Abstract
Endomicroscopy is an emerging imaging modality, that facilitates the acquisition of in vivo, in situ optical biopsies, assisting diagnostic and potentially therapeutic interventions. While there is a diverse and constantly expanding range of commercial and experimental optical biopsy platforms available, fibre-bundle endomicroscopy is currently the most widely used platform and is approved for clinical use in a range of clinical indications. Miniaturised, flexible fibre-bundles, guided through the working channel of endoscopes, needles and catheters, enable high-resolution imaging across a variety of organ systems. Yet, the nature of image acquisition though a fibre-bundle gives rise to several inherent characteristics and limitations necessitating novel and effective image pre- and post-processing algorithms, ranging from image formation, enhancement and mosaicing to pathology detection and quantification. This paper introduces the underlying technology and most prevalent clinical applications of fibre-bundle endomicroscopy, and provides a comprehensive, up-to-date, review of relevant image reconstruction, analysis and understanding/inference methodologies. Furthermore, current limitations as well as future challenges and opportunities in fibre-bundle endomicroscopy computing are identified and discussed.
Keywords: Fibre bundle endomicroscopy, confocal laser endomicroscopy, imaging, Image restoration, image analysis, image understanding
1. Introduction
The emergence of miniaturised optical-fibre based endoscopes has enabled real-time imaging, at cellular resolution, of tissues that were previously inaccessible through conventional endoscopy. Fibre-bundle endomicroscopy (FBEμ), the most prevalent endomicroscopy platform, has been clinically deployed for the acquisition of in vivo, in situ optical biopsies in a wide and ever-increasing range of organ systems predominantly in the gastrointestinal, urological and the respiratory tracts. Customarily, a coherent fibre bundle is guided through the working channel of an endoscope (or a needle) to a region of interest and intra-venous or topical dyes are employed to augment tissue fluorescence, enhancing the emitted signal of the imaged structure. Endomicroscopy has the potential to assist diagnostic and interventional procedures by aiding targeted sampling and increasing diagnostic yield and ultimately reducing the need for histopathological tissue biopsies and any associated delays. To date, the most widespread use of FBEμ (along with fluorescent dyes, such as fluorescein) is in the gastro-intestinal (GI) tract (Fugazza et al., 2016; Wallace and Fockens, 2009; Wang et al., 2015). In particular, FBEμ has been employed in the upper GI tract (East et al., 2016) to detect (i) structural changes in the oesophagus mucosa associated with squamous cell carcinoma and Barrett’s oesophagus, and (ii) polyps and neoplastic lesions as well as gastritis and metaplastic lesions in the stomach and duodenum. In the lower GI tract, fibre-endomicroscopy has been utilised to (i) detect colonic neoplasia (Su et al., 2013) and malignancy in colorectal polyps (Abu Dayyeh et al., 2015), as well as to (ii) assess the activity and relapse potential of Inflammatory Bowel Disease (IBD) (Rasmussen et al., 2015; Salvatori et al., 2012).
In pulmonology, the auto-fluorescence (at 488 nm) generated through the abundance of elastin and collagen has enabled the exploration of the distal pulmonary tract as well as the assessment of the respiratory bronchioles and alveolar gas exchanging units of the distal lung without the need for exogenous contrast agents. Clinical studies have demonstrated the ability of FBEμ to image a range of pathologies including (i) changes in cellularity in the alveolar space as indicator of acute lunge cellular rejection following lung transplantation (Yserbyt et al., 2014), (ii) cross-sectional and level of fluorescence changes in the alveolar structure in emphysema (Newton et al., 2012; Yserbyt et al., 2017), and (iii) elastic fibre distortion (Yserbyt et al., 2013) and neoplastic changes in epithelial cells (Fuchs et al., 2013; Thiberville et al., 20 07,2009) in bronchial mucosa.
Other clinical applications of FBEμ include (but are not limited to) imaging (i) structural epithelial changes observable in bladder neoplasia (Sonn et al., 2009) as well as upper tract urothelial carcinoma (Chen and Liao, 2014), (ii) pancreatobiliary strictures as well as in pancreatic cystic lesions (catheter and/or needle based endomicroscopy), detecting potential malignancy (Karia and Kahaleh, 2016; Smith et al., 2012), (iii) the oropharyngeal cavity, differentiating between healthy epithelium, squamous epithelium and squamous cell carcinoma (Abbaci et al., 2014), and (iv) brain tumours (surgical access), such as glioblastoma, providing immediate histological assessment of the brain-to-neoplasm interface and hence improving tumour resection (Mooney et al., 2014; Pavlov et al., 2016; Zehri et al., 2014). Furthermore, there has been an effort to develop molecularly targeted fluorescent probes, such as peptides (Burggraaf et al., 2015; Hsiung et al., 2008; Staderini et al., 2017), antibodies (Pan et al., 2014) and nanoparticles (Bharali et al., 2005), that can bind and amplify fluorescence in the presence of specific type of tumours (Hsiung et al., 2008; Khondee and Wang, 2013; Pan et al., 2014), inflammation (Avlonitis et al., 2013), bacteria (Akram et al., 2015b) and fibrogenesis (Aslam et al., 2015). Such fluorescent probes will give rise to molecular FBEμ, enhancing the imaging and the diagnostic capabilities of the technology and significantly augmenting utility.
The proliferation of probe-based confocal laser endomicroscopy (pCLE) in clinical practice, along with the emergence of novel, FBEμ architectures and molecularly targeted fluorescent probes necessitate the development of highly sensitive imaging platforms, as well as a range of custom, purpose specific image analysis and understanding methodologies that will assist the diagnostic process. This review provides a brief summary of the available endomicroscopic imaging platforms (Section 2), along with an overview of the state-of-the-art in fibre-bundle endomicroscopic (FBEμ) image computing methods, namely image reconstruction (Section 3), analysis (Section 4) and understanding/inference (Section 5). Owing to its more widespread dissemination, this review paper concentrates in FBEμ image computing. Yet, a small number of relevant image analysis and understanding techniques developed and assessed for other endomicroscopy platforms, offering viable solutions for FBEμ have also been included. Current limitations in FBEμ image computing as well as future challenges and opportunities are also identified and discussed (Section 6).
2. Technology overview
To date, four endomicroscopic imaging platforms, all exploiting different fundamental optical imaging technologies, have been commercialised for clinical use (NinePoint, Olympus, Pentax, Mauna Kea, Zeiss). While this review paper concentrates on fibre bundle based systems, a brief description of the currently available endomicroscopic imaging platforms, both commercial and research based, is provided.
NinePoint Medical (Bedford, Massachusetts, USA) developed the NVisionVLE platform, a Volumetric Laser Endomicroscopy (VLE) (Bouma et al., 2009; Vakoc et al., 2007; Yun et al., 2006) device that can acquire in-vivo, high-resolution (7 μm), volumetric data of a cavity (e.g. the gastro-intestinal tract) through a flexible, narrow diameter catheter (<2.8 mm). VLE combines principles of endoscopic Optical Coherence Tomography (OCT) (Tearney et al., 1997), along with Optical Frequency-Domain Imaging (OFDI) (Yun et al., 2003) to acquire a sequence of cross-sectional images 100-fold faster than conventional OCT, while maintaining the same resolution and contrast. Similar non-commercial OFDI technology has been employed by Tethered Capsule Endoscopy (TCE) to provide an imaging alternative for the gastro-intestinal tract (Gora et al., 2013).
Olympus Medical Systems Co. (Tokyo, Japan) developed a range of prototype endocytoscopes (Hasegawa, 2007), white-light, flexible, contact endoscopes that can image at cellular resolution (up to >1000x magnification). These, now discontinued, prototypes incorporated a miniaturised Charged-Coupled Device (CCD) sensor, the associated objective lenses and an adjacent light source at the distal end of the endoscope. All imaging at the endoscope/tissue contact layer was achieved via light scattering. Olympus provided a variety of options, from (i) full endoscope integration, to (ii) standalone, probes that could fit through a 3.7 mm endoscope work-ing channel (Ohigashi et al., 2006; Singh et al., 2010). An alternative non-commercial implementation, replacing the miniature sensor with a flexible, coherent fibre bundle enabling imaging at the proximal end of the fibre (Hughes et al., 2013), as well as a range of adaptations to achieve true reflectance endomicroscopy while avoiding back-reflections (Hughes et al., 2014; Liu et al., 2011; Sun et al., 2010) have been proposed.
Confocal laser endomicroscopy (CLE) employs a miniaturised optical fibre to acquire 2D images, predominantly fluorescent, across the examined tissue structure. Inspired from benchtop confocal microscopy (Minsky, 1988), a low-power laser signal (typically at 488 nm), focused to a single, finite point within the specimen, is scanned across a two-dimensional imaging plane, generating a 2D image commonly referred to as an optical section. Optical fibres are typically used for relaying light and may act as bidirectional pinholes, rejecting light outside the focal point, reducing the associated image blurring. There are currently, numerous experimental and two commercial CLE platforms with clinical utility, namely endoscope-based (eCLE) and probe-based (pCLE) endomicroscopy. A number of review papers (Jabbour et al., 2012; Oh et al., 2013) provide insight into the current CLE instrumentations. In brief, eCLE integrates a miniaturised confocal scanner into the distal tip of a device using a single core fibre (Delaney et al., 1994; Harris, 1992, 2003). Piezoelectric or electromagnetic actuators can be used to generate the 2D scanning pattern. The tissue signal generated at each individual (scanning) location is transferred through the single-core fibre to a detector and associated processing unit at the proximal end, where the image is accumulated and reconstructed after each complete scan. A clinical eCLE platform was developed by Pentax Medical (Tokyo Japan), integrating the confocal scanning facility into a conventional endoscope. This now discontinued device had a 12.8 mm diameter and enabled the acquisition of optical sections at a 500 × 500 μm field of view and 0.7 μm lateral resolution. Optiscan (Melbourne, Australia) has recently developed a pre-clinical eCLE platform, comprising of a 4 mm diameter standalone micro-endoscope with a < 0.5 μm lateral resolution (highest commercially available resolution) and a 475 × 475 μm field of view. The acquisition frame-rate in both devices is dependent on the associated acquisition aspect ratio and Z-stack depth (single vs multiple frames), with typically reported values ranging between 0.8 to 6 frames per second (for a single frame). Carl Zeiss Meditec (Jena, Germany) has recently developed a digital biopsy tool for neurosurgery (Leierseder, 2018) based on the underlying Optiscan eCLE technology (475 × 267 μm FOV at 488 nm excitation). A number of alternative, non-commercial experimental architectures utilising distal based scanning have been proposed, including (i) high speed imaging (Shi and Wang, 2010) via parallel distal scanning through a multi-core fibre; (ii) dual-axis imaging (Wang et al., 2003), separating the illumination and collection paths and enabling 2D (Liu et al., 2007), 3D (Ra et al., 2008) and multi-colour (Leigh and Liu, 2012) imaging capabilities with improved optical sectioning (axial resolution); and (iii) two-photon imaging (So et al., 20 00; Wu and Li, 2010), employing multiple, less energetic photons to induce transition of the imaged fluorescent structure to the desired excitation state, improving the resulting imaging resolution, penetration depth and reducing potential tissue photodamage.
Probe-based confocal laser endomicroscopy (pCLE) utilises a multicore imaging fibre-bundle for the acquisition of 2D optical en face sections of a tissue structure. Confocal scanning takes place at the proximal end of the fibre and is relayed by the fibre bundle. Each individual core within the bundle, often combined with a pinhole, rejects light outside the focal plane. Compared to eCLE, the optical setup for pCLE results in a substantially smaller distal endomicroscope as well as higher acquisition frame rates. In contrast, the imaging depth is fixed (and smaller) by the distal optics and the lateral resolution is determined (limited) by the inter-core distance of a particular multicore fibre bundle and distal optics design. Gmitro and Aziz (1993), Rouse et al. (2004) and Sabharwal et al. (1999) proposed an early implementation of pCLE with Dubaj et al. (2002) and Le Goualher et al. (2004b) introducing refinements for real-time confocal scanning in biomedical applications. Mauna Kea Technologies (Paris, France) developed the Cellvizio pCLE imaging platform along with a wide range of compatible multi-core fibre probes with diameters as small as 0.3 mm, field of views between 300 and 600 μm, and, excluding magnification from distal optics, approximate lateral resolution of 3.3 μm. Additional distal optics can be used to improve the imaging resolution to approximately 1 μm, at the expense of field of view (240 μm) and diameter (<3 mm). The probes’ miniature sizes enable use through the working channel of most commercially available endoscopes as well as some needles/catheters, while the relevant high data acquisition rate (>12 fps) enables real-time imaging of moving structures, resulting in Cellvizio (pCLE) being the most widely used endomicroscopy platform approved for clinical use. A rapidly growing volume of alternative, non-commercial experimental architectures is being developed, including (i) right-angle stage attachment for standard desktop confocal microscopes (ii) linescanning confocal endomicroscopy (Hughes and Yang, 2015, 2016), improving the acquisition frame rate without compromising substantially image quality; (iii) flexible and low-cost endomicroscopy architectures (Hong et al., 2016; Krstajić et al., 2016; Pierce et al., 2011; Shin et al., 2010), employing LED, widefield illumination; (iv) structured illumination endomicroscopy (Bozinovic et al., 2008; Ford et al., 2012b; Ford and Mertz, 2013), providing out-of-focus background rejection without beam scanning; (v) oblique back-illumination endomicroscopy (Ba et al., 2016; Ford et al., 2012a; Ford and Mertz, 2013), collecting phase-gradient images of thick scattering samples; and (vi) multi-spectral imaging (Bedard and Tkaczyk, 2012; Cha and Kang, 2013; Jean et al., 2007; Krstajic et al., 2016; Makhlouf et al., 2008; Rouse and Gmitro, 2000; Vercauteren et al., 2013; Waterhouse et al., 2016). These alternative architectures, along with pCLE can be grouped under the term Fibre-Bundle Endomicroscopy (FBEμ), which, due to its more widespread dissemination, is the technology primarily deliberated throughout this study.
3. Image reconstruction
The nature of image acquisition through coherent fibre bundles is a source of inherent limitations in FBEμ imaging. Coherent fibre bundles are comprised of multiple (>10.0 00) cores that (i) have variable size and shape, (ii) are irregularly distributed across the field of view, (iii) have variable light transition properties, including coupling efficiency and inter-core coupling spread, and (iv) have spatiotemporally variable auto-fluorescent (background) response at certain imaging wavelengths. Such properties directly limit the imaging capabilities of the technology. There has therefore been a substantial interest in the development of effective and efficient approaches to reconstruct FBEμ images, attempting to compensate for these inherent limitations. Table 1 provides an overview of the most relevant image reconstruction studies applicable to FBEμ, while Fig. 1 provides characteristic examples of the associated imaging limitations.
Table 1. Overview of reconstruction approaches for fibred endoscopic imaging.
| Topic | References | Methodology | Comments |
|---|---|---|---|
| Honeycomb effect & Fourier domain filters | (Dickens et al. 1998; Dickens et al. 1997,1999) | Manual band-reject filters with “high-boost” filter. | Simple to implement, and computationally efficient approaches that suppress the honeycomb structure. However, inherently susceptible to blurring the imaged structures. |
| Han et al. (2010) | Histogram equalisation with Gaussian low-pass filter. | ||
| Rupp et al. (2007) and Winter et al. (2006) | Low pass filter using alternative (circular, star-shaped), rotationally invariant kernels. | ||
| Lee and Han (2013b) | Gaussian based, notch reject filter, eliminating periodic, high-frequency components. | ||
| Ford et al. (2012b) | Iteratively blurring (low pass) cladding regions while maintaining core intensities. | ||
| Dumripatanachod and Piyawattanametha (2015) | Efficient with 2 1D top-hat filters (equivalent to square kernel). | ||
| Honeycomb effect & core interpolation | Elter et al. (2006), Le Goualher et al. (2004a) and Rupp et al. (2007,2009) | C0-2 continuous interpolation methods on irregular core lattice. | Simple and efficient approaches capable on maintaining the original core information. Successfully employed in clinical/commercial systems. |
| Zheng et al. (2017) | Enhancement of interpolated (bilinear) images using rotationally invariant Non-Local Means. | ||
| Winter et al. (2007) | Correcting for variable core PSF overlap over a colour-sensor’s Bayer pattern, suppressing false colour moiré patterns. | ||
| Honeycomb effect & image superimposition | Rupp et al. (2007) | Integrate the core locations of 4 shifted and aligned images, interpolate revised grid. | Capable of removing the honeycomb structure and increase the effective resolution of the acquired data. Developing real-time elastic registration approaches is a major challenge. |
| Kyrish et al. (2010), Lee and Han (2013a) and Lee et al. (2013) | Compounding images shifted (translation stage) with a range of predetermined patterns. | ||
| Cheon et al. (2014a), Cheon et al. (2014) and Vercauteren et al. (2005,2006) | Aligning (compensating for random movements) and combining consecutive frames. | ||
| Honeycomb effect & iterative reconstruction | Han and Yoon (2011) | Maximising the posterior probability in a Bayesian framework (Markov Random Fields). | Preliminary studies, successful at removing honeycomb structures. Not necessarily improve reconstruction error to interpolation. Computationally costly due to iterative nature |
| Liu et al. (2016) | l 1 norm minimisation (using iterative shrinkage thresholding - 1ST) in the wavelet domain. | ||
| Han et al. (2015) | Efficient, non-parametric iterative compressive sensing for inpainting cladding regions. | ||
| Variable coupling & background response | Ford et al. (2012b), Le Goualher et al. (2004a) and Zhong et al. (2009) | Affine intensity transform incorporating dark and bright-field information at each core. | Capable of supressing (in real time) the effect of spatio-temporally variable coupling and background response. Successfully employed in clinical/commercial systems. |
| Savoire et al. (2012) | Blind calibration exploring neighbouring core correlation to recursively (online) derive gain and offset coefficients in each core. | ||
| Vercauteren et al. (2013) | Multi-colour extension of Le Goualher et al. (2004a) dealing with geometric and chromatic distortions. | ||
| Cross coupling | Perperidis et al. (2017a) | Quantifying (and integrating into a linear model) cross coupling within fibre bundles. | Effective in supressing the effect of cross coupling. Computationally costly for real-time scenarios. |
| Karam Eldaly et al. (2018) | Deconvolution and image reconstruction, reducing the effect of inter-core coupling. |
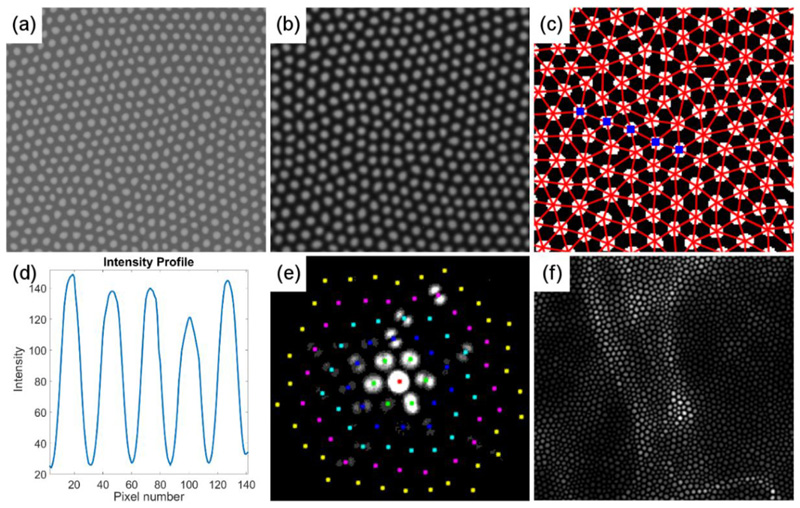
Fig. 1. Examples illustrating properties of coherent fibre bundles that limit the imaging capabilities of fibred endoscopy.
