Abstract
During cancer evolution constituent tumour cells compete under dynamic selection pressures. Phenotypic variation can be observed as intratumour heterogeneity, which is propagated by genome instability leading to mutations, somatic copy number alterations and epigenomic changes. TRACERx was set up in 2014 to observe the relationship between intratumour heterogeneity and patient outcome. By integrating multi-region sequencing of primary tumours with longitudinal sampling of a prospectively recruited patient cohort, cancer evolution can be tracked from early to late stage disease and through therapy. Here we review some of the key features of the studies and look to the future of the field.
Introduction
Over the last decade, the study of tumour evolution has expanded significantly, aided by the advent of technologies that enable rapid and relatively inexpensive sequencing of cancer exomes and genomes. This has shaped our understanding of tumour diversification and evolution, with genetic and non-genetic variation providing the basis for intratumour heterogeneity. By tracking both clonal (present within all tumour cells) and subclonal (present within a subset of tumour cells) mutations, tumours have been found to follow distinct evolutionary patterns. For instance, in some tumours there is evidence for a Darwinian pattern of evolution with selection of subclonal mutations, whereas in others there appears to be a neutral accumulation of mutations following a clonal driver event (1).
A key focus of research has been to link these observations to clinical outcomes, such as prognosis and treatment response, in order to understand the driving forces behind cancer progression, and to develop personalised approaches to cancer medicine. The national observational longitudinal studies, TRACERx (Tracking Cancer Evolution through Therapy) Lung (ClinicalTrials.gov Identifier: NCT01888601) and Renal (ClinicalTrials.gov Identifier: NCT03226886) have attempted to address this (2,3). Observations from the first 100 patients recruited to the TRACERx Lung and Renal studies have been published in a series of reports, with data collection now complete for the first 421 patients in the TRACERx Lung study. These studies involve sampling of primary, recurrent and subsequently progressive disease, as well as ‘liquid’ biopsies to isolate circulating tumour DNA (ctDNA) and circulating tumour cells (CTCs) from blood samples.
The TRACERx studies
The primary objectives of the TRACERx studies are to determine the extent of intratumour heterogeneity, establish its relationship with clinical outcome and the impact of adjuvant and other therapies on the cancer genomic landscape at recurrence.
Multi-region sampling and sequencing of surgically resected specimens in TRACERx allows for the detailed assessment of intratumour heterogeneity. However, the majority of sampling at relapse or progression, where clinically safe and possible, is limited to single-lesion biopsies. Invasive biopsies are avoided if the procedure is deemed to be high-risk, either due to the patient’s co-morbidities or the site of disease. ctDNA analysis in this context can capture genetic heterogeneity in a manner not limited to a single site of disease. Furthermore, co-recruitment into the Cancer Research UK (CRUK) funded national research autopsy study, PEACE (Posthumous Evaluation of Advanced Cancer Environment, ClinicalTrials.gov Identifier: NCT03004755), allows for extensive sampling of all metastatic disease sites in the post-mortem setting.
Whilst the benefit of retrospective analysis of data from significant resource projects such as The Cancer Genome Atlas and Pan Cancer Analysis of Whole Genomes cannot be overstated (4,5), large-scale longitudinal prospective studies of patients and their cancer genomes with clinical histories (case report forms), imaging and pathology analysis matched with paired recurrence specimens and ctDNA/CTC acquisitions are rare. Typically, genomic studies of cancer provide only a snapshot of the genomic landscape and seldom contextualise this temporally or spatially in a cohort of prospectively recruited patients in a real-world clinical setting with standard-of-care therapy to minimise confounding effects of treatment discordance across clinical sites. Furthermore, sampling, sequencing and processing protocols may be non-uniform, and the plethora of computational tools for interrogating genomic data may introduce technical inconsistencies and limit the reliability of subsequent retrospective analyses. Crucially, independent pathological review of specimens may not be performed, and clinical annotation is often absent or incomplete. Such studies frequently consider the cancer genome in isolation and in the absence of other features including its transcriptome or epigenome, as well as the immune-microenvironment, metabolome and microbiome. There is therefore an unmet scientific and clinical need for prospective, longitudinal evolutionary cohort studies of cancer ‘multi-omics’ with robust clinico-pathological correlates.
Here, we outline some of the lessons learnt from TRACERx longitudinal analyses, the importance of future such studies in other tumour types, and the questions outstanding in the field, offering a roadmap for cancer evolutionary studies in the coming decade.
Setting up a prospective longitudinal study in cancer evolution
TRACERx has galvanised a national research infrastructure linking surgical oncology, radiology, pathology, radiation and medical oncology disciplines, with trial specific procedures (TSPs) set up across multiple UK hospital sites to sample tumours with a view to performing detailed genomic and immunological analyses, alongside ctDNA and CTC analysis. Samples are tracked using a centralised database enabling sample oversight across all sites. Detailed case report forms are collected routinely from patients at each clinic visit recording their general medical and oncological history. In the adjuvant setting, patients are offered NICE-approved (National Institute for health and Care Excellence) standardised adjuvant protocols across all hospital sites according to disease stage, minimising the confounding variables associated with treatments differing between centres. This has established a unique clinical, pathological and scientific dataset. Collaborative efforts between the clinical trials centre, hospital trusts and research laboratories have been essential in overcoming some of the logistical challenges posed from sample collection to patient follow up.
Despite decades of research into non-small cell lung cancer (NSCLC), clinical outcomes for patients remain poor, with a 5-year relative survival of 26.5% reported in the November 2019 Surveillance, Epidemiology, and End Results Program (SEER) data submission (https://seer.cancer.gov/csr/1975_2017/). Moreover, significant numbers of patients with early-stage disease will go on to relapse despite surgery (long term survival is less than 50% following surgery (6)) and adjuvant chemotherapy. Although molecular driver events such as TP53 and KRAS mutations, as well as EGFR mutations or amplifications, and genome instability, are well-described in NSCLC (7), there remains an unmet need for insight into its biology and its relationship with the clinical course.
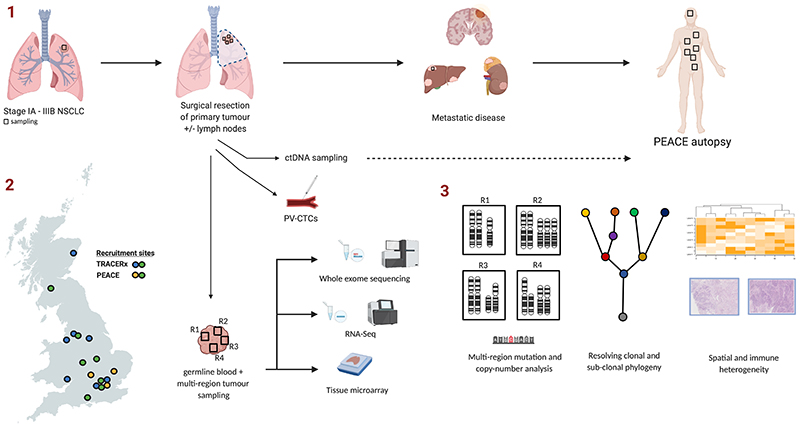
The structure of TRACERx Lung is illustrated in Figure 1. Following a diagnosis of early stage NSCLC, patients eligible for TRACERx Lung receive routine standard of care, undergoing surgery to resect their primary tumour with or without adjuvant chemotherapy depending on the disease stage. Multiple regions from the primary tumour (and in some cases lymph nodes) are obtained for analysis. Patients are followed up for at least 5 years and in patients with recurrent disease, tissue biopsies are obtained from the site of relapse where possible. TRACERx Lung has a target accrual of 842 patients from 14 hospital sites across the United Kingdom.
Figure 1. Outline of TRACERx Lung and PEACE autopsy studies.
1. Following a diagnosis of early stage non-small cell lung cancer, patients undergo surgery to resect their primary tumour (with adjuvant chemotherapy, depending on disease stage). Multiple regions from the primary tumour are sampled. In the event of recurrent disease, biopsies are obtained where possible. Patients that are enrolled in the PEACE study have multiple sites of disease sampled for analysis. Tumour samples are processed for whole exome sequencing, RNA-seq and tissue microarrays. 2. Recruitment sites for TRACERx and PEACE in the UK. 3. The core analysis of tumour samples involves calling clonal and subclonal mutation and copy number alterations, with construction of phylogenetic trees. Combining tissue microarray, RNA-seq data and other techniques has facilitated the study of immune and spatial heterogeneity. PV-CTC, pulmonary vein circulating tumour cells.
TRACERx Renal is a separate prospective multi-region study of primary tumours and paired metastatic lesions within 320 patients with clear cell renal cell carcinoma (ccRCC). Like NSCLC, surgery can be curative in early stage ccRCC, and survival rates are on average superior to those of NSCLC (exceeding 50%), but with significant heterogeneity in clinical outcome; approximately one-third of patients with localised ccRCC will relapse after surgical resection. Of note, surgery plays a significant role in the management of metastatic ccRCC, offering the chance to examine the primary tumour across all stages of disease, as well as access to paired metastatic disease. Previous studies had highlighted the common driver events of this disease, including loss of chromosome 3p which harbours four tumour suppressor genes: VHL, SETD2, BAP1 and PBRM1 all of which are frequently lost through mutation of the remaining allele. Significant intratumour heterogeneity of mutations and somatic copy number alterations (SCNAs) had also been described (8,9); TRACERx Renal aimed to explore associations between this intratumour heterogeneity and the diversity of clinical outcomes.
The target accrual of 842 in the Lung cohort was required to detect at least a 23% relative risk reduction and a 10% improvement in 5-year overall survival, comparing tumours with low and high intratumour heterogeneity and assuming a median disease-free survival of 30 months following surgery. For the TRACERx Renal study, a sample size of 320 was required to detect an association between intratumour heterogeneity and disease stage. Assuming a median overall disease survival of 15 months, this sample size was predicted to have enough power to detect a six-month difference in disease-free survival comparing low and high intratumour heterogeneity scores within stages I-III. Prospective studies of this size are essential to effectively integrate complex genomic and pathological data and make meaningful, clinically applicable conclusions in the context of standard-of-care clinical practice whilst mitigating statistical problems such as multiple testing that can plague small or retrospective analyses.
The dynamic fitness landscapes of different tumours throughout the disease course can share certain properties and be classified accordingly. Mapping evolutionary trajectories of the cancer genome throughout the disease course requires consideration of the myriad of ways in which tumours can adapt to selection pressures applied by a diverse tumour microenvironment. TRACERx uses multi-region whole exome sequencing (WES) to study the mutation and copy number landscape. This is complimented by RNA sequencing, T cell receptor sequencing, reduced-representation bisulfite sequencing, immunohistochemistry, fluorescent in-situ hybridisation, flow cytometry and imaging mass cytometry to reconcile mutational, copy number, epigenomic and transcriptomic heterogeneity in the context of the tumour and its immune microenvironment. Longitudinal blood sampling for CTCs and ctDNA allows for correlative genomic analysis between tissue and blood. Tissue microarrays are constructed for specialised immunohistochemistry assays and fresh frozen tumour samples are used for DNA and RNA extraction, improving the quality of downstream sequencing data relative to formalin-fixed, paraffin-embedded (FFPE) samples. Sequencing data is processed through a uniform bioinformatic pipeline. Published data are made available for clinical and scientific communities through a Data Access Committee overseen by the Cancer Research UK & University College London Cancer Trials Centre.
Patients who subsequently develop recurrent disease after initial curative surgery, where possible, undergo a biopsy to confirm relapse or to exclude a new cancer primary. Depending on the site of the disease, ease of access for biopsy, the performance status of the patient and associated co-morbidities, a decision can be made on the appropriateness of biopsy sampling. Despite the clinical indications for sampling in the relapse setting, all research biopsies are costed to support the interventional teams at the various hospital sites.
