Abstract
Contact-active antimicrobial polymer surfaces bear cationic charges and kill or deactivate bacteria by interaction with the negatively charged parts of their cell envelope (lipopolysaccharides, peptidoglycan, and membrane lipids). The exact mechanism of this interaction is still under debate. While cationic antimicrobial polymer surfaces can be very useful for short term applications, they lose their activity once they get contaminated by a sufficiently thick layer of adhering biomolecules or bacterial cell debris. This layer shields incoming bacteria from the antimicrobially active cationic surface moieties.
Besides discussing antimicrobial surfaces, this feature article focuses on recent strategies that were developed to overcome the contamination problem. This includes bifunctional materials with simultaneously presented antimicrobial and protein-repellent moieties; polymer surfaces which can be switched from an antimicrobial, cell-attractive to a cell-repellent state; polymer surfaces that can be regenerated by enzyme action; degradable antimicrobial polymers; and antimicrobial polymer surfaces with removable top layers.
Keywords: antimicrobial polymers, bioactive polymers, coatings, polymer surfaces, structure-property relationships
Figure For Toc_Abstract.
Introduction: Antimicrobial Resistance and Bacterial Infection in the Context of Medical Devices
Antimicrobial resistance
Antimicrobial resistance is a threat in modern healthcare and medicine. To put it in the words of the World Health Organization (WHO) ‘antibiotic resistance is no longer a prediction for the future; it is happening right now, across the world, and is putting at risk the ability to treat common infections in the community and hospitals. Without urgent, coordinated action, the world is heading towards a post-antibiotic era, in which common infections and minor injuries, which have been treatable for decades, can once again kill’.[1] Leading health organizations registered globally increasing numbers of antibiotic-resistant bacterial strains and a growing prevalence of resistant bacteria among pathogens in hospitals and the community.[1–4] Examples are Staphylococcus aureus that causes the majority of hospital-acquired bloodstream, skin and surgical site infections, strains of Escherichia coli that lead to severe urinary tract infections or bloody diarrhea (enterohemorrhagic E. coli, EHEC), and Pseudomonas aeruginosa, which causes bloodstream infections and pneumonia.[1] Strains of Enterococcus faecalis, commonly found in the gastrointestinal tract of humans, were associated with endocarditis, bloodstream infections, urinary tract infections, peritonitis and intra-abdominal abscesses.[4] In most of Europe, the resistance rate of E. faecalis against aminoglycoside-based antibiotics is now between 25% and 50%.[4] These reports on antibiotic resistance are already worrying, and become even more aggravating by the fact that we are running out of back-up antibiotics. Only few new classes of antibiotics, such as cyclic lipopeptides and oxazilidinones, have been discovered since the 1970s, and these have been associated with severe side effects, such as reduced bone marrow formation and neurotoxicity.
Infections in the context of medical devices
But how do bacterial infections come about in the first place? One important infection pathway in clinical settings is the contact between the human body and a medical device or implant, which may become an entrance door for bacteria. For example, single bacteria on urinary catheters can form a biofilm in less than 24 hours.[5] Patients that received a urinary catheter have an infection risk of 50% after 10 days, and 100% after 30 days.[6] As Busscher pointed out recently, ‘there is no such thing as a sterile operation theatre’:[7] even under the most rigorous hygiene protocols, contamination of implantation sites with single bacteria cannot be prevented. However, once these individual bacteria form colonies and are protected by the extracellular matrix of a biofilm, they are out of reach for our immune system, and antibiotics also fail to kill them. Studies have shown that an antibiotics concentration of up to 1000 times the regular dose is needed to eliminate bacteria encapsulated in biofilms.[8] Thus, materials which reduce bacterial infections associated with medical devices and implants would be highly welcome in the medical field.
Various kinds of polymer-based surfaces with and without intrinsic antimicrobial activity can be used to fight bacterial biofilms. They can be divided into “passive” materials that prevent protein adhesion (an initial step in biofilm formation), and “active” materials where the polymer itself is antimicrobial, or the matrix for another active ingredient. In the following two sections, we will discuss these two classes of polymers, as well as their shortcomings. We will then turn to strategies used to obtain antimicrobial polymer surfaces with longer-lasting or renewable antimicrobial activity. These include the combination of protein-repellent polymers with antimicrobial components, polymer surfaces with switchable properties, and the regeneration of antimicrobial surface activity, e.g. using enzymatic action or degradable polymers.
The Mechanism of Biofilm Formation, and Protein-repellent/”Antifouling” Polymer Surfaces
Biofilm formation
Biofilm formation is a problem in different and diverse situations. Prominent examples of biofilms are plaque on teeth, marine biofilms on ship hulls, sludge in water pipelines, and biofilms consisting of bacteria and yeasts on medical devices like urinary catheters.[9–11] A microbial biofilm typically consists of more than one species. These pathogens co-colonize surfaces to protect themselves jointly against biocides, external shear forces, or the immune system of a host organism.[5] Biofilm formation is initiated when a surface is immersed into a biological fluid. That surface is covered by proteins within seconds. The proteins form a so-called conditioning layer, to which all kinds of microorganisms can adhere and bind. In the following discussion, we will focus on bacteria. The initial binding of bacteria to the surface is reversible until the bacteria secrete adhesins (adhesive proteins), and thereby attach irreversibly to the substrate.[11, 12] The next step is proliferation and colony formation, followed by secretion of a thick peptidoglycan layer, the so-called extracellular matrix.[13] Quorum sensing and other kinds of chemical communication between the bacteria enable the cohabitation of various bacterial species inside a single extracellular matrix, and thus the maturation of the biofilm.[14] As the cells inside the biofilm proliferate further, pressure builds up inside the biofilm, and the extracellular matrix eventually ruptures. This enables planktonic bacteria and biofilm fragments to leave the biofilm and spread the infection further.[14]
Classification of “anti-fouling” polymer surfaces
There are a number of polymer-based materials that can prevent or at least slow down biofilm formation. Some of these materials target the first step of biofilm formation. They prevent the irreversible adhesion of proteins to the surface and are generally termed “anti(bio)fouling materials”. Unfortunately, this is an ill-defined expression, since it does not refer to proteins, but to biofouling in general, and in its common usage the molecule or organism whose adhesion is prevented by the “anti-fouling” coating is not specified. Terms like protein-repellent, bacteria-repellent, or zoosphore-repellent, are more precise and should be used instead when only specific material-species interactions are studied. We here use the word “anti-fouling” as a generic term for anti-adhesive, protein-repellent, or bacteria-repellent materials, or other materials that prevent the organism-specific interactions with a surface. “Anti-fouling” coatings are designed to prevent biofilm formation by keeping the interaction of the surface with its biological environment reversible. To do so, the adhesive forces between incoming bacteria, proteins or other biomolecules and the surface must be minimized. Most protein-repellent or “anti-fouling materials” fall into two main categories – so called non-fouling materials that have a low interfacial energy with water, and fouling-release materials that require a low amount of energy to remove adhered contaminations. For a bacterial cell or biomolecule, it is energetically not advantageous to settle on a nonfouling polymer coating because of the low interfacial energy with water of that material. By attaching to such a surface, only little enthalpy of adhesion is gained while entropy is lost, resulting in an unfavorable change in free energy. In the case of fouling-release coatings, the force needed to detach a microorganism from the surface is low enough to keep microorganism-surface binding reversible. Parameters such as surface roughness, hydrophobicity, surface charge, Lewis acidity, and stiffness also play a role in the adhesion process; however the exact role of each parameter and how it contributes to the overall surface properties is still under debate.[11] (Again, “nonfouling” and “fouling-release” are ill-defined terms. While they specify the mechanism by which contamination is suppressed, they do not define the potentially contaminating species.)
The best-studied polymeric non-fouling material is probably poly(ethylene glycol) (PEG), which has a particularly low interfacial energy with water (5 mJ m-2).[9] PEG is highly hydrophilic and, depending on its polymer architecture, swellable or soluble in water because it is an excellent hydrogen bond acceptor. It was shown that water forms a hydration layer near the PEG surface, which accounts for the low interfacial energy,[9] although other explanations for its protein-repellency also exist.[15, 16] While PEG has been the gold standard for protein-repellent materials for many years, it is not infinitely stable and prone to undergo oxidative degeneration.[17] Recently, polyzwitterions have attracted considerable attention as a potential substitute for PEG because of their comparatively high oxidative and hydrolytic stability.[18–21] Like many of the phospholipids that form the cell envelope of mammalian cells, polyzwitterions carry an equal amount of positive and negative charges, for example quaternary ammonium groups combined with phosphates/phosphonates (poly(phosphorylcholines)),[22–31] sulfone/sulfate groups (poly(sulfobetaines)),[32–36] or carboxylates (poly(carboxy-betaines)).[19, 21, 33–35, 37–41] Polyzwitterions are very hydrophilic and, due to their charged nature, they apparently bind even more water than PEG.[42] In addition to their protein-repellency, polyzwitterions are highly biocompatible and have been successfully used in a number of biomedical coatings including orthopedic implants.[42] Another class of non-fouling materials are weakly amphiphilic poly(meth)acrylate coatings containing esters and aromatic/aliphatic rings.[43] Although their mechanism of action is not yet fully understood, it was suggested that, besides phyico-chemical effects like hydrophilicity and molecular flexibility[44], molecular recognition of certain groups by bacteria could also contribute to their non-fouling properties.[45]
Other noteworthy materials that prevent biofilm formation are non-polar, fluorinated polymers such as Teflon, or poly(dimethylsiloxane) (PDMS)-based fouling-release materials. These have a low surface energy, but a high interfacial energy with water (for example, PDMS has an interfacial energy with water of 52 mJ m-2).[9] This makes them prone to protein adhesion, yet because of their low surface energy and/or modulus, only small forces (e.g. shear forces from hydrodynamic drag) are needed to remove contaminations.[9] However, when PDMS or Teflon adsorb lipids from biological fluids, these cannot be removed as easily. This process also changes the surface energy of the materials and enables protein adhesion and biofilm formation.[46]
The above described non-fouling or fouling-release materials were homogeneous surfaces. There have also been a few studies that investigated protein adhesion and biofilm formation on heterogeneous or “mixed” surfaces consisting of different polymers, e.g. PEG combined with fluorinated polymers.[47, 48] Some of these surfaces had excellently low protein adsorbance, and a low polymer-water interfacial energy (4 mJ m-2).[49] Recently, a triblock copolymer containing a combination of cationic and fluorinated moieties showed excellent resistance to bacterial biofilm formation.[50] Also, PDMS modified with zwitterionic polymer grafts were 50-70% less fouling than pure PDMS while retaining its mechanical properties.[26] Another noteworthy class of “anti-fouling” materials, probably of the fouling-release class, are the SLIPS surfaces by Aizenberg and co-workers,[51] which consist of nanoporous materials infiltrated with fluorinated liquids. Since we restricted ourselves to all-polymer based approaches, these will not discussed further. While the initial performance of many protein-repellent or “antifouling” coatings is good to excellent, many of them are not long term stable. Proteins may eventually adhere, either on attached lipids or solid debris (dust), which then initiates bacterial adhesion. Once this process is started, protein-repellent materials cannot defend themselves against bacterial colonization, which is problematic because even single bacteria can form a biofilm in less than 24 hours.[5] On catheters or medical implants, this can cause severe inflammatory reactions.
Antimicrobial Polymer Surfaces and Their Mechanism of Action
Classification of antimicrobial polymer surfaces
While “anti-fouling” polymer surfaces are passive and vulnerable once their defenses are breached, polymer-based antimicrobial surfaces actively interfere with bacterial proliferation. They can be roughly classified into two kinds of materials – leaching ones[5] and non-leaching, contact-active ones. Leaching polymer surfaces consist of a polymeric matrix that is used as a carrier material for the active ingredient. This matrix can be degradable or non-degradable. The architecture of the polymer matrix and the partition coefficient of the drug, i.e. its distribution between the polymer and the surrounding media, determine the usefulness of the system. Ideally, drug release should be linear to obtain materials with constant antimicrobial activity. Initial high doses (so-called “burst release”) are desirable in some cases, e.g. to kill bacteria on a freshly placed implant, but care must be taken that over-dosing and potentially toxic side effects are avoided. Many well-known biocides and disinfectants have been used in polymer-based leaching antimicrobial surfaces, for example silver, quaternary ammonium compounds, antibiotics, antimicrobial peptides, or triclosan.[5] In more sophisticated approaches, the active ingredient was covalently attached to the polymer matrix through a labile bond, and could be gradually released in a triggered or non-triggered way. Thus, leaching surfaces with highly active species like chlorite that would otherwise not be sufficiently stable were accessible.[5] Leaching polymer surfaces are quite popular because they can produce a high concentration of the antimicrobial agents locally. Their disadvantage is that they fail once the leaching component is exhausted and thus may even foster bacterial resistance when the leached drug doses become sub-lethal. Further, non-degradable leaching components like silver may contaminate the environment and accumulate in soils or waterways, where further resistance formation in environmental bacteria could occur. The mechanism for antimicrobial activity of leaching polymer surfaces depends on the component that is leached; these mechanisms and the leaching concept in general will not be discussed further in this paper.
Non-leaching polymer systems, where the polymers themselves are the antimicrobially active ingredient, are cationic and contact-killing. Their charge enables them to attract and “capture” negatively charged bacterial cells, and to further interact with the bacterial cell envelope. This polymer-membrane interaction apparently damages the bacterial membrane, prevents bacterial growth and can eventually kill the bacteria, but the exact mechanism is not yet fully understood.[7, 52] While the role of the cationic charge in this process is not questioned, the role of hydrophobicity, and the sequence of events that lead to cell death, is not clear. In particular, since cationic polymers immobilized on surfaces lack the degrees of freedom of polymers in solution, their respective interaction with bacterial cell envelopes and lipid membranes was considered to be quite different.[7] It was therefore doubted that surface-attached polymers could permeate bacterial cell walls and cell membranes in ways similar to solution-borne polymers.[7] The field of such contact-killing antimicrobial polymer surfaces has been extensively reviewed.[53–57] We here focus on work dedicated to understanding the mechanism of interaction of cationic antimicrobial polymer surfaces with bacteria and eukaryotic cells. When reading the original papers cited here, the reader is advised to pay close attention to how the surfaces were characterized. Many early reports are purely phenomenological and important physical parameters or surface topography were not/could not be studied in detail. Yet sometimes physical properties were used to interpret the data, which in some cases might have led to conclusions that need to be re-interpreted in the light of results obtained using more advanced techniques, and overall advanced knowledge of the field. This is not meant to belittle the contributions of those researchers; it is just how science evolves.