(a) Scanning Electron Microscopy (SEM) image of commercial coherent fibre bundle (FIGH-30-650S, Fujikura), along with (b) a uniform, flood illumination (520 nm) image of the same fibre bundle, using widefield endomicroscopy. The variable size and shape, as well as irregular distribution of the cores is apparent in both (a) and (b). (c) Binary masque and associated Delaunay triangulation of cores, identified within a uniformly illuminated image, similar to (b). (d) Intensity profile across the five cores highlighted in (c) illustrating the variations of coupling efficiency amongst different neighbouring cores. (e) Example inter-core coupling spread at 520 nm, as measured by Perperidis et al. (2017b). (f) Example raw widefield endomicroscopy image of auto-fluorescent alveoli structures from an ex-vivo, human lung. The imaged structured is heavily corrupted by the intrinsic characteristics of the imaging fibre bundle, highlighting the need for effective image reconstruction approaches. Images (a-b), (c) and (e) have been reproduced (cropped) from Figures 6, 7 and 8 respectively of the “Characterization and modelling of inter-core coupling in coherent fibre bundles" by Perperidis et al. (2017b) under the Creative Commons Attribution (CC BY) 4.0 International License (https://creativecommons.org/licenses/by/4.0).
3.1. Honeycomb effect
The most visually striking, and limiting artefact, arising from the transmission of the imaged scene through a coherent fibre bundle, is the so-called honeycomb effect. The honeycomb effect, illustrated in Fig. 1, is a consequence of the light being guided from the distal to the proximal end of the individual cores comprising the fibre-bundle but not through the surrounding cladding. Each core, while usually imaged across multiple pixels, contains intensity information on a single, discrete position within the imaged scene. Consequently, the resulting raw image data is a high-resolution rectangular matrix representation of a low-resolution, irregularly-sampled scene. Several studies have attempted to supress/remove the honeycomb effect in fibred endoscopy, generating continuous, high-resolution image sequences.
Throughout the years, a number of approaches employing band-pass filtering in the Fourier domain have been proposed (Dickens et al., 1998,Dickens et al., 1997,1999; Dumripatanachod and Piyawattanametha, 2015; Ford et al., 2012b; Han et al., 2010; Lee and Han, 2013b; Maneas et al., 2015; Rupp et al., 2007; Winter et al., 2006). Band-pass filters employing a range of different kernels, static and adaptive (derived from the core distribution across the bundle) were typically combined with a range of pre- and post-processing approaches to enhance the performance of suppressing the core honeycomb pattern. Band-pass filtering provides a simple and efficient approach to suppress/remove the honeycomb structure from fibred endoscopic images. However, given the irregularly distributed cores in most modern miniaturised fibrescopes, identifying suitable thresholds in the frequency domain that would remove the honeycomb effect (usually high frequency component) without blurring the underlying imaged structure (usually lower frequency component) can be inherently challenging.
In contrast to band-pass filtering, interpolating amongst the irregular core lattice effectively removes the undesired honeycomb structure while retaining the original image content at the core locations. To accurately identify the locations of each individual core, a uniformly illuminated calibration image is required. Local maxima and the Circular Hough Transforms (CHT) are amongst the wide range of off-the-shelf solutions for identifying core locations. Suggested interpolation methods (Elter et al., 2006; Le Goualher et al., 2004a; Rupp et al., 20 07,2009; Vercauteren et al., 2006) include (i) C° continuous nearest neighbour, triangulation-based and natural-neighbour based linear interpolations, (ii) C 1 continuous Clough-Tocher interpolation (Amidror, 2002) and a Bernstein-Bezier extension to natural neighbours (Farin, 1990), and (iii) C 2 continuous Radial Basis Functions (Amidror, 2002), b-spline approximation (Lee et al., 1997) and recursive Gaussian filter (Deriche, 1993) adaptation of Shepard’s interpolation. Zheng et al. (2017) attempted to refine the results of a bilinear interpolation using a rotationally invariant adaptation of Non-Local Means (NLM) filters. Moreover, Winter et al. (2007) proposed an extension of the core interpolation approach for single chip colour cameras, suppressing any false colour moiré patterns. While higher order continuity generated smoother images, a property that can be desirable in particular applications, the associated reconstruction accuracy was shown (Rupp et al., 2009) to be only marginally supe-rior to simple C 0 algorithms. On the other hand, for simple Voronoi Tessellation based approaches, all calculations could be performed once, at the calibration stage, generating look up tables to be employed during the subsequent image reconstruction task. Consequently, generating comparable results with less computational complexity makes such linear interpolation approaches more attractive candidates for real-time applications.
Superposition or compounding of spatially misaligned and partially decorrelated images is an approach that has been effective in the enhancement of medical ultrasound images (Perperidis et al., 2015; Rajpoot et al., 2009). In fibred endoscopy, movement of the fibre tip in successive frames accommodates the acquisition of information from the regions previously masked by the fibre cladding. Hence, by effectively aligning the imaged structures and combining a sequence of shifted frames (i) the honeycomb structure can be suppressed/eliminated, (ii) the imaging resolution can be increased. Numerous attempts have examined the effect of different shift patterns, altering the location of the core pattern with respect to the imaged structure, and superposition methods, such as deriving the average or maximum intensity of the aligned images on fibreoscopic images (Kyrish et al., 2010; Lee and Han, 2013a; Lee et al., 2013; Rupp et al., 2007). Alternatively, Cheon et al. (2014a, Cheon et al., 2014) and Vercauteren et al. (2005,2006) employed the random movements during data acquisition, as would be expected in a realistic clinical scenario, to create an enhanced composite image. The approach was first introduced as part an image mosaicing framework (see Section 4.1) with the main effort being placed in devising an accurate and efficient approach for the alignment of consecutive images. While small translational movements can be efficiently and potentially accurately estimated in real-time, in a realistic scenario with, image distortions as well as elastic and sometimes large structural deformations between consecutive frames, the accurate real-time alignment and compounding can be an eminently challenging task. Increased acquisition frame rate can potentially reduce the deformations between consecutive frames, making their effective alignment more realisable. However, increasing the acquisition frame rate can have detrimental effect in the signal to noise ratio and associated imaging limits of detection.
Recently, several more “sophisticated”, iterative methods have been proposed for the reconstruction of fibred endoscopic images and the removal of the associated fixed honeycomb pattern. Han and Yoon (2011) employed a Bayesian approximation algorithm to decouple the honeycomb effect. Liu et al. (2016), based on the empirical observation that natural images tend to be sparse in the wavelet domain, employed l 1 norm minimisation in the wavelet domain to remove the honeycomb pattern. Han et al. (2015) employed an efficient, non-parametric iterative compressive sensing technique for inpainting the cladding regions, without the need of any prior information with regards to the underlying core structure. Limited evaluation (on USAF resolution targets and some biological data) demonstrated the potential of such iterative approaches in image reconstruction, removing the honeycomb artefact as well as fibre bundle defects, while maintaining the spatial resolution and considerably increasing the image contrast and contrast to noise ratio (CNR). However, the current algorithm im-plementations are considered computationally expensive, making them unsuitable for real time applications. Nevertheless, accelerated, parallel processing though state-of-the-art Graphical Processing Units (GPU) could potentially enable the real-time implementation of such iterative approaches.
3.2. Variable coupling and background response
Coherent fibre bundles comprise of a large number cores, commonly in excess of 50 00. To reduce the effect of inter-core coupling, neighbouring cores tend to be of variable size and shape. A consequence of this core irregularity is the variable coupling efficiency observed across the fibre bundle. Furthermore, some imaging fibre bundles have exhibited an intrinsic, background auto-fluorescent response at certain imaging wavelengths (e.g. 488 nm). Auto-fluorescence, as with coupling efficiency, is also associated with the shape and size of each individual core. These innate fibre properties have a detrimental effect in imaging quality. Consequently, explicit calibration procedures have been developed in an attempt to supress their effect in fibred endoscopic imaging. Le Goualher et al. (2004a) proposed an off-line calibration process, utilising (i) an image of the fibre auto-fluorescent background (dark-field), as well as (ii) an image of a uniformly fluorescent medium (bright-field). More specifically, for every frame during data acquisition, geometric distortions caused by the resonant scanning mirrors were compensated and the intensity at each core location was normalised using an affine intensity transformation combining the dark and bright-field information. Ford et al. (2012b) and Zhoπg et al. (2009) extended the Le Goualher et al. (2004a) approach, introducing additional normalisation terms to partially compensate for camera bias, ambient background light and occasional system realignment. Vercauteren et al. (2013) adapted the off-line calibration approach in Le Goualher et al. (2004a) to deal with the distortion compensations (geometric and chromatic) for multi-colour acquisition. In particular, chromatic distortions were estimated and compensated by a symmetric and robust version of the Iterative Closest Point algorithm relying on orthogonal linear regression.
The aforementioned studies assumed constant gain (coupling efficiency) and offset (background auto-fluorescence) for each individual core. However, medium-dependent and slow time-varying coefficient deviations can introduce a static noise pattern on the acquired images. Savoire et al. (2012) explored the high correlation of signals between neighbouring cores to develop a blind on-line calibration process. For every core in the bundle, (i) linear regression on a temporal window estimated the relative gain and offset coefficients for the associated neighbouring core-pairs, (ii) regularised inversion derived the core’s actual gain and offset parameters. To compensate for slow time-varying coefficient changes, the process was performed recursively over temporal windows sufficiently large to enable a robust inversion process.
3.3. Inter-core coupling
Inter-core coupling is another limitation in coherent fibre bundles, resulting in blurring of the imaged structures and consequently a worsening in the associated limits of detection in FBEμ. Inter-core coupling has been studied both experimentally (Chen et al., 2008; Wood et al., 2017) and within the theoretical framework of coupled mode theory (Ortega-Quijano et al., 2010; Reichenbach and Xu, 2007; Wang and Nadkarni, 2014), providing (i) insights on the factors affecting cross talk, and (ii) solutions/recommendations for optimal design, selection and optimisation of fibre bundles. Yet, due to the trade-off between cross coupling and core density, cross coupling can be suppressed yet not eliminated through optimal fibre design. In a recent study, Perperidis et al. (2017a), introduced a novel approach for measuring, analysing and quantifying cross coupling within coherent fibre bundles, in a format that can be integrated into a linear model of the form v = Hu + w with v being the recorded image, u the original signal, H the convolution operator modelling the spread of light, and w an additive observation noise. Karam Eldaly et al. (2018) employed this linear model and demonstrated the potential of both optimisation-based and simulation-based approaches in reconstructing FBEμ data and reducing the effect of inter-core coupling. However, the computational requirements of the proposed methodology limit their current suitability in real-time clinical applications.
4. Image analysis
Analysis of the acquired data and quantification of the imaged structures and potential pathologies is an imperative component to the development of Computer Aided Diagnosis (CAD) systems. Such systems can capitalise on the real-time, optical biopsy capabilities of the technology. Yet, the underlying imaging technology, along with the nature of the clinical data acquisition, generating a steady stream of high resolution images with constricted Field of View (FOV), impose a series of inherent restrictions/challenges to the development of image analysis methodologies. To date, image analysis research for FBEμ can be broadly categorised into (i) mosaicing (Table 2), and (ii) quantification (Table 3) methods. However, the literature appears to be heavily unbalanced, concentrating predominantly on the task of mosaicing frame sequences to extend the associated FOV.
Table 2. Overview of mosaicing approaches for fibred endoscopic imaging.
| Topic | References | Methodology | Comments |
|---|---|---|---|
| Image based, real-time | Bedard et al. (2012) and Vercauteren et al. (2008) | Local rigid alignment through normalised cross correlation matching. | |
| Loewke et al. (2008) | Local rigid alignment through feature based optical flow refined via gradient descent on normalised cross correlation. | ||
| Certain assumptions and model simplifications required for achieving real time performance. | |||
| Image based, post-procedural | Vercauteren et al. (2005,2006) | Hierarchical framework of frame-to-reference transformations (on the original, sparsely sampled data) to derive a globally consistent rigid alignment, while compensating for motion distortions, elastic deformations. Global alignment seen as an estimation problem on a Lie group. | More complex models dealing with a range of local and global, rigid and elastic image transformations. Post-procedural approaches with real-time capacity compromised due to the underlying complex registration models. |
| Loewke et al. (2011,2007a) | Compensating for global (rigid) as well as local (elastic) transformations (including motion distortion. Fixed correspondence between images were replaced with a Gaussian Potential representing the amount of certainty in the registration. Global and local deformation potentials were maximised in an integrated optimisation problem | ||
| Hu et al. (2010) | Elastic registration of consecutive frames based on optical flow of robust image features (RANSAC strategy on Lucas-Kanade tracker. A Maximum a Posterior (MAP) estimation based image blending generated super-resolved images. | ||
| Image based, dynamic imaging | Mahé et al. (2015) | Dynamic mosaic obtained by solving a 3D Markov Random Field. Two-stage approach, of static mosaicing followed by stitching of the associated video segments. | Generating mosaics that maintain temporal information in the form of infinite loops. |
| External input based | Loewke et al. (2007b) | Initial rigid alignment using feedback from a robotic arm determining the five degree-of-freedom position/orientation of the fibre tip. | Actuators/sensors provide feedback on the scanning path improving the efficiency and/or the robustness of mosaicing. Hardware additions are limiting their suitability for endoscopic applications. |
| Vyas et al. (2015) | Initial rigid alignment using feedback from a six degree-of-freedom electromagnetic sensor positioned at the tip of the fibre-bundle. | ||
| Mahé et al. (2013) | Weak a-priori knowledge of the trajectory (spiral scan) used to derive spatio-temporal associations within the frame sequence. A hidden Markov model notation and a Viterbi algorithm was recovered the optimal frame associations, feeding a modification of the mosaicing algorithm by Vercauteren et al. (2006) to estimate the optimal transform. |
Table 3. Overview of quantification approaches for fibred endoscopic imaging.
| Organ (System) | Quantifying | References | Methodology | Comments |
|---|---|---|---|---|
| Circulatory | Red blood cell velocity. | Savoire et al. (2004) | Thresholding and line-fitting (M-estimators) translated (through trigonometry) to RBC velocity. | Inventive use of known and quantifiable artefact in raster scanning imaging systems for deriving physiological information. Preliminary results with uncertain clinical relevance. |
| Perchant et al. (2007) | ROI tracking and alignment through (i) scanning distortion compensation, and (ii) global affine registration, for blood velocity estimation through spatio-temporal correlation. | Feasibility study. Preliminary results with uncertain clinical relevance. |
||
| Oropharyngeal | Epithelial cells in vocal chords. | Mualla et al. (2014) | Watershed segmentation (borders) and local minima detection (location). | Empirical, ad-hoc approach employing off-the-shelf image analysis methods. Limited data can potentially lead to poor generalisation of the proposed methodology. |
| Gastro-intestinal | Intestinal crypts in Inflammatory Bowel Disease. (eCLE) | Couceiro et al. (2012) | Detecting (local maxima), segmenting (ellipse fitting on edge detection) and quantifying (number, connectivity). | Empirical, ad-hoc approaches employing off-the-shelf image analysis methods Heuristic parameter estimation, hard thresholds and limited data can potentially lead to poor generalisation of the proposed methodologies. |
| Intestinal crypts in colorectal polyps. | Prieto et al. (2016) | Contrast enhancement, thresholding (Otsu’s) and morphological filters (erosion, centre of mass, circularity). | ||
| Goblet cells in villi. (eCLE) | Boschetto et al. (2015a) | Detecting (matched filters), segmenting (Voronoi diagrams) cells and identifying (hard threshold) goblet cells within the villi. | ||
| Intestinal villi. (eCLE) | Boschetto et al. (2015b) | Detect via morphological filters (top-hat, morphological reconstruction and closing) and quad-tree decomposition. | ||
| Boschetto et al. (2016b) | Subdivide to superpixels, extract features and classify through Random Forests to generate a binary segmentation map. | Employing established data driven approaches with reasonable size of data, resulting on better generalisation potential. | ||
| Pulmonary | Alveoli sacs in mice distal lung. | Namati et al. (2008) | Segmenting (optimum separation thresholding) and quantifying (8-point connectivity) alveolar sacs. | Limited data and uncertain translatability to human alveoli sacks due to their large size relative to the limited field of view. |
| Stained mesenchymal stem cells in rat lungs. | Perez et al. (2017) | Contrast stretch, denoise (opening), threshold and count (connected component analysis). | Empirical, ad-hoc approach employing off-the-shelf image analysis methods. | |
| Stained bacteria in distal lung. | Karam Eldaly et al. (2018) | Outlier detection using a hierarchical Bayesian model along with a MCMC algorithm based on Gibbs sampler. | More elaborate approaches, adopting model-based and data-driven methodologies. | |
| Stained bacteria and cells in distal lung. | Seth et al., (2017, 2018) | Bacterial and cellular load using spatio-temporal template matching with a radial basis functions network. | They have potential for good generalisation and translation to clinical applications. |
4.1. Mosaicing
The miniaturisation of imaging fibre bundles in FBEμ constraints the effective field of view (potentially <500 μm) and thus limits sampling diversity, which have implications for navigation, target tissue identification and scene interpretation. To address these inherent limitations, multiple, partially overlapping frames acquired over time can be aligned and combined (stitched) into a single frame with extended field of view. The process has been referred to as image mosaicing. Over the years, there has been considerable research in the development of image mosaicing approaches (Ghosh and Kaabouch, 2016), employed in a range of applications, including endoscopic imaging (Bergen and Wittenberg, 2016). Yet, generic mosaicing approaches do not deal with the inherent properties and limitations of endomicroscopy as described in Vercauteren et al. (2006). Notably, FBEμ is a direct contact imaging technique. The interaction of a moving rigid fibre-bundle tip with soft tissue may result in non-linear deformations of the imaged structures. A model of probe-tissue interaction for FBEμ was proposed in Erden et al. (2013). Furthermore, in laser scanning based FBEμ platforms, an input frame does not represent a single point in time. Instead, each sampling point is acquired at a slightly different point in time, resulting in potential deformations when imaging fast moving objects. Finally, the imaged tissue structures are sampled through a sparse, irregularly distributed fibre bundle. Hence, due to these non-linear deformations, motion artefacts and irregular sampling of the input frames, there has been a substantial research interest in the development of custom mosaicing approaches optimised for endomicroscopic data. The proposed methodologies, some currently used in clinical practice, range from simple real-time, to more intricate post-procedural solutions, for either free-hand and/or robotically driven mosaicing platforms. Table 2 and Fig. 2 provide an overview of FBEμ mosaicing techniques and characteristic examples of the derived mosaics.
