Abstract
Membrane proteins perform a variety of functions, all crucially dependent on their orientation in the membrane. However, neither the exact number of transmembrane domains (TMDs) nor the topology of most proteins have been experimentally determined. Due to this most scientists rely primarily on prediction algorithms to determine topology and TMD assignments. Since these can give contradictory results, single-algorithm based predictions are unreliable. To map the extent of potential misanalysis we compared the predictions of nine algorithms on the yeast proteome, and find that they have little agreement when predicting TMD number and termini orientation. To view all predictions in parallel we created a webpage called TopologYeast: http://www.weizmann.ac.il/molgen/TopologYeast. Comparing each algorithm with experimental data, we also find poor agreement. Our analysis suggests that more systematic data on protein topology is required to increase the training sets for prediction algorithms and to have accurate knowledge of membrane protein topology.
Keywords: Transmembrane domains, Prediction algorithms, Yeast, membrane proteins, termini orientation
1. Introduction
It has been estimated in the literature that nearly 30% of all proteins in eukaryotic cells span membranes [1]. These proteins are essential for diverse functions such as transfer of molecules across membranes, signal transduction, organelle tethering and fusion, and a myriad of enzymatic activities. The structure, function and localization of such proteins is heavily affected by the number and location of their transmembrane domains (TMDs), as well as the orientation of each of their residues that together comprise the protein topology in the membrane. Hence, to better understand the function of any transmembrane protein it is essential to understand its topology.
2. Data Analysis
2.1. Experimental approaches to study TMD number and topology using systematic tools are essential for providing a whole-proteome view
A variety of biochemical techniques such as protease protection and cysteine scanning assays have been used for decades to map TMD number and topology in single proteins [2,3]. However, in recent years several experimental approaches have been published that enable the study of TMD number and protein membrane topology in a systematic manner on tens to hundreds of proteins, or even in entire proteomes. These have been extensively reviewed [2] but just a few examples include:
2.1.1. High sensitivity mass spectrometry (MS) coupled with protease protection assays
In short, peptides that are facing the cytosol will be sensitive to protease treatment and hence will be lost from MS analysis. This method was recently used to assess yeast mitochondrial protein membrane topology [4].
2.1.2. Genetic reporter tags to follow the termini of membrane proteins in living cells
For example, a Glycosylatable GFP (gGFP) tag that loses fluorescence when glycosylated in the lumen of the endoplasmic reticulum (ER) is a very powerful tool that has been developed for tracking orientation of a terminus for secretory proteins [5]. Another example for the use of reporter tags is the Suc2/His4C chimeric protein. The tag enables the following of glycosylation status (suggesting luminal orientation in the secretory pathway) since both the His4C and Suc2 peptides have N-linked glycosylation sites. Additionally, the His4C domain encodes for a histidinol dehydrogenase activity that converts histidinol to histidine, but only if located in the cytosol. This method was already successfully used to map the carboxy terminus (C’) orientation of hundreds of secretory proteins [6]. A third tagging approach was recently created based on a split Venus system [7]. A library was created with all yeast proteins containing one half of the split-Venus reporter on their amino terminus (N’). To determine termini orientation this library was mated with a strain expressing the other half of the Venus in the cytosol, and their ability to interact was assayed by complementation of the full Venus fluorophore and emission of fluorescence, suggesting that the N’ is facing the cytosol [8]
While more methods are starting to become available, such systematic analyses are usually only performed on one terminus of a protein, on subsets of proteins rather than on entire proteomes and often only in genetically pliable model organisms, such as yeast or bacteria, and not in other experimental systems. Hence we are still missing robust experimental data on topology at a proteome wide level for any organism.
2.2. Prediction algorithms based on different approaches and training sets show little overlap in their output on the presence and number of TMDs
While systematic experimental data on protein topology is slowly becoming available it is still not present for most organisms. In cases where experimental data is not available, prediction programs are widely used to estimate how many TMDs a protein has (TMD prediction), and the direction in which its N’ and C’ face (termini orientation). Over the years, many different prediction algorithms have been created, each following a different approach and using varying training datasets.
