Abstract
Skeletal muscle myofibers are large and elongated cells with multiple and evenly distributed nuclei. Nuclear distribution suggests that each nucleus influences a specific compartment within the myofiber and implies a functional role for nuclear positioning. Compartmentalization of specific mRNAs and proteins has been reported at the neuromuscular and myotendinous junctions, but mRNA distribution in non-specialized regions of the myofibers remains largely unexplored. We report that the bulk of mRNAs are enriched around the nucleus of origin and that this perinuclear accumulation depends on recently transcribed mRNAs. Surprisingly, mRNAs encoding large proteins – giant mRNAs – are spread throughout the cell and do not exhibit perinuclear accumulation. Furthermore, by expressing exogenous transcripts with different sizes we found that size contributes to mRNA spreading independently of mRNA sequence. Both these mRNA distribution patterns depend on microtubules and are independent of nuclear dispersion, mRNA expression level and stability, and the characteristics of the encoded protein. Thus, we propose that mRNA distribution in non-specialized regions of skeletal muscle is size selective to ensure cellular compartmentalization and simultaneous long-range distribution of giant mRNAs.
Keywords: Compartmentalization, mRNA distribution, mRNA size, mRNA transport, Multinucleation, Skeletal muscle
Introduction
Skeletal muscle is formed by bundles of myofibers: large and elongated multinucleated cells, which result from the fusion of hundreds of mononucleated myocytes (Abmayr and Pavlath, 2012). During development, nuclei undergo a series of tightly regulated migrations. Nuclei align at the center of the myofiber after myocyte fusion, but eventually spread and move to the cell periphery where they are anchored (Zhang et al., 2007). These tightly regulated movements depend initially on microtubules and later on desmin crosslinking and cell contraction (Cadot et al., 2012; Falcone et al., 2014; Gimpel et al., 2017; Metzger et al., 2012; Roman et al., 2017). Interestingly, nuclear distribution is not random in fully developed myofibers. Nuclei are distributed evenly along the cell, maximizing inter-nuclear spacing and minimizing transport distances (Bruusgaard et al., 2003). Exceptions to this nuclear spreading occur in specialized compartments of myofibers, such as the neuromuscular (Bruusgaard et al., 2003; Englander and Rubin, 1987; Merlie and Sanes, 1985) and myotendinous junctions (Bruusgaard et al., 2003), where a few nuclei accumulate.
Specific proteins and mRNAs also accumulate at these specialized regions. The myotendinous junction has higher levels of slow myosin heavy chain mRNA (Dix and Eisenberg, 1990) and membrane-associated proteins (Can et al., 2014), whereas the neuromuscular junction accumulates acetylcholine receptor (Merlie and Sanes, 1985) and Na+ channel mRNAs (Awad et al., 2001). The compartmentalization of acetylcholine receptor at this site results from the restricted expression of its mRNA by nuclei at the neuromuscular junction (Duclert and Changeux, 1995; Sanes and Lichtman, 1999). The observation that gene products are maintained in the vicinity of their nucleus of origin led to the idea of nuclear domains in myofibers (Hall and Ralston, 1989; Pavlath et al., 1989). This theory proposes that multinucleated myofibers are partitioned into regions under the influence of a single nucleus and its genetic products.
Although the relevance of nuclear localization for muscle function is not fully understood, proper nuclear positioning is important for skeletal muscle function in Drosophila (Metzger et al., 2012). In addition, central localization of nuclei is a distinct morphological change observed in a set of muscular disorders, including centronuclear myopathies (Biancalana et al., 2012; Jungbluth et al., 2008). It is yet to be defined whether nuclear positioning is a causal factor in the pathophysiology of muscle disorders, but according to the nuclear domain theory, defective nuclear dispersion should hinder the efficient distribution of nuclear products such as mRNA along the myofiber.
mRNA localization and transport mechanisms have been observed in multiple cell types, from yeast to mammalian cells (Mofatteh and Bullock, 2017). In large and polarized cells, such as neurons and oocytes, several mRNA localization mechanisms have been described. In mouse neurons, the localization of β-actin (ACTB) in dendrites is crucial for a rapid neuronal response upon stimulation. To match β-actin mRNA demands at dendrites, mRNA is transported via microtubules (Ma et al., 2011) as part of RNA-binding protein complexes and is translated upon stimulation (Buxbaum et al., 2014; Park et al., 2014). In the Drosophila oocyte, localization of specific mRNAs such as gurken (MacDougall et al., 2003; Tomancak et al., 1998), bicoid (Duncan and Warrior, 2002; Januschke et al., 2002) and oskar (Brendza et al., 2000; Krauss et al., 2009) is crucial to determine cell fate by establishing oocyte polarity through mRNA transport and anchoring mechanisms dependent on microtubules and actin, respectively.
In myofibers, mRNA localization and transport mechanisms are largely unknown. In non-specialized regions of the myofiber, mRNAs encoding costameric and sarcomeric proteins are localized at the site of protein localization (Fulton and Alftine, 1997). Additionally, the mRNAs encoding calsequestrin (CASQ1 and CASQ2) and DHPR (also known as CACNA1S) are enriched in the perinuclear region and sarcolemma in a striated pattern consistent with the protein localization at the sarcoplasmic reticulum and triads, respectively (Nissinen et al., 2005). The perinuclear enrichment and striated pattern of distribution is also observed for total mRNA (Nevalainen et al., 2013). However, there is a lack of comprehensive studies regarding mRNA localization and their transport mechanisms in myofibers, most likely due to the technical challenges imposed by their unique structure and dimension.
Here, we studied the local distribution of several mRNAs in non-specialized regions of differentiated skeletal myofibers. We observed that most mRNA is enriched around the nucleus of origin. Surprisingly, mRNAs that encode for large proteins are spread throughout the myofiber. We showed that large size of transcripts favors mRNA spreading in myofibers and confirmed this size-dependent distribution by expressing transcripts of various lengths. This spreading is independent of protein function or localization and of tissue specificity. We found that transcription is required for the enrichment of mRNA around the nucleus and that microtubules are involved in the distribution of both subsets of mRNA away from the nucleus.
Results
mRNA is enriched in the perinuclear region of differentiated skeletal muscle myofibers
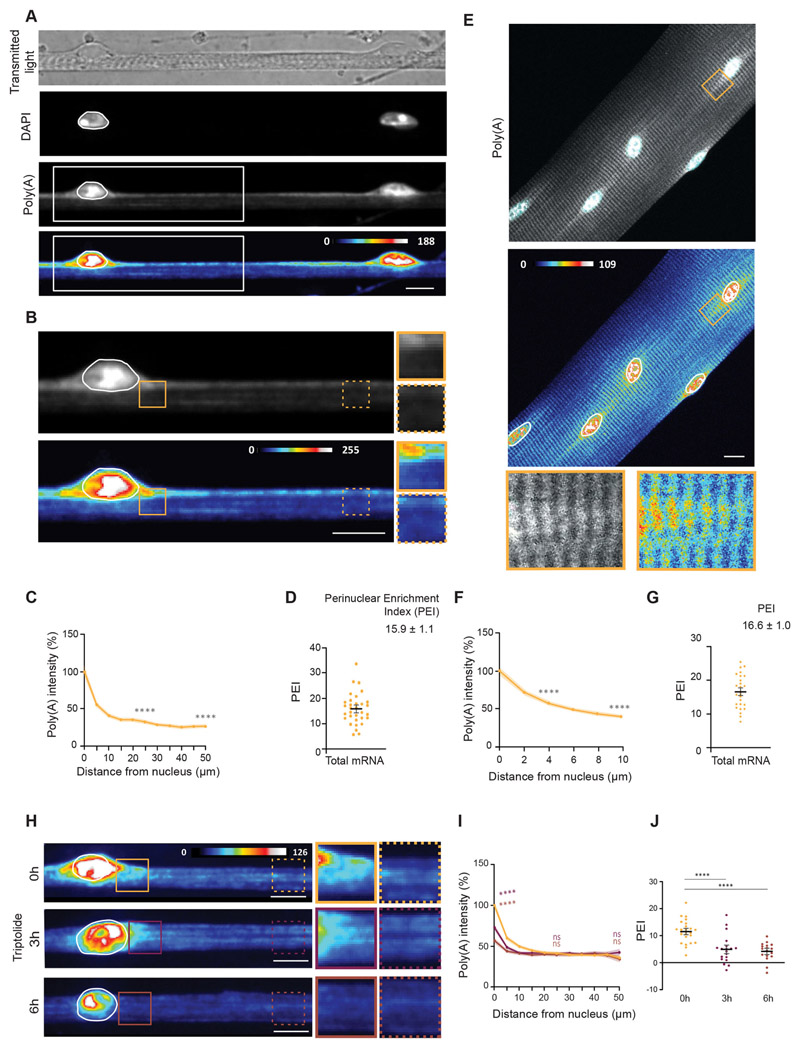
To determine the distribution of mRNA in non-specialized regions of skeletal myofibers, we used an in vitro system for primary myofiber differentiation previously established in our group (Falcone et al., 2014; Pimentel et al., 2017). Using this method, highly mature myofibers are formed in vitro in 6 days. These myofibers exhibit several hallmarks of skeletal muscle maturation, such as nuclear positioning at the periphery of the cell, transverse triad formation, sarcomeric striations and contractility. We performed fluorescence in situ hybridization (FISH) to detect total mRNA using a probe that recognizes mRNA poly(A) tails and observed an enrichment of mRNA in the perinuclear region of differentiated myofibers (Fig. 1A-C). RNase A treatment for 1 h showed that most of the poly(A) signal is specific (Fig. S1B-D). To quantify the perinuclear enrichment, we devised an mRNA perinuclear enrichment index (PEI) corresponding to the difference between the amount of mRNA close to the nucleus and the amount of mRNA away from the nucleus, expressed as a percentage of total mRNA (see Materials and Methods). The distribution was analyzed in a region of interest encompassing a myofiber segment of 50 μm from the nucleus, given that the mid-distance between two consecutive nuclei was on average 57.9 ±1.9 μm (mean±s.e.m.; Fig. S1A). We obtained an average PEI of 15.9±1.1 (mean±s.e.m.) for total mRNA (Fig. 1D). We observed a similar perinuclear enrichment of mRNA in ex vivo single myofibers, isolated from the extensor digitorum longus (EDL) muscle of adult mice and calculated an average PEI of 16.6±1.0 (Fig. 1E-G). Due to the higher proximity between nuclei in isolated myofibers, we analyzed a 10 μm concentric region of interest around each nucleus. Both these muscle models show that non-specialized nuclei accumulate mRNA in the perinuclear region.
Fig. 1. Newly transcribed mRNA is enriched in the perinuclear region in skeletal muscle myofibers.