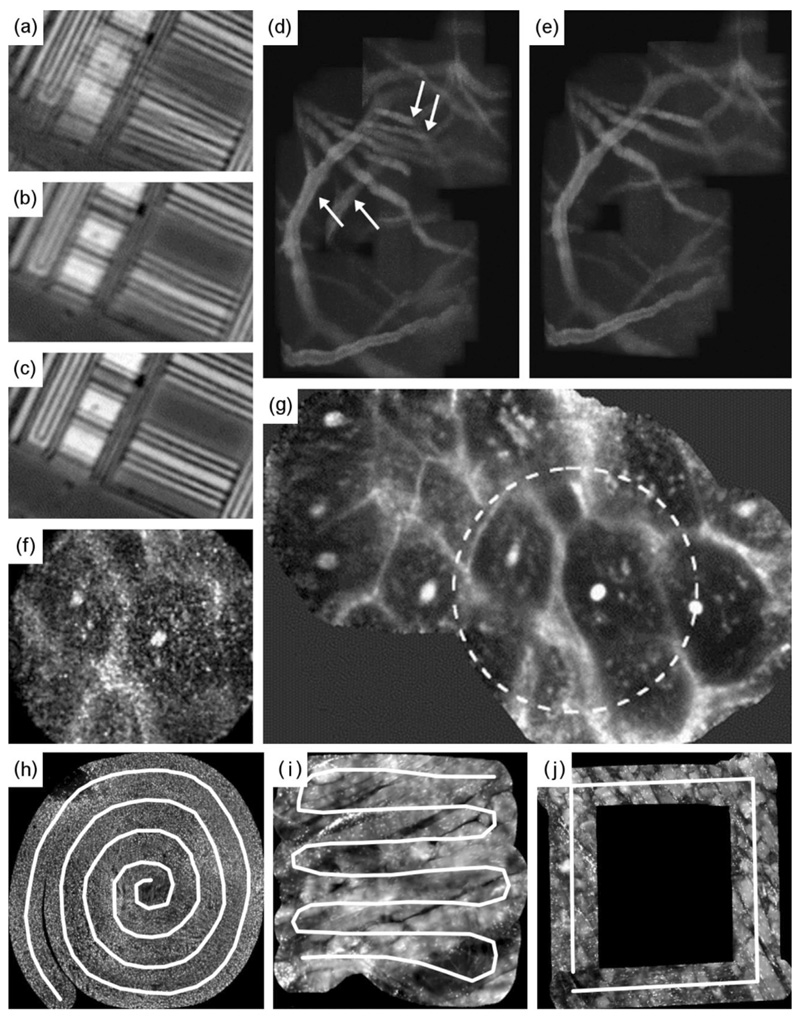
Fig. 2. Examples of mosaics employing a range of motion/deformation compensation algorithms as well as motorised acquisition path control.
(a-c) Mosaics of a silicon wafer using (a) local only, (b) local and motion distortion, and (c) global and motion distortion compensation; (d-e) Mosaics of mouse brain blood vessels using (d) local and (e) global motion and deformation compensation. While local, rigid frame alignment can generate mosaics in real-time, global and non-linear motion and deformation compensation is required for more accurate, continuous mosaics. (f-g) Global mosaic (circular ROI in (g) matching FOV in (f)) of human mouth mucosa where the accurate alignment and reconstruction can result in denoised and super-resolved images. (h-j) Customised and structured mosaic acquisition paths (such as spiral, raster and square scans). Images (a-c) and (f-g) reproduced and adapted (cropped/resized) with permission from Elsevier from Figures 18 and 22 respectively of the “Robust mosaicing with correction of motion distortions and tissue deformations for in vivo fibered microscopy” by Vercauteren et al. (2006). Images (d-e) are reproduced and adapted with permission from the Institute of Electrical and Electronics Engineers (IEEE) from Figure 9 of the “In Vivo Micro-Image Mosaicing” by Loewke et al. (2011). Image (h) is reproduced and adapted with permission from IEEE from Figure 3 of the “Conic-Spiraleur: A Miniature Distal Scanner for Confocal Microlaparoscope” by Erden et al. (2014). Image (i) is reproduced and adapted with permission from IEEE from Figure 13 of the “Building Large Mosaics of Confocal Edomicroscopic Images Using Visual Servoing” by Rosa et al. (2013). Image (j) is reproduced and adapted with permission from IEEE from Figure 6 of the “Understanding Soft-Tissue Behavior for Application to Microlaparoscopic Surface Scan” by Erden et al. (2013).
Early mosaicing approaches were post-procedural and addressed both rigid and elastic deformations (Fig. 2). Vercauteren et al. (20 05,20 06) were the first studies to identify the necessity for custom mosaicing approaches in endomicroscopy. Vercauteren et al. (2006) provided a hierarchical framework of frame-to-reference transformations (on the original, sparsely sampled data) to iteratively derive a globally consistent rigid alignment, while compensating for motion induced distortions, as well as for non-rigid deformations. Scattered data approximation was employed to reconstruct a continuous, regularly sampled image from the sparsely sampled inputs merged into a common reference. The proposed method, currently used as part of Mauna Kea’s post-procedural analysis software, was tested on phantom and in vivo data producing smooth mosaics with extended field of view and enhanced resolution (due to image reconstruction on partially overlapping, irregularly sampled images - see Superposition in Section 3.1). Loewke et al. (2007a) decomposed the problem into similar components, compensating for global (rigid) as well as local (elastic) transformations (incorporating the effect of motion distortions), maximising the certainty of the registration, both global and local, as an integrated optimisation problem. Averaging of overlapping pixels as well as multi-resolution pyramid blending were tested on both simulated as well as in vivo data, producing mosaics with smooth image transitions and sharp edges across the imaged structures. Finally, Hu et al. (2010) adopted a different approach, employing elastic registration of consecutive frames based on optical flow of robust image features and blending the mosaiced frames into a super-resolved image through a Maximum a Posterior (MAP) estimation technique. However, very limited FBEμ data (1 mosaic) were provided for the assessment of the technique.
The complex and descriptive models employed by the preceding methodologies have a direct effect on their computational requirements and consequently their real-time capability. Real-time mosaicing can provide much needed feedback, guiding the data acquisition process, ensuring a smooth continuous path over the desired region of interest (Fig. 2). Vercauteren et al. (2008) proposed an early real-time mosaicing algorithm, integrating translation and distortion (due to finite scanning speed) to a single rigid transformation (estimated through a fast, normalised cross correlation matching algorithm) followed from a simple “dead leaves” model for image blending. A very similar approach of aligning consecutive frames was employed by Bedard et al. (2012). Loewke et al. (2008) adopted a two-stage pair-wise registration between consecutive frames, (i) obtaining an initial translation estimate through optical flow on easily trackable features, and (ii) refining the es-timate through a gradient descent on cross correlation approach. A multi-resolution pyramid blending algorithm was also employed recombining overlapping regions to a composite image. To achieve real-time performance, these approaches needed to make certain assumptions and hence be subjected to a number of inherent limitations, such as the inability to compensate for global, accumulative alignment errors as well as any elastic deformations. A potential solution to these limitations, which has been adopted by both Loewke et al. (2011) and by Mauna Kea Technologies, is employ-ing a two-stage mosaicing strategy, including real-time mosaicing for live image acquisition, followed from a more accurate, postprocedural reconstruction.
Most mosaicing approaches for FBEμ have assumed a roughly static scene imaged with a moving field of view. However, this is not always the case in clinical applications with mosaicing removing dynamic information that can be of potential clinical use. He et al. (2010) proposed a method compensating for a range of movements, both operator induced as well as due to respiratory and cardiac motions, stabilising the field of view for improved monitoring of dynamic structural changes. In Mahé et al. (2015) dynamic video sequences on static mosaics were integrated, enabling the FOV extension without the associated loss of dynamic structural changes throughout the acquisition. A two-stage approach, of static mosaicing followed by stitching of the associated video segments was employed to reduce computational load. Visual artefacts at the seams across the mosaic were suppressed using a gradient-domain decomposition. Dynamic mosaics (infinite loops) from six organs (oesophagus, stomach, pancreas, bladder, biliary duct and colon) with various conditions were produced and clinically assessed by four experts. The produced visual summaries indicated higher level of consistency with the original data com-pared to static mosaicing.
Mosaicing techniques for FBEμ have, for the most part, concentrated in aligning and blending images with no a priori information on the acquisition trajectory, based exclusively on topology inference through the changes on the imaged structures. Such approaches call for large overlap amongst adjacent frames and, for effective, smooth results, can be computationally expensive. There has therefore been interest in incorporating such a priori trajectory information in the mosaicing process. Loewke et al. (2007b) utilised feedback from a robotic arm determining the five degree-of-freedom position and orientation of its end-effector (along with projective geometry) as an initial global rigid alignment amongst a frame sequence. Vyas et al. (2015) replaced the robotic arm with a six degree-of-freedom electromagnetic sensor positioned at the tip of the fibre-bundle in a proof-of-principle study. The positioning feedback from the sensor acted as a coarse global alignment followed from a fine tuning similar to Vercauteren et al. (2008). In Mahé et al. (2013) they used weak prior knowledge of the trajectory (spiral scan) to derive spatio-temporal associations within the frame sequence, linking overlapping frames from successive branches of the spiral scan and estimating optimal transforms similar to Vercauteren et al. (2006). While these approaches have been reported to improve the efficiency and/or the robustness of the mosaicing process, they require additional actuators/sensors at the tip of the fibre bundle, either to drive or to provide feedback on the scanning path. Such hardware additions are currently limiting their suitability for endoscopic applications, necessitating for future miniaturisation of the relevant technologies.
The previous studies described above dealt predominantly with free hand movements for producing an extended field of view through image mosaicing. While free-hand mosaicing is a very valuable tool, the ability to create customised scanning paths, ensuring a full imaging coverage of the region of interest, is also highly desirable. A number robotised distal scanning tips have been proposed (Erden et al., 2014; Rosa et al., 2013,2011; Zuo et al., 2015,2017b) facilitating customised and structured mosaic acquisition paths (such as spiral and raster scans) for FBEμ (Fig. 2). While a diverse set of architectures has been proposed, the plurality of solutions catered mostly for Minimal Invasive/Laparoscopic Surgery applications, with some prototypes suitable for endoscopic applications (Zuo et al., 2017a). Further miniaturisation is therefore imperative for facilitating robotically controlled scanning in endoscopy. In robotised scanning, the complex 3D surface of many of the examined structures, along with the lack of haptic feedback may result in loss of contact, or excessive contact/pressure between the fibre bundle and the imaged structure. Giataganas et al. (2015) created an adaptive probe mount that could maintain constant (low force magnitude) contact between the tissue and the imaging probe. Furthermore, the direct contact of the hard tip of the fibre bundle with the soft tissue can lead to tissue deformations, resulting in accumulative deviation off the desired path throughout a scan. There have been studies attempting to understand this soft tissue behaviour and provide feedback to the robotised scanner in an attempt to compensate for the anticipated path deviations. The feedback can be (i) through determining the loading-distance prior an automated scan and compensating respectively by adjusting the scan path (Erden et al., 2013), or (ii) through visual servoing (Rosa et al., 2013), estimating the imaged path in real-time through the mosaiced image data and adjusting accordingly to meet the desired scan path. While further research and development, especially in their miniaturisation is necessary, robotic scanning is anticipated to serve as a key milestone to the adoption of image mosaicing in a wide range of clinical endoscopic and laparoscopic procedures. Yet, the detailed discussion on such robotised scanning approaches is beyond the scope of this paper. Zuo and Yang (2017) has produced a thorough review on endomicroscopy for robot assisted intervention, providing details and discussion on a wide range of relevant studies.
4.2. Quantification
Aside from image mosaicing, there have been a limited number of image analysis studies for FBEμ images. These studies have predominantly concentrated on the detection and quantification of particles and structures that can act as indicators of pathological or physiological processes in the circulatory system, oropharyngeal, gastrointestinal and pulmonary tracts. For the most part, empirical, ad hoc observations, combined with simple, off-the-shelf image analysis approaches, have been employed. This section along with Table 3 and Fig. 3 provide a brief overview of the most relevant image analysis/quantification studies for fibred endomicro-scopic data.
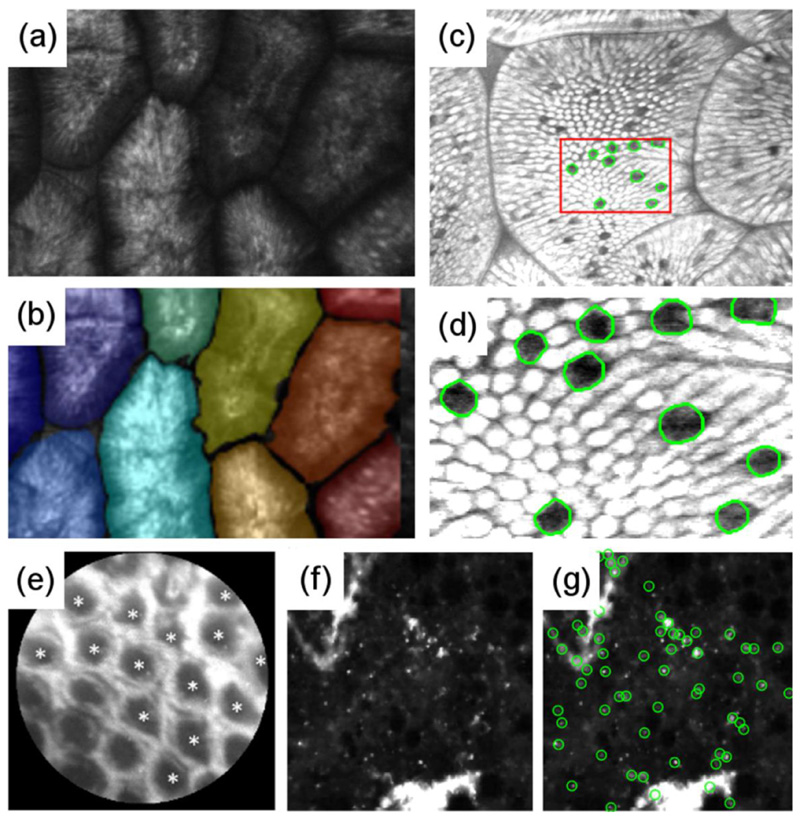
Fig. 3. Examples of image analysis performed on a range of organ systems.
(a-b) Segmentation of intestinal villi, and (c-d) detection of goblet cells within the villi. (e) Detection of intestinal crypts in colorectal polyps as a first step towards automated classification between benign and dysplastic epithelial tissue. (f-g) Detection of fluorescent stained bacteria, appearing as bright dots in (f), in the alveolar space of ovine distal lung. Images (a-b) are reproduced and adapted with permission from the Institute of Electrical and Electronics Engineers (IEEE) from Figures 1 and 4 respectively of the “Semiautomatic detection of villi in confocal endoscopy for the evaluation of celiac disease” by Boschetto et al. (2015b). Images (c-d) are reproduced and adapted with permission from IEEE from Figure 5 of the “Detection and density estimation of goblet cells in confocal endoscopy for the evaluation of celiac disease” by Boschetto et al. (2015a). Image (e) has been reproduced from Figure 8 of the “Quantitative analysis of ex vivo colorectal epithelium using an automated feature extraction algorithm for microendoscopy image data” by Prieto et al. (2016) under the Creative Commons Attribution (CC BY) 3.0 International License (https://creativecommons.org/licenses/by/3.0).
In the circulatory system, Savoire et al. (2004) proposed a method to estimate the velocity of Red Blood Cells (RBC) within micro-vessels from a single endomicroscopic frame, exploiting the skewing artefact introduced on fast moving RBC due to the relatively slow scanning speed of the vertical axis component (result-ing in circular RBCs appearing ellipsoidal). Perchant et al. (2007) developed algorithms to track and align a region of interest over consecutive frames for cell traffic analysis and blood velocity estimation. Huang et al. (2013) examined the variability in stained cardiac tissue structures imaged through FBEμ as a means for intraoperatively identifying nodal tissue in living rat hearts with potential application to neonatal open-heart surgery. In the orapharyngeal tract, Mualla et al. (2014) identified the borders and locations respectively of epithelial cells in the mucosa layer of vocal chords as the first step to analysing and quantifying structural changes.
In the gastrointestinal tract, Couceiro et al. (2012) developed a methodology that employed off-the-shelf algorithms for segmenting and quantifying intestinal crypts in endomicroscopic images as a potential indicator for Inflammatory Bowel Disease. Similarly, Prieto et al. (2016) employed crypt detection as a first step towards automated classification between benign and dysplastic epithelial tissue in colorectal polyps. Boschetto et al. (2015a,b,2016b) attempted to semi-automatically analyse and quantify fluorescent endomicroscopic images of the gastro-intestinal mucosa, as a first step to assist diagnosis and monitoring of coeliac Disease. Boschetto et al. (2016b,2015b) proposed methodologies for segmenting intestinal villi, while Boschetto et al. (2015a) proceeded in detecting and segmenting cells within the villi and differentiating between columnar and goblet cells of the epithelium.
In the pulmonary tract, Namati et al. (2008) analysed mice distal lung images and automatically quantified the number and size of alveolar sacs. Perez et al. (2017) applied a sequence of off-the-shelf image processing operations to count fluorescently labelled Mesenchymal Stem Cells injected into rat lungs, as a potential indicator for lung repair in radiation induced lung injury. Karam Eldaly et al. (2018) employed a fully unsupervised, hierarchical Bayesian approach for detecting bacteria labelled with a (green) fluorescent smart-probe (Akram et al., 2015a) within the, highly auto-fluorescent (also green) distal lung. The algorithm was an extension of McCool et al. (2016) for denoising along with outlier detection and removal in sparsely, irregularly sampled data. Such fully unsupervised approches offer a flexible and consistent methodology to deal with uncertainty in inference when limited amount of data or information is available. Seth et al. (2017, 2018) quantified bacterial and cellular load in the human lung adopting and adapting a learning-to-count (Arteta et al., 2014) approach, employing a multi-resolution, spatio-temporal template matching scheme using radial basis functions network.
5. Image understanding
Another component of the image computing pipeline is the higher-level understanding and exploitation of the acquired, reconstructed and sometimes processed data, in an attempt to extract clinically and biologically relevant information, and consequently guide the diagnostic process. Due to the nature of FBEμ data acquisition in a clinical setup, a large volume of continuous frame sequences is generated, sometimes surpassing 100 0 frames per video. These video sequences include uninformative/corrupted frames, off-target frames outside the examined anatomic structure and/or region of interest, as well as a range of on-target frames from healthy and pathological structures. This large, and sometimes very diverse, data volume acts as a major bottleneck in the analysis and quantification of the data, increasing the required human/computational resources, and potentially diluting the objectiveness of associated clinical procedure. The main body of FBEμ image understanding research to date can be broadly categorised into frame (i) classification (Tables 4–5 and Fig. 4), and (ii) content-based retrieval methods (Table 6).
