Abstract
Fibroblast growth factor receptor (FGFR) inhibitors like ponatinib and nintedanib are clinically approved for defined cancer patient cohorts but often exert dose-limiting adverse effects. Hence, we encapsulated FGFR inhibitors ponatinib, PD173074, and nintedanib into polylactic acid nanoparticles and liposomes to enable increased tumor accumulation/specificity and reduce side effects. Different methods of drug loading were tested and the resulting formulations compared regarding average size distribution as well as encapsulation efficiency. Appropriate encapsulation levels were achieved for liposomal preparations only. Nanoencapsulation resulted in significantly decelerated uptake kinetics in vitro with clearly decreased short-term (up to 72 hours) cytotoxicity at higher concentrations. However, in long-term clonogenic assays liposomal formations were equally or even more active as compared to the free drugs. Accordingly, in a FGFR inhibitor-sensitive murine osteosarcoma transplantation model (K7M2), only liposomal but not free ponatinib resulted in significant tumor growth inhibition (by 60.4%) at markedly reduced side effects.
Keywords: FGFR inhibitors, Nanoencapsulation, Liposomes, Drug delivery, Ponatinib, Nintedanib
Background
Fibroblast growth factor receptors (FGFRs) play an essential role in the regulation of cell survival, proliferation, migration and differentiation. FGFRs are a family of receptor tyrosine kinases (RTKs) exhibiting an extracellular immunoglobulin (Ig)-like ligand binding domain, a transmembrane domain and a cytoplasmic tyrosine kinase domain. Ligand binding induces receptor dimerization and transphosphorylation at critical tyrosine residues of the cytoplasmic receptor tail, leading to the activation of downstream-signaling effectors 1. Deregulation of FGFR signaling has been linked to several developmental syndromes, but represents also an important pathway for cancer initiation and progression 1. Since the beginning of this century, the development of small molecule kinase inhibitors, which target the ATP-binding pocket of the tyrosine kinase domain of RTKs (so-called “targeted therapy”) 2, has been a success story revolutionizing cancer treatment 3. The first FGFR inhibitor approved for cancer treatment was ponatinib (Figure 1). Ponatinib is a multi-kinase inhibitor 4, also targeting the ABL, SRC, platelet-derived growth factor receptor (PDGFR) and vascular endothelial growth factor receptor (VEGFR) families 5. End of 2012, this drug was approved for clinical use in ABL-positive chronic myeloid leukemia (CML) and later for acute lymphoblastic leukemia (ALL) 6. In 2014, nintedanib (Figure 1), a triple angiokinase inhibitor targeting the proangiogenic and pro-fibrotic pathways driven by FGFR, VEGFR and PDGFR 7, was approved for non-small-cell lung cancer (NSCLC) and idiopathic pulmonary fibrosis 8, 9. However, despite the specific targeting of oncogene-dependent cancer cells, the occurrence of severe side effects and rapid development of drug resistance, comparable to classical chemotherapy, are the major limitations for successful treatment with kinase inhibitors in the clinics. The major adverse effect of nintedanib observed in clinical studies, making discontinuation of treatment or dose reduction necessary, is reversible elevation in liver enzymes 10, 11. In the case of ponatinib, a clinical phase III trial could not be continued because of a high rate of severe arterial thrombotic events like life-threatening blood clots and severe narrowing of blood vessels 12. This resulted in a reduction of the maximum applicable dose for clinical routine 13. Additionally, also other adverse effects like skin rash, hypertension and abdominal pain are commonly observed for ponatinib 14, 15.
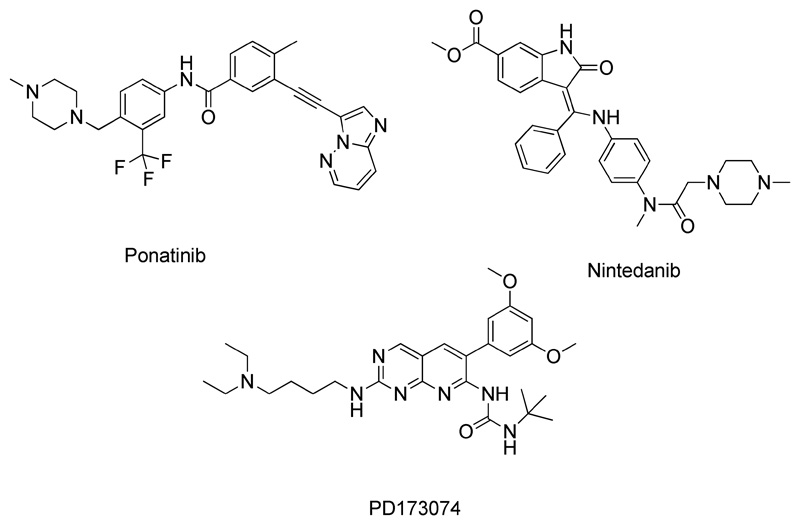
Figure 1. Chemical structures of the FGFR inhibitors investigated in this study.
Consequently, tools are urgently needed to eliminate these drawbacks and reduce adverse effects. In that respect, the encapsulation of anticancer kinase inhibitors into nanoparticulate drug formulations would be an ideal strategy, since it could enable increased tumor accumulation via the enhanced permeability and retention (EPR) effect 16 and - as a consequence - reduce side effects. The EPR effect allows nanoformulations to enrich in tumor tissue due to leaky blood vessels and an impaired lymphatic drainage system 17. Consequently, we investigated the possibility to encapsulate the multikinase inhibitors ponatinib and nintedanib as well as the more specific FGFR inhibitor PD17307418 which is currently in preclinical studies 19 into nanoformulations. On the one hand, poly(lactic acid) (PLA) nanoparticles were prepared, since PLA is biodegradable, biocompatible and approved as therapeutic drug carriers in humans by the US Food and Drug Administration (FDA) 20. On the other hand, we tested the encapsulation of these three drugs into liposomes by two different approaches: firstly, encapsulation by addition of the compounds to the lipid mixture 21 (reasonable for highly lipophilic compounds), and secondly, by the remote loading approach, which is one of the best established methods for the preparation of liposomes, with Doxil® (liposomal doxorubicin) as a clinically approved representative 22. The most promising nanoformulations were subsequently tested for their anticancer activity on a panel of FGFR-driven lung cancer cell lines in comparison to the free drugs. Furthermore, their uptake kinetics were investigated using flow cytometry measurements. Finally, the liposomal formulation of ponatinib was studied in vivo in a K7M2 tumor allograft model and compared to the free compound.
Methods
A detailed description of all methods can be found in the supplementary material section.
Very briefly: PLA polymeric nanoparticles as well as liposomes were synthesized loaded with three different FGFR inhibitors. The encapsulation efficiency, average size, PDI, zeta potential, stability and release kinetics were investigated. The most promising formulations were biologically investigated by MTT cytotoxicity assays, Western blot, ERK/AKT phosphorylation levels, cellular uptake via flow cytometry and in vivo studies.
Results
Polymeric nanoparticles - preparation and characterization
As a first approach nanoparticles of ponatinib, nintedanib and PD173074 (NP-ponatinib, NP-nintedanib, and NP-PD173074, respectively) were synthesized using the nanoprecipitation method 23 with the biocompatible and biodegradable PLA as polymer matrix. The encapsulation of the three drugs was performed by adding acetone solutions of PLA and the drug to an aqueous solution of the surfactant (Tween 80). After evaporation of the organic solvent, and adjustment of the volume to 1 mL under reduced pressure, non-encapsulated drug was removed by size exclusion chromatography (Sephadex G50). Subsequently, the nanoparticles were characterized regarding their size, polydispersity index (PDI) and encapsulation efficiency (EE). Dynamic light scattering (DLS) data showed (intensity-based) particle sizes between 96 – 147 nm with a PDI between 0.09 and 0.14, indicating very homogeneous nanoformulations (Table 1). The EE of the respective drugs was determined right after Sephadex purification of the nanoparticles by evaporation of the solvent, dissolution of the thin film in methanol and UV-Vis measurements. Unfortunately, the EE for all three drugs was very low with only 2 – 6 %.
