Chromosome segregation in bacteria is poorly understood outside some prominent model strains 1–5 and even less is known about how it is coordinated with other cellular processes. This is the case for the opportunistic human pathogen Streptococcus pneumoniae (the pneumococcus)6, lacking the Min and the nucleoid occlusion systems7 and possessing only an incomplete chromosome partitioning Par(A)BS system, in which ParA is absent8. The bacterial tyrosine-kinase (BY-Kinase9) CpsD that is required for capsule production was previously found to interfere with chromosome segregation10. Here, we identify a protein of unknown function that interacts with CpsD and drives chromosome segregation. RocS (Regulator of Chromosome Segregation) is membrane-bound and also interacts both with DNA and the chromosome partitioning protein ParB to properly segregate the origin of replication region to new daughter cells. In addition, we show that RocS interacts with the cell division protein FtsZ and hinders cell division. Altogether, this work reveals that RocS is the cornerstone of a nucleoid protection system ensuring proper chromosome segregation and cell division in coordination with the biogenesis of the protective capsular layer.
Previous studies have evidenced that ParB and SMC are involved, but not essential, in pneumococcal chromosome segregation8. Notably, individual or double deletion of parB and smc only lead to weak chromosome segregation defects, suggesting that other factors remain to be discovered. In line with this hypothesis, impaired autophosphorylation of the BY-kinase CpsD generated elongated cells with an aberrant nucleoid morphology10. CpsD is primarily described as a key regulator of the export and synthesis of the polysaccharide capsule, the main virulence factor of the pneumococcus, which is exclusively produced at the pneumococcal division septum10–13. To understand the potential relationship between capsule production and the chromosome biology, we first screened a yeast two-hybrid genomic library of a pneumococcal laboratory strain14 using CpsD or its membrane activator CpsC as baits. Indeed, the interaction between CpsD and CpsC mimics the behavior of BY-kinases found in proteobacteria15. Both CpsD and CpsC interacted with Spr0895, a protein with unknown function (Supplementary Fig. 1a). The interaction between Spr0895 and CpsD was confirmed in vitro and in vivo (Supplementary Fig. 1b-d). The spr0895 gene is conserved among Streptococcaceae (Supplementary Fig. 2) and is hereinafter referred to as rocS (Regulator of Chromosome Segregation) based on our observations below.
We first constructed a rocS deletion in the encapsulated virulent D39 strain and analyzed capsule production by immunofluorescence microscopy10. As observed for wild-type cells, capsule was detected over the entire surface of ΔrocS cells (Fig. 1a). Quantification of the fluorescent signal, together with the immuno-detection of the total fraction of capsule, revealed that capsule production and polymerization were not affected (Supplementary Fig. 3). However, although the cell shape of ΔrocS cells was not significantly altered, mutants displayed a growth defect with an increased generation time compared to wild-type cells (Supplementary Fig. 4). Surprisingly, when we looked at the DNA content of ΔrocS cells, we found that 13.9% of cells were anucleate (Fig. 1a,b). Deletion of rocS in the isogenic non-encapsulated mutant D39Δcps or the non-encapsulated laboratory R800 strain resulted in comparable fractions (15.4% and 15.7% respectively) of anucleate cells (Fig. 1b and Supplementary Fig. 5 and 6) indicating that nucleoid defects were not dependent on capsule production. Complementation of the ΔrocS D39 and R800 mutants with an ectopic copy of rocS (ΔrocS-PcomX-rocS) restored the wild-type phenotype with 1.5% and 1% of anucleate cells, respectively (Fig. 1b). By comparison, the deletion of parB or smc results in less than 4% of anucleate cells8. We therefore deleted parB or smc in the D39-ΔrocS-PcomX-rocS strain. Upon rocS induction, these mutants were as viable as the ΔrocS D39 mutant. However, the depletion of rocS induced an additive detrimental effect on cell viability (Supplementary Fig. 7). Consistently, we were unable to delete both rocS and either smc or parB, suggesting that RocS acts complementary with ParB and SMC in the pneumococcal chromosome biology.
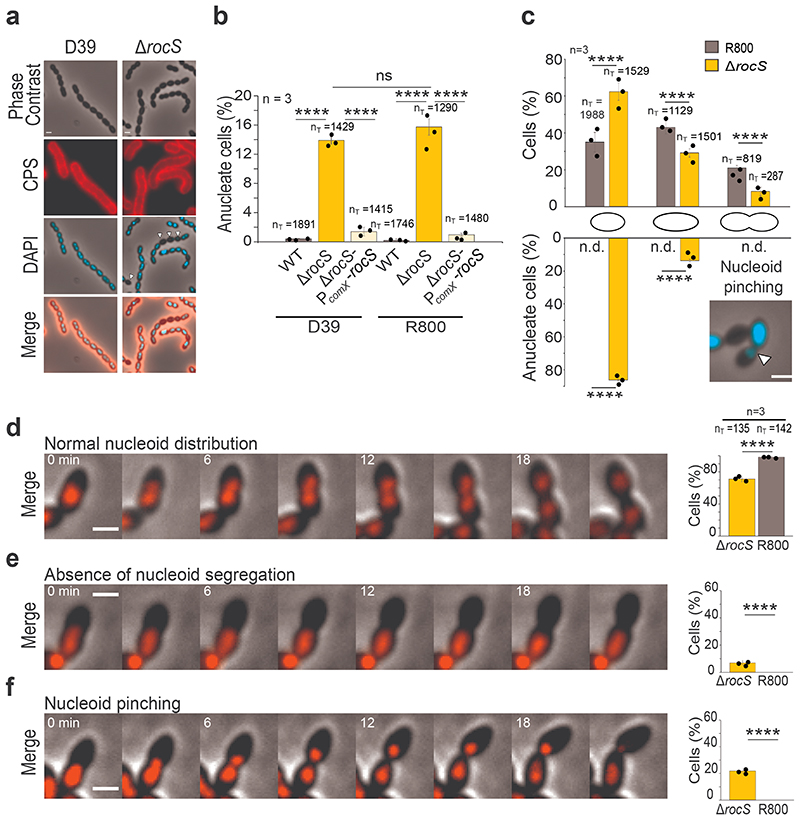
Figure 1. Impact of rocS deletion on capsule production and nucleoid distribution.
a. Detection of capsular polysaccharides (CPS) and DNA in D39 and ΔrocS cells. Phase contrast (grey), CPS (red), DAPI (blue) and merged images are shown. Arrowheads indicate anucleate cells. Images are representative of 3 experiments repeated independently. b. Percentage of anucleate cells in D39 and R800 (grey) strains, corresponding ΔrocS mutants (orange) and complemented strains (yellow). c. Percentage of anucleate cells in the course of the cell cycle. R800 (grey) and ΔrocS (orange) cells were sorted into three size groups (small, elongated and constricting cells) as a proxy for their progression in the cell cycle. The percentage of each group and the percentage of anucleate cells in each group are shown respectively in the upper and the lower bar chart. Arrowheads indicate chromosome pinching in constricting cells. n.d.= none detected. d-f. Still images from fluorescence time-lapse microscopy (Supplementary Video 1, 2 and 3) showing (d) a normal nucleoid segregation, (e) an absence of nucleoid segregation, or (f) a nucleoid pinching event during the cell division in WT (d) or ΔrocS cells (e and f) producing HlpA-mKate2. The percentage of each event (normal, absence or pinching) in WT and ΔrocS cells are shown in the corresponding bar chart. Scale bar, 1 μm. In b-f, nT indicates the number of cells analyzed from 3 independent experiments. Bar chart, with data points overlap, represents the mean ± SEM. Two-tailed P-values derived from two-population proportion tests for the following pairs of proportions: Panel b: ‘D39-WT’ vs ‘D39-ΔrocS’ (P<0.0001); ‘D39-ΔrocS’ vs ‘D39-ΔrocS-PcomX-rocS’ (P=2,49.10-12); ‘R800-WT’ vs ‘R800-ΔrocS’ (P<0.0001); ‘R800-ΔrocS’ vs ‘R800-ΔrocS-PcomX-rocS’ (P<0.0001); ‘D39-ΔrocS’ vs ‘R800-ΔrocS’ (P=0.158). Upper panel c: ‘R800-WT’ vs ‘R800-ΔrocS’ small cells (P<0.0001), elongated cells (P<0.0001) and constricting cells (P=7.29.10-12). Lower panel c: ‘R800-WT’ vs ‘R800-ΔrocS’ for small cells (P<0.0001) and elongated cells (P<0.0001). Panel d-f: ‘R800-WT’ vs ‘R800-ΔrocS’ (d) P=2.6.10-15, (e) P=8.2.10-15 and (f) P=3.45.10-5. **** P < 0.0001. ns P > 0.05.
