Abstract
Cell-free systems allow interference with gene expression processes without requiring elaborate genetic engineering procedures This makes it ideally suited for rapid prototyping of synthetic biological parts. Inspired by nature’s strategies for the control of gene expression via short antisense RNA molecules, we here investigated the use of small DNA (sDNA) for translational inhibition in the context of cell-free protein expression. We designed sDNA molecules to be complementary to the ribosome binding site (RBS) and the downstream coding sequence of targeted mRNA molecules. Depending on sDNA concentration and the promoter used for transcription of the mRNA, this resulted in a reduction of gene expression of targeted genes by up to 50-fold. We applied the cell-free sDNA technique (cf-sDNA) to modulate cell-free gene expression from the native T7 phage genome by suppressing the production of the major capsid protein of the phage. This resulted in a reduced phage titer, but at the same time drastically improved cell-free replication of the phage genome, which we utilized to amplify the T7 genome by more than 15,000-fold in a droplet-based serial dilution experiment. Our simple antisense sDNA approach extends the possibilities to exert translational control in cell-free expression systems, which should prove useful for cell-free prototyping of native phage genomes and also cell-free phage manipulation.
Introduction
Cell-free (CF) transcription-translation systems have become a standard tool in bottom-up synthetic biology1 where they are used for a wide variety of applications ranging from the elucidation of basic biological mechanisms2–4 to the rapid prototyping and screening of synthetic biological parts.5–7 Cell-free gene expression also plays a central role in the construction and study of minimal living systems.8, 9 Various attempts have been made to create synthetic cellular systems by encapsulating CF gene expression into cell-scale compartments.10, 11 In this context, important steps have been made towards autonomously replicating compartments such as the enclosed synthesis of the membrane constituents,12 and the concomitant growth of the compartments.13 Next to growth and division, autonomous propagation also requires the replication of the genetic material contained in the synthetic cells, which ultimately would confer the potential for Darwinian evolution.14
Genomic replication in this context has been mostly studied based on bacteriophages, which typically have their own – comparatively simple – replication machinery.15 More than 40 years ago the seminal Spiegelman experiment16 already demonstrated replication and evolution of the RNA genome of phage Qβ based on its own replicase. In order to generate a truly self-replicating system, however, it would be required to produce the replicase itself and also other phage components during the replication cycle. In the case of Qβ phage, this was hampered by the requirement for longer genome lengths and the appearance of short parasitic RNA sequences during replication. But partial self-replication of Qβ genes have been already shown.17 As an alternative, only quite recently the replication machinery of the □29 phage (containing a dsDNA genome and a more faithful DNA polymerase) has been utilized successfully to replicate genetic templates of up to 100 kilobase pairs (kbp).18
Apart from their fundamental interest as simple self-replicating systems, phages have provided a plethora of useful tools for molecular biology such as a wide range of DNA and RNA polymerases, ligases, and other enzymes. Moreover, in face of rising antibiotic resistances, the old idea to use bacteriophages for therapeutic applications receives increasingly more attention. Modification of natural bacteriophages could be used to improve their therapeutic value, e.g., via tail fiber engineering to alter their host range,19 repression of their lysogenic life cycle20 or by improving their ability to degrade biofilms.21
Conventional phage genome engineering lacks efficiency due to the limited time phages spend inside their host bacteria. For this reason, techniques such as the homologous recombination screening require up to 104 to 1010 phages.22 More advanced techniques employing CRISPR-based control systems have recently been shown to reduce these numbers.23 In vivo phage manipulation is additionally limited by the natural toxicity for their hosts, and the lack of efficient transfection methods for certain (large) phages and their host bacteria. For the production of viable phages, it is further important to keep essential phage genes unchanged.
Many of the limitations of phage manipulation can be easily overcome in a cell-free context – first of all, there is no requirement for the host bacterium, and thus neither toxicity nor transfection is an issue. Further, it is even possible to knock-down phage genes essential for the natural replication cycles, which provides new opportunities for phage manipulation.
In the present work, we took inspiration from bacterial gene regulation via small RNAs (sRNAs),24, 25 to specifically suppress the expression of phage genes without altering the genome itself. Importantly, in the cell-free context chemically unstable sRNA can be replaced by small single-stranded DNA molecules (sDNA), which can be easily added to the cell-free expression system from outside. Similar in function to bacterial sRNAs, we used antisense sDNA to target the ribosome binding sites (RBS) of specific mRNA transcripts, which resulted in a strong downregulation of translation of the corresponding proteins.
In order to demonstrate the viability of this approach, we first showed cf-sDNA regulation of CF expression of the fluorescent protein YPet. We then applied the technique to alter CF gene expression from the genome of bacteriophage T7. When targeting the gene encoding the major capsid protein, which is an essential component of the T7 capsid, the titer of the phages produced in the system strongly decreased, while phage genome replication increased eightfold. Using this capability, we were able to realize the first Spiegelman-type serial dilution experiment with phage T7 in vitro.