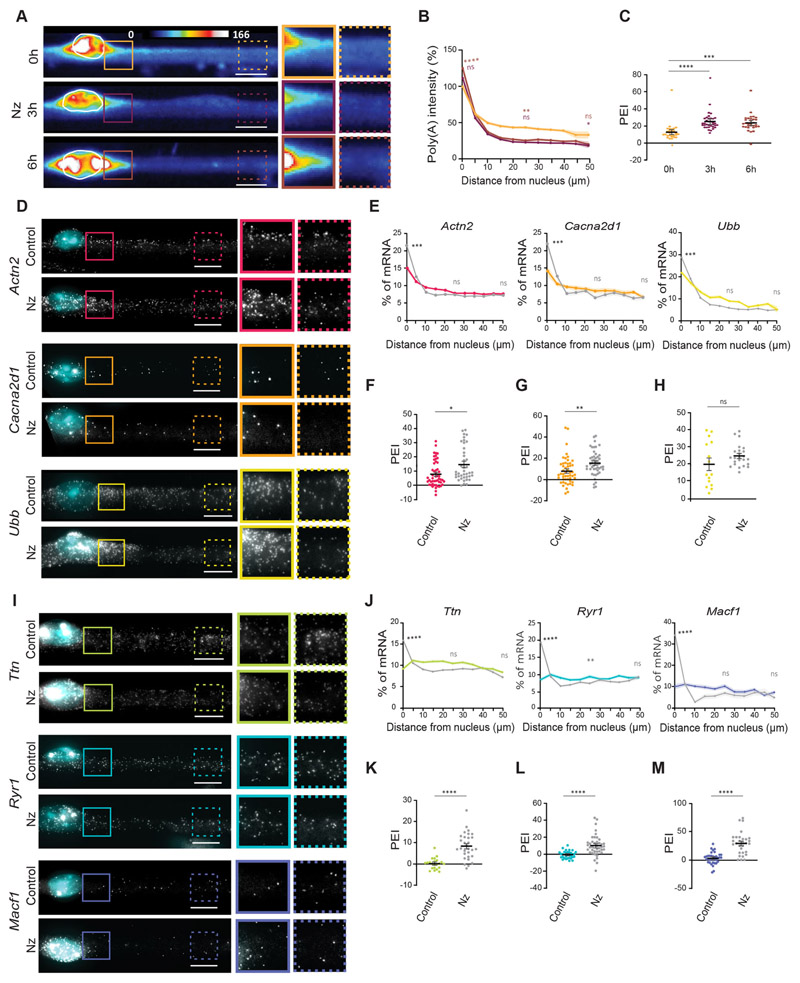
(A) Representative transmitted light, DAPI and FISH images, and FISH heatmap showing total mRNA in myofibers differentiated in vitro. The white line indicates a nucleus, determined using DAPI staining. White boxes indicate the region shown at 2× magnification in B. (B) Total mRNA FISH image and heatmap in the region highlighted in A. Images on the right show 1.5× magnifications of the perinuclear region (undashed box) and 50 μm away from the nucleus (dashed box). (C) Quantification of poly(A) intensity at different distances from the nucleus for myofibers differentiated in vitro, normalized to the average intensity close to the nucleus in each experiment. Mean±s.e.m. of 38 cell segments from three independent experiments. Statistical significance presented at 25 μm and 50 μm relative to 0 μm. (D) PEI, the percentage difference in total mRNA signal at 0 and 50 μm away from the nucleus. Mean±s.e.m. of 32 cell segments from three independent experiments. (E) Representative FISH image and heatmap of total mRNA in myofibers isolated from the EDL muscle of adult mice. A4× magnification of the highlighted area (box) is shown below, and nuclei are outlined in white. (F) Quantification of poly(A) intensity at different distances from the nucleus for myofibers isolated from the EDL muscle of adult mice, normalized to the average intensity close to the nucleus in each experiment. Mean±s.e.m. of 26 nuclei in five cells from two independent experiments. Statistical significance is presented at 4 μm and 10 μm relative to 0 μm. (G) PEI of total mRNA in myofibers isolated from the EDL muscle of adult mice. Mean ±s.e.m. of 26 nuclei in five cells from two independent experiments. (H) Representative FISH heatmaps of total mRNA distribution in in vitro myofibers treated for 3 and 6 h with 2 μMtriptolide. Nuclei are outlined in white. Images on the right show 1.5× magnifications of the perinuclear region (undashed box) and 50 μm away from the nucleus (dashed box). (I) Quantification of poly(A) intensity at different distances from the nucleus for myofibers treated with triptolide (color-coded as in H), normalized to the average intensity near the nucleus in untreated cells (0 h) of each experiment. Mean±s.e.m. of 33 (0 h), 35 (3 h) and 41 (6 h) cell segments, from four independent experiments. Statistical significance presented at 0 μm, 25 μm and 50 μm relative to untreated cells (0 h). (J) PEI of total mRNA in myofibers treated with triptolide for 0, 3 and 6 h. Mean±s.e.m. of 33 (0 h), 35 (3 h) and 41 (6 h) cell segments from four independent experiments. ****P<0.0001; ns, P>0.05 (one-way ANOVA with Tukey’s multiple comparisons test). FISH images are sum intensity projections and signal intensity is represented as heatmaps. Scale bars: 10 μm.
Perinuclear mRNA enrichment is modulated by newly transcribed mRNA
Non-specialized nuclei are transcriptionally active, as indicated by the strong nuclear signal of poly(A) FISH (Fig. 1A,E) and as previously described (Newlands et al., 1998; Nissinen et al., 2005). Thus, it is possible that the perinuclear mRNA enrichment is dependent on the accumulation of newly transcribed mRNAs from the closest nucleus. To test this hypothesis, we inhibited transcription using triptolide, an inhibitor of RNA polymerase I and II (Vispé et al., 2009), and performed FISH to detect total mRNA (Fig. 1H-J). We observed an overall reduction in the intensity of total mRNA 6 h after transcription inhibition, corresponding to 70.6% of the non-treated controls (Fig. S1E). This reduction occurred mostly in the perinuclear region, as depicted by the distribution profile of total mRNA 3 and 6 h after transcription inhibition (Fig. 1H,I). Thus, perinuclear mRNA accumulation is dependent on de novo transcription. To determine whether the perinuclear mRNA is transcribed by the nearest nucleus we co-cultured primary mouse myoblasts with human immortalized myoblasts (Fig. S1F-I). After differentiation, we identified chimeric myofibers containing a single human nucleus identifiable by its larger size and uniform DAPI staining (Fig. S1H). We used human- and mouse-specific single-molecule FISH (smFISH) probes targeting an mRNA encoding a Ca2+ channel (Cacna1s) to determine the spatial distribution of the mRNA relative to its nucleus of origin. We observed perinuclear accumulation of the human mRNA near the human nucleus and perinuclear accumulation of the mouse mRNA near the mouse nucleus (Fig. S1I). These results demonstrate that mRNA perinuclear enrichment is due to the accumulation of recently transcribed mRNAs from the nearest nucleus.
A subset of mRNAs are spread throughout the myofiber
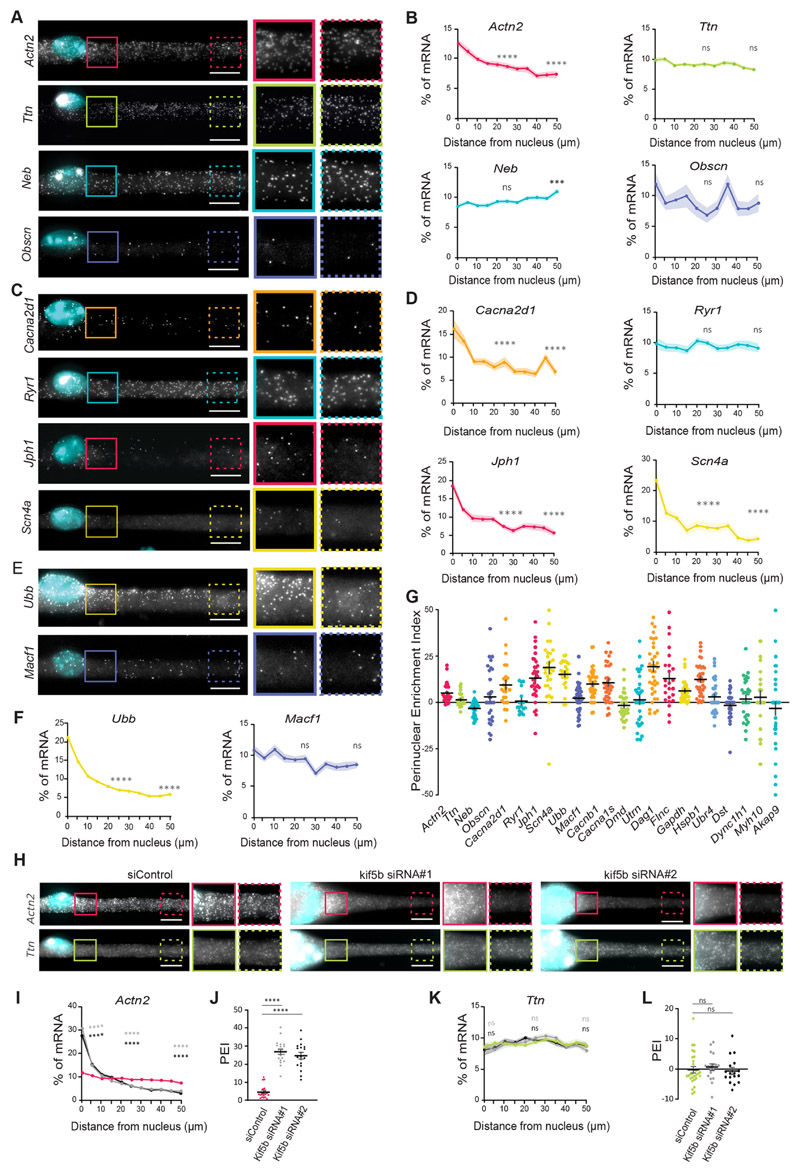
To determine whether the observed perinuclear enrichment of total mRNA is due to the accumulation of a particular subset of mRNAs, we performed smFISH to observe the spatial distribution of specific mRNA transcripts. We explored the distribution of mRNAs encoding proteins of sarcomeres, triads and costameres (Cretoiu et al., 2018) (characteristics of analyzed transcripts are summarized in Table S1).
We observed that mRNAs encoding sarcomeric proteins displayed different distribution profiles, as shown by their varying PEIs. Actn2 mRNAs presented the highest perinuclear enrichment (PEI=5.25±1.0; mean±s.e.m.) whereas Ttn (PEI=1.63±0.7), Neb (PEI=-3.03±0.7) and Obscn (PEI=3.17±2.7) were more evenly spread along the myofiber (Fig. 2A,B). This suggests that not all mRNAs exhibit a perinuclear enrichment, as we observed for total mRNA. Similarly, mRNAs encoding triad proteins Cacna2dl (PEI=9.55±2.5), Cacnb1 (PEI=10.1±1.2), Cacna1s (PEI=10.8 ±1.9), Jph1 (PEI=13.3±2.1) and Scn4a (PEI=19.0±3.1) accumulated in the perinuclear region, while Ryr1 mRNAs were more spread (PEI=0.7±1.7) (Fig. 2C,D; Fig. S2A,B). Dmd (dystrophin) and Utrn (utrophin) mRNAs, which encode costamere proteins, were also spread along the myofiber (Dmd, PEI=-1.4±1.3; Utrn, PEI=1.6±2.5; Fig. S2C,D). However, Dag1 (dystroglycan, PEI=19.6±2.6) and Flnc (filamin C, PEI=13.1±3.1) mRNAs, encoding sarcolemma proteins, displayed perinuclear enrichment. (Fig. S2C,D). These results show that a subset of mRNAs are not enriched in the perinuclear region, but instead are spread along the myofiber (PEI<5.0). These mRNA distribution patterns are observed for transcripts encoding proteins with different functions and localizations in the myofiber.
Fig. 2. A subset of mRNAs is spread throughout the myofiber independently of nuclear dispersion.
(A,C,E) Representative smFISH images of the distribution of Actn2, Ttn, Neb and Obscn (A); Cacna2d1, Ryr1, Jph1 and Scn4a (C); and Ubb and Macf1 (E) mRNAs. Images on the right show 1.5× magnifications of the perinuclear region (undashed boxes) and 50 μm away from the nucleus (dashed boxes) on the right. (B,D,F) Quantification of mRNA amount for the indicated mRNAs relative to distance to the nucleus (5 μm bins), normalized to total mRNA counts in each cell segment. Statistical significance is presented at 25 and 50 μm, comparing with 0 μm. Mean±s.e.m. of 28 (Actn2 and Ttn), 30 (Neb and Obscn), 24 (Cacna2d1 and Ryr1), 30 (Scn4a), 26 (Ubb) and 46 (Macf1) cell segments from three to four independent experiments. (G) Mean±s.e.m. PEI of the analyzed mRNAs. (H) Representative smFISH image of the distribution of Actn2 and Ttn mRNAs in cells transfected with control (siControl) and KIF5B siRNAs (kif5b siRNA#1 and kif5b siRNA#2). KIF5B depletion results in myofibers that exhibit nuclei aggregation. Images on the right for each condition show 1.5× magnifications of the perinuclear region (undashed boxes) and 50 μm away from the nuclei (dashed boxes). (I,K) Quantification of Actn2 (I) and Ttn (K) distribution in control and KIF5B-depleted myofibers, color-coded as in J and L, respectively. Mean±s.e.m. of 20 (siControl), 20 (siRNA#1) and 19 (siRNA#2) cell segments for Actn2 and 29 (siControl), 18 (siRNA#1) and 17 (siRNA#2) cell segments for Ttn mRNA from three independent experiments. Statistical significance presented at 0 μm, 25 μm and 50 μm relative to the control. (J,L) PEI of Actn2 (J) and Ttn (L) in control and KIF5B-depleted myofibers. Mean±s.e.m. of 20 (siControl), 20 (siRNA#1) and 19 (siRNA#2) cell segments for Actn2 and 29 (siControl), 18 (siRNA#1) and 17 (siRNA#2) cell segments for Ttn mRNA from three independent experiments. ****P<0.0001; ***P<0.001; ns, P>0.05 (one-way ANOVA with Tukey’s multiple comparisons test). smFISH images are maximum intensity projections. Scale bars: 10 μm.