To analyze the chromosome dynamics in the absence of RocS, we quantified the relative proportions of three size groups (small, elongated and constricting cells) for ΔrocS R800 cells (Fig. 1c). By comparison with the relative proportion observed for wild-type cells, we observed an increase in the number of small cells: 62.5% of ΔrocS cells displayed the morphology of rounded small cells while only 35% of wild-type cells harbored this morphology (Fig. 1c). Since the formation of mini-cells is usually associated with an aberrant localization of the divisome, we looked at its localization in ΔrocS cells using GFP-FtsA as a proxy. As observed for wild-type cells, GFP-FtsA localized at the division site at mid-cell in ΔrocS cells, suggesting that the localization of the division machinery was not affected in ΔrocS cells (Supplementary Fig. 8). Remarkably, the small cells constitute the large majority of the anucleate cells (86.3%) while elongated and constricting cells harbored asymmetric distribution of the nucleoid, suggesting that chromosome-pinching events occurred in ΔrocS cells (Fig. 1c). To confirm this, we followed the localization of the HlpA-mKate2 fusion, a pneumococcal histone-like protein16. As expected for wild-type cells, the chromosome duplicates at the early stage of the cell cycle and eventually splits into two parts that segregate to each daughter cell (Fig. 1d and Supplementary Video 1). In contrast, newly replicated chromosomes in ΔrocS cells were either not segregated (7%) (Fig. 1e and Supplementary Video 2), or partially segregated and eventually truncated by the newly forming septum (21.8%), a process also known as the guillotine effect17 (Fig. 1f and Supplementary Video 3). In the latter case, the signal of the truncated chromosome was ultimately degraded. In both cases, these aberrant chromosome-partitioning events led to the formation of small and anucleate cells. To test if chromosome replication was affected in the R800 ΔrocS mutant, we determined the ratio between the origin of replication (oriC) and the terminus region (ter) of the chromosome in exponentially growing cells 18 (Supplementary Fig. 9). As expected, we observed that wild-type cells displayed a characteristic mean ratio of 1.68 ± 0.28 whereas this ratio was close to 1 for a thermo-sensitive dnaA (encoding the replication initiator protein) mutant shifted to non-permissive temperature. The origin-to-terminus ratios of ΔrocS (1.67 ± 0.24) and complemented ΔrocS-PcomX-rocS (1.56 ± 0.24) cells were similar to that of wild-type cells, indicating that RocS is not involved in chromosome replication. Together, our results show that chromosome segregation rather than chromosome replication is severely affected in the absence of RocS.
To characterize the contribution of RocS to chromosome segregation, we next examined the localization of the origin of replication (oriC) during the cell cycle of wild-type and ΔrocS R800 cells (Fig. 2). We used a system based on the ectopic production of a fluorescent fusion of RepC, the ParB homolog of Enterococcus faecalis, and insertion of parSEf sites from E. faecalis near the pneumococcal oriC 19 (Fig. 2a). Neither expression of repC-gfp nor insertion of parSEf sites influenced the pneumococcal cell cycle as evidenced by wild-type growth kinetics and cell morphology (Supplementary Fig. 10). When produced, the RepC-GFP fusion formed diffraction-limited foci in the vicinity of oriC (Fig. 2b and Supplementary Fig. 10). As previously characterized20, oriC localized as a single focus located around mid-cell of nascent cells (Fig. 2b). The duplication of the focus was followed by rapid segregation of the two foci toward the center of each daughter cell where they remain as the cell elongate. Interestingly, new cycles of chromosome replication and segregation started early in the cell cycle, even before the completion of division, as attested by the 4.5% of nascent cells containing 2 foci and the 5% of cells at the later stage of the cell cycle containing 3 or 4 foci (Fig. 2b, c). By comparison, the subcellular localization of oriC throughout the cell cycle was strongly affected in the absence of RocS. After duplication, most of the two foci remained near mid-cell and did not segregate (Fig. 2b, c). On average, the spacing rate (distance between 2 foci of oriC in relation to the cell length) was significantly lower in ΔrocS cells (0.32±0.003) than in WT cells (0.47±0.003) (Fig. 2d). Furthermore, the proportion of cells with single foci was significantly higher in ΔrocS cells (47.6%) than in wild-type cells (23%) (Fig. 2c). Since chromosome replication was not affected in ΔrocS cells (Supplementary Fig. 9), this observation suggests that after replication, some oriC copies may be too close to be detected as separated foci in ΔrocS cells. Finally, we did not detect constricting cells containing 3 or 4 foci in ΔrocS cells (Fig. 2c). Thus, the two newly replicated chromosome origins segregate less efficiently in the absence of RocS, reflecting its crucial role in chromosome segregation.
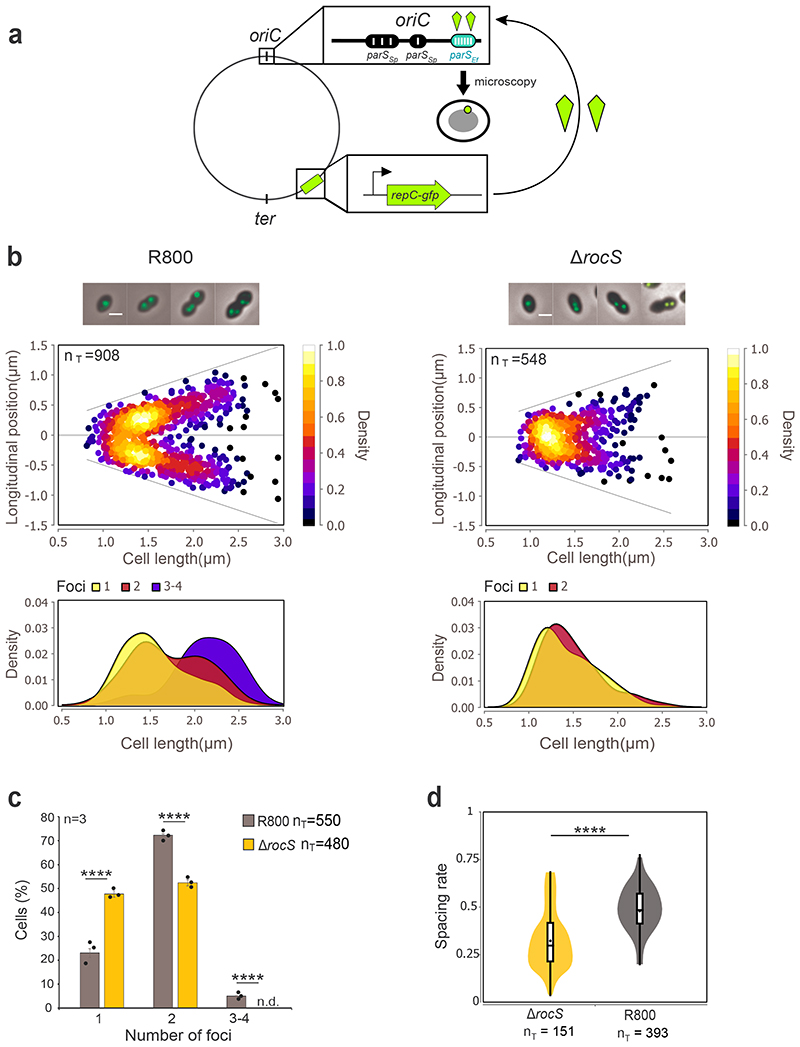
Figure 2. oriC segregation patterns in wild-type and ΔrocS cells.
a. Schematic representation of the Par system used to image the origin of replication (oriC). parS sequences from E. faecalis (parS Ef, blue oval) were inserted into the chromosome near the pneumococcal oriC while the parB homolog repC fused to gfp (RepC-GFP, green kite) is expressed ectopically under the control of the PcomX promoter. Upon loading of RepC-GFP onto parS Ef sites, the localization of oriC is followed by fluorescence microscopy (green dot). parS Sp indicates native pneumococcal parS sites. b. (upper panels) Localization heat maps of oriC (RepC-GFP) positions along the cell length in wild-type and ΔrocS R800 cells. Representative merged images between phase contrast and GFP fluorescence signal of cells with either 1, 2 or 3/4 foci are shown on the top. Scale bar, 1 μm. (lower panels) Kernel density plots of the cell length in relation to the number of foci in wild-type and ΔrocS R800 cells. c. Relative percentages of cells as a function of the number of oriC foci in WT (grey) and ΔrocS (orange) cells. Bar chart, with data points overlap, represents the mean ± SEM. Two-tailed P-values derived from a two-population proportion test for the following pairs of proportions: ‘R800-WT’ vs ‘R800-ΔrocS’ one foci (P<0.0001), two foci (P= 8.9.10-16), three foci (P=1,5.10-10). d. Measurements of the spacing rate (relative distance between 2 foci of oriC in relation to the cell length). Box indicates the 25th to 75th percentile and Whiskers indicate the minimum and the maximum. The mean and the median are indicated with a dot and a line in the box, respectively. Two-tailed P-value derived from a Mann-Whitney test between ‘R800-WT’ and ‘R800-ΔrocS’ is P = 7.9.10-9. **** P < 0.0001. nT indicates the number of cells analyzed. Experiments were performed in triplicate.