To assay how robust the different TMD prediction algorithms are, we used the protein coding genome of the yeast Saccharomyces cerevisiae (from here on called yeast) as a test case. The yeast proteome contains 5800 proteins (excluding dubious genes and pseudogenes) and has the most experimental data on protein topology and hence is a good model for such analyses. We compiled the predictions on the presence and number of TMDs predicted for all yeast proteins using all TMD prediction algorithms that could be used in batch running for the entire proteome and that gave topology predictions: HMMTOP [9], TMHMM [1], Topcons [10], Octopus [11], Philius [12], Polyphobius [13], Scampi [14], Spoctopus [15] and MEMSAT-SVM [16]. All nine algorithms are widely used and rely on different methods (such as Hidden Markov Models, Artificial Neural Networks, Support Vector Machines, Bayesian Networks or combinations of methods) (See Table 1 for more details) and are trained on several protein properties, such as chemical traits, distributions or predicted structures of amino acids (aa). Topcons combines the results from several algorithms (Octopus, Philius, PolyPhobius, Scampi and Spoctopus).
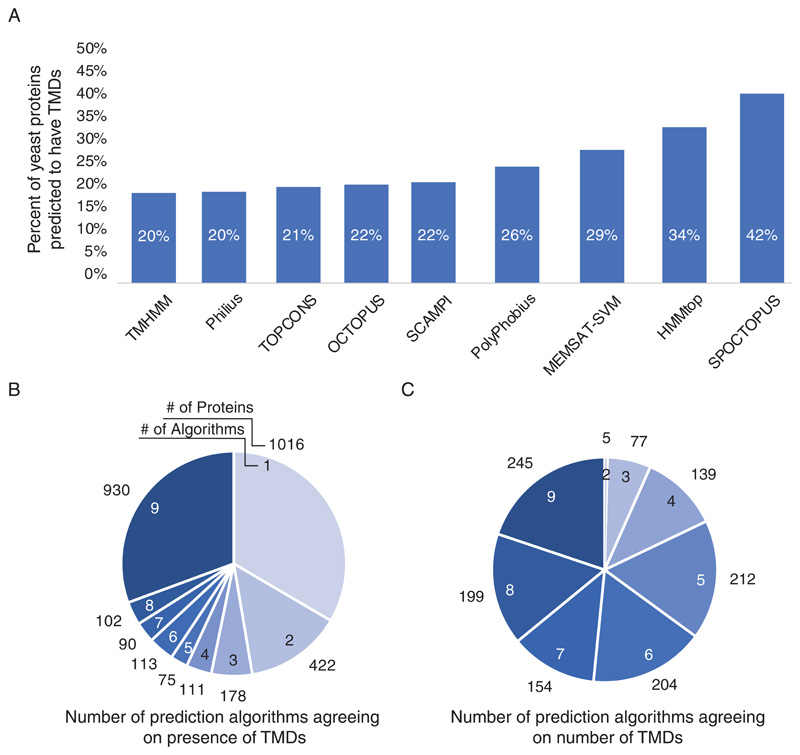
When viewing the results from the nine algorithms, it was immediately apparent that even in the simple question of whether a protein is membrane spanning or not, each of the prediction programs provides very different answers. The results of the nine algorithms varied between 20-42% of all proteins in the proteome being membrane spanning (Supplementary Table 1) (Figure 1A). Specifically, different algorithms gave conflicting assignments on whether even one TMD exists for over 2107 proteins (Supplementary Table 1). Only 2761 were consistently predicted to be soluble proteins and only 930 were consistently predicted by all 9 programs to have at least one TMD, although in many cases each program predicted a different number of TMDs (Figure 1B). When looking at the number of TMDs predicted for each protein we found that for only 245 proteins (~4.2% of the proteome) did all 9 programs agree (Figure 1C). This observation raises the concern that utilizing a single prediction algorithm most likely has a very low probability of really predicting whether a protein spans the membrane at all and if so, the number of times it does so.
