Abstract
Reports of long-lasting coronavirus disease 2019 (COVID-19) symptoms, the so-called ‘long COVID’, are rising but little is known about prevalence, risk factors or whether it is possible to predict a protracted course early in the disease. We analyzed data from 4,182 incident cases of COVID-19 in which individuals self-reported their symptoms prospectively in the COVID Symptom Study app1. A total of 558 (13.3%) participants reported symptoms lasting ≥28 days, 189 (4.5%) for ≥8 weeks and 95 (2.3%) for ≥12 weeks. Long COVID was characterized by symptoms of fatigue, headache, dyspnea and anosmia and was more likely with increasing age and body mass index and female sex. Experiencing more than five symptoms during the first week of illness was associated with long COVID (odds ratio = 3.53 (2.76-4.50)). A simple model to distinguish between short COVID and long COVID at 7 days (total sample size, n = 2,149) showed an area under the curve of the receiver operating characteristic curve of 76%, with replication in an independent sample of 2,472 individuals who were positive for severe acute respiratory syndrome coronavirus 2. This model could be used to identify individuals at risk of long COVID for trials of prevention or treatment and to plan education and rehabilitation services.
COVID-19 can manifest a wide severity spectrum from asymptomatic to fatal forms2. A further source of heterogeneity is symptom duration. Hospitalized patients are well recognized to have lasting dyspnea and fatigue in particular3, yet such individuals constitute only a small proportion of symptomatic COVID-19 (ref. 4). Few studies capture symptoms prospectively in the general population to ascertain with accuracy the duration of illness and the prevalence of long-lasting symptoms.
Here, we report a prospective observational cohort study of COVID-19 symptoms in 4,182 users of the COVID Symptom Study who reported testing positive for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and started logging on the app when feeling physically normal, enabling accurate determination of symptom onset (Methods)5,6. Symptom duration in these individuals was compared with that in age-, sex- and body mass index (BMI)-matched symptomatic controls who tested negative for COVID-19.
We then compared users with symptoms persisting over 28 d (LC28) to users with shorter duration of symptoms, that is, less than 10 d (short COVID). Our previous findings that clusters of symptoms predicted the need for acute respiratory support7 led us to hypothesize that persistent symptomatology in COVID-19 (long COVID) is associated with early symptom patterns that could be used for prediction.
Results
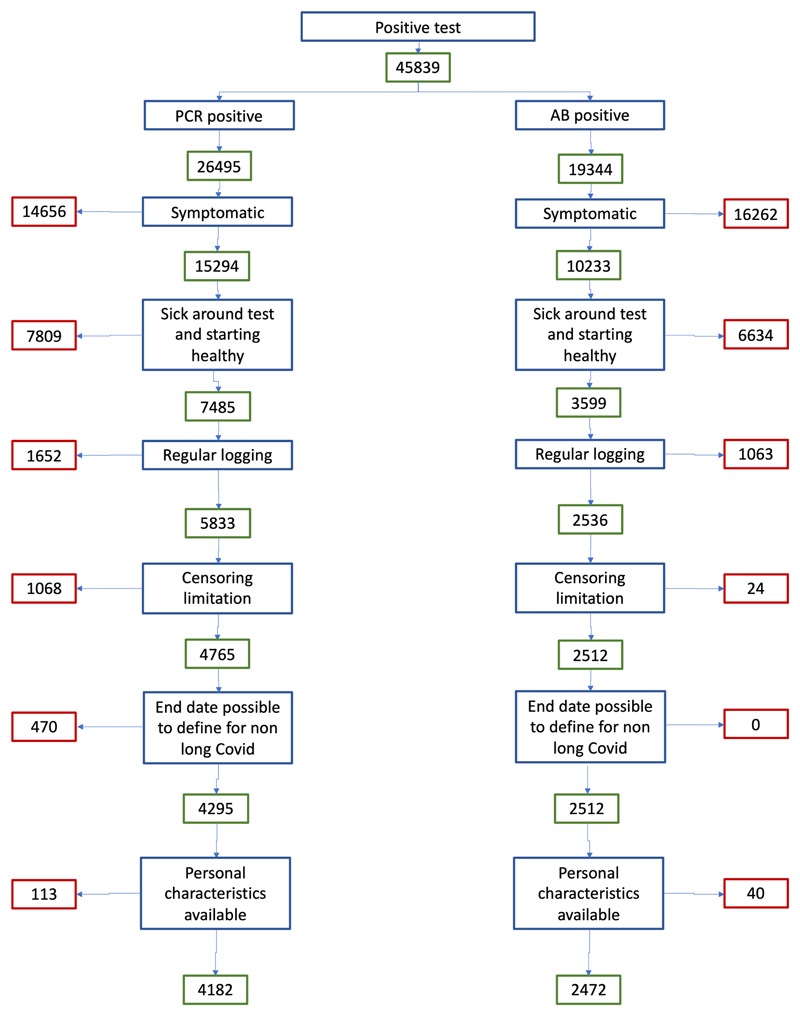
The COVID Symptom Study is a mobile application launched in response to the COVID-19 pandemic. Contributors to the app are prompted to provide daily information on their health status and symptoms, as well as results of any available COVID-19 test. Here we used data collected between 24 March 2020 (launch date in the United Kingdom) and 2 September 2020. During this time, 4,223,955 adults registered onto the app (mean age (standard deviation (s.d.)) 45.97 (15.8) years; 57% female), with the majority from the UK (88.2%), as well as the United States (7.3%) and Sweden (4.5%). From these, we selected 4,182 individuals who met the inclusion criteria to investigate the duration of persistent symptoms in COVID-19 (Extended Data Fig. 1).
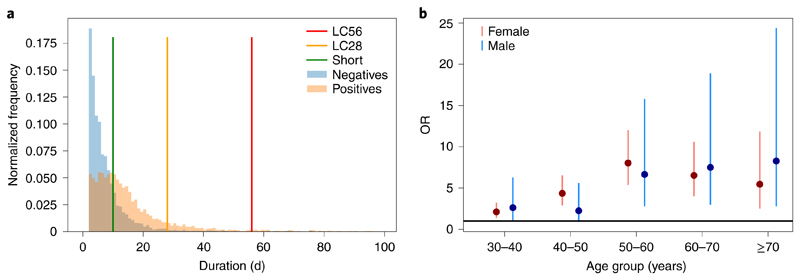
Figure 1 shows the duration of symptoms reported in the individuals who tested positive for COVID-19 overlaid on age-, sex- and BMI-matched symptomatic controls who tested negative for COVID-19. For controls, the median duration of symptoms was 5 d (3–9 d), with 2.4% reporting symptoms for ≥28 d (Fig. 1). For individuals who had a positive swab for COVID-19 (n = 4,182 from the UK, the US and Sweden), the overall median symptom duration was 11 (interquartile range (IQR), 6–19) days, with 558 (13.3%) people who met the LC28 definition (median (IQR), 41 (33–63) days). Of those, 189 (4.5%) met the definition for LC56 (duration ≥ 56 d) and 108 (2.6%) for LC84 (duration ≥ 84 d; all percentages were calculated with respect to the overall sample, n = 4,182). In contrast 1,591 (38.0%) individuals had short COVID (median (IQR) symptom duration, 6 (4–8) days). The proportions were comparable in all three countries (UK: 3,491, US: 218, Sweden: 473; LC28: UK 13.3%, US 16.1%, Sweden 12.1%; P = 0.35; LC56: UK 4.7%, US 5.5%, Sweden 2.5%; P=0.07). Supplementary Tables 1 and 2 report equivalent properties for groups that did not meet inclusion criteria and the effect of exclusion on the estimation of LC28 proportions. Supplementary Table 3 presents the population symptom reporting rate over the study period in those with positive tests, negative tests and without tests.
