Abstract
Very long chain fatty acids (VLCFAs) are essential building blocks for synthesis of the ceramides and sphingolipids. The first step in the fatty acid elongation cycle is catalysed by the 3-keto acyl-CoA synthases (in mammals, ELOVL elongases). Although ELOVLs are implicated in common diseases including insulin resistance, hepatic steatosis and Parkinson’s, their underlying molecular mechanisms are unknown. Here we report the structure of the human ELOVL7 elongase, which comprises an inverted transmembrane barrel surrounding a 35 Å long tunnel containing a covalently attached product analogue. The structure reveals the substrate binding sites in the narrow tunnel and an active site deep in the membrane. We demonstrate that chain elongation proceeds via an acyl-enzyme intermediate involving the second histidine in the canonical HxxHH motif. The unusual substrate-binding arrangement and chemistry suggest mechanisms for selective ELOVL inhibition, relevant for diseases where VLCFAs accumulate such as X-linked adrenoleukodystrophy.
Long and very long chain fatty acids (FAs) (LCFAs: 12C:20C and VLCFAs: >20C)1,2 are precursors for eicosanoid signalling molecules and for the synthesis of ceramides and sphingolipids3. These lipids perform many unique cellular functions, serving as structural components of biological membranes where they are essential for the myelin sheaths of nerves4,5, the skin permeability barrier6,7 and but also in retina8 and liver function4,9.
In eukaryotes, LCFAs produced in the cytosol are further elongated into VLCFAs through the action of four distinct endoplasmic reticulum (ER) membrane-embedded enzymes. The first, rate-limiting step of the FA elongation cycle is catalysed by 3-keto acyl-CoA synthases and consists of a condensation reaction between an acyl-CoA and malonyl-CoA to yield a 3-keto acyl-CoA (Fig. 1a). This product then undergoes a series of reduction and dehydration reactions catalysed by three other ER-resident enzymes to yield an acyl-CoA with two additional carbons in the acyl chain. There are two distinct families of fatty acid elongase condensing enzymes. In mammals10, yeast11,12, protists13,14 and other eukaryotes1, the condensation step is catalysed by the ELO elongases whereas plants and some protists primarily utilize unrelated 3-ketoacyl-CoA synthases (KCSs) that are homologous to Arabidopsis FA elongase 1 (FAE1) 1,15,16.
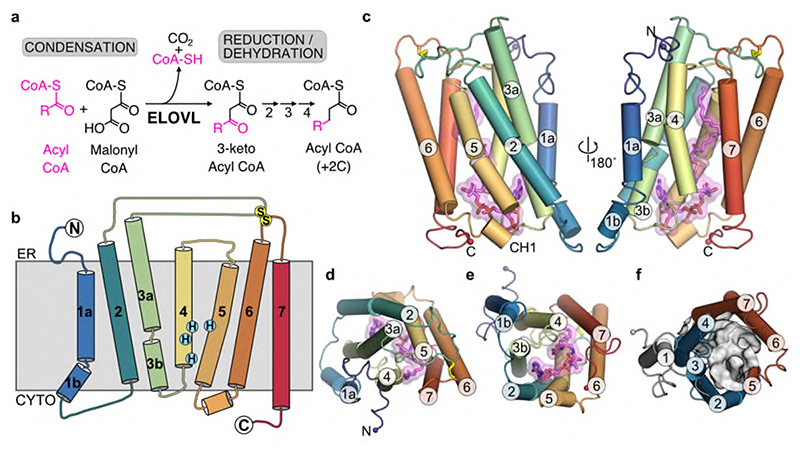
Fig. 1. Overall structure of ELOVL7.
a, FA elongase reaction sequence. b, Schematic representation of the seven transmembrane (TM) helix topology. c-e, Cartoon of ELOVL7 structure viewed c, parallel to the membrane plane and from either d, the ER or e, the cytoplasmic faces. f, ELOVL7 6-TM inverted barrel organisation viewed from the cytoplasmic face.
Mammals have seven elongation of very long chain fatty acids (ELOVL1-7) elongases, each with distinct preferences for acyl chain length and the degree of unsaturation3. Mutations in ELOVL elongases cause severe genetic diseases including Stargardt syndrome17 and spinocerebellar ataxia18,19. Mouse knockouts suggest ELOVL involvement in hepatic steatosis20 and insulin resistance21, and in particular ELOVL7 is implicated in cancer22–24, early-onset Parkinson’s disease25 and necroptosis26. However, very little is known about the molecular mechanisms underlying this key step in fatty acid and lipid synthesis by the ELOVLs.
In order to understand the molecular basis of acyl chain elongation by the ELOVL elongases, we purified and solved the crystal structure of human ELOVL7 in complex with a copurified, covalently bound 3-keto acyl-CoA and performed intact protein mass spectrometry (MS) experiments to probe its catalytic mechanism. The results obtained enabled us to propose a ping-pong type mechanism, achieved through the formation of an unusual acyl-imidazole intermediate upon reaction with the first acyl-CoA substrate.
Results
Structure of human ELOVL7
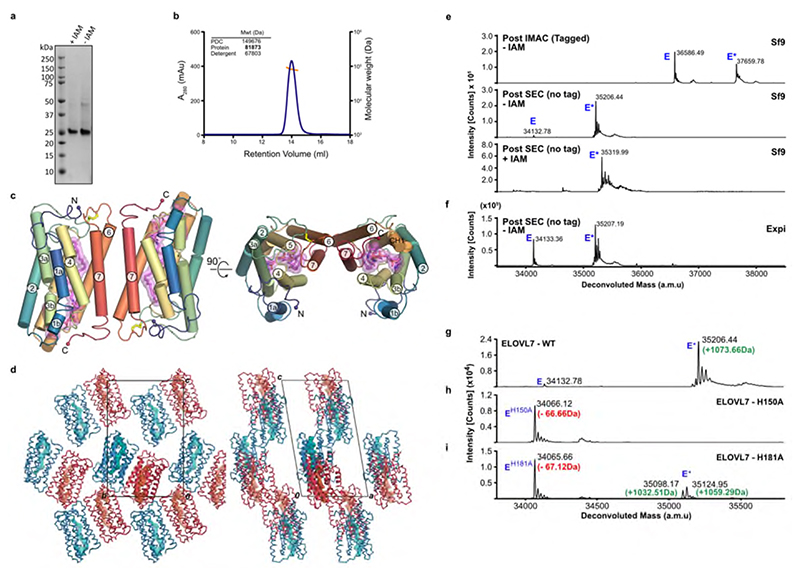
We have solved the structure of human ELOVL7 by X-ray crystallography to 2.6 Å resolution (see Methods, Table 1 and Extended Data Fig. 1a), revealing an inverted dimer with a small and unconserved interaction surface (870 Å2) in the crystals which was also observed in solution (Extended Data Fig. 1b-d).
Table 1. X-ray data collection, refinement & validation statistics.
| Hg peak | WT-native | WT-STARANISO | |
|---|---|---|---|
| Data collection | |||
| Space group | P21 | P21 | P21 |
| Cell dimensions | |||
| a, b, c (Å) | 62.34, 72.42, 111.68 | 63.23, 72.46, 112.04 | 63.23, 72.46, 112.04 |
| α, β, γ (°) | 90, 100.14, 90 | 90, 100.03, 90 | 90, 100.03, 90 |
| Resolution (Å) | 58-3.0 (3.18-3.0)* | 62-2.05 (2.10-2.05) | 62-2.05 (2.18-2.05) |
| R meas | 0.396 (2.516) | 0.161 (4.108) | 0.119 (0.839) |
| R pim | 0.093 (0.588) | 0.061 (1.534) | 0.062 (0.449) |
| I / σl | 6.2 (1.5) | 4.9 (0.4) | 8.3 (1.9) |
| CC1/2 | 0.996 (0.838) | 0.985 (0.473) | 0.987 (0.813) |
| Completeness (%) | 99.9 (99.6) | 99.5 (96.6) | 58.7 (17.5) |
| Redundancy | 17.6 (18) | 6.9 (6.9) | 6.9 (6.2) |
| Refinement | |||
| Resolution (Å) | 28.68-2.05 | ||
| No. reflections | 36690 | ||
| R work / R free | 0.2104 / 0.2261 | ||
| No. atoms | |||
| Protein | 4260 | ||
| Ligand/ion | 293 | ||
| Water | 112 | ||
| B-factors | |||
| Protein | 44 | ||
| Ligand/ion | 49 | ||
| Water | 37 | ||
| R.m.s deviations | |||
| Bond lengths (Å) | 0.008 | ||
| Bond angles (°) | 0.87 |
Values in parentheses are for highest-resolution shell.
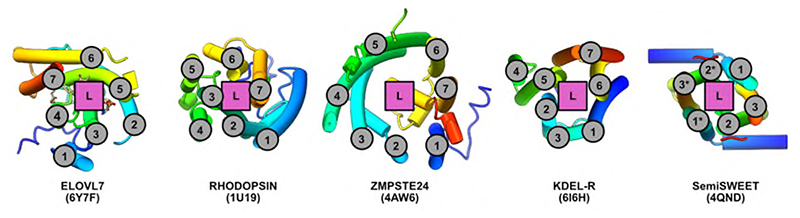
Overall ELOVL7 has seven transmembrane (TM) helices (TM1-TM7) with TM2-7 forming a six TM inverted barrel surrounding a narrow tunnel (Fig. 1b-f), formed by two units of three helices (TM2-4 and TM5-7) arranged in an antiparallel fashion. The two 3-helix units are assembled as an inverted repeat around the central tunnel (Fig. 1f) with TM1 lying against TM3/4, outside the barrel. This fold is unlike the GPCR 7TM fold and we did not find any six or seven TM protein structures with a similar fold (DALI27) (Extended Data Fig. 2). The 35 Å long central tunnel has a narrow (8-10 Å wide) opening on the cytoplasmic face of the protein and is sealed at its ER lumen end by the short buried loop between TM4-5, which connects the two halves of the barrel (Fig. 1b-e). This loop is separated from the ER lumen by the two disulphide-linked ER loops between TMs2-3 and TMs6-7.
