Abstract
HOTAIR is a lncRNA overexpressed in several epithelial cancers and strongly correlated with invasion. This lncRNA was proven a pivotal element of the epithelial-mesenchymal transition (EMT), a trans-differentiation process triggering metastasis. Snail, master inducer of EMT, requires HOTAIR to recruit EZH2 on specific epithelial target genes (i.e., HNF4α, E-cadherin and HNF1α) and cause their repression. Here we designed a HOTAIR deletion mutant form, named HOTAIR-sbid, including the putative Snail-binding domain but depleted of the EZH2 binding domain. HOTAIR-sbid acted as a dominant negative of the endogenous HOTAIR. In both murine and human tumor cells, HOTAIR-sbid impaired the ability of HOTAIR to bind Snail and, in turn, trigger H3K27me3/EZH2-mediated repression of Snail epithelial target genes. Notably, HOTAIR-sbid expression was proven to reduce cellular motility, invasiveness, anchorage-independent growth, and responsiveness to TGFβ-induced EMT. These data provide evidence on a lncRNA-based strategy to effectively impair the function of a master EMT-transcriptional factor.
Keywords: lncRNA, EZH2, PRC2, TGFβ, EMT
Introduction
Long noncoding (lnc) RNAs act as major players in physiological regulatory circuitries of the cell and are largely deregulated in different instances of human diseases, such as tumours (1,2). Specifically, HOTAIR (HOX Transcript Antisense Intergenic RNA, (3)) is a well-known lncRNA characterized as a scaffold for the PRC2 subunit EZH2 and able to act in trans to allow the establishment of a repressed chromatin state (4,5). This model of HOTAIR function was debated by considering promiscuous the specificity of the PRC2-RNA in vitro interactions (6,7), and even dispensable PRC2 for the HOTAIR-mediated repression of a luciferase reporter locus (8). However, there is no doubt that HOTAIR is an in vivo hallmark of poor prognosis and it impacts on epithelial tumour metastasis, by determining genome-wide PRC2 retargeting (4,9,10). Similarly, HOTAIR negative regulation significantly affects cancer cells migratory and invasive properties (11,12). HOTAIR-mediated recruitment of PRC2 to chromatin sites may imply more than direct RNA-DNA complementarity (4,13) and may conceivably involve the interaction with other partners. Notably, evidence was recently provided that, in the Epithelial to Mesenchymal Transition (EMT), HOTAIR bridges in specific chromatin sites the interaction between EZH2 and the “master” transcription factor (EMT-TF) Snail1 (Snail). In other words, Snail repressive function depends on its capacity to convey EZH2 to target sites by means of a direct interaction with HOTAIR (12,14).
Interestingly, these observations were validated for both murine and human HOTAIR. Of note EZH2 and Snail proteins are highly conserved in mouse (92.8% and 99.3% sequence identity, respectively) and human HOTAIR shares significant sequence similarity with its mouse homologue (55% identity), that is higher than the average 20% of human lncRNAs conserved in mouse (14). Here, in line with the growing effectiveness of RNA-based therapeutic approaches (15,16), an innovative anti-EMT strategy have been attempted. By means of an advanced computational tool, catRAPID (17), we estimated the binding potential of HOTAIR to Snail and EZH2 and an HOTAIR deletion mutant form (called HOTAIR-sbid, for Snail binding domain; nucleotides 620-845) was designed to correspond to the putative Snail-binding and depleted of the EZH2-binding domain. catRAPID method allows predictions of RNA-protein interactions through van der Waals, hydrogen bonding and secondary structure propensities of both protein and RNA sequences and identifies binding partners with an accuracy of 0.78 or higher (17,18).
HOTAIR-sbid has been functionally challenged in several model of tumoral and non-tumoral, murine and human, liver cells; this allowed to test its capacity (i) to interfere with the endogenous HOTAIR in Snail expressing cells (i.e. BW1J, Hep3B and HepG2 cell lines) and (ii) to prevent EMT induction by the niche factor TGFß in hepatocytes (i.e. D3 and E14 cell lines).
HOTAIR-sbid delivery in epithelial cancer cells was challenged as a functional and innovative lncRNA-based strategy in impairing Snail activity. Provided molecular evidence demonstrated that HOTAIR-sbid localizes in the nucleus and competes with endogenous wild type HOTAIR in Snail binding. This dominant negative function impairs the Snail-mediated recruitment of EZH2 to HNF4α, E-cadherin and HNF1α promoters, thus preventing their H3K27me3-mediated repression.
Functionally, HOTAIR-sbid was also demonstrated to cause a reduction of cell migration, invasion, anchorage-independent growth and, notably, to interfere with the responsiveness of non-tumorigenic hepatocytes to the niche factor TGFβ, main inducer of EMT.
Materials And Methods
Bioinformatic prediction
(i). Protein-RNA interaction prediction
Interactions were predicted using Global Score (18)overall binding propensity) and catRAPID fragments (19) identification of the binding sites). catRAPID estimates the binding propensity of protein-RNA pairs by combining secondary structure, hydrogen bonding and van der Waals contributions. As reported in a recent analysis of about half a million of experimentally validated interactions (20), the algorithm is able to separate interacting vs non-interacting pairs with an area under the ROC curve of 0.78. The Global Score and fragments calculations for the HOTAIR (NR_047528.1) and EZH2 (NP_031997.2) interaction are available at http://crg-webservice.s3.amazonaws.com/submissions/2019-12/234094/output/index.html?unlock=09cd6117fe; similarly, predictions for the interaction between HOTAIR and Snail (NP_035557.1) are at http://crg-webservice.s3.amazonaws.com/submissions/2019-12/233811/output/index.html?unlock=7d153ebbb3. The IgG control (BAX56602.1) interaction is calculated at http://crg-webservice.s3.amazonaws.com/submissions/2019-12/234361/output/index.html?unlock=45d56a80fd.
(ii). Structural conservation
Comparison of human and mouse HOTAIR sequences was performed by the Cluster W algorithm (21). We used CROSSalign (22,23), an algorithm based on Dynamic Time Warping (DTW), to check and evaluate the structural conservation between RNA molecules. The comparison between mouse (x-axis, NR_047528.1) and human (y-axis, NR_047517.1) HOTAIR structures are available at http://crg-webservice.s3.amazonaws.com/submissions/2019-12/233638/output/index.html?unlock=befdec937d. The lower the structural distance between the two RNAs, the closer is the line to the diagonal, as in the case of the 5’ (nucleotides 1-1000). The significance is assessed as in the original publications (22,23). Secondary structure prediction of RNA structure and secondary structure propensity was performed by CROSS soft constrains (23,24).
Cell culture conditions
Non-tumorigenic D3 and E14 hepatocytes were grown on collagen I-coated dishes in RPMI-1640 supplemented with 10% fetal bovine serum (Gibco Life Technology, Monza, Italy), 50 ng/ml epidermal growth factor, 30 ng/ml insulin-like growth factor II (PeproTech Inc., Rocky Hill, NJ, USA), 10 μg/ml insulin (Roche, Mannheim, Germany) and antibiotics. Where reported, cells were treated with 2,5 ng/ml TGFβl (PeproTech Inc.) for 24 h. Murine BW1J, human Hep3B and HepG2 cells were grown in DMEM supplemented with 10% fetal bovine serum (Gibco) and antibiotics. All cell lines were kindly provided by Dr. Alessandra Marchetti (BW1J, Hep3B and HepG2) and Prof. Laura Amicone from Sapienza University of Rome and used for the experiments three passages after thawing. All cell lines were tested for mycoplasma using the DAPI staining and the LookOut® Mycoplasma PCR Detection Kit (Merck MP0035). All cell lines were authenticated after thawing by morphology check, cell proliferation rate evaluation and species verification by PCR. Bacteria contamination was excluded.