Table 1. Data of the prepared polymeric and liposomal nanoformulationsa .
| Encapsulation efficiency [%] | PDI | Average size (nm) | |
|---|---|---|---|
| Ponatinib-Liposomes (L-ponatinib) |
92 | 0.15 | 122 |
| Ponatinib-Nanoparticles (NP-ponatinib) |
6 | 0.13 | 147 |
| Nintedanib-Liposomes | n.a. | 0.53 | 155 |
| Nintedanib-Liposomes (remote loading) (L-nintedanib) |
34 | 0.18 | 95 |
| Nintedanib-Nanoparticles (NP-nintedanib) |
5 | 0.09 | 96 |
|
PD173074-Liposomes (L-PD173074) |
23 | 0.17 | 113 |
|
PD173074-Nanoparticles (NP-PD173074) |
2 | 0.14 | 139 |
data of the most promising nanoformulation of each FGFR inhibitor written in bold
Liposomes - preparation and characterization
Due to the very low EE in case of the polymeric nanoparticles, encapsulation into liposomes was investigated. As liposomal building blocks DSPC/CHOL/DSPE-mPEG(2000) 55:40:5 mol/mol (DSPC = 1,2-distearoyl-sn-glycero-3-phosphocholine; CHOL = cholesterol; DSPE-mPEG(2000) = 1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N-[methoxy(polyethylene glycol)-2000)]] were used, well-known from already approved liposomal formulations 22. Several different methods for the preparation of liposomes are available influencing the resulting particle properties like size and EE. The so-called thin lipid film hydration method was the first described 24. Briefly, a thin film of lipids in organic solvents is formed by evaporation and rehydrated in aqueous solutions to form liposomes. The necessary size reduction and homogenization of the liposomes can be achieved using multiple extrusions through a polycarbonate membrane. However, both, the use of the Mini-Extruder from Avanti (Alabaster, AL, USA) and the Extruder from Northern Lipids (Vancouver, BC, Canada) resulted in EEs below 5 % (data not shown).
Therefore, we concentrated our next approaches on a different method of reducing the liposome size: ultrasonic homogenization with a microtip. Since the used drugs are highly lipophilic with calculated logP values of 2.38 for nintedanib, 5.01 for ponatinib and 5.11 for PD17307425, the drugs were added already at the beginning of the preparation to the lipid mixture. After hydration of the lipid-drug film with a 0.3 M (NH4)2SO4 solution and size-reduction by ultra-sonication, non-encapsulated drug was removed by size exclusion chromatography (Sephadex G50). This approach resulted in a PDI of 0.15 for liposomal ponatinib (L-ponatinib) and 0.17 for liposomal PD173074 (L-PD173074), indicating narrow size distributions (Table 1, Figures S1, S2). Average (intensity-based) sizes of 113 nm for L-PD173074 and 122 nm for L-ponatinib (Table 1) were obtained and the EE was high for L-ponatinib with 92 % and moderate for L-PD173074 with 23 %. In case of the slightly less lipophilic nintedanib, this method was unsuccessful, resulting in very inhomogeneous size distributions with a PDI of 0.53 (Table 1).
Another method for the encapsulation of compounds into liposomes is the so-called remote loading approach, which is for example applied in the preparation of Doxil® (encapsulated doxorubicin) as the most prominent FDA-approved representative 22. In Doxil®, doxorubicin is loaded via an ammonium sulfate gradient, with a lower intraliposomal pH compared to the extraliposomal solution 26. In order to ensure optimal remote loading by the ammonium sulfate gradient, the loaded molecules should have a logD at pH 7 in the range of –2.5 to 2 and a pK a ≤ 11 27. Therefore, the remote loading approach was tested for nintedanib, which has a calculated logD of 1.8 at pH 7 25 and a calculated pK a of 7.4 25. The remote loading liposomes were prepared by hydrating a thin lipid film with a 0.3 M (NH4)2SO4 solution and the size of the liposomes was reduced by ultra-sonication. To create the ammonium sulfate gradient, size exclusion chromatography (Sephadex G50) with phosphate buffered saline (PBS) at pH 7.4 as eluent was used. Subsequently, the water-soluble nintedanib-ethanesulfonate salt was mixed with the liposomes and stirred for 1.5 h at 65 °C and finally non-encapsulated drug was removed by another size exclusion column (Sephadex G50). The resulting liposomes had a PDI of 0.18, indicating a homogeneous size distribution (Figure S3), and a moderate EE of 34 %.
To check the reproducibility of the liposomes, between five to twelve different batches of the three most promising formulations (L-ponatinib and L-PD173074 by direct addition to the lipids and L-nintedanib by remote loading) were synthesized. The EE was well reproducible with values of 92 ± 7 % for L-ponatinib, 34 ± 10 % for L-nintedanib and 23 ± 1 % for L-PD173074. Also the average size and PDIs were highly reproducible with values of 122 ± 6 nm (PDI: 0.15 ± 0.04) for L-ponatinib, 98 ± 4 nm (PDI: 0.18 ± 0.04) for L-nintedanib and 112 ± 3 nm (PDI: 0.17 ± 0.02) for L-PD173074.
The lipid content in the liposomes was estimated with the Wako Phospholipids C assay, which revealed that more than 95 % of the used amount of lipids were still present in the final liposomal formulations, and the Sephadex purifications therefore only slightly reduced the overall yield.
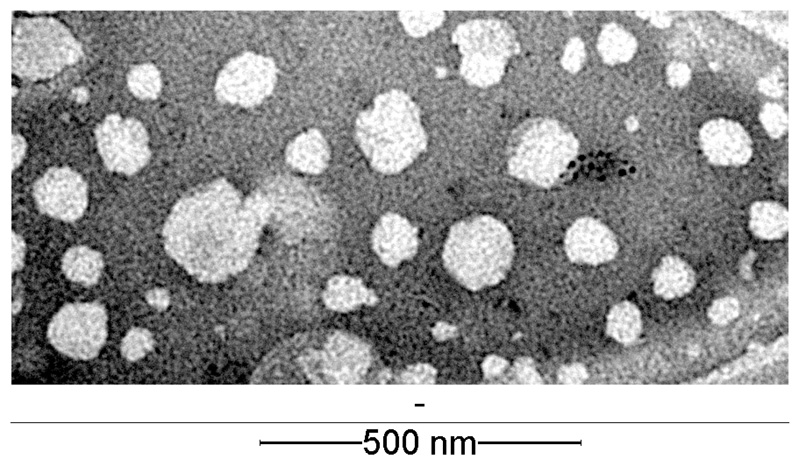
To investigate the morphology of the liposomes, negative stain transmission electron microscopy (TEM) measurements were performed. Figure 2 shows a representative image of L-PD173074, showing the almost spherical shapes of liposomes and a size slightly lower compared to the ~100 nm measured by DLS (representative TEM images of L-ponatinib and L-nintedanib can be found in Figures S4 and S5). This can likely be explained by the intensity-based DLS distributions, which can result in overestimated particle sizes. Consequently, we also calculated number-based size distributions, which revealed most abundant sizes at 80 nm for L-ponatinib, 75 nm for L-PD173074 and 71 nm for L-nintedanib (Figures S1–S3) in good agreement with the TEM images.
Figure 2. Transmission electron microscopy (TEM) image of L-PD173074 after negative staining with 2.5 % gadolinium acetate solution.
Finally, the electrostatic repulsion of the particle surfaces (zeta potential) was measured revealing slightly negative values for all liposomes at –2.6 ± 1 mV for L-ponatinib, –2.1 ± 1 mV for L-nintedanib and –2.1 ± 1 mV for L-PD173074, which is typical for PEGylated liposomes28, 29.
To check the stability of the liposomes, we performed DLS size and zeta potential measurements of the liposomal formulations after incubation at 37°C for 48 and 72 h. All parameters only slightly changed over time without any indications for degradation or agglomerations. For example the average size of L-ponatinib of 118 ± 1 nm (PDI: 0.12 ± 0.02) was stable with 117 ± 1 nm (PDI: 0.10 ± 0.01) after 48 h and 117 ± 1 nm (PDI: 0.09 ± 0.01) after 72 h. The zeta potential shifted from –2.0 ± 1 mV at time point zero to –2.5 ± 1 mV after 48 h and –2.4 ± 1 mV after 72 h at 37°C. The respective size distributions together with the measured values at the different times points for the liposomal formulations of all three inhibitors are shown in Figures S6–S8.
Drug release from liposomes
The drug release of the liposomal formulations L-ponatinib, L-PD173074 and L-nintedanib was investigated with two different approaches of the dialysis diffusion technique: firstly, the nanoformulations were introduced into a dialysis bag (molecular weight cut-off 14 kDa) and immersed into PBS. The release was measured for 48 h at 37°C by removing samples from the PBS solution and determining the drug amounts by fluorescence spectroscopy. This was possible as all three drugs showed distinct fluorescence properties when irradiated between 340–390 nm with emissions between 420–480 nm. This method resulted in very low release rates of 0.1 ± 0.05 % for L-ponatinib, 2.6 ± 0.2 % for L-nintedanib and 8.8 ± 1.6 % for L-PD173074, after 48 h (Figure 3).
