Abstract
Purpose
Tumor hypoxia fuels an aggressive tumor phenotype and confers resistance to anti-cancer treatments. We conducted a clinical trial to determine whether the antimalarial drug atovaquone, a known mitochondrial inhibitor, reduces hypoxia in non-small cell lung cancer (NSCLC).
Patients and methods
Patients with NSCLC scheduled for surgery were recruited sequentially into two cohorts: Cohort 1 received oral atovaquone at the standard clinical dose 750 mg twice-daily, whilst Cohort 2 did not. Primary imaging endpoint was change in tumor hypoxic volume (HV) measured by hypoxia PET-CT. Inter-cohort comparison of hypoxia gene expression signatures using RNA sequencing from resected tumors was performed.
Results
Thirty patients were evaluable for hypoxia PET-CT analysis, 15 per cohort. Median treatment duration was 12 days. Eleven (73.3%) atovaquone-treated patients had meaningful HV reduction with median change −28.0% [95% confidence interval (CI), −58.2 to −4.4]. In contrast, median change in untreated patients was +15.5% (95% CI, −6.5 to 35.5). Linear regression estimated the expected mean HV was 55% (95% CI, 24% to 74%) lower in Cohort 1 compared to Cohort 2 (p=0.004), adjusting for cohort, tumor volume and baseline HV. A key pharmacodynamic endpoint was reduction in hypoxia regulated genes, which were significantly downregulated in atovaquone-treated tumors. Data from multiple additional measures of tumor hypoxia and perfusion are presented. No atovaquone-related adverse events were reported.
Conclusions
This is the first clinical evidence that targeting tumor mitochondrial metabolism can reduce hypoxia and produce relevant anti-tumor effects at the mRNA level. Repurposing atovaquone for this purpose may improve treatment outcomes for NSCLC.
Keywords: Atovaquone, OXPHOS inhibitor, tumor hypoxia, FMISO PET-CT and NSCLC
Introduction
In contrast to normal tissues, hypoxia is a common finding in solid tumors resulting from imbalance between high oxygen demand and poor delivery (1). Principally through hypoxia-inducible factor signaling, hypoxia contributes significantly to tumorigenesis by directly contributing to the majority of cancer hallmarks, including sustained proliferation, immune system evasion, invasion and metastasis (2–4). Consequently, hypoxia results in a highly aggressive tumor phenotype.
Of further clinical relevance is that tumor hypoxia is a significant barrier to numerous anti-cancer treatments. In particular, hypoxia confers profound resistance to radiation treatment with up to three times the dose of radiation required to treat hypoxic tumors successfully (5), and is associated with inferior clinical outcomes following radiotherapy (6, 7). Hypoxia-related resistance to cytotoxic chemotherapy and immunotherapy is also well recognized (8, 9).
An emerging novel strategy to tackle hypoxia is to reduce tumor oxygen consumption rate (OCR) through metabolic reprogramming (10). Recently, the commonly prescribed and well-tolerated anti-malarial drug atovaquone has been discovered as a highly promising agent for this purpose. We reported previously that atovaquone reduces OCR in numerous cancer cell lines and alleviates hypoxia in xenograft models leading to tumor radiosensitization (11). Reduction in OCR occurs by inhibiting oxidative phosphorylation (OXPHOS) at complex III of the mitochondrial electron transport chain (ETC). Interestingly, besides causing hypoxia through oxygen consumption, tumor mitochondria are increasingly recognized as important mediators of drug resistance, invasion and metastasis (12).
However, demonstration that mitochondrial function can be modified in tumors using mitochondrial inhibitors has never previously been shown in the clinical setting. We therefore conducted a translational clinical trial to determine whether by using the pharmacodynamic endpoints of tumor hypoxia and gene expression analysis, we could prove the concept and effectiveness of atovaquone as a clinically useful mitochondrial inhibitor. Patients with NSCLC were chosen for this study given the urgent clinical need to improve treatment outcomes for this patient group, especially with regards to radiotherapy.
Non-invasive positron emission tomography computed tomography (PET-CT) imaging of tumor hypoxia is reliable and the most-accepted technique for measuring hypoxic change in patients (13–18) and was the primary method used. The most common radiolabeled tracers used for this purpose are nitroimidazole-based compounds such as [18F]-fluoromisonidazole (FMISO) and [18F]-fluoroazomycin arabinoside (FAZA), which under low oxygen tensions undergo irreversible intracellular enzymatic reduction. By utilizing such scans in this study, assessment of hypoxia in viable and therefore clinically relevant tumor regions was ensured.
Gene expression signatures measure the complex tumor transcriptome response to hypoxia (19) and were considered a key endpoint to demonstrate that reduction of hypoxia had a desired biological effect. Because development of validated and deliverable clinical hypoxia biomarkers is needed, putative circulating and tumor sample-based measures of hypoxia were also assessed. Assessment of changes in tumor perfusion and vasculature was also performed, thus ensuring a comprehensive assessment of tumor oxygenation.
We report the first clinical evidence that hypoxia can be reduced through targeting tumor metabolism and highlight atovaquone as a highly promising agent for combination with anti-cancer treatments in NSCLC.
Patients and Methods
Study design, patients and treatment
This was an open-label, non-randomized, two-cohort, window-of-opportunity study (NCT02628080) conducted at the Oxford Cancer and Haematology Centre (Oxford, UK). Ethical approval was obtained from National Research Ethics Service Committee South Central Oxford B (16/SC/0012). Trial conduct adhered to all regulatory requirements and was in full accordance with the provisions of the Declaration of Helsinki and Good Clinical Practice guidelines.
Eligible patients were aged ≥18 years, had a pathological or radiological diagnosis of NSCLC, were scheduled for surgical resection, had disease >2 cm in diameter and were Eastern Cooperative Oncology Group (ECOG) performance status 0-2. Patients were excluded if taking known ETC inhibitors. Complete eligibility criteria are provided in the trial protocol (Supplementary Materials and Methods). All patients provided written informed consent.
The trial was designed to recruit 30 patients sequentially into two equal-sized cohorts: Cohort 1 received atovaquone and Cohort 2 did not. All patients attended for two visits for evaluation of tumor hypoxia and clinical assessment. For Cohort 1, oral atovaquone (Wellvone 750 mg/5 mL micronized suspension, GlaxoSmithKline UK) was self-administered at a standard clinical dose of 750 mg twice-daily for 7-16 days between visits, as permitted by the surgical date. Patients were asked to take atovaquone together with fat-containing food to aid absorption. Shortly following the second visit, all patients underwent surgery with 500 mg/m2 oral pimonidazole hydrochloride (Hydroxyprobe-1, Natural Pharmacia International, USA) administered 16-24 hours before as an exogenous marker of hypoxia.
This study utilized multiple imaging and sample-based methods to assess tumor oxygenation with numerous pre-specified primary, secondary and exploratory endpoints. All endpoints are listed in the trial protocol and the trial schema is shown in Supplementary Fig. S1 (Supplementary Materials and Methods).
Tumor imaging outcomes
The primary imaging-related trial endpoint was change in tumor hypoxic volume (HV) measured using hypoxia PET-CT. Patients were imaged using GE Discovery 690 or 710 PET-CT scanners (GE Healthcare, USA). 370 MBq of FMISO (University of Cambridge, UK) or FAZA (University of Manchester, UK) was injected and immediately a 45-minute dynamic PET acquisition and then 10-minute acquisitions at 2 and 4 hours post-injection were obtained. CT was performed for localization and attenuation correction. The same tracer and scanner were mandated for both study visits per individual patient.
