Abstract
Neoadjuvant chemotherapy (NACT) may stimulate anti-cancer adaptive immune responses in high-grade serous ovarian cancer (HGSOC), but little is known about effects on innate immunity. Using omental biopsies from HGSOC, and omental tumors from orthotopic mouse HGSOC models that replicate the human tumor microenvironment, we studied the impact of platinum-based NACT on tumor-associated macrophages (TAMs). We found that chemotherapy reduces markers associated with alternative macrophage activation, while increasing expression of pro-inflammatory pathways, with evidence of inflammasome activation. Further evidence of a shift in TAM functions came from macrophage depletion via CSF1R inhibitors (CSF1Ri) in the mouse models. Although macrophage depletion in established disease had no impact on tumor weight or survival, CSF1Ri treatment after chemotherapy significantly decreased disease-free and overall survival. This decrease in survival was accompanied by significant inhibition of adaptive immune response pathways in the tumors. We conclude that chemotherapy skews the TAM population in HSGOC towards an anti-tumor phenotype that may aid adaptive immune responses, and therapies that enhance or sustain this during remission may delay relapse.
Introduction
Women with disseminated high-grade serous ovarian cancer (HGSOC) may be treated with neoadjuvant chemotherapy (NACT) before surgery, or receive upfront cytoreductive surgery followed by adjuvant chemotherapy. Because there is evidence that chemotherapy can stimulate host anti-tumor responses (1), we, and others, have used human tissues and blood samples to study this in a clinically relevant context. The known effects of NACT on the ovarian tumor microenvironment (TME) to-date can be summarized as follows: T-cell activation may be enhanced; T-cell, B-cell, and NK-cell densities can increase, whereas T regulatory cell (Treg) density decreases (2–5). Because a majority of HGSOC patients fail to respond to current immunotherapies (6,7), in spite prognostic correlation between T-cell densities and survival (4,8,9), novel immunotherapies that augment chemotherapy-induced immune stimulation could delay relapse, increase survival, and reduce the need for chemotherapy after surgical debulking. However, these studies have not investigated the effects of NACT on the abundant myeloid cell populations found in HGSOC that may provide novel targets after NACT and surgery. We previously reported that NACT leads to significant decreases in plasma of tumor-promoting inflammatory cytokines such as TNF, IL6, and IL8, with a concomitant increase in circulating IFNγ (3), but we did not investigate local changes in inflammatory cells such as tumor-associated macrophages (TAMs). Although TAMs are generally considered as tumor-promoting and immunosuppressive, they may have several phenotypic and functional states, some of which may aid anti-tumor immune responses (10).
The role of TAMs in HGSOC is not fully defined. There is no prognostic association with TAM density in biopsies as measured by the pan-macrophage marker CD68 (11), but higher densities of alternatively activated CD163+ macrophages have negative correlations (12). These results could be explained by our previous study of the pan-macrophage marker CD68 in stromal and malignant cell areas in pre-treatment primary tumor biopsies from 152 HGSOC patients (9). We found that overall CD68+ density does not have any association with patient survival; however, high stromal CD68+ density was significantly predictive of improved overall survival (9), suggesting that some TAM populations may aid anti-tumor immunity in HGSOC.
The aim of the work reported here, therefore, was to ask if NACT could modulate TAM populations in HGSOC. To understand how to exploit any immune activating effects of NACT, we also needed mouse models that recapitulate the molecular and cellular features of HGSOC. We previously described orthotopic syngeneic mouse models of HGSOC that show significant correlations with the TME of human tumors, in terms of their transcriptomes, host cell immune infiltrates, matrisome, vasculature, and tissue modulus (13). In this paper, we compared the effects of NACT in patient omental biopsies with the effects of chemotherapy on omental tumors in mouse models. We then asked if inhibiting TAM populations in the mouse models during remission, resulting from chemotherapy, would impact subsequent relapse. We conclude that chemotherapy in patients and mouse models decreases overall TAM populations, but those TAMs that remain may foster anti-tumor responses.
Materials and methods
Patients and samples
Tissues surplus to diagnostic and therapeutic requirements were collected along with clinical data under the Barts Gynae Tissue Bank HTA license number 12199 (REC no: 10/H0304/14 and 15/EE/0151). Patients gave written informed consent, and the study was approved by the East of England Cambridge UK national review board. Studies were conducted in accordance with the Declaration of Helsinki and the International Ethical Guidelines for Biomedical Research Involving Human Subjects. Samples were kindly donated by high-grade serous ovarian cancer (HGSOC) patients undergoing surgery at Barts Health NHS Trust and St. George’s University Hospitals NHS Foundation Trust. Omental metastases were collected prospectively before and after neoadjuvant chemotherapy (NACT) to include samples from stage 3/4 patients. Patients were treated with carboplatin (area under the curve [AUC] 5 mg per millilitre per minute) intravenously every three weekes. Paclitaxel standard treatment was at a dose of 175 mg/m2 intraveously every three weeks. Surgery was usually performed between 3 and 4 weeks after the last NACT. Human omental specimens (pre- and post-chemotherapy) were processed for flow cytometry, cell sorting, or immunohistochemistry within 5-7 hours from sample resection and were stored between 0-4˚C in PBS until processing.
Immunohistochemistry (IHC)
For the analysis of CD68 in tumors and stomal areas, the post-chemotherapy samples from interval cytoreduction surgery were contained in a tissue microarray (TMA)(3), whereas pre-chemotherapy samples were sections from diagnostic percutaneous core omental biopsies. This cohort consisted of twenty-six matched pre and post-NACT biopsy samples. Patient samples (pre- and post-NACT) were fixed in 10% formalin solution (HT501128, Sigma-Aldrich) and paraffin-embedded by the Pathology Department, Barts Cancer Institute. Sections were dewaxed, dehydrated, and incubated in Antigen Unmasking Solution (H-3300, Vector Laboratories) at 100˚C in a microwave for 20 minutes. Sections were incubated in 0.3% H2O2 in methanol for 10 minutes and in blocking buffer [2.5% bovine serum albumin (BSA; A4503, Sigma-Aldrich) and 2.5% goat serum (B15-035, PAA) in PBS] for 60 minutes at room temperature. Primary antibodies were applied in blocking buffer for 60 minutes at room temperature: CD68 (1:8000, M0814, Dako), PAX-8 (1:400, NBP1-32440 Novus), TREML4 (1:1100, ab204798, Abcam), cleaved caspase-3 (1:100, 9664S, Cell Signalling), F4/80 (1:100, MCA497, BioRad), and CD206 (1:100, MCA2235, BioRad). Washes were performed with PBS with 0.1% Tween-20 (PBST). Sections were incubated with Vector Impact Kit (H-4343, MP-63636, Vector Laboratories) for 1 hour at room temperature and washed three times in PBST before addition of DAB chromogen and counterstained with haematoxylin (GHS116, Sigma-Aldrich) for 20 seconds. Sections were imaged using the Panoramic digital slide scanner (3DHISTECH). Definiens® digital analysis software was used to quantify staining within cancer cell islands and stroma. The software-determined tumor and stromal separation for the entire tissue sample was reviewed with the oversight of Consultant Gynaecological Pathologist, Dr. Jacqueline McDermott (St. George’s University Hospitals NHS Foundation Trust). Size and number of human macrophage lakes across the entire tissue section, defining them as areas of continuous CD68+ staining >4000 μm2, 10,000 μm2, or >50,000 μm2 was measured in unmatched biopsies from nine patients pre- and twenty-three patients post-chemotherapy. For mouse sections, tumors analyzed for the presence of lakes using QuPath image analysis software (14). Total tumor area and individual lakes were manually annotated.
In vitro cell lines and macrophages
Human HGSOC cell lines were derived in our laboratory (G164, established in our laboratory from an omental metastasis of a patient with HGSOC), or kindly given by Professor David Bowtell (Peter MacCallum Cancer Centre, Melbourne, Australia; AOCS1, received in 2011). Both cell lines are described in (15) and were cultivated in in DMEM/F12 with Glutamax (cat.no 31331-093, Gibco) containing pen/strep (100 μg/mL; 15140-122, Gibco) supplemented with 4% human serum (H4522, Sigma-Aldrich) for G164 cells or 10% FBS (SV30160.03, lot. no. RXL35906, HyClone) and insulin, transferrin, sodium selenite (1X; 51300-044, Gibco) for AOCS1 cells. Cells were authenticated by STR sequencing with ATCC (135-XV) at the beginning and at the end of the project. 60577 and 30200 cell lines were established from tumors in genetic models generated by adenoviral transfection of the ovarian surface epithelium in FVB mice and are Tp53-/-, Brca-1-/- with inactivation of the Rb pathway (16). HGS2 is Tp53-/-, Pten-/- and Brca-2-/- and was generated from tumors derived from a genetic model established by Perets et al (17) that we backcrossed to B6 mice (13). Mouse cell lines 30200, 60577 (16), and HGS2 (13) were grown in DMEM/F12 with Glutamax with 4% FBS, pen/strep (100 μg/mL; 15140-122, Gibco) and insulin, transferrin, sodium selenite (1X; 51300-044, Gibco), murine epidermal growth factor (EGF; 0.2 μg/mL; E4127, Sigma), hydrocortisone (0.5 μg/mL; H0135, Sigma), and antibiotic-antimycotic (1X; 15240-062, Gibco). Cell lines were used within 5-6 passages from thawing. Routine testing for mycoplasma contamination using the MycoAlert PLUS Mycoplasma Detection Kit (cat. no. LT07-710, Lonza) has been consistently negative.
Human blood from healthy donors was obtained from leucocyte cones from the NHS Blood and Transplant service. Peripheral blood mononuclear cells (PBMCs) were isolated using Ficoll-Paque PLUS (17-1440-03 AG, GE Healthcare). Monocytes were isolated from PBMCs by magnetic isolation with CD14 microbeads (130-050-201, Miltenyi) on LS columns (130-042-401, Miltenyi) according to manufacturer’s instructions. Monocytes were differentiated to macrophages for seven days with M-CSF (100 ng/mL; 574806, Biolegend). Macrophages were polarized with IL4 (20 ng/mL; 200-04 Peprotech) and IL10 (20 ng/mL; 200-10, Peprotech) or IFNγ (10 ng/mL; 300-02 Peprotech), LPS (100 ng/mL; L2630, Sigma Aldrich) in the presence of M-CSF (100 ng/mL) for 72 hours.
Human and mouse HGSOC cell lines and human macrophages were stimulated with carboplatin (Hospira) or paclitaxel (Hospira), obtained from St. Bartholomew’s Hospital pharmacy, for 48 hours at the concentrations indicated in the legends. As a positive control for inflammasome activation, human macrophages were stimulated for 48 hours with LPS (200 ng/mL; LPS from E. coli O111:B4, tlrl-eblps, Invivogen) and nigericin (7.5 μM; tlrl-nig, Invivogen).
Viability assays
Human and mouse HGSOC cells were fixed after stimulation and stained in 70% ethanol (51976, Sigma-Aldrich) + 0.5% crystal violet (C0775, Sigma-Aldrich). Crystal violet was dissolved in 10% acetic acid (W200603, Sigma-Aldrich), and the optical density at 595nm (OD595) was quantified. Measurements were normalized to media only control analyzed in Prism v7.0. Human monocyte-derived macrophages were plated in 6-well plates at a density of 1×106 cells/well and stimulated with carboplatin (Hospira) or paclitaxel (Hospira) for 48 hours at the concentrations indicated in the legends. Cells were detatched using Cell Dissociation Buffer (Gibco, 13151014). Cells were washed and stained with FVD450 (1:500; 65-0863-14, eBioscience) for 25 minutes at 4˚C. After fixation, cells were analysed on a LSR Fortessa II flow cytometer (BD Biosciences). The results were analyzed using FlowJo v10.2 (Treestar Inc.). Measurements were normalized to media only control and analyzed in Prism v7.0.
