Abstract
Air-liquid interface (ALI) culture models currently represent a valid instrument to recreate the typical aspects of the respiratory tract in vitro in both healthy and diseased state. They can help reducing the number of animal experiments, therefore, supporting the 3R principle. This review discusses ALI cultures and co-cultures derived from immortalized as well as primary cells, which are used to study the most common disorders of the respiratory tract, in terms of both pathophysiology and drug screening. The article displays ALI models used to simulate inflammatory lung diseases such as chronic obstructive pulmonary disease (COPD), asthma, cystic fibrosis, lung cancer, and viral infections. It also includes a focus on ALI cultures described in literature studying respiratory viruses such as SARS-CoV-2 causing the global Covid-19 pandemic at the time of writing this review. Additionally, commercially available models of ALI cultures are presented. Ultimately, the aim of this review is to provide a detailed overview of ALI models currently available and to critically discuss them in the context of the most prevalent diseases of the respiratory tract.
Keywords: air-liquid interface, lung, 3D co-culture models, pulmonary administration, respiratory tract, SARS Cov-2
1. Introduction
Chronic respiratory inflammatory conditions observed in patients with asthma, cystic fibrosis and COPD are major causes for death and morbidity worldwide.[1] Besides, lung cancer and respiratory infections, including the global COVID-19 pandemic in the year 2020, are frequent causes of mortality. Therefore, more efficient treatment strategies are urgently sought for, particularly for diseases causing irreversible tissue destruction and loss of lung function. The complex cellular composition of the respiratory tract and its location at the air-liquid interface (ALI) hamper an accurate mimicking of the physiological situation. Various models have been applied in the past ranging from in vitro lung cell models to in vivo animal experiments.[2] In this field of research, animal models are routinely used since all in vitro models lack one or another aspect of lung anatomy or physiology, hampering validation of research results. Consequently, animal models are widely regarded as the sole reliable choice that is available. Yet, the anatomical differences between rodents and humans emphasize a substantial lack of functional homology regarding various biomolecules, drug deposition rates and localization of particulate drug delivery systems.[3] For example, the alveolar and the airway architecture show fundamental differences.[4,5] Mice only have 6-8 levels of branching airways while humans have up to 20 or more. Furthermore, mice do not have respiratory bronchioles comparable to humans, which are characterized by interruptions on their walls that project into the alveoli. They only have short terminal bronchioles opening straight into the alveolar ductules.[6] Therefore, interpretation of data derived from rodent models cannot easily be translated into human context. Furthermore, the strong support for the 3R principle (reduce, refine, replace) in experimental animal testing is constantly increasing.[7] These key facts have driven the development of alternative in vitro cell culture methods aimed at mimicking the respiratory tract. Hereby, ALI models derived from the field of inhalation toxicology have been described as the most promising approach.[8] The most important characteristic of ALI culture is that the apical surface of cells is exposed to air while the basal side is nourished by contact with liquid cell culture medium. This configuration allows cell differentiation towards a mucociliary phenotype, simulating in vivo conditions better than it is possible in conventional cell culture. ALI models hence allow to obtain relevant data of the respiratory tract since they can be constructed from human-derived cells and are therefore capable to model scenarios close to in vivo conditions.[9] Another advantage is that drugs administered as aerosols and particles are not diluted or changed structurally by contact with cell culture medium before they impact on the epithelium differently from submerged models. Furthermore, dosing can be exactly controlled in contrast to in vivo administration, resulting in a a better optimization of parameters in vitro and subsequently reduced amounts of experimental animals required in follow-up in vivo experiments. Besides, these lung models cannot only be used to help understand pathophysiological processes and perform drug screening. They also support the mechanistic understanding of the interaction of xenobiotics at the cellular level in healthy and diseased tissue, complementing findings gained from in vivo studies. However, for many applications, ALI mono-culture platforms fail to represent the cellular arrangement thoroughly, e.g. by lacking direct cell-cell interactions. Hence, co-culture models constituted of more than one cell type are widely being developed.[10] For many applications, they are beneficial over ordinary ALI culture models because they provide a morphology, function, and intercellular interactions with enhanced resemblance to physiological in vivo conditions.
This article focuses on the different types of ALI cell culture models resembling the human respiratory tract including the commonly used cell types and applications. Hereby, importance is given to models mimicking healthy as well as diseased states of the lung, e.g., in patients suffering from asthma, cystic fibrosis or COPD. Especially advanced systems using multiple cell types or even culturing cells with viruses or bacteria for pathogen-host interaction studies will be presented in detail.
2. Anatomical and cellular structure of the respiratory tract
The respiratory tract is part of the respiratory system that also includes parts of the central nervous system, the chest wall and the pulmonary circulation.[11] One can picture the respiratory tract as an upside-down tree with a complex network of bifurcations getting thinner and thinner with every branching step. Generally the respiratory tract can be divided into three main regions: (a) the extrathoracic (ET) region which includes the oral and nasal cavity, the pharyngeal and laryngeal tract to the trachea entrance, (b) the tracheobronchial (TB) or conducting region expanding from the trachea down to the terminal bronchioles, and (c) the alveolar (Al) region responsible for the gas exchange.[12] Within the airways, several structural and cellular mechanisms protect the organ against harmful materials and potential pathogens. First, a continuous layer of epithelial cells lines the entire respiratory tree. These cells form tight junction networks, building the specific structural integrity of the epithelial layer, and they are crucial for maintaining the normal functions of the respiratory system. Furthermore, the surfactant film coating the lower airways and the mucociliary escalator of the upper tract of the airways join forces to transport unwanted matter up the airways to be swallowed subsequently.[13] Also, a resident population of innate cells such as dendritic cells or macrophages inside and underneath the airway epithelium phagocytoses foreign material. [14,15]
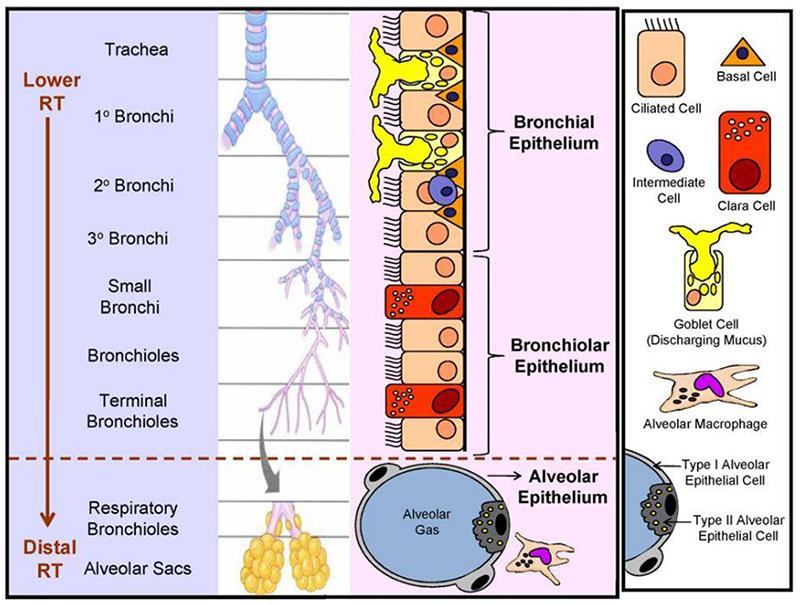
Cell types present in the different regions alter from the conducting to the respiratory part of the airways, accommodating the respective specific functions and defence mechanisms (Figure 1). The ET epithelium constitutes predominantly of ciliated columnar cells and goblet cells, while the TB tract is lined by a pseudostratified, columnar epithelium consisting of goblet cells, basal cells and ciliated cells, supporting the mucociliary clearance. Cuboidal ciliated cells and secretory Clara cells prevail in the epithelium of the bronchioles.[17] Apart from these tissue-specific epithelial cells, many non-epithelial migratory cells can be found such as mast cells, leukocytes and lymphocytes. The AI region further down the respiratory system is constituted of alveolar type I and II pneumocytes forming the alveolar epithelium. Type I pneumocytes account for about 95% of the internal lung surface and are mainly responsible for gas exchange, whereas type II cells mediate many other functions such as regulation of the lung surfactant system, the alveolar fluid content and secretion of antimicrobial and surface-active components. Besides, alveolar cells form the gas exchange barrier by attaching to endothelial cells via their basal membranes. Additionally, resident alveolar macrophages remove inhaled debris.
Figure 1.
Principle cell types found alongside the human respiratory tract varying in functions and defense mechanisms. Reproduced with permission.[16] Copyright 2010, Elsevier.
3. Cellular composition of ALI models
In vitro cell culture systems offer controllable, adaptable and reproducible models compared to in vivo and ex vivo systems. Numerous epithelial cell types have been grown at the ALI with the aim of mimicking distinct parts of the respiratory tract. Compared to submerged culture, differentiated cell morphology, altered biochemistry and response to tested materials have been described, showing good resemblance to the actual in vivo situation. ALI models can be established with primary material or with immortalized cell lines, both offering pros and cons. In general, primary cultures isolated directly from tissues represent a heterogenous population of several different types of epithelial cells. Primary cells from patient populations suffering from respiratory diseases such as cystic fibrosis,[18] asthma,[19] and COPD[20] have been isolated for research purposes. Moreover, primary epithelial cells were used to study virus-host-interactions.[21] Cells from different sections of the respiratory tract are cultured at ALI and can reproduce many features of the diseased state.[22] Each isolate, however, is unique, and therefore, it is impossible to completely reproduce it. Moreover, the isolation from normal human airway tissue comes with a limitated number of cells.[23] Epithelial immortalized cell lines, as presented in Table 1, have the advantage of homogenous clonality with less phenotypic differences compared to primary cells, resulting in more stable cultures with easier handling. This is the reason why immortalized human cell lines are very commonly utilized in ALI cultures of the respiratory tract. However, it is important to emphasize that due to the transformation process and clonality, they can potentially lack important molecules that are usually encountered in vivo. In general, primary epithelial cells display a better representation of the native microenvironment and in principle are optimal candidates for simulating in vivo conditions.
Table 1. Phenotypic characteristics of cell lines used in ALI cultures to mimic different parts of the respiratory tract.
| Cell type | Derivation | Phenotype | TEER [Ω ⋅ cm2] | Reference |
|---|---|---|---|---|
| RPMI 2650 | Nasal squamous cell carcinoma | Multilayered, non-ciliated, mucin expression, exhibiting tight junction formation | 41-270 | [28][27] |
| 16HBE14o- | Immortalized healthy tissue | Cuboidal monolayer, non-ciliated but microvilli present tight junction formation, transporter protein expression | ~ 250 | [34] |
| Calu-3 | Adenocarcinoma | Columnar monolayer, mucin expression, tight junction formation, microvilli formation | > 300 | [39][37][54] |
| BEAS-2B | Immortalized healthy tissue | Monolayer formation, cytokine secretion, antioxidant expression, no mucin secretion or tight junction formation | < 100 | [37] |
| A549 | Alveolar adenocarcinoma | Monolayer formation, membrane bound inclusion, alveolar type II-like, surfactant secretion, no tight junction formation | n.a. | [55][56][57] |
| NCI-H441 | Papillary adenocarcinoma | Polarized monolayer formation, alveolar type II-like, Clara cell-like | ~ 300 | [52] |
n.a. = not applicable
For the ET region of the respiratory system, only very few cell lines are suitable. The only immortalized cell line of human origin frequently used in nasal drug delivery research is the RPMI2650 cell line.[24] This epithelial cell line was obtained from an anaplastic squamous cell carcinoma of the human nasal septum and displays strong stability even after extended in vitro culturing without phenotype alteration exhibiting superior differentiation under ALI conditions.[25,26] RPMI2650 cells do not grow in polarized monolayers but in sheets of non-ciliated cells, and transepithelial electrical resistance (TEER) values range from 41-270 Ω ⋅ cm2.[27,28] Due to the lack of suitable immortalized cell lines of normal nasal tissue, many applications of drug delivery research use primary epithelial cells from nasal brushings or from nasal polyps.[29,30]
One of the most widely used human bronchial epithelial cell lines, 16HBE14o-, was established by transforming normal bronchial epithelial cells. They are used to mimic the TB region and have a cuboidal shape expressing tight junction proteins and develop proper TEER values.[31] While there are conflicting reports whether this cell line is ciliated or not, it has been shown that 16HBE14o- cells express several transport proteins.[32] Furthermore, although bronchial epithelial cells can typically be found at an air interface, these cells sometimes fail to polarize under ALI conditions.[33] The exact mechanism or reason for this is currently still unknown but improvements in culture conditions were shown to enable 16HBE14o- cell polarization at ALI, as it was shown for the RPMI 2650 cell line.[25,34] Apart from 16HBE14o-, another cell line obtained from healthy human epithelial cells, the BEAS-2B cell line, is also commonly used. It is particularly described in co-culture models to evaluate the influence of epithelial cells on co-cultured immune cells after exposition to tobacco smoke or diesel exhaust.[35,36] However, at the ALI, these cells do not appear to polarize, form tight junctions or produce mucus and only reach very low TEER values of < 100 Ω cm2.[37]
Another extensively used cell line mimicking bronchial cells is the Calu-3 cell line derived from a bronchial epithelial adenocarcinoma.[38] This cell line shows an excellent polarized monolayer formation at ALI together with high levels of tight junction proteins and mucus production.[39] Depending on the culture conditions, TEER values of Calu3 cells at ALI are usually bigger than 300 Ω cm2, sometimes even exceeding 1000 Ω cm2.[37] However, also for this cell line the data for cilia expression appear to be contradictory, which might be related to the number of cell passages.[38,40,41] Due to high stability, robustness, in vivo resemblance and easy culture, Calu-3 cells are often described as a suitable model for the respiratory epithelium.[42]
The most commonly used alveolar cell line mimicking the Al region is the A549 cell line from human pulmonary adenocarcinoma. Many studies suggest that these cells are unable to polarize and lack functional tight junctions, although they seem to express certain tight junction proteins such as Occludin and E-cadherin.[43] Despite these limitations, the A549 cell line contains multilamellar cytoplasmic inclusion bodies typically seen in human alveolar type II cells.[44] These cells additionally release surfactant to reduce surface tension, similarly to what is observed in vivo. [45,46] Therefore, A549 cells are still used in ALI co-culture models, mostly in presence of other epithelial cell lines to facilitate cell layer polarization but also together with immune cells and endothelial cells to mimic the alveolar barrier in the lung.[47–49] Besides A549, the NCI-H441 human alveolar cell line from lung adenocarcinoma has been utilized in several studies. This cell line has been described to have characteristics of both bronchiolar Clara cells and alveolar type II cells.[50,51] It can form polarized monolayers with TEER values of around 300 Ω cm2 and has mainly been employed to study the air-blood barrier in co-cultures with endothelial cells.[52,53]
In general, the complex regulation mechanisms of airway responses to allergens, pathogens and other antigens combined with the different cell types and cytokines present in the airways have a great influence on the microenvironment. Therefore, it is obvious that such a complex system cannot be mimicked by just one cell type, and reproducing this microenvironment as a field of research is advancing continuously. Depending on which part of the respiratory tract and, above all, which disease should be analyzed, there are multiple options for designing a suitable model.