Next, we followed the subcellular localization of RocS fused to the GFP. Expression and functionality of the GFP-RocS fusion is suitable for RocS localization studies as attested by wild-type growth kinetics, cell morphology, intracellular RocS level and a low level of anucleate R800 cells (3%) (Supplementary Fig. 11 and 12). By wide-field epifluorescence microscopy, the GFP-RocS fusion protein was shown to form one or two bright foci per cell that were mostly localized around mid-cell of small cells and that positioned toward the center of the daughter cell as cells elongate (Fig. 3a). However, when observed by total internal reflection fluorescence microscopy at relatively high frequency data acquisition, we also detected some highly dynamic but very faint foci with no specific localization during the cell cycle (Supplementary Fig. 13 and Supplementary Video 4). Using image averaging, we showed that the faint foci were homogeneously distributed all around the cell periphery. This suggested that the fain foci could represent small units of RocS diffusing at the cell membrane even if one cannot exclude that they could also be due to some degradation species of GFP-RocS (Supplementary Fig. 12). Interestingly, we observed that bright foci mostly co-localized with oriC (d<0.15μm; Supplementary Fig. 14), suggesting that only the bright foci might be involved in chromosome segregation. Supporting this, we detected that RocS interacts with the pneumococcal ParB protein both in vivo and in vitro (Supplementary Fig. 15). As ParB binds to 4 parS sites close to oriC 8, these data suggest that RocS acts together with ParB to allow proper chromosome segregation.
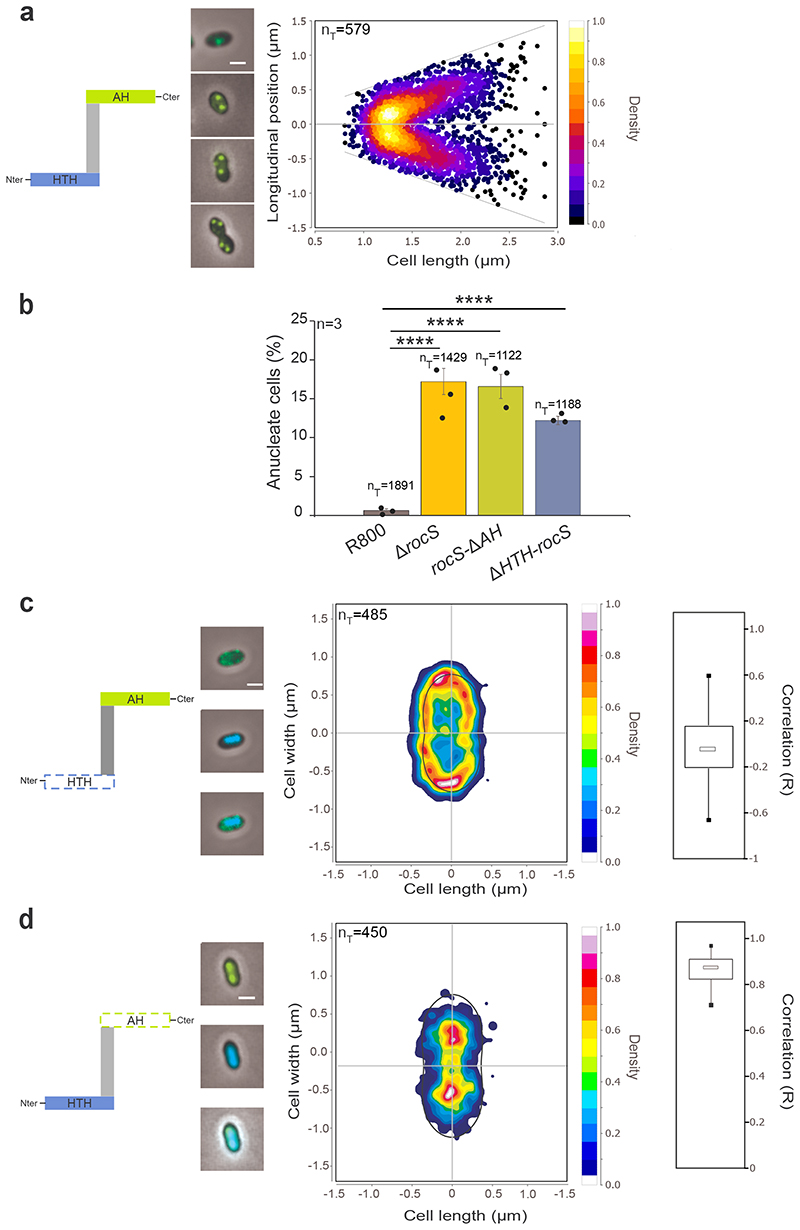
Figure 3. Localization of GFP-RocS and derivatives and impact on nucleoid localization.
Schematic representations of RocS and derivatives are shown on the left of panels a, c and d. a. Heat map representing the longitudinal localization of GFP-RocS as a function of the cell length in R800 cells. Representative merged images of cells with either 1, 2 or 3/4 foci are shown on the left. b. Relative percentage of anucleate cells for rocS-ΔAH and ΔHTH-rocS R800 strains. Bar chart, with data points overlap, represents the mean ± SEM. nT indicates the total number of cells analyzed from three independent experiments. Two-tailed P-values derived from a two-population proportion test for the following pairs of proportions: ‘R800-WT’ vs ‘R800-ΔrocS’ (P<0.0001), ‘R800-WT’ vs ‘R800-rocS-ΔAH’ (P<0.0001) and ‘R800-WT’ vs ‘R800-ΔHTH-rocS’ (P<0.0001). **** P<0.0001. c-d. Heat maps representing the 2-dimensional localization patterns of GFP-ΔHTH-RocS (c) and GFP-RocS-ΔAH (d) in R800 cells. Representative overlays of phase contrasts and, GFP or DAPI fluorescence signals, or both signals, are shown on the left. Scale bar, 1 μm. The distribution of the Pearson correlation coefficient (R), measured between the DAPI and GFP signals for each strain are shown as box (25th to 75th percentile) and whisker (minimum and maximum) plots on the right.
Bioinformatic analysis of the RocS sequence predicted the presence of a C-terminal membrane-binding amphipathic helix (AH) homologous to that of MinD of Escherichia coli 21 and an N-terminal helix-turn-helix domain (HTH, InterPro IPR000047) characteristic of DNA-binding proteins 22 (Supplementary Fig. 16). These two domains are required for the function of RocS in chromosome segregation as both ΔHTH-rocS and rocS-ΔAH R800 cells displayed growth and viability defects as well as an anucleate phenotype and cell shapes similar to ΔrocS R800 cells (Fig. 3c and Supplementary Fig. 17). In addition, deletion of either the AH or the HTH domain drastically altered the localization pattern of RocS (Fig. 3 c, d). The deletion of the N-terminal HTH domain resulted in the discontinuous redistribution of GFP-ΔHTH-RocS at the cell periphery. On the other hand, GFP-RocS-ΔAH co-localizes with the nucleoid in the pneumococcal cell (median R = 0.85, interquartile range = 0.83-0.92) (Fig. 3d) suggesting that RocS binds DNA via the HTH domain. ChIP-seq experiments (Supplementary Fig. 18) using a FLAG-RocS fusion protein (Supplementary Fig. 12) did not reveal any specific conserved DNA sequence targeted by RocS. We further showed that DNA binding was independent of the size, GC content and sequence of the DNA fragment (Supplementary Fig. 19a, b). Interestingly, analysis of the HTH domain of RocS indicates that it resembles that of regulators of the Lrp and MarR families22. Some members of these families, like LrpC from Bacillus subtilis, bind intrinsically curved sequences of DNA23. Therefore, RocS may recognize some topological features of the DNA. Finally, to confirm that the HTH of RocS is required and sufficient for DNA binding, we substituted the highly conserved glycine 15 residue of the HTH with a proline residue22 (Supplementary Fig. 16) and showed that DNA binding of RocS-G15P-ΔAH was nearly completely abolished (Supplementary Fig. 19c). Collectively, these data show that the C-terminal AH is required for the interaction of RocS with the membrane, whereas the N-terminal HTH domain mediates RocS DNA binding and that both domains are essential for RocS function.
