Abstract
Circulating proteins are vital in human health and disease and are frequently used as biomarkers for clinical decision-making or as targets for pharmacological intervention. Here we map and replicate protein quantitative trait loci (pQTL) for 90 cardiovascular proteins in over 30,000 individuals, resulting in 451 pQTLs for 85 proteins. For each protein we further perform pathway mapping to obtain trans-pQTL gene and regulatory designations. We substantiate these regulatory findings with orthogonal evidence for trans-pQTLs using mouse knock-down experiments (ABCA1, TRIB1) and clinical trial results (CCR2, CCR5), with consistent regulation. Finally we evaluate known drug targets, and suggest new target candidates or repositioning opportunities using Mendelian randomization. This identifies 11 proteins with causal evidence of involvement in human disease that have not previously been targeted, including (gene symbols) EGF, IL16, PAPPA, SPON1, F3, ADM, CASP8, CHI3L1, CXCL16, GDF15, and MMP12. Taken together these findings demonstrate the utility of large-scale mapping the genetics of the proteome, and provide a resource for future precision studies of circulating proteins in human health.
Proteins circulating in blood are derived from multiple organs and cell types, and consist of both actively secreted and passively leaked proteins. Plasma proteins are frequently used as biomarkers to diagnose and predict disease and have been of key importance for clinical practice and drug development for many decades.
Circulating proteins are attractive as potential drug targets as they can often be directly perturbed using conventional small molecules or biologics such as monoclonal antibodies1. However, a prerequisite for successful drug development is efficacy, which is predicated on the drug target playing a causal role in disease. One approach to clarifying causation is through Mendelian randomization (MR), which has successfully predicted the outcome of randomized controlled trials (RCT) for pharmacological targets such as PCSK9, LpPLA2 and NPC1L1, and is increasingly becoming a standard tool for triaging new drug targets2.
Recent technological developments of targeted proteomic methods have enabled hundreds to thousands of circulating proteins to be measured simultaneously in large studies3–6. This has paved the way for studies of genetic regulation of circulating proteins using genome-wide association studies (GWAS) for detection of protein quantitative trait loci (pQTL), some of which are referenced here3,4,7–9.
Here, we present a genome-wide meta-analysis of 90 cardiovascular-related proteins, many of which are established prognostic biomarkers or drug targets, measured using the Olink Proximity Extension Assay CVD-I panel 10 in 30,931 subjects across 14 studies. The identified pQTLs were combined with other sources of information to suggest new target candidates underpinned by insights into cis- and trans- regulation of protein levels and to evaluate past and present efforts to therapeutically modify the proteins analysed in the present investigation. We also show that protein-centric polygenic risk scores (PRS) can predict a substantial fraction of inter-individual variability in circulating protein levels, explaining a proportion of disease susceptibility attributable to specific biological pathways.
These are the first results to emerge from the SCALLOP consortium, a collaborative framework for pQTL mapping and biomarker analysis of proteins on the Olink platform (www.scallop-consortium.com).
Results
Genome-wide meta-analysis of 90 proteins reveals 467 independent genetic loci associated with plasma levels of 85 proteins
Ninety proteins in up to 21,758 participants from 13 cohorts passed quality control (QC) criteria and were available for GWAS meta-analysis [Supplementary Table 1]. We found a total of 401 pQTLs that were significant at a discovery P-value threshold conventional for GWAS (P<5x10-8) [Figure 1] [Supplementary Table 2]. Conditioning each of these primary pQTLs using the GCTA-COJO software, we identified an additional 144 proximal pQTLs that independently surpassed conventional genome-wide significance (P<5x10-8), termed as secondary pQTLs. We attempted to replicate the primary and secondary pQTLs in two independent studies (9,173 participants) whereupon the discovery and replication datasets were meta-analysed, leading to 315 primary pQTLs and 136 secondary pQTLs surpassing a Bonferroni corrected P-value (P<5.6x10-10). The discovery P-values were used for pQTLs absent in the replication dataset (nsnp=25) [Supplementary Table 2].
Figure 1. Chromosomal location of all associations discovered.
Cis-pQTLs are shown in red (bold) and trans-pQTLs in blue if they are significant at a conventional GWAS significance threshold of P<5x10-8. The gene annotations refer to the gene closest to the pQTL. A version of this figure with only loci selected according to the criteria for Mendelian randomization is available as [Extended figure 1].
Some proteins such as SCF, RAGE, PAPPA, CTSL1 and MPO showed association with more than nine primary pQTLs, but most proteins (22 of 85) were associated with 2 primary pQTLs. We also observed that some proteins were associated with multiple conditionally significant (secondary) pQTLs such as CCL-4 with 4 secondary signals, implicating complex genetic regulation of circulating CCL-4 at the CCL4 locus.
Analysis of trans-pQTLs suggests common mechanisms by which genetic variants affect plasma protein levels
A “best guess” causal gene for each of the CVD-I trans-pQTLs was assigned by a hierarchical approach based on analysis of protein-protein interactions (PPI), literature mining, genomic distance to gene and manual review of literature around the gene as well as the genomic context of the association signal. In total, 326 primary trans-pQTLs were assigned to unique genes and 30 trans-pQTLs were assigned more than one gene, with ABO, ST3GAL4, JMJD1C, SH2B3, ZFPM2 showing association with the levels of five or more CVD-I proteins [Extended Figure 2A and 2B] [Supplementary Table 2]. Extending this analysis to pQTLs from literature expanded the list of genes with five or more protein associations to include also KLKB1, GCKR, FUT2, TRIB1, SORT1 and F12 [Supplementary Table 4].
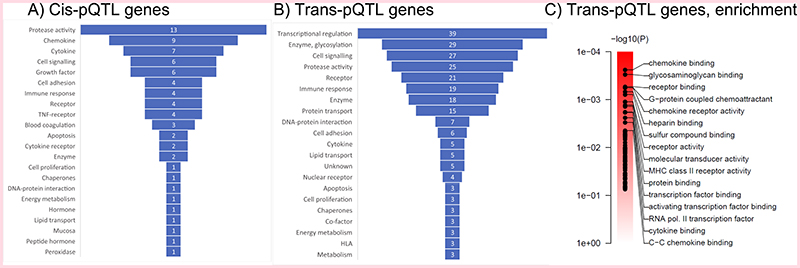
Gene ontology (GO) analysis of genes assigned to all significant trans-pQTLs showed functional enrichment for chemokine binding, glycosaminoglycan binding, receptor binding and G-protein coupled chemoattractant activity [Figure 2C]. A broader classification of genes assigned to both cis- and trans-pQTLs [Figure 2A, 2B] [Supplementary Table 2] using a wider set of tools (Online Methods) suggested that transcriptional regulation, post-translational modifications, such as glycation and sialylation, cell-signalling events, protease activity and receptor binding are potential common mechanisms by which trans-pQTLs influence circulating protein levels. The default gene calls and paths for the CVD-I trans-pQTLs based on PPI and literature mining can be visualised using the SCALLOP CVD-I network tool [Extended Figure 2C] whereas details on the classification of genes are available in the Online Methods, [Supplementary Information 1] and [Supplementary Table 3].
Figure 2. Classification of cis- and trans-pQTL genes.
A. The gene ontology label of all cis-pQTL genes, i.e. the protein-encoding genes. B. The gene-ontology label of all best-guess trans-pQTL genes. C. Gene set enrichment analysis of genes assigned to all significant trans-pQTLs, showing the top-gene sets from the Gene Ontology set Molecular Function.
Evidence of mRNA expression mediating associations with a third of cis pQTLs
We investigated the overlap of the CVD-I cis- and trans-pQTLs with expression quantitative trait loci (eQTL) by a combination of approaches and eQTL studies, including direct genetic lookups and colocalisation using PrediXcan 11 and SMR / HEIDI 12. For direct lookups, three studies were used: LifeLines-DEEP (whole blood), eQTLGen meta-analysis (whole blood and PBMCs) and GTEx (48 tissue types). Of 545 pQTLs from [Supplementary table 2], eQTL data were available for 434 SNP-transcript pairs, including 168 cis-pQTLs and 266 trans-pQTLs. Of these, 72 (43%) of cis-pQTLs had at least one corresponding eQTL (FDR<0.05) in any of the eQTL datasets investigated, implicating 42 of the 75 proteins with a cis-pQTL. At a more stringent eQTL p-value of P<5x10-8, the percentage with a corresponding eQTL was 26 %, similar to some previous reports 13–15 [Supplementary Table 5].