TRACERx patients who suffer disease relapse are, where sensitive and appropriate discussions allow, co-recruited into the PEACE study, allowing for sampling from all sites of disease in the post-mortem setting. The PEACE study has highlighted unique logistical challenges in relation to autopsies proceeding in a timely manner within National Health Service mortuary services and resources.
The COVID-19 pandemic has caused widespread disruption to cancer care and cancer clinical trials (10), with an inevitable impact on patient recruitment and follow up. Despite this, where possible funding bodies have supported research staff to persevere in delivering the research promised by studies such as TRACERx and PEACE.
Insights from TRACERx
Study Background
Intratumour heterogeneity has long been described in cancer and proposed as evidence for evolutionary processes at play in cancer development and progression. Early observations from Nowell, who described phenotypic heterogeneity within cancers, and Goldie, who postulated a link between genomic instability, tumour heterogeneity and drug resistance, supported a process of Darwinian clonal evolution in tumour biology (11,12).
Subsequently, retrospective studies from our group and others, prior to TRACERx, demonstrated that intratumour heterogeneity results from branched tumour evolution (8,13–17). Navin and colleagues used array comparative genomic hybridisation to illustrate differences between ploidy profiles of different tumours (13). Tumours showing ‘polygenomic’ profiles, indicating the presence of genomic heterogeneity, were common, and descent of different clones from a common ancestor was demonstrated. The advent of next-generation sequencing has also provided significant insight. For example, Greaves and colleagues demonstrated that acute lymphoblastic leukaemia evolves through branched, non-linear evolution producing distinct subclones (14). Serial transplantation in mouse models demonstrated that these different clones were functional and able to potentiate novel cancers. Our group reported intratumour heterogeneity within driver mutations for ccRCC, and suggested selection of subclonal mutations late in tumour evolution (8). In addition, subclonal driver mutations were also a predictor of poor prognosis in a longitudinal cohort of chronic lymphocytic leukaemia, and low frequency mutations present prior to treatment shown to contribute to resistance to targeted therapy (16,18).
Before TRACERx, it was understood that SCNAs contributed to intratumour heterogeneity (19). Exploration of the SCNA landscape across cancer types had demonstrated an association between the presence of SCNAs, genomic instability and poor clinical outcome, both in terms of survival and treatment resistance (20,21). However, the impact of the rate of acquisition of these events, reflecting the degree of ongoing chromosomal instability (CIN), upon clinical outcome was not known. In addition, whole-genome doubling events had been characterised across cancer types (22), but the relevance of this event to tumour evolution, as well as the extent to which CIN provides the variation that acts as the substrate for selection was unclear.
Studies of the prognostic or predictive relevance of intratumour heterogeneity had previously been limited to small, retrospective cohorts. TRACERx aimed to explore this question prospectively, attempting to address whether somatic heterogeneity might be associated with poor clinical outcome. The major findings are summarised in Table 1.
Table 1. Summary of key findings from TRACERx.
| Study | Summary | Tumour type | Samples | Key finding |
|---|---|---|---|---|
| Jamal-Hanjani et al, 2017 | Prospective analysis of intratumour heterogeneity | Non-small cell lung cancer (NSCLC) | Whole-exome sequencing (WES) of 327 biopsies from 100 patients | ITH of copy number events, but not mutations, is associated with adverse outcome |
| Turajlic et al, 2018 | Analysis of evolutionary trajectories | Clear-cell renal cell carcinoma (ccRCC) | 1,206 primary tumour regions from 101 patients | Different evolutionary subtypes, defined by features such as the clonality of driver mutations or aneuploidy, correlate with distinct clinical phenotypes |
| Turajlic et al, 2018 | Analysis of paired primary and metastasis | ccRCC | 575 primary and 335 metastatic biopsies from 100 patients | Genomic features, such as chromosomal complexity and loss of 9p, promote metastatic competence |
| Mitchell et al, 2018 | Multi-region analysis timing landmark events | ccRCC | Whole-genome sequencing of 95 biopsies from 33 patients | Loss of chromosome 3p, an important early event in ccRCC development, can occur in childhood and precede tumour development by 30-50 years. |
| Lopez et al, 2020 | Study of whole-genome duplication (WGD) events and deleterious events in tumour evolution | Pan-cancer | TRACERx 100 and TCGA cohorts | WGD is enriched in tumour types with a high rate of loss-of-heterozygosity and selected for when deleterious alterations are acquired at a high rate, suggesting WGD events may mitigate against the impact of deleterious alterations in tumour evolution. |
| Watkins et al, 2020 | Analysis of somatic copy-number alterations (SCNAs) in primary tumours and metastases that have undergone multi-region sampling | Pan-cancer | WES of 1,421 samples from 394 tumours, and 1,024 independent metastatic samples | SCNAs heterogeneity provides the substrate for ongoing tumour evolution; over one third of tumours show evidence of parallel evolution of SCNAs, and certain SCNAs recurred at high frequency subclonally, and in metastasis |
| Rosenthal et al, 2019 | Study of methods of immune evasion | NSCLC | WES, RNA-seq and tumour-infiltrating lymphocyte estimates from 258 regions of 88 tumours | Tumours experience strong selection pressures to evade immune surveillance early in tumour evolution, and develop heterogeneous methods of immune escape |
| Joshi et al, 2019 | Exploration of the T-cell receptor repertoire | NSCLC | 220 tumour regions, 64 nontumour lung tissue samples and 56 peripheral blood mononuclear cell samples from 72 patients | T-cell receptor heterogeneity varies between tumours, and is influenced regionally by genomic heterogeneity. TCRs present in blood samples can reflect tumoural T cell phenotype |
| Ghorani et al, 2020 | Study of the impact of tumour mutation burden (TMB) upon T cell differentiation | NSCLC | Flow cytometry of 63 tumour regions from 31 patients | TMB drives intratumoural T cell differentiation, and is associated with loss of progenitor-like and increased abundance in dysfunctional T-cell phenotypes; in NSCLC, this skewing is associated with poor clinical outcome. |
| Abduljabbbar et al, 2020 | Spatial histological analysis of lung cancer cohorts integrated with genomic and transcriptomic data | NSCLC | Multi-region histology from 100 TRACERx patients, and an independent cohort of 970 others. | Tumours with multiple immune-cold regions were associated with worse outcome, and genomic features influencing the anti-tumour immune response could be visualised histologically. |
| Abbosh et al, 2017 | Longitudinal ctDNA analysis throughout tumour evolution | NSCLC | 100 patients | Evolutionary dynamics of disease, including relapse and treatment resistance, could be profiled using ctDNA detection. |
| Chemi et al, 2019 | Analysis of the relationship between circulating tumour cells derived from the pulmonary vein (PV-CTCs) and disease relapse | NSCLC | 100 patients | The presence of PV CTCs is associated with an increased risk of relapse, and associated clones are more likely to be detected in metastasis than in the primary tumour. |
| Biswas et al, 2019 | Investigation of impact of intratumour heterogeneity upon biomarker utility | NSCLC | RNA-seq from 156 regions of 48 tumours | Intratumour heterogeneity renders gene expression biomarkers vulnerable to sampling bias, and a panel of clonally expressed genes can represent a robust biomarker in non-small cell lung cancer. |
Heterogeneity in SCNAs and clinical outcome: The TRACERx Lung 100 cohort
A multi-region sequencing analysis from the first 100 prospectively recruited patients was published in 2017 and sought to explore the clinical implications of intratumour heterogeneity (7). Survival analysis showed that heterogeneity of SCNAs (as defined by the proportion of these events that were present only in a subset of cancer cells), but not mutations, predicted for poor outcome. This heterogeneity, detected through multi-region tumour sampling, may reflect the dynamic nature of CIN, suggesting the clinical importance of ongoing CIN in tumour evolution. Furthermore, an analysis of the intratumour heterogeneity, and timing, of driver events revealed that most canonical driver mutations in NSCLC, such as in TP53, KRAS and EGFR were clonal, and occurred prior to a whole-genome duplication event. However, other events such as mutations in PIK3CA and chromatin modifying genes were more likely to be subclonal and to occur later in tumour evolution. Importantly, with the help of phased copy-number information leveraged from multi-region sequencing, parallel evolution (where somatic events affecting the same gene occurred independently within distinct subclones) was seen among different haplotypes in copy-number events affecting genomic segments containing driver genes. This analysis added explicit evolutionary context to earlier observations about the clinical impact of intratumour heterogeneity of driver events, and CIN (Figure 2).
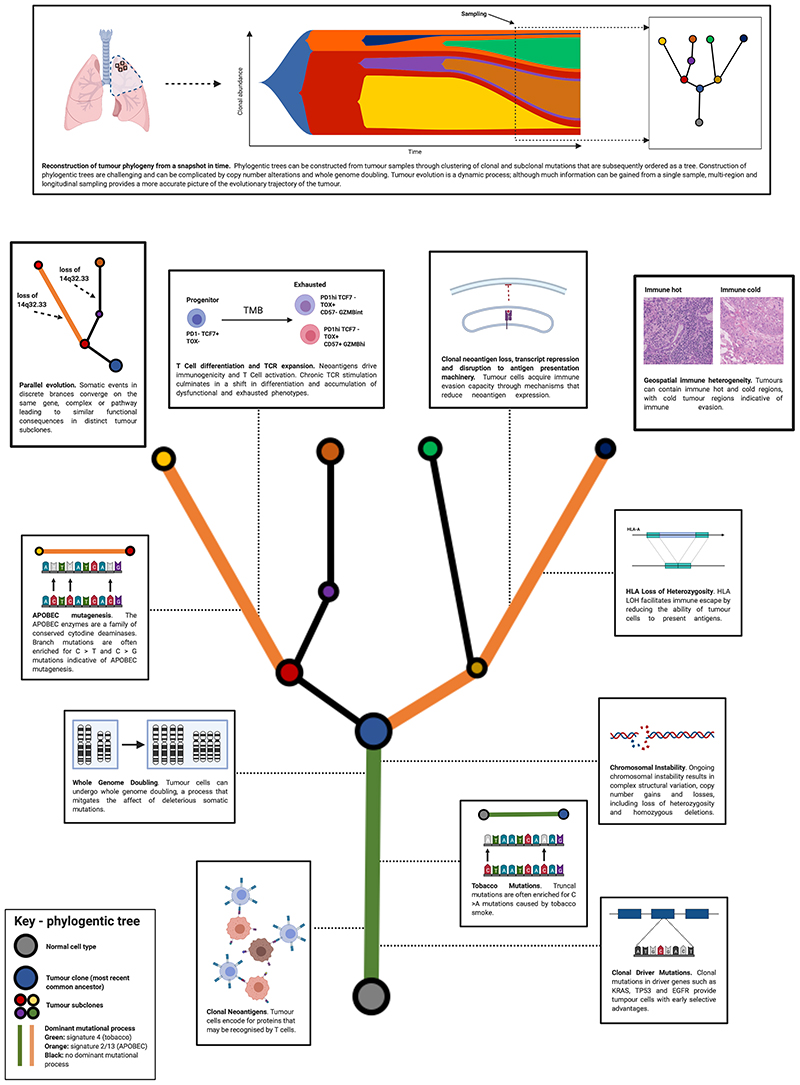
Figure 2. Tumour evolution in non-small cell lung cancer.
Evolutionary processes in non-small cell lung cancer are outlined. Top, subclonal dynamics over time can be represented by a fish plot, however a single sample in time only provides a snapshot. From this snapshot tumour phylogeny can be inferred. Bottom, evolutionary processes generating immune and genomic heterogeneity are described as part of a ‘tree’. Events that occur in the ‘trunk’ are clonal; i.e. they occur within every cell in the tumour. Through tumour evolution, subclones can emerge through selection; events that occur in these subclones are known as ‘branch’ events.