Results and Discussion
Repression of cell-free expression of a gene encoding a fluorescent protein
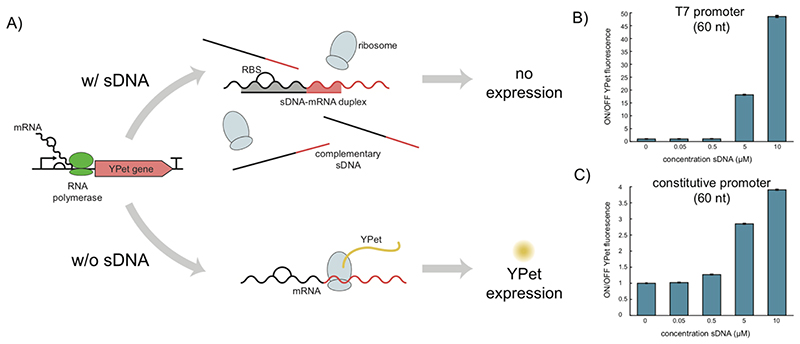
Control of gene expression levels via small RNA molecules is a widespread regulatory strategy in biology. Bacteria commonly use small RNAs, which are partially or completely complementary to mRNA (sub)-sequences. Hybridization of the sRNAs to their targets can modulate translation efficiency and mRNA stability – depending on the details, both enhancement and reduction are possible. One of the most straightforward regulatory mechanisms is based on the blockage of the ribosome binding site in the 5’ untranslated region of the mRNAs, which directly prevents translation initiation. We figured that the use of such short regulatory nucleic acids would be straightforward also in a cell-free context and allow the easy manipulation of gene expression reactions. As the stability of externally added RNA in extract-based cell-free systems is quite limited, we explored the use of short single-stranded DNA (sDNA) as cell-free translational repressors (Figure 1a).
Figure 1. Cell-free gene repression using small antisense DNA.
(A) Schematic overview of the working principle of the cf-sDNA technique. Following in vitro transcription of mRNA, sDNA already present in solution hybridizes with the targeted mRNA at the ribosome binding site and downstream sequence, which inhibits translational initiation and elongation. (B)+(C) Repression of YPet expression using different concentrations of sDNA as determined from YPet fluorescence. In (B) mRNA is transcribed from a T7 promoter while in (C) it is produced from an E.coli promoter (J23106). The ON/OFF YPet fluorescence is the ratio between the fluorescence of a negative control (0 μM sDNA, YPet fluorescence “ON”) and the fluorescence determined in samples with sDNA (YPet fluorescence “OFF”). Fluorescence values used are biological triplicates of fluorescence end levels after 13-14 h of incubation in the cell-free expression system. The given uncertainties are S.E., but they are hardly visible due to their small values.
To test the basic feasibility of this approach, we first investigated cf-sDNA mediated translational repression of mRNA for a fluorescent protein (YPet), which was transcribed from a plasmid in an E.coli -based cell extract.5, 26 Expression of YPet at 29°C was monitored via its fluorescence using a plate reader. The total length of the sDNA was 60 nucleotides (nt), which were complementary to the 8 nt long RBS and the following downstream sequence on the mRNA. Apart from blocking the RBS, sDNA-mRNA hybridization may also result in degradation of the mRNA by RNase H, which is also present in the cell extract.
We first studied the effect of the sDNA concentration and the promoter used for mRNA transcription on the repression efficiency by comparing the fluorescence end-levels of the corresponding CF transcription-translation reactions (Figures 1b & c). As expected, the repression strength increased with increasing concentrations of sDNA, while 0.05 μM did not show any effect when compared to the control. For both a T7 promoter (Figure 1b, Figure S1) and a constitutive E.coli promoter (J23106) (Figures 1c, Figure S2), repression was found to be highest at the maximum sDNA concentration used (10 μM). With roughly 50-fold repression compared to only 4-fold, the ON/OFF ratio achieved was much higher for the T7 promoter than for the constitutive promoter. Since sDNA and the ribosome compete for the same binding site on the mRNA, it is possible to gradually tune the repression strength of the targeted gene by changing the concentration of the sDNA.
We surmise that the different results for the two promoters is a consequence of the differing transcription rates of the RNA polymerases (RNAPs) used (20-90 nt/s for the E. coli RNAP vs. 240 nt/s for T7 RNAP). In contrast to the “slower” E.coli RNAP, T7 RNAP is expected to generate mRNA in excess, which creates an imbalance between available ribosomes and mRNAs and also increases sDNA-mRNA hybridization rates. The impact of batch-to-batch variations of the homemade cell extract can be seen in Figure S3. For comparison we also performed experiments with the commercially available cell extract myTXTL and PURExpress (Figure S4).