Figure 1. Comparison of the predictions from various algorithms on transmembrane domain presence and number.
(A) Bar graph showing the percent of proteins in the yeast genome predicted to be membrane spanning according to each of the prediction algorithms. (B) Pie chart showing the relative portion of prediction algorithms agreeing on presence of TMDs. Numbers inside pie chart represent the number of prediction programs that are in agreement. The numbers outside the pie chart represent the number of proteins falling into each category N=3037 (C) Pie chart showing the relative portion of prediction algorithms agreeing on the number of TMDs only in those proteins that were predicted to have more than one TMD by at least five programs. Numbers inside pie chart represent the number of prediction programs that are in agreement. The numbers outside the pie chart represent the number of proteins falling into each category N=1235
2.3. A new list of predicted tail-anchored proteins based on all TMD prediction algorithms pooled together suggests new proteins in this family
The location of TMDs across the aa sequence of a protein influences its structure and function. Often very extreme TMDs at either the N’ or C’ are harder to accurately predict. For example, a TMD at the very N’ may be confused with a highly hydrophobic signal peptide (SP) that does not form part of the mature protein but rather is only used to direct the protein into the secretory pathway and later cleaved off.
A single TMD at the C’ of a protein is the hallmark of tail-anchored (TA) proteins that are anchored to all intra-cellular membranes facing the cytosol thus enabling their cytosolic domains to carry out crucial functions. Due to their unique topology, distinct targeting and translocation pathways have evolved to cater for them. [17]
Despite intense research on TA proteins, the entire repertoire of yeast TA proteins has not been fully identified or verified. This may be due to the fact that any predictions to date only included a subset of prediction algorithms. Hence, we set out to predict all yeast TA proteins using the 9 algorithms mentioned above. We defined a TA protein as one that does not have any N’ targeting motif (SP or Mitochondrial Targeting Sequence (MTS)) [8] and harbours a single TMD at the C’ (no further than 80 aa from the last residue). Based on these criteria and agreement between at least 6 prediction algorithms (67% agreement), we predicted 78 proteins that could be defined as TA proteins with high confidence (Supplementary Table 2). Out of these 31 have previously been assigned as TA proteins [18,19,28–35,20–27] and 47 are newly predicted TA proteins (19 additional TA proteins were missed by our threshold and would have been captured had we lowered the threshold of high confidence). 198 additional proteins were predicted to have a TA by at least one prediction method. Hence many additional TA proteins may exist and this remains to be experimentally verified.
More generally our analysis shows that using several prediction algorithms in parallel raises the chance of capturing an entire topological family. 29 of the newly identified TA proteins are uncharacterized proteins with no known function. Since TA proteins perform important regulatory functions in cells it may give a clue to the functions of these new members of this topological family.
2.4. Termini orientation predictions show little robustness
Uncovering the membrane topology of an integral membrane protein is an essential step in determining its structural and functional properties. While assigning TMD presence and number is already fairly inaccurate (as discussed above), an additional complicated task is to define termini orientation, i.e. whether the termini are facing the cytosol (in) or the lumen of an organelle/ the extracellular space (out). To date, most assays for determining termini orientation at the single protein level rely on laborious biochemical techniques and not many systematic assays have been performed in organisms other than yeast. As a result, prediction algorithms are heavily utilized by protein researchers. As in the case of TMD prediction, termini orientation results can vary between one prediction algorithm to the other. Moreover, for some proteins matters are further complicated as their termini can reside at either side of the membrane while still supporting function [36].