4. ALI models in health and disease state
4.1. Respiratory viruses
It is estimated that 75% of all acute morbidities in developed countries are caused by acute respiratory diseases. The underlying reason for the majority of them are viruses. For the evaluation of virus-host interactions and the development of antiviral treatments, specific models capable of high-throughput screening in physiologically relevant conditions are required. In many other fields, animal experiments are used for this purpose but in virology, small animal models are often not suitable. Depending on the nature of the virus, some animal species are not susceptable to human viral infections or need virus adaption, thereby, potentially affecting viral pathogenicity. In many cases, the clinical state of the disease in humans is not reflected properly due to a lack of expression of specific human receptors. Therefore, it is advantageous to make efforts towards establishing advanced human in vitro models for a reliable analysis of virus-host interactions.
Immortalized human cell lines, such as Calu-3, are invaluable tools for the evaluation of virus replication cycles in lung epithelial cells.[58] However, the natural target cells of viruses in the respiratory tract are differentiated cells, whose characteristic features sometimes differ widely from immortalized cells. Therefore, the analysis of virus infections in continuous cell lines after lacks important aspects of the viral pathogenesis. In the last years, models composed of well-differentiated epithelial cells from airway tissue have been established to assess respiratory virus infections under more clinically relevant terms. The cells are cultured under ALI conditions forming a monolayered, polarized and differentiated epithelium.[59] This model closely resembles the airway epithelium in vivo regarding morphology and function, including mucus production and cilia movement.[60] For emerging respiratory viruses unable to proliferate in a traditional two-dimensional (2D) submerged cell culture due to the lack of expression of several entry factors, ALI models greatly facilitate virus isolation and characterization.[61,62] Ashraf et al. developed an ALI model to study the basic characteristics of human rhinovirus-C viruses (HRV–C).[63] This subtype of human rhinoviruses, which is considered the primary cause of the common cold, has been circulating unnoticed due to the failure of culturing under submerged conditions. The group developed a model for growing HRV-C in an ALI culture of differentiated human sinus epithelial cells characterized by a pseudostratified morphology, cilia and mucus producing goblet cells. Thus, they were not only able to analyze the characteristics of clinical HRV-C but also to compare the biological properties of different subtypes of rhinoviruses in the same cell culture system. Warner et al. strengthened the hypothesis that using physiologically relevant cell lines as well as a suitable cell culture model is fundamental. They used differentiated human airway epithelial cells cultured under ALI conditions to evaluate the replication and innate immunity of rhinoviruses. With their experiments they challenged older findings obtained from HeLa cells grown under conventional culture conditions.[64] Despite the potential advantages of primary cell-based models as discussed above, there are also some limitations. Ziegler et al. studied the susceptibility to the Epstein-Barr virus using an ALI model with primary bronchial epithelial cells.[65] Thereby they detected significant donor differences. Their results suggest a significant impact of host variables to the susceptibility in the nasopharynx together with the type of EBV infection (productive or non-productive). In conclusion, studying donor-dependent infection mechanisms as well as treatment responses, but also to improving the robustness and reproducibility of in vitro models for interpretable results represents a clinical need. Jonsdottir et al. established transgenic primary ALI cultures using lentiviral vectors aiming at allowing for more combinations for virus-host interactions in different cell types and species.[66] They hypothesized that transgenesis would enable the study of viral and/or host factors, relevant for respiratory virus infections. They also expected that studying interactions between the virus and cells engineered for targeted gene knockdown or overexpression would allow the elucidation of specific mechanisms involved in virus-host interactions. This model in fact offers the potential for translation to animal cells so that viral pathogenesis can also be studied in other species in the future.
Apart from testing different virus-host interactions, ALI models can also be utilized for the screening of different therapeutically relevant agents and their effectiveness on virus inactivation.[67] Especially for respiratory coronaviruses (CoV) an immediate unmet clinical need for broad-spectrum antiviral therapies was particularly emphasized by the 2020 COVID-19 pandemic. Originally, the importance of CoVs regarding human diseases was underrated and therefore, at the beginning of the pandemic, no vaccine or general therapy was available to treat CoV-induced disease in humans. However, some strains of the mainly zoonotic coronaviruses can enter new host species and spread there rapidly.[68] Both the Middle East respiratory syndrome CoV (MERS-CoV) and the severe acute respiratory syndrome CoV (SARS-CoV) have crossed the species barrier in the recent past, entered the human population and resulted in severe diseases. In 2019, a novel human infecting coronavirus (first provisionally named 2019-nCoV, later SARS-CoV-2) was first identified in Wuhan, in the Hubei province in China, and caused a worldwide pandemic, which is not yet under control by the time this article is written.[69] Scientists were able to rapidly isolate the virus from bronchial lavage fluid of patients suffering from SARS-CoV-2 mediated COVID-19 disease. They studied its biological characteristics using ALI models of primary human bronchial epithelial cells.[70,71] Hence, polarized ALI cultures represented a valuable and high–throughput tool for rapidly gaining information about infection, replication and pathogenesis of the new virus.[72] The thus obtained knowledge helped scientists and clinicians to decide upon suitable containment measures for the population. At the time of writing this article, two mRNA based vaccines against SARS-CoV-2 are approved in a few countries.[73,74] However, antiviral treatment options for patients with CoV infections are still very rare. Multiple therapeutic approaches are currently under development including commonly known antivirals, antibodies, interferons, vaccines and more recently also nucleic acid based therapeutics.[75–80] For the evaluation of this broad variety and number of new therapeutic entities, more physiologically relevant in vitro models are needed including not only well-differentiated primary cells but also co-cultures formed by more than a single cell type. One approach for a robust, high throughput in vitro screening platform is presented by Gard et al.[21] The group utilized an ALI cell-based model of human primary airway epithelial cells integrated into a high-throughput microfluidic platform (PREDICT96-ALI). This model can be used to study virus infections and has the potential to be used for fast and clinically relevant efficacy screening of different therapeutics.
Nonetheless, to this day, only very few co-culture models have been used for the analysis of virus-host interactions and possible therapies. Yoshikawa et al. cultured Calu-3 cells at the ALI to study the different functionalities of the apical and the basolateral domains in response to viral infection.[81] After virus inoculation of differentiated Calu-3 cells, the medium from both sides, apical and basolateral, was collected and incubated with dendritic cells or pulmonary macrophages in order to assess the potential of epithelial cytokines to modulate intrinsic factors of these cells. They showed an amplification of the early acute inflammatory response by both dendritic cells and pulmonary macrophages after infection of lung epithelial cells with SARS-CoV. In another study, a real ALI co-culture model of the human respiratory tract was established by Blom et al. using human bronchial epithelial cells (16HBE14o-cell line), human monocyte-derived dendritic cells and macrophage cultures.[82] Apart from establishing a reliable ALI co-culture, they aimed to study the interplay between those three different cell types as well as interactions with biomimetic nanocarriers such as liposomes and virosomes, which show a promising opportunity for vaccines and/or drug delivery systems for antiviral therapeutics. Both studies underline the need for advanced ALI co-culture models in the field of virology, not only with a differentiated epithelial layer but also in combination with different cell types that are present in the human lung tissue. With the implementation of such three-dimensional (3D) in vitro models, scientists will be able to gain improved insights into virus-host interactions and to obtain more reliable and translatable results regarding antiviral therapy, thereby, reducing animal experimentation to a minimum.
4.2. Cystic fibrosis
Cystic fibrosis (CF) represents one of the disorders involving the respiratory tract where ALI culture can help unveil the molecular processes of the disease and the search for new therapeutic approaches. In the Caucasian population cystic fibrosis is the most prevalent autosomal recessive disease, involving about 100,000 people worldwide. It is a result of mutations in the cystic fibrosis transmembrane conductance regulator (CFTR) gene, which encodes the transmembrane protein responsible for transport of chloride and bicarbonate ions across epithelial cells.[83] Despite being a monogenic disease, about 2,000 different mutations have been observed at the CFTR level, leading to different phenotypes and severity levels. The deletion of a phenylalanine in position 508 (ΔF508) is the most frequently encountered mutation, observed in about 70% of the CF population. The mutations result in a reduction of channel number, function or both, with severe consequences on the functionality of the affected tissues.[84] Although CF is a multi-organ disorder, the lung is the most affected. CFTR loss on the apical side of lung epithelium causes an imbalanced transport of ions and fluids across the cells, leading to unpaired mucociliary clearance, chronic inflammation and recurring bacterial infections. Respiratory failure certainly represents the primary cause of death and morbidity.[85]
After the discovery of the CFTR gene in 1989,[86] the understanding and treatment of CF greatly progressed in the past decades. This resulted in improved life quality and expectancy of the patients. The development of a mouse CFTR-knockout model helped identify many characteristics of CF. However, the CF mouse model lacked the development of spontaneous lung disease, which limited its use and pushed towards the development of alternative in vivo models in pigs and ferrets, and in vitro models.[87] In the last two decades polarized in vitro epithelial cultures have been fundamental for the progresses made in this field, especially for studying the role of CFTR in CF pathogenesis. Air-liquid interface culture systems are an ideal instrument for growing epithelial cells in vitro, since they allow the production of a differentiated airway epithelium including the main features found in vivo, particularly in terms of cellular differentiation, mucus secretion and barrier function.[88] For this purpose, both secondary and primary cell lines have been used. Among the immortalized cell lines, models using both CF, CFBE41o- cells, and non-CF cells, mostly 16HBE14o- and Calu-3, can be found. Calu-3 cells, in particular, are widely used due to their high transepithelial resistance, mucus secretion and high expression of CFTR protein [87]. Alternatively, CFBE41o- is a CF-immortalized cell line homozygous for the ΔF508 deletion, therefore simulating the CF epithelium with high accuracy.[89]
Despite the ease of use and accessibility, secondary cell lines are not representative of the great variety of scenarios found in CF epithelia, particularly in terms of CFTR variants. More appropriate patient-specific epithelial cell models need to be adopted. On this basis, primary cell lines are now considered the gold standard in CF research and are used for disease modeling as well as drug screening.[18] Primary human airway epithelial cells can be isolated from biopsy samples, lung explants or cadavers and, after an expansion step, they can be seeded on transwell filters and grown at ALI. This process allows the generation of a pseudostratified epithelium with mucociliary morphology displaying the key physiologic functions of CF cells.[90] Moreover, it is possible to assess CFTR channel conductance through an Ussing chamber or patch clamping.[91] Recently, primary nasal epithelial cells have also been explored for ALI studies. These cells can overcome one of the major drawbacks of primary lung cells, namely the limited accessibility associated to the invasive sampling methods. Nasal epithelial cells are obtained by non-invasive nasal brushing of patients and it was demonstrated that they show a polarization pattern well-correlating with primary lung cells. They share similar growth and structural characteristics and, remarkably, also an analogous CFTR expression.[92,93]
The development of polarized primary CF epithelia played a significant role in the progress achieved by precision medicine in the context of CF. In particular, they strongly supported the development of CFTR modulators.[94] This class of drugs directly modulates the defective CFTR channel. They function either as CFTR potentiators by opening the channel present at the cell surface or as CFTR correctors by enhancing the transport of the protein to the cell membrane.[95] These therapeutics were a milestone in the treatment of CF, since they exert their action directly on the primary cause of the disease. However, they are not accessible to the entire CF population since they are mutation specific, meaning that they are effective only on specific CFTR mutations. The in vitro investigation of these modulators on patient-derived cells helped to discriminate the efficiency of each drug on the different genotypes. The potentiator Ivacaftor, for example, was shown to work on the G551D mutation,[96] while the corrector Lumacaftor worked best on the ΔF508 mutation.[97] A wider study additionally showed the different effects of Lumacaftor on primary cells from patients with different genotypes, demonstrating the great potential of this airway model in the identification of patient-specific treatments.[98] Notably, Pranke et al. demonstrated that the efficiency of CFTR modulators could also be evaluated on nasal epithelium by testing the effect of two CFTR correctors, Lumacaftor and Tezacaftor on primary human bronchial and nasal cells. The results revealed only small response discrepancies in the two cell types, paving the way for using easily accessible nasal epithelial cells as predictors of CFTR modulators’ effectiveness.[99]
Apart from small molecule drugs, ALI models can also be exploited to evaluate the ability of macromolecule-based therapies, including siRNA to downregulate a target gene. In the context of CF, one example for a target is the epithelial sodium channel (ENaC), which is generally responsible for the transport of sodium. In CF, this channel is upregulated causing airway liquid depletion and thickened mucus. Manunta et al. were able to efficiently down-regulate ENaC expression on primary CF and non-CF cells grown in both submerged and ALI cultures. Interestingly, lower transfection efficiencies were observed in ALI cultures, reinforcing the importance of using advanced culture models to better mimic the in vivo situation.[100] This was confirmed by another study evidencing a similar behavior in CF cells cultured at ALI and in an in vivo mouse model in terms of dose regimen. In both systems, siRNA mediated a significantly higher ENaC knockdown after three repeated transfections compared to a single administration.[101]
Co-culture models, as already discussed, present a promising opportunity for further alignment between in vitro models and the complex in vivo situation. The addition of immune cells, in fact, greatly improves the imitation of the airway microenvironment.[88] In a study by d’Angelo et al., a triple co-culture system was proposed for testing the cellular internalization and pro-inflammatory effect of an ENaC-targeted siRNA therapy. This model comprised 16HBE14o- cells as well as monocyte-derived macrophages from human blood and dendritic cells. Interestingly, in this study the formulation was applied via nebulization through the Vitrocell Cloud system, thus increasing the resemblance to inhalation under in vivo conditions.[102]
Moreover, co-culture models have also contributed to shed light on another important aspect of CF, namely bacterial infection of the airways. Staphylococcus aureus and Pseudomonas aeruginosa are the bacterial strains mainly involved in lung infections of patients with CF. Several studies showed that it is possible to cultivate both S. aureus [103,104] and P. aeruginosa [105] on the apical side of polarized CF epithelial cells grown at ALI to observe the infection process, biofilm formation and inflammatory responses. In this model, the bacterial infection can therefore be performed under non-submerged conditions, providing a more natural condition for the infection process.[106]
Using a more sophisticated method, Yonker et al. developed a model of inflamed mucosa by co-culturing human airway basal cells and neutrophils. The group observed the migration of the latter in response to inflammation mediated by P. aeruginosa growth in the apical chamber of the Transwell® filter.[107] Moreover, co-culture of epithelial and bacterial cells can be very useful for testing the efficacy of antimicrobial agents. In case of P. aeruginosa, ALI co-cultures were adopted to evaluate the ability of antibiotics,[108] combination therapies [109] and antimicrobial peptides [110] to prevent bacterial infection while examining the consequences on human epithelial cells.