We finally questioned the biological role of the interaction between RocS and the tyrosine-autokinase CpsD (Supplementary Fig. 1). Previous findings showed that CpsD possesses a structural fold comparable to that of ParA proteins that usually assist ParB in chromosome segregation 10,24,25. Since ParA is absent in the pneumococcus7 and CpsD interacts directly with ParB, it was proposed that CpsD could act as a ParA-like protein10. Interestingly, this interaction is modulated by the autophosphorylation of CpsD and mimicking permanent phosphorylation of CpsD (CpsD-3YE) promotes capsule biogenesis and normal chromosome segregation by enabling ParB mobility10 (Fig. 4a). By contrast, defective autophosphorylation of CpsD (CpsD-3YF) not only impairs capsule production, but also reduces ParB mobility, inducing aberrant chromosome segregation and leading to cell elongation10 (Fig. 4b). By consequence, even in the absence of a conserved nucleoid occlusion system in the pneumococcus7, cell division appears to be blocked to protect the nucleoid against truncation by the newly forming septum when CpsD is not phosphorylated. To test whether RocS could be involved in this process, we deleted rocS in D39 strains mimicking either permanent or defective phosphorylation of CpsD (respectively ΔrocS-cpsD-3YE and ΔrocS-cpsD-3YF) and looked at the cell morphology, capsule production and DNA content. As expected, the deletion of rocS generated approximately 13% of anucleate cells in both cases (Fig. 4). Strikingly, while the deletion of rocS in the permanent phosphorylation cpsD-3YE mutant did not impact the cell morphology, the deletion of rocS suppressed the elongated phenotype of the defective phosphorylation cpsD-3YF mutant (Fig. 4). This property is specific to rocS as the deletion of parB in the defective phosphorylation cpsD-3YF mutant strain had only a modest impact on cell elongation (Fig. 4b). By contrast, overproducing RocS in the absence of CpsD also induced an elongated phenotype (Supplementary Fig. 20). This suggests that while the division block depends on the phosphorylation state of CpsD, the latter can be bypassed by the overexpression of rocS. By consequence, RocS, along with the CpsD phosphorylation level, blocks cell division to protect the nucleoid against truncation.
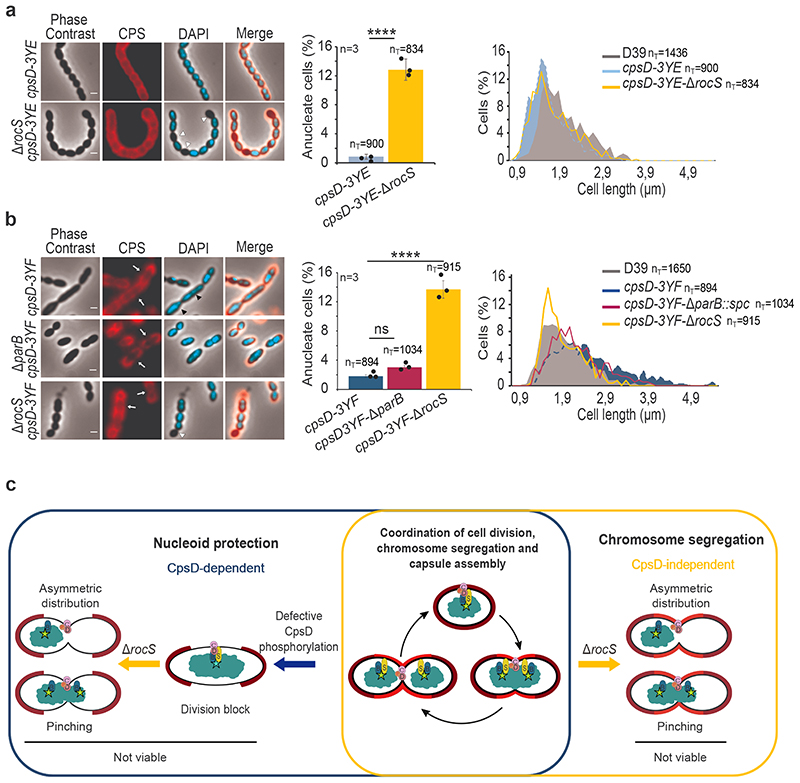
Figure 4. Deletion of rocS in phospho-ablative and phospho-mimetic CpsD mutants and model for the RocS nucleoid protection system.
Detection of CPS and DNA in (a) cpsD-3YE and cpsD-3YE-ΔrocS and (b) cpsD-3YF, cpsD-3YF- ΔparB and cpsD-3YF-ΔrocS. Phase contrast (grey), CPS (red), DAPI (blue) and merged images are shown on the left. White arrows show CPS production defects, white arrowheads show anucleate cells and black arrowheads show nucleoid segregation defects. Scale bar, 1 μm. The corresponding percentage of anucleate cells are shown as bar charts. Bar chart, with data points overlap, represents the mean ± SEM. Two-tailed P-values derived from a two-population proportion test for the following pairs of proportions:: ‘cpsD-3YE’ vs ‘cpsD-3YE-ΔrocS ‘(P<0.0001), ‘cpsD-3YF’ vs ‘cpsD-3YF-ΔrocS’ (P=1.9.10-13) and ‘cpsD-3YF’ vs ‘cpsD-3YF-ΔparB’ (P=1.2). ****: P < 0.0001. ns P >0.05. The corresponding distribution of the cell length are shown on the right as histograms. nT indicates the number of cells analyzed from 3 independent experiments and standard errors are indicated with error bars. c. Model for the nucleoid protection system coordinating capsule synthesis, chromosome segregation and cell division. Non-phosphorylated CpsD hinders both capsule synthesis and chromosome segregation inducing a division block. The deletion of rocS alleviates the division block and results in uncontrolled cell constriction with improper chromosome segregation (pinching and asymmetric distribution) leading to non-viable progeny. ParB, RocS, CpsD and its transmembrane activator CpsC are indicated by blue, yellow, brown and pink circles, respectively. Red “P” and the turquoise star indicate CpsD autophosphorylation and the oriC region, respectively. Capsule is shown in light (new capsule produced during cell division) and dark (inherit from the mother cell) red.
To get more insight into the interplay between RocS and CpsD, we looked at the co-localization between CpsD-mKate2 and GFP-RocS in D39. As expected, since CpsD localized exclusively at mid-cell throughout the cell cycle10, RocS co-localized with CpsD only at the early stage of the cell cycle (Supplementary Fig. 21). Since RocS migrates with oriC and thus with the nucleoid as the cell elongates, one can assume that RocS and CpsD can interact only when the nucleoid is not fully or properly segregated. Therefore, both the phosphorylation state of CpsD and the co-occurrence of RocS and CpsD at mid-cell could regulate the constriction and eventually block cell division when the nucleoid is not properly segregated. Interestingly, we found that RocS interacts with FtsZ in vitro (Supplementary Fig. 22) suggesting that this cell division block could result from a direct action of RocS on the Z-ring.
Our results suggest that RocS has two main roles during the pneumococcal cell cycle: (i) RocS, independently of CpsD, is required for proper chromosome partitioning and (ii) RocS, along with CpsD, regulate the constriction and eventually blocks the cell division to ensure proper capsule secretion and to protect the nucleoid against premature truncation (Fig. 4c). Typical nucleoid occlusion systems prevent the assembly of the FtsZ ring over the nucleoid26,27. However, FtsZ-rings were found to be properly positioned at the division septum in cpsD-3YF elongated cells10 indicating that the constriction rather than the assembly of the FtsZ-ring at mid-cell, was blocked by RocS. RocS constitutes therefore an authentic nucleoid protection system, which is mechanistically distinct from the typical nucleoid occlusion mechanisms. Cell elongation of the pneumococcus is not achieved by MreB-mediated lateral insertion of peptidoglycan, but rather organized by the Z-ring itself at the cell center 28. Preventing the assembly of the Z-ring over the nucleoid, like in rod-shaped bacteria, would thus hinder cell elongation and therefore the cell division of the pneumococcus. The latter, and probably all Streptococcaceae (Supplementary Fig. 2), have therefore evolved their own nucleoid protection system to avoid premature truncation of the nucleoid during cell division. Overall, our work demonstrate that RocS can be viewed as the cornerstone of a process connecting and coordinating capsule synthesis, chromosome segregation and cell division. The “raison d’être” of such a regulatory process coordinating capsule synthesis with cell cycle progression is likely to make sure that cells are covered by capsule at every step of the cell cycle in order to prevent detection by the human immune system.