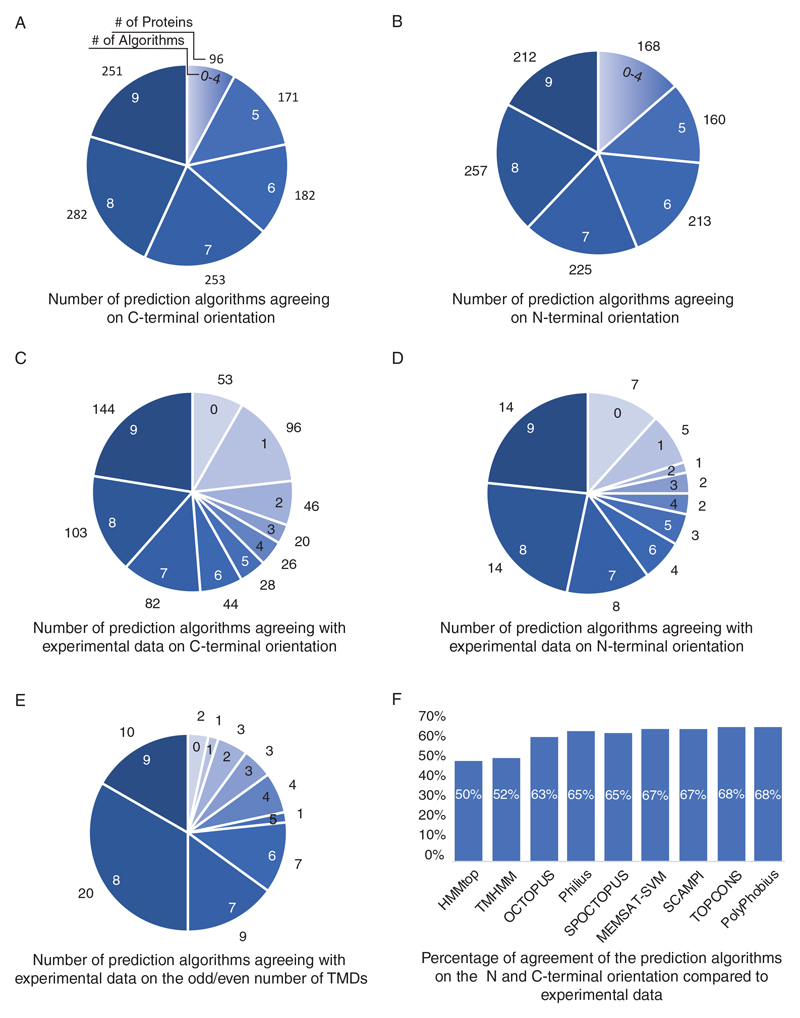
Indeed, looking at how our various prediction programs define an “in” orientation, we find very little agreement in their prediction for either N’ or C’ (Figure 2A, B). How can we know which one of the predictions is right? Or which prediction algorithm should be trusted in most instances? Studies for determining C’ orientation have, to date, been only performed on subsets of proteins and hence a simple systematic comparison was not possible. To collect as many datapoints to our comparisons we compiled the abovementioned data from the Suc2/His4C chimeric protein assays [6]. To these datapoints we added existing data, not originally intended for termini orientation assignments. Specifically, we added protein-protein interaction data from large datasets from three types of protein complementation assays (PCAs) performed with C’ tagging. In these assays one half of a reporter protein is attached to the C’ of a query protein and the other half of the reporter protein to the C’ of another. If one protein is cytosolic and one is spanning a membrane than the only way that the two tagged termini can interact and form the fully complemented assay protein (giving rise to a measurable phenotype) is if the tagged terminus is facing the cytosol. Hence this data can be used as a proxy for termini orientation even though the assay was not built for this matter but rather to assay protein-protein interactions.
Figure 2. Comparison of the predictions from various algorithms on termini orientation and protein topology.