Table 4. Overview of classification approaches for fibred endoscopic imaging employing traditional machine learning.
| Organ (System) | Classifying | References | Methodology | Comments |
|---|---|---|---|---|
| Pulmonary | Distal lung alveolar abnormalities. | Desir et al. (2012a), Désir et al. (2010,2012b), Hebert et al. (2012), Heutte et al. (2016), Koujan et al. (2018), Petitjean et al. (2009) and Saint-Réquier et al. (2009) |
Features: First Order Statistics, GLCMs, LBPs, SIFT, Scattering Transform, FREAK, ORB, Homomorphic filters, Structural Information (Canny and Sobel Edge Detectors), Sparse - Irregular LBPs, LQPs, HOGs, LDPs, Homogeneity, Spatial Frequency, Fractal Texture, Intensity, Wavelet and CNN Features. Classifiers: K-NN, SVMs, SVM-RFE, Gaussian Mixture Models, LDA, QDA, Random Forests, Generalised Linear Model, Gaussian Processes, Boosted Cascade of Classifiers, Neural Networks. Multiclass: One-vs-all and one-vs-one ECOCs, binary tree classification, Recursive SVM tree and Naïve Bayes. Other: Pruning trees for non-detection; feature selection (i.e. SDA, FSS and PCA) for dimensionality reduction; visual coding (Bag of Words, Sparse Coding and Fisher Kernel Coding) and classification on mosaics for enhanced classification performance. |
Simple and effective methodologies performing in most part binary classification. Results are positive indicating the potential strength of simple approaches in classifying endomicroscopic images. Primary limitations include (i) the limited scope of the classification, for example health VS pathological, when endomicroscopic sequences contain a plethora of frame classes, and (ii) the limited number of images used for training, testing and evaluation, making the proposed methodologies susceptible to a range of biases. |
| Informative frames within videos. | Leonovych et al. (2018) and Perperidis et al. (2016) | |||
| Cancerous nodules in airways and distal lung. | He et al. (2012), Rakotomamonjy et al. (2014) and Seth et al. (2016) | |||
| Gastro-intestinal | Oesophagus epithelial changes. | Ghatwary et al. (2017), Veronese et al. (2013) and Wu et al. (2017) | ||
| Intestinal adenocarcinoma. (eCLE) | Stefanescu et al. (2016) | |||
| Colorectal polyps. | André et al. (2012b) and Zubiolo et al. (2014) | |||
| Coeliac disease. (ecle) | Boschetto et al. (2016a) | |||
| Oropharyngeal | Pathological epithelium. | Jaremenko et al. (2015) and Vo et al. (2017) | ||
| Brain | Brain tumours (glioma and meningioma). | Kamen et al. (2016) and Wan et al. (2015) | ||
| Ovaries | Epithelial changes | Srivastava et al. (2005,2008) |
Table 5. Overview of classification approaches for fibred endoscopic imaging going beyond traditional machine learning.
| Organ (System) | Classifying | References | Methodology | Comments |
|---|---|---|---|---|
| Pulmonary | Cancerous nodules in airways. | Gil et al. (2017) | Unsupervised classification (compensating for limited data availability) using graph representation and community detection algorithms. | Early FBEμ classification approaches going beyond the traditional machine learning pipeline, exploring methods such as Convolutional Neural Networks (off the self as well as custom), transfer learning, unsupervised learning and multi-modal learning at a latent space. The results are very promising. Yet, more data, both in terms of numbers as well as in terms of diversity are necessary. Furthermore, custom solutions, taking into consideration the inherent FBEμ imaging properties, could further enhance the classification performance. |
| Gastro-intestinal | Oesophagus epithelial changes. | Hong et al. (2017) and Aubreville et al. (2017) | Custom CNN architecture for the multi-class frame classification. | |
| Oropharyngeal | Pathological epithelium. | Aubreville et al. (2017) | Full-training of LeNet-5 and shallow fine-tuning the Inception v3 (using the ImageNet database). | |
| Breast | Cancerous breast nodules. | Gu et al. (2017) | Multi-modal (FBEμ mosaics and histology) classification mapping the original features to a latent space for improved SVM performance. |
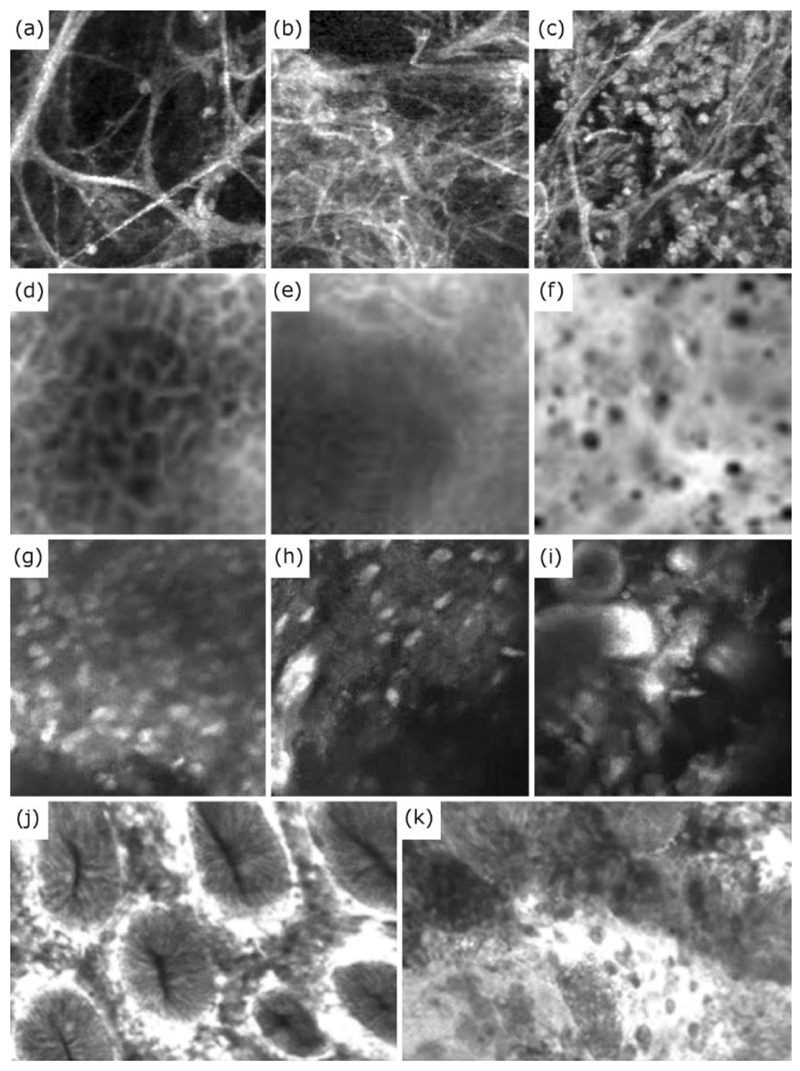
Fig. 4. Examples of structural changes observed in OEM images across variety of organ systems and conditions. These structural changes have been used to classify/detect a range of clinically relevant pathologies.
(a-c) Difference in tissue structure in the alveoli structures of the distal lung, indicating (a) healthy and (b) pathological elastin strands, as well as (c) alveoli sacs flooded with cells. (d-f) Difference between (d) healthy and (f) cancerous oral epithelium, along with (e) an example of oral epithelium with limited textural information where classification can be challenging. (g-i) Difference between (g-h) Glioblastoma and (i) Meningioma brain tumour images. (j-k) Difference between (j) healthy colon mucosa and (k) adenocarcinoma. Images (d-f) have been reproduced (cropped) from Figure 6 of the “Automatic Classification of Cancerous Tissue in Laserendomicroscopy Images of the Oral Cavity using Deep Learning” by Aubreville et al. (2017) under the Creative Commons Attribution (CC BY) 4.0 International License. Images (g-i) have been reproduced from Figure 3 of the “Automatic Tissue Differentiation Based on Confocal Endomicroscopic Images for Intraoperative Guidance in Neurosurgery” by Kamen et al. (2016) under CC BY 4.0. Images (j) and (k) have been reproduced (from Figures 2 and 3 respectively of the “Computer Aided Diagnosis for Confocal Laser Endomicroscopy in Advanced Colorectal Adenocarcinoma” by Ştefănescu et al. (2016) under CC BY 4.0 (https://creativecommons.org/licenses/by/4.0).
Table 6. Overview of image retrieval approaches for fibred endoscopic imaging.
| Topic | References | Methodology | Comments |
|---|---|---|---|
| Image retrieval through low-level visual features | André et al. (2009a) | Bag of Visual Words (k-means clustering) of multi-scale SIFT descriptors extracted from regularly distributed circular regions. | Thorough methodologies for image and video retrieval based solely on low-level information extracted from images. Due to lack of relevant ground truth, methodologies were evaluated as binary classification tasks (instead of retrieval). |
| André et al. (2009b) | Introduce (i) spatial information between local features by exploiting the co-occurrence matrix of their visual words (ii) temporal relationship across frames through mosaicing. | ||
| André et al. (2010) | Deriving visual words from individual frames and weighting the contributions of local regions through the relevant overlap rate derived during mosaicing. | ||
| André et al. (2012b,2011b) | Combining and clinically testing above approaches as a binary classification (kNN) between neoplastic/benign colonic epithelium. | ||
| André et al. (2011a) | (i) Generate the “perceived similarity” ground truth (manual assessment - Likert scale), and (ii) learn an adjusted similarity/distance metric (linear transform) for optimal mapping of video signatures (histograms of visual words). | First attempt to evaluate directly the performance of endomicroscopic video retrieval, through generating the perceived similarity of ground truth. | |
| Image retrieval combining low-level visual features with high-level semantic contex | André et al. (2012a,c) | Fisher-based approach transforming visual word histograms to 8 binary semantic concepts. Combine with adjusted similarity distance to improve “perceived similarity”. | Bridging the semantic gap between low-level visual features, extracted from the images, and high-level clinical knowledge, generated through human perception. |
| Watcharapichat (2012) | Gabor filter and Earth Mover’s Distance based retrieval enhanced through iterative “relevance feedback” and Isomap dimensionality reduction. | ||
| Tous et al. (2012) | Retrieval via (i) low-level, image-based features (LBPs & k-NN with Euclidian or Manhattan distances), (ii) high level key-word semantic descriptions (Apache Lucene search engine), and (iii) third party software compatibility through MPEG Query Format & JPEG Search standards. | ||
| Other image retrieval approaches | Kohandani Tafresh et al. (2014) | Semi-automated query adaptation of André et al. (2011b) via (i) temporal segmentation based on kinetic stability (Euclidean distance of SHIFT descriptors across consecutive frames), and (ii) manual selection of spatially stable segments. | Adaptations of André et al. (2011b) enhancing retrieval performance. |
| Gu et al. (2017) | Unsupervised, multimodal graph mining (i) deriving similar (cycle consistency) and dissimilar (geodesic distance) FBEμ and histology frame pairs, (ii) learning discriminative features in the associated latent space. |
5.1. Image classification
Classification of frames on pre-determined, clinically defined cohorts based on their content is currently the most investigated area of FBEμ image computing research. An abundance of studies have applied binary as well as multi-class classification on endomicroscopic images of a range of organ systems in an attempt to identify cancer in ovarian epithelium (Srivastava et al., 2005,2008), abnormalities in distal lung alveolar structures (Desir et al., 2012a; Désir et al., 2010,2012b; Hebert et al., 2012; Heutte et al., 2016; Koujan et al., 2018; Petitjean et al., 2009; Saint-Réquier et al., 2009), informative frames in brain (Izadyyazdanabadi et al., 2017; Izadyyazdanabadi et al., 2018) and pulmonary videos (Leonovych et al., 2018; Perperidis et al., 2016), cancerous nodules in the airways (Gil et al., 2017; He et al., 2012; Rakotomamonjy et al., 2014) and distal lung (Seth et al., 2016), pathological epithelium in the oropharyngeal cavity (Aubreville et al., 2017; Jaremenko et al., 2015; Vo et al., 2017), changes in oesophageal epithelium in cases of Barrett’s oesophagus (Ghatwary et al., 2017; Hong et al., 2017; Veronese et al., 2013; Wu et al., 2017), adenocarcinoma (Stefanescu et al., 2016), colorectal polyps (André et al., 2012b; Zubiolo et al., 2014) and celiac disease (Boschetto et al., 2016a) in intestinal epithelium, neoplastic tissue in breast nodules (Gu et al., 2017), as well as two types of common brain tumours, glioblastoma and meningioma (Kamen et al., 2016; Murthy et al., 2017; Wan et al., 2015). Methodologically, most of the aforementioned studies employed the same basic structure, defining a hand-crafted feature space descriptive of the underlying imaged structure, training a range of classifiers, to distinguish between pre-determined frame categories. For organs/structures that do not exhibit any auto-fluorescence at the imaging wavelengths, fluorescence dyes such as methylene blue and fluorescein, and molecular probes (He et al., 2012) were employed to generate the necessary fluorescent signal.
Commonly used feature descriptors include (i) first order image statistics, (ii) structural information through Skeletonisation, Sobel and Canny Edge Detectors, etc. (iii) Haralick’s texture parameters derived through gray Level Co-occurrence Matrices (GLCM), (iv) Local Binary Patterns (LBP) and their variation of Local Quinary Patterns (LQP), and (v) Scale Invariant Feature Transforms (SHIFT). Other less adopted descriptors employed as discriminative features include (i) spatial frequency based features extracted at Fourier domain (Srivastava et al., 2005,2008), (ii) fractal analysis (Stefanescu et al., 2016), (iii) Scattering transform (Rakotomamonjy et al., 2014; Seth et al., 2016), (iv) Fast Retina Keypoint (FREAK) (Wan et al., 2015), (v) oriented FAST and rotated BRIEF (ORB) (Wan et al., 2015), (vi) Histogram of orientated Gradients (HOG) (Gu et al., 2016; Vo et al., 2017), (vii) textons (Gu et al., 2016), (viii) Local Derivative Patterns (LDP) (Vo et al., 2017), as well as (ix) features extracted from Convolutional Neural Networks (CNN) prior to the fully connected layer employed for computing each class score (Gil et al., 2017; Vo et al., 2017). Leonovych et al. (2018) introduced Sparse Irregular Local Binary Patterns (SILBP), an adaptation of LBPs taking into consideration the sparse, irregular sampling imposed by the imaging fibre bundle on FBEμ images. Feature spaces combining two or more of the above descriptors are also frequent, with descriptors customarily extracted from the whole image, yet in some cases, regular or randomly distributed sub-windows/patches have been used, either on their own, or in conjunction to the whole image feature space.
A number of well-established classifiers have been assessed, including (i) k-Nearest Neighbours (kNN) (André et al., 2012b; Désir et al., 2010; Hebert et al., 2012; Saint-Réquier et al., 2009; Srivastava et al., 2005,2008), (ii) Linear and Quadratic Discriminant Analysis (LDA and QDA) (Leonovych et al., 2018; Srivastava et al., 2005,2008), (iii) Support Vector Machines (SVM) and their adaptation with Recursive Feature Elimination (SVM-RFE) (Désir et al., 2010, 2012b; Jaremenko et al., 2015; Leonovych et al., 2018; Petitjean et al., 2009; Rakotomamonjy et al., 2014; Saint-Réquier et al., 2009; Vo et al., 2017; Wan et al., 2015; Zubiolo et al., 2014), (iv) Random Forests (RF) and variants such as Extremely Randomised Trees (ET) (Desir et al., 2012a; Heutte et al., 2016; Jaremenko et al., 2015; Leonovych et al., 2018; Seth et al., 2016; Vo et al., 2017), (v) Gaussian Mixture Models (GMM) (He et al., 2012; Perperidis et al., 2016), (vi) Boosted Cascade of Classifiers (Hebert et al., 2012), (vii) Neural Networks (NN) (Stefanescu et al., 2016), (viii) Gaussian Processes Classifiers (GPC), and (ix) Lasso Generalised Linear Models (GLM) (Seth et al., 2016). Most studies employed leave-k-out and k-fold cross validation to assess the predictive capacity of the proposed methodology on limited, preannotated frames. In an attempt to enhance the classification performance and/or reduce the computational workload required for training and testing, some studies incorporated additional steps in the classification pipeline. In particular, feature selection (dimensionality reduction in feature space) such as Stepwise Discriminant Analysis (SDA), Forward Sequential Search (FSS), and Principal Component Analysis (PCA) were also used (Perperidis et al., 2016; Srivastava et al., 20 05,20 08) prior to the classification process. Furthermore, visual coding schemes, such as Bag-of-Words, Fisher Kernel Coding and Sparse Coding (Kamen et al., 2016; Vo et al., 2017; Wan et al., 2015), as well as reduction of non-detection, minimising the incorrectly classified images through rejection mechanisms (Desir et al., 2012a; Heutte et al., 2016), have been investigated.
Classification of endomicroscopic images has predominantly concentrated in binary cases, with a very limited number of studies having attempted multi-class classification (Boschetto et al., 2016a; Ghatwary et al., 2017; Hong et al., 2017; Koujan et al., 2018; Veronese et al., 2013; Wu et al., 2017; Zubiolo et al., 2014). To this end, Boschetto et al. (2016a) employed a multi-class Naïve Bayes classifier. Koujan et al. (2018) adopted the One-Versus-All (OVA) Error Correcting Output Codes (ECOC), a popular method (along with other ECOCs such as One-Versus-One and Ordinal) for multiclass classification using binary classifiers. Ghatwary et al. (2017) and Veronese et al. (2013) tackled the multi-class problem as a pre-determiπed sequence (tree) of binary classifications (through SVM), while Zubiolo et al. (2014) employed graph theory tools (minimum cut) to recursively estimate the optimal associated bipartitions (large SVM margin). Hierarchical (tree) binary classifications can potentially reduce the classification complexity from linear for OVA to logarithmic. Wu et al. (2017) improved the performance of multi-class classification performance incorporating unlabelled images through an adaptation of semi-supervised approach called Label Propagation method introduced by Zhou et al. (2003).
There have recently been some studies that do not follow the same basic structure of training a classifier on a hand-crafted feature space descriptive of the underlying imaged structures. Gil et al. (2017) proposed an unsupervised classification approach to compensate for the limited quantity of data available for training and testing decision support systems. The methodology used graph representation to codify feature space connectivity followed by community detection algorithms (Cazabet et al., 2010), representing space topology and detecting associated image communities. Gu et al. (2016) incorporated features extracted from endomicroscopy mosaics as well as associated histology images, to a supervised framework, mapping the original features to a latent space by maximising their semantic correlation. The derived latent features outperformed mono-modal features in binary classification (SVM) of breast cancer images. Furthermore, recent advances of Deep Learning architectures, such as Convolutional Neural Networks (CNN), have resulted in numerous powerful tools for binary or multi-class image classification, without the need for explicit definition of feature descriptors. Hong et al. (2017) proposed a custom CNN architecture with for the multi-class classification of epithelial changes in Barrett’s oesophagus. Aubreville et al. (2017) adopted and adapted two established CNN architectures for the detection of cancerous tissue in the oral cavity, (i) a patch-based classification based on full-training of LeNet-5 (Lecun et al., 1998), as well as (ii) a whole image classification based on shallow fine-tuning the Inception v3 network (Szegedy et al., 2016) pre-trained using ImageNet database (Deng et al., 2009). Similarly, Izadyyazdanabadi et al. (2017) fully-trained AlexNet (Krizhevsky et al., 2012) and GoogleNet (Szegedy et al., 2015) for the detection of diagnostic frames in brain endomicroscopy. Murthy et al. (2017) presented a novel multi-stage CNN, discarding images classified with high confidence at early stages, concentrating on more challenging images at subsequent, expert shallow networks. The proposed network demonstrated substantial improvement on traditional feature/classifier as well as CNN architectures when classifying (binary endomicroscopic brain tumour images. Izadyyazdanabadi et al., 2018) compared the classification performance amongst fully training CNNs from scratch against transfer learning through fine-tuning, shallow (fully connected layers) or deep (whole network), of pre-trained networks using conventional image databases such as ImageNet. Similar to Tajbakhsh et al. (2016), fine-tuning was found to be able to provide better or at least similar classification performance to training from scratch on limited medical image databases.
