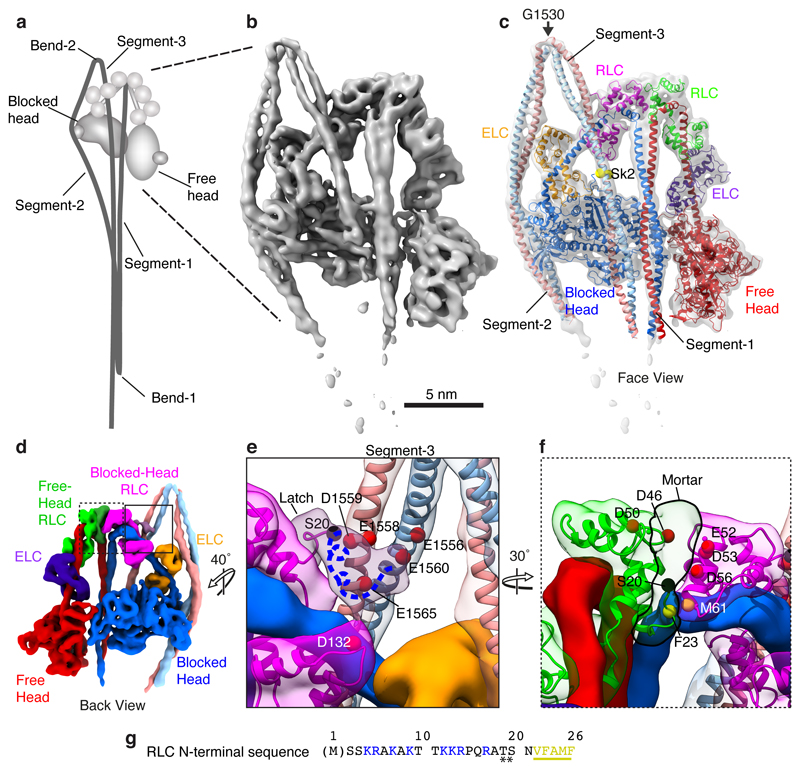
Figure 1. Structure of the heads region of shutdown SmM and contributions of the RLCs to the shutdown state.
a, Cartoon of the entire folded molecule showing head-head interaction (IHM), and path of the folded tail including its three segments and two bends. b, cryoEM density map of the heads region (face view) thresholded (0.369) to display secondary structure. c, Backbone depiction of pseudo-atomic model fitted into the density map with individual chains labelled. Residue G1530 is at the centre of bend-2; Sk2 denotes skip residue 2 (Q1592). d, Density map as in b (back view), segmented and coloured by chain as in c, boxed regions shown enlarged (e and f) to highlight additional density of the blocked-head RLC-N-terminal extension ‘latch’ (e) and free-head and RLC-N-terminal extension ‘mortar’ (f). Canonical RLCs shown translucent with fitted model. In e & f, the phosphorylatable serine (S20) is highlighted (black sphere). The approximate path of the remaining N-terminal extension (containing positively-charged residues (as shown in g) depicted as a blue dashed line. In e, a ring of negatively-charged residues in segment-3 is shown as red Cα spheres. In f, negatively-charged residues in RLCs that could interact with positively-charged residues in the N-terminal extension are shown as red Cα spheres; the hydrophobic interface between free-head and blocked-head RLCs is indicated by yellow spheres. g, Amino acid sequence of SmM RLC-N-terminal extension; basic residues highlighted blue; hydrophobic residues highlighted yellow. (Note: N-terminal Met is residue 1 in our numbering, thus residue numbers used in this paper are in some cases one higher than elsewhere in the literature.)