Subsequent insights into chromosomal instability and cancer evolution
Within the TRACERx Lung 100 cohort, 76% of tumours showed evidence of a whole-genome duplication (WGD) event, which is consistent with observations in other cancer types (22). Whilst WGD has been linked to poor prognosis and increased subclonal diversity, a biological rationale for the frequency with which this event is seen in cancer had not been defined. One hypothesis is that WGD might mitigate the negative impact of consecutive deleterious mutations that accumulate within a cancer cell lineage. Therefore, within the TRACERx and The Cancer Genome Atlas (TCGA) NSCLC cohorts, this was explored in greater detail (23). In cancers with extensive loss-of-heterozygosity (LOH) across the genome, where the negative impact of deleterious mutations would in theory be more marked, WGD was enriched, and in simulations, WGD appeared to be selected preferentially when deleterious alterations were acquired at a high rate. Negative selection against deleterious mutations in essential genes acquired prior to WGD within genomic segments of LOH was also demonstrated using dN/dS, a tool to measure selection adapted from evolutionary biology (24,25). These data suggest that WGD may be a selected event secondary to a “ratchet”-like phenomenon, mitigating the impact of the progressive accumulation of deleterious mutations in genes essential for lung cancer cell survival in regions of haploid LOH, brought about by excessive early CIN.
Although the finding in TRACERx that heterogeneity of SCNAs was associated with poor outcome suggests a role for CIN in fuelling subclonal selection, this was not explicitly demonstrated. In a subsequent pan-cancer analysis of multi-region data, enabling phasing of SCNAs, from 394 tumours, we explored this in greater detail revealing extensive heterogeneity of SCNAs in primary and metastatic tumours, reflective of ongoing CIN during tumour evolution (26). Importantly, in the majority of tumours, and in particular in those with evidence of a prior WGD event, the evolution of SCNAs was consistent with positive and negative selection of these events. Over one third of tumours showed evidence of one or more parallel evolutionary events, in which the same genes were affected by independent SCNAs within different subclones. Frequent focal subclonal SCNAs which were often subject to parallel evolution were revealed, including gains at 5p15.33 and 8q24.1, containing TERT and MYC respectively. In addition, in cases with both multi-region sampling of the primary and metastasis, subclonal copy-number events in the primary were found in all metastases, and in a combined analysis of paired and unpaired primary and metastatic samples, certain SCNAs that were frequently subclonal in primary tumours, such as those affecting CCND1 and MYC, were found to be significantly enriched within metastases in certain cancer types. This work builds upon observations from the TRACERx Lung 100 cohort, and supports the importance of ongoing CIN in tumour evolution.
Clinical correlates of cancer evolutionary subtypes
In TRACERx Renal, the evolutionary dynamics of ccRCC were shown to precede the development of a tumour mass. A multi-region whole-genome sequencing analysis of 33 patients with ccRCC enabled inference of the timing of landmark events in the development of this tumour (27). 3p loss, a near-ubiquitous event in this disease, was shown in some cases to precede the growth of a clinically detectable tumour by 30-50 years, revealing this event as an attractive potential therapeutic target to reduce the incidence of this tumour.
The first interim analysis of the TRACERx Renal cohort defined seven evolutionary subtypes of ccRCC (3), that follow distinct trajectories following 3p loss and VHL inactivation, suggesting that in many cases cancer evolution is deterministic. Tumours characterised by multiple clonal driver mutations had evidence of early fixation of high levels of chromosomal complexity and limited ongoing evolution; BAP1-driven tumours had similarly high levels of chromosomal complexity but with BAP1 as the sole additional driver; clonal expansion of PBRM1-mutant populations led to frequent fixation of additional driver events resulting in extensive intratumour heterogeneity, subclonal chromosomal complexity and evidence of frequent parallel evolution events; VHL monodriver tumours showed a paucity of additional drivers and minimal chromosomal complexity; and a subtype that in spite of 3p loss remained VHL-wildtype with cryptic early chromosomal complexity. Evolutionary features underpin heterogeneous clinical behaviour ranging from an absence of disease progression to indolent oligometastatic disease and rapid disease progression and death. Indeed, certain measures of intratumour heterogeneity were able to account for this variation in clinical behaviour. A survival analysis demonstrated that tumours with extensive chromosomal complexity and minimal intratumour heterogeneity of driver mutations were associated with poor outcome, whilst those with significant subclonal diversity had a more attenuated pattern of progression. Excellent outcomes were observed in tumours that lacked both diversity and chromosomal complexity.
Defining the clinical behaviours of tumours with different evolutionary trajectories remains a key scientific question across tumour types. Whilst subclonal selection may be a feature of some cancers, others may acquire driver events at an early stage in tumour evolution and show little or no evidence of subclonal selection (28–30). Understanding the extent of subclonal selection from a biopsy at a single time point is challenging. Moreover, heterogeneity in the sampling and tissue processing (single vs. multi-region, FFPE vs. fresh frozen samples), sequencing and analytical methods and a lack of detailed clinical annotation can confound attempts to draw conclusions across studies about the relationships between evolutionary subtypes and clinical outcome. To address this problem, a uniform protocol accompanied by robust clinico-pathological information is required.
Cancer evolution and metastasis
TRACERx Renal explored the evolutionary dynamics of metastasis, in particular attempting to establish the extent to which subclonal selection during branched tumour evolution drives this process. In ccRCC, lymphatic and haematogenous dissemination can cause spread to the lungs, liver, adrenal glands, brain and bones, as well as a local intravascular growth called a tumour thrombus; of the 98 patients included in this analysis, tumour thrombus was sampled in 25, lymph nodes sampled in 24, lesions of the adrenal glands in 20, bone in 17, lung in 13, and liver in 3 (31). Previous studies analysed cohorts of matched primary-metastasis retrospectively, whilst TRACERx Renal prospectively explored the evolutionary basis of metastasis (32). This has enabled the detailed comparison of primary tumour clones that go on to subsequently seed metastasis, and those that seemingly fail to metastasise, revealing a selection for clones harbouring extensive chromosomal complexity, as measured by Weighted Genome Integrity Index (wGII) scores (33,34), in metastatic seeding.
In this analysis, driver mutations were not enriched or depleted within metastases (32). However, copy-number loss of 9p, containing the tumour suppressor CDKN2A, as well as 14q, was significantly overrepresented within metastatic lesions whilst existing mostly as a subclonal event within the corresponding primary tumours. 9p loss was associated with an aggressive disease phenotype and reduced progression-free and overall survival. Of descriptive note, the study also revealed a small number of cases with pancreatic metastases, with lower chromosomal complexity when compared to other metastatic sites, which were associated with latent metastases at periods of 15 years post primary diagnosis. This highlights not only the role that evolutionary subtypes play in determining clinical outcome, but also that subtypes may favour different metastatic patterns.
These observations were supported by sampling of multiple metastases in the PEACE study; in one patient who experienced rapid disease progression, a clone showing 9p and 14q loss seeded all 13 metastatic sites; whilst in another patient showing attenuated progression, indolent metastases to the pancreas lacked 9p and 14q loss, whereas subsequent multi-site distant metastases had acquired these critical subclonal SCNAs.
Tumour evolution and the immune microenvironment
Tumour evolution and immune escape
The immune microenvironment plays a key role in shaping an evolving tumour; with positive selection favouring cancer cells that acquire immune evasion mechanisms (Figure 2). These include mutations and loss of heterozygosity at HLA loci, disruption of antigen presentation machinery and negative selection of neoantigens, which can be mediated by DNA copy number loss, transcript repression, epigenetic silencing and loss of neoantigen expressing clones. Clinical and experimental data suggest that functional anti-tumour immunity exerts a strong selection pressure on evolving tumour subclones (35–37).
HLA class I genes (HLA-A, HLA-B, HLA-C), located on the short arm of chromosome 6, encode cell surface proteins that present peptides to CD8+ cytotoxic T cells as part of the class I major histocompatibility complex (MHC). These genes are highly polymorphic, thus creating diversity in immune responses at the population level. Loss of HLA class I genes reduces the ability of tumour cells to present antigens, providing a mechanism to escape immune surveillance (38). Beta-2 microglobulin is also crucial for MHC class I binding and is located on the long arm of chromosome 15. The tool LOHHLA (Loss Of Heterozygosity in Human Leucocyte Antigen) was developed (39) in TRACERx to help quantify allele-specific HLA copy-number from bulk sequencing data, which can be challenging given the polymorphic nature of the HLA locus. In TRACERx Lung, HLA loss of heterozygosity (HLA LOH) occurred in 40% of tumours, of which in 65% it was observed as a subclonal (branching) event. Due to the mono-allelic loss of an HLA gene, there is a reduction (but not abrogation) of peptide presentation to tumour infiltrating cytotoxic T cells; no tumour harboured loss of all 6 HLA class I alleles.
HLA LOH occurred as a focal loss (defined in this case as a non-arm event) with a higher than expected frequency, suggesting that this event is subject to positive selection in tumour evolution. In four cases, loss of the same HLA allele occurred as separate branch events in the same tumour, indicating parallel evolution that converges on HLA loss. HLA LOH was associated with the expansion of potentially antigenic mutations; in lung adenocarcinoma, subclonal non-synonymous mutations were increased in tumours where HLA LOH had occurred. In tumour regions without HLA LOH, subclonal mutation burden was higher if HLA LOH had occurred in other regions of the same tumour, suggesting that increased mutational burden may drive selection pressure for HLA LOH. Furthermore, in lung adenocarcinomas, subclonal mutations were enriched for APOBEC mutational signatures (Signature 2 and Signature 13) in tumours with HLA LOH. Furthermore, a trend towards enrichment of HLA LOH in brain metastasis was observed, suggesting that this event may be permissive for metastasis and/or associated with later stage disease. In the study, tumours with clonal HLA LOH were associated with high immune infiltration and significantly elevated levels of PD-L1 staining by immunohistochemistry, indicating that HLA LOH is a prevalent mechanism of immune evasion in tumours where immune activity is high, reinforcing the concept that this even occurs in response to high immune selection pressure (39).
The TRACERx Lung study has provided a platform to explore the role of neoantigen evolution in immune escape. In 2016, we found in NSCLC that tumours with a high clonal neoantigen burden were associated with increased overall survival (40). Building on previous work, clonal neoantigen burden was associated with high tumour immune infiltrate and increased PD-L1 expression. Moreover, clonal neoantigen burden was associated with clinical response to the checkpoint inhibitor pembrolizumab, however, this association was not significant if the neoantigen burden was subclonal. In some cases, subclonal neoantigens induced by cytotoxic chemotherapy were enriched in patients where response to checkpoint inhibition was poor. The concept that clonal neoantigens may be particularly immunogenic has since been supported by preclinical models (41) as well as data from patients with metastatic melanoma (42) and has lead to the development of clonal neoantigen targeting adoptive T cell therapies (ClinicalTrials.gov Identifier: NCT04032847).
The intra- and intertumour heterogeneity of immune infiltration was characterised by integrating RNA-sequencing data with tumour infiltrating lymphocyte estimates in 258 regions from 88 TRACERx lung tumours (43). In this cohort, 28% of tumours had uniformly high levels of immune infiltration and 43% had uniformly low levels of immune infiltration, with disparate levels of infiltration in the remaining 28%. In lung adenocarcinoma, low immune infiltration was associated with increased subclonal diversity. This association between immune evasion and subclonal complexity is suggestive of an evolving tumour that has escaped immune surveillance.