5.2. Image retrieval
While a less prolific research area to the closely related image classification, a number of studies have developed Content Based Image Retrieval (CBIR) frameworks for endomicroscopic data. Unlike image classification, that groups images to a number of predetermined (trained) classes, CBIR methods search a database to find (and return) the images that are most similar (based to some image extracted feature set) to a given (query) image. In an early attempt, André et al. (2009a) adapted the Bag of Visual Words (BVW) approach of Sivic and Zisserman (2008) to endomicroscopic images, containing discriminative texture information (SIFT) extracted across a regular grid of overlapping disks at various scales (radius). André et al. (2009b) introduced to the retrieval process (i) spatial relationship between local features by exploiting the co-occurrence matrix of the visual words labelling the local features in each image, as well as (ii) temporal relationship between successive frames in a video sequence, by including image mosaics projecting the temporal dimension onto an extended field of view. In an attempt to avoid computationally costly non-rigid deformations required for a robust mosaic image, André et al. (2010) proposed a video retrieval approach named Bag of Overlap-Weighted Visual Words (BOWVW). BOWVW computed independently the BVW signatures from individual frames within a video sub-sequence, as per André et al. (2009a), and weighted the asso-ciated contributions (frame overlap rate) of their individual dense local regions to a single signature for the sub-sequence. The subsequence signatures were then incorporated (normalised sum) to a single signature for a whole video. The aforementioned studies were compared and combined to a single, integrated video retrieval approach (André et al., 2011b). However, due to the challenging task of generating ground truth for the evaluation of content based retrieval, the proposed methodology was evaluated as a binary classification task between neoplastic and benign epithelium in Colonic Polyps (André et al., 2012b).
In an attempt to address the challenging evaluation of true retrieval performance (André et al., 2011a) (i) developed a tool for generating the perceived similarity ground truth, enabling the direct evaluation of endoscopic video retrieval, and (ii) employed this ground truth information by employing a similarity distance learning technique to derive an optimal mapping of video signatures, improving the discrimination of similar video pairs. Another challenge in retrieval systems is bridging the “semantic gap” between the (sometimes conflicting) low-level visual features, extracted computationally from the images, and high-level clinical knowledge, generated through human perception. In clinical practice, new data are usually interpreted through similarity-based reasoning, combining both visual features and semantic concepts. André et al. (2012a,c) defined 8 mid-level binary semantic concepts that were either present or not in a colonic endomicroscopic video sequence. A Fisher-based approach was utilised to estimate the expressive power of each of the visual words (estimated as per André et al., 2011b) to each of these 8 semantic concepts. The derived semantic signatures were found to be informative and consistent with the low-level visual features, providing some relevant semantic translation, more familiar to the clinicians’ own language, of the visual retrieval outputs. In a separate attempt to alleviate the semantic gap, Watcharapichat (2012) proposed an interactive approach that the user has the ability to provide “relevance feedback” on the previously retrieved content, enabling the system to iteratively improve upon the search results. The feedback was combined on Isomap dimensionality reduction for improved performance and efficiency. Tous et al. (2012) developed a multi-media retrieval software enabling querying via low-level, image-based features as well as high level key-word semantic descriptions. The software ensured compatibility with third party applications through interface compliance with the MPEG Query Format (ISO/IEC 15938-12:2008) and JPEG Search (ISO/IEC 24800) standards.
In an attempt to improve the retrieval performance of André et al. (2011b), Kohandani Tafresh et al. (2014) introduced a simple and efficient semi-automated approach allowing clinicians to create more meaningful queries than unprocessed endomicro-scopic video sequences. The approach automatically temporally segmented endomicroscopic video sequence based on kinematic stability assessment, with informative sub-segments assumed spatially stable. Then, the clinician could manually select stable subsequences of interest generating a new augmented query video, leading to more reproducible and consistent retrieval results. Gu et al. (2017) proposed Unsupervised Multimodal Graph Mining (UMGM), a framework mining the latent similarity amongst endomicroscopic mosaics and histology patches for enhanced CBIR performance. While an extension of Gu et al. (2016), UMGM employed graph-based analysis over a large collection of histology patches without supervised information (matching pairs), minimising latent space distance between similar pairs while maximising the distance between dissimilar pairs.
6. Limitations and opportunities
Fibre bundle based endomicroscopy (FBEμ) offers several enabling capabilities for diagnostic and interventional procedures in a range of clinical indications. The literature to date has established a solid understanding of the limitations inherent to imaging through coherent fibre bundles, making substantial progress in terms of associated image computing methodologies. Characteristic examples of concentrated research effort have been (i) compensating for the honeycomb effect through the irregular, sparse sampling introduced along the coherent fibre bundle, and (ii) extending the limited field of view, a direct consequence of the fibre bundle miniaturisation for guidance through an endoscope’s working channel, through mosaicing spatially adjacent frames. Yet, FBEμ is a still a fledging imaging technology with tremendous potential for improvement assuming the research/technical challenges can be overcome. Throughout this review the following major image computing challenges/opportunities have been identified.
6.1. Image reconstruction
Image reconstruction research has concentrated predominantly in compensating for the honeycomb effect on raw FBEμ images, a consequence of the sparse, irregular sampling through the coherent fibre bundle. Yet, even straightforward approaches such as bilinear interpolation between the cores, as currently used in clinical practice (Cellvizio, Mauna Kea Technologies), have been found to generate satisfactory results, with subsequent improvements perceived as predominantly aesthetic. In contrast, very limited research has been performed compensating for other inherent artefacts known to have limiting effect on the imaging capabilities of the technology, such as (i) variable coupling and background response (due to irregularities amongst cores physical properties) and (ii) inter-core coupling across neighbouring cores. Optimal solutions to these problems can have a direct impact on the imaging signal to noise ratio, contrast and potentially the spatial resolution (computationally supressing cross coupling can conceivably enable smaller inter-core distances). Furthermore, with the notable exception of the work by Vercauteren et al. (2013), there have been no studies on multi-colour data acquisition, investigating and compensating for the effect of the aforementioned coupling/background artefacts along with other inherent limitations such as spectral mixing. Such enhanced imaging capabilities are of paramount importance into the advancing molecular endomicroscopy which has stringent requirements in terms of light detection (preferably at multiple wavelengths), especially when imaging small targets such as bacteria superimposed upon highly fluorescent background structures. Finally, existing reconstruction approaches tend to concentrate on a single limitation in FBEμ imaging, intrinsic to the coherent fibre bundle characteristics, either ignoring or downplaying the relevance of other limitations in the reconstructed images. In real-world applications this is rarely the case. There is therefore scope for the development of a unified image reconstruction methodology that compensates for a range of limitations, including but not limited to irregular sampling, varying coupling efficiency and inter-core coupling along with additional challenges introduced in multi-spectral acquisition such as chromatic aberrations and spectral mixing. This is of greater importance to widefield FBEμ, where poor sectioning already reduces limits of detection and subsequently the imaging capabilities of the technology. The emergence of deep-imaging (Wang, 2016), employing data-driven deep learning (Convolutional Neural Networks) for image formation/reconstruction from raw, irregularly sampled data, is expected to generate tremendous opportunities in biomedical imaging in general and FBEμ in extension. Ravì et al. (2018) for example has recently demonstrated a deep-learning based superresolution pipeline for FBEμ. Yet, this direction, while very promising, will eventually lead to additional challenges regarding the need for large amounts of carefully chosen, and meaningful “gold-standard” data to form the basis of the learning and inference processes. Furthermore, convolution filers have been the cornerstone of state-of-the-art deep learning approaches for classical regularly sampled images. Yet, FBEμ images are sparsely and irregularly sampled through a coherent fibre bundle subsequently reconstructed to a regularly sampled image, potentially introducing uncertainty to the image reconstruction process. There is therefore scope for developing novel deep-learning architectures applied directly on the irregularly sampled data.
6.2. Pathology detection and quantification
There is a substantial body of work on the classification of frames into clinically relevant groupings, based predominantly on the binary classification between healthy and pathological frames over a range of organ systems and associated pathologies. Generating hand-crafted feature descriptors and training a binary or multiclass classifier has been shown to generate reliable results in parsing videos and detecting abnormalities in endomicroscopic frame sequences. Yet, there has been very limited work on the semantic segmentation and subsequent pathology detection and quantification for FBEμ frames and mosaics. IIzadyyazdanabadi et al. (2018) for example proposed a weakly supervised CNN architecture for localising brain tumours in eCLE images. Pathology quantification will be imperative to any viable Computer-aided detection (CADe) and Computer-aided diagnosis (CADx) system. Furthermore, the existing image quantification studies have primarily adopted, empirical, ad hoc methodologies along with heuristic parameter estimation using hard thresholds, tested on very limited data. As a result, this can lead to poor generalisation as well as limited clinical utility. There is therefore an opportunity and need for the development of customised and robust methods that analyse and quantify the contents of FBEμ images, which when combined with state of the art detection and classification approaches, can identify and quantify pathology as the cornerstone for invaluable CAD systems. Ultimately, in certain clinical applications, pathology detection and quantification will also be aided by targeted molecular imaging agents.
6.3. Integration
To date, the tasks of image reconstruction, analysis and understanding have been dealt with independently, with notable exceptions the works of Hu et al. (2010), Ravì et al. (2018) and Vercauteren et al. (2006) that employ image mosaicing techniques to generate a super-resolved reconstructed image. Consequently, image reconstruction has been optimised primarily for user expe-rience. While user experience in a clinical setting is an extremely important factor, contributing to the success of the endomicroscopic procedure through effective guidance and on-target sampling, it is not necessarily a primary concern during the automated detection and quantification of pathology. Moreover, most studies have employed unimodal information derived exclusively from endomicroscopic images, with a small number of multimodal attempts integrating histological (Gu et al., 2017,2016), demographic and clinical (Seth et al., 2016) information in the decision-making pipeline. Yet, endomicroscopy (predominantly due to limited FOV and guidance capabilities) is unlikely to be used as a stand-alone tool in the clinical workflow. FBEμ will be integrated as part of a multimodality approach consolidating imaging across a range of scales, from organ level (radiology) to cellular level (microscopy), along with other clinically relevant information. There is therefore scope for (i) incorporating multi-modal information in the decision-making algorithms, and (ii) integrating the reconstruction, analysis and understanding of endomicroscopic images to novel unified frameworks with joint loss functions, optimised for the task in question such as identifying and quantifying pathology.
6.4. Data availability
Recent developments in Convolutional Neural Networks (CNNs) have acted as vehicle to substantial advances to image analysis and understanding across an ever-increasing range of areas, including medical imaging, with applications in image reconstruction, classification, segmentation and registration. Yet, to date there have been just a limited number studies employing Convolutional Neutral Networks for the classification and retrieval on FBEμ frames and mosaics. Instead, image understanding tasks has been tackled predominantly through traditional machine learning pathways, defining hand-crafted feature descriptors and subsequently training a binary or multi-class classifier on this feature set. A key con-straint in the effective adaptation and adoption of the technology (CNNs) has been, to a large extent, the limited data and associated annotations available. In particular, the plurality of FBEμ classification/retrieval studies have employed limited data, ranging between 100 and 200 annotated frames for combined training, validation and testing, with several studies using datasets of less than 100 frames. Furthermore, the available data have for the most part been acquired from a single clinical site and many times from a single operator, introducing potential bias and hindering the ability of the proposed methodologies for widespread generalisation. Similarly, there is often a lack of a gold reference standard and manual annotations can be weak, demonstrating large inter- and intraoperator variability. In tasks such as image restoration and analysis, the proposed methodologies of assessment have been constrained to simple simulated data, test targets, and in some cases to a very limited number of biological samples. There is therefore a need for the development of (i) large data repositories, containing a diverse collection of frames sequences acquired from different operators at multiple sites across the world, with easy access for the endomicroscopy research community, (ii) associated manual annotations, ideally from multiple operators with varied level of expertise, with quantifiable inter- and intra-operator variability. Providing standardised annotation tools, available alongside the data depositories can further enhance the consistency and robustness of these annotations.
6.5. Real-time capability
In much of the FBEμ image computing literature to date, the proposed methodologies have limited or no capacity for real-time application. Given the potential for FBEμ to perform in vivo, in situ assessment, at microscopic level (optical biopsies), the lack of realtime capability impairs the clinical application for such algorithms. There is therefore a necessity to design and test methodologies, from the ground up, with particular consideration for their real-time potential under pragmatic computational resources (at the time of testing and near future) for the intended clinical application.
7. Conclusions
Fibre bundle based endomicroscopy (FBEμ) is a relatively new medical imaging modality. Yet, the real time, microscopic imaging capabilities, commonly referred to as optical biopsy, make FBEμ a very promising diagnostic and monitoring tool, particularly when combined in the future with molecular imaging agents. Imaging through a miniaturised coherent fibre bundle, typically guided to the region of interest through the working channel of an endo-scope, imposes a number of inherent limitations to the technology. These limitations have motivated a diverse and ever-growing area of research for tailored image computing solutions. To date, considerable progress has been made in (i) image reconstruction, compensating for the honeycomb introduced by the coherent fibre bundle, (ii) extending the limited field of view through mosaicing adjacent frames, and (iii) classifying frames amongst two or more clinically relevant categories. However, there are still significant research challenges and opportunities remain for FBEμ to realise its full clinical potential.
Acknowledgements
Funding: This work was supported by the Engineering and Physical Sciences Research Council (EPSRC, United Kingdom) [EP/K03197X/1 and NS/A00 0050/1], as well as the Wellcome Trust [203145Z/16/Z and 203148/Z/16/Z].
Footnotes
Declaration of Competing Interest
Professor Vercauteren is a shareholder of Mauna Kea Technologies (Paris, France). Professor Dhaliwal is founder and shareholder of Edinburgh Molecular Imaging (Edinburgh, UK) and has in the past received funds for travel and meeting attendance from Mauna Kea Technologies (Paris, France).
Contributor Information
Antonios Perperidis, Email: Antonios.Perperidis@gmail.com.
Kevin Dhaliwal, Email: Kev.Dhaliwal@ed.ac.uk.
Stephen McLaughlin, Email: S.McLaughlin@hw.ac.uk.
Tom Vercauteren, Email: Tom.Vercauteren@kcl.ac.uk.