HOTAIR-sbid cloning and cell transfection
Murine HOTAIR sequence (NR_047528.1; nucleotides 620-845), obtained from NCBI (http: (https://www.ncbi.nlm.nih.gov/), was artificially synthesized, verified by sequencing and cloned (NheI-XbaI) in the pCDNA 3 vector.
BW1J, D3 and E14 cells were transfected with Lipofectamine LTX Reagent (Invitrogen, San Diego, CA, USA), according to the manufacturer’s protocol. Equal amounts of DNA (pCDNA3 empty vector or pCDNA3/HOTAIR-sbid)) were used. BW1J cells were analysed 48 h post-transfection; D3 and E14 cells were diluted 24h post-transfection, treated with TGFβ 30h post-transfection and collected after 24h as in (14). Hep3B and HepG2 cells were transfected with FuGENE® HD Transfection Reagent (Promega, Madison, WI, USA), according to the manufacturer’s protocol, diluted 24h post-transfection and collected at 48h post-transfection.
RNA immunoprecipitation
RIP was performed as reported in (25) starting from 1 mg of cleared lysate. Immunoprecipitated RNA was reverse transcribed for reverse transcription and real-time polymerase chain reaction (RT-qPCR) amplifications. List of primers is reported in Table S1. Primary antibodies for IP were: anti-EZH2 39901 (Active motif), anti-Snail AF3639 (R&D systems) and as negative control Normal Rabbit IgG 12370 (Millipore) or Normal Goat IgG AB-108-C (R&D systems).
Co-immunoprecipitation
Cells were lysed in 50 mM Tris-HCl (pH 8.0), 150 mM NaCl, 5 mM EGTA (pH 8.0), 50 mM NaF (pH 8.0), 10% glycerol, 1.5 mM MgCl2, 1% Triton X-100 containing protease and phosphatase inhibitors (complete EDTA-free; Roche Applied Science, Mannheim Germany) and protein concentrations determined by Bradford method. One milligram of cell lysates, after preclearing with protein A-sepharose (GE Healthcare, Little Chalfont, Buckinghamshire, UK), was incubated with 5 μg of anti-EZH2 39901 (Active motif), anti-Snail AF3639 (R&D systems) and as negative control Normal Rabbit IgG 12370 (Millipore) or Normal Goat IgG AB-108-C (R&D systems). The complexes were incubated for 3 h with protein A-sepharose. Immune complexes were washed, eluted and denatured in Laemmli buffer. Proteins from either cell lysates or immunoprecipitation were resolved on sodium dodecyl sulphate-polyacrylamide gel electrophoresis and transferred to nitrocellulose membrane (162-0115; Bio-Rad Laboratories). Blots were probed with primary anti-EZH2 05-1319 (Millipore Corp.) or anti-Snail L70G2 (Cell Signaling Technology Inc., Danvers, MA, USA) and immune complexes were detected with horseradish peroxidase-conjugated species-specific secondary antiserum (Bio-Rad Laboratories), followed by enhanced chemiluminescence reaction (Bio-Rad Laboratories).
RNA extraction, reverse transcription, quantitative polymerase chain reaction
RNAs were extracted by ReliaPrep™ RNA Tissue Miniprep System (Promega, Madison, WI, USA) and reverse transcribed with iScriptTM c-DNA Synthesis Kit (Bio-Rad Laboratories Inc., Hercules, CA, USA). Quantitative polymerase chain reaction (RT-qPCR) analyses were performed according to MIQE guidelines (26). cDNAs were amplified by qPCR reaction using GoTaq qPCR Master Mix (Promega, Madison, WI, USA). Relative amounts, obtained with 2(-ΔCt) method, were normalized with respect to the housekeeping gene 18S rRNA (mouse) or L32 (human). For primer details see Table S1.
Protein extraction and western blot analysis
Cells were lysed in Laemmli buffer, subsequently the proteins were resolved on sodium dodecyl sulphate-polyacrylamide gel electrophoresis and transferred to nitrocellulose membrane 0.45um (162-0115; Bio-Rad Laboratories, Hercules, CA). The following primary antibodies were used for immunoblotting: α-HNF4α (SC-6556 Santa Cruz Biotechnology, Inc., CA), α-HNF1α (NBP1-33596 Novus Biological), α-E-CADHERIN (610182 BD transduction laboratories, Franklin Lakes, New Jersey), α-SNAIL (L70G2, Cell Signaling Technology, Danvers, MA), α-FIBRONECTIN (F0916 Sigma, Merck Darmstadt Germany), α-VIMENTIN (V6389 Sigma, Merck Darmstadt Germany), and α-GAPDH (MAB-374 Millipore Corp., Bedford, MA), used as a loading control. The immune complexes were detected with horseradish peroxidase-conjugated species-specific secondary antiserum: (α-Rabbit 172-1019 and α-Mouse 170-6516 Bio-Rad Laboratories, Hercules, CA), α-Goat (705-036-147, Jackson Immuno Research, Cambridge UK) then by enhanced chemiluminescence reaction (Bio-Rad Laboratories, Hercules, CA). Densitometric analysis of protein expression was performed by using the Fiji-Image J image processing package.
Statistical analysis
Paired t-test and GraphPad Prism version 8.00 (GraphPad software, San Diego, CA, USA; http://www.graphpad.com) were used. A P-value (P) <0.05 was considered statistically significant (*P<0.05; **P<0.01 and ***P<0.001). Data were obtained from independent experiments and expressed as means ±s.e.m.
Chromatin immunoprecipitation (ChIP) analysis
ChIP analysis was performed as reported previously (14) by using 5 μg rabbit α-SNAIL (R&D system), α-EZH2 (Active motif) or the negative control normal rabbit immunoglobulin (IgG) (Millipore Corp., Bedford, MA) or normal goat immunoglobulin (IgG) (R&D system). 5 ng of immunoprecipitated DNA and the relative controls were used as templates for real-time qPCR analysis, performed in triplicate. qPCR analysis of the immunoprecipitated samples and of the negative controls (IgG) were both normalized to total chromatin input and expressed as percentage of Input (% Input). Histone ChIP analysis was performed by using 5 μg of the specific antibody (H3K27me3; 07-449; Millipore Corp., Bedford, MA) or of the negative control normal rabbit IgG (Millipore Corp., Bedford, MA), as reported previously (14). The DNA was extracted with phenol-chloroform, precipitated with ethanol and resuspended in 50μl of water, then used in the downstream qPCR analyses (primer pairs details are listed in Table S2).
Scratch Assay
Cell lines were maintained in culture medium (as above) until reaching 100% confluence, then shifted to serum-depleted culture medium to inhibit cell proliferation, as in (27); a scratch wound was created on the cell layer using a micropipette tip. Micrographs were taken at time 0, 24 or 48 h after the scratch. Cell-devoid areas at time 0 and 24 or 48 h after the scratch were quantified through Fiji-Image J image processing package.
Invasion assay
For transwell invasion assays, 8 μm pore 24-well cell culture plates (Corning Inc, NY, USA), coated with type I collagen (0.1 mg/ml; Upstate Biotechnology, CA, USA) were used. Equal numbers of cells were plated in the upper chamber in serum-free medium, in the lower chamber DMEM medium was supplemented with 20% FBS as a chemoattractant. Cells were fixed with 100% MetOH, stained with Giemsa’s solution, and manually counted in three random microscopic fields. Three independent experiments for each cell line were performed.
Anchorage-independent growth assay
For colony formation assay the same number of cells were plated on 6-cm cell culture dishes in 7,5 ml of soft-agar plating medium. Colonies were evaluated after two weeks. The experiments were performed three times.