To further explore the interplay between neoantigens and the immune response, we focused on neoantigen-directed mechanisms of immune escape, a term that encompasses both neoantigen depletion and disruption to antigen presentation. Potential mechanisms of neoantigen depletion that were analysed included DNA copy-number loss, neoantigen transcript repression, epigenetic silencing of neoantigen sequences and T cell mediated elimination of neoantigen expressing clones. In 43 of 88 TRACERx lung tumours, there was evidence of clonal neoantigen copy-number loss, and this was more likely to occur in regions of low immune infiltration. In highly immune infiltrated tumours where HLA alleles were intact, there was a depletion in expressed predicted neoantigenic transcripts (relative to non-neoantigenic transcripts), thus proposing neoantigen transcript repression as an alternative mechanism of immune escape. Irrespective of immune infiltration, in consistently expressed genes essential for lung cancer survival, there was a reduction in neoantigens. Furthermore, there was a decrease in immunoediting (defined here as a reduction in the ratio of observed-to-expected neoantigens) (44) from clonal to subclonal neoantigens in immune cold tumours. This suggests that these tumours once-contained an active immune microenvironment that became cold. Poorly expressed genes predicted to contain neoantigens, exhibited an increase in promoter hypermethylation (compared with expressed genes containing neoantigens and the same genes without neoantigenic mutations), suggesting that promoter hypermethylation is an additional mechanism in neoantigen silencing.
Disruption to antigen presentation encompassing HLA LOH and mutations affecting the MHC complex, HLA enhanceosome or peptide generation were found in 56% of lung adenocarcinomas and 78% of squamous cell carcinomas. By combining immune infiltration with these immune evasion mechanisms, the capacity for a tumour to evade the immune system was quantified. For tumours with low immune evasion capacity; uniformly high immune infiltrate with no evidence of DNA immunoediting and no disruption to antigen presentation, there was a significant improvement in disease free survival. In a multivariate model, both low immune evasion capacity and clonal neoantigen burden were predictors of disease-free survival, however subclonal neoantigen burden was not. The diversity of immune escape mechanisms likely reflects the strength of selection pressures exerted by the immune system and highlights an important consideration for immunotherapeutic design.
Intratumour heterogeneity and the T cell landscape
In the complex evolutionary arms race of tumour immunity the immune system drives selection of evolving tumour subclones, and reciprocally, the tumour mutational landscape may drive T cell activation, clonal expansion and differentiation. The expansion and spatial diversity of intratumoural TCRs were analysed (45) by sequencing the α-chain and β-chain T cell receptor (TCR) repertoires from 220 tumour regions and 119 matched (non-tumour lung or blood) samples from 72 patients in the TRACERx Lung 100 cohort. The most expanded intratumoural TCRs represented a higher proportion of the total TCR repertoire compared with those expanded in non-tumour lung samples. Furthermore, the number of expanded TCRs correlated with non-synonymous mutations in tumour regions, consistent with ongoing neoantigen specific T cell responses.
There were marked differences in TCR intratumour heterogeneity between tumours, whereby some tumours had a diverse regional TCR expression pattern and for others this pattern was more homogenous. TCRs were therefore defined as ubiquitous or regional depending on their spatial distribution throughout multi-region tumour specimens and, interestingly, this correlated with the number of clonal non-synonymous and subclonal non-synonymous mutations, respectively; implying that ubiquitous TCRs may recognize clonal neoantigens. Interestingly, expanded TCR CDR3 sequences formed highly related clusters according to amino acid composition; with regional TCR clustering far less prevalent than ubiquitous TCR clustering. There were also more unique DNA sequences encoding each ubiquitous expanded TCR when compared with regional TCR or random sampling, indicative of antigen driven convergent recombination. The positive association between ubiquitous TCRs and a CD8+ Th1 transcriptional phenotype suggests that a cytotoxic T cell response is linked with the presence of ubiquitous intratumoural TCRs. Clonal neoantigen reactive T cells, and cells bearing an exhausted phenotype also tended to harbour expanded TCR sequences.
Furthermore, expanded, ubiquitous TCR sequences in the tumour were preferentially found in peripheral T cell clones, and TCRs in the blood displayed both contraction and expansion throughout the clinical course of disease, suggesting that longitudinal tracking of specific subsets of TCRs in the blood may in future provide a method of monitoring dynamic intratumoral immune surveillance.
Tumour mutational landscape and T cell differentiation
T cells respond to antigen stimulation by activation, proliferation and differentiation, manifesting in redistribution of progenitor-progeny subsets (46). The relationship between tumour mutational burden (TMB), and T cell differentiation was explored in the TRACERx Lung 100 cohort (47). TMB is known to predict response to immune checkpoint blockade, yet tumour specific T cells often appear dysfunctional, suggesting neoantigens could both drive immunogenicity but also fuel chronic TCR stimulation that culminates in a shift in differentiation and accumulation of dysfunctional phenotypes. High dimensional flow cytometry analysis of multi-region TILs revealed highly diverse populations of CD4 and CD8 progenitor-like and dysfunctional tumour infiltrating lymphocytes. Ghorani and Reading then integrated these data with paired WES data from the TRACERx Lung 100 cohort, unveiling that TMB correlated with T cell subsets exhibiting evidence of antigen engagement; including dysfunctional T cells with high PD-1 expression, PD-1 expressing subsets with features of terminally differentiated effector memory cells (CD57, granzyme B and eomesodermin expression), and regulatory T cells with high co-inhibitory receptor expression. Conversely, T cell clusters lacking PD-1 with bystander or progenitor-like characteristics correlated negatively with TMB, implying a neoantigen-driven intra-tumoral differentiation process. Consistent with this, progenitor and dysfunctional T cell subsets shared TCR sequences and dysfunctional T cells correlating with TMB were phenotypically and transcriptomically similar to neoantigen multimer reactive T cells. The progenitor and dysfunctional subsets were additionally shown to express canonical stem (TCF7) and exhaustion (TOX), transcription factors, respectively by flow cytometry and scRNAseq. This putative program of neoantigen driven differentiation skewing was also characterised in bulk tumour RNA-seq by identifying a gene signature that mapped loss of transcription factors TCF7 and LEF1 (TL-DS). The TL-DS signature correlated with TMB in tumours and poor clinical outcomes in TRACERx patients with lung adenocarcinoma, but not squamous cell carcinoma. When applied to TCGA datasets in a multivariable model, the signature was associated with adverse survival outcomes in 6 different tumour types, including NSCLC, raising the possibility that in the absence of immunotherapy, chronic neoantigen stimulation could fuel loss of progenitor T cells and gain of dysfunctional subsets, ultimately precipitating fatal immune failure.
Importantly, the burden of clonal, but not subclonal mutations correlated with the loss of progenitor and gain of dysfunctional subsets, corroborating the observation that clonal neoantigens preferentially elicit immunoreactivity and suggesting that clonal neoepitopes may generate PD-1+ cells that could serve as a substrate for immunotherapies targeting the PD-1 axis (40,48). These data highlight that neoantigens may represent a double-edged sword; triggering protective immunity that can be de-repressed by checkpoint blockade but also serving as a co-factor for T cell dysfunction. More work is needed to better understand neoantigen driven T cell differentiation across tumour types.
Geospatial variability of the immune microenvironment and immune evasion
By developing a deep-learning pipeline utilising multi-region histology samples from the TRACERx Lung 100 and The Leicester Archival Thoracic Tumour Investigatory (LATTICe-A) cohorts (49), Yuan and colleagues integrated tumour morphological data with WES and RNA-sequencing data (50) to understand how the spatial organisation of stroma and immune cells impact evolutionary processes in immune escape. Following topological classification of immune hot and cold regions, the authors demonstrated high intratumour variability of immune infiltrate that did not correlate with stage. Comparing hot and cold immune tumour regions, pairs of immune cold regions of the same tumour shared more subclonal mutations than pairs of hot regions in lung adenocarcinoma but not squamous cell carcinoma, indicating that these cold regions may share a recent common ancestor with the capacity for immune evasion. Tumours with more than one cold region were at significantly higher risk of relapse compared with those with one or less. In lung adenocarcinoma but not squamous cell carcinoma, regions with intact HLA alleles correlated with increased geometric complexity of the cancer-stroma interface compared with those regions with HLA LOH. The proximity of lymphocytes to cancer cells was also quantified, with clonal neoantigen burden correlating with increased stromal lymphocytes. The study demonstrated that the association between geospatial immune configuration and mechanisms of cancer cell immune evasion are likely disease specific, and as we continue to refine complex imaging technology and integrate this with multi-omic data, these features will become clearer.
The work to identify immune escape mechanisms and geospatial variation in the tumour microenvironment in the TRACERx cohort complemented work on pre-invasive squamous cell lung cancer (51). Here, WES was combined with gene expression profiling, genome-wide methylation and image analysis using the deep learning approach in AdbulJabbar et al (50). By comparing regressive and progressive carcinoma in situ (CIS) lesions, key immune-evasive mechanisms found in progressive CIS lesions were identified. Regressive CIS lesions harbour a greater concentration of CD8+ T cells, and are associated with a relative increase in most immune cell types except macrophages. Progressive lesions had a higher mutational burden and also contained more genomic aberrations in genes involved in MHC Class I antigen presentation and hypermethylation at the HLA locus. There was also an inverse correlation between fibroblast TGF- beta response gene signature and the gradient between tumour and adjacent stroma TIL scores supporting previous observations of a fibroblast driven, TGF-beta mediated T cell exclusion response.
Taken together, data from the TRACERx study has provided insights into the interface between the immune system and the evolving tumour (Figure 2). Non-small cell lung cancer cells have an array of immune evasion mechanisms that may be selected for in the face of a hostile and diverse tumour microenvironment. Clonal neoantigens appear to drive T cell immune-reactivity and where clonal neoantigen burden is high, immune infiltration is often high. A strong CD8+ response is characterised by ubiquitous T cell receptor tumour antigen reactivity, which shows evidence of diverse clonotypes that converge on specific neoantigens. Consistent neoantigenic stimuli may result in a differential skewing of dysfunctional T cell subtypes, some with high PD-1 expression. HLA LOH is a prevalent mechanism for immune escape when infiltration is high, but as the tumour further develops capacity for immune invasion, the immune-stromal landscape becomes more heterogenous, with immune cold regions that may have been historically hot associated with copy number driven loss of clonal neoantigens, intratumour neoantigen heterogeneity, and stromal-mediated T cell exclusion. Tumours with high immune evasion capacity and heterogenous or low immune infiltration are associated with poor outcomes.
Translational developments in TRACERx
Findings from the TRACERx Lung study are being translated into clinical application with a view to demonstrating the clinical utility of its discoveries in improving methods of early detection, prognostication and therapeutic intervention in NSCLC.
Clinical utility of circulating biomarkers
The development of minimally invasive circulating biomarkers such as ctDNA and CTCs aims to improve approaches to early detection of relapse through minimal residual disease (MRD) monitoring and predictors of clinical outcome. Identification of patients with NSCLC at risk of relapse post-surgery may help identify those who would benefit from adjuvant therapies and potentially avoid unnecessary treatments and associated toxicities in those patients with a lower risk of disease recurrence. The prospective recruitment and longitudinal design inherent in TRACERx provided a platform to address this question. In creating bespoke multiplex PCR panels to detect clonal and subclonal single nucleotide variants (SNVs) in plasma, NSCLC relapse was profiled through ctDNA detection in the TRACERx Lung 100 cohort (52). By profiling preoperative plasma samples, the determinants of ctDNA detection were identified. Necrotic tumours were more likely to release ctDNA; with non-adenocarcinoma subtype, lymphovascular invasion and high Ki67 proliferation index independent predictors of ctDNA detection. Clonal SNVs were detected in all ctDNA positive patients, and the plasma variant allele frequency correlated with tumour size. We utilised a panel of between 25-30 variants per patient to detect and characterise recurrent disease. Mutations in ctDNA were detected in 13 out of 14 patients before or at confirmed radiological relapse with a median lead time of 70 days prior to imaging confirmed recurrence. By mapping ctDNA detected variants at recurrence back to the primary tumour WES data, we were able to identify the ancestral subclones leading to relapse. Finally, metastatic subclones were retrospectively tracked through ctDNA analysis in a patient who had subsequently participated in the PEACE study, using a panel of 20 clonal SNVs with 4-15 subclonal SNVs per metastatic subclone. Using this 103-SNV panel, ctDNA was detected 151 days post-surgery (315 days prior to clinical relapse), with 18 out of 20 shared subclonal SNVs (present in 3 of 9 metastatic clonal clusters) and two private subclonal SNVs detectable in ctDNA following clinical relapse on day 466. This bespoke approach is utilised as part of the Signatera assay (53).