References
- Abbaci M, Breuskin I, Casiraghi O, De Leeuw F, Ferchiou M, Temam S, Laplace-Builhé C. Confocal laser endomicroscopy for non-invasive head and neck cancer imaging: a comprehensive review. Oral Oncol. 2014;50:711–716. doi: 10.1016/j.oraloncology.2014.05.002. [DOI] [PubMed] [Google Scholar]
- Abu Dayyeh BK, Thosani N, Konda V, Wallace MB, Rex DK, Chauhan SS, Hwang JH, Komanduri S, Manfredi M, Maple JT, Murad FM, Siddiqui UD, Banerjee S. ASGE technology committee systematic review and metaanalysis assessing the asge pivi thresholds for adopting real-time endoscopic assessment of the histology of diminutive colorectal polyps. Gastrointest Endosc. 2015;81:502.e501–502.e516. doi: 10.1016/j.gie.2014.12.022. [DOI] [PubMed] [Google Scholar]
- Akram AR, Avlonitis N, Lilienkampf A, Perez-Lopez AM, McDonald N, Chankeshwara SV, Scholefield E, Haslett C, Bradley M, Dhaliwal K. A labelled-ubiquicidin antimicrobial peptide for immediate in situ optical detection of live bacteria in human alveolar lung tissue. Chem Sci. 2015a;6:6971–6979. doi: 10.1039/c5sc00960j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akram AR, Avlonitis N, Lilienkampf A, Perez-Lopez AM, McDonald N, Chankeshwara SV, Scholefield E, Haslett C, Bradley M, Dhaliwal K. A labelled-ubiquicidin antimicrobial peptide for immediate in situ optical de-tection of live bacteria in human alveolar lung tissue. Chem Sci. 2015b doi: 10.1039/c5sc00960j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amidror I. Scattered data interpolation methods for electronic imaging systems: a survey. J Electron Imaging. 2002;11:157–176. [Google Scholar]
- André B, Vercauteren T, Ayache N. Content-based retrieval in endomicroscopy: toward an efficient smart atlas for clinical diagnosis; MICCAI International Workshop on Medical Content-Based Retrieval For Clinical Decision Support; Toronto, Canada. Springer Berlin Heidelberg; 2012a. pp. 12–23. [Google Scholar]
- André B, Vercauteren T, Buchner AM, Krishna M, Ayache N, Wallace MB. Software for automated classification of probe-based confocal laser endomicroscopy videos of colorectal polyps. World J Gastroenterol. 2012b;18:5560–5569. doi: 10.3748/wjg.v18.i39.5560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- André B, Vercauteren T, Buchner AM, Wallace MB, Ayache N. Endomicroscopic video retrieval using mosaicing and visualwords; IEEE International Symposium on Biomedical Imaging: From Nano to Macro; Rotterdam, Netherlands. 2010. pp. 1419–1422. [Google Scholar]
- André B, Vercauteren T, Buchner AM, Wallace MB, Ayache N. Retrieval evaluation and distance learning from perceived similarity between endomicroscopy videos; International Conference on Medical Image Computing and Computer-Assisted Intervention; Toronto, Canada. Springer Berlin Heidelberg; 2011a. pp. 297–304. [DOI] [PubMed] [Google Scholar]
- André B, Vercauteren T, Buchner AM, Wallace MB, Ayache N. A smart atlas for endomicroscopy using automated video retrieval. Med Image Anal. 2011b;15:460–476. doi: 10.1016/j.media.2011.02.003. [DOI] [PubMed] [Google Scholar]
- André B, Vercauteren T, Buchner AM, Wallace MB, Ayache N. Learning semantic and visual similarity for endomicroscopy video retrieval. IEEE Trans Med Imaging. 2012c;31:1276–1288. doi: 10.1109/TMI.2012.2188301. [DOI] [PubMed] [Google Scholar]
- André B, Vercauteren T, Perchant A, Buchner AM, Wallace MB, Ayache N. Endomicroscopic image retrieval and classification using invariant visual features; IEEE International Symposium on Biomedical Imaging: From Nano to Macro; Boston, USA. 2009a. pp. 346–349. [Google Scholar]
- André B, Vercauteren T, Perchant A, Buchner AM, Wallace MB, Ayache N. Introducing space and time in local feature-based endomicroscopic image retrieval; MICCAI International Workshop on Medical Content-Based Retrieval for Clinical Decision Support; London, UK. Springer Berlin Heidelberg; 2009b. pp. 18–30. [Google Scholar]
- Arteta C, Lempitsky V, Noble JA, Zisserman A. In: Fleet D, Pajdla T, Schiele B, Tuytelaars T, editors. Interactive object counting; Computer Vision - ECCV 2014: 13th European Conference; Zurich, Switzerland. September 6-12, 2014; Springer International Publishing; 2014. pp. 504–518. Proceedings, Part III, Cham. [Google Scholar]
- Aslam T, Miele A, Chankeshwara SV, Megia-Fernandez A, Michels C, Akram AR, McDonald N, Hirani N, Haslett C, Bradley M, Dhaliwal K. Optical molecular imaging of lysyl oxidase activity - detection of active fibrogenesis in human lung tissue. Chem Sci. 2015;6:4946–4953. doi: 10.1039/c5sc01258a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aubreville M, Knipfer C, Oetter N, Jaremenko C, Rodner E, Denzler J, Bohr C, Neumann H, Stelzle F, Maier A. Automatic classification of cancerous tissue in laserendomicroscopy images of the oral cavity using deep learning. Sci Rep. 2017;7:11979. doi: 10.1038/s41598-017-12320-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avlonitis N, Debunne M, Aslam T, McDonald N, Haslett C, Dhaliwal K, Bradley M. Highly specific, multi-branched fluorescent reporters for analysis of human neutrophil elastase. Org Biomol Chem. 2013;11:4414–4418. doi: 10.1039/c3ob40212f. [DOI] [PubMed] [Google Scholar]
- Ba C, Palmiere M, Ritt J, Mertz J. Dual-modality endomicroscopy with co-registered fluorescence and phase contrast. Biomed Opt Express. 2016;7:3403–3411. doi: 10.1364/BOE.7.003403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedard N, Quang T, Schmeler K, Richards-Kortum R, Tkaczyk TS. Realtime video mosaicing with a high-resolution microendoscope. Biomed Opt Express. 2012;3:2428–2435. doi: 10.1364/BOE.3.002428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedard N, Tkaczyk TS. Snapshot spectrally encoded fluorescence imaging through a fiber bundle. J Biomed Opt. 2012;17:080508. doi: 10.1117/1.JBO.17.8.080508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergen T, Wittenberg T. Stitching and surface reconstruction from endo-scopic image sequences: a review of applications and methods. IEEE J Biomed Health Inform. 2016;20:304–321. doi: 10.1109/JBHI.2014.2384134. [DOI] [PubMed] [Google Scholar]
- Bharali DJ, Klejbor I, Stachowiak EK, Dutta P, Roy I, Kaur N, Bergey EJ, Prasad PN, Stachowiak MK. Organically modified silica nanoparticles: a nonviral vector for in vivo gene delivery and expression in the brain. Proc Natl Acad Sci U S A. 2005;102:11539–11544. doi: 10.1073/pnas.0504926102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boschetto D, Claudio GD, Mirzaei H, Leong R, Grisan E. Automatic classification of small bowel mucosa alterations in celiac disease for confocal laser endomicroscopy; SPIE Medical Imaging; San Diego, USA. 2016a. p. 6. [Google Scholar]
- Boschetto D, Mirzaei H, Leong RWL, Grisan E. Detection and density estimation of goblet cells in confocal endoscopy for the evaluation of celiac disease; 37th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC); Milan, Italy. 2015a. pp. 6248–6251. [DOI] [PubMed] [Google Scholar]
- Boschetto D, Mirzaei H, Leong RWL, Grisan E. Superpixel-based auto-matic segmentation of villi in confocal endomicroscopy; IEEE-EMBS International Conference on Biomedical and Health Informatics; Las Vegas, NV, USA. 2016b. pp. 168–171. [Google Scholar]
- Boschetto D, Mirzaei H, Leong RWL, Tarroni G, Grisan E. Semiautomatic detection of villi in confocal endoscopy for the evaluation of celiac disease; 37th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC); Milan, Italy. 2015b. pp. 8143–8146. [DOI] [PubMed] [Google Scholar]
- Bouma BE, Yun SH, Vakoc BJ, Suter MJ, Tearney GJ. Fourier-domain optical coherence tomography: recent advances toward clinical utility. Curr Opin Biotechnol. 2009;20:111–118. doi: 10.1016/j.copbio.2009.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozinovic N, Ventalon C, Ford TN, Mertz J. Fluorescence endomicroscopy with structured illumination. Opt Express. 2008;16:8016–8025. doi: 10.1364/oe.16.008016. [DOI] [PubMed] [Google Scholar]
- Burggraaf J, Kamerling IMC, Gordon PB, Schrier L, de Kam ML, Kales AJ, Bendiksen R, Indrevoll B, Bjerke RM, Moestue SA, Yazdanfar S, et al. Detection of colorectal polyps in humans using an intravenously administered fluorescent peptide targeted against c-Met. Nat Med. 2015;21:955–961. doi: 10.1038/nm.3641. [DOI] [PubMed] [Google Scholar]
- Cazabet R, Amblard F, Hanachi C. Detection of overlapping communities in dynamical social networks; IEEE Second International Conference on Social Computing; Minneapolis, USA. 2010. pp. 309–314. [Google Scholar]
- Cha J, Kang JU. Video-rate Multicolor Fiber-Optic Microscopy; Imaging and Applied Optics; Arlington, Virginia. 2013. IM4E.4 [Google Scholar]
- Chen SP, Liao JC. Confocal laser endomicroscopy of bladder and upper tract urothelial carcinoma: a new era of optical diagnosis? Curr Urol Rep. 2014;15:437. doi: 10.1007/s11934-014-0437-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Reichenbach KL, Xu C. Experimental and theoretical analysis of core-to-core coupling on fiber bundle imaging. Opt Express. 2008;16:21598–21607. doi: 10.1364/oe.16.021598. [DOI] [PubMed] [Google Scholar]
- Cheon GW, Cha J, Kang JU. Random transverse motion-induced spatial compounding for fiber bundle imaging. Opt Lett. 2014a;39:4368–4371. doi: 10.1364/OL.39.004368. [DOI] [PubMed] [Google Scholar]
- Cheon GW, Cha J, Kang JU. Spatial compound imaging for fiber-bundle optic microscopy; Proceedings of SPIE 8938, Optical Fibers and Sensors for Medical Diagnostics and Treatment Applications; 2014. p. 8938.893811 [Google Scholar]
- Couceiro S, Barreto JP, Freire P, Figueiredo P. Description and Classification of Confocal Endomicroscopic Images for the Automatic Diagnosis of Inflammatory Bowel Disease. Springer, MLMI; 2012. pp. 144–151. [Google Scholar]
- Delaney PM, Harris MR, King RG. Fiber-optic laser scanning confocal microscope suitable for fluorescence imaging. Appl Opt. 1994;33:573–577. doi: 10.1364/AO.33.000573. [DOI] [PubMed] [Google Scholar]
- Deng J, Dong W, Socher R, Li LJ, Li K, Fei-Fei L. ImageNet: a large-scale hierarchical image database; IEEE Conference on Computer Vision and Pattern Recognition; Miami, USA. 2009. pp. 248–255. [Google Scholar]
- Deriche R. Recursively Implementating the Gaussian and Its Derivatives. INRIA, Sophia Antipolis; France: 1993. p. 24. [Google Scholar]
- Desir C, Petitjean C, Heutte L, Salaun M, Thiberville L. Classification of endomicroscopic images of the lung based on random subwindows and extra-trees. IEEE Trans Biomed Eng. 2012a;59:2677–2683. doi: 10.1109/TBME.2012.2204747. [DOI] [PubMed] [Google Scholar]
- Désir C, Petitjean C, Heutte L, Thiberville L. Using a priori knowledge to classify in vivo images of the lung; 6th International Conference on Intelligent Computing; Changsha, China. Springer Berlin Heidelberg; 2010. pp. 207–212. [Google Scholar]
- Désir C, Petitjean C, Heutte L, Thiberville L, Salaün M. An SVM-based distal lung image classification using texture descriptors. Comput Med Imaging Graph. 2012b;36:264–270. doi: 10.1016/j.compmedimag.2011.11.001. [DOI] [PubMed] [Google Scholar]
- Dickens MM, Bornhop DJ, Mitra S. Removal of optical fiber interference in color micro-endoscopic images; Proceedings of the 11th IEEE Symposium on Computer-Based Medical Systems; 1998. pp. 246–251. [Google Scholar]
- Dickens MM, Houlne MP, Mitra S, Bornhop DJ. Soft computing method for the removal of pixelation in microendoscopic images; Proceedings of SPIE 3165, Applications of Soft Computing; 1997. pp. 186–194. [Google Scholar]
- Dickens MM, Houlne MP, Mitra S, Bornhop DJ. Method for depixelating micro-endoscopic images. Opt Eng. 1999;38:1836–1842. [Google Scholar]
- Dubaj V, Mazzolini A, Wood A, Harris M. Optic fibre bundle contact imaging probe employing a laser scanning confocal microscope. J Microsc. 2002;207:108–117. doi: 10.1046/j.1365-2818.2002.01052.x. [DOI] [PubMed] [Google Scholar]
- Dumripatanachod M, Piyawattanametha W. A fast depixelation method of fiber bundle image for an embedded system; 2015 8th Biomedical Engineering International Conference (BMEiCON); 2015. pp. 1–4. [Google Scholar]
- East JE, Vleugels JL, Roelandt P, Bhandari P, Bisschops R, Dekker E, Hassan C, Horgan G, Kiesslich R, Longcroft-Wheaton G, Wilson A, et al. Advanced endoscopic imaging: European society of gastrointestinal endoscopy (ESGE) technology review. Endoscopy. 2016;48:1029–1045. doi: 10.1055/s-0042-118087. [DOI] [PubMed] [Google Scholar]
- Elter M, Rupp S, Winter C. Physically motivated reconstruction of fiberscopic images; 18th International Conference on Pattern Recognition (ICPR’06); 2006. pp. 599–602. [Google Scholar]
- Erden MS, Rosa B, Boularot N, Gayet B, Morel G, Szewczyk J. Conic-Spiraleur: a miniature distal scanner for confocal microlaparoscope. IEEE/AsMe Trans Mechatron. 2014;19:1786–1798. [Google Scholar]
- Erden MS, Rosa B, Szewczyk J, Morel G. Understanding soft-tissue behavior for application to microlaparoscopic surface scan. IEEE Trans Biomed Eng. 2013;60:1059–1068. doi: 10.1109/TBME.2012.2234748. [DOI] [PubMed] [Google Scholar]
- Farin G. Surfaces over Dirichlet tessellations. Comput Aided Geom Des. 1990;7:281–292. [Google Scholar]
- Ford TN, Chu KK, Mertz J. Phase-gradient microscopy in thick tissue with oblique back-illumination. Nat Methods. 2012a;9:1195–1197. doi: 10.1038/nmeth.2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford TN, Lim D, Mertz J. Fast optically sectioned fluorescence hilo endomicroscopy. J Biomed Opt. 2012b;17:0211051–0211057. doi: 10.1117/1.JBO.17.2.021105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford TN, Mertz J. Video-rate imaging of microcirculation with single-exposure oblique back-illumination microscopy. J Biomed Opt. 2013;18:066007. doi: 10.1117/1.JBO.18.6.066007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs FS, Zirlik S, Hildner K, Schubert J, Vieth M, Neurath MF. Confocal laser endomicroscopy for diagnosing lung cancer in vivo . Eur Respir J. 2013;41:1401–1408. doi: 10.1183/09031936.00062512. [DOI] [PubMed] [Google Scholar]
- Fugazza A, Gaiani F, Carra MC, Brunetti F, Levy M, Sobhani I, Azoulay D, Catena F, de’Angelis GL, de’Angelis N. Confocal laser endomicroscopy in gastrointestinal and pancreatobiliary diseases: a systematic review and metaanalysis. Biomed Res Int. 2016;2016:31. doi: 10.1155/2016/4638683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghatwary N, Ahmed A, Ye X, Jalab H. Automatic grade classification of barretts esophagus through feature enhancement; SPIE Medical Imaging; Orlando, United States. 2017. p. 8. [Google Scholar]
- Ghosh D, Kaabouch N. A survey on image mosaicing techniques. J Vis Commun Image Represent. 2016;34:1–11. [Google Scholar]
- Giataganas P, Hughes M, Yang GZ. Force adaptive robotically assisted endomicroscopy for intraoperative tumour identification. Int J Comput Assist Radiol Surg. 2015;10:825–832. doi: 10.1007/s11548-015-1179-0. [DOI] [PubMed] [Google Scholar]
- Gil D, Ramos-Terrades O, Minchole E, Sanchez C, de Frutos NC, Diez-Ferrer M, Ortiz RM, Rosell A. Computer Assisted and Robotic Endoscopy and Clinical Image-Based Procedures. Springer International Publishing; Quebec, Canada: 2017. Classification of confocal endomicroscopy patterns for diagnosis of lung cancer; pp. 151–159. [Google Scholar]
- Gmitro AF, Aziz D. Confocal microscopy through a fiber-optic imaging bundle. Opt Lett. 1993;18:565–567. doi: 10.1364/ol.18.000565. [DOI] [PubMed] [Google Scholar]
- Gora MJ, Sauk JS, Carruth RW, Gallagher KA, Suter MJ, Nishioka NS, Kava LE, Rosenberg M, Bouma BE, Tearney GJ. Tethered capsule endomicroscopy enables less-invasive imaging of gastrointestinal tract microstructure. Nat Med. 2013;19:238–240. doi: 10.1038/nm.3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Y, Vyas K, Yang J, Yang GZ. Med Image Comput Comput Assist Interv. Springer International Publishing; Quebec City, Canada: 2017. Unsupervised feature learning for endomicroscopy image retrieval; pp. 64–71. [Google Scholar]
- Gu Y, Yang J, Yang GZ. Multi-view multi-modal feature embedding for endomicroscopy mosaic classification; IEEE Conference on Computer Vision and Pattern Recognition Workshops (CVPRW); Las Vegas, USA. 2016. pp. 1315–1323. [Google Scholar]
- Han J, Lee J, Kang JU. Pixelation effect removal from fiber bundle probe based optical coherence tomography imaging. Opt Express. 2010;18:7427–7439. doi: 10.1364/OE.18.007427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, Yoon SM. Depixelation of coherent fiber bundle endoscopy based on learning patterns of image prior. Opt Lett. 2011;36:3212–3214. doi: 10.1364/OL.36.003212. [DOI] [PubMed] [Google Scholar]
- Han J, Yoon SM, Yoon G. Decoupling structural artifacts in fiber optic imaging by applying compressive sensing. Optik. 2015;126:2013–2017. [Google Scholar]
- Harris MR. Scanning Confocal Microscope Including a Single Fibre for Transmitting Light to and Receiving Light from an Object. USA: 1992. [Google Scholar]
- Harris MR. Scanning Microscope With Miniature Head. Optiscal Ltd; USA: 2003. [Google Scholar]
- Hasegawa N. Magnifying Image Pickup Unit for an Endoscope, an Endoscope for in vivo Cellular Observation that uses it, and Endoscopic, in vivo Cellular Observation Methods. Olympus Corporation; 2007. [Google Scholar]
- He T, Xue Z, Lu K, Alvarado MV, Wong ST. SPIE Medical Imaging. SPIE; San Diego, USA: 2012. IntegriSense molecular image sequence classification using gaussian mixture model. [Google Scholar]
- He T, Xue Z, Xie W, Wong S, Wong K, Alvarado MV, Wong STC. In: Liao H, Edwards PJE, Pan X, Fan Y, Yang GZ, editors. A motion correction algorithm for microendoscope video computing in image-guided intervention; Medical Imaging and Augmented Reality: 5th International Workshop, MIAR 2010; September 19-20, 2010; Springer Berlin Heidelberg; 2010. pp. 267–275. Proceedings, Berlin, Heidelberg. [Google Scholar]
- Hebert D, Désir C, Petitjean C, Heutte L, Thiberville L. Detection of pathological condition in distal lung images; 9th IEEE International Symposium on Biomedical Imaging (ISBI); Barcelona, Spain. 2012. pp. 1603–1606. [Google Scholar]
- Heutte L, Petitjean C, Desir C. Pruning trees in random forests for minimis-ing non detection in medical imaging. In: Chen CH, editor. Handbook of Pattern Recognition and Computer Vision. fifth ed. World Scientific Publishing; 2016. [Google Scholar]
- Hong J, Park By, Park H. Convolutional neural network classifier for distinguishing Barrett’s esophagus and neoplasia endomicroscopy images; Annual International Conference of the IEEE Engineering in Medicine and Biology Society; Seogwipo, South Korea. 2017. pp. 2892–2895. [DOI] [PubMed] [Google Scholar]
- Hong X, Nagarajan VK, Mugler DH, Yu B. Smartphone microendoscopy for high resolution fluorescence imaging. J Innov Opt Health Sci. 2016;9:1650046 [Google Scholar]