Mouse experiments
Mouse experiments were performed under the licence PBE3719B3 in accordance with Animals (Scientific Procedures) Act 1986 and with the approval of our Institutional Ethics committee. Seven week-old female FVB and six week-old C57BL/6 mice were purchased from Charles River (UK). Mice received 1×107 cells (60577, 30200 or HGS2) injected intraeritoneally (i.p.) in 300 μL PBS. Mice were treated with carboplatin (20 mg/kg), paclitaxel (10mg/kg), or the combination of carboplatin (20 mg/kg, Hospira) + paclitaxel (10 mg/kg, Hospira), both from the pharmacy at St. Bartholomew’s Hospital, London. Carboplatin and paclitaxel were administered to mice i.p. in 300 μL volume starting 21 days (60577), 84 days (30200), or 49 days (HGS2) post-cell injection. Vehicle-treated controls received either 0.9% NaCl or cremophor (C5135, Sigma-Aldrich): ethanol (51976, Sigma-Aldrich) (1:1) dissolved in 0.9% NaCl. Mice were assessed daily and weighed twice weekly. The survival endpoint for mice was defined as a change in general health; specifically, 15% body weight loss over 72 hours or 20% over any time period, inability to ambulate, hunched posture, or difficulty breathing, as well as signs of ascites or palpable tumors exceeding an estimated size of 1.2 cm diameter. In survival experiments, assessment of mice was made by the same individual to limit inter-observer variability. In the majority of cases, survival determinations were made by a trained animal technician who was not directly involved in the experimental design. AZD7507 was obtained from AstraZeneca and was dissolved in 0.5% (w/v) methyl cellulose (M7027, Sigma-Aldrich) and 0.1% (v/v) Tween-80 (P4780, Sigma-Aldrich) in water and administered at 100 mg/kg twice daily by oral gavage, 5 days/week in the 60577 and 30200 model, starting as indicated in the figures and legends. BLZ945 (HY-12768, MedChemTronica) was dissolved in 10% sulfobutylether-b-cyclodextrin (HY-17031, MedChemTronica) in water and administered at 200 mg/kg once daily by oral gavage, 5 days/week in the 60577 model, starting at 70 days post-injection for 63 days.
Flow cytometry and cell sorting
Samples were minced and incubated with collagenase D (1 mg/mL; 11088866001, Roche) and DNAse I (25 mg/mL; D4513, Sigma-Aldrich) in 5% FBS RPMI (R8758, Sigma-Aldrich) media for 30 minutes under agitation at 37˚C, filtered through a 70μm strainer (352350, Falcon), followed by red blood cell lysis (555899, BD Biosciences) for 5 minutes. Mouse omental samples were digested in HBSS (9374543, Gibco) supplemented with collagenase (2 mg/mL; C9263, Sigma) and DNAase I for 20 minutes at 37˚C and filtered through a 70μm cell strainer. Cells were washed and resuspended in FACS buffer (PBS + 2% heat-inactivated FBS + 2 mM EDTA) and blocked for 15 minutes in Trustain blocker (1:200; 101319, Biolegend). The cell suspension was stained in FACS buffer for 30 minutes at 4˚C using the antibodies specified in Supplementary Table S1. Mouse and human cells were washed and stained with FVD450 or FVD506 (1:500; 65-0863- 14, 65-0866-14, eBioscience) for 25 minutes at 4˚C. After fixation in 1:1 10% formalin:FACS buffer for 10 minutes at 4˚C, fixative was washed out and cells were analysed on a LSR Fortessa II flow cytometer (BD Biosciences). The results were analyzed using FlowJo v10.2 (Treestar Inc.). Mean fluorescence intensity (MFI) was normalized according to this formula: SI=[MFI(positive)-MFI(negative)]/[2*SD(negative)]. SI=staining index, SD=standard deviation.
For sorting, dissociated cells were stained in FACS buffer for 30 minutes at 4˚C. Cells were stained using the lineage antibodies listed for flow cytometry diluted 1:100. Cells were stained with DAPI (2.5 μg/mL; 422801, Biolegend) immediately prior to sorting. FITC-HLA-DR+CD14+ cells were sorted using a FACSAria II cell sorter (BD Bioscience), lysed in RLT buffer (1015750, Qiagen) with 1% β-mecaptoethanol (M-6250, Sigma-Aldrich) and stored at -80˚C. 0.25-2×106 cells were sorted per sample, and cell purities of >96% were obtained.
RNA extraction and sequencing
Mouse tumor RNA was isolated from frozen omental tumors from the 60577 model, treated with and without AZD7507 after carboplatin, collected at end-point, as decribed in (13) with RNeasy Mini Kit (74104, Quiagen) according to manufacturer’s instructions. Three vehicle and three AZD7507 treated tumors were processed and analyzed. Human macrophage RNA was extracted from the sorted TAM population (HLA-DR+CD14+Lin- cells) using RNeasy Micro Kit (74004, Qiagen) according to manufacturer instructions. Five pre- and seven post-NACT unmatched samples were processed and analysed. RNA purity and integrity were assessed using a Nanodrop and an RNA NanoChip (5067-1511, Agilent) and RIN (RNA integrity number) values were ≥5.6 for mouse RNA and ≥8.6 for human TAM RNA.
RNA sequencing (RNA-seq) was carried out at the Wellcome Trust Centre for Human Genetics. Library preparation was carried out using ribosomal depletion, and sequencing was performed with HiSeq4000 for human samples and NovaSeq6000 for mouse samples, with 150bp read-length, paired-end, strand specific and a coverage of, on average, 54 million reads for human samples and 68 million for mouse. Raw reads were aligned to the reference genome GRCh37 (hg19) or GRCm38 (mm10). The number of reads aligned to the exonic region of each gene were counted using htseq-count (18) based on the Ensembl annotation. Only genes that achieved at least one read count per million reads (CPM) in at least 25% of the samples were kept. This led to 16,253 filtered genes, in total, for human and 16,344 for mouse. Conditional quantile normalization (cqn)(19) was performed counting for gene length and GC content, and log2 transformed RPKM expression matrices were generated. RNA-seq data have been deposited in Gene Expression Omnibus (GEO) under the accession numbers GSE158739 (human data) and GSE158812.
Differential gene expression and pathway analysis
Differential gene expression analysis was performed using edgeR and limma R packages (20,21). For human samples, the generalized linear model with a 0+group+batch design was used. For mouse samples, voom normalization and the linear model with a 0+group design was used. Gene set enrichment analysis (GSEA) was performed using the publicly available bioinformatics platform GenePattern (https://cloud.genepattern.org/, v3.9.9 and v3.9.11)(22) with GSEA pre-ranked (23) for Canonical Pathways (c2.cp.v6.2.symbols.gmt for human; c2.cp.v7.0.symbols.gmt for mouse data), and Gene Ontology Biological Processes (c5.go.bp.v7.2.symbols.gmt for human; c5.bp.v7.0.symbols.gmt for mouse data). Single sample GSEA (Fig. 7A) was performed using the R package GSVA (24).
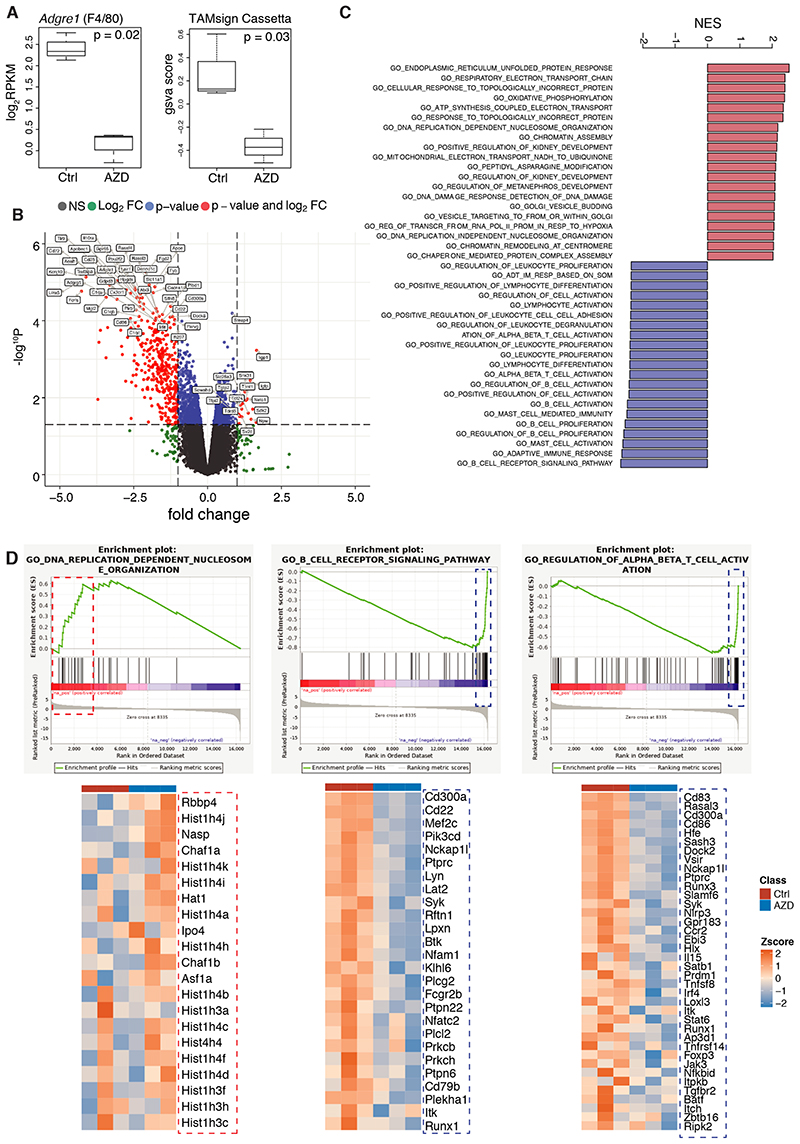
Figure 7. Transcriptomics of in vivo AZD7507-treated ovarian mouse model tumors.
(A) Boxplots illustrating expression of Adgre1 and the GSVA scores of TAM signature (24) in control (Ctrl) and AZD7507 (AZD)-treated samples. p values correspond to Bonferroni-Hochberg adjusted p value and Student’s t-test, respectively; n=3 per group. Boxplots illustrate median with interquartile range (first and third quartile) with whiskers indicating the minimum and maximum. (B) Volcano plot showing significantly differentially expressed genes in red (p<0.05); gene symbols of top differentially expressed genes are indicated. (C) Barplot illustrating top enriched canonical pathways in GSEA of post- vs pre-treatment (FDR<0.05). (D) GSEA enrichment plots of DNA replication-dependent nucleosome organization, B-cell receptor signaling pathway, regulation of T-cell activation, and heatmaps of the core enrichment genes for these pathways.
Western blotting
Human macrophages (stimulated as indicated in the “In vitro cell line and macrophage” section) were lysed with a RIPA buffer (R0278, Sigma-Alrich) containing 1:10 complete mini EDTA protease inhibitor (11836153001, Roche) and 1:100 phosphatase inhibitor cocktail (P5726, Sigma-Alrich). The protein concentration in the extracts was calculated using a BCA assay. Cell extracts (25 μg) were run on a NuPAGE Novex 4% to 12% Bis-Tris Gel (1.5 mm, NP0335BOX, Invitrogen) and transferred to a nylon membrane. The membrane was blocked overnight (4°C in PBS with 0.1% Tween and 5% milk powder) and probed using: cleaved caspase-3 (1:1000, 9664S, Cell signaling), gasdermin E (GSDME; 1:1000, ab215191, Abcam), and β-actin (1:2000, A1978, Sigma). A rabbit or mouse horseradish peroxidase-conjugated secondary antibody (NXA931, NA9340V, GE Healthcare) incubation allowed visualization using ECL Prime (RPN2232, GE Healthcare) or the Luminata Forte (WBLUF0500, Millipore) and the Amersham Imager 600 (GE Healthcare).
TUNEL (terminal deoxynucleotidyl transferase dUTP nick end labelling) assay
The Novus Biologicals APO-BRDU-IHC reagent kit (cat no. NBP2-31164, Novus Biologicals) was used in accordance with the manufacturer’s instructions following the staining protocol for paraffin embedded tissue. 4μm sections of FFPE tissue were cut by the Bart’s Cancer Institute Pathology Department. Human HGSOC sections post chemotherapy known to contain macrophage lakes were stained. Kit positive controls, in addition to a tissue positive control section, produced by incubating sections for 30 minutes with 1μg/mL DNAse in 1xPBS + 1 mM MgSO4, were stained. Sections were de-paraffinized in xylene (cat. no. X/0100/17, Fisher Scientific), rehydrated through an ethanol (cat. no. E/0650DF/17, Fisher Scientific) series (100%, 90%, 80%, 70%), and washed in PBS. Slides were then incubated with proteinase K solution for 20 minutes at room temperature. Slides were then washed in PBS, followed by endogenous peroxidase blocking by incubating slides with 3% hydrogen peroxide in methanol for 5 minutes at room temperature and then washing in PBS. Slides where then equilibrated in reaction buffer. Following addition of labelling reaction (reaction buffer, TdT enzyme, Br-dUTP and distilled water), the sections were incubated in a humidified chamber for 1.5 hours at 37˚C. The slides were washed in 1x PBS followed by incubation with Blocking buffer for 10 minutes at room temperature. Slides were then incubated with biotinylated BrDU antibody for 1.5 hours at room temperature, washed in PBS, and incubated with 1x conjugate solution for 30 minutes at room temperature. After washing in PBS, the slides were incubated with DAB for 15 minutes, and counterstained with methyl green. Slides were then mounted (cat. no. 06522, Sigma-Aldrich) and imaged on an Axiophot microscope (Zeiss).