A similar phylogenetic approach to MRD detection is now being implemented in the clinical trial setting. MERMAID-1 is a phase III trial (ClinicalTrials.gov Identifier: NCT04385368), in collaboration with ArcherDx and AstraZeneca. The trial aims to assess the efficacy of the PD-L1 inhibitor durvalumab in combination with chemotherapy in patients with resected stage II-III NSCLC. Following resection of the primary tumour, MRD detection through bespoke ctDNA panels will be used in the assessment of disease-free and overall survival. Furthermore, using the same approach, Powles and colleagues have demonstrated that patients with muscle-invasive urothelial cancer who are ctDNA positive post cystectomy may benefit from atezolizumab (54). This analysis was done as part of the IMvigor010 trial; the Signatera assay utilising a bespoke panel of 16 somatic mutations (55,56).
Using the circulating tumour cell (CTC) capture platform CellSearch, CTCs taken from the pulmonary vein (PVT-CTCs) of patients from the TRACERx Lung 100 cohort were enumerated at surgery (57). CTCs were detected at a threshold of at least 1 PV-CTC in 7.5mL of blood in 48% of patients. A high PV-CTC count (> 7 PV-CTCs per 7.5mL blood) was an independent predictor for lung cancer relapse. Each doubling of PV-CTC count was associated with lung cancer specific relapse indicating a continuous relationship between PV-CTCs and clinical outcome. WES data from three primary tumour regions was compared to single cell sequencing data from 6 single PV-CTCs (three of which were found to be EpCAM and cytokeratin expressing circulating epithelial cells - CECs) and the metastatic relapse biopsy (10 months after surgery) in a single patient. They demonstrated a higher number of shared mutations between the PV-CTC and the metastatic sample compared with the PV-CTC and primary sample, with phylogenetic reconstruction demonstrating that the PV-CTC and metastatic sample came from the same branch. PV-CTCs taken at the time of surgery shared a common progenitor with the metastasis that was detected 10 months later, indicating that early dissemination of PV-CTCs may result in metastatic seeding.
Prognostic signatures and therapeutic stratification
Prognostic gene expression signatures have been utilised to predict clinical outcome in patients with a variety of cancer types, however, the interpretability of these biomarkers may be confounded by sampling bias driven by intratumour heterogeneity (58–60). By combining multi-region RNA sequencing from 48 patients in the TRACERx Lung 100 cohort with TCGA and the Uppsala NSCLC datasets (61–63), the impact of RNA intratumour heterogeneity gene expression signatures was explored in lung adenocarcinoma (64). We demonstrated regional discordance of prognostic scores between biopsies from the same tumour, thus highlighting the vulnerability of such scoring approaches to sampling bias. By stratifying genes into quadrants based on intra- and intertumour heterogeneity, and selecting a subset of genes that were expressed with low intratumour heterogeneity but high intertumour heterogeneity, we generated a 23-gene prognostic signature termed ORACLE (Outcome Risk Associated Clonal Lung Expression biomarker).
As a clonal expression biomarker, the ORACLE score was associated with outcome in the Uppsala cohort, in a multivariate model that adjusted for TNM stage, adjuvant treatment status, age, performance status, smoking history, gender and Ki67 staining. The genes that were stratified in the low intratumour heterogeneity but high intertumour heterogeneity quadrant (Q4) were enriched in proliferation pathways, such as nucleosome assembly and mitotic prometaphase. These genes retain transcriptional stability despite ongoing CIN, and are subject to clonal copy-number gains early in tumour evolution.
ORACLE was able to classify patients with stage I lung adenocarcinoma into low and high-risk groups. RNA intratumour heterogeneity also correlated strongly with subclonal copy-number diversity, indicating that ongoing chromosomal instability is a strong driver of transcriptomic heterogeneity. Finally, by comparing genes identified in each quadrant using this selection approach to genes with pan-cancer prognostic scores from Prediction of Clinical Outcomes from Genomic Profiles (PRECOG) dataset, we found genes in Q4 were significantly enriched for prognostic genes in 19 of 39 malignancies. This approach to gene expression profiling may improve refinement of prognostication including response to therapy.
From the bench to clinical trials
Whilst TRACERx aims to establish the relationship between intratumour heterogeneity and clinical outcome, we are yet to understand the relevance of clonality in therapeutically targeting actionable driver events. DARWIN II (Deciphering Antitumour Response and Resistance With INtratumour Heterogeneity, ClinicalTrials.gov Identifier: NCT02314481) is a multi-arm non-randomised phase II trial which aims to investigate the role of predicted neoantigens and intratumour heterogeneity on response to anti PD-L1 therapy, as well as outcomes in patients treated with targeted therapies against clonal or subclonal actionable alterations (Figure 3). Patients with relapsed disease in the TRACERx Lung study or those with available multi-region sequencing data from a primary NSCLC are eligible. Patients are stratified to specific therapies depending on their targetable alteration; BRAFV600 (vemurafenib), ALK/RET translocation (alectinib), and HER2 amplification (trastuzumab emtansine). All other patients are consented to the PD-L1 inhibitor atezolizumab. Sampling before and after treatment, including at autopsy, will facilitate the study of resistance to immune- and targeted therapies. Longitudinal monitoring of predictive biomarkers of response and resistance may also guide future patient stratification and combination therapies. Recruitment to this trial is ongoing.
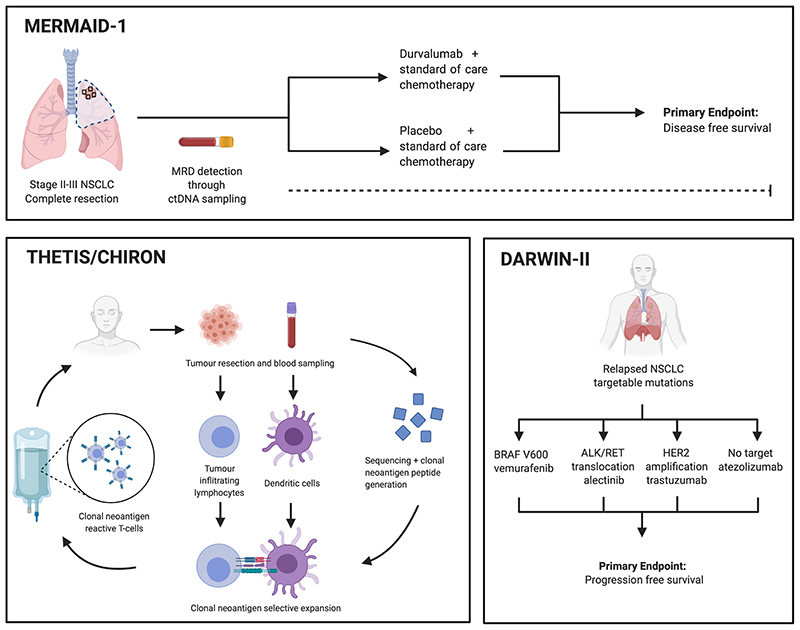
Figure 3. Clinical trials in tumour evolution.
Outlined are key trials that have been associated with the TRACERx project. Top, the MERMAID-1 trial incorporates bespoke ctDNA panels as MRD detection, following on from the work by Abbosh et al (52). Left, the THETIS and CHIRON trials are first-in-man studies that look at the efficacy of autologous clonal neoantigen reactive T cells in non-small cell lung cancer and melanoma. Clonal neoantigens are identified following tumour sequencing. T cells are subsequently expanded with neo-antigenic peptides and returned to the patient. Right, the DARWIN-II trial investigates the impact of intra-tumour heterogeneity in targeted therapy response in relapsed non small cell lung cancer, combining data from primary tumours sampled in the TRACERx Lung project.
Despite recent improvements in NSCLC clinical outcomes with PD-1/PD-L1 checkpoint inhibitors, 45-50% of patients with metastatic disease do not achieve an optimal response with standard of care first line chemotherapy plus PD-1/PD-L1 combinations, and almost 70% patients experience disease progression or die within 12 months of treatment (65,66). In this setting, second-line therapy is often associated with minimal clinical benefit. With evidence that clonal neoantigens drive T cell reactivity, the first-in-human open-label, multicentre phase I/IIa trial to characterise the safety and clinical activity of autologous clonal neoantigen targeting T cells (cNeT) was announced in 2019. Developed by Achilles Therapeutics, the CHIRON and THETIS trials (ClinicalTrials.gov Identifier: NCT03997474 and NCT04032847) investigate the safety and clinical activity of a therapeutic product derived from autologous tumour-infiltrating lymphocytes (TILs) primed to recognise patient-specific tumour clonal neoantigen epitopes in NSCLC and melanoma respectively (Figure 3).
Future plans to address unanswered questions
Our understanding of cancer evolution has improved significantly over the last decade. Here, we outline outstanding questions in the field and areas of unmet clinical need that could be addressed in future studies.
The role of chromosomal instability in the disease course
SCNAs provide a heterogenous substrate for ongoing tumour evolution, and TRACERx has highlighted the relationship between heterogeneity driven by SCNAs and adverse clinical outcome. Future studies should aim to highlight specific copy-number events that may be prerequisites for certain evolutionary trajectories. Moreover, there is an ongoing effort to characterise the processes that generate SCNAs and drive CIN, understand cancer cell tolerance of this, and define their relationship with driver events. Developing the necessary datasets and tools required to measure the rate of CIN might help shed light on its exact role in the natural history of a tumour.
Non-genetic variation and tumour evolution
Cancers may exploit transcriptomic and epigenomic variation during their evolution (67,68). This can provide a tumour with alternate mechanisms of immune evasion as well as fuelling adaptive evolution in response to selective pressures, such as targeted therapies (69). Understanding the evolutionary context of this non-genetic variation, and assessing epigenetic and transcriptomic heterogeneity, in both treated and untreated cancers must be addressed and will require a repertoire of studies using a variety of bulk and single-cell sequencing technologies.
The anti-tumour immune response as a therapeutic target
Understanding which neoantigens stimulate an effective anti-tumour response is critical. Neoantigens that are clonal, highly expressed and dissimilar to ‘self’ with appropriate mitigation from HLA loss or copy-number loss tend to stimulate a more effective anti-tumour immune response (70). Moreover, neoantigens that are formed via frameshift mutations are more likely to be recognised by multiple HLA alleles. Understanding and leveraging high-quality neoantigens for treatment stratification promises to enhance immunotherapeutic approaches, including adoptive T cell therapies and cancer vaccines (71). Evolving selection pressures exerted by the immune response that affect the transitions between pre-invasive to invasive and early stage to disseminated disease may also have profound clinical relevance in the context of early detection and cancer progression.
Understanding clinical correlates of cancer evolutionary trajectories
A key objective of TRACERx has been to understand the relationship between intratumour heterogeneity and clinical outcome. In TRACERx Renal, tumour subtypes with distinct evolutionary trajectories were described associated with distinct clinical outcomes (3,32). Expanding such analyses to large cohorts of cancers of other types and integrating data from pre-invasive lesions, primary tumours, recurrences and distant metastases may reveal novel insights into the evolutionary dynamics of cancer and support optimal disease classification, facilitate new screening and treatment strategies. Longitudinal sampling, with liquid biopsies to study ctDNA as well as tissue biopsies at relapse or autopsy, may help to understand the impact of treatment on cancers, as well as the development and evolution of therapy-resistant subclones. It is hoped that this might improve our ability to better classify early stage disease and to predict the likely clinical course of the disease.
Functional and translational validation of observations
Ascribing functional weight to observations made in bulk sequencing studies is crucially important. Advanced technologies such as patient-derived organoid systems and genetically engineered mouse models will help validate these findings, including further elucidation of the importance of order in somatic events, quantification of non-genetic heterogeneity and refined modelling of the immune interface in tumour evolution (72,73). In addition, complex cellular imaging can provide additional spatial resolution of intratumour heterogeneity and the tumour microenvironment (74).