ELOVL7 copurifies with a bound 3-keto acyl-CoA
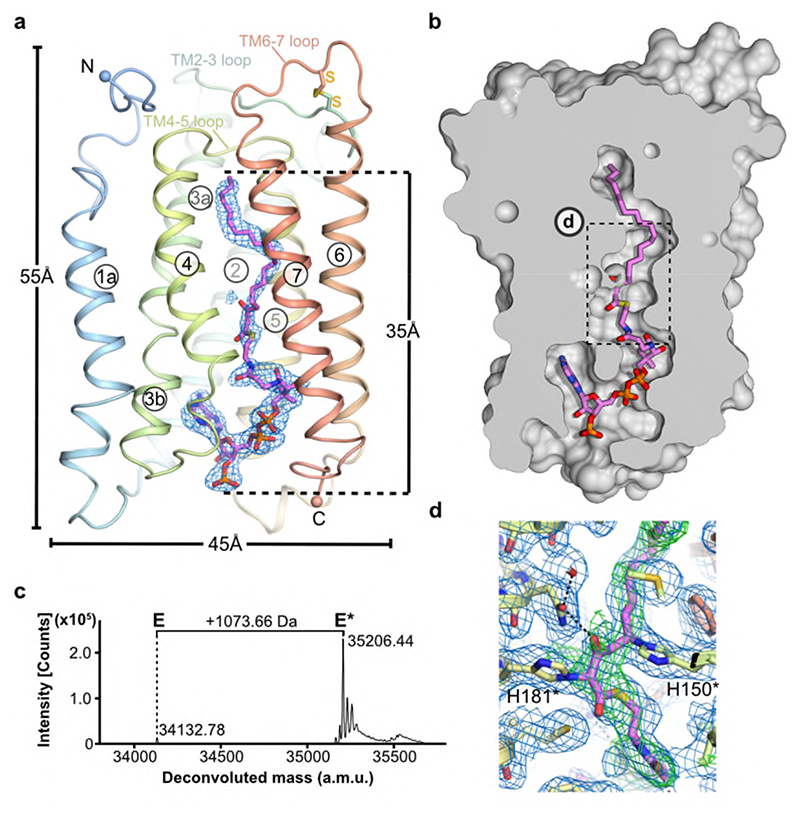
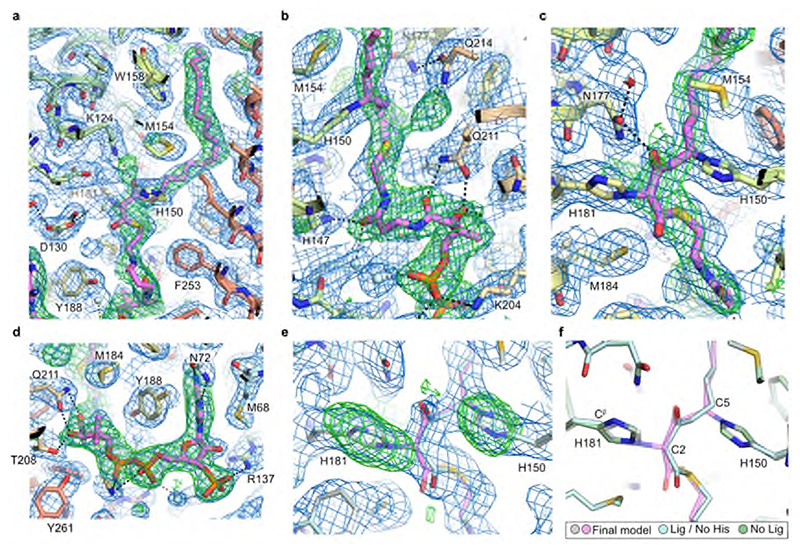
Surprisingly, the electron density maps clearly showed a long hooked density extending from the closed ER end of the tunnel to the cytoplasmic surface of the protein. This electron density is consistent with there being a bound acyl-CoA within the active site tunnel spanning its entire length (Fig. 2a,b and Extended Data Fig. 3). Denaturing intact mass spectrometry analysis of protein samples taken during purification showed two species: a form with the expected mass of the purified protein construct (34132.78 Da) and a modified form with a mass adduct of 1073.66 Da (Fig. 2c). This covalent mass adduct was observed both with protein obtained from over-expression in Spodoptera frugiperda (Sf9) insect cells or mammalian (Expi293F) cells (Extended Data Fig. 1e,f). The form with the mass adduct was more stable during purification, so that following size exclusion chromatography (SEC), it represented >90% of the purified protein (Fig. 2c). Based on the shape of the electron density within the central tunnel and the observed mass adduct, we have modelled a covalently bound 3-keto eicosanoyl(C20)-CoA (Mw 1074.02 Da), an analogue of the reaction product which co-purified with ELOVL7. The electron density clearly shows that the bound 3-keto C20-CoA is covalently attached to the imidazole rings of both H150 and H181 (Fig. 2d and Extended Data Fig. 3); the C5 carbon of the 3-keto C20-CoA is attached to the N epsilon (Nε) of H150 and the C2 carbon atom is attached to the Nε of H181.
Fig. 2. Heterologously expressed ELOVL7 is covalently bound to a 3-keto acyl-CoA.
a, Cartoon showing the covalently bound 3-keto eicosanoyl-CoA along with the FoFc omit electron density map (blue mesh, contoured at 3σ). b, Cutaway molecular surface representation showing the extent of the enclosed central active site tunnel. c, Intact mass analysis of Sf9 purified protein (E) highlighting adduct species (E*; +1073.66Da). Data shown from one experiment. Similar adduct peaks were observed for all purifications tested (n=3). d, Electron density in region around covalent linkages to active site histidines. Final BUSTER 2mFo-DFc (blue mesh, contoured at 1sigma) and omit mFo-DFc (green mesh, contoured at 2.5sigma) electron density maps are overlaid on the final model.
The substrate binding site lies within an extended tunnel
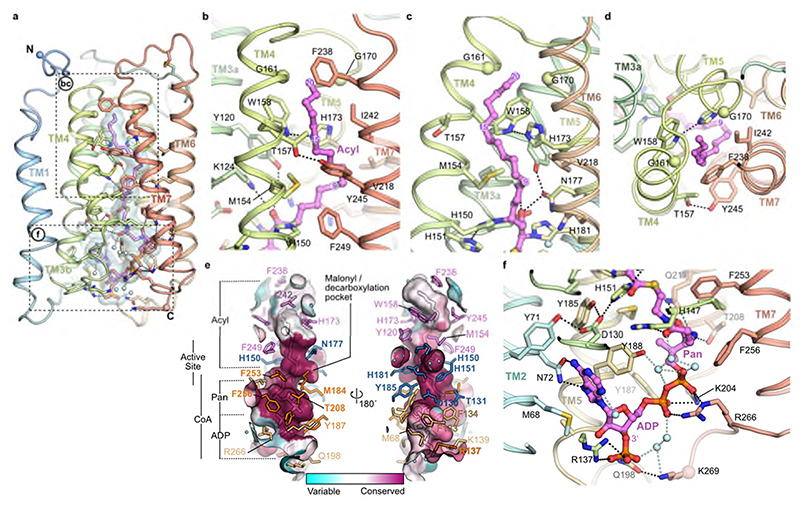
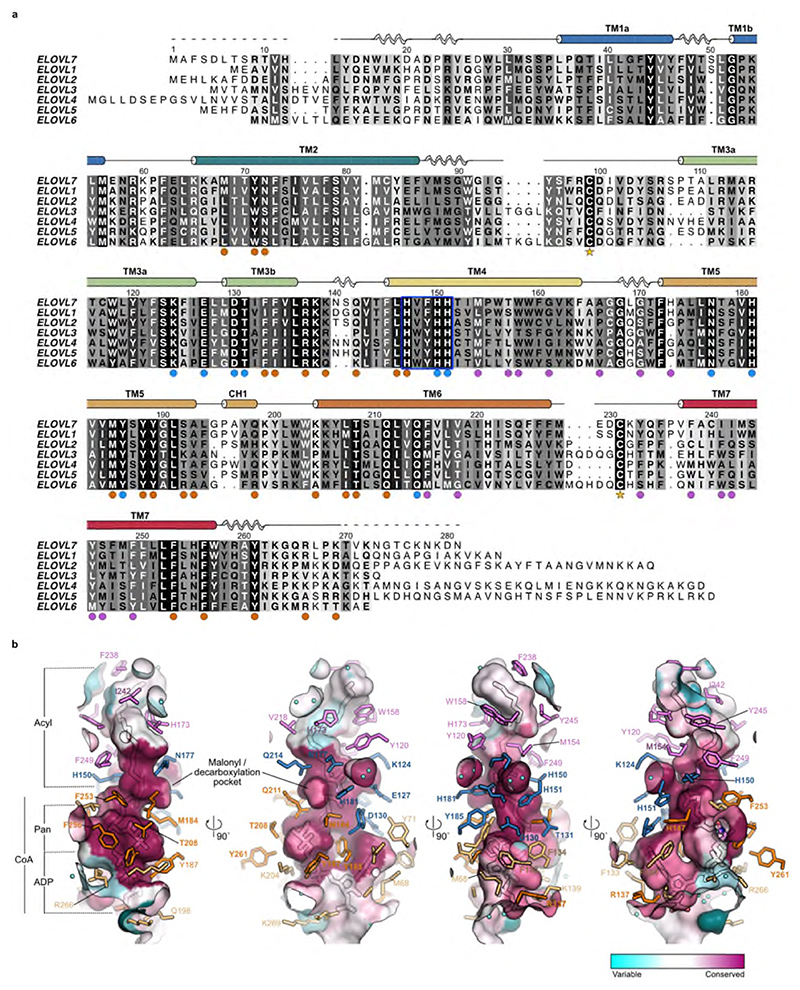
The covalently bound 3-keto acyl-CoA seen in the crystal structure conveniently marks out the binding sites for both the acyl and CoA components of the substrates and products (Fig. 3) and reveals the architecture of the active site (Fig. 4). The acyl chain binding site is located at the upper, occluded end of the tunnel and is terminated by very short TM4-5 loop (Fig. 3a-d). It is lined predominantly by hydrophobic sidechains from TM4 and TM7 (M154, W158, G161, G170, F238, I242 and Y245), and is curved, allowing both unsaturated and saturated acyl chains to bind (Fig. 3b-d). The tunnel is kinked in two positions, around M154 at the C9 carbon of the acyl chain and adjacent to T157 and H173 at the C15 acyl carbon (Fig. 3b,c). These locations correspond to Δ6 and Δ12 sites of unsaturation in fatty acid substrates. The acyl chain of the 3-keto acyl-CoA seen in the structure has 20 ordered carbons and it does not occupy the entire pocket. There is space for two additional carbons, suggesting that 3-keto acyl-CoA products with up to 22 carbons could be accommodated. While ELOVL7 has been reported to have a preference for C18-CoA substrates, it can accept substrates with carbon chains up to C20 in length3,28 which is consistent with the observed length of the acyl chain binding site in the structure. The seven ELOVL isoenzymes have different preferences for acyl chain length and number of double bonds3, and sequence alignments of the human ELOVL elongases suggest that the residues lining the acyl chain binding site are not well conserved (Fig. 3e; Extended Data Fig. 4), reflecting their abilities to accommodate different acyl chains. Differences in particular in the TM6-TM7 loop and at the N-terminal region of TM7 (Extended Data Fig. 4a) are likely to impact the size and shape of the acyl binding tunnel. Previous studies with yeast ELOs demonstrated that changes in this region were able to influence fatty acid chain length29.
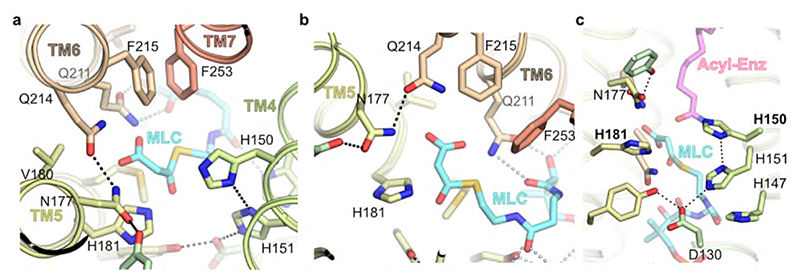
Fig. 3. Acyl chain and CoA binding sites.
a, Overview of the central acyl-CoA binding tunnel. b-d, Acyl chain binding pocket. e, Conservation of active site tunnel. Molecular surface representation is coloured by amino acid conservation score calculated by CONSURF49 analysis of a diverse set of ELOV1-7 family members. The various subregions of the tunnel are indicated (ADP / Pan from CoA and Acyl chain). Amino acid residues that form the binding tunnel are coloured according to region (pink, acyl; blue, catalytic site; orange CoA binding). f, Cytoplasmic-facing CoA 3’-phospho ADP/pantothenic (Pan) binding pocket.
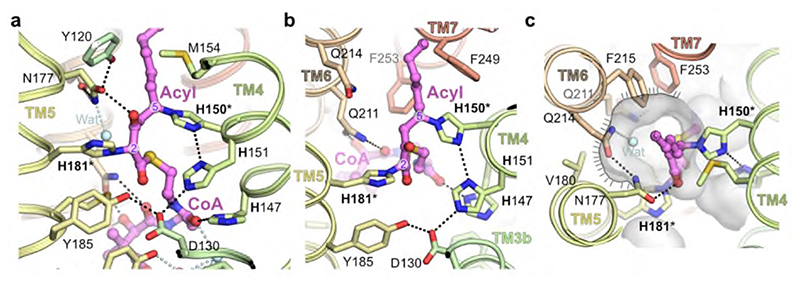
Fig. 4. Catalytic site around the HxxHH motif.
a,b, Orthogonal views of central active site. c, Putative malonyl binding site adjacent to catalytic histidines. Molecular surface is shown in transparent grey highlighting the putative binding site for malonyl moiety of second substrate (indicated with hatched line). In the crystal structure this is filled by a water molecule (Wat). Covalently modified histidines (H150/H181) are highlighted by asterisks.