ELISA
Ascites from the 60577 model at endpoint was collected and centrifuged in a refrigerated centrifuge at 1300 rpm for 5 minutes and stored at -20˚C. ELISAs for IL1ß were performed using the Mouse IL1ß Duoset (DY401, R&D) with undiluted ascites according to manufacturer’s instructions. Concentration was extrapolated from a standard curve. Abs450 was read using a BMG Labtech FLUOstar Optima reader (Labtech).
Proximity ligation assay (PLA)
Human macrophages were grown on coverslips and fixed in 10% formal saline (HT501128, Sigma-Aldrich) for 10 minutes at room temperature, washed with PBS, and stored at 4˚C. Tissue sections from paraffin-embedded tumors underwent antigen retrieval similarly to the immunohistochemistry protocol described. Samples were permeabilized in 0.5% Triton-X100 (T8787, Sigma-Aldrich.) in PBS for 10 minutes at room temperature. Specimens were then washed, blocked in 5% BSA (A4503, Sigma-Aldrich, or 1% BSA and 2% FCS for tissue) in PBS for 60 minutes at room temperature. Specimens were incubated with primary antibodies NLRP3 (1:50) and ASC (1:50)(AG-20B-0014-C100, AG-25B-0006-C100, AdipoGen) overnight at 4°C, followed by washing three times in 0.1% Triton-X100 in PBS for 10 minutes. Samples were then incubated with PLA probes (1:5; DUO92002, DUO92004, Sigma- Aldrich) for 60 minutes at room temperature (90 minutes for tissue slides), followed by washing three times with 0.1% triton-X100 for 5 minutes. Ligation reaction was applied at 37°C in a humidity chamber for 30 minutes (1 hour for tissue slides). After washing twice in 0.1% triton X100 for 3 minutes each, polymerization reaction was applied at 37°C in a humidity chamber for 100 minutes in the dark (overnight for tissue slides) and washed three times for 5 minutes. Samples were then stained with DAPI (1 μg/mL; 40043, Biotium) for 3 minutes (30 minutes for tissue slides), followed by three 5-minute washes in PBS and mounted using FluorSave reagent (345789, Calbiochem). Imaging was done on a LSM 710 confocal microscope. Approximately 30 cells were imaged using the 63x objective under oil immersion. LSM files were exported and analysed in Fiji v1.2/ImageJ software. For analysis of cells, regions of interest were drawn around each cell. The “find maxima” process was used to isolate the green specs individual points and counted. For the analysis of mouse tumors, 10 fields for each tumor were analyzed. Signal was thresholded to remove background, and the process “analyse particles” with a size=4-20 pixels was used to count. For the analysis of human tumors, 6-10 fields were analyzed and positive signal was counted manually.
Statistics
All data are expressed as the mean of the individual experiments ± the standard error of the mean (SEM) unless otherwise specified. Differences were considered significant at p < 0.05, using a Student’s t-test, ANOVA test followed by Bonferroni post-test, or non-parametric test as appropriate, performed with the statistical analysis software Graphpad Prism 7/8. P values are specified. If not specified, t-test or ANOVA were applied. For survival curves, Log-rank (Mantel-Cox) p value is shown.
Results
NACT changes TAM populations in HGSOC omental biopsies
We first used a cohort of twenty-six matched pre and post-NACT omental biopsy samples, for which, we had previously studied T- and B-cell infiltrates and activity (2,3). Using IHC, we stained the sections for the pan-macrophage marker CD68 and quantified staining within malignant cell islands and stromal areas. Because CD68+ cells often have multiple cytoplasmic extensions, we quantified CD68 staining as a percentage of the malignant cell area, the stromal area, or the overall TMA core/tissue area. Supplementary Fig. S1A shows a representative analysis of a single TMA core, illustrating tumor and stroma separation and CD68 staining. No difference in CD68+ area between pre-treatment and post-NACT biopsies was observed when the entire sample was analysed (Fig. 1A-B). However, NACT-treatment caused a significant reduction in the mean CD68+ area in the malignant cell regions (7.88% pre-treatment vs. 1.01% post-NACT p<0.0001). The stromal CD68+ area was increased after NACT in four of the patient samples (Fig. 1A–B), suggesting that CD68+ cells may not be uniformly distributed after treatment.
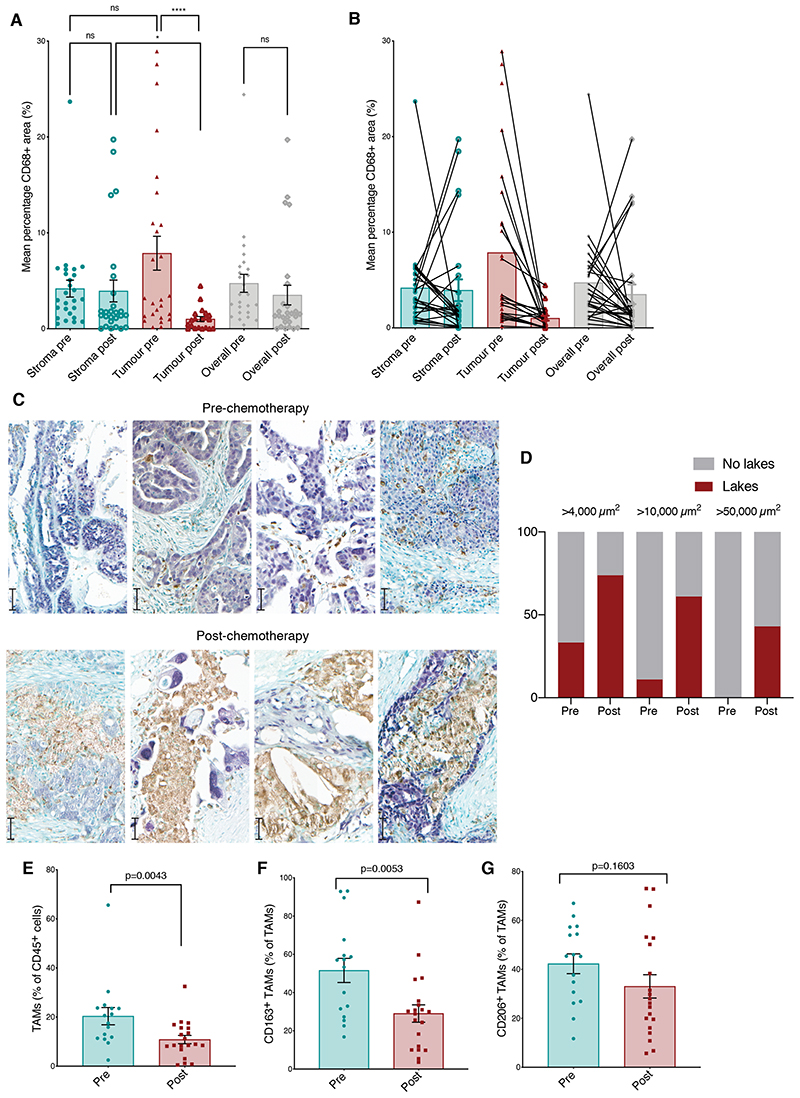
Figure 1. Effect of NACT on TAMs populations.
(A, B) Mean percentage area of CD68 staining pre- and post-chemotherapy within stromal areas, malignant cell areas, and the biopsy overall was quantified by Definiens analysis in matched pre and post-chemotherapy human HGSOC biopsy samples from the omentum. (A) Data presented from twenty-six matched pre and post-NACT biopsy samples. (B) Data presented indicating matched sample pairs. ****p<0.0001, *p<0.05. Wilcoxon test was applied for matched analyses and Mann-Whitney test for unmatched analyses. (C) Representative images of dual color IHCfor four pre- and four post-chemotherapy human HGSOC metastases showing malignant cells (anti-PAX8, purple) and myeloid cells (anti-CD68, brown). Scale bars: 50μm. (D) Quantification of ‘macrophage lakes’ within human HGSOC omental metastases as areas of continuous CD68 staining greater than 4,000 μm2, 10,000 μm2 and 50,000 μm2. Data represent quantification of unmatched biopsies from nine patients pre- and twenty-three patients post-chemotherapy. Fisher’s exact test p-values are shown. (E-G) Flow cytometry analysis of HGSOC omental metastases from sixteen patients treated with upfront surgery (green) and twenty patients having received NACT (red). (E) Frequency of TAMs as a percentage of CD45+ cells. Frequency of TAMs positive for (F) CD163 and (G) CD206 staining. Mann-Whitney test was applied. Error bars represent SEM.
We, therefore, used dual color IHC to investigate the distribution of CD68+ cells in tissue sections rather than the smaller TMAs in a different unmatched cohort of nine pre-treatment and twenty-three post NACT patients. We observed large stromal clusters of CD68+ cells surrounding or in close proximity to viable-appearing malignant cells (Fig. 1C). There were significantly more macrophage lakes within post-NACT tissue sections compared to pre-chemotherapy (Fig. 1D). Because we previously reported that high stromal CD68+ cell densities in pre-treatment primary tumor biopsies from 226 HGSOC patients had a significant positive association with overall survival (OS)(9), this suggested that some TAM populations in HGSOC may aid host anti-tumor responses. We, therefore, looked for correlations between CD68+ density and OS. In patients for whom we had follow-up data, OS was greater in patients with high compared to low CD68+ areas following chemotherapy (log-rank p=0.05), and a near-significant trend (p=0.08) to improved survival associated with a high CD68+ area within the stroma before chemotherapy (Supplementary Fig. S1B-C). Taken with our published data (9), the survival curves shown here, albeit on a small cohort of patients, suggest that high stromal TAM density may confer positive influence on OS and that TAMs within the stromal and malignant cell areas of the HGSOC TME may have different functions and impacts on cancer growth. In summary, results show that chemotherapy depletes macrophages from the malignant cell islands of the HGSOC TME and alters the overall phenotype of the residual macrophages which are largely resident in the stroma.
TAM phenotypes pre-treatment and post-NACT
We next asked if NACT affected TAM phenotype. Using flow cytometry in sixteen omental samples from patients having upfront surgery and twenty unmatched samples from patients receiving interval reduction surgery post-NACT, we defined TAMs as CD45+Lin-CD14+HLA-DR+ cells, excluding CD3+, CD19+, CD20+, CD56+, CD66b+, and Siglec8+ cells (Supplementary Fig. S1D). In support of the data from Fig. 1A–B, we found a significant reduction in the number of TAMs, as a percentage of CD45+ leukocytes, in the post-NACT group compared to biopsies from patients receiving upfront surgery (Fig. 1E). There was also a decrease in CD163+ TAMs, considered a marker of alternatively activated macrophages (Fig. 1F, Supplementary Fig. S1E) and associated with poorer prognosis in HGSOC (12). The mean fluorescence intensity (MFI) of CD163 was also lower in the post-NACT cohort (Supplementary Fig. S1F). Although differences in numbers of CD206+ TAMs were not significant, there was a reduction in cell surface expression (Fig. 1G, Supplementary Fig. S1G). The MFI for HLA-DR was unchanged, although it showed a trend for decreased expression (Supplementary Fig. S1H). The data thus far show that 3-4 weeks after the last dose of neoadjuvant platinum and taxane chemotherapy, TAM density was reduced in the malignant cell areas of the omental TME, with a decrease in cell surface markers associated with alternatively activated macrophages.
NACT alters the transcriptome of TAMs in omental tumors
To further investigate changes in macrophage cell function and activity after NACT, we used RNA-seq to study the transcriptomes of CD45+HLA-DR+Lin-CD14+ cells sorted as a bulk population, from five pre-treatment and seven post-NACT unmatched omental HGSOC biopsies, obtaining between 250,000-750,000 cells per sample. Transcript expression of a total of 16,253 genes were quantified, of which 12,614 were protein-coding genes. Supplementary Fig. S2A demonstrates high expression of macrophage genes relative to lineage genes of other cell types. Pre- and post-NACT samples segregated by unsupervised clustering with some pre- and post-samples interspersed between the two groups, indicating discrete features in transcriptomes of the macrophages with some heterogeneity (Supplementary Fig. S2B).