Early work on contact-killing antimicrobial polymer surfaces
The study of contact-active antimicrobial polymer surfaces began with pioneering work on surface-attached poly(phosphonium) salts from the group of Endo, which paved the way for future generations.[58] In these studies, surfaces with different phosphonium content were obtained by varying reaction times. The surfaces with the highest phosphonium content had the best activity and quantitatively killed E. coli and S. aureus bacteria. This was interpreted as an effect of increasing charge density. Since surface charge or coating thickness were not directly measured, this trend might also be the result of a better surface coverage due to longer reaction times. In scanning electron microscopy images, the deformation of bacteria by these surfaces was clearly visible, which supposedly indicated membrane damage.[58] Toxicological data of the polymers was not reported, however from the styrene-based monomer structure it can be inferred that these surfaces must have been hydrophobic and thus potentially toxic.
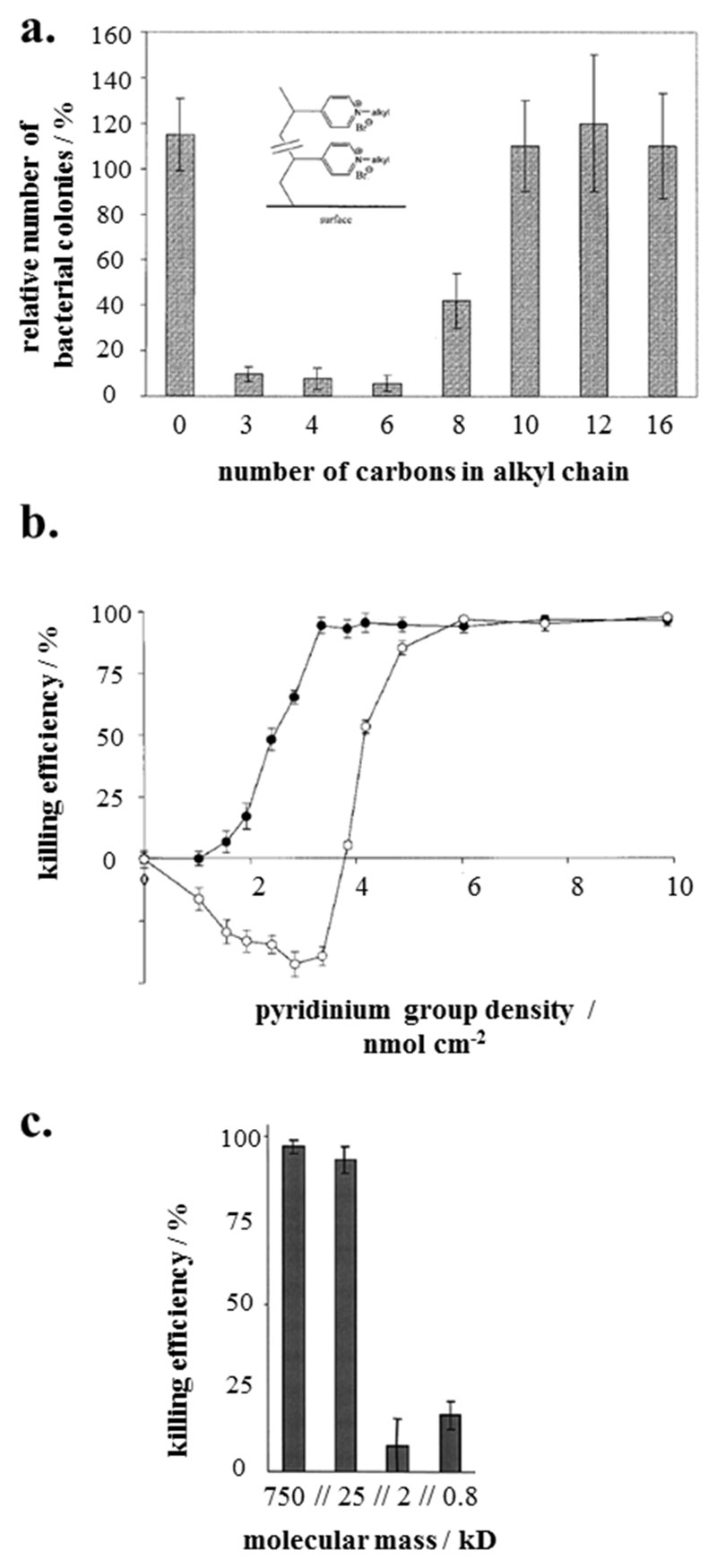
Further groundbreaking work on antimicrobial polymer surfaces by Tiller and Klibanov focused on poly(vinyl pyridinium)-based polymers that were surface-immobilized by various grafting-onto methods.[59–64] The effect of changes in hydrophobicity on the antimicrobial activity of the materials was investigated. Polymer surfaces with short alkyl chain lengths on the pyridinium ring (propyl to hexyl) had higher antimicrobial activity than longer ones (Figure 1a.).[59]
Figure 1.
a. Effect of alkyl chain length on the antimicrobial properties. The percentage of colony forming units is plotted against the number of carbon atoms per alkyl chain.[59] b. Effect of the density of the surface-attached pyridinium groups (determined by titration) on the antimicrobial activity. Killing efficiency is plotted versus pyridinium group density. For airborne bacteria (black symbols), the minimum charge density for quantitative killing was 3 nmol cm-2, for the water-borne bacteria (open symbols), it was about 6 nmol cm-2;[60, 62] c. Effect of molecular weight of poly(ethylene imine) on bactericidal activity.[59] Adapted with permission from a. Proc. Natl. Acad. Sci. U. S. A. 2001,[59]; b., c.: John Wiley and Sons, 2002 and 2003.[60, 62]
When correlating the number of pyridinium groups on the surfaces with antimicrobial activity (Figure 1b),[60] a sigmoidal relationship was obtained. This suggests that there is a certain charge threshold after which addition of further charge does not improve the antimicrobial activity.[60] Klibanov and coworkers also found that in some cases, high molecular weight polymers were more antimicrobially active than low molecular weight polymers (Figure 1c).[59, 62] In the light of these results, they postulated that long polyelectrolyte chains grafted onto a substrate can act on bacteria like a needle that would pierce a balloon (“hole-poking”[54]), i.e. that the chains would permeate and burst bacterial membranes.[54, 59–62, 65, 66] While the discussed experimental results are flawless, this mechanistic interpretation merits careful (re-)consideration. Polyelectrolyte chains are not spike-like rigid rods under physiological conditions, but collapse to coils,[67] unless they are densely grafted polymer-brushes (which seems not to the be the case here). It is difficult to visualize how such structures would “poke”. Additionally, recent studies showed that 2-3 nm thin antimicrobial polymer brushes, much too short to permeate and pierce bacterial cell walls, were highly bactericidal.[68] An alternative interpretation would be that Klibanov’s surfaces with high molecular weight chains had a better surface coverage than those with lower molecular weight ones, or that these long, not densely-grafted surface-attached polymer chains could interact more easily with the bacterial membranes than shorter chains, for example by adhering to the outer envelope of the bacteria. Both alternatives would explain their higher antimicrobial activity without the need of a minimum molecular weight for antimicrobial activity, which was not found in other reports.[68, 69] In any case, the uncontested achievement of Klibanov’s team was that they identified hydrophobicity and charge density as tools to tune the antimicrobial activity of cationic polymer surfaces and thus provided valuable guidelines for the design of antimicrobial polymers.
Mechanistic considerations
Additional work by Kügler on poly(vinyl pyridinium)-based polymers confirmed the correlation between charge density and antimicrobial activity, however they found no simple correlation between layer thickness and charge density.[70] As to mechanisms, the authors argue that the exchange of the divalent counterions from inside the bacterial membrane against the polyelectrolyte surface as a “counterion” would be lethal to the bacteria, because Mg2+ and Ca2+ stabilize bacterial membranes.[70] This is in line with results found for cationic polymers in solution by Tirrell.[71] Tiller and Klibanov had also speculated that, in addition to their “hole-poking” mechanism, the replacement of the divalent cations ‘might be itself sufficient for a lethal outcome’.[59] Notably, such an effect would not require direct interaction between polymer chains and the bacterial membrane (see discussion below). Russell and Matyjaszewski investigated a cationic polymer surface with a two dimensional charge density gradient. They also found a correlation between surface charge density and antimicrobial activity of their polymer surfaces.[72, 73] Interestingly, the authors argued that the killing mechanism of antimicrobial surfaces may not only depend on factors like charge density, but also on surface architecture, which is very plausible: densely grafted, brush-like surfaces and loosely packed surfaces with long chains will not interact in the same way with bacterial membranes.[73, 74] This aspect could explain many discrepancies and contradictory results in this field.
Ober and coworkers found for cationic poly(pyridinium) surfaces with hydrophobic alkyl or fluoroalkyl groups[69] that the antimicrobial activity did not depend on the molecular weight, but on the composition of the top few nanometers of the surface.[69] In the light of the careful analysis of these surfaces, these conclusions are very convincing. Chen and coworkers investigated quaternary ammonium polymers grafted onto a surface by sum frequency generation vibrational spectroscopy[75] and found that the charged polymer parts segregated to the air-polymer interface, and that the exact structure of that interface depended on the alkyl residue of the ammonium group.[75] Both studies indicated that there is also a hydrophobic component to the antimicrobial action of surface bound polymers. Recently, Chan-Park presented antimicrobial hydrogels made from poly(l-lysine)[76] and alkylated, quaternized chitosan that had excellent antimicrobial activity.[77] As mechanism of action, they proposed that the highly cationic hydrogel can “suck” negatively charged lipids out of bacterial membranes through electrostatic interactions (“anion sponge”)[77] A similar argument was also suggested by Tiller, who postulated the idea that cationic surfaces can act as “lipid sponges”.[78] There is a simulation experiment in Chan-Park’s paper that suggested such a mechanism.[79] Experimental evidence for the “anion sponge” theory is proposed in the work of Tiller[78] and in a more recent study.[80] Both groups demonstrated that cationic polymer surfaces can irreversibly adsorb negatively charged phospholipids, and are not antimicrobially active after that interaction.[78] These works indeed confirm that negatively charged phospholipids adsorb on polycationic surface-attached polymers significantly more than charge-neutral phospholipids do,[78] and that the relative thickness increase due to lipid adsorption of these surface-attached polymer networks with different cross-linking density is inversely proportional to their degree of cross-linking.[80] This is perfectly in line with results from work on polyelectrolyte-surfactant complexes formed in solution and in gels,[81, 82] and the known inverse dependency of swellability on cross-linking of surface-attached polymer networks.[83] However, it is less clear which conclusions to draw from these studies for the “anion sponge”/”lipid sponge” theory. In these model experiments, there is direct, unshielded contact between phospholipids, most of them possibly forming liposomes, and the surface-attached polycationic network. Flowing liposomes across surfaces is a well-known method to form physisorbed lipid monolayers.[84] However, such surface-liposome interaction is assumed to break the entire vesicle - the higher the curvature of the liposomes, the easier they break. Cationic charges will most likely facilitate the rupture of these vesicles. Thus, an alternative explanation for the above results is that the entire liposomes break when in contact with the polycationic surfaces, and that the phospholipids then irreversibly bind to the surfaces. Another argument for the “anion sponge” theory was the observation that surfaces consisting of surface-tethered hydrophilic poly(ethyloxazolines) with a cationic end-group behave differently than the above described polycationic surfaces when in contact with anions – on the hydrophilic poly(ethyloxazolines), no loss of antimicrobial activity upon treatment with SDS and negative phospholipids was observed, i.e. these surfaces are still antimicrobially active after washing.[78] This could indicate that SDS and negatively charged phospholipids bind reversibly to these surfaces, or do not bind at all. We follow the argumentation of Russell and coworkers[73] and propose that this different behavior compared to the highly charged surfaces described before is a surface architecture effect. The poly(ethyloxazolines) present their end-groups in the same way as proteins and other biomolecules are presented when conjugated to spacers,[74] where the spacer hydrophilicity and the overall lower segment density of the surface ensure bioavailability of the presented molecule or drug. Additionally, in this particular case of cationic end-groups, there will no polyelectrolyte effect - due to the flexibility of the 100 repeat-unit long spacer, each cationic charge can avoid the other, i.e. will move away from the other charges to a distance larger than the Bjerrum length, so that counterion condensation will not be present. Each charged end group thus acts separately on the anionic counterions, and these counterion can therefore be much more easily replaced, e.g. by washing. Therefore, these surfaces would still be antimicrobially active after treatment with SDS or phospholipid. Thus, even though these experiments do not prove the “anion sponge” theory, they are excellent model experiments to study the strength of the electrostatic interactions of different polycationic surfaces.
In addition to the above presented argument that the anion complexation experiments cannot unambiguously prove the “anion sponge” theory because the whole liposome breaks when brought in contact with the polycationic surfaces, one also has to remember that pure liposome models fail to capture the entire structure of bacterial membranes, which also has been observed in other experiments.[85] In reality, the bacterial membrane is protected by a 20-80 nm thick, highly cross-linked peptidoglycan layer (for Gram-positive bacteria), or by a lipopolysaccharide (LPS) layer and an additional outer lipid membrane (for Gram-negative bacteria). Thus, it is a well-organized separate compartment of the bacterial cell and has a distance from the surface-attached polycationic network of 20 to 80 nm. (Since the peptidoglycan layer and the lipopolysaccharide layer are also polyanionic, this raises the question why an external polycationic network would not form a polyelectrolyte complex with these species, and even if it did not, how would it reach the phospholipid membrane, and pull out negatively charged lipids from that membrane through the thick, polyanionic network surrounding it? Additionally, while lateral motion of lipids in membranes is easy, pulling out a lipid from an intact membrane would require a lot more energy. The typical binding free energy of a phospholipid in a membrane is about 80–100 kJ/mol, as determined experimentally and by MD simulations.[86, 87] This is a lot, compared to lateral motion of liquids within the membrane. Also, if one assumes that the kinetics of a lipid leaving the membrane can be compared to a lipid flip-flop (which is plausible), it appears that it would take 100 times longer for this event than a 5 nm lateral diffusion of that lipid.[88] Thus, it seems more likely that anionic lipids in bacteria would cluster in the presence of an electric field. This in turn would cause line tension in the membrane at the phase domains, which would destabilize the bacterial membrane sufficiently to cause leakage. In addition to these considerations, a mechanism according to the “anion sponge” theory would require even more than the above estimated 80–100 kJ/mol, because additional energy would be required to sufficiently flatten out the bacteria to enable close contact with the surfaces.[89, 90]
That same argument applies to many other theories that rely on direct interaction between the polycationic network and the bacterial membrane - it is just as difficult to picture lipids coming out of the membrane, as it is to imagine individual polymer chains reptating or “poking” through the polyanionic bacterial cell envelope. However, the charming feature of the “anion sponge” theory is that it can be re-interpreted to imply that the charge present on polycationic surfaces might have a long range effect on the bacterial membrane that is not based on direct contact between the membrane and the surface. For example, the contact with the polycationic surface might cause changes in the structure of the peptidoglycan or LPS layer. Such an effect could be based on changes in the local charge distribution of peptidoglycan or LPS, their mechanical properties or their integrity. All of the above could in turn affect the lipid organization of the bacterial membrane underneath, or its charge distribution or fluidity. Any of these effects would make the lipid membranes much more fragile and prone to damage, even without direct polymer-membrane interaction. At the same time, such a mechanism would still account for the well-known membrane damage effects observed when using live-dead stains on bacteria in contact with polycationic antimicrobial surfaces. Thus, it may be worthwhile to think about the mechanism of action of these surfaces in terms of signal transduction rather than in terms of direct interaction. However, this is purely speculative so far and needs to be further investigated.