Co-localisation analysis of CVD-I cis-pQTLs and mRNA levels was performed in selected tissues from the GTEx project by first imputing mRNA expression of the CVD-I protein-encoding transcripts using the PrediXcan11 algorithm in one of the SCALLOP CVD-I cohorts (IMPROVE), and then testing imputed mRNA levels for association with CVD-I plasma protein levels using linear regression. Twenty-six of the 90 CVD-I proteins were associated with their corresponding mRNA transcript (FDR<0.05) in at least one of the 20 GTEx tissues investigated [Extended Figure 3]. All 26 proteins were among the 42 proteins found to also be an eQTL by direct lookups. Proteins CCL4, CD40, CHI3L1, CSTB and IL-6RA all associated with their corresponding transcript across five or more tissues whereas proteins ST2 and RAGE showed significant association exclusively in lung, and CTSD exclusively in skeletal muscle.
To further investigate if the CVD-I protein pQTLs overlap with eQTLs, we used the SMR/HEIDI methods12, using data from the Consortium for the Architecture of Gene Expression (CAGE) study. SMR/HEIDI tests the hypothesis that there is a single variant affecting protein and gene expression (pleiotropy or causality), with the alternative hypothesis being that protein and gene expression are affected by two distinct variants. In total, 125 associations between 96 genes and 54 proteins were identified at an experiment-wise SMR test significance level (PSMR<0.05/8558) and a stringent HEIDI test threshold (PHEIDI > 0.01) [Supplementary Table 6], of which 23.2 % were in cis-pQTL regions, such as IL-8 and U-PAR. The 96 genes were located in 74 loci, suggesting that pleiotropic associations between protein and mRNA expression were present for 18.4 % of significant and suggestive primary loci using SMR / HEIDI.
A minor proportion of cis-acting pQTLs are in high linkage-disequilibrium with non-synonymous coding variants
“Pseudo-pQTLs” caused by epitope effects, i.e. differential assay recognition depending on presence of protein-altering variants, is a theoretical possibility for cis-pQTLs and likely dependent on the method of protein quantification 4,16. To evaluate the potential for pseudo-pQTLs among the CVD-I pQTLs, we investigated presence of protein-altering variants for sentinel variants or variants in high linkage disequilibrium with a sentinel variant. Of the 90 proteins, 85 had at least one pQTL, including 12 with only cis-pQTLs, 10 with only trans-pQTLs and 63 with both cis- and trans-pQTLs. Of the 170 primary or secondary cis-pQTLs for 75 proteins, 20 cis-pQTLs for 18 proteins had a sentinel variant in high linkage disequilibrium (LD; R2>0.9) with a protein-altering variant, which suggests potential to affect assay performance [Supplementary Table 2].
Orthogonal evidence supports causal gene to protein relationships for a subset of the CVD-I trans-pQTLs
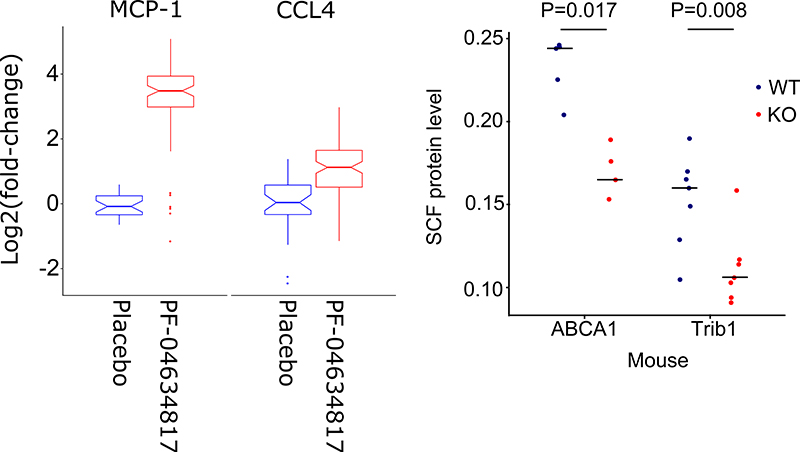
Of the 326 trans-pQTLs identified, eight were assigned to gene products targeted by compounds or antibodies that have been in clinical development [Supplementary Table 7]. Assuming that trans- pQTLs represent causal relationships between gene variants and proteins, we hypothesized that the downstream CVD-I proteins associated with CVD-I trans-pQTL genes would be modulated on therapeutic modification of the gene product. Support for this hypothesis was obtained by previous work showing that circulating FABP4 is upregulated upon treatment with glitazones (PPARG inhibitors)17; that circulating IL-6 is increased after treatment with tociluzumab18 (IL6R inhibitor) and that circulating TNF-R2 is decreased upon infliximab (TNFA inhibitor) treatment in patients with Crohn’s disease19, which supports CVD-I trans-pQTLs for these proteins. Along these lines, we present novel evidence from a clinical trial supporting our observations that a CCR5 variant is a trans-pQTL for plasma CCL-4 and a variant in CCR2 is a trans-pQTL for plasma MCP-1 [Supplementary table 2]. CCR5 and CCR2 are targeted in combination by the small-molecule dual-inhibitor PF-04634817 20. To test whether dual inhibition of CCR5 and CCR2 resulted in a change of circulating CCL-4 and MCP-1 respectively, we measured these proteins in 350 type 2 diabetes patients in a randomized, double-blind, placebo-controlled phase-II trial evaluating the efficacy of PF-04634817 in diabetic nephropathy (NCT01712061). In addition, we also measured known or suspected ligands of CCR5 and CCR2, including CCL-3, CCL-5 (RANTES) and CCL-8, and 5 additional proteins that were present on the Olink CVD-I panel, and for which assays were readily available. Compared to placebo, we observed a 9.25-fold increase in circulating MCP-1 levels (p < 0.0001) and a 2.11-fold increase in circulating CCL4 levels (p < 0.0001) at week 12 [Figure 3A]. An alternative ligand for CCR-2; CCL-8 did not change following exposure to PF-04634817, and neither did other CCR-5 ligands, such as CCL-5 (RANTES) and CCL-3. Moreover, EN-RAGE, FGF-23, KIM-1, myoglobin and TNFR-2 were unchanged following PF-04634817 exposure [Extended Figure 4]. We conclude that CVD-I trans-pQTLs at CCR5 and CCR2 were concordant with the effects of PF-04634817 in human.
Figure 3. Clinical trial in humans and knock down experiment in mice corresponds to trans pQTL effects.
A) In humans treated with a small molecule dual-inhibitor of CCR5 and CCR2 (PF-04634817) the induction of MCP-1 and CCL4, mirrors the observed CVD-I trans-pQTLs. Box plots elements are according to standards for box-and-whisker diagrams. B) In mice, knockdown of ABCA1 or TRIB1 resulted in decreased circulating SCF levels mirroring CVD-I trans-pQTLs for SCF. Shown in the plot are SCF levels of individual mice represented by circles (wild-type in blue and transgenic mice in red) and the median level per group. P-value is calculated using a two-sided T-test.
Two of the genes implicated by CVD-I trans-pQTLs, ABCA1 and TRIB1 for circulating SCF levels, were also investigated in the mouse. Mice with liver-specific or whole-body knockdown of ABCA1 21 and TRIB1 22 respectively showed decreased plasma levels of SCF compared to matched wild-type controls [Figure 3B], concordant with the human CVD-I trans-pQTLs.