We next sought to understand whether either of the two distribution patterns was exclusive to muscle-specific mRNAs. We therefore analyzed the distribution of ubiquitously expressed mRNAs: Ubb (ubiquitin B), Macf1 (microtubule-actin crosslinking factor 1), Gapdh (glyceraldehyde 3-phosphate dehydrogenase), Hspb1 (Heat shock protein β1), Ubr4 (E3 ubiquitin-protein ligase UBR4), Dst (dystonin), Dync1h1 (cytoplasmic dynein 1 heavy chain 1), Myh10 (myosin-10) and Akap9 (A-kinase anchor protein 9). Similarly to muscle-specific mRNAs, some ubiquitously expressed mRNAs accumulated in the perinuclear region, whereas others were more spread (Fig. 2E–G; Fig. S2E,F). This shows that non-muscle mRNAs can also exhibit both distribution patterns.
Differential distribution of mRNA is independent of nuclear dispersion
Nuclei are dispersed evenly along myofibers through a mechanism dependent on the crosslinking of microtubules by kinesin-1 (Metzger et al., 2012). To test whether nuclear dispersion influences the distribution patterns of mRNAs, we induced clustering of nuclei by kinesin-1 (KIF5B) siRNA-mediated knockdown (Fig. S3A). We analyzed mRNAs enriched in the perinuclear region (Actn2 and Cacna2d1) and spread along the myofiber (Ryr1 and Ttn) upon KIF5B knockdown. We found that both mRNA distribution patterns were present upon KIF5B knockdown, although we observed an increase of the PEI for Actn2 and Cacna2d1 when compared to control (Fig. 2H–L; Fig. S3B–F). Given our data on transcription inhibition (Fig. 1H–J), the increase in PEI of mRNAs enriched in the perinuclear region is most likely due to the clustering of mRNA-producing nuclei. An alternative hypothesis is that KIF5B is implicated in the transport of Actn2 and Cacna2d1 mRNAs away from the nuclei, since KIF5B has been previously implicated in mRNA transport (Brendza et al., 2000). However, we can conclude that nuclear dispersion is not required for the observed mRNA distribution patterns (perinuclear accumulated versus spread).
Transcript size contributes to mRNA spreading
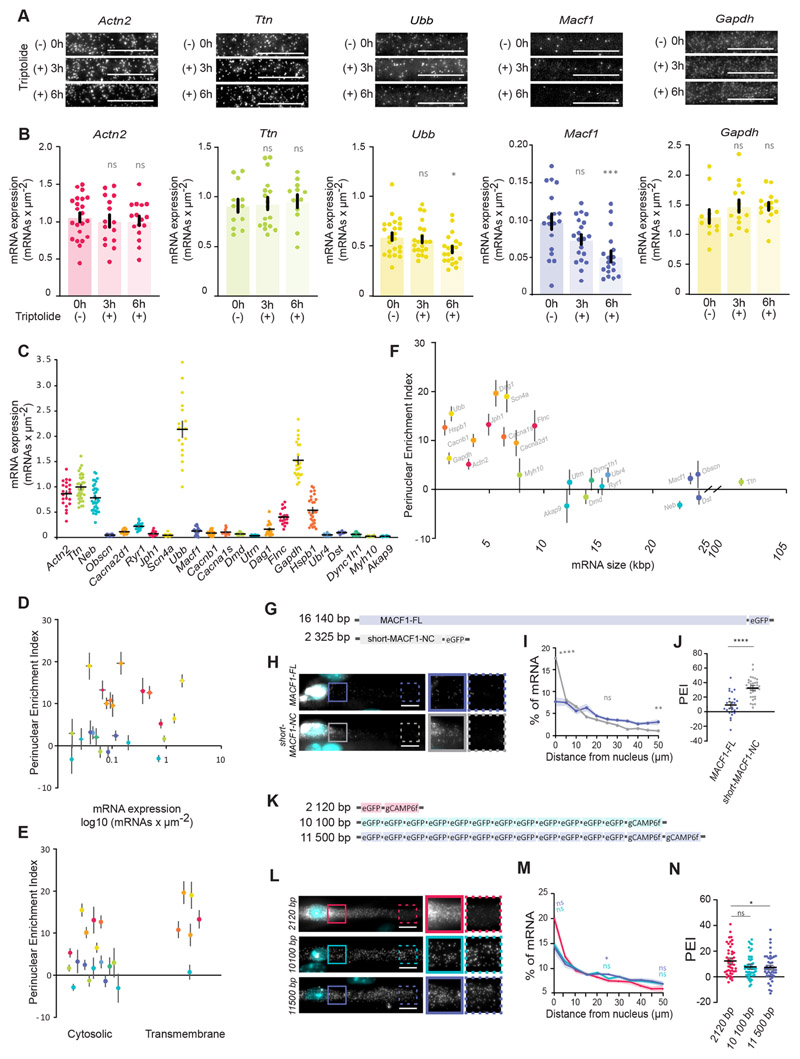
A possible explanation for the two types of mRNA distribution pattern (perinuclear accumulated versus spread) could result from different kinetics of mRNA synthesis and degradation. mRNAs with shorter half-life may be enriched near their nucleus of origin, whereas long-lived mRNAs could be able to reach farther. In addition, highly expressed mRNAs could be enriched in the perinuclear region.
We measured the stability of spread and perinuclear-accumulated mRNAs by blocking transcription for 3 and 6 h and quantified the number of mRNA molecules. We used smFISH because our in vitro culture contains different mononucleated cell types (myoblasts, myofibroblasts) and myofibers at different stages of differentiation (Pimentel et al., 2017; Roman et al., 2018). We observed that Actn2, Ttn and Gapdh were stable during this time, indicating that the half-life of these mRNAs is longer than 6 h in in vitro myofibers (Fig. 3A,B). In contrast, Ubb and Macf1 mRNA levels decreased to 78% and 52%, respectively, after 6 h of transcription inhibition (Fig. 3B). Given that Macf1 transcripts were spread along the myofiber but had a shorter half-life than Actn2, Gapdh and Ubb, differences in mRNA stability do not explain their differential distributions. We also measured the expression levels of all analyzed mRNAs (Fig. 3C) and we did not observe a correlation between mRNA expression levels and the corresponding PEI (Fig. 3D), indicating that perinuclear enrichment (PEI>5.0) is not due to the high expression level of certain mRNAs. Of note, the lack of correlation does not exclude the involvement of these or other factors in mRNA distribution. It is possible that several mechanisms are in place to orchestrate the differential distribution of mRNA in skeletal muscle. Additionally, we considered whether the mRNA distribution patterns identified could be related to differences in the proteins they encode (cytosolic versus transmembrane), but we found that mRNAs encoding cytosolic or transmembrane proteins could display both distribution patterns (Fig. 3E). Some of the largest human genome mRNAs encode muscle-specific proteins, such as titin and nebulin (Meyer and Wright, 2013). By plotting the predicted mRNA size of the analyzed mRNAs against the PEI, we observed a correlation between mRNA size and the type of distribution (Fig. 3F). Transcripts that displayed perinuclear enrichment (PEI>5) were those with a regular size below 10,000 basepairs (bps). However, most mRNAs that were spread through the myofiber (PEI<5) were those with predicted sizes above 10,000 bp and that encode large proteins. We refer to these transcripts as ‘giant mRNAs’. Myh10, with PEI=2.91±3.4 (mean±s.e.m.) and mRNA size of 7783 bp was the only exception from the pool of 23 tested transcripts.
Fig. 3. mRNA size contributes to mRNA spreading.
(A) Representative smFISH images of Actn2, Ttn, Ubb, Macf1 and Gapdh mRNA 3 h and 6 h after 2 μM triptolide treatment in myofibers differentiated in vitro. (B) Quantification of Actn2, Ttn, Ubb, Macf1 and Gapdh mRNA number per μm2 between two adjacent nuclei, measured by smFISH, after 3 h and 6 h of 2 μM triptolide treatment. Mean±s.e.m. of 22, 15, 16 (Actn2); 12, 14, 12 (Gapdh); 12, 17, 12 (Ttn); 18, 20, 18 (Macf1); and 23, 22, 21 (Ubb) cell segments in 2-3 independent experiments for 0 h, 3 h and 6 h treatments, respectively. (C) Quantifications of transcript number per μm2 (mRNA expression level) measured by smFISH for all the mRNAs in this study. Mean±s.e.m. of 22 (Actn2), 33 (Ttn), 35 (Neb), (Obscn), 19 (Cacna2d1), 22 (Ryr1), 21 (Jph1), 22 (Scn4a), 17 (Ubb), 23 (Macf1), 29 (Cacnb1), 10 (Cacna1s), 22 (Dmd),13 (Utrn), 18 (Dag1), 16 (Flnc), 19 (Gapdh), 27 (Hspb1), 15 (Ubr4), 19 (Dst), 14 (Dync1h1), 16 (Myh10) and 28 (Akap9) cell segments in four to seven independent experiments. (D-F) Scatter plots illustrating the relationship between mean±s.e.m. PEI of each mRNA (warm colors for perinuclear-enriched and cold colors for spread mRNAs) and mRNA expression levels (D), characteristics of mRNA-encoded protein (E) and predicted mRNA size (F). (G) Schematic representation of full-length MACF1 (MACF1-FL) and shorter N- and C-terminal (short-MACF1-NC) eGFP fusion. (H) Representative smFISH images of MACF1-FL and short-MACF1-NC mRNAs. Images on the right show 1.5× magnifications of the perinuclear region (undashed box) and 50 μm away from the nucleus (dashed box). (I) Quantification of MACF1-FL and MACF1-NC mRNA distribution color-coded as in H. Mean±s.e.m. of 27 (MACF1-FL) and 37 (short-MACF1-NC) cell segments from five independent experiments. Statistical significance presented at 0 μm, 25 μm and 50 μm relative to MACF1-FL. (J) PEI of MACF1-FL and short-MACF1-NC mRNA. Mean±s.e.m. of 27 (MACF1-FL) and 37 (short-MACF1-NC) cell segments from five independent experiments. (K) Schematic representation of combinations of eGFP and GCaMP6f with different transcript sizes. (L) Representative smFISH images of eGFP and GCaMP6f mRNAs. Images on the right show 1.5× magnifications of the perinuclear region (undashed box) and 50 μm away from the nucleus (dashed box). (M) Quantification of eGFP and GCaMP6f mRNAs distribution color-coded as in L. Mean±s.e.m. of 45 (2120 bp), 50 (10,100 bp) and 45 (11,500 bp) cell segments from six independent experiments. Statistical significance presented at 0 μm, 25 μm and 50 μm. (N) PEI of eGFP and GCaMP6f mRNAs of the indicated transcript sizes. Mean±s.e.m. of 45 (2120 bp), 50 (10,100 bp) and 45 (11,500 bp) cell segments from six independent experiments. ****P<0.0001; ***P<0.001; **P<0.01; *P<0.05; ns, P>0.05 (one-way ANOVA with Tukey’s multiple comparisons test and two-tailed, unpaired Student’s t-test for comparisons between two experimental conditions). 95% confidence interval. Scale bars: 10 μm.