Results
Bioinformatic prediction of HOTAIR functional domains
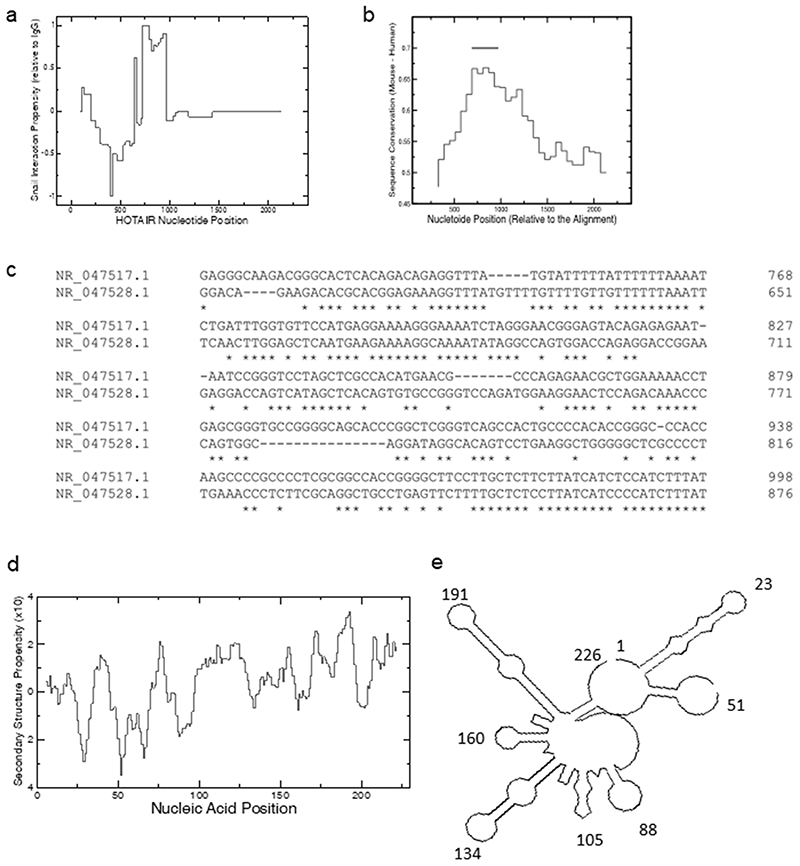
A direct interaction between HOTAIR and EZH2 was recently reported (14), in accordance with previous studies indicating the binding between the D1 domain of HOTAIR and PRC2 components (4,28). In agreement with this evidence, catRAPID Global Score (18) predicts an interaction between murine HOTAIR and EZH2, propensity of 0.98, on a scale ranging from 0 to 1, where 0 indicates no RNA-binding ability and 1 is strong affinity for the interaction. Interestingly, similar binding scores were found for the interaction between murine HOTAIR and Snail, propensity of 0.91, while the negative control (Ig gamma chain C region, IgG) does not show significant interaction propensity. The interactions of EZH2 and Snail are both predicted in the 5’ of HOTAIR, with the EZH2 binding (nucleotides 1-500) slightly upstream Snail (nucleotides 500-1000). Then, by using catRAPID fragments (19), we further investigated on the Snail binding site of HOTAIR. The algorithm identifies a region (henceforth called HOTAIR-sbid), located at nucleotides 620-845, where the interaction is predicted to occur with high specificity (Figure 1 A). The human HOTAIR lncRNA shares significant sequence similarity (55% identity) with its mouse homologue, which is higher than the average similarity between human and mouse lncRNAs (29). Interestingly, nucleotides 620-845, predicted by catRAPID to interact with Snail, show the highest sequence conservation between human and mouse HOTAIR (Figure 1 B, C).
Figure 1. Bioinformatic analysis of HOTAIR interaction propensity and conservation.
(a) Prediction of HOTAIR interaction propensity with Snail. The score is normalized relatively to the control IgG. (b) Sequence conservation between human and mouse HOTAIR. The most conserved region in HOTAIR corresponds to Snail binding site predicted by catRAPID (black bar). The positions are relative to the alignments between the two sequences. (c) Comparison of human and mouse HOTAIR sequences in the HOTAIR-sbid region by Cluster W algorithm. (d) Double stranded content of HOTAIR-sbid. The CROSS algorithm was employed to predict the secondary structure propensity. (e) Secondary structure prediction of HOTAIR-sbid by CROSS soft constrains in RNA structure.
In agreement with the sequence conservation, CROSSalign (22,23) predicts that the secondary structures of both human and mouse HOTAIR are highly similar in the region at nucleotides 620-845 (Supplementary Figure S1), thus providing additional evidence that HOTAIR-sbid has a potential functional role. Indeed, the structure contains several double stranded regions, which are indicative of protein interactions (30) (Figure 1 D, E).
Overall, these analyses predict the existence of a Snail-interacting HOTAIR domain conserved in mouse and human.
HOTAIR-sbid displays a biochemical dominant negative function on human HOTAIR
Based on the above described bioinformatics analysis, a HOTAIR-sbid mutant (nucleotides 620-845), including the putative highly-specific Snail-binding sequence and depleted of the EZH2-binding region, was designed, cloned in a plasmid vector (see methods) and expressed in the various experimental cell models described below. Firstly, the cellular distribution of HOTAIR-sbid was explored, taking into account that the nuclear localization would be the primary determinant for the expected functions of this mutant: as shown in Supplementary Figure S2 (A, B), cellular fractionation assay provided evidence about the HOTAIR-sbid nuclear enrichment. Then, its capacity to form a complex (i) with Snail, (ii) EZH2 and (iii) to compete with endogenous HOTAIR was tested. To discriminate between the endogenous HOTAIR and HOTAIR-sbid, this analysis was performed in human Hep3B sparse cultures. Note that low confluency in vitro may act as a stressful condition triggering hepatoma cells dedifferentiation. Hep3B, indeed, when grown at low density, express both HOTAIR and Snail and, in turn, repress the Snail target genes HNF4α, HNF1α and E-cadherin, mimicking in vivo mesenchymal, invasive tumor cells. On the other hand, dense hepatoma cultures, as differentiated tumor cells, maintain epithelial markers and repress Snail as well as HOTAIR (see (31) and Supplementary Figure S3). Thus, different confluence of this in vitro cell model allows to mimic different steps of epithelial tumorigenesis.
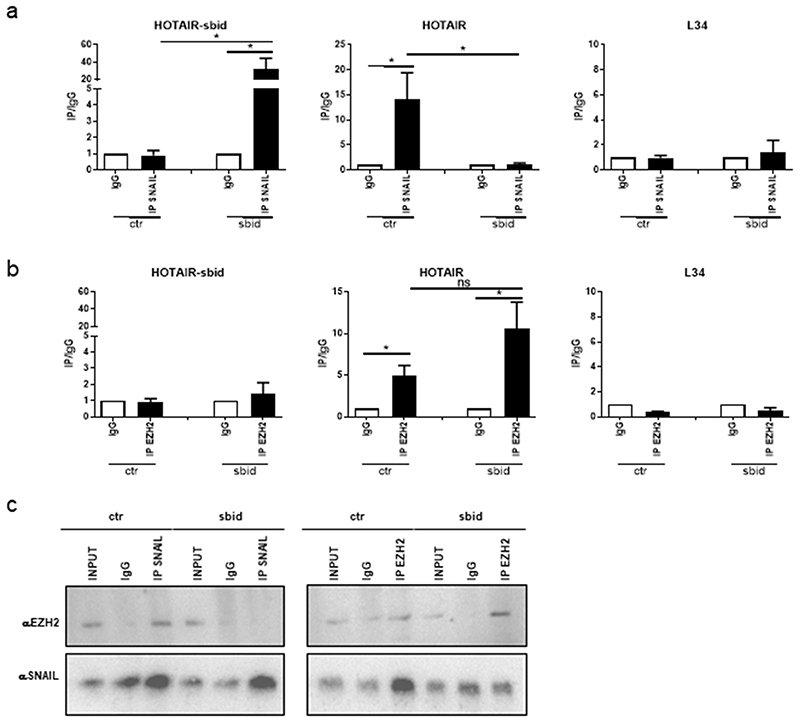
RIP assays, performed on Hep3B extracts from sparse cultures, highlighted that (i) Snail interacts with both endogenous HOTAIR and HOTAIR-sbid (Figure 2 A); (ii) EZH2 interacts with endogenous HOTAIR and not with HOTAIR-sbid (Figure 2 B); and most importantly, (iii) HOTAIR-sbid impairs the interaction of endogenous HOTAIR with Snail (while HOTAIR binding to EZH2 is retained) (Figure 2 A, B). Finally, as shown by reciprocal Snail and EZH2 co-immunoprecipitation experiments (Figure 2 C), HOTAIR-sbid impairs the interaction between Snail and EZH2.
