837–849. doi: 10.1101/gr.131169.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst J, Kellis M. ChromHMM: automating chromatin-state discovery and characterization. Nature methods. 2012;9:215–216. doi: 10.1038/nmeth.1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fellmann C, Hoffmann T, Sridhar V, Hopfgartner B, Muhar M, Roth M, Lai DY, Barbosa IA, Kwon JS, Guan Y, et al. An optimized microRNA backbone for effective single-copy RNAi. Cell reports. 2013;5:1704–1713. doi: 10.1016/j.celrep.2013.11.020. [DOI] [PubMed] [Google Scholar]
- Frank SR, Parisi T, Taubert S, Fernandez P, Fuchs M, Chan HM, Livingston DM, Amati B. MYC recruits the TIP60 histone acetyltransferase complex to chromatin. EMBO Rep. 2003;4:575–580. doi: 10.1038/sj.embor.embor861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs G, Hollander D, Voichek Y, Ast G, Oren M. Cotranscriptional histone H2B monoubiquitylation is tightly coupled with RNA polymerase II elongation rate. Genome Res. 2014;24:1572–1583. doi: 10.1101/gr.176487.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs M, Gerber J, Drapkin R, Sif S, Ikura T, Ogryzko V, Lane WS, Nakatani Y, Livingston DM. The p400 complex is an essential E1A transformation target. Cell. 2001;106:297–307. doi: 10.1016/s0092-8674(01)00450-0. [DOI] [PubMed] [Google Scholar]
- Fujinaga K, Irwin D, Huang Y, Taube R, Kurosu T, Peterlin BM. Dynamics of human immunodeficiency virus transcription: P-TEFb phosphorylates RD and dissociates negative effectors from the transactivation response element. Molecular and cellular biology. 2004;24:787–795. doi: 10.1128/MCB.24.2.787-795.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabay M, Li Y, Felsher DW. MYC activation is a hallmark of cancer initiation and maintenance. Cold Spring Harbor perspectives in medicine. 2014;4 doi: 10.1101/cshperspect.a014241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlach JM, Furrer M, Gallant M, Birkel D, Baluapuri A, Wolf E, Gallant P. PAF1 complex component Leo1 helps recruit Drosophila Myc to promoters. Proceedings of the National Academy of Sciences of the United States of America. 2017;114:E9224–E9232. doi: 10.1073/pnas.1705816114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gothe HJ, Bouwman BAM, Gusmao EG, Piccinno R, Petrosino G, Sayols S, Drechsel O, Minneker V, Josipovic N, Mizi A, et al. Spatial Chromosome Folding and Active Transcription Drive DNA Fragility and Formation of Oncogenic MLL Translocations. Mol Cell. 2019;75:267–283.:e212. doi: 10.1016/j.molcel.2019.05.015. [DOI] [PubMed] [Google Scholar]
- Gregersen LH, Svejstrup JQ. The Cellular Response to Transcription-Blocking DNA Damage. Trends Biochem Sci. 2018;43:327–341. doi: 10.1016/j.tibs.2018.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudjonsson T, Altmeyer M, Savic V, Toledo L, Dinant C, Grofte M, Bartkova J, Poulsen M, Oka Y, Bekker-Jensen S, et al. TRIP12 and URB5 suppress spreading of chromatin ubiquitylation at damaged chromosomes. Cell. 2012;150:697–709. doi: 10.1016/j.cell.2012.06.039. [DOI] [PubMed] [Google Scholar]