Figure 3. Release profiles of L-ponatinib, L-nintedanib and L-PD173074 over 48 h with the dialysis bag diffusion technique in PBS at 37°C (data from two independent experiments).
Since especially ponatinib and PD173074 are highly lipophilic and consequently possess extremely low solubility in PBS at pH 7.4, reference experiments with the free inhibitors only were not possible. However, this would be important as at least some amounts of the drugs could be stuck at the cellulose membrane of the dialysis bags. Therefore, a second method was applied using Float-A-Lyzer Tubes with an 8–10 kDa molecular weight cut-off. The tubes were filled with the liposomal formulations and immersed into PBS for 48 h at 37°C. These tubes enable the determination of the remaining amount of drug inside the liposomes30 and consequently a possible interaction of the released inhibitor and the dialysis tube would not influence the measured drug concentration. This approach revealed that after 48 h only 0.8 ± 0.6 % for L-ponatinib, 16 ± 3.9 % for L-nintedanib, and 2.2 ± 1.5 % for L-PD173074 have been released. Thus, although the two methods show slightly different values, overall these results indicate a very stable entrapment of ponatinib, PD173074 and nintedanib into the liposomes.
Cytotoxicity of liposomal drug formulations in FGFR1-driven lung cancer cells
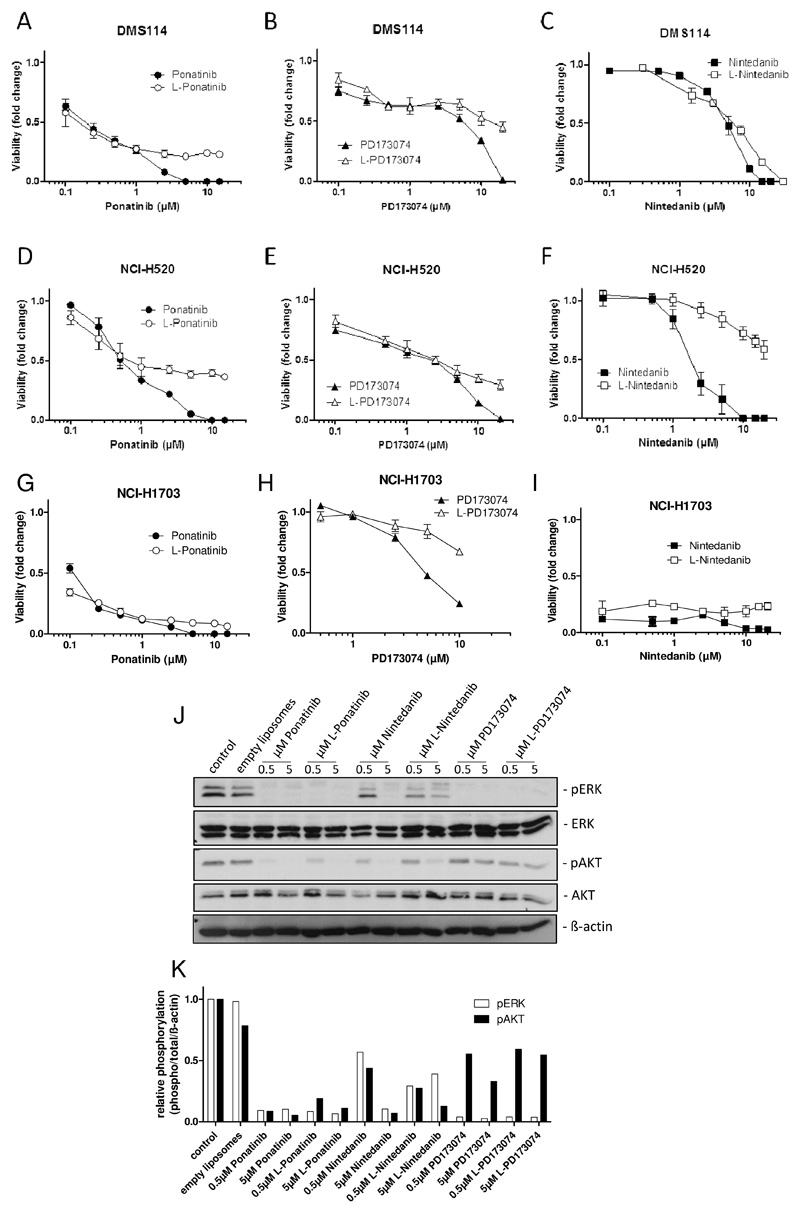
To test the cytotoxic potentials of the liposomal formulations in comparison to the free drugs, viability assays were performed using the FGFR1-driven lung cancer cell lines DMS114, NCI-H520 and NCI-H170331, 32. Treatment with liposomal drug formulations showed similar to slightly reduced cytotoxic activity as depicted by the moderately elevated IC50 values in comparison to the free drugs with the exception of L-nintedanib, which was found to be distinctly less active in NCI-H520 cells (Table 2).
Table 2. IC50 values (in μmol/L) of FGFR inhibitors and their respective liposomal formulations in FGFR1-driven lung cancer cell lines.
| Drug | DMS114 (IC50 ± SD) | Fold change | NCI-H520 (IC50 ± SD) | Fold change | NCI-H1703 (IC50 ± SD) | Fold change |
|---|---|---|---|---|---|---|
| Ponatinib | 0.51 ± 0.22 | 0.53 ± 0.24 | 0.23 ± 0.03 | |||
| L-Ponatinib | 0.62 ± 0.44 | 1.2 | 0.38 ± 0.14 | 0.7 | 0.27 ±0.09 | 1.2 |
| Nintedanib | 3.79 ± 1.26 | 2.12 ± 1.06 | 0.23 ± 0.01 | |||
| L-Nintedanib | 4.48 ± 2.65 | 1.2 | 16.4 ± 7.20 | 7.7 | 0.33 ± 0.05 | 1.4 |
| PD173074 | 2.97 ± 3.75 | 2.27 ± 0.01 | 6.33 ± 1.89 | |||
| L-PD173074 | 7.13 ± 9.24 | 2.4 | 2.04 ± 1.05 | 0.9 | 15.23 ± 0.82 | 2.4 |
Accordingly, the resulting fold change in cytotoxicity of liposomal as compared to the free drugs ranged between 0.7 and 7.7 (Table 2). Interestingly, the activity of the liposomal drugs was virtually identical to that of the respective free drugs at low concentrations below and around the respective IC50 values. In contrast, at high concentrations, the free drugs exerted profoundly stronger cytotoxic potentials as compared to their liposomal counterparts.
This was illustrated by distinctly flatter viability dose-response curves e.g. in L-ponatinib- or L-PD173074-treated DMS114 and NCI-H520 cells in comparison to the respective free drugs at concentrations above the IC50 values (Figure 4A–F). This indicates that upon a certain threshold concentration, a plateau was reached above which a further increase of liposomal drug administration did not result in further increased cytotoxicity under two-dimensional cell culture conditions. However, in the highly sensitive NCI-H1703 cells comparable differences were observed for L-PD173074, while no major differences were observed for L-ponatinib and L-nintedanib as compared to the respective free drugs also at high doses (Figure 4G–I). The high sensitivity of NCI-H1703 cells towards FGFR inhibition by ponatinib and nintedanib likely explains why the toxicity of liposomal drugs is not reduced in comparison to the free drugs. Furthermore, we wanted to evaluate our novel FGFR inhibitor formulations with respect to their inhibitory potentials on oncogenic FGFR downstream signaling cascades. 24 hours drug exposure of NCI-H520 cells revealed a strong inhibitory effect of liposomal drugs on ERK and AKT phosphorylation - major indicators of FGFR1 downstream signaling to the MAPK and the PI3K/AKT pathway, respectively. All three liposomal inhibitor formulations were virtually comparably or moderately less efficient on MAPK and PI3K pathway inhibition as their respective free drugs at both tested concentrations of 0.5 and 5 μM (Figure 4J–K). Interestingly, PD173074 both as free and as liposomal drug was less efficiently inhibiting the AKT phosphorylation as compared to nintedanib and ponatinib, while MAPK inhibition detected by ERK phosphorylation was comparable for all three compounds. Hence, in the cell culture situation liposomal formulations of the investigated FGFR inhibitors exhibited comparable to slightly reduced potency with respect to FGFR1-downstream pathway inhibition and cytotoxicity was only marginally reduced in comparison to the respective free drugs. Only at higher concentrations distinct differences were observed.