4.3. Asthma
Asthma is a chronic lung illness with more than 300 million affected people worldwide, associated with a growing burden for healthcare systems of both industrialized and developing countries. The most prominent feature of asthma is a generalized inflammation of the upper airways resulting in recurrent episodes of coughing, wheezing, chest tightness and dyspnoea.[111] In addition to a severe symptomatology, chronic inflammation has also serious consequences on lung structure and functionality, especially in terms of airway hyperresponsiveness, obstruction and remodeling as well as mucus hypersecretion.[112] The mechanisms behind the inflammatory status rely on inappropriate immunological responses to common inhaled allergens, which trigger cytokine secretion by T helper 2 (Th2) immune cells. The sustained release of inflammatory cytokines mediates a perpetuated inflammatory state.[113] Apart from Th2 cells, other immune cells such as eosinophils, mast cells and dendritic cells are involved, defining asthma as an immunological disorder.[114] Indeed, asthma involves several cell types including epithelial cells, which represent the first line defense and regulators of immune responses against the environmental factors. Activated epithelial cells secrete cytokines and chemokines that recruit dendritic cells, which are responsible for the following coordination of the inflammatory cascade. Airway epithelium functionality, however, is altered in the disease state.[115] Apart from the infiltration of immune cells, bronchial biopsies of asthmatic patients point out structural changes of airway epithelium in response to chronic inflammation. In addition to the increased number of mucus-secreting goblet cells and enhanced angiogenesis, they also display a remodeled, damaged epithelium missing tight and adherent junctions with consequent loss of apicobasal polarity.[116] This condition is strictly correlated to the breakdown of the defense mechanisms observed in asthma.
The alterations of the airway epithelium make the development of suitable experimental models essential to comprehend the ongoing immunological and structural modifications and to identify possible therapeutic targets. Mouse models of allergic asthma have been widely used to investigate asthma driving mechanisms and to screen therapeutic agents. Nonetheless, they present anatomical as well as immunological discrepancies with human airways. As described above, mouse lungs differ in terms of lobar structure and branching pattern of the bronchi.[117,118] Moreover, mice lack bronchodilatory nerves and the ability to cough, along with a different pattern of mediators secreted by mast cells.[119] The complex human in vitro and ex vivo systems now available embody a valid support to the well-established in vivo models. Epithelial cells grown at ALI can be used as a powerful tool to mimic the asthmatic epithelium in vitro with high similarity to the in vivo situation. The simplest way to reproduce the polarized epithelium at ALI for asthma studies is adopting common epithelial cell lines of the respiratory tract such as 16HBE14o-, BEAS-2B and Calu-3. They are easily accessible and relatively simple to grow on Transwell® supports. Additionally, they express tight junctions, making them suitable for barrier function studies.[120] Stewart et al. evaluated different immortalized cell lines in comparison to primary bronchial epithelial cells in terms of TEER values and marker expression. They observed that Calu3 cells featured a similar expression pattern of ZO-1, E-Cadherin and MUC5AC to primary cells, although lacking a localized expression of β-tubulin.[37] Nonetheless, secondary cell lines lack the genetic features and the structural changes observed in asthmatic epithelium. The improvements achieved in primary cell extraction from bronchial biopsies allowed their implementation in asthma research as ex vivo model of airway epithelia. Primary cells from asthmatic patients show marked differences from the ones obtained from healthy individuals due to the increased secretion of inflammatory cytokines and mucus production. Furthermore, they retain a less differentiated phenotype with diminished capability to repair injuries.[121] Additionally, they have disrupted tight junctions that lower TEER values and increase permeability.[122] ALI culture of asthmatic primary cells can therefore be considered a suitable model to depict the morphologic and inflammatory imbalances caused by chronic inflammation. Notably, these models retain the phenotypic differences typical of the disease state. Gras et al. confirmed that reconstituted bronchial epithelium from mild and severe asthma patients retains a trend in inflammatory marker expression and mucus production that varies in correlation to the severity of asthma.[123] Thanks to the high resemblance to the real-life condition, ALI culture of primary epithelial cells of the respiratory tract has been a valuable tool for the identification of novel drug targets and screening of alternative therapeutic options. They were, for instance, used for analyzing the role of transforming growth factor-β (TGF-β) [124] and histocompatibility antigen G (HLA-G) [125] in airway remodeling. Moreover, in vitro cultures of primary human cells from asthmatic patients are also exploited to detect novel subsets of asthmatic patients, as the recently identified interleukin-6-driven asthmatic group.[126] As mentioned above, this model system plays a crucial role in the preclinical investigation of novel therapeutic agents and for understanding the mechanisms of action of therapies already in use. β2-adrenoreceptor agonists, for example, together with corticosteroids are first choices in the step therapy approach for the management of asthma symptomatology. Holden et al. cultured primary cells as well as BEAS-2B cells at ALI as a simplified version of the respiratory tract to investigate the effect on inflammatory response of the β2-agonists in epithelial cells in combination with corticosteroids.[127] Potential anti-inflammatory agents were also screened using ALI culture. It was confirmed that one molecule inhibited nuclear factor κ-B2 (IKK2i) leading to a reduction of inflammatory mediators in corticosteroid unresponsive epithelial cells,[128] while an src-family kinase inhibitor was able to reduce TNF-α release, a key inflammatory mediator, and improved barrier properties of severe asthmatic ALI cultures.[129] Sexton et al. tested if a human monoclonal antibody inhibited a serine protease from the tissue kallikrein family in primary cultures of bronchial epithelial cells, which induces bronchoconstriction and mucus hypersecretion in the airways. The monoclonal antibody reduced the mucus secretion and the inflammatory burden. The results from ALI cultures showed that antibody treatment restored kallikrein expression and reduced mucus sexretion. In vivo studies in a sheep asthma model emphasized that the monoclonal antibody decreased bronchoconstriction and hyperresponsiveness as well.[130]
As already discussed in the context of cystic fibrosis, nasal primary epithelial cells offer a promising alternative to bronchial primary cells thanks to their higher availability and effortless extraction procedure. A study comparing primary asthmatic nasal cells with non-asthmatic and bronchial cells demonstrated that the former express higher levels of typical mediators which are commonly encountered in asthmatic patients, such as vascular endothelial growth factor (VEGF) and TGF-β. This observation strengthened the hypothesis that nasal epithelial cells are a suitable surrogate for primary lung epithelial cells.[131] Healey et al. utilized nasal epithelial cells from asthmatic donors in the preclinical testing of an siRNA-based therapy aimed at downregulating STAT-6 expression in epithelial cells, a gene involved in bronchial inflammation of asthma.[132] Similarly, Bequignon et al. investigated the ability of a monoclonal antibody to bind the neonatal Fc receptor in human nasal epithelial cells as a potential administration route in asthma-related chronic rhinosinusitis.[30] Despite the advantages offered by this alternative source of primary cells, it is important to consider that they have some intrinsic differences in comparison to the bronchial ones that might affect the reliability of the results. It is therefore important to keep in mind the implications of choosing one or the other source of cells.
The asthma research field has greatly benefited from ALI culture of epithelial cells. However, simple monoculture models cannot represent the complex cellular network typical of the disease state. Asthma is a multicellular disease involving epithelial cells as well as several immune cells. Co-culture systems more closely represent the complexity encountered in asthmatic lungs since they bring together two or three different cell types on the same Transwell® filter. Co-cultures for asthma-related studies generally present lung epithelial cells, primary or immortalized, in the apical chamber of a Transwell® to form a polarized epithelium, while the basolateral chamber hosts a subtype of immune cells including dendritic cells,[133] eosinophils,[134] or T cells.[135] A co-culture formed by primary epithelial cells and T cells was used by Wawrzyniak et al. to study the role of T cell secreted cytokines and histone deacetylases on the integrity of the epithelial barrier integrity. They demonstrated a beneficial effect on barrier integrity after treatment with a histone deacetylase inhibitor.[135] Co-cultures were also used to study airway remodeling in asthma, a typical feature of the disease. Haghi et al. developed an airway remodeling model by growing primary bronchial epithelial cells on the apical chamber and airway smooth muscle cells on the basolateral one.[136] In another study, Reeves et al. developed a co-culture of primary human bronchial epithelial cells derived from asthmatic children and human lung fibroblasts to study the fibroblast-myofibroblast transition. They showed that it is possible to restore the healthy condition by using a monoclonal antibody inhibiting TGF-β, a central factor involved in airway remodeling.[137] Triple co-cultures would be an even more exemplary model of the asthmatic environment. However, studies exploiting this method are infrequent. A recent study by Paplinska-Goryca et al. provides a triple co-culture model formed by primary epithelial cells, dendritic cells and macrophages that might be helpful for better understanding the pathogenesis of asthma thanks to the higher complexity of the system.[138]
4.4. Lung cancer
Lung cancer is the most prevalent cause of cancer-related deaths, both in men and also in women.[139] Therapeutic advancements of the last ten years are barely reflected in the small decline of lung cancer mortality. One reason is the histologic diversity of lung cancer including the three most common types: adenocarcinoma, squamous cell carcinoma and small cell carcinoma as well as several less frequent types such as adenosquamous carcinoma and large cell neuroendocrine carcinoma. Due to this high variability, it is necessary to examine the exact molecular mechanisms of each cellular type to further improve therapy options. Hereby, animal models are an important standard tool. Apart from known drawbacks such as high costs, species differences and limited availability, ethical issues regarding the use of animals in tumor research are controversially discussed.[140] Therefore, also in this field extensive research has been conducted in the last years to establish reliable and physiologically relevant in vitro cell culture models, thereby, reducing the number of animals for experiments in tumor research and drug development. Since conventional 2D cell cultures are not capable of mimicking the complex architecture and microenvironment of lung cancer in vivo, 3D cell cultures and co-cultures contribute greatly to the knowledge of tumor cell pathophysiology and also for anti-tumor drug discovery.[141]
Often, ALI systems are used to study the exact mechanisms of malignant transformations of different cell types because of their resemblance to the human lung physiology.[142,143] ALI systems also serve studies about how a specific signaling pathway can potentially be inhibited.[144] Horie et al. established a 3D co-culture model at the ALI using A549 cells and lung cancer-associated fibroblasts.[145] They found that fibroblasts enhanced A549 cell invasion into collagen gels, showing their tumor-promoting role through the production of instructive signals including growth factors and chemokines. Due to its aggressive nature and high mortality, it is especially important to improve the early detection and chemoprevention of lung cancer. However, the pathobiology of early stages is poorly understood. To this purpose, Correia et al. engineered an ALI culture of immortalized human bronchial epithelial cells overlaid on a fibroblast containing collagen layer to induce the activation of oncogenes.[146] They confirmed that deregulation of oncogene SOX2, under proper in vivo mimicking circumstances, leads to bronchial dysplasia. These findings are important steps forward towards the development of therapeutics used for primary and secondary chemoprevention.[144]
The most frequently found 3D in vitro tumor models are lung cancer spheroids constituted of one or more cell types.[147,148] Tumor spheroids are cell constructs that have self-organized to exhibit a 3D structure resembling the cancerous state. It was shown that genes expressed in 3D spheroids, especially the ones responsible for aggressive tumor growth, recapitulate the in vivo phenotype better than the respective 2D models.[149] Also, three dimensional models hold the potential to guide personalized medicine in the future, clearly demonstrating their superiority over conventional models.[150] However, particularly in lung cancer research, spheroid models have one critical disadvantage. Many of them lack air exposure and therefore do not reflect the physiological environment correctly. Subsequently, these cultures are unsuited models for testing the efficacy of aerosolized drugs. Meenach et al. compared lung tumor spheroids in air- as well as liquid-interface culture for treatment with paclitaxel-containing PEGylated phospholipid microparticles in form of a dry powder.[151] For the cultivation at ALI conditions, A549 cells were seeded on collagen coated transmembrane inserts in a 24-well plate and then incubated for 24 h under submerged conditions. Subsequently, the medium on the apical side was discarded and spheroid formation was evident after 9 days. The group showed that IC50 values of paclitaxel can differ substantially between treatment of a tumor spheroid grown under submerged conditions and another one cultured at ALI conditions. Moreover, they stated that the data from ALI evaluation were reported in μg/dose, in contrary to the usual μM results, thereby the patient dose in terms of mg/kg via direct inhalation of the drug can be easily determined. Gupta et al. developed a high-throughput model growing A549 lung adenocarcinoma cells as 3D spheroids at the air-liquid interface.[152] They found that, due to limited drug diffusion, the cytotoxic effect of paclitaxel in A549 ALI spheroids was only noticeable on the outer layer of the cell complex. However, the co-administration of a tumor penetrating peptide enhanced paclitaxel penetration depth. These results emphasize the importance of air-grown three-dimensional in vitro models not only to characterize tumor growth and microenvironment of different cell types, but also to gain a deeper knowledge of the resistance mechanisms in lung cancer. Such models strongly improve efficacy and success of the screening process of chemotherapeutics and drug combinations and serve as a first step before conducting expensive in vivo pre-clinical or clinical studies.
Apart from spheroids, other ALI models have been developed. Movia et al. used a multilayered cell culture of A549 cells to study four anti-cancer drugs delivered by a clinical nebulizer as liquid aerosol.[153] Their results clearly demonstrated the advantages of this model: the biological complexity due to 3D architecture, closer resemblance to the patient status by modeling the multidrug resistance (MDR) observed in human patients, and applicability of aerosol administration methods due to ALI conditions. In a subsequent study, the group incorporated human fibroblasts into their multilayered cell culture (MCC) model to assess their role in MDR.[154] Indeed, cancer cell-fibroblast crosstalk led to a higher MDR than the one found in an ALI multilayered monoculture. These studies emphasize that a monoculture, even if it is three-dimensional and under ALI conditions, still does not reflect the in vivo conditions sufficiently. Therefore, it is crucial to integrate several other key factors, such as immune cells, extracellular matrix and genetic variability, into the in vitro platform in further studies. Zhang et al. assessed the impact of tumor microenvironment on bone marrow mesenchymal stem cells.[155] Conclusively, they found that an in vitro ALI lung cancer A549 microenvironment might induce stem cells to undergo alterations in cell proliferation, morpholgy, cytoskeleton, karyotype and migration ability. This study is an important example for the versatility and flexibility of advanced ALI in vitro models. In lung cancer research, they can be used for the evaluation of various therapeutic approaches by simply incorporating key factors of interest.
Due to the versatile nature of lung cancer and the increasing development of multidrug resistance in tumors, new and innovative therapy options are constantly needed to improve therapeutic efficacy. In the last decade, nanotechnology emerged as a promising alternative and/or addition to conventional treatment strategies. When using these nanocarriers in pulmonary administration, the lung can be used as port of entry limiting systemic distribution and avoiding first pass metabolism.[156,157] Many attempts have been described in the literature to improve the delivery of already approved chemotherapeutics, such as paclitaxel or doxorubicin, by using nanocarriers.[158–160] Furthermore, nanocarriers are often used to encapsulate new therapeutic entities for lung cancer treatments such as nucleic acids,[161] and also for co-delivery of different therapeutic agents.[162]
The huge variety and number of combinations of delivery systems with already existing or new active pharmaceutical ingredients requires testing in more complex and physiologically relevant in vitro models to assess and compare efficacy. By using 3D co-culture systems scientist can make reliable statements about penetration, efficacy, toxicity and other characteristics of drug formulations, thereby reducing the amount of animal experiments drastically. Conclusively, the development of these advanced cell culture systems is crucial because in combination with organ-on-a-chip models and simulation approaches they could potentially lead the way to animal-free research.