Methods
Strains and growth conditions
Strains used in this study are listed in the Table S1. Streptococcus pneumoniae R800, D39Δcps 29 and D39 and derivatives were cultivated at 37°C in C+Y medium or Todd-Hewitt Yeast (THY) broth at pH 7.4.
Cell growth curves were monitored in JASCO V-630-BIO-spectrophotometer and the optical density was read automatically every 10 min. Escherichia coli XL1-B strain30 was used for cloning and E. coli BL2131 for overproduction of CpsC/D, RocS, RocS-ΔAH, ParB and FtsZ. E. coli strains were grown in lysogeny broth (LB) supplemented with appropriate antibiotic. Growth was monitored by optical density (OD) readings at 550 nm or 600 nm for S. pneumoniae or E. coli strains, respectively.
Construction of plasmids and strains
Gene modifications (gfp, mkate2 and flag fusions, knock-out and domain deletion) in S. pneumoniae were achieved by homologous recombination using the two-step procedure based on a bicistronic kan-rpsL cassette called Janus32 and constructed at their native chromosomal locus. They are thus expressed under the control of the native promoter and represent the only source of the protein.
ΔrocS D39, ΔrocS R800, ΔrocS-Δsmc R800 and ΔrocS-ΔparB R800 strains were complemented ectopically for rocS expression using the strategy described by33 using the competence inducible system of Streptococcus thermophilus. The ComS-inducible comR DNA fragment was introduced between the treR and amiF loci of both strains. Then, the rocS copy under the control of the comX promoter was inserted between the cpsN and cpsO genes in R800 or at the bgaA locus in D39 strains.
For constructing the system for tagging ori, we used the parS sites and the ParB homologue RepC fused to GFP from Enteroccocus faecali 19. The parS sites were inserted between thmA and IS1167 loci near the pneumococcal origin of replication. Then, the repC-gfp under the control of the promoter of the comX gene of Streptococcus thermophilus were used by PCR and inserted between the cpsN and cpsO genes in the R800 strain. repC-gfp expression was induced with 1 μM of ComS.
To construct the thermo-sensitive dnaA R800 mutated strain, we PCR amplified the dnaA(T1193C) mutated gene of the D39 thermo-sensitive mutant described in Kjos et al. 16. The DNA fragment was then transformed in the R800 strain and cells were plated at 30°C. After overnight growth, colonies were resuspended in THY and cultured again on plates at either 30°C or 40°C. The mutation in dnaA was checked by DNA sequencing in clones growing at 30°C but not at 40°C.
For the construction of plasmids overproducing RocS-ΔAH-6His or native FtsZ, we PCR amplified DNA fragments coding for either RocS from Met1 to Gln150 or FtsZ from Met1 to Arg419, respectively, using chromosomal DNA from S. pneumoniae R800 as template. The obtained rocS or ftsZ DNA fragments were cloned between either the NdeI and PstI or the NdeI and HindIII cloning sites of pT7-734. Site-directed mutagenesis of glycine 15 to proline of RocS was performed by PCR using the plasmid pT7.7-rocSΔAH (Table S1) as a template. The other plasmids used in this study are described in Table S1.
The oligonucleotides used for all construction are listed in Table S2. Plasmids and pneumococcal strains were verified by DNA sequencing to verify error-free PCR amplification.
Protein purification
Purification of the chimera 6His-CpsC/D and ParB-6His was performed as described previously10. To purify RocS-ΔAH-6His, E. coli BL21 were used and cultured at 37 °C in LB medium. At OD600 = 0.6, 1mM IPTG was added and cells culture were continued for 3 h at 37 °C. Cells were then harvested by centrifugation and resuspended in buffer A (Tris-Hcl 25 mM, pH 7.5; NaCl 1 M, imidazole 10 mM; glycerol 10%) containing 10 mg mL-1 of lysozyme, 1 μg mL–1 of protease inhibitor (Roche Diagnostics). After sonication and centrifugation, the supernatant was loaded on to a Ni-NTA agarose resin (Qiagen) and extensively washed with buffer A containing 20 mM imidazole. RocS-6His was eluted with buffer B (Tris-Hcl 25 mM, pH 7.5; NaCl 300 mM, imidazole 300 mM; glycerol 10%). Pure fractions were pooled and dialyzed against buffer C (HEPES 50 mM, pH 7.5 or Tris pH 7,5 25 mM ; NaCl 150 mM, glycerol 10%).
To purify FtsZ, E. coli BL21 were used and cultured at 37 °C in LB medium. At OD600 = 0.6, 1mM IPTG was added and cells culture were continued for 2 h at 37 °C. Cells were then harvested by centrifugation and resuspended in buffer D (Tris-Hcl 50 mM, pH 8; KCl 50 mM, EDTA 1 mM) containing 10 mg mL-1 of lysozyme, 1 μg mL–1 of protease inhibitor (Roche Diagnostics) and 1 μg mL–1 DNase-RNase (Sigma). After sonication and centrifugation, Ammonium sulfate was added to the supernatant at 4°C to a final concentration of 30% and stirred for 30 min. The mixture was then centrifuged at 25,000 x g for 30 min, and the pellet was retained, resuspended in buffer D and the solution was dialyzed against Buffer D for 4h at 4°C. The supernatant was then applied to a HiTrap Q HP column (GE Healthcare). After extensive washing, the protein was eluted with a gradient of 0 to 50% of buffer E (buffer D + KCl 1M).
Peak fractions containing FtsZ were pooled and concentrated in Amicon filters (10 kDa cutoff). The concentrated lysate was further injected into a GE-Hiload 16/600 superdex 200 size exclusion chromatography column. The FtsZ protein peaks were collected in buffer D and analyzed on SDS PAGE. Homogenous fractions were collected and concentrated as mentioned above. The final buffer was 50 mM Tris.HCl pH8, 200 mM KCl, 1 mM EDTA, 10% glycerol. Protein concentrations were determined using a Coomassie assay protein dosage reagent (Uptima) and proteins were then aliquoted and frozen at -80C.
Co-immunoprecipitation and immunoblot analysis
For co-immunoprecipitation, cultures of S. pneumoniae cells were grown at 37°C in C+Y medium until OD550nm = 0.3. Cells pellets were incubated at 30°C for 30 min in buffer A (Tris-HCl 0.1 M, pH 7.5; MgCl2 2 mM, Sucrose 1 M, 6 mg mL-1 of DNase I and RNase A, 1 μg mL–1 of protease inhibitor). After centrifugation at 4°C, the pellet was resuspended in buffer B (Tris-Hcl 0.1 M, pH 7.5; EDTA 1 mM, 0.1% Triton, 6 mg mL-1 of DNase I and RNase A, 1 μg mL–1 of protease inhibitor) and incubated 15 min at room temperature before being harvested by centrifugation. The supernatant was then incubated with Dynabeads (Invitrogen) coupled with 20 μg anti-FLAG or anti-GFP antibodies and incubated for 2 hour at 4°C. After extensive wash with buffer C (Tris-Hcl 10 mM, pH 7.5, EDTA 0.5 mM, 0.1% Triton, NaCl 150 mM, 1 μg mL–1 of protease inhibitor), Protein-bounded bead were eluted with SDS-PAGE loading buffer at 95°C for 10 min and analyzed by SDS-PAGE and immunoblotting using either a rabbit anti-GFP antibody at 1/10,000 (AMS Biotechnology), the anti-FLAG antibody at 1/1,000 (Sigma) or the anti-mKate2 (1/3000) antibody (Invitrogen).
For immunoblot analysis, S. pneumoniae cells were resuspended in TE-buffer (25 mM Tris-HCl pH 7.5, 1 mM EDTA) supplemented with protease and phosphatase inhibitor cocktail II (Sigma-Aldrich) and lysed by sonication. 25 μg of crude extracts were analyzed by SDS-PAGE, electrotransferred onto a polyvinylidene difluoride membrane and incubated with either rabbit anti-RocS at 1/5000 (produced by Eurogentec with purified RocS-ΔAH-6His), rabbit-anti-enolase polyclonal antibody at 1/50000035 or rabbit anti-serotype 2 CPS polyclonal antibody at 1/2,000 (Statens serum Institute). A goat anti-rabbit polyclonal antibody horseradish peroxidase (HRP) conjugated (Biorad) was used at 1/5000 to reveal immunoblots.