Fig. 1. Distribution of disease duration and age effect on duration.
a, Distribution of symptom duration in COVID-19. The colored bars indicate the limits to define short, LC28 and LC56 disease duration. The y axis represents the normalized frequency of symptom duration; 2.4% of negative controls and 3.3% of individuals with COVID-19 reported symptoms for ≥28d. b, ORs and 95% CIs of LC28 for each age decile compared to the 20- to 30-year-old age group when considering LC28 versus short COVID (1,516 females and 633 males). For males aged 20-30 years (n = 117), the proportion who had LC28 was 4.5%, compared with 5.6% of females in same age range (n = 357).
Table 1 summarizes the descriptive characteristics of the study population stratified by symptom and disease duration. LC28 was significantly associated with age, rising from 9.9% in the individuals aged 18–49 years to 21.9% in those aged ≥70 years (P < 0.0005), with an escalation in odds ratio (OR) by age decile (Fig. 1b and Supplementary Table 4). LC28 disproportionately affected women (14.9%) compared with men (9.5%), although not in the older age group (≥70 years). Long COVID affected all socioeconomic groups, as assessed using the Index of Multiple Deprivation (IMD; Extended Data Fig. 2). Individuals with long COVID were more likely to have required hospital assessment (Table 1). Asthma was the only preexisting condition significantly associated with LC28 (OR = 2.14 (95% confidence interval (CI) 1.55–2.96); Extended Data Fig. 3).
Table 1. Characteristics of individuals with COVID-19 by symptom duration, compared to age-, sex- and BMI-matched app users who tested negative for COVID-19.
| Positive PCR test | Matched negative sample | |||||
|---|---|---|---|---|---|---|
| Short (<10 d) | LC28 (≥28 d) (including LC56) | LC56 (≥56 d) | Intermediate (≥10 d < 28 d | Overall | ||
| Number | 1,591 | 558 | 189 | 1,915 | 4,182 | 4,182 |
| UK/SE/US (numbers; %) | 1,365/139/87; 85.8/8.7/5.5 | 466/57/35; 83.5/10.2/6.3 | 165/12/12; 87.3/6.3/6.3 | 1,558/271/86; 81.4/14.2/4.5 | 3,491/473/218; 83.5/11.3/5.2 | 3,882/131/169; 92.8/3.1/4.1 |
| Male (%) | 32.7 | 20.3*** | 16.9* | 27.9 | 28.5 | 28.5 |
| Age, years (median, IQR) | 38 (29-49) | 50 (39-57)*** | 52 (43-59)*** | 43 (33-53) | 42 (32-53) | 42 (32-53) |
| Age group (18-49/50-69/>70) (numbers; %) | 1,122/331/38; 75.3/22.2/2.5 | 259/262/24; 47.5/48.1/4.4 | 69/96/11; 39.2/54.5/6.3 | 1,293/594/28; 67.5/31.0/1.5 | 2,627/1,195/96; 62.8/28.6/2.3 | 2,821/1,264/97; 67.5/30.2/2.3 |
| Obese (%) | 23.8 | 27.6* | 26.5 | 277*** | 26.3 | 26.4 |
| BMI (kg/m2) (median, IQR) | 25.5 (22.7-29.7) | 26.1 (23.3-30.5) | 25.9(23.3-30.5) | 26.2 (23.2-30.7)*** | 25.9(23.3-30.3) | 25.9 (23.0-30.3) |
| Asthma (%) | 7.7 | 15.8*** | 18.0*** | 10.0* | 10.0 | 13.7 |
| Lung disease (%) | 12.8 | 16.5** | 15.9 | 13.3 | 13.6 | 13.7 |
| Diabetes (%) | 3.0 | 3.9 | 5.8* | 2.6 | 2.9 | 2.8 |
| Heart disease (%) | 1.7 | 3.2** | 4.8** | 1.6 | 1.9 | 1.7 |
| Kidney disease (%) | 0.5 | 0.9 | 0.5 | 0.4 | 0.6 | 0.6 |
| IMD (median decile, IQR) | 7(4-9) | 7(5-9) | 7(5-9) | 7(4-9)* | 7(4-9) | 7(5-9)*** |
| IMD quintilesa (numbers; %) | 64/75/334/132/634 5.2/6.1/27.0/10.7/51.2 | 23/23/86/49/240 5.5/5.5/20.4/11.6/57.0 | 10/9/26/18/88 6.6/6.0/17.2/11.9/58.3 | 155/246/310/334/397 10.7/17.1/21.5/23.2/27.5 | 158/194/830/363/1653 4.9/6.1/26.0/11.4/51.7 | 118/193/895/376/2057 3.2/5.3/24.6/10.3/56.5 |
| Visit to hospital (%) | 7.0 | 31.5*** | 43.9*** | 14.3*** | 13.9 | 4.1 |
| Number of symptoms in the first week (median, IQR) | 5(3-7) | 7(5-9)*** | 7(5-9)*** | 6(4-8)*** | 6(4-8) | 3 (2-4)*** |
Asterisks indicate statistically significant differences compared to short COVID; *P<0.1, **P<0.05 and ***P<0.01. Comparisons were performed with respect to the ‘short duration’ within the positive group. Matched negatives were compared to the overall positive population. Two-sided Mann-Whitney U tests were performed for continuous variables, and chi-squared tests were performed when comparing proportions. UK, United Kingdom; SE, Sweden; US, United States of America
IMD information is available only for app users from the UK who entered a complete postcode.