Differential expression analysis revealed 858 protein-coding genes with different expression post-NACT vs. pre-chemotherapy TAMs (Supplementary Table S2). Of these, 81 were upregulated and 777 downregulated post-NACT (Fig. 2A–B). The gene that showed the greatest fold-change increase in expression was TREML4 (triggering receptor expressed on myeloid cells-like 4), a positive regulator of TLR7 and other TLR signaling (25) that is upregulated in M1-polarized macrophages (26)(Fig. 2B). FGFR2 (fibroblast growth factor receptor 2) was among the genes showing the greatest fold-change decrease in expression after chemotherapy and was of interest considering the reported role of FGF2 in polarizing TAMs to a tumor-promoting phenotype (27).
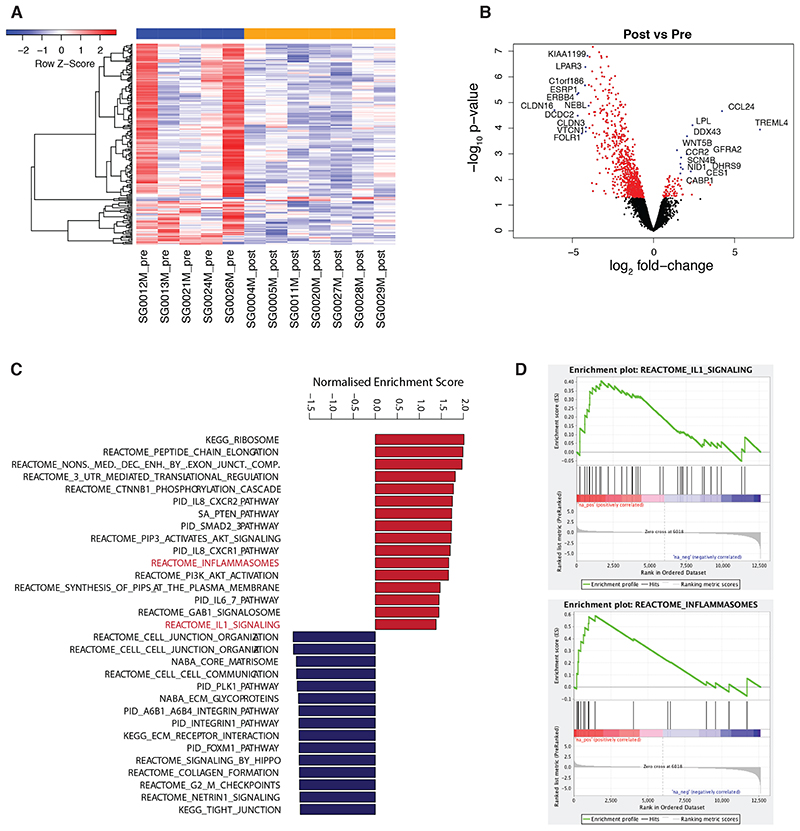
Figure 2. Transcriptomics of TAMs from pre and post-chemotherapy biopsies.
(A) Heatmap of differentially expressed genes (p < 0.05). (B) Volcano plot showing significantly differentially expressed genes in red (p<0.05). Gene symbols of top differentially expressed genes are indicated. (C) Barplot illustrating top enriched canonical pathways in GSEA of post- vs pre-treatment (p<0.05). (D) GSEA enrichment plots of the inflammasome and IL1 signaling pathways. Data correspond to n=5 pre- and n=7 post-NACT unmatched samples.
Gene set enrichment analysis (GSEA) for canonical pathways and gene ontology biological processes (Fig. 2C, Supplementary Table S3A-B) showed that amongst the top twenty significantly upregulated pathways in the post-NACT samples were activation of AKT/PI3K signaling, IL6 signaling, IL8 signaling, activation of the inflammasome, and IL1β signaling (Fig. 2C–D). We also observed upregulation of lipid metabolism processes, suggesting altered lipid profiles in post-chemotherapy TAMs (Supplementary Table S3B). The top significantly downregulated pathways in the post-NACT TAMs were extracellular matrix (ECM) formation and remodeling, ECM interactions and signaling, and cell cycle and proliferation pathways. This reduction in ECM pathway genes is of particular interest and may suggest a decrease in cells analogous to the subset of pro-fibrotic TAMs found in pancreatic cancer biopsies and mouse models (28). In summary, bulk RNA-seq data of CD45+HLA-DR+Lin-CD14+ cells from HGSOC omental metastases showed that 3-4 weeks after NACT, there was an increase in expression of inflammatory pathways, evidence of activation of the inflammasome, and a decrease in markers and pathways associated with tumor-promoting TAMs. This also suggested that pyroptosis, a form of programmed cell death characterized by cell membrane lysis and the release of the proinflammatory cytokines IL1β and IL18 (29), may be occurring in macrophages post-chemotherapy.
NACT activates the inflammasome in TAMs
Because the RNA-seq data provided evidence of macrophage cell death after NACT, we re-analyzed the flow cytometry data for evidence of cytotoxicity, finding a significant increase in non-viable TAMs as a percentage of all TAMs in post-NACT samples compared to pre-treatment samples (Fig. 3A). To study this further, we conducted in vitro viability assays on monocytes isolated from normal PBMCs differentiated to classically activated (M1) or alternatively activated (M2) phenotypes in vitro (see Methods) and treated with doses of chemotherapy that were toxic for malignant HGSOC cells. The cells were then treated with carboplatin at different concentrations for 48 hours, and cell viability and phenotype were assessed by flow cytometry. Macrophages were killed by carboplatin at concentrations in the range 50-200μM (Fig. 3B). This was within the range of IC50 value for the two human HGSOC cell lines tested (Fig. 3C). The IC50 was lower for M1/classically activated macrophages (93μM) than M2-polarized (203μM) macrophages. Paclitaxel was also toxic for macrophages at similar doses to human HGSOC cell lines (Supplementary Fig. S3A-B).
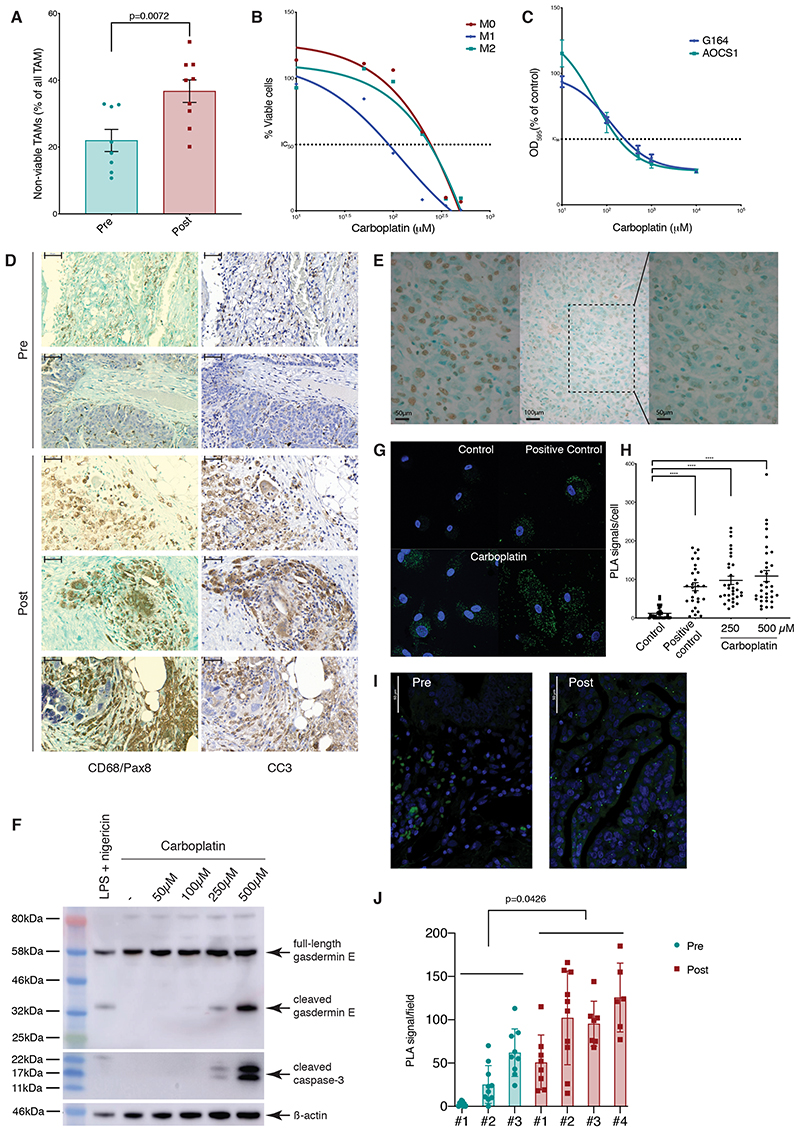
Figure 3. Effect of NACT on inflammasome activation.
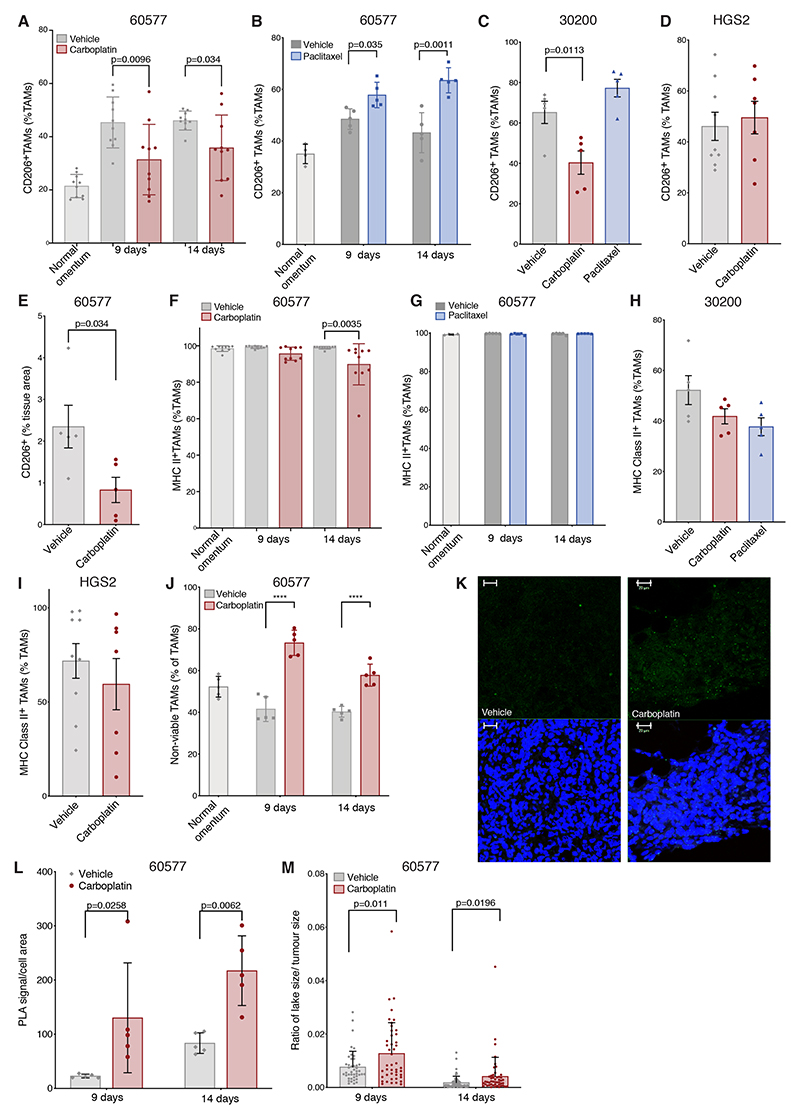
(A) Flow cytometry analysis of human HGSOC omental metastases pre- and post-chemotherapy showing non-viable (FVD+) TAMs (Lin−HLA-DR+) as percentage of total TAMs in 8 (green) and 9 patients (red). (B) Human CD14+ monocytes, polarized to M1 (LPS/IFNγ), M2 (IL4/IL10), or M0 phenotypes, were treated with varying concentrations of carboplatin for 48 hours. Viable cells as percentage of single cells, normalized to media only control for each condition are shown from one experiment representative of two. IC50 values: M0=201.7μM, M1=93.5μM, and M2=203.3μM. (C) AOCS1 and G164 cells were treated with different concentrations of carboplatin for 48 hours. OD595 measurements for crystal violet staining are presented as average of three experiments, normalized to media-only control. IC50 values: G164=98.63μM, AOCS1=45.1μM. (D) Representative staining of five HGSOC omental biopsies pre- and post-chemotherapy from consecutive sections. Left: CD68 (brown), PAX8 (purple), green counterstaining. Right: cleaved caspase-3 (brown), blue counterstaining. Scale bar: 200μM. (E) Representative TUNEL staining within ‘macrophage lakes’ post-chemotherapy. Left: positive control, tissue incubated with DNAase prior to TUNEL staining. Center: post-chemotherapy HGSOC biopsy showing an region within a ‘macrophage lake’, x20 magification. Right: x40 magnification of the shaded area of the center image. Scale bar 50, 100, and 50 μM. (F) Western blot showing cleaved gasdermin E and caspase-3 in lysates from M2 macrophages treated with varying concentrations of carboplatin for 24 hours or LPS/nigericin as a positive control. Data relate to a single experiment. (G) PLA staining for ASC/NLRP3 interaction in M2-differentiated macrophages. Media only control, LPS/nigericin treated cells (positive control), cells treated with carboplatin 250μM (bottom left) and 500μM (bottom right) for 24 hours. Green specks indicate NLRP3 inflammasome complexes. Scale bar: 25μM. (H) Quantification of PLA signal per cell for one experiment. One-way ANOVA test was applied; ****p<0.0001. (I, J) PLA staining for ASC/NLRP3 interaction performed on pre- and post-chemotherapy HGSOC omental biopsies. Green specks indicate NLRP3 inflammasome complexes. Scale bar 50 μM. (J) Quantification of PLA signal from 3 pre- and 4 post-chemotherapy biopsies. P-value from the t-test comparing averages of pre- and post-chemotherapy is shown. Error bars represent SEM.