The CoA binding site lies at the open, cytoplasmic end in the tunnel, with the CoA almost completely buried within the core of the enzyme (95% of the 1400 Å2 ligand surface area is buried). The 3’-phospho-ADP/pantothenate portion of the CoA adopts a ‘U’ shaped conformation wrapped around the sidechain of Y188 (Fig. 3f). The adenine ring is inserted in a narrow cleft between TM2, 3 and 5. N72 on TM2 forms hydrogen bonds to both N5 and N6 on the adenine base and D130 on TM3b provides a third hydrogen bond to the base (Fig. 3f). The 3’ phosphate projects towards the solvent and forms a salt bridge with R137. The diphosphate forms salt bridges with K204 and R266 and a hydrogen bond with the sidechain of Y187. The pantothenate (Pan) moiety follows a helical path and is hydrogen bonded to the sidechains of T208, Q211 and H147. The cysteamine is elongated and sits in a hydrophobic pocket surrounded by M184 and F253. Together these extensive interactions ensure a highly specific binding site for the CoA portion of the substrates.
The catalytic site is surrounded by conserved histidines
The catalytic site of ELOVL elongases lies at the centre of the membrane and the residues lining this section of the tunnel are highly conserved across all seven elongases (Fig. 3e). Previous studies on the ELO family have identified a number of conserved sequence motifs including a HxxHH histidine-box that are essential for activity29,30. A site directed mutagenesis study in the ELOVL homologue EloA from the social amoeba Dictyostelium discoideum identified a series of residues that were essential for activity (equivalent to K124, D130, H150, H151, N177, H181 and Q211 in ELOVL7)30 and all of these residues lie within the active site.
In the structure, the 3-keto acyl-CoA thioester sits just below a triad of histidines – H150 and H151 from the conserved 147HxxHH151 motif at the N-terminal end of TM4 are positioned on one side of the active site and H181 on TM5 lies on the other side directly opposite to H150 (Fig. 4). All three histidines of the HxxHH motif lie on the same face of TM4 and interact with the bound 3-keto acyl-CoA (Fig. 4ab). Although this type of histidine motif is sometimes associated with metal ion binding, for example in fatty acyl desaturases31, the electron density maps show no indication of bound metal ions and the geometry is not suitable for metal ion binding, indicating that metal-assisted catalysis is unlikely.
The sidechains of H150 and H181 are covalently attached to the bound 3-keto acyl-CoA. H150 is bound to the C5 carbon via the imidazole ring N epsilon and H181 is bound to the C2 carbon between the two carbonyl groups. The 3-keto group is hydrogen-bonded to the sidechains of K124 and N177. Adjacent to the sidechain of H181 is a side pocket off the main tunnel between TM5 and TM6. This shallow pocket is lined by the sidechains of N177, V180, H181, M184, Q211, Q214, F215 and F253 (Fig. 4c). Modelling suggests that this pocket probably accommodates the malonyl portion of the second substrate malonyl-CoA after the acyl-CoA has bound. Hereafter we refer to this sub-region of the active site as the ‘malonyl / decarboxylation pocket’.
Fatty acid extension proceeds via a ping-pong type mechanism
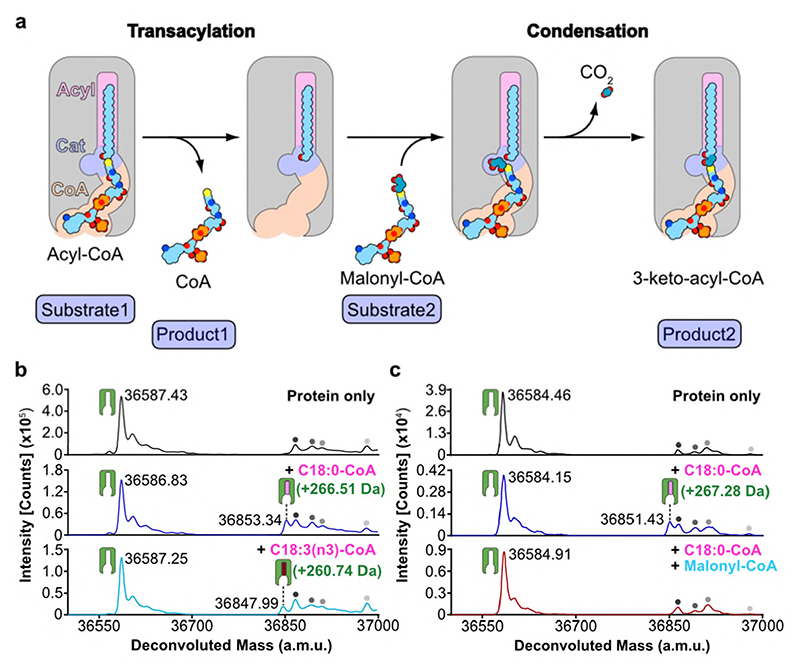
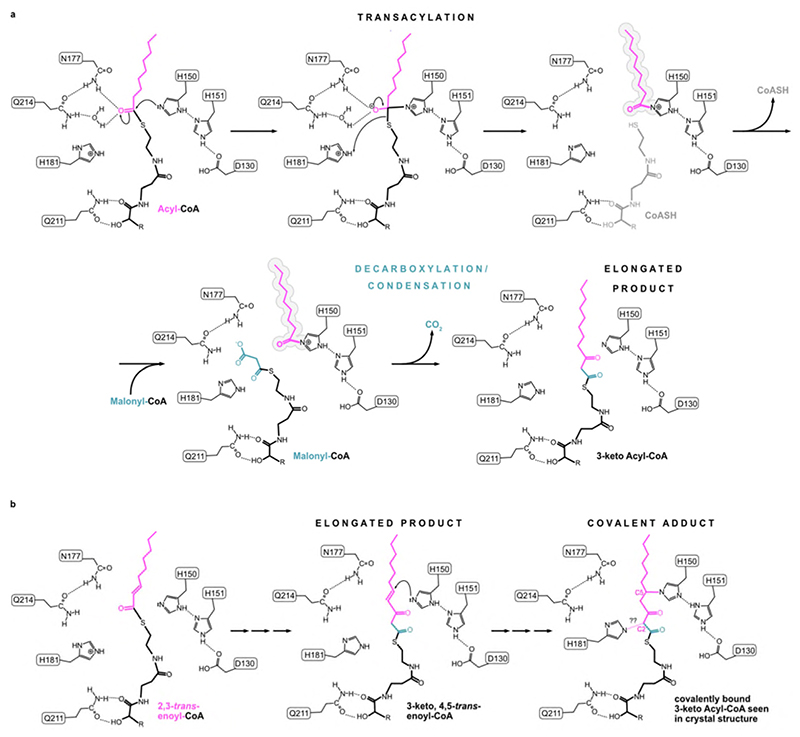
Acyl transferase reactions generally involve either a ping-pong or a ternary complex mechanism. In the first, the two substrates bind one after the other and a covalent intermediate is involved, whereas in the second both substrates bind at the same time and the acyl group is transferred directly from one substrate to the other. In ELOVL7 the narrow substrate binding tunnel does not provide sufficient space for both substrates to bind at the same time (Fig. 2b), excluding a ternary complex mechanism. The narrow tunnel is however consistent with a ping-pong type mechanism, where the first product must leave before the second substrate binds (Fig. 5a). Therefore, we propose that in the first step of the reaction the acyl-CoA binds, with the FA tail occluded at the closed ER end of the tunnel and the CoA moiety near the entrance. Then the CoA is cleaved and can dissociate, leaving behind the buried acyl chain. In the second step, the malonyl-CoA binds at the more accessible CoA binding site, with the malonyl unit in a side pocket in the central active site. Decarboxylation and carbon-carbon bond formation will then yield the 3-keto acyl-CoA product. The entry and exit route for acyl-CoAs and 3-keto acyl-CoAs is likely to involve lateral movement of the hydrophobic acyl chain directly into the membrane, between two TM helices, rather than having the acyl chain move along the whole of the tunnel, parts of which are hydrophilic. The structure suggests that this entry/exit portal lies between TM4 and TM7, between the two 3TM halves of the barrel (Fig. 1f).
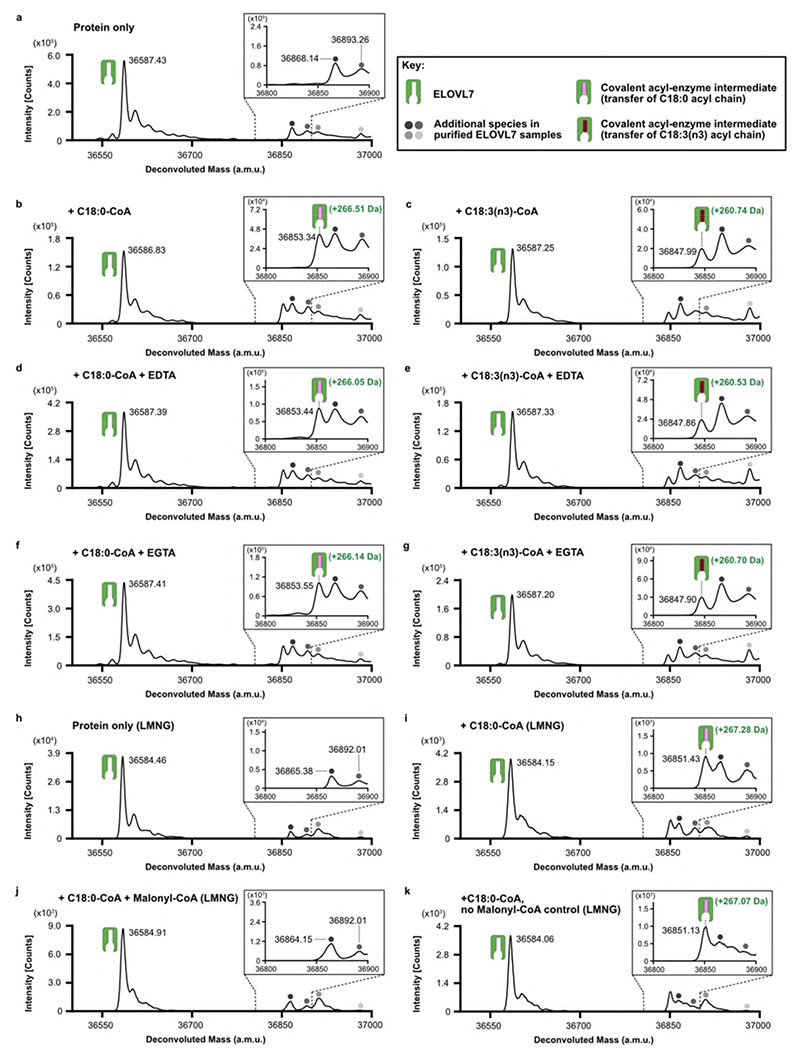
Fig. 5. Proposed ping-pong mechanism and evidence for a covalent acyl-enzyme intermediate.
a, Schematic outlining proposed ELOVL ping-pong mechanism. b-c, Intact mass analysis of ELOVL7 in presence of b, acyl-CoA substrates demonstrating transacylation to form an acyl-enzyme intermediate and c, acyl-CoA followed by malonyl-CoA. Intact mass analysis of protein only (black), protein + C18:0 acyl-CoA (blue), protein + C18:3 acyl-CoA (cyan) and protein + C18:0 acyl-CoA + malonyl-CoA (red). Peaks are indicated as follows: protein (green icon), acyl enzyme intermediate (green icon + pink/red oblong), background species present in all traces (grey circles). (n=2 biological repeats, see Supplementary Figures 1-2 for replicate traces; Supplementary Table 2 for experimental and theoretical masses and mass errors).