Mendelian randomization analysis revealed 25 CVD-1 proteins causal for complex traits with strong evidence
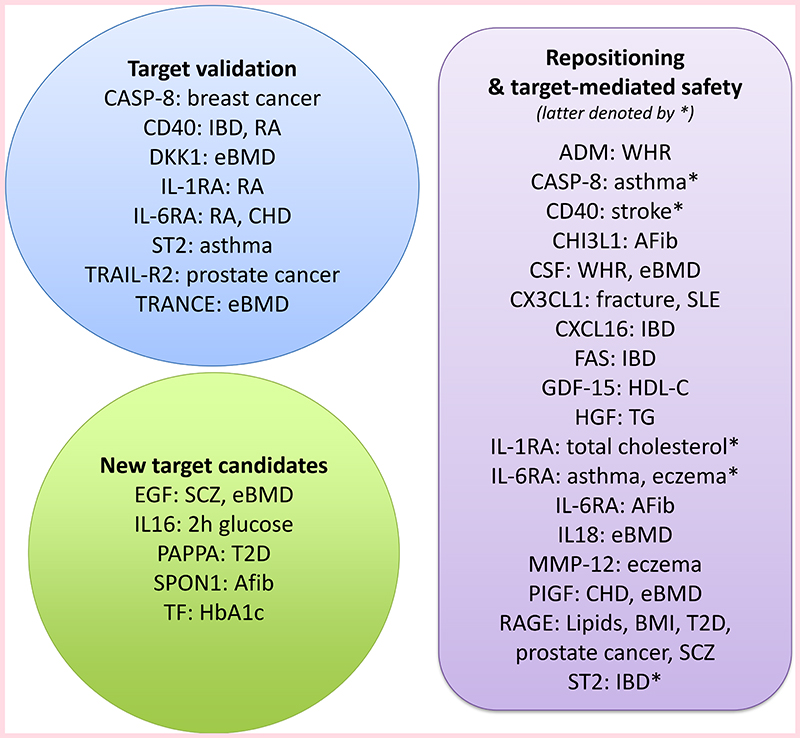
To identify potential causal disease pathways indexed by proteins, we conducted an MR analysis of 85 proteins across 38 outcomes. 25 proteins showed strong evidence of causality for at least one disease or phenotype and an additional 24 proteins showed intermediate evidence of causality. [Figure 4A] [Extended Figure 7] [Supplementary Figure 1]. Using open-source information (clinicaltrials.gov) (www.ebi.ac.uk/chembl/) (www.drugbank.ca/) (www.opentargets.org) and Clarivate Integrity (integrity.clarivate.com), we identified records on past or present clinical drug development programs for 14 of the 25 proteins, all of which have been in phase 2 trials or later [Supplementary Table 7]. Of the 14 proteins, seven proteins were targeted for an indication different from the phenotype implicated by our MR analysis. Eleven of the 25 proteins have never been targeted in clinical trials, but may provide new promising target candidates for indications closely related to the traits in the MR analysis.
Figure 4. Main findings of Mendelian randomization analysis.
A. Heatmap of Mendelian randomization analyses of 38 complex traits. ICD-10 chapter of indication and clinical trial stage indicated for each target B. Forest plot showing CVD-I proteins with strong evidence of causality in the Mendelian randomization analysis. Drug development abbreviations: PC: pre-clinical, Ph1: Phase 1, Ph2: Phase 2, Ph3: Phase 3, post-MA: post-marketing authorisation. ICD-10 chapters of disease: A-B: infectious and parasitic; C-D: neoplasms; D: blood and immune; E: endocrine, nutritional and metabolic; F: mental and behavioural; G: nervous system; H: eye, adnexa, ear and mastoid; I: circulatory system; J: respiratory system; K: digestive system; L: skin and subcutaneous tissue; M: musculoskeletal and connective tissue; N: genitourinary; O: pregnancy, childbirth, puerperium; P: perinatal; Q: congenital, deformations and chromosomal; R: clinical and lab findings; S-T: injury, poisoning; U: provisional assignment (new diseases unknown aetiology); V-Y: external causes; Z: health status & health services
Several published MR findings were confirmed, including that IL6RA variants associated with higher circulating levels of interleukin-6 (IL-6) and soluble IL6-RA were associated with lower risk of coronary heart disease (CHD), rheumatoid arthritis (RA) and atrial fibrillation but higher risks of atopy, such as asthma and eczema23. We also replicated previous findings suggesting a causal contribution of IL-1ra to rheumatoid arthritis (RA) but an inverse causal relationship with cholesterol levels 24, and a protective role of genetically higher MMP-12 against stroke 4,25.
Some novel MR observations included higher levels of CD40 protein and increased risk of RA, higher MMP-12 and increased risk of eczema, and higher TRAIL-R2 proteins levels and prostate cancer. Further, Dkk-1 has been targeted by a humanised monoclonal antibody (DKN-01) in clinical trials for advanced cancer (NCT01457417, NCT02375880), and was in our study causally linked to higher risk of bone fractures and lower risk of estimated bone mineral density (eBMD). In addition, strong evidence for protective roles of PLGF in CHD, CASP-8 in breast cancer and ST2 in asthma was observed. RAGE was causally linked to several traits, including lower body mass index (BMI) and a corresponding lower risk of type 2 diabetes (T2D), higher total cholesterol and triglycerides and higher risk of prostate cancer and schizophrenia. A small molecule brain penetrant RAGE inhibitor was tested in a phase 2 trial of Alzheimer’s disease (NCT00566397), but was stopped early for futility. We saw no strong signal for Alzheimer’s disease (or vascular disease) in our MR analysis. Our findings identify potential target-mediated effects across multiple other complex phenotypes that might manifest in beneficial and/or harmful effects on patients receiving RAGE-modifying therapies.
We also collated observational evidence for 23 of the 50 protein-trait pairs identified as causal in the MR analysis [supplementary table 10]. The direction of effect inferred from observational studies was concordant with the effect direction from MR estimates for 12 pairs.
Heritability analysis and polygenic risk scores (PRS) demonstrates large differences in genetic architecture
We calculated SNP-heritability contributed by the major reported loci (major loci hSNP 2, any pQTL included in [supplementary table 2]), as well as additional genome-wide SNP-heritability (polygenic hSNP 2) for each protein included in the entire SCALLOP CVD-I meta-analysis. We observed a large range of different genetic architectures: Differences in magnitude of the genetic component (hSNP 2) ranged from 0.01 (EGF) to 0.46 (IL-6RA). Differences in the contribution from non-genome-wide significant SNPs ranged from essentially monogenic (e.g. IL-6RA) to others showing considerable locus heterogeneity with genetic contributions originating entirely from a polygenic background with no single dominating locus (e.g. PDGF-B and Galanin) [Figure 5].
Figure 5. SNP heritability and variance explained by genetics.
A. SNP-Heritability in the SCALLOP consortium discovery cohorts stratified by contributions major loci (light red) and polygenic effects (dark red). In the independent MDC cohort, additional variability explained by adding major loci (light blue) and polygenic risk scores (dark blue). Significance was reported according to the LDSC algorithm (blue) or a linear regression model (red). B. Differences in how protein levels are affected by polygenic (non-genome-wide significant) loci vs major loci, shown for both the SCALLOP consortium discovery cohorts as hSNP2 and for the MDC cohort as variability explained.
In addition, we calculated the out of sample variance explained in the independent Malmo Diet and Cancer (MDC) study (N=4,678) both for genome-wide significant loci (major loci V.E.PRS), as well as additional variance explained by adding PRS (polygenic V.E.PRS) [Figure 5]. The protein PRS’ applied in the MDC study for 11 proteins exceeded 10 % of variance explained (V.E.PRS) and the PRS’ for another 14 proteins exceeded 5 % of variance explained, suggesting that the genetic contribution to inter-individual variability of CVD-I protein levels is considerable.
A polygenic risk score for circulating ST2 levels shows a dose-response relationship with asthma
Since circulating ST2 showed strong evidence of causation in asthma and inflammatory bowel disease (IBD) and the polygenic V.E.PRS model for ST2 explained nearly 20 % of its variance, we attempted to quantify the effect of the ST2 polygenic V.E.PRS on circulating ST2 levels in the MDC study, and risk of asthma and IBD in 337,484 unrelated White British subjects in the UK Biobank. The range of circulating ST2 across 11 categories of the ST2 PRS in MDC was nearly 1.2 standard deviations [Figure 6A]. Corroborating the Mendelian randomization analysis, the ST2 PRS showed a strong negative dose-response relationship with risk of asthma (p=1.2x10-8) and a positive trend for risk of IBD (p=0.13) [Figure 6B and 6C]. Overlaying the linear trends for ST2 levels, asthma and IBD using meta-regression, an increase in the PRS equivalent to a 1 standard deviation higher circulating ST2, corresponded to a 8.6 % (95%CI 3.8%, 13.2%; P=0.004) reduction in the relative risk of asthma and a 4.3 % (95%CI −3.8%, 13.0%; P=0.263) increase in the relative risk of IBD [extended Figure 8].
Figure 6. Mendelian randomization using polygenic risk scores.
A. Association of a polygenic risk score (PRS) with ST2 levels in the independent MDC cohort. B. Association of the ST2 PRS with asthma in the UK-biobank. B. Association of the ST2 PRS with inflammatory bowel disease (IBD) in the UK-biobank. The ST2 PRS was divided into 11 quantiles, with the middle group (quantile number 6) as the reference category. Effect estimates are presented as quantile-specific mean differences (ST2) and odds ratios (asthma and IBD) relative to the reference category.
