Abstract
The diverse number of neurons in the cerebral cortex are generated during development by neural stem cells lining the ventricle, and they continue maturing postnatally. Dynamic chromatin regulation in these neural stem cells is a fundamental determinant of the emerging property of the functional neural network, and the chromatin remodellers are critical determinants of this process. Chromatin remodellers participate in several steps of this process from proliferation, differentiation, migration leading to complex network formation which forms the basis of higher-order functions of cognition and behaviour. Here we review the role of these ATP-dependent chromatin remodellers in cortical development in health and disease and highlight several key mouse mutants of the subunits of the complexes which have revealed how the remodelling mechanisms control the cortical stem cell chromatin landscape for expression of stage-specific transcripts. Consistent with their role in cortical development, several putative risk variants in the subunits of the remodelling complexes have been identified as the underlying causes of several neurodevelopmental disorders. A basic understanding of the detailed molecular mechanism of their action is key to understating how mutations in the same networks lead to disease pathologies and perhaps pave the way for therapeutic development for these complex multifactorial disorders.
Keywords: NuRD complex, BAF (mSWI/SNF) complex, ISWI complex, INO80 complex, Corticogenesis, Neurodevelopmental disorders
1. Introduction
The cerebral cortex is the domain of all higher-order functions in the brain, including motor and sensory activities, cognition and social behaviour. Neurulation commences with the establishment of a neural plate from a single sheet of neuroepithelial cells (NECs) that eventually close to give rise to the neural tube (Gotz and Huttner, 2005). These NECs divide symmetrically to self-renew and expand the pool, and at the onset of neurogenesis at embryonic day (E)10.5, give rise to apical radial glia cells (aRGs) (Fig. 1) (Tuoc et al., 2014). aRGs are bipolar cells that line the ventricular zone (VZ) of the neocortex and extend processes towards both the apical and basal surfaces. Similar to NECs, aRGs divide both symmetrically for pool expansion as well as asymmetrically to self-renew and form a differentiating daughter cell (Tuoc et al., 2014). aRGs can directly give rise to neurons upon asymmetric division (direct neurogenesis) or can give rise to basal intermediate progenitors (bIPs) or basal radial glia (bRGs), that line the subventricular zone (SVZ), which then differentiate to form neurons (indirect neurogenesis) (Georgala et al., 2011; Gotz and Huttner, 2005; Govindan and Jabaudon, 2017; Jabaudon, 2017; Molyneaux et al., 2007; Sessa et al., 2008; Tuoc et al., 2014). bRGs are found in both the lateral and medial cortex of mice, albeit at lower proportions as compared to gyren-cephalic mammals. bRGs have been further characterised into separate populations based on the expression of Hopx, a marker for human bRGs (Wang 2011; Vaid 2018). bIPs are fate restricted with a limited self-renewal capacity (Hevner, 2019; Kalebic and Huttner, 2020). Both aRGs and bIPs give rise to neurons, that populate the cortical plate (CP) to form the six-layered neocortex (Agirman et al., 2017; Budday et al., 2015; Florio and Huttner, 2014; Leone et al., 2008; Lodato and Arlotta, 2015; Molyneaux et al., 2007; Mukhtar and Taylor, 2018). For a detailed review on mammalian corticogenesis refer to Florio and Huttner (2014); and Lodato and Arlotta (2015). The neocortex is formed in an inside out fashion, beginning with the formation of layer VI. New-born neurons migrate past the early-born neurons and occupy their positions in the growing cortical plate to create newer layers (Fig. 1). As corticogenesis progresses, aRGs also give rise to astrocytes and oligodendrocytes, which mature postnatally (Guillemot, 2007; Martynoga et al., 2012). An interesting feature of mammalian nervous system is that the same set of progenitors give rise to both neurons and glial cells. Neurogenesis precedes gliogenesis, and this transition from neuron to glia production is termed as the neuron-glia cell fate switch (Gao 2014; Mukhtar and Taylor, 2018).
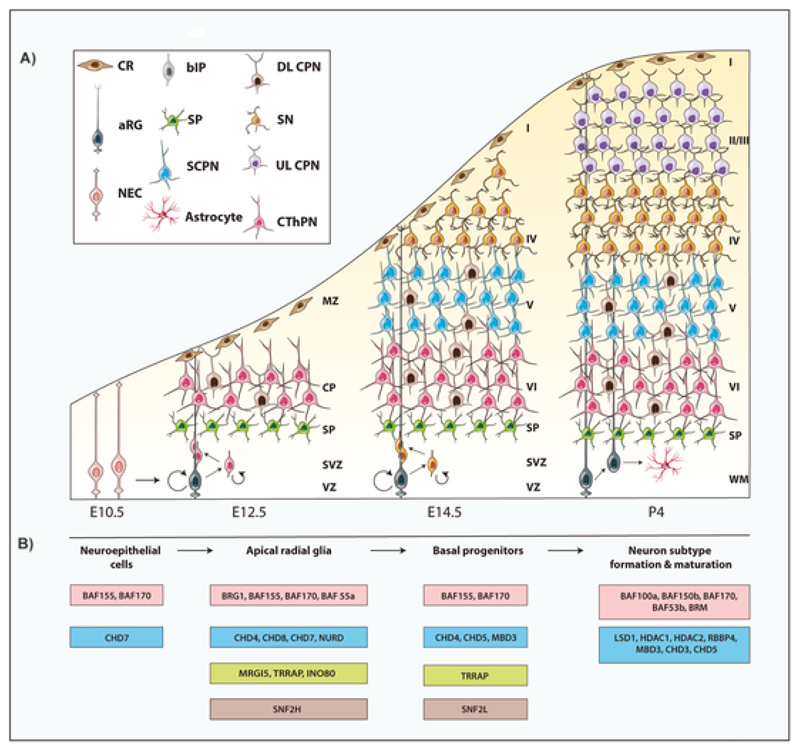
Fig. 1. Snapshot of cortical development and the chromatin remodelling complexes involved in its formation.
Excitatory projection neurons emerge from the progenitors in the dorsal telencephalon which line the ventricle. At early embryonic ages, the dorsal neuroep-ithelium consists of NECs which divide both symmetrically for pool expansion and asymmetrically to give rise to aRGs in the VZ. aRGs self-amplify by symmet-ric division and also give rise to bIPs, bRGs in the SVZ (indirect neurogenesis) and neurons (direct neurogenesis) by asymmetric division. bIPs and bRGs can also self-renew and produce neurons that fill the CP. The CR cells in the MZ do not originate from the cortical progenitor zone. The various PNs of the neocor-tex are generated in a temporally timed manner in an inside out fashion with layer VI generated first followed by V,IV, III and II as development progresses. The newly generated PNs use the aRGs or bRGs as guidance to migrate and occupy distinct positions in the cortical plate. Chromatin remodellers involved in the various steps of cortical development are depicted. Abbreviations: Neuroepithelial precursors cells (NE or NECs), aRGs-apical radial glia, bIPs-basal inter-mediate progenitors, bRGs-basal radial glia, VZ-ventricular zone, SVZ-subventricular zone, SP-subplate, CP-cortical plate, MZ – marginal zone, WM-white matter, CThPN-corticothalamic projection neuron of layer 6, PN-projection neuron, SCPN-subcerebral projection neuron of layer 5, DL CPN-deep layer callosal projection neuron of layer VI and V, UL CPN-upper layer projection neuron of layers II and III, SN – stellate neuron of layer IV.
Corticogenesis involves expression of transcriptional waves of proliferative (P), neurogenic (Ng) and neuronal (N) transcripts sequentially. These P-type, Ng-type and N-type transcripts display enriched localisation in the VZ, SVZ and CP respectively. The P-type transcripts are the first to be repressed as development progresses (Telley et al., 2016) This sequential expression and repression of transcripts at different stages of corticogenesis require dynamic chromatin remodelling to promote/inhibit chromatin accessibility.
Chromatin accessibility is a dynamic and changing process occurring during cortical development wherein nucleosomes are rearranged to bring about gene regulation. A recent study showed that predicted regulatory elements in accessible chromatin at different stages of human cortical development display motifs for various transcription factors known to regulate corticogenesis at that stage (Markenscoff-Papadimitriou et al., 2020). PAX6 motifs on open regulatory elements were observed at 14 gestational weeks (gw), and PBX1 motifs were observed at 19gw. Similarly, the deep layers displayed motifs for TBR1, ETV1, ETV2; whereas the upper layers showed motifs for LHX2, LHX5, BRN1 and BRN2. This accessibility of motifs at the regulatory elements is essential for transcription factor binding. Transcription factors can then recruit chromatin remodelling complexes to regulate gene expression. For example, the transcription factor LHX2 interacts with NuRD subunits, and specific subunits bind to distal LHX2 binding regions in the genome to repress layer V formation during layer VI neurogenesis (Muralidharan et al., 2017).
Chromatin Remodellers are translocases that alter the chromatin structure to enable access to transcriptional machinery (Clapier et al., 2017). Chromatin remodelling complexes consist of multiple protein subunits. They are ATP-driven and contain a single core subunit belonging to the Sucrose non-fermentable (SNF2) family that acts as an ATPase. Based on the sequence similarity and domain structures of the ATPase subunits, these complexes have been classified into four sub-families - Nucleosome Remodelling and Deacetylase (NuRD), BRG1-or BRM Associated Factors (BAF or SWItch/Sucrose non-fermentable - SWI/SNF), Imitation SWItch (ISWI) and Inositol 80 (INO80) (Hargreaves and Crabtree, 2011; Ho and Crabtree, 2010; Hota and Bruneau, 2016). The subunits of these complexes are expressed in the different zones of the developing cortical primordium (Sokpor et al., 2018) and thus regulate several critical steps of corticogenesis starting from progenitor proliferation to neuronal maturation (Fig. 1).
A key feature of the mammalian genome is the highly compact arrangement of chromatin structure in the form of nucleosomes. This compaction accommodated the genome expansion that resulted from gene duplication and a relative increase in protein-coding and noncoding regulatory elements. In comparison to yeast and flies, mammals underwent only a small increase in the total number of genes, in contrast, the amount of regulatory DNA expanded substantially in the mammalian genome (Alfert et al., 2019; Hajheidari et al., 2019). This led to the proposal that the function of a limited number of proteincoding genes is diversified by establishing a pattern of gene expression mediated by transcriptional regulation.
Therefore, chromatin regulation at different levels, namely DNA methylation, histone modification, and nucleosome remodelling evolved to compact DNA and regulate gene expression (Hajheidari et al., 2019; Ho and Crabtree, 2010; Wu et al., 2009).
The chromatin modifiers have thus expanded and diversified their function. For example, SWI/SNF has changed from a monomorphic complex with a role in transcriptional activation in yeast to a dimorphic complex in invertebrates with two ATPase paralogs that have evolved to activate and repress gene expression transcriptionally. In the vertebrates, SWI/SNF complexes are polymorphic with two ATPase paralogs and multiple variants for most of the associated subunits, permitting a combinatorial assembly of the complex (Hajheidari et al., 2019; Ho and Crabtree, 2010; Wu et al., 2009). The chromodomain helicase DNA-binding (CHD) proteins have diverged and acquired new functions to act through multi-subunit complexes such as NuRD. The ISWI family consists of 2 ATPases, conserved from yeast to mammals. The INO80 complex has acquired new subunits such as YY1, NFRKB, and Uch37 over the course of evolution (Conaway and Conaway, 2009).
Hence, the emergence of ATP dependent chromatin remodelling complexes in parallel with multicellularity and morphological complexity was a quintessential step in the evolutionary trajectory of eukaryotes.
2. Composition of the chromatin remodelling complexes
2.1. Core constituents of the NuRD complex
The NuRD complex possesses dual enzymatic activities of - ATPase and deacetylase. The core components of the complex are CHD3/4/5, HDAC1 and HDAC2, MBD2/3, MTA1/2/3, RBBP4 and RBBP7, GATAD2A/B and LSD1(KDM1A) (Fig. 2); (Kolla et al., 2015; Tong et al., 1998; Wade et al., 1998; Wang et al., 2009; Xue et al., 1998; Zhang et al., 1998, 1999). The chromodomain helicase DNA-binding (CHD) proteins 3, 4 and 5 are ATPases, and form the catalytic core in independent NuRD complexes (Tong et al., 1998; Wade et al., 1998; Xue et al., 1998; Zhang et al., 1998; Nitarska et al., 2016). The PHD fingers of the chromodomain proteins interact with histone H3 tails (Mansfield et al., 2011; Musselman et al., 2009). The Histone deacetylases 1 and 2 (HDAC1 and HDAC2) provide the second catalytic activity to the complex. HDAC1 and HDAC2 belong to class I of histone deacetylases (Gregoretti et al., 2004) and deacetylate both histone and non-histone proteins (Seto and Yoshida, 2014). MBD2 and MBD3 belong to the methyl cytosine-guanosine (CpG)-binding domain (MBD) protein family (Le Guezennec et al., 2006). MBD3 preferentially occupies methylated CpG islands, whereas MBD2 displays localisation on both methylated and unmethylated CpG islands (Cramer et al., 2014). Metastasis associated genes 1 and 2 (MTA1/2) act as scaffolds for the NuRD com-plex and help in complex assembly (Alqarni et al., 2014). The retinoblastoma-binding proteins RBBP4 and RBBP7 are histone chaperones (Murzina et al., 2008). RBBP4 interacts with MTA1 for recruiting both itself and RBBP7 into the NuRD complex (Alqarni et al., 2014). The GATA zinc finger domain-containing D2A and D2B (GATAD2A and GATAD2B) proteins are also known as p66α and p66β, respectively. p66 contains two conserved regions - the CR1 and CR2 domains. The CR1 domain interacts with MBD3, and the CR2 domain is necessary for targeting both p66 and MBD3 to specific nuclear loci (Feng et al., 2002). Both p66α and p66β contain sumoylation sites, and mutants for these sites affect p66α-HDAC1 and p66β-RBBP7 interactions in fibroblasts (Gong et al., 2006). LSD1, lysine specific demethylase is one of the newest additions to the NuRD complex and provides histone demethylase activity (Wang et al., 2009).
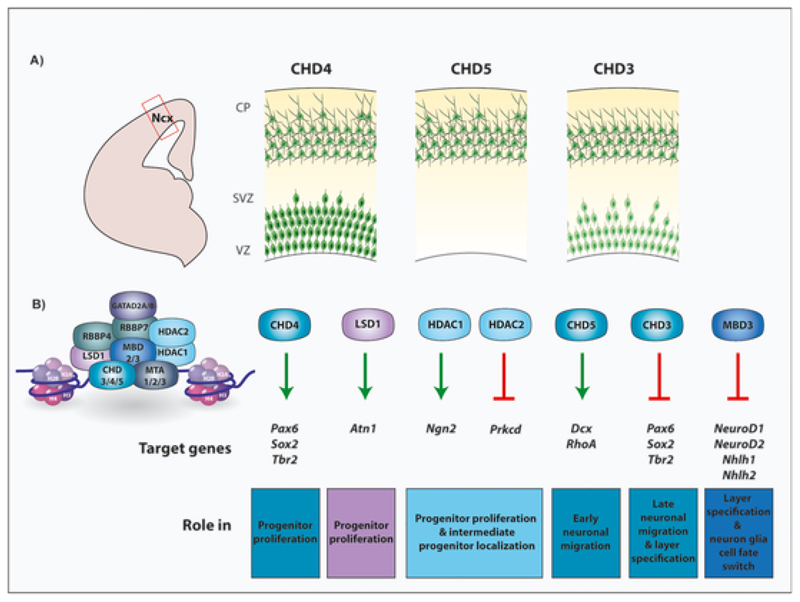
Fig. 2. Members of the NuRD complex mediate different aspects of cortical development.
Diagram illustrating expression pattern of the NuRD ATPases CHD4, CHD5 and CHD3 at the peak of cortical neurogenesis (E15.5) in mice. CHD4 is expressed by the VZ progenitors as well as the post-mitotic neurons of the cortical plate. CHD5 is primarily expressed in the cortical plate. CHD3 exhibits differential expression, with higher expression in the post-mitotic neurons and low expression in the progenitors (top) (Nitarska et al., 2016). Graphical representation of the NuRD subunits, their target genes and their roles in corticogenesis. Deletion of these subunits leads to defects in various steps of corticogenesis. CHD4 positively regulates Pax6, Sox2 and Tbr2 to enhance progenitor proliferation. LSD1 activates its effector ATN1 by binding to the LBAL locus downstream of the Atn1 gene, to maintain the progenitor pool during corticogenesis. HDAC1/2 temporally controls localisation of intermediate progenitors, by activating the HDAC1 target gene Ngn2. CHD5 regulates the expression of neuronal migration genes Dcx and RhoA and promotes early radial migration. CHD3 inhibits the expression of the progenitor genes Pax6, Sox2 and Tbr2 and plays a role in both late migration of neurons as well as layer specification. MBD3 plays a role in both layer specification and neuron-glia cell fate switch. It represses the expression of neurogenic factors Neurod1, Neurod2, Nhlh1 and Nhlh2 at the beginning of gliogenesis (bottom) (LBAL: LSD1 binding site at the Atn1 locus).
2.2. Core constituents of the BAF complex/mSWI/SNF complex
The BAF complexes comprise of up to 15 subunits containing a single catalytic core of either BRM or BRG1 with ATPase activity and exhibit dynamicity in terms of subunit composition at different stages of development in various tissues. BAF45 subunits have PHD fingers which facilitate the interaction with histone tails, wherein BAF45c is known to bind to monomethylated H3K4 and BAF45d interacts with both monomethylated H3K4 and crotonylated H3K14 in vitro (Ho et al., 2019).
BAF47 and BAF250a/b have a role in promoting DNA binding at the regulatory sequences (Allen et al., 2015; Ho et al., 2019).
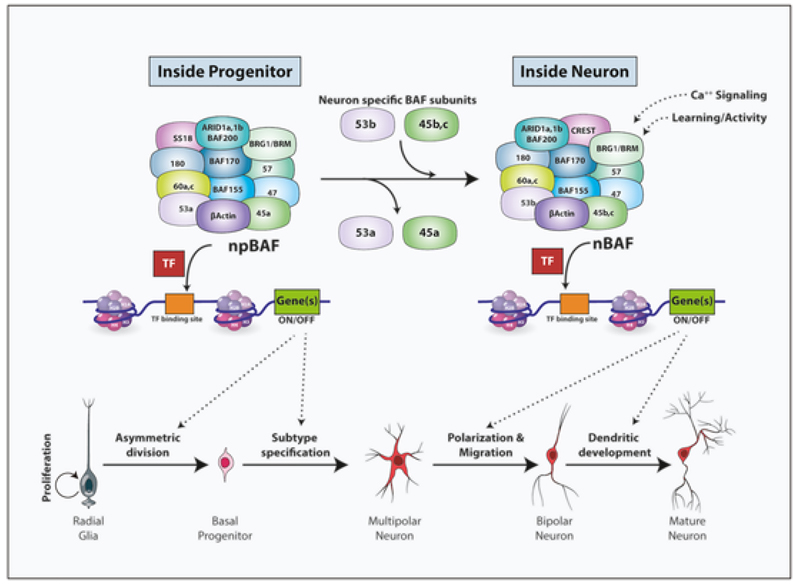
The esBAF (embryonic stem cell BAF), npBAF (neural progenitor-specific BAF) and nBAF (neuron-specific BAF) are the three predominantly studied canonical BAF complexes that play regulatory roles in embryonic development, neural progenitors and neurons respectively (Fig. 3) (Alfert et al., 2019; Ho et al., 2009; Son and Crabtree, 2014).
Fig. 3. Differential roles of progenitor and neuron-specific BAF complexes in neurogenesis.
The BAF complexes in the progenitors (npBAF) and neurons (nBAF) share multiple subunits but display a few stage-specific subunits. The progenitor specific subunits BAF53a, BAF45a and SS18 in the npBAF are exchanged for the neuronal-specific subunits BAF53b, BAF45b/c and CREST in the nBAF. The npBAF complex interacts with transcription factors such as PAX6 to regulate gene expression during symmetric and asymmetric progenitor division and neuronal subtype specification. The nBAF, which acts in neurons controls steps of neuronal maturation including polarisation, migration and dendritogenesis.
Multiple variants of BAF subunits are encoded by gene families (DEAD/H Helicase family), but these are mutually exclusive, and only a single variant from each family is recruited in a complex. BAF180, BAF60, BAF250, BAF47, BCL7, β-actin and BAF155 are a few of the core subunits which are present in both npBAF and nBAF. BAF45a/d, SS18 and BAF53a are specific to npBAF of progenitors and are replaced by BAF45b/c, CREST and BAF53b respectively in nBAF of neurons (Alfert et al., 2019; Son and Crabtree, 2014; Staahl et al., 2013) (Fig. 3).
2.3. Core constituents of the ISWI complex
The mammalian homologues of the ISWI proteins are the SNF2H and the SNF2L chromatin remodelling proteins encoded by the Smarca5 and Smarca1 genes, respectively (Aihara et al., 1998; Lazzaro and Picketts, 2001). ISWI proteins are ATPase motor proteins. The catalytic ATPase activity is at the N -terminal of the ISWI protein, whereas the C-terminal HAND-SANT-SLIDE (HSS) domain mediates its interaction with the linker DNA and the histone H4 tails. The autoregulatory motifs AutoN and NegC-confer specificity (Clapier and Cairns, 2012; Yan et al., 2016). The ATP-mediated activity of the motor proteins of this complex help in the translocation of the DNA along nucleosomes, thereby leading to the formation of equally distanced nucleosomes (Corona et al., 1999; Ito et al., 1997). The SNF2H or the SNF2L are the catalytic subunits of the 8 mammalian ISWI complexes whose compositions vary depending on the interacting partners namely the BAZ (bromodomain adjacent to PHD zinc finger) domain-containing proteins and may also include partner proteins containing AT hooks or DNA-binding homeobox containing proteins and the different transcription and chromatin remodelling factors (DDT) domains for DNA binding (Table 1) (Goodwin and Picketts, 2018).
Table 1.
| ISWI Complex | ISWI Protein | Other partner proteins |
|---|---|---|
| CERF | SNF2L | CECR2 |
| NURF | SNF2L | BPTF, RBBP4, RBBP7 |
| ACF | SNF2H | ACF-1 |
| CHRAC | SNF2H | ACF-1, CHRAC15, CHRAC17 |
| NoRC | SNF2H | TIP5 |
| WICH | SNF2H | WSTF |
| SNF2H-Cohesin | SNF2H | RAD21, SMC1/2/3, SA1/SA2 and NuRD complex proteins |
| RSF | SNF2H | RSF-1 |
The 8 complexes are ACF (ATP-utilising chromatin assembly and remodelling factor), CHRAC (chromatin assembly complex), WICH (WSTF-ISWI chromatin remodelling complex), RSF (remodelling and spacing factor), CERF (CECR2-containing remodelling factor), NoRC (nucleolar remodelling complex), NURF (Nucleosome remodelling factor complex) and SNF2H-cohesin complex (Table 1). Each of the complexes performs a particular function and has been identified in distinct cell types (Goodwin and Picketts, 2018; Manelyte and Langst, 2013).