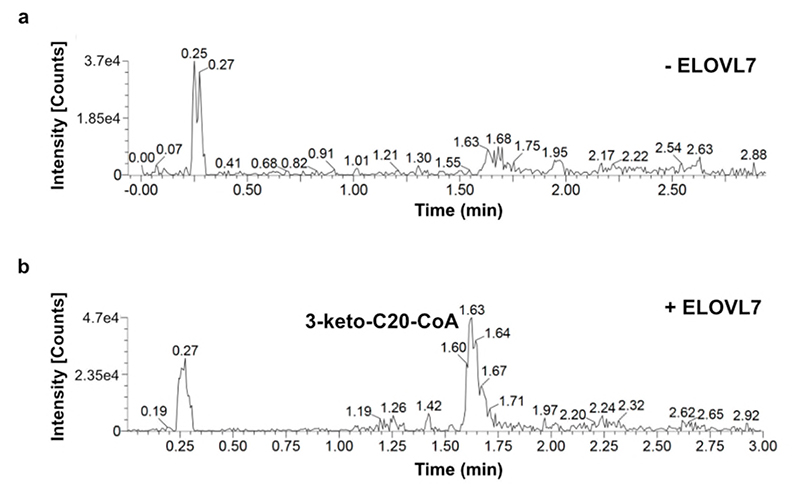
To investigate the mechanism in more detail, we monitored the ELOVL7-catalysed elongation reaction using mass spectrometry. Although the unmodified enzyme is unstable and readily precipitates during purification, we were able to confirm that detergent-purified ELOVL7 converts the substrates stearoyl-CoA (C18:0) and malonyl-CoA to give the product 3-keto-eicosanoyl (C20)-CoA (Extended Data Fig. 5a,b), indicating that although ~90% of the final purified enzyme is covalently modified by the 3-keto acyl-CoA and therefore non-functional, the remaining unmodified protein is active.
A key feature of a ping-pong type reaction mechanism is the formation of a substituted enzyme intermediate prior to the binding of the second substrate. The formation of such a covalent acyl-enzyme intermediate at the end of the first step was confirmed by denaturing intact mass spectrometry analysis of the enzyme after incubation with an acyl-CoA substrate (C18:0-CoA). We observed a 266.51 Da adduct (Fig. 5b and Extended Data Fig. 6a,b), consistent with the transfer of a C18:0-acyl group to the protein. Similarly, incubation with the unsaturated C18:3(n3)-CoA substrate also resulted in the formation of a 260.74 Da adduct (Fig.5b; Extended Data Fig 6c). Addition of EDTA or EGTA did not interfere with covalent modification of the protein by either substrate, supporting the hypothesis that metal ions are not required for catalysis (Extended Data Fig. 6d-g).
The sequential nature of the reaction was demonstrated by initial incubation of the enzyme with the first substrate (C18:0-CoA), which gave rise to the acyl-intermediate species (Fig. 5c and Extended Data Fig. 6h-i), followed by further incubation with the second substrate malonyl-CoA. Analysis of the mixture at the end of the reaction shows a complete loss of the deconvoluted mass peak corresponding to the adduct intermediate, consistent with the reaction having gone to completion (Fig. 5c and Extended Data Fig. 6j).
Catalysis via an acyl-imidazole intermediate involving H150
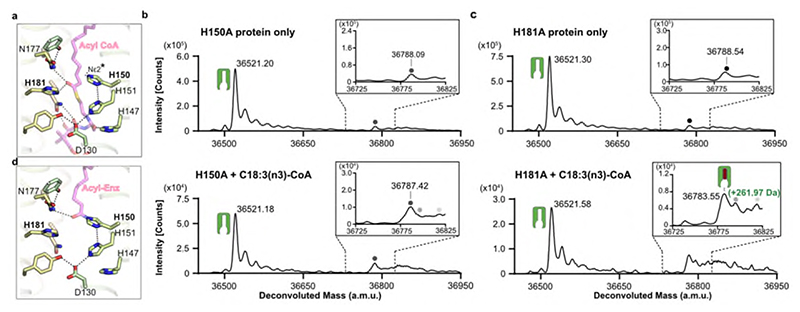
Many fatty acid synthases and other acyltransferases require the formation of acyl-enzyme intermediates with either thio- or oxy-ester covalent linkages32, usually involving cysteine or serine33 residues. However, there are no cysteines or serines in or near the ELOVL active site. Instead, the active site is surrounded by several histidines that have been shown to be essential for activity including the two histidines (H150 / H181) that are covalently bound to the 3-keto C20-CoA in the crystal structure.
Based on the structure, we hypothesised that H150 was a candidate for the role as the primary nucleophile. The nucleophilicity of H150 is likely enhanced by a bonding network involving H151, D130 and Y185 that ensures that the Nε atom is unprotonated (Fig. 4ab; Fig.6a). A H150A mutation gave protein that was not modified upon incubation with C18:3(n3)-CoA (Fig. 6b) (we were unable to use C18:0-CoA as substrate due to the presence of background peaks in the MS traces). In contrast, incubation of the H181A protein with C18:3(n3)-CoA gave a covalent adduct of 261.97 Da (Fig. 6c). The catalytic behaviour of the His mutants was also reflected in the adduct species observed in the over-expressed protein. For the H150A mutation, we were not able to detect the presence of any adducts whereas for the H181A mutation we detected both unmodified protein and adduct species of around 1030-1060Da. (Extended Data Fig. 1h,i). Together, these results are consistent with the H150 sidechain being the site where the fatty acyl unit is covalently attached in the first step of the elongation reaction.
Fig. 6. Covalent acyl-enzyme intermediate is formed upon substrate reaction at H150.
a, Structural model for acyl-CoA binding. b-c, LC-ESI-MS intact mass analysis of mutant proteins upon incubation at 37°C for 2h. b, H150A mutant protein incubated in the absence of substrate (upper panel) or in the presence of 100 μM C18:3(n3)-CoA (lower panel). No acyl-enzyme intermediate formation could be detected with the H150A mutant protein. c, H181A mutant protein incubated in the absence of substrate (upper) or in the presence of 100μM C18:3(n3)-CoA (lower). Expected mass shift upon reaction with C18:3(n3)-CoA: +260.42 Da. The presence of a background peak at ~36788 Da precluded testing with C18:0-CoA. All intact mass experiments were repeated twice with similar results (n=2 biological repeats, see Supplementary Figure 3 for replicate traces; Supplementary Table 2 for experimental and theoretical masses and mass errors). Peaks are indicated as follows: protein (green icon), acyl enzyme intermediate (green icon + pink/red oblong), background species present in all traces (grey circles). d, Structural model for acyl-imidazole H150 intermediate. Ligand complexes are models based on the product analogue crystal structure. The unprotonated imidazole nitrogen of H150 involved in the transacylation step is highlighted.
Using the ELOVL7 structure with the covalently bound 3-keto acyl-CoA as a guide, we modelled the positions of each substrate and product during the reaction, indicating how the transacylation reaction could proceed (Fig. 6a,d). Once the first substrate, the fatty acyl-CoA has bound, then H150 is ideally placed to act as a nucleophile to attack the thioester carbonyl of the fatty acyl-CoA. The sidechains of N177 and H181, together with a water molecule (which hydrogen bonds to N177 and Q214), are suitably positioned to stabilise the oxyanion prior to release of CoA. This transient tetrahedral intermediate would then collapse back to the carbonyl, ejecting the CoA and completing the transacylation step of the reaction (Fig. 6d; Extended Data Fig. 7a).
Covalent acyl adducts to histidines are uncommon, but they are not entirely without precedent: HlyC, the enzyme that activates the Escherichia coli toxin prohemolysin, catalyses acyl transfer via a histidine32,34 and the enzymes of the complement system, C3 and C4, utilise a histidine for internal acyl transfer35,36. In addition, artificial enzyme systems have been created in which a nucleophilic histidine is acylated to form a covalent acyl-imidazole intermediate, confirming that such additions are possible37–39.
Condensation of acyl-enzyme intermediate with malonyl-CoA
In the second ‘condensation’ step of the reaction, malonyl-CoA is combined with the bound acyl chain through a Claisen-like condensation. This reaction requires malonyl-CoA binding followed by decarboxylation and carbon-carbon bond formation either in a concerted or sequential manner, resulting in the formation of the 3-keto acyl-CoA product that is elongated by 2 carbon units. Initially malonyl-CoA would bind to the CoA binding site, and modelling suggests that the malonyl unit would be positioned so that the carboxylate would lie within the malonyl / decarboxylation pocket at the side of the tunnel between TM5 and TM6 (Fig. 4c). This shallow pocket is lined by a semicircle of amide groups from N177, Q211 and Q214, as well as H181, which are all ideally placed to interact with the malonyl group and could promote decarboxylation (Fig. 7a,b). Such roles are supported by mutational studies of the equivalent residues in EloA which gave either inactive enzyme (H181, N177 and Q211 (ELOVL7 numbering)), or enzyme with 10% of WT activity (Q214)30. However, the precise roles of the residues lining the decarboxylation pocket and the interactions that promote malonyl-CoA decarboxylation will require further investigation. Following decarboxylation, the resulting nucleophilic enolate carbanion would be well-placed to react with the acyl intermediate on H150 (Fig. 7c), to form the 3-keto acyl-CoA product which would then be released from the enzyme.
Fig. 7. Model for malonyl-CoA binding at the decarboxylation site.
a, Putative structural model for malonyl-CoA (MLC) binding within the conserved decarboxylation side pocket off the main tunnel. b, Conserved sidechains recognise malonyl carboxylate (viewed looking from H150). c, Composite structural model of second condensation reaction step with bound acy-limidazole intermediate and malonyl-CoA.
Formation of the 3-keto acyl-CoA adduct in the structure
Interestingly, in the crystal structure the sidechain of H150 is covalently linked to the C5 of the bound 3-keto acyl-CoA. A covalent bond at this position would not be expected with the known substrates or products. This species likely represents a ‘dead-end’ product complex, from a side reaction with an abnormal substrate, that has become irreversibly bound to the enzyme. We speculate that this crosslink could have been the result of ELOVL7’s conjugate addition of the nucleophilic imidazole of H150 to an unsaturated acyl-CoA substrate or breakdown product, with an adjacent double bond in the acyl chain (e.g., 2,3-trans-enoyl-CoA), thus allowing the formation of a displaced acyl-imidazole covalent bond. For example, such a reaction could occur if over-expressed ELOVL7 accepted a 2,3-trans-enoyl-CoA as the first substrate, instead of the usual acyl-CoA and elongation occurred as normal via the proposed acyl-imidazole intermediate. However, on formation of the 3-keto product, H150 could react with the adjacent unsaturated carbon-carbon bond to give rise to the observed C5 linkage (Extended Data Fig. 7b). Acyl-CoAs with a double bond in this location are formed both during the FA elongation cycle and during breakdown of VLCFAs in the peroxisomes.
The other unusual feature of the trapped covalent 3-keto-adduct is the second covalent linkage from C2 to H181. This bond is not as easily rationalised as the bond from H150 to C5 but is clearly seen in the electron density. Possibly this unusual bond between the nucleophilic tau-N of histidine and the nucleophilic alpha-carbon of the 3-keto ester is the result of an uncharacterised rearrangement in the active site or trapping of an oxidised product.