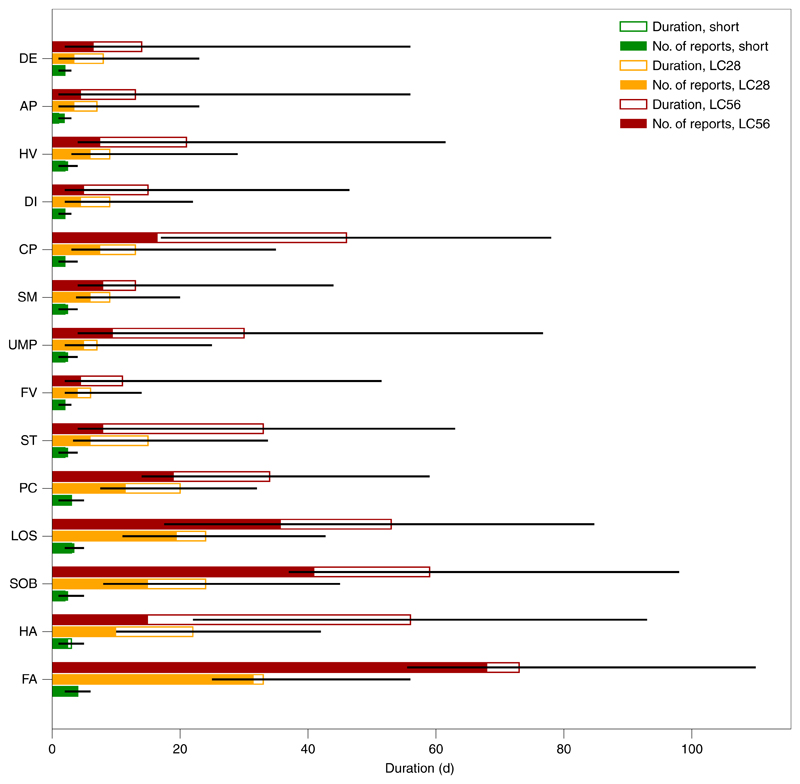
Fatigue (97.7%) and intermittent headaches (91.2%) were the most commonly reported symptoms in the individuals with LC28, followed by anosmia and lower respiratory symptoms (Fig. 2 and Supplementary Table 5). Free-text additional symptoms were more commonly reported by individuals with LC28 (81%) compared to short COVID (45%), and cardiac symptoms (for example, palpitations and tachycardia; LC28 6.1%; short COVID 0.5%; P < 0.0005), concentration or memory issues (4.1% versus 0.2%; P < 0.0005), tinnitus and earache (3.6% versus 0.2%; P < 0.0005) and peripheral neuropathy symptoms (pins and needles and numbness; 2% versus 0.5%; P = 0.004) disproportionately reported in those with LC28. Most of these additional symptoms were reported for the first time 3–4 weeks after symptom onset.
Fig. 2. Symptoms by short, LC28 and LC56 disease duration.
Each symptom is ordered from top to bottom by increasing frequency of occurrence. For short (n = 1,591), LC28 (n = 558) and LC56 (n = 189) disease durations, the median duration of report is represented by the total (hollowed) bar height and associated IQR is represented by the black line. The filled bars represent the number of times a report has been given. For both duration and the number of reported days of symptoms, the x axis reflects the number of days. This highlights the differences in the symptoms in terms of their intermittence throughout the course of the disease. DE, delirium; AP, abdominal pain; HV, hoarse voice; DI, diarrhea; CP, chest pain; SM, skipped meals; UMP, unusual muscle pains; FV, fever; ST, sore throat; PC, persistent cough; LOS, loss of smell; SOB, shortness of breath; HA, headache; FA, fatigue.
We found two main patterns of symptomatology within LC28: individuals reporting exclusively fatigue, headache and upper respiratory complaints (shortness of breath, sore throat, persistent cough and loss of smell) and those with additional multisystem complaints, including ongoing fever and gastroenterological symptoms (Extended Data Fig. 4). In the individuals with LC28, ongoing fever (OR 2.16 (CI 1.50–3.13)) and skipped meals (OR 2.52 (CI 1.74–3.65)) were associated with hospital assessment. Details of the frequency of symptoms persisting beyond 28 and 56 d after symptom onset are provided in Supplementary Table 6.
Individuals with LC28 were more likely than those with a duration of <28 days to report symptom relapses (16.0% versus 8.4%; P < 0.0005). By comparison, in the matched group of individuals who tested negative for SARS-CoV-2, symptom relapse was reported in 11.5% of individuals, and relapse was longer in the LC28 group (median (IQR), 9 (5–18) versus 6 (4–10) days).
We explored how to estimate risk of LC28 among individuals who tested positive for COVID-19 using only data available early in the disease course (first week of symptoms). Individuals who reported more than five symptoms in the first week (the median number reported) were significantly more likely to go on to experience LC28, (OR 3.95 (CI 3.10–5.04)). This strong risk factor was predictive in both sexes and in all age groups (Extended Data Fig. 5).
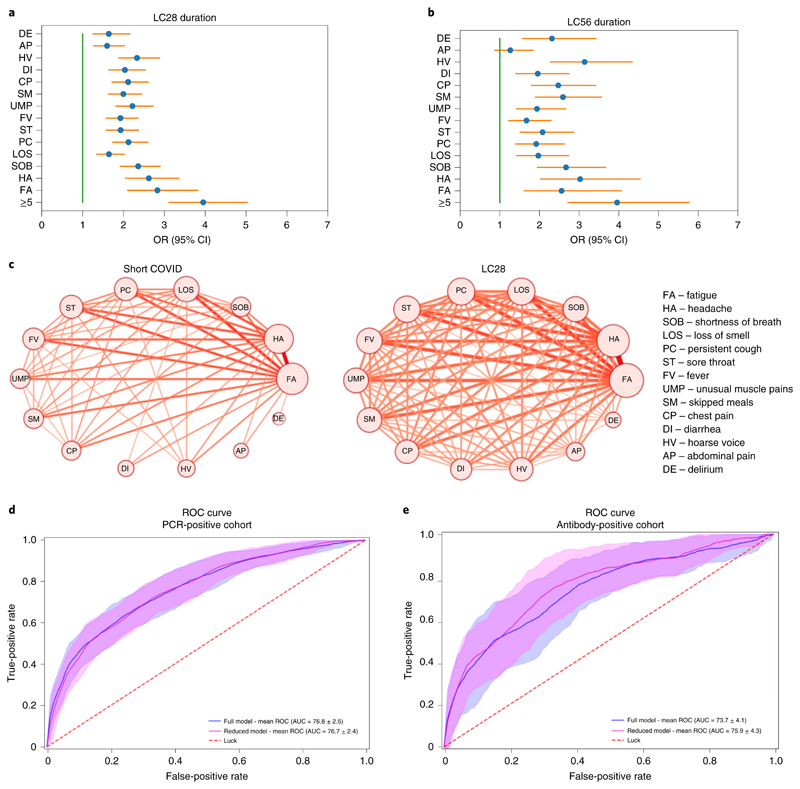
The five symptoms experienced during the first week that were most predictive of LC28 in the individuals with COVID-19 were: fatigue (OR 2.83 (CI 2.09–3.83)), headache (OR 2.62 (2.04–3.37)), dyspnea (OR 2.36 (CI 1.91–2.91), hoarse voice (OR 2.33 (1.88–2.90)) and myalgia (OR 2.22 (1.80–2.73); Fig. 3). Similar patterns were observed in both sexes. In adults aged over 70 years, loss of smell (which was generally less common in this age group) was the most predictive symptom of long COVID (OR 7.35 (CI 1.58–34.22)) before fever (OR 5.51 (CI 1.75–17.36) and hoarse voice (OR 4.03 (CI 1.21–13.42; Extended Data Fig. 5). Co-occurrence plots of symptoms in short COVID versus LC28 further illustrate early multisymptom involvement in long COVID (Fig. 3c).