Influence of sDNA length on translation repression
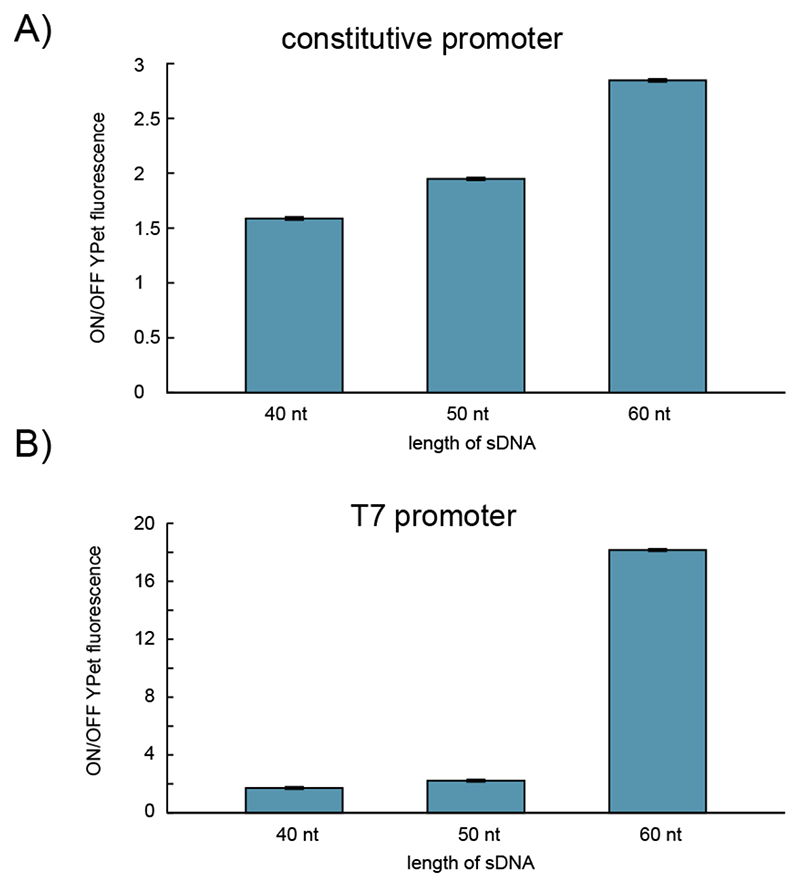
We next studied the efficiency of translational repression by cf-sDNA as a function of length. To this end, we tested 40, 50 and 60 nt long sDNAs at a concentration of 5 μM each, both for the constitutive and the T7 promoter (Figure 2). For both promoters, translational repression clearly increased with the length of the sDNA. This is consistent with the higher thermodynamic stability of longer sDNA-mRNA duplexes, which shifts the equilibrium away from ribosomes initiating translation at the RBS. Apart from this, longer sDNA-mRNA stretches will be less likely displaced by ribosomes, and will be more frequently attacked by RNaseH. Further, longer sDNA has a higher chance of finding accessible sequence regions in mRNAs rich in secondary structure. Surprisingly, however, a reduction of sDNA length from 60 nt to 50 nt reduces repression strength in the case of the T7-transcribed mRNA to the much lower level of the E.coli RNAP-transcribed molecules. An analysis of the minimal free energy of the RNA-DNA dimers by using the Vienna RNA Suite27 and NUPACK.
Figure 2. Repression efficiency of 5 μM sDNA with different lengths for a YPet gene controlled.
(A) by an E.coli promoter (J23106) and (B) by a T7 promoter. The ON/OFF YPet fluorescence is the ratio between the fluorescence of a negative control (0 μM sDNA, YPet fluorescence “ON”) and the fluorescence measured in samples with sDNA (YPet fluorescence “OFF”). Fluorescence values used are technical triplicates of fluorescence end levels after 13-14 h in the cell-free expression system. The given uncertainties are S.E., but they are hardly visible due to their small values.
Gene knockdown in a native phage genome
We moved on to apply the cf-sDNA strategy in the context of cell-free phage manipulation. In conventional approaches, the function of specific phage genes is probed via the generation of corresponding knockout variants of the genome, for which it is necessary to alter the phage genome. Although analogous techniques used for mammalian cells have evolved quickly in the past, bacteriophages still require laborious screening experiments.22 As it has been shown that various E.coli phages can be produced from their genomes in vitro, new opportunities have opened up that allow cell-free engineering and manipulation of the bacteriophages. For instance, the T7 phage can replicate its genome with only three phage proteins (gp5, gp4 and gp2.5) and one host protein (thioredoxin)28 without the T7 RNAP. After a vast in vitro production of gp5, gp4 and gp2.5 these proteins should be able to replicate the phage genome. In this context, cf-sDNA could provide a simple and straightforward approach towards in vitro knock-down studies.