Further work is required to clarify whether ELOVL7 can accept an enoyl-CoA as a first substrate and whether it could lead to covalent modification of the enzyme. 2,3-trans-enoyl-CoAs are the substrates of the reductase which carries out the final step in the elongation cycle and this metabolite may accumulate during ELOVL7 over-expression. Previous reports suggest the FA elongation cycle enzymes are able to form a multi-enzyme complex3,29,40 which may facilitate substrate channelling and would potentially prevent ELOVLs forming such catalytically dead-end complexes under normal circumstances.
Discussion
The structure of ELOVL7 reveals a narrow extended tunnel which is involved in binding and extension of acyl-CoAs. Previous studies have highlighted a series of histidine residues that are essential for ELOVL enzyme activity and these are clustered halfway along the membrane-embedded tunnel, at the midpoint of the bilayer, where they are poised for catalysis. Through a combination of mutagenesis and intact protein MS analysis, our results show that the elongation reaction proceeds through a stable acyl-imidazole intermediate formed between the substrate acyl-CoA and the second His in the ELOVL HxxHH motif. A second group of sidechains previously demonstrated to be required for elongase activity (N177, H181, Q211, and Q214)30 lie adjacent to the modified histidine, where they could play a role in the decarboxylation of malonyl-CoA, the second substrate. The resulting enolate nucleophile would attack the acyl-enzyme intermediate giving rise to the 3-keto acyl-CoA product.
Both ELOs and FAE1-type KCSs found in plants and some protists transfer a two-carbon unit from malonyl-CoA to a fatty acyl-CoA through a decarboxylating Claisen-like condensation. However, ELOs and KCSs share no sequence or structure similarities. The structure of ELOVL7 reveals a unique TM architecture that encloses a central catalytic tunnel. In contrast, KCSs are predicted to be anchored to the membrane by one or two N-terminal TM helices and their soluble catalytic domain shares sequence similarity to ketoacyl-ACP synthase IIIs (KASIII) and polyketide synthases15. As with these soluble condensing enzymes, KCSs possess a Cys-His-Asn catalytic triad and the conserved cysteine is thought to act as a nucleophile, forming a covalent acyl-enzyme intermediate upon reaction with an acyl-CoA substrate41,42. ELOs do not possess a recognisable catalytic triad and therefore their mechanism of acyl chain transfer has remained elusive. Our structural and biochemical data provide novel insights into the catalytic mechanism of the ELO family of elongases, demonstrating that these enzymes use their conserved HxxHH motif to generate a hydrogen-bonding network which activates a histidine residue to act as the nucleophile in the first step of the reaction. To our knowledge, this finding represents the first example of a nucleophilic histidine involved in acyl transfer within the context of fatty acid metabolism.
Failure of the VLCFA elongation process through ELOVL mutations leads to loss of ceramides and sphingolipids required for myelin, skin barrier and retinal function. Mutations in the ELOVL4 gene cause Stargardt disease-317, ichthyosis, intellectual disability, spastic quadriplegia43, and spinocerebellar ataxia 34 (SCA34)18,44. ELOVL5 mutations could cause spinocerebellar ataxia 38 (SCA38)19. Elovl3 knockout mice suffer severely from hair loss and have an imbalance in the lipid species of the sebum45,46; and mouse knockout studies of ELOVL5 / ELOVL6 suggest associations with hepatic steatosis20 and obesity-induced insulin resistance21. ELOVL7, the most recently discovered ELOVL elongase, is associated with prostate22,23 and gynaecological24 cancer and early onset Parkinson’s disease25. ELOVL7 knockdown reduces cell death and membrane permeabilization during necroptosis, a form of programmed cell death26. The unusual architecture, active site and chemistry revealed by our ELOVL elongase structure provides exciting new avenues for design of modulators of ELOVLs. These may be of value for patients with X-linked adrenoleukodystrophy (X-ALD), as they are unable to break down VLCFAs, leading to accumulation in cells and further elongation by ELOVL147. X-ALD affects the adrenal cortex and nervous system, leading to adrenocortical insufficiency and myelopathy, with progressive demyelination of nerves in severe cases. The formulation known as Lorenzo’s oil has been shown to alleviate symptoms of X-ALD and is reported to inhibit ELOVL148. The design of specific ELOVL elongase inhibitors may offer a route to reduction of VLCFAs, thus providing a novel approach to therapy for this devastating disease.
Methods
Cloning
The Homo sapiens ELOVL7 gene, encoding the ELOVL7 protein (Uniprot ID: A1L3X0) was cloned into the baculovirus transfer vector pFB-CT10HF-LIC (available from The Addgene Nonprofit Plasmid Repository) for expression in Sf9 cells (Thermo-Fisher Scientific, Cat no. 11496015) using the primers shown in Extended Data Table 1. A C-terminal TEV-cleavable His10-FLAG tag was fused to the protein for purification. For mammalian expression, the same construct was cloned into the pHTBV1.1-LIC baculovirus transfer vector (The BacMam vector backbone (pHTBV1.1), kindly provided by Professor Frederick Boyce, Massachusetts General Hospital, Cambridge, MA and adapted for ligation independent cloning in-house for expression in Expi293F cells (Thermo-Fisher Scientific, Cat no.A14527) which similarly confers a TEV cleavable C-terminal His10-FLAG tag.
The mutagenesis was performed using Q5® Site-Directed Mutagenesis Kit (NEB) according to manufacturer’s instructions using primers listed in Extended Data Table 1.
Protein Production
For both insect and mammalian expression, baculoviral DNA was produced by transposition of DH10Bac with either the ELOVL7-pFB-CT10HF-LIC or ELOVL7-pHTBV1.1-LIC transfer vectors, which were then used to transfect Sf9 cells to produce baculovirus for transduction using the transfection reagent Insect GeneJuice® (MerckMillipore). The virus amplification was performed by infecting mid-log Sf9 cells (2x106 cells ml-1) grown in Sf-900II™ media with 2% fetal bovine serum. For large scale protein production, Sf9 cells at the density of 2x106 cell ml-1 were infected with 5 ml of P2 (second passage) recombinant baculovirus in Sf-900™ II in 2 L roller bottles (Biofil) and incubated for 72 h at 27°C. Cells were harvested by centrifugation at 900 x g for 15 mins, washed with phosphate buffered saline (PBS), and pelleted again prior to flash freezing in liquid N2, then stored at -80 °C for further use.
For mammalian (Expi293F) expression, 1 L of Expi293F cell cultures (2 x 106 cells ml-1) in Freestyle 293™ Expression Medium (Thermo-Fisher) were transduced with 30 ml of P3 baculovirus (third passage) in the presence of 5 mM sodium butyrate in a 2 L roller bottle (Biofil). Cells were grown in a humidity controlled orbital shaker for 48 hours at 37 °C with 8% CO2 before being harvested using the same process as for insect cells.
Protein Purification
All the following steps were performed at 4°C unless otherwise indicated. Cell pellets were resuspended in lysis buffer (50 mM HEPES-NaOH, pH 7.5, 500 mM NaCl, 5% v/v glycerol, 1 mM TCEP-NaOH, Roche protease inhibitor cocktail EDTA-free) at the ratio of 50 ml / L equiv. original cell culture. The resuspension was then passed twice through an EmulsiFlex-C5 homogenizer (Avestin) at 10000 psi. Membrane proteins were extracted from the cell lysate with 1% w/v octyl glucose neopentyl glycol (OGNG; Generon, Cat. No. NG311) / 0.1% cholesteryl hemisuccinate tris salt (CHS; Sigma-Aldrich, Cat. No. C6512) and rotated for 2 h. Cell debris was removed by centrifugation at 35,000 x g for 1 h. The supernatant was supplemented with 5 mM imidazole pH 8.0 before incubation with Co2+ charged TALON resin (Clontech) for 1 h on a rotator (1 ml resin slurry per L original culture volume). The Talon resin was collected by centrifugation at 700 x g for 5 mins and washed with 30 column volumes of washing buffer (50 mM HEPES-NaOH, pH 7.5, 500 mM NaCl, 1 mM TCEP-NaOH, 0.12% w/v OGNG / 0.012% w/v CHS and 20 mM imidazole pH 8.0) before the target protein was eluted with elution buffer (washing buffer supplemented with 250 mM imidazole pH 8.0). The eluted protein was then exchanged into lysis buffer supplemented with 0.15% w/v OGNG / 0.015% w/v CHS by passing over a pre-equilibrated Sephadex PD-10 desalting column (GE Life Sciences). TEV protease was added to the desalted protein at a weight ratio of 1:10 for overnight tag cleavage. The His-tagged TEV protease was removed by a second Talon resin binding step for 1 h, and the flow through was collected and concentrated to <1ml using a 100 kDa molecular weight cutoff (MWCO) Vivaspin 20 centrifugal concentrator (GE Life Sciences) and further purified by size exclusion chromatography on a Superdex 200 10/300 Increase GL column (GE Healthcare) in size exclusion chromatography (SEC) buffer (20 mM HEPES-NaOH, pH 7.5, 200 mM NaCl, 1 mM TCEP-NaOH, 0.08% w/v OGNG/ 0.008% w/v CHS). The H150A and H181A mutated protein were expressed and purified in the same way as the wild type protein. In addition, we attempted to produce a H151A version of the protein, but this variant proved to be too unstable to purify.
For structural studies, the flow through from the reverse Talon step was incubated with 50 mM iodoacetamide (IAM) (Merck Millipore) for 20 mins at room temperature. IAM was removed by passing the reaction mixture down a PD-10 desalting column prior to concentration. After SEC, fractions containing the highest concentration of ELOVL7 were pooled and concentrated to 12-25 mg/ml using a 100 kDa MWCO Vivaspin 20 centrifugal concentrator.
Crystallization
Initial protein crystals were grown at 4°C in condition E10 of the MemGold2-ECO Screen (Molecular Dimensions; 0.05 M Na-acetate pH 4.5, 0.23 M NaCl, 33 % v/v polyethylene glycol (PEG) 400) in 3-well sitting-drop crystallisation plates (SwissCi) with 150 nl drops and 2:1 and 1:1 protein to reservoir ratios. Crystals appeared after 4-7 days and grew to full size within 3-4 weeks. Two microlitre hanging drops were set up in 24-well XRL plates (Molecular Dimensions) at protein to reservoir ratios of 2.5:1, 2:1 and 1.5:1. The best crystals grew in 0.1 M Na-acetate pH 4.5, 0.23 M NaCl, 34-38% v/v PEG400 using the IAM-modified protein at a concentration of 5-8 mg/ml and were harvested after 12-14 days. Crystals could also be grown for the unalkylated protein but were limited in size and poorly reproducible despite using seeding. Prior to vitrification, crystals were sequentially transferred to mother liquor solutions with an increasing amount of PEG400 to a final concentration of PEG400 of 46% v/v over 10-15 mins. For heavy atom derivatisation, crystals were looped into drops containing reservoir solution supplemented with 10 mM mercury chloride and soaked for 10 mins. Hg-soaked crystals were then treated to the same PEG400 escalation strategy using Hg-free solutions before being vitrified in liquid nitrogen.
Data Collection
All crystals were screened and X-ray diffraction data were collected on the I24 microfocus beamline at the Diamond Light Source (Didcot, UK) from vitrified crystals at 100 K. Multiple data sweeps were collected from crystals using 0.2° oscillation and a beam-size of 20 μm x 20 μm. Crystals were translated by at least 25 μm between collection areas. Native data were collected at a wavelength of 0.9686 Å. The high resolution native dataset (WT-native) was assembled from four overlapping 100° wedges of data collected from separate volumes of a single crystal. 960° of anomalous data were also collected from a mercury-derivatised crystal at a wavelength close to the Hg LIII edge (λ = 0.992 Å) in a single pass (Hg peak).