2.4. Core constituents of the INO80 complex
The INO80/SWR remodeller family consists of 3 complexes, namely INO80, SNF2-related CBP activator protein (SRCAP) and P400/TIP60 (Gerhold and Gasser, 2014). The complexes are named based on the core ATPase subunit, and share the RUVB helicases – RVB1 and RVB2 and actin related proteins (ARPs). The RUVB subunits RVB1 and RVB2 are ATP-dependent helicases, that unwind DNA (Gerhold and Gasser, 2014). Actin and ARPs are involved in histone recognition, with different ARP subunits displaying differential preferences (Willhoft and Wigley, 2020). The ARP4 subunit which is shared between the complexes, interacts with unmodified and phosphorylated H2A, whereas ARP8 of the INO80 complex interacts with H3/H4 tetramers (Willhoft and Wigley, 2020). The Yin Yang 1(YY-1) subunit of INO80 is a transcription factor and helps target INO80 to gene promoters (Cai et al., 2007). KAT5, previously known as TIP60, is a lysine acetyltransferase and transformation/transcription domain-associated protein (TRRAP) is a co-factor of KATs/HATs (Tapias et al., 2014). The UCH37 protein encoded by the UCHL5 gene is a deubiquitinating enzyme (Burgie et al., 2012). Functions of many subunits belonging to this complex family have not been well characterised.
3. Role of the chromatin remodellers in the developing dorsal telencephalon
The interplay of morphogens and TFs determines the specification of the dorsal neural fate in the developing telencephalon, which goes on to form the neocortex and hippocampus (Tole and Hebert, 2013). Loss of critical subunits of the BAF complex affects this crucial initial step of telencephalic specification.
FoxG1-Cre, which is expressed in the entire telencephalon (dorsal and ventral - from E8.5) (Hebert and McConnell, 2000), mediated removal of both BAF155 and BAF170 resulted in complete loss of cortex and head structures regions suggesting a very early and crucial role for BAF complexes in neural fate specification and perhaps the proliferation of NECs (Bachmann et al., 2016; Narayanan et al., 2015; Nguyen et al., 2016). SRG3 – mouse homologue of human BAF155, heterozygote mutants show exencephaly (defect in neural tube closure leading to the brain growing outside the skull) (Kim et al., 2001).
After dorsal telencephalon specification, the various subunits of the remodelling complexes play key roles in several steps of corticogenesis in order to develop a functional neural network (Fig. 1B). Deletion of the subunits affects major steps of cortical development including progenitor proliferation, progenitor maintenance, bIP generation and localisation, differentiation, neuronal subtype specification, migration, neuron-glia cell fate switch and maturation as delineated below.
3.1. Progenitor proliferation, bIP generation and neuron differentiation
All stem cells, namely NECs, aRGs, aIPs, bIPs, bRGs in the developing cortex are capable of proliferation by symmetric divisions. The intermediate progenitors divide symmetrically fewer times than the aRGs. Thus, progenitor proliferation is important to sustain the stem cell pool required to generate the entire cortex and to produce the proper/correct number of neurons. Asymmetric divisions ensure the generation of bIPs from aRGs and differentiation of neurons from bIPs and aRGs.
3.1.1. Members of the NuRD complex
The different members of the NuRD complex which are expressed in the VZ and show similar role in regulation of the PAX6+ aRGs are HDAC1&2, CHD4, LSD1 and MBD3. HDAC1 and HDAC2 are expressed in the VZ progenitors during neurogenesis. HDAC2 also shows enhanced expression in post-mitotic neurons in the cortical plate. CHD4 appears early in development around E12.5 in the VZ, with lower expression around E18.5. LSD1 is ubiquitously expressed in the developing cortex, (Fuentes et al., 2012). MBD3 is expressed in the VZ/SVZ and cortical plate, albeit at lower levels as compared to other NuRD subunits (Sokpor et al., 2018).
Central Nervous system (CNS) specific deletion of either Hdac1 or Hdac2 did not produce any overt brain phenotypes owing to genetic redundancy. Deletion of both resulted in prenatal and postnatal death depending on whether an CNS-specific early (Nestin-Cre) or late acting Cre (hGFAP-Cre) was used (Hagelkruys et al., 2014; Montgomery et al., 2009). The brains were smaller in size, and the cortical plate was reduced in thickness due to decreased proliferation and increased cell death (Hagelkruys et al., 2014). Brain-specific complete deletion of Hdac 1 and a single allele of Hdac2 (Hdac 1-/- and Hdac2 +/-) resulted in no defect. However, deletion of both the Hdac2 alleles and a single allele of Hdac 1 (Hdac 1+/- and Hdac2-/-) resulted in reduced proliferation in the cortex and cerebellum eventually leading to neonatal death. Thus, a single allele of Hdac2 is sufficient to rescue the essential brain development functions of HDAC1 (Hagelkruys et al., 2014). A similar phenotype was observed in heterozygous knock-in mice generated with an amino acid substitution of histidine to alanine (Hdac 1-H141A; Hdac2-H142A) which codes for an inactive protein. The Hdac 1 knock-in mice display normal development, whereas the Hdac2 knock-in mice show severe nervous system defects, including a reduction in the cortical and cerebellar sizes due to a dominant-negative effect. A global increase in the acetylation marks is also observed in the Hdac2 knock-in mice brain consistent with its role as a histone deacetylase (Hagelkruys etal., 2016).
Cortex specific deletion (Emx1-Cre) of Hdac1/2 at the onset of neurogenesis using Emx1-Cre precluded the embryonic and postnatal lethality observed in the other studies and enabled a study of the functions of the two deacetylases specifically in the developing cortical primordium (Tang et al., 2019). The double mutant resulted in a smaller brain and a significant reduction in cortical thickness. The mutant displays a thinner VZ and reduced number of PAX6 + aRGs which are distributed across the entire cortex and later undergo apoptosis (Fig. 4).
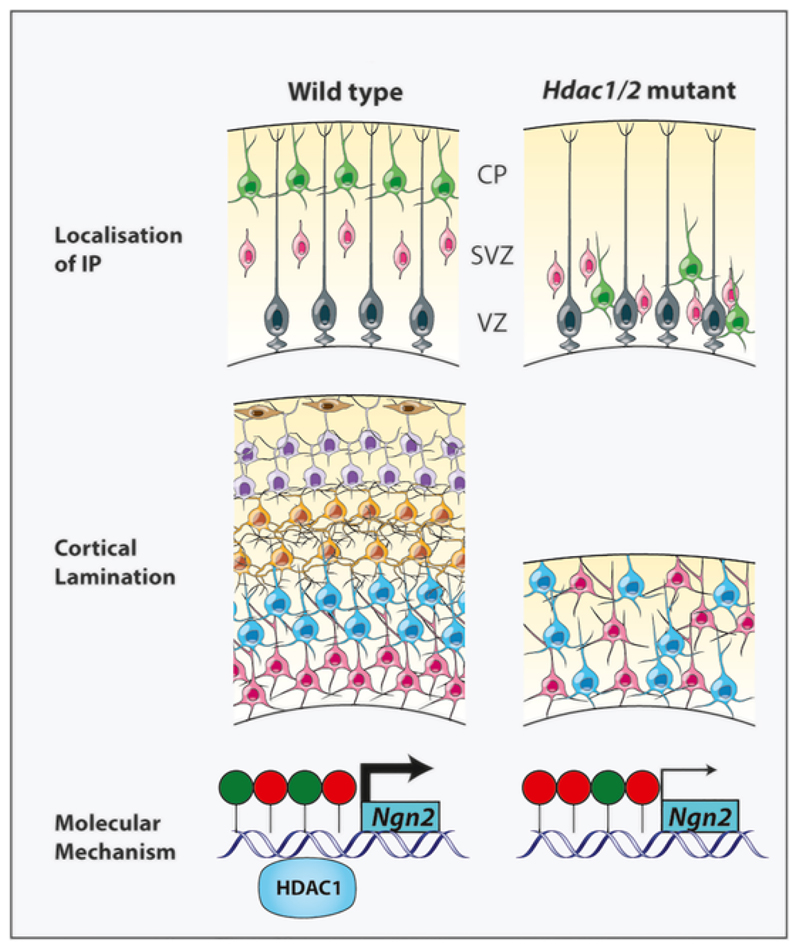
Fig. 4. Regulation of Intermediate progenitor localisation by Hdac1/2 in the cortex.
Deletion of Hdac1/2 at E10.5 leads to accumulation of intermediate progenitors in the cortical VZ (top). Analysis at E16.5 shows that the mutant cortex is thinner, and does not display upper layer neurons (middle). Ngn2 is a target and downstream effector of HDAC1. HDAC1 binds to the Ngn2 promoter, which also displays the H3K9 acetylation (green sphere) and H3K27 trimethylation (red spheres) marks. In the mutant cortex, the H3K9ac mark is reduced, whereas the H2K27me3 mark is increased, thereby repressing NGN2 expression (bottom) (Tang et al., 2019).
Thus, HDAC1 and HDAC2 have partially redundant roles in the proliferation of PAX6+ aRGs in the developing cortex and that HDAC2 has a more specific role in progenitor proliferation.
The intermediate progenitor (IP) cells are generated from apical radial glia and are classified into two subtypes, based on the molecular expression pattern. IPs located in the VZ are bipolar and divide away from the apical surface. IPs localised in the subventricular zone are multipolar and express neuronal differentiation markers such as NEUROD1. Both subtypes express the classical IP marker TBR2 and differentiating IPs produce glutamatergic neurons that occupy all cortical layers (Hevner, 2019).
Besides aRG proliferation, HDAC1/2 temporally regulate the positioning of bIPs in the developing cortex. Emx1 -Cre mediated deletion of HDAC1/2 causes mislocalisation of bIPs to the VZ. These ectopic bIPs undergo apical cell division, display precocious cell cycle exit and are prone to differentiation. Newly postmitotic TBR1 + neurons generated by these mutant IPs also mislocalise in the VZ. The mislocalisation affects cortical lamination at E16.5 as seen by reduced upper layers (UL) neurons and mispositioned deep layer (DL)neurons. HDAC1 occupies the Ngn2 promoter, which has both the H3K9ac active mark and the H3K27me3 repressive mark (Fig. 4). In the mutants, NGN2 expression is diminished coinciding with a reduction in the active mark and an increase in the repressive mark. NGN2 overexpression in HDAC1 and HDAC2 knockdown rescues the ectopic IP localisation, whereas NGN2 knockdown recapitulates the ectopic IP positioning. NGN2 is thus a downstream effector of HDAC1 and HDAC2 in their function of maintaining proper IP localisation. Interestingly, deletion of HDAC1 and HDAC2 post E13.5 still causes reduced cortical thickness but does not affect bIP localisation, suggesting a temporal window for their role in bIP localisation (Tang et al., 2019).
Nestin-Cre mediated deletion of Chd4 results in death at birth in mice. The Chd4 mutant mice have a smaller brain, with reduced cortical thickness. Mutant progenitors precociously exit the cell cycle and fail to differentiate, and later become apoptotic. The number of TBR2+ intermediate progenitors is reduced in the mutants, with decreased proliferation. CHD4 occupies the promoters of Sox2, Pax6 and Tbr2 at higher levels in progenitors as compared to postmitotic neurons, and the expression of these transcription factors is downregulated in the Chd4 mutants, providing a mechanism for its action (Fig. 2B). CHD4 is therefore required for bIP pool maintenance, and Chd4 deletion thus leads to depletion of the bIP pool leading to defective cortical lamination with a specific reduction in the numbers of neurons of the upper layers (Nitarska et al., 2016).
shRNA mediated knockdown of LSD1 in rat cortex or Nestin-Cre mediated deletion in LSD1 conditional mutants leads to precocious differentiation of progenitors into neurons. This defect is due to LSD1’s role in positively regulating the proliferation of aRGs in the ventricular zone. The knockdown brains show fewer SOX2 + and TBR2 + progenitor cells and a corresponding increase in the number of TUJ1 + immature neurons. Further Lsd1 knockdown leads to a migration defect wherein the cells are stuck in the intermediate zone (IZ). LSD1 mediates this function via its direct downstream effector Atrophin1. Interestingly the authors note that H3K4me2 marks increase in the absence of LSD1 on the LBAL (LSD1 binding site at Atrophin1) locus. This seems to be counterintuitive to the reported activatory function of LSD1 which is mediated by the removal of the dimethyl H3K9 instead of the H3K4. The authors conclude that it is perhaps the first report of LSD1 mediated H3K4 methylation leading to gene activation. Atrophin1 (Atn1) is both necessary like LSD1 and sufficient in progenitor proliferation in the aRGs. LSD1 thus maintains the cortical progenitor pool by regulating Atn1 by binding to the LBAL locus (Zhang et al., 2014).
CNS specific deletion of Mbd3 using Nestin-Cre yields a small cortex and a reduction in the number of TBR2 + bIPS. Decreased asymmetric division of aRGs suggests a role for MBD3 in bIPs generation. An opposing function to that of MBD3 in the progenitor comes from Suppressor of Mek null - Smek 1/2. SMEK1/2 polyubiquitinates MBD3 and prevents the repressive binding of MBD3, MTA1, HDAC1/2 (NuRD subunits) to neuronal and proneuronal genes. This leads to progenitors exiting the cell cycle and becoming postmitotic. Smek1/2 null mutants have decreased TUJ1+, same phenotype as overexpression of MBD3. Loss of MBD3 rescued the defective neurogenesis in Smek1/2 mutant. Thus, SMEK1/2 targets MBD3 for degradation at the onset of neurogenesis (Moon et al., 2017).
The Holo-NuRD complex is formed by an interaction of the HDAC and CHD remodeller subcomplexes, mediated by the MBD protein (Basu et al., 2020; Spruijt et al., 2020). MBD3 is expressed in the VZ, SVZ and a subpopulation of post-mitotic neurons (Knock et al., 2015), but is degraded at the onset of neurogenesis (Moon et al., 2017) and is essential for gliogenesis (Knock et al., 2015). MBD2 null mice are viable and show behavioural defects, whereas MBD3 null mice are embryonic lethal (Hendrich et al., 2001). MBD3 perhaps is redundant for MBD2 in the brain, or MBD2 deletion alone is not sufficient to cause a cellular phenotype, indicating the need for a detailed characterisation of MBD2 function in cortical development. All these together suggest that deletion of the MBD2/3 genes should be sufficient to disrupt the Holo-NuRD complex in the neocortex. A complete and comprehensive picture on the function of the NuRD complex could be obtained by deletion of the MBD2/3 genes spatio-temporally in the neocortex, helping identify its functions both in progenitors and post-mitotic cells. Mutants of HDAC1/2, CHD4, LSD1 and MBD3 exhibit a reduction in the number of Tbr2+ bIP cells, suggesting that this cellular phenotype is governed by chromatin remodelling through the NuRD complex. And indeed the functioning of the NuRD complex could be hindered by deletion of the HDAC1/2, MBD3 and CHD4 proteins, as observed by their positioning within the Holo-NuRD complex (Basu et al., 2020). The multiple subunits of the NuRD complex namely HDACs, LSD1, CHDs and RBBPs are known to function individually or as part of other complexes. HDAC1 and HDAC2 are part of the NuRD, SIN3 (switch-independent 3), Mi-DAC (mitotic deacetylase), CoREST (co-repressor of REST), MIER (mesoderm induction early response) and RERE (arginine-glutamic acid dipeptide repeats) complexes (Millard et al., 2017). LSD1 is known to associate with the CoREST complex (Yang et al., 2006). CHDs are known to function both individually as well as part of multimeric complexes like NuRD and ChAHP (CHD4, ADNP, HP1γ) (Nitarska et al., 2016; Tabar et al., 2021). RBBP4 and RBBP7 are a part of both the NuRD complex and NuRF subcomplex of ISWI (Goodwin and Picketts, 2018; Kolla et al., 2015).
Besides, the distinctive developmental and lineage specific expression of the different subunits themselves, it raises the possibility that the complex does not function as holo-NuRD but rather the subunits perhaps come together in a context/site specific manner to dynamically regulate the cortical genome. Sequential ChIP-sequencing will verify the co-occurrence of multiple chromatin regulators on the same gene loci and will clearly be a way forward to understand the function of the NuRD complex in its entirety and its subunits individually.
3.1.2. Members of the BAF complex
The npBAF complex, required to maintain self-renewal and proliferation of neural stem cells, has a different combination of subunits as compared to nBAF that is important for neuron subtype specificity, neuronal maturation, terminal differentiation and dendritic development (Ho and Crabtree, 2010; Sokpor et al., 2017; Son and Crabtree, 2014) (Fig. 3). Both these complexes contain either BRG1 or BRM as the ATPase which is expressed throughout mouse development and in both progenitors and postmitotic neurons (Olave, 2002; Sokpor et al., 2018).
npBAF contains high levels of BAF155 and low levels of BAF170 and vice versa for nBAF (Narayanan et al., 2018; Staahl et al., 2013; Tuoc et al., 2013a, 2013b). As the neurons exit mitosis, subunits BAF53a, BAF45a/d and SS18 of npBAF are switched with BAF53b, BAF45b/c and CREST (Son and Crabtree, 2014; Staahl et al., 2013). The stoichiometry of composition of BAF170 and BAF155 in the two BAF complexes is crucial for modulating the switch between direct and indirect neurogenesis by controlling euchromatin structure (Tuoc et al., 2013a). Consistent with their role, expression of BAF45a and BAF53a is restricted to neural progenitors, while BAF45b, BAF45c and BAF53b are expressed in post-mitotic neurons in the cortex (Lessard et al., 2007). BAF170 and BAF155 are expressed in apical progenitors between E12.5–14.5. At the peak of upper layer neurogenesis (E15.5), they show complementary expression pattern with BAF170 being absent from VZ progenitors and expressed in the cortical plate (Tuoc et al., 2013a).
The mouse mutants for these various subunits divulge the functional role of each of these subunits in maintaining the progenitors in the developing cortical primordium.
Nestin-Cre mediated deletion of BRG1 ATPase subunit leads to a reduced cortex (Lessard et al., 2007; Matsumoto et al., 2006). aRGs show defective proliferation and exit cell cycle to give rise to postmitotic neurons (Lessard et al., 2007).
BRG1 containing npBAF regulates aRG proliferation by activating Notch signalling pathway and repressing Sonic hedgehog signalling pathway by transcriptional regulation of key components of these pathways such as Patched, Olig1, and Jagged1 (Lessard et al., 2007).
BAF45a, BAF53a and SS18 are a part of npBAF and have characterised roles in aRG proliferation. BAF45a and BAF53a are both necessary and sufficient for aRG proliferation, whereas SS18 knockdown in cultures affects the self-renewal capacity of progenitors (Braun et al., 2021; Lessard et al., 2007; Staahl et al., 2013). Mechanistically, BAF53a containing npBAF regulates the chromatin accessibility of genes required for neural progenitor proliferation and cell cycle maintenance by opposing the PRC2 mediated repression. In the absence of BAF53a in these mutants, there is an increase in the levels of PRC2 mediated H3K27me3 repressive marks on cell cycle genes such as Ccdn1, Cdk2 (Braun et al., 2021). The switch from BAF53a to BAF53b in BAF complex in post-mitotic neurons is known to be regulated by miR-9 and miR-124 (Yoo et al., 2009). In progenitors, REST and its co-repressors restrict the expression of these miRNAs that have a binding site in the 3’UTR of Baf53a (Conaco et al., 2006). Upon differentiation, the expression of these miRNA is derepressed in neurons which in turn promote the substitution of BAF53b in nBAF complex by silencing Baf53a expression. Prolonged expression of BAF53a leads to increased progenitor proliferation (Yoo et al., 2009).
For indirect neurogenesis, aRGs undergoes asymmetric division to give rise to bIPs, which then differentiate into neurons. This is a crucial step as it increases the final number of neurons by increasing the progenitor pool and is also considered as the step which contributed towards increased cortical size in primates. BAF complexes regulate this crucial step in corticogenesis. The differential combinatorial stoichiometry of the BAF complex allows for such distinct regulations of the chromatin landscape in the different cells and expression of cell-type specific genes (Wang et al., 1996).
BAF170 and BAF155 have complementary roles in the cortex.
Deletion of BAF170 from E13.5 cortical progenitors leads to increased IPs with no alteration to Pax6 + aRGs and a larger cortex. Overexpression of BAF170 had an opposite effect of reduced IPs and reduction in cortical thickness and volume. Mechanistically BAF170 competes with BAF155 to recruit PAX6/REST repressor to PAX6 target genes Tbr2, Cux1, and Tle1 which regulate the production of bIPs and late cortical progenitors (Tuoc et al., 2013a, 2013b).
Emx1-Cre mediated knockout of BAF155 in cortex leads to increased conversion of aRGs to bRGS by delamination and depletion of bIPs. BAF155 regulates the production of bIPs by positively regulating the expression of PAX6 dependent genes like CEP4 one of the key players of cytoskeleton remodelling and is reported to regulate progenitor delamination (Narayanan et al., 2018).
To analyse the role for BAF170 in regulating bRGs production, the authors overexpressed BAF170 in E14.5 cortex which also leads to in-creased expression of PAX6 and TBR2 + bRGs in the IZ. This phenotype was reverted by the overexpression of both BAF155 and BAF 170 (Narayanan et al., 2018). Thus, BAF155 and BAF170 have opposing roles in bIPs generation and maintenance. BAF155 works in concert with PAX6 whereas BAF170 has an opposite role. These studies delineate how competing chromatin remodeller subunits balance the generation of bIPs population at specific developmental time points in the developing cortex. Very recently, RBM15-RNA-binding motif protein 15 has been shown to regulate the expression of BAF155 in the cortex. RBM15 has been reported to regulate the progenitor delamination (bRGs) by modulating the stability of Baf155 mRNA. METTL3 (methyltransferase-like 3), is a core catalytic component of the N6 - methyladenosine (m6A) RNA methylation machinery, together with RBM15, affect the stability of BAF155. Baf155 mRNAs have three m6A methylation sites, RBM15 activates Baf155 mRNA degradation by promoting m6A methylation. Ablation of RBM15 leads to an increased expression of BAF155, whereas, RBM15 overexpression phenocopies Baf155cKO mutants (Xie et al., 2019).
A cortex specific deletion of BAF155 and BAF170 at E10.5 resulted in a thin cortex with severely affected progenitor proliferation and complete absence of the BAF complex (Narayanan et al., 2015). A global reduction in active chromatin marks H3K9Ac and increase in repressive chromatin marks H3K27me2 and H3K27me3 was seen in the mutants. BAF155 and BAF170 are critical subunits required to maintain the integrity of the BAF complex and the absence of which leads to the dissolution of the complex (Narayanan et al., 2015; Sohn et al., 2007).
Deletion of both BAF155 and BAF 170 at later stages of corticogenesis using hGFAP-cre (E15.5 onwards in the dorsal telencephalon) leads to genome wide increase in H3K4me2 active marks and specific upregulation of proliferation pathways and associated genes and Wnt/β-catenin signaling (Nguyen H et al., 2018). This was confirmed by a concurrent increase in proliferation of PAX6+, SOX2+ aRGs. These mutants also showed suppression of neuronal differentiation related genes seen concomitantly with increased H3K27me3 globally. There was a significantly reduced number of TBR2+ IPs, and late born SATB2 +, BRN2+ neurons in dorsal and lateral pallium of the mutants.