Four-hour hypoxia PET-CT images were analyzed using Hermes Hybrid Viewer software (Hermes Medical Solutions AB, Sweden). Tumors were outlined by an experienced PET radiologist (R.E.M.). Blood in the descending aorta was outlined to provide the background mean standardized uptake value (SUVmean). To determine the HV, voxel-by-voxel SUVs were divided by background SUVmean to provide tumor-to-blood ratio (TBR) values for each tumor voxel and TBR ≥1.4 voxels were classified as hypoxic, as previously described (15). Volumes of hypoxic voxels pre- and post-atovaquone were compared and ≥10% reduction was defined as a meaningful reduction, as previously published (14) (further explained in Supplementary Materials and Methods).
Secondary imaging endpoints were change in tumor perfusion using dynamic hypoxia PET, perfusion CT (pCT) and dynamic contrast-enhanced (DCE) MRI. Non-prespecified T2* map and T1 shMOLLI MRI analysis were also performed to investigate changes in tumor venous oxygenation and biochemistry, respectively. Methodology for all these imaging techniques is provided in Supplementary Materials and Methods.
Tumor sample and circulating marker outcomes
Resected tumors were formalin-fixed and paraffin-embedded (FFPE) using a standard clinical protocol. Inter-cohort comparison of immunohistochemistry for the exogenous hypoxia marker pimonidazole was a co-primary trial endpoint and performed using, whenever possible, three entire tumor cross sections. Immunohistochemical staining for endothelial markers CD31 and CD146, as markers of vascular density, was also performed (Supplementary Materials and Methods).
Transcriptomic analysis for Buffa (20), Hallmark (21), Harris (22) and Winter (23) hypoxia signatures was conducted from the same tumor regions with OXPHOS, glycolysis, reactive oxygen species (ROS), angiogenesis, VEGF, epithelial-to-mesenchymal transition (EMT), integrated stress response and STAT3 signaling pathway signatures also studied. Genes in each signature are listed in Supplementary Tables S1-S4. RNA isolation was performed using RecoverAll Total Nucleic Acid Isolation Kit for FFPE (Invitrogen, USA), as per manufacturer’s instructions (Supplementary Materials and Methods). Paired end library preparations and RNA 3’ end sequencing (HiSeq4000 75bp) was performed by Oxford Genomics Centre, Wellcome Centre for Human Genetics, UK. FastQC was used to evaluate quality of sequencing reads at both base and read levels (24). Reads with low-quality bases (phred score <20) were removed. Filtered reads were aligned to the human reference genome (hg19) using STAR (25). Gene expression was quantified using RSEM (26) and normalized counts were logged base 2.
Change in plasma levels of vascular endothelial growth factor (VEGF), carbonic anhydrase IX (CAIX) and osteopontin were additional co-primary endpoints and change in plasma microRNA-210 (miR-210) was a secondary endpoint (Supplementary Materials and Methods). Circulating levels of these hypoxic cellular response proteins have been shown to correlate with other measures of tumor hypoxia and thus represent putative hypoxia measures, whilst also functioning as negative prognostic biomarkers for patients treated with radiotherapy (27–31).
Atovaquone plasma levels were also measured on the morning of the second trial visit, with no time interval mandated between plasma sampling and preceding dose as treatment duration was sufficient to achieve steady state levels (Supplementary Materials and Methods).
Clinical assessment
Adverse event (AE) monitoring occurred until surgery. AEs were graded according to National Cancer Institute’s Common Terminology Criteria v4.0. Given the established excellent safety profile of atovaquone, only grade ≥3 AEs or grade ≥2 atovaquone-related AEs were reportable.
Evaluability and statistical analysis
Primary analyses were on a modified intention-to-treat basis. Hypoxia PET-CT and plasma marker analyses included patients with interpretable data from both visits. Primary imaging analysis using hypoxia PET-CT excluded patients with insufficient baseline HV to reliably measure change (predefined as <1.5 mL). Patients were included for pimonidazole analysis if pimonidazole was administered 16-24 hours prior to surgery.
Statistical analyses used Stata v15.0 (StataCorp, USA) and R v3.6.3 (Comprehensive R Archive Network, https://CRAN.R-projct.org).
Percentage change in HV, plasma markers and other measures were calculated between trial visits for each patient. The binomial method was used to derive 95% CIs for median changes within cohorts. For primary imaging analyses, a linear regression model was used to assess mean difference between cohorts at the second imaging visit adjusted for cohort, baseline measure and tumor volume with values log-transformed to improve model validity. Spearman’s coefficients were reported for correlation analysis. Statistical tests were performed at the 5% level.
For RNA sequencing (RNAseq), normalized read count data for all samples were processed by Limma-Voom method to identify differentially expressed genes (32). Gene Set Enrichment Analysis (GSEA) was carried out using gene sets from the Molecular Signatures Database (MSigDB) v7.1 (21,33). GSEA3.0.0 was used with default settings (33). Gene set variation analysis (GSVA) and single sample gene set enrichment analysis (ssGSEA) was carried out using the GSVA Bioconductor package (34). sigQC protocol was used to evaluate signature applicability and compute signature summary scores (35). Heatmaps were created using the Pheatmap R package (https://CRAN.R-project.org/package=pheatmap). Inter-cohort differences in expression were compared using independent samples t-test for hypoxia signature analysis and Wilcoxon rank-sum test for all other expression analysis (equal variances).
Results
From May 2016 to August 2018, 46 patients were recruited, 23 per cohort. Cohorts were well-balanced in key clinical characteristics, except for male predominance in untreated patients (Table 1). CONSORT diagrams are shown in Supplementary Figs. S1 and S2. Median length of atovaquone treatment between imaging timepoints was 12 [interquartile range (IQR): 7-13] days and the median length prior to tumor resection was 14 (IQR: 12-15) days. Median atovaquone plasma concentration was 62.3 μM (IQR: 50.2-94.2), with all patients achieving a level shown preclinically to reduce tumor hypoxia (11) (Supplementary Fig. S3). Median length between visits for untreated patients was 13 (IQR: 6-13) days.
Table 1. Clinical characteristics of patients at baseline.
| Characteristic | Atovaquone cohort n=23 | No atovaquone cohort n=23 |
|---|---|---|
| Age (yr) | 69.7 (58.5, 74.1) | 68.6 (62.4, 71.4) |
| Sex | ||
| Male | 11 (47.8) | 17 (73.9) |
| Female | 12 (52.2) | 6 (26.1) |
| Ethnicity | ||
| White | 23 (100) | 23 (100) |
| Smoking Status | ||
| Never smoker | 0 (0.0) | 2 (8.7) |
| Ex-smoker | 22 (95.7) | 21 (91.3) |
| Current smoker | 1 (4.4) | 0 (0.0) |
| ECOG performance status | ||
| 0 | 16 (69.6) | 16 (69.6) |
| 1 | 7 (30.4) | 7 (30.4) |
| Histologya | ||
| Adenocarcinoma | 15 (65.2) | 11 (47.8) |
| Squamous cell carcinoma | 7 (30.4) | 8 (34.8) |
| Adenosquamous carcinoma | 1 (4.4) | 0 (0.0) |
| Large cell | 0 (0.0) | 1 (4.4) |
| Giant cell carcinoma | 0 (0.0) | 1 (4.4) |
| Pleomorphic carcinoma | 0 (0.0) | 1 (4.4) |
| Not otherwise specified | 0 (0.0) | 1 (4.4) |
| TNM stagea | ||
| T1b | 0 (0.0) | 1 (4.4) |
| T1c | 2 (8.7) | 1 (4.4) |
| T2vis | 1 (4.4) | 2 (8.7) |
| T2a | 3 (13.0) | 4 (17.4) |
| T2b | 5 (21.7) | 3 (13.0) |
| T3 | 7 (30.4) | 5 (21.7) |
| T4 | 5 (21.7) | 7 (30.5) |
| N0 | 13 (56.5) | 12 (52.2) |
| N1 | 7 (30.4) | 8 (34.8) |
| N2 | 3 (13.0) | 3 (13.0) |
| M0 | 23 (100.0) | 22 (95.7) |
| M1b | 0 (0.0) | 1 (4.4) |
| Tumor maximum diameter (mm) | 45 (33, 60) | 44 (29, 60) |
| Hemoglobin (g/dL) | 13.2 (12.0, 14.3) | 13.9 (12.9, 14.7) |
Abbreviations: ECOG, Eastern Cooperative Oncology Group; TNM, tumor, node and metastasis.