Unmerged scaled data from the xia2.dials automated processing pipeline50 was truncated and merged using AIMLESS (CCP451,52). ELOVL7 crystals are monoclinic and contain two copies of the enzyme in each asymmetric unit. All diffraction data were highly anisotropic and limited to between 3.4 - 4.5 Å resolution in the worse direction and 2.05 – 3 Å in the best direction. Anisotropic resolution limits for the WT-Native data along each of the three principal directions as defined by AIMLESS were 3.5 Å, 2.05 Å, 2.65 Å and 3.24 Å, 2.05 Å, 2.22 Å based on Mn (I/sd(i)) > 2 and CC1/2 > 0.5 respectively. Data truncated in AIMLESS with isotropic resolution limits was used for phasing and initial cycles of model building. The high resolution native dataset (nominal overall resolution 2.6 Å based on Mn (I/sd(I)) > 2 limit as defined by AIMLESS) was further processed with STARANISO (v2.3.2553) to the maximum resolution of 2.05 Å and used for refinement (see Table 1 for details).
Phasing
Phasing was carried out in PHENIX54 using SIRAS with the Hg-peak data and a 3Å isomorphous lower resolution native dataset. Two Hg2+ sites were located with phenix.hyss using data to 4.5 Å. The resulting 3 Å phased electron density map had clear protein density allowing the identification of the NCS relationship between the two ELOVL7 molecules in the asymmetric unit. After two-fold averaging using RESOLVE55, the resultant map was of sufficient quality to manually model all the TM helices. Initial phases were further improved by cross-crystal averaging with a non-isomorphous, less anisotropic and slightly higher resolution dataset (unit cell dimensions 64.14 Å x 71.93 Å x 111.74 Å β = 106.7°) using DMMULTI56. The resultant map was of excellent quality and the majority of the structure could be built automatically with BUCCANEER57.
Model Building and Refinement
The BUCCANEER-built model was manually rebuilt in COOT58 and refined using BUSTER (version 2.10.3, 29-NOV-201959). The model was refined with LSSR restraints60 and a single TLS group was refined for each protein chain. Ligand dictionaries were generated using GRADE (Global Phasing Ltd). All data to 2.05 Å from the STARANISO processing was used in refinement despite the low completeness and truncation to lower resolutions resulted in a refined model with worse geometry. The final model comprises residues 16-269 (chain B, 14-269), a covalently bound 3-keto-CoA acyl lipid, four OGNG detergent molecules and 112 solvent molecules. None of the resolved cysteines appear to be modified with IAM. Data collection and refinement statistics are reported in Table 1. The covalent bonds between H150 / H181 and the bound 3-keto-acyl-CoA have been modelled as N-C linkages. While the proposed conjugate addition involving H150 will involve its nucleophilic Nε2 atom, it is not possible to distinguish CH from N in the electron density maps and so the H181 unusual linkage could also be via the histidine CE1.
Molecular models for the complexes with substrate, covalently bound acyl-imidazole intermediate, malonyl-CoA and 3-keto-eicosanoyl product were built in COOT based on the crystal structure using simple manual placement and correction of any steric clashes.
Denaturing Intact Mass Spectrometry
The denaturing intact mass spectrometry measurements were performed using an Agilent 1290 Infinity LC System in-line with an Agilent 6530 Accurate-Mass Q-TOF LC/MS (Agilent Technologies Inc.). The solvent system was consisted of 0.1% Optima™ LC/MS grade formic acid (Fisher Chemical) in HPLC electrochemical grade water (Fisher Chemical) (solvent A) and 0.1% formic acid in Optima™ LC/MS grade methanol (Fisher Chemical) (solvent B). Typically, 1-2 μg of protein sample was diluted to 60 μl with 30% methanol in 0.1% formic acid. 60 μl of sample was injected onto a ZORBAX StableBond 300 C3 column (Agilent Technologies Inc.) by an auto sampler. The flowrate of the LC system was set to 0.5 ml/min. 30% of solvent B was applied in the beginning and the sample elution was initiated by a linear gradient from 30% to 95% of solvent B over 7 min. 95% B was then applied for 2 min, followed by 2 min equilibration with 30% B. The mass spectrometer was in positive ion, 2 GHz detector mode and spectra were recorded with capillary, fragmentor and collision cell voltages of 4000 V, 250 V and 0 V, respectively. The drying gas was supplied at 350°C with flow rate of 12 l/min and nebulizer at 60 psi. The data was acquired from 100-3200 m/z. Data analysis was performed using MassHunter Qualitative Analysis Version B.07.00 (Agilent) software.
Assignment of the deconvoluted mass peak to the unmodified state of the protein was achieved by identifying a -89.62 Da mass shift relative to the theoretical mass of the purified protein. This corresponds to the loss of the initiator methionine, followed by acetylation of the new N-terminus (theoretical mass shift of -89.16 Da). Further characterisation of adduct formation and IAM modification was carried out by analysing the deconvoluted mass shift from the unmodified protein peak to those corresponding to covalently modified forms of the protein. This enabled the determination of a mass shift of +1073.66 Da for the copurified adduct, and the observation that 1, 2 or 3 sites can be simultaneously modified by IAM (predicted +57.07 Da per site modified).
In order to trap the covalent acyl-enzyme intermediate, the purified, tagged, wild-type ELOVL7 protein at 1.5 mg/mL (obtained after the desalting step that followed IMAC elution) was incubated with 100 μM C18:0-CoA (Avanti Polar Lipids, Cat. No. 870718) or C18:3(n3)-CoA (Avanti Polar Lipids, Cat. No. 870732) for 2 hours at 37 °C, in the presence and absence of 1 mM EDTA, 1 mM EGTA, or 100 μM Malonyl-CoA (Sigma-Aldrich, Cat. No. M4263). The reaction was terminated by dilution into 30% methanol in 0.1% formic acid, as described above. Covalent acyl-enzyme intermediate formationwas identified by monitoring the presence of a mass shift upon incubation with the substrate, corresponding to the addition of the substrate acyl chain through attachment of the histidine imidazole to the thioester carbonyl, resulting in thioester cleavage and loss of CoA (predicted +266.47 Da upon reaction with C18:0-CoA; +260.42 Da upon reaction with C18:3(n3)-CoA). The site of covalent modification was probed by carrying out this experiment with the His150Ala and His181Ala mutants, which allowed identification of His150 as the nucleophile involved in covalent intermediate formation.
In order to probe the sequential nature of the reaction, protein was solubilised and purified in 0.002% (w/v) LMNG as the unmodified form of the enzyme is more stable in this detergent than in OGNG/CHS. The protein (2 mg/mL) was incubated with 100 μM C18:0-CoA for 15 minutes at 37 °C, and any precipitated protein at the end of this incubation was removed by centrifugation at 21,000 x g for 30 minutes. The presence of a covalent acyl-enzyme intermediate at this stage was monitored as described above. Next, 200 μM malonyl-CoA (Sigma-Aldrich, Cat. No. M4263) was added to the reaction and incubated for 3h at 4 °C. Subsequent denaturing intact mass spectrometry analysis of the reaction mixture revealed the loss of the covalent acyl-imidazole intermediate deconvoluted mass peak. Control samples taken after incubation with C18:0 CoA were also incubated for 3h at 4° C in the absence of malonyl-CoA (Extended Data Fig. 6k). All intact mass experiments were carried out at least twice using distinct biological samples. See Supplementary information file for replicate traces and mass data.
Product formation detected by mass spectrometry
To follow the enzymatic reaction, a small molecule mass spectrometry experiment was performed using LC-MS with a Waters system equipped with a Waters 2545 binary gradient module, a Waters SQ Detector 2, Waters 2489 UV/visible detector, and a Waters 2424 ELS detector. Masslynx 4.0 software by Waters (Beverly, MA) was applied for data processing. Analytical separation of malonyl-CoA, C18:0-CoA and the 3-keto-eicosanoyl (C20:0)-CoA reaction product was carried out on a Phenomenex Kinetex 5 μm EVO C18 column (100 mm × 3.0 mm, 100 Å) using a flow rate of 2 mL/min in a 3 min gradient elution, with 20 μL injections. The mobile phase was a mixture of 93% H2O, 5% acetonitrile, and 2% of 0.5 M ammonium acetate adjusted to pH 6 with glacial acetic acid (solvent A) and 18% H2O, 80% acetonitrile, and 2% of 0.5 M ammonium acetate adjusted to pH 6 with glacial acetic acid (solvent B). Gradient elution was as follows: 95:5 (A/B) 0.35 min, 95:5 (A/B) to 5:95 (A/B) over 1 min, 5:95 (A/B) over 0.75 min, and then reversion back to 95:5 (A/B) over 0.1 min and 95:5 (A/B) over 0.8 min. A blank injection of MeOH (20 μL) was included between runs to avoid any carry-over from the previous sample.
Samples were analysed by mass spectrometry in negative ESI single ion scans set for mass ions of 852.4 m/z (ESI-) (malonyl-CoA, Sigma-Aldrich, Cat. No. M4263), 1032.9 m/z (ESI-) (C18:0-CoA, Sigma-Aldrich, Cat. No. S0802) and 1074.9 m/z (ESI-) (3-keto-eicosanoyl-CoA). The retention times for these compounds were 0.23, 1.60 and 1.63 mins, respectively. The capillary voltage was set to 3.00 kV, the cone voltage was set to 30 V. Nitrogen was used as desolvation gas at flow rate of 500 L/h. Desolvation temperature was set to 250 °C. Experiments were carried out with two distinct protein purifications giving similar results.
Size exclusion chromatography with multiangle light scattering (SEC-MALS)
SEC-MALS analysis was performed by injecting 100 μl of purified protein at 1 mg/mL onto a Superdex 200 10/300 Increase GL column (GE Healthcare), pre-equilibrated in 20 mM HEPES pH 7.5, 200 mM NaCl, 0.1% w/v OGNG / 0.01% w/v CHS, using an OMNISEC RESOLVE system (Malvern). Light scattering and refractive index changes were monitored using an integrated OMNISEC REVEAL multi-detector module (Malvern), and the data was analysed using OMNISEC 5.1 software. To enable calculation of the protein molecular weight within the protein-detergent complex, the detergent dn/dc was experimentally determined by generating a standard curve of refractive index peak area vs. detergent concentration. The slope of this curve, corresponding to the detergent dn/dc, was 0.127 ml/g. The protein dn/dc was assumed to be 0.185 ml/g.
Extended Data
Extended Data Fig. 1. Properties of purified ELOVL7.
a, SDS-PAGE gel of purified ELOVL7. Data shown is from a single purification but similar results were observed for multiple purifications / modifications. b, SEC profile and MALS analysis showing that OGNG-solubilised protein exists as a dimer in solution. Experiment carried out for tag-cleaved ELOVL7 purifications with and without IAM modification. Similar results were seen for each sample. c, Representation of head-to-tail dimer present in the crystal. d, ELOVL7 head-to-tail dimer packing within the crystal lattice. e, f, Intact mass analysis of ELOVL7 protein at various stages during purification. Deconvoluted mass spectra are shown for ELOVL7 protein purified from (e) Sf9 (n=3 or 4) and (f) Expi293F cells (n=2). For protein purified after expression in insect cells, the samples are shown after immobilized metal affinity chromatography (IMAC), after cleavage of the tag and size exclusion chromatography (SEC) and after treatment with iodoacetamide (IAM). The expected mass of the untagged ‘native’ enzyme (E) based on the sequence is 34222.38 Da. The observed mass peaks (Sf9, 34132.78 Da; Expi 34133.36 Da) correspond to the loss of the N-terminal methionine (-131.20 Da) and acetylation of the resulting new N-terminus (+42.04 Da). All samples were run in their reduced state. The modified material (E*) appears as an adduct with an average mass shift of +1073.6 Da. The addition of 113.55 Da upon treatment with IAM suggests modification of two cysteine residues. g-i, Deconvoluted intact mass spectra for the untagged g, WT (n=3), h, H150A (n=2) and i, H181A mutants (n=2). The expected mass decrease of a His-to-Ala mutation is 66.06 Da. Evidence of in vivo modification (E*) is observed for the H181A mutant but not for H150A. For intact mass experiments, theoretical and experimental masses along with mass errors are given in Supplementary Table 1.