Disruption of npBAF subunits - BAF45a, BAF53a, SS18, BAF155 and BAF170 leads to progenitor proliferation defects.
3.1.3. Members of the ISWI complex
The ISWI ATPases SNF2L and SNF2H are both expressed early during neurogenesis in the developing cortex. As neurogenesis proceeds, the levels of SNF2L and SNF2H increase and decrease respectively. The Snf2l gene also encodes a brain-specific transcript of 3.7 kb, which starts peaking postnatally (Lazzaro and Picketts, 2001). The complementary expression pattern of Snf2h and Snf2l in the developing cortex (Banting et al., 2005, Lazarro and Picketts, 2001) is suggestive of the fact that the chromatin remodelling activities of the two ISWI proteins could be required at particular times in specific regions performing distinct functions in cortical development. Snf2h is abundantly expressed in the cortical progenitors during development whereas Snf2l is expressed in newly postmitotic neurons (Alvarez-Saavedra et al., 2014; Lazzaro and Picketts, 2001).
Interestingly, when the ATP-binding motif of SNF2L was deleted conditionally the resulting mutant (Ex6DEL) showed increased cortical thickness (Yip et al., 2012). The brain to body mass was also increased by 1.4-fold. The increased cortex size was due to increased proliferation of TBR2+ bIPs resulting in changes in cortical lamination and the timing of neurogenesis was also altered. Mechanistically, SNF2L bound to the FoxG1 locus and regulated its expression. Transcription factor, FOXG1 positively regulates basal progenitor expansion in the developing cortex (Kumamoto et al., 2013; Martynoga et al., 2005). FOXG1 was indeed downstream to SNF2L and necessary for IP maintenance as evident by the rescue of the bIP proliferation and the increased brain size phenotype when the Ex6Del mutants were crossed with Foxg1 heterozygous mice to reduce the FOXG1 dosage. Thus, SNF2L represses FoxG1 expression and thereby keeps the bIP numbers in the cortex at adequate levels and regulates the timing of neurogenesis in the cortex (Yip et al., 2012).
Whereas a cortex specific deletion of the Snf2h leads to an opposite phenotype of reduced TBR2+ IPCs and a shrunken cortex. The decreased TBR2+ bIPs resulted in decreased callosal projection neurons leading to partial corpus callosum agenesis. Snf2h depletion leads to altered axonal targeting of the corticothalamic projection neurons and neurons that project across the corpus callosum (Alvarez-Saavedra et al., 2019). This role of Snf2h is consistent with the study where the loss of Snf2h causes proliferation defects observed in the cerebellum granule neuron progenitors (Alvarez-Saavedra et al., 2014).
These results suggest a critical role for Snf2h in maintaining progenitor proliferation and contrasts with the role of Snf2l – the other ISWI mammalian protein thereby suggesting that the two play distinct roles in the developing cortical primordium and merits further detailed analysis on their molecular mechanisms.
BAZ1 or WSTF target genes identified by ChIP-seq from human fetal neural progenitors and neurons derived from them, distinguished its role in both (Lalli et al., 2016). WSTF regulated stem cell renewal genes in the progenitors vs neural differentiation and synaptic genes in the neurons. Further haploinsufficent levels of WSTF dramatically affected the differentiation of the progenitors into neurons and the progenitors remained proliferative. Transcriptomic profiling of these cells also showed an upregulation of mitotic genes alongwith Wnt-activated receptor genes. Thus WSTF function in progenitors is to regulate the balance between proliferation vs neuron generation.
3.1.4. Members of INO80/SWR1 complex
INO80 is expressed in the forebrain from E11.5 with enhanced expression in the cortical VZ and is observed postnatally as well (Elsen et al., 2018; Keil et al., 2020). INO80 deletion from progenitors using Emx1-Cre yields a smaller brain size (Keil et al., 2020). Accumulation of DNAdouble-strand breaks in the progenitors leads to increased apoptosis and loss of aRGs and bIPs in the mutant medial cortex. The cortical lamination is disrupted only in the medial cortex, where Emx1-Cre excises Ino80 specifically from progenitors undergoing symmetric division. The INO80 function in the symmetric progenitors is through its role in DNA repair, but not chromatin remodelling providing an insight into the multifunctional aspects of remodeller subunits. Interestingly, the transcription factor YY1 of the INO80 complex plays an opposite role and is important for neuronal differentiation. Knockdown of YY1 leads to an increase in the progenitors and a concurrent decrease in the number of post-mitotic neurons (Knauss et al., 2018).
MRG15, a subunit of the P400/TIP60 complex, exhibits weak expression in the VZ/SVZ progenitors and enhanced expression in the post-mitotic neurons in the cortical plate at E14.5 (Sokpor et al., 2018). It plays a role in both progenitor proliferation and neuronal differentiation (Chen et al., 2009, 2011). Mrg15 null mice show a thinner neural tube at E10.5 due to increased apoptosis. Mutant neurosphere cultures are smaller in size and show reduced passaging capacity due to proliferation defects. Mutant neurospheres resist differentiation, with an overall reduction in the number of neurons but not glia.
TRRAP is expressed in all cells at E14.5 but displays the highest expression in basal progenitors consistent with its role in progenitors (Telley et al., 2016).
CNS specific mutants of Trrap, another subunit of the P400/TIP60 complex, also show proliferation defects. The mutant brain size and cortical thickness are reduced. Neural progenitors in the mutant show reduced proliferation and precocious differentiation into neurons. Mutant aRGs and bIPs show localisation defects, with a greater number of aRGs differentiating into bIPs and neurons. This phenotype in the mutant is rescued by overexpressing cyclins A2 and B1 (Tapias et al., 2014).
Though the various subunits have overlapping expression patterns in the developing cortical primordium, their roles seem to be in regulating very context-specific aspects of the progenitor proliferation, bIP generation and neuron differentiation thereby highlighting the dynamic nature of the associations leading to temporal control of corticogenesis.
3.2. Neuronal subtype specification
The projection neurons within the cortical layers show diversity in terms of morphology, molecular identity, location, function and synaptic properties (Lodato and Arlotta, 2015). Neuronal subtypes are specified temporally, guided by the spatiotemporal expression of signalling proteins and transcription factors (Molyneaux et al., 2007). Proper specification of neuronal subtypes is essential to form a functional cortex (Mukhtar and Taylor, 2018).
3.2.1. Members of the NuRD complex in neuronal subtype specification
Transcription factors in concert with chromatin remodellers play a crucial role in neuronal subtype specification. In the developing mice brain, Satb2 mutant shows an absence of corpus callosum, thicker anterior commissure, and UL migration and genetic programming defects. SATB2 represses CTIP2 expression in the UL neurons by employing HDAC1 and MTA2 on the matrix attachment region (MAR) for the Ctip2 gene locus. In the absence of SATB2, HDAC1, and MTA2 interaction with the Ctip2 locus is reduced, and CTIP2 is expressed in the UL neurons which now start projecting subcortically (Fig. 5) (Alcamo et al., 2008; Britanova et al., 2008). Ectopic CTIP2 expression in the developing UL neurons is also observed in Ski null mutants. SKI, a transcriptional regulator, interacts with SATB2 and helps recruit HDAC1 to the CTIP2 locus. In the absence of SKI, only MTA2 is recruited to the Ctip2 locus, and SATB2-HDAC1 interaction is reduced (Fig. 5) (Baranek et al., 2012). CTIP2 is thus a downstream regulator of SKI and SATB2, and NuRD subunits are recruited to suppress ectopic CTIP2 expression in the UL neurons (Baranek et al., 2012; Britanova et al., 2008). Postnatally, there is an increase in the number of CTIP2 and SATB2 co-expressing neurons in the somatosensory cortex. The protein LMO4 competes with SATB2 for HDAC1 in postnatal neurons, thus disrupting the NuRD complex and de-repressing CTIP2 expression (Harb et al., 2016).
Fig. 5. Interaction between transcription factors and NuRD.
In cortical progenitors, LHX2 putatively recruits HDAC2, LSD1 and RBBP4 and represses FEZF2 and SOX11 expression by removing active marks from their gene loci. The sub-cerebral projection neuron identity of layer V is hence suppressed during layer VI formation. In the absence of LHX2, the NuRD subunits possibly cannot interact with the gene loci, thus retaining the active marks and precocious formation of layer V sub-cerebral projection neurons (Muralidharan et al., 2017). In post-mitotic neurons, transcription factor SATB2 and transcription regulator SKI repress CTIP2 expression in callosal projection neurons by recruiting HDAC1 and MTA2 to the matrix attachment region (MAR) upstream of the Ctip2 gene locus. In the Ski mutant, HDAC1 is not recruited to the MAR locus whereas, in the Satb2 mutant, the SKI-NuRD complex is not recruited to the MAR locus. CTIP2 expression is thus facilitated in the Ski and Satb2 mutants, providing a subcerebral projection identity to the neurons (Britanova et al., 2008; Baranek et al., 2012).
Several transcription factors have been characterised, and well-studied as postmitotic determinants of neuronal subtype specification namely TBR1 (Bedogni et al., 2010; Han et al., 2011; McKenna et al., 2011), SOX5 (Kwan et al., 2008; Lai et al., 2008), SOX4 and SOX11 (Shim et al., 2012) and FEZF2 (McKenna et al., 2011), SATB2 (Britanova et al., 2008), BRN1 and BRN2 (Dominguez et al., 2013; Sugitani et al., 2002), but a progenitor specific mechanism was not known. LHX2 is expressed in the progenitors at the time of deep layer neurogenesis but is not detected in the postmitotic DL neurons. Postnatally LHX2 expression comes up in the UL neurons but not in deep layers suggesting a possible role in deep layer neurogenesis. Cortex-specific deletion of Lhx2 using the Emx1-Cre line alters the DL cortical lamination. Mutant cortices exhibit an expansion of layer V at the expense of layer VI, coinciding with an increase and decrease in the number of CTIP+ and TBR1+ neurons, respectively. The expression of layer V markers FEZF2 and SOX11 is enhanced in these mutants within a day of LHX2 removal, with an increase in the active marks H3K4me3 and H3K9ac at the transcription start sites or LHX2 binding regions on the Fezf2 and Sox11 gene loci. LHX2 acts by interacting with NuRD subunits HDAC2, RBBP4 and LSD1, which occupy distal LHX2 binding regions and transcription start sites of the Fezf2 and Sox11 gene loci and repress their expression during layer VI specification (Figure) (Muralidharan et al., 2017).
A similar progenitor specific mechanism was shown in the CNS specific Mbd3 deletion mutants exhibiting disrupted cortical lamination. The CTIP2+ CSMNs is expanded in the mutant cortex, which also exhibits intermingling of TBR1, CTIP2 and SATB2 positive cells, with no distinction in the layers (Knock et al., 2015).
In-depth mechanistic analysis exists for few factors where a combination of biochemical techniques combined with genome-wide binding and mouse genetics have established a clear connection starting with gene regulation to phenotypic outcomes. The above studies have elegantly delineated the role of transcription factors and chromatin remodellers on specific gene loci to make epigenetic changes leading to altered gene expression. Cortical lamination defects as seen in the mutants above signify the importance of chromatin remodellers in fine tuning gene expression during cortical development. Further studies to dissect the role of these remodellers in controlling gene expression via epigenetic changes would enhance our understanding of cortical development.
3.2.2. Members of the BAF complex in neuronal subtype specification
Close relatives, Ctip1 and Ctip2 have complementary expression pattern and roles in the developing cortex.
CTIP2/BCL11b/BAF100b is highly expressed in subcerebral projection neurons and cortical spinal motor neurons of layer V (Wiegreffe et al., 2015; Leid et al., 2004) and forms a part of l5-nBAF complex (Narayanan and Tuoc, 2014). Ctip2 null mice fail to project corticospinal neurons to the spinal cord (Arlotta et al., 2005). FEZF2 is critical for the formation of sub-cerebral projection neurons (Molyneaux et al., 2005) and CTIP2 is a downstream effector of FEZF2 in guiding the axonal tracts to sub-cortical regions (Chen et al., 2008). As previously discussed, its expression is regulated by NuRD subunits, giving an example of how these remodellers perhaps cross-regulate each other.
Whereas CTIP1/BAF100a/BCL11a, highly expressed in Layer VI–CThPN and Layers II-IV, plays a cell-autonomous role in establishing somatosensory area identity and projection neuron subtype identity in developing cortex. Emx1-Cre mediated deletion of Baf100a showed an abnormal expansion of subcerebral projection neurons at the expense corticothalamic and deep-layer callosal neurons resulting in the formation of an expanded layer V (Woodworth et al., 2016). It controls somatosensory area patterning by suppressing the expression of motor specific genes in sensory areas (Greig et al., 2016). Ctip1 depletion results in abnormal differentiation of layer IV neurons and causes a subset of them to project to motor thalamic nuclei instead of sensory thalamic nuclei. Further Ctip1 deletion affects the differentiation of Layer IV granule neurons which affects the integration of thalamocortical inputs to form proper barrel fields. Thus, CTIP1 is a crucial post-mitotic determinant of area identity in the cortex and ensures the specification of somatosensory cortex.
3.3. Neuronal migration
Newborn neurons generated in the ventricular and subventricular zones migrate radially towards the basal surface to occupy their positions in the cortical layers. Early born neurons migrate by somal translocation, whereas the late-born neurons migrate by multipolar migration up to the IZ. These multipolar cells then become bipolar and migrate further guided by radial glia fibres. As the cortex is formed in an inside-out fashion, late-born neurons migrate past the early-born neurons. This neuronal migration is essential for proper cortical lamination, and defects can lead to cortical malformations (Buchsbaum and Cappello, 2019).
CHD3 and CHD5 display enhanced expression post E15.5 with elevated levels in the cortical plate (Knockdown of CHD5 at E13.5 leads to accumulation of cells in the IZ, whereas a majority of these cells reach the cortical plate in control. The mutant cells fail to transition into bipolar cells and remain multipolar but express the UL markers SATB2 and CUX1, forming an ectopic layer below the cortical plate. CHD5 occupies the promoters of RhoA and Dcx, which are known to regulate neuronal migration and mutant cells exhibit reduced levels of both. Knockdown of CHD3 at E13.5 leads to retention of cells in the deep layers, with multipolar identity. A majority of these mutant cells expressed TBR1 and SOX5, and only a few expressed UL markers BRN2 and CUX1. CHD3 shows higher occupancy on Sox2, Pax6 and Tbr2 promoters in post-mitotic neurons as compared to neural progenitors, and acts to repress them. Ectopic early expression of CHD3 at E13.5 also causes downregulation of protein expression. CHD5 is thus essential for radial migration, whereas CHD3 is necessary for both radial migration as well as layer specification (Fig. 2) (Nitarska et al., 2016). CHD3 is directly activated by TBR1 and TBR2, which supports its expression and function later in neuronal development (Elsen et al., 2018).
A contrasting study by Egan et al., 2013 showed that CHD5 knockdown at E14.5 leads to progenitor accumulation in the VZ/SVZ as these cells fail to undergo terminal differentiation (Egan et al., 2013).
The HDAC2 protein contains two cysteine residues which can be S-nitrosylated on treatment with BDNF or S-nitrocysteine, thus promoting chromatin remodelling. Mutations of the cysteine residues prevent HDAC2 S-nitrosylation (Nott et al., 2008). Electroporation of E14.5 mice cortices with mutated Hdac2 leads to accumulation of cells in the IZ, whereas these cells reach the cortical plate in the controls. The BAF complex protein BRM is a target of S-nitrosylated HDAC2, and its expression is reduced in mutant HDAC2 cells. Overexpression of BRM in mutant cells rescues the neuronal migration phenotype. S-nitrosylation of HDAC2 is hence necessary for neuronal migration and the BAF complex subunit BRM is its downstream regulator. This study thus identified the interaction between subunits of 2 different remodelling complexes – BAF and NuRD (Nott et al., 2013).
Deletion of Ctip1 at E14.5 leads to delayed migration of UL neurons and the mutant neurons remain multipolar. UL neurons exhibit specific molecular markers but impaired dendritic morphology. These mutant neurons undergo apoptosis postnatally, giving rise to a thinner cortex and CC. CTIP1 acts by directly repressing the signalling molecule SE-MA3C. Overexpression of SEMA3C recapitulates the Ctip1 mutant phenotype and knockdown in Ctip1 mutants rescues the impaired migration (Wiegreffe et al., 2015).
3.4. Neuritogenesis
Neuritogenesis and synapse formation are the building blocks to form a mature neural circuitry. Dendritic size and topology both affect firing (van Elburg et al., 2010).
NuRD complex subunits are expressed throughout development, and thus their roles extend to regulate maturation of neurons (Nott et al., 2008).
LSD1 has four known isoforms – LSD1, LSD1-2a, LSD1-2a/8a and LSD1-8a, wherein the 8a isoforms are neuron-specific (Zibetti et al., 2010).
Knockdown of the neuron-specific LSD1 isoforms in rat cortical cultures causes defects in neuritogenesis, with a reduced number of neurite branches, width and length. Overexpression of the neuro-specific LSD1 isoforms has the opposite effect with increased neuritogenesis (Zibetti et al., 2010). The phosphorylated amino acid threonine within the neuron-specific 8a exon of Lsd1 is necessary for neuritogenesis. A phosphodefective mutant phenocopied Lsd1 knockdown phenotype, whereas a demethylase activity inactivating mutant rescued the phenotype in a phospho-defective mutant (Toffolo et al., 2014). The neuro-specific LSD1 isoforms are crucial for neuronal maturation.
S-Nitrosylation of HDAC2 promotes its dissociation from the chromatin, thus facilitating the acetylation of histones H3 and H4. Disrupting this nitrosylation by mutating the two cysteine residues leads to a reduction in the total and average dendritic length in cortical cultures. Short interfering RNA mediated silencing of HDAC2 causes the opposite effect with an increase in the average dendritic length and number of branch points per neuron. These dendritic phenotypes are rescued in the presence of wildtype HDAC2. Thus, HDAC2 S-nitrosylation plays a role in dendritic length and branching (Nott et al., 2008).
Early on cell-intrinsic mechanism drive corticogenesis and later stages of development, namely migration, dendritic maturation are both driven by cell-intrinsic and extrinsic input activity cues. Calcium signalling plays a significant role in dendritic maturation. The down-stream trans-activator of the Calcium signalling is Calcium responsive trans-activator (CREST) and is both a necessary and sufficient regulator of this process.
CREST expression is detected around E18.5 in post-mitotic neurons of the developing cortex, peaks at postnatal P1 and starts to decline after P10 (Aizawa et al., 2004). CREST regulates dendritic architecture and branching both during pre-natal (Staahl et al., 2013) and postnatal development (Aizawa et al., 2004). Absence of CREST and BAF53b impairs calcium-dependent dendritic outgrowth and branching in the cortex (Aizawa et al., 2004; Wu et al., 2007). Mechanistically, BAF53b helps in recruiting nBAF containing CREST to the promoters of key players in axon growth such as Ephexin1, Gap43. Moreover, knocking down other neuron-specific subunits of nBAF BRG, BAF57, or BAF45b had similar defects in dendritic morphogenesis (Wu et al., 2007). These highlight the importance of the combinatorial chemistry of the nBAF complex in regulating dendritogenesis.
ARID1B (BAF250b) knockdown leads to dendritic arborization defects in the mice cortex (Ka et al., 2016). shRNA mediated knockdown of Arid1b leads to both apical and basal dendritic defects. Apical dendritic length and number are reduced, with fewer dendrites reaching the pial surface. Basal dendrites in the knockdown are shorter and extend only laterally, as opposed to a lateral and towards VZ orientation of control neurons. The number and length of spines are reduced, leading to impaired excitatory and inhibitory functions. The expression of CFOS, ARC and neurite associated genes (Stmn22, Gprin11 and Gap43) is reduced in the knockdown, and overexpression of C-FOS or ARC partially rescues the apical dendritic phenotype.
3.5. Glial fate specification
Astrocytes are essential for neural circuit formation, and play key roles in synaptogenesis (Fossati et al., 2020). Oligodendrocytes are myelinating cells that help propagate action potentials through saltatory conduction (Freeman and Rowitch, 2013; Parras et al., 2020).
Neurogenic mechanisms are actively repressed during gliogenic phase to prevent prolonged neuronal generation in the developing cortex (Hirabayashi and Gotoh, 2010; Miller and Gauthier, 2007; Yao et al., 2016).
MBD3 plays a crucial role in the neuron-glia cell fate switch. Nestin- Cre mediated deletion of Mbd3 leads to enhanced and prolonged-expression of the neurogenic factors NEUROD1, NEUROD2, NHLH1 and NHLH2 in the mutant cortex. Concurrently, the expression of the glial markers GFAP, NEUROD4 and S100B is significantly reduced. MBD3 occupies the gene loci of all four factors and potentially acts by repressing the expression of these genes at the onset of gliogenesis (Knock et al., 2015). MBD3 knockdown cultures also causes an increase and a corresponding decrease in the number of TUJ1+ neurons and GFAP + astrocytes, respectively. A similar reduction in GFAP + glial cells lining the VZ is recapitulated in tamoxifen driven Emx1-Cre mediated deletion of MBD3 in mice. MBD3 occupies gene loci of the neurogenic genes Neurog1, Fezf2, Lef1 and Tcfap2c in gliogenic NPCs, but not in neurogenic NPCs. Deletion of Mbd3 before the gliogenic phase in cortical cultures, leads to enhanced expression of FEZF2 and TCFAP2C and a reduced expression of GFAP (Tsuboi et al., 2018). Thus, MBD3 functions to keep neurogenic genes repressed in gliogenic NPCs to ensure glia production.
In the adult brain, HDAC1 is primarily expressed in glial cells, and HDAC2 is expressed in the neurons (MacDonald and Roskams, 2008; Sokpor et al., 2018).
HDAC1 and HDAC2 play opposing roles in oligodendrogenesis. Knockdown of HDAC1 or HDAC2 in oligodendrocyte precursor cells causes reduced and increased formation of oligodendrocytes, respectively (Egawa et al., 2019). HDAC2 is essential for preventing precocious oligodendrocyte differentiation and acts by inhibiting expression of SOX10, which is required for terminal differentiation (Castelo-Branco et al., 2014). Hdac1/2 knockout in OLIG1 expressing cells leads to failure in oligodendrocyte differentiation. In the absence of HDAC1/2, the Wnt signalling protein B-Catenin sequesters the oligodendrogenesis protein TCF7L2 (Ye et al., 2009).