- Guo J, Li T, Schipper J, Nilson KA, Fordjour FK, Cooper JJ, Gordan R, Price DH. Sequence specificity incompletely defines the genome-wide occupancy of Myc. Genome Biol. 2014;15:482. doi: 10.1186/s13059-014-0482-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamperl S, Bocek MJ, Saldivar JC, Swigut T, Cimprich KA. Transcription-Replication Conflict Orientation Modulates R-Loop Levels and Activates Distinct DNA Damage Responses. Cell. 2017;170:774–786.:e719. doi: 10.1016/j.cell.2017.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidelberger JB, Voigt A, Borisova ME, Petrosino G, Ruf S, Wagner SA, Beli P. Proteomic profiling of VCP substrates links VCP to K6-linked ubiquitylation and c-Myc function. EMBO Rep. 2018;19 doi: 10.15252/embr.201744754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herold S, Kalb J, Buchel G, Ade CP, Baluapuri A, Xu J, Koster J, Solvie D, Carstensen A, Klotz C, et al. Recruitment of BRCA1 limits MYCN-driven accumulation of stalled RNA polymerase. Nature. 2019;567:545–549. doi: 10.1038/s41586-019-1030-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho JW, Jung YL, Liu T, Alver BH, Lee S, Ikegami K, Sohn KA, Minoda A, Tolstorukov MY, Appert A, et al. Comparative analysis of metazoan chromatin organization. Nature. 2014;512:449–452. doi: 10.1038/nature13415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang CH, Lujambio A, Zuber J, Tschaharganeh DF, Doran MG, Evans MJ, Kitzing T, Zhu N, de Stanchina E, Sawyers CL, et al. CDK9-mediated transcription elongation is required for MYC addiction in hepatocellular carcinoma. Genes & development. 2014;28:1800–1814. doi: 10.1101/gad.244368.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutterer C, Eickhoff J, Milbradt J, Korn K, Zeittrager I, Bahsi H, Wagner S, Zischinsky G, Wolf A, Degenhart C, et al. A novel CDK7 inhibitor of the Pyrazolotriazine class exerts broad-spectrum antiviral activity at nanomolar concentrations. Antimicrob Agents Chemother. 2015;59:2062–2071. doi: 10.1128/AAC.04534-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iglewicz B, Hoaglin DC. How to Detect and Handle Outliers. (ASQC Quality Press); 1993. [Google Scholar]
- Jacquet K, Fradet-Turcotte A, Avvakumov N, Lambert JP, Roques C, Pandita RK, Paquet E, Herst P, Gingras AC, Pandita TK, et al. The TIP60 Complex Regulates Bivalent Chromatin Recognition by 53BP1 through Direct H4K20me Binding and H2AK15 Acetylation. Mol Cell. 2016;62:409–421. doi: 10.1016/j.molcel.2016.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaenicke LA, von Eyss B, Carstensen A, Wolf E, Xu W, Greifenberg AK, Geyer M, Eilers M, Popov N. Ubiquitin-Dependent Turnover of MYC Antagonizes MYC/PAF1C Complex Accumulation to Drive Transcriptional Elongation. Mol Cell. 2016;61:54–67. doi: 10.1016/j.molcel.2015.11.007. [DOI] [PubMed] [Google Scholar]
- Ju BG, Lunyak VV, Perissi V, Garcia-Bassets I, Rose DW, Glass CK, Rosenfeld MG. A topoisomerase IIbeta-mediated dsDNA break required for regulated transcription. Science. 2006;312:1798–1802. doi: 10.1126/science.1127196. [DOI] [PubMed] [Google Scholar]
- Kalkat M, Resetca D, Lourenco C, Chan PK, Wei Y, Shiah YJ, Vitkin N, Tong Y, Sunnerhagen M, Done SJ, et al. MYC Protein Interactome Profiling Reveals Functionally Distinct Regions that Cooperate to Drive Tumorigenesis. Mol Cell. 2018;72:836–848.:e837. doi: 10.1016/j.molcel.2018.09.031. [DOI] [PubMed] [Google Scholar]
- Kelstrup CD, Young C, Lavallee R, Nielsen ML, Olsen JV. Optimized fast and sensitive acquisition methods for shotgun proteomics on a quadrupole orbitrap mass spectrometer. J Proteome Res. 2012;11:3487–3497. doi: 10.1021/pr3000249. [DOI] [PubMed] [Google Scholar]