Data shown as patient number with percentage or mean with interquartile range.
Histology and staging are following resection, unless this was not performed, in which case diagnostic histology and imaging were used, respectively.
Tumor hypoxia imaging analysis
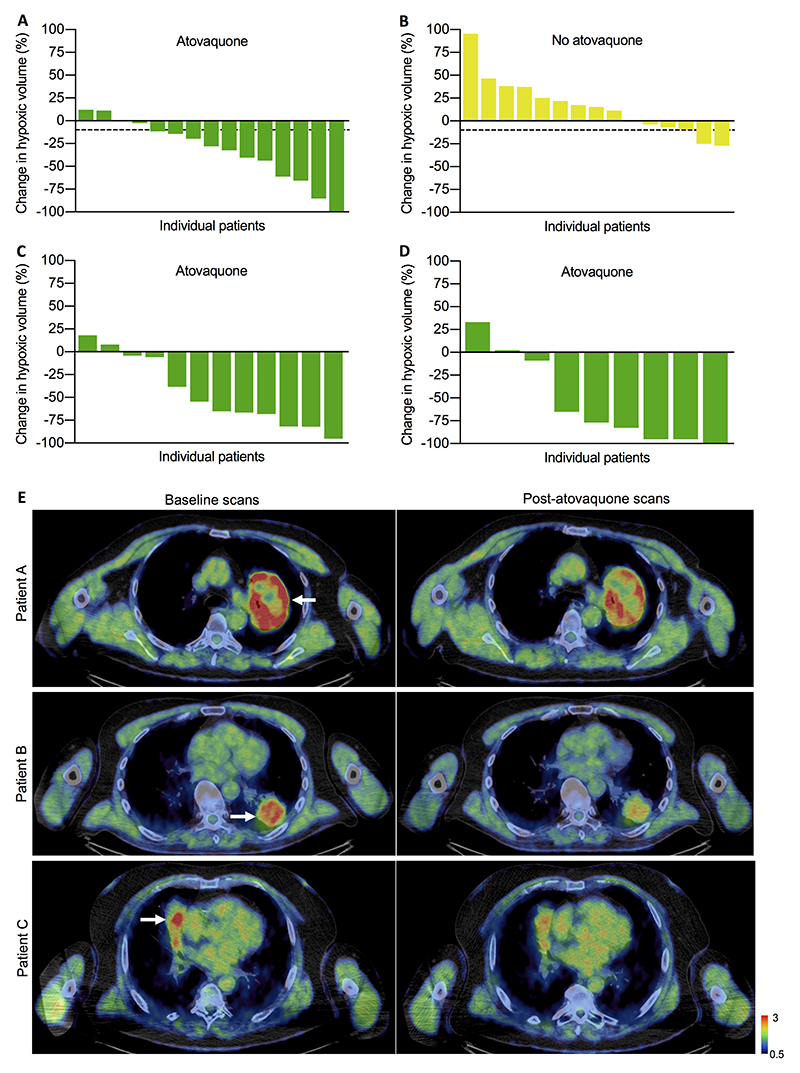
Thirty patients were evaluable for assessing change in HV using hypoxia PET-CT, 15 per cohort. Thirteen patients were deemed unevaluable due to insufficient baseline HV (predefined as <1.5mL) and one patient had uninterpretable scan images. Eleven (73.3%) atovaquone-treated patients had >10% reduction in HV from baseline and the median change was −28.0% (95% CI, −58.2 to −4.4). In contrast, median change in untreated patients was +15.5% (95% CI, −6.5 to 35.5) with >10% reduction observed in only two (13.3%) patients. Estimated geometric mean HV was 55% (95% CI, 24% to 74%) lower at the second visit for atovaquone-treated patients compared to untreated patients (p=0.004), after adjusting for baseline HV and tumor volume. HV changes per individual patient are shown in Figs. 1A and 1B and summarized in Table 2.
Figure 1. Change in tumor HV measured by hypoxia PET-CT.
Waterfall plots of percentage change in HV from baseline for A) atovaquone-treated and B) untreated patients using TBR ≥1.4 to define HV. More than 10% reduction in HV represents a meaningful reduction and is indicated by the dashed line. Changes in HV using TBR C) ≥1.6 and D) ≥1.8 are shown for atovaquone-treated patients with sufficient HV (≥1.5 mL) to reliably measure change. E) Representative FMISO PET-CT scans for three patients (A, B and C) at baseline and post-atovaquone treatment (standardized uptake value scale 0.5-3). Arrows on the pre-treatment scans indicate tumor location. Areas with high tracer uptake (red) represent hypoxic regions. Treatment length between scans was 14 days for patients A and C, and 13 days for patient B. The reduction in HV from baseline for patients A, B and C was 33%, 61% and 66%, respectively.
Table 2. Summary of 4-hour hypoxia PET-CT results.
| Atovaquone cohort n=15 | No atovaquone cohort n=15 | |||
|---|---|---|---|---|
| Tumor volume (mL) | HV (mL) | Tumor volume (mL) | HV (mL) | |
| Baseline scan | 59.3 (30.7, 179.5) | 28.9 (5.4, 50.8) | 43.7 (20.6, 80.7) | 11.4 (2.6, 34.0) |
| Pre-surgery scan | 53.7 (28.5, 186.6) | 13.7 (1.7, 37.4) | 43.8 (23.1, 79.3) | 13.1 (3.2, 32.7) |
| Change from baseline | +4.0% (−2.8, 6.2) | −28.0% (−58.2, −4.4) | +2.8% (−1.7, 14.1) | +15.5% (−6.5, 35.5) |
| Treatment estimate for HV | 0.45 (0.26, 0.76) | |||
Median values with interquartile range for scan visit data and median percentage change from baseline with 95% confidence interval.
Treatment estimate is geometric mean of hypoxic volume (HV) in atovaquone-treated group in relation to untreated group and is equivalent to 55% (95% CI: 24% to 74%) reduction in the atovaquone-treated group. Treatment estimate was adjusted for baseline tumor volume and hypoxic volume with values log-transformed.
All patients had FMISO PET-CT, except for one patient in the atovaquone treatment cohort who had FAZA PET-CT.
Volumes of baseline tumor hypoxia did vary in both cohorts, however this did not appear to affect the degree or direction of response. On review of individual patient hypoxia PET-CT data (Supplementary Table S1), the four atovaquone-treated patients who did not have a meaningful reduction in HV had a range of small (24.6 mL), medium (59.3 mL and 45.9 mL) and large (366.9 mL) HVs at baseline. Similarly, in the two untreated patients in whom HV decreased, one had significant baseline HV (44.8 mL) whilst the other had very little (3.5 mL).
Given the reduction in HV in atovaquone-treated patients, unplanned analysis using higher hypoxia PET tracer uptake thresholds to define hypoxia, thus measuring more intensely hypoxic regions, was performed. Using TBR ≥1.6 and ≥1.8 to define tumor hypoxic voxels, even more marked reduction following treatment was observed in patients whose tumors contained sufficient (>1.5 mL) HV at this level to reliably measure change (Figs. 1C and 1D).
Representative example hypoxia PET-CT images before and after atovaquone treatment are shown in Fig. 1E.