Fig. 3. Prediction of long COVID compared with short COVID and illustration of multi-system presentation.
a,b, Symptom correlates of long COVID for LC28 (n = 558; a) and LC56 (n = 189; b) compared to short COVID (n = 1,591) with correction for age and sex. Error bars indicate the 95% CI for the ORs. c, Co-occurrence network of symptom pairs in which nodes represent symptoms, the frequency of symptoms corresponds to the size of the node, and the likelihood of symptom pair co-occurrence is represented by the weight of the edges linking them. Edges representing a co-occurrence of less than 10% were removed. d, ROC curve of the cross-validated full and reduced models on the PCR cohort. e, ROC curve when training on the whole PCR cohort of short and LC28 (n = 2,149) and testing on the antibody-positive cohort (n = 1,440 short COVID and n = 165 LC28) for the full (blue) and reduced (magenta) models. Random predictive probability is indicated by the dashed red line.
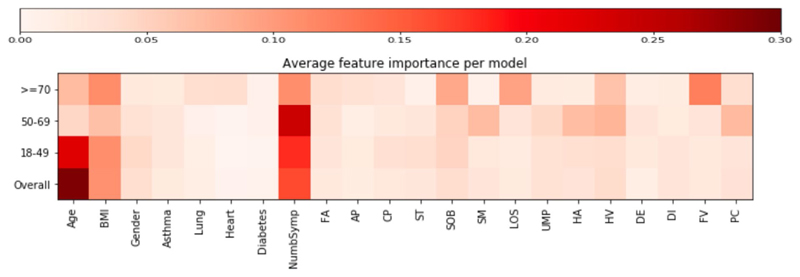
We created random forest prediction models using a combination of symptom reporting during the first week, personal characteristics and comorbidities. Using all features, the average AUC-ROC (area under the curve of the receiver operating characteristic curve) was 76.8% (s.d. 2.5; Fig. 3d) in the classification between short COVID and LC28. The strongest predictor was increasing age (29.2%) followed by the number of symptoms during the first week (16.3%). Feature importance was relatively similar across age-specific models. However, in participants aged over 70 years, features such as fever, anosmia and comorbidities were important, and could be early warning signals in older adults (Extended Data Fig. 6).
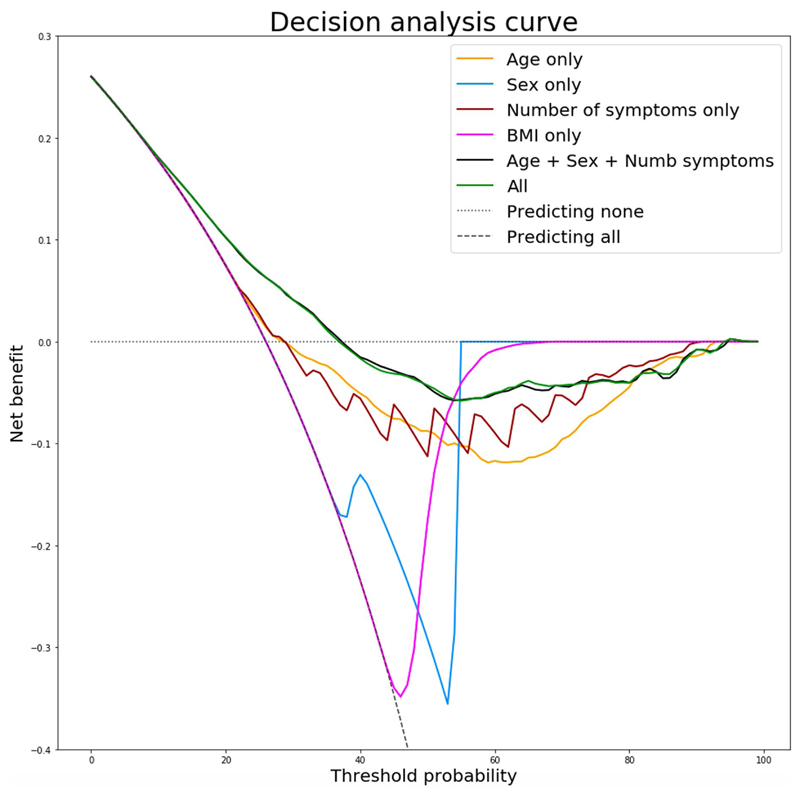
To create a model usable in healthcare settings, we simplified the prediction model to include only symptom number in the first week with age and sex in a logistic regression model, obtaining an AUC-ROC of 76.7% (s.d. 2.4) (Fig. 3d), for which the calibration slope had a median of 0.99 (IQR 0.92–1.13). When optimizing the balance between false positives and false negatives, we obtained a specificity of 73.4% (s.d. 9.7) and a sensitivity of 68.7% (s.d. 9.9). Specificity, sensitivity, positive predictive value and negative predictive value at different thresholds are presented in Supplementary Table 7. A comparison of decision analysis curves between other simple prediction models highlighted the superiority of this approach (Extended Data Fig. 7).
Key predictive findings of our analysis were validated in an independent dataset of 2,412 individuals who reported testing positive for antibodies (but without a positive PCR result) to SARS-CoV-2 from 2 weeks after symptom onset where, again, more than five different symptoms in the first week of illness was the strongest predictor (OR 4.60 (95% CI 3.28–6.46)). The simple prediction model was similarly predictive of LC28 in the antibody group, with an AUC-ROC of 75.9% (s.d. 4.3%) and median calibration slope of 1.09 (0.85–1.63; Fig. 3e).
Discussion
While this study provides insights into the clinical presentation of long COVID, there are limitations and any generalization should be considered carefully. Our study was limited by being confined to app contributors, rather than a representative sample of the population. App users were disproportionately female, and those over 70 years of age were underrepresented, which could increase or decrease our estimate of the prevalence and duration of long COVID. Caution is needed in the interpretation of associations found in smaller population subgroups. Swab test results were self-reported and were all assumed to be from PCR with reverse transcription (RT–PCR), as antigen tests were not available at the time. Applying a weighting to make the cohort representative of the UK population (Methods), the estimated proportion of people experiencing symptomatic COVID-19 who went on to suffer long COVID was similar: 14.5%, 5.1% and 2.2% for 4-, 8- and 12-week durations, respectively. Although estimates could be inflated due to PCR testing that was restricted to those who were more severely unwell early in the pandemic in the UK, or if regular logging or test results encouraged a systematic bias in symptom reporting, long-COVID prevalence in the current study could also be underestimated if individuals with prolonged symptoms were more likely to stop logging symptoms on the app. Our participant selection criteria were chosen to identify cases and disease onset with confidence. Demographics of excluded groups with upper and lower bounds for estimates given each exclusion criteria (Supplementary Tables 1 and 2) and basal symptom reporting (Supplementary Table 3), suggest that our estimates are likely to be conservative. We had insufficient numbers to explore risk factors for disease lasting longer than 2 months, and were unable to analyze the impact of ethnicity due to incomplete data. Further, due to the use of very regular assessment, we could not find any other external dataset for external validation. In addition, the list of symptoms on the app, while comprehensive, is not necessarily exhaustive, although analysis of the free-text responses allowed us to highlight other symptoms present in long COVID, such as cardiac and neurological manifestations. With emerging evidence of ongoing myocardial inflammation8,9 associated with COVID-19, this calls for specific studies of cardiac and neurological longer-term sequelae of COVID-19.