Figure 4. Reduced cytotoxicity of liposomal as compared to free FGFR inhibitors.
A-I. Viability of DMS114 (A-C), NCI-H520 (D-F) and NCI-H1703 cells (G-I) in response to increasing concentrations of free or liposomal ponatinib (A, D, G), PD173074 (B, E, H) or nintedanib (C, F, I) was analyzed by MTT assay after 72 hours drug exposure. J. Expression/phosphorylation of FGFR1 downstream signaling proteins was analyzed by Western blot in NCI-H520 cells treated with the indicated concentrations of free or liposomal ponatinib, nintedanib or PD173074 for 24 hours. ß-actin was used as loading control. One representative dataset out of three independent experiments is shown. K. Representative densitometric quantification of ERK and AKT phosphorylation levels depicted in (J) was performed using ImageJ software. Values are normalized to total ERK/AKT and to ß-actin protein levels and are given relative to the untreated control.
Liposomal FGFR inhibitors exhibit decelerated cellular uptake kinetics
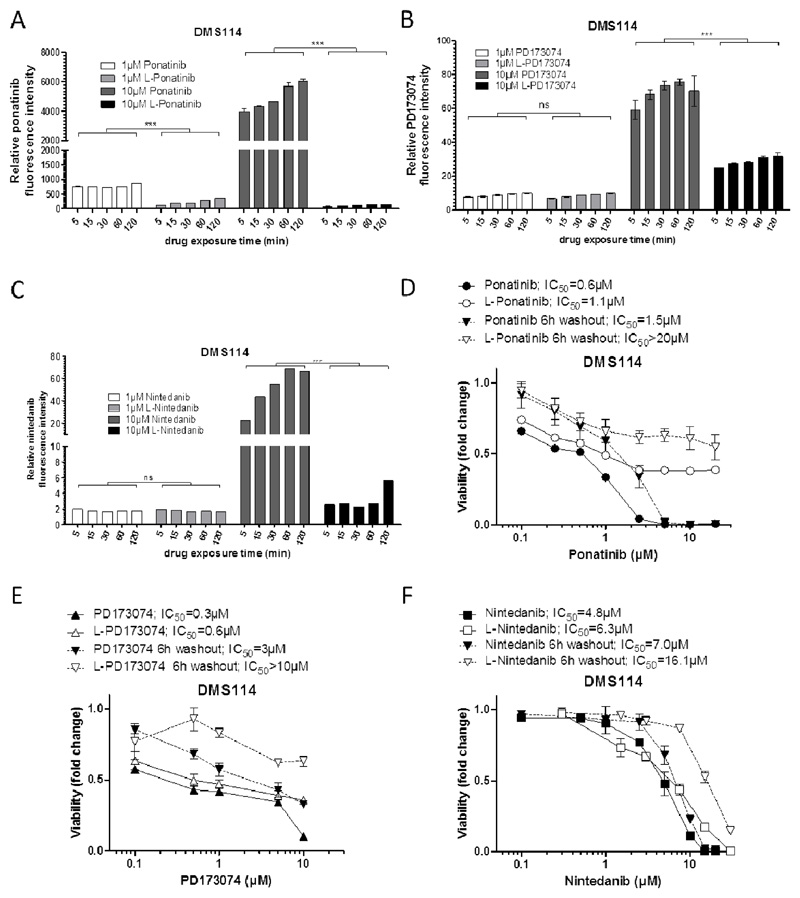
Consequently, we were interested whether liposomal drug formulation has an impact on cellular uptake kinetics and whether this effect may account for the strong differences in activity at higher concentrations in comparison to the respective free drugs. To this end, we exploited the intrinsic fluorescence activities of ponatinib, PD173074 and nintedanib 33 to compare drug uptake levels by flow cytometry. Interestingly, at lower concentration (1 μM), we did not observe differences in the time-dependent increase of intracellular drug levels in all investigated cell lines. However, in case of ponatinib a significantly accelerated uptake was observed for the free drug as compared to the liposomal formulation in all cell lines (Figure 5 A, Figure S9 A, D). Strikingly, however, administration of 10 μM of the liposomal formulations of all three drugs led to markedly lower intracellular accumulation levels as compared to the respective free drugs (Figure 5 A–C, Figure S9). In addition, at least for ponatinib, no difference in drug uptake kinetics was observed between 1 μM and 10 μM of the liposomal formulation for the duration of the experiment (240 min), indicating that this short-time scale depicts a dose-independent limiting threshold for drug uptake. These reduced intracellular accumulation levels of 10 μM liposomal drugs (38.8-, 2.3- and 14.2-fold lower in DMS114 cells, 6.2-, 2.4- and 17.3-fold in NCI-H1703 cells and 2.9-, 1.9- and 11.2-fold in NCI-H520 cells for L-ponatinib, L-PD173074 and L-nintedanib after 2 hours, respectively) correlate with the observations from cell viability assays (see Figure 4), where a distinct difference in the activity between liposomal vs. respective free drugs was observed at concentrations higher than 1 μM. To further investigate the time dependency of drug accumulation, we monitored ponatinib as compared to L-ponatinib uptake in NCI-H1703 cells up to 72 hours with sub-toxic drug doses (Figure S9G). These results showed that longer drug exposure times do result in dose-dependent ponatinib accumulation also as liposomal formulation. Still, decreased levels of intracellular L-ponatinib were apparent after up to 72 hours of drug exposure. However, it is likely that upon long-term exposure, these reduced drug levels are still sufficient to induce the same extent of cytotoxicity as did free ponatinib. In addition, we investigated uptake kinetics in non-cancer cells. To this end, we used the non-malignant murine, immortalized NIH-3T3 derived fibroblast lineage 3T3-L1. Generally, intracellular drug levels at the indicated time points were lower as compared to those determined in cancer cells (compare Figure 5A and S9A/D). While again no major differences in uptake kinetics of L-ponatinib as compared to free ponatinib were detectable at low doses (0.1 μM and 1 μM), a significant decrease was found at 10 μM (Figure S9H). This reduction, however, was relatively moderate as compared to the malignant cell counterparts.
Figure 5. Liposomal drug formulation decreases cellular uptake kinetics.
A-C. Time- and dose-dependent intracellular accumulation of liposomal (L) or free ponatinib (A), PD173074 (B) and nintedanib (C) in DMS114 cells was measured by flow cytometry. Ponatinib and PD173074 were detected using the 405 nm laser and the 450/40 nm bandpass emission filter, nintedanib was analyzed using 488 nm laser and the 530/30 nm bandpass emission filter. *** p<0.001, two-way ANOVA, Bonferroni post-test. ns, non-significant. Statistical significance levels are indicated by asterisks and include testing of each time-point between free and liposomal drugs for the given concentrations. D-F. Viability of DMS114 cells in response to different exposure times (6 and 72 hours) of free or liposomal ponatinib (D), PD173074 (E) and nintedanib (F) was analyzed by MTT assay after 72 hours.
Subsequently, we wanted to find out whether the slower uptake kinetics of the liposomal ponatinib, PD173074 and nintedanib have a time-dependent influence on their cytotoxic potential. We, thus, performed cytotoxicity assays in which cells were exposed to drugs for 6 hours and viability was determined following a 72 hours drug-free follow-up phase (Figure 5D–F). Indeed, the fold change of liposomal vs. free drugs was distinctly stronger in 6 hours-treated DMS114 cells as compared to those treated for the full 72 hours (Figure 5D–F, >13.3-fold vs 1.8-fold for ponatinib, >3.3-fold vs. 2.0-fold for PD173074, and 2.3-fold vs 1.3-fold for nintedanib, respectively). Together, these results demonstrate that exposure of lung cancer cells to FGFR inhibitors loaded into liposomes decelerated drug uptake kinetics which also may explain the reduced cytotoxic activity at higher concentrations. While these effects were clearly observable for both DMS114 and NCI-H520 cells, the cytotoxic activities of free versus liposomal drugs were comparable in NCI-H1703 cells. This might be a consequence of the exceptionally high sensitivity of this cell line towards FGFR inhibition, resulting in liposomal drug uptake over 72 hours to be sufficient to induce the same extent of cell death as exerted by the respective free drugs. To compensate the decelerated uptake kinetics of liposomal drugs, we additionally performed clonogenic assays allowing long-term drug exposure in vitro. Indeed, exposure of cells for 168 hours with rising ponatinib concentrations led to equal or even slightly enhanced toxicity profiles of liposomal as compared to the respective free drugs. Especially NCI-H1703 cells were even slightly more sensitive to L-ponatinib as compared to free ponatinib also at high concentrations in this experimental setting (Figure S10). This suggests that the cytotoxicity of liposomally encapsulated FGFR TKIs, after an initial latency period, rises between 72 and 168 hours to the same potency as the respective free drugs.