To test whether mRNA size influences mRNA distribution in myofibers, we transiently expressed eGFP-tagged full-length MACF1 (MACF1-FL; 16,140 bp) and a shorter MACF1 N- and C-terminal fusion (short-MACF1-NC; 2325 bp) (Wu et al., 2008) (Fig. 3G–J). Using eGFP smFISH probes we determined the localization of the exogenous mRNAs. MACF1-FL mRNA was evidently more spread, whereas short-MACF1-NC was more enriched around the nucleus, with a PEI 3.5-fold higher (Fig. 3J). These data suggest that transcript size might contribute to mRNA distribution. To understand whether the role of transcript size on mRNA distribution is independent of the transcript sequence, we expressed constructs encoding a variable number of eGFP and GCaMP6f repeats in myofibers and analyzed their mRNA distribution (Fig. 3K–N). We found that the distribution of the shorter eGFP–GCaMP6f mRNA (2110 bp) resulted in higher perinuclear accumulation in comparison to mRNAs containing 12 repeats of eGFP and one or two GCaMP6f sequences (10,100 bp and 11,500 bp, respectively; Fig. 3K–N). Overall, these data suggest that mRNA distribution in skeletal muscle myofibers might be influenced by the size of the transcripts in a sequence-independent manner.
mRNA distribution patterns are maintained regardless of translation
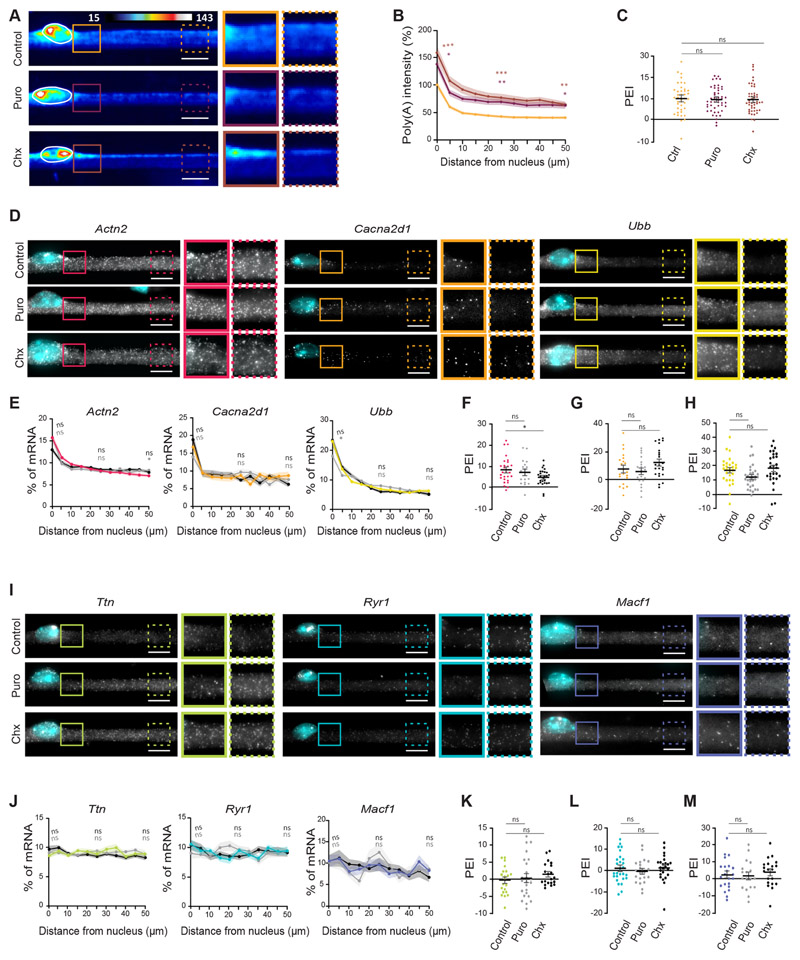
Translation-dependent and -independent pathways have been described to localize transcripts encoding transmembrane and cytosolic proteins to the endoplasmic reticulum (Cui et al., 2015; Cui and Palazzo, 2012; Jagannathan et al., 2014; Voigt et al., 2017). Accordingly, it has been observed that the perinuclear enrichment of transferrin receptor mRNA in muscle is translation dependent (Ralston et al., 1997). We observed an accumulation of ribosomal RNA in the perinuclear region (Fig. S4A–C), suggesting a perinuclear enrichment of translational machinery. To understand whether active translation determines the distribution of mRNA, we inhibited translation using both puromycin and cycloheximide. We observed no effect on total mRNA distribution (Fig. 4A–C), although translation inhibition led to an increase in the level of total mRNA (Fig. S4D). To further investigate the effects of translation inhibition, we performed smFISH on a subset of six transcripts encoding cytoplasmic and transmembrane proteins representative of normal size (Actn2, Cacna2d1 and Ubb) and giant mRNAs (Ttn, Ryr1 and Macf1) (Fig. 4D–M). The perinuclear accumulation of normal-size mRNAs was in general maintained (Fig. 4D–H). However, cycloheximide induced a significant decrease in the PEI of Actn2 (Fig. 4F) and puromycin slightly decreased the PEI of Ubb (Fig. 4H). The distribution of the giant mRNAs tested remained unchanged after translation inhibition (Fig. 4I–M). Similarly to total mRNA, translation inhibition altered the levels of some mRNAs (Fig. S4E,F). These results suggest that translation is not a determinant for the differential distribution of normal-size and giant mRNAs. However, our analysis does not exclude the possibility of a larger impact on the distribution of other transcripts.
Fig. 4. mRNA distribution patterns are maintained upon translation inhibition.
(A) Representative FISH heatmaps of total mRNA in myofibers treated for 6 h with 200 μg/ml puromycin (Puro) or 20 μg/ml cycloheximide (Chx). Images on the right show 1.5× magnifications of the perinuclear region (undashed boxes) and 50 μm away from the nucleus (dashed boxes). (B) Quantification of poly(A) intensity at different distances from the nucleus in control, puromycin- and cycloheximide-treated cells (color-coded as in A), normalized to the average intensity near the nucleus in untreated cells of each experiment. Mean±s.e.m. of 40 (control), 48 (Puro) and 47 (Chx) cell segments from three independent experiments. Statistical significance presented at 0 μm, 25 μm and 50 μm relative to control. (C) PEI of total mRNA in control myofibers (Ctrl) and myofibers treated for6 h with puromycin or cycloheximide. Mean±s.e.m. of 40, 48 and 47 cell segments, respectively, from three independent experiments. (D) Representative smFISH images of the distribution of Actn2, Cacna2d1 and Ubb mRNA in myofibers treated for 6 h with puromycin or cycloheximide. Images on the right show 1.5× magnifications of the perinuclear region (undashed boxes) and 50 μm away from the nucleus (dashed boxes). (E) Quantification of Actn2, Cacna2d1 and Ubb mRNA amount relative to distance to the nucleus in control, puromycin- and cycloheximide-treated cells (color-coded as in F–H). Mean±s.e.m. of 28, 19 and 26 (Actn2); 21, 21 and 23 (Cacna2d1); and 29, 30 and 28 (Ubb) cell segments, respectively, from three independent experiments. Statistical significance presented at 0 μm, 25 μm and 50 μm relative to control. (F-H) PEI of Actn2 (F), Cacna2d1 (G) and Ubb (H) mRNA in control, puromycin and cycloheximide-treated cells. Mean±s.e.m. of 28, 19 and 26 (Actn2); 21, 21 and 23 (Cacna2d1); and 29, 30 and 28 (Ubb) cell segments, respectively, from three independent experiments. (I) Representative smFISH images of the distribution of Ttn, Ryr1 and Macf1 mRNA in myofibers treated for 6 h with puromycin and cycloheximide. Images on the right show 1.5× magnifications of the perinuclear region (undashed boxes) and 50 μm away from the nucleus (dashed boxes). (J) Quantification of Ttn, Ryr1 and Macf1 mRNA amount relative to distance to the nucleus in control and puromycin and cycloheximide-treated cells (color-coded as in K–M). Mean±s.e.m. of 22, 24 and 20 (Ttn); 27, 21 and 24 (Ryr1); and 22, 18 and 20 (Macf1) cell segments, respectively, from three independent experiments. Statistical significance presented at 0 μm, 25 μm and 50 μm relative to control. (K–M) PEI of Ttn (K), Ryr1 (L) and Macf1 (M) mRNA in control and puromycin- and cycloheximide-treated cells. Mean ±s.e.m. of 22, 24 and 20 (Ttn); 27, 21 and 24 (Ryr1); and 22, 18 and 20 (Macf1) cell segments, respectively, from three independent experiments. ***P<0.001; **P<0.01; *P<0.05; ns, P>0.05 (one-way ANOVA with Tukey’s multiple comparisons test). smFISH images are maximum intensity projections. Scale bars: 10 μm.
Microtubules are involved in mRNA distribution away from the nucleus of origin
mRNA distribution is often mediated by the cytoskeleton, particularly by microtubules (Mofatteh and Bullock, 2017). In skeletal muscle, microtubules nucleate from the nuclear envelope and Golgi (Bugnard et al., 2005; Gimpel et al., 2017; Tassin et al., 1985), and are mostly organized parallel to the longitudinal axis of the myofiber.
To test the involvement of the microtubule cytoskeleton in mRNA distribution, we treated myofibers with nocodazole, which induces microtubule depolymerization (Fig. S5A). We observed a progressive accumulation in total mRNA in the perinuclear region after 3 h and 6 h of nocodazole treatment (Fig. 5A–C), suggesting that mRNA could be transported away from the nucleus of origin via microtubules. Microtubule depolymerization did not affect the total amount of mRNA, indicating that it does not exert a significant effect in overall mRNA transcription and/or degradation (Fig. S5B). Using smFISH, we investigated whether the effect of microtubule depolymerization on mRNA distribution was specific to giant mRNAs, which are spread throughout the myofiber. We analyzed a diverse panel of six mRNAs, including the largest and the smallest transcripts in our probe set (Ttn and Ubb, respectively). Nocodazole treatment for 6 h induced a general decrease in the total cytoplasmic levels (Fig. S5C,D) and perinuclear accumulation of both normal-size (Actn2, Cacna2d1 and Ubb; Fig. 5D–H) and giant mRNAs (Ttn, Ryr1 and Macf1; Fig. 5I–M), resulting in an increase in PEI, more evident in giant mRNAs. Nocodazole treatment did not affect the localization of sarcoplasmic reticulum (stained for RYR1), endocytic pathway organelles such as the Golgi (stained for giantin, also known as GOLGB1) and lysosomes/late endosomes (stained for LAMP1), or mitochondria (stained for HSP60, also known as HSPD1) (Fig. S5E–G). This suggests that the perinuclear accumulation of mRNA is not coupled to the localization of these organelles.
Fig. 5. mRNA distribution away from the nucleus of origin is perturbed by microtubule depolymerization.
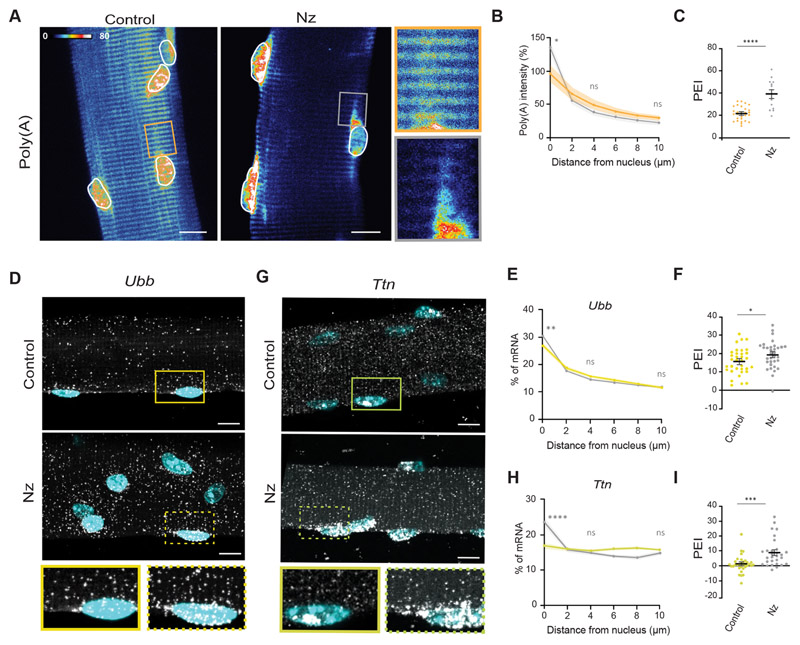
(A) Representative FISH heatmaps of total mRNA in myofibers treated for 3 h and 6 h with 4 μg/ml nocodazole (Nz). Images on the right show 1.5× magnifications of the perinuclear region (undashed box) and 50 μm away from the nucleus (dashed box). (B) Quantification of poly(A) intensity at different distances from the nucleus (color-coded as in A), normalized to the average intensity near the nucleus in the 0 h (untreated) cells of each experiment. Mean±s.e.m. of 42-58 cell segments from four independent experiments. (C) PEI of total mRNA in myofibers treated for 3 h and 6 h with nocodazole. Mean±s.e.m. of 42-58 cell segments from four independent experiments. (D) Representative smFISH images of the distribution of Actn2, Cacna2d1 and Ubb mRNA in myofibers treated for 6 h with nocodazole. Images on the right show 1.5× magnifications of the perinuclear region (undashed box) and 50 μm away from the nucleus (dashed box). (E) Quantification of Actn2, Cacna2d1 and Ubb mRNA amount relative to distance to the nucleus in control and nocodazole-treated cells (color-coded as in F–H). Mean±s.e.m. of 51 and 40 (Actn2), 51 and 49 (Cacna2d1) and 22 and 36 (Ubb) cell segments, respectively, from three independent experiments. (F–H) PEI of Actn2 (F), Cacna2d1 (G) and Ubb (H) mRNA in control and nocodazole-treated cells. Mean±s.e.m. of 51 and 40 (Actn2), 51 and 49 (Cacna2d1) and 22 and 36 (Ubb) cell segments, respectively, from three independent experiments. (I) Representative smFISH images of the distribution of Ttn, Ryr1 and Macf1 mRNA in myofibers treated for 6 h with nocodazole. Images on the right show 1.5× magnifications of the perinuclear region (undashed box) and 50 μm away from the nucleus (dashed box). (J) Quantification of Ttn, Ryr1 and Macf1 mRNA amount relative to distance to the nucleus in control and nocodazole-treated cells (color-coded as in K–M). Mean±s.e.m. of 28 and 46 (Ttn), 32 and 41 (Ryr1) and 36 and 30 (Macf1) cell segments, respectively, from three independent experiments. (K-M) PEI of Ttn (K), Ryr1 (L) and Macf1 (M) mRNA in control and nocodazole-treated cells. Mean±s.e.m. of 28 and 46 (Ttn), 32 and 41 (Ryr1) and 36 and 30 (Macf1) cell segments, respectively, from three independent experiments. ****P<0.0001; ***P<0.001; **P<0.01; *P<0.05; ns, P>0.05 (two-tailed, unpaired Student’s t-test for comparisons between different conditions and the respective untreated control). smFISH images are maximum intensity projections. Scale bars: 10 μm.