At the population level, it is critical to quantify the burden of long COVID to assess its impact on the healthcare system and appropriately distribute resources. In our study, prospective logging of a wide range of symptoms allowed us to conclude that the proportion of people with symptomatic COVID-19 who experience prolonged symptoms is considerable, and relatively stable across three countries with different cultures. Whether looking at a 4-week or an 8-week threshold for defining long duration, those experiencing long COVID were consistently older, more likely to be female, and more likely to have required hospital assessment than in the group reporting symptoms for a short period of time. Those going on to experience LC28 had multisystem disease from the start, supporting the need for holistic care10. While asthma was not reported as a factor of risk for hospitalization in some studies11, its association with long COVID (LC28) warrants further investigation. Analysis of the pathophysiological drivers underlying the risk factors for long COVID identified here is a critical next step.
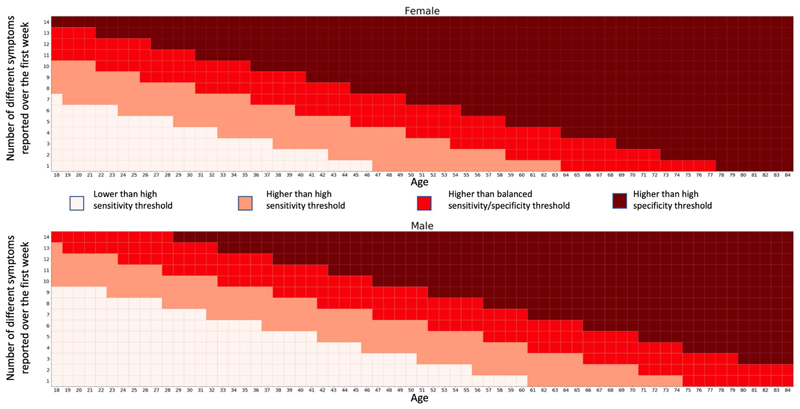
We found that early disease features were predictive of duration. With only three features—the number of symptoms in the first week, age and sex—we built a model designed to separate short (<10 d) and long (≥28 d) duration of COVID-19. The model generalized with the same performance to the population that reported antibody testing. This information could feature in targeted education material for both affected individuals and healthcare providers, and we present typical nomograms for use in clinical settings (Extended Data Fig. 8), with model results at different thresholds depending on whether high sensitivity, specificity or a balanced model is required (Supplementary Table 7). Moreover, this method could help determine at-risk groups and be used to target early intervention trials and clinical service developments to support rehabilitation in primary and specialist care12 to alleviate long COVID and facilitate timely recovery.
Online content
Any methods, additional references, Nature Research reporting summaries, source data, extended data, supplementary information, acknowledgements, peer review information; details of author contributions and competing interests; and statements of data and code availability are available at https://doi.org/10.1038/s41591-021-01292-y.
Methods
Ethical approval
All subscribers provided informed consent for use of their data for COVID-19 research. In the UK, the app and study were approved by King’s College London (KCL) ethics committee (REMAS no. 18210, review reference LRS-19/20–18210). In Sweden, ethical approval for the study was provided by the central ethics committee (DNR no. 2020–01803). In the US, this study was approved by the Partners Human Research Committee (protocol no. 2020P000909).
Dataset
Data used in this study were acquired through the COVID Symptom Study app, a mobile health application developed by Zoe Global with input from physicians and scientists at KCL, Massachusetts General Hospital, and Lund and Uppsala Universities5,6. The app, which collects data on personal characteristics and enables prospective logging of symptoms, was launched in the UK, the US and Sweden between 24 March 2020 (UK) and 30 April 2020 (Sweden), and rapidly reached over 4 million users from the community. App users were asked to report their health status daily, and any incident COVID-19 test (both undertaking of the test and its result). Questions on the app are appended in Supplementary Table 9. The current study focused on 4,182 users who reported testing positive for SARS-CoV-2 by PCR swab test with symptom onset between 25 March 2020 and 30 June 2020, for whom the date of symptom onset matched clinically with the date of test, and in whom duration of symptoms could be estimated (see Supplementary Fig. 1 for a flowchart of study inclusion). We repeated the analyses in an independent subgroup of 2,412 app users who reported a positive test result for antibodies to SARS-CoV-2 at least 2 weeks after symptom onset, but without swab test results (Supplementary Fig. 1).
To understand how the duration and relapse rate compared to a similar population not suffering from COVID-19, we selected an additional matched sample from all app users who met the study inclusion criteria but who tested negative by PCR swab test, and, for each individual with COVID-19, we chose the individual from the negative group with the smallest Euclidean distance based on sex, age and BMI13.
Definitions
Symptoms considered when determining disease duration were: abdominal pain, chest pain, sore throat, shortness of breath, fatigue, hoarse voice, delirium, diarrhea, skipped meals, fever, persistent cough, unusual muscle pains, loss of smell and headache.
Onset of disease was defined as the first day of reporting at least one symptom and a sum of symptoms being nonzero for more than 1 d.
Disease end was defined as the last day of symptom reporting before reporting as healthy for the next consecutive 7 d, or the last day of reporting with fewer than five symptoms before ceasing use of the app. For included participants who had ceased using the app and whose cumulative number of symptoms were fewer than five, disease end was considered as the last log.
Relapse was defined as two or more days of symptoms (minimum of one symptom) within a 7-d window after 1 week of healthy logging, if initial symptoms were temporally close to a positive swab test.
Long COVID was defined as symptoms that persisted for more than 4 weeks (28 d, LC28), more than 8 weeks (56 d, LC56) or more than 12 weeks (LC84) between symptom onset and end, while short duration was defined as an interval of less than 10 d between symptom onset and end, without a subsequent relapse (short COVID).
Inclusion and exclusion criteria
To be included in the subsequent analysis, users of the COVID Symptoms Study app were selected based on the following criteria: age ≥ 18 years; BMI greater than 15 and less than 55, a positive SARS-CoV-2 swab test (PCR) confirming the diagnosis of COVID-19; disease onset between 14 d before and 7 d after the test date, and before the 30 June 2020 (to limit right censoring).