Dynamic hypoxia PET results indicated reduction in tumor hypoxia (k5) following treatment: −11.8% (95% CI: −23.8 to 6.8) vs 11.4% (95% CI: −25.1 to 22.6) for atovaquone-treated and untreated patients, respectively (Table 3). Complete 4-hour and dynamic hypoxia PET-CT data obtained for each patient is provided in Supplementary Table S1.
Table 3. Summary of dynamic hypoxia PET-CT results.
| Atovaquone cohort n=13 | No atovaquone cohort n=13 | |||||
|---|---|---|---|---|---|---|
| v B (%) | K1 (mL/min/g) | k 5 (1/min) | v B (%) | K1 (mL/min/g) | k 5 (1/min) | |
| Baseline scan | 3.7 (2.0, 5.7) | 0.42 (0.36, 0.54) | 0.0038 (0.0027, 0.0050) | 2.1 (0.1, 4.7) | 0.29 (0.27, 0.47) | 0.0044 (0.0038, 0.0062) |
| Pre-surgery scan | 4.9 (2.7,5.1) | 0.43 (0.36,0.50) | 0.0037 (0.0029, 0.0069) | 2.8 (0.1, 4.6) | 0.37 (0.24, 0.42) | 0.0046 (0.0033, 0.0058) |
| Change from baseline | +6.8% (−16.9, 142.2) | −5.7 (−23.3, 15.3) | −11.8% (−23.8, 6.8) | +7.4% (−7.6, 43.5) | −10.4% (−29.3, 6.9) | +11.4% (−25.1, 22.6) |
Median values with interquartile range for scan visit data and median percentage change from baseline with 95% confidence interval.
All patients had FMISO PET-CT, except one patient in the atovaquone treatment cohort who had FAZA PET-CT.
For patients with complete MRI datasets, increase in T2* was observed in five (42%) of 12 atovaquone-treated patients in keeping with increased venous oxygenation, whilst no such change was observed in any untreated patients (Supplementary Figs. S4A and S4B).
Tumor tissue hypoxia transcriptomic and immunohistochemical analysis
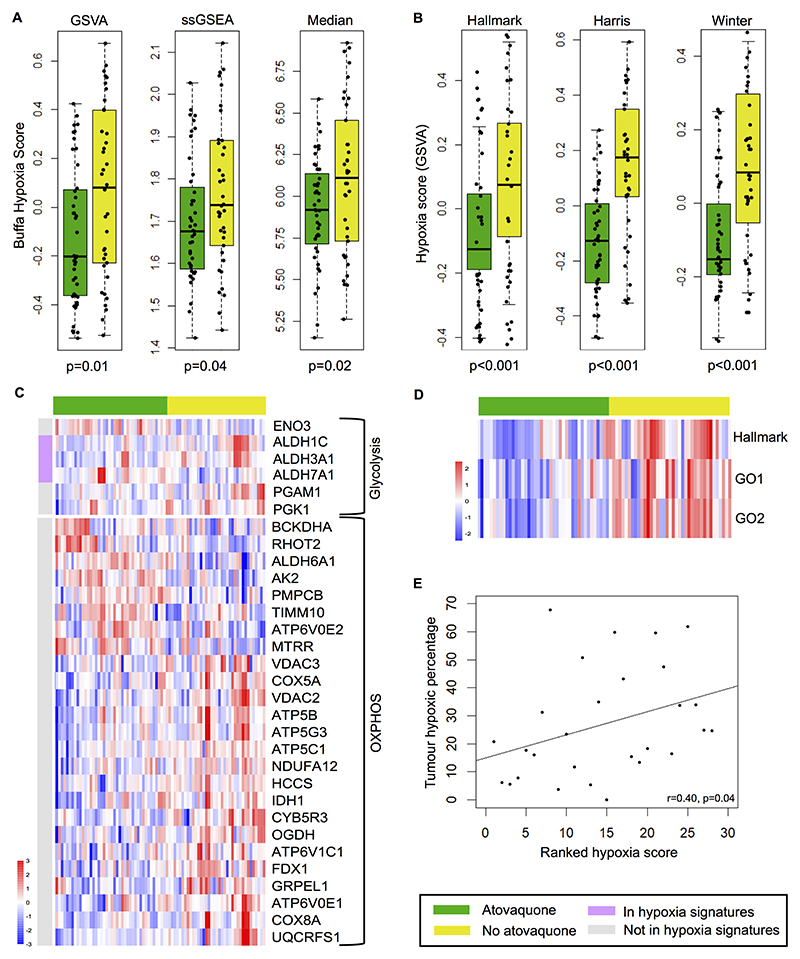
Hypoxia transcriptomic analysis revealed significant differences between atovaquone-treated and untreated tumors. Expression of the Buffa hypoxia signature was lower in treated tumors regardless of whether GSVA (p=0.01), ssGSEA (p=0.04) or median (p=0.02) summarization scores were used (Fig. 2A). This was confirmed by inter-cohort comparison of GSVA analysis using Hallmark (p<0.001), Harris (p<0.001) and Winter (p<0.001) hypoxia signatures (Fig. 2B and Supplementary Fig. S5). GSEA confirmed all four hypoxia gene signatures were significantly enriched in non-treated patients with normalized enrichment score (NES) 1.58 to 2.07, p<0.05 in all cases. Expression of OXPHOS and glycolysis genes was altered (Fig. 2C) and a striking lower expression of genes activated in response to ROS in the group of patients treated with atovaquone was observed (Fig. 2D). Although it may seem paradoxical, it is well known that ROS production is enhanced in hypoxia (36), therefore, a lower level of ROS in atovaquone-treated patients may be a direct consequence of increased oxygen levels. Hypoxia signature validity was confirmed by assessing expression metrics (Supplementary Fig. S6) and correlation with hypoxia PET-CT (Fig. 2E). The Buffa signature correlated with hypoxic tumor percentage (Spearman’s rho=0.40, p=0.04), with significance maintained in a mixed-effects linear model accounting for replicate RNAseq samples (effect size=0.62, p=0.03).
Figure 2. Altered expression of tumor hypoxia- and metabolism-related genes in atovaquone-treated patients as measured by RNAseq.
A) Box and whisker plots of the Buffa hypoxia gene signature expression score in atovaquone-treated and untreated tumor samples using GSVA, ssGSEA and median summarization methods. Each data point represents an expression score obtained from tissue from an entire cross section of tumor. B) GSVA scores for Hallmark, Harris and Winter hypoxia gene signatures. Median values, interquartile range and absolute range (minus outliers) are shown with independent sample t-test significance provided. C) Heatmaps illustrating differentially expressed KEGG glycolysis and oxidative phosphorylation (OXPHOS) genes between treatment groups. Genes present in any of the four hypoxia signatures are indicated by the pink bar. D) Heatmap showing lower expression of ROS associated genes in atovaquone-treated patients using Hallmark and Gene Ontology (GO1 and GO2) ROS signature GSVA scores. E) Positive correlation between Buffa GSVA hypoxia score and hypoxic tumor percentage as measured by hypoxia PET-CT.
Fifteen patients in Cohort 1 and 11 patients in Cohort 2 were evaluable for tumor pimonidazole analysis. Pimonidazole scores were comparable between cohorts, in keeping with equivalent HV tumor percentage for both cohorts at the pre-surgery visit (Supplementary Figs. S7 and S8 and Table S2). Significant positive correlations were observed between pimonidazole score and HV percentage defined using the higher PET tracer uptake thresholds (Supplementary Fig. S9).
Circulating hypoxia markers
Changes in plasma levels of all four putative markers of tumor hypoxia could be assessed in only 10 patients as VEGF was undetectable in nine patients, CAIX was undetectable in one patient and miR-210 was unmeasurable in 15 patients due to sample hemolysis (Supplementary Fig. S10 and Tables S3-6). No agreement was observed between individual markers or between any marker and hypoxia PET-CT or pimonidazole immunohistochemistry.