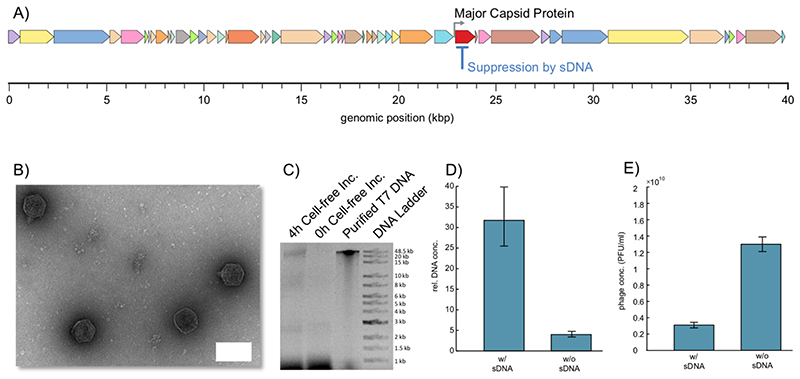
To demonstrate this capability, we here used cf-sDNA to suppress translation of the major capsid protein of T7 phage (Figure 3a). The gene of interest is controlled by a T7 promoter and is terminated by a T7 terminator (T7-T phi), which naturally makes it a suitable candidate for our approach. Further, the major capsid protein is not encoded in a polycistronic region of the genome. In the experiments, we first supplied the T7 phage genome (without sDNA) to the CF expression system and measured phage expression after 4 h at 29 °C using a plaque assay, which resulted in phage concentrations of about 3×109 PFU/mL to 1.3×1010 PFU/mL. Additionally, transmission electron microscopy (TEM) was used to check the formation of the phages (Figure. 3b). In negative control experiments – either without DNA or without cell extract – no phages where detected. Upon addition of deoxy-nucleoside triphosphates (dNTPs) to the expression system containing the phage genome, also replication of the T7 DNA could be detected after 4h of incubation using agarose gel electrophoresis (AGE)(Figure 3c).
Figure 3. Repressing the gene of the major capsid protein of phage T7.
(A) Schematic illustrating of the full genome of the T7 phage with its encoded genes. (B) Transmission electron micrograph of active T7 phages formed in the cell-free system after 4 h of incubation at 29 °C in the absence of sDNA. Scale bar: 50 nm. (C) The presence of the T7 phage genome in the cell-free system at the start (0h) and after 4 h of incubation in the cell-free expression system at 29 °C was checked using agarose gel electrophoresis. Purified T7 DNA was taken from a column-purified T7 genome extracted from a phage stock. The visible bands in the range from 1 kb to 1.5 kb result from the cell-free system, while the bands below 0.5 kb are residual DNA fragments from the phage DNA preparation. The high molecular weight band after 4h indicates successful replication of the full-length phage genome. (D) Relative concentration of the phage DNA incubated in the cell extract used with and without complementary sDNA. The relative concentration of the genomic DNA was determined via qPCR. (E) Concentration of active phages in PFU/mL measured utilizing a plaque assay for samples treated with and without sDNA. The given uncertainties are S.E.
We then targeted the major capsid gene for knockdown with a 60 nt long sDNA covering its RBS and downstream sequence. As the major capsid protein is essential for the assembly of the phage capsids, we expected a profound effect of its knockdown on gene expression in the CF system. We quantified the replication yield of the T7 genome with and without sDNA via qPCR. To this end, we targeted the region between gene locations 8426 and 9007 by the qPCR primers. The calibration curve for the DNA concentration calculation was measured beforehand using purified phage DNA (Figure S7) In the control sample without sDNA, the DNA concentration showed a 4-fold increase, whereas in the sample containing sDNA the DNA replication increased by more than 30-fold (Figure 3d). We also tested how the repression of the major capsid protein affected production of active phages and found that the phage titer roughly decreased by a factor of four (Figure 3e).
These results demonstrate that cf-sDNA technique can be successfully used to alter gene expression levels of a native phage without genome engineering. The reduction in phage titer is easily understood due to the essential role of the major capsid assembly. The concomitant increase in DNA replication can be explained through the higher amount of DNA available for replication, since packaging of the genome into the phage capsid is impaired by the knockdown. Furthermore, since the RBS is blocked, the ribosomes are not sequestered by the mRNA and might be free for the translation of more DNA replication proteins.
T7 genome replication in serial dilution experiments
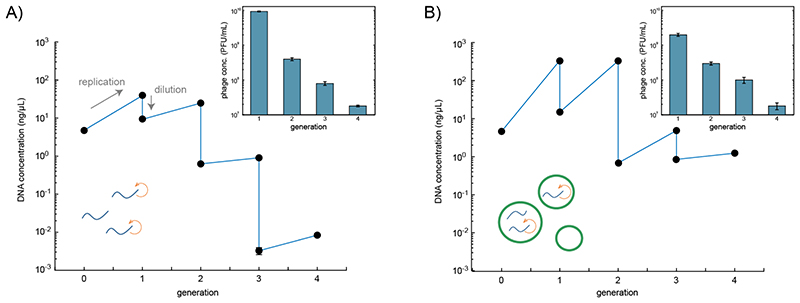
Encouraged by the strong improvement in genome replication caused by the sDNA-mediated repression of the major capsid protein, we next tested whether one could perform multiple replication rounds for the T7 phage genome in a serial dilution setting similar to the original Spiegelman experiment.16 To this end, phage DNA (0.5 nM) was incubated in the cell-free expression system supplemented with dNTPs (5 μM) at 29 °C for 4 h, and diluted afterwards by transferring 3 % (v/v) of the sample into freshly prepared cell extract, followed by another incubation round at 29 °C for 4 h (Figure 4a). Before each dilution step, the DNA concentration was quantified using qPCR as described above. Further, the replication of the full genome was again verified using AGE (Figure S8). As expected, the DNA concentration increased due to replication in each generation. Typical for serial dilution experiments, however, the amplification per generation decreased with each dilution step.