Yeast-two hybrid
The yeast two hybrid genetic screens were carried out using a mating strategy as described previously14,36. Construction of the pGBDU-cpsD and the pGBDU-cpsC bait plasmids and expressing CpsD fused to the DNA-binding domain of Gal4 (BD) was described in10. This plasmid was introduced in the PJ69-4(α) haploid strain. This strain was then mated with PJ69-4 haploid(α) strain harboring a library of pGAD plasmids expressing genomic fragments of S. pneumoniae R6 in fusion with the GAL4 activating domain (AD)14. Potential binary interactions were selected by the ability of the yeast diploids to grow on synthetic media agar SC–LUH lacking Leucine (L) and Uracil (U) to select for maintenance of plasmids pGAD and pGBDU, respectively, as well as histidine (H), to selects for the interaction37. Additionally, binary interactions were tested by a matrix-based approach by mating haploid cells expressing BD-CpsD, with haploid cells of complementary mating type expressing the AD-prey protein fusions RocS50-163, RocS, CpsC and CpsD. Diploids were first selected onto –LU media and further tested for interacting phenotypes (i.e. ability to grow on SC–LUH selective agar plates) to reveal binary interactions between bait and prey proteins.
Preparation and analysis of CPS
CPS were prepared as previously described10. Briefly, S. pneumoniae cultures were grown until OD550nm = 0.3, washed once with PBS and resuspended in buffer A (Tris-HCL 50nM, pH 7.4; sucrose 20%; MgSO4 50 nM). The solution was then supplemented with 400 units of mutanolysin and 6 μg/μl of DNase and RNase and incubated overnight at room temperature. After centrifugation at 16,000 x g for 20 min at 4 °C, pellets were resuspended in the same volume of buffer A. 10 μL of the mixture were then mixed with 5 μl of buffer B (Tris-HCl 50 mM, pH 8.0; EDTA 50 mM; Tween20 0.5%; Triton X100 0.5%) and 20 μg of proteinase K, incubated 30 min at 37°C and analyzed by SDS-PAGE and immunoblotting.
Microscopy techniques
Cells were grown until OD550nm = 0.1. For immunofluorescence microscopy, cells were mixed with the rabbit-serotype 2 CPS polyclonal antibody (Statens Serum Institute) at 1/1,000, washed and then incubated with the anti-rabbit Dylight-549 antibody (Jackson ImmunoResearch) at 1/2,000. After a last wash with PBS, CPS were imaged and fluorescence intensity was measured as described previously10.
For DAPI staining, 10 μl of S. pneumoniae cell culture were mixed with 1 μl of DAPI at 2 μg/μl (Molecular Probes) and incubated 5 min at room temperature. For mKate2 and GFP fluorescence imaging, cells were spotted on pads made of 1.5% agarose in C+Y medium at 37°C as previously described in38.
Slides were visualized with a Nikon TiE microscope fitted with an Orca-CMOS Flash4 V2 camera with a 100 × 1.45 objective. Images were collected using NIS-Elements (Nikon). For TIRF experiments, data acquisition was done every 100 ms to 2 sec. Images were analyzed using the software ImageJ (http://rsb.info.nih.gov/ij/) and the plugin MicrobeJ39.
Diffraction-limited foci of RepC-GFP or GFP-RocS were detected using the feature/spot detection option in MicrobeJ. This option combines spatial 2D filtering (Median Filter) and 2D local maxima algorithm to localize single fluorescent maxima in each detected cell. Each maximum was then fit to a single peak or a multi peak 2D Gaussian curve, to determine their amplitude, their FWHM (Full width at half Maximum) and their coordinates at the subpixel resolution. Maxima were finally filtered based on the goodness of the fit and their amplitude. Their sub-cellular localizations were automatically computed for each associated particle.
Microscale thermophoretic analysis
Microscale thermophoresis40 was used to test the interaction of RocS-AH with the chimera CpsC/D, ParB and FtsZ. Binding experiments were carried out with a Monolith NT.115 Series instrument (Nano Temper Technologies GMBH). RocS-ΔAH was labeled with the red dye NT-647. Briefly, sample containing 50 nM of labeled RocS-ΔAH-6His and increasing concentrations of 6His-CpsC/D (from 275 pM to 9 μM) or ParB-6His (from 427 pM to 14 μM) or FtsZ (from 686 pM to 22.5 μM) were loaded on K023 Monolith NT.115 hydrophobic capillaries and thermophoresis was measured for 30 s at 25°C. Each measurement was made in triplicate. Experiments were carried out at 25°C in 10mM HEPES pH 7.5, 150mM NaCl and 0.05% Tween-20. Analysis was performed with the Monolith software. Affinity KD was quantified by analyzing the change in normalized fluorescence (FNorm = fluorescence after thermophoresis/initial fluorescence) as a function of the concentration of the titrated 6His-CpsC/D or ParB-6His proteins.
oriC-ter ratio determination by real-time qPCR
Genomic DNA was extracted using the DNA maxima Kit (Qiagen). Real-time qPCR was performed as described previously18. Briefly, each 20 μl sample consisted of 8.8 ng of DNA, 0.6 pmol of each primer (Table S2), and 10 μl of the 2x SYBR Green Supermix (Bio-Rad). Amplification was performed on an iQ5 Real-Time PCR Detection System (Bio-Rad). To find amplification efficiencies, Monte Carlo simulations were performed in R. Average Ct-values and their corresponding standard deviations were used to simulate 10,000 new sets of Ct-values that were used to compute the amplification efficiencies for each set. From that population of possible efficiencies, averages and standard deviations were derived. Analysis of the real-time qPCR experiments for oriC-ter ratio determination was performed using the 2-ΔΔCT method 41, with the important difference that the earlier found amplification efficiencies were used to determine the fold-change per cycle, instead of assuming it to equal 2. As a reference, cells with an assumed oriC-ter ratio of 1 were used. For that, a thermo-sensitive dnaA-mutant (dnaA-T1193C) was grown at 30°C until an OD600 of 0.05. Then, cells were transferred to non-permissive temperature (40°C) and incubated for 1 hour, followed by harvesting and isolation of chromosomal DNA. Uncertainties in oriC-ter ratios were also determined by Monte Carlo simulations.
Bioinformatic analyses
For the phylogenetic analysis, homologues of RocS were retrieved using iterative BLASTP from BLAST package 2.2.6 against a local database containing 4466 prokaryotic complete proteomes retrieved from NCBI ftp (ftp://ftp.ncbi.nlm.nih.gov/). The Spr0895 amino acid sequence (NP_358489.1) was used as first seed. Protein sequences detected as homologues were aligned with MAFFT v7.123b42 and used to build an HMM profile with HMMER v3.1b143. The profile was then used to query the local database with HMMSEARCH from the HMMER package. Plasmid sequences were removed from the analysis. Phylogeny of Lactobacillales has been inferred from a supermatrix of ribosomal proteins. One strain per family was selected to represent each family in Lactobacillales and a sequence of one species of Listeriaceae was added to root the tree. The sequences were aligned using MAFFT (L-INS-I option) and trimmed with BMGE-1.1 (option BLOSUM30)44. The evolution model was chosen using BIC criteria and the phylogeny was inferred using PhyML45 (LG+I+F+G4, 8 sequences, 6219 positions).
Secondary structure predictions of RocS were obtained using PSIPRED 46. The helical representation of RocS and MinD of Escherichia coli was made using http://www.tcdb.org/progs/?tool=pepwheel.
ChIP-Seq and data analysis
The protocol for immunoprecipitation of FLAG-tagged RocS was largely performed as described by Minnen et al.8 and was performed in duplicate. Specifically, cells were pre-cultured in acid C+Y (pH 6.8) and grown until OD600=0.2. Cells were then diluted 1:50 in acid C+Y, to a final volume of 250 mL and grown until OD600 0.20. Then, 25 mL of fixation buffer (11% formaldehyde; 5 mM NaOH; 50 mM Tris, pH 8.0; 100 mM NaCl; 0.5 mM EGTA; 1 mM EDTA) was added, the culture was mixed by inversion and incubated at RT for 30 min. Formaldehyde was quenched by the addition of 92 mL of 1M Tris pH 8 and 10 min incubation at RT. First, cells were spun down at 5000g for 12 min at 4°C and washed in 20 mL ice-cold PBS. Secondly, cells were spun down at 5000g for 12 min at 4°C and washed in 10 mL ice-cold PBS. Thirdly, cells were spun down at 5000g for 12 min at 4°C and washed in 1 mL ice-cold PBS. Finally, cells were spun down at 11000g for 2 min at 4°C, supernatant was removed and the pellet was snap-frozen in liquid nitrogen and stored at -80°C.