Tumor perfusion and vascular analysis
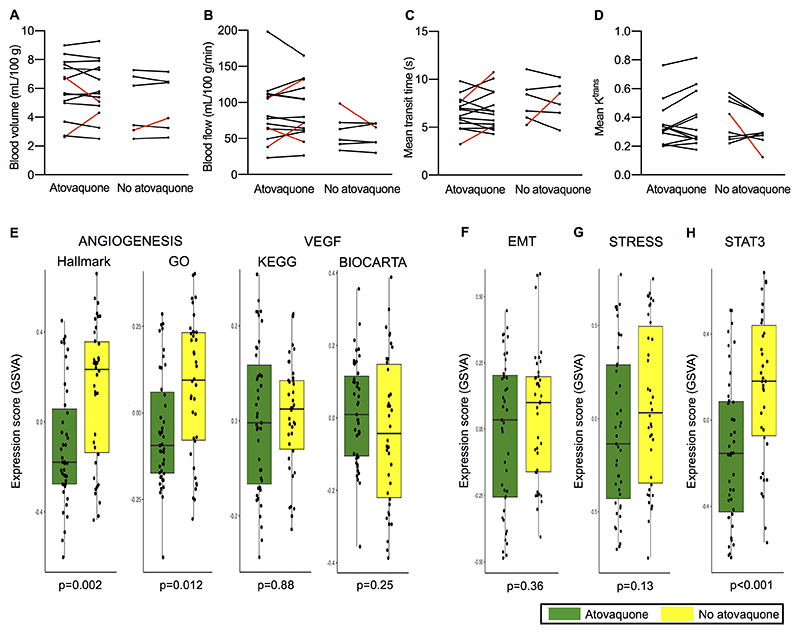
Chronic hypoxia is known to stimulate sustained angiogenesis. As a result, tumor vascularization becomes abnormal leading to impaired tumor perfusion and consequently restricted delivery of oxygen, thus reinforcing hypoxia (37). We investigated whether reversal of hypoxia by atovaquone may improve tumor perfusion through vascular normalization. Using pCT, no meaningful changes in tumor blood volume (Fig. 3A), blood flow (Fig. 3B) or blood mean transit time (Fig. 3C) were observed following treatment, therefore suggesting that hypoxic reduction was not due to increased tumor perfusion. pCT data obtained for each patient is provided in Supplementary Table S7. A number of patients were unevaluable for pCT imaging analyses, with the most common reason being uninterpretable scan images due to variations in the rate of contrast injection as measured by the analysis input function and significant interference of signal form adjacent structures such as heart and great vessels (n=4). DCE-MRI Ktrans (Fig. 3D), as well as dynamic PET-derived vB and K1 (Table 3) were also not altered for either cohort signifying that tumor perfusion was unchanged. We also analyzed the endothelial markers CD31 and CD146 by immunohistochemistry in order to assess a potential decrease in vessel density as indicative of vascular normalization (37). In agreement with the lack of increased perfusion, the endothelial marker analysis did not show a lower vessel density in atovaquone-treated patients, with in fact a trend for the reverse effect observed (Supplementary Fig. S11). In addition, MRI T1 shMOLLI analysis indicated that no changes in tumor biochemistry occurred to suggest significant alteration in cellularity either (Supplementary Figs. S4C and S4D), in keeping with preclinical data showing that as monotherapy atovaquone does not have direct cytotoxic effects (11).
Figure 3. Effect of atovaquone on tumor perfusion and gene expression signatures.
Change in pCT-derived A) blood volume, B) blood flow and C) mean transit time, as well as D) DCE MRI-derived Ktrans for atovaquone-treated and untreated patients. The few cases where a meaningful change in imaging parameter was observed is indicated by red lines. Only data for patients with complete scan data sets and evaluable for primary imaging analysis are shown. E-H) Box and whisker plots showing GSVA scores in atovaquone-treated and untreated tumor samples. E) Angiogenesis-related gene signatures: Hallmark and Gene Ontology (GO) angiogenesis signature scores, and KEGG and BIOCARTA VEGF signaling pathway expression scores. F) EMT signature. G) Integrated stress response (STRESS) genes. H) STAT3-induced gene expression. Each data point represents an expression score obtained from tissue from an entire cross section of tumor. The median value, interquartile range and absolute range (minus outliers) is shown with Wilcoxon rank-sum test significance provided.
Despite the reported absence of an improvement in vascular function, RNAseq analysis revealed a significantly lower expression of angiogenesis signature genes in atovaquone-treated patients: Hallmark Angiogenesis (p=0.002) and Gene Ontology Angiogenesis (p=0.012) (Fig. 3E). Although there is no significant overlap between the hypoxia and angiogenesis signatures (Supplementary Fig. S12) many of the genes in the angiogenesis signature have been linked to the hypoxia response (38–40). Preclinical and clinical evidence has demonstrated that inhibition of the VEGF angiogenic pathway effectively increases tumor oxygenation linked to features of vascular normalization (37). Of note, we did not find differences in the expression of VEGF signaling signatures between untreated and atovaquone-treated patients: KEGG VEGF (p=0.88) and BIOCARTA VEGF (p=0.25) (Fig. 3E).
Altogether, these results suggest that atovaquone treatment reduces angiogenic signaling in NSCLC patients, but this has no impact on vessel functionality, at least within the timeframe of the study.
EMT, integrated stress response and STAT3
Upon OXPHOS inhibition, cancer cells are able to shift to a more glycolytic metabolism for energy production. Since increased glycolysis has been linked to enhanced tumor aggressiveness (41), we investigated whether atovaquone was able to promote epithelial-to-mesenchymal transition (EMT) by analyzing a previously published gene expression signature (42). As shown in Fig. 3F, the expression of the EMT signature was similar in untreated and atovaquone-treated patients (p=0.36), suggesting that atovaquone does not induce a more aggressive tumor phenotype.
Aside from inhibiting OXPHOS, atovaquone has recently been shown to modulate two signaling pathways in cancer cells. In preclinical models of acute myeloid leukemia (AML), atovaquone consistently upregulated four genes involved in the integrated stress response (REDD1, ATF3, CHOP, and CHAC1) (43), suggesting that atovaquone is able to induce a state of cellular stress, which may have antitumoral effects. In contrast to these pre-clinical findings, we found that these four stress response genes were not induced following atovaquone treatment (p=0.13) (Fig. 3G). A gene signature of STAT3 activation was previously used to demonstrate that atovaquone inhibited STAT3 function in multiple myeloma models (44). Using the same signature, we also found that atovaquone treatment strongly diminished the expression of STAT3-induced genes (p<0.001) (Fig. 3H).
Safety and tolerability
In keeping with the established favorable safety profile of atovaquone, only one AE was reported (Grade 3 hypertension) in the atovaquone treatment cohort and was deemed unrelated to treatment.
Discussion
This study demonstrates that the mitochondrial inhibitor atovaquone reduces tumor hypoxia in patients with NSCLC, therefore providing the first clinical confirmation for the strategy of targeting tumor metabolism to alter the tumor microenvironment in this way.