Figure 4.
Serial transfer experiments with T7 DNA in cell extract. In every generation the phage DNA was incubated for 4 h at 29 °C in freshly supplied cell extract, followed by a dilution to 3 % (v/v) of its original amount. The DNA concentration of each sample was measured by qPCR at the beginning of the incubation and after 4 h of incubation. Insets: concentration of active T7 phages determined using a plaque assay. All data points represent technical triplicates. In (A) incubation was carried out in bulk solution, while in (B) cell extract and phage DNA were incubated in picoliter-sized water-in-oil droplets. After four dilution steps the theoretical amplification is 80-fold in (A) and 15,698-fold in (B). The given uncertainties are S.E.
To explore the influence of encapsulation within cell-scale compartments on DNA replication efficiency, we enclosed the phage genome and the cell-free system inside water-in-oil emulsion droplets formed by manual shaking.29 After 4 h of incubation at 29 °C the emulsion was broken and its contents mixed. As before, DNA amplification was quantified via qPCR and AGE, and 3 % of the mixture were combined with fresh cell extract and encapsulated for another replication round.
As in the bulk experiment, the replication efficiency is lower in the later generations of the serial dilution experiments. However, instead of only a ≈ 10-fold increase in DNA concentration per generation in bulk, the encapsulated samples showed a 100-fold increase of DNA concentration in the first two generations. After four generations the DNA concentrations in droplets were still 100-fold larger than the bulk concentrations. The plaque assays carried out after each generation showed a reduction in the concentration of active phages from generation to generation (Figure 4 insets) for both the bulk and droplet experiment. The phage titers determined for these two experiments were almost the same, except for the first generation where in droplets nearly one order of magnitude less active phages were produced.
The global reduction of replicated DNA from generation to generation might be caused by the accumulation of mutations in genes required for DNA replication or the appearance of short parasitic DNA sequences, which are replicated faster than the full genome. Another explanation could be the accumulation of inhibitory products, which would lead to decreased polymerase expression, etc. All mechanisms reduce the production of essential proteins required for DNA replication.
When phages reproduce in bacteria, parasitic sequences get lost after lysis or result in less active or inactive phages, whose number will diminish under evolutionary competition. In serial dilution experiments, evolutionary pressure is reduced, and thus parasitic sequences can accumulate over time and finally poison the system. As a consequence, replication of the full phage genome is reduced. This hypothesis is also supported by our observation that the reduction in DNA concentration over four generations is considerably less for the compartmentalized replication reaction than in bulk, where parasitic DNA sequences can spread over the whole system more rapidly.
Conclusion
In summary, we were able to show that small antisense DNA molecules (sDNA) complementary to the RBS and coding region of an mRNA can be used to suppress gene expression in a cell-free expression system, analogously to the action of sRNAs in bacteria, to RNAi in eucaryotes and similarly to CRISPRi. Translational repression by sDNA is highly sequence specific and orthogonal as it relies on complementary base pairing of a 60 nt long DNA strand.
Based on externally added, synthetic DNA, cf-sDNA provides a quick and simple approach to control gene expression outputs in vitro. Protein expression levels can be tuned via the sDNA concentration, which potentially could be used for the fine-tuning of in vitro gene networks.
As shown in this work, cf-sDNA could be a particularly useful tool for the cell-free manipulation of phage genomes. Phage manipulation is laborious and is complicated by the huge variety of existing bacteriophages. We have demonstrated that sDNA allows to tune the CF expression of phage proteins in a rapid and straightforward manner. Systematic application of cf-dsDNA in this context could help to improve the functional annotation of the proteomes of newly found phages drastically. At the moment, cf-sDNA is still limited to the control of monocistronic genes, however. Using cf-sDNA repression, we were also able to strongly improve in vitro replication of a 40 kbp long phage genome in a serial dilution experiment, enabling one of the first “Spiegelman-type” experiments with phage T7. Because of the high processivity of T7 DNA polymerase and the low replication error rate of about 15×10-6 bases/genome/replication, the T7 DNA replication system could be an interesting module for the realization of synthetic cells. As shown here, repression of the major capsid protein via cf-sDNA provides a simple approach to harness its capabilities.
Methods
Transcription translation reaction (TX-TL)
For the generation of crude S30 cell extract a BL21-Rosetta 2(DE3) mid-log phase culture was bead-beaten with 0.1 mm glass beads in a Minilys homogenizer (Peqlab, Germany) as described in Sun et all.26 The composite buffer contained 50 mM HEPES(pH 8), 1.5 mM ATP and GTP, 0.9 mM CTP and UTP, 0.2 mg/mL tRNA, 26 mM coenzyme A, 0.33 mM NAD, 0.75 mM cAMP, 68 mM folinic acid, 1 mM spermidine, 30 mM PEP, 1 mM DTT and 2 % PEG-8000, 6 mM Mg-glutamat 80mM K-glutamat and amino acids.