Next, we investigated the role of microtubules in mRNA distribution using myofibers fully differentiated in vivo. We isolated and cultured ex vivo single muscle fibers from the EDL of adult mice and treated them with nocodazole for 9 h (Fig. S5H). Consistent with our in vitro results, nocodazole-treated ex vivo myofibers displayed total mRNA enrichment in the perinuclear region, resulting in an increase in PEI (Fig. 6A–C). To analyze single-molecule mRNA distribution, we performed hybridization chain reaction (HCR)-based signal-amplified smFISH in isolated single fibers. The amplification step in this smFISH method increases sensitivity and thus enables single transcript analysis throughout the entire myofiber. Disruption of the microtubule network in isolated myofibers also led to the accumulation of Ttn (giant mRNA; Fig. 6G–I) and a milder accumulation of Ubb (normal-size mRNA) transcripts (Fig. 6D–F) around the nucleus, similar to our results in vitro. Overall, these results indicate that microtubules are involved in the distribution of both normal-size and giant mRNAs in myofibers.
Fig. 6. mRNA distribution in ex vivo isolated fibers is perturbed by microtubule depolymerization.
(A) Representative FISH heatmap of total mRNA distribution in isolated myofibers treated for 9 h with 4 μg/ml nocodazole (Nz), compared with the untreated control. Images on the right show 3× magnifications of the highlighted regions (boxes). Nuclei are outlined in white. (B) Quantification of mean±s.e.m. poly(A) intensity at different distances from the nucleus in nocodazole-treated isolated myofibers (color-coded as in C), normalized to the average intensity close to the nucleus in the control (untreated) of each experiment. (C) PEI of total mRNA in control and nocodazole-treated myofibers. Mean±s.e.m. of 28 nuclei from six cells for the control and 14 nuclei from four cells for the nocodazole-treated sample from two independent experiments. (D) Representative smFISH images of Ubb mRNA distribution in control and nocodazole-treated isolated myofibers. Images beneath show 1.5× magnifications of the region around the single nuclei indicated by boxes. (E) Quantification of mean±s.e.m. Ubb mRNA amount relative to the distance to the nucleus (2 μm bins) in control and nocodazole-treated isolated myofibers (color-coded as in F). (F) PEI of Ubb mRNA in control and nocodazole-treated myofibers. Mean±s.e.m. of 29 and 34 nuclei, respectively, from 5-6 cells from three independent experiments. (G) Representative smFISH images of Ttn mRNA distribution in control and nocodazole-treated isolated myofibers. Images beneath show 1.5× magnifications of the region around the single nuclei indicated by boxes. (H) Quantification of mean±s.e.m. Ttn mRNA amount relative to the distance to the nucleus (2 μm bins) in control and nocodazole-treated isolated myofibers (color-coded as in I). (I) PEI of Ttn mRNA in control and nocodazole-treated myofibers. Mean±s.e.m. of 34 and 37 nuclei, respectively, from 5-6 cells from two independent experiments. ****P<0.0001; ***P<0.001; **P<0.01; *P<0.05; ns, P>0.05 (two-tailed unpaired Student’s t-test for comparisons between different conditions and the respective untreated control at 0 μm). smFISH images are maximum intensity projections. Scale bars: 10 μm.
Discussion
In this work, we performed a detailed study concerning the distribution of mRNA in non-specialized regions of skeletal muscle. We showed that the bulk of mRNAs are enriched in the perinuclear region due to the accumulation of newly transcribed mRNAs from the closest nucleus. Additionally, we identified a subset of transcripts encoding some of the biggest mammalian proteins that are uniformly distributed throughout the myofiber. Large size favors the spreading of mRNAs in skeletal muscle in a sequence-independent manner. Finally, we showed that the microtubule cytoskeleton is involved in the distribution of these mRNAs away from the nucleus of origin, regardless of mRNA size.
Besides being the largest cells in the body, myofibers are also structurally unique: triads and sarcomeres are packed in the cytoplasm in numerous repeats of the same structures (Cretoiu et al., 2018). Thus, skeletal muscle constitutes an interesting system to study long-range mRNA transport for local protein delivery. Although this is a long-standing subject of interest, the improvement of RNA-imaging technologies and in vitro systems only recently enabled comprehensive studies in these complex cells. Most of the preceding reports have shown that gene products are restricted to the region surrounding the nucleus (Hall and Ralston, 1989; Pavlath et al., 1989; Ralston et al., 1997). This has led to the general acceptance of the nuclear domain theory, stating that each nucleus can influence a limited volume of cytoplasm. Here we show that de novo transcription is required for the accumulation of mRNAs in the perinuclear area.
Recent studies have shown the existence of different populations of nuclei with specific transcription signatures within myofibers (Dos Santos et al., 2020; Kim et al., 2020; Petrany et al., 2020). These differences could allow for tailored local responses to either intra- or extra-cellular stimuli. The fact that nuclei near the perimysium have a specific transcriptional profile (Kim et al., 2020) is one of the examples implying that the compartmentalization within myofibers goes beyond the classical examples of the myotendinous and neuromuscular junctions. The existence of different nuclei populations argues for cellular compartmentalization that can only be achieved if the distribution of genetic products is limited. In the case of the multinucleated fungus Ashbya gossypii (also known as Eremothecium gossypii), the heterogeneity of cyclin transcripts near each nucleus allows for cell-cycle asynchrony within the shared cytoplasm (Dundon et al., 2016; Lee et al., 2013). Our observation that mRNAs are enriched in the perinuclear region of non-specialized nuclei indicates that specific nuclei transcription profiles in myofibers can result in cell compartmentalization by maintaining transcripts in the vicinity of the nucleus that transcribes these transcripts. Thus, the area around each nucleus in non-specialized regions of the multinucleated myofiber is compartmentalized, similarly to the neuromuscular and myotendinous junction regions.
In contrast, we also found that giant mRNAs are evenly spread throughout the myofiber, instead of accumulating in the perinuclear region. Importantly, we found that the dispersion of nuclei throughout the myofiber is not involved in the establishment of the two types of mRNA distribution that we identified (spread versus perinuclear accumulated). Intrinsic properties of the mRNA are likely to dictate mRNA distribution in skeletal muscle. By analyzing the distribution of exogenous mRNAs with size up to 12 kbp, we determined that the size of a transcript affects its distribution regardless of the nucleotide sequence. The relation between mRNA size and cellular localization is deeply understudied. Interestingly, a positive correlation between the size of β-actin mRNA and its distribution in chicken embryo fibroblasts has been previously observed (Yamagishi et al., 2009). This is most likely dependent on the properties of the cytoplasm, because an identical pattern is observed using dextrans of different molecular masses. Similarly, we observed that transcripts with high molecular mass are spread in relation to the nuclei. It is probable that a uniform mRNA distribution is not exclusive to this subset of transcripts (Kann and Krauss, 2019); however, it seems reasonable to expect a more efficient distribution of mRNAs encoding very large proteins to compensate for their constrained diffusion. In fact, mRNA accumulation in stress granules is correlated with the size of the coding region and UTRs (Khong et al., 2017; Moon et al., 2019). How mRNA size could regulate localization remains unknown. Giant mRNA transport could be enhanced in myofibers through more effective binding to motors or binding to specific RNA-binding proteins. Supporting this hypothesis, it has been shown that the RNA-binding protein FMR1 preferentially interacts with long transcripts, promoting their translation (Greenblatt and Spradling, 2018). The transport of mRNA to distribute it along the myofiber would be energetically more efficient than distributing each protein. It would also allow for a faster local response to stimuli, such as muscle damage, similarly to the local translation mechanisms described in neurons (Holt and Schuman, 2013). However, we do not exclude that the distribution of normal-size mRNAs can also be actively regulated. We assessed the role of translation in normal-size and giant mRNA distribution and we observed only slight effects, but those were restricted to normal-size mRNAs. It is possible that translation could also contribute to the retention of mRNAs in the perinuclear region.
We show that microtubules contribute to mRNA distribution away from the nucleus. In differentiated skeletal muscle, there is a polarization of microtubules relative to nuclei, given that the nuclear envelope acts as a microtubule nucleator (Bugnard et al., 2005). A recent report in cardiomyocytes has shown that microtubules are required for mRNA localization and proper translation during cardiac muscle hypertrophy (Scarborough et al., 2021). This indicates that microtubule-dependent mRNA distribution is conserved in other muscle types. We observed that microtubule depolymerization has a larger impact on giant mRNAs than on normal-size mRNAs that are enriched in the perinuclear region. These data support the model that giant mRNA distribution could be more efficient, spreading these transcripts throughout the myofiber. It is possible that mRNAs are directly transported along microtubules, but also that microtubules are required for mRNA distribution through indirect effects such as mRNA anchoring, or mRNA hitchhiking on vesicles or organelles (Kugler and Lasko, 2009; Salogiannis and Reck-Peterson, 2017). Our data also does not exclude the possible role of passive mRNA distribution through diffusion, followed by differential anchoring according to size. A detailed characterization of the mechanisms involved in the microtubule-dependent effect on the distribution of both normal-size and giant mRNAs will be crucial to clarify these hypotheses.
The roles for mRNA localization in mammalian cells are being gradually unraveled (Cioni et al., 2018; Herbert and Costa, 2019; Holt and Bullock, 2009). Although many described mechanisms are transcript-specific, an understanding of how this process can be generally regulated is arising (Moor et al., 2017; Zappulo et al., 2017). In mononucleated cells, there is a tight regulation between transcription levels and the size of the cytoplasm (Padovan-Merhar et al., 2015). If mRNA concentration is also regulated in multinucleated cells, this process must be more complex due to the shared cytoplasm. The perinuclear accumulation of the bulk of mRNAs could allow for such regulation to take place in each nuclear compartment.
Materials and Methods
In vitro primary myofibers
All procedures using animals were approved by the institutional ethics committee and followed the guidelines of the National Research Council Guide for the care and use of laboratory animals.