4. Role of chromatin remodellers in neurodevelopmental disorders (NDDs)
Neurodevelopmental Disorders (NDDs) are complex conditions arising due to defects in nervous system development. These conditions affect major aspects of brain function, including speech, cognition, motor skills, language and behaviour (Celen et al., 2017; Homma et al., 2019; Kruszka et al., 2019; Lalli et al., 2016). Childhood NDDs (diagnosis in infancy or childhood) include Intellectual disability (ID), Autism spectrum disorder (ASD), attention-deficit/hyperactivity disorder (ADHD) whereas adolescent NDDs (diagnosis in adolescent age) include Schizophrenia and bipolar disorder (Morris-Rosendahl and Crocq, 2020; Stein et al., 2020). The emerging hypothesis in the field is that these conditions which show a wide spectrum of symptoms share genetic and environmental risks factors which lead to overlapping disease mechanisms affecting brain development (Owen et al., 2011; Singh et al., 2017). A comprehensive list of human NDDs caused by mutations in the chromatin remodeller complex subunits with their clinical features is summarised in Table 2. Chromatin regulation plays a critical role and is a fundamental determinant of the generation of the cerebral cortex (Telley et al., 2019). Indeed the long list of mutations in chromatin remodellers comes as no surprise and suggests that chromatin remodelling seems to be at the root of these disorders. Genetic mutations in the remodellers can have a functional impact on their protein activity, which then leads to a cellular impact on neural function, leading to the formation of a dysfunctional neural circuitry ultimately affecting human be-haviour. In this section we discuss known mechanisms of chromatin dysregulation caused by mutations in the subunits of the 4 families of ATP dependent chromatin remodelling complexes followed by a description of overlapping cellular/behavioural phenotypes leading to NDDs.
Table 2.
| Family | Subunit | Syndrome/NDD | Mutations | Clinical brain Phenotypes | References | |
|---|---|---|---|---|---|---|
| Gene | Protein | |||||
| NuRD | CHD3 | CHD3 | Snyders Blok-Campeau Syndrome | Missense, nonsense, frameshift, deletion, splice-site variant | ID, DD, delayed myelination, macrocephaly/microcephaly | Drivas et al. (2020); Snijders Blok et al., 2018 |
| CHD4 | CHD4 | Sifrim-Hitz-Weiss syndrome (SIHIWES) | Missense, in-frame deletions, nonsense | ID, DD, macrocephaly, hydrocephalus, thin corpus callosum, | Farnung et al. (2020); Weiss et al. (2020) | |
| CHD5 | CHD5 | Autism-like | Missense | DD, cerebral palsy, speech impairment | http://www.mygene2.org [03 October 2020 accessed] | |
| MBD2/3 | MBD2/3 | Autism | SNPs | Cukier et al. (2009); Li et al. (2005) | ||
| HDAC1/2 | HDAC1/2 | Schizophrenia, Angelman syndrome | – | ID, DD | Jamal et al. (2017); Schroeder et al. (2017); Sharma et al. (2008) | |
| GATAD2B | GATAD2B | GATAD2B-associated neurodevelopmental disorder (GAND) | Deletion, frameshift, nonsense, splice-site, missense | ID, DD, macrocephaly, thin corpus callosum, widened CSF spaces, hypomyelination | de Ligt et al. (2012); Hamdan et al. (2014); Luo et al, 2017; Shieh et al. (2020); Ueda et al. (2019); Willemsen et al. (2013) | |
| KDM1A | KDM1A | Kabuki and KBG-like syndrome | Missense, nonsense, splice-variant, nonsense | ID, DD, thin corpus callosum, delayed myelination, cerebellar defects | Chong et al., 2015; Rauch et al. (2012); Tunovic et al. (2014) | |
| BAF | ARIDla | BAF250A | Coffin Siris Syndrome | Nonsense, frameshift indel, missense | CNS structural abnormalities | Bramswig et al. (2017); Tsurusaki et al. (2012); Wieczorek et al. (2013) |
| SMARCA4 | BRG1 | |||||
| SMARCE1 | BAF57 | |||||
| ARID2 | BAF200 | |||||
| SMARCA2 | BRM | Coffin Siris syndrome, Nicolaides-Baraitser syndrome, Schizophrenia | Partial deletion, missense, intronic alteration, duplication | CNS structural abnormalities, ID, microcephaly, behavioural problems and seizures, delusions, thought disorder, cognitive dysfunction | Ejaz et al. (2016); Miyake et al., 2014; Sousa et al. (2014); Wolff et al., 2012 | |
| SMARCB1 | BAF47 | Coffin Siris Syndrome, Kleefstra syndrome | In-frame deletion, missense | CNS structural abnormalities, ID, behavioural anomalies | Kleefstra et al., 2012; Santen et al., 2013; Tsurusaki et al. (2012); Wieczorek et al., 2013 | |
| ARIDlb | BAF250B | Coffin Siris Syndrome, ID, Hirschsprung’s disease, Schizophrenia, Autism, 6q25 Microdeletion Syndrome | Missense, nonsense, frameshift indel, translocation, microdeletion, interstitial deletion | CNS structural abnormalities, developmental delay, epilepsy, and hypoplasia of the corpus callosum and cerebellum, delusions, thought disorder, cognitive dysfunction, defects in social and communication skills, restricted and repetitive behaviour, corpus callosum agenesis | Backx et al. (2011); Halgren et al. (2012); Santen et al., 2013; Takenouchi et al. (2016), Ronzoni et al. (2016) | |
| SMARCD1 | BAF60 | Coffin Siris Syndrome, Nicolaides-Baraitser syndrome | Missense, nonsense | CNS structural abnormalities, ID, microcephaly, behavioural problems and seizures | Machol et al. (2019); Nixon et al., 2019 | |
| SMARCC1 | BAF155 | Autism | Missense | Defects in social and communication skills, restricted and repetitive behaviour | Neale et al. (2013); O’Roak et al. (2012) | |
| PBRM | BAF180a | Autism | Missense | |||
| ACTL6b | BAF53b | Autism | Missense | Defects in social and communication skills, restricted and repetitive behaviour | Bell et al. (2019); Wenderski et al. (2020) | |
| ACTL6b | BAF53b | DECAM syndrome | – | Developmental delay, Epileptic encephalopathy, Cerebral atrophy and abnormal Myelination | Bell et al. (2019); Yuksel et al. (2019) | |
| BCL11a | BAF100a | Autism, Schizophrenia, p15–16.1 Microdeletion syndrome, Intellectual Disability | Microdeletion, missense, frameshift | Defects in social and communication skills, restricted and repetitive behaviour, delusions, thought disorder, cognitive dysfunction, developmental delay, hearing and visual impairment and corpus callosum agenesis | Basak et al. (2015); Bagheri et al. (2016); Chen et al. (2020); De Rubeis et al. (2014), Korenke et al. (2020) | |
| SMARCC2 | BAF170 | Coffin Siris Syndrome, Nicolaides-Baraitser syndrome like, Autism | Splice site mutation | CNS structural abnormalities, ID, microcephaly, behavioural problems and seizures, defects in social and communication skills, restricted and repetitive behaviour | Machol et al. (2019) | |
| ISWI | SMARCA1 | SNF2L | Coffin-Siris like syndrome; Schizophrenia | Nonsense, missense | ID, microcephaly with cortical atrophy, spasticity, dysmorphic features | Homann et al. (2016); Karaca et al. (2015) |
| BAZ1A | ACF-1 | Intellectual Disability | Missense | ID, corpus callosum agenesis | Zaghlool et al. (2016) | |
| BAZ1B | WSTF | Williams Syndrome | Microdeletion | Anxiety, ADHD | Lalli et al. (2016) | |
| BPTF | BPTF | Unnamed syndrome; Silver-Russell syndrome | Nonsense, missense, frameshift, CNV deletion, splice-variant | ID, DD, microcephaly, dysmorphic facial features; pre- and postnatal growth retardation | Stankiewicz et al. (2017); Wakeling et al. (2017) | |
| RAD21 | RAD21 | Cornelia de Lange (CdL) | LOF | Microcephaly, holoprosencephaly | Goel 2020; Kruszka et al. (2019) | |
| SMC1 | SMC1 | Cornelia de Lange syndrome and holoprosencephaly | Nonsense, missense | Holoprosencephaly | Kruszka et al. (2019) | |
| SMC3 | SMC3 | ID and holoprosencephaly | Missense, nonsense, in-frame deletion/duplication | Holoprosencephaly | Kruszka et al. (2019) | |
| STAG2 | SA2 | STAG2-related X-linked Intellectual Deficiency and holoprosencephaly | LOF | Moderate ID, language and hearing defects, developmental delay, microcephaly, characteristic facial features, holoprosencephaly | Kruszka et al. (2019); Soardi et al. (2017) | |
| INO80/SWR1 | INO80 | Intellectual Disability and Global developmental delay | – | ID, DD, microcephaly | Alazami et al. (2015) | |
| SRCAP | Floating Harbor Syndrome | Nonsense | ID, ADHD, speech delay, cranio-facial features | Greenberg et al. (2019); Homma et al. (2019); Zhang et al. (2019) | ||
| TIP60/KAT5 | Unnamed syndrome | Missense | ID, DD, corpus callosum agenesis/dysgenesis, characteristic facial features | Humbert et al. (2020) | ||
| Other CHDs | CHD1 | CHD1 | Pilarowski-Bj ornsson Syndrome, 5q15-q21.2 deletion, ASD | Missense, deletion | Speech apraxia, DD, autism, craniofacial dysmorphism, motor delay | Neale et al. (2013); Pilarowski et al. (2018); Zepeda-Mendoza et al., 2019 |
| CHD2 | CHD2 | Developmental Epileptic Encephalopathy, Epilepsy with myoclonicatonic epilepsy, ASD | Nonsense, missense, deletion | Photosensitivity, myoclonic seizures, ID | Angione et al. (2019); Carvill et al. (2013); Neale et al. (2013); Trivisano et al. (2015) | |
| CHD7 | CHD7 | CHARGE Syndrome, ASD | Missense, nonsense, frameshift, microdeletions, translocation, intronic SNV | Developmental delay, hearing impairment, defects in social and communication skills, restricted and repetitive behaviour | Johnson et al. (2006); O’Roak et al., 2012; Vissers et al. (2004); Zhang et al. (2020) | |
| CHD8 | CHD8 | 14q11.2 microdeletion, Zahir Friedman syndrome, Schizophrenia | de novo deletion, Microdeletion, CNVs, missense, frameshift, nonsense, single amino acid deletions, duplications, point mutations | Developmental delay, cognitive impairment, dysmorphic features, ASD/DD/ID, macrocephaly, speech defects, defects in social and communication skills, restricted and repetitive behaviour, hallucinations, persecutory delutions and catatonic behaviors | Bernier et al. (2014); Kimura et al., (2016); McCarthy et al., (2014); Merner et al., 2016; Stessman et al. (2017); Talkowski et al., 2012; Yasin et al. (2019); Yasin et al., 2020; Zahir et al., 2007 | |
The mutations in these subunit genes can disrupt protein function in the following ways:
4.1. Disruption of protein binding to histones or DNA
Sifrim-Hitz-Weiss syndrome (SIHIWES) is caused by mutations within the CHD4 gene locus. Variants located within the PHD and chromodomains affect both the ATPase and nucleosome remodelling activities of CHD4 (Weiss et al., 2020). A protein modelling study showed that a few of these mutations affect the binding of CHD4 to H3 or DNA, or disrupt binding of Zn2+ to CHD4, thus affecting its activity (Farnung et al., 2020). CHD4 is important for maintenance of bIP pool (Nitarska et al., 2016) and mutations could possibly lead to depletion of the progenitor pool and perhaps be causative of the thin corpus callosum seen in the patients.
4.2. Disruption of protein-protein interaction
GATAD2B-associated neurodevelopmental disorder (GAND) is caused due to mutations in the GATAD2B gene. The GATAD2B protein contains 2 conserved regions termed CR1 and CR2, and all identified missense mutations lie within these regions. Mutations located in the CR1 and CR2 domain impaired the ability of GATAD2B to interact with MBD2/MBD3 or CHDs respectively (Shieh et al., 2020).
4.3. Truncated/catalytically impaired enzyme activity
Patients with de novo mutations in LSD1 exhibit ID and DD, with Kabuki and KBG like syndrome (Chong et al., 2015). Biochemical analysis of three variants – E379K, D556G and Y761H showed that the protein is structurally stable, but enzymatically impaired coupled to a reduced half-life (Pilotto et al., 2016). Chromosome translocation mutation in ARID1B is implicated in a patient with ID and corpus callosum agenesis. De novo translocation between chromosome chr4 (q25.3) and chr14 (q13.2) results in formation of a fusion transcript of ARID1B and MRPP3. This fusion transcript is suggested to contribute to disease phenotypes likely by haploinsufficiency (Backx et al., 2011).
Further studies on downstream targets which are mis-regulated due to these mutations and alteration in protein activity will reveal how these mutations affect neurobiological pathways important for cortical development. This will be crucial to establish a cause-and-effect relationship between the mutations and the altered cellular neural development outcomes leading to clinical symptoms in patients with NDDs.
4.3.1. Altered cellular and behavioural phenotypes –
The various steps of neural development are critical for the formation of a functional neural circuitry leading to behaviour. Defects in the neural development processes as outlined below could alter the behavioural outcome manifesting as a clinical disease symptom in patients with NDDs.
Progenitor proliferation defect-Williams syndrome is caused due to microdeletion of ~28 genes within chromosome 7, including BAZ1B which encodes the WSTF protein of the ISWI complex. ChIP-seq analysis of genes bound by WSTF in human NSCs showed occupancy on genes implicated in self-renewal and neurogenesis. Transcriptomic analysis of WSTF haploinsufficient neuronal cultures showed bound genes to be misregulated and mirror the transcriptomic profiling from patients with Williams syndrome containing the microdeletion including overactivation of WNT-activated receptor genes. Knockdown of BAZ1B inhibits neural progenitor differentiation and cells remain proliferative. Thus, WSTF is crucial for regulating the progenitor proliferation and its haploinsufficiency changes the balance between progenitor proliferation and differentiation leading to reduced neuronal output (Lalli et al., 2016).
Neuronal cell death-Mutations in the BPTF gene cause a yet unnamed syndrome. ID, DD, speech, and motor delay have been reported in these patients (Table 2). Zebrafish bptf mutant embryos exhibit a smaller head size, and increased apoptosis. These mutants also showed craniofacial patterning defects, with an increase in the angle of the ceratohyal cartilage. The mutants further showed increased apoptosis but not changed proliferation suggesting that microcephaly could possibly be because of increased neuronal cell death (Stankiewicz et al., 2017).
Dendritic impairments-De novo mutation in the CHD5 gene locus has been reported on MyGene2 website in a (http://www.mygene2.org [03 October 2020 accessed]). Genes implicated in ASD are affected in Chd5 null mice, and in vitro cortical neurons exhibit dendritic impairments. These mice are neophobic - prefer familiar spaces and objects, and show social communication defects. These correlate with known autistic behaviours (Pisansky et al., 2017).
Recessive mutations in ACTL6B (BAF53b) have been implicated as a cause of autism. Patient iPSCs showed that ACTL6B is not included in the nBAF complex due to protein destability. Patients display CC hypoplasia and a reduction in white matter, which is recapitulated in a null mutant of Actl6b. These null mutants also exhibit characteristic autistic behaviours of hyperactivity and social interaction defects with memory impairment. The non-neuronal isoform ACTL6A is recruited in the mutant nBAF, and chromatin-complex interactions are impaired. Patient mutations in ACTL6B rerouted dendrites in the fly olfactory nervous system (Wenderski et al., 2020). ACTL6B regulates dendritic morphogenesis in cortical development (Aizawa et al., 2004; Wu et al., 2007) and thus the autistic behaviours could be a result of the impaired dendritogenesis, CC hypoplasia and reduction in white matter.
Mutations in ACTL6B (BAF53b) have been identified to cause Developmental delay, epileptic encephalopathy, Cerebral atrophy and abnormal Myelination (DECAM) syndrome (Yuksel et al., 2019). Loss of dendrites and a large nucleus are observed in both ACTL6B knockout in control iPSC derived neuronal cultures and patient iPSC derived neuronal cultures with mutations in ACTL6B (Bell et al., 2019)).
Mutations in ARID1b are frequently implicated in autism spectrum disorders and non-syndromic ID (Halgren et al., 2012). Loss of function mutations in these patients are suspected to contribute to the disease. ARID1b loss of function leads to dendritic defects as it is part of the nBAF complex which functions in postmitotic neurons. Patients with de novo deletions in ARID1B/BAF250b have severe speech impairment, and varying degrees of dysmorphic facies (Celen et al., 2017). Arid1b heterozygous mice exhibit impaired learning, memory, social interaction and showed anxiety like behaviour. Mutant littermates displayed growth impairment consistent with clinical features of ASD patients (Jung et al., 2017; Shibutani et al., 2017).
Synapse formation defects - RNAi induced deletion of simj, the fly ortholog of the GATAD2A and GATAD2B genes causes habituation delay and synapse formation defects - reduced synaptic area, lower number of branches and branch points in Drosophila (Willemsen et al., 2013).
CSS is commonly associated with mutations in multiple members of the BAF complex, including mutations in the SMARCA2 (BRM), SMAR-CB1 (BAF47), SMARCA4 (BRG1), SMARCE1 (BAF57), ARID1A (BAF250a) and ARID1B genes (BAF250b) (Tsurusaki et al., 2012). Mutation in SMARCD1/BAF60a is associated with syndromic NDDs which share overlapping clinical features of CSS and Nicolaides-Baraitser syndrome (NCBRS) (Table 2). Studies in Drosophila show that Bap60 (Drosophila ortholog of BAF60a) is required for regulation of genes for synaptic development and circuit formation involved in learning and memory (Nixon et al., 2019).
Corpus Callosum defects-The corpus callosum is a white matter tract which serves to connect the two cerebral hemispheres. Callosal projection neurons of the cortex form this tract, and hence impairments in corticogenesis affect CC formation. Corpus callosum agenesis is seen as a common clinical feature in many patients with mutations in different chromatin remodeller subunits, namely LSD1, BAF250b, BAF200a and GATAD2a and TIP60.
These patients also show developmental delay and Intellectual disability. This suggests that there might exist a correlation between CC agenesis perhaps caused by defective callosal projection neurons subtype identity leading to ID and DD. Indeed, mouse mutant of BCl11a shows partial agenesis of the CC (Woodworth et al., 2016) and in a separate study, BCl11a mice heterozygotes mimicking BCL11a haploinsufficiency show ID like symptoms (Dias et al., 2016).
A missense mutation within the BAZ1A (ACF-1) gene has been reported to cause ID. The missense mutation causes a change in the conserved amino acid phenylalanine to cysteine. Global RNA seq analysis of patient sample showed enrichment for genes involved in nervous system development, and 25 differentially expressed transcripts that cause ID (Zaghlool et al., 2016). Another patient with BAZ1A and NBAS (Neuroblastoma amplified sequence) mutations displayed corpus callosum agenesis (Weitensteiner et al., 2018).
Myelination defects-Another commonly observed feature in NDDs is myelination defect seen in patients with mutations in CHD3, GATAD2 and LSD1. The myelin sheath is formed of oligodendrocytes in the CNS. A proper balance in the expression of chromatin remodeller subunits is essential for oligodendrocyte formation, as discussed earlier for HDAC1 and HDAC2. A recent review discusses in detail the role of chromatin remodellers in oligodendrogenesis (Parras et al., 2020). Perhaps some of these mutations lead to defective olidendrocyte generation, and this needs to be probed further at the molecular level in mice and in vitro iPSC derived model systems.
Behavioural deficits-Single nucleotide polymorphisms (SNPs) within the MBD2 and MBD3 gene loci have been observed in autistic individuals (Cukier et al., 2009; Li et al., 2005). Rodent studies have shown that MBD2 deficient animals display abnormal maternal behaviour, whereas high maternal licking/grooming females show higher MBD2 expression in hippocampal CA1 and DG (Hendrich et al., 2001; Weaver et al., 2014). Human genes associated with ASD are altered in Mbd2 null mice, suggesting that MBD2 is an upstream regulator of ASD-associated genes. These mice also show behavioural defects, including memory impairment, reduced social-interaction and increased anxiety. Mice with MBD2 knockdown in adulthood also display similar behaviour (Lax et al., 2019). These phenotypes have been previously established in other rodent models of ASD (Edalatmanesh et al., 2013; Sosa-Diaz et al., 2014).
SMARCA2/BRM has been implicated in Schizophrenia in the Japanese population (Koga et al., 2009). A single risk variant allele showed nuclear localisation defect, which hampers protein functions. Smarca2 null mice show impaired social interaction, a behaviour commonly observed in SZ.
Cohesin mutations cause chromatin architecture defects, leading to transcriptional changes. Smc3 knockout using Nestin-Cre or Tau-Cre shows that these mutant mice exhibit phenotypes associated with CdL syndrome (Cornelia de Lange syndrome. Behavioural analysis showed that low cohesin expression causes high anxiety in these mice (Fujita et al., 2017).
The chromodomain group of proteins are ATPases, that can each function individually as a chromatin remodeller. Several NDDs have mutations within the CHD group of genes (Table 2). We summarise a few known causal pathological mechanisms.
CHD1-Pilarowski–Bjornsson Syndrome (PILBOS) is characterised by speech apraxia, DD, autism and facial dysmorphic features, with patients displaying missense mutations. All these mutations displaced highly conserved amino acids in the CHD1 protein. Fibroblasts from a single patient with R618Q variant showed a global increase in the H3K27me3 mark, suggesting closed chromatin to be plausible cause for chromatin mysregulation (Pilarowski et al., 2018). Xenopus laevis embryo morphants of Chd1 display narrower faces and smaller mouths, with reduction in craniofacial cartilages. Increased apoptosis in the head coupled to a reduced expression of the neural crest marker gene tfap-2α is observed in these morphants. The study suggests towards a possible control of CHD1 expression through retinoic acid signals, but further studies need to be conducted to delineate the role of the signalling molecule in CHD1 expression (Wyatt et al., 2021).
CHD2-Mutations in CHD2 causes Developmental Epileptic Encephalopathy-Zebrafish larvae morphants of chd2 display both developmental and behavioural anomalies, with epileptiform discharges, similar to those observed in mouse model of epilepsy (Suls et al., 2013).
CHD8-Loss of function mutations in CHD8 are associated with ASD, intellectual disability and macrocephaly (Bernier et al., 2014). CHD8 has several key roles in neurodevelopment beginning from proliferation of progenitors, dendritic arborization, to neurotransmission and synapse development as studied from loss of function mouse models (Hoffmann and Spengler, 2021). The role of pathogenic mutations in CHD8 is not well understood. To mimic the disease mutations, mice with heterozygous frameshift deletion in Chd8 display cognitive deficits, increased brain regional volume and perturbed expression of other ASD associated genes and biological pathways consistent with the clinical traits of ASD patients carrying CHD8 mutations (Gompers et al., 2017). Katayama et al., reported that haploinsufficiency of Chd8 led to characteristic ASD like behaviours such as anxiety, repetitive behaviour and altered social behaviour in mice (Katayama et al., 2016).
Knockdown of CHD8 in human NSCs led to dysregulated expression of ASD risk genes involved in neurodevelopment. These NSCs also showed defects in proliferation and differentiation (Cotney et al., 2015). In a similar study using hiPSC derived cerebral organoids, CRISPR/Cas9 mediated disruption of CHD8 led to upregulated expression of ASD and SZ risk genes indicating the role of CHD8 in neurodevelopmental pathophysiology (Wang et al., 2017).