Figure 2. HOTAIR-sbid interferes with the SNAIL/HOTAIR/EZH2 complex.
(a) RIP assays with goat polyclonal anti-SNAIL or preimmune IgG on Hep3B cells expressing the HOTAIR-sbid (sbid) or the empty vector (Ctr). (b) RIP assays with rabbit polyclonal anti-EZH2 or preimmune IgG on Hep3B cells expressing the HOTAIR-sbid (sbid) or the empty vector (ctr). RNA levels in immunoprecipitates (IP) and IgG were determined by qRT-PCR. HOTAIR-sbid, endogenous HOTAIR lncRNA and, as negative control, ribosomal L34 RNA, were reported as IP/IgG. Data are reported as means ±s.e.m. of four independent experiments. (c) Co - immunoprecipitation of Snail and EZH2. Immunoprecipitations with goat polyclonal anti-SNAIL, rabbit polyclonal anti-EZH2 and the relative preimmune IgG were performed on protein extracts from Hep3B cells expressing the HOTAIR-sbid (sbid) or the empty vector (Ctr) as a control. Immunoblots were performed using mouse anti-Snail and mouse anti-EZH2 antibodies.
Overall, these results indicate as HOTAIR-sbid displays a biochemical dominant negative function on human endogenous HOTAIR.
HOTAIR-sbid displays an in vivo dominant negative function on endogenous HOTAIR.
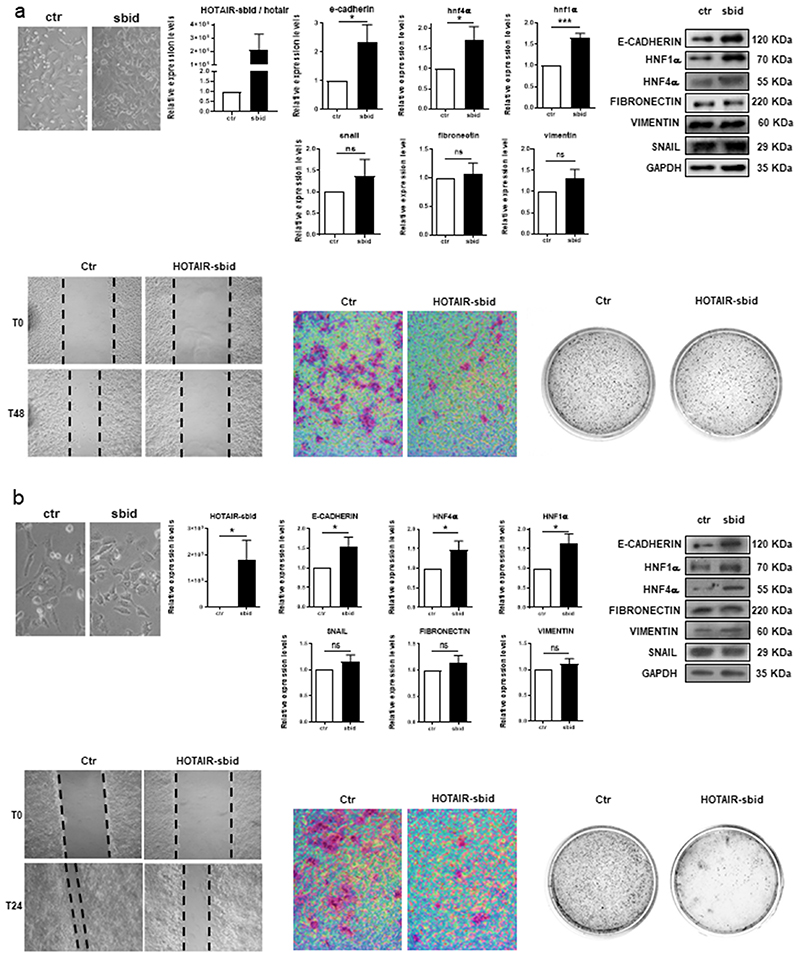
We then aimed at investigating the HOTAIR-sbid functional role in Snail-expressing human and murine cells where it was previously demonstrated the Snail/HOTAIR/EZH2-mediated repression of HNF4α, HNF1α and E-cadherin (14). In murine BW1J (Figure 3 A and Supplementary Figure S4), human Hep3B (Figure 3 B and Supplementary Figure S4) and HepG2 hepatoma cell lines (Supplementary Figure S5), HOTAIR-sbid impaired HOTAIR-mediated (i) repression at both RNA and protein levels of epithelial genes previously characterized as Snail-targets (32–34), (ii) both migration ability and (iii) invasion properties (see also Supplementary Figure S6) (assays performed in absence of cellular proliferation; see also, Supplementary Figure S7), and (iv) anchorage-independent growth. However, the expression of mesenchymal genes (i.e. Snail, Fibronectin and Vimentin) did not resulted significantly modulated nor at RNA or at protein level (Figure 3A, 3B and Supplementary Figure S4).
Figure 3. HOTAIR-sbid interferes with the SNAIL/HOTAIR/EZH2-mediated functions.
(a) Analysis of BW1J cells expressing HOTAIR-sbid (sbid) or the empty vector as a control (ctr) as indicated. Top left: phase contrast micrographs; top center: RT-qPCR analysis for the indicated epithelial (e-cadherin, hnf4α, hnf1α) and mesenchymal (snail, vimentin and fibronectin) genes; the values are calculated by the 2(-ΔCt) method, expressed as fold of expression versus the control (arbitrary value=1) and shown as mean±s.e.m. Statistically significant differences are reported (*P<0.05, ***P<0.001) for six independent experiments; top right: Western blot analysis (WB) for E-CADHERIN, HNF1α, HNF4α, FIBRONECTIN, VIMENTIN, SNAIL on protein extracts. GAPDH was used as a loading control. All the experiments have been performed four times and WB images represent one indicative experiment of the independent ones. Densitometric analysis of protein expression relative to the independent experiments are shown in Supplementary Figure S6. Bottom left: Scratch assay at the indicted time. Bottom centre: invasion assay; Bottom right: anchorage independent growth analysis in soft agar assay. Quantification of migration and invasion analysis are reported in Supplementary Figure S8. (b) Analysis of Hep3B cells expressing HOTAIR-sbid (sbid) or the empty vector as a control (ctr) as in (a). Data are relative to six (RT-qPCR) or seven (WB) independent experiments.