Extended Data Fig. 2. TM helix topology of ELOVL7.
TM helical topology of ELOVL7 is compared with other six and seven membered TM bundles. TM helices are numbered and location of substrate/ligand site marked. Underlying cartoon representations of each structure are coloured from blue to red from the N- to C-termini respectively. PDB accession codes are shown in parentheses.
Extended Data Fig. 3. Electron density clearly shows covalently bound 3-keto-eicosanoyl-CoA.
a-d, Electron density running along the catalytic tunnel. Final BUSTER 2mFo-DFc (blue mesh, contoured at 1σ) and omit mFo-DFc (green mesh, contoured at 2.5σ) electron density maps are overlaid on the final model. e, Comparison of a test refinement in which the imidazole groups of H150 and H181 were removed from the model (grey carbon protein atoms / palecyan ligand carbon atoms) and the final model (palecyan protein carbons / violet ligand carbon atoms). The BUSTER 2mFo-DFc (blue mesh, contoured at 1σ) and mFo-DFc (green mesh, contoured at 3σ) maps for the refined histidine-truncated model / unlinked acyl-CoA are shown. f, Comparison of various refined models (green carbons - protein only; palecyan - protein without H150/181 sidechain plus ligand; grey/violet - final model with covalently attached ligand).
Extended Data Fig. 4. Sequence alignment and active site conservation of human ELOVL family members.
a, Sequence alignment of human ELOVL1-7. The conserved histidine box (147HxxHH151) is highlighted by a blue box. Filled circles below alignment indicate residues with a proposed catalytic role (blue) and residues interacting with either the CoA (orange) or acyl (plum) portion of the substrate. Cysteines that form the disulphide bridge (C99-C231) between the TM2/3 and TM6/7 loops are indicated by stars. b, Conservation of active site tunnel. Molecular surface representation is coloured by amino acid conservation score calculated by CONSURF analysis49 of a diverse set of ELOVL1-7 family members. The various subregions of the tunnel are indicated (ADP / Pan from CoA and Acyl chain). Amino acid residues that form the binding tunnel are coloured according to region (pink, acyl; blue, catalytic site; orange CoA binding).
Extended Data Fig. 5. WT ELOVL7 activity.
a,b Activity of residual WT enzyme on incubation with stearoyl-CoA (C18:0) and malonyl-CoA. Selected ion recording is shown for a, reaction mixture without added enzyme and b, reaction mixture after 3hr incubation with ELOVL7 enzyme. The ion peak at 1.61 mins corresponds to the expected 3-keto-eicosanoyl (C20)-CoA product of the elongation reaction. This experiment was carried out with two biological repeats with similar results.
Extended Data Fig. 6. Identification of a covalent acyl-enzyme intermediate of ELOVL7.
Purified, tagged, wildtype ELOVL7 was incubated in the presence and absence of known substrates and metalchelating agents prior to LC-ESI-MS intact mass analysis. a-g, Deconvoluted intact mass spectra for ELOVL7 incubated for 2h at 37°C. a, in the absence of substrates. b, ELOVL7 incubated with 100μM C18:0-CoA. Expected mass addition for acyl intermediate upon reaction with C18:0-CoA: +266.47 Da. c, ELOVL7 incubated with 100μM C18:3(n3)-CoA. Expected mass addition for acyl intermediate upon reaction with C18:3(n3)-CoA: +260.42 Da. d-e, ELOVL7 incubated with d, 100μM C18:0-CoA or e, 100μM C18:3(n3)-CoA in the presence of 1mM EDTA. f-g ELOVL7 incubated with f, 100μM C18:0-CoA or g, 100μM C18:3(n3)-CoA in the presence of 1mM EGTA. h-k, Sequential reaction of LMNG-purified ELOVL7 with C18:0-CoA and malonyl-CoA. h, LMNG-purified ELOVL in the absence of substrates. i, ELOVL7 incubated with 100μM C18:0-CoA. j, Purified ELOVL7 initially incubated with 100μM C18:0-CoA, followed by incubation with 200 μM malonyl-CoA. Addition of the second substrate leads to loss of the acyl-enzyme intermediate peak, consistent with the reaction having gone to completion. k, control ELOVL7 sample taken after incubation with C18:0-CoA was further incubated in the absence of malonyl-CoA, showing that covalent intermediate loss only occurs in the presence of malonyl-CoA. All experiments were repeated independently twice with similar results (n=2 biological repeats, see Supplementary Figure 1 and Figure 2 for replicate traces. See Supplementary Table 2 for theoretical and experimental masses and mass errors).
Extended Data Fig. 7. Proposed ping-pong reaction mechanism for ELOVL7.
a, Transacylation step with acyl chain of first substrate being transferred to H150. In the second step, malonyl-CoA binds and undergoes decarboxylation and a condensation reaction to form the elongated 3-keto product. b, Proposed reaction steps leading to C-N covalent adduct with H150 seen in crystal structure. In this scenario a 2,3-trans-enoyl-CoA serves as the first substrate (left) leading to the 3-keto,4,5-trans-enoyl-CoA ‘product’ (middle) which subsequently crosslinks to H150 via a conjugate addition reaction of H150 (right). The nature of the reaction that leads to H181 crosslinking to the C2 atom of the 3-keto-acyl-CoA is not clear.
Extended Data Table 1. Mutagenesis primer sequences.
| Primer purpose | Primer sequence (5’ to 3’) | |
|---|---|---|
| WT ELOVL7 | Forward | TTAAGAAGGAGATATACTATGGCCTTCAGTGATCTTAC |
| Reverse | GATTGGAAGTAGAGGTTCTCTGCATTATCTTTGTTTTTGCAAG | |
| H150A | Forward | TCATGTATTCGCTCATACCATCATGCCG |
| Reverse | AGGAAAGTCACTTGGCTATTTTTC | |
| H151A | Forward | TGTATTCCATGCTACCATCATGCC |
| Reverse | TGAAGGAAAGTCACTTGG | |
| H181A | Forward | TACAGCTGTAGCTGTAGTCATGTATTCC |
| Reverse | TTTAGAAGGGCATGGAATG |
Supplementary Material
Acknowledgements
L.N., A.C.W.P., G.F.R., A.C., V.C., S.R.B., D.S., T.M., L.S., S.M.M.M., N.B.-B., E.P.C. were members of the SGC, a registered charity (number 1097737) that receives funds from AbbVie, Bayer Pharma AG, Boehringer Ingelheim, the Canada Foundation for Innovation, Genome Canada, Janssen, Merck KGaA, Merck & Co., Novartis, the Ontario Ministry of Economic Development and Innovation, Pfizer, São Paulo Research Foundation-FAPESP and Takeda, as well as the Innovative Medicines Initiative Joint Undertaking ULTRA-DD grant 115766 and the Wellcome grant no 106169/Z/14/Z. T.C.P. is supported by a Wellcome PhD studentship (102164/B/15/Z). A.Q. is supported by Wellcome grant no 202892/Z16/Z. The authors would like to thank Diamond Light Source for beam time (BAG proposal mx19301), and the I24 beamline staff for assistance with beam time, crystal screening and data collection. We thank Oliver Smart and Clemens Vonrhein at Global Phasing Ltd for assistance with generating covalent ligand restraints. We acknowledge the use of the UCSF Chimera package from the Resource for Biocomputing, Visualisation, and Informatics at the University of California, San Francisco (supported by NIGMS P41-GM103311). We are grateful to the Membrane Protein Laboratory (Wellcome grant ref (202892/Z16/Z)) at the Research Complex at Harwell for access to SEC-MALS equipment and assistance with these experiments.
Footnotes
Author Contributions
L.N. purified protein, carried out biophysical characterisations, assisted by V.C. and D.S., and obtained crystals that diffracted to beyond 4 Å resolution. L.N. and A.C.W.P. collected X-ray diffraction data and solved and built the structure. T.C.P. purified protein samples, performed mass spectrometry adduct analysis and SEC-MALS experiments. A.Q. provided access to equipment and assisted in SEC-MALS. G.F.R. assisted with design and execution of the rapid fire mass spectrometry activity assay, supervised by P.E.B.. T.M. assisted with mass spectrometry methods optimisation. S.R.B. and A.C. were involved in the early stages of the project, including design of constructs, optimisation of the protein purification, production and screening of initial crystals and initial MS studies. J.D.L. assisted with early stages of the project, including testing constructs and providing materials. Constructs were screened for expression by L.S. and large scale insect cell expressions were produced by S.M.M.M., supervised by N.B.B. Data were analysed and the paper was written by L.N., T.C.P., A.C.W.P. G.F.R., P.E.B. and E.P.C.. E.P.C. supervised all aspects of the project.
Competing Interests
The authors declare no competing interests.
Data Availability
Atomic coordinates and structure factors for the reported crystal structure are deposited in the PDB under accession code 6Y7F.