(A) The number of prediction algorithms agreeing on C-terminal orientation. Numbers inside pie chart represent the number of prediction programs that are in agreement. The numbers outside the pie chart represent the number of proteins falling into each category N=1235 (B) The number of prediction algorithms agreeing on N-terminal orientation. Numbers inside pie chart represent the number of prediction programs that are in agreement. The numbers outside the pie chart represent the number of proteins falling into each category N=1235 (C) Pie chart showing the number of prediction algorithms agreeing with experimental data on C-terminal orientation. Numbers inside pie chart represent the number of prediction programs that are in agreement. The numbers outside the pie chart represent the number of proteins falling into each category N=642 (D) Pie chart showing the number of prediction algorithms agreeing with experimental data on N-terminal orientation. Numbers inside pie chart represent the number of prediction programs that are in agreement. The numbers outside the pie chart represent the number of proteins falling into each category N=60 (E) Pie chart showing the relative number of prediction algorithms that agree with experimental data on whether TMD number is even or odd. Numbers inside pie chart represent the number of prediction programs that are in agreement. The numbers outside the pie chart represent the number of proteins falling into each category N=60 (F) Bar graph showing the percentage of agreement with complete topology (termini orientation and odd/even number of TMDs) between the various prediction and the experimental data N=60
The DHFR PCA reporter confers resistance to the cytostatic drug methotrexate [37]. We found 3 studies that carried out such analysis and from them deduced C’ orientation for 430 yeast proteins [37–39]. A second PCA approach is based on the split-ubiquitin system [40]. In this approach the protein interaction enables cleavage of a ubiquitin releasing a transcription factor activating transcription of the HIS3 reporter gene and enabling growth in medium lacking histidine. We compiled the data from ten different studies that utilized this system and deduced C’ orientation for an additional 210 proteins [40–49]. Finally, we compiled the data from three different studies that utilized the split Venus approach on C’ tagged proteins and compiled information on an additional 112 proteins [50–52]. Most proteins were only represented in one of these datasets thus there is minimal overlap. In total, we compiled the topology assignments from 16 PCA experiments performed on C’ tags, spanning a total of 15,843 independent interaction data points (Supplementary Table 3). Comparing the experimental data from the C’ topology and PCA experiments with the topology predictors enabled us to ask which ones correctly assigned termini orientation (Figure 2C).
Since N’ orientation was systematically assayed for yeast TMD proteins we compared the experimental data [8,53] to the predictions (Figure 2D).
For both termini it seems like nearly half of all proteins did not show agreement with most algorithms (Figure 2C,D). This makes it difficult to deteremine a specific algorithm that outperforms the rest.
Finally, to assess which prediction algorithm is the most accurate at the level of whole protein topology (orientation of termini and number of TMDs) we compared the outputs of all prediction algorithms to the experimental data on protein topology discussed above. We coupled the C’ and N’ orientation experimental data to determine whether the number of TMDs should be odd or even (if both termini are facing the same direction TMD number should be even) (Figure 2E) as well as whole protein topology (Figure 2F). We then compared this to the various algorithms focusing on 1014 yeast proteins predicted to have a TMD by at least 5 algorithms. In general, most TMD prediction programs had a similar, non-satisfactory chance of compatibility with the experimental data, though each one with a different roster of proteins (Figure 2E, F) (Supplementary Table 2).
Taking the N’ and C’ termini prediction comparisons together we can suggest that either all algorithms do not predict termini orientation well or that the experimental/computational analyses that were done to date to define terminus orientation are not satisfactory. Regardless, creating more systematic datasets on experimental evidence is rapidly needed to assess the superiority of any of the algorithms, or to increase the learning sets of prediction algorithms enabling them to increase their accuracy and sensitivity of prediction.
3. Conclusions and outlook
Our study of 9 widely utilized prediction algorithms suggests that, at present, prediction programs still fall short of providing a strong platform for studying protein TMD number and topology. Additionally, our analyses suggest that in cases where experimental proof does not exist, a hybrid computational and experimental prediction might be the best approach for prediction of TMD number and topology.