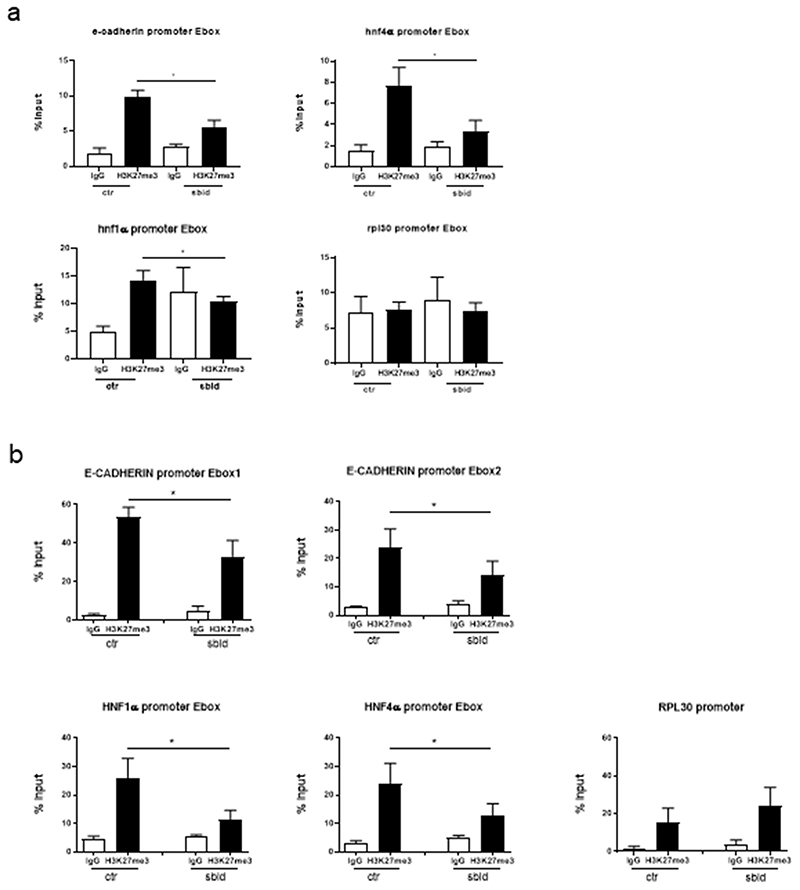
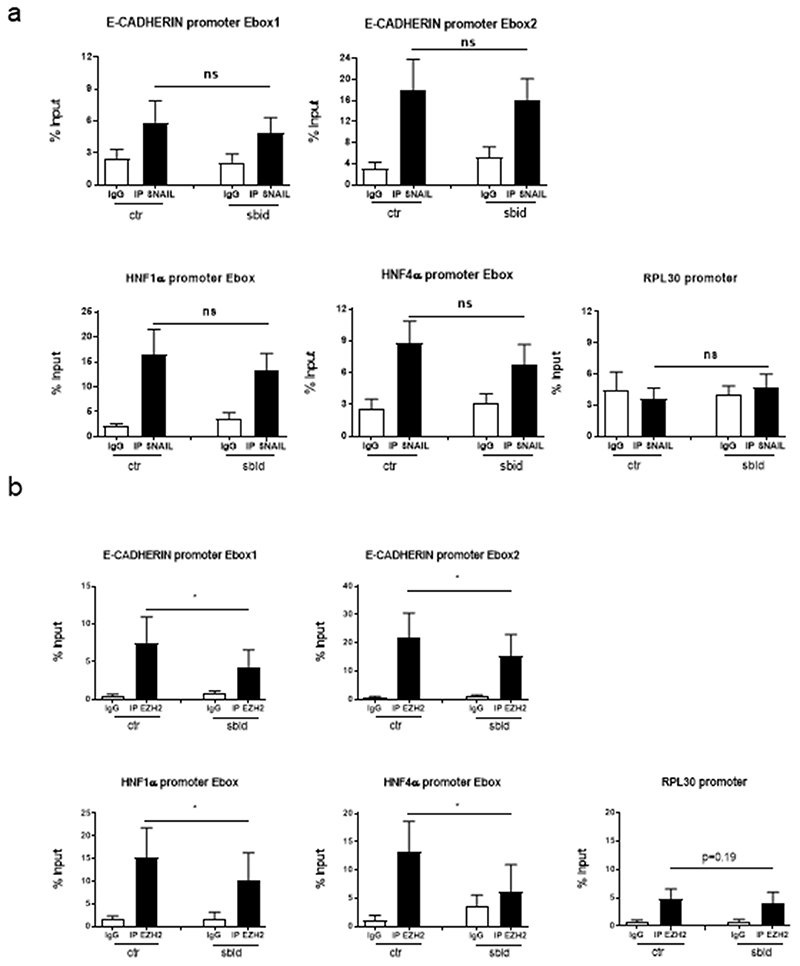
Mechanistically, ChIP assays on HNF4α, E-cadherin and HNF1α promoters’ Snail-binding sites highlighted a significant decrease of H3K27me3 (Figure 4 A, B). Furthermore, the direct recruitment of both Snail and EZH2 on the same promoter regions was assessed; as shown in Figure 5 A and B, while Snail recruitment appeared not affected, EZH2 binding was impaired by HOTAIR-sbid.
Figure 4. HOTAIR-sbid interferes with the SNAIL/HOTAIR/EZH2-mediated promoters H3K27me3.
qPCR analysis of ChIP assays with anti-H3K27me3 antibody (H3K27me3) and, as control, normal rabbit IgG (IgG) on chromatin from BW1J cells (a) and Hep3B (b) expressing HOTAIR-sbid (sbid) or the empty vector as a control (ctr). Data show the enrichment of H3K27 trimethylation on the Snail consensus binding sites on the murine promoters of HNF1α, HNF4α and E-cadherin. Rpl30 promoter was used as a negative control. Values derived from five independent experiments are reported as means ± s.e.m. and expressed as the percentage of the Input chromatin (% Input). Statistically significant differences are reported (*P<0.05).
Figure 5. HOTAIR-sbid interferes with the EZH2 recruitment on epithelial genes promoters.
qPCR analysis of ChIP assays with anti-SNAIL (a) or anti-EZH2 (b) antibodies and, as control, normal goat IgG (IgG) (a) or normal rabbit IgG (IgG) (b) on chromatin from Hep3B cells expressing HOTAIR-sbid (sbid) or the empty vector as a control (ctr). Data show the recruitment of SNAIL or EZH2 on the Snail consensus binding sites on the human promoters of HNF1α, HNF4α and E-cadherin. Rpl30 promoter was used as a negative control. Values derived from four independent experiments are reported as means ±s.e.m. and expressed as the percentage of the Input chromatin (% Input). Statistically significant differences are reported (*P<0.05).
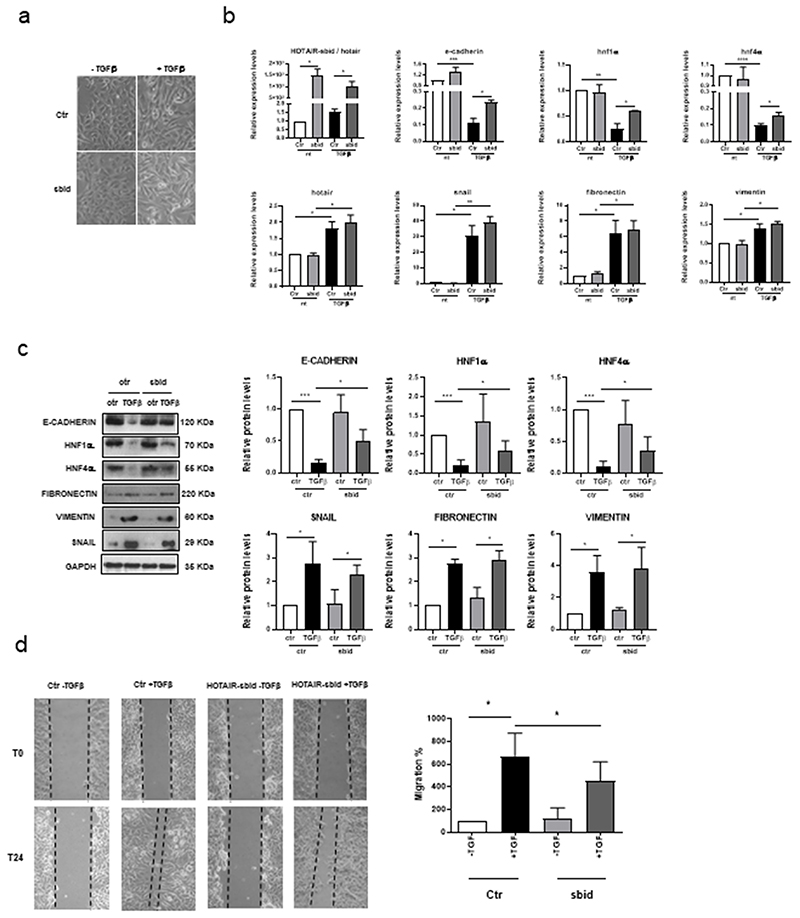
Finally, HOTAIR-sbid functional role was tested in two hepatocyte cell models (D3 and E14 cell lines (35),(36)) suitable to study the TGFβ-mediated EMT (in both cellular systems Snail and HOTAIR are induced in response to this niche factor (14)). As shown in Figure 6 A–D and Supplementary Figure S8 A–D, in both cell line HOTAIR-sbid impaired the TGFβ-induced (i) acquisition of the mesenchymal morphology, (ii) repression of epithelial genes (i.e. HNF4α, E-cadherin and HNF1α and (iii) acquisition of migratory properties. On the other hand, in line with previous findings achieved by HOTAIR silencing (14), mesenchymal genes (i.e. Snail, Fibronectin and Vimentin) were found upregulated (Figure 6 B, C and Supplementary Figure S8 B, C).