References
- 1.Leonard AE, Pereira SL, Sprecher H, Huang Y-S. Elongation of long-chainfatty acids. Progress in Lipid Research. 2004;43:36–54. doi: 10.1016/s0163-7827(03)00040-7. [DOI] [PubMed] [Google Scholar]
- 2.Sassa T, Kihara A. Metabolism of very long-chain Fatty acids: genes and pathophysiology. Biomol Ther (Seoul) 2014;22:83–92. doi: 10.4062/biomolther.2014.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ohno Y, et al. ELOVL1 production of C24 acyl-CoAs is linked to C24 sphingolipid synthesis. Proc Natl Acad Sci U S A. 2010;107:18439–44. doi: 10.1073/pnas.1005572107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Imgrund S, et al. Adult ceramide synthase 2 (CERS2)-deficient mice exhibit myelin sheath defects, cerebellar degeneration, and hepatocarcinomas. J Biol Chem. 2009;284:33549–60. doi: 10.1074/jbc.M109.031971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Isokawa M, Sassa T, Hattori S, Miyakawa T, Kihara A. Reduced chain length in myelin sphingolipids and poorer motor coordination in mice deficient in the fatty acid elongase Elovl1. FASEB Bioadv. 2019;1:747–759. doi: 10.1096/fba.2019-00067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sassa T, et al. Impaired epidermal permeability barrier in mice lacking elovl1, the gene responsible for very-long-chain fatty acid production. Mol Cell Biol. 2013;33:2787–96. doi: 10.1128/MCB.00192-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li W, et al. Depletion of ceramides with very long chain fatty acids causes defective skin permeability barrier function, and neonatal lethality in ELOVL4 deficient mice. Int J Biol Sci. 2007;3:120–8. doi: 10.7150/ijbs.3.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harkewicz R, et al. Essential role of ELOVL4 protein in very long chain fatty acid synthesis and retinal function. J Biol Chem. 2012;287:11469–80. doi: 10.1074/jbc.M111.256073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pewzner-Jung Y, et al. A critical role for ceramide synthase 2 in liver homeostasis: II.insights into molecular changes leading to hepatopathy. J Biol Chem. 2010;285:10911–23. doi: 10.1074/jbc.M109.077610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jakobsson A, Westerberg R, Jacobsson A. Fatty acid elongases in mammals: their regulation and roles in metabolism. Prog Lipid Res. 2006;45:237–49. doi: 10.1016/j.plipres.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 11.Toke DA, Martin CE. Isolation and Characterization of a Gene Affecting Fatty Acid Elongation in Saccharomyces cerevisiae. Journal of Biological Chemistry. 1996;271:18413–18422. doi: 10.1074/jbc.271.31.18413. [DOI] [PubMed] [Google Scholar]
- 12.Oh C-S, Toke DA, Mandala S, Martin CE. ELO2 and ELO3, Homologues of the Saccharomyces cerevisiae ELO1 Gene, Function in Fatty Acid Elongation and Are Required for Sphingolipid Formation. Journal of Biological Chemistry. 1997;272:17376–17384. doi: 10.1074/jbc.272.28.17376. [DOI] [PubMed] [Google Scholar]
- 13.Lee SH, Stephens JL, Englund PT. A fatty-acid synthesis mechanism specialized for parasitism. Nature Reviews Microbiology. 2007;5:287–297. doi: 10.1038/nrmicro1617. [DOI] [PubMed] [Google Scholar]
- 14.Ramakrishnan S, et al. Apicoplast and Endoplasmic Reticulum Cooperate in Fatty Acid Biosynthesis in Apicomplexan Parasite Toxoplasma gondii*. Journal of Biological Chemistry. 2012;287:4957–4971. doi: 10.1074/jbc.M111.310144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haslam TM, Kunst L. Extending the story of very-long-chain fatty acid elongation. Plant Sci. 2013;210:93–107. doi: 10.1016/j.plantsci.2013.05.008. [DOI] [PubMed] [Google Scholar]
- 16.Venegas-Calerón M, Beaudoin F, Sayanova O, Napier JA. Co-transcribed Genes for Long Chain Polyunsaturated Fatty Acid Biosynthesis in the Protozoon Perkinsus marinus Include a Plant-like FAE1 3-Ketoacyl Coenzyme A Synthase. Journal of Biological Chemistry. 2007;282:2996–3003. doi: 10.1074/jbc.M607051200. [DOI] [PubMed] [Google Scholar]
- 17.Zhang K, et al. A 5-bp deletion in ELOVL4 is associated with two related forms of autosomal dominant macular dystrophy. Nat Genet. 2001;27:89–93. doi: 10.1038/83817. [DOI] [PubMed] [Google Scholar]
- 18.Cadieux-Dion M, et al. Expanding the clinical phenotype associated with ELOVL4 mutation: study of a large French-Canadian family with autosomal dominant spinocerebellar ataxia and erythrokeratodermia. JAMA Neurol. 2014;71:470–5. doi: 10.1001/jamaneurol.2013.6337. [DOI] [PubMed] [Google Scholar]
- 19.Di Gregorio E, et al. ELOVL5 mutations cause spinocerebellar ataxia 38. Am J Hum Genet. 2014;95:209–17. doi: 10.1016/j.ajhg.2014.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moon YA, Hammer RE, Horton JD. Deletion of ELOVL5 leads to fatty liver through activation of SREBP-1c in mice. J Lipid Res. 2009;50:412–23. doi: 10.1194/jlr.M800383-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matsuzaka T, et al. Crucial role of a long-chain fatty acid elongase, Elovl6, in obesity-induced insulin resistance. Nat Med. 2007;13:1193–202. doi: 10.1038/nm1662. [DOI] [PubMed] [Google Scholar]
- 22.Tamura K, et al. Novel lipogenic enzyme ELOVL7 is involved in prostate cancer growth through saturated long-chain fatty acid metabolism. Cancer Res. 2009;69:8133–40. doi: 10.1158/0008-5472.CAN-09-0775. [DOI] [PubMed] [Google Scholar]
- 23.Tolkach Y, et al. Signatures of Adverse Pathological Features, Androgen Insensitivity and Metastatic Potential in Prostate Cancer. Anticancer Res. 2015;35:5443–51. [PubMed] [Google Scholar]
- 24.Zhang X, Wang Y. Identification of hub genes and key pathways associated with the progression of gynecological cancer. Oncol Lett. 2019;18:6516–6524. doi: 10.3892/ol.2019.11004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li G, et al. Association of GALC, ZNF184, IL1R2 and ELOVL7 With Parkinson’s Disease in Southern Chinese. Front Aging Neurosci. 2018;10:402. doi: 10.3389/fnagi.2018.00402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Parisi LR, et al. Membrane Disruption by Very Long Chain Fatty Acids during Necroptosis. ACS Chem Biol. 2019;14:2286–2294. doi: 10.1021/acschembio.9b00616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Holm L, Rosenstrom P. Dali server: conservation mapping in 3D. Nucleic Acids Res. 2010;38:W545–9. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Naganuma T, Sato Y, Sassa T, Ohno Y, Kihara A. Biochemical characterization of the very long-chain fatty acid elongase ELOVL7. FEBS Letters. 2011;585:3337–3341. doi: 10.1016/j.febslet.2011.09.024. [DOI] [PubMed] [Google Scholar]
- 29.Denic V, Weissman JS. A molecular caliper mechanism for determining very long-chain fatty acid length. Cell. 2007;130:663–77. doi: 10.1016/j.cell.2007.06.031. [DOI] [PubMed] [Google Scholar]
- 30.Hernandez-Buquer S, Blacklock BJ. Site-directed mutagenesis of a fatty acid elongase ELO-like condensing enzyme. FEBS Lett. 2013;587:3837–42. doi: 10.1016/j.febslet.2013.10.011. [DOI] [PubMed] [Google Scholar]
- 31.Wang H, et al. Crystal structure of human stearoyl-coenzyme A desaturase in complex with substrate. Nat Struct Mol Biol. 2015;22:581–5. doi: 10.1038/nsmb.3049. [DOI] [PubMed] [Google Scholar]
- 32.Röttig A, Steinbüchel A. Acyltransferases in bacteria. Microbiology and molecular biology reviews : MMBR. 2013;77:277–321. doi: 10.1128/MMBR.00010-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Paiva P, Sousa SF, Ramos MJ, Fernandes PA. Understanding the Catalytic Machinery and the Reaction Pathway of the Malonyl-Acetyl Transferase Domain of Human Fatty Acid Synthase. ACS Catalysis. 2018;8:4860–4872. [Google Scholar]
- 34.Trent MS, Worsham LMS, Ernst-Fonberg ML. HlyC, the Internal Protein Acyltransferase That Activates Hemolysin Toxin: Roles of Various Conserved Residues in Enzymatic Activity As Probed by Site-Directed Mutagenesis. Biochemistry. 1999;38:9541–9548. doi: 10.1021/bi9905617. [DOI] [PubMed] [Google Scholar]
- 35.Dodds AW, Ren XD, Willis AC, Law SK. The reaction mechanism of the internal thioester in the human complement component C4. Nature. 1996;379:177–9. doi: 10.1038/379177a0. [DOI] [PubMed] [Google Scholar]
- 36.Gadjeva M, et al. The covalent binding reaction of complement component C3. J Immunol. 1998;161:985–90. [PubMed] [Google Scholar]
- 37.Elliott RJ, Bennet AJ, Braun CA, MacLeod AM, Borgford TJ. Active-site variants of Streptomyces griseus protease B with peptide-ligation activity. Chem Biol. 2000;7:163–71. doi: 10.1016/s1074-5521(00)00086-7. [DOI] [PubMed] [Google Scholar]
- 38.Bolon DN, Mayo SL. Enzyme-like proteins by computational design. Proc Natl Acad Sci U S A. 2001;98:14274–9. doi: 10.1073/pnas.251555398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Burke AJ, et al. Design and evolution of an enzyme with a non-canonical organocatalytic mechanism. Nature. 2019;570:219–223. doi: 10.1038/s41586-019-1262-8. [DOI] [PubMed] [Google Scholar]
- 40.Naganuma T, Kihara A. Two modes of regulation of the fatty acid elongase ELOVL6 by the 3-ketoacyl-CoA reductase KAR in the fatty acid elongation cycle. PLoS One9. 2014:e101823. doi: 10.1371/journal.pone.0101823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ghanevati M, Jaworski JG. Active-site residues of a plant membrane-bound fatty acid elongase beta-ketoacyl-CoA synthase, FAE1 KCS. Biochim Biophys Acta. 2001;1530:77–85. doi: 10.1016/s1388-1981(00)00168-2. [DOI] [PubMed] [Google Scholar]
- 42.Ghanevati M, Jaworski JG. Engineering and mechanistic studies of the Arabidopsis FAE1 β-ketoacyl-CoA synthase, FAE1 KCS. European Journal of Biochemistry. 2002;269:3531–3539. doi: 10.1046/j.1432-1033.2002.03039.x. [DOI] [PubMed] [Google Scholar]
- 43.Aldahmesh MA, et al. Recessive mutations in ELOVL4 cause ichthyosis, intellectual disability, and spastic quadriplegia. Am J Hum Genet. 2011;89:745–50. doi: 10.1016/j.ajhg.2011.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Giroux JM, Barbeau A. Erythrokeratodermia with ataxia. Arch Dermatol. 1972;106:183–8. [PubMed] [Google Scholar]
- 45.Herron BJ, Bryda EC, Heverly SA, Collins DN, Flaherty L. Scraggly, a new hair loss mutation on mouse chromosome 19. Mamm Genome. 1999;10:864–9. doi: 10.1007/s003359901105. [DOI] [PubMed] [Google Scholar]
- 46.Westerberg R, et al. Role for ELOVL3 and fatty acid chain length in development of hair and skin function. J Biol Chem. 2004;279:5621–9. doi: 10.1074/jbc.M310529200. [DOI] [PubMed] [Google Scholar]
- 47.Engelen M, Kemp S, Poll-The B-T. X-Linked Adrenoleukodystrophy: Pathogenesis and Treatment. Current Neurology and Neuroscience Reports. 2014;14:486. doi: 10.1007/s11910-014-0486-0. [DOI] [PubMed] [Google Scholar]
- 48.Sassa T, Wakashima T, Ohno Y, Kihara A. Lorenzo’s oil inhibits ELOVL1 and lowers the level of sphingomyelin with a saturated very long-chain fatty acid. J Lipid Res. 2014;55:524–30. doi: 10.1194/jlr.M044586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Glaser F, et al. ConSurf: identification of functional regions in proteins by surfacemapping of phylogenetic information. Bioinformatics. 2003;19:163–4. doi: 10.1093/bioinformatics/19.1.163. [DOI] [PubMed] [Google Scholar]
- 50.Winter G, et al. DIALS: implementation and evaluation of a new integration package. Acta Crystallographica Section D. 2018;74:85–97. doi: 10.1107/S2059798317017235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Winn MD, et al. Overview of the CCP4 suite and current developments. Acta Crystallographica Section D. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Evans P. Scaling and assessment of data quality. Acta Crystallographica Section D-Biological Crystallography. 2006;62:72–82. doi: 10.1107/S0907444905036693. [DOI] [PubMed] [Google Scholar]
- 53.Tickle IJ, et al. STARANISO. Global Phasing Ltd; Cambridge, UK: 2018. [Google Scholar]
- 54.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallographica Section D-Biological Crystallography. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Terwilliger T. SOLVE and RESOLVE: automated structure solution, density modification and model building. Journal of Synchrotron Radiation. 2004;11:49–52. doi: 10.1107/s0909049503023938. [DOI] [PubMed] [Google Scholar]
- 56.Cowtan KD, Zhang KYJ, Main P. DM/DMMULTI software for phase improvement by density modification. International Union of Crystallography; 2012. [Google Scholar]
- 57.Cowtan K. The Buccaneer software for automated model building. 1. Tracing protein chains. Acta Crystallographica Section D. 2006;62:1002–1011. doi: 10.1107/S0907444906022116. [DOI] [PubMed] [Google Scholar]
- 58.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallographica Section D-Biological Crystallography. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bricogne G, et al. BUSTER version 2.10.3. Global Phasing Ltd; Cambridge, UK: 2017. [Google Scholar]
- 60.Smart OS, et al. Exploiting structure similarity in refinement: automated NCS and target-structure restraints in BUSTER. Acta Crystallogr D Biol Crystallogr. 2012;68:368–80. doi: 10.1107/S0907444911056058. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Atomic coordinates and structure factors for the reported crystal structure are deposited in the PDB under accession code 6Y7F.