We next looked for evidence of TAM death in the ‘macrophage lakes’ in human HGSOC sections with dual-color IHC for CD68 and PAX8 and in consecutive sections, cleaved caspase-3 (CC3) as a marker of apoptosis (Fig. 3D). There was clear overlap of CC3 and CD68 staining patterns, most notably within post-chemotherapy ‘macrophage lakes’ (Fig. 1C). Despite the fact that CC3 is frequently regarded as a marker of apoptosis, assessment of cell and nuclear morphology suggested that the CC3+ areas were not apoptotic (Fig. 3D). The distribution of the staining was also unusual for apoptosis; CC3-positivity is normally seen within individual cells rather than large clusters of cells. In keeping with the morphological appearances of these cells, they were not TUNEL-positive (Fig. 3E).
Published data suggest that caspase-3 can cleave gasdermin E to activate pyroptosis following chemotherapy drug treatment (30), either directly by inducing cell lysis or indirectly by triggering membrane pore-mediated potassium efflux that in turn activates the classical NLRP3/ASC/caspase-1 inflammasome and pyroptosis (31). Consistent with this, we observed caspase-3 and gasdermin E cleavage in macrophages following carboplatin treatment (Fig. 3F). To test if cleavage of gasdermin E could activate the inflammasome, we treated differentiated macrophages in the presence of 250μM and 500μM carboplatin and performed a proximity ligation assay (PLA) to test for the interaction of NLRP3 (NOD-, LRR-, and pyrin domain-containing 3) and ASC (apoptosis-associated speck-like protein containing a CARD)(Fig. 3G), as a readout of inflammasome activation, finding a significant increase in the number of PLA signals/cell in treated macrophages (Fig. 3G–H). We concluded that carboplatin treatment resulted in formation of the NLRP3 inflammasome complex, which was most likely triggered by gasdermin E-mediated potassium efflux. Therefore, we then used the PLA assay to look for NLRP3 inflammasome activation in HGSOC tissue sections pre- and post- NACT. Complex formation was detected more frequently in post-treatment biopsies (Fig. 3I–J).These results support the pathway analysis of the RNA-sequencing data. Collectively, these data provide evidence of inflammasome activation in human macrophages in response to carboplatin treatment.
Chemotherapy alters TAM numbers in mouse models of HGSOC
We then asked whether we could see similar effects of chemotherapy in mouse models of HGSOC that recapitulate many aspects of the human omental tumor microenvironment (13). We used three of these syngeneic orthotopic murine models, 60577, 30200 and HGS2, that harbor the genetic features of some human HGSOCs and recapitulate key features of the human TME, although there are distinct differences between them (13)(see Methods). When injected i.p., mice develop tumors within the omentum, as well as other peritoneal sites. We first assessed the sensitivity of the 60577, 30200, and HGS2 mouse cell lines to carboplatin and paclitaxel in vitro (Supplementary Fig. S4A-B). IC50 values were similar between the three cell lines and within the range described for two human ovarian cancer cell lines (Fig. 3C).
In all in vivo experiments, mice were treated once peritoneal disease was established (approximately 3, 10, and 7 weeks after i.p. injection of cells for 60577, 30200, and HGS2, respectively). As shown in survival experiments (Fig. 4A–C), established peritoneal tumors from all three models showed some therapeutic response to three doses of carboplatin given weekly, in a regime to mimic the platinum component of NACT in the patients. However, there were obvious differences between the models. 60577 tumors were the most sensitive to carboplatin treatment, with extension of survival in excess of 20 weeks, whereas in 30200 and HGS2 tumors, the difference in median survival was about three weeks. Omental weights were used as a surrogate measure of tumor burden as previously described (13). Nine and fourteen days after the start of treatment, when mice had received two doses of carboplatin, there was a significant reduction in omental weight in mice bearing 60577 tumors (Supplementary Fig. S4C). Two doses of paclitaxel as a single agent did not have appreciable effects on omental weight in 60577-bearing mice (Supplementary Fig. S4D). There was a significant effect on omental weight 48 hours after the third dose of treatment in carboplatin-treated 30200 tumors, but again paclitaxel had no activity (Supplementary Fig. S4E). A trend for a reduction in tumor weight was observed in HGS2 tumors treated with three doses of carboplatin, but the difference was not significant (Supplementary Fig. S4F), suggesting that this model is the least sensitive to this chemotherapeutic treatment.
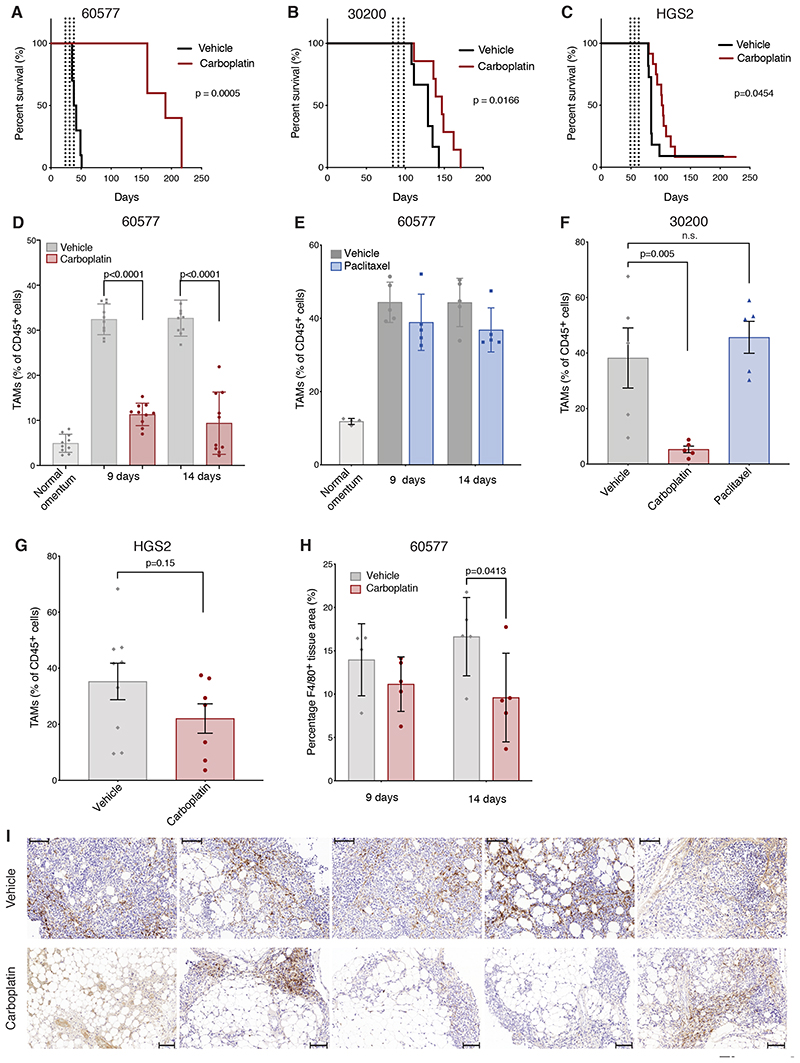
Figure 4. Effect of chemotherapy on TAMs in murine models of HGSOC.
(A-C) Kaplan-Meier curves comparing mice treated three times with carboplatin via i.p. injections (red) or vehicle (black) once weekly. Dashed lines indicate treatments. (A) 60577 injected mice. Treatment was started at 21 days. One experiment of two is shown; n=10 (vehicle), n=5 (carboplatin). (B) 30200 injected mice. Treatment was started at 84 days. N= 6 (vehicle), n=7 (carboplatin). (C) HGS2 injected mice. Treatment was started at 49 days. N=10 (vehicle), n=11 (carboplatin). (D-G) Flow cytometry analysis of omental tumors treated with carboplatin, paclitaxel, vehicle alone, or healthy omental controls. TAM frequency is expressed as a percentage of the CD45+ population. (D) Two independent 60577 experiments; n=9/10 vehicle-, 10 carboplatin-treated mice per time point at 9 and 14 days after the first dose of carboplatin (two doses administered); 20 healthy omenta pooled into 10. (E) One experiment; 5 paclitaxel-treated, 5 vehicle-treated mice per time point at 9 and 14 days after the first dose of paclitaxel; 10 healthy omenta pooled into 4. (F) One 30200 experiment; 5 mice per group; 16 days after the first dose, following three doses. (G) Two HGS2 experiments pooled together 21 days after the first dose, following three doses, 9 and 7 mice, respectively. (H, I) F4/80 staining quantified by Definiens digital analysis expressed as percentage positive area of the biopsy of 60577 omental tumors, from one experiment; 5 carboplatin-treated mice, 4-5 vehicle-treated mice, at the indicated time points following two doses of carboplatin treatment. Scale bar: 100 μM. Error bars represent SEM.
We next used flow cytometry to study macrophage cell populations in the omental tumors, defining murine TAMs as CD45+Ly6C-Ly6G-F4/80+CD11b+ cells (Supplementary Fig. S4G). The number of these cells, as a percentage of CD45+ cells, was significantly increased in the omental tumors compared to normal omentum from age-matched mice in the 60577 model (Fig. 4D–E). In established omental tumors, we found a significant decline in TAMs, as a percentage of CD45+ cells, two days after both the first and second weekly dose of carboplatin in the 60577 model (Fig. 4D, Supplementary Fig. S4H) but not after paclitaxel (Fig. 4E). The same decline was seen after the third weekly dose of carboplatin in the 30200 model but not after paclitaxel (Fig. 4F). There was also a decline in TAMs with the HGS2 model at the same time point, but this did not reach significance (Fig. 4G). The decline in TAMs shown by the flow cytometry data were confirmed in carboplatin-treated 60577 tumors using IHC for the macrophage marker F4/80 (Fig. 4H–I).
Chemotherapy alters TAM phenotypes in mouse models of HGSOC
We next asked if there was a change in phenotype of the murine TAMs after carboplatin treatment. Because there was no reliable commercially available antibody for murine CD163 at the time these experiments were conducted, we used CD206 as a marker of alternatively activated macrophages. CD206+ TAMs were significantly increased in established 60577 omental tumors compared to age-matched normal mouse omentum (Fig. 5A). CD206+ TAMs (as a percentage of the TAM population) decreased in carboplatin- compared to vehicle-treated 60577 omental tumors after the first and second dose (Fig. 5A, Supplementary Fig. S4H), whereas there was an increase in paclitaxel-treated tumors (Fig. 5B). We also found significant reductions in CD206+ TAMs in the 30200 model after three doses of carboplatin but not paclitaxel (Fig. 5C). The finding was, however, not replicated in the HGS2 model (Fig. 5D). The CD206 data were confirmed by IHC in 60577 omental tumors (Fig. 5E). Because MHC Class II was not one of our positive gating criteria in the mouse tumor experiments as HLA-DR had been in the patient samples, we were able to measure the frequency of MHC Class II-expressing TAMs (Fig. 5F–I). There were significant reductions in the 60577 model at the indicated time points with carboplatin but not with paclitaxel and no significant differences were seen in 30200 and HGS2 models.