It is not always unambiguous to establish a linear cause and effect relationship between the genetic mutations, the underlying altered cellular neural development phenotype and the clinical symptoms in NDDs. Reasons include, the dynamic expression and function of the subunits of the chromatin remodelling complexes throughout development and perhaps postnatally as well, makes it hard to pinpoint the exact stage at which the mutation affects, the polygenic nature of these disorders, the effect of environment in exacerbating the symptoms, to name a few.
One can adequately model aspects of these disorders using a human-specific model system such iPSCs-derived neurons from patients to capture the genetics of the disease and to understand human disease pathological mechanisms.
5. Concluding remarks and future perspectives
Genome organization is crucial for transcriptional regulation and is an emerging property of its function. Dynamic spatio-temporal gene expression programs determine neural cell outcomes during cortical development (Yuan et al., 2020). The timed and cell-type specific expression of transcripts requires specific chromatin configurations which are mediated individually by the different subunits of the chromatin remodelling complexes. Thus, each subunit has a unique function and contributes towards a dynamic genome which is a crucial driver of the process of proliferation and differentiation in the neural cells. Such a cell intrinsic mechanism propels cortical development initially and at later stages acts in concert with extrinsic signaling to make a functional cortical network (Telley et al., 2019). Given several putative risk variants have been identified in the very same chromatin remodellers a basic understanding of their mode of action is imperative to create a framework to study neuropathological mechanisms in NDDs. With the advent of mouse genetics and functional genomics the last several decades have revealed a mechanistic understanding of this process.
Key concepts which have emerged reveal the dynamicity of the process. For instance, the NuRD subunits display varying levels of expression in the developing mouse neocortex (Fuentes et al., 2012; MacDonald and Roskams, 2008; Nitarska et al., 2016; Sokpor et al., 2018). Some like LSD1 is ubiquitously expressed whereas CHDs have distinct zonal expression. It is not clear whether they function as holo-NuRD complex or perhaps have site-specific genome occupancy and function depending on the complex they are a part of at a given location and time. Disruption of NuRD subunits affects all steps of corticogenesis, right from progenitor proliferation to the neuron-glia cell fate switch. Progenitor proliferation defects are observed in mutants of HDAC1/2, CHD4, LSD1 and MBD3 (Hagelkruys et al., 2014, 2016; Moon et al., 2017; Nitarska et al., 2016; Tang et al., 2019; Zhang et al., 2014). Recruitment of HDACs, MTA2, RBBP4 and LSD1 by transcription factors at gene loci is important for regulating neuronal subtype specification (Britanova et al., 2008; Muralidharan et al., 2017). CHD3, CHD5 and S-nitrosylated HDAC2 have been shown to be important for neuronal migration (Nitarska et al., 2016; Nott et al., 2013). Phosphorylated LSD1 and S-nitrosylated HDAC2 play a role in neuritogenesis, exemplifying how post-translational modifications of these remodellers are necessary in cortical development. Disruption of these modifications in both HDAC2 and LSD1 led to impaired neuritogensis (Nott et al., 2008; Toffolo et al., 2014). The neuron-glia cell fate switch is affected in mutants of MBD3 and HDAC1/2 (Egawa et al., 2019; Knock et al., 2015; Tsuboi et al., 2018; Ye et al., 2009).
In the case of BAF complex, cell-type specific expression of different subunit isoforms enables switching and diverse combinatorial assembly of the complex thereby giving rise to the heterogeneous spatiotemporal pattern of gene expression in a tissue-specific manner (Ho and Crabtree, 2010; Sokpor et al., 2018; Wang et al., 1996; Yoo and Crabtree, 2009). Similar to the NuRD subunits, disruption of BAF subunits also affects all steps of corticogenesis. Mutants of BRG1, BAF45a, BAF53a and SS18 exhibit progenitor proliferation defects, as these subunits form an essential part of the npBAF complex (Son and Crabtree, 2014; Staahl et al., 2013; Lessard et al., 2007). However, each of these components seem to control aRG proliferation through different downstream effectors; illustrating how a multimeric complex modulates progenitor proliferation. A stoichiometric ratio of BAF170 and BAF155 is necessary for the controlled production of bIPs. The transcription factors CTIP1 and CTIP2, which form an essential part of the BAF complex, are involved in neuronal subtype specification. ARID 1b and the nBAF subunits – CREST, BAF53b and BAF45b are necessary for regulating dendritogenesis. Thus the combinatorial chemistry of these remodellers enable a probabilistic genome, one as recently proposed (Misteli 2020) which renders it dynamic, allows for quick changes and fine tuning of gene expression in the different progenitor and post mitotic zones of the developing neocortex.
Much attention has been given to the NuRD and the BAF complexes and their role in cortical development. Though the roles of members of the INO80/SWR and ISWI complexes in cortical development has not been well characterised, various studies indicate that the different subunits display varying expression patterns in the developing cortex.
SRCAP and TIP60 show enhanced expression in neurons (Elsen et al., 2018), as compared to INO80 which is enriched in the cortical VZ, suggesting a differential functional role of the INO80 complexes in progenitors and neurons similar to the CHDs (Nitarska et al., 2016). The expression of the cohesins SMC1 and SMC3 peaks in the embryonic brain at E15.5, post which it decreases gradually and peaks again post-natally at day 5 (Fujita et al., 2017). ACF1, CHRAC15 and CHRAC17 show high expression in the aRGs, followed by bIPs and post-mitotic neurons (Telley et al., 2016), suggesting a putative role of the ACF and CHRAC complexes in cortical progenitors. TIP5 and CECR2 are expressed in both progenitors and neurons but are enriched in the bIPs (Elsen et al., 2018; Telley et al., 2016). RSF1 and the three cohesins RAD21, SA1 and SA2 are expressed in all layers, but enriched in the progenitors (Telley et al., 2019). WSTF and BPTF display high expression in the aRGs and moderate expression in the bIPs (Elsen et al., 2018; Telley et al., 2019). More studies using biochemical and genomics approaches and animal models are necessary to unravel the roles of the ISWI and INO80 subunits in chromatin remodelling during corticogenesis. A complete picture of chromatin control of cortical development will emerge once the molecular underpinnings of the function of the above members of the chromatin remodelling complexes is deciphered.
ChIP-seq has been the mainstay to uncover protein-DNA interactions. This method, though very useful to study histone modifications, TF and co-factor binding, is time-consuming and requires a large number of cells (1-10million cells) and is not performed under native conditions. Newer techniques in the field of genomics have paved the way for in-depth characterisation of capturing the dynamicity of protein-chromatin interactions efficiently for low cell numbers. Cleavage Under Targets and Tagmentation (CUT & Tag) is an enzyme tethering based in situ method to capture chromatin accessibility and TF binding sites in the genome with more efficiency. This strategy uses a Tn5 transposase loaded with sequencing adapters to integrate them at the site of cleavage, within the nuclei, thus reducing the efforts of sequence library creation (Kaya-Okur et al., 2019). Cleavage Under Targets and Release Using Nuclease’ (CUT&RUN) is an in-situ method that employs TF-specific antibodies to interact with target protein (TF) in intact cells, following which protein A-MNase fragments the DNA bound by the TFs and these cleaved fragments diffuse out of nucleus. DNA extracted from the supernatant is used for paired end sequencing (Skene and Henikoff, 2017).
Chromatin integration labelling (ChIL) is an immunoprecipitation free method, wherein DNA sequences interacting with TFs and histone modifications are captured by antibody directed cleavage by Tn5 transposase. Cells are fixed and labelled with a primary antibody, a secondary antibody conjugated to ChIL DNA probe containing T7 promoter sequence, primer sequence for sequencing library preparation and Tn5 transposase binding site is used to integrate the ChIL probe into the genome. The integrated sequence is amplified by T7 RNA polymerase by in situ transcription and transcribed RNA is used for sequencing library preparation (Harada et al., 2019).
Assay for Transposase Accessible Chromatin (ATAC-seq) with high-throughput sequencing uses hyperactive Tn5 transposases loaded with sequencing adaptors. Transposase cleaves DNA in the region of open chromatin and tags adaptors to create DNA fragments, which can be used for sequencing (Buenrostro et al., 2013; Sun et al., 2019).
Recently, ATAC-seq was used to reveal accessible chromatin regulatory regions in human brain development and further used to map genetic risk for disease (Trevino et al., 2020).
The limitations to ChIP seq such as the requirement of high input cell size, low mapping resolution and low signal to noise ratio are overcome by these latest techniques. These techniques populate peaks at low sequencing depth with low background noise and high reproducibility with low cell number samples. Besides each of these techniques have their own advantages for example ChIL and CUT &Tag have an advantage of using them for single cell profiling. Given the subunits of the chromatin remodelling complexes have spatio-temporal functions these epigenomic profiling techniques would allow for high resolution of genome-dynamics in the different neural cells across developmental timelines. Such datasets would establish molecular signatures in successive generations of progenitors, neurons and glia.
Chromatin Remodelling complexes play a significant role in cortical development, and mutations in their subunits are perhaps causative of NDDs. Some recent advances in the field could be useful in establishing cause to effect relationships. Hi-coupled MAGMA utilises Hi-C datasets to interpret gene-SNP association and is useful to predict the effect of distal element SNPs on a gene in NDDs (Sey et al., 2020). Engineered chromatin remodelling proteins (E-ChRPs) that were shown to manipulate nucleosome positioning can be employed to study the functional effects of incorrect nucleosome positioning (Donovan et al., 2019).
A majority of the NDDs are polygenic, and the phenotypes observed are due to a combined effect rather than due to a single gene. It is therefore essential to study these NDDs using newer human models – pluripotent stem cells and organoids. Three-dimensional organoids generated from pluripotent stem cells and the advent of gene manipulation techniques such as CRISPR editing have led to a paradigm shift in our understanding of disease mechanisms. These technologies facilitate the creation of disease-associated mutations in cultured iPSC/ESCs and correcting mutations in patient-derived iPSC (Hotta and Yamanaka, 2015). Further advancements in these approaches such as in vivo genomic and RNA base editing (Sinnamon et al., 2020; Yeh et al., 2018), and combining multiple isogenic iPSC lines with various mutations associated with disease and differentiating them into neural lineage allow for better understanding of complex polygenic disorders (Cederquist et al., 2020).
Acknowledgements
We thank Disha Chauhan, PhD (The Visual Stories Studio) for making the scientific illustrations in this review. BM acknowledges support from a DBT/Wellcome Trust India Alliance: Intermediate Career Fellowship (IA/I/19/1/504288), DST-SERB: Start up research grant (SRG/2020/001612), DBT-Har Gobind Khorana Innovative Young Biotechnologist Award and institutional support from inStem by the Department of Biotechnology, Government of India.
Footnotes
Uncited references
Bornelov et al., 2018, Santen and Aten, 2013
Declaration of competing interest
none.
References
- Agirman G, Broix L, Nguyen L. Cerebral cortex development: an outside-in perspective. FEBS Lett. 2017;591:3978–3992. doi: 10.1002/1873-3468.12924. [DOI] [PubMed] [Google Scholar]
- Aihara T, Miyoshi Y, Koyama K, Suzuki M, Takahashi E, Monden M, Nakamura Y. Cloning and mapping of SMARCA5 encoding hSNF2H, a novel human homologue of Drosophila ISWI. Cytogenet Cell Genet. 1998;81:191–193. doi: 10.1159/000015027. [DOI] [PubMed] [Google Scholar]
- Aizawa H, Hu SC, Bobb K, Balakrishnan K, Ince G, Gurevich I, Cowan M, Ghosh A. Dendrite development regulated by CREST, a calcium-regulated transcriptional activator. Science. 2004;303:197–202. doi: 10.1126/science.1089845. [DOI] [PubMed] [Google Scholar]
- Alazami AM, Patel N, Shamseldin HE, Anazi S, Al-Dosari MS, Alzahrani F, Hijazi H, Alshammari M, Aldahmesh MA, Salih MA, Faqeih E, et al. Accelerating novel candidate gene discovery in neurogenetic disorders via whole-exome sequencing of prescreened multiplex consanguineous families. Cell Rep. 2015;10:148–161. doi: 10.1016/j.celrep.2014.12.015. [DOI] [PubMed] [Google Scholar]
- Alcamo EA, Chirivella L, Dautzenberg M, Dobreva G, Farinas I, Grosschedl R, McConnell SK. Satb2 regulates callosal projection neuron identity in the developing cerebral cortex. Neuron. 2008;57:364–377. doi: 10.1016/j.neuron.2007.12.012. [DOI] [PubMed] [Google Scholar]
- Alfert A, Moreno N, Kerl K. The BAF complex in development and disease. Epigenet Chromatin. 2019;12(19) doi: 10.1186/s13072-019-0264-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen MD, Freund SM, Zinzalla G, Bycroft M. The SWI/SNF subunit INI1 contains an N-terminal winged helix DNA binding domain that is a target for mutations in schwannomatosis. Structure. 2015;23:1344–1349. doi: 10.1016/j.str.2015.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alqarni SS, Murthy A, Zhang W, Przewloka MR, Silva AP, Watson AA, Lejon S, Pei XY, Smits AH, Kloet SL, Wang H, et al. Insight into the architecture of the NuRD complex: structure of the RbAp48-MTA1 subcomplex. J Biol Chem. 2014;289:21844–21855. doi: 10.1074/jbc.M114.558940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez-Saavedra M, De Repentigny Y, Lagali PS, Raghu Ram EV, Yan K, Hashem E, Ivanochko D, Huh MS, Yang D, Mears AJ, Todd MA, et al. Snf2h-mediated chromatin organization and histone H1 dynamics govern cerebellar morphogenesis and neural maturation. Nat Commun. 2014;5:4181. doi: 10.1038/ncomms5181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez-Saavedra M, Yan K, De Repentigny Y, Hashem LE, Chaudary N, Sarwar S, Yang D, Ioshikhes I, Kothary R, Hirayama T, Yagi T, et al. Snf2h drives chromatin remodeling to prime upper layer cortical neuron development. Front Mol Neurosci. 2019;12(243) doi: 10.3389/fnmol.2019.00243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angione K, Eschbach K, Smith G, Joshi C, Demarest S. Genetic testing in a cohort of patients with potential epilepsy with myoclonic-atonic seizures. Epilepsy Res. 2019;150:70–77. doi: 10.1016/j.eplepsyres.2019.01.008. [DOI] [PubMed] [Google Scholar]
- Arlotta P, Molyneaux BJ, Chen J, Inoue J, Kominami R, Macklis JD. Neuronal subtype-specific genes that control corticospinal motor neuron development in vivo. Neuron. 2005;45:207–221. doi: 10.1016/j.neuron.2004.12.036. [DOI] [PubMed] [Google Scholar]
- Bachmann C, Nguyen H, Rosenbusch J, Pham L, Rabe T, Patwa M, Sokpor G, Seong RH, Ashery-Padan R, Mansouri A, Stoykova A, et al. mSWI/SNF (BAF) complexes are indispensable for the neurogenesis and development of embryonic olfactory epithelium. PLoS Genet. 2016;12:e1006274. doi: 10.1371/journal.pgen.1006274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backx L, Seuntjens E, Devriendt K, Vermeesch J, Van Esch H. A balanced translocation t(6;14)(q25.3;q13.2) leading to reciprocal fusion transcripts in a patient with intellectual disability and agenesis of corpus callosum. Cytogenet Genome Res. 2011;132:135–143. doi: 10.1159/000321577. [DOI] [PubMed] [Google Scholar]
- Bagheri H, Badduke C, Qiao Y, Colnaghi R, Abramowicz I, Alcantara D, Dunham C, Wen J, Wildin RS, Nowaczyk MJ, Eichmeyer J, et al. Identifying candidate genes for 2p15p16.1 microdeletion syndrome using clinical, genomic, and functional analysis. JCI Insight. 2016;1:e85461. doi: 10.1172/jci.insight.85461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banting GS, Barak O, Ames TM, Burnham AC, Kardel MD, Cooch NS, Davidson CE, Godbout R, McDermid HE, Shiekhattar R. CECR2, a protein involved in neurulation, forms a novel chromatin remodeling complex with SNF2L. Hum Mol Genet. 2005;14:513–524. doi: 10.1093/hmg/ddi048. [DOI] [PubMed] [Google Scholar]
- Baranek C, Dittrich M, Parthasarathy S, Bonnon CG, Britanova O, Lanshakov D, Boukhtouche F, Sommer JE, Colmenares C, Tarabykin V, Atanasoski S. Protooncogene Ski cooperates with the chromatin-remodeling factor Satb2 in specifying callosal neurons. Proc Natl Acad Sci U S A. 2012;109:3546–3551. doi: 10.1073/pnas.1108718109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basak A, Hancarova M, Ulirsch JC, Balci TB, Trkova M, Pelisek M, Vlckova M, Muzikova K, Cermak J, Trka J, Dyment DA, et al. BCL11A deletions result in fetal hemoglobin persistence and neurodevelopmental alterations. J Clin Invest. 2015;125:2363–2368. doi: 10.1172/JCI81163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu S, Shukron O, Ponjavic A, Parruto P, Boucher W, Zhang W, Reynolds N, Lando D, Shah D, Sober LH, Jartseva A, et al. 2020. [DOI]
- Bedogni F, Hodge RD, Elsen GE, Nelson BR, Daza RA, Beyer RP, Bammler TK, Rubenstein JL, Hevner RF. Tbr1 regulates regional and laminar identity of postmitotic neurons in developing neocortex. Proc Natl Acad Sci U S A. 2010;107:13129–13134. doi: 10.1073/pnas.1002285107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell S, Rousseau J, Peng H, Aouabed Z, Priam P, Theroux JF, Jefri M, Tanti A, Wu H, Kolobova I, Silviera H, et al. Mutations in ACTL6B cause neurodevelopmental deficits and epilepsy and lead to loss of dendrites in human neurons. Am J Hum Genet. 2019;104:815–834. doi: 10.1016/j.ajhg.2019.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernier R, Golzio C, Xiong B, Stessman HA, Coe BP, Penn O, Witherspoon K, Gerdts J, Baker C, Vulto-van Silfhout AT, Schuurs-Hoeijmakers JH, et al. Disruptive CHD8 mutations define a subtype of autism early in development. Cell. 2014;158:263–276. doi: 10.1016/j.cell.2014.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bornelov S, Reynolds N, Xenophontos M, Gharbi S, Johnstone E, Floyd R, Ralser M, Signolet J, Loos R, Dietmann S, Bertone P, et al. Thenucleosome remodeling and deacetylation complex modulates chromatin structure at sites of active transcription to fine-tune gene expression. Mol Cell. 2018;71:56–72.:e54. doi: 10.1016/j.molcel.2018.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bramswig NC, Caluseriu O, Ludecke HJ, Bolduc FV, Noel NC, Wieland T, Surowy HM, Christen HJ, Engels H, Strom TM, Wieczorek D, et al. Heterozygosity for ARID2 loss-of-function mutations in individuals with a Coffin-Siris syndrome-like phenotype. Hum Genet. 2017;136:297–305. doi: 10.1007/s00439-017-1757-z. [DOI] [PubMed] [Google Scholar]
- Braun SMG, Petrova R, Tang J, Krokhotin A, Miller EL, Tang Y, Panagiotakos G, Crabtree GR. BAF subunit switching regulates chromatin accessibility to control cell cycle exit in the developing mammalian cortex. Genes Dev. 2021;35:335–353. doi: 10.1101/gad.342345.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britanova O, de Juan Romero C, Cheung A, Kwan KY, Schwark M, Gyorgy A, Vogel T, Akopov S, Mitkovski M, Agoston D, Sestan N, et al. Satb2 is a postmitotic determinant for upper-layer neuron specification in the neocortex. Neuron. 2008;57:378–392. doi: 10.1016/j.neuron.2007.12.028. [DOI] [PubMed] [Google Scholar]
- Buchsbaum IY, Cappello S. Neuronal migration in the CNS during development and disease: insights from in vivo and in vitro models. Development. 2019:146. doi: 10.1242/dev.163766. [DOI] [PubMed] [Google Scholar]
- Budday S, Steinmann P, Kuhl E. Physical biology of human brain development. Front Cell Neurosci. 2015;9(257) doi: 10.3389/fncel.2015.00257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods. 2013;10(12):1213–1218. doi: 10.1038/nmeth.2688. 10, 1213-1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgie SE, Bingman CA, Soni AB, Phillips GN., Jr Structural characterization of human Uch37. Proteins. 2012;80:649–654. doi: 10.1002/prot.23147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Y, Jin J, Yao T, Gottschalk AJ, Swanson SK, Wu S, Shi Y, Washburn MP, Florens L, Conaway RC, Conaway JW. YY1 functions with INO80 to activate transcription. Nat Struct Mol Biol. 2007;14:872–874. doi: 10.1038/nsmb1276. [DOI] [PubMed] [Google Scholar]
- Carvill GL, Heavin SB, Yendle SC, McMahon JM, O’Roak BJ, Cook J, Khan A, Dorschner MO, Weaver M, Calvert S, Malone S, et al. Targeted resequencing in epileptic encephalopathies identifies de novo mutations in CHD2 and SYNGAP1. Nat Genet. 2013;45:825–830. doi: 10.1038/ng.2646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castelo-Branco G, Lilja T, Wallenborg K, Falcao AM, Marques SC, Gracias A, Solum D, Paap R, Walfridsson J, Teixeira AI, Rosenfeld MG, et al. Neural stem cell differentiation is dictated by distinct actions of nuclear receptor corepressors and histone deacetylases. Stem Cell Reports. 2014;3:502–515. doi: 10.1016/j.stemcr.2014.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cederquist GY, Tchieu J, Callahan SJ, Ramnarine K, Ryan S, Zhang C, Rittenhouse C, Zeltner N, Chung SY, Zhou T, Chen S, et al. A multiplex human pluripotent stem cell platform defines molecular and functional subclasses of autism-related genes. Cell Stem Cell. 2020;27:35–49.:e36. doi: 10.1016/j.stem.2020.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celen C, Chuang JC, Luo X, Nijem N, Walker AK, Chen F, Zhang S, Chung AS, Nguyen LH, Nassour I, Budhipramono A, et al. Arid1b haploinsufficient mice reveal neuropsychiatric phenotypes and reversible causes of growth impairment. Elife. 2017:6. doi: 10.7554/eLife.25730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B, Wang SS, Hattox AM, Rayburn H, Nelson SB, McConnell SK. The Fezf2-Ctip2 genetic pathway regulates the fate choice of subcortical projection neurons in the developing cerebral cortex. Proc Natl Acad Sci USA. 2008;105:11382–11387. doi: 10.1073/pnas.0804918105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Cao H, Kaufmann T, Westlye LT, Tost H, Meyer-Lindenberg A, Schwarz E. Identification of reproducible BCL11A alterations in schizophrenia through individual-level prediction of coexpression. Schizophr Bull. 2020 doi: 10.1093/schbul/sbaa047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Pereira-Smith OM, Tominaga K. Loss of the chromatin regulator MRG15 limits neural stem/progenitor cell proliferation via increased expression of thep21 Cdk inhibitor. Stem Cell Res. 2011;7:75–88. doi: 10.1016/j.scr.2011.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Takano-Maruyama M, Pereira-Smith OM, Gaufo GO, Tominaga K. MRG15, a component of HAT and HDAC complexes, is essential for proliferation and differentiation of neural precursor cells. J Neurosci Res. 2009;87:1522–1531. doi: 10.1002/jnr.21976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong JX, Yu J-H, Lorentzen P, Park KM, Jamal SM, Tabor HK, Rauch A, Saenz MS, Boltshauser E, Patterson KE, Nickerson DA, et al. Gene discovery for Mendelian conditions via social networking: de novo variants in KDM1A cause developmental delay and distinctive facial features. Genet Med. 2015;18:788–795. doi: 10.1038/gim.2015.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clapier CR, Cairns BR. Regulation of ISWI involves inhibitory modules antagonized by nucleosomal epitopes. Nature. 2012;492:280–284. doi: 10.1038/nature11625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clapier CR, Iwasa J, Cairns BR, Peterson CL. Mechanisms of action and regulation of ATP-dependent chromatin-remodelling complexes. Nat Rev Mol Cell Biol. 2017;18:407–422. doi: 10.1038/nrm.2017.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conaco C, Otto S, Han J-J, Mandel G. Reciprocal actions of RECT and a microRNA promote neuronal identity. Proc Natl Acad Sci U S A. 2006;103:2422–2427. doi: 10.1073/pnas.0511041103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conaway RC, Conaway JW. The INO80 chromatin remodeling complex in transcription, replication and repair. Trends Biochem Sci. 2009;34:71–77. doi: 10.1016/j.tibs.2008.10.010. [DOI] [PubMed] [Google Scholar]