4.5. COPD
Chronic obstructive pulmonary disease (COPD), together with asthma, is considered one of the chronic respiratory diseases displaying the highest impact on healthcare systems worldwide. Based on the World Health Organization report, it affects more than 250 million people around the world, with more than 90% of mortality in low- and middle-income countries.[163] COPD is triggered by persistent exposure to toxic gases and particles, where cigarette smoke exposure was identified as a central risk factor. Tobacco smoke, in fact, mediates an abnormal chronic inflammatory status resulting in severe consequences on lung structure and functionality.[164] COPD affects small airways, lung parenchyma as well as larger airways, and it is characterized by a progressive obstruction of the airways that ultimately leads to lung failure. The clinical manifestations of COPD can be grouped into two major subsets, chronic bronchitis and emphysema, which affect large and distal airways, respectively. Chronic bronchitis is distinguished by chronic inflammation and remodeling of the large airways together with mucus secretion, while emphysema shows a progressive destruction of airway walls as well as loss of alveolar cells, which consequently impairs gas exchange.[165] The abnormal inflammatory response typical of COPD is linked to an enhanced presence of inflammatory cells in the airways, with neutrophils, macrophages and CD8+ T cells playing a prominent role. These cells secrete cytokines and chemokines including TNF-α, IL-1β and IL-8, which mediate a perpetuated inflammatory condition.[166] Similarly to asthma, bronchial epithelial cells retain a central function also in COPD. While at physiological condition airway epithelium acts as a defensive barrier towards external agents, the alterations of the homeostatic environment driven by toxic agents cause severe modifications of epithelium structure and functionality. This is reflected particularly in terms of reduced mucociliary clearance and increased permeability to external factors. Therefore, lung epithelium faces increased permeability, reduced cilia beat ability as well as decreased mucus clearance. Moreover, in this imbalanced status, epithelial cells are not only the target of inflammatory mediators, but they also secrete cytokines and chemokines that perpetuate and worsen the inflammatory status. The increased secretion of TGF-beta and EGF, for example, is directly linked to fibrosis and mucus secretion.[167]
Considering the central role played by epithelial cells in COPD, ALI models can grant deeper understanding of the disease driving mechanisms as well as identification of novel therapeutic options. COPD is directly correlated to lung exposure to toxic pollutants such as tobacco smoke or diesel exhaust. Therefore, ALI cultures represent a straightforward tool mimicking the in vivo lung environment of the disease. Several studies have shown that after growing and differentiating cells at ALI, they can be exposed to cigarette smoke to obtain an in vitro system incorporating most of the effects observed also in vivo. Shamberger et al. demonstrated that cigarette smoke exposure of healthy human primary bronchial epithelial cells alters their differentiation and functionality. Apart from impairing the epithelial barrier integrity, it also affected cellular differentiation, resulting in an elevated number of mucus-secreting cells while the number of ciliated cells was decreased. These changes caused a mucus-rich lung environment.[168] Indeed, mucus hypersecretion is one of the main features of COPD. Culturing epithelial cells at ALI is essential to investigate this distinct mucus hypersecretion trait of the disease [169] along with reduced mucus clearance by ciliated cells.[170] The effect of tobacco smoke on epithelial barrier integrity has also been explored in terms of tight junction loosening [171] and airway remodeling.[172] Both factors are crucial for the pathophysiology of the disease and, similarly to the ones described above, their understanding was expanded by ALI cultures. Cells grown under this condition, in fact, form differentiated epithelia with different cellular subsets, such as ciliated cells, and can even secrete mucus, a condition not reproducible under submerged cultures. Recently, a new manufacturing method was established to grow primary small airway epithelial cells at the ALI. Small airways are the part of the airways mainly affected by chronic bronchitis. Gindele et al. showed that once primary cells from COPD patients were grown at ALI and exposed to cigarette smoke, their behavior well correlated to the in vivo conditions in terms of barrier integrity, mucus secretion and cellular differentiation, making them a suitable tool for further COPD treatment studies.[173] The effect of cigarette smoke on barrier integrity is reflected also in an increased susceptibility to microbial infections.[174] Co-culture of epithelial cells grown at ALI and bacteria were developed along with exposure to cigarette smoke. Amatngalim et al. grew primary epithelial cells from COPD and non-COPD subjects at ALI and studied the different response to Haemophilus influenzae after exposure to cigarette smoke. They observed that antibacterial activity was lower in primary cells from COPD patients and suppressed after cigarette smoke exposure.[175]
To further improve the translatability of ALI-based systems in COPD research, efforts have been made towards the advancement of exposure systems for tobacco smoke. Azzopardi et al. used an aerosol exposure chamber to uniformly expose lung epithelial cells grown at ALI and used it to study the consequences on cellular viability and cytokine release after the aerosolization of tobacco smoke. This system allowed the investigation of tobacco effect on cells with various exposure regimens and exposure times that well correlate with the real life parameters.[176] In another study, primary COPD epithelial cells were exposed to diesel exhaust, another toxic agent responsible for triggering the disease, using a Vitrocell nebulization system. Instead of using suspended particles, this exposure system allowed to reproduce in vitro exposure conditions similar to the ones observed in everyday life.[177] As discussed above, co-culture models can fill the rift between in vitro and in vivo models thanks to more advanced cellular complexity.
In COPD research, several studies have exploited co-culture models to study the cellular networks involved in the disease. Ladjemi et al. developed a co-culture model formed by primary epithelial cells from COPD patients and B-cells. They used this system to test how the bronchial epithelium influenced the humoral response in the lung. Specifically, they observed the effect of Interleukin 6 (IL-6) secreted by epithelial cells on immunoglobulin A (IgA) secretion by B cells, which is increased in COPD patients.[178] In another study, co-culture of respiratory epithelial cells and lung fibroblasts was established to understand the mechanisms behind airway remodeling and inflammation in COPD. The authors observed a stronger interleukin1 α (IL-1α) mediated inflammation in co-cultures exposed to cigarette extract, confirming the connection between smoke and inflammation.[179] An additional co-culture model was established between primary small airway epithelial cells and macrophages to analyze the epithelial wound injury mechanisms in respiratory diseases.[180] COPD co-culture models could therefore represent a useful tool to gain a deeper knowledge of the driving mechanisms of the disease as well as for in vitro screening of potential therapeutic agents. So far, most of the treatment-related studies were carried out using ALI cultures involving only a single cell line, mostly epithelial cells. The mucus-secreting cell line Calu-3, for example, was utilized to test the ability of simvastatin, a drug mediating reduced mucus secretion of epithelial cells of COPD patients.[181] Calu-3 cells were also exposed to cigarette smoke extract to examine the ability of the antimicrobial peptide Cathelicidin LL-37 to prevent the disruption of tight junctions. This effect was expected to reverse the impaired activity of the epithelium typical of COPD.[182] Another study investigated the impact of Roflumilast, a phosphodiesterase inhibitor, on mucociliary clearance impairment. A Vitrocell nebulizer was used to nebulize cigarette smoke on primary human bronchial epithelial cells pre-treated with Rofumilast. An Ussing chamber was used to determine the recovery of mucociliary activity.[183] Primary cell-based ALI cultures were also used to test the potential of monoclonal antibody candidates to improve the COPD phenotype. ALI cells treated with an anti-TGF-β monoclonal antibody reversed the progressive de-differentiation of the epithelium typical of the disease.[184] Monoclonal antibodies directed towards IL-1α and IL-1β decreased cigarette smoke-mediated airway inflammation in primary human bronchial epithelial cells.[185] Taken together, ALI models of mono- and co-culture have been used in a variety of pharmacological and pharmacotherapeutic studies trying to understand and treat COPD better.
5. Commercially available ALI models
As described above in detail, there is an unmet urgent clinical need for standardized 3D in vitro ALI models of the respiratory tract to study toxicology and for efficient drug screening. Therefore, several companies have specialized in the fabrication of ALI models mimicking the morphology of healthy and diseased human tissue. Two of these models, available under the brand names MucilAir™ (Epithelix) and EpiAirway™ (MatTek Corporation), are made of primary human epithelial cells which are freshly isolated from nasal or bronchial biopsies.[186] They can accurately reproduce the biophysiology of human airway epithelia comprising a functional mucociliary system and the secretion of mucus.[187] It was shown that both, MucilAir™ and EpiAirway™ models, express tight and adherent junction proteins, as well as functional ABC drug efflux transporters.[188,189] Hoffmann et al. showed similar permeability of 30 model substances when comparing MucilAir™ to nasal and bronchial epithelium in human tissue.[190] These findings confirm the match of these ALI models with major features of a normal human nasal and bronchial epithelium.[191] Hence, these models are widely used for toxicology studies, drug efficacy and formulation screening.[192–195]
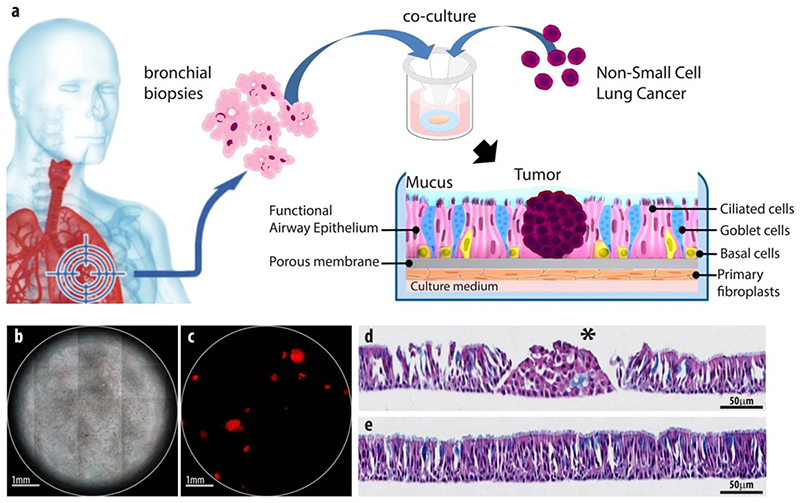
With two other models expressing a different phenotype, EpiAlveolar™ and SmallAir™, scientists are additionally able to analyze the impact of different therapeutics on the lower respiratory tract.[197,198] The two leading companies in this field, MatTek Corporation and Epithelix, recently also developed co-cultures of ALI epithelial cells and fibroblasts (EpiAirway™ FT, MucilAir™ HF) for advanced analysis of cell interaction and better mimicking in vivo conditions.[199,200] These commercial models open various possibilities for further modifications with other cell types, bacteria and viruses depending on the application. Outlaw et al. infected cells of the EpiAirway™ model with SARS CoV-2 to study the efficacy of a lipopeptide as virus entry inhibitor (Figure).[201] As shown by Signer et al., MucilAir™ can readily be infected with different respiratory coronaviruses in order to evaluate the effect of new antiviral agents.[202] Another example to show the modularity of these systems is given by Mas et al. (Epithelix).[203,204] They combined a functional respiratory epithelium, primary lung fibroblasts and proliferating tumor nodules from a KRAS mutated Non-small Cell Lung Cancer cell line. After confirming the biological relevance of this brand named OncoCilAir™ model regarding the in vivo situation, the group showed reduced growth of tumors when treated with MEK inhibitors and the standard anticancer agent docetaxel. Subsequently, this model has been used in a variety of other studies including specific tumor targeting and multi organ chips.[205,206]
In general, organ-on-a-chip devices present the possibility to create artificial tissue microenvironments simulating those conditions found in in vivo. This can also be used for mimicking lung functions involving a complex structure and fluid and solid mechanical stress.[207,208] Zamprogno et al. established a lung-on-a-chip model to reconstitute the lung alveolar barrier. They used a hexagonal gold mesh with a suspended stretchable membrane to culture alveolar epithelial cells in ALI conditions mimicking physiological lung movement.[209] Huh et al. developed a microfluidic system replicating a functional unit of the living human lung. [210] Therefore, they constructed a compartmentalized microchannel system consisting of two chambers separated by a mesoporous elastomeric membrane. Human alveolar epithelial cells were seeded into the chambers and cultured at ALI including pulmonary microvascular endothelial cells. With this model, the group was able to simulate physiological breathing motions by stretching the mesoporous membrane. Although these particular techniques and execution are not yet optimized, there is already a microfluidic based ALI lung model on the market. The company SynVivo developed a device containing a co-culture of epithelial cells embedded into vasculature comprised of endothelial cells. Hereby, tight junctions are formed, functional cilia are built, and airway tubules form and transport mucus.[211] These examples including many more that can be found in literature demonstrate that lung-on-a-chip models are valid tools in pharmaceutical development.[212]
6. Aerosolization systems
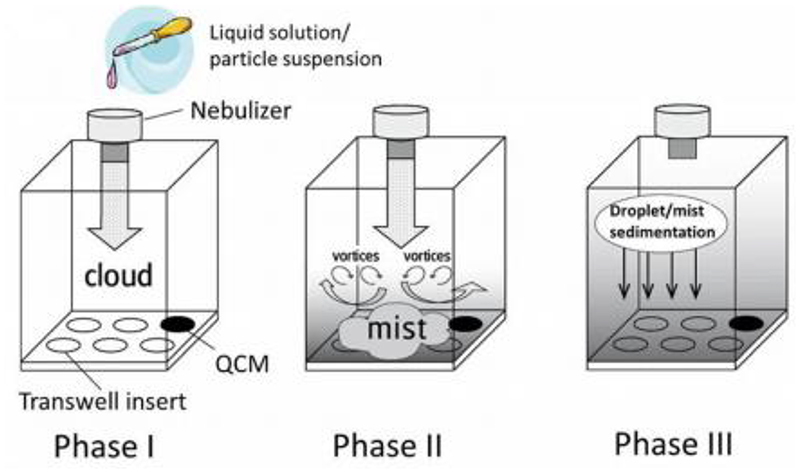
All the models described above demonstrate the possibility to mimic the exceptionally complex nature of the lung epithelium in vitro using ALI cultures. However, so far, the lack of high-throughput technology to obtain dosimetrically accurate aerosol-to-cell drug delivery hampers the development of aerosolized inhalable therapeutics. Lenz et al. investigated the ALICE-CLOUD system, an aerosol-to-cell exposure system with a vibrating mesh nebulizer developed for the use in standard multi-well plates.[214] The same group found that aerosolized drug delivery with the ALICE system results in a ca. 8 μm thin liquid layer, which is about 1000-fold lower than typical media heights under submerged cell culture conditions.[215] Therefore, it resembles the clinical conditions in the bronchial regime making aerosolized drug delivery to ALI cells crucial for biokinetic studies. This system can be combined with co-culture systems to examine the potential effect of particles in the lung using a sophisticated in vitro model.[216] Vitrocell Systems were among the first companies to offer a commercial version of this system. An interesting application of a tetra-culture in combination with the Vitrocell system was investigated by Klein et al. regarding the effects of particle deposition on the lung. [216] In this model, four different cell lines (epithelial cells, macrophages, mast cells and endothelial cells) were grown on the same Transwell® insert demonstrating the potential offered such ALI systems. Cells were grown on both sides of the insert to mimic in vivo cellular distribution (Figure 5).