Figure 5. Effect of chemotherapy on murine TAM phenotype.
(A-D) Flow cytometry data showing the effect of carboplatin and paclitaxel on CD206+ TAM percentage of the total TAM population in 60577, 30200, and HGS2 models. (A) Two independent 60577 experiment; n=9/10 vehicle-, 10 carboplatin-treated mice at 9 and 14 days after the first dose of carboplatin (two doses administered), 20 healthy omenta pooled into 10. (B) One experiment; 5 paclitaxel-treated, 5 vehicle-treated mice at 9 and 14 days after the first dose of paclitaxel (two doses administered); 10 healthy omenta pooled into 4. (C) One 30200 experiment; 5 mice per group, 16 days after the first dose, following three doses. (D) Two HGS2 experiments pooled together; 21 days after the first dose, following three doses, 9 and 7 mice, respectively. (E) Quantification of CD206 staining by Definiens digital analysis in 60577 omental tumors; from one experiment; 5 carboplatin-treated mice, 5 vehicle-treated mice, at 14 days following the start of treatment. (F-I) Flow cytometry data showing the effect of carboplatin and paclitaxel on MHCII+ TAM percentage of the total TAM population in 60577, 30200s and HGS2 models in the same experiments described in A-D. (J) Flow cytometry analysis omental tumors treated with carboplatin, vehicle, or healthy omental controls. Non-viable TAM frequency is expressed as a percentage of the total TAM population from one experiment; 5 carboplatin-treated mice, 4-5 vehicle-treated mice per time point; 10 healthy omenta pooled to 5. (K) Representative examples of PLA staining for ASC/NLRP3 interaction on sections of 60577 tumors treated with vehicle or carboplatin for the indicated times. Small green specks indicate NLRP3 inflammasome complexes. Scale bar 20 μM (L) Quantification of PLA signal shown in (K); 5 tumors for each time point and treatment. P-value from ANOVA comparing averages pre- and post-chemotherapy is shown. (M) Quantification of macrophage lakes in control and carboplatin-treated mice, 9 and 14 days from the start of the treatment. Each dot corresponds to the ratio of the area of a lake versus the total tumor area. Error bars represent SEM.
The data suggest that carboplatin, but not paclitaxel, results in loss of TAMs from the TME of murine HGSOC sensitive to carboplatin and changes their phenotype in a way that largely replicates the human data we described. In line with the patient data, we found significantly more non-viable TAMs in carboplatin-treated compared to control tumors at all time points in 60577 tumors (Fig. 5J). There was also evidence of inflammasome activation after NACT in the 60577 model, with a positive signal for ASC-NLRP3 complex formation observed at higher frequency in carboplatin-treated tumors (Fig. 5K–L) and increased size of macrophage ‘lakes’ 14 days after carboplatin treatment (Fig. 5M). Thus, data so far show that chemotherapy treatment of omental metastases, either platinum/taxane NACT in human HGSOC or single-agent carboplatin in murine HGSOC tumors, decreases the number of TAMs, with a decreased positivity for alternative activation markers and an increase in non-viable TAMs in carboplatin-susceptible models. We also presented evidence that the mechanism of action of carboplatin involves inflammasome activation.
Inhibition of TAM populations after chemotherapy decreases disease-free survival
Because we concluded that chemotherapy depleted TAMs in both murine and human HGSOC at the same time as augmenting TAM populations that may aid host anti-tumor responses, we hypothesized that targeting TAMs after chemotherapy may modulate disease-free survival (DFS). We tested this hypothesis in the 60577 model where the mice relapsed 20 weeks after three doses of chemotherapy. We speculated that if chemotherapy increases anti-tumor TAMs, depletion of TAM populations post-chemotherapy would decrease DFS, whereas it may have an opposite effect later during remission. First, we asked if alternatively activated TAMs were increased in relapsed tumors. We compared TAMs from omental tumors that reached humane endpoint in control untreated groups and in mice that had relapsed after chemotherapy. TAM numbers were similar in untreated tumors and relapsed tumors (Fig. 6A), but there was a significant increase CD206+ TAMs (Fig. 6B), with no change in numbers with high MHC expression (Fig. 6C), suggesting that there may be more alternatively activated TAMs at relapse.
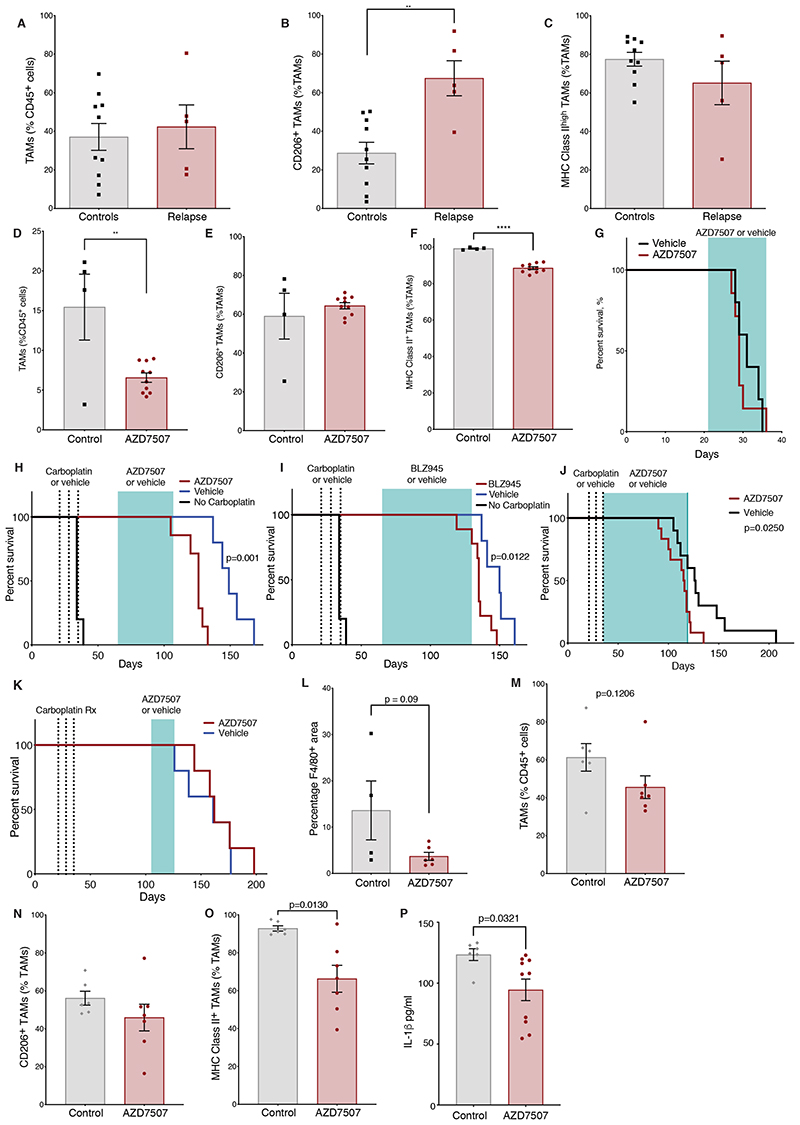
Figure 6. Effect of TAMs depletion in combination with chemotherapy in vivo.
(A-C) Flow cytometry analysis of omental tumors from 60577 model treated with carboplatin and relapsed (relapse) or vehicle-treated tumor (controls); 5 mice (relapse) and 10 mice (control) sacrificed at end point. Frequency of TAMs as percentage of CD45+ cells (A). Frequency of CD206+ TAMs and (B) MHC Class II+ TAMs (C) expressed as percentage of the TAM population. (D-F) Flow cytometry analysis of 60577 omental tumors treated with AZD7507 for 21 days or vehicle alone; from one experiment with 10 AZD7507-treated and 4 vehicle-treated mice. Treatment started 21 days after cell injection and continued for 12 days. (D) TAM frequency as percentage of the CD45+ cells, (E) CD206+ TAMs, and (F) MHC Class II+ TAMs as percentage of the TAM population. (G) Kaplan Meier curves showing survival of mice treated with vehicle or AZD7507 starting 21 days after 60577 cell injection until end-point from one experiment, n=5 vehicle, n=7 AZD7507. (H, I) Kaplan Meier curves showing 60577-injected AZD7507- (H, J) or BLZ945- (I) treated mice, vehicle-treated controls, and mice not treated with chemotherapy. Carboplatin treatments (dashed lines) and AZD7507, BLZ945 (green shading) treatment are indicated, starting four weeks after chemotherapy (H, I) or immediately after (J). Data from one experiment is shown (n=5 vehicle, n=7 AZD7507 in H, n=5 vehicle, n=9 BLZ945 in I, n=10 vehicle, n=12 AZD7507 in J). (K), Kaplan Meier curves showing 60577-injected AZD7507-treated mice (red line) and vehicle-treated controls (blue line). Carboplatin treatments (dashed lines) and AZD7507 or vehicle treatment (green shading) are indicated. Data from one experiment are shown (n=5 vehicle, n=5 for AZD7507); differences are non-significant. (L) Quantification of F4/80 staining by Definiens analysis in relapsed tumors from the experiment shown in H. F4/80 staining expressed as percentage positive area of the biopsy. (M-O) Flow cytometry analysis of 60577 omental tumors treated with AZD7507 started immediately after carboplatin treatment. Data relate to a single experiment with 10 AZD7507-treated mice and 10 vehicle-treated mice. Treatment continued for 84 days. (M) TAM frequency as a percentage of the CD45+ cells, (N) CD206+ TAMs and (O) MHC Class II+ TAMs as percentage of the TAM population. (P) ELISA of IL1β in ascities fluid of mice from the experiment described in K-M. Error bars represent SEM.
To test the above-stated hypothesis, we treated mice with two orally available inhibitors of the pan-macrophage receptor CSF1R (CSF1Ri), AZD7507 and BLZ945, both selective and potent inhibitors of CSF1R kinase activity (32,33) in order to inhibit TAM recruitment to murine tumors (the reported biochemical IC50 values are 3nM for AZD7507 and 1nM BLZ945; both have negligible activity against other kinases).
Using AZD7507, we confirmed that this agent depleted TAM in established 60577 omental tumors (Fig. 6D). AZD7507 did not change the frequency of TAMs expressing CD206 (Fig. 6E), but there was a small but significant decrease in the frequency of MHCII-expressing TAMs (Fig. 6F). In spite of the decrease in macrophage cell number and change in phenotype, treatment of established 60577 tumors had no impact on mouse survival when the CSF1Ri was given as a single agent (Fig. 6G). The CFS1Ri BLZ945 also depleted TAMs in established 60577 tumors without a significant impact on tumor weight after 4 days treatment (Supplementary Fig. S5A-B). Treatment of mice with AZD7507 as a single agent also led to a significant reduction of CD4+ cells and CD19+ B cells infiltrating the tumors (Supplementary Fig. S5C-D). A similar effect was seen with treatment with BLZ945 (Supplementary Fig. S5E-F).
Having shown that these agents could deplete TAMs in untreated tumors, we treated cohorts of 60577 mice with three doses of carboplatin and then with AZD507, BLZ945, or appropriate vehicle controls to see if this would influence time to relapse and overall survival (OS). In support of our hypothesis, long term treatment with these two inhibitors significantly decreased mouse survival (p=0.0006 and p=0.012 for AZD7507 and BLZ945, respectively, 30 days after the end of carboplatin)(Fig. 6H–I). In another experiment, we started treatment with AZD7507 immediately after the end of carboplatin treatment and again observed a significant decrease in DFS and OS (p=0.025)(Fig. 6J). However, if AZD7507 started later, 10 weeks after carboplatin treatment ceased, there was no impact on DFS (Fig. 6K). This provides further evidence that carboplatin induced changes in TAM populations towards an anti-tumor phenotype. These data also suggest that TAM populations persist in the TME after the end of chemotherapy.