Figure 6. HOTAIR-sbid interferes with the SNAIL/HOTAIR/EZH2-mediated functions in EMT.
D3 treated with TGFβ (+TGFβ) or left untreated (-TGFβ) and expressing HOTAIR-sbid or the empty vector as a control (Ctr). (a) Phase contrast micrographs; (b) RT–qPCR analysis for the indicated epithelial (e-cadherin, hnf1α, hnf4α), mesenchymal (snail, fibronectin and vimentin) genes and for HOTAIR and HOTAIR-sbid. The values are calculated by the 2(-ΔCt) method, expressed as fold of expression versus the control (arbitrary value=1) and shown as mean ±s.e.m. Statistically significant differences are reported (*P<0.05, **P<0.01) for three independent experiments. (c) Left: Western blot analysis (WB) for E-CADHERIN, HNF1α, HNF4α, FIBRONECTIN, VIMENTIN, SNAIL protein extracts. GAPDH was used as a loading control. All the experiments have been performed three times and WB images represent one indicative experiment of the independent ones. Right: Densitometric analysis of protein expression relative to the independent experiments. (d) Left: Scratch assay at the indicted time; Right: quantification of migration abilities of four independent experiments.
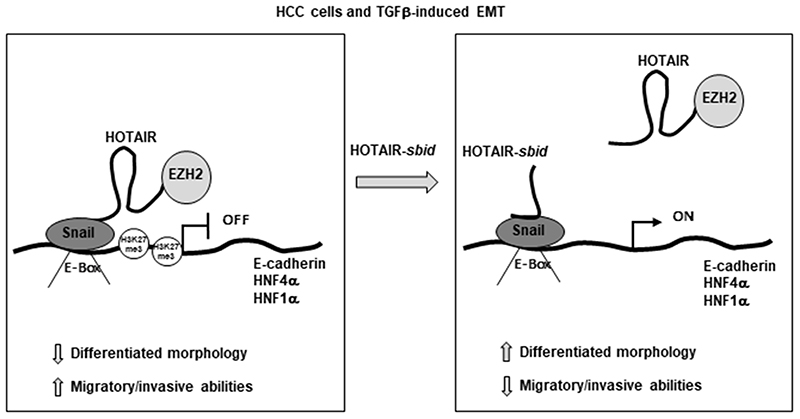
Overall, these results indicate as HOTAIR-sbid (i) displays a dominant negative activity on Snail repressive function and (ii) significantly affects cellular EMT outcome, both in terms of epithelial program repression and migratory abilities (Figure 7).
Figure 7. Schematic representation of HOTAIR-sbid activity in cancer cells and in TGFβ-induced EMT.
Discussion
The main result of this research was the design and the validation of a RNA molecule with dominant negative function on the HOTAIR-mediated Snail repressive activity (on pivotal epithelial genes, i.e. HNF4α, HNF1α and E-cadherin), thus allowing the rescue of a more differentiated phenotype of hepatocellular carcinoma cells and the protection from the TGFβ-induced EMT of non-tumorigenic hepatocytes. Specifically, we designed the deletion mutant HOTAIR-sbid that competes with the wild type endogenous HOTAIR form. Our computational approach, previously applied to other lncRNAs, such as SAMMSON (37) and XIST (18), was instrumental to define the precise binding region for Snail. The mutant was found to impair the assembly of the Snail/HOTAIR/EZH2 functional complex that controls the execution of the Snail-mediated EMT (14).
The EMT reversible trans-differentiation program is essential for the dissemination of malignant epithelial tumours by conferring stem properties, motility and, finally, ability to metastasize (for review, (38)). HOTAIR acts as a “mesenchymal” gene positively regulated in TGFβ-induced EMT and epistatic with respect to the EMT-TF Snail by controlling its repressive function in the chromatin context (14). Consistently, this lncRNA promotes tumorigenesis and metastasis formation in different epithelial cancers, including HCC: its increased expression causes the acquisition of metastatic properties while its knockdown impairs migration, invasion ability and EMT in carcinoma cells (4,9,10,39,40). Coherently, HOTAIR repression is required in the maintenance of the epithelial differentiated state (12). In HCC, HOTAIR was shown to act also at post-transcriptional level (for review, (41)) by sponging several miRs (e.g. miR-29b, thus enhancing DNMT3b (42) involved in EMT/MET dynamics (43), or miR-23p-3p, targeting the EMT-TF ZEB1 (40,44)). All this body of evidence identifies this lncRNA as a suitable candidate in therapies counteracting epithelial tumour progression.
Over the last years much effort has been concentrated on the setting of RNA-based therapeutics, mimicking or antagonizing endogenous RNA functions (e.g. the delivery of antisense oligonucleotides for mRNAs, interfering RNAs, in vitro transcribed mRNAs or oligonucleotides aptamers) (15,16). In particular for the liver, one of the more achievable organs, several siRNA-based approaches, with lower off-target effects than conventional therapies, have been attempted (45); for example approaches have been challenged by using molecules chemically modified or loaded into lipid particles and nanoparticles (46). Nevertheless, since miRNA-mimics may present drawbacks as multiple targets (summarized in (47)), lncRNA-based strategies appear promising.
We validated for the first time, to our knowledge, a dominant negative-based RNA strategy to interfere with a pivotal function of a lncRNA. Specifically, HOTAIR-sbid function was proven dominant negative against that of the endogenous HOTAIR, in chromatin context of both murine and human cell lines; following our computational design, HOTAIR-sbid was shown to interact with Snail but not with the general chromatin modifier EZH2. More interestingly, the endogenous HOTAIR was shown to maintain the capacity to interact with EZH2 while its ability to bind to Snail was hampered by HOTAIR-sbid; in turn, as highlighted by Co-IP analysis, the interaction between Snail and EZH2 was impaired. At the epigenetic level, competition between endogenous HOTAIR and exogenous HOTAIR-sbid, inhibiting the formation of the tripartite complex Snail/HOTAIR/EZH2, causes the decrease of H3K27me3 levels on E-cadherin, HNF1α and HNF4α promoter regions and, consequently, the rescue of their expression. It should be remembered that HNF1α and HNF4α in hepatic cells control cell differentiation and maintenance of the epithelial phenotype, acting on thousands of target genes, also by repression of the mesenchymal program (34).