Exclusion criteria were: individuals who started app reporting when already unwell; users reporting as exclusively healthy throughout the study period; users with gaps of more than 7 d after an unhealthy report who did not report any hospital assessment (to allow for gaps due to hospitalization). In addition, individuals reporting symptoms for fewer than 28 d but who reported more than five symptoms at their last log were excluded, as accurate symptom duration could not be ascertained.
Inclusion and exclusion criteria were similar for the matched sample of individuals with a negative COVID-19 test, which differed only on the result of their RT–PCR test.
To assess the impact of the different exclusion criteria on rates of LC28, we show the lower and upper bounds of these proportions according to lower and upper bound assumptions on duration (Supplementary Table 4). We also provide an estimation of the proportion of LC28 when accounting for a possible rate of false negatives ranging from 2% to 30% based on the distribution estimated from the matched negative sample. Supplementary Table 5 presents the demographics of the different excluded groups.
Statistical testing and modeling
Data collected prospectively until 2 September 2020 were included, to allow sufficient time to ascertain duration. We used univariable and multivariable logistic regression to assess symptoms associated with short and long COVID, respectively, adjusting for sex and age, using Statsmodels v0.11.1 in Python 3.7. Separate models were fitted to subgroups stratified by sex and age (18–49, 50–69 and >70 years). For analysis of relapse, existence and duration of relapse were compared between the LC28 group and the whole control sample, using a Mann–Whitney U test.
We used a k-modes clustering analysis to investigate whether there was evidence of different subtypes of long COVID, using k-mode package v0.10.2. The number of ideal symptom clusters was obtained via a silhouette analysis with Dice distance metrics. Differences between LC28 and short COVID were visualized using a co-occurrence network (NetworkX), applying a 10% threshold to remove rare edges to aid visualization.
Finally, to create a predictive model for long COVID LC28, we used sklearn v0.22.2.post1, training random forest classifiers using stratified repeated cross-validation (ten times, five folds) with a grid search for hyperparameter estimation including, as features, information available during the first week of illness, reported comorbidities (asthma, lung disease, heart disease, kidney disease and diabetes) and personal characteristics (BMI, age and sex). In addition to a global consideration of the studied sample population, separate models stratified by age were also entrained using a similar cross-validation setting (hyperparameter search and stratified sampling). After running the cross-validation for each model structure (50 times), the feature importance was averaged across the different repeated folds. To create a simplified linear model, we applied a Lasso least angle regression information criterion with Bayesian information criterion for feature selection.
This resulted in a model that included only age, sex and the number of symptoms experienced during the first week. Using only these three features, a logistic regression model was then assessed with the same stratification and cross-validation.
To assess performance on the test dataset (antibody positive), cross-validation was also performed to obtain an indication of the variability in performance using models that were trained on the whole PCR-positive sample. For the reduced logistic regression model, the score was given by the following formula:
S = 0.259503 × NumberSymptoms + 0.055457 × age – 0.633310 × sex – 3.20 (where sex is encoded as 1 – female/2 – male)
Where ‘NumberSymptoms’ corresponds to the sum of different symptoms experienced over the first week among the list of 14 symptoms reported on daily logs. This score was then transformed to a probability using the formula: 1/(1 + exp(–score))
Software code and packages
The following packages in Python 3.7 were used for the analyses performed in this study: numpy v1.16.4, pandas v0.25.0, Statsmodels v0.11.1, k-mode v0.10.2, NetworkX v2.3, scipy v1.3.1, sklearn v0.22.2.post1 and exetera (https://github.com/KCL-BMEIS/ExeTera/).
Matching with negative cases
The negative cases were selected using the same inclusion rules and were matched to the positive cases using the minimum Euclidean distance between the vectors of features created by age, BMI and sex applying a Hungarian matching algorithm. The sex feature was multiplied by 100 to ensure balance between feature strength.
To assess the impact of possible false negatives in the estimate of prevalence of LC28, for both extremes of the expected proportion of false-negative results (2% and 29%), we randomly sampled (100 times) individuals from the matched sample and adjusted the estimate of LC28 according to the mean proportion of LC28 obtained during the random sampling.
Rebalancing to UK population demographics
Lastly, rebalancing with respect to the UK population was performed by re-weighting the age and sex proportions of LC28 in the studied sample by those of the UK population using census data from 2018. The weighting per age group is described in Supplementary Table 8.
Ascertainment of parameters
The wording of the questions on the app during registration and description of symptom presentation is available in Supplementary Table 9. Specific comments regarding changes and interpretation are in square brackets.
IMD deciles were calculated within each country in the UK as an indicator of area-based socioeconomic status using the postcode of the app contributors. Deciles were collapsed to quintiles in the figures. The IMD was downloaded from the following relevant government sources, using the most recently available IMD at the time of analysis: England (2019): https://www.gov.uk/government/statistics/english-indices-of-deprivation/; Scotland (2016): https://www2.gov.scot/Topics/Statistics/SIMD/; Wales (2019): https://statswales.gov.wales/Catalogue/Community-Safety-and-Social-Inclusion/Welsh-Index-of-Multiple-Deprivation/WIMD-2019/.
Reporting Summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Extended Data
Extended Data Fig. 1. Study inclusion criteria.
Individuals reporting symptoms for at most 1 day were considered for the purpose of this analysis to be asymptomatic. We further excluded users who joined the app already unhealthy, for which the onset of disease was not calculable. Of the remainder, we excluded those who only reported intermittent unhealthy report and restricted to individuals reporting prospective symptoms at least once a week over the course of the disease. The left side of the diagram represents the inclusion flowchart for individuals reporting a positive swab test while the right side reflects the inclusion flowchart for individuals with antibody positive test only.
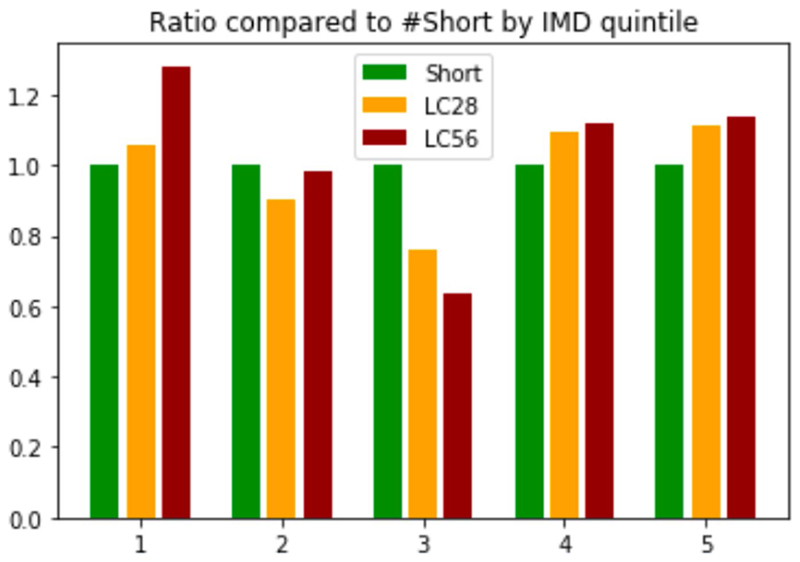
Extended Data Fig. 2. IMD ratio compared to short-COVID.