We also studied the TAM populations when mice reached humane endpoint in the experiment shown in Fig. 6H and 6J. For mice shown in Fig. 6H, there was a trend for a decrease in density of F4/80+ cells in the 60577 omental tumors by IHC (Fig. 6L). Flow cytometry of endpoint tumors from the AZD7507 experiment shown in Fig. 6J similarly showed a decline in F4/80+ macrophages, but this was not significant (Fig. 6M). Within the remaining TAMs, CD206+ cells did not decrease significantly in AZD7507-treated tumors (Fig. 6N). There was, however, a significant and pronounced decrease in MHC Class II+ TAMs (Fig. 6O, Supplementary Fig. S5G) after long-term AZD7507 treatment, suggesting that antigen presentation was decreased. Because there was evidence that chemotherapy induced inflammatory cell death in both human and mouse tumors, we assayed IL1β as a marker of inflammasome activation in ascites from the AZD7507 experiment (Fig. 6J) at end point and found a significant decrease in IL1ß (Fig. 6P). We also tested the effects of AZD7507 post-carboplatin therapy in the 30200 model, which had less sensitivity to three doses of chemotherapy, and there was no effect on DFS or OS (Supplementary Fig. S5H).
In summary, although CSF1R inhibitors were able to decrease F4/80+ cells in established omental tumors and those that had relapsed after chemotherapy, our experiments suggested that this did not augment host anti-tumor responses. Indeed, single-agent CSF1Ri treatment also depleted cells of the adaptive immune system in the omental tumors, and some schedules decreased DFS and OS when given in an attempt to prevent relapse. We conclude that long-term depletion of CSF1R+ cells was preventing potential anti-tumor actions of myeloid cells that had been induced by the carboplatin.
CSF1Ri treatment reprograms the microenvironment of relapsing tumors
We next conducted RNA-seq on omental biopsies from AZD7507 or vehicle-treated tumors at end point (Fig. 6J). Principal component analysis illustrated differences in the transcriptomes of the vehicle- compared to AZD7507-treated tumors (Supplementary Fig. S6A). Continued suppression of myeloid cells was confirmed with a significant decrease of F4/80 (Adgre1) in the AZD7507-treated group (Fig. 7A, left panel). Expression of murine orthologs of the human TAM signature determined by Cassetta et al (34) were also significantly decreased in the AZD7507-treated tumors (Fig. 7A, right panel). Differential expression analysis of AZD7507- versus vehicle-treated tumors revealed 352 genes (p-value<0.05 and log2FC>|1|), of which 336 genes were downregulated in AZD7507-treated tumors and 16 upregulated (Fig. 7B, Supplementary Table S4). The top downregulated genes included innate and adaptive immune genes, such as Tlr9, Cx3cr1, Il10ra, Cd72, C1qb, Cd83, Irf8, Ifi207, and Slc11a1, as well as expected downregulation of Csf1r. The top upregulated genes included the interferon inducible GTPase Iigp1, malignant cell-related genes such as Snx31 and Neto1, and adhesion molecules such as Sdk2. Gene-set enrichment analysis (GSEA) showed significant decreases in B- and T-cell activation in CSF1Ri-treated tumors and an upregulation of malignant cell processes such as developmental pathways, DNA damage response and DNA replication (Fig. 7C–D, Supplementary Table S5). Fig. 7D illustrates core enrichment genes in the GSEA pathways relating to the upregulation of DNA replication pathways and the downregulation of T and B cell activation. These results point to an important role of macrophages post-treatment in the activation of adaptive immunity.
Discussion
We found that TAM populations in omental samples from patients and mouse orthotopic models were altered by carboplatin chemotherapy, both in terms of density and phenotype. Because we found that pan-macrophage depletion starting shortly after chemotherapy in the mouse models reduced DFS and adaptive immunity, we concluded that chemotherapy enhanced the anti-tumor actions of TAMs and their ability to support adaptive immune responses.
A majority of patients with HGSOC have a good response to initial chemotherapy but, even with the use of more targeted treatments such as PARP inhibitors in an adjuvant setting (35), many patients will eventually relapse with increasingly drug-resistant disease. The work described here, taken together with published investigations of the adaptive immune response in this context, suggests that therapies that can enhance this nascent chemotherapy-induced immune response could increase DFS and even reduce the amount of chemotherapy given. As a majority of TAMs are thought to be tumor-promoting, one approach would be to reduce TAM recruitment to post-chemotherapy micrometastases. There are several antibody and small-molecule approaches to macrophage depletion currently in early-phase clinical trials (10), but our model would suggest that this could be detrimental because it would also reduce TAMs capable of fostering anti-tumor immune responses. On the basis of our current results, and our previously published data showing stimulation of adaptive immunity after NACT in HGSOC (2,3) as well as a positive associaton between stromal TAM density and survival (9), we suggest that macrophage-repolarizing approaches may maintain and even enhance the immune-stimulatory effects of chemotherapy in HGSOC and other cancers.
We do not know why only the stromal macrophages are associated with a better prognosis. It is known that both ECM components and stiffness of the matrix can influence the phenotype of macrophages (reviewed in (36)). High infiltration of macrophages at the invasive front of colorectal cancer sections correlates with a better response to chemotherapy (37). Similarly, in human pancreatic ductal adenocarcinoma, a high density of TAMs at the tumor–stroma interface positively predicts responsiveness to gemcitabine adjuvant chemotherapy (38). Stromal macrophages may have functions distinct from those in tumor islands, and/or may be predictive of response, rather than simply prognosis.
We observed differences in the tissue localization of CD68+ macrophages in HGSOC sections three weeks following NACT compared to untreated samples. In chemotherapy-treated samples, we frequently observed large clusters of macrophages (macrophage ‘lakes’) often in close proximity to or surrounding islands of viable malignant cells. In contrast, pre-chemotherapy samples, macrophages were scattered in the stroma or malignant cell islands. A fibro-inflammatory response including infiltrating macrophages and ‘foam’ cells has been described as a tissue response to chemotherapy within omental HGSOC metastases (39). Indeed, assessment of these infiltrates form a part of the chemotherapy response score (CRS) and are associated with a favourable response to chemotherapy when associated with a concomitant tumour cell response (39).
There are a number of limitations to our work. Post-chemotherapy changes in the TME are dynamic but we studied just one time point – 3-4 weeks after the last treatment cycle in our patients. However, because we were able to replicate the effects of chemotherapy in the HGSOC mouse models, we could study dynamic changes. Also, we focused on a single disease site, the omentum, because diagnostic omental biopsies are readily available to match biopsies taken at post-NACT surgery from the same patient, and our mouse cell lines develop large omental tumor deposits (13). However, other groups have confirmed our initial studies on the effects of NACT on T- and B-cell responses in other HGSOC disease sites (5). We should also note that heterogeneity of TAM populations and the effect of chemotherapy in the different TAM subsets is an important factor in understanding the dynamics of TAMs, and single-cell RNA-seq will be valuable in future analysis. We studied platinum and taxane treatments individually, which of course was not possible in the patients, and further study of combination treatments in the mice are warranted.
The NLRP3 inflammasome is a multi-protein complex that mediates maturation and release of cytokines such as IL1β, as well as initiating pyroptosis. We found evidence for increased NLRP-3 inflammasome activation in carboplatin-treated macrophages and post-NACT mouse and human tumors as a result of chemotherapy-induced, caspase-3-mediated activation of gasdermin E. Gasdermin E in turn forms pores on the plasma membrane that may either directly cause pyroptosis, or potassium efflux that in turn activates the NLRP3-inflammasome. Once formed, the inflammasome promotes the maturation of IL1β and other cytokines, along with the induction of pyroptosis. Activation of the inflammasome and gasdermin E-induced pyroptosis in the tumor microenvironment have been reported to have a critical role in the activation of anti-tumor immunity in some cancer models (40,41).
Our data support a model, at least in vitro, whereby carboplatin induces caspase-3 dependent cleavage of gasdermin E that in turn forms pore in the plasma membrane. Similarly to that described in (30), we observed cleaved caspase-3-positive/TUNEL-negative macrophages, and NLRP3 inflammasome formation, in biopsies from carboplatin-treated patients. Because NLRP3-inflammasome formation is known to mediate caspase-1 auto-proteolytic activation and IL1β cleavage and release, we hypothesize that this also occurs in macrophages. We do not know whether this exclusively happens in macrophages or also in cancer cells. Because we observed PLA-positive cells that morphologically looked like tumor cells, it is possible that inflammasome activation also occurs in malignant cells.
Pathways associated with IL1, IL6, and IL8 signaling were enhanced in the post-NACT human TAM RNA-seq data. The actions of these inflammatory cytokines in cancer is likely to be context dependent – in early disease, neutralizing these inflammatory cytokines may be beneficial, but acute and high levels of these inflammatory cytokines generated by the dying malignant cells after chemotherapy may help stimulate longer-lasting adaptive immune responses as our results suggest. In terms of anti-tumor actions of IL1ß, a study highlights a critical role for myeloid cells in the stimulation of immunogenic cell death after oxaliplatin and mitoxantrone chemotherapy in mouse cancer models. The mechanism of this involves myeloid cell PTEN-promoting inflammasome activation and production of IL1β. They were also able to find evidence for this mechanism in breast cancer patients treated with anthracycline-based adjuvant chemotherapy (42). However, there is also abundant pre-clinical evidence that inflammatory cytokines such as IL1ß, IL6 and IL8 are targets for cancer therapy, rather than being involved in host anti-tumor responses (43–45). In the CANTOS trial of 10,061 patients with cardiovascular disease, treatment with the IL1ß antibody canakinumab reduces incidence of lung cancer in a dose-dependent manner (46). One interpretation of this result is that the patients (many of whom were smokers) had undetected early lung cancers and that in this context, chronic IL1β production was promoting these nascent tumors. However, these important findings need to be replicated in further clinical trials.
We believe that the mouse models of treatment and relapse that we have described here will allow us to ask if neutralizing inflammatory cytokines, inflammasome inhibitor drugs, or therapies that repolarize TAMs will prolong or reduce remission or act together with chemo- or immunotherapies.
Supplementary Material
Synopsis.
Neoadjuvant chemotherapy is shown to reduce the density of tumor-associated macrophages (TAMs). In addition, chemotherapy reduces M2-related markers and increases proinflammatory signaling in TAMs, including inflammasome activation, thus, highlighting an opportunity to leverage this to improve anti-tumor responses.
Acknowledgments
We would like to thank the patients for donating samples, doctors and nurses at St. Bartholomew’s Gynaecological Cancer Centre and St. George University Hospital for their support. We would also like to thank the Barts Gynae Tissue Bank and the BCI Flow Cytometry Facility which is funded by a CORE SERVICE GRANT at Barts Cancer Institute (Core Award C16420/A18066). In particular, we thank Dr. Rebecca Pike for her advice and support in flow cytometry and cell sorting. This work was funded by Cancer Research UK Programme Grants C587/A16354, C587/A25714 (CB, EM, AL, PK, FB); Cancer Research UK Clinical Bursary A21222, Wellcome Trust Clinical Reseach Training Fellowship 201118/Z/16/Z (OH); Well Being of Women RTF1013, ELS906 (SE); AA was funded by Barts Charity Grant CIF9035B.
Footnotes
Authors’ disclosures
FB is on the Scientific Advisory Board of Verseau Therapeutics Inc. and has acted as a consultant for Novartis Pharma Canada Inc. STB is an AstraZeneca employee and shareholder.
Authors’ contributions
OH Study design, execution of experiments, analysis of data, manuscript writing; CB Study design, execution of experiments, manuscript writing; EM Study design, analysis of data, manuscript writing; AL, CP Execution of experiments; PK, SE Study design, execution of experiments; SB, STB Provision of study material, critically revising the manuscript; AA Study design, manuscript writing; DB Study design, critically revising the manuscript; FB Study design, manuscript writing.