Longitudinal, prospective and comprehensive clinical studies to augment cancer research
TRACERx is the product of a national, comprehensive, multi-disciplinary research infrastructure that links a diverse range of clinical specialties and scientific disciplines, made possible through the collaborative nature and uniform clinical care adherent to strict standard of care guidelines provided by a national health service. Studies of cancer evolution should endeavour to incorporate comprehensive clinical information with detailed genomic and immunological analyses (Table 2). Establishing a sufficiently powered prospectively recruited cohort with robust sampling methods and detailed clinical annotation is important. Furthermore, considering the need for longitudinal sampling from early to late stage disease, with the incorporation of research autopsies, can help establish a dataset encompassing the disease course in its entirety.
Table 2. A template for studies of tumour evolution.
| Feature | Benefit |
|---|---|
| Multi-centre | Involvement of different centres of helps with timely recruitment of a large cohort enabling analysis of sufficiently power for statistically meaningful observations. |
| Prospectively recruited | Prospective analyses are critical when making meaningful and robust inferences about clinical factors such as outcome. |
| Multiple samples | Analysis of different samples over time, such as through sampling at relapse or post-mortem or ctDNA analysis, and space, using multi-region sequencing, can help to elucidate the evolutionary context of cellular events. |
| Uniform sampling method | It is critical that studies define a method for sampling, processing and sequencing that is adhered to across sites to reduce technical inconsistencies. |
| Independent pathology review | Reduces impact of heterogeneous interpretation of histopathological specimen |
| Multi-omics | Enables capturing of greater degree of tumour variation, including non-genetic events and microenvironmental alterations. |
| Detailed clinical annotation | Robust protocols for inputting clinical data reduces missing data. This is critical for prospectively defined variables of interest, but also for retrospective analysis, such as when examining the impact of clinic-pathological variables or environmental exposures. |
| Reproducible bioinformatic pipeline | Transparent, portable and easily replicable analytical approaches improve efficiency and collaborative approaches to the study of tumour evolution. |
Maintaining a comprehensive and reproducible bioinformatic pipeline that is adaptable to different tissue types and sampling time points is crucial, with novel software and technological developments making portable analytical frameworks a reality. Finally, multiple samples within individual patients, whether they be from different regions of the same tumour, relapse, metastasis, from liquid biopsies or taken at autopsy, are likely to provide granularity to the inferences we can make regarding tumour evolution.
Outside of NSCLC and ccRCC, numerous studies have used multi-region or longitudinal sampling to address questions pertaining to intratumour heterogeneity and tumour evolution (75–81). Large, prospective, longitudinal clinical cohorts however are rare. The Glioma Longitudinal Analysis Consortium (GLASS) is a multi-institutional initiative that aims to profile longitudinal molecular trajectories of gliomas over several time points through treatment (82). Studies such as this are essential and are powered to address key, treatment-focussed questions in the field. Developing large prospective clinical studies in other tumour types that integrate multi-omic approaches with longitudinal follow up will build on observations made to date.
TRACERx has helped to shed light on the evolutionary forces at work within lung and renal tumours. Longitudinal and multi-region sampling has helped to explore this at the molecular level, and some key observations are being tested in the clinic. However, many questions remain unanswered, and further studies will be critical to understanding the clinical relevance of evolutionary processes within cancer and the way in which we can leverage these to benefit patients.
Statement of Significance.
Cancers evolve and adapt to environmental challenges such as immune surveillance and treatment pressures. The TRACERx studies were set up in 2014 to track cancer evolution in a clinical setting, through primary disease to recurrence. Through multi-region and longitudinal sampling, evolutionary processes have been detailed in the tumour and the immune microenvironment in non-small cell lung cancer and clear cell renal cell carcinoma. TRACERx has revealed the potential therapeutic utility of targeting clonal neoantigens and ctDNA detection in the adjuvant setting as a minimal residual disease detection tool, primed for translation into clinical trials.
Acknowledgements
We would like to thank Tom Watkins, Alex Frankell, Rachel Rosenthal, Chris Abbosh, Yinyin Yuan and Kroopa Joshi for their thoughtful input and review of the manuscript. The fish plot was created using the fishplot R package from https://github.com/chrisamiller/fishplot/. Figures were created with the BioRender software at BioRender.com. CB and JRMB contributed equally to this work.
Footnotes
Conflicts of Interests
CS is Royal Society Napier Research Professor. This work was supported by the Francis Crick Institute that receives its core funding from Cancer Research UK (FC001169), the UK Medical Research Council (FC001169), and the Wellcome Trust (FC001169). CS is funded by Cancer Research UK (TRACERx, PEACE and CRUK Cancer Immunotherapy Catalyst Network), Cancer Research UK Lung Cancer Centre of Excellence, the Rosetrees Trust, Butterfield and Stoneygate Trusts, NovoNordisk Foundation (ID16584), Royal Society Professorship Enhancement Award (RP/EA/180007), the NIHR BRC at University College London Hospitals, and the CRUK-UCL Centre, Experimental Cancer Medicine Centre, and the Breast Cancer Research Foundation (BCRF). This research is supported by a Stand Up To Cancer‐LUNGevity-American Lung Association Lung Cancer Interception Dream Team Translational Research Grant (Grant Number: SU2C-AACR-DT23-17). Stand Up To Cancer is a division of the Entertainment Industry Foundation. Research grants are administered by the American Association for Cancer Research, the Scientific Partner of SU2C. CS receives funding from the European Research Council (ERC) under the European Union’s Seventh Framework Programme (FP7/2007-2013) Consolidator Grant (FP7-THESEUS-617844), European Commission ITN (FP7-PloidyNet 607722), an ERC Advanced Grant (PROTEUS) from the European Research Council under the European Union’s Horizon 2020 research and innovation programme (grant agreement No. 835297), and Chromavision from the European Union’s Horizon 2020 research and innovation programme (grant agreement 665233). CS acknowledges grant support from Pfizer, AstraZeneca, Bristol Myers Squibb, Roche-Ventana, Boehringer-Ingelheim, Archer Dx Inc (collaboration in minimal residual disease sequencing technologies) and Ono Pharmaceutical, is an AstraZeneca Advisory Board member and Chief Investigator for the MeRmaiD1 clinical trial, has consulted for Pfizer, Novartis, GlaxoSmithKline, MSD, Bristol Myers Squibb, Celgene, AstraZeneca, Illumina, Amgen, Genentech, Roche-Ventana, GRAIL, Medicxi, Bicycle Therapeutics, and the Sarah Cannon Research Institute, has stock options in Apogen Biotechnologies, Epic Bioscience, GRAIL, and has stock options and is co-founder of Achilles Therapeutics. CS holds European patents relating to assay technology to detect tumour recurrence (PCT/GB2017/053289); to targeting neoantigens (PCT/EP2016/059401), identifying patent response to immune checkpoint blockade (PCT/EP2016/071471), determining HLA LOH (PCT/GB2018/052004), predicting survival rates of patients with cancer (PCT/GB2020/050221), identifying patients who respond to cancer treatment (PCT/GB2018/051912), a US patent relating to detecting tumour mutations (PCT/US2017/28013) and both a European and US patent related to identifying insertion/deletion mutation targets (PCT/GB2018/051892).
ST is funded by Cancer Research UK (grant reference number C50947/A18176), the National Institute for Health Research (NIHR) Biomedical Research Centre at the Royal Marsden Hospital and Institute of Cancer Research (grant reference number A109), the Kidney and Melanoma Cancer Fund of The Royal Marsden Cancer Charity, The Rosetrees Trust (grant reference number A2204), Ventana Medical Systems Inc (grant reference numbers 10467 and 10530), the National Institute of Health U01 Award and Melanoma Research Alliance. ST has received speaking fees from Roche, Astra Zeneca, Novartis and Ipsen. ST has the following patents filed: Indel mutations as a therapeutic target and predictive biomarker PCTGB2018/051892 and PCTGB2018/051893 and Clear Cell Renal Cell Carcinoma Biomarkers P113326GB.
MJH has received funding from Cancer Research UK, National Institute for Health Research, Rosetrees Trust, UKI NETs and NIHR University College London Hospitals Biomedical Research Centre. MJH is a member of the Scientific Advisory Board and Steering Committee for Achilles Therapeutics.
Figure Declaration
These figures are unpublished original works, created by CB for the express purpose of publication in this Cancer Discovery article. CB holds an institutional Biorender license through the Francis Crick Institute.