Very recently, Mei, Busscher and Lootjens compared the antibacterial activity of quaternary ammonium compounds in solution and on surfaces using atomic force microscopy.[7] When surface-immobilized S. epidermidis was exposed to quaternary ammonium polymers in solution, the bacteria wrinkled, disintegrated, and eventually detached from the substrate.[7] On a surface covered with the same quaternary ammonium polymer, no wrinkling of the bacteria was visible, indicating that the interaction between the bacteria and the surface was confined to their contact zone. (This is in contrast to early results by Endo using scanning electron microscopy,[58] see above.) However, the bacteria were strongly flattened on that surface, and the adhesive force between the bacteria and the cationic quaternary ammonium polymer surface was about 100 times higher than between bacteria and an untreated glass surfaces.[7] Likewise, it was 25 times higher than between bacteria and a cationic α-poly(l-lysine) surface.[7] (One of the reasons for the latter, besides structural difference of the polymer architecture, could be that the quaternary ammonium polymer is a strong polyelectrolyte, while the weak polyelectrolyte α-poly(l-lysine) is only partially protonated under physiological conditions.) The authors argued that high electrostatic forces between bacteria and cationic polymer surfaces may prevent the bacteria from detaching during proliferation, so that they eventually die. This alternative mechanism would also not require a direct interaction between the polymers and the membrane, but solely the electrostatic interaction between surface and cell envelope. It convincingly explains the experimental fact that there is a charge threshold for antimicrobial activity, even though it does not explain why hydrophobicity affects antimicrobial activity. However, if Chen’s and Obers findings[69, 75] are considered in this context, one can assume that the presence of hydrophobic groups may affect the surface charge density, and thus indirectly also the antimicrobial activity. (It should also be noted that in the above presented study, the authors speculated about removal of lipids from the membrane as part of the mechanism, but no direct experimental evidence of this was presented.[7])
Two other recent studies discuss structure-property relationships and mechanistic aspects of low molecular weight antimicrobial polymers that have been spin-coated onto solid substrates.[91, 92] Because these polymers are insoluble in aqueous media, the authors assume that they also stay surface-immobilized during the experiments. Even when assuming that this is correct, and no macroscopic delamination or leaching of the polymers happens, these materials cannot be compared to surface-attached polymer coatings. First, they are low molecular weight polymers and therefore have significantly higher degrees of freedom when in contact with bacteria than surface-attached polymers: The physical properties of the latter are governed by the equilibrium between the elastic, ionic and mixing contributions to the free enthalpy.[52] For the non-surface-attached low molecular weight polymers, there is no such elastic contribution. Thus, the data in these studies,[91, 92] while interesting by itself, cannot be used to draw conclusions about the mechanism of surface-attached polymers, but rather describes an in-between state between solution-borne polymers and surface-attached polymers.
Studies on surface-attached antimicrobial poly(oxonorbornenes)
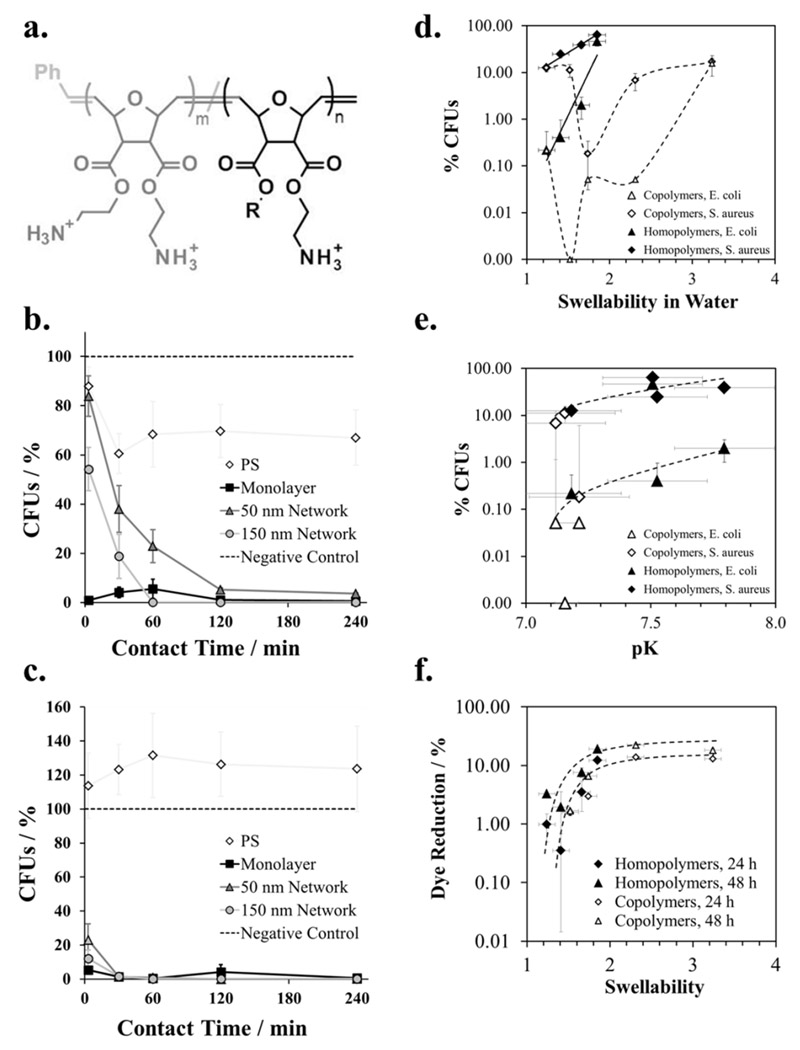
Because each of the above discussed studies investigated a specific and limited set of parameters for different systems, we recently devised a set of experiments designed to correlate several structural parameters of antimicrobial polymer surfaces with antimicrobial activity for the same type of material, namely poly(oxonorborenenes).[52] These provide a simple synthetic platform where parameters like polymer layer thickness, charge density and hydrophobicity can be systematically changed. To fully characterize these materials, we analyzed the following structural parameters: layer thickness (by ellipsometry), surface topography (by atomic force microscopy), surface zeta potential and acid constant as parameters related to the surface charge (, determined by electrokinetic measurements), and parameters related to the surface hydrophobicity (swellability, measured by surface plasmon resonance spectroscopy; surface energy, determined by the Zisman method). The polymer surfaces studied were surface-attached polymer networks made from facially amphiphilic, cationic poly(oxonorbornenes), and had constant cross-linker content. They contained amphiphilic repeat units with one variable hydrophobic R group and one hydrophilic charged group, and an optional repeat unit with two charged groups but no hydrophobic groups (Figure 2a).[52]
Figure 2.
a. Chemical structure of the polymers used to make surface-attached polymer networks. Homopolymer series: amphiphilic repeat units only (m = 0, R = Methyl to Butyl); Copolymer series: R = butyl, m = 0, 0.1, 0.5, 0.9 and 1, respectively. b) Antimicrobial activity against S. aureus. Surviving colony forming units (CFUs, in %) are plotted vs. contact time for homopolymers with R = butyl and different layer thickness.[93] c) Antimicrobial activity against E. coli. Surviving colony forming units (CFUs, in %) are plotted vs. contact time for homopolymers with R = butyl and different layer thickness.[93] d) Correlation of antimicrobial activity of homopolymers and copolymers against E. coli and S. aureus after t = 30 min with swellability. Surviving colony forming units (CFUs, in %) are plotted vs. swellability in water.[52] e) Correlation of antimicrobial activity of homopolymers and copolymers against E. coli and S. aureus after t = 30 min with pK.[52] f) Correlation of cell toxicity with swellability for homopolymers and copolymers. Alamar Blue dye reduction (a measure of metabolic activity) of immortalized human keratinocytes is plotted versus swellability in water.[52, 93] Copyright: b.,c.: Al-Ahmad et al., 2014;[93] d-f. Zou et al., [52] published by The Royal Society of Chemistry, 2015.
First, the layer thickness was increased from ~ 3 nm (polymer monolayer) to 50 nm and 150 nm (networks) while the polymer structure was the same (m = 0, R = butyl, Mn = 180,000 g mol-1).[93] The antimicrobial activity of all three samples against E. coli and S. aureus was about the same after 4 hours of contact time, but the killing kinetics varied significantly with layer thickness (Figure 2b. and c.).[93] Surprisingly, both networks killed S. aureus more slowly than the monolayer, but no effect was found for E. coli. The reason for this is not yet understood, but surface coverage issues can be excluded as the monolayer is the foundation on which both networks are build (i.e. surface coverage cannot be worse for the networks than for the monolayer). It could be interpreted as either an adhesion effect or an effect of substrate modulus. For both bacteria, the 50 nm network killed more slowly than the 150 nm network. This finding indicates that the 150 nm network had either a higher surface area to contact and kill the bacteria, or an effect of the stiff silicon substrate on which the networks were built (such as modulus or limited swellability near the surface) could be sensed more strongly by the bacteria on the thinner network. In either case, the data demonstrates that “layer thickness” is a much more complicated parameter than anticipated and may contain combined effects of differences in surface architecture, mechanical properties and chemical properties. This would explain why there is not yet a consensus in the field about the effect of layer thickness or molecular weight on antimicrobial properties. It also stresses the importance to be sure to compare “apples to apples” in these kinds of studies, which is unfortunately more easily said than done in many cases.
Using the same system of surface-attached poly(oxonorbornene) networks, but this time with constant layer thickness (~ 150 nm) and constant cross-linking density, we investigated the effect of changes of hydrophobicity and charge density on antimicrobial activity, cell compatibility and physical properties.[52] The characterization data for homopolymers (m = 0 in Figure 2a) with a hydrophobicity gradient (R from Methyl to Butyl, Figure 2a), and for copolymers with a charge density gradient (composition from m = 0.0 to 0.1 and n = 1 to 0, respectively, Figure 2a) are shown in Table 1.
Table 1.
Physical characterization of surface-attached networks made from poly(oxonorbornene) homopolymers (m = 0) and copolymers (m ≠ 0). The dry layer thickness was measured by ellipsometry; the apparent surface tension was determined using the Zisman method; swellability ratios in water were determined by surface plasmon resonance spectroscopy; the maximum surface charge ζmax under acidic conditions, the surface charge under pseudo-physiological conditions (ζphys), the isoelectric point and the acid constant pK were obtained by electrokinetic measurements.[52] Copyright: Zou et al.,[52] published by The Royal Society of Chemistry, 2015.
| Dry Layer Thickness / nm | Apparent Surface Tension / mN m-1 | Swellability Ratio / H2O | ζmax / mV | Iso-electric point | ζphys / mV | pK | |
|---|---|---|---|---|---|---|---|
| Diamine | 157 ± 4 | 61.0 | 3.2 | 62 ± 3 | 7.5 ± 0.2 | 0 | 7.4 |
| Methyl | 147 ± 3 | 56.8 | 1.9 | 77 ±5 | 7.6 ± 0.2 | 7 | 7.4 |
| Ethyl | 143 ± 3 | 53.0 | 1.7 | 73 ± 4 | 7.9 ± 0.2 | 23 | 7.7 |
| Propyl | 149 ± 3 | 51.5 | 1.4 | 85 ± 2 | 7.8 ± 0.2 | 11 | 7.5 |
| Butyl | 153 ± 4 | 48.8 | 1.2 | 84 ± 1 | 7.3 ± 0.2 | -2 | 7.2 |
| B:D = 1:9 | 148 ± 3 | 58.0 | 2.3 | 50 ± 3 | 7.5 ± 0.2 | 1 | 7.2 |
| B:D = 5:5 | 158 ± 3 | 55.3 | 1.7 | 74 ± 3 | 7.6 ± 0.2 | 2 | 7.2 |
| B:D = 9:1 | 152 ± 4 | 53.5 | 1.5 | 88 ±3 | 7.5 ± 0.2 | -6 | 7.2 |
For the homopolymers, the swellability of the polymer networks in water increases systematically with shorter alkyl groups R. Also, their antimicrobial activity correlated with swellability (Figure 2d). This correlation did not hold for the copolymers (Figure 2d). However, a correlation of log (antimicrobial activity) with pK (determined from zeta potential measurements) was found for both homo- and copolymers (Figure 2e). Care must be taken not to over-interpret this data: small yet significant effects were considered, and the 16 data points contain 3 outliers. Nevertheless, the antimicrobial activity of most homopolymers and copolymers against E. coli scaled with pK and clustered in the lower half of the plot, while their antimicrobial activity against S. aureus clusters in the upper half of the plot and also scaled with pK (Figure 2e). This relative shift of the E. coli curve and the S. aureus curve reflects the different sensitivity of these bacteria to contact-active antimicrobial polymers. On the other hand, the maximum zeta potential of the surfaces under acidic conditions (which is proportional to the maximum number of chargeable groups on the surface) did not correlate with antimicrobial activity.[52] This is because the maximum number of chargeable groups is not necessarily the same as the number of charges present: It is well known from the polyelectrolyte literature that the number of actually charged groups on a polymer depends on the charge separation, and that charges cannot be closer to each other than the Bjerrum length.[67] This is discussed in more detail for this system elsewhere.[52] Thus, when swellability changes (which it does for the copolymer series) the distance between the chargeable groups is affected, which may or may not be balanced by counter ion condensation, depending on the surface architecture of the system investigated. pK, on the other hand, ranks the ability of the overall system to form surface charges under given environmental conditions and could thus be correlated with antimicrobial activity. On the other hand, a correlation between cell toxicity and swellability was observed for all polymer surfaces in this data set (homo- and copolymers, Figure 2f). Thus, while swellability alone is not sufficient to describe the effect of structural changes on antimicrobial activity, it still can be used as a single parameter to map cell compatibility.