Ratio of LC28 (n=558) and LC56 (n=189) vs short-COVID (n=1591) by Index of Multiple Deprivation (IMD) quintile.
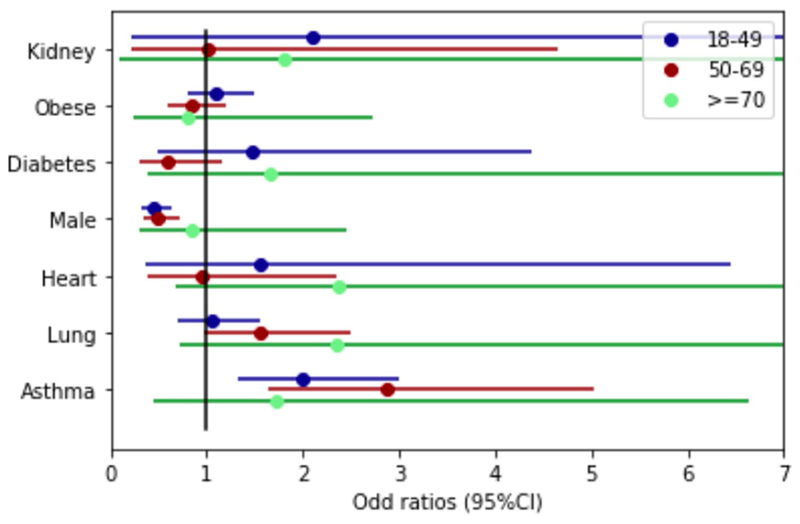
Extended Data Fig. 3. Odds ratio of LC28 per comorbidity.
Odds ratios and associated 95% confidence interval for the risk of developing Long Covid 28 for each comorbidity or risk factor, correcting for age and gender in each age group (18-49 n=1466, 50-69 n=621, >=70 n=62).
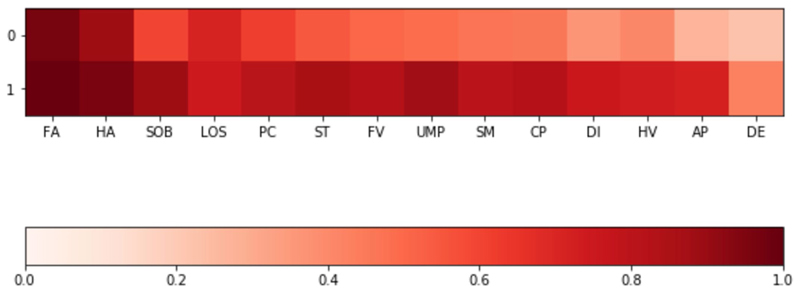
Extended Data Fig. 4. Symptom clustering in LC28.
Clustering of symptoms in the LC28 group, indicating a common strong higher airways component with fatigue, headache, and loss of smell for both groups; and a more multi system presentation for the second group. Colouring presents the frequency of reporting of a given symptom. Abbreviations: DE – delirium, AP – Abdominal Pain, HV – Hoarse Voice, DI – Diarrhoea, CP – Chest Pain, SM – skipped meals, UMP – Unusual Muscle pains, FV – Fever, ST – Sore Throat, PC – Persistent Cough, LOS – Loss of smell, SOB – Shortness of breath, HA – Headache, FA – Fatigue -.
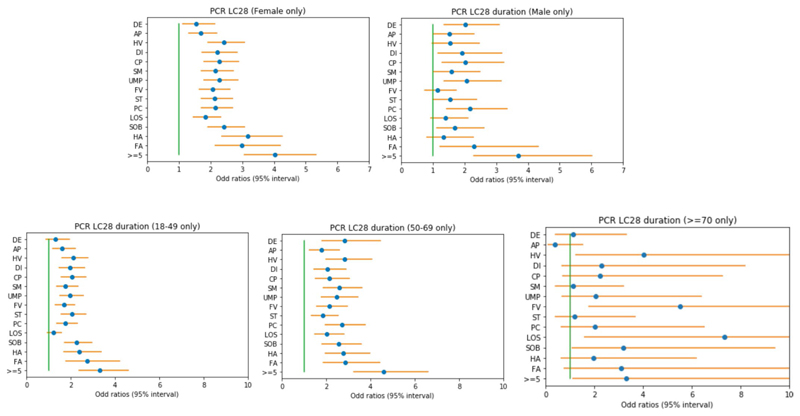
Extended Data Fig. 5. Odds ratios of LC28 per sex and age group.
Odds ratios and associated 95% confidence intervals of LC28 when presenting a given symptom during the first week compared t, correcting for age and gender (if necessary) in different subgroups female(a) (n=1516), male (b) (n=633), 18-49 (c) (n=1466), 50-69 (d) (n=621), >=70 (e) (n=62). Abbreviations: DE – delirium, AP – Abdominal Pain, HV – Hoarse Voice, DI – Diarrhoea, CP – Chest Pain, SM – skipped meals, UMP – Unusual Muscle pains, FV – Fever, ST – Sore Throat, PC – Persistent Cough, LOS – Loss of smell, SOB – Shortness of breath, HA – Headache, FA – Fatigue.
Extended Data Fig. 6. Comparison of feature importance.
Comparison of mean feature importance (proportion ranging from 0 to 1) for the cross-validated random forest models across the different age groups when considering personal characteristics and presented symptoms during the first week of the disease. Abbreviations – (Abbreviations DE – delirium, AP – Abdominal Pain, HV – Hoarse Voice, DI – Diarrhoea, CP – Chest Pain, SM – skipped meals, UMP – Unusual Muscle pains, FV – Fever, ST – Sore Throat, PC – Persistent Cough, LOS – Loss of smell, SOB – Shortness of breath, HA – Headache, FA – Fatigue).
Extended Data Fig. 7. Decision Analysis Curve.
Decision analysis curve comparing the final simple model to other models of simple logistic regression considering different feature associations.
Extended Data Fig. 8. Nomograms.
Example of nomograms that could be used to assess risk of developing LC28 based on 7 days of symptoms and corresponding table of sensitivity, specificity positive and negative predictive values at the different thresholds, given a prevalence of 13.3%. For a sensitive model, for example to apply further monitoring for the development of Long-COVID, the threshold between white and pink could be used, with a PPV of 34% and NPV of (98%), whereas more specific model, for example to recruit to trials to prevent Long-COVID, might use the dark red threshold, with a PPV of 60%, although some individuals who would go on to have Long-COVID would not be recruited (NPV 82%). Symptoms considered for the count: Fatigue – Headache – Shortness of breath – Fever – Persistent cough – Sore throat – Hoarse voice – Abdominal pain – Diarrhoea – Delirium – Chest pain – Loss of smell – Skipped meals – Unusual muscle pains.