- Hsiung PL, Hardy J, Friedland S, Soetikno R, Du CB, Wu AP, Sahbaie P, Crawford JM, Lowe AW, Contag CrH, Wang TD. Detection of colonic dysplasia in vivo using a targeted heptapeptide and confocal microendoscopy. Nat Med. 2008;14:454–458. doi: 10.1038/nm1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu M, Penney G, Rueckert D, Edwards P, Bello F, Figl M, Casula R, Cen Y, Liu J, Miao Z, Hawkes D. In: Liao H, Edwards PJE, Pan X, Fan Y, Yang GZ, editors. A robust mosaicing method with super-resolution for optical medical images; Medical Imaging and Augmented Reality: 5th International Workshop, MIAR 2010; Beijing, China. September 19-20, 2010; Springer Berlin Heidelberg; 2010. pp. 373–382. Proceedings, Berlin, Heidelberg. [Google Scholar]
- Huang C, Kaza AK, Hitchcock RW, Sachse FB. dentification of nodal tissue in the living heart using rapid scanning fiber-optics confocal microscopy and extracellular fluorophores. Circ Cardiovasc Imaging. 2013;6:739–746. doi: 10.1161/CIRCIMAGING.112.000121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes M, Chang TP, Yang GZ. Fiber bundle endocytoscopy. Biomed Opt Express. 2013;4:2781–2794. doi: 10.1364/BOE.4.002781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes M, Giataganas P, Yang GZ. Color reflectance fiber bundle endomicroscopy without back-reflections. J Biomed Opt. 2014;19:030501. doi: 10.1117/1.JBO.19.3.030501. [DOI] [PubMed] [Google Scholar]
- Hughes M, Yang GZ. High speed, line-scanning, fiber bundle fluorescence confocal endomicroscopy for improved mosaicking. Biomed Opt Express. 2015;6:1241–1252. doi: 10.1364/BOE.6.001241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes M, Yang GZ. Line-scanning fiber bundle endomicroscopy with a virtual detector slit. Biomed Opt Express. 2016;7:2257–2268. doi: 10.1364/BOE.7.002257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- IIzadyyazdanabadi M, Belykh E, Cavallo C, Zhao X, Gandhi S, Borba Moreira L, Eschbacher J, Nakaji P, Preul MC, Yang Y. Weakly-supervised learning-based feature localization in confocal laser endomicroscopy glioma images; International Conference on Medical Image Computing and Computer-Assisted Intervention; Springer Berlin Heidelberg; 2018. [Google Scholar]
- Izadyyazdanabadi M, Belykh E, Martirosyan NL, Eschbacher J, Nakaji P, Y Y, Preul MC. Improving utility of brain tumor confocal laser endomicroscopy: objective value assessment and diagnostic frame detection with con-volutional neural networks; SPIE Medical Imaging; 2017. p. 9. [Google Scholar]
- Izadyyazdanabadi M, Belykh E, Mooney MA, Martirosyan N, Eschbacher J, Nakaji P, Preul MC, Yang Y. Convolutional neural networks: ensemble modeling, fine-tuning and unsupervised semantic localization for intraoperative cle images. J Vis Commun Image Representation. 2018;54:10–20. [Google Scholar]
- Jabbour JM, Saldua MA, Bixler JN, Maitland KC. Confocal endomicroscopy: instrumentation and medical applications. Ann Biomed Eng. 2012;40:378–397. doi: 10.1007/s10439-011-0426-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaremenko C, Maier A, Steidl S, Hornegger J, Oetter N, Knipfer C, Stelzle F, Neumann H. Bild-verarbeitung Für Die Medizin. Springer Berlin Heidelberg; Lübeck, Germany: 2015. Classification of confocal laser endomicroscopic images of the oral cavity to distinguish pathological from healthy tissue; pp. 479–485. [Google Scholar]
- Jean F, Bourg-Heckly G, Viellerobe B. Fibered confocal spectroscopy and multicolor imaging system for in vivo fluorescence analysis. Opt Express. 2007;15:4008–4017. doi: 10.1364/oe.15.004008. [DOI] [PubMed] [Google Scholar]
- Kamen A, Sun S, Wan S, Kluckner S, Chen T, Gigler AM, Simon E, Fleischer M, Javed M, Daali S, Igressa A, Charalampaki P. Automatic tissue differentiation based on confocal endomicroscopic images for intraoperative guidance in neurosurgery. Biomed Res Int. 2016;2016:8. doi: 10.1155/2016/6183218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karam Eldaly A, Altmann Y, Perperidis A, Akram A, Dhaliwal K, McLaughlin S. Bacteria detection in fcfm using a bayesian approach. IEEE Trans Biomed Eng. 2018 [Google Scholar]
- Karam Eldaly A, Altmann Y, Perperidis A, Krstajic N, Choudhary TR, Dhaliwal K, McLaughlin S. Deconvolution and Restoration of Optical Endomicroscopy Images. IEEE Trans Comput Imag. 2018;4(2):194–205. [Google Scholar]
- Karia K, Kahaleh M. A review of probe-based confocal laser endomicroscopy for pancreaticobiliary disease. Clin Endosc. 2016;49:462–466. doi: 10.5946/ce.2016.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khondee S, Wang TD. Progress in molecular imaging in endoscopy and endomicroscopy for cancer imaging. J Healthc Eng. 2013;4:1–22. doi: 10.1260/2040-2295.4.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohandani Tafresh M, Linard N, Andre B, Ayache N, Vercauteren T. Med Image Comput Comput Assist Interv. Springer International Publishing; Boston, USA: 2014. Semi-automated query construction for content-based endomicroscopy video retrieval; pp. 89–96. [DOI] [PubMed] [Google Scholar]
- Koujan MR, Ahsan A, McCool P, Westerfeld J, Wilson D, Dhaliwal K, McLaugh-lin S, Perperidis A. Multi-class classification of pulmonary endomicroscopic images; IEEE International Symposium on Biomedical Imaging (ISBI); Washington D.C., USA. 2018. pp. 1574–1577. [Google Scholar]
- Krizhevsky A, Sutskever I, Hinton GE. ImageNet classification with deep convolutional neural networks; International Conference on Neural Information Processing Systems; Lake Tahoe, USA. 2012. pp. 1097–1105. [Google Scholar]
- Krstajic N, Akram AR, Choudhary T, McDonald N, Tanner MG, Pedretti E, Dalgarno PA, Scholefield E, Girkin JM, Moore A, Bradley M, Dhaliwal K. Two-color widefield fluorescence microendoscopy enables multiplexed molecular imaging in the alveolar space of human lung tissue. J Biomed Opt. 2016;21:046009. doi: 10.1117/1.JBO.21.4.046009. [DOI] [PubMed] [Google Scholar]
- Kyrish M, Kester R, Richards-Kortum R, Tkaczyk T. Improving spatial reso-lution of a fiber bundle optical biopsy system. Proc SPIE. 2010;7558:755807. doi: 10.1117/12.842744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Goualher G, Perchant A, Genet M, Cavé C, Viellerobe B, Berier F, Abrat B, Ayache N. In: Barillot C, Haynor DR, Hellier P, editors. Towards optical biopsies with an integrated fibered confocal fluorescence microscope; Proceedings of the Medical Image Computing and Computer-Assisted Intervention; Berlin, Heidelberg. Springer Berlin Heidelberg; 2004a. pp. 761–768. [Google Scholar]
- Le Goualher G, Perchant A, Genet M, Cavé C, Viellerobe B, Berier F, Abrat B, Ayache N. In: Barillot C, Haynor DR, Hellier P, editors. Towards optical biopsies with an integrated fibered confocal fluorescence microscope; Medical Image Computing and Computer-Assisted Intervention - MICCAI 2004: 7th International Conference; Saint-Malo, France. September 26-29, 2004; Springer Berlin Heidelberg; 2004b. pp. 761–768. Proceedings, Part II, Berlin, Heidelberg. [Google Scholar]
- Lecun Y, Bottou L, Bengio Y, Haffner P. Gradient-based learning applied to document recognition. Proc IEEE. 1998;86:2278–2324. [Google Scholar]
- Lee C, Han J. Elimination of honeycomb patterns in fiber bundle imaging by a superimposition method. Opt Lett. 2013a;38:2023–2025. doi: 10.1364/OL.38.002023. [DOI] [PubMed] [Google Scholar]
- Lee CY, Cha YM, Han JH. Imaging and Applied Optics. Optical Society of America; Arlington, Virginia: 2013. Restoration method for fiber bundle microscopy using interpolation based on overlapping self-shifted images; JTu4A.23 [Google Scholar]
- Lee CY, Han JH. Integrated spatio-spectral method for efficiently suppress-ing honeycomb pattern artifact in imaging fiber bundle microscopy. Opt Commun. 2013b;306:67–73. [Google Scholar]
- Lee S, Wolberg G, Shin SY. Scattered data interpolation with multilevel B-splines. IEEE Trans Vis Comput Graph. 1997;3:228–244. [Google Scholar]
- Leierseder S. Laser+Photonics. AT-Fachverlag GmbH; Fellbach, Germany: 2018. Confocal Endomicroscopy During Brain surgery; pp. 76–79. [Google Scholar]
- Leigh SY, Liu JTC. Multi-color miniature dual-axis confocal microscope for point-of-care pathology. Opt Lett. 2012;37:2430–2432. doi: 10.1364/OL.37.002430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonovych O, Koujan MR, Akram A, Westerfeld J, Wilson D, Dhaliwal K, McLaughlin S, Perperidis A. Medical Image Understanding and Analysis (MIUA) Springer; Southampton, UK: 2018. Texture descriptors for classifying sparse, irregularly sampled optical endomicroscopy images. [Google Scholar]
- Liu JTC, Mandella MJ, Ra H, Wong LK, Solgaard O, Kino GS, Piyawattanametha W, Contag CH, Wang TD. Miniature near-infrared dual-axes confocal microscope utilizing a two-dimensional microelectromechanical systems scanner. Opt Lett. 2007;32:256–258. doi: 10.1364/ol.32.000256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Huang Y, Kang JU. Dark-field illuminated reflectance fiber bundle endoscopic microscope. J Biomed Opt. 2011;16:046003. doi: 10.1117/1.3560298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Zhang L, Kirby M, Becker R, Qi S, Zhao F. Iterative l1-min algo-rithm for fixed pattern noise removal in fiber-bundle-based endoscopic imaging. J Opt Soc Am A. 2016;33:630–636. doi: 10.1364/JOSAA.33.000630. [DOI] [PubMed] [Google Scholar]
- Loewke K, Camarillo D, Piyawattanametha W, Breeden D, Salisbury K. Real-time image mosaicing with a hand-held dual-axes confocal microscope. Proc SPIE. 2008:68510F–68519. [Google Scholar]
- Loewke K, Camarillo D, Piyawattanametha W, Mandella M, Contag C, Thrun S, Salisbury J. In vivo micro-image mosaicing. IEEE Trans Biomed Eng. 2011;58:159–171. doi: 10.1109/TBME.2010.2085082. [DOI] [PubMed] [Google Scholar]
- Loewke K, Camarillo D, Salisbury K, Thrun S. Deformable image mosaicing for optical biopsy, computer vision, 2007; ICCV 2007. IEEE 11th International Conference; 2007a. pp. 1–8. [Google Scholar]
- Loewke KE, Camarillo DB, Jobst CA, Salisbury JK. Real-time image mosaicing for medical applications. Stud Health Technol Inform. 2007b;125:304–309. [PubMed] [Google Scholar]
- Mahé J, Linard N, Tafreshi MK, Vercauteren T, Ayache N, Lacombe F, Cuingnet R. In: Navab N, Hornegger J, Wells WM, Frangi A, editors. Motion-Aware mosaicing for confocal laser endomicroscopy; Medical Image Computing and Computer-Assisted Intervention - MICCAI 2015: 18th International Conference; Munich, Germany. October 5-9, 2015; Springer International Publishing; 2015. pp. 447–454. Proceedings, Part I, Cham. [Google Scholar]
- Mahé J, Vercauteren T, Rosa B, Dauguet J. In: Mori K, Sakuma I, Sato Y, Barillot C, Navab N, editors. A viterbi approach to topology inference for large scale endomicroscopy video mosaicing; Medical Image Computing and Computer-Assisted Intervention - MICCAI 2013: 16th International Conference; Nagoya, Japan. September 22-26, 2013; Springer Berlin Heidelberg; 2013. pp. 404–411. Proceedings, Part I, Berlin, Heidelberg. [DOI] [PubMed] [Google Scholar]
- Makhlouf H, Gmitro AF, Tanbakuchi AA, Udovich JA, Rouse AR. Multispectral confocal microendoscope for in vivo and in situ imaging. J Biomed Opt. 2008;13:044016. doi: 10.1117/1.2950313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maneas E, dos Santos GS, Deprest J, Wimalasundera R, David AL, Vercauteren T, Ourselin S. In: Guang-Zhong Yang AD, editor. Adaptive filtering of fibre-optic fetoscopic images for fetal surgery; The Hamlyn Symposium on Medical Robotics; 2015. [Google Scholar]
- McCool P, Altmann Y, Perperidis A, McLaughlin S. Robust Markov random field outlier detection and removal in subsampled images; 2016 IEEE Statistical Signal Processing Workshop (SSP); 2016. pp. 1–5. [Google Scholar]
- Minsky M. Memoir on inventing the confocal scanning microscope. Scanning. 1988;10:128–138. [Google Scholar]
- Mooney MA, Zehri AH, Georges JF, Nakaji P. Laser scanning confocal endomicroscopy in the neurosurgical operating room: a review and discussion of future applications. Neurosurg Focus. 2014;36:E9. doi: 10.3171/2013.11.FOCUS13484. [DOI] [PubMed] [Google Scholar]
- Mualla F, Schöll S, Bohr C, Neumann H, Maier A. Epithelial cell detection in endomicroscopy images of the vocal folds; Proceedings of the International Multidisciplinary Microscopy Congress; Antalya, Turkey. Springer International Publishing; 2014. pp. 201–205. [Google Scholar]
- Murthy VN, Singh V, Sun S, Bhattacharya S, Chen T, Comaniciu D. Cascaded deep decision networks for classification of endoscopic images; SPIE Medical Imaging; 2017. p. 15. [Google Scholar]
- Namati E, Thiesse J, Ryk JD, Mclennan G. In vivo assessment of alveolar morphology using a flexible catheter-based confocal microscope. IET Comput Vis. 2008;2:228–235. [Google Scholar]
- Newton RC, Kemp SV, Yang GZ, Elson DS, Darzi A, Shah PL. Imaging parenchymal lung diseases with confocal endomicroscopy. Respir Med. 2012;106:127–137. doi: 10.1016/j.rmed.2011.09.009. [DOI] [PubMed] [Google Scholar]
- Oh G, Chung E, Yun SH. Optical fibers for high-resolution in vivo microen-doscopic fluorescence imaging. Opt Fiber Technol. 2013;19:760–771. [Google Scholar]
- Ohigashi T, Kozakai N, Mizuno R, Miyajima A, Murai M. Endocytoscopy: novel endoscopic imaging technology for in-situ observation of bladder cancer cells. J Endourol. 2006;20:698–701. doi: 10.1089/end.2006.20.698. [DOI] [PubMed] [Google Scholar]
- Ortega-Quijano N, Fanjul-Vélez F, Arce-Diego JL. Optical crosstalk influence in fiber imaging endoscopes design. Opt Commun. 2010;283:633–638. [Google Scholar]
- Pan Y, Volkmer JP, Mach KE, Rouse RV, Liu JJ, Sahoo D, Chang TC, Metzner TJ, Kang L, van de Rijn M, Skinner EC, et al. Endoscopic molecular imaging of human bladder cancer using a CD47 antibody. Sci Transl Med. 2014;6:260ra148. doi: 10.1126/scitranslmed.3009457. [DOI] [PubMed] [Google Scholar]
- Pavlov V, Meyronet D, Meyer-Bisch V, Armoiry X, Pikul B, Dumot C, Beuriat PA, Signorelli F, Guyotat J. Intraoperative probe-based confocal laser endomicroscopy in surgery and stereotactic biopsy of low-grade and high-grade gliomas: a feasibility study in humans neurosurg. Clin N Am. 2016;79:604–612. doi: 10.1227/NEU.0000000000001365. [DOI] [PubMed] [Google Scholar]
- Perchant A, Vercauteren T, Oberrietter F, Savoire N, Ayache N. Region tracking algorithms on laser scanning devices applied to cell traffic analysis; 4th IEEE International Symposium on Biomedical Imaging: From Nano to Macro; Arlington, USA. 2007. pp. 260–263. [Google Scholar]
- Perez JR, Ybarra N, Chagnon F, Serban M, Lee S, Seuntjens J, Lesur O, El Naqa I. Tracking of mesenchymal stem cells with fluorescence endomicroscopy imaging in radiotherapy-induced lung injury. Sci Rep. 2017;7:40748. doi: 10.1038/srep40748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perperidis A, Akram A, Altmann Y, McCool P, Westerfeld J, Wilson D, Dhaliwal K, McLaughlin S. Automated detection of uninformative frames in pulmonary optical endomicroscopy (OEM) IEEE Trans Biomed Eng. 2016;64:87–98. doi: 10.1109/TBME.2016.2538084. [DOI] [PubMed] [Google Scholar]
- Perperidis A, Cusack D, White A, McDicken N, MacGillivray T, Anderson T. Temporal compounding: a novel implementation and its impact on quality and diagnostic value in echocardiography. Ultrasound Med Biol. 2015;41:1749–1765. doi: 10.1016/j.ultrasmedbio.2015.02.008. [DOI] [PubMed] [Google Scholar]
- Perperidis A, Parker H, Karam Eldaly A, Altmann Y, Thomson RR, Tanner MG, McLaughlin S. Characterisation and modelling of inter-core coupling in coherent fibre bundles. Opt Express In press. 2017a doi: 10.1364/OE.25.011932. [DOI] [PubMed] [Google Scholar]
- Perperidis A, Parker HE, Karam-Eldaly A, Altmann Y, Dhaliwal K, Thomson RR, Tanner MIG, McLaughlin S. Characterization and modelling of intercore coupling in coherent fiber bundles. Opt Express. 2017b;25:11932–11953. doi: 10.1364/OE.25.011932. [DOI] [PubMed] [Google Scholar]
- Petitjean C, Benoist J, Thiberville L, Salaün M, Heutte L. Classification of in-vivo endomicroscopic images of the alveolar respiratory system; IAPR Conference on Machine Vision Applications (MVA); Yokohama, JAPAN. 2009. pp. 471–474. [Google Scholar]
- Pierce M, Yu D, Richards-Kortum R. High-resolution fiber-optic microen-doscopy for in situ cellular imaging. J Vis Exp. 2011:2306. doi: 10.3791/2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prieto SP, Lai KK, Laryea JA, Mizell JS, Muldoon TJ. Quantitative analysis of ex vivo colorectal epithelium using an automated feature extraction algorithm for microendoscopy image data. J Med Imaging. 2016;3:024502. doi: 10.1117/1.JMI.3.2.024502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ra H, Piyawattanametha W, Mandella MJ, Hsiung PL, Hardy J, Wang TD, Contag CH, Kino GS, Solgaard O. Three-dimensional in vivo imaging by a handheld dual-axes confocal microscope. Opt Express. 2008;16:7224–7232. doi: 10.1364/oe.16.007224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakotomamonjy A, Petitjean C, Salaün M, Thiberville L. Scattering features for lung cancer detection in fibered confocal fluorescence microscopy images. Artif Intell Med. 2014;61:105–118. doi: 10.1016/j.artmed.2014.05.003. [DOI] [PubMed] [Google Scholar]
- Rasmussen DN, Karstensen JG, Riis LB, Brynskov J, Vilmann P. Confocal laser endomicroscopy in inflammatory bowel disease - a systematic review. J Crohn’s Colitis. 2015;9:1152–1159. doi: 10.1093/ecco-jcc/jjv131. [DOI] [PubMed] [Google Scholar]
- Rajpoot K, Noble JA, Grau V, Szmigielski C, Becher H. Multiview RT3D echocardiography image fusion; International Conference on Functional Imaging and Modeling of the Heart (FIMH 2009); 2009. pp. 134–143. [Google Scholar]
- Ravì D, Szczotka AB, Ismail Shakir D, Pereira SP, Vercauteren T. Effective deep learning training for single-image super-resolution in endomicroscopy exploiting video-registration-based reconstruction. Int J Comput Ass Rad Sur. 2018;13(6):917–924. doi: 10.1007/s11548-018-1764-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichenbach KL, Xu C. Numerical analysis of light propagation in image fibers or coherent fiber bundles. Opt Express. 2007;15:2151–2165. doi: 10.1364/oe.15.002151. [DOI] [PubMed] [Google Scholar]
- Rosa B, Erden MS, Vercauteren T, Herman B, Szewczyk J, Morel G. Building large mosaics of confocal edomicroscopic images using visual servoing. IEEE Trans Biomed Eng. 2013;60(4):1041–1049. doi: 10.1109/TBME.2012.2228859. [DOI] [PubMed] [Google Scholar]
- Rosa B, Herman B, Szewczyk J, Gayet B, Morel G. Laparoscopic optical biopsies: in vivo robotized mosaicing with probe-based confocal endomicroscopy; 2011 IEEE/RSJ International Conference on Intelligent Robots and Systems; 2011. pp. 1339–1345. [Google Scholar]
- Rouse AR, Gmitro AF. Multispectral imaging with a confocal microendo-scope. Opt Lett. 2000;25:1708–1710. doi: 10.1364/ol.25.001708. [DOI] [PubMed] [Google Scholar]
- Rouse AR, Kano A, Udovich JA, Kroto SM, Gmitro AF. Design and demonstration of a miniature catheter for a confocal microendoscope. Appl Opt. 2004;43:5763–5771. doi: 10.1364/ao.43.005763. [DOI] [PubMed] [Google Scholar]