- Corona DF, Längst G, Clapier CR, Bonte EJ, Ferrari S, Tamkun JW, Becker PB. ISWI is an ATP-dependent nucleosome remodeling factor. Mol Cell. 1999;3:239–245. doi: 10.1016/s1097-2765(00)80314-7. [DOI] [PubMed] [Google Scholar]
- Cotney J, Muhle RA, Sanders SJ, Liu L, Willsey AJ, Niu W, Liu W, Klei L, Lei J, Yin J, Reilly SK, et al. The autism-associated chromatin modifier CHD8 regulates other autism risk genes during human neurodevelopment. Nat Commun. 2015;6:6404. doi: 10.1038/ncomms7404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cramer JM, Scarsdale JN, Walavalkar NM, Buchwald WA, Ginder GD, Williams DC., Jr Probing the dynamic distribution of bound states for methylcytosine-binding domains on DNA. J Biol Chem. 2014;289:1294–1302. doi: 10.1074/jbc.M113.512236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cukier HN, Rabionet R, Konidari I, Rayner-Evans MY, Baltos ML, Wright HH, Abramson RK, Martin ER, Cuccaro ML, Pericak-Vance MA, Gilbert JR. Novel variants identified in methyl-CpG-binding domain genes in autistic individuals. Neurogenetics. 2009;11:291–303. doi: 10.1007/s10048-009-0228-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- deLigt J, Willemsen MH, van Bon BW, Kleefstra T, Yntema HG, Kroes T, Vulto-van Silfhout AT, Koolen DA, de Vries P, Gilissen C, del Rosario M, et al. Diagnostic exome sequencing in persons with severe intellectual disability. N Engl J Med. 2012;367:1921–1929. doi: 10.1056/NEJMoa1206524. [DOI] [PubMed] [Google Scholar]
- De Rubeis S, He X, Goldberg AP, Poultney CS, Samocha K, Cicek AE, Kou Y, Liu L, Fromer M, Walker S, Singh T, et al. Synaptic, transcriptional and chromatin genes disrupted in autism. Nature. 2014;515:209–215. doi: 10.1038/nature13772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dias C, Estruch SB, Graham SA, McRae J, Sawiak SJ, Hurst JA, Joss SK, Holder SE, Morton JE, Turner C, Thevenon J, et al. BCL11A haploinsufficiency causes an intellectual disability syndrome and dysregulates transcription. Am J Hum Genet. 2016;99:253–274. doi: 10.1016/j.ajhg.2016.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez MH, Ayoub AE, Rakic P. POU-III transcription factors (Brn1, Brn2, and Oct6) influence neurogenesis, molecular identity, and migratory destination of upper-layer cells of the cerebral cortex. Cerebr Cortex. 2013;23:2632–2643. doi: 10.1093/cercor/bhs252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donovan DA, Crandall JG, Banks OGB, Jensvold ZD, Truong V, Dinwiddie D, McKnight LE, McKnight JN. Engineered chromatin remodeling proteins for precise nucleosome positioning. Cell Rep. 2019;29:2520–2535.:e2524. doi: 10.1016/j.celrep.2019.10.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drivas TG, Li D, Nair D, Alaimo JT, Alders M, Altmuller J, Barakat TS, Bebin EM, Bertsch NL, Blackburn PR, Blesson A, et al. A second cohort of CHD3 patients expands the molecular mechanisms known to cause Snijders Blok-Campeau syndrome. Eur J Hum Genet. 2020;28:1422–1431. doi: 10.1038/s41431-020-0654-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edalatmanesh MA, Nikfarjam H, Vafaee F, Moghadas M. Increased hippocampal cell density and enhanced spatial memory in thevalproic acid rat model of autism. Brain Res. 2013;1526:15–25. doi: 10.1016/j.brainres.2013.06.024. [DOI] [PubMed] [Google Scholar]
- Egan CM, Nyman U, Skotte J, Streubel G, Turner S, O’Connell DJ, Rraklli V, Dolan MJ, Chadderton N, Hansen K, Farrar GJ, et al. CHD5 is required for neurogenesis and has a dual role in facilitating gene expression and polycomb gene repression. Dev Cell. 2013;26:223–236. doi: 10.1016/j.devcel.2013.07.008. [DOI] [PubMed] [Google Scholar]
- Egawa N, Shindo A, Hikawa R, Kinoshita H, Liang AC, Itoh K, Lok J, Maki T, Takahashi R, Lo EH, Arai K. Differential roles of epigenetic regulators in the survival and differentiation of oligodendrocyte precursor cells. Glia. 2019;67:718–728. doi: 10.1002/glia.23567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ejaz R, Babul-Hirji R, Chitayat D. The evolving features of Nicolaides-Baraitser syndrome - a clinical report of a 20-year follow-up. Clin Case Rep. 2016;4:351–355. doi: 10.1002/ccr3.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsen GE, Bedogni F, Hodge RD, Bammler TK, MacDonald JW, Lindtner S, Rubenstein JLR, Hevner RF. The epigenetic factor landscape of developing neocortex is regulated by transcription factors Pax6--> Tbr2--> Tbr1. Front Neurosci. 2018;12:571. doi: 10.3389/fnins.2018.00571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farnung L, Ochmann M, Cramer P. Nucleosome-CHD4 chromatin remodeler structure maps human disease mutations. Elife. 2020:9. doi: 10.7554/eLife.56178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Q, Cao R, Xia L, Erdjument-Bromage H, Tempst P, Zhang Y. Identification and functional characterization of thep66/p68 components of the MeCP1 complex. Mol Cell Biol. 2002;22:536–546. doi: 10.1128/mcb.22.2.536-546.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florio M, Huttner WB. Neural progenitors, neurogenesis and the evolution of the neocortex. Development. 2014;141:2182–2194. doi: 10.1242/dev.090571. [DOI] [PubMed] [Google Scholar]
- Fossati G, Matteoli M, Menna E. Astrocytic factors controlling synaptogenesis: a team play. Cells. 2020;9 doi: 10.3390/cells9102173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman MR, Rowitch DH. Evolving concepts of gliogenesis: a look way back and ahead to the next 25 years. Neuron. 2013;80:613–623. doi: 10.1016/j.neuron.2013.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuentes P, Ca novas J, Berndt FA, Noctor SC, Kukuljan M. Co REST/LSD1 control the development of pyramidal cortical neurons. Cerebr Cortex. 2012;22:1431–1441. doi: 10.1093/cercor/bhr218. [DOI] [PubMed] [Google Scholar]
- Fujita Y, Masuda K, Bando M, Nakato R, Katou Y, Tanaka T, Nakayama M, Takao K, Miyakawa T, Tanaka T, Ago Y, et al. Decreased cohesin in the brain leads to defective synapse development and anxiety-related behavior. J Exp Med. 2017;214:1431–1452. doi: 10.1084/jem.20161517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgala PA, Manuel M, Price DJ. The generation of superficial cortical layers is regulated by levels of the transcription factor Pax6. Cerebr Cortex. 2011;21:81–94. doi: 10.1093/cercor/bhq061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerhold CB, Gasser SM. INO80 and SWR complexes: relating structure to function in chromatin remodeling. Trends Cell Biol. 2014;24:619–631. doi: 10.1016/j.tcb.2014.06.004. [DOI] [PubMed] [Google Scholar]
- Goel H, Parasivam G. Another case of holoprosencephaly associated with RAD21 loss-of-function variant. Brain. 2020;143:e64. doi: 10.1093/brain/awaa173. [DOI] [PubMed] [Google Scholar]
- Gompers AL, Su-Feher L, Ellegood J, Copping NA, Riyadh MA, Stradleigh TW, Pride MC, Schaffler MD, Wade AA, Catta-Preta R, Zdilar I, et al. Germline Chd8 haploinsufficiency alters brain development in mouse. Nat Neurosci. 2017;20:1062–1073. doi: 10.1038/nn.4592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Z, Brackertz M, Renkawitz R. SUMO modification enhances p66-mediated transcriptional repression of the Mi-2/NuRD complex. Mol Cell Biol. 2006;26:4519–4528. doi: 10.1128/MCB.00409-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin LR, Picketts DJ. The role of ISWI chromatin remodeling complexes in brain development and neurodevelopmental disorders. Mol Cell Neurosci. 2018;87:55–64. doi: 10.1016/j.mcn.2017.10.008. [DOI] [PubMed] [Google Scholar]
- Gotz M, Huttner WB. The cell biology of neurogenesis. Nat Rev Mol Cell Biol. 2005;6:777–788. doi: 10.1038/nrm1739. [DOI] [PubMed] [Google Scholar]
- Govindan S, Jabaudon D. Coupling progenitor and neuronal diversity in the developing neocortex. FEBS Lett. 2017;591:3960–3977. doi: 10.1002/1873-3468.12846. [DOI] [PubMed] [Google Scholar]
- Greenberg RS, Long HK, Swigut T, Wysocka J. Single amino acid change underlies distinct roles of H2A.Z subtypes in human syndrome. Cell. 2019;178:1421–1436.:e1424. doi: 10.1016/j.cell.2019.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregoretti IV, Lee YM, Goodson HV. Molecular evolution of the histone deacetylase family: functional implications of phylogenetic analysis. J Mol Biol. 2004;338:17–31. doi: 10.1016/j.jmb.2004.02.006. [DOI] [PubMed] [Google Scholar]
- Greig LC, Woodworth MB, Greppi C, Macklis JD. Ctip1 controls acquisition of sensory area identity and establishment of sensory input fields in the developing neocortex. Neuron. 2016;90:261–277. doi: 10.1016/j.neuron.2016.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillemot F. Cell fate specification in the mammalian telencephalon. Prog Neurobiol. 2007;83:37–52. doi: 10.1016/j.pneurobio.2007.02.009. [DOI] [PubMed] [Google Scholar]
- Hagelkruys A, Lagger S, Krahmer J, Leopoldi A, Artaker M, Pusch O, Zezula J, Weissmann S, Xie Y, Schofer C, Schlederer M, et al. A single allele of Hdac2 but not Hdac1 is sufficient for normal mouse brain development in the absence of its paralog. Development. 2014;141:604–616. doi: 10.1242/dev.100487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagelkruys A, Mattes K, Moos V, Rennmayr M, Ringbauer M, Sawicka A, Seiser C. Essential nonredundant function of the catalytic activity of histone deacetylase 2 in mouse development. Mol Cell Biol. 2016;36:462–474. doi: 10.1128/MCB.00639-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajheidari M, Koncz C, Bucher M. Chromatin evolution-key innovations underpinning morphological complexity. Front Plant Sci. 2019;10:454. doi: 10.3389/fpls.2019.00454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halgren C, Kjaergaard S, Bak M, Hansen C, El-Schich Z, Anderson CM, Henriksen KF, Hjalgrim H, Kirchhoff M, Bijlsma EK, Nielsen M, et al. Corpus callosum abnormalities, intellectual disability, speech impairment, and autism in patients with haploinsufficiency of ARID1B. Clin Genet. 2012;82:248–255. doi: 10.1111/j.1399-0004.2011.01755.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamdan FF, Srour M, Capo-Chichi JM, Daoud H, Nassif C, Patry L, Massicotte C, Ambalavanan A, Spiegelman D, Diallo O, Henrion E, et al. De novo mutations in moderate or severe intellectual disability. PLoS Genet. 2014;10:e1004772. doi: 10.1371/journal.pgen.1004772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han W, Kwan KY, Shim S, Lam MM, Shin Y, Xu X, Zhu Y, Li M, Sestan N. TBR1 directly represses Fezf2 to control the laminar origin and development of the corticospinal tract. Proc Natl Acad Sci U S A. 2011;108:3041–3046. doi: 10.1073/pnas.1016723108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada A, Maehara K, Handa T, Arimura Y, Nogami J, Hayashi-Takanaka Y, Shirahige K, Kurumizaka H, Kimura H, Ohkawa Y. A chromatin integration labelling method enables epigenomic profiling with lower input. Nat Cell Biol. 2019;21:287–296. doi: 10.1038/s41556-018-0248. [DOI] [PubMed] [Google Scholar]
- Harb K, Magrinelli E, Nicolas CS, Lukianets N, Frangeul L, Pietri M, Sun T, Sandoz G, Grammont F, Jabaudon D, et al. Area-specific development of distinct projection neuron subclasses is regulated by postnatal epigenetic modifications. Elife. 2016;5:e09531. doi: 10.7554/eLife.09531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hargreaves DC, Crabtree GR. ATP-dependent chromatin remodeling: genetics, genomics and mechanisms. Cell Res. 2011;21:396–420. doi: 10.1038/cr.2011.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert JM, McConnell SK. Targeting of cre to the Foxg1 (BF-1) locus mediates loxP recombination in the telencephalon and other developing head structures. Dev Biol. 2000;222:296–306. doi: 10.1006/dbio.2000.9732. [DOI] [PubMed] [Google Scholar]
- Hendrich B, Guy J, Ramsahoye B, Wilson VA, Bird A. Closely related proteins MBD2 and MBD3 play distinctive but interacting roles in mouse development. Genes Dev. 2001;15:710–723. doi: 10.1101/gad.194101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hevner RF. Intermediate progenitors and Tbr2 in cortical development. J Anat. 2019;235:616–625. doi: 10.1111/joa.12939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirabayashi Y, Gotoh Y. Epigenetic control of neural precursor cell fate during development. Nat Rev Neurosci. 2010;11:377–388. doi: 10.1038/nrn2810. [DOI] [PubMed] [Google Scholar]
- Ho L, Crabtree GR. Chromatin remodelling during development. Nature. 2010;463:474–484. doi: 10.1038/nature08911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho L, Jothi R, Ronan JL, Cui K, Zhao K, Crabtree GR. An embryonic stem cell chroma tin remodeling complex, esBAF, is an essential component of the core pluripotency transcriptional network. Proc Natl Acad Sci U S A. 2009;106:5187–5191. doi: 10.1073/pnas.0812888106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho PJ, Lloyd SM, Bao X. Unwinding chromatin at the right places: how BAF is targeted to specific genomic locations during development. Development. 2019;146 doi: 10.1242/dev.178780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann A, Spengler D. Chromatin remodeler CHD8 in autism and brain development. J Clin Med. 2021:10. doi: 10.3390/jcm10020366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homann OR, Misura K, Lamas E, Sandrock RW, Nelson P, McDonough SI, DeLisi LE. Whole-genome sequencing in multiplex families with psychoses reveals mutations in the SHANK2 and SMARCA1 genes segregating with illness. Mol Psychiatr. 2016;21:1690–1695. doi: 10.1038/mp.2016.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homma TK, Freire BL, Honjo R, Dauber A, Funari MFA, Lerario AM, Albuquerque EVA, Vasques GA, Bertola DR, Kim CA, Malaquias AC, et al. Growth and clinical characteristics of children with floating-harbor syndrome: analysis of current original data and a review of the literature. Horm Res Paediatr. 2009;92:115–123. doi: 10.1159/000503782. [DOI] [PubMed] [Google Scholar]
- Hota SK, Bruneau BG. ATP-dependent chromatin remodeling during mammalian development. Development. 2016;143:2882–2897. doi: 10.1242/dev.128892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotta A, Yamanaka S. From genomics to gene therapy: induced pluripotent stem cells meet genome editing. Annu Rev Genet. 2015;49:47–70. doi: 10.1146/annurev-genet-112414-054926. [DOI] [PubMed] [Google Scholar]
- Humbert J, Salian S, Makrythanasis P, Lemire G, Rousseau J, Ehresmann S, Garcia T, Alasiri R, Bottani A, Hanquinet S, Beaver E, et al. De novo KAT5 variants cause a syndrome with recognizable facial dysmorphisms, cerebellar atrophy, sleep disturbance, and epilepsy. Am J Hum Genet. 2020;107:564–574. doi: 10.1016/j.ajhg.2020.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T, Bulger M, Pazin MJ, Kobayashi R, Kadonaga JT. ACF, an ISWI-containing and ATP-utilizing chromatin assembly and remodeling factor. Cell. 1997;90:145–155. doi: 10.1016/s0092-8674(00)80321-9. [DOI] [PubMed] [Google Scholar]
- Jabaudon D. Fate and freedom in developing neocortical circuits. Nat Commun. 2017;8 doi: 10.1038/ncomms16042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamal I, Kumar V, Vatsa N, Shekhar S, Singh BK, Sharma A, Jana NR. Rescue of altered HDAC activity recovers behavioural abnormalities in a mouse model of Angelman syndrome. Neurobiol Dis. 2017;105:99–108. doi: 10.1016/j.nbd.2017.05.010. [DOI] [PubMed] [Google Scholar]
- Johnson D, Morrison N, Grant L, Turner T, Fantes J, Connor JM, Murday V. Confirmation of CHD7 as a cause of CHARGE association identified by mapping a balanced chromosome translocation in affected monozygotic twins. J Med Genet. 2006;43:280–284. doi: 10.1136/jmg.2005.032946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung EM, Moffat JJ, Liu J, Dravid SM, Gurumurthy CB, Kim WY. Arid1b haploinsufficiency disrupts cortical interneuron development and mouse behavior. Nat Neurosci. 2017;20:1694–1707. doi: 10.1038/s41593-017-0013-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ka M, Chopra DA, Dravid SM, Kim WY. Essential roles for ARID1B in dendritic arborization and spine morphology of developing pyramidal neurons. J Neurosci. 2016;36:2723–2742. doi: 10.1523/JNEUROSCI.2321-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalebic N, Huttner WB. Basal progenitor morphology and neocortex evolution. Trends Neurosci. 2020 doi: 10.1016/j.tins.2020.07.009. [DOI] [PubMed] [Google Scholar]
- Karaca E, Harel T, Pehlivan D, Jhangiani SN, Gambin T, Coban Akdemir Z, Gonzaga-Jauregui C, Erdin S, Bayram Y, Campbell IM, Hunter JV, et al. Genes that affect brain structure and function identified by rare variant analyses of mendelian neurologic disease. Neuron. 2015;88:499–513. doi: 10.1016/j.neuron.2015.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katayama Y, Nishiyama M, Shoji H, Ohkawa Y, Kawamura A, Sato T, Suyama M, Takumi T, Miyakawa T, Nakayama KI. CHD8 haploinsufficiency results in autistic-like phenotypes in mice. Nature. 2016;537:675–679. doi: 10.1038/nature19357. [DOI] [PubMed] [Google Scholar]
- Kaya-Okur HS, Wu SJ, Codomo CA, Pledger ES, Bryson TD, Henikoff JG, Ahmad K, Henikoff S. CUT&Tag for efficient epigenomic profiling of small samples and single cells. Nat Commun. 2019;10(1930) doi: 10.1038/s41467-019-09982-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keil JM, Doyle DZ, Qalieh A, Lam MM, Funk OH, Qalieh Y, Shi L, Mohan N, Sorel A, Kwan KY. Symmetric neural progenitor divisions require chromatin-mediated homologous recombination DNA repair by Ino80. Nat Commun. 2020;11:3839. doi: 10.1038/s41467-020-17551-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JK, Huh SO, Choi H, Lee KS, Shin D, Lee C, Nam JS, Kim H, Chung H, Lee HW, Park SD, et al. Srg3, a mouse homolog of yeast SWI3, is essential for early embryogenesis and involved in brain development. Mol Cell Biol. 2001;21:7787–7795. doi: 10.1128/MCB.21.22.7787-7795.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura H, Wang C, Ishizuka K, Xing J, Takasaki Y, Kushima I, Aleksic B, Uno Y, Okada T, Ikeda M, Mori D, et al. Identification of a rare variant in CHD8 that contributes to schizophrenia and autism spectrum disorder susceptibility. Schizophr Res. 2016;178:104–106. doi: 10.1016/j.schres.2016.08.023. [DOI] [PubMed] [Google Scholar]
- Kleefstra T, Kramer JM, Neveling K, Willemsen MH, Koemans TS, Vissers LE, Wissink-Lindhout W, Fenckova M, van den Akker WM, Kasri NN, Nillesen WM, et al. Disruption of an EHMT1-associated chromatin-modification module causes intellectual disability. Am J Hum Genet. 2012;91:73–82. doi: 10.1016/j.ajhg.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knauss JL, Miao N, Kim SN, Nie Y, Shi Y, Wu T, Pinto HB, Donohoe ME, Sun T. Long noncoding RNA Sox2ot and transcription factor YY1 co-regulate the differentiation of cortical neural progenitors by repressing Sox2. Cell Death Dis. 2018;9:799. doi: 10.1038/s41419-018-0840-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knock E, Pereira J, Lombard PD, Dimond A, Leaford D, Livesey FJ, Hendrich B. Themethyl binding domain 3/nucleosome remodelling and deacetylase complex regulates neural cell fate determination and terminal differentiation in the cerebral cortex. Neural Dev. 2015;10:13. doi: 10.1186/s13064-015-0040-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koga M, Ishiguro H, Yazaki S, Horiuchi Y, Arai M, Niizato K, Iritani S, Itokawa M, Inada T, Iwata N, Ozaki N, et al. Involvement of SMARCA2/BRM in the SWI/SNF chromatin-remodeling complex in schizophrenia. Hum Mol Genet. 2009;18:2483–2494. doi: 10.1093/hmg/ddp166. [DOI] [PubMed] [Google Scholar]
- Kolla V, Naraparaju K, Zhuang T, Higashi M, Kolla S, Blobel GA, Brodeur GM. The tumour suppressor CHD5 forms a NuRD-type chromatin remodelling complex. Biochem J. 2015;468:345–352. doi: 10.1042/BJ20150030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korenke GC, Schulte B, Biskup S, Neidhardt J, Owczarek-Lipska M. A Novel de novo Frameshift Mutation in the BCL11A Gene in a Patient with Intellectual Disability Syndrome and Epilepsy. Mol Syndromol. 2020;11:135–140. doi: 10.1159/000508566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruszka P, Berger SI, Casa V, Dekker MR, Gaesser J, Weiss K, Martinez AF, Murdock DR, Louie RJ, Prijoles EJ, Lichty AW, Brouwer OF, et al. Cohesin complex-associated holoprosencephaly. Brain. 2019;142:2631–2643. doi: 10.1093/brain/awz210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumamoto T, Toma K, Gunadi, McKenna WL, Kasukawa T, Katzman S, Chen B, Hanashima C. Foxg1 coordinates the switch from nonradially to radially migrating glutamatergic subtypes in the neocortex through spatiotemporal repression. Cell Rep. 2013;3:931–945. doi: 10.1016/j.celrep.2013.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwan KY, Lam MMS, Krsnik Z, Kawasawa YI, Veronique Lefebvre V, Šestan N. SOX5 postmitotically regulates migration, postmigratory differentiation, and projections of subplate and deep-layer neocortical neurons. Proc Natl Acad Sci U S A. 2008;105:16021–16026. doi: 10.1073/pnas.0806791105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai T, Jabaudon D, Molyneaux BJ, Azim E, Arlotta P, Menezes JR, Macklis JD. SOX5 controls the sequential generation of distinct corticofugal neuron subtypes. Neuron. 2008;57:232–247. doi: 10.1016/j.neuron.2007.12.023. [DOI] [PubMed] [Google Scholar]
- Lalli MA, Jang J, Park JH, Wang Y, Guzman E, Zhou H, Audouard M, Bridges D, Tovar KR, Papuc SM, Tutulan-Cunita AC, et al. Haploinsufficiency of BAZ1B contributes to Williams syndrome through transcriptional dysregulation of neurodevelopmental pathways. Hum Mol Genet. 2016;25:1294–1306. doi: 10.1093/hmg/ddw010. [DOI] [PubMed] [Google Scholar]
- Lax E, DoCarmo S, Enuka Y, Sapozhnikov DM, Welikovitch LA, Mahmood N, Rabbani SA, Wang L, Britt JP, Hancock WW, Yarden Y, et al. 2019. [DOI] [PMC free article] [PubMed]
- Lazzaro MA, Picketts DJ. Cloning and characterization of the murine Imitation Switch (ISWI) genes: differential expression patterns suggest distinct developmental roles for Snf2h and Snf2l. J Neurochem. 2001;77:1145–1156. doi: 10.1046/j.1471-4159.2001.00324.x. [DOI] [PubMed] [Google Scholar]