Due to the high diversity and disparity between programs we have decided to create an easy platform to simplify the visualisation of the available data (predictions as well as experimental) utilized in this manuscript. To this end, we have integrated these data into a database, called TopologYeast, available for viewing at http://www.weizmann.ac.il/molgen/TopologYeast Our findings make a strong argument for the importance of further gathering of topology information using systematic approaches for entire genomes. High-throughput approaches and the resulting large data sets are increasingly becoming available, but currently only a small portion of the information that they contain is utilized to understand and explore complicated biological phenomena. We believe that many questions, such as termini orientation and TMD number, can be calculated from some of these data. More generally, any additional information will be of wide use to the community of scientists working to understand protein functions.
Supplementary Material
Acknowledgements
We would like to thank Ines Castro, Emma Fenech and Nir Fluman for critical reading of the manuscript. We would like to thank Dr. Jaime Prilusky for custom scripting. The Schuldiner lab is supported through a Peroxisystem ERC CoG (646604), a Volkswagen foundation “Life” grant and an SFB 1190 “Gates and contact sites” grant. MS is an incumbent of the Dr. Gilbert Omenn and Martha Darling Professorial Chair in Molecular Genetics.
Footnotes
Conflict of interest:
The authors declare no conflict of interest.
References
- [1].Krogh A, Larsson B, von Heijne G, Sonnhammer EL. J Mol Biol. 2001;305:567. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- [2].Lee H, Kim H. Biochem Biophys Res Commun. 2014;453:268. doi: 10.1016/j.bbrc.2014.05.111. [DOI] [PubMed] [Google Scholar]
- [3].Bogdanov M, Zhang W, Xie J, Dowhan W. Methods. 2005;36:148. doi: 10.1016/j.ymeth.2004.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Morgenstern M, Stiller SB, Lübbert P, Peikert CD, Dannenmaier S, Drepper F, Weill U, Höß P, Feuerstein R, Gebert M, Bohnert M, et al. Cell Rep. 2017;19:2836. doi: 10.1016/j.celrep.2017.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Lee H, Min J, von Heijne G, Kim H. Biochem Biophys Res Commun. 2012;427:780. doi: 10.1016/j.bbrc.2012.09.138. [DOI] [PubMed] [Google Scholar]
- [6].Kim H, Melén K, Osterberg M, von Heijne G. Proc Natl Acad Sci U S A. 2006;103:11142. doi: 10.1073/pnas.0604075103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Jin L, Baker B, Mealer R, Cohen L, Pieribone V, Pralle A, Hughes T. J Neurosci Methods. 2011;199:1. doi: 10.1016/j.jneumeth.2011.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Weill U, Yofe I, Sass E, Stynen B, Davidi D, Natarajan J, Ben-Menachem R, Avihou Z, Goldman O, Harpaz N, Chuartzman S, et al. Nat Methods. 2018;15:617. doi: 10.1038/s41592-018-0044-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Tusnady GE, Simon I. Bioinformatics. 2001;17:849. doi: 10.1093/bioinformatics/17.9.849. [DOI] [PubMed] [Google Scholar]
- [10].Bernsel A, Viklund H, Hennerdal A, Elofsson A. Nucleic Acids Res. 2009;37:W465. doi: 10.1093/nar/gkp363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Viklund H, Elofsson A. Bioinformatics. 2008;24:1662. doi: 10.1093/bioinformatics/btn221. [DOI] [PubMed] [Google Scholar]