References
- 1.Galluzzi L, Buque A, Kepp O, Zitvogel L, Kroemer G. Immunological Effects of Conventional Chemotherapy and Targeted Anticancer Agents. Cancer Cell. 2015;28(6):690–714. doi: 10.1016/j.ccell.2015.10.012. [DOI] [PubMed] [Google Scholar]
- 2.Montfort A, Pearce O, Maniati E, Vincent BG, Bixby L, Bohm S, et al. A Strong B-cell Response Is Part of the Immune Landscape in Human High-Grade Serous Ovarian Metastases. Clin Cancer Res. 2017;23(1):250–62. doi: 10.1158/1078-0432.CCR-16-0081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bohm S, Montfort A, Pearce OM, Topping J, Chakravarty P, Everitt GL, et al. Neoadjuvant Chemotherapy Modulates the Immune Microenvironment in Metastases of Tubo-Ovarian High-Grade Serous Carcinoma. Clin Cancer Res. 2016;22(12):3025–36. doi: 10.1158/1078-0432.CCR-15-2657. [DOI] [PubMed] [Google Scholar]
- 4.Lo CS, Sanii S, Kroeger DR, Milne K, Talhouk A, Chiu DS, et al. Neoadjuvant Chemotherapy of Ovarian Cancer Results in Three Patterns of Tumor-Infiltrating Lymphocyte Response with Distinct Implications for Immunotherapy. Clin Cancer Res. 2017;23(4):925–34. doi: 10.1158/1078-0432.CCR-16-1433. [DOI] [PubMed] [Google Scholar]
- 5.Jimenez-Sanchez A, Cybulska P, Mager KL, Koplev S, Cast O, Couturier DL, et al. Unraveling tumor-immune heterogeneity in advanced ovarian cancer uncovers immunogenic effect of chemotherapy. Nat Genet. 2020 doi: 10.1038/s41588-020-0630-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Odunsi K. Immunotherapy in ovarian cancer. Ann Oncol. 2017;28(suppl_8):viii1–viii7. doi: 10.1093/annonc/mdx444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bast RC, Jr, Matulonis UA, Sood AK, Ahmed AA, Amobi AE, Balkwill FR, et al. Critical questions in ovarian cancer research and treatment: Report of an American Association for Cancer Research Special Conference. Cancer. 2019;125(12):1963–72. doi: 10.1002/cncr.32004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang L, Conejo-Garcia JR, Katsaros D, Gimotty PA, Massobrio M, Regnani G, et al. Intratumoral T cells, recurrence, and survival in epithelial ovarian cancer. The New England journal of medicine. 2003;348(3):203–13. doi: 10.1056/NEJMoa020177. [DOI] [PubMed] [Google Scholar]
- 9.Montfort A, Owen S, Piskorz AM, Supernat A, Moore L, Al-Khalidi S, et al. Combining measures of immune infiltration shows additive effect on survival prediction in high-grade serous ovarian carcinoma. Br J Cancer. 2020 doi: 10.1038/s41416-020-0822-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mantovani A, Marchesi F, Malesci A, Laghi L, Allavena P. Tumour-associated macrophages as treatment targets in oncology. Nat Rev Clin Oncol. 2017;14(7):399–416. doi: 10.1038/nrclinonc.2016.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Milne K, Kobel M, Kalloger SE, Barnes RO, Gao D, Gilks CB, et al. Systematic analysis of immune infiltrates in high-grade serous ovarian cancer reveals CD20, FoxP3 and TIA-1 as positive prognostic factors. PLoS One. 2009;4(7):e6412. doi: 10.1371/journal.pone.0006412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yuan X, Zhang J, Li D, Mao Y, Mo F, Du W, et al. Prognostic significance of tumor-associated macrophages in ovarian cancer: A meta-analysis. Gynecol Oncol. 2017;147(1):181–7. doi: 10.1016/j.ygyno.2017.07.007. [DOI] [PubMed] [Google Scholar]
- 13.Maniati E, Berlato C, Gopinathan G, Heath O, Kotantaki P, Lakhani A, et al. Mouse Ovarian Cancer Models Recapitulate the Human Tumor Microenvironment and Patient Response to Treatment. Cell reports. 2020;30(2):525–40.:e7. doi: 10.1016/j.celrep.2019.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bankhead P, Loughrey MB, Fernandez JA, Dombrowski Y, McArt DG, Dunne PD, et al. QuPath: Open source software for digital pathology image analysis. Sci Rep. 2017;7(1):16878. doi: 10.1038/s41598-017-17204-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tamura N, Shaikh N, Muliaditan D, Soliman TN, McGuinness JR, Maniati E, et al. Specific Mechanisms of Chromosomal Instability Indicate Therapeutic Sensitivities in High-Grade Serous Ovarian Carcinoma. Cancer Res. 2020;80(22):4946–59. doi: 10.1158/0008-5472.CAN-19-0852. [DOI] [PubMed] [Google Scholar]
- 16.Szabova L, Bupp S, Kamal M, Householder DB, Hernandez L, Schlomer JJ, et al. Pathway-specific engineered mouse allograft models functionally recapitulate human serous epithelial ovarian cancer. PLoS One. 2014;9(4):e95649. doi: 10.1371/journal.pone.0095649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Perets R, Wyant GA, Muto KW, Bijron JG, Poole BB, Chin KT, et al. Transformation of the fallopian tube secretory epithelium leads to high-grade serous ovarian cancer in Brca;Tp53;Pten models. Cancer Cell. 2013;24(6):751–65. doi: 10.1016/j.ccr.2013.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Anders S, Pyl PT, Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31(2):166–9. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hansen KD, Irizarry RA, Wu Z. Removing technical variability in RNA-seq data using conditional quantile normalization. Biostatistics. 2012;13(2):204–16. doi: 10.1093/biostatistics/kxr054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rerat M, Pouchan C. Dynamic scalar and tensor polarizabilities of the 2 (1)P and 2 (3)P states of He. Phys Rev A. 1994;49(2):829–32. doi: 10.1103/physreva.49.829. [DOI] [PubMed] [Google Scholar]
- 22.Reich M, Liefeld T, Gould J, Lerner J, Tamayo P, Mesirov JP. GenePattern 2.0. Nat Genet. 2006;38(5):500–1. doi: 10.1038/ng0506-500. [DOI] [PubMed] [Google Scholar]
- 23.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. PNAS. 2005;43:15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hanzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC bioinformatics. 2013;14:7. doi: 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ramirez-Ortiz ZG, Prasad A, Griffith JW, Pendergraft WF, 3rd, Cowley GS, Root DE, et al. The receptor TREML4 amplifies TLR7-mediated signaling during antiviral responses and autoimmunity. Nat Immunol. 2015;16(5):495–504. doi: 10.1038/ni.3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gonzalez-Cotto M, Guo L, Karwan M, Sen SK, Barb J, Collado CJ, et al. TREML4 Promotes Inflammatory Programs in Human and Murine Macrophages and Alters Atherosclerosis Lesion Composition in the Apolipoprotein E Deficient Mouse. Frontiers in immunology. 2020;11:397. doi: 10.3389/fimmu.2020.00397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Im JH, Buzzelli JN, Jones K, Franchini F, Gordon-Weeks A, Markelc B, et al. FGF2 alters macrophage polarization, tumour immunity and growth and can be targeted during radiotherapy. Nat Commun. 2020;11(1):4064. doi: 10.1038/s41467-020-17914-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhu Y, Herndon JM, Sojka DK, Kim KW, Knolhoff BL, Zuo C, et al. Tissue-Resident Macrophages in Pancreatic Ductal Adenocarcinoma Originate from Embryonic Hematopoiesis and Promote Tumor Progression. Immunity. 2017;47(2):323–38.:e6. doi: 10.1016/j.immuni.2017.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xue Y, Enosi Tuipulotu D, Tan WH, Kay C, Man SM. Emerging Activators and Regulators of Inflammasomes and Pyroptosis. Trends Immunol. 2019;40(11):1035–52. doi: 10.1016/j.it.2019.09.005. [DOI] [PubMed] [Google Scholar]
- 30.Wang Y, Gao W, Shi X, Ding J, Liu W, He H, et al. Chemotherapy drugs induce pyroptosis through caspase-3 cleavage of a gasdermin. Nature. 2017;547(7661):99–103. doi: 10.1038/nature22393. [DOI] [PubMed] [Google Scholar]
- 31.Kovacs SB, Miao EA. Gasdermins: Effectors of Pyroptosis. Trends Cell Biol. 2017;27(9):673–84. doi: 10.1016/j.tcb.2017.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Scott DA, Dakin LA, Daly K, Del Valle DJ, Diebold RB, Drew L, et al. Mitigation of cardiovascular toxicity in a series of CSF-1R inhibitors, and the identification of AZD7507. Bioorg Med Chem Lett. 2013;23(16):4591–6. doi: 10.1016/j.bmcl.2013.06.031. [DOI] [PubMed] [Google Scholar]
- 33.Pyonteck SM, Akkari L, Schuhmacher AJ, Bowman RL, Sevenich L, Quail DF, et al. CSF-1R inhibition alters macrophage polarization and blocks glioma progression. Nat Med. 2013;19(10):1264–72. doi: 10.1038/nm.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cassetta L, Fragkogianni S, Sims AH, Swierczak A, Forrester LM, Zhang H, et al. Human Tumor-Associated Macrophage and Monocyte Transcriptional Landscapes Reveal Cancer-Specific Reprogramming, Biomarkers, and Therapeutic Targets. Cancer Cell. 2019;35(4):588–602.:e10. doi: 10.1016/j.ccell.2019.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moore K, Colombo N, Scambia G, Kim BG, Oaknin A, Friedlander M, et al. Maintenance Olaparib in Patients with Newly Diagnosed Advanced Ovarian Cancer. N Engl J Med. 2018;379(26):2495–505. doi: 10.1056/NEJMoa1810858. [DOI] [PubMed] [Google Scholar]
- 36.Yang M, McKay D, Pollard JW, Lewis CE. Diverse Functions of Macrophages in Different Tumor Microenvironments. Cancer Res. 2018;78(19):5492–503. doi: 10.1158/0008-5472.CAN-18-1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Malesci A, Bianchi P, Celesti G, Basso G, Marchesi F, Grizzi F, et al. Tumor-associated macrophages and response to 5-fluorouracil adjuvant therapy in stage III colorectal cancer. Oncoimmunology. 2017;6(12):e1342918. doi: 10.1080/2162402X.2017.1342918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Di Caro G, Cortese N, Castino GF, Grizzi F, Gavazzi F, Ridolfi C, et al. Dual prognostic significance of tumour-associated macrophages in human pancreatic adenocarcinoma treated or untreated with chemotherapy. Gut. 2016;65(10):1710–20. doi: 10.1136/gutjnl-2015-309193. [DOI] [PubMed] [Google Scholar]
- 39.Bohm S, Faruqi A, Said I, Lockley M, Brockbank E, Jeyarajah A, et al. Chemotherapy Response Score: Development and Validation of a System to Quantify Histopathologic Response to Neoadjuvant Chemotherapy in Tubo-Ovarian High-Grade Serous Carcinoma. J Clin Oncol. 2015 doi: 10.1200/JCO.2014.60.5212. [DOI] [PubMed] [Google Scholar]
- 40.Ghiringhelli F, Apetoh L, Tesniere A, Aymeric L, Ma Y-Y, Ortiz C, et al. Activation of the NLRP3 Inflammasome in Dendritic Cells Induces IL-1beta-dependent Adaptive Immunity Against Tumors. Nat Med. 2009;15:1170–8. doi: 10.1038/nm.2028. [DOI] [PubMed] [Google Scholar]
- 41.Zhang Z, Zhang Y, Xia S, Kong Q, Li S, Liu X, et al. Gasdermin E suppresses tumour growth by activating anti-tumour immunity. Nature. 2020;579(7799):415–20. doi: 10.1038/s41586-020-2071-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huang Y, Wang H, Hao Y, Lin H, Dong M, Ye J, et al. Myeloid PTEN promotes chemotherapy-induced NLRP3-inflammasome activation and antitumour immunity. Nat Cell Biol. 2020;22(6):716–27. doi: 10.1038/s41556-020-0510-3. [DOI] [PubMed] [Google Scholar]
- 43.Apte RN, Voronov E. Immunotherapeutic approaches of IL-1 neutralization in the tumor microenvironment. J Leukoc Biol. 2017;102(2):293–306. doi: 10.1189/jlb.3MR1216-523R. [DOI] [PubMed] [Google Scholar]
- 44.Van Gorp H, Lamkanfi M. The emerging roles of inflammasome-dependent cytokines in cancer development. EMBO Rep. 2019;20(6) doi: 10.15252/embr.201847575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Taniguchi K, Karin M. IL-6 and related cytokines as the critical lynchpins between inflammation and cancer. Seminars in immunology. 2014;26(1):54–74. doi: 10.1016/j.smim.2014.01.001. [DOI] [PubMed] [Google Scholar]
- 46.Ridker PM, MacFadyen JG, Thuren T, Everett BM, Libby P, Glynn RJ, et al. Effect of interleukin-1beta inhibition with canakinumab on incident lung cancer in patients with atherosclerosis: exploratory results from a randomised, double-blind, placebo-controlled trial. Lancet. 2017 doi: 10.1016/S0140-6736(17)32247-X. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.