Achievements and challenges in the field of contact-killing antimicrobial polymer surfaces
For contact-killing antimicrobial polymer surfaces, polymer charge and hydrophobicity have been identified as the leading parameters that affect antimicrobial activity, while layer thickness and molecular weight of the polymer seem to matter only when they cause differences in surface coverage or modulus. Their mode of action is still under debate, yet it seems that these materials vary so much in their relative chemistry and architecture that there is no single explanation for their activity. Finally, it should be mentioned that in spite of their promising antimicrobial properties, most cationic contact-active polymer surfaces suffer from a fundamental problem: their charge, i.e. the same feature that makes them active, also causes protein adhesion and accumulation of debris of killed bacteria on the surface. If a surface manages to kill all bacteria present in a given application, this may not be problematic. However, once the surface is sufficiently covered and deactivated, any incoming bacteria can irreversibly settle and proliferate on these contaminations. Consequently, for many applications, antimicrobial activity alone is not enough. In the next sections of this contribution, we therefore discuss how we and others have developed alternative approaches to obtain more robust, long-term stable antimicrobial polymer surfaces.
Materials with Combined Antimicrobial Activity and Protein-repellency
Concept
One popular approach to make antimicrobial polymer surfaces more robust and long-lived is to combine them with protein-repellent polymers. The key idea behind this material design is that the resulting polymer surface will interfere with two of the early steps of biofilm formation. First, the protein-repellent component will slow down the rate of protein adsorption; second, the antimicrobial component will kill bacteria that come near the surface or attach to it. Additionally, the protein-repellent component will slow down the contamination of the surfaces with cell debris. There are several ways to combine antimicrobial and protein-repellent action in a single material, some of which have been summarized in a recent review by Chen.[94] In principle, leaching and non-leaching materials can be used as antimicrobial components and be combined with the respective protein-repellent or “anti-fouling” moieties. The effectiveness of the resulting materials depends on the intrinsic bioactivity of its components, and on the method by which these have been combined, i.e. whether they are sufficiently bioavailable on the resulting bifunctional surface.
Leaching materials
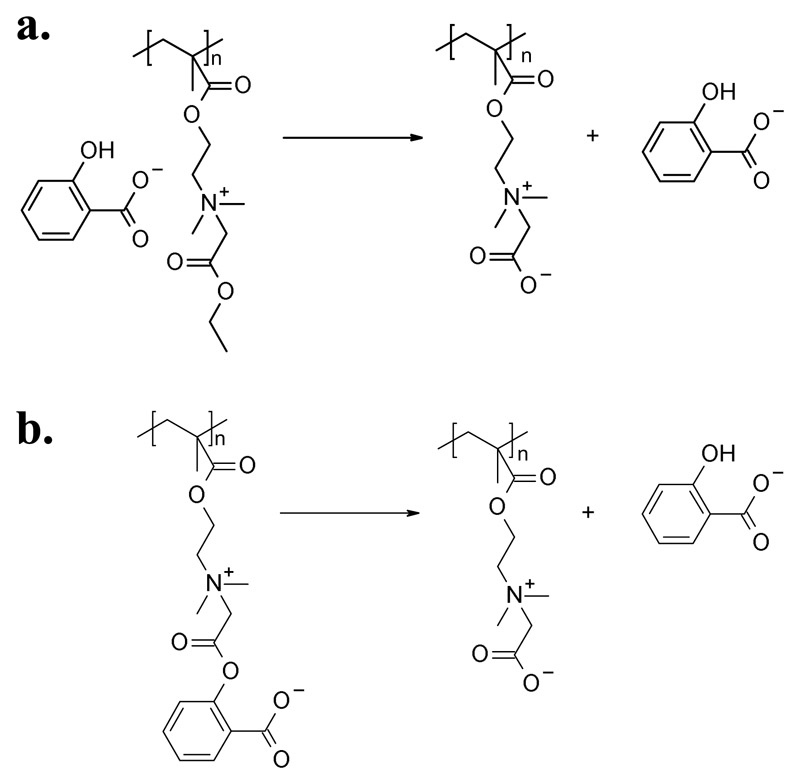
The easiest way to combine antimicrobial action and protein-repellency in a single material is embedding the antimicrobial component (antibiotics, antimicrobial peptides, or bactericidal heavy metals like silver) inside a protein-repellent polymer matrix.[95–97] Care must be taken in this approach that the release of the active component is well controlled in order to avoid toxic effects or sublethal doses.[5] Two conceptually beautiful and ingenious materials based on the leaching concept were developed by Jiang and coworkers (Figure 3a.). A surface-attached polycationic polymer hydrogel was loaded with the intrinsically antibiotic salicylate as counterion.[98] Upon hydrolysis of the ester groups of the polymer hydrogel, its charge switched from polycationic to polyzwitterionic and thus protein-repellent. This simultaneously released the salicylate counterion. The system showed a one log reduction in protein adsorption and a three log reduction of growth of E. coli and S. epidermidis.[98] The drawback of the system was that the switching worked only once. Next, the hydrogel was re-designed so that it was initially a polyzwitterionic material, and the salicylate became part of the hydrolyzable ester group.[99] When this system was hydrolyzed, the initial polyzwitterion was transformed into another polyzwitterion and also released the antimicrobial salicylate (Figure 3b.).[99] The resulting material had excellent antimicrobial activity, though not quantitative resistance to bacterial adhesion. At the same time, the salicylate release could be controlled through the hydrolysis kinetics. [99]
Figure 3.
a) Surface-attached polymer hydrogel made from a polycation with a salicylate counterion. Upon hydrolysis of its ester group, the polycation became polyzwitterionic, and the antimicrobial salicylate was released.[98] b) In a similar system, the salicylate was covalently attached to the polyzwitterionic hydrogel through the ester group. Upon hydrolysis, the salicylate was released, but the polymer hydrogel remained polyzwitterionic and thus protein-repellent.[99]
Non-leaching materials
Other systems consist of protein-repellent carrier polymers to which the antimicrobial components are covalently attached by a hydrophilic, protein-repellent spacer molecule. As shown in the field of bioconjugation, such hydrophilic spacers lead to high bioavailability of the active compound, combined with low unspecific protein adhesion.[74] There are various examples of antibiotics that have thus been surface-immobilized using PEG spacers.[100, 101] Other polymers and surface architectures were also used to present covalently attached antimicrobial components, e.g. antimicrobial peptides or quaternary ammoinium compounds.[102–106]
In yet another approaches to obtain bifunctional materials, the antimicrobial polymer is the active component, not just the matrix, and is combined with a protein-repellent polymer. We recently grafted protein-repellent polyzwitterions covalently onto antimicrobial poly(oxonorbornenes).[107] The resulting system was fully protein-repellent and antimicrobially active against E. coli and S. aureus bacteria. Using a hydrogel made from cross-linked PEG and a cationic polycarbonate, Yang obtained a material with excellent antimicrobial activity.[108] Ye et al. reported surface-attached polymer brushes with a core-shell architecture containing polyzwitterions at the inside and cationic polymers at the outside, which were grafted onto a synthetic membrane. The resulting material moderately reduced the adsorption of the protein BSA and E. coli and was moderately antimicrobial.[109] Another system consisting of hydrophilic poly(hydroxylethyl methacrylate) and the natural polymer chitosan had moderate antimicrobial activity, but good protein-repellency.[110] As one of the few materials that contained hydrophobic protein-repellent materials in combination with an antimicrobial polymer, a bifunctional membrane from antibiofouling poly(vinylidene fluoride) and antimicrobial poly[2-(N,N-dimethylamino)ethyl methacrylate] was described; however, this material was only protein-repellent when it was non-protonated, and in this state it was not antibacterial.[19]
There were also attempts to make simultaneously antimicrobial and protein-repellent material using the layer-by-layer (LbL) approach, in which alternating layers of polyanions and antimicrobial polycations were applied onto a surface.[111–114] The idea behind this application of LbL is that the two polymers do not form a stack of discrete polymer layers; the individual polymers rather undergo mixing. Thus, the surface would contain sufficient cationic polymer patches to be antimicrobial, but these would be neutralized by patches of the anionic polymer so that the material is overall protein-repellent. The data from the above cited LbL studies and our own work[114] indicate that it is very difficult to obtain LbL materials that are antimicrobially active, protein-repellent, and at the same time stable under physiological conditions. In some cases, this was solved by covalent cross-linking of the two polymers involved.[115] The team of Therien-Aubin combined a stable LbL base layer consisting of poly(allylamine hydrochloride) and poly(styrene sulfonate) with protein-repellent poly(allyl glycidyl ether brushes), onto which micro-patterned polycationic, antimicrobial and polyzwitterionic patches with a spacing of 2-25 μm were attached.[116] This qualitatively reduced protein adhesion and bacterial adhesion. A dependency of these properties on the spacing of the micro-pattern was not studied.
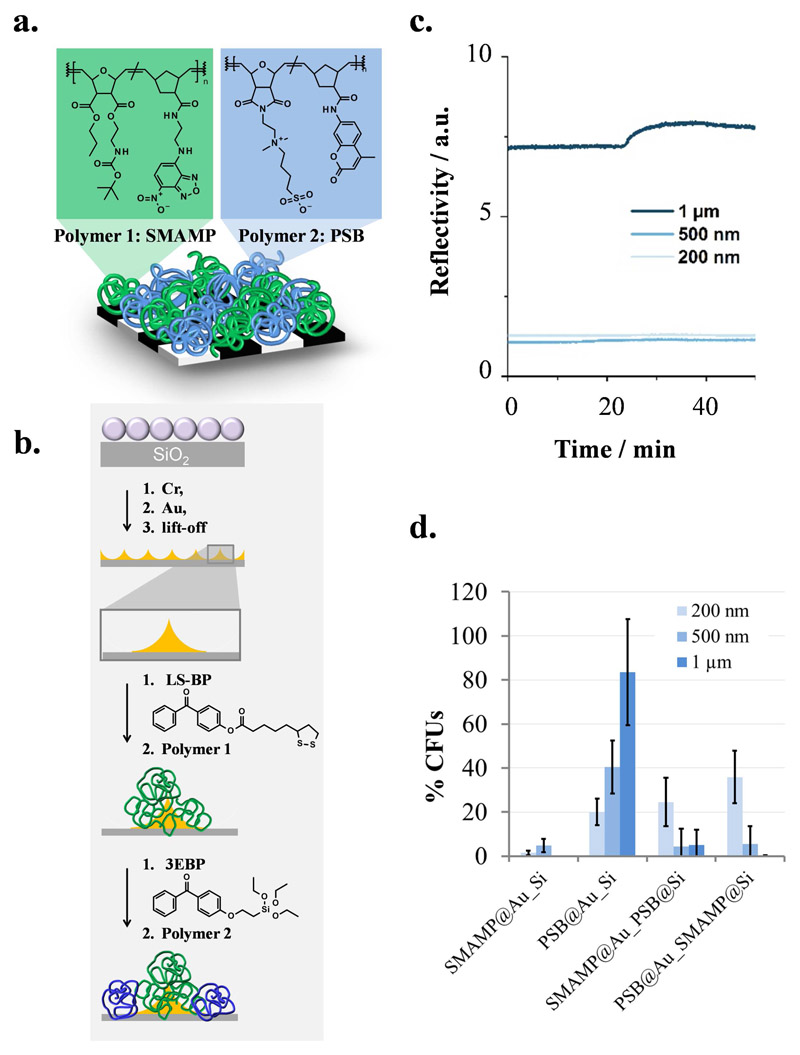
Judging from the above described systems, the leaching approach is quite successful to obtain materials with dual activity. It has, however, all the drawbacks of leaching systems described above (risk of bacterial resistance, contamination of the environment, allergic reactions, failure when the active component has completely leached). The above described non-leaching systems, on the other hand, all suffered from the problem that it is very difficult to simultaneously present enough antimicrobial moieties on the surface to be efficient against bacteria, and at the same time resist protein adsorption. Thus, for such systems the surface architecture must be precisely controlled; otherwise, either antimicrobial activity or protein-repellency is compromised. We therefore designed a material to systematically investigate structure-property relationships for combined materials made from antimicrobial and protein-repellent polymers. The target material consists of surface-attached polymer patches made from antimicrobial poly(oxonorbornene) SMAMPs and protein-resistant polymer poly(sulfobetaines) (PSB, Figure 4a.). These were immobilized side by side, like the black and white fields of a chess board, on a surface having a chemical contrast of gold islands on a silicon wafer. The surface functionalization process is described elsewhere.[117] In short, the gold islands on silicon were created by colloid lithography and site-selectively functionalized with the two bioactive polymers using linker molecules with orthogonal reactivity (Figure 4b). Since it was expected that a size range from a few hundreds of nanometers to a few micrometers would particularly affect the biological activity of our materials,[118–120] the size of the antimicrobial and protein-repellent patches was varied from 200 nm to 1 μm. The site-selective immobilization of one polymer on silicon, and the other on gold, was confirmed by fluorescence spectroscopy and atomic force microscopy. The protein-adsorption of two bifunctional materials with different spacing (SMAMP on islands, PSB on background and vice versa) was tested using surface plasmon resonance spectroscopy. The data shows that the materials were strongly protein-repellent for all spacings. For the 200 nm and 500 nm spacing, this effect was quantitative, while slight protein adhesion (less than 1%) was observed for the 1 μm structures (Figure 4c.). In the antimicrobial activity tests, it was found that the antimicrobial activity depended strongly on the spacing: the activity of the 200 nm structures was compromised, while the 1 μm structures were quantitatively active against E. coli (Figure 4d.)