- Kim J, Guermah M, McGinty RK, Lee JS, Tang Z, Milne TA, Shilatifard A, Muir TW, Roeder RG. RAD6-Mediated transcription-coupled H2B ubiquitylation directly stimulates H3K4 methylation in human cells. Cell. 2009;137:459–471. doi: 10.1016/j.cell.2009.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Woo AJ, Chu J, Snow JW, Fujiwara Y, Kim CG, Cantor AB, Orkin SH. A Myc network accounts for similarities between embryonic stem and cancer cell transcription programs. Cell. 2010;143:313–324. doi: 10.1016/j.cell.2010.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SY, Herbst A, Tworkowski KA, Salghetti SE, Tansey WP. Skp2 regulates Myc protein stability and activity. Mol Cell. 2003;11:1177–1188. doi: 10.1016/s1097-2765(03)00173-4. [DOI] [PubMed] [Google Scholar]
- Kim W, Bennett EJ, Huttlin EL, Guo A, Li J, Possemato A, Sowa ME, Rad R, Rush J, Comb MJ, et al. Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol Cell. 2011;44:325–340. doi: 10.1016/j.molcel.2011.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein IA, Resch W, Jankovic M, Oliveira T, Yamane A, Nakahashi H, Di Virgilio M, Bothmer A, Nussenzweig A, Robbiani DF, et al. Translocation-capture sequencing reveals the extent and nature of chromosomal rearrangements in B lymphocytes. Cell. 2011;147:95–106. doi: 10.1016/j.cell.2011.07.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knobel PA, Belotserkovskaya R, Galanty Y, Schmidt CK, Jackson SP, Stracker TH. USP28 is recruited to sites of DNA damage by the tandem BRCT domains of 53BP1 but plays a minor role in double-strand break metabolism. Molecular and cellular biology. 2014;34:2062–2074. doi: 10.1128/MCB.00197-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzine F, Gupta A, Baranello L, Wojtowicz D, Ben-Aissa K, Liu J, Przytycka TM, Levens D. Transcription-dependent dynamic supercoiling is a short-range genomic force. Nat Struct Mol Biol. 2013;20:396–403. doi: 10.1038/nsmb.2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kress TR, Sabo A, Amati B. MYC: connecting selective transcriptional control to global RNA production. Nat Rev Cancer. 2015;15:593–607. doi: 10.1038/nrc3984. [DOI] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nature methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lans H, Hoeijmakers JHJ, Vermeulen W, Marteijn JA. The DNA damage response to transcription stress. Nature reviews. 2019;20:766–784. doi: 10.1038/s41580-019-0169-4. [DOI] [PubMed] [Google Scholar]
- Lin CY, Loven J, Rahl PB, Paranal RM, Burge CB, Bradner JE, Lee TI, Young RA. Transcriptional amplification in tumor cells with elevated c-Myc. Cell. 2012;151:56–67. doi: 10.1016/j.cell.2012.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzin F, Benary U, Baluapuri A, Walz S, Jung LA, von Eyss B, Kisker C, Wolf J, Eilers M, Wolf E. Different promoter affinities account for specificity in MYC-dependent gene regulation. eLife. 2016;5 doi: 10.7554/eLife.15161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madabhushi R, Gao F, Pfenning AR, Pan L, Yamakawa S, Seo J, Rueda R, Phan TX, Yamakawa H, Pao PC, et al. Activity-Induced DNA Breaks Govern the Expression of Neuronal Early-Response Genes. Cell. 2015;161:1592–1605. doi: 10.1016/j.cell.2015.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnaghi P, D’Alessio R, Valsasina B, Avanzi N, Rizzi S, Asa D, Gasparri F, Cozzi L, Cucchi U, Orrenius C, et al. Covalent and allosteric inhibitors of the ATPase VCP/p97 induce cancer cell death. Nat Chem Biol. 2013;9:548–556. doi: 10.1038/nchembio.1313. [DOI] [PubMed] [Google Scholar]
- Michalski A, Damoc E, Lange O, Denisov E, Nolting D, Muller M, Viner R, Schwartz J, Remes P, Belford M, et al. Ultra high resolution linear ion trap Orbitrap mass spectrometer (Orbitrap Elite) facilitates top down LC MS/MS and versatile peptide fragmentation modes. Mol Cell Proteomics. 2012;11:O111 013698. doi: 10.1074/mcp.O111.013698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosimann C, Hausmann G, Basler K. Parafibromin/Hyrax activates Wnt/Wg target gene transcription by direct association with beta-catenin/Armadillo. Cell. 2006;125:327–341. doi: 10.1016/j.cell.2006.01.053. [DOI] [PubMed] [Google Scholar]