Dynabeads™ Protein G (Invitrogen) were prepared according to the manufacturer’s instructions and loaded with 10 μg of anti-FLAG antibody. Cell pellets were resuspended in 2 mL ice-cold lysis buffer (50 mM Hepes-KOH, pH 7.55; 140 mM NaCl; 1 mM EDTA; 1% Triton X-100; 0.1% sodium deoxycholate; 1 mM PMSF; protease inhibitor cocktail; 100 mg/mL RNase) and transferred to a 5 mL round-bottom tube. Samples were sonicated on ice for twice 10 x 30 sec on a Sonics Vibracell VCX130 with 65% amplitude. Samples were then split into 200 μL whole-cell extract (WCE, stored at -20°C) and 800 μL for immunoprecipitation. The latter fractions were incubated for 2-4 hours at 4°C on a rotating wheel. Supernatant was removed on a magnet. The beads were washed three times 5 min, shaking at 800 rpm at RT. The first wash was performed with 1 mL lysis buffer, the second wash with 1 mL lysis buffer with extra NaCl (500 mM final concentration), and the third wash with 1 mL wash buffer (10 mM Tris, pH 8.0; 250 mM LiCl; 1 mM EDTA; 0.5% NP-40; 0.5% sodium deoxycholate; 1 mM PMSF). Supernatant was removed and beads were resuspended in 520 μL TES buffer. WCE samples were thawed and combined with 300 μL TES buffer and 20 μL of 10% SDS. To elute DNA, both WCEs and immunoprecipitates (IPs) were incubated overnight on a shaker at 65°C. On a magnet, the DNA-containing supernatant was transferred to a fresh tube.
To the DNA samples, 1 μL phenol per μL of sample was added, followed by vortexing and centrifugation at 11000g for 5 min. The DNA-containing layer was then added to 1 μL chloroform per 1 μL of sample, followed by vortexing and centrifugation at 11000g for 5 min. The DNA-containing layer was transferred to a fresh tube and 1 μL of glycogen (Roche) and 40 μL of 3M NaOAc (pH 5.3) were added. After mixing, 1 mL of pure ethanol was added and tubes were incubated for 20 min at -20°C, followed by centrifugation for 15 min at 4°C. The pellets were resuspended in 100 μL TE (pH 8.0) and incubated for 15 min at 65°C. DNA fragmentation was verified on an agarose gel.
GATC Biotech performed further library preparation and sequencing on an Illumina HiSeq with 50 nt single-end reads. Due to an insufficient amount of material in one of the immunoprecipitate samples, we collected data on 2 WCE samples and 1 IP sample.
Sequencing reads were mapped to the S. pneumoniae R6 genome using Bowtie247. Visualization and peak calling was performed with SeqMonk (https://www.bioinformatics.babraham.ac.uk/projects/seqmonk/"). Although no significant enrichment was detected by SeqMonk, we selected the 6 most intense peaks and extracted the 500 nucleotides surrounding the respective maximums. Motif enrichment analysis was then performed using MEME-ChIP (https://www.ncbi.nlm.nih.gov/pubmed/21486936).
Electrophoretic mobility shift assay (EMSA)
EMSA were carried out by incubating different concentrations of purified protein RocS-ΔAH-6His or RocS-G15P-ΔAH-6His (0; 5; 10; 15 μM) with 50 ng of DNA in the following buffer (500mM Tris-HCl pH 8.8, 50mM MgSO4). DNA fragments of different length and percentage of GC content were PCR amplified (pUC18, gfp or genomic DNA of Pseudomonas aeruginosa PA7) using primers listed in Table S2. Reactions were incubated for 15 min at 37 °C before being loaded on 1% agarose gels. Gels were stained with ethidium bromide and imaged with UV light.
Supplementary Material
Acknowledgments
Work on the Grangeasse lab is supported by grants from the CNRS, the University of Lyon, the Agence National de la Recherche (ANR-10-BLAN-1303-01 and ANR-15-CE32-0001-01), the Region Auvergne-Rhône-Alpes (financial support for C.M. and P.S.G.), the “Fondation pour la Recherche Médicale” (financial support for N.D. (ING20150532637) and C.M. (FDT20170437272)) and the Bettencourt-Schueller Foundation. Work in the Veening lab is supported by the Swiss National Science Foundation (project grant 31003A_172861), a JPIAMR grant (50-52900-98-202) from the Netherlands Organization for Health Research and Development (ZonMW) and ERC consolidator grant 771534-PneumoCaTChER. We thank Stéphanie Ravaud for help in RocS structural predictions, Andrew Fenton (University of Sheffield, UK) for providing us with the D39Δcps strain and Keith Weaver (University of South Dakota) for providing us with the pAD1 plasmid. We acknowledge the contribution of the Protein Science of the “SFR Biosciences Gerland-Lyon Sud (UMS344/US8)”.
Footnotes
Author contributions
C.G. directed the study. C.M. conducted the experiments of cell imaging and analyses with A.D., genetics with J.N., protein purification and western blot analysis with J.P.L, C.F. and S.N.N. C.M. and N.D. implemented the oriC localization system. J.P.L. performed microscale thermophoresis experiments. C.M. and J.S. performed oriC/ter ratio and ChIP-Seq experiments. M.F.N.G. performed yeast two-hybrid experiments. P.S.G. performed phylogeny analyses. All authors designed and analyzed the data. C.G. and J.W.V. wrote the manuscript and all authors edited the manuscript.
Competing interests
The authors declare no competing financial interests.
Data availability
The data that support the findings of this study are available from the corresponding author upon request. The ChIP-seq data were deposited at NCBI SRA (accession number PRJNA511435) and GEO (accession number GSE129717).
References
- 1.Toro E, Shapiro L. Bacterial chromosome organization and segregation. Cold Spring Harb Perspect Biol. 2010;2:a000349. doi: 10.1101/cshperspect.a000349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reyes-Lamothe R, Nicolas E, Sherratt DJ. Chromosome replication and segregation in bacteria. Annu Rev Genet. 2012;46:121–143. doi: 10.1146/annurev-genet-110711-155421. [DOI] [PubMed] [Google Scholar]
- 3.Wang X, Montero Llopis P, Rudner DZ. Organization and segregation of bacterial chromosomes. Nat Rev Genet. 2013;14:191–203. doi: 10.1038/nrg3375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Badrinarayanan A, Le TB, Laub MT. Bacterial chromosome organization and segregation. Annu Rev Cell Dev Biol. 2015;31:171–199. doi: 10.1146/annurev-cellbio-100814-125211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bohm K, et al. Novel Chromosome Organization Pattern in Actinomycetales-Overlapping Replication Cycles Combined with Diploidy. mBio. 2017;8 doi: 10.1128/mBio.00511-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grangeasse C. Rewiring the Pneumococcal Cell Cycle with Serine/Threonine- and Tyrosine-kinases. Trends Microbiol. 2016;24:713–724. doi: 10.1016/j.tim.2016.04.004. [DOI] [PubMed] [Google Scholar]
- 7.Pinho MG, Kjos M, Veening JW. How to get (a)round: mechanisms controlling growth and division of coccoid bacteria. Nat Rev Microbiol. 2013;11:601–614. doi: 10.1038/nrmicro3088. [DOI] [PubMed] [Google Scholar]
- 8.Minnen A, Attaiech L, Thon M, Gruber S, Veening JW. SMC is recruited to oriC by ParB and promotes chromosome segregation in Streptococcus pneumoniae. Mol Microbiol. 2011;81:676–688. doi: 10.1111/j.1365-2958.2011.07722.x. [DOI] [PubMed] [Google Scholar]
- 9.Grangeasse C, Nessler S, Mijakovic I. Bacterial tyrosine kinases: evolution, biological function and structural insights. Philos Trans R Soc Lond B Biol Sci. 2012;367:2640–2655. doi: 10.1098/rstb.2011.0424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nourikyan J, et al. Autophosphorylation of the Bacterial Tyrosine-Kinase CpsD Connects Capsule Synthesis with the Cell Cycle in Streptococcus pneumoniae. PLoS Genet. 2015;11:e1005518. doi: 10.1371/journal.pgen.1005518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Morona JK, Morona R, Miller DC, Paton JC. Mutational Analysis of the Carboxy-Terminal (YGX)(4) Repeat Domain of CpsD, an Autophosphorylating Tyrosine Kinase Required for Capsule Biosynthesis in Streptococcus pneumoniae. J Bacteriol. 2003;185:3009–3019. doi: 10.1128/JB.185.10.3009-3019.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yother J. Capsules of Streptococcus pneumoniae and other bacteria: paradigms for polysaccharide biosynthesis and regulation. Annu Rev Microbiol. 2011;65:563–581. doi: 10.1146/annurev.micro.62.081307.162944. [DOI] [PubMed] [Google Scholar]