In vitro myofibers were differentiated as previously described (Pimentel et al., 2017) from primary myoblasts isolated from 5–7-day-old C57BL/6 mice. Briefly, muscles were dissected, digested mechanically and incubated for 90 min at 37°C with agitation in collagenase type V (Sigma-Aldrich, St Louis, MO, USA) and dispase II (Invitrogen, Waltham, MA, USA) in DPBS (Gibco, Carlsbad, CA, USA). After centrifugation, cells were pre-plated for 4 h to allow for fibroblast adherence. The remaining cells were collected, centrifuged and plated at a density of 200,000 cells/ml in 20% FBS (Eurobio Scientific, Les Ulis, France) IMDM with Glutamax (Gibco) in μ-dish 35 mm dishes (Ibidi, Gräfelfing, Germany) pre-coated with 1% Matrigel (Corning Inc., Corning, NY, USA) in IMDM with Glutamax (Gibco). After 3 days of proliferation, medium was replaced by 10% horse serum (Invitrogen, Waltham, MA, USA) IMDM with Glutamax supplemented with 100 ng/ml recombinant agrin (R&D Systems, Minneapolis, MN, USA) to induce differentiation. An upper coating of 50% Matrigel in 10% horse serum IMDM with Glutamax was added the following day. All experiments were performed 6 days after differentiation induction.
Ex vivo isolated single myofibers
Ex vivo isolated single myofibers were obtained from EDL muscle as previously described (Rosenblatt et al., 1995). EDL muscle was explanted from adult (3–6-months-old) C57BL/6 mice, regardless of gender, and digested in 0.2% collagenase type I (Sigma-Aldrich) in DMEM (Gibco) for 90 min at 37°C. Mechanical dissociation of myofibers was performed using a thin Pasteur pipette and followed under a stereomicroscope. Single fibers were collected and fixed immediately or plated in Ibidi dishes followed by 50% Matrigel coating in IMDM with Glutamax. Plated myofibers were maintained in 10% horse serum IMDM with Glutamax for the duration of the experiment.
Human and mouse myoblast co-culture
Human immortalized myoblasts C8220, a kind gift from Vincent Mouly (Institute of Myology, Paris, France), were cultivated in Skeletal Muscle Cell Growth Medium (Promocell, Heidelberg, Germany) and kept at 20–60% confluence to avoid committing the cells to differentiation, as previously described (Mamchaoui et al., 2011). Cells were trypsinized using TrypLE Express (Gibco), centrifuged for 5 min at 350 g and mixed with mouse myoblasts at plating, at a ratio of 1:10 cells. Several human to mouse cell ratios were tested, and this was the most favorable to obtain a few myofibers with only one human nucleus. The nuclei were distinguished by the uniform (human) or punctuate (mouse) DAPI staining. Mouse nuclei appear punctuate due to higher fluorescence in A/T-rich regions in DNA, which are absent in human DNA (Blau et al., 1983).
Plasmids and transfections
For kinesin-1 (KIF5B) depletion, in vitro cells were transfected with 20 nM KIF5B siRNA#1 and siRNA#2 (ID s68781 and s68782, respectively; Thermo Fisher Scientific, Waltham, MA) 5 h prior to differentiation induction, as previously described (Pimentel et al., 2017), using RNAiMAX (Life Technologies, Carlsbad, CA) following the manufacturer’s instructions. Silencer Select Negative Control No. 1 (Thermo Fisher Scientific, #4390843) was used as a control.
For exogenous expression of MACF1-FL, short-MACF1-NC and eGFP–GCaMP6f constructs, cells were transfected at the time of differentiation induction for 16 h with 2 μg of the appropriate plasmid using 2 μl of Lipofectamine 3000 (Life Technologies) according to manufacturer’s instructions. MACF1-FL and short-MACF1-NC were a kind gift from the Elaine Fuchs lab (Rockefeller University, New York, NY, USA). In order to create the EGFP–GCaMP6f constructs, the GCaMP6f stop codon in the p1×GCaMP6f-N1 vector (modified from p2×EGFP-N1, Addgene plasmid #122162) was replaced by a BspEI site using site-directed mutagenesis (SDM). A 12×EGFP cassette was directionally cloned into the SDM-resulting backbone p1×GCaMP6f(mut)-N1, generating a plasmid with one GCaMP6f and 12 repetitions of EGFP. This process was repeated to introduce two repetitions of concatemeric GCaMP6f and 12 repetitions of concatemeric EGFPs in the backbone. p2×EGFP-N1 and p12×EGFP-N1 were deposited at Addgene by Georg Mayr (Addgene plasmids #122162 and #122172, respectively) (Böhm et al., 2017).
Drug treatments
To inhibit transcription, 2 μM triptolide (Sigma-Aldrich) was added to in vitro myofibers for 3 h and 6 h. To inhibit translation, 200 μg/ml puromycin (Sigma-Aldrich) or 20 μg/ml cycloheximide (Sigma-Aldrich) was added for 6 h. To depolymerize microtubules, 4 μg/ml nocodazole (Sigma-Aldrich) was used for 3 h and/or 6 h. In ex vivo isolated single myofibers, 4 μg/ml nocodazole was added for 9 h.
Reverse transcription quantitative PCR for siRNA validation
After transfection, total RNA was extracted from in vitro myofibers at differentiation day 6 using TRIzol Reagent (Thermo Fisher Scientific) according to the manufacturer’s instructions. RNA yield and purity was assessed using a Nanodrop 2000 apparatus. cDNA was synthesized using the High capacity RNA-to-cDNA kit (Applied Biosystems, Foster City, CA, USA) following the manufacturer’s instructions. Reverse transcription quantitative PCR (RT-qPCR) was performed using Power SYBR Green PCR MasterMix (Alfagene) according to the manufacturer’s instructions and using forward and reverse primers at 0.25 μM and 1:20 cDNA dilution. Amplification of kif5b was performed using primers 5’-TACAGACCAAGAGAAGAGCAGG-3’ (forward) and 5’-AGTCT-GAAGTTCTTTTGCCACG-3’ (reverse). As a control, housekeeping gene Hprt was amplified using the primers 5’-GTTAAGCAGTACAG-CCCCAAA-3’ (forward) and 5’-AGGGCATATCCAACAACAAACTT-3’ (reverse). RNA extraction, cDNA synthesis and RT-qPCRs were performed three times, and relative transcription levels were determined using the ΔCt method.
smFISH and total mRNA FISH
In vitro differentiated myofibers were washed once with RNase-free PBS (Ambion, Austin, TX, USA) and fixed in 3.7% formaldehyde (Sigma-Aldrich) for 10 min at room temperature. Myofibers were washed in PBS and permeabilized overnight in 70% ethanol. After washing for 5 min with 10% formamide (Ambion) in 2× saline sodium citrate buffer (SSC, Sigma-Aldrich), myofibers were incubated overnight at 37°C in 10% formamide, 1% dextran sulfate (Sigma-Aldrich) in 2× SSC containing RNA probes coupled to Qasar 570 or Qasar 670 at a concentration of 1.25 μM and 2.50 μM, respectively. mRNA probes were designed to align to the coding sequence of the mRNA of interest using the Stellaris probe designer (https://www.biosearchtech.com/stellaris-designer; Biosearch Technologies, CA, USA). The Stellaris probe set sequences are provided in Table S2. To stain total mRNA, in primary myofibers or isolated single fibers, we used a poly(dT) probe with 60 nucleotides conjugated at the 5’ end with Alexa Fluor 488 at 5 nM (Eurofins Scientific, Luxembourg City, Luxembourg). As control for poly(A) intensity signal, we added 1 mg/ml ribonuclease A (RNase-A; Sigma-Aldrich) to in vitro myofibers post fixation for 1 h at 37° C. Myofibers were washed twice in 10% formamide in 2× SSC for 30 min at 37°C and incubated with DAPI (Sigma-Aldrich) during the second wash. Finally, myofibers were washed in 2× SSC for 5 min and covered in Vectashield Antifade Mounting Medium (Vector Laboratories, Burlingame, CA, USA). For smFISH, image acquisition was performed on a Zeiss Cell Observer wide-field inverted microscope using a 63× Plan-Apochromat oil objective (NA=1.4). Digital images were acquired by a cooled Axiocam 506m camera upon excitation with a Zeiss HXP 120 metal halide light source using 1000 ms exposure time. For total mRNA FISH, image acquisition was performed on a Zeiss Cell Observer inverted CSU-X1 spinning-disk microscope. Digital images were acquired by an Evolve 512 EMCCD camera through a 40× Plan-Apochromat oil objective (NA=1.4). All samples were imaged soon after RNA labeling. Cell regions including two consecutive nuclei in the field of view were chosen based on the striations seen using transmitted light and the position of nuclei at the cell periphery.
Hybridization chain reaction-based signal-amplified smFISH
For single mRNA detection in ex vivo isolated single myofibers, HCR-based signal-amplified smFISH was performed according to the manufacturer’s instructions. Ex vivo single myofibers were fixed and permeabilized as described for in vitro myofibers in the section above. Probes were designed to bind to the coding sequence of the mRNAs of interest by Molecular Instruments (Los Angeles, CA). A set of probes comprises multiple probe pairs that bind to the mRNA of interest in different subregions and contain an HCR initiator split between the probe pair. The HCR initiator triggers a chain reaction once incubated with DNA hairpins coupled to a fluorescent molecule that stores energy for self-assembly. Myofibers were washed twice in 2× SSC and incubated for 30 min at 37°C with pre-heated hybridization buffer (Molecular Instruments). Then, myofibers were incubated overnight at 37°C with hybridization buffer containing 10 nM of the probe set of interest. Myofibers were washed four times for 5 min at 37°C with pre-heated wash buffer (Molecular Instruments) and twice with 0.1% Tween 20 (Sigma-Aldrich). For HCR amplification, DNA hairpins were snap cooled by heating for 90 s at 95°C and cooled down to room temperature for 30 min. Myofibers were incubated with amplification buffer (Molecular Instruments) for 30 min, followed by a 45 min incubation in the dark with amplification buffer containing 1 μM DNA hairpins labelled with Alexa Fluor 546 or Alexa Fluor 647. After DAPI staining, fibers were washed five times in 5× SSC containing 0.1% Tween 20 and covered in Vectashield Antifade Mounting Medium. Image acquisition was performed on a Zeiss LSM 880 confocal inverted microscope using a 63× Plan-Apochromat oil Objective (NA=1.4) and GaSP detectors.
Immunofluorescence
In vitro or isolated single myofibers were fixed for 10 min in 4% paraformaldehyde (Science Services GmbH, Munich, Germany) and washed twice in PBS. Myofibers were permeabilized with 0.5% Triton X-100 (Sigma-Aldrich) in PBS for 5 min and washed with PBS. After incubation for 30 min in blocking solution [5% BSA (Sigma-Aldrich) and 10% goat serum (Sigma-Aldrich) in PBS], primary antibodies were diluted in 0.1% saponin (Sigma-Aldrich), 5% BSA and 10% goat serum in PBS and incubated overnight at 4°C. Myofibers were washed three times for 5 min in PBS. Secondary antibodies and DAPI were incubated for 60 min in 0.1% saponin, 5% BSA and 10% goat serum in PBS. After washing three times for 5 min in PBS, cells were covered in Fluoromount-G mounting medium (Southern Biotech, Birmingham, AL, USA). Image acquisition was performed on a Zeiss LSM 880 confocal inverted microscope using a 63× Plan-Apochromat oil Objective (NA=1.4) and GaSP detectors.
Monoclonal anti-tyrosinated α-tubulin (YL1/2) produced in rat (ECACC, UK) was used diluted at 1:50. Mouse monoclonal anti-acetylated tubulin antibody from Sigma-Aldrich (T7451) was diluted at 1:200. Mouse monoclonal anti-RYR1 (#MA3-925; Thermo Fisher Scientific) was diluted at 1:300. Rabbit polyclonal anti-giantin (#909701; BioLegend, San Diego, CA, USA) was diluted at 1:1000. Rat monoclonal anti-LAMP1 (ab25245; Abcam, Cambridge, UK) was diluted at 1:200. Mouse monoclonal anti-Hsp60 (#611562; BD Biosciences, Franklin Lanes, NJ, USA) was diluted at 1:200. Donkey anti-mouse IgG secondary antibody Alexa Fluor 488 (Thermo Fisher Scientific), goat anti-rat IgG secondary antibody Alexa Fluor 555 (Thermo Fisher Scientific) and goat anti-rabbit IgG Alexa Fluor 647 (Thermo Fisher Scientific) were used diluted 1:400.