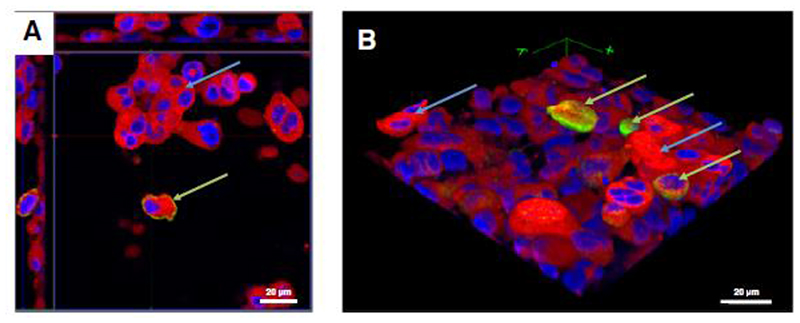
Figure 5.
Z-stack image series for the analysis of THP-1 macrophage and HMC-1 cell distribution in the tetra-culture system of the apical compartment, analyzed via CLSM. Cell membranes are stained with cell mask deep red dye (red), and cell nuclei are stained with DAPI (blue); Macrophage-like cells are counterstained with an anti-CD11b-antibody. A: x–y orthoslice. B: 3D image of the tetra-culture based on the the z-stack from A. THP-1 (green arrows) and HMC-1 (blue arrows) cells are situated on top of the epithelial cells. EA.hy 926 cells were not present in the 3D reconstruction.[216] Reproduced with permission. Copyright 2013, BioMed Central.
Another example of a commercially available aerosolization system is CULTEX from Cultex® Technology. The company did not only develop an exposure system but also a computer-controlled long-term cultivation system.[217,218] Using these two modules together allows prolonged ALI cultivation of normal human bronchial epithelial cells for a period of 38 days exhibiting in vivo-like differentiation characteristics for inhalation toxicological studies. The computer-controlled system can operate independently, reducing the risk of contamination and eliminating process variability. With the CULTEX aerosolization system, the particle or gas exposure takes place for 15-60 min under humid atmosphere at 37°C. The device was extensively validated for the inhalation of airborne particles by Tsoutsoupoulos et al..[219] After testing the aerosolization of 24 different substances, the results demonstrated the device to be robust, transferable and predictive for in vitro screening. Other exposure systems with a limited number of users have also been described.[220] Additionally, nebulizers can be used as well with the drawback of uncertain dose deposition after direct nebulization on top of ALI cultures. Many of the systems described in the literature need further modification for standardised particle deposition and dosimetry.[221–224]
7. Conclusion
In the last years, the need for alternatives to in vivo models to reduce, refine and replace (3R) animal experiments increased drastically. Furthermore, anatomical differences between commonly used laboratory animals, such as mice or rats, and humans lead to a significant lack of functional homology, especially regarding the respiratory tract. Modeling this part of the human body in vitro requires multiple considerations in order to simulate in vivo pathophysiology as closely as possible. In this review, different in vitro air-liquid interface cultures mimicking the human respiratory tract were described in detail. The most frequently utilized cell lines representing different areas of the respiratory tract were discussed together with the possibilities of replacing these with human-derived primary cells. Besides, different approaches to mimic diseases of the human respiratory tract, such as asthma, COPD or viral infections, were discussed. These can be of great use in gaining deeper understanding of disease pathophysiology and in high throughput drug screening to find new therapeutic options. Yet, the recreation of such a complex microenvironment in vitro using epithelial cells in combination with other cell types, such as immune cells, bacteria or even viruses, is particularly challenging leaving room for continuous progression in this field of research. Moreover, the broad variety of culture methods, cell sources and exposure setups requires further evaluation of robustness, complexity, reproducibility and ease-of-use of the in vitro setup. Until now there is no golden standard cell model. However, a few companies have started commercializing validated and standardized ALI models from primary human respiratory epithelium. In conclusion, the use of the ALI culture technique, especially in co-culture models, has the potential to result in significant advances in the development of more physiologically relevant tissue models for drug discovery and disease modelling, thereby reducing the number of experimental animals required in lung research.
Figure 2.
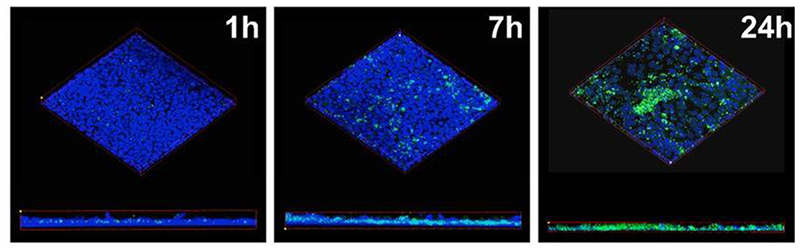
Polarized cell monolayers after infection with GFP-expressing S. aureus USA100 (green) and consecutively fixed and stained with Hoechst (blue) for confocal imaging at distinct time points. [104] Reproduced with permission. Copyright 2018, mSphere.
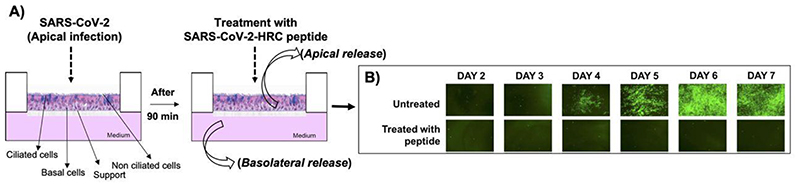
Figure 3. Cholesterol-conjugated peptides derived from SARS-CoV-2 block the SARS-CoV-2-mNeonGreen viral spread in human airway epithelial cells (HAE).
(A) Infection of HAE cells with SARS-CoV-2 (2,000 PFU/well for a multiplicity of infection of ~0.02) for 90 min with subsequent addition of SARS-CoV-2 peptide. Collection of liquid from the apical or basolateral surfaces daily. (B) Fluorescent virus is presented at the indicated days with or without peptide treatment. Adapted with permission.[196] Copyright 2020, American Society for Microbiology.
Figure 4. The OncoCilAir™ model.
(a) Scheme of lung cancer invasion of a healthy human airway modelled by OncoCilAir™ tissue. (b) Phase contrast and (c) fluorescence images of a human respiratory epithelium with EGFR tumor nodules (mRFP labeled) at the ALI. (d + e) Haematoxylin eosin histological staining of differentiated functional region of the airway (e) and with an array of non-polarized tumor cells (d, star) invading the epithelium (d). Reproduced with permission.[213] Copyright 2017, Nature.
Figure 6.
Scheme of nebulization and sedimentation process of various solutions or suspensions using the VITROCELL® Cloud 6 system with a quartz crystal microbalance (QCM). Phase 1: emission of droplet cloud from nebulizer. Phase 2: emitted cloud is transformed into a fine mist of droplets, which is distributed uniformely filling the chamber from bottom up. Phase 3: droplet deposition onto the cells via sedimentation. Reproduced with permission.[225]. Copyright 2020, BioMed Central.
Acknowledgements
This project has received funding from the European Research Council (ERC) (Grant agreement ERC-2014-StG637830).
Footnotes
Domizia Baldassi and Bettina Gabold contributed equally to this work.
The authors have declared no conflicts of interest for this article.
References
- [1].Wang H, Naghavi M, Allen C, Barber RM, Bhutta ZA, Carter A, Casey DC, Charlson FJ, Chen AZ, Coates MM, Coggeshall M, et al. Lancet. 2016;388:1459–1544. doi: 10.1016/S0140-6736(16)31012-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Bérubé K, Aufderheide M, Breheny D, Clothier R, Combes R, Duffin R, Forbes B, Gaça M, Gray A, Hall I, Kelly M, et al. In Vitro models of inhalation toxicity and disease: The report of a FRAME workshop. ATLA Altern to Lab Anim. 2009:89–141. [PubMed] [Google Scholar]
- [3].Spits H, Villaudy J. Nat Biotechnol. 2019;37:1129–1130. doi: 10.1038/s41587-019-0269-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Shapiro SD. Am J Respir Crit Care Med. 2006;174:1171–1173. doi: 10.1164/rccm.2609001. [DOI] [PubMed] [Google Scholar]
- [5].Ware LB. Am J Physiol Cell Mol Physiol. 2008;294:L149-L150. doi: 10.1152/ajplung.00472.2007. [DOI] [PubMed] [Google Scholar]
- [6].Bal HS, Goshal NG. Lab Anim. 1988;22:76–82. doi: 10.1258/002367788780746539. [DOI] [PubMed] [Google Scholar]
- [7].Van Patter LE, Blattner C. Soc Anim. 2020;28:171–190. [Google Scholar]
- [8].Lacroix G, Koch W, Ritter D, Gutleb AC, Larsen ST, Loret T, Zanetti F, Constant S, Chortarea S, Rothen-Rutishauser B, Hiemstra PS, et al. Appl Vitr Toxicol. 2018;4:91–106. doi: 10.1089/aivt.2017.0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Dvorak A, Tilley AE, Shaykhiev R, Wang R, Crystal RG. Am J Respir Cell Mol Biol. 2011;44:465–473. doi: 10.1165/rcmb.2009-0453OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Chary A, Serchi T, Moschini E, Hennen J, Cambier S, Ezendam J, Blömeke B, Gutleb AC. ALTEX. 2019;36:403–418. doi: 10.14573/altex.1901241. [DOI] [PubMed] [Google Scholar]
- [11].Person A, Mintz ML. Anatomy and Physiology of the Respiratory Tract. In: Mintz ML, editor. Disord Respir Tract Common Challenges Prim Care. Humana Press; Totowa, NJ: 2006. pp. 11–15. [Google Scholar]
- [12].Leslie KO, Wick MR. Pract Pulm Pathol A Diagnostic Approach A Vol Pattern Recognit Ser. 2017:1–13. [Google Scholar]
- [13].Schürch S, Gehr P, Im Hof V, Geiser M, Green F. Respir Physiol. 1990;80:17–32. doi: 10.1016/0034-5687(90)90003-h. [DOI] [PubMed] [Google Scholar]
- [14].Holt PG, Schon-Hegrad MA, Mcmenamin PG. Int Rev Immunol. 1990;6:139–149. doi: 10.3109/08830189009056625. [DOI] [PubMed] [Google Scholar]
- [15].Lehnert BE. Environ Health Perspect. 1992;97:17–46. doi: 10.1289/ehp.929717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].BéruBé K, Prytherch Z, Job C, Hughes T. Toxicology. 2010;278:311–318. doi: 10.1016/j.tox.2010.04.004. [DOI] [PubMed] [Google Scholar]
- [17].McDowell EM, Barrett LA, Glavin F, Harris CC, Trump BF. J Natl Cancer Inst. 1978;61:539–549. [PubMed] [Google Scholar]
- [18].Awatade NT, Wong SL, Hewson CK, Fawcett LK, Kicic A, Jaffe A, Waters SA. Front Pharmacol. 2018;9:1–11. doi: 10.3389/fphar.2018.01429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Papazian D, Würtzen PA, Hansen SWK. Int Arch Allergy Immunol. 2016;170:1–21. doi: 10.1159/000445833. [DOI] [PubMed] [Google Scholar]
- [20].Gindele JA, Kiechle T, Benediktus K, Birk G, Brendel M, Heinemann F, Wohnhaas CT, Leblanc M, Zhang H, Barel YS, Cr RG, et al. Sci Rep. 2020;10:1–17. doi: 10.1038/s41598-020-63345-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Gard AL, Maloney R, Cain BP, Miller CR, Luu RJ, Coppeta JR, Liu P, Wang JP, Azizgolshani H, Fezzie RF, Balestrini JL, et al. BioRxiv. 2020:2020.05.23.112797 [Google Scholar]
- [22].Pezzulo AA, Starner TD, Scheetz TE, Traver GL, Tilley AE, Harvey BG, Crystal RG, McCray PB, Zabner J. Am J Physiol - Lung Cell Mol Physiol. 2011;300:25–31. doi: 10.1152/ajplung.00256.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Rothen-Rutishauser B, Blank F, Mühlfeld C, Gehr P. Expert Opin Drug Metab Toxicol. 2008;4:1075–1089. doi: 10.1517/17425255.4.8.1075. [DOI] [PubMed] [Google Scholar]
- [24].Kreft ME, Lasi E, Kristan K. Pharm Res. 2015;32:665–679. doi: 10.1007/s11095-014-1494-0. [DOI] [PubMed] [Google Scholar]
- [25].Mercier C, Perek N, Delavenne X. Eur J Drug Metab Pharmacokinet. 2018;43:13–24. doi: 10.1007/s13318-017-0426-x. [DOI] [PubMed] [Google Scholar]
- [26].Sibinovska N, Simon Ž, Kristan K. Eur J Pharm Biopharm. 2019;145:85–95. doi: 10.1016/j.ejpb.2019.10.008. [DOI] [PubMed] [Google Scholar]
- [27].Reichl S, Becker K. J Pharm Pharmacol. 2012;64:1621–1630. doi: 10.1111/j.2042-7158.2012.01540.x. [DOI] [PubMed] [Google Scholar]
- [28].Kreft ME, Lasi E, Kristan K. Pharm Res. 2014;32:665–79. doi: 10.1007/s11095-014-1494-0. [DOI] [PubMed] [Google Scholar]
- [29].Schlachet I, Sosnik A. ACS Appl Mater Interfaces. 2019;11:21360–21371. doi: 10.1021/acsami.9b04766. [DOI] [PubMed] [Google Scholar]
- [30].Bequignon E, Dhommée C, Angely C, Thomas L, Bottier M, Escudier E, Isabey D, Coste A, Louis B, Papon JF, Gouilleux-Gruart V. Int J Mol Sci. 2019;20:1379. doi: 10.3390/ijms20061379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Forbes B. Pharm Sci Technol Today. 2000;3:18–27. doi: 10.1016/s1461-5347(99)00231-x. [DOI] [PubMed] [Google Scholar]
- [32].Ehrhardt C, Kneuer C, Laue M, Schaefer UF, Kim K, Lehr C. Pharm Res. 2003;20:545–51. doi: 10.1023/a:1023230328687. [DOI] [PubMed] [Google Scholar]
- [33].Ehrhardt C, Kneuer C, Fiegel J, Hanes J, Friedrich U. Cell Tissue Res. 2002;308:391–400. doi: 10.1007/s00441-002-0548-5. [DOI] [PubMed] [Google Scholar]
- [34].Forbes B, Shah A, Martin GP, Lansley AB. Int J Pharm. 2003;257:161–167. doi: 10.1016/s0378-5173(03)00129-7. [DOI] [PubMed] [Google Scholar]
- [35].Bleck B, Tse DB, Jaspers I, Curotto MA, Lafaille D, Reibman J, Bleck B, Tse DB, Jaspers I, De Lafaille AC, Reibman J. J Immunol. 2006;176:7431–7437. doi: 10.4049/jimmunol.176.12.7431. [DOI] [PubMed] [Google Scholar]
- [36].Mayer AK, Bartz H, Fey F, Schmidt LM, Dalpke AH. Eur J Immunol. 2008;38:1689–1699. doi: 10.1002/eji.200737936. [DOI] [PubMed] [Google Scholar]
- [37].Stewart CE, Torr EE, Mohd Jamili NH, Bosquillon C, Sayers I. J Allergy. 2012;2012:1–11. doi: 10.1155/2012/943982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Patel J, Pal D, Vangala V, Gandhi M, Mitra AK. Pharm Res. 2002;19:1696–1703. doi: 10.1023/a:1020761514471. [DOI] [PubMed] [Google Scholar]
- [39].Grainger CI, Greenwell LL, Lockley DJ, Martin GP, Forbes B. Pharm Res. 2006;23:1482–1490. doi: 10.1007/s11095-006-0255-0. [DOI] [PubMed] [Google Scholar]
- [40].Fiegel J, Ehrhardt C, Schaefer UF, Lehr C, Hanes J. Pharm Res. 2003;20:788–796. doi: 10.1023/a:1023441804464. [DOI] [PubMed] [Google Scholar]
- [41].Shen B, Finkbeiner WE. Am J Physiol. 1994;266:493–501. doi: 10.1152/ajplung.1994.266.5.L493. [DOI] [PubMed] [Google Scholar]