Further effort should be made to extend current analysis to a broader number of genes known to be target of HOTAIR (48). Functionally, HOTAIR-sbid alters mouse and human HCC cells behaviour: it significantly impairs the migratory capacity, the invasion abilities and anchorage-independent growth of both cell lines. Most importantly, in two non-tumorigenic cellular models, previously validated to respond to TGFβ with an EMT/MET dynamic (i.e. D3 and E14 cell lines (14,49)), HOTAIR-sbid affects cellular morphology, migration and repression of master epithelial genes (see Figure 6 and Supplementary Figure S8). However, HOTAIR-sbid does not impair the TGFβ-mediated mesenchymal gene expression, further suggesting the relevance of the previously reported Snail independent TGFβ-induced response (50).
Collectively, these data represent an early promise in the control of tumour progression and in the rescue of the epithelial phenotype of cancer cells. Moreover, they open new perspectives for the future analysis of the effectiveness of this strategy also in vivo where HOTAIR-sbid could counteract different aspect of tumour progression. Overall, this innovative approach, based on the use of a dominant negative form of a lncRNA normally expressed in tumour cells, could represent one additional step in the use of highly specific RNA therapeutics.
Supplementary Material
Significance.
This study defines an innovative RNA-based strategy to interfere with a pivotal function of the tumor-related lncRNA HOTAIR, comprising a dominant negative mutant that was computationally designed and impairs epithelial-to-mesenchymal transition.
Acknowledgements
We thank Prof. Andres Ramos (UCL) for critical reading and suggestions.
The research leading to these results has been supported by European Research Council (RIBOMYLOME_309545 and ASTRA_855923), the H2020 projects IASIS_727658 and INFORE_825080, the Spanish Ministry of Economy and Competitiveness BFU2017-86970-P as well as the collaboration with Peter St. George-Hyslop financed by the Wellcome Trust (to G.G.T.); by Associazione Italiana per la Ricerca sul Cancro (AIRC) IG 18843, FFO2019 grant from CIB (Consorzio Italiano per le Biotecnologie) and Sapienza University of Rome RG11916B6A9C42C7 (to M.T.); by Sapienza University of Rome RM116154BE5E14B2 and RM11916B6A80C2CF (to C.C.).
Abbreviations
- EMT
Epithelial to Mesenchymal Transition
- MET
Mesenchymal to Epithelial Transition
- TGFβ
transforming growth factor β
- lncRNAs
long non-coding RNAs
- EZH2
Enhancer of Zeste Homolog 2
- HCC
hepatocellular carcinoma
- PRC2
Polycomb Repressive Complex 2
- H3K27me3
tri-methylation of histone H3 lysine 27
- HNF4α
hepatocyte nuclear factor 4 alpha
- HNF1 α
hepatocyte nuclear factor 1 alpha
- RT-qPCR
reverse-transcription and quantitative polymerase chain reaction
- qPCR
quantitative polymerase chain reaction
- ChIP
Chromatin Immunoprecipitation
- RIP
RNA immunoprecipitation
Footnotes
Conflict of interests
The authors declare no potential conflicts of interest.
References
- 1.Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annual review of biochemistry. 2012;81:145–66. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huarte M. The emerging role of lncRNAs in cancer. Nature medicine. 2015;21:1253–61. doi: 10.1038/nm.3981. [DOI] [PubMed] [Google Scholar]
- 3.Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–23. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–6. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, et al. Long noncoding RNA as modular scaffold of histone modification complexes. Science (New York, NY. 2010;329:689–93. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davidovich C, Zheng L, Goodrich KJ, Cech TR. Promiscuous RNA binding by Polycomb repressive complex 2. Nature structural & molecular biology. 2013;20:1250–7. doi: 10.1038/nsmb.2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cifuentes-Rojas C, Hernandez AJ, Sarma K, Lee JT. Regulatory interactions between RNA and polycomb repressive complex 2. Molecular cell. 2014;55:171–85. doi: 10.1016/j.molcel.2014.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Portoso M, Ragazzini R, Brencic Z, Moiani A, Michaud A, Vassilev I, et al. PRC2 is dispensable for HOTAIR-mediated transcriptional repression. The EMBO journal. 2017;36:981–94. doi: 10.15252/embj.201695335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Geng YJ, Xie SL, Li Q, Ma J, Wang GY. Large intervening non-coding RNA HOTAIR is associated with hepatocellular carcinoma progression. The Journal of international medical research. 2011;39:2119–28. doi: 10.1177/147323001103900608. [DOI] [PubMed] [Google Scholar]
- 10.Yang Z, Zhou L, Wu LM, Lai MC, Xie HY, Zhang F, et al. Overexpression of long non-coding RNA HOTAIR predicts tumor recurrence in hepatocellular carcinoma patients following liver transplantation. Annals of surgical oncology. 2011;18:1243–50. doi: 10.1245/s10434-011-1581-y. [DOI] [PubMed] [Google Scholar]
- 11.Geng YJ, Xie SL, Li Q, Ma J, Wang GY. Large intervening non-coding RNA HOTAIR is associated with hepatocellular carcinoma progression. The Journal of international medical research. 39:2119–28. doi: 10.1177/147323001103900608. [DOI] [PubMed] [Google Scholar]
- 12.Battistelli C, Sabarese G, Santangelo L, Montaldo C, Gonzalez FJ, Tripodi M, et al. The lncRNA HOTAIR transcription is controlled by HNF4alpha-induced chromatin topology modulation. Cell death and differentiation. 2019;26:890–901. doi: 10.1038/s41418-018-0170-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kalwa M, Hanzelmann S, Otto S, Kuo CC, Franzen J, Joussen S, et al. The lncRNA HOTAIR impacts on mesenchymal stem cells via triple helix formation. Nucleic acids research. 2016;44:10631–43. doi: 10.1093/nar/gkw802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Battistelli C, Cicchini C, Santangelo L, Tramontano A, Grassi L, Gonzalez FJ, et al. The Snail repressor recruits EZH2 to specific genomic sites through the enrollment of the lncRNA HOTAIR in epithelial-to-mesenchymal transition. Oncogene. 2017;36:942–55. doi: 10.1038/onc.2016.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lieberman J. Tapping the RNA world for therapeutics. Nature structural & molecular biology. 2018;25:357–64. doi: 10.1038/s41594-018-0054-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Setten RL, Rossi JJ, Han SP. Author Correction: The current state and future directions of RNAi-based therapeutics. Nat Rev Drug Discov. 2020;19:290. doi: 10.1038/s41573-019-0027-2. [DOI] [PubMed] [Google Scholar]
- 17.Agostini F, Zanzoni A, Klus P, Marchese D, Cirillo D, Tartaglia GG. catRAPID omics: a web server for large-scale prediction of protein-RNA interactions. Bioinformatics. 2013;29:2928–30. doi: 10.1093/bioinformatics/btt495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cirillo D, Blanco M, Armaos A, Buness A, Avner P, Guttman M, et al. Quantitative predictions of protein interactions with long noncoding RNAs. Nat Methods. 2016;14:5–6. doi: 10.1038/nmeth.4100. [DOI] [PubMed] [Google Scholar]