Reverse Mendelian randomization identifies widespread causal relationships, where complex phenotypes affects CVD-I proteins
To investigate whether genetic susceptibility (liability) to complex disease and phenotypes causally alter circulating levels of CVD-I proteins, we also performed MR using 38 complex phenotypes (including continuous risk factors, such as adiposity and clinical outcomes, such as T2D) as exposure and CVD-I protein levels as outcomes. All CVD-I proteins were causally altered by at least one complex phenotype. BMI and estimated glomerular filtration rate (eGFR) causally affected 32 and 29 of the 85 tested proteins respectively [Figure 7A] [Extended Figure 7] [Supplementary Figure 2]. BMI seemed to causally affect protein levels in both positive and negative directions, whereas only REN (renin) was causally decreased with genetically higher eGFR. In an effort to elucidate whether these estimates were recapitulated in simple observational analyses, we compared effect estimates from linear regression analyses of associations of BMI and eGFR with each respective CVD-I protein in one of the participating study cohorts (IMPROVE). The correlation between the observational and MR estimates were high for BMI (R=0.78), and more modest for eGFR (R=0.50) [Figure 7B–C].
Figure 7. Mendelian randomization with proteins as outcome.
A. Heatmap showing the causal estimates of 38 complex traits on CVD-I protein levels. B. Correlation between beta-values for association between body mass index and circulating levels of CVD-I proteins in the IMPROVE cohort, and causal estimates from the Mendelian randomization analysis of body mass index genetic liability on same CVD-I proteins. C. Same as B but for estimated glomerular filtration rate.
Discussion
Using a meta-analysis approach including >30,000 individuals, we identified and replicated 315 primary and 136 secondary pQTLs for 85 circulating proteins to yield new insights for translational studies and drug development. Our study demonstrates that pQTLs can be harnessed to enhance evaluation of therapeutic hypotheses for protein targets, and to support those hypotheses with basic insights into potential protein regulatory pathways and biomarker strategies. However, we also observed large differences between proteins in relation to genetic architecture, suggesting that the relative strength to apply these strategies is likely protein-dependent.
Our pQTL-based framework was developed to address several key challenges associated with drug development, including a) mapping of protein regulatory pathways, b) identification of new target candidates c) repositioning of drugs, d) target-associated safety and e) matching of target mechanisms to patients by protein biomarkers or genetic PRS’ [Figure 8].
Figure 8. Protein-trait relationships that support target validation, repositioning, target-mediated safety and new candidates for drug development.
For more information, see data presented in [Supplementary Table 7].
The mapping of trans-pQTLs, which typically have smaller effects on protein levels [Extended Figure 9], was aided by the large SCALLOP discovery sample size, yielding on average 4 independent pQTLs per protein. A causal gene was assigned for each trans-pQTL to generate hypotheses that can be further tested using in vitro or in vivo perturbation experiments. The robustness of causal gene assignments for a few selected trans-pQTLs was demonstrated using samples from a randomised controlled trial testing a dual small-molecular inhibitor of the protein products of assigned genes (CCR5, CCR2) and transgenic mice with liver-specific knockdown of assigned genes (ABCA1, TRIB1). Although further studies will be needed for orthogonal validation of most of the genes assigned from the CVD-I trans-pQTLs, several of the implicated genes have previously been identified as regulators of some of the CVD-I proteins including CASP1 26, NLRC4 26 and GSDMD 27 for IL-18, FLT1 28 for PLGF, ADAM17 29 for TNFR1 and SLC34A1 30 for FGF-23 [Supplementary Table 2].
Further, we attempted to estimate the proportion of pQTLs that were likely to be driven by effects on mRNA expression, using multiple eQTL approaches and datasets. The lowest estimate was obtained with SMR/HEIDI, suggesting that 18.4 % of pQTLs were also eQTLs whereas direct look-up and co-localisation analysis using PrediXcan yielded estimates between 26 % - 29 %. We conclude that the majority of pQTLs identified for the CVD-I proteins were not explained by eQTLs.
Clinical-stage targeting with any drug modality was reported for 35 of the 90 proteins on the Olink CVD-I panel [Supplementary Table 7]. Our MR analysis identified 11 proteins with causal evidence of involvement in human disease that have not previously been targeted. Among those, four proteins were causal for a disease phenotype and did not show strong evidence of inverse causality with another phenotype (increasing specificity for intended indication), including CHI3L1 and SPON1 for atrial fibrillation and PAPPA for type-2 diabetes. Strong causal evidence was also identified for proteins targeted in phase-2 or later development. The MR evidence was concordant with drug indications for several protein targets but for some also suggested alternative indications or that monitoring of target-associated safety might be warranted. Monoclonal antibodies that block the CD40 ligand binding to CD40 – a critical element in T cell activation – have been shown to have positive clinical effects in patients with autoimmune diseases; but increased risk of thromboembolism precluded further clinical development31. These observations from clinical trials are in line with our findings that genetically lower levels of CD40 are associated with lower risk of RA, but higher risk of stroke. There are ongoing efforts to modify CD40L antibodies to retain efficacy while avoiding thromboembolism 31. However, our results suggest that decreasing circulating CD40 levels may have target-mediated beneficial effects on RA risk, while increasing the risk of ischemic stroke, i.e. that the increased risk of thromboembolism (manifest as stroke) is an on-target adverse effect. TRAIL-R2 is a key receptor for TRAIL, which has been shown to selectively drive tumour cells into apoptosis. Therefore, considerable effort to agonise TRAIL-R2 for treating cancers has been made in the past years32. We demonstrated that increased circulating TRAIL-R2 is protective against prostate cancer, which may suggest that this cancer type should be investigated in clinical trials evaluating the efficacy of TRAIL-R2 agonists.
Biomarkers can be broadly classified as generic biomarkers for disease risk or prognosis, or as biomarkers reflecting the activity of specific disease processes or biology. Biomarkers that enable matching of target mechanisms to patient subgroups with greater than average benefit from treatment are enablers of precision medicine. We showed that CCR2/CCR5 small-molecule inhibition modulated circulating levels of CCL-4 and MCP-1, which may suggest that trans-pQTLs can guide selection of exploratory biomarkers to monitor the efficacy of target mechanisms. We also identified multiple complex traits causally affecting circulating protein levels. For example, eGFR and BMI causally influenced over 1/3 of the CVD-I proteins, suggesting that future biomarker studies should consider these traits as potential confounders. Moreover, the causal phenotype-to-protein associations may represent pathway-related causality to the complex phenotype of interest; or alternatively, ‘reverse causality’ which might pose an opportunity to evaluate implicated proteins as surrogate biomarkers for efficacy in interventional trials 33. We found that higher BMI causally lowered RAGE, while higher circulating levels of RAGE were causally linked to a lower risk of T2D. Thus, developing a hypothetical therapeutic to increase RAGE might represent a mechanism by which it is possible to off-set the risk of T2D arising from the global increases in obesity.
Protein-centric PRS’ may allow stratification of individuals with genetic propensity for high circulating protein levels. Only 10 % of the protein-centric PRS’ explained 10 % or more of the protein variance in the independent replication cohort, including ST2, a prognostic biomarker for heart failure34. ST2 showed evidence of inverse causality in asthma and positive causality in IBD. By constructing a genome-wide polygenic risk score for ST2 levels from the MDC study, applying it to the UK Biobank and comparing asthma and IBD prevalence across eleven quantiles of the ST2 PRS, estimated the magnitude of ST2 increase required to decrease the risk of asthma to similar levels as individuals in the highest ST2 PRS category. Such use of PRS for proteins may be expanded to other disease endpoints and may be of use in precision medicine, to guide which patients may obtain most benefit from drugs that pharmacologically alter individual proteins.
In conclusion, our findings provide a comprehensive toolbox for evaluation and exploitation of therapeutic hypothesis and precision medicine approaches in complex disease. Such approaches provide an excellent opportunity to rejuvenate the drug development pipeline for new treatments.
Online Methods
Selection of proteins
Proteins for the Olink PEA CVD-I panel were selected by mining the literature for protein biomarkers associated with cardiovascular risk or prognosis in human observational studies and in animal models and by bringing in protein biomarker suggestions from leading cardiovascular disease researchers 10. The list of proteins curated from these sources was then pruned down based on availability of high-quality antibodies and relative abundance of the proteins in human plasma.