Liposomal but not free ponatinib is active against a highly tumorigenic murine osteosarcoma model in vivo
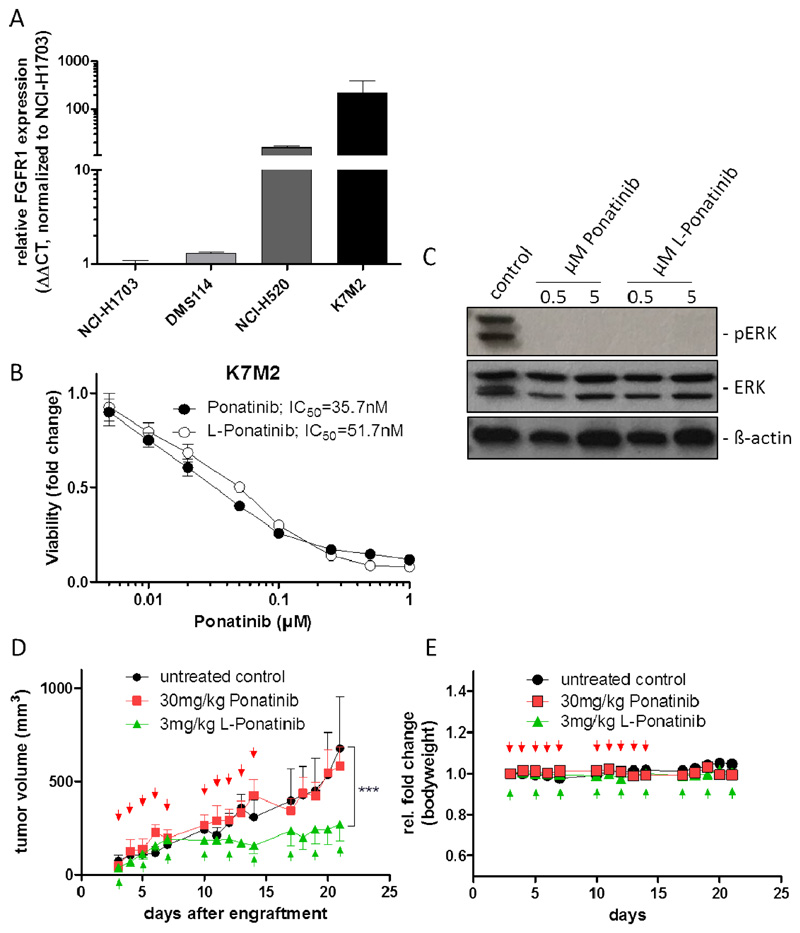
The “targeting” strategy of liposomal formulation of the investigated FGFR inhibitors yielded the expected and anticipated retardation of cellular drug uptake and a concomitant slight decrease in short-term (72 hours) cytotoxicity in our cell culture-based analyses. We, thus, aimed at testing whether these effects - together with the commonly observed prolonged plasma half-life of liposomal as compared to free drugs - might turn out to be beneficial for the tolerability as well as the long-term treatment efficacy against tumors in vivo. In an initial study, we administered free ponatinib orally (30 mg/kg) and L-ponatinib intravenously (3 mg/kg) to SCID mice in order to evaluate systemic tolerability. Notably in these experiments, oral gavage of free ponatinib resulted in severe cutaneous adverse effects in all mice, while animals treated with L-ponatinib showed no side effects and no signs of distress (Figure S11). This is an interesting finding, as ponatinib has been shown to exhibit a maximum serum concentration of 316 ng/mL after oral administration of 30 mg/kg 34. Presuming a 100% bioavailability, the 3 mg/kg intravenously injected L-ponatinib translates into distinctly higher serum concentrations of the liposomal (30 μg/mL) as compared to the free drug. As a next step, we tested the tumorigenicity of our three human FGFR inhibitor-sensitive cell models in SCID mice. However, all three cell lines proved to be non-tumorigenic or unstably growing as subcutaneous xenografts and were thus not suitable for the testing of our new drug formulation. However, subsequent studies revealed that the murine K7M2 osteosarcoma cell model exhibits very high FGFR1 expression levels even as compared to the FGFR1-amplified NCI-H520 cell line (Figure 6A). In line, we found K7M2 cells to be exquisitely sensitive towards both free and liposomal ponatinib in cell culture, exhibiting comparable IC50 values in the lower nanomolar range (Figure 6B). Concomitantly, the effect on the MAPK pathway activity was very potent, as illustrated by strong inhibition of ERK phosphorylation at both tested concentrations (Figure 6C). Thus, to evaluate the long-term treatment efficacy of L-ponatinib, syngeneic tumors from this murine model were used. As shown in Figure 6D, repeated application of L-ponatinib (3 mg/kg, i.v.) to K7M2-bearing mice resulted in a marked and significant tumor growth retardation (60.4% growth reduction at day 21 post-engraftment). In contrast, free ponatinib, given orally at a standard dose of 30 mg/kg, failed to be active (Figure 6D). Importantly, the liposomal drug formulation was well tolerated and did not affect animal body weight or behavior (Figure 6E). This indicates that the liposomal formulation is able to enhance both activity as well as tolerability of ponatinib in vivo.
Figure 6. Liposomal ponatinib is active in a murine osteosarcoma allograft model.
A. FGFR1 mRNA expression levels of NCI-H1703, DMS114, NCI-H520 and K7M2 cells was analyzed by qRT-PCR. Data were normalized to the housekeeping gene ACTB and are given relative to NCI-H1703 cells. B. Viability of K7M2 cells exposed to increasing concentrations of free or liposomal (L) ponatinib was analyzed by MTT assay after 72 hours drug exposure. C. Expression/phosphorylation of FGFR1 downstream signaling proteins was analyzed by Western blot in K7M2 cells treated with the indicated concentration of free or liposomal ponatinib for 1 hour. ß-actin was used as loading control. D. Efficacy of indicated doses of liposomal and free ponatinib on K7M2 subcutaneous allograft growth in BALB/c mice. Days of treatment are indicated by the arrows. *** p < 0.001, two-way ANOVA, Bonferroni post-test. The asterisk indicates statistical significance of L-ponatinib compared to both free ponatinib and the untreated control. E. Body weights of K7M2-engrafted BALB/c mice was assessed as indicator of therapeutic adverse effects. Treatment days are indicated by arrows.
Discussion
Liposomal drug formulations are an established and very important tool to improve the efficacy and tolerability of anticancer drugs. This is illustrated by the approval of liposomal formulations of doxorubicin (Doxil®, Myocet® and Lipodox®), daunorubicin (DaunoXome®), vincristine (Marqibo®) and mifamurtide (Mepact®) 35, 36. Additionally, many other liposomal formulations for classical chemotherapeutics, like e.g. of cisplatin and paclitaxel, are currently under clinical evaluation 35, 36. However, in case of the large class of targeted anticancer agents, clinical approvals of liposomal preparations are so far missing and only a number of preclinical studies have been reported 37. These include for instance the clinically used tyrosine kinase inhibitors (TKIs) sunitinib, imatinib, gefitinib and cabozantinib 38–42, but also experimental compounds such as the pan-kinase inhibitor staurosporine or the VEGFR2 inhibitor apatinib 43, 44. In addition, some studies have addressed nanoparticle-based co-delivery of cytotoxic drugs with a TKI like paclitaxel with lapatinib 45 and doxorubicin with erlotinib 30. With respect to cancer nanomedicine, FGFRs have been frequently used as targets for tumor-selective delivery strategies of chemotherapeutics-loaded nanoparticles 46–49. In contrast, regarding FGFRs as therapeutic target per se, solely one very recent publication for ponatinib encapsulated into polymeric micelles with and without a Janus kinase 2 (JAK2) inhibitor is available 50.