- 19.Cirillo D, Agostini F, Klus P, Marchese D, Rodriguez S, Bolognesi B, et al. Neurodegenerative diseases: quantitative predictions of protein-RNA interactions. RNA. 2013;19:129–40. doi: 10.1261/rna.034777.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lang B, Armaos A, Tartaglia GG. RNAct: Protein-RNA interaction predictions for model organisms with supporting experimental data. Nucleic acids research. 2019;47:D601–D6. doi: 10.1093/nar/gky967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thompson JD, Higgins DG, Gibson TJ. Clustal-W -Improving the Sensitivity of Progressive Multiple Sequence Alignment through Sequence Weighting, Position-Specific Gap Penalties and Weight Matrix Choice. Nucleic acids research. 1994;22:4673–80. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Delli Ponti R, Armaos A, Marti S, Tartaglia GG. A Method for RNA Structure Prediction Shows Evidence for Structure in lncRNAs. Front Mol Biosci. 2018;5:111. doi: 10.3389/fmolb.2018.00111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Delli Ponti R, Marti S, Armaos A, Tartaglia GG. A high-throughput approach to profile RNA structure. Nucleic acids research. 2017;45:e35. doi: 10.1093/nar/gkw1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reuter JS, Mathews DH. RNAstructure: software for RNA secondary structure prediction and analysis. Bmc Bioinformatics. 2010;11 doi: 10.1186/1471-2105-11-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dahm GM, Gubin MM, Magee JD, Techasintana P, Calaluce R, Atasoy U. Method for the isolation and identification of mRNAs, microRNAs and protein components of ribonucleoprotein complexes from cell extracts using RIP-Chip. J Vis Exp. 2012 doi: 10.3791/3851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem. 2009;55:611–22. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- 27.Magistri P, Battistelli C, Strippoli R, Petrucciani N, Pellinen T, Rossi L, et al. SMO Inhibition Modulates Cellular Plasticity and Invasiveness in Colorectal Cancer. Front Pharmacol. 2017;8:956. doi: 10.3389/fphar.2017.00956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Somarowthu S, Legiewicz M, Chillon I, Marcia M, Liu F, Pyle AM. HOTAIR forms an intricate and modular secondary structure. Molecular cell. 2015;58:353–61. doi: 10.1016/j.molcel.2015.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hezroni H, Koppstein D, Schwartz MG, Avrutin A, Bartel DP, Ulitsky I. Principles of long noncoding RNA evolution derived from direct comparison of transcriptomes in 17 species. Cell Rep. 2015;11:1110–22. doi: 10.1016/j.celrep.2015.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.de Groot NS, Armaos A, Grana-Montes R, Alriquet M, Calloni G, Vabulas RM, et al. RNA structure drives interaction with proteins. Nat Commun. 2019;10 doi: 10.1038/s41467-019-10923-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Conacci-Sorrell M, Simcha I, Ben-Yedidia T, Blechman J, Savagner P, Ben-Ze’ev A. Autoregulation of E-cadherin expression by cadherin-cadherin interactions: the roles of beta-catenin signaling, Slug, and MAPK. J Cell Biol. 2003;163:847–57. doi: 10.1083/jcb.200308162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nature reviews. 2014;15:178–96. doi: 10.1038/nrm3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cicchini C, Filippini D, Coen S, Marchetti A, Cavallari C, Laudadio I, et al. Snail controls differentiation of hepatocytes by repressing HNF4alpha expression. J Cell Physiol. 2006;209:230–8. doi: 10.1002/jcp.20730. [DOI] [PubMed] [Google Scholar]
- 34.Santangelo L, Marchetti A, Cicchini C, Conigliaro A, Conti B, Mancone C, et al. The stable repression of mesenchymal program is required for hepatocyte identity: a novel role for hepatocyte nuclear factor 4alpha. Hepatology. 2011;53:2063–74. doi: 10.1002/hep.24280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Amicone L, Spagnoli FM, Spath G, Giordano S, Tommasini C, Bernardini S, et al. Transgenic expression in the liver of truncated Met blocks apoptosis and permits immortalization of hepatocytes. The EMBO journal. 1997;16:495–503. doi: 10.1093/emboj/16.3.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bisceglia F, Battistelli C, Noce V, Montaldo C, Zammataro A, Strippoli R, et al. TGFbeta Impairs HNF1alpha Functional Activity in Epithelial-to-Mesenchymal Transition Interfering With the Recruitment of CBP/p300 Acetyltransferases. Front Pharmacol. 2019;10:942. doi: 10.3389/fphar.2019.00942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vendramin R, Verheyden Y, Ishikawa H, Goedert L, Nicolas E, Saraf K, et al. SAMMSON fosters cancer cell fitness by concertedly enhancing mitochondrial and cytosolic translation. Nature structural & molecular biology. 2018;25:1035–46. doi: 10.1038/s41594-018-0143-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dongre A, Weinberg RA. New insights into the mechanisms of epithelial-mesenchymal transition and implications for cancer. Nature reviews. 2019;20:69–84. doi: 10.1038/s41580-018-0080-4. [DOI] [PubMed] [Google Scholar]
- 39.Zhang Z, Gao Z, Rajthala S, Sapkota D, Dongre H, Parajuli H, et al. Metabolic reprogramming of normal oral fibroblasts correlated with increased glycolytic metabolism of oral squamous cell carcinoma and precedes their activation into carcinoma associated fibroblasts. Cell Mol Life Sci. 2020;77:1115–33. doi: 10.1007/s00018-019-03209-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gong X, Zhu Z. Long Noncoding RNA HOTAIR Contributes to Progression in Hepatocellular Carcinoma by Sponging miR-217-5p. Cancer Biother Radiopharm. 2020 doi: 10.1089/cbr.2019.3070. [DOI] [PubMed] [Google Scholar]
- 41.Wu L, Zhang L, Zheng S. Role of the long non-coding RNA HOTAIR in hepatocellular carcinoma. Oncol Lett. 2017;14:1233–9. doi: 10.3892/ol.2017.6312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yu F, Chen B, Dong P, Zheng J. HOTAIR Epigenetically Modulates PTEN Expression via MicroRNA-29b: A Novel Mechanism in Regulation of Liver Fibrosis. Mol Ther. 2017;25:205–17. doi: 10.1016/j.ymthe.2016.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cicchini C, de Nonno V, Battistelli C, Cozzolino AM, De Santis Puzzonia M, Ciafre SA, et al. Epigenetic control of EMT/MET dynamics: HNF4alpha impacts DNMT3s through miRs-29. Biochim Biophys Acta. 2015;1849:919–29. doi: 10.1016/j.bbagrm.2015.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang T, He X, Chen A, Tan K, Du X. LncRNA HOTAIR contributes to the malignancy of hepatocellular carcinoma by enhancing epithelial-mesenchymal transition via sponging miR-23b-3p from ZEB1. Gene. 2018;670:114–22. doi: 10.1016/j.gene.2018.05.061. [DOI] [PubMed] [Google Scholar]
- 45.Burnett JC, Rossi JJ. RNA-based therapeutics: current progress and future prospects. Chem Biol. 2012;19:60–71. doi: 10.1016/j.chembiol.2011.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kaczmarek JC, Kowalski PS, Anderson DG. Advances in the delivery of RNA therapeutics: from concept to clinical reality. Genome Med. 2017;9:60. doi: 10.1186/s13073-017-0450-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Grijalvo S, Alagia A, Jorge AF, Eritja R. Covalent Strategies for Targeting Messenger and Non-Coding RNAs: An Updated Review on siRNA, miRNA and antimiR Conjugates. Genes (Basel) 2018;9 doi: 10.3390/genes9020074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chu C, Qu K, Zhong FL, Artandi SE, Chang HY. Genomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactions. Molecular cell. 2011;44:667–78. doi: 10.1016/j.molcel.2011.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Noce V, Battistelli C, Cozzolino AM, Consalvi V, Cicchini C, Strippoli R, et al. YAP integrates the regulatory Snail/HNF4alpha circuitry controlling epithelial/hepatocyte differentiation. Cell Death Dis. 2019;10:768. doi: 10.1038/s41419-019-2000-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cicchini C, Laudadio I, Citarella F, Corazzari M, Steindler C, Conigliaro A, et al. TGFbeta-induced EMT requires focal adhesion kinase (FAK) signaling. Exp Cell Res. 2008;314:143–52. doi: 10.1016/j.yexcr.2007.09.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.