Figure 4.
a. Cartoon illustration of the target material: polymer patches made from green-fluorescent, antimicrobial poly(oxonorbornene) SMAMP and blue-fluorescent, protein-repellent poly(oxonorbornene) PSB immobilized on a structured substrate. b. Surface fabrication process: a layer of polystyrene colloids on silicon was used as lithographic mask for the evaporation of chromium (adhesive layer) and gold. After liftoff, a gold-on-silicon pattern was obtained. The gold islands were functionalized with the gold-selective molecule LS-BP, which was used to immobilize polymer 1 by a UV-triggered reaction between that polymer and the benzophenone group of the linker. The silicon background was functionalized with the silane 3EBP, which was then used to immobilize polymer 2 (also through its benzophenone group). c) Fibrinogen adhesion on PSB@SiO2_SMAMP@Au with a spacing of 200 nm, 500 nm and 1 μm, respectively, studied by surface plasmon resonance spectroscopy (kinetics mode). d) Antimicrobial activity (% CFUs) of structured monofunctional (SMAMP@Au_Si and PSB@Au_Si, polymer attached to the gold islands) and bifunctional surfaces (SMAMP@Au_PSB@Si and PSB@Au_SMAMP@Si) with spacings of 200 nm, 500 nm and 1 μm, respectively, against E. coli.[117] Reprinted/adapted with permission from ACS Biomat. Sci. Eng. 2017, ASAP; Copyright (2017) American Chemical Society.
The data shows why it is so difficult to obtain combined antimicrobial and protein-repellent properties simultaneously in a single material using contact-active polymers: if the antimicrobial patches are too small, the material cannot interact sufficiently with the bacterial cells to damage or kill them, and the antimicrobial activity is compromised. If the patch size is increased, the antimicrobial activity is restored to 95-100% CFU reduction because the SMAMP patches become once again accessible to the bacteria. However, the antimicrobial patches become also more easily accessible for proteins and thus protein-adhesive. While a “perfect” material with 100% protein-repellency and 100% antimicrobial activity could not be obtained, the material with PSB on the islands and SMAMP in the background at a spacing of 1 μm quantitatively killed E. coli and reduced protein adhesion by 99.4%. It thus exceeded the state-of-the-art in simultaneously active antimicrobial and protein-repellent materials. While it is fundamentally interesting and important to understand these structure-property relationships, open questions remain. For example, the long-term stability of and biofilm formation on these materials in biological fluids need to be studied in more detail, and the surface fabrication process needs to be simplified to become more attractive for medical applications.
Regeneration of Antimicrobial Activity of Polymer Surfaces
The problem that cationic antimicrobial surfaces become inactive when they are contaminated by debris of dead bacteria or by other biomolecules was considered quite early in the development of these materials. For example, in their studies on poly(vinyl-N-alkylpyridinium) polymers, which are among the first reports of contact-killing antimicrobial polymers, the authors mention this drawback.[59, 60] They also assert that these surfaces ‘could be rejuvenated simply by periodic washings’ with a detergent.[60] This was shown by Klibanov and coworkers, who treated fabrics impregnated these polymers with soap for 12 h at 50°C, and demonstrated that their antimicrobial activity could be regenerated to at least 95% of its initial value by washing.[121] Even though this value was further reduced after each washing cycle,[121] it indicates that the removal of bacterial debris is difficult, yet not impossible. The reduction of antimicrobial activity is most likely associated with adsorption of anionic detergent, which was (using SDS as an example) reported to irreversibly adhere to other polycationic surfaces.[78] Work from the area of stimulus-responsive polymers also demonstrated that the removal of cell debris from a surface is not trivial. For example, surfaces that can be switched from adhesive to non-adhesive were reported. Chen’s mixed polymer brushes made from N,N-dimethylaminoethyl methacrylate and 3-acrylamidobenzene-1-boronic acid were cationic/adhesive at pH 4.5. A buffer wash at pH 9 neutralized the acid and removed up to 92% of the adhered S. aureus bacteria.[122] Self-assembled monolayers made from cationic and anionic surfactants could be switched from adhesive to non-adhesive by an external potential.[123] Both these systems required an external stimulus (pH or potential change) to switch their properties, and were not intrinsically antimicrobial.
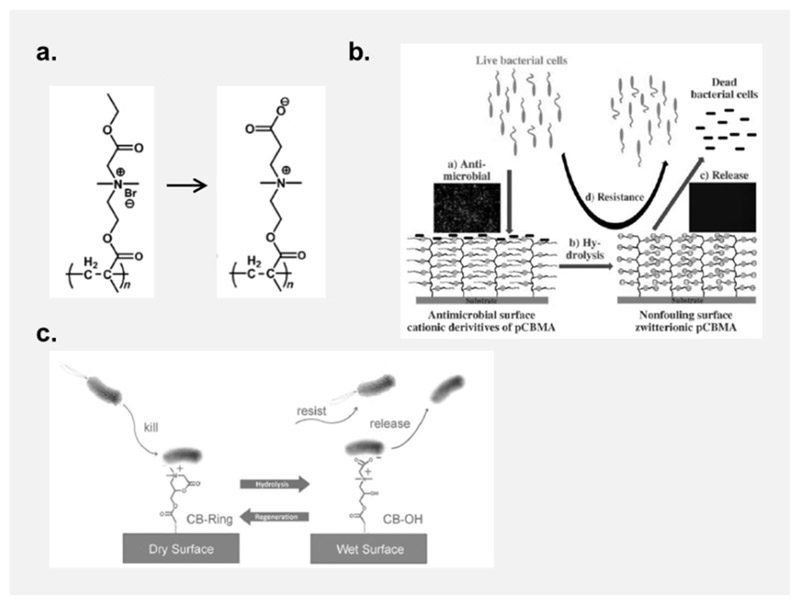
Jiang and coworkers presented the first self-cleaning surfaces that were switchable from a cationic, antimicrobial to a zwitterionic, protein-repellent state.[124] The cationic quaternary ammonium part of these polymers contained one residue that could be hydrolyzed from an ester to a carboxylate, so that the net charge of the surface changed to neutral upon hydrolysis (Figure 5a. and b.).[124] While the idea is beautiful, hydrolysis of the ester groups required high pH values, and the switching process only worked once, and in one direction. A reversible system was presented by the same team using a N,N-dimethyl-2-morpholinone polymer that was cationic in the ring-closed form, and zwitterionic in the ring-opened form (Figure 5c.).[125] As a dry surface, the polycationic material was strongly antimicrobial, but hydrolyzed to the protein-repellent polyzwitterionic form when in contact with aqueous media. The system could be regenerated into the cationic, antimicrobial state several times without degradation or loss of properties.[125] While the regeneration conditions are still not ideal for practical applications, this paper is an important milestone. It demonstrates that adhered bacteria can in principle be removed from cationic polymer surfaces in the early stages of biofilm formation. Meanwhile, this principle has been imitated in other systems.[126, 127]
Figure 5.
Charge-switching polymer surfaces: a. An antimicrobial quaternary ammonium polymer (QA) becomes zwitterionic when its ester group is hydrolyzed. [124] b. While the cationic QA surface is antibacterial and adhesive, the debris of dead bacteria is released when the surface becomes zwitterionic. This surface also repels further incoming bacteria.[124] c. A cationic morpholine-based polymer surface can be switched to a zwitterionic state by hydrolysis, yielding a bacteria-repellant and resistant material. Unlike the material presented in a. and b., this process is reversible.[125] Copyright: Reproduced with permission by a, b: Wiley-VCH Verlag GmbH & Co. KGaA, 2008; c: Wiley-VCH Verlag GmbH & Co. KGaA, 2012.
Chen presented an interesting approach based on an LbL material, where the polyanionic species contained adamantyl as a supramolecular motif.[128] This surface was loaded with a low molecular weight biocide immobilized on a corresponding cyclodextrin supramolecular motif. When exposed to bacteria, the cationic biocide killed them. Washing with the surfactant sodium dodecyl sulfate broke the supramolecular interactions and removed the biocide together with the attached bacteria.[128] The system could then be re-loaded with new biocide, and the process could be repeated several times without loss of activity.[128] Zhao and coworkers also used the supramolecular motif of adamantyl and cyclodextrin interaction, but this time to assemble mixed brushes from poly(N-isopropylacrylamide) (PNIPAM) and poly[(methacryloyloxy)ethyl]trimethylammonium chloride] on a silicon surface.[129] Using the thermo-responsive properties of PNIPAM, the material reversibly killed about 96% of S. aureus bacteria at 37°C, and released between 80-90% of the dead bacteria at 4°C.[129] Lopez et al. combined PNIPAM with oligo(phenylene-ethynylene) that was antimicrobial upon UV-A irradiation. Above the LCST, the material was antimicrobial, killing up to 62% bacteria. Below the LCST, the material was protein-repellent and released up to 62% of attached dead bacteria.[94, 130] When the system was additionally loaded with low molecular weight quaternary ammonium salts, the killing efficiency against E. coli and S. aureus was between 80-85%, and it released 65-80% of the attached bacteria.[131] Similar effects were obtained with release systems consisting of nano-patterned PNIPAM brushes and quaternary ammonium salts,[132] or PNIPAM with lysozyme.[133] While these materials are conceptually very interesting, the remaining question is how practical this temperature dependent property is for real-life applications.
Chen also presented an approach that fits into the context of surface regeneration, which was based on an enzyme immobilized on a polymer surface.[134] This surface was active due to the presence of the enzyme lysozyme, which degraded attached E. coli bacteria. The bacterial debris and the enzyme could be removed from that surface by washing with 1-4 M NaCl. After-reloading the surfaces with lysozyme, the surface activity was regenerated.[134] In the context of regeneration by washing, Yin and coworkers presented moderately dense block copolymer brushes with a polycationic core and a polyzwitterionic shell.[135] These polymers killed 76% of S. aureus bacteria and about 95% of E. coli in the dry state. 73% of adhered S. aureus bacteria and 90% of adhered E. coli bacteria could be released in the wet state from these bifunctional materials.[135] It was assumed that the dry material exposed its antimicrobial core, while the swollen material consisted of a polyzwitterionic corona that covered that shell, and thus prevented bacterial adhesion. As discussed before, an inverse surface architecture (polyzwitterionic core and polycationic shell) was presented by Elimelech,[109] however removal of bacteria was not discussed for this material.
Since it is so difficult to make the perfectly antimicrobial, infinitely protein-repellent surface, why not make an antimicrobial surface that can rejuvenate instead? This principle is applied in all kinds of surfaces that are erodible, for example using degradable polymers. Since intrinsically antimicrobial, degradable polymers are rare,[136–138] most degradation concepts rely on leaching from an inactive degradable polymer matrix, e.g. cellulose, cellulose acetate, poly(lactic acid) (PLA),[139, 140] poly(glycolic acid) (PGA), and PLA-PGA copolymers,[141] chitosan[142–144], poly(ε-caprolactone),[145] poly(anhydride esters),[146] or various layer-by-layer assemblies.[11, 147–149] Unlike the situation in non-degradable matrices, the leaching of the active ingredient in these materials does not only occur by passive diffusion, but is assisted by degradation of the matrix. This approach has the advantage that the degradation kinetics can be used to influence the release kinetics. A contact-active, degradable polymer surface has been presented by Bieser et al.,[150] who grafted antimicrobial N,N-dimethyldodecylammonium to cellulose and showed that the material obtained was degradable in the presence of the enzyme cellulase, and thus “self-polishing”.[151] The antimicrobial activity could be regenerated to 80% of its initial value.[150] Nottelet and coworkers presented a poly(lactide) surface onto which an antimicrobial polymer had been grafted by Huisgen addition. While the self-regeneration aspect of this material was not studied, the material had a 5 log reduction of bacterial growth of E. coli and S. aureus compared to untreaded PLA and could thus be a promising candidate in the quest for non-leaching degradable antimicrobial materials.[140]
The general problem with degradable polymer coatings is that the erosion process is difficult to control, as many degradable polymers are semi-crystalline and have different degradation kinetics in their crystalline and amorphous regions. Thus, the roughness of such coatings increases continuously during degradation.[152] which may facilitate bacterial adhesion. To avoid this problem, we designed a material that can erode in a defined and controlled way. It consists of a multi-stack of alternating antimicrobial and degradable polymer layers and was designed to shed its layers sequentially, like a reptile shedding its skin (Figure 6a.).[153] The key idea of the design is that the top antimicrobial layer of the material is removed by disintegrating the degradable layer underneath, and not the antimicrobial layer itself. The morphology of the emerging layer is dialed in during the build-up of the material, and is retrieved at each shedding event.
Figure 6.