Concerning the liposomal stability, relatively similar values were reported in these studies for the different encapsulated drugs. In case of the liposomal sunitinib formulation, 18 % of sunitinib were released after 48 h 39. Comparably, for the liposomal imatinib, gefitinib and cabozantinib formulations between ~20 and 40 % release after 48 h were reported, respectively 40–42. For the drug combination studies of classical chemotherapeutic agents and TKIs, slightly faster release kinetics (~ 70% after 48 h) compared to the nanoformulations of only a single TKI were observed 30, 45. In comparison, our liposomal formulations of ponatinib, PD173074 and nintedanib showed very high stability with respect to size, PDI and zeta potential and the release kinetics with <10% in 48 h (only in case of nintedanib using the Float-A-Lyzer tubes 16% release after 48 h were measured). The EE of the liposomal formulations of sunitinib, imatinib and cabozantinib were reported to be between 80 to 90% 39, 40, 42. In contrast, for gefitinib a broad range from 20 to 90% EE, depending on the lipids used for preparation, were observed 41. The EE of our nintedanib and PD173074 liposomes was moderate with 34 ± 10 and 23 ± 1 %, respectively, and only for ponatinib a very high EE of 92 ± 7 % could be achieved. This indicates that the EE strongly depends on the exact chemical nature and physico-chemical properties of the encapsulated inhibitor. Notably, in the recently reported polymeric micelles of ponatinib the EE was extremely low with only around 3% 50. It is worth mentioning that also in this work encapsulation into polymeric nanoparticles was very ineffective with EE only between 2 and 6 %. The significantly higher EE into liposomes compared to the polymeric nanoparticles might be due to stronger interaction of the lipid bilayer with the very lipophilic drugs compared to polylactic acid of the polymeric micelles/nanoparticles. Hence, encapsulation into liposomes seems to be of great advantage to achieve high-loaded nanoformulations.
In terms of cytotoxicity studies under two-dimensional cell culture conditions, only two of the above-mentioned publications with liposomal formulations of tyrosine kinase inhibitors reported viability data as compared to respective free drugs. On the one hand, liposomal gefitinib exerted strongly reduced activity compared to free gefitinib (with a fold change between 15–20) 41 while, on the other hand, liposomal cabozantinib exhibited a much stronger cytotoxicity compared to the free inhibitor (fold change between 0.02 to 0.18) 42. Usually, PEGylated liposomal formulations should reduce the biological activity compared to the free drugs 51–53. As such, so-called “stealth liposomes” hamper endocytic uptake by macrophages 54, 55. Our liposomal formulations exhibited a double-edged behavior on a panel of FGFR-driven lung cancer cell lines. At low concentrations similar activities compared to the free drugs were observed, but at concentrations higher than 1 μM the activity of the liposomal formulations was distinctly lower. To further investigate this effect, the uptake kinetics were investigated by flow cytometry exploiting the intrinsic fluorescence properties of the three FGFR inhibitors (33 and unpublished data). Indeed, quite similar uptake dynamics (at least for nintedanib and PD173074) were observed at 1 μM, while the one of ponatinib was slower in case of the nanoformulations. In contrast, the uptake was strongly reduced compared to the free drugs at 10 μM for all three TKIs in line with the lower cytotoxic activity. Concerning NCI-H1703 cells, the very strong sensitivity of this cell line towards FGFR inhibition may account for the unaltered cytotoxicity profiles of liposomal as compared to free drugs even despite reduced intracellular uptake levels. Importantly, longer drug exposure times (168 hours) led to a comparable activity profile between free and liposomal ponatinib in vitro. This observation may reflect the desired temporary detargeting effect of liposomal drug encapsulation, leading to an initial latency phase (up to 72 hours at higher concentrations), but eventually to an assimilation of anticancer activity compared to the free drug at later time points.
Finally, the liposomal formulation with the highest EE (liposomal ponatinib) was investigated in vivo against the K7M2 osteosarcoma allograft in BALB/c mice. K7M2 cells express high levels of FGFR1 and exhibit exquisite sensitivity against FGFR inhibitors in vitro. Off-target toxicity represents a major obstacle in systemic anticancer therapy 37. In case of molecularly-designed TKIs, this is based on expression of target RTKs also in normal tissues 56. Due to the high conservation of tyrosine kinase domains, TKIs often exhibit non-exclusive specificity profiles. This also applies for ponatinib, which – besides Abl kinase and FGFR - also targets VEGFR and PDGFR family members 57. These receptors are highly expressed in endothelial cells and, as such, are crucial factors in the maintenance of blood vessel integrity. Consequently, ponatinib administration has been observed to pose a considerable risk for vascular toxicity 58, 59. In the clinical situation, commonly observed ponatinib adverse effects include cutaneous toxicity such as non-specific rash or dryness 14. Moreover, several cases of pityriasis rubra pilaris-like dermatitis as well as folliculocentric, ichthyosiform and seborrhoeic eruptions have been documented in patients 60. The skin-associated side-effects observed in our study in mice treated with free ponatinib likely recapitulated these clinical symptoms. The absence of such side effects of liposomal ponatinib demonstrates that a nanoparticulate formulation is highly favorable with respect to its safety profile as compared to the free drug. We assume that this improved toxicity profile is a consequence of a reduced non-specific biodistribution. Proof of concept of this assumption and in-depth analysis of differences in pharmacokinetics such as tissue distribution as well as histopathological effects between free and liposomal ponatinib is pending in a current follow-up study.
This excellent tolerability is especially interesting in the light of the fact that intravenous administration of liposomal ponatinib was predicted to yield much higher plasma concentrations as compared to the orally administered free drug 34. In addition, liposomal drug dose in absolute numbers was ten-fold lower than that of free ponatinib (3mg/kg versus 30mg/kg), yet only this formulation resulted in significant antitumor activity. Hence, the lower amount of L-ponatinib seems to more specifically target the tumor as compared to oral free ponatinib.
In conclusion, in the present study we have demonstrated that stable liposomal formulations of FGFR inhibitors can be generated and -at least in case of ponatinib-significantly improve therapeutic efficacy and decrease side effects in vivo. Consequently, the presented liposomal formulation of ponatinib might represent a possible strategy to reduce the toxicity and increase the antitumor efficacy of ponatinib in the clinical routine.
Supplementary Material
Acknowledgements
We thank Ao. Univ.-Prof. Lars Gille from the University of Veterinary Medicine of Vienna for helpful discussions. We also thank Dr. Irena Herbacek from the Medical University of Vienna for flow cytometry analysis and Gerhard Zeitler for devoted animal care. Electron microscopy measurements were performed at the “Core Facility of Cell Imaging and Ultrastructure Research” of the University of Vienna.
Funding organizations
This work was supported by the Mahlke geb. Obermann-Stiftung of the University of Vienna FA526009 (to B. Keppler) and the Austrian Science Fund (FWF) grant P28853 (to C. Kowol) and P30105 (to W. Berger).
Abbreviations
- ALL
acute lymphoblastic leukemia
- CML
chronic myeloid leukemia
- DLS
dynamic light scattering
- EPR
enhanced permeability and retention
- EE
encapsulation efficiency
- FGFR
fibroblast growth factor receptor
- NSCLC
non-small-cell lung cancer
- PDGFR
platelet-derived growth factor receptor
- PDI
polydispersity index
- PLA
polylactic acid
- TEM
transmission electron microscopy
- VEGFR
vascular endothelial growth factor receptor
Footnotes
Conflicts of interest
The authors declare no conflicts of interest.