As an energy source in this buffer phosphoenolpyruvate (PEP) was utilized instead of 3-phosphoglyceric acid (3-PGA). All components were stored at −80 °C before usage. A single cell-free reaction consisted of 42 % (v/v) composite buffer, 25 % (v/v) DNA plus additives and 33 % (v/v) S30 cell extract. For ATP regeneration 13.3 mM maltodextrin and 1 U of T7 RNA polymerase (NEB, M0251S) were added to the cell-free reaction mix. All measurements took place at 29 °C with 2 nM of plasmid unless indicated otherwise.
Phage assembly
Phages were assembled according to Rustad et al.30 with the following adjustments. Phage DNA was mixed with a TX-TL system based on cell extract, an energy solution and an amino acid solution as described in Sun et al.26 with one amendment in Buffer B (6 mM Mg-glutamate, 100 mM K-glutamate, 3 mM DTT, 1.5 mM each amino acid except leucine, 1.25 mM leucine, 50 mM HEPES, 1.5 mM ATP and GTP, 0.9 mM CTP and UTP, 0.2 mg/ml tRNA, 0.26 mM CoA, 0.33 mM NAD, 0.75 mM cAMP, 0.068 mM folinic acid, 1 mM spermidine, 30 mM 3-PGA, 4 % PEG-8000, 6 mM Mg-glutamat 80mM K-glutamat and amino acids).5 For 6 reactions à 13 μL, 2.5 μL PEG 8000 (36 % w/v), 4 μL dNTPs (25 mM), 0.8 μL ATP (500 mM), 37.5 μL Buffer B, 2 μL GamS (150 μM), 28.5 μL TX-TL and 1.6 μL DNA (10 nM) were mixed with nuclease free water to a final volume of 80 μL. All constituents were mixed (except DNA), chilled for 5 min on ice, followed by the addition of DNA. This 13 μL assembly mix was incubated for 4 h at 29 °C to generate the phages.
Transmission electron microscopy
The vesicle solution was adsorbed on glow discharged formvar-supported carbon-coated Cu400 TEM grids (FCF400-CU, Science Services, Munich, Germany) for 2 min, followed by negative staining using a 2 % aqueous uranyl formate solution with 25 mM sodium hydroxide for 45 s. Afterwards the grid was dried and stored under vacuum for 30 min. Imaging was carried out using a Philips CM100 transmission electron microscope at 100 kV. For image acquisition an AMT 4 mega-pixel CCD camera was used and imaging was performed at a magnification between ×8,900 and ×15,500. For image processing the plugin Scale Bar Tools for Microscopes for Java-based software ImageJ was used.
Fluorescence measurements
Cell-free expression and transcription was characterized via plate reader measurements, with the corresponding filter sets for the fluorescence (BMG FLUOstar Optima) using 15 mL reaction volumes in 384-well plates.
qPCR
For the quantification of the amplification of the T7 DNA the samples were flash frozen in liquid nitrogen and stored at -80 °C until all samples were collected. The qPCR reactions were performed on a BioRad IQ5 instrument by cycling 1x 1 min 95 °C, 45x 30 s 95 °C and 15 s 60 °C, 1x melt curve 55-95 °C. The reactions were prepared with 7 μL of 1:100 diluted cell extract sample containing the DNA and LunaScript Universal MasterMix 2x (New England Biolabs) in white PCR stripes with flat lid (AB-1191, ThermoFisher). Three technical replicates were recorded for each sample. The design for the specific primers for the recorded gene can be found in the supporting information (SI section DNA sequences). The cycle threshold (ct) values were calculated by the intersection of the fluorescence curve from the DNA dye at 20 % of the maximum intensity of the brightest sample. From the ct value the concentration was determined based on a beforehand measured calibration curve (Figure S7).
Droplet generation
FC-40 oil (Sigma Aldrich, # F9755) with 2 % (w/w) PFPE/PEG-surfactant (Raindance Technologies) were used to create the droplets by shaking (Figure S6).29 After the reaction, droplets were broken by adding perfluorooctanol (PFO; 370533, Sigma) to the droplets (five volumes of PFO to one volume of the aqueous droplet contents).
T7 stock preparation
A single plaque of the T7 phage was picked to be incubated at 37 °C and 250 rpm in a rotary shaker with the corresponding host-bacteria at an OD of 0.4 at 600 nm. The sample was incubated for approximately 2 h until the lysis cleared the solution. The solution was then centrifuged at 5000 rcf at 4 °C for 5 minutes and the supernatant containing the phages was mixed with 10 % w/v PEG 8000 and 1 M NaCl for precipitation of the phages. After storage of the phages at 4 °C overnight, the sample was centrifuged for 15 minutes at 4 °C at 7000 rcf. After discarding the supernatant, the precipitated phages were resuspended in phage buffer (1x PBS, 1 mM MgCl2, 1 mM MgSO4), followed by a filtration with a 0.45 μm sized filter. The resulting titer was measured by a plaque assay.
Plaque assay
For the plaque-assay 0.5 % agarose NZCYM medium was melted, split into 4 mL aliquots and stored in a water bath at 48 °C. Separately, the assembled phages were diluted 102-108-fold in phage buffer (1x PBS, 1mM MgCl2, 1mM MgSO4). 100 μL of each dilution was mixed with an equal volume of corresponding host bacterium overnight culture. This mixture was added to the 0.5 % agarose NZCYM medium aliquots and poured on a 1 % NZCYM agar plate (Figure S9). After the suspension had solidified at room temperature, the plates were incubated at 37 °C until plaques became visible.