Image analysis for spatial distribution measurements
All images were processed in Fiji (Schindelin et al., 2012). Intensity color-coded images were obtained by applying the ‘royal’ lookup table (LUT). For total mRNA, z-stacks of the entire myofiber height were converted to sum intensity projections. Poly(A) signal intensity of the z-stacks was quantified for a linescan from the nucleus to 50 μm away, spanning the width of the cell. The average background intensity was subtracted, and the values obtained were normalized to the average intensity near the nuclei in the control of each independent experiment. The average of all normalized intensities along the 50 μm myofiber segment was plotted against distance in multiples of 5 μm.
For smFISH analysis in in vitro myofibers, z-stacks of the entire myofiber height were converted to maximum intensity projections. Nucleus area was defined by creating a mask based on the threshold of the DAPI signal, and a mask of each cell area was determined manually. Cells were divided into bins of 5 μm from the edge of the nearest nucleus to 50 μm away. The number of fluorescent spots corresponding to single mRNAs was quantified in each bin, after detection using the open source ThunderSTORM plug-in for ImageJ (Ovesný et al., 2014). Each bin was plotted as the percentage relative to the total mRNA in the cell segment.
In ex vivo isolated single myofibers, z-stacks of the entire myofiber height were converted to sum and maximum intensity projections for total mRNA and smFISH analysis, respectively. Nuclei areas were defined by creating a mask based on the threshold of the DAPI signal. The area around each nucleus was divided into bins of 2 μm by defining concentric regions of interest with 2 μm radius increments, restricted to the area inside the cell with a manually determined mask. mRNA amount was quantified from the bin closest to the nucleus to 10 μm away, given that the nuclear distances are shorter when compared with those of in vitro myofibers. Total mRNA was quantified by measuring the total intensity of poly(A) signal in each concentric bin, and single mRNAs were detected using the open source ThunderSTORM plug-in for ImageJ.
Perinuclear enrichment index and mRNA expression calculation
The mRNA perinuclear enrichment index (PEI) corresponds to the percentage difference between the amount of mRNA in the region closest to the nucleus and the amount of mRNA in the region furthest away from the nucleus, as follows: .
For in vitro differentiated myofibers, the PEI was calculated between the segment bins at 0–5 μm and 45–50 μm to the nucleus. For ex vivo isolated myofibers, given that the nuclear distances are shorter, the PEI was calculated using the concentric bins with radius 0–2 μm and 8–10 μm away from the nucleus periphery.
To calculate mRNA expression and degradation levels in the in vitro myofiber cytoplasm, the total number of mRNAs in a maximum intensity projection of cell segments between two adjacent nuclei was divided by the area (mRNAs μm−2).
Data representation and statistics
All graphs were plotted using GraphPad Prism (San Diego, CA, USA) and formatted in Adobe Illustrator (Adobe Inc., San Jose, CA, USA). Statistical tests were performed using GraphPad Prism. For mRNA distribution histograms, a one-way analysis of covariance (ANOVA) was performed and the significance in the middle bin and the last bin was compared with the closest bin to the nucleus, applying Tukey’s multiple comparisons test. For mRNA distribution in different experimental conditions, a Student’s t-test or one-way ANOVA was performed comparing the region closest to the nucleus, 25 μm and 50 μm away. For mRNA amount, a one-way ANOVA with Tukey’s multiple comparisons test was applied between conditions. Statistical significance is represented as follows: ****P<0.0001; ***P<0.001; **P<0.01; *P<0.05; ns, no significance. Error bars, or shading in mRNA distribution graphs, represent the s.e.m. For each set of results, n is indicated in the figure legend and represents the number of cell segments or the number of nuclei.
Supplementary Material
Acknowledgements
We thank all members of the Edgar Gomes Laboratory, Claudio Franco Laboratory and Bruno Cadot for discussions; Graciano Leal and Katharina Hennig for support in in vitro myofiber culture; Sara Ferreira and the Animal Facility staff at iMM for support in C57BL/6 mice colony maintenance; António Temudo, Ana Nascimento, Aida Lima and José Rino, from the Bioimaging facility at iMM, for imaging assistance; the Elaine Fuchs Laboratory for providing the MACF1 constructs; and the Molecular and Transgenic Tools Platform of Champalimaud Foundation for cloning advice and services.
Funding
This work was supported by the European Research Council (617676, 810207); a European Molecular Biology Organization Installation Grant (2758); AFM-Telethon (21661); Fundaçao para a Ciência e a Tecnologia (PTDC/BIA-CEL/31075/2017); and Congento LISBOA-01-0145-FEDER-022170, co-financed by Fundação para a Ciência e a Tecnologia and Lisboa2020, under the PORTUGAL2020 agreement (European Regional Development Fund). H.P. (PD/BD/128287/2017), M.R.P. (SFRH/BD/52227/2013) and C.S. (2020.04642.BD) were supported by fellowships from the Fundação para a Ciência e a Tecnologia.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: H.P., M.R.P., W.R., E.R.G.; Methodology: H.P., M.R.P.; Validation: H.P., M.R.P., E.R.G.; Formal analysis: H.P., M.R.P., C.S.; Investigation: H.P., M.R.P., C.S.; Resources: H.P., M.R.P., W.R.; Data curation: L.M.O., A.P.; Writing - original draft: H.P., M.R.P.; Writing - review & editing: A.P., W.R., E.R.G.; Visualization: H.P., M.R.P.; Supervision: E.R.G.; Project administration: H.P., M.R.P., E.R.G.; Funding acquisition: E.R.G.
Peer review history
The peer review history is available online at https://journals.biologists.com/jcs/article-lookup/doi/10.1242/jcs.256388
References
- Abmayr SM, Pavlath GK. Myoblast fusion: lessons from flies and mice. Development (Cambridge, England) 2012;139:641–656. doi: 10.1242/dev.068353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awad SS, Lightowlers RN, Young C, Chrzanowska-Lightowlers ZM, Lomo T, Slater CR. Sodium channel mRNAs at the neuromuscular junction: distinct patterns of accumulation and effects of muscle activity. The J Neurosci. 2001;21:8456–8463. doi: 10.1523/JNEUROSCI.21-21-08456.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biancalana V, Beggs AH, Das S, Jungbluth H, Kress W, Nishino I, North K, Romero NB, Laporte J. Clinical utility gene card for: Centronuclear and myotubular myopathies. Eur J Hum Genet. 2012;20:1101. doi: 10.1038/ejhg.2012.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blau HM, Chiu C-P, Webster C. Cytoplasmic activation of human nuclear genes in stable heterocaryons. Cell. 1983;32:1171–1180. doi: 10.1016/0092-8674(83)90300-8. [DOI] [PubMed] [Google Scholar]
- Böhm J, Thavaraja R, Giehler S, Nalaskowski MM. A set of enhanced green fluorescent protein concatemers for quantitative determination of nuclear localization signal strength. Anal Biochem. 2017;533:48–55. doi: 10.1016/j.ab.2017.06.015. [DOI] [PubMed] [Google Scholar]
- Brendza RP, Serbus LR, Duffy JB, Saxton WM. Afunction for kinesin I in the posterior transport of oskar mRNA and Staufen protein. Science. 2000;289:2120–2122. doi: 10.1126/science.289.5487.2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruusgaard JC, Liestøl K, Ekmark M, Kollstad K, Gundersen K. Number and spatial distribution of nuclei in the muscle fibres of normal mice studied in vivo. J Physiol. 2003;551:467–478. doi: 10.1113/jphysiol.2003.045328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugnard E, Zaal KJM, Ralston E. Reorganization of microtubule nucleation during muscle differentiation. Cell Motility. 2005;60:1–13. doi: 10.1002/cm.20042. [DOI] [PubMed] [Google Scholar]
- Buxbaum AR, Wu B, Singer RH. Single β-actin mRNA detection in neurons reveals a mechanism for regulating its translatability. Science. 2014;343:419–422. doi: 10.1126/science.1242939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadot B, Gache V, Vasyutina E, Falcone S, Birchmeier C, Gomes ER. Nuclear movement during myotube formation is microtubule and dynein dependent and is regulated by Cdc42, Par6 and Par3. EMBO Rep. 2012;13:741–749. doi: 10.1038/embor.2012.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Can T, Faas L, Ashford DA, Dowle A, Thomas J, O’Toole P, Blanco G. Proteomic analysis of laser capture microscopy purified myotendinous junction regions from muscle sections. Proteome Sci. 2014;12:25. doi: 10.1186/1477-5956-12-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cioni J-M, Koppers M, Holt CE. Molecular control of local translation in axon development and maintenance. Curr Opin Neurobiol. 2018;51:86–94. doi: 10.1016/j.conb.2018.02.025. [DOI] [PubMed] [Google Scholar]
- Cretoiu D, Pavelescu L, Duica F, Radu M, Suciu N, Cretoiu SM. Myofibers. In: Xiao J, editor. Muscle Atrophy. Springer; 2018. pp. 23–46. [DOI] [PubMed] [Google Scholar]
- Cui XA, Palazzo AF. Visualization of endoplasmic reticulum localized mRNAs in mammalian cells. J Vis Exp. 2012;70:e50066. doi: 10.3791/50066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui XA, Zhang H, Ilan L, Liu AX, Kharchuk I, Palazzo AF. mRNA encoding Sec61β, a tail-anchored protein, is localized on the endoplasmic reticulum. J Cell Sci. 2015;128:3398–3410. doi: 10.1242/jcs.168583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dix DJ, Eisenberg BR. Myosin mRNA accumulation and myofibrillogenesis at the myotendinous junction of stretched muscle fibers. J Cell Biol. 1990;111:1885–1894. doi: 10.1083/jcb.111.5.1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dos Santos M, Backer S, Saintpierre B, Izac B, Andrieu M, Letourneur F, Relaix F, Sotiropoulos A, Maire P. Single-nucleus RNA-seq and FISH identify coordinated transcriptional activity in mammalian myofibers. Nat Commun. 2020;11:5102. doi: 10.1038/s41467-020-18789-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duclert A, Changeux JP. Acetylcholine receptor gene expression at the developing neuromuscular junction. Physiol Rev. 1995;75:339–368. doi: 10.1152/physrev.1995.75.2.339. [DOI] [PubMed] [Google Scholar]
- Duncan JE, Warrior R. The cytoplasmic dynein and kinesin motors have interdependent roles in patterning the Drosophila oocyte. Curr Biol. 2002;12:1982–1991. doi: 10.1016/S0960-9822(02)01303-9. [DOI] [PubMed] [Google Scholar]
- Dundon SER, Chang S-S, Kumar A, Occhipinti P, Shroff H, Roper M, Gladfelter AS. Clustered nuclei maintain autonomy and nucleocytoplasmic ratio control in a syncytium. Mol Biol Cell. 2016;27:2000–2007. doi: 10.1091/mbc.E16-02-0129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Englander LL, Rubin LL. Acetylcholine receptor clustering and nuclear movement in muscle fibers in culture. J Cell Biol. 1987;104:87–95. doi: 10.1083/jcb.104.1.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falcone S, Roman W, Hnia K, Gache V, Didier N, Lainé J, Auradé F, Marty I, Nishino I, Charlet-Berguerand N, et al. N-WASP is required for Amphiphysin-2/BIN1-dependent nuclear positioning and triad organization in skeletal muscle and is involved in the pathophysiology of centronuclear myopathy. EMBO Mol Med. 2014;6:1455–1475. doi: 10.15252/emmm.201404436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulton AB, Alftine C. Organization of protein and mRNA for titin and other myofibril components during myofibrillogenesis in cultured chicken skeletal muscle. Cell Struct Funct. 1997;22:51–58. doi: 10.1247/csf.22.51. [DOI] [PubMed] [Google Scholar]