- Le Guezennec X, Vermeulen M, Brinkman AB, Hoeijmakers WA, Cohen A, Lasonder E, Stunnenberg HG. MBD2/NuRD and MBD3/NuRD, two distinct complexes with different biochemical and functional properties. Mol Cell Biol. 2006;26:843–851. doi: 10.1128/MCB.26.3.843-851.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leid M, Ishmael JE, Avram D, Shepherd D, Fraulob V, Dolle P. CTIP1 and CTIP2 are differentially expressed during mouse embryogenesis. Gene Expr Patterns. 2004;4:733–739. doi: 10.1016/j.modgep.2004.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leone DP, Srinivasan K, Chen B, Alcamo E, McConnell SK. The determ ination of projection neuron identity in the developing cerebral cortex. Curr Opin Neurobiol. 2008;18:28–35. doi: 10.1016/j.conb.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lessard J, Wu JI, Ranish JA, Wan M, Winslow MM, Staahl BT, Wu H, Aebersold R, Graef IA, Crabtree GR. An essential switch in subunit composition of a chromatin remodeling complex during neural development. Neuron. 2007;55:201–215. doi: 10.1016/j.neuron.2007.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Yamagata T, Mori M, Yasuhara A, Momoi MY. Mutation analysis of methyl-CpG binding protein family genes in autistic patients. Brain Dev. 2005;27:321–325. doi: 10.1016/j.braindev.2004.08.003. [DOI] [PubMed] [Google Scholar]
- Lodato S, Arlotta P. Generating neuronal diversity in the mammalian cerebral cortex. Annu Rev Cell Dev Biol. 2015;31:699–720. doi: 10.1146/annurev-cellbio-100814-125353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo X, Zou Y, Tan B, Zhang Y, Guo J, Zeng L, Zhang R, Tan H, Wei X, Hu Y, Zheng Y, et al. Novel GATAD2B loss-of-function mutations cause intellectual disability in two unrelated cases. J Hum Genet. 2017;62:513–516. doi: 10.1038/jhg.2016.164. [DOI] [PubMed] [Google Scholar]
- MacDonald JL, Roskams AJ. Histonedeacetylases 1 and 2 areexpressed at distinct stages of neuroglial development. Dev Dynam. 2008;237:2256–2267. doi: 10.1002/dvdy.21626. [DOI] [PubMed] [Google Scholar]
- Machol K, Rousseau J, Ehresmann S, Garcia T, Nguyen TTM, Spillmann RC, Sullivan JA, Shashi V, Jiang YH, Stong N, Fiala E, et al. Expanding the spectrum of BAF-related disorders: de novo variants in SMARCC2 cause a syndrome with intellectual disability and developmental delay. Am J Hum Genet. 2019;104:164–178. doi: 10.1016/j.ajhg.2018.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manelyte L, Langst G. Chromatin Remodelers and Their Way of Action. Chromatin Remodelling. 2013 [Google Scholar]
- Mansfield RE, Musselman CA, Kwan AH, Oliver SS, Garske AL, Davrazou F, Denu JM, Kutateladze TG, Mackay JP. Plant homeodomain (PHD) fingers of CHD4 are histone H3-binding modules with preference for unmodified H3K4 and methylated H3K9. J Biol Chem. 2011;286:11779–11791. doi: 10.1074/jbc.M110.208207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markenscoff-Papadimitriou E, Whalen S, Przytycki P, Thomas R, Binyameen F, Nowakowski TJ, Kriegstein AR, Sanders SJ, State MW, Pollard KS, Rubenstein JL. A chromatin accessibility atlas of the developing human telencephalon. Cell. 2020;182:754–769.:e718. doi: 10.1016/j.cell.2020.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martynoga B, Drechsel D, Guillemot F. Molecular control of neurogenesis: a view from the mammalian cerebral cortex. Cold Spring Harb Perspect Biol. 2012:4. doi: 10.1101/cshperspect.a008359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martynoga B, Morrison H, Price DJ, Mason JO. Foxg1 is required for specification of ventral telencephalon and region-specific regulation of dorsal telencephalic precursor proliferation and apoptosis. Dev Biol. 2005;283:113–127. doi: 10.1016/j.ydbio.2005.04.005. [DOI] [PubMed] [Google Scholar]
- Matsumoto S, Banine F, Struve J, Xing R, Adams C, Liu Y, Metzger D, Chambon P, Rao MS, Sherman LS. Brg1 is required for murine neural stem cell ma intenance and gliogenesis. Dev Biol. 2006;289:372–383. doi: 10.1016/j.ydbio.2005.10.044. [DOI] [PubMed] [Google Scholar]
- McCarthy SE, Gillis J, Kramer M, Lihm J, Yoon S, Berstein Y, Mistry M, Pavlidis P, Solomon R, Ghiban E, Antoniou E, et al. De novo mutations in schizophrenia implicate chromatin remodeling and support a genetic overlap with autism and intellectual disability. Mol Psychiatr. 2014;19:652–658. doi: 10.1038/mp.2014.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna WL, Betancourt J, Larkin KA, Abrams B, Guo C, Rubenstein JL, Chen B. Tbr1 and Fezf2 regulate alternate corticofugal neuronal identities during neocortical development. J Neurosci. 2011;31:549–564. doi: 10.1523/JNEUROSCI.4131-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merner N, Forgeot d’Arc B, Bell SC, Maussion G, Peng H, Gauthier J, Crapper L, Hamdan FF, Michaud JL, Mottron L, Rouleau GA, et al. A de novo frameshift mutation in chromodomain helicase DNA-binding domain 8 (CHD8): a case report and literature review. Am J Med Genet. 2016;170A:1225–1235. doi: 10.1002/ajmg.a.37566. [DOI] [PubMed] [Google Scholar]
- Millard CJ, Watson PJ, Fairall L, Schwabe JWR. Targeting class I histone deacetylases in a “complex”. Environment Trends Pharmacol Sci. 2017;38:363–377. doi: 10.1016/j.tips.2016.12.006. [DOI] [PubMed] [Google Scholar]
- Miller FD, Gauthier AS. Timing is everything: making neurons versus glia in the developing cortex. Neuron. 2007;54:357–369. doi: 10.1016/j.neuron.2007.04.019. [DOI] [PubMed] [Google Scholar]
- Miyake N, Tsurusaki Y, Matsumoto N. Numerous BAF complex genes are mutated in Coffin-Siris syndrome. Am J Med Genet C Semin Med Genet. 2014;166C:257–261. doi: 10.1002/ajmg.c.31406. [DOI] [PubMed] [Google Scholar]
- Molyneaux BJ, Arlotta P, Hirata T, Hibi M, Macklis JD. Fezl is required for the birth and specification of corticospinal motor neurons. Neuron. 2005;47:817–831. doi: 10.1016/j.neuron.2005.08.030. [DOI] [PubMed] [Google Scholar]
- Molyneaux BJ, Arlotta P, Menezes JR, Macklis JD. Neuronal subtype specification in the cerebral cortex. Nat Rev Neurosci. 2007;8:427–437. doi: 10.1038/nrn2151. [DOI] [PubMed] [Google Scholar]
- Montgomery RL, Hsieh J, Barbosa AC, Richardson JA, Olson EN. Histone deacetylases 1 and 2 control the progression of neural precursors to neurons during brain development. J Neurosci. 2009;106:7876–7881. doi: 10.1073/pnas.0902750106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon BS, Yun HM, Chang WH, Steele BH, Cai M, Choi SH, Lu W. Smek promotes corticogenesis through regulating Mbd3’s stability and Mbd3/NuRD complex recruitment to genes associated with neurogenesis. PLoS Biol. 2017;15:e2001220. doi: 10.1371/journal.pbio.2001220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris-Rosendahl DJ, Crocq MA. Neurodevelopmental disorders-the history and future of a diagnostic concept. Dialogues Clin Neurosci. 2020;22:65–72. doi: 10.31887/DCNS.2020.22.1/macrocq. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhtar T, Taylor V. Untangling cortical complexity during development. J Exp Neurosci. 2018;12:1179069518759332. doi: 10.1177/1179069518759332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muralidharan B, Khatri Z, Maheshwari U, Gupta R, Roy B, Pradhan SJ, Karmodiya K, Padmanabhan H, Shetty AS, Balaji C, Kolthur-Seetharam U, et al. LHX2 interacts with the NuRD complex and regulates cortical neuron subtype determinants Fezf2 and Sox11. J Neurosci. 2017;37:194–203. doi: 10.1523/JNEUROSCI.2836-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murzina NV, Pei XY, Zhang W, Sparkes M, Vicente-Garcia J, Pratap JV, McLaughlin SH, Ben-Shahar TR, Verreault A, Luisi BF, Laue ED. Structural basis for the recognition of histone H4 by the histone-chaperone RbAp46. Structure. 2008;16:1077–1085. doi: 10.1016/j.str.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musselman CA, Mansfield RE, Garske AL, Davrazou F, Kwan AH, Oliver SS, O’Leary H, Denu JM, Mackay JP, Kutateladze TG. Binding of the CHD4 PHD2 finger to histone H3 is modulated by covalent modifications. Biochem J. 2009;423:179–187. doi: 10.1042/BJ20090870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanan R, Pham L, Kerimoglu C, Watanabe T, Castro Hernandez R, Sokpor G, Ulmke PA, Kiszka KA, Tonchev AB, Rosenbusch J, Seong RH, et al. Chromatin remodeling BAF155 subunit regulates thegenesis of basal progenitors in developing cortex. iScience. 2018;4:109–126. doi: 10.1016/j.isci.2018.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanan R, Pirouz M, Kerimoglu C, Pham L, Wagener RJ, Kiszka KA, Rosenbusch J, Seong RH, Kessel M, Fischer A, Stoykova A, et al. Loss of BAF (mSWI/SNF) complexes causes global transcriptional and chromatin state changes in forebrain development. Cell Rep. 2015;13:1842–1854. doi: 10.1016/j.celrep.2015.10.046. [DOI] [PubMed] [Google Scholar]
- Narayanan R, Tuoc TC. Roles of chromatin remodeling BAF complex in neural differentiation and reprogramming. Cell Tissue Res. 2014;356:575–584. doi: 10.1007/s00441-013-1791-7. [DOI] [PubMed] [Google Scholar]
- Neale BM, Kou Y, Liu L, Ma’ayan A, Samocha KE, Sabo A, Lin CF, Stevens C, Wang LS, Makarov V, Polak P, et al. Patterns and ra tes of exonic de novo mutations in autism spectrum disorders. Nature. 2013;485:242–245. doi: 10.1038/nature11011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen H, Sokpor G, Pham L, Rosenbusch J, Stoykova A, Staiger JF, Tuoc T. Epigenetic regulation by BAF (mSWI/SNF) chromatin remodeling complexes is indispensable for embryonic development. Cell Cycle. 2016;15:1317–1324. doi: 10.1080/15384101.2016.1160984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitarska J, Smith JG, Sherlock WT, Hillege MM, Nott A, Barshop WD, Vashisht AA, Wohlschlegel JA, Mitter R, Riccio A. A functional switch of NuRD chromatin remodeling complex subunits regulates mouse cortical development. Cell Rep. 2016;17:1683–1698. doi: 10.1016/j.celrep.2016.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nixon KCJ, Rousseau J, Stone MH, Sarikahya M, Ehresmann S, Mizuno S, Matsumoto N, Miyake N, Study DDD, Baralle D, McKee S, et al. A syndromic neurodevelopmental disorder caused by mutations in SMARCD1, a core SWI/SNF subunit needed for context-dependent neuronal gene regulation in flies. Am J Hum Genet. 2019;104:596–610. doi: 10.1016/j.ajhg.2019.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nott A, Nitarska J, Veenvliet JV, Schacke S, Derijck AA, Sirko P, Muchardt C, Pasterkamp RJ, Smidt MP, Riccio A. S-nitrosylation of HDAC2 regulates the expression of the chromatin-remodeling factor Brm during radial neuron migration. Proc Natl Acad Sci U S A. 2013;110:3113–3118. doi: 10.1073/pnas.1218126110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nott A, Watson PM, Robinson JD, Crepaldi L, Riccio A. S-Nitrosylation of histone deacetylase 2 induces chromatin remodelling in neurons. Nature. 2008;455:411–415. doi: 10.1038/nature07238. [DOI] [PubMed] [Google Scholar]
- O’Roak BJ, Vives L, Girirajan S, Karakoc E, Krumm N, Coe BP, Levy R, Ko A, Lee C, Smith JD, Turner EH, et al. Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature. 2012;485:246–250. doi: 10.1038/nature10989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olave I. Identification of a polymorphic, neuron-specific chromatin remodeling complex. Genes & Development. 2002;16:2509–2517. doi: 10.1101/gad.992102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen MJ, O’Donovan MC, Thapar A, Craddock N. Neurodevelopmental hypothesis of schizophrenia. Br J Psychiatry. 2011;198:173–175. doi: 10.1192/bjp.bp.110.084384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parras C, Marie C, Zhao C, Lu QR. Chromatin remodelers in oligodendroglia. Glia. 2020;68:1604–1618. doi: 10.1002/glia.23837. [DOI] [PubMed] [Google Scholar]
- Pilarowski GO, Vernon HJ, Applegate CD, Boukas L, Cho MT, Gurnett CA, Benke PJ, Beaver E, Heeley JM, Medne L, Krantz ID, et al. Missense variants in the chromatin remodeler CHD1 are associated with neurodevelopmental disability. J Med Genet. 2018;55:561–566. doi: 10.1136/jmedgenet-2017-104759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilotto S, Speranzini V, Marabelli C, Rusconi F, Toffolo E, Grillo B, Battaglioli E, Mattevi A. LSD1/KDM1A mutations associated to a newly described form of intellectual disability impair demethylase activity and binding to transcription factors. Hum Mol Genet. 2016;25:2578–2587. doi: 10.1093/hmg/ddw120. [DOI] [PubMed] [Google Scholar]
- Pisansky MT, Young AE, O’Connor MB, Gottesman, Bagchi A, Gewirtz JC. Mice lacking the chromodomain helicase DNA-binding 5 chromatin remodeler display autism-like characteristics. Transl Psychiatry. 2017;7:e1152. doi: 10.1038/tp.2017.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauch A, Wieczorek D, Graf E, Wieland T, Endele S, Schwarzmayr T, Albrecht B, Bartholdi D, Beygo J, Di Donato N, Dufke A, et al. Rangeof genetic mutations associated with severe non-syndromic sporadic intellectual disability: an exome sequencing study. Lancet. 2012;380:1674–1682. doi: 10.1016/s0140-6736(12)61480-9. [DOI] [PubMed] [Google Scholar]
- Ronzoni L, Tagliaferri F, Tucci A, Baccarin M, Esposito S, Milani D. Interstitial 6q25 microdeletion syndrome: ARID1B is the key gene. Am J Med Genet. 2016;170A:1257–1261. doi: 10.1002/ajmg.a.37553. [DOI] [PubMed] [Google Scholar]
- Santen G, Aten E, Vulto-van Silfhout A, Pottinger C, van Bon B, van Minderhout I, Snowdowne R, van der Lans C, Boogaard M, Linssen M, Vijfhuizen L, et al. Coffin-siris syndrome and the BAF complex: genotype-phenotype study in 63 patients. Hum Mutat. 2013;16(19):2509–2517. doi: 10.1101/gad.992102. [DOI] [PubMed] [Google Scholar]
- Schroeder FA, Gilbert TM, Feng N, Taillon BD, Volkow ND, Innis RB, Hooker JM, Lipska BK. Expression of HDAC2 but not HDAC1 transcript is reduced in dorsolateral prefrontal cortex of patients with schizophrenia. ACS Chem Neurosci. 2017;8:662–668. doi: 10.1021/acschemneuro.6b00372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessa A, Mao CA, Hadjantonakis AK, Klein WH, Broccoli V. Tbr2 directs conversion of radial glia into basal precursors and guides neuronal amplification by indirect neurogenesis in the developing neocortex. Neuron. 2008;60:56–69. doi: 10.1016/j.neuron.2008.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seto E, Yoshida M. Erasers of histone acetylation: the histone deacetylase enzymes. Cold Spring Harb Perspect Biol. 2014;6:a018713. doi: 10.1101/cshperspect.a018713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sey NYA, Hu B, Mah W, Fauni H, McAfee JC, Rajarajan P, Brennand KJ, Akbarian S, Won H. A computational tool (H-MAGMA) for improved prediction of brain-disorder risk genes by incorporating brain chromatin interaction profiles. Nat Neurosci. 2020;23:583–593. doi: 10.1038/s41593-020-0603-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma RP, Grayson DR, Gavin DP. Histone deactylase 1 expression is increased in the prefrontal cortex of schizophrenia subjects: analysis of the National Brain Databank microarray collection. Schizophr Res. 2008;98:111–117. doi: 10.1016/j.schres.2007.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibutani M, Horii T, Shoji H, Morita S, Kimura M, Terawaki N, Miyakawa T, Hatada I. Arid1b haploinsufficiency causes abnormal brain gene expression and autism-related behaviors in mice. Int J Mol Sci. 2017;18 doi: 10.3390/ijms18091872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shieh C, Jones N, Vanle B, Au M, Huang AY, Silva APG, Lee H, Douine ED, Otero MG, Choi A, Grand K, et al. GATAD2B-associated neurodevelopmental disorder (GAND): clinical and molecular insights into a NuRD-related disorder. Genet Med. 2020;22:878–888. doi: 10.1038/s41436-019-0747-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim S, Kwan KY, Li M, Lefebvre V, Sestan N. Cis-regulatory control of corticospinal system development and evolution. Nature. 2012;486:74–79. doi: 10.1038/nature11094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh T, Walters JTR, Johnstone M, Curtis D, Suvisaari J, Torniainen M, Rees E, Iyegbe C, Blackwood D, McIntosh AM, Kirov G, et al. The contribution of rare variants to risk of schizophrenia in individuals with and without intellectual disability. Nat Genet. 2017;49:1167–1173. doi: 10.1038/ng.3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinnamon JR, Kim SY, Fisk JR, Song Z, Nakai H, Jeng S, McWeeney SK, Mandel G. In vivo repair of a protein underlying a neurological disorder by programmable RNA editing. Cell Rep. 2020;32:107878. doi: 10.1016/j.celrep.2020.107878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skene PJ, Henikoff S. An efficient targeted nuclease strategy for high-resolution mapping of DNA binding sites. Elife. 2017;6 doi: 10.7554/eLife.21856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijders Blok L, Rousseau J, Twist J, Ehresmann S, Takaku M, Venselaar H, Rodan LH, Nowak CB, Douglas J, Swoboda KJ, Steeves MA, et al. CHD3 helicase domain mutations cause a neurodevelopmental syndrome with macrocephaly and impaired speech and language. Nat Commun. 2018;9 doi: 10.1038/s41467-018-06014-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soardi FC, Machado-Silva A, Linhares ND, Zheng G, Qu Q, Pena HB, Martins TMM, Vieira HGS, Pereira NB, Melo-Minardi RC, Gomes CC, et al. Familial STAG2 germline mutation defines a new human cohesinopathy. NPJ Genom Med. 2017;2:7. doi: 10.1038/s41525-017-0009-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohn DH, Lee KY, Lee C, Oh J, Chung H, Jeon SH, Seong RH. SRG3 interacts directly with the major components of the SWI/SNF chromatin remodeling complex and protects them from proteasomal degradation*. J Biol Chem. 2007;282:10614–10624. doi: 10.1074/jbc.M610563200. [DOI] [PubMed] [Google Scholar]
- Sokpor G, Castro-Hernandez R, Rosenbusch J, Staiger JF, Tuoc T. ATP-dependent chromatin remodeling during cortical neurogenesis. Front Neurosci. 2018;12:226. doi: 10.3389/fnins.2018.00226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokpor G, Xie Y, Rosenbusch J, Tuoc T. Chromatin remodeling BAF (SWI/SNF) complexes in neural development and disorders. Front Mol Neurosci. 2017;10:243. doi: 10.3389/fnmol.2017.00243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Son EY, Crabtree GR. The role of BAF (mSWI/SNF) complexes in mammalian neural development. Am J Med Genet C Semin Med Genet. 2014;166C:333–349. doi: 10.1002/ajmg.c.31416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sosa-Diaz N, Bringas ME, Atzori M, Flores G. Prefrontal cortex, hippocampus, and basolateral amygdala plasticity in a rat model of autism spectrum. Synapse. 2014;68:468–473. doi: 10.1002/syn.21759. [DOI] [PubMed] [Google Scholar]
- Sousa SB, Hennekam RC. Nicolaides-Baraitser Syndrome International, C, 2014. Phenotype and genotype in Nicolaides-Baraitser syndrome. Am J Med Genet C Semin Med Genet. 2014;166C:302–314. doi: 10.1002/ajmg.c.31409. [DOI] [PubMed] [Google Scholar]
- Spruijt CG, Grawe C, Kleinendorst SC, Baltissen MPA, Vermeulen M. Cross-linking mass spectrometry reveals the structural topology of peripheral NuRD subunits relative to the core complex. FEBS J. 2020 doi: 10.1111/febs.15650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staahl BT, Tang J, Wu W, Sun A, Gitler AD, Yoo AS, Crabtree GR. Kinetic analysis of npBAF to nBAF switching reveals exchange of SS18 with CREST and integration with neural developmental pathways. J Neurosci. 2013;33:10348–10361. doi: 10.1523/JNEUROSCI.1258-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stankiewicz P, Khan TN, Szafranski P, Slattery L, Streff H, Vetrini F, Bernstein JA, Brown CW, Rosenfeld JA, Rednam S, Scollon S, et al. Haploinsufficiency of the chromatin remodeler BPTF causes syndromic developmental and speech delay, postnatal microcephaly, and dysmorphic features. Am J Hum Genet. 2017;101:503–515. doi: 10.1016/j.ajhg.2017.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein DJ, Szatmari P, Gaebel W, Berk M, Vieta E, Maj M, deVries YA, Roest AM, de Jonge P, Maercker A, Brewin CR, et al. Mental, behavioral and neurodevelopmental disorders in the ICD-11: an international perspective on key changes and controversies. BMC Med. 2020;18:21. doi: 10.1186/s12916-020-1495-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stessman HA, Xiong B, Coe BP, Wang T, Hoekzema K, Fenckova M, Kvarnung M, Gerdts J, Trinh S, Cosemans N, Vives L, et al. Targeted sequencing identifies 91 neurodevelopmental-disorder risk genes with autism and developmental-disability biases. Nat Genet. 2017;49:515–526. doi: 10.1038/ng.3792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugitani Y, Nakai S, Minowa O, Nishi M, Jishage K, Kawano H, Mori K, Ogawa M, Noda T. Brn-1 and Brn-2 share crucial roles in the production and positioning of mouse neocortical neurons. Genes Dev. 2002;16:1760–1765. doi: 10.1101/gad.978002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suls A, Jaehn JA, Kecskes A, Weber Y, Weckhuysen S, Craiu DC, Siekierska A, Djemie T, Afrikanova T, Gormley P, von Spiczak S, et al. De novo loss-of-function mutations in CHD2 cause a fever-sensitive myoclonic epileptic encephalopathy sharing features with Dravet syndrome. Am J Hum Genet. 2013;93:967–975. doi: 10.1016/j.ajhg.2013.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Miao N, Sun T. Detect accessible chromatin using ATAC-sequencing, from principle to applications. Hereditas. 2019;156 doi: 10.1186/s41065-019-0105-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabar MS, Giardina C, Feng YJ, Francis H, Sani HM, Low JKK, Mackay JP, Bailey CG, Rasko JEJ. 2021. [DOI]
- Takenouchi T, Yoshihashi H, Sakaguchi Y, Uehara T, Honda M, Takahashi T, Kosaki K, Miyama S. Hirschsprung disease as a yet undescribed phenotype in a patient with ARID1B mutation. Am J Med Genet. 2016;170:3249–3252. doi: 10.1002/ajmg.a.37861. [DOI] [PubMed] [Google Scholar]
- Talkowski ME, Rosenfeld JA, Blumenthal I, Pillalamarri V, Chiang C, Heilbut A, Ernst C, Hanscom C, Rossin E, Lindgren AM, Pereira S, et al. Sequencing chromosomal abnormalities reveals neurodevelopmental loci that confer risk across diagnostic boundaries. Cell. 2012;149:525–537. doi: 10.1016/j.cell.2012.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang T, Zhang Y, Wang Y, Cai Z, Lu Z, Li L, Huang R, Hagelkruys A, Matthias P, Zhang H, et al. HDAC1 and HDAC2 regulate intermediate progenitor positioning to safeguard neocortical development. Neuron. 2019;101:1117–1133.:e1115. doi: 10.1016/j.neuron.2019.01.007. [DOI] [PubMed] [Google Scholar]
- Tapias A, Zhou ZW, Shi Y, Chong Z, Wang P, Groth M, Platzer M, Huttner W, Herceg Z, Yang YG, Wang ZQ. Trrap-dependent histone acetylation specifically regulates cell-cycle gene transcription to control neural progenitor fate decisions. Cell Stem Cell. 2014;14:632–643. doi: 10.1016/j.stem.2014.04.001. [DOI] [PubMed] [Google Scholar]
- Telley L, Agirman G, Prados J, Amberg N, Fievre S, Oberst P, Bartolini G, Vitali I, Cadilhac C, Hippenmeyer S, Nguyen L, et al. Temporal patterning of apical progenitors and their daughter neurons in the developing neocortex. Science. 2019;364 doi: 10.1126/science.aav2522. http://genebrowser.unige.ch/telagirdon/ [DOI] [PubMed] [Google Scholar]
- Telley L, Govindan S, Prados J, Stevant I, Nef S, Dermitzakis E, Dayer A, Jabaudon D. Sequential transcriptional waves direct the differentiation of newborn neurons in the mouse neocortex. Science. 2016;351:1443–1446. doi: 10.1126/science.aad8361. http://genebrowser.unige.ch/science2016/ [DOI] [PubMed] [Google Scholar]
- Toffolo E, Rusconi F, Paganini L, Tortorici M, Pilotto S, Heise C, Verpelli C, Tedeschi G, Maffioli E, Sala C, Mattevi A, Battaglioli E. Phosphorylation of neuronal Lysine-Specific Demethylase 1LSD1/KDM1A impairs transcriptional repression by regulating interaction with CoREST and histone deacetylases HDAC1/2. J Neurochem. 2014;128:603–616. doi: 10.1111/jnc.12457. [DOI] [PubMed] [Google Scholar]
- Tole S, Hebert J. Telencephalon Patterning. 2013.