Supplementary Material
The Supporting Information is available free of charge on the ACS Publications website at DOI: Plasmid, DNA, RNA and primer sequences used in this study; fluorescence plate reader data; microscopy image; qPCR calibration curve; agarose gel and agar plate images; free energy values for DNA-RNA dimerization (PDF)
Acknowledgment
Funded by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) – Project-ID 364653263 – TRR 235 and by the European Research Council (grant No. 694410, project AEDNA). We would like to thank Lukas Oesinghaus for providing help with the evaluation of the qPCR Data. We thank Jean-Paul Pirnay, Maya Merabishvili and Yves Briers for helpful discussions.
Footnotes
Author Contributions
K.V. and T.P. designed research. K.V. performed research, E.F. and S.v.S. assisted in cell extract preparation. K.V., F.C.S., and T.P. analyzed data. K.V., F.C.S., and T.P. wrote the manuscript.
Notes
A patent application has been filed by the authors’ employer, the Technical University of Munich. Invitris has an option to negotiate a license to the patent rights. The authors declared no other potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
References
- 1.Laohakunakorn N, Grasemann L, Lavickova B, Michielin G, Shahein A, Swank Z, Maerkl SJ. Bottom-Up Construction of Complex Biomolecular Systems With Cell-Free Synthetic Biology. Front Bioeng Biotechnol. 2020;8:431. doi: 10.3389/fbioe.2020.00213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moore SJ, MacDonald JT, Wienecke S, Ishwarbhai A, Tsipa A, Aw R, Kylilis N, Bell DJ, McClymont DW, Jensen K, Polizzi KM, et al. Rapid acquisition and model-based analysis of cell-free transcription-translation reactions from nonmodel bacteria. Proc Natl Acad Sci USA. 2018;115(19):E4340. doi: 10.1073/pnas.1715806115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wandera KG, Collins SP, Wimmer F, Marshall R, Noireaux V, Beisel CL. An enhanced assay to characterize anti-CRISPR proteins using a cell-free transcription-translation system. Methods. 2020;172:42–50. doi: 10.1016/j.ymeth.2019.05.014. [DOI] [PubMed] [Google Scholar]
- 4.Yim SS, Johns NI, Park J, Gomes ALC, McBee RM, Richardson M, Ronda C, Chen SP, Garenne D, Noireaux V, Wang HH. Multiplex transcriptional characterizations across diverse bacterial species using cell-free systems. Mol Syst Biol. 2019;15(8):e50762. doi: 10.15252/msb.20198875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Garamella J, Marshall R, Rustad M, Noireaux V. The All E. coli TX-TL Toolbox 2.0: A Platform for Cell-Free Synthetic Biology. ACS Synth Biol. 2016;5(4):344–355. doi: 10.1021/acssynbio.5b00296. [DOI] [PubMed] [Google Scholar]
- 6.Niederholtmeyer H, Sun ZZ, Hori Y, Yeung E, Verpoorte A, Murray RM, Maerkl SJ. Rapid cell-free forward engineering of novel genetic ring oscillators. eLife. 2015;4:e09771. doi: 10.7554/eLife.09771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sun ZZ, Yeung E, Hayes CA, Noireaux V, Murray RM. Linear DNA for Rapid Prototyping of Synthetic Biological Circuits in an Escherichia coliBased TX-TL Cell-Free System. ACS Synth Biol. 2014;3(6):387–397. doi: 10.1021/sb400131a. [DOI] [PubMed] [Google Scholar]
- 8.Schwille P, Spatz J, Landfester K, Bodenschatz E, Herminghaus S, Sourjik V, Erb TJ, Bastiaens P, Lipowsky R, Hyman A, Dabrock P, et al. MaxSynBio: Avenues Towards Creating Cells from the Bottom Up. Angew Chem. 2018;57(41):13382–13392. doi: 10.1002/anie.201802288. [DOI] [PubMed] [Google Scholar]
- 9.Vogele K, Pirzer T, Simmel FC. Genetically Encoded Membranes for Bottom-Up Biology. ChemSystemsChem. 2019;46:2543 [Google Scholar]
- 10.Adamala KP, Martin-Alarcon DA, Guthrie-Honea KR, Boyden ES. Engineering genetic circuit interactions within and between synthetic minimal cells. Nat Chem. 2016:1–66. doi: 10.1038/nchem.2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Noireaux V, Libchaber A. A vesicle bioreactor as a step toward an artificial cell assembly. Proc Natl Acad Sci USA. 2004;101(51):17669–17674. doi: 10.1073/pnas.0408236101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Scott A, Noga MJ, de Graaf P, Westerlaken I, Yildirim E, Danelon C. Cell-Free Phospholipid Biosynthesis by Gene-Encoded Enzymes Reconstituted in Liposomes. PLOS ONE. 2016;11(10):e0163058. doi: 10.1371/journal.pone.0163058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vogele K, Frank T, Gasser L, Goetzfried MA, Hackl MW, Sieber SA, Simmel FC, Pirzer T. Towards synthetic cells using peptide-based reaction compartments. Nat Commun. 2018;9(1) doi: 10.1038/s41467-018-06379-8. 3862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Matsumura S, Kun Á, Ryckelynck M, Coldren F, Szilágyi A, Jossinet F, Rick C, Nghe P, Szathmáry E, Griffiths AD. Transient compartmentalization of RNA replicators prevents extinction due to parasites. Science. 2016;354(6317):1293–1296. doi: 10.1126/science.aag1582. [DOI] [PubMed] [Google Scholar]