- 13.Henriques MX, Rodrigues T, Carido M, Ferreira L, Filipe SR. Synthesis of capsular polysaccharide at the division septum of Streptococcus pneumoniae is dependent on a bacterial tyrosine kinase. Mol Microbiol. 2011;82:515–534. doi: 10.1111/j.1365-2958.2011.07828.x. [DOI] [PubMed] [Google Scholar]
- 14.Mirouze N, Claverys J-P, Noirot P Université Paul, S. Identification du produit d’un gène tardif impliqué dans la régulation de la compétence et dans le processing de l’ADN lors de la transformation naturelle chez S pneumoniae. 2007 [s.n.] [Google Scholar]
- 15.Bechet E, et al. Tyrosine-kinases in bacteria: from a matter of controversy to the status of key regulatory enzymes. Amino Acids. 2009;37:499–507. doi: 10.1007/s00726-009-0237-8. [DOI] [PubMed] [Google Scholar]
- 16.Kjos M, Veening JW. Tracking of chromosome dynamics in live Streptococcus pneumoniae reveals that transcription promotes chromosome segregation. Mol Microbiol. 2014;91:1088–1105. doi: 10.1111/mmi.12517. [DOI] [PubMed] [Google Scholar]
- 17.Yamanaka K, Ogura T, Niki H, Hiraga S. Identification of two new genes, mukE and mukF, involved in chromosome partitioning in Escherichia coli. Mol Gen Genet. 1996;250:241–251. doi: 10.1007/BF02174381. [DOI] [PubMed] [Google Scholar]
- 18.Slager J, Kjos M, Attaiech L, Veening JW. Antibiotic-induced replication stress triggers bacterial competence by increasing gene dosage near the origin. Cell. 2014;157:395–406. doi: 10.1016/j.cell.2014.01.068. [DOI] [PubMed] [Google Scholar]
- 19.Francia MV, Weaver KE, Goicoechea P, Tille P, Clewell DB. Characterization of an active partition system for the Enterococcus faecalis pheromone-responding plasmid pAD1. J Bacteriol. 2007;189:8546–8555. doi: 10.1128/JB.00719-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van Raaphorst R, Kjos M, Veening JW. Chromosome segregation drives division site selection in Streptococcus pneumoniae. Proc Natl Acad Sci U S A. 2017;114:E5959–E5968. doi: 10.1073/pnas.1620608114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou H, Lutkenhaus J. Membrane binding by MinD involves insertion of hydrophobic residues within the C-terminal amphipathic helix into the bilayer. J Bacteriol. 2003;185:4326–4335. doi: 10.1128/JB.185.15.4326-4335.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aravind L, Anantharaman V, Balaji S, Babu MM, Iyer LM. The many faces of the helix-turn-helix domain: transcription regulation and beyond. FEMS Microbiol Rev. 2005;29:231–262. doi: 10.1016/j.femsre.2004.12.008. [DOI] [PubMed] [Google Scholar]
- 23.Tapias A, Lopez G, Ayora S. Bacillus subtilis LrpC is a sequence-independent DNA-binding and DNA-bending protein which bridges DNA. Nucleic Acids Res. 2000;28:552–559. doi: 10.1093/nar/28.2.552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leipe DD, Wolf YI, Koonin EV, Aravind L. Classification and evolution of P-loop GTPases and related ATPases. J Mol Biol. 2002;317:41–72. doi: 10.1006/jmbi.2001.5378. [DOI] [PubMed] [Google Scholar]
- 25.Gerdes K, Howard M, Szardenings F. Pushing and pulling in prokaryotic DNA segregation. Cell. 2010;141:927–942. doi: 10.1016/j.cell.2010.05.033. [DOI] [PubMed] [Google Scholar]
- 26.Wu LJ, Errington J. Coordination of cell division and chromosome segregation by a nucleoid occlusion protein in Bacillus subtilis. Cell. 2004;117:915–925. doi: 10.1016/j.cell.2004.06.002. [DOI] [PubMed] [Google Scholar]
- 27.Bernhardt TG, de Boer PA. SlmA, a nucleoid-associated, FtsZ binding protein required for blocking septal ring assembly over Chromosomes in E. coli. Mol Cell. 2005;18:555–564. doi: 10.1016/j.molcel.2005.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fleurie A, et al. Interplay of the serine/threonine-kinase StkP and the paralogs DivIVA and GpsB in pneumococcal cell elongation and division. PLoS genetics. 2014;10:e1004275. doi: 10.1371/journal.pgen.1004275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fenton AK, Mortaji LE, Lau DT, Rudner DZ, Bernhardt TG. CozE is a member of the MreCD complex that directs cell elongation in Streptococcus pneumoniae. Nat Microbiol. 2016;2:16237. doi: 10.1038/nmicrobiol.2016.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bullock WO, Fernandez JM, short JM. a high efficiency plasmid transforming recA Escherichia coli strain with beta-galactosidase selection. Biotechniques. 1987;5:376. [Google Scholar]
- 31.Studier FW, Moffatt BA. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol. 1986;189:113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- 32.Sung CK, Li H, Claverys JP, Morrison DA. An rpsL cassette, janus, for gene replacement through negative selection in Streptococcus pneumoniae. Appl Environ Microbiol. 2001;67:5190–5196. doi: 10.1128/AEM.67.11.5190-5196.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Berg KH, Biornstad TJ, Straume D, Havarstein LS. Peptide-regulated gene depletion system developed for use in Streptococcus pneumoniae. J Bacteriol. 2011;193:5207–5215. doi: 10.1128/JB.05170-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cortay JC, et al. In vitro asymmetric binding of the pleiotropic regulatory protein, FruR, to the ace operator controlling glyoxylate shunt enzyme synthesis. J Biol Chem. 1994;269:14885–14891. [PubMed] [Google Scholar]
- 35.Fleurie A, et al. Mutational dissection of the S/T-kinase StkP reveals crucial roles in cell division of Streptococcus pneumoniae. Mol Microbiol. 2012;83:746–758. doi: 10.1111/j.1365-2958.2011.07962.x. [DOI] [PubMed] [Google Scholar]
- 36.Marchadier E, et al. An expanded protein-protein interaction network in Bacillus subtilis reveals a group of hubs: Exploration by an integrative approach. Proteomics. 2011;11:2981–2991. doi: 10.1002/pmic.201000791. [DOI] [PubMed] [Google Scholar]
- 37.James P, Halladay J, Craig EA. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.de Jong IG, Beilharz K, Kuipers OP, Veening JW. Live Cell Imaging of Bacillus subtilis and Streptococcus pneumoniae using Automated Time-lapse Microscopy. J Vis Exp. 2011 doi: 10.3791/3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ducret A, Quardokus EM, Brun YV. MicrobeJ, a tool for high throughput bacterial cell detection and quantitative analysis. Nat Microbiol. 2016;1:16077. doi: 10.1038/nmicrobiol.2016.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jerabek-Willemsen M, Wienken CJ, Braun D, Baaske P, Duhr S. Molecular interaction studies using microscale thermophoresis. Assay Drug Dev Technol. 2011;9:342–353. doi: 10.1089/adt.2011.0380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 42.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Eddy SR. A new generation of homology search tools based on probabilistic inference. Genome Inform. 2009;23:205–211. [PubMed] [Google Scholar]
- 44.Criscuolo A, Gribaldo S. BMGE (Block Mapping and Gathering with Entropy): a new software for selection of phylogenetic informative regions from multiple sequence alignments. BMC Evol Biol. 2010;10:210. doi: 10.1186/1471-2148-10-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guindon S, et al. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 46.Jones DT. Protein secondary structure prediction based on position-specific scoring matrices. J Mol Biol. 1999;292:195–202. doi: 10.1006/jmbi.1999.3091. [DOI] [PubMed] [Google Scholar]
- 47.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Slager J, Aprianto R, Veening JW. Deep genome annotation of the opportunistic human pathogen Streptococcus pneumoniae D39. Nucleic Acids Res. 2018;46:9971–9989. doi: 10.1093/nar/gky725. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon request. The ChIP-seq data were deposited at NCBI SRA (accession number PRJNA511435) and GEO (accession number GSE129717).