Supplementary Material
Supplementary information The online version contains supplementary material available at https://doi.org/10.1038/s41591-021-01292-y.
Acknowledgements
Zoe Global provided in-kind support for all aspects of building, running and supporting the app and service to all users worldwide. Support for this study was provided by the National Institute for Health Research (NIHR)-funded Biomedical Research Centre based at Guy’s and St Thomas’ (GSTT) NHS Foundation Trust. This work was supported by the UK Research and Innovation London Medical Imaging & Artificial Intelligence Centre for Value-Based Healthcare. Investigators also received support from the Wellcome Trust, Medical Research Council (MRC), British Heart Foundation (BHF), Alzheimer’s Society, European Union, NIHR, COVID-19 Driver Relief Fund (CDRF) and the NIHR-funded BioResource, Clinical Research Facility and Biomedical Research Centre (BRC) based at GSTT NHS Foundation Trust in partnership with KCL. S.O. was supported by the French government, through the 3IA Côte d’Azur Investments in the Future project managed by the National Research Agency (ANR) with the reference number ANR-19-P3IA-0002. A.T.C. was supported by a Stuart and Suzanne Steele MGH Research Scholar Award. C.M. was funded by the Chronic Disease Research Foundation and by the MRC AimHy project grant. L.H.N., D.A.D., A.D.J. and A.T.C. were supported by the Massachusetts Consortium on Pathogen Readiness (MassCPR) and M. Schwartz and L. Schwartz. J.M. was partially supported by the European Commission Horizon 2020 program (H2020-MSCA-IF-2015-703787). The work performed on the Swedish study was supported by grants from the Swedish Research Council, Swedish Heart-Lung Foundation and the Swedish Foundation for Strategic Research (LUDC-IRC 15-0067).
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Author contributions
Obtained funding: C.J.S., S.O., E.L.D. and T.S. Study design and development of concept: C.H.S. and C.J.S. Data collection: C.J.S., J.C.P., P.W.F., A.T.C., T.S., R.D. and J.W. Data analysis: C.H.S., B.M., T.V., M.S.G., R.C.B., K.K., M.A., L.S.C., E.M., M.M., M.J.C., A.M., S.G., R.D., L.H.N., D.A.D., C.M.A., A.D.J., J.M., N.T., T.F. and M.F.G. Study coordination: C.J.S., S.O., T.S., T.F., M.F.G., P.W.F. and J.W. Writing the manuscript: C.H.S., C.J.S., R.S.P., F.M.K.W., E.L.D. and C.M. All authors reviewed and revised the final manuscript.
Competing interests
Zoe Global codeveloped the app pro bono for noncommercial purposes. Investigators received support from the Wellcome Trust, the MRC/BHF, European Union, French government, Alzheimer’s Society, NIHR, CDRF and the NIHR-funded BioResource, Clinical Research Facility and BRC based at GSTT NHS Foundation Trust in partnership with KCL. R.D., J.W., J.C.P., A.M. and S.G. work for Zoe Global, and T.S. and P.W.F. are consultants to Zoe Global. L.H.N., D.A.D., J.M., P.W.F. and A.T.C. previously participated as investigators on a diet study unrelated to this work that was supported by Zoe Global. C.H.S., M.S.G., E.M., K.K., M.A., L.S.C., M.M., T.V., M.J.C. and S.O. declare no competing interests.
Peer review information Nature Medicine thanks Lauren Collins and the other, anonymous, reviewers for their contributions to the peer review of this work. Jennifer Sargent was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Reprints and permissions information is available at www.nature.com/reprints.
Data availability
Data used in this study are available to bona fide researchers through UK Health Data Research at https://web.www.healthdatagateway.org/dataset/fddcb382-3051-4394-8436-b92295f14259/. Source data are provided with this paper.
Code availability
Code for data extraction is available at https://github.com/KCL-BMEIS/ExeTera/.
References
- 1.Menni C, et al. Real-time tracking of self-reported symptoms to predict potential COVID-19. Nat Med. 2020 doi: 10.1038/s41591-020-0916-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wiersinga WJ, Rhodes A, Cheng AC, Peacock SJ, Prescott HC. Pathophysiology, transmission, diagnosis and treatment of coronavirus disease 2019 (COVID-19): a review. J Am Med Assoc. 2020;324:782–793. doi: 10.1001/jama.2020.12839. [DOI] [PubMed] [Google Scholar]
- 3.Sheehy LM. Considerations for postacute rehabilitation for survivors of COVID-19. JMIR Public Health Surveill. 2020;6:e19462. doi: 10.2196/19462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alwan NA. A negative COVID-19 test does not mean recovery. Nature. 2020;584:170. doi: 10.1038/d41586-020-02335-z. [DOI] [PubMed] [Google Scholar]
- 5.Menni C, et al. Real-time tracking of self-reported symptoms to predict potential COVID-19. Nat Med. 2020;26:1037–1040. doi: 10.1038/s41591-020-0916-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Drew DA, et al. Rapid implementation of mobile technology for real-time epidemiology of COVID-19. Science. 2020 doi: 10.1126/science.abc0473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sudre CH, et al. Symptom clusters in COVID-19: a potential clinical prediction tool from the COVID Symptom study app. medRxiv. 2020 doi: 10.1101/2020.06.12.20129056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mitrani RD, Dabas N, Goldberger JJ. COVID-19 cardiac injury: implications for long-term surveillance and outcomes in survivors. Heart Rhythm. 2020 doi: 10.1016/j.hrthm.2020.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Padda I, Khehra N, Jaferi U, Parmar MS. The neurological complexities and prognosis of COVID-19. SN Compr Clin Med. 2020 doi: 10.1007/s42399-020-00527-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.National Institute for Health Research. Living with COVID-19. 2020 doi: 10.3310/themedreview_41169. [DOI] [Google Scholar]
- 11.Robinson LB, et al. COVID-19 severity in hospitalized patients with asthma: a matched cohort study. J Allergy Clin Immunol Pract. 2021;9:497–500. doi: 10.1016/j.jaip.2020.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Greenhalgh T, Knight M, A’Court C, Buxton M, Husain L. Management of post-acute COVID-19 in primary care. Br Med J. 2020;370:m3026. doi: 10.1136/bmj.m3026. [DOI] [PubMed] [Google Scholar]
- 13.Spiel C, et al. A Euclidean distance-based matching procedure for nonrandomized comparison studies. Eur Psychol. 2008;13:180–187. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary information The online version contains supplementary material available at https://doi.org/10.1038/s41591-021-01292-y.
Data Availability Statement
Data used in this study are available to bona fide researchers through UK Health Data Research at https://web.www.healthdatagateway.org/dataset/fddcb382-3051-4394-8436-b92295f14259/. Source data are provided with this paper.
Code for data extraction is available at https://github.com/KCL-BMEIS/ExeTera/.