- Gimpel P, Lee YL, Sobota RM, Calvi A, Koullourou V, Patel R, Mamchaoui K, Nédélec F, Shackleton S, Schmoranzer J, et al. Nesprin-1α-dependent microtubule nucleation from the nuclear envelope via Akap450 is necessary for nuclear positioning in muscle cells. Curr Biol. 2017;27:2999–3009.:e9. doi: 10.1016/j.cub.2017.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenblatt EJ, Spradling AC. Fragile X mental retardation 1 gene enhances the translation of large autism-related proteins. Science. 2018;361:709–712. doi: 10.1126/science.aas9963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall ZW, Ralston E. Nuclear domains in muscle cells. Cell. 1989;59:771–772. doi: 10.1016/0092-8674(89)90597-7. [DOI] [PubMed] [Google Scholar]
- Herbert SP, Costa G. Sending messages in moving cells: mRNA localization and the regulation of cell migration. Essays Biochem. 2019;63:595–606. doi: 10.1042/EBC20190009. [DOI] [PubMed] [Google Scholar]
- Holt CE, Bullock SL. Subcellular mRNA localization in animal cells and why it matters. Science. 2009;326:1212–1216. doi: 10.1126/science.1176488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt CE, Schuman EM. The central Dogma decentralized: new perspectives on RNA function and local translation in neurons. Neuron. 2013;80:648–657. doi: 10.1016/j.neuron.2013.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagannathan S, Reid DW, Cox AH, Nicchitta CV. De novo translation initiation on membrane-bound ribosomes as a mechanism for localization of cytosolic protein mRNAs to the endoplasmic reticulum. RNA (NewYork, NY) 2014;20:1489–1498. doi: 10.1261/rna.045526.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Januschke J, Gervais L, Dass S, Kaltschmidt JA, Lopez-Schier H, St Johnston D, Brand AH, Roth S, Guichet A. Polar transport in the Drosophila oocyte requires Dynein and Kinesin I cooperation. Curr Biol. 2002;12:1971–1981. doi: 10.1016/S0960-9822(02)01302-7. [DOI] [PubMed] [Google Scholar]
- Jungbluth H, Wallgren-Pettersson C, Laporte J. Centronuclear (myotubular) myopathy. Orphanet J Rare Dis. 2008;3:26. doi: 10.1186/1750-1172-3-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kann AP, Krauss RS. Multiplexed RNAscope and immunofluorescence on whole-mount skeletal myofibers and their associated stem cells. Development. 2019;146:dev179259. doi: 10.1242/dev.179259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khong A, Matheny T, Jain S, Mitchell SF, Wheeler JR, Parker R. The stress granule transcriptome reveals principles of mRNA accumulation in stress granules. Mol Cell. 2017;68:808–820.:e5. doi: 10.1016/j.molcel.2017.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M, Franke V, Brandt B, Lowenstein ED, Schöwel V, Spuler S, Akalin A, Birchmeier C. Single-nucleus transcriptomics reveals functional compartmentalization in syncytial skeletal muscle cells. Nat Commun. 2020;11:6375. doi: 10.1038/s41467-020-20064-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krauss J, López de Quinto S, Nüsslein-Volhard C, Ephrussi A. Myosin-V regulates oskar mRNA localization in the Drosophila oocyte. Curr Biol. 2009;19:1058–1063. doi: 10.1016/j.cub.2009.04.062. [DOI] [PubMed] [Google Scholar]
- Kugler J-M, Lasko P. Localization, anchoring and translational control of oskar, gurken, bicoid and nanos mRNA during Drosophila oogenesis. Fly. 2009;3:15–28. doi: 10.4161/fly.3.1.7751. [DOI] [PubMed] [Google Scholar]
- Lee CH, Zhang H, Baker AE, Occhipinti P, Borsuk ME, Gladfelter AS. Protein aggregation behavior regulates cyclin transcript localization and cell-cycle control. Dev Cell. 2013;25:572–584. doi: 10.1016/j.devcel.2013.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma B, Savas JN, Yu M-S, Culver BP, Chao MV, Tanese N. Huntingtin mediates dendritic transport of β-actin mRNA in rat neurons. Sci Rep. 2011;1:140. doi: 10.1038/srep00140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDougall N, Clark A, MacDougall E, Davis I. Drosophila gurken (TGFα) mRNA localizes as particles that move within the oocyte in two dynein-dependent steps. Dev Cell. 2003;4:307–319. doi: 10.1016/S1534-5807(03)00058-3. [DOI] [PubMed] [Google Scholar]
- Mamchaoui K, Trollet C, Bigot A, Negroni E, Chaouch S, Wolff A, Kandalla PK, Marie S, Di Santo J, StGuily JL, et al. Immortalized pathological human myoblasts: towards a universal tool for the study of neuromuscular disorders. Skeletal Muscle. 2011;1:34. doi: 10.1186/2044-5040-1-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merlie JP, Sanes JR. Concentration of acetylcholine receptor mRNA in synaptic regions of adult muscle fibres. Nature. 1985;317:66–68. doi: 10.1038/317066a0. [DOI] [PubMed] [Google Scholar]
- Metzger T, Gache V, Xu M, Cadot B, Folker ES, Richardson BE, Gomes ER, Baylies MK. MAP and kinesin-dependent nuclear positioning is required for skeletal muscle function. Nature. 2012;484:120–124. doi: 10.1038/nature10914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer LC, Wright NT. Structure of giant muscle proteins. Frontier Physiol. 2013;4:368. doi: 10.3389/fphys.2013.00368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mofatteh M, Bullock SL. SnapShot: subcellular mRNA localization. Cell. 2017;169:178.:e1. doi: 10.1016/j.cell.2017.03.004. [DOI] [PubMed] [Google Scholar]
- Moon SL, Morisaki T, Khong A, Lyon K, Parker R, Stasevich TJ. Multicolour single-molecule tracking of mRNA interactions with RNP granules. Nat Cell Biol. 2019;21:162–168. doi: 10.1038/s41556-018-0263-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moor AE, Golan M, Massasa EE, Lemze D, Weizman T, Shenhav R, Baydatch S, Mizrahi O, Winkler R, Golani O, et al. Global mRNA polarization regulates translation efficiency in the intestinal epithelium. Science. 2017;357:1299–1303. doi: 10.1126/science.aan2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nevalainen M, Kaakinen M, Metsikkö K. Distribution of mRNA transcripts and translation activity in skeletal myofibers. Cell Tissue Res. 2013;353:539–548. doi: 10.1007/s00441-013-1659-x. [DOI] [PubMed] [Google Scholar]
- Newlands S, Levitt LK, Robinson CS, Karpf ABC, Hodgson VRM, Wade RP, Hardeman EC. Transcription occurs in pulses in muscle fibers. Genes Dev. 1998;12:2748–2758. doi: 10.1101/gad.12.17.2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nissinen M, Kaisto T, Salmela P, Peltonen J, Metsikko K. Restricted distribution ofmRNAs encoding a sarcoplasmic reticulum or transverse tubule protein in skeletal myofibers. J Histochem Cytochem. 2005;53:217–227. doi: 10.1369/jhc.4A6431.2005. [DOI] [PubMed] [Google Scholar]
- Ovesný M, Křížek P, Borkovec J, Švindrych Z, Hagen GM. ThunderSTORM: a comprehensive ImageJ plug-in for PALM and STORM data analysis and super-resolution imaging. Bioinformatics. 2014;30:2389–2390. doi: 10.1093/bioinformatics/btu202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padovan-Merhar O, Nair GP, Biaesch AG, Mayer A, Scarfone S, Foley SW, Wu AR, Churchman LS, Singh A, Raj A. Single mammalian cells compensate for differences in cellular volume and DNA copy number through independent global transcriptional mechanisms. Mol Cell. 2015;58:339–352. doi: 10.1016/j.molcel.2015.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park HY, Lim H, Yoon YJ, Follenzi A, Nwokafor C, Lopez-Jones M, Meng X, Singer RH. Visualization of dynamics of single endogenous mRNA labeled in live mouse. Science. 2014;343:422–424. doi: 10.1126/science.1239200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlath GK, Rich K, Webster SG, Blau HM. Localization of muscle gene products in nuclear domains. Nature. 1989;337:570–573. doi: 10.1038/337570a0. [DOI] [PubMed] [Google Scholar]
- Petrany MJ, Swoboda CO, Sun C, Chetal K, Chen X, Weirauch MT, Salomonis N, Millay DP. Single-nucleus RNA-seq identifies transcriptional heterogeneity in multinucleated skeletal myofibers. Nat Commun. 2020;11 doi: 10.1038/s41467-020-20063-w. 6374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pimentel MR, Falcone S, Cadot B, Gomes ER. In Vitro Differentiation of Mature Myofibers for Live Imaging. J Vis Exp. 2017;119:e55141. doi: 10.3791/55141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralston E, McLaren RS, Horowitz JA. Nuclear domains in skeletal myotubes: the localization of transferrin receptor mrna is independent of its half-life and restricted by binding to ribosomes. Exp Cell Res. 1997;236:453–462. doi: 10.1006/excr.1997.3753. [DOI] [PubMed] [Google Scholar]
- Roman W, Martins JP, Carvalho FA, Voituriez R, Abella JVG, Santos NC, Cadot B, Way M, Gomes ER. Myofibril contraction and crosslinking drive nuclear movement to the periphery of skeletal muscle. Nat Cell Biol. 2017;19:1189–1201. doi: 10.1038/ncb3605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roman W, Martins JP, Gomes ER. Local arrangement of fibronectin by myofibroblasts governs peripheral nuclear positioning in muscle cells. Dev Cell. 2018;46:102–111.:e6. doi: 10.1016/j.devcel.2018.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenblatt JD, Lunt AI, Parry DJ, Partridge TA. Culturing satellite cells from living single muscle fiber explants. In Vitro Cell Dev Biol Animal. 1995;31:773–779. doi: 10.1007/BF02634119. [DOI] [PubMed] [Google Scholar]
- Salogiannis J, Reck-Peterson SL. Hitchhiking: a non-canonical mode of microtubule-based transport. Trends Cell Biol. 2017;27:141–150. doi: 10.1016/j.tcb.2016.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanes JR, Lichtman JW. Development of the vertebrate neuromuscular junction. Annu Rev Neurosci. 1999;22:389–442. doi: 10.1146/annurev.neuro.22.1.389. [DOI] [PubMed] [Google Scholar]
- Scarborough EA, Uchida K, Vogel M, Erlitzki N, Iyer M, Phyo SA, Bogush A, Kehat I, Prosser BL. Microtubules orchestrate local translation to enable cardiac growth. Nat Commun. 2021;12 doi: 10.1038/s41467-021-21685-4. 1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tassin AM, Maro B, Bornens M. Fate of microtubule-organizing centers during myogenesis in vitro. J Cell Biol. 1985;100:35–46. doi: 10.1083/jcb.100.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomancak P, Guichet A, Zavorszky P, Ephrussi A. Oocyte polarity depends on regulation of gurken by Vasa. Development. 1998;125:1723–1732. doi: 10.1242/dev.125.9.1723. [DOI] [PubMed] [Google Scholar]
- Vispe S, DeVries L, Créancier L, Besse J, Bréand S, Hobson DJ, Svejstrup JQ, Annereau J-P, Cussac D, Dumontet C, et al. Triptolide is an inhibitor of RNA polymerase I and II-dependent transcription leading predominantly to down-regulation of short-lived mRNA. Mol Cancer Ther. 2009;8:2780–2790. doi: 10.1158/1535-7163.MCT-09-0549. [DOI] [PubMed] [Google Scholar]
- Voigt F, Zhang H, Cui XA, Triebold D, Liu AX, Eglinger J, Lee ES, Chao JA, Palazzo AF. Single-Molecule Quantification of Translation-Dependent Association of mRNAs with the Endoplasmic Reticulum. Cell Reports. 2017;21:3740–3753. doi: 10.1016/j.celrep.2017.12.008. [DOI] [PubMed] [Google Scholar]
- Wu X, Kodama A, Fuchs E. ACF7 regulates cytoskeletal-focal adhesion dynamics and migration and has ATPase activity. Cell. 2008;135:137–148. doi: 10.1016/j.cell.2008.07.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagishi M, Shirasaki Y, Funatsu T. Size-dependent accumulation of mRNA at the leading edge of chicken embryo fibroblasts. Biochem Biophys Res Commun. 2009;390:750–754. doi: 10.1016/j.bbrc.2009.10.043. [DOI] [PubMed] [Google Scholar]
- Zappulo A, van den Bruck D, Ciolli Mattioli C, Franke V, Imami K, McShane E, Moreno-Estelles M, Calviello L, Filipchyk A, Peguero-Sanchez E, et al. RNA localization is a key determinant of neurite-enriched proteome. Nat Commun. 2017;8:583. doi: 10.1038/s41467-017-00690-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Xu R, Zhu B, Yang X, Ding X, Duan S, Xu T, Zhuang Y, Han M. Syne-1 and Syne-2 play crucial roles in myonuclear anchorage and motor neuron innervation. Development. 2007;134:901–908. doi: 10.1242/dev.02783. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.