References
- 1.Turajlic S, Sottoriva A, Graham T, Swanton C. Resolving genetic heterogeneity in cancer. Nat Rev Genet. 2019;20:404–16. doi: 10.1038/s41576-019-0114-6. [Internet]. Available from: http://www.nature.com/articles/s41576-019-0114-6. [DOI] [PubMed] [Google Scholar]
- 2.Jamal-Hanjani M, Hackshaw A, Ngai Y, Shaw J, Dive C, Quezada S, et al. Tracking Genomic Cancer Evolution for Precision Medicine: The Lung TRACERx Study. PLoS Biol. 2014;12:e1001906. doi: 10.1371/journal.pbio.1001906. [Internet] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Turajlic S, Xu H, Litchfield K, Rowan A, Horswell S, Chambers T, et al. Deterministic Evolutionary Trajectories Influence Primary Tumor Growth: TRACERx Renal. Cell. 2018;173:595–610.:e11. doi: 10.1016/j.cell.2018.03.043. [Internet]. Available from: https://linkinghub.elsevier.com/retrieve/pii/S0092867418303751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McLendon R, Friedman A, Bigner D, Van Meir EG, Brat DJ, Mastrogianakis GM, et al. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–8. doi: 10.1038/nature07385. [Internet]. Available from: http://www.nature.com/articles/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Campbell PJ, Getz G, Korbel JO, Stuart JM, Jennings JL, Stein LD, et al. Pan-cancer analysis of whole genomes. Nature. 2020;578:82–93. doi: 10.1038/s41586-020-1969-6. [Internet]. Available from: http://www.nature.com/articles/s41586-020-1969-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Subotic D, Van Schil P, Grigoriu B. Optimising treatment for post-operative lung cancer recurrence. Eur Respir J. 2016;47:374–8. doi: 10.1183/13993003.01490-2015. [Internet] [DOI] [PubMed] [Google Scholar]
- 7.Jamal-Hanjani M, Wilson GA, McGranahan N, Birkbak NJ, Watkins TBK, Veeriah S, et al. Tracking the Evolution of Non-Small-Cell Lung Cancer. N Engl J Med. 2017;376:2109–21. doi: 10.1056/NEJMoa1616288. [Internet] [DOI] [PubMed] [Google Scholar]
- 8.Gerlinger M, Rowan AJ, Horswell S, Larkin J, Endesfelder D, Gronroos E, et al. Intratumor Heterogeneity and Branched Evolution Revealed by Multiregion Sequencing. N Engl J Med. 2012;366:883–92. doi: 10.1056/NEJMoa1113205. [Internet] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gerlinger M, Horswell S, Larkin J, Rowan AJ, Salm MP, Varela I, et al. Genomic architecture and evolution of clear cell renal cell carcinomas defined by multiregion sequencing. Nat Genet. 2014;46:225–33. doi: 10.1038/ng.2891. [Internet]. Available from: http://www.nature.com/articles/ng.2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bailey C, Black JRM, Swanton C. Cancer Research: The Lessons to Learn from COVID-19. Cancer Discov. 2020;10:1263–6. doi: 10.1158/2159-8290.CD-20-0823. [Internet] [DOI] [PubMed] [Google Scholar]
- 11.Nowell P. The clonal evolution of tumor cell populations. Science (80-) 1976;194:23–8. doi: 10.1126/science.959840. [Internet] [DOI] [PubMed] [Google Scholar]
- 12.Goldie JH, Coldman AJ. The Genetic Origin of Drug Resistance in Neoplasms: Implications for Systemic Therapy. Cancer Res. 1984;44:3643–53. [PubMed] [Google Scholar]
- 13.Navin N, Krasnitz A, Rodgers L, Cook K, Meth J, Kendall J, et al. Inferring tumor progression from genomic heterogeneity. Genome Res. 2010;20:68–80. doi: 10.1101/gr.099622.109. [Internet] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Anderson K, Lutz C, van Delft FW, Bateman CM, Guo Y, Colman SM, et al. Genetic variegation of clonal architecture and propagating cells in leukaemia. Nature. 2011;469:356–61. doi: 10.1038/nature09650. [Internet]. Available from: http://www.nature.com/articles/nature09650. [DOI] [PubMed] [Google Scholar]
- 15.Nik-Zainal S, Van Loo P, Wedge DC, Alexandrov LB, Greenman CD, Lau KW, et al. The Life History of 21 Breast Cancers. Cell. 2012;149:994–1007. doi: 10.1016/j.cell.2012.04.023. [Internet]. Available from: https://linkinghub.elsevier.com/retrieve/pii/S0092867412005272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Landau DA, Carter SL, Stojanov P, McKenna A, Stevenson K, Lawrence MS, et al. Evolution and Impact of Subclonal Mutations in Chronic Lymphocytic Leukemia. Cell. 2013;152:714–26. doi: 10.1016/j.cell.2013.01.019. [Internet]. Available from: https://linkinghub.elsevier.com/retrieve/pii/S0092867413000718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marusyk A, Tabassum DP, Altrock PM, Almendro V, Michor F, Polyak K. Non-cell-autonomous driving of tumour growth supports sub-clonal heterogeneity. Nature. 2014;514:54–8. doi: 10.1038/nature13556. [Internet]. Available from: http://www.nature.com/articles/nature13556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bozic I, Reiter JG, Allen B, Antal T, Chatterjee K, Shah P, et al. Evolutionary dynamics of cancer in response to targeted combination therapy. Elife. 2013;2 doi: 10.7554/eLife.00747. [Internet]. Available from: https://elifesciences.org/articles/00747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McGranahan N, Burrell RA, Endesfelder D, Novelli MR, Swanton C. Cancer chromosomal instability: therapeutic and diagnostic challenges. EMBO Rep. 2012;13:528–38. doi: 10.1038/embor.2012.61. [Internet] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Carter SL, Eklund AC, Kohane IS, Harris LN, Szallasi Z. A signature of chromosomal instability inferred from gene expression profiles predicts clinical outcome in multiple human cancers. Nat Genet. 2006;38:1043–8. doi: 10.1038/ng1861. [Internet]. Available from: http://www.nature.com/articles/ng1861. [DOI] [PubMed] [Google Scholar]
- 21.Lee AJX, Endesfelder D, Rowan AJ, Walther A, Birkbak NJ, Futreal PA, et al. Chromosomal Instability Confers Intrinsic Multidrug Resistance. Cancer Res. 2011;71:1858–70. doi: 10.1158/0008-5472.CAN-10-3604. [Internet] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zack TI, Schumacher SE, Carter SL, Cherniack AD, Saksena G, Tabak B, et al. Pan-cancer patterns of somatic copy number alteration. Nat Genet. 2013;45:1134–40. doi: 10.1038/ng.2760. [Internet]. Available from: http://www.nature.com/articles/ng.2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.López S, Lim EL, Horswell S, Haase K, Huebner A, Dietzen M, et al. Interplay between whole-genome doubling and the accumulation of deleterious alterations in cancer evolution. Nat Genet. 2020;52:283–93. doi: 10.1038/s41588-020-0584-7. [Internet]. Available from: http://www.nature.com/articles/s41588-020-0584-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kryazhimskiy S, Plotkin JB. The Population Genetics of dN/dS. Gojobori T, editor. PLoS Genet. 2008;4:e1000304. doi: 10.1371/journal.pgen.1000304. [Internet] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martincorena I, Roshan A, Gerstung M, Ellis P, Van Loo P, McLaren S, et al. High burden and pervasive positive selection of somatic mutations in normal human skin. Science (80-) 2015;348:880–6. doi: 10.1126/science.aaa6806. [Internet] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Watkins TBK, Lim EL, Petkovic M, Elizalde S, Birkbak NJ, Wilson GA, et al. Pervasive chromosomal instability and karyotype order in tumour evolution. Nature. 2020;587:126–32. doi: 10.1038/s41586-020-2698-6. [Internet]. Available from: http://www.nature.com/articles/s41586-020-2698-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mitchell TJ, Turajlic S, Rowan A, Nicol D, Farmery JHR, O’Brien T, et al. Timing the Landmark Events in the Evolution of Clear Cell Renal Cell Cancer: TRACERx Renal. Cell. 2018;173:611–623.:e17. doi: 10.1016/j.cell.2018.02.020. [Internet]. Available from: https://linkinghub.elsevier.com/retrieve/pii/S0092867418301648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Williams MJ, Werner B, Barnes CP, Graham TA, Sottoriva A. Identification of neutral tumor evolution across cancer types. Nat Genet. 2016;48:238–44. doi: 10.1038/ng.3489. [Internet]. Available from: http://www.nature.com/articles/ng.3489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Notta F, Chan-Seng-Yue M, Lemire M, Li Y, Wilson GW, Connor AA, et al. A renewed model of pancreatic cancer evolution based on genomic rearrangement patterns. Nature. 2016;538:378–82. doi: 10.1038/nature19823. [Internet]. Available from: http://www.nature.com/articles/nature19823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Baca SC, Prandi D, Lawrence MS, Mosquera JM, Romanel A, Drier Y, et al. Punctuated Evolution of Prostate Cancer Genomes. Cell. 2013;153:666–77. doi: 10.1016/j.cell.2013.03.021. [Internet]. Available from: https://linkinghub.elsevier.com/retrieve/pii/S0092867413003437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bianchi M, Sun M, Jeldres C, Shariat SF, Trinh Q-D, Briganti A, et al. Distribution of metastatic sites in renal cell carcinoma: a population-based analysis. Ann Oncol. 2012;23:973–80. doi: 10.1093/annonc/mdr362. [Internet]. Available from: https://linkinghub.elsevier.com/retrieve/pii/S0923753419346551. [DOI] [PubMed] [Google Scholar]
- 32.Turajlic S, Xu H, Litchfield K, Rowan A, Chambers T, Lopez JI, et al. Tracking Cancer Evolution Reveals Constrained Routes to Metastases: TRACERx Renal. Cell. 2018;173:581–594.:e12. doi: 10.1016/j.cell.2018.03.057. [Internet]. Available from: https://linkinghub.elsevier.com/retrieve/pii/S0092867418303891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chin SF, Teschendorff AE, Marioni JC, Wang Y, Barbosa-Morais NL, Thorne NP, et al. High-resolution aCGH and expression profiling identifies a novel genomic subtype of ER negative breast cancer. Genome Biol. 2007;8:R215. doi: 10.1186/gb-2007-8-10-r215. [Internet] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Burrell RA, McClelland SE, Endesfelder D, Groth P, Weller M-C, Shaikh N, et al. Replication stress links structural and numerical cancer chromosomal instability. Nature. 2013;494:492–6. doi: 10.1038/nature11935. [Internet]. Available from: http://www.nature.com/articles/nature11935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Galon J. Type, Density, and Location of Immune Cells Within Human Colorectal Tumors Predict Clinical Outcome. Science (80-) 2006;313:1960–4. doi: 10.1126/science.1129139. [Internet] [DOI] [PubMed] [Google Scholar]
- 36.Charoentong P, Finotello F, Angelova M, Mayer C, Efremova M, Rieder D, et al. Pan-cancer Immunogenomic Analyses Reveal Genotype-Immunophenotype Relationships and Predictors of Response to Checkpoint Blockade. Cell Rep. 2017;18:248–62. doi: 10.1016/j.celrep.2016.12.019. [Internet]. Available from: https://linkinghub.elsevier.com/retrieve/pii/S2211124716317090. [DOI] [PubMed] [Google Scholar]
- 37.Milo I, Bedora-Faure M, Garcia Z, Thibaut R, Périé L, Shakhar G, et al. The immune system profoundly restricts intratumor genetic heterogeneity. Sci Immunol. 2018;3:eaat1435. doi: 10.1126/sciimmunol.aat1435. [Internet] [DOI] [PubMed] [Google Scholar]
- 38.Algarra I, Garc=a-Lora A, Cabrera T, Ruiz-Cabello F, Garrido F. The selection of tumor variants with altered expression of classical and nonclassical MHC class I molecules: implications for tumor immune escape. Cancer Immunol Immunother. 2004;53 doi: 10.1007/s00262-004-0517-9. [Internet] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McGranahan N, Rosenthal R, Hiley CT, Rowan AJ, Watkins TBK, Wilson GA, et al. Allele-Specific HLA Loss and Immune Escape in Lung Cancer Evolution. Cell. 2017;171:1259–1271.:e11. doi: 10.1016/j.cell.2017.10.001. [Internet]. Available from: https://linkinghub.elsevier.com/retrieve/pii/S0092867417311856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McGranahan N, Furness AJS, Rosenthal R, Ramskov S, Lyngaa R, Saini SK, et al. Clonal neoantigens elicit T cell immunoreactivity and sensitivity to immune checkpoint blockade. Science (80-) 2016;351:1463–9. doi: 10.1126/science.aaf1490. [Internet] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gejman RS, Chang AY, Jones HF, DiKun K, Hakimi AA, Schietinger A, et al. Rejection of immunogenic tumor clones is limited by clonal fraction. Elife. 2018;7 doi: 10.7554/eLife.41090. [Internet]. Available from: https://elifesciences.org/articles/41090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wolf Y, Bartok O, Patkar S, Eli GB, Cohen S, Litchfield K, et al. UVB-Induced Tumor Heterogeneity Diminishes Immune Response in Melanoma. Cell. 2019;179:219–235.:e21. doi: 10.1016/j.cell.2019.08.032. [Internet]. Available from: https://linkinghub.elsevier.com/retrieve/pii/S0092867419309511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rosenthal R, Cadieux EL, Salgado R, Bakir MAl, Moore DA, Hiley CT, et al. Nature. Vol. 567. Nature Publishing Group; 2019. Neoantigen-directed immune escape in lung cancer evolution; pp. 479–85. [Internet]. Available from: http://www.nature.com/articles/s41586-019-1032-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rooney MS, Shukla SA, Wu CJ, Getz G, Hacohen N. Molecular and Genetic Properties of Tumors Associated with Local Immune Cytolytic Activity. Cell. 2015;160:48–61. doi: 10.1016/j.cell.2014.12.033. [Internet]. Available from: https://linkinghub.elsevier.com/retrieve/pii/S0092867414016390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Joshi K, de Massy MR, Ismail M, Reading JL, Uddin I, Woolston A, et al. Nat Med. Vol. 25. Springer US; 2019. Spatial heterogeneity of the T cell receptor repertoire reflects the mutational landscape in lung cancer; pp. 1549–59. [Internet] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reading JL, Gálvez-Cancino F, Swanton C, Lladser A, Peggs KS, Quezada SA. The function and dysfunction of memory CD8 + T cells in tumor immunity. Immunol Rev. 2018;283:194–212. doi: 10.1111/imr.12657. [Internet] [DOI] [PubMed] [Google Scholar]