- [42].Braakhuis HMAU, He RAU, Vandebriel RJAU, Gremmer ERAU, Zwart EAU, Vermeulen JPAU, Fokkens PAU, Boere JAU, Gosens IAU, Cassee FRAU. JoVE. 2020:e61210 [Google Scholar]
- [43].Caraballo Juan C, Yshii Cecilia, Westphal Whitney, Moninger Thomas, Comellas Alejandro P. Respirology. 2011;16:340–349. doi: 10.1111/j.1440-1843.2010.01910.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Lieber M, Smith B, Szakal A, Nelson-rees W, Todaro G. Int J Cancer. 1976;70:62–70. doi: 10.1002/ijc.2910170110. [DOI] [PubMed] [Google Scholar]
- [45].Blank F, Rothen-Rutishauser BM, Schurch S, Gehr P. J Aerosol Med. 2006;19:392–405. doi: 10.1089/jam.2006.19.392. [DOI] [PubMed] [Google Scholar]
- [46].Öhlinger K, Kolesnik T, Meindl C, Gallé B, Absenger-Novak M, Kolb-Lenz D, Fröhlich E. Toxicol Vitr. 2019;60:369–382. doi: 10.1016/j.tiv.2019.06.014. [DOI] [PubMed] [Google Scholar]
- [47].Wang Y, Adamcakova-dodd A, Steines BR, Jing X, Salem AK, Thorne PS. NanoImpact. 2020;18:100215. doi: 10.1016/j.impact.2020.100215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Cappellini F, Di Bucchianico S, Karri V, Latvala S, Malmlöf M, Kippler M, Elihn K, Hedberg J, Wallinder IO, Gerde P, Karlsson HL. Nanomaterials. 2020;10:618. doi: 10.3390/nano10040618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Lehmann AD, Daum N, Bur M, Lehr CM, Gehr P, Rothen-Rutishauser BM. Eur J Pharm Biopharm. 2011;77:398–406. doi: 10.1016/j.ejpb.2010.10.014. [DOI] [PubMed] [Google Scholar]
- [50].Zhang L, Whitsett JA, Stripp BR. Biochim Biophys Acta. 1997;1350:359–367. doi: 10.1016/s0167-4781(96)00180-7. [DOI] [PubMed] [Google Scholar]
- [51].Rehan VK, Torday JS, Peleg S, Gennaro L, Vouros P, Padbury J, Rao DS, Reddy GS. Mol Genet Metab. 2002;76:46–56. doi: 10.1016/s1096-7192(02)00022-7. [DOI] [PubMed] [Google Scholar]
- [52].Woollhead AM, Baines DL. J Biol Chem. 2006;281:5158–5168. doi: 10.1074/jbc.M509947200. [DOI] [PubMed] [Google Scholar]
- [53].Papritz M, Pohl C, Wübbeke C, Moisch M, Hofmann H, Iris M, Thiermann H, James C, Kehe K. Toxicol Appl Pharmacol. 2010;245:361–369. doi: 10.1016/j.taap.2010.04.002. [DOI] [PubMed] [Google Scholar]
- [54].Sibinovska N, Žakelj S, Roškar R, Kristan K. Int J Pharm. 2020;585:119484. doi: 10.1016/j.ijpharm.2020.119484. [DOI] [PubMed] [Google Scholar]
- [55].Rothen-rutishauser B, Blank F, Mühlfeld C, Gehr P. Expert Opin Drug Metab Toxicol. 2008;4:1075–1090. doi: 10.1517/17425255.4.8.1075. [DOI] [PubMed] [Google Scholar]
- [56].Blank F, Rothen-Rutishauser BM, Schurch S, Gehr P. J Aerosol Med. 2006;19:392–405. doi: 10.1089/jam.2006.19.392. [DOI] [PubMed] [Google Scholar]
- [57].Öhlinger K, Kolesnik T, Meindl C, Gallé B, Absenger-Novak M, Kolb-Lenz D, Fröhlich E. Toxicol Vitr. 2019;60:369–382. doi: 10.1016/j.tiv.2019.06.014. [DOI] [PubMed] [Google Scholar]
- [58].Josset L, Menachery VD, Gralinski LE, Agnihothram S, Sova P, Carter VS, Yount BL, Graham RL, Baric RS, Katzea MG. MBio. 2013;4:1–11. doi: 10.1128/mBio.00165-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Qian Z, Travanty EA, Oko L, Edeen K, Berglund A, Wang J, Ito Y, Holmes KV, Mason RJ. Am J Respir Cell Mol Biol. 2013;48:742–748. doi: 10.1165/rcmb.2012-0339OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Gray TE, Guzman K, Davis CW, Abdullah LH, Nettesheim P. Am J Respir Cell Mol Biol. 1996;14:104–112. doi: 10.1165/ajrcmb.14.1.8534481. [DOI] [PubMed] [Google Scholar]
- [61].Sungnak W, Huang N, Bécavin C, Berg M, Queen R, Litvinukova M, Talavera-López C, Maatz H, Reichart D, Sampaziotis F, Worlock KB, et al. Nat Med. 2020;26:681–687. doi: 10.1038/s41591-020-0868-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Abo KM, Ma L, Matte T, Huang J, Alysandratos KD, Werder RB, Mithal A, Lou Beermann M, Lindstrom-Vautrin J, Mostoslavsky G, Ikonomou L, et al. BioRxiv. 2020:2020.06.03.132639 [Google Scholar]
- [63].Ashraf S, Brockman-schneider R, Bochkov YA, Pasic TR, Gern JE. Virology. 2013;436:143–149. doi: 10.1016/j.virol.2012.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Warner SM, Wiehler S, Michi AN, Proud D. 2019. pp. 1–13. [DOI] [PMC free article] [PubMed]
- [65].Ziegler P, Bai Y, Tian Y, Abrahamsson S, Green A, Moore J, Lee SE, Myerburg MM, Park HJ, Tang KW, Shair KHY. BioRxiv. 2020:2020.08.31.272096 [Google Scholar]
- [66].Jonsdottir HR, Marti S, Geerts D, Rodriguez R, Thiel V, Dijkman R. Viruses. 2019;11 doi: 10.3390/v11080747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Jonsdottir HR, Dijkman R. Virol J. 2016;13:1–9. doi: 10.1186/s12985-016-0479-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].De Wit E, Van Doremalen N, Falzarano D, Munster VJ. Nat Rev Microbiol. 2016;14:523–534. doi: 10.1038/nrmicro.2016.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Wang C, Horby PW, Hayden FG, Gao GF. Lancet. 2020;395:470–473. doi: 10.1016/S0140-6736(20)30185-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, Zhao X, Huang B, Shi W, Lu R, Niu P, et al. N Engl J Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Lu R, Zhao X, Li J, Niu P, Yang B, Wu H, Wang W, Song H, Huang B, Zhu N, Bi Y, et al. Lancet. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Ravindra NG, Alfajaro MM, Gasque V, Wei J, Filler RB, Huston NC, Wan H, Szigeti-Buck K, Wang B, Montgomery RR, Eisenbarth SC, et al. BioRxiv Prepr Serv Biol. 2020:2020.05.06.081695 [Google Scholar]
- [73].Polack FP, Thomas SJ, Kitchin N, Absalon J, Gurtman A, Lockhart S, Perez JL, Pérez Marc G, Moreira ED, Zerbini C, Bailey R, et al. N Engl J Med. 2020 [Google Scholar]
- [74].Jackson LA, Anderson EJ, Rouphael NG, Roberts PC, Makhene M, Coler RN, McCullough MP, Chappell JD, Denison MR, Stevens LJ, Pruijssers AJ, et al. N Engl J Med. 2020;383:1920–1931. doi: 10.1056/NEJMoa2022483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Sheahan TP, Sims AC, Graham RL, Menachery VD, Gralinski LE, Case JB, Leist SR, Pyrc K, Feng JY, Trantcheva I, Bannister R, et al. Sci Transl Med. 2017;9 doi: 10.1126/scitranslmed.aal3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Jiang L, Wang N, Zuo T, Shi X, Poon KMV, Wu Y, Gao F, Li D, Wang R, Guo J, Fu L, et al. Sci Transl Med. 2014;6:1–10. doi: 10.1126/scitranslmed.3008140. [DOI] [PubMed] [Google Scholar]
- [77].Lundin A, Dijkman R, Bergström T, Kann N, Adamiak B, Hannoun C, Kindler E, Jónsdóttir HR, Muth D, Kint J, Forlenza M, et al. PLoS Pathog. 2014;10 doi: 10.1371/journal.ppat.1004166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Shalhoub S, Farahat F, Al-Jiffri A, Simhairi R, Shamma O, Siddiqi N, Mushtaq A. J Antimicrob Chemother. 2015;70:2129–2132. doi: 10.1093/jac/dkv085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Liu R, Wang J, Shao Y, Wang X, Zhang H, Shuai L, Ge J, Wen Z, Bu Z. Antiviral Res. 2018;150:30–38. doi: 10.1016/j.antiviral.2017.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].de Wilde AH, Wannee KF, Scholte FEM, Goeman JJ, ten Dijke P, Snijder EJ, Kikkert M, van Hemert MJ. J Virol. 2015;89:8318–8333. doi: 10.1128/JVI.01029-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Yoshikawa T, Hill T, Li K, Peters CJ, Tseng CTK. J Virol. 2009;83:3039–3048. doi: 10.1128/JVI.01792-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Blom RAM, Erni ST, Krempaská K, Schaerer O, Van Dijk RM, Amacker M, Moser C, Hall SRR, Von Garnier C, Blank F. PLoS One. 2016;11:1–25. doi: 10.1371/journal.pone.0163539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Boucher R. Adv Drug Deliv Rev. 2002;54:1359–1371. doi: 10.1016/s0169-409x(02)00144-8. [DOI] [PubMed] [Google Scholar]
- [84].Ratjen F, Bell SC, Rowe SM, Goss CH, Quittner AL, Bush A. Nat Rev Dis Prim. 2015;1:15010. doi: 10.1038/nrdp.2015.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Stoltz DA, Meyerholz DK, Welsh MJ. N Engl J Med. 2015;372:351–362. doi: 10.1056/NEJMra1300109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Riordan JR, Rommens JM, Kerem BS, Alon NOA, Rozmahel R, Grzelczak Z, Zielenski J, Lok SI, Plavsic N, Chou JL, Drumm ML, et al. Science (80-) 1989;245:1066–1073. doi: 10.1126/science.2475911. [DOI] [PubMed] [Google Scholar]
- [87].Rosen BH, Chanson M, Gawenis LR, Liu J, Sofoluwe A, Zoso A, Engelhardt JF. J Cyst Fibros. 2018;17:S28-S34. doi: 10.1016/j.jcf.2017.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Castellani S, Di Gioia S, di Toma L, Conese M. Anal Cell Pathol. 2018;2018 doi: 10.1155/2018/3839803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Ehrhardt C, Collnot EM, Baldes C, Becker U, Laue M, Kim KJ, Lehr CM. Cell Tissue Res. 2006;323:405–415. doi: 10.1007/s00441-005-0062-7. [DOI] [PubMed] [Google Scholar]
- [90].Scott WO, Randell H, Leslie Fulcher M, Olsen JC. Methods Mol Biol. 2011;2:285–310. doi: 10.1007/978-1-61779-120-8_18. [DOI] [PubMed] [Google Scholar]
- [91].Mou H, Brazauskas K, Rajagopal J. Pediatr Pulmonol. 2015;50:S14-S23. doi: 10.1002/ppul.23233. [DOI] [PubMed] [Google Scholar]
- [92].Schögler A, Blank F, Brügger M, Beyeler S, Tschanz SA, Regamey N, Casaulta C, Geiser T, Alves MP. Respir Res. 2017;18:1–10. doi: 10.1186/s12931-017-0706-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Brewington JJ, Filbrandt ET, LaRosa FJ, Moncivaiz JD, Ostmann AJ, Strecker LM, Clancy JP. JCI Insight. 2018;3:1–14. doi: 10.1172/jci.insight.99385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Stacey ETZ, Martiniano L, Sagel Scott D. Curr Opin Pediatr. 2016;28:312–317. doi: 10.1097/MOP.0000000000000351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Lopes-Pacheco M. Front Pharmacol. 2016;7:1–20. doi: 10.3389/fphar.2016.00275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Van Goor F, Hadida S, Grootenhuis PDJ, Burton B, Cao D, Neuberger T, Turnbull A, Singh A, Joubran J, Hazlewood A, Zhou J, et al. Proc Natl Acad Sci U S A. 2009;106:18825–18830. doi: 10.1073/pnas.0904709106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Van Goor F, Hadida S, Grootenhuis PDJ, Burton B, Stack JH, Straley KS, Decker CJ, Miller M, McCartney J, Olson ER, Wine JJ, et al. Proc Natl Acad Sci U S A. 2011;108:18843–18848. doi: 10.1073/pnas.1105787108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Awatade NT, Uliyakina I, Farinha CM, Clarke LA, Mendes K, Solé A, Pastor J, Ramos MM, Amaral MD. EBioMedicine. 2015;2:147–153. doi: 10.1016/j.ebiom.2014.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Pranke IM, Hatton A, Simonin J, Jais JP, Le Pimpec-Barthes F, Carsin A, Bonnette P, Fayon M, Stremler-Le Bel N, Grenet D, Thumerel M, et al. Sci Rep. 2017;7:1–11. doi: 10.1038/s41598-017-07504-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Manunta MDI, Tagalakis AD, Attwood M, Aldossary AM, Barnes JL, Munye MM, Weng A, McAnulty RJ, Hart SL. Sci Rep. 2017;7:1–12. doi: 10.1038/s41598-017-00662-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Tagalakis AD, Munye MM, Ivanova R, Chen H, Smith CM, Aldossary AM, Rosa LZ, Moulding D, Barnes JL, Kafetzis KN, Jones SA, et al. Thorax. 2018;73:847–856. doi: 10.1136/thoraxjnl-2017-210670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].d’Angelo I, Costabile G, Durantie E, Brocca P, Rondelli V, Russo A, Russo G, Miro A, Quaglia F, Petri-Fink A, Rothen-Rutishauser B, et al. J Aerosol Med Pulm Drug Deliv. 2017;31:jamp.2017.1364. doi: 10.1089/jamp.2017.1364. [DOI] [PubMed] [Google Scholar]
- [103].Mitchell G, Grondin G, Bilodeau G, Cantin AM, Malouin F. Infect Immun. 2011;79:3541–3551. doi: 10.1128/IAI.00078-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Kiedrowski MR, Gaston JR, Kocak BR, Coburn SL, Lee S, Pilewski JM, Myerburg MM, Bomberger JM. MSphere. 2018;3:1–17. doi: 10.1128/mSphere.00341-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Michelle Di Paola CMK, Park Amber J, Ahmadi Saumel, Roach Elyse J, Wu Yu-Sheng, Struder-Kypke Michaela, Lam Joseph S, Bear Christine E. MBio. 2017;8:1–14. doi: 10.1128/mBio.02073-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Hasan S, Sebo P, Osicka R. FEBS J. 2018;285:4343–4358. doi: 10.1111/febs.14582. [DOI] [PubMed] [Google Scholar]
- [107].Yonker LM, Mou H, Chu KK, Pazos MA, Leung H, Cui D, Ryu J, Hibbler RM, Eaton AD, Ford TN, Falck JR, et al. Sci Rep. 2017;7:1–12. doi: 10.1038/s41598-017-08567-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Halldorsson S, Gudjonsson T, Gottfredsson M, Singh PK, Gudmundsson GH, Baldursson O. Am J Respir Cell Mol Biol. 2010;42:62–68. doi: 10.1165/rcmb.2008-0357OC. [DOI] [PubMed] [Google Scholar]
- [109].Moreau-Marquis S, O’Toole GA, Stanton BA. Am J Respir Cell Mol Biol. 2009;41:305–313. doi: 10.1165/rcmb.2008-0299OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Lashua LP, Melvin JA, Deslouches B, Pilewski JM, Montelaro RC, Bomberger JM. J Antimicrob Chemother. 2016;71:2200–2207. doi: 10.1093/jac/dkw143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].GA Network. The Global Asthma Report 2018. 2018 http://globalasthmareport.org/burden/burden.php.