- Moyal L, Lerenthal Y, Gana-Weisz M, Mass G, So S, Wang SY, Eppink B, Chung YM, Shalev G, Shema E, et al. Requirement of ATM-dependent monoubiquitylation of histone H2B for timely repair of DNA double-strand breaks. Mol Cell. 2011;41:529–542. doi: 10.1016/j.molcel.2011.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhar M, Ebert A, Neumann T, Umkehrer C, Jude J, Wieshofer C, Rescheneder P, Lipp JJ, Herzog VA, Reichholf B, et al. SLAM-seq defines direct gene-regulatory functions of the BRD4-MYC axis. Science. 2018;360:800–805. doi: 10.1126/science.aao2793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murr R, Loizou JI, Yang YG, Cuenin C, Li H, Wang ZQ, Herceg Z. Histone acetylation by Trrap-Tip60 modulates loading of repair proteins and repair of DNA double-strand breaks. Nature cell biology. 2006;8:91–99. doi: 10.1038/ncb1343. [DOI] [PubMed] [Google Scholar]
- Nakamura K, Kato A, Kobayashi J, Yanagihara H, Sakamoto S, Oliveira DV, Shimada M, Tauchi H, Suzuki H, Tashiro S, et al. Regulation of homologous recombination by RNF20-dependent H2B ubiquitination. Mol Cell. 2011;41:515–528. doi: 10.1016/j.molcel.2011.02.002. [DOI] [PubMed] [Google Scholar]
- Nie Z, Hu G, Wei G, Cui K, Yamane A, Resch W, Wang R, Green DR, Tessarollo L, Casellas R, et al. c-Myc Is a Universal Amplifier of Expressed Genes in Lymphocytes and Embryonic Stem Cells. Cell. 2012;151:68–79. doi: 10.1016/j.cell.2012.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira DV, Kato A, Nakamura K, Ikura T, Okada M, Kobayashi J, Yanagihara H, Saito Y, Tauchi H, Komatsu K. Histone chaperone FACT regulates homologous recombination by chromatin remodeling through interaction with RNF20. Journal of cell science. 2014;127:763–772. doi: 10.1242/jcs.135855. [DOI] [PubMed] [Google Scholar]
- Olson CM, Jiang B, Erb MA, Liang Y, Doctor ZM, Zhang Z, Zhang T, Kwiatkowski N, Boukhali M, Green JL, et al. Pharmacological perturbation of CDK9 using selective CDK9 inhibition or degradation. Nat Chem Biol. 2018;14:163–170. doi: 10.1038/nchembio.2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlando DA, Chen MW, Brown VE, Solanki S, Choi YJ, Olson ER, Fritz CC, Bradner JE, Guenther MG. Quantitative ChIP-Seq normalization reveals global modulation of the epigenome. Cell reports. 2014;9:1163–1170. doi: 10.1016/j.celrep.2014.10.018. [DOI] [PubMed] [Google Scholar]
- Orthwein A, Noordermeer SM, Wilson MD, Landry S, Enchev RI, Sherker A, Munro M, Pinder J, Salsman J, Dellaire G, et al. A mechanism for the suppression of homologous recombination in G1 cells. Nature. 2015;528:422–426. doi: 10.1038/nature16142. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Peter S, Bultinck J, Myant K, Jaenicke LA, Walz S, Muller J, Gmachl M, Treu M, Boehmelt G, Ade CP, et al. Tumor cell-specific inhibition of MYC function using small molecule inhibitors of the HUWE1 ubiquitin ligase. EMBO molecular medicine. 2014;6:1525–1541. doi: 10.15252/emmm.201403927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poli J, Gerhold CB, Tosi A, Hustedt N, Seeber A, Sack R, Herzog F, Pasero P, Shimada K, Hopfner KP, et al. Mec1, INO80, and the PAF1 complex cooperate to limit transcription replication conflicts through RNAPII removal during replication stress. Genes & development. 2016;30:337–354. doi: 10.1101/gad.273813.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popov N, Wanzel M, Madiredjo M, Zhang D, Beijersbergen R, Bernards R, Moll R, Elledge SJ, Eilers M. The ubiquitin-specific protease USP28 is required for MYC stability. Nature cell biology. 2007;9:765–774. doi: 10.1038/ncb1601. [DOI] [PubMed] [Google Scholar]