- [12].Reynolds SM, Käll L, Riffle ME, Bilmes JA, Noble WS. PLoS Comput Biol. 2008;4:e1000213. doi: 10.1371/journal.pcbi.1000213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Kall L, Krogh A, Sonnhammer ELL. Bioinformatics. 2005;21:i251. doi: 10.1093/bioinformatics/bti1014. [DOI] [PubMed] [Google Scholar]
- [14].Bernsel A, Viklund H, Falk J, Lindahl E, von Heijne G, Elofsson A. Proc Natl Acad Sci U S A. 2008;105:7177. doi: 10.1073/pnas.0711151105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Viklund H, Bernsel A, Skwark M, Elofsson A. Bioinformatics. 2008;24:2928. doi: 10.1093/bioinformatics/btn550. [DOI] [PubMed] [Google Scholar]
- [16].Nugent T, Jones DT. BMC Bioinformatics. 2009;10:159. doi: 10.1186/1471-2105-10-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Aviram N, Schuldiner M. J Cell Sci. 2017;130:4079. doi: 10.1242/jcs.204396. [DOI] [PubMed] [Google Scholar]
- [18].Albright CF, Orlean P, Robbins PW. Proc Natl Acad Sci U S A. 1989;86:7366. doi: 10.1073/pnas.86.19.7366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Lewis MJ, Pelham HR. Cell. 1996;85:205. doi: 10.1016/s0092-8674(00)81097-1. [DOI] [PubMed] [Google Scholar]
- [20].Becherer KA, Rieder SE, Emr SD, Jones EW. Mol Biol Cell. 1996;7:579. doi: 10.1091/mbc.7.4.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Elgersma Y, Kwast L, van den Berg M, Snyder WB, Distel B, Subramani S, Tabak HF. EMBO J. 1997;16:7326. doi: 10.1093/emboj/16.24.7326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Spang A, Courtney I, Grein K, Matzner M, Schiebel E. J Cell Biol. 1995;128:863. doi: 10.1083/jcb.128.5.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Ungermann C, von Mollard GF, Jensen ON, Margolis N, Stevens TH, Wickner W. J Cell Biol. 1999;145:1435. doi: 10.1083/jcb.145.7.1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Youker RT, Walsh P, Beilharz T, Lithgow T, Brodsky JL. Mol Biol Cell. 2004;15:4787. doi: 10.1091/mbc.E04-07-0584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Ballensiefen W, Ossipov D, Schmitt HD. J Cell Sci. 1998;111(Pt 1):1507. doi: 10.1242/jcs.111.11.1507. [DOI] [PubMed] [Google Scholar]
- [26].Nichols BJ, Ungermann C, Pelham HR, Wickner WT, Haas A. Nature. 1997;387:199. doi: 10.1038/387199a0. [DOI] [PubMed] [Google Scholar]
- [27].Banfield DK, Lewis MJ, Pelham HR. Nature. 1995;375:806. doi: 10.1038/375806a0. [DOI] [PubMed] [Google Scholar]
- [28].Hay JC, Scheller RH. Curr Opin Cell Biol. 1997;9:505. doi: 10.1016/s0955-0674(97)80026-9. [DOI] [PubMed] [Google Scholar]
- [29].Burri L, Varlamov O, Doege CA, Hofmann K, Beilharz T, Rothman JE, Söllner TH, Lithgow T. Proc Natl Acad Sci U S A. 2003;100:9873. doi: 10.1073/pnas.1734000100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Kagiwada S, Hosaka K, Murata M, Nikawa J, Takatsuki A. J Bacteriol. 1998;180:1700. doi: 10.1128/jb.180.7.1700-1708.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Sommer T, Jentsch S. Nature. 1993;365:176. doi: 10.1038/365176a0. [DOI] [PubMed] [Google Scholar]
- [32].Esnault Y, Feldheim D, Blondel MO, Schekman R, Képès F. J Biol Chem. 1994;269:27478. [PubMed] [Google Scholar]
- [33].Holthuis JCM. EMBO J. 1998;17:113. doi: 10.1093/emboj/17.1.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Rossi G, Salminen A, Rice LM, Brünger AT, Brennwald P. J Biol Chem. 1997;272:16610. doi: 10.1074/jbc.272.26.16610. [DOI] [PubMed] [Google Scholar]