- [112].Kudo M, Ishigatsubo Y, Aoki I. Front Microbiol. 2013;4:1–16. doi: 10.3389/fmicb.2013.00263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Wegmann M. Expert Rev Mol Diagn. 2009;9:85–100. doi: 10.1586/14737159.9.1.85. [DOI] [PubMed] [Google Scholar]
- [114].Barnes PJ. Nat Rev Immunol. 2018;18:454–466. doi: 10.1038/s41577-018-0006-6. [DOI] [PubMed] [Google Scholar]
- [115].Lambrecht BN, Hammad H. Nat Med. 2012;18:684–692. doi: 10.1038/nm.2737. [DOI] [PubMed] [Google Scholar]
- [116].Barnes PJ. Nat Rev Immunol. 2008;8:183–192. doi: 10.1038/nri2254. [DOI] [PubMed] [Google Scholar]
- [117].Ware LB. Am J Physiol Cell Mol Physiol. 2008;294:L149-L150. doi: 10.1152/ajplung.00472.2007. [DOI] [PubMed] [Google Scholar]
- [118].Bal HS, Goshal NG. Lab Anim. 1988;22:76–82. doi: 10.1258/002367788780746539. [DOI] [PubMed] [Google Scholar]
- [119].Blume C, Davies DE. Eur J Pharm Biopharm. 2013;84:394–400. doi: 10.1016/j.ejpb.2012.12.014. [DOI] [PubMed] [Google Scholar]
- [120].Blume C, Davies DE. Eur J Pharm Biopharm. 2013;84:394–400. doi: 10.1016/j.ejpb.2012.12.014. [DOI] [PubMed] [Google Scholar]
- [121].Papazian D, Würtzen PA, Hansen SWK. Int Arch Allergy Immunol. 2016;170:1–21. doi: 10.1159/000445833. [DOI] [PubMed] [Google Scholar]
- [122].Xiao C, Puddicombe SM, Field S, Haywood J, Broughton-Head V, Puxeddu I, Haitchi HM, Vernon-Wilson E, Sammut D, Bedke N, Cremin C, et al. J Allergy Clin Immunol. 2011;128 doi: 10.1016/j.jaci.2011.05.038. [DOI] [PubMed] [Google Scholar]
- [123].Gras D, Bourdin A, Vachier I, De Senneville L, Bonnans C, Chanez P. J Allergy Clin Immunol. 2012;129:1259–1266.:e1. doi: 10.1016/j.jaci.2012.01.073. [DOI] [PubMed] [Google Scholar]
- [124].Hackett TL, Warner SM, Stefanowicz D, Shaheen F, Pechkovsky DV, Murray LA, Argentieri R, Kicic A, Stick SM, Bai TR, Knight DA. Am J Respir Crit Care Med. 2009;180:122–133. doi: 10.1164/rccm.200811-1730OC. [DOI] [PubMed] [Google Scholar]
- [125].Carlini F, Picard C, Garulli C, Piquemal D, Roubertoux P, Chiaroni J, Chanez P, Gras D, Di Cristofaro J. Front Immunol. 2017;8:1–11. doi: 10.3389/fimmu.2017.00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Jevnikar Z, Östling J, Ax E, Calvén J, Thörn K, Israelsson E, Öberg L, Singhania A, Lau LCK, Wilson SJ, Ward JA, et al. J Allergy Clin Immunol. 2019;143:577–590. doi: 10.1016/j.jaci.2018.05.026. [DOI] [PubMed] [Google Scholar]
- [127].Holden NS, Rider CF, Bell MJ, Velayudhan J, King EM, Kaur M, Salmon M, Giembycz MA, Newton R. Br J Pharmacol. 2010;160:410–420. doi: 10.1111/j.1476-5381.2010.00708.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Woodman LB, Wan WYH, Milone R, Grace K, Sousa A, Williamson R, Brightling CE. Chest. 2013;143:1656–1666. doi: 10.1378/chest.12-1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Hardyman MA, Wilkinson E, Martin E, Jayasekera NP, Blume C, Swindle EJ, Gozzard N, Holgate ST, Howarth PH, Davies DE, Collins JE. J Allergy Clin Immunol. 2013;132:665–675.:e8. doi: 10.1016/j.jaci.2013.03.005. [DOI] [PubMed] [Google Scholar]
- [130].Sexton DJ, Chen T, Martik D, Kuzmic P, Kuang G, Chen J, Nixon AE, Zuraw BL, Forteza RM, Abraham WM, Wood CR. Biochem J. 2009;422:383–392. doi: 10.1042/BJ20090010. [DOI] [PubMed] [Google Scholar]
- [131].Lopez-Guisa JMDJ, Powers C, File D, Cochrane E, Jimenez N. J Allergy Clin Immunol. 2012;129:990–7. doi: 10.1016/j.jaci.2011.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Healey GD, Evans N, Hopkin JM, Davies G, Walker W. J Cell Mol Med. 2013;17:356–364. doi: 10.1111/jcmm.12014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Gras D, Martinez-Anton A, Bourdin A, Garulli C, De Senneville L, Vachier I, Vitte J, Chanez P. Eur Respir J. 2017;49:1–11. doi: 10.1183/13993003.02399-2016. [DOI] [PubMed] [Google Scholar]
- [134].Jiao D, Wong CK, Tsang MSM, Chu IMT, Liu D, Zhu J, Chu M, Lam CWK. Sci Rep. 2017;7:1–13. doi: 10.1038/s41598-017-02085-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Wawrzyniak P, Wawrzyniak M, Wanke K, Sokolowska M, Bendelja K, Rückert B, Globinska A, Jakiela B, Kast JI, Idzko M, Akdis M, et al. J Allergy Clin Immunol. 2017;139:93–103. doi: 10.1016/j.jaci.2016.03.050. [DOI] [PubMed] [Google Scholar]
- [136].Haghi M, Hittinger M, Zeng Q, Oliver B, Traini D, Young PM, Huwer H, Schneider-Daum N, Lehr CM. Mol Pharm. 2015;12:2625–2632. doi: 10.1021/acs.molpharmaceut.5b00124. [DOI] [PubMed] [Google Scholar]
- [137].Reeves SR, Kolstad T, Lien TY, Herrington-Shaner S, Debley JS. Respir Res. 2015;16:1–12. doi: 10.1186/s12931-015-0185-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138].Paplinksa-Goryca M, Misiukiewicz-Stepien P, Nejman-Gryz P, Proboszcz M, Mlacki M, Gorska K, Krenke R. Clin Immunol. 2019:108421. doi: 10.1016/j.clim.2020.108421. [DOI] [PubMed] [Google Scholar]
- [139].Siegel RL, Miller KD, Jemal A. CA Cancer J Clin. 2020;70:7–30. doi: 10.3322/caac.21590. [DOI] [PubMed] [Google Scholar]
- [140].Stevens JL, Baker TK. Drug Discov Today. 2009;14:162–167. doi: 10.1016/j.drudis.2008.11.009. [DOI] [PubMed] [Google Scholar]
- [141].Weigelt B, Ghajar CM, Bissell MJ. Adv Drug Deliv Rev. 2014;69–70:42–51. doi: 10.1016/j.addr.2014.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [142].Jakiela B, Gielicz A, Plutecka H, Hubalewska M, Mastalerz L, Bochenek G, Soja J, Januszek R, Musial J, Sanak M. Prostaglandins Other Lipid Mediat. 2013;106:116–123. doi: 10.1016/j.prostaglandins.2013.05.001. [DOI] [PubMed] [Google Scholar]
- [143].Shaykhiev R, Zuo WL, Chao IW, Fukui T, Witover B, Brekman A, Crystal RG. Proc Natl Acad Sci U S A. 2013;110:12102–12107. doi: 10.1073/pnas.1303058110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [144].Lee J, Ryu SH, Kang SM, Chung WC, Gold KA, Kim ES, Hittelman WN, Hong WK, Koo JS. Cancer Prev Res. 2011;4:1306–1315. doi: 10.1158/1940-6207.CAPR-10-0364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [145].Horie M, Saito A, Mikami Y, Ohshima M, Morishita Y, Nakajima J, Kohyama T, Nagase T. Biochem Biophys Res Commun. 2012;423:158–163. doi: 10.1016/j.bbrc.2012.05.104. [DOI] [PubMed] [Google Scholar]
- [146].Correia LL, Johnson JA, McErlean P, Bauer J, Farah H, Rassl DM, Rintoul RC, Sethi T, Lavender P, Rawlins EL, Littlewood TD, et al. Am J Respir Crit Care Med. 2017;195:1494–1508. doi: 10.1164/rccm.201510-2084OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [147].Zanoni M, Piccinini F, Arienti C, Zamagni A, Santi S, Polico R, Bevilacqua A, Tesei A. Sci Rep. 2016;6:1–11. doi: 10.1038/srep19103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [148].Kim M, Mun H, Sung CO, Cho EJ, Jeon HJ, Chun SM, Jung DJ, Shin TH, Jeong GS, Kim DK, Choi EK, et al. Nat Commun. 2019:10. doi: 10.1038/s41467-019-11867-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [149].Han K, Pierce SE, Li A, Spees K, Anderson GR, Seoane JA, Lo YH, Dubreuil M, Olivas M, Kamber RA, Wainberg M, et al. Nature. 2020;580 doi: 10.1038/s41586-020-2099-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [150].Di Liello R, Ciaramella V, Barra G, Venditti M, Della Corte CM, Papaccio F, Sparano F, Viscardi G, Iacovino ML, Minucci S, Fasano M, et al. ESMO Open. 2019;4:1–6. doi: 10.1136/esmoopen-2019-000536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [151].Meenach SA, Tsoras AN, McGarry RC, Mansour HM, Hilt JZ, Anderson KW. Int J Oncol. 2016;48:1701–1709. doi: 10.3892/ijo.2016.3376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [152].Gupta SK, Torrico Guzmán EA, Meenach SA. Int J Cancer. 2017;141:2143–2153. doi: 10.1002/ijc.30913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [153].Movia D, Bazou D, Volkov Y, Prina-mello A. Sci Rep. 2018:1–19. doi: 10.1038/s41598-018-31332-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [154].Movia D, Bazou D, Prina-mello A. BMC Cancer. 2019:1–21. doi: 10.1186/s12885-019-6038-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [155].Zhang YM, Zhang ZM, Guan QL, Liu YQ, Wu ZW, Li JT, Su Y, Yan CL, Luo YL, Qin J, Wang Q, et al. Exp Ther Med. 2017;14:2983–2991. doi: 10.3892/etm.2017.4909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [156].Gagnadoux F, Hureaux J, Vecellio L, Urban T, Le Pape A, Valo I, Montharu J, Leblond V, Boisdron-Celle M, Lerondel S, Majoral C, et al. J Aerosol Med Pulm Drug Deliv. 2008;21:61–70. doi: 10.1089/jamp.2007.0656. [DOI] [PubMed] [Google Scholar]
- [157].Lee WH, Loo CY, Traini D, Young PM. Asian J Pharm Sci. 2015;10:481–489. [Google Scholar]
- [158].Joshi N, Shirsath N, Singh A, Joshi KS, Banerjee R. Sci Rep. 2014;4 doi: 10.1038/srep07085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [159].Rosière R, Van Woensel M, Gelbcke M, Mathieu V, Hecq J, Mathivet T, Vermeersch M, Van Antwerpen P, Amighi K, Wauthoz N. Mol Pharm. 2018;15:899–910. doi: 10.1021/acs.molpharmaceut.7b00846. [DOI] [PubMed] [Google Scholar]
- [160].Almuqbil RM, Heyder RS, Bielski ER, Durymanov M, Reineke JJ, da Rocha SRP. Mol Pharm. 2020 doi: 10.1021/acs.molpharmaceut.0c00083. [DOI] [PubMed] [Google Scholar]
- [161].Mehta A, Dalle Vedove E, Isert L, Merkel OM. Pharm Res. 2019:36. doi: 10.1007/s11095-019-2665-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [162].Zhang C, Zhang S, Zhi D, Zhao Y, Cui S, Cui J. Colloids Surfaces A Physicochem Eng Asp. 2020;585:124054 [Google Scholar]
- [163].Brandsma CA, Van den Berge M, Hackett TL, Brusselle G, Timens W. J Pathol. 2019 doi: 10.1002/path.5364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [164].Marin L, Colombo P, Bebawy M, Young PM, Traini D. Expert Opin Drug Deliv. 2011;8:1205–1220. doi: 10.1517/17425247.2011.588697. [DOI] [PubMed] [Google Scholar]
- [165].Tuder RM, Petrache I. J Clin Invest. 2012;122:2749–2755. doi: 10.1172/JCI60324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [166].MacNee W. Proc Am Thorac Soc. 2005;2:258–266. doi: 10.1513/pats.200504-045SR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [167].Gao W, Li L, Wang Y, Zhang S, Adcock IM, Barnes PJ, Huang M, Yao X. Respirology. 2015;20:722–729. doi: 10.1111/resp.12542. [DOI] [PubMed] [Google Scholar]
- [168].Schamberger AC, Staab-weijnitz CA, Mise-racek N, Eickelberg O. Sci Rep. 2015:1–9. doi: 10.1038/srep08163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [169].Haswell LE, Hewitt K, Thorne D, Richter A, Gaça MD. Toxicol Vitr. 2010;24:981–987. doi: 10.1016/j.tiv.2009.12.019. [DOI] [PubMed] [Google Scholar]
- [170].Gohy S, Carlier FM, Fregimilicka C, Detry B, Lecocq M, Ladjemi MZ, Verleden S, Hoton D, Weynand B, Bouzin C, Pilette C. Sci Rep. 2019:1–12. doi: 10.1038/s41598-019-54292-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [171].Meiners S, Eickelberg O, Schamberger AC, Mise N, Jia J, Genoyer E, Ali O. Am J Respir Cell Mol Biol. 2014;50:1040–1052. doi: 10.1165/rcmb.2013-0090OC. [DOI] [PubMed] [Google Scholar]