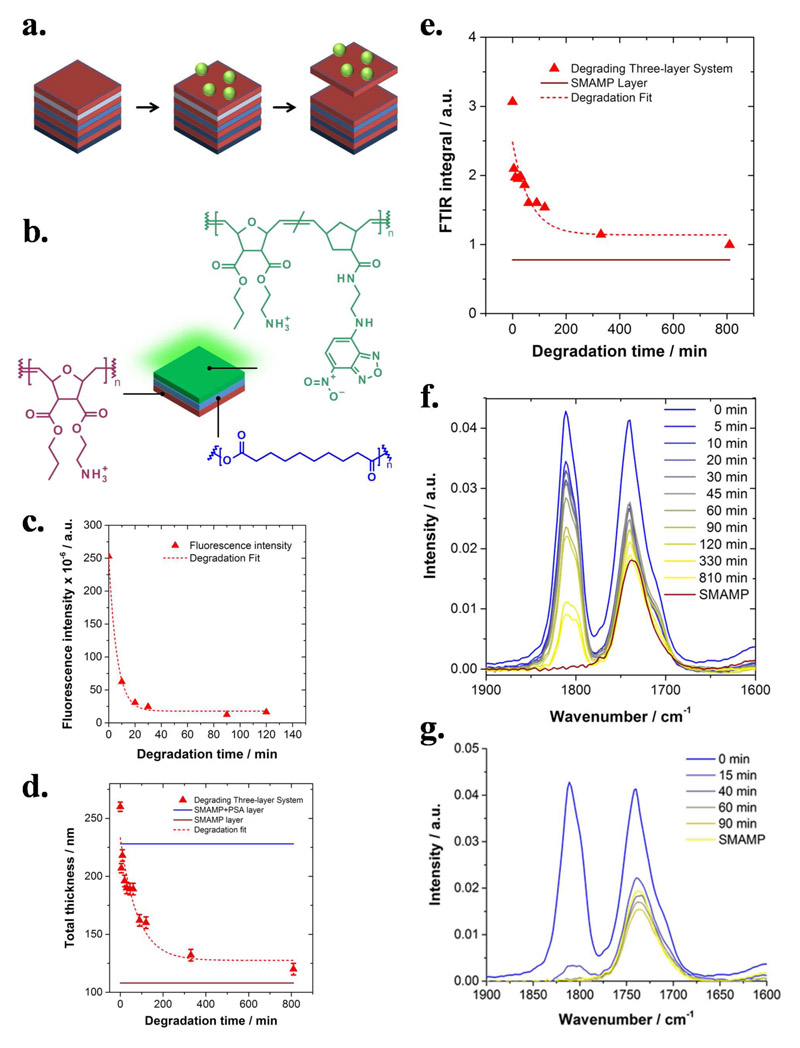
a. Cartoon illustration of the target material, a polymer thin film multi-stack consisting of alternating layers of antimicrobial (red) and degradable polymers (blue). Once the top antimicrobial layer is contaminated, the layer underneath (light blue) is degraded, so that the top layer is detached. Thus, the system can shed its outer layer like a reptile shedding its skin, and a fresh antimicrobial layer is revealed. Because the degradable layers have different degradation kinetics (illustrated by different shades of blue), the process should be repeatable several times. b. The proof-of-concept stack consists of an antimicrobial bottom layer made from a surface-attached SMAMP polymer network (red), a poly(sebacic anhydride) (PSA) interlayer (blue), and a 2-(4-nitro-2,1,3-benzoxadiazol-7-yl) (NBD) labelled antimicrobial SMAMP network as the top layer (green). c.-f. Degradation studies of the three-layer system in 0.1 M HCl: c. Fluorescence intensity versus degradation time (imaging time = 1 s; the intensity at t = 0 min was calculated from the fit data; the fit function was Intensity = y 0 + A · e −k1·tdeg, with y 0 = 17, A = 235, and k 1 = 0.16 min-1); d. Thickness determined by ellipsometry versus degradation time. The straight lines in the image mark the initial thickness of the SMAMP (red) and SMAMP + PSA layers (blue), respectively. The data was fitted using Thickness = y 0 + A · e −k1·(tdeg-t0), with y 0 = 128 nm, A = 106 nm, t 0 = −0.33 min, and k 1 = 0.12 min-1. e. Integrated FTIR intensity of the carbonyl region versus degradation time (straight line = initial thickness of the SMAMP layer). The data was fitted using Intensity = y 0 + A · e −k1·(tdeg-t0), with y 0 = 1.14, A = 1.33, t 0 = 0.52 min and k 1 = 0.16 min-1); f. FTIR spectra of the carbonyl region at defined time points. g. Degradation studies by FTIR of the three-layer system in 3 M HCl. The SMAMP bottom layer before degradation is included as reference (yellow curve).[153] (b. is reprinted with permission from ACS Macro Lett. 2015, 4, 1337. Copyright (2015) American Chemical Society.)
At first glance, this idea resembles the concept of sacrificial layers frequently used in microsystems fabrication.[154] The difference is that sacrificial layers are dissolved in a particular solvent for removal, not degraded. Since polymer degradation is a phenomenon with distinct kinetics and depends on the polymer structure, this allows better control over the shedding event. By choosing polymers with different degradation rates for the individual degradable layers, this should enable selective and sequential layer shedding. The selective shedding of discrete layers from a polymer thin film multi-stack has not been successfully reported in the literature before, but a similar idea has been followed in the context of LbL-systems.[155] In this context, it was not possible to selectively shed layers, because LbL assemblies do not consist of discrete alternating layers, but of interpenetrating anionic and cationic domains.[155, 156] This gave two design rules for our concept: first, it was necessary to avoid strong electrostatic attraction between adjacent polymer layers, and second, one should use significantly thicker layers to obtain a “pan-cake stack” rather than a “scrambled egg” morphology.
A proof-of-concept of our idea was realized in a three-layer-system consisting of an antimicrobial, surface-attached poly(oxanorbornene) SMAMP network as the bottom layer (Figure 6b., red), a degradable poly(sebacic anhydride) (PSA) inter-layer (Figure 6b, blue), and a green fluorescent antimicrobial poly(oxanorbornene) SMAMP network as the top layer (Figure 6b, green).[157] The build-up of this system was studied using ellipsometry, fluorescence microscopy and FTIR spectroscopy. Since degradation of the material in HEPES buffer (“pseudo-physiological” conditions) was slow and led to undesired side-reactions, the degradation was studied in 0.1 M HCl using fluorescence microscopy, ellipsometry, and FTIR spectroscopy, which were used to determine the thickness of the system and the rate constant of degradation, respectively. FTIR and ellipsometry showed that the material did not fall back to the original thickness of the first SMAMP layer, suggesting incomplete layer shedding, or that fragments of the removed layers migrated into the bottom SMAMP network. However, the rate constants of degradation determined by fluorescence microscopy, ellipsometry and FTIR spectroscopy were in good agreement (see caption of Figure 6). Fluorescence measurements monitored only the shedding of the top layer of the system, while FTIR measured the presence or absence of the entire material on the surface. The fact that the three rate constants matched means that the top fluorescent layer disappeared at the same speed as the PSA layer vanished. This is a clear indication that the system degraded at a speed dictated by the PSA layer, and did not delaminate in an uncontrolled way.
Complete degradation of the proof-of-concept system was observed in 3 M HCl (Figure 6g.). FTIR spectra of this system showed that the peak of the asymmetric anhydride stretching vibration from the PSA layer at about 1815 cm-1 vanished completely and the intensity of the ester peak from the SMAMP fell below the level of the initial SMAMP single layer (Figure 6g, yellow line). The latter is possibly an indication of a small amount of ester hydrolysis in the SMAMP layer. Antimicrobial activity assays demonstrated that the regenerated SMAMP system was fully antimicrobially active.[153] Thus, it was demonstrated that the layer shedding approach is a novel concept that might be useful to regenerate the surface functionality of contaminated surfaces. As with the other approaches presented above, it still has to be proven that this concept holds under practical considerations and in real life applications. Potential applications of this technology, once it has sufficiently matured, are surfaces that are too hard to reach to just wipe them clean, e.g. deep sea sensors, or medical devices where surface regeneration could extend the lifetime of the device, e.g. the inner lumen of urinary catheters. To become truly useful, such a system should be able to shed sequentially and selectively under physiological conditions.
Concluding Remarks
In this paper, we addressed the yet not fully understood mechanism of action of antimicrobial polymer surface, and the central problem for the long-term activity of these materials – their inability to remain antimicrobially active when they become covered by biomolecules and bacterial debris. We presented several creative and fundamentally interesting approaches to deal with this problem, among them bifunctional polymer surfaces made from protein-repellent and antimicrobial components, switchable polymer surfaces, surfaces regenerated by enzyme action, erodible polymer surfaces, and polymer surfaces that can shed discrete layers to regenerate antimicrobial activity. While many of the presented materials are academically interesting and increase our fundamental understanding of the interaction of bacteria with material surfaces, they still have one common problem – most of them are too complicated for practical applications. Let’s come back to the initially sketched scenario of bacterial resistance and biofilm formation on medical devices. What the medical field needs to successfully fight biofilms in these settings are polymer coatings that are long-term stable and can be easily attached to the surfaces of real-life devices (which are non-planar and not as ideally smooth as laboratory model surfaces). Also, such long-term stable materials should be usable without additional washing or regeneration procedures – otherwise, acceptance by healthcare practitioners and/or patient compliance cannot be expected. Additionally, these surfaces need to be sterilizable without degradation of the material. They must have a sufficiently long shelf-life, and must be stable against oxidation and hydrolysis under the conditions of use. We have recently developed a material that might bring us one step closer to fulfill all of these criteria – a protein-repellent and simultaneously antimicrobial polyzwitterion.[158] We also demonstrated that we could surface-attach this material to a commercially available porous polyurethane wound dressing. [158]
In conclusion, even though most of the above presented materials need to be re-engineered to become useful for medical applications, their development and study taught scientists a lot about the underlying problem – interaction of bacteria with polymer surfaces. It is hard to predict how the field will evolve. With “static” polymer surfaces (i.e. those that do not change properties during time of use) we currently try to extend the surface lifetimes by making them more inert to protein adhesion, or by increasing their antimicrobial activity. However, this is just a race of bacterial adhesion vs. detachment, which the bacteria will eventually win. The future might therefore lie in the development of “dynamic” polymer surfaces, which can actively respond to contamination without the need of an operator to switch or steer their properties. While this is still more science fiction than science, we believe that even with the “static” approaches, long-term stable polymer surfaces that perform well under physiological conditions are within reach. We will therefore continue the development of these materials to contribute to the solution of an important problem in modern medicine and healthcare – the infection of medical devices with bacteria.
Acknowledgements
Funding of this work by the German Research Foundation (Emmy-Noether-Program, LI1714/5-1), the Ministry for Science, Research and Art of the State of Baden-Württemberg, Germany (GenMik I + II graduate school), the Freiburg Institute for Advanced Studies (FRIAS) of the Albert-Ludwigs-Universität Freiburg, Germany and the European Research Council (ERC-StG REGENERATE) is gratefully acknowledged.
Author Biography
 Karen Lienkamp studied Chemistry at the University of Cambridge and the Freie Universität Berlin. She obtained her Ph.D. in Physical Chemistry from the Johannes Gutenberg-Universität Mainz for work on biomimetic cylindrical polyelectrolyte brushes. After a PostDoc at the University of Massachusetts, Amherst, USA, she started her own research group as a Junior Fellow of the Freiburg Institute for Advanced Studies (FRIAS) and the Department of Microsystems Engineering (IMTEK) at the Albert-Ludwigs-Universität in Freiburg. She was awarded an Emmy-Noether-Grant from the German Research Foundation (DFG), and a Starting Grant from the European Research Council (ERC). She obtained the Venia Legendi in 2017. Her research interests are functional polymers, hierarchical polymer architectures, and biomimetic materials.
Karen Lienkamp studied Chemistry at the University of Cambridge and the Freie Universität Berlin. She obtained her Ph.D. in Physical Chemistry from the Johannes Gutenberg-Universität Mainz for work on biomimetic cylindrical polyelectrolyte brushes. After a PostDoc at the University of Massachusetts, Amherst, USA, she started her own research group as a Junior Fellow of the Freiburg Institute for Advanced Studies (FRIAS) and the Department of Microsystems Engineering (IMTEK) at the Albert-Ludwigs-Universität in Freiburg. She was awarded an Emmy-Noether-Grant from the German Research Foundation (DFG), and a Starting Grant from the European Research Council (ERC). She obtained the Venia Legendi in 2017. Her research interests are functional polymers, hierarchical polymer architectures, and biomimetic materials.
References
- [1].World Health Organization (WHO) Report on global surveillance of antimicrobial resistance. http://www.who.int/mediacentre/factsheets/fs194/en/, 2014/11/24.
- [2].Center for Disease Control. http://www.cdc.gov/mrsa/, 2014/11/17.
- [3].European Center for Disease Prevention and Control. http://ecdc.europa.eu/en/activities/surveillance/EARS-Net/database/Pages/database.aspx, 2014/11/17.
- [4].European Antimicrobial Resistance Surveillance Network (EARS-Net) Antimicrobial resistance surveillance in Europe 2013. www.ecdc.europa.eu/en/publications/layouts/forms/Publication_DispForm.aspx?List=4f55ad51-4aed-4d32-b960-af70113dbb90&ID=1205, 2014/11/24.