- Puc J, Kozbial P, Li W, Tan Y, Liu Z, Suter T, Ohgi KA, Zhang J, Aggarwal AK, Rosenfeld MG. Ligand-dependent enhancer activation regulated by topoisomerase-I activity. Cell. 2015;160:367–380. doi: 10.1016/j.cell.2014.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao X, Liu Y, Prada ML, Mohan AK, Gupta A, Jaiswal A, Sharma M, Merisaari J, Haikala HM, Talvinen K, et al. URB5 Is Coamplified with MYC in Breast Tumors and Encodes an Ubiquitin Ligase That Limits MYC-Dependent Apoptosis. Cancer research. 2020;80:1414–1427. doi: 10.1158/0008-5472.CAN-19-1647. [DOI] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahl PB, Lin CY, Seila AC, Flynn RA, McCuine S, Burge CB, Sharp PA, Young RA. c-Myc regulates transcriptional pause release. Cell. 2010;141:432–445. doi: 10.1016/j.cell.2010.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez F, Ryan DP, Gruning B, Bhardwaj V, Kilpert F, Richter AS, Heyne S, Dundar F, Manke T. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 2016;44:W160–165. doi: 10.1093/nar/gkw257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabo A, Kress TR, Pelizzola M, de Pretis S, Gorski MM, Tesi A, Morelli MJ, Bora P, Doni M, Verrecchia A, et al. Selective transcriptional regulation by Myc in cellular growth control and lymphomagenesis. Nature. 2014;511:488–492. doi: 10.1038/nature13537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L, Shao N, Liu X, Nestler E. ngs.plot: Quick mining and visualization of next-generation sequencing data by integrating genomic databases. BMC Genomics. 2014;15:284. doi: 10.1186/1471-2164-15-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh S, Szlachta K, Manukyan A, Raimer HM, Dinda M, Bekiranov S, Wang YH. Pausing sites of RNA polymerase II on actively transcribed genes are enriched in DNA double-stranded breaks. The Journal of biological chemistry. 2020;295:3990–4000. doi: 10.1074/jbc.RA119.011665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith T, Heger A, Sudbery I. UMI-tools: modeling sequencing errors in Unique Molecular Identifiers to improve quantification accuracy. Genome Res. 2017;27:491–499. doi: 10.1101/gr.209601.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sondalle SB, Longerich S, Ogawa LM, Sung P, Baserga SJ. Fanconi anemia protein FANCI functions in ribosome biogenesis. Proceedings of the National Academy of Sciences of the United States of America; 2019. pp. 2561–2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soucek L, Whitfield JR, Sodir NM, Masso-Valles D, Serrano E, Karnezis AN, Swigart LB, Evan GI. Inhibition of Myc family proteins eradicates KRas-driven lung cancer in mice. Genes & development. 2013;27:504–513. doi: 10.1101/gad.205542.