- [172].Heijink IH, De Bruin HG, Dennebos R, Jonker MR, Noordhoek JA, Brandsma C, Van Den Berge M. Eur Respir J. 2016;48:504–515. doi: 10.1183/13993003.01541-2015. [DOI] [PubMed] [Google Scholar]
- [173].Gindele JA, Kiechle T, Benediktus K, Birk G, Brendel M, Heinemann F, Wohnhaas CT, Leblanc M, Zhang H, Barel YS, Cr RG, et al. 2020. pp. 1–17. [DOI] [PMC free article] [PubMed]
- [174].Comer DM, Kidney JC, Ennis M, Elborn JS. Eur Respir J. 2013;41:1058–1067. doi: 10.1183/09031936.00063112. [DOI] [PubMed] [Google Scholar]
- [175].Amatngalim GD, Schrumpf A, Henic A. J Innate Immun. 2017:359–374. doi: 10.1159/000455193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [176].Azzopardi D, Haswell LE, Foss-smith G, Hewitt K, Asquith N, Corke S, Phillips G. Toxicol Vitr. 2015;29:1720–1728. doi: 10.1016/j.tiv.2015.06.016. [DOI] [PubMed] [Google Scholar]
- [177].Zarcone MC, Van Schadewijk A, Duistermaat E, Hiemstra PS, Kooter IM. 2017. pp. 1–11. [DOI] [PMC free article] [PubMed]
- [178].Ladjemi MZ, Lecocq M, Weynand B, Bowen H, Gould HJ, Van Snick J, Detry B, Pilette C. Eur Respir J n d. :980–993. doi: 10.1183/09031936.00063914. [DOI] [PubMed] [Google Scholar]
- [179].Osei ET, Noordhoek JA, Hackett TL, Spanjer AIR, Postma DS, Timens W, Brandsma C, Heijink IH. Eur Respir J n d. :359–369. doi: 10.1183/13993003.01911-2015. [DOI] [PubMed] [Google Scholar]
- [180].Gindele JA, Mang S, Pairet N, Christ I, Gantner F, Lamb DJ. PLoS One. 2017:1–15. doi: 10.1371/journal.pone.0184386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [181].Marin L, Traini D, Bebawy M, Colombo P, Buttini F, Haghi M, Xin H, Young P. Eur J Pharm Biopharm. 2013;84:566–572. doi: 10.1016/j.ejpb.2013.01.021. [DOI] [PubMed] [Google Scholar]
- [182].Tatsuta M, Kan-o K, Ishii Y, Yamamoto N, Ogawa T, Fukuyama S, Ogawa A, Fujita A, Nakanishi Y, Matsumoto K. Respir Res. 2019;20:1–14. doi: 10.1186/s12931-019-1226-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [183].Schmid A, Baumlin N, Ivonnet P, Dennis JS, Campos M, Krick S. Respir Res. 2015:1–11. doi: 10.1186/s12931-015-0294-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [184].Gohy ST, Hupin C, Fregimilicka C, Detry BR, Bouzin C, Chevronay HG, Lecocq M, Weynand B, Ladjemi MZ, Pierreux CE, Birembaut P, et al. Eur Respir J n d. :1258–1272. doi: 10.1183/09031936.00135814. [DOI] [PubMed] [Google Scholar]
- [185].Lamb DJ, Bucher H, Mang S, Keck M, Schiele F, Wittenbrink M, Fuchs K, Jung B, Erb KJ, Peter D. Pulm Pharmacol Ther. 2017;44:96–105. doi: 10.1016/j.pupt.2017.03.008. [DOI] [PubMed] [Google Scholar]
- [186].Reus AA, Maas WJM, Jansen HT, Constant S, Staal YCM, Van Triel JJ, Kuper CF. Toxicol Vitr. 2014;28:258–264. doi: 10.1016/j.tiv.2013.10.025. [DOI] [PubMed] [Google Scholar]
- [187].Huang S, Wiszniewski L, Constant S, Roggen E. Toxicol Vitr. 2013;27:1151–1156. doi: 10.1016/j.tiv.2012.10.010. [DOI] [PubMed] [Google Scholar]
- [188].Mercier C, Jacqueroux E, He Z, Hodin S, Perek N, Boudard D, Delavenne X, Constant S. Eur J Pharm Biopharm. 2019;139:186–196. doi: 10.1016/j.ejpb.2019.04.002. [DOI] [PubMed] [Google Scholar]
- [189].Rotoli BM, Barilli A, Visigalli R, Ferrari F, Frati C, Lagrasta CA, Di Lascia M, Riccardi B, Puccini P, Asta VD. 1:1–14. doi: 10.3390/ijms21093190. nd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [190].Hoffmann W, Gradinaru J, Farcal L, Caul-Futy M, Huang S, Wiszniewski L, Parissis N, Morath S, Fortaner S, Cole T, Reginato E, et al. Appl Vitr Toxicol. 2018;4:139–148. [Google Scholar]
- [191].Furubayashi T, Inoue D, Nishiyama N, Tanaka A. 2020.
- [192].Iskandar AR, Xiang Y, Frentzel S, Talikka M, Leroy P, Kuehn D, Guedj E, Martin F, Mathis C, Ivanov NV, Peitsch MC, et al. Toxicol Sci. 2015;147:207–221. doi: 10.1093/toxsci/kfv122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [193].Kooter IM, Gröllers-Mulderij M, Duistermaat E, Kuper F, Schoen ED. Toxicol Vitr. 2017;44:339–348. doi: 10.1016/j.tiv.2017.07.006. [DOI] [PubMed] [Google Scholar]
- [194].Sancey L, Sabido O, He Z, Rossetti F, Guignandon A, Bin V, Coll JL, Cottier M, Lux F, Tillement O, Constant S, et al. J Nanobiotechnology. 2020:1–18. doi: 10.1186/s12951-020-00683-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [195].Mistry A, Bowen LE, Dzierlenga MW, Hartman JK, Slattery SD. Toxicol Vitr. 2020;69:104968. doi: 10.1016/j.tiv.2020.104968. [DOI] [PubMed] [Google Scholar]
- [196].Outlaw M, Victor K, Bovier Francesca T, Mears Megan C, Cajmat Maria N, Zhu Yun, Lin Michelle J, Addetia Amin, Lieberman Nicole AP, Peddu Vikas, Xie Xuping, et al. MBio. 2020;11:1–14. doi: 10.1128/mBio.01935-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [197].Huang S, Boda B, Vernaz J, Ferreira E, Wiszniewski L, Constant S. Eur J Pharm Biopharm. 2017;118:68–72. doi: 10.1016/j.ejpb.2016.12.006. [DOI] [PubMed] [Google Scholar]
- [198].Barosova H, Maione AG, Septiadi D, Sharma M, Haeni L, Balog S, O’Connell O, Jackson GR, Brown D, Clippinger AJ, Hayden P, et al. ACS Nano. 2020;14:3941–3956. doi: 10.1021/acsnano.9b06860. [DOI] [PubMed] [Google Scholar]
- [199].Bolmarcich J, Jackson GR, Oldach J, Bachelor M, Wilbert S, Kenney T, Wright CD, Hayden PJ. 2017;XX:1–15. [Google Scholar]
- [200].De Matos Raphael S, Vuilleumier Jeremy, Mas Christophe, Constant Samuel, Staedler Davide. Gerber-Lemaire. 2019:31659–31669. doi: 10.1039/c9ra05299b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [201].Outlaw M, Victor K, Bovier Francesca T, Mears Megan C, Cajmat Maria N, Zhu Yun, Lin Michelle J, Addetia Amin, Lieberman Nicole AP, Peddu Vikas, Xie Xuping, et al. Moscon. 2020;11:1–14. [Google Scholar]
- [202].Signer J, Jonsdottir HR, Albrich WC, Strasser M, Züst R, Ryter S, Ackermann-Gäumann R, Lenz N, Siegrist D, Suter A, Schoop R, et al. Virol J. 2020;17:136. doi: 10.1186/s12985-020-01401-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [203].Mas C, Boda B, CaulFuty M, Huang S, Wiszniewski L, Constant S. J Biotechnol. 2015;205:111–119. doi: 10.1016/j.jbiotec.2015.01.012. [DOI] [PubMed] [Google Scholar]
- [204].Kilin V, Mas C, Constant S, Wolf JP, Bonacina L. Sci Rep. 2017;7:1–10. doi: 10.1038/s41598-017-16628-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [205].Schimek K, Frentzel S, Luettich K, Bovard D, Rütschle I, Boden L, Rambo F, Erfurth H, Dehne E, Winter A, Marx U, Hoeng J. 2020:1–13. doi: 10.1038/s41598-020-64219-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [206].Sancey L, Sabido O, He Z, Rossetti F, Guignandon A, Bin V, Coll JL, Cottier M, Lux F, Tillement O, Constant S, et al. J Nanobiotechnology. 2020:1–18. doi: 10.1186/s12951-020-00683-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [207].Dellaquila A, Thomée EK, McMillan AH, Lesher-Pérez SC. Lung-on-a-chip platforms for modeling disease pathogenesis. in: Organ-on-a-Chip Eng Microenviron Saf Effic Test. 2019:133–180. [Google Scholar]
- [208].Shrestha J, Razavi Bazaz S, Aboulkheyr Es H, Yaghobian Azari D, Thierry B, Ebrahimi Warkiani M, Ghadiri M. Crit Rev Biotechnol. 2020;40:213–230. doi: 10.1080/07388551.2019.1710458. [DOI] [PubMed] [Google Scholar]
- [209].Zamprogno P, Wüthrich S, Achenbach S, Thoma G, Stucki JD, Hobi N, Schneider-Daum N, Lehr CM, Huwer H, Geiser T, Schmid RA, et al. Commun Biol. 2021;4:168. doi: 10.1038/s42003-021-01695-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [210].Huh D, Matthews BD, Mammoto A, Montoya-Zavala M, Hsin HY, Ingber DE. Science (80-) 2010;328 doi: 10.1126/science.1188302. 1662 LP – 1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [211].Liu Z, Mackay S, Gordon DM, Anderson JD, Haithcock DW, Garson CJ, Tearney GJ, Solomon GM, Pant K, Prabhakarpandian B, Rowe SM, et al. Biomed Opt Express. 2019;10:5414–5430. doi: 10.1364/BOE.10.005414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [212].Kızılkurtlu AA, Polat T, GBA, Akpek* A. Curr Pharm Des. 2018;24:5386–5396. doi: 10.2174/1381612825666190208122204. [DOI] [PubMed] [Google Scholar]
- [213].Kilin V, Mas C, Constant S, Wolf JP, Bonacina L. Sci Rep. 2017;7:1–10. doi: 10.1038/s41598-017-16628-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [214].Lenz A, Stoeger T, Cei D, Schmidmeir M, Semren N. Am J Respir Cell Mol Biol. 2014;51:526–535. doi: 10.1165/rcmb.2013-0479OC. [DOI] [PubMed] [Google Scholar]
- [215].Schmid O, Jud C, Umehara Y, Mueller D, Bucholski A, Gruber F, Denk O, Egle R, Petri-Fink A, Rothen-Rutishauser B. J Aerosol Med Pulm Drug Deliv. 2017;30:411–424. doi: 10.1089/jamp.2016.1361. [DOI] [PubMed] [Google Scholar]
- [216].Klein SG, Serchi T, Hoffmann L, Blömeke B, Gutleb AC. Part Fibre Toxicol. 2013;10 doi: 10.1186/1743-8977-10-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [217].Tsoutsoulopoulos A, Gohlsch K, Möhle N, Breit A, Hoffmann S, Krischenowski O, Mückter H, Gudermann T, Thiermann H, Aufderheide M, Steinritz D. Toxicol Vitr. 2019;58:245–255. doi: 10.1016/j.tiv.2019.03.020. [DOI] [PubMed] [Google Scholar]
- [218].Aufderheide M, Förster C, Beshay M, Branscheid D, Emura M. Exp Toxicol Pathol. 2016;68:77–87. doi: 10.1016/j.etp.2015.10.001. [DOI] [PubMed] [Google Scholar]
- [219].Tsoutsoulopoulos A, Gohlsch K, Möhle N, Breit A, Hoffmann S, Krischenowski O, Mückter H, Gudermann T, Thiermann H, Aufderheide M, Steinritz D. Toxicol Vitr. 2019;58:245–255. doi: 10.1016/j.tiv.2019.03.020. [DOI] [PubMed] [Google Scholar]
- [220].Polk WW, Sharma M, Sayes CM, Hotchkiss JA, Clippinger AJ. Part Fibre Toxicol. 2016;13:20. doi: 10.1186/s12989-016-0131-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [221].Ji J, Hedelin A, Malmlöf M, Kessler V, Seisenbaeva G, Gerde P, Palmberg L. PLoS One. 2017;12:e0170428. doi: 10.1371/journal.pone.0170428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [222].Upadhyay S, Palmberg L. Toxicol Sci. 2018;164:21–30. doi: 10.1093/toxsci/kfy053. [DOI] [PubMed] [Google Scholar]
- [223].Jeannet N, Fierz M, Kalberer M, Burtscher H, Geiser M. Nanotoxicology. 2015;9:34–42. doi: 10.3109/17435390.2014.886739. [DOI] [PubMed] [Google Scholar]
- [224].Ihalainen M, Jalava P, Ihantola T, Kasurinen S, Uski O, Sippula O, Hartikainen A, Tissari J, Kuuspalo K, Lähde A, Hirvonen M-R, et al. Aerosol Sci Technol. 2019;53:133–145. [Google Scholar]
- [225].Ding Y, Weindl P, Lenz AG, Mayer P, Krebs T, Schmid O. Part Fibre Toxicol. 2020;17:1–20. doi: 10.1186/s12989-020-00376-w. [DOI] [PMC free article] [PubMed] [Google Scholar]