Intra- and inter-plate coefficients of variation (CV) of the CVD-I panel are available from Olink Proteomics AB (https://www.olink.com/resources-support/document-download-center/). In addition, we calculated the inter-plate coefficient of variation using data from a pooled plasma sample in one of the participating cohorts -the IMPROVE study. The mean inter-plate CV was averaged across proteins was 16.6 %, (range 11 % -26 %) [Supplementary Table 1].
Cohorts and data collection
Summary statistics from GWAS of Olink CVD-I proteins were obtained from 13 cohorts of European ancestry. The details of all study cohorts are shown in [Supplementary Table 9]. Together the cohorts included a total of 21,758 individuals; although the average per-protein sample size was 17,747, since not all proteins passed quality control (QC) in all cohorts. Each cohort provided data imputed to 1000 Genomes Project phase 3 reference or later or to the Haplotype Reference Consortium (HRC) reference, which resulted in the testing of 21.4M SNPs. Because imputation schemes varied by cohort, this resulted in an average of 20.3M SNPs under investigation for each protein.
Each cohort applied quality control measures for call rate filters, sex mismatch, population outliers, heterozygosity and cryptic relatedness as documented in [Supplementary Table 8]. Prior to running the genetic analyses, NPX values of proteins (on the log2 scale) were rank-based inverse normal transformed and/or standardised to unit variance, thus avoiding potential Olink batch-differences between cohorts. Genetic analyses were conducted using additive model regressions, with adjustment for population structure and study-specific parameters [Supplementary Table 8]. Forest plots of cohort-specific effects are available for all significant and suggestive pQTLs using the online tool. Each contributing cohort uploaded the resulting summary statistics in a standardized format using a secure computational cluster provided by Neic Tryggve (https://neic.no/tryggve/). All meta-analysis was performed in duplicate at two different research centres using completely separate bioinformatic pipelines (L.F. and S.G.).
Data cleaning and meta-analysis
A per-protein filtering threshold of >80% samples above the Olink detection limit was applied to each cohort, leaving data on 90 of the 92 proteins to be analysed. The remaining files had an average of 3% missing samples (per cohort statistics available in [Supplementary Table 8]). Minor allele frequencies were compared with those reported in 1000 Genomes EUR. A per-SNP filter was applied based on imputation quality level (at default setting for respective imputation algorithm) and minor allele count (at least 10 alleles per cohort). This resulted in the omission of 10% of the SNPs. Finally, meta-analysis was performed using METAL (2011-03-25) 35, applying the inverse-variance weighted approach (i.e. the STDERR option). Throughout the manuscript, P-values from this test are reported as-is, with multiple testing burden handled through appropriate thresholds. Cis-pQTLs were defined as a signal within 1 Mb of the gene encoding the protein and all other signals were defined as trans-pQTLs. See [extended figure 5] for flow chart overview of meta analysis.
Replication analyses
We sought to replicate the findings in the Malmö Diet and Cancer (MDC) population-based cohort with 4,678 individuals, and in the Swedish Mammography Cohort Clinical (SMCC, part of the Swedish national research infrastructure SIMPLER described at www.simpler4health.se) population-based study of 4,495 women. In MDC, genotypes were imputed to the Haplotype Reference Consortium reference (HRC Unlimited v1.0.1) and data were analysed using linear regression in EPACTS 3.3.0 (linear Wald test). The genotypes in SMCC were measured using Illumina’s Global Screening Array and were imputed up to HRC v1.1 and 1000G phase3 (v5), and linear regressions of rank-based inverse-normal transformed protein values adjusting for age, storage time, and PC1-15 were performed using PLINK v2 (4 Mar 2019).
Conditional and joint association analysis
To identify secondary signals at the 401 loci reported in [Supplementary table 2], we performed analyses conditioning on the primary signal using conditional-joint analysis in GCTA (version 1.26.0) 36,37. The Stanley cohort was chosen as an ancestrally well-matched LD-reference cohort. Meta-analysis summary data were processed with filtering for MAF (0.01) and r2 (<0.001) to ensure that secondary association signals identified were not driven by LD with the primary signal. See [Extended figure 6] for a flow chart of signal selection criteria.
Cross-reference of pQTLs with other complex traits
For each pQTL association, we searched PubMed and the EBI GWAS catalogue (URL: https://www.ebi.ac.uk/gwas/ : November 2018) for published SNPs with any complex trait within 10kb or having an LD of r2>= 0.85.
Comparison between eQTLs and pQTL
To identify eQTL that corresponded to each pQTL, we used three independent eQTL studies: LifeLines-DEEP 38, GTEx39 and eQTLGen40. Each SNP-protein pQTL pair was first converted to SNP-gene pairs using Olink platform protein identification and the gene annotation of Ensembl v91. Then, the significance of eQTLs for these SNP-gene pairs was assessed in three eQTL datasets, using two different cut-offs: a stringent genome-wide significance threshold (P<5x10-8) and a nominal significance of P<0.05.
In the eQTL dataset of LifeLines-DEEP, individual-level whole blood RNA-seq, protein and genotype data were available. This allowed for a direct comparison of the concordance of blood eQTLs and pQTLs. To do so, we re-tested eQTL associations for all pQTL pairs, using a previously published pipeline 41. The resulting eQTLs were considered genome-wide significant if it passed the permutation-based FDR <0.05 level, or to be nominally significant if the P-value was < 0.05.
In the eQTL datasets of GTEx v7 and eQTL-Gen, we did not have access to individual level data. Thus, the comparisons were conducted using publicly available eQTL results. In these datasets, we considered an eQTL genome-wide significant if it was within the reported genome-wide significant list, and nominally significant if it had a nominal P-value < 0.05. Altogether, if one pQTL pair had at least one significant eQTL effect in any dataset irrespective of allelic direction it was considered an overlapping pQTL-eQTL pair.
Expression SMR analysis
We performed an SMR and HEIDI (heterogeneity in dependent instruments) analysis12 to identify the expression levels of genes that were associated with protein abundance through pleiotropy using pQTL summary statistics from this study and cis-eQTL summary data from published studies42,43.
The eQTL summary data used in the SMR analysis were from the Consortium for the Architecture of Gene Expression (CAGE), comprising 38,624 normalized gene expression probes and ~8 million SNPs from 2,765 blood samples. The eQTL effects were in standard deviation (SD) units of expression levels. We excluded the gene probes in the major histocompatibility complex (MHC) region and included only the gene probes with at least one cis-eQTL at P<5×10-8 (a basic assumption of SMR), resulting in 9,538 gene expression probes.
The SMR test uses a SNP instrument (i.e., the top associated eQTL) to detect association between two phenotypes (i.e., gene and protein in this case). The HEIDI test utilises LD between the SNP instrument and other SNPs in the cis-region to distinguish whether the association identified by the SMR test is driven by a set of shared genetic variants between two traits (pleiotropic or causal model) or distinct sets of variants in LD (linkage model)12. Only the associations that surpassed the genome-wide significance level of the SMR test (P SMR < 0.05 / m with m being the number of SMR tests) and were not rejected by the HEIDI test (P HEIDI > 0.01) were reported as significant.
PrediXcan and transcript-wide association of CVD-I protein levels
Imputation of gene expression was performed in the IMPROVE study. After standard quality control, genotypes were pre-phased using Eagle2, and then subsequently imputed by minimac4 using the 1000 Genomes reference. A filter on RSQ 0.8 and minor allele frequency 0.01 was set on the imputed genotypes prior to prediction with PrediXcan, which used 44 tissue models based on GTEx v7.
Using protein data collected on the CVD-I chip in the same individuals, the associations between protein levels in plasma and the predicted expression of their respective coding gene across 20 tissues (from the PrediXcan model) were modelled by a linear model in R. False discovery rate were estimated based on Q-values (using the R package qvalue). In total, 64 genes in one to 18 tissues were tested for associations between protein levels and predicted expression. Heatmaps were constructed (using the pheatmap package in R) for any gene with a significant association (FDR<0.05) in at least one tissue.
Systems Biology
Two sets of network analysis were performed, one using the protein-protein interaction (PPI) data from the inBio Map™ (InWeb_InBioMap) and one using significant associations from text-mining (TM). These two networks each had 13,033 and 14,635 nodes, respectively; and 147,882 and 193,777 edges, respectively. In both setups, the shortest path between any of the cis-gene intermediaries to the protein was identified; altogether 12,436 pairs were compared. Of the 372 trans-pQTL associations reported in [Supplementary Table 2], 335 associations had both cis-gene intermediaries and plasma protein in the network allowing their analysis. The likelihood of a path arising by chance was calculated by permutation sampling, using 1,000,000 random networks were generated with a conserved degree distribution. A new algorithm was developed for de novo random network generation, which generated random networks with a nearly conserved degree distribution in a feasible time-frame. Further details are available in [Supplementary Information 1].