- [5].Hetrick EM, Schoenfisch MH. Chem Soc Rev. 2006;35:780. doi: 10.1039/b515219b. [DOI] [PubMed] [Google Scholar]
- [6].Dennis GM, Paul AT. Emerg Infect Dis. 2001;7:342. [Google Scholar]
- [7].Asri LATW, Crismaru M, Roest S, Chen Y, Ivashenko O, Rudolf P, Tiller JC, van der Mei HC, Loontjens TJA, Busscher HJ. Adv Fund Mater. 2014;24:346. [Google Scholar]
- [8].Díaz C, Miñán A, Schilardi PL, de Mele Fernández Lorenzo M. Int J Antimicrob Agents. 2012;40:221. doi: 10.1016/j.ijantimicag.2012.05.012. [DOI] [PubMed] [Google Scholar]
- [9].Krishnan S, Weinman CJ, Ober CK. J Mater Chem. 2008;18:3405 [Google Scholar]
- [10].Hucknall A, Kim D-H, Rangarajan S, Hill RT, Reichert WM, Chilkoti A. Adv Mater. 2009;21:1968. doi: 10.1002/adma.200803125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Lichter JA, Van VKJ, Rubner MF. Macromolecules. 2009;42:8573 [Google Scholar]
- [12].Hall-Stoodley L, Costerton JW, Stoodley P. Nat Rev Microbiol. 2004;2:95. doi: 10.1038/nrmicro821. [DOI] [PubMed] [Google Scholar]
- [13].Stewart PS, William CJ. Lancet. 2001;358:135. doi: 10.1016/s0140-6736(01)05321-1. [DOI] [PubMed] [Google Scholar]
- [14].Gupta P, Sarkar S, Das B, Bhattacharjee S, Tribedi P. Arch Microbiol. 2016;198:1. doi: 10.1007/s00203-015-1148-6. [DOI] [PubMed] [Google Scholar]
- [15].Jeon SI, Lee JH, Andrade JD, De Gennes PG. J Colloid Interface Sci. 1991;142:149. [Google Scholar]
- [16].Jeon SI, Andrade JD. J Colloid Interface Sci. 1991;142:159. [Google Scholar]
- [17].Konradi R, Pidhatika B, Muehlebach A, Textor M. Langmuir. 2008;24:613. doi: 10.1021/la702917z. [DOI] [PubMed] [Google Scholar]
- [18].Ladd J, Zhang Z, Chen S, Hower JC, Jiang S. Biomacromolecules. 2008;9:1357. doi: 10.1021/bm701301s. [DOI] [PubMed] [Google Scholar]
- [19].Zhang Z, Vaisocherova H, Cheng G, Yang W, Xue H, Jiang S. Biomacromolecules. 2008;9:2686. doi: 10.1021/bm800407r. [DOI] [PubMed] [Google Scholar]
- [20].Cheng G, Li G, Xue H, Chen S, Bryers JD, Jiang S. Biomaterials. 2009;30:5234. doi: 10.1016/j.biomaterials.2009.05.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Jiang S, Cao Z. Adv Mater. 2010;22:920. doi: 10.1002/adma.200901407. [DOI] [PubMed] [Google Scholar]
- [22].Lewis AL. Colloids Surf, B. 2000;18:261. doi: 10.1016/s0927-7765(99)00152-6. [DOI] [PubMed] [Google Scholar]
- [23].Iwata R, Suk-In P, Hoven VP, Takahara A, Akiyoshi K, Iwasaki Y. Biomacromolecules. 2004;5:2308. doi: 10.1021/bm049613k. [DOI] [PubMed] [Google Scholar]
- [24].Rose SF, Okere S, Hanlon GW, Lloyd AW, Lewis AL. J Mater Sci: Mater Med. 2005;16:1003. doi: 10.1007/s10856-005-4755-y. [DOI] [PubMed] [Google Scholar]
- [25].Hirota K, Murakami K, Nemoto K, Miyake Y. FEMS Microbiol Lett. 2005;248:37. doi: 10.1016/j.femsle.2005.05.019. [DOI] [PubMed] [Google Scholar]
- [26].Goda T, Konno T, Takai M, Moro T, Ishihara K. Biomaterials. 2006;27:5151. doi: 10.1016/j.biomaterials.2006.05.046. [DOI] [PubMed] [Google Scholar]
- [27].Goda T, Konno T, Takai M, Ishihara K. Colloids Surf, B. 2007;54:67. doi: 10.1016/j.colsurfb.2006.09.006. [DOI] [PubMed] [Google Scholar]
- [28].Iwata R, Satoh R, Iwasaki Y, Akiyoshi K. Colloids Surf, B. 2008;62:288. doi: 10.1016/j.colsurfb.2007.10.018. [DOI] [PubMed] [Google Scholar]
- [29].Bertal K, Shepherd J, Douglas CWI, Madsen J, Morse A, Edmondson S, Armes SP, Lewis A, MacNeil S. J Mater Sci. 2009;44:6233 [Google Scholar]
- [30].Kitano K, Inoue Y, Matsuno R, Takai M, Ishihara K. Colloids Surf, B. 2009;74:350. doi: 10.1016/j.colsurfb.2009.08.004. [DOI] [PubMed] [Google Scholar]
- [31].Yoshimoto K, Hirase T, Madsen J, Armes SP, Nagasaki Y. Macromol Rapid Commun. 2009;30:2136. doi: 10.1002/marc.200900484. [DOI] [PubMed] [Google Scholar]
- [32].Zhang Z, Chen S, Chang Y, Jiang S. J Phys Chem B. 2006;110:10799. doi: 10.1021/jp057266i. [DOI] [PubMed] [Google Scholar]
- [33].Zhang Z, Chao T, Chen S, Jiang S. Langmuir. 2006;22:10072. doi: 10.1021/la062175d. [DOI] [PubMed] [Google Scholar]
- [34].Cheng G, Zhang Z, Chen S, Bryers JD, Jiang S. Biomaterials. 2007;28:4192. doi: 10.1016/j.biomaterials.2007.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Colak S, Tew GN. Macromolecules. 2008;41:8436 [Google Scholar]
- [36].Garg G, Chauhan GS, Gupta R, Ahn JH. J Colloid Interface Sci. 2010;344:90. doi: 10.1016/j.jcis.2009.12.016. [DOI] [PubMed] [Google Scholar]
- [37].Zhang Z, Chen S, Jiang S. Biomacromolecules. 2006;7:3311. doi: 10.1021/bm060750m. [DOI] [PubMed] [Google Scholar]
- [38].Vaisocherova H, Yang W, Zhang Z, Cao Z, Cheng G, Piliarik M, Homola J, Jiang S. Anal Chem. 2008;80:7894. doi: 10.1021/ac8015888. [DOI] [PubMed] [Google Scholar]
- [39].Tada S, Inaba C, Mizukami K, Fujishita S, Gemmei-Ide M, Kitano H, Mochizuki A, Tanaka M, Matsunaga T. Macromol Biosci. 2009;9:63. doi: 10.1002/mabi.200800150. [DOI] [PubMed] [Google Scholar]
- [40].Yang W, Xue H, Li W, Zhang J, Jiang S. Langmuir. 2009;25:11911. doi: 10.1021/la9015788. [DOI] [PubMed] [Google Scholar]
- [41].Gao C, Li G, Xue H, Yang W, Zhang F, Jiang S. Biomaterials. 2010;31:1486. doi: 10.1016/j.biomaterials.2009.11.025. [DOI] [PubMed] [Google Scholar]
- [42].Lowe S, O’Brien-Simpson NM, Connal LA. Polymer Chemistry. 2015;6:198. [Google Scholar]
- [43].Hook AL, Chang C-Y, Yang J, Luckett J, Cockayne A, Atkinson S, Mei Y, Bayston R, Irvine DJ, Langer R, Anderson DG, et al. Nat Biotechnol. 2012;30:868. doi: 10.1038/nbt.2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Sanni O, Chang C-Y, Anderson DG, Langer R, Davies MC, Williams PM, Williams P, Alexander MR, Hook AL. Advanced healthcare materials. 2015;4:695. doi: 10.1002/adhm.201400648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Hook AL, Chang C-Y, Yang J, Atkinson S, Langer R, Anderson DG, Davies MC, Williams P, Alexander MR. Adv Mater. 2013;25:2542. doi: 10.1002/adma.201204936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Toes GJ, van Muiswinkel KW, van Oeveren W, Suurmeijer AJH, Timens W, Stokroos I, van den Dungen JJAM. Biomaterials. 2001;23:255. doi: 10.1016/s0142-9612(01)00103-x. [DOI] [PubMed] [Google Scholar]
- [47].Weinman CJ, Finlay JA, Park D, Paik MY, Krishnan S, Sundaram HS, Dimitriou M, Sohn KE, Callow ME, Callow JA, Handlin DL, et al. Langmuir. 2009;25:12266. doi: 10.1021/la901654q. [DOI] [PubMed] [Google Scholar]
- [48].Gudipati CS, Finlay JA, Callow JA, Callow ME, Wooley KL. Langmuir. 2005;21:3044. doi: 10.1021/la048015o. [DOI] [PubMed] [Google Scholar]
- [49].Krishnan S, Ayothi R, Hexemer A, Finlay JA, Sohn KE, Perry R, Ober CK, Kramer EJ, Callow ME, Callow JA. Langmuir. 2006;22:5075. doi: 10.1021/la052978l. [DOI] [PubMed] [Google Scholar]
- [50].Park D, Finlay JA, Ward RJ, Weinman CJ, Krishnan S, Paik M, Sohn KE, Callow ME, Callow JA, Handlin DL, Willis CL, et al. ACS Appl Mater Interfaces. 2010;2:703. doi: 10.1021/am900748v. [DOI] [PubMed] [Google Scholar]
- [51].Wong TS, Kang SH, Tang SK, Smythe EJ, Hatton BD, Grinthal A, Aizenberg J. Nature. 2011;477:443. doi: 10.1038/nature10447. [DOI] [PubMed] [Google Scholar]
- [52].Zou P, Laird D, Riga EK, Deng Z, Perez-Hernandez H-R, Guevara-Solarte DL, Steinberg T, Al-Ahmad A, Lienkamp K. Journal of Materials Chemistry B: Materials for Biology and Medicine. 2015;3:6224. doi: 10.1039/c5tb00906e. [DOI] [PubMed] [Google Scholar]
- [53].Siedenbiedel F, Tiller JC. Polymers. 2012;4:46. [Google Scholar]
- [54].Klibanov AM. J Mater Chem. 2007;17:2479 [Google Scholar]
- [55].Kenawy E-R, Worley SD, Broughton R. Biomacromolecules. 2007;8:1359. doi: 10.1021/bm061150q. [DOI] [PubMed] [Google Scholar]
- [56].Timofeeva L, Kleshcheva N. Appl Microbiol Biotechnol. 2011;89:475. doi: 10.1007/s00253-010-2920-9. [DOI] [PubMed] [Google Scholar]
- [57].Ferreira L, Zumbuehl A. J Mater Chem. 2009;19:7796 [Google Scholar]
- [58].Kanazawa A, Ikeda T, Endo T. J Polym Sci, Part A: Polym Chem. 1993;31:1467 [Google Scholar]
- [59].Tiller JC, Liao C-J, Lewis K, Klibanov AM. Proc Natl Acad Sci U S A. 2001;98:5981. doi: 10.1073/pnas.111143098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Tiller JC, Lee SB, Lewis K, Klibanov AM. Biotechnol Bioeng. 2002;79:465. doi: 10.1002/bit.10299. [DOI] [PubMed] [Google Scholar]
- [61].Lin J, Tiller JC, Lee SB, Lewis K, Klibanov AM. Biotechnol Lett. 2002;24:801. [Google Scholar]
- [62].Lin J, Qiu S, Lewis K, Klibanov AM. Biotechnol Bioeng. 2003;83:168. doi: 10.1002/bit.10651. [DOI] [PubMed] [Google Scholar]
- [63].Park D, Wang J, Klibanov AM. Biotechnol Prog. 2006;22:584. doi: 10.1021/bp0503383. [DOI] [PubMed] [Google Scholar]
- [64].Milovic NM, Wang J, Lewis K, Klibanov AM. Biotechnol Bioeng. 2005;90:715. doi: 10.1002/bit.20454. [DOI] [PubMed] [Google Scholar]
- [65].Lin J, Qiu S, Lewis K, Klibanov AM. Biotechnol Prog. 2002;18:1082. doi: 10.1021/bp025597w. [DOI] [PubMed] [Google Scholar]
- [66].Lewis K, Klibanov AM. Trends Biotechnol. 2005;23:343. doi: 10.1016/j.tibtech.2005.05.004. [DOI] [PubMed] [Google Scholar]
- [67].Dautzenberg H. Polyelectrolytes : formation, characterization, and application. Hanser Publishers; Hanser/Gardner, Munich; New York; Cincinnati: 1994. [Google Scholar]
- [68].Madkour AE, Dabkowski JM, Nusslein K, Tew GN. Langmuir. 2009;25:1060. doi: 10.1021/la802953v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Krishnan S, Ward RJ, Hexemer A, Sohn KE, Lee KL, Angert ER, Fischer DA, Kramer EJ, Ober CK. Langmuir. 2006;22:11255. doi: 10.1021/la061384v. [DOI] [PubMed] [Google Scholar]
- [70].Kugler R, Bouloussa O, Rondelez F. Microbiology. 2005;151:1341. doi: 10.1099/mic.0.27526-0. [DOI] [PubMed] [Google Scholar]
- [71].Takigawa DY, Tirrell DA. Makromolekulare Chemie, Rapid Communications. 1985;6:653. [Google Scholar]
- [72].Sang B Lee, Richard R Koepsel, Scott W Morley, Matyjaszewski K, Sun Y, Alan J Russell. Biomacromolecules. 2004;5:877. [Google Scholar]
- [73].Murata H, Koepsel RR, Matyjaszewski K, Russell AJ. Biomaterials. 2007;28:4870. doi: 10.1016/j.biomaterials.2007.06.012. [DOI] [PubMed] [Google Scholar]
- [74].Goddard JM, Hotchkiss JH. Prog Polym Sci. 2007;32:698. [Google Scholar]
- [75].Ye S, Majumdar P, Chisholm B, Stafslien S, Chen Z. Langmuir. 2010;26:16455. doi: 10.1021/la1001539. [DOI] [PubMed] [Google Scholar]
- [76].Zhou C, Li P, Qi X, Sharif ARM, Poon YF, Cao Y, Chang MW, Leong SSJ, Chan-Park MB. Biomaterials. 2011;32:2704. doi: 10.1016/j.biomaterials.2010.12.040. [DOI] [PubMed] [Google Scholar]
- [77].Avery CW, Palermo EF, McLaughlin A, Kuroda K, Chen Z. Anal Chem. 2011;83:1342. doi: 10.1021/ac1025804. [DOI] [PubMed] [Google Scholar]
- [78].Bieser AM, Tiller JC. Macromol Biosci. 2011;11:526. doi: 10.1002/mabi.201000398. [DOI] [PubMed] [Google Scholar]
- [79].Li P, Poon YF, Li W, Zhu H-Y, Yeap SH, Cao Y, Qi X, Zhou C, Lamrani M, Beuerman RW, Kang E-T, et al. Nature Materials. 2011;10:149. doi: 10.1038/nmat2915. [DOI] [PubMed] [Google Scholar]
- [80].Gao J, White EM, Liu Q, Locklin J. ACS Appl Mater Interfaces. 2017;9:7745. doi: 10.1021/acsami.6b14940. [DOI] [PubMed] [Google Scholar]
- [81].Langevin D. Adv Colloid Interface Sci. 2009;147–148:170. doi: 10.1016/j.cis.2008.08.013. [DOI] [PubMed] [Google Scholar]
- [82].Hansson P. Current Opinion in Colloid & Interface Science. 2006;11:351. [Google Scholar]
- [83].Toomey R, Freidank D, Rühe J. Macromolecules. 2004;37:882. [Google Scholar]