- Rupp S, Elter M, Winter C. Improving the accuracy of feature extraction for flexible endoscope calibration by spatial super resolution; 29th Annual In-ternational Conference of the IEEE Engineering in Medicine and Biology Society; 2007. pp. 6565–6571. [DOI] [PubMed] [Google Scholar]
- Rupp S, Winter C, Elter M. Evaluation of spatial interpolation strategies for the removal of comb-structure in fiber-optic images; Annual International Conference of the IEEE Engineering in Medicine and Biology Society; 2009. pp. 3677–3680. [DOI] [PubMed] [Google Scholar]
- Sabharwal YS, Rouse AR, Donaldson LT, Hopkins MF, Gmitro AF. Slit-scanning confocal microendoscope for high-resolution in vivo imaging. Appl Opt. 1999;38:7133–7144. doi: 10.1364/ao.38.007133. [DOI] [PubMed] [Google Scholar]
- Saint-Réquier A, Lelandais B, Petitjean C, Désir C, Heutte L, Salaün M, Thiberville L. Characterization of endomicroscopic images of the distal lung for computer-aided diagnosis; 5th International Conference on Intelligent Computing; Ulsan, South Korea. 2009. pp. 994–1003. [Google Scholar]
- Salvatori F, Siciliano S, Maione F, Esposito D, Masone S, Persico M, De Palma GD. Confocal laser endomicroscopy in the study of Colonic Mucosa in IBD patients: a review. Gastroenterology Research and Practice. 2012;2012:6. doi: 10.1155/2012/525098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savoire N, Andre B, Vercauteren T. Proceedings of the Medical Image Computing and Computer Assisted Intervention. 2012. Online blind calibration of non-uniform photodetectors: application to endomicroscopy; pp. 639–646. [DOI] [PubMed] [Google Scholar]
- Savoire N, Le Goualher G, Perchant A, Lacombe F, Malandain G, Ayache N. Measuring blood cells velocity in microvessels from a single image: ap-plication to in vivo and in situ confocal microscopy; 2nd IEEE International Symposium on Biomedical Imaging (ISBI): Nano to Macro; 2004. pp. 456–459. [Google Scholar]
- Seth S, Akram A, McCool P, Westerfeld J, Wilson D, McLaughlin S, Dhaliwal K, Williams C. Assessing the utility of autofluorescence-based pulmonary optical endomicroscopy to predict the malignant potential of solitary pulmonary nodules in humans. Sci Rep. 2016;6:31372. doi: 10.1038/srep31372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seth S, Akram AR, Dhaliwal K, Williams CKI. In: Valdés Hernández M, González-Castro V, editors. Estimating bacterial load in fcfm imaging; Medical Image Understanding and Analysis: 21st Annual Conference, MIUA 2017; Edinburgh, UK. July 11-13, 2017; Springer International Publishing; 2017. pp. 909–921. Proceedings, Cham. [Google Scholar]
- Seth S, Akram AR, Dhaliwal K, Williams CKI. Estimating bacterial and cellular load in fcfm imaging. J Imaging. 2018;4:11. [Google Scholar]
- Shi Y, Wang L. Photonics Asia 2010. SPIE; Beijin, China: 2010. Fast confocal endomicroscopy based on multi-fiber parallel scanning; p. 6. [Google Scholar]
- Shin D, Pierce MC, Gillenwater AM, Williams MD, Richards-Kortum RR. A fiber-optic fluorescence microscope using a consumer-grade digital camera for in vivo cellular imaging. PLoS ONE. 2010;5:e11218. doi: 10.1371/journal.pone.0011218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R, Mei SLCY, Tam W, Raju D, Ruszkiewicz A. Real-time histology with the endocytoscope. World J Gastroenterol. 2010;16:5016–5019. doi: 10.3748/wjg.v16.i40.5016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivic J, Zisserman A. Efficient visual search for objects in videos. Proc IEEE. 20008;96:548–566. [Google Scholar]
- Smith I, Kline PE, Gaidhane M, Kahaleh M. A review on the use of confocal laser endomicroscopy in the bile duct. Gastroenterol Res Pract. 2012;2012:5. doi: 10.1155/2012/454717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- So PTC, Dong CY, Masters BR, Berland KM. Two-Photon excitation fluorescence microscopy. Annu Rev Biomed Eng. 2000;2:399–429. doi: 10.1146/annurev.bioeng.2.1.399. [DOI] [PubMed] [Google Scholar]
- Sonn GA, Jones SE, Tarin TV, Du CB, Mach KE, Jensen KC, Liao JC. Optical biopsy of human bladder neoplasia with in vivo confocal laser endomicroscopy. J Urol. 2009;182:1299–1305. doi: 10.1016/j.juro.2009.06.039. [DOI] [PubMed] [Google Scholar]
- Srivastava S, Rodriguez JJ, Rouse AR, Brewer MA, Gmitro AF. Analysis of confocal microendoscope images for automatic detection of ovarian cancer; IEEE International Conference on Image Processing; 2005. [Google Scholar]
- Srivastava S, Rodríguez JJ, Rouse AR, Brewer MA, Gmitro AF. Computer-aided Identification of Ovarian Cancer in Confocal Microendoscope Images. SPIE. 2008 doi: 10.1117/1.2907167. [DOI] [PubMed] [Google Scholar]
- Staderini M, Megia-Fernandez A, Dhaliwal K, Bradley M. Peptides for optical medical imaging and steps towards therapy. Bioorg Med Chem. 2017 doi: 10.1016/j.bmc.2017.09.039. [DOI] [PubMed] [Google Scholar]
- Stefanescu D, Streba C, Cârţână ET, Saftoiu A, Gruionu G, Gruionu LG. Computer aided diagnosis for confocal laser endomicroscopy in advanced colorectal adenocarcinoma. PLoS ONE. 2016;11:e0154863. doi: 10.1371/journal.pone.0154863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su P, Liu Y, Lin S, Xiao K, Chen P, An S, He J, Bai Y. Efficacy of confocal laser endomicroscopy for discriminating colorectal neoplasms from non-neoplasms: a systematic review and meta-analysis. Colorectal Dis. 2013;15:e1–e12. doi: 10.1111/codi.12033. [DOI] [PubMed] [Google Scholar]
- Sun J, Shu C, Appiah B, Drezek R. Needle-compatible single fiber bundle image guide reflectance endoscope. J Biomed Opt. 2010;15:040502–040503. doi: 10.1117/1.3465558. [DOI] [PubMed] [Google Scholar]
- Szegedy C, Liu W, Jia Y, Sermanet P, Reed S, Anguelov D, Erhan D, Vanhoucke V, Rabinovich A. Going deeper with convolutions; IEEE Conference on Computer Vision and Pattern Recognition; 2015. pp. 1–9. [Google Scholar]
- Szegedy C, Vanhoucke V, Ioffe S, Shlens J, Wojna Z. Rethinking the inception architecture for computer vision; IEEE Conference on Computer Vision and Pattern Recognition; Las Vegas, USA. 2016. pp. 2818–2826. [Google Scholar]
- Tajbakhsh N, Shin JY, Gurudu SR, Hurst RT, Kendall CB, Gotway MB, Liang J. Convolutional neural networks for medical image analysis: full training or fine tuning? IEEE Trans Med Imaging. 2016;35:1299–1312. doi: 10.1109/TMI.2016.2535302. [DOI] [PubMed] [Google Scholar]
- Tearney GJ, Brezinski ME, Bouma BE, Boppart SA, Pitris C, Southern JF, Fujimoto JG. In vivo endoscopic optical biopsy with optical coherence to-mography. Science. 1997;276:2037. doi: 10.1126/science.276.5321.2037. [DOI] [PubMed] [Google Scholar]
- Thiberville L, Moreno-Swirc S, Vercauteren T, Peltier E, Cavé C, Bourg Heckly G. In vivo imaging of the bronchial wall microstructure using fibered confocal fluorescence microscopy. Am J Respir Crit Care Med. 2007;175:22–31. doi: 10.1164/rccm.200605-684OC. [DOI] [PubMed] [Google Scholar]
- Thiberville L, Salaün M, Lachkar S, Dominique S, Moreno-Swirc S, Vever-Bizet C, Bourg-Heckly G. In vivo confocal fluorescence endomicroscopy of lung cancer. J Thorac Oncol. 2009;4:S48–S51. doi: 10.1513/pats.200902-009AW. [DOI] [PubMed] [Google Scholar]
- Tous R, Delgado J, Zinkl T, Toran P, Alcalde G, Goetz M, Ferrer-Roca O. The anatomy of an optical biopsy semantic retrieval system. IEEE Multimed. 2012;19:16–27. [Google Scholar]
- Vakoc BJ, Shishko M, Yun SH, Oh WY, Suter MJ, Desjardins AE, Evans JA, Nishioka NS, Tearney GJ, Bouma BE. Comprehensive esophageal mi-croscopy by using optical frequency-domain imaging (with video) Gastrointest Endosc. 2007;65:898–905. doi: 10.1016/j.gie.2006.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vercauteren T, Doussoux F, Cazaux M, Schmid G, Linard N, Durin MA, Gharbi H, Lacombe F. Endoscopic Microscopy. VIII-2013. SPIE; 2013. Multicolor probe-based confocal laser endomicroscopy: a new world for in vivo and real-time cellular imaging, spie bios-spie photonics west, bios. [Google Scholar]
- Vercauteren T, Meining A, Lacombe F, Perchant A. Real time autonomous video image registration for endomicroscopy: fighting the compromises; SPIE BiOS; 2008. p. 8. [Google Scholar]
- Vercauteren T, Perchant A, Malandain G, Pennec X, Ayache N. Robust mosaicing with correction of motion distortions and tissue deformations for in vivo fibered microscopy. Med Image Anal. 2006;10:673–692. doi: 10.1016/j.media.2006.06.006. [DOI] [PubMed] [Google Scholar]
- Vercauteren T, Perchant A, Pennec X, Ayache N. In: Duncan JS, Gerig G, editors. Mosaicing of confocal microscopic in vivo soft tissue video sequences; Medical Image Computing and Computer-Assisted Intervention - MICCAI 2005: 8th International Conference; Palm Springs, CA, USA. October 26-29, 2005; Springer Berlin Heidelberg; 2005. pp. 753–760. Proceedings, Part I, Berlin, Heidelberg. [DOI] [PubMed] [Google Scholar]
- Veronese E, Grisan E, Diamantis G, Battaglia G, Crosta C, Trovato C. Hybrid patch-based and image-wide classification of confocal laser endomicroscopy images in barrett’s esophagus surveillance; IEEE International Symposium on Biomedical Imaging; San Francisco, USA. 2013. pp. 362–365. [Google Scholar]
- Vo K, Jaremenko C, Bohr C, Neumann H, Maier A. Bildverarbeitung Für Die Medizin. Springer; Berlin Heidelberg: 2017. Automatic classification and pathological staging of confocal laser endomicroscopic images of the vocal cords; pp. 312–317. [Google Scholar]
- Vyas K, Hughes M, Yang GZ. Electromagnetic tracking of handheld high-resolution endomicroscopy probes to assist with real-time video mosaicking; SPIE BiOS; 2015. p. 8. [Google Scholar]
- Wallace MB, Fockens P. Probe-Based confocal laser endomicroscopy. Gastroenterology. 2009;136:1509–1513. doi: 10.1053/j.gastro.2009.03.034. [DOI] [PubMed] [Google Scholar]
- Wan S, Sun S, Bhattacharya S, Kluckner S, Gigler A, Simon E, Fleischer M, Charalampaki P, Chen T, Kamen A. Med Image Comput Comput Assist Interv. Springer International Publishing; Munich, Germany: 2015. Towards an efficient computational framework for guiding surgical resection through intra-operative endo-microscopic pathology; pp. 421–429. [Google Scholar]
- Wang G. A perspective on deep imaging. IEEE Access. 2016;4:8914–8924. [Google Scholar]
- Wang J, Nadkarni SK. The influence of optical fiber bundle parameters on the transmission of laser speckle patterns. Opt Express. 2014;22:8908–8918. doi: 10.1364/OE.22.008908. [DOI] [PubMed] [Google Scholar]
- Wang KK, Carr-Locke DL, Singh SK, Neumann H, Bertani H, Galmiche JP, Arsenescu RI, Caillol F, Chang KJ, Chaussade S, Coron E, et al. Use of probe-based confocal laser endomicroscopy (pCLE) in gastrointestinal applications a consensus report based on clinical evidence. United Eur Gastroenterol J. 2015;3:230–254. doi: 10.1177/2050640614566066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang TD, Mandella MJ, Contag CH, Kino GS. Dual-axis confocal microscope for high-resolution in vivo imaging. Opt Lett. 2003;28:414–416. doi: 10.1364/ol.28.000414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watcharapichat P. Image Processing and Classification Algorithm to Detect Cancerous Cells Morphology When Using In-Vivo Probe-Based Confocal Laser Endomicroscopy For the Lower Gastrointestinal Tract. Department of Computing. Imperial College; London, UK: 2012. p. 65. [Google Scholar]
- Waterhouse DJ, Joseph J, Neves AA, di Pietro M, Brindle KM, Fitzgerald RC, B SE. Design and validation of a near-infrared fluorescence endoscope for detection of early esophageal malignancy using a targeted imaging probe; SPIE BiOS; San Francisco, CA, USA. 2016. p. 9. [DOI] [PubMed] [Google Scholar]
- Winter C, Rupp S, Elter M, Munzenmayer C, Gerhauser H, Wittenberg T. Automatic adaptive enhancement for images obtained with fiberscopic endoscopes. IEEE Trans Biomed Eng. 2006;53:2035–2046. doi: 10.1109/TBME.2006.877110. [DOI] [PubMed] [Google Scholar]
- Winter C, Zerfaß T, Elter M, Rupp S, Wittenberg T. In: Ayache N, Ourselin S, Maeder A, editors. Physically motivated enhancement of color images for fiber endoscopy; Medical Image Computing and Computer-Assisted Intervention - MICCAI 2007: 10th International Conference; Brisbane, Australia. October 29 - November 2, 2007; Springer Berlin Heidelberg; 2007. pp. 360–367. Proceedings, Part II, Berlin, Heidelberg. [DOI] [PubMed] [Google Scholar]
- Wood H, Harrington K, Stone JM, Birks TA, Knight JC. Quantitative characterization of endoscopic imaging fibers. Opt Express. 2017;25:1985–1992. doi: 10.1364/OE.25.001985. [DOI] [PubMed] [Google Scholar]
- Wu H, Tong L, Wang MD. Improving multi-class classification for endomicroscopic images by semi-supervised learning; EEE EMBS International Conference on Biomedical & Health Informatics; Orlando, USA. 2017. pp. 5–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Li X. Two-photon fluorescence endomicroscopy. Ch. 32. In: Costa N, Cartaxo A, editors. Advances in Lasers and Electro Optics. InTech; Rijeka: 2010. [Google Scholar]
- Yserbyt J, Dooms C, Decramer M, Verleden GM. Acute lung allograft rejection: diagnostic role of probe-based confocal laser endomicroscopy of the respiratory tract. J Heart Lung Transplant. 2014;33:492–498. doi: 10.1016/j.healun.2014.01.857. [DOI] [PubMed] [Google Scholar]
- Yserbyt J, Dooms C, Janssens W, Verleden GM. Endoscopic advanced imaging of the respiratory tract: exploring probe-based confocal laser endomicroscopy in emphysema. Thorax. 2017 doi: 10.1136/thoraxjnl-2016-209746. [DOI] [PubMed] [Google Scholar]
- Yserbyt J, Dooms C, Ninane V, Decramer M, Verleden G. Perspectives using probe-based confocal laser endomicroscopy of the respiratory tract. Swiss Med Wkly. 2013:143. doi: 10.4414/smw.2013.13764. [DOI] [PubMed] [Google Scholar]
- Yun SH, Tearney GJ, de Boer JF, Iftimia N, Bouma BE. High-speed optical frequency-domain imaging. Opt Express. 2003;11:2953–2963. doi: 10.1364/oe.11.002953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun SH, Tearney GJ, Vakoc BJ, Shishkov M, Oh WY, Desjardins AE, Suter MJ, Chan RC, Evans JA, Jang IK, Nishioka NS, de Boer JF, Bouma BE. Comprehensive volumetric optical microscopy in vivo. Nat Med. 2006;12:1429–1433. doi: 10.1038/nm1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zehri AH, Ramey W, Georges JF, Mooney MA, Martirosyan NL, Preul MC, Nakaji P. Neurosurgical confocal endomicroscopy: a review of contrast agents, confocal systems, and future imaging modalities. Surg Neurol Int. 2014:5. doi: 10.4103/2152-7806.131638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Z, Cai B, Kou J, Liu W, Wang Z. In: Chen W, Hosoda K, Menegatti E, Shimizu M, Wang H, editors. A honeycomb artifacts removal and super resolution method for fiber-optic images; Intelligent Autonomous Systems 14: Proceedings of the 14th International Conference IAS-14; Cham. Springer International Publishing; 2017. pp. 771–779. [Google Scholar]
- Zhong W, Celli JP, Rizvi I, Mai Z, Spring BQ, Yun SH, Hasan T. In vivo high-resolution fluorescence microendoscopy for ovarian cancer detection and treatment monitoring. Br J Cancer. 2009;101:2015–2022. doi: 10.1038/sj.bjc.6605436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou D, Bousquet O, Lal TN, Weston J, Scholkopf B. Learning with local and global consistency; Proceedings of the 16th International Conference on Neural Information Processing Systems; Whistler, British Columbia, Canada. MIT Press; 2003. pp. 321–328. [Google Scholar]
- Zubiolo A, Malandain G, André B, Debreuve E. A recursive approach for multiclass support vector machine application to automatic classification of endomicroscopic videos; International Conference on Computer Vision Theory and Applications; Lisbon, Portugal. 2014. pp. 441–447. [Google Scholar]
- Zuo S, Hughes M, Seneci C, Chang TP, Yang GZ. Toward intraopera-tive breast endomicroscopy with a novel surface-scanning device. IEEE Trans Biomed Eng. 2015;62:2941–2952. doi: 10.1109/TBME.2015.2455597. [DOI] [PubMed] [Google Scholar]
- Zuo S, Hughes M, Yang GZ. A balloon endomicroscopy scanning device for diagnosing barrett’s oesophagus; 2017 IEEE International Conference on Robotics and Automation (ICRA); 2017a. pp. 2964–2970. [Google Scholar]
- Zuo S, Hughes M, Yang GZ. Flexible robotic scanning device for intraoperative endomicroscopy in mis. IEEE/ASME Trans Mechatron. 2017b;22:1728–1735. [Google Scholar]
- Zuo S, Yang GZ. Endomicroscopy for computer and robot assisted intervention. IEEE Rev Biomed Eng. 2017:1–1. doi: 10.1109/RBME.2017.2686483. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Recent developments in Convolutional Neural Networks (CNNs) have acted as vehicle to substantial advances to image analysis and understanding across an ever-increasing range of areas, including medical imaging, with applications in image reconstruction, classification, segmentation and registration. Yet, to date there have been just a limited number studies employing Convolutional Neutral Networks for the classification and retrieval on FBEμ frames and mosaics. Instead, image understanding tasks has been tackled predominantly through traditional machine learning pathways, defining hand-crafted feature descriptors and subsequently training a binary or multi-class classifier on this feature set. A key con-straint in the effective adaptation and adoption of the technology (CNNs) has been, to a large extent, the limited data and associated annotations available. In particular, the plurality of FBEμ classification/retrieval studies have employed limited data, ranging between 100 and 200 annotated frames for combined training, validation and testing, with several studies using datasets of less than 100 frames. Furthermore, the available data have for the most part been acquired from a single clinical site and many times from a single operator, introducing potential bias and hindering the ability of the proposed methodologies for widespread generalisation. Similarly, there is often a lack of a gold reference standard and manual annotations can be weak, demonstrating large inter- and intraoperator variability. In tasks such as image restoration and analysis, the proposed methodologies of assessment have been constrained to simple simulated data, test targets, and in some cases to a very limited number of biological samples. There is therefore a need for the development of (i) large data repositories, containing a diverse collection of frames sequences acquired from different operators at multiple sites across the world, with easy access for the endomicroscopy research community, (ii) associated manual annotations, ideally from multiple operators with varied level of expertise, with quantifiable inter- and intra-operator variability. Providing standardised annotation tools, available alongside the data depositories can further enhance the consistency and robustness of these annotations.