References
- 1.Wesche J, Haglund K, Haugsten EM. Fibroblast growth factors and their receptors in cancer. Biochem J. 2011;437:199–213. doi: 10.1042/BJ20101603. [DOI] [PubMed] [Google Scholar]
- 2.Stephens P, Edkins S, Davies H, Greenman C, Cox C, Hunter C, et al. A screen of the complete protein kinase gene family identifies diverse patterns of somatic mutations in human breast cancer. Nat Genet. 2005;37:590–2. doi: 10.1038/ng1571. [DOI] [PubMed] [Google Scholar]
- 3.Wu P, Nielsen TE, Clausen MH. FDA-approved small-molecule kinase inhibitors. Trends Pharmacol Sci. 2015;36:422–439. doi: 10.1016/j.tips.2015.04.005. [DOI] [PubMed] [Google Scholar]
- 4.Huang WS, Metcalf CA, Sundaramoorthi R, Wang Y, Zou D, Thomas RM, et al. Discovery of 3-[2-(imidazo[1,2-b]pyridazin-3-yl)ethynyl]-4-methyl-N-{4-[(4-methylpiperazin-1-y l)methyl]-3-(trifluoromethyl)phenyl}benzamide (AP24534), a potent, orally active pan-inhibitor of breakpoint cluster region-abelson (BCR-ABL) kinase including the T315I gatekeeper mutant. J Med Chem. 2010;53:4701–19. doi: 10.1021/jm100395q. [DOI] [PubMed] [Google Scholar]
- 5.Hoy SM. Ponatinib: a review of its use in adults with chronic myeloid leukaemia or Philadelphia chromosome-positive acute lymphoblastic leukaemia. Drugs. 2014;74:793–806. doi: 10.1007/s40265-014-0216-6. [DOI] [PubMed] [Google Scholar]
- 6.Gainor JF, Chabner BA. Ponatinib: Accelerated Disapproval. Oncologist. 2015;20:847–848. doi: 10.1634/theoncologist.2015-0253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Roth GJ, Binder R, Colbatzky F, Dallinger C, Schlenker-Herceg R, Hilberg F, et al. Nintedanib: from discovery to the clinic. J Med Chem. 2015;58:1053–63. doi: 10.1021/jm501562a. [DOI] [PubMed] [Google Scholar]
- 8.Takeda M, Okamoto I, Nakagawa K. Clinical development of nintedanib for advanced non-small-cell lung cancer. Ther Clin Risk Manag. 2015;11:1701–6. doi: 10.2147/TCRM.S76646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fala L. Ofev (Nintedanib): First Tyrosine Kinase Inhibitor Approved for the Treatment of Patients with Idiopathic Pulmonary Fibrosis. Am Health Drug Benefits. 2015;8:101–104. [PMC free article] [PubMed] [Google Scholar]
- 10.Reck M, Kaiser R, Eschbach C, Stefanic M, Love J, Gatzemeier U, et al. A phase II double-blind study to investigate efficacy and safety of two doses of the triple angiokinase inhibitor BIBF 1120 in patients with relapsed advanced non-small-cell lung cancer. Ann Oncol. 2011;22:1374–81. doi: 10.1093/annonc/mdq618. [DOI] [PubMed] [Google Scholar]
- 11.Okamoto I, Kaneda H, Satoh T, Okamoto W, Miyazaki M, Morinaga R, et al. Phase I safety, pharmacokinetic, and biomarker study of BIBF 1120, an oral triple tyrosine kinase inhibitor in patients with advanced solid tumors. Mol Cancer Ther. 2010;9:2825–33. doi: 10.1158/1535-7163.MCT-10-0379. [DOI] [PubMed] [Google Scholar]
- 12.Lipton JH, Chuah C, Guerci-Bresler A, Rosti G, Simpson D, Assouline S, et al. Ponatinib versus imatinib for newly diagnosed chronic myeloid leukaemia: an international, randomised, open-label, phase 3 trial. Lancet Oncol. 17:612–621. doi: 10.1016/S1470-2045(16)00080-2. [DOI] [PubMed] [Google Scholar]
- 13.Dorer DJ, Knickerbocker RK, Baccarani M, Cortes JE, Hochhaus A, Talpaz M, et al. Impact of dose intensity of ponatinib on selected adverse events: Multivariate analyses from a pooled population of clinical trial patients. Leuk Res. 2016;48:84–91. doi: 10.1016/j.leukres.2016.07.007. [DOI] [PubMed] [Google Scholar]
- 14.Cortes JE, Kim DW, Pinilla-Ibarz J, le Coutre P, Paquette R, Chuah C, et al. A phase 2 trial of ponatinib in Philadelphia chromosome-positive leukemias. N Engl J Med. 2013;369:1783–96. doi: 10.1056/NEJMoa1306494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Price KE, Saleem N, Lee G, Steinberg M. Potential of ponatinib to treat chronic myeloid leukemia and acute lymphoblastic leukemia. Onco Targets Ther. 2013;6:1111–1118. doi: 10.2147/OTT.S36980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Portney NG, Ozkan M. Nano-oncology: drug delivery, imaging, and sensing. Anal Bioanal Chem. 2006;384:620–30. doi: 10.1007/s00216-005-0247-7. [DOI] [PubMed] [Google Scholar]
- 17.Maeda H, Nakamura H, Fang J. The EPR effect for macromolecular drug delivery to solid tumors: Improvement of tumor uptake, lowering of systemic toxicity, and distinct tumor imaging in vivo. Adv Drug Deliv Rev. 2013;65:71–9. doi: 10.1016/j.addr.2012.10.002. [DOI] [PubMed] [Google Scholar]
- 18.Mohammadi M, Froum S, Hamby JM, Schroeder MC, Panek RL, Lu GH, et al. Crystal structure of an angiogenesis inhibitor bound to the FGF receptor tyrosine kinase domain. Embo j. 1998;17:5896–904. doi: 10.1093/emboj/17.20.5896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nguyen PT, Tsunematsu T, Yanagisawa S, Kudo Y, Miyauchi M, Kamata N, et al. The FGFR1 inhibitor PD173074 induces mesenchymal-epithelial transition through the transcription factor AP-1. Br J Cancer. 2013;109:2248–2258. doi: 10.1038/bjc.2013.550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mishra B, Patel BB, Tiwari S. Colloidal nanocarriers: a review on formulation technology, types and applications toward targeted drug delivery. Nanomedicine. 2010;6:9–24. doi: 10.1016/j.nano.2009.04.008. [DOI] [PubMed] [Google Scholar]
- 21.Sharma A, Sharma US. Liposomes in drug delivery: Progress and limitations. Int J Pharm. 1997;154:123–140. [Google Scholar]
- 22.Barenholz Y. Doxil(R)--the first FDA-approved nano-drug: lessons learned. J Control Release. 2012;160:117–34. doi: 10.1016/j.jconrel.2012.03.020. [DOI] [PubMed] [Google Scholar]
- 23.Fessi H, Puisieux F, Devissaguet JP, Ammoury N, Benita S. Nanocapsule formation by interfacial polymer deposition following solvent displacement. Int J Pharm. 1989;55:R1–R4. [Google Scholar]
- 24.Bangham AD, De Gier J, Greville GD. Osmotic properties and water permeability of phospholipid liquid crystals. Chem Phys Lipids. 1967;1:225–246. [Google Scholar]
- 25.(ACD/Labs) Software V11.02. Advanced Chemistry Development, Inc; 2017. [Google Scholar]
- 26.Barenholz Y. Liposome application: problems and prospects. Curr Opin Colloid Interface Sci. 2001;6:66–77. [Google Scholar]
- 27.Zucker D, Marcus D, Barenholz Y, Goldblum A. Liposome drugs’ loading efficiency: A working model based on loading conditions and drug’s physicochemical properties. J Control Release. 2009;139:73–80. doi: 10.1016/j.jconrel.2009.05.036. [DOI] [PubMed] [Google Scholar]
- 28.Nakamura K, Yamashita K, Itoh Y, Yoshino K, Nozawa S, Kasukawa H. Comparative studies of polyethylene glycol-modified liposomes prepared using different PEG-modification methods. Biochimica et Biophysica Acta (BBA)-Biomembranes. 2012;1818:2801–2807. doi: 10.1016/j.bbamem.2012.06.019. [DOI] [PubMed] [Google Scholar]
- 29.Garbuzenko O, Zalipsky S, Qazen M, Barenholz Y. Electrostatics of PEGylated micelles and liposomes containing charged and neutral lipopolymers. Langmuir. 2005;21:2560–2568. doi: 10.1021/la0479105. [DOI] [PubMed] [Google Scholar]