- 47.Ghorani E, Reading JL, Henry JY, de Massy MR, Rosenthal R, Turati V, et al. The T cell differentiation landscape is shaped by tumour mutations in lung cancer. Nat Cancer. 2020;1:546–61. doi: 10.1038/s43018-020-0066-y. [Internet]. Available from: http://www.nature.com/articles/s43018-020-0066-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thommen DS, Koelzer VH, Herzig P, Roller A, Trefny M, Dimeloe S, et al. A transcriptionally and functionally distinct PD-1+ CD8+ T cell pool with predictive potential in non-small-cell lung cancer treated with PD-1 blockade. Nat Med. 2018;24:994–1004. doi: 10.1038/s41591-018-0057-z. [Internet]. Available from: http://www.nature.com/articles/s41591-018-0057-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Moore DA, Sereno M, Das M, Baena Acevedo JD, Sinnadurai S, Smith C, et al. In situ growth in early lung adenocarcinoma may represent precursor growth or invasive clone outgrowth—a clinically relevant distinction. Mod Pathol. 2019;32:1095–105. doi: 10.1038/s41379-019-0257-1. [Internet]. Available from: http://www.nature.com/articles/s41379-019-0257-1. [DOI] [PubMed] [Google Scholar]
- 50.AbdulJabbar K, Raza SEA, Rosenthal R, Jamal-Hanjani M, Veeriah S, Akarca A, et al. Geospatial immune variability illuminates differential evolution of lung adenocarcinoma. Nat Med. 2020;26:1054–62. doi: 10.1038/s41591-020-0900-x. [Internet]. Available from: http://www.nature.com/articles/s41591-020-0900-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pennycuick A, Teixeira VH, AbdulJabbar K, Raza SEA, Lund T, Akarca AU, et al. Immune Surveillance in Clinical Regression of Preinvasive Squamous Cell Lung Cancer. Cancer Discov. 2020;10:1489–99. doi: 10.1158/2159-8290.CD-19-1366. [Internet] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Abbosh C, Birkbak NJ, Wilson GA, Jamal-Hanjani M, Constantin T, Salari R, et al. Nature. Vol. 545. Nature Publishing Group; 2017. Phylogenetic ctDNA analysis depicts early-stage lung cancer evolution; pp. 446–51. [Internet]. Available from: http://www.nature.com/articles/nature22364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bratman SV, Yang SYC, Iafolla MAJ, Liu Z, Hansen AR, Bedard PL, et al. Personalized circulating tumor DNA analysis as a predictive biomarker in solid tumor patients treated with pembrolizumab. Nat Cancer. 2020;1:873–81. doi: 10.1038/s43018-020-0096-5. [Internet]. Available from: http://www.nature.com/articles/s43018-020-0096-5. [DOI] [PubMed] [Google Scholar]
- 54.Powles T, Assaf ZJ, Davarpanah N, Hussain M, Oudard S, Gschwend JE, et al. 10 - Clinical outcomes in post-operative ctDNA-positive muscle-invasive urothelial carcinoma (MIUC) patients after atezolizumab adjuvant therapy. ESMO Immuno-Oncology Virtual Congress 2020 (9-12 December) Ann Oncol. 2020;31((suppl)) [Google Scholar]
- 55.Hussain MHA, Powles T, Albers P, Castellano D, Daneshmand S, Gschwend J, et al. IMvigor010: Primary analysis from a phase III randomized study of adjuvant atezolizumab (atezo) versus observation (obs) in high-risk muscle-invasive urothelial carcinoma (MIUC) J Clin Oncol. 2020;38:5000–5000. doi: 10.1200/JCO.2020.38.15_suppl.5000. [Internet] [DOI] [Google Scholar]
- 56.Christensen E, Birkenkamp-Demtröder K, Sethi H, Shchegrova S, Salari R, Nordentoft I, et al. Early Detection of Metastatic Relapse and Monitoring of Therapeutic Efficacy by Ultra-Deep Sequencing of Plasma Cell-Free DNA in Patients With Urothelial Bladder Carcinoma. J Clin Oncol. 2019;37:1547–57. doi: 10.1200/JCO.18.02052. [Internet] [DOI] [PubMed] [Google Scholar]
- 57.Chemi F, Rothwell DG, McGranahan N, Gulati S, Abbosh C, Pearce SP, et al. Pulmonary venous circulating tumor cell dissemination before tumor resection and disease relapse. Nat Med. 2019;25:1534–9. doi: 10.1038/s41591-019-0593-1. [Internet]. Available from: http://www.nature.com/articles/s41591-019-0593-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gyanchandani R, Lin Y, Lin H-M, Cooper K, Normolle DP, Brufsky A, et al. Intratumor Heterogeneity Affects Gene Expression Profile Test Prognostic Risk Stratification in Early Breast Cancer. Clin Cancer Res. 2016;22:5362–9. doi: 10.1158/1078-0432.CCR-15-2889. [Internet] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gulati S, Martinez P, Joshi T, Birkbak NJ, Santos CR, Rowan AJ, et al. Systematic Evaluation of the Prognostic Impact and Intratumour Heterogeneity of Clear Cell Renal Cell Carcinoma Biomarkers. Eur Urol. 2014;66:936–48. doi: 10.1016/j.eururo.2014.06.053. [Internet]. Available from: https://linkinghub.elsevier.com/retrieve/pii/S0302283814006277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Boutros PC. The path to routine use of genomic biomarkers in the cancer clinic. Genome Res. 2015;25:1508–13. doi: 10.1101/gr.191114.115. [Internet] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hammerman PS, Voet D, Lawrence MS, Voet D, Jing R, Cibulskis K, et al. Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012;489:519–25. doi: 10.1038/nature11404. [Internet]. Available from: http://www.nature.com/articles/nature11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Collisson EA, Campbell JD, Brooks AN, Berger AH, Lee W, Chmielecki J, et al. Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511:543–50. doi: 10.1038/nature13385. [Internet]. Available from: http://www.nature.com/articles/nature13385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Djureinovic D, Hallström BM, Horie M, Mattsson JSM, La Fleur L, Fagerberg L, et al. Profiling cancer testis antigens in non-small-cell lung cancer. JCI Insight. 2016;1 doi: 10.1172/jci.insight.86837. [Internet]. Available from: https://insight.jci.org/articles/view/86837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Biswas D, Birkbak NJ, Rosenthal R, Hiley CT, Lim EL, Papp K, et al. Nat Med. Vol. 25. Springer US; 2019. A clonal expression biomarker associates with lung cancer mortality; pp. 1540–8. [Internet] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gandhi L, Rodríguez-Abreu D, Gadgeel S, Esteban E, Felip E, De Angelis F, et al. Pembrolizumab plus Chemotherapy in Metastatic Non-Small-Cell Lung Cancer. N Engl J Med. 2018;378:2078–92. doi: 10.1056/NEJMoa1801005. [Internet] [DOI] [PubMed] [Google Scholar]
- 66.Paz-Ares L, Luft A, Vicente D, Tafreshi A, Gümüş M, Mazières J, et al. Pembrolizumab plus Chemotherapy for Squamous Non-Small-Cell Lung Cancer. N Engl J Med. 2018;379:2040–51. doi: 10.1056/NEJMoa1810865. [Internet] [DOI] [PubMed] [Google Scholar]
- 67.McDonald OG, Li X, Saunders T, Tryggvadottir R, Mentch SJ, Warmoes MO, et al. Epigenomic reprogramming during pancreatic cancer progression links anabolic glucose metabolism to distant metastasis. Nat Genet. 2017;49:367–76. doi: 10.1038/ng.3753. [Internet]. Available from: http://www.nature.com/articles/ng.3753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Calabrese C, Davidson NR, Demircioğlu D, Fonseca NA, He Y, Kahles A, et al. Genomic basis for RNA alterations in cancer. Nature. 2020;578:129–36. doi: 10.1038/s41586-020-1970-0. [Internet]. Available from: http://www.nature.com/articles/s41586-020-1970-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bell CC, Gilan O. Principles and mechanisms of non-genetic resistance in cancer. Br J Cancer. 2020;122:465–72. doi: 10.1038/s41416-019-0648-6. [Internet]. Available from: http://www.nature.com/articles/s41416-019-0648-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.McGranahan N, Swanton C. Neoantigen quality, not quantity. Sci Transl Med. 2019;11:eaax7918. doi: 10.1126/scitranslmed.aax7918. [Internet] [DOI] [PubMed] [Google Scholar]
- 71.De Mattos-Arruda L, Vazquez M, Finotello F, Lepore R, Porta E, Hundal J, et al. Neoantigen prediction and computational perspectives towards clinical benefit: recommendations from the ESMO Precision Medicine Working Group. Ann Oncol. 2020;31:978–90. doi: 10.1016/j.annonc.2020.05.008. [Internet]. Available from: https://linkinghub.elsevier.com/retrieve/pii/S0923753420398240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yuki K, Cheng N, Nakano M, Kuo CJ. Organoid Models of Tumor Immunology. Trends Immunol. 2020;41:652–64. doi: 10.1016/j.it.2020.06.010. [Internet] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.LaFave LM, Kartha VK, Ma S, Meli K, Del Priore I, Lareau C, et al. Epigenomic State Transitions Characterize Tumor Progression in Mouse Lung Adenocarcinoma. Cancer Cell. 2020;38:212–228.:e13. doi: 10.1016/j.ccell.2020.06.006. [Internet]. Available from: https://linkinghub.elsevier.com/retrieve/pii/S153561082030310X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ali HR, Jackson HW, Zanotelli VRT, Danenberg E, Fischer JR, Bardwell H, et al. Imaging mass cytometry and multiplatform genomics define the phenogenomic landscape of breast cancer. Nat Cancer. 2020;1:163–75. doi: 10.1038/s43018-020-0026-6. [Internet]. Available from: http://www.nature.com/articles/s43018-020-0026-6. [DOI] [PubMed] [Google Scholar]
- 75.Hu Z, Li Z, Ma Z, Curtis C. Nat Genet. Vol. 52. Springer US; 2020. Multi-cancer analysis of clonality and the timing of systemic spread in paired primary tumors and metastases; pp. 701–8. [Internet] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Morita K, Wang F, Jahn K, Hu T, Tanaka T, Sasaki Y, et al. Nat Commun. Vol. 11. Springer US; 2020. Clonal evolution of acute myeloid leukemia revealed by high-throughput single-cell genomics; p. 5327. [Internet] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yates LR, Knappskog S, Wedge D, Farmery JHR, Gonzalez S, Martincorena I, et al. Genomic Evolution of Breast Cancer Metastasis and Relapse. Cancer Cell. 2017;32:169–184.:e7. doi: 10.1016/j.ccell.2017.07.005. [Internet]. Available from: https://linkinghub.elsevier.com/retrieve/pii/S1535610817302970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Araf S, Wang J, Korfi K, Pangault C, Kotsiou E, Rio-Machin A, et al. Genomic profiling reveals spatial intra-tumor heterogeneity in follicular lymphoma. Leukemia. 2018;32:1261–5. doi: 10.1038/s41375-018-0043-y. [Internet]. Available from: http://www.nature.com/articles/s41375-018-0043-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Barry P, Vatsiou A, Spiteri I, Nichol D, Cresswell GD, Acar A, et al. The Spatiotemporal Evolution of Lymph Node Spread in Early Breast Cancer. Clin Cancer Res. 2018;24:4763–70. doi: 10.1158/1078-0432.CCR-17-3374. [Internet] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Spiteri I, Caravagna G, Cresswell GD, Vatsiou A, Nichol D, Acar A, et al. Ann Oncol. Vol. 30. Elsevier Masson SAS; 2019. Evolutionary dynamics of residual disease in human glioblastoma; pp. 456–63. [Internet] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Landau DA, Sun C, Rosebrock D, Herman SEM, Fein J, Sivina M, et al. Nat Commun. Vol. 8. Springer US; 2017. The evolutionary landscape of chronic lymphocytic leukemia treated with ibrutinib targeted therapy; p. 2185. [Internet] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Barthel FP, Johnson KC, Varn FS, Moskalik AD, Tanner G, Kocakavuk E, et al. Nature. Vol. 576. Springer US; 2019. Longitudinal molecular trajectories of diffuse glioma in adults; pp. 112–20. [Internet] [DOI] [PMC free article] [PubMed] [Google Scholar]