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun XX, He X, Yin L, Komada M, Sears RC, Dai MS. The nucleolar ubiquitin-specific protease USP36 deubiquitinates and stabilizes c-Myc. Proceedings of the National Academy of Sciences of the United States of America; 2015. pp. 3734–3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasselli L, Xi Y, Zheng W, Tennen RI, Odrowaz Z, Simeoni F, Li W, Chua KF. SIRT6 deacetylates H3K18ac at pericentric chromatin to prevent mitotic errors and cellular senescence. Nat Struct Mol Biol. 2016;23:434–440. doi: 10.1038/nsmb.3202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesi A, de Pretis S, Furlan M, Filipuzzi M, Morelli MJ, Andronache A, Doni M, Verrecchia A, Pelizzola M, Amati B, et al. An early Myc-dependent transcriptional program orchestrates cell growth during B-cell activation. EMBO Rep. 2019:e47987. doi: 10.15252/embr.201947987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Oss SB, Cucinotta CE, Arndt KM. Emerging Insights into the Roles of the Paf1 Complex in Gene Regulation. Trends Biochem Sci. 2017;42:788–798. doi: 10.1016/j.tibs.2017.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Oss SB, Shirra MK, Bataille AR, Wier AD, Yen K, Vinayachandran V, Byeon IL, Cucinotta CE, Heroux A, Jeon J, et al. The Histone Modification Domain of Paf1 Complex Subunit Rtf1 Directly Stimulates H2B Ubiquitylation through an Interaction with Rad6. Mol Cell. 2016;64:815–825. doi: 10.1016/j.molcel.2016.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkitaraman AR. Cancer suppression by the chromosome custodians, BRCA1 and BRCA2. Science. 2014;343:1470–1475. doi: 10.1126/science.1252230. [DOI] [PubMed] [Google Scholar]
- von der Lehr N, Johansson S, Wu S, Bahram F, Castell A, Cetinkaya C, Hydbring P, Weidung I, Nakayama K, Nakayama KI, et al. The F-Box Protein Skp2 Participates in c-Myc Proteosomal Degradation and Acts as a Cofactor for c-Myc-Regulated Transcription. Mol Cell. 2003;11:1189–1200. doi: 10.1016/s1097-2765(03)00193-x. [DOI] [PubMed] [Google Scholar]
- Vos SM, Farnung L, Boehning M, Wigge C, Linden A, Urlaub H, Cramer P. Structure of activated transcription complex Pol II-DSIF-PAF-SPT6. Nature. 2018a;560:607–612. doi: 10.1038/s41586-018-0440-4. [DOI] [PubMed] [Google Scholar]
- Vos SM, Farnung L, Urlaub H, Cramer P. Structure of paused transcription complex Pol II-DSIF-NELF. Nature. 2018b;560:601–606. doi: 10.1038/s41586-018-0442-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walz S, Lorenzin F, Morton J, Wiese KE, von Eyss B, Herold S, Rycak L, Dumay-Odelot H, Karim S, Bartkuhn M, et al. Activation and repression by oncogenic MYC shape tumour-specific gene expression profiles. Nature. 2014;511:483–487. doi: 10.1038/nature13473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen H, Li Y, Xi Y, Jiang S, Stratton S, Peng D, Tanaka K, Ren Y, Xia Z, Wu J, et al. ZMYND11 links histone H3.3K36me3 to transcription elongation and tumour suppression. Nature. 2014;508:263–268. doi: 10.1038/nature13045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu G, Paige JS, Jaffrey SR. Global analysis of lysine ubiquitination by ubiquitin remnant immunoaffinity profiling. Nat Biotechnol. 2010;28:868–873. doi: 10.1038/nbt.1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan WX, Mirzazadeh R, Garnerone S, Scott D, Schneider MW, Kallas T, Custodio J, Wernersson E, Li Y, Gao L, et al. BLISS is a versatile and quantitative method for genome-wide profiling of DNA double-strand breaks. Nat Commun. 2017;8:15058. doi: 10.1038/ncomms15058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Zaugg K, Mak TW, Elledge SJ. A role for the deubiquitinating enzyme USP28 in control of the DNA-damage response. Cell. 2006;126:529–542. doi: 10.1016/j.cell.2006.06.039. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The ChIP-Rx, BLISS and RNA sequencing data are deposited at the GEO (Gene Expression Omnibus) database (GEO accession: GSE150217). Original image data have been deposited to Mendeley Data: 10.17632/n4rr8ck4w3.1