Hypoxia PET-CT was the primary method used to measure tumor HV with a rapid reduction observed in the majority of atovaquone-treated patients. The increase in median HV observed in the non-treated cohort, indicating a worsening of tumor hypoxia over time, made the reduction in treated patients all the more striking. Since this imaging modality is the most accepted method for measuring hypoxic change in the clinical setting (13, 16, 17), our findings provide the best available evidence that atovaquone is a clinically useful hypoxia modifier. Furthermore, as the nitroimidazole-based hypoxia PET tracers used in this study accumulate only in viable tissue at oxygen levels below the threshold at which hypoxia-related radioresistance becomes apparent (45), reduction in uptake should translate into improved tumor radiation response. In support of this, clinical studies have repeatedly demonstrated that high uptake of such tracers functions as a negative prognostic biomarker for patients treated with radiation (7, 46). Our observation that the most pronounced reduction was in the most intensely hypoxic, and thus radiobiologically resistant tumor regions, provides particular impetus for developing atovaquone as a novel tumor radiosensitizer.
Confirmation of the pharmacodynamic effect of hypoxia reduction by the reduced expression of key hypoxia-related tumor gene signatures not only further establishes atovaquone as a hypoxia modifier, but also demonstrates the alteration in oxygenation was of biological relevance. Furthermore, transcriptomic analysis revealed altered expression of OXPHOS and glycolysis pathways, as well as decreased ROS signature expression, in keeping with effects on tumor metabolism more widely. The downregulation of ROS-induced genes in the tumors of patients treated with atovaquone is in agreement with higher oxygen levels, as it is well established that ROS production is enhanced under hypoxia (36).
Preclinical evidence suggests that the mechanism of action by which atovaquone reduces tumor hypoxia is through decreased tumor oxygen consumption (11). It is possible, however, that reversal of tumor hypoxia could cause a normalization of the vasculature, through a decrease in angiogenic signaling, and consequently improve tumor perfusion, which in turn could contribute to tumor oxygenation (37). Our observation that atovaquone treatment was not associated with changes in tumor perfusion, as assessed by multimodality perfusion imaging, strongly suggest that vascular normalization does not contribute to the hypoxia alleviation reported in NSCLC patients treated with atovaquone. This was further supported by the lack of decreased vessel density, considered a feature of vascular normalization (37), in atovaquone-treated patients. Therefore, the decrease in hypoxia is likely the consequence of increased oxygenation due to diminished oxygen consumption.
RNAseq analysis revealed reduced angiogenic signaling in atovaquone-treated patients. The absence of improved perfusion with atovaquone treatment may indicate that the reported decrease in angiogenic signaling does not suffice or requires a longer time to cause a significant vascular normalization. In addition, the fact that VEGF signaling, regarded as one of the main drivers of tumor angiogenesis (37), remained unaltered with atovaquone treatment may also partly explain the lack of substantial vascular normalization and increased perfusion, despite an overall decrease in angiogenic signaling.
Atovaquone did not lead to an increased expression in genes involved in EMT, suggesting that treatment did not make the tumors more aggressive. This is important because of the possibility that the inhibition of OXPHOS and subsequent increased glycolysis could lead to increased tumor aggressiveness and metastasis (41). Our study precluded long-term follow-up, therefore, we are unable to assess whether incidence of metastasis or other clinical outcomes differed between cohorts, and these are important endpoints for future clinical testing of this agent as an anti-cancer treatment.
Aside from modulating metabolism, atovaquone has recently been shown to inhibit STAT3 signaling and upregulate integrated stress response genes, as demonstrated in multiple myeloma and AML models, respectively (43,44). Whilst we did not see a change in the expression of stress response genes, we did find a decrease in genes affected by STAT3 signaling. Interestingly, unlike the above-mentioned studies in hematological malignancies, our preclinical studies did not show an anti-tumor effect of atovaquone as single agent (11). In this current study, we did not observe a reduction in tumor volume with atovaquone treatment, but it is feasible this may occur with longer treatment duration.
Although this study demonstrates hypoxia alleviation in NSCLC, preclinical evidence demonstrates that atovaquone inhibits OCR in a wide range of tumor cell lines (11) and therefore it is expected this effect would occur in patients with different tumors types. Since hypoxia confers resistance to radiation, chemotherapy and immunotherapy (5, 7–9), co-administration with numerous such well-established treatments widens clinical applicability further.
Several FDA-approved drugs inhibit OXPHOS through off-target effects and have been proposed clinical hypoxia modifiers (10), of which metformin is most-studied. This mitochondrial complex I inhibitor (47) reduces OCR in cancer cell lines and ameliorates hypoxia in xenografts, leading to improved tumor radiation response (11, 48, 49). Although metformin has been shown to alter tumor metabolism in breast cancer patients (50), its effect on tumor oxygenation in patients has not been reported. Importantly, concentrations required to reduce tumor OCR in preclinical models are higher than clinically achievable. In keeping with this, a recent phase II study combining metformin with chemoradiotherapy in NSCLC failed to improve outcomes (51). In contrast, all patients in our study had atovaquone plasma levels at or above that known to reduce tumor OCR in preclinical experiments, with levels observed to be in-keeping with that stated in the Summary of Product Characteristics for the oral micronized preparation used in this study (52).
Favorable toxicity is paramount when developing radiosensitizers since standard (chemo)radiotherapy regimes are already significantly toxic. The established excellent safety profile of atovaquone, as was observed in this study, further highlights this drug as highly promising for this purpose. As atovaquone is safe, FDA-approved and inexpensive, repurposing this agent as an anti-cancer treatment has the potential for rapid adoption into routine clinical practice, whilst helping to contribute to more sustainable healthcare systems.
Our study had limitations. Firstly, this was a non-randomized study and, although sizable for a translational study, patient numbers were relatively small. For this reason, it was decided not to investigate the mutational status of these tumors as the patient numbers would be too small to meaningfully assess whether certain mutations could be associated with response to atovaquone. Secondly, as baseline tumor tissue was not collected, assessment of change in hypoxia signatures and immunohistochemistry per patient was not possible. For pimonidazole immunohistochemistry, analysis was impeded due to surgery occurring outside the narrow time-window required following pimonidazole administration for a number of patients. For evaluable patients, comparable scores between cohorts was in keeping with hypoxia PET-CT-derived HV equivalence prior to surgery, with correlation between these measures observed. As atovaquone-treated patients appeared to have more baseline HV, the lack of difference in pimonidazole staining may in fact represent additional evidence for hypoxia modification.
Validated and deliverable clinical hypoxia biomarkers are needed to enable selection of patients with hypoxic tumors for treatment, and the challenge of identifying such markers was apparent in this trial. Putative circulating hypoxia markers previously shown to correlate with radiotherapy outcomes in NSCLC (28) were often undetectable or provided conflicting results. Despite difficulties correlating hypoxia measures, observed agreement between hypoxia PET-CT and gene expression signatures suggests a transcriptomic approach may be a particularly promising future strategy. Agreement between the different hypoxia signature scores and correlation with hypoxia PET-CT was also observed, supporting signature validity as hypoxia biomarkers in NSCLC.
Here we provide the first clinical report that tumor hypoxia can be reduced by targeting tumor metabolism. Repurposing atovaquone for this purpose has the potential to improve the outcomes of numerous established treatments in NSCLC, as well as in other tumor types. In light of these highly promising findings, we have initiated phase I clinical testing combining this agent with chemoradiation in NSCLC.
Supplementary Material
Statement of Translational Relevance.
The antimalarial drug atovaquone, a known inhibitor of mitochondrial oxygen consumption by targeting OXPHOS, rapidly reduces tumor hypoxia in patients with NSCLC. This is the first clinical report that targeting mitochondrial metabolism is an effective strategy to alter the tumor microenvironment in this way. As tumor hypoxia is a well-recognized barrier to radiation, chemotherapy and immunotherapy, combining this agent with such treatments may improve outcomes for many patients. In NSCLC, this is particularly important as novel radiosensitizers are needed to improve radiation outcomes. Atovaquone reduces oxygen consumption in many tumor cell lines, therefore, hypoxia modification is expected in patients with various tumor types. As atovaquone is FDA-approved and well-tolerated, repurposing this agent as an anti-cancer treatment has the potential for rapid adoption into clinical practice. Utilizing established as well as putative tumor hypoxia measures, findings presented contribute to development of clinically useful biomarkers to enable patient selection for hypoxia-reducing therapies.