Assignment of cis-intermediary genes
To assign the most plausible causal gene for each of the CVD-I trans-pQTLs we applied a hierarchical approach based on analysis of InWeb_InBioMap PPI, TM, and genomic distance between gene and lead variant at each locus. Results were then manually reviewed by literature, gene expression analysis (proteinatlas.org) and published pQTLs which led to the re-assignment of 52 genes. The algorithmic gene assignment was overruled or complemented for instances when the assigned gene was different from the gene assigned by multiple prior studies [Supplementary table 4]. Gene Ontology analysis of most plausible genes was performed using the DAVID bioinformatics tools and the GO MF gene set definition, with default settings. The Panther pathway tool, Uniprot and the Human Protein Atlas were used to classify the genes according to basic functional class (see URLs).
Human in-vivo validation of trans-pQTLs
PF-04634817 is a competitive dual inhibitor of CCR2 and CCR5 receptors. In the recent B1261007 study, (ClinicalTrials.gov Identifier: NCT01712061), samples were collected from subjects with diabetic nephropathy and treated with PF-04634817 for 12 weeks. CCL-2 (MCP-1) was measured in serum by ELISA at Eurofins (The Netherlands). CCL4 (MIP-1b) and CCL-8 were measured in plasma using Luminex assays (Bio-Rad, Berkeley, CA). CCL5 (RANTES), was measured in plasma as part of a multi-analyte panel at Myriad Rules Based Medicine (Austin, TX).
Mouse in-vivo validation of trans-pQTLs
Plasma from transgenic- and matched control mice were randomised on a PCR plate. The samples included five mice with targeted deletion of hepatocyte ABCA121 together with five matched control mice, three mice with whole-body TRIB122 knockdown and three controls and four mice with liver-specific knockdown of TRIB1 and four matched controls. Protein levels of stem cell factor (SCF) was measured using the Olink PEA Mouse exploratory panel according to the manufacturer’s instruction (Olink Proteomics, Uppsala, Sweden). The plasma levels of SCF were normalised against average protein concentrations using information on an additional 91 proteins. TRIB1 whole-body and liver-specific mice were analysed jointly as were the respective wild-type controls. The median plasma levels of SCF were compared using the Mann-Whitney U test for unpaired samples.
Mendelian randomization
To study the causal effects of the protein on selected disease outcomes, we performed two-sample Mendelian randomization analyses. We used between-study heterogeneity to guide the instrumental variable selection. In the presence of between-study heterogeneity (P-het<9x10-5), variants had to surpass a Bonferroni-corrected p-value threshold in discovery (P<5.6x10-10) and show nominal significance (P<0.05) in the replication studies (9,173 individuals), with directionally concordant beta coefficients. In the absence of between-study heterogeneity we included variants showing conventional genome-wide significance (P<5x10-8) in a meta-analysis of the discovery and replication datasets. From these, we created two sets of instrumental variables (IVs) for each of the 85 proteins with variants reaching multiple testing-corrected significance in our discovery GWAS: (a) cis IVs including one or more independent variants (LD r2=0.001 within ±1Mb of the transcript boundaries of the gene encoding the protein); and (b) pan IVs including all independent (LD r2=0) variants associated with the protein, i.e. combining cis and trans pQTLs. The per-allelic beta coefficients from the main GWAS analyses were used as weights in the IVs. For the outcomes, we obtained the relevant SNP-to-trait summary statistics from publicly-available GWAS as outcomes [Supplementary Table 9]. When lead variants from our main GWAS were not available in these summary statistics, we replaced them with proxies (LD r2>0.85). For each individual SNP-protein and SNP-outcome association, we generated an instrumental variable Wald ratio estimate, with standard errors obtained using the delta method. When the instrument included more than one SNP, summary IV estimates were generated by combining individual SNP Wald estimates by inverse-variance weighted fixed-effect meta-analysis. We report associations with a Benjamini-Hochberg false discovery rate (FDR) ≤ 5%, applied separately to summary estimates from cis-pQTL and pan-pQTL IVs, using pooled estimates for all 38 diseases. We graded the evidence of causality using a framework outlined in [Extended Figure 7], using the following categories: strong (cis-IV estimate FDR≤ 5%); intermediate (pan-IV estimate FDR≤ 5% with: (i) no heterogeneity between cis-IV estimate and pan-IV estimate; and (ii) no evidence of the MR estimate being unduly influenced by a trans-pQTL in leave-one-out analysis); or weak (pan-IV estimate FDR≤ 5% but: no cis-pQTL IV available; heterogeneity between cis- and all-IVs; or evidence of undue influence by a trans-pQTL). Heterogeneity between pan-IV and cis-IV estimates were calculated using Cochran’s Q tests, with P<0.05 denoting evidence against the null hypothesis, and applying a Bonferroni adjustment for multiple testing. Mendelian randomization was conducted in duplicate by two separate analysts and analyses were performed in Stata (StataCorp, Texas, USA) version 13.3 using the mrivests, metan and multproc commands and R. Of the 2437 IV estimates derived using cis-pQTL instruments across the 85 proteins and 38 outcome traits, the IV estimates of 50 protein-to-disease associations met the FDR≤5% (corresponding to an uncorrected P≤1.1x10-3). Of the 3044 IV estimates composed using all pQTL instruments, 281 IV estimates met FDR≤ 5% (corresponding to P≤ 4.7x10-3; [Figure 4A]).
Heritability analyses
We estimated the total SNP-heritability (hSNP 2) for the plasma level of each protein from the summary statistics of each individual GWAS by summing the contributions from two independent partitions of the SNPs: primary major loci and polygenic background. We defined the variance explained by primary major loci (major loci hSNP 2) as the sum of the estimated variance explained (2*β2*f*(1-f)), where f is the minor allele frequency, and owing to the fact that the phenotypic variance has been standardized across lead SNPs indexing all primary genome-wide significant loci. We used LDSC regression44 to estimate the contribution of the polygenic background (polygenic hSNP 2) for each protein, which we define as the contribution of all loci not indexed by a genome-wide significant lead SNP. LDSC regression is known to perform poorly when large effect, major genes are present, as it was derived under the assumption of a simple polygenic genetic architecture44. To account for this and avoid double counting the variance explained by major loci through LD surrogates, prior to estimating the LDSC regression polygenic hSNP 2, we censored all SNPs within 10 Mb of genome-wide significant lead SNPs for all primary loci.
Polygenic risk score calculation
Polygenic risk scores were derived using LDpred algorithm45, which adjusts the effect of each SNP allele for those of other SNP alleles in linkage disequilibrium (LD) with it, and also takes into account the likelihood of a given allele to have a true effect according to a user-defined parameter, which we used as all 7 default LDpred-settings, with values from 1 through 1x10-5. The algorithm was directed to use HapMap3 SNPs that had a minor allele frequency >0.05, Hardy-Weinberg equilibrium P>1e-05 and imputation score >0.95. Variance explained in the independent MDC-study was tested according to a step-wise model, first including non-genetic covariates, then additional variability explained by adding SNPs from genome-wide significant SNPs (major loci V.E.PRS), and then additional variability explained by adding the 7 LDpred-derived scores as additional covariates (polygenic V.E.PRS).
ST2 polygenic risk score for asthma and inflammatory bowel disease in the UK biobank
Prior to analysis subjects who were not White British (based on self-reported ancestry in combination with genetic PCA) in the maximum unrelated subset were filtered out. All bi-allelic SNPs with MAF >= 1% and MaCH rsq >= 0.8 were kept. The Z-score transformed LDpred PRS (wt2) for ST2 was calculated as described for MDC in 337,484 White British UK Biobank participants. Association with asthma and IBD were tested using logistic regression adjusting for age, sex, PC1-10, genotype batch using either the continuous PRS or the PRS quantile-bins as predictors. The UK Biobank protocol has been described previously46 and is available online (https://www.ukbiobank.ac.uk). The genotype quality control (QC), phasing, and imputation was performed centrally and has been previously described 47. Outcomes (defined based on self-reported data at baseline and/or the inpatient and death registry [including primary and secondary causes as well as prevalent and incident disease]) Asthma: Self-reported touchscreen (6152), self-reported nurse interview (20002), or ICD-10 “J45”. Conflicting self-reported results set to missing unless “J45” was reported. Inflammatory bowel disease: nurse interview (20002) or ICD-10 K50-K52.