- Tong JK, Hassig CA, Schnitzler GR, Kingston RE, Schreiber SL. Chromatin deacetylation by an ATP-dependent nucleosome remodelling complex. Nature. 1998;395:917–921. doi: 10.1038/27699. [DOI] [PubMed] [Google Scholar]
- Trevino AE, Sinnott-Armstrong N, Andersen J, Yoon SJ, Huber N, Pritchard JK, Chang HY, Greenleaf WJ, Pasca SP. Chromatin accessibility dynamics in a model of human forebrain development. Science. 2020;367 doi: 10.1126/science.aay1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trivisano M, Striano P, Sartorelli J, Giordano L, Traverso M, Accorsi P, Cappelletti S, Claps DJ, Vigevano F, Zara F, Specchio N. CHD2 mutations are a rare cause of generalized epilepsy with myoclonic-atonic seizures. Epilepsy Behav. 2015;51:53–56. doi: 10.1016/j.yebeh.2015.06.029. [DOI] [PubMed] [Google Scholar]
- Tsuboi M, Kishi Y, Yokozeki W, Koseki H, Hirabayashi Y, Gotoh Y. Ubiquitination-independent repression of PRC1 targets during neuronal fate restriction in the developing mouse neocortex. Dev Cell. 2018;47:758–772.:e755. doi: 10.1016/j.devcel.2018.11.018. [DOI] [PubMed] [Google Scholar]
- Tsurusaki Y, Okamoto N, Ohashi H, Kosho T, Imai Y, Hibi-Ko Y, Kaname T, Naritomi K, Kawame H, Wakui K, Fukushima Y, et al. Mutations affecting components of the SWI/SNF complex cause Coffin-Siris syndrome. Nat Genet. 2012;44:376–378. doi: 10.1038/ng.2219. [DOI] [PubMed] [Google Scholar]
- Tunovic S, Barkovich J, Sherr EH, Slavotinek AM. Denovo ANKRD11 and KDM1A gene mutations in a male with features of KBG syndrome and Kabuki syndrome. Am J Med Genet. 2014;164A:1744–1749. doi: 10.1002/ajmg.a.36450. [DOI] [PubMed] [Google Scholar]
- Tuoc TC, Boretius S, Sansom SN, Pitulescu ME, Frahm J, Livesey FJ, Stoykova A. Chromatin regulation by BAF170 controls cerebral cortical size and thickness. Dev Cell. 2013a;25:256–269. doi: 10.1016/j.devcel.2013.04.005. [DOI] [PubMed] [Google Scholar]
- Tuoc TC, Narayanan R, Stoykova A. BAF chromatin remodeling complex: cortical size regulation and beyond. Cell Cycle. 2013b;12:2953–2959. doi: 10.4161/cc.25999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuoc TC, Pavlakis E, Tylkowski MA, Stoykova A. Control of cerebral size and thickness. Cell Mol Life Sci. 2014;71:3199–3218. doi: 10.1007/s00018-014-1590-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda K, Yanagi K, Kaname T, Okamoto N. A novel mutation in the GATAD2B gene associated with severe intellectual disability. Brain Dev. 2019;41:276–279. doi: 10.1016/j.braindev.2018.10.003. [DOI] [PubMed] [Google Scholar]
- van Elburg RA, van Ooyen A. Impact of dendritic size and dendritic topology on burst firing in pyramidal cells. PLoS Comput Biol. 2010;6:e1000781. doi: 10.1371/journal.pcbi.1000781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vissers LE, van Ravenswaaij CM, Admiraal R, Hurst JA, deVries BB, Janssen IM, van der Vliet WA, Huys EH, de Jong PJ, Hamel BC, Schoenmakers EF, et al. Mutations in a new member of the chromodomain gene family cause CHARGE syndrome. Nat Genet. 2004;36:955–957. doi: 10.1038/ng1407. [DOI] [PubMed] [Google Scholar]
- Wade PA, Jones PL, Vermaak D, Wolffe AP. A multiple subunit Mi-2 histone deacetylase from Xenopus laevis cofractionates with an associated Snf2 superfamily ATPase. Curr Biol. 1998;8:843–846. doi: 10.1016/S0960-9822(98)70328-8. [DOI] [PubMed] [Google Scholar]
- Wakeling EL, Brioude F, Lokulo-Sodipe O, O’Connell SM, Salem J, Bliek J, Canton AP, Chrzanowska KH, Davies JH, Dias RP, Dubern B. Diagnosis and management of Silver-Russell syndrome: first international consensus statement. Nat Rev Endocrinol. 2017;13:105–124. doi: 10.1038/nrendo.2016.138. [DOI] [PubMed] [Google Scholar]
- Wang P, Mokhtari R, Pedrosa E, Kirschenbaum M, Bayrak C, Zheng D, Lachman HM. CRISPR/Cas9-mediated heterozygous knockout of the autism gene CHD8 and characterization of its transcriptional networks in cerebral organoids derived from iPS cells. Mol Autism. 2017;8:11. doi: 10.1186/s13229-017-0124-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Xue Y, Zhou S, Kuo AA, Cairns BR, Crabtree GR. Diversity and specialization of mammalian SWI/SNF complexes. Genes Dev. 1996;10:2117–2130. doi: 10.1101/gad.10.17.2117. [DOI] [PubMed] [Google Scholar]
- Wang Y, Zhang H, Chen Y, Sun Y, Yang F, Yu W, Liang J, Sun L, Yang X, Shi L, Li R, et al. LSD1 is a subunit of the NuRD complex and targets the metastasis programs in breast cancer. Cell. 2009;138:660–672. doi: 10.1016/j.cell.2009.05.050. [DOI] [PubMed] [Google Scholar]
- Weaver IC, Hellstrom IC, Brown SE, Andrews SD, Dymov S, Diorio J, Zhang TY, Szyf M, Meaney MJ. The methylated-DNA binding protein MBD2 enhances NGFI-A (egr-1)-mediated transcriptional activation of the glucocorticoid receptor. Philos Trans R Soc Lond B Biol Sci. 2014;369 doi: 10.1098/rstb.2013.0513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss K, Lazar HP, Kurolap A, Martinez AF, Paperna T, Cohen L, Smeland MF, Whalen S, Heide S, Keren B, Terhal P, et al. TheCHD4-related syndrome: a comprehensive investigation of the clinical spectrum, genotype-phenotype correlations, and molecular basis. Genet Med. 2020;22:389–397. doi: 10.1038/s41436-019-0612-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weitensteiner V, Zhang R, Bungenberg J, Marks M, Gehlen J, Ralser DJ, Hilger AC, Sharma A, Schumacher J, Gembruch U, Merz WM, et al. Exome sequencing in syndromic brain malformations identifies novel mutations in ACTB, and SLC9A6, and suggests BAZ1A as a new candidate gene. Birth Defects Res. 2018;110:587–597. doi: 10.1002/bdr2.1200. [DOI] [PubMed] [Google Scholar]
- Wenderski W, Wang L, Krokhotin A, Walsh JJ, Li H, Shoji H, Ghosh S, George RD, Miller EL, Elias L, Gillespie MA, et al. Loss of theneural-specific BAF subunit ACTL6B relieves repression of early response genes and causes recessive autism. Proc Natl Acad Sci U S A. 2020;117:10055–10066. doi: 10.1073/pnas.1908238117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieczorek D, Bogershausen N, Beleggia F, Steiner-Haldenstatt S, Pohl E, Li Y, Milz E, Martin M, Thiele H, Altmuller J, Alanay Y, et al. A comprehensive molecular study on Coffin-Siris and Nicolaides-Baraitser syndromes identifies a broad molecular and clinical spectrum converging on altered chromatin remodeling. Hum Mol Genet. 2013;22:5121–5135. doi: 10.1093/hmg/ddt366. [DOI] [PubMed] [Google Scholar]
- Wiegreffe C, Simon R, Peschkes K, Kling C, Strehle M, Cheng J, Srivatsa S, Liu P, Jenkins NA, Copeland NG, Tarabykin V, et al. Bcl11a (Ctip1) controls migration of cortical projection neurons through regulation of Sema3c. Neuron. 2015;87:311–325. doi: 10.1016/j.neuron.2015.06.023. [DOI] [PubMed] [Google Scholar]
- Willemsen MH, Nijhof B, Fenckova M, Nillesen WM, Bongers EM, Castells-Nobau A, Asztalos L, Viragh E, van Bon BW, Tezel E, Veltman JA, et al. GATAD2B loss-of-function mutations cause a recognisable syndrome with intellectual disability and are associated with learning deficits and synaptic undergrowth in Drosophila. J Med Genet. 2013;50:507–514. doi: 10.1136/jmedgenet-2012-101490. [DOI] [PubMed] [Google Scholar]
- Willhoft O, Wigley DB. INO80 and SWR1 complexes: the non-identical twins of chromatin remodelling. Curr Opin Struct Biol. 2020;61:50–58. doi: 10.1016/j.sbi.2019.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff D, Endele S, Azzarello-Burri S, Hoyer J, Zweier M, Schanze I, Schmitt B, Rauch A, Reis A, Zweier C. In-frame deletion and missense mutations of the C-terminal helicase domain of SMARCA2 in three patients with nicolaides-baraitser syndrome. Mol Syndromol. 2012;2:237–244. doi: 10.1159/000337323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodworth MB, Greig LC, Liu KX, Ippolito GC, Tucker HO, Macklis JD. Ctip1 regulates the balance between specification of distinct projection neuron subtypes in deep cortical layers. Cell Rep. 2016;15:999–1012. doi: 10.1016/j.celrep.2016.03.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu JI, Lessard J, Crabtree GR. Understanding the words of chromatin regulation. Cell. 2009;136:200–206. doi: 10.1016/j.cell.2009.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu JI, Lessard J, Olave IA, Qiu Z, Ghosh A, Graef IA, Crabtree GR. Regulation of dendritic development by neuron-specific chromatin remodeling complexes. Neuron. 2007;56:94–108. doi: 10.1016/j.neuron.2007.08.021. [DOI] [PubMed] [Google Scholar]
- Wyatt BH, Raymond TO, Lansdon LA, Darbro BW, Murray JC, Manak JR, Dickinson AJG. Using an aquatic model, Xenopus laevis, to uncover the role of chromodomain 1 in craniofacial disorders. Genesis. 2021;59:e23394. doi: 10.1002/dvg.23394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y, Castro-Hernandez R, Sokpor G, Pham L, Narayanan R, Rosenbusch J, Staiger JF, Tuoc T. RBM15 modulates the function of chromatin remodeling factor BAF155 through RNA methylation in developing cortex. Mol Neurobiol. 2019;56:7305–7320. doi: 10.1007/s12035-019-1595-1. [DOI] [PubMed] [Google Scholar]
- Xue Y, Wong J, Moreno GT, Young MK, Côté J, Wang W. NURD, a novel complex with both ATP-dependent chromatin-remodeling and histone deacetylase activities. Mol Cell. 1998;2:851–861. doi: 10.1016/S1097-2765(00)80299-3. [DOI] [PubMed] [Google Scholar]
- Yan L, Wang L, Tian Y, Xia X, Chen Z. Structure and regulation of the chromatin remodeller ISWI. Nature. 2016;540:466–469. doi: 10.1038/nature20590. [DOI] [PubMed] [Google Scholar]
- Yang M, Gocke CB, Luo X, Borek D, Tomchick DR, Machius M, Otwinowski Z, Yu H. Structural basis for CoREST-dependent demethylation of nucleosomes by the human LSD1 histone demethylase. Mol Cell. 2006;23:377–387. doi: 10.1016/j.molcel.2006.07.012. [DOI] [PubMed] [Google Scholar]
- Yao B, Christian KM, He C, Jin P, Ming GL, Song H. Epigenetic mechanisms in neurogenesis. Nat Rev Neurosci. 2016;17:537–549. doi: 10.1038/nrn.2016.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasin H, Gibson WT, Langlois S, Stowe RM, Tsang ES, Lee L, Poon J, Tran G, Tyson C, Wong CK, Marra MA, et al. A distinct neurodevelopmental syndrome with intellectual disability, autism spectrum disorder, characteristic facies, and macrocephaly is caused by defects in CHD8. J Hum Genet. 2019;64:271–280. doi: 10.1038/s10038-019-0561-0. [DOI] [PubMed] [Google Scholar]
- Yasin H, Zahir FR. Chromodomain helicase DNA-binding proteins and neurodevelopmental disorders. Journal of Translational Genetics and Genomics. 2020 doi: 10.20517/jtgg.2020.30. [DOI] [Google Scholar]
- Ye F, Chen Y, Hoang T, Montgomery RL, Zhao XH, Bu H, Hu T, Taketo MM, van Es JH, Clevers H, Hsieh J, et al. HDAC1 and HDAC2 regulate oligodendrocyte differentiation by disrupting the beta-catenin-TCF interaction. Nat Neurosci. 2009;12:829–838. doi: 10.1038/nn.2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh WH, Chiang H, Rees HA, Edge ASB, Liu DR. In vivo base editing of post-mitotic sensory cells. Nat Commun. 2018;9:2184. doi: 10.1038/s41467-018-04580-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yip DJ, Corcoran CP, Alvarez-Saavedra M, DeMaria A, Rennick S, Mears AJ, Rudnicki MA, Messier C, Picketts DJ. Snf2l regulates Foxg1-dependent progenitor cell expansion in the developing brain. Dev Cell. 2012;22:871–878. doi: 10.1016/j.devcel.2012.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo AS, Crabtree GR. ATP-dependent chromatin remodeling in neural development. Curr Opin Neurobiol. 2009;19:120–126. doi: 10.1016/j.conb.2009.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo AS, Staahl BT, Chen L, Crabtree GR. MicroRNA-mediated switching of chromatin-remodelling complexes in neural development. Nature. 2009;460:642–646. doi: 10.1038/nature08139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan W, Ma S, Brown JR, Kim K, Murek V, Trastulla L, Meissner A, Lodato S, Shetty A, Levin JZ, Buenrostro JD, et al. 2020. [DOI] [PMC free article] [PubMed]
- Yuksel Z, Yazol M, Gumus E. Pathogenic homozygous variations in ACTL6B cause DECAM syndrome: developmental delay, Epileptic encephalopathy, Cerebral Atrophy, and abnormal Myelination. Am J Med Genet. 2019;179:1603–1608. doi: 10.1002/ajmg.a.61210. [DOI] [PubMed] [Google Scholar]
- Zaghlool A, Halvardson J, Zhao JJ, Etemadikhah M, Kalushkova A, Konska K, Jernberg-Wiklund H, Thuresson AC, Feuk L. A role for the chromatin-remodeling factor BAZ1A in neurodevelopment. Hum Mutat. 2016;37:964–975. doi: 10.1002/humu.23034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahir F, Firth HV, Baross A, Delaney AD, Eydoux P, Gibson WT, Langlois S, Martin H, Willatt L, Marra MA, Friedman JM. Novel deletions of 14q11.2 associated with developmental delay, cognitive impairment and similar minor anomalies in three children. J Med Genet. 2007;44:556–561. doi: 10.1136/jmg.2007.050823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zepeda-Mendoza C, Goodenberger ML, Kuhl A, Rice GM, Hoppman N. Familial segregation of a 5q15-q21.2 deletion associated with facial dysmorphism and speech delay. Clin Case Rep. 2019;7:1154–1160. doi: 10.1002/ccr3.2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Xu D, Yuan L, Sun Y, Xu Z. Epigenetic regulation of Atrophin1 by lysine-specific demethylase 1 is required for cortical progenitor maintenance. Nat Commun. 2014;5:5815. doi: 10.1038/ncomms6815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Chen S, Qin H, Yuan H, Pi Y, Yang Y, Huang H, Li G, Sun Y, Wang Z, Ma H, et al. Novel genotypes and phenotypes among Chinese patients with Floating-Harbor syndrome. Orphanet J Rare Dis. 2019;14(144) doi: 10.1186/s13023-019-1111-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R, He H, Yuan B, Wu Z, Wang X, Du Y, Chen Y, Qiu Z. An intronic variant of CHD7 identified in autism patients interferes with neuronal differentiation and development. Research Square Preprint. 2020 doi: 10.21203/rs.3.rs-58118/v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, LeRoy G, Seelig H-P, Lane WS, Reinberg D. The dermatomyositis-specific autoantigen Mi2 is a component of a complex containing histone deacetylase and nucleosome remodeling activities. Cell. 1998;95:279–289. doi: 10.1016/S0092-8674(00)81758-4. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Ng H-H, Erdjument-Bromage H, Tempst P, Bird A, Reinberg D. Analysis of the NuRD subunits reveals a histone deacetylase core complex and a connection with DNA methylation. Genes Dev. 1999;13:1924–1935. doi: 10.1101/gad.13.15.1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zibetti C, Adamo A, Binda C, Forneris F, Toffolo E, Verpelli C, Ginelli E, Mattevi A, Sala C, Battaglioli E. Alternative splicing of the histone demethylase LSD1/KDM1 contributes to the modulation of neurite morphogenesis in the mammalian nervous system. J Neurosci. 2010;30:2521–2532. doi: 10.1523/JNEUROSCI.5500-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]