Acknowledgements
We thank all patients and their families for taking part and the staff at Oxford University Hospitals National Health Service Foundation Trust. This trial was sponsored by the University of Oxford. Trial management was provided by the Oncology Clinical Trials Office at the University of Oxford as part of the UKCRC Oxford Clinical Trials Research Unit. We thank Alix Hampson for assistance with measurement of plasma hypoxia marker and atovaquone levels.
Funding
This work was funded by the Howat Foundation (C5255/A25069). Additional support was obtained from CRUK & EPSCRC Cancer Imaging Centre in Oxford (C5255/A16466), CRUK/MRC Oxford Institute for Radiation Oncology (C5255/A23755) and the NIHR Oxford Experimental Cancer Medicine Centre. D.R.M. was funded by a NIHR/HEE Clinical Lectureship (ICA-CL-2016-02-009). P.S.M. was supported by the Jean Shanks Foundation/Pathological Society of Great Britain & Ireland Clinical Research Training Fellowship. J.A.B. was supported by the EPSRC/MRC Centre for Doctoral Training in Systems Approaches to Biomedical Science (EP/G037280/1) and the EPSRC Impact Acceleration Account (EP/R511742/1). G.S.H. was supported by a CRUK Clinician Scientist Fellowship (C34326/A19590).
Footnotes
Conflict of interest disclosure statement
The authors have no conflicts of interest to declare
References
- 1.Carmeliet P, Jain RK. Angiogenesis in cancer and other diseases. Nature. 2000;407:249–57. doi: 10.1038/35025220. [DOI] [PubMed] [Google Scholar]
- 2.Petrova V, Annicchiarico-Petruzzelli M, Melino G, Amelio I. The hypoxic tumour microenvironment. Oncogenesis. 2018;7:10. doi: 10.1038/s41389-017-0011-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.LaGory EL, Giaccia AJ. The Ever Expanding Role of HIF in Tumour and Stromal Biology. Nat Cell Biol. 2016;18:356–65. doi: 10.1038/ncb3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rankin EB, Nam J-M, Giaccia AJ. Hypoxia: Signaling in the Metastatic Cascade. Trends Cancer. 2016;2:295–304. doi: 10.1016/j.trecan.2016.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gray LH, Conger AD, Ebert M, Hornsey S, Scott OCA. The Concentration of Oxygen Dissolved in Tissues at the Time of Irradiation as a Factor in Radiotherapy. Br J Radiol. 1953;26:638–48. doi: 10.1259/0007-1285-26-312-638. [DOI] [PubMed] [Google Scholar]
- 6.Nordsmark M, Bentzen SM, Rudat V, Brizel D, Lartigau E, Stadler P, et al. Prognostic value of tumor oxygenation in 397 head and neck tumors after primary radiation therapy. An international multi-center study. Radiother Oncol. 2005;77:18–24. doi: 10.1016/j.radonc.2005.06.038. [DOI] [PubMed] [Google Scholar]
- 7.Eschmann S-M, Paulsen F, Reimold M, Dittmann H, Welz S, Reischl G, et al. Prognostic impact of hypoxia imaging with 18F-misonidazole PET in non-small cell lung cancer and head and neck cancer before radiotherapy. J Nucl Med. 2005;46:253–60. [PubMed] [Google Scholar]
- 8.Walsh JC, Lebedev A, Aten E, Madsen K, Marciano L, Kolb HC. The Clinical Importance of Assessing Tumor Hypoxia: Relationship of Tumor Hypoxia to Prognosis and Therapeutic Opportunities. Antioxid Redox Signal. 2014;21:1516–54. doi: 10.1089/ars.2013.5378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li Y, Patel SP, Roszik J, Qin Y. Hypoxia-Driven Immunosuppressive Metabolites in the Tumor Microenvironment: New Approaches for Combinational Immunotherapy. Front Immunol. 2018;9:1591. doi: 10.3389/fimmu.2018.01591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ashton TM, McKenna WG, Kunz-Schughart LA, Higgins GS. Oxidative Phosphorylation as an Emerging Target in Cancer Therapy. Clin Cancer Res. 2018;24:2482–90. doi: 10.1158/1078-0432.CCR-17-3070. [DOI] [PubMed] [Google Scholar]
- 11.Ashton TM, Fokas E, Kunz-Schughart LA, Folkes LK, Anbalagan S, Huether M, et al. The anti-malarial atovaquone increases radiosensitivity by alleviating tumour hypoxia. Nat Commun. 2016;7:12308. doi: 10.1038/ncomms12308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Porporato PE, Filigheddu N, Pedro JMB-S, Kroemer G, Galluzzi L. Mitochondrial metabolism and cancer. Cell Res. 2018;28:265–80. doi: 10.1038/cr.2017.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fleming IN, Manavaki R, Blower PJ, West C, Williams KJ, Harris AL, et al. Imaging tumour hypoxia with positron emission tomography. Br J Cancer. 2015;112:238–50. doi: 10.1038/bjc.2014.610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McGowan DR, Skwarski M, Bradley KM, Campo L, Fenwick JD, Gleeson FV, et al. Buparlisib with thoracic radiotherapy and its effect on tumour hypoxia: A phase I study in patients with advanced non-small cell lung carcinoma. Eur J Cancer. 2019;113:87–95. doi: 10.1016/j.ejca.2019.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koh WJ, Rasey JS, Evans ML, Grierson JR, Lewellen TK, Graham MM, et al. Imaging of hypoxia in human tumors with [F-18]fluoromisonidazole. Int J Radiat Oncol Biol Phys. 1992;22:199–212. doi: 10.1016/0360-3016(92)91001-4. [DOI] [PubMed] [Google Scholar]
- 16.Okamoto S, Shiga T, Yasuda K, Ito YM, Magota K, Kasai K, et al. High Reproducibility of Tumor Hypoxia Evaluated by 18F-Fluoromisonidazole PET for Head and Neck Cancer. J Nucl Med. 2013;54:201–7. doi: 10.2967/jnumed.112.109330. [DOI] [PubMed] [Google Scholar]
- 17.Grkovski M, Schwartz J, Rimner A, Schöder H, Carlin SD, Zanzonico PB, et al. Reproducibility of 18F-fluoromisonidazole intratumour distribution in non-small cell lung cancer. EJNMMI Res. 2016;6:79. doi: 10.1186/s13550-016-0210-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Salem A, Asselin M-C, Reymen B, Jackson A, Lambin P, West CML, et al. Targeting Hypoxia to Improve Non–Small Cell Lung Cancer Outcome. J Natl Cancer Inst. 2018;110:14–30. doi: 10.1093/jnci/djx160. [DOI] [PubMed] [Google Scholar]
- 19.Harris BHL, Barberis A, West CML, Buffa FM. Gene Expression Signatures as Biomarkers of Tumour Hypoxia. Clin Oncol. 2015;27:547–60. doi: 10.1016/j.clon.2015.07.004. [DOI] [PubMed] [Google Scholar]
- 20.Buffa FM, Harris AL, West CM, Miller CJ. Large meta-analysis of multiple cancers reveals a common, compact and highly prognostic hypoxia metagene. Br J Cancer. 2010;102:428–35. doi: 10.1038/sj.bjc.6605450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liberzon A, Birger C, Thorvaldsdóttir H, Ghandi M, Mesirov JP, Tamayo P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015;1:417–25. doi: 10.1016/j.cels.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harris AL. Hypoxia — a key regulatory factor in tumour growth. Nat Rev Cancer. 2002;2:38–47. doi: 10.1038/nrc704. [DOI] [PubMed] [Google Scholar]