Meta-regression analysis for ST2 PRS, asthma and IBD
We estimated the per-quantile and per-SD associations of the weighted PRS for ST2 (MDC study) on risks of asthma and IBD (UK Biobank) by taking the quantile associations with ST2, asthma and IBD and conducting meta-regression analyses whereby the dependent variable was the quantile-specific logOR and corresponding SE of asthma or IBD and the independent variable was the quantile specific beta coeffient for ST2. This was conducted using the “metareg” package in STATA SE v13.1 (Statacorp, USA). Plots from the metaregression are presented in [Extended Figure 8].
Observational evidence
Observational evidence for the CVD-I proteins showing strong evidence of causality in Mendelian randomization was collated from literature or by de-novo analysis in the IMPROVE cohort [supplementary table 10]. To identify evidence from literature, we searched for the protein name or aliases in combination with the implicated trait trait/disease in PubMed. For clinical outcome traits, only those reported as “significant” by the paper were included, and the table provides the directional information provided. For quantitative outcome traits, standardised betas and p-values are reported.
Supplementary Material
Acknowledgements
Secure computing was supported by NeIC Tryggve, which is the Nordic collaboration for sensitive data funded by NeIC and ELIXIR nodes of participating countries.
Sources of Funding for SMCC, part of the national research infrastructure SIMPLER. We acknowledge the national research infrastructure SIMPLER (the Swedish Infrastructure for Medical Population-based Life-course and Environmental Research) for provisioning of facilities and support. SIMPLER receives funding through the Swedish Research Council under the grant no 2017-00644. This study was also supported by additional grants from the Swedish Research Council (grants no 2017-06100; no 2015-05997 and no 2015-03257), from the Swedish Research Council for Health, Working Life and Welfare (FORTE grant no 2017-00721) and Stiftelsen Olle Engkvist Byggmästare (grant no 2017/49)
S Lubitz is supported by NIH grant 1R01HL139731 and American Heart Association 18SFRN34250007.
The Orkney Complex Disease Study (ORCADES) was supported by the Chief Scientist Office of the Scottish Government (CZB/4/276, CZB/4/710), a Royal Society URF to J.F.W., the MRC Human Genetics Unit quinquennial programme “QTL in Health and Disease”, Arthritis Research UK and the European Union framework program 6 EUROSPAN project (contract no. LSHG-CT-2006-018947). DNA extractions were performed at the Edinburgh Clinical Research Facility. We would like to acknowledge the invaluable contributions of the research nurses in Orkney, the administrative team in Edinburgh and the people of Orkney.
MAK is supported by a Senior Research Fellowship from the National Health and Medical Research Council (NHMRC) of Australia (APP1158958). He also has a research grant from the Sigrid Juselius Foundation, Finland
AB was supported by a Wellcome PhD training fellowship for clinicians (204979/Z/16/Z), the Edinburgh Clinical Academic Track (ECAT) programme
J. G. Smith and the genotyping of MPP-RES was supported by grants from the Swedish Heart-Lung Foundation (2016-0134 and 2016-0315), the Swedish Research Council (2017-02554), the European Research Council (ERC-STG-2015-679242), the Crafoord Foundation, Skåne University Hospital, the Scania county, governmental funding of clinical research within the Swedish National Health Service, a generous donation from the Knut and Alice Wallenberg foundation to the Wallenberg Center for Molecular Medicine in Lund, and funding from the Swedish Research Council (Linnaeus grant Dnr 349-2006-237, Strategic Research Area Exodiab Dnr 2009-1039) and Swedish Foundation for Strategic Research (Dnr IRC15-0067) to the Lund University Diabetes Center.
The study of the LifeLines-DEEP cohort is supported by the Netherlands Heart Foundation CVON grant 2018-27 to JF and AZ, Netherlands Organization for Scientific Research (NWO-Vidi grant 864.13.013 to JF, 016.178.056 to AZ, 917.14.374 to LF, Veni grant 194.006 to DZ, gravitation grant ExposomeNL to AZ, gravitation 024.003.001 to JF), European Research Council (ERC starting grant 715772 to AZ, 637640 to LF), LF also receives financial support from Oncode Institute.
We would like to thank Professor John Parks at Wake Forest School of Medicine, Winston-Salem, NC and Professor Daniel Rader at Perelman School of Medicine at the University of Pennsylvania, Philadelphia, PA for their kind donations of samples from transgenic mice and controls. This research has been conducted using the UK Biobank Resource under Application Number 13721.
Footnotes
Author Contributions
La.F, SG, QW, DHH, ÅKH, DVZ, EF, EMD, EI, AM contributed to meta-analysis. La.F, ÅKH, DVZ, YW, JRG, YC, AC, FM, EF, Lu.F, TQ, RW, HJW, JY, AM contributed to functional analysis. La.F, SG, QW, GDS, TP, TQ, JY, LW, ASB, MVH, EI, AM contributed to Mendelian randomization. JP, NE, SEB, TB, ADB, St.E, AK, MAK, SHC, JD, Sö.E, CF, Lu.F, PF, VG, Ch.H, AH, ÅJ, PKJ, LL, CML, SL, EMD, MM, APM, RM, MWN, OP, BP, EP, JS, PS, UV, HJW, AZ, JÄ, JF, GS, TE, Ca.H, UG, ML, Ag.S, JFW, LW, ASB, EI, AM contributed to cohort level analysis. BS, LM, AM contributed to mouse experiments. KP, JDG, JL, WZ, AQ, AM contributed to clinical trials. La.F, ÅKH, An.S, JRG, FM, EF, AI, TW, AM contributed to other downstream analysis. La.F, SG, ÅKH, GE, CF, OM, KM, PMN, JN, MOM, MS, AM contributed to replication analysis. La.F, SG, QW, MVH, EI, AM contributed to writing. La.F, SG, QW, ASB, MVH, EI, AM contributed to project planning. All authors gave final approval to publish.
Competing Interests Statement
The other authors declare no competing interests
Data availability
The full summary statistics of the Olink CVD-I protein GWAS have been deposited at the SCALLOPCVD-I online resource, allowing access to interactive SCALLOP-CVD-I tools and unrestricted download access for secondary analyses. Additionally, a full copy has been deposited at https://doi.org/10.5281/zenodo.2615265 for long-term retention, as well as with GWAS catalog. A copy of the polygenic scores have been deposited at the PGS catalog.