- 15.Weigel C, Seitz H. Bacteriophage replication modules. FEMS Microbiol Rev. 2006;30(3):321–381. doi: 10.1111/j.1574-6976.2006.00015.x. [DOI] [PubMed] [Google Scholar]
- 16.Mills DR, Peterson RL, Spiegelman S. An extracellular Darwinian experiment with a self-duplicating nucleic acid molecule. Proc Natl Acad Sci USA. 1967;58(1):217. doi: 10.1073/pnas.58.1.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mizuuchi R, Ichihashi N. Sustainable replication and coevolution of cooperative RNAs in an artificial cell-like system. Nat Ecol Evol. 2018;2(10):1654–1660. doi: 10.1038/s41559-018-0650-z. [DOI] [PubMed] [Google Scholar]
- 18.Libicher K, Hornberger R, Heymann M, Mutschler H. In vitro self-replication and multicistronic expression of large synthetic genomes. Nat Commun. 2020;11(1):904. doi: 10.1038/s41467-020-14694-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ando H, Lemire S, Pires DP, Lu TK. Engineering Modular Viral Scaffolds for Targeted Bacterial Population Editing. Cell Syst. 2015;1(3):187–196. doi: 10.1016/j.cels.2015.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dedrick RM, Guerrero-Bustamante CA, Garlena RA, Russell DA, Ford K, Harris K, Gilmour KC, Soothill J, Jacobs-Sera D, Schooley RT, Hatfull GF, et al. Engineered bacteriophages for treatment of a patient with a disseminated drug-resistant Mycobacterium abscessus. Nat Med. 2019;25(5):730–733. doi: 10.1038/s41591-019-0437-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lu TK, Collins JJ. Dispersing biofilms with engineered enzymatic bacteriophage. Proc Natl Acad Sci USA. 2007;104(27):11197. doi: 10.1073/pnas.0704624104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pires DP, Cleto S, Sillankorva S, Azeredo J, Lu TK. Genetically Engineered Phages: a Review of Advances over the Last Decade. Microbiol Mol Biol Rev. 2016;80(3):523. doi: 10.1128/MMBR.00069-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kiro R, Shitrit D, Qimron U. Efficient engineering of a bacteriophage genome using the type I-E CRISPR-Cas system. RNA Biol. 2014;11(1):42–44. doi: 10.4161/rna.27766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Villa JK, Su Y, Contreras LM, Hammond MC. Synthetic Biology of Small RNAs and Riboswitches. Microbiol Spectr. 2018;6(3) doi: 10.1128/microbiolspec.rwr-0007-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wassarman KM. Small RNAs in Bacteria. Proc Natl Acad Sci USA. 2002;109(2):141–144. doi: 10.1016/s0092-8674(02)00717-1. [DOI] [PubMed] [Google Scholar]
- 26.Sun ZZ, Hayes CA, Shin J, Caschera F, Murray RM, Noireaux V. Protocols for implementing an Escherichia coli based TX-TL cell-free expression system for synthetic biology. J Vis Exp. 2013;(79):e50762. doi: 10.3791/50762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gruber AR, Lorenz R, Bernhart SH, Neuböck R, Hofacker IL. The Vienna RNA Websuite. Nucleic Acids Res. 2008;36(suppl_2):W70–W74. doi: 10.1093/nar/gkn188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hamdan SM, Richardson CC. Motors, Switches, and Contacts in the Replisome. Annu l Rev Biochem. 2009;78(1):205–243. doi: 10.1146/annurev.biochem.78.072407.103248. [DOI] [PubMed] [Google Scholar]
- 29.Weitz M, Kim J, Kapsner K, Winfree E, Franco E, Simmel FC. Diversity in the dynamical behaviour of a compartmentalized programmable biochemical oscillator. Nat Chem. 2014;6(4):295–302. doi: 10.1038/nchem.1869. [DOI] [PubMed] [Google Scholar]
- 30.Rustad M, Eastlund A, Marshall R, Jardine P, Noireaux V. Synthesis of Infectious Bacteriophages in an E. coli-based Cell-free Expression System. J Vis Exp. 2017;(126):e56144. doi: 10.3791/56144. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The Supporting Information is available free of charge on the ACS Publications website at DOI: Plasmid, DNA, RNA and primer sequences used in this study; fluorescence plate reader data; microscopy image; qPCR calibration curve; agarose gel and agar plate images; free energy values for DNA-RNA dimerization (PDF)