- 23.Winter SC, Buffa FM, Silva P, Miller C, Valentine HR, Turley H, et al. Relation of a Hypoxia Metagene Derived from Head and Neck Cancer to Prognosis of Multiple Cancers. Cancer Res. 2007;67:3441–9. doi: 10.1158/0008-5472.CAN-06-3322. [DOI] [PubMed] [Google Scholar]
- 24.Andrews S. FastQC: A Quality Control Tool for High Throughput Sequence Data. Available at http://www.bioinformatics.babraham.ac.uk/projects/fastqc/
- 25.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dunst J, Stadler P, Becker A, Kuhnt T, Lautenschläger C, Molls M, et al. Tumor hypoxia and systemic levels of vascular endothelial growth factor (VEGF) in head and neck cancers. Strahlenther Onkol. 2001;177:469–73. doi: 10.1007/pl00002428. [DOI] [PubMed] [Google Scholar]
- 28.Ostheimer C, Bache M, Güttler A, Kotzsch M, Vordermark D. A pilot study on potential plasma hypoxia markers in the radiotherapy of non-small cell lung cancer. Strahlenther Onkol. 2014;190:276. doi: 10.1007/s00066-013-0484-1. [DOI] [PubMed] [Google Scholar]
- 29.İlie M, Mazure NM, Hofman V, Ammadi RE, Ortholan C, Bonnetaud C, et al. High levels of carbonic anhydrase IX in tumour tissue and plasma are biomarkers of poor prognostic in patients with non-small cell lung cancer. Br J Cancer. 2010;102:1627–35. doi: 10.1038/sj.bjc.6605690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jiang M, Li X, Quan X, Li X, Zhou B. Clinically Correlated MicroRNAs in the Diagnosis of Non-Small Cell Lung Cancer: A Systematic Review and Meta-Analysis. BioMed Res Int. 2018;2018:5930951. doi: 10.1155/2018/5930951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Świtlik WZ, Karbownik MS, Suwalski M, Kozak J, Szemraj J. Serum miR-210-3p as a Potential Noninvasive Biomarker of Lung Adenocarcinoma: A Preliminary Study. Genet Test Mol Biomark. 2019;23:353–8. doi: 10.1089/gtmb.2018.0275. [DOI] [PubMed] [Google Scholar]
- 32.Law CW, Chen Y, Shi W, Smyth GK. voom: precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 2014;15:R29. doi: 10.1186/gb-2014-15-2-r29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hänzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics. 2013;14:7. doi: 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dhawan A, Barberis A, Cheng W-C, Domingo E, West C, Maughan T, et al. Guidelines for using sigQC for systematic evaluation of gene signatures. Nat Protoc. 2019;14:1377–400. doi: 10.1038/s41596-019-0136-8. [DOI] [PubMed] [Google Scholar]
- 36.Hernansanz-Agustín P, Choya-Foces C, Carregal-Romero S, Ramos E, Oliva T, Villa-Piña T, et al. Na + controls hypoxic signalling by the mitochondrial respiratory chain. Nature. 2020;586:287–91. doi: 10.1038/s41586-020-2551-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Goel S, Duda DG, Xu L, Munn LL, Boucher Y, Fukumura D, et al. Normalization of the vasculature for treatment of cancer and other diseases. Physiol Rev. 2011;91:1071–121. doi: 10.1152/physrev.00038.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kanehisa M. Toward understanding the origin and evolution of cellular organisms. Protein Sci. 2019;28:1947–51. doi: 10.1002/pro.3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kanehisa M, Sato Y, Furumichi M, Morishima K, Tanabe M. New approach for understanding genome variations in KEGG. Nucleic Acids Res. 2019;47:D590–5. doi: 10.1093/nar/gky962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kanehisa M, Goto S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Georgakopoulos-Soares I, Chartoumpekis DV, Kyriazopoulou V, Zaravinos A. EMT Factors and Metabolic Pathways in Cancer. Front Oncol. 2020;10:499. doi: 10.3389/fonc.2020.00499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Byers LA, Diao L, Wang J, Saintigny P, Girard L, Peyton M, et al. An Epithelial–Mesenchymal Transition Gene Signature Predicts Resistance to EGFR and PI3K Inhibitors and Identifies Axl as a Therapeutic Target for Overcoming EGFR Inhibitor Resistance. Clin Cancer Res. 2013;19:279–90. doi: 10.1158/1078-0432.CCR-12-1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stevens AM, Xiang M, Heppler LN, Tošić I, Jiang K, Munoz JO, et al. Atovaquone is active against AML by upregulating the integrated stress pathway and suppressing oxidative phosphorylation. Blood Adv. 2019;3:4215–27. doi: 10.1182/bloodadvances.2019000499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xiang M, Kim H, Ho VT, Walker SR, Bar-Natan M, Anahtar M, et al. Gene expression–based discovery of atovaquone as a STAT3 inhibitor and anticancer agent. Blood. 2016;128:1845–53. doi: 10.1182/blood-2015-07-660506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Krohn KA, Link JM, Mason RP. Molecular Imaging of Hypoxia. J Nucl Med. 2008;49:129S–148S. doi: 10.2967/jnumed.107.045914. [DOI] [PubMed] [Google Scholar]
- 46.Rischin D, Hicks RJ, Fisher R, Binns D, Corry J, Porceddu S, et al. Prognostic significance of [18F]-misonidazole positron emission tomography-detected tumor hypoxia in patients with advanced head and neck cancer randomly assigned to chemoradiation with or without tirapazamine: a substudy of Trans-Tasman Radiation Oncology Group Study 98.02. J Clin Oncol. 2006;24:2098–104. doi: 10.1200/JCO.2005.05.2878. [DOI] [PubMed] [Google Scholar]
- 47.Owen MR, Doran E, Halestrap AP. Evidence that metformin exerts its anti-diabetic effects through inhibition of complex 1 of the mitochondrial respiratory chain. Biochem J. 2000;348:607–14. [PMC free article] [PubMed] [Google Scholar]
- 48.Zannella VE, Dal Pra A, Muaddi H, McKee TD, Stapleton S, Sykes J, et al. Reprogramming metabolism with metformin improves tumor oxygenation and radiotherapy response. Clin Cancer Res. 2013;19:6741–50. doi: 10.1158/1078-0432.CCR-13-1787. [DOI] [PubMed] [Google Scholar]
- 49.De Bruycker S, Vangestel C, Van den Wyngaert T, Pauwels P, Wyffels L, Staelens S, et al. 18F-Flortanidazole Hypoxia PET Holds Promise as a Prognostic and Predictive Imaging Biomarker in a Lung Cancer Xenograft Model Treated with Metformin and Radiotherapy. J Nucl Med. 2019;60:34–40. doi: 10.2967/jnumed.118.212225. [DOI] [PubMed] [Google Scholar]
- 50.Lord SR, Cheng W-C, Liu D, Gaude E, Haider S, Metcalf T, et al. Integrated Pharmacodynamic Analysis Identifies Two Metabolic Adaption Pathways to Metformin in Breast Cancer. Cell Metab. 2018;28:679–88. doi: 10.1016/j.cmet.2018.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tsakiridis T, Hu C, Skinner HD, Santana-Davila R, Lu B, Erasmus JJ, et al. Initial reporting of NRG-LU001 ( NCT02186847), randomized phase II trial of concurrent chemoradiotherapy (CRT) +/- metformin in locally advanced Non-Small Cell Lung Cancer (NSCLC) J Clin Oncol. 2019;37:8502. [Google Scholar]
- 52.GlaxoSmithKline UK. Wellvone 750 mg/5ml oral suspension Summary of Product Characteristics. Available form https://www.medicines.org.uk/emc/product/952/smpc.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.