References
- 1.Chames P, Van Regenmortel M, Weiss E, Baty D. Therapeutic antibodies: successes, limitations and hopes for the future. Br J Pharmacol. 2009;157:220–233. doi: 10.1111/j.1476-5381.2009.00190.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Holmes MV, Ala-Korpela M, Smith GD. Mendelian randomization in cardiometabolic disease: challenges in evaluating causality. Nat Rev Cardiol. 2017;14:577–590. doi: 10.1038/nrcardio.2017.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Folkersen L, et al. Mapping of 79 loci for 83 plasma protein biomarkers in cardiovascular disease. PLoS Genet. 2017;13:e1006706. doi: 10.1371/journal.pgen.1006706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sun BB, et al. Genomic atlas of the human plasma proteome. Nature. 2018;558:73–79. doi: 10.1038/s41586-018-0175-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Williams SA, et al. Plasma protein patterns as comprehensive indicators of health. Nat Med. 2019;25:1851–1857. doi: 10.1038/s41591-019-0665-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lehallier B, et al. Undulating changes in human plasma proteome profiles across the lifespan. Nat Med. 2019;25:1843–1850. doi: 10.1038/s41591-019-0673-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Enroth S, Johansson A, Enroth SB, Gyllensten U. Strong effects of genetic and lifestyle factors on biomarker variation and use of personalized cutoffs. Nat Commun. 2014;5:4684. doi: 10.1038/ncomms5684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Emilsson V, et al. Co-regulatory networks of human serum proteins link genetics to disease. Science. 2018;361:769–773. doi: 10.1126/science.aaq1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Melzer D, et al. A genome-wide association study identifies protein quantitative trait loci (pQTLs) PLoS Genet. 2008;4:e1000072. doi: 10.1371/journal.pgen.1000072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Assarsson E, et al. Homogenous 96-plex PEA immunoassay exhibiting high sensitivity, specificity, and excellent scalability. PLoS One. 2014;9:e95192. doi: 10.1371/journal.pone.0095192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gamazon ER, et al. A gene-based association method for mapping traits using reference transcriptome data. Nat Genet. 2015;47:1091–1098. doi: 10.1038/ng.3367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhu Z, et al. Integration of summary data from GWAS and eQTL studies predicts complex trait gene targets. Nat Genet. 2016;48:481–487. doi: 10.1038/ng.3538. [DOI] [PubMed] [Google Scholar]
- 13.Sun W, et al. Common Genetic Polymorphisms Influence Blood Biomarker Measurements in COPD. PLoS Genet. 2016;12:e1006011. doi: 10.1371/journal.pgen.1006011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chick JM, et al. Defining the consequences of genetic variation on a proteome-wide scale. Nature. 2016;534:500–505. doi: 10.1038/nature18270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhernakova DV, et al. Individual variations in cardiovascular-disease-related protein levels are driven by genetics and gut microbiome. Nat Genet. 2018;50:1524–1532. doi: 10.1038/s41588-018-0224-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Solomon T, et al. Identification of Common and Rare Genetic Variation Associated With Plasma Protein Levels Using Whole-Exome Sequencing and Mass Spectrometry. Circ Genom Precis Med. 2018;11:e002170. doi: 10.1161/CIRCGEN.118.002170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cabre A, et al. Fatty acid binding protein 4 is increased in metabolic syndrome and with thiazolidinedione treatment in diabetic patients. Atherosclerosis. 2007;195:e150–158. doi: 10.1016/j.atherosclerosis.2007.04.045. [DOI] [PubMed] [Google Scholar]
- 18.Nishimoto N, et al. Mechanisms and pathologic significances in increase in serum interleukin-6 (IL-6) and soluble IL-6 receptor after administration of an anti-IL-6 receptor antibody, tocilizumab, in patients with rheumatoid arthritis and Castleman disease. Blood. 2008;112:3959–3964. doi: 10.1182/blood-2008-05-155846. [DOI] [PubMed] [Google Scholar]
- 19.Gustot T, et al. Profile of soluble cytokine receptors in Crohn’s disease. Gut. 2005;54:488–495. doi: 10.1136/gut.2004.043554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gale JD, et al. Effect of PF-04634817, an Oral CCR2/5 Chemokine Receptor Antagonist, on Albuminuria in Adults with Overt Diabetic Nephropathy. Kidney Int Rep. 2018;3:1316–1327. doi: 10.1016/j.ekir.2018.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bashore AC, et al. Targeted Deletion of Hepatocyte Abca1 Increases Plasma HDL (High-Density Lipoprotein) Reverse Cholesterol Transport via the LDL (Low-Density Lipoprotein) Receptor. Arterioscler Thromb Vasc Biol. 2019;39:1747–1761. doi: 10.1161/ATVBAHA.119.312382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Burkhardt R, et al. Trib1 is a lipid- and myocardial infarction-associated gene that regulates hepatic lipogenesis and VLDL production in mice. J Clin Invest. 2010;120:4410–4414. doi: 10.1172/JCI44213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rosa M, et al. A Mendelian randomization study of IL6 signaling in cardiovascular diseases, immune-related disorders and longevity. NPJ Genom Med. 2019;4:23. doi: 10.1038/s41525-019-0097-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Interleukin 1 Genetics, C. Cardiometabolic effects of genetic upregulation of the interleukin 1 receptor antagonist: a Mendelian randomisation analysis. Lancet Diabetes Endocrinol. 2015;3:243–253. doi: 10.1016/S2213-8587(15)00034-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mahdessian H, et al. Integrative studies implicate matrix metalloproteinase-12 as a culprit gene for large-artery atherosclerotic stroke. J Intern Med. 2017;282:429–444. doi: 10.1111/joim.12655. [DOI] [PubMed] [Google Scholar]
- 26.Kaplanski G. Interleukin-18: Biological properties and role in disease pathogenesis. Immunol Rev. 2018;281:138–153. doi: 10.1111/imr.12616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Heilig R, et al. The Gasdermin-D pore acts as a conduit for IL-1beta secretion in mice. Eur J Immunol. 2018;48:584–592. doi: 10.1002/eji.201747404. [DOI] [PubMed] [Google Scholar]
- 28.Autiero M, et al. Role of PlGF in the intra- and intermolecular cross talk between the VEGF receptors Flt1 and Flk1. Nat Med. 2003;9:936–943. doi: 10.1038/nm884. [DOI] [PubMed] [Google Scholar]
- 29.Dri P, et al. TNF-Induced shedding of TNF receptors in human polymorphonuclear leukocytes: role of the 55-kDa TNF receptor and involvement of a membrane-bound and non-matrix metalloproteinase. J Immunol. 2000;165:2165–2172. doi: 10.4049/jimmunol.165.4.2165. [DOI] [PubMed] [Google Scholar]
- 30.Tenenhouse HS, Sabbagh Y. Novel phosphate-regulating genes in the pathogenesis of renal phosphate wasting disorders. Pflugers Arch. 2002;444:317–326. doi: 10.1007/s00424-002-0839-4. [DOI] [PubMed] [Google Scholar]
- 31.Xie JH, et al. Engineering of a novel anti-CD40L domain antibody for treatment of autoimmune diseases. J Immunol. 2014;192:4083–4092. doi: 10.4049/jimmunol.1303239. [DOI] [PubMed] [Google Scholar]
- 32.de Miguel D, Lemke J, Anel A, Walczak H, Martinez-Lostao L. Onto better TRAILs for cancer treatment. Cell Death Differ. 2016;23:733–747. doi: 10.1038/cdd.2015.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Holmes MV, Davey Smith G. Can Mendelian Randomization Shift into Reverse Gear? Clin Chem. 2019;65:363–366. doi: 10.1373/clinchem.2018.296806. [DOI] [PubMed] [Google Scholar]
- 34.McCarthy CP, Januzzi JL., Jr Soluble ST2 in Heart Failure. Heart Fail Clin. 2018;14:41–48. doi: 10.1016/j.hfc.2017.08.005. [DOI] [PubMed] [Google Scholar]
- 35.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet. 2011;88:76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang J, et al. Conditional and joint multiple-SNP analysis of GWAS summary statistics identifies additional variants influencing complex traits. Nat Genet. 2012;44:369–375.:S361-363. doi: 10.1038/ng.2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tigchelaar EF, et al. Cohort profile: LifeLines DEEP, a prospective, general population cohort study in the northern Netherlands: study design and baseline characteristics. BMJ Open. 2015;5:e006772. doi: 10.1136/bmjopen-2014-006772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Consortium GT, et al. Genetic effects on gene expression across human tissues. Nature. 2017;550:204–213. doi: 10.1038/nature24277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Võsa Urmo. Unraveling the polygenic architecture of complex traits using blood eQTL metaanalysis. bioRxv. 2018 Oct 19; [Google Scholar]
- 41.Westra HJ, et al. Systematic identification of trans eQTLs as putative drivers of known disease associations. Nat Genet. 2013;45:1238–1243. doi: 10.1038/ng.2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lloyd-Jones LR, et al. The Genetic Architecture of Gene Expression in Peripheral Blood. Am J Hum Genet. 2017;100:371. doi: 10.1016/j.ajhg.2017.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McRae AF, et al. Identification of 55,000 Replicated DNA Methylation QTL. Scientific Reports. 2018;8 doi: 10.1038/s41598-018-35871-w. Article number: 17605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bulik-Sullivan BK, et al. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet. 2015;47:291–295. doi: 10.1038/ng.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vilhjalmsson BJ, et al. Modeling Linkage Disequilibrium Increases Accuracy of Polygenic Risk Scores. Am J Hum Genet. 2015;97:576–592. doi: 10.1016/j.ajhg.2015.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sudlow C, et al. UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 2015;12:e1001779. doi: 10.1371/journal.pmed.1001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bycroft C, et al. The UK Biobank resource with deep phenotyping and genomic data. Nature. 2018;562:203–209. doi: 10.1038/s41586-018-0579-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The full summary statistics of the Olink CVD-I protein GWAS have been deposited at the SCALLOPCVD-I online resource, allowing access to interactive SCALLOP-CVD-I tools and unrestricted download access for secondary analyses. Additionally, a full copy has been deposited at https://doi.org/10.5281/zenodo.2615265 for long-term retention, as well as with GWAS catalog. A copy of the polygenic scores have been deposited at the PGS catalog.