- [84].Lis M, Dorner F, Tew GN, Lienkamp K. Langmuir. 2016;32:5946. doi: 10.1021/acs.langmuir.6b00230. [DOI] [PubMed] [Google Scholar]
- [85].Lienkamp K, Kumar K-N, Som A, Nuesslein K, Tew GN. Chem--Eur J. 2009;15:11710. doi: 10.1002/chem.200802558. [DOI] [PubMed] [Google Scholar]
- [86].Tieleman DP, Marrink S-J. J Am Chem Soc. 2006;128:12462. doi: 10.1021/ja0624321. [DOI] [PubMed] [Google Scholar]
- [87].Cevc G, Marsh D. Phospholipid Bilayers: Physical Principles and Methods. John Wiley & Sons; New York: 1987. [Google Scholar]
- [88].Berg JM, Tymoczko JL, Stryer L. Section 12.6, Lipids and Many Membrane Proteins Diffuse Rapidly in the Plane of the Membrane. In: Freeman WH, editor. Biochemistry. 5th. New York: 2002. [Google Scholar]
- [89].larikov DD, Kargar M, Sahari A, Russel L, Gause KT, Behkam B, Ducker WA. Biomacromolecules. 2014;15:169. doi: 10.1021/bm401440h. [DOI] [PubMed] [Google Scholar]
- [90].Kargar M, Pruden A, Ducker WA. Journal of Materials Chemistry B: Materials for Biology and Medicine. 2014;2:5962. doi: 10.1039/c4tb00835a. [DOI] [PubMed] [Google Scholar]
- [91].Hoque J, Akkapeddi P, Yadav V, Manjunath GB, Uppu DSSM, Konai MM, Yarlagadda V, Sanyal K, Haldar J. ACS Appl Mater Interfaces. 2015;7:1804. doi: 10.1021/am507482y. [DOI] [PubMed] [Google Scholar]
- [92].Altay E, Yapaoz MA, Keskin B, Yucesan G, Eren T. Colloids Surf B Biointerfaces. 2015;127:73. doi: 10.1016/j.colsurfb.2015.01.020. [DOI] [PubMed] [Google Scholar]
- [93].Al-Ahmad A, Zou P, Guevara-Solarte DL, Hellwig E, Steinberg T, Lienkamp K. PLoS One. 2014:e111357. doi: 10.1371/journal.pone.0111357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Yu Q, Wu Z, Chen H. Acta Biomater. 2015;16:1. doi: 10.1016/j.actbio.2015.01.018. [DOI] [PubMed] [Google Scholar]
- [95].Jewrajka SK, Haldar S. Polym Compos. 2011;32:1851 [Google Scholar]
- [96].Ho CH, Tobis J, Sprich C, Thomann R, Tiller JC. Adv Mater. 2004;16:957. [Google Scholar]
- [97].Hu R, Li G, Jiang Y, Zhang Y, Zou J-J, Wang L, Zhang X. Langmuir. 2013;29:3773. doi: 10.1021/la304708b. [DOI] [PubMed] [Google Scholar]
- [98].Cheng G, Xue H, Li G, Jiang S. Langmuir. 2010;26:10425. doi: 10.1021/la101542m. [DOI] [PubMed] [Google Scholar]
- [99].Mi L, Jiang S. Biomaterials. 2012;33:8928. doi: 10.1016/j.biomaterials.2012.09.011. [DOI] [PubMed] [Google Scholar]
- [100].Aumsuwan N, Heinhorst S, Urban MW. Biomacromolecules. 2007;8:3525. doi: 10.1021/bm700803e. [DOI] [PubMed] [Google Scholar]
- [101].Aumsuwan N, McConnell MS, Urban MW. Biomacromolecules. 2009;10:623. doi: 10.1021/bm8013473. [DOI] [PubMed] [Google Scholar]
- [102].Gao G, Lange D, Hilpert K, Kindrachuk J, Zou Y, Cheng JTJ, Kazemzadeh-Narbat M, Yu K, Wang R, Straus SK, Brooks DE, et al. Biomaterials. 2011;32:3899. doi: 10.1016/j.biomaterials.2011.02.013. [DOI] [PubMed] [Google Scholar]
- [103].Laloyaux X, Fautre E, Blin T, Purohit V, Leprince J, Jouenne T, Jonas AM, Glinel K. Adv Mater. 2010;22:5024. doi: 10.1002/adma.201002538. [DOI] [PubMed] [Google Scholar]
- [104].Glinel K, Jonas AM, Jouenne T, Leprince J, Galas L, Huck WTS. Bioconjugate Chem. 2009;20:71. doi: 10.1021/bc800280u. [DOI] [PubMed] [Google Scholar]
- [105].Salwiczek M, Qu Y, Gardiner J, Strugnell RA, Lithgow T, McLean KM, Thissen H. Trends Biotechnol. 2014;32:82. doi: 10.1016/j.tibtech.2013.09.008. [DOI] [PubMed] [Google Scholar]
- [106].Fik CP, Konieczny S, Pashley DH, Waschinski CJ, Ladisch RS, Salz U, Bock T, Tiller JC. Macromol Biosci. 2014;14:1569. doi: 10.1002/mabi.201400220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Zou P, Hartleb W, Lienkamp K. J Mater Chem. 2012;22:19579 [Google Scholar]
- [108].Liu SQ, Yang C, Huang Y, Ding X, Li Y, Fan WM, Hedrick JL, Yang Y-Y. Adv Mater. 2012;24:6484. doi: 10.1002/adma.201202225. [DOI] [PubMed] [Google Scholar]
- [109].Ye G, Lee J, Perreault F, Elimelech M. ACS Appl Mater Interfaces. 2015;7:23069. doi: 10.1021/acsami.5b06647. [DOI] [PubMed] [Google Scholar]
- [110].Yang WJ, Cai T, Neoh K-G, Kang E-T, Dickinson GH, Teo SL-M, Rittschof D. Langmuir. 2011;27:7065. doi: 10.1021/la200620s. [DOI] [PubMed] [Google Scholar]
- [111].Decher G. Science. 1997;277:1232 [Google Scholar]
- [112].Wong SY, Li Q, Veselinovic J, Kim B-S, Klibanov AM, Hammond PT. Biomaterials. 2010;31:4079. doi: 10.1016/j.biomaterials.2010.01.119. [DOI] [PubMed] [Google Scholar]
- [113].Wong SY, Han L, Timachova K, Veselinovic J, Hyder MN, Ortiz C, Klibanov AM, Hammond PT. Biomacromolecules. 2012;13:719. doi: 10.1021/bm201637e. [DOI] [PubMed] [Google Scholar]
- [114].Dorner F, Malek-Luz A, Saar JS, Bonaus S, Al-Ahmad A, Lienkamp K. Macromol Chem Phys. 2016;217:2154 [Google Scholar]
- [115].Yang WJ, Pranantyo D, Neoh K-G, Kang E-T, Teo SL-M, Rittschof D. Biomacromolecules. 2012;13:2769. doi: 10.1021/bm300757e. [DOI] [PubMed] [Google Scholar]
- [116].Ma W, Rahaman MS, Therien-Aubin H. Journal of Water Reuse and Desalination. 2015;5:326. [Google Scholar]
- [117].Vöhringer Maria, Hartleb Wibke, Lienkamp K. 2017 doi: 10.1021/acsbiomaterials.7b00140. submitted. [DOI] [PubMed] [Google Scholar]
- [118].Diaz C, Salvarezza RC, Fernandez Lorenzo de Mele MA, Schilardi PL. ACS Appl Mater Interfaces. 2010;2:2530. doi: 10.1021/am100313z. [DOI] [PubMed] [Google Scholar]
- [119].Diaz C, Schilardi PL, dos Santos Claro PC, Salvarezza RC, Fernandez Lorenzo de Mele MA. ACS Appl Mater Interfaces. 2009;1:136. doi: 10.1021/am8000677. [DOI] [PubMed] [Google Scholar]
- [120].Diaz C, Schilardi PL, Salvarezza RC, Fernandez Lorenzo de Mele M. Langmuir. 2007;23:11206. doi: 10.1021/la700650q. [DOI] [PubMed] [Google Scholar]
- [121].Mukherjee K, Rivera JJ, Klibanov AM. Appl Biochem Biotechnol. 2008;151:61. doi: 10.1007/s12010-008-8151-1. [DOI] [PubMed] [Google Scholar]
- [122].Xiong X, Wu Z, Yu Q, Xue L, Du J, Chen H. Langmuir. 2015;31:12054. doi: 10.1021/acs.langmuir.5b02002. [DOI] [PubMed] [Google Scholar]
- [123].Pranzetti A, Mieszkin S, Iqbal P, Rawson FJ, Callow ME, Callow JA, Koelsch P, Preece JA, Mendes PM. Adv Mater. 2013;25:2181. doi: 10.1002/adma.201204880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Cheng G, Xue H, Zhang Z, Chen S, Jiang S. Angew Chem, Int Ed. 2008;47:8831. doi: 10.1002/anie.200803570. [DOI] [PubMed] [Google Scholar]
- [125].Cao Z, Mi L, Mendiola J, Ella-Menye J-R, Zhang L, Xue H, Jiang S. Angew Chem, Int Ed. 2012;51:2602. doi: 10.1002/anie.201106466. [DOI] [PubMed] [Google Scholar]
- [126].Cao B, Tang Q, Li L, Humble J, Wu H, Liu L, Cheng G. Advanced Healthcare Materials. 2013;2:1096. doi: 10.1002/adhm.201200359. [DOI] [PubMed] [Google Scholar]
- [127].Cao B, Li L, Wu H, Tang Q, Sun B, Dong H, Zhe J, Cheng G. Chem Commun (Cambridge, U K) 2014;50:3234. doi: 10.1039/c3cc48878k. [DOI] [PubMed] [Google Scholar]
- [128].Wei T, Zhan W, Cao L, Hu C, Qu Y, Yu Q, Chen H. ACS Appl Mater Interfaces. 2016;8:30048. doi: 10.1021/acsami.6b11187. [DOI] [PubMed] [Google Scholar]
- [129].Shi Z-Q, Cai Y-T, Deng J, Zhao W-F, Zhao C-S. ACS Appl Mater Interfaces. 2016;8:23523. doi: 10.1021/acsami.6b07397. [DOI] [PubMed] [Google Scholar]
- [130].Yu Q, Ge W, Atewologun A, Stiff-Roberts AD, López GP. Colloids and Surfaces B: Biointerfaces. 2015;126:328. doi: 10.1016/j.colsurfb.2014.12.043. [DOI] [PubMed] [Google Scholar]
- [131].Yu Q, Ge W, Atewologun A, Lopez GP, Stiff-Roberts AD. Journal of Materials Chemistry B. 2014;2:4371. doi: 10.1039/c4tb00566j. [DOI] [PubMed] [Google Scholar]
- [132].Yu Q, Cho J, Shivapooja P, Ista LK, López GP. ACS applied materials & interfaces. 2013;5:9295. doi: 10.1021/am4022279. [DOI] [PubMed] [Google Scholar]
- [133].Yu Q, Ista LK, Lopez GP. Nanoscale. 2014;6:4750. doi: 10.1039/c3nr06497b. [DOI] [PubMed] [Google Scholar]
- [134].Tan M, Wang H, Wang Y, Chen G, Yuan L, Chen H. Journal of Materials Chemistry B. 2014;2:569. doi: 10.1039/c3tb21358g. [DOI] [PubMed] [Google Scholar]
- [135].Yan S, Luan S, Shi H, Xu X, Zhang J, Yuan S, Yang Y, Yin J. Biomacromolecules. 2016;17:1696. doi: 10.1021/acs.biomac.6b00115. [DOI] [PubMed] [Google Scholar]
- [136].Nederberg F, Zhang Y, Tan JPK, Xu K, Wang H, Yang C, Gao S, Guo XD, Fukushima K, Li L, Hedrick JL, et al. Nat Chem. 2011;3:409. doi: 10.1038/nchem.1012. [DOI] [PubMed] [Google Scholar]
- [137].Engler AC, Tan JPK, Ong ZY, Coady DJ, Ng VWL, Yang YY, Hedrick JL. Biomacromolecules. 2013;14:4331. doi: 10.1021/bm401248t. [DOI] [PubMed] [Google Scholar]
- [138].Mizutani M, Palermo EF, Thoma LM, Satoh K, Kamigaito M, Kuroda K. Biomacromolecules. 2012;13:1554. doi: 10.1021/bm300254s. [DOI] [PubMed] [Google Scholar]
- [139].Martinez-Abad A, Ocio MJ, Lagaron JM. J Appl Polym Sci. 2014;131:41001/1 [Google Scholar]
- [140].El Habnouni S, Lavigne J-P, Darcos V, Porsio B, Garric X, Coudane J, Nottelet B. Acta Biomater. 2013;9:7709. doi: 10.1016/j.actbio.2013.04.018. [DOI] [PubMed] [Google Scholar]
- [141].McClanahan JR, Peyyala R, Mahajan R, Montelaro RC, Novak KF, Puleo DA. Int J Antimicrob Agents. 2011;38:530. doi: 10.1016/j.ijantimicag.2011.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [142].Zhou C-E, Kan C-w. Cellulose. 2014;21:2951 [Google Scholar]
- [143].Kilicarslan M, Gumustas M, Yildiz S, Baykara T. Current drug delivery. 2014;11:98. doi: 10.2174/15672018113109990055. [DOI] [PubMed] [Google Scholar]
- [144].Joerger RD, Sabesan S, Visioli D, Urian D, Joerger MC. Packaging Technology & Science. 2009;22:125. [Google Scholar]
- [145].Dave RN, Joshi HM, Venugopalan VP. Antimicrob Agents Chemother. 2011;55:845. doi: 10.1128/AAC.00477-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [146].Johnson ML, Uhrich KE. J Biomed Mater Res, Part A. 2009;91A:671. doi: 10.1002/jbm.a.32288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [147].Pavlukhina S, Zhuk I, Mentbayeva A, Rautenberg E, Chang W, Yu X, van de Belt-Gritter B, Busscher HJ, van der Mei HC, Sukhishvili SA. NPG Asia Materials. 2014;6:e121 [Google Scholar]
- [148].Shukla A, Fleming KE, Chuang HF, Chau TM, Loose CR, Stephanopoulos GN, Hammond PT. Biomaterials. 2010;31:2348. doi: 10.1016/j.biomaterials.2009.11.082. [DOI] [PubMed] [Google Scholar]
- [149].Pavlukhina S, Lu Y-M, Patimetha A, Libera M, Sukhishvili S. Biomacromolecules. 2010;11:3448. doi: 10.1021/bm100975w. [DOI] [PubMed] [Google Scholar]
- [150].Bieser AM, Thomann Y, Tiller JC. Macromol Biosci. 2011;11:111. doi: 10.1002/mabi.201000306. [DOI] [PubMed] [Google Scholar]
- [151].Wu Y, Qin Y, Yuan M, Li L, Chen H, Cao J, Yang J. Polym Adv Technol. 2014;25:948. [Google Scholar]
- [152].Shakesheff KM, Davies MC, Domb A, Jackson DE, Roberts CJ, Tendler SJB, Williams PM. Macromolecules. 1995;28:1108 [Google Scholar]
- [153].Dorner F, Boschert D, Schneider A, Hartleb W, Al-Ahmad A, Lienkamp K. ACS Macro Lett. 2015;4:1337. doi: 10.1021/acsmacrolett.5b00686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [154].Ziaie B, Baldi A, Lei M, Gu Y, Siegel RA. Adv Drug Delivery Rev. 2004;56:145. doi: 10.1016/j.addr.2003.09.001. [DOI] [PubMed] [Google Scholar]
- [155].Lynn DM. Adv Mater. 2007;19:4118 [Google Scholar]
- [156].Hammond PT. AIChE J. 2011;57:2928 [Google Scholar]
- [157].Dorner F. Degradierbare antimikrobielle Multilagen. Albert-Ludwigs-Universität; Freiburg: 2015. [Google Scholar]
- [158].Kurowska M, Eickenscheidt A, Guevara-Solarte DL, Widyaya VT, Marx F, Al-Ahmad A, Lienkamp K. Biomacromolecules. 2017;18:1373. doi: 10.1021/acs.biomac.7b00100. [DOI] [PubMed] [Google Scholar]