- [35].Beilharz T, Egan B, Silver PA, Hofmann K, Lithgow T. J Biol Chem. 2003;278:8219. doi: 10.1074/jbc.M212725200. [DOI] [PubMed] [Google Scholar]
- [36].Nasie I, Steiner-Mordoch S, Gold A, Schuldiner S. J Biol Chem. 2010;285:15234. doi: 10.1074/jbc.M110.108746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Tarassov K, Messier V, Landry CR, Radinovic S, Serna Molina MM, Shames I, Malitskaya Y, Vogel J, Bussey H, Michnick SW, Molina MM, et al. Science. 2008;320:1465. doi: 10.1126/science.1153878. [DOI] [PubMed] [Google Scholar]
- [38].Schlecht U, Miranda M, Suresh S, Davis RW, St Onge RP. Proc Natl Acad Sci U S A. 2012;109:9213. doi: 10.1073/pnas.1204952109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Messier V, Zenklusen D, Michnick SW. Cell. 2013;153:1080. doi: 10.1016/j.cell.2013.04.035. [DOI] [PubMed] [Google Scholar]
- [40].Miller JP, Lo RS, Ben-Hur A, Desmarais C, Stagljar I, Noble WS, Fields S. Proc Natl Acad Sci U S A. 2005;102:12123. doi: 10.1073/pnas.0505482102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Hruby A, Zapatka M, Heucke S, Rieger L, Wu Y, Nussbaumer U, Timmermann S, Dünkler A, Johnsson N. J Cell Sci. 2011;124:35. doi: 10.1242/jcs.077065. [DOI] [PubMed] [Google Scholar]
- [42].Mo C, Bard M. Biochim Biophys Acta. 2005;1737:152. doi: 10.1016/j.bbalip.2005.11.002. [DOI] [PubMed] [Google Scholar]
- [43].Eckert JH, Johnsson N. J Cell Sci. 2003;116:3623. doi: 10.1242/jcs.00678. [DOI] [PubMed] [Google Scholar]
- [44].Yan A, Lennarz WJ. Glycobiology. 2005;15:1407. doi: 10.1093/glycob/cwj026. [DOI] [PubMed] [Google Scholar]
- [45].Chavan M, Yan A, Lennarz WJ. J Biol Chem. 2005;280:22917. doi: 10.1074/jbc.M502858200. [DOI] [PubMed] [Google Scholar]
- [46].Möckli N, Deplazes A, Hassa PO, Zhang Z, Peter M, Hottiger MO, Stagljar I, Auerbach D. Biotechniques. 2007;42:725. doi: 10.2144/000112455. [DOI] [PubMed] [Google Scholar]
- [47].Xue X, Lehming N. J Mol Biol. 2008;379:212. doi: 10.1016/j.jmb.2008.03.069. [DOI] [PubMed] [Google Scholar]
- [48].Labedzka K, Tian C, Nussbaumer U, Timmermann S, Walther P, Müller J, Johnsson N. J Cell Sci. 2012;125:4103. doi: 10.1242/jcs.105320. [DOI] [PubMed] [Google Scholar]
- [49].Gulati S, Balderes D, Kim C, Guo ZA, Wilcox L, Area-Gomez E, Snider J, Wolinski H, Stagljar I, Granato JT, Ruggles KV, et al. FASEB J. 2015;29:4682. doi: 10.1096/fj.14-264796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Sung M-K, Lim G, Yi D-G, Chang YJ, Bin Yang E, Lee K, Huh W-K. Genome Res. 2013;23:736. doi: 10.1101/gr.148346.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Pu J, Ha CW, Zhang S, Jung JP, Huh W-K, Liu P. Protein Cell. 2011;2:487. doi: 10.1007/s13238-011-1061-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Gallina I, Colding C, Henriksen P, Beli P, Nakamura K, Offman J, Mathiasen DP, Silva S, Hoffmann E, Groth A, Choudhary C, et al. Nat Commun. 2015;6:6533. doi: 10.1038/ncomms7533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Yofe I, Weill U, Meurer M, Chuartzman S, Zalckvar E, Goldman O, Ben-Dor S, Schütze C, Wiedemann N, Knop M, Khmelinskii A, et al. Nat Methods. 2016;13:371. doi: 10.1038/nmeth.3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.