- 30.Morton SW, Lee MJ, Deng ZJ, Dreaden EC, Siouve E, Shopsowitz KE, et al. A nanoparticle-based combination chemotherapy delivery system for enhanced tumor killing by dynamic rewiring of signaling pathways. Sci Signal. 2014;7:ra44. doi: 10.1126/scisignal.2005261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weiss J, Sos ML, Seidel D, Peifer M, Zander T, Heuckmann JM, et al. Frequent and focal FGFR1 amplification associates with therapeutically tractable FGFR1 dependency in squamous cell lung cancer. Sci Transl Med. 2010;2:62.:ra93. doi: 10.1126/scitranslmed.3001451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Englinger B, Lotsch D, Pirker C, Mohr T, van Schoonhoven S, Boidol B, et al. Acquired nintedanib resistance in FGFR1-driven small cell lung cancer: role of endothelin-A receptor-activated ABCB1 expression. Oncotarget. 2016;7:50161–50179. doi: 10.18632/oncotarget.10324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Englinger B, Kallus S, Senkiv J, Heilos D, Gabler L, van Schoonhoven S, et al. Intrinsic fluorescence of the clinically approved multikinase inhibitor nintedanib reveals lysosomal sequestration as resistance mechanism in FGFR-driven lung cancer. J Exp Clin Cancer Res. 2017;36:122. doi: 10.1186/s13046-017-0592-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gozgit JM, Wong MJ, Moran L, Wardwell S, Mohemmad QK, Narasimhan NI, et al. Ponatinib (AP24534), a multitargeted pan-FGFR inhibitor with activity in multiple FGFR-amplified or mutated cancer models. Mol Cancer Ther. 2012;11:690–9. doi: 10.1158/1535-7163.MCT-11-0450. [DOI] [PubMed] [Google Scholar]
- 35.Wicki A, Witzigmann D, Balasubramanian V, Huwyler J. Nanomedicine in cancer therapy: Challenges, opportunities, and clinical applications. J Control Release. 2015;200:138–157. doi: 10.1016/j.jconrel.2014.12.030. [DOI] [PubMed] [Google Scholar]
- 36.Anselmo AC, Mitragotri S. Nanoparticles in the clinic. Bioeng Transl Med. 2016;1:10–29. doi: 10.1002/btm2.10003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shi J, Kantoff PW, Wooster R, Farokhzad OC. Cancer nanomedicine: progress, challenges and opportunities. Nat Rev Cancer. 2017;17:20–37. doi: 10.1038/nrc.2016.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu P, Nielsen TE, Clausen MH. Small-molecule kinase inhibitors: an analysis of FDA-approved drugs. Drug Discov Today. 2016;21:5–10. doi: 10.1016/j.drudis.2015.07.008. [DOI] [PubMed] [Google Scholar]
- 39.Maitani Y, Saito H, Seishi Y, Iwase Y, Yamauchi T, Higashiyama K, et al. A combination of liposomal sunitinib plus liposomal irinotecan and liposome co-loaded with two drugs enhanced antitumor activity in PC12-bearing mouse. J Drug Target. 2012;20:873–82. doi: 10.3109/1061186X.2012.723215. [DOI] [PubMed] [Google Scholar]
- 40.Fan Y, Du W, He B, Fu F, Yuan L, Wu H, et al. The reduction of tumor interstitial fluid pressure by liposomal imatinib and its effect on combination therapy with liposomal doxorubicin. Biomaterials. 2013;34:2277–88. doi: 10.1016/j.biomaterials.2012.12.012. [DOI] [PubMed] [Google Scholar]
- 41.Zhou X, Yung B, Huang Y, Li H, Hu X, Xiang G, et al. Novel liposomal gefitinib (L-GEF) formulations. Anticancer Res. 2012;32:2919–23. [PubMed] [Google Scholar]
- 42.Kulkarni AA, Vijaykumar VE, Natarajan SK, Sengupta S, Sabbisetti VS. Sustained inhibition of cMET-VEGFR2 signaling using liposome-mediated delivery increases efficacy and reduces toxicity in kidney cancer. Nanomedicine. 2016;12:1853–1861. doi: 10.1016/j.nano.2016.04.002. [DOI] [PubMed] [Google Scholar]
- 43.Mukthavaram R, Jiang P, Saklecha R, Simberg D, Bharati IS, Nomura N, et al. High-efficiency liposomal encapsulation of a tyrosine kinase inhibitor leads to improved in vivo toxicity and tumor response profile. Int J Nanomedicine. 2013;8:3991–4006. doi: 10.2147/IJN.S51949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Song Z, Lin Y, Zhang X, Feng C, Lu Y, Gao Y, et al. Cyclic RGD peptide-modified liposomal drug delivery system for targeted oral apatinib administration: enhanced cellular uptake and improved therapeutic effects. Int J Nanomedicine. 2017;12:1941–1958. doi: 10.2147/IJN.S125573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ravar F, Saadat E, Kelishadi PD, Dorkoosh FA. Liposomal formulation for co-delivery of paclitaxel and lapatinib, preparation, characterization and optimization. J Liposome Res. 2016;26:175–87. doi: 10.3109/08982104.2015.1070174. [DOI] [PubMed] [Google Scholar]
- 46.Cai L, Wang X, Wang W, Qiu N, Wen J, Duan X, et al. Peptide ligand and PEG-mediated long-circulating liposome targeted to FGFR overexpressing tumor in vivo. Int J Nanomedicine. 2012;7:4499–510. doi: 10.2147/IJN.S32817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xiao W, Chen X, Yang L, Mao Y, Wei Y, Chen L. Co-delivery of doxorubicin and plasmid by a novel FGFR-mediated cationic liposome. Int J Pharm. 2010;393:119–26. doi: 10.1016/j.ijpharm.2010.04.018. [DOI] [PubMed] [Google Scholar]
- 48.Chen X, Wang X, Wang Y, Yang L, Hu J, Xiao W, et al. Improved tumor-targeting drug delivery and therapeutic efficacy by cationic liposome modified with truncated bFGF peptide. J Control Release. 2010;145:17–25. doi: 10.1016/j.jconrel.2010.03.007. [DOI] [PubMed] [Google Scholar]
- 49.Terada T, Mizobata M, Kawakami S, Yamashita F, Hashida M. Optimization of tumor-selective targeting by basic fibroblast growth factor-binding peptide grafted PEGylated liposomes. J Control Release. 2007;119:262–70. doi: 10.1016/j.jconrel.2007.01.018. [DOI] [PubMed] [Google Scholar]
- 50.Mu C-F, Xiong Y, Bai X, Sheng Y-J, Cui J. Codelivery of Ponatinib and SAR302503 by Active Bone-Targeted Polymeric Micelles for the Treatment of Therapy-Resistant Chronic Myeloid Leukemia. Mol Pharm. 2017;14:274–283. doi: 10.1021/acs.molpharmaceut.6b00872. [DOI] [PubMed] [Google Scholar]
- 51.Gabizon AA, Tzemach D, Horowitz AT, Shmeeda H, Yeh J, Zalipsky S. Reduced toxicity and superior therapeutic activity of a mitomycin C lipid-based prodrug incorporated in pegylated liposomes. Clin Cancer Res. 2006;12:1913–20. doi: 10.1158/1078-0432.CCR-05-1547. [DOI] [PubMed] [Google Scholar]
- 52.Horowitz AT, Barenholz Y, Gabizon AA. In vitro cytotoxicity of liposome-encapsulated doxorubicin: dependence on liposome composition and drug release. Biochim Biophys Acta. 1992;1109:203–209. doi: 10.1016/0005-2736(92)90084-y. [DOI] [PubMed] [Google Scholar]
- 53.Song H, Zhang J, Han Z, Zhang X, Li Z, Zhang L, et al. Pharmacokinetic and cytotoxic studies of pegylated liposomal daunorubicin. Cancer Chemother Pharmacol. 2006;57:591–8. doi: 10.1007/s00280-005-0076-6. [DOI] [PubMed] [Google Scholar]
- 54.Immordino ML, Dosio F, Cattel L. Stealth liposomes: review of the basic science, rationale, and clinical applications, existing and potential. Int J Nanomedicine. 2006;1:297–315. [PMC free article] [PubMed] [Google Scholar]
- 55.Hioki A, Wakasugi A, Kawano K, Hattori Y, Maitani Y. Development of an in vitro drug release assay of PEGylated liposome using bovine serum albumin and high temperature. Biol Pharm Bull. 2010;33:1466–70. doi: 10.1248/bpb.33.1466. [DOI] [PubMed] [Google Scholar]
- 56.Broekman F, Giovannetti E, Peters GJ. Tyrosine kinase inhibitors: Multi-targeted or single-targeted? World J Clin Oncol. 2011;2:80–93. doi: 10.5306/wjco.v2.i2.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gentile C, Martorana A, Lauria A, Bonsignore R. Kinase Inhibitors in Multitargeted Cancer Therapy. Curr Med Chem. 2017;24:1671–1686. doi: 10.2174/0929867324666170112112734. [DOI] [PubMed] [Google Scholar]
- 58.Damrongwatanasuk R, Fradley MG. Cardiovascular Complications of Targeted Therapies for Chronic Myeloid Leukemia. Curr Treat Options Cardiovasc Med. 2017;19:24. doi: 10.1007/s11936-017-0524-8. [DOI] [PubMed] [Google Scholar]
- 59.Yilmaz M, Jabbour E. Tyrosine Kinase Inhibitors Early in the Disease Course: Lessons From Chronic Myelogenous Leukemia. Semin Oncol. 2015;42:876–86. doi: 10.1053/j.seminoncol.2015.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Alloo A, Sheu J, Butrynski JE, DeAngelo DJ, George S, Murphy GF, et al. Ponatinib-induced pityriasiform, folliculocentric and ichthyosiform cutaneous toxicities. Br J Dermatol. 2015;173:574–7. doi: 10.1111/bjd.13692. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.